- 1Department of Biochemistry, Faculty of Science, University of Geneva, Geneva, Switzerland
- 2School of Pharmaceutical Sciences, University of Geneva, CMU, Geneva, Switzerland
- 3Institute of Pharmaceutical Sciences of Western Switzerland, University of Geneva, CMU, Geneva, Switzerland
- 4ACCESS Screening Platform, NCCR Chemical Biology, Faculty of Science, University of Geneva, Geneva, Switzerland
- 5Mycology Group, Research Department Plant Protection, Agroscope, Nyon, Switzerland
Introduction: Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis, remains a serious threat to human health worldwide and the quest for new anti-tubercular drugs is an enduring and demanding journey. Natural products (NPs) have played a significant role in advancing drug therapy of infectious diseases.
Methods: This study evaluated the suitability of a high-throughput infection system composed of the host amoeba Dictyostelium discoideum (Dd) and Mycobacterium marinum (Mm), a close relative of Mtb, to identify anti-infective compounds. Growth of Dd and intracellular Mm were quantified by using luminescence and fluorescence readouts in phenotypic assays. The system was first benchmarked with a set of therapeutic anti-Mtb antibiotics and then used to screen a library of biotransformed stilbenes.
Results: The study confirmed both efficacy of established antibiotics such as rifampicin and bedaquiline, with activities below defined anti-mycobacterium susceptibility breakpoints, and the lack of activity of pyrazinamide against Mm. The screening revealed the promising anti-infective activities of trans-δ-viniferins and in particular of two compounds 17 and 19 with an IC50 of 18.1 μM, 9 μM, respectively. Both compounds had no activity on Mm in broth. Subsequent exploration via halogenation and structure-activity relationship studies led to the identification of derivatives with improved selectivity and potency. The modes of action of the anti-infective compounds may involve inhibition of mycobacterial virulence factors or boosting of host defense.
Discussion: The study highlights the potential of biotransformation and NP-inspired derivatization approaches for drug discovery and underscores the utility of the Dd-Mm infection system in identifying novel anti-infective compounds.
Introduction
Tuberculosis, an infectious disease caused by Mycobacterium tuberculosis (Mtb), has tragically claimed the lives of nearly 30 million people across the globe over the past decade, as reported by the World Health Organization (WHO) (Bagcchi, 2023). The tuberculous mycobacteria within the Mtb complex pose a difficult challenge for drug targeting, as these intracellular pathogens reside inside a vacuole in macrophages. Adding to this complexity, the intricate and waxy mycobacteria cell wall serves as an impermeable barrier to effective antibiotic treatment (Alderwick et al., 2015). The quest for new anti-tubercular drugs is an enduring and demanding journey further complicated by Mtb’s ability to enter a metabolically dormant state. Indeed, latent infections are often accompanied by non-heritable antibiotic tolerance, hindering the efficacy of conventional antibiotics and presenting a complicated challenge in disease control (Lewis, 2007). Adding to the gravity of the situation is the swift emergence of Multi-Drug-Resistant (MDR) and Extensively-Drug-Resistant (XDR) strains, which demand prolonged antibiotic treatment associated with severe side effects, high costs, and a limited chance of cure (Dheda and Migliori, 2012). Regrettably, despite the escalating threat of antibiotic resistance, the implementation of new antibiotics had been declining. However, thanks to a world-wide effort centered in universities, much progress has recently been achieved with the identification of a substantial number of drug candidates and newly approved drugs with novel mechanisms of action (Koul et al., 2007; Makarov et al., 2009; Lu et al., 2019; Alsayed and Gunosewoyo, 2023). Consequently, the development of new regimens for the treatment of drug-susceptible, MDR, and XDR Mtb infections are now conceivable. In fact, in 2019, the first 6-month regimen for the treatment of multidrug-resistant (MDR) and extensively drug-resistant (XDR) TB was approved, consisting of just three drugs with two novel mechanisms of action: bedaquiline, pretomanid, and linezolid (Koul et al., 2008; Conradie et al., 2020). Despite this success, a continuation of the effort to “fill the pipeline” is necessary.
In the pursuit of new anti-Mtb compounds, two primary approaches have been embraced: targeted drug discovery and whole-cell screening. While the main drawback of targeted drug discovery is that the selected target may not be essential in vivo, phenotypic screenings have exhibited more promise, giving rise to compounds such as SQ109 which is in clinical trial phase II (Reddy et al., 2010), bedaquiline (Andries et al., 2005), or delamanid (Matsumoto et al., 2006). Phenotypic screens in whole-cell assays offer numerous advantages over target-based drug discovery, including the identification of novel targets, probing multiple targets simultaneously or detection and exclusion of compounds with low membrane permeability. The fact that 40 years passed before bedaquiline was discovered as a new first-in-class drug (Mahajan, 2013), emphasizes the need for new targets and the importance of phenotypic assays.
Even though SQ109, bedaquiline and delamanid were discovered by phenotypic whole-cell screens on mycobacteria in broth, in vitro screening neglects that Mtb is an intracellular, vacuolar or cytosolic pathogen. Consequently, screening approaches should better take into account mycobacteria’s infection biology by making use of dedicated host-pathogen systems. Recent efforts shifted their focus towards a variety of in cellulo infection models, encompassing both cells of animal and non-animal origin. The experimentally versatile Amoebae – Mycobacterium marinum (Mm) infection models provide robust and ethically compliant platforms for investigating mycobacterial pathogenicity (reviewed in Cardenal-Munoz et al., 2018; Dunn et al., 2018; Guallar-Garrido and Soldati, 2024). Crucially, both Mtb and Mm employ highly similar strategies to establish an infection by hindering phagosome maturation and avoiding killing in macrophages and amoebae, while Mm offers experimental advantages, such as faster growth and being an organism requiring biosafety level 2 infrastructure (Ramakrishnan, 2004; Tobin and Ramakrishnan, 2008). On the host side, two main amoebae systems, Acanthamoeba castellanii (Kicka et al., 2014; Diop et al., 2018, 2019) and Dictyostelium discoideum (Dd), are commonly used to study the infectious process caused by various pathogenic bacteria, including Salmonella, Mycobacteria, Legionella, or Pseudomonas (Solomon et al., 2003; Steinert and Heuner, 2005; Hagedorn and Soldati, 2007; Dunn et al., 2018). Both amoebae are professional phagocytes employing evolutionary conserved strategies to capture and kill bacteria. Similarly to an Mtb infection, after uptake of Mm in macrophages or amoebae, the phagosome becomes a Mycobacteria-Containing Vacuole (MCV), an active interface between the host cell and the mycobacteria, being at the crossroads of host machineries such as membrane damage repair, autophagy, oxidative burst and lysosomal fusion, and virulence factors of the pathogen, such as type VII secretion systems and their effectors. Eventually, the MCV loses its integrity, allowing Mm to access the host cytosol (Cardenal-Munoz et al., 2017; Dunn et al., 2018; Lopez-Jimenez et al., 2018). Amoebae systems and especially Dd offer experimental advantages over animal derived cellular models mainly due to low maintenance cost and ease of genetic manipulation, rendering them ideal platforms for high-throughput approaches. The Dd-Mm system is a powerful 3R model system. The 3R principles were coined in the 1950s and stand for Replacement, Reduction and Refinement of animal experiments (Russell and Burch, 1959).
Direct screening inside an infected host allows to rapidly exclude compounds that are toxic to the host or have poor pharmacokinetics, thereby significantly contributing to decrease the usually high attrition rate in drug screens (Huszár et al., 2020). Most importantly, such systems open the possibility to identify host-targeting compounds that boost cell-autonomous defence and bacteria killing. This strategy termed adjunctive or host-directed therapy (HDT) is of particular interest as a strategy to circumvent resistant strains such as MDR-Mtb (Gries, 2024). For example, bedaquiline has been shown to activate macrophages in addition to its antibiotic effect (Giraud-Gatineau et al., 2020). The drug’s concentration in lipid droplets of the host cell (Greenwood et al., 2019) might play a substantial role in its mode of action and emphasizes the need of considering host biology when searching for new drugs. Besides unlocking host targets, host-pathogen screening systems also have the potential to guide the way to targets within bacterial effectors, which are specific for the intracellular life of mycobacteria (Gries et al., 2020). Compounds with such an activity have been identified by investigating hits from a host-pathogen screen, which were inactive in a screen on mycobacteria (Rybniker et al., 2015).
Even with the amoeba-Mm infection model as a valuable screening tool, the choice of chemical space to screen is crucial to success. In this regard, natural products (NPs) and their derivatives have played a significant role throughout history in advancing drug therapy, with notable contributions in fields such as cancer and infectious diseases (Newman and Cragg, 2016, 2020). The hegemony of NPs in drug discovery was severely challenged in the 1990s, with a major shift towards synthetic and combinatorial chemistry. However, this shift did not deliver the expected success (David et al., 2015). Recent advances in “omics” techniques, computational methods, and innovative screening models have revitalized the field of phenotypic screens in complex systems, and opened up new avenues of exploration (Rossiter et al., 2017; Wolfender et al., 2019). Despite this advance, several challenges in NP drug discovery remain, such as access to novel biological material, intraspecies variability and supply (Kingston, 2011; David, 2018). An alternative approach that takes advantage of the aforementioned advance is the generation of new NP-like compounds based on NP scaffolds. Such a strategy can take advantage of the use of enzymes to generate derivatives of known NPs that are not or not yet found in nature. Biotransformation has a number of advantages over classical organic chemistry. Reactions can take place under mild conditions, near neutral pH, ambient temperatures and atmospheric pressure, and protection of certain functional groups is often not required.
Based on this strategy, we recently presented the use of an enriched enzyme fraction secreted by the fungus Botrytis cinerea (“enzymatic secretome”) to transform NPs, such as stilbenes, into more complex structures with improved biological activities against fungi, breast cancer, bacteria and viruses (Gindro et al., 2017; Righi et al., 2020; Huber et al., 2022a; Zwygart et al., 2023). This approach yielded a small library of more than 70 compounds distributed in six scaffolds. Since stilbene derivatives have been reported to have potential anti-TB activity in various in vitro assays (Suarez et al., 2017; Reinheimer et al., 2018; Peraman et al., 2021), this library was subjected to an anti-infective screening workflow in the Dd-Mm infection model. The goal of this proof-of-concept study was to develop and benchmark the Dd-Mm system to high-throughput capacity in order to screen a focused library of natural products derivatives for anti-infective activities, which theoretically include both antivirulence and host defence-boosting activities that will be disentangled in future studies.
Materials and methods
Generation of a stilbene dimer library by chemoenzymatic synthesis
The initial screening library was generated by chemoenzymatic synthesis for a previous study on Wnt pathway inhibition (Huber et al., 2022a). Briefly, compounds 1–4 (99% purity) were purchased from Biopurify Phytochemicals Ltd. (Chengdu, Sichuan, China), and dimers 5–54 were obtained by enzymatic radical dimerization of 1 and 2 using the enzymatic secretome of Botrytis cinerea Pers (Gindro et al., 2017). Each compound was isolated by semi-preparative high-resolution liquid chromatography (semi-preparative-HPLC) and characterized by nuclear magnetic resonance (NMR), high-resolution mass spectrometry (HRMS) and ultraviolet (UV) spectroscopy. Details on the synthesis, isolation and characterization of the compounds can be found in their original reference (Huber et al., 2022a).
Isolation of trans-δ-viniferin enantiomers
Both enantiomers of the trans-δ-viniferin derivatives 11, 17, 19, and 21 were previously isolated for their antibacterial evaluation against Staphylococcus aureus (Huber et al., 2023). This was achieved by using a chiral stationary phase in HPLC. Each enantiomer was characterized by electronic circular dichroism (ECD) and optical rotation to assess its absolute configuration. Details of the isolation and characterization of these enantiomers can be found in the original reference (Huber et al., 2022b; Huber, 2023).
Generation of trans-δ-viniferin derivatives
The collection of trans-δ-viniferin derivatives 55–94 was generated for a previous study on antibacterial activity against S. aureus (Huber et al., 2023). Several strategies were employed, including (1) light-induced double-bond isomerization, (2) O-methylation of the phenolic moieties, (3) oxidation of the dihydrobenzofuran ring to benzofuran, (4) radical dimerization of other stilbene monomers and (5) oxidative halogenation. Each compound was isolated by semi-preparative HPLC and characterized by NMR, HRMS and UV spectroscopy. Details of the synthesis, isolation and characterization of the compounds can be found in their original reference (Huber, 2023).
