- 1Graduate School of Horticulture, Chiba University, Matsudo, Japan
- 2Regional Environment Conservation Division, National Institute for Environmental Studies, Tsukuba, Japan
Iodate reductase (Idr) gene cluster (idrABP1P2) is involved in bacterial iodate (IO3−) respiration under anaerobic conditions. Putative idr gene clusters are present in both anaerobic and aerobic bacteria; however, the specific physiological roles of idr genes in aerobic bacteria remain unclear. Therefore, in this study, three marine aerobic bacteria with putative idr gene clusters (Roseovarius azorensis, Notoacmeibacter marinus, and Aliiroseovarius sediminilitoris) were grown in the presence of iodate to determine whether they can reduce iodate to iodide (I−). All tested bacteria almost completely reduced 2 mM iodate under static conditions but only reduced 0.1–0.5 mM iodate under shaking conditions. Moreover, the washed cell suspension of R. azorensis reduced iodate only when the cells were pre-grown statically in the presence of iodate. Transcriptional analysis revealed that the expression levels of idrA, idrB, idrP1, and idrP2 genes were upregulated in R. azorensis when the cells were grown statically in the presence of iodate. Specifically, idrA expression was induced by 0.1 μM iodate and was up to 14-fold higher compared to that of the non-iodate control. These results suggest that marine aerobic bacteria reduce iodate under oxygen-limited conditions, and that this capacity is induced by environmentally relevant levels of iodate in seawater. Our results suggest that marine aerobic bacteria contribute to iodide production in marine surface waters, thereby affecting the global iodine cycling and ozone budget.
1 Introduction
In marine environments, iodine mainly exists in inorganic forms, such as iodate (IO3−) and iodide (I−) ions (Wong, 1991; Luther, 2023). These inorganic iodine species are ubiquitously distributed in seawater at concentrations of 0.4–0.5 μM. Iodate is thermodynamically more stable in oxidized seawater (Sillen, 1961). However, iodide-rich water sources, such as the oxygen minimum zone (OMZ) of Arabian Sea (Farrenkopf et al., 1997; Farrenkopf and Luther, 2002) and hypersaline anoxic brine in the Orca Basin, Gulf of Mexico (Wong et al., 1985), are also observed. High concentrations of iodide are also observed in surface water (Tian and Nicolas, 1995; Campos et al., 1996; Tian et al., 1996; Moriyasu et al., 2023). Moriyasu et al. (2023) recently reported a hot zone of iodide comprising up to 50% of the total iodine concentration in tropical surface waters.
Iodide in marine surface water plays an important role in atmospheric iodine photochemistry and has a significant impact on the global ozone (O3) budget (Carpenter et al., 2021). Iodide in seawater reacts with gaseous ozone to form volatile I2 and hypoiodous acid (Jones et al., 2010; Carpenter et al., 2013). These volatile iodine compounds undergo photolysis in the atmosphere, releasing iodine atoms (I), which react with ozone to form the IO radical that plays a key role in the formation of cloud condensation nuclei, thus impacting the regional climate (O’Dowd et al., 2002; Saiz-Lopez et al., 2012). The reaction between iodide and ozone at the sea surface is the primary source of atmospheric iodine and the primary sink for atmospheric ozone (MacDonald et al., 2014). Atmospheric iodine reaches the land via wet (rainfall) and dry deposition, where it is taken up by animals and plants (Fuge and Johnson, 2015). Dietary iodine deficiency causes endemic goiter and congenital hypothyroidism (Chaker et al., 2022; Dijck-Brouwer et al., 2022). Therefore, iodide levels in marine surface water significantly influence the public health.
As the iodide concentration in seawater often correlates with chlorophyll fluorescence (Moriyasu et al., 2023; Jones et al., 2024), many studies investigated iodate reduction by microalgae (phytoplankton). Many microalgal cultures are capable of iodate reduction, including growth-associated and senescence-associated reduction (Wong et al., 2002; Chance et al., 2007; Bluhm et al., 2010; van Bergeijk et al., 2016; Hepach et al., 2020). Nitrate reductase is suggested as the key enzyme catalyzing iodate reduction (Tsunogai and Sase, 1969); however, some studies suggest the involvement of other enzymes in this process (Waite and Truesdale, 2003; Mok et al., 2018; Hepach et al., 2020). Recently, bacterial iodate reduction has attracted considerable attention (Yeager et al., 2017). Some bacteria act as iodate-respiring prokaryotes and dissimilatorily reduce iodate as a terminal electron acceptor (Yamazaki et al., 2020; Reyes-Umana et al., 2022a). Iodate-respiring bacteria, such as Pseudomonas sp. SCT and Denitromonas sp. IR-12, possess gene cluster idrABP1P2 in their genome that forms an operon-like structure. Transcription of the idrABP1P2 cluster is specifically upregulated under iodate-respiring conditions (Yamazaki et al., 2020). Notably, idrA knockout mutant of Denitromonas sp. IR-12 does not grow in the presence of iodate (Reyes-Umana et al., 2022a). The idrA gene encodes the catalytic subunit of a novel dimethyl sulfoxide reductase family protein containing a molybdenum cofactor and [3Fe–4S] cluster. The idrB gene encodes the electron-transfer subunit containing the [2Fe–2S] cluster. Both idrP1 and idrP2 encode di-heme proteins closely related to cytochrome c peroxidase, but their functions remain unclear.
The idrABP1P2 cluster is widely distributed in the genomes of anaerobic, facultative anaerobic, and aerobic bacteria (Yamazaki et al., 2020; Reyes-Umana et al., 2022a). The idrABP1P2 cluster is present in various aerobic Alphaproteobacteria isolated from surface seawater (Supplementary Figure S1). Alphaproteobacteria are predominantly found together with Gammaproteobacteria and Cyanobacteria in the surface waters of Mediterranean Sea, Black Sea, South Atlantic Ocean, and Northwestern Pacific Ocean (Haro-Moreno et al., 2018; Cabello-Yeves et al., 2021; Coutinho et al., 2021; Shih et al., 2023). In this study, three aerobic marine Alphaproteobacteria with the idrABP1P2 cluster in their genomes, namely Roseovarius azorensis (Rajasabapathy et al., 2014), Notoacmeibacter marinus (Huang et al., 2017), and Aliiroseovarius sediminilitoris (Park et al., 2015), were grown with iodate under various conditions to determine their iodate reduction capacity. R. azorensis was selected as the representative bacterium, and the effects of environmental factors on its iodate-reducing ability and transcription of idrABP1P2 genes were investigated. Furthermore, iodate concentration necessary to induce idrA transcription was examined and whether this concentration was comparable to the iodine levels in seawater was assessed. The possible physiological roles of iodate reduction by aerobic marine bacteria were also discussed.
