- 1College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 2Sichuan Key Laboratory of Animal Epidemic Disease and Human Health, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
Porcine circovirus 4 (PCV4) was first identified in 2019, categorized within the genus Circovirus in the family Circoviridae. To date, the virus has not been isolated from clinical samples. Meanwhile, many aspects of the biology and pathogenic mechanisms of PCV4 infection remain unknown. In this study, PCV4 was successfully rescued from an infectious clone. We utilized a PCV4 virus stock derived from this infectious clone to intranasally inoculate 4-week-old specific-pathogen-free piglets to evaluate PCV4 pathogenesis. The rescued PCV4 was capable of replicating in both PK-15 cells and piglets, with the virus detectable in nearly all collected samples from the challenge groups. Pathological lesions and PCV4-specific antigens were observed in various tissues and organs, including the lungs, kidneys, lymph nodes, spleen, and liver, in the inoculated piglets. Additionally, the levels of pro-inflammatory cytokines in the serum of the PCV4-inoculated group were significantly elevated compared to the control group, indicating that the induced inflammatory response may contribute to tissue damage associated with PCV4 infection. These findings offer new insights into the pathogenesis and inflammatory responses associated with PCV4-related diseases.
1 Introduction
Porcine circovirus (PCV) belongs to the genus Circovirus in the family Circoviridae. Prior to 2019, only three genotypes, namely PCV1, PCV2, and PCV3, had been documented worldwide (Allan et al., 1998; Hamel et al., 1998; Rodríguez-Arrioja et al., 2000; Lekcharoensuk et al., 2004; Phipps et al., 2007; Krakowka et al., 2008). PCV1 was a non-pathogenic viral particle (Gagnon et al., 2010; Ma et al., 2011; Saha et al., 2011). PCV2 is the predominant pathogen responsible for PCVAD, characterized by immunosuppression leading to clinical signs (Krakowka et al., 2008; Becskei et al., 2010; Doster et al., 2010; Henriksson et al., 2011; Resendes et al., 2011). In October 2016, PCV3 was detected in the USA (Wang et al., 2020). Subsequently, PCV3 has been detected in countries such as Poland, Japan, and South Korea (Kwon et al., 2017; Stadejek et al., 2017; Hayashi et al., 2018; Deng et al., 2023). In 2019, PCV4, was identified in farmed pigs, in Hunan Province, China (Zhang et al., 2020). It was also reported in several provinces and cities in China and South Korea (Tian et al., 2021; Hu et al., 2022; Wang et al., 2022; Chen et al., 2023; Fu et al., 2024). Extensive research has indicated that PCV2 and PCV3 cause a series of serious diseases and syndromes in swine farms, posing a threat to the health of pigs and the economic efficiency of the farming industry (Mora-Díaz et al., 2020; Yang et al., 2022). However, the pathogenicity and harmfulness of PCV4 have not been fully clarified. As a newly discovered virus, the inability to isolate PCV4 from clinical samples significantly hampers the assessment of its pathogenicity, the development of effective prevention and control strategies, and the creation of vaccines against it.
Infectious clones are typically created by inserting the viral genome into a plasmid vector, which can then be transcribed into RNA or directly used to produce infectious virus particles in host cells (Fenaux et al., 2002; Park et al., 2016; Jiang et al., 2019; Niu et al., 2022; Shakir et al., 2023). This approach allows detailed investigation of viral functions, including gene expression, replication mechanisms, and host interactions. For viruses that are difficult to isolate, we often employ the method of infectious clones. In 2002, Fenaux et al. (2002) reported the first construction and use of an infectious molecular DNA clone of PCV2. This molecular clone was used for direct in vivo transfection of pigs to characterize the disease and associated pathological lesions of PCV2 infection. Jiang et al. (2019) have recently reported that PDNS-like disease can be reproduced in pigs infected with a cloned PCV3 virus in 2019.
In this study, we utilized the predominantly endemic strain in the Southwest of China to construct the pSK recombinant plasmid and we developed the rescued virus expression vector pSK-PCV4 (Xu et al., 2022c). Pathological lesions and PCV4-specific antigens were detected in various tissues and organs, including the lungs, kidneys, lymph nodes, spleen, and liver. Additionally, there was a notable increase in pro-inflammatory cytokines and chemokines in the serum. These results contribute to the assessment of PCV4 infectivity and pathogenicity, providing valuable insights for the prevention, treatment, and vaccine development for related diseases.
2 Materials and methods
2.1 Plasmid construction and virus rescuing
The full-length genome of PCV4 was synthesized based on the sequence of PCV4/SC-GA2022ABTC (OP497960.1) published by our laboratory. A molecular DNA clone of the PCV4 genome, flanked by Sac I and Hind III restriction enzyme sites at the 5′ and 3′ ends respectively, was inserted into the pBluescript SK+ vector, which had been predigested with the same enzymes to create the recombinant plasmid pSK-PCV4. Sequencing of the recombinant plasmid confirmed the absence of errors introduced during PCR amplification.
When the cells reached approximately 70% confluency, 250 ng of recombinant plasmid was transfected into the cells using Lipofectamine 3,000 reagent (Thermo Fisher Scientific, Waltham, MA, United States). The cells were additionally treated with 300 mM D-glucosamine for 30 min at 24 h after transfection. Then, the old medium was discarded and the cells was incubated with DMEM containing 2% FBS.
Seventy-two hours after transfection, the cells were subjected to three successive freeze–thaw cycles and named F0. Inoculate the F0 into freshly cultured PK-15 cells and perform successive passaging within the PK-15 cell line. The supernatants of virus culture medium were collected from generations F0 to F6, respectively. Total viral DNA was extracted from the collected samples using the DNA/RNA Extraction Kit (Vazyme, Nanjing, China). PCV4 genomic copies were quantified using a validated TB Green II-based real-time quantitative PCR (qPCR) assay, following the protocol described before (Xu et al., 2022b).
2.2 Identification of rescued PCV4
The generated PCV4 virus stock was then serially passaged in PK15 cells, and the titer of the PCV4 virus stock was determined as described previously for PCV2 and PCV3 titration and expressed as the 50% tissue culture infective dose (TCID50)/ml (Fenaux et al., 2002; Jiang et al., 2019). To confirm the virus was successfully rescued, the F6 virus was used to infect PK15 cells, and the cells were subjected to an immunofluorescence assay after 72 h.
Simultaneously, the infected PK15 cells were washed twice with pre-cooled phosphate-buffered saline (PBS, pH7.4) after removing the culture medium. Subsequently, 200 μL of RIPA cell lysis buffer was added to each well of a 6-well plate and incubated on ice for 20 min. Following incubation, all lysed samples were scraped off with a sterile cell spatula and transferred to pre-cooled 1.5 mL centrifuge tubes. The supernatant was then centrifuged at 12,000 rpm at 4°C for 10 min to isolate total cellular proteins. The resulting supernatant constituted the total cellular protein extract. The protein level of the rescued virus was further assessed through western blot analysis of the Cap protein from the F6 generation virus, using a mouse anti-PCV4 Cap polyclonal antibody developed in our laboratory as the primary antibody. For a detailed description of the preparation process, refer to the Supplementary material S1.