Antibacterial assay
Mm WT expressing the lux operon (luxCDABE) under the hsp60 promoter (Andreu et al., 2010; Arafah et al., 2013) was cultured for 24 h prior to the experiment at 32°C under shaking conditions in 10 mL 7H9 broth (Becton Dickinson, Difco Middlebrook 7H9) containing 0.2% glycerol (Sigma Aldrich), 10% OADC (Becton Dickinson) and 0.05% tyloxapol (Sigma Aldrich). The Mm culture was centrifuged at 300 RPM for 1 min to sediment large bacterial clumps. The OD600 of the supernatant was then measured to determine and subsequently adjust the bacterial density to 3.75*105 bacteria per ml in 7H9 medium. Each well of a 384-well plate (Interchim FP-BA8240) was filled with 20 μL of the bacterial suspension (7.5*103 bacteria), and 0.2 μL of the dissolved compounds were added in a 1:100 dilution to each well to a final vehicle concentration of 0.5% ethanol. Each compound was tested in technical duplicates, and at least 24 technical replicates of control conditions were included: a vehicle control (VC) with a final concentration of 0.5% ethanol and rifabutin at a final concentration of 10 μM as a positive control. The plates were sealed with a gas-impermeable membrane (H769.1, Carl Roth), briefly centrifuged and bacterial growth was monitored using an Agilent BioTek H1 plate reader, an Agilent BioTek BioStack plate stacker, and luminescence was recorded over 72 h at 32°C with readings taken every hour. For every assay plate the robust Z’-factor was calculated using the vehicle control and the positive control. Each plate included 4 positive controls (Rifabutin) and 12 vehicle control wells (0.5% EtOH). The average Z’-factor was calulated as a mean of 3 biological replicates.
Anti-infective assay
Dd Ax2(ka) expressing mCherry at the act5 locus (Paschke et al., 2019) was cultured in 10 cm culture dishes (Falcon) in HL5-C medium (Formedium). Mm M strain expressing the bacterial lux operon (luxCDABE) was cultured at 32°C under shaking conditions in 7H9 broth (Becton Dickinson, Difco Middlebrook 7H9) containing 0.2% glycerol (Sigma Aldrich), 10% OADC (Becton Dickinson) and 0.05% tyloxapol (Sigma Aldrich). The day before the experiment 107 amoebae were plated in a 10 cm Petri dish. On the day of the experiment, infection was performed at MOI 25 by spinoculation in HL5-C and subsequently washing off extracellular bacteria (Mottet et al., 2021). Then, the cell suspension of infected amoebae was detached from the culture dish and resuspended in HL5-C with 5 U/mL penicillin and 5 μg/mL of streptomycin (Gibco) to inhibit extracellular growth of bacteria. The density of infected cells was quantified using a Countess (Thermo Fisher Scientific), and 1 × 104 cells in 20 μL of HL5-C with penicillin and streptomycin were seeded into each well of a 384-well plate (Interchim FP-BA8240). Test compounds resuspended in 50% ethanol were tested in technical triplicates and added in a 1:100 dilution to a final vehicle control concentration of 0.5% ethanol. The plates were sealed (H769.1, Carl Roth), briefly centrifuged and fluorescence and luminescence were monitored using an Agilent BioTek H1 plate reader and an Agilent BioTek BioStack plate stacker in a temperature-controlled environment (set to 24°C) over 72 h with readings taken every hour.
For both assays, growth curves were obtained by measuring the luminescence and fluorescence as a proxy for bacterial growth and host growth, respectively, for 72 h with time-points taken every hour. The “normalized residual growth” was computed by calculating the area under the curve (AUC, trapezoid method) and normalization to the vehicle control (0.5% ethanol, set at 1) and a baseline curve (set at 0). The baseline curve was calculated by taking the median of the first measurement of all wells in a plate and extrapolating it over the full time course. The threshold for hit detection was arbitrarily fixed at a cut-off of normalized residual growth <0.5. Normalized values were averaged over technical and biological replicates (all experiments at least n = 3 and N = 3) before estimating the IC50. For IC50 estimation, a robust 4PL regression was used, constraining the top value to 1 and, if necessary, the bottom value to –2. This was performed using GraphPad Prism (Version 8.0.1). The MIC was determined as the lowest experimental concentration, where the averaged normalized residual growth plus one standard deviation was ≤0. For every assay plate the robust Z’-factor was calculated using the vehicle control and the positive control. Each plate included 24 positive controls (Rifabutin) and 60 vehicle control wells (0.5% EtOH). The average Z’-factor was calulated as a mean of 3 biological replicates.
Readouts of the assays and terminology
To classify the readouts of the antibacterial and infection assays, we coined terms that encompass all the states/populations identified. The growth of Mm in broth is simply defined as “Mm-broth.” The situation is slightly more complex for the infection assay. Our standard protocol leads to the robust and reproducible infection of about 50% of the amoebae with approximately 1–5 bacteria per cell at time zero (Trofimov et al., 2018; Mottet et al., 2021; Bagcchi, 2023). Therefore, “Dd-inf” refers to the monitoring of the mixed infected and un-infected Dd population, which evolves through the infection course. The situation is similar for the Mm populations. At time zero, the number of extracellular Mm is negligeable, but during infection the plate reader assay does not distinguish the intracellular from the extracellular Mm signal (the latter resulting from multiple release mechanisms: exocytosis, host-cell lysis and ejection). Therefore,” Mm-inf” refers to the monitoring of the intracellular and extracellular mixed Mm populations during infection.
Anti-infective assay with high-content microscopy
The procedure described here was optimized from Mottet et al. (2021). To monitor both the host and pathogen during the course of infection in a quantitative manner, Dd Ax2(ka) cells expressing mCherry knocked-in at the act5 locus (Paschke et al., 2019) were infected as described above with Mm WT stably expressing GFP (Msp:12-GFP). As described above, the day before the experiment, 107 amoebae were plated in a 10 cm Petri dish. On the day of the experiment, infection was performed, as described above, in HL5-C at MOI 25 by spinoculation. This was followed by eight washes in HL5-C to clear a maximum of non-internalized bacteria. The infected amoebae were detached mechanically and resuspended in HL5-C with 0.75 U/mL penicillin and 0.75 μg/mL of streptomycin (Gibco). Compounds were diluted in HL5-C to a 10x stock of the final assay concentration and pre-plated in 20 μL into the respective wells of a 96-well μ-plate (iBIDI 96-well μ-plate, 89,626). Subsequently, 7,500 cells per well were added in a volume of 180 μL. Every condition has been plated in at least three technical replicates and at least three biological replicates. The 96-well plate was sealed with a gas permeable seal (4titude, 4ti-0516/96) and centrifuged briefly.
Microscopy images were acquired at 25°C with a 40X plan Apo objective at selected time points over 48 h on the ImageXpress Micro C® automated microscope (Molecular Devices) in wide field mode. Focus was obtained automatically by autofocusing on the well bottom and plate bottom, at every well and every time point. In a grid of 4 × 5, 20 images per well, corresponding to a range of 500–6,000 cells, have been taken using the Texas-Red channel for Dd expressing mCherry and FITC channel for Mm expressing GFP in each experiment. Image analysis was performed using MetaXpress Custom Module editor software. Objects were effectively segmented by size and intensity. More precisely, amoebae were segmented using a “find blob module” using the Texas-Red channel. To segment the bacteria, the FITC image was first subjected to a light deconvolution with a top hat filter and then segmented using a “find blob module.” Intracellular bacteria and infected amoebae were determined by associating the mask of the amoebae and the bacteria. Several parameters such as intracellular and extracellular bacterial area sum, amoebae area sum, averaged and integrated fluorescence intensity and feature counts of bacteria and amoeba were extracted. The growth curves for the area sum of uninfected amoebae, infected amoebae and intracellular bacteria of independent biological replicates were averaged and plotted using GraphPad Prism (Version 8.0.1).
Results
Benchmarking the Dd-Mm high-throughput system
The high-throughput Dictyostelium discoideum - Mycobacterium marinum (Dd-Mm) infection model was benchmarked by monitoring dose–response curves (DRCs) for seven antibiotics used in TB chemotherapy (Figure 1). First, we used an anti-infective assay to measure the intracellular growth of a bioluminescent Mm strain (Mm-inf) inside Dd cells expressing mCherry, allowing to also measure in parallel the growth of the host amoebae (Dd-inf). Second, we used an antibiotic assay measuring the growth of Mm in broth (Mm-broth) (see definitions in Materials and methods). In both assays, the luminescence serves as a proxy for Mm mass and metabolic activity. The bacteria or the infected amoebae population were plated in 384-well plates, treated with descending concentrations of each antibiotic and monitored for 72 h using a plate reader. The average robust Z’-factor of the respective benchmarking experiments was 0.73 for Mm-inf and 0.61 for Mm-broth. The antibiotics rifampicin, isoniazid, ethambutol, bedaquiline, ethionamide and rifabutin exhibited sterilizing activity in both assays at their respective maximal tested concentration (Figures 1A–I). In addition, we observed a slight pro-infective effect of isoniazid and pyrazinamide between 0.4 and 3.3 μM (Figures 1E,J), but the latter showed otherwise no activity against Mm (Figure 1J). Moreover, rifampicin and ethambutol showed dramatically different activities in infection and in broth with MIC of 10 and 1.1 μM, respectively for rifampicin (Figure 1D), and MIC of 30 and 3.3 μM, respectively for ethambutol (Figure 1F). Rifabutin and ethionamide also exhibited moderate differences of activity on Mm-inf and Mm-broth. Both rifampicin and rifabutin affected amoebae growth slightly at the highest tested concentrations, but, as illustrated for rifampicin in Figure 1C, did not prohibit Dd from reaching a plateau in its growth phase.
