- 1College of Animal Science and Technology, Qingdao Agricultural University, Qingdao, China
- 2College of Animal Science and Technology, Jilin Agricultural University, Changchun, China
Probiotics are one of the management tools to improve the host’s healthy microbiota. The positive effects of probiotics on host health are species-specific, so probiotics isolated from host’s own gut may be most beneficial. Many of the metabolites (e.g., short-chain fatty acids, bacteriocins, and hydrogen peroxide) produced by Lactobacillus johnsonii have specific inhibitory profiles against invading pathogens. In this study, we isolated L. johnsonii GJ231 from the intestinal tract of healthy female beagles. The genome size of 1.763 M encoded a total of 1,691 predicted genes. Many carbohydrate-active enzymes responsible for carbohydrate degradation and the production of short-chain fatty acids were also predicted. The metabolic profile of short-chain fatty acids in L. johnsonii GJ231 was determined using LC–MS/MS. The bacteriocin-producing gene bacteriocin (lactacin F) in L. johnsonii GJ231 was also predicted. In vitro, experiments demonstrated that GJ231 can thrive in weak acids, 0.3% bile salts, and artificial gastrointestinal fluid models. It was tolerant of to high temperatures up to 70°C, was non- hemolytic, inhibited pathogenic bacteria, and had a high antioxidant capacity. In vivo safety experiments conducted in mice revealed that oral administration of GJ231 not only had no toxic side effect but also increased their antioxidant capacity. In conclusion, combining the above test results, which collectively demonstrate that canine-derived L. johnsonii GJ231 was a non-pathogenic, acid-tolerant and bile-salt-tolerant probiotic strain that inhibits pathogenic bacteria and improves host antioxidant function. This may make it a promising candidate for the development of innovative functional foods for pets.
1 Introduction
Dogs and cats have been companions to humans for thousands of years (Grześkowiak et al., 2015). Nowadays, pet owners consider their pets as family members, so the health of pets is a major concern (Redfern et al., 2017). The health of companion animals depends on their gut microbiota (Grześkowiak et al., 2015). Pet owners and veterinarians have typically used antibiotics to treat or prevent animal diseases. However, the use of antibiotics can lead to gut microbiota disruption, and overuse can lead to a rapid increase in antibiotic residue and resistance (Ma et al., 2021; Shao et al., 2021). Close contact between pets and their owners can lead to the transfer of bacterial resistance, resulting in harm to both parties as a result (Scarborough et al., 2021; El-Razik et al., 2023). Therefore, there is a considerable need to develop new products to replace antibiotics.
In recent years, there has been a growing interest in the use of probiotics as an alternative to antibiotics for the prevention and treatment of bacterial infections, in particular Lactobacillus and Bidobacterium (Wan et al., 2019). Probiotics are “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Hill et al., 2014). Probiotics such as lactic acid bacteria (LAB) can inhibit the adhesion and activity of pathogenic bacteria, modulate immunity, improve intestinal barrier integrity, and produce organic acids as well as antimicrobial compounds (La Fata et al., 2018; Sanders et al., 2019). Lactobacillus johnsonii, a type of LAB, has been shown to improve antioxidant capacity, alleviate colitis, and improve hypercholesterolemia (Kim et al., 2006; Jia et al., 2022; Yoon et al., 2023).
Probiotic candidates isolated from host’s own gut may be most effective, as the co-evolution of gut microbes with the host can better prepare the strain to colonize and thrive in the host’s gastrointestinal tract (Garcia-Gutierrez et al., 2019; Johnson et al., 2023). A previous study reported that L. johnsonii strains were specific to their hosts (Buhnik-Rosenblau et al., 2012). Whole-genome sequencing, aided by the rapid advancement of sequencing technology, is becoming essential for studying microorganisms, presenting a new technique for investigating the probiotic potential and possible pathogenic features of microbes (Tyagi et al., 2022). In this study, we isolated L. johnsonii strain GJ231 from the gastrointestinal tract of healthy beagles. A comprehensive analysis of the probiotic potential and safety of strain GJ231 was conducted using whole-genome sequencing, along with in vivo and in vitro tests, demonstrating the strain’s potential for use in companion animals.
2 Materials and methods
2.1 Sample collection
Animals were acquired from the Jimo Pet Base of Qingdao Agricultural University. Six well-grown and healthy 6-month-old female beagles with an average weight of 5.45 ± 0.36 kg were selected. All test animals were not given probiotics or antibiotics in the past 2 months. A cotton swab was dipped into a small amount of saline and used for sampling. Samples were promptly transported to the laboratory.
2.2 Strain isolation and identification
Experimental samples were subjected to gradient dilution (10−3–10−7), and 50 μL of each dilution was coated onto Man, Rogosa, and Sharpe (MRS) agar plates (Haibo, China). The plates were incubated inverted at 37°C for 48–72 h. Single colonies with lysogenic rings were selected and transferred to MRS broth. After 2–3 passages, glycerol (25.0%) was added as a preservative, and cultures were stored for future use. Bacterial DNA was extracted with a Bacterial Genome Kit (Tiangen, China). Subsequently, polymerase chain reaction (PCR) was performed using the primers 16S-27F (5’-AGAGTTTGATCCTGGCTCAG-3′) and 16S-1492R (5’-TACGGCTACCTTGTTACGACTT-3′) (Wang et al., 2018) and the resulting products were sent to Qingke Biotech (Beijing, China) for sequencing. The sequencing results were compared with other sequence data at National Center for Biotechnology Information, and a phylogenetic tree (Neighbor Joining) was reconstructed using Mega 7.0 (Sudhir Kumar, King Abdulaziz University, Saudi Arabia) (Kumar et al., 2016).
L. johnsonii GJ231 used in this study has been conserved at the China Center for Type Culture Collection (CCTCC M2023981).
2.3 Probiotic potential
2.3.1 Growth and acid production curves of strains
GJ231 cultured for 18–20 h was inoculated into MRS broth at 37°C with an inoculum amount of 2.5% (v/v) and then incubated for 0, 1, 2, 3, 6, 9, 12, 15, 18, 21, 24, 27, 30, 36, or 48 h, respectively. The optical density at 600 nm (OD600) was measured at each time point, while the pH value was determined at different time points using a pH detector (Shanghai INESA, China).
2.3.2 Antimicrobial ability
Cultures in the exhibiting stationary phase after 18 h of incubation (1 × 108 CFU/mL) and the bacterial suspension (BS) was centrifuged at 4°C, 10,621 × g (10,000 rpm in Centrifuge 5430R) for 10 min. The cell-free supernatant (CFS) was collected, and the remaining bacterial pellet (BP) was resuspended in the same volume of PBS. Similarly, cultures of pathogenic bacteria (Escherichia coli ATCC 25922; Staphylococcus aureus ATCC 25923; Salmonella enterica subsp. Enterica serovar Typhimurium ATCC 14028; Pseudomonas aeruginosa ATCC 27853; Listeria monocytogenes ATCC 19115) in the stationary phase of growth were adjusted to a concentration approximately 1 × 107 CFU/mL. A two-layer method was adopted using Oxford cups on Tryptone Soy Agar (TSA) (Solarbio, China), with 100 μL of CFS, BS, BP, or CFS7.0 (CFS was adjusted to pH 7.0) added in each well. The wells were then incubated at 37°C for 24 h, and the inhibition zone diameter (IZD) was determined.
2.3.3 High temperature tolerance
The method described by Zhao et al. (2023) was followed with minor modifications. Briefly, GJ231 bacterial solution cultured for 18–20 h was placed into a water bath heated to 37, 50, 60, 70, or 80°C for 5 min, respectively. Then was quickly transferred to ice for 30–60 s. The number of bacteria in 100 μL of the solution was then calculated by plate counting. The survival rate was calculated using the following formula:
Where Tinitial1 and Ttreatment1 are the number of bacteria (log CFU/mL) before and after treatment, respectively.
2.3.4 Resistance to gastrointestinal environmental
The method described by Gilliland and Walker (1990) was followed with minor modifications. Briefly, GJ231 bacterial suspension cultured for 18–20 h were centrifuged at 4°C, 10,621 × g for 10 min and the supernatant was discarded. The bacterial cells were washed three times with PBS (pH 7.0). The cells were then resuspended in 0.1 and 0.3% (w/v) bile salt solutions, and the 0, 1, 2, and 4 h incubations were taken for counting using dilution plate counting method. Similarly, bacterial cultures from GJ231 were cultured until reaching the stationary phase, and samples were taken at 1.5 h or 2 h intervals after resuspension in gastric or intestinal fluids. The remaining steps were the same as those for pre-bile salt tolerance treatment. For the treatment with gastrointestinal fluid, samples were initially resuspended in gastric fluid for 1.5 h, followed by resuspension in intestinal fluid for 2 h before being sampled and counted. All cultures were incubated at 37°C, and survival rates were calculated using Equation 1.
2.3.5 Antioxidant test
Ascorbic acid was used as a positive reference and a DPPH free radical scavenging capacity assay kit (Solarbio, China) was operated. The absorbance was measured at 515 nm (Cheng et al., 2021). Calculated using Equation 2.
(2) Asample1 is the absorbance of the sample; Acontrol1 is the absorbance of the mixture to sample and anhydrous ethanol; Ablank1 is the absorbance of the mixture of the extract solution and working solution.
A working solution was made by mixing 7 mM ABTS and 2.45 mM potassium persulfate and after incubation for 12 h away from light. Add 20 μL of sample to 200 μL of working solution and mix well. Next, the reaction was carried out for 20 min at room temperature and protected from light, and the absorbance was measured at 734 nm. As a positive control, ascorbic acid was utilized. Calculated using Equation 3.
(3) Asample2 is the absorbance of a mixture of sample and working solution; Acontrol2 is the absorbance of the mixture of deionized water and sample; Ablank2 is the absorbance of the mixture of deionized water and working solution.
O2- scavenging capacity was determined using the Superoxide Anion Scavenging Capacity Kit (Solarbio, China). The OD value was measured at 560 nm. Calculated using Equation 4 (Zhao et al., 2023).
(4) Asample3 is the absorbance of a mixture of sample and working solution; Ablank3 is the absorbance of the mixture of deionized water and working solution.
2.3.6 Autoaggregation activity
Cultures during the stationary phase were washed three times with PBS (pH 7.0), and the OD600 (A0) of the bacterial solution was determined. The bacterial solution was vortexed for 25–30 s and then incubated at 37°C for 8 h. The OD600 (A1) was then determined. Calculated using Equation 5.
2.3.7 Cell surface hydrophobicity
GJ231 was cultured at 37°C for 18–20 h and then centrifuged at 4°C at 10,621 × g for 10 min. Cells were washed three times with PBS (pH 7.0) and resuspended until the OD600 was approximately 0.25 ± 0.05 (A2). An equal volume of xylene and chloroform (1,1, v/v) was then added, and samples were vortexed for 120 s before incubating at 37°C for 3 h. The OD600 of the aqueous phase was then measured (A3). Calculated using Equation 6.
2.3.8 Hemolytic activity
A culture of GJ231 was streaked on the blood agar base (Oxoid, Germany) containing 5.0% (v/v) sheep blood, which were incubated at 37°C for 20–24 h (Nataraj et al., 2023). Blank blood plates without inoculation were used as a negative control (data not shown), and Staphylococcus aureus (ATCC 25923) was used as a positive control.
2.4 Complete genome sequencing analysis
GJ231 bacterial pellet and genomic DNA were extracted using the SDS extraction method (Wang et al., 2023c). Libraries were prepared using an SQK-LSK110 kit (Oxford, United Kingdom) following the manufacturer’s instruction manual using 1.0 g DNA. After sample purification and quality control, small fragment libraries were generated using a Universal Plus DNA Library Prep Kit for MGI V2 (Vazyme, China). The libraries underwent quality checks and were sequenced using the Nanopore PromethION and Illumina NovaSeq 6,000 platforms. Reads were assembled using Unicycler (Version: 0.5.0) software, plasmids were identified using PlasFlow software, and the assembled genome was used for coding gene prediction using Prokka (Version: 1.14.6) software (Seemann, 2014). Functional elements, genomic functions, database annotation, and websites were listed in Supplementary Table S1. Thresholds for carbohydrase annotation were set at an E-value of <1e-18 and coverage of >35.0%. Pathogen–host interaction thresholds were set at an E-value of <1e-5, and virulence factor annotation was performed using the VFDB database with a coverage of >40.0% (Wang et al., 2023b).
Potential gene clusters for natural product biosynthesis in GJ231 were predicted using the antiSMASH database. In addition, the bacteriocin production gene cluster was predicted using the Bagel4 database. The amino acid sequences of bacteriocin lactacin F (lafA and lafx), which have the highest sequence similarity, were submitted to the SWISS-MODEL web server to generate protein models (Wang et al., 2024).
