- Department of Hematology, Nanjing Drum Tower Hospital, Affiliated Hospital of Medical School, Nanjing University, Nanjing, China
Background: Primary immune thrombocytopenia (ITP) is an immune-mediated hematologic disorder characterized by a reduction in platelet count, increasing the risk of bleeding. Recent studies have indicated a close association between alterations in gut microbiota and the development of ITP. However, the mechanisms by which gut microbiota influence the occurrence and progression of ITP through plasma metabolites remain poorly understood. Evidence suggests extensive interactions between gut microbiota and plasma metabolites, implying a potential role for gut microbiota in influencing ITP through alterations in plasma metabolites, which requires further investigation.
Methods: In this study, summarized GWAS data (including 211 gut microbiota taxa, 1,400 plasma metabolites or ratios, and an ITP patient cohort) were retrieved from the MiBioGen and GWAS Catalog databases. Using a two-sample Mendelian randomization (MR) approach, we screened gut microbiota and plasma metabolites potentially causally related to ITP. We further identified plasma metabolites serving as mediators through which gut microbiota affect ITP and calculated the strength of the mediation effect. To ensure result stability, we primarily used the inverse variance weighted (IVW) method as the main judgment index. We also utilized MR Egger and inverse variance weighted methods to detect heterogeneity in the results, and employed MR-Egger and MR-PRESSO methods to assess the presence of pleiotropy.
Results: Though two-sample MR analysis, 8 gut microbiota taxa were found to have causal relationships with ITP. After excluding six plasma metabolites with pleiotropy, 39 plasma metabolites were found to be causally related to ITP (P < 0.05). Eleven plasma metabolites were identified as having causal relationships between gut microbiota and plasma metabolites. Finally, using the delta method, it was calculated that Sphingomyelin levels (8.0%, 95%CI: 0.9% to 11.5%, P = 0.047) and Glucose-to-mannose ratio (6.5%, 95%CI: 0.7% to 9.5%, P = 0.039) are intermediates for Intestinimonas influencing ITP, while Bilirubin (Z,Z) to etiocholanolone glucuronide ratio (5.6%, 95%CI: 4.7% to 6.9%, P = 0.043) is an intermediate for Senegalimassilia influencing ITP.
Conclusion: Gut microbiota can influence the development of ITP through changes in plasma metabolites. Sphingomyelin levels, Glucose-to-mannose ratio, and Bilirubin (Z,Z) to etiocholanolone glucuronide ratio are newly discovered intermediates through which gut microbiota influence ITP, providing potential indicators and targets for clinical diagnosis and treatment. This study highlights the intricate relationship between gut microbiota and plasma metabolites in the context of ITP, suggesting new avenues for clinical diagnosis and treatment.
Introduction
Primary immune thrombocytopenia (ITP) is an autoimmune disorder characterized by a reduced platelet count, leading to increased bleeding and bruising (Rodeghiero, 2023). The incidence of ITP is approximately 3.3 per 100,000 adults annually, with a slightly higher prevalence in females and individuals over 60 years of age (Bussel et al., 2023). The pathogenesis of ITP is primarily due to the immune system erroneously targeting and destroying platelets. Autoantibodies, particularly those against platelet glycoproteins such as GPIIb/IIIa and GPIb/IX, play a central role in this process (Semple et al., 2020). In addition to antibody-mediated destruction, T-cells and the accumulation of certain metabolites may directly or indirectly lyse platelets or impair megakaryocyte function, leading to reduced platelet production (Malik et al., 2023; Zhang et al., 2022). Known risk factors for ITP include viral infections, certain medications, and genetic predispositions, as indicated by familial cases and associations with specific HLA types (Cines, 2023).
Emerging research highlights the role of gut microbiota in the pathogenesis of ITP. Dysbiosis, or imbalance in the gut microbiota, may influence immune responses and contribute to the development and progression of ITP (Wang et al., 2021; Liu et al., 2020; Wang et al., 2023). However, the precise mechanisms underlying the influence of gut microbiota on ITP remain unclear and warrant further investigation.
Metabolites play a significant role in the pathogenesis of autoimmune diseases, including ITP (Zhang et al., 2022; Yang and Cong, 2021). Recent research has shown that plasma metabolic disturbances can influence immune cell function and contribute to the dysregulation seen in autoimmune conditions (Fernández-Ochoa et al., 2020). In autoimmune diseases, metabolites such as short-chain fatty acids (SCFAs), amino acids, and lipid mediators can modulate immune responses, either exacerbating or alleviating inflammation (Rasouli-Saravani et al., 2023; Mondanelli et al., 2019). In the context of ITP, studies have identified specific metabolites that may be involved in disease progression and severity (Wen et al., 2022). However, the precise roles of these metabolites in ITP remain elusive. The gut microbiota, a complex community of microorganisms residing in the gastrointestinal tract, significantly impacts the host's metabolic landscape through the production and modulation of various metabolites (Honda and Littman, 2016). For instance, gut microbiota ferments dietary fibers to produce SCFAs such as acetate, propionate, and butyrate, which play critical roles in maintaining gut barrier integrity, modulating immune responses, and providing energy sources for colonic cells. These SCFAs can enter the bloodstream, influencing systemic metabolic processes and exerting anti-inflammatory effects crucial in managing autoimmune diseases (Du et al., 2022). Additionally, gut bacteria modify primary bile acids into secondary bile acids, which regulate host metabolism and immune function by interacting with receptors like the farnesoid X receptor and G protein-coupled bile acid receptor 1 (Sepe et al., 2016). Gut microbiota also metabolizes tryptophan into various metabolites that modulate immune responses and intestinal health. In the context of lipid metabolism, gut bacteria influence sphingolipid levels, which are involved in cell signaling and immune regulation, and fatty acid profiles, impacting systemic inflammation and immune function (Rooks and Garrett, 2016). These findings suggest a complex interaction between microbial communities and host metabolism in the pathogenesis of autoimmune diseases. Therefore, it is reasonable to speculate that causal relationships may exist between gut microbiota, plasma metabolites, and ITP. Our study aims to elucidate these potential associations and identify specific metabolites that could serve as valuable tools for early diagnosis and potential clinical treatment.
Mendelian randomization (MR), an approach that utilizes genetic variants as instrumental variables (IVs), is instrumental in establishing causal relationships between exposures and clinical outcomes while controlling for confounders and mitigating reverse causation bias (Davies et al., 2018). Increasing evidence supports the utility of human genetic data related to gut microbial characteristics in clinical investigations, thereby positioning MR as a powerful tool to infer causal links between gut microbiota and ITP. In this study, we employed a two-step MR analysis and mediation analyses using summary statistics from the most extensive and current genome-wide association studies (GWAS) of the gut microbiota, plasma metabolites, and ITP. These analyses aimed to elucidate the intricate associations between these variables, providing valuable insights into their causal relationships.
Methods
Study design
The study mainly conducted two steps of analyses as described in Figure 1: Step 1: the analysis of causal effects of 196 gut microbiota taxa on ITP; Step 2: analysis of candidate plasma metabolites as the mediation bridged the gut microbiota and ITP. In brief, the causal relationships between 1,400 plasma metabolites and ITP were analyzed. Then, the plasma metabolites positively associated with ITP were further screened for causal relationships with gut microbiota. Finally, the mediated effect of plasma metabolites was calculated. Mendelian randomization is based on three core assumptions: (1) the IVs are closely associated with the exposure factors. In our study, we ensured a strong correlation between genetic IVs and exposure factors (gut microbiota, plasma metabolites, or ITP) by removing linkage disequilibrium and strict correlation; (2) IVs are not associated with confounding factors. To ensure this, we used two sensitivity analysis methods (MR-PRESSO regression and MR Egger regression) to exclude the influence of confounding factors on the reliability of the results; (3) IVs do not affect the outcome directly, and it can only affect outcome via the exposure. All genetic instrumental variables were verified to have no association with the outcomes (Bowden and Holmes, 2019).
