- 1Hubei Key Laboratory of Biological Resources Protection and Utilization, Hubei Minzu University, Enshi, China
- 2College of Biological and Food Engineering, Hubei Minzu University, Enshi, China
- 3College of Life Science, Baicheng Normal University, Baicheng, China
Selenium (Se) is a vital trace element integral to numerous biological processes in both plants and animals, with significant impacts on soil health and ecosystem stability. This review explores how soil microorganisms facilitate Se transformations through reduction, oxidation, methylation, and demethylation processes, thereby influencing the bioavailability and ecological functions of Se. The microbial reduction of Se compounds, particularly the conversion of selenate and selenite to elemental Se nanoparticles (SeNPs), enhances Se assimilation by plants and impacts soil productivity. Key microbial taxa, including bacteria such as Pseudomonas and Bacillus, exhibit diverse mechanisms for Se reduction and play a substantial role in the global Se cycle. Understanding these microbial processes is essential for advancing soil management practices and improving ecosystem health. This review underscores the intricate interactions between Se and soil microorganisms, emphasizing their significance in maintaining ecological balance and promoting sustainable agricultural practices.
1 Introduction
Se is a vital trace element with significant roles in both geochemistry and biological systems (Sharma et al., 2015; Wang et al., 2022e). In soil ecosystems, microorganisms are crucial in regulating the availability and cycling of Se, which in turn influences its geochemical behavior and bioavailability (Wells and Stolz, 2020). This trace element is an essential component of critical enzymes involved in oxidation–reduction reactions that maintain cellular homeostasis in various organisms (Wells et al., 2021). These processes contribute to antioxidant defenses and reduce oxidative stress (Ekumah et al., 2021). By participating in nutrient cycling, Se not only impacts the metabolic activities of soil microorganisms and plant growth but also contributes to biodiversity and ecosystem stability by modulating the survival and reproduction of various organisms across trophic levels (Graves et al., 2019; Wang et al., 2022e). In addition to its ecological importance, Se is crucial for human health, supporting antioxidative, immunomodulatory, and anticarcinogenic functions, as well as playing a role in detoxification processes (Mehri, 2020; Zhao et al., 2023). Deficiencies in Se can lead to a range of health issues, including compromised immune function, thyroid dysfunction, and cardiovascular diseases (Rahman et al., 2019; Crespo et al., 2024). Therefore, the diverse roles of Se have profound implications for both ecosystem function and human health.
In soil, Se mainly exists as selenate, selenite, or organic Se compounds (methylated compounds, selenoamino acids, selenoproteins, and their derivatives) (Pyrzyńska, 1996; Ullah et al., 2019). Microorganisms regulate the form and availability of Se through reduction and oxidation processes (Srivastava et al., 2023), which are essential for maintaining soil health and productivity (Gómez-Gómez et al., 2019). For instance, some microorganisms reduce Se from higher oxidation states to more accessible forms for plants, while others oxidize organic Se into inorganic forms, facilitating its cycling within the ecosystem (Fischer et al., 2020).
Microbially mediated Se cycling not only affects soil content and plant uptake but also has broader implications for ecosystem services and food chain dynamics (Khanna et al., 2023). As Se accumulates in food chains, microbial processes such as reduction, methylation, and demethylation regulate its conversion into bioavailable or less toxic forms. These processes influence the movement of Se between soil, plants, and animals, ultimately affecting the ecological balance, species survival, and overall health of entire ecosystems (Wang et al., 2022e). Given the significant role of microorganisms in Se cycling and ecosystem functioning, we provide a brief overview of the current state of knowledge on microbial processes involved in Se transformations and their ecological implications.
2 Microbial transformation and utilization of Se
In soil, Se predominantly occurs in two soluble inorganic forms: selenate (SeO42−, Se6+) and selenite (SeO32−, Se4+) (Sharma and Sharma, 2021). Additionally, Se is present as part of organic compounds, including selenocysteine, selenomethionine, and selenocysteine, which are important for its biological cycling (Petrović, 2021). Microorganisms play a vital role in transforming inorganic Se compounds into insoluble elemental Se (Se0), which can be filtered out of drainage water (Zhou et al., 2020). Additionally, they reduce Se compounds such as selenate, converting them into more easily absorbed organic forms such as methyl selenocysteine (Liao et al., 2024). These organic forms are readily absorbed by plants, thereby entering the food chain. Microbial communities exhibit variability in Se bioavailability, using various mechanisms and metabolic pathways to detect and utilize Se. The four known biological transformations of Se—reduction (both assimilatory and dissimilatory), oxidation, methylation, and demethylation—play a crucial role in Se cycling within ecosystems (Figure 1) (Khanna et al., 2023; Chilala et al., 2024).
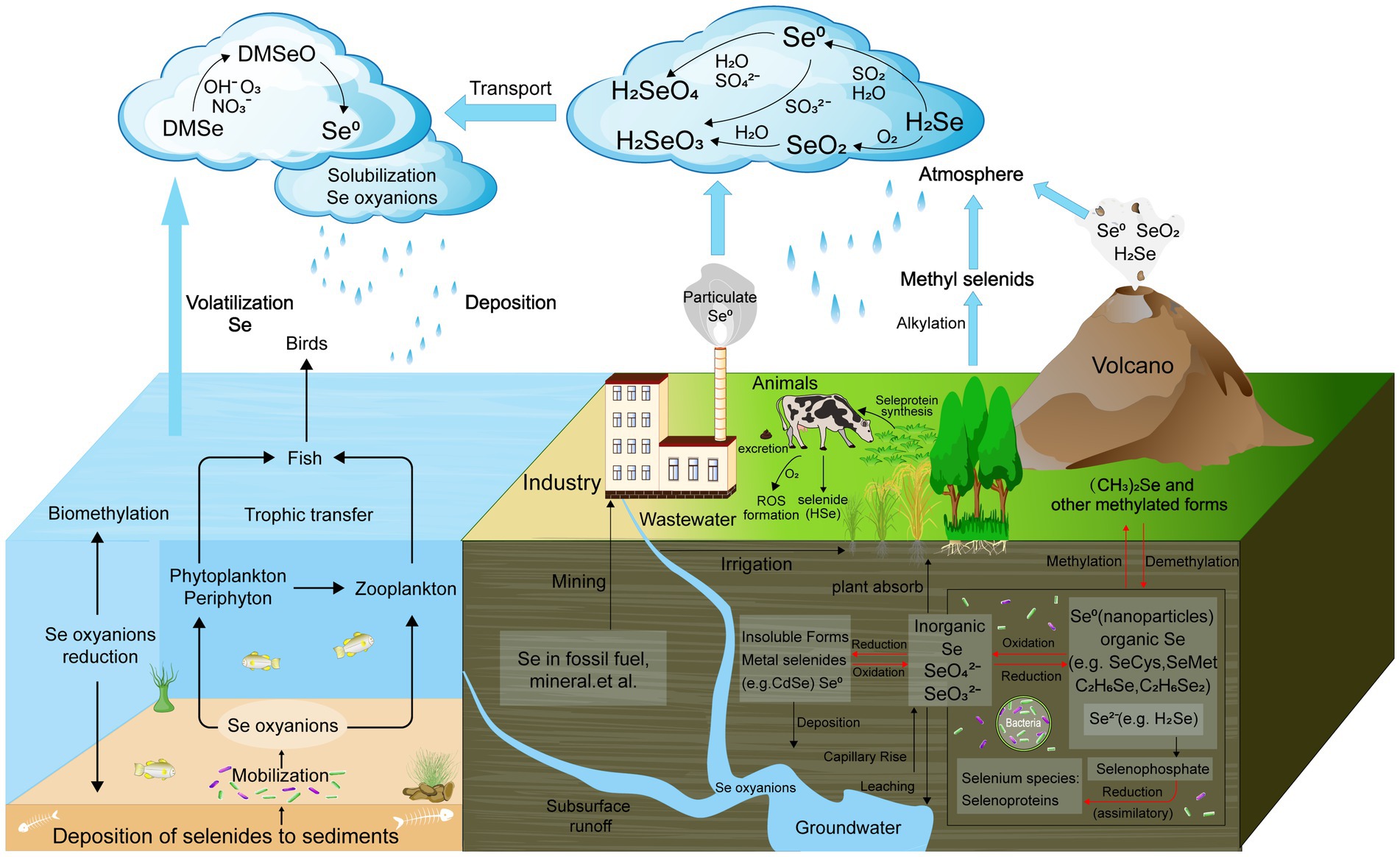
Figure 1. Schematic biogeochemical behavior of Se in the ecosystem. The four known biological transformations of Se—reduction, oxidation, methylation, and demethylation are marked with red.
2.1 Se-reducing bacteria
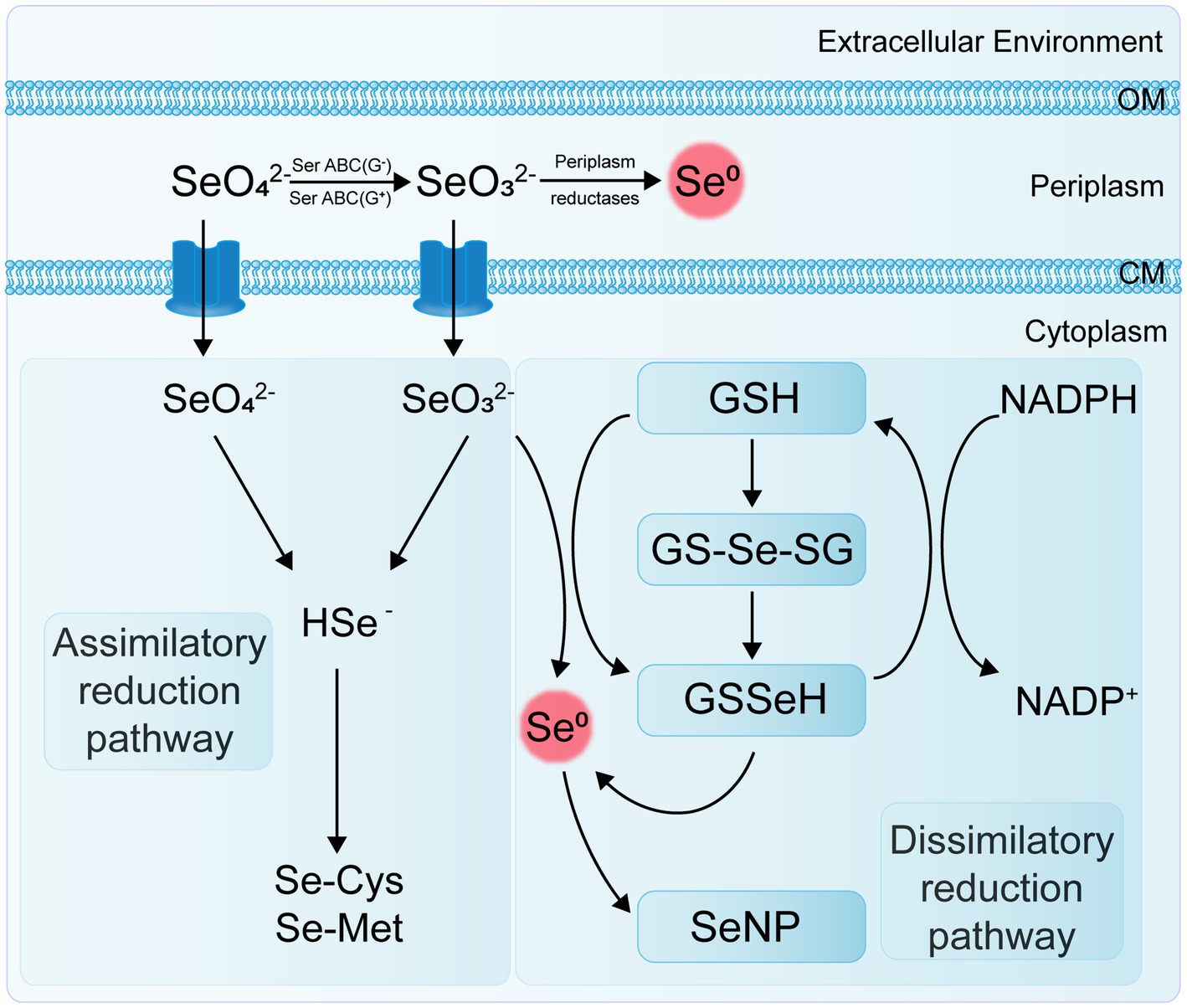
Se-reducing bacteria are microorganisms capable of reducing inorganic Se compounds from higher to lower oxidation states. These Se reduction processes can be categorized into two main types: assimilatory reduction and dissimilatory reduction (Figure 2). Assimilatory reduction involves converting Se(VI) or Se(IV) into Se-containing amino acids and selenoproteins. In contrast, dissimilatory reduction converts Se(VI) and Se(IV) into SeNPs or selenides. Both processes promote Se cycling and enhance its bioavailability in soil ecosystems.
