- 1Engineering Research Center of Southwest Animal Disease Prevention and Control Technology, Ministry of Education of the People’s Republic of China, Chengdu, China
- 2Key Laboratory of Animal Disease and Human Health of Sichuan Province, Chengdu, China
- 3International Joint Research Center for Animal Disease Prevention and Control of Sichuan Province, Chengdu, China
- 4Institute of Veterinary Medicine and Immunology, Sichuan Agricultural University, Chengdu, China
- 5Research Center of Avian Disease, College of Veterinary Medicine, Sichuan Agricultural University, Chengdu, China
- 6Sinopharm Yangzhou VAC Biological Engineering Co., Ltd., Yangzhou, China
Alphaherpesvirus is a widespread pathogen that causes diverse diseases in humans and animals and can severely damage host health. Alphaherpesvirus particles comprise a DNA core, capsid, tegument and envelope; the tegument is located between the nuclear capsid and envelope. According to biochemical and proteomic analyses of alphaherpesvirus particles, the tegument contains at least 24 viral proteins and plays an important role in the alphaherpesvirus life cycle. This article reviews the important role of tegument proteins and their interactions during the viral life cycle to provide a reference and inspiration for understanding alphaherpesvirus infection pathogenesis and identifying new antiviral strategies.
1 Introduction
Herpesviruses are a class of double-stranded DNA viruses with a tegument structure (Davison et al., 2009). According to the different biological characteristics and genetic structures of viruses, the International Committee on Taxonomy of Viruses (ICTV) has divided Herpesviridae into three subfamilies: Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae (Gatherer et al., 2021). At present, ICTV has classified Alphaherpesvirinae a into 49 viruses in 5 genera, common type includes herpes simplex virus type-1/2 (HSV-1/2), duck plague virus (DPV), pseudorabies virus (PRV), varicella-zoster virus (VZV), equine herpesvirus (EHV), bovine herpesvirus (BHV-1) and Marek’s disease virus (MDV)and so on (International Committee on Taxonomy of Viruses Executive Committee, 2020).
The complete virion of all herpesviruses consists of four parts: the core containing the double-stranded DNA genome, the capsid, the tegument and the envelope. The presence of a complex tegument between the viral capsid and the envelope is typical of herpesvirus morphology, and this layer contains many proteins (Table 1), which can be divided into three groups: inner tegument proteins closely related to the nucleocapsid, outer tegument proteins that contact the capsule membrane and “intermediate” tegument proteins, which are unique to the Alphaherpesvirinae. Intermediate tegument proteins are among the most abundant tegument proteins and act as the central hub of the protein interaction network (Wolfstein et al., 2006; Radtke et al., 2010).
Herpesviruses are dependent on host cells for proliferation, and the virus enters host cells through endocytosis and membrane fusion (Mercer et al., 2010). When virions infect cells, viral glycoproteins mediate the fusion of the viral envelope and cell membrane (Johnson and Baines, 2011). Subsequently, the viral nucleocapsid is released into the cytoplasm, and the viral DNA migrates to the nucleus through the nuclear pore and begins to be replicated, initiating transcription of the viral genome (Liashkovich et al., 2011). The capsid assembles in the nucleus, and the genome is packaged into it (Heming et al., 2017). Next, the virions encased in the primary envelope enter the inner nuclear membrane (INM) through budding and then fuse with the outer nuclear membrane to undergo de-envelopment (Bigalke and Heldwein, 2016). The viral capsid is released into the cytosol, where the virus then acquires the tegument and the secondary envelope to reach maturation, and the progeny virus is discharged from the cell by exocytosis or via the endoplasmic reticulum system (Crump, 2018).
Tegument proteins and their interactions play important roles in the viral life cycle, including but not limited to targeting the movement of virion components into and out of the nucleus, recruitment of molecular motors during movement into and out of the cell, regulation of viral and host gene and protein expression, and assembly of virions during nuclear egress (Kelly et al., 2009). They also provide a scaffold for the assembly of the virion, creating a network of interactions that connect the capsid and the viral envelope proteins (Guo et al., 2010). This article reviews the important functions of tegument proteins and their interactions at various stages of the viral life cycle, with an emphasis on tegument proteins from the Alphaherpesvirinae, in particular HSV-1 PRV and VZV, since these represent the most extensively studied.
2 Function of alphaherpesvirus tegument proteins at various stages of the virus life cycle
2.1 Virus adsorption, invasion and decapsidation
Herpesvirus attachment to cell surfaces is a relatively nonspecific process based on electrical charge and is achieved by reversible binding of viral envelope glycoproteins (gC and gB in Alphaherpesvirinae) to heparan sulfate proteoglyca(HSPG) and chondroitin sulfate proteoglycan(CSPG) (Spear and Longnecker, 2003). These initial entry steps rely on interactions between glycoproteins, primarily gB, gC, and gD, as well as host cell receptors and do not directly require tegument proteins (Spear, 2004). The virus particles entering the cell undergo different degrees of degeneration, expansion, and cleavage, and some virus components are released; subsequently, most of the viral tegument proteins are dissociated, and their respective functions are then exerted. This dissociation process occurs mainly via the viral kinases pUS3 and pUL13 (Morrison et al., 1998). The remaining tegument proteins are connected to the nucleocapsid and participate in the transport of microtubule-dependent cytokinetic proteins to the nucleocapsid. The HSV-1 inner tegument proteins pUL36 and pUL37 interact with the cellular motor proteins kinesin and dynein, respectively, and these interactions lead to bidirectional or hopping movement of the capsid along the microtubule (Döhner et al., 2002; Douglas et al., 2004; Antinone and Smith, 2010; Radtke et al., 2010; Kramer and Enquist, 2013). For PRV, VP1/2 (pUL36 homologous protein) interacts with the Dynein intermediate chain (DIC) and the large subunit p150 of the dynactin-activating protein to transport cargo through a network of microtubules (Bremner et al., 2009). However, in HSV-1, the pUL37 protein interacts with dynetin DIC during the early stage of infection. This pUL37-DIC interaction is synchronized with DIC phosphorylation in infected cells (Musarrat et al., 2021). Since the kinetic protein and kinetic protein motor complex cooperate in retrocellular transport, the binding of pUL37 to the kinetic protein may stabilize the movement of the entire virus, facilitating capsid transport to the nucleus within the cell. In addition, pUL34 can interact with DYNC1/1a (Ye et al., 2000; Martínez-Moreno et al., 2003). The inner tegument proteins participate in the recruitment of molecular motors, thus enabling transport of the viral capsid along microtubules.
In this process, major structural components of the HSV-1 tegument are phosphorylated upon entry into infected cells; for example, the phosphorylation of VP13/14 and VP22 mediates their dissociation from virions in vitro, and the phosphorylation of VP1/2 may be necessary for its interaction with the remaining tegument (Morrison et al., 1998).
2.2 Viral gene transcription and DNA replication
The viral DNA entering the nucleus is cycled to stimulate the transcription and translation of the viral immediate early (IE) gene under the action of the transcriptional activator VP16, and after translation, the IE product activates and regulates the transcription of the early (E) gene. The translated E protein products activate and regulate the transcription of late (L) genes, which are mainly structural proteins, in a cascade reaction (Simmen et al., 1997; Liu et al., 2015). Therefore, the tegument protein VP16 is a key promoter of this process. VP16 works with host cytokine 1 (HCF-1) and octamer transcription factor-1 (Oct-1/POU2F1) to form a complex (VIC) that activates transcription of the immediate early rise gene and is critical for initiating the lytic replication of HSV-1 (Shaw et al., 1995). After the entry of HSV-1 into cells, HCF-1 interacts with VP16 through six Kelch domains on its N-terminus (amino acid sequence 3-455 aa) and mediates the nuclear input of VP16 (Shaw et al., 1995). After entering the nucleus, the host protein Oct-1 binds to the HFC-1 and VP16 complexes via its POU-specific domain (POUS) and recognizes five HSV-1 IE genes (ICP0, ICP4, ICP22, ICP27, ICP27). The VP16 reaction sequence (TAATGARAT) in the promoter activates its transcription by recruiting lysine-specific demethylase 1 (LSD1) to demethylate the histone bound to the IE promoter (Liang et al., 2009; Zhou et al., 2010; Roizman and Whitley, 2013). Transcription of the IE gene then activates the remaining viral gene, initiating the viral cleavage replication cycle. HSV-1 gene promoters include binding sites for host cell and viral proteins. These sequences include multiple GC-rich sequences or GC boxes in the promoters of the IE and E genes that bind to the cellular transcription factor specific protein 1(Sp1) (Jones and Tjian, 1985; Jones et al., 1985). Sp1 is a universally expressed zinc finger transcription factor with a recognized role in regulating viral and cellular gene transcription (Kaczynski et al., 2003). It can promote HSV-1 transcription in conditions or contexts in which the activity or expression of viral transactivators like VP16 or ICP0 is limited (Sodroski et al., 2024). Following acute infection, BoHV-1 establishes lifelong latency in sensory neurons present in trigeminal ganglia (TG) and unknown cells in pharyngeal tonsil (El-Mayet et al., 2017). In the process, the immediate early transcription unit 1 (IEtu1) promoter drives expression of bICP0 and bICP4, The IEtu1 promoter contains two glucocorticoid receptor (GR) responsive elements (GREs) that are transactivated by activated GR. GC-rich motifs, including consensus binding sites for Sp1, are in the IEtu1 promoter sequences (El-Mayet and Jones, 2024a,b). Members of the KLF family, including Krüppel-like factor 15 (KLF15), belong to the Sp1 transcription factor family (Bieker, 2001; Kaczynski et al., 2003). Members of the KLF family bind GC- or Ca- rich motifs, including some containing consensus Sp1 binding sites (Bieker, 2001). Since BoHV-1 and HSV-1 are GC-rich genomes, KLF15 may stimulate certain viral promoters. Under stress stimulation, GRs and KLF15 form a feed-forward loop that stimulates viral gene expression and productive infection following stressful stimuli. (Mangan and Alon, 2003; Sasse et al., 2013, 2015; El-Mayet et al., 2017). New studies have demonstrated that ICP27 enhancer sequences were transactivated by GR and KLF15. Mutation of a consensus Sp1 binding site within ICP27 enhancer sequences impaired transactivation by GR and KLF15 (Ostler and Jones, 2021).
There are also many tegument proteins that play important roles in this process. In DPV, pUL14 interacts with VP16 via a nonclassical nuclear localization signal (NLS) sequence, facilitating the translocation of VP16 to the nucleus (Li et al., 2016). This process is necessary for effective nuclear transport of VP16 (Yamauchi et al., 2008). VP1/2 (pUL36) is the largest tegument protein of HSV-1, and the interaction between VP16 and VP1/2 induces the transcription of the IE gene (Svobodova et al., 2012). VP16 interacts with the virion host shutoff protein (VHS), which is encoded by the UL41 gene, in this process. Previous studies have shown that during viral replication, HSV-1 VHS induces the accumulation of viral proteins and gradually reduces the production of host proteins (Matis and Kúdelová, 2001). This phenomenon, known as “host shut down”, is achieved through VHS activity, and its structural role is strictly related to endoribonuclease activity, which is responsible for host mRNA degradation and viral mRNA turnover (Smiley et al., 2001). VP16 stimulates IE gene transcription, and VHS inhibits host protein synthesis and induces accelerated turnover of cellular and viral mRNAs, while VHS has been shown to play a role in the transition from IE gene to E gene expression by actively degrading IE gene mRNA (Everly et al., 2002; Esclatine et al., 2004; Taddeo and Roizman, 2006). In this model, another protein, VP22, is also thought to play an important role in transcriptional activation because both VHS and VP22 can bind to VP16 (Smibert et al., 1994; Elliott et al., 1995; O’Regan et al., 2007b); therefore, it has been proposed that these three proteins form the trimer complex needed to neutralize the RNase activity of VHS and that in the absence of VP22 or VP16, VHS eventually degrades viral mRNA in an unrestricted manner, resulting in a complete shutdown of viral protein synthesis (Sciortino et al., 2007; Duffy et al., 2009; Mbong et al., 2012b). In addition, pUL47 can deliver a portion of VHS to the nucleus. In transfected cells, VHS with mutations in the nuclear export signal (NES) steadily degraded mRNA but did not in infected cells, suggesting that pUL47 largely blocked viral mRNA degradation by VHS to help regulate this process (Shu et al., 2013). VP16 triggers the cascade reaction by regulating IE gene transcription, and the subsequent expression of the IE proteins ICP0 and ICP4 can also enhance the effective expression of the HSV-1 gene (Cai and Schaffer, 1992; Mossman and Smiley, 1999). ICP0 is a viral E3 ubiquitin ligase that targets restrictive host cell proteins for degradation and promotes IE, E, and L protein expression (Cai and Schaffer, 1992; Mossman and Smiley, 1999; Boutell et al., 2002; Orzalli et al., 2012). Both VP16 and ICP0 promote the removal of nucleosomes and restrictive heterochromatin deposited on the viral genome, thereby increasing viral DNA accessibility and subsequent gene expression (Hagglund and Roizman, 2004; Herrera and Triezenberg, 2004; Cliffe and Knipe, 2008). ICP4 is a viral transcription factor necessary for the stable expression of the E and L genes (Watson and Clements, 1980; DeLuca and Schaffer, 1985). Moreover, ICP4 can bind to specific or nonspecific sequences in the viral promoter while recruiting universal RNA polymerase II (Pol II) to HSV-1 DNA to increase the transcription of most viral genes (DeLuca and Schaffer, 1985; Sodroski et al., 2024), and VP22 appears to promote ICP4 synthesis after transcription (Okada et al., 2018).