2.5 Safety evaluation in vivo
Forty Kunming white mice (from Jinan Pengyue Experimental Animal Breeding Co., Ltd.) were randomly divided into two groups, with an equal number of males and females in each group. The oral administration experiment began after 7 d of acclimatization with unrestricted access to food and water, and a room temperature of 25°C. The control group (CK) was administered 0.1 mL of saline daily via oral gavage for 28 d, while the GJ231 group was administered an equal volume of GJ231 bacterial solution (1 × 109 CFU/mL) daily via oral gavage for 28 d. Throughout the study, mice were monitored daily for diarrhea, and their body weights and food intake were recorded weekly. Before the end of the experiment, the mice were fasted for 12 h, rendered unconscious with 1.0% (w/v) sodium pentobarbital (50 mg/kg), and blood was collected from the aorta. Blood samples were centrifuged at 825 × g (3,000 rpm, using a centrifuge 5,702 Eppendorf, Germany) at 4°C for 15 min. The serum was collected and assayed for biochemical markers using a kit (Nanjing, China). Subsequently, the hearts, livers, spleens, kidneys, and thymuses of the mice were observed and collected. The organs of 10 mice in each group were randomly selected and added to 10 mL of sterile saline (1.0 g/mL). The organs were homogenized and crushed, and 0.1 mL of the homogenate was spread evenly on MRS plates. The plates were then incubated at 37°C for 48 h for observation.
2.6 Short chain fatty acids
Short-chain fatty acids in the supernatant of strain GJ231 were determined using LC–MS/MS. Briefly, 30 to 40 mg of frozen faecal samples were placed into a 1.5 mL centrifuge tube. Then, 1 mL of 50% acetonitrile (ACN, Fisher Chemical, United States) was added, followed by 2 to 3 metal grinding beads. The sample was processed in an E6618 tissue grinder (Beyotime, China) for 1 min at 60 Hz and then centrifuged at 15871 × g (13,000 rpm in a Centrifuge 5,430 Eppendorf, Germany) for 10 min at 4°C. One hundred microliters of supernatant was taken and diluted proportionally to 10 mg of sample per 1.8 mL of 50% ACN solution. The mixture was vortexed and shaken (Scilogex, USA) for 30 s and then centrifuged at 15871 × g for 30 s. Twenty microliters of the supernatant was aspirated, and 10 μL of 200 mM 3-nitrophenylhydrazine-HCl (3-NPH-HCl, Sigma Aldrich, United States) was added separately. Ten microlitres of 200 mM N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide-HCl (EDC-HCl, Sigma Aldrich, United States), 80 μL of 50% ACN, 50 μL of 7% pyridine (Sigma–Aldrich, United States), and 1 μL of isotope internal standard solution (Toronto Research Chemicals, Canada) were vortexed and shaken for 3 min, derivatized in a constant-temperature water bath at 40°C for 30 min and centrifuged at 15871 × g for 1 min at 4°C. Finally, 20 μL of the reaction solution after derivatization was added to 280 μL of 50% ACN, vortexed for 30 s, centrifuged at 15871 × g for 10 min at 4°C, aspirated into the injection vial, and then subjected to LC–MS/MS analysis (LC-30 HPLC, SCIEX QTRAP 5500 mass spectrometry, Phenomenex: Kinetex C18, 2.6 μm 100 × 3.00 mm, column temperature: 40°C, flow rate: 0.7 mL/min).
2.7 Statistical analysis
Data were expressed as the mean ± standard error of mean (SEM). Statistical significance was determined using t-tests and one-way analysis of variance in GraphPad Prism 8.3.0, with statistical significance set at a level of p < 0.05.
2.8 Data availability
The whole-genome sequence data has been deposited under accession number PRJNA1028141 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1028141) (17-Oct-2023).
3 Results
3.1 Identification of GJ231
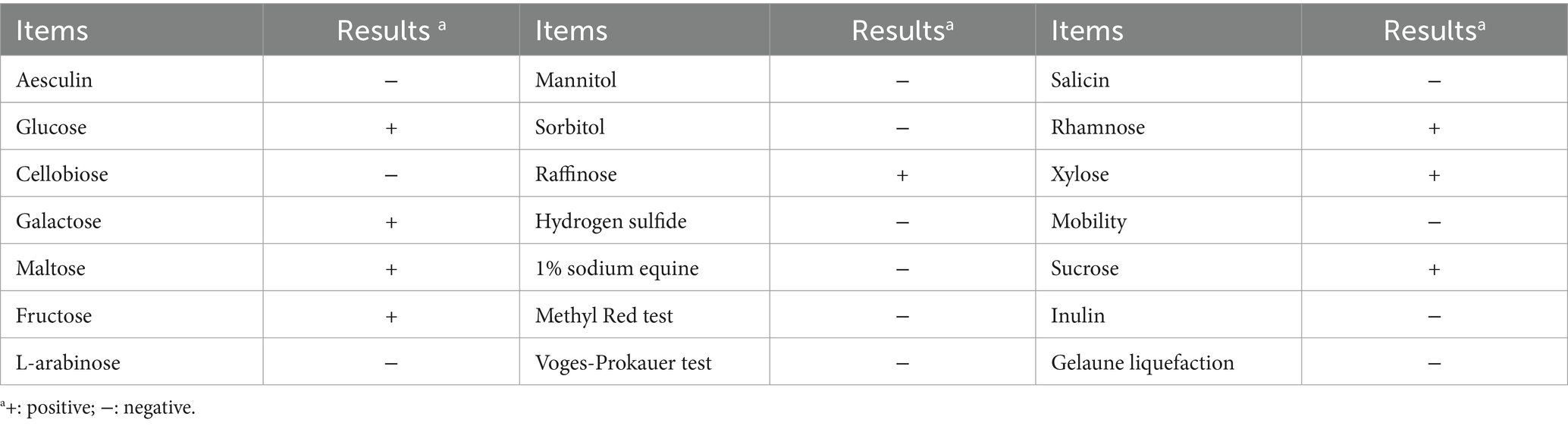
The results of physiological and biochemical experiments on strain GJ231 were shown in Table 1. Strain GJ231 could utilize glucose, galactose, maltose, fructose, raffinose, rhamnose, xylose and sucrose. The colony morphology of strain GJ231 on MRS agar plates was raised, creamy white, and spherical or ovoid (Figure 1A). After Gram staining, bacteria appeared under a microscope as purple, rod-shaped cells with rounded ends, indicating that they are Gram-positive (Figure 1B). By reconstructing an evolutionary tree, we determined that GJ231 shared a nucleotide similarity of 99% with L. johnsonii N6-2 (Figure 1C). We therefore concluded that GJ231 represented L. johnsonii.
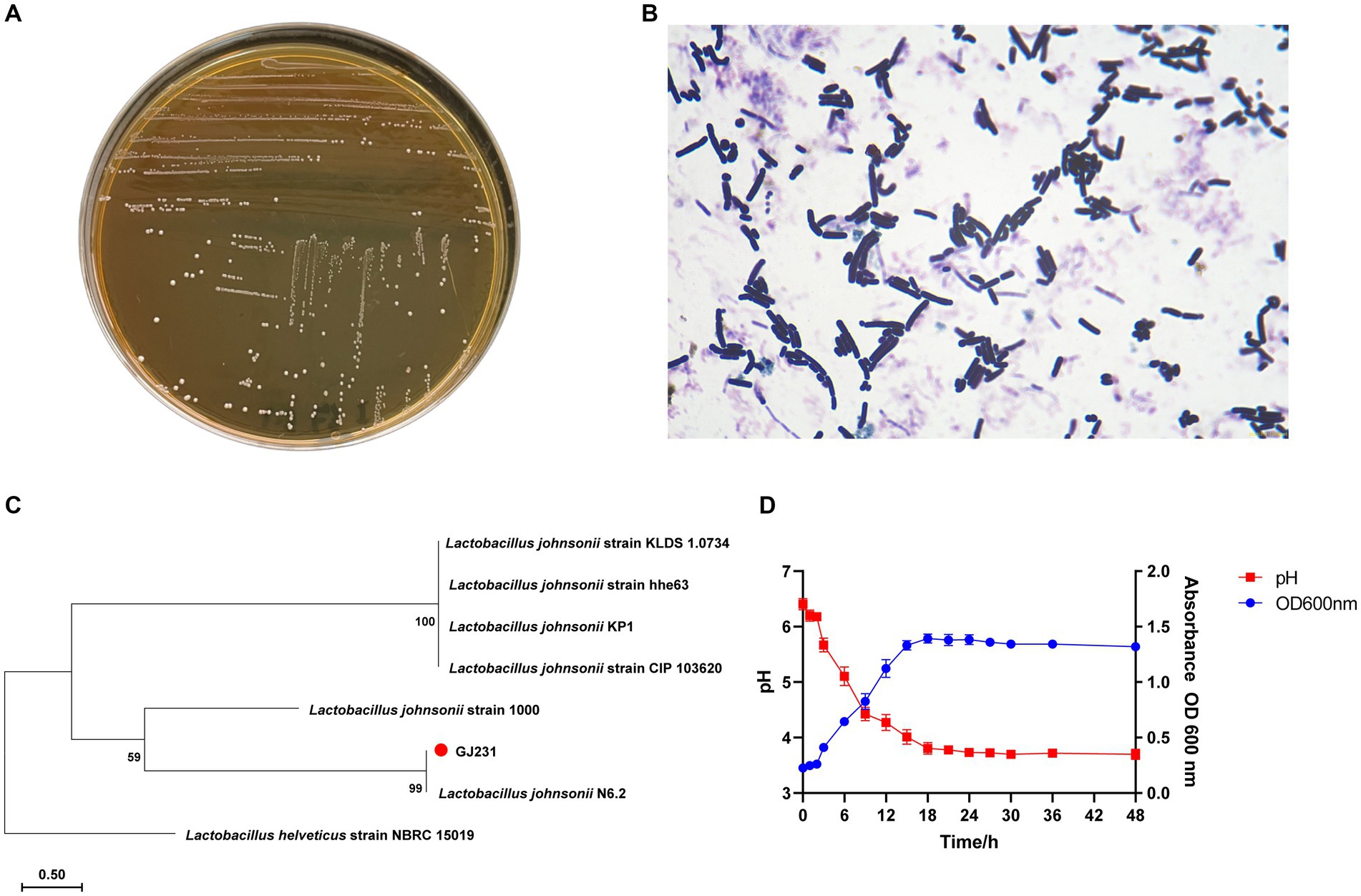
Figure 1. Morphology, staining and phylogenetic tree of strain GJ231. (A) Morphology of strain GJ231, (B) gram staining results of strain GJ231, (C) neighbor-joining phylogenetic tree of strain GJ231 (Lactobacillus helveticus as an exogenous species) and (D) growth and acid-producing curve.
3.2 Growth and acid production curve
L. johnsonii GJ231 was in the lag period from 0 to 3 h, entered the exponential phase of growth after 3 h and reached the stationary phase at 18 h. In addition, the strain had a good capacity for acid production (Figure 1D).
3.3 Antimicrobial ability
The bacteriostatic activity of GJ231 against five pathogenic bacteria (Figure 2; Supplementary Table S2) was observed, with the most significant bacteriostatic effect observed against L. monocytogenes, followed by P. aeruginosa, S. aureus, Salmonella, and E. coli. However, there was no significant difference in the inhibitory effects of BS or CFS against the same pathogenic bacteria (p > 0.05). When the pH of the CFS was adjusted to pH 7.0, the inhibitory effect disappeared, suggesting that organic acids may play the primary inhibitory role in the CFS of GJ231. Furthermore, the BP of GJ231 had no inhibitory effect on pathogenic bacteria.
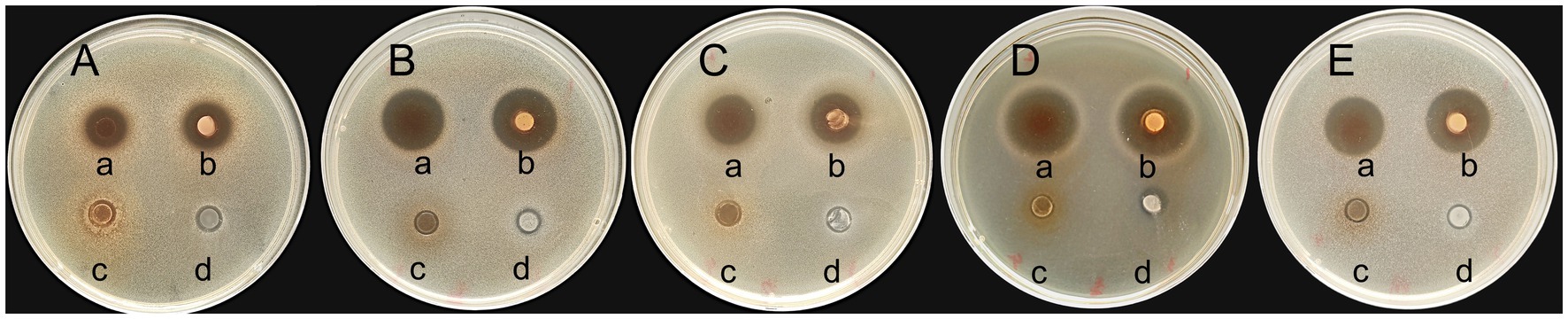
Figure 2. The inhibitory effects of GJ231 against pathogenic indicator bacteria. (A) Escherichia coli; (B) Staphylococcus aureus; (C) Salmonella; (D) Listeria monocytogenes; and (E) Pseudomonas aeruginosa. In each TSA agar, (a) added the cell-free supernatant of GJ231; (b) added the bacterial suspension of GJ231; (c) added the cell-free supernatant (pH = 7.0) of GJ231; (d) added the bacterial pellet.