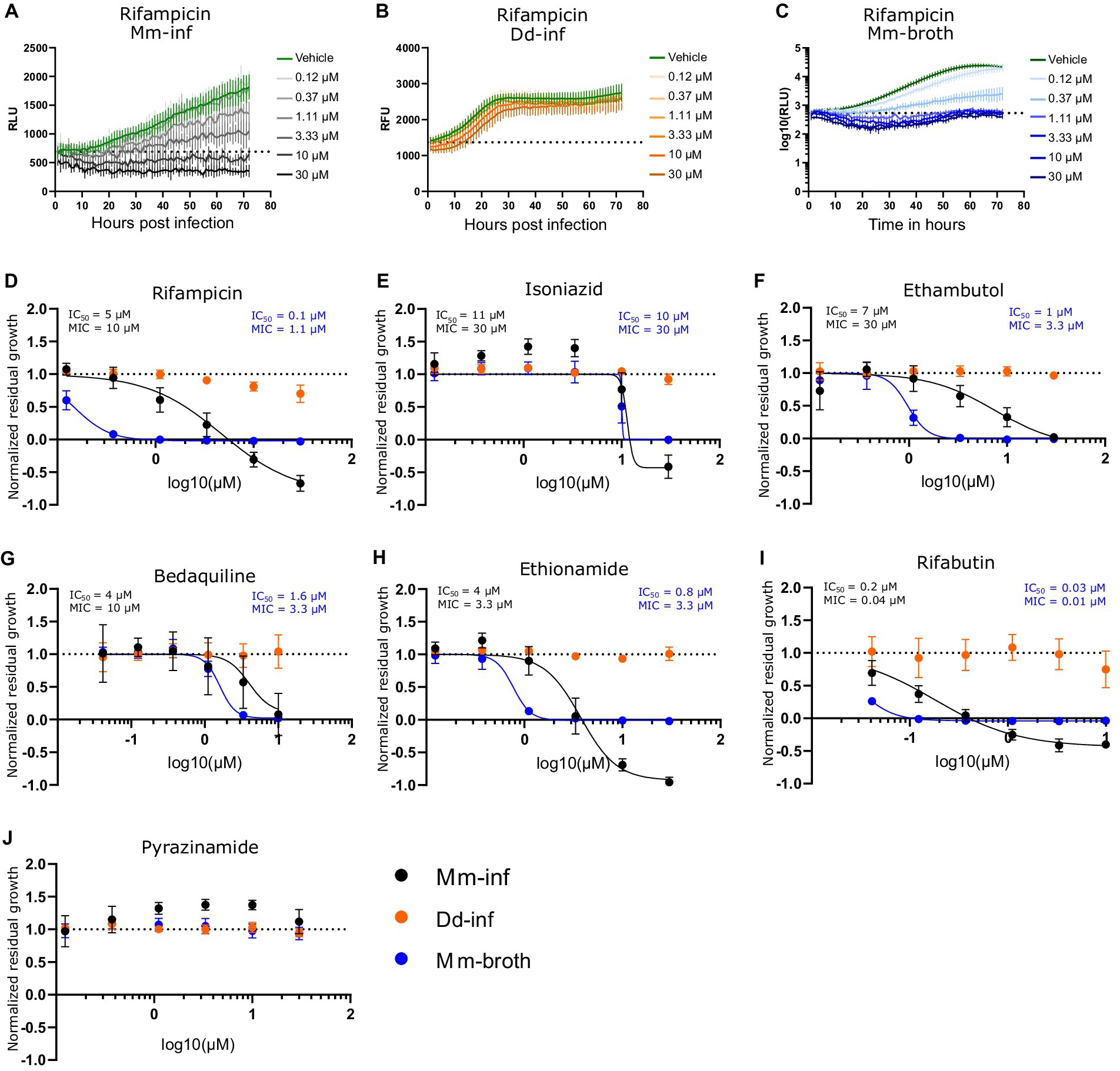
Figure 1. Dose response curves of benchmarked antibiotics A selection of antibiotics used for treatment of a TB infection were benchmarked in the Dd-Mm high-throughput system, in infection and in broth. In panels (A–C), 72 h growth curves of Mm in infection (Mm-inf), Dd in infection (Dd-inf) and Mm in broth (Mm-broth), respectively, at different concentrations of rifampicin are shown. The dashed line represents the median over the first measurements of all wells of the respective experiment. On the y-axis are random luminescence units (RLU) or random fluorescence units (RFU). The former were log10 transformed in (C). The corresponding dose–response curve of Rifampicin is shown in (D). The dose–response curves depicted in panel (E) isoniazid, (F) (ethambutol), (G) (bedaquiline), (H) (ethionamide), (I) (rifabutin) and (J) (pyrazinamide) were created from growth curves in analogous manner. Black points show Mm-inf, orange points Dd-inf and blue points Mm-broth. Log10 of the test concentration in μM is shown on the x-axis. A dashed line at y = 1 represents the normalized residual growth of the vehicle control. Depicted are means of at least three biological replicates and the respective standard deviations. Black and blue text inlays show the MIC and IC50 of the anti-infective or the antibacterial assay, respectively.
Primary screening of the stilbene monomer and dimer library in a high-throughput and dual readout growth inhibition assay
Next, we selected a library of compounds obtained through chemoenzymatic dimerization allowing structure activity relationship (SAR) investigations, while being relatively diverse. The compound structures can be classified into five scaffolds: trans-δ-viniferin, pallidol, leachianol, restrytisol and acyclic dimer (or labruscol). Each scaffold is derived from a different coupling reaction, as described in our previous article (Huber et al., 2022a). Restrytisol, leachianol and acyclic dimer scaffolds are further classified according to their relative stereochemistry. Two parameters explain the diversity of compounds obtained in each scaffold: First, the starting materials: resveratrol, pterostilbene, or a mixture of the two, which lead to hydroxylated and O-methylated derivatives, and second, the cosolvent. The leachianol and acyclic dimer scaffolds are the only ones capable of incorporating a solvent molecule during their synthesis by a solvolysis reaction, i.e., a nucleophilic attack of the alcohol on an sp2 carbon of an intermediate. This explains the large number of members in these two series. In addition to these 50 stilbene dimers, four stilbene monomers, which were used in generating dimers or derivatives of the initial library were included in the library (resveratrol, pterostilbene, pinostilbene and isorhapontigenin, Figure 2A).
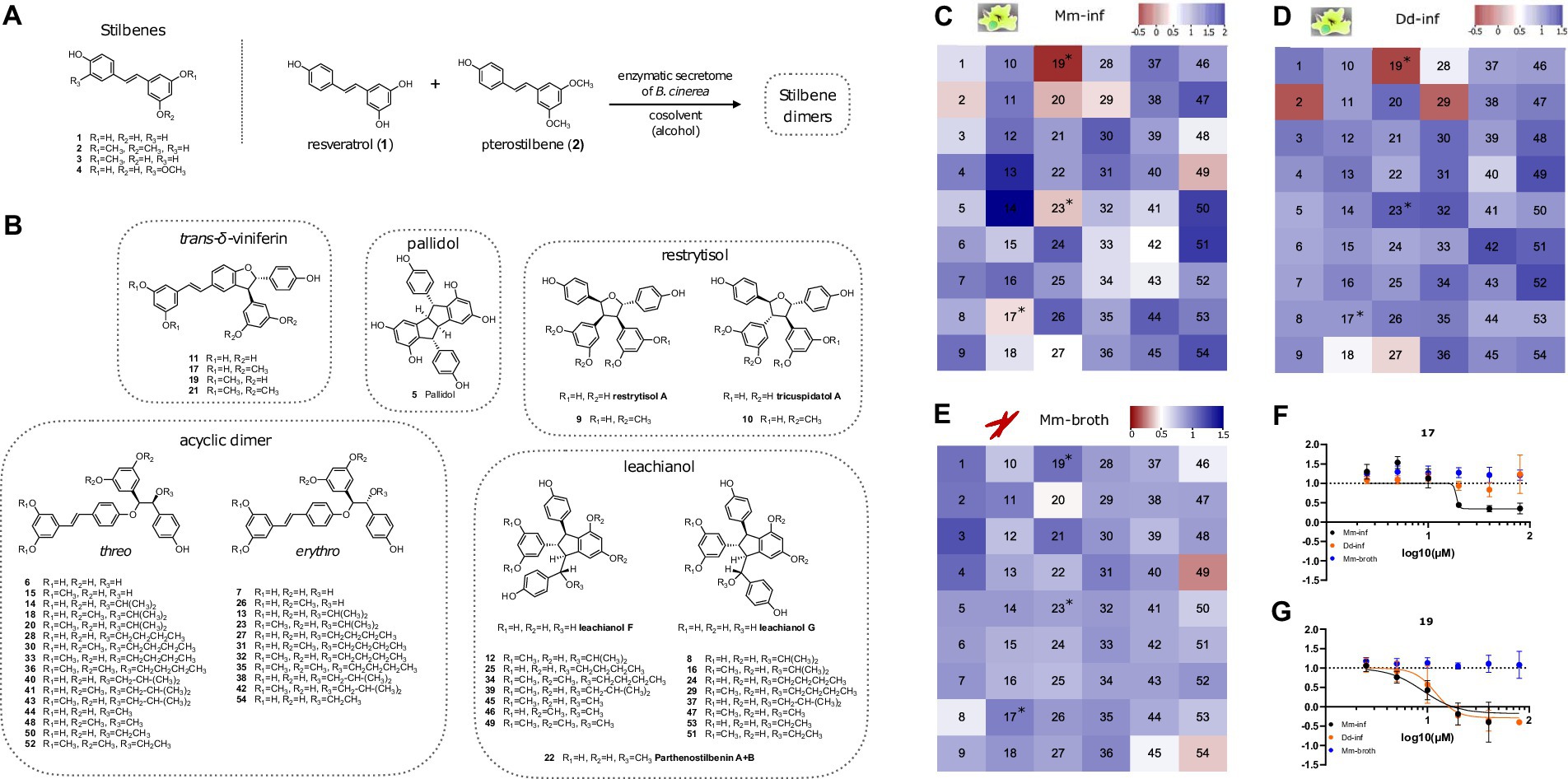
Figure 2. Overview of compound structures in the library and results of the screening. (A) Stilbene monomers (1–4) and biotransformation reaction with the enzymatic secretome of B. cinerea. (B) Structures of the stilbene dimers obtained by biotransformation of resveratrol and pterostilbene (5–54). Heatmaps in (C–E) color code normalized residual growth of Mm-inf (C), Dd-inf (D) and Mm-broth (E) under treatment with compounds at 20 μM each. Values below the cut-off of 0.5 are displayed in shades of red, while values above the cut-off are displayed in shades of blue. Numbers in the heatmap correspond to the compounds described in panels (A,B). Compounds meeting the cut-off in Mm in infection, but not in broth (i.e., Mm-inf ≤ 0.5 and Mm-broth ≥0.5) after a validation in a dose–response curve are marked with an asterisk in all three heatmaps. (F,G) Depict these dose–response curves of 17 and 19, respectively, of which both are analogs of the same scaffold, the trans-δ-viniferins. The corresponding dose–response curve for 23 can be found in Supplementary Figure S1. The log10 of the concentration in μM is shown on the x-axis; a dashed line at y = 1 represents the normalized residual growth of the vehicle control. Black points show Mm-inf, orange points Dd-inf and blue points Mm-broth. Shown are means of at least three biological replicates and the respective standard deviations. Note that in (G), for visual clarity the highest dosage (80 μM, normalized residual growth of –2.2) was omitted for Mm-inf.
Screening directly for anti-infective activity is more stringent than screening first for antibiotic activity on Mm in broth, since the latter compounds standardly suffer from a high attrition rate due to poor pharmacokinetics. We aim at identifying anti-infective compounds, which are defined by reduced intracellular Mm growth (Mm-inf). We focus on anti-infective compounds that are ineffective on Mm in broth (Mm-broth), so-called strict anti-infective compounds, since this implies a potentially novel mode of action specific to mycobacterial infection biology. While this is the main objective, we also take into account the effect of a compound on amoeba growth (Dd-inf) and aim at identifying compounds with a high selectivity against the pathogen. For all three aspects, the hit threshold of growth reduction by at least half was applied. We subjected the library to the anti-infective and antibacterial high-throughput assays, analogously to the procedure applied in Figure 1. The hit cut-off for any readout was set at 50% of residual growth. This allowed a hit classification for each compound in the library and for each readout: Mm-inf, Dd-inf and Mm-broth. The average robust Z’-factors for this screen were 0.71 for Mm-inf and 0.74 for Mm-broth.
One of the four monomers 1–4, pterostilbene (2) showed anti-infective activity (0.28 of normalized residual growth in Mm-inf), but was inactive on Mm in broth (Supplementary Figure S1B). However, this compound exhibited a strong Dd growth inhibition (–0.16 of normalized residual growth of Dd-inf), which is a possible indirect cause for its activity on Mm. Pterostilbene was tested in a DRC, revealing that activity on Dd-inf occurred already at lower concentrations than activity on Mm during infection (IC50 estimate of 11.1 and 19.8 μM respectively, Supplementary Figure S1B and Supplementary Table S1), weakening a possible causal link between activity on Dd and Mm during infection. The other monomers were also tested in a DRC, confirming the lack of activity of resveratrol (1, Supplementary Figure S1A) and isorhapontigenin (4, Supplementary Figure S1D) even at high concentrations, while pinostilbene (3, Supplementary Figure S1C) was active on Mm-inf, at high concentrations (>20 μM).
Several stilbene dimers were hits on Mm-inf, in particular 17 (with a normalized residual growth of 0.34), 19 (–0.32), 20 (0.20), 23 (0.23), 27 (0.47), 29 (0.35), 42 (0.47), and 49 (0.19) (Figure 2C and Supplementary Table S1). In broth, three of these dimers had an antibacterial activity with normalized residual growth below 0.5: 20 (0.47), 49 (0.27) and 54 (0.38) (Figure 2E), whereas 17, 19, 23, 27, 29 and 42 were shown to be anti-infectives without antibiotic activity. Among these six compounds, 19 (normalized residual growth of –0.22), 27 (0.32) and 29 (–0.10) were also inhibitors of Dd growth (Figure 2D and Supplementary Table S1). The other three dimers, 17 (normalized residual growth of 0.91), 23 (1.1) and 42 (1.2) were inactive on Dd (Figure 2D and Supplementary Table S1).
