
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CORRECTION article
Front. Microbiol., 25 June 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1441327
This article is a correction to:
Patterns of within-host spread of Chlamydia trachomatis between vagina, endocervix and rectum revealed by comparative genomic analysis
 Sandeep J. Joseph1‡
Sandeep J. Joseph1‡ Sankhya Bommana2‡
Sankhya Bommana2‡ Noa Ziklo2
Noa Ziklo2 Mike Kama3
Mike Kama3 Deborah Dean2,4,5,6,7,8*†‡
Deborah Dean2,4,5,6,7,8*†‡ Timothy D. Read9*†‡
Timothy D. Read9*†‡A corrigendum on
Patterns of within-host spread of Chlamydia trachomatis between vagina, endocervix and rectum revealed by comparative genomic analysis
by Joseph, S. J., Bommana, S., Ziklo, N., Kama, M., Dean, D., and Read, T. D. (2023). Front. Microbiol. 14:1154664. doi: 10.3389/fmicb.2023.1154664
In the published article, there was an error in Figure 2 as published. Some of the colors in the “ompA genotype” column of the heatmap did not match the true genotype of strain in the phylogenetic tree. The corrected Figure 2 and its caption appear below.
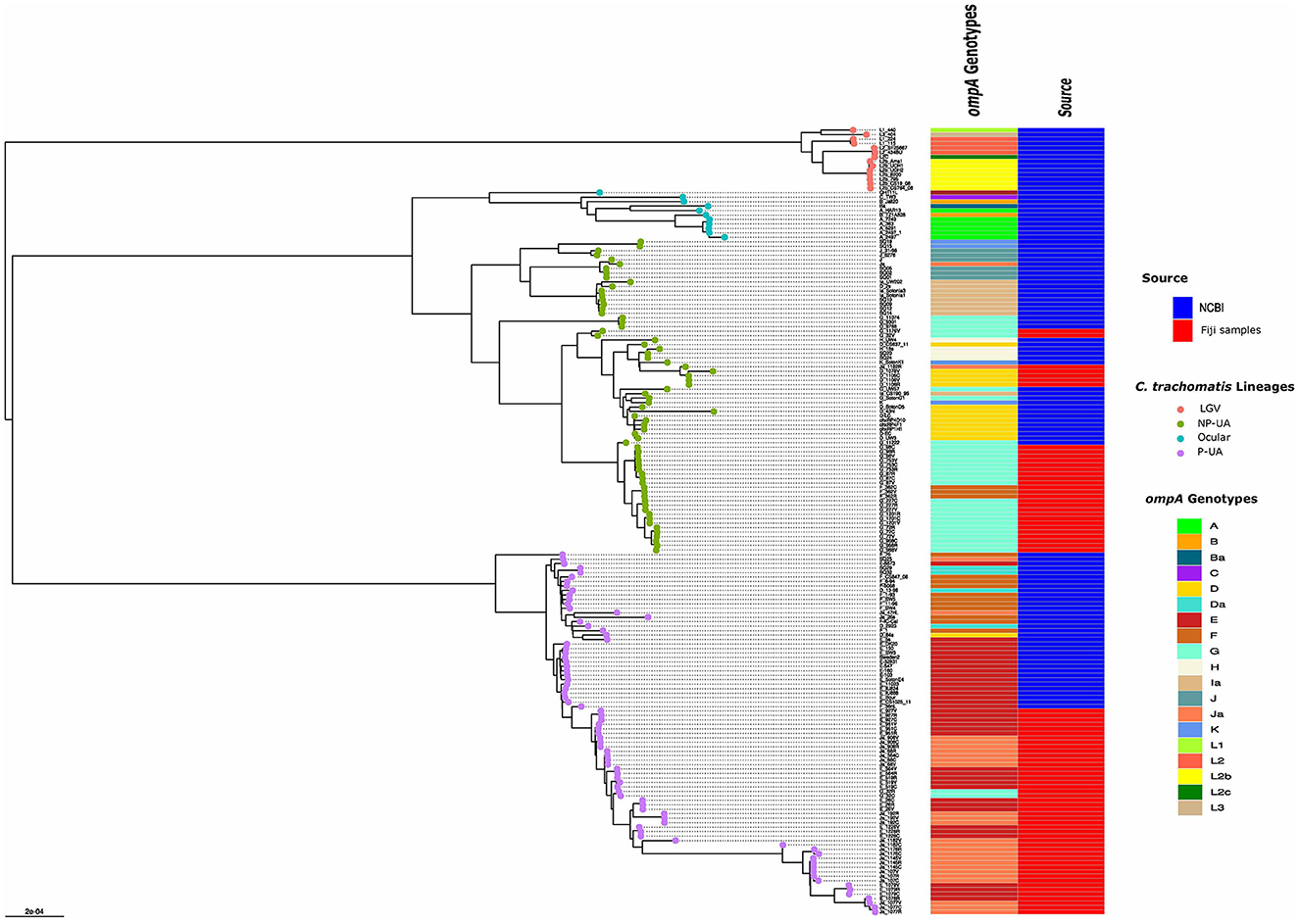
Figure 2. Global phylogeny with clade designations. The global phylogeny of high-quality C. trachomatis Fiji genomes plus selected complete C. trachomatis reference and clinical genomes representing global diversity from the National Center for Biotechnology Information (NCBI). Sample names are < ompA genotype > - < participant ID > - < body site code, where C = endocervix, R = rectum and V = vagina >. The round tips are colored by the 4 clade designations [LGV, ocular, prevalent- urogenital and anorectal (P-UA), non-prevalent urogenital and anorectal (NP-UA)]. The first column to the right of the tree denotes the ompA genotype with code at the lower right; the second column represents the source of the genomes from NCBI or the Fijian samples.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: Chlamydia trachomatis, single nucleotide polymorphisms, single variable polymorphisms, sexually transmitted diseases, chlamydiae
Citation: Joseph SJ, Bommana S, Ziklo N, Kama M, Dean D and Read TD (2024) Corrigendum: Patterns of within-host spread of Chlamydia trachomatis between vagina, endocervix and rectum revealed by comparative genomic analysis. Front. Microbiol. 15:1441327. doi: 10.3389/fmicb.2024.1441327
Received: 30 May 2024; Accepted: 10 June 2024;
Published: 25 June 2024.
Edited and reviewed by: Axel Cloeckaert, Institut National de recherche pour l'agriculture, l'alimentation et l'environnement (INRAE), France
Copyright © 2024 Joseph, Bommana, Ziklo, Kama, Dean and Read. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deborah Dean, ZGVib3JhaC5kZWFuQHVjc2YuZWR1; Timothy D. Read, dHJlYWRAZW1vcnkuZWR1
†These authors have contributed equally to this work
‡ORCID: Sandeep J. Joseph orcid.org/0000-0003-0697-2487
Sankhya Bommana orcid.org/0000-0002-4469-7925
Deborah Dean orcid.org/0000-0002-4490-1746
Timothy D. Read orcid.org/0000-0001-8966-9680
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.