- 1College of Animal Science and Technology, Chongqing Key Laboratory of Herbivore Science, Southwest University, Chongqing, China
- 2Tengda Animal Husbandry Co., Ltd., Chongqing, China
- 3Chongqing Academy of Animal Science, Chongqing, China
The growth rate of young ruminants has been associated with production performance in later life, with recent studies highlighting the importance of rumen microbes in supporting the health and growth of ruminants. However, the specific role of rumen epithelium bacteria and microbiota-host interactions in influencing the early life growth rate of ruminants remains poorly understood. In this study, we investigated the rumen fermentation pattern, microbiota characteristics, and global gene expression profiles of the rumen epithelium in 6-month-old goats with varying growth rates. Our results showed that goats with high average daily gain (HADG) exhibited higher rumen propionate concentrations. Goats with low average daily gain (LADG) had the higher relative abundances of rumen epithelium bacteria genera U29-B03 and Quinella, while exhibiting a lower relative abundance of Lachnospiraceae UCG-009. In the rumen fluid, the relative abundances of bacteria genus Alloprevotella were lower and Desulfovibrio were higher in LADG goats compared to HADG goats. Additionally, the relative abundance of fungal genus Symmetrospora was lower in LADG goats compared to HADG goats. Transcriptome analysis showed that 415 genes were differentially expressed between LADG and HADG goats, which were enriched in functions related to cell junction and cell adhesion, etc. Correlation analysis revealed that rumen epithelium bacteria genera UCG-005 and Candidatus Saccharimonas were negatively associated, while Lachnospiraceae NK3A20 group and Oscillospiraceae NK4A214 group were positively associated with average daily gain (ADG) and genes related to barrier function. The rumen fluid bacteria genus Alloprevotella was positively correlated, while Desulfovibrio was negatively correlated with rumen propionate and ammoniacal nitrogen (NH3-N) concentrations, as well as genes related to barrier function and short chain fatty acids (SCFAs) transport. In summary, our study reveals that the higher ruminal fermentation efficiency, improved rumen epithelial barrier functions, and enhanced SCFAs transport in HADG goats could be attributed to the rumen microbiota, particularly the rumen epithelium bacteria, such as Lachnospiraceae and Oscillospiraceae NK4A214 group.
Introduction
The rumen constitutes a complex natural ecosystem that harbors diverse microbes responsible for biomass degradation, which provide nutrients for the host’s physiological needs. In return, the rumen provides a unique environment characterized by anaerobic conditions, stable temperature, and high osmotic pressure for the rumen microbes (Kamra, 2005). Emerging evidence suggests that the stable mutualistic relationship between rumen and its microbial inhabitants is pivotal for the growth, development (Malmuthuge et al., 2019; Jize et al., 2022), feed efficiency (Fonseca et al., 2023), and mitigation of acidosis (Li et al., 2019) in ruminants. However, these studies predominantly focused on rumen fluid-associated bacteria, which are more sensitive to the changes in environmental conditions (Liu et al., 2016) and differ from rumen epithelium bacteria (Pei et al., 2010). Rumen epithelium bacteria, directly adhering to the epithelium, likely engage in more interactions with the host. Moreover, fungi are immensely important for degrading ligno-carbohydrate complex in rumen (Bhagat et al., 2023). Yin et al. (2023) identified several fungal biomarkers in both rumen and rectum that were associated with growth rate in lambs. However, our understanding of the functional roles of rumen fungi remains limited and further investigations are required.
Early-life growth rate significantly influences later-life production performance. The pre-weaning and pre-pubertal average daily gain (ADG) has been reported to be correlated with milk production (Soberon and Van Amburgh, 2013; Gelsinger et al., 2016) and fertility (Schubach et al., 2019; Costa et al., 2021) in ruminants. There is a growing interest in exploring key microbes that affect the health and growth performance of ruminant animals. Yin et al. (2023) identified several microbes in both the rumen and the rectum associated with post-weaning ADG in lambs. The ADG-related rumen fluid microbiota, including the Prevotellaceae family, Streptococcus, and Candidatus Saccharimonans, is believed to contribute to nutrient metabolism functions and short chain fatty acids (SCFAs) production in dairy goats (Wang et al., 2023). Nevertheless, the role of rumen epithelium bacteria and microbiota-host interactions in influencing the early-life growth performance of goats remains unclear.
The objective of the present study was to investigate the relationships between ruminal microbiota and early-life growth performance in ruminants, and to further explore the underlying mechanisms involved in the microbiota-host interactions at transcriptome level. Here, 6-month-old goats with different ADG, raised under identical conditions, were selected as model animals to assess their rumen fermentation patterns, rumen microbiota characteristics, and global gene expression profiles of the rumen epithelium. The findings of this study may contribute to the development of novel strategies aimed at improving the growth rate of young ruminants, thereby potentially enhancing production performance in later life.
Materials and methods
Ethics statement
All animal experimental and protocols were approved by the Institutional Animal Care and Use Committee of Southwest University, Beibei, China (approval IACUC-20220630-04).
Animal experiment design
A total of 49 female Dazu black kids from Tengda Animal Husbandry Co., Ltd. (Chongqing, China) were used for the experiment. Birth weights of all goat kids were recorded immediately after birth and they were weaned at 2 months of age. The goats were fed with a total mixed ration (TMR) ad libitum twice daily at 0700 and 1,600, consisting of corn grain (475 g/kg), soybean meal (40 g/kg), alfalfa hay (450 g/kg), NaCl (5 g/kg), dicalcium phosphate (10 g/kg), sodium bicarbonate (10 g/kg), and a trace minerals and vitamins supplement (10 g/kg). The diet was antibiotic-free, meeting the recommendations of the Feeding Standard of Meat-producing Sheep and Goats of China, NY/T 816–2004 (Ministry of Agriculture, MOA, PRC, 2004).
At approximately 6 months of age (183.4 ± 6.2 days), the goats were weighed before morning feeding and then slaughtered after anesthesia. The ADG was calculated as the weight gain from birth to 6 months of age divided by the number of days. The average ADG for all 49 goats was 86.8 ± 10.9 g/day (Supplementary Table S1), after which goats were ranked based on individual ADG. The top 10% of goats were designated as the high ADG group (HADG, 99.8 ± 12.8 g/day), while the bottom 10% were designated as the low ADG group (LADG, 63.6 ± 3.94 g/day).
Dry matter intake was continuously measured for 2 days before slaughtering (844 ± 28 g/day for LADG goats and 857 ± 37 g/day for HADG goats, p = 0.55). The rumen fluid was filtered using a four-layer cheesecloth, and pH values were measured with a portable pH meter (PHB-4, Rex Instrument, Shanghai, China). Rumen epithelium samples were collected from the ventral sac of each goat immediately after slaughter and rinsed with 0.01 M phosphate-buffered saline (PBS) buffer. All samples were promptly immersed in liquid nitrogen and then stored at −80°C for subsequent analysis.
The SCFAs and NH3-N analysis
The rumen fluid was centrifuged for 10 min at 10,000 × g, and the supernatants were filtered with a membrane (0.22 μm). Acetate, propionate, and butyrate concentrations were determined using high-performance liquid chromatography (KC-811 column, Shodex; mobile phase, 3 mM perchloric acid; flow rate, 1.0 mL/min; temperature: 50°C). Ammonia-N (NH3-N) concentration was determined using the phenol-hypochlorite colorimetric method.
Rumen microbiota analysis
Total microbial DNA was extracted from rumen fluid and epithelial samples using the E.Z.N.A.® stool DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to the manufacturer’s protocol. The quality of the DNA was determined by 1% agarose gel electrophoresis. Bacterial 16S rRNA gene fragments (V3-V4) were amplified with primers 338F (5’-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′), and fungi ITS rRNA gene was amplified with primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3′) using an ABI GeneAmp® 9,700 PCR thermocycler (ABI, CA, United States). The amplicons were paired-end sequenced using the Illumina PE250 platform (Illumina, San Diego, USA). The raw sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) database (Accession Number: PRJNA1099693). The resulting sequences were filtered using FASTP (version 0.19.6) and merged using FLASH (version 1.2.11). The amplicon sequence variant (ASV), obtained using DADA2 in QIIME2 (version 2020.2), were assigned to taxonomies using SILVA database (version 138) and UNITE database (version 8.0) for bacteria and fungi, respectively. The alpha diversity indices, including Chao1, ACE, Sobs, Shannon, and Simpson, were analyzed using Mothur (version 1.30.2). Principal coordinate analysis (PCoA) was performed based on weighted UniFrac distances and significance was determined using ANOSIM with 999 permutations.
Transcriptome analysis
Total RNA was extracted from rumen epithelium using Trizol reagent (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol. RNA quality was determined by a 5,300 (Bioanalyser Agilent, Palo Alto, CA, United States) and high-quality RNA samples were sent to Majorbio Biotech (Shanghai, China) for commercial library preparation and sequencing on the Illumina Novaseq 6,000. The DEGs (differential expression genes) between groups were analyzed using DESeq2. Genes with |log2FC| ≥ 1 and FDR < 0.05 were considered as differentially expressed. The gene ontology (GO) functional-enrichment analyses were performed by Goatools to identify significantly enriched GO terms at a Bonferroni-corrected p-value ≤0.05. RNA sequencing data were deposited in the NCBI’s Gene Expression Omnibus under the accession number PRJNA1101717.
