
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 10 July 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1432883
This article is part of the Research TopicInsights in Infectious Agents and Disease: 2023/2024View all 34 articles
 Giorgia Fabrizio1
Giorgia Fabrizio1 Francesca Sivori2
Francesca Sivori2 Ilaria Cavallo2
Ilaria Cavallo2 Mauro Truglio2
Mauro Truglio2 Luigi Toma3
Luigi Toma3 Francesca Sperati4
Francesca Sperati4 Massimo Francalancia2
Massimo Francalancia2 Francisco Obregon2
Francisco Obregon2 Luisa Pamparau2
Luisa Pamparau2 Daniela Kovacs5
Daniela Kovacs5 Fulvia Pimpinelli2*
Fulvia Pimpinelli2* Enea Gino Di Domenico1*
Enea Gino Di Domenico1*Sodium hypochlorite (NaOCl) is widely recognized for its broad-spectrum antimicrobial efficacy in skin wound care. This study investigates the effectiveness of NaOCl against a range of bacterial and fungal isolates from pressure ulcer (PU) patients.
We analyzed 20 bacterial isolates from PU patients, comprising carbapenem-resistant Klebsiella pneumoniae (CRKP), multidrug-resistant Acinetobacter baumannii (MDRAB), methicillin-resistant Staphylococcus aureus (MRSA), methicillin-susceptible Staphylococcus aureus (MSSA), along with 5 Candida albicans isolates. Antibiotic resistance profiles were determined using standard susceptibility testing. Whole-genome sequencing (WGS) was employed to identify antimicrobial resistance genes (ARGs) and disinfectant resistance genes (DRGs). Genetic determinants of biofilm formation were also assessed. The antimicrobial activity of NaOCl was evaluated by determining the minimum inhibitory concentration (MIC) and the minimal biofilm eradication concentration (MBEC) for both planktonic and biofilm-associated cells.
CRKP and MDRAB showed resistance to fluoroquinolones and carbapenems, while MRSA exhibited resistance to β-lactams and levofloxacin. MSSA displayed a comparatively lower resistance profile. WGS identified significant numbers of ARGs in CRKP and MDRAB, with fewer DRGs compared to MRSA and MSSA. All isolates possessed genes associated with fimbriae production and adhesion, correlating with pronounced biofilm biomass production. NaOCl demonstrated substantial antimicrobial activity against both planktonic cells and biofilms. The MIC90 for planktonic bacterial cells was 0.125 mg/mL, and the MBEC90 ranged from 0.225 to 0.5 mg/mL. For planktonic C. albicans, the MIC90 was 0.150 mg/mL, and the MBEC90 was 0.250 mg/mL.
These results highlight the challenge in treating biofilm-associated infections and underscore the potential of NaOCl as a robust antimicrobial agent against difficult-to-treat biofilm infections at concentrations lower than those typically found in commercial disinfectants.
Pressure ulcers (PUs) represent a global problem in hospitalized patients and a significant burden to healthcare systems. Recognized as largely preventable, PUs are integral to hospital safety protocols, aiming to enhance patient quality of life and reduce hospitalization durations (Kottner et al., 2019). Globally, approximately 12% of adult hospitalized patients experience pressure injuries, with the prevalence of those acquired in hospitals ranging from 7 to 9% (Li et al., 2020; Wang et al., 2024). Most PUs fail to progress beyond the inflammatory phase, and infection often impacts wound healing (Goldberg and Diegelmann, 2020). PUs are predisposed to microbial colonization and at high risk for multidrug-resistant (MDR) organism infections (Norman et al., 2016; Di Domenico et al., 2017; Binsuwaidan et al., 2023). Among MDR pathogens, ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, Enterobacter sp.) pathogens have caused a significant concern due to their resistance against antibiotics (Vale De Macedo et al., 2021). Complications related to treating infected PUs are not limited to the presence of MDR organisms. Increasing evidence suggests biofilm accounts for a major virulence factor in ulcers’ pathogenesis (Cavallo et al., 2024). Indeed, PUs offer an ideal environment for biofilm formation due to the reduced host immune response and the presence of necrotic tissues that promote microbial adhesion (Siddiqui and Bernstein, 2010; Zhao et al., 2013; Cavallo et al., 2024). Bacteria embedded in a mature biofilm matrix are highly tolerant to conventional antimicrobial treatments. As a result, biofilm bacteria are difficult to eradicate since they require a minimum inhibitory concentration (MIC) impossible to reach in vivo due to the side effects and the toxicity of the drugs at renal and/or hepatic levels (Percival et al., 2015; Di Domenico et al., 2022). In response to the escalating challenge of antibiotic resistance, which is often exacerbated by indiscriminate systemic therapy, the British Society for Antimicrobial Chemotherapy (BSAC) and the European Wound Management Association (EWMA) recommend prioritizing the use of non-antibiotic antimicrobials for treating infected wounds (Lipsky et al., 2016). Topical antiseptics are preferred over topical antibiotics because they are broader in their spectrum of activity and less likely to cause allergic reactions. The ideal antiseptic for managing an infected skin wound should combine therapeutic efficacy with tissue tolerability (Kramer et al., 2018). Sodium hypochlorite (NaOCl) solutions are considered among the first choices for skin wound care due to their effectiveness against a wide range of planktonic and biofilm microorganisms, their role in promoting physiological tissue repair, and their anti-inflammatory actions (Ciccia et al., 2018; Lineback et al., 2018; Myltykbayeva et al., 2020). The NaOCl has longed to be used as an antiseptic for patients on dialysis and to irrigate wounds and burns (Bruch, 2006; Cruz et al., 2006; Alvarez et al., 2010). The NaOCl is a cheaper antiseptic than chlorhexidine, slightly volatile and less flammable than alcohol, less vulnerable to deactivation by blood and serum protein than povidone-iodine, and less toxic than povidone-iodine (Alvarez et al., 2010). Besides, NaOCl has never been correlated with bacterial contamination and hospital outbreaks. The NaOCl has a broad spectrum of action and is an isotonic and physiological solution that does not cause any osmotic stress toward damaged tissues (Mourad and Hobro, 2020). Furthermore, due to the positive activity/tolerability ratio, the 0.05% NaOCl solution has to be considered one of the reference solutions for the disinfection of damaged skin (Maniscalco et al., 2023).
The present study was designed to comprehensively investigate the antimicrobial resistance patterns and biofilm production capabilities of ESKAPE pathogens and Candida albicans isolates derived from PUs. Specifically, it assessed the efficacy of NaOCl in combating these pathogens, focusing on its role against planktonic cells and disrupting biofilm structures. The findings are intended to contribute to a deeper understanding of biofilm resilience mechanisms and inform more effective treatment approaches for managing infections associated with PUs.
From the Microbial Strain Repository of the Laboratory of Microbiology and Virology, San Gallicano Dermatological Institute, Rome, a total of 25 consecutive bacterial isolates, including five carbapenem-resistant K. pneumoniae (CRKP), five multidrug-resistant A. baumannii (MDRAB), five methicillin-resistant (MRSA) and five methicillin-susceptible S. aureus (MSSA), and five C. albicans from patients with infected PUs admitted between August 2022 and July 2023 were included in the study. Antimicrobial susceptibility testing was assessed by the BD PhoenixTM automated microbiology system (Becton Dickinson Diagnostic Systems, Sparks, MD, United States). Multidrug-resistant (MDR) bacteria were defined according to the European Centre for Disease Prevention and Control (ECDC) criteria.1 The antifungal susceptibility testing was performed utilizing Sensititre YeastOne YO10 AST Plate (Thermo Fisher Scientific, United States) and following manufacturers’ instructions.
PUs were identified as localized skin and underlying tissue lesions caused by compression between bony prominences and external surfaces. When a patient presented with multiple ulcers, all ulcers exhibiting local signs of infection were cultured, and the stage of the most severe ulcer was assigned to the case. Infected PUs were categorized based on specific guidelines (Livesley and Chow, 2002). Ethical approval was secured from the Central Ethics Committee I.R.C.C.S. Lazio, Rome (Protocol 8,993—05.07.2022, trial registry number 1733/22), adhering to the Helsinki Declaration.
The minimum inhibitory concentration (MIC) of NaOCl against the selected pathogens was established through plate counting to determine the CFU/ml. A standard inoculum of 5 × 105 colony-forming units (CFU)/mL was utilized. NaOCl was diluted in serial two-fold increments in cation-adjusted Mueller–Hinton broth (MHB). The MIC90 was identified as the lowest antibiotic concentration, resulting in a 90% reduction in bacterial survival compared to the untreated control (Sivori et al., 2022). All experiments were performed in triplicate.
The multiple antibiotic resistance (MAR) index for each isolate was determined using the following equation:
Where a represents the number of antibiotics that the isolate resists, and b denotes the total number of antibiotics tested against the isolate (Krumperman, 1983).
