- 1Department of Animal Science, The University of Tennessee, Knoxville, TN, United States
- 2Department of Genome Science and Technology, The University of Tennessee, Knoxville, TN, United States
Introduction: The rising prevalence of Extended-Spectrum Beta-Lactamase (ESBL)-producing Klebsiella species (spp.) poses a significant threat to human and animal health and environmental safety. To address this pressing issue, a comprehensive study was undertaken to elucidate the burden and dissemination mechanisms of ESBL-Klebsiella spp. in dairy cattle farms.
Methods: Fifty-seven Klebsiella species were isolated on CHROMagar™ ESBL plates and confirmed with MADLI-TOF MS and whole genome sequenced from 14 dairy farms.
Results and discussion: Six families of beta-lactamase (bla) (blaCTX−M, blaSHV, blaTEM, blaOXY, blaOXA, and blaSED) were detected in ESBL-Klebsiella spp. genomes. Most (73%) of isolates had the first three types of beta-lactamase genes, with blaSHV being the most frequent, followed by blaCTX−M. Most (93%) isolates harbored two or more bla genes. The isolates were genotypically MDR, with 26 distinct types of antibiotic resistance genes (ARGs) and point mutations in gyrA, gyrB, and parC genes. The genomes also harbored 22 different plasmid replicon types, including three novel IncFII. The IncFII and Col440I plasmids were the most frequent and were associated with blaCTXM−27 and qnrB19 genes, respectively. Eighteen distinct sequence types (STs), including eight isolates with novel STs of K. pneumoniae, were detected. The most frequently occurring STs were ST353 (n = 8), ST469 (n = 6), and the novel ST7501 (n = 6). Clusters of ESBL-Klebsiella strains with identical STs, plasmids, and ARGs were detected in multiple farms, suggesting possible clonal expansion. The same ESBL variant was linked to identical plasmids in different Klebsiella STs in some farms, suggesting horizontal spread of the resistance gene. The high burden and dual spread mechanism of ESBL genes in Klebsiella species, combined with the emergence of novel sequence types, could swiftly increase the prevalence of ESBL-Klebsiella spp., posing significant risks to human, animal, and environmental health. Immediate action is needed to implement rigorous surveillance and control measures to mitigate this risk.
1 Introduction
Klebsiella species belongs to the genus Klebsiella in the family of Enterobacteriaceae, and the bacteria naturally reside in the digestive tracts of healthy animals and humans (Martin and Bachman, 2018). Klebsiella spp. is identified as one of the most concerning bacteria involved in carrying and disseminating genes mediating resistance to the highest priority and critically important classes of antibiotics (HPCIA), such as third-generation cephalosporins (3GCs) (Boucher et al., 2009; Navon-Venezia et al., 2017). Studies conducted in human health settings documented that Klebsiella spp. is one of the most effective bacteria in disseminating extended-spectrum beta-lactamase (ESBL) genes, which confer resistance to 3GC. The bacteria transfer ESBL genes vertically to their subsequent generation and horizontally by acting as donors via mobile genetic elements (MGEs) (Woodford and Livermore, 2011; Wyres and Holt, 2018).
Klebsiella spp. is recognized as one of the most frequent causes of community and hospital-associated infections caused by extended-spectrum beta-lactamase (ESBL)-producing bacteria in Europe (Navon-Venezia et al., 2017) and the US (CDC, 2019). The US Centers for Disease Control and Prevention (CDC) recently reported a 50% increase in ESBL-producing Enterobacteriaceae infections over six years from 2012 to 2017 (CDC, 2019). Surprisingly, nearly half of these cases were community-associated and had no prior history of healthcare exposure. This rapid increase has led the CDC to designate ESBL-producing Enterobacteriaceae, including Klebsiella spp., as a “serious threat” to public health (CDC, 2019). However, as almost all previous studies on ESBL-producing Klebsiella spp. were focused on humans (Tadesse et al., 2012; Davis and Price, 2016; Martin and Bachman, 2018), the cause for the rapid increase in ESBL-producing Enterobacteriaceae infections in community settings is not yet understood.
As a common colonizer of dairy cattle’s gastrointestinal tract (GIT) and one of the causative agents of coliform mastitis, Klebsiella spp. is frequently exposed to beta-lactam antibiotics, especially to 3GCs, which is often used for prophylactic and therapeutic purposes in dairy cattle. Previous studies showed that the prevalence of ceftiofur, a veterinary 3GC-resistant Enterobacteriaceae, is significantly higher in dairy cattle than in humans and other food animals, such as pigs and chickens (Tadesse et al., 2012). A recent molecular study indicated a similarity between K. pneumoniae isolates from animals and humans, suggesting inter-species transmission of the bacteria (Davis and Price, 2016).
The widespread presence of Klebsiella spp. in the GIT of dairy cattle, combined with the frequent use of ceftiofur, a 3GC, in dairy farms, may render dairy cattle a potential reservoir for ESBL-producing Klebsiella species (Gelalcha et al., 2022). However, this bacterium has been largely overlooked and understudied in dairy cattle farms. As a result, information on the burden of ESBL-Klebsiella spp. on dairy farms and the possible mechanisms of its transmission within and between farms is unknown.
Without a detailed understanding of the burden and mechanisms of the spread of these bacteria within and between farms, it is challenging to effectively control its transmission from dairy farms to humans through different routes. Thus, we hypothesized that dairy cattle may serve as a potential reservoir for ESBL-producing Klebsiella spp. and ESBL genes that spread vertically and horizontally in the dairy production system. Therefore, this study aimed to identify ESBL genes and other co-carried resistance genes in Klebsiella species and understand how these resistance genes spread through clonal expansion or horizontal transfer.
2 Materials and methods
2.1 Sample collection and laboratory analyses
We conducted a cross-sectional study in 14 East Tennessee dairy farms. We obtained isolates from various sources, including rectal fecal samples using sterile rectal gloves from dairy cows (n = 424) and calves (n = 84) and farm environmental samples of manure (n = 30), feed (n = 15), and water (n = 19). Feed and water samples were pooled from multiple feeding and watering troughs on the farm and placed in sterile containers. Manure samples were pooled from different farm pens and placed in sterile collection tubes. All samples were transported to the laboratory in coolers to maintain appropriate temperature and prevent bacterial growth. The samples were processed and plated directly on CHROMagar™ ESBL (DRG International, Inc., Springfield, NJ, USA) as described in Gelalcha et al. (2023a). Presumptive ESBL-Klebsiella spp. were confirmed using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) as described in our previous publications (Gelalcha et al., 2023a,b). Chromosomal DNA was extracted from all MALDI-TOF-confirmed ESBL-Klebsiella spp. using Qiagen MagAttract HMW DNA kit (Qiagen, Germantown, MD, USA) as described in Gelalcha et al. (2023b). The genomic DNA libraries were prepared using the Nextera XT DNA Library prep kit from Illumina Corporation (Illumina Inc., San Diego, CA, USA), following the manufacturer’s instructions. Whole genome sequencing (WGS) was conducted on Klebsiella isolates (n = 56) using Illumina NovaSeq PE250 at the University of Tennessee Genomics Core (Knoxville, TN, USA). This study was approved by the University of Tennessee’s Institutional Animal Care and Use Committee (IACUC) Registration Number: 2782-0720.
2.2 Statistical data analyses
The collected raw data was entered into Microsoft Excel for Windows 10 (Microsoft Corp., Redmond, WA, USA) before being transferred to SPSS (SPSS Statistics for Windows, Version 27.0, IBM Corp, Armonk, NY, USA) for statistical analysis. Descriptive (frequency) statistics were used to summarize the collected data.
2.3 Whole genome sequence data processing and analyses
The whole genome sequence data analysis involved demultiplexing of the sequence data, followed by quality assessment using FastQC v0.11.9 (Babraham Bioinformatics, 2024) and multiQC v1.14 (Ewels et al., 2016). Trimmomatic v0.39 was used to remove adapters, filter, and trim raw reads that had low-quality (Bolger et al., 2014). Preprocessed reads of each isolate were then subjected to de novo assembly using the default parameters of the SPAdes genome assembler v3.15.5 (Bankevich et al., 2012). BBMap (version 39.06) was used to filter contigs shorter than 500 bp.1 QUAST (v5.2.0) was used to generate basic genome assembly statistics (Mikheenko et al., 2018). For downstream analysis, PlasmidSPAdes modes were used to assemble only the plasmids. Several web-based online bioinformatics tools were used to analyze the genetic characteristics of bacterial isolates. To confirm bacterial spp. KmerFinder 3.22 (Hasman et al., 2014; Larsen et al., 2014; Clausen et al., 2018) and https://pubmlst.org/bigsdb?db=pubmlst_rmlst_seqdef_kiosk were used. Antibiotic resistance genes (ARG) and mutations conferring resistance were detected using3 (Zankari et al., 2017; Bortolaia et al., 2020) and CARD4 (Alcock et al., 2020). Plasmids associated with ARGs were identified using plasmid finder5 (Camacho et al., 2009; Carattoli et al., 2014), and their sequence types were characterized using pMLST 2.06. Mobile genetic elements (including plasmids) associated with ARGs were identified using MobileElementFinder7 (Camacho et al., 2009; Zankari et al., 2012; Joensen et al., 2014). The ESBL-Klebsiella spp. sequence type (ST) was determined based on seven housekeeping genes (gapA, infB, mdh, pgi, phoE, rpoB, and tonB) as described in Diancourt et al. (2005). The ST of the Klebsiella spp. were assigned using tools at the Center for Genomic epidemiology,8 and the Klebsiella Pasteur MLST sequence database.9 The combination of the best matching allele was selected to determine the sequence type of each Klebsiella spp.
Phylogenetic analyses of the ESBL-Klebsiella species isolates were automated using GToTree (version 1.8.6) (Lee, 2019). The GToTree default parameters were used to build the tree based on nucleotide sequence alignment using “Bacteria” genes. GCF 000240185.1 K. pneumoniae HS11286 was used as the reference genome. Interactive Tree of Life (iTOL version 6) was used to visualize the tree interactively online.