2.1.1 Microbial assimilatory reduction of Se
The end products of the microbial assimilatory reduction of Se are Se-containing amino acids and selenoproteins, produced through specific and non-specific pathways. The primary Se-containing amino acids are selenocysteine (Sec or SeCys, U, the 21st amino acid) and selenomethionine (SeMet, the 22nd amino acid). The biosynthesis of SeCys is specific and specialized (Kieliszek et al., 2020), while SeMet can non-specifically replace methionine in protein synthesis. Se is an essential element for various organisms across almost all bacterial phyla. Bacteria capable of Se assimilation are widely distributed, with at least one-third of over 5,200 sequenced bacterial species possessing Se assimilation capabilities (Zhang et al., 2022b). Two key enzymes identified in the Se assimilation process in eukaryotes, bacteria, and archaea are selenocysteine synthase (SelA) and selenophosphate synthetase (SelD) (Hoffman et al., 2023). The latest research found that bacterial SelA, a homo-decameric enzyme composed of a pentamer of dimers, has a unique and highly symmetrical arrangement not seen in Archaea and Eukarya (Serrão et al., 2024). SelD interacts directly with the complex formed by SelA and SelC to supply the necessary selenophosphate for synthesizing Sec-tRNASec (Scortecci et al., 2020). Additionally, selenocysteine-specific transfer RNA (tRNASec) and the selenocysteine insertion sequence (SECIS) are important factors involved in microbial Se assimilation (Wells et al., 2021; Manta et al., 2022).
2.1.2 Microbial dissimilatory reduction of Se
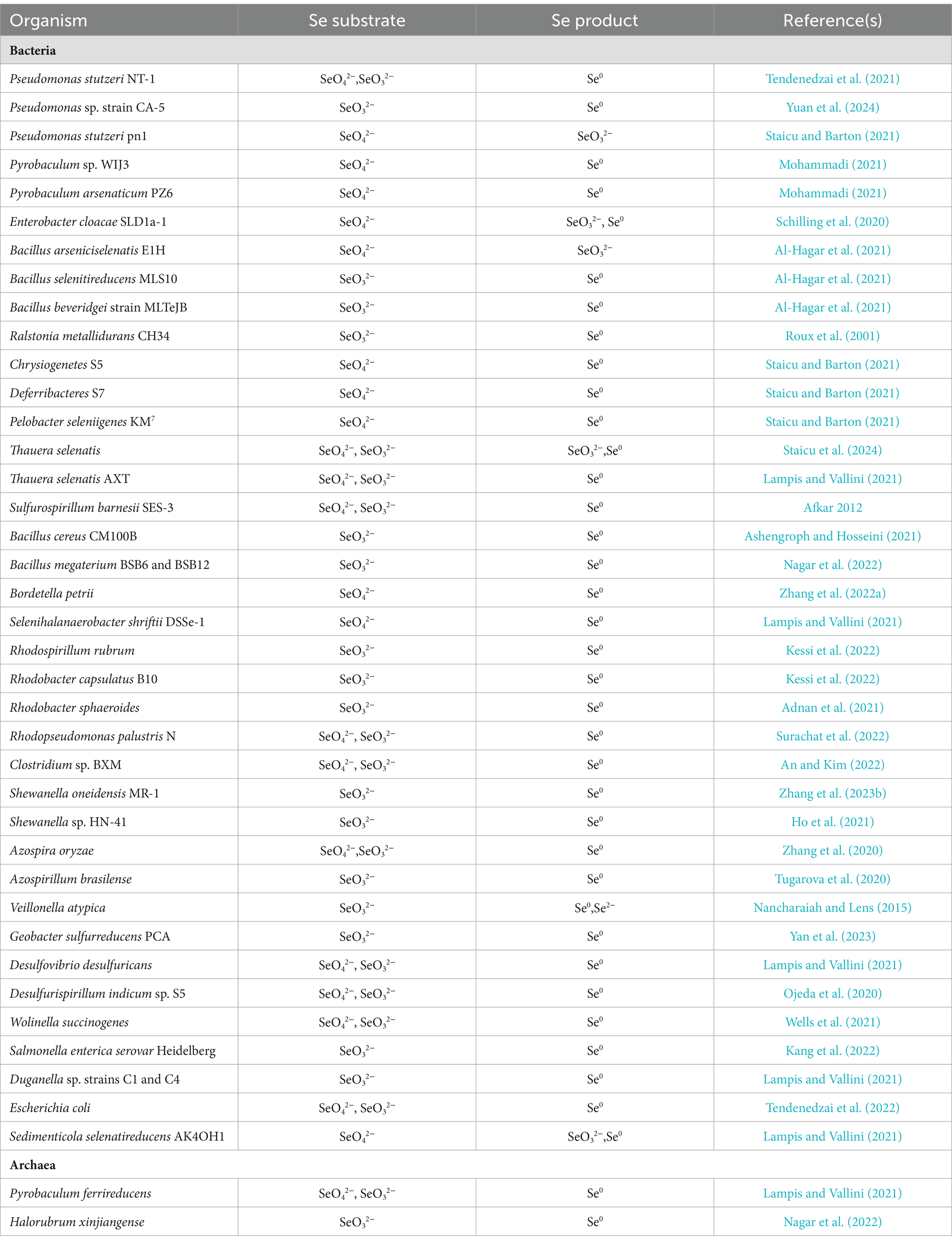
The dissimilatory reduction of Se(VI) and Se(IV) by bacteria occurs widely in soil, sediment, and aquatic environments, typically producing SeNPs under both anaerobic and aerobic conditions. Various bacteria, including Escherichia coli, Rhodospirillum rubrum, Rhodobacter sphaeroides, Bacillus subtilis, Pseudomonas fluorescens, and Bacillus licheniformis, have been identified as capable of reducing selenate or selenite to SeNPs (Table 1). Ojeda et al. (2020) summarized several Se(IV) reduction pathways, including thiol group-mediated reduction reactions, the thioredoxin-thioredoxin reductase system, siderophore-mediated reduction reactions, sulfide-mediated reduction reactions, and dissimilatory reduction. Bacteria and fungi with thiol groups utilize these groups to facilitate Se reduction, while some microorganisms reduce selenate to Se compounds, thereby participating in the Se cycle.
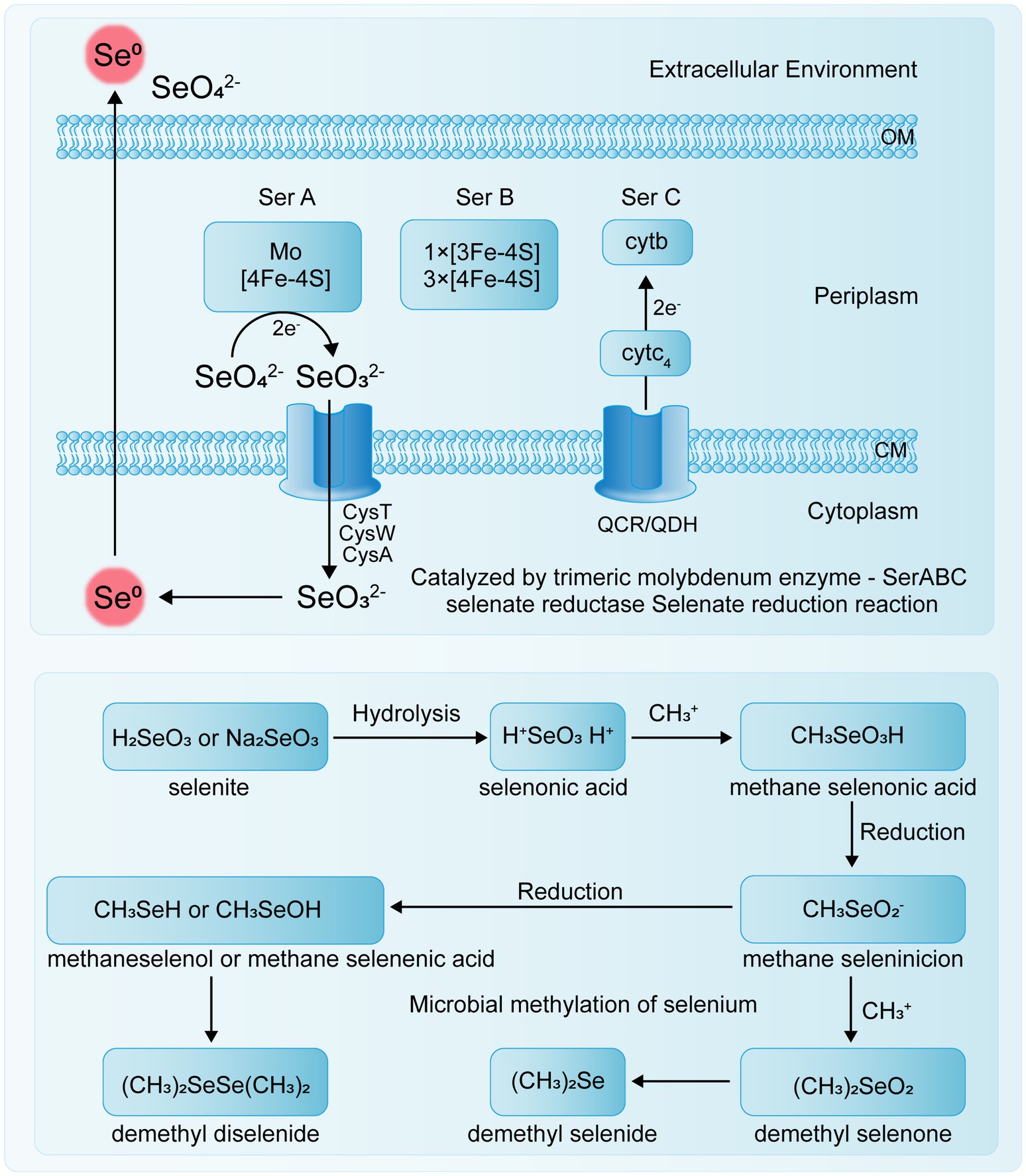
Transposon mutagenesis genetic analysis has revealed that the selenate reductase in E. coli is a molybdoenzyme and that the reduction of selenite to Se0 and the reduction of selenate to selenite are two independent processes (Bébien et al., 2002). Kuroda et al. (2011) also found that the strain B. selenatarsenatis SF-1 independently reduces selenate to selenite through anaerobic respiration. This process is facilitated by a selenate reductase complex, with the operon SrdBCA displaying typical features of a membrane-bound and molybdopterin-containing oxidoreductase capable of encoding a respiratory selenate reductase complex. To date, a specific selenate reductase has only been purified from the Gram-negative β-proteobacterium Thauera selenatis, which uses selenate as the terminal electron acceptor for anaerobic respiration (Mohapatra et al., 2022). However, the biological mechanisms and enzymes involved in other selenate and selenite reduction processes remain to be characterized.
The genus Clostridium includes strains of anaerobic bacteria capable of reducing Se. These bacteria typically inhabit anoxic or microaerophilic environments, utilizing thiol groups or other reducing agents to convert selenate to selenides (Figueiredo et al., 2020). The genus Desulfovibrio comprises sulfate-reducing bacteria (SRB) that usually thrive in environments rich in organic matter and sulfur. These bacteria also possess the capability to reduce Se, using reducing agents such as hydrogen sulfide to transform selenate into selenides (Gao et al., 2023). Both archaeal and bacterial domains can use selenate and selenite as terminal electron acceptors. Under anaerobic conditions, they reduce soluble selenate and selenite to insoluble elemental Se through dissimilatory reduction (Xu et al., 2023). For example, Se-reducing bacteria can use carbon sources and electron donors to reduce Se oxyanions to elemental Se (Sinharoy and Lens, 2022); Enterobacter cloacae SLD1a-1 reduces selenate in the periplasmic space to SeNPs using a membrane-bound Se (VI) reductase, with SeO₄2− as an alternative electron acceptor, and rapidly expels the nanoparticles (Escobar-Ramírez et al., 2021).