2.3 Animal experiment
Six 4-week-old specific pathogen-free piglets purchased from Wanjiahao in Meishan, Sichuan, were randomly divided into two groups: control group and PCV4 inoculation group. Each group of three piglets was housed in a separate room and provided with sterile food and water ad libitum. Before inoculation, all piglets tested negative for PCV2, PCV3, PRV, CSFV, PEDV, JEV and PRRSV by qPCR or RT-qPCR. After a one-week acclimation period, the PCV4-inoculated group received an nasal injection with 1 × 105.93 TCID50/ml of the PCV4, while the control group was similarly injected with PBS. After 28 days, all piglets were humanely euthanized.
2.4 Clinical signs
All pigs were weighed daily, and relative weight gain were calculated weekly. Rectal temperatures were recorded daily. Clinical observations were documented each day.
2.5 Sample collection
Blood samples were collected from all pigs every 3 days and placed in serum separation tubes. The blood was centrifuged at 2000 rpm for 10 min. The serum was stored at −80°C for future use.
At day 28, necropsies were performed, and samples of the heart, liver, spleen, lungs, kidneys, tonsils, and lymph nodes were collected into separate bags and stored at −80°C.
2.6 Assessment of viremia and distribution of PCV4 in various organs
PCV4 viral loads in tissues and sera were measured by qPCR described previously (Xu et al., 2022b). Samples underwent a 10-fold dilution in PBS, were then vortexed at 4°C, and subsequently centrifuged at a speed of 12,000 rpm for a duration of 10 min. The extracted DNA was counted and stored at −80°C. Results were analyzed using Bio-Rad CFX Manager 4.1 (Bio-Rad Laboratories, Shanghai, Co., Ltd.).
2.7 HE and IHC of rescued PCV4-inoculated piglets
At the end of the 28-day experiment, gross pathological lesions were observed. The same tissue samples were then fixed in 4% paraformaldehyde, dehydrated through a graded ethanol series, and embedded in paraffin. Subsequently, the samples were sectioned into 4 μm thick slices and stained with hematoxylin and eosin (H&E) for histopathological examination under an optical microscope.
For IHC, Firstly, tissue sections are deparaffinized in xylene and rehydrated through graded alcohols to water. Antigen retrieval is performed by heating the sections in a citrate buffer solution. The slides are then cooled and rinsed with PBS. To block endogenous peroxidase activity, the sections are incubated with hydrogen peroxide. The mouse anti-PCV4 Cap polyclonal antibody is then applied to the sections and incubated at 4°C overnight. After rinsing with PBS, the sections are incubated with an HRP-conjugated goat anti-mouse IgG (Bioss, Beijing, Co., Ltd.). Following this, a chromogen such as DAB (Servicebio, Wuhan, Co., Ltd.) is added to visualize the antibody–antigen complex, resulting in a brown precipitate. The stained sections are then examined under a microscope to assess the localization and intensity of the antigen expression.
2.8 ELISA of cytokine
The serum levels of IL-1β, IL-6, IL-10, IL-12, IFN-γ, and TNF-α at 0, 7, 14, 21, and 28 days were measured using an ELISA assay (Jianglai Biotechnology, Shanghai, Co., Ltd.).
2.9 Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.5.1 (GraphPad Software, San Diego, CA, United States). Data were analyzed using two-way ANOVA followed by t-tests to determine statistical significance, with a p-value of less than 0.05 considered statistically significant.
3 Results
3.1 Generation of PCV4 infectious stock
The full-length genome of PCV4 was synthesized based on the sequence of PCV4/SC-GA2022ABTC (OP497960.1) published by our laboratory. The synthesized PCV4 genome was ligated into the pSK vector to produce the PCV4 clone (Figure 1A). To obtain PCV4 viral particles, PCV4 infectious clones were transfected into PK-15 cells and passaged continuously. The viral load of PCV4 was monitored by qPCR at each passage, confirming the presence of PCV4 genomic DNA in every passage, indicative of viral genome replication within the cells (Figure 1B). Throughout the experimental period, no discernible cytopathic effects (CPE) were observed in transfected PK-15 cells. IFA results revealed that the PCV4 Cap protein within cells infected by the sixth generation of the virus exhibited significant reactivity with mouse anti-PCV4 Cap polyclonal antibodies. Relative to the control group, the widespread distribution of PCV4 within the cells was prominently evident, with a predominance observed in the cytoplasm compared to the nucleus, underscoring the successful rescue of PCV4 and its replication within the cellular environment (Figure 1C). The viral titer of the PCV4 stock after six passages was quantified as 105.93 TCID50/ml at 72 h post-infection. Furthermore, Western blot analysis detected a distinct 27 kD PCV4 Cap protein band in cell cultures collected from the sixth generation, contrasting with the absence of bands in the control group, indicative of expression of the viral Cap gene within the cells (Figure 1D).
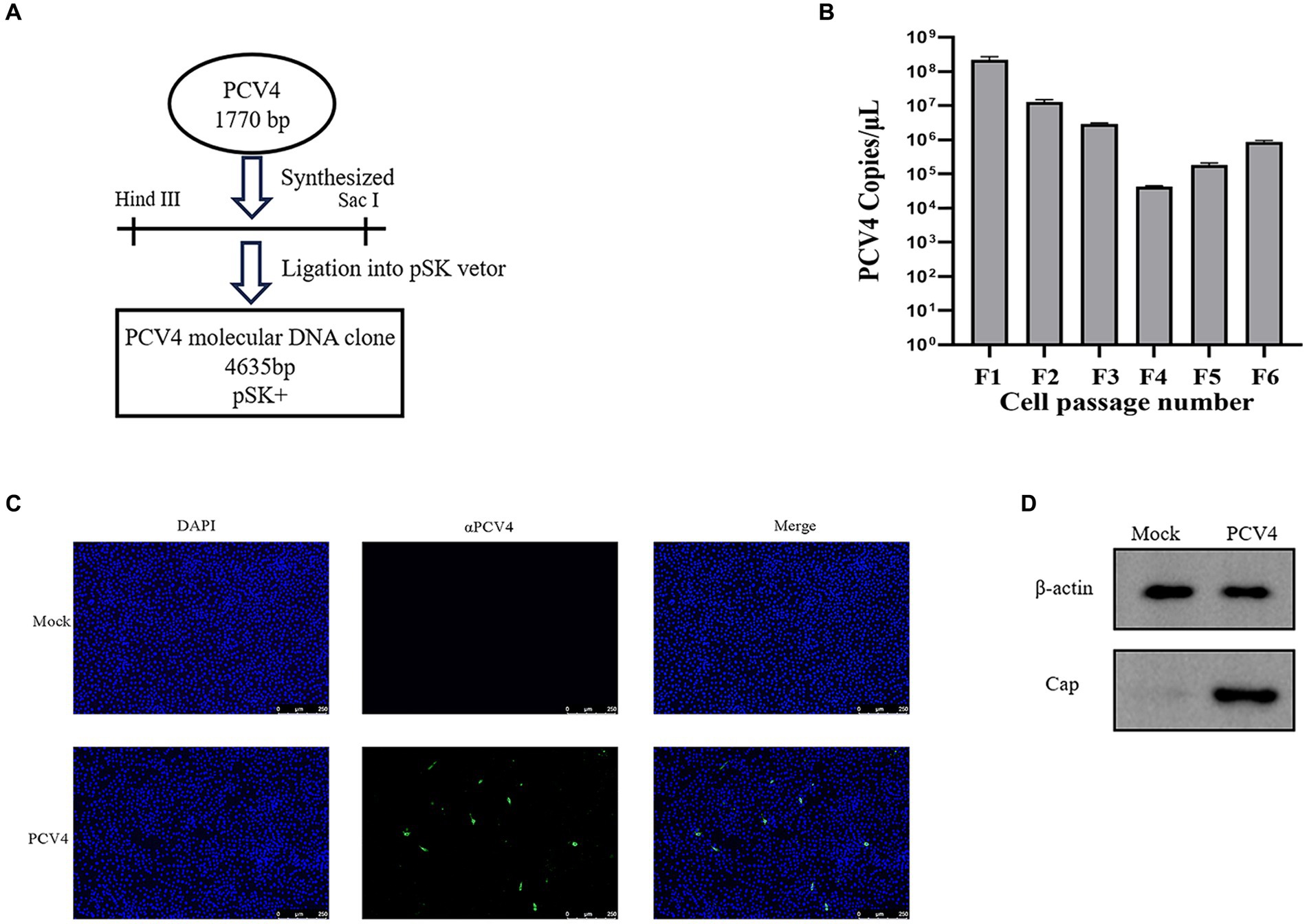
Figure 1. PCV4 infectious clone. (A) Construction of an infectious PCV4 molecular DNA clone. (B) The copy numbers of each passage. (C) Immunofluorescence analysis of PCV4-infected cells. Cells were stained with mouse anti-PCV4 Cap polyclonal antibody (1:100), followed by FITC-labeled goat anti-mouse IgG (green, 1:100). The nuclei were stained with DAPI (blue, 1:500). (D) Extract the total protein from the sixth-generation viral fluid for Western blot (WB) analysis.
