
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol., 18 July 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1434987
Mycotoxins are secondary metabolites produced during the growth, storage, and transportation of crops contaminated by fungi and are physiologically toxic to humans and animals. Aflatoxin, zearalenone, deoxynivalenol, ochratoxin, patulin, and fumonisin are the most common mycotoxins and can cause liver and nervous system damage, immune system suppression, and produce carcinogenic effects in humans and animals that have consumed contaminated food. Physical, chemical, and biological methods are generally used to detoxify mycotoxins. Although physical methods, such as heat treatment, irradiation, and adsorption, are fast and simple, they have associated problems including incomplete detoxification, limited applicability, and cause changes in food characteristics (e.g., nutritive value, organoleptic properties, and palatability). Chemical detoxification methods, such as ammonification, ozonation, and peroxidation, pollute the environment and produce food safety risks. In contrast, bioenzymatic methods are advantageous as they achieve selective detoxification and are environmentally friendly and reusable; thus, these methods are the most promising options for the detoxification of mycotoxins. This paper reviews recent research progress on common mycotoxins and the enzymatic principles and mechanisms for their detoxification, analyzes the toxicity of the degradation products and describes the challenges faced by researchers in carrying out enzymatic detoxification. In addition, the application of enzymatic detoxification in food and feed is discussed and future directions for the development of enzymatic detoxification methods are proposed for future in-depth study of enzymatic detoxification methods.
Mycotoxins are secondary metabolites produced by certain fungi that infect crops (grains, nuts, and fruits) and pose a threat to humans and animals (Xu et al., 2022). Humans are exposed to mycotoxin either by directly consuming mycotoxin-present crops or indirectly by ingesting through the consumption of livestock or poultry fed contaminated crops (Abraham et al., 2022a). Short-term consumption of highly mycotoxin-contaminated food/feed can be fatal to humans and animals, while prolonged exposure to threshold doses can result in liver and nervous system damage, immunosuppression and chronic diseases with long-term carcinogenic effects (Yu and Pedroso, 2023). At present, the detoxification of mycotoxins is commonly achieved by physical, chemical, and biological methods. Physical detoxification methods, such as thermal treatment, irradiation, and adsorption, are fast and simple, but may result in incomplete detoxification; however, their applicability is limited due to potential impacts on food characteristics (e.g., nutritive value, organoleptic properties, and palatability) (Haidukowski et al., 2019; Murugesan et al., 2021; Raesi et al., 2022; Zhang et al., 2022; Jing et al., 2023; Romero-Sánchez et al., 2024). Chemical detoxification methods, such as ozonation, peroxidation, and ammoniation achieve high detoxification efficiencies but are associated with environmental degradation, the generation of hazardous by-products, the use of potentially toxic reagents, and significant costs (Afsah-Hejri et al., 2020; Shen and Singh, 2022a,b; Demirci et al., 2023). Newly developed physicochemical detoxification methods, such as cold plasma technology (Gavahian and Khaneghah, 2020), pulsed light sterilization technology (Wang L. et al., 2022; Li et al., 2023b), and the application of novel nanomaterials (Lin et al., 2022), also affect food characteristics and are costly. Enzymatic detoxification involves the use of biological enzymes to reduce or remove mycotoxins. Compared with traditional physical and chemical detoxification methods, enzymatic detoxification is performed under mild conditions without nutrient loss or secondary contamination, while being highly efficient, versatile, highly specific, and low in toxicity.
To date, more than 400 mycotoxins have been identified that belong to different categories and originate from different fungi, the most common of which are aflatoxin, zearalenone deoxynivalenol, ochratoxin, patulin, and fumonisin. Mycotoxins not only contaminate food production systems, causing enormous losses to the global economy, but are also passed on to humans through the food chain, posing a major threat to human health. The directional flow direction of mycotoxin contaminants in food is shown in Figure 1. Complete elimination becomes a challenge once mycotoxins are present. The most effective approach to preventing mycotoxin contamination is to intervene before fungal infections occur and before plants produce mycotoxins (Jouany, 2007).
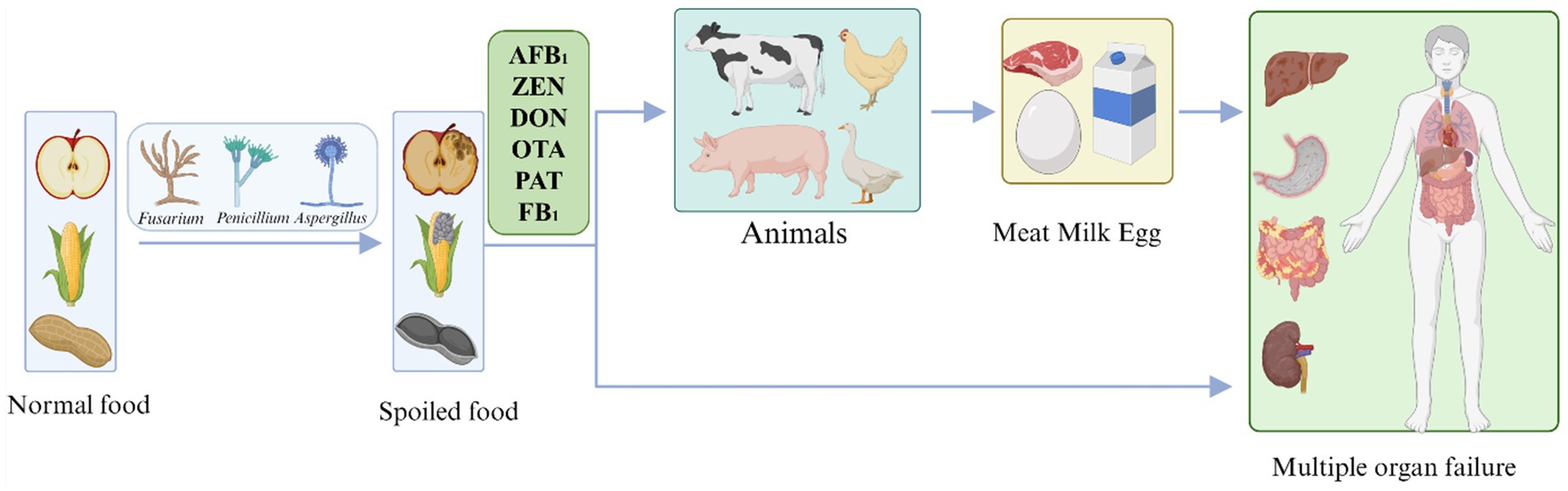
Figure 1. The flow direction of mycotoxins in foods. AFB1, Aflatoxin B1; ZEN, Zearalenone; DON, Deoxynivalenol; OTA, Ochratoxin; PAT, Patulin; and FB1, Fumonisin B1.
Cultivate healthy seeds of disease- and pest-resistant varieties that can resist fungal diseases and prevent fungal invasion. Use crop rotation, appropriate irrigation and fertilization, fungicides and disinfection are used during the growing season to reduce the population of toxigenic fungi. Timely harvesting of mature crops, prompt drying to control moisture content, and storage and transport in a dry environment. Protecting the external structure of seeds and grains minimizes the risk of fungal contamination. High-risk components of mycotoxins can be removed by removing diseased plants, discarding damaged grains, and thoroughly washing grain soil (Jouany, 2007; Ogunade et al., 2018; Matumba et al., 2021; Nada et al., 2022). Governments can use publicity and education to raise people’s awareness of the toxic hazards of mycotoxins and to encourage good hygiene habits in their daily lives. Countries should also implement stricter guidelines for mycotoxin levels in cereals and develop relevant legislation policies to minimize the threat of mycotoxins to humans.
To our knowledge, existing reviews do not comprehensively evaluate the types of mycotoxin detoxification enzymes or their degradation mechanisms. And only a few reviews have been published to date to systematize them (Ben Taheur et al., 2019; Lyagin and Efremenko, 2019; Venkatesh and Keller, 2019; Abdi et al., 2021; Abraham et al., 2022a; Wang Y. et al., 2022; Sun H. et al., 2023). This review provides a detailed summary of the principles and mechanisms underlying the bioenzymatic detoxification of mycotoxins that are prevalent in major crops, analyzes the toxicity of the degradation products, and discusses their potential application in the detoxification of mycotoxins in food and feed. Through this review, it is hoped that the understanding of mycotoxin detoxification enzymes will be more thorough and will provide a theoretical and practical basis for future research and applications related to the biological detoxification of mycotoxins in the food and feed industries.
Aflatoxin (AF), a derivative of difurocoumarin, is produced by Aspergillus fungi and is typically found in corn, peanuts, rice, nuts, and other crops infected with Aspergillus flavus; these infections result from long-term storage, poor storage conditions, and poor sanitation during processing (Aristil et al., 2020). Among the various types of aflatoxins, the most common are aflatoxin B1 (AFB1), aflatoxin B2 (AFB2), aflatoxin G1 (AFG1), and aflatoxin G2 (AFG2) (Figure 2A; Du et al., 2023).
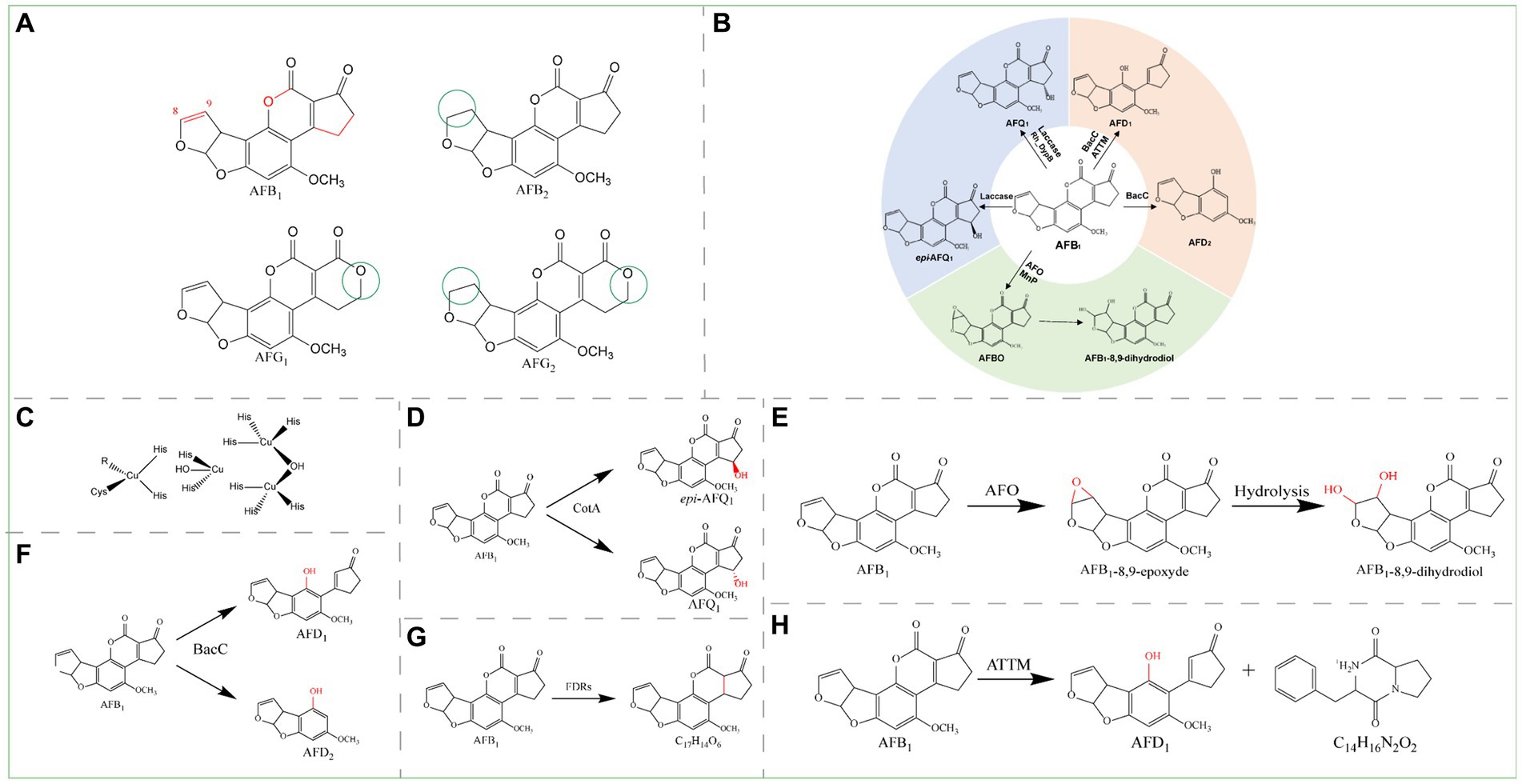
Figure 2. (A) Structural formula of AFB1 and its derivatives. The red group is the detoxifying enzyme action group and the green group shows the difference between AFB1 derivatives and them. AFB1, Aflatoxin B1; AFB2, Aflatoxin B2; AFG1, Aflatoxin G1; AFG2, Aflatoxin G2. (B) Main degradation and transformation products of AFB1. (C) Structure of the active center of laccase. (D) Detoxification of AFB1 by CotA laccase. (E) Degradation of AFB1 by AFO. (F) Degradation of AFB1 by BacC. (G) Degradation of AFB1 by FDRs. (H) Degradation of AFB1 by ATTM.
Aflatoxin B1 is the most common, most toxic, and most thoroughly researched class of mycotoxins and was classified by the World Health Organization as a group 1 carcinogen in 1993 (Janik et al., 2020). Liver cancer is likely to occur after long-term exposure to AFB1 (Do et al., 2020), and 28% of liver cancer cases worldwide are caused by AFB1 (Liu and Wu, 2010; Liu et al., 2012). For food safety reasons, more than 100 countries and regions have established detailed limit standards for different types of food. For example, the European Union (EU) has set a limit of 2 μg/kg AFB1 for all cereals and cereal products (Commission Regulation (EU), 2010). The mechanism by which AFB1 causes liver cancer involves the oxidation of AFB1 in the liver by cytochrome P450 3A4 (CYP3A4) in microsomes; this process generates the reactive metabolite AFB1-8,9-epoxide (AFBO), which covalently binds to nucleic acids and leads to liver cancer. A study in poultry revealed that the toxin is metabolized mainly by cytochrome P450 1A2 (CYP1A2) and P450 3A4 (CYP3A4) and causes numerous mutations, particularly in the p53 tumor suppressor gene (Ülger et al., 2020). The main toxic groups of AFB1 include the C8-C9 double bond on the difuran ring, the lactone ring in the coumarin structure, and the pentanone ring (Figure 2A). There are three main pathways by which toxic groups can be targeted to degrade AFB1: the first pathway involves the C8-C9 double bond on the difuran ring in the toxin structure, the second pathway involves lactone ring modification or ring opening, and the third pathway involves hydroxylation and reduction of the pentenone ring. The main degradation and transformation products of AFB1 include aflatoxin Q1 (AFQ1), epi-aflatoxin Q1 (epi-AFQ1), aflatoxin D1 (AFD1), aflatoxin D2 (AFD2), and AFBO. Due to the instability of AFBO, AFB1-8,9-dihydrodiol is eventually generated (Figure 2B). AFB1 toxicity can be reduced by the epoxidation of C8-C9, hydroxylation of C3, or epoxidation and modification of the lactone ring. Oxidoreductases, lactonucleases, and other degrading enzymes can catalyze the conversion of these groups, thereby achieving enzymatic detoxification.
Oxidoreductases, such as laccases, oxidases, reductases, and peroxidases, degrade AFB1 by different mechanisms. Degradation of AFB1 by laccases involves hydroxylation of the pentanone ring, aflatoxin oxidase involves oxidation of the furan ring, reductases involve reduction of the double bond of the coumarin lactone ring and the α, β-unsaturated ester bond between the lactone ring and the pentanone ring, and peroxidases involve hydroxylation of the pentanone ring and oxidation of the furan ring by mechanisms similar to those of laccase and aflatoxin oxidase, respectively.
Laccases are a very important class of oxidoreductases that can degrade AFB1. As an extracellular multicopper oxidase, laccase can catalyze the oxidation of aromatic amines, phenols, carboxylic acids, and other heterocyclic compounds to less toxic or nontoxic compounds; the structure of its active center is shown in Figure 2C. Laccase can oxidize AFB1 indirectly through a mediator or directly (Okwara et al., 2021); in practical applications, direct oxidation is a more feasible route. In indirect oxidation, the mediator is first oxidized by laccase, and then the oxidized mediator can oxidize the substrate. Through this system, laccases can overcome the limitation of redox potential and expand the range of substrates. However, copper clusters in laccases directly oxidize the substrate through free radicals generated during direct oxidation.
With acetosyringone (AS) as the mediator, Ery4 laccase can completely remove 0.1 μg/mL AFB1 in vitro after 1 h and likely converts the compounds into AFQ1, epi-AFQ1, AFB1-8,9-dihydrodiol or AFB1-dialdehyde, AFB2a, and AFM1 (Loi et al., 2023). With methyl syringate (MS) as a mediator, the BsCotA laccase from Bacillus subtilis oxidizes AFB1 to AFQ1; in this process, the toxicity of the product is significantly reduced, and the conversion efficiency depends on the dosages of laccase and the mediator (Wang et al., 2024). In another experiment, CotA laccase from Bacillus licheniformis ANSB821 was expressed in E. coli, AFB1 was directly converted to AFQ1 and epi-AFQ1 effectively in the absence of redox mediators (Figure 2D), and the degradation rate reached 96% within 12 h at pH 8.0 and 37°C (Guo et al., 2020). Compared with CotA laccase, the Q441A mutant exhibited better pH stability, better thermal stability and higher degradation efficiency (Liu et al., 2023a). The recombinant laccase Lac3 from Trametes sp. C30 broke the double bond of the furan ring and lactone ring in the coumarin moiety and degraded AFB1 into C16H22O4, C14H16N2O2, C7H12N6O, and C24H30O6 with a degradation rate of 91%; in addition, the toxicity of the degradation products was significantly reduced. Studies in mouse HepG2 cells have shown that Lac3-degraded AFB1 reduces hepatocyte apoptosis, histopathological lesions in the liver and kidney, oxidative stress, and inflammation (Liu et al., 2021). Recombinant fmb-rL103 laccase (2.5 mM) from Bacillus vallismortis can degrade AFB1 (10 ng/mL) at 37°C and pH 7.0 with a degradation rate greater than 50% (Bian et al., 2022). However, the degradation mechanism remains uncertain.