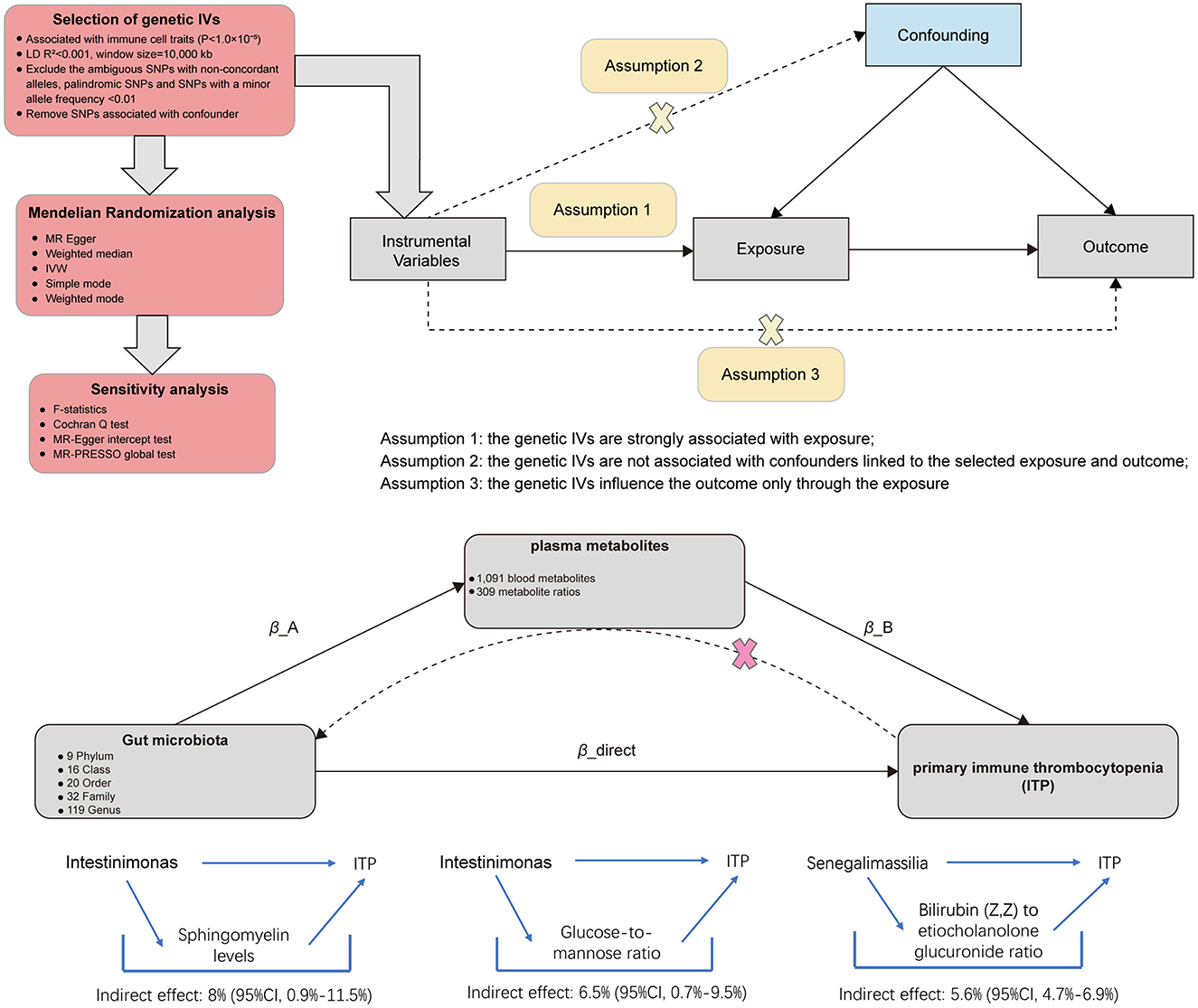
Figure 1. Diagrams illustrating associations examined in this study. The total effect between gut microbiota and primary immune thrombocytopenia (ITP). β_total is the total effect using genetically predicted gut microbiota as exposure and ITP as outcome. β_reverse is the total effect using genetically predicted ITP as exposure and gut microbiota as outcome. The total effect was decomposed into: (i) indirect effect using a two-step approach (where β_A is the total effect of gut microbiota on plasma metabolites, and β_B is the effect of plasma metabolites on ITP) and the product method (β_A × β_B) and (ii) direct effect (β_direct = β_total – β_A × β_B). Proportion mediated was the indirect effect divided by the total effect.
Data sources
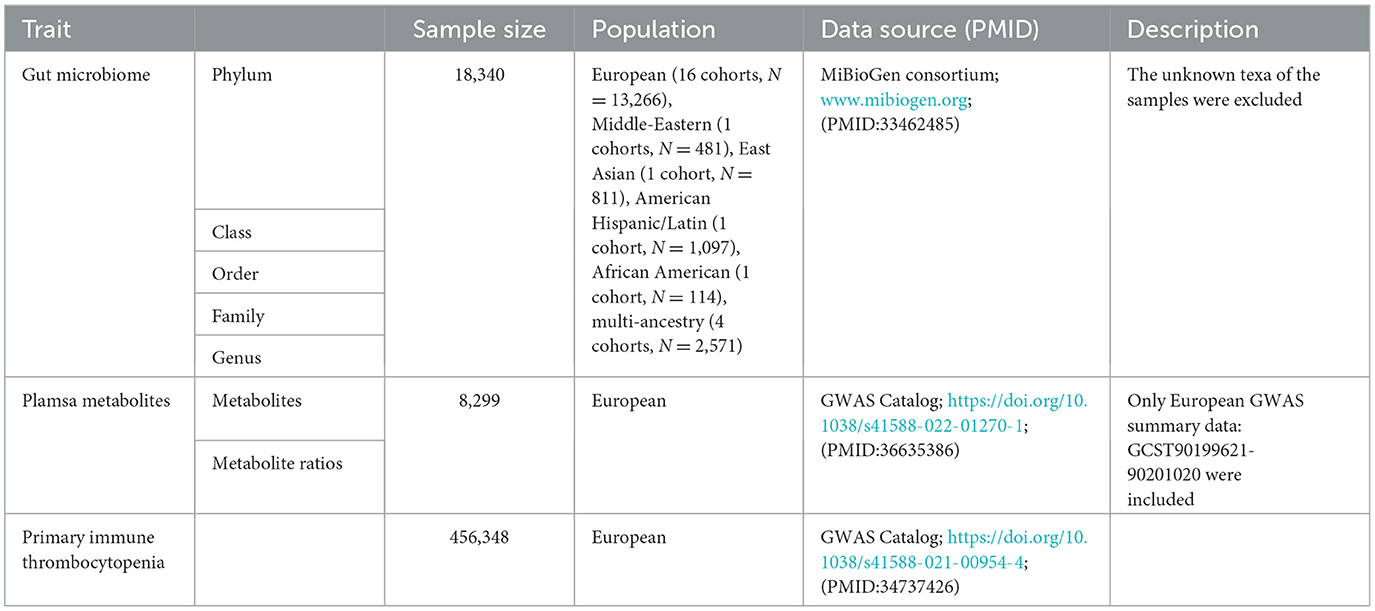
MiBioGen consortium designed and processed genome-wide genotypes and 16S fecal microbiome data from 18,340 individuals mainly from European background and the latest GWAS summary data included a total of 211 gut microbiota taxa (Kurilshikov et al., 2021). To enhancing the interpretability and scientific rigor of our findings, we deliberately excluded unknown microbial stains. So, a total of 196 microbiota taxa (119 genera, 32 families, 20 orders, 16 classes, and 9 phyla) were included in this study. The genetic data for metabolomics (comprising 1,091 metabolites and 309 metabolite ratios) from 8,299 unrelated European ancestry individuals were derived from the study by Yiheng Chen et al. and included in the GWAS Catalog (https://www.ebi.ac.uk/gwas/diagram) (Chen et al., 2023). The GWAS summary data of ITP from 456,348 European ancestry was acquired from the study by Jiang et al. (2021) and also included in the GWAS Catalog. Additional details are shown in Table 1. All GWAS data are from different consortia or organizations, and thus there is no overlapping sample.
