- 1Laboratory of Cultivation and Protection for Non-Wood Forest Trees, Central South University of Forestry and Technology, Changsha, China
- 2Key Laboratory of Forest Bio-resources and Integrated Pest Management for Higher Education in Hunan Province, Central South University of Forestry and Technology, Changsha, China
- 3State Key Laboratory of North China Crop Improvement and Regulation, College of Horticulture, Hebei Agricultural University, Baoding, China
- 4Hunan Tianhua Tea-oil Technology, Changsha, China
Introduction: Camellia anthracnose is caused by multiple Colletotrichum species, resulting in severe yield losses of oil-tea Camellia. Colletotrichum fructicola is one of the major anthracnose pathogens of oil-tea Camellia worldwide. However, developing unique molecular markers for the rapid and accurate detection of Colletotrichum fructicola from diverse Colletotrichum species, as well as early monitoring and effective control of the disease, remains largely unexplored.
Methods: C. fructicola-specific genes were obtained using a BLAST search of the sequences of predicted genes in C. fructicola against the genome sequences of Colletotrichum fungal pathogens. In this study, Colletotrichum fructicola-specific molecular markers were developed for rapid and accurate detection of C. fructicola among Camellia anthracnose causing fungal pathogens.
Results: Using genomic DNA-based end-point PCR and qPCR, three C. fructicola-specific genes with the ability to distinguish C. fructicola from other oil-tea Camellia anthracnose-related Colletotrichum species, including Colletotrichum camelliae, Colletotrichum gloeosporioides, and Colletotrichum siamense, and oil-tea Camellia fungal pathogens belonging to the genus Neopestalotiopsis, Pestalotiopsis, and Alternaria, were validated as molecular markers. In addition, these three molecular markers were highly sensitive to detecting C. fructicola using DNA extracted from the inoculated leaves of oil-tea Camellia.
Discussion: These findings enable us to rapidly and uniquely detect the Camellia anthracnose disease caused by Colletotrichum fructicola, which will equip farmers with an effective tool for monitoring Camellia anthracnose disease in the field and taking timely control measurements in advance.
1 Introduction
Anthracnose caused by Colletotrichum species is a devastating disease on oil-tea Camellia oleifera (Wang et al., 2020; Chen et al., 2022). The Colletotrichum pathogens, including Colletotrichum camelliae, Colletotrichum fructicola, Colletotrichum siamense, Colletotrichum aenigma and Colletotrichum gloeosporioides, have been reported to infect the buds, fruits and leaves of oil-tea Camellia oleifera in southern China (Jin et al., 2009; Chen et al., 2022, 2023). With the expanded cultivation of oil-tea Camellia in China, epidemics of anthracnose disease increasingly occur, resulting in reduced annual production of oil tea ranging from 10 to 30% (Qing et al., 2009). C. fructicola is the most predominantly identified species in diseased leaves of oil tea, followed by C. camelliae (Li et al., 2016). C. fructicola that was first isolated from diseased Coffea arabica in northern Thailand is a dominant pathogen of oil-tea anthracnose, belonging to the C. gloeosporioides complex (Prihastuti et al., 2009). As a member of Colletotrichum genus that is ranked among the top 10 plant pathogenic fungi (Dean et al., 2012), C. fructicola has the ability to cause anthracnose, bitter rot, and leaf spot diseases on a wide range of woody or herbaceous plants worldwide (Cannon et al., 2012; Fu et al., 2019; Li et al., 2021, 2022; Martin et al., 2021; Evallo et al., 2022; Zhao et al., 2022). The Chinese native commercial shrub oil-tea Camellia is threatened by C. fructicola because the pathogen can produce a large number of conidia spores, which are primary initial and secondary infection resources and can be easily disseminated to neighboring healthy blossoms, young fruit or leaves by rain splash, wind-driven rain and human conducted cultivation activities (De Silva et al., 2017; Chen et al., 2021).
Distinguishing specific pathogen species can facilitate the selection of an adequate disease management strategy (Cai et al., 2011). Quick and accurate detection and differentiation of different Colletotrichum species is essential in monitoring the oil-tea Camellia anthracnose epidemic to control the spread of the disease. Diagnosing plant diseases routinely depends on symptom observation and fulfillment of Koch's postulates in combination with morphological and molecular characterization (Damm et al., 2010; Liu et al., 2016). Currently, lab-based molecular identification and quantification of fungal pathogens using qPCR and multilocus phylogenetic analyses based on DNA sequences of the ribosomal internal transcribed spacer regions, calmodulin, β-tubulin, actin, chitin synthase-encoding genes and glyceraldehyde-3-phosphate dehydrogenase facilitate the differentiation of individual Colletotrichum species (Chen et al., 2022). For instance, multilocus phylogenetic analysis based on multiple gene sequences has been used as a supplement to morphological analysis to reliably and accurately identify C. fructicola within the C. gloeosporioides complex, including morphologically and physiologically identical 22 species plus one subspecies (Weir et al., 2012; Giblin et al., 2018; Grammen et al., 2019; Guarnaccia et al., 2021). In addition, based on multilocus phylogenetic analysis in combination with morphological characterization, 106 Colletotrichum isolates obtained from leaves of Camellia sinensis with anthracnose symptoms in the tea-growing regions of China have been identified to belong to 11 Colletotrichum species (Wang et al., 2016). The identification of 232 isolates collected from diseased leaves and fruits of oil-tea Ca. oleifera based on morphology and multilocus phylogenetic analyses found that they belong to five species, including C. camelliae, C. fructicola, C. siamense, C. aenigma, and C. gloeosporioides (Wang et al., 2020). Although multi-gene phylogenetic analysis coupled with morphological characterization has been successfully used to differentiate C. fructicola from multiple Colletotrichum species, the identification process is time-consuming, labor-intensive, and cost-effective. Therefore, the development of a single molecular marker for fast and accurate detection of C. fructicola from complex Colletotrichum species is required.
In this study, BLAST search of the predicted coding gene sequences of C. fructicola against the genomes of three anthracnose Colletotrichum species, including Colletotrichum camelliae, Colletotrichum gloeosporioides, and Colletotrichum siamense, was conducted to identify C. fructicola-unique coding sequences. Specific primers based on C. fructicola-unique coding sequences were developed and examined for the detection of C. fructicola using DNA-based end-point PCR and qPCR. Finally, three primer sets with high accuracy are obtained to detect and differentiate C. fructicola from other oil-tea Camellia anthracnose-related fungal species using end-point PCR and qPCR with DNA extracted from the diseased plant tissue.