Aflatoxin oxidase (AFO) was the first enzyme discovered that could degrade AFB1 (Cao et al., 2011). AFO has no obvious similarity to any known oxidase, and its protein structure suggests that it is a new aflatoxin oxidase that is different from reported oxidases such as laccase; it is therefore listed separately. AFO is an intracellular enzyme that acts on the AFB1 difuran ring and oxidizes it to AFBO. Due to the instability of AFBO, AFBO is further hydrolyzed to AFB1-8,9-dihydrodiol (Figure 2E) (Wu et al., 2015).
The reductase BacC from Bacillus subtilis UTB1, which catalyzes the degradation of AFB1, may catalyze the reduction of the double bond of the lactone ring within the coumarin moiety. The ester bond then undergoes hydrolysis to yield carboxylic acid, and AFD1 is produced through decarboxylation. Alternatively, after hydrolysis, the derivative AFD2 can be produced through cleavage of the bond with the ciclopeptone ring (Figure 2F). BacC contributes to the reduction of aflatoxin levels in pistachio fruits: treatment of pistachio nuts with B. subtilis UTB1 incubated at 30°C for 7 days reduced aflatoxin levels by 69.1% (Afsharmanesh et al., 2018).
F420H2-Dependent reductases A and B (FDR-A and FDR-B) from Mycobacterium smegmatis were previously found to catalyze the reduction of α, β-unsaturated ester bonds between the lactone and pentanone rings (Figure 2G) (Taylor et al., 2010). Subsequently, it was found that MSMEG_5998 from the FDR family of Mycobacterium smegmae exhibits greater degradation activity toward AFB1 in the pH range of 7.0–10.0, and compared with native enzymes, the thioredoxin Trx-linked MSMEG_5998 recombinant enzyme exhibits greater degradation activity in vitro. In a HepG2 cell culture model, Trx-linked MSMEG_5998 can reduce AFB1-induced cytotoxicity in HepG2 cells by ameliorating DNA damage and p53-mediated apoptosis (Li et al., 2019).
Heterologous expression with the manganese peroxidase PhcMnp in Kluyveromyces lactis generated a product that exhibited a good ability to degrade AFB1. The degradation rate reached 75.71% at 40°C and pH 4.5 for 40 h, and the degradation product was identified as AFB1-8,9-dihydrodiol. Peanut samples contaminated with AFB1 were treated, and the recombinant strain degraded more than 90% of the AFB1 in the peanut samples (Xia et al., 2022).
The recombinant type B dye decolorizing peroxidase (Rh_DypB) can degrade AFB1 to AFQ1 and numerous low molecular mass compounds in vitro. After reacting with 0.1 U/mL Rh_DypB and 0.1 mM H2O2 for 96 h, the degradation rate of AFB1 reached 96% (Loi et al., 2020). The dye decolorizing peroxidase type B (DypB) can effectively degrade AFB1 under the action of Mn2+ (Mangini et al., 2024).
Ery4 laccase cannot degrade AFB1 without a redox mediator; in contrast, MnP requires H2O2, and Dyp requires H2O2 and Mn2+ as cofactors to degrade AFB1. However, due to the potential toxicity and high cost of the redox mediators, H2O2 and Mn2+, these mechanisms are not practical for food and feed applications. Therefore, laccases without redox mediators, such as CotA laccase, Lac3 laccase, and fmb-103 laccase, and oxidoreductases without cofactors are advantageous for the detoxification of mycotoxins in food and feed.
Lactonases are a family of enzymes that degrade AFB1 by destroying the AFB1 lactone ring, and opening the lactone ring can effectively reduce the toxicity of AFB1. Certain Bacillus species possess the aiiA gene, which encodes N-acyl-homoserine lactone (AHL) lactonase; this lactonase is responsible for AFB1 degradation and cleaves the lactone ring by targeting the lactone bond. This enzymatic hydrolysis mechanism is based on an AHL with a structure similar to that of AFB1, which also possesses a lactone ring. In a toxicity test with Artemia salina, these Bacillus strains were able to significantly reduce the toxicity of AFB1 after incubation at 30°C for 48 h (González Pereyra et al., 2019). The highest rate of AFB1 degradation by ATTM lactonase from Bacillus megaterium HNGDA6 was 86.78% at 80°C and pH 8.5, and the degradation products were AFD1 (C16H14O5) and C14H16N2O2. The former product results from the decarboxylation of the lactone ring-opening form of AFB1, whereas the other three toxicity sites (the C8-C9 double bond of the difuran ring, the lactone ring, and the pentanone ring) are disrupted (Figure 2H) (Cheng et al., 2023).
Pantoea aflatoxin degradation enzyme (PADE1) is an AFB1-degrading enzyme from Pantoea sp. T6 that was identified as an outer membrane protein A and showed the highest degradation rate of AFB1 (48.5%) at 40°C and pH 7.0 (Xie et al., 2019). However, the Pseudomonas AFB1-degrading enzyme (PADE2) from Pseudomonas aeruginosa M19 showed the greatest AFB1 degrading activity at 65°C and pH 6.0. Cu2+ and Fe3+ strongly enhanced the activity, whereas Ca2+ and Zn2+ strongly inhibited the activity (Song et al., 2019). The genetically engineered Trametes versicolor AFB1-degrading enzyme (TV-AFB1D) converted 67.4% of AFB1 to other compounds within 5 h at 32°C and pH 7.0. Recombinant TV-AFB1D engineered Saccharomyces cerevisiae strains effectively degrade AFB1 in contaminated rice (Yang P. et al., 2022).
The above three types of enzymes (oxidoreductases, lactonases, and degrading enzymes) can degrade AFB1, but only a few studies have investigated the degradation and transformation mechanism of enzymes and identified the structure of degradation products; some intermediate degradation products and the catalytic mechanisms of enzymes remain unknown. More detoxification enzymes and degradation products should be identified to clarify the biological detoxification mechanisms of these enzymes. The detoxifying enzymes of AFB1 are summarized in Table 1.
Zearalenone (ZEN) is a toxic compound with a resorcylic acid lactone structure produced by Fusarium fungi (Figure 3A). This toxin is present in moldy grains, such as barley, corn, and wheat, and typically results from improper postharvest storage or processing (Bouajila et al., 2022). ZEN is a non-steroidal estrogen mycotoxin that interferes with the endocrine system and exerts toxic effects on the reproductive system of animals. ZEN can cause adverse conditions such as non-infectious abortion, an abnormal estrous cycle, and abnormal embryo development (Yang et al., 2020; Yadav et al., 2021). ZEN has five structural derivatives: α/β-zearalenol (α/β-ZOL), α/β-zearalanol (α/β-ZAL), and zearalanone (ZAN) (Figure 3A). ZEN and its derivatives possess chemical structures similar to those of the animal endogenous estrogen β-estradiol; thus ZEN and its derivatives exhibit toxicity through interactions with the receptors of β-estradiol in animal cells, and compared with ZEN, α-ZAL, and α-ZOL exhibit greater cytotoxicity (Jouany, 2007).
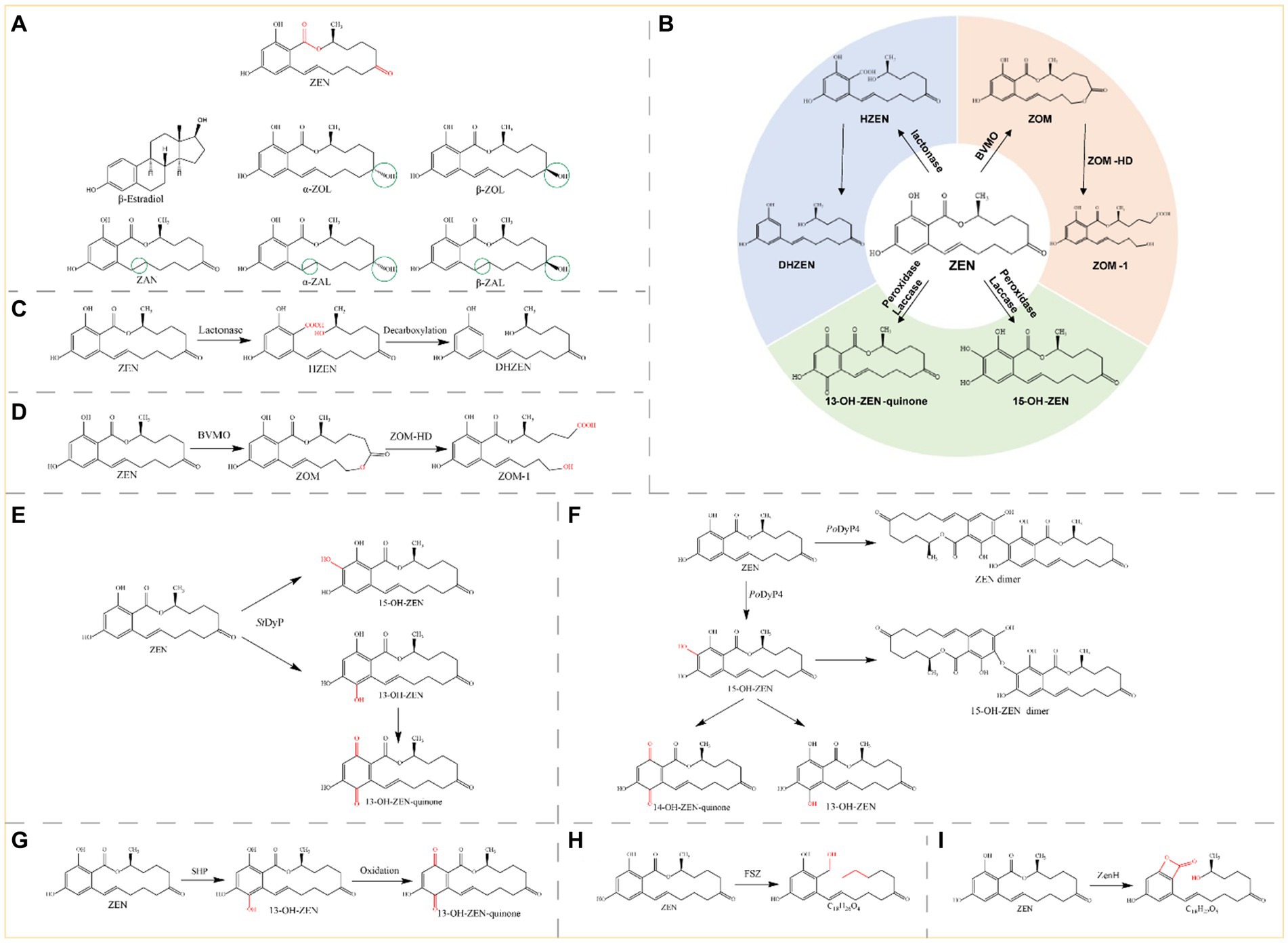
Figure 3. (A) Structural formula of ZEN and its derivatives. The red group is the detoxifying enzyme action group and the green group shows the difference between ZEN derivatives and them. ZEN, Zearalenone; β-estradiol; α-ZOL, α-Zearalenol; β-ZOL, β-Zearalenol; ZAN, Zearalanone; α-ZAL, α-Zearalanol; β-ZAL, β-Zearalanol. (B) Main degradation and transformation products of ZEN. (C) Degradation of ZEN by lactonase. (D) Degradation by monooxygenase and carboxylester hydrolase. (E) Degradation of ZEN by StDyP. (F) Degradation of ZEN by PoDyP4. (G) Degradation of ZEN by SHP. (H) Degradation of ZEN by FSZ. (I) Degradation of ZEN by ZenH.
Therefore, although ZEN is the most abundant toxin in moldy grains, the toxicity of both the ZEN molecule and its derivatives should be emphasized. The main toxic group of ZEN is the lactone ring (Figure 3A), and the known pathways of ZEN biodegradation involve hydrolysis of the lactone ring, oxidation of the ketone group (or hydrolysis followed by the addition of oxygen), and hydroxylation of the aromatic groups. The main degradation products of ZEN include DHZEN, ZOM-1, 15-OH-ZEN, and 13-OH-ZEN-quinone (Figure 3B). Lactonases can hydrolyze the ester bond on the lactone ring of zearalenone, and the toxicity of the hydrolyzed product is significantly reduced or even eliminated. Monooxygenases and carboxylester hydrolases destroy the C6 ketone group of ZEN, and the metabolite ZOM-1 loses its estrogenic activity. Peroxidases and laccases can hydroxylate the aromatic ring on ZEN, the resulting products are transformed into 13-OH-ZEN-quinone and 15-OH-ZEN, respectively, and the estrogenic biotoxicity of both products is significantly reduced. Lactonases, oxidoreductases and other novel enzymes detoxify ZEN by different mechanisms, generating different products.
Zearalenone lactone hydrolase (ZHD) degrades ZEN by disrupting the ZEN lactone structure, thereby achieving ZEN detoxification. Lactonase first hydrolyses ZEN to hydrolyzed ZEN (HZEN), which is further decarboxylated to form decarboxylated HZEN (DHZEN) (Figure 3C).
Takahashi-Ando et al. (2002) cloned and expressed a lactonohydrolase from Clonostachys rosea IFO 7063 in Escherichia coli and were the first to report the mechanism by which the lactonohydrolase ZHD101 promotes ZEN degradation and detoxification. The ZEN hydrolase RmZHD from Rhinocladiella mackenziei degraded ZEN through proton transfer and nucleophilic-substituted ring opening to form a hydroxyl product (Zhou et al., 2020). The amino acid sequences of ZLHY-6 from Clonostachys rosea 31535 and ZHD101 exhibited high identity (98.3%). The optimum temperature and pH ranges for ZLHY-6 to ZEN were 25–41°C and 6.5–10, respectively. By optimizing ZLHY-6, irreversible degradation of ZEN was achieved, with a degradation rate of up to 96.31%. The ZEN in crude oil was reduced from 1257.3 to 13 μg/kg after detoxification with ZLHY-6 during crude oil refining (Chang et al., 2020). PR-ZHD from Clonostachys rosea GRZ7 was successfully heterologously expressed in Penicillium canescens strain PCA-10, and ZEN was completely removed from the model solutions after 3 h at pH 8.5 and 30°C. The addition of PR-ZHD (8 mg/g of dried grain) to flour samples prepared from the infected ZEN contaminated grain (approximately 16 μg/g) completely decontaminated the samples after an overnight exposure (Shcherbakova et al., 2020). The BAMF_RS30125 gene from Bacillus amyloliquefaciens H6 was heterologously expressed in E. coli, and the recombinant purified protein ZTE138 was obtained. The degradation rates of ZEN were 34.20 and 59.79% at 36 and 72 h, respectively (Xu et al., 2020). ZHD607 from Phialophora americana, a mesophilic lactone hydrolase, was heterologously expressed in Pichia pastoris and showed optimum activity at 35°C and pH 8.0. Two ZHD607 mutants, ZHDM1 and I160Y, showed 2.9- and 3.4-fold greater ZEN degradation, respectively, than did ZHD607 (Yu et al., 2020). ZENG from Gliocladium roseum MA918 was identified as a highly efficient degrading enzyme. The recombinant ZENG showed maximum activity at pH 7.0 and 38°C (> 70% degradation at ZEN concentrations of 10–80 μg/mL). This is the first report of a neutral ZEN-degrading lactonase that can degrade ZEN and its derivatives (α-ZOL and α-ZAL) (Zhang et al., 2020). In addition, the half-life (t1/2) of the thermostable ZENG mutant S162P/S220R was 36.8-fold greater than that of the wild-type enzyme at 55°C, and the melting temperature (Tm) was 8.2°C higher (Fang et al., 2022). ZenA showed high degradation activity for ZEN, converting ZEN to HZEN within the first 10 min of the reaction, and HZEN was partially decarboxylated to DHZEN during the 2 h of the reaction. The metabolism of ZEN in the reticulorumen of dairy cows was studied and the concentration of ZEN in the reticulorumen was significantly reduced by ZenA administration (Gruber-Dorninger et al., 2021). ZHD-P from Trichoderma aggressivum exhibited 97% amino acid sequence identity with ZHD101, and the specific activity of the heterologously expressed and purified ZHD-P was 1.55-fold greater than that of ZHD101. The optimum temperature and pH for ZEN degradation were 45°C and 7.5–9.0, respectively, and the degradation activity of ZHD-P increased with the addition of Ca2+, K+, Mg2+, Mn2+, and Na+ (Chen S. et al., 2021). ZHD_LD from Exophiala spinifera exhibited 60.15% amino acid identity with ZHD101, and the purified recombinant ZHD_LD showed high ZEN degradation activity under optimum reaction conditions (50°C and pH 9.0) (Gao et al., 2022). The novel lactonase Zhd11B from Fonsecaea monophora is a neutral lactone hydrolase with a broad substrate spectrum that can efficiently hydrolyze ZEN and its more toxic derivatives (α-ZAL and β-ZAL). Through cap domain swapping, the activities of ZEN, α-ZAL and β-ZAL were further improved by 1.5-, 1.6-, and 2.9-fold, respectively (Jiang et al., 2022). Zhd11D from Phialophora attinorum, which has the highest degradation rate of ZEN at pH 8.0 and 35°C, increased enzyme activity by 1.5-fold and thermostability by 2-fold (40°C) by fusing a segment of the amphiphilic short peptide S1 at the N-terminus of Zhd11D. In addition, S1-Zhd11D showed a higher rate of hydrolysis of ZEN than Zhd11D in peanut oil (Wang Z. et al., 2023).
Most zearalenone-degrading lactonases are temperature sensitive and can only maintain high activity within a narrow temperature range. Therefore, resolving the structure of lactonases and improving their temperature stability represent future research directions.
Monooxygenases, peroxidases, and laccases are oxidoreductases that degrade ZEN. Baeyer-Villiger monooxygenases (BVMOs) and carboxylester hydrolases (ZOM-HDs) from Apiotrichum mycotoxinivorans function together to achieve detoxification (Mascotti et al., 2015). First, a BVMO enzyme catalyzes the insertion of an oxygen atom between C5 and C6, thus transforming the cyclic ketone to a zearalenone oxidized metabolite (ZOM). Next, ZOM-HD is recruited to hydrolyze ZOM, breaking the ester bond at C6 to produce a non-estrogen compound (ZOM-1) (Figure 3D) (Sun et al., 2020).