2 Materials and methods
2.1 Growth conditions and culture media
R. azorensis KCTC 32421T (Rajasabapathy et al., 2014), N. marinus KCTC 52427T (Huang et al., 2017), and A. sediminilitoris KCTC 23959T (Park et al., 2015) were purchased from Korean Collection for Type Cultures (Jeonbuk, Korea). They were routinely subcultured aerobically at 30°C on the marine agar 2216 medium (Becton Dickinson, Sparks, MD). For iodate reduction, the cells were cultured in a 100 mL Erlenmeyer flask containing 25 mL of marine broth 2216 (Becton Dickinson) supplemented with 2 mM potassium iodate (Fujifilm Wako Chemicals, Osaka, Japan). The flask was sealed with a silicone septum and incubated at 30°C under static or shaking conditions at 180 rpm.
2.2 Iodate reduction by washed cell suspension
R. azorensis was grown under static or shaking conditions in the presence or absence of 2 mM iodate as described above. After 3 days, the cells were collected and washed twice with the washing buffer containing 20 mM Tris-HCl (pH 8.0), 330 mM NaCl, 30 mM MgCl2·6H2O, 13 mM MgSO4·7H2O, 12 mM CaCl2·2H2O, and 7 mM KCl. The washed cells were resuspended in the same buffer to yield an optical density at 600 nm (OD600) of 1.0. The washed cell suspension (20 mL) was dispensed into the 100 mL Erlenmeyer flask or 60 mL serum bottle under N2 gas stream. Glucose (5 mM) and iodate (1 mM) were added to the cell suspension as the electron donor and acceptor, respectively. The flask was sealed with a silicone septum and incubated aerobically with shaking as described above. The serum bottle was sealed with a thick butyl rubber stopper and aluminum cap and incubated anaerobically under static conditions.
2.3 Preparation of crude cell extract and enzyme assay
To prepare the crude extract, the cells were grown statically with 2 mM iodate for 3 days, collected, washed, and resuspended in the washing buffer. The cells were disrupted via sonication (Q500 Sonicator; Qsonica, Newtown, CT, United States), followed by centrifugation (17,000 × g, 30 min, 4°C) to remove the cell debris, and the supernatant was used as the crude enzyme. Then, iodate reductase (Idr) activity was assayed using a spectrophotometer with reduced methyl viologen (MV) as an electron donor, as previously described (Yamazaki et al., 2020). The detection limit of the enzyme assay was approximately 0.1 U mg−1.
2.4 Electrophoresis, activity staining, and liquid chromatography-tandem mass spectrometry analyses
Electrophoresis was performed in two steps to separate the proteins present in the crude extract (Yamazaki et al., 2020). First, non-denaturing gel electrophoresis was performed at 4°C using 10% polyacrylamide gel in 25 mM Tris-glycine buffer (pH 8.3), excluding both the reducing agent and SDS. Following non-denaturing electrophoresis, the gel was incubated in a nitrogen atmosphere with 20 mM Tris-HCl (pH 6.8) containing 0.3 mM MV, 12 mM iodate, and 10 mM dithionite. Then, the clear band (active band) was excised from the gel, boiled with 2% SDS and 5% 2-mercaptoethanol for 5 min for complete denaturation, and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). A Protein Molecular Weight Marker (Takara, Otsu, Japan) was used as the standard. Proteins were visualized by staining with Coomassie brilliant blue R-250, and the selected bands were excised, trypsin-digested, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis, as previously described (Shevchenko et al., 1996).
2.5 Transcriptional analysis of idr genes
All manipulations were performed as previously described (Yamazaki et al., 2020). RNA was obtained from the static and shaking cultures in the presence (0.1–1,000 μM) or absence of iodate. Cells in the late exponential growth phase were collected, and pellets were stored at −80°C. Total RNA was extracted using the RNeasy Miniprep kit (QIAGEN, Hilden, Germany) and treated with the TURBO DNA-free kit (Ambion, Carlsbad, CA, United States) to remove the residual DNA. cDNA was synthesized using the SuperScript First-Strand Synthesis System for reverse transcription-polymerase chain reaction (RT-PCR; Invitrogen, Carlsbad, CA, United States). Quantification of idrA, idrB, idrP1, and idrP2 in cDNA samples was performed using quantitative PCR (qPCR). The 16S rRNA gene was quantified in the cDNA samples to normalize the qRT-PCR data. As shown in Supplementary Table S3, new primers were designed for qPCR analysis. SYBR Green detection was performed for qPCR assays using the StepOnePlus instrument (Applied Biosystems). Standard curves were plotted using serial dilutions of cDNA prepared from cells grown in iodate. The slopes of standard curves were used to calculate PCR efficiency according to the following equation:
E values for all genes and primer pairs ranged from 95 to 105%, with R2 values >0.99.
2.6 Analytical technique
Iodate and iodide concentrations were determined using the ion chromatograph IC-2010 (Tosoh, Tokyo, Japan), as previously described (Yamazaki et al., 2020). The detection limits of iodate and iodide were 3 and 5 μM, respectively.
3 Results
3.1 Iodate reduction by marine aerobic bacterial cultures
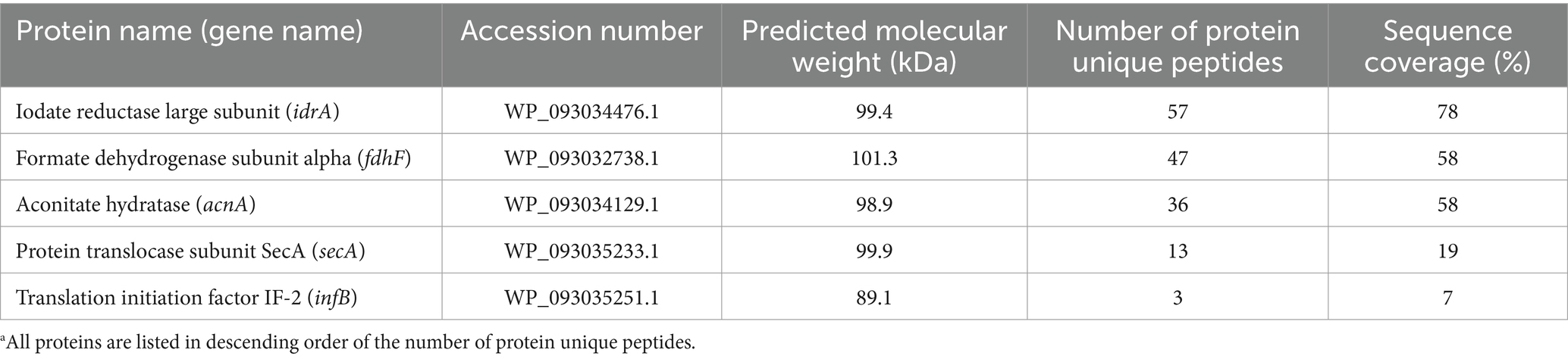
R. azorensis, N. marinus, and A. sediminilitoris were grown in the presence of 2 mM iodate under static and shaking conditions. Under static conditions, all bacteria reduced 2 mM iodate to iodide almost completely within 3 days (Figures 1, 2; Supplementary Figures S2, S3). However, the bacteria only reduced 0.1–0.5 mM iodate under shaking conditions despite much faster growth than that under static conditions. We attempted to grow these bacteria under anaerobic conditions in serum bottles filled with N2 gas. However, they could not grow or reduce iodate under these conditions (data not shown).