3.2 Clinical signs induced by PCV4 infection
In the PCV4-inoculated group, one of the three piglets first exhibited clinical symptoms such as loss of appetite and diarrhea at 17 days post-infection (dpi). However, no noticeable clinical symptoms were observed in the other piglets inoculated with PCV4. The marked pathological changes in multiple organs of the PCV4-inoculated piglets indicate its pathogenicity in piglets. Rectal temperatures were measured every day. Piglets in the PCV4 inoculated group exhibited varying degrees of elevated body temperature (Figure 2A). Following PCV4 inoculation, weight gain in the PCV4 group piglets decreased after 21 dpi and continued to decline until the end of the experiment (Figure 2B). As expected, the control group showed no significant clinical symptoms.
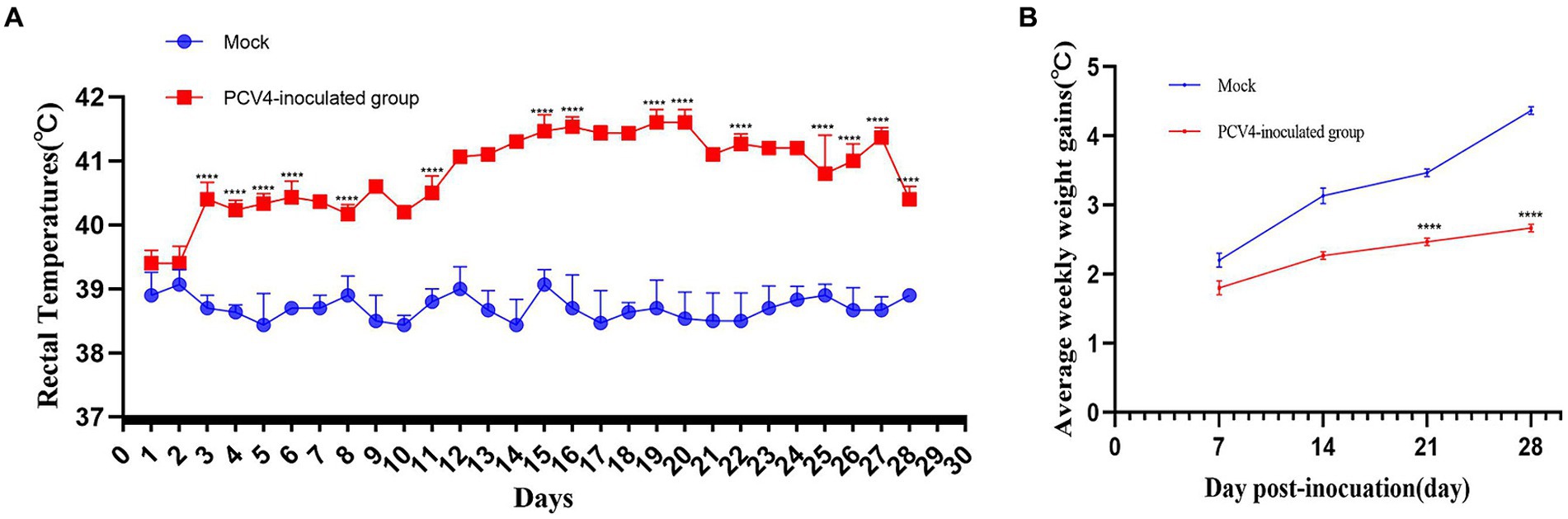
Figure 2. (A) Rectal temperature kinetics of the 4-week-old PCV4-inoculated piglets. (B) Average weekly weight gains of the 4-week-old PCV4-inoculated piglets following inoculation. * Means p < 0.05, ** means p < 0.01, *** means p < 0.005, **** means p < 0.0001 when comparing the PCV4-inoculted group with the control group.
3.3 Assessment of viral loads in sera and organs post-dissection
Throughout the study, all pigs in the control group tested negative for PCV4 in serum samples. In contrast, pigs in the infection group exhibited a significant increase in viremia starting at 7 dpi, peaking at 21 dpi (Figure 3A).
