- 1Department of Research and Development, Center for Industrialization of Agricultural and Livestock Microorganisms, Jeongeup-si, Republic of Korea
- 2School of Biological Sciences and Technology, Chonnam National University, Gwangju, Republic of Korea
Global climate change poses a significant threat to plant growth and crop yield and is exacerbated by environmental factors, such as drought, salinity, greenhouse gasses, and extreme temperatures. Plant growth-promoting rhizobacteria (PGPR) help plants withstand drought. However, the mechanisms underlying PGPR–plant interactions remain unclear. Thus, this study aimed to isolate PGPR, Bacillus megaterium strains CACC109 and CACC119, from a ginseng field and investigate the mechanisms underlying PGPR-stimulated tolerance to drought stress by evaluating their plant growth-promoting activities and effects on rice growth and stress tolerance through in vitro assays, pot experiments, and physiological and molecular analyses. Compared with B. megaterium type strain ATCC14581, CACC109 and CACC119 exhibited higher survival rates under osmotic stress, indicating their potential to enhance drought tolerance. Additionally, CACC109 and CACC119 strains exhibited various plant growth-promoting activities, including phosphate solubilization, nitrogen fixation, indole-3-acetic acid production, siderophore secretion, 1-aminocyclopropane-1-carboxylate deaminase activity, and exopolysaccharide production. After inoculation, CACC109 and CACC119 significantly improved the seed germination of rice (Oryza sativa L.) under osmotic stress and promoted root growth under stressed and non-stressed conditions. They also facilitated plant growth in pot experiments, as evidenced by increased shoot and root lengths, weights, and leaf widths. Furthermore, CACC109 and CACC119 improved plant physiological characteristics, such as chlorophyll levels, and production of osmolytes, such as proline. In particular, CACC109- and CACC119-treated rice plants showed better drought tolerance, as evidenced by their higher survival rates, greater chlorophyll contents, and lower water loss rates, compared with mock-treated rice plants. Application of CACC109 and CACC119 upregulated the expression of antioxidant-related genes (e.g., OsCAT, OsPOD, OsAPX, and OsSOD) and drought-responsive genes (e.g., OsWRKY47, OsZIP23, OsDREB2, OsNAC066, OsAREB1, and OsAREB2). In conclusion, CACC109 and CACC119 are promising biostimulants for enhancing plant growth and conferring resistance to abiotic stresses in crop production. Future studies should conduct field trials to validate these findings under real agricultural conditions, optimize inoculation methods for practical use, and further investigate the biochemical and physiological responses underlying the observed benefits.
1 Introduction
Global population growth and extreme climate change pose important agricultural challenges that may cause insufficient food supply worldwide (FAO, 2017). Rice (Oryza sativa L.), a crucial staple feeding nearly half of the global population, is grown on around 163 million hectares of land worldwide (Fukagawa and Ziska, 2019; Shahbandeh, 2024). Despite its importance, rice productivity is affected by abiotic and biotic stresses. Drought and high salinity are abiotic stresses that severely affect rice growth, development, and production. Specifically, drought is considered the most important constraint to rice production, affecting approximately 19–23 million hectares of rice fields (Kumar, 2017).
Under drought, plants exhibit changes in various properties, including physiological (e.g., photosynthesis, osmotic balance, transpiration, leaf water content, and stomatal water transmission activity), morphological (e.g., leaf area, leaf number, root length, leaf aging, maturation, and growth stage), and biochemical (e.g., antioxidant content, chlorophyll content, proline accumulation, hormonal content, and secondary metabolites) (Anjum et al., 2011; Sharma et al., 2012; Haworth et al., 2013; Ammar et al., 2015; Conesa et al., 2016; Tiwari et al., 2021; Yang et al., 2021). Although these mechanisms enable plants to minimize water loss and usage, they can adversely affect plant growth and development, consequently reducing yield (Nakabayashi and Saito, 2015).
With the urgent need to support crop growth to meet food demands amid limited water resources, interest in enhancing drought tolerance and discovering sustainable solutions to related food security challenges has increased. The acquisition of essential nutrients under drought is a significant challenge for plants, often limiting their growth and productivity. Plant growth-promoting rhizobacteria (PGPR) represent a wide variety of beneficial soil bacteria that colonize the rhizosphere and plant roots, and can grow in, on, or around plant tissues. PGPR can increase nutrient availability and uptake, directly and indirectly supporting plant growth and resilience under drought (Backer et al., 2018). Under drought, PGPR play pivotal roles in nutrient solubilization and uptake. These bacteria solubilize phosphorus and other essential nutrients, making them more accessible to plants, while also improving water content and potential, thereby enhancing mineral quality and grain yield (Creus et al., 2004; Richardson and Simpson, 2011; Hardoim et al., 2015). Nitrogen fixation by PGPR contributes to soil nitrogen enrichment, providing an additional nitrogen source for plants. This process is essential for nitrogen acquisition in nutrient-poor, dry soils (Hardoim et al., 2015). Siderophore production by PGPR is another vital mechanism that enhances nutrient acquisition under drought. Siderophores are iron-chelating compounds that microbes secrete to sequester iron from the soil environment, making it more accessible to plants. This process is especially critical under drought, where nutrient availability is limited (Sharma and Johri, 2003; Ferreira et al., 2019). In addition, PGPR can improve drought tolerance by stimulating the production of drought-resistant compounds, such as bacterial exopolysaccharides (EPS), 1-aminocyclopropane-1-carboxylate (ACC) deaminase, volatile organic compounds (VOCs), and phytohormones, such as abscisic acid (ABA), ethylene (ET), and indole-3-acetic acid (IAA) (Vejan et al., 2016; Vurukonda et al., 2016; Tiepo et al., 2018). The production of EPS by PGPR helps retain soil moisture and stabilize soil structure, further improving the water holding capacity and nutrient availability in the soil, thereby enhancing plant growth and alleviating drought (Vurukonda et al., 2016). ACC deaminase is a PGPR-produced enzyme that can modulate plant ET levels. ET is a phytohormone associated with stress responses, and high levels of ET can inhibit plant growth. ACC deaminase lowers ET levels by breaking down its precursor, ACC, thereby mitigating stress effects and promoting root elongation and nutrient uptake under drought conditions (Glick et al., 1998; Mayak et al., 2004; Vurukonda et al., 2016). VOCs emitted by soil microorganisms also enhance the ability of plants to cope with abiotic stresses, such as drought, by inducing system resistance. VOCs such as acetoin and 2,3-butanediol improve plant growth and drought resistance by modulating stress-related pathways and enhancing water use efficiency (Ryu et al., 2003; Vejan et al., 2016). Phytohormones, including ABA and ET, are critical in plant response to drought. ABA plays central roles in regulating stomatal closure, reducing water loss, and activating stress-responsive genes. Microbial interactions can influence ABA levels, enhancing the drought response of plant. While ET is involved in the regulation of growth and stress responses, its overproduction can be detrimental. Microbial modulation of ET levels via ACC deaminase activity helps maintain a balance, promoting plant growth and stress tolerance (Glick et al., 1998; Cutler et al., 2010; Cohen et al., 2015; Vurukonda et al., 2016). PGPR produce plant growth-promoting (PGP) hormones such as auxins that enhance root growth and nutrient acquisition. The best-known auxin, IAA, modifies the plant root architecture by increasing root length, root branching, and total root surface area resulting in improved water and nutrient uptake and enhanced cellular defense against water stress (Glick, 2012; Vurukonda et al., 2016). These studies indicate the promising potential of PGPR in enhancing drought tolerance.
Plants initiate specific responses that reprogram genetic, molecular, and physiological processes to mitigate the adverse effects of drought on their growth and productivity. Many plants achieve stress resistance through PGPR-mediated activation of various stress-responsive genes. Plants inoculated with PGPR under drought stress exhibit distinct gene expression profiles compared with non-inoculated plants (Saakre et al., 2017; Kaushal, 2019). For example, inoculation of sugarcane (Saccharum spp.) with Gluconacetobacter diazotrophicus PAL5 under drought activates ABA-dependent signaling genes, enhancing drought tolerance (Vargas et al., 2014). Colonization of plants by PGPR strains can markedly alter gene expression in response to drought. In rice, treatment with Pseudomonas fluorescens (Pf1) activates genes such as Hsp20, bZIP1, and COC1 involved in the ABA signaling pathway, PKDP encoding a protein kinase, and AP2-EREBP associated with developmental and stress defense pathways (Saakre et al., 2017). Similarly, inoculation of Arabidopsis thaliana with GAP-P45 alters the expression patterns of genes related to polyamine biosynthesis (ADC, AIH, CPA, SPDS, SPMS, and SAMDC), leading to increased polyamine levels and reduced osmotic stress during drought (Sen et al., 2018). In wheat, PGPR strains such as Arthrobacter protophormiae SA3, Bacillus subtilis LDR2, and Dietzia natronolimnaea STR1 enhance drought and salinity tolerance by modifying the expression of genes and transcription factors related to the ET signaling pathway (Barnawal et al., 2019). In soybean, inoculation with Pseudomonas simiae AU under drought conditions results in the upregulation of genes encoding transcription factors (DREB/EREB), osmoprotectants (P5CS, GOLS), and water transporters (PIP, TIP), leading to improved drought tolerance compared with control plants (Vaishnav and Choudhary, 2019). Li et al. (2020) found that treating drought-stressed wheat with a cell filtrate from Streptomyces pactum Act12 improves growth by upregulating genes related to water deficit resistance, including those involved in cell wall expansion (EXPA2 and EXPA6), proline accumulation (P5CS), and stomatal closure (SnRK) (Li et al., 2020). Additionally, application of Bacillus subtilis GOT9 to A. thaliana and Brassica campestris enhances drought tolerance and survival by hyper-inducing various genes responsible for improved drought tolerance phenotypes (Woo et al., 2020). Inoculation with Pseudomonas putida (RA) in chickpea plants under drought confers drought tolerance by differentially expressing genes involved in transcription activation (DREB1A and NAC1), stress response (LEA and DHN), reactive oxygen species (ROS) scavenging (CAT, APX, and GST), ET biosynthesis (ACO and ACS), salicylic acid (PR1), and jasmonate (MYC2) signaling (Tiwari et al., 2021). Similarly, potted rice plants inoculated with Bacillus endophyticus PB3, Bacillus altitudinis PB46, and Bacillus megaterium PB50 exhibit significant drought tolerance, particularly with strain PB50, by upregulating genes such as LEA, SNAC1, HSP70, RAB16B, and bZIP23, along with increased osmolyte accumulation (Devarajan et al., 2021). Recently, Omara et al. (2022) have demonstrated that inoculating rice plants with B. megaterium, Pseudomonas azotoformans, and Rhizobium sp. enhances growth and drought tolerance by altering the expression of genes related to growth and stress response (COX1, AP2-EREBP, GRAM, NRAMP6, NAM, GST, and DHN) and expansion (EXP1, EXP2, and EXP3) (Omara et al., 2022).