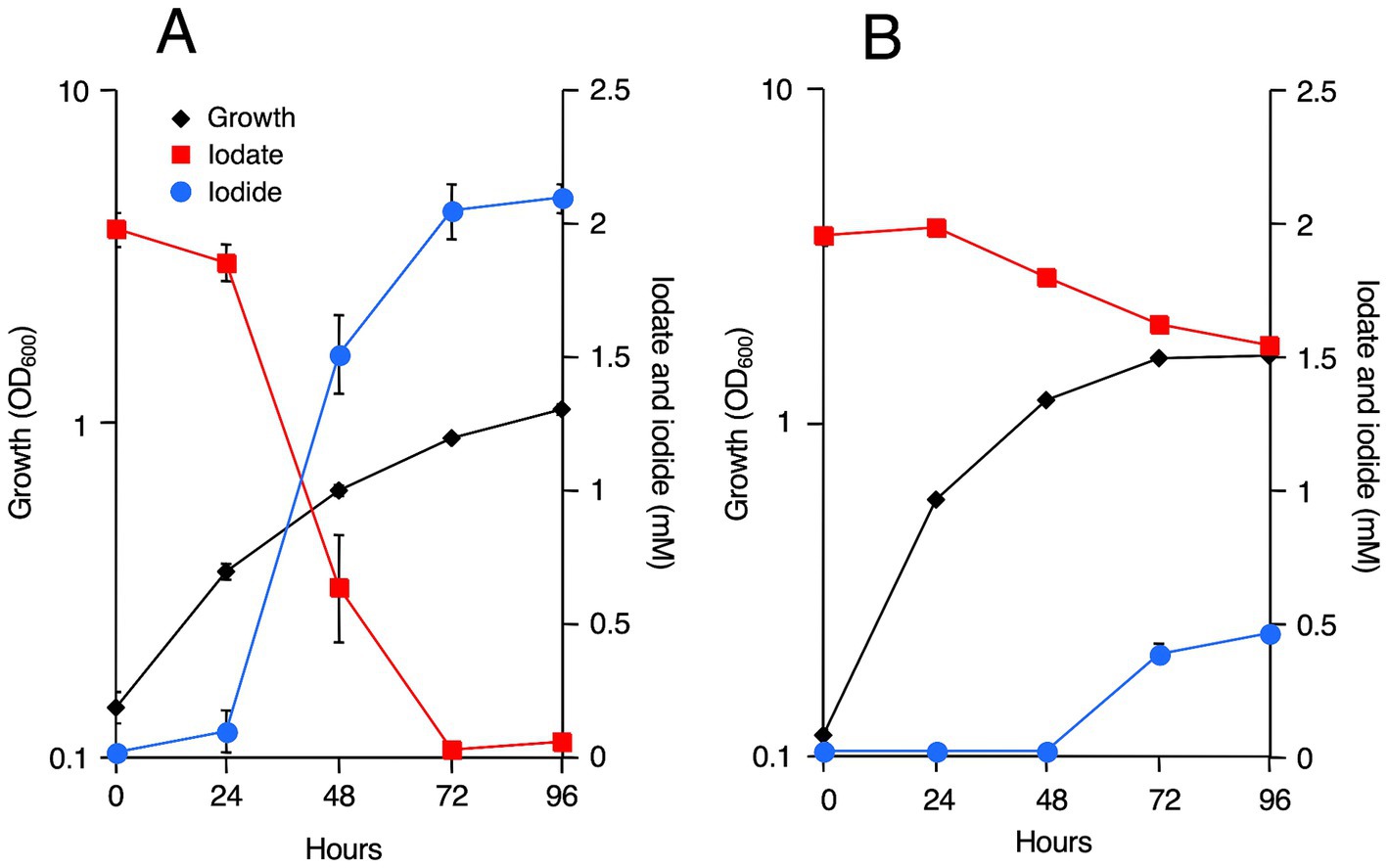
Figure 1. Iodate reduction by Roseovarius azorensis in marine broth 2216 under static (A) and shaking (B) conditions. All experiments were performed in triplicate, and error bars represent the standard deviations. Absence of bars indicates that the error is too small to be denoted by symbols.
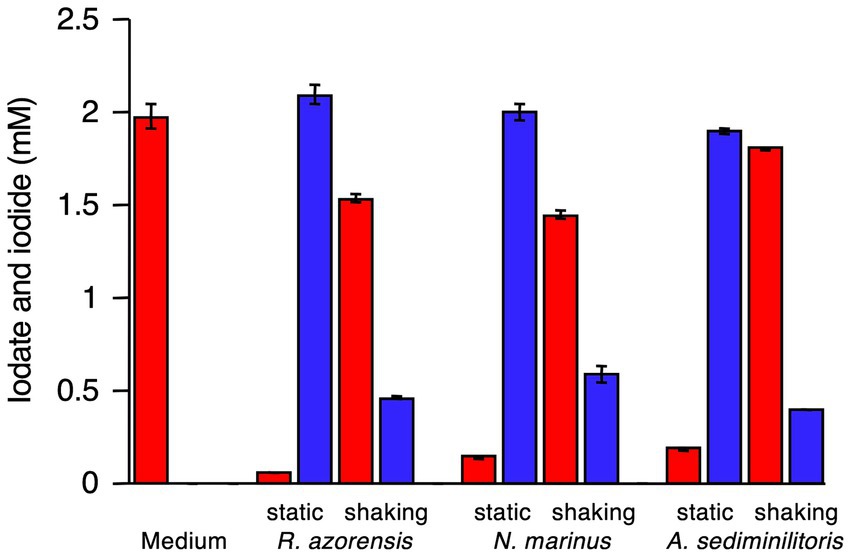
Figure 2. Iodate reduction by R. azorensis, Notoacmeibacter marinus, and Aliiroseovarius sediminilitoris in marine broth 2216 under static and shaking conditions. Iodate (red) and iodide (blue) concentrations after cultivation for 96 h are shown. Data from an uninoculated medium used as a control are also shown. All experiments were performed in triplicate, and error bars represent the standard deviations.