Whole-genome sequencing (WGS) was performed on all bacterial strains included in this study. DNA for whole-genome sequencing (WGS) was extracted using QIAsymphony DSP Virus/Pathogen Kits (Qiagen, Hilden, Germany) following the manufacturer’s protocols. The quality of the DNA reads was enhanced by trimming using FastP v0.23.4, and the sequences were assembled with SPAdes v3.15.5 (Bankevich et al., 2012; Chen et al., 2018). These sequences were annotated using Prokka v1.14.6. A pan-genome analysis was conducted using Roary v3.13.0 (Page et al., 2015). Predictions for antibiotic resistance genes were performed utilizing the Comprehensive Antibiotic Resistance Database (CARD) v3.2.8 and the Resistance Gene Identifier (RGI) tool, specifically focusing on “perfect” and “strict” matches to high-quality reference sequences, with a threshold of 97% identity for inclusion (Alcock et al., 2023). Virulence factors were determined by using Blastn against the Virulence Factor Database (VFDB), requiring at least 80% coverage and 90% identity (Chen et al., 2016; Cavallo et al., 2022). The abundance of Clusters of Orthologous Groups (COG) categories was assessed using eggNOG-mapper v2.1.12 aligned with the eggNOG database v5.0, under default settings (Cantalapiedra et al., 2021).
Biofilm formation was quantified using crystal violet (CV) to assess biomass 24 h post-incubation. Sterile 96-well polystyrene plates were inoculated with 200 μL of a bacterial or yeast cell suspension (105 CFU/mL) in MHB and incubated at 37°C for 24 h without agitation. Following incubation, the medium was discarded, and the wells were washed three times with 200 μL of sterile distilled water. The plates were then air-dried for 45 min before the adherent cells were stained with 200 μL of 0.1% CV solution for 15 min. Excess stain was removed by washing the wells three times with 200 μL of sterile distilled water. The stain absorbed by the biofilm-forming cells was solubilized in 200 μL of an ethanol-acetone mixture (4:1 ratio), and the absorbance of each well was measured at 570 nm (OD570) using a Multiskan SkyHigh spectrophotometer (Thermo Fisher Scientific, Ohio, United States). All experiments were conducted in triplicate and repeated independently at least three times (Di Domenico et al., 2018; Truglio et al., 2024).
Cell suspensions diluted to approximately 5 × 105 CFU/mL were used to inoculate a 96-well polystyrene flat-bottom plate with 100 μL of MHB for biofilm cultivation. After a 24-h incubation at 37°C, the wells were washed twice with sterile deionized water and resuspended in 100 μL of MHB. The biofilms were then thoroughly scraped, and viable cells were quantified by serial dilution and plating on blood agar for bacterial cultures or Sabouraud’s dextrose agar for C. albicans to determine CFU/ml.
Cell suspensions, diluted to approximately 5 × 105 CFU/mL, were utilized to inoculate a 96-well polystyrene flat-bottom plate with 100 μL of MHB for biofilm cultivation. After incubation at 37°C for 24 h, the wells were rinsed with 0.45% saline solution to eliminate nonadherent cells. Subsequently, the cells were resuspended in 100 μL of MHB containing serial dilutions of NaOCl. The plate was then incubated for an additional 20 h at 37°C. Following this incubation period, the contents of the wells were aspirated. Each well was washed twice with sterile deionized water, and the cells were resuspended in 100 μL of MHB. The biofilms were thoroughly scraped, and the total number of viable cells was quantified by serial dilution and plating on blood agar plates for bacterial cultures or Sabouraud’s dextrose agar for C. albicans to determine CFU. MBEC90 values were established as the lowest concentrations of NaOCl, resulting in a 90% reduction in cell viability within the biofilms relative to the untreated control.
The metabolic activity of planktonic and biofilm isolates was determined by a resazurin-based assay for bacterial cells or 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl) -5-[(phenylamino) carbonyl]-2H- tetrazolium hydroxide (XTT) reduction assay for yeast cells as described previously (Di Domenico et al., 2018; Sivori et al., 2022). All experiments were conducted in triplicate and repeated independently at least three times.
For biofilms that had already formed, eDNA quantification was carried out by first adding 100 μL of TE buffer to each well, followed by 100 μL of a freshly prepared PicoGreen solution (consisting of 1 μL PicoGreen dye diluted in 199 μL TE buffer). The wells containing PicoGreen were then incubated for 5 min. Subsequently, fluorescence intensity was measured using a Wallace Victor 3, 1,420 Multicolor fluorescence plate reader (PerkinElmer), with settings at excitation 485 nm and emission 535 nm, and a 0.1-s integration time. This procedure was replicated across three biological replicates for each strain. A standard curve was generated using Lambda DNA (Invitrogen Molecular Probes) for each assay run (Sivori et al., 2022).
Biofilms were grown on μ-Slide (Ibidi, Gräfelfing, Germany) inoculated with ~1 × 105 cells in 500 μL of fresh MH medium and incubated for 48 h at 37°C. The culture medium was changed after 24 h of biofilm growth. Then, biofilms were treated with different concentrations of NaOCl in a fresh MH medium and incubated for an additional 22 h at 37°C. According to supplier specifications, biofilms were stained using the LIVE/DEAD BacLight Bacterial Viability Kit (Life Technologies, New York, NY, United States) and examined with an Axio Observer inverted fluorescence microscope equipped with an Apotome system (Carl Zeiss International, Oberkochen, Germany). Data was analyzed using ZEN 3.2 (blue edition) software (Truglio et al., 2024). All the images are available in Supplementary Figure S1.
We reported the categorical variables through absolute and relative frequencies, whereas the continuous variables through median and means with standard deviations (SD). Kolmogorov–Smirnov normality test was calculated for all the continuous variables and the Kruskal-Wallis test was used to evaluate the differences between groups. Since we are dealing with multiple comparisons, the Bonferroni correction was applied. The correlation between variables was investigated using the Spearman correlation coefficient. A univariate regression linear analysis was conducted to investigate the relationship between variables. Statistical analyses were performed with GraphPad Prism 10.0 (GraphPad Software, Inc., San Diego, CA, United States). Differences were considered statistically significant for p-values of p < 0.05 (*), p < 0.01 (**), and p < 0.001 (***).
Bacterial isolates comprised five carbapenem-resistant Klebsiella pneumoniae (CRKP), five multidrug-resistant Acinetobacter baumannii (MDRAB), five methicillin-resistant Staphylococcus aureus (MRSA), and five methicillin-susceptible Staphylococcus aureus (MSSA). Antibiotic susceptibility patterns are summarized in Figures 1A,B.
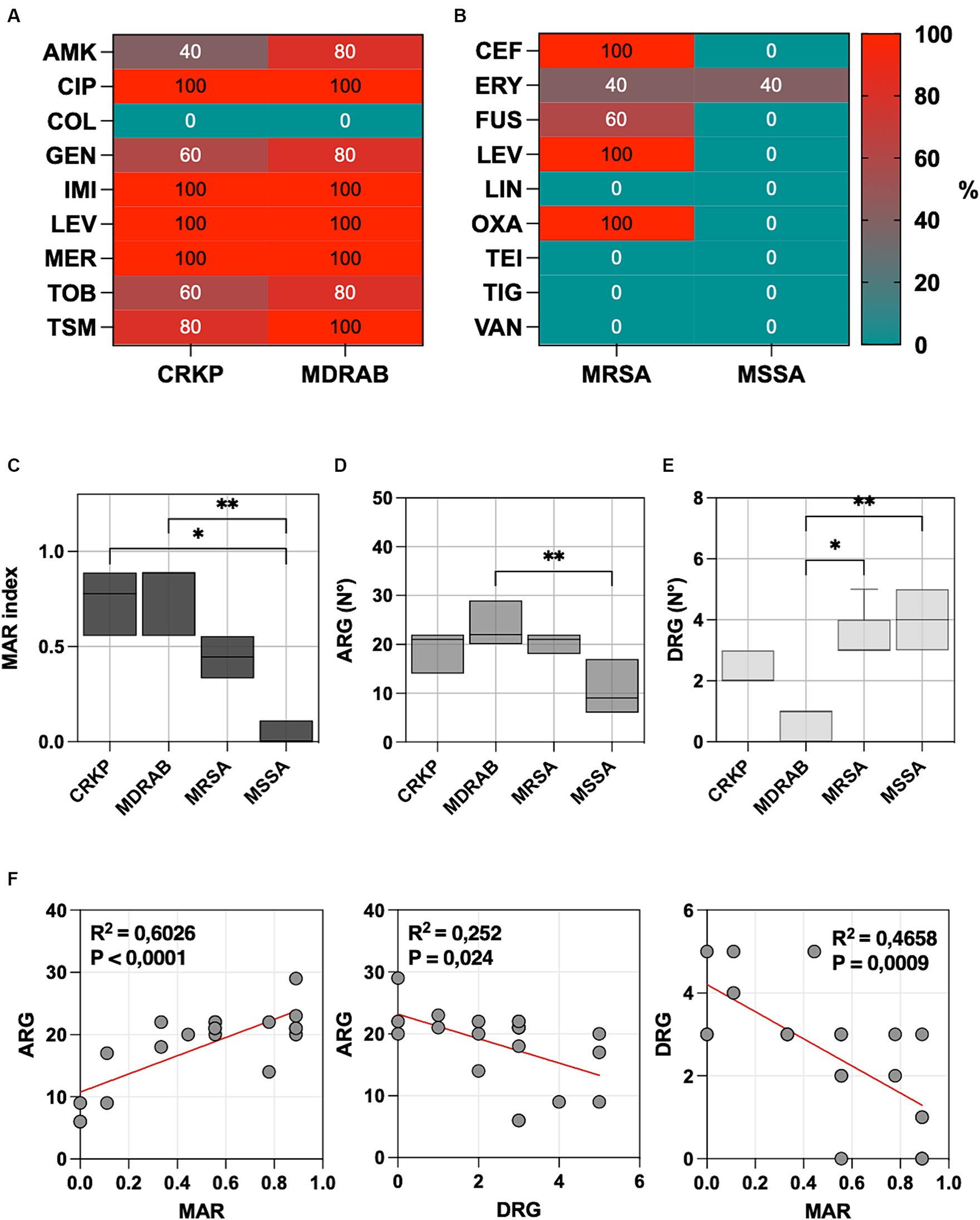
Figure 1. (A) The percentage of carbapenem-resistant Klebsiella pneumoniae (CRKP), multidrug-resistant Acinetobacter baumannii (MDRAB), tested non-susceptible or resistant to amikacin (AMK), ciprofloxacin (CIP), colistin (COL), gentamicin (GEN), imipenem (IMI), levofloxacin (LEV), meropenem (MER), tobramycin (TOB) and trimethoprim-sulfamethoxazole (TSM). (B) The percentage of methicillin-resistant Staphylococcus aureus (MRSA) and methicillin-susceptible S. aureus (MSSA) tested non-susceptible or resistant to ceftaroline (CEF), daptomycin (DAP), erythromycin (ERY), fusidic acid (FUS), levofloxacin (LEV), linezolid (LIN), oxacillin (OXA), teicoplanin (TEI), tigecycline (TIG), and vancomycin (VAN). (C) Multiple antibiotic resistance (MAR) index box plots, (D) predictions of antibiotic resistance genes (ARG), and (E) disinfectant resistance genes (DRG) using CARD v3.2.8. (F) Linear regression analysis illustrates the correlation among the MAR index, ARGs, and DRGs. Significance was assessed by the Kruskal Wallis statistic test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
All CRKP and MDRAB isolates were resistant to fluoroquinolones (ciprofloxacin and levofloxacin) and carbapenems (imipenem and meropenem), with colistin being the sole effective agent. MRSA displayed resistance to ceftaroline, levofloxacin, and oxacillin, while MSSA showed a generally lower resistance profile, with an 80% resistance noted only to erythromycin. The MIC values for each strain and the different antibiotics are provided in Supplementary Table S1.