Bacteria, similar to plants, are also affected by hydric stress during water scarcity. Therefore, identifying and selecting drought-tolerant bacteria is valuable for developing new biostimulants suitable for water-deficient regions (García et al., 2017). Panax ginseng is a perennial herb of significant medicinal importance in Korea (Kim, 2018). Ginseng stands out among crops due to the high commercial value of its roots, and maintaining healthy root development is essential throughout its 4–6-year cultivation period (Durairaj et al., 2018; Ding et al., 2022). However, ginseng cultivation is frequently challenged by biotic factors, such as microbial pathogens and pests, and abiotic factors, including drought, salinity, and temperature (Foyer et al., 2016), leading to changes in microbial communities. Notably, a close relationship exists between changes in microbial communities and edaphic factors in soils where ginseng is cultivated (Punja et al., 2008; Li et al., 2018). Ginseng roots and the surrounding soil harbor a diverse microbial community, including PGPR (Ji et al., 2021). However, the specific mechanisms underlying PGPR–plant interactions and their optimization for sustainable agricultural practices remain unclear.
Thus, this study aimed to investigate the effects of bacterial strains isolated from the rhizosphere and soil of ginseng plants growing in a drought-stressed field on the growth and drought resistance of rice and elucidate the underlying mechanisms. Two bacterial strains, Bacillus megaterium CACC109 and CACC119, were identified for their osmotic stress tolerance and PGP traits. Various parameters, including seed germination, shoot and root growth, physiological properties, osmolyte production, expression of antioxidant-related genes and drought-responsive genes (DRGs), and soil composition, were compared between rice treated with these strains and control plants. We hypothesized that rice plants inoculated with bacterial strains isolated from the rhizosphere under drought would exhibit enhanced growth and improved drought resistance due to the beneficial impact of the bacteria on plant metabolism.
2 Materials and methods
2.1 Isolation and characterization of rhizosphere bacteria
Ginseng roots and soil were collected from a drought-stressed ginseng field (36°8′15.1″N, 127°28′3.0″E) in South Korea. The roots were washed in sterile distilled water to remove bulk soil and then transferred to new sterilized bottles containing 100 mL of 2-(N-morpholino)ethanesulfonic acid (MES) buffer (pH 5.7). The mixture was shaken at 200 rpm for 30 min at 27°C. Subsequently, the roots were carefully removed, and the shaken solution was stored as the rhizosphere sample. The rhizosphere solution was then centrifuged at 500 rpm for 5 min at 27°C, and the resulting supernatant was transferred to a new 50 mL conical tube and centrifuged at 8000 rpm for 15 min. The supernatant was discarded, and the resulting pellet was resuspended in 30 mL of sterile water. Bacteria were isolated using the agar plate method in combination with serial dilutions. Rhizosphere samples were diluted with sterile distilled water at a ratio of 1:10, and each dilution (0.1 mL) was cultured on Luria–Bertani (LB) broth at 30°C for 2 days. Each colony was selected and streaked onto LB agar for purification (Choi et al., 2020). The various bacterial isolates obtained from the samples were designated as “CACC” (Cialm Agricultural Cultures Collection), followed by their respective isolation numbers. The isolated bacterial cells were then grown in LB medium and stored at −80°C.
The isolated bacteria were identified based on 16S rRNA gene sequences. They were cultured in LB broth at 30°C in a shaker at 200 rpm, and the cells were collected by centrifugation at 5,000 rpm for 10 min and 4°C. Genomic DNA was extracted using the ExiPrep™ Plus Bacteria Genomic DNA Kit (BIONEER, Daejeon, South Korea) following the manufacturer’s protocols. The 16S rRNA gene was amplified from the genomic DNA using the universal primers 518F (CCAGCAGCCGCGGTAATAC) and 805R (GACTACCAGGGTATCTAATC), followed by a BLAST search in the National Center for Biotechnology Information (NCBI) database to identify sequence homologies.
2.1.1 Phylogenetic analysis of Bacillus spp.
The full-length 16S rRNA gene sequences from the bacterial strains were compared with the sequences available in the NCBI database.1 The sequences were aligned using Multiple Sequence Comparison by Log-Expectation (Edgar, 2004). A phylogenetic tree was constructed using the neighbor-joining method, and the relative stability of the branches was evaluated by bootstrap analysis of 1,000 datasets using MEGA11 software (Tamura et al., 2021).
2.1.2 Phosphate solubilization
To quantify phosphate solubilization, bacterial strains were inoculated into a National Botanical Research Institute Phosphorus (NBRIP) medium consisting of 10 g glucose, 5 g MgCl2∙6H2O, 0.25 g MgSO4∙7H2O, 0.2 g KCl, 0.1 g (NH4)2SO4, and 5 g tricalcium phosphate as the sole P source in 1 L of distilled water (Olsen et al., 1954). Cultured bacterial strains (0.3 mL) were inoculated into 30 mL of NBRIP medium in Erlenmeyer flasks. Controls were prepared using the NBRIP medium only. After incubation for 7 days at 30°C and 150 rpm, the culture medium was centrifuged to quantify soluble phosphorus using the MARS 6 microwave instrument from CEM (Kamp-Lintfort, Germany). For the digestion, 5 mL of the supernatant was transferred to a 110 mL vessel, and 15 mL of 70% nitric acid solution was added. The mixture was subjected to a temperature–time ramp, reaching a final temperature of 180°C within 15 min, and this temperature was maintained for 20 min to ensure complete acid digestion. Following digestion, the 70% nitric acid was evaporated with heat, and the residue was diluted with 3% nitric acid solution for analysis. The diluted sample was analyzed using inductively coupled plasma mass spectrometry (ICP-MS) on an iCAP-RQ instrument (Thermo Fisher Scientific, Waltham, MA, United States) equipped with an ASX-560 autosampler (TELEDYNE, Englewood, CO, United States). The analysis utilized the 31P isotope for phosphorus quantification. A kinetic energy discrimination mode with helium gas was employed to mitigate interference from 14N16OH+ polyatomic ions (Fleischer et al., 2014).
2.1.3 Nitrogen fixation
Molecular nitrogen fixation was assessed using a nitrogen-free medium (Nfb) and the phenate method to detect NH4-N production (Ahmad et al., 2013). A bacterial colony was inoculated into 25 mL of Nfb and cultured at 30°C for 7 days. The culture medium was centrifuged at 4°C and 8,000 rpm for 20 min to obtain a supernatant. The supernatant was then diluted 10 times into 25 mL portions and added with 1 mL of phenol, 1 mL of sodium nitroprusside, and 2.5 mL of oxidizing solution. The mixed solution was incubated for 1 h, and the absorbance was measured at 640 nm to quantify NH4-N production using an ammonium standard solution (0.1, 0.2, 0.5, and 1 mg N/L) for calibration. Distilled water was used as a blank.
2.1.4 IAA production
The colorimetric determination of IAA was performed as previously described (Meudt and Gaines, 1967). The bacterial strains were cultured in DF medium [2 g (NH4)2SO4, 1 g KH2PO4, 15 g Na2HPO4·7H2O, 0.2 g MgSO4·7H2O, 1 mg FeSO4·7H2O, 10 μg H3BO3,11 μg MnSO4·H2O, 125 μg ZnSO4·7H2O, 78 μg CuSO4·5H2O, and 17 μg Na2MoO4·2H2O per liter] supplemented with 0.5 g tryptophan and then incubated at 30°C and 180 rpm for 5 days. After centrifugation at 5000 rpm for 20 min, 1 mL of the supernatant was mixed with 2 mL of modified Salkowski reagent (50 mL of perchloric acid and 1 mL of 0.05 M FeCl3) and left in the dark at 24°C for 30 min. The optical density (OD) was measured at 530 nm using microplate reader (SPARK, TECAN, Männedorf, Switzerland), and the IAA concentration was calculated from a standard curve prepared using IAA solutions ranging from 0 to 100 ng/mL.
2.1.5 Siderophore production
Siderophore production was assessed using King’s B broth (20 g protease peptone, 10 mL glycerol, 1.5 g KH2PO4, and 1.5 g MgSO4·7H2O in 1 L). The medium was iron-deficient to induce siderophore synthesis. For quantitative analysis, 0.4 mL of the bacterial cultures were inoculated into 40 mL of King’s B broth and incubated at 30°C for 4 days as previously described (Nagarajkumar et al., 2004; Cherif-Silini et al., 2016). After centrifugation at 5000 rpm for 30 min, the supernatant was adjusted to pH 2.9, mixed with an equal volume of ethyl acetate, shaken for 30 min, and then recentrifuged at 5,000 rpm for 30 min. The upper ethyl acetate layer was collected and mixed with Hathway reaction solution (1 mL of 0.1 M FeCl3 in 0.1 N HCl added to 100 mL distilled water plus 1 mL of potassium ferricyanide). After 20 min, the absorbance was measured at 700 nm using microplate reader (SPARK, TECAN). Siderophore production was quantified based on the measurements, and a standard curve was prepared using dihydroxybenzoic acid.
2.1.6 ACC deaminase production
The ACC deaminase activity was evaluated using a previously described method (Misra and Chauhan, 2020), which measures α-ketobutyrate production resulting from ACC cleavage by ACC deaminase. Briefly, to measure ACC deaminase production, 20 μL of 0.5 M ACC was added to 0.2 mL of cell pellets obtained by inoculating the isolated strain into DF salts minimal medium containing 3 mM ACC as the sole nitrogen source [replacing (NH4)2SO4]. The reaction was allowed to proceed for 15 min. After the addition of 0.56 M HCl and 2,4-dinitrophenylhydrazine reagent (0.2% 2,4-dinitrophenylhydrazine in 2 M HCl), the mixture was stirred for 30 min and then added with 2 mL of 2 N NaOH. Absorbance was measured at 540 nm, and ACC deaminase activity was quantified by comparing the absorbance with a standard curve of known α-ketobutyrate concentrations.
2.1.7 Bacterial growth and exopolysaccharide formation
Bacterial growth was evaluated at different water potentials of 0, −0.15, and −0.49 MPa, which were generated using LB liquid medium supplemented with polyethylene glycol (PEG) 6,000 at concentrations of 0, 10, and 20%, respectively. The selected strains were inoculated into the medium at an initial OD of 0.01 and then cultured at 30°C with shaking at 180 rpm for 48 h. Bacterial growth was monitored by measuring the OD600nm using microplate reader (SPARK, TECAN). Growth at 0 MPa was considered 100%, whereas growth at −0.15 and −0.49 MPa was expressed as a percentage relative to that at 0 MPa.