3.2 Iodate reduction by the washed cell suspension of Roseovarius azorensis
R. azorensis was selected as the representative bacterium and grown under static and shaking conditions in the presence or absence of iodate. Washed cell suspensions were prepared from four types of cultures and incubated with iodate aerobically (in Erlenmeyer flasks with shaking) or anaerobically (in serum bottles filled with N2 gas). As shown in Figure 3, cells pre-grown under shaking conditions with or without iodate did not reduce iodate, regardless of the incubation conditions (aerobic or anaerobic). Similarly, cells pre-grown under static conditions without iodate did not reduce iodate, regardless of the incubation conditions. Notably, cells pre-grown under static conditions with iodate rapidly reduced iodate and produced iodide under anaerobic conditions (Figure 3). However, the same cells did not reduce iodate under aerobic conditions. These results suggest that the iodate-reducing capacity is induced only when R. azorensis is grown statically in the presence of iodate and that the induced cells can reduce iodate only under anaerobic conditions, but not under aerobic conditions.
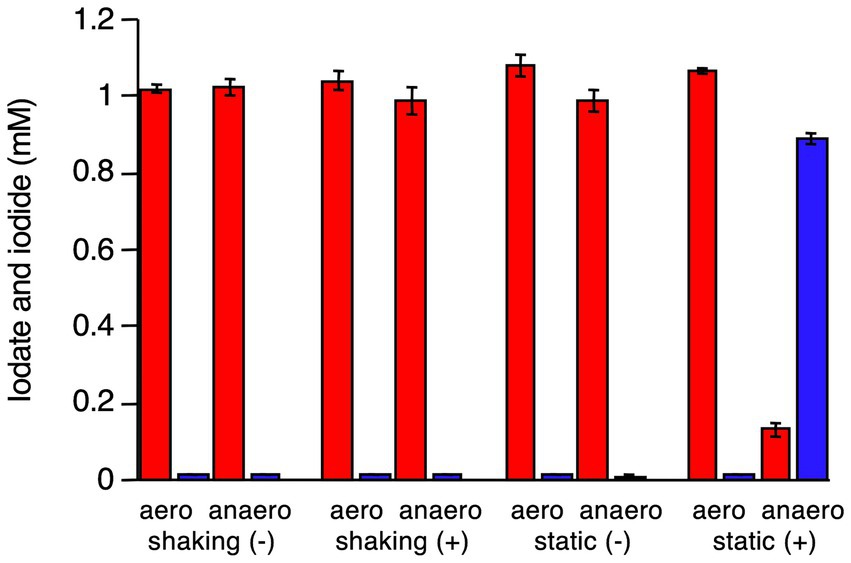
Figure 3. Iodate reduction by the washed cell suspension of R. azorensis under various conditions. Cells were pre-grown under shaking and static conditions with (+) or without (−) iodate. Then, the washed cells were incubated aerobically (aero) or anaerobically (anaero). Iodate (red) and iodide (blue) concentrations in washed cells after incubation for 8 h are shown. All experiments were performed in triplicate, and error bars represent the standard deviations.
3.3 Activity and identification of Idr
R. azorensis was grown under static conditions in the presence of iodate, and Idr activity in the crude extract was determined using reduced MV as an electron donor. However, significant Idr activity of more than 0.1 U mg−1 was not detected in this study (data not shown). To visualize the Idr activity directly on a polyacrylamide gel, the crude extract was subjected to SDS-PAGE under non-denaturing conditions. The gel was stained for Idr activity under anaerobic conditions using reduced MV as an electron donor. A clear band was observed in the gel (Figure 4A), excised, and subjected to SDS-PAGE under denaturing conditions. As shown in Figure 4B, multiple bands with apparent molecular weights of 44–100 kDa were observed after Coomassie brilliant blue staining. A band with a molecular weight of approximately 100 kDa was excised, trypsin-digested, and subjected to liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. From this band, 24 proteins with apparent molecular weights of 9–101 kDa were identified (Supplementary Table S1). One protein with accession number WP_093034476.1 showed the highest sequence coverage of 78% and highest number of unique protein peptides (Table 1). BLASTP analysis revealed that this protein was a homolog of IdrA in Pseudomonas sp. SCT, suggesting its involvement in iodate reduction in R. azorensis.
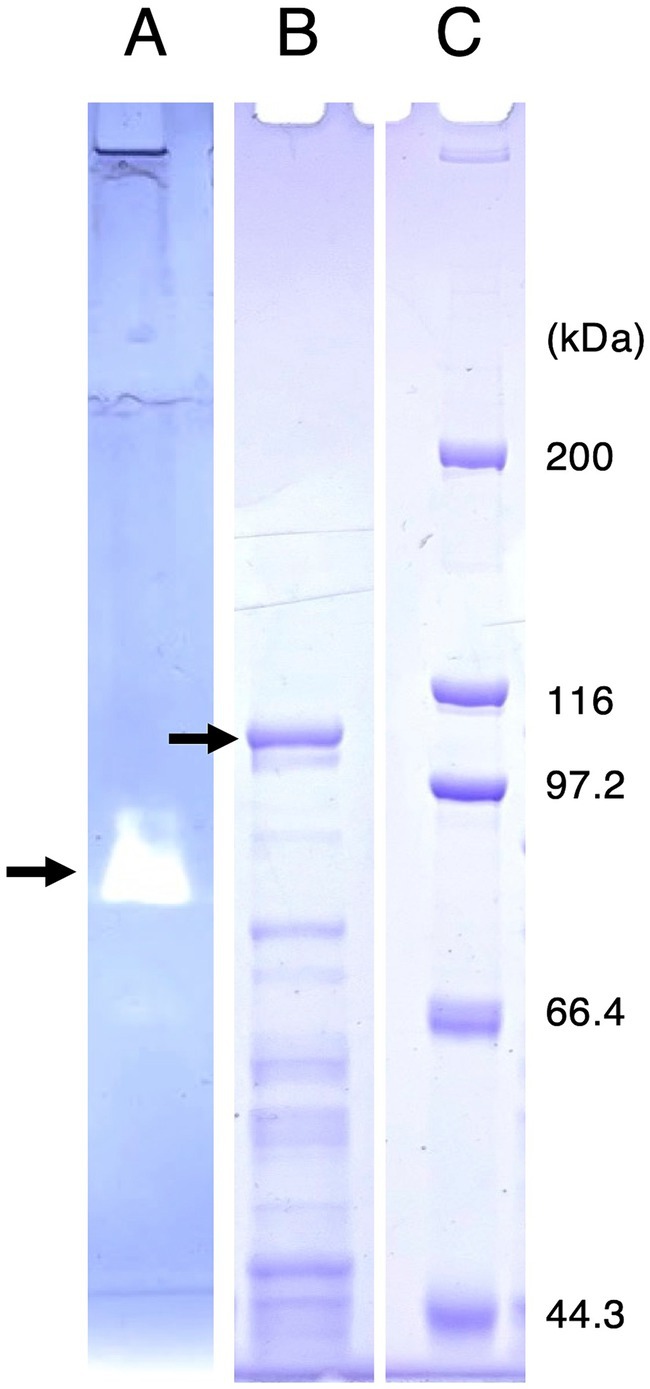
Figure 4. (A) Activity staining of iodate reductase (Idr) protein. Roseovarius azorensis crude extract was subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) at 4°C under non-denaturing conditions (without SDS and reducing agents). After electrophoresis, the gel was stained anaerobically with methyl viologen and iodate as the electron donor and acceptor, respectively. A clear band (arrow) represents the active band. (B) Separation of the excised active band under completely denaturing conditions (100°C, 5 min with SDS and 2-mercaptoethanol). The gel was stained with Coomassie brilliant blue R-250. A band with a molecular weight of approximately 100 kDa (arrow) was excised, trypsin-digested, and subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. (C) Standard marker proteins. Note that (B,C) show images of the same SDS-PAGE gel.
