- 1Antibiotics Research and Re-evaluation Key Laboratory of Sichuan Province, Sichuan Industrial Institute of Antibiotics, School of Pharmacy, Chengdu University, Chengdu, China
- 2State Key Laboratory of Agricultural Microbiology, Hubei Hongshan Laboratory, College of Life Science and Technology, Huazhong Agricultural University, Wuhan, China
Introduction: Limosilactobacillus reuteri effectively colonizing the gut, secretes antimicrobial compounds and strengthens immune system function. Considering these health benefits, increasing its stress assessments efficiency could improve its commercial viability.
Methods: In this work, the resistance of L. reuteri FP41 to acid, bile salts, and freeze-drying was examined.
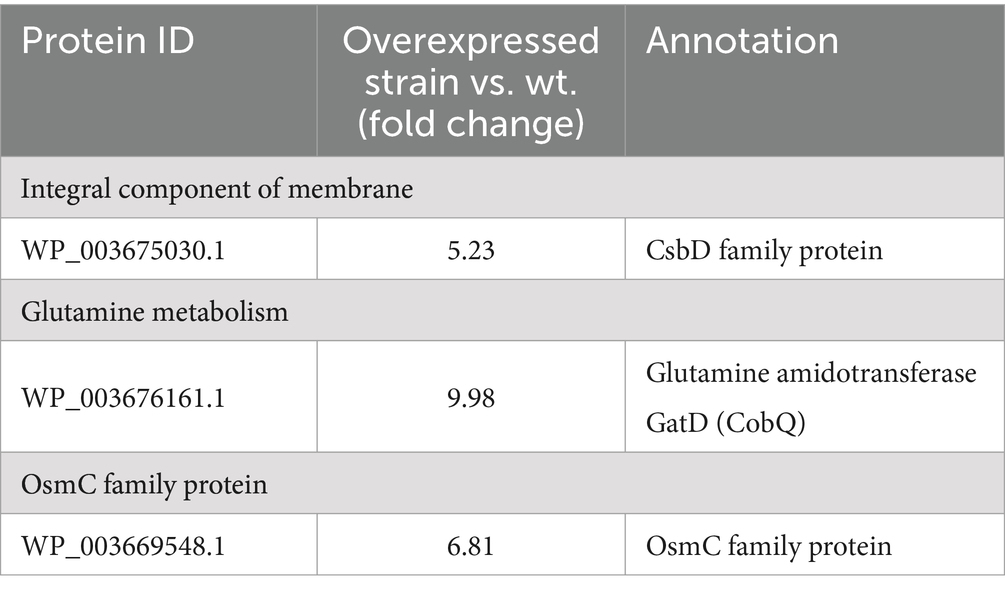
Results: The findings showed that strain FP41 demonstrated a strong resistance to acid/bile salt stresses. The transcriptome revealed a significant up-regulation of various stress response genes, including those related to membrane integrity, glutamine metabolism, OsmC family protein, ABC transporters, and chaperonin. Subsequent research demonstrated that overexpression of three stress response-specific proteins, including glutamate decarboxylase GatD, osmotically induced bacterial protein OsmC, and membrane protein component CsbD, significantly increased the survival rate of L. reuteri Z204 under acid/bile salts stress. Notably, overexpression of the OsmC, CsbD, and GatD proteins also enhanced the survival of L. reuteri after freeze-drying.
Discussion: The development of a unique cross-protection method is highlighted in this study, that might significantly increase cellular resistance to acid, bile salts, and cold stresses. This finding could significantly impact the way that L. reuteri is employed in industrial manufacturing processes.
Introduction
Probiotics are known as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host.” Through consumption of fermented foods or powdered probiotic supplements, lactic acid bacteria (LAB), a type of bacteria that can produce significant levels of lactic acid, are commonly employed as probiotics to improve immunity and promote health (Daba and Elkhateeb, 2020). Bacteria are exposed to a variety of extreme environmental conditions throughout the production and utilization of LAB, including osmotic pressure, temperature, oxidation stress, and starvation (Mbye et al., 2020). Bacteria encounter challenges in the gastrointestinal tract due to low pH and high osmotic pressure caused by stomach acid and bile salts. Probiotics, on the other hand, need to overcome these stresses in order to colonize and function effectively in the human intestine. The phospholipids and proteins in the cell membranes of bacteria in the gastrointestinal system can be disrupted by exposure to bile salts, which can impair their ability to survive and disturb their cellular homeostasis (Ruiz et al., 2013). The favored approach for long-term preservation in strain resource centers and production industries is now freeze-drying due to its more effective long-term cell survival rate and ease of preservation, transportation, and application (Liu Y. et al., 2022). Bacteria are exposed to multi-stresses throughout the freeze-drying process, including cold, desiccation, crystallization, osmotic stress and others. These stresses have the ability to alter the structure of cell membrane and cellular physiology, which may lower bacterial activity (Gao et al., 2022). But its understanding of how exactly LAB responds to these stressors is still limited.
Extreme environmental stimuli usually have an impact on a wide range of biological functions in bacteria. Adaptive mechanisms, including alterations to protein expression, can be induced by sublethal environmental stresses, enabling them to withstand harsh environments (Mbye et al., 2020). For example, the expression of stress response proteins HSPs, under harsh settings may enhance the probiotics’ ability to withstand heat during food processing and boost their survival rate after freeze-drying (Liu et al., 2015; Chen et al., 2017). It is now commonly recognized that lactic acid rapidly permeates the cytoplasmic membrane, dissociates into protons to cause intracellular acidification, and ultimately causes damage to DNA and proteins, all of which negatively affect LAB (Shabala et al., 2006). The ATP synthase subunit beta and chaperone protein DnaK was used to against the acid stress in L. pentosus (Pérez Montoro et al., 2018). Modulation membrane fluidity, protecting and repairing macromolecules, strengthening the proton pump, controlling enzyme and metabolic activity, and consuming an excessive amount of cytoplasmic protons can all help reduce these negative effects (Gao et al., 2022). LAB possesses several established bile salts tolerance mechanisms, including stress protein synthesis, alterations to cell membranes, and bile salts metabolism (Bagon et al., 2021; Ayyash et al., 2021). It’s worth noting that resistance to certain stresses as well as unrelated stresses that improve overall tolerance for multiple environmental stresses can cross-protect probiotic survival in LAB (Gao et al., 2022). Accordingly, in order to minimize stress-related damage and improve survival rates, it is crucial to gain a thorough understanding of how LAB responds to environmental stresses. It is essential for the screening and selection of candidate probiotic strains (Mbye et al., 2020).
Almost all vertebrates and other animals have the heterotypic fermentative bacterium Limosilactobacillus reuteri in the gastrointestinal tracts, which serves a variety of health-beneficial functions for its host (Mu et al., 2018). Crucial steps towards expanding L. reuteri’s industrial application include investigating its stress response system and enhancing its adaptability through genetic modification. In this study, we utilized transcriptome analysis to identify the critical genes that L. reuteri FP41, a highly viable and resistant bacterium isolated from the intestine of a healthy pig, responds to acid/bile salt stress. By assessing the survival rate under the acid/salts stress after these observed stress-related genes were overexpressed, the mechanism underlying L. reuteri’s resistance to these stresses was studied. Furthermore, the identified genes were investigated for their potential role in L. reuteri’s cross-protection mechanism and freeze-drying tolerance. This work provides new insights into the processes by which L. reuteri responds to environmental stress and establish a framework for future research on the utilization of cross-protection mechanisms in stress response.
Materials and methods
Strains, cultural conditions, and transformation
L. reuteri strains FP41 (MGSC70001), FP42, and Z204 were isolated from the feces of pigs (The fecal samples were gradient-diluted with sterile water, covered with MRS Plates, and single colonies were chosen and repeatedly emphasized before being screened and identified.), L. reuteri strains ATCC 23272 was obtained from ATCC. The DH5α strain of Escherichia coli is used for gene cloning. E. coli strains was cultured in LB medium with or without 100 μg/mL Ampicillin in order to screen positive clones (Coolaber, Beijing, CN). The strains of L. reuteri ATCC23272, FP41, FP42, and Z204 were cultured at 37°C in MRS medium.
The construction of overexpression plasmids was based on pMG36e. These genes were amplified from L. reuteri strains FP41 genomic DNA using Phanta DNA Polymerase (Vazyme, Nanjing, China), and the primers listed in Supplementary Table S1. The restriction enzyme XbaI/NotI was used to digest the osmC, csbD, and cobQ gene segments that were produced using PCR and plasmid pMG36e over night at 37°C. The digested products were subsequently transformed into DH5a after being exposed to T4 ligase for 2 hours at 22°C. After an overnight culture, transformants were chosen, PCR analysis and sequence were used to confirm positive clones. Plasmids from positive clones were extracted and being electroporated into L. reuteri 204. For L. reuteri strains’ electroporation (Liu et al., 2023; Guo et al., 2023), 500 ng plasmids were added to 80 μL competent cells and electroporated at 2,500 V, 25 μF, and 200 Ω. The cuvette was immediately put in an ice bath for 5 min after electroporation and filled with prechilled modified MRS liquid medium supplemented with 171.15 g/L sucrose (Sinopharm, Beijing, CN). Following a 6 h incubation at 37°C, the cells were centrifuged and the remaining bacterial solution was resuspended in 100 μL. The resuspended solution was spread on an agar plate containing MRS medium with 5 μg/mL erythromycin (Coolaber, Beijing, CN), and incubated at 37°C for 2 days.
