
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 13 November 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1434554
 Qiannan Li1,2
Qiannan Li1,2 Lianfu Yang1,2
Lianfu Yang1,2 Ting Zhu1,2
Ting Zhu1,2 Xiyv Yv1,2
Xiyv Yv1,2 Boxin Zhang1,2
Boxin Zhang1,2 Hongzhe Li2
Hongzhe Li2 Junjie Hao2
Junjie Hao2 Lei Zhang1,2
Lei Zhang1,2 Pengzhang Ji1,2
Pengzhang Ji1,2 Jiahong Dong1,2*
Jiahong Dong1,2*Background: Disporopsis, a member of the Liliaceae family and a perennial herb, is predominantly cultivated in southwestern and southeastern China. Its rhizome, referred to as Zhugenqi, serves as a traditional Chinese medicinal herb for the treatment of bone injuries. However, viral diseases have emerged as a significant challenge in the cultivation of Disporopsis.
Objective: The aim of this study was to identify and characterize viruses present in diseased samples of Disporopsis spp. using high-throughput sequencing (HTS) and reverse transcription-polymerase chain reaction (RT-PCR) to enhance the understanding of the virome associated with Disporopsis and to inform diagnostic and control strategies for viral diseases in this plant.
Methods: Diseased samples of Disporopsis spp. were subjected to HTS and RT-PCR for virus identification. A total of five viruses were detected, including three novel viruses and two known viruses. The novel viruses were provisionally named Disporopsis chlorotic stripe virus (DCSV), Disporopsis pernyi-associated partitivirus (DaPTV), and Disporopsis pernyi-associated lispi-like virus (DaLV). Sequence identity and phylogenetic analyses were performed to confirm the novelty and taxonomic placement of these viruses.
Results: DCSV exhibited polyprotein sequence identities ranging from 47.6% to 83.6% with other potyviruses, with the highest identity (83.6%) shared with Polygonatum kingianum virus 5 (PKgV5). DaLV shared an amino acid sequence identity of 34.59% with maize suscal virus (MSV), and DaPTV shared an identity of 76.18–85.10% with Paris alphapartitivirus (ParAPV). Phylogenetic analyses supported the potential classification of the three novel viruses as new members of their respective genera. Two isolates of polygonatum mosaic-associated virus 1 (PMaV1) were identified in Disporopsis for the first time, showing divergences of 96.33% and 98.86% from existing isolates. RT-PCR analysis of 67 Disporopsis field samples collected from four cities in China revealed that more than half of the samples tested positive for at least one of the five viruses. PMaV1 and DaLV were the most prevalent, detected in 22 and 34 out of the 67 samples, respectively. Other viruses were detected at low rates and/or had limited distribution.
Conclusion: This study provides insights into the virome infecting Disporopsis and offers valuable information for the diagnosis and control of viral diseases in this plant. The identification of five viruses, including three potential new members of their respective genera, contributes to the understanding of the viral threats to Disporopsis cultivation.
Plants belonging to be genus Disporopsis Hance mainly grow in China and several countries in Southeast Asia such as Laos, Myanmar, Thailand and Vietnam (Ghorbani et al., 2011; Sydara et al., 2014; Nguyen et al., 2019; Sarapan et al., 2023). The rhizome or the entire plant is used as a traditional remedy for easing thirst, nourishing Yin, and moisturizing the body by minority groups in southwest China (Long and Li, 2004; Zeng et al., 2023). Pharmacological studies have shown that the rhizome of Disporopsis spp. is rich in bioactive substances such as steroidal saponins, rutin, luteolin, quercetin, and betulinic acid, which have antioxidant, antimicrobial, anti-inflammatory, antibiosis, and anticancer properties (Lin et al., 2014; Wang et al., 2015). The rhizome extracts also have biological control effects on Pseudoperonospora cubensis, Phytophthora infestans and Podosphaera xanthii (Zhu et al., 2018; Han et al., 2022).
High-throughput sequencing (HTS), or deep sequencing techniques, have significantly helped to advances in discovery, diagnostics, and evolutionary studies in virosphere (Roossinck et al., 2015; Roossinck, 2017; Bernardo et al., 2018; Zhang et al., 2018). Many novel and known viruses have been identified in plants, including both wild and cultivated varieties, through the use of high-throughput sequencing (HTS). This is particularly true for medicinal plants such as wild citrus (Kreuze et al., 2009; Zhang and Wang, 2020), greenhood orchids (Pterostylidinae) (Bolger et al., 2014), Paris polyphylla (Grabherr et al., 2011), Mallotus japonicus (Chen and Chen, 2002), and Aconitum carmichaelii (Yang, 2022). The obtained comprehensive genomic data has enabled the virus’ evolution, phytopathology, and epidemiology, as well as disease management strategies, to be further studied (Kreuze et al., 2009).
With the development and industrialization of Chinese herbal medicine, the Disporopsis planting areas have been expanded. However, plant diseases have become an important bottleneck to Disporopsis production (Zhang and Wang, 2020). In recent years, Disporopsis plant leaves displaying virus-like disease symptoms, such as chlorosis, leaf rolling and yellowing, have also been discovered during field surveys. However, there have not yet been any reports about Disporopsis-infecting viruses. In this study, we used HTS in combination with RT-PCR to identify Disporopsis-infecting viruses in China. Three novel Disporopsis-infecting viruses belonging to the genera Potyvirus, Alphapartivirus and Lispiviridae, two different isolates belonging to polygonatum mosaic-associated virus 1 and one isolate of Paris mosaic necrosis virus were identified and characterized. The resulting virus genome information for Disporopsis plants have also been made available for future investigations on the evolution, genetic diversity and epidemiology of Disporopsis-infecting viruses.
Entire plants of Disporopsis were collected from four regions in China, including Kunming city in the Yunnan province, Wenchuan city in the Sichuan province, Enshi city in the Hubei province, and Chongqing city. Additionally, the disease incidence in the field was estimated and recorded.
A total of 67 samples displaying virus-like or unusual symptoms were collected, including 13 D. pernyi samples from Kunming, 19 D. pernyi samples from Wenchuan, 15 D. fuscopicta samples from Enshi, and 20 Disporopsis sp. seedling samples (species was not identified) from Chongqing. The Disporopsis samples exhibited disease symptoms like leaf stripe, leaf-roll, mottling, and chlorosis. The plants were transplanted into a greenhouse at the Yunnan University of Chinese Medicine for later sampling and testing. Part of the diseased leaves were stored at −80°C. Samples were labeled with the plant species name, symptoms, collection location, and collection date (Supplementary Table S1).
Seedlings of Nicotiana tabacum var. K326 were obtained through seed germination. Seedlings of Disporopsis spp. and Polygonatum kingianum were purchased from plantations in Kunming in the Yunnan province and Wenchuan county in the Sichuan province, and they were grown in a growth chamber with a photoperiod of 25°C for 16 h (day)/18°C for 8 h (/night). Mechanical virus inoculation was carried out as follows: the inoculum was prepared by grinding infected leaf tissue with a mortar and pestle in phosphate buffer (mixing 8.0 mL of 1.0 mol-L−1 NaH2PO4 and 42 mL of 1 mol-L−1 Na2HPO4 solution); the leaves of disease-free plants were then lightly dusted with carborundum, and the virus solution was rubbed onto the leaves using a cotton swab. For mock-inoculated control plants, leaves were rubbed with the phosphate buffer alone. The symptomatic and mock-inoculated leaves were then collected at 7 dpi and stored at −80°C for the RT-PCR assay.
The samples from Kunming, Yunnan province, exhibiting chlorotic stripe symptoms were sent to OE Biotech Co., Ltd. (Shanghai, China) for HTS. The total RNA was extracted using TRIzol (Ambion, Hillsboro, Oregon, United States) following the manufacturer’s instructions. Ribosomal RNA was depleted using the RiboZero kit (Illumina, San Diego, CA, United States) according to the manufacturer’s instructions. cDNA libraries were synthesized using the TruSeq Stranded Total RNA Kit with Ribo-Zero Gold (Illumina). The obtained cDNA libraries were paired-end deep sequenced reads using the Illumina HiSeq 2500. Raw data (raw reads) were processed using Trimmomatic by removing the reads containing ploy-N and the low-quality reads (Bolger et al., 2014). After removing the adaptor sequences, the clean reads were assembled into expressed sequence tag contigs and de novo assembled into transcripts using Trinity 2.4 version in paired-end mode (Grabherr et al., 2011). For subsequent analysis, the longest transcript was designated as an unigene based on the similarity and length of the sequence. The unigenes were functionally annotated by querying them against the NCBI nonredundant (NR), SwissProt, and Clusters of orthologous groups for eukaryotic complete genomes (KOG) databases using Blastx with a threshold E-value of 10−5.