The Klebsiella species genomes have been submitted to the Pasteur Institute bacterial isolate Genome Sequence Database BIGSdb_20240715165555_3499682_34488.
3 Results
3.1 Microbiological and whole genome sequence (WGS) result of Klebsiella species
Using MALDI-TOF MS, we identified 57 Klebsiella spp. consisting of four distinct species from 14 farms (Supplementary Table 1). These include K. pneumoniae (n = 52), K. variicola (2 isolates), K. oxytoca (2), and K. aerogenes (1).
Of the 57 Klebsiella isolates identified, 56 were subjected to WGS. The MALDI-TOF MS test misidentified two K. pneumoniae isolates as K. variicola, later confirmed by WGS analysis. Therefore, the total number of K. pneumoniae isolates sequenced was 53. Two isolates initially identified as K. oxytoca by MALDI-TOF MS from the same farm were later determined to be K. michiganensis through WGS analysis. One K. michiganensis genome was contaminated with the E. coli genome (43%) and excluded from further downstream WGS analysis. Thus, further genomic analysis was carried out on the remaining 55 Klebsiella spp (53 K. pneumoniae and one from K. michiganensis and K. aerogenes). Summary statistics related to Klebsiella species genome assembly, such as coverage, number of contigs, N50, and others, were given in Supplementary Figure 1.
3.2 Genetic determinants of resistance to beta-lactam antibiotics
All sequenced isolates had one or more kinds of ESBL-encoding genes. This study detected six families of beta-lactamase genes from the ESBL-Klebsiella spp. genomes. These include blaCTX−M, blaSHV, blaTEM, blaOXY, blaOXA, and blaSED. The majority (73%; 40/55) of the isolates harbored three types of beta-lactamase genes, namely balCTX−M, blaSHV, and blaTEM, in their genomes. Only five isolates carried a single kind of beta-lactamase gene, four of which had blaSHV−2, and the remaining one had blaCTX−M. The remaining ten isolates had two beta-lactamase genes in their genomes. In this manuscript, we will focus only on ESBL variants and inhibitor-resistant beta-lactamase genes in each family.
Interestingly, thirteen isolates of K. pneumonia had inhibitor-resistant blaSHV genes, either blaSHV−56 or blaSHV−26 (Table 1). In addition, inhibitor-resistant blaTEM variant beta-lactamases, including blaTEM−33, blaTEM−34, blaTEM−35, blaTEM−36, and blaTEM−122, were each detected in five separate isolates.
Regarding distribution within isolates, blaSHV was the most frequent and was detected in 53 isolates, followed by blaCTX−M and blaTEM, which were detected in the genomes of 51 and 40 isolates, respectively. Furthermore, blaSHV exhibited the highest level of diversity with 30 identified variants. However, only 40% (n = 12) of blaSHV variants were ESBL. In most cases (41 out of 53 isolates), multiple variants of blaSHV were found clustered together on the same contigs within the genome of an isolate, creating a resistance island.
Of the 51 isolates containing blaCTX−M, only three had more than one variant of the blaCTX−M gene, while 48 had only one variant. However, two Klebsiella isolated from the same farm harbored six distinct variants of the blaCTX−M genes per genome (Table 1). These variants include blaCTX−M−27, blaCTX−M−146, blaCTX−M−32, blaCTX−M−61, blaCTX−M−138, and blaCTX−M−1. All these genes were located on the same contigs except for blaCTX−M−27. In this study, blaCTX−M−27 was the most frequent blaCTX−M allele, followed by blaCTX−M−1 and blaCTX−M−65 (Table 1). Among the two rare Klebsiella spp. reported in this study, K. michiganensis had blaCTX−M−1 and blaOXY−1−7 genes, while K. aerogenes had blaCTX−M−65.
Among 24 different variants of blaTEM, the narrow spectrum blaTEM−1B was detected in 97.6% (40/41) of Klebsiella isolates carrying this gene, making it the most prevalent variant. Only two isolates possessed blaTEM−1c. The remaining 22 blaTEM variants were found in just one Klebsiella isolates each. Among the detected blaTEM, only three of them, blaTEM−16, blaTEM−29, and blaTEM−163, were categorized as ESBL variants. One isolate harbored seventeen blaTEM variants, including four inhibitor-resistant beta-lactamases (blaTEM−33, blaTEM−34, blaTEM−35, and blaTEM−36) and fourteen other variants (indicated with the superscript “e” in Table 1) on a contig. Similarly, other Klebsiella isolates from the same farm harbored nine blaTEM variants, all co-located on the same contigs (the genes are indicated with the superscript “f” in Table 1).
3.3 Detection of horizontally transferable resistance genes in ESBL-Klebsiella isolates
Apart from beta-lactamase genes, Klebsiella spp. harbored resistance genes against eight different classes of antibiotics in their genomes (Table 2). All 55 Klebsiella isolates were genotypically multidrug resistant, carrying resistance genes to as high as seven classes of antibiotics in their genomes. In addition to the beta-lactamase genes, about 26 resistance genes mediating resistance to about eight classes of antibiotics were detected. These include resistance genes against (fluoro)quinolones, macrolides, aminoglycosides, folate pathway inhibitors, tetracycline, chloramphenicol, lincosamides, and phosphonic acids.
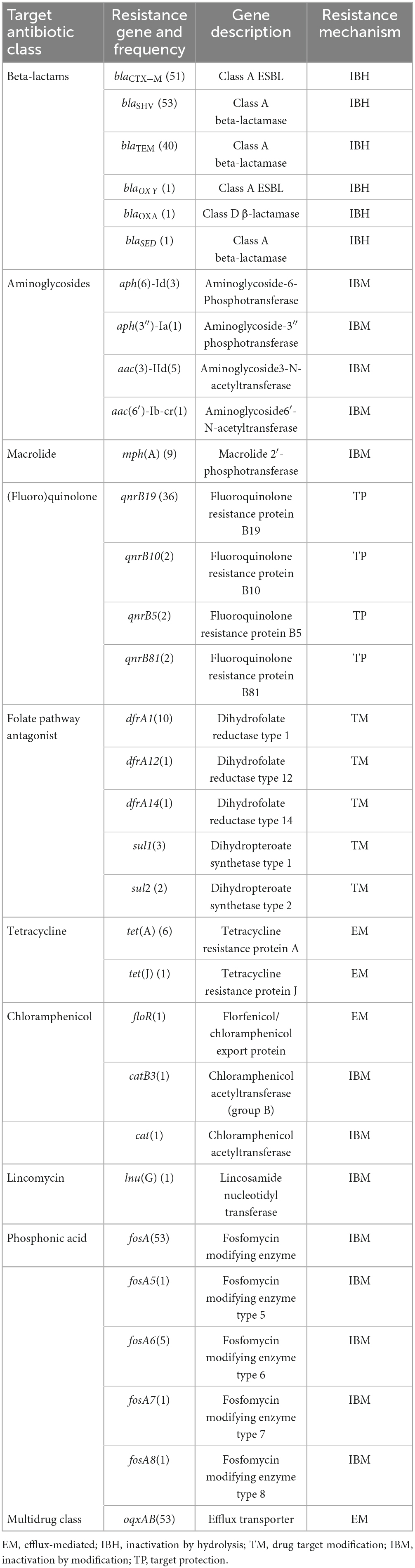
Table 2. Horizontally transferable resistance genes co-harbored with beta-lactamase genes in Klebsiella species.
Multiple variants of resistance genes against specific classes of antibiotics were detected, such as five variants of each of the fosfomycin and sulfonamide resistance genes and four variants of each of the aminoglycoside and quinolone resistance genes. Among these genes, the most frequently occurring ones were the fosfomycin resistance gene (fosA) and multidrug efflux pump encoding genes (oqxAB), present in the genome of 53 isolates. The plasmid-mediated fluoroquinolone resistance gene, qnrB19, was detected in 36 Klebsiella isolates.
Isolates from the same farm exhibited similar resistance gene patterns. For instance, all isolates from Farm B had the same resistance pattern, with respect to the blaCTX−M variant type (blaCTX−M−1), the presence of macrolide resistance gene mph(A), and sulfonamide resistance gene (dfrA1). Similarly, all isolates from Farm J had blaSHV−2 genes (no blaCTX−M or dfrA1). In contrast, identical sequence types between farms showed the same resistance pattern. For example, ST353 detected in three farms (Farm A, B, and J) carried the same gene variants, including blaCTX−M−1, mph(A), blaSHV, dfrA1, qnrB19, and fosA.
The most frequent chromosomal mutations detected in ESBL-Klebsiella genomes were DNA gyrases (gyrA and gyrB) and topoisomerase IV (parC). Multiple mutations that mediate fluoroquinolone resistance, gyrA, and parC were detected in all 55 Klebsiella spp. genomes.
3.4 Plasmid replicon types and their association with resistance genes
From the WGS data of 55 ESBL-Klebsiella isolates obtained from six farms, 22 different plasmid replicon types were identified. On average, each ESBL-Klebsiella isolate harbored five different kinds of replicons, with a minimum of two and a maximum of nine replicon types. Isolates with similar genotypes tend to have similar numbers and replicon types of plasmids. The most frequently detected replicon types in this study were IncF family plasmids (IncFII, IncFIA, and FIB) and Col plasmids such as Col440I, Col(IMGS31), Col(MG828), ColRNAI, Col(pHAD28), ColpVC, Col(BS512), and Col156. Additionally, a limited number of isolates were found to harbor broad-host-range plasmids, including IncH plasmids (such as ncHI2A and IncHI2), IncN, and IncR.
Most (96.4%) of the Klebsiella isolates’ genomes harbored the Col440I plasmid. Among the narrow host range, IncF family plasmids, including FII, IncFIA, and FIB replicon types, were highly prevalent, with up to four subtypes of these plasmids present in each isolate. Among the IncF family plasmids, the IncFII plasmid replicon types were the most frequently detected (89%), followed by IncFIB (85%). Notably, all IncFII plasmid replicon types were found to have the same sequence type, ST F2: A−: B−.