The Multiple Antibiotic Resistance (MAR) index for each isolate was determined by dividing the number of antibiotics to which the isolate was resistant by the total number of antibiotics tested (Figure 1C). The highest MAR index was observed in MDRAB (median 0.889, range 0.556–0.889), followed by CRKP (median 0.778, range 0.556–0.889), and MRSA (median 0.444, range 0.333–0.556). MSSA isolates (median 0.000, range 0.000–0.111) had a significantly lower MAR index compared to MDRAB (p = 0.001) and CRKP (p = 0.014), but not to MRSA.
Whole-genome sequencing (WGS) identified three CRKP isolates as sequence type (ST) 307 and two as ST501. In MDRAB, four isolates were classified as ST2 and one as ST139. MRSA predominantly belonged to ST22 (n = 4), with a single ST5 isolate. MSSA demonstrated greater diversity, with ST398 being the most frequent (n = 3), followed by single isolates of ST5 and ST7.
All CRKP strains harbored the SHV β-lactamase genes responsible for extended-spectrum β-lactamases (ESBLs). Specifically, SHV-11 and SHV-28 were present in two strains, while SHV-1 was detected in one strain. Additionally, two strains possessed the OXA-1 gene, contributing to ESBL resistance. Notably, all the CRKP strains tested positive for the carbapenem-hydrolyzing β-lactamase KPC gene. In one positive isolate for SHV, OXA-1, and KPC, the ompK37 gene, which confers reduced permeability to β-lactams, was also identified. Furthermore, all CRKP isolates harbored 4 to 6 quinolone resistance genes.
In MDRAB, class D oxacillinase (OXA) resistance genes were the most commonly identified. Three isolates were found to have OXA-23. All isolates also carried other OXA genes, such as two OXA66, two OXA82, and one OXA545. Significantly, the MDRAB strains were positive for the multidrug efflux pump genes adeIJK.
All MRSA isolates resistant to levofloxacin carried the gyrA and parC genes, which are known to confer resistance to quinolones. As expected, the mecA gene conferring resistance to oxacillin was detected in all MRSA isolates. In MSSA, erythromycin ribosome methylase genes (ermA or ermC) were identified in 2 isolates.
Predictions of antibiotic resistance genes (ARGs) and disinfectant resistance genes (DRGs) showed that MDRAB exhibited the highest level of ARGs, with a median number of 22 genes (range 20–29), followed by CRKP (21 genes, range 14–22), MRSA (21 genes, range 18–22), and MSSA (9 genes, range 6–17), as reported in Figure 1D. Conversely, DRGs were more abundant in MSSA (4 genes, range 3–5), followed by MRSA (3 genes, range 3–5), CRKP (2 genes, range 2–3), and MDRAB (0 genes, range 0–1), as indicated in Figure 1E.
All CRKP isolates contain kpnEF genes, which confer resistance to broad-spectrum disinfectants. The qacEΔ1 gene, linked to antiseptic resistance, appears in 40% of CRKP and MDRAB. In S. aureus, the NorC efflux pump, crucial for resistance to various disinfectants, is universally present. MRSA showed a median of 3 genes related to resistance in disinfectant. In particular, all the isolates were positive for arlR and mgrA, which modulates the expression of norA and norB. Similarly, MgrA regulates the expression of a multitude of genes, including the regulation of several multidrug efflux pumps, including NorA, NorB, and NorC. The other efflux pump linked to resistance to disinfectant is SdrM, which has been detected in 3 MSSA and one MRSA strain.
Linear regression analysis (Figure 1F), plotting the number of MAR index and ARG, showed a positive correlation (p < 0.0001). Conversely, a negative correlation was observed between ARG and DRG (p = 0.0240) and DRG and AMR (p = 0.0009).
WGS analysis revealed the widespread occurrence of genes responsible for adhesion and biofilm formation. In particular, CRKP harbored the mrkABCDF gene cluster, encoding type 3 fimbriae mediating adhesion to abiotic surfaces and biofilm formation and crucial for persistence in the nosocomial setting.
MDRAB isolates consistently harbored genes for producing pili and other surface structures that facilitate adherence and biofilm development. Genes such as bap for biofilm-associated protein and csuABCDE for chaperone-usher pili assembly were detected in 83.3% of MDRAB isolates, indicating a common biofilm-forming strategy.
In S. aureus, both MSSA and MRSA strains showed the presence of the icaADBC operon, which is instrumental in intercellular adhesion and biofilm polysaccharide synthesis. Additionally, fibronectin-binding protein genes fnbA and fnbB were ubiquitously present, indicating a potential mechanism for cellular adhesion in diverse environmental niches. Clumping factor genes clfA and clfB, along with the collagen adhesion gene cna and the biofilm-associated sasG, were also identified, highlighting the multifaceted approach of S. aureus in establishing infection and chronicity.
Accordingly, CRKP, MDRAB, and S. aureus (both MSSA and MRSA) exhibited comparable levels of biofilm formation, the total number of biofilm cells expressed as CFU/ml, and the metabolic activity of the biofilm cells (MAB) as reported in Figures 2A–C. Interestingly, the amount of extracellular DNA (eDNA) within the biofilm matrix (Figure 2D) was significantly higher in MRSA isolates compared to CRKP (p = 0.0395), MDRAB (p = 0.0302), and MSSA (p = 0.0005). Fluorescence microscopy revealed that all isolates formed biofilms with thicknesses ranging from 25 to 45 μm, displaying typical maturation features such as cell aggregates and water channels (Figure 2E and Supplementary Figure S1).