The total RNA was extracted using the Eastep® Super Total RNA Extraction Kit according to the manufacturer’s instructions. Specific primers for each virus were designed based on the transcriptome contigs (Table 1) and were then used in RT-PCR to amplify the corresponding viral fragments. RT-PCR was conducted using a PrimeScriptTM One-step RT-PCR Kit ver.2 (TaKaRa Biotechnology Co., Ltd., Dalian, China). The 25 μL RT-PCR reaction contained 3 μL of total RNAs, 0.5 μL of each primer (10 μM), 12.5 μL of 2 × 1 Reaction Mix, 1 μL of Enzyme Mix and 7.5 μL of distilled water. The thermal cycling conditions were 1 cycle of reverse transcription at 50°C for 30 min and denaturation at 94°C for 2 min, 35 cycles of amplification at 94°C for 30 s, annealing temperature for 30 s and 72°C for 60 s, and a final extension of 72°C for 10 min. Sanger sequencing of the target fragment clones was subsequently performed to verify the accuracy of the new and known viruses identified by HTS. The complete genome sequence of Polygonatum mosaic virus 1-Disporopsis 1 (PMaV1-D1) was obtained by assembling the Sanger sequenced RT-PCR and 5′ RACE amplified reads, and the partial genome sequences of the DCSV, DaLV, and PMaV1-D2 isolates were assembled from the RT-PCR amplified sequenced reads. The viral full-length primers were designed based on the allele sequences of each target virus (new virus or known virus) obtained in this study (Supplementary Tables S2–S5). Based on a modified 5′RACE method by Chen and Chen (2002) and Yang (2022), the first-strand cDNA was first synthesized by reverse transcription using super M-MuLV Reverse Transcriptase from Sangong Bioengineering Co. (Shanghai). The total RNA was converted into cDNA in a 5 μL reaction that consisted of 1 μL total RNA, 1 μL the 3′-specific primer ZHM1, 1 μL of dNTPs, and 2 μL DEPC. The reaction was incubated at 65°C for 5 min and then left on ice for 2 min. Next 2 μL 5 × Super M-MLV Buffer, 1 μL of Solution I, 0.5 μL of RNase Inhibitor, 0.5 μL of Super M-MLV Reverse, and 1 μL of DEPC were mixed with the cDNA solution to form a 10-μL system solution, and the reaction was incubated at 42°C for 60 min and then 80°C for 15 min. Subsequently, a PCR was performed using the cDNA and the 3′-specific primer ZHM1. The 50 μL reaction contained 0.5 μL of cDNA, 0.5 μL of Ex Taq, 0.5 μL of 5′-end specific primers, 0.5 μL of ZHM1, 0.5 μL of dNTPs, and 38 μL of DEPC. The thermal cycling conditions were as follows: 2 min at 94°C, 35 cycles of 30 s at 94°C, 30 s at TM, and 72°C for 60 s, and finally, an extension period of 72°C for 10 min. The products were recovered, cloned and Sanger sequenced as described above.
The sequence identities of the isolate viruses were analyzed alongside sequences of published viruses in the NCBI database using DNAMAN vers.7.0 (LynnonBiosoft, United States). Open reading frames (ORFs) were predicted using ORF Finder on the NCBI website.1 The phylogenetic relationships among the amino acid sequences were analyzed in Mega X. The phylogenetic trees were constructed using the maximum likelihood algorithm and LG model with 1,000 bootstrap replicates (Kumar et al., 2018).
The virus-infected Disoropsis plants displayed the following symptoms: chlorotic stripes along the leaf vein, top leaf rolling, and mottle and mosaic symptoms on leaves (Figure 1). Four viruses except DaPTV were detected in (A), DSCV and DpLV were detected in (B), all viruses were detected in (C), DpLV and DaPTV were detected in (D,E) and DCSV and PMaV1-D1 were detected in (F). The most severe symptoms appeared between June and September in Kunming. No symptoms were observed on healthy plants.

Figure 1. Typical symptoms of virus diseases on leaves of Disporopsis plants: (A) chlorotic stripe, (B) leafroll, rugose and mottle, (C) mottle, (D) yellow, (E) mild mottle, (F) rugose. (A,B) D. longifolia from Yunnan Province, (C–F).
HTS was performed on the leaves from one Disporopsis pernyi plant that showed chlorotic stripe symptoms in Kunming (E102°49′16″, N24°50′9″). A total of 24.28 M raw reads were obtained, with a total length of 2,201,943 bp and an average GC content of 61.14%. After quality trimming and size filtering, approximately 24.06 M clean reads remained. Subsequently, the clean reads were de novo assembled into 3,532 contigs and annotated by using BLASTx to search the GenBank database and identify the potential viral agents. Next, seven contigs were mapped to viral genome sequences in GenBank (Table 2). One 9,685 nucleotide long contig had a high amino acid sequence identity of 82% with Polygonatum kingianum virus 5 (GenBank accession number: MN873571.1), which belonged to the genus Potyvirus. Two of 9,635 nt and 8,863 nt contigs had high aa sequence identities of more than 93% with polygonatum mosaic-associated virus (PMaV1, GenBank accession number: OP380926.1), One contig of 7,531 nt in length had a high amino acid sequence identity of 34.59% with the ORF4-encoded protein of maize suscal virus (GenBank accession number: MZ270532.1), which belonged to the Lispiviridae family. Lastly, two contigs of 2010 nt and 1873 nt in length had high amino acid sequence identities with RdRp and CP of Paris alphapartitivirus (GenBank accession number: OL960006.1 and OL960007.1). RT-PCR assays and Sanger sequencing further confirmed the HTS results. The viruses in the HTS-tested D. pernyi included three novel viruses and two PMaV1 isolates. Three novel viruses were temporarily named Disporopsis chlorotic stripe virus (DCSV), Disporopsis pernyi associated partitivirus (DaPTV) and Disporopsis pernyi associated lispi-like virus (DaLV), respectively.
The near complete genomic sequences of Disporopsis chlorotic stripe virus (DCSV, GenBank accession number: PP691760) was determined to be 9,640 nt long using RT-PCR, excluding the 3′-terminal poly(A) tail. The genome organization of DCSV was identical to that of other members of the Potyvirus genus in the Potyviridae family and contained a large ORF encoding a polyprotein of 3,090 aa residues. The 5′ and 3′ untranslated regions (UTR) were 136 nt and 231 nt long, respectively. Nine highly conserved proteolytic cleavage sites in the polyprotein were identified by comparing them with the consensus protease recognition motifs from other potyviruses. Ten putative mature proteins were identified from these cleavage sites: P1 (321 aa), HC-Pro (458 aa), P3 (351 aa), 6K1 (52 aa), CI (589 aa), 6K2 (53 aa), VPg (193 aa), NIa-Pro (243 aa), NIb (516 aa) and CP (314 aa) (Figure 2). The small ORF PIPO within the P3 of potyviruses was also identified due to the presence of GAAAAAAA in the DCSV genome sequence (2,932–2,939 nt).
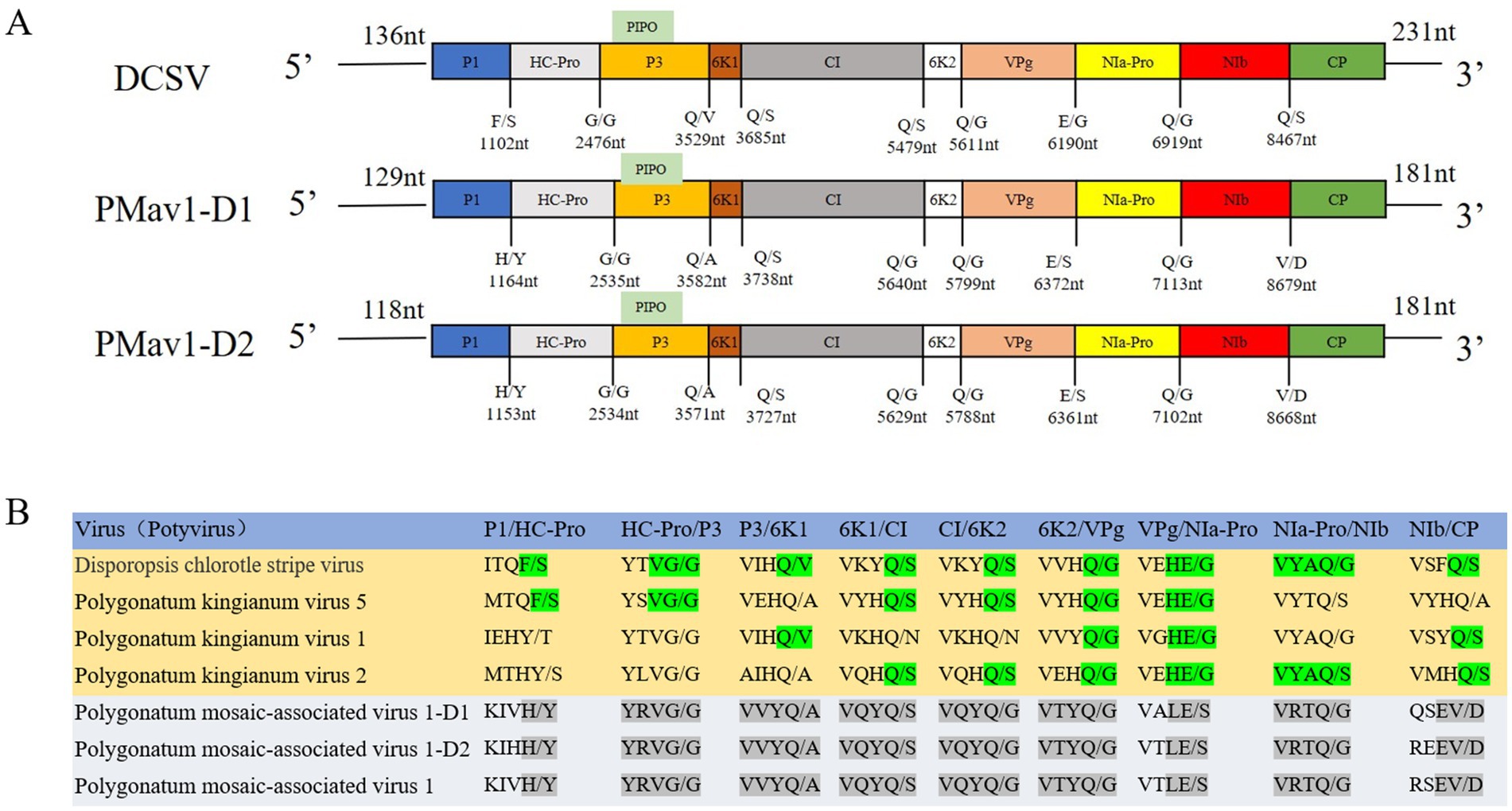
Figure 2. (A) The cleavage sites of DCSV, PMaV1-D1, and PMaV1-D2, along with their corresponding proteins. These sites were sequentially cleaved into nine mature proteins, including P1, HC-Pro, P3, 6K1, CI, 6K2, VPg, NIa-Pro, NIb, CP. (B) PMaV1 isolates protein cleavage sites and corresponding proteins.