The StDyP gene from Streptomyces thermocarboxydus was expressed in E. coli, and StDyP was able to slightly degrade ZEN; the process could be accelerated in the presence of Mn2+ and 1-hydroxybenzotriazole, and the degradation products were 15-OH-ZEN and 13-OH-ZEN. However, 13-OH-ZEN is unstable and autooxidizes to 13-OH-ZEN-quinone (Figure 3E) (Qin et al., 2021b).
The spore CotA laccase from Bacillus licheniformis was heterologously expressed in E. coli and could directly oxidize ZEN; moreover, the addition of the redox mediators 2,2′-azino-bis-(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) and AS promoted the degradation of ZEN by the CotA laccase. Proliferation assays in human breast cancer MCF-7 cells showed that CotA laccase and CotA laccase-mediator systems eliminated the estrogenicity of ZEN. Additionally, the immobilization of CotA laccase onto chitosan microspheres resulted in greater thermal stability than that of free CotA laccase. The rates of ZEN removal in corn meal by free and immobilized CotA laccase were 70 and 90%, respectively (Guo et al., 2022).
PoDyP4, a dye-decolorizing peroxidase from Pleurotus ostreatus, was expressed in E. coli. It almost completely degraded 1 mM ZEN within 2 h at pH 6.0 and 40°C with good pH and temperature stability. ZEN can be detoxified by hydroxylation, oxidation, and polymerization reactions (Figure 3F) (Ding et al., 2024).
Soybean hull peroxidase (SHP) extracted from soybean hulls degraded 95% of the ZEN in 100 μM H2O2 buffer when added in a stepwise manner within 1 h. The degradation of ZEN by SHP in maize steep liquor, maize flour, wheat flour, and rice flour was 85, 60, 66, and 54%, respectively, (Figure 3G) (Guo et al., 2024).
Several novel ZEN-degrading enzymes have also been identified. The ZEN-degrading enzyme FSZ from Aspergillus niger ZEN-S-FS10 is a novel enzyme (with less than 10% homology to ZHD101 and RmZHD) that degrades ZEN at a rate of 75–80% (pH 7.0, 28°C) and produces C18H26O4 as the degradation product (Figure 3H). FSZ can function at 28–38°C and pH 2.0–7.0 and can degrade ZEN derivatives (α-ZAL, β-ZOL, and ZAN), and the degradation rates of ZEN and ZAN are greater than those of α-ZAL and β-ZOL. The degradation rate of FSZ was 78.43% after incubation with corn flour containing 1.0 μg/mL ZEN for 28 h, which showed a good degradation effect (Ji et al., 2022).
ZenH is a novel enzyme from Aeromicrobium sp. HA that can degrade ZEN and exhibits the highest similarity (21.52% homology) to the lactonase ZHD607 from Phallophora americana. ZenH degrades ZEN into C18H22O5 (Figure 3I), which is an isomer of ZEN. Treatment of ZEN-contaminated corn meal with ZenH resulted in ZEN degradation rates ranging from 75.7 to 85.3% (Hu et al., 2023).
In conclusion, studies on the biodegradation and transformation mechanism of ZEN were carried out relatively later than studies on AFB1, so the degradation and transformation mechanism of ZEN by detoxification enzymes has not been explored. The detoxifying enzymes of ZEN are summarized in Table 2.
Deoxynivalenol (DON), also known as vomitoxin, is a class of sesquiterpenoid metabolites produced by Fusarium fungi and is among the typical trichothecene mycotoxins (Figure 4A).
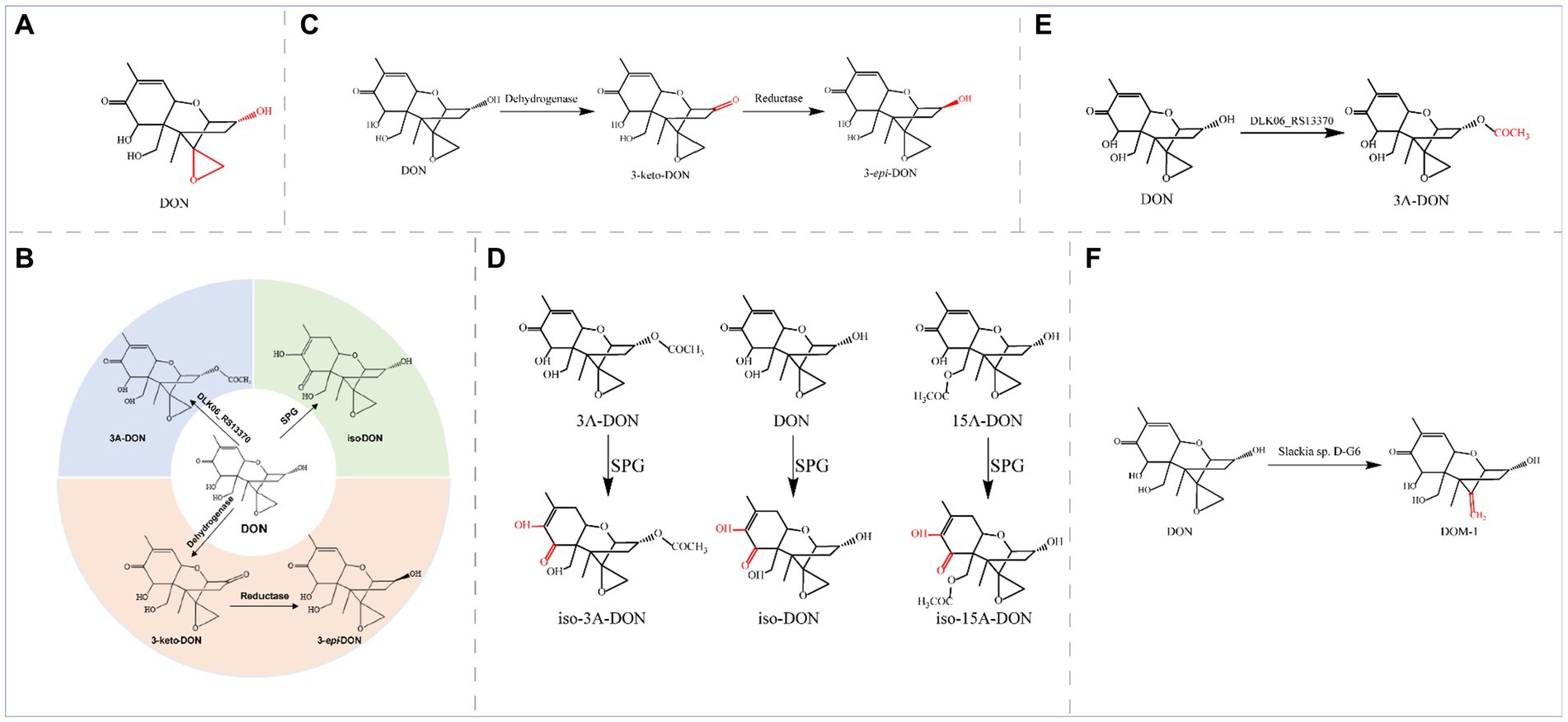
Figure 4. (A) Structural formula of DON and its corresponding detoxification enzyme action groups. The red group is the detoxifying enzyme action group. (B) Main degradation and transformation products of DON. (C) Degradation of DON by dehydrogenase and reductase. (D) Degradation of DON and its derivatives by SPG. (E) Degradation of DON by the enzyme encoded by DLK06_RS13370. (F) Degradation of DON by D-G6.
This toxin is mainly found in grains, such as wheat, corn, and barley, that are contaminated with Fusarium and potentially jeopardize the health of both humans and animals (Mishra et al., 2020; Tu et al., 2023). High concentrations of deoxynivalenol can cause decreased appetite, digestive system problems, and immune system damage (Gao Y. et al., 2020; Chen et al., 2023). In severe cases, deoxynivalenol can also result in teratogenicity, carcinogenicity, and immunotoxicity (Yao and Long, 2020). Different animals exhibit different sensitivities to DON, with pigs being particularly sensitive to DON (Holanda and Kim, 2021). The main toxic groups of DON are the C12-13 epoxy ring and the OH group at C3 (Figure 4A), and the degradation and transformation of these two groups are crucial for biological detoxification. The enzymatic elimination of DON can oxidize the OH group at C3 in DON to generate the corresponding ketone group, 3-keto-DON. Alternatively, isomerization of the C3 atom can generate the epimer of DON (3-epi-DON) or transform a low-toxicity 3-keto-DON intermediate to 3-epi-DON, which is almost nontoxic; iso-DON can also be generated through isomerization. These methods can significantly reduce the physiological toxicity of DON. The main degradation and transformation products of DON are as follows (Figure 4B): dehydrogenases oxidize DON to 3-keto-DON, and then reductases isomerize 3-keto-DON to 3-epi-DON; glyoxalase isomerizes DON to iso-DON; and acyltransferases convert DON to 3-acetyl-deoxynivalenol (3-ADON).
Oxidoreductases coordinate detoxification, and dehydrogenases and reductases catalyze the conversion of DON through a two-step sequential reaction. In this reaction, the dehydrogenases first oxidize DON to 3-keto-DON and then the reductases reduce 3-keto-DON to the final product, 3-epi-DON, to epimerize the C3 atom (Figure 4C).
The OH group is oxidized and isomerized at C3 by two DON detoxification enzymes from Devosia mutans 17-2-E-8, a pyrroloquinoline quinone (PQQ)-dependent dehydrogenase (DepA) and an NADPH-dependent aldo-keto reductase (DepB) (He et al., 2016; Carere et al., 2018a). Specifically, DepA oxidizes DON to 3-keto-DON (Yang H. et al., 2022), DepB reduces 3-keto-DON to 3-epi-DON (Carere et al., 2018b), and the intermediate product 3-keto-DON is an order of magnitude less toxic than DON; in contrast, the final product (3-epi-DON) is virtually nontoxic. Two highly active DON detoxifying enzymes are also present in Devosia strain D6-9 and catalyze sequential reactions; specifically, the quinone-dependent DON dehydrogenase QDDH oxidizes DON to 3-keto-DON, and two NADPH-dependent aldo-keto reductases (AKR13B2 and AKR6D1) convert 3-keto-DON to 3-epi-DON (He et al., 2020). Deoxynivalenol dehydrogenase (DDH) from Pelagibacterium halotolerans ANSP101 shares 57.55% sequence identity with the dehydrogenase DepA from Devosia, which oxidizes DON to 3-keto-DON using phenazine methosulfate (PMS), dichlorophenolindophenol (DCPIP), or pyrroloquinoline quinone (PQQ) as the hydrogen acceptor. The TDDH mutant can degrade DON over a wide pH range (6.0–11.0) and temperature range (20–45°C) in the presence of the same hydrogen receptor and possesses a greater ability to degrade DON (Qin et al., 2022). DepBRleg from Rhizobium leguminosarum, a member of the new aldo-keto reductase family AKR18, is also an NADPH-dependent DON detoxification enzyme. DepBRleg converts 3-keto-DON to 3-epi-DON and DON in diastereomeric ratios of 67.2 and 32.8%, respectively (Abraham et al., 2022b). Sorbose dehydrogenase (SDH) from Ketogulonicigenium vulgare Y25 can effectively convert DON to 3-keto-DON, a substance in which DON loses four hydrogen atoms; the SDH mutants F103L and F103A were more effective, with 5- and 23-fold increases in Vmax, respectively (Li D. et al., 2023). SDH and its mutants exhibit excellent thermal stability and activity over wide pH and temperature ranges, with potential applications in detoxifying DON. YoDDH, a new DON-degrading enzyme from Youhaiella tibetensis, showed the highest degradation activity of DON at 40°C and pH 4.5 in the presence of Ca2+ and PQQ, and YoDDH ultimately degraded 73% of DON (100 μM) when incubated under optimum conditions for 3 h (Shi et al., 2024).
Transferases can catalyze the detoxification of DON by transferring specific groups. The specialized glyoxalase I from Gossypium hirsutum (SPG) exhibits the highest degradation activity at pH 9.0 and 55°C; in addition, SPG can reduce the toxicity of DON by isomerization to transfer the C8 carbonyl to C7 and double bonds from C9-C10 to C8-C9 to generate the less toxic iso-DON. Furthermore, SPG recognizes the DON derivatives 15A-DON and 3A-DON; generating iso-15A-DON and iso-3A-DON, respectively (Figure 4D). The SPGY62A mutant was more efficient at detoxifying DON, 3A-DON, and 15A-DON (the catalytic activity of the enzyme increased by more than 70%) (Huang et al., 2020; Hu et al., 2022).
Acinetobacter pittii S12, which was isolated and characterized from soil samples, also degraded DON with a high degradation rate of 78.32%. The DLK06_RS13370 gene, a pivotal gene for DON detoxification in the strain, was expressed in E. coli, and the resulting recombinant protein exhibited acyltransferase activity; this enzyme converted a hydroxyl group into an acetyl moiety, thereby converting DON to 3-ADON (Figure 4E) (Liu et al., 2023b).
Some strains can also de-epoxidize C12-C13, such as the novel DON detoxifying bacterium, Slackia sp. D-G6, isolated from chicken intestines, in which D-G6 de-epoxidizes DON into a nontoxic form called DOM-1 (Figure 4F) (Gao X. et al., 2020).
The detoxifying enzymes of DON are summarized in Table 3.
Ochratoxins are a group of isocoumarin derivative compounds produced mainly by Aspergillus and Penicillium fungi. There are three main groups of ochratoxins: ochratoxin A (OTA), ochratoxin B (OTB), and ochratoxin C (OTC) (Figure 5A). Among these toxins, OTA is the most common, ubiquitous, and virulent mycotoxin in the ochratoxin family (Mwabulili et al., 2023).
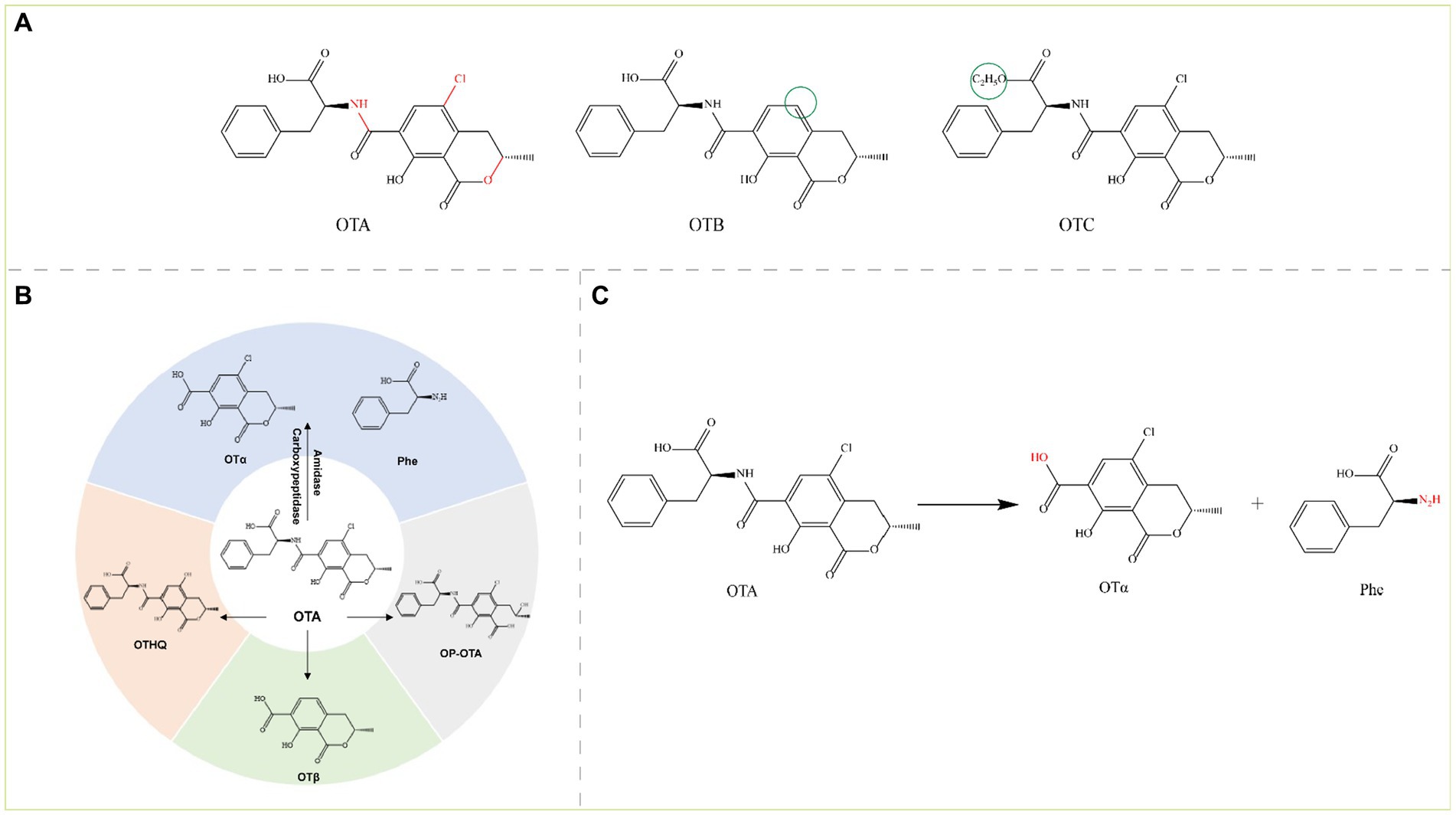
Figure 5. (A) Structural formula of OTA and its derivatives. The red group is the detoxifying enzyme action group and the green group shows the difference between OTA derivatives and them. OTA, Ochratoxin A; OTB, Ochratoxin B; and OTC, Ochratoxin C. (B) Main degradation transformation products of OTA. (C) Degradation of OTA by carboxypeptidase.
The ochratoxin skeleton consists of a dihydroisocoumarin group linked to phenylalanine by an amide bond, and OTA and OTC also contain a p-chlorophenol structure (Freire et al., 2020). OTA is commonly found in grains (wheat, maize, barley, oats, rye, rice, etc.) contaminated by Aspergillus and Penicillium. OTA is a potent nephrotoxin and is also hepatotoxic, carcinogenic, teratogenic, genotoxic, immunotoxic, and embryotoxic (Kumar et al., 2020; Gu et al., 2021). The main toxic groups of OTA are amide bonds, isocoumarin rings, lactone rings, and chlorine atoms, and the toxins are detoxified by several reactions, including hydrolysis of amide bonds, hydroxylation, ring opening of the lactone ring, and dichlorination. These reactions are accomplished mainly with enzymes such as amidases, peptidases (aminopeptidases and carboxypeptidases) and lipases. Currently, the production of ochratoxin α (OTα) and L-β-phenylalanine (Phe) by the hydrolysis of amide bonds is the most important mechanism for the degradation and detoxification of OTA. However, other pathways can lead to the detoxification of OTA such as dechlorination to OTB and further degradation to ochratoxin β (OTβ), hydroxylation of the isocoumarin ring to ochratoxin hydroquinone (OTHQ), and lactone ring-opening to lactone-opened OTA (OP-OTA) (OP-OTA is more toxic than OTA) (Figure 5B) (Wu et al., 2011).