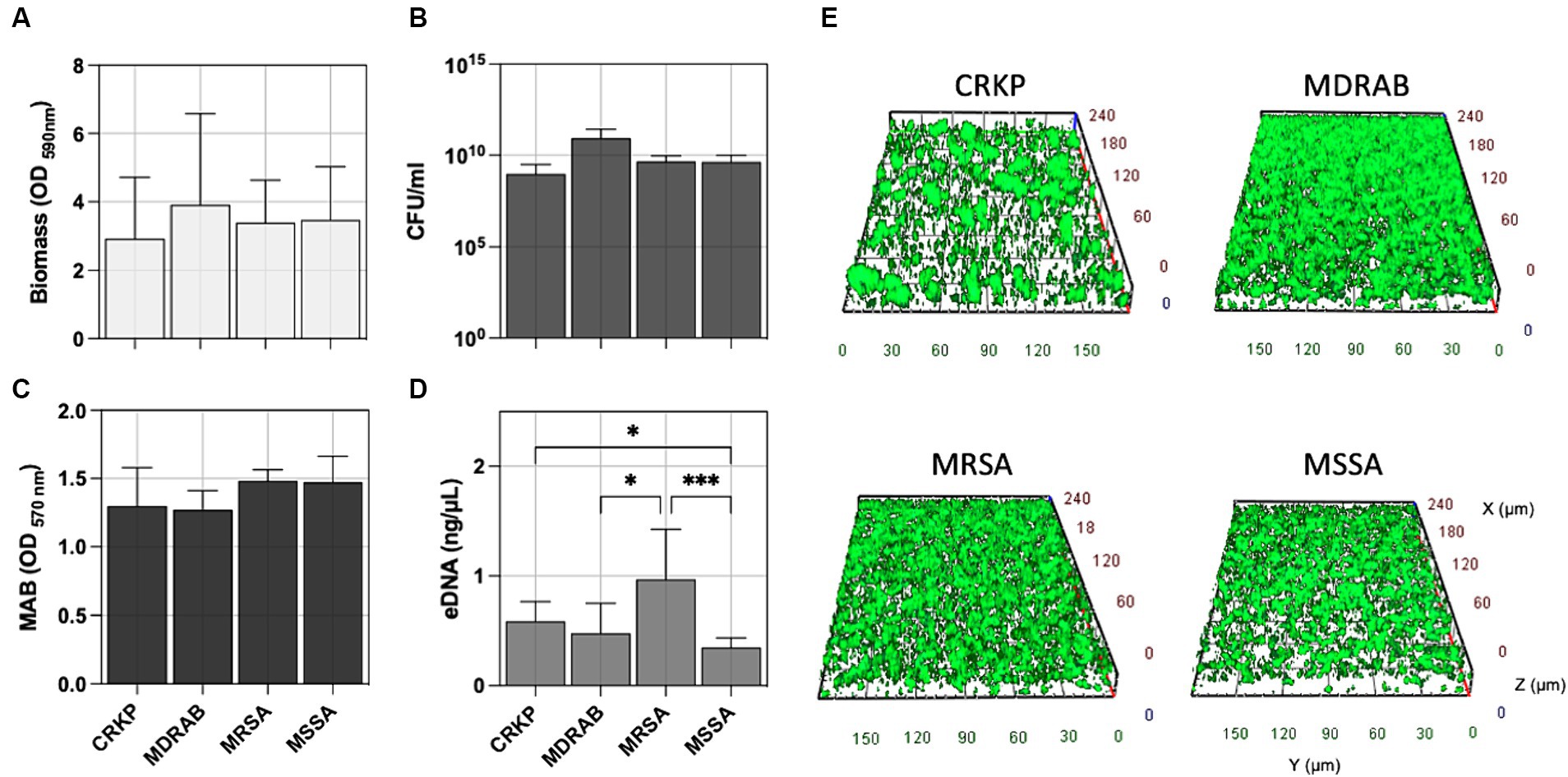
Figure 2. Comparative biofilm analysis of carbapenem-resistant Klebsiella pneumoniae (CRKP), multidrug-resistant Acinetobacter baumannii (MDRAB), methicillin-resistant Staphylococcus aureus (MRSA), and methicillin-susceptible S. aureus (MSSA) was quantitatively assessed by evaluating (A) the biomass using crystal violet (CV) staining, (B) the count of biofilm-forming cells (CFU/ml), (C) metabolic activity of biofilm cell (MAB) expressed as the area under the growth curve by the resazurin assay, and (D) the amount of extracellular DNA (eDNA) in the biofilm matrix. (E) Representative fluorescence microscopy images of biofilm formation for the indicated bacterial species stained with the Live/Dead BacLight kit after 24 h of incubation at 37°C. Computerized 3D reconstruction (X, Y, and Z axis) of the biofilm layers (in μm). Significance was assessed by the Kruskal Wallis statistic test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
The bacterial pathogens were exposed to sodium hypochlorite (NaOCl) to evaluate its antimicrobial and antibiofilm efficacy. Survival fractions of cells were assessed in comparison to untreated control strains, with NaOCl concentrations ranging from 0.125 to 0.275 mg/mL (Figure 3A). The minimum inhibitory concentration (MIC90), defined as the lowest concentration needed to inhibit 90% of planktonic bacterial growth relative to controls, was determined. For planktonic isolates, the MIC90 was 0.175 mg/mL for CRKP and 0.125 mg/mL for MDRAB, MRSA, and MSSA (Figure 3B). Biofilm-forming isolates were further tested to determine the minimum biofilm eradication concentration (MBEC90), which is the lowest concentration of NaOCl needed to eradicate 90% of the biofilm biomass. The MBEC90 values were found to be 0.250 mg/mL for CRKP, 0.125 mg/mL for MDRAB, 0.225 mg/mL for MRSA, and 0.150 mg/mL for MSSA (Figure 3B). Overall, the MBEC90 values were significantly higher (p < 0.001) than the corresponding MIC90 values, indicating a greater resistance of biofilm-associated cells to NaOCl treatment.
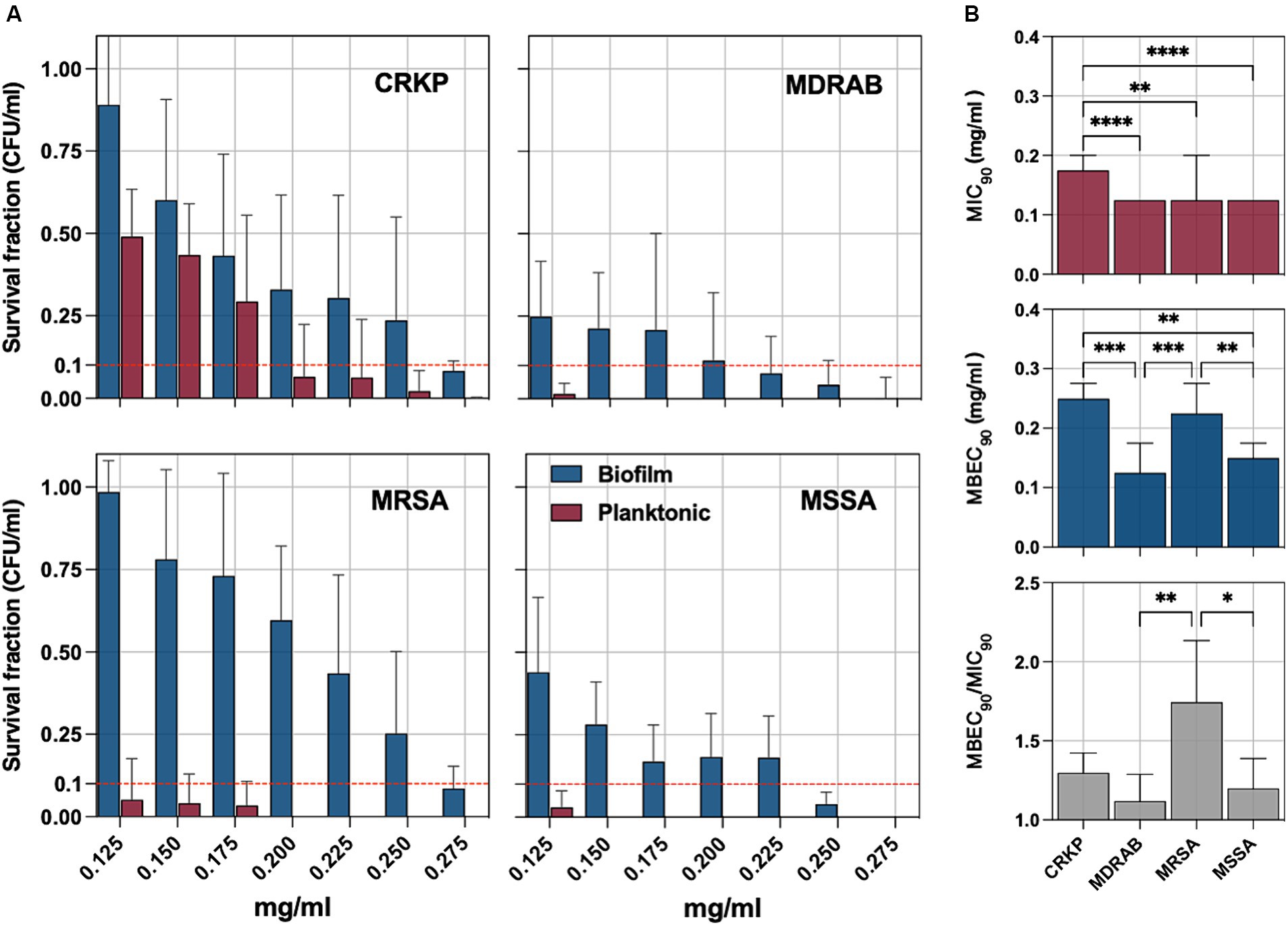
Figure 3. (A) The antimicrobial and antibiofilm activity of an electrolytic sodium hypochlorite solution (NaOCl) against carbapenem-resistant Klebsiella pneumoniae (CRKP), multidrug-resistant Acinetobacter baumannii (MDRAB), methicillin-resistant Staphylococcus aureus (MRSA), and methicillin-susceptible S. aureus (MSSA) is displayed through survival cell fractions compared to untreated control strains at concentrations ranging from 0.125 to 0.275 mg/mL. (B) The minimum inhibitory concentration (MIC90) is the lowest concentration (mg/ml) needed to inhibit 90% of planktonic bacterial growth relative to controls and the minimum biofilm eradication concentration (MBEC90). The antimicrobial and antibiofilm activity of NaOCl is demonstrated through the survival of bacterial cell fractions, compared to untreated control strains, at concentrations ranging from 0.125 to 0.275 mg/mL. The minimum inhibitory concentration (MIC90) and the minimum biofilm eradication concentration (MBEC90) are defined as the lowest NaOCl concentrations required to inhibit 90% of planktonic and biofilm bacterial growth, respectively, compared to untreated controls. The MBEC90/MIC90 ratio was used to quantify the biofilm tolerance to NaOCl for all tested strains. Significance was assessed by the Kruskal Wallis statistic test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
The MBEC90/MIC90 ratio, indicating the fold increase in the NaOCl dose needed to inhibit or kill cells in biofilms compared to planktonic growth, was used to quantify biofilm tolerance. The MBEC90/MIC90 ratio was significantly higher in the MRSA group compared to the MSSA (p = 0,0286) and MDRAB (p = 0,0014) but not CRKP (Figure 3B).
