- 1Department of Plant Pathology, Faculty of Agriculture, Kasetsart University, Bangkok, Thailand
- 2Department of Plant Science, Faculty of Science, Mahidol University, Bangkok, Thailand
- 3Forest Pathology Research Laboratory, Department of Forest Sciences, Faculty of Agriculture and Forestry, University of Helsinki, Helsinki, Finland
Rigidoporus microporus, the causative agent of the white root rot disease of rubber trees, poses a significant threat to natural rubber production worldwide. Understanding the molecular mechanisms facilitating its pathogenicity would be crucial for developing effective disease management strategies. The pathogen secretes effector proteins, which play pivotal roles in modulating host immune responses and infection. In this study, in silico analyses identified 357 putative secreted effector proteins from the R. microporus genome. These were then integrated into previous RNA-seq data obtained in response to rubber tree latex exposure. Annotation of putative effectors suggested the abundance of proteins in several families associated with the virulence of R. microporus, especially hydrophobin proteins and glycoside hydrolase (GH) proteins. The contribution of secreted effectors to fungal pathogenicity was discussed, particularly in response to rubber tree latex exposure. Some unknown highly expressed effectors were predicted for the protein structures, revealing their similarity to aminopeptidase, ubiquitin ligase, spherulin, and thaumatin protein. This integrative study further elucidates the molecular mechanism of R. microporus pathogenesis and offers alternative targets for developing control strategies for managing white root rot disease in rubber plantations.
1 Introduction
The white root rot disease (WRD) of rubber tree (Hevea brasiliensis), one of the elite crops worldwide, leads to a serious problem especially in the first few years after planting because they can affect water and nutrient uptake, and hence reduce growth and yield of natural rubber (Mohammed et al., 2014). This disease has been suggested to rank high as a major rubber tree catastrophe in many rubber tree planting countries, particularly in Thailand, Malaysia, Indonesia, Sri Lanka, Nigeria, and Cote d’Ivoire, as well as in several other countries (According to CABI compendium).1 The disease is caused by the Basidiomycete fungal pathogen Rigidoporus microporus (Fr.) Overeem., which can persist on dead root debris for a long time before reaching the living roots and switching to the pathogenic phase (Wattanasilakorn et al., 2012). The symptoms of infected rubber trees manifest in the form of leaf discoloration of leaves (e.g., yellowish brown), early flowering, and fruiting off-season. However, the observation at the initial stage of disease based on these aboveground symptoms requires a trained mind as it was hardly discriminated from symptoms of drought or other abiotic stresses. The only obvious aboveground sign of WRD fungus is the production of the orange basidiocarp with a whitish margin at the lower stem trunk (Omorusi, 2012), which is, unfortunately, considered a severe and late event of the WRD, by which time the whole rubber tree root may already be damaged.
Insights into the molecular responses of rubber trees against R. microporus were reported recently by a number of authors. Once the rubber tree root has been infected and colonized by fungus, the systemic molecular responses in the rubber tree can be observed in the roots (Siddiqui et al., 2017), stem (Oghenekaro et al., 2016a; Janket et al., 2022), and leaves (Woraathasin et al., 2017; Longsaward et al., 2023). Several rubber tree defense-related proteins were noticed as the alleviated mechanisms for tolerating WRD disease. Defects in photosynthesis after being infected by R. microporus may have contributed to the leaf discoloration and growth retardation of rubber trees.
Understanding the mechanisms of how the R. microporus pathogen causes WRD would highly contribute to disease management. Identifying and understanding as many potential virulence factors of the fungal pathogen will help to facilitate the selection of suitable strategies and tools to combat them. Rubber tree roots colonized by R. microporus will exhibit observable fungal mycelia that grow firmly attached to the root surface. The woody root then turns creamer and softer due to the activity of cell wall-degrading enzymes released by R. microporus to promote its further invasion of plant tissue (Omorusi, 2012). Delignification, a process driven by key enzymes including lignin peroxidases, manganese peroxidases, and laccases, has been reported as one of the fungal virulence that causes root rot symptoms (Paleaz et al., 1995). From the analysis of de novo transcriptome, R. microporus expresses peroxidases, laccases, aldo-keto reductases, oxidases, and oxidoreductases for depolymerization of rubber lignin during the saprotrophic growth into the rubber wood (Oghenekaro et al., 2016b). Among these, the laccase was proven to have enzyme activity correlated to the pathogenicity of R. microporus, the higher virulence isolates showed higher laccase activity (Suwandi, 2007).
Recent breakthrough technologies allow the exploration of the genome sequence from several species, including the WRD fungus R. microporus. According to Oghenekaro et al. (2020), the genome analysis in this fungal pathogen revealed a larger number of cellulose-degrading enzymes, than the hemicellulose-and pectin-degrading enzymes. Several protein candidates related to fungal virulence were annotated. Interestingly, there is evidence showing that the rubber tree latex is unable to suppress fungal growth but, conversely, promotes the growth of R. microporus with the help of supportive latex degradation genes. This highlights the aggressiveness of R. microporus against the host’s basal defenses. Although several small secreted proteins (SSPs) were proposed as effector proteins due to their upregulation when co-culturing with other competitive fungal species (Oghenekaro et al., 2020), none of the studies have systemically identified the putative effector proteins from the R. microporus fungus.
Effectors were recently defined as molecules that influence the interaction between organisms in a way that benefits the one who releases or uses them (Todd et al., 2022). This expands the classical definition in plant pathology, where effectors are traditionally referred to as molecules from pathogens that manipulate the host’s cellular functions and structures to support their infection and trigger plant defenses (Kamoun, 2006). According to the Zig-Zag model (Jones and Dangl, 2006), pathogens employ effectors to bypass the basal pattern-triggered immunity (PTI) expressed by the host. Microbial pathogens have developed various effectors as sole strategy to interfere with several molecular functions of their hosts including (i) proteins (Alfano and Collmer, 2004; Stergiopoulos and de Wit, 2009), (ii) small RNAs (Weiberg et al., 2013; Wang et al., 2017), and (iii) secondary metabolites (Friesen et al., 2008; Stergiopoulos et al., 2013; Collemare et al., 2019) especially polyketide derivatives (Hassani et al., 2022). Effectors are crucial to the virulence of pathogens and largely contribute to the pathogenesis in suppressing or avoiding the host immune systems. Fungal effectors can be divided based on their pathogenic functions into biotrophic effectors, which facilitate colonization in the host via suppressing plant immunity (Uhse and Djamei, 2018), and necrotrophic effectors that highly disrupt host cells such as phytotoxins and cell wall-degrading enzymes (Kim et al., 2016; Shao et al., 2021; Samperna et al., 2022). The hemibiotrophic fungi on the other hand could use either biotrophic or necrotrophic effectors as adaptive modes for their lifestyles (Kabbage et al., 2015). Therefore, the information on effectors utilized by particular pathogens and the understanding of their function could benefit the effective disease management of crop plants or forest trees.
Identification of all effectors in an organism, a so-called effectoromics analysis, provides a broader understanding of fungal virulence and lifestyle (Todd et al., 2022). However, the identification of the effectorome is highly challenging from the determination of how effectors should be defined, so it influences the elucidation and identification methods. Regarding effector proteins, the taxonomic distribution of effector proteins is limited and they contain very low or no sequence similarity to other protein families or other organisms. The lack of homology among effectors leads to a fewer number of effector protein families. To date, the effectors can also be defined as the (i) canonical effector proteins that are usually cysteine-rich proteins of less than 300 amino acid residues with the presence of signal peptide, and their genes are highly expressed during the interaction with the host or other competitive microbes, and (ii) the non-canonical effector proteins that implemented the well-known conserved domains and motifs from other known pathogenic effector regardless of the protein size and protein secretion (Todd et al., 2022).
To explore the effectoromic in the WRD fungus, the secreted proteins were in silico screened from the total proteins in the R. microporus genome (Oghenekaro et al., 2020). Thereafter, the secreted protein candidates were predicted for the effector characteristics in this study, through the Effector P prediction tool and the search for effector-conserved motifs. Several protein families identified as potential effector proteins were compared to the other relative fungal species and also discussed with the available transcriptome dataset of genes expressed in response to latex, a rubber tree basal defense. In addition, some of the key R. microporus virulent effectors with low sequence homology were predicted for their structures. The integrative information of R. microporus-secreted effectors would accelerate the applications for effective disease management in future, especially the chemical or biofungicide targeting of these virulence factors of WRD fungus. Moreover, the information would benefit in prioritizing effector candidates from R. microporus for further characterization in the lab.
2 Materials and methods
2.1 Effector identification
The fungal proteome, acquired from the JGI genome portal of Rigidoporus microporus ED310 v 1.0 (Oghenekaro et al., 2020) (accessed on November 20, 2023),2 was used for the in silico identification of effector candidates using steps of machine learning-based tools available online. To identify potential fungal effectors, the 10,917 proteins of R. microporus were subjected to secreted protein prediction by Signal P 6.0 (Teufel et al., 2022) (accessed on November 21, 2023).3 Then, the secreted protein candidates were screened for effector potential by (i) Effector P 3.0 (accessed on November 21, 2023)4 (Sperschneider and Dodds, 2022) and (ii) a search for effector-conserved motifs using Find Individual Motif Occurrence (FIMO) v. 5.5.5 (accessed on January 31, 2024)5 (Grant et al., 2011). In this study, the predicted apoplastic effectors and cytoplasmic effectors from Effector P as well as the secreted proteins with effector-conserved motifs were considered as effectors of the R. microporus fungus.
2.2 Domain analysis of secreted proteins
The secreted protein candidates were retrieved for their related information from the MycoCosm database,6 i.e., the amino acid sequences, protein ID, transcript ID, domain-associated protein family (PFam), and GO terms. All candidates were re-predicted for the protein domain via HMMscan (accessed on 15 January 2024)7 (Potter et al., 2018) in order to resolve those candidates without annotation information from MycoCosm database.
2.3 Analysis of secreted protein expression from the transcriptome data
The expression of secreted protein candidates identified here was retrieved from the transcriptomic analysis of R. microporus in response to natural latex treatment. The transcriptome data in NCBI BioProject 4977868 were previously analyzed for differentially expressed genes (DEGs) by comparing three replicates of pure R. microporus culture supplemented with rubber wood to three replicates of pure R. microporus culture supplemented with rubber wood or rubber latex (Oghenekaro et al., 2020). The protein candidates in this study were screened from the list of DEGs and their expression data were used to discuss the potential molecular mechanisms of the fungi during interaction with the rubber tree.
2.4 Prediction of uncharacterized effectors
The structural-based prediction of highly expressed effectors that were previously considered unknown proteins was carried out through the D-I-TASSER server.9 The server predicted the secondary-and 3D structures, ligand binding sites, enzyme activity, and GO terms from the template protein in the PDB database. The protein structure was visualized by UCSF Chimera v. 1.15.