The broad host range plasmid IncN was detected in all nine K. pneumoniae with identical STs obtained from three farms (Farms A, B, and G) and K. michiganensis strain obtained from Farm L. The IncN plasmid was identified as ST1 in all these isolates and was primarily linked with blaCTXM−1. On the other hand, the IncH plasmids (IncHI2A and IncHI2) were detected in only three Klebsiella isolates with identical genotypes (ST111), which carried blaCTXM−65. The IncN plasmid was not detected in the remaining 45 isolates obtained from the other farms. The IncR plasmids were identified in only two K. pneumoniae isolates in a single farm but were not associated with any of the resistance genes identified in this study. Three Novel IncF RSTs (due to novel FIA allele) were detected in three Klebsiella isolates obtained from the same farm. Two of them have the same nearest Sequence type, K4: A−: B−, and the ST closest to the third novel allele was K7: A−: B−.
3.5 Association between resistance genes and mobile genetic elements
Most isolates (88.5%; 46/52) containing blaCTX−M genes were found to have these genes associated with mobile genetic elements (MGEs). In 80% (37/46) of the isolates, blaCTX−M was associated with plasmids, while in the remaining 20%, it was associated with insertion sequences (IS) of different types. In 67.4% (31/46) of the isolates, the blaCTX−M−27 and blaTEM−1B genes co-occurred and were associated with epidemic resistance plasmids, IncFII, which had identical plasmid sequence types, STF2: A−: B−. In six isolates obtained from three different farms, the genes blaCTX−M−1 and mph(A) were both associated with broad host range plasmids of the IncN type, which were subtyped as ST1. The gene blaCTX−M−65, often detected in ST111, was associated with IncHI2 ST3, IS102, or IS26. Other resistance genes, such as aph(6)-Id, aph(3′′)-Ib, and sul2, were also co-harbored with different variants of blaCTX−M on the same MGEs. Similarly, other resistance genes were often associated with MGEs (for example, qnrB19 was associated withol440I plasmids in 73% of the isolates harboring this resistance gene). Additionally, the tet(A) gene was often found to be associated with broad host range plasmids of the IncHI2A type with ST3 (Table 3).
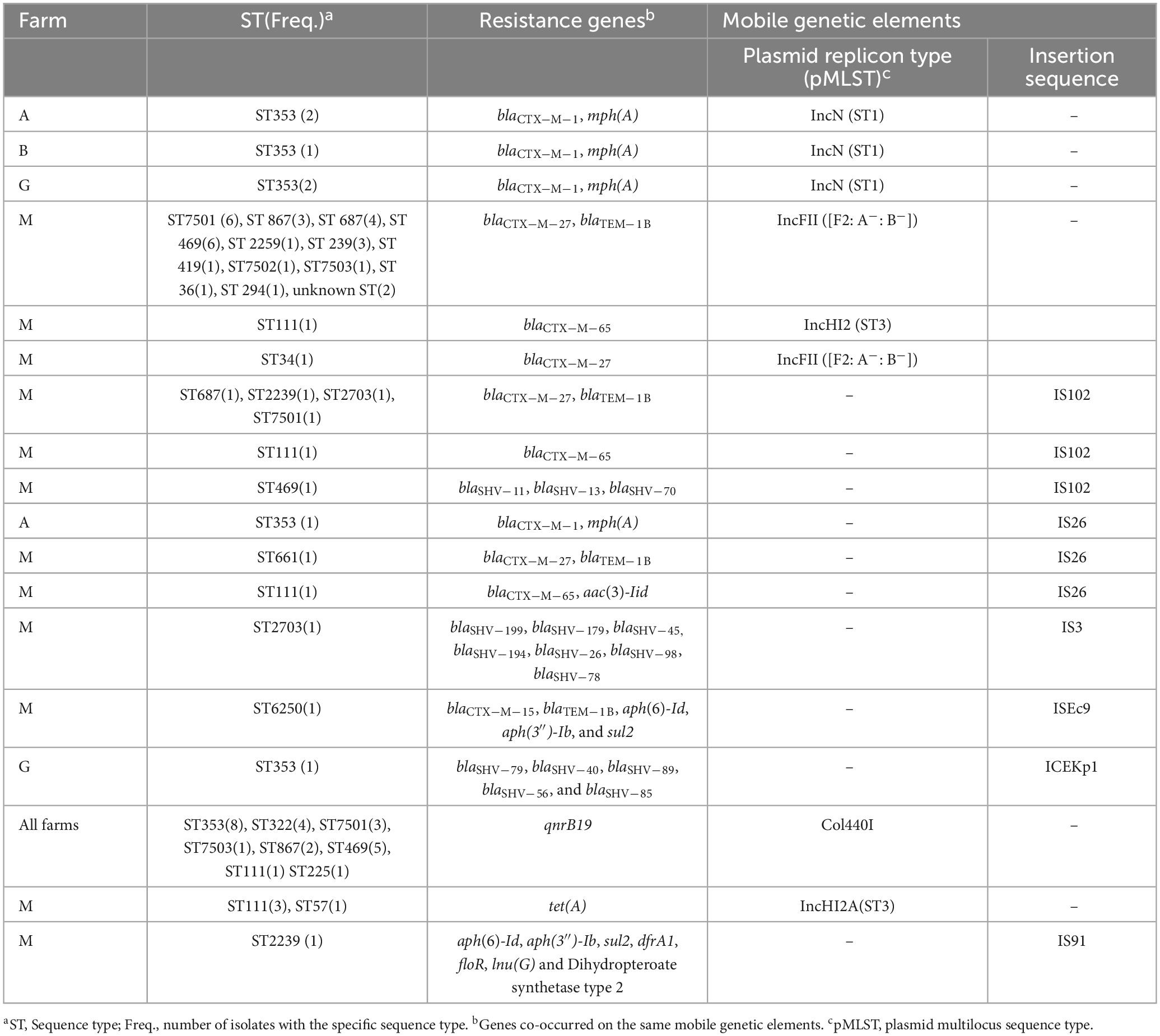
Table 3. Association between Klebsiella species sequence types, resistance genes, and mobile genetic elements.
3.6 Genetic diversity and possible spread mechanisms of ESBL-Klebsiella species in the dairy farms
Eighteen distinct sequence types (STs) of K. pneumoniae were identified, including three novel STs (ST7501, ST7502, and ST7503) detected in eight samples. The most frequently occurring STs were ST353 (n = 8), ST469 (n = 6), the novel ST7501 (n = 6), ST687 (n = 5), ST322 (n = 4), ST111 (n = 4), ST2239 (n = 4), ST867 (n = 3), and ST225 (n = 2). The other seven STs were observed only once, including ST7502, ST7503, ST34, ST6250, ST2703, ST36, ST294, ST661, and ST419. The ST K. aerogenes was identified as ST57. The ST of K. michiganensis was unknown and could not be assigned as the https://bigsdb.pasteur.fr database handles only the K. pneumoniae species complex. The remaining five K. pneumoniae isolates could not be assigned to specific sequence types because their genome assembly statistics did not meet the criteria set by the database, and alternative databases8 provided ambiguous results.
ESBL-producing Klebsiella spp. were found to spread both clonally through specific Klebsiella spp. STs, and horizontally, via plasmids between genetically unrelated Klebsiella spp. Some of the Klebsiella STs showed both within-farm and between-farm similarity. ST353, for example, was found in three different farms: three isolates, each from farms A and B, and two isolates from farm G. All these isolates carry the same ESBL variant (blaCTX−M−1) associated with plasmids with identical sequence types, IncN (ST1). Farms A and B are in the same county, but in different cities, and Farm G is in a different county.
On the other hand, some STs were only detected within a single farm. For instance, all four ST322 isolates were found exclusively in farm J and had the same blaSHV-2 variants. One of these STs was isolated from a water sample collected from water troughs.
Farm M had the largest number of sequenced Klebsiella isolates (76.4%; 42/55) and displayed the greatest diversity regarding Klebsiella STs. It contained 13 distinct sequence types of K. pneumoniae and ten unknown STs. The most common STs in this farm include the novel ST7501 (n = 6), ST469 (n = 6), and ST687 (n = 5), followed by ST111 and ST2239, each occurring four times in the farm. Despite the genetic diversity, most isolates in farm M harbored indistinguishable plasmid (e.g., IncFII STF2: A−: B−) associated with the same variants of ESBL (blaCTX−M−27). The same ESBL variant was also detected associated with the same plasmid sequence types among diverse STs of Klebsiella spp. A similar pattern was also observed for other resistance genes, such as qnrB19 and tet(A) (Table 3). In addition, the phylogenetic tree displayed several clusters of Klebsiella isolates within a farm, and some isolates from a different farm were also clustered together (Figure 1).