Under aerobic or microaerobic conditions, various bacterial strains can reduce selenate and selenite to elemental Se or selenides as a detoxification mechanism. Se-tolerant aerobic bacteria of the genus Bacillus (EU573774.1), isolated from Se-rich rhizosphere soil, reduce Se(IV) under aerobic conditions to produce amorphous α-Se0 nanospheres. This demonstrates the potential of this Bacillus isolate for neutralizing toxic Se(IV) anions in the environment (Hashem et al., 2021). The adsorption of selenite onto B. subtilis cells is mediated by thiol sites on the cell envelope, resulting in the formation of reduced organic Se compounds (e.g., R1S–Se–SR2) (Yu et al., 2018). The environmental bacterial isolate Stenotrophomonas maltophilia actively reduces SeO₄2− and SeO₃2− to red, amorphous Se0 and produces volatile alkyl selenides such as dimethyl selenide (DMSe), dimethyl selenyl sulfide (DMSeS), and dimethyl diselenide (DMDSe) (Lashani et al., 2023). The SeNPs synthesized by the strain S. maltophilia SeITE02 exhibit significant antibacterial and antibiofilm activity against various pathogens, including both reference strains and clinical isolates (Piacenza et al., 2023). Members of the genus Rhizobium can reduce selenite to Se0 under both aerobic and denitrifying conditions via molybdenum-containing proteins. However, they do not reduce selenate nor use selenite or selenate as terminal electron acceptors (Nikam et al., 2022).
Strain E1H (B. arsenicoselenatis) is an anaerobic bacterium characterized by spore-forming rods and grows by reducing Se(VI) to Se(IV). Strain MLS10 (B. selenitireducens) is a microaerophilic Se-reducing bacillus that grows by reducing Se(IV) to Se0. When co-cultured, these strains can completely reduce Se(VI) to Se0 (Boltz and Rittmann, 2021). Various phototrophic bacteria in redox steady states, such as Rhodospirillum rubrum, can also reduce Se salts and selenite to elemental Se, which is then excreted through the membrane and cell wall (Tugarova et al., 2020). The genus Selenihalanaerobacter consists of anaerobic bacteria specialized in Se utilization, capable of using selenate as a terminal electron acceptor. Through selenate reduction, these bacteria convert it to selenite and elemental Se, playing a significant role in Se cycling (Gonzalez-Gil et al., 2016; Yadav et al., 2023).
Different types of Se-reducing bacteria exhibit various mechanisms for Se reduction, and even a single bacterium may use multiple mechanisms. Current research indicates that enzymes involved in Se reduction include dehydrogenase I, sulfite reductase, nitrite reductase (NiR), and dimethyl sulfoxide reductase (Khanna et al., 2023). Reddy and Bandi (2022) discovered that the Se-reducing bacterium Pseudomonas seleniipraecipitans can reduce selenate to Se0 through nitrate reductase (NR) and that glutathione reductase and thioredoxin reductase may also be involved in the reduction of Se(IV). Selenomonas ruminantium reduces selenite to red elemental Se by incorporating it into selenocysteine, selenohomocysteine, selenomethionine, and selenoethionine (Hendawy et al., 2021). Ralstonia metallidurans CH34 (formerly known as Alcaligenes CH34) is a soil bacterium noted for its ability to grow in the presence of various heavy metals. It has been reported that strain CH34 can resist up to 6 mM selenite and, during the lag phase, adapts to selenite stress by reducing selenite to red elemental Se (Mohammadi and Baldwin, 2024).
The non-enzymatic reduction of Se(IV) primarily involves sulfide-mediated reduction and thiol compound-mediated reduction, such as by reduced glutathione (GSH). Studies have demonstrated that the abiotic reduction of selenite by glutathione produces reactive oxygen species (ROS). This indicates that glutathione is involved in the reduction of selenite to elemental Se, as mediated by Rhodospirillum rubrum and E. coli (Wang et al., 2022a). Additionally, Se reduction in some bacteria may be associated with extracellular secretions. For instance, in Pseudomonas aeruginosa strain JS-11, the metabolite pyocyanin-1-carboxylic acid (PCA) and known redox agents (such as NADH and NADH-dependent reductases) are responsible for reducing SeO₃2− to Se0 (Xue et al., 2024). These Se-reducing bacteria exhibit diverse ecological characteristics and physiological properties, and their distribution and activity in the soil significantly impact Se cycling and bioavailability. Research on these bacteria enhances our understanding of the biogeochemical cycle of Se and the functioning of soil ecosystems.
2.2 Se-oxidizing Bacteria
Se-oxidizing bacteria are microorganisms capable of oxidizing organic Se compounds or inorganic Se compounds from lower to higher oxidation states. These bacteria include heterotrophs, sulfur-oxidizing bacteria, and possibly fungi (Luo et al., 2022). They play a crucial role in soil and water environments, influencing the bioavailability and cycling of Se. Sarathchandra and Watkinson (1981) discovered that Bacillus megaterium, isolated from soil, could oxidize elemental Se in laboratory cultures to selenite and trace amounts of selenate (with selenite content <1%). This observation represents an important but previously unreported oxidation step in the biological Se cycle. To date, few studies have demonstrated that the oxidation of Se0 in soil is predominantly biological and occurs at a relatively slow rate. There is limited research on the types of Se-oxidizing bacteria, particularly pure cultures capable of oxidizing Se0, and the biosynthetic mechanisms of Se oxidation (Wasserman et al., 2021). This may be because microbial oxidation in the soil is partly limited by the adsorption of Se(IV) on the soil surface, and the process of Se0 being oxidized by microorganisms to higher oxidation states of Se is very slow (Wang et al., 2022a).
Additionally, studying the oxidation of Se0 by soil microorganisms is crucial for increasing the amount of bioavailable Se in soil, enhancing Se enrichment in plants, and ultimately supplementing Se in the human diet. Agrobacterium sp. strain T3F4 can oxidize Se0 to Se(IV), establish stable colonies in the soil, and dissolve soil-bound Se(IV), thereby enhancing Se bioavailability and promoting Se uptake in crops (Zhu et al., 2021). Guo et al. (2024a) demonstrated that Agrobacterium T3F4 enhances the absorption of Se by cabbage plants by oxidizing Se in the soil, thereby increasing its bioavailability. Four Se-oxidizing bacteria, namely Dyella spp. LX-1 and LX-66, and Rhodanobacter spp. LX-99 and LX-100, isolated from seleniferous soil, were involved in the oxidation of SeMet, SeCys2, selenourea, and Se0 to Se(IV) in pure cultures (Luo et al., 2022).
Some strains of the genus Thiobacillus can oxidize Se independently. They typically inhabit sulfur-rich environments and use sulfur oxidase to oxidize Se0 to selenate or other higher oxidation state Se compounds. Thiobacillus ASN-1 primarily produces Se(VI) rather than Se(IV) as the oxidation product of Se0. Intermediate metabolites from Se0 oxidation, such as volatile fatty acids, hydrogen, and methane, are utilized by heterotrophic Se(VI) reducers for Se(VI) detoxification (Li et al., 2022). Kushwaha et al. (2022) found that acetate, glucose, and sulfide promoted Se oxidation, confirming the involvement of chemoautotrophic and chemoheterotrophic species of Thiobacillus. Leptothrix sp. MnB-1 can oxidize Se0 to Se(VI) through complex mechanisms, though its enzymes and pathways have not yet been reported (Guo et al., 2023).
2.3 Microbial transport of Se
Some microorganisms actively absorb and transport Se through specific membrane proteins or carriers, acquiring Se from the environment or surrounding organic matter. Current research generally agrees that Se(VI) is transported via sulfate [S(VI)] transport channels, while Se(IV) can be transported through various mechanisms. Kinetic studies by Goff (2020) on the transport of selenite, selenate, and sulfate in wild-type E. coli K-12 indicate that K-12 shares the same transport system for Se and sulfate. Microorganisms produce specific enzymes, known as selenoenzymes, during the absorption and transport process, converting inorganic Se or organic Se compounds into forms that can be utilized by microbial cells (Chaudière, 2023). In yeast, Kieliszek et al. (2020) reported that the proton-coupled monocarboxylate transporter Jen1p in Saccharomyces cerevisiae transports selenite into the cell in a proton-dependent manner, similar to the transport mechanism for lactate. This facilitates high-affinity uptake of selenite and promotes its accumulation within the cell.
Lazard (2021) discovered that in S. cerevisiae, the uptake of selenite is influenced by the phosphate concentration in the growth medium. Both high- and low-affinity phosphate transport proteins are involved in this process. Under low-phosphate (Pi) conditions, the high-affinity transporter Pho84p primarily mediates selenite uptake. When phosphate is abundant, selenite is absorbed through low-affinity transporters, such as Pho87p, Pho90p, and Pho91p. This shift occurs because the high-affinity transport, which includes Pho84p and Pho89p, is upregulated by the phosphate signal transduction pathway (PHO) during Pi starvation. Pho84p operates most effectively at neutral to acidic pH, while Pho89p functions at alkaline pH. In contrast, the low-affinity transporters Pho87p, Pho90p, and Pho91p, which have a lower affinity for phosphate, are constitutively expressed but their activity is post-transcriptionally downregulated in low-phosphate conditions by Spl2p, a component of the PHO regulon.
The transport mechanism of Se0 in bacteria remains unclear, and specific selenite transport proteins have not been identified. Studies by Peng et al. (2021) indicate that E. coli K-12 can transport sulfate, selenate, and selenite via ABC transporters, but this system has a binding affinity for sulfate that is five times stronger than for selenate and 37 times stronger than for selenite. Se(VI) reductase is a trimeric complex with structural similarities to other enzymes, though it also exhibits some detailed differences. For example, in T. selenatis AXT, the Se(VI) reductase SerABC is located in the periplasmic space, exhibits high substrate specificity, and produces SeNPs that accumulate in the cytoplasm or extracellularly (Lampis and Vallini, 2021). The SerA subunit is a 96 kDa catalytic subunit containing a molybdenum cofactor, SerB is a 40 kDa iron–sulfur protein containing one [3Fe-4S] cluster and three [4Fe-4S] clusters, and SerC is a 23 kDa protein containing a heme b cofactor (Staicu and Barton, 2021). The genes encoding these three subunits, serA, serB, serC, and serD, form an operon, with serD encoding a system-specific molecular chaperone protein involved in the insertion of the molybdenum cofactor into SerA (Wang et al., 2022b). Electrons are transferred through quinol cytochrome c oxidoreductase (QCR) or quinol dehydrogenase (QDH) to cytochrome c4 in the periplasm, and then to the cytochrome b of the Se(VI) reductase complex, specifically to the SerC subunit of SerABC (Figure 2).
It has been reported that ABC transporters play a significant role in the mechanisms of Se uptake, metabolism, and tolerance in various plants (Jiao et al., 2022). For instance, in tea plants (Camellia sinensis), exposure to selenate increases the expression of ABCC8 and ABCG14, while selenite exposure upregulates ABCA2 and ABCC4 (Ren et al., 2022). In ryegrass, ABC5 and ABC11 are induced under high-dose selenite treatment (Chen et al., 2022). In the Se hyperaccumulator Stanleya pinnata, ABCC2, ABCF5, and ABCG40 are believed to be involved in vacuolar sequestration and the distribution of excess Se(VI) to mitigate toxicity (Rao et al., 2021).
Arbuscular mycorrhizal fungi (AMF) have the ability to absorb sulfate. Specifically, the sulfate transporters GBC38160.1 and GBC25943.1, along with the sulfate permease PKY50973.1 in Rhizophagus irregularis (an AMF species), as well as the sulfate transporters EDR02618.1 and EDR02177.1, and sulfate permeases EDR11271.1 and EDR00466.1 in the ectomycorrhizal fungus Laccaria bicolor, have been shown to enhance the uptake and transport of sulfate to the host plants. Given that Se and sulfur (S) belong to the same group (VI-A) in the periodic table, Se exhibits chemical properties that are highly similar to sulfur. Se can be absorbed in the form of selenate or selenite and is transported to the host plant via sulfur assimilation pathways, including sulfur transporters. This process leads to the biosynthesis of SeCys, SeMet, and various Se-homologs of sulfur-containing metabolites (Ye et al., 2020).