3.4 Complete genome sequencing and bioinformatic analysis
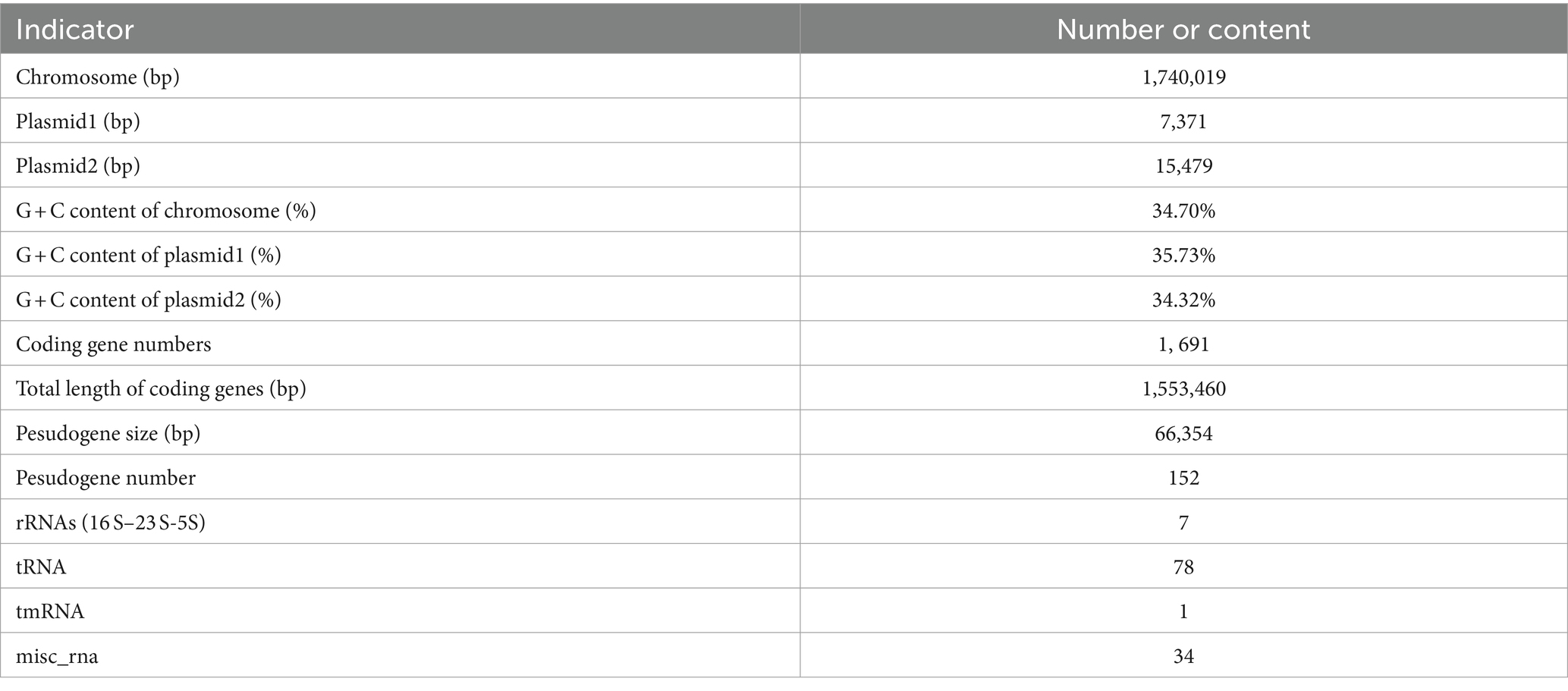
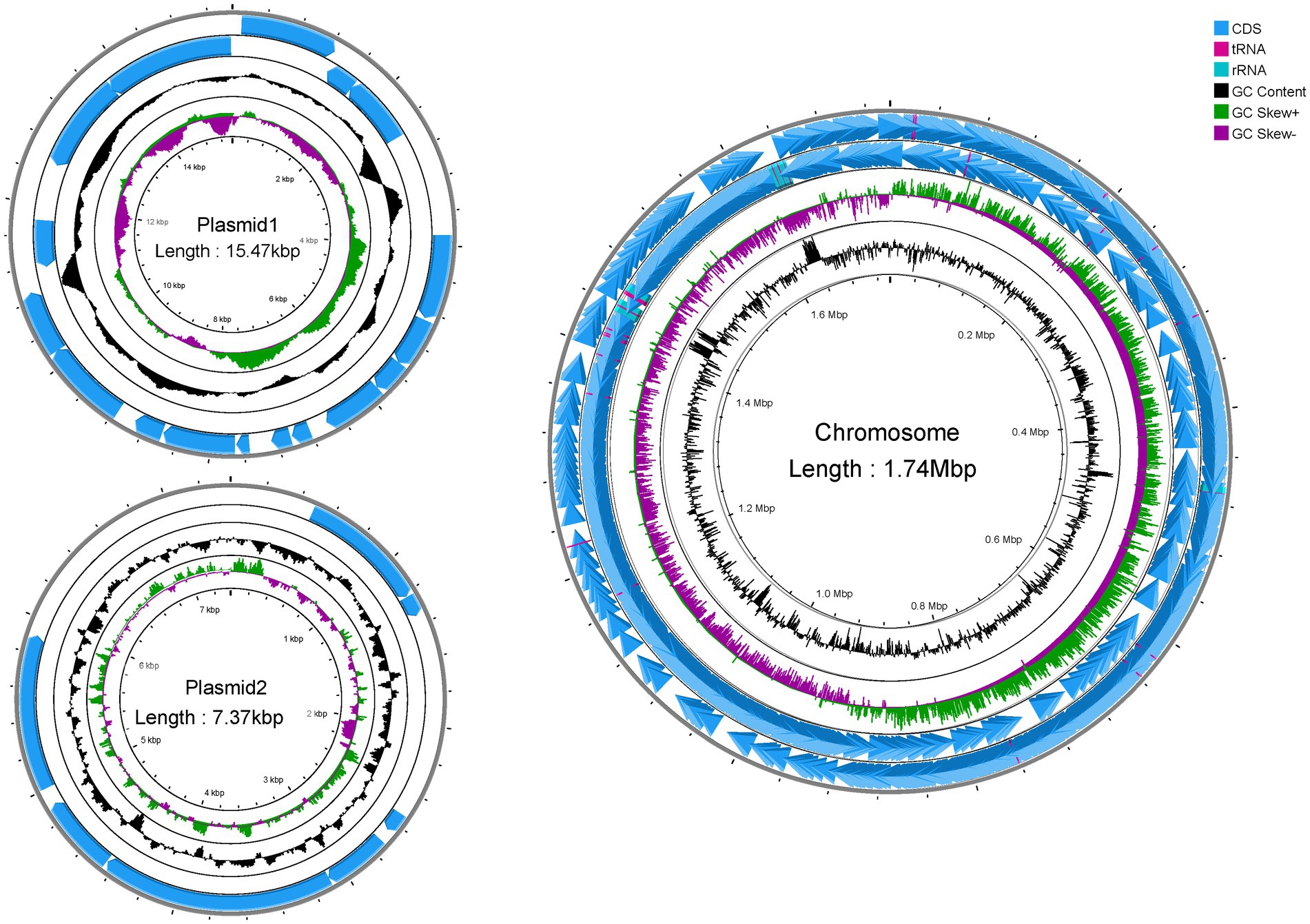
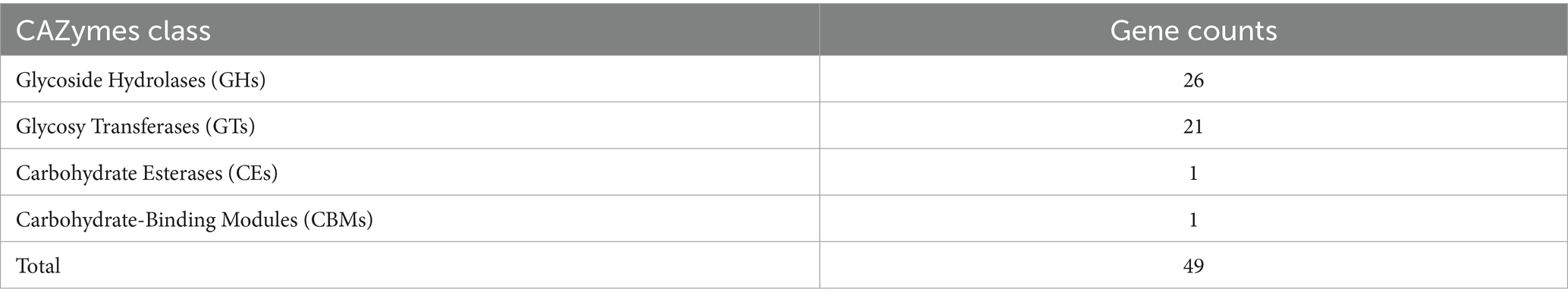
Genome-wide sequencing revealed that GJ231 contained one chromosome with a length of 1,740,019 bp and two plasmids; these had a G + C content of 34.70, 35.73, and 34.32%, respectively (Table 2; Figure 3). There were 1,691 coding sequences (CDSs) with a total length of 1,553,460 bp. Furthermore, we identified 152 pseudogenes, 7 rRNAs, and 78 tRNAs. Our analysis revealed three genomic islands (Supplementary Table S3), eight prophages (Supplementary Table S4), 189 repeated sequences totaling 17,652 bp in length (Supplementary Table S5), 17 insertions (Supplementary Table S6), two cytochromes P450 (Supplementary Table S7), and 83 virulence factors (Supplementary Table S8). In addition, there were 49 carbohydrate-active enzymes, with 26 being glycoside hydrolases and many being glycosyltransferases (Table 3). Moreover, the whole genome of GJ231 contained a 185-bp CRISPR-Cas sequence located on the chromosome (Supplementary Table S9). In addition, three resistance genes, namely InuA, ErmT, and ErmB, were predicted (Table 4).
Figure 4A showed a map of the coding genes in GJ231 annotated in the universal database. A total of 1,674 genes were predicted using the COG database (Figure 4B). Translation and carbohydrate metabolic processes, as well as the phosphoenolpyruvate-dependent glucose phosphotransferase system, were identified as highly active biological processes. We also determined that the plasma membrane, an essential cellular component, plays a crucial role in cellular function, while ATP binding, DNA binding, and metal ion binding were essential molecular functions (Figure 4C). KEGG annotation revealed 1,498 genes classified into 23 functional groups. These groups included global and overview maps (284 genes), carbohydrate metabolism (120 genes), and membrane transport (98 genes), as shown in Figure 4D. The database annotation results indicated that L. johnsonii GJ231 has a robust metabolic capacity and can adapt to multiple ecological niches.