Furthermore, eight highly conserved motifs were found in the polyprotein amino acid sequences. The high level of conservation among these sequences demonstrated that these proteins likely perform essential functions in viruses and therefore are retained over the course of viral evolution (Zhang et al., 2023). These include the 630PTK632 (aphid transmission) and 501FRNK504 motifs in HC-Pro; 1266GAVGSGKST1274 (NTP binding), 1355DECH1358 (potential helicase activity), and 1484VATNIIENGVTL1495 (potential helicase activity) motifs in CI, 2612GNNSGQPSTVVDNT2625 (RNA-dependent polymerase activity) and 2656GDD2658 (RNA-dependent polymerase activity) motifs in NIb, and 2858DAG2860 (aphid transmission) motif in CP (Figure 3) (Valli et al., 2007; Puli’uvea et al., 2017; Worrall et al., 2019; Zheng et al., 2008; Chung et al., 2008). The numbers in superscript represent the positions of the sequences along the DCSV genome.
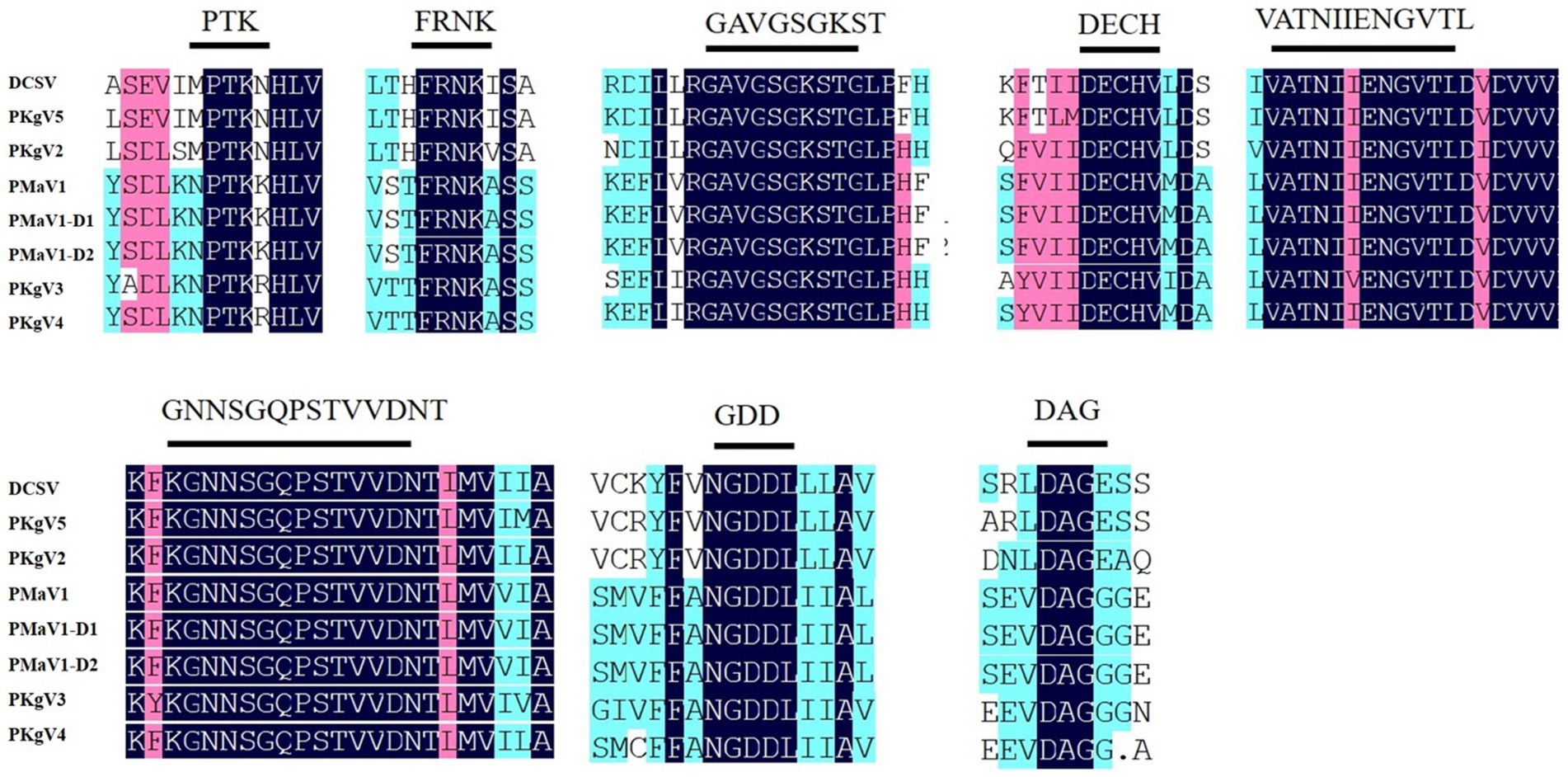
Figure 3. Conserved motifs among DCSV, PMaV1-D1, PMaV1-D2 and other potyviruses. These polyproteins contain the typical highly conserved potyviral motifs. The shaded black areas indicate conserved amino acid residues, respectively.
The genome sequences of DCSV and other potyviruses listed in Supplementary Tables S6, S7, S10, S11, except for PKgV5, shared nucleotide identities of 53.10–69.60% and amino acid identities of 48.10–75.70%. Five proteins, P1 (14.10–54.00%), HC-Pro (46.30–71.10%), P3 (25.50–64.10%) and 6K2 (41.50–58.50%), VPg (51.90–79.40%), had the amino acid identities of less than 80% with other potyviruses, other five proteins, 6K1 (42.30–82.70%), CI (55.70–80.80%), NIa-Pro (52.70–83.10%), NIb (58.30–83.40%) and CP (59.80–86.90%) had amino acid identities of more than 80% with other potyviruses. These values were all under the current species demarcation threshold (Supplementary Tables S6, S8) for the Potyviridae family (Shukla and Ward, 1989; Adams et al., 2005). Sequence alignment comparisons showed that the polyprotein and CP of DCSV had more than 82% amino acid identity with PKgV5, which is beyond the current species demarcation threshold (Supplementary Table S6). However, given that the genome sequence of PKgV5 was only confirmed using the assembled HTS contig data, without using RT-PCR, we propose that only DSCV is a novel potyvirus, and name it Disporopsis chlorotic stripe virus.
The phylogenetic analyses were performed using the deduced polyprotein sequences of DCSV and 42 other potyviruses available in GenBank (Figure 4). The maximum likelihood tree placed DCSV within the Potyvirus genus and into the TuMV subgroup. DCSV was most closely related to putative polyprotein sequences belonging to an unconfirmed Polygonatum kingianum virus 5 (PKgV5, QVN46485.1) and Polygonatum kingianum virus 2 (QIJ96720.1).