C. albicans is the main fungal species involved in pressure ulcer infection; accordingly, five isolates were included in the study. Figure 4A reports the antifungal susceptibility testing (AfST) data for the C. albicans clinical isolates. According to EUCAST clinical breakpoints, all the isolates were susceptible to amphotericin B, fluconazole, micafungin, and voriconazole. Resistance was observed in anidulafungin (40%), posaconazole (60%) and itraconazole (100%).
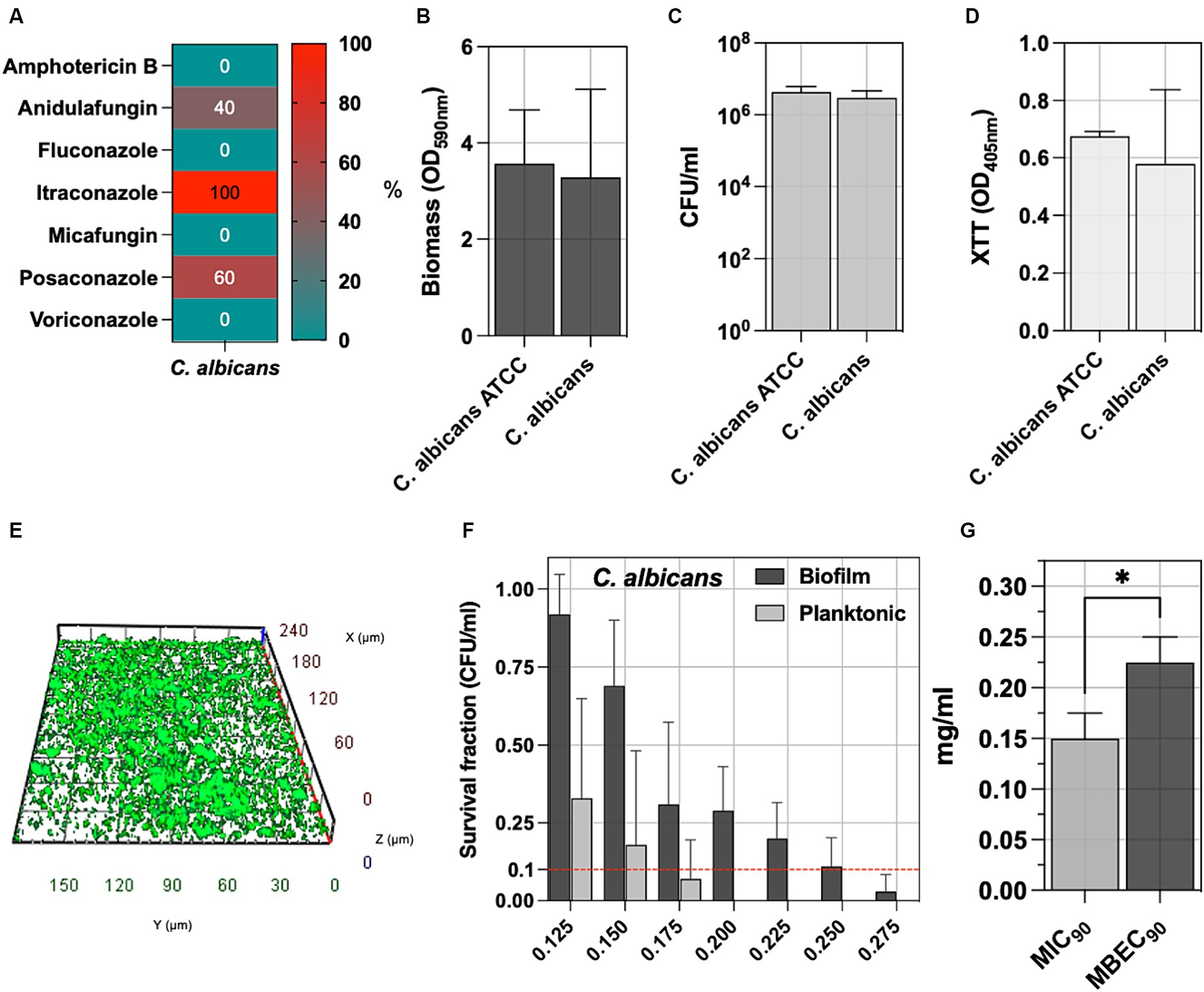
Figure 4. (A) The percentage of C. albicans tested non-susceptible or resistant according to EUCAST breakpoints to amphotericin B, anidulafungin, fluconazole, itraconazole, micafungin, posaconazole, voriconazole. (B) The biofilm of 5 C. albicans clinical isolates was assessed by measuring the biomass with CV staining, (C) the count of biofilm-forming cells (CFU/ml), and (D) the metabolic activity of the biofilm cells with XTT. (E) Representative fluorescence microscopy image stained with the Live/Dead BacLight kit after 24 h of incubation at 37°C. The C. albicans ATCC 2091, classified as a strong biofilm producer, was included as a control strain. (F) The antimicrobial and antibiofilm activity of NaOCl against C. albicans isolates was demonstrated through survival cell fractions, compared to untreated control strains, at concentrations ranging from 0.125 to 0.275 mg/mL. (G) The minimum inhibitory concentration (MIC90) and the minimum biofilm eradication concentration (MBEC90) are defined as the lowest NaOCl concentrations (mg/ml) required to inhibit 90% of planktonic and biofilm bacterial growth, respectively, compared to untreated controls. Significance was assessed by the Kruskal Wallis statistic test. *, p < 0.05; **, p < 0.01; ***, p < 0.001, ****, p < 0.0001.
Biofilm was assessed by measuring biomass production, counting the total number of biofilm cells expressed as CFU/ml, determining the metabolic activity of the biofilm cells (XTT), and comparing it with the reference C. albicans ATCC 2091 strain (Figures 4B–D). No significant differences were observed between C. albicans clinical isolates and the C. albicans ATCC 2091, classified as a strong biofilm producer strain (Di Domenico et al., 2018). Fluorescence microscopy reveals a well-defined architecture within the C. albicans biofilm, as evidenced by the consistent thickness between 20 to 35 μm. The three-dimensional analysis further highlights the presence of dense cellular clusters and heterogeneity of the biofilm matrix, underscoring the spatial organization (Figure 4E and Supplementary Figure S1).
The antifungal activity of NaOCl against planktonic and biofilm yeast cells, ranging from 0.125 to 0.275 mg/mL, was tested against C. albicans isolates (Figure 4F). Cell survival fractions were assessed in comparison to untreated control strains. NaOCl showed an MIC90 of 0.150 mg/mL (0.125–0.175 mg/mL) against planktonic C. albicans, while the MBEC90 was 0.225 mg/mL (0.175–0.250 mg/mL) (Figure 4G). The MBEC90/MIC90 ratio showed a median value of 1.43 (range 1.29–1.80) in the biofilm-mediated tolerance in C. albicans isolates (Figure 3H).
This study provides valuable insights into the in vitro efficacy of a NaOCl solution against clinical bacterial pathogens and C. albicans isolated from patients with PUs. The presence of strains such as CRKP, MDRAB, MRSA, and MSSA within PUs mirrors the findings of previous studies, which have similarly noted these pathogens as significant contributors to the complexity of wound infections (Li et al., 2021). The high levels of AMR observed in MDRAB and CRKP are alarming, given their role in healthcare-associated infections and the growing prevalence in PUs worldwide (Zhen et al., 2020). Similarly, MRSA presents significant concerns in managing PU infections due to its multidrug resistance profile correlated with suboptimal wound healing outcomes and extended durations of hospitalization (Binsuwaidan et al., 2023). In the current study, a MAR index greater than 0.4 indicates a high prevalence of multiple drug resistance among clinical isolates (Krumperman, 1983; Jain et al., 2021). The MAR index has been confirmed as an effective metric for assessing the risk associated with microbial isolates. Indeed, linear regression analysis delineates a statistically significant positive correlation between the MAR index and the number of ARGs. This relationship reinforces the utility of the MAR index not only as an essential epidemiological tool to monitor drug resistance but also as a predictor of the genetic propensity for resistance within bacterial populations in PUs (Krumperman, 1983; Jain et al., 2021).
WGS has refined our understanding of bacterial strains implicated in PU infections, revealing high-risk clones such as CRKP ST307 and ST501, which are known for their adaptability and prevalence in healthcare settings (Villa et al., 2017; Fasciana et al., 2019; Heiden et al., 2020). In MDRAB, four isolates were classified as ST2, the globally predominant ST. This ubiquity poses a significant challenge to infection control, underscoring the need for a concerted global response to effectively manage its spread (Cherubini et al., 2022). The ST22 was the most abundant detected in MRSA strains in four isolates. ST22 is particularly concerning due to its association with virulence factors such as Panton-Valentine leucocidin and toxic shock syndrome toxin 1 genes, which may enhance the ability to cause severe infections (Kaneko et al., 2023). Conversely, MSSA demonstrated a greater sequence type diversity than their Gram-negative counterparts and MRSA, with ST398 being the most frequent. The diversity observed in MSSA could reflect a broader range of ecological niches and host interactions, potentially impacting their pathogenicity and transmission dynamics (Vandendriessche et al., 2014).
