
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 18 July 2024
Sec. Systems Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1432049
Background: The gut microbiota and plasma metabolites play important roles in the progression of drug-induced liver injury (DILI). We investigated the causal associations between the gut microbiota, plasma metabolome, and DILI.
Methods: The summary data for gut microbiota (n = 18,340), plasma metabolome (n = 8,299), and DILI (n = 366,838) were obtained from the large genome-wide association studies. A two-sample Mendelian randomization was performed to explore the associations between the gut microbiota, plasma metabolome, and DILI. Additionally, a two-step Mendelian randomization was performed to explore the potential metabolites.
Results: Five taxa were causally associated with DILI, including Oscillospira [odds ratio (OR) = 2.257, 95% confidence interval (CI) = 1.110–4.590], Blautia (OR = 2.311, 95% CI = 1.010–5.288), Roseburia (OR = 2.869, 95% CI = 1.429–5.761), Fusicatenibacter (OR = 1.995, 95% CI = 1.024–3.890), and Prevotella 7 (OR = 1.549, 95% CI = 1.065–2.253). Moreover, 53 metabolites were causally associated with DILI. After mediation analysis, four taxa were found to affect DILI through five mediation metabolites. N6-carbamoylthreonyladenosine mediated the effect of Blautia on DILI. Acetylcarnitine mediated the effect of Fusicatenibacter on DILI. In addition, 4-cholesten-3-one mediated the effect of Prevotella 7 on DILI. Furthermore, 5,6-dihydrothymine levels and the salicylate-to-citrate ratio mediated the effect of Oscillospira on DILI.
Conclusion: We found that the gut microbiota could affect DILI through plasma metabolites, which could serve as potential biomarkers for risk stratification and elucidate underlying mechanisms for further investigation of DILI.
Drug-induced liver injury (DILI) is liver damage caused by various chemicals and drugs (European Association for the Study of the Liver et al., 2019), affecting 13%−15% of patients with acute liver failure in Western countries (European Association for the Study of the Liver et al., 2019). Compared to other etiologies, patients with DILI present worse transplant-free survival (European Association for the Study of the Liver et al., 2019). Therefore, it is imperative to investigate the occurrence and progression of DILI.
The gut microbiota has been reported to play a pivotal role in liver disease (Chu et al., 2023). However, epidemiological evidence on the associations between the gut microbiota and DILI is conflicting. For example, it has been reported that the abundance of Blautia decreased in mice with DILI compared to controls (Zhang et al., 2022), whereas the abundance of Blautia was reported to increase in mice with DILI compared to controls in other studies (Wang et al., 2022). Conflicting evidence on the association between Bacteroides and DILI was also reported (Chu et al., 2023). The causal effects of the gut microbiota on DILI and its underlying mechanisms remain unclear.
Interestingly, it has been found that the gut microbiota could affect DILI by modulating blood metabolites such as adrenic acid and aspartic acid (Zhao et al., 2022). Given the potential associations between the gut microbiota, blood metabolites, and DILI, we investigated the causal associations between the gut microbiota, plasma metabolome, and DILI, aiming to provide potential targets for risk stratification and to elucidate the underlying mechanisms for further investigation of DILI.
Mendelian randomization, an instrumental variable method utilizing single nucleotide polymorphisms (SNPs) as instruments, has been employed to elucidate the causal associations between two features. Mendelian randomization offers two advantages: mitigating bias resulting from confounding factors and addressing issues related to reverse causality (Emdin et al., 2017). Therefore, we performed a Mendelian randomization analysis to investigate the causal associations between the gut microbiota, plasma metabolome, and DILI.
The study design is shown in Figure 1. Initially, we identified SNPs as instrument variables and performed a univariable Mendelian randomization analysis to explore the causal effects of the gut microbiota and plasma metabolomes on DILI. Next, we implemented a two-step Mendelian randomization to identify potential metabolites as mediators in the associations between the gut microbiota and DILI. Our study was performed in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology Reporting Guidelines (Skrivankova et al., 2021) (Supplementary material).
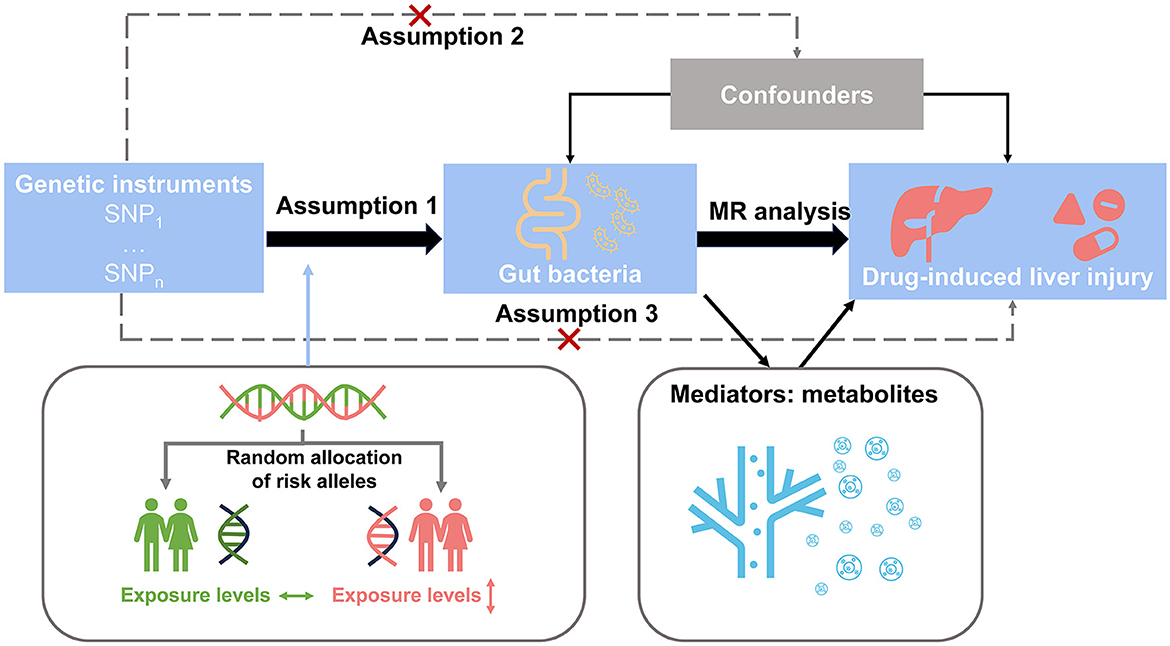
Figure 1. Study design overview. Step 1: We identified SNPs as instrument variables and performed univariable Mendelian randomization analysis to explore the causal effects of the gut microbiota and plasma metabolome on drug-induced liver injury. Step 2: We implemented two-step Mendelian randomization to identify potential metabolites as mediators in the associations between the gut microbiota and drug-induced liver injury.