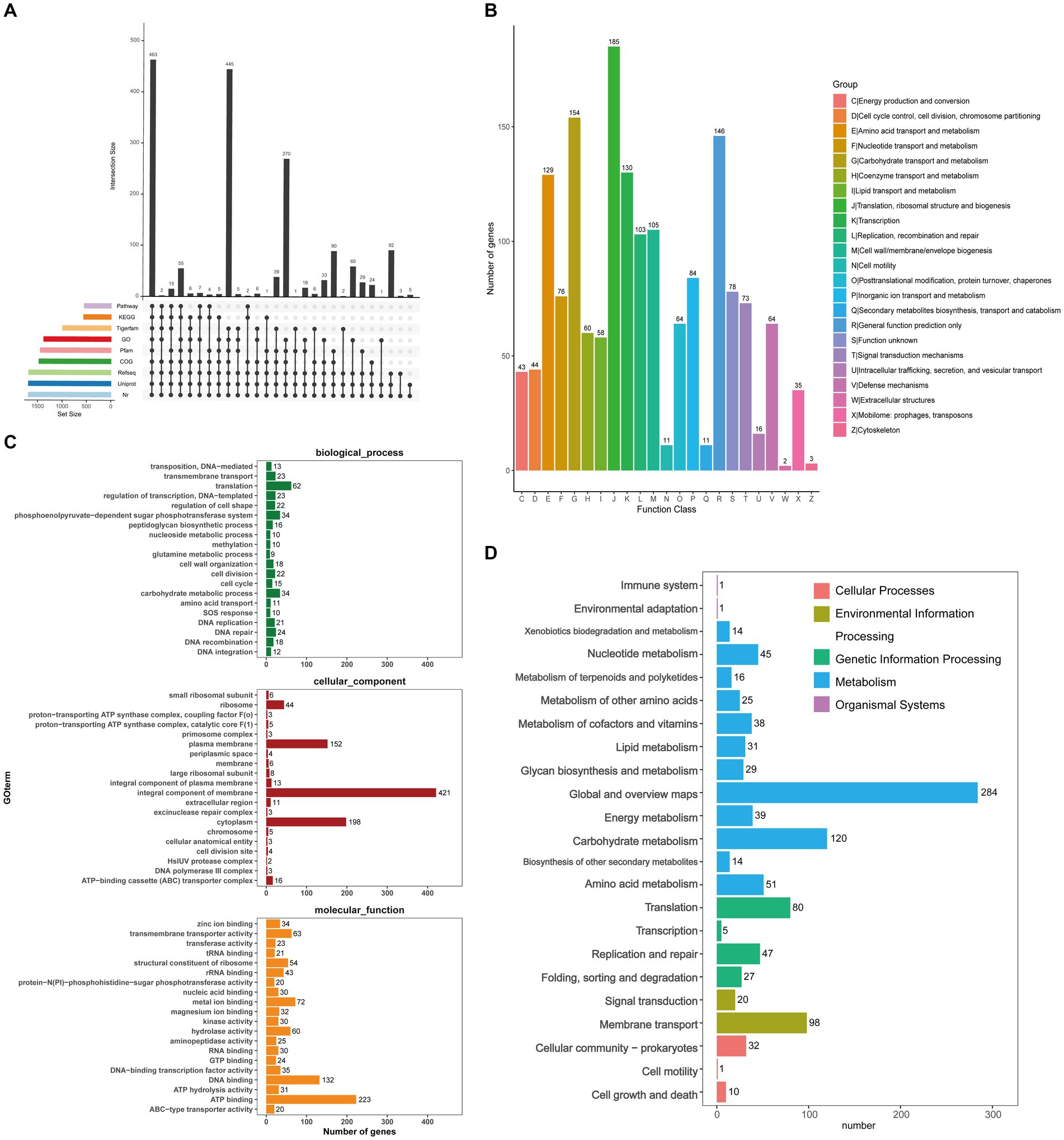
Figure 4. Prediction function of Lactobacillus johnsonii GJ231. (A) Coding gene database annotation statistics; (B) COG; (C) GO analysis and (D) KEGG pathways of proteins functional.
We annotated 537 pathogen–host interactions (Figure 5A). Furthermore, we identified 387 membrane transporter proteins, with primary active transporters being the most numerous at 164 (Figure 5B). The three most common structural domains identified were 65 ABC transporters (ABC_tran), 29 Major Facilitator Superfamily (MFS_1) domains, and 24 RecF/RecN/SMC_N terminal domains (SMC_N) (Figure 5C). The amino acid sequence database of non-redundant (NR) proteins was also analyzed, consistent with the results of 16S rRNA gene sequencing (Figure 5D). In addition, genes conferring resistance to acid, bile salts, high temperatures, and oxidative stress were also identified in the genome of GJ231 (Table 5).
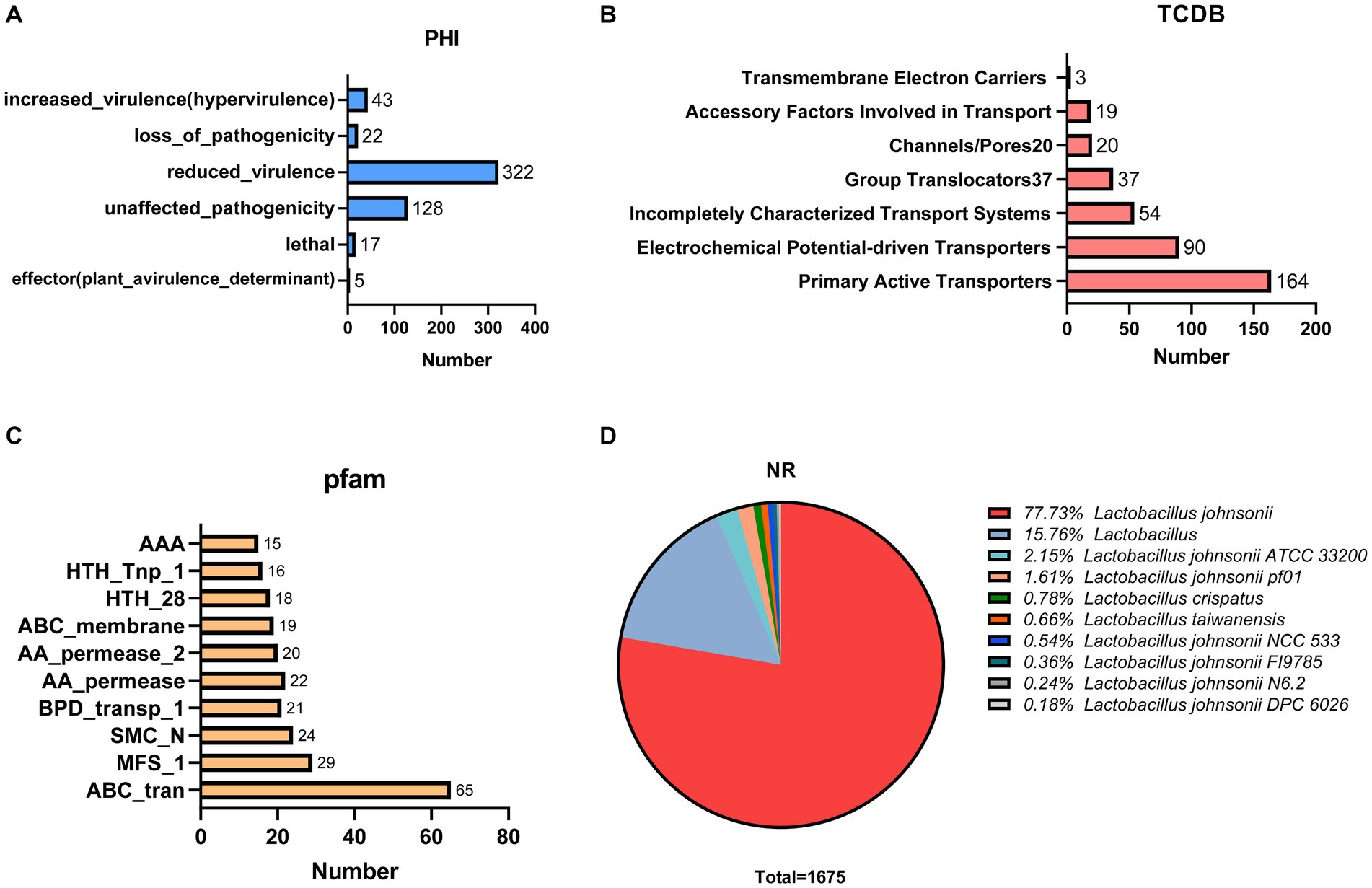
Figure 5. Proprietary database annotations of Lactobacillus johnsonii GJ231. (A) Pathogen host interactions annotations; (B) TCDB transporter protein; (C) PFAM domain and (D) NR annotations of Lactobacillus johnsonii GJ231.
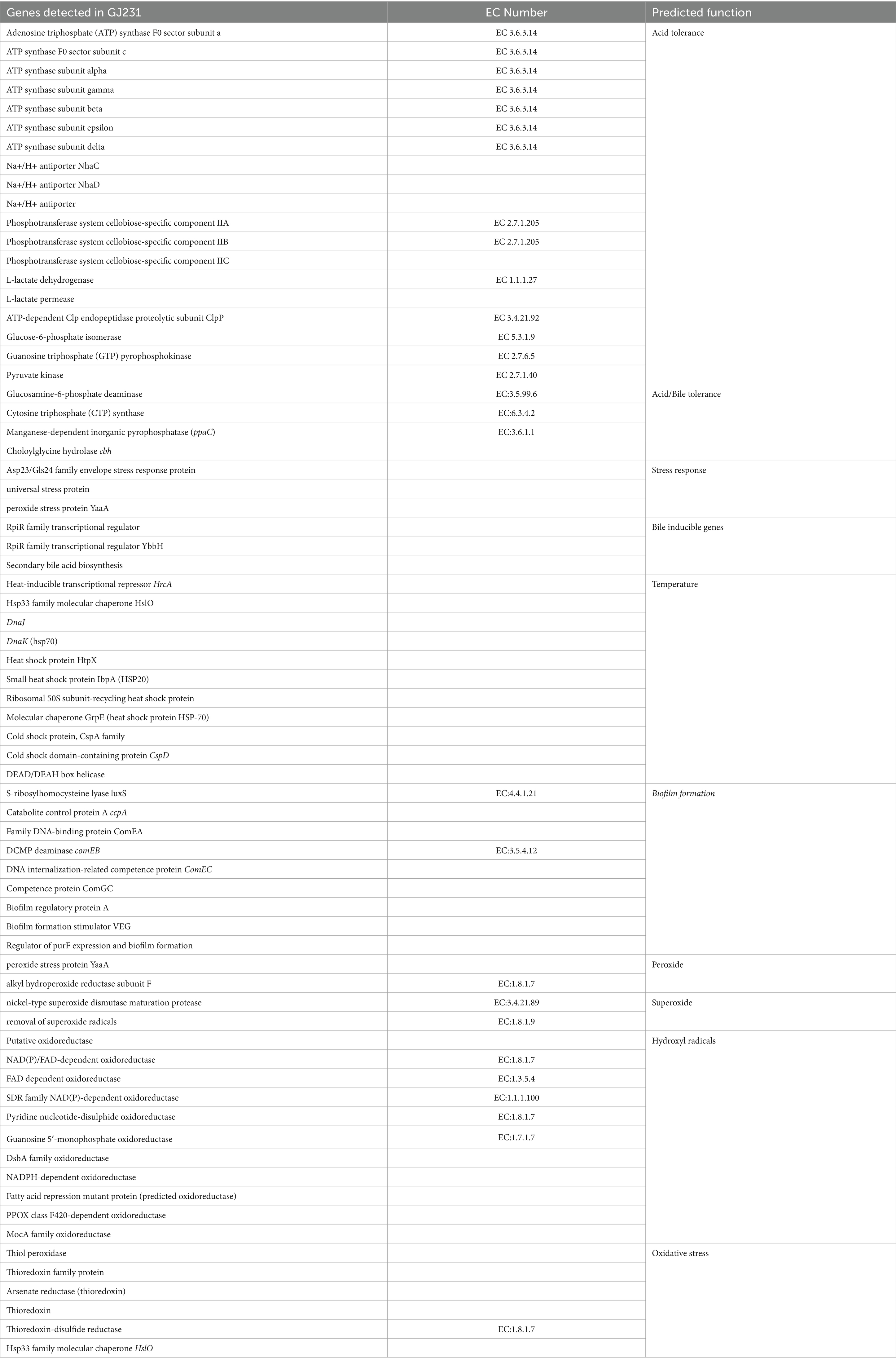
Table 5. Probiotic genes that render resistance to Lactobacillus johnsonii GJ231 in harsh gastric conditions.
We used the antiSMASH database to identify potential gene clusters for natural product biosynthesis in GJ231, and two potential products, gassericin E and gassericin T, were identified (Figures 6A–C). In addition, several potential bacteriocins were predicted in the GJ231 genome, such as gassericin, lactacin F (lafA and lafx), pediocin, and bacteriocin helveticin J (Figure 6D). Furthermore, the tertiary structure of the protein lactacin F (lafA and lafx) was modeled (Figure 6E).