Growth curves test
Every two hours, the OD600 value was measured while the 1% overnight-cultured L. reuteri was added to MRS liquid medium and incubated at 37°C. The culture was diluted and plated onto solid MRS media after a 12 h fermentation. After that, the plates were incubated for 48 h at 37°C to count the viable bacteria. The pH of MRS was adjusted to 2.0, 3.0, 4.0, 5.0, 6.0 and 6.8 to obtain the acid tolerance growth curves; on the other hand, the addition of pig bile salt (0.02, 0.04, 0.06, and 0.08%) to MRS generated the bile salt tolerance growth curves.
RNA library construction, quantification, and transcriptome analysis
The quality and the quantity of the extracted total RNA were determined by Nano Photometer spectrophotometer and Qubit 2.0 Fluorometer. RNA was examined for integrity by Agilent 2,100 bioanalyzer before being used as input material for cDNA library preparations.
HiScript III RT SuperMix for qPCR (Vazyme, Nanjing, CN) was used to reverse transcribed 1 μg of each total RNA sample in order to synthesis cDNA for RT-qPCR (Conesa et al., 2016). Utilizing the Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, CN), real-time qPCR was performed out in an ABI Real-Time PCR Instrument. Supplementary Table S1 lists the amplified targets’ primer sequences.
Using the RIBO-Zero kit, rRNA was extracted to enrich the mRNA, which was then fragmented and used as a template for cDNA synthesis with random hexamers for transcriptome analysis. The double-stranded cDNA was purified using AMPure XP Beads and the USER enzyme was used to remove the second strand containing Uracil. Using an Illumina HiSeq2000 instrument, the cDNA end was repaired, polyA tail was added, splice sequenced and length screened. Before aligning the raw data to the reference genome sequence of L. reuteri I5007, adapter sequences and low-quality bases were removed in order to obtain clean reads (Powell et al., 2012). For rapid genome-to-sequencing data comparison, the Burrows-Wheeler Transform (BWT) algorithm with Ferragina-Manzini (FM) indexes was used. The reads mapped to each gene were then counted using the HTSeq software, and per kilobase per million mapped reads (RPKM) was then calculated based on gene length and the number of mapped reads (Chang et al., 2009; McClure et al., 2013). We expressed transcript expression levels using RPKM in order to determine the levels of gene expression in different groups. Significant differences in gene expression were analyzed using the DESeq2 software (Kanehisa et al., 2004). To identify differentially expressed genes (DEGs), a p-value>0.05 and a log foldchange (logFC) < 2 were established as criteria. Transcriptome data were deposited in the SRA database under Accession number PRJNA1032396.
Acid tolerance test
The artificial gastric juice (Pedersen et al., 2004) contained: 3.50 g/L glucose, 2.05 g/L NaCl, 0.11 g/L CaCl2, 0.37 g/L KCl, 0.60 g/L KH2PO4, 0.05 g/L pig bile salt (Sinopharm, Beijing, CN); 8.30 g/L peptone (OXOID, United Kingdom), 0.10 g/L Lysozyme (Amresco, United States), 13.30 mg/L Pesin (Macklin, Shanghai, CN), and the pH was adjusted to 2.5 for sterilization and backup use. 1 mL of L. reuteri overnight cultures was added into 9 mL artificial gastric juice, and cultured in a 37°C incubator. Viable bacterial count was measured by sampling at 0, 1, and 3 h, respectively. The equation for survival rate is as follows: survival rate = number of bacteria after treatment / number of bacteria in 0 h before treatment * 100%.
Bile tolerance test
MRS media containing 0.1, 0.3 and 0.5% (w/v) of pig bile salt were prepared, respectively (Nithya and Halami, 2012). One ml of L. reuteri overnight cultures was inoculated into 9 mL of pig bile solution at different concentrations, and the mixture was then incubated at 37°C for 12 h. Samples were taken for dilution plate counting at the 0, 12 h. Viable bacteria count was measured. The equation for survival rate is as follows: survival rate = number of bacteria after treatment / number of bacteria in 0 h before treatment * 100%.
Freeze drying test
The bacteria were centrifuged for 10 min at 8, 000 g, 4°C. The bacterial pellet was collected and the supernatant was removed. The bacteria suspension was mixed with the protection agent solution (10% skimmed milk powder, 6% maltose, 1.5% mannitol and 0.8% glycine) at a volume ratio of 1:9 after the bacteria mud and water were mixed at a mass ratio of 1:1. 1 mL was taken and divided into the freeze-drying bottle once it was completely homogenous. The procedures for freeze drying were as follows: pre-freeze for 5 h at −80°C, followed by freeze drying for 12 h at −50°C and 0.25 mba of vacuum pressure. The equation for survival rate is as follows: survival rate = number of bacteria after treatment / number of bacteria in 0 h before treatment * 100%.
Proteome analysis
Proteins were extracted from the overnight cultured sample by lysing it in SDT buffer (4% SDS, 100 mM Tris–HCl, pH 7.6). After filter-aided sample preparation (FASP), protein digestion was performed. For peptide quantification (OD280), the resulting peptide digests from each sample were desalted using C18 Cartridges, lyophilized, and reconstituted in 40 μL of 0.1% (v/v) formic acid. Using a timsTOF Pro mass spectrometry system (Bruker) coupled to Nanoelute (Bruker), LC–MS/MS analysis was conducted. After loading the peptides onto a C18-reversed phase analytical column (Thermo Scientific Easy Column, 25 cm long, 75 μm inner diameter, resin particle size: 1.9 μm) with an initial composition of 95% buffer A (0.1% Formic acid in water), followed by separated using a linear gradient of buffer B (99.9% acetonitrile and 0.1% Formic acid) at a flow rate of 300 nL/min. With a 1.5 kV electrospray voltage applied, the mass spectrometer operated in positive ion mode. The TOF detector analyzed precursor ions and fragments within the mass range of m/z 100–1700. The timsTOF Pro was utilized in the parallel accumulation serial fragmentation (PASEF) (Meier et al., 2018) mode, and the subsequent parameters were followed for data collection: 1 MS and 10 MS/MS PASEF scans were performed with an ion mobility coefficient (1/K0) value ranging from 0.6 to 1.6 Vs cm2, and active exclusion was enabled with a 24 s release time. MaxQuant (Cox and Mann, 2008) version 1.6.14 software was used for combining and analyzing the MS raw data for each sample (Yu et al., 2004; Finn et al., 2016; Schwanhäusser et al., 2011; Cox et al., 2011). The MS data have been deposited to the ProteomeXchange Consortium via the iProX repository with the data set identifier IPX0007438000. The DEGs are listed in Supplementary Table S2.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 8 and SPSS. One-way analysis of variance (ANOVA) was used to evaluate the data, and then Tukey’s back testing was performed to determine the statistical difference between each group.
Results and discussion
Analysis of Limosilactobacillus reuteri growth characteristics and the acid/bile tolerance
As a probiotic, L. reuteri’s tolerance to low pH and high bile salts is vital for its survival in gastric juice and the intestines. The starting strains for further stress resistance studies were selected based on the growth characteristics and acid/bile tolerance of L. reuteri.
According to the findings, the strains of L. reuteri FP41, FP42, Z204 and ATCC 23272 showed the highest growth with OD600 values of 3.73, 2.83, 2.88 and 3.94, respectively (Figure 1A), and with the highest viable counts were 9.15 × 108 CFU/mL, 4.55 × 108 CFU/mL, 7.10 × 108 CFU/mL and 12.98 × 108 CFU/mL, respectively. These results indicate that, while growing similarly to type strain ATCC 23272, the candidate strain FP41’s growth curve was significantly better than FP42 and Z204. Strong acid tolerance was demonstrated by L. reuteri FP41, FP42, ATCC 23272 and Z204, as they were able to survive in gastric juice with a pH of 2.5 (Figure 1B), suggesting their advantages in passing through the human digestive tract (pH 1.5–7.5) (Fallingborg, 1999). Furthermore, FP41 and FP42 showed a significantly higher survival rate than ATCC 23272 and Z204 at 3 h upon acid treatment. Even at a concentrations of 0.5%, L. reuteri FP41, FP42 and ATCC23272 all shown good tolerance to bile salts (Figure 1C), suggesting their advantages in passing through the human small intestine (Begley et al., 2005). When treated with pig bile salts at concentration of 0.3 and 0.5%, FP41 had a significantly higher survival rate than ATCC23272, which is similar to acid tolerance. Furthermore, compared to other strains, Z204 showed a markedly lower bile salt tolerance, even at a bile salts concentration of just 0.1%. These findings led us to choose FP41 as the transcriptome analysis strain and Z204 as the host for the overexpression of stress proteins.