2 Materials and methods
2.1 Fungal isolates and cultivation
The oil-tea Camellia anthracnose-related fungal species were originally isolated from diseased leaves and stored at the Key Laboratory of National Forestry and Grassland Administration for Control of Diseases and Pests of South Plantation, Central South University of Forestry and Technology, Changsha, China (Li et al., 2016). Colletotrichum spp. [Colletotrichum camelliae, Colletotrichum gloeosporioides, Colletotrichum siamense and Colletotrichum fructicola (CFLH16)], together with three other fungal strains belonging to the genus Neopestalotiopsis, Pestalotiopsis and Alternaria, were recovered from −80°C glycerol stocks and cultured in potato dextrose broth (PDB) medium shaking at 150 rpm and temperature of 28°C for 5 days to produce conidia.
2.2 Colletotrichum frucitcola conidia inoculation
Conidia preserved at −80°C were first recovered on solid potato dextrose agar (PDA) plate for 5 days at 28°C, and then the hyphae were transferred to PDB medium shaking at 150 rpm and temperature of 28°C for another 5 days. C. fructicola conidia were harvested by filtering a 5-day-old fungal liquid culture through three layers of Miracloth (Merck Millipore, Darmstadt, Germany). After washing with sterile water three times, the conidia were resuspended to 105 spores mL−1 in sterile water with 0.01% (v/v) Tween-20 for oil-tea Camellia inoculation. Surface sterilized Camellia leaves were placed in a Petri dish with the abaxial side up, and a volume of 1 mL of this spore suspension was sprayed using a handheld sprayer on the plant leaves per Petri dish. The treated materials were then cultivated in Petri dishes to keep humid for up to 5 days at 28°C before being harvested for total genomic DNA extraction.
2.3 DNA extraction from fungal conidia and oil-tea Camellia leaves
Fungal conidia were individually isolated by filtrate 5-day-old fungal PDB culture through three layers of Miracloth, and were washed with sterile water three times. Healthy Ca. oleifera leaves were collected from the orchard in the campus of Central South University of Forestry and Technology, Changsha, China. The conidia samples and Ca. oleifera leaves were frozen in liquid nitrogen, and these samples were ground to fine powder by Mixer Mill MM400 (Retsch, Haan, Germany). The total genomic DNA was isolated using CTAB method. For 500 mg fungal or botanic material, or inoculated oil-tea Camellia leaves at 5 days with C. fructicola conidia 1 mL CTAB extraction buffer supplemented with 10 μL β-mercaptoethanol was added and then vortexed to mix the sample powder with CTAB thoroughly before incubation at 65°C for 1 h. These CTAB mixtures were then cooled at room temperature for 1 h and gently mixed with an equal volume of chloroform. The extraction buffers were centrifuged at 10,000 g for 10 min to isolate the supernatant individually. The supernatant was transferred to a new centrifuge tube with an equal volume of isopropanol to precipitate total genomic DNA by gently mixing the supernatant with isopropanol. The DNA was collected as a pellet with centrifugation at 10,000 g for 10 min, and the DNA pellet was washed twice with ice-cold 70% (v/v) ethanol before dissolving in 100 μL distilled water. The total genomic DNA was quantified using the Eppendorf BioPhotometer Plus (Eppendorf, Hamburg, Germany).
2.4 Sequence analysis and primer design
Genome sequences of Colletotrichum camelliae, Colletotrichum gloeosporioides, Colletotrichum siamense, and Colletotrichum fructicola were downloaded from the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/). All coding sequences of C. fructicola were aligned to the genomes of Colletotrichum camelliae, Colletotrichum gloeosporioides, and Colletotrichum siamense, respectively, using BLASTN with an e-value cutoff value of 1e−5. The coding sequence without any BLAST hits against other Colletotrichum species was selected, and the genes without intron were taken as the candidate markers for Colletotrichum fructicola identification. Primers targeting selected C. fructicola unique genes were designed using Primer5 software (Supplementary Table S1).
2.5 End-point PCR amplification
Each PCR reaction was performed in a 20 μL reaction mixture comprised of 1 μL DNA template, 1 μL of 10 μ mol/L forward and reverse primer (10 μM of each primer), 10 μL 2 × Es Taq MasterMix (Cwbio, Taizhou, Jiangsu, China), and 7 μL distilled water. The PCR program used to produce proper amplicons was: initial denaturation at 94°C for 2 min; 35 cycles of 94°C denaturations for 30 s, annealing at 69°C for 30 s, extension at 72°C for 10 s; followed by a final extension at 72°C for 2 min on a Mastercycler nexus (Eppendorf, Hamburg, Germany). The PCR products were examined by 2% agarose gel electrophoresis. This experiment was repeated three times, and the same results were obtained.
2.6 qPCR assay
qPCR was performed using a 7500 Real-time PCR System (Applied Biosystems, Foster City, CS, USA) with T5 Fast qPCR Mix (SYBR Green I) (Tsingke Biotechnology, Beijing, China), in a final volume of 20 μL containing 0.1 μM of each primer from the respective primer set, 1 μL genomic DNA. The real-time PCR amplification program consisted of an initial denaturation step at 95°C for 4 min, followed by 40 cycles of denaturation for 15 s at 95°C, annealing for 30 s at 69°C and extension for 30 s at 72°C. Melt curve analysis was carried out following amplification by continuously heating from 60 to 95°C, with the acquisition of fluorescence at 0.3°C intervals and a 15 s hold at each increment.
3 Results
3.1 Identification of C. fructicola-specific genes for primer design
To rapidly and simply differentiate C. fructicola from other Colletotrichum species, we conducted a BLAST search of coding sequences of C. fructicola against the genome sequences of C. camelliae, C. gloeosporioides, C. siamense, respectively. Nine hundred and sixty-five C. fructicola-specific gene-coding sequences were obtained (Figure 1). Based on the predicted gene function and annotation from NCBI, 115 C. fructicola-specific genes encoding proteins participating in various cellular processes and interactions with the environment that are essential for fungal survival and reproduction were selected from 965 C. fructicola specific genes (Figure 1). To design C. fructicola specific primers that can be used for genomic DNA-based PCR detection, 22 genes without intron were selected (Supplementary Figure S1). qPCR primer pairs were designed to target these C. fructicola-specific gene regions (Supplementary Table S1).