Genome-wide association study (GWAS) data sources are summarized in Table 1. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Declaration of Helsinki of 1975, as revised in 2008. All studies included in the cited genome-wide association studies had been approved by a relevant review board. All participants provided informed consent.
GWAS data on the gut microbiota were obtained from the MiBioGen consortium based on 18,340 participants, of which 78% were Europeans (Kurilshikov et al., 2021). The MiBioGen consortium analyzed the genome information and 16S fecal microbiome, which resulted in 211 taxa (131 genera, 35 families, 20 orders, 16 classes, and nine phyla). Only data from 131 genera were used in our study. Moreover, GWAS data on plasma metabolomes were obtained from 8,299 European individuals in the Canadian Longitudinal Study on Aging cohort, including 1,091 metabolites and 309 metabolite ratios (Chen et al., 2023). Furthermore, GWAS data for DILI were obtained from the FinnGen consortium, including 388 cases and 366,450 controls (Kurki et al., 2023), which were diagnosed based on the International Classification of Diseases (ICD10: K71). Refer to the cited papers for more detailed information on each GWAS data source. Overlapping individuals have negligible effects on power when selected instrument variables are strong enough (Pierce and Burgess, 2013).
SNPs for the gut microbiota and plasma metabolites were selected as instrument variables with a p-value (< 1 × 10−5). All SNPs were clumped for independent inheritance (R2 < 0.001, within 10 Mb). F-statistics were calculated to assess SNPs' validity, with a threshold of exceeding 10 (Pierce and Burgess, 2013).
A univariable Mendelian randomization analysis was performed to explore the causal effects of the gut microbiota and plasma metabolomes on DILI. Five methods [inverse-variance-weighted (IVW), MR-Egger, weighted median, simple mode, and weighted mode] were used to evaluate and validate causal effects, and assumptions and advantages are summarized in Supplementary Table S1. Inverse-variance-weighted was the primary method used. Cochran's Q statistic was calculated to evaluate heterogeneity. The MR-Egger intercept was performed to evaluate pleiotropy. Only a causal effect with inverse-variance-weighted p-value of < 0.05 and without heterogeneity and pleiotropy (corresponding p-value of >0.05) was identified as a significant effect. Moreover, based on the above criteria, we selected more stringent criteria with consistent directionality across the five methods to analyze the associations between plasma metabolomes and DILI. Furthermore, we used MetaboAnalyst 5.0 (https://www.metaboanalyst.ca/) to implement the enrichment analysis of identified metabolites. Furthermore, reverse causality between the gut microbiota and DILI was assessed using the above methods.
A two-step Mendelian randomization (Burgess et al., 2015) was used to explore the mediation roles of plasma metabolites in the causal effects of the gut microbiota on DILI. Based on the causal effects (βEO) of the gut microbiota on DILI, two causal effects were explored: the causal effects (βMO) of mediators (plasma metabolites) on DILI and the causal effects (βEM) of exposures (the gut microbiota) on mediators. βEM × βMO represented the mediation effect (βmediation). Where there was evidence that the gut microbiota affected plasma metabolites, which in turn affected DILI, we used the product of the coefficient method to evaluate mediation effects. The mediation proportion was calculated using the following formula: βmediation/βEO. Standard errors for mediation effects were derived using the delta method.
All tests were two-sided and performed using the TwoSampleMR (version 0.5.7) packages in R software (version 4.0.2). All IVW results were corrected for multiple testing using the false discovery rate (FDR) method. An FDR p-value of < 0.05 was identified as statistically significant. A p-value of < 0.05 was identified as nominally significant.
SNPs for each exposure and mediator were identified as genetic instruments based on the selection criteria. All SNP F-statistics exceeded 10, indicating enough validity (Supplementary Tables S2–S4).
As shown in Figure 2A, the causal effects of the gut microbiota on DILI were explored, and five taxa presented causal effects on DILI using the inverse-variance-weighted method without heterogeneity or pleiotropy. As shown in Figure 2C, these taxa were all associated with an increasing risk of DILI. Oscillospira (OR = 2.257, 95% CI = 1.110–4.590, p = 0.025), Blautia (OR = 2.311, 95% CI = 1.010–5.288, p = 0.047), and Roseburia (OR = 2.869, 95% CI = 1.429–5.761, p = 0.003) presented causal effects on the increasing risk of DILI, as well as Fusicatenibacter (OR = 1.995, 95% CI = 1.024–3.890, p = 0.043) and Prevotella 7 (OR = 1.549, 95% CI = 1.065–2.253, p = 0.022). Only nominally significant effects were detected rather than statistically significant effects. No reverse causality was detected in the above associations (Supplementary Table S5). Neither heterogeneity nor pleiotropy was detected in the above associations (Supplementary Table S6).