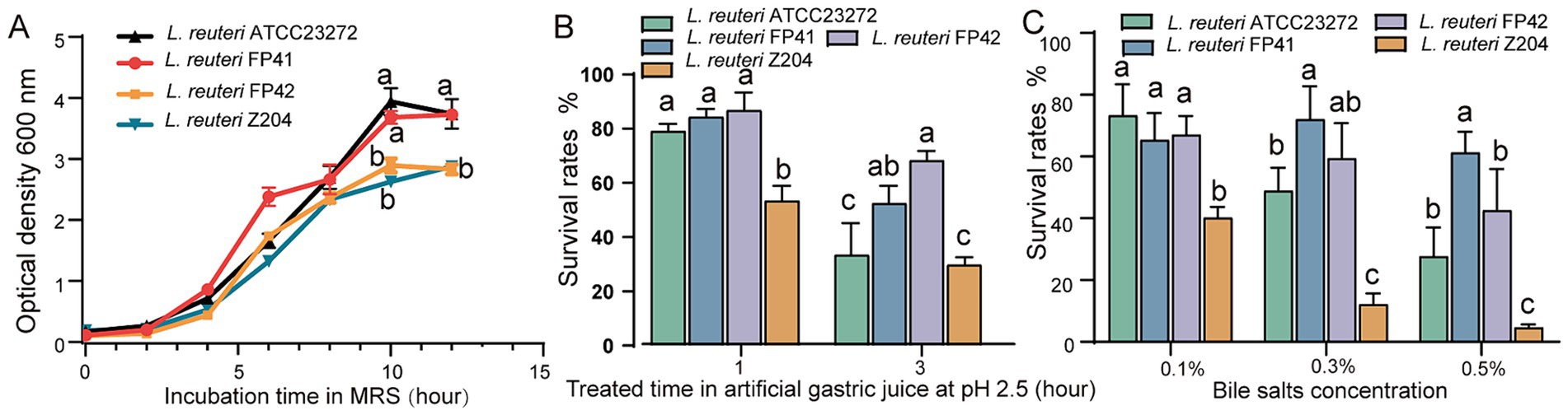
Figure 1. The growth characteristics and the acid/bile tolerance in L. reuteri. (A) Growth curves of FP41, FP42, Z204 and ATCC23272 in MRS based on absorbance at 600 nm. Survival rates of FP41, FP42, Z204 and ATCC23272 in pH 2.5 artificial gastric juice treatment (B) and different pig bile salts concentration treatment (C). Error bars: standard derivations of three independent experiments. Data with different superscript letters (a, b, and c) are significantly different (p < 0.05) according to one-way ANOVA followed by Tukey’s test.
Transcriptome sequencing of Limosilactobacillus reuteri acid/bile salts stress
The growth of L. reuteri FP41 was examined under various pH conditions or variable concentrations of bile salts in order to determine an appropriate sample period and treatment conditions. The results showed that growth was significantly slower at pH 4.0 than it was for the control group at pH 6.8, that adjusting the pH to 2.0 and 3.0 successfully inhibited the growth of bacteria (Figure 2A). Bile salts inhibited L. reuteri FP41 growth gradually; the greatest effect was observed at a concentration of 0.08% (Figure 2B). For RNA-sequencing, L. reuteri FP41 was thus cultivated in MRS at pH 4.0 or 0.08% bile salts concentration till OD600 = 1.0.
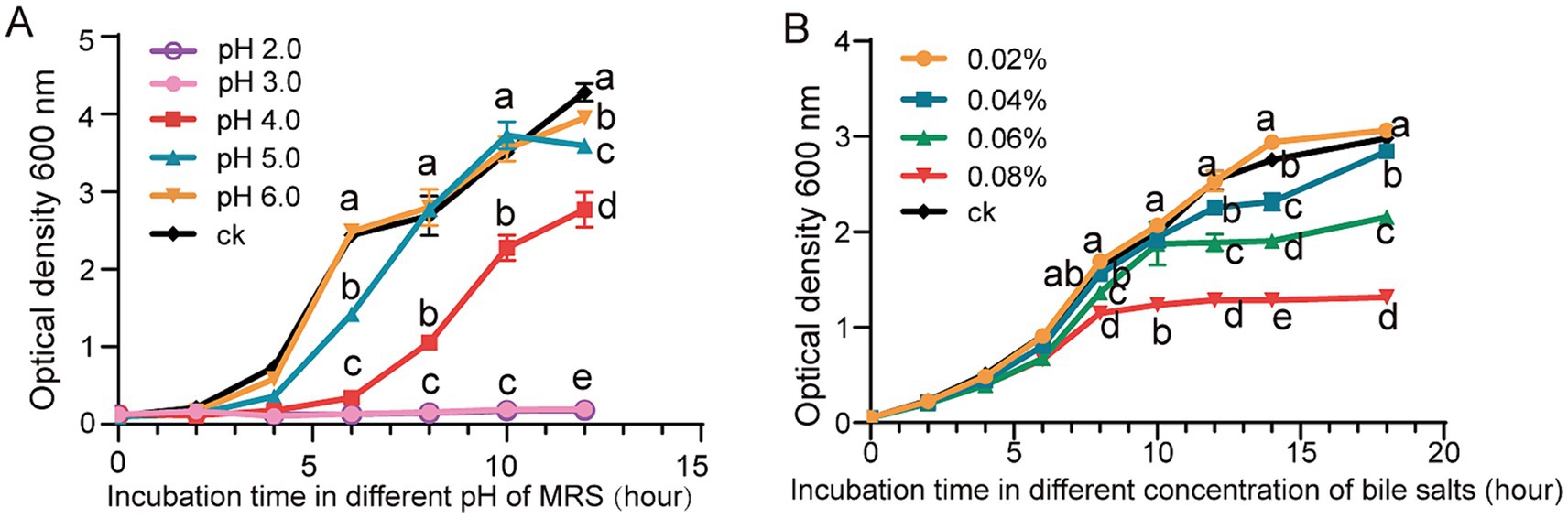
Figure 2. The growth characteristics and the acid/bile tolerance in L. reuteri. The growth curves of L. reuteri FP41 in MRS under different pH values (A) and different bile salts concentrations (B). Error bars: standard derivations of three independent experiments. Data with different superscript letters (a, b, c, d, and e) are significantly different (p < 0.05) according to one-way ANOVA followed by Tukey’s test.
The stress response genes expression in Limosilactobacillus reuteri under acid/bile salts
A total of 233 genes were significantly differentially expressed in the pH 4.0 group, of which 126 genes were up-regulated compared with the control group, and 607 genes were significantly differentially expressed in the bile salts stress group, of which 315 genes were up-regulated compared with the control group, according to transcriptome data analysis (Supplementary Table S1). Among the genes that have been significantly up-regulated are cytochrome, glutamine metabolism, OsmC family protein, ABC transporter system, and integral component of membranes (Table 1; Supplementary Table S1). Under stress of acid and bile salts, most genes were up-regulated concurrently, and the highest number of genes related to the membrane was seen. This finding aligns with earlier research indicating that probiotics largely alter cell membranes to enhance their ability to withstand acidic or bile salt environments (Ayyash et al., 2021; van Der Heide and Poolman, 2000). The cytoplasmic membrane is a major barrier to most solutes and is sensitive to stress (Spano and Massa, 2006), which is consistent with our findings indicating the upregulation of a significant number of genes related to membranes following treatment with acid/bile salt (Table 1; Supplementary Table S1). Particularly, responses to various stresses, such as heat, acid, oxidative stress, phosphate starvation, and salt stress, are mediated by the membrane protein CsbD (Han et al., 2017; Prágai and Harwood, 2002). In our study, we found that under acid/bile salts stress, CsbD was markedly upregulated (Table 1). Furthermore, in response to acid stress, bacteria can produce or consume ammonia by upregulating certain genes associated with glutamine metabolism (Lu et al., 2013). This is consistent with our findings, as glutamine metabolism-related gene gatD was similarly up-regulated (Table 1). These findings imply that L. reuteri might have an ammonia cycle to deal with acid stress, and that the ammonia molecule that results is directed at the MurT active site. Notably, under acid/bile salts stress, OsmC, a thiol-dependent organic hydroperoxide reductase (Ohr), showed significant up-regulation (Table 1). In Corynebacterium glutamicum (Si et al., 2019), as well as in peroxide metabolism and preservation against oxidative stress in mycobacteria (Saikolappan et al., 2011), the protein OsmC plays a critical protective role against organic hydroperoxides (OHPs) stress. Similar results were found in the transcriptome data (Supplementary Table S1), which is consistent with previous findings that ABC transporters are positively associated with acid tolerance in L. lactis (Zhu et al., 2019) and that chaperonin pumps significantly contribute to resistance against acid and bile salts stress (Gao et al., 2022; Zhang et al., 2017).