Tissue RNA extraction and real-time quantitative PCR
Total RNA was extracted from rumen epithelium using Trizol reagent (Tiangen Biotech, Beijing, China) according to the manufacturer’s protocol. RNA degradation and contamination were monitored through 1% agarose gels electrophoresis. RNA concentration and purity were examined using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, United States). The gene expression was detected by quantitative real-time PCR (RT-qPCR) using the 2−∆∆CT method. The primer sequences are listed in Supplementary Table S2. β-actin and GAPDH were used as the endogenous reference genes. All reactions were run in triplicate for each sample.
Statistical analysis
Statistical significance was assessed using an unpaired Student’s two-tailed t-test for average daily gain, ruminal pH, NH3-N, and SCFAs, or the one-way ANOVA with Tukey’s post hoc tests for alpha diversity of the bacterial community, using GraphPad Prism (version 9.2.0, GraphPad Software, San Diego, CA, United States). Results are expressed as means with the standard error of the means (SEM). A p-value of less than 0.05 was considered statistically significant. Correlations between rumen microbiota and growth performance, ruminal SCFAs and NH3-N, or gene expression were estimated by Spearman correlation analysis using the ‘Hmisc’ and ‘Corrplot’ packages in R (version 4.3.2).
Results
Ruminal pH, NH3-N, and short-chain fatty acids concentrations
HADG goats had higher rumen propionate concentrations (p < 0.05) and tended to have higher rumen NH3-N concentration (p = 0.07, Figure 1). However, no significant differences were found in rumen fluid pH, and ruminal acetate and butyrate concentrations between HADG and LADG goats.
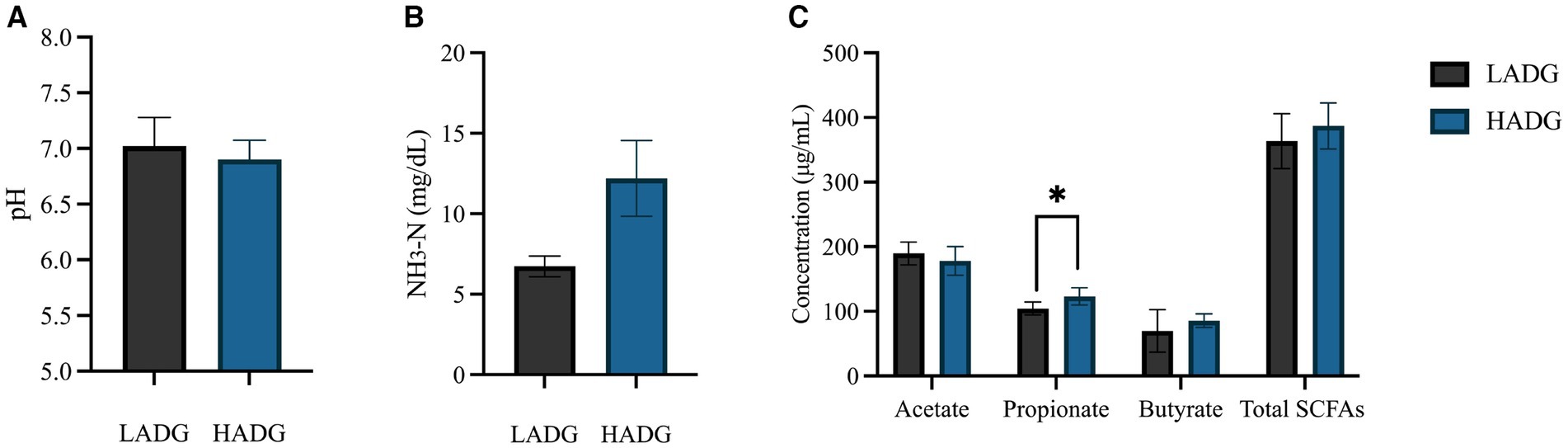
Figure 1. The ruminal pH (A), NH3-N (B), and short-chain fatty acids (SCFAs) concentrations (C) of low average daily gain (LADG) or high average daily gain (HADG) goats. * p < 0.05.
Microbiota composition of rumen fluid and epithelium
In the current study, 16 s and ITS gene sequencing techniques were used to determine the differences in rumen microbiota, especially for rumen epithelium bacteria, between HADG and LADG goats, aiming to explore the potential microbial targets for the development of novel intervention strategies to improve the growth performance of young ruminants.
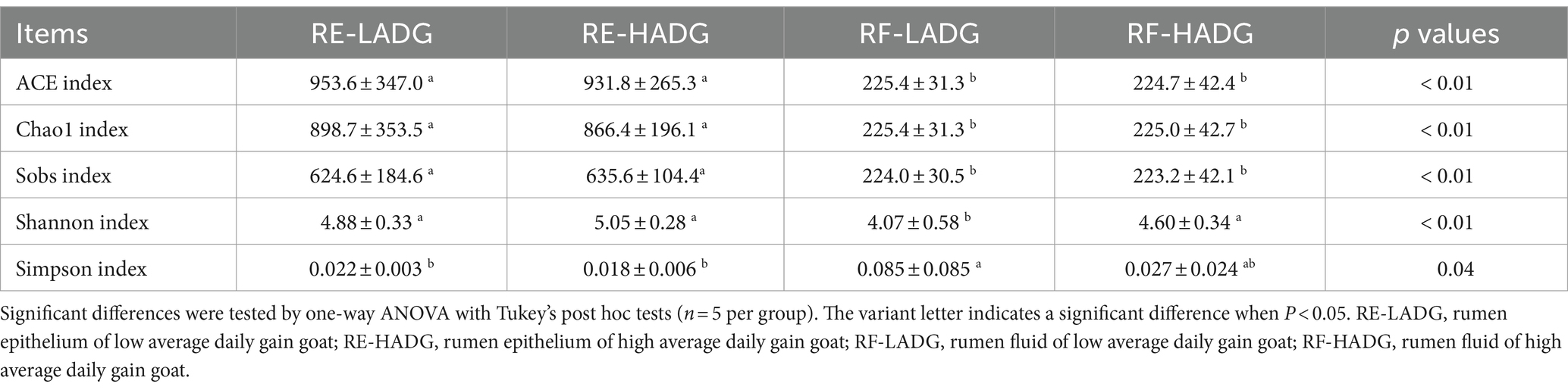
The alpha diversity of the bacterial community in rumen fluid and epithelium is shown in Table 1. Bacterial community richness indices (Chao1, Ace, and Sobs) were greater in rumen epithelium of both HADG and LADG goats compared with rumen fluid of the same groups (p < 0.01). Nonetheless, no significant difference was observed in bacterial community composition between HADG and LADG goats in either rumen fluid or epithelium. In rumen fluid, the bacterial diversity of HADG was higher than that of LADG (p < 0.01), as indicated by Shannon indices.
As shown in Figure 2A, bacterial communities of rumen epithelium were well separated from those in the rumen fluid, with 52.25 and 15.57% of the variations explained by principal component 1 (PC1) and PC2, respectively. The rumen fluid bacterial composition of LADG goats was separated from the HADG goats (Figure 2C). However, the rumen epithelium bacterial composition was similar between LADG and HADG goats (Figure 2B).
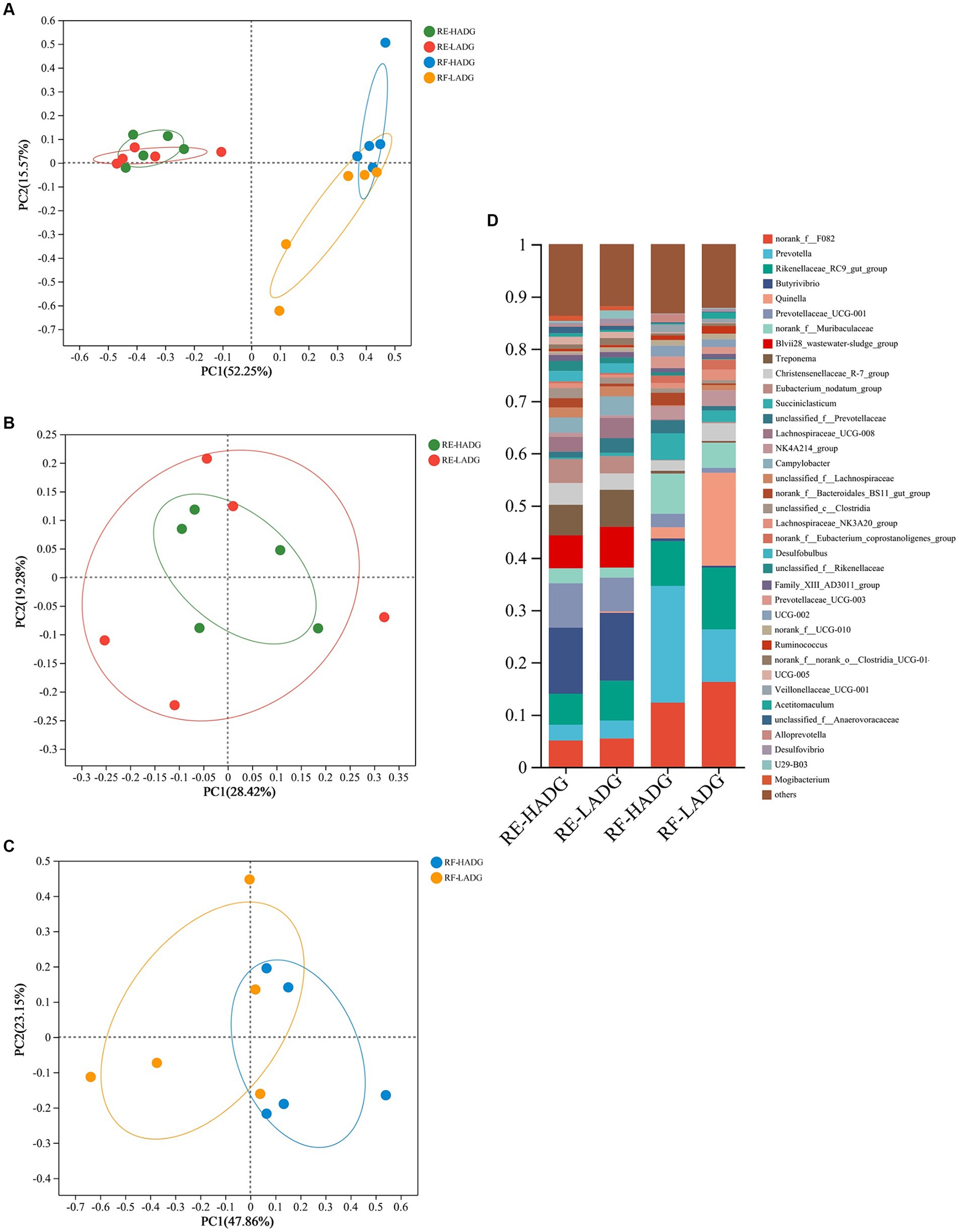
Figure 2. Principal coordinate analysis (PCoA) of bacterial compositional profiles of all samples (A), rumen epithelium samples (B), and rumen fluid samples (C) between LADG and HADG goats, based on weighted UniFrac distances. Abundant genera (D) in the rumen bacteria of LADG or HADG goats. Data are shown as means. RE-LADG, rumen epithelium of low average daily gain goat; RE-HADG, rumen epithelium of high average daily gain goat; RF-LADG, rumen fluid of low average daily gain goat; RF-HADG, rumen fluid of high average daily gain goat.