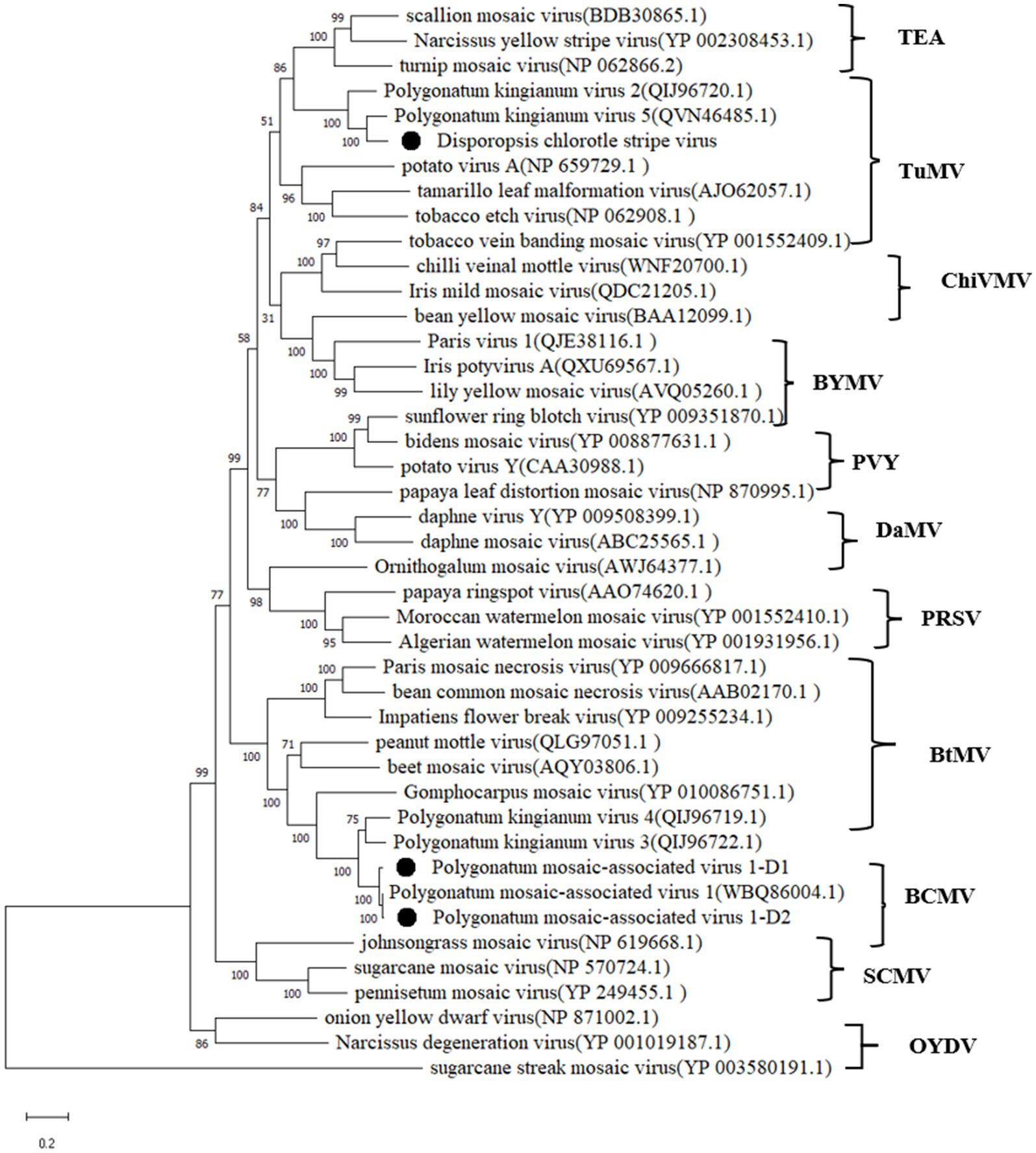
Figure 4. Maximum-likelihood tree based on the deduced polyprotein sequences of DCSV, PMaV1-D1, PMaV1-D2 and representative members of the genus Potyvirus. Each of the subgroups is indicated by abbreviation of the representative virus. Bootstrap analysis was applied using 1,000 bootstrap replicates. The scale bar representing a genetic distance of 0.2. Sugarcane streak mosaic virus, a member of the genus Poacevirus, was used as an outgroup.
PMaV1 is a potyvirus that was first identified in Polygonatum cyrtonema Hua (Li et al., 2023). In this study, two PMaV1 isolates were detected in Disporopsis, and were named PMaV1-Disporopsis 1 (GenBank accession number: PP691759) and PMaV1-Disporopsis 2 (GenBank accession number: PP691758), respectively. The complete genome sequences were determined to be 9,637 nt and 9,626 nt in length, excluding the 3′-terminal poly(A) tail, using RT-PCR. PMaV1-Disporopsis 1 and PMaV1-Disporopsis 2 encoded a polyprotein of 3,109 aa and 3,084 aa, respectively. The 5′and 3′-UTRs of PMaV1-Disporopsis 1 were 129 nt and 181 nt. The 5′ and 3′-UTRs of PMaV1-Disporopsis 2 were 118 nt and 181 nt (Figure 2). The polyproteins of the two PMaV1 isolates had 96.33 and 98.86% identity with those of a PMaV1 isolated from Polygonatum cyrtonema, respectively. All three PMaV1 isolates shared the same conserved motifs (Li et al., 2023). The size difference in the polyproteins of the two Disporopsis isolates of PMaV1 was due to the deletion of the CP N-terminal. The CP of PMaV1-Disporopsis 1 and Disporopsis 2 consisted of 260 aa and 183aa residues, respectively (Supplementary Tables S8, S10, S11). Nine other proteins within the two isolates were identical in size (Figure 2). Moreover, PIPO ORF was identified due to the presence of GAAAAAA in PMaV1-Disporopsis 1 (nt positions 2,992–2,998) and PMaV1-Disporopsis 2 (nt positions 2,981–2,987), respectively (Figure 2).
Paris mosaic necrosis virus (a potyvirus) was first reported in Paris polyphylla var. yunnanensis, which belongs to the Liliaceae family, and subsequently detected in Polygonatum kingianum (Lan et al., 2018; Hu et al., 2022). PMNV was also detected in four D. pernyi samples from Kunming using RT-PCR with the virus specific primer pairs targeting NIb, CP, and the 3′-UTR sequence of PMNV. The RT-PCR products were cloned and sequenced. Sequence analysis showed that the CP gene of the four PMNV Disporopsis isolates was 813 nt long and shared 86.4–92.4% and 90.4–95.8% of their nucleotide and amino acid sequences, respectively, with PMNV-cn isolate from P. polyphylla var. yunnanensis.
A 7,531 nt contig was highly homologous to the RdRp of maize suscal virus (GenBank accession number: MZ270532), with a query coverage of 80.00% and an amino acid identity of 34.59%, as well as to other lispi-like viruses with a query coverage of about 50.00% and amino acid identity of about 28% (Supplementary Table S9). Sequence analysis showed that the RdRP of Disporopsis pernyi-associated lispi-like virus possessed the conserved motifs: Mononegavirales RNA-dependent RNA polymerase (pfam00946) and Mononegavirales mRNA-capping region V (pfam14318) using NCBI Conserved Domain Search2 (Wang et al., 2023). The phylogenetic tree based on the RdRP from lispi-like viruses showed that Disporopsis pernyi associated lispi-like virus was clustered in a subbranch with maize suscal virus (Figure 5). Using the specific primer pairs derived from this contig (Supplementary Figure S1), we detected this lispi-like virus in 32 Disporopsis samples using RT-PCR, there by indicating that this virus (putatively named Disporopsis pernyi lispi-like virus, GenBank accession number: PP803564) occurred frequently in Disporopsis.

Figure 5. Maximum-likelihood tree based on the deduced polyprotein sequences of Disporopsis pernyi lispi-like virus representative members of the family Lispiviridae. Each of the subgroups is indicated by abbreviation of the representative virus. Bootstrap analysis was applied using 1,000 bootstrap replicates. The scale bar representing a genetic distance of 0.5.
RNA1 and RNA2 of Disporopsis pernyi-associated partitivirus (DaPTV, GenBank accession number: PP803563 and PP803565) were determined to be 2,010 nt and 1,873 nt in length. The genome structure of DaPTV was identical to that of other alphapartitiviruses (Figure 6). RNA1 encoded an RdRp of 586aa (190–1,950 nt) and RNA2 encoded a CP of 490 aa (169–1,641 nt), Both RNA1 and RNA2 had high amino acid sequence identities of 85 and 76.1% with those of Paris alphapartitivirus (GenBank accession number: OL960006.1 and OL960007.1) (Table 3). These percent values were all under the current species demarcation threshold (≤ 90% aa sequence identity in the RdRP, and ≤80% aa sequence identity in the CP) (Vainio et al., 2018) but within the Alphapartitivirus genus (Figure 7). Thus, we suggest that DaPTV is a new member of the Alphapartitivirus genus. To confirm the presence of the virus in Disporopsis, RT-PCR was conducted using specific primer sets (as listed in Table 1) that target RNA1 and RNA2. The sequences obtained from the RT-PCR products were then analyzed and confirmed to match the virus’s identity. DaPTV-RNA1 primers were used to detect 13 samples from Kunming, 9 samples were found to have bands, which proved the existence of DaPTV (Figure 8).