Se-absorbing bacteria primarily include Fibrobacter succinogenes, Ruminococcus albus, Streptococcus spp., Lactobacillus spp., and Prevotella ruminicola. The transport and absorption mechanisms for Se(IV) in these bacteria differ from those for S(VI) or Se(VI) and are not influenced by the presence of S(VI) or Se(VI). In Selenomonas, selenite is incorporated into selenocysteine, selenoethionine, selenohomocysteine, and selenomethionine, and is also reduced to Se0 (Hendawy et al., 2021). The soil bacterium Pseudomonas putida KT2440 has been demonstrated to reduce and transport selenite under aerobic conditions through central metabolic processes, sulfur metabolism, and oxidative stress responses, leading to the production of SeNPs. Genes such as sucA, D2HGDH, and PP_3148 indicate that the metabolism of 2-oxoglutarate and glutamate plays a crucial role in the conversion of selenite to elemental Se. Conversely, mutations affecting sulfite reductase activity significantly impair the bacteria’s ability to reduce selenite.
Additionally, genes related to sulfur metabolism (ssuEF, sfnCE, sqrR, sqr, and pdo2) and stress response (gqr, lsfA, ahpCF, and sadI) have been identified as key players in the selenite conversion process (Avendaño et al., 2023). In the photosynthetic bacterium Rhodobacter sphaeroides, selenite enters the cytoplasm mediated by a polyol transporter from red algae, but similar polyhydroxy compound transport systems have not been reported in other bacteria (Wang et al., 2022a). Additionally, Padhi and Chatterjee (2023) studied the phenotypic characteristics and selenite consumption in DedA mutants of Ralstonia metallidurans, indicating that DedA is involved in the transport and absorption of selenite.
2.4 Microbial methylation and demethylation of Se
Studies have shown that some microorganisms possess the ability to methylate Se, converting inorganic or organic Se compounds into methylated Se compounds, such as methyl selenate (Figure 3) (Ojeda et al., 2020). Because the resulting methylated Se compounds are often volatile, this process serves as a detoxification mechanism for soil environments and has important applications in the remediation of Se-contaminated sites. Yang et al. (2024) reviewed the mechanisms by which environmental microorganisms methylate and demethylate Se compounds, indicating that microbial methylation of reduced Se oxyanions is a potentially effective detoxification process.
For instance, Rhodocyclus tenuis and Rhodospirillum rubrum can utilize Se(VI) to form DMSe and DMDSe in photoautotrophic processes. Additionally, R. tenuis can produce DMSe from selenides (Pal et al., 2022). Studies have shown that if the initial Se species are Se(IV), Se(VI), or Se(0), the Se methylation process necessarily involves both reduction and methylation steps, with different microorganisms using various pathways for Se methylation (Genchi et al., 2023). In soil, inorganic Se compounds can be converted by some microorganisms into volatile methylated substances (DMSe, DMDSe, and dimethyl selenone or methyl selenite), first undergoing Se methylation followed by Se reduction (Petrović, 2021). In some bacteria, such as Corynebacterium sp., Se is first reduced and then methylated (Garg et al., 2020).
Several enzymes associated with Se methylation have been identified (Table 2). Thiopurine methyltransferase (TPMT) is a monomeric, cytoplasmic S-adenosylmethionine (SAM)-dependent methyltransferase that plays a crucial role in thiopurine metabolism. It catalyzes the S-methylation of aromatic and heterocyclic thiol compounds, including 6-mercaptopurine (6-MP) and thiopurine, and also facilitates the methylation of sulfur, Se, and tellurium elements (Mu et al., 2024). Bacterial thiopurine methyltransferase (bTPMT) plays a crucial role in the methylation of seleno compounds, contributing to the detoxification process by converting reactive Se species into less toxic, volatile forms such as DMSe. Ranjard et al. (2003) discovered that bTPMT and thiocoraline methyltransferase are capable of methylating selenite and (methyl) selenocysteine into DMSe and DMDSe through a homologous protein, MmtA, in Pseudomonas sp. Hsa.28 and E. coli cells. Alonso-Fernandes et al. (2023) demonstrated that the SAM-dependent methyltransferase encoded by the TehB gene in E. coli is a highly potent detoxification protein. This enzyme can detoxify oxyanions derived from the chalcogen elements tellurium and Se without differentiating between selenite and tellurite oxyanions.
To date, methanogenic bacteria, iron-reducing bacteria, and SRB are the most commonly identified microorganisms involved in demethylation processes (Du et al., 2019). Studies have shown that Se can reduce net methylation of mercury (Hg) by inhibiting SRB activity, suggesting that Se may either promote Hg demethylation or inhibit methylation by other microorganisms (Guo et al., 2024b). Microbial-mediated methylation and demethylation of Hg occur during the conversion of sulfate (SO42−) to sulfide (S2−) (Wang et al., 2022c). Consequently, the transformation of SeO42− to Se2− may similarly occur during this process, as the reactions involving Se and S share several common enzymes (Chen and Li, 2023).
2.5 Case studies of Se and other related microorganisms
2.5.1 Symbiotic relationships enhancing Se uptake
The interactions between Se and microorganisms have garnered significant attention in the fields of environmental microbiology and ecotoxicology. Research indicates that certain mycorrhizal fungi can form symbiotic relationships with plants, facilitating the uptake and utilization of Se through symbiotic cooperation, thereby enhancing plant Se uptake from the soil (Li et al., 2020). Li et al. (2021) isolated 14 Se-tolerant endophytic bacteria from the Cardamine hupingshanensis in the Se-rich region of Enshi, China. These bacteria belong to 11 different genera and exhibit tolerance to various concentrations of selenite and selenate. Additionally, a study found that the mycorrhizal fungus Glomus intraradices can enhance Se uptake in leguminous plants, thereby increasing Se accumulation in the plants (Liu et al., 2024).
2.5.2 Effects of Se on soil microbial communities
Several studies have explored the effects of Se on the structure and function of soil microbial communities. Kang et al. (2024) found that the addition of Se significantly alters the community structure of soil bacteria and fungi, leading to changes in the abundance and diversity of specific microbial strains. Rosenfeld et al. (2018) indicated that bacterial community richness and diversity decrease significantly with increased Se levels, suggesting that Se contamination shapes communities in favor of Se-tolerant microorganisms. Additional studies have focused on the impact of Se on microbial metabolism and growth. While Se is an essential trace element for microorganisms, high concentrations can be toxic, adversely affecting microbial metabolism and growth. In soils with elevated Se concentrations, Se-reducing bacteria can be utilized to convert highly toxic inorganic Se compounds into less toxic forms through bioremediation, thereby mitigating the toxic effects of Se in soil or water.
3 The critical role of microorganisms in Se cycling and soil remediation
3.1 Microbial mechanisms in Se cycling
Microorganisms are central to the biogeochemical cycling of Se in soil ecosystems, primarily through their ability to mediate redox transformations, methylation, and volatilization processes. These microbial activities convert Se between different oxidation states, such as SeO₄2− and SeO₃2−, and into Se0, or volatile methylated forms. These transformations are crucial for regulating Se mobility, bioavailability, and toxicity in soils. For instance, Se-reducing bacteria can reduce selenate to elemental Se, a less toxic and insoluble form, thereby preventing the accumulation of harmful Se in the environment. Conversely, certain bacteria oxidize reduced Se forms back to SeO₄2−, contributing to its recycling within the ecosystem. Additionally, microorganisms such as fungi and algae can methylate Se, converting it into volatile compounds such as DMSe, which are released into the atmosphere, thus removing Se from the soil and participating in the global Se cycle (Figure 1).
3.2 Microbial remediation of Se-contaminated soils
With the extensive use of Se compounds in agricultural and industrial production, increasing amounts of Se (mainly SeO₄2− and SeO₃2−) are being released into the environment, posing threats to soil safety and the natural environment. Currently, approximately 2,700 tons of Se are produced annually, but only approximately 15% is recovered (Gebreeyessus and Zewge, 2019). Therefore, there is an urgent need for efficient, cost-effective, and environmentally friendly Se remediation technologies to mitigate the environmental impact of Se and enable its recycling. Bioremediation, especially microbial remediation, offers unique advantages in cost control and recovery efficiency, making it a highly regarded approach in the study of Se pollution remediation technologies. Consequently, microbial remediation technologies are increasingly researched and applied in the treatment of Se-contaminated soils.
The accumulation and volatilization of Se by algae in surface water are important parts of the biogeochemical cycle of Se (Zhang et al., 2023a). Liu et al. (2019) demonstrated that adding six types of algae to an artificial wetland water treatment system can enhance Se removal efficiency. This method significantly reduces the risk of Se accumulation in ecotoxic forms by promoting Se volatilization. This study found that the most effective algae for Se removal was the Chlorella vulgaris strain (Cv). Cv primarily accumulated Se in the form of selenomethionine (a precursor to volatile Se) and Se0. Within 72 h, Cv removed 96% of Se (provided as selenate), with up to 61% volatilized into the atmosphere.
Selenate-respiring bacteria, such as T. selenatis, have been used in laboratory-scale bioreactor systems to treat refinery wastewater containing selenite. In these systems, T. selenatis and a significant population of denitrifying bacteria reduce selenite present in the water, along with selenite formed from the reduction of selenate. The results showed a 95% reduction in Se oxyanion content in wastewater with an initial Se concentration of 3.4 mg/L (Sinharoy and Lens, 2020).
In practical applications, T. selenatis has been used to treat Se-containing wastewater in a medium-scale bioreactor system in the Panoche Water District of California’s San Joaquin Valley. After reduction by T. selenatis in the reactor, the concentration of Se oxyanions (selenate and selenite) in the discharge water was reduced by 98%. Analysis of the reactor effluent indicated that 97.9% of the oxidized Se was converted to insoluble but recoverable Se0. The small Se0 particles produced by reduction were also recovered using a flocculant (Nalmet 8,072), with 91 to 96% of the total recovered Se being Se0 (Arias-Paić et al., 2022). The selenate-reducing bacterium Bacillus sp. SF-1, using lactate as an electron donor, can remove 41.8 mg-Se/L of selenate and excess lactate from wastewater. Strain SF-1 converts selenate to selenite and then to elemental Se through anaerobic respiration. When the cell retention time in the bioreactor was relatively short at 2.9 h, the selenate was effectively converted entirely to selenite. With a longer cell retention time, the soluble Se (selenate and selenite) was successfully reduced to non-toxic elemental Se, with a production rate of 0.45 mg/L/h of elemental Se (Baggio, 2020). Otsuka and Yamashita (2020) added Pseudomonas stutzeri NT-I to a 256-liter medium-scale bioreactor, utilizing its ability to reduce oxidized Se to elemental Se. This approach proved effective in detoxifying soluble Se in smelting wastewater containing Se.