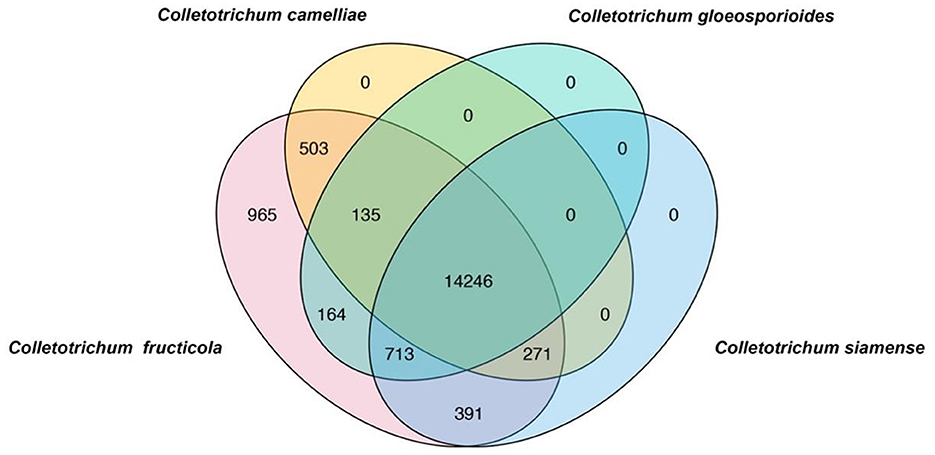
Figure 1. Venn diagrams showing the unique genes in different Colletotrichum species. BLAST search was conducted using all coding sequences of C. fructicola against the genome sequences of Colletotrichum camelliae, Colletotrichum gloeosporioides, and Colletotrichum siamense. C. fructicola-specific genes are indicated by pink.
3.2 Validation of C. fructicola primers specificity by end-point PCR
To evaluate the specificity of the designed C. fructicola primer sets, the genomic DNA extracted from four oil-tea Camellia pathogenic Colletotrichum spp. and the other three fungal genera isolated from diseased leaves of oil-tea Camellia anthracnose were used as the template for end-point PCR amplification with different primer sets, respectively. Among the 22 C. fructicola-specific primer pairs, end-point PCR amplification with three primer sets targeting v012077 (putative AC transposase), v013293 (nonribosomal peptide synthetase dtxS1), and v004178 (acyltransferase easC) produced specific bands with the expected sizes of 215, 243, and 208 bp, respectively, while no amplicons were generated from C. camelliae, C. gloeosporioides, C. siamense, Neopestalotiopsis, Pestalotiopsis and Alternaria (Figure 2). In addition, we optimized the annealing temperature of each primer pair and found that end-point PCR with the annealing temperature at 69°C produced the single amplicon of the target sequences (Figure 2). Taken together, these findings indicated that primer pairs v012077F/R, v013293F/R, and v004178F/R are specific for C. fructicola identification from the examined oil-tea Camellia anthracnose fungal pathogens.
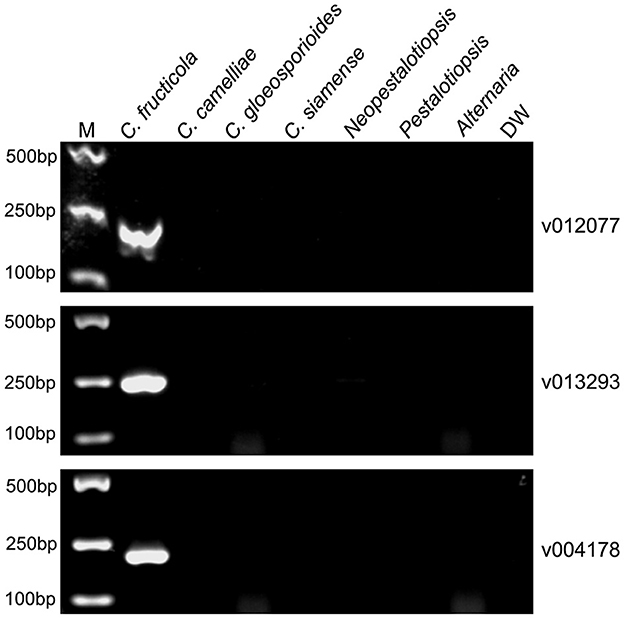
Figure 2. PCR amplification using three C. fructicola-specific primer sets to determine their presence in different Camellia anthracnose-related fungal pathogens. End-point PCR with primer pairs targeting different C. fructicola genes v012077, v013293, and v004178 was performed, respectively. The genomic DNAs of each oil-tea Camellia anthracnose-related fungal pathogens were used as templates. M: DL2000 Marker. DW: distilled water.
3.3 Determining the specificity and efficiency of C. fructicola-specific primers using qPCR assay
To assess the amplification specificity of v012077, v013293, and v004178 primers using qPCR assay, the melting curves were generated with the genomic DNA extracted from C. fructicola conidia. As shown in Figure 3A, the tested primer sets generated specific amplicons in qPCR assay, as evidenced by the appearance of a single peak in the melting curve in each sample. The amplification product of v012077 was specific, with a single peak around 83.7°C in the melting curve (Figure 3A). A single peak around 85.8°C in the melting curve of v013293 amplicons was observed in each sample, representing the specific amplification of this fragment (Figure 3B). The melting curve of v004178 products showed a single peak at 89.3°C (Figure 3C). These results suggested that the designed primer pairs targeting C. fructicola genes v012077, v013293, and v004178 can amplify specific PCR products using DNA-based qPCR assay. In addition, we performed qPCR assay using a set of 10-fold serially diluted C. fructicola genomic DNA to validate the primer efficiency. The amplification with v012077, v013293, and v004178 primer pairs produced a linear relationship between all diluted DNA templates and Ct values with a high correlation coefficient (R2 = 0.979–0.994) (Figures 3D–F). The high R2 value indicated that the Ct values are reliable predictors of the number of copies of the target gene detected in the qPCR assay. Overall, these results indicated that three C. fructicola-specific primer pairs were suitable for the sensitive detection of the target genes using genomic DNA-based qPCR.