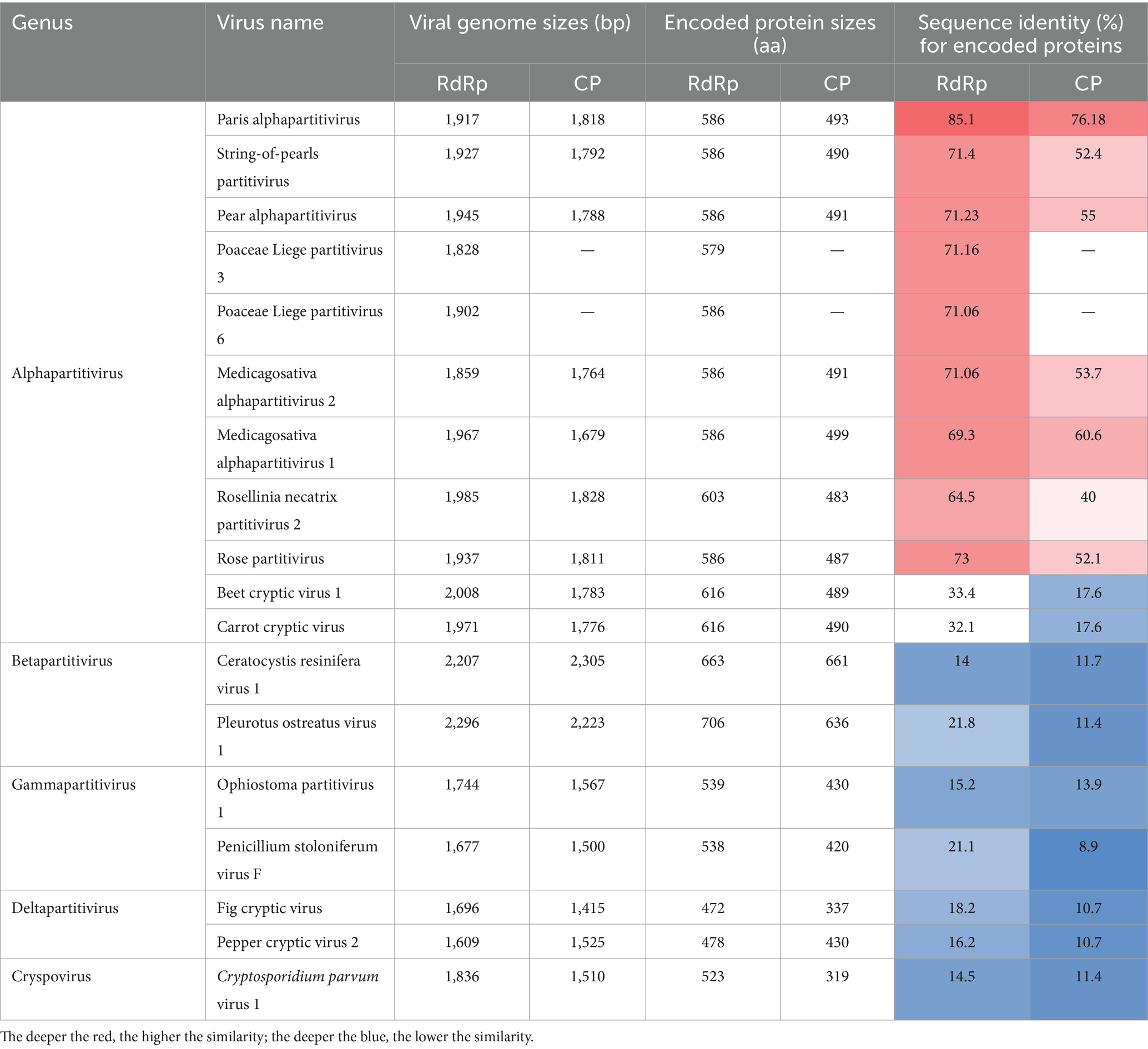
Table 3. Amino acid (aa) sequence identities of Disporopsis pernyi associated partitivirus and members of the family Partitiviridae.
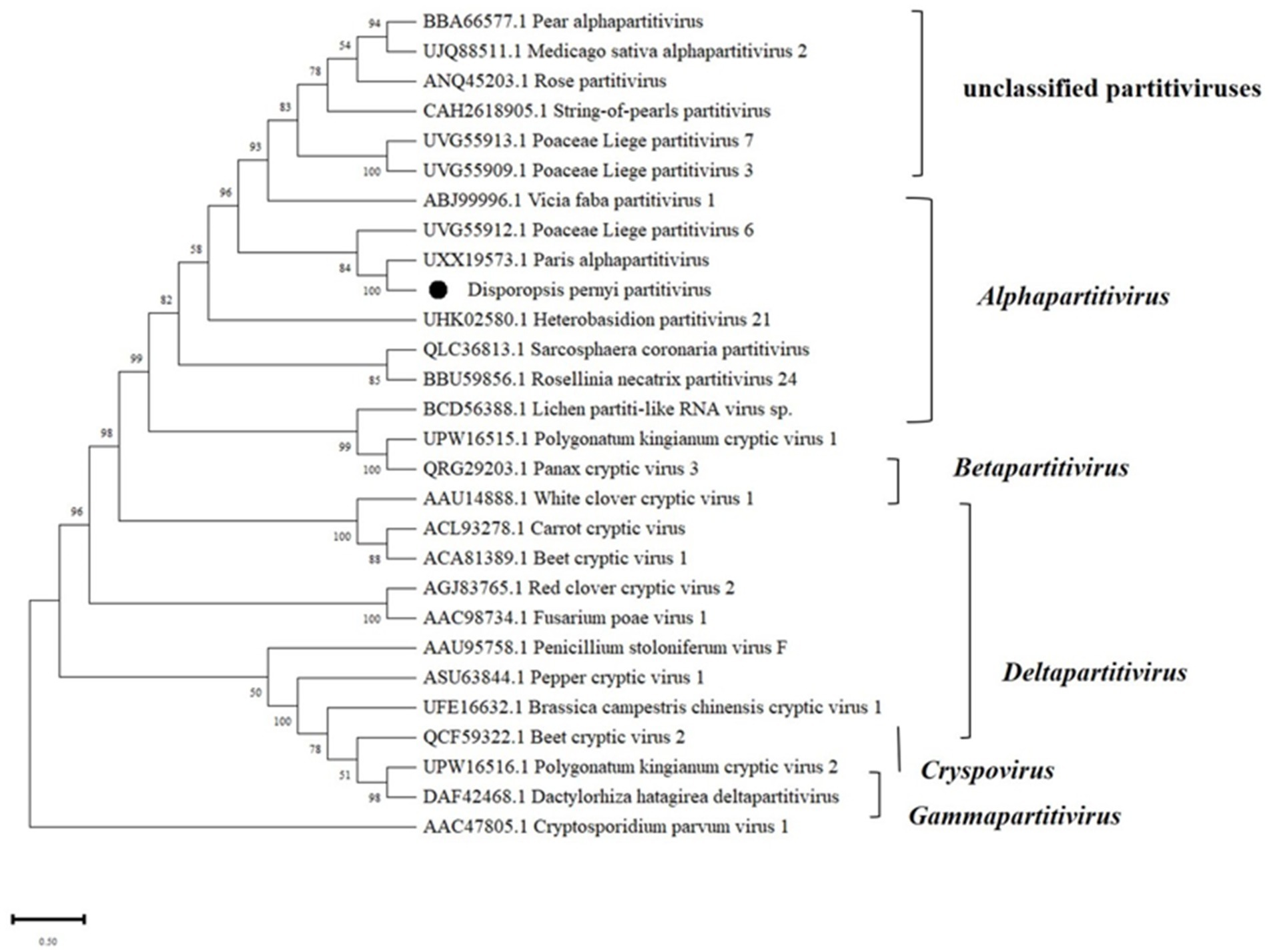
Figure 7. Maximum likelihood phylogenetic tree of amino acid sequences of the RdRp of representative members of the family Partitiviridae. Bootstrap probabilities for each branch node were estimated using 1,000 replicates. Phylogenetic tree was constructed using MEGA X software. Cryptosporidium parvum virus 1 was used as outgroup. Viral genome sequences were downloaded from NCBI viral genome database (http://www.ncbi.nlm.nih.gov/genome/viruses/). The scale bar representing a genetic distance of 0.5.