In these highly virulent STs, the genetic determinants of resistance to frontline antibiotics underscore the pathogens’ capacity to withstand conventional antibiotic therapy and the need for alternative treatment strategies. Accordingly, topical antiseptics, including NaOCl, are favored over antibiotics due to rising drug resistance and are recommended as first-line treatment for infected skin lesions (McCullough and Carlson, 2014). Specifically, NaOCl has been demonstrated to be active against biofilms by denaturing proteins in the matrix and inhibiting major enzymatic functions in bacterial cells. Although NaOCl disinfectants are effective against biofilms at concentrations lower than those typically found in commercial products, the use of sub-lethal concentrations of some sodium-containing disinfectants could promote the formation of biofilms on environmental surfaces (Barnes and Greive, 2013; Cincarova et al., 2016; Lineback et al., 2018).
Our study demonstrated that CRKP exhibited significantly higher MIC90 values for NaOCl than the other isolates. For CRKP, the higher MIC90 and MBEC90 values compared to the other pathogens could be partially explained by the presence of kpnEF genes, which are involved in broad-spectrum antimicrobial resistance and clinically relevant disinfectants (Srinivasan and Rajamohan, 2013). In addition, two of the CRKP strains also have the qacEΔ1 gene that confers multiple resistance to antibiotics and antiseptics in Gram-negative bacteria (Kucken et al., 2000). The diminished efficacy of NaOCl may result from the activity of these efflux pumps actively removing disinfectants from the cell, thereby reducing the disinfectant’s ability to achieve the desired antimicrobial effect. This would require higher concentrations of NaOCl to inhibit or eradicate CRKP, as reflected in the MIC90 and MBEC90 values. MDRAB exhibited a low frequency of genes related to disinfectants. In particular, two isolates have the qacEΔ1 gene and one the amvA gene (Rajamohan et al., 2010; Short et al., 2021). Accordingly, MDRAB showed a lower MIC90 and MBEC90 to CRKP despite exhibiting a comparable biomass level.
For MRSA, the presence of regulatory systems like ArlRS and MgrA and efflux pumps like NorC may indicate a sophisticated system of resistance that operates beyond just preventing the entrance of antiseptics and could involve regulatory pathways and interactions with the cell’s stress responses (Liang et al., 2005; Truong-Bolduc et al., 2005; Costa et al., 2024).
The observed MBEC90 values for MSSA, which are lower than those for MRSA, imply a differential efficacy of the MSSA efflux pump system in the presence of NaOCl or point to inherent structural or compositional variations in MSSA biofilms that may render them more susceptible to NaOCl-mediated disinfection (McCarthy et al., 2015; Oliva et al., 2021). Indeed, in MSSA, the most prevalent efflux pump linked to resistance to disinfectant is SdrM, which shares 23% identity with the S. aureus MDR efflux pumps NorB (Yamada et al., 2006). Additionally, the biofilm matrix composition, particularly the eDNA content, could account for variations in disinfection susceptibility. The significantly higher eDNA content in MRSA biofilms compared to MSSA might confer a greater structural integrity or protection against NaOCl, considering that eDNA is a known contributor to biofilm stability and antimicrobial resistance (McCarthy et al., 2015; Sivori et al., 2022). Hence, while MSSA and MRSA biofilms demonstrate similar thicknesses and morphological maturation, the divergent eDNA concentrations suggest a structural disparity with potential implications for disinfection strategies.
Indeed, our findings demonstrated that the MBEC90/MIC90 ratio was significantly higher in the MRSA group compared to the MSSA and MDRAB groups but not when compared to CRKP. This observation suggests the possible contribution of specific elements within the biofilm matrix to the increased tolerance observed. Notably, matrix characterization revealed that MRSA strains have a higher level of eDNA content within their biofilm matrix. This higher eDNA content may play a crucial role in limiting the activity of NaOCl, as eDNA can contribute to the structural integrity and protective function of the biofilm, potentially impeding the penetration and efficacy of antimicrobial agents like NaOCl (Sivori et al., 2022). Further studies are warranted to elucidate the exact mechanisms by which eDNA influences biofilm resilience and to explore potential strategies to counteract this protective effect.
The efficacy of NaOCl against both planktonic and biofilm-forming microbial isolates, as reported in this study, highlights its significant antimicrobial and antibiofilm potential. Notably, the MIC90 values, ranging from 0.125 mg/mL for MDRAB, MRSA, and MSSA to 0.175 mg/mL for CRKP, underline the effectiveness of NaOCl in inhibiting bacterial growth at concentrations considerably lower than those typically found in commercial disinfectants. These findings are consistent with prior studies indicating that sodium hypochlorite disinfectants can effectively reduce biofilms (DeQueiroz and Day, 2007; Tiwari et al., 2018; Koti et al., 2024). This aligns with previous findings that noted that NaOCl and hydrogen peroxide disinfectants destroy both the biofilm matrix and the bacteria cells within, establishing them as superior anti-biofilm agents (DeQueiroz and Day, 2007; Lineback et al., 2018; Tiwari et al., 2018). Nevertheless, the MBEC90 values are markedly higher than MIC90, suggesting that NaOCl requires increased concentrations while effectively disrupting established biofilms. This observation is also consistent with past research, suggesting that biofilms pose significant challenges to disinfectant efficacy due to their dense cellular matrices and protective barriers (Gilbert et al., 2001; Mitchell et al., 2015).
A previous study raises critical points about the potential for sub-lethal concentrations of sodium-containing disinfectants to inadvertently promote biofilm formation (Cincarova et al., 2016). This phenomenon represents a paradoxical risk, where disinfectant use intended to control pathogen spread instead enhances their protective niches. Specifically, our quantification of biofilm cells (CFU/ml) did not indicate increased biofilm production for our isolates under sub-lethal concentrations of NaOCl. However, we cannot exclude the possibility of increased biomass production. Indeed, our study’s focus on measuring the bactericidal effects via cell counts offers a direct evaluation of antibacterial activity rather than biomass estimation alone.
C. albicans, showed susceptibility to amphotericin B, fluconazole, micafungin, and voriconazole, confirming their efficacy as initial treatments in clinical settings. Conversely, resistance to anidulafungin, posaconazole, and itraconazole raises concerns about emerging antifungal resistance (Kessler et al., 2022). NaOCl displayed differential efficacy against planktonic and biofilm forms of C. albicans . While NaOCl effectively inhibits planktonic cells at low concentrations, higher concentrations are needed to eradicate biofilms, as evidenced by higher MBEC 90 compared to MIC 90 values, reflecting biofilms’ inherent resistance (Short et al., 2021). Importantly, the consistency in biofilm structure across the clinical isolates and the reference strain indicates that NaOCl must overcome a common, robust defense mechanism to be effective. The concentration of NaOCl required for biofilm eradication implies that it could play a role in disinfection protocols aimed at preventing the spread of C. albicans, particularly in healthcare environments. This positions NaOCl as a potentially valuable agent in the disinfection arsenal against C. albicans in both planktonic and biofilm states as confirmed by the low MBEC90/MIC90 ratio.
This study provides valuable insights but has limitations affecting its generalizability. The modest sample size of 25 isolates may not fully reflect the diversity of resistance patterns or biofilm capabilities seen in broader clinical contexts. Additionally, while WGS offers detailed genetic perspectives on resistance mechanisms, the functional impact on clinical outcomes is still speculative. The findings on biofilm susceptibility to NaOCl, derived from in vitro single-species cultures, might not accurately mimic the complex conditions in PU infections. These factors underscore the necessity for extended research with larger and more varied samples and more comprehensive antimicrobial evaluations to bridge the gap between laboratory and clinical settings. Overall, while these limitations highlight the need for further research, the study demonstrates that the efficacy of NaOCl against both planktonic and biofilm-forming microbial isolates occurs at concentrations significantly lower than the 0.5 mg/mL typically found in commercial disinfectant products (Maniscalco et al., 2023). This indicates that even at diluted concentrations, NaOCl is effective in targeting and controlling these clinically relevant microbes, reinforcing its valuable role in infection control and disinfection protocols.
The application of NaOCl demonstrated a potent antimicrobial and antibiofilm activity, though it was markedly more effective against planktonic than biofilm-embedded cells. The low MBEC90/MIC90 ratio suggests that the biofilm matrix is poorly effective in protecting the bacterial and C. albicans isolates from NaOCl.
The observed susceptibility of bacterial pathogens and C. albicans to NaOCl, both in planktonic and biofilm states, suggests that NaOCl could be a broad-spectrum agent applicable in a multi-pathogen context, reducing the microbial burden and promoting PU’s healing (Serena et al., 2022). This distinction between MIC90 and MBEC90 values points to the resilience of biofilm architectures and emphasizes the need for higher concentrations of NaOCl for biofilm eradication of microbial isolates from PUs.
Interestingly, the correlation analyses reveal a nuanced interaction in CRKP and MRDAB between biofilm biomass, cell count, and DRGs, which may inform future studies aiming to disrupt biofilm formation or enhance disinfectant penetration in the biofilm matrix.
Ultimately, our study supports the use of electrolytic NaOCl solutions as a potential antiseptic at concentrations considerably lower than those typically found in commercial disinfectants.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
GF: Writing – original draft, Writing – review & editing. FS: Writing – original draft, Writing – review & editing. IC: Writing – original draft, Writing – review & editing. MT: Writing – original draft, Writing – review & editing. LT: Writing – original draft, Writing – review & editing. FS: Writing – original draft, Writing – review & editing. MF: Writing – original draft, Writing – review & editing. FO: Writing – original draft, Writing – review & editing. LP: Writing – original draft, Writing – review & editing. DK: Writing – original draft, Writing – review & editing. FP: Writing – original draft, Writing – review & editing. ED: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Italian Ministry of Health (RC 2024). This publication was supported by an unconditional grant by Angelini Pharma S.p.A.