To replicate efficiently, herpesviruses have also evolved numerous sophisticated protective mechanisms. For example, dUTPases are known to catalyze the hydrolysis of dUTP to dUMP and pyrophosphate and to play a role in the accurate replication of DNA genomes by preventing the misincorporation of dUTP into replicated DNA (Sedwick et al., 1986; Vértessy and Tóth, 2009). The HSV-1 dUTPase (vdUTPase) is encoded by the pUL50 gene and is conserved throughout Herpesviridae (Preston and Fisher, 1984; McGeehan et al., 2001). In infected cells, pUS3 phosphorylates serine 187 of dUTPase, and this phosphorylation promotes viral replication by regulating the optimal enzyme activity of dUTPase (Kato et al., 2014c). When serine 187 was replaced by alanine, the virulence of recombinant HSV-1 in the central nervous system of mice was significantly reduced, but it had no effect on viral replication or pathogenicity in the eyes or vagina of mice after ocular or vaginal inoculation, respectively (Kato et al., 2014b). Moreover, this enhancement of vdUTPase activity compensates for the low dUTPase activity in the host cell, resulting in efficient viral replication (Kato et al., 2014a). In addition, HSV-1 ICP34.5 can interact with proliferating nuclear antigens and is also important for regulating viral DNA replication (Brown et al., 1997; Harland et al., 2003); the direct interaction of BHV-1 pUL49 with histones or of HSV-1 pUL49 with TAF-1 appears to regulate nucleosome formation and may play an unknown role in early viral replication (Ren et al., 2001; van Leeuwen et al., 2003).
2.3 Nuclear egress and envelopment
The herpesvirus capsid is assembled in the nucleus and undergoes a two-step process to cross the nuclear membrane. First, the capsid becomes encapsulates in the INM with the help of the nuclear egress complex (NEC) protein pUL31/34. Second, the perinuclear envelope virion (PEV) fuses with the outer nuclear membrane (ONM) to transport the capsid into the cytoplasm. Once inside the cytoplasm, the capsid is re-encapsulated in the Golgi/trans-Golgi apparatus, where it produces mature virions (Roller and Johnson, 2021). NEC is a heterodimer complex of pUL31 and pUL34 that is essential for herpesvirus nucleation, where pUL34 acts as a type II membrane protein that tethers the NEC to the INM and exposes pUL31 to the karyoplasm (Mettenleiter et al., 2013). The pUL31 protein of HSV-1 relies on physical interaction with the pUL34 protein for stability in infected cells (Ye et al., 2000), and pUL31 is also necessary for the localization of the pUL34 protein to the nucleus, especially the INM (Yamauchi et al., 2001). During this process, the protein kinase pUS3 is recruited to the thin nuclear layer to phosphorylate and dissociate the NEC, thereby promoting capsid budding and nucleation (Mou et al., 2007, 2009). The phosphorylation of pUL31 and pUL34 determines the nature of nuclear envelope deformation, while the viral protein pUL21 regulates the phosphorylation of the two NEC components (Muradov et al., 2021); pUL47 colocalizes with pUL31, pUL34, and pUS3 in the nuclear membrane and forms high-order complexes with these viral proteins in HSV-1-infected cells. The localization of pUL47, pUL31, and pUL34 on the nuclear membrane is regulated by pUS3 kinase activity (Kato et al., 2011; Deng et al., 2022). Moreover, pUL47 may promote the recruitment of the nucleocapsid by interacting with the pUL17-pUL25-pUL31 complex, thereby upregulating the primary capsular membrane of the nucleocapsid to stabilize the binding between the capsid and the pUL34/pUL31 complex at the nuclear membrane, which is necessary for the formation of an effective primary envelope of the nucleocapsid during HSV-1 nucleation (Liu et al., 2014). In addition to pUS3, pUL13 also acts as a phosphokinase that phosphorylates pUS3 and thus controls the nuclear edge localization of pUL31 and pUL34 (Kato et al., 2006; Cano-Monreal et al., 2009). It has also been suggested that the pUL16 and pUL21 proteins work together to promote capsid nucleation (Gao et al., 2017).
The nucleocapsid buds into the perinuclear space at a specific site in the INM, obtains the primary envelope, fuses with the ENM and is released into the cytoplasm, completing the de-envelopment process. Moreover, the immature virions obtain more tegument proteins (mainly outer tegument proteins) in the cytoplasm and then reach the Golgi apparatus, where secondary envelopment occurs, and the final envelope is obtained by budding (Guo et al., 2010). In the literature, tegument proteins are typically divided conceptually into “inner tegument proteins”, which are more closely associated with the capsid, “outer tegument proteins”, which are more closely associated with the capsule membrane, and “intermediate tegument proteins”, which are located between these two types of proteins (Hogue, 2021). Multiple protein interactions are involved in the process of secondary envelopment. Currently, a widely accepted model is that the viral glycoprotein embedded in the Golgi membrane recruits the outer tegument protein components, while the viral capsid is surrounded by the inner tegument protein (Owen et al., 2015).
VP22 (pUL49) is a central hub of tegument-membrane interactions. VP22 (pUL49) has been reported to bind the cytoplasmic tails of gE, gM, and gD, and BHV-1 VP22 can also interact directly with gN, a glycoprotein encoded by the pUL49.5 gene, without relying on gM (Fuchs et al., 2002b; Elliott et al., 2005; Farnsworth et al., 2007; O’Regan et al., 2007a; Stylianou et al., 2009; Maringer et al., 2012; Pannhorst et al., 2018). VP22 (pUL49) can also bind to pUL16 (Starkey et al., 2014), and pUL16 in turn interacts with pUL21, pUL11, and gE (Farnsworth et al., 2007; Harper et al., 2010; Meckes et al., 2010; Han et al., 2011; Chadha et al., 2012) to provide additional contact with the membrane. Furthermore, VP22 may also bind directly to cell membranes via alkaline residues that bind to acidic cytophospholipids (Brignati et al., 2003). VP16 (pUL48) is another central hub of tegument-membrane interactions and has been reported to bind to gH (Gross et al., 2003), although this interaction may not be conserved (Omar et al., 2013).
In addition, there are several important tegument-membrane interactions, such as those involving pUL51 and pUL11, which appear to be conserved, and they can directly bind the membrane by acyl modification [pUL51 is palmitoacylated (Nozawa et al., 2003); additionally, pUL11 is both myristoylated and palmitoylated (MacLean et al., 1989; Loomis et al., 2001)]. Because the “intermediate tegument protein” type is specific to the alphaherpesviruses, the above membrane contact model involving gD and gE (Roller and Johnson, 2021) is also specific to the alphaherpesvirus subfamily, indicating that the modification of pUL51 and pUL11 is important for f virion release among the entire herpesvirus family.
Furthermore, VP22 and VP16 are also at the core of the tegument protein interaction network. The correct assembly of ICP0 (Maringer and Elliott, 2010), ICP4 (Elliott et al., 2005), and pUL47 (Che et al., 2013; Sucharita et al., 2021) into virions requires the participation of VP22; VP22 is also required for the effective redistribution of VP16, ICP0, ICP4, ICP27, and the cellular protein Hsc-70 into the cytoplasm of infected cells, and the double-leucine motif of VP22 is critical for the regulation of this process (Tanaka et al., 2012). VP22 assembly itself involves two determinants, the C-terminal domain and the internal VP16 interaction domain, both of which are necessary for VP22 to effectively recruit to the virus assembly site (Hafezi et al., 2005), but this regulation is independent of the interaction between the two (O’Regan et al., 2007b). Similarly, pUL16 also facilitates the assembly of VP22, and VP22 assembly is even more dependent on pUL16 than on the other binding proteins, gE and VP16 (O’Regan et al., 2007b). In HSV-1, pUL16, pUL11, and pUL21 form a complex. pUL16 binds to the capsid before it reaches the Golgi apparatus, promoting capsid maturation. pUL11 binds to the nuclear membrane and binds to pUL16, thereby increasing the chances of pUL16 binding to the capsid, and in the absence of pUL11, the ability of pUL16 to bind to the capsid is reduced by 70% (Meckes et al., 2010). Then, pUL11 accumulates in the Golgi apparatus. After the nucleocapsid is successfully transported to the Golgi apparatus, pUL11 and pUL21 are involved in the germination and maturation of virions through interactions (Loomis et al., 2006; Meckes et al., 2010). Finally, pUL11 is involved in the release of the virus into the extracellular environment via direct contact of pUL11 with the membrane; similarly, in the absence of the UL11 gene, empty capsids and DNA-containing capsids accumulate on the nuclear membrane, and infected viruses are rarely released (Baines et al., 1995) In PRV, pUL21 is required for the effective incorporation of pUL46, pUL49 and pUS3 into mature virions (Roller and Johnson, 2021).
pUL36 (VP1/2) is necessary for the successful assembly of VP16 onto the capsid (Ko et al., 2010); pUL36, in turn, interacts with pUL37, and both require each other to be incorporated into the virus particles (Trus et al., 1992; Fossum et al., 2009; Krautwald et al., 2009; Kelly et al., 2012). The pUL37-targeted secondary envelopment of HSV-1 requires pUL36 but does not require a capsid (Desai et al., 2001). This finding can be explained by the interaction of pUL37 with pUL46 and pUL36 with VP16 (Trus et al., 1992; Steiner et al., 2007; Krautwald et al., 2009) because these outer tegument proteins accumulate at the site of secondary envelopment. The N-terminal region of HSV-1 pUL36 has the binding sites of pUL37 and VP16, and the C-terminal region has two binding regions of the outer capsid protein pUL25. Thus, HSV-1 pUL36 serves as a linker between the capsid and other tegument proteins (Cohen et al., 1980; Kelly et al., 2012). The absence of UL36 or pUL37 prevents the acquisition of a large amount of tegument in the cytoplasm, leading to the accumulation of uncoated capsids in the cytoplasm.
pUL51, pUL7, and pUL14 are also conserved in alphaherpesviruses, and the loss of these proteins causes misassembly of the secondary envelope (Nozawa et al., 2002; Fuchs et al., 2005; Klupp et al., 2005; Albecka et al., 2017). Both pUL7 and pUL14 can bind pUL51, possibly forming a three-part complex. During HSV-1 infection, pUL51 forms a complex with pUL7 and is primarily responsible for recruiting pUL7 to the cytoplasmic membrane and the tegument of the virion. In addition, the pUL7-pUL51 complex regulates adhesion stability and maintains cell morphology during infection, but the specific mechanism involved needs further study.
3 Other aspects
3.1 Tegument proteins are involved in innate immune escape
Because herpesvirus infection is lifelong, the virus must evade the host’s immune response during latent infection to prevent its elimination by the host (Whitley et al., 1998). Most tegument proteins are involved in viral innate immune escape (Yang et al., 2019). For example, the recently reported DPV pUL41 selectively downregulates IRF7 expression by reducing its mRNA accumulation, thereby inhibiting the DNA-sensing pathway (Gao et al., 2022); PRV pUL13 inhibits the expression of RIG-I and MDA5 by inhibiting the activation of the transcription factor NF-κB. pUL13 can interact with the CDN domain of STING and recruit RNF5 to promote its ubiquitination and degradation (Kong et al., 2022). Notably, VP22, one of the major tegument proteins, mediates a different mechanism by which other viral proteins participate in immune escape. VP22 accumulates and effectively destroys preformed cGAS-DNA complexes both in vitro and in cells. This inhibition relies on DNA-induced liquid phase separation of viral proteins rather than direct interaction with cGAS (Xu et al., 2021). There are also tegument protein interactions involved in these processes; for example, among antiviral mechanisms, PKR is a typical interferon-stimulated gene (ISG) protein (Duffy et al., 2009; Mbong et al., 2012a). Pennisi et al. (2020) reported for the first time that ph-PKR accumulation occurs in different cell lines via a VHS-dependent mechanism and that a lack of VHS RNAse activity affects the negative regulation of ph-PKR mediated by HSV-1. That is, VHS can affect mRNA accumulation during viral infection, allowing HSV-1 to control PKR. In addition, the authors demonstrated for the first time that both serine/threonine kinase proteins pUS3/pUL13 control ph-PKR accumulation during HSV-1 replication and that pUS3 regulates the phosphorylation of PKR by interacting with VHS (Pennisi et al., 2021).