The production of EPS (total biopolymers) at different water potentials (0, −0.15, and −0.49 MPa) was determined in broth cultures of the same strains. After inoculation into nutrient broth supplemented with 5% glucose and incubation at 30°C and 120 rpm for 40 h, the supernatant was collected by centrifugation at 12,000 rpm for 20 min at 4°C. Acetone was added to the supernatant in a ratio of 1:2 (v:v), and the mixture was allowed at 4°C for 24 h to precipitate the EPS. The precipitate was filtered through filter paper and dried at 24°C, and the weight of the filter paper was measured. The blank was measured using untreated filter paper for value calculation. Each treatment was performed in triplicate in two biological experiments, including control samples inoculated with B. megaterium ATCC14581.
2.1.8 Whole genome sequencing, gene prediction, and annotation
The genomes of CACC109 and CACC119 were assembled de novo from PacBio sequencing data by CJ Bioscience, Inc. (South Korea). Contigs were assembled and circularized using SMRT Link (Pacific Biosciences, Menlo Park, CA, United States) and Circlator 1.4.0 (Sanger Institute, Essex, United Kingdom). Gene prediction and functional annotation were performed using the EzBioCloud genome database, Prodigal 2.6.2 (CDSs) (Hyatt et al., 2010), tRNAscan-SE 1.3.1 (tRNA) (Schattner et al., 2005), and Rfam 12.0, (rRNA and other non-coding RNAs) (Nawrocki et al., 2015). CRISPR detection was conducted using PilerCR 1.06 (Edgar, 2007) and CRT 1.2 (Bland et al., 2007). Functional classification was performed using EggNOG 4.5.2 Additional annotation was achieved by comparing the predicted CDSs with the Swissprot (The UniProt Consortium, 2015), KEGG (Kanehisa et al., 2014), and SEED databases (Overbeek, 2005) using the UBLAST program (Edgar, 2010).
2.2 Effect of B. megaterium CACC109 and CACC119 on drought tolerance
2.2.1 Effects of bacterial strains on rice germination and growth under osmotic-stressed and non-stressed conditions
Germination and seedling growth assays were performed on rice seeds to evaluate the efficacy of the isolated bacterial strains in improving osmotic stress tolerance. Saechungmu rice (Oryza sativa L.) seeds were surface sterilized with 2% sodium hypochlorite and 70% ethanol and then washed multiple times with distilled water. The seeds were filled to the 20 mL mark in a 50 mL conical tube and either 1 × 107 CFU (colony-forming unit)/mL of bacterial suspension or 0.85% saline (mock) was added up to the 40 mL mark, ensuring the seeds were completely submerged. The seeds were then gently shaken for 8 h. Twenty seeds from each treatment were evenly distributed among three Petri dishes each containing Murashige & Skoog (MS) medium (DUCHEFA BIOCHEMIE, The Netherlands) with different water potentials (0, −0.15, and −0.49 MPa) achieved with PEG 6000. Germination was carried out at 27°C in the dark for 5 days, and then the germination rate, root length, and growth parameters were measured. The germination rate was calculated by dividing the number of germinated seeds by the total number of seeds and expressed as a ratio (Islam et al., 2016).
A similar assay was conducted using an iso-osmotic solution of mannitol to assess the effect of the isolated bacterial strains on osmotic stress tolerance. Seeds treated with bacteria or mock solution were germinated in a dark box at 27°C for 3 days. Uniformly germinated seeds were then transferred to MS agar supplemented with 0, 150, and 200 mM mannitol and grown under a 12-h light–dark cycle at 27°C for 3 days. Seedling root length was measured after 3 days. Each assay consisted of 10 seeds arranged in Petri dishes, with three replicates per treatment, and securely sealed with Parafilm.
2.2.2 Experimental design for plant growth and drought study
To study the effects of bacterial strains on rice growth and various physiological, molecular, and biochemical responses under drought and non-drought conditions, Saechungmu rice seeds were germinated at 27°C in the dark for 3 days. Uniformly germinated seedlings, 5 or 30, were selected and planted in regular or Wagner pots, respectively, filled with autoclaved potting soil for rice plants (Punong, Gyeongbuk, South Korea). At 7 days post-planting, the plants were treated weekly for 4 weeks with either 5 mL of 1 × 107 CFU/mL B. megaterium CACC109, 1 × 107 CFU/mL B. megaterium CACC119, or 0.85% saline (mock) per plant. A 1,000-fold dilution of Hyponex fertilizer was applied to the plants at 10 days post-planting. The plants were maintained in a growth chamber with a constant temperature of 27°C, a 12-h light–dark cycle, and approximately 70% relative humidity. After 6 weeks of growth, watering was halted for half of the mock-, CACC109-, and CACC119-treated rice plants, while the remaining plants continued to grow under well-watered conditions, serving as the control group. Soil moisture content was monitored daily using a moisture analyzer MA37 (Sartorius, Göttingen, Germany) to ensure a consistent water retention rate of 20–40% throughout the drought.
Chlorophyll content was assessed by measuring 20 leaves from mock-, CACC109-, and CACC119-treated independent plants in both the well-watered and water-stressed groups using a SPAD-502 meter (Konica Minolta, Tokyo, Japan). The height, weight, and water retention of 20 plants from each treatment group were assessed 15 or 37 days after inducing drought in the well-watered and water-stressed groups. Each plant, including its roots, was weighed to determine the fresh weight, followed by drying at 65°C for 3 days and subsequent weighing to obtain the dry weight. Water retention for each plant was calculated as the difference between the fresh and dry weights.
For gene expression analysis, leaf samples were collected from the mock-, CACC109-, and CACC119-treated plants grown under well-watered and water-stressed conditions at three time intervals (early, 10 days; middle, 20 days; and late, 35 days post-drought). For each treatment group, four biological replicates were sampled, with each biological replicate being a pool of three leaf tissues from independent plants. The collected samples were immediately flash-frozen in liquid nitrogen and stored at −80°C.
2.2.3 Estimation of proline content
Total proline accumulation in the leaf samples was quantified as previously described (Bates et al., 1973). Briefly, 150 mg of leaf samples were ground with liquid nitrogen and homogenized in 2 mL of 3% sulfosalicylic acid. After centrifugation at 12,000 rpm for 10 min, the supernatant was transferred to a new tube. Then, 0.5 mL of the supernatant was mixed with 0.5 mL of ninhydrin and 0.5 mL of glacial acetic acid in a 15 mL conical tube. The mixture was boiled for 45 min and immediately cooled on ice for 30 min. The samples were mixed with 1.5 mL of toluene and centrifuged at 700 rpm for 5 min to separate the total proline. Total proline content was measured at 520 nm against a blank using a standard curve prepared with proline.
2.2.4 Total RNA extraction and gene expression analysis
Total RNA from leaf tissue was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, United States). The RNA was treated with DNase I (Promega, Madison, WI, United States), cleaned using UltraPure™ Phenol:Chloroform:Isoamyl Alcohol (25, 24:1, v/v) (Thermo Fisher Scientific), and checked for quality and quantity using NANODROP ONE (Thermo Fisher Scientific). For each line, cDNA was synthesized from 2 μg of total RNA using M-MLV reverse transcriptase (Promega). Quantitative PCR analyses were conducted on a CFX96 real-time PCR system (Bio-Rad Laboratories, Hercules, CA, United States) using SYBR Green Supermix (containing hot-start iTaq DNA polymerase, dNTPs, MgCl2, SYBR® Green I dye, enhancers, and stabilizers from Bio-Rad Laboratories) and gene-specific primers. Samples were prepared in triplicate, with a total reaction volume of 20 μL containing the following components: 2 μL of cDNA, 1 μL of each primer (forward and reverse, at 10 pM each), 10 μL of SYBR Green Supermix, and 6 μL of double-distilled water. Amplification reactions were initiated with a denaturation step at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, 60°C for 10 s, and 72°C for 20 s. Melting curves were recorded over a temperature range of 65°C to 95°C in 0.5°C increments. Rice Actin served as an endogenous control for normalization. Expression levels were determined using the comparative cycle threshold (Ct) method. Details of the quantitative reverse transcription polymerase chain reaction (qRT-PCR) primers used are listed in Supplementary Table S1.
2.2.5 Analysis of chemical properties of soil
Soil samples (200 g each) were collected from each pot after the drought stress experiment. The samples were air-dried at room temperature, crushed to pass through a 2 mm mesh sieve, and thoroughly mixed. The pH was measured in distilled water at a ratio of 1:5 (soil sample: distilled water) using a glass electrode pH meter. Exchangeable cations from the soil were extracted with 0.05 N NH4OAc (pH 7.0) and determined using iCAP™ RQplus ICP-MS (Thermo Fisher Scientific). Available phosphoric acid was extracted following the Lancaster method and determined using iCAP™ RQplus ICP-MS (Thermo Fisher Scientific).
2.3 Statistical analysis
Statistical analyses were performed using the JMP Pro 16 software package. Two-way analyses of variance (ANOVA) with Tukey’s honest significant difference (HSD) test were used to assess the statistical significance of the bacterial growth, EPS production, seed germination, plant height, root length, chlorophyll content, proline production, and gene expression assay. Students’ t-test was used to compare significant differences in plant growth-promoting traits (phosphate solubility, nitrogen fixation, IAA productivity, siderophore productivity, and ACC deaminase activity), plant weight, water retention, survival rate under salinity stress, and soluble organic components in soil. The significance level was determined at a p < 0.05.
3 Results
3.1 PGPR isolated and screened from the ginseng rhizosphere
Five ginseng rhizosphere soil samples were collected from ginseng cultivation fields in South Korea to isolate PGPR with osmotic stress tolerance. A total of 31 isolates were obtained from the samples and identified based on their 16S rRNA gene sequences (Supplementary Table S2). Subsequently, the isolates were screened for osmotic stress tolerance in LB medium supplemented with PEG 6000. Two isolates exhibited better growth than the B. megaterium type strain ATCC14581 under osmotic stress (Figure 1A), indicating their efficacy as stress-tolerant isolates. Therefore, these strains were selected for further studies.