3.3 Influence of microbial Se cycling on plant growth
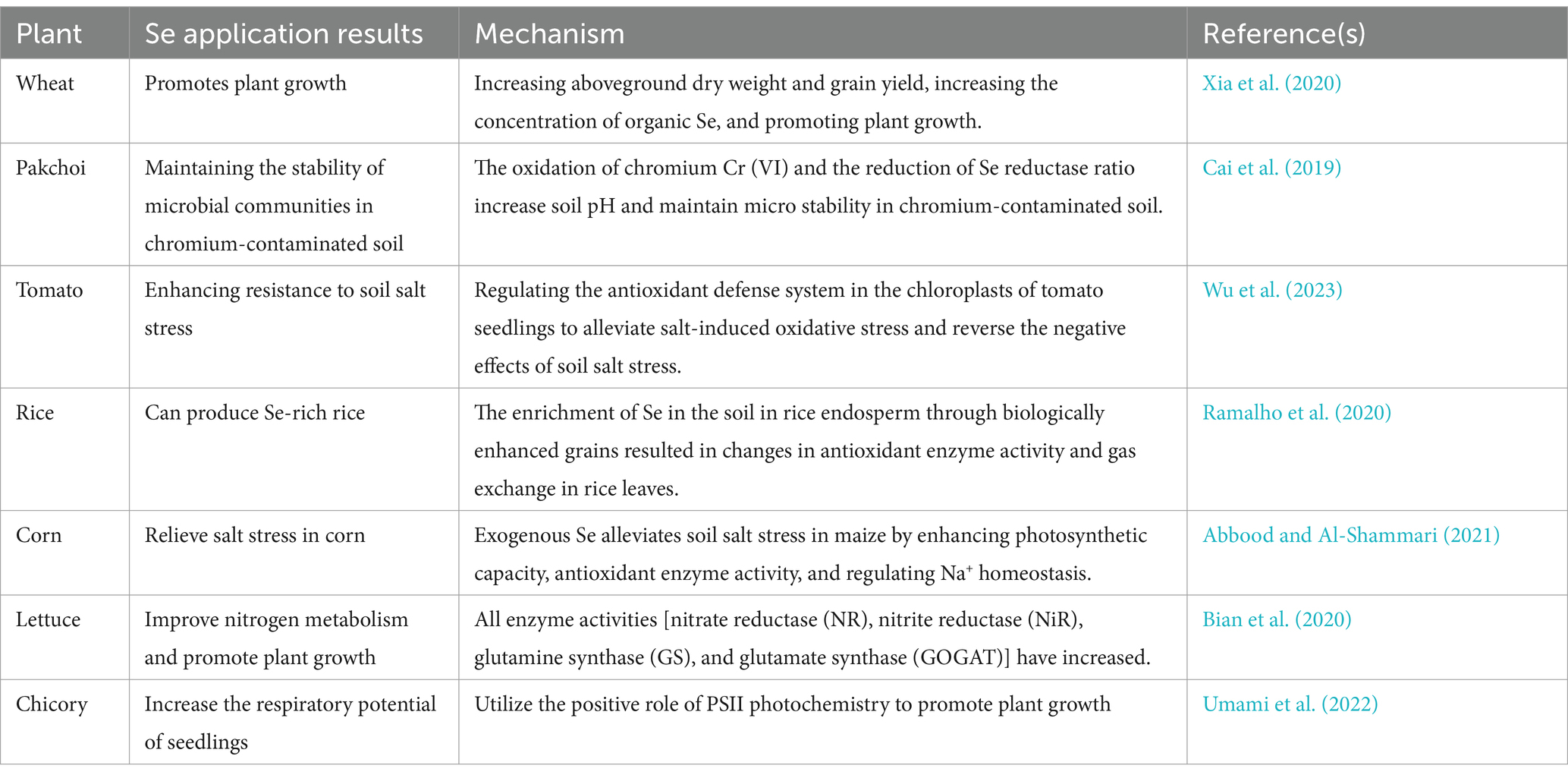
Microbial transformations of Se significantly influence its availability and form, directly affecting plant uptake and health. The form of Se in soil, such as selenides or Se0, cannot be directly absorbed and utilized by plants (Wang et al., 2022e). Microorganisms convert these inorganic forms into more bioavailable compounds, such as selenate and selenite, which plants can readily assimilate. This microbial activity is crucial for enabling plants to benefit from Se, promoting their growth, nutritional quality, and resistance to environmental stress. Se plays multiple roles in plants, including promoting growth, enhancing resistance, and improving crop quality (Table 3).
Studies have shown that the presence of Se-transforming microbes can increase the concentration of essential Se-containing compounds in plants, leading to improved antioxidant enzyme activities and enhanced tolerance to abiotic stresses. Bian et al. (2020) analyzed various enzyme activities—NR, NiR, glutamine synthetase (GS), and glutamate synthase (GOGAT)—in lettuce when different doses and forms (selenate and selenite) of Se were applied. They found that all enzyme activities increased, particularly with the application of selenite. These results indicated that appropriate concentrations of Se could promote the accumulation of starch in plant chloroplasts, benefiting plant growth. Similar conclusions were reached in studies on chicory (Umami et al., 2022) and potatoes (Li et al., 2023) when Se was applied.
Appropriate amounts of Se enhance the activities of several key enzymes, including superoxide dismutase, glutathione reductase, dehydroascorbate reductase, monodehydroascorbate reductase, glutathione peroxidase, and thioredoxin reductase. These enzymes play a crucial role in restoring photosynthetic responses and enzyme activity in the electron transport chain of stressed plants (Liu et al., 2022). The addition of suitable amounts of Se can increase the biomass and chlorophyll content of wheat seedlings as well as their antioxidant capacity. Various Se treatments have been shown to increase the content of antioxidant compounds (anthocyanins, flavonoids, and phenolic compounds) and the activities of antioxidant enzymes (peroxidase and catalase). Optimal Se supplementation can reduce the production of free radicals, decrease lipid peroxidation in membranes, promote biomass accumulation, mitigate chloroplast damage, and increase chlorophyll content (Lanza and Dos Reis, 2021). Das et al. (2022) observed that lower concentrations of Se supplementation improved the growth and arsenic resistance of crops such as rice seedings. Applying Se to rice seedings increased the activity of metallothioneins (MT), thiols, and glutathione-S-transferase (GST), thereby reducing oxidative damage caused by arsenic.
3.3.1 Impact of Se cycling on soil biodiversity and ecological function
Microbial processes are crucial in driving Se cycling within soil ecosystems, significantly influencing soil biodiversity and ecological functions (Wang et al., 2022d). Through reduction, methylation, and volatilization, microorganisms convert Se into various chemical forms, impacting its bioavailability and toxicity (Figure 1). These transformations play a key role in detoxifying Se, converting harmful forms such as selenate into less toxic elemental Se or volatile compounds such as DMSe, which can be released from the soil (Moulick et al., 2024).
The presence of Se in soil can also shape microbial community structures by selecting Se-tolerant species, which may lead to shifts in biodiversity and ecosystem function. Liu et al. (2024) found that Se could successfully decrease soil Cd availability and relieve the harm of Cd in wheat by modifying rhizosphere soil microbial communities. Rosenfeld et al. (2018) suggested that Se contamination shapes communities in favor of Se-tolerant microorganisms. While these shifts can enhance the soil’s ability to cope with Se stress, they may also disrupt the balance of microbial communities, potentially affecting other ecosystem processes.
Furthermore, microbial Se cycling is interconnected with other biogeochemical cycles, such as those of sulfur, carbon, and manganese, further complicating its impact on soil health (Rosenfeld et al., 2020; Li et al., 2022). Understanding the role of microorganisms in Se cycling is vital for developing strategies to manage soil health, particularly in Se-impacted environments. Effective management of these microbial processes can enhance soil resilience, protect biodiversity, and support sustainable agricultural practices.
4 Future perspectives and conclusion
4.1 Future research directions
To effectively utilize Se-reducing or Se-oxidizing microorganisms, future research should focus on the following areas: (1) enhancing microbial resilience and efficiency: screening or engineering microbial strains with high Se reduction capabilities, tolerance to harsh environmental conditions and high Se concentrations, and ensuring ecological safety; (2) optimizing microbial remediation techniques: accelerate microbial processes and establish efficient methods for recovering Se reduction products to make remediation more time-effective; (3) understanding molecular interactions: elucidate the molecular mechanisms of microbial influence on Se redox reactions and their regulation by environmental factors; (4) exploring microbial Se interactions in diverse ecosystems: investigate microbial Se interactions across various, including extreme and human-impacted, environments to understand their ecological roles; (5) developing innovative technologies: create new strategies that leverage microbial Se interactions for improved agricultural productivity and effective remediation of contaminated soils and water; (6) expanding knowledge of Se metabolism: discover and characterize Se-oxidizing bacteria and the enzymatic processes governing Se transformations for a comprehensive understanding of Se metabolism; (7) characterizing microbial roles in Se dynamics: clarify how microorganisms affect soil Se transformations and plant Se uptake to better manage Se cycling in ecosystems; and (8) balancing reduction and oxidation processes: study both Se reduction and the less-explored oxidation processes to develop effective and balanced Se management strategies. By addressing these research directions, the goal is to develop more effective tools and strategies for managing Se in the environment, with profound implications for both environmental science and agriculture. This integrated approach will help mitigate Se-related environmental challenges while enhancing the sustainability of agricultural practices.
5 Conclusion
This review highlights the crucial role of soil microorganisms in the Se cycle, encompassing reduction, oxidation, methylation, and transport processes. These microbial activities are essential for regulating Se bioavailability and mitigating its toxicity, thereby maintaining soil health and ecological balance. The interactions between microorganisms and Se not only enhance soil fertility but also contribute to the resilience of ecosystems in Se-contaminated environments. Understanding these processes is vital for developing effective soil management practices and bioremediation strategies that promote sustainable agricultural and environmental outcomes. Future research should focus on elucidating the molecular mechanisms of microbial Se transformation and exploring novel microbial strains to optimize Se cycling in diverse ecosystems.
Author contributions
ZJ: Writing – original draft. ZW: Writing – review & editing. YZ: Writing – review & editing. MP: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the Natural Science Foundation of Hubei Province (2022CFB674 and 2023AFB484), the Open Fund of Hubei Key Laboratory of Biological Resources Protection and Utilization (Hubei Minzu University) (No. KYPT012408), and the Enshi Science and Technology Plan Project (Young Talent Project) (No. D20220071).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbood, H. Y., and Al-Shammari, A. A. H. (2021). Effect of spraying with Nano selenium and salicylic acid in reducing salt stress and growth and yield of corn (Zea Mays L.). NVEO Nat. Volatiles Essent. Oils J. NVEO, 8, 1903–1922.
Adnan, F., Jalil, A., Ahmed, T., Rahman, A., Dawood, N., Haider, G., et al. (2021). TRAP transporter TakP: a key player in the resistance against selenite-induced oxidative stress in Rhodobacter sphaeroides. Microbiol. Res. 252:126828. doi: 10.1016/j.micres.2021.126828
Afkar, E. (2012). Localization of the dissimilatory arsenate reductase in Sulfurospirillum barnesii strain SeS-3. Am. J. Agric. Biol. Sci. 7, 97–105.
Al-Hagar, O. E., Abol-Fotouh, D., Abdelkhalek, E. S., Elsoud, M. M. A., and Sidkey, N. (2021). Bacillus niabensis OAB2: outstanding bio-factory of selenium nanoparticles. Mater. Chem. Phys. 273:125147. doi: 10.1016/j.matchemphys.2021.125147
Alonso-Fernandes, E., Fernández-Llamosas, H., Cano, I., Serrano-Pelejero, C., Castro, L., Díaz, E., et al. (2023). Enhancing tellurite and selenite bioconversions by overexpressing a methyltransferase from Aromatoleum sp. CIB. Microb. Biotechnol. 16, 915–930. doi: 10.1111/1751-7915.14162
Amin, B. H., Ahmed, H. Y., El Gazzar, E. M., and Badawy, M. M. (2021). Enhancement the mycosynthesis of selenium nanoparticles by using gamma radiation. Dose-Response 19:155932582110593. doi: 10.1177/15593258211059323
An, T., and Kim, Y.-K. (2022). Effect of selenium and tungsten on cell growth and metabolite production in syngas fermentation using “Clostridium autoethanogenum”. J. Biotechnol. 356, 60–64. doi: 10.1016/j.jbiotec.2022.07.004
Anusiya, G., Gowthama Prabu, U., Yamini, N., Sivarajasekar, N., Rambabu, K., Bharath, G., et al. (2021). A review of the therapeutic and biological effects of edible and wild mushrooms. Bioengineered 12, 11239–11268. doi: 10.1080/21655979.2021.2001183
Arias-Paić, M., Tsuchihashi, R., Gress, A., Miller, D., Papendick, J., and Kennedy, A. M. (2022). Treatment of selenium-laden agricultural drainage water using a full-scale bioreactor. J. Environ. Eng. 148:04022014. doi: 10.1061/(ASCE)EE.1943-7870.0001990
Ashengroph, M., and Hosseini, S.-R. (2021). A newly isolated Bacillus amyloliquefaciens SRB04 for the synthesis of selenium nanoparticles with potential antibacterial properties. Int. Microbiol. 24, 103–114. doi: 10.1007/s10123-020-00147-9
Avendaño, R., Muñoz-Montero, S., Rojas-Gätjens, D., Fuentes-Schweizer, P., Vieto, S., Montenegro, R., et al. (2023). Production of selenium nanoparticles occurs through an interconnected pathway of Sulphur metabolism and oxidative stress response in Pseudomonas putida KT2440. Microb. Biotechnol. 16, 931–946. doi: 10.1111/1751-7915.14215
Baggio, G. (2020). Investigation on bacterial selenite reduction to elemental selenium by Bacillus mycoides SeITE01 and Stenotrophomonas maltophilia SeITE02 through spectroscopic and metabolomics analyses, with characterization of biogenic selenium nanoparticles (bio-SeNPs). Verona: Universita degli studi di Verona.
Bébien, M., Kirsch, J., Méjean, V., and Verméglio, A. (2002). Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148, 3865–3872. doi: 10.1099/00221287-148-12-3865
Bian, Z.-H., Bo, L., Cheng, R.-F., Yu, W., Tao, L., and Yang, Q.-C. (2020). Selenium distribution and nitrate metabolism in hydroponic lettuce (Lactuca sativa L.): effects of selenium forms and light spectra. J. Integr. Agric. 19, 133–144. doi: 10.1016/S2095-3119(19)62775-9
Boltz, J. P., and Rittmann, B. E. (2021). Microbial ecology of selenium-respiring bacteria. in Environmental technologies to treat selenium pollution: Principles and engineering, (London: IWA Publishing), 145–170.