3 Results
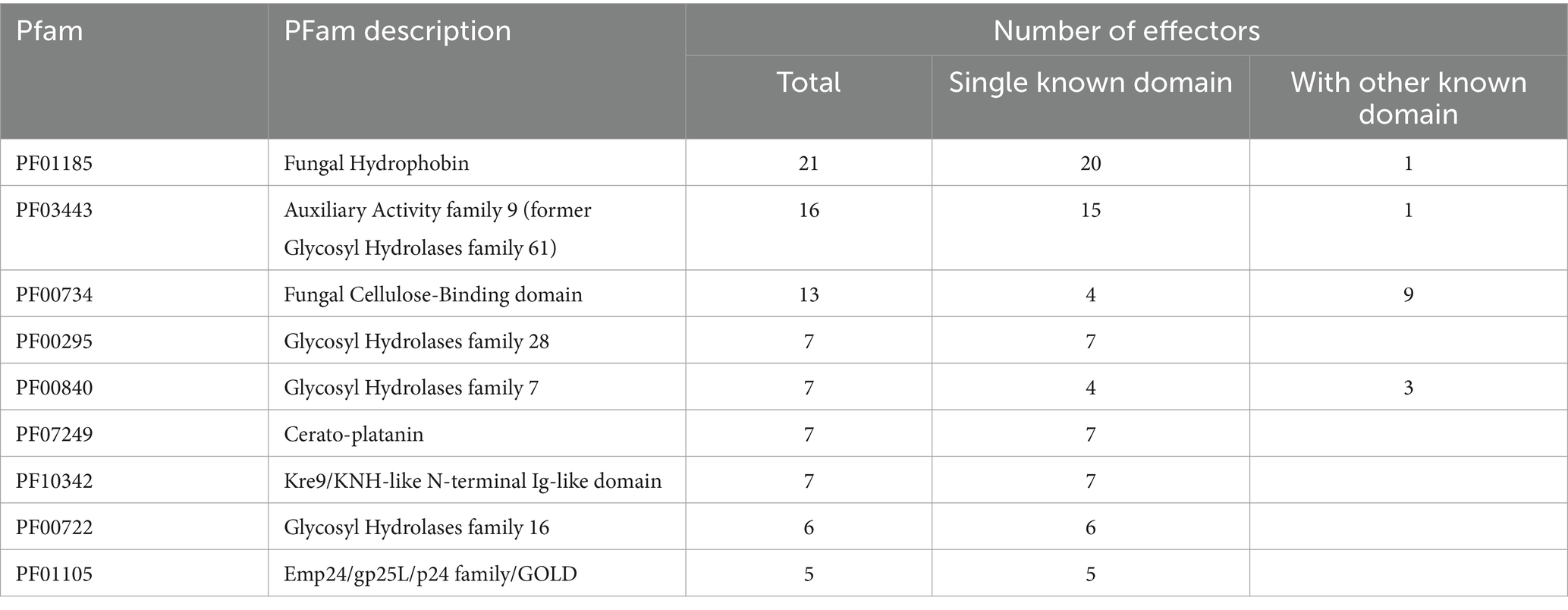
In this study, the candidate proteins potentially acting as effectors of the pathogenic fungus R. microporus were analyzed using machine learning-based tools available online. A total of 10,917 unique proteins reported in the R. microporus genome were assessed for the secretory feature by Signal P v. 6.0, revealing 720 proteins with high-potential signal peptides (Supplementary Table S1). These secreted proteins were then scrutinized for their effector characteristics by Effector P 3.0 (Supplementary Table S1). Among the 720 secreted proteins, Effector P predicted 183 as apoplastic effectors, 53 as cytoplasmic effectors, 10 proteins as both apoplastic and cytoplasmic effector features, while the remaining 474 candidates were categorized as non-effector-secreted proteins (Supplementary Table S2; Figure 1A).
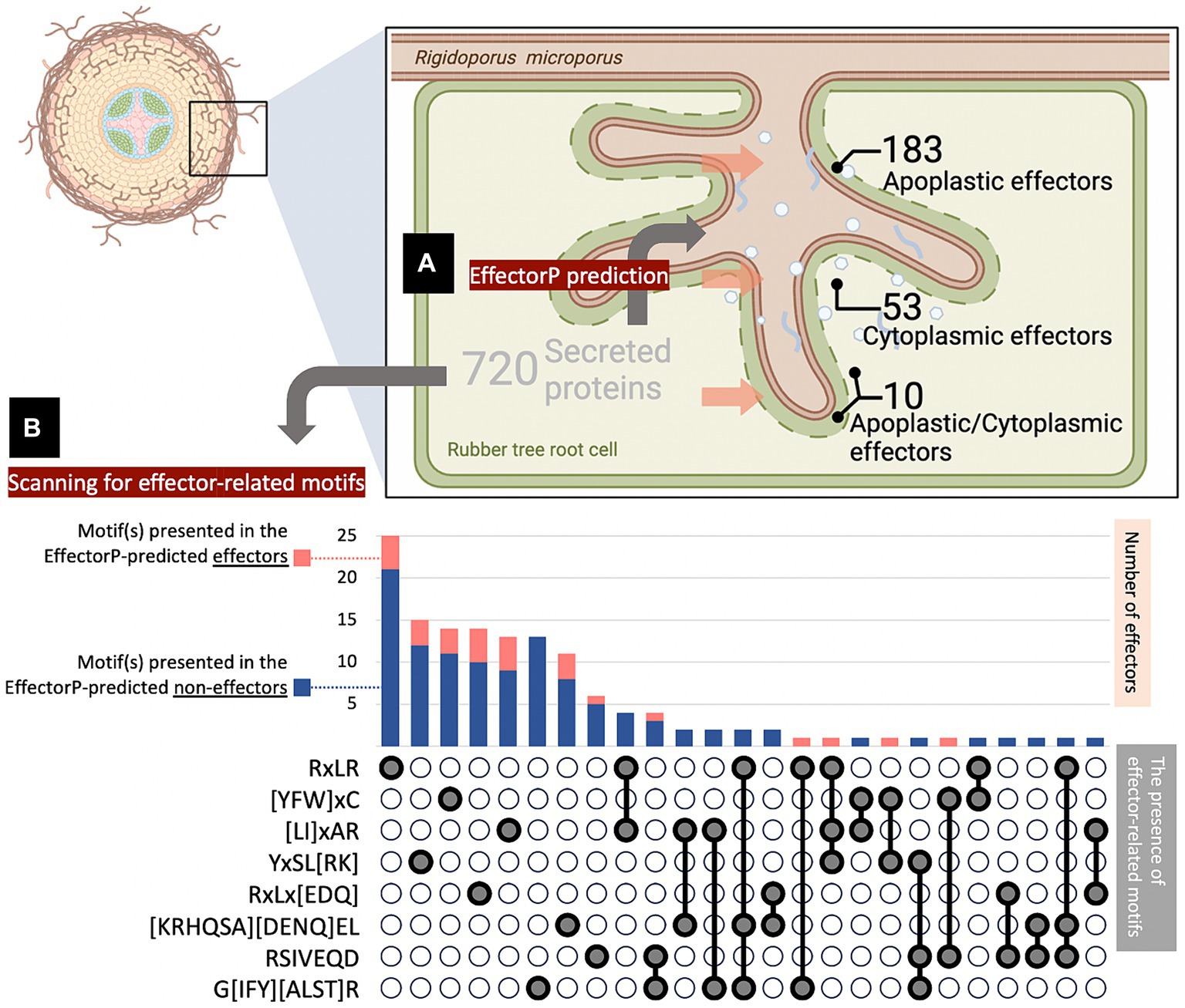
Figure 1. Number of effector candidates identified in silico from the Rigidoporus microporus fungus. A total of 720 proteins were predicted as secreted proteins, of which 246 effectors were predicted by the Effector P tool, and another 111 effectors were predicted by FIMO analysis of effector-conserved motifs. (A) Effector P predicted 183 apoplastic effectors, 53 cytoplasmic effectors, 10 apoplastic/cytoplasmic effectors, and 474 non-effectors. The penetration of fungus into the root was designed by BioRender. (B) The number of effectors with conserved motifs was investigated by FIMO analysis. The presence of one-, two-, or three-motifs on individual effectors was indicated in gray below the stacked chart, which shows the number of effector proteins predicted as effectors by Effector P (pink) and those overlooked by Effector P (navy). Each of the selected motifs was searched across the 720 secreted protein sequences identified in this study, using FIMO analysis with an E-value cut-off of 0.0001 (Supplementary Table S4).
To expand the effector identification, well-known motifs conserved in fungal effectors were focused and used as criteria to determine the effector-likeliness of secreted proteins from R. microporus. The presence of effector-related motifs in the candidate sequences was addressed using the FIMO analysis tool. Consequently, the analysis identified additional effectors containing these selected motifs (Figure 1B; Supplementary Table S4). This included 27 effector proteins predicted by Effector P and another 111 additional proteins that were overlooked by Effector P prediction (Supplementary Table S3). As a result, a total of 357 effector proteins have been identified as the effectorome of R. microporus in this study comprising 246 effectors predicted by Effector P and 111 effectors containing conserved fungal effector motifs (Figure 1).
The information associated with the secreted proteins and effectors identified in this study was obtained from the fungal genomic resource of R. microporus. The information includes annotation of Gene Ontology (GO) terms, Classification of Protein Families by Pfam and InterPro entry, Kyoto Encyclopedia of Genes and Genomes (KEGG), Carbohydrate-Active Enzymes (CAZy), Conserved Protein Domain Family (KOG), Transporter Classification Database (TCDB), and Peptidase database (MEROPS) (Supplementary Tables S2, S3). Among the 720 secreted proteins, 463 have been annotated with at least one of the aforementioned categories (64.31%), while 257 remain unannotated (35.69%), of which 141 were identified as effectors (Figure 2A). Consequently, further analysis was conducted on the protein re-annotation of these effectors and other secreted proteins using hmmscan for protein domain identification (Supplementary Tables S2, S3). This process improved the annotation, matching 8 previously unannotated effectors and 1 other unannotated secreted protein to Pfam, accounting for 1% of the secretome (Figure 2A).