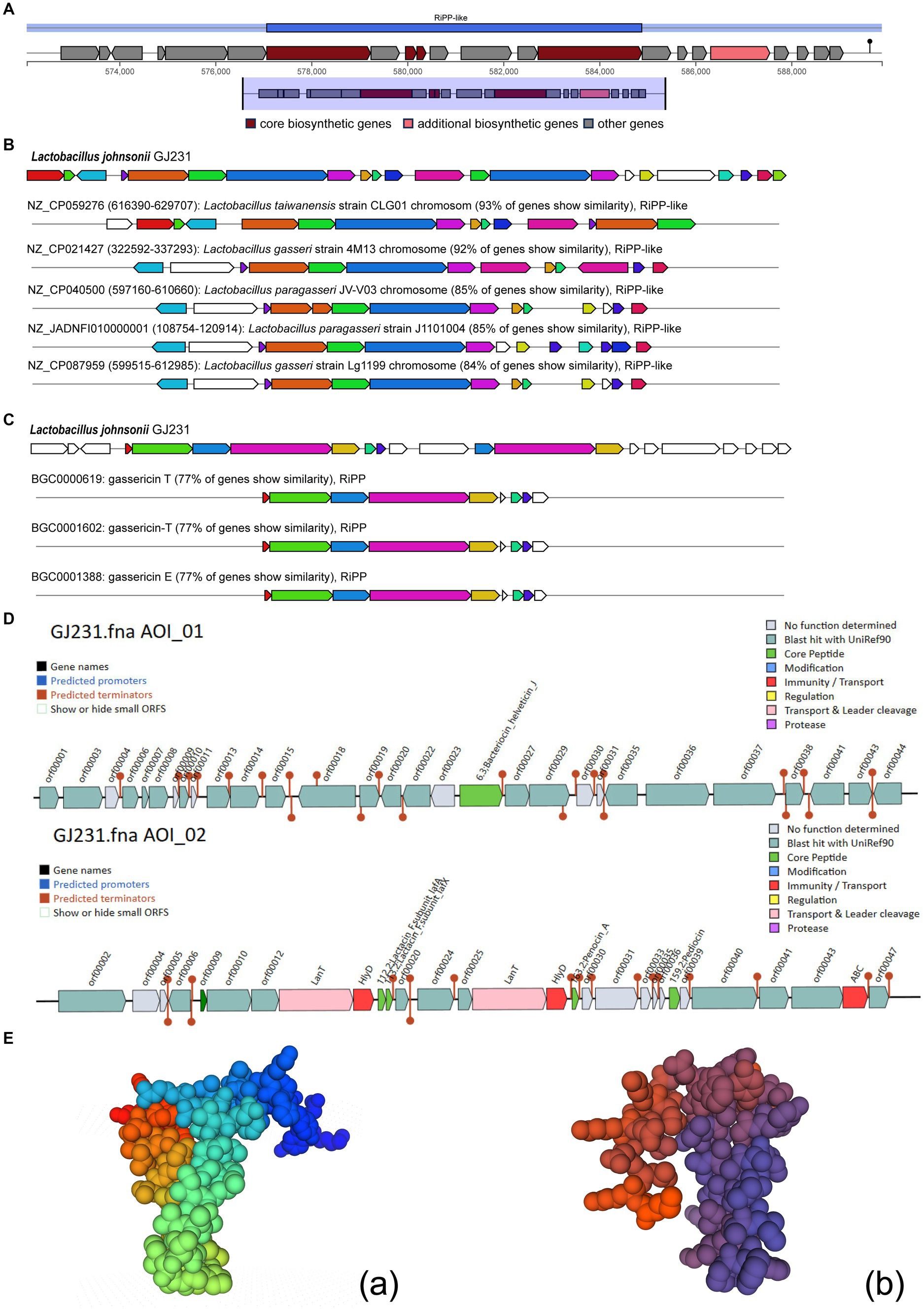
Figure 6. Bacteriocin prediction of Lactobacillus johnsonii GJ231. (A) Region of RiPP-like; (B) Cluster Blast; (C) Known Cluster Blast; (D) bacteriocin prediction by BAGEL 4 and (E) Protein tertiary structure prediction of Lactobacillus johnsonii GJ231.
3.5 Evaluation of the probiotic potential
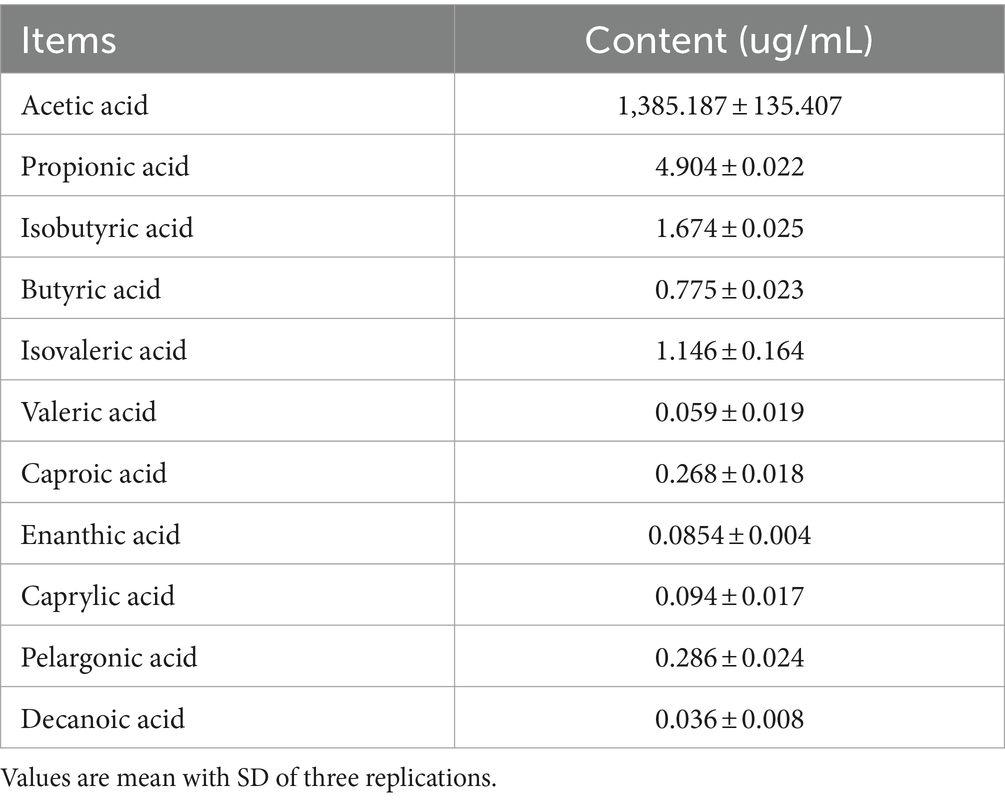
Strain GJ231 tolerated high temperatures well, with over 60.0% survival after treatment at 70°C and approximately 40.0% survival after treatment at 80°C (Figure 7A). The survival rate was still greater than 85.0% with 0.1% bile salt treatment and greater than 80.0% with 0.3% bile salt treatment (Figures 7B,C). Moreover, the survival rate of strain GJ231 after treatment with gastric fluid was almost 80.0%, the attrition rate of strain GJ231 after treatment with intestinal fluid was very low, and the survival rate after co-treatment with gastrointestinal fluid was about 79.0% (Figure 7D). In addition, strain GJ231 exhibited significantly lower DPPH radical scavenging compared with ABTS and O2- scavenging (Figure 7E; P < 0.01). Adhesion of the strain is also an important indicator of in vitro probiotic properties. Strain GJ231 exhibited 50.0% autoaggregation activity and greater than 60.0% cell surface hydrophobicity (Figure 7F). Furthermore, GJ231 tested negative for hemolysis (Figure 7Gb), while Staphylococcus aureus exhibited β-hemolysis (Figure 7Gb). LC–MS/MS results showed that the metabolite (short-chain fatty acids) was mainly acetic acid at 1304.31 μg/mL (Table 6).
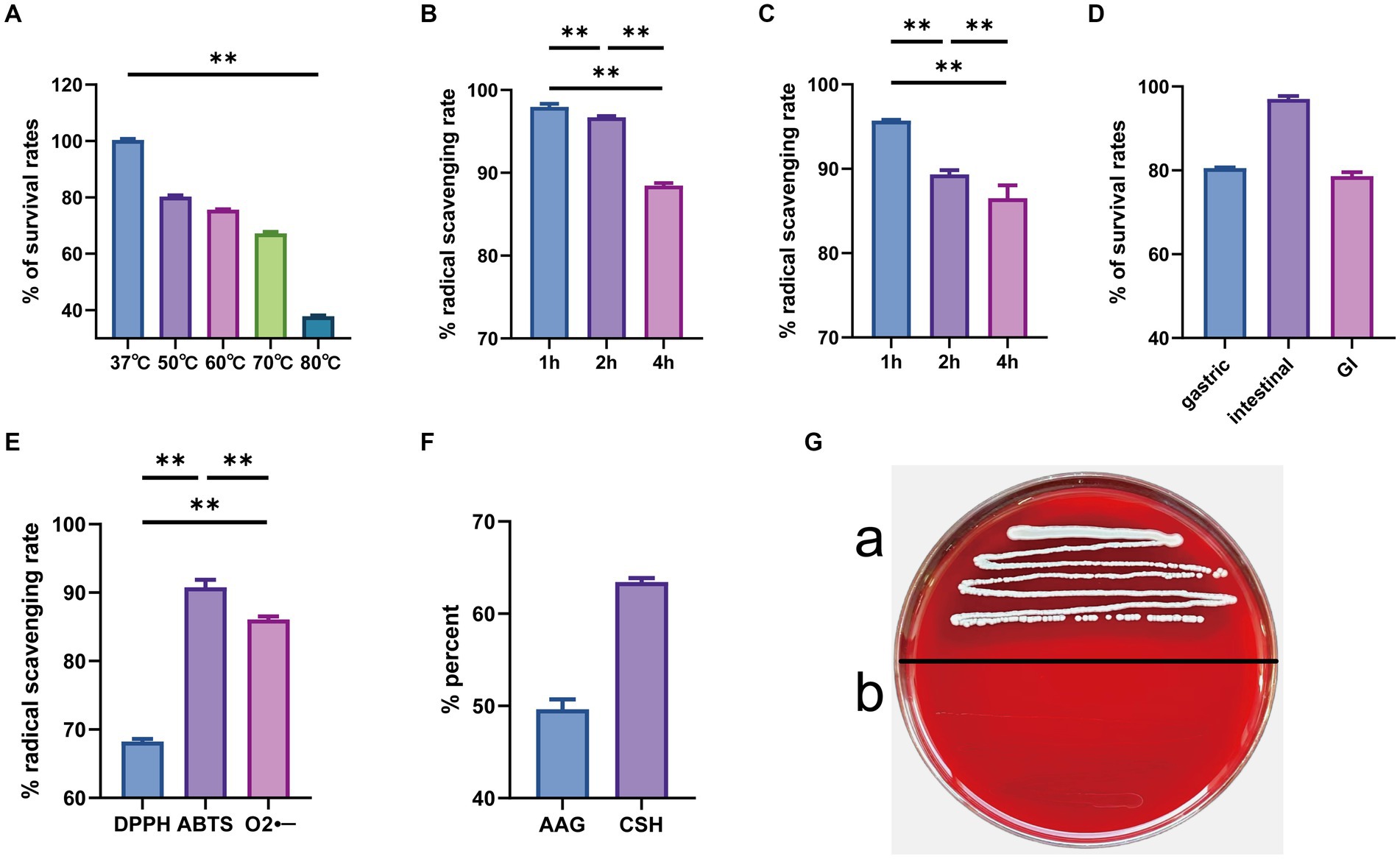
Figure 7. In vitro evaluation of probiotic potential. (A) High temperature resistance; (B) 0.1% bile salt resistance; (C) 0.3% bile salt resistance; (D) gastrointestinal tract models; (E) radical scavenging rate; (F) autoaggregation ability and cell surface hydrophobicity; (G) hemolysis tests (a. Staphylococcus aureus and b. GJ231). **p < 0.01 indicate differences between different types of treatment of GJ231.
3.6 Safety evaluation in vivo
Safety experiments for the oral administration of GJ231 in mice were conducted. Throughout the oral experiment, all mice were active and did not develop diarrhea, die, or exhibit any other signs of illness. No significant pathological changes were observed in the various organs during autopsy. The body weight, average daily gain, heart coefficient, liver coefficient, spleen coefficient, and kidney coefficient of mice orally administered with GJ231 were not significantly different from those of the CK group (Figures 8A–F; P > 0.05). The thymus coefficient of mice orally administered with GJ231 was significantly higher than that of the CK group (Figure 8G; P < 0.01). The liver and kidney function indexes of mice were also measured. There was no significant difference in aspartate aminotransferase (AST) and alanine aminotransferase (ALT) levels in the oral GJ231 group compared with those in the CK group (Figures 9A,B; P > 0.05). However, the blood urea nitrogen (BUN) and malondialdehyde (MDA) contents were significantly lower than those in the CK group (Figures 9C,D; P < 0.01). In addition, the total antioxidant capacity (T-AOC) and superoxide dismutase (SOD) levels in mice orally administered with the GJ231 group were significantly higher than those in the CK group (Figures 9E,F; P < 0.01). However, the catalase (CAT) and Glutathione peroxidase (GSH-PX) levels did not show a significant difference compared with those in the CK group (Figures 9G,H; P > 0.05).