3.4 Transcriptional analysis of idr gene cluster
Expression levels of idrA, idrB, idrP1, and idrP2 relative to those of 16S rRNA gene were quantified and compared using RNA extracted from cells grown under static and shaking conditions in the presence or absence of iodate. As shown in Figure 5 and Supplementary Table S2, expression patterns of these four genes were very similar. Highest expression was observed in the cells grown statically with iodate and lowest expression was observed in the cells grown under shaking conditions without iodate. The presence of iodate induced idr gene expression by 8.0–14-fold under shaking growth conditions and by 59–102-fold under static growth conditions (Supplementary Table S2). Static growth conditions induced idr gene expression by 5.7–12-fold in the absence of iodate and by 43–61-fold in the presence of iodate. Therefore, transcription of the idrABP1P2 cluster is upregulated when R. azorensis is grown statically in the presence of iodate.
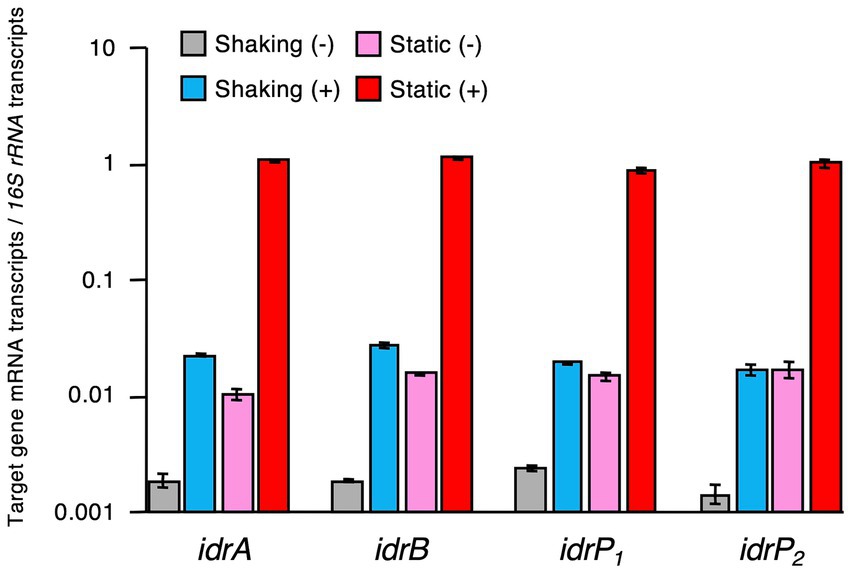
Figure 5. Expression levels of iodate reductase (idr) genes relative to those of 16S rRNA gene in R. azorensis cells grown under various conditions. R. azorensis cells were grown under shaking conditions without (gray) or with (cyan) iodate and under static conditions without (pink) or with (red) iodate. Values represent the ratio of the relative quantity of idrABP1P2 transcripts to that of 16S rRNA gene transcripts. All values represent the mean values obtained from triplicate experiments, and bars represent the standard deviations.
Next, we determined the effect of iodate on idrA expression. R. azorensis was grown under static conditions in the absence or presence of 0.1–1,000 μM iodate. As shown in Figure 6, idrA expression was detected at 0.1 μM iodate and was up to 14-fold higher than that in the non-iodate control.
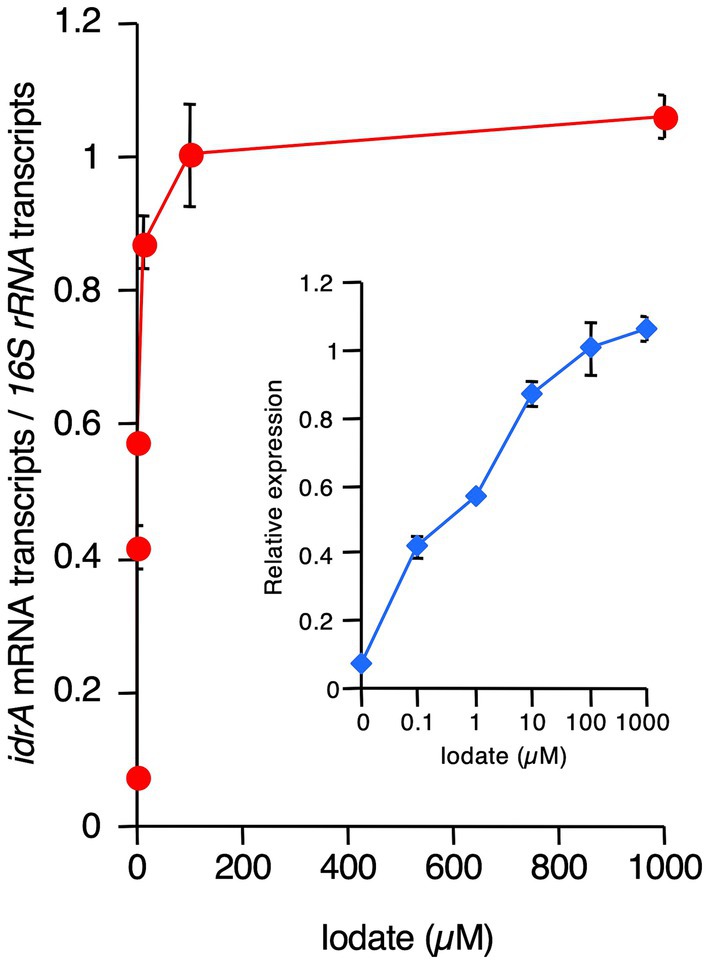
Figure 6. Effects of increasing concentrations of iodate on idrA expression levels in R. azorensis. Expression of idrA gene relative to that of 16S rRNA gene was determined as a function of increasing iodate concentrations in R. azorensis cultures. The bacterium was grown statically in the absence (0 μM) or presence of the indicated concentrations of iodate. All values represent the mean values obtained from triplicate experiments, and bars indicate the standard deviations. Absence of bars indicates that the error is too small to be denoted by symbols. Inlet shows the same graph, but with the X-axis displayed in the logarithmic scale.