We found that the Butyrivibrio (12.6% in HADG goats and 12.9% in LADG goats), Prevotellaceae UCG-001 (8.5% in HADG goats and 6.5% in LADG goats), Blvii28 wastewater-sludge group (6.3% in HADG goats and 7.3% in LADG goats), Rikenellaceae RC9 gut group (5.9% in HADG goats and 7.6% in LADG goats), Treponema (5.9% in HADG goats and 7.1% in LADG goats), and norank_f_F082 (5.0% in HADG goats and 5.4% in LADG goats) were predominant genera in the rumen epithelium (Figure 2D). In contrast, the Prevotella (12.6% in HADG goats and 12.9% in LADG goats), norank_f_F082 (12.3% in HADG goats and 16.2% in LADG goats), Rikenellaceae RC9 gut group (8.6% in HADG goats and 11.8% in LADG goats), Quinella (2.1% in HADG goats and 17.9% in LADG goats), norank_f_Muribaculaceae (7.8% in HADG goats and 4.8% in LADG goats), Succiniclasticum (5.1% in HADG goats and 2.2% in LADG goats) were predominant genus in the rumen fluid.
In rumen epithelium, the relative abundances of genera U29-B03 and Quinella were higher, while Lachnospiraceae_UCG-009, norank_f_norank_o_WCHB1-41, and Lachnospiraceae_FCS020_group were lower in LADG goats compared with HADG goats (p < 0.05, Figure 3A). Additionally, the relative abundance of unclassified_c_Clostridia tended to be lower (p = 0.06), and Desulfovibrio tended to be higher (p = 0.09) in rumen epithelium of LADG goats compared with HADG goats. In rumen fluid, the relative abundances of genera Alloprevotella and unclassified_o_Bacteroidales were lower, and Desulfovibrio and U23-B03 were higher in LADG goats compared with HADG goats (p < 0.05, Figure 3B). Moreover, the relative abundance of norank_f_Bacteroidales_BS11_gut_group tended to be lower (p = 0.06), and Quinella (p = 0.09) tended to be higher in LADG goats compared with HADG goats.
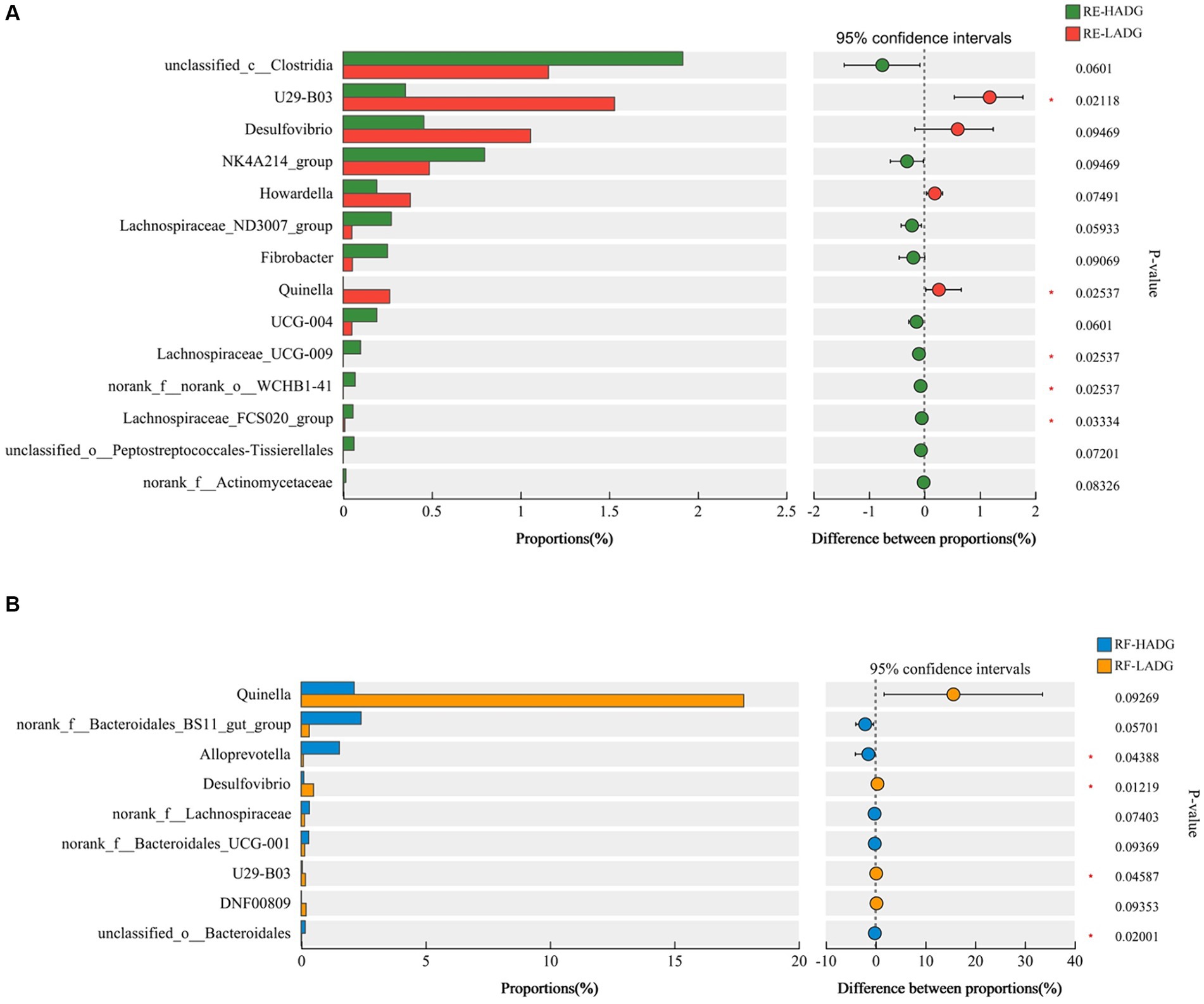
Figure 3. Differential enrichment of bacterial genera in rumen epithelium samples (A) and rumen fluid samples (B) between LADG and HADG goats, based on Wilcoxon rank-sum test. Data are shown as means. RE-LADG, rumen epithelium of low average daily gain goat; RE-HADG, rumen epithelium of high average daily gain goat; RF-LADG, rumen fluid of low average daily gain goat; RF-HADG, rumen fluid of high average daily gain goat.
To explore whether rumen fungi were related to the growth performance, we analyzed the fungal compositions of rumen fluid in LADG and HADG goats (Figure 4). The Saccharomyces (42.6% in HADG goats and 34.8% in LADG goats), Cladosporium (25.7% in HADG goats and 8.1% in LADG goats), Wallemia (3.0% in HADG goats and 10.3% in LADG goats), and Monascus (4.4% in HADG goats and 6.0% in LADG goats) were predominant genera in the rumen fluid. Alpha diversity analysis showed that the rumen fluid fungal composition of LADG goats was not clearly separated from the HADG goats. However, the relative abundance of genus Symmetrospora was lower in LADG goats compared with HADG goats (p < 0.05). Additionally, the relative abundance of genus unclassified_f_Cladosporiaceae tended to be lower (p = 0.06), and Aspergillus (p = 0.06) tended to be higher in LADG goats compared with HADG goats.