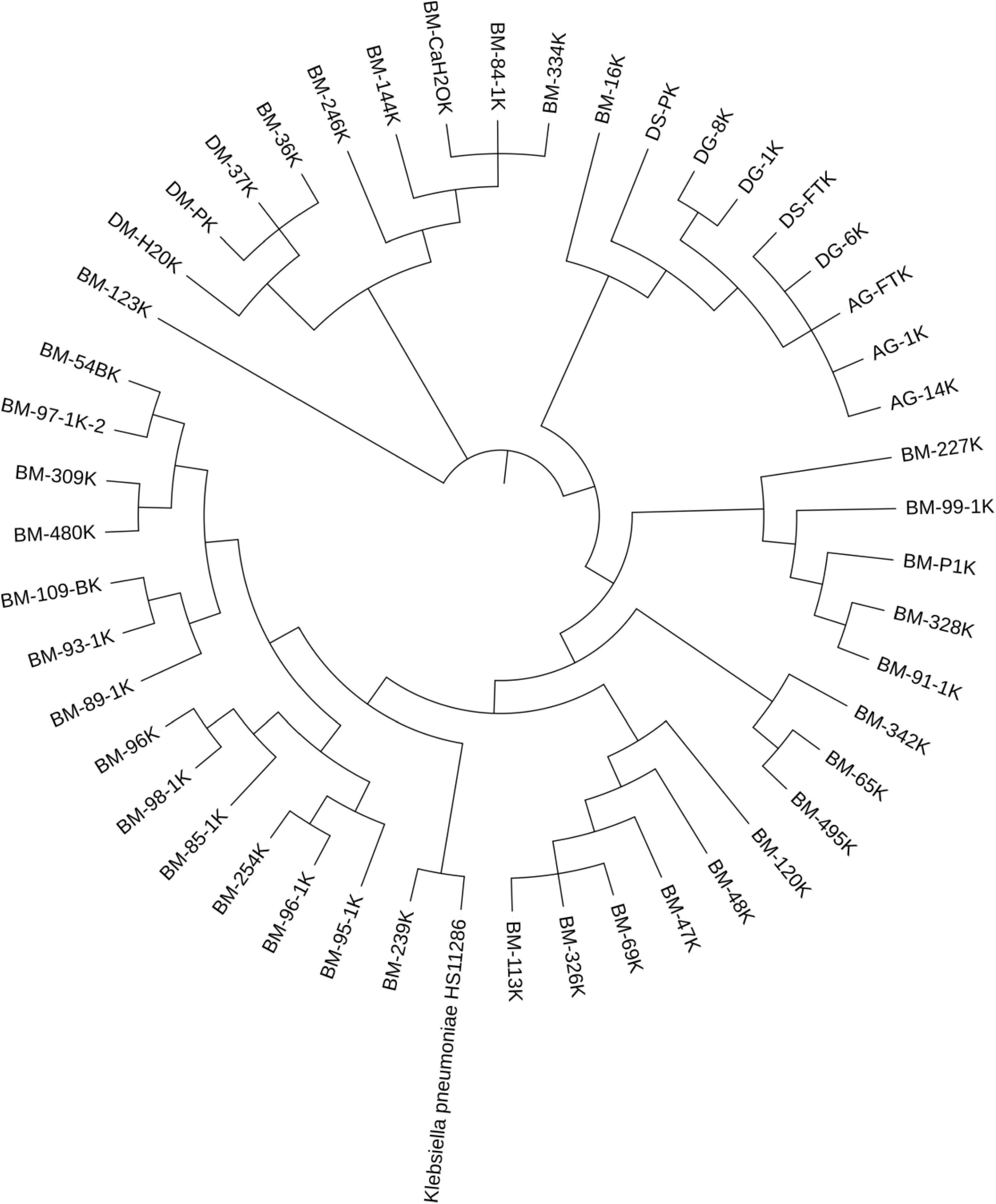
Figure 1. Phylogenetic tree of ESBL-Klebsiella pneumoniae. Isolates sharing the same first two letters were collected from the same farm. The letters indicate the farm and county, while the numbers represent the individual animal’s ID within the farm (except manure, feed, and water samples). Most of the isolates were obtained from the same farm, and they showed multiple clustering, with isolates in each cluster having identical sequence types.
4 Discussion
The detection of at least one ESBL-encoding gene in all sequenced Klebsiella spp. generally concurs with the CHROMagar™ ESBL screening result, which shows the high sensitivity and specificity of the chromogenic agar. In this study, detecting six distinct beta-lactamase gene families in Klebsiella genomes indicates that these bacteria have an arsenal of genes to inactivate beta-lactam antibiotics and can serve as a reservoir of these genes (Navon-Venezia et al., 2017; Wyres and Holt, 2018). This is particularly problematic given that these resistance genes can transfer from Klebsiella spp. to other pathogenic bacterial species that cause life-threatening infections in humans and animals that are difficult to treat with available antibiotics (Wyres and Holt, 2018). This finding agrees with the previous report that indicated Klebsiella spp. is a “key source and trafficker” of resistance genes, including ESBL genes (Navon-Venezia et al., 2017; Wyres and Holt, 2018). A similar level of ESBL burden was reported from Klebsiella spp. isolated from a food animal in China (Wu et al., 2016).
Among the 30 blaSHV variants, The ESBL variants such as blaSHV−27, −13, and −70 occurred frequently, and blaSHV−27 occurred alone, whereas the remaining three co-occurred, suggesting they were relatively widespread among the Klebsiella isolates in the farms. We did not find previous reports of blaSHV ESBL variants from Klebsiella spp. in US dairy farms. However, a study conducted in Oklahoma in 2005 detected blaSHV−1 from K. pneumoniae isolated from cattle farms. It is unclear whether these were beef or dairy farms (Kim et al., 2005). In addition, blaSHV variants have been reported in Klebsiella spp, isolated from humans in the US (Yigit et al., 2001; Paterson et al., 2003). Some blaSHV variants identified in this study, such as blaSHV−11, have previously been reported in E. coli and K. pneumoniae isolated from dairy farms in Japan (Ohnishi et al., 2013). In contrast, others, like blaSHV−2, have been reported in E. coli isolated from beef cattle in Canada (Cormier et al., 2016).
The identification of the blaCTX−M gene as the second most prevalent ESBL gene in the Klebsiella genomes (93%) supports the recent studies that showed this gene family is rapidly disseminating among ESBL-producing Enterobacteriaceae (Zhao and Hu, 2013; Bevan et al., 2017; Irrgang et al., 2017; Afema et al., 2018).
Eight blaCTX−M gene variants were detected in this study most of which were reported previously from other members of Enterobacteriaceae, mainly from E. coli isolates from US dairy farms. For instance, blaCTX−M−1 was reported from K. pneumoniae isolates from mastitis milk in Iowa and New York (Yang et al., 2019; Zheng et al., 2022). The blaCTX−M−1 was also detected from E. coli and Salmonella isolates retrieved from dairy cattle fecal samples obtained from Ohio, Washington, Texas, and the Southwestern region of the US, suggesting that it is a common blaCTX−M variant in dairy farms (Dunne et al., 2000; Cummings et al., 2009; Taylor et al., 2020; Carey et al., 2022). Due to selective pressure, it is possible that K. pneumoniae acquired this gene through horizontal transfer from E. coli or other sources.
Similarly, blaCTX−M−15, blaCTX−M−27, blaCTX−M−32, and blaCTX−M−65 were reported from E. coli isolated from fecal samples in different parts of the US, including Washington, Texas, and the Southwestern region of the country (Donaldson et al., 2006; Cummings et al., 2009; Afema et al., 2018; Taylor et al., 2020; Carey et al., 2022). The blaCTX−M−61, blaCTX−M−146, and blaCTX−M−138 detected in this study were not previously reported from US dairy farms. In addition, except blaCTX−M−1, all other blaCTX−M variants were not reported previously from Klebsiella isolates from the US dairy farms.
Among the detected blaCTX−M genes, blaCTX−M−27 was the most prevalent (detected in 67% of the Klebsiella spp. genomes) but limited to a single farm, whereas blaCTX−M−1 was relatively widespread and detected in five farms. Unlike this study, an earlier study reported that blaCTX−M−15 is the dominant variant, followed by blaCTX−M−27 in ESBL-E. coli isolates from US dairy farms (Afema et al., 2018). However, blaCTX−M−15, which was detected only in one Klebsiella isolate in the current study, was reported as the most prevalent blaCTX−M variant detected in Klebsiella isolates from humans in different parts of the US (Castanheira et al., 2014), and elsewhere (Bonnet, 2004; Calbo and Garau, 2015). In addition, blaCTX−M−15 was also reported from ESBL-Klebsiella pneumoniae in France (Haenni et al., 2014), ESBL-K. ozaena in Italy (Stefani et al., 2014) and ESBL-K. oxytoca in Egypt (Ahmed and Shimamoto, 2011) that were isolated from dairy cows.
In this study, blaTEM was another frequently detected beta-lactamase gene. Most blaTEM variants detected in this study have not been functionally characterized to determine whether ESBL or non-ESBL (Table 1). The ESBL detected in this study include blaTEM −16, −29, and −163. The narrow-spectrum blaTEM−1B variant was found in 95% of ESBL-Klebsiella isolates carrying the blaTEM gene, but further research is needed to determine if it provides a selective advantage to the host bacteria.
Most Klebsiella isolates harbored only one blaTEM variant, except two from the same farm, which had 17 and 9 variants each on the same contig, likely due to horizontal gene transfer or accumulation of mutation due to persistent exposure to antibiotics, raising concerns about the spread of these genes via MGEs (Carattoli, 2013).
To our knowledge, no previous studies in the US reported the blaTEM gene from Klebsiella spp. in dairy farms. However, a study from Pennsylvania reported blaTEM in E. coli, but the variant specification was not provided (Donaldson et al., 2006). Various blaTEM gene was also reported from K. pneumoniae isolates from humans in the US (Yigit et al., 2001; Paterson et al., 2003). An extensive European review revealed that several studies documented blaTEM variants different from those observed in the current study in E. coli isolates from dairy cattle (Dantas Palmeira and Ferreira, 2020). The lack of previous reports of blaTEM and blaSHV genes in Klebsiella isolates from US dairy farms likely reflects limited research rather than their actual absence, highlighting the need for ongoing surveillance of bacterial resistance in food production environments.
The rare β-lactamase genes, blaOXA−1 and blaSED−1 were detected in K. pneumoniae, whereas blaOXY 1−7 was in K. michiganensis. To our knowledge, this is the first study to identify these resistance genes in Klebsiella spp. from US dairy farms. The blaSED−1 gene naturally occurs in Citrobacter sedlakii (Petrella et al., 2001; Naas et al., 2017). Thus, K. pneumoniae may have acquired this gene via horizontal gene transfer from C. sedlakii in the animal GIT. K. michiganensis is a recently described novel member of the K. oxytoca complex (Saha et al., 2013; Yang et al., 2022). The detection of blaOXY 1−7 variants of ESBL aligns with previous studies that reported that K. oxytoca inherently carries blaOXY, a chromosomally encoded class A beta-lactamase gene (Fournier et al., 1994). However, plasmid-associated blaOXY was recently reported from K. oxytoca in Spain (Gonzalez-Lopez et al., 2009). This gene was not associated with plasmid or any other MGEs in this study, suggesting it is a chromosomal beta-lactamase gene.