3.2 Tegument proteins regulate autophagy pathways
Autophagy is a typical degradation pathway in which cytoplasmic contents are encapsulated in cell membranes called autophagosomes. Subsequently, autophagosomes can fuse with endosomal organelles to form diplosomes, which can be transported within the cell and fused with lysosomes to form degraded autophagolysosomes (Morishita and Mizushima, 2019; McShane and Selbach, 2022; Zhang, 2022). Uninfected cells typically exhibit low autophagy activity, but autophagy is strongly upregulated in response to cellular stress signals, which may include viral infection processes. Autophagy was originally thought to be antiviral because it can lead to lysosomal degradation of viral proteins and particles (Hogue, 2021). Some herpesviruses have strategies to regulate autophagy, and many rely on cellular autophagy mechanisms to successfully complete their life cycle (Yin et al., 2017; Xu et al., 2018). Examples include pUL21 (Ma et al., 2023)and VP16 (Ming et al., 2022)for PRV and ICP34.5 (Tallóczy et al., 2006; Ripa et al., 2022), pUS11 (Lussignol and Esclatine, 2017) and ICP0 (Liu et al., 2021; Waisner et al., 2023) for HSV-1. ICP34.5 is the first-discovered anti-autophagy viral protein of herpesviruses (Tallóczy et al., 2002). The ability to control autophagy by interacting with BECN1 is necessary for HSV-1 neurovirulence (Orvedahl et al., 2007); pUL21, a recently discovered herpesvirus protein associated with autophagy, triggers cGAS degradation through Tollip-mediated selective autophagy, thereby inhibiting innate immunity (Ma et al., 2023); and the host E3 ligase TRIM23, which induces autophagy to limit HSV-1 replication (and that of additional viruses), triggers proteasomal degradation of ICP0 through ubiquitination of K11 and K48 interactions (Liu et al., 2021). The major autophagy adaptor proteins Sequestosome 1 and Optineurin are downregulated in the early stages of HSV-1 infection (Waisner and Kalamvoki, 2019). In PRV, pUSP14 inhibition promotes the binding of K63-linked polyubiquitin chains in VP16 to K168 and subsequent degradation of VP16 via SQSTM1/p62-mediated selective autophagy (Ming et al., 2022). pUS11 has been shown to protect HeLa cells from heat and astrosporin-induced apoptosis and counteract autophagy responses (Javouhey et al., 2008; Lussignol et al., 2013); it also regulates the cleavage of caspase-8, the initiator of mammalian exogenous or death receptor-mediated apoptosis (Musarra-Pizzo et al., 2022).
However, the genomes of some herpesviruses do not encode the viral proteins ICP34.5 or pUS11, two known autophagy-associated proteins (Heinz et al., 2021); examples include DPV and VZV, although they belong to the same alphaherpesvirus subfamily as HSV-1 and PRV. Whether there is a different pattern in the relationship between DPV or VZV and autophagy remains unclear. However, as mentioned above, most of the viral proteins that have been reported to be involved in the autophagy pathway are tegument proteins, which led us to wonder whether there are other tegument proteins that play a role in the autophagy pathway.
3.3 Role of tegument proteins in the ESCRT mechanism
Like many cystal virus families (Votteler and Sundquist, 2013; Hurley, 2015; Scourfield and Martin-Serrano, 2017; Barnes and Wilson, 2019), herpesviruses utilize the ESCRT mechanism required for transport to construct and drive the rupture of their cystal membrane (Pawliczek and Crump, 2009; Kharkwal et al., 2014, 2016; Barnes and Wilson, 2019). The ESCRT machinery manipulates organelle membranes in various normal cellular processes by depositing components of ESCRT-III, a spring-like filament that drives planar bilayer deformation into vesicles and tubules (Alonso et al., 2016; Olmos and Carlton, 2016; Christ et al., 2017; Scourfield and Martin-Serrano, 2017; McCullough et al., 2018). HSV-1 can bypass the normal cellular pathway of ESCRT complex assembly (Pawliczek and Crump, 2009; Christ et al., 2017; Barnes and Wilson, 2019, 2020) by polymerizing ESCRT-III filaments directly on the surface of the cortex-binding capsid to support capsule deformation (Butt et al., 2020). When budding is complete, the ESCRT-III filaments contract their neck membranes to undergo shearing, and this process is accompanied by the disassembly and removal of ESCRT-III via hexameric VPS4 (McCullough et al., 2015, 2018; Christ et al., 2017). There are several tegument proteins involved in linking ESCRT mechanisms. For example, arginine clusters in the HSV-1 pUL34 disordered domain mediate interactions with the linker protein ALIX of ESCRT-III, which results in the recruitment of the INM by the ESCRT-III mechanism for effective primary membrane deposition (Arii et al., 2022). The helix-transhelical conformation of HSV-1 pUL51 is similar to that of the ESCRT-III component CHMP4B, which forms ESCRT-III-like filaments, and pUL51 bends the membrane in the cell, directly promoting membrane shearing during viral assembly. The interaction of pUL51 with pUL7 is conserved in all three herpesvirus subfamilies, and the pUL7 protein regulates the state of pUL51 during this process, controlling its ability to clip the capsule membrane at the correct time (Butt et al., 2020). Mutation of the glutamate 233 residue in the active site of the VPS4 ATPase produces the VPS4-EQ allele, which binds ATP but does not hydrolyze ATP (Sun et al., 2017). Vps4-eq has a major negative effect because hexamers containing a mixture of wild-type VPS4 and VPS4-EQ subunits can bind to ESCRT-III filaments but cannot drive their breakdown, irreversibly “locking” the VPS4 hexamer to the bud neck and preventing cutting (Davies et al., 2010; Han et al., 2015; Sun et al., 2017; Han and Hill, 2019). Thus, the expression of VPS4-EQ blocks the completion of HSV-1 and PRV envelopment (Crump et al., 2007; Kharkwal et al., 2014) and arrests the HSV-1 capsid in a partially membrane-closed state (Crump et al., 2007; Kharkwal et al., 2016). The absence of pUL36 reduces the number of complexes containing the HSV-1 capsid VPS4-EQ by approximately 70% (Barnes and Wilson, 2019), suggesting that pUL36 plays an important role in capsid recruitment to the ESCRT-III/Vps4 deposition site (Kharkwal et al., 2016). However, while pUL36 is necessary for the assembly of multiple outer tegument proteins and capsular proteins into capsular granules (Vittone et al., 2005; Mijatov et al., 2007; Ko et al., 2010; Svobodova et al., 2012; Koenigsberg and Heldwein, 2017, 2018), its role in capside-ESCRt-II-VPS4 assembly may be indirect (Kharkwal et al., 2016). In addition, HSV-1 VP1/2 (pUL36) has been shown to deubiquitinate the ESCRT-I protein TSG101 (Caduco et al., 2013; Calistri et al., 2015).
4 Conclusion
Herpesvirus tegument proteins play an important role in almost every part of the viral life cycle, especially that of alphaherpesviruses (Figure 1), perhaps because of their unique “intermediate tegument proteins” that are involved in viral particle transport and morphogenesis (Mocarski, 2007). After the virus enters the cell, tegument proteins participate in the dissociation of virus components, nuclear transport of the capsid and docking with the nuclear pore. After the virus enters the nucleus, these proteins regulate mRNA transcription, DNA replication and protein translation. In the later stage of infection, tegument proteins participate in capsid nucleation and primary capsule formation and form extensive and powerful tegument-tegument interactions and tegument-membrane interaction networks during secondary envelopment, which are responsible for virion morphogenesis (Figure 2). It can be assumed that antiviral drugs targeting and blocking conserved tegument protein interactions, such as the interactions between pUL36 and pUL37 or between VP16 and VP22, will be highly important. There is some redundancy in the function of tegument proteins overall, so that particles are still assembled, encapsulated and released from infected cells even after most of the genes encoding structural proteins have been deleted. Because of their redundancy, untangling the tegument protein interactions involved in these complex multistep processes is a challenge, and the use of more advanced techniques, such as liquid chromatography–mass spectrometry (LS-MS), high-resolution confocal microscopy, and bimolecular fluorescence complementarity (BiFC) detection, will help elucidate the protein–protein interactions of tegument proteins in more detail.
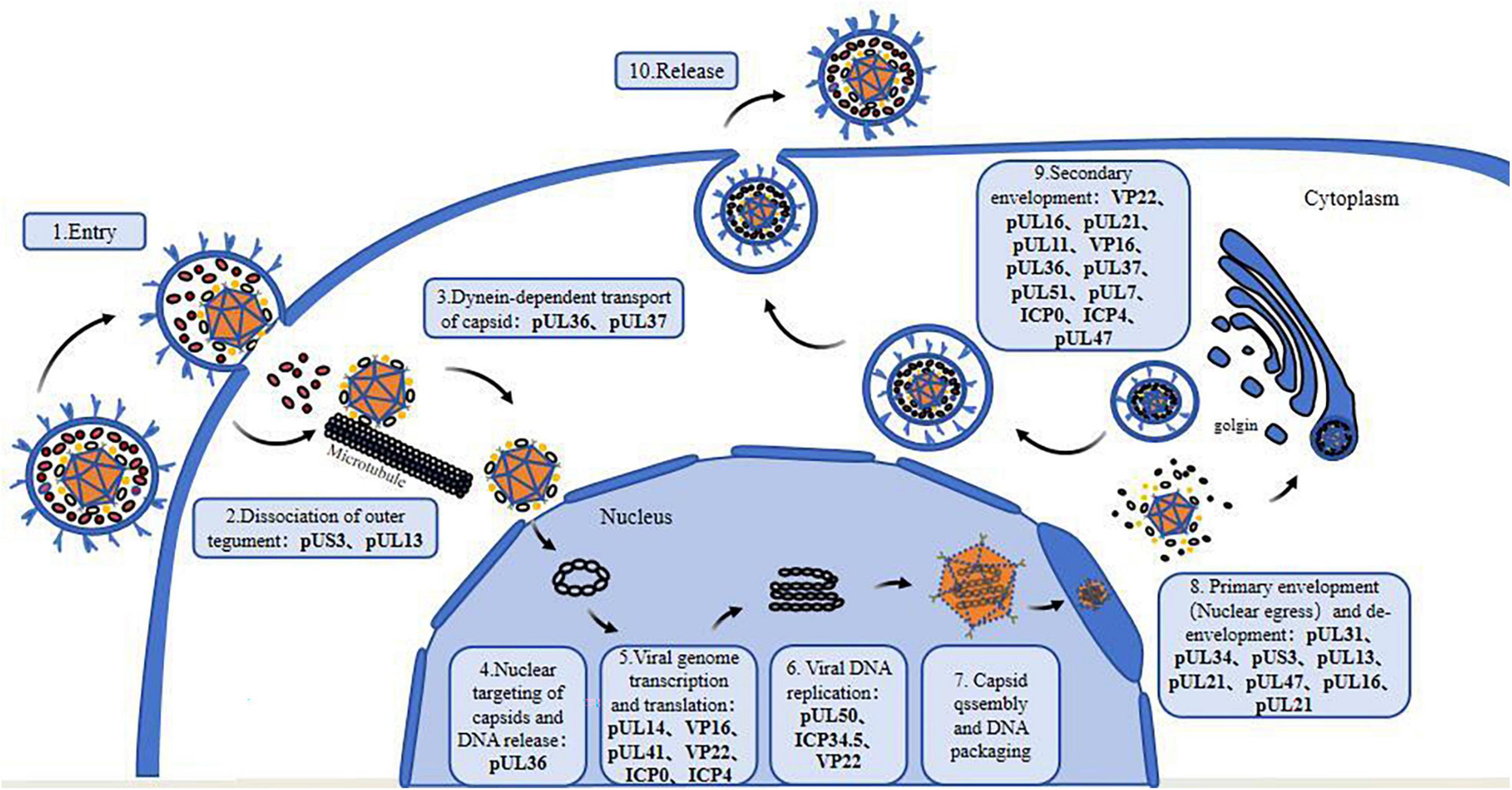
Figure 1. Tegument proteins involved in the life cycle of alphaherpesviruses. 1. Viral entry into the host cell; 2. Dissociation of outer tegument proteins; 3. Dynein-dependent transport of capsid; 4. Nuclear targeting of the capsid and viral DNA release; 5. Transcription and translation of the viral genome; 6. Viral DNA replication; 7. Nucleocapsid formation and viral DNA packaging; 8. Primary envelopment (nuclear egress) and de-envelopment; 9. Secondary envelopment; 10. Maturation and release of virions.