All six anti-infective dimers (17, 19, 23, 27, 29, and 42) were subsequently validated in the anti-infective assay in a DRC. All of them exhibited a dose-dependent activity, but only 17, 19, and 23 passed again the 0.5 cut-off for Mm-inf at 20 μM (Figures 2F,G and Supplementary Figure S1E). A high anti-infective activity was confirmed for 19 (IC50 = 9 μM, Supplementary Table S1), but was seen as potentially correlated to a high activity on Dd growth (IC50 = 12.1 μM, see Supplementary Table S1). For 17, a lower maximal effect on Mm-inf was observed (normalized residual growth of 0.3 for 17 compared to –0.2 for 19), but also a much lower activity on Dd growth (IC50 = 18.1 μM for Mm-inf, an estimate for Dd-inf was not possible due to the shape of the curve, Figure 2F and Supplementary Table S1) indicating a better selectivity of 17 toward the bacteria compared to 19. Both compounds did not show any growth inhibition of Mm-broth. Regarding compound 23, activity at 20 μM was confirmed by a DRC with an IC50 of 10.4 μM and no activity on Dd growth (Supplementary Figure S1E).
Surprisingly, few compounds (13, 14, 50, 51, and 54) had a pro-infective activity and enhanced Mm growth during infection with a normalized residual growth value greater than 1.5 (Figure 2C and Supplementary Table S1). This feature was exclusively identified during infection and not in broth. In the scope of this study, we focused on conditions that limit Mm growth, therefore, these compounds were not further explored.
Both 17 and 19 belong to the trans-δ-viniferin scaffold, while 23 is an acyclic dimer. Besides 17 and 19, which bear two O-methyl groups in a di-O-methylated ring, 11 and 21 also belong to the trans-δ-viniferin scaffold (Figure 2B), but with either no (11) or four (21) O-methyl groups. Both 11 and 21 showed no activity at 20 μM on Mm-inf with normalized residual growth of 1.1 and 1.4, respectively, or in broth with values of 1. On the acyclic dimers side, compounds 13 and 14 are both closely related to 23 but have no O-methyl groups and are inactive in infection (1.7, 2.0 respectively) and in broth (0.85, 0.90 respectively) (Supplementary Table S1).
In summary, the primary screen revealed 20 and 49 to be active both against extracellular and intracellular bacteria, whereas 2, 17, 19, and 23 were validated anti-infectives without antibiotic properties, partly with activity against Dd (2 and 19), and 54 was exclusively active against Mm-broth. Pterostilbene (2) is a monomer, while 17 and 19 both belong to the trans-δ-viniferin scaffold and 23 is an acyclic dimer. Both, trans-δ-viniferin and the acyclic dimer families contain very similar but inactive molecules that differ in the number and position of O-methyl groups. Based on these different activity patterns of the trans-δ-viniferins we subsequently focused on exploring this set of analogs.
High-content microscopy characterization of two hit compounds
After identifying and validating the trans-δ-viniferin derivatives 17 and 19 as anti-infectives without antibiotic activity, they were further validated using time-lapse high-throughput and high-content microscopy. While the plate reader assay provided aggregated data of a partially infected cell population, High-content microscopy allowed us to focus exclusively on infected cells in the population. Both uninfected Dd and Dd infected with Mm were treated with 20, 10 or 2.5 μM of 17 or 19 and the infection was monitored by collecting microscopy images at five time points for up to 48 h. The top concentration of 20 μM was chosen to be consistent with the primary screen whereas the dilutions were chosen to observe an effect similar to the vehicle control at the lowest dose. Mm and Dd were both segmented (Figures 3A,B) and the area of Dd (Figures 3C,D), infected Dd (i.e., Dd overlapping with a bacterium, Figures 3E,F) and the area of intracellular bacteria, (i.e., Mm overlapping with Dd, Figures 3G,H) were quantified. For all quantified parameters, a dose-dependency was observed for 17 and 19. The growth of uninfected Dd was more affected by 19 than by 17 (Figures 3C,D), while the growth of infected Dd (Figures 3E,F) and intracellular Mm (Figures 3G,H) showed the same pattern. Overall, the growth of intracellular Mm and infected Dd was almost completely inhibited by 19, also the growth of uninfected Dd was significantly reduced. For 17, this effect was weaker and, in addition, the growth restriction of uninfected Dd was less pronounced than the effect on intracellular Mm and infected Dd. In summary, validation of the activity of 17 and 19 using high-content microscopy confirmed that 17 was more selective for the intracellular pathogen over the host, although the overall activity was lower than that of 19.
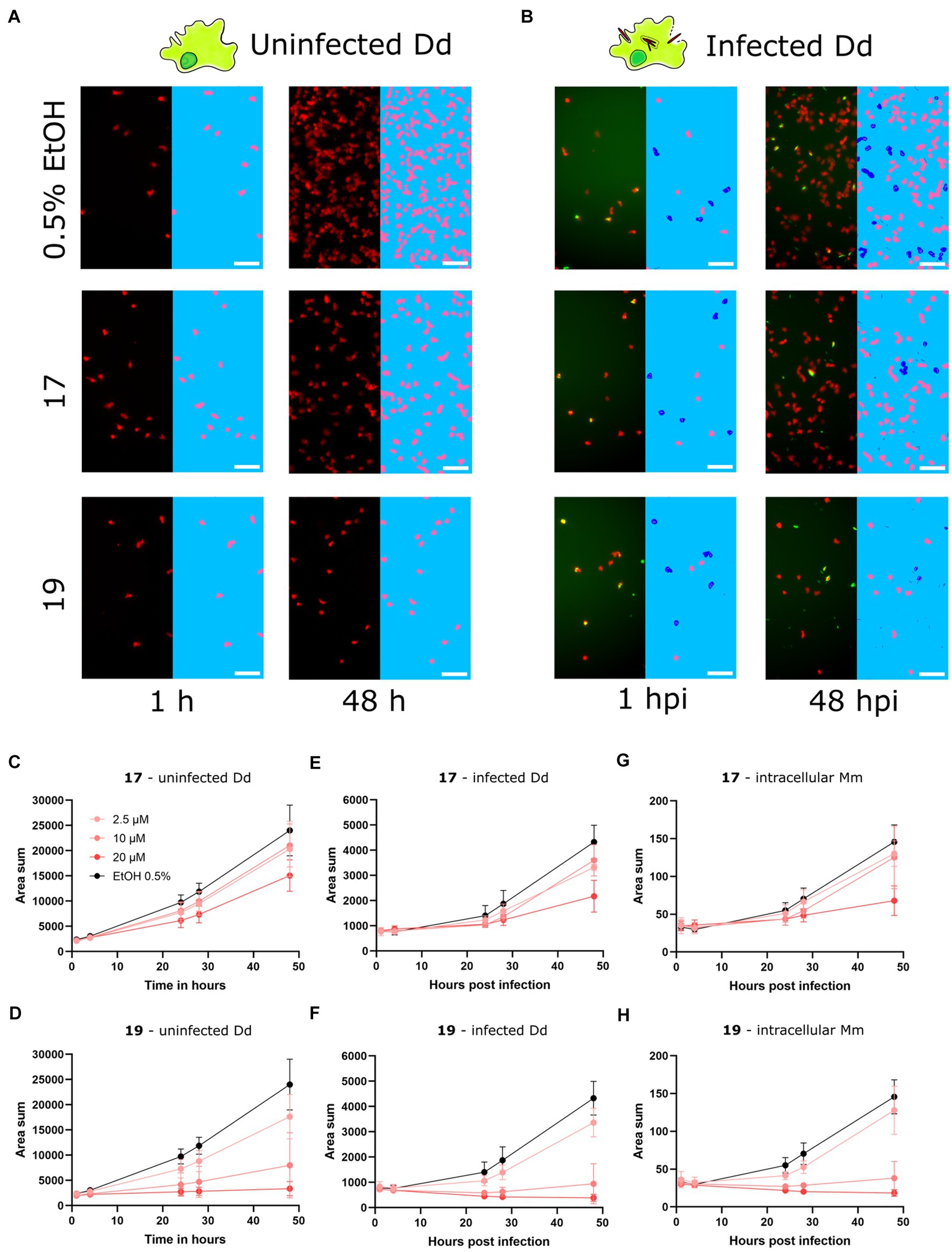
Figure 3. Characterization of primary hits with high-content microscopy Panels (A,B) show high-content microscopy panels illustrating the effects in infected and uninfected amoebae under treatment with 17 and 19 at 20 μM or 0.5% ethanol as the vehicle control at 1 and 48 h post infection (hpi). The left parts of the panels show the microscopy image, while the right parts of the panels show the corresponding segmentation. Uninfected Dd are shown in pink, infected Dd are shown in dark blue, intracellular bacteria are shown in white and extracellular bacteria are shown in black. Quantifications over the full time course and at concentrations of 2.5, 10, and 20 μM of 17 and 19 are depicted in (C–H). (C,D) Show the segmented area of uninfected Dd over time. (E,F) Show the segmented area of Dd determined to be infected by Dd-Mm area overlap, over time. (G,H) Show the segmented area of Mm determined to be intracellular by Dd-Mm area overlap, over time. Data points represent the average of three independent biological replicates, error bars are SDs.
After validating 17 and 19 as anti-infective compounds against Mm, we wondered if an interaction between 17 and established antibiotics could be determined. We tested this compound for synergy with rifampicin and isoniazid in a checkerboard assay and additionally tested for synergy between rifampicin and isoniazid. In brief, we did not find striking synergy but rather the pro-infective effect of 17 at low dosages (see Figure 2F) dominated the antibiotic effect of both drugs (between 1.25 and 10 μM of 17, see Supplementary Figures S3C,E).
Evaluation of trans-δ-viniferin derivatives
After validating the activity of 17 and 19 by high-content microscopy, their structure–activity relationship was further investigated, as 17 and 19 showed different activity and selectivity profiles despite having very similar structures. First, the impact of their absolute configuration was investigated. Indeed, the stilbene dimers (5–54) are all obtained as racemic mixtures by phenoxy radical coupling. Since enantiomers often show different activities, the previously separated enantiomers of the four trans-δ-viniferin derivatives (Supplementary Figure S2A, 11, 17, 19, 21) (Huber et al., 2022b) were evaluated for their anti-Mm activity during infection. The results, shown in Supplementary Figures S2B–D, indicated similar activity for both enantiomers of 17 and 19. Both enantiomers of 21 remained inactive, while a slight difference was observed between the enantiomers of 11. The latter result has not been further investigated due to the low level of activity observed.
Next, a series of derivatives of trans-δ-viniferin was generated and tested. These derivatives were obtained by light isomerization of the double bond (compounds 55–58, Figure 4A), by O-methylation of the phenolic moieties (compounds 59–64, Figure 4B) and by oxidation to generate benzofuran derivatives (compounds 66–69, Figure 4C). A methoxylated trans-δ-viniferin derivative (65) was also obtained by radical coupling of isorhapontigenin (4, Figure 4D). Cis isomerization and oxidation were performed to change the general shape of the molecule, the former reorienting a diphenol ring, and the latter making the structure planar. On the other hand, O-methylation allowed to study in detail the influence of the number and position of O-methyl groups, whose importance has been highlighted in the primary screen by the difference in activity between compounds 11 (no O-methyl group, inactive), 17 and 19 (two O-methyl groups, active), and 21 (four O-methyl groups, inactive).