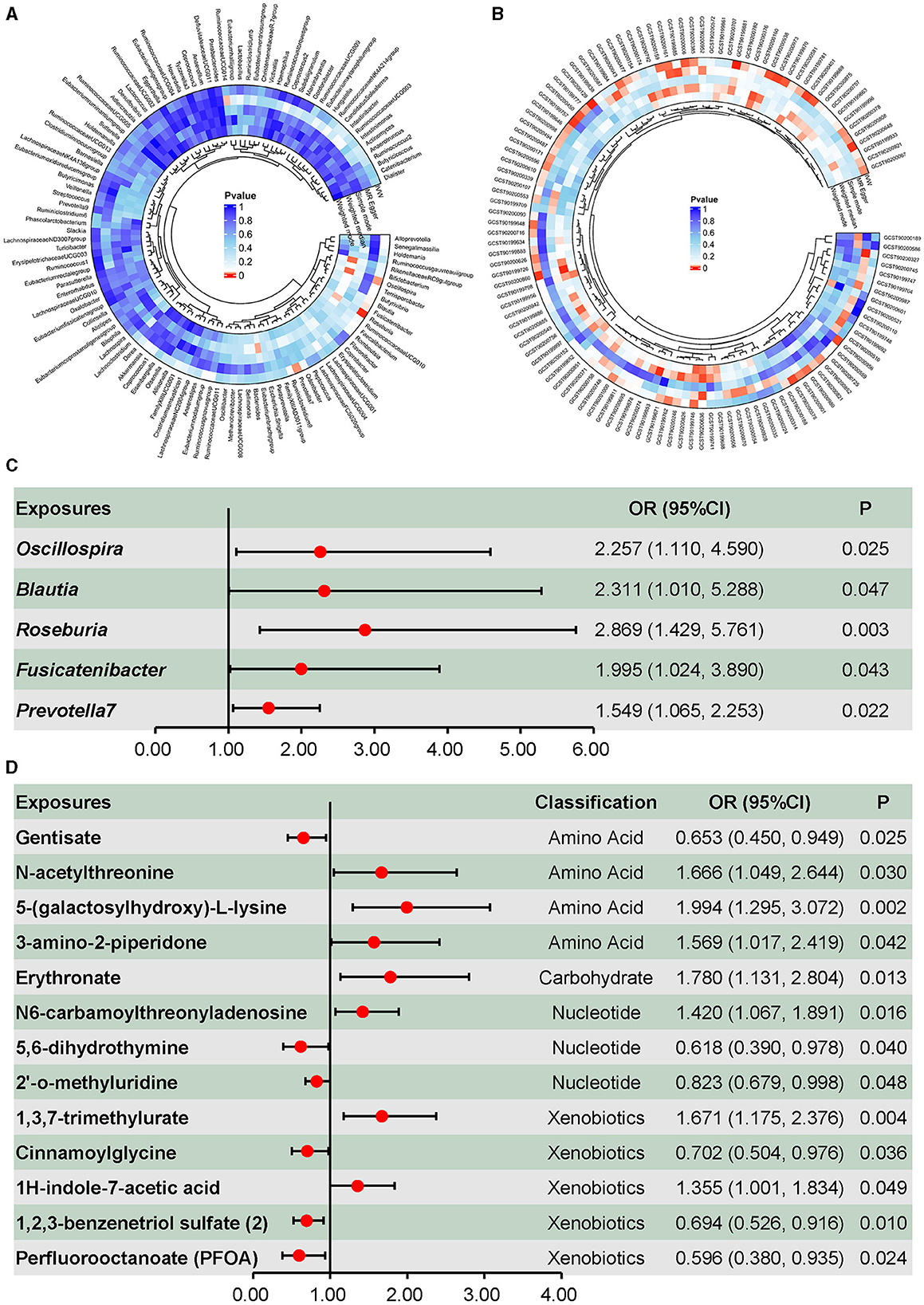
Figure 2. Causal effects of the gut microbiota and plasma metabolites on drug-induced liver injury. (A) Screening of the causal effects of the gut microbiota on drug-induced liver injury. (B) Screening of the causal effects of plasma metabolites on drug-induced liver injury. (C) Causal effects of the gut microbiota on drug-induced liver injury. (D) Causal effects of plasma amino acids, carbohydrate, nucleotides, and xenobiotics on drug-induced liver injury.
Metabolites associated with DILI are summarized in Figure 2B, in which 53 metabolites were identified using the inverse-variance-weighted method and with persistent directionality across all five methods, without heterogeneity or pleiotropy. Among these 53 metabolites, there were 30 known metabolites, including amino acids, carbohydrates, nucleotides, xenobiotics, and lipids. Causal associations between DILI and four amino acids, one carbohydrate, three nucleotides, and five xenobiotics are shown in Figure 2D, including seven positive and six negative causal effects on DILI. Moreover, 17 lipids presented causal effects on DILI, including five positive and 12 negative effects (Figure 3A). Based on known metabolites, enrichment analysis indicated that they were enriched in the beta-oxidation pathways of very long-chain fatty acids and ascorbate and aldarate metabolism (Figures 3B, C).