In addition to beta-lactamase genes, the genomes of ESBL-Klebsiella isolates carry resistance genes for up to seven classes of antibiotics. This finding concurs with previous studies conducted in humans, animals, and environmental samples that showed ESBL-producing Enterobacteriaceae carried multiple resistance genes to structurally unrelated classes of antibiotics (Kim et al., 2005; Cantón and Coque, 2006; Tacao et al., 2014; CDC, 2019; Peirano and Pitout, 2019; Gelalcha et al., 2022; Gelalcha and Kerro Dego, 2022). Like previous studies (Kim et al., 2005; Taylor et al., 2020; Carey et al., 2022), genes mediating resistance to CIAs, such as aminoglycoside modifying genes, macrolide modifying genes, and plasmid-mediated quinolone resistance genes (PMQR), were detected in many isolates. The use of 3GC (ceftiofur), fluoroquinolones (enrofloxacin), and macrolides (erythromycin) in dairy calves might result in the co-selection and spread of these resistance genes in Enterobacteriaceae in dairy farms (Carey et al., 2022).
The detection of these ARGs against CIAs in many isolates is concerning, as the horizontal transfer of these genes from commensal Klebsiella spp. to other enteric pathogens may impede the treatment of infections caused by these pathogens (Leclercq, 2002; Poirel et al., 2012; Zhang et al., 2021). The PMQR, qnrB, was found in 36 (66%) isolates, making it the most frequent ARG to CIAs. This finding is consistent with previous studies that reported frequent co-occurrence of ESBL and fluoroquinolone resistance genes in Enterobacteriaceae (Paterson et al., 2000; Wener et al., 2010; Al-Assil et al., 2013).
The most frequently detected ARGs on the Klebsiella genome with the ESBL gene were the fosfomycin resistance gene (fosA) and a multidrug efflux pump (oqxAB) gene, each present in 53 (96%) of the isolates. Since its first report in 2004 in E. coli isolates from swine in Europe (Cantón and Coque, 2006; Gelalcha and Kerro Dego, 2022), studies have shown that the oqxAB gene is increasing among Enterobacteriaceae (Perez et al., 2013; Wong et al., 2015). According to an international report on ESBL-producing K. pneumoniae, the oqxAB gene was found in about 88% of isolates from various hospitals in different countries, including the US (Perez et al., 2013). In a separate study of K. pneumoniae clinical isolates from Ohio, oqxAB was also detected in most isolates (Perez et al., 2013). The co-occurrence of the oqxAB and ESBL genes has significant veterinary and human health implications. This gene is often associated with MGE and encodes a multidrug efflux pump that confers resistance to multiple classes of antibiotics, including disinfectants (Li et al., 2019).
The high prevalence of the fosA gene, which confers resistance to fosfomycin in Klebsiella spp., is worrisome. Fosfomycin is a promising broad-spectrum antibiotic used in the US to treat MDR Gram-negative organisms such as ESBL-producing Enterobacteriaceae and carbapenem-producing K. pneumoniae (Sastry and Doi, 2016). This resistance gene is considered rare in the US and was reported seven years ago from a clinical ESBL-E. coli from a woman patient in Pennsylvania (Alrowais et al., 2015). Thus, detecting fosfomycin resistance genes in 96% of Klebsiella spp. found in the GIT of dairy cattle and the farm environment is concerning because the same genes may spread to human pathogenic strains through MGEs, or the bacteria may spread to humans via the food chain or direct contact with carrier animals (Arca et al., 1997; Hou et al., 2013; Wang et al., 2017). Studies showed that fosA genes are relatively common in Eastern Asia, particularly China. They were detected from ESBL-E. coli isolates from food animals such as dairy cattle and pets (Hou et al., 2012, 2013; Wang et al., 2017). More investigation is required to determine the origin of these genes in commensal Klebsiella spp. since fosfomycin is not used in US dairy farms.
Apart from horizontally transferable genes, multiple chromosomal mutations that affect quinolone resistance-determining regions (QRDRs) (gyrA, gyrB, parC) were detected. All 55 Klebsiella spp. genomes had QRDRs, including gyrA and parC, the primary resistance mechanisms to (fluoro)quinolones in Gram-negative bacteria, including Klebsiella spp (Geetha et al., 2020).
This study found that ESBL-Klebsiella isolates had diverse plasmids, with an average of five replicon types per isolate. In addition, this study also identified three novel plasmid STs of the IncF family (FIA replicon types). The discovery of new plasmid STs can help understand the spread and evolution of ARGs or plasmids and control the spread of antibiotic resistance in different environments (Villa et al., 2010; Shin et al., 2012).
In this study, both narrow-host range or epidemic resistance plasmids (ERPs) of the IncF family (IncFII, IncFIA, and FIB) and broad-host-range plasmids (IncHI2A, IncHI2, IncN, and IncR) were detected. Previous studies also reported similar replicon types of plasmids in ESBL-E. coli isolated from dairy farms in the US (Afema et al., 2018). After the Col440I plasmid, the conjugative narrow host range, or ERPs of IncF families, were highly prevalent (seen in 89% of the Klebsiella genomes), with up to four subtypes of this plasmid present in each isolate. The high prevalence of ERPs in Klebsiella spp. is concerning as previous studies showed that these families of plasmids could easily acquire resistance genes and rapidly spread throughout Enterobacteriaceae, particularly among related genotypes (Carattoli, 2009; Mathers et al., 2015).
In this study, the IncFII replicon type with ST F2: A−: B− was most frequent and was mainly associated with two co-occurring beta-lactamase genes, blaCTX−M−27 and blaTEM−1B, in over 56% (31/55) of sequenced Klebsiella isolates. Interestingly, these IncFII replicon type-beta-lactamase variant combinations were detected in isolates obtained from the same farm, suggesting that this specific subtype of plasmid may contribute to the spread of the two ARGs in Klebsiella on the farm. A previous study from the US dairy farm also reported that IncF plasmid families were associated with blaCTX−M−27 in E. coli (Afema et al., 2018).
The study identified the IncN plasmid ST1 linked to blaCTXM−1− in six clonally related K. pneumoniae isolates from three farms and one K. michiganensis strain from a fourth farm. In six Klebsiella isolates, balCTX−M−1 co-occurred with the mph(A) gene on the identical plasmids, IncN ST1. Similarly, a previous study from New York dairy farms reported the co-occurrence of blaCTX−M−1 and mph(A) on IncN plasmids in Klebsiella spp. isolated from cows with mastitis (Yang et al., 2019). The presence of the balCTX−M−1 gene and other ARGs on identical plasmids can facilitate the emergence and persistence of MDR through co-selection (Tyson et al., 2019). In addition, unlike the present study, a previous study on US dairy cattle reported IncN plasmid in association with other blaCTX−M genes (e.g., blaCTXM−15, 27, and −65) in E. coli (Afema et al., 2018). Another broad-host-range plasmid, IncHI2(ST3), was associated with blaCTXM−65 and/or tet(A) genes in five K. pneumoniae isolates. Previous studies also reported that the IncHI2A plasmid is associated with multiple kinds of resistance genes in Enterobacteriaceae isolated from animals and humans and is responsible for inter-species transmission of ARGs (Garcia Fernandez et al., 2007; Wyrsch et al., 2019).
In this study, 75% of qnrB19 detected in Klebsiella genome was associated with small Col440I plasmids in all the farms, suggesting its widespread occurrence. This concurs with a recent study from Germany that reported frequent association of the qnrB gene with Col440I plasmids in ESBL-producing E. coli isolated from livestock (Juraschek et al., 2022). The wide distribution of qnrB-carrying Col440I plasmids among different STs of Klebsiella spp. and other members of Enterobacteriaceae suggests that they may play a crucial role in maintaining and spreading qnrB19 genes within Enterobacteriaceae. However, further investigation is necessary to understand the mechanisms responsible for the Col440I plasmid spread and maintenance among Enterobacteriaceae even in the absence of selection pressure, as most Klebsiella isolates in this study were obtained from adult dairy cattle that do not use (fluoro)quinolones.
In this study, in addition to plasmids, variants of ESBL co-occurred with other ARGs such as aac(3)-Iid, aph(6)-Id, aph(3′′)-Ib, sul2 associated with other MGEs such as Insertion sequences (IS), and Integrative Conjugative Element (ICEKp1). Previous studies widely reported the co-occurrence of ESBL and other unrelated ARGs (Lin et al., 2008; Tacao et al., 2014; Farzand et al., 2019; Astanheira and Bradford, 2021). This will facilitate the mobility of diverse ARG genes and the spread of MDR among the bacterial population. For instance, in three Klebsiella genomes, one or more ESBL variants co-occurred with mph(A) or aac(3)-Iid on the same contigs with IS26. This has an important implication on the epidemiology of resistance genes, as previous studies reported that IS26 helps to capture and mobilize drug resistance genes found in both chromosomes (Roy Chowdhury et al., 2015, 2018) and plasmids (Cain and Hall, 2012; Garcia et al., 2016). Studies also showed that IS26 also aids in plasmid stability and persistence by facilitating the deletion of plasmid backbone sequences whose expression is burdensome for the cell (Porse et al., 2016).
The final objective of this study was to determine the genetic relatedness and spread mechanism of ESBL-producing Klebsiella spp. within and between farms. This study demonstrated that the ESBL-producing Klebsiella spp. spreads through two modes. These include (1) clones or STs with specific Klebsiella spp.-plasmid-ESBL gene variant combinations were disseminated widely within and between farms, (2) plasmids harboring ESBL genes were detected in dissimilar Klebsiella spp. STs suggest possible horizontal transfer of the genes that might contribute to the successful spread of the bacteria in the study farms.