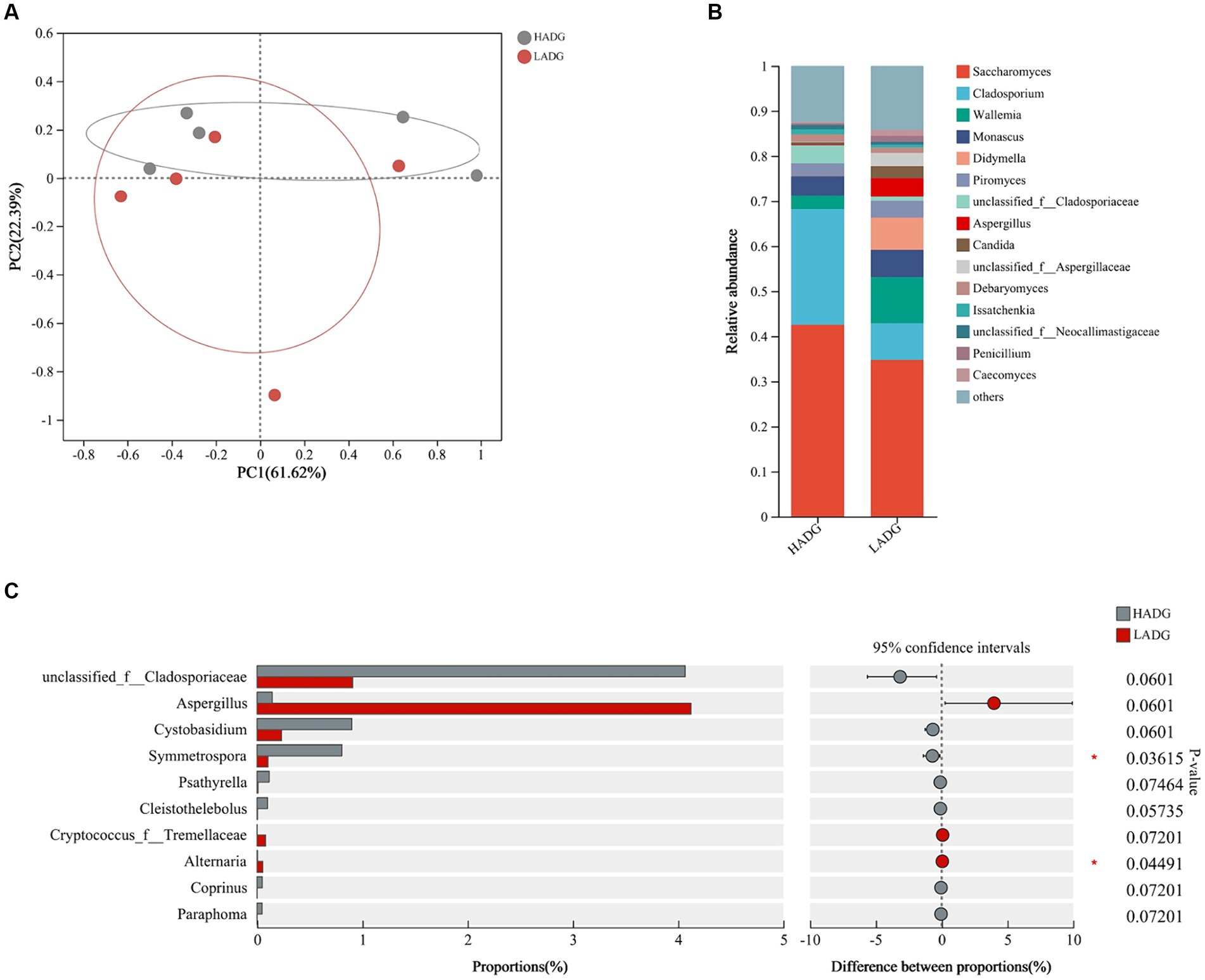
Figure 4. Principal coordinate analysis (PCoA) of fungal compositional profiles of rumen fluid samples between LADG and HADG goats, based on weighted UniFrac distances (A). Abundant genera in the rumen fungi of LADG or HADG goats (B). Differential enrichment of fungus genera of rumen epithelium samples (C) between LADG and HADG goats, based on Wilcoxon rank-sum test. Data are shown as means. n = 5 per group. LADG, low average daily gain goats; HADG, high average daily gain goats.
RNA-Seq analysis of the rumen epithelium
Principal components analysis (PCA) did not reveal strong clustering, with 34.41 and 14.76% variations explained by PC1 and PC2, respectively (Figure 5A). Further analysis of differentially expressed genes (DEGs) showed that 168 DEGs were upregulated, while 147 DEGs were downregulated in HADG goats compared with LADG goats (Figure 5B). 65 GO terms, including cell adhesion, cell junction, positive regulation of vascular endothelial growth factor production, regulation of developmental process, positive regulation of immune system process, etc., were identified (Figure 5C). Specifically, three DEGS, SPON2, SCARF2, VNN1, were upregulated and POSTN was downregulated, which were involved in regulation of cell adhesion, in HADG goats compared with LADG goats. Three upregulated genes (CDH24, ITGB2, APOE) and one downregulated gene (ACTN2) were enriched in the GO terms of cell junction in HADG goats. The ARNT and CCBE1 involved in regulation of vascular endothelial growth factor production were upregulated in HADG goats. Five upregulated genes (GPR4, CTSK, MDK, SNAI1, TMEM119, CYP126B1) and 1 downregulated gene (FRZB) were enriched in the GO terms of regulation of developmental process in HADG goats. Additionally, three genes (ITGAM, STXBP1, and PLVAP) involved in regulation of immune system process were upregulated in HADG goats.
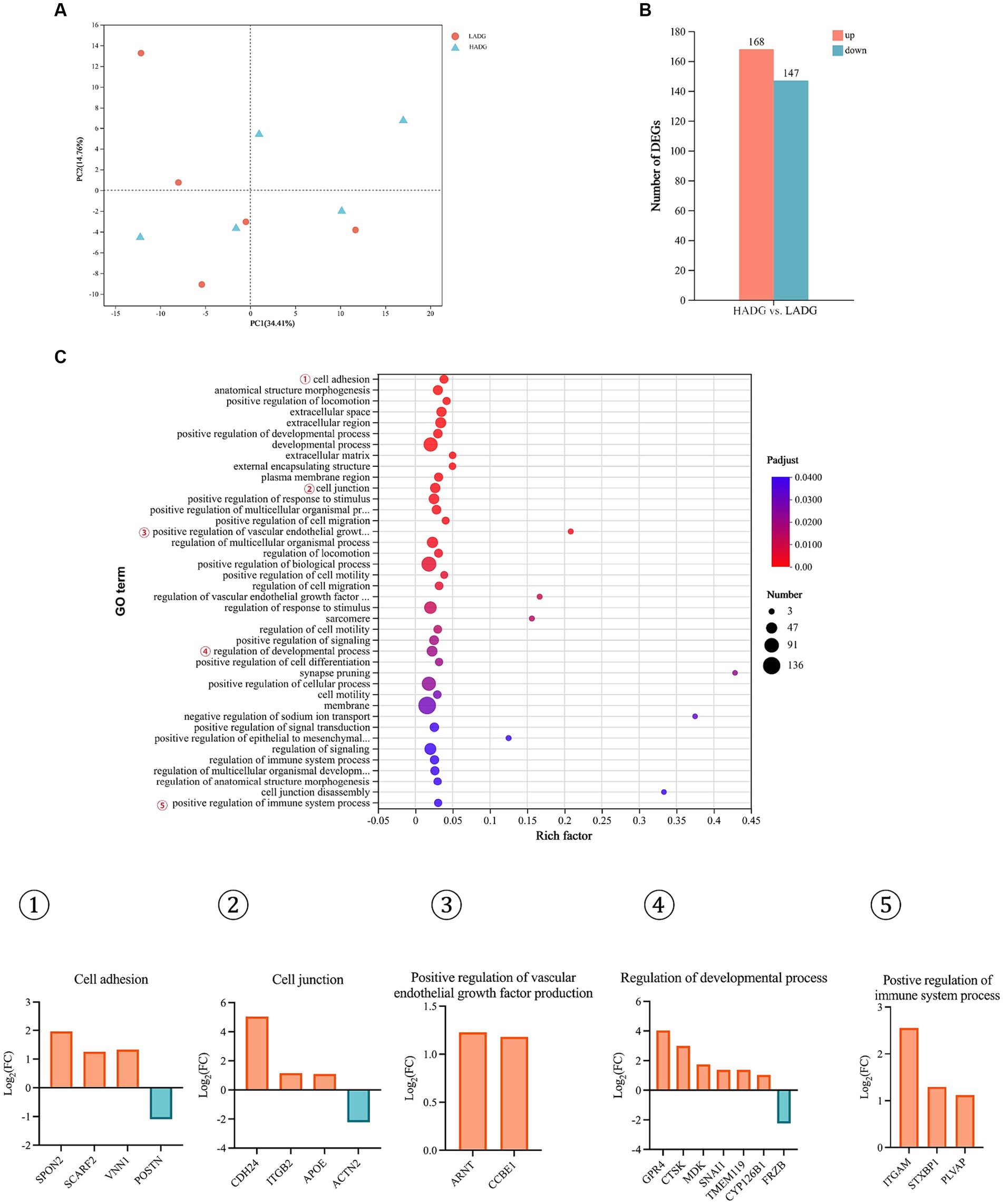
Figure 5. RNA-seq analysis of the rumen epithelium of HADG and LADG goats. Principal component analysis (PCA) of gene expressions in rumen epithelium between LADG and HADG goats (A). The number of differentially expressed genes (DEGs) between HADG and LADG goats (B). The GO pathways significantly enriched in the DEGs in rumen epithelium between LADG and HADG goats (C). n = 5 per group. LADG, low average daily gain goats; HADG, high average daily gain goats.