Carboxypeptidase detoxifies the amide bond of OTA, which hydrolyzes OTA to the virtually nontoxic OTα and the nontoxic Phe (Figure 5C); OTα is almost a thousand times less toxic than OTA. Cleavage (hydrolysis) of OTA amide bonds by carboxypeptidases is the main pathway for OTA detoxification.
Pitout (1969) discovered that carboxypeptidase A (CPA) from bovine pancreas, which was the first enzyme found to degrade OTA, could degrade OTA. In a study on CPA in contaminated wheat flour, researchers showed that CPA from three different sources (animal, vegetable, and microbial) could degrade OTA, and the best results were observed with microbial sources (Kupski et al., 2018). However, the poor thermal stability of native CPA limits its application; therefore, to produce efficient catalytic CPAs with in vitro stability, researchers have mostly genetically engineered the CPA gene and performed heterologous expression. Compared with those of other OTA degradation enzymes, the catalytic mechanism and crystal structure of CPA are well established. Lu Xiong constructed a mature CPA (M-CPA) without a propeptide or signal peptide and expressed it in P. pastoris. M-CPA degraded OTA up to 93.36%, with an optimum pH of 8 and temperature of 40°C. M-CPA could effectively degrade OTA in red wine (Xiong et al., 2020). CPA can be used to detoxify OTA in contaminated foods and feeds, and it shows good potential for various applications. Carboxypeptidase Y (CPY), produced by S. cerevisiae, is an analogous enzyme to CPA and breaks down OTA to the less toxic OTα; however, the native CPY is also defective, with very low specific activity and a very slow OTA hydrolysis reaction (only 52% of the OTA was converted to OTα in 5 days) (Abrunhosa et al., 2010). In recent years, researchers have identified several other carboxypeptidases, such as the carboxypeptidase cp4 gene from Lysobacter sp. CW239, which is expressed in E. coli and can degrade OTA to produce OTα; however, the OTA degradation rate of rCP4 after 24 h was only 36.8% (Wei W. et al., 2020). Moreover, the carboxypeptidase DacA gene from Bacillus subtilis CW14 was expressed in E. coli, and recombinant DacA converted OTA to OTα with a degradation rate of 71.3% within 24 h at 30°C and pH 7.0 (Xu et al., 2021). The carboxypeptidase BnOTase2 from the Brevundimonas naejangsanensis ML17 strain could hydrolyze OTA to OTα and OTB to OTβ with a degradation rate of 100% (Peng et al., 2022, 2023). In addition, AfOTH from Alcaligenes faecalis subsp. phenolicus DSM 16503T exhibited both amidase and carboxypeptidase dual activity, with higher activity at pH 6.0–9.0 and 30–60°C (Sánchez-Arroyo et al., 2024).
Amidohydrolases are also a typical class of OTA-degrading enzymes that produce OTα by hydrolyzing amide bonds. The novel microbial ochratoxin amidase gene from Aspergillus niger was cloned and homologously expressed in Aspergillus niger, and the recombinant protein ochratoxinase (OTase) showed optimum activity at pH 6.0 and 66°C; compared with carboxypeptidase A and Y, OTase hydrolyzed OTA more efficiently (Dobritzsch et al., 2014). The novel degrading enzyme N-acyl-L-amino acid amidohydrolase (AfOTase) from A. faecalis DSM 16503 belongs to the peptidase family M20, and recombinant AfOTase (rAfOTase) efficiently degrades OTA to OTα at an optimum temperature and pH of 50°C and 6.5, respectively (Zhang et al., 2019). The amidohydrolase ADH3 from Stenotrophomonas acidaminiphila is the most potent OTA detoxification enzyme reported to date; ADH3 hydrolyzes OTA to OTα and Phe, and recombinant ADH3 (1.2 μg/mL) expressed in E. coli completely degrades 50 μg/L OTA in 90 s. However, several hours are needed for other OTA-degrading enzymes to fully degrade OTA (Luo et al., 2022). The catalytic activity of the rationally modified S88E mutant expressed in P. pastoris increased by 3.7-fold (Dai et al., 2023). The salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans DSM 6986T (PsSDO) is a versatile metalloenzyme with both dioxygenase and amide hydrolase catalytic activities; through its amidase activity, this enzyme can hydrolyze the amide bond of OTA to produce OTα and L-β-phenylalanine (Sánchez-Arroyo et al., 2023). Chr1-3858681-3267, a metallo-dependent amidohydrolase from the aerobic gram-negative bacterium Stenotrophomonas sp. 043-1a, also degrades OTA (Gonaus et al., 2023).
Lipases and Nudix family hydrolases can also detoxify OTA. The Nudix hydrolase Nh-9 gene from the Bacillus velezensis IS-6 strain was expressed in E. coli, the recombinant Nh-9 enzyme was purified to degrade 1.0 μg/mL OTA by 68% within 24 h at 37°C and pH 7.0, and OTA was detoxified to OTα; Fe2+ and Cu2+ enhanced the degradation of OTA (Jahan et al., 2023). Amano lipase A from Aspergillus niger (ANL) and porcine pancreas lipase (PPL) hydrolyzed OTA, but the enzymes exhibited different degrees of hydrolysis; ANL completely degraded OTA and OTB after 3 and 10 h, respectively, whereas PPL failed to completely degrade OTA (only 43% of OTA was degraded after 25 h) but completely degraded OTB within 9 h (Santos et al., 2023).
Among the above OTA degradation and transformation pathways, the degradation pathway with OTα as the end product is recognized as the most effective degradation and detoxification pathway. The reported detoxification enzymes for OTA are summarized in Table 4.
Patulin (PAT) is a toxic polyketide lactone compound produced primarily by Aspergillus, Penicillium, and Byssochlamys fungi (Figure 6A) (Mahato et al., 2021).
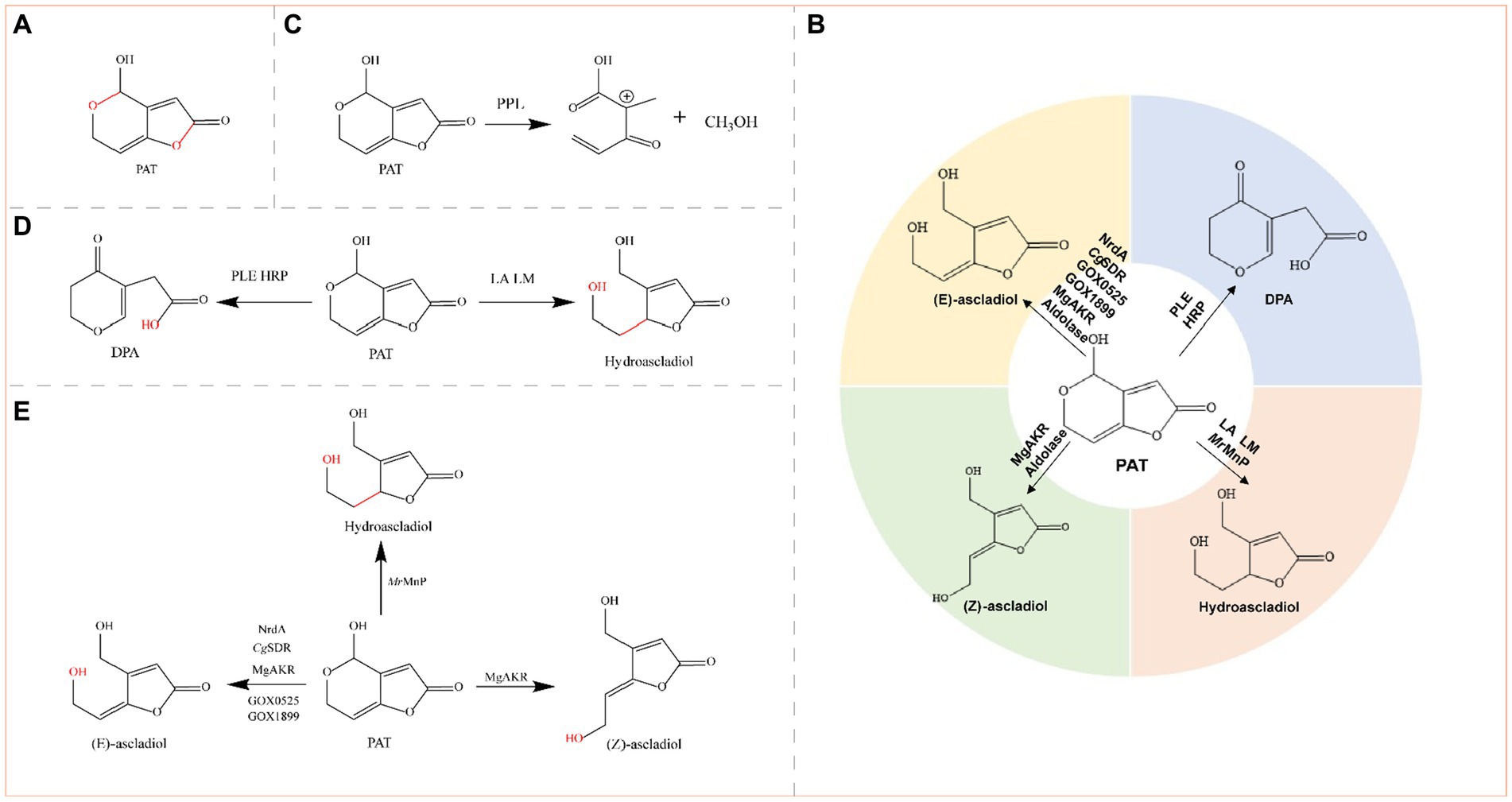
Figure 6. (A) Structural formula of PAT (patulin) and its corresponding detoxification enzyme action groups. The red group is the detoxifying enzyme action group. (B) Main degradation transformation products of PAT. (C) Degradation of PAT by PPL. (D) Possible degradation of PAT by PLE, HRP, LA, and LM. (E) Degradation of PAT by oxidoreductases.
Patulin is produced primarily in rotten fruits such as apples, pears, blueberries, cherries, peaches, plums, strawberries, and mulberries. Most human intake of PAT occurs via apples and apple products because rotten apples contain relatively high concentrations of PAT (Zheng et al., 2021). Animal experiments have shown that PAT affects a variety of organs, including the gut, liver, and kidneys (Wei C. et al., 2020). The main toxic groups of PAT are hemiacetal and lactone rings (Figure 6A). PAT can be reduced to E-ascladiol by short-chain dehydrogenases or the aldo-keto reductase family of enzymes, which can be converted to Z-ascladiol catalyzed by cellular sulfhydryl compounds, both of which are virtually nontoxic (Sekiguchi et al., 1983). E-ascladiol can also be reduced by manganese peroxidase to hydroascladiol, a much less toxic compound. Degradation of PAT to E-ascladiol appears to be more prevalent, but certain esterases and peroxidases convert PAT to deoxypatulinic acid (DPA), which is significantly less toxic than PAT (Tannous et al., 2017). Overall, the detoxification of PAT is achieved by the conversion of PAT to less toxic degradation products such as DPA, (E)-ascladiol, (Z)-ascladiol, and hydroascldiol by lipases, transferases, oxidoreductases, and aldolases (Figure 6B).
Lipase detoxifies PAT by destroying its lactone and hemiacetal rings. PPL was used to degrade PAT in pear juice at a dose of 0.02 g/mL, and was effective at 40°C within 24 h; the maximum degradation rate was 61.38% and no decrease in the quality or nutrition of the pear juice was observed (Figure 6C) (Xiao et al., 2019). The efficiency of the enzyme is greatly increased at lower PAT concentrations, so it is important to control PAT contamination before it spreads. The efficiency of the enzyme can also be improved if an immobilized PPL enzyme is applied (Yan et al., 2024).
The lipase RL12 from Ralstonia sp. strain SL312, 100 μg/mL RL12, degraded more than 80% of the PAT in apple juice within 24 h at an optimum pH of 7.5 and a temperature of 37°C. In addition, the nutritional and sensory qualities of apple juice are improved by the enzymatic processes (He et al., 2022).
Commercial enzymes are now available for the degradation of PAT in fruit juices. For example, esterase from porcine liver (PLE), peroxidase from horseradish (HRP), lipase A from Aspergillus niger (LA) and lipase M from Mucor javanicus (LM) degrade 97.8, 53.2, 76.3, and 68.2% of PAT in apple juice, respectively, and the effects of the four enzymes on the quality of apple juice are essentially within acceptable limits. Among these enzymes, PLE and HRP might degrade PAT by breaking its lactone ring, generating DPA as a degradation product; LA and LM might degrade PAT by destroying its hemiacetal structure, generating hydroascladiol as the degradation product (Figure 6D) (Liu X. et al., 2023).
The main transferase enzymes that exert detoxifying effects on PAT are phosphoribosyltransferase and methyltransferase, but their detoxification mechanisms and degradation products are unknown and should be further explored. The orotate phosphoribosyltransferase (OPRTase) from Rhodotorula mucilaginosa was reported to effectively degrade PAT in apple juice at 25°C. OPRTase (0.15 g/L) degraded 1 mg/L PAT, and the final PAT degradation rate was greater than 80% (1 mg/L) (Tang et al., 2019). PcCRG1 from Pichia caribbica, an S-adenosylmethionine-dependent methyltransferase, degraded PAT from 20 μg/mL to undetectable levels within 72 h. The PcCRG1 enzyme was also found to effectively degrade PAT in apple juice (Wang K. et al., 2019).
Oxidoreductases degrade PAT to either hydroascladiol or ascladiol (E-ascladiol or Z-ascladiol); manganese peroxidases degrade PAT to hydroascladiol and ribonucleoside diphosphate reductases, short-chain dehydrogenase/reductases (SDRs), and aldo-keto reductases degrade PAT to ascladiol (Figure 6E).
Ribonucleoside diphosphate reductase (NrdA) from Enterobacter cloacae subsp. TT-09 converts PAT to E-ascladiol (Xing et al., 2020). The short-chain dehydrogenase/reductase (SDR) gene CgSDR, which was cloned from Candida guilliermondii, was heterologously expressed in E. coli. The resulting recombinant CgSDR (150 μg/mL) with NADPH dependence converted 80% of the PAT in apple juice to E-ascladiol, and the biodegradation process did not affect the quality of the apple juice (Xing et al., 2021). After CgSDR was covalently linked to dopamine/polyethyleneimine-codeposited magnetic Fe3O4 particles for immobilization, the thermal and storage stabilities of the enzyme, proteolysis resistance, and reusability were greatly improved (Xing et al., 2023). The NADPH-dependent short-chain dehydrogenases GOX0525 and GOX1899 from Gluconobacter oxydans ATCC 621, both of which belong to the short-chain dehydrogenase/reductase (SDR) family, completely converted PAT to E-ascladiol within 24 h (Chan et al., 2022). MrMnP from Moniliophthora roreri was the first manganese peroxidase found to degrade PAT efficiently; the MrMnP gene was heterologously expressed in P. pastoris, and MrMnP produced the most rapid degradation of PAT in the malonate/Mn2+ system. At 0.5 U/mL, MrMnP completely removed 5 mg/L PAT within 5 h and eliminated up to 95% of the PAT in apple juice after 24 h (Wang S. et al., 2022). The novel NADPH-dependent PAT aldo-keto reductase MgAKR from Meyerozyma guilliermondii was expressed in E. coli, and MgAKR converted PAT to ascladiol. At an optimum reaction temperature of 16°C and pH 6.0, 88% of the PAT (5 μg/mL) in fresh pear juice was degraded by MgAKR (300 μg/mL) within 4 h, and the biodegradation process did not affect the quality of the fresh pear juice (Zhang et al., 2024).
Aldolase degrades PAT to form ascladiol, and aldolase from R. mucilaginosa degrades 2 mg/L PAT in apple juice at 0.7 mg/mL. The optimum pH and temperature were 7.0 and 25°C, respectively, but the enzyme was stable within the pH range of 5.5–7.0 and from 4 to 25°C. In addition, there was no significant difference in the quality parameters or volatile components of apple juice before and after degradation, and the purified aldolase cloned and recombinantly expressed in E. coli degraded more than 99% of the PAT (Yang et al., 2021; Li et al., 2022b).
Compared with its degradation by transferases, the degradation of PAT by lipases, oxidoreductases and aldolases has been more thoroughly studied; however, the degradation mechanism and products of PAT by transferases are unclear and should be further explored. Some of the above enzymes synergized product processing with PAT degradation and did not reduce the product quality during the mycotoxin degradation process, leading to an ideal state. The enzymes that detoxify PAT are summarized in Table 5.
Fumonisin is a water-soluble metabolite produced by Fusarium fungi, such as Fusarium verticillioides and Fusarium proliferatum (Rudyk et al., 2023). The fumonisins include fumonisin B1 (FB1), fumonisin B2 (FB2), fumonisin B3 (FB3), fumonisin B4 (FB4), fumonisin A1 (FA1), and fumonisin A2 (FA2), of which FB1 and FB2 are more toxic (Figure 7A); in contrast, several other toxins have very low concentrations of contaminants and are less toxic (Ben Taheur et al., 2019). FB1 is the most abundant and toxic variant of naturally contaminated maize, accounting for 70–80% of the total fumonisins. Recent studies have focused on the detoxification of FB1; other toxins are less frequently reported.