4 Discussion
All three strains used in this study reduced 2 mM iodate almost completely under static conditions, but not under shaking conditions (Figures 1, 2; Supplementary Figures S2, S3). This was expected as the proteins encoded by idrA and idrB genes contain [3Fe–4S] and [2Fe–2S] clusters, respectively (Yamazaki et al., 2020). Fe–S clusters in metalloproteins are highly sensitive to molecular oxygen due to the decreased bioavailability of ferric irons and direct oxidation of these clusters by reactive oxygen species (Boyd et al., 2014). Experiments with washed cell suspension clearly showed that static growth conditions were necessary to confer iodate-reducing capacity to R. azorensis cells (Figure 3). Even though the cells exhibited iodate-reducing capacity, they did not reduce iodate under aerobic conditions, suggesting that enzyme Idr is oxygen-sensitive. Furthermore, transcriptomic analysis revealed that the idrABP1P2 cluster was highly upregulated under static conditions (Figure 5). These results suggest that oxygen-limited conditions are essential for both iodate reduction by Idr and biosynthesis of Idr.
In addition to oxygen, iodate is another environmental factor significantly affecting iodate reduction by marine aerobic bacteria (Figures 3, 5). Induction of Idr and upregulation of idrABP1P2 cluster expression by iodate have been observed in various iodate-reducing bacteria, such as Pseudomonas sp. SCT and Azoarcus sp. DN11 (Yamazaki et al., 2020; Sasamura et al., 2023). Therefore, iodate may be a universal inducer of the idr gene cluster in bacteria. Here, transcription of idrA was stimulated at iodate concentrations >0.1 μM (Figure 6). As the average concentration of total dissolved iodine in seawater is 0.45 μM (Wong, 1991) and iodate is the more stable form of iodine in seawater, transcription of idr gene cluster and biosynthesis of Idr proteins may be induced by environmental levels of iodate in seawater. Thus, aerobic bacteria may actually reduce iodate in marine surface waters at relatively low levels of oxygen.
Where are such conditions present in the well-oxygenated surface seawater? Although we did not use any devises for measurement of dissolved oxygen concentration in our culture medium, a recent paper by Huang et al. (2021) reported that dissolved oxygen concentrations in bacterial shaking and static cultures are approximately 8 mg L−1 and 1 mg L−1, respectively. Considering that typical dissolved oxygen concentrations in surface seawater are 5 to 8 mg L−1 (Wright et al., 2012), our shaking cultures successfully mimicked surface water environment. However, dissolved oxygen concentrations in surface seawater are sometimes very low, for example 1 to 3 mg L−1 in the eastern tropical North Pacific (Hoogakker et al., 2018). In addition, the interior of organic-rich aggregates or detritus could potentially be suitable micro-habitats for aerobic iodate-reducing bacteria. Heterotrophic aerobic bacteria associated with particulate organic matter may reduce iodate after active degradation and assimilation of organic compounds, which is combined with the rapid consumption of dissolved oxygen in the micro-habitats. Such local and transient hypoxic environments may be hot spots of iodate reduction in marine surface waters. If the associated particulate organic matter is microalgal remains, the iodate reduction observed during algal senescence may partly be due to heterotrophic aerobic bacteria (Bluhm et al., 2010; Hepach et al., 2020).
Putative dissolved oxygen concentration in our static cultures (1 mg L−1) was similar to those observed in OMZs, which occur in the Pacific Ocean, the Atlantic Ocean, and the Arabian Sea (Wright et al., 2012). Thus, it may be possible that aerobic bacteria also contribute to iodide production in these oxygen-depleted marine waters. It was reported that Alphaproteobacteria are predominant together with Gammaproteobacteria in OMZs of the eastern tropical South Pacific (Stevens and Ulloa, 2008) and the Arabian Sea (Bandekar et al., 2018). In addition, Kalvelage et al. (2015) found that Alphaproteobacterial terminal respiratory oxidase genes and transcripts are abundant in the metagenomes and metatranscriptomes of OMZs off Peru and Chile, suggesting that microaerobic respiration is a major mode of organic matter degradation in these OMZs. Furthermore, metagenomic analysis of the eastern tropical Pacific and the Arabian Sea OMZs actually identified idrA genes (Reyes-Umana et al., 2022a). These results suggest the possibility that aerobic iodate-reducing bacteria are producing iodide from iodate in various OMZs together with anaerobic iodate-respiring bacteria. Considering that total iodine concentrations in OMZs are sometimes significantly higher than those in surface seawater (Chapman, 1983), and that iodide is predominantly observed over iodate in these oxygen-depleted waters (Farrenkopf et al., 1997; Farrenkopf and Luther, 2002), aerobic bacteria might play a significant role in iodine cycling in deep waters.
Physiological mechanisms by which marine aerobic bacteria reduce iodate remain unclear. As these bacteria were unable to grow anaerobically with iodate as the terminal electron acceptor (data not shown), it may not be an anaerobic respiratory process. As iodate reduction only occurs under oxygen-limited conditions, iodate may be used as an electron sink to dispose the excess reducing power instead of oxygen. This iodate-reducing capacity may aid aerobic bacteria in maintaining their metabolism in iodate-rich seawater, even under oxygen-limited conditions, and might prevent them from losing viability. Considering that the idrABP1P2 cluster is acquired by horizontal gene transfer (Reyes-Umana et al., 2022a, 2022b), iodate reduction should have an advantage for aerobic bacteria.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
KK: Data curation, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing, Formal analysis. SY: Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. SA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by JSPS KAKENHI (grant number 20H02896).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1446596/full#supplementary-material
References
Bandekar, M., Ramaiah, N., Jain, A., and Meena, R. M. (2018). Seasonal and depth-wise variations in bacterial and archaeal groups in the Arabian Sea oxygen minimum zone. Deep-Sea Res. II: Top. Stud. Oceanogr. 156, 4–18. doi: 10.1016/j.dsr2.2017.12.015
Bluhm, K., Croot, P., Wuttig, K., and Lochte, K. (2010). Transformation of iodate to iodide in marine phytoplankton driven by cell senescence. Aquat. Biol. 11, 1–15. doi: 10.3354/ab00284
Boyd, E. S., Thomas, K. M., Dai, Y., Boyd, J. M., and Outten, F. W. (2014). Interplay between oxygen and Fe–S cluster biogenesis: insights from the Suf pathway. Biochemistry 53, 5834–5847. doi: 10.1021/bi500488r
Cabello-Yeves, P. J., Callieri, C., Picazo, A., Mehrshad, M., Haro-Moreno, J. M., Roda-Garcia, J. J., et al. (2021). The microbiome of the Black Sea water column analyzed by shotgun and genome centric metagenomics. Environ. Microbiome 16:5. doi: 10.1186/s40793-021-00374-1
Campos, M. L. A. M., Farrenkopf, A. M., Jickells, T. D., and Luther, G. W. III (1996). A comparison of dissolved iodine cycling at the Bermuda Atlantic Time-series Station and Hawaii Ocean Time-series Station. Deep-Sea Res. II: Top. Stud. Oceanogr. 43, 455–466. doi: 10.1016/0967-0645(95)00100-X
Carpenter, L. J., Chance, R. J., Sherwen, T., Adams, T. J., Ball, S. M., Evans, M. J., et al. (2021). Marine iodine emissions in a changing world. Proc. Math. Phys. Eng. Sci. 477:20200824. doi: 10.1098/rspa.2020.0824
Carpenter, L. J., MacDonald, S. M., Shaw, M. D., Kumar, R., Saunders, R. W., Parthipan, R., et al. (2013). Atmospheric iodine levels influenced by sea surface emissions of inorganic iodine. Nat. Geosci. 6, 108–111. doi: 10.1038/ngeo1687
Chaker, L., Razvi, S., Bensenor, I. M., Azizi, F., Pearce, E. N., and Peeters, R. P. (2022). Hypothyroidism. Nat. Rev. Dis. Primers 8:30. doi: 10.1038/s41572-022-00357-7
Chance, R., Malin, G., Jickells, T., and Baker, A. R. (2007). Reduction of iodate to iodide by cold water diatom cultures. Mar. Chem. 105, 169–180. doi: 10.1016/j.marchem.2006.06.008
Chapman, P. (1983). Changes in iodine speciation in the Benguela Current upwelling system. Deep-Sea Res. 30:1247–1259.