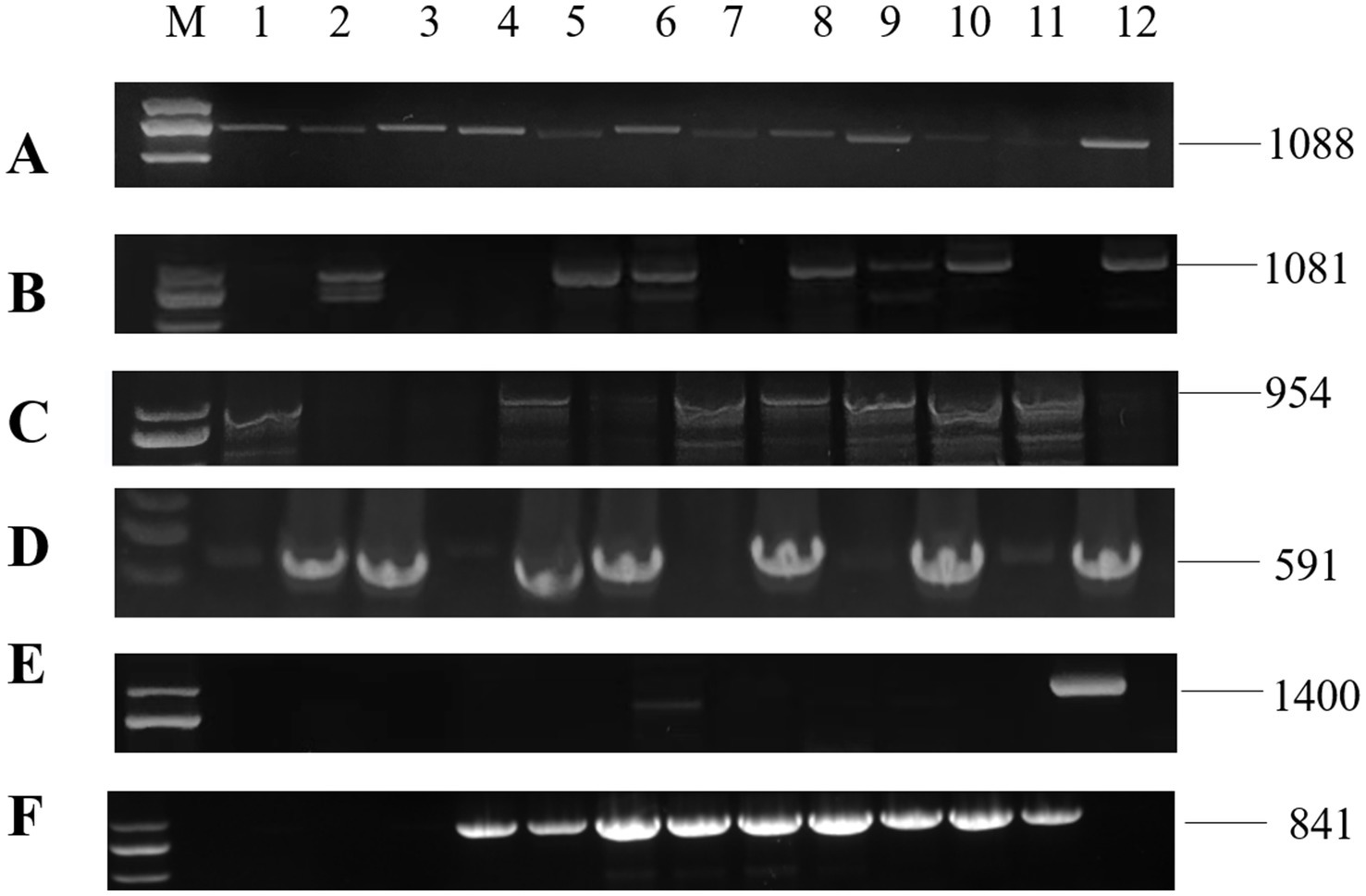
Figure 8. RT-PCR detection for viruses in diseased plants of Disporopsis: (A) DCSV, (B) PMaV1-D1, (C) PMaV1-D2, (D) DpLV, (E) PMNV, (F) DaPTV, Lanes (M) DNA marker, (D-KM1-12) diseased samples.
In order to test the infectivity of DCSV, PMaV1 and PMNV in Disporopsis, the viruses were extracted from three virus-infected leaves exhibiting chlorotic and yellow stripe, and mottle symptoms and were mechanically inoculated into virus-free seedlings of Disporopsis spp., Polygonatum kingianum and Nicotiana tabacum var. K326. At 7 days post-inoculation (dpi), the leaves of the inoculated Disporopsis plants displayed the characteristic chlorotic stripe symptoms, and DCSV, PMaV1, and PMNV were detected using RT-PCR. The leaves of the P. kingianum plants displayed the characteristic yellow and necrotic spot symptoms, and DCSV and PMaV1 were detected using RT-PCR (Figure 9). The leaves of the N. tabacum var. K326 plants displayed crinkling and mild mottle symptoms, and DCSV and PMNV were detected using RT-PCR. Thus the results indicated that DCSV, PMaV1, and PMNV could effectively infect Disporopsis through the mechanical inoculation (Figure 9; Supplementary Figure S2).
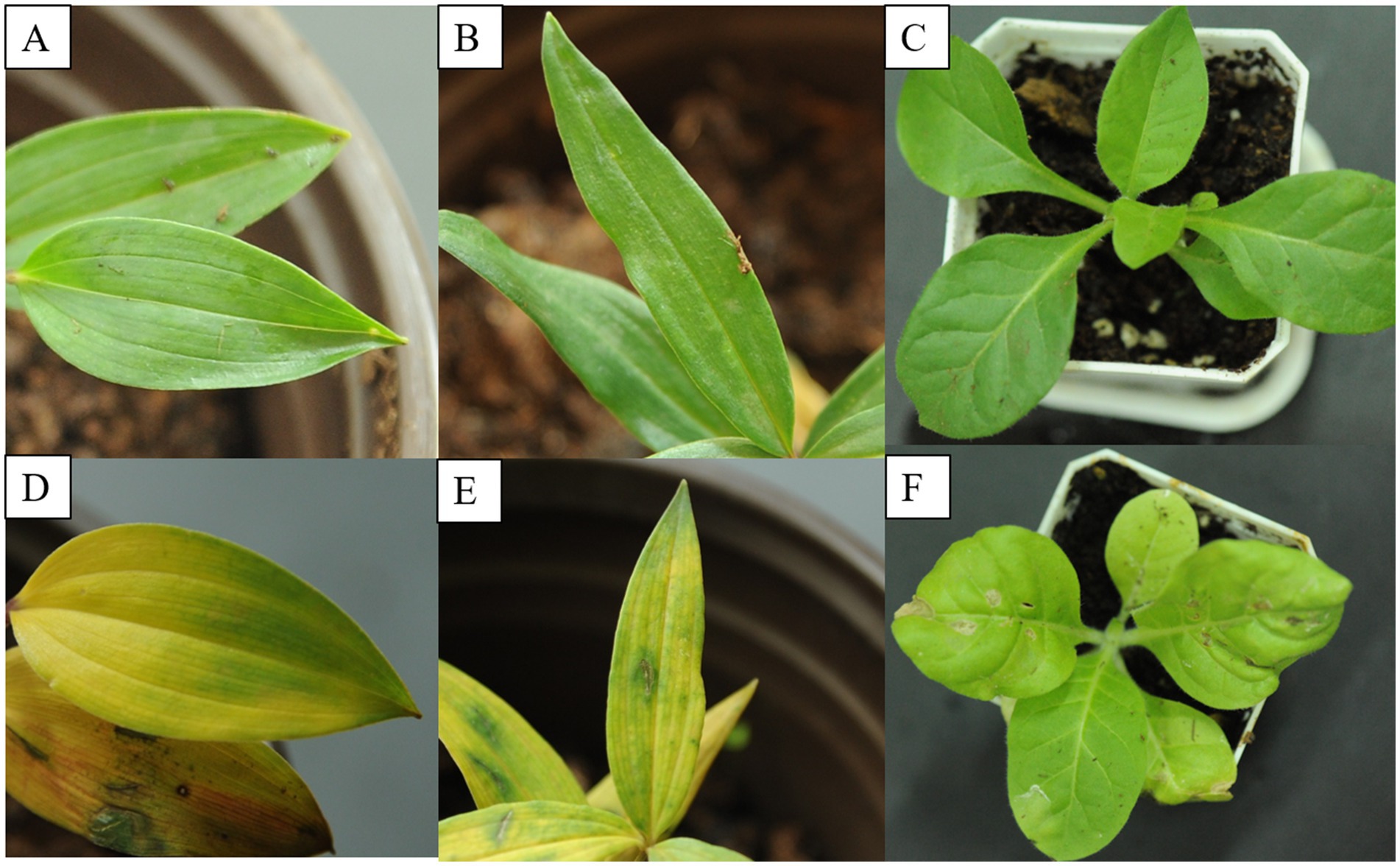
Figure 9. Infectivity of DCSV, PMaV1 and PMNV. (A–C) Represents virus-free seedling of D. pernyi, P. kingianum and N. tabacum var. K326, respectively. (D–F) Indicates the symptomatic leaves of D. pernyi, P. kingianum and N. tabacum var. K326, respectively.