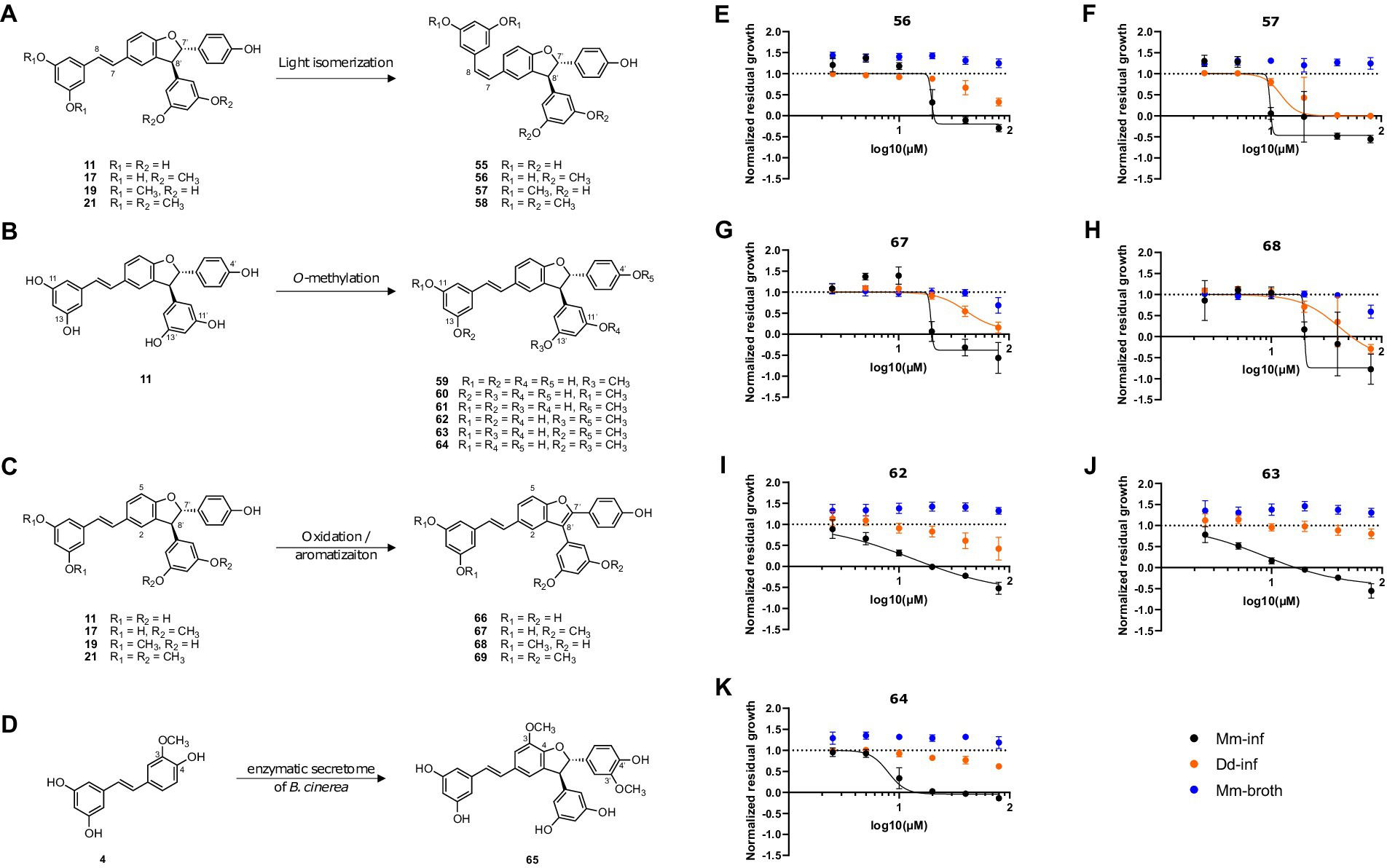
Figure 4. Synthesis of trans-δ-viniferin derivatives and corresponding structure activity relationship panel (A) shows light-isomerization leading to cis isomers 55–58. (B) Shows O-methylation of 11 leading to mono-O-methylated derivatives 59–61 and di-O-methylated derivatives 62–64. (C) Shows oxidation resulting in planar structures with a benzofuran moiety, 66–69. (D) Shows a radical coupling reaction of isorhapontigenin (4) leading to a dimer with two added methoxy groups (65) compared to trans-δ-viniferin (11). Panels (E–K) show dose–response curves of normalized residual growth of the aforementioned derivatives. (E,F) Show cis isomers 56 and 57, (G,H) benzofuran derivatives of 17 and 19, 67 and 68, respectively. (I–K) Show di-O-methylated derivatives 62, 63, and 64. Black points show Mm-inf, orange points Dd-inf and blue points Mm-broth; Log10 of the concentration in μM is shown on the x-axis. A dashed line at y = 1 represents the normalized residual growth of the vehicle control. Depicted are means of at least three biological replicates and the respective SDs.
Among the derivatives, the two chemical reactions that change the global shape of the molecules were first investigated. In the case of a specific protein target, such a change is expected to strongly influence the activity. The cis isomers (Figure 4A) showed a very close activity to the parent compounds [Mm-inf IC50 = 18.9 μM for 17, 19.7 μM for 56 (cis isomer); 9.0 μM for 19, 9.8 μM for 57 (cis isomer)] (Figure 4E, F). Similarly, the cis isomers 55 and 58 of the inactive trans-δ-viniferin 11 and 21, respectively, were also inactive at 20 μM (Supplementary Table S1). For the benzofuran derivatives, the change in activity was again weak for 17 (19.6 μM for 67, Figure 4G and Supplementary Table S1), but a decrease in activity was observed for 19 (20.1 μM for 68, Supplementary Table S1 and Figure 4H). The benzofuran derivative of 21 (69) remained inactive (Supplementary Figure S1G and Supplementary Table S1), while the corresponding derivative of 11 (66) showed nonspecific activity at higher dose than 17 and 19 (Mm-inf IC50 estimates of 29.7 μM, Dd-inf IC50 estimates of 26.2 μM; normalized residual growth of Mm-inf at 20 μM of 0.55) (Supplementary Figure S1F). A direct comparison with the DRC of 11 was not made because 11 was considered inactive at 20 μM in the primary screen (normalized residual growth of Mm-inf at 20 μM of 1.08). The complete IC50 data are given in Supplementary Table S1.
The data from the primary screen already showed that the number and position of O-methyl groups was important for the activity and selectivity of the compounds. Indeed, 17 and 19 which have both two O-methyl groups, showed different activity and selectivity in our assay. This could be due to the distinct positions of these groups. The impact of O-methyl groups was further supported by the decreased anti-infective activities of 11 (no O-methyl group) and 21 (four O-methyl groups). The importance of the O-methyl groups was therefore systematically investigated.
First, mono-O-methylated derivatives (59–61) were found to have weaker activities (less than 50% growth reduction of Mm-inf at 20 μM) and were therefore not investigated further (Supplementary Table S1).On the other hand, the di-O-methylated derivatives 62–64 showed an improved activity on Mm-inf (Figures 4I–K) compared to 17 and 19. Compound 62, showed a stronger maximal effect than 17 (–0.6 and 0.4, respectively), and its IC50 was slightly improved compared to 17 (13.4 and 18.9 μM, respectively, Figure 4I). Also 63 and 64 showed an improved maximal effect (−0.4 for 63 and 0 for 64), and the IC50 improved by about a factor of two, compared to 17 (8.6 and 8.0 μM for 63 and 64, respectively, Supplementary Table S1). Moreover, 62, 63, and 64 showed a reduced activity on the host during infection compared to 19, while maintaining a similar effect on Mm-inf (lowest measured value of Dd-inf 0.42, 0.8 and 0.62, respectively; −0.40 for 19). Notably, none of the modifications induced a gain of activity against Mm in broth, confirming the anti-infective property of the scaffold. On the other hand, compound 65, corresponding to a dimethoxylated derivative of 11 (keeping the free phenol groups) did not reduce Mm growth during infection, but rather showed pro-infective activity at 20 μM and was therefore disregarded (Supplementary Figure S1).
After identifying 62, 63, and 64 as trans-δ-viniferin derivatives with superior potency and selectivity over the initial screening hits 17 and 19, the activity profiles of the two most selective compounds 63 and 64 were validated using high-content microscopy (Figure 5). Similar to Figure 3, uninfected and infected Dd were treated with 20, 10, and 2.5 μM of 63 or 64 and cells were monitored for 48 h. Uninfected Dd, infected Dd and intracellular Mm were segmented (Figures 5A,B) treated with compounds and/or the vehicle. Quantification of uninfected Dd confirmed only a slight growth inhibition at 20 μM for 63 and 64 (Figures 5C,D), whereas the infection was well controlled by treatment with each of the compounds (Figures 5E–H).
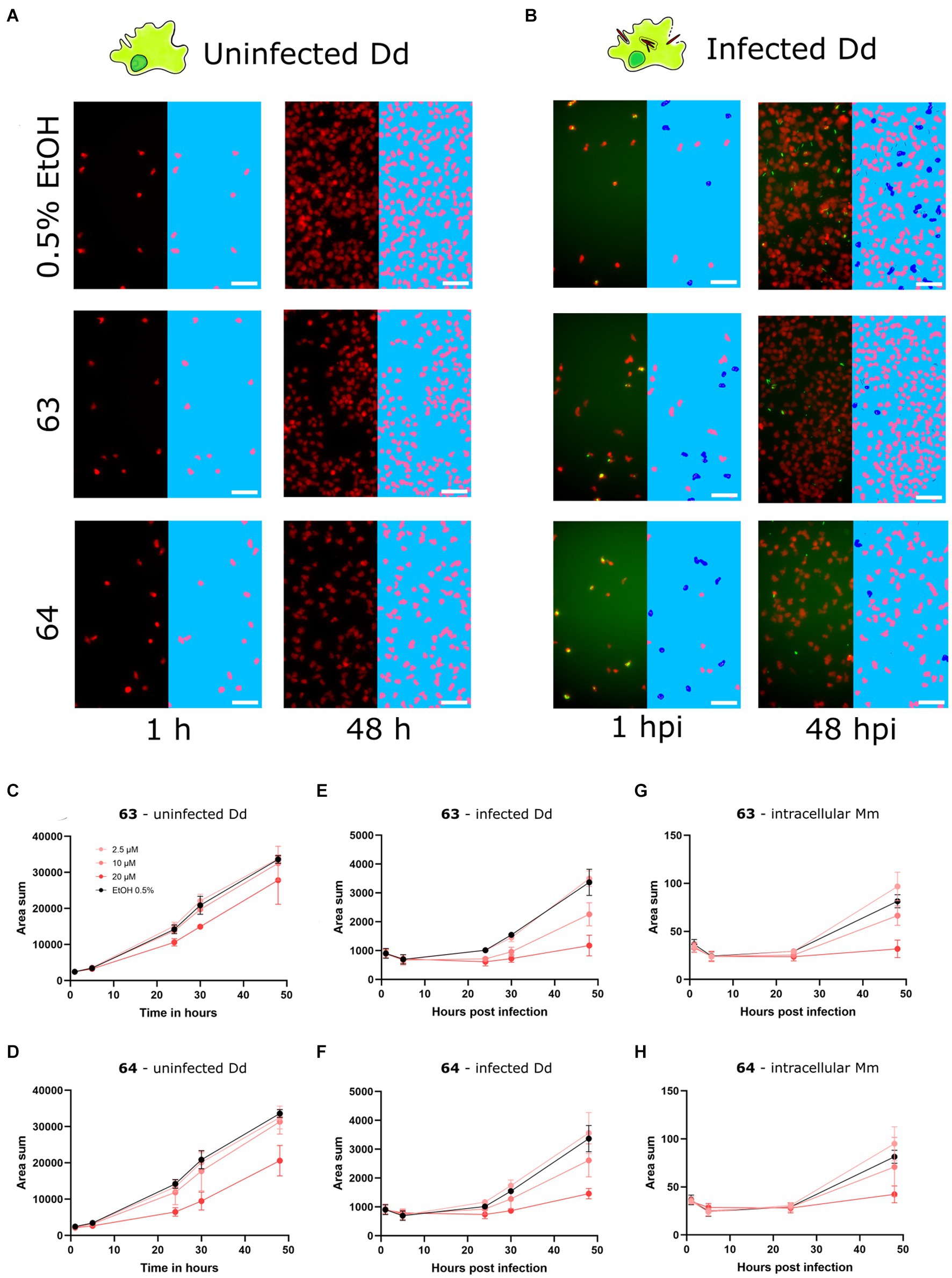
Figure 5. Characterization of improved derivatives with high-content microscopy Panels (A,B) show panels from high-content microscopy, illustrating effects in infected and uninfected amoebae under treatment with 63 and 64 at 20 μM or 0.5% ethanol as the vehicle control at 1 and 48 hpi. The left part of the panels shows the microscopy image, while the right part of the panels shows the segmented cells. Uninfected Dd are shown in pink, infected Dd are shown in dark blue, intracellular bacteria are shown in white and extracellular bacteria are shown in black. Quantifications over the full time-course at concentrations of 5, 10 and 20 μM of 63 and 64 are depicted in (C–H). (C,D) Show the segmented area of uninfected Dd over time. (E,F) Show the segmented area of Dd determined to be infected by Dd-Mm area overlap, over time. (G,H) Show the segmented area of Mm determined to be intracellular by Dd-Mm area overlap, over time. Data points represent the average of three independent biological replicates, error bars are SDs.