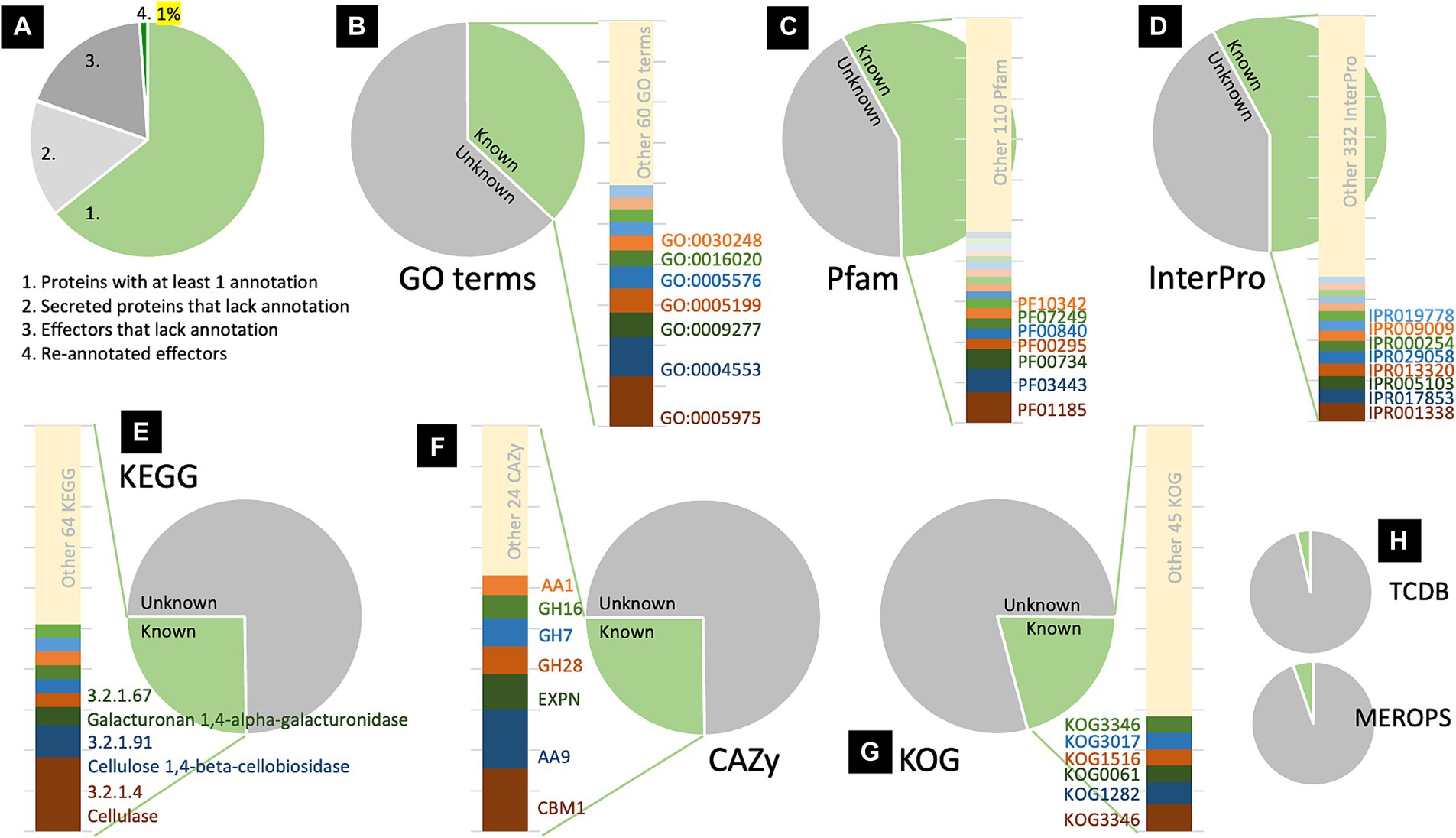
Figure 2. Analysis of protein information revealed the proportion of annotated and unannotated effector proteins of Rigidoporus microporus identified in this study. Approximately 64% of secreted proteins were annotated to at least one annotation based on the dataset of the R. microporus genome in the MycoCosm database (A). The effector proteins are analyzed for their GO terms (B), Pfam accessions (C), InterPro entries (D), KEGG (E), CAZy (F), KOG (G), as well as TCDB and MEROPS (H). The proportion of each known annotation was plotted as a stacked chart.
To investigate the annotation profiles of R. microporus effectorome, the information of 357 effectors was thoroughly analyzed across each of the 8 annotation tools (Figures 2B–H). The datasets from Pfam and InterPro were more informative for the R. microporus effectorome than GO terms and other annotations (Figure 2). Figures 2C,D showed that 42% of the effector proteins were associated with known protein families. The results revealed that a total of 128 PFam domains and 346 InterPro entries are associated with 206 and 207 effectors, respectively. However, 151 effectors still lack the matched Pfam domain annotations by hmmscan. Among the annotated domains, the 9 most frequently encountered Pfam domains in R. microporus effectors are listed (Table 1), highlighting the prevalence of domains such as fungal hydrophobin domain (PF01185), auxiliary activity family 9 domain (PF03443), fungal cellulose binding domain (PF00734), and others.
The transcriptomic data of R. microporus change upon the treatment of rubber tree latex, and based on the experiment by Oghenekaro et al. (2020), it was utilized to analyze effector candidates potentially involved in the rubber tree-R. microporus interaction. Among the 357 identified effectors, 319 were differentially expressed while 38 showed no significant changes upon latex treatment (Supplementary Tables S2, S3). Table 2 illustrates the significant differential expression rank of effector candidates with some abundant domains outlined in Table 1 and Figure 2. Using the threshold at a 1.5-fold difference, the 17 candidates with significantly lower expression levels and the 14 with significantly higher expression levels are listed. The results highlight the prominent roles of effectors containing fungal hydrophobin domains, which exhibited robust responses of R. microporus to rubber tree latex.
Table 2. List of the differentially expressed Rigidoporus microporus effectors that significantly change (more than 1.5 Log2Fold) following the treatment with rubber tree latex with the low expression level (A) and the high expression level (B) based on the transcriptome data reported by Oghenekaro et al. (2020).
The results showed 7 of the 14 highest expressed effectors with no annotation to any types of proteins, including the proteins 882769, 872712, 467301, 942071, 941140, 89931, and 948612 (Table 2B). To understand more about how some of these unknown latex-responsive effectors could contribute to the resilience of the pathogen to plant defense, the protein structural-based prediction was then performed by the D-I-TASSER server (Figure 3). Additional information on five previously uncharacterized effector proteins, including 882769, 872712, 942071, 941140, and 89931, was uncovered (Table 3).
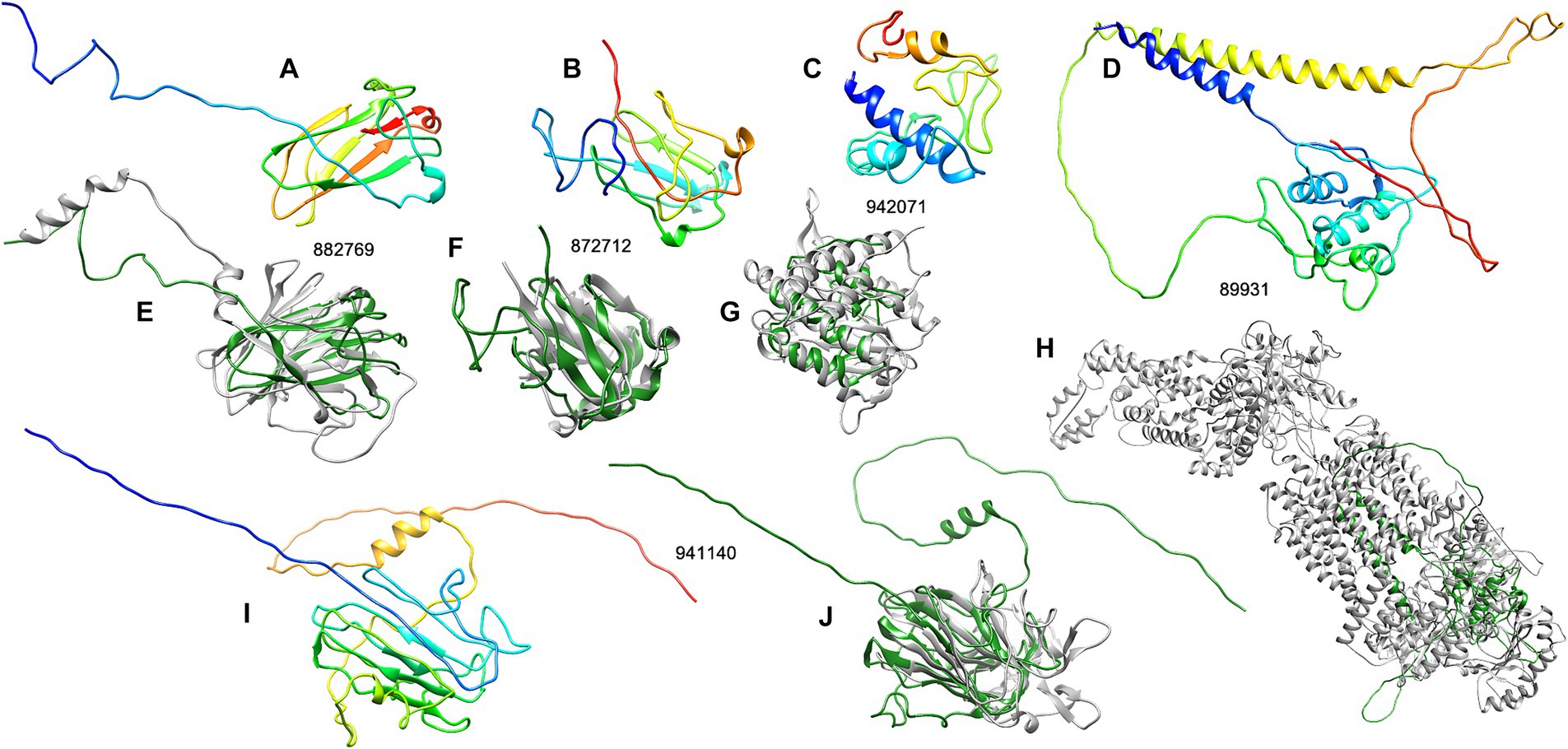
Figure 3. The structure analysis of five unknown R. microporus effectors whose expression was highly induced upon the interaction with rubber tree latex. The 3D structures of each candidate effector were predicted by the D-I-TASSER server (A–C,I) and compared with the most relevant template structure (E–H,J) including effector protein 882769 (A) compared with the PDB: 8PD4A (E), effector protein 872712 (B) compared with the PDB: 1HDFA (F), effector protein 942071 (C) compared with the PDB: 5JQKA2 (G), effector protein 89931 (D) compared with the PDB: 7TBWA (H), and effector protein 941140 (I) compared with the PDB: 2AHNA (J). The color range of the 3D structures represents the N-terminal (blue) to the C-terminal (red) of each candidate effector (A–C,I). The structural comparison showed the similarity of effector (green) and the PDB template (gray) (E–H,J).