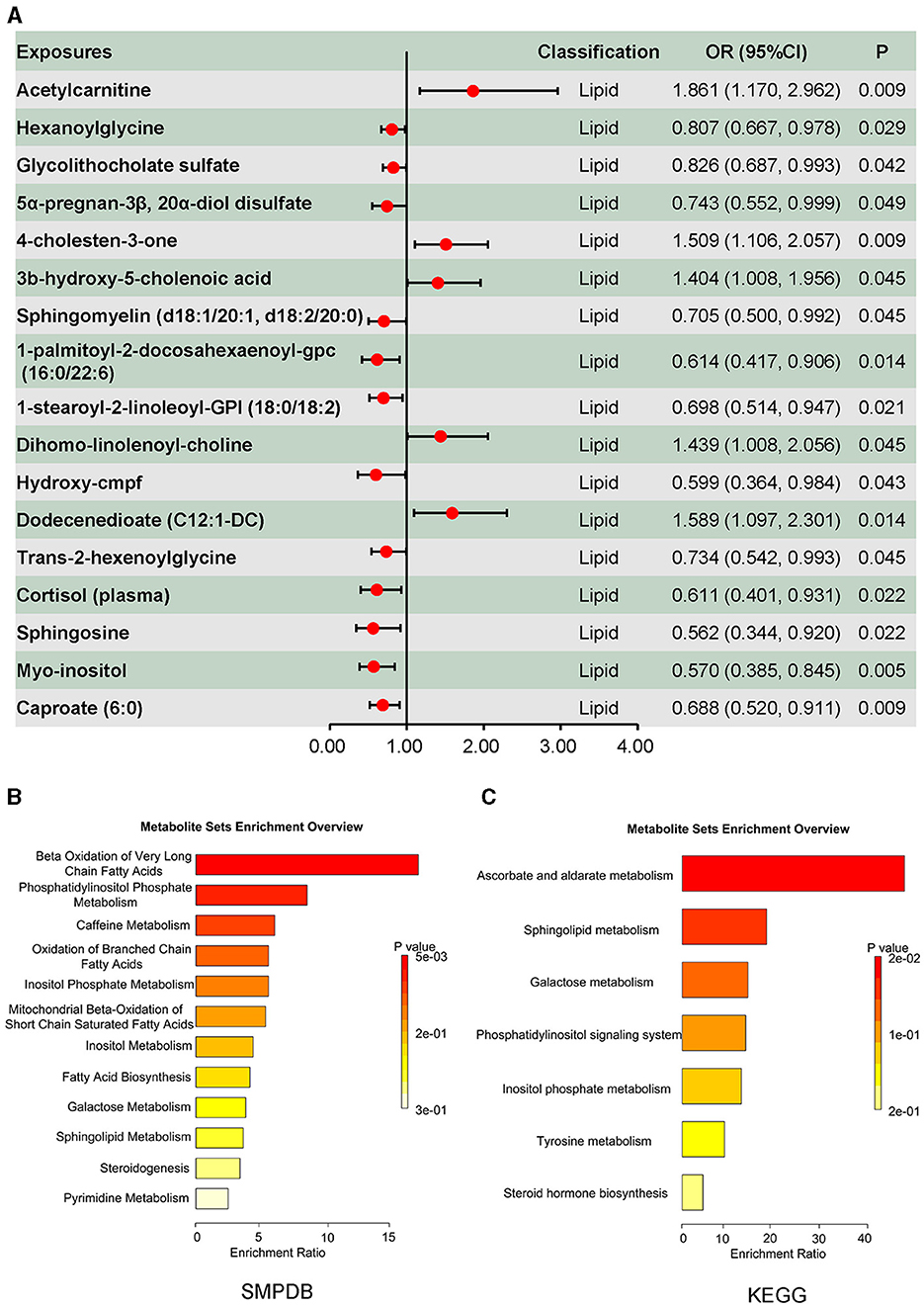
Figure 3. Causal effects of plasma lipid on drug-induced liver injury, and the enrichment analysis results of the causal effect of plasma metabolites on drug-induced liver injury. (A) Causal effects of plasma lipid on drug-induced liver injury. (B) Enrichment analysis results of the causal effect of plasma metabolites on drug-induced liver injury based on Small Molecule Pathway Database (SMPDB). (C) Enrichment analysis results of the causal effect of plasma metabolites on drug-induced liver injury based on the KEGG database.
Furthermore, 14 metabolite ratios and nine unknown metabolites were associated with the DILI (Table 2). Only nominally significant effects were detected rather than statistically significant effects. Neither heterogeneity nor pleiotropy was detected in the above associations (Supplementary Table S6).