Overall, the confirmation of a reduced effect of 63 and 64 on the growth of Dd compared to 19, while having a similar potency as 19, and higher than 17, against Mm in infection render these two derivatives promising hits for further investigation. Interestingly, these four compounds are all trans-δ-viniferin isomers with two O-methyl groups, the only difference being their positions: both methoxy groups on a single ring for 17 and 19, or on two different rings for 63 and 64. The di-O-methylated ring connected to the conjugated double bond system in 19 seemed to contribute to activity against Dd, whereas the presence of at least one mono-O-methylated ring was beneficial both for maximum effect and selectivity. On the other hand, the other derivatives (cis isomers and benzofuran derivatives) did not allow to draw clear conclusions about the structure–activity relationship. The cis isomerization did not affect the activity of any of the parent compounds (11, 17, 19, and 21), while the oxidation to benzofuran resulted in either no or only a small change (11, 17, 21) or a decrease in activity (19).
In addition to the aforementioned modifications, we also halogenated compounds 11, 17, 19, and 21 with chlorine, bromine and iodine at different positions (Supplementary Table S1 and Supplementary Figure S4) and subsequently performed preliminary experiments of these derivatives in 96 well plates in infection (n = 3, N = 2) and in broth (n = 3, N = 1). The experiment in broth confirmed no gain of antibiotic activity of any analog (Supplementary Table S1). The experiments in infection also showed no gain of activity but revealed several inactive analogs. Although no completely consistent pattern emerged, it appeared that halogenation at the diphenol rings had the most impact on activity. In particular, double halogenation, e. g. in the case of the inactive analogs of 17, 77 (di-brominated in positions 10 and 14) and 80 (di-chlorinated in positions 10 and 14) (Supplementary Table S1), decreases the predicted pka of the hydroxyl groups from approximately 9 to almost 7, rendering them partly deprotonated in the cell. The two inactive analogs of 19, 83, and 89 also show at least one halogenation at the diphenol ring, whereas halogenation on the di-O-methylated ring, such as in 85 and 86 did not decrease activity (Supplementary Table S1).
Discussion
In this study, we first benchmarked the high-throughput Dd-Mm infection system and subsequently deployed it in a phenotypic screen on a small library of natural product derivatives. We found trans-δ-viniferins to be an anti-infective scaffold, with activity specific to Mm in infection. In particular, compounds 17 and 19 stood out, which we subsequently explored by halogenation and further SAR on stereochemistry, planarity and O-methylation. Although we did not find striking and consistent patterns in this SAR, we confirmed anti-infective activity and higher potency for three O-methylated derivatives, 62, 63, and 64.
During benchmarking the Dd-Mm system, we observed no activity of pyrazinamide both in infection and in broth (Figure 1J). This is in accordance with Mm being known to be naturally resistant to pyrazinamide (Aubry et al., 2000; Finkelstein and Oren, 2011). Resistance to isoniazid has also been reported (Aubry et al., 2000; Lorian, 2005), and in both assays isoniazid had the highest observed MIC (30 μM or 4.1 μg/mL). The National Committee for Clinical Laboratory Standards (NCCLS) defined breakpoints of antibacterial susceptibility for MIC90 from clinical isolates. Breakpoints for Mm were defined for rifabutin (2 μg/mL), ethambutol (4 μg/mL) (Woods and Clinical and Laboratory Standards Institute, 2011) and ethionamide (5 μg/mL) (Lorian, 2005). MICs observed in this study are all below the respective breakpoints, with the exception of ethambutol in infection (30 μM or 6.1 μg/mL). Indeed, ethambutol and rifampicin showed a large difference of activity between infection and growth in broth. Such a pattern was reported for rifampicin in Raw264.7 (Brodin et al., 2010) and THP-1 macrophages, due to export from the host cell via P-gp, but specifically not for ethambutol (Hartkoorn et al., 2007). Dd possesses an array of ABC transporters and is known for rapid drug export (Anjard et al., 2002; Miranda et al., 2013), emphasizing its stringency as a drug screening model in regard to pharmacokinetics.
After screening a library of stilbene dimers in the Dd-Mm system, we detected mostly anti-infective activities in contrast to classical antibiotic compounds, characterized by no activity in broth throughout the tested concentration range. Subsequently, we investigated the scaffold via targeted SAR and found derivatives with improved selectivity and maximal effect. This demonstrates not only the power of the Dd-Mm system as a 3R infection model, but also the value of exploring natural chemistry by novel approaches of derivatization. The observed anti-infective phenotype might be caused by accumulation inside Dd, which we speculate is unlikely, since Dd is known to rapidly export xenobiotics through its array of ABC transporters (Anjard et al., 2002; Miranda et al., 2013), as it seems to be the case for rifampicin. An anti-infective phenotype is thus likely to be caused by an antivirulence or host-directed defence booster mode of action (Gries, 2024).
Previous work has identified host-directed compounds in mammalian cell systems, and some have advanced to clinical studies (Huang et al., 2024). Several studies have shed light on the potential of tyrosine kinase inhibitors for TB treatment. For example, therapeutic doses of imatinib have proven effective in enhancing phagosome acidification and maturation, leading to a reduction in intracellular Mtb survival, both in vitro and in infected mice (Napier et al., 2011; Bruns et al., 2012). Another HDT approach involves metformin, a medication commonly prescribed for type 2 diabetes, which activates AMP-activated protein kinase. Metformin not only reduces the bacterial burden but also improves lung pathology in both mice and humans by enhancing autophagy and increasing the production of reactive oxygen species (Singhal et al., 2014). An emerging avenue focuses on inhibiting Mtb’s intracellular acquisition of fatty acids. Tetrahydrolipstatin (THL), which is recognized as an inhibitor of pancreatic lipase and marketed as a weight loss aid, shows promise as an anti-Mtb compound. Although the precise mechanism of its action is only partially understood, it is believed that THL depletes essential lipids from the intracellular environment, which are critical for Mtb’s survival (Parker et al., 2009).
Members of the group of compounds with a trans-δ-viniferin scaffold (11, 17, 19, 21.) exerted the strongest anti-infective effect in our assay. Members of this family are phytoalexins found in various plants, including grapevines (Vitis vinifera L.), and they act as defense compounds produced by plants in response to stress, infection, or other environmental challenges, including microbial pathogens (Langcake and Pryce, 1976). Phenolic compounds such as stilbenes and the trans-δ-viniferins presented here have many reported biological activities against bacteria (Langcake and Pryce, 1976; Vestergaard and Ingmer, 2019; Righi et al., 2020), but also diverse positive effects on mammalian cells (Rimando et al., 2005; Qureshi et al., 2012; Li et al., 2016; Ren et al., 2018).
In a recent study, investigating essential requirements for infection of Mm in Dd via Transposon sequencing (Tn-Seq), we highlighted genes and pathways that might act as targets for antivirulence compounds (Lefrançois et al., 2024). Members of lipid metabolism are prominent candidates, such as genes required for phthiocerol dimycocerosates (PDIMs) synthesis (Quigley et al., 2017) or the Mce operons, which are associated with lipid transport (Klepp et al., 2022).
The host membrane might also be altered in a way that makes it less sensitive to the bacteria membrane damaging machinery, for example by interfering with cholesterol in the MCV membrane. Cholesterol has been discussed to play a role in membrane damage, bacterial entry and also during infection of other intracellular, vacuolar pathogens (Gatfield and Pieters, 2000; Howe and Heinzen, 2006; De Jonge et al., 2007). In addition, in Dd a knock-out of vacuolins, the functional homologues of the mammalian flotillins which organize membrane microdomains rich in sterols, lead to intracellular growth attenuation of Mm (Bosmani et al., 2021).
Since we did not observe a striking shift in IC50 upon manipulating the shape of the trans-δ-viniferins 17 and 19 via planarisation (benzofuran derivatives) and cis-isomerisation of the double bond, we speculate that the mechanism of action is unlikely to follow the conventional pattern of selective binding to a catalytic pocket on a protein but may indeed rely on interference with cellular membranes, as discussed above.
However, trans-δ-viniferins have been reported as Wnt modulators, which in turn is hypothesized to be a target for pharmaceutical autophagy induction (Huber et al., 2022a). Interestingly, autophagy is also discussed as a central process of cell autonomous defence against Mtb and Mm (Aylan et al., 2023; Golovkine et al., 2023).
Since trans-δ-viniferins show activities in many biological systems, this class of compounds might be described as pan assay interference compounds (PAINs) (Baell, 2016). PAINs are compounds that in principle can be developed into drugs, but due to their promiscuous target interactions in a less systematic manner and thus at a higher cost. For example, catecholamine drugs contain catechols, which are considered a classical feature of a PAIN (Baell, 2016). Other structural features of PAINs, such as a flexible three-dimensional shape and reactive side groups are present in trans-δ-viniferins. Nevertheless, we observed striking specific activity on intracellular Mm for 17, 62, 63, and 64. In addition, current developments in pharmaceutical research try to embrace drugs with multiple targets, facilitated by advancing techniques to decipher complicated mechanisms of action, possibly allowing to progress putative PAINs more systematically, in the future.
A few structures were also found to enhance Mm growth during infection, but not in broth. During infection, 13, 14, 50, 51, 54 resulted in a normalized residual growth value greater than 1.5 (Figure 1C and Supplementary Table S1). Compounds that promote growth during infection, so-called pro-infectives may also be of fundamental interest. Although these compounds are generally ignored in most screenings and have limited value in drug discovery, they could be potential discovery tools to understand the mechanisms restricting intracellular mycobacterial survival (Subhash and Sundaramurthy, 2021). In fact, such compounds might act by disabling cell-autonomous defences and thus might enhance our understanding of innate immune mechanisms. As our main objective in this study is to focus on compounds that limit Mm growth, we did not further explore pro-infective compounds.
In summary, we emphasize the value of using nature inspired methods to derivatize simple scaffolds to bioactive compounds. Additionally, we demonstrated the effectiveness and potential of the Dd-Mm infection system for drug discovery, a platform fully compliant with the 3R model. This could be considered an added value to the drug discovery pipeline, offering an affordable and ethically sound system that complements the primary efforts in Mtb drug discovery. A remaining question is the validation of hit compounds in the Mm-Zebrafish model to confirm their efficacy in a more complex vertebrate model system.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
JN: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. RH: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft, Writing – review & editing. SV: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. DM: Data curation, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. LM: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. KG: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. EQ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. TS: Conceptualization, Data curation, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. NH: Conceptualization, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. We thank the Swiss National Science Foundation for providing financial support for this project, which aims to improve natural products chemical biodiversity for drug discovery by fungal secretome assisted biotransformation (grant 205321_182438/1 EFQ and KG). This work has also been supported by grants awarded to TS: A SystemsX grant (HostPathX), an SNF Sinergia grant (CRSII5_189921) and two SNF grants (310030_169386, 310030_188813). JN was supported by a Swiss Government Excellence PhD Scholarship.
Acknowledgments
We acknowledge Richard Fish from the Zebrafish Core Facility, Department of Genetic Medicine and Development, Faculty of Medicine, CMU, Geneva, Switzerland, for developing the zebrafish infection assay with us and performing initial experiments. We thank Emilie Michellod from the Mycology Group, Research Department Plant Protection, Agroscope, for the generation of the enzymatic secretome of Botrytis cinerea used on the biotransformation reactions. Furthermore, we thank Camille-Léa Dubuis for initial experiments with the library.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1439814/full#supplementary-material
Abbreviations
Dd, Dictyostelium discoideum; Mm, Mycobacterium marinum; Mtb, Mycobacterium tuberculosis; TB, Tuberculosis; SAR, structure–activity relationship; NP, natural product; WHO, World Health Organization; MDR, multi-drug-resistant; IC50, half maximal inhibitory concentration; DRC, dose–response curve; AUC, area under the curve; VC, vehicle control; ECD, electronic circular dichroism; NMR, nuclear magnetic resonance; HRMS, high-resolution mass spectrometry; HDT, host-directed therapy; MCV, mycobacteria-containing vacuole.
