
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 03 July 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1439009
Background: The emergence and spread of multidrug-resistant Staphylococcus aureus strains demonstrates the urgent need for new antimicrobials. Xanthorrhizol, a plant-derived sesquiterpenoid compound, has a rapid killing effect on methicillin-susceptible strains and methicillin-resistant strains of S. aureus achieving the complete killing of staphylococcal cells within 2 min using 64 μg/mL xanthorrhizol. However, the mechanism of its action is not yet fully understood.
Methods: The S. aureus cells treated with xanthorrhizol were studied using optical diffraction tomography. Activity of xanthorrhizol against the wild-type and mscL null mutant of S. aureus ATCC 29213 strain was evaluated in the time-kill assay. Molecular docking was conducted to predict the binding of xanthorrhizol to the SaMscL protein.
Results: Xanthorrhizol treatment of S. aureus cells revealed a decrease in cell volume, dry weight, and refractive index (RI), indicating efflux of the cell cytoplasm, which is consistent with the spontaneous activation of the mechanosensitive MscL channel. S. aureus ATCC 29213ΔmscL was significantly more resistant to xanthorrhizol than was the wild-type strain. Xanthorrhizol had an enhanced inhibitory effect on the growth and viability of exponentially growing S. aureus ATCC 29213ΔmscL cells overexpressing the SaMscL protein and led to a noticeable decrease in their viability in the stationary growth phase. The amino acid residues F5, V14, M23, A79, and V84 were predicted to be the residues of the binding pocket for xanthorrhizol. We also showed that xanthorrhizol increased the efflux of solutes such as K+ and glutamate from S. aureus ATCC 29213ΔmscL cells overexpressing SaMscL. Xanthorrhizol enhanced the antibacterial activity of the antibiotic dihydrostreptomycin, which targets the MscL protein.
Conclusion: Our findings indicate that xanthorrhizol targets the SaMscL protein in S. aureus cells and may have important implications for the development of a safe antimicrobial agent.
The sesquiterpenoid compound xanthorrhizol, isolated from the rhizomes of Curcuma xanthorrhiza Roxb. (Zingiberaceae), exhibits broad-spectrum antibacterial activity, mainly against gram-positive bacteria such as S. aureus, Streptococcus mutans, Actinomyces viscosus, Porphyromonas gingivalis, Bacillus cereus, Listeria monocytogenes, and Streptococcus salivarius (Hwang et al., 2000a; Lee et al., 2008); against the yeast Candida albicans (Hwang et al., 2000b); and against various filamentous fungi (Rukayadi and Hwang, 2007). Xanthorrhizol has shown limited activity against gram-negative species (Mata et al., 2001; Lee et al., 2008; Oon et al., 2015). Mutation of the imp (lptD) gene significantly increased the susceptibility of E. coli cells to xanthorrhizol (Mata et al., 2001), which made it possible to identify two potential targets for xanthorrhizol in E. coli: tRNA-specific adenosine deaminase (TadA) and enoyl-ACP reductase (FabI) (Yogiara et al., 2015, 2020). In addition to its antimicrobial activity, xanthorrhizol also exhibits anticancer, anti-inflammatory, antioxidant and other biological activities without detectable toxic side effects (Choi et al., 2005; Chung et al., 2007; Oon et al., 2015). An inhibitory effect of xanthorrhizol was detected against severe acute respiratory syndrome (SARS) coronavirus 2 (SCoV2), as well as against other related coronaviruses such as SARS-CoV-1 (SCoV1) and the human coronavirus that causes the common cold (Kim et al., 2021).
A time-killing kinetic study showed that after 4 h of incubation with 8 μg/mL xanthorrhizol (1xMIC), no viable S. aureus cells were found (Lee et al., 2008). This rapid killing activity of xanthorrhizol has previously been shown against S. mutans (Hwang et al., 2000a), and electron microscopy analysis revealed disintegration of S. mutans cells, indicating that xanthorrhizol disrupted membrane structure or induced cell rupture (Kim et al., 2008). This effect is similar to that of osmotic downward shock, where the cell membrane ruptures, releasing intracellular content and resulting in cell death due to the large difference in osmotic strength between the cell and the environment (Chure et al., 2018). In response to changes in membrane tension and to avoid cell membrane damage, regulated efflux of water and small intracellular molecules occurs though transmembrane protein channels (Martinac et al., 1987; Chure et al., 2018). However, mechanosensitive channels not only protect bacteria from osmotic forces and prevent the selective release of cytoplasmic proteins under conditions of hypoosmotic shock (Kouwen et al., 2009a) but are also thought to play a role in the uptake and efflux of antibiotics (Iscla et al., 2014; Wray et al., 2016).
The S. aureus has the mechanosensitive channel of large conductance MscL (Liu et al., 2009; Dorwart et al., 2010) and one MscS type transporter, homologous to both ykuT/mscT (34.30%) and yfkC/mscC (35.50%) mechanosensitive channels of small conductance in B. subtilis (Hoffmann et al., 2008; Casey and Sleator, 2021). This surprisingly low presence of mechanosensitive channel nomenclature in S. aureus might be explained by the fact that hypoosmotic shock occurs relatively rarely in such an ecological niche as human skin (Casey and Sleator, 2021). The mechanosensitive channel MscL seems to be a very attractive target for antibiotic therapy due to its presence in almost all bacterial species but not in mammals, and it is highly conserved (Iscla et al., 2015). Some compounds have been shown to target mechanosensitive channel MscL in S. aureus (SaMscL) (Kouwen et al., 2009b; Iscla et al., 2015; Wray et al., 2019b, 2020, 2021, 2022; Blount and Iscla, 2020; Wang and Blount, 2022).
In the present study, we found that the natural antimicrobial agent xanthorrhizol acts by targeting the SaMscL mechanosensitive channel to inhibit the exponential growth of S. aureus cells, reduce the viability of S. aureus stationary cultures in a MscL-dependent manner, and enhance the anti-MscL activity of the antibiotic dihydrostreptomycin (Wray et al., 2016), which allows the use of xanthorrhizol as a potential antimicrobial agent.
The strains and plasmids used in this study are listed in Table 1. The LB medium for E. coli and tryptic soy broth (TSB) for S. aureus strain cultivation were purchased from Difco (San Jose, USA). The following concentrations of antibiotics were used: ampicillin, 100 μg/mL; erythromycin, 20 μg/mL; and kanamycin, 25 μg/mL. Dihydrostreptomycin sulfate (Sigma Aldrich St. Louis, MO) was used at concentrations ranging from 8 to 50 μg/mL. Xanthorrhizol was purchased from Cayman Chemical (Ann Arbor, Michigan, United States), and purified xanthorrhizol was also provided by Professor Jae-Kwan Hwang (Department of Biotechnology, Yonsei University, Seoul, Republic of Korea).
To measure the MIC values, a standard microdilution method was used according to a previously described procedure (Wiegand et al., 2008), with some modifications. Briefly, overnight cultures of the S. aureus strains grown in Mueller Hinton broth were reinoculated in the same fresh medium. When the cultures reached an OD600 of 0.5, the cells were adjusted to a final desired inoculation concentration of 5 × 105 cfu/ml. The xanthorrhizol concentrations ranged from 1 to 1,024 μg/mL, prepared via serial twofold dilution. The MIC was determined as the lowest concentration of the antimicrobial compound at which the bacteria did not demonstrate visible growth. This experiment was carried out in duplicate.
To measure the 3D RI tomogram of individual bacterial cells, optical diffraction tomography (ODT) was performed using a Mach–Zehnder interferometer equipped with a digital micromirror device (Oh et al., 2020). S. aureus cells were cultivated in TSB medium to a final OD600 of 0.5 and prepared for ODT as previously described (Oh et al., 2020). To perform bacterial imaging, the S. aureus cells were resuspended in TSB medium and divided into two parts: one was supplemented with xanthorrhizol at a final concentration of 64 μg/mL, and the other was supplemented with DMSO. Ten microlitres of the resuspended solution was placed between two coverslips and then measured with an optical imaging system for 30 min at 5-min intervals. All the measurements were performed at room temperature (25°C). The 3D rendered image was acquired using commercial software (TomoStudioTM, Tomocube Inc., Daejeon, Republic of Korea), which more clearly defines the overall refractive index (RI) distribution of bacteria. Quantitative analysis of cell parameters was performed as previously described (Oh et al., 2020).
Bacteria grown overnight in TSB medium were diluted 400-fold in fresh TSB medium, incubated at 37°C to a final OD600 of 0.5 and then diluted 10-fold. The cells were split into two groups and treated with xanthorrhizol (32–64 μg/mL) or with DMSO at a concentration equal to that used for drug-treated bacteria. The cells were then cultivated at 37°C for 1 or 5 h. Samples were taken after 37 s, 1.25, 2.5, 5, 10, and 30 min, and 1, 2 3, and 5 h; then, the samples were serially diluted 1:10 with sterile PBS containing 1 mM EDTA and 0.2% Kolliphor P188 (Sigma Aldrich, St. Louis, MO) and plated on solid TSA medium for colony counting. Kolliphor P188 was used to inactivate xanthorrhizol for live cell detection at each test point. The values represent the means of three independent experiments. The error bars indicate the standard errors.
The structure of EcMscL is not available in PDB. Therefore, the sequence of EcMscL was downloaded from UniProtKB. Structure of the M. tuberculosis MscL (PDB Code 2OAR) was used as the template for homology modeling (Wray et al., 2016). The homology model of EcMscL was built using Prime in Schrödinger Suite 2021-2 (Schrödinger, LLC, 2021a). Protein structures were prepared using the Protein Preparation Wizard. All missing loops were filled using Prime and implemented in Maestro version 2021-2 (Schrödinger, LLC, 2021b). Hydrogen atoms were added to the complex structure at pH 7.4, and their positions were optimized using PROPKA in Maestro. Restrained energy minimization was performed with an OPLS4 force field within 0.3 Å root mean square deviation (RMSD). Molecular docking was conducted to predict the binding poses of using the Glide (Maestro version 2021–2). Xanthorrhizol was docked with extra precision (XP) using Maestro’s Glide on the constructed homology model.