Relative gene expression of the rumen epithelium
The relative mRNA expression levels associated with tight junction, SCFAs transport, and inflammation are shown in Figure 6. The results demostrated that the relative expressions of ZO-1 (p = 0.01), Occludin (p = 0.02), NHE-2 (p = 0.03), and NHE-3 (p = 0.03) were higher in the rumen epithelium of HADG goats compared with LADG goats. Furthermore, the relative expressions of Claudin-1 (p = 0.10), Claudin-4 (p = 0.09), and MCT-4 (p = 0.08) tended to increase in the rumen epithelium of HADG goats compared with LADG goats. However, the relative mRNA expressions related to inflammation did not show significant differences between the two groups.
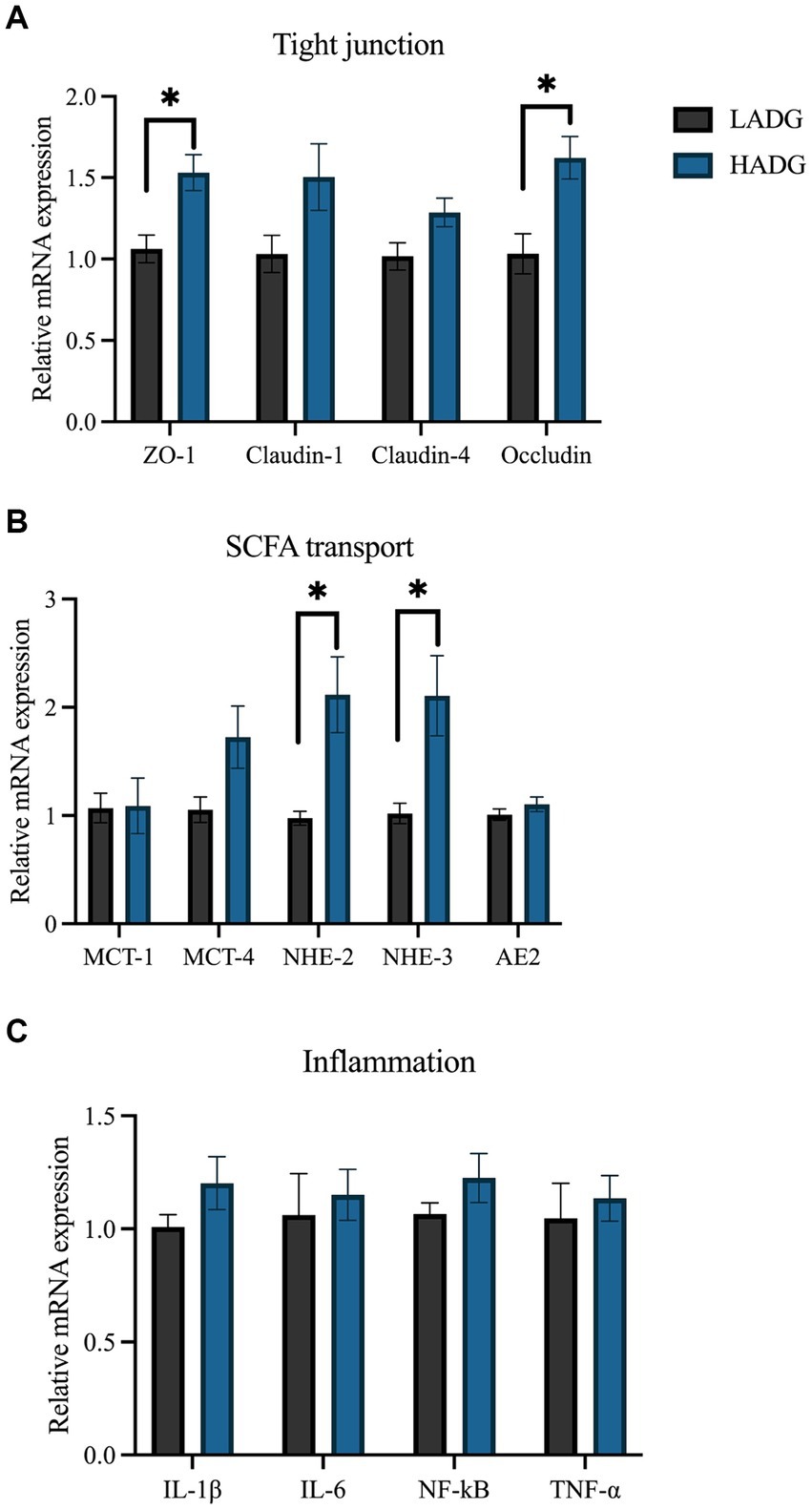
Figure 6. RT-PCR analyses of the relative mRNA expression of genes associated with tight junction (A), short-chain fatty acids (SCFAs) transport (B), and inflammation (C). The values represent the mean ± SD (n = 5 per group). *p < 0.05.
Correlation analyses of the rumen microbiota, growth performance, rumen SCFAs and NH3-N concentrations, or gene expression levels
The Spearman correlation analysis revealed that the relative abundances of genera UCG-005 and Candidatus Saccharimonas in rumen epithelium were negatively associated with ADG, and relative expressions of ZO-1 and Occludin (p < 0.05, Figure 7). The relative abundances of genera Lachnospiraceae NK3A20 group and Oscillospiraceae NK4A214 group in the rumen epithelium were positively correlated with ADG and the relative expressions of ZO-1 and Occludin (p < 0.05). Additionally, the relative abundances of genera norank_f_norank_o_Clostridia_UCG-014 and Desulfovibrio in rumen epithelium were negatively associated with the relative expressions of ZO-1 and Occludin (p < 0.05), while the relative abundance of genus Oscillospiraceae NK4A214 group in the rumen epithelium was positively associated with the relative expressions of MCT-4, NHE-2, NHE-3, and AE2 (p < 0.05).
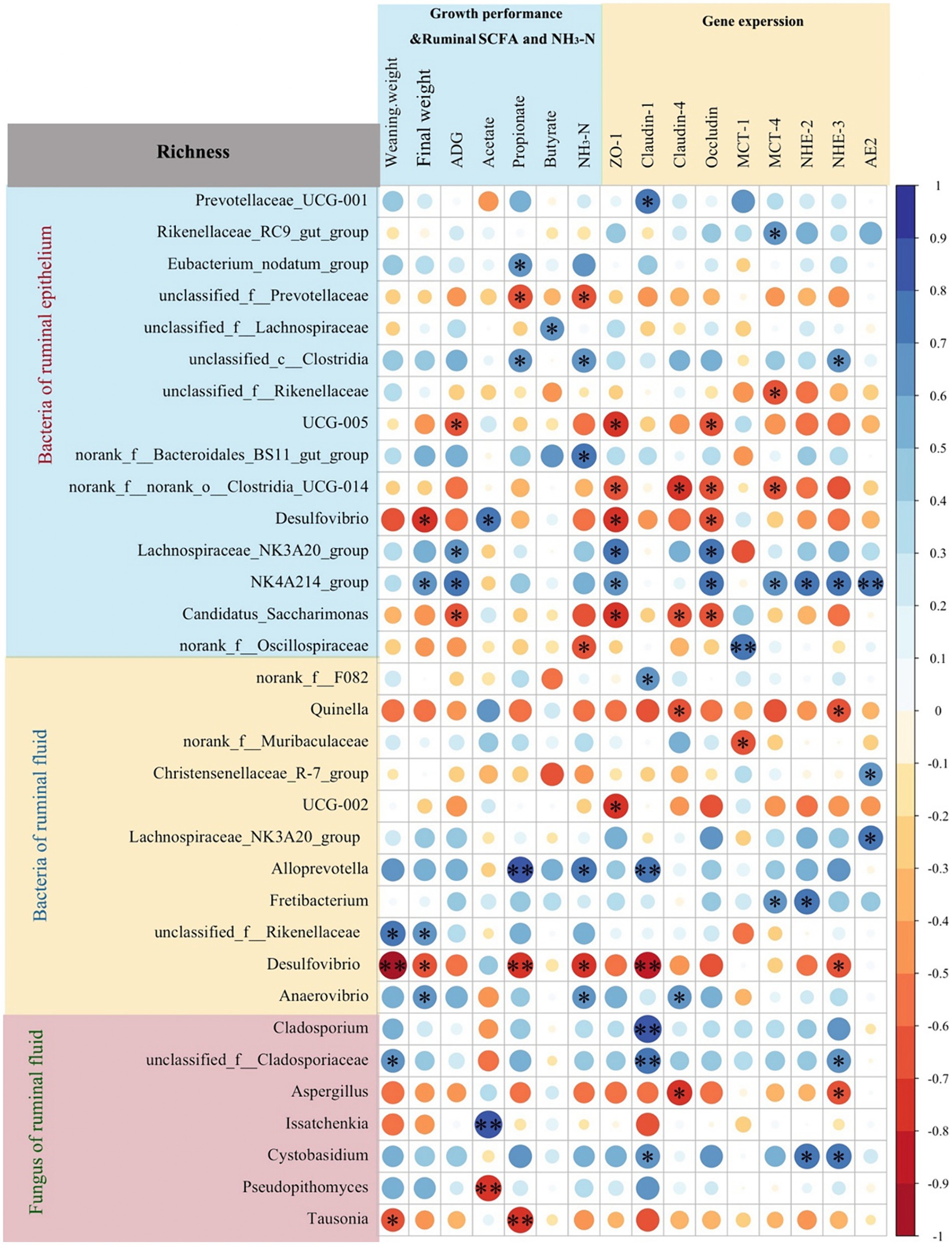
Figure 7. Spearman correlation analysis between rumen microbiota richness (fungi and bacteria of rumen fluid, and bacteria of rumen epithelium) and growth performance, ruminal short-chain fatty acids (SCFAs) and NH3-N concentrations, or gene expression levels. *p < 0.05, **p < 0.01.