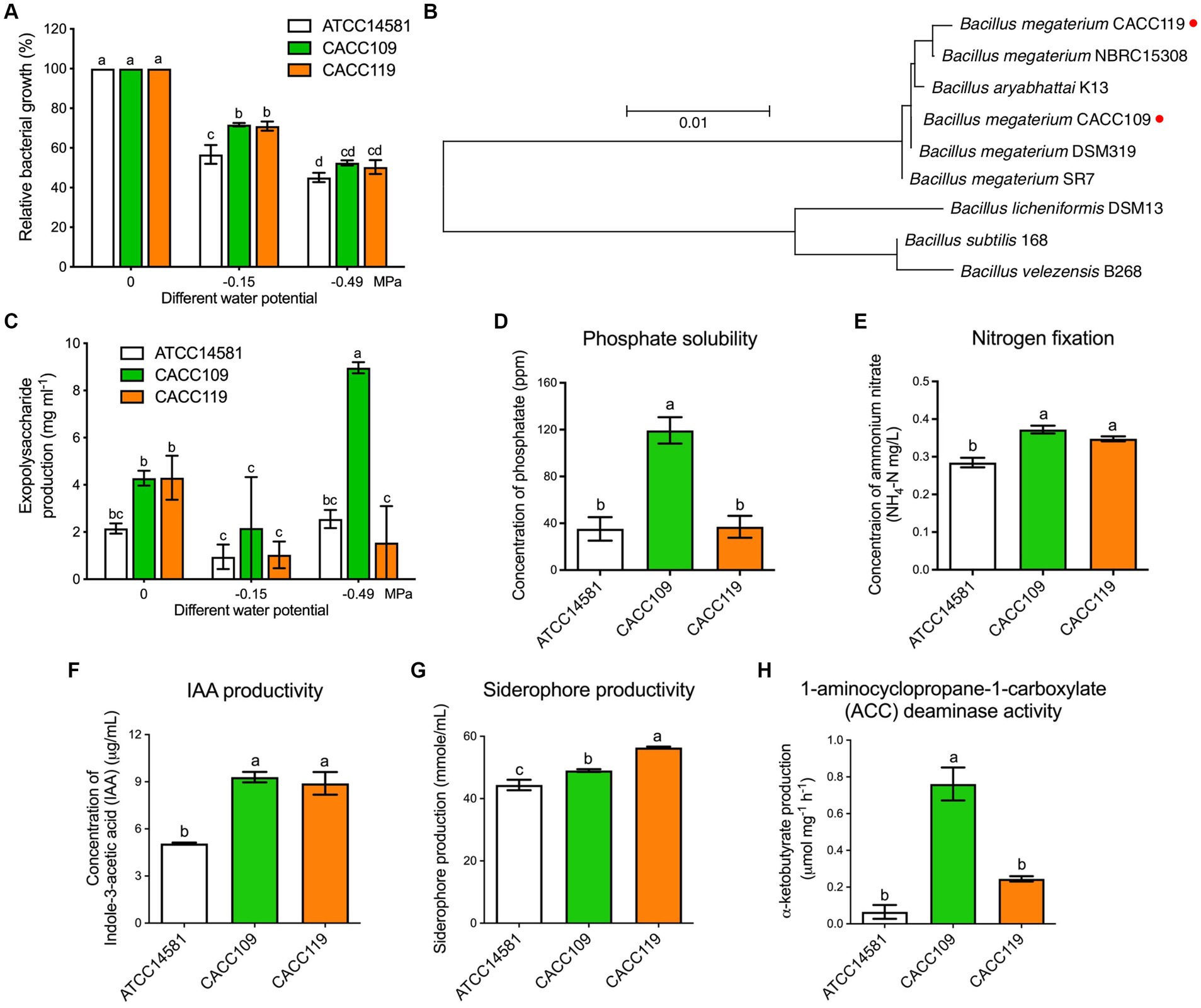
Figure 1. Isolation, characterization, and plant growth-promoting properties of water stress-tolerant Bacillus megaterium strains CACC109 and CACC119. (A) Relative growth rates of B. megaterium strains CACC109 and CACC119 in vitro at different water potentials. Growth was measured 24 h after incubation. Growth at 0 MPa was converted to 100%, and the growth values at −0.15 and −0.49 MPa were expressed as a percentage relative to that at 0 MPa. Data are expressed as the mean ± SD of three biological replicates. Different letters indicate significant differences among the bacterial strains in the various water potentials (p < 0.05, two-way ANOVA and Tukey’s test). B. megaterium ATCC14581 was used as a control. (B) Evolutionary relationships of CACC109 and CACC119 with other known Bacillus strains. Sequences were aligned through “Clustal W” using MEGA 11. The maximum-likelihood method with the Tamura-Nei model was used to construct the phylogenetic tree based on 16S rRNA gene nucleotide sequences. Bootstrapped consensus tree derived from 1,000 replicates. Evolutionary analysis was performed in MEGA 11. Red dots indicate the positions of CACC109 and CACC119. (C) EPS production of CACC109 and CACC119 in vitro at different water potentials 40 h after inoculation. Bars represent the mean ± SE of three biological replicates. Different letters indicate significant differences among the bacterial strains in the various water potentials (p < 0.05, two-way ANOVA and Tukey’s test). (D) Phosphate solubilization, (E) nitrogen fixation, (F) indole-3-acetic acid (IAA) production, (G) siderophore production, and (H) 1-aminocyclopropane-a-carboxylate (ACC) deaminase activity. Data in (D–H) are expressed as mean ± SD of three biological replicates. Different letters indicate significant differences (p < 0.05, Student’s t-test). B. megaterium ATCC14581 type strain was used as a control. B. megaterium ATCC14581 was used as a control. Accession numbers: Bacillus megaterium CACC119 (OR915158), Bacillus megaterium NBRC15308 (CP009920), Bacillus aryabhattai K13 (CP024035), Bacillus megaterium CACC109 (CP117689), Bacillus megaterium DSM319 (CP001982), Bacillus megaterium SR7 (CP022674), Bacillus licheniformis DSM13 (AE017333), Bacillus subtilis 168 (CP053102), Bacillus velezensis B268 (CP053764).
Identification of these two isolates by 16S rRNA gene sequencing revealed that they belonged to B. megaterium and thus were designated as B. megaterium CACC109 and CACC119. The obtained sequences of CACC109 and CACC119 were submitted to NCBI GenBank under accession numbers CP117689 and OR915158, respectively. A phylogenetic analysis was conducted using 16S rRNA gene sequences. The sequences of CACC109 and CACC119, along with their closest relatives from NCBI GenBank, were used to construct a phylogenetic tree with the maximum-likelihood method. The analysis showed that CACC109 was most closely related to B. megaterium DSM319, whereas CACC119 was most closely related to B. megaterium NBRC15308 (Figure 1B).
The EPS production of the bacterial strains subjected to different levels of osmotic stress induced by PEG6000 was quantified. Under non-osmotic stress, CACC109 and CACC119 produced 4.28 and 4.3 mg mL−1 EPS, respectively, which were significantly higher than the 2.15 mg mL−1 EPS produced by ATCC14581 (Figure 1C). However, EPS production decreased under −0.15 MPa osmotic stress. Remarkably, CACC109 demonstrated the highest EPS production, reaching 8.96 mg mL−1 at −0.49 MPa, whereas CACC119 exhibited substantially lower levels than CACC109 and similar levels to ATCC14581 (Figure 1C).
Analysis of the PGP properties of osmotic-tolerant strains CACC109 and CACC119 revealed that CACC109 had a significant ability to solubilize phosphate, with a maximum soluble P2O5 concentration of 119 μg/mL, whereas CACC119 did not (Figure 1D). In terms of nitrogen fixation, CACC109 and CACC119 produced 0.372 and 0.347 μg/mL NH4, respectively (Figure 1E). In terms of IAA production, CACC109 and CACC119 produced significantly higher concentrations of indole compounds than ATCC14581 (Figure 1F). Although both strains tested positive for siderophore production, CACC119 exhibited significantly greater production of siderophores than CACC109 (Figure 1G). Meanwhile, CACC109 exhibited significant ACC deaminase activity, whereas CACC119 did not (Figure 1H).
3.2 Effects of CACC109 and CACC119 on seed germination and root growth
The effects of CACC109 and CACC119 on the germination and root growth of rice seeds were examined under osmotic-stressed and non-stressed conditions. Under non-stressed conditions (0 MPa), the CACC109-, CACC119-, ATCC14581-, and mock-treated seeds showed identical germination ratios, with the maximum germination observed on day three (Figure 2A). At −0.15 MPa, no significant differences in germination rates were observed among the treatment groups (Figure 2A). However, the germination rates of the CACC109- and CACC119-treated seeds were significantly higher than those of the mock- and ATCC14581-treated lines at −0.49 MPa, indicating that both PGPR strains supported seed germination in rice under osmotic stress challenge (Figure 2A). In addition, the CACC109- and CACC119-treated seeds had significantly greater root growth than the mock- and ATCC14581-treated seeds under osmotic-stressed and non-stressed conditions (Figure 2B).