In-frame deletion of the mscL gene was conducted using the pHoss1 suicide plasmid (Abdelhamed et al., 2015) and overlap extension PCR with the primers mL1 (5′-GATATCGGATCCATATGACGTCGACGTAGAGCACCCATACCTAACG-3′) and mL4 (5′-TTTCTCACGTAATAAATCCATTACACTCAACCTCTCTTT-3′) for amplification of the 1 kb upstream fragment and mL3 (5′-AAAGAGAGGTTGAGTGTAATGGATTTATTACGTGAGAAA-3′) and mL2 (5′-GTTTGAATCATTAGATCCCATGGGAAATGGAAATGTCGAGGCTG-3′) for amplification of the 1 kb downstream fragment of the mscL gene. Then, both PCR fragments were used as templates for the second PCR with the mL1 and mL2 primers, and the resulting 2 kb PCR fragment was inserted between the SalI/NcoI restriction sites of pHoss1 to generate the pHoss1/ΔmscL plasmid. The pHoss1/ΔmscL plasmid was subsequently transformed into the methylation-deficient strain E. coli ET12567. Demethylated plasmid pHoss1/ΔmscL DNA was delivered into the S. aureus ATCC 29213 cells by electroporation. Electrocompetent S. aureus cells were prepared and electroporated as previously described (Schneewind and Missiakas, 2014). The transformed cells were recovered in TSB medium at 30°C for 3 h and then incubated on TSA solid medium supplemented with erythromycin (20 μg/mL) for 48 h at 30°C. One erythromycin-resistant clone was selected and incubated for 24 h in TSB medium supplemented with erythromycin (20 μg/mL) at 42°C. Then, 1 μl of the cell culture was diluted 1,000-fold with fresh TSB (without Ery) and incubated for 24 h at 30°C, which was repeated twice. Thereafter, the culture was reinoculated into fresh TSB, incubated for 8 h at 42°C, serially diluted and plated onto solid TSA supplemented with anhydrotetracycline (2.5 μg/mL). After 2 days of cultivation at 30°C, clones sensitive to erythromycin were selected and checked for loss of the mscL gene by PCR.
The B. subtilis/E. coli shuttle vector pBE-S (Takara Bio, Inc., Shiga, Japan) with an origin of replication derived from pUB110 was used to construct a protein expression vector suitable for S. aureus. The SpeI/HindIII fragment of pBE-S was replaced with the NheI/HindIII fragment from the plasmid pHT01 carrying the lacI gene and the IPTG-inducible Pgrac promoter to generate the pBEP expression vector. The mscL gene was amplified from the genomic DNA of S. aureus ATCC 29213 using the primers mL5 (5′-CAATTAAAGGAGGAAGGATCCATGTTAAAAGAATTCAAAGAG-3′) and mL6 (5′-GATGTCTAGACTGCAGGTCGACTTTTTTCTCACGTAATAAATC-3′) and inserted into the BamHI/SalI sites of the pBEP plasmid in frame with a C-terminal 6x-histidine tag to construct pBEP-SaMscL (Supplementary Figure S1). The pBEP and pBEP-SaMscL plasmids were transferred to the S. aureus ATCC 29213ΔmscL mutant strain, and IPTG-induced SaMscL protein expression was confirmed by SDS–PAGE and Western blotting using Histidine (C-term) tag (6XHis) -AP Mouse Monoclonal Antibody (Thermo Fisher Scientific, Waltham, MA, United States) (Supplementary Figure S1). Site-directed mutagenesis of the mscL gene to introduce the substitutions E4C, F5C, and F8C was performed using the forward primers SauC4, SauC5 and SauC8 listed in Supplementary Table S1 and the reverse primer mL6. The SaMscL mutants V14C, M23C, F73C, A77C, F78C, A79C, V84C, and F85C were constructed through overlap extension PCR using a QuickChange II-E Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, United States) with the primer pairs shown in Supplementary Table S1. Changes in the sequence are underlined.
Overnight cultures of wild-type S. aureus ATCC 29213 and its mscL null mutant grown in TSB medium were diluted 400-fold in the same medium and incubated with vigorous shaking for 1.5 h at 37°C. Thereafter, 1 μL of cell suspension was added to 99 μL of xanthorrhizol-enriched TSB medium prepared by twofold dilution in a 96-well plate in 3 replicates. The plates were incubated in an Infinite 200 Pro microplate reader (Tecan Austria GmbH, Grödig, Austria) at 37°C for 16 h. The OD600 was measured every 10 min.
The MIC curves for xanthorrhizol against the exponentially growing S. aureus ATCC 29213ΔmscL mutant strain with controlled expression of the SaMscL protein were obtained with some modifications. Overnight cultures of S. aureus ATCC 29213ΔmscL harboring the pBEP and pBEP-SaMscL plasmids grown in TSBKan25 medium were diluted 100-fold in fresh medium and incubated for 30 min at 37°C. Expression was then induced with 1 mM IPTG, and the cells were grown for 1 h. One microlitre of cell suspension was added to 99 μL of xanthorrhizol-enriched TSBKan25IPTG1mM medium prepared by twofold dilution in a 96-well plate in 3 replicates and incubated in an Infinite 200 Pro microplate reader (Tecan Austria GmbH, Grödig, Austria) at 37°C for 16 h. The OD600 was measured every 10 min.
The viability of exponentially growing S. aureus ATCC 29213ΔmscL cultures harboring the empty vector plasmid pBEP or pBEP-SaMscL was studied 1 h after IPTG induction. Overnight cultures grown in TSBKan25 medium were diluted 1/100 in the same medium, cultivated for 30 min under vigorous aeration at 37°C and induced for 1 h with 1 mM IPTG. After this, the cells were washed with PBS, diluted in PBS to a final concentration of approximately 106 CFU/mL, and treated with 12.8 μg/mL xanthorrhizol. Samples were collected at 5, 10, 20, and 40 min and 1 h, serially diluted 1:10 with sterile PBS containing 1 mM EDTA and 0.2% Kolliphor P188 (Sigma Aldrich, St. Louis, MO) and plated on solid TSA medium for colony counting.
Overnight cultures of S. aureus ATCC 29213 and its mscL null mutant grown in TSB were treated with 64 μg/mL xanthorrhizol for 6 h at 37°C. Sterile DMSO was used in the control experiment. Afterwards, the cultures were diluted 20-fold (Wray et al., 2020) with sterile PBS containing 1 mM EDTA and 0.2% Kolliphor P188 (Sigma Aldrich St. Louis, MO, United States), serially diluted 1:10 in the same buffer and plated on solid TSA medium for colony counting.
S. aureus ATCC 29213ΔmscL cells harboring the pBEP and pBEP-SaMscL plasmids were induced with 1 mM IPTG at an OD600 of 0.3 and incubated overnight at 37°C. Overnight cultures were divided into two parts: one was treated with 64 μg/mL xanthorrhizol, and sterile DMSO was added to the other. After 6 h of incubation at 37°C, samples were taken and processed as described above.
Sample preparation for the glutamate assay was performed as previously described (Wray et al., 2020), with some modifications. Briefly, overnight cultures of the S. aureus ATCC 29213ΔmscL mutant strain carrying the plasmids pBEP and pBEP-SaMscL were diluted 100-fold in fresh TSBKan25 medium, incubated at 37°C and induced with 1 mM IPTG for 1 h after reaching an OD600 of 0.25. Afterwards, each culture was divided into two parts: one was treated with 25 μg/mL xanthorrhizol, and sterile DMSO was added to the other. The culture was incubated for 17 h at 37°C, after which the final OD600 was recorded. For each sample, 6 mL of overnight culture was pelleted, and then the pellets were resuspended in TSB medium, adjusting the volume for the final OD600, and sonicated for 2 min. Glutamate measurements were performed using the EnzyChrom Glutamate Assay Kit (BioAssay Systems, Hayward, CA, United States) in a 96-well plate with 3 replicates, and the OD at 565 nm was read in an Infinite 200 Pro microplate reader (Tecan Austria GmbH, Grödig, Austria) at 25°C. The amount of intracellular K+ was measured in the same samples using the Potassium (K) Turbidimetric Assay Kit (MyBioSource, San Diego, CA, United States) as described in the manual in a 96-well plate; 3 replicates of the OD at 450 nm were read in an Infinite 200 Pro microplate reader (Tecan Austria GmbH, Grödig, Austria) at 25°C. Viability assays were performed simultaneously.
All experiments were performed in triplicate, and the mean ± SD values are displayed in each graph. The significance of differences between mean values of two measured parameters was assessed using a 2-tailed, paired Student’s t test with unequal variances. Differences were considered significant when the p value was < 0.05.
The MIC of xanthorrhizol was determined to be 64 μg/mL for both the MRSA strain ATCC 33592 and the MSSA strain ATCC 29213. Xanthorrhizol at this concentration (1xMIC) was used for the time-kill assay against ATCC 33592 cells (Figure 1A). Samples taken at 1 min and 15 s and later after xanthorrhizol treatment showed no living cells (Figure 1A). We also found no living cells 5 h after exposure to 1 × MIC xanthorrhizol (data not shown).