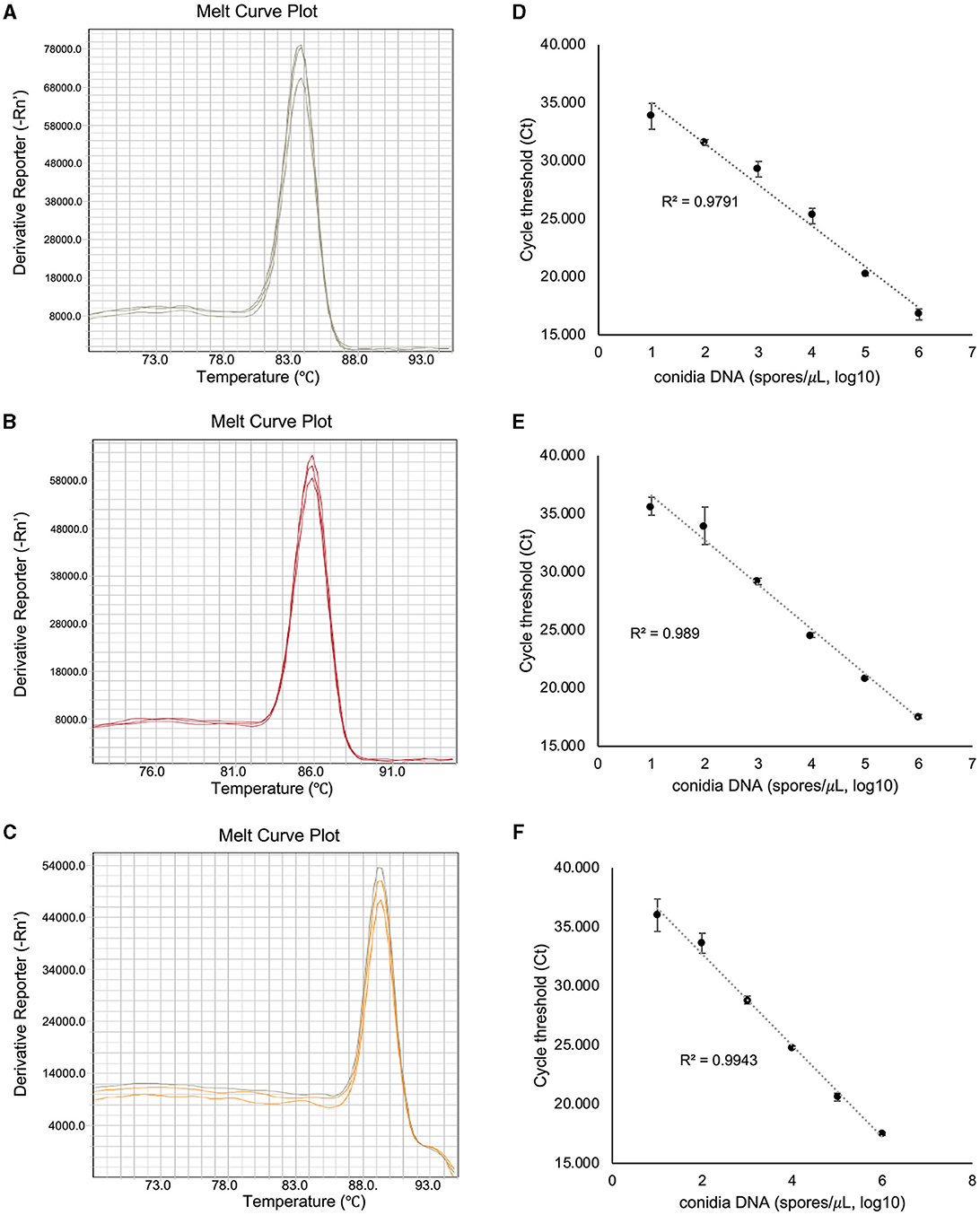
Figure 3. Evaluation of the amplification specificity and efficiency of C. fructicola-specific primers by qPCR. Melting curve analysis showing a single peak in each qPCR reaction with non-specific products was detected in C. fructicola v012077 (A), v013293 (B), and v004178 (C), DNA extracted from C. fructicola conidia was used as the template for amplification. The primer efficiencies (R2) of C. fructicola v012077 (D), v013293 (E), and v004178 (F) primers were analyzed with a serial dilution of genomic DNA extracted from fungal conidia as templates. Error bars are standard deviations.
3.4 Validating the sensitivity of C. fructicola-specific primers
To determine the sensitivity of C. fructicola v012077, v013293, and v004178 primer pairs for the direct detection of C. fructicola, end-point PCR and qPCR were performed with a set of 10-fold serially diluted C. fructicola genomic DNA that mixed with Ca. oleifera DNA as templates. The oil-tea Camellia TUB primers were included as a positive control to show the presence of plant genomic DNA (Cao et al., 2023), and a single band of TUB amplicon was produced in each sample (Figure 4). The unique PCR products with the expected sizes of 215, 243, and 208 bp were observed in amplicons using the template DNA extracted from 104 to 106 spores, respectively. In addition, the sensitivity of the qPCR assay to detect C. fructicola was evaluated. Table 1 showed that the sensitivity of qPCR reached 1 spore (0.063 pg DNA of C. fructicola) for the v012077 primer set and 10 spores (0.63 pg DNA) for v013293 and v004178 primer pairs (Ct value < 35). These results indicated that three C. fructicola-specific primer pairs can be used to steadily detect C. fructicola by end-point PCR and qPCR without effects by Camellia DNA.