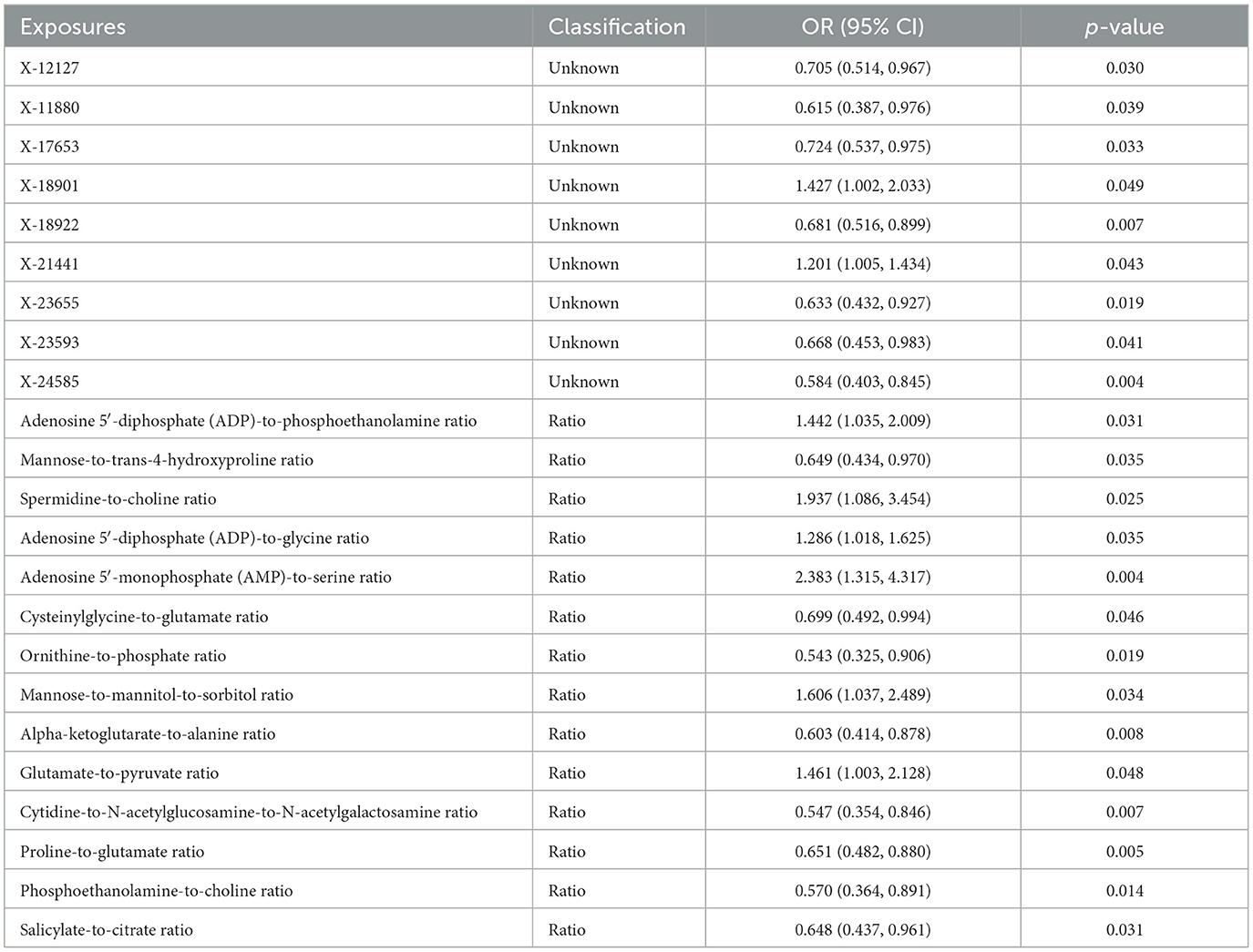
Table 2. Causal effects of plasma metabolite ratios and unknown metabolites on drug-induced liver injury.
A two-step Mendelian randomization was implemented based on the identified gut taxa and plasma metabolites to explore the mediation roles of plasma metabolites in the effects of the gut microbiota on DILI. As shown in Figure 4A, five mediation pathways were identified. N6-carbamoylthreonyladenosine mediated 10.03% (95% CI = 0.00%−20.53%, Figure 4B) of the effect of Blautia on DILI. Acetylcarnitine mediated 14.62% (95% CI = 0.00%−31.41%, Figure 4B) of the effect of Fusicatenibacter on DILI. In addition, 4-cholesten-3-one mediated 3.61% (95% CI = 0.00%−7.99%, Figure 4B) of the effect of Prevotella 7 on DILI. Moreover, 5,6-dihydrothymine levels and the salicylate-to-citrate ratio mediated 10.20% (95% CI = 0.00%−23.42%, Figure 4B) and 8.35% (95% CI = 0.00%−19.60%, Figure 4B) of the effect of Oscillospira on DILI, respectively. Detailed information for mediation analysis is summarized in Supplementary Table S7. These findings indicated that the gut microbiota could affect DILI via plasma metabolites.
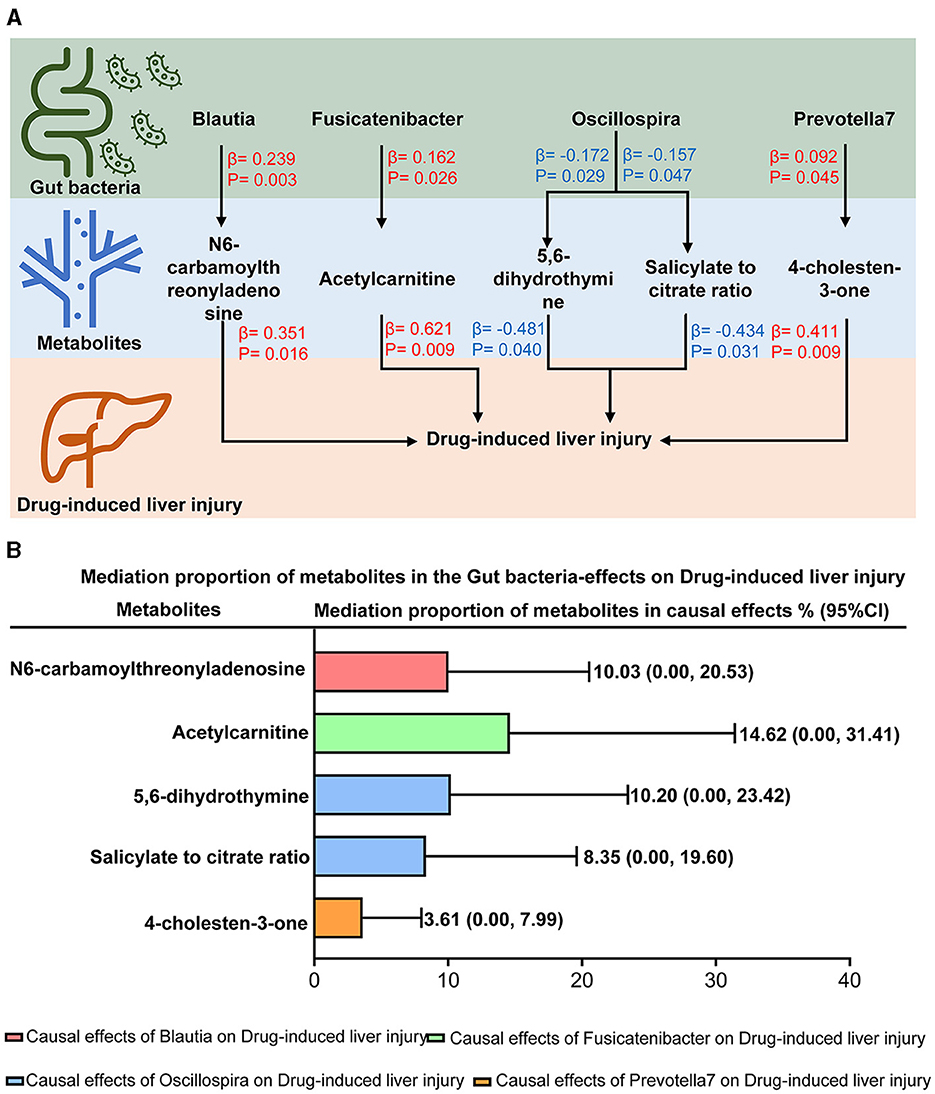
Figure 4. Mediation effects of plasma metabolites on the causal effects of the gut microbiota on drug-induced liver injury. (A) The mediation mode of plasma metabolites on the causal effects of the gut microbiota -on drug-induced liver injury in the mediation analysis. β indicated the causal effects using inverse-variance-weighted method (p < 0.05). Red-β and blue-β signified positive and negative effects, respectively. (B) Mediation proportion of plasma metabolites on the effects of the gut microbiota on drug-induced liver injury.