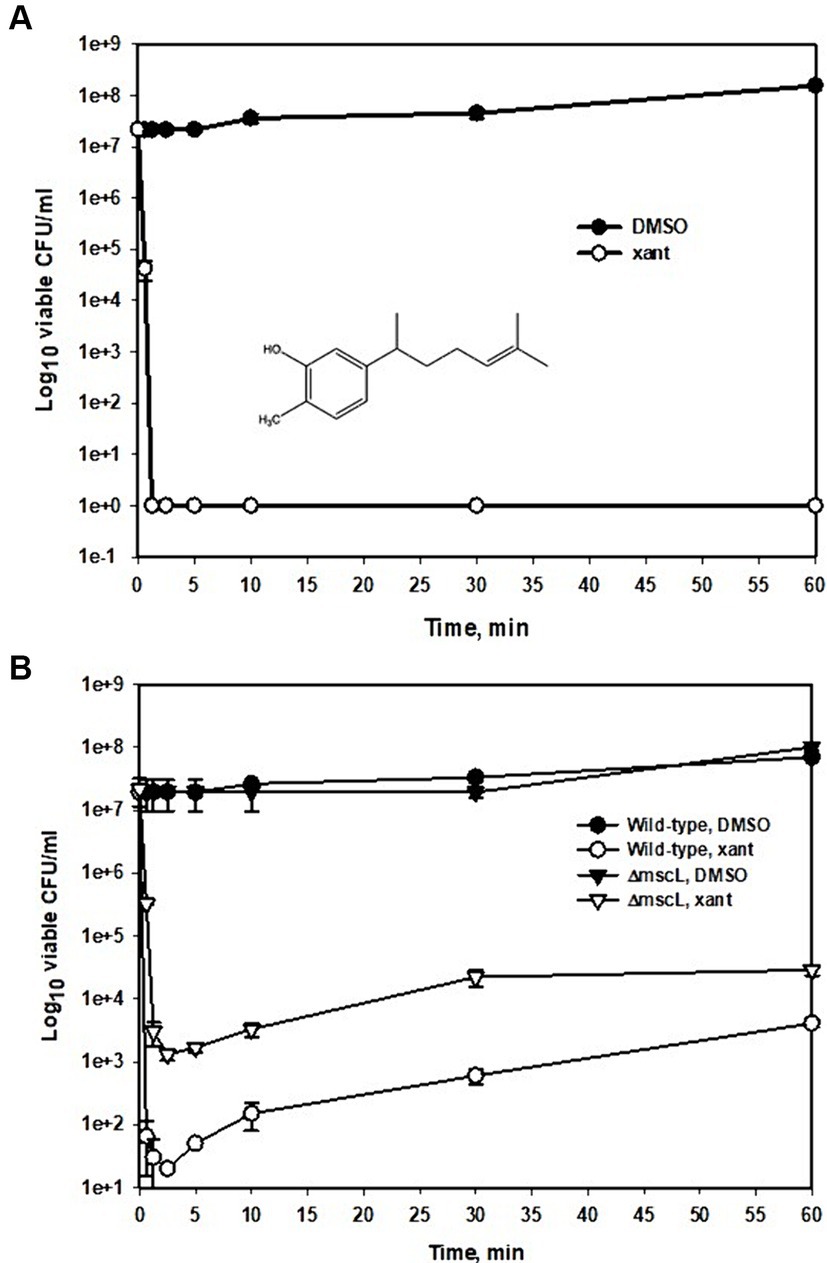
Figure 1. Time-kill kinetic curves of xanthorrhizol against MRSA ATCC 33592 or against MSSA ATCC 29213 and ATCC 29213ΔmscL. Exponentially growing cultures of ATCC 33592 (A) or ATCC 29213 and its mscL null mutant (B) were exposed to 64 μg/mL (A) and 32 μg/mL (B) xanthorrhizol and incubated at 37°C for 1 h. Error bars represent standard deviations from three independent experiments.
Since the MRSA strain ATCC 33592 is resistant to many antibiotics used in genetic experiments, such as erythromycin (100 μg/mL), kanamycin (25 μg/mL), and tetracycline (30 μg/mL) (our unpublished data), we plotted a time–kill curve for the MSSA strain ATCC 29213 (Figure 1B) using 0.5 × MIC xanthorrhizol for a more detailed study of its action against S. aureus. This strain also exhibited high sensitivity to xanthorrhizol when the number of living cells decreased from 2 × 107 CFU/ml to 65 CFU/mL during the first 37 s of incubation with xanthorrhizol and remained at the same level until 5 min after treatment (Figure 1B). From the 10th minute, the number of living cells in the experimental group began to increase, possibly because the time-kill assay was carried out under conditions conducive to growth, and from the 30th minute, the difference between the number of living cells treated with xanthorrhizol and those not treated decreased from 5 and a half orders of magnitude to 4 after 1 h of incubation, amounting to approximately 4 × 103 CFU/ml (Figure 1B).
To investigate changes in individual cells of S. aureus ATCC 29213 treated with xanthorrhizol, we measured 3D RI tomograms of bacteria using optical diffraction tomography (ODT) (Oh et al., 2020) and analyzed the resulting images. We visualized the dynamics of S. aureus cells in growth medium supplemented with or without xanthorrhizol (1xMIC) for 5 min (Figure 2A). As shown in Figure 2A, DMSO-treated control cells appeared healthy during the observation period. In contrast, S. aureus cells treated with xanthorrhizol over time began to look like “shadows” when cytoplasmic components were released, resulting in empty cell membranes (Figure 2A). In addition, between 20 and 25 min of exposure to xanthorrhizol, the RI value suddenly decreased (Figure 2B; Supplementary Table S2), which could be the time point at which bacterial lysis was initiated, since a decrease in the RI value of bacteria indicates the efflux of cell cytoplasm (Oh et al., 2020). This result is consistent with the cytoplasmic concentration data (Figure 2C; Supplementary Table S2), which showed a rapid decrease between 20 and 25 min of observation, which means that after bacterial lysis, the internal contents were released outside the cell wall, resulting in a decrease in the cytoplasmic concentration (Oh et al., 2020). Other morphological characteristics of bacterial growth under xanthorrhizol treatment, such as cell volume and cellular dry mass, also demonstrated a reduction in values starting from the 15th minute (Figures 2D,E; Supplementary Table S2). Moreover, the formation of “shadow cells,” as shown in Figure 2A, was found to be consistent over time, with a decrease in cell volume and dry mass for xanthorrhizol-treated cells. In DMSO-treated control S. aureus cells, the RI and cytoplasm concentration remained unchanged during the observation period (Figures 2B,C; Supplementary Table S2), and the cell volume and cellular dry mass increased (Figures 2D,E; Supplementary Table S3), indicating continuous growth.
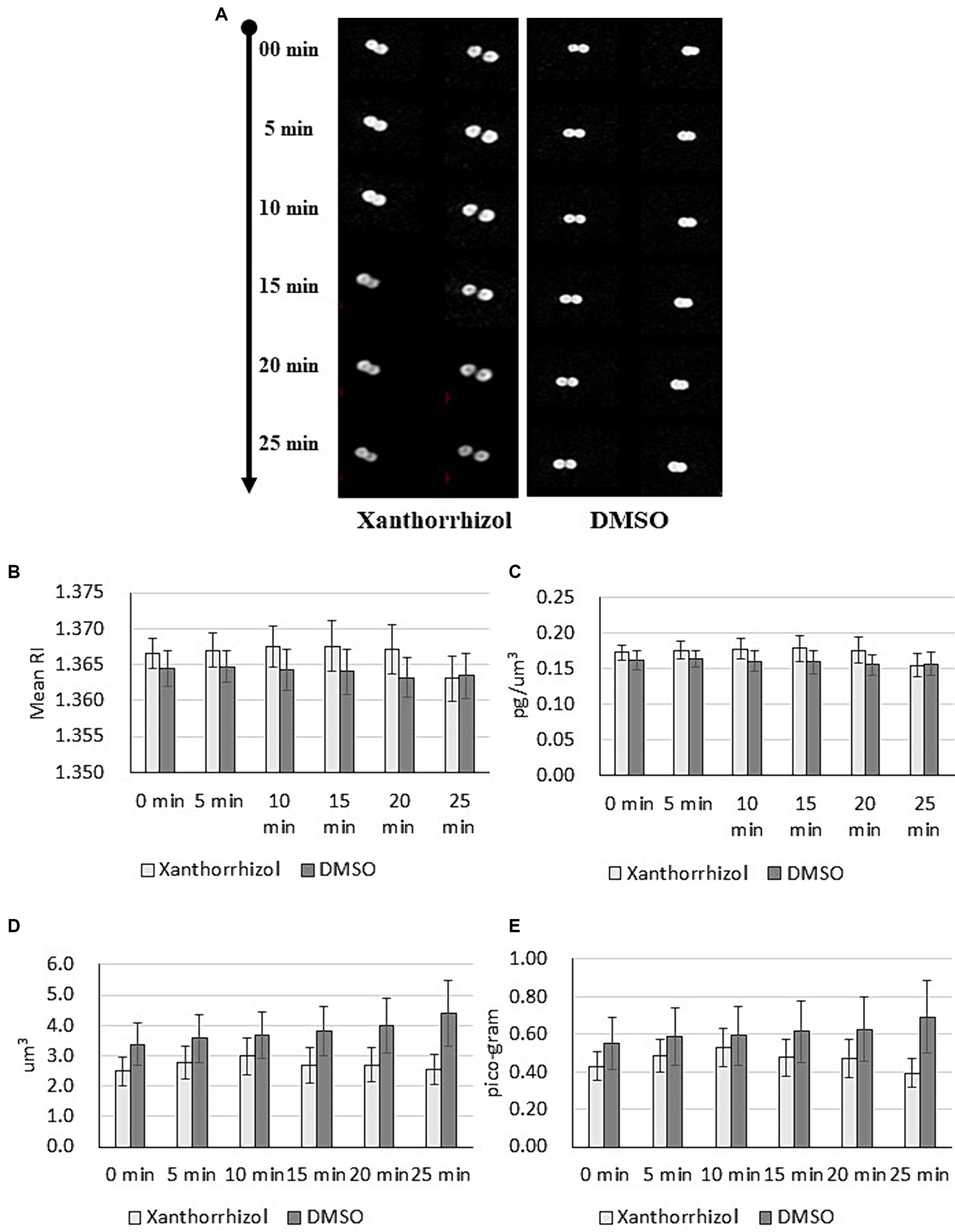
Figure 2. Maximum RI projection images (A) and quantitative analysis results (B–E) for S. aureus ATCC 29213 cells treated with xanthorrhizol or DMSO over time. The mean RI (B), cytoplasm concentration (C), cell volume (D) and cellular dry mass (E) for the control (n = 11) and xanthorrhizol (n = 12) groups were determined over time. The error bars indicate the standard errors.
Thus, the morphological changes found in xanthorrhizol-treated S. aureus cells are consistent with the spontaneous activation of MscL, resulting in the loss of solutes and osmolytes and hence a decrease in the size of S. aureus.
To determine whether the xanthorrhizol susceptibility of S. aureus was dependent on the MscL mechanosensitive channel, we constructed a S. aureus ATCC 29213 mutant lacking the mscL gene and compared the results of a time-kill assay of this mutant with those of the wild-type strain (Figure 1B). The number of living cells of the mscL null mutant after 37 s of exposure to xanthorrhizol was 2 orders of magnitude greater than that of the wild-type strain. The difference in live cell numbers between the wild type and the ΔmscL mutant then began to decrease, reaching approximately 1 order of magnitude after 1 h of xanthorrhizol treatment (Figure 1B).
We tested the effect of xanthorrhizol on the growth of S. aureus ATCC 29213 cells expressing the SaMscL channel at the endogenous level or under IPTG induction in dose dependence experiments (Figure 3A). The naturally expressed protein SaMscL appears to be involved in the xanthorrhizol-induced growth inhibition of wild-type S. aureus ATCC 29213 since the OD600 values of overnight cultures of this strain grown in the presence of 25 and 50 μg/mL xanthorrhizol were 34 and 28% lower, respectively than those of the ΔmscL mutant (Figure 3A). This effect was enhanced when the SaMscL protein was overexpressed in the S. aureus ATCC 29213ΔmscL mutant strain (Figure 3A). The OD600 values of IPTG-induced S. aureus ATCC 29213ΔmscL (pBEP-SaMscL) cells were 2.5- and 100-fold lower than those of cells carrying the empty vector in the presence of 25 μg/mL and 50 μg/mL xanthorrhizol (Figure 3A).