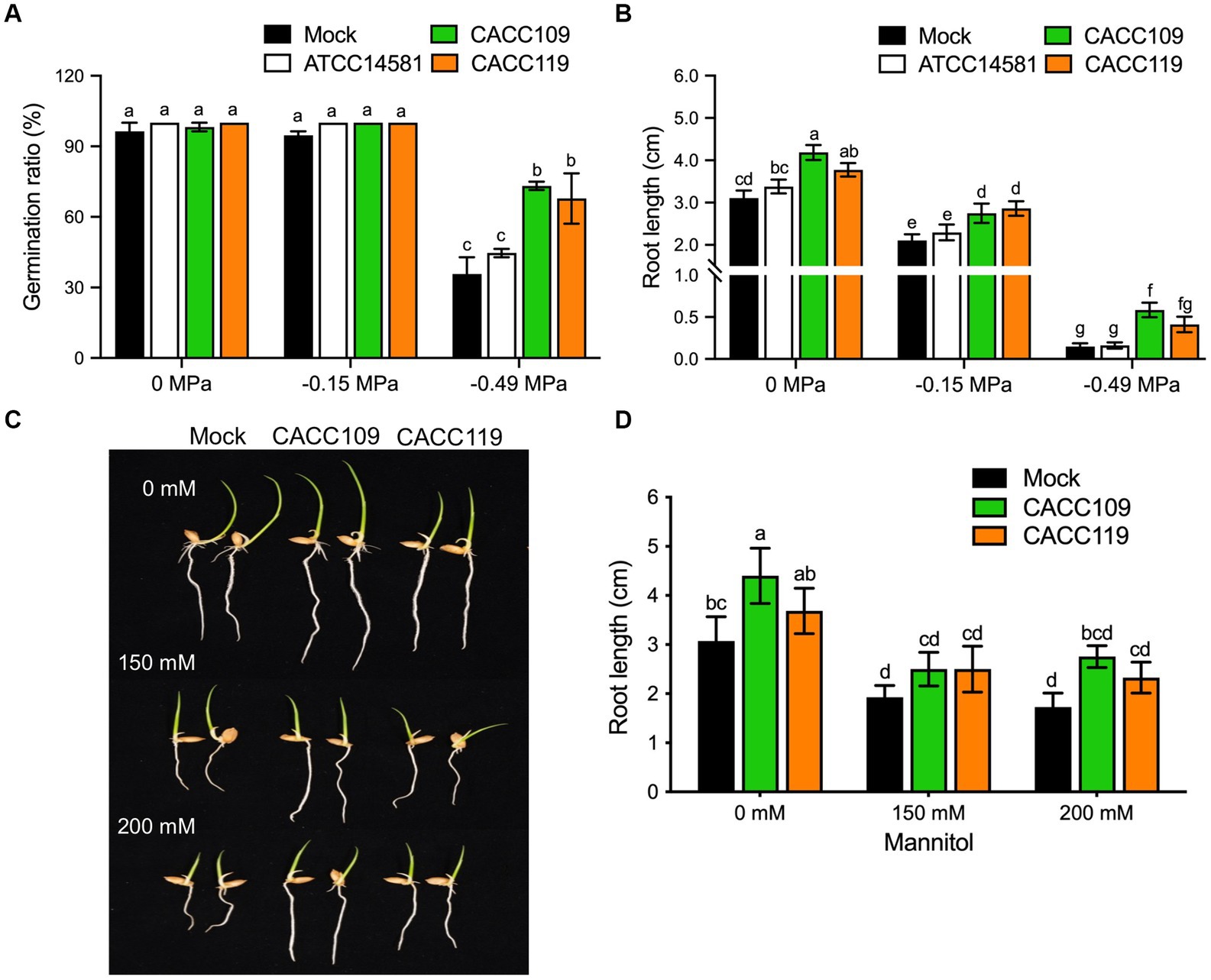
Figure 2. Effects of CACC109 and CACC119 on the osmotic stress resistance in rice seedlings. (A) Germination rate and (B) root length in rice seeds treated with mock, CACC109, or CACC119 at different water potentials. (C) Root growth of rice seedlings treated with mock, CACC109, or CACC119 grown in Murashige & Skoog (MS) media containing 0, 150, and 200 mM mannitol for 3 days. (D) Root length in (C) (n > 30). In (A,B), seeds were soaked in mock, CACC109, or CACC119 for 8 h and incubated at 30°C for 5 days under different water potentials created with PEG6000. In (C,D), for the mannitol assays, rice seeds were soaked in mock, CACC109, or CACC119 for 10 h, germinated in the magenta box, and similar-sized 3-day-old seedlings were transferred to MS media containing 0, 150, and 200 mM mannitol. Images and measurements were taken 3 days after growth on the MS medium. Assays were performed using 1 × 107 CFU/mL of CACC109 and CACC119. Data represent the mean ± SE of three independent experiments. Different letters indicate significant differences among the treatments in the various osmotic stresses (p < 0.05, two-way ANOVA and Tukey’s test).
To validate the effects of CACC109 and CACC119 on rice root growth, we germinated rice seeds and selected evenly germinated seedlings. The seedlings were then exposed to different concentrations of mannitol to induce osmotic stress (Claeys et al., 2014). The root lengths of the mock-, CACC109-, and CACC119-treated seedlings gradually decreased with increasing osmotic stress. This effect is similar to the observed effects of PEG-induced osmotic stress. In the absence of osmotic stress, the CACC109- and CACC119-treated seedlings had significantly longer roots than the mock-treated seedlings (Figures 2C,D). When exposed to 150 or 200 mM mannitol, the CACC109- and CACC119-treated seedlings had significantly longer roots than the mock-treated seedlings, indicating that both CACC strains promoted root growth of the rice seedlings under normal and drought conditions (Figures 2C,D). Collectively, these data indicated that CACC109 and CACC119 enhanced osmotic stress tolerance, promoted seed germination, and facilitated root development in rice seedlings.
3.3 Effects of CACC109 and CACC119 on plant growth and drought resistance
We investigated the effects of PGPR CACC109 and CACC119 on rice growth and drought stress resilience. Under well-watered conditions, the CACC109- and CACC119-treated rice plants had significantly greater shoot and root growth than the mock-treated plants (Figures 3A–D). Treatment with CACC109 and CACC119 notably increased the rice leaf width (Figures 3E,F). The CACC109- and CACC119-treated plants exhibited 64 and 40% higher shoot weight, respectively, than the mock-treated plants (Figure 3G). Compared with the mock treatment, CACC109 considerably increased root biomass due to axial and lateral root development, whereas CACC119 had a less pronounced effect (Supplementary Figure S1). These results indicated that CACC application significantly promoted plant growth in rice.
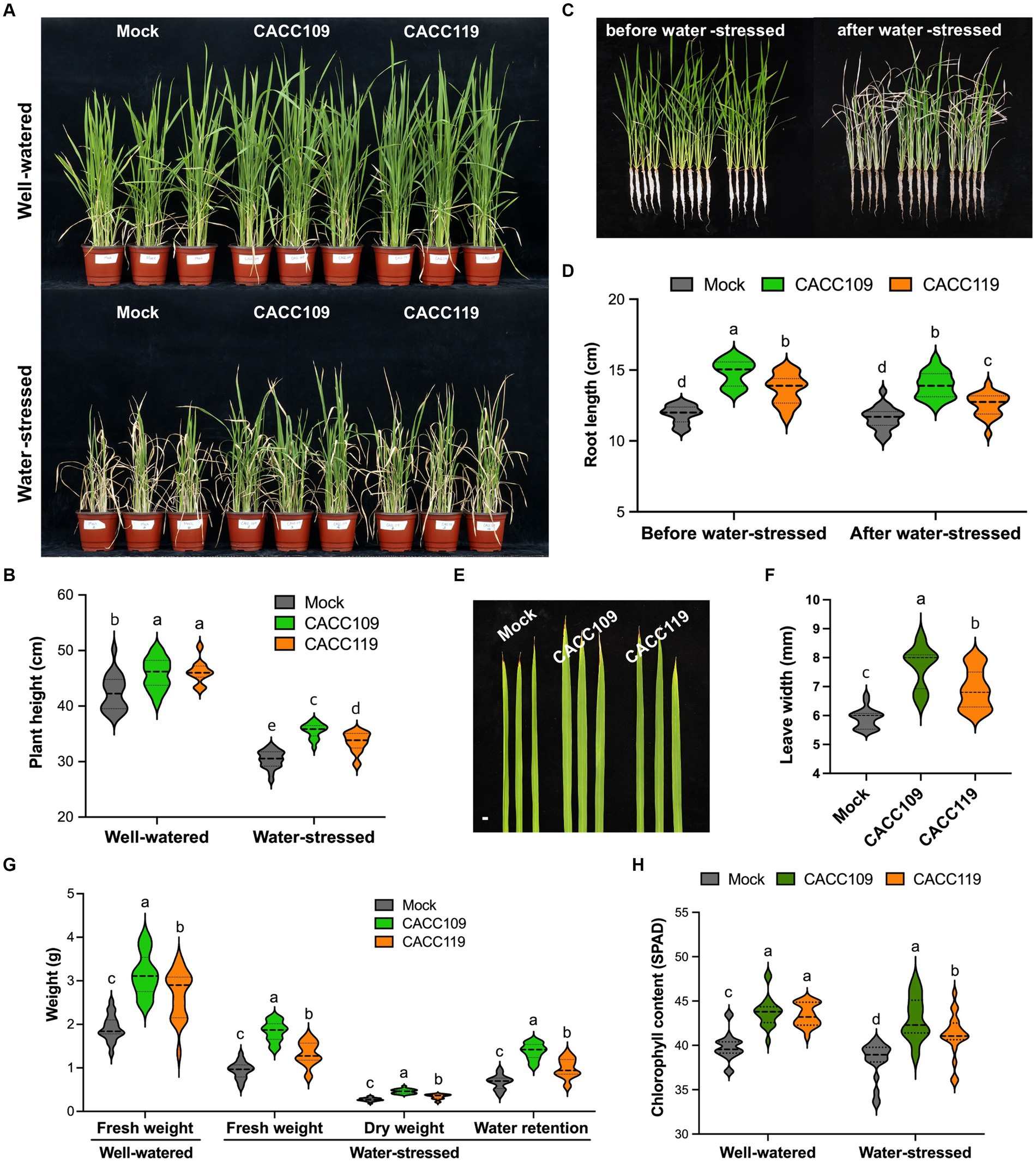
Figure 3. Growth promotion and drought resilience in rice by CACC109 and CACC119. (A) Rice plants treated with mock, CACC109, or CACC119 under well-watered and water-stressed conditions. The 6-week-old mock-, CACC109-, or CACC119-treated plants (25 plants for each line) were subjected to drought with 20–40% soil water potential for 15 days. (B) Height of rice plants treated with mock, CACC109, or CACC119 under well-watered and water-stressed conditions. (C) Aboveground shoot and root lengths of rice plants treated with mock, CACC109, or CACC119 before water stress (6 weeks old) or after water stress (15 days under drought). (D) Root length in (C). (E) Flag leaf morphology of 7-week-old rice plants treated with mock, CACC109, or CACC119 under well-watered conditions, bar = 5 mm. (F) Leaf width in (E). (G) Fresh weight of mock-, CACC109-, or CACC119-treated rice plants grown under well-watered conditions for 8 weeks. Fresh weight, dry weight, and water retention (fresh weight – dry weight) of mock-, CACC109-, or CACC119-treated rice plants after 15 days of drought. (H) Chlorophyll content of mock-, CACC109-, or CACC119-treated rice plants grown under well-watered or water-stressed conditions. In (B–H), values are means ± SD (n = 20, individual plants). Different letters indicate significant differences (p < 0.05, two-way ANOVA and Tukey’s test in B,D,H; Student’s t-test in F,G). Control plants (mock) were treated with the same amount of saline solution as a negative control. Chlorophyll and height were measured on mature leaves. The results presented here are representative of three independent experiments.
After 15 days of water stress, all mock-treated rice plants displayed typical severe dehydration symptoms with significant wilting, and only a few leaves remained green. By contrast, the leaves of most CACC109-treated plants appeared less dehydrated and remained green. The CACC119-treated plants exhibited an intermediate drought stress phenotype relative to the mock- and CACC109-treated plants (Figure 3A). Additionally, water stress caused growth retardation in the mock- and PGPR-treated plants, although this effect was more pronounced in the mock-treated plants (Figures 3A–D). Under drought, the CACC109- and CACC119-treated plants had greater shoot weights than the mock-treated plants (Figure 3G). Under water-stressed conditions, the CACC109-treated plants showed significantly higher shoot dry weight than the mock-inoculated plants. However, no significant difference in shoot dry weight was found between the CACC119- and mock-treated plants (Figure 3G). Drought is typically characterized by decreased water and chlorophyll contents (Bhandari et al., 2023). The CACC109- and CACC119-treated plants had higher water retention than the mock-treated plants (Figure 3G). After 10 days of drought, the chlorophyll content in the mock-treated plants decreased by 7%, whereas those in the CACC109- and CACC119-treated plants decreased by only 3 and 3.5%, respectively, compared with their respective controls (Figure 3H). These data were consistent with the phenotypes of rice plants under drought in each treatment group. Taken together, these results indicated that CACC109 and CACC119 significantly improved the growth and the drought tolerance of rice plants.