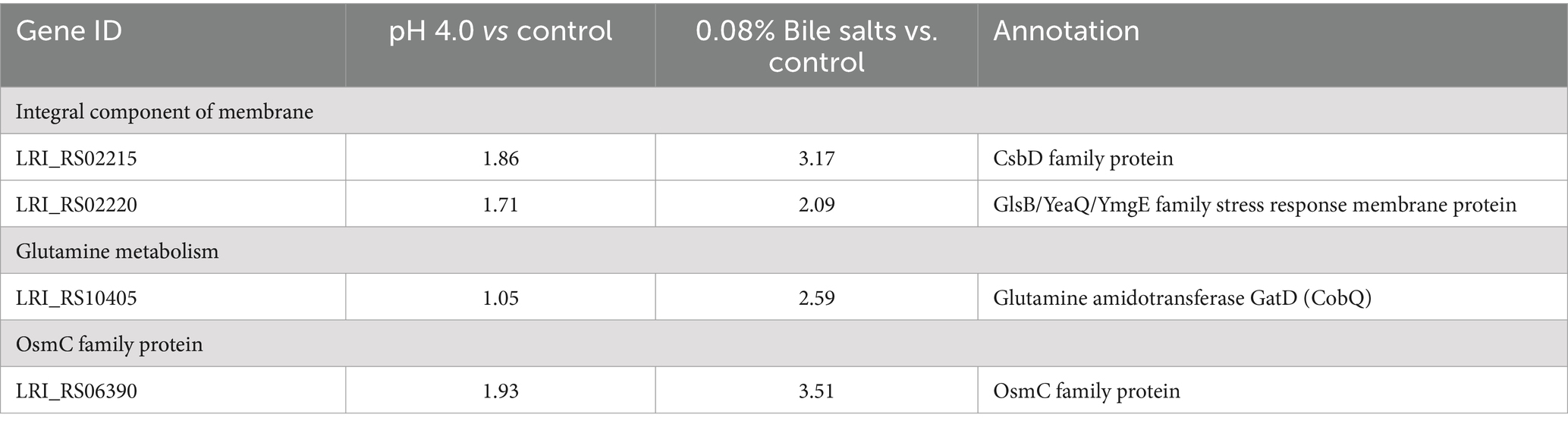
Table 1. Transcriptional changes (log2-fold) of genes response to acid/bile salts stress in L. reuteri FP41 according to the transcriptome data.
The construction of the overexpression plasmid
To validate the transcriptome results and demonstrate the protective role of these up-regulated genes against acid, bile salts and vacuum freeze-drying stress, overexpress the genes csbD, osmC, and gatD in L. reuteri Z204. These gene segments were cloned at the immediate downstream of the p32 promoter for overexpression in an E. coli-L. reuteri shuttle vector pMG36e (Figure 3A). Primer osmC-F/R, csbD-F/R, and gatD-F/R enabled the successful amplification of DNA fragments corresponding to three genes: osmC, csbD, and gatD, respectively (Figure 3B). These genes were effectively integrated into the pMG36e expression vector, transformed into DH5α, and the positive clone plasmids pMG36e-gatD/osmC/csbD were obtained by verifying the transformants using the universal 36-F/R primer (Figure 3C). These recombinant plasmids were then electroporated into L. reuteri Z204 in order to overexpress. Universal 36-F/R primer was used to confirm the existence of positive recombinant over-expressing bacteria (Figure 3D).
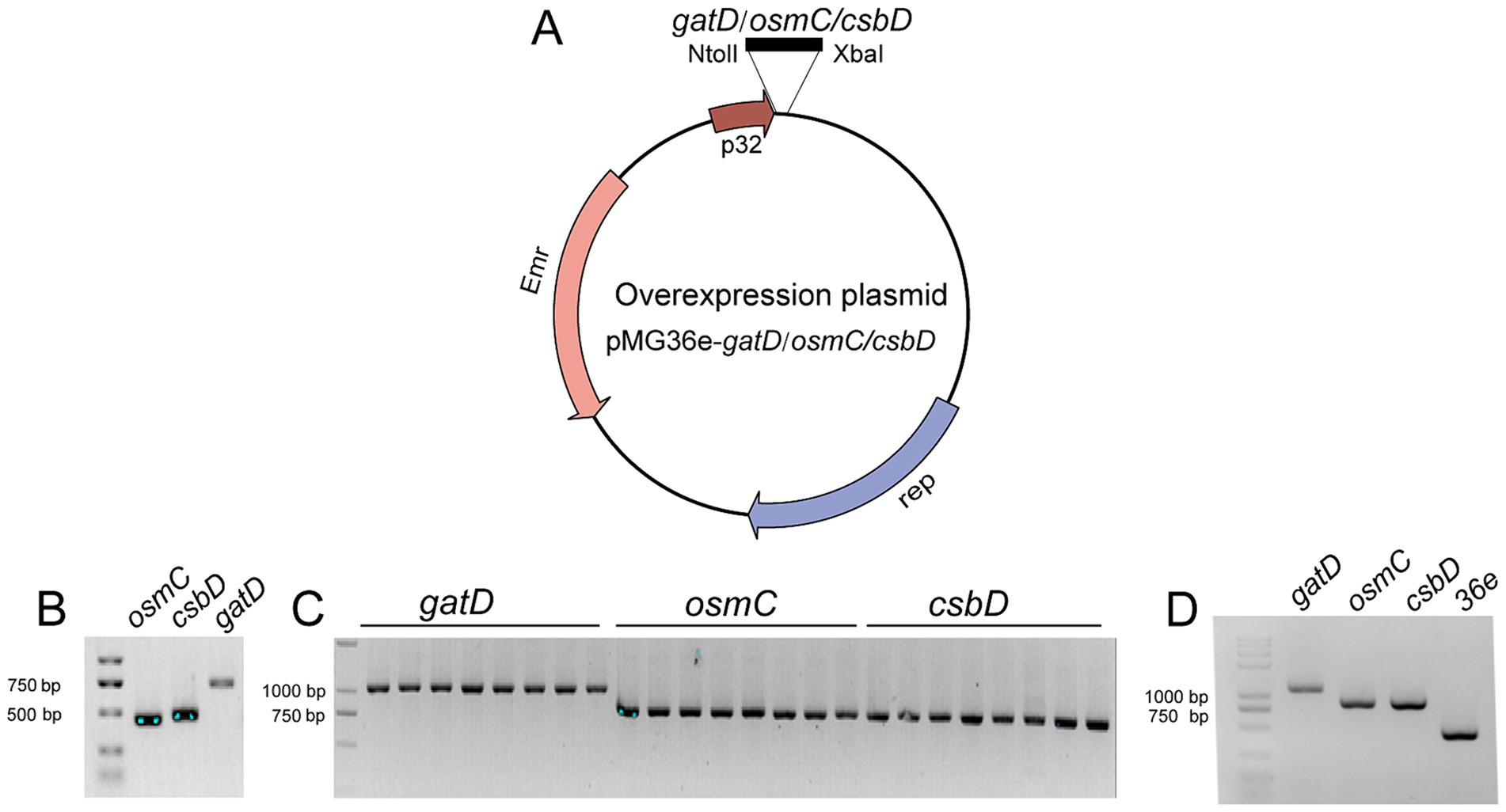
Figure 3. Schematic representation of the construction of the overexpression plasmid. (A) The plasmid map of pMG36e-gatD/osmC/csbD. (B) PCR amplification of these genes from L. reuteri strains FP41 genomic DNA. The plasmid in the single colonies of the transformants in E. coli (C) and in L. reuteri (D).
To confirm the overexpression of genes encoding OsmC, CsbD, and GatD proteins, we chose to measure the transcription level of osmC, csbD, and gatD genes by RT-qPCR to determine whether they were overexpressed (p < 0.0001, p = 0.0096, p = 0.0059) (Figure 4), and further validated their expression at the protein level through TOF-MS analysis (Table 2). Based on the RT-qPCR and proteome results, all three genes were overexpressed in the overexpressed strain.