In this study, we systemically investigated the causal effects of the gut microbiota and blood metabolites on DILI. We revealed that five taxa and 53 metabolites were causally associated with DILI. Further analysis showed that four taxa affected DILI via five mediation metabolites, which could serve as potential biomarkers for risk stratification and as underlying mechanisms for further investigation of DILI.
Blautia and Fusicatenibacter positively affected DILI by modulating plasma N6-carbamoylthreonyladenosine and acetylcarnitine levels, respectively. A previous study found that the abundance of the Balutia genus increased in mice with methotrexate-induced liver injury (Wang et al., 2022), whereas it decreased in mice with triclosan-induced liver injury (Zhang et al., 2022). Our result supported the hypothesis that Blautia was positively associated with DILI. Besides, we found that Blautia could positively affect DILI via modulating plasma N6-carbamoylthreonyladenosine levels. Similarly, we revealed that Fusicatenibacter exerted a positive causal effect on DILI by modulating plasma acetylcarnitine levels. Fusicatenibacter was reported to increase in patients with hepatic cirrhosis and portal vein thrombosis (Huang et al., 2023; Sato et al., 2023). Our result further investigated the role of Fusicatenibacter in DILI (Alotaibi et al., 2016). Interestingly, we found that Fusicatenibacter positively affects DILI through increasing acetylcarnitine levels, which is the underlying mechanism that needs to be explored in future studies.
Prevotella 7 exerted a positive effect on DILI via modulating the plasma 4-cholesten-3-one level. Based on the urine bacteria analysis, the abundance of Prevotella was reported to be increased in patients with anti-tuberculosis drug-induced hepatotoxicity (Wu et al., 2022). Moreover, a previous study found that Prevotella was enriched in mice fed a Western diet, resulting in aggravated carbon tetrachloride-induced liver injury (Yang et al., 2021). Increasing plasma 4-cholesten-3-one levels were reported as a biomarker for non-alcoholic fatty liver disease risk (Pang et al., 2022). Our study indicated the new role of 4-cholesten-3-one due to increased Prevotella 7 and the underlying mechanism of DILI.
Oscillospira positively affected DILI by modulating plasma 5,6-dihydro thymine levels and the salicylate-to-citrate ratio. Observational studies on the associations between Oscillospira and liver disease are conflicting. It was reported that Oscillospira increased in fructose-treated mice, activating the lipopolysaccharide-dependent proinflammatory Toll-like receptor 4/NLRP3 inflammasome pathway (Mastrocola et al., 2018). However, previous studies found that Oscillospira decreased in patients with DILI compared to those at risk of nonalcoholic fatty liver disease and controls (Rodriguez-Diaz et al., 2022). Our study supported the evidence that Oscillospira exerts a positive effect on DILI. It has been found that 5,6-dihydrothymine is associated with DNA damage and repair (Park et al., 2011), whereas there is no evidence of an association between 5,6-dihydrothymine and liver disease. Our study revealed the underlying mechanism that Oscillospira exerted a positive effect on DILI via modulating plasma 5,6-dihydrothymine levels. Besides, although salicylate was reported as a hepatotoxic drug (Doi and Horie, 2010), updated evidence showed the preventative effect of sodium salicylate nanoparticles on cisplatin-mediated hepatotoxicity (Alkhalaf et al., 2023). Furthermore, patients with DILI present with serious mitochondrial damage, resulting in the dysfunction of citric acid (Sanyal et al., 2001; Chella Krishnan et al., 2018; Fromenty, 2020). Our study showed that the salicylate-to-citrate ratio protected the liver from hepatoxicity via the above mechanisms, whereas Oscillospira presented a positive effect on DILI by weakening the protective effect of salicylate on the citrate ratio.
Roseburia exerted a positive effect on DILI. A previous study found that Roseburia increased in mice with d-galactosamine/lipopolysaccharide-induced acute liver injury (Liu et al., 2022), which aligned with our result. However, Roseburia was reported to decrease in mice with carbon tetrachloride-induced liver injury (Yang et al., 2021), though this difference could be attributed to the differences between mouse models and human patients. Moreover, we found no mediating metabolites in the effect of Roseburia on DILI, indicating that Roseburia could directly affect hepatoxicity.