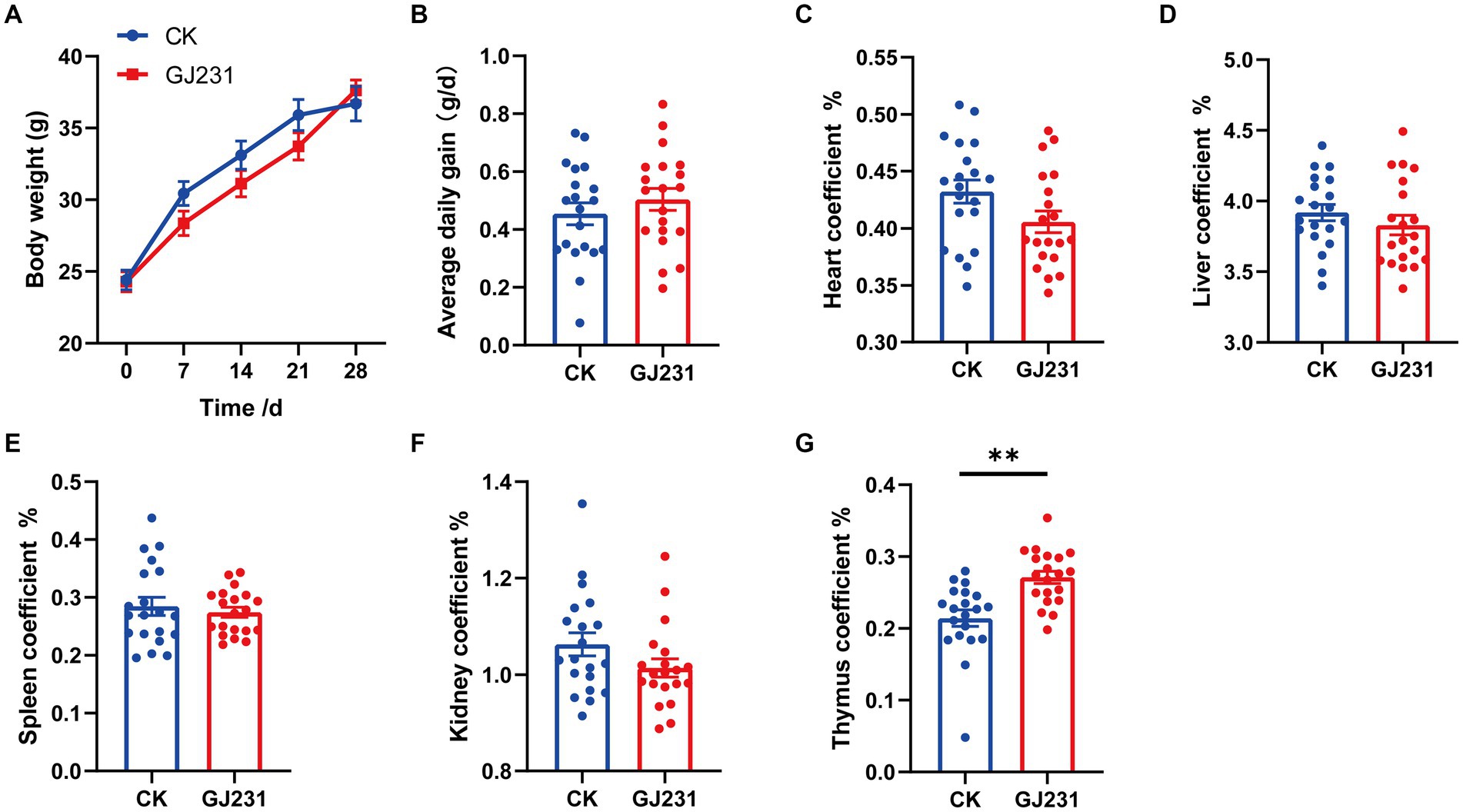
Figure 8. Effect of oral administration of Lactobacillus johnsonii GJ231 on organ indices in mice. (A) Body weight; (B) average daily gain; (C) heart coefficient; (D) liver coefficient; (E) spleen coefficient; (F) kidney coefficient and (G) thymus coefficient in mice. **p < 0.01 indicate differences between different types of treatment of Lactobacillus johnsonii GJ231, n = 20.
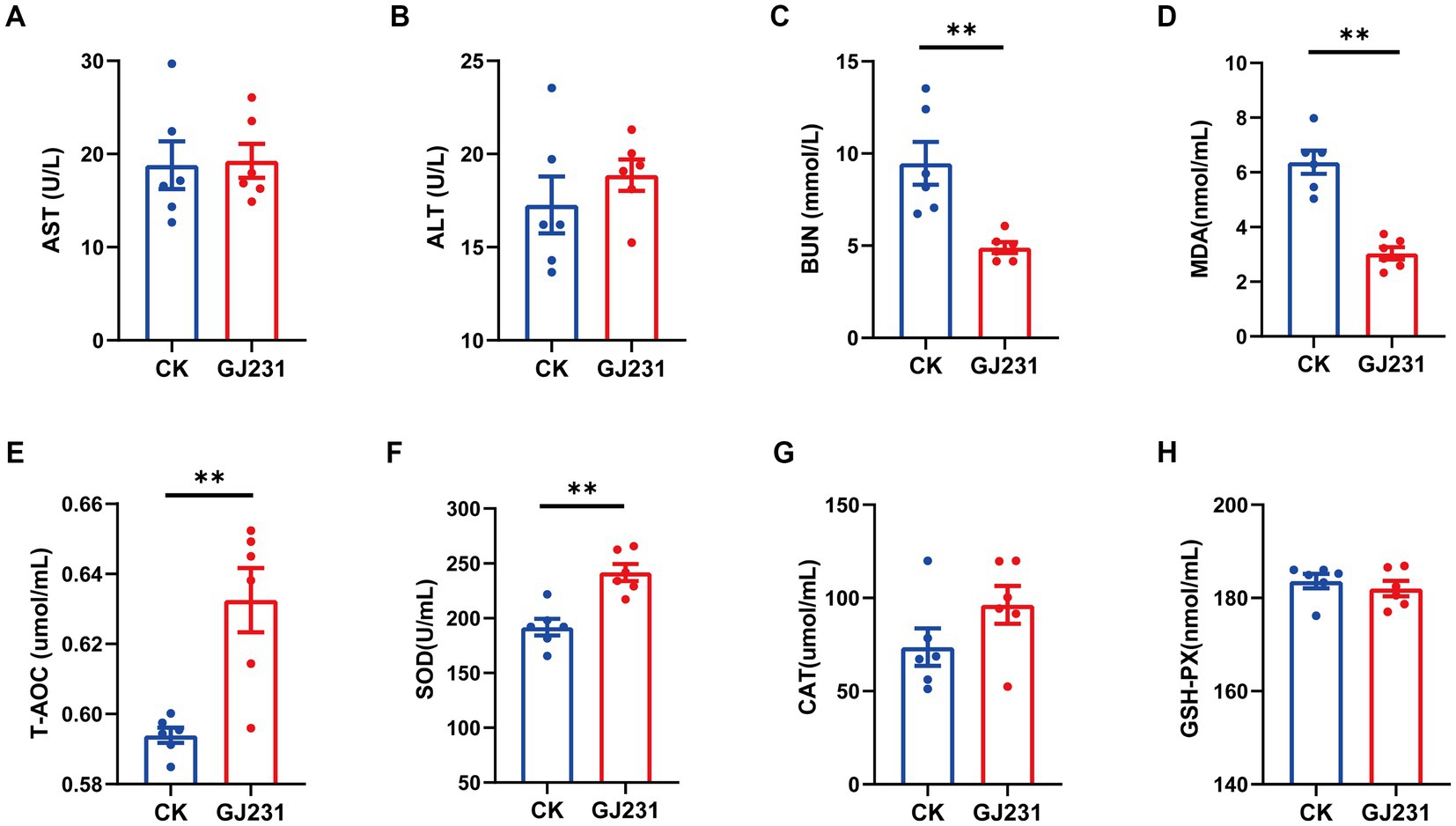
Figure 9. Effect of oral administration of Lactobacillus johnsonii GJ231 on serum indices in mice. (A) Aspartate amino transferase; (B) alanine aminotransferase; (C) blood urea nitrogen; (D) malondialdehyde; (E) total antioxidant capacity; (F) superoxide dismutase; (G) catalase; and (H) glutathione peroxidase in mice. **p < 0.01 indicate differences between different types of treatment of Lactobacillus johnsonii GJ231, n = 6.
4 Discussion
Evaluating probiotic strains by combining phenotypic characterization and whole-genome sequencing analysis provides more comprehensive information about their potential biological properties than using one method of evaluation alone and is one of the most effective methods for exploring high-quality probiotic resources (Evanovich et al., 2019). In this study, a strain GJ231 from the healthy beagles was evaluated by a combination of whole genome sequencing and in vitro and in vivo experiments.
Whole-genome sequencing can analyze the complete genetic information of a strain at the gene level, providing a fast and accurate method for evaluating the function and safety of probiotics (Umanets et al., 2023). In this study, the whole-genome sequencing of strain GJ231 revealed that the strain had a genome size of 1.763 M with one chromosome and a G + C content of 34.70%, which was consistent with previous reports (Chen et al., 2023). Compared with Lactiplantibacillus plantarum and Lactobacillus rhamnosus, L. johnsonii has a smaller genome and lower G + C content, indicating a high degree of variation among different species of Lactobacillus to facilitate better adaptation to the environment (Jarocki et al., 2018). Understanding how carbohydrates are processed by the gut microbiota can help us investigate the influence of dietary carbohydrates on host health, as carbohydrate-active enzymes play a crucial role in host nutrient metabolism. The GJ231 genome contains 49 CAZymes: 26 glycoside hydrolase (GH, 53.06%) genes, 21 glycosyl transferase (GT, 42.86%) genes, one carbohydrate esterase (CE, 2.04%) gene, and one carbohydrate-binding module (CBM, 2.04%). It is worth noting that GJ231 contains numerous genes encoding GHs and GTs, which catalyze the transfer of sugars to specific receptors and play a crucial role in forming surface structures recognized by the host immune system. GHs catalyze the cleavage of glycosidic bonds, releasing abundant energy. Both GHs and GTs can help the host to resist the invasion and adhesion of potential pathogenic bacteria and their toxins to the intestinal epithelium (Mazmanian et al., 2008; Becerra et al., 2015). In addition, the presence of CRISPR-Cas was detected in the GJ231 genome, indicating that strain GJ231 may inhibit horizontal transfer of virulence or antibiotic resistance genes and has the capacity to resist foreign genetic elements (phage, plasmids, and insertion sequences) (Kim et al., 2021).
Transporter Classification Database (TCDB) is a database that categorizes membrane transport proteins (Saier et al., 2006). Transporter proteins are essential for bacterial life activities as they are involved in the uptake of nutrients and defense against endogenous and environmental stresses (Piepenbreier et al., 2017). The most prominent transporters in the GJ231 genome were Primary Active Transporters (164, 42.38%) followed by Electrochemical Potential-driven Transporters (90, 23.26%). Furthermore, the most PFAM-annotated proteins in strain GJ231 were ABC transporter proteins (65), which were involved in critical host transport processes and play critical roles in host defense and antibiotic resistance (Feng et al., 2020). The ABC transporter proteins in GJ231 may have similar functions. Moreover, when the GJ231 genome was annotated using the NR Database, the highest number of genes was annotated to L. johnsonii, consistent with the results of 16S rRNA gene sequencing.
The bacteriostatic effects of LAB have been widely reported (Adetoye et al., 2018; Mani-López et al., 2022). LAB can inhibit the growth of pathogenic bacteria through metabolites such as organic acids, extracellular polysaccharides, and bacteriocins. By decreasing intracellular pH and enabling the accumulation of ionized forms of organic acids in the cytoplasm, organic acids, especially lactic and acetic acids, exercise bactericidal action (van Zyl et al., 2020). The results of this study showed that a BS of GJ231 and the CFS were bacteriostatic against five pathogenic bacteria. We hypothesized that the organic acids were responsible for bacteriostatic activity of the CFS, as the bacteriostatic effect disappeared when the pH was changed. Therefore, we examined the organic acid content in the supernatant of strain GJ231 and the results showed a high content of acetic acid. It was noteworthy that the GJ231 genome also contained clusters of secondary metabolic genes, such as gassericin E and gassericin T, as well as gene encoding bacteriocins including gassericin, lactacin F, pediocin, and bacteriocin helveticin J. Chen et al. (2023) predicted that bacterioncin helveticin J was present in every bacteriocin-containing strain of L. johnsonii. Lactacin F, a two-component class II peptide bacteriocin, has been reported to reduce inflammation in patients with inflammatory bowel disease (Allison et al., 1994; Miri et al., 2022). Bacteriocin is one of the critical factors in the ability of probiotics to inhibit pathogens in the gastrointestinal tract (GIT). The antimicrobial activity of bacteriocins provides bacteriocin-producing probiotic strains with a competitive advantage in the complex GIT environment. These exciting phenomena will be studied more in depth in future research.