3.4 Evaluation of CACC109 and CACC119 on drought tolerance in rice
Additional pot experiments were conducted to evaluate the effects of PGPR CACC109 and CACC119 on the drought tolerance in rice plants. These studies were conducted using a large number of rice plants grown in Wagner pots to mimic conditions in rice fields. We supplied CACC109 and CACC119 in the same manner as in the small-pot test and carefully maintained water levels throughout the experiment to simulate rice field conditions. To investigate the interaction between plants and PGPR, we collected leaf tissues from the CACC109-, CACC119-, and mock-treated plants under non-stressed and water-stressed conditions at different stages of drought (early, 10 days; middle, 20 days; and late, 35 days) to examine physiological traits and gene expression.
After 37 days of water deprivation, all mock-treated plants exhibited severe drought symptoms, characterized by significant wilting and a few leaves remaining green (Figure 4A), whereas the CACC109-treated plants appeared less dehydrated and remained green. The CACC119-treated plants exhibited an intermediate drought stress phenotype compared with the mock- and CACC109-treated plants (Figure 4A). Similar results were observed in these experiments, showing that the application of CACC109 and CACC119 increased the total chlorophyll content, fresh weight, and water retention in the rice plants under drought (Figures 4B,C). The weights of the drought-stressed plants were categorized (Figure 4D). In the mock treatment group, 69.27% of the plants weighed less than 0.75 g, whereas 26.13% weighed within 0.75–0.99 g, with only 4.6% weighing over 1.0 g. By contrast, 26.53% of the CACC109-treated plants weighed within 0.5–0.749 g, 26.64% within 0.75–0.99 g, and 46.83% exceeded 1 g. Meanwhile, 41.42% of the CACC119-treated plants weighed less than 0.749 g, 49.29% weighed within 0.75–0.99 g, and 9.29% exceeded 1 g (Figure 4D). Overall, these data confirmed that CACC109 and CACC119 enhanced the drought tolerance in rice plants.
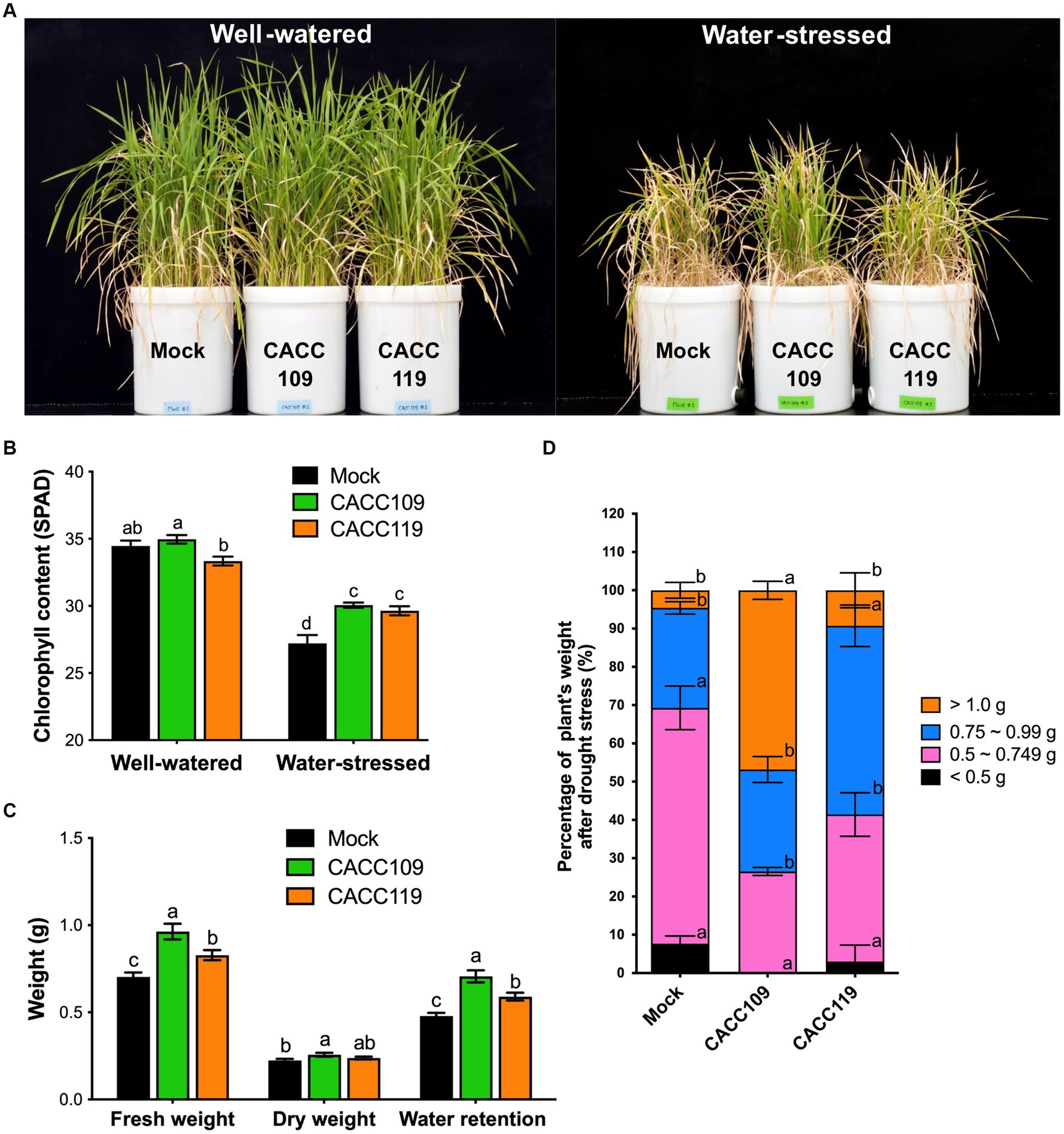
Figure 4. Enhanced drought tolerance in rice plants treated with CACC109 and CACC119. (A) Representative images of rice plants treated with mock, CACC109, or CACC119 under both well-watered and water-stressed conditions. (B) The chlorophyll content of rice plants grown under well-watered or water-stressed conditions. (C) Fresh weight, dry weight, and water retention (fresh weight-dry weight) of rice plants under drought. (D) Individual fresh weights of rice under water-stressed conditions. In (B–D), values are means ± SD (n = 30, individual plants). Different letters represent significant differences (p < 0.05, two-way ANOVA and Tukey’s test in B; Student’s t-test in C,D). Control plants (mock) were treated with equal volumes of saline solution as negative control. The soil water content in the water-stressed group throughout the drought stress period was determined using a moisture analyzer MA37 and calculated as a percentage of field capacity. The experiment was performed two times.
3.5 Enhanced proline accumulation and gene expression by CACC109 and CACC119 under drought stress
Plants under drought and high salinity often accumulate proline to enhance stress adaptation (Hayat et al., 2012). We evaluated the proline levels in the leaf tissues from the CACC109-, CACC119-, and mock-treated plants under well-watered and water-stressed conditions. No significant difference in proline content was found between the PGPR- and mock-treated plants under normal growth conditions (Figures 5A,B). All plants subjected to water stress displayed significantly higher proline concentrations than the well-watered control group (Figures 5A,B), indicating that the drought treatment was successful. Interestingly, at the early stage of water stress, the CACC109- and CACC119-treated plants had significantly higher leaf tissue proline concentrations than the mock-treated plants (Figure 5A). As the water stress continued, all plants accumulated more proline, but the mock- and CACC119-treated plants showed significantly higher proline accumulation than the CACC109-treated plants (Figure 5B). This result indicated that the mock- and CACC119-treated plants experienced relatively stronger water stress intensity than the CACC109-treated plants.
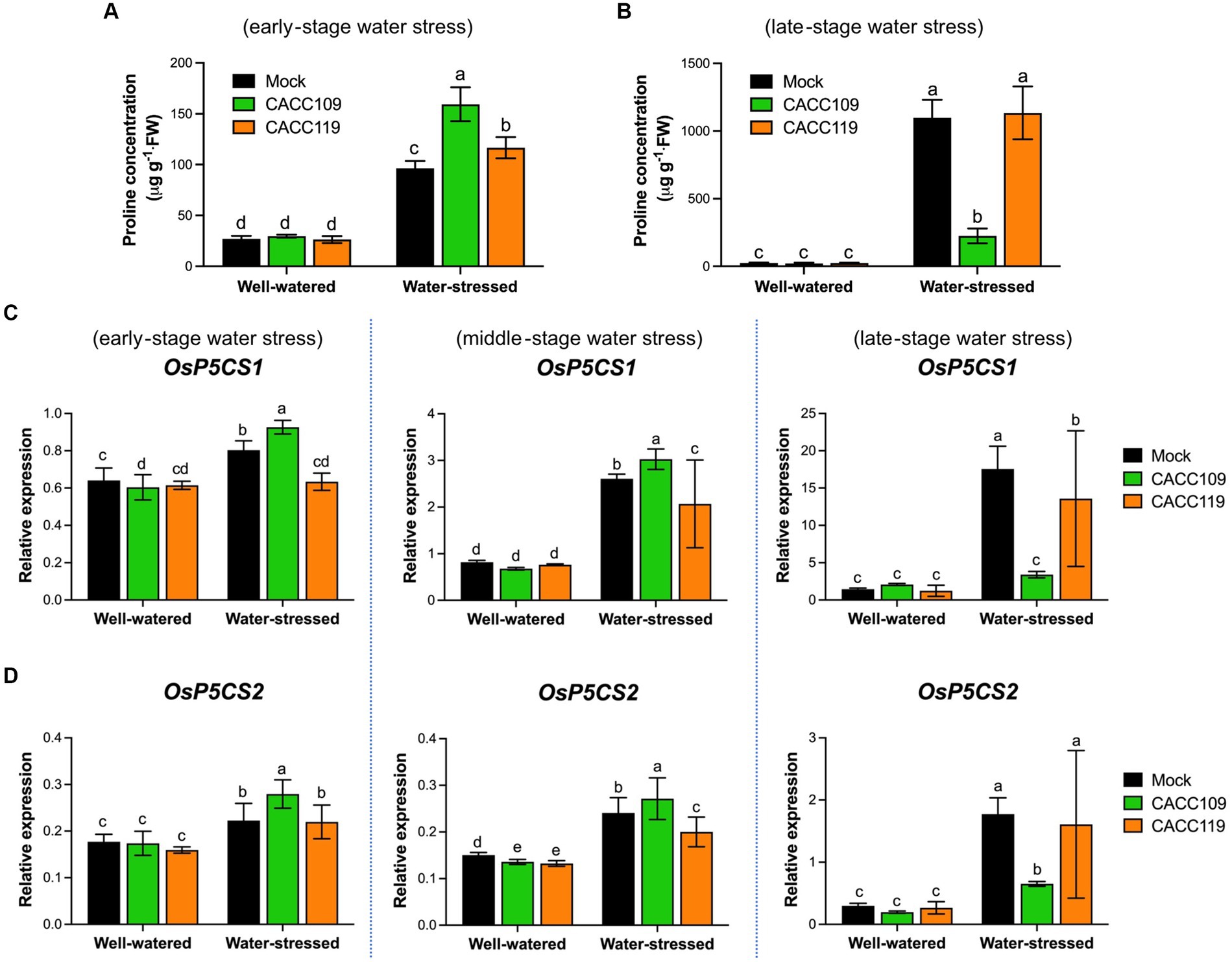
Figure 5. Effects of CACC109 and CACC119 on drought-induced proline production. (A,B) Proline content measured in the leaves of mock-, CACC109-, and CACC119-treated plants with or without drought stress in accordance with the method described by (Lugtenberg and Kamilova, 2009). Proline levels were quantified with four biological replicates at (A) 10 days and (B) 35 days post-drought. (C,D) Expression levels of pyrroline-5-carboxylate synthetase 1 (P5CS1) and P5CS2 in rice leaves grown with mock, CACC109, or CACC119 under well-watered and water-stressed conditions. P5CS catalyzes the first step of proline biosynthesis from glutamate. Six-week-old plants were subjected to drought by maintaining soil water potential at 20–40% for 37 days. Leaf tissues were collected from three independent plants treated with mock, CACC109, or CACC119 in each of the two pots under well-watered and water-stressed conditions at 10 (early), 20 (middle), and 35 (late) days after drought treatment. The expression levels of OsP5CS1 and OsP5CS2 were estimated by quantitative real-time PCR, and OsActin was used as an internal control for normalization. Values are mean ± SD of two independent experiments (p < 0.05, two-way ANOVA and Tukey’s test).