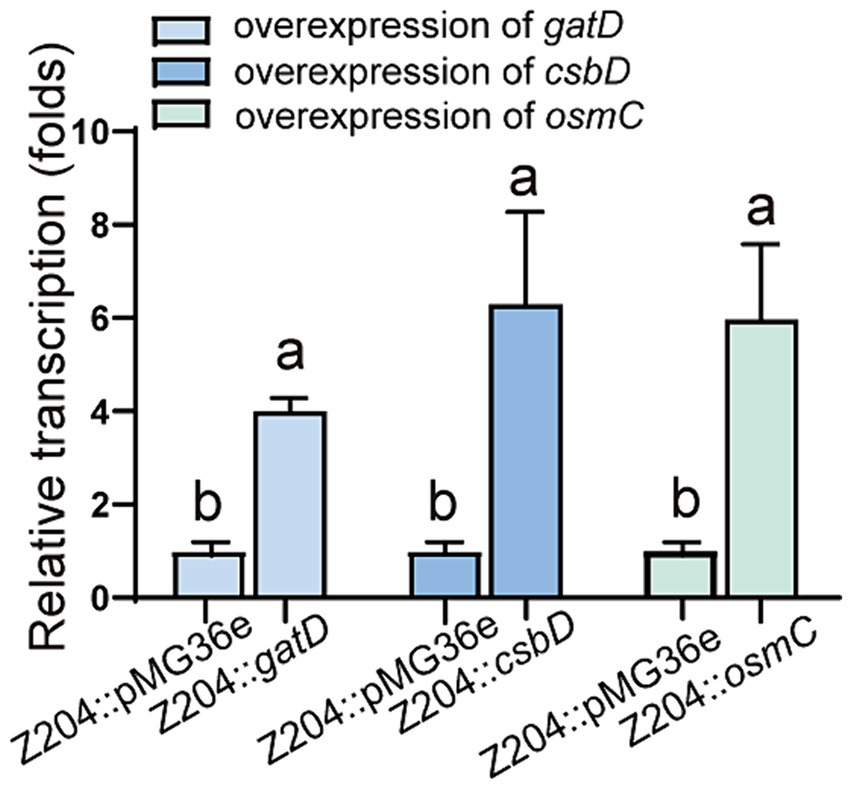
Figure 4. The mRNA levels of GatD, CsbD, and OsmC in the L. reuteri Z204 overexpressed strain measured by RT-qPCR.
Functional validation of stress resistance genes under acid, bile salts, and vacuum freeze-drying stress
Limosilactobacillus reuteri strains overexpressing CsbD protein (Z204::csbD) were assessed for survival rates in artificial gastric juice, bile salts medium, and vacuum freeze-drying conditions. The overexpression of CsbD protein significantly enhanced L. reuteri Z204’s resistance to acid (Figure 5A), bile salts (Figure 5B), and its viability after freeze-drying (Figure 5C) in comparison to the control group Z204::36e strain. These findings imply that the membrane protein CsbD may act as a global regulator of L. reuteri’s cross-protection against acid, bile salts, and low-temperature treatment by modulating the membrane composition to provide cross-protection against low-temperature stress (Meneghel et al., 2017).
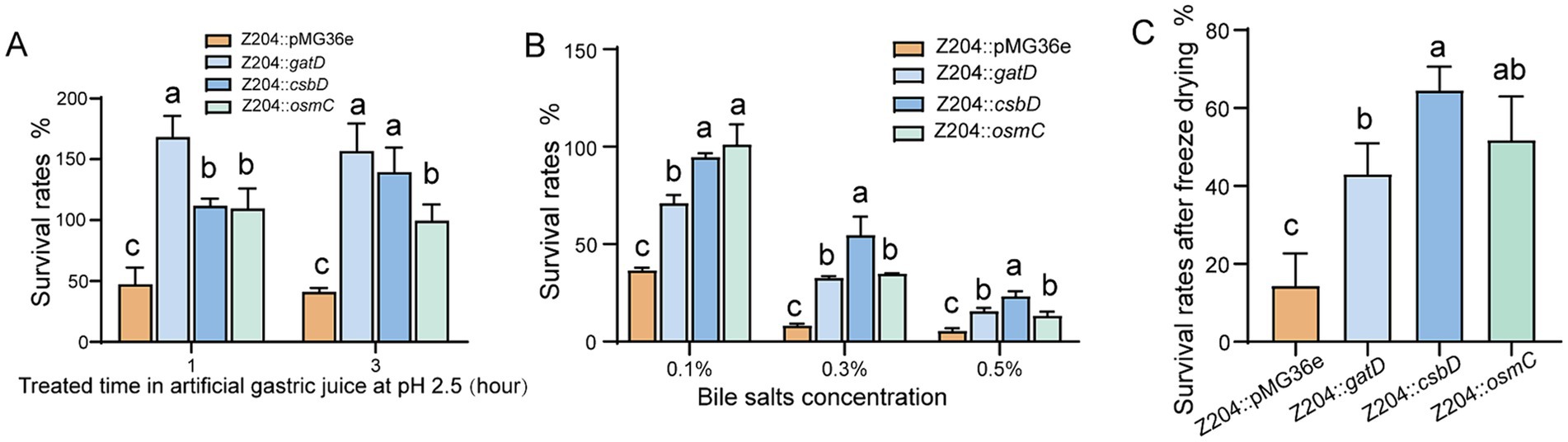
Figure 5. The L. reuteri Z204 overexpressed strains survival rates under the three stress conditions. The L. reuteri Z204 overexpressed strains survival rates under the artificial gastric juice stress (A), bile salts stress (B), and vacuum freeze-drying (C). Error bars: standard derivations of three independent experiments. Data with different superscript letters (a, b, and c) are significantly different (p < 0.05) according to one-way ANOVA followed by Tukey’s test.
We experimented with artificial gastric juice, bile salts medium, and vacuum freeze-drying on L. reuteri strains overexpressing GatD. According to our research, overexpression of the GatD protein enhances L. reuteri’s resistance to acid tolerance (Figure 5A), increases its bile salts tolerance (Figure 5B), and improves its survival rate after freeze-drying (Figure 5C). These results imply that the glutamine amidotransferase GatD acts as a global regulator, providing L. reuteri with cross-protection against acid, bile salts, and low temperatures. Proton neutralization may be the mechanism by which GatD enhances resistance to salts and low-temperature drying stress; this mechanism is comparable to the glutamine-dependent acid tolerance mechanism that has previously been described (Lu et al., 2013).
It’s interesting to note that L. reuteri Z204 demonstrated enhanced acid resistance (Figure 5A), bile salts resistance (Figure 5B), and survival rate after freeze-drying (Figure 5C) upon overexpression of OsmC protein, which is involved in peroxide metabolism and oxidative stress defense (Si et al., 2019; Lyu et al., 2016). Our results may provide support to the hypothesis that OsmC, an osmotically generated bacterial protein, acts as a general regulator to provide L. reuteri with cross-protection against acid, bile salts, and low-temperature.
Discussion
Immune homeostasis and physiology in the human gut are positively impacted by Lactic acid bacteria (Finamore et al., 2019; Meng et al., 2020). Furthermore, they protect the host by limiting the binding of pathogenic microbes and maintaining the balance between gut microbiota and lymphocytes (Riaz Rajoka et al., 2017). In order to influence host immunity and health, LAB are currently developed as fermented foods or probiotic powder (Daba and Elkhateeb, 2020; Wang et al., 2015; Wang et al., 2020). A key factor in establishing the characteristics of probiotics is their viability prior to technological processing and use (Bisutti and Stephan, 2020). Therefore, in order to prevent stress-related damage and increase the survival rate of linked products, it is crucial to comprehend the stress response and molecular control mechanism of LAB under various environmental conditions. Environmental harm may be lessened by LAB’s physiological and metabolic responses (Guan and Liu, 2020). Cross-protection, the ability to withstand both related and unrelated stresses, is a feature of adaptations seen under mild stress that can increase overall tolerance to different environmental pressures (Louesdon et al., 2014). Furthermore, it has the ability to stimulate cells within the global response signal, providing protection against external stressors (Papadimitriou et al., 2016). In order to resist multiple environmental stress conditions, also known as cross-protection (Gao et al., 2022), LAB exhibits numerous adaptive mechanisms by modulating their gene expression and signaling pathways, including up-regulation of response proteins, accumulation of suitable solutes, and regulation of membrane lipids (Zhen et al., 2020; Kulkarni et al., 2018). In the present study, overexpression of the membrane protein CsbD, the osmosis-induced bacterial protein OsmC, and the acid-related protein glutamate decarboxylase GatD, respectively, increased the acid tolerance (Figure 5A), bile salt tolerance (Figure 5B), and freeze-drying ability (Figure 5C) of L. reuteri. Through the expression of particular stress resistance proteins, the cross-protective characteristics of L. reuteri were shown to increase overall tolerance to numerous stimuli.