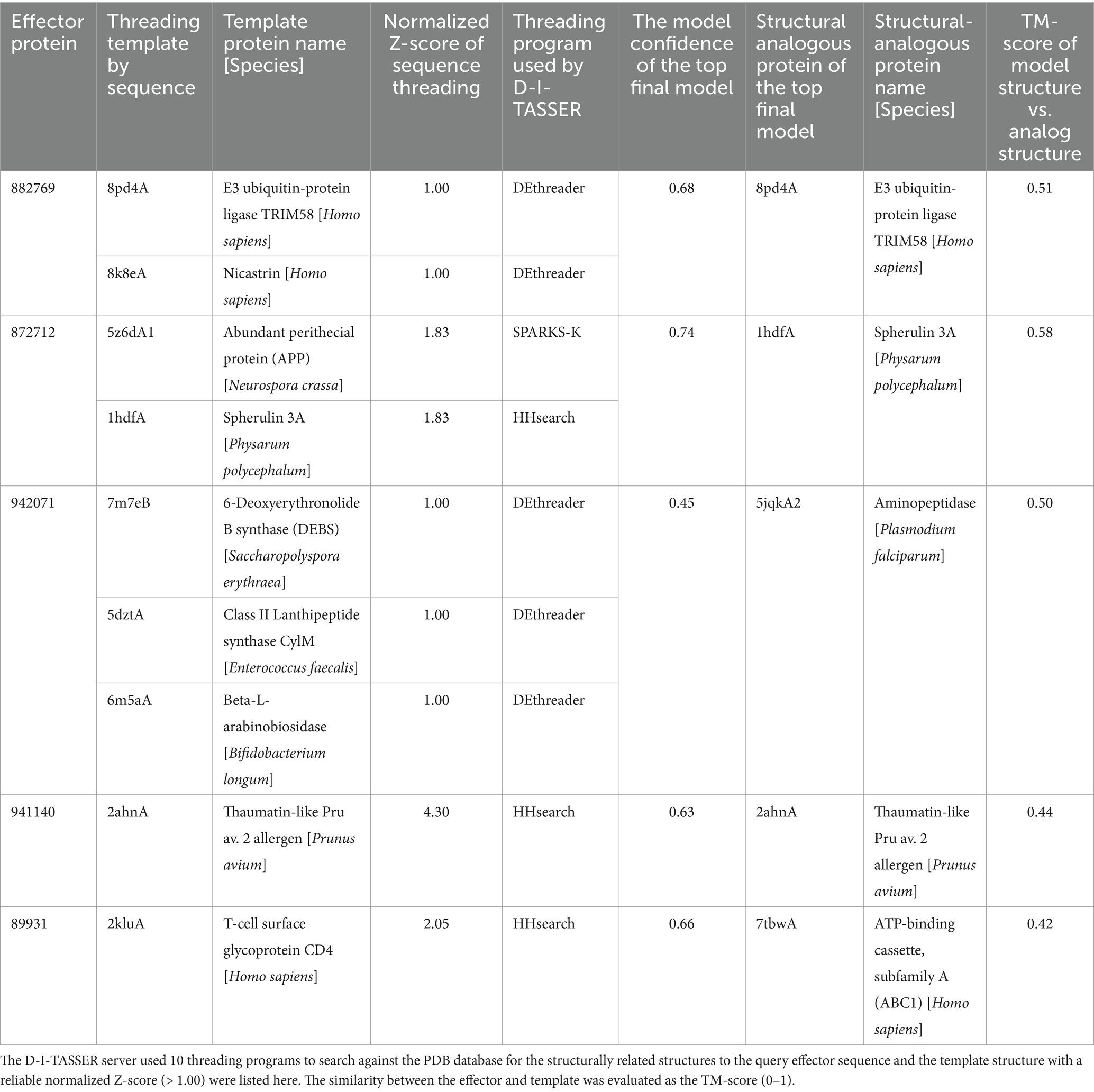
Table 3. The structurally related proteins in PDB that were selected as templates for the prediction of unknown R. microporus effector protein structures by the D-I-TASSER server.
The three effectors, including proteins 882769, 872712, and 942071, are structurally related to the E3 ubiquitin ligase protein, spherulin protein, and aminopeptidase protein, respectively, with reliable similarity scores (Table 3). The visualization of the structural comparison showed the likeliness of the known protein despite their low sequence similarities to any of the associated PFam accessions (Figures 3E–G). Meanwhile, the central domain of protein 941140 showed a structural similarity to the thaumatin-like allergen of sweet cherry (Table 3), but the structure of this effector exhibited the extended N-and C-terminal further than the allergen structure (Figure 3J). The predicted structure of effector 89931 was highly unique (Figure 3D) and, however, distinct from the closest structural template ATP-binding cassette structure (Figure 3H).
4 Discussion
Effector proteins of phytopathogenic fungi play vital roles during invasive growth, especially those secreted proteins with enzyme activity capable of altering host structural barriers and interfering with cellular immune components. Analyses of bioinformatic data by the currently developed tools, especially the machine-learning-based algorithms, have allowed the more feasible effectoromic studies extended from the wet lab in profiling the secreted proteomics and metabolomics of fungal pathogens. The identification could be done computationally by analyzing the conserved sequences among previously proven effectors and then predicting the likelihood of query sequences from the genome (Todd et al., 2022). The knowledge from the effectoromics study has been proposed to lead to alternative disease controls by targeting the virulent factors of particular pathogens (Vleeshouwers et al., 2011; Lanver et al., 2017; Lin et al., 2023). Although the molecular basis of WRD-rubber tree interactions has been studied (Oghenekaro et al., 2016a; Oghenekaro et al., 2016b; Oghenekaro et al., 2020; Janket et al., 2022; Longsaward et al., 2023), none of the previous research focused on the effectorome in R. microporus fungus.
The effector proteins are predicted and characterized using the Signal P server (Teufel et al., 2022) and the Effector P server (Sperschneider and Dodds, 2022). Employing the Signal P and Effector P predictions, the effectors of R. microporus were in silico analyzed, which led to the identification of 246 protein candidates as secreted effector proteins (Figure 1). The secreted effectors from plant pathogenic fungi are proposed to reach the extracellular space and then target the host components in the apoplast, or enter the cytoplasm to combat the intracellular defense of the host. Using the criteria of Rocafort et al. (2020), predicted by the Effector P server, R. microporus effectors could mainly be categorized into 183 apoplastic, 53 cytoplasmic, and 10 apoplastic/cytoplasmic effectors.
Effector-related motifs such as a well-known RxLR motif, which is renowned for mediating the entry of oomycetes and fungi into host cells (Kale et al., 2010; Sonah et al., 2016; Whisson et al., 2016), and the effector-conserved Y/F/WxC motif (Godfrey et al., 2010; Seong and Krasileva, 2023; Cao et al., 2023) were documented. Effector-related motifs were sometimes also used for the identification of non-canonical effectors. Noncanonical effectors are effectors with contrary characteristics such as containing more than 400 amino acids (Bohnert et al., 2004), few cysteine residues (Gout et al., 2006), and no signal peptide (Liu et al., 2014). These are normally submerged under the in silico identification of effector based on the canonical characters. In Pucinia graminis, Zhao et al. (2020) searched the effector-related motifs to identify the novel effector candidates and found 719 RxLR effectors, 138 Y/F/WxC effectors, and other groups of effectors related to other specific motifs. The RxLR motif-containing effectors of Phytophthora kernoviae have been identified and shown to contribute to pathogenicity (Wang et al., 2021). In this study, several effector-related motifs were identified in the sequence of secreted R. microporus proteins, including those predicted by Effector P through in silico analysis. An additional 111 effectors were further identified from the R. microporus secretome that had been overlooked by Effector P prediction. The most frequently found motif is the RxLR motif, which is presented in 33 effector candidates (Figure 1B). This includes 25 effectors containing only the RxLR effector-conserved motif and 8 effectors with the combination of RxLR and other effector-conserved motif(s). Among them, 1 effector in Emp24/gp25L/p24 family/GOLD family (PF01105), and another effector protein ID 915929 are contained RxLR. The later effector protein ID 915929 consists of the domain of tetratricopeptide-like repeats (PF13414) and DNAJ domain (PF00226), and it interestingly has 3 effector-related motifs including RxLR, [LI]xAR, and YxSL[RK] motifs (Supplementary Table S2). However, the relationship between these identified motifs and the effector functions in R. microporus pathogenicity still requires further experimental investigation. Future studies should prioritize candidate effectors with conserved motif(s) to better understand their roles in pathogenicity.
The localization where the effector protein from R. microporus was predicted to function (Figure 1A) may somehow be related to the role of the effector during the pathogenicity of WRD. Supplementary Table S2 in this study notes the identified R. microporus effectors with the characteristics of apoplastic effectors, cytoplasmic effectors, and versatile ones. Regardless of the uncharacterized effectors that are unable to be categorized into any protein family, several annotated protein families of R. microporus effectors showed specificity to the localization. The key cytoplasmic effectors are those effector proteins in families PF01105 (n = 5), PF00085 (n = 3), etc. (Supplementary Table S2). The five cytoplasmic effectors with the domain of PF01105 are in the Emp24/gp25L/p24 family/GOLD family, which function for the secretion via the Golgi apparatus (Anantharaman and Aravind, 2002). The GOLD domain in the Pb257 effector protein was recently proven to be crucial for the virulence of Plasmodiophora brassicae in killing the plant cells and causing the clubroot symptom (Yang et al., 2024). Moreover, the three cytoplasmic effectors with thioredoxin domain (PF00085) are noted. In a soilborne fungal pathogen Verticillium dahlia, thioredoxins were suggested as a key secreted virulent factor contributing to the scavenging of defensive ROS from host plants (Tian et al., 2023). However, most cytoplasmic effectors in R. microporus showed decreased expression upon latex treatments (Supplementary Table S2). The effector protein ID 942071 is the only uncharacterized cytoplasmic effector that showed higher transcription (2.055-fold) in response to latex treatment (Table 2B; Supplementary Table S2), and it was predicted to be structurally closest to the aminopeptidase protein (Table 3; Figure 3G). The aminopeptidase-like effector from R. microporus may be crucial for cleaving the peptide from rubber tree latex and facilitating fungal pathogen growth upon latex treatment, as experimentally proven by Oghenekaro et al. (2020). This resembles the virulent peptidases from several plant pathogenic fungi such as peptidases from Zymoseptoria tritici and relatives (Krishnan et al., 2018), and two secreted peptidases from Fusarium oxysporum, FoAPY1 (Qian et al., 2022) and FolAsp (Wang et al., 2023).
Apoplastic effectors secreted by fungal pathogens are crucial for the pathogenicity of particular diseases as they can be detected by the host apoplastic immune response, and consequently trigger the disease symptoms, and/or, can bypass the plant defense by deactivating the protective molecules in apoplast (Chen et al., 2023). A total of 183 secreted proteins in the R. microporus genome were predicted to be apoplastic effectors (Figure 1A; Supplementary Table S2), although most of them remain uncharacterized and unknown. The annotated apoplastic effectors in WRD fungus largely consist of the abundant protein families listed in Table 1, except the earlier mentioned cytoplasmic effectors in family PF01105.
It is worth highlighting the effector proteins in the hydrophobin family (PF01185) as one of the major effectors of R. microporus (n = 21) because of their abundance in the genome (n = 28) and their differential expression in response to the latex application (Table 2). Hydrophobin proteins are important proteins located on the surface of fungi, contributing not only to the fungal pathogenicity but also to the growth and formation of the fruiting body (Karlsson et al., 2007; Mgbeahuruike et al., 2012). In the pathogenicity of filamentous fungal pathogens, hydrophobins are reported to be involved in appressorium formation and mediated attachment to the host surface and subsequent infection (Linder et al., 2005). In the biological control activity, hydrophobins were found to be the major expressed proteins in Trichoderma atroviride during the interaction with plants and with the Rhizoctonia solani pathogen (Guzman-Guzman et al., 2017). These suggest their importance in the virulence of fungi. In the WRD pathosystem, Oghenekaro et al. (2016b) explored the transcription of R. microporus genes during the invasion of rubber tree wood. Over 2.5-fold higher abundance of two transcripts encoding hydrophobins (CL996.Contig2 and Unigene3334), as well as the 145-fold lower abundance of another hydrophobin (CL2382.Contig2) were observed in R. microporus when growing on the rubber wood-supplemented media. Accordingly, the integration of the identified effectors with transcriptomic data here revealed the dual pattern of hydrophobin expression upon the response to rubber tree latex (Table 2). In total, four R. microporus hydrophobins (Table 2A) showed sharp downregulation lower than 2-fold compared to the latex-free control. Meanwhile, a triple of hydrophobin proteins (Table 2B) showed high upregulation. Interestingly, one of the top upregulated effectors is the hydrophobin protein with collagen triple helix repeat domain (PF01391) whose protein ID is 884303 and its increased expression was 2.6 fold upon latex treatment (Table 2B). These datasets suggest some hydrophobins as R. microporus apoplastic effectors that are being suppressed by rubber tree latex, and some redundant hydrophobins as another set of effectors to compensate for the virulence of R. microporus.