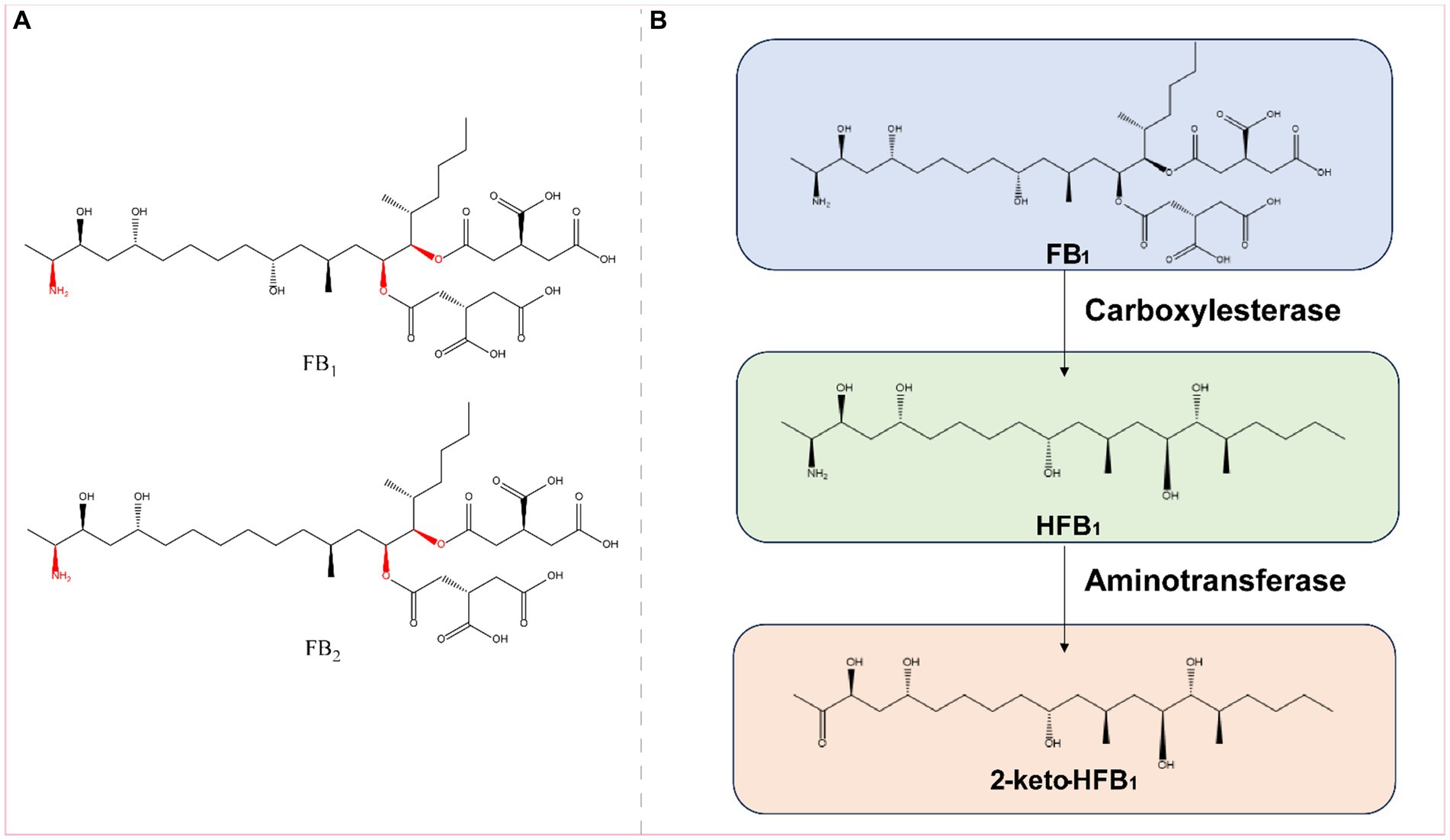
Figure 7. (A) Structural formula of FB1 (fumonisin B1) and its corresponding detoxification enzyme action groups. The red group is the detoxifying enzyme action group. (B) Main degradation transformation products of FB1.
Fumonisin B1 is found primarily on grains such as corn and rice that are contaminated with Fusarium. Exposure to FB1 can cause varying degrees of damage to the nervous, respiratory, digestive, and reproductive systems (Gao et al., 2023). FB1 is toxic to the liver, kidneys, and mammalian cells (Chen J. et al., 2021). The main toxic groups of FB1 are the C14- and C15-tricarboxylic acid groups and the C2-amino group (Figure 7A). FB1 can be detoxified by the synergistic action of carboxylesterases and aminotransferases, and this detoxification process is divided into two steps: in the first step, the C-14 and C-15 tricarboxylic acid groups are removed in the presence of carboxylesterases, converting FB1 to hydrolyzed FB1 (HFB1); in the second step, HFB1 is converted to 2-keto-HFB1 in the presence of aminotransferases (Figure 7B) (Blackwell et al., 1999; Heinl et al., 2010, 2011).
The carboxylesterases involved in FB1 detoxification include the fumonisin esterases FumD and FumDSB. Among these enzymes, FumD is a commercial enzyme used in the animal feed industry that effectively reduces FB1 in maize and maize products, and the detoxification of FB1 by FumD is specific and irreversible (Alberts et al., 2019, 2021). A novel carboxylesterase from the bacterium Sphingomonadales FumDSB was expressed in E. coli, and under ideal conditions, FumDSB degraded FB1 up to 100%. In addition, FumDSB showed high degradation activity, excellent pH stability, good thermal stability at 30–40°C and activity over a wide pH range (6.0–9.0); thus, FumDSB is well suited for use under physiological conditions in animals and can be developed as a promising food and feed additive. FumDSB can alleviate the inflammatory response induced by FB1 in growing pigs when it is added to the feed (Li et al., 2021; Liu et al., 2022).
After FB1 is hydrolyzed to HFB1 by carboxylesterases, it combines with aminotransferases and continues to remove C2-amino acids. The aminotransferase FumIS from Sphingopyxis sp. MTA144 and the aminotransferase FumIB from Bacterium ATCC 55552 efficiently removed C2-amino acids from HFB1. Feed enzymes can be used to degrade naturally antinutritive substances in agricultural products or to improve the nutritional value of feed. FumI may be suitable for future use as a feed enzyme (Hartinger et al., 2010, 2011). Yue Wang reported three fumonisin detoxifying aminotransferases, FumTSTA, FumUPTA, and FumPHTA, that share 61–74% sequence identity with FumIS and FumIB and exhibit good pH and thermal stabilities (Wang Y. et al., 2023).
Because the detoxification of FB1 requires the sequential action of carboxylesterases and aminotransferases, Kailin Li generated the fusion enzyme FUMDI by connecting the carboxylesterase gene (fumD) with the aminotransferase gene (fumI) through PCR. Researchers subsequently expressed the enzyme in P. pastoris, in which it almost completely degraded 5 μg/mL FB1 in 24 h. The experiments demonstrated that the fusion enzyme FUMDI and its degradation products were not toxic to human gastric epithelial cells (Li K. et al., 2022). The detoxification enzymes of FB1 are summarized in Table 6.
There are also several mycotoxins, such as sterigmatocystin, gliotoxin, and citrinin, which are difficult to analyze and are less common in nature; therefore, enzymatic degradation by these mycotoxins is less studied and reported at present.
Sterigmatocystin (STE) (Figure 8A), a polyketide mycotoxin produced mainly by fungi (such as Aspergillus versicolor and Aspergillus nidulans), is a carcinogenic, teratogenic, and mutagenic toxin that contaminates foods such as maize, peanuts, rice, and wheat. In Aspergillus strains, STE is a direct precursor (O-methylsterigmatocystin) of AFB1 and AFG1 and can be rapidly converted to both substances (Zingales et al., 2020).
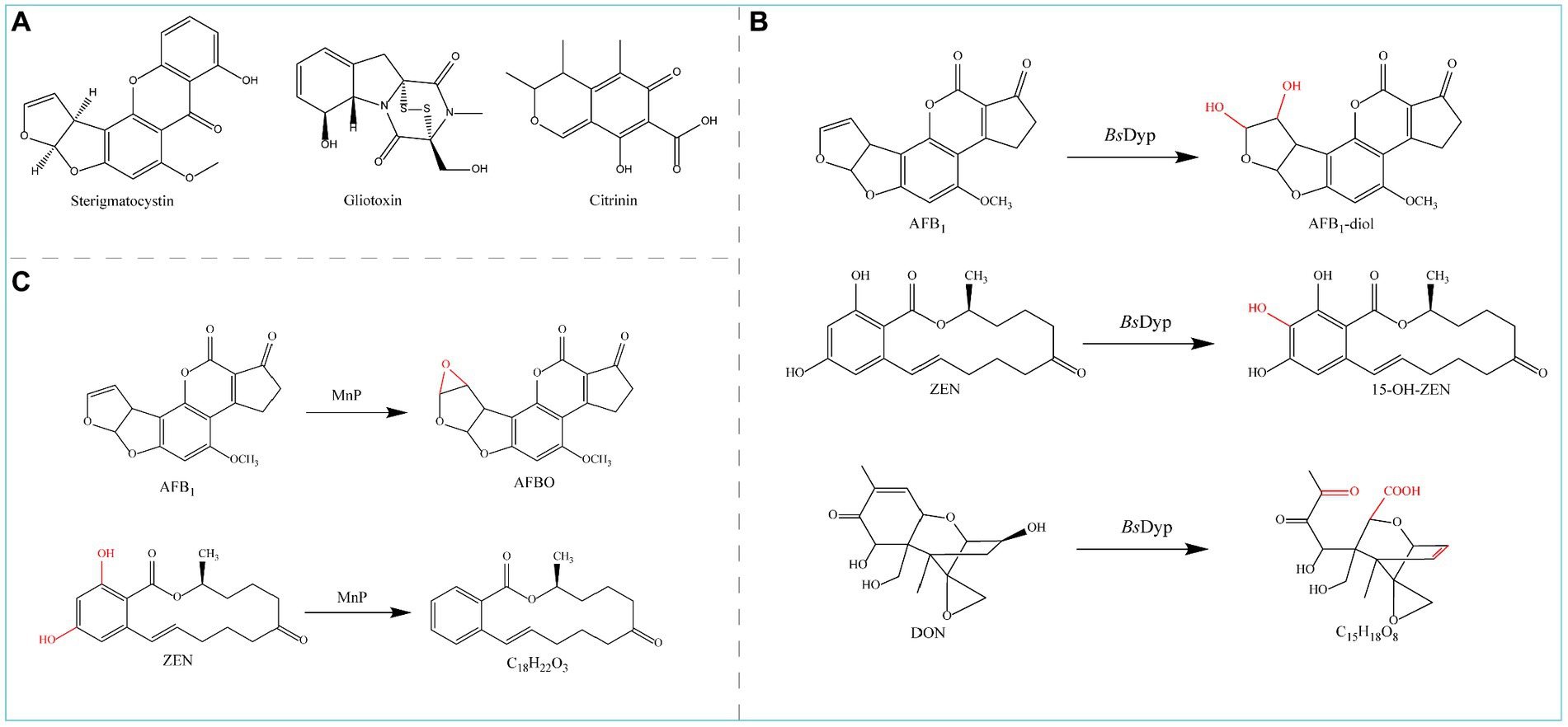
Figure 8. (A) Chemical structural formula of Sterigmatocystin, Gliotoxin, and Citrinin. (B) Degradation of AFB1 and ZEN by MnP. (C) Degradation of AFB1, ZEN, and DON by BsDyP.
Gliotoxin (Figure 8A), a mycotoxin produced by Aspergillus fumigatus, can cause immunosuppression, genotoxicity, cytotoxicity, and apoptosis (Pena et al., 2015). Gliotoxin is synthesized by the gliotoxin oxidase GliT and can therefore be reverse degraded via its synthetic pathway (Dolan et al., 2017).
Citrinin (CTN) (Figure 8A) is a nephrotoxic compound often produced by Penicillium, Aspergillus, and Monascus-contaminated seeds and foodstuffs (Flajs and Peraica, 2009; Wu et al., 2022). After R. borbori PS45 and E. cloacae PS21 were cultured on mineral media supplemented with 5 ppm citrinin, minimal citrinin degradation residues were observed (Kanpiengjai et al., 2016).
The research examples listed in part 2 (common mycotoxins and corresponding enzymes used for detoxification) are mostly based on the degradation of one mycotoxin by one enzyme. However, foods and feeds often contain a variety of mycotoxins, and multiple mycotoxins may have synergistic and superimposed toxicological effects on humans and animals. An ideal and economical detoxification pathway to address multiple mycotoxin contaminants is the application of one or several enzymes to degrade multiple mycotoxins; in addition, multiple detoxification enzymes can be simultaneously expressed by the same host bacterium to enhance the effect of detoxification and improve the efficiency of detoxification.
Manganese peroxidase (MnP) can simultaneously degrade four major mycotoxins, AFB1, ZEN, DON, and FB1, in the presence of a dicarboxylic acid malonate; MnP converts AFB1 to AFBO and ZEN to C18H22O4 (Figure 8B) and detoxifies DON and FB1, although the degradation products are unknown (Wang et al., 2019b).
The commercial peroxidase (POD) enzyme (Armoracia rusticana) degraded both OTA and ZEN in a model solution and beer with respective degradation rates of 27.0 and 64.9% for OTA and ZEN in the model solution and 4.8 and 10.9% in the beer after 360 min (De Oliveira Garcia et al., 2020).
The dye-decolorizing peroxidase-encoding gene BsDyP from Bacillus subtilis SCK6 was expressed in E. coli, and the purified recombinant BsDyP effectively degraded different types of mycotoxins such as AFB1, ZEN, and DON in the presence of Mn2+; the degradation products were AFB1-diol, 15-OH-ZEN, and C15H18O8 (Figure 8C), respectively, leading to significantly reduced toxicity (Qin et al., 2021a).
The CotA laccase gene from Bacillus licheniformis ZOM-1 was expressed in E. coli. CotA can degrade Alternaria toxin alternariol (AOH) in addition to the oxidative degradation of ZEN and AFB1. The optimum temperature of the CotA laccase was 80°C, and the pH was approximately 9.0 (Sun F. et al., 2023).
Lac-W, a laccase from Weizmannia coagulans 36D1, directly degrades AFB1, ZEN, DON, T-2 toxin, FB1, and OTA in the absence of redox mediators, and the degradation effect decreases in the following order: AFB1 > ZEN > DON > T-2 toxin > FB1 > OTA. The optimum conditions for AFB1 degradation by Lac-W are 30°C at pH 9.0, and the degradation products are not toxic to intestinal porcine epithelial cells (Hao et al., 2023).
The lactonase gene AttM, which can degrade AFB1, was expressed in E. coli; in addition to degrading AFB1, AttM also degraded OTA and ZEN at pH 8.5 and 80°C (Cheng et al., 2023).
Some enzymes can degrade both AFB1 and ZEN mycotoxins. BsCotA laccase degraded AFB1 by 98.0% and ZEN by 100.0% in the presence of the mediator methyl syringate (Wang et al., 2019a). Lac-2 laccase from the fungus Pleurotus pulmonarius was expressed in P. pastoris, and recombinant Lac-2 efficiently degraded ZEN at pH 4.0–8.0 and 40–60°C. Both Lac2-ABTS and Lac2-AS were effective at degrading ZEN, and Lac2-AS was effective at degrading AFB1 at pH 7.0 and 37°C (99.82% degradation after 1 h) (Song et al., 2021). The StMCO gene was cloned form Streptomyces thermocarboxydus and expressed in E. coli, and the recombinant laccase StMCO directly degraded AFB1 and ZEN to AFQ1 and 13-OH-ZEN-quinone, respectively, which can be significantly accelerated in the presence of the mediator ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonate)) (Qin et al., 2021c). The genes from Acinetobacter nosocomialis Y1 Porin and Peroxiredoxin degraded AFB1 and ZEN, producing AFD1 and α/β-ZAL as the degradation products, and the recombinant proteins exhibited a high degradation rate of both toxins (Adegoke et al., 2023).
The toxicity of mycotoxins is usually attributed to specific functional groups in their chemical structures; AFB1 and OTA both consist of a coumarin moiety, while the primary structures of AFB1, ZEN, OTA, and PAT are based on lactone rings. Some toxins contain the same toxic group, and certain degrading enzymes have strong group specificity but broader substrate specificity; thus, these enzymes can degrade a variety of mycotoxins with specific toxic groups. Molecular docking simulations have shown that enzymes with broad substrate specificity have substrate-binding pockets large enough to allow substrates of different structures to enter the active site of the enzyme, thereby catalyzing the degradative transformation of multiple functional groups of different mycotoxins. Therefore, the future development of enzymes that can catalyze multiple mycotoxins by expanding the substrate specificity of enzymes through genetic engineering will provide ideas for future mycotoxin detoxification.
In addition, studies on the detoxification of various mycotoxins by recombinant fusion enzyme technology have increased; in this technology, two or more single genes are combined by PCR. For example, zearalenone hydrolase (ZHD) and carboxypeptidase (CP) combine to form zearalenone hydrolase-carboxypeptidase (ZHDCP) for the detoxification of OTA and ZEN. The ZHDCP enzyme completely detoxified OTA (100%) within 30 min at pH 7.0 and 30°C, degraded ZEN by 60% in 48 min at pH 7.0 and 35°C and completely detoxified ZEN in 2 h (Azam et al., 2019). Another recombinant fusion enzyme for mycotoxin detoxification was ZPF1, which was formed by linking zearalenone hydrolase and manganese peroxidase using the linker peptide GGGGS; ZPF1 was successfully secreted and expressed in a newly constructed food-grade recombinant strain of K. lactis GG799 (pKLAC1-ZPF1) and degraded AFB1 and ZEN at high rates. Thus, this enzyme has potential for application in the food industry (Xia et al., 2021).
For the expression of fusion enzymes, it is important to select host bacteria that are not harmful to humans. E. coli, a type of opportunistic pathogen, is sometimes unsuitable for the expression of fungal enzymes, especially those used in the food industry. In contrast, we can choose food-safe yeast as a host bacterium, which is a suitable organism for the production of food enzymes.
These enzymes do not follow the conventional approach in which one enzyme degrades one specific mycotoxin. The enzymes that can simultaneously degrade multiple mycotoxins are summarized in Table 7.
Microbiological detoxification of mycotoxins relies mainly on fungi and bacteria, which will be briefly described using the examples of S. cerevisiae and lactic acid bacteria (LAB). S. cerevisiae grows rapidly and is suitable for use in agricultural production. LAB has immunomodulatory properties that prevent the germination of mold spores, and can be used as a biological food preservative (Shetty and Jespersen, 2006). The microbiological degradation mechanism is mainly by enzymatic and degradative sorption (Abdi et al., 2021). Some strains of S. cerevisiae from West African fermented foods bind AFB1 in significant amounts, up to a maximum concentration of 20 μg/mL of bound AFB1. Both viable and non-viable cells are effective aflatoxin binders and non-viable cells bound higher levels of the toxin than viable cells (Shetty et al., 2007). Lactiplantibacillus plantarum is a facultative heterofermentative LAB, 17 out of 33 LAB from plants showed removal of ZEN from the liquid medium, with L. plantarum BCC 47723 isolated from wild spider flower pickle (Pag-sian-dorng) showing the highest removal (23%) (Adunphatcharaphon et al., 2021).