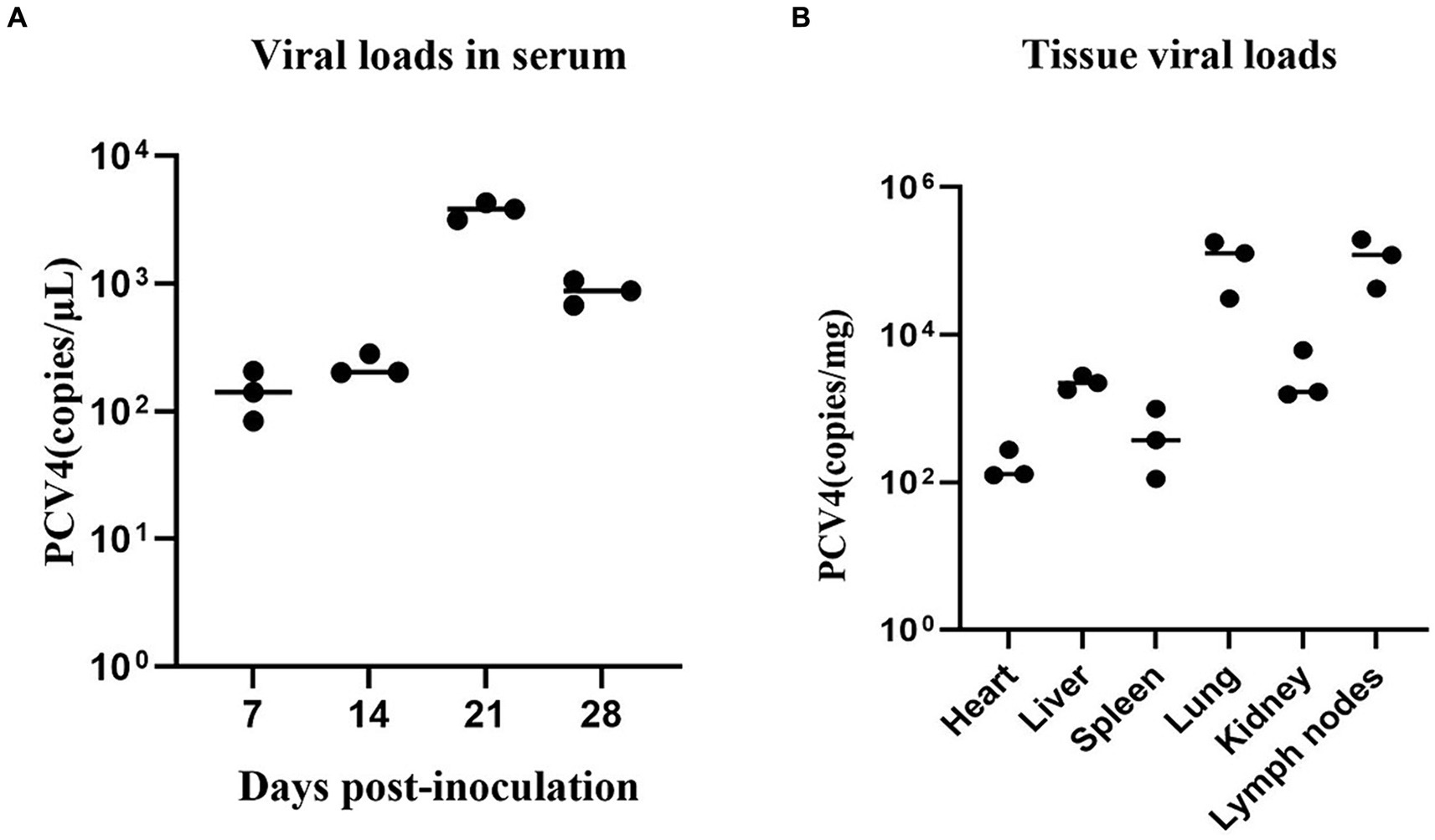
Figure 3. (A) PCV4 loads in sera from 4-week-old. PCV4-inoculated piglets were quantitatively assayed by qPCR. (B) Viral load of 4-week-old piglets in different tissues. * Means p < 0.05, ** means p < 0.01, *** means p < 0.005, **** means p < 0.0001 when comparing the PCV4-inoculted group with the control group.
As shown in Figure 3B, PCV4 was detected in the heart, liver, spleen, lungs, kidney, and lymph nodes using qPCR. The detected genomic copies indicated that the lungs and lymph nodes had the highest PCV4 copy numbers. No PCV4 infection was observed in the brain, small intestine, testes, or ovaries.
3.4 Histopathological lesions by PCV4 infection
Histopathological analysis revealed significant changes in the group inoculated with PCV4 Hepatocytes displayed mild vacuolar degeneration, with significant bile stasis evident between hepatocytes and bile ducts. In the spleen, there was a reduction in white pulp cells, accompanied by degeneration and necrosis of reticular cells in the red pulp. Lung tissues exhibited structural damage, characterized by alveolar wall rupture and degeneration, leading to shedding of alveolar epithelial cells into the alveolar cavity. Additionally, alveolar expansion and lymphocyte infiltration were observed. Renal tubules displayed moderate vacuolar degeneration of epithelial cells, along with tubular lumen narrowing and mesangial cell proliferation. Lymph nodes exhibited localized atrophy, with blurred boundaries between the cortex and medulla. The cortical lymphoid follicles were atrophied and reduced in number, with increased lymphocyte apoptosis/necrosis and prominent pigment deposition (Figure 4).
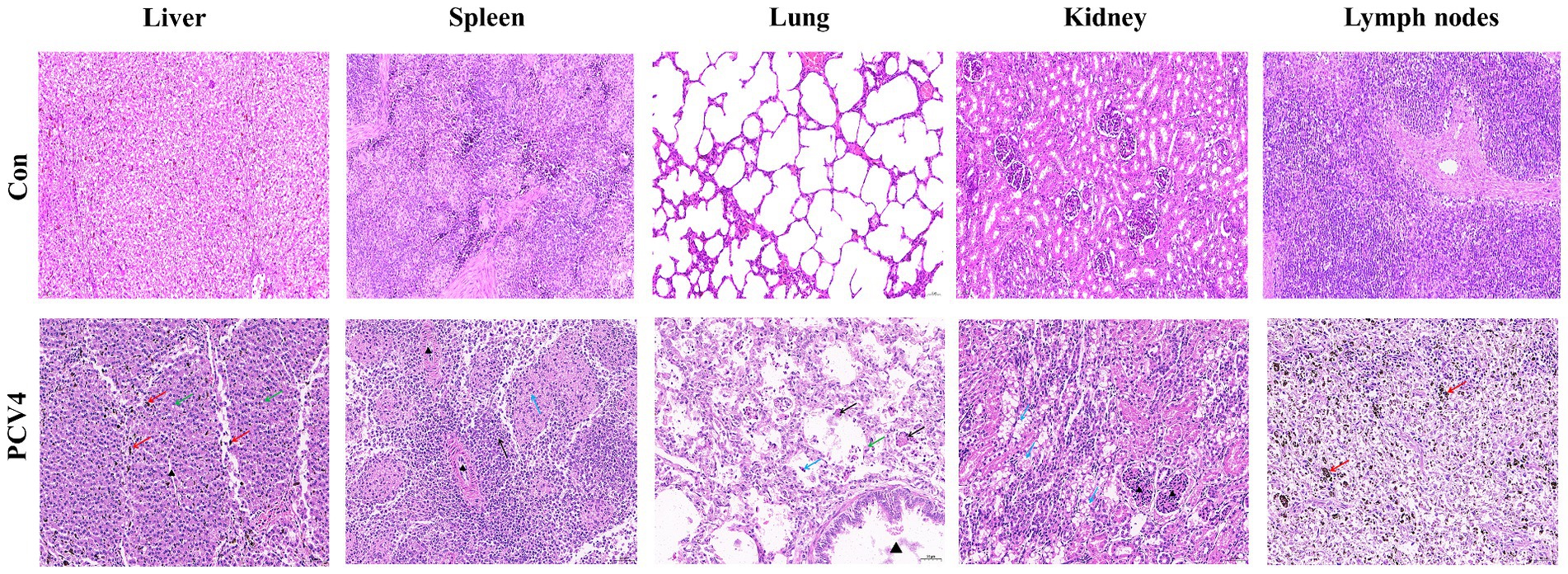
Figure 4. Histopathological lesions of organs in the 4-week-old PCV4-inoculated piglets. Bars, 50 μm.