The secreted fungal effectors seem to largely target the host structural barrier, especially the cell-wall-related carbohydrate macromolecules. The high number of R. microporus effectors containing the fungal cellulose binding domain (PF00734) are identified in this study as another group of apoplastic effectors (Supplementary Table S2) as well as the effectors presenting conserved motifs (Supplementary Table S3). These effectors are often associated with glycoside hydrolases (GH) domains and have been annotated within the CAZy database, which categorizes carbohydrate-hydrolyzing enzymes. Several studies on fungal effectorome analysis have utilized the CAZy annotation as a classification tool for effectors. For instance, it has been applied in studies of mycorrhizal populations (Kohler et al., 2015; Martin et al., 2016), endophytic fungi in the genus Hymenoscyphus (Rafiqi et al., 2023), and the palm dieback pathogen (Rafiqi et al., 2022), where their effectors comprised a significant portion of GHs and auxiliary activity (AA) enzymes. In this study, analysis of the CAZy information for R. microporus effectors revealed CBM, GHs, and AAs, as some of the most frequently observed activities (Figure 1F). GH is a large protein superfamily consisting of several classes of GH protein families that exhibit enzymatic cleavage on specific glycosidic bonds of various polysaccharides (Shrivasrava, 2020). Classes of GH proteins that have been considered are mostly cell-wall-degrading enzymes that are extensively mentioned to be important proteins that contribute to the pathogenicity of fungal pathogens in disrupting the host cell structure (Lopez et al., 2018; Bradley et al., 2022). Here, several proteins belonging to the formerly-named GH class 61 (GH61), which is now designated auxiliary activity 9 (AA9) protein (PF03443), were identified as apoplastic effectors of R. microporus (Table 1). The expression profile of AA9 proteins is mostly shown to be suppressed upon latex treatment (Supplementary Table S2), except for the IDs 925354 and 872163 which, by contrast, showed the highest latex-responsive expression (Table 2B). The AA9 proteins are lytic polysaccharide monooxygenase (LPMO) commonly responsible for the cleavage of crystalline cellulose (Vaaje-Kolstad et al., 2010; Levasseur et al., 2013). They were the highly expressed candidates of R. microporus during the saprotrophic invasion on rubber wood (Oghenekaro et al., 2016b) as well as other saprotrophic fungi (Wymelenberg et al., 2011; Raffaello et al., 2014; MacDonald et al., 2011). On the other hand, the biotrophic pathogen Ustilago maydis lacks this class of LPMO protein (Figure 4), which aligns with previous evolutionary comparisons showing that U. maydis has lost its LPMO, GH6, and GH7 enzymes (Kohler et al., 2015). Furthermore, GH7 is the hydrolyzing enzyme cleaving the non-crystalline cellulose at the beta-1,4-glycosidic bond at the ends of the chain (Medie et al., 2012). GH7 was recognized as the family with the highest transcript abundance among other GH families during the analysis of R. microporus-rubber tree wood interaction. It was observed that a gene encoding GH7 protein (CL2079.Contig4) had a 71-fold increased expression and the two AA9 proteins (CL374.Contig3, CL374.Contig4) had more than 16-fold increased transcripts (Oghenekaro et al., 2016b). In response to latex, the expression of these two GHs was mostly suppressed, while some of them were induced (Table 2).
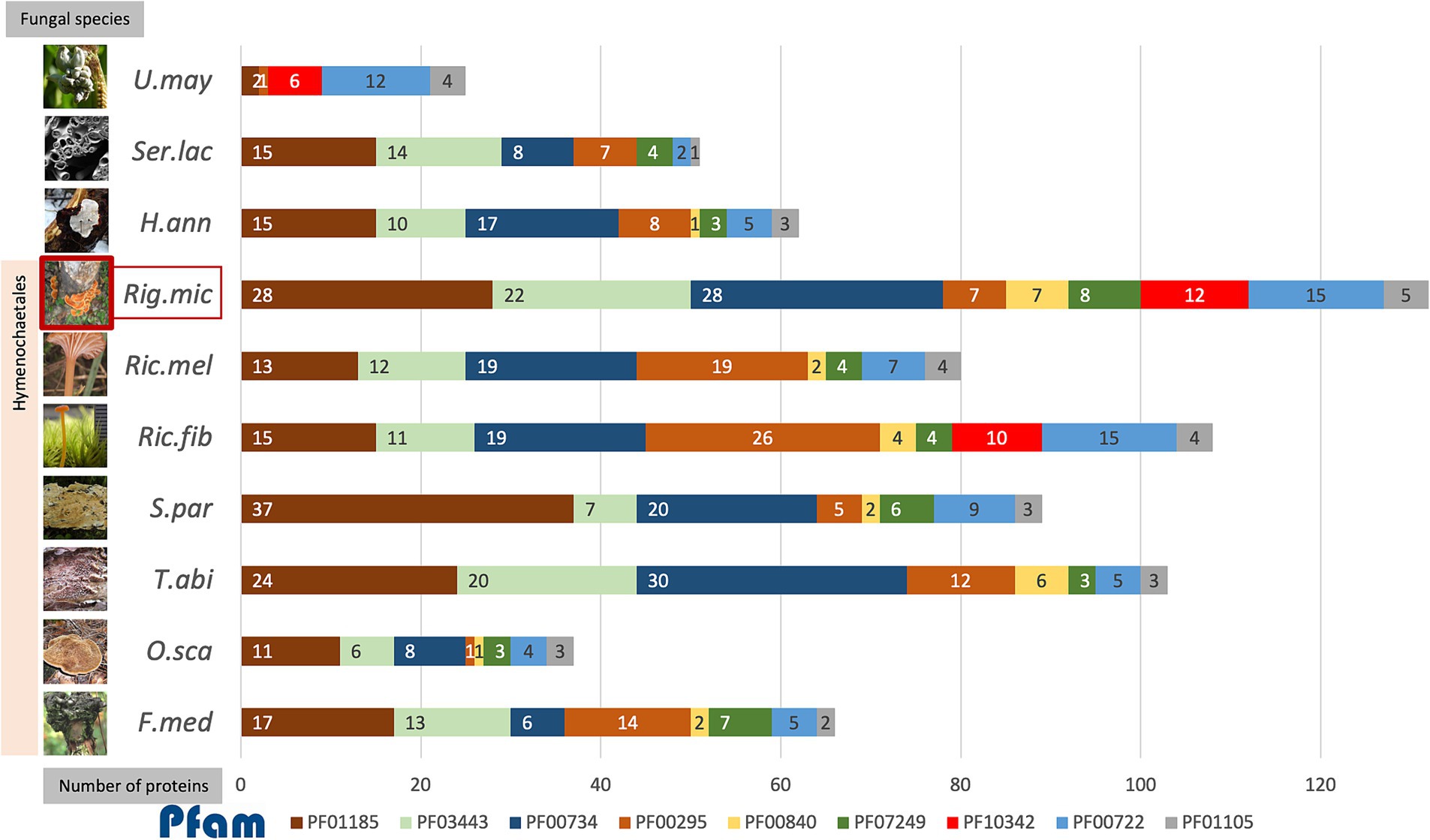
Figure 4. The accumulative bar chart showed the number of proteins encoding each of the top 9 protein families identified as effectors in R. microporus (Rig.mic) compared to other representative species in the order Hymenochaetales, including Fomitiporia mediterranea (F.med), Onnia scaura (O.sca), Trichaptum abietinum (T.abi), Schizopora paradoxa (S.par), Rickenella fibula (Ric.fib), and Rickenella mellea (Ric.mel), as well as the white rot pathogen Heterobasidion annosum (H.ann), brown rot fungus Serpula lacrymans (Ser.lac), and the biotrophic pathogen Ustilago maydis (U.may). The data on the number of proteins in the fungal genome belonging to each Pfam was collected from the Multigene Clusters (MCL) of the R. microporus genome in the MycoCosm database (based on the running Rigmic1-comparative.1858) and of the 3 other pathogens H.ann, Ser.lac, and U.may individually. The fungal images were retrieved from the description of each fungus in the MycoCosm database.
The aforementioned scenario illustrates the potential interaction, in which the WRD fungus largely utilizes several GHs as virulent factors for invading rubber trees and the rubber tree alleviates the rotting by the activity of its latex. When comparing among R. microporus-related fungal species in the order Hymenochaetales, the AA9 family (PF03443), GH7 proteins (PF00840), and the fungal cellulose binding domain (PF00734) contain more individual protein members in R. microporus and the powerful saprotrophic white rotter Trichaptum abietinum than in other species (Figure 4). The high candidate numbers of these proteins in WRD fungus and the rotter T. abietinum further support the potential contribution of two GH proteins to the evolutionary shift in the Hymenochaetales order, allowing these two fungi to cause rot symptoms on infected trees. Interestingly, several effector-related protein families of R. microporus contain a higher number of protein candidates compared to those in the white rot fungal pathogen H. annosum (Figure 4).
A high number of proteins belonging to the GH16 family (PF00722) were annotated in R. microporus, the bryophilous-associated fungus Rickenella fibula, as well as in the biotrophic pathogen U. maydis (Figure 4). The GH16 protein showed activity not only with the terrestrial polysaccharides but also with the marine polysaccharides (Viborg et al., 2019). As mentioned by Bradley et al. (2022), an example of GH16 from Botrytis cinerea, BcCrh1 protein, has been studied and proposed for its role in responsibility for infection and cell death induction on the host (Bi et al., 2021). Several GH16 proteins identified as apoplastic effectors of R. microporus were slightly triggered upon latex treatment (Supplementary Table S2). Notably, effector proteins containing Kre9/KNH-like N-terminal Ig-like domain (PF10342) were found only in R. microporus, Rickenella fibula, and U. maydis, but lacking in other representative Hymenochaetales species (Figure 4). They are mostly upregulated when challenged with the latex. The Kre9_KHN proteins were membrane-located proteins involved in the biosynthesis of fungal cell walls and the fruiting body formation (Szeto et al., 2007). The evidence to show the relevance of Kre9/KNH-like-domain-containing protein to the fungal pathogenicity is lacking, but their presence in R. microporus rather than most of other Hymenochaetales species together with the latex-responsive expression suggests the support of latex on the fungal growth as observed in Oghenekaro et al. (2020). This will facilitate the progressive increase of fungal mycelia and, consequently more rubber tree roots would be severely infected.