References
Alderwick, L. J., Harrison, J., Lloyd, G. S., and Birch, H. L. (2015). The mycobacterial Cell Wall--peptidoglycan and arabinogalactan. Cold Spring Harb. Perspect. Med. 5:a021113. doi: 10.1101/cshperspect.a021113
Alsayed, S. S. R., and Gunosewoyo, H. (2023). Tuberculosis: pathogenesis, current treatment regimens and new drug targets. Int. J. Mol. Sci. 24:5202. doi: 10.3390/ijms24065202
Andreu, N., Zelmer, A., Fletcher, T., Elkington, P. T., Ward, T. H., Ripoll, J., et al. (2010). Optimisation of Bioluminescent Reporters for Use with Mycobacteria. PLoS ONE 5:e10777. doi: 10.1371/journal.pone.0010777
Andries, K., Verhasselt, P., Guillemont, J., Göhlmann, H. W. H., Neefs, J.-M., Winkler, H., et al. (2005). A Diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307, 223–227. doi: 10.1126/science.1106753
Anjard, C., Loomis, W. F., and Dictyostelium Sequencing, C. (2002). Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot. Cell 1, 643–652. doi: 10.1128/ec.1.4.643-652.2002
Arafah, S., Kicka, S., Trofimov, V., Hagedorn, M., Andreu, N., Wiles, S., et al. (2013). Setting up and monitoring an infection of Dictyostelium discoideum with mycobacteria. Methods Mol. Biol. 983, 403–17. doi: 10.1007/978-1-62703-302-2_22
Aubry, A., Jarlier, V., Escolano, S., Truffot-Pernot, C., and Cambau, E. (2000). Antibiotic susceptibility pattern of Mycobacterium marinum. Antimicrob. Agents Chemother. 44, 3133–3136. doi: 10.1128/AAC.44.11.3133-3136.2000
Aylan, B., Bernard, E. M., Pellegrino, E., Botella, L., Fearns, A., Athanasiadi, N., et al. (2023). ATG7 and ATG14 restrict cytosolic and phagosomal Mycobacterium tuberculosis replication in human macrophages. Nat. Microbiol. 8, 803–818. doi: 10.1038/s41564-023-01335-9
Baell, J. B. (2016). Feeling Nature’s PAINS: natural products, natural product drugs, and Pan assay interference compounds (PAINS). J. Nat. Prod. 79, 616–628. doi: 10.1021/acs.jnatprod.5b00947
Bagcchi, S. (2023). WHO’s global tuberculosis report 2022. Lancet Microbe 4:e20. doi: 10.1016/S2666-5247(22)00359-7
Bosmani, C., Perret, A., Leuba, F., Guého, A., Hanna, N., and Soldati, T. (2021). Disruption of vacuolin microdomains in the host Dictyostelium discoideum increases resistance to Mycobacterium marinum-induced membrane damage and infection. Microbiology. doi: 10.1101/2021.11.16.468763
Brodin, P., Poquet, Y., Levillain, F., Peguillet, I., Larrouy-Maumus, G., Gilleron, M., et al. (2010). High content phenotypic cell-based visual screen identifies Mycobacterium tuberculosis Acyltrehalose-containing glycolipids involved in phagosome remodeling. PLoS Pathog. 6:e1001100. doi: 10.1371/journal.ppat.1001100
Bruns, H., Stegelmann, F., Fabri, M., Döhner, K., van Zandbergen, G., Wagner, M., et al. (2012). Abelson tyrosine kinase controls phagosomal acidification required for killing of Mycobacterium tuberculosis in human macrophages. J. Immunol. 189, 4069–4078. doi: 10.4049/jimmunol.1201538
Cardenal-Munoz, E., Arafah, S., Lopez-Jimenez, A. T., Kicka, S., Falaise, A., Bach, F., et al. (2017). Mycobacterium marinum antagonistically induces an autophagic response while repressing the autophagic flux in a TORC1-and ESX-1-dependent manner. PLoS Pathog. 13:e1006344. doi: 10.1371/journal.ppat.1006344
Cardenal-Munoz, E., Barisch, C., Lefrancois, L. H., Lopez-Jimenez, A. T., and Soldati, T. (2018). When Dicty met Myco, a (not so) romantic story about one Amoeba and its intracellular pathogen. Front. Cell. Infect. Microbiol. 7:529. doi: 10.3389/fcimb.2017.00529
Conradie, F., Diacon, A. H., Ngubane, N., Howell, P., Everitt, D., Crook, A. M., et al. (2020). Treatment of highly drug-resistant pulmonary tuberculosis. N. Engl. J. Med. 382, 893–902. doi: 10.1056/NEJMoa1901814
David, B. (2018). New regulations for accessing plant biodiversity samples, what is ABS? Phytochem. Rev. 17, 1211–1223. doi: 10.1007/s11101-018-9573-1
David, B., Wolfender, J.-L., and Dias, D. A. (2015). The pharmaceutical industry and natural products: historical status and new trends. Phytochem. Rev. 14, 299–315. doi: 10.1007/s11101-014-9367-z
De Jonge, M. I., Pehau-Arnaudet, G., Fretz, M. M., Romain, F., Bottai, D., Brodin, P., et al. (2007). ESAT-6 from Mycobacterium tuberculosis dissociates from its putative chaperone CFP-10 under acidic conditions and exhibits membrane-lysing activity. J. Bacteriol. 189, 6028–6034. doi: 10.1128/JB.00469-07
Dheda, K., and Migliori, G. B. (2012). The global rise of extensively drug-resistant tuberculosis: is the time to bring back sanatoria now overdue? Lancet 379, 773–775. doi: 10.1016/S0140-6736(11)61062-3
Diop, E. A., Queiroz, E. F., Kicka, S., Rudaz, S., Diop, T., Soldati, T., et al. (2018). Survey on medicinal plants traditionally used in Senegal for the treatment of tuberculosis (TB) and assessment of their antimycobacterial activity. J. Ethnopharmacol. 216, 71–78. doi: 10.1016/j.jep.2017.12.037
Diop, E. H. A., Queiroz, E. F., Marcourt, L., Kicka, S., Rudaz, S., Diop, T., et al. (2019). Antimycobacterial activity in a single-cell infection assay of ellagitannins from Combretum aculeatum and their bioavailable metabolites. J. Ethnopharmacol. 238:111832. doi: 10.1016/j.jep.2019.111832
Dunn, J. D., Bosmani, C., Barisch, C., Raykov, L., Lefrançois, L. H., Cardenal-Muñoz, E., et al. (2018). Eat prey, live: Dictyostelium discoideum as a model for cell-autonomous defenses. Front. Immunol. 8:1906. doi: 10.3389/fimmu.2017.01906
Finkelstein, R., and Oren, I. (2011). Soft tissue infections caused by marine bacterial pathogens: epidemiology, diagnosis, and management. Curr. Infect. Dis. Rep. 13, 470–477. doi: 10.1007/s11908-011-0199-3
Gatfield, J., and Pieters, J. (2000). Essential role for cholesterol in entry of mycobacteria into macrophages. Science 288, 1647–1651. doi: 10.1126/science.288.5471.1647
Gindro, K., Schnee, S., Righi, D., Marcourt, L., Nejad Ebrahimi, S., Codina, J. M., et al. (2017). Generation of antifungal stilbenes using the enzymatic Secretome of Botrytis cinerea. J. Nat. Prod. 80, 887–898. doi: 10.1021/acs.jnatprod.6b00760
Giraud-Gatineau, A., Coya, J. M., Maure, A., Biton, A., Thomson, M., Bernard, E. M., et al. (2020). The antibiotic bedaquiline activates host macrophage innate immune resistance to bacterial infection. eLife 9:e55692. doi: 10.7554/eLife.55692
Golovkine, G. R., Roberts, A. W., Morrison, H. M., Rivera-Lugo, R., McCall, R. M., Nilsson, H., et al. (2023). Autophagy restricts Mycobacterium tuberculosis during acute infection in mice. Nat. Microbiol. 8, 819–832. doi: 10.1038/s41564-023-01354-6
Greenwood, D. J., Dos Santos, M. S., Huang, S., Russell, M. R. G., Collinson, L. M., Mac Rae, J. I., et al. (2019). Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science 364, 1279–1282. doi: 10.1126/science.aat9689
Gries, R. (2024). Deciphering novel modes of action of Lead compounds with antibacterial or anti virulence activity against Mycobacterium tuberculosis. Universität Köln. doi: 10.1038/s42003-024-06620-9
Gries, R., Sala, C., and Rybniker, J. (2020). Host-directed therapies and anti-virulence compounds to address anti-microbial resistant tuberculosis infection. Appl. Sci. 10:2688. doi: 10.3390/app10082688
Guallar-Garrido, S., and Soldati, T. (2024). Exploring the relevance of the Dictyostelium discoideum-Mycobacterium marinum infection model for tuberculosis research. Dis. Model. Mech. 17:dmm050698. doi: 10.1242/dmm.050698
Hagedorn, M., and Soldati, T. (2007). Flotillin and RacH modulate the intracellular immunity of Dictyostelium to Mycobacterium marinum infection. Cell. Microbiol. 9, 2716–2733. doi: 10.1111/j.1462-5822.2007.00993.x
Hartkoorn, R. C., Chandler, B., Owen, A., Ward, S. A., Bertel Squire, S., Back, D. J., et al. (2007). Differential drug susceptibility of intracellular and extracellular tuberculosis, and the impact of P-glycoprotein. Tuberculosis 87, 248–255. doi: 10.1016/j.tube.2006.12.001
Howe, D., and Heinzen, R. A. (2006). Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell. Microbiol. 8, 496–507. doi: 10.1111/j.1462-5822.2005.00641.x
Huang, X., Lowrie, D. B., Fan, X.-Y., and Hu, Z. (2024). Natural products in anti-tuberculosis host-directed therapy. Biomed. Pharmacother. 171:116087. doi: 10.1016/j.biopha.2023.116087
Huber, R. (2023). Improving the chemical diversity of natural products for drug discovery through biotransformation by Fungi. Université de Genève. Available at: https://archive-ouverte.unige.ch/unige:173990 (Accessed January 24, 2024)
Huber, R., Koval, A., Marcourt, L., Héritier, M., Schnee, S., Michellod, E., et al. (2022a). Chemoenzymatic synthesis of original stilbene dimers possessing Wnt inhibition activity in triple-negative breast Cancer cells using the enzymatic Secretome of Botrytis cinerea Pers. Front. Chem. 10:881298. doi: 10.3389/fchem.2022.881298
Huber, R., Marcourt, L., Héritier, M., Luscher, A., Guebey, L., Schnee, S., et al. (2023). Generation of potent antibacterial compounds through enzymatic and chemical modifications of the trans-δ-viniferin scaffold. Sci. Rep. 13:15986. doi: 10.1038/s41598-023-43000-5
Huber, R., Marcourt, L., Quiros-Guerrero, L.-M., Luscher, A., Schnee, S., Michellod, E., et al. (2022b). Chiral separation of stilbene dimers generated by biotransformation for absolute configuration determination and antibacterial evaluation. Front. Chem. 10:912396. doi: 10.3389/fchem.2022.912396
Huszár, S., Chibale, K., and Singh, V. (2020). The quest for the holy grail: new antitubercular chemical entities, targets and strategies. Drug Discov. Today 25, 772–780. doi: 10.1016/j.drudis.2020.02.003
Kicka, S., Trofimov, V., Harrison, C., Ouertatani-Sakouhi, H., McKinney, J., Scapozza, L., et al. (2014). Establishment and validation of whole-cell based fluorescence assays to identify anti-mycobacterial compounds using the Acanthamoeba castellanii - Mycobacterium marinum host-pathogen system. PLoS One 9:e87834. doi: 10.1371/journal.pone.0087834
Kingston, D. G. I. (2011). Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 74, 496–511. doi: 10.1021/np100550t
Klepp, L. I., Sabio, Y., Garcia, J., and Bigi, F. (2022). Mycobacterial MCE proteins as transporters that control lipid homeostasis of the cell wall. Tuberculosis 132:102162. doi: 10.1016/j.tube.2021.102162
Koul, A., Dendouga, N., Vergauwen, K., Molenberghs, B., Vranckx, L., Willebrords, R., et al. (2007). Diarylquinolines target subunit c of mycobacterial ATP synthase. Nat. Chem. Biol. 3, 323–324. doi: 10.1038/nchembio884
Koul, A., Vranckx, L., Dendouga, N., Balemans, W., Van Den Wyngaert, I., Vergauwen, K., et al. (2008). Diarylquinolines are bactericidal for dormant mycobacteria as a result of disturbed ATP homeostasis. J. Biol. Chem. 283, 25273–25280. doi: 10.1074/jbc.M803899200
Langcake, P., and Pryce, R. J. (1976). The production of resveratrol by Vitis vinifera and other members of the Vitaceae as a response to infection or injury. Physiol. Plant Pathol. 9, 77–86. doi: 10.1016/0048-4059(76)90077-1
Lefrançois, L. H., Nitschke, J., Wu, H., Panis, G., Prados, J., Butler, R. E., et al. (2024). Temporal genome-wide fitness analysis of Mycobacterium marinum during infection reveals the genetic requirement for virulence and survival in amoebae and microglial cells. mSystems, e01326–23. doi: 10.1128/msystems.01326-23
Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56. doi: 10.1038/nrmicro1557
Li, C. L., Santhanam, B., Webb, A. N., Zupan, B., and Shaulsky, G. (2016). Gene discovery by chemical mutagenesis and whole-genome sequencing in Dictyostelium. Genome Res. 26, 1268–1276. doi: 10.1101/gr.205682.116
Lopez-Jimenez, A. T., Cardenal-Munoz, E., Leuba, F., Gerstenmaier, L., Barisch, C., Hagedorn, M., et al. (2018). The ESCRT and autophagy machineries cooperate to repair ESX-1-dependent damage at the Mycobacterium-containing vacuole but have opposite impact on containing the infection. PLoS Pathog. 14:e1007501. doi: 10.1371/journal.ppat.1007501
Lorian, V. (2005). Antibiotics in laboratory medicine. Philadelphia, USA: Lippincott Williams & Wilkins.