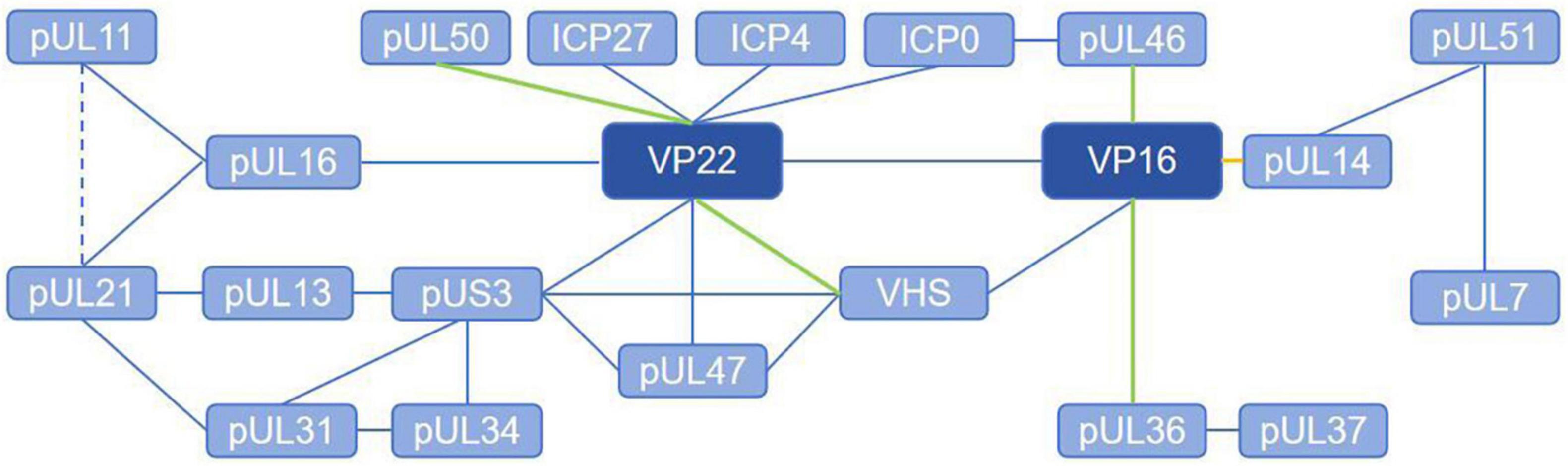
Figure 2. Tegument proteins interaction network of alphaherpesviruses. The solid line represents the direct interaction and the dashed line represents the brief interaction. The blue lines indicate that the interaction is verified with co-ip, the green lines indicate that the interaction is verified with GST-Pulldown, and the yellow lines indicate that the interaction is verified with BiFC.
Due to the important functions of tegument proteins and their interactions in the viral life cycle, further elucidation of their functions and mechanisms will provide important reference data for the search for therapeutic targets and vaccine development.
Author contributions
YC: Writing – original draft, Investigation, Methodology. MW: Funding acquisition, Writing – review and editing, Methodology. AC: Funding acquisition, Writing – review and editing, Supervision. WZ: Funding acquisition, Writing – review and editing. QY: Methodology, Writing – review and editing. BT: Conceptualization, Writing – review and editing. XO: Supervision, Writing – review and editing. JH: Methodology, Writing – review and editing. YW: Formal analysis, Writing – review and editing. SZ: Data curation, Writing – review and editing. DS: Resources, Writing – review and editing. YH: Formal analysis, Writing – review and editing. XZ: Supervision, Writing – review and editing. ZW: Formal analysis, Writing – review and editing. DZ: Methodology, Writing – review and editing. RJ: Supervision, Writing – review and editing. SC: Supervision, Writing – review and editing. ML: Methodology, Writing – review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by grants from the National Natural Science Foundation of China (32373049), the China Agriculture Research System of MOF and MARA (CARS-42-17), and the Program Sichuan Veterinary Medicine and Drug Innovation Group of China Agricultural Research System (SCCXTD-2020-18).
Conflict of interest
WZ was employed by Sinopharm Yangzhou VAC Biological Engineering Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Afroz, S., Garg, R., Fodje, M., and den Hurk, S. (2018). The major tegument protein of bovine herpesvirus 1, VP8, interacts with DNA damage response proteins and induces apoptosis. J. Virol. 92:18. doi: 10.1128/jvi.00773-18
Albecka, A., Owen, D. J., Ivanova, L., Brun, J., Liman, R., Davies, L., et al. (2017). Dual function of the pUL7-pUL51 tegument protein complex in herpes simplex virus 1 infection. J. Virol. 91:16. doi: 10.1128/jvi.02196-16
Alonso, Y. A. M., Migliano, S. M., and Teis, D. (2016). ESCRT-III and Vps4: A dynamic multipurpose tool for membrane budding and scission. FEBS J. 283, 3288–3302. doi: 10.1111/febs.13688
Antinone, S. E., and Smith, G. A. (2010). Retrograde axon transport of herpes simplex virus and pseudorabies virus: A live-cell comparative analysis. J. Virol. 84, 1504–1512. doi: 10.1128/jvi.02029-09
Arii, J., Takeshima, K., Maruzuru, Y., Koyanagi, N., Nakayama, Y., Kato, A., et al. (2022). Role of the arginine cluster in the disordered domain of herpes simplex virus 1 UL34 for the recruitment of ESCRT-III for viral primary envelopment. J. Virol. 96:e0170421. doi: 10.1128/jvi.01704-21
Baines, J. D., Jacob, R. J., Simmerman, L., and Roizman, B. (1995). The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69, 825–833. doi: 10.1128/jvi.69.2.825-833.1995
Barnes, J., and Wilson, D. W. (2019). Seeking closure: How do herpesviruses recruit the cellular ESCRT apparatus? J. Virol. 93:19. doi: 10.1128/jvi.00392-19
Barnes, J., and Wilson, D. W. (2020). The ESCRT-II subunit EAP20/VPS25 and the Bro1 domain proteins HD-PTP and BROX are individually dispensable for herpes simplex virus 1 replication. J. Virol. 94, 19. doi: 10.1128/jvi.01641-19
Bieker, J. J. (2001). Krüppel-like factors: Three fingers in many pies. J. Biol. Chem. 276, 34355–34358. doi: 10.1074/jbc.R100043200
Bigalke, J. M., and Heldwein, E. E. (2016). Nuclear exodus: Herpesviruses lead the way. Annu. Rev. Virol. 3, 387–409. doi: 10.1146/annurev-virology-110615-042215
Black, D., Ritchey, J., Payton, M., and Eberle, R. (2014). Role of the virion host shutoff protein in neurovirulence of monkey B virus (Macacine herpesvirus 1). Virol. Sin. 29, 274–283. doi: 10.1007/s12250-014-3495-x
Boutell, C., Sadis, S., and Everett, R. D. (2002). Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76, 841–850. doi: 10.1128/jvi.76.2.841-850.2002
Bremner, K. H., Scherer, J., Yi, J., Vershinin, M., Gross, S. P., and Vallee, R. B. (2009). Adenovirus transport via direct interaction of cytoplasmic dynein with the viral capsid hexon subunit. Cell Host Microbe 6, 523–535. doi: 10.1016/j.chom.2009.11.006
Brignati, M. J., Loomis, J. S., Wills, J. W., and Courtney, R. J. (2003). Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J. Virol. 77, 4888–4898. doi: 10.1128/jvi.77.8.4888-4898.2003
Brown, S. M., MacLean, A. R., McKie, E. A., and Harland, J. (1997). The herpes simplex virus virulence factor ICP34.5 and the cellular protein MyD116 complex with proliferating cell nuclear antigen through the 63-amino-acid domain conserved in ICP34.5, MyD116, and GADD34. J. Virol. 71, 9442–9449. doi: 10.1128/jvi.71.12.9442-9449.1997
Butt, B. G., Owen, D. J., Jeffries, C. M., Ivanova, L., Hill, C. H., Houghton, J. W., et al. (2020). Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex. Elife 9:e53789. doi: 10.7554/eLife.53789
Caduco, M., Comin, A., Toffoletto, M., Munegato, D., Sartori, E., Celestino, M., et al. (2013). Retraction. Tsg101 interacts with herpes simplex virus 1 VP1/2 and is a substrate of VP1/2 ubiquitin-specific protease domain activity. J. Virol. 87:6537. doi: 10.1128/jvi.00725-13
Cai, W., and Schaffer, P. A. (1992). Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66, 2904–2915. doi: 10.1128/jvi.66.5.2904-2915.1992
Calistri, A., Munegato, D., Toffoletto, M., Celestino, M., Franchin, E., Comin, A., et al. (2015). Functional interaction between the ESCRT-I component TSG101 and the HSV-1 tegument ubiquitin specific protease. J. Cell Physiol. 230, 1794–1806. doi: 10.1002/jcp.24890
Cano-Monreal, G. L., Wylie, K. M., Cao, F., Tavis, J. E., and Morrison, L. A. (2009). Herpes simplex virus 2 UL13 protein kinase disrupts nuclear lamins. Virology 392, 137–147. doi: 10.1016/j.virol.2009.06.051
Chadha, P., Han, J., Starkey, J. L., and Wills, J. W. (2012). Regulated interaction of tegument proteins UL16 and UL11 from herpes simplex virus. J. Virol. 86, 11886–11898. doi: 10.1128/jvi.01879-12
Chadha, P., Sarfo, A., Zhang, D., Abraham, T., Carmichael, J., Han, J., et al. (2017). Domain Interaction Studies of Herpes Simplex Virus 1 Tegument Protein UL16 reveal its interaction with mitochondria. J. Virol. 91:e1995-19. doi: 10.1128/jvi.01995-16
Che, X., Oliver, S. L., Reichelt, M., Sommer, M. H., Haas, J., Roviš, T. L., et al. (2013). ORF11 protein interacts with the ORF9 essential tegument protein in varicella-zoster virus infection. J. Virol. 87, 5106–5117. doi: 10.1128/jvi.00102-13
Christ, L., Raiborg, C., Wenzel, E. M., Campsteijn, C., and Stenmark, H. (2017). Cellular functions and molecular mechanisms of the ESCRT membrane-scission machinery. Trends Biochem. Sci. 42, 42–56. doi: 10.1016/j.tibs.2016.08.016
Chuard, A., Courvoisier-Guyader, K., Rémy, S., Spatz, S., Denesvre, C., and Pasdeloup, D. (2020). The tegument protein pUL47 of Marek’s disease virus is necessary for horizontal transmission and is important for expression of glycoprotein gC. J. Virol. 95:e1645–20. doi: 10.1128/jvi.01645-20
Cliffe, A. R., and Knipe, D. M. (2008). Herpes simplex virus ICP0 promotes both histone removal and acetylation on viral DNA during lytic infection. J. Virol. 82, 12030–12038. doi: 10.1128/jvi.01575-08
Cohen, G. H., Ponce, de Leon, M., Diggelmann, H., Lawrence, W. C., Vernon, S. K., et al. (1980). Structural analysis of the capsid polypeptides of herpes simplex virus types 1 and 2. J. Virol. 34, 521–531. doi: 10.1128/jvi.34.2.521-531.1980
Coller, K. E., and Smith, G. A. (2008). Two viral kinases are required for sustained long distance axon transport of a neuroinvasive herpesvirus. Traffic 9, 1458–1470. doi: 10.1111/j.1600-0854.2008.00782.x
Crump, C. (2018). Virus assembly and egress of HSV. Adv. Exp. Med. Biol. 1045, 23–44. doi: 10.1007/978-981-10-7230-7_2
Crump, C. M., Yates, C., and Minson, T. (2007). Herpes simplex virus type 1 cytoplasmic envelopment requires functional Vps4. J. Virol. 81, 7380–7387. doi: 10.1128/jvi.00222-07
Davies, B. A., Azmi, I. F., Payne, J., Shestakova, A., Horazdovsky, B. F., Babst, M., et al. (2010). Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol. Biol. Cell 21, 3396–3408. doi: 10.1091/mbc.E10-06-0512
Davison, A. J., Eberle, R., Ehlers, B., Hayward, G. S., McGeoch, D. J., Minson, A. C., et al. (2009). The order Herpesvirales. Arch. Virol. 154, 171–177. doi: 10.1007/s00705-008-0278-4
DeLuca, N. A., and Schaffer, P. A. (1985). Activation of immediate-early, early, and late promoters by temperature-sensitive and wild-type forms of herpes simplex virus type 1 protein ICP4. Mol. Cell Biol. 5, 1997–2008. doi: 10.1128/mcb.5.8.1997-2008.1985
Deng, L., Wan, J., Cheng, A., Wang, M., Tian, B., Wu, Y., et al. (2022). Duck plague virus US3 protein kinase phosphorylates UL47 and regulates the subcellular localization of UL47. Front. Microbiol. 13:876820. doi: 10.3389/fmicb.2022.876820
Deng, L., Wang, M., Cheng, A., Yang, Q., Wu, Y., Jia, R., et al. (2020). The pivotal roles of US3 protein in cell-to-cell spread and virion nuclear egress of duck plague virus. Sci. Rep. 10:7181. doi: 10.1038/s41598-020-64190-2
Desai, P., Sexton, G. L., McCaffery, J. M., and Person, S. (2001). A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75, 10259–10271. doi: 10.1128/jvi.75.21.10259-10271.2001
Döhner, K., Wolfstein, A., Prank, U., Echeverri, C., Dujardin, D., Vallee, R., et al. (2002). Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell 13, 2795–2809. doi: 10.1091/mbc.01-07-0348
Douglas, M. W., Diefenbach, R. J., Homa, F. L., Miranda-Saksena, M., Rixon, F. J., Vittone, V., et al. (2004). Herpes simplex virus type 1 capsid protein VP26 interacts with dynein light chains RP3 and Tctex1 and plays a role in retrograde cellular transport. J. Biol. Chem. 279, 28522–28530. doi: 10.1074/jbc.M311671200
Du, X., Zhou, D., Zhou, J., Xue, J., Wang, G., and Cheng, Z. (2022). Marek’s disease virus serine/threonine kinase Us3 facilitates viral replication by targeting IRF7 to block IFN-β production. Vet. Microbiol. 266:109364. doi: 10.1016/j.vetmic.2022.109364
Duffy, C., Mbong, E. F., and Baines, J. D. (2009). VP22 of herpes simplex virus 1 promotes protein synthesis at late times in infection and accumulation of a subset of viral mRNAs at early times in infection. J. Virol. 83, 1009–1017. doi: 10.1128/jvi.02245-07
Dunn, L. E. M., and Baines, J. D. (2023). Herpes simplex virus 1 immediate early transcription initiation, pause-release, elongation, and termination in the presence and absence of ICP4. J. Virol. 97:e0096023. doi: 10.1128/jvi.00960-23
Elliott, G., Hafezi, W., Whiteley, A., and Bernard, E. (2005). Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79, 9735–9745. doi: 10.1128/jvi.79.15.9735-9745.2005
Elliott, G., Mouzakitis, G., and O’Hare, P. (1995). VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69, 7932–7941. doi: 10.1128/jvi.69.12.7932-7941.1995
El-Mayet, F. S., and Jones, C. (2024a). A cell cycle regulator, E2F2, and glucocorticoid receptor cooperatively transactivate the bovine alphaherpesvirus 1 immediate early transcription unit 1 promoter. J. Virol. 98:0042324. doi: 10.1128/jvi.00423-24