The extent of the two modes of ESBL transmission appeared to be different between the farms. In four farms (Farm A, B, G, and J), the spread of the ESBL genes within the farms might be primarily attributed to clonal expansion. This was evident by clusters of genetically similar isolates with the same STs-ESBL gene variant-plasmid type combinations and the same chromosomal mutations in QRDRs, indicating the clonal transmission of specific resistant Klebsiella clones throughout the dairy herd. For example, all K. pneumoniae from farms A, B, and G were identified as ST353, and they all carry blaCTX−M−1 on the plasmid, IncN (ST1). The detection of the identical ST of K. pneumoniae harboring the same ESBL variant on indistinguishable plasmids among dairy cattle on the same farm indicates potential animal-to-animal transmission of the bacteria or exposure to a common source (Podder et al., 2014).
The detection of the same ESBL-producing K. pneumoniae STs, with the same ESBL genes and plasmid profile, in three separate farms, suggests that there may be a clonal spread of the bacteria between the farms through feed, contaminated vehicles, movement of dairy cattle, and humans as described in previous studies (Davis et al., 2015; Medhanie et al., 2016; Afema et al., 2018). However, further study is needed to determine whether the spread of the bacteria is due to clonal dissemination via the mechanisms mentioned above or if there is a unique adaptation between the bacterial STs, the plasmid, and the ESBL gene.
Similarly, in Farm J, all Klebsiella isolates were identified as ST322 and harbored the same blaSHV−2 variants. The isolates also carried identical epidemic resistance plasmids, IncFIIK (K7: A−: B−), and novel alleles for IncFIA replicon types, suggesting the possible clonal spread of the ESBL-producing Klebsiella spp. in the farm. Since one of the isolates in this farm was obtained from a water sample taken from the water troughs, the animals on the farm might have independently acquired the same ST of K. pneumoniae by consuming the contaminated water from the same trough.
In farm M, evidence of clonal and horizontal spread of blaCTX−M genes was observed. However, horizontal spread appeared to be the primary factor in disseminating blaCTX−M genes on the farm. This was supported by identifying the same predominant blaCTX−M variants, specifically the blaCTX−M−27 gene, associated with the same epidemic-resistant plasmid, IncFII (ST F2: A−: B−).
About 31 (74%) of Klebsiella spp. Obtained from this farm, The three novel STs, the eight distinct STs, and five Unknown STs contained blaCTX−M−27 associated with IncFII (ST F2: A−: B−) plasmids. This indicates a potential horizontal spread of this ESBL variant among diverse Klebsiella spp. genotypes in the farm. This concurs with the previous studies that showed narrow host range plasmids of the IncF group can acquire resistance genes and quickly spread among specific members of Enterobacteriaceae (Carattoli, 2009; Mathers et al., 2015). In addition, some Klebsiella spp. with identical STs (e.g., the novel ST7501, n = 6; ST469, n = 6; ST 687, n = 4; and ST239, n = 3) had the same blaCTX−M−27 on IncFII (ST F2: A−: B−), suggesting a possible clonal spread of these STs within the animals in the farm.
5 Conclusion
The study identified six families of beta-lactamase genes and 26 distinct types of horizontally transferable antibiotic resistance genes. Multiple mutations in fluoroquinolone resistance regions were detected in all isolates. Most Klebsiella spp. carried numerous variants of β-lactamase and other ARGs, suggesting that these bacteria are a reservoir of resistance genes. The Klebsiella genomes harbored 22 different plasmid replicon types, with IncFII and Col440I plasmids being the most frequently detected associated with blaCTX−M and qnrB19, respectively. Most farms had genetically identical ESBL-K. pneumoniae strains carrying the same plasmids and ESBL genes, indicating clonal expansion and animal-to-animal transmission. Additionally, identical isolates from two farms in the same county suggest the possible inter-farm spread of the bacteria. The detection of identical ESBL variants with the same plasmid subtype in different Klebsiella STs suggests a possible horizontal spread of ESBL genes via plasmid transfer. These dual mechanisms of the spread of ESBL genes in Klebsiella spp. underscore the potential for a rapid increase in the prevalence of ESBL-Klebsiella spp. Given Klebsiella’s role as a reservoir for antibiotic resistance genes, robust strategies are needed to combat antibiotic resistance spread to reduce the potential animal and human health risks. Furthermore, the identification of novel STs of ESBL-K. pneumoniae and novel STs of epidemic resistance plasmids in dairy farms underscore the importance of monitoring dairy farms for emerging antibiotic-resistant pathogens.
Data availability statement
The assembled sequence data has now been submitted to the Pasteur Institute Bacterial Isolate Genome Sequence Database and can be accessed at https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_isolates&page=submit&submission_id=BIGSdb_20240715165555_3499682_34488&view=1 BIGSdb_20240715165555_3499682_34488.
Ethics statement
The animal study was approved by the University of Tennessee’s Institutional Animal Care and Use Committee (IACUC) Registration Number: 2782-0720. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BG: Conceptualization, Writing – review & editing, Investigation, Methodology, Writing – original draft. RM: Formal Analysis, Writing – review & editing. AG: Investigation, Writing – review & editing. OKD: Conceptualization, Supervision, Writing – review & editing.
Funding
The authors declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We thank East Tennessee dairy farmers for volunteering to participate in this study. We thank the Institut Pasteur teams for the curation and maintenance of BIGSdb-Pasteur databases at http://bigsdb.pasteur.fr/.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1439363/full#supplementary-material
Supplementary Figure 1 | Summary statistics of Klebsiella species genome assembly.
Footnotes
- ^ https://sourceforge.net/projects/bbmap/
- ^ https://cge.food.dtu.dk/services/kmerfinder/
- ^ https://cge.food.dtu.dk/services/resfinder/
- ^ https://card.mcmaster.ca/analyze/rgi
- ^ https://cge.food.dtu.dk/services/plasmidfinder/
- ^ https://cge.food.dtu.dk/services/pmlst/
- ^ https://cge.food.dtu.dk/services/mobileelementfinder/
- ^ https://cge.food.dtu.dk/services/mlst/
- ^ https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_klebsiella_seqdef&page=sequencequery
References
Afema, J., Ahmed, S., Besser, T., Jones, L., Sischo, W., and Davis, M. (2018). Molecular epidemiology of dairy cattle-associated Escherichia coli carrying blaCTX-M genes in washington state. Appl. Environ. Microbiol. 84:e2430–17. doi: 10.1128/AEM.02430-17
Ahmed, A., and Shimamoto, T. (2011). Molecular characterization of antimicrobial resistance in Gram-negative bacteria isolated from bovine mastitis in Egypt. Microbiol. Immunol. 55, 318–327. doi: 10.1111/j.1348-0421.2011.00323.x
Al-Assil, B., Mahfoud, M., and Hamzeh, A. (2013). Resistance trends and risk factors of extended spectrum β-lactamases in Escherichia coli infections in Aleppo, Syria. Am. J. Infect. Control 41, 597–600. doi: 10.1016/j.ajic.2012.09.016
Alcock, B., Raphenya, A., Lau, T., Tsang, K., Bouchard, M., Edalatmand, A., et al. (2020). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 48, D517–D525. doi: 10.1093/nar/gkz935
Alrowais, H., McElheny, C., Spychala, C., Sastry, S., Guo, Q., Butt, A., et al. (2015). Fosfomycin resistance in Escherichia coli, Pennsylvania, USA. Emerg. Infect. Dis. 21, 2045–2047. doi: 10.3201/eid2111.150750
Arca, P., Reguera, G., and Hardisson, C. (1997). Plasmid-encoded fosfomycin resistance in bacteria isolated from the urinary tract in a multicentre survey. J. Antimicrob. Chemother. 40, 393–399. doi: 10.1093/jac/40.3.393
Astanheira, M., and Bradford, P. (2021). Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 3:dlab092. doi: 10.1093/jacamr/dlab092
Babraham Bioinformatics (2024). FastQC: A quality control tool for high throughput sequence data 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed July 20, 2024).