The cytoplasmic membrane is the main barrier for the majority of solutes and a major target of stress injury (Spano and Massa, 2006), research has demonstrated that sigmaB transcription factors triggered by metabolic stress control the membrane protein component csbD protein, which is a general stress response protein resistant to oxidative stress, protein denaturation, and osmotic stress (Prágai and Harwood, 2002; Akbar et al., 1999). In addition, the csbD protein was overexpressed in Methyocystis sp. strain SC2 (Han et al., 2017) and Group B Streptococcus (GBS) (Jia et al., 2023) in response to bile salt stress. Among these, in Group B Streptococcus (GBS), csbD promotes the transcription of several ABC transporter genes to excrete bile salts upon detecting bile salt stress. This enhances bacterial survivability in bile salts (Jia et al., 2023). As shown by our results, which showed that a large number of membrane proteins were up-regulated upon acid or bile salts stress (Table 1; Supplementary Table S1). L. reuteri Z204’s resistance to acid (Figure 5A) and bile salts (Figure 5B), as well as its survival rates after freeze-drying (Figure 5C), may be enhanced by overexpressing the membrane protein component CsbD protein. These results appear to indicate the hypothesis that the membrane protein CsbD functions as a global regulator of L. reuteri’s cross-protection against treatment with acid, bile salts, and low-temperature. This might be the consequence of either the regulation of global metabolic stress activators or the regulation of transport genes by csbD proteins. Altering the composition of the membrane, which can provide cross-protection against low-temperature stress may also have an impact on another cause (Meneghel et al., 2017). Many LAB improve stress resistance by altering the lipid composition of cytoplasmic membranes with various pretreatments (Papadimitriou et al., 2016). In order to prevent protons influx into cells from entering cells, LAB tends to increase the stiffness and density of the plasma membrane during acid stress, such as by modifying the content of fatty acids (van Der Heide and Poolman, 2000; Broadbent et al., 2010). By producing more unsaturated fatty acids, cyclic fatty acids, and branched-chain fatty acids, L. acidophilus has been shown to improve membrane fluidity under lactose starvation and display better tolerance to low-temperature stress (Wang et al., 2011).
The genes for glutamate decarboxylase (GAD), a pyridoxal phosphate (PLP)-dependent enzyme, are found in some E. coli and LAB strains (Barrett et al., 2012; Mazzoli and Pessione, 2016). By catalyzing the α-decarboxylation of L-glutamate or glutamate to produce gamma-aminobutyric acid (GABA), which helps bacteria in dealing with acid stress, GAD plays a crucial part in the glutamate-dependent acid resistance system (De Biase and Pennacchietti, 2012; Liu S. et al., 2022). Our findings showed that the glutamate decarboxylase GatD was up-regulated in response to acid stress (Table 1), which is consistent with a previous study’s finding that the GDA is crucial for the bacteria’s acid resistance system (De Biase and Pennacchietti, 2012). Otherwise, according to our findings, overexpression of the acid-associated protein glutamate decarboxylase GatD might increase L. reuteri’s bile salts tolerance (Figure 5B), survival rate after freeze-drying (Figure 5C), as well as its ability to withstand acid (Figure 5A). It’s interesting to note that the strain treated with artificial gastric fluid grew better than the untreated strain in the acid tolerance test of the overexpressed GatD strain used in this work. This phenomenon could be caused by: despite the fact that the properties of GADs differ significantly between LAB species and strains, most LAB GADs are significantly more active in acidic environments than in neutral pH (Cui et al., 2020). Consequently, the GatD overexpression strain’s glutamate decarboxylase GatD was able to deacid L-glutamate or glutamate into GABA, which gave the strain more energy for growth than it would have without treatment (Mazzoli and Pessione, 2016). As a consequence, the acid-treated strain grew more better than the untreated strain. According to these results, L. reuteri’s tolerance to acid, bile salts, and low-temperature is controlled globally by the glutamate decarboxylase GatD, which acts as a global regulator to cross-protection the acid, bile and low-temperature tolerance. By altering the proton content in the cells, the GatD may be able to improve resistance to salts and low-temperature drying stress.
Oxidative stress is brought on by the large-scale generation of reactive oxygen species (ROS) by LAB during fermentation. ROS (superoxide and hydroxyl radicals) can have a deleterious effect on microbial components as proteins, lipids, DNA, and RNA during oxidative stress (Larasati et al., 2018). Thermus thermophilus (Rehse et al., 2004), Thermococcus kodakarensis KOD1 (Park et al., 2008), E. coli (Shin et al., 2004; Lesniak et al., 2003), Mycobacterium tuberculosis and M. smegmatis (Saikolappan et al., 2011), have all been found to contain the OsmC protein, which has hydroperoxide peroxidase properties and can shield microorganisms from oxidative damage by lowering peroxide substrates. Furthermore, Thermococcus kodakarensis KOD1 overexpressed OsmC in order to fend off oxidative damage brought on by high osmotic pressure when exposed to salt (Park et al., 2008). Interestingly, both acid and bile salt stressors were shown to increase the expression of the osmotically induced bacterial protein OsmC in this study (Table 1). Additionally, L. reuteri Z204’s resistance to acid (Figure 5A), bile salts (Figure 5B), and freeze-drying (Figure 5C) could all be enhanced by overexpression of the OsmC protein. These results seem to verify the idea that OsmC, an osmotically induced bacterial protein, functions as a general regulator of L. reuteri cross-protection against oxidative stress exposure to acid, bile salts, and low-temperature.
Understanding the adaptive response mechanism under adverse conditions is crucial for optimizing the performance of LAB during production and downstream application (Derunets et al., 2024). Theoretically, new strategies to increase the survival rate of LAB under different stresses can be developed with a thorough grasp of the molecular mechanisms of environmental stress adaptation in lactic acid bacteria and appropriate protective measures based on the stress response mechanisms. In addition, a methodical comprehension of the “stressome” of LAB and the development of strains that enhance tolerance via cross-protection—particularly alterations in critical enzyme activities—will offer valuable new approaches for the focused screening or molecular modification of stress-resistant LABS in the fields of environmental and industrial protection (Gao et al., 2022). In this case, creating an acid-tolerant module made up of the transcription factor of the AR system (gadE), a periplasmic protein chaperone (hdeB), and two reactive oxygen scavenging enzymes (sodB and katE) may increase the growth robustness and productivity of industrial E. coli in mildly acidic environments (Yao et al., 2022). Enhancing the stress resistance of LAB through pre-adaptation (Bisson et al., 2023; Bommasamudram et al., 2023), genetic modification of strains (Yao et al., 2022), and modification of culture conditions (Hao et al., 2021) are particularly essential for decreasing production costs, stabilizing product quality, and enhancing the survival and vitality of strains used in industrial production. This will help us make the most use of lactic acid bacteria as starter cultures and probiotics in industrial production.