El-Mayet, F. S., and Jones, C. (2024b). Specificity protein 1 (Sp1) and glucocorticoid receptor (GR) stimulate bovine alphaherpesvirus 1 (BoHV-1) replication and cooperatively transactivate the immediate early transcription unit 1 promoter. J. Virol. 98:e0143623. doi: 10.1128/jvi.01436-23
El-Mayet, F. S., Harrison, K. S., and Jones, C. (2021). Regulation of Krüppel-like factor 15 expression by herpes simplex virus type 1 or bovine herpesvirus 1 productive infection. Viruses 13:148. doi: 10.3390/v13061148
El-Mayet, F. S., Sawant, L., Thunuguntla, P., and Jones, C. (2017). Combinatorial effects of the glucocorticoid receptor and krüppel-like transcription factor 15 on bovine herpesvirus 1 transcription and productive infection. J. Virol. 91:17. doi: 10.1128/jvi.00904-17
Esclatine, A., Taddeo, B., Evans, L., and Roizman, B. (2004). The herpes simplex virus 1 UL41 gene-dependent destabilization of cellular RNAs is selective and may be sequence-specific. Proc. Natl. Acad. Sci. U.S.A. 101, 3603–3608. doi: 10.1073/pnas.0400354101
Everly, D. N. Jr., Feng, P., Mian, I. S., and Read, G. S. (2002). mRNA degradation by the virion host shutoff (Vhs) protein of herpes simplex virus: Genetic and biochemical evidence that Vhs is a nuclease. J. Virol. 76, 8560–8571. doi: 10.1128/jvi.76.17.8560-8571.2002
Fan, D., Wang, M., Cheng, A., Jia, R., Yang, Q., Wu, Y., et al. (2020). The role of VP16 in the life cycle of alphaherpesviruses. Front. Microbiol. 11:1910. doi: 10.3389/fmicb.2020.01910
Fan, W. H., Roberts, A. P., McElwee, M., Bhella, D., Rixon, F. J., and Lauder, R. (2015). The large tegument protein pUL36 is essential for formation of the capsid vertex-specific component at the capsid-tegument interface of herpes simplex virus 1. J. Virol. 89, 1502–1511. doi: 10.1128/jvi.02887-14
Farnsworth, A., Wisner, T. W., and Johnson, D. C. (2007). Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J. Virol. 81, 319–331. doi: 10.1128/jvi.01842-06
Feutz, E., McLeland-Wieser, H., Ma, J., and Roller, R. J. (2019). Functional interactions between herpes simplex virus pUL51, pUL7 and gE reveal cell-specific mechanisms for epithelial cell-to-cell spread. Virology 537, 84–96. doi: 10.1016/j.virol.2019.08.014
Fossum, E., Friedel, C. C., Rajagopala, S. V., Titz, B., Baiker, A., Schmidt, T., et al. (2009). Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog. 5:e1000570. doi: 10.1371/journal.ppat.1000570
Fuchs, W., Granzow, H., Klopfleisch, R., Klupp, B. G., Rosenkranz, D., and Mettenleiter, T. C. (2005). The UL7 gene of pseudorabies virus encodes a nonessential structural protein which is involved in virion formation and egress. J. Virol. 79, 11291–11299. doi: 10.1128/jvi.79.17.11291-11299.2005
Fuchs, W., Granzow, H., Klupp, B. G., Kopp, M., and Mettenleiter, T. C. (2002a). The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76, 6729–6742. doi: 10.1128/jvi.76.13.6729-6742.2002
Fuchs, W., Klupp, B. G., Granzow, H., Hengartner, C., Brack, A., Mundt, A., et al. (2002b). Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76, 8208–8217. doi: 10.1128/jvi.76.16.8208-8217.2002
Fuchs, W., Ziemann, K., Teifke, J. P., Werner, O., and Mettenleiter, T. C. (2000). The non-essential UL50 gene of avian infectious laryngotracheitis virus encodes a functional dUTPase which is not a virulence factor. J. Gen. Virol. 81, 627–638. doi: 10.1099/0022-1317-81-3-627
Gao, J., Finnen, R. L., Sherry, M. R., Le Sage, V., and Banfield, B. W. (2020). Differentiating the roles of UL16, UL21, and Us3 in the nuclear egress of herpes simplex virus capsids. J. Virol. 94:20. doi: 10.1128/jvi.00738-20
Gao, J., Hay, T. J. M., and Banfield, B. W. (2017). The product of the herpes simplex virus 2 UL16 gene is critical for the egress of capsids from the nuclei of infected cells. J. Virol. 91:e350–17. doi: 10.1128/jvi.00350-17
Gao, L., Liu, R., Yang, F., Li, X., Liu, C., Qi, X., et al. (2022). Duck enteritis virus inhibits the cGAS-STING DNA-sensing pathway to evade the innate immune response. J. Virol. 96:e0157822. doi: 10.1128/jvi.01578-22
Gatherer, D., Depledge, D. P., Hartley, C. A., Szpara, M. L., Vaz, P. K., Benkő, M., et al. (2021). ICTV virus taxonomy profile: Herpesviridae 2021. J. Gen. Virol. 102:1673. doi: 10.1099/jgv.0.001673
Gershburg, S., Geltz, J., Peterson, K. E., Halford, W. P., and Gershburg, E. (2015). The UL13 and US3 protein kinases of herpes simplex virus 1 cooperate to promote the assembly and release of mature, infectious virions. PLoS One 10:e0131420. doi: 10.1371/journal.pone.0131420
Gross, S. T., Harley, C. A., and Wilson, D. W. (2003). The cytoplasmic tail of Herpes simplex virus glycoprotein H binds to the tegument protein VP16 in vitro and in vivo. Virology 317, 1–12. doi: 10.1016/j.virol.2003.08.023
Guo, H., Shen, S., Wang, L., and Deng, H. (2010). Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1, 987–998. doi: 10.1007/s13238-010-0120-0
Hafezi, W., Bernard, E., Cook, R., and Elliott, G. (2005). Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J. Virol. 79, 13082–13093. doi: 10.1128/jvi.79.20.13082-13093.2005
Hagglund, R., and Roizman, B. (2004). Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J. Virol. 78, 2169–2178. doi: 10.1128/jvi.78.5.2169-2178.2004
Han, H., and Hill, C. P. (2019). Structure and mechanism of the ESCRT pathway AAA+ ATPase Vps4. Biochem. Soc. Trans. 47, 37–45. doi: 10.1042/bst20180260
Han, H., Monroe, N., Votteler, J., Shakya, B., Sundquist, W. I., and Hill, C. P. (2015). Binding of substrates to the central pore of the Vps4 ATPase is autoinhibited by the microtubule interacting and trafficking (MIT) domain and activated by MIT interacting motifs (MIMs). J. Biol. Chem. 290, 13490–13499. doi: 10.1074/jbc.M115.642355
Han, J., Chadha, P., Meckes, D. G. Jr., Baird, N. L., and Wills, J. W. (2011). Interaction and interdependent packaging of tegument protein UL11 and glycoprotein e of herpes simplex virus. J. Virol. 85, 9437–9446. doi: 10.1128/jvi.05207-11
Harland, J., Dunn, P., Cameron, E., Conner, J., and Brown, S. M. (2003). The herpes simplex virus (HSV) protein ICP34.5 is a virion component that forms a DNA-binding complex with proliferating cell nuclear antigen and HSV replication proteins. J. Neurovirol. 9, 477–488. doi: 10.1080/13550280390218788
Harper, A. L., Meckes, D. G. Jr., Marsh, J. A., Ward, M. D., Yeh, P. C., Baird, N. L., et al. (2010). Interaction domains of the UL16 and UL21 tegument proteins of herpes simplex virus. J. Virol. 84, 2963–2971. doi: 10.1128/jvi.02015-09
He, T., Wang, M., Cheng, A., Yang, Q., Jia, R., Wu, Y., et al. (2020). Duck enteritis virus pUL47, as a late structural protein localized in the nucleus, mainly depends on residues 40 to 50 and 768 to 777 and inhibits IFN-β signalling by interacting with STAT1. Vet. Res. 51:135. doi: 10.1186/s13567-020-00859-w
He, T., Wang, M., Cheng, A., Yang, Q., Jia, R., Wu, Y., et al. (2021). DPV UL41 gene encoding protein induces host shutoff activity and affects viral replication. Vet. Microbiol. 255:108979. doi: 10.1016/j.vetmic.2021.108979
Heinz, J., Kennedy, P. G. E., and Mogensen, T. H. (2021). The role of autophagy in varicella zoster virus infection. Viruses 13:1053. doi: 10.3390/v13061053
Heming, J. D., Conway, J. F., and Homa, F. L. (2017). Herpesvirus Capsid Assembly and DNA Packaging. Adv. Anat. Embryol. Cell Biol. 223, 119–142. doi: 10.1007/978-3-319-53168-7_6
Herrera, F. J., and Triezenberg, S. J. (2004). VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 78, 9689–9696. doi: 10.1128/jvi.78.18.9689-9696.2004
Hogue, I. B. (2021). Tegument assembly, secondary envelopment and exocytosis. Curr. Issues Mol. Biol. 42, 551–604. doi: 10.21775/cimb.042.551
International Committee on Taxonomy of Viruses Executive Committee (2020). The new scope of virus taxonomy: Partitioning the virosphere into 15 hierarchical ranks. Nat. Microbiol. 5, 668–674. doi: 10.1038/s41564-020-0709-x
Jambunathan, N., Chouljenko, D., Desai, P., Charles, A. S., Subramanian, R., Chouljenko, V. N., et al. (2014). Herpes simplex virus 1 protein UL37 interacts with viral glycoprotein gK and membrane protein UL20 and functions in cytoplasmic virion envelopment. J. Virol. 88, 5927–5935. doi: 10.1128/jvi.00278-14
Jarosinski, K. W., and Osterrieder, N. (2010). Further analysis of Marek’s disease virus horizontal transmission confirms that U(L)44 (gC) and U(L)13 protein kinase activity are essential, while U(S)2 is nonessential. J. Virol. 84, 7911–7916. doi: 10.1128/jvi.00433-10
Javouhey, E., Gibert, B., Arrigo, A. P., Diaz, J. J., and Diaz-Latoud, C. (2008). Protection against heat and staurosporine mediated apoptosis by the HSV-1 US11 protein. Virology 376, 31–41. doi: 10.1016/j.virol.2008.02.031
Johnson, D. C., and Baines, J. D. (2011). Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394. doi: 10.1038/nrmicro2559
Jones, K. A., and Tjian, R. (1985). Sp1 binds to promoter sequences and activates herpes simplex virus ‘immediate-early’ gene transcription in vitro. Nature 317, 179–182. doi: 10.1038/317179a0
Jones, K. A., Yamamoto, K. R., and Tjian, R. (1985). Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell 42, 559–572. doi: 10.1016/0092-8674(85)90113-8
Kaczynski, J., Cook, T., and Urrutia, R. (2003). Sp1- and Krüppel-like transcription factors. Genome Biol. 4:206. doi: 10.1186/gb-2003-4-2-206
Kato, A., Liu, Z., Minowa, A., Imai, T., Tanaka, M., Sugimoto, K., et al. (2011). Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J. Virol. 85, 9599–9613. doi: 10.1128/jvi.00845-11
Kato, A., Oda, S., Watanabe, M., Oyama, M., Kozuka-Hata, H., Koyanagi, N., et al. (2018). Roles of the Phosphorylation of Herpes Simplex Virus 1 UL51 at a Specific Site in Viral Replication and Pathogenicity. J. Virol. 92:e1035–18. doi: 10.1128/jvi.01035-18
Kato, A., Tsuda, S., Liu, Z., Kozuka-Hata, H., Oyama, M., and Kawaguchi, Y. (2014c). Herpes simplex virus 1 protein kinase Us3 phosphorylates viral dUTPase and regulates its catalytic activity in infected cells. J. Virol. 88, 655–666. doi: 10.1128/jvi.02710-13
Kato, A., Shindo, K., Maruzuru, Y., and Kawaguchi, Y. (2014b). Phosphorylation of a herpes simplex virus 1 dUTPase by a viral protein kinase, Us3, dictates viral pathogenicity in the central nervous system but not at the periphery. J. Virol. 88, 2775–2785. doi: 10.1128/jvi.03300-13
Kato, A., Hirohata, Y., Arii, J., and Kawaguchi, Y. (2014a). Phosphorylation of herpes simplex virus 1 dUTPase upregulated viral dUTPase activity to compensate for low cellular dUTPase activity for efficient viral replication. J. Virol. 88, 7776–7785. doi: 10.1128/jvi.00603-14
Kato, A., Yamamoto, M., Ohno, T., Tanaka, M., Sata, T., Nishiyama, Y., et al. (2006). Herpes simplex virus 1-encoded protein kinase UL13 phosphorylates viral Us3 protein kinase and regulates nuclear localization of viral envelopment factors UL34 and UL31. J. Virol. 80, 1476–1486. doi: 10.1128/jvi.80.3.1476-1486.2006
Kelly, B. J., Diefenbach, E., Fraefel, C., and Diefenbach, R. J. (2012). Identification of host cell proteins which interact with herpes simplex virus type 1 tegument protein pUL37. Biochem. Biophys. Res. Commun. 417, 961–965. doi: 10.1016/j.bbrc.2011.12.044
Kelly, B. J., Fraefel, C., Cunningham, A. L., and Diefenbach, R. J. (2009). Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res. 145, 173–186. doi: 10.1016/j.virusres.2009.07.007
Kew, C., Lui, P. Y., Chan, C. P., Jin, D. Y., and Kok, K. H. (2016). Inhibition of RIG-I-dependent innate immunity by herpes simplex virus type I Us11 protein. Hong Kong Med. J. 22, 46–47.