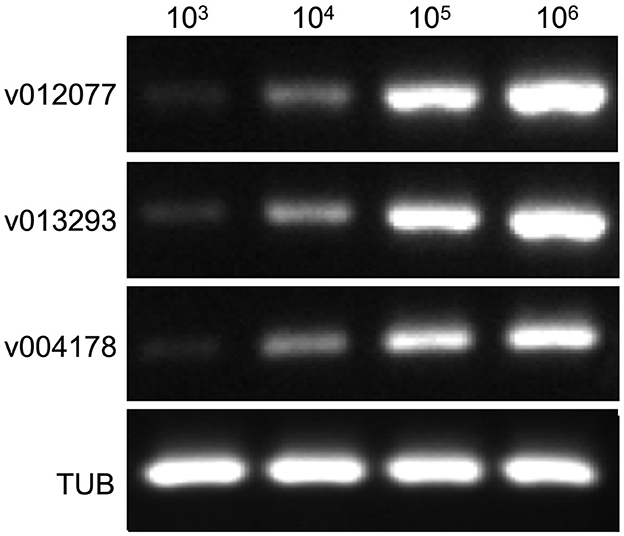
Figure 4. Determining the detection sensitivity of C. fructicola-specific primers via end-point PCR using C. fructicola and Ca. oleifera genomic DNA mixture. End-point PCR amplifications were performed with serial diluted C. fructicola conidial total DNA samples which were supplemented with oil-tea Camellia leaf genomic DNA. Oil-tea Camellia TUB was used as the plant inner control to indicate the plant genomic DNA in each reaction. M: DL2000 Marker. v012077, v013293, and v004178.
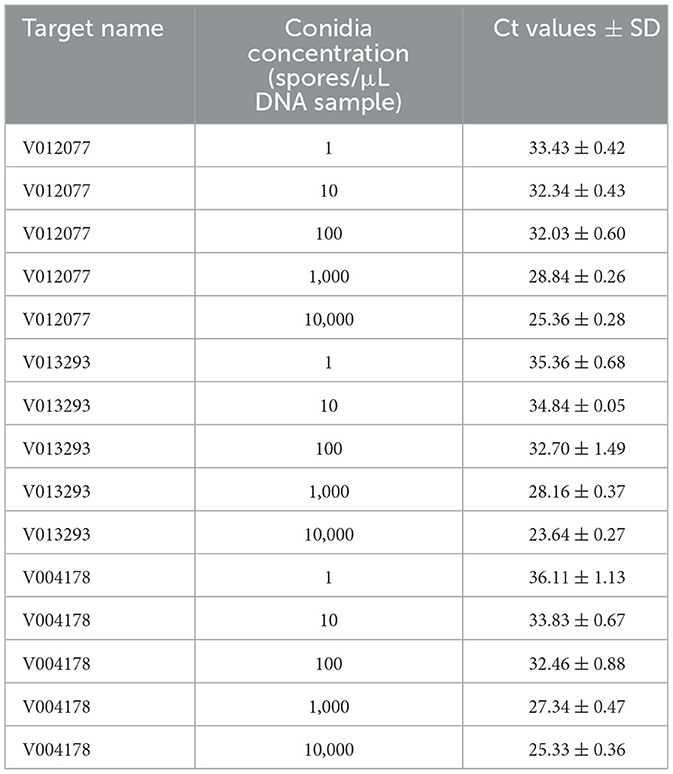
Table 1. Detection sensitivity of C. fructicola-specific primers using DNA extracted from a spore suspension with concentration ranging from 1 to 104 spores/μL by qPCR.
3.5 Detection of C. fructicola in inoculated oil-tea Camellia leaves using qPCR
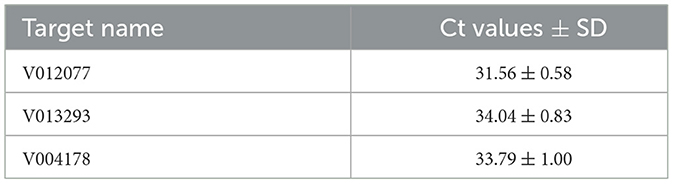
To validate the practical application of v012077, v013293, and v004178 primer pairs in directly detecting C. fructicola in Ca. oleifera leaves, the lower surface of detached Ca. oleifera leaves was inoculated with conidia (105 spores/mL conidial suspension) by spraying. Leaf samples were harvested 5 days post inoculation for DNA extraction, and qPCR was then conducted to detect C. fructicola. As shown in Table 2, qPCR analysis using v012077, v013293, and v004178 primer sets can detect C. fructicola with Ct values 31.56, 34.04, and 33.79, respectively. The primer set of v012077 produced the lowest Ct value, indicating that the v012077 primer set is more sensitive than the other two primer sets. These results demonstrated the three C. fructicola-specific molecular markers based on qPCR technique can be used for direct detection of C. fructicola in inoculated oil-tea Camellia leaves, which can be used for practical surveillance of this pathogen.
4 Discussion
C. fructicola is a versatile pathogen of the C. gloeosporioides complex, of which over 100 plant species have been reported as its hosts and the host list is continuing to grow (Bragard et al., 2021). As the predominant causal agent of oil-tea Camellia anthracnose, C. fructicola poses a significant threat to the oil-tea industry. Therefore, accurate detection and specific identification of C. fructicola are urgently required to safeguard oil-tea Camellia cultivation. The goal of this study was to develop molecular markers suitable for specific detection and identification of C. fructicola from oil-tea Camellia. In this work, we developed three C. fructicola-specific molecular markers to reliably detect C. fructicola in inoculated oil-tea Camellia leaves through DNA-based end-point PCR and qPCR approaches. We also determined the specificity of these three molecular markers in distinguishing C. fructicola from other oil-tea Camellia anthracnose-isolated Colletotrichum species, as well as from oil-tea Camellia-related fungal pathogenic genus Neopestalotiopsis, Pestalotiopsis, and Alternaria. This study provided a rapid and straightforward method to reliably distinguish C. fructicola from C. gloeosporioides complex, as well as from other oil-tea Camellia anthracnose-related fungal species in China.