Genetic manipulation based on the factors and signaling pathways regulating bacterial stress response has drawn special interest among molecular mechanisms of how LAB responds to stressful situations. In this study, the efficacy of L. reuteri’s resistance to acid resistance, bile salts resistance, or freeze-drying was improved by overexpressing csbD, osmC, and gatD. Overall, GatD was most successful in enhancing acid tolerance, whereas CsbD was most effective in improving bile salts tolerance. Furthermore, it was found that CsbD was the most successful in increasing freeze-drying tolerance, followed by OsmC. These findings enhance our understanding of the molecular mechanism adopted by L. reuteri in response to acid, bile salts, and freeze-drying stress. In order to promote global cross-protection, these findings suggest a novel practical strategy to improve resistance to acid, bile salts resistance, or freeze-drying stress function. They also show that LAB respond to environmental stress and that cross-protection strategies can increase resistance to extremely high levels of environmental stress. This establishes the framework for further investigations on the LAB stress mechanisms that provide cross-protection.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
ZL: Writing – review & editing, Writing – original draft, Funding acquisition. XZ: Writing – original draft, Methodology, Investigation, Data curation. HB: Writing – review & editing, Visualization, Software, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32100054 to ZL).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1437803/full#supplementary-material
References
Akbar, S., Lee, S. Y., Boylan, S. A., and Price, C. W. (1999). Two genes from Bacillus subtilis under the sole control of the general stress transcription factor sigmaB. Microbiology 145, 1069–1078. doi: 10.1099/13500872-145-5-1069
Ayyash, M. M., Abdalla, A. K., AlKalbani, N. S., Baig, M. A., Turner, M. S., Liu, S. Q., et al. (2021). Invited review: characterization of new probiotics from dairy and nondairy products-insights into acid tolerance, bile metabolism and tolerance, and adhesion capability. J. Dairy Sci. 104, 8363–8379. doi: 10.3168/jds.2021-20398
Bagon, B. B., Valeriano, V. D. V., Oh, J. K., Pajarillo, E. A. B., Lee, J. Y., and Kang, D. K. (2021). Exoproteome perspective on the bile stress response of Lactobacillus johnsonii. Proteomes 9:10. doi: 10.3390/proteomes9010010
Barrett, E., Ross, R. P., O'Toole, P. W., Fitzgerald, G. F., and Stanton, C. (2012). γ-Aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 113, 411–417. doi: 10.1111/j.1365-2672.2012.05344.x
Begley, M., Gahan, C. G. M., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Bisson, G., Maifreni, M., Innocente, N., and Marino, M. (2023). Application of pre-adaptation strategies to improve the growth of probiotic lactobacilli under food-relevant stressful conditions. Food Funct. 14, 2128–2137. doi: 10.1039/D2FO03215E
Bisutti, I. L., and Stephan, D. (2020). Influence of fermentation temperature and duration on survival and biocontrol efficacy of Pseudomonas fluorescens Pf153 freeze-dried cells. J. Appl. Microbiol. 128, 232–241. doi: 10.1111/jam.14458
Bommasamudram, J., Kumar, P., Kapur, S., Sharma, D., and Devappa, S. (2023). Development of Thermotolerant lactobacilli cultures with improved probiotic properties using adaptive laboratory evolution method. Probiotics Antimicrob. Proteins 15, 832–843. doi: 10.1007/s12602-021-09892-3
Broadbent, J. R., Larsen, R. L., Deibel, V., and Steele, J. L. (2010). Physiological and transcriptional response of Lactobacillus casei ATCC 334 to acid stress. J. Bacteriol. 192, 2445–2458. doi: 10.1128/JB.01618-09
Chang, T. H., Huang, H. D., Wu, L. C., Yeh, C. T., Liu, B. J., and Horng, J. T. (2009). Computational identification of riboswitches based on RNA conserved functional sequences and conformations. RNA 15, 1426–1430. doi: 10.1261/rna.1623809
Chen, M. J., Tang, H. Y., and Chiang, M. L. (2017). Effects of heat, cold, acid and bile salt adaptations on the stress tolerance and protein expression of kefir-isolated probiotic Lactobacillus kefiranofaciens M1. Food Microbiol. 66, 20–27. doi: 10.1016/j.fm.2017.03.020
Conesa, A., Madrigal, P., Tarazona, S., Gomez-Cabrero, D., Cervera, A., McPherson, A., et al. (2016). A survey of best practices for RNA-seq data analysis. Genome Biol. 17:13. doi: 10.1186/s13059-016-0881-8
Cox, J., and Mann, M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372. doi: 10.1038/nbt.1511
Cox, J., Neuhauser, N., Michalski, A., Scheltema, R. A., Olsen, J. V., and Mann, M. (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805. doi: 10.1021/pr101065j
Cui, Y., Miao, K., Niyaphorn, S., and Qu, X. (2020). Production of gamma-aminobutyric acid from lactic acid Bacteria: a systematic review. Int. J. Mol. Sci. 21:995. doi: 10.3390/ijms21030995
Daba, G. M., and Elkhateeb, W. A. (2020). Bacteriocins of lactic acid bacteria as biotechnological tools in food and pharmaceuticals: current applications and future prospects. Biocatal. Agric. Biotechnol. 28:101750. doi: 10.1016/j.bcab.2020.101750
De Biase, D., and Pennacchietti, E. (2012). Glutamate decarboxylase-dependent acid resistance in orally acquired bacteria: function, distribution and biomedical implications of the gadBC operon. Mol. Microbiol. 86, 770–786. doi: 10.1111/mmi.12020
Derunets, A. S., Selimzyanova, A. I., Rykov, S. V., Kuznetsov, A. E., and Berezina, O. V. (2024). Strategies to enhance stress tolerance in lactic acid bacteria across diverse stress conditions. World J. Microbiol. Biotechnol. 40:126. doi: 10.1007/s11274-024-03905-3
Fallingborg, J. (1999). Intraluminal pH of the human gastrointestinal tract. Dan. Med. Bull. 46, 183–196.
Finamore, A., Roselli, M., Donini, L., Brasili, D. E., Rami, R., Carnevali, P., et al. (2019). Supplementation with Bifidobacterium longum Bar33 and Lactobacillus helveticus Bar13 mixture improves immunity in elderly humans (over 75 years) and aged mice. Nutrition 63–64, 184–192. doi: 10.1016/j.nut.2019.02.005
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Gao, X., Kong, J., Zhu, H., Mao, B., Cui, S., and Zhao, J. (2022). Lactobacillus, Bifidobacterium and Lactococcus response to environmental stress: mechanisms and application of cross-protection to improve resistance against freeze-drying. J. Appl. Microbiol. 132, 802–821. doi: 10.1111/jam.15251
Guan, N., and Liu, L. (2020). Microbial response to acid stress: mechanisms and applications. Appl. Microbiol. Biotechnol. 104, 51–65. doi: 10.1007/s00253-019-10226-1
Guo, Q., Yan, Y., Zhang, Z., Xu, B., Bangash, H. L., Sui, X., et al. (2023). Developing the Limosilactobacillus reuteri chassis through an endogenous programmable endonuclease-based genome editing tool. ACS Synth. Biol. 12, 3487–3496. doi: 10.1021/acssynbio.3c00450
Han, D., Link, H., and Liesack, W. (2017). Response of Methylocystis sp. strain SC2 to salt stress: physiology, global transcriptome, and amino acid profiles. Appl. Environ. Microbiol. 83:e00866-17. doi: 10.1128/AEM.00866-17
Hao, F., Fu, N., Ndiaye, H., Woo, M. W., Jeantet, R., and Chen, X. D. (2021). Thermotolerance, survival, and stability of lactic acid Bacteria after spray drying as affected by the increase of growth temperature. Food Bioprocess Technol. 14, 120–132. doi: 10.1007/s11947-020-02571-1
Jia, L., Yuan, C., Pan, F., Zhou, X., Fan, H., and Ma, Z. (2023). CsbD, a novel group B streptococcal stress response factor that contributes to bacterial resistance against environmental bile salts. J. Bacteriol. 205:e0044822. doi: 10.1128/jb.00448-22
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277D–2280D. doi: 10.1093/nar/gkh063
Kulkarni, S., Haq, S. F., Samant, S., and Sukumaran, S. (2018). Adaptation of Lactobacillus acidophilus to thermal stress yields a Thermotolerant variant which also exhibits improved survival at pH 2. Probiotics Antimicrob. Proteins 10, 717–727. doi: 10.1007/s12602-017-9321-7
Larasati, B. A., Panunggal, B., Afifah, D. N., Anjani, G., and Rustanti, N. (2018). Total lactic acid bacteria, antioxidant activity, and acceptance of synbiotic yoghurt with red ginger extract (Zingiberofficinale var. rubrum). IOP Conf. Ser. Earth Environ. Sci. 116:012037. doi: 10.1088/1755-1315/116/1/012037
Lesniak, J., Barton, W. A., and Nikolov, D. B. (2003). Structural and functional features of the Escherichia coli hydroperoxide resistance protein OsmC. Protein Sci. 12, 2838–2843. doi: 10.1110/ps.03375603
Liu, L., Helal, S. E., and Peng, N. (2023). CRISPR-Cas-based engineering of probiotics. Biodes. Res. 5:0017. doi: 10.34133/bdr.0017
Liu, H. Y., Roos, S., Jonsson, H., Ahl, D., Dicksved, J., Lindberg, J. E., et al. (2015). Effects of Lactobacillus johnsonii and Lactobacillus reuteri on gut barrier function and heat shock proteins in intestinal porcine epithelial cells. Physiol. Rep. 3:e12355. doi: 10.14814/phy2.12355