There are approximately 400 known mycotoxins that contaminate food, feed, fruits, and crops and cause a range of physiological toxicity in humans and other animals. For safety purposes, many countries have imposed mandatory thresholds for mycotoxin levels. The most important step in controlling mycotoxins is prevention, i.e., preventing fungal contamination and the production of mycotoxins. However, mycotoxin contamination often cannot be prevented completely throughout the growth, storage, and transportation of crops as well as the production, processing and preservation of food; thus, a detoxification method must be developed to treat mycotoxin-contaminated foods (raw materials) and eliminate or reduce toxicity to humans and animals. Mycotoxins can be detoxified by physical, chemical and biological methods. Biological enzymatic methods are advantageous over physical and chemical methods in terms of their selectivity, safety and environmental friendliness. However, these methods face the following challenges: (1) enzymatic detoxification mostly reduces the toxicity of mycotoxins, as the products of enzyme degradation are often less toxic but still exhibit physiological toxicity; (2) the processing and use of fungal-contaminated raw materials are sometimes affected after treatment with enzymatic detoxification processes; (3) at present, only a few types of mycotoxins and their degradation enzymes and degradation mechanisms have been thoroughly studied, and some mycotoxin degradation enzymes or degradation mechanisms remain unclear; (4) information on the structures of mycotoxin degradation products and their physiological toxicities is lacking, and more research is needed to improve knowledge of mycotoxin degradation products; and (5) relevant mycotoxin detection technology and screening, optimization and safety assessment of biodegradation enzymes still need to be improved. In this paper, we reviewed the main enzymes used to detoxify mycotoxins and the mechanisms underlying detoxification. Based on these findings, several aspects of the native enzymes used for mycotoxin detoxification, including their instability, low activity, and inhibitory activity against products or substrates, should be improved. In the past 10 years, genetic engineering technology has been widely used for heterologous mycotoxin detoxification enzyme expression and engineering of modifications to enhance the detoxification effect of enzymes to expand their application. Additionally, studying enzyme immobilization has become an important method for improving the efficiency of enzymes in the application of detoxification enzymes. Studies on the structural biology of existing detoxification enzymes have revealed the mechanism by which enzymes detoxify mycotoxins, and potential detoxification enzymes can be better explored through the inclusion of bioinformatics. As a green detoxification method, the technology used for the enzymatic detoxification of mycotoxins is maturing; in addition, the industrialization process is accelerating, and the application fields are expanding. In the future, research on the enzymatic detoxification of mycotoxins should focus on the development of high-efficiency enzymes, the development of multienzyme cascades and synergistic degradation systems, and the development of detoxification enzymes and production systems that can be used directly in foods and feeds, are particularly important.
ML: Data curation, Formal analysis, Investigation, Validation, Writing – original draft, Writing – review & editing. XZ: Investigation, Resources, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. HL: Data curation, Formal analysis, Investigation, Writing – original draft. YZ: Data curation, Formal analysis, Investigation, Writing – review & editing. WX: Conceptualization, Supervision, Visualization, Writing – review & editing. WF: Supervision, Visualization, Writing – review & editing. PS: Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Shandong Province (grant number ZR2022MC159).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdi, M., Asadi, A., Maleki, F., Kouhsari, E., Fattahi, A., Ohadi, E., et al. (2021). Microbiological detoxification of mycotoxins: focus on mechanisms and advances. IDDT 21, 339–357. doi: 10.2174/1871526520666200616145150
Abraham, N., Chan, E. T. S., Zhou, T., and Seah, S. Y. K. (2022a). Microbial detoxification of mycotoxins in food. Front. Microbiol. 13:957148. doi: 10.3389/fmicb.2022.957148
Abraham, N., Schroeter, K. L., Zhu, Y., Chan, J., Evans, N., Kimber, M. S., et al. (2022b). Structure–function characterization of an aldo–keto reductase involved in detoxification of the mycotoxin, deoxynivalenol. Sci. Rep. 12:14737. doi: 10.1038/s41598-022-19040-8
Abrunhosa, L., Paterson, R., and Venâncio, A. (2010). Biodegradation of ochratoxin A for food and feed decontamination. Toxins 2, 1078–1099. doi: 10.3390/toxins2051078
Adegoke, T. V., Yang, B., Tian, X., Yang, S., Gao, Y., Ma, J., et al. (2023). Simultaneous degradation of aflatoxin B1 and zearalenone by Porin and Peroxiredoxin enzymes cloned from Acinetobacter nosocomialis Y1. J. Hazard. Mater. 459:132105. doi: 10.1016/j.jhazmat.2023.132105
Adunphatcharaphon, S., Petchkongkaew, A., and Visessanguan, W. (2021). In vitro mechanism assessment of zearalenone removal by plant-derived Lactobacillus plantarum BCC 47723. Toxins 13:286. doi: 10.3390/toxins13040286
Afsah-Hejri, L., Hajeb, P., and Ehsani, R. J. (2020). Application of ozone for degradation of mycotoxins in food: a review. Comp. Rev. Food Sci. Food Safe 19, 1777–1808. doi: 10.1111/1541-4337.12594
Afsharmanesh, H., Perez-Garcia, A., Zeriouh, H., Ahmadzadeh, M., and Romero, D. (2018). Aflatoxin degradation by Bacillus subtilis UTB1 is based on production of an oxidoreductase involved in bacilysin biosynthesis. Food Control 94, 48–55. doi: 10.1016/j.foodcont.2018.03.002
Alberts, J. F., Davids, I., Moll, W.-D., Schatzmayr, G., Burger, H.-M., Shephard, G. S., et al. (2021). Enzymatic detoxification of the fumonisin mycotoxins during dry milling of maize. Food Control 123:107726. doi: 10.1016/j.foodcont.2020.107726
Alberts, J., Schatzmayr, G., Moll, W.-D., Davids, I., Rheeder, J., Burger, H.-M., et al. (2019). Detoxification of the fumonisin mycotoxins in maize: an enzymatic approach. Toxins 11:523. doi: 10.3390/toxins11090523
Aristil, J., Venturini, G., Maddalena, G., Toffolatti, S. L., and Spada, A. (2020). Fungal contamination and aflatoxin content of maize, moringa and peanut foods from rural subsistence farms in South Haiti. J. Stored Prod. Res. 85:101550. doi: 10.1016/j.jspr.2019.101550
Azam, M. S., Yu, D., Liu, N., and Wu, A. (2019). Degrading ochratoxin a and zearalenone mycotoxins using a multifunctional recombinant enzyme. Toxins 11:301. doi: 10.3390/toxins11050301
Ben Taheur, F., Kouidhi, B., Al Qurashi, Y. M. A., Ben Salah-Abbès, J., and Chaieb, K. (2019). Review: biotechnology of mycotoxins detoxification using microorganisms and enzymes. Toxicon 160, 12–22. doi: 10.1016/j.toxicon.2019.02.001
Bian, L., Zheng, M., Chang, T., Zhou, J., and Zhang, C. (2022). Degradation of aflatoxin B1 by recombinant laccase extracellular produced from Escherichia coli. Ecotoxicol. Environ. Saf. 244:114062. doi: 10.1016/j.ecoenv.2022.114062
Blackwell, B. A., Gilliam, J. T., Savard, M. E., David Miller, J., and Duvick, J. P. (1999). Oxidative deamination of hydrolyzed fumonisin B1 (AP1) by cultures of Exophiala spinifera. Nat. Toxins 7, 31–38. doi: 10.1002/(SICI)1522-7189(199902)7:1<31::AID-NT36>3.0.CO;2-W
Bouajila, A., Lamine, M., Hamdi, Z., Ghorbel, A., and Gangashetty, P. (2022). A nutritional survey of local barley populations based on the mineral bioavailability, fatty acid profile, and geographic distribution of fusarium species and the mycotoxin zearalenone (ZEN). Agronomy 12:916. doi: 10.3390/agronomy12040916
Cao, H., Liu, D., Mo, X., Xie, C., and Yao, D. (2011). A fungal enzyme with the ability of aflatoxin B1 conversion: purification and ESI-MS/MS identification. Microbiol. Res. 166, 475–483. doi: 10.1016/j.micres.2010.09.002
Carere, J., Hassan, Y. I., Lepp, D., and Zhou, T. (2018a). The enzymatic detoxification of the mycotoxin deoxynivalenol: identification of DepA from the DON epimerization pathway. Microb. Biotechnol. 11, 1106–1111. doi: 10.1111/1751-7915.12874
Carere, J., Hassan, Y. I., Lepp, D., and Zhou, T. (2018b). The identification of DepB: an enzyme responsible for the final detoxification step in the deoxynivalenol epimerization pathway in Devosia mutans 17-2-E-8. Front. Microbiol. 9:1573. doi: 10.3389/fmicb.2018.01573
Chan, E. T. S., Zhu, Y., Li, X.-Z., Zhou, T., and Seah, S. Y. K. (2022). Characterization of two dehydrogenases from Gluconobacter oxydans involved in the transformation of patulin to ascladiol. Toxins 14:423. doi: 10.3390/toxins14070423
Chang, X., Liu, H., Sun, J., Wang, J., Zhao, C., Zhang, W., et al. (2020). Zearalenone removal from corn oil by an enzymatic strategy. Toxins 12:117. doi: 10.3390/toxins12020117
Chen, S., Pan, L., Liu, S., Pan, L., Li, X., and Wang, B. (2021). Recombinant expression and surface display of a zearalenone lactonohydrolase from Trichoderma aggressivum in Escherichia coli. Protein Expr. Purif. 187:105933. doi: 10.1016/j.pep.2021.105933
Chen, J., Wei, Z., Wang, Y., Long, M., Wu, W., and Kuca, K. (2021). Fumonisin B1: mechanisms of toxicity and biological detoxification progress in animals. Food Chem. Toxicol. 149:111977. doi: 10.1016/j.fct.2021.111977
Chen, J., Zhang, X., He, Z., Xiong, D., and Long, M. (2023). Damage on intestinal barrier function and microbial detoxification of deoxynivalenol: a review. J. Integr. Agric. S2095311923004355. doi: 10.1016/j.jia.2023.11.038
Cheng, S., Wu, T., Zhang, H., Sun, Z., Mwabulili, F., Xie, Y., et al. (2023). Mining lactonase gene from aflatoxin B1-degrading strain Bacillus megaterium and degrading properties of the recombinant enzyme. J. Agric. Food Chem. 71, 20762–20771. doi: 10.1021/acs.jafc.3c05725
Commission Regulation (EU) (2010). No 165/2010 of 26 February 2010 amending Regulation (EC) No 1881/2006 setting maximum levels for certain contaminants in foodstuffs as regards aflatoxins. Text with EEA relevance.
Dai, L., Niu, D., Huang, J.-W., Li, X., Shen, P., Li, H., et al. (2023). Cryo-EM structure and rational engineering of a superefficient ochratoxin A-detoxifying amidohydrolase. J. Hazard. Mater. 458:131836. doi: 10.1016/j.jhazmat.2023.131836
De Oliveira Garcia, S., Sibaja, K. V. M., Nogueira, W. V., Feltrin, A. C. P., Pinheiro, D. F. A., Cerqueira, M. B. R., et al. (2020). Peroxidase as a simultaneous degradation agent of ochratoxin A and zearalenone applied to model solution and beer. Food Res. Int. 131:109039. doi: 10.1016/j.foodres.2020.109039
Demirci, A. S., Tirpanci Sivri, G., Tunc, M., and Mutlu, S. (2023). Detoxification of unshelled hazelnut artificially contaminated with aflatoxins by gaseous ozone. Food Measure 17, 3515–3522. doi: 10.1007/s11694-023-01886-9
Ding, S., Lin, C., Xiao, Q., Feng, F., Wang, J., Zhang, X., et al. (2024). Effective degradation of zearalenone by dye-decolorizing peroxidases from Pleurotus ostreatus and its metabolic pathway and toxicity analysis. Sci. Total Environ. 908:168500. doi: 10.1016/j.scitotenv.2023.168500
Do, T. H., Tran, S. C., Le, C. D., Nguyen, H.-B. T., Le, P.-T. T., Le, H.-H. T., et al. (2020). Dietary exposure and health risk characterization of aflatoxin B1, ochratoxin A, fumonisin B1, and zearalenone in food from different provinces in northern Vietnam. Food Control 112:107108. doi: 10.1016/j.foodcont.2020.107108
Dobritzsch, D., Wang, H., Schneider, G., and Yu, S. (2014). Structural and functional characterization of ochratoxinase, a novel mycotoxin-degrading enzyme. Biochem. J. 462, 441–452. doi: 10.1042/BJ20140382
Dolan, S. K., Bock, T., Hering, V., Owens, R. A., Jones, G. W., Blankenfeldt, W., et al. (2017). Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus. Open Biol. 7:160292. doi: 10.1098/rsob.160292
Du, Q., Zhang, W., Xu, N., Jiang, X., Cheng, J., Wang, R., et al. (2023). Efficient and simultaneous removal of aflatoxin B1, B2, G1, G2, and zearalenone from vegetable oil by use of a metal–organic framework absorbent. Food Chem. 418:135881. doi: 10.1016/j.foodchem.2023.135881
Fang, Y., Huang, Z., Xu, W., Wang, C., Sun, Y., Zhang, W., et al. (2022). Efficient elimination of zearalenone at high processing temperatures by a robust mutant of Gliocladium roseum zearalenone lactonase. Food Control 142:109222. doi: 10.1016/j.foodcont.2022.109222
Flajs, D., and Peraica, M. (2009). Toxicological properties of Citrinin. Arch. Ind. Hyg. Toxicol. 60, 457–464. doi: 10.2478/10004-1254-60-2009-1992
Freire, L., Braga, P. A. C., Furtado, M. M., Delafiori, J., Dias-Audibert, F. L., Pereira, G. E., et al. (2020). From grape to wine: fate of ochratoxin A during red, rose, and white winemaking process and the presence of ochratoxin derivatives in the final products. Food Control 113:107167. doi: 10.1016/j.foodcont.2020.107167
Gao, H., Lu, D., Xing, M., Xu, Q., and Xue, F. (2022). Excavation, expression, and functional analysis of a novel zearalenone-degrading enzyme. Folia Microbiol. 67, 633–640. doi: 10.1007/s12223-022-00967-4
Gao, Z., Luo, K., Zhu, Q., Peng, J., Liu, C., Wang, X., et al. (2023). The natural occurrence, toxicity mechanisms and management strategies of fumonisin B1: a review. Environ. Pollut. 320:121065. doi: 10.1016/j.envpol.2023.121065
Gao, Y., Meng, L., Liu, H., Wang, J., and Zheng, N. (2020). The compromised intestinal barrier induced by mycotoxins. Toxins 12:619. doi: 10.3390/toxins12100619
Gao, X., Mu, P., Zhu, X., Chen, X., Tang, S., Wu, Y., et al. (2020). Dual function of a novel bacterium, Slackia sp. D-G6: detoxifying deoxynivalenol and producing the natural estrogen analogue, equol. Toxins 12:85. doi: 10.3390/toxins12020085
Gavahian, M., and Khaneghah, A. M. (2020). Cold plasma as a tool for the elimination of food contaminants: recent advances and future trends. Crit. Rev. Food Sci. Nutr. 60, 1581–1592. doi: 10.1080/10408398.2019.1584600
Gonaus, C., Wieland, L., Thallinger, G. G., and Prasad, S. (2023). Ochratoxin a degrading enzymes of Stenotrophomonas sp. 043-1a. FEMS Microbiol. Lett. 370:fnad028. doi: 10.1093/femsle/fnad028
González Pereyra, M. L., Martínez, M. P., and Cavaglieri, L. R. (2019). Presence of aiiA homologue genes encoding for N-acyl homoserine lactone-degrading enzyme in aflatoxin B1-decontaminating Bacillus strains with potential use as feed additives. Food Chem. Toxicol. 124, 316–323. doi: 10.1016/j.fct.2018.12.016
Gruber-Dorninger, C., Faas, J., Doupovec, B., Aleschko, M., Stoiber, C., Höbartner-Gußl, A., et al. (2021). Metabolism of zearalenone in the rumen of dairy cows with and without application of a zearalenone-degrading enzyme. Toxins 13:84. doi: 10.3390/toxins13020084
Gu, K., Ryu, D., and Lee, H. J. (2021). Ochratoxin A and its reaction products affected by sugars during heat processing. Food Chem. 348:129038. doi: 10.1016/j.foodchem.2021.129038
Guo, Y., Qin, X., Tang, Y., Ma, Q., Zhang, J., and Zhao, L. (2020). CotA laccase, a novel aflatoxin oxidase from Bacillus licheniformis, transforms aflatoxin B1 to aflatoxin Q1 and epi-aflatoxin Q1. Food Chem. 325:126877. doi: 10.1016/j.foodchem.2020.126877