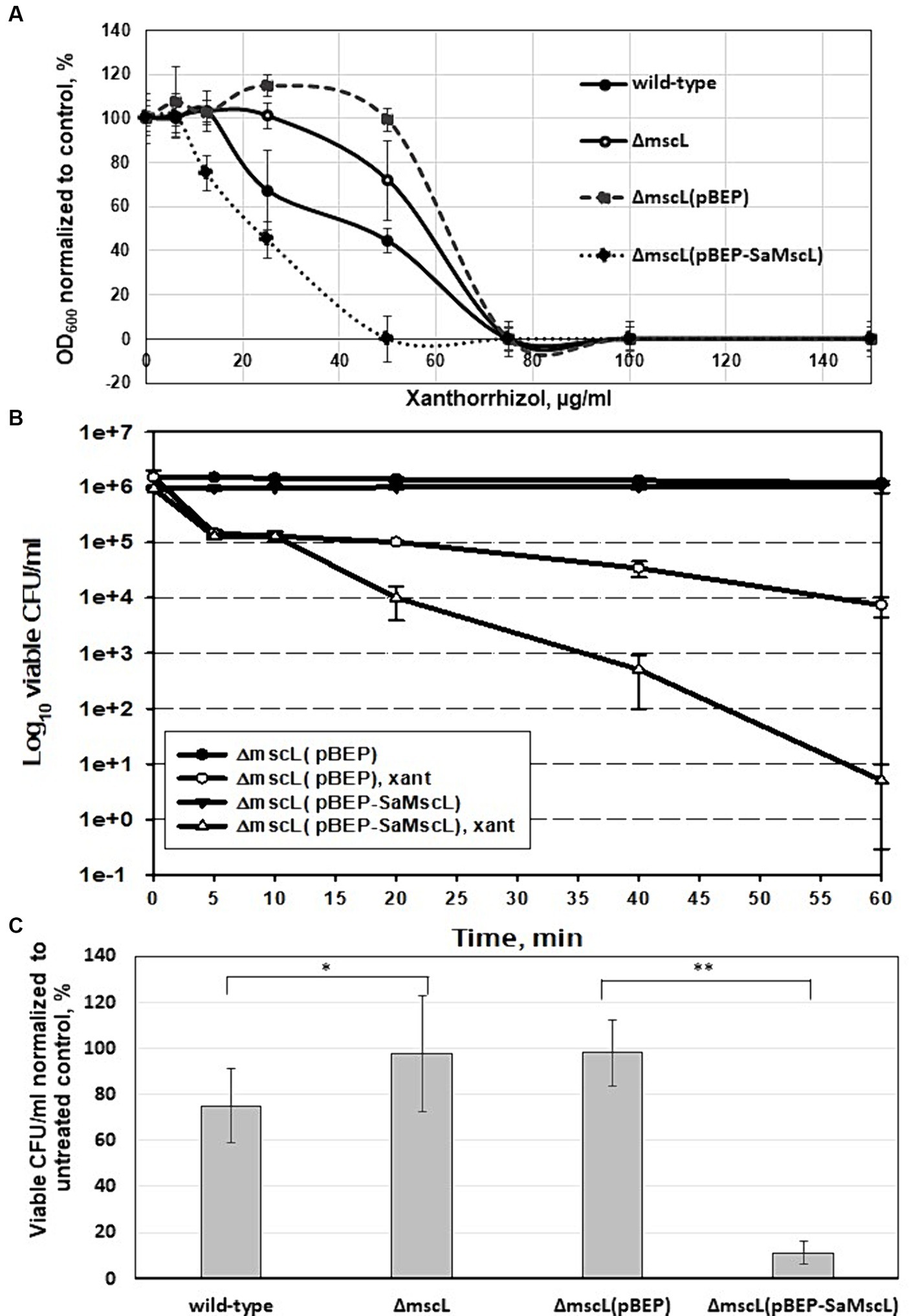
Figure 3. Effect of SaMscL protein deficiency or overproduction on the viability of S. aureus ATCC 29213 and S. aureus ATCC 29213ΔmscL in the presence of xanthorrhizol. Growth inhibition by increasing concentrations of xanthorrhizol was shown for exponentially growing S. aureus ATCC 29213, S. aureus ATCC 29213ΔmscL, S. aureus ATCC 29213ΔmscL (pBEP) and S. aureus ATCC 29213ΔmscL (pBEP-SaMscL) cultures (A). The decrease in growth is reflected as the percentage of the OD600 of the treated cultures to that of the untreated control. The error bars indicate the standard errors. Time-kill kinetic curves of xanthorrhizol against exponentially growing S. aureus ATCC 29213ΔmscL cells carrying the plasmids pBEP or pBEP-SaMscL were obtained 1 h after IPTG induction (B), as described in the Materials and Methods section. The viability of stationary-phase cultures of S. aureus ATCC 29213, S. aureus ATCC 29213ΔmscL, S. aureus ATCC 29213ΔmscL (pBEP) and S. aureus ATCC 29213ΔmscL (pBEP-SaMscL) was studied after treatment with xanthorrhizol for 6 h (C), where *p < 0.01 (n = 3); **p < 0.002 (n = 6) by a 2-tailed, paired Student’s t test.
Additionally, accelerated cell death was demonstrated in exponentially growing S. aureus ATCC 29213ΔmscL cells overexpressing the SaMscL protein compared to empty vector-containing cells treated with 12.8 μg/mL xanthorrhizol (1/5 MIC) (Figure 3B). Therefore, after 1 h of incubation with xanthorrhizol, the number of living S. aureus ATCC 29213ΔmscL (pBEP-SaMscL) cells was found to be 3 orders of magnitude lower than that of S. aureus ATCC 29213ΔmscL (pBEP) cells (Figure 3B).
Previous studies have shown that some antimicrobial agents can significantly reduce the viability of “dormant”/stationary-phase S. aureus cells in a MscL-dependent manner (Wray et al., 2019a, 2020). We tested the viability of S. aureus ATCC 29213 and its ΔmscL mutant cells that reached the stationary phase and were then exposed to 1xMIC xanthorrhizol for 6 h. As shown in Figure 3C, the ΔmscL mutant was more resistant to xanthorrhizol and exhibited close to 100% survival, in contrast to wild-type cells, the viability of which was only 75%. Overexpression of SaMscL in S. aureus ATCC 29213ΔmscL (pBEP-SaMscL) resulted in a sharp decrease in viability (Figure 3C), with only 11% of the cells surviving after xanthorrhizol treatment, in contrast to cells carrying the empty plasmid, 98% of which survived. Thus, the ability of xanthorrhizol to kill stationary-phase S. aureus cells in a MscL-dependent manner may have important medical significance, since S. aureus biofilms found in infection sites (Moormeier and Bayles, 2017) are difficult to treat. Xanthorrhizol has previously been shown to have strong activity against S. mutans biofilms (Rukayadi and Hwang, 2006), possibly due to its action against MscL.
To provide further evidence of the effectiveness of xanthorrhizol against the SaMscL channel, we tested whether xanthorrhizol could increase the potency of dihydrostreptomycin (DHS), a cidal antibiotic, in targeting E. coli MscL and its orthologues from B. subtilis and S. aureus expressed in the E. coli ΔmscL mutant (Iscla et al., 2014; Wray et al., 2016). The MIC of DHS was determined to be 16 μg/mL for wild-type S. aureus ATCC 29213 and 32 μg/mL for S. aureus ATCC 29213ΔmscL. To obtain more detailed information on the activity of DHS against S. aureus, we studied the growth inhibition of exponentially growing cells of the ATCC 29213 strain and its ΔmscL mutant in a dose dependence experiment. As shown in Figure 4A, the OD600 values of the wild-type and ΔmscL overnight cultures were 85 and 91%, respectively, compared to those of the untreated cells at a DHS concentration of 8 μg/mL. However, starting at a DHS concentration of 10 μg/mL, the wild-type OD600 decreased sharply to 24% and subsequently reached 1% at 20 μg/mL DHS, in contrast to the ΔmscL mutant, which retained the same level of viability at DHS concentrations ranging from 10 to 20 μg/mL (Figure 4A). The results of this experiment clearly confirmed that DHS targets SaMscL.