Genetic instrumental variables (IVs) selection
For acquiring sufficient IVs, the displaying statistical significance of single nucleotide polymorphisms (SNPs) was set at P-value <1.0 × 10−5 (Malik et al., 2023). We extracted pertinent details including the chromosome (CHR), genomic location, effect allele (EA), other allele (OA), effect allele frequency (EAF) (if available), effect sizes (β), standard error (SE), P-value, and sample size (N). To ensure independence among the selected SNPs, a linkage disequilibrium (LD) threshold of r2 <0.001 was established, utilizing reference panel data from 1,000 Genomes Project Europeans samples (phase 3). This threshold facilitated the retention of independent SNPs with the lowest P-values (Bowden et al., 2015). Last, we calculated the explained variance (R2) and the F-statistic method was employed for SNP screening, calculated by dividing β by the square of the standard error, with a cut-off value set at 10 (Burgess et al., 2017). Subsequently, the identified SNPs were scrutinized using PhenoScanner V2 to identify potential confounding variables and confounders, including age, sex, race, and other diseases (Kamat et al., 2019).
MR analysis
Primary analysis
We performed MR analysis in R software (version 4.4.3, http://www.r-project.org) with “Two-Sample MR” package (version 0.5.6) (Broadbent et al., 2020). In order to ascertain the causal effects of gut microbiota and plasma metabolites on ITP, we conducted two-sample MR analysis separately. The inverse variance weighted (IVW) approach served as the primary analysis method, while the Wald ratios test was utilized for features containing only one IV (Burgess et al., 2013). MR results were reported as odds ratios (ORs) along with their corresponding 95% confidence intervals (CIs). Statistical significance was determined when the P-value of the inverse variance weighted (IVW) method was <0.05. The exposure factors with no sufficient SNPs for harmonization were excluded.
Reverse causality analysis
To assess reverse directional causation effects between gut microbiota and primary immune thrombocytopenia (ITP), we considered ITP as the “exposure” and gut microbiota associated with ITP as the “outcome.” SNPs significantly associated with ITP (P < 1 × 10−5) were selected as instrumental variables (IVs).
Sensitivity analysis
The “MR_PRESSO” package was employed for multiplicity testing (Ong and MacGregor, 2019). Cochran's Q test was conducted to assess the heterogeneity of each SNP (Cohen et al., 2015). Leave-one-out analysis was performed to assess the influence of each SNP on the overall results (Burgess and Thompson, 2017). Furthermore, MR-PRESSO regression and MR-Egger regression were utilized to examine potential horizontal pleiotropy effects (Verbanck et al., 2018).
Mediation analysis
In the two-sample analysis, gut microbiota and plasma metabolites showing significant causal effects on ITP were selected for mediation analysis. We investigated whether gut microbiota had a causal effect on plasma metabolites, which in turn had causal effects on ITP. The percentage mediated by the mediating effect was calculated by dividing the indirect effect by the total effect. 95% confidence intervals were computed using the “RMediation” package. Results with P < 0.05 and mediation percentages between 0% and 100% were considered statistically meaningful (Tofighi and MacKinnon, 2011).
Results
Causal effects of gut microbiota on ITP
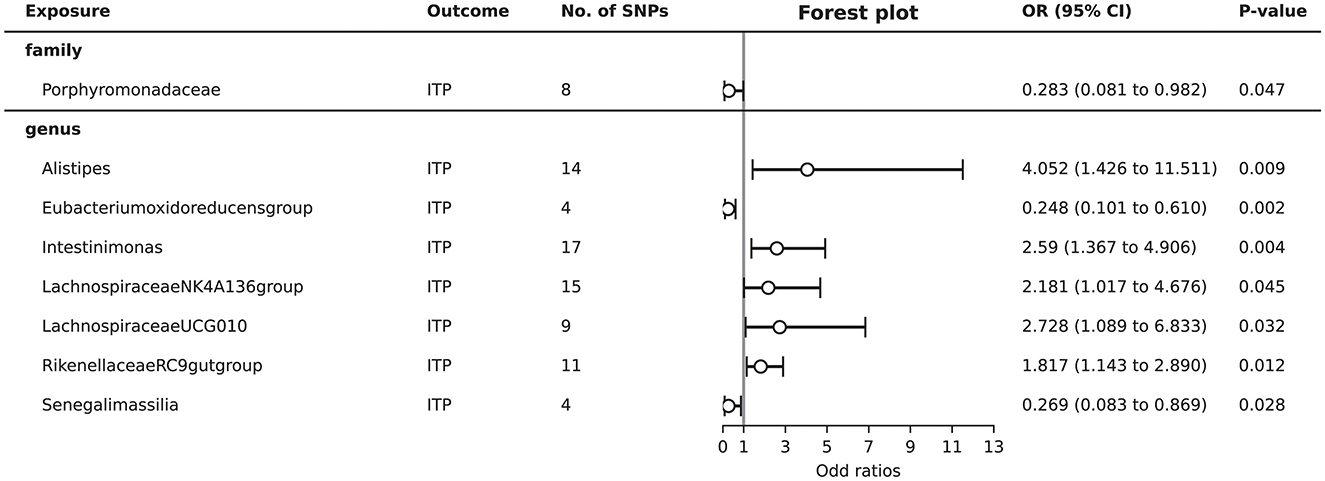
Total of eight gut microbiota (including one family, seven genera) were associated with ITP (Supplementary Table S3, Figure 2). Detailed 2,182 SNPs information for included microbiota is shown in Supplementary Table S1.
As shown in Figure 2, MR analysis suggested that genetic prediction of five gut microbiota (genus Alistipes, genus Intestinimonas, genus Lachnospiraceae NK4A136 group, genus Lachnospiraceae UCG010, and genus Rikenellaceae RC9 gut group) was associated with an increased risk of ITP. The genus Alistipes (OR = 4.052, 95%CI = 1.426–11.511, P = 0.009), genus Intestinimonas (OR = 2.590, 95%CI = 1.367–4.906, P = 0.004), genus Lachnospiraceae NK4A136 group (OR = 2.181, 95%CI = 1.017–4.676, P = 0.045), genus Lachnospiraceae UCG010 (OR = 2.728, 95%CI = 1.089–6.833, P = 0.032), and genus Rikenellaceae RC9 gut group (OR = 1.817, 95%CI = 1.143–2.890, P = 0.012) significantly increased the risk of ITP.