In rumen fluid, the relative abundance of Quinella had a negative correlation with the relative expressions of claudin-4 and NHE-3 (p < 0.05). Notably, the relative abundances of Alloprevotella in rumen fluid had a positive correlation with rumen propionate (p < 0.01) and NH3-N (p < 0.05) concentrations, and the relative expressions of Claudin-1 (p < 0.01). Conversely, the relative abundance of Desulfovibrio in rumen fluid had a negative correlation with weaning weight (p < 0.01), rumen propionate (p < 0.01) and NH3-N (p < 0.05) concentrations, and the relative expressions of ZO-1 (p < 0.01) and NHE-3 (p < 0.05).
Furthermore, the relative abundances of fungal genera Cladosporium (p < 0.01), unclassified_f_Cladosporiaceae (p < 0.01), and Cystobasidium (p < 0.05) showed positive correlations with the relative expressions of Claudin-1. The relative of fungal genus Issatchenkia were positively (p < 0.01), and Pseudopithomyces were negatively (p < 0.01) associated with rumen acetate concentration.
Discussion
We observed distinct rumen fermentation patterns between LADG and HADG goats, as reflected by lower rumen propionate and NH3-N concentrations in LADG goats. Propionate, a crucial precursor for gluconeogenesis in ruminants, is known to consume hydrogen and contribute to higher energy utilization efficiency and reduced methane formation in the rumen (Moss et al., 2000; Goopy et al., 2011). Shabat et al. (2016) further demonstrated that efficient ruminants exhibit higher ruminal propionate concentrations. Moreover, the tendency of higher rumen NH3-N concentration observed in HADG goats may favor microbial protein production. These findings suggest that the rumen microbiota of HADG goats likely had higher fermentation efficiency, supporting the higher ADG of these goats compared with LADG goats.
While most studies have focused on the rumen fluid associated bacteria, it is crucial to acknowledge the distinct microbiota residing in the rumen epithelium (Pei et al., 2010). Malmuthuge et al. (2019) emphasized the inadequacy of studies only based only on rumen contents in capturing the complete rumen microbiome. In our current study, alpha diversity analysis revealed higher bacterial diversity and richness in rumen epithelium compared to rumen fluid. Notably, Butyrivibrio and Prevotellaceae UCG-001 were predominant in rumen epithelium, contrasting with Prevotella and norank_f_F082, which dominated the rumen fluid. However, it is worth noting that diversity indices were reported to be higher in rumen digesta than in mucosal tissue in pre-weaning calves (Malmuthuge et al., 2014), likely due to the differences in growth stage and diet between weaned and pre-weaned ruminants.
The higher bacterial diversity and richness imply a more intricate microbial population and complex host-bacteria interactions within the rumen epithelium. Previous studies have highlighted the involvement of ruminal epithelium bacteria in various crucial processes such as oxygen scavenging, tissue recycling, urea digestion, and anatomic and functional development of rumen (Cheng et al., 1979; Jiao et al., 2015). In our study, we observed that LADG goats had higher abundances of the genera U29-B03 and Quinella in the rumen epithelium compared to HADG goats. While U29-B03, a member of the Rikenellaceae family, has been reported to positively influence SCFAs production (Du et al., 2021), it is worth noting that Wang et al. (2023) found an unclassified Rikenellaceae to be positively correlated with the acetate levels but negatively correlated with growth performance traits in dairy goats. Additionally, Yi et al. (2022) reported a negative correlation between Quinella abundance and propionate concentration. When enriched, Quinella cells, were found to predominantly ferment glucose into lactate, with minimal production of acetate, propionate, and CO2 (Brough et al., 1970). Furthermore, genera U29-B03 and Quinella abundances were higher in the rumen fluid of LADG goats compared with HADG goats in the current study. Thus, these findings suggest that the genera U29-B03 and Quinella may be the key bacteria influencing the fermentation pattern, which could partly explain the lower propionate concentration observed in LADG goats.
The genus Desulfovibrio comprises the predominant sulfate-reducing bacteria in the rumen (Drewnoski et al., 2014; Zhao et al., 2020). Hydrogen sulfide gas (H2S) is the principal terminal metabolite produced by Desulfovibrio via the dissimilatory sulfate reduction pathway (Rey et al., 2013). In our study, the abundance of Desulfovibrio was found to be higher in both the rumen epithelium and fluid in LADG goats compared with HADG goats. Although the relationship between ruminal Desulfovibrio and the production performance of ruminants remains unclear, overgrowth of Desulfovibrio is known to correlate with several human intestinal and extra-intestinal diseases (Singh et al., 2023). Wu et al. (2021) found that a high sulfur diet increases the ruminal abundance of Desulfovibrio and compromises the integrity and barrier function of the rumen epithelium. According to the Spearman correlation analysis, the abundance of Desulfovibrio was negatively correlated with ruminal propionate and NH3-N concentrations, as well as with the expressions of epithelial tight junction-related genes. Based on these findings, we speculated that Desulfovibrio may play a critical role in influencing the growth performance of LADG goats, but further investigations are warranted to clarify the ecological functions of Desulfovibrio in the rumen.
In our study, we observed lower abundances of the genus Alloprevotella in the rumen fluid and several members of family Lachnospiraceae (e.g., Lachnospiraceae_ND3007_group and Lachnospiraceae_UCG-009) in the rumen epithelium of LADG goats compared with HADG goats. Alloprevotella, belonging to the family Prevotellaceae, has the capacity to produce moderate amounts of acetate and significant amounts of succinic acid (Downes et al., 2013). Our findings revealed a positive correlation between propionate concentration in the rumen and Alloprevotella, consistent with previous research by Fan et al. (2020). Furthermore, Xie et al. (2022) proposed that species within the Lachnospiraceae family may play a key role in carbohydrate metabolism and influence feed efficiency in dairy cows. In our study, genera within Lachnospiraceae showed positive correlation with ADG, ruminal butyrate concentration, and the expression of genes related to epithelial tight junction. These results suggest that Alloprevotella and Lachnospiraceae could potentially serve as probiotics, enhancing fermentation efficiency and promoting host health.
We further explored the differences in fungal composition to gain a comprehensive understanding of the rumen microbiota between LADG and HADG goats. We observed a lower abundance of genus Symmetrospora and a tendency towards higher abundances of the genera unclassified_f_Cladosporiaceae and Aspergillus in LADG goats. Symmetrospora foliicola has previously been associated with body weight in lambs at 90 days of age (Yin et al., 2023). Moreover, several mycotoxins, such as aflatoxins and ochratoxin A, can be produced by Aspergillus fungi and may disrupt ruminal functions (Loh et al., 2020). We also observed a negative correlation between the abundance of Aspergillusand and the mRNA expression of Claudin-4, suggesting that Aspergillus may impair the integrity of the ruminal barrier.
The rumen epithelium serves as a critical site for host-bacteria interactions, acting as the first line of defense against pathogens and facilitating nutrient uptake to support the development and growth of host (Petri et al., 2018). Transcriptome analysis in our study revealed differential expression of 415 genes between LADG and HADG goats, with enrichment in cell junction (CDH24, ITGB2, APOE, and ACTN2), cell adhesion (SPON2, SCARF2, VNN1, and POSTN), and others. The tight junction barrier of rumen epithelium plays a pivotal role in preventing microbial invasion and the entry of harmful substances, which is crucial for the growth and health of ruminants (Aschenbach et al., 2019). Our findings indicated lower expression levels of CDH24, ZO-1, and Occludin in the rumen epithelium of LADG goats. Additionally, PLVAP was demonstrated to play a critical role in endothelial barrier function and intestinal homeostasis (Stan et al., 2012). Consistent with our results, studies on monogastric animals have shown lower expression of tight junction proteins in low-birth-weight piglets (Huang et al., 2021; Wang et al., 2023). Furthermore, several bacteria inhabiting the rumen epithelium were significantly associated with the mRNA expression of ZO-1 and Occludin in our study, suggesting that these bacteria may influence the growth performance by regulating the rumen epithelial barrier.
In the current study, the transcriptome analysis showed that cell adhesion related genes, such as SPON2, VNN1, and CTSK, were upregulated in HADG goats. The SPON2 encodes mind in, which binds to bacteria and their components, acting as an opsonin (He et al., 2004). VNN1, which encodes pantetheinase, is involved in coenzyme A metabolism, lipid metabolism, and energy production (Bartucci et al., 2019). Salzano et al. (2023) suggested that the increased VNN1 expression in rumen epithelium of buffaloes fed a green feed diet was associated with the anti-inflammatory activities. Moreover, the gene CTSK, involved in the regulation of developmental processes, has been shown to maintain the normal composition of intestinal microbiota (Sina et al., 2013). However, the functions of these DEGs in ruminants are not well understood, and further research is needed to confirm these findings.