We thank those who encouraged us to keep sippin’ on straight chlorine.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1432883/full#supplementary-material
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920
Alvarez, J. A., Macias, J. H., Macias, A. E., Rodríguez, E., Muñoz, J. M., Mosqueda, J. L., et al. (2010). Povidone-iodine against sodium hypochlorite as skin antiseptics in volunteers. Am. J. Infect. Control 38, 822–825. doi: 10.1016/j.ajic.2010.05.019
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barnes, T. M., and Greive, K. A. (2013). Use of bleach baths for the treatment of infected atopic eczema. Australas. J. Dermatol. 54, 251–258. doi: 10.1111/ajd.12015
Binsuwaidan, R., Khan, M. A., Alzahrani, R. H., Aldusaymani, A. M., Almallouhi, N. M., Alsabti, A. S., et al. (2023). Prevalence of multidrug-resistant and ESBL-producing bacterial pathogens in patients with chronic wound infections and spinal cord injury admitted to a tertiary care rehabilitation hospital. Antibiotics 12:1587. doi: 10.3390/antibiotics12111587
Bruch, M. K. (2006). “Toxicity and safety of topical sodium hypochlorite” in Contributions to nephrology. eds. C. Ronco and G. J. Mishkin (Basel: Karger), 24–38.
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, Orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 38, 5825–5829. doi: 10.1093/molbev/msab293
Cavallo, I., Sivori, F., Mastrofrancesco, A., Abril, E., Pontone, M., Di Domenico, E. G., et al. (2024). Bacterial biofilm in chronic wounds and possible therapeutic approaches. Biology 13:109. doi: 10.3390/biology13020109
Cavallo, I., Sivori, F., Truglio, M., De Maio, F., Lucantoni, F., Cardinali, G., et al. (2022). Skin dysbiosis and Cutibacterium acnes biofilm in inflammatory acne lesions of adolescents. Sci. Rep. 12:21104. doi: 10.1038/s41598-022-25436-3
Chen, L., Zheng, D., Liu, B., Yang, J., and Jin, Q. (2016). VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 44, D694–D697. doi: 10.1093/nar/gkv1239
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Cherubini, S., Perilli, M., Segatore, B., Fazii, P., Parruti, G., Frattari, A., et al. (2022). Whole-genome sequencing of ST2 A. baumannii causing bloodstream infections in COVID-19 patients. Antibiotics 11:955. doi: 10.3390/antibiotics11070955
Ciccia, M., Chakrokh, R., Molinazzi, D., Zanni, A., Farruggia, P., and Sandri, F. (2018). Skin antisepsis with 0.05% sodium hypochlorite before central venous catheter insertion in neonates: a 2-year single-center experience. Am. J. Infect. Control 46, 169–172. doi: 10.1016/j.ajic.2017.08.012
Cincarova, L., Polansky, O., Babak, V., Kulich, P., and Kralik, P. (2016). Changes in the expression of biofilm-associated surface proteins in Staphylococcus aureus food-environmental isolates subjected to sublethal concentrations of disinfectants. Biomed. Res. Int. 2016, 1–12. doi: 10.1155/2016/4034517
Costa, F. G., Mills, K. B., Crosby, H. A., and Horswill, A. R. (2024). The Staphylococcus aureus regulatory program in a human skin-like environment. MBio 15:e0045324. doi: 10.1128/mbio.00453-24
Cruz, D. N., Ocampo, C., Brendolan, A., Menara, G., Corradi, V., De Cal, M., et al. (2006). “Effectiveness of sodium hypochlorite in the prevention of catheter related infections” in Contributions to nephrology. eds. C. Ronco and G. J. Mishkin (Basel: Karger), 97–102.
DeQueiroz, G. A., and Day, D. F. (2007). Antimicrobial activity and effectiveness of a combination of sodium hypochlorite and hydrogen peroxide in killing and removing Pseudomonas aeruginosa biofilms from surfaces. J. Appl. Microbiol. 103, 794–802. doi: 10.1111/j.1365-2672.2007.03299.x
Di Domenico, E. G., Cavallo, I., Guembe, M., Prignano, G., Gallo, M. T., Bordignon, V., et al. (2018). The clinical biofilm ring test: a promising tool for the clinical assessment of biofilm-producing Candida species. FEMS Yeast Res. 18:foy025. doi: 10.1093/femsyr/foy025
Di Domenico, E., Farulla, I., Prignano, G., Gallo, M., Vespaziani, M., Cavallo, I., et al. (2017). Biofilm is a major virulence determinant in bacterial colonization of chronic skin ulcers independently from the multidrug resistant phenotype. Int J Mol Sci 18:1077. doi: 10.3390/ijms18051077
Di Domenico, E. G., Oliva, A., and Guembe, M. (2022). The current knowledge on the pathogenesis of tissue and medical device-related biofilm infections. Microorganisms 10:1259. doi: 10.3390/microorganisms10071259
Fasciana, T., Gentile, B., Aquilina, M., Ciammaruconi, A., Mascarella, C., Anselmo, A., et al. (2019). Co-existence of virulence factors and antibiotic resistance in new Klebsiella pneumoniae clones emerging in south of Italy. BMC Infect. Dis. 19:928. doi: 10.1186/s12879-019-4565-3
Gilbert, P., Das, J. R., Jones, M. V., and Allison, D. G. (2001). Assessment of resistance towards biocides following the attachment of microorganisms to, and growth on, surfaces. J. Appl. Microbiol. 91, 248–254. doi: 10.1046/j.1365-2672.2001.01385.x
Goldberg, S. R., and Diegelmann, R. F. (2020). What makes wounds chronic. Surg. Clin. N. Am. 100, 681–693. doi: 10.1016/j.suc.2020.05.001
Heiden, S. E., Hübner, N.-O., Bohnert, J. A., Heidecke, C.-D., Kramer, A., Balau, V., et al. (2020). A Klebsiella pneumoniae ST307 outbreak clone from Germany demonstrates features of extensive drug resistance, hypermucoviscosity, and enhanced iron acquisition. Genome Med. 12:113. doi: 10.1186/s13073-020-00814-6
Jain, P., Bepari, A. K., Sen, P. K., Rafe, T., Imtiaz, R., Hossain, M., et al. (2021). High prevalence of multiple antibiotic resistance in clinical E. coli isolates from Bangladesh and prediction of molecular resistance determinants using WGS of an XDR isolate. Sci. Rep. 11:22859. doi: 10.1038/s41598-021-02251-w
Kaneko, H., Yanagi, Y., Otake, S., Sato, M., Saito, T., and Nakaminami, H. (2023). The emerging threat of methicillin-resistant Staphylococcus aureus (MRSA) clone ST22-PT, carrying both Panton–valentine leucocidin and toxic shock syndrome toxin 1 genes. J. Antimicrob. Chemother. 78, 1023–1027. doi: 10.1093/jac/dkad039
Kessler, S. Q. S., Lang, P. M., Dal-Pizzol, T. S., and Montagner, F. (2022). Resistance profiles to antifungal agents in Candida albicans isolated from human oral cavities: systematic review and meta-analysis. Clin. Oral Invest. 26, 6479–6489. doi: 10.1007/s00784-022-04716-2
Koti, K., Rodas-Gonzalez, A., Nadon, C., McAllister, T., Yang, X., and Narváez-Bravo, C. (2024). Evaluating disinfectant efficacy on mixed biofilms comprising Shiga toxigenic Escherichia coli, lactic acid bacteria, and spoilage microorganisms. Front. Microbiol. 15:1360645. doi: 10.3389/fmicb.2024.1360645
Kottner, J., Cuddigan, J., Carville, K., Balzer, K., Berlowitz, D., Law, S., et al. (2019). Prevention and treatment of pressure ulcers/injuries: the protocol for the second update of the international clinical practice guideline 2019. J. Tissue Viability 28, 51–58. doi: 10.1016/j.jtv.2019.01.001
Kramer, A., Dissemond, J., Kim, S., Willy, C., Mayer, D., Papke, R., et al. (2018). Consensus on wound antisepsis: update 2018. Skin Pharmacol. Physiol. 31, 28–58. doi: 10.1159/000481545
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170. doi: 10.1128/aem.46.1.165-170.1983
Kucken, D., Feucht, H.-H., and Kaulfers, P.-M. (2000). Association of qacE and qacE Δ1 with multiple resistance to antibiotics and antiseptics in clinical isolates of gram-negative bacteria. FEMS Microbiol. Lett. 183, 95–98. doi: 10.1016/S0378-1097(99)00636-9
Li, Z., Lin, F., Thalib, L., and Chaboyer, W. (2020). Global prevalence and incidence of pressure injuries in hospitalised adult patients: a systematic review and meta-analysis. Int. J. Nurs. Stud. 105:103546. doi: 10.1016/j.ijnurstu.2020.103546
Li, Z., Xie, J., Yang, J., Liu, S., Ding, Z., Hao, J., et al. (2021). Pathogenic characteristics and risk factors for ESKAPE pathogens infection in burn patients. IDR 14, 4727–4738. doi: 10.2147/IDR.S338627
Liang, X., Zheng, L., Landwehr, C., Lunsford, D., Holmes, D., and Ji, Y. (2005). Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J. Bacteriol. 187, 5486–5492. doi: 10.1128/JB.187.15.5486-5492.2005