Cerato-platanin proteins (PF07249) are one of the conserved effector protein families in fungi (Gaderer et al., 2014; Luti et al., 2020), which were previously recognized for their role in the production of phytotoxin and triggering the systemic acquired resistance (SAR) and hypersensitive responses (HR) in plants (Frias et al., 2014; Frias et al., 2011). The proteins in this family have been suggested as crucial effectors utilized by fungal pathogens (Vasina et al., 2016; Pan et al., 2018; Zhang et al., 2020; Khairi et al., 2022). In the necrotrophic fungus Sclerotinia sclerotiorum, its cerato-platanin SsCP1 protein interacts with the pathogenesis-related protein 1 (PR-1) of Nicotiana benthamiana and contributes to its virulence (Yang et al., 2018). In R. microporus, Maiden et al. (2024) reported the stable expression of a gene encoding RmCP, a cerato-platanin homolog, during the infection into rubber trees. The analysis here identified 7 cerato-platanins (Table 1), which is the highest number compared to the number of cerato-platanins identified in other representative species in Hymenochaetales (Figure 4). Overall, the R. microporus effectors belonging to Cerato-platanin were upregulated against latex treatment, except for the slightly lowered ID 137475 (Supplementary Table S2).
Moreover, some of the identified unknown effectors of R. microporus that showed a high level of differential expression response to the rubber tree latex (Table 2B) were predicted for their tertiary structures by The I-TASSER server (Table 3; Figure 3). These key unknown effectors are proposed here to be the structural relatives of known proteins, noted as the E3 ubiquitin ligase-like protein (ID 882769), spherulin-like protein (ID 872712), aminopeptidase-like protein (ID 942071), and thaumatin-like protein (ID 941140).
Previously, E3 ubiquitin ligase activity was mentioned to be used by fungal pathogens to interfere with the proteasome of host plants and facilitate pathogenicity (Duplan and Rivas, 2014). The ubiquitination is a key process in regulating the secretion of fungal effectors, such as in the rice blast Magnaporthe oryzae (Wang et al., 2022). The predicted structure of the highly upregulated effector ID 882769 showed a similarity toward the E3 ubiquitin ligase (Figure 3E). Moreover, the spherulin 3A protein from the slime mold Physarum polycephalum was the template and the structural-related protein to the effector ID 872712 (Table 3; Figure 3F). Though the spherulin protein was also identified in the grape canker fungus Botryosphaeria parva (UniProt ID: R1EXI5), there is limited information on how spherulin proteins participate in the pathogenicity of plant fungal pathogens. However, spherulin is a starvation-related protein in fungi that exhibits the conserved H[I/T]HPRATEI to the germin-like proteins in plants. They are both considered members of cupin superfamily proteins (Dunwell et al., 2004). Another identified R. microporus effector containing RxLR motif (ID 888205) also belongs to the cupin family, although its expression was slightly lower. Therefore, the relatedness of the spherulin-like effector from R. microporus to the fungal virulence may resemble the pathogenicity mediated by a cupin-domain-containing effector named FoCupin1 from Fusarium oxysporum f. sp. cubense (Yan et al., 2022). Further functional investigation of these effectors regarding the effects of rubber tree latex on the fungal proteasome, secretome, as well as the protein interaction to the host immune merits to be investigated.
Thaumatin (PF00314) is well-known as an antifungal protein in plants in the PR-5 family (de Jesus-Pires et al., 2020). Because of the antifungal activity or cleaving of the beta-1,3-glucanase, the thaumatin-like proteins in R. microporus effector identified in this study (Supplementary Tables S2, S3) may not likely contribute to the pathogenicity of the WRD. It is most likely to contribute to the mycoparasitic activity in competing with other fungal organisms. In fungi, a thaumatin-like protein (TLP) was recently identified in Rhizoctonia solani and is being suggested as an elicitor protein triggering the defense of maize plants (Liu et al., 2024). Although a thaumatin-like protein was predicted as one of the upregulated R. microporus effectors, another thaumatin (ID 862870) was the most downregulated effector (Table 2A). The compensation of thaumatin expression in R. microporus upon latex treatment was also documented (Supplementary Table S2).
5 Conclusion
In conclusion, the dataset of putative secreted effectors from the white root rot fungus Rigidoporus microporus was explored for the first time based on the characteristics predicted by the combination of Signal P and Effector P servers, as well as the presence of fungal-conserved effector motifs. The cytoplasmic effectors and apoplastic effectors of R. microporus were categorized despite several of them being unknown effectors. Most of the known effectors belong to hydrophobins and GH, which facilitate invasive growth and the degradation of rubber tree cell walls. The dataset of the response toward the latex application was integrated and underlined the crucial groups of effectors which may be utilized by R. microporus to cause the WRD in rubber trees (Figure 5). Taken together, the in silico identification of effectors opens up the opportunity for experimental validation of the several prioritized effector candidates. It would also benefit the development of novel strategies for managing the WRD of rubber trees in future.
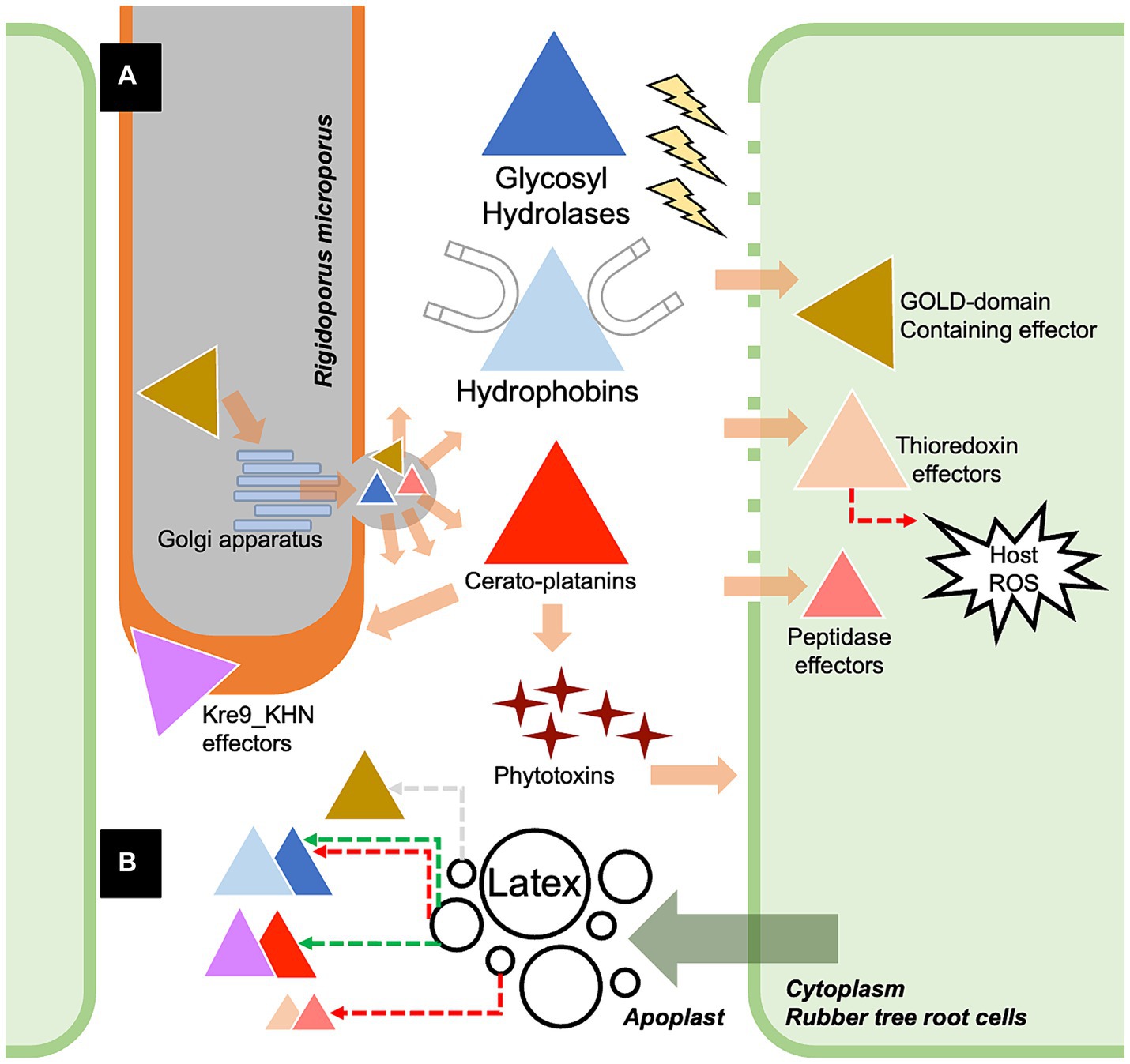
Figure 5. The proposed scenario showing the partial molecular pathogenicity of white root rot disease mediated by the identified effectors of Rigidoporus microporus fungus in this study (A) and the effects of rubber tree latex on the expression of some effectors (B). Briefly, R. microporus exhibits GOLD-domain-containing effector proteins that may facilitate the secretion function via the Golgi apparatus, further promoting the release of fungal effectors toward the rubber tree cells. There are also several classes of glycoside hydrolases (GHs) proteins that are the apoplastic effectors that cleave the cell wall components of rubber trees. The hydrophobins are the common apoplastic effectors of R. microprus that also destroy the host cell wall and enable the attachment of fungus to the rubber tree’s cell surface. Furthermore, the hyphal-enhancing and phytotoxins-producing cerato-platanin proteins are another group of apoplastic effectors of R. microporus. GOLD-domain-containing effectors, thioredoxin effectors, and peptidase effectors are proposed as crucial cytoplasmic effectors of R. microporus, which interfere with the molecular defense of rubber trees. Although the latex released by the rubber tree showed suppressive activity against these fungal effectors, some of them could be induced by the latex based on the experiment by Oghenekaro et al. (2020). The fungal effectors from R. microporus are illustrated as triangles with different colors corresponding to each of the containing protein domains. The functional locations of effectors were demonstrated based on the prediction in Supplementary Table S2. The colors of dashed arrows indicate the downregulation (red) or upregulation (green) of effectors or the plant defense.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
RL: Conceptualization, Data curation, Formal analysis, Visualization, Writing – original draft. UV: Conceptualization, Writing – review & editing. ZW: Conceptualization, Writing – review & editing. FA: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Research Council of Finland (Grant no. 353365 to FOA) and Open access funded by Helsinki University Library.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1439454/full#supplementary-material
Footnotes
1. ^https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.47610
2. ^https://genome.jgi.doe.gov/portal/Rigmic1/Rigmic1.download.html
3. ^https://services.healthtech.dtu.dk/services/SignalP-6.0/
4. ^https://effectorp.csiro.au/
5. ^https://meme-suite.org/meme/tools/fimo
6. ^https://mycocosm.jgi.doe.gov/Rigmic1/Rigmic1.home.html
7. ^https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan
References
Alfano, J. R., and Collmer, A. (2004). Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42, 385–414. doi: 10.1146/annurev.phyto.42.040103.110731
Anantharaman, V., and Aravind, L. (2002). The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 3:research0023. doi: 10.1186/gb-2002-3-5-research0023
Bi, K., Scalschi, L., Jaisawal, N., Mengiste, T., Fried, R., Belen Sanz, A., et al. (2021). The Botrytis cinerea Crh1 transglycosylase is a cytoplasmic effector triggering plant cell death and defense response. Nat. Commun. 12:2166. doi: 10.1038/s41467-021-22436-1
Bohnert, H. U., Fudal, I., Dioh, W., Tharreau, D., Notteghem, J. L., and Lebrun, M. H. (2004). A putative polyketide synthase/peptide synthetase from Magnaporthe gisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513. doi: 10.1105/tpc.104.022715
Bradley, E. L., Okmen, B., Doehlemann, G., Henrissat, B., Bradshaw, R. E., and Mesarich, C. H. (2022). Secreted glycoside hydrolase proteins as effectors and invasion patterns of plant-associated fungi and oomycetes. Front. Plant Sci. 13:853106. doi: 10.3389/fpls.2022.853106