Colletotrichum pathogens isolated from the leaves of Ca. oleifera anthracnose belong to Colletotrichum gloeosporioides complex (CGSC), and they are morphologically and physiologically identical or similar (Cannon et al., 2012; Weir et al., 2012). Therefore, the predominant oil-tea Camellia anthracnose pathogen C. fructicola cannot be reliably identified based on morphological characteristics. Since these Ca. oleifera pathogenic Colletotrichum species are genetically related, multilocus gene sequencing analysis is compulsory for C. fructicola identification (Li et al., 2016; Giblin et al., 2018; Grammen et al., 2019; Guarnaccia et al., 2021). Molecular markers for C. fructicola quick detection and identification with a single PCR procedure are urgently required. We developed three C. fructicola-specific molecular markers (v012077, v013293, and v004178) that are capable of distinguishing C. fructicola from other examined Colletotrichum species with genomic DNA-based end-point PCR and qPCR assays (Figures 2, 3). Previous studies showed that PCR primers targeting ribosomal internal transcribed spacer (ITS), calmodulin (CAL), β-tubulin (TUB2), actin, chitin synthase-encoding genes and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) have been developed (Chen et al., 2022). The ribosomal internal transcribed spacer regions have been extensively used to design Colletotrichum species primers, such as C. acutatum detection with ribosomal internal transcribed spacer regions based primers from strawberry leaves and C. abscissam/C. gloeosporioides from Citrus leaves (Debode et al., 2009; Pereira et al., 2019). Besides these commonly used Colletotrichum identification regions, a cutinase gene-derived primer pair was used for indiscriminately distinguishing C. gloeosporioides complex, including C. fructicola, C. gloeosporioides, C. aenigma and C. siamense, from strawberry anthracnose (Yang et al., 2022). However, the high sequence similarity between these phylogenetic markers among species of C. gloeosporioides complex hindered the detection accuracy of these molecular markers and the development of C. fructicola-specific molecular markers. Reclassification of C. fructicola into C. chrysophilum has been reported by adding additional loci sequences to construct phylogenic tree. For example, C. fructicola isolated from diseased peach in the United States has been re-identified as C. chrysophilum by using eight loci analyses, including Actin (ACT), APN2 (DNA lyase gene, adjacent to MAT1-2-1 gene), ApMAT (the intergenic region between APN2 and partial sequence of the mating-type gene MAT1-2-1), CAL, GAPDH, GS (glutamine synthetase), ITS, and TUB2 (Khodadadi et al., 2020). Astolfi et al. (2022) reported that C. fructicola originally identified from glomerella leaf spot on apple in Southern Brazil and Uruguay was reclassified as C. chrysophilum by using APN2, ApMAT, CAL, and GS sequences in addition to original GAPDH, GD, and TUB2 sequences (Astolfi et al., 2022). These recent findings suggest that increasing the loci for the classification of Colletotrichum species is essential although the majority of identification and classification of Colletotrichum species causing anthracnose worldwide depends on the number of loci ranging from 3 to 5 (De Aguiar Carraro et al., 2022; Marins et al., 2022; Vieira et al., 2022). The C. fructicola isolate used in this study was originally isolated from the leaves of tea-oil Camellia anthracnose in China using four widely used loci sequences, including ITS, CL, GS, and GD (Li et al., 2016). C. chrysophilum has been increasingly reported to be responsible for anthracnose in America and Europe, while no studies reported that C. chrysophilum causes anthracnose in Asia, especially in China, on oil-tea trees (Chen et al., 2022; Castillo-Cabrera et al., 2023). We cannot exclude the possibility that the identified and reported Colletotrichum species in Asian countries, especially from oil-tea anthracnose, can be reclassified by increasing the used loci sequences. Revaluating the classification of the reported Colletotrichum species causing anthracnose will be urgently required in a near future.
High sensitivity is a crucial feature of C. fructicola molecular markers used for practical applications. TaqMan real-time PCR targeting the ApMat gene has a detection threshold of 1 ng C. fructicola genomic DNA (He et al., 2023). A species-specific hydrolysis probe real-time PCR assay that combines CHLAD and FRLAD primer-probe sets improves the detection sensitivity to 5 pg fugnal genomic DNA in apple (McHenry and Aćimović, 2024). The detection threshold of C. fructicola using a two-step nested PCR assay in strawberry reaches 1 pg genomic DNA (Chung et al., 2022). Our study achieved a comparable detection limit of 0.63 pg DNA with qPCR assays using v013293 and v004178 primer pairs, and the v012077 primed qPCR detected as little as 0.063 pg C. fructicola genomic DNA. The high sensitivity of the C. fructicola molecular markers developed in this work enables us to detect the presence of C. fructicola fungal pathogen with a single qPCR assay.
Our results showed that end-point PCR and qPCR with primer pairs targeting three genes v012077, v013293, and v004178 are able to generate specific amplicons from C. fructicola-oil-tea Camellia genomic DNA mixture (Figure 4, Table 1). Therefore, these findings rule out the possibility that DNA extracted from the leaves of oil-tea Camellia may interfere with the specificity of C. fructicola-specific primers for detecting the pathogen. Previously, we identified a TUB primer pair that can be used in various oil-tea Camellia species to normalize qPCR results for the relative quantification of fungal biomass in planta (Cao et al., 2023). These three C. fructicola-specific primer sets, in combination with the oil-tea Camellia TUB primer, can be potentially applied for practical quantitative evaluation of C. fructicola in oil-tea Camellia orchards regardless of the cultivated tree cultivar/species. As C. fructicola endangers a broad list of economically important host plants (Cannon et al., 2012; Fu et al., 2019; Li et al., 2021, 2022; Martin et al., 2021; Evallo et al., 2022; Zhao et al., 2022), the application of these C. fructicola-specific primers in the detection of the pathogen from other vulnerable plants for quarantine can be included in future works. Moreover, these C. fructicola-specific primers, in combination with suitable plant report genes, have the potential to be used as an efficient tool to study the dynamics of C. fructicola growth in the cognate host, which is essential for in vivo fundamental studies of C. fructicola-host plant interactions.
In conclusion, we developed three C. fructicola-specific molecular markers that can accurately distinguish C. fructicola from other oil-tea Camellia anthracnose isolated fungal pathogens, including from these closely related fungal species belonging to C. gloeosporioides complex. The C. fructicola-specific molecular markers based on end-point PCR and qPCR assays can be used for rapid and accurate detection and identification of this fungal pathogen from inoculated leaves of oil-tea Camellia, thus providing valuable tools for Camellia anthracnose monitoring caused by C. fructicola. We are confident in the sanitary applications of these three C. fructicola-specific molecular markers in oil-tea Camellia nurseries and orchards.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LC: Writing – original draft. KS: Writing – original draft. YL: Writing – original draft. XX: Writing – original draft. XS: Writing – original draft. WD: Writing – original draft. CW: Writing – original draft. LM: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the General Project of Hunan Natural Science Foundation (2023JJ30990 and 2024JJ5628), Changsha Outstanding Innovative Youth Fund (kq2306014), the Natural Science Foundation of China (31901314), and Major Science Project of Hunan Education Department (22A0182).