Our study provided potential biomarkers for risk stratification and elucidated the underlying mechanisms for further investigation of DILI. DILI is the most common cause of acute liver failure. The outcome would be fatal for these patients without liver transplantation (European Association for the Study of the Liver et al., 2019). Moreover, ~20% of DILI patients would progress into chronicity, which would result in cirrhosis, liver failure, death, and/or liver transplantation (Lo Re et al., 2015). However, owing to the limitations of existing predictive models in terms of predictive effects and clinical feasibility, accurately predicting chronic and fatal DILI remains difficult. Our study identified the causal associations between the gut microbiota, blood metabolites, and DILI, which could serve as potential biomarkers to predict the outcomes of DILI patients. Furthermore, the mechanism of DILI is involved in hepatic metabolism and excretion of the DILI-causative agent, leading to cellular stress, cell death, activation of an adaptive immune response, and a failure to adapt (Andrade et al., 2019). Our study revealed that the gut microbiota could affect DILI via plasma metabolites, which could be the underlying mechanism for further investigation of DILI.
There are several strengths in our study. First, we used Mendelian randomization analysis to explore the associations between the gut microbiota, plasma metabolome, and toxic liver injury, which could mitigate bias from confounding factors and address issues related to reverse causality (Emdin et al., 2017). Second, we revealed that the gut microbiota could affect DILI via plasma metabolites, which could serve as potential biomarkers for risk stratification and as underlying mechanisms for further investigation of DILI. Third, we assessed causal associations in the European population to avoid bias arising from population structure.
Limitations need to be considered when interpreting our results. First, we used less stringent criteria (1 × 10−5) to identify instrument variables for partial exposures. However, it was reassuring that F-statistics exceeded 10, indicating that the weak instrument bias should be minimal (Pierce and Burgess, 2013). Second, the study populations were limited to those with European ancestry. Therefore, our results should be cautiously interpreted across non-European populations.
We found causal associations between the gut microbiota, blood metabolites, and DILI, which could serve as potential biomarkers to predict the outcomes of DILI patients. Furthermore, we revealed that the gut microbiota could affect DILI via plasma metabolites, which could be an underlying mechanism of DILI worthy of further investigation.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
This is a secondary analysis based on summary statistics from existing, published studies. The ethical approval and informed consent have been obtained by all original studies. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
HF: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft. SZ: Methodology, Writing – review & editing. SS: Methodology, Writing – review & editing, Formal analysis, Investigation. QX: Writing – review & editing, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Nos. 82070604, 82270618, and 82300686). The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors thank all investigators for sharing these data.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1432049/full#supplementary-material
DILI, drug-induced liver injury; FDR, false discovery rate; GWAS, genome-wide association study; SMPDB, small molecule pathway database; SNPs, single nucleotide polymorphisms.
Alkhalaf, M., Mohamed, N. A., and El-Toukhy, S. E. (2023). Prophylactic consequences of sodium salicylate nanoparticles in cisplatin-mediated hepatotoxicity. Sci. Rep. 13:10045. doi: 10.1038/s41598-023-35916-9
Alotaibi, S. A., Alanazi, A., Bakheet, S. A., Alharbi, N. O., and Nagi, M. N. (2016). Prophylactic and therapeutic potential of acetyl-L-carnitine against acetaminophen-induced hepatotoxicity in mice. J. Biochem. Mol. Toxicol. 30, 5–11. doi: 10.1002/jbt.21733
Andrade, R. J., Chalasani, N., Björnsson, E. S., Suzuki, A., Kullak-Ublick, G. A., Watkins, P. B., et al. (2019). Drug-induced liver injury. Nat. Rev. Dis. Primers 5:58. doi: 10.1038/s41572-019-0105-0
Burgess, S., Daniel, R. M., Butterworth, A. S., and Thompson, S. G. (2015). Network Mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int. J. Epidemiol. 44, 484–495. doi: 10.1093/ije/dyu176
Chella Krishnan, K., Kurt, Z., Barrere-Cain, R., Sabir, S., Das, A., Floyd, R., et al. (2018). Integration of multi-omics data from mouse diversity panel highlights mitochondrial dysfunction in nonalcoholic fatty liver disease. Cell Syst. 6, 103–115.e107. doi: 10.1016/j.cels.2017.12.006
Chen, Y., Lu, T., Pettersson-Kymmer, U., Stewart, I. D., Butler-Laporte, G., Nakanishi, T., et al. (2023). Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 55, 44–53. doi: 10.1038/s41588-022-01270-1
Chu, H. K., Ai, Y., Cheng, Z. L., Yang, L., and Hou, X. H. (2023). Contribution of gut microbiota to drug-induced liver injury. Hepatobiliary Pancreat. Dis. Int. 22, 458–465. doi: 10.1016/j.hbpd.2023.06.008
Doi, H., and Horie, T. (2010). Salicylic acid-induced hepatotoxicity triggered by oxidative stress. Chem. Biol. Interact. 183, 363–368. doi: 10.1016/j.cbi.2009.11.024