Bosu, S., Rajamohan, N., Rajasimman, M., Raut, N., and Vasseghian, Y. (2023). Hydrogen based membrane bioreactor for environmental remediation of pollutants-review on applications and mechanism. Int. J. Hydrog. Energy 48, 6546–6559. doi: 10.1016/j.ijhydene.2021.12.136
Cai, M., Hu, C., Wang, X., Zhao, Y., Jia, W., Sun, X., et al. (2019). Selenium induces changes of rhizosphere bacterial characteristics and enzyme activities affecting chromium/selenium uptake by pak choi (Brassica campestris L. ssp. Chinensis Makino) in chromium contaminated soil. Environ. Pollut. 249, 716–727. doi: 10.1016/j.envpol.2019.03.079
Chaudière, J. (2023). Biological and catalytic properties of selenoproteins. Int. J. Mol. Sci. 24:10109. doi: 10.3390/ijms241210109
Chen, X., and Li, B. (2023). How nature incorporates sulfur and selenium into bioactive natural products. Curr. Opin. Chem. Biol. 76:102377. doi: 10.1016/j.cbpa.2023.102377
Chen, Q., Yu, L., Chao, W., Xiang, J., Yang, X., Ye, J., et al. (2022). Comparative physiological and transcriptome analysis reveals the potential mechanism of selenium accumulation and tolerance to selenate toxicity of Broussonetia papyrifera. Tree Physiol. 42, 2578–2595. doi: 10.1093/treephys/tpac095
Chilala, P., Skalickova, S., and Horky, P. (2024). Selenium status of southern Africa. Nutrients 16:975. doi: 10.3390/nu16070975
Crespo, L., Lucena, B. S., Martínez, F., Mozzi, F., and Pescuma, M. (2024). Selenium bioactive compounds produced by beneficial microbes. Adv. Appl. Microbiol. 126, 63–92.
Das, S., Majumder, B., and Biswas, A. K. (2022). Selenium alleviates arsenic induced stress by modulating growth, oxidative stress, antioxidant defense and thiol metabolism in rice seedlings. Int. J. Phytoremediation 24, 763–777. doi: 10.1080/15226514.2021.1975639
Du, H., Ma, M., Igarashi, Y., and Wang, D. (2019). Biotic and abiotic degradation of methylmercury in aquatic ecosystems: a review. Bull. Environ. Contam. Toxicol. 102, 605–611. doi: 10.1007/s00128-018-2530-2
Ekumah, J.-N., Ma, Y., Akpabli-Tsigbe, N. D. K., Kwaw, E., Ma, S., and Hu, J. (2021). Global soil distribution, dietary access routes, bioconversion mechanisms and the human health significance of selenium: a review. Food Biosci. 41:100960. doi: 10.1016/j.fbio.2021.100960
Escobar-Ramírez, M. C., Castañeda-Ovando, A., Pérez-Escalante, E., Rodríguez-Serrano, G. M., Ramírez-Moreno, E., Quintero-Lira, A., et al. (2021). Antimicrobial activity of se-nanoparticles from bacterial biotransformation. Fermentation 7:130. doi: 10.3390/fermentation7030130
Figueiredo, G. G. O., Lopes, V. R., Romano, T., and Camara, M. C. (2020). “Clostridium” in Beneficial microbes in agro-ecology (Netherlands: Elsevier), 477–491.
Fischer, S., Krause, T., Lederer, F., Merroun, M. L., Shevchenko, A., Hübner, R., et al. (2020). Bacillus safensis JG-B5T affects the fate of selenium by extracellular production of colloidally less stable selenium nanoparticles. J. Hazard. Mater. 384:121146. doi: 10.1016/j.jhazmat.2019.121146
Gao, P., Zhang, X., Huang, X., Chen, Z., Marietou, A., Holmkvist, L., et al. (2023). Genomic insight of sulfate reducing bacterial genus Desulfofaba reveals their metabolic versatility in biogeochemical cycling. BMC Genomics 24:209. doi: 10.1186/s12864-023-09297-2
Garg, N., Bharti, A., Sharma, A., Saroy, K., Cheema, A., and Bisht, A. (2020). “Prokaryotic and eukaryotic microbes: potential tools for detoxification and bioavailability of metalloids” in Metalloids in plants: advances and future prospects, (New Jersey: John Wiley & Sons), 149–183.
Gebreeyessus, G. D., and Zewge, F. (2019). A review on environmental selenium issues. SN Appl. Sci. 1:55. doi: 10.1007/s42452-018-0032-9
Genchi, G., Lauria, G., Catalano, A., Sinicropi, M. S., and Carocci, A. (2023). Biological activity of selenium and its impact on human health. Int. J. Mol. Sci. 24:2633. doi: 10.3390/ijms24032633
Goff, J. L. (2020). The role of microbial sulfur metabolism in biogeochemical cycling of tellurium and selenium. New Jersey: Rutgers The State University of New Jersey, School of Graduate Studies.
Gómez-Gómez, B., Pérez-Corona, T., Mozzi, F., Pescuma, M., and Madrid, Y. (2019). Silac-based quantitative proteomic analysis of Lactobacillus reuteri CRL 1101 response to the presence of selenite and selenium nanoparticles. J. Proteome 195, 53–65. doi: 10.1016/j.jprot.2018.12.025
Gonzalez-Gil, G., Lens, P. N., and Saikaly, P. E. (2016). Selenite reduction by anaerobic microbial aggregates: microbial community structure, and proteins associated to the produced selenium spheres. Front. Microbiol. 7:179111. doi: 10.3389/fmicb.2016.00571
Graves, S. D., Liber, K., Palace, V., Hecker, M., Doig, L. E., and Janz, D. M. (2019). Effects of selenium on benthic macroinvertebrates and fathead minnow (Pimephales promelas) in a boreal lake ecosystem. Ecotoxicol. Environ. Saf. 182:109354. doi: 10.1016/j.ecoenv.2019.06.037
Guo, P., Du, H., Zhao, W., Xiong, B., Wang, M., He, M., et al. (2024b). Selenium-and chitosan-modified biochars reduce methylmercury contents in rice seeds with recruiting Bacillus to inhibit methylmercury production. J. Hazard. Mater. 465:133236. doi: 10.1016/j.jhazmat.2023.133236
Guo, J., Luo, X., Zhang, Q., Duan, X., Yuan, Y., and Zheng, S. (2024a). Contributions of selenium-oxidizing bacteria to selenium biofortification and cadmium bioremediation in a native seleniferous cd-polluted sandy loam soil. Ecotoxicol. Environ. Saf. 272:116081. doi: 10.1016/j.ecoenv.2024.116081
Guo, Q., Ye, J., Zeng, J., Chen, L., Korpelainen, H., and Li, C. (2023). Selenium species transforming along soil–plant continuum and their beneficial roles for horticultural crops. Horticul. Res. 10:uhac270. doi: 10.1093/hr/uhac270
Hashem, A. H., Abdelaziz, A. M., Askar, A. A., Fouda, H. M., Khalil, A. M., Abd-Elsalam, K. A., et al. (2021). Bacillus megaterium-mediated synthesis of selenium nanoparticles and their antifungal activity against Rhizoctonia solani in faba bean plants. J. Fungi 7:195. doi: 10.3390/jof7030195
Hendawy, A. O., Sugimura, S., Sato, K., Mansour, M. M., Abd El-Aziz, A. H., Samir, H., et al. (2021). Effects of selenium supplementation on rumen microbiota, rumen fermentation, and apparent nutrient digestibility of ruminant animals: a review. Fermentation 8:4. doi: 10.3390/fermentation8010004
Ho, C. T., Nguyen, T.-H., Lam, T.-T., Le, D.-Q., Nguyen, C. X., Lee, J. H., et al. (2021). Biogenic synthesis of selenium nanoparticles by Shewanella sp. HN-41 using a modified bioelectrochemical system. Electron. J. Biotechnol. 54, 1–7. doi: 10.1016/j.ejbt.2021.07.004
Hoffman, K. S., Chung, C. Z., Mukai, T., Krahn, N., Jiang, H.-K., Balasuriya, N., et al. (2023). Recoding UAG to selenocysteine in Saccharomyces cerevisiae. RNA 29, 1400–1410. doi: 10.1261/rna.079658.123
Hoyos, B. S., Hernandez-Tenorio, F., Miranda, A. M., Villanueva-Mejía, D. F., and Sáez, A. A. (2023). Systematic analysis of genes related to selenium bioaccumulation in microalgae: a review. Biology 12:703. doi: 10.3390/biology12050703
Jiao, L., Zhang, L., Zhang, Y., Wang, R., Lu, B., and Liu, X. (2022). Transcriptome analysis provides new insight into the distribution and transport of selenium and its associated metals in selenium-rich rice. Environ. Pollut. 301:118980. doi: 10.1016/j.envpol.2022.118980
Kang, Y., Ming, J., Fu, W., Long, L., Wen, X., Zhang, Q., et al. (2024). Selenium fertilizer improves microbial community structure and diversity of Rhizospheric soil and selenium accumulation in tomato plants. Commun. Soil Sci. Plant Anal. 55, 1430–1444. doi: 10.1080/00103624.2024.2315931
Kang, R., Wang, W., Liu, Y., Huang, S., Xu, J., Zhao, L., et al. (2022). Dietary selenium sources alleviate immune challenge induced by Salmonella Enteritidis potentially through improving the host immune response and gut microbiota in laying hens. Front. Immunol. 13:928865. doi: 10.3389/fimmu.2022.928865
Kessi, J., Turner, R. J., and Zannoni, D. (2022). Tellurite and selenite: how can these two oxyanions be chemically different yet so similar in the way they are transformed to their metal forms by bacteria? Biol. Res. 55:17. doi: 10.1186/s40659-022-00378-2
Khanna, K., Kumar, P., Ohri, P., and Bhardwaj, R. (2023). Harnessing the role of selenium in soil–plant-microbe ecosystem: Ecophysiological mechanisms and future prospects. Plant Growth Regul. 100, 197–217. doi: 10.1007/s10725-022-00830-z
Kieliszek, M., Bierla, K., Jiménez-Lamana, J., Kot, A. M., Alcántara-Durán, J., Piwowarek, K., et al. (2020). Metabolic response of the yeast Candida utilis during enrichment in selenium. Int. J. Mol. Sci. 21:5287. doi: 10.3390/ijms21155287
Kuroda, M., Yamashita, M., Miwa, E., Imao, K., Fujimoto, N., Ono, H., et al. (2011). Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J. Bacteriol. 193, 2141–2148. doi: 10.1128/JB.01197-10
Kushwaha, A., Goswami, L., Lee, J., Sonne, C., Brown, R. J., and Kim, K.-H. (2022). Selenium in soil-microbe-plant systems: sources, distribution, toxicity, tolerance, and detoxification. Crit. Rev. Environ. Sci. Technol. 52, 2383–2420. doi: 10.1080/10643389.2021.1883187
Lampis, S., and Vallini, G. (2021). Microbial reduction of selenium oxyanions: energy-yielding and detoxification reactions. in Environmental technologies to treat selenium pollution: Principles and engineering, (London: IWA Publishing), 101–143.