Genetic prediction of three gut microbiota (family Porphyromonadaceae, genus Eubacterium oxidoreducens group, and genus Senegalimassilia) was associated with a decreased risk of ITP. The family Porphyromonadaceae (OR = 0.283, 95%CI = 0.081–0.982, P = 0.047), genus Eubacterium oxidoreducens group (OR = 0.248, 95%CI = 0.101–0.610, P = 0.002), and genus Senegalimassilia (OR = 0.269, 95%CI = 0.083–0.869, P = 0.028) significantly decreased the risk of ITP.
According to the MR-Egger regression intercept approach and MRPRESSO analysis, horizontal pleiotropy was observed in the family Oxalobacteraceae, genus Eggerthella, genus ErysipelotrichaceaeUCG003, genus Oscillibacter, genus Peptococcus, genus Anaerofilum, and genus Ruminiclostridium5 in the MR study. The Cochran's Q tests indicated heterogeneity in the MR results for the family Actinomycetaceae, genus Actinomyces, genus Anaerofilum, genus Butyricimonas, genus Eubacterium brachy group, genus Eubacterium rectale group, genus Eubacterium ventriosum group, genus Hungatella, genus Olsenella, genus Ruminiclostridium5, and genus Veillonella. These findings do not affect the identification of the eight gut microbiota with a causal relationship with ITP in the MR analysis, ensuring the reliability of the results (Supplementary Table S2).
Reverse causal effects of ITP on gut microbiota
As shown in Supplementary Table S6, ITP has causal effect on 7 of 8 gut microbiota [family Porphyromonadaceae (OR = 1.024, 95%CI = 1.013–1.036, P < 0.001), genus Alistipes (OR = 1.029, 95%CI = 1.021–1.038, P < 0.001), genus Eubacterium oxidoreducens group (OR = 0.964, 95%CI = 0.948–0.980, P < 0.001), genus LachnospiraceaeNK4A136group (OR = 1.018, 95%CI = 1.009–1.026, P < 0.001), genus LachnospiraceaeUCG010 (OR = 0.986, 95%CI = 0.976–0.995, P = 0.003), genus RikenellaceaeRC9gutgroup (OR = 0.938, 95%CI = 0.918–0.959, P < 0.001), and genus Senegalimassilia (OR = 0.964, 95%CI = 0.943–0.986, P = 0.001)] identified with causal effect on ITP. Detailed 335 SNPs information for reverse causal analyses is shown in Supplementary Table S4.
In the MR-Egger regression intercept and MRPRESSO analysis, horizontal pleiotropy was detected for the associations between ITP and genus Alistipes, genus Eubacterium oxidoreducens group, and family Porphyromonadaceae. Cochran's Q tests indicated that the causal relationships between ITP and family Porphyromonadaceae, genus Senegalimassilia, and genus Senegalimassilia were influenced by heterogeneity. Based on these MR results, we identified reverse causal confounding between ITP and genus Lachnospiraceae NK4A136 group, genus Lachnospiraceae UCG010, and genus Rikenellaceae RC9 gut group, leading to their exclusion from subsequent mediation analysis (Supplementary Table S5).
Causal effects of plasma metabolites on ITP
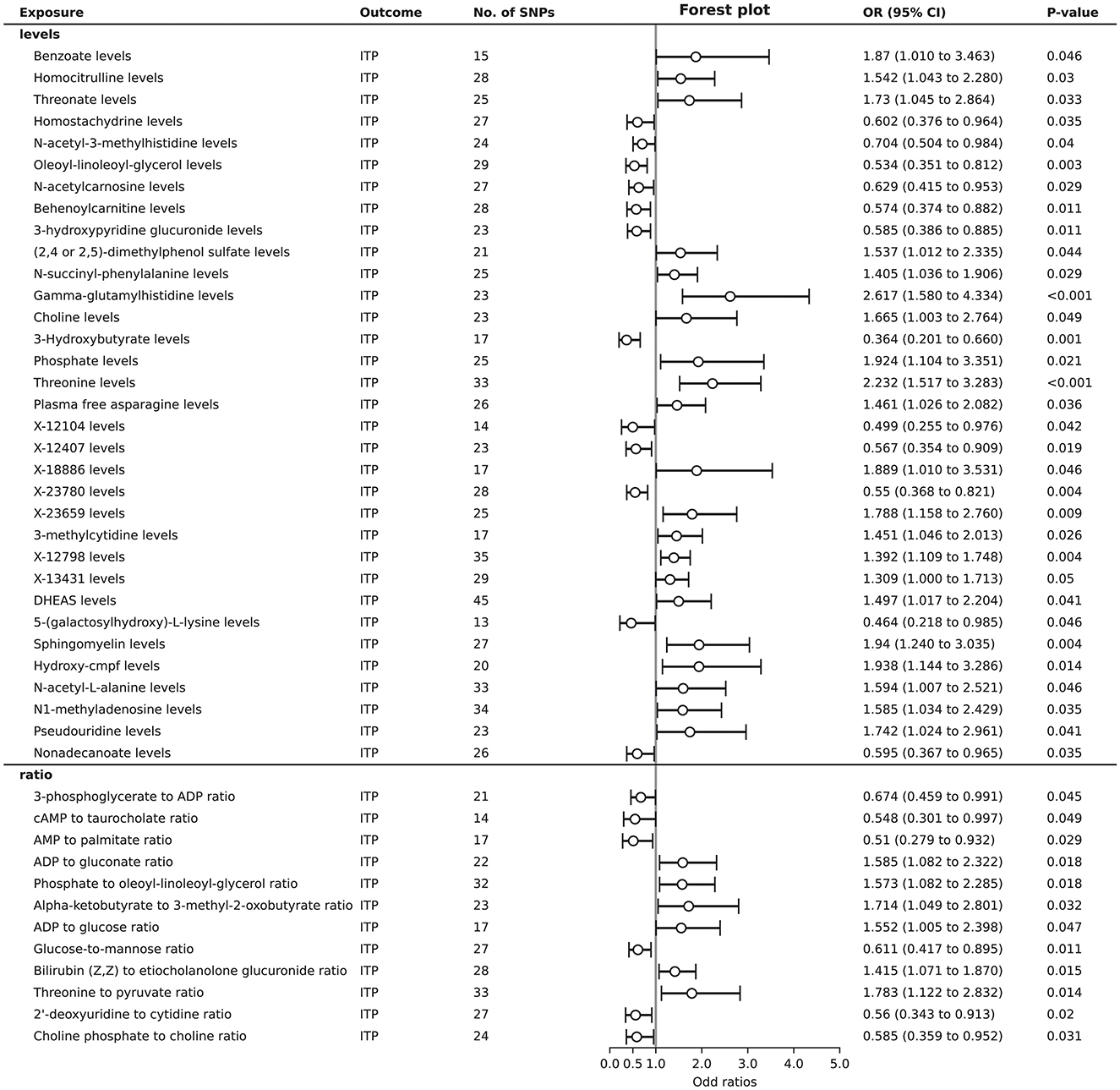
A total of 45 plasma metabolites were found to be associated with ITP. Among the levels of plasma metabolites, 21 showed a positive causal relationship with ITP, while 12 exhibited a negative causal relationship. Regarding the ratios of plasma metabolites, elevated levels of 6 ratios were identified as risk factors for ITP, whereas elevated levels of another 6 ratios were identified as protective factors (Figure 3). Detailed 1,124 SNPs information for MR analysis of 45 plasma metabolites on ITP is shown in Supplementary Table S7.