Lu, X., Williams, Z., Hards, K., Tang, J., Cheung, C.-Y., Aung, H. L., et al. (2019). Pyrazolo[1,5-a]pyridine inhibitor of the respiratory cytochrome bcc complex for the treatment of drug-resistant tuberculosis. ACS Infect. Dis. 5, 239–249. doi: 10.1021/acsinfecdis.8b00225
Mahajan, R. (2013). Bedaquiline: first FDA-approved tuberculosis drug in 40 years. Int. J. App. Basic Med. Res. 3, 1–2. doi: 10.4103/2229-516X.112228
Makarov, V., Manina, G., Mikusova, K., Möllmann, U., Ryabova, O., Saint-Joanis, B., et al. (2009). Benzothiazinones kill Mycobacterium tuberculosis by blocking arabinan synthesis. Science 324, 801–804. doi: 10.1126/science.1171583
Matsumoto, M., Hashizume, H., Tomishige, T., Kawasaki, M., Tsubouchi, H., Sasaki, H., et al. (2006). OPC-67683, a nitro-Dihydro-Imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med. 3:e466. doi: 10.1371/journal.pmed.0030466
Miranda, E. R., Zhuchenko, O., Toplak, M., Santhanam, B., Zupan, B., Kuspa, A., et al. (2013). ABC transporters in Dictyostelium discoideum development. PLoS One 8:e70040. doi: 10.1371/journal.pone.0070040
Mottet, M., Bosmani, C., Hanna, N., Nitschke, J., Lefrançois, L. H., and Soldati, T. (2021). Novel single-cell and high-throughput microscopy techniques to monitor Dictyostelium discoideum-Mycobacterium marinum infection dynamics. Methods Mol. Biol. 2314, 183–203. doi: 10.1007/978-1-0716-1460-0_7
Napier, R. J., Rafi, W., Cheruvu, M., Powell, K. R., Zaunbrecher, M. A., Bornmann, W., et al. (2011). Imatinib-sensitive tyrosine kinases regulate mycobacterial pathogenesis and represent therapeutic targets against tuberculosis. Cell Host Microbe 10, 475–485. doi: 10.1016/j.chom.2011.09.010
Newman, D. J., and Cragg, G. M. (2016). Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661. doi: 10.1021/acs.jnatprod.5b01055
Newman, D. J., and Cragg, G. M. (2020). Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 83, 770–803. doi: 10.1021/acs.jnatprod.9b01285
Parker, S. K., Barkley, R. M., Rino, J. G., and Vasil, M. L. (2009). Mycobacterium tuberculosis Rv3802c encodes a phospholipase/thioesterase and is inhibited by the antimycobacterial agent tetrahydrolipstatin. PLoS One 4:e4281. doi: 10.1371/journal.pone.0004281
Paschke, P., Knecht, D. A., Williams, T. D., Thomason, P. A., Insall, R. H., Chubb, J. R., et al. (2019). Genetic engineering of Dictyostelium discoideum cells based on selection and growth on Bacteria. J. Vis. Exp. e58981. doi: 10.3791/58981-v
Peraman, R., Meka, G., Chilamakuru, N. B., Kutagulla, V. K., Malla, S., Ashby, C. R., et al. (2021). Novel stilbene scaffolds efficiently target Mycobacterium tuberculosis nucleoid-associated protein, HU. New J. Chem. 45, 10683–10692. doi: 10.1039/D0NJ05947A
Quigley, J., Hughitt, V. K., Velikovsky, C. A., Mariuzza, R. A., El-Sayed, N. M., and Briken, V. (2017). The Cell Wall lipid PDIM contributes to Phagosomal escape and host cell exit of Mycobacterium tuberculosis. MBio 8. doi: 10.1128/mBio.00148-17
Qureshi, A. A., Guan, X., Reis, J. C., Papasian, C. J., Jabre, S., Morrison, D. C., et al. (2012). Inhibition of nitric oxide and inflammatory cytokines in LPS-stimulated murine macrophages by resveratrol, a potent proteasome inhibitor. Lipids Health Dis. 11:76. doi: 10.1186/1476-511X-11-76
Ramakrishnan, L. (2004). Using Mycobacterium marinum and its hosts to study tuberculosis. Curr. Sci. 86, 82–92.
Reddy, V. M., Einck, L., Andries, K., and Nacy, C. A. (2010). In vitro interactions between new antitubercular drug candidates SQ109 and TMC207. Antimicrob. Agents Chemother. 54, 2840–2846. doi: 10.1128/AAC.01601-09
Reinheimer, C., Büttner, D., Proschak, E., Bode, H. B., Kempf, V. A. J., and Wichelhaus, T. A. (2018). Anti-tubercular activity of a natural stilbene and its synthetic derivatives. GMS Infect. Diseases 6:Doc01. doi: 10.3205/ID000036
Ren, G., Rimando, A. M., and Mathews, S. T. (2018). AMPK activation by pterostilbene contributes to suppression of hepatic gluconeogenic gene expression and glucose production in H4IIE cells. Biochem. Biophys. Res. Commun. 498, 640–645. doi: 10.1016/j.bbrc.2018.03.035
Righi, D., Huber, R., Koval, A., Marcourt, L., Schnee, S., Le Floch, A., et al. (2020). Generation of stilbene antimicrobials against multiresistant strains of Staphylococcus aureus through biotransformation by the enzymatic Secretome of Botrytis cinerea. J. Nat. Prod. 83, 2347–2356. doi: 10.1021/acs.jnatprod.0c00071
Rimando, A. M., Nagmani, R., Feller, D. R., and Yokoyama, W. (2005). Pterostilbene, a new agonist for the peroxisome proliferator-activated receptor α-isoform, lowers plasma lipoproteins and cholesterol in Hypercholesterolemic hamsters. J. Agric. Food Chem. 53, 3403–3407. doi: 10.1021/jf0580364
Rossiter, S. E., Fletcher, M. H., and Wuest, W. M. (2017). Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 117, 12415–12474. doi: 10.1021/acs.chemrev.7b00283
Russell, W. M. S., and Burch, R. L. (1959). The principles of humane experimental technique. London: Methuen.
Rybniker, J., Vocat, A., Sala, C., Busso, P., Pojer, F., Benjak, A., et al. (2015). Lansoprazole is an antituberculous prodrug targeting cytochrome bc1. Nat. Commun. 6:7659. doi: 10.1038/ncomms8659
Singhal, A., Jie, L., Kumar, P., Hong, G. S., Leow, M. K.-S., Paleja, B., et al. (2014). Metformin as adjunct antituberculosis therapy. Sci. Transl. Med. 6:263ra159. doi: 10.1126/scitranslmed.3009885
Solomon, J. M., Leung, G. S., and Isberg, R. R. (2003). Intracellular replication of Mycobacterium marinum within Dictyostelium discoideum: efficient replication in the absence of host coronin. Infect. Immun. 71, 3578–3586. doi: 10.1128/iai.71.6.3578-3586.2003
Steinert, M., and Heuner, K. (2005). Dictyostelium as host model for pathogenesis. Cell. Microbiol. 7, 307–314. doi: 10.1111/j.1462-5822.2005.00493.x
Suarez, M., Valencia, J., Cadena, C., Maiti, R., Datta, C., Puerto, G., et al. (2017). Diarylethenes display in vitro anti-TB activity and are efficient hits targeting the Mycobacterium tuberculosis HU protein. Molecules 22:1245. doi: 10.3390/molecules22081245
Subhash, N., and Sundaramurthy, V. (2021). Advances in host-based screening for compounds with intracellular anti-mycobacterial activity. Cell. Microbiol. 23:e13337. doi: 10.1111/cmi.13337
Tobin, D. M., and Ramakrishnan, L. (2008). Comparative pathogenesis of Mycobacterium marinum and Mycobacterium tuberculosis. Cell. Microbiol. 10, 1027–1039. doi: 10.1111/j.1462-5822.2008.01133.x
Trofimov, V., Kicka, S., Mucaria, S., Hanna, N., Ramon-Olayo, F., Del Peral, L. V., et al. (2018). Antimycobacterial drug discovery using mycobacteria-infected amoebae identifies anti-infectives and new molecular targets. Sci. Rep. 8:3939. doi: 10.1038/s41598-018-22228-6
Vestergaard, M., and Ingmer, H. (2019). Antibacterial and antifungal properties of resveratrol. Int. J. Antimicrob. Agents 53, 716–723. doi: 10.1016/j.ijantimicag.2019.02.015
Wolfender, J.-L., Litaudon, M., Touboul, D., and Queiroz, E. F. (2019). Innovative omics-based approaches for prioritisation and targeted isolation of natural products – new strategies for drug discovery. Nat. Prod. Rep. 36, 855–868. doi: 10.1039/C9NP00004F
G. L. Woods , and Clinical and Laboratory Standards Institute (Eds.) (2011). Susceptibility testing of mycobacteria, nocardiae and other aerobic actinomycetes: Approved standard. 2nd Edn. Wayne, Pa: CLSI.
Keywords: anti-infectives, natural products, phenotypic screening, Mycobacterium marinum , Dictyostelium discoideum , stilbene derivatives
Citation: Nitschke J, Huber R, Vossio S, Moreau D, Marcourt L, Gindro K, Queiroz EF, Soldati T and Hanna N (2024) Discovery of anti-infective compounds against Mycobacterium marinum after biotransformation of simple natural stilbenes by a fungal secretome. Front. Microbiol. 15:1439814. doi: 10.3389/fmicb.2024.1439814
Edited by:
Maria Rosalia Pasca, University of Pavia, ItalyReviewed by:
Ben Gold, Weill Cornell Medicine, United StatesArshad Rizvi, Emory University, United States
Copyright © 2024 Nitschke, Huber, Vossio, Moreau, Marcourt, Gindro, Queiroz, Soldati and Hanna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thierry Soldati, VGhpZXJyeS5Tb2xkYXRpQHVuaWdlLmNo; Nabil Hanna, TmFiaWwuTmFubmFAdW5pZ2UuY2g=
 Jahn Nitschke
Jahn Nitschke Robin Huber
Robin Huber Stefania Vossio
Stefania Vossio Dimitri Moreau
Dimitri Moreau Laurence Marcourt
Laurence Marcourt Katia Gindro
Katia Gindro Emerson F. Queiroz
Emerson F. Queiroz Thierry Soldati
Thierry Soldati Nabil Hanna
Nabil Hanna