3.5 Tissue distribution of PCV4 antigen
Immunohistochemical staining for PCV4 antigen was performed on the lungs, liver, kidney, spleen, and lymph nodes of piglets. The tissues from control piglets were negative for PCV4 antigen. In the liver, cells within the hepatic sinusoids exhibited a marked positive reaction. In the lungs, both the alveolar walls and the exfoliated cells within the alveolar spaces showed strong positive signals for the PCV4 antigen. In the spleen, the positive reactions were mainly distributed in the red pulp, with a few positive reactions observed in the lymphocytes of the white pulp. In the lymph nodes, clear positive reactions were detected in the cortical necrotic areas and around some local lymphoid follicles. In the kidney, there was a slight positive reaction within the epithelial cells of the renal tubules (Figure 5).
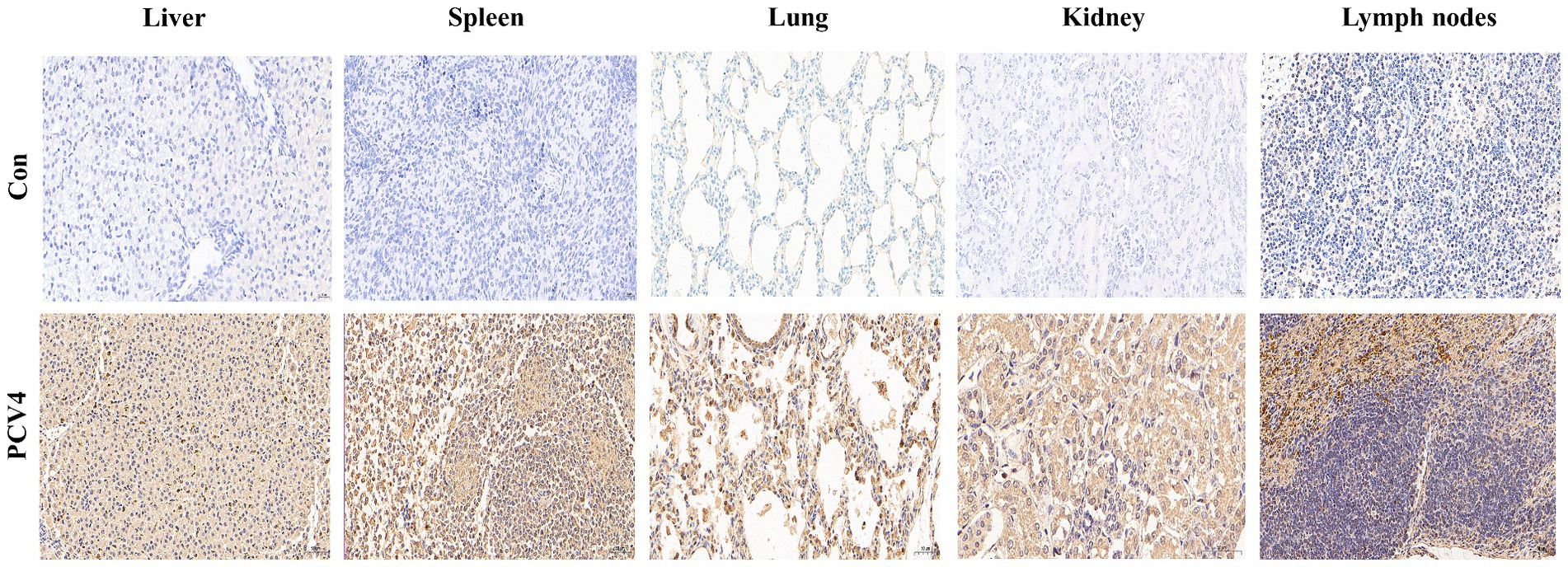
Figure 5. Immunohistochemical staining of organs and tissues of the 4-week-old piglets inoculated with PCV4.
3.6 The expression levels of cytokine in PCV4 infected piglets
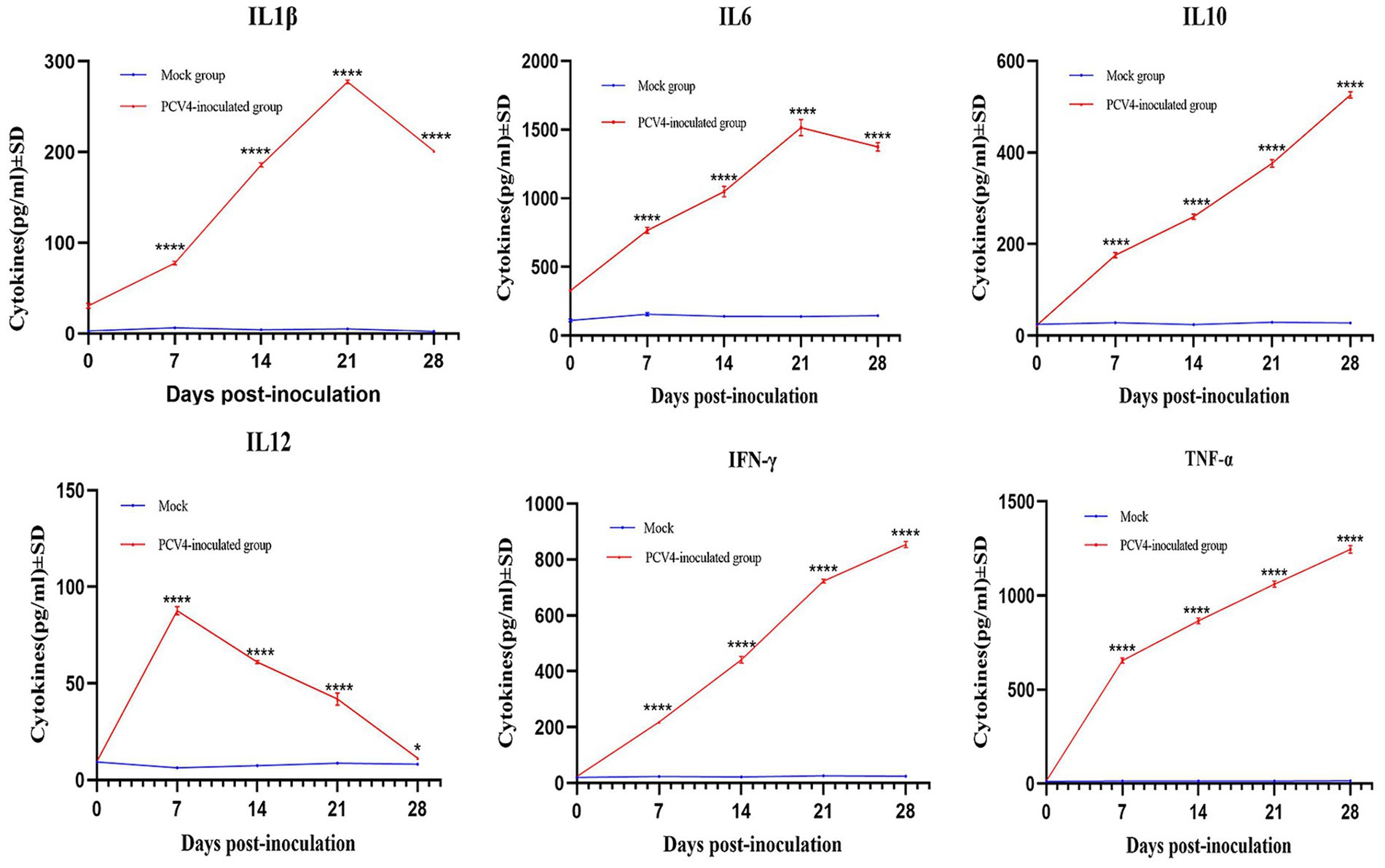
To further confirm that PCV4 infection induces inflammatory responses in piglets, we analyzed the expression levels of cytokines in the serum. As shown in Figure 6, compared to the control group, the expression levels of IL-1β, IL-6, and TNF-α in the PCV4-inoculated group continuously increased during the infection period, peaking at 21 days. The expression level of IL-10 and IFN-γ in the PCV4-inoculated group gradually increased throughout the infection period. In contrast, the expression level of IL-12 was low and continuously decreased during the infection period.