The specific primer pairs (Table 1) that were designed based on the contig sequences obtained with HTS were used in RT-PCR and Sanger sequencing to detect the virus presence in 67 Disporopsis samples. Four viruses were detected: DCSV, the two PMaV1 isolates, DaLV and DaPTV. In addition, we detected Paris mosaic necrosis virus (PMNV), which has been reported to infect Paris sp. and Polygonatum sp. with the specific primer pair that targets its CP. The expected PCR products were recovered and sequenced (Figure 8). The results showed DaLV has the highest incidence rate and was detected in 31 out of 67 samples, followed by PMaV1 Disporopsis 2 isolate in 15 out of 67. PMNV was only detected in four samples from Kunming. Among the 19 samples from the Wenchuan botanical garden in the Sichuan province, only one sample was infected by DaLV, no virus in other 18 samples. All 13 samples from Kunming were infected by DaLV. Moreover, DaPTV was detected in 11 out of 67 samples, including nine samples from Kunming and two samples from Chongqing. Mixed infection was prevalent in the samples from Kunming, with 12 out of 13 being mixed infected. Six viruses including DCSV, two PMaV1 isolate, PMNV, DaLV and DaPTV were detected in 1/13. PMaV1 and DaLV were predominant viruses in the tested samples and were detected in 22 and 34 out of 67 samples, respectively (Table 4; Supplementary Table S12).
Disporopsis spp. have been used as medicinal plants in certain Asian countries (Sydara et al., 2014; Nguyen et al., 2019; Sarapan et al., 2023; Grabherr et al., 2011). The species belonging to the Disporopsis Hance genus include D. pernyi, D. aspersa, D. fuscopicta, D. longifolia, D. jinfushanensis, and D. undulata are native to China (Liang and Tamura, 2000). In the Yunnan and Guizhou provinces in southwest China, minority groups, such as those belonging to the Hani and Miao ethnicities, use the dried rhizome of Disporopsis spp. to cure pustules, cough, asthma and pneumonia (Grabherr et al., 2011; Tian, 2013). HTS provides a powerful tool for detecting the presence of known viruses and discovering novel plant viruses (Roossinck et al., 2015). So far, no viruses have been reported in Disporopsis. This study, used HTS to characterize the virus species in Disporopsis pernyi from Kumming displaying chlorotic stripe symptoms. The analysis of the virome revealed that there were five viruses, including three novel and two known virus species in this single D. pernyi sample. Based on the genome sequence, the three novel viruses were tentatively named Disporopsis chlorotic stripe virus (DCSV), Disporopsis pernyi-associated partitivirus (DaPTV), and Disporopsis pernyi-associated lispi-like virus (DaLV), belonging to Potyvirus, Alphapartitivirus and Lispiviridae, respectively. The two known viruses: Polygontanum mottle associated virus 1 (PMaV1) and Paris mosaic necrosis virus (PMNV), were both first reported in Disporopsis in this study. Two distinct strains of PMaV1, named PMaV1-Disporopsis 1 and PMaV1-Disporopsis 2, were identified within the Disporopsis genus. These strains were distinguished from one another by analyzing their genomic sequences.
To confirm that these five viruses were effectively capable of infecting Disporopsis, we homogenated samples exhibiting different viral symptoms, such as chlorotic stripes and yellow necrotic spots, and inoculated the viruses mechanically into virus-free seedlings of D. pernyi. The RT-PCR results confirmed that DCSV, PMaV1 and PMNV could infect D. pernyi using mechanical inoculation methods, while DaPTV and DaLV could not.
The phylogenetic analysis based on the RdRp amino acid sequences showed that DaLV clustered closely with maize suscal lispivirus and formed a distinct clade. A Blastp search revealed that DaLV and maize suscal lispivirus (accession number: UWX11517.1) shared the highest amino acid sequence identity (34.59%). Both DaLV and MSV shared relatively low identities (28.0%) with sequences belonging to the lispivirids and that were less than the genus-level threshold, thereby suggesting that DaLV may belong to a new genus with in Lispiviridae of the order Mononegavirales. Unfortunately, only the RdRp fragment of the DaLV genome could be obtained from the HTS data in this study. However, DaLV was detected in 32 out of 67 Disporopsis samples from the Yunnan province, Sichuan province, Hubei province and Chongqian city, thereby suggesting that DaLV frequently infects Disporopsis. Most members of this family have been identified within arthropod hosts, and these viruses are only known from their genome sequences (Li et al., 2023). In our experimental of mechanical inoculation, DaLV was not detected from the inoculated plants, indicating that DaLV might have a higher effective transmission mode in nature than artificial inoculation. Thus, further research is needed to sequence and characterize the full-length genome of DaLV, clarify other biological features, i.e., host range, vector, pathogenicity and virus-host interaction.
The genome structure of DaPTV was identical to those of alphapartitiviruses (Figure 7). Thus, we suggest that DaPTV is a member of the genus Alphapartitivirus. RT-PCR was carried out with primer sets (Table 1) were designed to target different regions of RNA1 and RNA2 in order to confirm the presence of this virus in Disporopsis, and the sequences of the RT-PCR products confirmed that they belonged to DaPTV (Figure 8). Furthermore, the phylogenetic analysis based on the RdRp and CP amino acid sequences indicated that DaPTV clustered closely with alphapartitiviruses that had been identified in plants like Paris polyphylla and Poaceae liege. DaPTV shared 87 and 78.19% amino acid sequence identities and 99–100% coverage with RdRp and CP of Paris alphapartitivirus (Table 3), which are below the species-level identity threshold of 90%. Therefore, this suggests that DaPTV may be a new alphapartitivirus. Using the specific primers targeting DaPTV, the virus was detected in 11 out of 67 Disporopsis samples, thereby indicating that DaPTV is prevalent in Disporopsis. For partivirids, bisegmented double-stranded (ds) RNA viruses without known natural vectors, their host range includes plants, fungi, and protozoa with persistent infections. The partitiviruses have hence sometimes been called cryptic viruses, or cryptoviruses, especially in the case of plant partitiviruses which generally have the asymptomatic infections in the host organisms, or have few, if any, deleterious effects on host cells (Nibert and Schiff, 2014). Plant partitiviruses were transmitted by seed (Vainio et al., 2018). Thus, further research is needed to provide the supporting evidence for partitivirus-host interactions and host effects,
An analysis on the incidence of viruses among 67 Disporopsis samples revealed that 46 plants out of the 67 were infected with at least one virus. PMaV1 and DaLV were the most predominantly, found viruses and were detected in 22 and 34 out of 67 samples, respectively. Mixed infections were also common, particularly in samples from Kunming.
Aside from mechanical transmission, members of the Potyvirus genus are usually transmitted by aphids or seed in nature (Atreya et al., 1991). The three potyviruses (DCSV, PMaV1, and PMNV) in this study may have been transmitted by aphids, agricultural practices, or contaminated seeds to Disporopsis from crops or weeds grown in the same or nearby fields. The viruses may also have traveled across farms and even regions on contaminated seeds. Further study is required to understand the epidemiology of these potyviruses and the importance of the transmission vectors and intermediate hosts on the virus’ pathogenicity.
This study has successfully identified and characterized three novel viruses that infect Disoropsis plants based on their genomic features or infectivity, and it provides a foundation for understanding the impact of these viruses on plant health and for developing future management strategies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PP691760, https://www.ncbi.nlm.nih.gov/genbank/, PP691759, https://www.ncbi.nlm.nih.gov/genbank/, PP691758, https://www.ncbi.nlm.nih.gov/genbank/, PP803564, https://www.ncbi.nlm.nih.gov/genbank/, PP803563, https://www.ncbi.nlm.nih.gov/genbank/, PP803565.
QL: Writing – original draft, Validation, Data curation, Writing – review & editing. LY: Validation, Writing – review & editing. TZ: Validation, Writing – review & editing. XY: Data curation, Writing – review & editing. BZ: Validation, Writing – review & editing. HL: Investigation, Resources, Writing – review & editing. JH: Investigation, Resources, Writing – review & editing. LZ: Investigation, Resources, Writing – review & editing. PJ: Investigation, Resources, Writing – review & editing. JD: Data curation, Funding acquisition, Investigation, Methodology, Resources, Supervision, Writing – review & editing, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by High Level Talents Project of Yunnan University of Chinese Medicine (2019YZG09), Yunnan Provincial Science and Technology Department-Applied Basic Research Joint Special Funds of Yunnan University of Chinese Medicine (202101AZ070001-011) and Wang Yuan Chao Expert Workstation in Yunnan Province (202305AF150018), Yunnan Provincial Science and Technology Department (202304BI090004).