In the sensitivity analysis results for 45 plasma metabolites, Cochran's Q-test showed no evidence of heterogeneity in the causal relationship between these SNPs. The results of the MR-Egger regression intercept indicated potential horizontal pleiotropy for threonate levels, behenoylcarnitine (C22) levels, X-23780 levels, the phosphate to oleoyl-linoleoyl-glycerol (18:1 to 18:2) (Bussel et al., 2023) ratio, nonadecanoate (19:0) levels, and the 2′-deoxyuridine to cytidine ratio. Conversely, the MR-PRESSO test did not identify significant pleiotropy. To maximize the discovery of plasma metabolites potentially mediating the causal link between gut microbiota and ITP, we included the levels or ratios of these 45 plasma metabolites in the subsequent analysis (Supplementary Table S8).
Causal effects of gut microbiota on plasma metabolites associated to ITP
After harmonization, there were 2,836 SNPs were filtered for further MR analysis (Supplementary Table S10). A total of 12 causal relationships were identified between the 8 gut microbiota and 45 plasma metabolites associated with ITP (Supplementary Table S12). Specifically, the genus Alistipes was found to have a protective effect on the plasma levels of hydroxy-cmpf (OR = 0.780, 95% CI = 0.638–0.953, P = 0.015) and (2,4 or 2,5)-dimethylphenol sulfate (OR = 0.724, 95% CI = 0.578–0.907, P = 0.005), while it was a risk factor for N-acetyl-L-alanine levels (OR = 1.341, 95% CI = 1.118–1.608, P = 0.002). The genus Eubacterium oxidoreducens group was identified as a risk factor for the ratio of threonine to pyruvate (OR = 1.227, 95% CI = 1.080–1.394, P = 0.002). The genus Intestinimonas was a risk factor for sphingomyelin levels (OR = 1.122, 95% CI = 1.008–1.249, P = 0.035) and a protective factor for the 2′-deoxyuridine to cytidine ratio (OR = 0.870, 95% CI = 0.767–0.987, P = 0.030) and the glucose-to-mannose ratio (OR = 0.882, 95% CI = 0.789-0.986, P = 0.027). The family Porphyromonadaceae may promote the levels of sphingomyelin (OR = 1.265, 95% CI = 1.009–1.585, P = 0.042) and X-13431 (OR = 1.263, 95% CI = 1.003–1.590, P = 0.047), while inhibiting the levels of phosphate (OR = 0.772, 95% CI = 0.615–0.970, P = 0.026) and X-23659 (OR = 0.795, 95% CI = 0.639–0.989, P = 0.039). The genus Senegalimassilia was negatively associated with the ratio of bilirubin (Z,Z) to etiocholanolone glucuronide (OR = 0.810, 95% CI = 0.692–0.949, P = 0.009) (Figure 4).
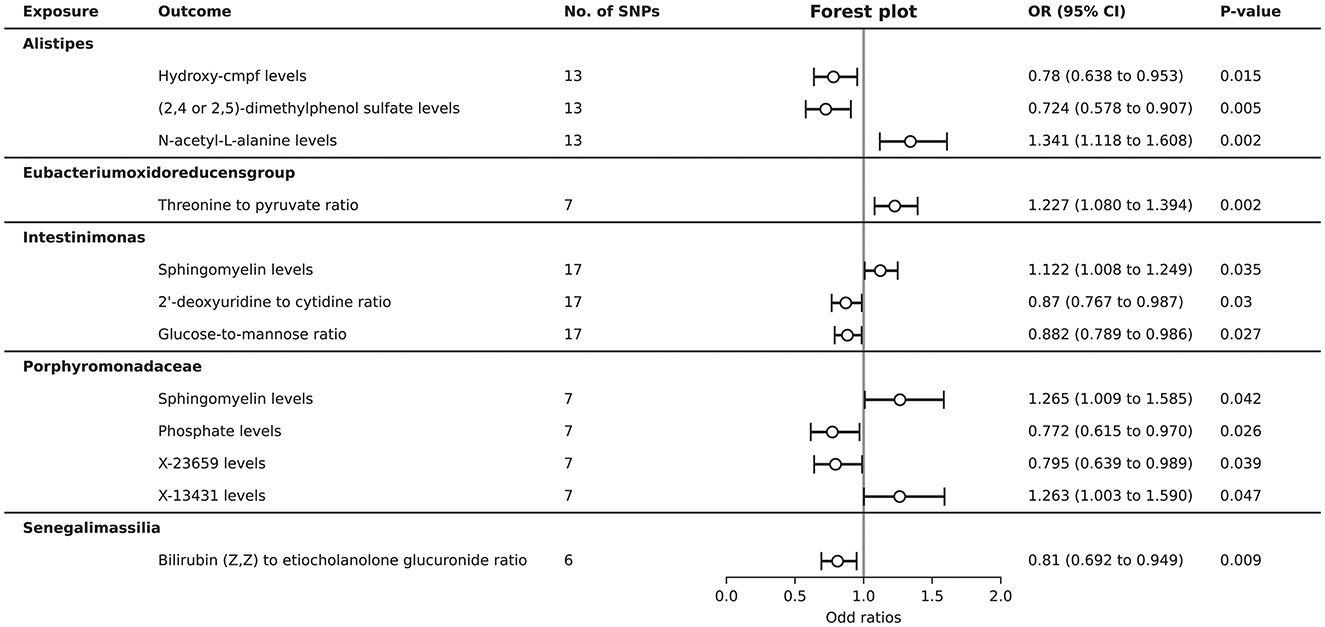
Figure 4. Forest plot to visualize the causal effects of gut microbiota on plasma metabolites associated with ITP.
Although seven pairs of causal relationships between the included gut microbiota and plasma metabolites did not pass the sensitivity analysis, the MR-Egger regression intercept approach and the MRPRESSO analysis indicated that heterogeneity and genetic pleiotropy did not bias the significant results (Supplementary Table S11).
Mediation analysis of gut microbiota on ITP via plasma metabolites
Based on the above analysis, gut microbiota and plasma metabolites both have causal effects on ITP. It appears that plasma metabolites mediate the pathway between gut microbiota and ITP. Using the delta method, we identified Sphingomyelin levels (β = 0.076, 95% CI = 0.003–0.183, P = 0.047) and the Glucose-to-mannose ratio (β = 0.062, 95% CI = 0.002–0.151, P = 0.039) as mediators in the effect of genus Intestinimonas on ITP. Additionally, the Bilirubin (Z,Z) to etiocholanolone glucuronide ratio (β = −0.073, 95% CI = −0.171–−0.007, P = 0.043) was identified as a mediator in the effect of genus Senegalimassilia on ITP. In terms of mediation proportion, Sphingomyelin levels and the Glucose-to-mannose ratio accounted for 8% (95% CI = 0.9%−11.5%) and 6.5% (95% CI = 0.7%−9.5%) of the effect of genus Intestinimonas on ITP, respectively. The Bilirubin (Z,Z) to etiocholanolone glucuronide ratio accounted for 5.6% (95% CI = 4.7%−6.9%) of the effect of genus Senegalimassilia on ITP (Table 2).