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A., Dvorkin, M., Kulikov, A., et al. (2012). SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bevan, E., Jones, A., and Hawkey, P. (2017). Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Bolger, A., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bonnet, R. (2004). Growing group of extended-spectrum beta-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 48, 1–14. doi: 10.1128/AAC.48.1.1-14.2004
Bortolaia, V., Kaas, R., Ruppe, E., Roberts, M., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Boucher, H., Talbot, G., Bradley, J., Edwards, J., Gilbert, D., Rice, L., et al. (2009). Bad bugs, no drugs: No ESKAPE! An update from the infectious diseases society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Cain, A., and Hall, R. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Calbo, E., and Garau, J. (2015). The changing epidemiology of hospital outbreaks due to ESBL-producing Klebsiella pneumoniae: The CTX-M-15 type consolidation. Future Microbiol. 10, 1063–1075. doi: 10.2217/fmb.15.22
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: Architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421
Cantón, R., and Coque, T. (2006). The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9, 466–475. doi: 10.1016/j.mib.2006.08.011
Carattoli, A. (2009). Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 2227–2238. doi: 10.1128/AAC.01707-08
Carattoli, A. (2013). Plasmids and the spread of resistance. Int. J. Med. Microbiol. 303, 298–304. doi: 10.1016/j.ijmm.2013.02.001
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Larsen, M., Lund, O., Villa, L., et al. (2014). in silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Carey, A., Capik, S., Giebel, S., Nickodem, C., Pineiro, J., Scott, H., et al. (2022). Prevalence and profiles of antibiotic resistance genes mph(A) and qnrB in extended-spectrum β-lactamase (ESBL)-producing Escherichia coli isolated from dairy calf feces. Microorganisms 10:411. doi: 10.3390/microorganisms10020411
Castanheira, M. F., Krause, K., Jones, R., and Sader, H. (2014). Contemporary diversity of β-lactamases among Enterobacteriaceae in the nine U.S. census regions and ceftazidime-avibactam activity tested against isolates producing the most prevalent β-lactamase groups. Antimicrob. Agents Chemother. 58, 833–838. doi: 10.1128/AAC.01896-13
Clausen, P., Aarestrup, F., and Lund, O. (2018). Rapid and precise alignment of raw reads against redundant databases with KMA. BMC Bioinform. 19, 307. doi: 10.1186/s12859-018-2336-6
Cormier, A., Chalmers, G., McAllister, T., Cook, S., Zaheer, R., Scott, H., et al. (2016). Extended-spectrum-cephalosporin resistance genes in Escherichia coli from beef cattle. Antimicrob. Agents Chemother. 60, 1162–1163. doi: 10.1128/AAC.02516-15
Cummings, K., Warnick, L., Alexander, K., Cripps, C., Grohn, Y., McDonough, P., et al. (2009). The incidence of salmonellosis among dairy herds in the northeastern United States. J. Dairy Sci. 92, 3766–3774. doi: 10.3168/jds.2009-2093
Dantas Palmeira, J., and Ferreira, H. (2020). Extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae in cattle production - a threat around the world. Heliyon 6:e03206. doi: 10.1016/j.heliyon.2020.e03206
Davis, G., and Price, L. B. (2016). Recent research examining links among Klebsiella pneumoniae from food, food animals, and human extraintestinal infections. Curr. Environ. Health Rep. 3, 128–135. doi: 10.1007/s40572-016-0089-9
Davis, M., Sischo, W., Jones, L., Moore, D., Ahmed, S., Short, D., et al. (2015). Recent emergence of Escherichia coli with cephalosporin resistance conferred by blaCTX-M on washington state dairy farms. Appl. Environ. Microbiol. 81, 4403–4410. doi: 10.1128/AEM.00463-15
Diancourt, L., Passet, V., Verhoef, J., Grimont, P., and Brisse, S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43, 4178–4182. doi: 10.1128/JCM.43.8.4178-4182.2005
Donaldson, S., Straley, B., Hegde, N., Sawant, A., DebRoy, C., and Jayarao, B. (2006). Molecular epidemiology of ceftiofur-resistant Escherichia coli isolates from dairy calves. Appl. Environ. Microbiol. 72, 3940–3948. doi: 10.1128/AEM.02770-05
Dunne, E., Fey, P., Kludt, P., Reporter, R., Mostashari, F., Shillam, P., et al. (2000). Emergence of domestically acquired ceftriaxone-resistant Salmonella infections associated with AmpC β-lactamase. JAMA 284, 3151–3156. doi: 10.1001/jama.284.24.3151
Ewels, P., Magnusson, M., Lundin, S., and Kaller, M. (2016). MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. doi: 10.1093/bioinformatics/btw354
Farzand, R., Rajakumar, K., Zamudio, R., Oggioni, M., Barer, M., and O’Hare, H. (2019). ICEKp2: Description of an integrative and conjugative element in Klebsiella pneumoniae, co-occurring and interacting with ICEKp1. Sci. Rep. 9:13892. doi: 10.1038/s41598-019-50456-x
Fournier, B., Arlet, G., Lagrange, P., and Philippon, A. (1994). Klebsiella oxytoca: Resistance to aztreonam by overproduction of the chromosomally encoded β-lactamase. FEMS Microbiol. Lett. 116, 31–36. doi: 10.1111/j.1574-6968.1994.tb06671.x
Garcia Fernandez, A., Cloeckaert, A., Bertini, A., Praud, K., Doublet, B., Weill, F., et al. (2007). Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51, 4177–4180. doi: 10.1128/AAC.00603-07
Garcia, V., Garcia, P., Rodriguez, I., Rodicio, R., and Rodicio, M. (2016). The role of IS26 in evolution of a derivative of the virulence plasmid of Salmonella enterica serovar Enteritidis which confers multiple drug resistance. Infect. Genet. Evol. 45, 246–249. doi: 10.1016/j.meegid.2016.09.008
Geetha, P., Aishwarya, K., Mariappan, S., and Sekar, U. (2020). Fluoroquinolone Resistance in clinical isolates of Klebsiella pneumoniae. J. Lab. Phys. 12, 121–125. doi: 10.1055/s-0040-1716478
Gelalcha, B., Agga, G., and Kerro Dego, O. (2022). “Antimicrobial usage for the management of mastitis in the USA: Impacts on antimicrobial resistance and potential alternative approaches,” in Mastitis in dairy cattle, sheep and goats, ed. O. Kerro Dego (London: Intech Open), 1–21. doi: 10.5772/intechopen.101533
Gelalcha, B., and Kerro Dego, O. (2022). Extended-spectrum β-lactamases producing Enterobacteriaceae in the USA dairy cattle farms and implications for public health. Antibiotics 11:1313. doi: 10.3390/antibiotics11101313
Gelalcha, B., Gelgie, A., and Kerro Dego, O. (2023a). Prevalence and antimicrobial resistance profile of extended-spectrum beta-lactamase-producing E. coli in east tennessee dairy farms. Front. Vet. Sci. Sec. Vet. Infect. Dis. 10:1260433. doi: 10.3389/fvets.2023.1260433
Gelalcha, B., Mohammed, R., Gelgie, A., and Kerro Dego, O. (2023b). Molecular epidemiology and pathogenomics of extended-spectrum β-lactamase producing- Escherichia coli and - Klebsiella pneumoniae isolates from bulk tank milk in Tennessee, USA. Front. Microbiol. 14:1283165. doi: 10.3389/fmicb.2023.1283165
Gonzalez-Lopez, J., Coelho, A., Larrosa, M., Lavilla, S., Bartolome, R., and Prats, G. (2009). First detection of plasmid-encoded blaOXY beta-lactamase. Antimicrob. Agents Chemother. 53, 3143–3146. doi: 10.1128/AAC.01473-08
Haenni, M. C., Métayer, V., Bour, M., Signol, E., Madec, J., and Gay, E. (2014). Comparative prevalence and characterization of ESBL-producing Enterobacteriaceae in dominant versus subdominant enteric flora in veal calves at slaughterhouse, France. Vet. Microbiol. 171, 321–327. doi: 10.1016/j.vetmic.2014.02.023
Hasman, H., Saputra, D., Sicheritz-Ponten, T., Lund, O., Svendsen, C., Frimodt-Moller, N., et al. (2014). Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J. Clin. Microbiol. 52, 139–146. doi: 10.1128/JCM.02452-13
Hou, J., Huang, X., Deng, Y., He, L., Yang, T., Zeng, Z., et al. (2012). Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from Pets in China. Antimicrob. Agents Chemother. 56, 2135–2138. doi: 10.1128/AAC.05104-11
Hou, J., Yang, X., Zeng, Z., Lv, L., Yang, T., Lin, D., et al. (2013). Detection of the plasmid-encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J. Antimicrob. Chemother. 68, 766–770. doi: 10.1093/jac/dks465
Irrgang, A., Falgenhauer, L., Fischer, J., Ghosh, H., Guiral, E., Guerra, B., et al. (2017). CTX-M-15-producing E. coli isolates from food products in germany are mainly associated with an IncF-Type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbiol. 8:2318. doi: 10.3389/fmicb.2017.02318
Joensen, K., Scheutz, F., Lund, O., Hasman, H., Kaas, R., Nielsen, E., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Juraschek, K., Malekzadah, J., Malorny, B., Kasbohrer, A., Schwarz, S., Meemken, D., et al. (2022). Characterization of qnrB-carrying plasmids from ESBL- and non-ESBL-producing Escherichia coli. BMC Genom. 23:365. doi: 10.1186/s12864-022-08564-y
Kim, S., Wei, C., Tzou, Y., and An, H. (2005). Multidrug-resistant Klebsiella pneumoniae isolated from farm environments and retail products in Oklahoma. J. Food Prot. 68, 2022–2029. doi: 10.4315/0362-028X-68.10.2022
Larsen, M., Cosentino, S., Lukjancenko, O., Saputra, D., Rasmussen, S., Hasman, H., et al. (2014). Benchmarking of methods for genomic taxonomy. J. Clin. Microbiol. 52, 1529–1539. doi: 10.1128/JCM.02981-13
Leclercq, R. (2002). Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 34, 482–492. doi: 10.1086/324626
Lee, M. (2019). GToTree: A user-friendly workflow for phylogenomics. Bioinformatics 35, 4162–4164. doi: 10.1093/bioinformatics/btz188
Li, J., Zhang, H., Ning, J., Sajid, A., Cheng, G., Yuan, Z., et al. (2019). The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob. Resist. Infect. Control 8:44. doi: 10.1186/s13756-019-0489-3
Lin, T., Lee, C., Hsieh, P., Tsai, S., and Wang, J. (2008). Characterization of integrative and conjugative element ICEKp1-associated genomic heterogeneity in a Klebsiella pneumoniae strain isolated from a primary liver abscess. J. Bacteriol. 190, 515–526. doi: 10.1128/JB.01219-07