4 Discussion
PCV4, as a newly virus, which has a genome of 1770 nucleotides and encodes key replication and capsid proteins, has been reported across numerous Chinese provinces and detected in various species, indicating potential cross-species transmission (Tian et al., 2021;Wang et al., 2022; Wu et al., 2022; Xu et al., 2022c; Xu et al., 2023).
Currently, most research on PCV4 is limited to epidemiological investigations and cross-species transmission studies (Hou et al., 2022; Xu et al., 2022c). Although PCV4 was first identified in 2019, retrospective studies indicate that cases of PCV4 infection existed in China as early as 2008. The exact pathogenic mechanisms of PCV4 remain unclear due to the inability to successfully isolate the virus, posing challenges for disease prevention.
Fenaux et al. (2002) constructed an infectious molecular clone of PCV2 and demonstrated its infectivity by directly injecting the cloned PCV2 plasmid DNA into the liver and lymph nodes of pigs. This approach employed in vitro-constructed recombinant plasmids and an in vivo transfection system to elucidate the structural and functional relationships of PCV2 genes. Through this method, the roles of various regions or genes of PCV2 in viral replication and pathogenesis within the host were elucidated. PCV3, rescued from infectious clones, can induce typical clinical signs in piglets, such as PMWS and PDNS, respectively (Jiang et al., 2019). The clinical signs induced by these rescued viruses closely resemble those observed in field cases, indicating that infectious clones are effective for rescuing PCVs (Fenaux et al., 2002; Jiang et al., 2019). In this study, akin to the methodology employed for PCV2, PCV3 and PCV4 infectious clones, we utilized the PSK vector (Niu et al., 2022). Specifically, we employed the sequence of the SC-GA2022ABTC strain (belonging to PCV4c), which was initially identified in the southwestern region of China and is characterized by a distinctive amino acid sequence (239V for the Rep protein, and 27N, 28R, and 212M for the Cap protein) (Xu et al., 2022c).
We collected supernatants from each viral passage and quantified PCV4 viral load using qPCR. qPCR results indicated high copy numbers in the F1 and F2 generations, potentially attributable to residual transfected plasmid. Meanwhile, IHC and WB results demonstrated that the Cap protein continued to be expressed after multiple passages, confirming the successful rescue of the PCV4 infectious clone strain in this study. Constructing an infectious clone of PCV4 is advantageous as it allows for the generation of a biologically pure and homogeneous infectious virus stock, essential for definitive characterization of the disease and pathological lesions associated with PCV4 infection in piglets.
To assess the pathogenicity of the rescued PCV4 infectious clone strain and its capacity to elicit immune responses, we inoculated intranasally 4-week-old piglets with the rescued virus. The experimental results indicated that viremia was detectable as early as 7 dpi, reaching peak at 21 dpi. Previous studies have shown that Severe lymphoid depletion t in lymphoid tissues are the characteristic histological lesions of the PCV2 and PCV3 (Beach and Meng, 2012; Segalés, 2012). HE staining showed significant lymphocyte infiltration and increased apoptosis/necrosis of lymphocytes in piglets inoculated with the PCV4 infectious clone strain. This suggests that PCV4 infection compromised the pigs’ immune systems, leading to immunosuppression. IHC revealed a substantial presence of PCV4-positive cells across various tissues and organs in the inoculated piglets. These observations suggest that the tissue lesions caused by PCV4 are linked not only to viral replication within the host but also to the virus-induced inflammatory response. Furthermore, PCV4 was detected in several organs, including the heart, liver, spleen, lungs, kidneys, and lymph nodes. This indicates that PCV4 exhibits tissue tropism. The genomic copy numbers detected in different tissues indicated that the lungs and lymph nodes contained the highest PCV4 copy numbers. Previous studies have demonstrated that PCV2 and PCV3 can replicate their viral genomes more efficiently within lymph nodes (Phipps et al., 2007; Krakowka et al., 2008; Mora-Díaz et al., 2020), making these nodes the most common sites for PCV3 investigation and detection. Our research indicates that PCV4 may replicate more effectively in both lymph nodes and lungs.
Pigs infected with PCV4 exhibited milder clinical signs and no piglets died during the study period. This suggests that, under laboratory conditions, the pathogenicity of PCV4 may be relatively low. This is different from the infection of PCV2. Histopathological lesions in multiple tissues and organs similar to those of PMWS were reproduced with the PCV2 molecular DNA clone as well as with the infectious virus prepared in vitro from the molecular DNA clone. Besides, piglets that were inoculated with PCV3 alone and been reported in cases of natural infection with the virus presented characteristics of PDNS-like clinical signs and lesions. The clinical symptoms and severity of PCV4-induced disease may be influenced by several factors, including the immune status of the pig population, and co-infection with other porcine pathogens.
Pro-inflammatory cytokines play a crucial role in the development and maintenance of inflammation, potentially leading to multi-organ damage. To confirm that PCV4 infection triggers an inflammatory response, we measured cytokine (IL-1β, IL-6, IFN-γ and TNF-α) expression in pig serum. IL-1β is expressed in various tissues and cells, especially in macrophages within lymphoid organs. IL-6 is not a specific marker for infection, as its levels can increase in response to tissue injury or inflammatory stimuli. TNF-α is a key cytokine involved in inflammation and immune regulation, promoting the production of other cytokines and immune responses. The levels of IL-1β, IL-6, IFN-γ and TNF-α in the PCV4-inoculated group remained elevated throughout the infection period. Same infection as PCV2 and PCV3, upregulation of pro-inflammatory cytokines appears to play an important role in PCV4 pathogenesis (Meng, 2013; Chen et al., 2023).