Kharkwal, H., Smith, C. G., and Wilson, D. W. (2014). Blocking ESCRT-mediated envelopment inhibits microtubule-dependent trafficking of alphaherpesviruses in vitro. J. Virol. 88, 14467–14478. doi: 10.1128/jvi.02777-14
Kharkwal, H., Smith, C. G., and Wilson, D. W. (2016). Herpes simplex virus capsid localization to ESCRT-VPS4 complexes in the presence and absence of the large tegument protein UL36p. J. Virol. 90, 7257–7267. doi: 10.1128/jvi.00857-16
Kim, D., Cianfrocco, M. A., Verhey, K. J., and Smith, G. A. (2024). The HSV-1 pUL37 protein promotes cell invasion by regulating the kinesin-1 motor. Proc. Natl. Acad. Sci. U.S.A. 121:e2401341121. doi: 10.1073/pnas.2401341121
Kim, I. J., Chouljenko, V. N., Walker, J. D., and Kousoulas, K. G. (2013). Herpes simplex virus 1 glycoprotein M and the membrane-associated protein UL11 are required for virus-induced cell fusion and efficient virus entry. J. Virol. 87, 8029–8037. doi: 10.1128/jvi.01181-13
Klupp, B. G., Granzow, H., Klopfleisch, R., Fuchs, W., Kopp, M., Lenk, M., et al. (2005). Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79, 3831–3840. doi: 10.1128/jvi.79.6.3831-3840.2005
Ko, D. H., Cunningham, A. L., and Diefenbach, R. J. (2010). The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84, 1397–1405. doi: 10.1128/jvi.01721-09
Koenigsberg, A. L., and Heldwein, E. E. (2017). Crystal structure of the N-terminal half of the traffic controller UL37 from herpes simplex virus 1. J. Virol. 91:e1244–17. doi: 10.1128/jvi.01244-17
Koenigsberg, A. L., and Heldwein, E. E. (2018). The dynamic nature of the conserved tegument protein UL37 of herpesviruses. J. Biol. Chem. 293, 15827–15839. doi: 10.1074/jbc.RA118.004481
Kong, Z., Yin, H., Wang, F., Liu, Z., Luan, X., Sun, L., et al. (2022). Pseudorabies virus tegument protein UL13 recruits RNF5 to inhibit STING-mediated antiviral immunity. PLoS Pathog. 18:e1010544. doi: 10.1371/journal.ppat.1010544
Koyanagi, N., Kato, A., Takeshima, K., Maruzuru, Y., Kozuka-Hata, H., Oyama, M., et al. (2018). Regulation of herpes simplex virus 2 protein kinase UL13 by phosphorylation and its role in viral pathogenesis. J. Virol. 92:18. doi: 10.1128/jvi.00807-18
Kramer, T., and Enquist, L. W. (2013). Directional spread of alphaherpesviruses in the nervous system. Viruses 5, 678–707. doi: 10.3390/v5020678
Krautwald, M., Fuchs, W., Klupp, B. G., and Mettenleiter, T. C. (2009). Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J. Virol. 83, 3389–3396. doi: 10.1128/jvi.02090-08
Li, F., Zhang, Y., Chen, S., Wang, M., Jia, R., Zhu, D., et al. (2016). Identification of the nuclear localization signal region of duck enteritis virus UL14 and its interaction with VP16. Intervirology 59, 187–196. doi: 10.1159/000452711
Li, S., Liu, S., Dai, Z., Zhang, Q., Xu, Y., Chen, Y., et al. (2021). The UL16 protein of HSV-1 promotes the metabolism of cell mitochondria by binding to ANT2 protein. Sci. Rep. 11:14001. doi: 10.1038/s41598-021-93430-2
Liang, Y., Vogel, J. L., Narayanan, A., Peng, H., and Kristie, T. M. (2009). Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat. Med. 15, 1312–1317. doi: 10.1038/nm.2051
Liashkovich, I., Hafezi, W., Kühn, J. M., Oberleithner, H., and Shahin, V. (2011). Nuclear delivery mechanism of herpes simplex virus type 1 genome. J. Mol. Recogn. 24, 414–421. doi: 10.1002/jmr.1120
Liu, C., Cheng, A., Wang, M., Chen, S., Jia, R., Zhu, D., et al. (2015). Duck enteritis virus UL54 is an IE protein primarily located in the nucleus. Virol. J. 12:198. doi: 10.1186/s12985-015-0424-z
Liu, X., Acharya, D., Krawczyk, E., Kangas, C., Gack, M. U., and He, B. (2021). Herpesvirus-mediated stabilization of ICP0 expression neutralizes restriction by TRIM23. Proc. Natl. Acad. Sci. U.S.A. 118:2113060118. doi: 10.1073/pnas.2113060118
Liu, X., Matrenec, R., Gack, M. U., and He, B. (2019). Disassembly of the TRIM23-TBK1 complex by the Us11 protein of herpes simplex virus 1 impairs autophagy. J. Virol. 93:e497–19. doi: 10.1128/jvi.00497-19
Liu, Z., Kato, A., Shindo, K., Noda, T., Sagara, H., Kawaoka, Y., et al. (2014). Herpes simplex virus 1 UL47 interacts with viral nuclear egress factors UL31, UL34, and Us3 and regulates viral nuclear egress. J. Virol. 88, 4657–4667. doi: 10.1128/jvi.00137-14
Loomis, J. S., Bowzard, J. B., Courtney, R. J., and Wills, J. W. (2001). Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75, 12209–12219. doi: 10.1128/jvi.75.24.12209-12219.2001
Loomis, J. S., Courtney, R. J., and Wills, J. W. (2006). Packaging determinants in the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 80, 10534–10541. doi: 10.1128/jvi.01172-06
Lu, X., Huang, C., Zhang, Y., Lin, Y., Wang, X., Li, Q., et al. (2017). The Us2 gene product of herpes simplex virus 2 modulates NF-κB activation by targeting TAK1. Sci. Rep. 7:8396. doi: 10.1038/s41598-017-08856-4
Lussignol, M., and Esclatine, A. (2017). Herpesvirus and Autophagy: “All right, everybody be cool, this is a robbery! Viruses 9:372. doi: 10.3390/v9120372
Lussignol, M., Queval, C., Bernet-Camard, M. F., Cotte-Laffitte, J., Beau, I., Codogno, P., et al. (2013). The herpes simplex virus 1 Us11 protein inhibits autophagy through its interaction with the protein kinase PKR. J. Virol. 87, 859–871. doi: 10.1128/jvi.01158-12
Ma, W., Wang, H., and He, H. (2019). Bovine herpesvirus 1 tegument protein UL41 suppresses antiviral innate immune response via directly targeting STAT1. Vet. Microbiol. 239:108494. doi: 10.1016/j.vetmic.2019.108494
Ma, Z., Bai, J., Jiang, C., Zhu, H., Liu, D., Pan, M., et al. (2023). Tegument protein UL21 of alpha-herpesvirus inhibits the innate immunity by triggering CGAS degradation through TOLLIP-mediated selective autophagy. Autophagy 19, 1512–1532. doi: 10.1080/15548627.2022.2139921
MacLean, C. A., Clark, B., and McGeoch, D. J. (1989). Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70, 3147–3157. doi: 10.1099/0022-1317-70-12-3147
Mangan, S., and Alon, U. (2003). Structure and function of the feed-forward loop network motif. Proc. Natl. Acad. Sci. U.S.A. 100, 11980–11985. doi: 10.1073/pnas.2133841100
Maringer, K., and Elliott, G. (2010). Recruitment of herpes simplex virus type 1 immediate-early protein ICP0 to the virus particle. J. Virol. 84, 4682–4696. doi: 10.1128/jvi.00126-10
Maringer, K., Stylianou, J., and Elliott, G. (2012). A network of protein interactions around the herpes simplex virus tegument protein VP22. J. Virol. 86, 12971–12982. doi: 10.1128/jvi.01913-12
Martínez-Moreno, M., Navarro-Lérida, I., Roncal, F., Albar, J. P., Alonso, C., Gavilanes, F., et al. (2003). Recognition of novel viral sequences that associate with the dynein light chain LC8 identified through a pepscan technique. FEBS Lett. 544, 262–267. doi: 10.1016/s0014-5793(03)00516-7
Matis, J., and Kúdelová, M. (2001). Early shutoff of host protein synthesis in cells infected with herpes simplex viruses. Acta Virol. 45, 269–277.
Mbong, E. F., Woodley, L., Dunkerley, E., Schrimpf, J. E., Morrison, L. A., and Duffy, C. (2012a). Deletion of the herpes simplex virus 1 UL49 gene results in mRNA and protein translation defects that are complemented by secondary mutations in UL41. J. Virol. 86, 12351–12361. doi: 10.1128/jvi.01975-12
Mbong, E. F., Woodley, L., Frost, E., Baines, J. D., and Duffy, C. (2012b). Deletion of UL21 causes a delay in the early stages of the herpes simplex virus 1 replication cycle. J. Virol. 86, 7003–7007. doi: 10.1128/jvi.00411-12
McCullough, J., Clippinger, A. K., Talledge, N., Skowyra, M. L., Saunders, M. G., Naismith, T. V., et al. (2015). Structure and membrane remodeling activity of ESCRT-III helical polymers. Science 350, 1548–1551. doi: 10.1126/science.aad8305
McCullough, J., Frost, A., and Sundquist, W. I. (2018). Structures, functions, and dynamics of ESCRT-III/vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 34, 85–109. doi: 10.1146/annurev-cellbio-100616-060600
McGeehan, J. E., Depledge, N. W., and McGeoch, D. J. (2001). Evolution of the dUTPase gene of mammalian and avian herpesviruses. Curr. Protein Pept. Sci. 2, 325–333. doi: 10.2174/1389203013380964
McShane, E., and Selbach, M. (2022). Physiological functions of intracellular protein degradation. Annu. Rev. Cell Dev. Biol. 38, 241–262. doi: 10.1146/annurev-cellbio-120420-091943
Meckes, D. G. Jr., Marsh, J. A., and Wills, J. W. (2010). Complex mechanisms for the packaging of the UL16 tegument protein into herpes simplex virus. Virology 398, 208–213. doi: 10.1016/j.virol.2009.12.004
Mercer, J., Schelhaas, M., and Helenius, A. (2010). Virus entry by endocytosis. Annu. Rev. Biochem. 79, 803–833. doi: 10.1146/annurev-biochem-060208-104626
Mettenleiter, T. C., Müller, F., Granzow, H., and Klupp, B. G. (2013). The way out: What we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 15, 170–178. doi: 10.1111/cmi.12044
Mijatov, B., Cunningham, A. L., and Diefenbach, R. J. (2007). Residues F593 and E596 of HSV-1 tegument protein pUL36 (VP1/2) mediate binding of tegument protein pUL37. Virology 368, 26–31. doi: 10.1016/j.virol.2007.07.005
Ming, S. L., Zhang, S., Wang, Q., Zeng, L., Zhou, L. Y., Wang, M. D., et al. (2022). Inhibition of USP14 influences alphaherpesvirus proliferation by degrading viral VP16 protein via ER stress-triggered selective autophagy. Autophagy 18, 1801–1821. doi: 10.1080/15548627.2021.2002101
Mocarski, E. S. (2007). “Comparative analysis of herpesvirus-common proteins,” in Human herpesviruses: Biology, therapy, and immunoprophylaxis, eds A. Arvin, G. Campadelli-Fiume, E. Mocarski, P. S. Moore, B. Roizman, R. Whitley, et al. (Cambridge: Cambridge University Press).