Guo, Y., Tang, Y., Zhang, L., Liu, Y., Ma, Q., and Zhao, L. (2024). Enzymatic characterization and application of soybean hull peroxidase as an efficient and renewable biocatalyst for degradation of zearalenone. Int. J. Biol. Macromol. 260:129664. doi: 10.1016/j.ijbiomac.2024.129664
Guo, Y., Wang, Y., Liu, Y., Ma, Q., Ji, C., and Zhao, L. (2022). Detoxification of the mycoestrogen zearalenone by Bacillus licheniformis spore CotA laccase and application of immobilized laccase in contaminated corn meal. LWT 163:113548. doi: 10.1016/j.lwt.2022.113548
Haidukowski, M., Casamassima, E., Cimmarusti, M. T., Branà, M. T., Longobardi, F., Acquafredda, P., et al. (2019). Aflatoxin B1-adsorbing capability of Pleurotus eryngii mycelium: efficiency and modeling of the process. Front. Microbiol. 10:1386. doi: 10.3389/fmicb.2019.01386
Hao, W.-B., Gu, X., Yu, X., Zhao, Y., Li, C., Jia, M., et al. (2023). Laccase lac-W detoxifies aflatoxin B1 and degrades five other major mycotoxins in the absence of redox mediators. Environ. Pollut. 338:122581. doi: 10.1016/j.envpol.2023.122581
Hartinger, D., Heinl, S., Schwartz, H. E., Grabherr, R., Schatzmayr, G., Haltrich, D., et al. (2010). Enhancement of solubility in Escherichia coli and purification of an aminotransferase from Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B1. Microb. Cell Factories 9:62. doi: 10.1186/1475-2859-9-62
Hartinger, D., Schwartz, H., Hametner, C., Schatzmayr, G., Haltrich, D., and Moll, W.-D. (2011). Enzyme characteristics of aminotransferase FumI of Sphingopyxis sp. MTA144 for deamination of hydrolyzed fumonisin B1. Appl. Microbiol. Biotechnol. 91, 757–768. doi: 10.1007/s00253-011-3248-9
He, J. W., Hassan, Y. I., Perilla, N., Li, X.-Z., Boland, G. J., and Zhou, T. (2016). Bacterial epimerization as a route for deoxynivalenol detoxification: the influence of growth and environmental conditions. Front. Microbiol. 7:572. doi: 10.3389/fmicb.2016.00572
He, Q., Liang, J., Zhao, Y., Yuan, Y., Wang, Z., Gao, Z., et al. (2022). Enzymatic degradation of mycotoxin patulin by an extracellular lipase from Ralstonia and its application in apple juice. Food Control 136:108870. doi: 10.1016/j.foodcont.2022.108870
He, W.-J., Shi, M.-M., Yang, P., Huang, T., Zhao, Y., Wu, A.-B., et al. (2020). A quinone-dependent dehydrogenase and two NADPH-dependent aldo/keto reductases detoxify deoxynivalenol in wheat via epimerization in a Devosia strain. Food Chem. 321:126703. doi: 10.1016/j.foodchem.2020.126703
Heinl, S., Hartinger, D., Thamhesl, M., Schatzmayr, G., Moll, W.-D., and Grabherr, R. (2011). An aminotransferase from bacterium ATCC 55552 deaminates hydrolyzed fumonisin B1. Biodegradation 22, 25–30. doi: 10.1007/s10532-010-9371-y
Heinl, S., Hartinger, D., Thamhesl, M., Vekiru, E., Krska, R., Schatzmayr, G., et al. (2010). Degradation of fumonisin B1 by the consecutive action of two bacterial enzymes. J. Biotechnol. 145, 120–129. doi: 10.1016/j.jbiotec.2009.11.004
Holanda, D. M., and Kim, S. W. (2021). Mycotoxin occurrence, toxicity, and detoxifying agents in pig production with an emphasis on deoxynivalenol. Toxins 13:171. doi: 10.3390/toxins13020171
Hu, Y., Li, H., Min, J., Yu, Y., Liu, W., Huang, J.-W., et al. (2022). Crystal structure and biochemical analysis of the specialized deoxynivalenol–detoxifying glyoxalase SPG from Gossypium hirsutum. Int. J. Biol. Macromol. 200, 388–396. doi: 10.1016/j.ijbiomac.2022.01.055
Hu, J., Wang, G., Hou, M., Du, S., Han, J., Yu, Y., et al. (2023). New hydrolase from Aeromicrobium sp. HA for the biodegradation of zearalenone: identification, mechanism, and application. J. Agric. Food Chem. 71, 2411–2420. doi: 10.1021/acs.jafc.2c06410
Huang, J.-Q., Fang, X., Tian, X., Chen, P., Lin, J.-L., Guo, X.-X., et al. (2020). Aromatization of natural products by a specialized detoxification enzyme. Nat. Chem. Biol. 16, 250–256. doi: 10.1038/s41589-019-0446-8
Jahan, I., Tai, B., Ma, J., Hussain, S., Du, H., Guo, L., et al. (2023). Identification of a novel Bacillus velezensis IS-6 nudix hydrolase Nh-9 involved in ochratoxin a detoxification by transcriptomic profiling and functional verification. J. Agric. Food Chem. 71, 10155–10168. doi: 10.1021/acs.jafc.3c01910
Janik, E., Niemcewicz, M., Ceremuga, M., Stela, M., Saluk-Bijak, J., Siadkowski, A., et al. (2020). Molecular aspects of mycotoxins—a serious problem for human health. IJMS 21:8187. doi: 10.3390/ijms21218187
Ji, J., Yu, J., Xu, W., Zheng, Y., Zhang, Y., and Sun, X. (2022). Isolation and mechanistic characterization of a novel zearalenone-degrading enzyme. Food Secur. 11:2908. doi: 10.3390/foods11182908
Jiang, T., Wang, M., Li, X., Wang, H., Zhao, G., Wu, P., et al. (2022). The replacement of main cap domain to improve the activity of a ZEN lactone hydrolase with broad substrate spectrum. Biochem. Eng. J. 182:108418. doi: 10.1016/j.bej.2022.108418
Jing, G., Wang, Y., Wu, M., Liu, W., Xiong, S., Yu, J., et al. (2023). Photocatalytic degradation and pathway from mycotoxins in food: a review. Food Rev. Int. 40, 276–292. doi: 10.1080/87559129.2023.2166062
Jouany, J. P. (2007). Methods for preventing, decontaminating and minimizing the toxicity of mycotoxins in feeds. Anim. Feed Sci. Technol. 137, 342–362. doi: 10.1016/j.anifeedsci.2007.06.009
Kanpiengjai, A., Mahawan, R., Lumyong, S., and Khanongnuch, C. (2016). A soil bacterium rhizobium borbori and its potential for citrinin-degrading application. Ann. Microbiol. 66, 807–816. doi: 10.1007/s13213-015-1167-1
Kumar, P., Mahato, D. K., Sharma, B., Borah, R., Haque, S., Mahmud, M. M. C., et al. (2020). Ochratoxins in food and feed: occurrence and its impact on human health and management strategies. Toxicon 187, 151–162. doi: 10.1016/j.toxicon.2020.08.031
Kupski, L., Queiroz, M. I., and Badiale-Furlong, E. (2018). Application of carboxypeptidase a to a baking process to mitigate contamination of wheat flour by ochratoxin A. Process Biochem. 64, 248–254. doi: 10.1016/j.procbio.2017.09.006
Li, Y., Cai, R., Fu, C., Qi, L., Yuan, Y., Yue, T., et al. (2023). Degradation of patulin in apple juice by pulsed light and its effect on the quality. Food Bioprocess Technol. 16, 870–880. doi: 10.1007/s11947-022-02978-y
Li, N., Cui, R., Zhang, F., Meng, X., and Liu, B. (2022). A novel enzyme from Rhodotorula mucilaginosa aldolase: isolation, identification and degradation for patulin in apple juice. Process Biochem. 116, 148–156. doi: 10.1016/j.procbio.2022.03.001
Li, C.-H., Li, W.-Y., Hsu, I.-N., Liao, Y.-Y., Yang, C.-Y., Taylor, M. C., et al. (2019). Recombinant aflatoxin-degrading F420H2-dependent reductase from Mycobacterium smegmatis protects mammalian cells from aflatoxin toxicity. Toxins 11:259. doi: 10.3390/toxins11050259
Li, D., Liang, G., Mu, P., Lin, J., Huang, J., Guo, C., et al. (2023). Improvement of catalytic activity of sorbose dehydrogenase for deoxynivalenol degradation by rational design. Food Chem. 423:136274. doi: 10.1016/j.foodchem.2023.136274
Li, Z., Wang, Y., Liu, Z., Jin, S., Pan, K., Liu, H., et al. (2021). Biological detoxification of fumonisin by a novel carboxylesterase from Sphingomonadales bacterium and its biochemical characterization. Int. J. Biol. Macromol. 169, 18–27. doi: 10.1016/j.ijbiomac.2020.12.033
Li, K., Yu, S., Yu, D., Lin, H., Liu, N., and Wu, A. (2022). Biodegradation of fumonisins by the consecutive action of a fusion enzyme. Toxins 14:266. doi: 10.3390/toxins14040266
Lin, X., Yu, W., Tong, X., Li, C., Duan, N., Wang, Z., et al. (2022). Application of nanomaterials for coping with mycotoxin contamination in food safety: from detection to control. Crit. Rev. Anal. Chem. 54, 355–388. doi: 10.1080/10408347.2022.2076063
Liu, Y., Chang, C.-C. H., Marsh, G. M., and Wu, F. (2012). Population attributable risk of aflatoxin-related liver cancer: systematic review and meta-analysis. Eur. J. Cancer 48, 2125–2136. doi: 10.1016/j.ejca.2012.02.009
Liu, Y., Guo, Y., Liu, L., Tang, Y., Wang, Y., Ma, Q., et al. (2023a). Improvement of aflatoxin B1 degradation ability by Bacillus licheniformis CotA-laccase Q441A mutant. Heliyon 9:e22388. doi: 10.1016/j.heliyon.2023.e22388
Liu, Q., Huang, L., Cui, Z., Qiao, B., Li, F., and Wang, C. (2022). FumDSB can alleviate the inflammatory response induced by fumonisin B1 in growing pigs. Food Additiv Contamin Part A 39, 1619–1633. doi: 10.1080/19440049.2022.2100490
Liu, Y., Mao, H., Woldemariam Yohannes, K., Wan, Z., Cao, Y., Tron, T., et al. (2021). Degradation of aflatoxin B1 by a recombinant laccase from Trametes sp. C30 expressed in Saccharomyces cerevisiae: a mechanism assessment study in vitro and in vivo. Food Res. Int. 145:110418. doi: 10.1016/j.foodres.2021.110418
Liu, X., Wang, L., Wang, S., Cai, R., Yue, T., Yuan, Y., et al. (2023). Detoxification of patulin in apple juice by enzymes and evaluation of its degradation products. Food Control 145:109518. doi: 10.1016/j.foodcont.2022.109518
Liu, Y., and Wu, F. (2010). Global burden of aflatoxin-induced hepatocellular carcinoma: a risk assessment. Environ. Health Perspect. 118, 818–824. doi: 10.1289/ehp.0901388
Liu, Y., Xu, L., Shi, Z., Wang, R., Liu, Y., Gong, Y., et al. (2023b). Identification of an Acinetobacter pittii acyltransferase involved in transformation of deoxynivalenol to 3-acetyl-deoxynivalenol by transcriptomic analysis. Ecotoxicol. Environ. Saf. 263:115395. doi: 10.1016/j.ecoenv.2023.115395
Loi, M., De Leonardis, S., Ciasca, B., Paciolla, C., Mulè, G., and Haidukowski, M. (2023). Aflatoxin B1 degradation by Ery4 laccase: from in vitro to contaminated corn. Toxins 15:310. doi: 10.3390/toxins15050310
Loi, M., Renaud, J. B., Rosini, E., Pollegioni, L., Vignali, E., Haidukowski, M., et al. (2020). Enzymatic transformation of aflatoxin B1 by Rh_DypB peroxidase and characterization of the reaction products. Chemosphere 250:126296. doi: 10.1016/j.chemosphere.2020.126296
Luo, H., Wang, G., Chen, N., Fang, Z., Xiao, Y., Zhang, M., et al. (2022). A superefficient ochratoxin a hydrolase with promising potential for industrial applications. Appl. Environ. Microbiol. 88:e0196421. doi: 10.1128/AEM.01964-21
Lyagin, I., and Efremenko, E. (2019). Enzymes for detoxification of various mycotoxins: origins and mechanisms of catalytic action. Molecules 24:2362. doi: 10.3390/molecules24132362
Mahato, D. K., Kamle, M., Sharma, B., Pandhi, S., Devi, S., Dhawan, K., et al. (2021). Patulin in food: a mycotoxin concern for human health and its management strategies. Toxicon 198, 12–23. doi: 10.1016/j.toxicon.2021.04.027
Mangini, V., Rosini, E., Caliandro, R., Mangiatordi, G. F., Delre, P., Sciancalepore, A. G., et al. (2024). DypB peroxidase for aflatoxin removal: new insights into the toxin degradation process. Chemosphere 349:140826. doi: 10.1016/j.chemosphere.2023.140826
Mascotti, M. L., Lapadula, W. J., and Juri Ayub, M. (2015). The origin and evolution of Baeyer—Villiger monooxygenases (BVMOs): an ancestral family of flavin monooxygenases. PLoS One 10:e0132689. doi: 10.1371/journal.pone.0132689
Matumba, L., Namaumbo, S., Ngoma, T., Meleke, N., De Boevre, M., Logrieco, A. F., et al. (2021). Five keys to prevention and control of mycotoxins in grains: a proposal. Glob. Food Sec. 30:100562. doi: 10.1016/j.gfs.2021.100562
Mishra, S., Srivastava, S., Dewangan, J., Divakar, A., and Kumar Rath, S. (2020). Global occurrence of deoxynivalenol in food commodities and exposure risk assessment in humans in the last decade: a survey. Crit. Rev. Food Sci. Nutr. 60, 1346–1374. doi: 10.1080/10408398.2019.1571479
Murugesan, P., Brunda, D. K., Moses, J. A., and Anandharamakrishnan, C. (2021). Photolytic and photocatalytic detoxification of mycotoxins in foods. Food Control 123:107748. doi: 10.1016/j.foodcont.2020.107748
Mwabulili, F., Xie, Y., Li, Q., Sun, S., Yang, Y., and Ma, W. (2023). Research progress of ochratoxin a bio-detoxification. Toxicon 222:107005. doi: 10.1016/j.toxicon.2022.107005
Nada, S., Nikola, T., Bozidar, U., Ilija, D., and Andreja, R. (2022). Prevention and practical strategies to control mycotoxins in the wheat and maize chain. Food Control 136:108855. doi: 10.1016/j.foodcont.2022.108855
Ogunade, I. M., Martinez-Tuppia, C., Queiroz, O. C. M., Jiang, Y., Drouin, P., Wu, F., et al. (2018). Silage review: mycotoxins in silage: occurrence, effects, prevention, and mitigation. J. Dairy Sci. 101, 4034–4059. doi: 10.3168/jds.2017-13788
Okwara, P. C., Afolabi, I. S., and Ahuekwe, E. F. (2021). Application of laccase in aflatoxin B1 degradation: a review. IOP Conf. Ser. Mater. Sci. Eng. 1107:012178. doi: 10.1088/1757-899X/1107/1/012178
Pena, G. A., Monge, M. P., González Pereyra, M. L., Dalcero, A. M., Rosa, C. A. R., Chiacchiera, S. M., et al. (2015). Gliotoxin production by Aspergillus fumigatus strains from animal environment. Micro-analytical sample treatment combined with a LC-MS/MS method for gliotoxin determination. Mycotoxin Res 31, 145–150. doi: 10.1007/s12550-015-0225-7
Peng, M., Zhang, Z., Xu, X., Zhang, H., Zhao, Z., and Liang, Z. (2023). Purification and characterization of the enzymes from Brevundimonas naejangsanensis that degrade ochratoxin a and B. Food Chem. 419:135926. doi: 10.1016/j.foodchem.2023.135926
Peng, M., Zhao, Z., and Liang, Z. (2022). Biodegradation of ochratoxin a and ochratoxin B by Brevundimonas naejangsanensis isolated from soil. Food Control 133:108611. doi: 10.1016/j.foodcont.2021.108611
Pitout, M. J. (1969). The hydrolysis of ochratoxin a by some proteolytic enzymes. Biochem. Pharmacol. 18, 485–491. doi: 10.1016/0006-2952(69)90224-X
Qin, X., Su, X., Tu, T., Zhang, J., Wang, X., Wang, Y., et al. (2021a). Enzymatic degradation of multiple major mycotoxins by dye-decolorizing peroxidase from Bacillus subtilis. Toxins 13:429. doi: 10.3390/toxins13060429
Qin, X., Xin, Y., Su, X., Wang, X., Wang, Y., Zhang, J., et al. (2021b). Efficient degradation of zearalenone by dye-decolorizing peroxidase from Streptomyces thermocarboxydus combining catalytic properties of manganese peroxidase and laccase. Toxins 13:602. doi: 10.3390/toxins13090602
Qin, X., Xin, Y., Zou, J., Su, X., Wang, X., Wang, Y., et al. (2021c). Efficient degradation of aflatoxin B1 and zearalenone by laccase-like multicopper oxidase from Streptomyces thermocarboxydus in the presence of mediators. Toxins 13:754. doi: 10.3390/toxins13110754
Qin, X., Zhang, J., Liu, Y., Guo, Y., Tang, Y., Zhang, Q., et al. (2022). A quinoprotein dehydrogenase from Pelagibacterium halotolerans ANSP101 oxidizes deoxynivalenol to 3-keto-deoxynivalenol. Food Control 136:108834. doi: 10.1016/j.foodcont.2022.108834
Raesi, S., Mohammadi, R., Khammar, Z., Paimard, G., Abdalbeygi, S., Sarlak, Z., et al. (2022). Photocatalytic detoxification of aflatoxin B1 in an aqueous solution and soymilk using nano metal oxides under UV light: kinetic and isotherm models. LWT 154:112638. doi: 10.1016/j.lwt.2021.112638
Romero-Sánchez, I., Gracia-Lor, E., and Madrid-Albarrán, Y. (2024). Aflatoxin detoxification by thermal cooking treatment and evaluation of in vitro bioaccessibility from white and brown rice. Food Chem. 436:137738. doi: 10.1016/j.foodchem.2023.137738
Rudyk, H., Tomaszewska, E., Arciszewski, M. B., Muszyński, S., Tomczyk-Warunek, A., Dobrowolski, P., et al. (2023). Histomorphometrical changes in intestine structure and innervation following experimental fumonisins intoxication in male Wistar rats. Pol. J. Vet. Sci. 23, 77–88. doi: 10.24425/pjvs.2020.132751