5 Conclusion
PCV4, rescued from infectious clones, demonstrates the ability to replicate both in vitro and in vivo, leading to mild pathological lesions in piglets. This study substantiates the role of PCV4 in subclinical infections, which are marked by mild multisystemic inflammation and lymphocyte depletion in immune organs. These findings offer new insights into the pathogenesis and inflammatory responses associated with PCV4-related diseases, which are instrumental in developing effective prevention and control strategies against PCV4. Further research is crucial to understand and elucidate the mechanisms of interaction between PCV4 and its host, particularly in terms of immune evasion and pathogenesis.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Sichuan Agricultural University Animal Ethical and welfare Committee [license number: 20220283]. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LC: Writing – review & editing, Writing – original draft. L-SD: Writing – original draft, Validation, Supervision. TX: Writing – original draft, Validation, Supervision. S-YL: Writing – original draft, Conceptualization. Y-RA: Writing – original draft, Methodology. LZ: Writing – original draft, Conceptualization. Z-WX: Writing – review & editing, Writing – original draft, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by Technology Plan (Grant No. 2022YFN0007), the Chongqing Municipal Technology Innovation and Application Development Project (No. cstc2021jscx-dxwt BX0007), the Basic construction task of experimental pig resource development and innovation team (NCTIP-XD1C14), the Porcine Major Science and Technology Project of Sichuan Science and Technology Plan (No. 2021ZDZX0010-3), Key Research and Development Program in rural areas of Sichuan Science and Technology Department (Grant No. 2020YFN0147), Agricultural Industrial Technology System of Sichuan Provincial Department of Agriculture (Grant No. CARS-SVDIP), and Key Research and Development Project of Sichuan Science (2023YFN0021).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1443119/full#supplementary-material
References
Allan, G. M., McNeilly, F., Kennedy, S., Daft, B., Clarke, E. G., Ellis, J. A., et al. (1998). Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J. Vet. Diagn. Invest. 10, 3–10. doi: 10.1177/104063879801000102
Beach, N. M., and Meng, X. J. (2012). Efficacy and future prospects of commercially available and experimental vaccines against porcine circovirus type 2 (PCV2). Virus Res. 164, 33–42. doi: 10.1016/j.virusres.2011.09.041
Becskei, Z., Aleksić-Kovačević, S., Rusvai, M., Balka, G., Jakab, C., Petrović, T., et al. (2010). Distribution of porcine circovirus 2 cap antigen in the lymphoid tissue of pigs affected by postweaning multisystemic wasting syndrome. Acta Vet. Hung. 58, 483–498. doi: 10.1556/AVet.58.2010.4.9
Chen, D., Zhang, L., and Xu, S. (2023). Pathogenicity and immune modulation of porcine circovirus 3. Front. Vet. Sci. 10:1280177. doi: 10.3389/fvets.2023.1280177
Chen, X. M., Zhao, Y. Y., Liu, X. C., Han, Y. Y., Zhang, Y. H., Hou, C. Y., et al. (2023). Molecular detection and genetic characteristics of a novel porcine circovirus (porcine circovirus 4) and porcine reproductive and respiratory syndrome virus in Shaanxi and Henan provinces of China. Comp. Immunol. Microbiol. Infect. Dis. 98:102009. doi: 10.1016/j.cimid.2023.102009
Deng, H., Zhu, S., Zhu, L., Jian, Z., Zhou, Y., Li, F., et al. (2023). Histopathological changes and inflammatory response in specific pathogen-free (SPF) with porcine circovirus type 3 infection. Animals (Basel) 13:530. doi: 10.3390/ani13030530
Doster, A. R., Subramaniam, S., Yhee, J. Y., Kwon, B. J., Yu, C. H., Kwon, S. Y., et al. (2010). Distribution and characterization of IL-10-secreting cells in lymphoid tissues of PCV2-infected pigs. J. Vet. Sci. 11, 177–183. doi: 10.4142/jvs.2010.11.3.177
Fenaux, M., Halbur, P. G., Haqshenas, G., Royer, R., Thomas, P., Nawagitgul, P., et al. (2002). Cloned genomic DNA of type 2 porcine circovirus is infectious when injected directly into the liver and lymph nodes of pigs: characterization of clinical disease, virus distribution, and pathologic lesions. J. Virol. 76, 541–551. doi: 10.1128/jvi.76.2.541-551.2002
Fu, P. F., Wang, Y. H., Liu, G., Wang, D. M., Huang, W. W., Guo, D. Q., et al. (2024). First molecular detection and genetic characterization of porcine circovirus 4 in the Gansu Province of China. PLoS One 19, 19:e0293135. doi: 10.1371/journal.pone.0293135
Gagnon, C. A., Music, N., Fontaine, G., Tremblay, D., and Harel, J. (2010). Emergence of a new type of porcine circovirus in swine (PCV): a type 1 and type 2 PCV recombinant. Vet. Microbiol. 144, 18–23. doi: 10.1016/j.vetmic.2009.09.072
Hamel, A. L., Lin, L. L., and Nayar, G. P. (1998). Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72, 5262–5267. doi: 10.1128/JVI.72.6.5262-5267.1998
Hayashi, S., Ohshima, Y., Furuya, Y., Nagao, A., Oroku, K., Tsutsumi, N., et al. (2018). First detection of porcine circovirus type 3 in Japan. J. Vet. Med. Sci. 80, 1468–1472. doi: 10.1292/jvms.18-0079
Henriksson, S., Blomström, A. L., Fuxler, L., Fossum, C., Berg, M., and Nilsson, M. (2011). Development of an in situ assay for simultaneous detection of the genomic and replicative form of PCV2 using padlock probes and rolling circle amplification. Virol. J. 8:37. doi: 10.1186/1743-422X-8-37
Hou, C. Y., Zhang, L. H., Zhang, Y. H., Cui, J. T., Zhao, L., Zheng, L. L., et al. (2022). Phylogenetic analysis of porcine circovirus 4 in Henan Province of China: a retrospective study from 2011 to 2021. Transbound. Emerg. Dis. 69, 1890–1901. doi: 10.1111/tbed.14172. Epub 2021 Jun 11
Hu, X., Ding, Z., Li, Y., Chen, Z., and Wu, H. (2022). Serum investigation of antibodies against porcine circovirus 4 rep and cap protein in Jiangxi Province, China. Front. Microbiol. 13:944679. doi: 10.3389/fmicb.2022.944679
Jiang, H., Wang, D., Wang, J., Zhu, S., She, R., Ren, X., et al. (2019). Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J. Virol. 93, e02045–e02018. doi: 10.1128/JVI.02045-18
Krakowka, S., Hartunian, C., Hamberg, A., Shoup, D., Rings, M., Zhang, Y., et al. (2008). Evaluation of induction of porcine dermatitis and nephropathy syndrome in gnotobiotic pigs with negative results for porcine circovirus type 2. Am. J. Vet. Res. 69, 1615–1622. doi: 10.2460/ajvr.69.12.1615