Mohnke, J., Stark, I., Fischer, M., Fischer, P. M., Schlosser, A., Grothey, A., et al. (2022). pUL36 deubiquitinase activity augments both the initiation and the progression of lytic herpes simplex virus infection in IFN-primed cells. J. Virol. 96:e0096322. doi: 10.1128/jvi.00963-22
Morishita, H., and Mizushima, N. (2019). Diverse cellular roles of autophagy. Annu. Rev. Cell. Dev. Biol. 35, 453–475. doi: 10.1146/annurev-cellbio-100818-125300
Morrison, E. E., Wang, Y. F., and Meredith, D. M. (1998). Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J. Virol. 72, 7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998
Mossman, K. L., and Smiley, J. R. (1999). Truncation of the C-terminal acidic transcriptional activation domain of herpes simplex virus VP16 renders expression of the immediate-early genes almost entirely dependent on ICP0. J. Virol. 73, 9726–9733. doi: 10.1128/jvi.73.12.9726-9733.1999
Mou, F., Forest, T., and Baines, J. D. (2007). US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81, 6459–6470. doi: 10.1128/jvi.00380-07
Mou, F., Wills, E., and Baines, J. D. (2009). Phosphorylation of the U(L)31 protein of herpes simplex virus 1 by the U(S)3-encoded kinase regulates localization of the nuclear envelopment complex and egress of nucleocapsids. J. Virol. 83, 5181–5191. doi: 10.1128/jvi.00090-09
Muradov, J. H., Finnen, R. L., Gulak, M. A., Hay, T. J. M., and Banfield, B. W. (2021). pUL21 regulation of pUs3 kinase activity influences the nature of nuclear envelope deformation by the HSV-2 nuclear egress complex. PLoS Pathog. 17:e1009679. doi: 10.1371/journal.ppat.1009679
Musarra-Pizzo, M., Pennisi, R., Lombardo, D., Velletri, T., and Sciortino, M. T. (2022). Direct cleavage of caspase-8 by herpes simplex virus 1 tegument protein US11. Sci. Rep. 12:12317. doi: 10.1038/s41598-022-15942-9
Musarrat, F., Chouljenko, V., and Kousoulas, K. G. (2021). Cellular and viral determinants of HSV-1 entry and intracellular transport towards nucleus of infected cells. J. Virol. 95:20. doi: 10.1128/jvi.02434-20
Nozawa, N., Daikoku, T., Koshizuka, T., Yamauchi, Y., Yoshikawa, T., and Nishiyama, Y. (2003). Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77, 3204–3216. doi: 10.1128/jvi.77.5.3204-3216.2003
Nozawa, N., Daikoku, T., Yamauchi, Y., Takakuwa, H., Goshima, F., Yoshikawa, T., et al. (2002). Identification and characterization of the UL7 gene product of herpes simplex virus type 2. Virus Genes 24, 257–266. doi: 10.1023/a:1015332716927
Oda, S., Arii, J., Koyanagi, N., Kato, A., and Kawaguchi, Y. (2016). The Interaction between herpes simplex virus 1 tegument proteins UL51 and UL14 and its role in virion morphogenesis. J. Virol. 90, 8754–8767. doi: 10.1128/jvi.01258-16
Okada, A., Suganuma, S., Badr, Y., Omatsu, T., Mizutani, T., Ohya, K., et al. (2018). Decreased expression of the immediate early protein, ICP4, by deletion of the tegument protein VP22 of equine herpesvirus type 1. J. Vet. Med. Sci. 80, 311–315. doi: 10.1292/jvms.17-0380
Olmos, Y., and Carlton, J. G. (2016). The ESCRT machinery: New roles at new holes. Curr. Opin. Cell Biol. 38, 1–11. doi: 10.1016/j.ceb.2015.12.001
Omar, O. S., Simmons, A. J., Andre, N. M., Wilson, D. W., and Gross, S. T. (2013). Pseudorabies virus and herpes simplex virus type 1 utilize different tegument-glycoprotein interactions to mediate the process of envelopment. Intervirology 56, 50–54. doi: 10.1159/000339467
O’Regan, K. J., Murphy, M. A., Bucks, M. A., Wills, J. W., and Courtney, R. J. (2007b). Incorporation of the herpes simplex virus type 1 tegument protein VP22 into the virus particle is independent of interaction with VP16. Virology 369, 263–280. doi: 10.1016/j.virol.2007.07.020
O’Regan, K. J., Bucks, M. A., Murphy, M. A., Wills, J. W., and Courtney, R. J. (2007a). A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358, 192–200. doi: 10.1016/j.virol.2006.08.024
Orvedahl, A., Alexander, D., Tallóczy, Z., Sun, Q., Wei, Y., Zhang, W., et al. (2007). HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 1, 23–35. doi: 10.1016/j.chom.2006.12.001
Orzalli, M. H., DeLuca, N. A., and Knipe, D. M. (2012). Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. U.S.A. 109, E3008–E3017. doi: 10.1073/pnas.1211302109
Ostler, J. B., and Jones, C. (2021). Stress induced transcription factors transactivate the herpes simplex virus 1 infected cell protein 27 (ICP27) transcriptional enhancer. Viruses 13:296. doi: 10.3390/v13112296
Owen, D. J., Crump, C. M., and Graham, S. C. (2015). Tegument assembly and secondary envelopment of alphaherpesviruses. Viruses 7, 5084–5114. doi: 10.3390/v7092861
Pannhorst, K., Wei, H., Yezid, H., He, J., and Chowdhury, S. I. (2018). Bovine herpesvirus 1 U(L)49.5 interacts with gM and VP22 to ensure virus cell-to-cell spread and virion incorporation: Novel role for VP22 in gM-independent U(L)49.5 virion incorporation. J. Virol. 92:e240–18. doi: 10.1128/jvi.00240-18
Parameswaran, P., Payne, L., Powers, J., Rashighi, M., and Orzalli, M. H. (2024). A viral E3 ubiquitin ligase produced by herpes simplex virus 1 inhibits the NLRP1 inflammasome. J. Exp. Med. 221:518. doi: 10.1084/jem.20231518
Pawliczek, T., and Crump, C. M. (2009). Herpes simplex virus type 1 production requires a functional ESCRT-III complex but is independent of TSG101 and ALIX expression. J. Virol. 83, 11254–11264. doi: 10.1128/jvi.00574-09
Pennisi, R., Musarra-Pizzo, M., Lei, Z., Zhou, G. G., and Sciortino, M. T. (2020). VHS, in the list of citations. US3 and UL13 viral tegument proteins are required for Herpes Simplex Virus-Induced modification of protein kinase R. Sci. Rep. 10:5580. doi: 10.1038/s41598-020-62619-2
Pennisi, R., Musarra-Pizzo, M., Lei, Z., Zhou, G. G., and Sciortino, M. T. (2021). Author correction: VHS, US3 and UL13 viral tegument proteins are required for Herpes Simplex Virus-Induced modification of protein kinase R. Sci. Rep. 11:7503. doi: 10.1038/s41598-021-87273-0
Preston, V. G., and Fisher, F. B. (1984). Identification of the herpes simplex virus type 1 gene encoding the dUTPase. Virology 138, 58–68. doi: 10.1016/0042-6822(84)90147-8
Radtke, K., Kieneke, D., Wolfstein, A., Michael, K., Steffen, W., Scholz, T., et al. (2010). Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. doi: 10.1371/journal.ppat.1000991
Rémy, S., Blondeau, C., Le Vern, Y., Lemesle, M., Vautherot, J. F., and Denesvre, C. (2013). Fluorescent tagging of VP22 in N-terminus reveals that VP22 favors Marek’s disease virus (MDV) virulence in chickens and allows morphogenesis study in MD tumor cells. Vet. Res. 44:125. doi: 10.1186/1297-9716-44-125
Ren, X., Harms, J. S., and Splitter, G. A. (2001). Bovine herpesvirus 1 tegument protein VP22 interacts with histones, and the carboxyl terminus of VP22 is required for nuclear localization. J. Virol. 75, 8251–8258. doi: 10.1128/jvi.75.17.8251-8258.2001
Richards, A. L., Sollars, P. J., Pitts, J. D., Stults, A. M., Heldwein, E. E., Pickard, G. E., et al. (2017). The pUL37 tegument protein guides alpha-herpesvirus retrograde axonal transport to promote neuroinvasion. PLoS Pathog. 13:e1006741. doi: 10.1371/journal.ppat.1006741
Ripa, I., Andreu, S., López-Guerrero, J. A., and Bello-Morales, R. (2022). Interplay between autophagy and herpes simplex virus type 1: ICP34.5, one of the main actors. Int. J. Mol. Sci. 23:643. doi: 10.3390/ijms232113643
Roizman, B., and Whitley, R. J. (2013). An inquiry into the molecular basis of HSV latency and reactivation. Annu. Rev. Microbiol. 67, 355–374. doi: 10.1146/annurev-micro-092412-155654
Roller, R. J., and Johnson, D. C. (2021). Herpesvirus nuclear egress across the outer nuclear membrane. Viruses 13:2356. doi: 10.3390/v13122356
Sarfo, A., Starkey, J., Mellinger, E., Zhang, D., Chadha, P., Carmichael, J., et al. (2017). The UL21 tegument protein of herpes simplex virus 1 is differentially required for the syncytial phenotype. J. Virol. 91:e1161–17. doi: 10.1128/jvi.01161-17
Sasse, S. K., Mailloux, C. M., Barczak, A. J., Wang, Q., Altonsy, M. O., Jain, M. K., et al. (2013). The glucocorticoid receptor and KLF15 regulate gene expression dynamics and integrate signals through feed-forward circuitry. Mol. Cell. Biol. 33, 2104–2115. doi: 10.1128/mcb.01474-12
Sasse, S. K., Zuo, Z., Kadiyala, V., Zhang, L., Pufall, M. A., Jain, M. K., et al. (2015). Response element composition governs correlations between binding site affinity and transcription in glucocorticoid receptor feed-forward loops. J. Biol. Chem. 290, 19756–19769. doi: 10.1074/jbc.M115.668558
Sciortino, M. T., Taddeo, B., Giuffrè-Cuculletto, M., Medici, M. A., Mastino, A., and Roizman, B. (2007). Replication-competent herpes simplex virus 1 isolates selected from cells transfected with a bacterial artificial chromosome DNA lacking only the UL49 gene vary with respect to the defect in the UL41 gene encoding host shutoff RNase. J. Virol. 81, 10924–10932. doi: 10.1128/jvi.01239-07
Scourfield, E. J., and Martin-Serrano, J. (2017). Growing functions of the ESCRT machinery in cell biology and viral replication. Biochem. Soc. Trans. 45, 613–634. doi: 10.1042/bst20160479
Sedwick, W. D., Brown, O. E., and Glickman, B. W. (1986). Deoxyuridine misincorporation causes site-specific mutational lesions in the lacI gene of Escherichia coli. Mutat. Res. 162, 7–20. doi: 10.1016/0027-5107(86)90066-7
Shahin, F., Raza, S., Yang, K., Hu, C., Chen, Y., Chen, H., et al. (2017). Bovine herpesvirus 1 tegument protein UL21 plays critical roles in viral secondary envelopment and cell-to-cell spreading. Oncotarget 8, 94462–94480. doi: 10.18632/oncotarget.21776
Shaw, P., Knez, J., and Capone, J. P. (1995). Amino acid substitutions in the herpes simplex virus transactivator VP16 uncouple direct protein-protein interaction and DNA binding from complex assembly and transactivation. J. Biol. Chem. 270, 29030–29037. doi: 10.1074/jbc.270.48.29030
Shu, M., Taddeo, B., and Roizman, B. (2013). The nuclear-cytoplasmic shuttling of virion host shutoff RNase is enabled by pUL47 and an embedded nuclear export signal and defines the sites of degradation of AU-rich and stable cellular mRNAs. J. Virol. 87, 13569–13578. doi: 10.1128/jvi.02603-13
Simmen, K. A., Newell, A., Robinson, M., Mills, J. S., Canning, G., Handa, R., et al. (1997). Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J. Virol. 71, 3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997
Smibert, C. A., Popova, B., Xiao, P., Capone, J. P., and Smiley, J. R. (1994). Herpes simplex virus VP16 forms a complex with the virion host shutoff protein vhs. J. Virol. 68, 2339–2346. doi: 10.1128/jvi.68.4.2339-2346.1994
Smiley, J. R., Elgadi, M. M., and Saffran, H. A. (2001). Herpes simplex virus vhs protein. Methods Enzymol. 342, 440–451. doi: 10.1016/s0076-6879(01)42565-1
Sodroski, C. N., Oh, H. S., Chou, S. F., and Knipe, D. M. (2024). Sp1 facilitates continued HSV-1 gene expression in the absence of key viral transactivators. mBio 15:e0347923. doi: 10.1128/mbio.03479-23
Spear, P. G. (2004). Herpes simplex virus: Receptors and ligands for cell entry. Cell Microbiol 6, 401–410. doi: 10.1111/j.1462-5822.2004.00389.x