Conflict of interest
CW was employed by Hunan Tianhua Tea-oil Technology.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1442922/full#supplementary-material
Supplementary Figure S1 | Prediction of introns in selected C. fructicola unique genes.
Supplementary Table S1 | Primers targeting selected genes for C. fructicola identification.
References
Astolfi, P., Velho, A. C., Moreira, V., Mondino, P. E., Alaniz, S. M., and Stadnik, M. J. (2022). Reclassification of the main causal agent of glomerella leaf spot on apple into Colletotrichum chrysophilum in Southern Brazil and Uruguay. Phytopathology 112, 1825–1832. doi: 10.1094/PHYTO-12-21-0527-SC
Bragard, C., Dehnen-Schmutz, K., Di Serio, F., Gonthier, P., Jacques, M.-A., Miret, J. A. J., et al. (2021). Pest categorisation of Colletotrichum fructicola. EFSA J. 19:6803. doi: 10.2903/j.efsa.2021.6803
Cai, L., Giraud, T., Zhang, N., Begerow, D., Cai, G., and Shivas, R. G. (2011). The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers. 50, 121–133. doi: 10.1007/s13225-011-0127-8
Cannon, P. F., Damm, U., Johnston, P. R., and Weir, B. S. (2012). Colletotrichum – current status and future directions. Stud. Mycol. 73, 181–213. doi: 10.3114/sim0014
Cao, L., Sun, X., Dong, W., Ma, L., and Li, H. (2023). Detection and quantification of anthracnose pathogen Colletotrichum fructicola in cultivated tea-oil Camellia species from southern China using a DNA-based qPCR assay. Plant Dis. 107, 363–371. doi: 10.1094/PDIS-04-22-0901-RE
Castillo-Cabrera, J., Pacheco-Esteva, M. C., Ramos-Caballero, B., Mora-Romero, G. A., Leyva-Madrigal, K. Y., and Vásquez-López, A. (2023). First Report of Colletotrichum chrysophilum causing anthracnose on nance (Byrsonima crassifolia) in Mexico. Plant Dis. 108:209. doi: 10.1094/PDIS-05-23-0941-PDN
Chen, X., Chen, X., Tan, Q., Mo, X., Liu, J., and Zhou, G. (2022). Recent progress on harm, pathogen classification, control and pathogenic molecular mechanism of anthracnose of oil-tea. Front. Microbiol. 13:918339. doi: 10.3389/fmicb.2022.918339
Chen, X., He, Y., Wang, Z., Niu, A., Xue, Y., Zhou, D., et al. (2023). Research progress and management strategies of fungal diseases in Camellia oleifera. Front. Microbiol. 14:1215024. doi: 10.3389/fmicb.2023.1215024
Chen, X., Jiang, L., Bao, A., Liu, C., Liu, J., and Zhou, G. (2021). Molecular characterization, pathogenicity and biological characterization of Colletotrichum species associated with anthracnose of Camellia yuhsienensis Hu in China. Forests 12:1712. doi: 10.3390/f12121712
Chung, P. C., Wu, H. Y., Chen, Y. C., Hung, T. H., and Chung, C. L. (2022). Development of a nested PCR assay for detecting Colletotrichum siamense and Colletotrichum fructicola on symptomless strawberry plants. PLoS ONE 17:e0270687. doi: 10.1371/journal.pone.0270687
Damm, U., Baroncelli, R., Cai, L., Kubo, Y., O'Connell, R., Weir, B., et al. (2010). Colletotrichum: species, ecology and interactions. IMA Fungus 1, 161–165. doi: 10.5598/imafungus.2010.01.02.08
De Aguiar Carraro, T., Lichtemberg, P. D. S. F., Michailides, T. J., Miranda Borges, M. I., Pereira, W. V., and May De Mio, L. L. (2022). Identification and characterization of Colletotrichum species associated with anthracnose on persimmon in Brazil. Fungal Biol. 126, 235–249. doi: 10.1016/j.funbio.2021.12.003
De Silva, D. D., Crous, P. W., Ades, P. K., Hyde, K. D., and Taylor, P. W. J. (2017). Life styles of Colletotrichum species and implications for plant biosecurity. Fungal Biol. Rev. 31, 155–168. doi: 10.1016/j.fbr.2017.05.001
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Debode, J., Van Hemelrijckb, W., Baeyena, S., Creemersb, P., Heungensa, K., and Maes, M. (2009). Quantitative detection and monitoring of Colletotrichum acutatum in strawberry leaves using real-time PCR. Plant Pathol. 58, 504–514. doi: 10.1111/j.1365-3059.2008.01987.x
Evallo, E., Taguiam, J. D., and Balendres, M. A. (2022). Colletotrichum fructicola associated with fruit anthracnose of persimmon. J. Phytopathol. 170, 194–201. doi: 10.1111/jph.13068
Fu, M., Crous, P. W., Bai, Q., Zhang, P. F., Xiang, J., Guo, Y. S., et al. (2019). Colletotrichum species associated with anthracnose of Pyrus spp. in China. Persoonia 42, 1–35. doi: 10.3767/persoonia.2019.42.01
Giblin, F. R., Tan, Y. P., Mitchell, R., Coates, L. M., Irwin, J. A. G., and Shivas, R. G. (2018). Colletotrichum species associated with pre-and post-harvest diseases of avocado and mango in eastern Australia. Aust. Plant Pathol. 47, 269–276. doi: 10.1007/s13313-018-0553-0
Grammen, A., Wenneker, M., Van Campenhout, J., Pham, K. T. K., Van Hemelrijck, W., Bylemans, D., et al. (2019). Identification and pathogenicity assessment of Colletotrichum isolates causing bitter rot of apple fruit in Belgium. Eur. J. Plant Pathol. 153, 47–63. doi: 10.1007/s10658-018-1539-z
Guarnaccia, V., Martino, I., Gilardi, G., Garibaldi, A., and Gullino, M. L. (2021). Colletotrichum spp. causing anthracnose on ornamental plants in northern Italy. J. Plant Pathol. 103, 127–137. doi: 10.1007/s42161-020-00684-2
He, J., Sun, M. L., Li, D. W., Zhu, L. H., Ye, J. R., and Huang, L. (2023). A real-time PCR for detection of pathogens of anthracnose on Chinese fir using TaqMan probe targeting ApMat gene. Pest Manag. Sci. 79, 980–988. doi: 10.1002/ps.7260
Jin, A. X., Zhou, G. Y., and Li, H. (2009). Progress, problem and prospect of oil camellia anthracnose (Colletotrichum gloeosporioides) research. Forest Pest Dis. 28, 27–30.