Lanza, M. G. D. B., and Dos Reis, A. R. (2021). Roles of selenium in mineral plant nutrition: ROS scavenging responses against abiotic stresses. Plant Physiol. Biochem. 164, 27–43. doi: 10.1016/j.plaphy.2021.04.026
Lashani, E., Moghimi, H., Turner, R. J., and Amoozegar, M. A. (2023). Selenite bioreduction by a consortium of halophilic/halotolerant bacteria and/or yeasts in saline media. Environ. Pollut. 331:121948. doi: 10.1016/j.envpol.2023.121948
Lazard, M. (2021). Selenium metabolism and toxicity in the yeast Saccharomyces cerevisiae. Med. Res. Arch. 9, 1–13. doi: 10.18103/mra.v9i9.2550
Li, J., Awasthi, M. K., Xing, W., Liu, R., Bao, H., Wang, X., et al. (2020). Arbuscular mycorrhizal fungi increase the bioavailability and wheat (Triticum aestivum L.) uptake of selenium in soil. Ind. Crop. Prod. 150:112383. doi: 10.1016/j.indcrop.2020.112383
Li, S., Chen, H., Jiang, S., Hu, F., Xing, D., and Du, B. (2023). Selenium and nitrogen fertilizer management improves potato root function, photosynthesis, yield and selenium enrichment. Sustain. For. 15:6060. doi: 10.3390/su15076060
Li, L., Zhang, B., Li, L., and Borthwick, A. G. (2022). Microbial selenate detoxification linked to elemental sulfur oxidation: independent and synergic pathways. J. Hazard. Mater. 422:126932. doi: 10.1016/j.jhazmat.2021.126932
Li, Q., Zhou, S., and Liu, N. (2021). Diversity of endophytic bacteria in Cardamine hupingshanensis and potential of culturable selenium-resistant endophytes to enhance seed germination under selenate stress. Curr. Microbiol. 78, 2091–2103. doi: 10.1007/s00284-021-02444-6
Liang, X., Perez, M. A. M.-J., Nwoko, K. C., Egbers, P., Feldmann, J., Csetenyi, L., et al. (2019). Fungal formation of selenium and tellurium nanoparticles. Appl. Microbiol. Biotechnol. 103, 7241–7259. doi: 10.1007/s00253-019-09995-6
Liao, Q., Li, A.-M., Xing, Y., Liang, P.-X., Jiang, Z.-P., Liu, Y.-X., et al. (2024). Selenobacteria-mediated se transformation and uptake involving the unique genetic code. Front. Plant Sci. 15:1392355. doi: 10.3389/fpls.2024.1392355
Liu, F., Huang, J.-C., Zhou, C., Gao, W., Xia, S., He, S., et al. (2019). Development of an algal treatment system for selenium removal: effects of environmental factors and post-treatment processing of se-laden algae. J. Hazard. Mater. 365, 546–554. doi: 10.1016/j.jhazmat.2018.11.017
Liu, H., Wang, H., Nie, Z., Tao, Z., Peng, H., Shi, H., et al. (2024). Combined application of arbuscular mycorrhizal fungi and selenium fertilizer increased wheat biomass under cadmium stress and shapes rhizosphere soil microbial communities. BMC Plant Biol. 24:359. doi: 10.1186/s12870-024-05032-5
Liu, H., Xiao, C., Qiu, T., Deng, J., Cheng, H., Cong, X., et al. (2022). Selenium regulates antioxidant, photosynthesis, and cell permeability in plants under various abiotic stresses: a review. Plan. Theory 12:44. doi: 10.3390/plants12010044
Luo, X., Wang, Y., Lan, Y., An, L., Wang, G., Li, M., et al. (2022). Microbial oxidation of organic and elemental selenium to selenite. Sci. Total Environ. 833:155203. doi: 10.1016/j.scitotenv.2022.155203
Manta, B., Makarova, N. E., and Mariotti, M. (2022). The selenophosphate synthetase family: a review. Free Radic. Biol. Med. 192, 63–76. doi: 10.1016/j.freeradbiomed.2022.09.007
Mehri, A. (2020). Trace elements in human nutrition (II)–an update. Int. J. Prev. Med. 11:2. doi: 10.4103/ijpvm.IJPVM_48_19
Nancharaiah, Y. V., and Lens, P. N. L. (2015). Ecology and biotechnology of selenium-respiring bacteria. Microbiol. Mol. Biol. Rev. 79, 61–80. doi: 10.1128/mmbr.00037-14
Mohammadi, E. (2021). Kinetics of total dissolved selenium removal in a chemostat. Vancouver: University Of British Columbia.
Mohammadi, E., and Baldwin, S. A. (2024). Effect of acetate and methanol on the kinetics of total dissolved selenium removal in a chemostat. J. Chem. Technol. Biotechnol. 99, 1325–1341. doi: 10.1002/jctb.7629
Mohapatra, D. P., Robinson, K. A., Huang, F., Kirpalani, D., and Loewen, M. C. (2022). Insights into increasing Selenate reductase enzyme activity in the presence of nitrogen-doped graphite electrodes for selenium effluent treatment. Water 14:931. doi: 10.3390/w14060931
Moulick, D., Mukherjee, A., Das, A., Roy, A., Majumdar, A., Dhar, A., et al. (2024). Selenium–An environmentally friendly micronutrient in agroecosystem in the modern era: An overview of 50-year findings. Ecotoxicol. Environ. Saf. 270:115832. doi: 10.1016/j.ecoenv.2023.115832
Mu, H., Ye, L., and Wang, B. (2024). Detailed resume of S-methyltransferases: categories, structures, biological functions and research advancements in related pathophysiology and pharmacotherapy. Biochem. Pharmacol. 226:116361. doi: 10.1016/j.bcp.2024.116361
Nagar, D., Braganca, J. M., and Furtado, I. J. (2022). “Extremophilic Bacteria and Archaea in the valorization of metalloids: arsenic, selenium, and tellurium” in Enzymes in the valorization of waste (Florida: CRC Press), 65–80.
Nikam, P. B., Salunkhe, J. D., Minkina, T., Rajput, V. D., Kim, B. S., and Patil, S. V. (2022). A review on green synthesis and recent applications of red nano selenium. Results Chem. 4:100581. doi: 10.1016/j.rechem.2022.100581
Ojeda, J. J., Merroun, M. L., Tugarova, A. V., Lampis, S., Kamnev, A. A., and Gardiner, P. H. (2020). Developments in the study and applications of bacterial transformations of selenium species. Crit. Rev. Biotechnol. 40, 1250–1264. doi: 10.1080/07388551.2020.1811199
Otsuka, O., and Yamashita, M. (2020). Selenium recovery from wastewater using the selenate-reducing bacterium Pseudomonas stutzeri NT-I. Hydrometallurgy 197:105470. doi: 10.1016/j.hydromet.2020.105470
Padhi, Y., and Chatterjee, S. (2023). XdfA, a novel membrane-associated DedA family protein of Xanthomonas campestris, is required for optimum virulence, maintenance of magnesium, and membrane homeostasis. MBio 14, e01361–e01323. doi: 10.1128/mbio.01361-23
Pal, A., Bhattacharjee, S., Saha, J., Sarkar, M., and Mandal, P. (2022). Bacterial survival strategies and responses under heavy metal stress: a comprehensive overview. Crit. Rev. Microbiol. 48, 327–355. doi: 10.1080/1040841X.2021.1970512
Peng, J.-J., Yue, S.-Y., Fang, Y.-H., Liu, X.-L., and Wang, C.-H. (2021). Mechanisms affecting the biosynthesis and incorporation rate of selenocysteine. Molecules 26:7120. doi: 10.3390/molecules26237120
Petrović, M. (2021). Selenium: widespread yet scarce, essential yet toxic. ChemTexts 7:11. doi: 10.1007/s40828-021-00137-y
Piacenza, E., Sule, K., Presentato, A., Wells, F., Turner, R. J., and Prenner, E. J. (2023). Impact of biogenic and Chemogenic selenium nanoparticles on model eukaryotic lipid membranes. Langmuir 39, 10406–10419. doi: 10.1021/acs.langmuir.3c00718
Pyrzyńska, K. (1996). Speciation analysis of some organic selenium compounds. A review. Analyst 121, 77R–83R. doi: 10.1039/AN996210077R
Rahman, M. M., Hossain, K. F. B., Banik, S., Sikder, M. T., Akter, M., Bondad, S. E. C., et al. (2019). Selenium and zinc protections against metal-(loids)-induced toxicity and disease manifestations: a review. Ecotoxicol. Environ. Saf. 168, 146–163. doi: 10.1016/j.ecoenv.2018.10.054
Ramalho, J. C., Roda, F. A., Pessoa, M. F., Reboredo, F. H., Pais, I. P., Ndayiragije, A., et al. (2020). Selenium agronomic biofortification in rice: improving crop quality against malnutrition. in Future rice demand (Singapore: Springer), 179–203.
Ranjard, L., Nazaret, S., and Cournoyer, B. (2003). Freshwater bacteria can methylate selenium through the thiopurine methyltransferase pathway. Appl. Environ. Microbiol. 69, 3784–3790. doi: 10.1128/aem.69.7.3784-3790.2003
Rao, S., Yu, T., Cong, X., Lai, X., Xiang, J., Cao, J., et al. (2021). Transcriptome, proteome, and metabolome reveal the mechanism of tolerance to selenate toxicity in Cardamine violifolia. J. Hazard. Mater. 406:124283. doi: 10.1016/j.jhazmat.2020.124283
Reddy, B., and Bandi, R. (2022). “Synthesis of selenium nanoparticles by using microorganisms and Agri-based products” in Agri-waste and microbes for production of sustainable nanomaterials (Netherlands: Elsevier), 655–683.
Ren, H., Li, X., Guo, L., Wang, L., Hao, X., and Zeng, J. (2022). Integrative transcriptome and proteome analysis reveals the absorption and metabolism of selenium in tea plants [Camellia sinensis (L.) O. Kuntze]. Front. Plant Sci. 13:848349. doi: 10.3389/fpls.2022.848349
Rosenfeld, C. E., James, B. R., and Santelli, C. M. (2018). Persistent bacterial and fungal community shifts exhibited in selenium-contaminated reclaimed mine soils. Appl. Environ. Microbiol. 84, e01394–e01318. doi: 10.1128/AEM.01394-18
Rosenfeld, C. E., Sabuda, M. C., Hinkle, M. A., James, B. R., and Santelli, C. M. (2020). A fungal-mediated cryptic selenium cycle linked to manganese biogeochemistry. Environ. Sci. Technol. 54, 3570–3580. doi: 10.1021/acs.est.9b06022
Roux, M., Sarret, G. R., Pignot-Paintrand, I., Fontecave, M., and Coves, J. (2001). Microbial interaction with metals and metalloids. Appl. Environ. Microbiol. 67, 769–773. doi: 10.1128/AEM.67.2.769-773.2001
Ruiz-Fresneda, M. A., Morales-Hidalgo, M., Povedano-Priego, C., Jroundi, F., Hidalgo-Iruela, J., Cano-Cano, M., et al. (2024). Unlocking the key role of bentonite fungal isolates in tellurium and selenium bioremediation and biorecovery: implications in the safety of radioactive waste disposal. Sci. Total Environ. 912:169242. doi: 10.1016/j.scitotenv.2023.169242
Saghafi, D., Bagherifam, S., Hatami, M., and Lajayer, B. A. (2020). The role of microbes in detoxification and availability of metalloids.” in Metalloids in plants: advances and future prospects, (New Jersey: John Wiley & Sons), 65–93.
Santelli, C. M., Sabuda, M. C., and Rosenfeld, C. E. (2023). Time-resolved examination of fungal selenium redox transformations. ACS Earth and Space Chemistry 7, 960–971. doi: 10.1021/acsearthspacechem.2c00288
Sarathchandra, S., and Watkinson, J. (1981). Oxidation of elemental selenium to selenite by Bacillus megaterium. Science 211, 600–601.
Schilling, K., Basu, A., Wanner, C., Sanford, R. A., Pallud, C., Johnson, T. M., et al. (2020). Mass-dependent selenium isotopic fractionation during microbial reduction of seleno-oxyanions by phylogenetically diverse bacteria. Geochim. Cosmochim. Acta 276, 274–288. doi: 10.1016/j.gca.2020.02.036
Scortecci, J. F., Serrao, V. H. B., Fernandes, A. F., Basso, L. G., Gutierrez, R. F., Araujo, A. P. U., et al. (2020). Initial steps in selenocysteine biosynthesis: the interaction between selenocysteine lyase and selenophosphate synthetase. Int. J. Biol. Macromol. 156, 18–26. doi: 10.1016/j.ijbiomac.2020.03.241
Serrão, V. H. B., Minari, K., Pereira, H. D. M., and Thiemann, O. H. (2024). Bacterial selenocysteine synthase structure revealed by single-particle cryoEM. Curr. Res. Struct. Biol. 7:100143. doi: 10.1016/j.crstbi.2024.100143
Sharma, V. K., McDonald, T. J., Sohn, M., Anquandah, G. A., Pettine, M., and Zboril, R. (2015). Biogeochemistry of selenium. A review. Environ. Chem. Lett. 13, 49–58. doi: 10.1007/s10311-014-0487-x
Sharma, S., and Sharma, A. K. (2021). Selenium distribution and chemistry in water and soil. in Selenium contamination water, (New Jersey: John Wiley & Sons), 20–38.