In our study, the mRNA expressions of NHE-2 and NHE-3 were found to be higher in HADG goats. The Na+/H+ exchanger (NHE) proteins regulate the intracellular pH of rumen epithelium by transporting H+ to the lumen or extracellular space and taking up Na+ into epithelial cells (Schlau et al., 2012). Increased NHE activities are known to lower the local pH near epithelial cells, thereby enhancing the uptake of the undissociated form of SCFAs via simple diffusion (Graham et al., 2007). In addition, the presence of the genus Oscillospiraceae NK4A214 group in the rumen epithelium was positively associated with the relative expressions of MCT-4, NHE-2, NHE-3, and AE2. Previously classified in the family Ruminococcaceae, Oscillospiraceae NK4A214 group has been linked to improving growth and lactation performance in ruminants (Tong et al., 2018; Huang et al., 2021; Wang et al., 2023). Consequently, Oscillospiraceae NK4A214 group could serve as a potential probiotic to enhance growth or production by mediating SCFAs transport-related genes in the rumen epithelium. However, Oscillospiraceae NK4A214 group remains uncultured, and its exact mechanism of action in rumen remains unclear, warranting further investigation.
Conclusion
In summary, our study reveals that HADG goats exhibit higher ruminal fermentation efficiency, improved rumen epithelial barrier functions, and enhanced SCFAs transport, all of which support the development and growth of goats. These differences can be attributed to the rumen microbiota, particularly to the rumen epithelium bacteria (Figure 8). Thus, our findings offer a deeper understanding of the association between rumen microbiota and growth performance, and identify several potential microbial targets for the development of novel intervention strategies to improve the growth performance of young ruminants.
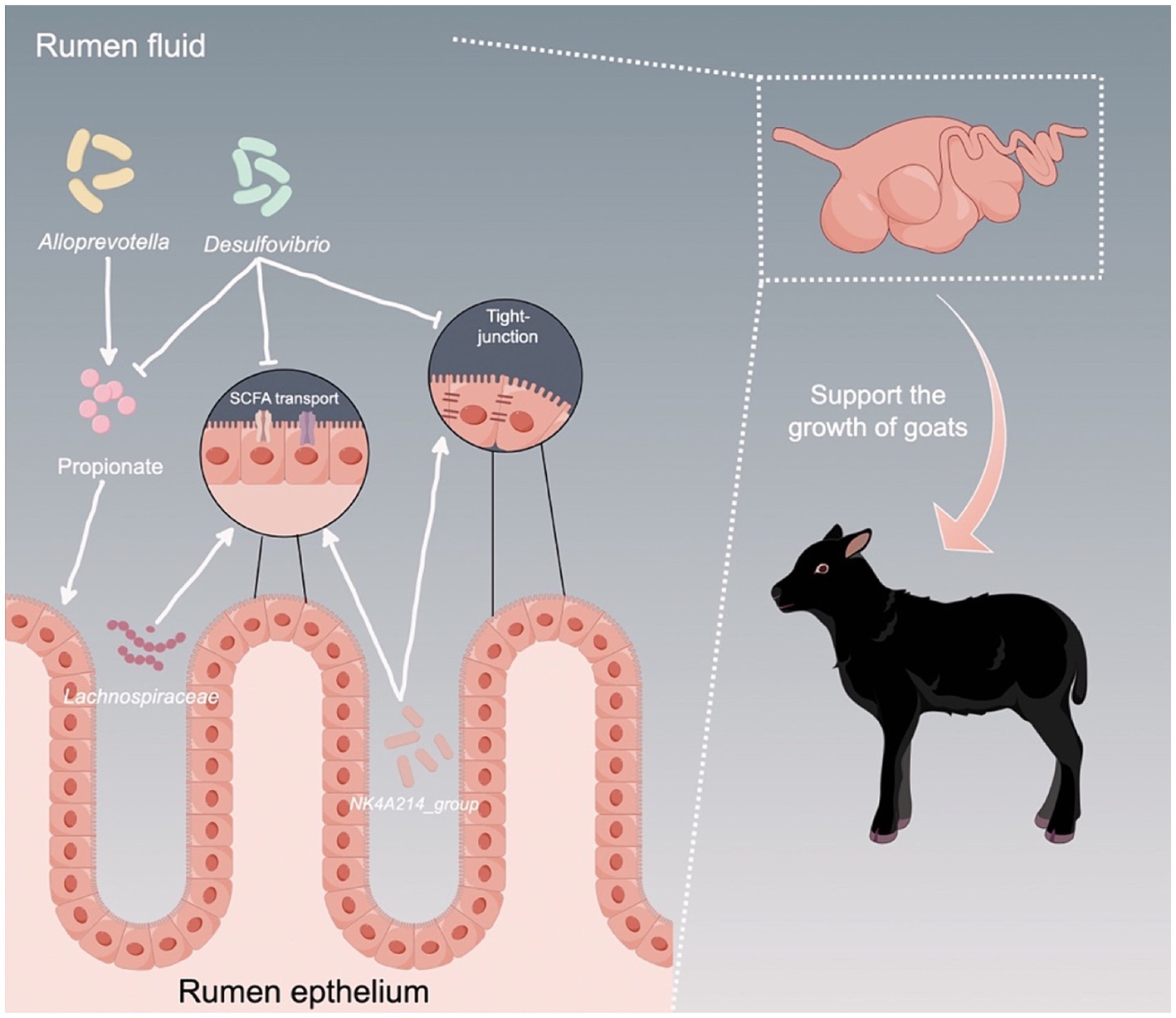
Figure 8. Proposed model of the interactions between ruminal microbiota and the host to support the growth of young ruminants. Arrows indicates upregulation, while T bars indicate downregulation.
Data availability statement
The raw sequencing reads of the rumen microbiota, along with the RNA sequencing data, were deposited into the NCBI Sequence Read Archive (SRA) database and NCBI’s Gene Expression Omnibus under the accession numbers PRJNA1099693 and PRJNA1101717, respectively.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of Southwest University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
JC: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. XZ: Methodology, Writing – original draft, Writing – review & editing. XC: Methodology, Writing – original draft. BW: Methodology, Writing – original draft. YF: Funding acquisition, Investigation, Resources, Writing – original draft. SS: Investigation, Resources, Writing – original draft. DG: Investigation, Resources, Writing – original draft. DH: Investigation, Resources, Writing – review & editing. YS: Investigation, Resources, Writing – original draft. XD: Investigation, Methodology, Writing – original draft. YZ: Conceptualization, Funding acquisition, Writing – review & editing. ZZ: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (32202686), Key project of Chong Qing Natural Science Foundation (grant number. cstc2020jcyj-zdxmX0005), The Strategic Collaboration Project jointly funded by Chongqing Municipal People's Government and Chinese Academy of Agricultural Sciences, and Chongqing Municipal Training Program of Innovation and Entrepreneurship for Undergraduates (grant number X202310635055).
Acknowledgments
The authors appreciated the Figdraw (www.figdraw.com) for their assistance in drawing figure.
Conflict of interest
DG and DH were employed by the Tengda Animal Husbandry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1445223/full#supplementary-material
References
Aschenbach, J. R., Zebeli, Q., Patra, A. K., Greco, G., Amasheh, S., and Penner, G. B. (2019). Symposium review: the importance of the ruminal epithelial barrier for a healthy and productive cow. J. Dairy Sci. 102, 1866–1882. doi: 10.3168/jds.2018-15243
Bartucci, R., Salvati, A., Olinga, P., and Boersma, Y. L. (2019). Vanin 1: its physiological function and role in diseases. Int. J. Mol. Sci. 20:3891. doi: 10.3390/ijms20163891
Bhagat, N. R., Kumar, S., Kumari, R., and Bharti, V. K. (2023). A review on rumen anaerobic fungi: current understanding on carbohydrate fermentation and roughages digestion in ruminants. Appl. Biochem. Microbiol. 59, 231–249. doi: 10.1134/S0003683823030043
Brough, B. E., Reid, T., and Howard, B. (1970). The biochemistry of the rumen bacterium" Quin's oval" —part 1. N. Z. J. Sci. 13, 570–575.