Coutinho, F. H., von Meijenfeldt, F. A. B., Walter, J. M., Haro-Moreno, J. M., Lopéz-Pérez, M., van Verk, M. C., et al. (2021). Ecogenomics and metabolic potential of the South Atlantic Ocean microbiome. Sci. Total Environ. 765:142758. doi: 10.1016/j.scitotenv.2020.142758
Dijck-Brouwer, D. A. J., Muskiet, F. A. J., Verheesen, R. H., Schaafsma, G., Schaafsma, A., and Geurts, J. M. W. (2022). Thyroidal and extrathyroidal requirements for iodine and selenium: a combined evolutionary and (patho)physiological approach. Nutrients 14:3886. doi: 10.3390/nu14193886
Farrenkopf, A. M., and Luther, G. W. III (2002). Iodine chemistry reflects productivity and denitrification in the Arabian Sea: evidence for flux of dissolved species from sediments of western India into the OMZ. Deep-Sea Res. II: Top. Stud. Oceanogr. 49, 2303–2318. doi: 10.1016/S0967-0645(02)00038-3
Farrenkopf, A. M., Luther, G. W. III, Truesdale, V. W., and van der Weijden, C. H. (1997). Sub-surface iodide maxima: evidence for biologically catalyzed redox cycling in Arabian Sea OMZ during the SW intermonsoon. Deep-Sea Res. II: Top. Stud. Oceanogr. 44, 1391–1409. doi: 10.1016/S0967-0645(97)00013-1
Fuge, R., and Johnson, C. C. (2015). Iodine and human health, the role of environmental geochemistry and diet, a review. Appl. Geochem. 63, 282–302. doi: 10.1016/j.apgeochem.2015.09.013
Haro-Moreno, J. M., López-Pérez, M., de la Torre, J. R., Picazo, A., Camacho, A., and Rodriguez-Valera, F. (2018). Fine metagenomic profile of the Mediterranean stratified and mixed water columns revealed by assembly and recruitment. Microbiome 6:128. doi: 10.1186/s40168-018-0513-5
Hepach, H., Hughes, C., Hogg, K., Collings, S., and Chance, R. (2020). Senescence as the main driver of iodide release from a diverse range of marine phytoplankton. Biogeosciences 17, 2453–2471. doi: 10.5194/bg-17-2453-2020
Hoogakker, B. A. A., Lu, Z., Umling, N., Jones, L., Zhou, X., Rickaby, R. E. M., et al. (2018). Glacial expansion of oxygen-depleted seawater in the eastern tropical Pacific. Nature 562, 410–413. doi: 10.1038/s41586-018-0589-x
Huang, Z., Guo, F., Lai, Q., and Shao, Z. (2017). Notoacmeibacter marinus gen. nov., sp. nov., isolated from the gut of a limpet and proposal of Notoacmeibacteraceae fam. nov. in the order Rhizobiales of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol. 67, 2527–2531. doi: 10.1099/ijsem.0.001951
Huang, Y., Lu, Z., Jiang, T., Zeng, Y., Zeng, Y., and Chen, B. (2021). Oxygen availability affects the synthesis of quorum sensing signal in the facultative anaerobe Novosphingomonas pentaromativorans US6-1. Appl. Microbiol. Biotechnol. 105, 1191–1201. doi: 10.1007/s00253-021-11089-1
Jones, C. E., Hornsby, K. E., Sommariva, R., Dunk, R. M., von Glasow, R., McFiggans, G., et al. (2010). Quantifying the contribution of marine organic gases to atmospheric iodine. Geophys. Res. Lett. 37, 1–6. doi: 10.1029/2010GL043990
Jones, M. R., Chance, R., Bell, T., Jones, O., Loades, D. C., May, R., et al. (2024). Iodide, iodate & dissolved organic iodine in the temperate coastal ocean. Front. Mar. Sci. 11: 1277595.