The authors thank Fang Xiang Li Qi Garden at the YUCM for plant base of Disporopsis plants. The authors also thank Prof. Furong Xu for providing the greenhouse to maintain Disporopsis plant samples which was tested through HTS.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1434554/full#supplementary-material
Adams, M. J., Antoniw, J. F., and Fauquet, C. M. (2005). Molecular criteria for genus and species discrimination within the family Potyviridae. Arch. Virol. 150, 459–479. doi: 10.1007/s00705-004-0440-6
Atreya, P. L., Atreya, C. D., and Pirone, T. P. (1991). Amino acid substitutions in the coat protein result in loss of insect transmissibility of a plant virus. Proc. Natl. Acad. Sci. U.S.A. 88, 7887–7891. doi: 10.1073/pnas.88.17.7887
Bernardo, P., Charles-Dominique, T., Barakat, M., Ortet, P., Fernandez, E., Filloux, D., et al. (2018). Geometagenomics illuminates the impact of agriculture on the distribution and prevalence of plant viruses at the ecosystem scale. ISME J. 12, 173–184. doi: 10.1038/ismej.2017.155
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chen, J., and Chen, J. (2002). Determination of genome sequence of potyviruses by degenerated PCR and RACE methods. Chin. J. Virol. 4, 371–374. (in Chinese)
Chung, B. Y., Miller, W. A., Atkins, J. F., and Firth, A. E. (2008). An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U.S.A. 105, 5897–5902. doi: 10.1073/pnas.0800468105
Ghorbani, A., Langenberger, G., Feng, L., and Sauerborn, J. (2011). Ethnobotanical study of medicinal plants utilised by Hani ethnicity in Naban River Watershed National Nature Reserve, Yunnan, China. J. Ethnopharmacol. 134, 651–667. doi: 10.1016/j.jep.2011.01.011
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Han, L., Zhao, W., Li, A., Zhou, B., Zhang, J., and Wu, W. (2022). Antifungal activity of l-azetidine-2-carboxylic acid isolated from Disporopsis aspera rhizomes against Podosphaera xanthii. Pest Manag. Sci. 78, 1946–1952. doi: 10.1002/ps.6812
Hu, J., Que, K., and Mu, L. (2022). First report of Paris mosaic necrosis virus infecting Polygonatum kingianum. Chin. Acta Phytopathol Sin. (in Chinese). 52, 712–714. doi: 10.13926/j.cnki.apps.000596
Kreuze, J. F., Perez, A., Untiveros, M., Quispe, D., Fuentes, S., Barker, I., et al. (2009). Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: a generic method for diagnosis, discovery and sequencing of viruses. Virology 388, 1–7. doi: 10.1016/j.virol.2009.03.024
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lan, P., Zhao, J., Zhou, Y., Li, Y., Shen, D., Liao, Q., et al. (2018). Complete genome sequence of Paris mosaic necrosis virus, a distinct member of the genus Potyvirus. Arch. Virol. 163, 787–790. doi: 10.1007/s00705-017-3649-x
Li, Q., Zhang, B., Hu, J., Zhang, L., Ji, P., and Dong, J. (2023). Complete genome sequence of polygonatum mosaic-associated virus 1, a novel member of the genus Potyvirus in China. Arch. Virol. 168:42. doi: 10.1007/s00705-022-05665-7
Liang, S.-Y., and Tamura, M. N. (2000). “Disporopsis Hance” in Flora of China. eds. Z. Y. Wu and P. H. Raven (Beijing: Science Press), 232–234.
Lin, Q., Zhang, L., Yang, D., and Chunjie, Z. (2014). Contribution of phenolics and essential oils to the antioxidant and antimicrobial properties of Disporopsis pernyi (Hua) Diels. J. Med. Food 17, 714–722. doi: 10.1089/jmf.2013.2905
Long, C. L., and Li, R. (2004). Ethnobotanical studies on medicinal plants used by the red-headed Yao people in Jinping, Yunnan province, China. J. Ethnopharmacol. 90, 389–395. doi: 10.1016/j.jep.2003.10.021
Nibert, M. L., and Schiff, L. A. (2014). Reovirus RNA packaging and morphogenesis. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins. 975–1002.
Nguyen, T. S., Xia, N. H., Chu, T. V., and Sam, H. V. (2019). Ethnobotanical study on medicinal plants in traditional markets of Son La province, Vietnam. For. Soc. 3:171. doi: 10.24259/fs.v3i2.6005
Puli’uvea, C., Khan, S., Chang, W. L., Valmonte, G., Pearson, M. N., and Higgins, C. M. (2017). First complete genome sequence of vanilla mosaic strain of dasheen mosaic virus isolated from the Cook Islands. Arch. Virol. 162, 591–595. doi: 10.1007/s00705-016-3133-z
Roossinck, M. J. (2017). Deep sequencing for discovery and evolutionary analysis of plant viruses. Virus Res. 239, 82–86. doi: 10.1016/j.virusres.2016.11.019
Roossinck, M. J., Martin, D. P., and Roumagnac, P. (2015). Plant virus metagenomics: advances in virus discovery. Phytopathology 105, 716–727. doi: 10.1094/PHYTO-12-14-0356-RVW
Sarapan, A., Hodkinson, T. R., and Suwanphakdee, C. (2023). Assessment of morphological, anatomical and palynological variation in the medicinal plant Disporopsis longifolia Craib (Asparagaceae) for botanical quality control. Plants 12:259. doi: 10.3390/plants12020259
Shukla, D. D., and Ward, C. W. (1989). Structure of potyvirus coat proteins and its application in the taxonomy of the potyvirus group. Adv. Virus Res. 36, 273–314. doi: 10.1016/S0065-3527(08)60588-6
Sydara, K., Xayvue, M., Souliya, O., Elkington, B. G., and Soejarto, D. D. (2014). Inventory of medicinal plants of the Lao People’s Democratic Republic: a mini review. J. Med. Plant Res. 8, 1262–1274. doi: 10.5897/JMPR2014.5534
Tian, X. X. (2013). China Miao enthnicity medicine. Guiyang: Guizhou Scientific and Technological Press.
Vainio, E. J., Chiba, S., Ghabrial, S. A., Maiss, E., Roossinck, M., Sabanadzovic, S., et al. (2018). ICTV virus taxonomy profile: Partitiviridae. J. Gen. Virol. 99, 17–18. doi: 10.1099/jgv.0.000985
Valli, A., López-Moya, J. J., and García, J. A. (2007). Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. J. Gen. Virol. 88, 1016–1028. doi: 10.1099/vir.0.82402-0
Wang, J., Chitsaz, F., Derbyshire, M. K., Gonzales, N. R., Gwadz, M., Lu, S., et al. (2023). The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388. doi: 10.1093/nar/gkac1096
Wang, Y., Li, S., Han, D., Meng, K., Wang, M., and Zhao, C. (2015). Simultaneous determination of rutin, luteolin, quercetin, and betulinic acid in the extract of Disporopsis pernyi (Hua) Diels by UPLC. J. Anal. Chem. 2015:130873. doi: 10.1155/2015/130873
Worrall, E. A., Hayward, A. C., Fletcher, S. J., and Mitter, N. (2019). Molecular characterization and analysis of conserved potyviral motifs in bean common mosaic virus (BCMV) for RNAi-mediated protection. Arch. Virol. 164, 181–194. doi: 10.1007/s00705-018-4065-6
Yang, L. (2022). Molecular identification and construction of infectious cDNA clone of citrus yellow spot virus. Southwest University.
Zeng, Q., Luo, H., Xu, Z., Duan, P., Liu, X., and Tan, M. (2023). Determination of heavy metals in Rhizoma Bambusiculata by inductively coupled plasma mass spectrometry. Chin. J. Ethnomed. Ethnopharm. 32, 22–27. (in Chinese)
Zhang, Y. Z., Shi, M., and Holmes, E. C. (2018). Using metagenomics to characterize an expanding virosphere. Cell 172, 1168–1172. doi: 10.1016/j.cell.2018.02.043
Zhang, F., Su, X., Zhang, S., Wang, M., Wang, T., Zheng, X., et al. (2023). Genomic characterization and seed transmission of a novel unclassified partitivirus infecting Polygonatum kingianum Coll. et Hemsl. Heliyon 9:e16719. doi: 10.1016/j.heliyon.2023.e16719
Zhang, Q., and Wang, Y. (2020). A new Colletotrichum species from Disporopsis fuscopicta in Yunnan province (in China). Chin. J. Fungal Res. 18, 330–334. doi: 10.13341/j.jfr.2020.8011
Zheng, L., Wayper, P. J., Gibbs, A. J., Fourment, M., Rodoni, B. C., and Gibbs, M. J. (2008). Accumulating variation at conserved sites in potyvirus genomes is driven by species discovery and affects degenerate primer design. PLoS One 3:e1586. doi: 10.1371/journal.pone.0001586
Keywords: Disporopsis, high-throughput sequencing, virome, potyvirus, lispivirus, partitivirus
Citation: Li Q, Yang L, Zhu T, Yv X, Zhang B, Li H, Hao J, Zhang L, Ji P and Dong J (2024) Detection and discovery of plant viruses in Disporopsis through high-throughput sequencing. Front. Microbiol. 15:1434554. doi: 10.3389/fmicb.2024.1434554
Received: 21 May 2024; Accepted: 23 October 2024;
Published: 13 November 2024.
Edited by:
Wei Wei, Agricultural Research Service (USDA), United StatesReviewed by:
R. Vinoth Kumar, SRM Institute of Science and Technology, IndiaCopyright © 2024 Li, Yang, Zhu, Yv, Zhang, Li, Hao, Zhang, Ji and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiahong Dong, ZG9uZ2pobkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.