Cheng, K., McCowan, R., and Costerton, J. (1979). Adherent epithelial bacteria in ruminants and their roles in digestive tract function. Am. J. Clin. Nutr. 32, 139–148. doi: 10.1093/ajcn/32.1.139
Costa, A., Boselli, C., and De Marchi, M. (2021). Effect of body weight and growth in early life on the reproductive performances of Holstein heifers. Agriculture 11:159. doi: 10.3390/agriculture11020159
Downes, J., Dewhirst, F. E., Tanner, A. C., and Wade, W. G. (2013). Description of Alloprevotella rava gen. Nov., sp. nov., isolated from the human oral cavity, and reclassification of Prevotella tannerae Moore et al. 1994 as Alloprevotella tannerae gen. Nov., comb. nov. Int. J. Syst. Evol. Microbiol. 63, 1214–1218. doi: 10.1099/ijs.0.041376-0
Drewnoski, M., Pogge, D., and Hansen, S. (2014). High-sulfur in beef cattle diets: a review. J. Anim. Sci. 92, 3763–3780. doi: 10.2527/jas.2013-7242
Du, M., Yang, C., Liang, Z., Zhang, J., Yang, Y., Ahmad, A. A., et al. (2021). Dietary energy levels affect carbohydrate metabolism-related bacteria and improve meat quality in the longissimus thoracis muscle of yak (Bos grunniens). Front. Vet. Sci. 8:718036. doi: 10.3389/fvets.2021.718036
Fan, Q., Wanapat, M., Yan, T., and Hou, F. (2020). Altitude influences microbial diversity and herbage fermentation in the rumen of yaks. BMC Microbiol. 20, 1–13. doi: 10.1186/s12866-020-02054-5
Fonseca, P., Lam, S., Chen, Y., Waters, S., Guan, L., and Cánovas, A. (2023). Multi-breed host rumen epithelium transcriptome and microbiome associations and their relationship with beef cattle feed efficiency. Sci. Rep. 13:16209. doi: 10.1038/s41598-023-43097-8
Gelsinger, S., Heinrichs, A., and Jones, C. (2016). A meta-analysis of the effects of preweaned calf nutrition and growth on first-lactation performance. J. Dairy Sci. 99, 6206–6214. doi: 10.3168/jds.2015-10744
Goopy, J. P., Woodgate, R., Donaldson, A., Robinson, D. L., and Hegarty, R. (2011). Validation of a short-term methane measurement using portable static chambers to estimate daily methane production in sheep. Anim. Feed. Sci. Tech. 166, 219–226. doi: 10.1016/j.anifeedsci.2011.04.012
Graham, C., Gatherar, I., Haslam, I., Glanville, M., and Simmons, N. L. (2007). Expression and localization of monocarboxylate transporters and sodium/proton exchangers in bovine rumen epithelium. Am. J. Physiol. Regul. Integr. Comp. Physiol. 292, R997–R1007. doi: 10.1152/ajpregu.00343.2006
He, Y. W., Li, H., Zhang, J., Hsu, C. L., Lin, E., Zhang, N., et al. (2004). The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat. Immunol. 5, 88–97. doi: 10.1038/ni1021
Huang, C., Ge, F., Yao, X., Guo, X., Bao, P., Ma, X., et al. (2021). Microbiome and metabolomics reveal the effects of different feeding systems on the growth and ruminal development of yaks. Front. Microbiol. 12:682989. doi: 10.3389/fmicb.2021.682989
Jiao, J., Huang, J., Zhou, C., and Tan, Z. (2015). Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microb. 81, 3502–3509. doi: 10.1128/AEM.00203-15
Jize, Z., Zhuoga, D., Xiaoqing, Z., Na, T., Jiacuo, G., Cuicheng, L., et al. (2022). Different feeding strategies can affect growth performance and rumen functions in Gangba sheep as revealed by integrated transcriptome and microbiome analyses. Front. Microbiol. 13:908326. doi: 10.3389/fmicb.2022.908326
Li, W., Gelsinger, S., Edwards, A., Riehle, C., and Koch, D. (2019). Transcriptome analysis of rumen epithelium and meta-transcriptome analysis of rumen epimural microbial community in young calves with feed induced acidosis. Sci. Rep. 9:4744. doi: 10.1038/s41598-019-40375-2
Liu, J. H., Zhang, M. L., Zhang, R. Y., Zhu, W. Y., and Mao, S. Y. (2016). Comparative studies of the composition of bacterial microbiota associated with the ruminal content, ruminal epithelium and in the faeces of lactating dairy cows. Microb. Biotechnol. 9, 257–268. doi: 10.1111/1751-7915.12345
Loh, Z. H., Ouwerkerk, D., Klieve, A. V., Hungerford, N. L., and Fletcher, M. T. (2020). Toxin degradation by rumen microorganisms: a review. Toxins 12:664. doi: 10.3390/toxins12100664
Malmuthuge, N., Griebel, P. J., and Guan, L. L. (2014). Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl. Environ. Microb. 80, 2021–2028. doi: 10.1128/AEM.03864-13
Malmuthuge, N., Liang, G., and Guan, L. L. (2019). Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 20, 1–16. doi: 10.1186/s13059-019-1786-0
Moss, A. R., Jouany, J. P., and Newbold, J. (2000). Methane production by ruminants: its contribution to global warming. Ann. Zootech. 49, 231–253. doi: 10.2527/1995.7382483x
Pei, C. X., Mao, S. Y., Cheng, Y. F., and Zhu, W. Y. (2010). Diversity, abundance and novel 16S rRNA gene sequences of methanogens in rumen liquid, solid and epithelium fractions of Jinnan cattle. Animal 4, 20–29. doi: 10.1017/S1751731109990681
Petri, R., Kleefisch, M., Metzler-Zebeli, B., Zebeli, Q., and Klevenhusen, F. (2018). Changes in the rumen epithelial microbiota of cattle and host gene expression in response to alterations in dietary carbohydrate composition. Appl. Environ. Microb. 84, e00384–e00318. doi: 10.1128/AEM.00384-18
Rey, F. E., Gonzalez, M. D., Cheng, J., Wu, M., Ahern, P. P., and Gordon, J. I. (2013). Metabolic niche of a prominent sulfate-reducing human gut bacterium. Proc. Natl. Acad. Sci. 110, 13582–13587. doi: 10.1073/pnas.1312524110
Salzano, A., Fioriniello, S., D’Onofrio, N., Balestrieri, M. L., Aiese Cigliano, R., Neglia, G., et al. (2023). Transcriptomic profiles of the ruminal wall in Italian Mediterranean dairy buffaloes fed green forage. BMC Genomics 24:133. doi: 10.1186/s12864-023-09215-6
Schlau, N., Guan, L., and Oba, M. (2012). The relationship between rumen acidosis resistance and expression of genes involved in regulation of intracellular pH and butyrate metabolism of ruminal epithelial cells in steers. J. Dairy Sci. 95, 5866–5875. doi: 10.3168/jds.2011-5167
Schubach, K. M., Cooke, R. F., Brandão, A. P., Schumaher, T. F., Pohler, K. G., Bohnert, D. W., et al. (2019). Impacts of postweaning growth rate of replacement beef heifers on their reproductive development and productivity as primiparous cows. J. Anim. Sci. 97, 4171–4181. doi: 10.1093/jas/skz262
Shabat, S. K. B., Sasson, G., Doron-Faigenboim, A., Durman, T., Yaacoby, S., Berg Miller, M. E., et al. (2016). Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 10, 2958–2972. doi: 10.1038/ismej.2016.62
Sina, C., Lipinski, S., Gavrilova, O., Aden, K., Rehman, A., Till, A., et al. (2013). Extracellular cathepsin K exerts antimicrobial activity and is protective against chronic intestinal inflammation in mice. Gut 62, 520–530. doi: 10.1136/gutjnl-2011-300076
Singh, S. B., Carroll-Portillo, A., and Lin, H. C. (2023). Desulfovibrio in the gut: the enemy within? Microorganisms 11:1772. doi: 10.3390/microorganisms11071772
Soberon, F., and Van Amburgh, M. (2013). Lactation biology symposium: the effect of nutrient intake from milk or milk replacer of preweaned dairy calves on lactation milk yield as adults: a meta-analysis of current data. J. Anim. Sci. 91, 706–712. doi: 10.2527/jas.2012-5834
Stan, R. V., Tse, D., Deharvengt, S. J., Smits, N. C., Xu, Y., Luciano, M. R., et al. (2012). The diaphragms of fenestrated endothelia: gatekeepers of vascular permeability and blood composition. Dev. Cell 23, 1203–1218. doi: 10.1016/j.devcel.2012.11.003
Tong, J., Zhang, H., Yang, D., Zhang, Y., Xiong, B., and Jiang, L. (2018). Illumina sequencing analysis of the ruminal microbiota in high-yield and low-yield lactating dairy cows. PLoS One 13:e0198225. doi: 10.1371/journal.pone.0198225
Wang, D., Chen, L., Tang, G., Yu, J., Chen, J., Li, Z., et al. (2023). Multi-omics revealed the long-term effect of ruminal keystone bacteria and the microbial metabolome on lactation performance in adult dairy goats. Microbiome 11:215. doi: 10.1186/s40168-023-01652-5
Wu, H., Li, Y., Meng, Q., and Zhou, Z. (2021). Effect of high sulfur diet on rumen fermentation, microflora, and epithelial barrier function in steers. Animals 11:2545. doi: 10.3390/ani11092545
Xie, Y., Sun, H., Xue, M., and Liu, J. (2022). Metagenomics reveals differences in microbial composition and metabolic functions in the rumen of dairy cows with different residual feed intake. Anim. Microbiome 4:19. doi: 10.1186/s42523-022-00170-3
Yi, S., Dai, D., Wu, H., Chai, S., Liu, S., Meng, Q., et al. (2022). Dietary concentrate-to-forage ratio affects rumen bacterial community composition and metabolome of yaks. Front. Nutr. 9:927206. doi: 10.3389/fnut.2022.927206
Yin, X., Duan, C., Ji, S., Tian, P., Ju, S., Yan, H., et al. (2023). Average daily gain in lambs weaned at 60 days of age is correlated with rumen and rectum microbiota. Microorganisms 11:348. doi: 10.3390/microorganisms11020348
Keywords: microbes, rumen epithelium, transcriptome, growth rate, goat
Citation: Chen J, Zhang X, Chang X, Wei B, Fang Y, Song S, Gong D, Huang D, Sun Y, Dong X, Zhao Y and Zhao Z (2024) Multi-omics analysis reveals the effects of host-rumen microbiota interactions on growth performance in a goat model. Front. Microbiol. 15:1445223. doi: 10.3389/fmicb.2024.1445223
Edited by:
Jie Yin, Hunan Agricultural University, ChinaReviewed by:
Limei Lin, Nanjing Agricultural University, ChinaLifeng Dong, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Chen, Zhang, Chang, Wei, Fang, Song, Gong, Huang, Sun, Dong, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongquan Zhao, emhvbmdxdWFuemhhb0AxMjYuY29t; Juncai Chen, anVuY2FpLmNoZW5AaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Juncai Chen
Juncai Chen Xiaoli Zhang1†
Xiaoli Zhang1† Yawang Sun
Yawang Sun Xianwen Dong
Xianwen Dong Yongju Zhao
Yongju Zhao