Lineback, C. B., Nkemngong, C. A., Wu, S. T., Li, X., Teska, P. J., and Oliver, H. F. (2018). Hydrogen peroxide and sodium hypochlorite disinfectants are more effective against Staphylococcus aureus and Pseudomonas aeruginosa biofilms than quaternary ammonium compounds. Antimicrob. Resist. Infect. Control 7:154. doi: 10.1186/s13756-018-0447-5
Lipsky, B. A., Dryden, M., Gottrup, F., Nathwani, D., Seaton, R. A., and Stryja, J. (2016). Antimicrobial stewardship in wound care: a position paper from the British Society for Antimicrobial Chemotherapy and European wound management association. J. Antimicrob. Chemother. 71, 3026–3035. doi: 10.1093/jac/dkw287
Livesley, N. J., and Chow, A. W. (2002). Infected pressure ulcers in elderly individuals. Clin. Infect. Dis. 35, 1390–1396. doi: 10.1086/344059
Maniscalco, R., Mangano, G., De Joannon, A. C., Vergassola, M., Zucchi, S., Mannello, F., et al. (2023). Effect of sodium hypochlorite 0.05% on MMP-9 extracellular release in chronic wounds. J Clin Med 12:3189. doi: 10.3390/jcm12093189
McCarthy, H., Rudkin, J. K., Black, N. S., Gallagher, L., O’Neill, E., and O’Gara, J. P. (2015). Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 5:1. doi: 10.3389/fcimb.2015.00001
McCullough, M., and Carlson, G. W. (2014). Dakin’s solution: historical perspective and current practice. Ann. Plast. Surg. 73, 254–256. doi: 10.1097/SAP.0b013e3182a634f7
Mitchell, K. F., Zarnowski, R., Sanchez, H., Edward, J. A., Reinicke, E. L., Nett, J. E., et al. (2015). Community participation in biofilm matrix assembly and function. Proc. Natl. Acad. Sci. USA 112, 4092–4097. doi: 10.1073/pnas.1421437112
Mourad, K. A., and Hobro, S. (2020). Developing chlorine-based antiseptic by electrolysis. Sci. Total Environ. 709:136108. doi: 10.1016/j.scitotenv.2019.136108
Myltykbayeva, Z., Kovaleva, G., Mukhitdinov, A., Omarova, S., and Nadirov, R. (2020). In vivo comparison of chlorine-based antiseptics versus alcohol antiseptic for surgical hand antisepsis. Scientifica 2020, 1–6. doi: 10.1155/2020/3123084
Norman, G., Dumville, J. C., Moore, Z. E., Tanner, J., Christie, J., and Goto, S. (2016). Antibiotics and antiseptics for pressure ulcers. Cochrane Database Syst. Rev. 2017:CD011586. doi: 10.1002/14651858.CD011586.pub2
Oliva, A., Stefani, S., Venditti, M., and Di Domenico, E. G. (2021). Biofilm-related infections in gram-positive Bacteria and the potential role of the long-acting agent Dalbavancin. Front. Microbiol. 12:749685. doi: 10.3389/fmicb.2021.749685
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Percival, S. L., Suleman, L., Vuotto, C., and Donelli, G. (2015). Healthcare-associated infections, medical devices and biofilms: risk, tolerance and control. J. Med. Microbiol. 64, 323–334. doi: 10.1099/jmm.0.000032
Rajamohan, G., Srinivasan, V. B., and Gebreyes, W. A. (2010). Molecular and functional characterization of a novel efflux pump, AmvA, mediating antimicrobial and disinfectant resistance in Acinetobacter baumannii. J. Antimicrob. Chemother. 65, 1919–1925. doi: 10.1093/jac/dkq195
Serena, T. E., Serena, L., Al-Jalodi, O., Patel, K., and Breisinger, K. (2022). The efficacy of sodium hypochlorite antiseptic: a double-blind, randomised controlled pilot study. J. Wound Care 31, S32–S35. doi: 10.12968/jowc.2022.31.Sup2.S32
Short, F. L., Liu, Q., Shah, B., Clift, H. E., Naidu, V., Li, L., et al. (2021). The Acinetobacter baumannii disinfectant resistance protein, AmvA, is a spermidine and spermine efflux pump. Commun. Biol. 4:1114. doi: 10.1038/s42003-021-02629-6
Siddiqui, A. R., and Bernstein, J. M. (2010). Chronic wound infection: facts and controversies. Clin. Dermatol. 28, 519–526. doi: 10.1016/j.clindermatol.2010.03.009
Sivori, F., Cavallo, I., Kovacs, D., Guembe, M., Sperduti, I., Truglio, M., et al. (2022). Role of extracellular DNA in Dalbavancin activity against methicillin-resistant Staphylococcus aureus (MRSA) biofilms in patients with skin and soft tissue infections. Microbiol Spectr. 10:e0035122. doi: 10.1128/spectrum.00351-22
Srinivasan, V. B., and Rajamohan, G. (2013). KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-Spectrum antimicrobial resistance. Antimicrob. Agents Chemother. 57, 4449–4462. doi: 10.1128/AAC.02284-12
Tiwari, S., Rajak, S., Mondal, D. P., and Biswas, D. (2018). Sodium hypochlorite is more effective than 70% ethanol against biofilms of clinical isolates of Staphylococcus aureus. Am. J. Infect. Control 46, e37–e42. doi: 10.1016/j.ajic.2017.12.015
Truglio, M., Sivori, F., Cavallo, I., Abril, E., Licursi, V., Fabrizio, G., et al. (2024). Modulating the skin mycobiome-bacteriome and treating seborrheic dermatitis with a probiotic-enriched oily suspension. Sci Rep. 14:2722. doi: 10.1038/s41598-024-53016-0
Truong-Bolduc, Q. C., Dunman, P. M., Strahilevitz, J., Projan, S. J., and Hooper, D. C. (2005). MgrA is a multiple regulator of two new efflux pumps in Staphylococcus aureus. J. Bacteriol. 187, 2395–2405. doi: 10.1128/JB.187.7.2395-2405.2005
Vale De Macedo, G. H. R., Costa, G. D. E., Oliveira, E. R., Damasceno, G. V., Mendonça, J. S. P., Silva, L. D. S., et al. (2021). Interplay between ESKAPE pathogens and immunity in skin infections: an overview of the major determinants of virulence and antibiotic resistance. Pathogens 10:148. doi: 10.3390/pathogens10020148
Vandendriessche, S., Vanderhaeghen, W., Larsen, J., De Mendonca, R., Hallin, M., Butaye, P., et al. (2014). High genetic diversity of methicillin-susceptible Staphylococcus aureus (MSSA) from humans and animals on livestock farms and presence of SCCmec remnant DNA in MSSA CC398. J. Antimicrob. Chemother. 69, 355–362. doi: 10.1093/jac/dkt366
Villa, L., Feudi, C., Fortini, D., Brisse, S., Passet, V., Bonura, C., et al. (2017). Diversity, virulence, and antimicrobial resistance of the KPC-producing Klebsiella pneumoniae ST307 clone. Microb. Genom. 3:e000110. doi: 10.1099/mgen.0.000110
Wang, I., Walker, R. M., Gillespie, B. M., Scott, I., Sugathapala, R. D. U. P., and Chaboyer, W. (2024). Risk factors predicting hospital-acquired pressure injury in adult patients: an overview of reviews. Int. J. Nurs. Stud. 150:104642. doi: 10.1016/j.ijnurstu.2023.104642
Yamada, Y., Hideka, K., Shiota, S., Kuroda, T., and Tsuchiya, T. (2006). Gene cloning and characterization of SdrM, a chromosomally-encoded multidrug efflux pump, from Staphylococcus aureus. Biol. Pharm. Bull. 29, 554–556. doi: 10.1248/bpb.29.554
Zhao, G., Usui, M. L., Lippman, S. I., James, G. A., Stewart, P. S., Fleckman, P., et al. (2013). Biofilms and inflammation in chronic wounds. Adv. Wound Care 2, 389–399. doi: 10.1089/wound.2012.0381
Zhen, X., Stålsby Lundborg, C., Sun, X., Gu, S., and Dong, H. (2020). Clinical and economic burden of Carbapenem-resistant infection or colonization caused by Klebsiella pneumoniae, Pseudomonas aeruginosa, Acinetobacter baumannii: a multicenter study in China. Antibiotics 9:514. doi: 10.3390/antibiotics9080514
Keywords: NaOCl, biofilm, Acinetobacter baumannii, Klebsiella pneumoniae, Staphylococcus aureus, Candida albicans, sodium hypochlorite solution, skin, ESKAPE
Citation: Fabrizio G, Sivori F, Cavallo I, Truglio M, Toma L, Sperati F, Francalancia M, Obregon F, Pamparau L, Kovacs D, Pimpinelli F and Di Domenico EG (2024) Efficacy of sodium hypochlorite in overcoming antimicrobial resistance and eradicating biofilms in clinical pathogens from pressure ulcers. Front. Microbiol. 15:1432883. doi: 10.3389/fmicb.2024.1432883
Received: 14 May 2024; Accepted: 25 June 2024;
Published: 10 July 2024.
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Kurt Henry Piepenbrink, University of Nebraska-Lincoln, United StatesCopyright © 2024 Fabrizio, Sivori, Cavallo, Truglio, Toma, Sperati, Francalancia, Obregon, Pamparau, Kovacs, Pimpinelli and Di Domenico. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Enea Gino Di Domenico, ZW5lYS5kaWRvbWVuaWNvQHVuaXJvbWExLml0; Fulvia Pimpinelli, ZnVsdmlhLnBpbXBpbmVsbGlAaWZvLml0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.