Khodadadi, F., González, J. B., Martin, P. L., Giroux, E., Bilodeau, G. J., Peter, K. A., et al. (2020). Identification and characterization of Colletotrichum species causing apple bitter rot in New York and description of C. noveboracense sp. nov. Sci. Rep. 10:11043. doi: 10.1038/s41598-020-66761-9
Li, C., Tanga, B., Cao, S., Bao, Y., Sun, W., Zhao, Y., et al. (2021). Biocontrol ability and action mechanism of dihydromaltophilin against Colletotrichum fructicola causing anthracnose of pear fruit. Pest Manag. Sci. 77, 1061–1069. doi: 10.1002/ps.6122
Li, H., Zhou, G.-Y., Liu, J.-A., and Xu, J. (2016). Population genetic analyses of the fungal pathogen Colletotrichum fructicola on tea-oil trees in China. PLoS ONE 11:e0156841. doi: 10.1371/journal.pone.0156841
Li, W., Ran, F., Long, Y., Mo, F., Shu, R., and Yin, X. (2022). Evidences of Colletotrichum fructicola causing anthracnose on Passiflora edulis Sims in China. Pathogens 11:6. doi: 10.3390/pathogens11010006
Liu, F., Tang, G., Zheng, X., Li, Y., Sun, X., Qi, X., et al. (2016). Molecular and phenotypic characterization of Colletotrichum species associated with anthracnose disease in peppers from Sichuan Province, China. Sci. Rep. 6:32761. doi: 10.1038/srep32761
Marins, É. F. C., da Silva, M. J. D. S., da Silva, J. L., Silva-Cabral, J. R. A., Costa, J. F. D. O., Feij,ó, F. M., et al. (2022). Colletotrichum species associated with sugarcane red rot in Brazil. Fungal Biol. 126, 290–299. doi: 10.1016/j.funbio.2022.02.002
Martin, P. L., Krawczyk, T., Khodadadi, F., Acimovic, S. G., and Peter, K. A. (2021). Bitter rot of apple in the mid-atlantic United States: causal species and evaluation of the impacts of regional weather patterns and cultivar susceptibility. Phytopathology 111, 966–981. doi: 10.1094/PHYTO-09-20-0432-R
McHenry, D. J., and Aćimović, S. G. (2024). New species-specific real-time PCR assays for colletotrichum species causing bitter rot of apple. Microorganisms 12:878. doi: 10.3390/microorganisms12050878
Pereira, W. V., Bertolini, E., Cambra, M., and Massola Junior, N. S. (2019). Multiplex real-time PCR for detection and quantification of Colletotrichum abscissum and C. gloeosporioides on Citrus leaves. Eur. J. Plant Pathol. 155, 1047–1059. doi: 10.1007/s10658-019-01831-x
Prihastuti, H., Cai, L., Chen, H., McKenzie, E. H. C., and Hyde, K. D. (2009). Characterization of Colletotrichum species associated with coffee berries in northern Thailand. Fungal Divers. 39, 89–109.
Qing, A. X., Zhou, G. Y., and Li, H. (2009). Recent advances in tea-oil tree anthracnose research. Chin. For. Pathol. 28, 27–31.
Vieira, W. A. D. S., Veloso, J. S., Silva, A. C. D., Nunes, A. D. S., Doyle, V. P., Castlebury, L. A., et al. (2022). Elucidating the Colletotrichum spp. diversity responsible for papaya anthracnose in Brazil. Fungal Biol. 126, 623–630. doi: 10.1016/j.funbio.2022.08.001
Wang, Y., Chen, J. Y., Xu, X., Cheng, J., Zheng, L., Huang, J., et al. (2020). Identification and characterization of Colletotrichum species associated with anthracnose disease of Camellia oleifera in China. Plant Dis. 104, 474–482. doi: 10.1094/PDIS-11-18-1955-RE
Wang, Y. C., Hao, X. Y., Wang, L., Xiao, B., Wang, X. C., and Yang, Y. J. (2016). Diverse Colletotrichum species cause anthracnose of tea plants (Camellia sinensis (L.) O. Kuntze) in China. Sci. Rep. 6:35287. doi: 10.1038/srep35287
Weir, B. S., Johnston, P. R., and Damm, U. (2012). The Colletotrichum gloeosporioides species complex. Stud. Mycol. 73, 115–180. doi: 10.3114/sim0011
Yang, J., Duan, K., Liu, Y., Song, L., and Gao, Q.-H. (2022). Method to detect and quantify colonization of anthracnose causal agent Colletotrichum gloeosporioides species complex in strawberry by real-time PCR. J. Phytopathol. 170, 326–336. doi: 10.1111/jph.13082
Keywords: Colletotrichum fructicola, oil-tea Camellia, molecular markers, pathogen detection, end-point PCR, qPCR
Citation: Cao L, Shi K, Liu Y, Xie X, Sun X, Dong W, Wang C and Ma L (2024) Identification of specific genes as molecular markers for rapid and accurate detection of oil-tea Camellia anthracnose pathogen Colletotrichum fructicola in China. Front. Microbiol. 15:1442922. doi: 10.3389/fmicb.2024.1442922
Received: 03 June 2024; Accepted: 05 August 2024;
Published: 26 August 2024.
Edited by:
Ravinder Kumar, Indian Agricultural Research Institute (ICAR), IndiaReviewed by:
Gerardo Vázquez-Marrufo, Universidad Michoacana de San Nicolás de Hidalgo, MexicoYin Song, Northwest A&F University, China
Copyright © 2024 Cao, Shi, Liu, Xie, Sun, Dong, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingxue Cao, dDIwMTkyNDQ1QGNzdWZ0LmVkdS5jbg==; Lisong Ma, bWFsaXNvbmdAaGViYXUuZWR1LmNu
†These authors have contributed equally to this work
 Lingxue Cao
Lingxue Cao Kailin Shi3†
Kailin Shi3† Lisong Ma
Lisong Ma