Sánchez-Arroyo, A., Plaza-Vinuesa, L., De Las Rivas, B., Mancheño, J. M., and Muñoz, R. (2024). Structural and functional analysis of the key enzyme responsible for the degradation of ochratoxin a in the Alcaligenes genus. Int. J. Biol. Macromol. 267:131342. doi: 10.1016/j.ijbiomac.2024.131342
Sánchez-Arroyo, A., Plaza-Vinuesa, L., Rivas, B. D. L., Mancheño, J. M., and Muñoz, R. (2023). The salicylate 1,2-dioxygenase from Pseudaminobacter salicylatoxidans DSM 6986T is a bifunctional enzyme that inactivates the mycotoxin ochratoxin a by a novel amidohydrolase activity. Int. J. Biol. Macromol. 237:124230. doi: 10.1016/j.ijbiomac.2023.124230
Santos, J., Castro, T., Venâncio, A., and Silva, C. (2023). Degradation of ochratoxins A and B by lipases: a kinetic study unraveled by molecular modeling. Heliyon 9:e19921. doi: 10.1016/j.heliyon.2023.e19921
Sekiguchi, J., Shimamoto, T., Yamada, Y., and Gaucher, G. M. (1983). Patulin biosynthesis: enzymatic and nonenzymatic transformations of the mycotoxin (E)-ascladiol. Appl. Environ. Microbiol. 45, 1939–1942. doi: 10.1128/aem.45.6.1939-1942.1983
Shcherbakova, L., Rozhkova, A., Osipov, D., Zorov, I., Mikityuk, O., Statsyuk, N., et al. (2020). Effective zearalenone degradation in model solutions and infected wheat grain using a novel heterologous lactonohydrolase secreted by recombinant Penicillium canescens. Toxins 12:475. doi: 10.3390/toxins12080475
Shen, M.-H., and Singh, R. K. (2022a). Decomposing aflatoxins in peanuts using advanced oxidation processes by UV and H2O2. Food Bioprocess Technol. 15, 1647–1657. doi: 10.1007/s11947-022-02844-x
Shen, M.-H., and Singh, R. K. (2022b). Detoxifying aflatoxin contaminated peanuts by high concentration of H2O2 at moderate temperature and catalase inactivation. Food Control 142:109218. doi: 10.1016/j.foodcont.2022.109218
Shetty, P. H., Hald, B., and Jespersen, L. (2007). Surface binding of aflatoxin B1 by Saccharomyces cerevisiae strains with potential decontaminating abilities in indigenous fermented foods. Int. J. Food Microbiol. 113, 41–46. doi: 10.1016/j.ijfoodmicro.2006.07.013
Shetty, P. H., and Jespersen, L. (2006). Saccharomyces cerevisiae and lactic acid bacteria as potential mycotoxin decontaminating agents. Trends Food Sci. Technol. 17, 48–55. doi: 10.1016/j.tifs.2005.10.004
Shi, Y., Xu, W., Ni, D., Zhang, W., Guang, C., and Mu, W. (2024). Identification and application of a novel deoxynivalenol-degrading enzyme from Youhaiella tibetensis. Food Chem. 435:137609. doi: 10.1016/j.foodchem.2023.137609
Song, Y., Wang, Y., Guo, Y., Qiao, Y., Ma, Q., Ji, C., et al. (2021). Degradation of zearalenone and aflatoxin B1 by Lac2 from Pleurotus pulmonarius in the presence of mediators. Toxicon 201, 1–8. doi: 10.1016/j.toxicon.2021.08.003
Song, J., Zhang, S., Xie, Y., and Li, Q. (2019). Purification and characteristics of an aflatoxin B1 degradation enzyme isolated from Pseudomonas aeruginosa. FEMS Microbiol. Lett. 366:fnz034. doi: 10.1093/femsle/fnz034
Sun, H., He, Z., Xiong, D., and Long, M. (2023). Mechanisms by which microbial enzymes degrade four mycotoxins and application in animal production: a review. Anim. Nutr. 15, 256–274. doi: 10.1016/j.aninu.2023.09.003
Sun, J., Xia, Y., and Ming, D. (2020). Whole-genome sequencing and bioinformatics analysis of apiotrichum mycotoxinivorans: predicting putative zearalenone-degradation enzymes. Front. Microbiol. 11:1866. doi: 10.3389/fmicb.2020.01866
Sun, F., Yu, D., Zhou, H., Lin, H., Yan, Z., and Wu, A. (2023). CotA laccase from Bacillus licheniformis ZOM-1 effectively degrades zearalenone, aflatoxin B1 and alternariol. Food Control 145:109472. doi: 10.1016/j.foodcont.2022.109472
Takahashi-Ando, N., Kimura, M., Kakeya, H., Osada, H., and Yamaguchi, I. (2002). A novel lactonohydrolase responsible for the detoxification of zearalenone: enzyme purification and gene cloning. Biochem. J. 365, 1–6. doi: 10.1042/bj20020450
Tang, H., Li, X., Zhang, F., Meng, X., and Liu, B. (2019). Biodegradation of the mycotoxin patulin in apple juice by Orotate phosphoribosyltransferase from Rhodotorula mucilaginosa. Food Control 100, 158–164. doi: 10.1016/j.foodcont.2019.01.020
Tannous, J., Snini, S. P., El Khoury, R., Canlet, C., Pinton, P., Lippi, Y., et al. (2017). Patulin transformation products and last intermediates in its biosynthetic pathway, E- and Z-ascladiol, are not toxic to human cells. Arch. Toxicol. 91, 2455–2467. doi: 10.1007/s00204-016-1900-y
Taylor, M. C., Jackson, C. J., Tattersall, D. B., French, N., Peat, T. S., Newman, J., et al. (2010). Identification and characterization of two families of F420H2-dependent reductases from mycobacteria that catalyse aflatoxin degradation. Mol. Microbiol. 78, 561–575. doi: 10.1111/j.1365-2958.2010.07356.x
Tu, Y., Liu, S., Cai, P., and Shan, T. (2023). Global distribution, toxicity to humans and animals, biodegradation, and nutritional mitigation of deoxynivalenol: a review. Comp. Rev. Food Sci. Food Safe 22, 3951–3983. doi: 10.1111/1541-4337.13203
Ülger, T. G., Uçar, A., Çakıroğlu, F. P., and Yilmaz, S. (2020). Genotoxic effects of mycotoxins. Toxicon 185, 104–113. doi: 10.1016/j.toxicon.2020.07.004
Venkatesh, N., and Keller, N. P. (2019). Mycotoxins in conversation with bacteria and fungi. Front. Microbiol. 10:403. doi: 10.3389/fmicb.2019.00403
Wang, X., Bai, Y., Huang, H., Tu, T., Wang, Y., Wang, Y., et al. (2019a). Degradation of aflatoxin B1 and zearalenone by bacterial and fungal laccases in presence of structurally defined chemicals and complex natural mediators. Toxins 11:609. doi: 10.3390/toxins11100609
Wang, Y., Chen, Y., Jiang, L., and Huang, H. (2022). Improvement of the enzymatic detoxification activity towards mycotoxins through structure-based engineering. Biotechnol. Adv. 56:107927. doi: 10.1016/j.biotechadv.2022.107927
Wang, X., Cui, L., Liu, M., Qi, Z., Luo, H., Huang, H., et al. (2024). Theoretical insights into the mechanism underlying aflatoxin B1 transformation by the BsCotA-methyl syringate system. Ecotoxicol. Environ. Saf. 272:116049. doi: 10.1016/j.ecoenv.2024.116049
Wang, L., Liu, X., Cai, R., Ge, Q., Zhao, Z., Yue, T., et al. (2022). Detoxification of ochratoxin a by pulsed light in grape juice and evaluation of its degradation products and safety. Innovative Food Sci. Emerg. Technol. 78:103024. doi: 10.1016/j.ifset.2022.103024
Wang, Z., Luo, F., Jiang, S., Selvaraj, J. N., Zhou, Y., and Zhang, G. (2023). Biochemical characterization and molecular modification of a zearalenone hydrolyzing enzyme Zhd11D from Phialophora attinorum. Enzym. Microb. Technol. 170:110286. doi: 10.1016/j.enzmictec.2023.110286
Wang, X., Qin, X., Hao, Z., Luo, H., Yao, B., and Su, X. (2019b). Degradation of four major mycotoxins by eight manganese peroxidases in presence of a dicarboxylic acid. Toxins 11:566. doi: 10.3390/toxins11100566
Wang, Y., Sun, J., Zhang, M., Pan, K., Liu, T., Zhang, T., et al. (2023). Detoxification of fumonisins by three novel transaminases with diverse enzymatic characteristics coupled with carboxylesterase. Food Secur. 12:416. doi: 10.3390/foods12020416
Wang, S., Wang, X., Penttinen, L., Luo, H., Zhang, Y., Liu, B., et al. (2022). Patulin detoxification by recombinant manganese peroxidase from Moniliophthora roreri expressed by Pichia pastoris. Toxins 14:440. doi: 10.3390/toxins14070440
Wang, K., Zheng, X., Yang, Q., Zhang, H., Apaliya, M. T., Dhanasekaran, S., et al. (2019). S-adenosylmethionine-dependent methyltransferase helps Pichia caribbica degrade patulin. J. Agric. Food Chem. 67, 11758–11768. doi: 10.1021/acs.jafc.9b05144
Wei, W., Qian, Y., Wu, Y., Chen, Y., Peng, C., Luo, M., et al. (2020). Detoxification of ochratoxin a by Lysobacter sp. CW239 and characteristics of a novel degrading gene carboxypeptidase cp4. Environ. Pollut. 258:113677. doi: 10.1016/j.envpol.2019.113677
Wei, C., Yu, L., Qiao, N., Zhao, J., Zhang, H., Zhai, Q., et al. (2020). Progress in the distribution, toxicity, control, and detoxification of patulin: a review. Toxicon 184, 83–93. doi: 10.1016/j.toxicon.2020.05.006
Wu, Q., Dohnal, V., Huang, L., Kuca, K., Wang, X., Chen, G., et al. (2011). Metabolic pathways of ochratoxin A. CDM 12, 1–10. doi: 10.2174/138920011794520026
Wu, Y.-Z., Lu, F.-P., Jiang, H.-L., Tan, C.-P., Yao, D.-S., Xie, C.-F., et al. (2015). The furofuran-ring selectivity, hydrogen peroxide-production and low Km value are the three elements for highly effective detoxification of aflatoxin oxidase. Food Chem. Toxicol. 76, 125–131. doi: 10.1016/j.fct.2014.12.004
Wu, J., Yang, C., Yang, M., Liang, Z., Wu, Y., Kong, X., et al. (2022). The role of ER stress and ATP/AMPK in oxidative stress meditated hepatotoxicity induced by citrinin. Ecotoxicol. Environ. Saf. 237:113531. doi: 10.1016/j.ecoenv.2022.113531
Xia, Y., He, R., Sun, Y., Zhou, H., Gao, M., Hu, X., et al. (2022). Food-grade expression of manganese peroxidases in recombinant kluyveromyces lactis and degradation of aflatoxin B1 using fermentation supernatants. Front. Microbiol. 12:821230. doi: 10.3389/fmicb.2021.821230
Xia, Y., Wu, Z., He, R., Gao, Y., Qiu, Y., Cheng, Q., et al. (2021). Simultaneous degradation of two mycotoxins enabled by a fusion enzyme in food-grade recombinant Kluyveromyces lactis. Bioresour. Bioprocess. 8:62. doi: 10.1186/s40643-021-00395-1
Xiao, Y., Liu, B., Wang, Z., Han, C., Meng, X., and Zhang, F. (2019). Effective degradation of the mycotoxin patulin in pear juice by porcine pancreatic lipase. Food Chem. Toxicol. 133:110769. doi: 10.1016/j.fct.2019.110769
Xie, Y., Wang, W., and Zhang, S. (2019). Purification and identification of an aflatoxin B1 degradation enzyme from Pantoea sp. T6. Toxicon 157, 35–42. doi: 10.1016/j.toxicon.2018.11.290
Xing, M., Chen, Y., Dai, W., He, X., Li, B., and Tian, S. (2023). Immobilized short-chain dehydrogenase/reductase on Fe3O4 particles acts as a magnetically recoverable biocatalyst component in patulin bio-detoxification system. J. Hazard. Mater. 448:130986. doi: 10.1016/j.jhazmat.2023.130986
Xing, M., Chen, Y., Li, B., and Tian, S. (2021). Characterization of a short-chain dehydrogenase/reductase and its function in patulin biodegradation in apple juice. Food Chem. 348:129046. doi: 10.1016/j.foodchem.2021.129046
Xing, M., Li, B., Chen, Y., and Tian, S. (2020). Ribonucleoside diphosphate reductase plays an important role in patulin degradation by Enterobacter cloacae subsp. dissolvens. J. Agric. Food Chem. 68, 5232–5240. doi: 10.1021/acs.jafc.0c01613
Xiong, L., Peng, M., Zhao, M., and Liang, Z. (2020). Truncated expression of a carboxypeptidase a from bovine improves its enzymatic properties and detoxification efficiency of ochratoxin a. Toxins 12:680. doi: 10.3390/toxins12110680
Xu, X., Pang, M., Liu, J., Wang, Y., Wu, X., Huang, K., et al. (2021). Genome mining reveals the genes of carboxypeptidase for OTA-detoxification in Bacillus subtilis CW14. Int. J. Biol. Macromol. 186, 800–810. doi: 10.1016/j.ijbiomac.2021.07.085
Xu, L., Sun, X., Wan, X., Li, H., Yan, F., Han, R., et al. (2020). Identification of a Bacillus amyloliquefaciens H6 thioesterase involved in zearalenone detoxification by transcriptomic analysis. J. Agric. Food Chem. 68, 10071–10080. doi: 10.1021/acs.jafc.0c03954
Xu, H., Wang, L., Sun, J., Wang, L., Guo, H., Ye, Y., et al. (2022). Microbial detoxification of mycotoxins in food and feed. Crit. Rev. Food Sci. Nutr. 62, 4951–4969. doi: 10.1080/10408398.2021.1879730
Yadav, R., Yadav, P., Singh, G., Kumar, S., Dutt, R., and Pandey, A. (2021). Non infectious causes of abortion in livestock animals-a review. Int. J. Livest. Res. 11, 1–13. doi: 10.5455/ijlr.20201031015650
Yan, X., Chen, K., Jia, H., Zhao, Q., Du, G., Guo, Q., et al. (2024). Infiltration of porcine pancreatic lipase into magnetic hierarchical mesoporous UiO-66-NH2 metal–organic frameworks for efficient detoxification of patulin from apple juice. Food Chem. 431:137172. doi: 10.1016/j.foodchem.2023.137172
Yang, Q., Pang, B., Solairaj, D., Hu, W., Legrand, N. N. G., Ma, J., et al. (2021). Effect of Rhodotorula mucilaginosa on patulin degradation and toxicity of degradation products. Food Additiv Contamin Part A 38, 1427–1439. doi: 10.1080/19440049.2021.1923821
Yang, C., Song, G., and Lim, W. (2020). Effects of endocrine disrupting chemicals in pigs. Environ. Pollut. 263:114505. doi: 10.1016/j.envpol.2020.114505
Yang, P., Xiao, W., Lu, S., Jiang, S., Jiang, S., Chen, J., et al. (2022). Characterization of a Trametes versicolor aflatoxin B1-degrading enzyme (TV-AFB1D) and its application in the AFB1 degradation of contaminated rice in situ. Front. Microbiol. 13:960882. doi: 10.3389/fmicb.2022.960882
Yang, H., Yan, R., Li, Y., Lu, Z., Bie, X., Zhao, H., et al. (2022). Structure–function analysis of a quinone-dependent dehydrogenase capable of deoxynivalenol detoxification. J. Agric. Food Chem. 70, 6764–6774. doi: 10.1021/acs.jafc.2c01083
Yao, Y., and Long, M. (2020). The biological detoxification of deoxynivalenol: a review. Food Chem. Toxicol. 145:111649. doi: 10.1016/j.fct.2020.111649
Yu, J., and Pedroso, I. R. (2023). Mycotoxins in cereal-based products and their impacts on the health of humans, livestock animals and pets. Toxins 15:480. doi: 10.3390/toxins15080480
Yu, X., Tu, T., Luo, H., Huang, H., Su, X., Wang, Y., et al. (2020). Biochemical characterization and mutational analysis of a lactone hydrolase from Phialophora americana. J. Agric. Food Chem. 68, 2570–2577. doi: 10.1021/acs.jafc.9b05853
Zhang, Z., Xu, W., Wu, H., Zhang, W., and Mu, W. (2020). Identification of a potent enzyme for the detoxification of zearalenone. J. Agric. Food Chem. 68, 376–383. doi: 10.1021/acs.jafc.9b06223
Zhang, H., Yang, C., Du, J., Huang, J., He, J., and Zhang, W. (2022). Efficient and safe detoxification of aflatoxin B1 in peanut oil by synergistic modification of montmorillonite with histidine and acid. LWT 170:114056. doi: 10.1016/j.lwt.2022.114056
Zhang, H., Zhang, Y., Yin, T., Wang, J., and Zhang, X. (2019). Heterologous expression and characterization of a novel ochratoxin a degrading enzyme, N-acyl-L-amino acid amidohydrolase, from Alcaligenes faecalis. Toxins 11:518. doi: 10.3390/toxins11090518
Zhang, Y., Zhao, Q., Ngolong Ngea, G. L., Godana, E. A., Yang, Q., and Zhang, H. (2024). Biodegradation of patulin in fresh pear juice by an aldo-keto reductase from Meyerozyma guilliermondii. Food Chem. 436:137696. doi: 10.1016/j.foodchem.2023.137696
Zheng, X., Wei, W., Zhou, W., Li, H., Rao, S., Gao, L., et al. (2021). Prevention and detoxification of patulin in apple and its products: a review. Food Res. Int. 140:110034. doi: 10.1016/j.foodres.2020.110034
Zhou, J., Zhu, L., Chen, J., Wang, W., Zhang, R., Li, Y., et al. (2020). Degradation mechanism for zearalenone ring-cleavage by zearalenone hydrolase RmZHD: a QM/MM study. Sci. Total Environ. 709:135897. doi: 10.1016/j.scitotenv.2019.135897
Keywords: mycotoxins, enzymes, detoxification, degradation, fungi, mechanism
Citation: Liu M, Zhang X, Luan H, Zhang Y, Xu W, Feng W and Song P (2024) Bioenzymatic detoxification of mycotoxins. Front. Microbiol. 15:1434987. doi: 10.3389/fmicb.2024.1434987
Received: 19 May 2024; Accepted: 08 July 2024;
Published: 18 July 2024.
Edited by:
Pierina Visciano, University of Teramo, ItalyReviewed by:
Ye Tian, Chinese Academy of Sciences (CAS), ChinaCopyright © 2024 Liu, Zhang, Luan, Zhang, Xu, Feng and Song. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Song, c29uZ3BlbmdAbGN1LmVkdS5jbg==; c29uZ3BlbmdAenltZXJjay5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.