Probiotics are widely used in pets to prevent or combat diseases as they improve overall health. With the increasing variety of probiotic strains, the safety evaluation of probiotics has become a popular issue (Ishibashi and Yamazaki, 2001; Wang et al., 2023a). Hemolytic strains may cause sepsis in the host. Therefore, non- hemolytic bacteria are considered essential for exceptional probiotics. GJ231 was non- hemolytic on blood agar plates. In addition, the non- hemolytic enterotoxin (Nhe) and hemolysin BL (Hbl) genes were not identified in the GJ231 genome, which aligns with the strain’s lack of hemolytic activity. A potential risk for antibiotic resistance was the possibility of vertical or horizontal transfer. The GJ231 genome contains three antibiotic resistance genes: InuA, ErmT, and ErmB. Although there was a risk of gene transfer in GJ231, these genes are localized to genomic DNA rather than plasmid DNA, and therefore the risk was reduced. In addition, we annotated GJ231 with CRISPR-Cas, which prevents the horizontal transfer of virulence or antibiotic resistance genes, reducing the risk of gene transfer (Saidumohamed and Ganapathy Bhat, 2021). Analysis of potential metabolites in strain GJ231 did not reveal the presence of any toxic products.
After ingestion, bacteria are immediately exposed to the dual stresses of weak acids and bile salts. Therefore, acid and bile salt tolerance are essential criteria for selecting probiotics. Because the strong acid tolerance of LAB is not universal, strain-by-strain testing is required (Kailasapathy and Chin, 2000). Strain GJ231 exhibited both acid tolerance and bile salt tolerance. At weak acids, ATP synthase hydrolyses ATP to produce a transmembrane ionic gradient, which facilitates bacterial tolerance to strong acids (Deckers-Hebestreit and Altendorf, 1996). The GJ231 genome contains genes encoding ATP synthase and sodium antiporter proteins, as well as genes for L-lactate dehydrogenase, L-lactate permease, ATP-dependent Clp, and the phosphotransferase system. All of these components have mechanisms for providing resistance to weak acids and can work together to maintain pH homeostasis (Liang et al., 2023). Choloylglycine hydrolase (CBH), which hydrolyses the amide bond between glycine and bile acids, and inorganic pyrophosphatase, which affects the activity of bile acid coenzymes, were identified in the genome of L. johnsonii GJ231. Their presence implied a potential mechanism for the bile salt tolerance of strain GJ231 (Da et al., 1987; Lakshmi Ragavan and Das, 2021). The GJ231 genome also contained a variety of genes related to heat and stress tolerance, which echoed the results of the in vitro test. The heat-inducible transcriptional repressor HrcA and chaperone proteins (HslO, DnaJ, DnaJ, HtpX, HSP20, and HSP70) can help maintain the structure of proteins by preventing misfolding (Gladysheva et al., 2022; Chen et al., 2024). In addition, HSP had the ability to modulate the immune response (Huang et al., 2019). Under hyperosmotic conditions, the synergistic action of chaperone proteins (dnaK and dnaJ) encoded by the GJ231 genome contributes to repairing damage to the cellular macromolecular machinery (Schröder et al., 1993). In vitro assays revealed the excellent antioxidant capacity of strain GJ231. GJ231 also contained various oxidative stress-related enzymes scavenging of superoxide, peroxide, and hydroxyl radicals. These enzymes play a vital role in the antioxidant system and are closely related to the strain’s resistance to oxidative stress (Ezraty et al., 2017).
In vivo experiments are the primary method used to evaluate the safety of probiotics (Fan et al., 2023). A previous study reported that after administering L. delbrueckii subsp. lactis CIDCA to mice, no differences in body weight, feed intake, or organ indices were found in the group with CIDCA (de Jesus et al., 2022). The liver is an important organ responsible for metabolism and detoxification (Cui et al., 2023). In this study, the oral administration of GJ231 did not have a significant effect on body weight, food intake, or liver function (AST and ALT) in mice. This demonstrating that oral administration of GJ231 had no toxic effect on mice. However, we found the oral administration of GJ231 an increased thymic index in mice. The development of the thymus as a major lymphoid organ is vital for adaptive immunity (Kernfeld et al., 2018; Chen et al., 2019). SOD is the first line of defense against oxidative damage and plays a crucial role in oxidative stress and T-AOC is an oxidative stress marker (Zhao et al., 2021; Zhou et al., 2023). MDA is a byproduct of lipid peroxidation and has been utilized as a biomarker to indicate oxidative stress in organisms. In this study, the levels of serum T-AOC and SOD were significantly increased, while the levels of serum MDA was notably decreased in mice after administering GJ231. It suggested that the oral administration of GJ231 improved the mice’s ability to handle oxidative stress.
Probiotics have been reported to promote health and well-being in healthy dogs, e.g., as an adjunct product for the treatment of atopic dermatitis in dogs (Marsella, 2009; Kim et al., 2015) and to reduce the incidence of diarrhea (Nixon et al., 2019; Shmalberg et al., 2019). However, some studies have shown that supplemental feeding of E. faecium NCIB 10415 (isolated from the feces of healthy dogs) had no positive effect on the health status of dogs (Vahjen and Männer, 2003). Therefore, more studies are needed to discover the negative and positive effects of probiotics (canine or non-canine origin) on healthy and sick dogs to recycle probiotic products in dogs. Therefore, a strain with good probiotic potential (Lactobacillus johnsonii GJ231) was obtained in this study; larger studies are needed to explore the beneficial significance of the strain Lactobacillus johnsonii GJ231 at a broader level for prescription pet food and functional pet food development and/or improvement of canine intestinal health.
5 Conclusion
We isolated L. johnsonii GJ231 from the intestinal tract of healthy beagles. In vitro experiments revealed that GJ231 was tolerant of acid, bile salts, and heat. It was also capable of inhibiting the growth of various pathogens, had excellent free-radical scavenging activity, exhibited high levels of adherence, and possessed genes with the potential to biosynthesize a variety of bacteriocins. In conclusion, L. johnsonii GJ231 is a promising probiotic candidate for functional pet foods.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by the Laboratory Animal Ethics Committee of Qingdao Agricultural University (Approved No. DWKJ2023121901 Qingdao, China). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MZ: Data curation, Methodology, Visualization, Writing – original draft. YL: Data curation, Methodology, Visualization, Writing – original draft. YZ: Data curation, Methodology, Visualization, Writing – original draft. GL: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The manuscript was supported by the High-level Talents Fund of Qingdao Agricultural University (1121021) to GL.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1437036/full#supplementary-material
References
Adetoye, A., Pinloche, E., Adeniyi, B. A., and Ayeni, F. A. (2018). Characterization and anti-salmonella activities of lactic acid bacteria isolated from cattle faeces. BMC Microbiol. 18:96. doi: 10.1186/s12866-018-1248-y
Allison, G. E., Fremaux, C., and Klaenhammer, T. R. (1994). Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J. Bacteriol. 176, 2235–2241. doi: 10.1128/jb.176.8.2235-2241.1994
Becerra, J. E., Yebra, M. J., and Monedero, V. (2015). An l-Fucose operon in the probiotic Lactobacillus rhamnosus GG is involved in adaptation to gastrointestinal conditions. Appl. Environ. Microbiol. 81, 3880–3888. doi: 10.1128/AEM.00260-15
Buhnik-Rosenblau, K., Matsko-Efimov, V., Jung, M., Shin, H., Danin-Poleg, Y., and Kashi, Y. (2012). Indication for co-evolution of Lactobacillus johnsonii with its hosts. BMC Microbiol. 12:149. doi: 10.1186/1471-2180-12-149
Chen, D., Miao, Z., Peng, M., Xing, H., Zhang, H., and Teng, X. (2019). The co-expression of circRNA and mRNA in the thymuses of chickens exposed to ammonia. Ecotoxicol. Environ. Saf. 176, 146–152. doi: 10.1016/j.ecoenv.2019.03.076
Chen, D., Shen, F., Liu, J., Tang, H., Teng, X., Yang, F., et al. (2024). Luteolin enhanced antioxidant capability and induced pyroptosis through NF-κB/NLRP3/Caspase-1 in splenic lymphocytes exposure to ammonia. Sci. Total Environ. 919:170699. doi: 10.1016/j.scitotenv.2024.170699
Chen, K., Zhou, X., Zhao, J., Ross, R. P., Stanton, C., Chen, W., et al. (2023). Comparative genomics of Lactobacillus johnsonii reveals extensive intraspecific genetic variation. Food Biosci. 56:103190. doi: 10.1016/j.fbio.2023.103190
Cheng, M., Kong, R., Zhang, R., Wang, X., Wang, J., and Chen, M. (2021). Effect of glyoxal concentration on the properties of corn starch/poly(vinyl alcohol)/carvacrol nanoemulsion active films. Ind. Crop. Prod. 171:113864. doi: 10.1016/j.indcrop.2021.113864
Cui, J., Hao, Z., Zhou, Q., Qiu, M., Liu, Y., Liu, Y., et al. (2023). Chlorpyrifos induced autophagy and mitophagy in common carp livers through AMPK pathway activated by energy metabolism disorder. Ecotoxicol. Environ. Saf. 258:114983. doi: 10.1016/j.ecoenv.2023.114983
Da, V., Am, B., and Es, K. (1987). Bile acid: CoASH ligases from guinea pig and porcine liver microsomes. Purification and characterization. J. Biol. Chem. 262:5360-5. Available at: https://pubmed.ncbi.nlm.nih.gov/3558396/.