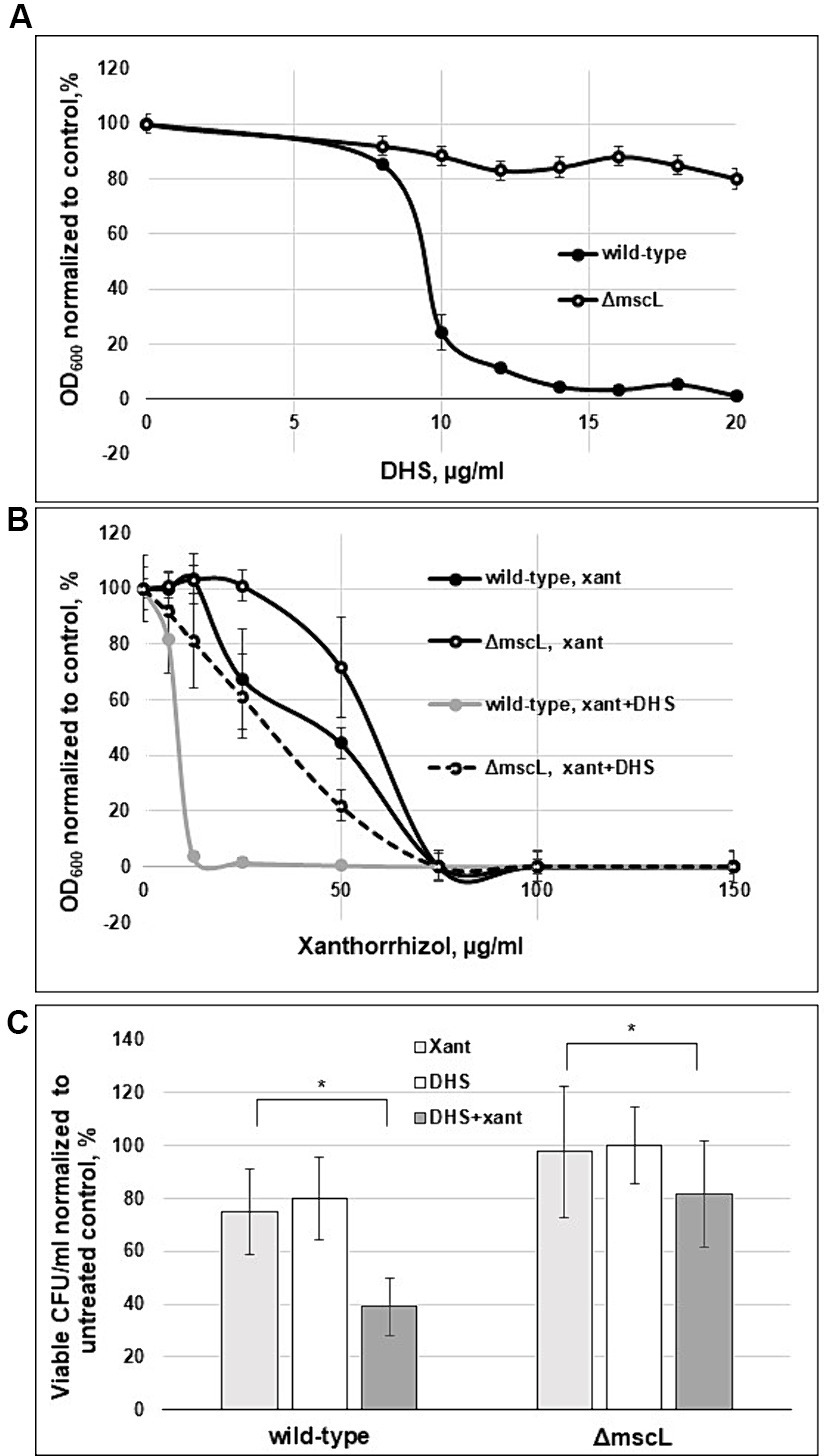
Figure 4. Xanthorrhizol increases the potency of DHS in a MscL-dependent manner. The concentration-dependent effects of DHS (A) and xanthorrhizol in the presence of DHS (8 μg/mL) are shown (B) for exponentially growing S. aureus ATCC 29213 and S. aureus ATCC 29213ΔmscL cells. The decrease in growth is reflected as the percentage of the OD600 of the treated cultures to that of the untreated control. The error bars indicate the standard errors. The viability of stationary-phase cultures of S. aureus ATCC 29213 and S. aureus ATCC 29213ΔmscL (C) was studied after treatment with 50 μg/mL DHS or a combination of DHS (50 μg/mL) and xanthorrhizol (64 μg/mL) for 6 h, where *p < 0.01 (n = 3); *p < 0.05 (n = 3) by a 2-tailed, paired Student’s t test.
We then tested whether xanthorrhizol could increase the potency of the subthreshold concentration of DHS. Exponentially growing cells of the ATCC 29213 strain and its ΔmscL mutant were cultivated in the presence of 8 μg/mL DHS and various concentrations of xanthorrhizol (Figure 4B). After combined treatment with DHS and xanthorrhizol, the growth of the wild-type strain greatly decreased compared with that after treatment with DHS or xanthorrhizol alone (Figures 4A,B). Compared with the wild-type strain, the ΔmscL mutant was more resistant to the simultaneous action of DHS and xanthorrhizol (Figure 4B). However, its viability was also lower with the combination of DHS and xanthorrhizol than with each antimicrobial agent used separately (Figures 4A,B).
We found a similar effect after combinatorial treatment of stationary-phase S. aureus ATCC 29213 and S. aureus ATCC 29213ΔmscL cells, wherein the viability of both strains decreased by approximately half or 11% for the wild-type strain or the ΔmscL mutant, respectively, compared to that in the separate treatment with DHS or xanthorrhizol (Figure 4C). Interestingly, the stationary-phase S. aureus ATCC 29213 cells were more resistant to DHS, and to observe a decrease in viability, we increased the concentration of DHS from 16 μg/mL (1 × MIC) to 50 μg/mL. In this experiment, the ΔmscL mutant also exhibited increased resistance to both antimicrobials in combination or separately (Figure 4C).
The data obtained here showed that xanthorrhizol increases the likelihood of DHS passing through the SaMscL channel, followed by a decrease in the MIC for DHS. However, a visible decrease in the viability of mutant ΔmscL cells after combined treatment with DHS and xanthorrhizol may indicate that SaMscL is not a unique pathway for entry into S. aureus cells.
Previous studies have shown that treatment of E. coli cells overexpressing the McsL channel with some antimicrobial agents increased the efflux of K+ and glutamate from the cytoplasm due to inappropriate gating of MscL (Iscla et al., 2014; Wray et al., 2020). We tested the K+ and glutamate levels in IPTG-induced S. aureus ATCC 29213ΔmscL (pBEP-SaMscL) cells treated with xanthorrhizol. As shown in Figure 5, the relative amount of K+ decreased by 19% and that of glutamate decreased by 30% after treatment with xanthorrhizol compared to 2 and 6%, respectively, in S. aureus ATCC 29213ΔmscL cells carrying the empty vector. Thus, the flux data obtained are consistent with the idea that SaMscL is gated in vivo in response to xanthorrhizol treatment.
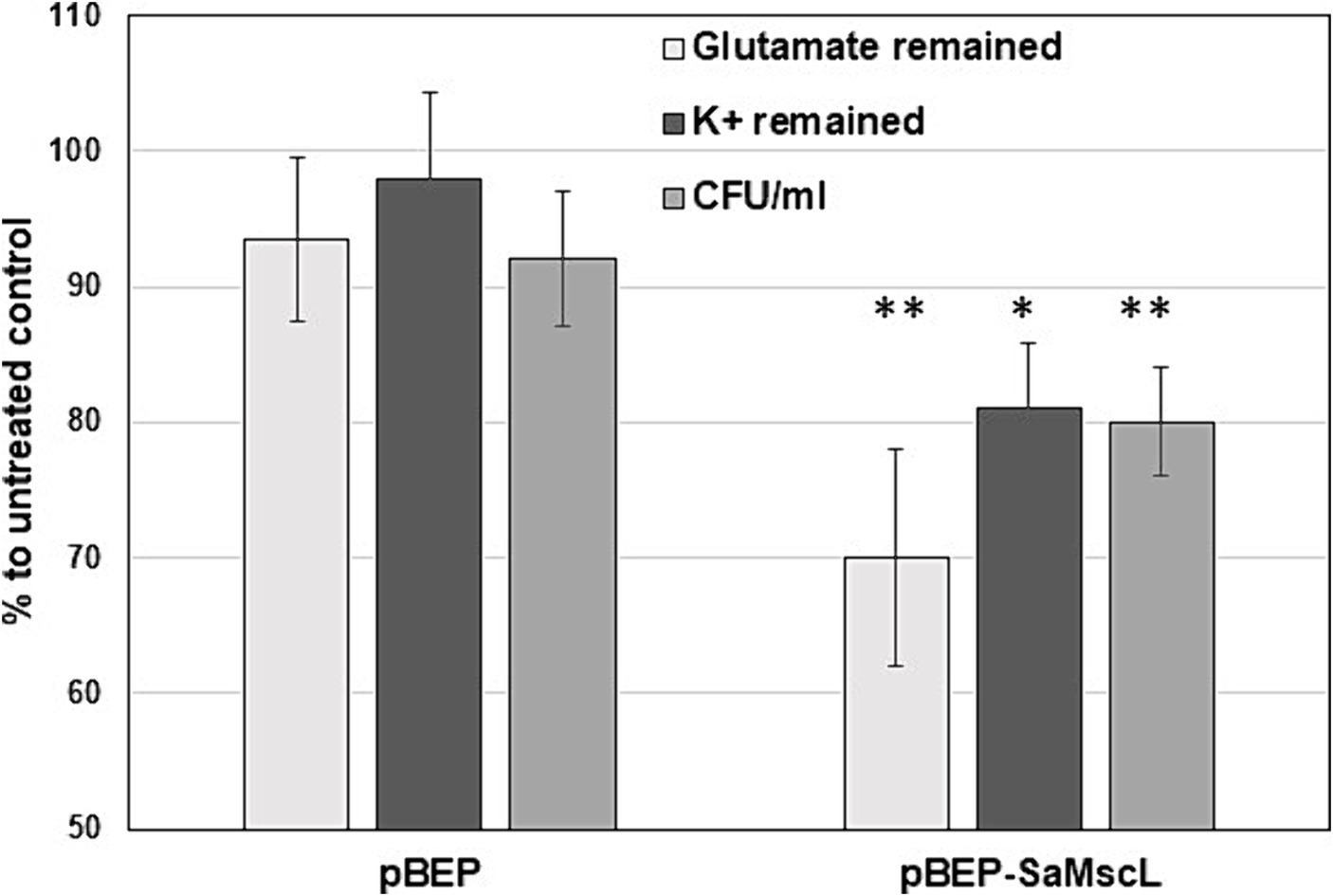
Figure 5. The decrease in the steady-state levels of glutamate and K+ in S. aureus ATCC 29213ΔmscL cells treated with xanthorrhizol is associated with the SaMscL protein The glutamate and K+ levels in S. aureus ATCC 29213ΔmscL cells harboring the empty expression vector pBEP or pBEP-SaMscL after overnight incubation with xanthorrhizol are shown as percentages of those in the untreated controls. Error bars reflect standard deviations from three independent measurements for glutamate, **p < 0.002; for K+, *p < 0.01 by a 2-tailed, paired Student’s t test. The viability of the bacterial cells was measured at the same time and is displayed as a percentage of the viability of the untreated controls. Error bars represent standard deviations from three independent measurements, **p < 0.002 by a 2-tailed, paired Student’s t test.
Previously, successful attempts to determine the binding sites for some antibacterial agents in MscL by comparison to the E. coli channel (EcMscL) have been described using a library of cysteine mutants (Wray et al., 2016, 2019b, 2020). Based on molecular modeling of EcMscL (Wray et al., 2019b), we identified two amino acid residues, F7 and K97, that possibly contribute to the binding of xanthorrhizol (Figures 6A,B). Sequence alignment between the MscL orthologues from E. coli and S. aureus showed that F5 and K85 in SaMscL are analogs of F7 and K97 from EcMscL (Figure 6C). To determine other amino acids involved in xanthorrhizol binding, we checked several residues from SaMscL (E4, F5, F8, V14, M23, F73, A77, F78, A79, V84, and K85). The reason for choosing the amino acid residues mentioned above was that their counterparts in EcMscL have been shown or suspected to be involved in binding to other antimicrobials (Wray et al., 2019b, 2020, 2022). Single substitutions of the aforementioned amino acid residues for cysteine were introduced into SaMscL, a cysteine-free protein, and the expression plasmids pBEP-SaMscL carrying each mutated mscL gene were transformed into the S. aureus ATCC 29213ΔmscL strain. Exponential- and stationary-phase S. aureus ATCC 29213ΔmscL (pBEP-SaMsc) cells overexpressing cysteine mutants of SaMscL were treated with xanthorrhizol at 1/5 × MIC or 1 × MIC, respectively, as described in the Materials and Methods section.