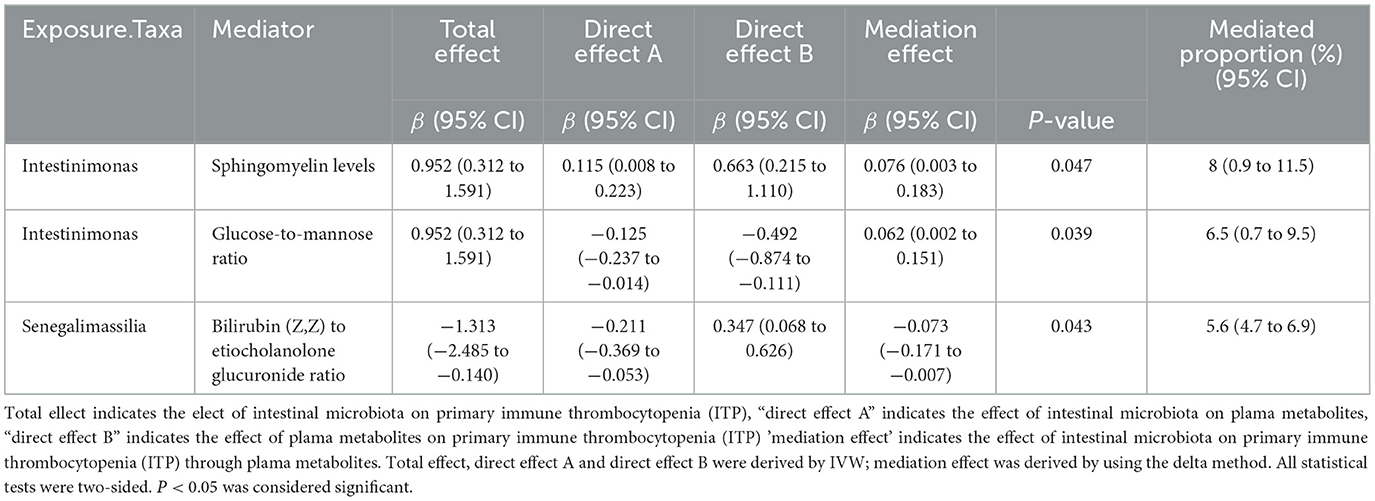
Table 2. The mediation effect of gut microbiota on primary immune thrombocytopenia (ITP) via plama metabolites.
Discussion
This study is the first to identify plasma mediators influenced by the gut microbiota in immune thrombocytopenia (ITP) from a big data perspective. We conducted a large-scale two-sample Mendelian randomization analysis using summary data from the MiBioGen consortium GWAS meta-analysis, alongside plasma metabolites and ITP data summarized from relevant literature, to identify plasma metabolites mediating the causal relationship between gut microbiota and ITP. Our Mendelian randomization analysis revealed a unidirectional causal relationship between five gut microbiota taxa (family Porphyromonadaceae, genus Alistipes, genus Eubacterium oxidoreducens group, genus Intestinimonas, and genus Senegalimassilia) and ITP. Among the 45 plasma metabolites with a causal relationship with ITP, sphingomyelin levels, glucose-to-mannose ratio, and bilirubin (Z,Z) to etiocholanolone glucuronide ratio mediated the effects of genus Intestinimonas and genus Senegalimassilia on ITP.
Host-microbiota interactions are crucial for host physiology and disease phenotypes, with dysbiosis potentially promoting disease development through alterations in pathogens and their associated metabolites (Elsouri et al., 2021; Opoku et al., 2022). Current evidence suggests bidirectional communication between gut microbiota composition and function and the occurrence and progression of Immune Thrombocytopenia (ITP) (Li et al., 2023). As an example, one group reported antibodies against platelets were identified on one female patient's platelets who developed thrombocytopenia after two fecal microbial transplantations (Malnick et al., 2015). In our study, among identified 8 gut microbiota taxa causally linked to ITP, genus Lachnospiraceae NK4A136 group, genus Lachnospiraceae UCG010, and genus Rikenellaceae RC9 gut group, exhibited bidirectional causal relationships with ITP. Among the microbiota promoting ITP, we identified genus Alistipes within the phylum Bacteroidetes, consistent with previous reports of a decreased Firmicutes/Bacteroidetes ratio in ITP (Yu et al., 2022). Additionally, the causal relationship between genus Intestinimonas within the family Lachnospiraceae and ITP parallels findings by Dongmei Guo, which demonstrated a causal link between Lachnospiraceae and ITP based on different outcome cohorts (Guo et al., 2023). In terms of protective factors against ITP development, family Porphyromonadaceae and genus Eubacterium oxidoreducens group were identified as significant protective factors. Although their roles in ITP have not been previously reported, the protective effects of specific microbiota within the family Porphyromonadaceae against autoimmune diseases are well-established (Mao et al., 2023). Moreover, some discrepancies between our results and previous Mendelian randomization studies on the causal relationship between gut microbiota and ITP may be attributed to the heterogeneity in ITP etiology and differences in the outcome cohort data utilized (Guo et al., 2023; Jiang et al., 2024).
Metabolites are intermediate or end products of metabolic reactions, with plasma metabolite levels influenced by various factors such as gut microbiota and medications (Lavelle and Sokol, 2020). Changes in plasma metabolites can affect disease outcomes and serve as potential therapeutic targets (Yoon et al., 2021). Using GC-MS technology, the potential associations between ITP and plasma metabolites have been gradually uncovered (Zhang et al., 2022); however, systematic studies on their causal relationships are lacking. Through Mendelian randomization analysis, this study newly identified 45 plasma metabolites with potential causal links to ITP. Fujii et al. (2021) discovered that defects in sphingomyelin synthase 1 lead to thrombocytopenia, which aligns with our finding that Sphingomyelin level is a protective factor for ITP. Importantly, we identified that genus Intestinimonas promotes plasma Sphingomyelin levels, thereby protecting against ITP from the perspective of gut microbiota. Additionally, an increase in mannose in platelets has been observed in ITP patients (Ramírez-López et al., 2021); our study validated that the Glucose-to-mannose ratio acts as a negative regulatory mediator in the effect of genus Intestinimonas on ITP. In summary, the reported associations between metabolites and ITP are consistent with the causal relationships identified in our study.
Investigations into the gut microbiota and metabolites indicate that their changes are closely related to the clinical characteristics of ITP (Wang et al., 2023). For example, it has been reported that among ITP patients receiving treatment, the increased abundance of Pseudomonas in the gut suggests a mechanism for good prognosis (Rui et al., 2023). Enzymes produced by Pseudomonas, such as alkaline protease and elastase, have been shown to play a role in inhibiting the activity of neutrophils and natural killer cells, while also inhibiting lymphocyte proliferation by proteolytic hydrolysis of IL-2, ultimately improving ITP (Theander et al., 1988). Likely, our found plasma metabolites causally associated to a certain of gut microbiota are potential biomarkers reflecting the severity of ITP during disease progression. On the other hand, there seems to be a bidirectional relationship between the composition of the microbiota and treatment in ITP patients. For example, the genus Lachnospiraceae is clearly present in the gut microbiota of ITP and RA patients (Wu et al., 2016). In recent years, probiotics and prebiotics have been recommended for the treatment of various diseases, including autoimmune diseases. Thereby, our explored three gut microbiota-plasma metabolites-ITP relationship are ideal candidate probiotics and prebiotics hopefully utilized in the treatment of ITP.