Kalvelage, T., Lavik, G., Jensen, M. M., Revsbech, N. P., Löscher, C., Schunck, H., et al. (2015). Aerobic microbial respiration in oceanic oxygen minimum zones. PLoS One 10:e0133526. doi: 10.1371/journal.pone.0133526
Luther, G. W. III (2023). Review on the physical chemistry of iodine transformations in the oceans. Front. Mar. Sci. 10:1085618. doi: 10.3389/fmars.2023.1085618
MacDonald, S. M., Gómez Martín, J. C., Chance, R., Warriner, S., Saiz-Lopez, A., Carpenter, L. J., et al. (2014). A laboratory characterization of inorganic iodine emissions from the sea surface: dependence on oceanic variables and parameterization for global modeling. Atmos. Chem. Phys. 14, 5841–5852. doi: 10.5194/acp-14-5841-2014
Mok, J. K., Toporek, Y. J., Shin, H.-D., Lee, B. D., Lee, M. H., and DiChristina, T. J. (2018). Iodate reduction by Shewanella oneidensis does not involve nitrate reductase. Geomicrobiol J. 35, 570–579. doi: 10.1080/01490451.2018.1430189
Moriyasu, R., Bolster, K. M., Hardisty, D. S., Kadko, D. C., Stephens, M. P., and Moffett, J. W. (2023). Meridional survey of the Central Pacific reveals iodide accumulation in equatorial surface waters and benthic sources in the abyssal plain. Global Biogeochem. Cycles 37:e2021. doi: 10.1029/2021GB007300
O’Dowd, C. D., Jimenez, J. L., Bahreini, R., Flagan, R. C., Seinfeld, J. H., Hämeri, K., et al. (2002). Marine aerosol formation from biogenic iodine emissions. Nature 417, 632–636. doi: 10.1038/nature00775
Park, S., Park, J.-M., Kang, C.-H., and Yoon, J.-H. (2015). Aliiroseovarius pelagivivens gen. nov., sp. nov., isolated from seawater, and reclassification of three species of the genus Roseovarius as Aliiroseovarius crassostreae comb. nov., Aliiroseovarius halocynthiae comb. nov. and Aliiroseovarius sediminilitoris comb. nov. Int. J. Syst. Evol. Microbiol. 65, 2646–2652. doi: 10.1099/ijs.0.000315
Rajasabapathy, R., Mohandass, C., Dastager, S. G., Liu, Q., Khieu, T.-N., Son, C. K., et al. (2014). Roseovarius azorensis sp. nov., isolated from seawater at Espalamaca, Azores. Antonie Van Leeuwenhoek 105, 571–578. doi: 10.1007/s10482-013-0109-9
Reyes-Umana, V., Henning, Z., Lee, K., Barnum, T. P., and Coates, J. D. (2022a). Genetic and phylogenetic analysis of dissimilatory iodate-reducing bacteria identifies potential niches across the world’s oceans. ISME J. 16, 38–49. doi: 10.1038/s41396-021-01034-5
Reyes-Umana, V., Kretschmer, J., and Coates, J. D. (2022b). Isolation of a dissimilatory iodate-reducing Aromatoleum sp. from a freshwater creek in the San Francisco Bay area. Front. Microbiol. 12:804181. doi: 10.3389/fmicb.2021.804181
Saiz-Lopez, A., Plane, J. M. C., Baker, A. R., Carpenter, L. J., von Glasow, R., Martín, J. C. G., et al. (2012). Atmospheric chemistry of iodine. Chem. Rev. 112, 1773–1804. doi: 10.1021/cr200029u
Sasamura, S., Ohnuki, T., Kozai, N., and Amachi, S. (2023). Iodate respiration by Azoarcus sp. DN11 and its potential use for removal of radioiodine from contaminated aquifers. Front. Microbiol. 14:1162788. doi: 10.3389/fmicb.2023.1162788
Shevchenko, A., Wilm, M., Vorm, O., and Mann, M. (1996). Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858. doi: 10.1021/ac950914h
Shih, C.-Y., Chen, S.-Y., Hsu, C.-R., Chin, C.-H., Chiu, W.-C., Chang, M.-H., et al. (2023). Distinctive microbial community and genome structure in coastal seawater from a human-made port and nearby offshore island in northern Taiwan facing the northwestern Pacific Ocean. PLoS One 18:e0284022. doi: 10.1371/journal.pone.0284022
Sillen, L. G. (1961). “The physical chemistry of seawater” in Oceanography. ed. M. Sears (Washington, DC: American Association for the Advancement of Science), 549–581.
Stevens, H., and Ulloa, O. (2008). Bacterial diversity in the oxygen minimum zone of the eastern tropical South Pacific. Environ. Microbiol. 10, 1244–1259. doi: 10.1111/j.1462-2920.2007.01539.x
Tian, R. C., Marty, J. C., Nicolas, E., Chiavérini, J., Ruiz-Ping, D., and Pizay, M. D. (1996). Iodine speciation: a potential indicator to evaluate new production versus regenerated production. Deep-Sea Res. I: Oceanogr. Res. Pap. 43, 723–738. doi: 10.1016/0967-0637(96)00023-4
Tian, R. C., and Nicolas, E. (1995). Iodine speciation in the northwestern Mediterranean Sea: method and vertical profile. Mar. Chem. 48, 151–156. doi: 10.1016/0304-4203(94)00048-I
Tsunogai, S., and Sase, T. (1969). Formation of iodide-iodine in the ocean. Deep-Sea Res. 16, 489–496.
van Bergeijk, S. A., Hernández, L., Zubía, E., and Cañavate, J. P. (2016). Iodine balance, growth and biochemical composition of three marine microalgae cultured under various inorganic iodine concentrations. Mar. Biol. 163:107. doi: 10.1007/s00227-016-2884-0
Waite, T. J., and Truesdale, V. W. (2003). Iodate reduction by Isochrysis galbana is relatively insensitive to de-activation of nitrate reductase activity—are phytoplankton really responsible for iodate reduction in seawater? Mar. Chem. 81, 137–148. doi: 10.1016/S0304-4203(03)00013-6
Wong, G. T. F., Piumsomboon, A. U., and Dunstan, W. M. (2002). The transformation of iodate to iodide in marine phytoplankton cultures. Mar. Ecol. Prog. Ser. 237, 27–39. doi: 10.3354/meps237027
Wong, G. T. F., Takayanagi, K., and Todd, J. F. (1985). Dissolved iodine in waters overlying and in the Orca Basin, Gulf of Mexico. Mar. Chem. 17, 177–183. doi: 10.1016/0304-4203(85)90072-6
Wright, J. J., Konwar, K. M., and Hallam, S. J. (2012). Microbial ecology of expanding oxygen minimum zones. Nat. Rev. Microbiol. 10, 381–394. doi: 10.1038/nrmicro2778
Yamazaki, C., Kashiwa, S., Horiuchi, A., Kasahara, Y., Yamamura, S., and Amachi, S. (2020). A novel dimethylsulfoxide reductase family of molybdenum enzyme, Idr, is involved in iodate respiration by Pseudomonas sp. SCT. Environ. Microbiol. 22, 2196–2212. doi: 10.1111/1462-2920.14988
Keywords: iodate reduction, iodide production, marine aerobic bacteria, Roseovarius azorensis, idrABP1P2
Citation: Kine K, Yamamura S and Amachi S (2024) Iodate reduction by marine aerobic bacteria. Front. Microbiol. 15:1446596. doi: 10.3389/fmicb.2024.1446596
Edited by:
Partha Basu, Indiana University–Purdue University Indianapolis, United StatesReviewed by:
Michael Wells, Louisiana Tech University, United StatesXiyang Dong, Third Institute of Oceanography of the Ministry of Natural Resources, China
Copyright © 2024 Kine, Yamamura and Amachi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Seigo Amachi, YW1hY2hpQGZhY3VsdHkuY2hpYmEtdS5qcA==
 Ken Kine1
Ken Kine1 Shigeki Yamamura
Shigeki Yamamura Seigo Amachi
Seigo Amachi