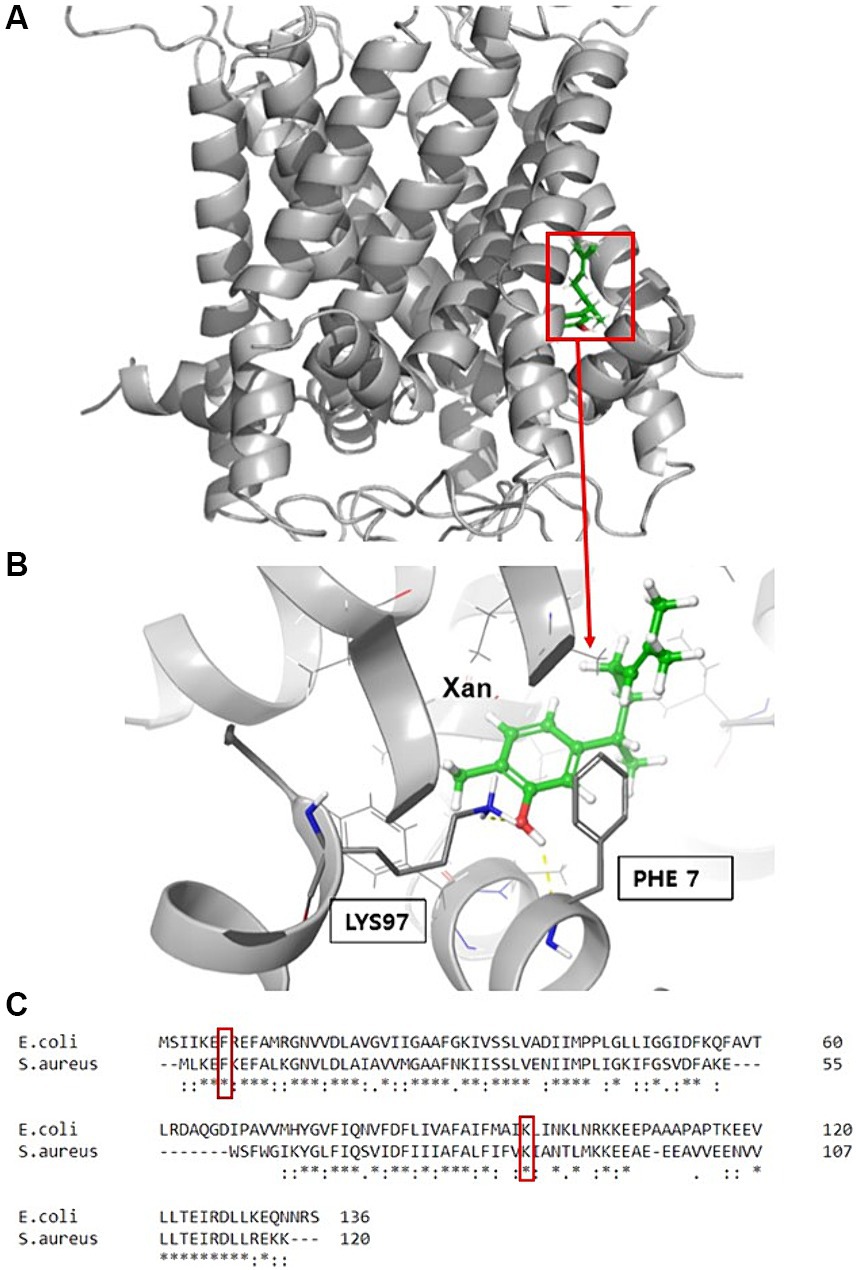
Figure 6. Molecular modeling of EcMscL with the docking site for xanthorrhizol. Xanthorrhizol binding pocket (A) in the EcMscL complex and an enlarged view of xanthorrhizol relative to the positions of residues F7 and K97 (B). Sequence alignment of the MscLs from E. coli and S. aureus with the corresponding residues (C).
Based on the viability data for exponentially growing cells, the SaMscL cysteine mutants could be divided into three groups. Group 1 included the V14C, M23C, F73C, A77C, A79C, V84C, and K85C mutants, which showed almost the same viability in the presence of xanthorrhizol as cells carrying the empty vector (Table 2; Figure 7A). The mutant from group 2, E4C, was as sensitive to xanthorrhizol as cells carrying wild-type SaMscL (Table 2; Figure 7B). Group 3, consisting of mutants F5C, F8C and F78C, demonstrated moderate tolerance/sensitivity to xanthorrhizol, with a decrease in the number of living cells after 1 h of incubation with xanthorrhizol of 3–4 orders of magnitude, compared to 1–2 orders of magnitude in group 1 and 5 orders of magnitude in group 2 (Table 2; Figure 7B).
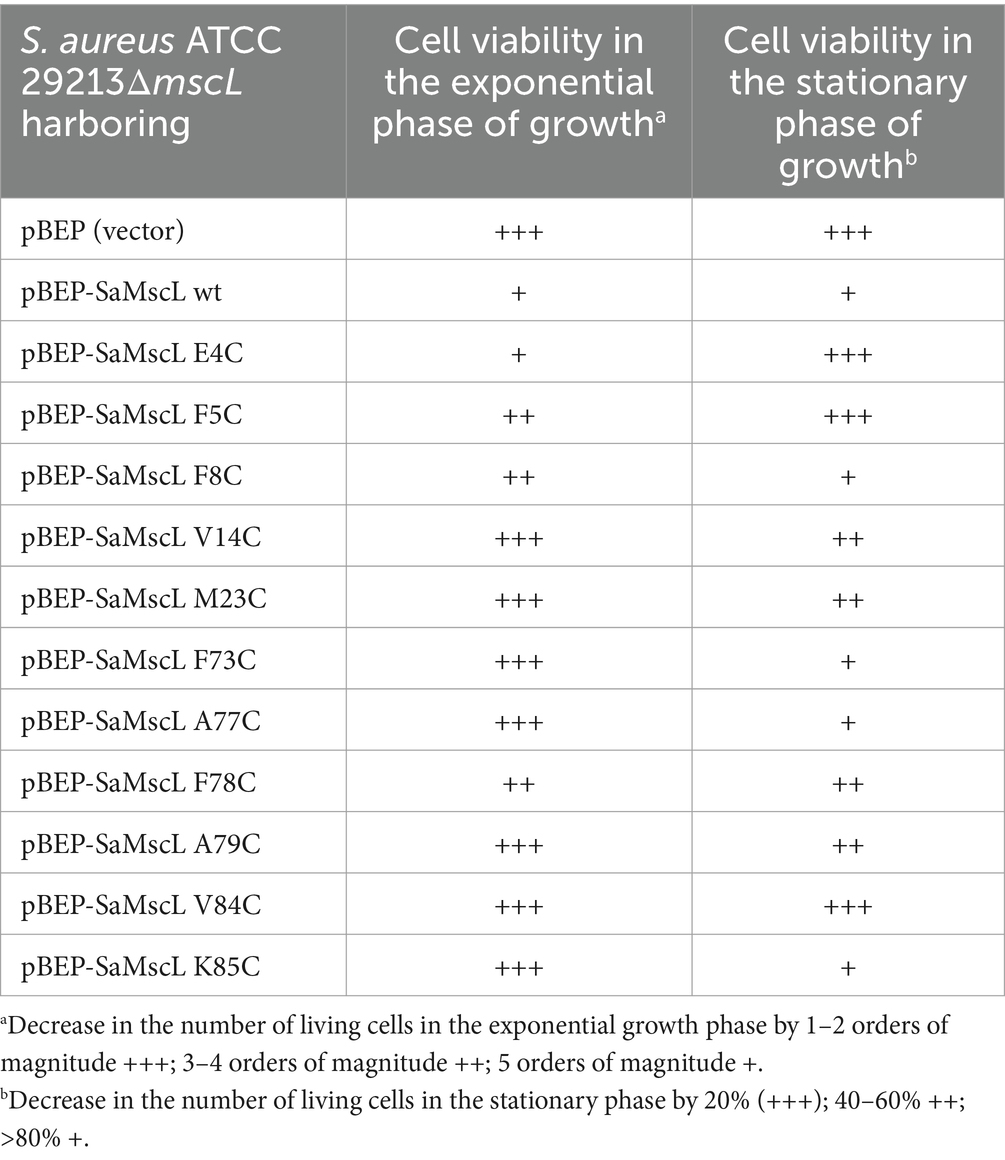
Table 2. Comparative analysis of single amino acid substitutions in the SaMscL protein affecting its tolerance to xanthorrhizol.