Siddharthan, S., Thangaraj, S., Paulraj, S., RajaMohmed, B., Rakkamuthu, K., Dharmaraj, V., et al. (2024). Myco-remediation of selenium contaminated environment and future prospects: An overview. Environ. Qual. Manag. 33, 869–877. doi: 10.1002/tqem.22159
Sinharoy, A., and Lens, P. N. (2020). Biological removal of selenate and selenite from wastewater: options for selenium recovery as nanoparticles. Curr. Pollut. Rep. 6, 230–249. doi: 10.1007/s40726-020-00146-4
Sinharoy, A., and Lens, P. N. (2022). Biological selenate and selenite reduction by waste activated sludge using hydrogen as electron donor. J. Environ. Manag. 319:115745. doi: 10.1016/j.jenvman.2022.115745
Srivastava, P., Sachan, K., Baskar, P., Saikanth, D., Lytand, W., Kumar, R., et al. (2023). Soil microbes expertly balancing nutrient demands and environmental preservation and ensuring the delicate stability of our ecosystems-a review. Int. J. Plant Soil Sci. 35, 989–1000. doi: 10.9734/ijpss/2023/v35i183363
Staicu, L. C., and Barton, L. L. (2021). Selenium respiration in anaerobic bacteria: does energy generation pay off? J. Inorg. Biochem. 222:111509. doi: 10.1016/j.jinorgbio.2021.111509
Staicu, L. C., Dziurzyński, M., Wόjtowicz, P. J., and Gόrecki, A. (2024). “The impact of selenium on an Archaea-dominated, methanogenic granular sludge consortium” in Geomicrobiology: Natural and anthropogenic settings (Switzerland: Springer), 33–49.
Surachat, K., Kantachote, D., Deachamag, P., and Wonglapsuwan, M. (2022). In silico genomic analysis of Rhodopseudomonas palustris strains revealed potential biocontrol agents and crop yield enhancers. Biol. Control 176:105085. doi: 10.1016/j.biocontrol.2022.105085
Telo da Gama, J. (2023). The role of soils in sustainability, climate change, and ecosystem services: challenges and opportunities. Ecologies 4, 552–567. doi: 10.3390/ecologies4030036
Tendenedzai, J. T., Brink, H. G., and Chirwa, E. M. (2021). Formation of elemental selenium nanoparticles (SeNPs) from the reduction of selenite (SeO32-) by a pure culture of Pseudomonas Stutzeri NT-i. Chem. Eng. Trans. 86, 193–198. doi: 10.3303/CET2186033
Tendenedzai, J. T., Chirwa, E. M., and Brink, H. G. (2022). Enterococcus spp. cell-free extract: An abiotic route for synthesis of selenium nanoparticles (SeNPs), their characterisation and inhibition of Escherichia coli. Nano 12:658. doi: 10.3390/nano12040658
Tugarova, A. V., Mamchenkova, P. V., Khanadeev, V. A., and Kamnev, A. A. (2020). Selenite reduction by the rhizobacterium Azospirillum brasilense, synthesis of extracellular selenium nanoparticles and their characterisation. New Biotechnol. 58, 17–24. doi: 10.1016/j.nbt.2020.02.003
Ullah, H., Liu, G., Yousaf, B., Ali, M. U., Irshad, S., Abbas, Q., et al. (2019). A comprehensive review on environmental transformation of selenium: recent advances and research perspectives. Environ. Geochem. Health 41, 1003–1035. doi: 10.1007/s10653-018-0195-8
Umami, N., Rahayu, E., Suhartanto, B., Agus, A., and Rahman, M. (2022). Selenium application in improving chicory (Cichorium intybus) productivity and quality. Trop. Animal Sci. J. 45, 337–347. doi: 10.5398/tasj.2022.45.3.337
Wang, J., Dai, J., Chen, G., and Jiang, F. (2022c). Role of sulfur biogeochemical cycle in mercury methylation in estuarine sediments: a review. J. Hazard. Mater. 423:126964. doi: 10.1016/j.jhazmat.2021.126964
Wang, Z., Huang, W., and Pang, F. (2022e). Selenium in soil–plant-microbe: a review. Bullet. Environ. Contaminat. Toxicol. 108, 167–181. doi: 10.1007/s00128-021-03386-2
Wang, F.-F., Liu, G.-P., Zhang, F., Li, Z.-M., Yang, X.-L., Yang, C.-D., et al. (2022b). Natural selenium stress influences the changes of antibiotic resistome in seleniferous forest soils. Environ. Microb. 17:26. doi: 10.1186/s40793-022-00419-z
Wang, D., Rensing, C., and Zheng, S. (2022a). Microbial reduction and resistance to selenium: mechanisms, applications and prospects. J. Hazard. Mater. 421:126684.doi: 10.1016/j.jhazmat.2021.126684
Wang, Y., Shi, X., Huang, X., Huang, C., Wang, H., Yin, H., et al. (2022d). Linking microbial community composition to farming pattern in selenium-enriched region: potential role of microorganisms on se geochemistry. J. Environ. Sci. 112, 269–279. doi: 10.1016/j.jes.2021.05.015
Wasserman, N. L., Schilling, K., Johnson, T. M., and Pallud, C. (2021). Selenium isotope shifts during the oxidation of selenide-bearing minerals. ACS Earth Space Chem. 5, 1140–1149. doi: 10.1021/acsearthspacechem.1c00036
Wells, M., Basu, P., and Stolz, J. F. (2021). The physiology and evolution of microbial selenium metabolism. Metallomics 13:mfab024. doi: 10.1093/mtomcs/mfab024
Wells, M., and Stolz, J. F. (2020). Microbial selenium metabolism: a brief history, biogeochemistry and ecophysiology. FEMS Microbiol. Ecol. 96:fiaa209. doi: 10.1093/femsec/fiaa209
Wu, H., Fan, S., Gong, H., and Guo, J. (2023). Roles of salicylic acid in selenium-enhanced salt tolerance in tomato plants. Plant Soil 484, 569–588. doi: 10.1007/s11104-022-05819-1
Xia, Q., Yang, Z., Shui, Y., Liu, X., Chen, J., Khan, S., et al. (2020). Methods of selenium application differentially modulate plant growth, selenium accumulation and speciation, protein, anthocyanins and concentrations of mineral elements in purple-grained wheat. Front. Plant Sci. 11:1114. doi: 10.3389/fpls.2020.01114
Xu, R., Kolton, M., Tao, W., Sun, X., Su, P., Huang, D., et al. (2023). Anaerobic selenite-reducing bacteria and their metabolic potentials in se-rich sediment revealed by the combination of DNA-stable isotope probing, metagenomic binning, and metatranscriptomics. J. Hazard. Mater. 457:131834. doi: 10.1016/j.jhazmat.2023.131834
Xue, S.-J., Zhang, X.-T., Li, X.-C., Zhao, F.-Y., Shu, X., Jiang, W.-W., et al. (2024). Multi-pathways-mediated mechanisms of selenite reduction and elemental selenium nanoparticles biogenesis in the yeast-like fungus Aureobasidium melanogenum I15. J. Hazard. Mater. 470:134204. doi: 10.1016/j.jhazmat.2024.134204
Yadav, P., Pandey, S., and Dubey, S. K. (2023). Selenite bioreduction with concomitant green synthesis of selenium nanoparticles by a selenite resistant EPS and siderophore producing terrestrial bacterium. Biometals 36, 1027–1045. doi: 10.1007/s10534-023-00503-y
Yan, S., Cheng, K. Y., Bohu, T., Morris, C., Lomheim, L., Yang, I., et al. (2023). Microbial community dynamics in a feedback-redox controlled bioreactor process that enabled sequential selenate, nitrate and sulfate removal, and elemental selenium recovery. J. Water Process Eng. 54:103881. doi: 10.1016/j.jwpe.2023.103881
Yang, X., Mao, K., Chang, C., Wang, J., Wu, Q., Shao, Y., et al. (2024). Interactions of selenium and mercury in soil–plant systems: characterizations, occurrences, and mechanisms. Crit. Rev. Environ. Sci. Technol., 1–19. doi: 10.1080/10643389.2024.2332135
Ye, Y., Qu, J., Pu, Y., Rao, S., Xu, F., and Wu, C. (2020). Selenium biofortification of crop food by beneficial microorganisms. J. Fungi 6:59. doi: 10.3390/jof6020059
Yu, Q., Boyanov, M. I., Liu, J., Kemner, K. M., and Fein, J. B. (2018). Adsorption of selenite onto Bacillus subtilis: the overlooked role of cell envelope sulfhydryl sites in the microbial conversion of se (IV). Environ. Sci. Technol. 52, 10400–10407. doi: 10.1021/acs.est.8b02280
Yuan, Z., Fang, Y., He, M., Wang, X., and Pan, F. (2024). An optimized method for the bioremediation of se pollution and the biosynthesis of se nanoparticles with sodium selenite-reducing Bacteria Stenotrophomonas sp. EGS12. J. Biobaased Mater. Bioenergy 18, 810–818. doi: 10.1166/jbmb.2024.2428
Zambonino, M. C., Quizhpe, E. M., Jaramillo, F. E., Rahman, A., Santiago Vispo, N., Jeffryes, C., et al. (2021). Green synthesis of selenium and tellurium nanoparticles: current trends, biological properties and biomedical applications. Int. J. Mol. Sci. 22:989. doi: 10.3390/ijms22030989
Zayed, A., Pilon-Smits, E., deSouza, M., Lin, Z.-Q., and Terry, N. (2020). “Remediation of selenium-polluted soils and waters by phytovolatilization” in Phytoremediation of contaminated soil and water (CRC Press), 61–83.
Zhang, G., Chen, X., Li, F., Que, W., Qian, J., Fang, J., et al. (2023a). Effects of environmental factors on selenite volatilization by freshwater microalgae. Sci. Total Environ. 854:158539. doi: 10.1016/j.scitotenv.2022.158539
Zhang, Y., Jin, J., Huang, B., Ying, H., He, J., and Jiang, L. (2022b). Selenium metabolism and selenoproteins in prokaryotes: a bioinformatics perspective. Biomol. Ther. 12:917. doi: 10.3390/biom12070917
Zhang, X., Wang, L., Zeng, T., Liu, Y., Wang, G., Liu, J., et al. (2022a). The removal of selenite and cadmium by immobilized biospheres: efficiency, mechanisms and bacterial community. Environ. Res. 211:113025. doi: 10.1016/j.envres.2022.113025
Zhang, Z., Xiong, Y., Chen, H., and Tang, Y. (2020). Understanding the composition and spatial distribution of biological selenate reduction products for potential selenium recovery. Environ. Sci. 6, 2153–2163. doi: 10.1039/D0EW00376J
Zhang, X., Zhong, M., Zhou, R., Qin, W., and Si, Y. (2023b). Se (IV) reduction and extracellular biosynthesis of Nano-se (0) by Shewanella oneidensis MR-1 and Shewanella putrefaciens. Process Biochem. 130, 481–491. doi: 10.1016/j.procbio.2023.05.016
Zhao, L., Liu, M., Sun, H., Yang, J.-C., Huang, Y.-X., Huang, J.-Q., et al. (2023). Selenium deficiency-induced multiple tissue damage with dysregulation of immune and redox homeostasis in broiler chicks under heat stress. Sci. China Life Sci. 66, 2056–2069. doi: 10.1007/s11427-022-2226-1
Zhou, X., Yang, J., Kronzucker, H. J., and Shi, W. (2020). Selenium biofortification and interaction with other elements in plants: a review. Front. Plant Sci. 11:586421. doi: 10.3389/fpls.2020.586421
Zhu, D., Niu, Y., Fan, K., Zhang, F., Wang, Y., Wang, G., et al. (2021). Selenium-oxidizing Agrobacterium sp. T3F4 steadily colonizes in soil promoting selenium uptake by pak choi (Brassica campestris). Sci. Total Environ. 791:148294. doi: 10.1016/j.scitotenv.2021.148294
Keywords: selenium, soil, microorganisms, biogeochemical cycling, selenium reduction
Citation: Jiang Z, Wang Z, Zhao Y and Peng M (2024) Unveiling the vital role of soil microorganisms in selenium cycling: a review. Front. Microbiol. 15:1448539. doi: 10.3389/fmicb.2024.1448539
Edited by:
Cui Li, Northwestern Polytechnical University, ChinaReviewed by:
Claudia Muñoz-Villagrán, University of Santiago, ChileRisheng Xu, Northwestern Polytechnical University, China
Copyright © 2024 Jiang, Wang, Zhao and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mu Peng, cGVuZ211MTAyNUBob3RtYWlsLmNvbQ==
 Zhihui Jiang
Zhihui Jiang Zhiyong Wang1,2
Zhiyong Wang1,2 Mu Peng
Mu Peng