Our study exploring the relationships between gut microbiota, plasma metabolites, and ITP encountered several limitations. Firstly, the generalizability of our findings is constrained by the predominantly European ancestry of our sample, which may not accurately reflect the genetic and lifestyle diversity influencing gut microbiota in different populations. Additionally, the use of 16S rRNA gene sequencing provides taxonomic insights only up to the genus level, lacking the depth of species-level analysis that metagenomic sequencing could offer. The associations observed between certain microbiota and ITP risk are preliminary, and given the complexity of interactions within the gut microbiome and with ITP, these findings should be interpreted with caution. Moreover, our approach based on Mendelian randomization (MR) may not fully capture the intricate, potentially non-linear relationships between gut microbiota and ITP. It's necessary to perform prospective cohort studies to help further validate these causal associations. The current limitations of available GWAS datasets for ITP also hinder our ability to conduct reliable replication studies using diverse GWAS data, which is crucial for establishing definitive causal relationships. The relatively small sample size of existing ITP GWAS datasets reduces the power to detect true causal effects of certain exposures, potentially increasing the risk of false-negative results. Therefore, future research requires larger GWAS datasets to validate our findings and further elucidate the complex role of gut microbiota in the pathogenesis of ITP. Lastly, the hypothesis-driven nature of MR emphasizes the detection of biologically plausible causal relationships. Thus, our exploratory work prioritizes biologically meaningful associations that should be further investigated in larger datasets in future studies.
To our knowledge, this is the first comprehensive study to evaluate the causal relationships between gut microbiota, plasma metabolites, and ITP. We have identified a significant role for gut microbiota and serum metabolites in the pathogenesis and progression of ITP in the host. These findings underscore the necessity of further exploring the mechanisms underlying the interactions between gut microbiota and ITP and deeper mechanisms based on individual-level data and experimental studies will be further explored to gain a more nuanced understanding. Moreover, our results provide novel insights that may inform the development of microbiota-based therapies and plasma metabolite-targeted interventions for ITP.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
YH: Writing – original draft. CZ: Writing – original draft. KS: Writing – review & editing, Conceptualization. XD: Writing – review & editing. BC: Writing – review & editing, Supervision, Formal analysis.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the Natural Science Foundation of Jiangsu Province (BK20210028).
Acknowledgments
The authors thank the investigators of the original studies for sharing the GWAS summary statistics.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1447729/full#supplementary-material
Abbreviations
OR, Odd ratio; SNP, Single nucleotide polymorphisms; ITP, Primary immune thrombocytopenia; GWAS, Genome-wide association studies; MR, Mendelian randomization; IVW, Inverse variance weighted; SCFA, Short-chain fatty acid; IV, Instrumental variable; CHR, Chromosome; EA, Effect allele; OA, Other allele; EAF, Effect allele frequency; β, Effect size; SE, Standard error; N, Sample size.
References
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Bowden, J., and Holmes, M. V. (2019). Meta-analysis and Mendelian randomization: a review. Res. Synth. Methods 10, 486–496. doi: 10.1002/jrsm.1346
Broadbent, J. R., Foley, C. N., Grant, A. J., Mason, A. M., and Staley, J. R. (2020). MendelianRandomization v0.5.0: updates to an R package for performing Mendelian randomization analyses using summarized data. Wellcome Open Res. 5:252. doi: 10.12688/wellcomeopenres.16374.2
Burgess, S., Butterworth, A., and Thompson, S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. doi: 10.1002/gepi.21758
Burgess, S., Small, D. S., and Thompson, S. G. (2017). A review of instrumental variable estimators for Mendelian randomization. Stat. Methods Med. Res. 26, 2333–2355. doi: 10.1177/0962280215597579
Burgess, S., and Thompson, S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389. doi: 10.1007/s10654-017-0255-x
Bussel, J. B., Hou, M., and Cines, D. B. (2023). Management of primary immune thrombocytopenia in pregnancy. N. Engl. J. Med. 389, 540–548. doi: 10.1056/NEJMra2214617
Chen, Y., Lu, T., Pettersson-Kymmer, U., Stewart, I. D., Butler-Laporte, G., Nakanishi, T., et al. (2023). Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 55, 44–53. doi: 10.1038/s41588-022-01270-1
Cines, D. B. (2023). Pathogenesis of refractory ITP: overview. Br. J. Haematol. 203, 10–16. doi: 10.1111/bjh.19083
Cohen, J. F., Chalumeau, M., Cohen, R., Korevaar, D. A., and Khoshnood, B. (2015). Cochran's Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J. Clin. Epidemiol. 68, 299–306. doi: 10.1016/j.jclinepi.2014.09.005
Davies, N. M., Holmes, M. V., and Davey Smith, G. (2018). Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362:k601. doi: 10.1136/bmj.k601
Du, H. X., Yue, S. Y., Niu, D., Liu, C., Zhang, L. G., Chen, J., et al. (2022). Gut microflora modulates Th17/Treg cell differentiation in experimental autoimmune prostatitis via the short-chain fatty acid propionate. Front. Immunol. 13:915218. doi: 10.3389/fimmu.2022.915218
Elsouri, K., Arboleda, V., Heiser, S., and Kesselman, M. M. (2021). Microbiome in rheumatoid arthritis and celiac disease: a friend or foe. Cureus 13:e15543. doi: 10.7759/cureus.15543
Fernández-Ochoa, Á., Brunius, C., Borrás-Linares, I., Quirantes-Piné, R., Cádiz-Gurrea, M. d. L., Consortium, P. C., et al. (2020). Metabolic disturbances in urinary and plasma samples from seven different systemic autoimmune diseases detected by HPLC-ESI-QTOF-MS. J. Proteome Res. 19, 3220–3229. doi: 10.1021/acs.jproteome.0c00179
Fujii, Y., Taniguchi, M., Nagaya, S., Ueda, Y., Hashizume, C., Watanabe, K., et al. (2021). A novel mechanism of thrombocytopenia by PS exposure through TMEM16F in sphingomyelin synthase 1 deficiency. Blood Adv. 5, 4265–4277. doi: 10.1182/bloodadvances.2020002922
Guo, D., Chen, Q., Wang, G., Li, C., and FinnGen consortium. (2023). Causal relationship between gut microbiota and immune thrombocytopenia: a Mendelian randomization study of two samples. Front. Microbiol. 14:1190866. doi: 10.3389/fmicb.2023.1190866
Honda, K., and Littman, D. R. (2016). The microbiota in adaptive immune homeostasis and disease. Nature 535, 75–84. doi: 10.1038/nature18848
Jiang, C., Deng, S., Ma, X., Song, J., Li, J., Yuan, E., et al. (2024). Mendelian randomization reveals association of gut microbiota with Henoch-Schonlein purpura and immune thrombocytopenia. Int. J. Hematol. 120, 50–59. doi: 10.1007/s12185-024-03777-1