Emdin, C. A., Khera, A. V., and Kathiresan, S. (2017). Mendelian randomization. JAMA 318, 1925–1926. doi: 10.1001/jama.2017.17219
European Association for the Study of the Liver Clinical Practice Guideline Panel, and EASL Governing Board representative. (2019). EASL clinical practice guidelines: drug-induced liver injury. J. Hepatol. 70, 1222–1261. doi: 10.1016/j.jhep.2019.02.014
Fromenty, B. (2020). Alteration of mitochondrial DNA homeostasis in drug-induced liver injury. Food Chem. Toxicol. 135:110916. doi: 10.1016/j.fct.2019.110916
Huang, X. Y., Zhang, Y. H., Yi, S. Y., Lei, L., Ma, T., Huang, R., et al. (2023). Potential contribution of the gut microbiota to the development of portal vein thrombosis in liver cirrhosis. Front. Microbiol. 14:1217338. doi: 10.3389/fmicb.2023.1217338
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Kurki, M. I., Karjalainen, J., Palta, P., Sipilä, T. P., Kristiansson, K., Donner, K. M., et al. (2023). FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518. doi: 10.1038/s41586-022-05473-8
Liu, S., Yin, R., Yang, Z., Wei, F., and Hu, J. (2022). The effects of rhein on D-GalN/LPS-induced acute liver injury in mice: results from gut microbiome-metabolomics and host transcriptome analysis. Front. Immunol. 13:971409. doi: 10.3389/fimmu.2022.971409
Lo Re, V. 3rd, Haynes, K., Forde, K. A., Goldberg, D. S., Lewis, J. D., Carbonari, D. M., et al. (2015). Risk of acute liver failure in patients with drug-induced liver injury: evaluation of Hy's law and a new prognostic model. Clin. Gastroenterol. Hepatol. 13, 2360–2368. doi: 10.1016/j.cgh.2015.06.020
Mastrocola, R., Ferrocino, I., Liberto, E., Chiazza, F., Cento, A. S., Collotta, D., et al. (2018). Fructose liquid and solid formulations differently affect gut integrity, microbiota composition and related liver toxicity: a comparative in vivo study. J. Nutr. Biochem. 55, 185–199. doi: 10.1016/j.jnutbio.2018.02.003
Pang, Y., Kartsonaki, C., Lv, J., Millwood, I. Y., Fairhurst-Hunter, Z., Turnbull, I., et al. (2022). Adiposity, metabolomic biomarkers, and risk of nonalcoholic fatty liver disease: a case-cohort study. Am. J. Clin. Nutr. 115, 799–810. doi: 10.1093/ajcn/nqab392
Park, Y., Li, Z., Cloutier, P., Sanche, L., and Wagner, J. R. (2011). DNA damage induced by low-energy electrons: conversion of thymine to 5,6-dihydrothymine in the oligonucleotide trimer TpTpT. Radiat. Res. 175, 240–246. doi: 10.1667/RR2381.1
Pierce, B. L., and Burgess, S. (2013). Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 178, 1177–1184. doi: 10.1093/aje/kwt084
Rodriguez-Diaz, C., Taminiau, B., García-García, A., Cueto, A., Robles-Díaz, M., Ortega-Alonso, A., et al. (2022). Microbiota diversity in nonalcoholic fatty liver disease and in drug-induced liver injury. Pharmacol. Res. 182:106348. doi: 10.1016/j.phrs.2022.106348
Sanyal, A. J., Campbell-Sargent, C., Mirshahi, F., Rizzo, W. B., Contos, M. J., Sterling, R. K., et al. (2001). Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120, 1183–1192. doi: 10.1053/gast.2001.23256
Sato, S., Iino, C., Chinda, D., Sasada, T., Tateda, T., Kaizuka, M., et al. (2023). Effect of liver fibrosis on oral and gut microbiota in the Japanese general population determined by evaluating the fibroscan-aspartate aminotransferase score. Int. J. Mol. Sci. 24:13470. doi: 10.3390/ijms241713470
Skrivankova, V. W., Richmond, R. C., Woolf, B. A. R., Yarmolinsky, J., Davies, N. M., Swanson, S. A., et al. (2021). Strengthening the reporting of observational studies in epidemiology using mendelian randomization: the STROBE-MR statement. JAMA 326, 1614–1621. doi: 10.1001/jama.2021.18236
Wang, C., Zhao, S., Xu, Y., Sun, W., Feng, Y., Liang, D., et al. (2022). Integrated microbiome and metabolome analysis reveals correlations between gut microbiota components and metabolic profiles in mice with methotrexate-induced hepatoxicity. Drug Des. Devel. Ther. 16, 3877–3891. doi: 10.2147/DDDT.S381667
Wu, S., Wang, M., Zhang, M., and He, J. Q. (2022). Metabolomics and microbiomes for discovering biomarkers of antituberculosis drugs-induced hepatotoxicity. Arch. Biochem. Biophys. 716:109118. doi: 10.1016/j.abb.2022.109118
Yang, L., Li, Y., Wang, S., Bian, X., Jiang, X., Wu, J., et al. (2021). Western diet aggravated carbon tetrachloride-induced chronic liver injury by disturbing gut microbiota and bile acid metabolism. Mol. Nutr. Food Res. 65:e2000811. doi: 10.1002/mnfr.202000811
Zhang, P., Zheng, L., Duan, Y., Gao, Y., Gao, H., Mao, D., et al. (2022). Gut microbiota exaggerates triclosan-induced liver injury via gut-liver axis. J. Hazard. Mater. 421:126707. doi: 10.1016/j.jhazmat.2021.126707
Keywords: toxic liver disease, drug-induced liver disease, gut microbiota, plasma metabolome, Mendelian randomization, liver disease
Citation: Fu H, Zhao S, Song S and Xie Q (2024) Gut microbiota causally affects drug-induced liver injury via plasma metabolites: a Mendelian randomization study. Front. Microbiol. 15:1432049. doi: 10.3389/fmicb.2024.1432049
Received: 15 May 2024; Accepted: 28 June 2024;
Published: 18 July 2024.
Edited by:
Jinbo Xiong, Ningbo University, ChinaReviewed by:
Kendall S. Stocke, University of Louisville, United StatesCopyright © 2024 Fu, Zhao, Song and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Xie, eGllcWluZ3JqaEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.