We also investigated the time-course expression patterns of the proline biosynthesis genes Delta 1-Pyrroline-5-carboxylate synthetase 1 and 2 (P5CS1 and P5CS2) in the CACC109-, CACC119-, and mock-treated plants grown under well-watered and water-stressed conditions. As expected, no significant differences in the transcript levels of P5CS1 and P5CS2 were found among the treatment groups under well-watered conditions (Figures 5C,D). The expression of P5CS1 and P5CS2 was upregulated by water stress in all treated rice plants. Specifically, the expression levels of P5CS1 and P5CS2 were higher in the CACC109-treated plants than in the mock- and CACC119-treated plants at the early and middle stages of water stress but lower at the late stage of water stress (Figures 5C,D). The observed trends in the transcript levels of P5CS1 and P5CS2 were consistent with proline accumulation. These data suggested that rice produced proline in response to water stress and that inoculation with the PGPR further improved stress tolerance in rice plants by enhancing proline accumulation.
3.6 Effects of CACC109 and CACC119 on the expression of genes responsible for eliminating reactive oxygen species
Under drought, reactive oxygen species (ROS) trigger defense mechanisms associated with ABA and Ca2+ flux (Mahmood et al., 2019). However, excessive ROS accumulation leads to cell death through oxidative damage, emphasizing the necessity of ROS-detoxifying systems for plant resilience to drought.
We analyzed the time-course expression of antioxidant-related genes, such as catalase (CAT), superoxide dismutase (SOD), peroxidase (POD), ascorbate peroxidase (APX), and glutathione peroxidase (GPX), in the mock-, CACC109-, and CACC119-treated plants under well-watered and water-stressed conditions. Under non-stressed conditions, no significant differences in the expression of antioxidant-related genes were found among the treated plants (Figure 6). However, under water stress, the expression levels of these genes were significantly higher in the mock-, CACC109-, and CACC119-treated plants than in their respective non-stressed controls (Figure 6). Specifically, during drought, the levels of OsCAT and OsPOD were significantly higher in the CACC109- and CACC119-treated plants than in the mock-treated controls (Figures 6A,B). At the early stage of water stress, OsAPX expression was higher in the mock- and CACC109-treated plants than in the CACC119-treated plants. As the stress continued, OsAPX expression became higher in the CACC109-treated plants than in the mock- and CACC119-treated plants. However, in the late stages of stress, OsAPX expression was notably higher in the mock- and CACC119-treated plants than in the CACC109-treated plants (Figure 6C). The expression levels of OsSOD and OsGPX were higher in the CACC109-treated plants than in the mock- and CACC119-treated plants at the early and middle stages but lower at the late stage of drought (Figures 6D,E).
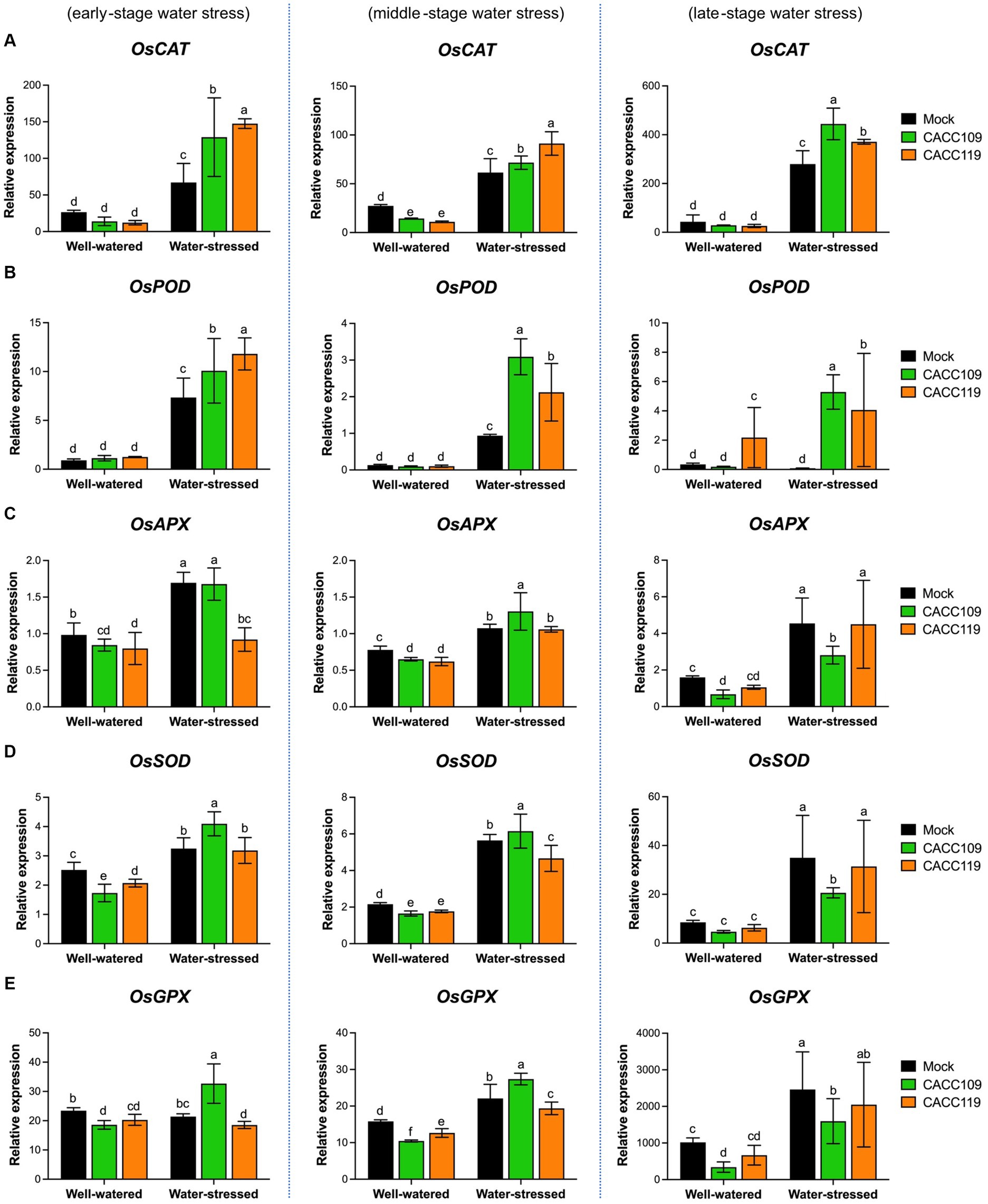
Figure 6. Effects of CACC109 and CACC119 on the expression of antioxidant-related genes in rice leaves. Expression of (A) OsCAT (catalase), (B) OsPOD (peroxidase), (C) OsAPX (ascorbate peroxidase), (D) OsSOD (superoxide dismutase), and (E) OsGPX (glutathione peroxidase) in mock-, CACC109-, and CACC119-treated plants grown under well-watered or water-stressed conditions. Six-week-old plants were subjected to drought stress by maintaining soil water potential at 20–40% for 37 days. Leaf tissue was collected from three independent plants treated with mock, CACC109, or CACC119 in each of the two pots under well-watered and water-stressed conditions at 10 (early), 20 (middle), and 35 (late) days after drought treatment. Data were normalized by the comparative cycle threshold method with OsActin as the internal control and presented as relative expression. Values are mean ± SD of two independent experiments. Different letters indicate significant differences (p < 0.05, two-way ANOVA and Tukey’s test).
Antioxidant activity was significantly higher in the PGPR-treated plants under water stress than in those under control conditions. Additionally, the expression of antioxidant-related genes was modulated below a certain threshold because of the enhanced stress tolerance mediated by the PGPR.
3.7 Effects of CACC109 and CACC119 on the expression of DRGs
The above results suggest that CACC109 and CACC119 positively affect plant responses to drought. Drought triggers a significant reprogramming of gene expression (Shinozaki and Yamaguchi-Shinozaki, 2006). To gain insights into the molecular mechanisms underlying PGPR-induced drought tolerance, we analyzed the expression of ABA-dependent [e.g., OsbZIP23 (basic region/leucine zipper motif), OsNAC066 (NAM, ATAF1/2, and CUC2), OsAREBs (ABA-responsive element binding protein)] and ABA-independent DRGs [e.g., OsWRKY47, OsDREB2s (dehydration-responsive element binding protein), and OsERD1 (early response to dehydration1)] in the mock-, CACC109-, and CACC119-treated plants with or without water stress. Under well-watered conditions, no significant differences in the expression of all DRGs were found between the leaf tissues from the mock- and PGPR-treated plants (Figures 7, 8). The expression levels of all DRGs were significantly higher in the mock-, CACC109-, and CACC119-treated plants under water stress than in their well-watered counterparts, indicating that all plants were affected by drought (Figures 7, 8).