Cao, Y., Kummel, F., Logemann, E., Gebauer, J. M., Lawson, A. W., Yu, D., et al. (2023). Structural polymorphisms within a common powdery mildew effector scaffold as a driver of coevolution with cereal immune receptors. PNAS 120:e2307604120. doi: 10.1073/pnas.2307604120
Chen, X. R., Wang, Y., Kale, S. D., Fang, Y., and Srivastava, V. (2023). Editorial: Apoplastic effectors – what roles do they play in plant-pathogen interactions? Front. Microbiol. 14:1149771. doi: 10.3389/fmicb.2023.1149771
Collemare, J., O’Connell, R., and Lebrun, M. H. (2019). Nonproteinaceous effectors: the terra incognita of plant-fungal interactions. New Phytol. 223, 590–596. doi: 10.1111/nph.15785
de Jesus-Pires, C., Ferreira-Neto, J. R. C., Bezerra-Neto, J. P., Kido, E. A., de Olivera Silva, R. L., Pandolfi, V., et al. (2020). Plant thaumatin-like proteins: function, evolution and biotechnological applications. Curr. Protein Pept. Sci. 21, 36–51. doi: 10.2174/1389203720666190318164905
Dunwell, J. M., Purvis, A., and Khuri, S. (2004). Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17. doi: 10.1016/j.phytochem.2003.08.016
Duplan, V., and Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5:42. doi: 10.3389/fpls.2014.00042
Frias, M., Brito, N., Gonzalez, M., and Gonzalez, C. (2014). The phytotoxic activity of the cerato-platanin BcSpl1 resides in a two-peptide motif in the protein surface. Mol. Plant Pathol. 15, 342–351. doi: 10.1111/mpp.12097
Frias, M., Gonzalez, C., and Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. doi: 10.1111/j.1469-8137.2011.03802.x
Friesen, T. L., Faris, J. D., Solomon, P. S., and Oliver, R. P. (2008). Host-specific toxins: effectors of necrotrophic pathogenicity. Cell. Microbiol. 10, 1421–1428. doi: 10.1111/j.1462-5822.2008.01153.x
Gaderer, R., Bonazza, K., and Seidl-Seiboth, V. (2014). Cerato-platanins: a fungal family with intriguing properties and application potential. Appl. Microbiol. Biotechnol. 98, 4795–4803. doi: 10.1007/s00253-014-5690-y
Godfrey, D., Bohlenius, H., Pedersen, C., Zhang, Z., Emmersen, J., and Thordal-Christensen, H. (2010). Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11:317. doi: 10.1186/1471-2164-11-317
Gout, L., Fudal, I., Kuhn, M. L., Blaise, F., Ekert, M., Cattolico, L., et al. (2006). Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol. Microbiol. 60, 67–80. doi: 10.1111/j.1365-2958.2006.05076.x
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Guzman-Guzman, P., Aleman-Duarte, M. I., Delaye, L., Herrera-Estrella, A., and Olmedo-Monfil, V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18:16. doi: 10.1186/s12863-017-0481-y
Hassani, M. A., Oppong-Danquah, E., Feurtey, A., Tasdemir, D., and Stukenbrock, E. H. (2022). Differential regulation and production of secondary metabolites among isolates of the fungal wheat pathogen Zymoseptoria tritici. Appl. Environ. Microbiol. 88:e0229621. doi: 10.1128/aem.02296-21
Janket, J., Pengnoo, A., Kongsawadworakul, P., and Viboonjun, U. (2022). Proteomic analysis of rubber trees uncovers a systemic response to white root rot disease. Plant Omics 15, 37–47. doi: 10.21475/POJ.15.01.22.p3756
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kabbage, M., Yarden, O., and Dickman, M. B. (2015). Pathogenic attributes of Sclerotinia sclerotiorum: switching from a biotrophic to necrotrophic lifestyle. Plant Sci. 233, 53–60. doi: 10.1016/j.plantsci.2014.12.018
Kale, S. D., Gu, B., Capelluto, D. G. S., Dou, D., Feldman, E., Rumore, A., et al. (2010). External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142, 284–295. doi: 10.1016/j.cell.2010.06.008
Kamoun, S. (2006). A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44, 41–60. doi: 10.1146/annurev.phyto.44.070505.143436
Karlsson, M., Stenlid, J., and Olson, A. (2007). Two hydrophobin genes from the conifer pathogen Heterobasidion annosum are expressed in aerial hyphae. Mycologia 99, 227–231. doi: 10.3852/mycologia.99.2.227
Khairi, M. H. F., Nor Muhammad, N. A., Bunawan, H., Abdul Murad, A. M., and Ramzi, A. B. (2022). Unveiling the core effector proteins of oil palm pathogen Ganoderma boninense via pan-secretome analysis. J. Fungi 8:793. doi: 10.3390/jof8080793
Kim, K. T., Jeon, J., Choi, J., Cheong, K., Song, H., Choi, G., et al. (2016). Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 7:186. doi: 10.3389/fpls.2016.00186
Kohler, A., Kuo, A., Nagy, L. G., Morin, E., Barry, K. W., Buscot, F., et al. (2015). Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat. Genet. 47, 410–415. doi: 10.1038/ng.3223
Krishnan, P., Ma, X., McDonald, B. A., and Brunner, P. C. (2018). Widespread signatures of selection for secreted peptidases in a fungal plant pathogen. BMC Evol. Biol. 18:7. doi: 10.1186/s12862-018-1123-3
Lanver, D., Tollot, M., Schweizer, G., Presti, L. L., Reissmann, S., Ma, L. S., et al. (2017). Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15, 409–421. doi: 10.1038/nrmicro.2017.33
Levasseur, A., Drula, E., Lombard, V., Coutinho, P. M., and Henrissat, B. (2013). Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6:41. doi: 10.1186/1754-6834-6-41
Lin, X., Jia, Y., Heal, R., Prokchorchick, M., Sindalovskaya, M., Olave-Achury, A., et al. (2023). Solanum americanum genome-assisted discovery of immune receptors that detect potato late blight pathogen effectors. Nat. Genet. 55, 1579–1588. doi: 10.1038/s41588-023-01486-9
Linder, M. B., Szilvay, G. R., Nakari-Setala, T., and Penttila, M. E. (2005). Hydrophobins: the protein-amphiphiles of filamentous fungi. FEMS Microbiol. Rev. 29, 877–896. doi: 10.1016/j.femsre.2005.01.004
Liu, J., Feng, S., Liu, T., Mao, Y., Shen, S., Liu, Y., et al. (2024). Molecular characterization revealed the role of thaumatin-like proteins of Rhizoctonia solani AG4-JY in inducing maize disease resistance. Front. Microbiol. 15:1377726. doi: 10.3389/fmicb.2024.1377726
Liu, T., Song, T., Zhang, X., Yuan, H., Su, L., Li, W., et al. (2014). Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5:4686. doi: 10.1038/ncomms5686
Longsaward, R., Pengnoo, A., Kongsawadworakul, P., and Viboonjun, U. (2023). A novel rubber tree PR-10 protein involved in host-defense response against the white root rot fungus Rigidoporus microporus. BMC Plant Biol. 23:157. doi: 10.1186/s12870-023-04119-3
Lopez, D., Ribeiro, S., Label, P., Fumanal, B., Venisse, J. S., Kohler, A., et al. (2018). Genome-wide analysis of Corynespora cassicola leaf fall disease putative effectors. Front. Microbiol. 9:276. doi: 10.3389/fmicb.2018.00276
Luti, S., Sella, L., Quarantin, A., Pazzagli, L., and Baccelli, I. (2020). Twenty years of research on cerato-platanin family proteins: clues, conclusions, and unsolved issues. Fungal Biol. Rev. 34, 13–24. doi: 10.1016/j.fbr.2019.10.001
MacDonald, J., Doering, M., Canam, T., Gong, Y., Guttman, D. S., Campbell, M. M., et al. (2011). Transcriptomic responses of the softwood-degrading white-rot fungus Phanerochaete carnosa during growth on coniferous and deciduous wood. Appl. Environ. Microbiol. 77, 3211–3218. doi: 10.1128/AEM.02490-10
Maiden, N. A., Atan, S., Syd Ali, N., Ahmad, K., and Wong, M. Y. (2024). The cerato-platanin gene, rmcp, from Rigidoporus microporus was stably expressed during infection of Hevea brasiliensis. J. Rubber Res. 27, 373–382. doi: 10.1007/s42464-024-00253-7
Martin, F., Kohler, A., Murat, C., Veneault-Fourrey, C., and Hibbett, D. S. (2016). Unearthing the roots of ectomycorrhizal symbiosis. Nat. Rev. Microbiol. 14, 760–773. doi: 10.1038/nrmicro.2016.149
Medie, F. M., Davies, G. J., Drancourt, M., and Henrissat, B. (2012). Genome analyses highlight the different biological roles of cellulases. Nat. Rev. Microbiol. 10, 227–234. doi: 10.1038/nrmicro2729
Mgbeahuruike, A. C., Karlsson, M., and Asiegbu, F. O. (2012). Differential expression of two hydrophobins (Pgh1 and Pgh2) from the biocontrol fungus Phlebiopsis gigantea. Fungal Biol. 116, 620–629. doi: 10.1016/j.funbio.2012.03.001
Mohammed, C. L., Rimbawanto, A., and Page, D. E. (2014). Management of basidiomycete root-and stem-rot diseases in oil palm, rubber and tropical hardwood plantation crops. Forest Pathol. 44, 428–446. doi: 10.1111/efp.12140
Oghenekaro, A. O., Kovalchuk, A., Raffaello, T., Camarero, S., Gressler, M., Henrissat, B., et al. (2020). Genome sequencing of Rigidoporus microporus provides insights on genes important for wood decay, latex tolerance and interspecific fungal interactions. Sci. Rep. 10:5250. doi: 10.1038/s41598-020-62150-4
Oghenekaro, A. O., Omorusi, V. I., and Asiegbu, F. O. (2016a). Defense-related gene expression of Hevea brasiliensis clones in response to the white root rot pathogen Rigidoporus microporus. Forest Pathol. 46, 318–326. doi: 10.1111/efp.12260
Oghenekaro, A. O., Raffaello, T., Kovalchuk, A., and Asiegbu, F. O. (2016b). De novo transcriptomic assembly and profiling of Rigidoporus microporus during saprotrophic growth on rubber wood. BMC Genomics 17:234. doi: 10.1186/s12864-016-2574-9
Omorusi, V. I. (2012). “Effects of white root rot disease on Hevea brasiliensis (Mull. Arg.) – challenges and control approach” in Plant science. eds. N. K. Dhal and S. C. Sahu (IntechOpen).