Jiang, L., Zheng, Z., and Fang, H. (2021). A generalized linear mixed model association tool for biobank-scale data. Nat. Genet. 53, 1616–1621. doi: 10.1038/s41588-021-00954-4
Kamat, M. A., Blackshaw, J. A., Young, R., Surendran, P., Burgess, S., Danesh, J., et al. (2019). PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics 35, 4851–4853. doi: 10.1093/bioinformatics/btz469
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Lavelle, A., and Sokol, H. (2020). Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 17, 223–237. doi: 10.1038/s41575-019-0258-z
Li, X., Zhang, M., He, L., Zhou, J., Shen, P., Dai, W., et al. (2023). Gut microbiota alterations in children and their relationship with primary immune thrombocytopenia. Front. Pediatr. 11:1213607. doi: 10.3389/fped.2023.1213607
Liu, C., Cheng, L., Ji, L., Li, F., Zhan, Y., Wu, B., et al. (2020). Intestinal microbiota dysbiosis play a role in pathogenesis of patients with primary immune thrombocytopenia. Thromb. Res. 190, 11–19. doi: 10.1016/j.thromres.2020.03.012
Malik, A., Sayed, A. A., Han, P., Tan, M. M., Watt, E., Constantinescu-Bercu, A., et al. (2023). The role of CD8+ T-cell clones in immune thrombocytopenia. Blood 141, 2417–2429. doi: 10.1182/blood.2022018380
Malnick, S. D., Oppenheim, A., and Melzer, E. (2015). Immune thrombocytopenia caused by fecal microbial transplantation in a patient with severe recurrent clostridium difficile infection. J. Clin. Gastroenterol. 49, 888–889. doi: 10.1097/MCG.0000000000000404
Mao, D., Tao, B., Sheng, S., Jin, H., Chen, W., Gao, H., et al. (2023). Causal effects of gut microbiota on age-related macular degeneration: a mendelian randomization study. Invest. Ophthalmol. Vis. Sci. 64:32. doi: 10.1167/iovs.64.12.32
Mondanelli, G., Iacono, A., Carvalho, A., Orabona, C., Volpi, C., Pallotta, M. T., et al. (2019). Amino acid metabolism as drug target in autoimmune diseases. Autoimmun. Rev. 18, 334–348. doi: 10.1016/j.autrev.2019.02.004
Ong, J. S., and MacGregor, S. (2019). Implementing MR-PRESSO and GCTA-GSMR for pleiotropy assessment in Mendelian randomization studies from a practitioner's perspective. Genet. Epidemiol. 43, 609–616. doi: 10.1002/gepi.22207
Opoku, Y. K., Asare, K. K., Ghartey-Quansah, G., Afrifa, J., Bentsi-Enchill, F., Ofori, E. G., et al. (2022). Intestinal microbiome-rheumatoid arthritis crosstalk: the therapeutic role of probiotics. Front. Microbiol. 13:996031. doi: 10.3389/fmicb.2022.996031
Ramírez-López, A., Román, M. T. Á., Manzano, E. M., Acuña, P., Arias-Salgado, E. G., Salces, M. M., et al. (2021). The importance of platelet glycoside residues in the haemostasis of patients with immune thrombocytopaenia. J. Clin. Med. 10:1661. doi: 10.3390/jcm10081661
Rasouli-Saravani, A., Jahankhani, K., Moradi, S., Gorgani, M., Shafaghat, Z., Mirsanei, Z., et al. (2023). Role of microbiota short-chain fatty acids in the pathogenesis of autoimmune diseases. Biomed. Pharmacother. 162:114620. doi: 10.1016/j.biopha.2023.114620
Rodeghiero, F. (2023). Recent progress in ITP treatment. Int. J. Hematol. 117, 316–330. doi: 10.1007/s12185-022-03527-1
Rooks, M. G., and Garrett, W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Rui, X., Fu, Y., Cai, J., Zhang, Y., Fu, Q., He, C., et al. (2023). Gut microbiota were altered with platelet count and red blood cell count in immune thrombocytopenia patients with different treatments. Front. Cell. Infect. Microbiol. 13:1168756. doi: 10.3389/fcimb.2023.1168756
Semple, J. W., Rebetz, J., and Maouia, A. (2020). An update on the pathophysiology of immune thrombocytopenia. Curr. Opin. Hematol. 27, 423–429. doi: 10.1097/MOH.0000000000000612
Sepe, V., Renga, B., Festa, C., Finamore, C., Masullo, D., Carino, A., et al. (2016). Investigation on bile acid receptor regulators. Discovery of cholanoic acid derivatives with dual G-protein coupled bile acid receptor 1 (GPBAR1) antagonistic and farnesoid X receptor (FXR) modulatory activity. Steroids 105, 59–67. doi: 10.1016/j.steroids.2015.11.003
Theander, T. G., Kharazmi, A., Pedersen, B. K., Christensen, L. D., Tvede, N., Poulsen, L. K., et al. (1988). Inhibition of human lymphocyte proliferation and cleavage of interleukin-2 by Pseudomonas aeruginosa proteases. Infect. Immun. 56, 1673–1677. doi: 10.1128/iai.56.7.1673-1677.1988
Tofighi, D., and MacKinnon, D. P. (2011). RMediation: an R package for mediation analysis confidence intervals. Behav. Res. Methods 43, 692–700. doi: 10.3758/s13428-011-0076-x
Verbanck, M., Chen, C. Y., and Neale, B. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Wang, H., Bi, H., Yang, M., Wang, X., Song, C., Yang, W., et al. (2023). Intestinal flora altered and correlated with interleukin-2/4 in patients with primary immune thrombocytopenia. Hematology 28:2277501. doi: 10.1080/16078454.2023.2277501
Wang, Y., Liu, F., Zhang, G., Su, Y., Sun, X., Chen, Q., et al. (2021). Gut microbiome alterations and its link to corticosteroid resistance in immune thrombocytopenia. Sci. China Life Sci. 64, 766–783. doi: 10.1007/s11427-020-1788-2
Wen, S., Wang, Z., Feng, J., Yang, Y., Lin, X., Huang, H., et al. (2022). NMR-based metabolomics identify metabolic change in spleen of idiopathic thrombocytopenic purpura patients. Metabolites 12:565. doi: 10.3390/metabo12060565
Wu, X., He, B., Liu, J., Feng, H., Ma, Y., Li, D., et al. (2016). Molecular Insight into Gut Microbiota and Rheumatoid Arthritis. Int. J. Mol. Sci. 17:431. doi: 10.3390/ijms17030431
Yang, W., and Cong, Y. (2021). Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 18, 866–877. doi: 10.1038/s41423-021-00661-4
Yoon, N., Jang, A. K., Seo, Y., and Jung, B. H. (2021). Metabolomics in autoimmune diseases: focus on rheumatoid arthritis, systemic lupus erythematous, and multiple sclerosis. Metabolites 11:812. doi: 10.3390/metabo11120812
Yu, X., Zheng, Q., He, Y., Yu, D., Chang, G., Chen, C., et al. (2022). Associations of gut microbiota and fatty metabolism with immune thrombocytopenia. Front. Med. 9:810612. doi: 10.3389/fmed.2022.810612
Keywords: primary immune thrombocytopenia, plasma metabolites, gut microbiota, Mendelian randomization, mediation analyses
Citation: Hong Y, Zhang C, Shen K, Dong X and Chen B (2024) Genetically predicted plasma metabolites mediate the causal relationship between gut microbiota and primary immune thrombocytopenia (ITP). Front. Microbiol. 15:1447729. doi: 10.3389/fmicb.2024.1447729
Received: 03 July 2024; Accepted: 07 October 2024;
Published: 28 October 2024.
Edited by:
Arezoo Mirzaei, Isfahan University of Medical Sciences, IranReviewed by:
Davood Mansury, Isfahan University of Medical Sciences, IranMahtab Hadadi, Isfahan University of Medical Sciences, Iran
Copyright © 2024 Hong, Zhang, Shen, Dong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing Chen, Y2hlbmJpbmdfbmp1QDEyNi5jb20=; Xiaoqing Dong, cWdzd25zQDEyNi5jb20=
†These authors have contributed equally to this work
 Yang Hong
Yang Hong Cuilin Zhang†
Cuilin Zhang† Xiaoqing Dong
Xiaoqing Dong Bing Chen
Bing Chen