Martin, R., and Bachman, M. (2018). Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front. Cell Infect. Microbiol. 8:4. doi: 10.3389/fcimb.2018.00004
Mathers, A., Peirano, G., and Pitout, J. (2015). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Medhanie, G., Pearl, D., McEwen, S., Guerin, M., Jardine, C., Schrock, J., et al. (2016). On-farm starling populations and other environmental and management factors associated with the presence of cefotaxime and ciprofloxacin resistant E. coli among dairy cattle in Ohio. Prev. Vet. Med. 134, 122–127. doi: 10.1016/j.prevetmed.2016.10.006
Mikheenko, A., Prjibelski, A., Saveliev, V., Antipov, D., and Gurevich, A. (2018). Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34, i142–i150. doi: 10.1093/bioinformatics/bty266
Naas, T., Oueslati, S., Bonnin, R., Dabos, M., Zavala, A., Dortet, L., et al. (2017). β-lactamase database (BLDB) - structure and function. J. Enzym. Inhib. Med. Ch. 32, 917–919. doi: 10.1080/14756366.2017.1344235
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Ohnishi, M. O., Esaki, H., Harada, K., Sawada, T., Murakami, M., Marumo, K., et al. (2013). Herd prevalence of Enterobacteriaceae producing CTX-M-type and CMY-2 β-lactamases among Japanese dairy farms. J. Appl. Microbiol. 115, 282–289. doi: 10.1111/jam.12211
Paterson, D., Hujer, K., Hujer, A., Yeiser, B., Bonomo, M., Rice, L., et al. (2003). Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: Dominance and widespread prevalence of SHV- and CTX-M-type beta-lactamases. Antimicrob. Agents Chemother. 47, 3554–3560. doi: 10.1128/AAC.47.11.3554-3560.2003
Paterson, D., Mulazimoglu, L., Casellas, J., Ko, W., Goossens, H., Von Gottberg, A., et al. (2000). Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin. Infect. Dis. 30, 473–478. doi: 10.1086/313719
Peirano, G., and Pitout, J. (2019). Extended-spectrum β-lactamase-producing Enterobacteriaceae: Update on molecular epidemiology and treatment options. Drugs 79, 1529–1541. doi: 10.1007/s40265-019-01180-3
Perez, F., Rudin, S., Marshall, S., Coakley, P., Chen, L., Kreiswirth, B., et al. (2013). OqxAB, a quinolone and olaquindox efflux pump, is widely distributed among multidrug-resistant Klebsiella pneumoniae isolates of human origin. Antimicrob. Agents Chemother. 57, 4602–4603. doi: 10.1128/AAC.00725-13
Petrella, S., Clermont, D., Casin, I., Jarlier, V., and Sougakoff, W. (2001). Novel class A β-lactamase Sed-1 from Citrobacter sedlakii: Genetic diversity of β-lactamases within the Citrobacter genus. Antimicrob. Agents Chemother. 45, 2287–2298. doi: 10.1128/AAC.45.8.2287-2298.2001
Podder, M., Rogers, L., Daley, P., Keefe, G., Whitney, H., and Tahlan, K. (2014). Klebsiella species associated with bovine mastitis in Newfoundland. PLoS One 9:e106518. doi: 10.1371/journal.pone.0106518
Poirel, L., Cattoir, V., and Nordmann, P. (2012). Plasmid-mediated quinolone resistance; interactions between human, animal, and environmental ecologies. Front. Microbiol. 3:24. doi: 10.3389/fmicb.2012.00024
Porse, A., Schonning, K., Munck, C., and Sommer, M. (2016). Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873. doi: 10.1093/molbev/msw163
Roy Chowdhury, P., Charles, I., and Djordjevic, S. P. (2015). A role for Tn6029 in the evolution of the complex antibiotic resistance gene loci in genomic island 3 in enteroaggregative hemorrhagic Escherichia coli O104:H4. PLoS One 10:e0115781. doi: 10.1371/journal.pone.0115781
Roy Chowdhury, P., McKinnon, J., Liu, M., and Djordjevic, S. (2018). Multidrug resistant uropathogenic Escherichia coli ST405 with a novel, composite IS26 transposon in a unique chromosomal location. Front. Microbiol. 9:3212. doi: 10.3389/fmicb.2018.03212
Saha, R., Farrance, C., Verghese, B., Hong, S., and Donofrio, R. (2013). Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr. Microbiol. 66, 72–78. doi: 10.1007/s00284-012-0245-x
Sastry, S., and Doi, Y. (2016). Fosfomycin: Resurgence of an old companion. J. Infect. Chemother. 22, 273–280. doi: 10.1016/j.jiac.2016.01.010
Shin, J., Choi, M., and Ko, K. (2012). Replicon sequence typing of IncF plasmids and the genetic environments of blaCTX-M-15 indicate multiple acquisitions of blaCTX-M-15 in Escherichia coli and Klebsiella pneumoniae isolates from South Korea. J. Antimicrob. Chemother. 67, 1853–1857. doi: 10.1093/jac/dks143
Stefani, S., Giovanelli, I., Anacarso, I., Condo, C., Messi, P., de Niederhausern, S., et al. (2014). Prevalence and characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae in food-producing animals in Northern Italy. New Microbiol. 37, 551–555.
Tacao, M., Moura, A., Correia, A., and Henriques, I. (2014). Co-resistance to different classes of antibiotics among ESBL-producers from aquatic systems. Water Res. 48, 100–107. doi: 10.1016/j.watres.2013.09.021
Tadesse, D., Zhao, S., Tong, E., Ayers, S., Singh, A., Bartholomew, M., et al. (2012). Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950-2002. Emerg. Infect. Dis. 18, 741–749. doi: 10.3201/eid1805.111153
Taylor, E., Ossa-Trujillo, C., Vinasco, J., Jordan, E. R., Buitrago, G. J. A., and Hagevoort, K. N. (2020). Use of critically important antimicrobial classes early in life may adversely impact bacterial resistance profiles during adult years: Potential co-selection for plasmid-borne fluoroquinolone and macrolide resistance via extendedspectrum β-lactam use in dairy cattle. Lett. Appl. Microbiol. 72, 220–224. doi: 10.1111/lam.13419
Tyson, G., Li, C., Hsu, C., Bodeis-Jones, S., and McDermott, P. (2019). Diverse fluoroquinolone resistance plasmids from retail meat E. coli in the United States. Front. Microbiol. 10:2826. doi: 10.3389/fmicb.2019.02826
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wang, X., Dong, Z., Schwarz, S., Zhu, Y., Hua, X., Zhang, Y., et al. (2017). Plasmids of diverse Inc groups disseminate the fosfomycin resistance gene fosA3 among Escherichia coli Isolates from pigs, chickens, and dairy cows in Northeast China. Antimicrob. Agents Chemother. 61, e859–17. doi: 10.1128/AAC.00859-17
Wener, K., Schechner, V., Gold, H. S., Wright, S. B., and Carmeli, Y. T. (2010). Treatment with fluoroquinolones or with β-lactam-beta-lactamase inhibitor combinations is a risk factor for isolation of extended-spectrum-β-lactamase-producing Klebsiella species in hospitalized patients. Antimicrob. Agents Chemother. 54, 2010–2016. doi: 10.1128/AAC.01131-09
Wong, M., Chan, E., and Chen, S. (2015). Evolution and dissemination of OqxAB-like efflux pumps, an emerging quinolone resistance determinant among members of Enterobacteriaceae. Antimicrob. Agents Chemother. 59, 3290–3297. doi: 10.1128/AAC.00310-15
Woodford, N. T., and Livermore, D. (2011). Multiresistant Gram-negative bacteria: The role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. doi: 10.1111/j.1574-6976.2011.00268.x
Wu, H., Wang, M., Liu, Y., Wang, X., Wang, Y., Lu, J., et al. (2016). Characterization of antimicrobial resistance in Klebsiella species isolated from chicken broilers. Int. J. Food Microbiol. 232, 95–102. doi: 10.1016/j.ijfoodmicro.2016.06.001
Wyres, K., and Holt, K. (2018). Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically important bacteria. Curr. Opin. Microbiol. 45, 131–139. doi: 10.1016/j.mib.2018.04.004
Wyrsch, E., Reid, C., DeMaere, M., Liu, M., Chapman, T., Chowdhury, P., et al. (2019). Complete sequences of multiple-drug resistant IncHI2 ST3 plasmids in Escherichia coli of porcine origin in Australia. Front. Sustain. Food Syst. 3:18. doi: 10.3389/fsufs.2019.00018
Yang, J., Long, H., Hu, Y., Feng, Y., McNally, A., and Zong, Z. (2022). Klebsiella oxytoca complex: Update on taxonomy, antimicrobial resistance, and virulence. Clin. Microbiol. Rev. 35:e0000621. doi: 10.1128/CMR.00006-21
Yang, Y., Higgins, C., Rehman, I., Galvao, K., Brito, I., Bicalho, M., et al. (2019). Genomic diversity, virulence, and antimicrobial resistance of Klebsiella pneumoniae strains from cows and humans. Appl Environ Microbiol. 85, e2654–18. doi: 10.1128/AEM.02654-18
Yigit, H., Queenan, A., Anderson, G., Domenech-Sanchez, A., Biddle, J., Steward, C., et al. (2001). Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 45, 1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001
Zankari, E., Allesoe, R., Joensen, K., Cavaco, L., Lund, O., and Aarestrup, F. (2017). PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 72, 2764–2768. doi: 10.1093/jac/dkx217
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, X., Li, Q., Lin, H., Zhou, W., Qian, C., Sun, Z., et al. (2021). High-level aminoglycoside resistance in human clinical Klebsiella pneumoniae complex isolates and characteristics of armA-carrying IncHI5 plasmids. Front. Microbiol. 12:636396. doi: 10.3389/fmicb.2021.636396
Zhao, W., and Hu, Z. (2013). Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 39, 79–101. doi: 10.3109/1040841X.2012.691460
Keywords: Klebsiella pneumoniae, beta-lactamase genes, dairy farm, genetic diversity, plasmids, transmission, whole genome sequence
Citation: Gelalcha BD, Mohamed RI, Gelgie AE and Kerro Dego O (2024) Molecular epidemiology of extended-spectrum beta-lactamase-producing-Klebsiella species in East Tennessee dairy cattle farms. Front. Microbiol. 15:1439363. doi: 10.3389/fmicb.2024.1439363
Received: 27 May 2024; Accepted: 30 July 2024;
Published: 24 September 2024.
Edited by:
Kristina Kadlec, Independent researcher, Wunstorf, GermanyReviewed by:
Salome N. Seiffert, Zentrum für Labormedizin (ZLM), SwitzerlandDong Chan Moon, Korea National Institute of Health, Republic of Korea
Copyright © 2024 Gelalcha, Mohamed, Gelgie and Kerro Dego. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oudessa Kerro Dego, b2tlcnJvZGVAdXRrLmVkdQ==
 Benti D. Gelalcha
Benti D. Gelalcha Ruwaa I. Mohamed
Ruwaa I. Mohamed Aga Edema Gelgie
Aga Edema Gelgie Oudessa Kerro Dego
Oudessa Kerro Dego