Paleaz, F., Martinez, M. J., and Martinez, A. T. (1995). Screening of 68 species of basidiomycetes for enzymes involved in lignin degradation. Mycol. Res. 99, 37–42. doi: 10.1016/S0953-7562(09)80313-4
Pan, Y., Wei, J., Yao, C., Reng, H., and Gao, Z. (2018). SsSm1, a cerato-platanin family protein, is involved in the hyphal development and pathogenic process of Sclerotinia sclerotiorum. Plant Sci. 270, 37–46. doi: 10.1016/j.plantsci.2018.02.001
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., and Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46, W200–W204. doi: 10.1093/nar/gky448
Qian, H., Song, L., Wang, L., Wang, B., and Liang, W. (2022). The secreted FoAPY1 peptidase promotes fusarium oxysporum invasion. Front. Microbiol. 13:1040302. doi: 10.3389/fmicb.2022.1040302
Raffaello, T., Chen, H., Kohler, A., and Asiegbu, F. O. (2014). Transcriptomic profiles of Heterobasidion annosum under abiotic stresses and during saprotrophic growth in bark, sapwood and heartwood. Environ. Microbiol. 16, 1654–1667. doi: 10.1111/1462-2920.12321
Rafiqi, M., Jelonek, L., Diouf, A. M., Mbaye, A. L., Rep, M., and Diarra, A. (2022). Profile of the in silico secretome of the palm dieback pathogen, fusarium oxysporum f. sp. albedinis, a fungus that puts natural oases at risk. PLoS One 17:e0260830. doi: 10.1371/journal.pone.0260830
Rafiqi, M., Kosawang, C., Peers, J. A., Jelonek, L., Yvanne, H., McMullan, M., et al. (2023). Endophytic fungi related to the ash dieback causal agent encode signatures of pathogenicity on European ash. IMA Fungus 14:10. doi: 10.1186/s43008-023-00115-8
Rocafort, M., Fudal, I., and Mesarich, C. H. (2020). Apoplastic effector proteins of plant-associated fungi and oomycetes. Curr. Opin. Plant Biol. 56, 9–19. doi: 10.1016/j.pbi.2020.02.004
Samperna, S., Masi, M., Vurro, M., Evidente, A., and Marra, M. (2022). Cyclopaldic acid, the main phytotoxic metabolite of Diplodia cupressi, induces programmed cell death and autophagy in Arabidopsis thaliana. Toxins 14:474. doi: 10.3390/toxins14070474
Seong, K., and Krasileva, K. V. (2023). Prediction of effector protein structures from fungal phytopathogens enables evolutionary analyses. Nat. Microbiol. 8, 174–187. doi: 10.1038/s41564-022-01287-6
Shao, D., Smith, D. L., Kabbage, M., and Roth, M. G. (2021). Effectors of plant necrotrophic fungi. Front. Plant Sci. 12:687713. doi: 10.3389/fpls.2021.687713
Shrivasrava, S. (2020). “Introduction to glycoside hydrolases: classification, identification and occurrence” in Industrial applications of glycoside hydrolases. ed. S. Shrivastava (Singapore: Springer), 3–84.
Siddiqui, N., Middleton, C., Ribeiro, C., Atan, S., and Di Cola, A. (2017). Gal-based proteomic study for differential expression of Hevea brasiliensis root proteins in response to infection by soil fungus Rigidoporus microporus. Acta Hortic. 1152, 229–234. doi: 10.17660/ActaHortic.2017.1152.31
Sonah, H., Deshmukh, R. K., and Belanger, R. R. (2016). Computational prediction of effector proteins in fungi: opportunities and challenges. Front. Plant Sci. 7:00126. doi: 10.3389/fpls.2016.00126
Sperschneider, J., and Dodds, P. N. (2022). EffectorP 3.0: prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant-Microbe Interact. 35, 146–156. doi: 10.1094/MPMI-08-21-0201-R
Stergiopoulos, I., Collemare, J., Mehrabi, R., and de Wit, P. J. G. M. (2013). Phytotoxic secondary metabolites and peptides produced by plant pathogenic Dothideomycetes fungi. FEMS Microbiol. Rev. 37, 67–93. doi: 10.1111/j.1574-6976.2012.00349.x
Stergiopoulos, I., and de Wit, P. J. G. M. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. doi: 10.1146/annurev.phyto.112408.132637
Suwandi, S. (2007). “Mode of dispersal and variation in population of white root rot fungus Rigidoporus microporus as revealed by mycelial incompatibility” in International workshop on white root disease of Hevea rubber. Eds. S. Pawirosoemardjo, B. Setyawan, H. Suryaningtyas, and M. Supriadi (IRRDB), 68–75. Available at: https://repository.unsri.ac.id/7261/2/Pages_from_Proceedings_of__International_Workshop_on_White_Root_Disease_Hevea_Rubber.pdf
Szeto, C. Y. Y., Leung, G. S., and Kwan, H. S. (2007). Le.MAPK and its interacting partner, Le.DRMIP, in fruiting body development in Lentinula edodes. Gene 393, 87–93. doi: 10.1016/j.gene.2007.01.030
Teufel, F., Armenteros, J. J. A., Johansen, A. R., Gislason, M. H., Pihl, S. I., Tsirigos, K. D., et al. (2022). SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 40, 1023–1025. doi: 10.1038/s41587-021-01156-3
Tian, L., Zhuang, J., Li, J. J., Zhu, H., Klosterman, S. J., Dai, X. F., et al. (2023). Thioredoxin VdTrx1, an unconventional secreted protein, is a virulence factor in Verticillium dahlia. Front. Microbiol. 14:1130468. doi: 10.3389/fmicb.2023.1130468
Todd, J. N. A., Carreon-Anguiano, K. G., Islas-Flores, I., and Canto-Canche, B. (2022). Fungal effectoromics: a world in constant evolution. Int. J. Mol. Sci. 23:13433. doi: 10.3390/ijms232113433
Uhse, S., and Djamei, A. (2018). Effectors of plant-colonizing fungi and beyond. PLoS Pathog. 14:e1006992. doi: 10.1371/journal.ppat.1006992
Vaaje-Kolstad, G., Westereng, B., Horn, S. J., Liu, Z., Zhai, H., Sørlie, M., et al. (2010). An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222. doi: 10.1126/science.1192231
Vasina, D. V., Pavlov, A. R., and Koroleva, O. V. (2016). Extracellular proteins of Trametes hirsute st. 072 induced by copper ions and a lignocellulose substrate. BMC Microbiol. 16:106. doi: 10.1186/s12866-016-0729-0
Viborg, A. H., Terrapon, N., Lombard, V., Michel, G., Czjzek, M., Henrissat, B., et al. (2019). A subfamily roadmap of the evolutionary diverse glycoside hydrolase family 16 (GH16). J. Biol. Chem. 294, 15973–15986. doi: 10.1074/jbc.RA119.010619
Vleeshouwers, V. G. A. A., Raffaele, S., Vossen, J. H., Champouret, N., Oliva, R., Segretin, M. E., et al. (2011). Understanding and exploiting late blight resistance in the age of effectors. Annu. Rev. Phytopathol. 49, 507–531. doi: 10.1146/annurev-phyto-072910-095326
Wang, S., Vetukuri, R. R., Kushwaha, S. K., Hedley, P. E., Morris, J., Studholme, D. J., et al. (2021). Haustorium formation and a distinct biotrophic transcriptome characterize infection of Nicotiana benthamiana by the tree pathogen Phytophthora kernoviae. Mol. Plant Pathol. 22, 954–968. doi: 10.1111/mpp.13072
Wang, M., Weiberg, A., Dellota, E. Jr., Yamane, D., and Jin, H. (2017). Botrytis small RNA Bc-SiR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 14, 421–428. doi: 10.1080/15476286.2017.1291112
Wang, Y., Yang, N., Zheng, Y., Yue, J., Bhadauria, V., Peng, Y. L., et al. (2022). Ubiquitination in the rice blast fungus Magnaporthe oryzae: from development and pathogenicity to stress responses. Phytopathol. Res. 4:1. doi: 10.1186/s42483-021-00106-w
Wang, C., Zheng, Y., Liu, Z., Qian, Y., Li, Y., Yang, L., et al. (2023). The secreted FolAsp aspatic protease facilitates the virulence of fusarium oxysporum f. sp. lycopersici. Front. Microbiol. 14:1103418. doi: 10.3389/fmicb.2023.1103418
Wattanasilakorn, S., Sdoodee, S., Nualsri, C., and Chuenchit, S. (2012). Screening of rubber (Hevea brasiliensis Muell. Arg.) rootstocks for the white root rot disease resistance. J. Agr. Tech. 8, 2385–2395.
Weiberg, A., Wang, M., Lin, F. M., Zhao, H., Zhang, Z., Kaloshian, I., et al. (2013). Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123. doi: 10.1126/science.1239705
Whisson, S. C., Boevink, P. C., Wang, S., and Birch, P. R. (2016). The cell biology of late blight disease. Curr. Opin. Microbiol. 34, 127–135. doi: 10.1016/j.mib.2016.09.002
Woraathasin, N., Nakkanong, K., and Nualsri, C. (2017). Cloning and expression analysis of HbPR-1b and HbPR-3 in Hevea brasiliensis during inoculation with Rigidoporus microporus. Pak. J. Biol. Sci. 20, 233–243. doi: 10.3923/pjbs.2017.233.243
Wymelenberg, A. V., Gaskell, J., Mozuch, M., BonDurant, S. S., Sabat, G., Ralph, J., et al. (2011). Significant alteration of gene expression in wood decay fungi Postia placenta and Phanerochaete chrysosporium by plant species. Appl. Environ. Microbiol. 77, 4499–4507. doi: 10.1128/AEM.00508-11
Yan, T., Zhou, X., Li, J., Li, G., Zhao, Y., Wang, H., et al. (2022). FoCupin1, a Cupin_1 domain-containing protein, is necessary for the virulence of fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 13:1001540. doi: 10.3389/fmicb.2022.1001540
Yang, H., Liao, R., Jin, C., Feng, X., Zhang, Y., Wang, X., et al. (2024). The conserved GOLD domain in the Plasmodiophora brassicae effector Pb257 is required for triggering cell death and root swelling. Phytopathol. Res. 6:18. doi: 10.1186/s42483-024-00236-x
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217, 739–755. doi: 10.1111/nph.14842
Zhang, Z., Li, Y., Luo, L., Hao, J., and Li, J. (2020). Characterization of cmcp gene as a pathogenicity factor of Ceratocytis manginecans. Front. Microbiol. 11:1824. doi: 10.3389/fmicb.2020.01824
Keywords: effectorome, Rigidoporus microporus, rubber tree, virulence, white root rot disease
Citation: Longsaward R, Viboonjun U, Wen Z and Asiegbu FO (2024) In silico analysis of secreted effectorome of the rubber tree pathogen Rigidoporus microporus highlights its potential virulence proteins. Front. Microbiol. 15:1439454. doi: 10.3389/fmicb.2024.1439454
Edited by:
Jian Chen, Jiangsu University, ChinaReviewed by:
Maryam Rafiqi, Independent Researcher, Richmond, United KingdomChristopher Brian Lawrence, Virginia Tech, United States
Copyright © 2024 Longsaward, Viboonjun, Wen and Asiegbu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fred O. Asiegbu, ZnJlZC5hc2llZ2J1QGhlbHNpbmtpLmZp
 Rawit Longsaward
Rawit Longsaward Unchera Viboonjun
Unchera Viboonjun Zilan Wen
Zilan Wen Fred O. Asiegbu
Fred O. Asiegbu