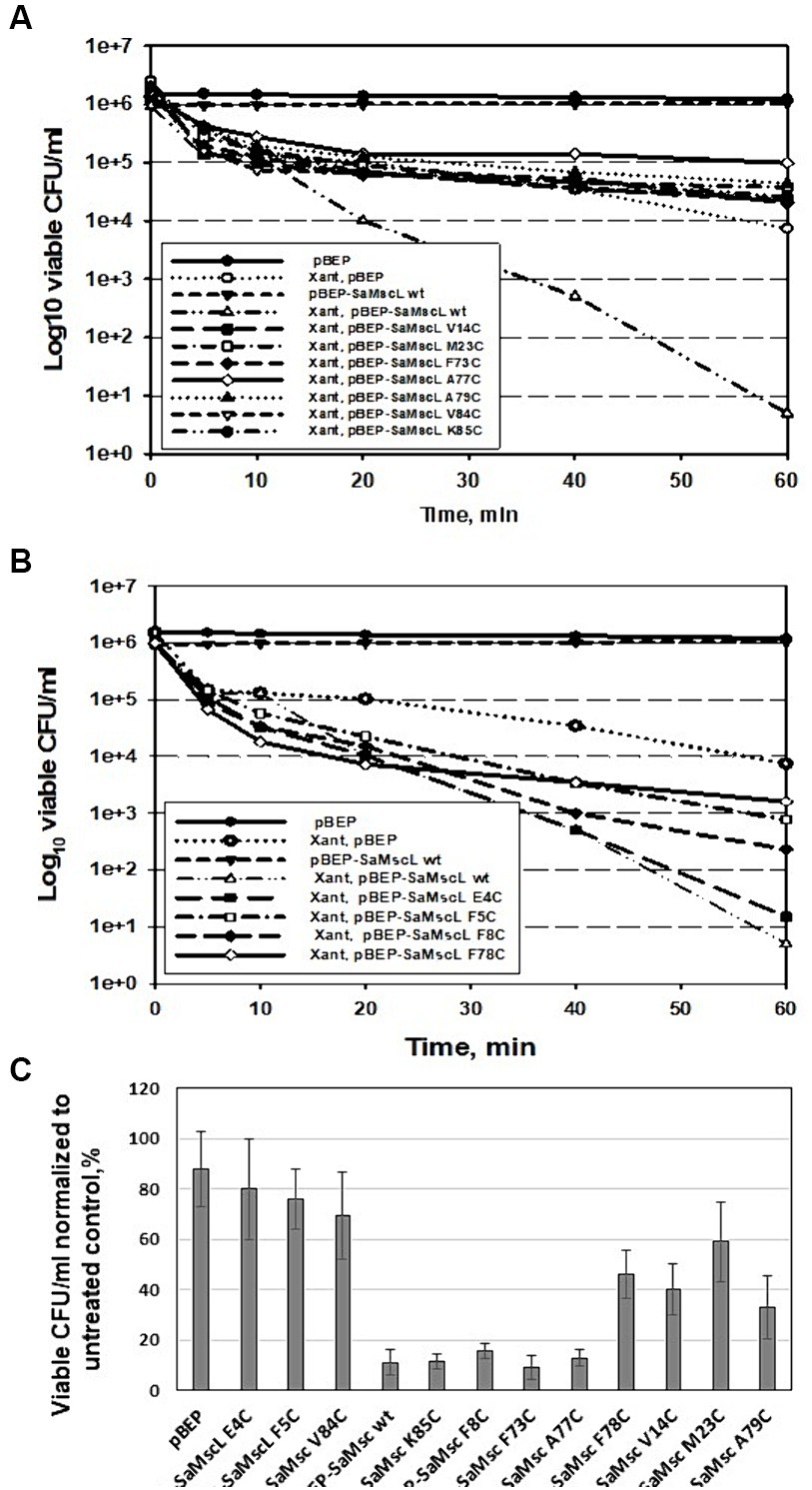
Figure 7. Detection of key amino acid residues involved in the interaction of SaMscL with xanthorrhizol. Time-kill kinetic curves of xanthorrhizol against exponentially growing S. aureus ATCC 29213ΔmscL cells carrying a plasmid (pBEP) with or without mutated mscL genes (A,B). The viability of stationary-phase cultures of S. aureus ATCC 29213ΔmscL carrying the empty vector or expressing wild-type and mutants of SaMscL was studied after treatment with 64 μg/mL xanthorrhizol for 6 h (C).
The viability of stationary-phase cells was found to be different from that of exponentially growing cells. Therefore, group 1 consisted of the E4C, F5C, and V84C mutants, whose tolerance to xanthorrhizol was almost the same as that of cells carrying the empty vector (78, 80, and 69.5%, respectively) (Table 2; Figure 7C). The composition of group 2 was changed to include the mutants F8C, F73C, A77C, and K85C, which exhibited xanthorrhizol sensitivity similar to that of cells carrying wild-type SaMscL (Table 2; Figure 7C). Group 3, with moderate tolerance/sensitivity to xanthorrhizol in the stationary phase, consisted of the mutants V14C, M23V, F78C, and A79C, with cell viabilities of 33–59% (Table 2; Figure 7C).
By comparing viability data from experiments using exponential- and stationary-phase S. aureus cells, the amino acid residues F5, V14, M23, A79, and V84 were predicted to be the residues for the binding of xanthorrhizol.
In the present paper, we report that xanthorrhizol is active against MRSA and MSSA strains at therapeutic concentrations (Figure 1). The undoubted advantage of xanthorrhizol (1 × MIC) is its rapid killing effect on MRSA, which prevents the formation of resistant clones. The 3D RI tomogram of individual staphylococcal cells showed morphological changes after treatment with xanthorrhizol, which are consistent with spontaneous activation of SaMscL, such as a reduction in size and the emergence of “shadow cells” as a result of the loss of solutes and osmolytes (Figure 2). Here, we showed that the efficacy of xanthorrhizol against S. aureus was, at least partially, associated with the mechanosensitive channel SaMscL. The results obtained in experiments with S. aureus ATCC 29213, lacking the mscL gene, with or without SaMscL overexpression, demonstrated that the natural food-borne compound xanthorrhizol targets SaMscL (Figure 3). Our results showed that single cysteine substitutions of many amino acid residues in the SaMscL protein led to a significant decrease in the antimicrobial activity of xanthorrhizol, especially in exponentially growing cells, indicating multiple binding sites for xanthorrhizol with the SaMscL protein. This may explain the rapid killing effect of xanthorrhizol against methicillin-sensitive and MRSA strains during the first 10 min of incubation with xanthorrhizol.
Xanthorrhizol enhanced the antimicrobial effect of DHS against the SaMscL channel in a MscL-dependent manner (Figure 4). A similar effect has been previously described for other antimicrobial compounds, such as 011A and K05, which enhance the effectiveness of DHS, kanamycin, tetracycline and ampicillin against S. aureus and M. smegmatis, suggesting their potential use as antibiotic adjuvants (Wray et al., 2019a, 2020). The authors hypothesized that modulation of MscL gating caused by the above-mentioned compounds may specifically permeabilize the membrane with subsequent facilitation of antibiotic uptake into the cytoplasm. These compounds have been classified as a group that directly affects mechanosensitive channels (Sidarta et al., 2022). Xanthorrhizol apparently belongs to this group, which is confirmed by the identification of amino acid residues that promote its binding to the SaMscL channel. However, a visible decrease in the viability of mutant ΔmscL cells after combined treatment with DHS and xanthorrhizol may indicate that SaMscL is not a unique pathway for entry into S. aureus cells. This assumption is supported by our findings that a mutant strain of S. aureus lacking the mscL gene remained sensitive to xanthorrhizol, but at concentrations higher than those observed for the wild-type strain (Figures 3A, 4B). Xanthorrhizol appears to share some of its antimicrobial properties with curcumin, which also limits bacterial growth in a MscL-dependent manner and increases the activity of MscL channels, possibly due to changes in biophysical properties of the membrane, for example by its thinning, since the direct binding site for curcumin has not been identified (Wray et al., 2021; Lane and Pliotas, 2023). Curcumin, a flavonoid polyphenol isolated from rhizome of turmeric (Curcuma longa L.) (Teow et al., 2016), which is active against both gram-negative and gram-positive bacteria, but its activity against S. aureus is less than that of xanthorrhizol (Teow et al., 2016; our unpublished data).
Based on the results presented in this study, we hypothesize that xanthorrhizol, an antimicrobial agent of plant origin, may be preferable to artificially synthesized compounds for antibacterial therapy. Xanthorrhizol is a multitarget antibiotic that has been evolutionarily adapted to effectively kill gram-positive bacteria, many of which are harmful to humans, but not gram-negative bacteria. The selective activity of xanthorrhizol is especially important for the human body since most gut-resident E. coli cells prevent colonization by pathogenic microflora and produce vitamins K and B12, which are essential for the blood coagulation process and red blood cell formation (Cepas and Soto, 2020). On the other hand, some E. coli species are known to cause infections of the intestines or urinary tract, which necessitates their elimination. To solve this problem, xanthorrhizol has been shown to have good antimicrobial effects against four gram-negative bacteria (E. coli, Salmonella enterica serovar Typhi, S. enterica serovar Typhimurium, and Vibrio cholerae) in combination with polymyxin B nonapeptide and some food-grade antimicrobial agents (Kim et al., 2019).
In sum, we found here that the SaMscL channel is required for xanthorrhizol-dependent decrease in growth and viability of S. aureus cells. Treatment with xanthorrhizol increases the effectiveness of another MscL-targeting antibiotic DHS, and also results in potassium and glutamate fluxes, which is consistent with the opening of the SaMscL channel by xanthorrizol. Our findings thus demonstrate that the SaMscL protein may truly be a target for xanthorrhizol, which provides the opportunity to use this agent as a potential antibiotic against S. aureus. The high level of conservation of the mechanosensitive channel MscL among bacterial species can be used to further improve the function of xanthorrhizol and to develop a safe antimicrobial agent for use both against gram-positive pathogens and in combination with other agents against gram-negative pathogens.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
EM: Formal analysis, Investigation, Methodology, Validation, Writing – original draft, Writing – review & editing. JK: Methodology, Software, Writing – original draft, Writing – review & editing. HJ: Formal analysis, Methodology, Software, Writing – review & editing. KN: Conceptualization, Writing – original draft, Writing – review & editing. J-GP: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Research Foundation of Korea (NRF) Grant (NRF-2017 M3A9E4077291) and GenoFocus internal grant.
We thank Dr. Kee-Hang Lee (Tomocube Inc., Daejeon, Republic of Korea) for optical diffraction tomography of the S. aureus cells.