Kwon, T., Yoo, S. J., Park, C. K., and Lyoo, Y. S. (2017). Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 207, 178–180. doi: 10.1016/j.vetmic.2017.06.013
Lekcharoensuk, P., Morozov, I., Paul, P. S., Thangthumniyom, N., Wajjawalku, W., and Meng, X. J. (2004). Epitope mapping of the major capsid protein of type 2 porcine circovirus (PCV2) by using chimeric PCV1 and PCV2. J. Virol. 78, 8135–8145. doi: 10.1128/JVI.78.15.8135-8145.2004
Ma, H., Shaheduzzaman, S., Willliams, D. K., Gao, Y., and Khan, A. S. (2011). Investigations of porcine circovirus type 1 (PCV1) in vaccine-related and other cell lines. Vaccine 29, 8429–8437. doi: 10.1016/j.vaccine.2011.07.123
Meng, X. J. (2013). Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system. Annu. Rev. Anim. Biosci. 1, 43–64. doi: 10.1146/annurev-animal-031412-103720
Mora-Díaz, J., Piñeyro, P., Shen, H., Schwartz, K., Vannucci, F., Li, G., et al. (2020). Isolation of PCV3 from perinatal and reproductive cases of PCV3-associated disease and in vivo characterization of PCV3 replication in CD/CD growing pigs. Viruses 12:219. doi: 10.3390/v12020219
Niu, G., Zhang, X., Ji, W., Chen, S., Li, X., Yang, L., et al. (2022). Porcine circovirus 4 rescued from an infectious clone is replicable and pathogenic in vivo. Transbound. Emerg. Dis. 69, e1632–e1641. doi: 10.1111/tbed.14498
Park, J. S., Kim, C. K., Um, J. H., Ju, Y. R., Lee, Y. S., Choi, Y. K., et al. (2016). Development of infectious clones of a wild-type Korean rabies virus and evaluation of their pathogenic potential. Virus Res. 223, 122–130. doi: 10.1016/j.virusres.2016.07.004
Phipps, S., Lam, C. E., Mahalingam, S., Newhouse, M., Ramirez, R., Rosenberg, H. F., et al. (2007). Eosinophils contribute to innate antiviral immunity and promote clearance of respiratory syncytial virus. Blood 110, 1578–1586. doi: 10.1182/blood-2007-01-071340
Resendes, A. R., Majó, N., VandenIngh, T. S., Mateu, E., Domingo, M., Calsamiglia, M., et al. (2011). Apoptosis in postweaning multisystemic wasting syndrome (PMWS) hepatitis in pigs naturally infected with porcine circovirus type 2 (PCV2). Vet. J. 189, 72–76. doi: 10.1016/j.tvjl.2010.06.018
Rodríguez-Arrioja, G. M., Segalés, J., Balasch, M., Rosell, C., Quintant, J., Folch, J. M., et al. (2000). Serum antibodies to porcine circovirus type 1 and type 2 in pigs with and without PMWS. Vet. Rec. 146, 762–764. doi: 10.1136/vr.146.26.762
Saha, D., Lefebvre, D. J., Ducatelle, R., Doorsselaere, J. V., and Nauwynck, H. J. (2011). Outcome of experimental porcine circovirus type 1 infections in mid-gestational porcine foetuses. BMC Vet. Res. 7:64. doi: 10.1186/1746-6148-7-64
Segalés, J. (2012). Porcine circovirus type 2 (PCV2) infections: clinical signs, pathology and laboratory diagnosis. Virus Res. 164, 10–19. doi: 10.1016/j.virusres.2011.10.007
Shakir, S., Zaidi, S. S., Hashemi, F. S. G., Nyirakanani, C., and Vanderschuren, H. (2023). Harnessing plant viruses in the metagenomics era: from the development of infectious clones to applications. Trends Plant Sci. 28, 297–311. doi: 10.1016/j.tplants.2022.10.005
Stadejek, T., Woźniak, A., Miłek, D., and Biernacka, K. (2017). First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound. Emerg. Dis. 64, 1350–1353. doi: 10.1111/tbed.12672
Tian, R. B., Zhao, Y., Cui, J. T., Zheng, H. H., Xu, T., Hou, C. Y., et al. (2021). Molecular detection and phylogenetic analysis of porcine circovirus 4 in Henan and Shanxi provinces of China. Transbound. Emerg. Dis. 68, 276–282. doi: 10.1111/tbed.13714
Wang, Y., Noll, L., Lu, N., Porter, E., Stoy, C., Zheng, W., et al. (2020). Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound. Emerg. Dis. 67, 1284–1294. doi: 10.1111/tbed.13467
Wang, Y., Yan, S., Ji, Y., Yang, Y., Rui, P., Ma, Z., et al. (2022). First identification and phylogenetic analysis of porcine circovirus type 4 in Fur animals in Hebei, China. Animals (Basel) 12:3325. doi: 10.3390/ani12233325
Wu, H., Hou, C., Wang, Z., Meng, P., Chen, H., and Cao, H. (2022). First complete genomic sequence analysis of porcine circovirus type 4 (PCV4) in wild boars. Vet. Microbiol. 273:109547. doi: 10.1016/j.vetmic.2022.109547
Xu, T., Chen, X. M., Fu, Y., Ai, Y., Wang, D. M., Wei, Z. Y., et al. (2022a). Cross-species transmission of an emerging porcine circovirus (PCV4): first molecular detection and retrospective investigation in dairy cows. Vet. Microbiol. 273:109528. doi: 10.1016/j.vetmic.2022.109528
Xu, T., Chen, L., Huang, B. Z., Zhu, L., Sun, X. G., Lai, S. Y., et al. (2023). The first dog-origin porcine circovirus type 4 complete genomic sequence have high homology with that of pig-derived strains. Front. Microbiol. 14:1121177. doi: 10.3389/fmicb.2023.1121177
Xu, T., Hou, C. Y., Zhang, Y. H., Li, H. X., Chen, X. M., Pan, J. J., et al. (2022b). Simultaneous detection and genetic characterization of porcine circovirus 2 and 4 in Henan province of China. Gene 808:145991. doi: 10.1016/j.gene.2021.145991
Xu, T., You, D., Wu, F., Zhu, L., Sun, X. G., Lai, S. Y., et al. (2022c). First molecular detection and genetic analysis of porcine circovirus 4 in the southwest of China during 2021–2022. Front. Microbiol. 13:1052533. doi: 10.3389/fmicb.2022.1052533
Yang, Y., Xu, T., Wen, J., Yang, L., Lai, S., Sun, X., et al. (2022). Prevalence and phylogenetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) in the southwest of China during 2020-2022. Front. Vet. Sci. 9:1042792. doi: 10.3389/fvets.2022.1042792
Keywords: porcine circovirus 4 (PCV4), pathogenicity, rescue, immune response, infection model
Citation: Chen L, Deng L-S, Xu T, Lai S-Y, Ai Y-R, Zhu L and Xu Z-W (2024) Rescue and characterization of PCV4 infectious clones: pathogenesis and immune response in piglets. Front. Microbiol. 15:1443119. doi: 10.3389/fmicb.2024.1443119
Edited by:
Peirong Jiao, South China Agricultural University, ChinaReviewed by:
Naidong Wang, Hunan Agricultural University, ChinaPu Sun, Chinese Academy of Agricultural Sciences, China
Copyright © 2024 Chen, Deng, Xu, Lai, Ai, Zhu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Wen Xu, YWJ0Y3h6d0AxMjYuY29t; Ling Zhu, YWJ0Y3psNzJAMTI2LmNvbQ==
†These authors share first authorship
 Lan Chen1
†
Lan Chen1
† Ling Zhu
Ling Zhu Zhi-Wen Xu
Zhi-Wen Xu