Liu, S., Wen, B., Du, G., Wang, Y., Ma, X., Yu, H., et al. (2022). Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH. Food Chem. 403:134436. doi: 10.1016/j.foodchem.2022.134436
Liu, Y., Zhang, Z., and Hu, L. (2022). High efficient freeze-drying technology in food industry. Crit. Rev. Food Sci. Nutr. 62, 3370–3388. doi: 10.1080/10408398.2020.1865261
Louesdon, S., Charlot-Rougé, S., Juillard, V., Tourdot-Maréchal, R., and Béal, C. (2014). Osmotic stress affects the stability of freeze-dried Lactobacillus buchneri R1102 as a result of intracellular betaine accumulation and membrane characteristics. J. Appl. Microbiol. 117, 196–207. doi: 10.1111/jam.12501
Lu, P., Ma, D., Chen, Y., Guo, Y., Chen, G. Q., Deng, H., et al. (2013). L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 23, 635–644. doi: 10.1038/cr.2013.13
Lyu, C., Hu, S., Huang, J., Luo, M., Lu, T., Mei, L., et al. (2016). Contribution of the activated catalase to oxidative stress resistance and γ-aminobutyric acid production in Lactobacillus brevis. Int. J. Food Microbiol. 238, 302–310. doi: 10.1016/j.ijfoodmicro.2016.09.023
Mazzoli, R., and Pessione, E. (2016). The neuro-endocrinological role of microbial glutamate and GABA signaling. Front. Microbiol. 7:1934. doi: 10.3389/fmicb.2016.01934
Mbye, M., Baig, M. A., AbuQamar, S. F., El-Tarabily, K. A., Obaid, R. S., Osaili, T. M., et al. (2020). Updates on understanding of probiotic lactic acid bacteria responses to environmental stresses and highlights on proteomic analyses. Compr. Rev. Food Sci. Food Saf. 19, 1110–1124. doi: 10.1111/1541-4337.12554
McClure, R., Balasubramanian, D., Sun, Y., Bobrovskyy, M., Sumby, P., Genco, C. A., et al. (2013). Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 41:e140. doi: 10.1093/nar/gkt444
Meier, F., Brunner, A. D., Koch, S., Koch, H., Lubeck, M., Krause, M., et al. (2018). Online parallel accumulation-serial fragmentation (PASEF) with a novel trapped ion mobility mass spectrometer. Mol. Cell. Proteomics 17, 2534–2545. doi: 10.1074/mcp.TIR118.000900
Meneghel, J., Passot, S., Cenard, S., Réfrégiers, M., Jamme, F., and Fonseca, F. (2017). Subcellular membrane fluidity of Lactobacillus delbrueckii subsp. bulgaricus under cold and osmotic stress. Appl. Microbiol. Biotechnol. 101, 6907–6917. doi: 10.1007/s00253-017-8444-9
Meng, D., Sommella, E., Salviati, E., Campiglia, P., Ganguli, K., Djebali, K., et al. (2020). Indole-3-lactic acid, a metabolite of tryptophan, secreted by Bifidobacterium longum subspecies infantis is anti-inflammatory in the immature intestine. Pediatr. Res. 88, 209–217. doi: 10.1038/s41390-019-0740-x
Mu, Q., Tavella, V. J., and Luo, X. M. (2018). Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 9:757. doi: 10.3389/fmicb.2018.00757
Nithya, V., and Halami, P. M. (2012). Evaluation of the probiotic characteristics of Bacillus species isolated from different food sources. Ann. Microbiol. 63, 129–137. doi: 10.1007/s13213-012-0453-4
Papadimitriou, K., Alegria, A., Bron, P. A., de Angelis, M., Gobbetti, M., Kleerebezem, M., et al. (2016). Stress physiology of lactic acid Bacteria. Microbiol. Mol. Biol. Rev. 80, 837–890. doi: 10.1128/MMBR.00076-15
Park, S. C., Pham, B. P., van Duyet, L., Jia, B., Lee, S., Yu, R., et al. (2008). Structural and functional characterization of osmotically inducible protein C (OsmC) from Thermococcus kodakaraensis KOD1. Biochim. Biophys. Acta 1784, 783–788. doi: 10.1016/j.bbapap.2008.02.002
Pedersen, C., Jonsson, H., Lindberg, J. E., and Roos, S. (2004). Microbiological characterization of wet wheat distillers' grain, with focus on isolation of lactobacilli with potential as probiotics. Appl. Environ. Microbiol. 70, 1522–1527. doi: 10.1128/AEM.70.3.1522-1527.2004
Pérez Montoro, B., Benomar, N., Caballero Gómez, N., Ennahar, S., Horvatovich, P., Knapp, C. W., et al. (2018). Proteomic analysis of Lactobacillus pentosus for the identification of potential markers involved in acid resistance and their influence on other probiotic features. Food Microbiol. 72, 31–38. doi: 10.1016/j.fm.2017.11.006
Powell, S., Szklarczyk, D., Trachana, K., Roth, A., Kuhn, M., Muller, J., et al. (2012). eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289. doi: 10.1093/nar/gkr1060
Prágai, Z., and Harwood, C. R. (2002). Regulatory interactions between the pho and σB-dependent general stress regulons of Bacillus subtilis. Microbiology 148, 1593–1602. doi: 10.1099/00221287-148-5-1593
Rehse, P. H., Ohshima, N., Nodake, Y., and Tahirov, T. H. (2004). Crystallographic structure and biochemical analysis of the Thermus thermophilus osmotically inducible protein C. J. Mol. Biol. 338, 959–968. doi: 10.1016/j.jmb.2004.03.050
Riaz Rajoka, M. S., Shi, J., Zhu, J., Shao, D., Huang, Q., Yang, H., et al. (2017). Capacity of lactic acid bacteria in immunity enhancement and cancer prevention. Appl. Microbiol. Biotechnol. 101, 35–45. doi: 10.1007/s00253-016-8005-7
Ruiz, L., Margolles, A., and Sánchez, B. (2013). Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 4:396. doi: 10.3389/fmicb.2013.00396
Saikolappan, S., Das, K., Sasindran, S. J., Jagannath, C., and Dhandayuthapani, S. (2011). OsmC proteins of mycobacterium tuberculosis and Mycobacterium smegmatis protect against organic hydroperoxide stress. Tuberculosis (Edinb.) 91, S119–S127. doi: 10.1016/j.tube.2011.10.021
Schwanhäusser, B., Busse, D., Li, N., Dittmar, G., Schuchhardt, J., Wolf, J., et al. (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. doi: 10.1038/nature10098
Shabala, L., Mcmeekin, T., Budde, B., and Siegumfeldt, H. (2006). Listeria innocua and Lactobacillus delbrueckii subsp. bulgaricus employ different strategies to cope with acid stress. Int. J. Food Microbiol. 110, 1–7. doi: 10.1016/j.ijfoodmicro.2006.01.026
Shin, D. H., Choi, I. G., Busso, D., Jancarik, J., Yokota, H., Kim, R., et al. (2004). Structure of OsmC Fromescherichia coli: a salt-shock-induced protein. Acta Crystallogr. D Biol. Crystallogr. 60, 903–911. doi: 10.1107/S0907444904005013
Si, M., Su, T., Chen, C., Wei, Z., Gong, Z., and Li, G. (2019). OsmC in Corynebacterium glutamicum was a thiol-dependent organic hydroperoxide reductase. Int. J. Biol. Macromol. 136, 642–652. doi: 10.1016/j.ijbiomac.2019.06.047
Spano, G., and Massa, S. (2006). Environmental stress response in wine lactic acid bacteria: beyond Bacillus subtilis. Crit. Rev. Microbiol. 32, 77–86. doi: 10.1080/10408410600709800
van Der Heide, T., and Poolman, B. (2000). Glycine betaine transport in Lactococcus lactis is osmotically regulated at the level of expression and translocation activity. J. Bacteriol. 182, 203–206. doi: 10.1128/JB.182.1.203-206.2000
Wang, Y., Delettre, J., Corrieu, G., and Béal, C. (2011). Starvation induces physiological changes that act on the cryotolerance of Lactobacillus acidophilus RD758. Biotechnol. Prog. 27, 342–350. doi: 10.1002/btpr.566
Wang, J., Tang, H., Zhang, C., Zhao, Y., Derrien, M., Rocher, E., et al. (2015). Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 9, 1–15. doi: 10.1038/ismej.2014.99
Wang, C., Zhang, C., Li, S., Yu, L., Tian, F., Zhao, J., et al. (2020). Effects of probiotic supplementation on dyslipidemia in type 2 diabetes mellitus: a meta-analysis of randomized controlled trials. Foods 9:1540. doi: 10.3390/foods9111540
Yao, X., Liu, P., Chen, B., Wang, X., Tao, F., Lin, Z., et al. (2022). Synthetic acid stress-tolerance modules improve growth robustness and lysine productivity of industrial Escherichia coli in fermentation at low pH. Microb. Cell Factories 21:68. doi: 10.1186/s12934-022-01795-4
Yu, C. S., Lin, C. J., and Hwang, J. K. (2004). Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 13, 1402–1406. doi: 10.1110/ps.03479604
Zhang, W., Guo, H., Cao, C., Li, L., Kwok, L. Y., Zhang, H., et al. (2017). Adaptation of Lactobacillus casei Zhang to gentamycin involves an alkaline shock protein. Front. Microbiol. 8:2316. doi: 10.3389/fmicb.2017.02316
Zhen, N., Zeng, X., Wang, H., Yu, J., Pan, D., Wu, Z., et al. (2020). Effects of heat shock treatment on the survival rate of Lactobacillus acidophilus after freeze-drying. Food Res. Int. 136:109507. doi: 10.1016/j.foodres.2020.109507
Keywords: Limosilactobacillus reuteri , acid stress, bile salts stress, freeze-drying, cross-protection
Citation: Liu Z, Zhao X and Bangash HI (2024) Expression of stress responsive genes enables Limosilactobacillus reuteri to cross-protection against acid, bile salt, and freeze-drying. Front. Microbiol. 15:1437803. doi: 10.3389/fmicb.2024.1437803
Edited by:
Konstantinos Papadimitriou, Agricultural University of Athens, GreeceReviewed by:
E. Emma Tymczyszyn, National University of Quilmes, ArgentinaJuan Moreira, University of Arkansas, United States
Anastasios Nikolaou, Democritus University of Thrace, Greece
Copyright © 2024 Liu, Zhao and Bangash. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenzhen Liu, bGl1emhlbnpoZW4wNDEwQG91dGxvb2suY29t
 Zhenzhen Liu
Zhenzhen Liu Xiao Zhao
Xiao Zhao Hina Iqbal Bangash2
Hina Iqbal Bangash2