EM and J-GP were employed by GenoFocus Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1439009/full#supplementary-material
Abdelhamed, H., Lawrence, M. L., and Karsi, A. (2015). A novel suicide plasmid for efficient gene mutation in Listeria monocytogenes. Plasmid 81, 1–8. doi: 10.1016/j.plasmid.2015.05.003
Blount, P., and Iscla, I. (2020). Life with bacterial mechanosensitive channels, from discovery to physiology to pharmacological target. Microbiol. Mol. Biol. Rev. 84, e00055–e00019. doi: 10.1128/MMBR.00055-19
Casey, D., and Sleator, R. D. (2021). A genomic analysis of osmotolerance in Staphylococcus aureus. Gene 767:145268. doi: 10.1016/j.gene.2020.145268
Cepas, V., and Soto, S. M. (2020). Relationship between virulence and resistance among gram-negative bacteria. Antibiotics 9:719. doi: 10.3390/antibiotics9100719
Choi, M. A., Kim, S. H., Chung, W. Y., Hwang, J. K., and Park, K. K. (2005). Xanthorrhizol, a natural sesquiterpenoid from Curcuma xanthorrhiza, has an anti-metastatic potential in experimental mouse lung metastasis model. Biochem. Biophys. Res. Commun. 326, 210–217. doi: 10.1016/j.bbrc.2004.11.020
Chung, W. Y., Park, J. H., Kim, M. J., Kim, H. O., Hwang, J. K., Lee, S. K., et al. (2007). Xanthorrhizol inhibits 12-O-tetradecanoylphorbol-13-acetate-induced acute inflammation and two-stage mouse skin carcinogenesis by blocking the expression of ornithine decarboxylase, cyclooxygenase-2 and inducible nitric oxide synthase through mitogen-activated protein kinases and/or the nuclear factor-κB. Carcinogenesis 28, 1224–1231. doi: 10.1093/carcin/bgm005
Chure, G., Lee, H. J., Rasmussen, A., and Phillip, R. (2018). Connecting the dots between mechanosensitive channel abundance, osmotic shock, and survival at single-cell resolution. J. Bacteriol. 200, e00460–e00418. doi: 10.1128/JB.00460-18
Dorwart, M. R., Wray, R., Brautigam, C. A., Jiang, Y., and Blount, P. (2010). S.aureus MscL is a pentamer in vivo but of variable stoichiometries in vitro: implications for detergent solubilized membrane proteins. PLoS Biol. 8:e1000555. doi: 10.1371/journal.pbio.1000555
Hoffmann, T., Boiangiu, C., Moses, S., and Bremer, E. (2008). Responses of Bacillus subtilis to Hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74, 2454–2460. doi: 10.1128/AEM.01573-07
Hwang, J. K., Shim, J. S., Baek, N. I., and Pyun, Y. R. (2000a). Xanthorrhizol: a potential antibacterial agent from Curcuma xanthorrhiza against Streptococcus mutans. Planta Med. 66, 196–197. doi: 10.1055/s-0029-1243135
Hwang, J. K., Shim, J. S., and Pyun, Y. R. (2000b). Antibacterial activity of xanthorrhizol from Curcuma xanthorrhiza against oral pathogens. Fitoterapia 71, 321–323. doi: 10.1016/s0367-326x(99)00170-7
Iscla, I., Wray, R., Blount, P., Larkins-Ford, J., Conery, A. L., Ausubel, F. M., et al. (2015). A new antibiotic with potent activity targets MscL. J. Antibiot. 68, 453–462. doi: 10.1038/ja.2015.4
Iscla, I., Wray, R., Wei, S., Posner, B., and Blount, P. (2014). Streptomycin potency is dependent on MscL channel expression. Nat. Commun. 5, 1–7. doi: 10.1038/ncomms5891
Kim, M., Cho, H., Ahn, D. G., Jung, H. G., Seo, H. Y., Kim, J. S., et al. (2021). In vitro replication inhibitory activity of xanthorrhizol against severe acute respiratory syndrome coronavirus 2. Biomedicines 9:1725. doi: 10.3390/biomedicines9111725
Kim, J. E., Kim, H. E., Hwang, J.-K., Lee, H. J., Kwon, H. K., and Kim, B. I. (2008). Antibacterial characteristics of Curcuma xanthorrhiza extract on Streptococcus mutans biofilm. J. Microbiol. 46, 228–232. doi: 10.1007/s12275-007-0167-7
Kim, M. S., Kim, H.-R., Kim, H., Choi, S.-K., Kim, C.-H., Hwang, J.-K., et al. (2019). Expansion of antibacterial spectrum of xanthorrhizol against Gram-negatives in combination with PMBN and food-grade antimicrobials. J. Microbiol. 57, 405–412. doi: 10.1007/s12275-019-8511-2
Kouwen, T. R. H. M., Antelmann, H., van der Ploeg, R., Denham, E. L., Hecker, M., and van Dijl, J. M. (2009a). MscL of Bacillus subtilis prevents selective release of cytoplasmic proteins in a hypotonic environment. Proteomics 9, 1033–1043. doi: 10.1002/pmic.200800483
Kouwen, T. R. H. M., Trip, E. N., Denham, E. L., Sibbald, M. J. J. B., Dubois, J.-Y. F., and van Dijl, J. M. (2009b). The large mechanosensitive channel MscL determines bacterial susceptibility to the bacteriocin sublancin 168. Antimicrob. Agents Chemother. 53, 4702–4711. doi: 10.1128/AAC.00439-09
Lane, B. J., and Pliotas, C. (2023). Approaches for the modulation of mechanosensitive MscL channel pores. Front. Chem. 11:1162412. doi: 10.3389/fchem.2023.1162412
Lee, L. Y., Shim, J. S., Rukayadi, Y., and Hwang, J. K. (2008). Antibacterial activity of xanthorrhizol isolated from Curcuma xanthorrhiza Roxb. against foodborne pathogens. J. Food Prot. 71, 1926–1930. doi: 10.4315/0362-028x-71.9.1926
Liu, Z., Gandhi, C. S., and Rees, D. C. (2009). Structure of a tetrameric MscL in an expanded intermediate state. Nature 461, 120–124. doi: 10.1038/nature08277
Martinac, B., Buechner, M., Delcour, A. H., Adler, J., and Kung, C. (1987). Pressure-sensitive ion channel in Escherichia coli. Proc. Natl. Acad. Sci. USA 84, 2297–2301. doi: 10.1073/pnas.84.8.2297
Mata, R., Martinez, E., Bye, R., Morales, G., Singh, M. P., Janso, J. E., et al. (2001). Biological and mechanistic activities of xanthorrhizol and 4-(1′,5′- dimethylhex-4′-enyl)-2-methylphenol isolated from Iostephane heterophylla. J. Nat. Prod. 64, 911–914. doi: 10.1021/np010076o
Moormeier, D. E., and Bayles, K. W. (2017). Staphylococcus aureus biofilm: a complex developmental organism. Mol. Microbiol. 104, 365–376. doi: 10.1111/mmi.13634
Oh, J., Ryu, J. S., Lee, M., Jung, J., Han, S., Chung, H. J., et al. (2020). Three-dimensional label-free observation of individual bacteria upon antibiotic treatment using optical diffraction tomography. Biomed. Opt. Express 11, 1257–1267. doi: 10.1364/BOE.377740
Oon, S. F., Nallappan, M., Tee, T. T., Shohaimi, S., Kassim, N. K., Sa’ariwijaya, M. S. F., et al. (2015). Xanthorrhizol: a review of its pharmacological activities and anticancer properties. Cancer Cell Int. 15, 1–15. doi: 10.1186/s12935-015-0255-4
Rukayadi, Y., and Hwang, J. K. (2006). In vitro activity of xanthorrhizol against Streptococcus mutans biofilms. Lett. Appl. Microbiol. 42, 400–404. doi: 10.1111/j.1472-765X.2006.01876.x
Rukayadi, Y., and Hwang, J. K. (2007). In vitro antimycotic activity of xanthorrhizol isolated from Curcuma xanthorrhiza Roxb. against opportunistic filamentous fungi. Phytother. Res. 21, 434–438. doi: 10.1002/ptr.2092
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual. 2nd Edn. New York: Cold Spring Harbor Laboratory Press.
Schneewind, O., and Missiakas, D. (2014). Genetic manipulation of Staphylococcus aureus. Curr. Protoc. Microbiol. 32:9C.3.1-9C.3.19. doi: 10.1002/9780471729259.mc09c03s32
Sidarta, M., Baruah, L., and Wenzel, M. (2022). Roles of bacterial mechanosensitive channels in infection and antibiotic susceptibility. Pharmaceuticals 15:770. doi: 10.3390/ph15070770
Teow, S. Y., Liew, K., Ali, S. A., Khoo, A. S. B., and Peh, S. C. (2016). Antibacterial action of curcumin against Staphylococcus aureus: a brief review. J. Trop. Med. 2016, 1–10. doi: 10.1155/2016/2853045
Wang, J., and Blount, P. (2022). Feeling the tension: the bacterial mechanosensitive channel of large conductance as a model system and drug target. Curr. Opin. Physio. 31:100627. doi: 10.1016/j.cophys.2022.100627
Wiegand, I., Hilpert, K., and Hancock, R. E. W. (2008). Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 3, 163–175. doi: 10.1038/nprot.2007.521
Wray, R., Herrera, N., Iscla, I., Wang, J., and Blount, P. (2019a). An agonist of the MscL channel affects multiple bacterial species and increases membrane permeability and potency of common antibiotics. Mol. Microbiol. 112, 896–905. doi: 10.1111/mmi.14325
Wray, R., Iscla, I., and Blount, P. (2021). Curcumin activation of a bacterial mechanosensitive channel underlies its membrane permeability and adjuvant properties. PLoS Pathog. 17:e1010198. doi: 10.1371/journal.ppat.1010198
Wray, R., Iscla, I., Gao, Y., Li, H., Wang, J., and Blount, P. (2016). Dihydrostreptomycin directly binds to, modulates, and passes through the MscL channel pore. PLoS Biol. 14:e1002473. doi: 10.1371/journal.pbio.1002473
Wray, R., Iscla, I., Kovacs, Z., Wang, J., and Blount, P. (2019b). Novel compounds that specifically bind and compounds that specifically bind and modulate MscL: insights into channel gating mechanisms. FASEB J. 33, 3180–3189. doi: 10.1096/fj.201801628R
Wray, R., Wang, J., Blount, P., and Iscla, I. (2022). Activation of a bacterial mechanosensitive channel, MscL, underlies the membrane permeabilization of dual-targeting antibacterial compounds. Antibiotics 11:970. doi: 10.3390/antibiotics11070970
Wray, R., Wang, J., Iscla, I., and Blount, P. (2020). Novel MscL agonists that allow multiple antibiotics cytoplasmic access activate the channel through a common binding site. PLoS One 15:e0228153. doi: 10.1371/journal.pone.0228153
Yogiara, K., Kim, D., Hwang, J.-K., and Pan, J.-G. (2015). Escherichia coli ASKA clone library harboring tRNA-specific adenosine deaminase (tadA) reveals resistance towards xanthorrhizol. Molecules 20, 16290–16305. doi: 10.3390/molecules200916290
Keywords: food-grade antimicrobial, xanthorrhizol, MscL, Staphylococcus aureus , mutants
Citation: Mordukhova EA, Kim J, Jin H, No KT and Pan J-G (2024) The efficacy of the food-grade antimicrobial xanthorrhizol against Staphylococcus aureus is associated with McsL channel expression. Front. Microbiol. 15:1439009. doi: 10.3389/fmicb.2024.1439009
Received: 27 May 2024; Accepted: 18 June 2024;
Published: 03 July 2024.
Edited by:
Miklos Fuzi, Independent Researcher, Seattle, WA, United StatesReviewed by:
Paul Blount, University of Texas Southwestern Medical Center, United StatesCopyright © 2024 Mordukhova, Kim, Jin, No and Pan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jae-Gu Pan, amdwYW5AZ2Vub2ZvY3VzLmNvbQ==; amdwYW5Aa3JpYmIucmUua3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.