de Jesus, L. C. L., de Jesus Sousa, T., Coelho-Rocha, N. D., Profeta, R., Barroso, F. A. L., Drumond, M. M., et al. (2022). Safety evaluation of Lactobacillus delbrueckii subsp. lactis CIDCA 133: a health-promoting Bacteria. Probiot. Antimicrob. Proteins 14, 816–829. doi: 10.1007/s12602-021-09826-z
Deckers-Hebestreit, G., and Altendorf, K. (1996). The F0F1-type ATP synthases of bacteria: structure and function of the F0 complex. Ann. Rev. Microbiol. 50, 791–824. doi: 10.1146/annurev.micro.50.1.791
El-Razik, K. A. A., Ibrahim, E. S., Arafa, A. A., Hedia, R. H., Younes, A. M., and Hasanain, M. H. (2023). Molecular characterization of tetracycline and vancomycin-resistant Enterococcus faecium isolates from healthy dogs in Egypt: a public health threat. BMC Genomics 24:610. doi: 10.1186/s12864-023-09708-4
Evanovich, E., de, P. J., and Guerreiro, J. F. (2019). Comparative genomic analysis of Lactobacillus plantarum: an overview. Int. J. Genom. 2019:e4973214. doi: 10.1155/2019/4973214
Ezraty, B., Gennaris, A., Barras, F., and Collet, J.-F. (2017). Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 15, 385–396. doi: 10.1038/nrmicro.2017.26
Fan, X., Jiang, X., Guo, Y., Zhang, T., Zeng, X., Wu, Z., et al. (2023). In vitro and in vivo evaluation of the safety of Levilactobacillus brevis CGMCC1.5954 with probiotic potential based on tri-generation whole genome sequencing and animal studies. Food Biosci. 53:102654. doi: 10.1016/j.fbio.2023.102654
Feng, Z., Liu, D., Wang, L., Wang, Y., Zang, Z., Liu, Z., et al. (2020). A putative efflux transporter of the ABC family, YbhFSR, in Escherichia coli functions in tetracycline efflux and Na+(Li+)/H+ transport. Front. Microbiol. 11:556. doi: 10.3389/fmicb.2020.00556
Garcia-Gutierrez, E., Mayer, M. J., Cotter, P. D., and Narbad, A. (2019). Gut microbiota as a source of novel antimicrobials. Gut Microbes 10, 1–21. doi: 10.1080/19490976.2018.1455790
Gilliland, S. E., and Walker, D. K. (1990). Factors to consider when selecting a culture of Lactobacillus acidophilus as a dietary adjunct to produce a Hypocholesterolemic effect in humans. J. Dairy Sci. 73, 905–911. doi: 10.3168/jds.S0022-0302(90)78747-4
Gladysheva, I. V., Cherkasov, S. V., Khlopko, Y. A., and Plotnikov, A. O. (2022). Genome characterization and probiotic potential of Corynebacterium amycolatum human vaginal isolates. Microorganisms 10:249. doi: 10.3390/microorganisms10020249
Grześkowiak, Ł., Endo, A., Beasley, S., and Salminen, S. (2015). Microbiota and probiotics in canine and feline welfare. Anaerobe 34, 14–23. doi: 10.1016/j.anaerobe.2015.04.002
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document. The international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Huang, H., Jiao, X., Xu, Y., Han, Q., Jiao, W., Liu, Y., et al. (2019). Dietary selenium supplementation alleviates immune toxicity in the hearts of chickens with lead-added drinking water. Avian Pathol. 48, 230–237. doi: 10.1080/03079457.2019.1572102
Ishibashi, N., and Yamazaki, S. (2001). Probiotics and safety. Am. J. Clin. Nutr. 73, 465S–470S. doi: 10.1093/ajcn/73.2.465s
Jarocki, P., Podleśny, M., Krawczyk, M., Glibowska, A., Pawelec, J., Komoń-Janczara, E., et al. (2018). Complete genome sequence of Lactobacillus rhamnosus pen, a probiotic component of a medicine used in prevention of antibiotic-associated diarrhoea in children. Gut Pathogens 10:5. doi: 10.1186/s13099-018-0235-z
Jia, D.-J.-C., Wang, Q.-W., Hu, Y.-Y., He, J.-M., Ge, Q.-W., Qi, Y.-D., et al. (2022). Lactobacillus johnsonii alleviates colitis by TLR1/2-STAT3 mediated CD206+ macrophagesIL-10 activation. Gut Microbes 14:2145843. doi: 10.1080/19490976.2022.2145843
Johnson, A., Miller, E. A., Weber, B., Figueroa, C. F., Aguayo, J. M., Johny, A. K., et al. (2023). Evidence of host specificity in Lactobacillus johnsonii genomes and its influence on probiotic potential in poultry. Poult. Sci. 102:102858. doi: 10.1016/j.psj.2023.102858
Kailasapathy, K., and Chin, J. (2000). Survival and therapeutic potential of probiotic organisms with reference to Lactobacillus acidophilus and Bifidobacterium spp. Immunol. Cell Biol. 78, 80–88. doi: 10.1046/j.1440-1711.2000.00886.x
Kernfeld, E. M., Genga, R. M., Neherin, K., Magaletta, M. E., Xu, P., and Maehr, R. (2018). A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity 48, 1258–1270.e6. doi: 10.1016/j.immuni.2018.04.015
Kim, H. S., Jeong, S. G., Ham, J. S., Chae, H. S., Lee, J. M., and Ahn, C. N. (2006). Antioxidative and probiotic properties of Lactobacillus gasseri NLRI-312 isolated from Korean infant feces. Asian Australas. J. Anim. Sci. 19, 1335–1341. doi: 10.5713/ajas.2006.1335
Kim, H., Rather, I. A., Kim, H., Kim, S., Kim, T., Jang, J., et al. (2015). A double-blind, placebo controlled-trial of a probiotic strain Lactobacillus sakei Probio-65 for the prevention of canine atopic dermatitis. J. Microbiol. Biotechnol. 25, 1966–1969. doi: 10.4014/jmb.1506.06065
Kim, E., Yang, S.-M., Kim, D., and Kim, H.-Y. (2021). Complete genome sequencing and comparative genomics of three potential probiotic strains, Lacticaseibacillus casei FBL6, Lacticaseibacillus chiayiensis FBL7, and Lacticaseibacillus zeae FBL8. Front. Microbiol. 12:794315. doi: 10.3389/fmicb.2021.794315
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
La Fata, G., Weber, P., and Mohajeri, M. H. (2018). Probiotics and the gut immune system: indirect regulation. Probiot. Antimicrob. Proteins 10, 11–21. doi: 10.1007/s12602-017-9322-6
Lakshmi Ragavan, M., and Das, N. (2021). Effective probiotic delivery: current trends and future. Perspectives 497–517. doi: 10.1016/B978-0-12-822909-5.00029-0
Liang, X., Dai, N., Yang, F., Zhu, H., Zhang, G., and Wang, Y. (2023). Molecular identification and safety assessment of the potential probiotic strain Bacillus paralicheniformis HMPM220325 isolated from artisanal fruit dairy products. Food Funct. 15, 747–765. doi: 10.1039/D3FO04625G
Ma, F., Xu, S., Tang, Z., Li, Z., and Zhang, L. (2021). Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosafety Health 3, 32–38. doi: 10.1016/j.bsheal.2020.09.004
Mani-López, E., Arrioja-Bretón, D., and López-Malo, A. (2022). The impacts of antimicrobial and antifungal activity of cell-free supernatants from lactic acid bacteria in vitro and foods. Compr. Rev. Food Sci. Food Saf. 21, 604–641. doi: 10.1111/1541-4337.12872
Marsella, R. (2009). Evaluation of Lactobacillus rhamnosus strain GG for the prevention of atopic dermatitis in dogs. Am. J. Vet. Res. 70, 735–740. doi: 10.2460/ajvr.70.6.735
Mazmanian, S. K., Round, J. L., and Kasper, D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453, 620–625. doi: 10.1038/nature07008
Miri, S. T., Sotoodehnejadnematalahi, F., Amiri, M. M., Pourshafie, M. R., and Rohani, M. (2022). Comparison of the prevalence of bacteriocin encoding genes in Lactobacillus spp. isolated from fecal samples of healthy volunteers. IBD-patient and IBD-recovered. Iran J. Microbiol. 14, 219–226. doi: 10.18502/ijm.v14i2.9191
Nataraj, B. H., Kumari, M., Nagpal, R., Ali, S. A., and Behare, P. V. (2023). Safety evaluation of indigenous probiotic Limosilactobacillus fermentum NCDC 400 using whole genome sequences and in vitro approaches. Food Biosci. 56:103101. doi: 10.1016/j.fbio.2023.103101
Nixon, S. L., Rose, L., and Muller, A. T. (2019). Efficacy of an orally administered anti-diarrheal probiotic paste (pro-Kolin advanced) in dogs with acute diarrhea: a randomized, placebo-controlled, double-blinded clinical study. J. Vet. Intern. Med. 33, 1286–1294. doi: 10.1111/jvim.15481
Piepenbreier, H., Fritz, G., and Gebhard, S. (2017). Transporters as information processors in bacterial signalling pathways. Mol. Microbiol. 104, 1–15. doi: 10.1111/mmi.13633
Redfern, A., Suchodolski, J., and Jergens, A. (2017). Role of the gastrointestinal microbiota in small animal health and disease. Vet. Rec. 181:370. doi: 10.1136/vr.103826
Saidumohamed, B. E., and Ganapathy Bhat, S. (2021). Indian oil sardine (Sardinella longiceps) gut derived Bacillus safensis SDG14 with enhanced probiotic competence for food and feed applications. Food Res. Int. 150:110475. doi: 10.1016/j.foodres.2021.110475
Saier, M. H., Tran, C. V., and Barabote, R. D. (2006). TCDB: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 34, D181–D186. doi: 10.1093/nar/gkj001
Sanders, M., Merenstein, D., Reid, G., Gibson, G., and Rastall, R. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Scarborough, R., Hardefeldt, L., Browning, G., and Bailey, K. (2021). Pet owners and antibiotics: knowledge, opinions, expectations, and communication preferences. Antibiotics 10:1326. doi: 10.3390/antibiotics10111326
Schröder, H., Langer, T., Hartl, F. U., and Bukau, B. (1993). DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. EMBO J. 12, 4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shao, Y., Wang, Y., Yuan, Y., and Xie, Y. (2021). A systematic review on antibiotics misuse in livestock and aquaculture and regulation implications in China. Sci. Total Environ. 798:149205. doi: 10.1016/j.scitotenv.2021.149205
Shmalberg, J., Montalbano, C., Morelli, G., and Buckley, G. J. (2019). A randomized double blinded placebo-controlled clinical trial of a probiotic or metronidazole for acute canine diarrhea. Front. Vet. Sci. 6:163. doi: 10.3389/fvets.2019.00163
Tyagi, A., Sharma, C., Srivastava, A., Naveen Kumar, B. T., Pathak, D., and Rai, S. (2022). Isolation, characterization and complete genome sequencing of fish pathogenic Aeromonas veronii from diseased Labeo rohita. Aquaculture 553:738085. doi: 10.1016/j.aquaculture.2022.738085
Umanets, A., Surono, I. S., and Venema, K. (2023). I am better than I look: genome based safety assessment of the probiotic Lactiplantibacillus plantarum IS-10506. BMC Genomics 24:518. doi: 10.1186/s12864-023-09495-y
Vahjen, W., and Männer, K. (2003). The effect of a probiotic Enterococcus faecium product in diets of healthy dogs on bacteriological counts of Salmonella spp., Campylobacter spp. and Clostridium spp. in faeces. Arch. Tierernahr. 57, 229–233. doi: 10.1080/0003942031000136657
van Zyl, W. F., Deane, S. M., and Dicks, L. M. T. (2020). Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 12:1831339. doi: 10.1080/19490976.2020.1831339
Wan, M. L. Y., Forsythe, S. J., and El-Nezami, H. (2019). Probiotics interaction with foodborne pathogens: a potential alternative to antibiotics and future challenges. Crit. Rev. Food Sci. Nutr. 59, 3320–3333. doi: 10.1080/10408398.2018.1490885
Wang, Q., Li, W., Liu, H., Tan, B., Dong, X., Chi, S., et al. (2023a). The isolation, identification, whole-genome sequencing of Clostridium butyricum LV1 and its effects on growth performance, immune response, and disease-resistance of Litopenaeus vannamei. Microbiol. Res. 272:127384. doi: 10.1016/j.micres.2023.127384
Wang, Y., Li, A., Zhang, H., Zhang, L., and Li, J. (2018). Probiotic potential of Leuconostoc pseudomesenteroides and Lactobacillus strains isolated from yaks. Front. Microbiol. 9:2987. doi: 10.3389/fmicb.2018.02987
Wang, H., Qiu, Y., Chen, S., Zhao, Y., Wu, Y., and Wang, Y. (2024). Whole-genome sequencing of a protease-producing strain isolated from fermented golden pompano and molecular docking of the protease to fish proteins. Food Biosci. 57:103598. doi: 10.1016/j.fbio.2024.103598
Wang, Z., Su, J., Ali, A., Gao, Z., Zhang, R., Li, Y., et al. (2023c). Microbially induced calcium precipitation driven by denitrification: performance, metabolites, and molecular mechanisms. J. Environ. Manag. 338:117826. doi: 10.1016/j.jenvman.2023.117826
Wang, Y., Yang, H., Mu, G., and Wu, X. (2023b). Safety evaluation and complete genome analysis emphasis on extracellular polysaccharide of two strains of Limosilactobacillus fermentum MWLf-4 and Lactipiantibacillus plantarum MWLp-12 from human milk. Food Biosci. 51:102356. doi: 10.1016/j.fbio.2023.102356
Yoon, H., Lee, Y., Park, H., Kang, H.-J., Ji, Y., and Holzapfel, W. H. (2023). Lactobacillus johnsonii BFE6154 ameliorates diet-induced hypercholesterolemia. Probiot. Antimicrob. Proteins 15, 451–459. doi: 10.1007/s12602-021-09859-4
Zhao, M., Zhang, Y., Li, Y., Liu, K., Zhang, C., and Li, G. (2023). Complete genome sequence and probiotic properties of Pediococcus acidilactici CLP03 isolated from healthy Felis catus. Probiot. Antimicrob. Proteins. doi: 10.1007/s12602-023-10187-y
Zhao, H., Zhang, R., Yan, X., and Fan, K. (2021). Superoxide dismutase nanozymes: an emerging star for anti-oxidation. J. Mater. Chem. B 9, 6939–6957. doi: 10.1039/d1tb00720c
Keywords: Lactobacillus johnsonii, beagle, whole genome sequencing, probiotic, functional food
Citation: Zhao M, Li Y, Zhang Y and Li G (2024) Genomic analysis and functional properties of Lactobacillus johnsonii GJ231 isolated from healthy beagles. Front. Microbiol. 15:1437036. doi: 10.3389/fmicb.2024.1437036
Edited by:
Baichuan Deng, South China Agricultural University, ChinaReviewed by:
Yun Ji, China Agricultural University, ChinaXiaohua Teng, Northeast Agricultural University, China
Copyright © 2024 Zhao, Li, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guangyu Li, dGNzbGd5QDEyNi5jb20=
†These authors have contributed equally to this work
 Mengdi Zhao
Mengdi Zhao Yueyao Li1†
Yueyao Li1† Guangyu Li
Guangyu Li