Spear, P. G., and Longnecker, R. (2003). Herpesvirus entry: An update. J. Virol. 77, 10179–10185. doi: 10.1128/jvi.77.19.10179-10185.2003
Starkey, J. L., Han, J., Chadha, P., Marsh, J. A., and Wills, J. W. (2014). Elucidation of the block to herpes simplex virus egress in the absence of tegument protein UL16 reveals a novel interaction with VP22. J. Virol. 88, 110–119. doi: 10.1128/jvi.02555-13
Steiner, I., Kennedy, P. G., and Pachner, A. R. (2007). The neurotropic herpes viruses: Herpes simplex and varicella-zoster. Lancet Neurol. 6, 1015–1028. doi: 10.1016/s1474-4422(07)70267-3
Stylianou, J., Maringer, K., Cook, R., Bernard, E., and Elliott, G. (2009). Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83, 5204–5218. doi: 10.1128/jvi.00069-09
Sucharita, S., Zhang, K., and van Drunen Littel-van den Hurk, S. (2021). VP8, the major tegument protein of bovine herpesvirus-1, is partially packaged during early tegument formation in a VP22-dependent manner. Viruses 13:1854. doi: 10.3390/v13091854
Sun, S., Li, L., Yang, F., Wang, X., Fan, F., Yang, M., et al. (2017). Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution. Nat. Commun. 8:16064. doi: 10.1038/ncomms16064
Svobodova, S., Bell, S., and Crump, C. M. (2012). Analysis of the interaction between the essential herpes simplex virus 1 tegument proteins VP16 and VP1/2. J. Virol. 86, 473–483. doi: 10.1128/jvi.05981-11
Taddeo, B., and Roizman, B. (2006). The virion host shutoff protein (UL41) of herpes simplex virus 1 is an endoribonuclease with a substrate specificity similar to that of RNase A. J. Virol. 80, 9341–9345. doi: 10.1128/jvi.01008-06
Tallóczy, Z., Jiang, W., Virgin, H. W., Leib, D. A., Scheuner, D., Kaufman, R. J., et al. (2002). Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc. Natl. Acad. Sci. U.S.A. 99, 190–195. doi: 10.1073/pnas.012485299
Tallóczy, Z., Virgin, H. W., and Levine, B. (2006). PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2, 24–29. doi: 10.4161/auto.2176
Tanaka, M., Kato, A., Satoh, Y., Ide, T., Sagou, K., Kimura, K., et al. (2012). Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence. J. Virol. 86, 5264–5277. doi: 10.1128/jvi.06913-11
Tang, S., Patel, A., and Krause, P. R. (2019). Hidden regulation of herpes simplex virus 1 pre-mRNA splicing and polyadenylation by virally encoded immediate early gene ICP27. PLoS Pathog. 15:e1007884. doi: 10.1371/journal.ppat.1007884
Trapp-Fragnet, L., Courvoisier, K., Rémy, S., Pape, G. L., Loustalot, F., and Denesvre, C. (2019). Identification of Marek’s disease virus VP22 tegument protein domains essential for virus cell-to-cell spread, nuclear localization, histone association and cell-cycle arrest. Viruses 11:537. doi: 10.3390/v11060537
Trus, B. L., Newcomb, W. W., Booy, F. P., Brown, J. C., and Steven, A. C. (1992). Distinct monoclonal antibodies separately label the hexons or the pentons of herpes simplex virus capsid. Proc. Natl. Acad. Sci. U.S.A. 89, 11508–11512. doi: 10.1073/pnas.89.23.11508
van Gent, M., Chiang, J. J., Muppala, S., Chiang, C., Azab, W., Kattenhorn, L., et al. (2022). The US3 kinase of herpes simplex virus phosphorylates the RNA sensor RIG-I To suppress innate immunity. J. Virol. 96:e0151021. doi: 10.1128/jvi.01510-21
van Leeuwen, H., Okuwaki, M., Hong, R., Chakravarti, D., Nagata, K., and O’Hare, P. (2003). Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J. Gen. Virol. 84, 2501–2510. doi: 10.1099/vir.0.19326-0
Vértessy, B. G., and Tóth, J. (2009). Keeping uracil out of DNA: Physiological role, structure and catalytic mechanism of dUTPases. Acc. Chem. Res. 42, 97–106. doi: 10.1021/ar800114w
Vittone, V., Diefenbach, E., Triffett, D., Douglas, M. W., Cunningham, A. L., and Diefenbach, R. J. (2005). Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79, 9566–9571. doi: 10.1128/jvi.79.15.9566-9571.2005
Votteler, J., and Sundquist, W. I. (2013). Virus budding and the ESCRT pathway. Cell Host Microbe 14, 232–241. doi: 10.1016/j.chom.2013.08.012
Waisner, H., and Kalamvoki, M. (2019). The ICP0 protein of herpes simplex virus 1 (HSV-1) downregulates major autophagy adaptor proteins sequestosome 1 and optineurin during the early stages of HSV-1 infection. J. Virol. 93:e1258–19. doi: 10.1128/jvi.01258-19
Waisner, H., Lasnier, S., Suma, S. M., and Kalamvoki, M. (2023). Effects on exocytosis by two HSV-1 mutants unable to block autophagy. J. Virol. 97:e0075723. doi: 10.1128/jvi.00757-23
Wan, J., Li, F., Wang, M., Cheng, A., Tian, B., Yang, Q., et al. (2022). The protein encoded by the duck plague virus UL14 gene regulates virion morphogenesis and affects viral replication. Poult. Sci. 101:101863. doi: 10.1016/j.psj.2022.101863
Watson, R. J., and Clements, J. B. (1980). A herpes simplex virus type 1 function continuously required for early and late virus RNA synthesis. Nature 285, 329–330. doi: 10.1038/285329a0
Whitley, R. J., Kimberlin, D. W., and Roizman, B. (1998). Herpes simplex viruses. Clin. Infect. Dis. 26, 541–553; quiz554–545. doi: 10.1086/514600
Wolfstein, A., Nagel, C. H., Radtke, K., Döhner, K., Allan, V. J., and Sodeik, B. (2006). The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7, 227–237. doi: 10.1111/j.1600-0854.2005.00379.x
Wu, L., Wang, M., Cheng, A., Tian, B., Huang, J., Wu, Y., et al. (2023). Duck plague virus tegument protein vp22 plays a key role in the secondary envelopment and cell-to-cell spread. Vet. Res. 54:60. doi: 10.1186/s13567-023-01191-9
Xu, C., Wang, M., Song, Z., Wang, Z., Liu, Q., Jiang, P., et al. (2018). Pseudorabies virus induces autophagy to enhance viral replication in mouse neuro-2a cells in vitro. Virus Res. 248, 44–52. doi: 10.1016/j.virusres.2018.02.004
Xu, G., Liu, C., Zhou, S., Li, Q., Feng, Y., Sun, P., et al. (2021). Viral tegument proteins restrict cGAS-DNA phase separation to mediate immune evasion. Mol. Cell 81:2823–2837.e2829. doi: 10.1016/j.molcel.2021.05.002
Yamauchi, Y., Kiriyama, K., Kubota, N., Kimura, H., Usukura, J., and Nishiyama, Y. (2008). The UL14 tegument protein of herpes simplex virus type 1 is required for efficient nuclear transport of the alpha transinducing factor VP16 and viral capsids. J. Virol. 82, 1094–1106. doi: 10.1128/jvi.01226-07
Yamauchi, Y., Shiba, C., Goshima, F., Nawa, A., Murata, T., and Nishiyama, Y. (2001). Herpes simplex virus type 2 UL34 protein requires UL31 protein for its relocation to the internal nuclear membrane in transfected cells. J. Gen. Virol. 82, 1423–1428. doi: 10.1099/0022-1317-82-6-1423
Yan, K., Liu, J., Guan, X., Yin, Y. X., Peng, H., Chen, H. C., et al. (2019). The Carboxyl Terminus Of Tegument Protein pUL21 contributes to pseudorabies virus neuroinvasion. J. Virol. 93:e2052–18. doi: 10.1128/jvi.02052-18
Yan, Z., Yue, J., Zhang, Y., Hou, Z., Li, D., Yang, Y., et al. (2024). Pseudorabies virus VHS protein abrogates interferon responses by blocking NF-κB and IRF3 nuclear translocation. Virol. Sin. S1995-820X(24)00078-6. doi: 10.1016/j.virs.2024.05.009
Yang, L., Shen, B., Wang, M., Cheng, A., Yang, Q., Wu, Y., et al. (2021). The intracellular domain of duck plague virus glycoprotein E affects UL11 protein incorporation into viral particles. Vet. Microbiol. 257:109078. doi: 10.1016/j.vetmic.2021.109078
Yang, L., Wang, M., Cheng, A., Yang, Q., Wu, Y., Huang, J., et al. (2022). Features and functions of the conserved herpesvirus tegument protein UL11 and its binding partners. Front. Microbiol. 13:829754. doi: 10.3389/fmicb.2022.829754
Yang, L., Wang, M., Cheng, A., Yang, Q., Wu, Y., Jia, R., et al. (2019). Innate immune evasion of alphaherpesvirus tegument proteins. Front. Immunol. 10:2196. doi: 10.3389/fimmu.2019.02196
Ye, G. J., Vaughan, K. T., Vallee, R. B., and Roizman, B. (2000). The herpes simplex virus 1 U(L)34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74, 1355–1363. doi: 10.1128/jvi.74.3.1355-1363.2000
Ye, R., Su, C., Xu, H., and Zheng, C. (2017). Herpes simplex virus 1 ubiquitin-specific protease UL36 abrogates NF-κB activation in DNA sensing signal pathway. J. Virol. 91:16. doi: 10.1128/jvi.02417-16
Yin, H. C., Zhao, L. L., Li, S. Q., Niu, Y. J., Jiang, X. J., Xu, L. J., et al. (2017). Autophagy activated by duck enteritis virus infection positively affects its replication. J. Gen. Virol. 98, 486–495. doi: 10.1099/jgv.0.000696
Yin, H., Li, Z., Zhang, J., Huang, J., Kang, H., Tian, J., et al. (2020). Construction of a US7/US8/UL23/US3-deleted recombinant pseudorabies virus and evaluation of its pathogenicity in dogs. Vet. Microbiol. 240:108543. doi: 10.1016/j.vetmic.2019.108543
Zhang, H. (2022). The genetics of autophagy in multicellular organisms. Annu. Rev. Genet. 56, 17–39. doi: 10.1146/annurev-genet-022422-095608
Zhang, J., Zhao, J., Xu, S., Li, J., He, S., Zeng, Y., et al. (2018). Species-specific deamidation of cGAS by herpes simplex virus UL37 protein facilitates viral replication. Cell Host Microbe 24:234–248.e235. doi: 10.1016/j.chom.2018.07.004
Zhang, L., Huang, F., Liu, J., Xu, Y., Miao, Y., Yuan, Y., et al. (2021). HSV-1-encoded ICP0 degrades the host deubiquitinase BRCC36 to antagonize interferon antiviral response. Mol. Immunol. 135, 28–35. doi: 10.1016/j.molimm.2021.03.027
Zhang, R., Xu, A., Qin, C., Zhang, Q., Chen, S., Lang, Y., et al. (2017). Pseudorabies virus dUTPase UL50 induces lysosomal degradation of type i interferon receptor 1 and antagonizes the alpha interferon response. J. Virol. 91:e1148–17. doi: 10.1128/jvi.01148-17
Zhao, N., Wang, F., Kong, Z., and Shang, Y. (2022). Pseudorabies virus tegument protein UL13 suppresses RLR-mediated antiviral innate immunity through regulating receptor transcription. Viruses 14:465. doi: 10.3390/v14071465
Zhou, G., Te, D., and Roizman, B. (2010). The CoREST/REST repressor is both necessary and inimical for expression of herpes simplex virus genes. mBio 2:e313–10. doi: 10.1128/mBio.00313-10
Keywords: alphaherpesvirus, tegument protein, life cycle, interaction, innate immune escape, autophagy, ESCRT
Citation: Cui Y, Wang M, Cheng A, Zhang W, Yang Q, Tian B, Ou X, Huang J, Wu Y, Zhang S, Sun D, He Y, Zhao X, Wu Z, Zhu D, Jia R, Chen S and Liu M (2024) The precise function of alphaherpesvirus tegument proteins and their interactions during the viral life cycle. Front. Microbiol. 15:1431672. doi: 10.3389/fmicb.2024.1431672
Received: 12 May 2024; Accepted: 20 June 2024;
Published: 02 July 2024.
Edited by:
Rahul K. Suryawanshi, Gladstone Institutes, United StatesReviewed by:
Mengmeng Zhao, Foshan University, ChinaFouad S. El-mayet, Oklahoma State University, United States
Copyright © 2024 Cui, Wang, Cheng, Zhang, Yang, Tian, Ou, Huang, Wu, Zhang, Sun, He, Zhao, Wu, Zhu, Jia, Chen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anchun Cheng, Y2hlbmdhbmNodW5AdmlwLjE2My5jb20=
†These authors have contributed equally to this work
 Yuxi Cui
Yuxi Cui Mingshu Wang
Mingshu Wang Anchun Cheng
Anchun Cheng Wei Zhang6
Wei Zhang6 Qiao Yang
Qiao Yang Bin Tian
Bin Tian Xumin Ou
Xumin Ou Juan Huang
Juan Huang Shaqiu Zhang
Shaqiu Zhang Di Sun
Di Sun Dekang Zhu
Dekang Zhu Shun Chen
Shun Chen Mafeng Liu
Mafeng Liu