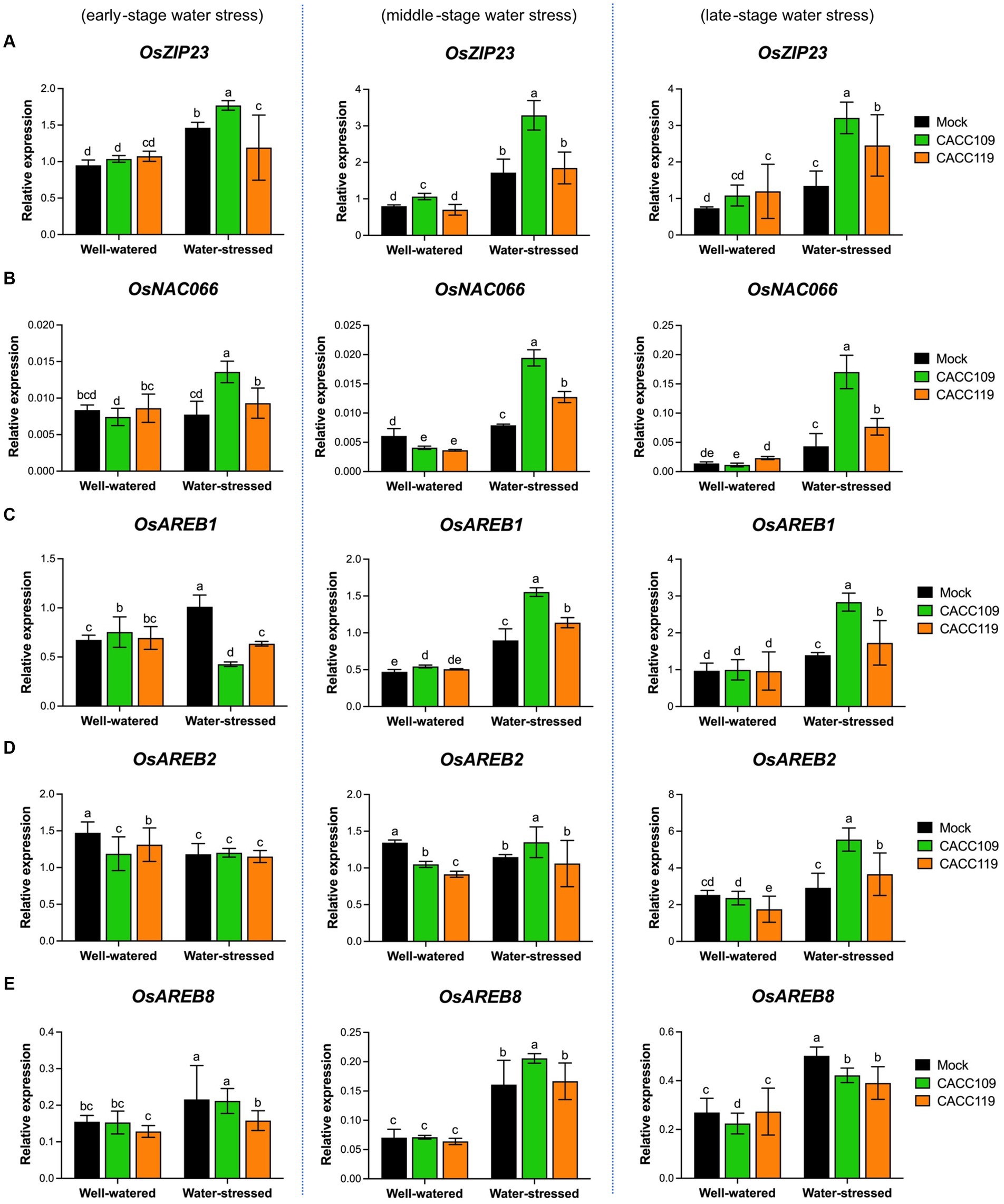
Figure 7. Effects of CACC109 and CACC119 on the expression of ABA-dependent drought-responsive genes in rice leaves. Expression of (A) OsZIP23, (B) OsNAC066, (C) OsAREB1, (D) OsAREB2, and (E) OsAREB8 in mock-, CACC109-, and CACC119-treated plants grown under well-watered or water-stressed conditions. Six-week-old plants were subjected to drought stress by maintaining soil water potential at 20–40% for 37 days. Leaf tissues were collected from three independent plants treated with mock, CACC109, or CACC119 in each of the two pots under well-watered and water-stressed conditions at 10 (early), 20 (middle), and 35 (late) days after drought treatment. The expression levels of drought-responsive genes were estimated by quantitative real-time PCR, and OsActin was used as an internal control for normalization. Values are mean ± SD of two independent experiments. Different letters indicate significant differences (p < 0.05, two-way ANOVA and Tukey’s test).
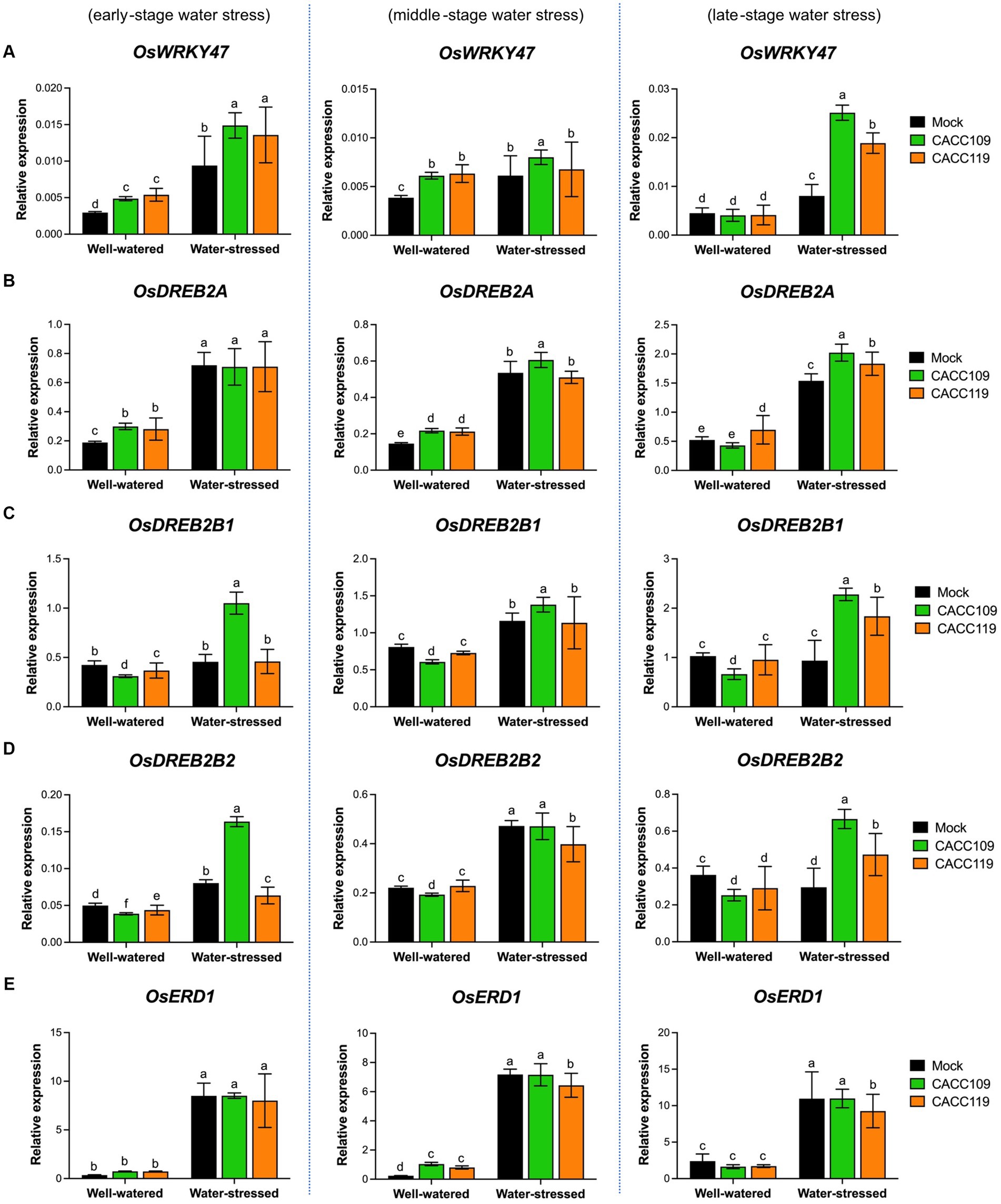
Figure 8. Effects of CACC109 and CACC119 on the expression of ABA-independent drought-responsive genes in rice leaves. Expression of (A) OsWRKY47, (B) OsDREB2A, (C) OsDREB2B1, (D) OsDREB2B2, and (E) OsERD1 in mock-, CACC109-, and CACC119-treated plants grown under well-watered or water-stressed conditions. Six-week-old plants were subjected to drought by maintaining soil water potential at 20–40% for 37 days. Leaf tissues were collected from three independent plants treated with mock, CACC109, or CACC119 in each of the two pots under well-watered and water-stressed conditions at 10 (early), 20 (middle), and 35 (late) days after drought treatment. Expression levels of drought-responsive genes were estimated by quantitative real-time PCR, and OsActin was used as an internal control for normalization. Values are mean ± SD of two independent experiments. Different letters indicate significant differences (p < 0.05, two-way ANOVA and Tukey’s test).
Analysis of the expression of ABA-dependent DRGs showed that during water stress, OsZIP23 expression peaked in the CACC109-treated plants, whereas the CACC119-treated plants exhibited notably higher OsZIP23 expression than the mock-treated plants at the late stage (Figure 7A). Meanwhile, OsNAC066 expression was highest in the CACC109-treated plants, followed by the CACC119-treated plants, and then the control plants at all time points after drought (Figure 7B). At the early stage of water stress, the expression levels of OsAREB1 and OsAREB2 were similar or lower in the PGPR-treated plants than in the mock-treated plants. However, as the stress progressed to the middle and late stages, the expression of these genes became significantly higher in the CACC109- and CACC119-treated plants than in the mock-treated plants (Figures 7C,D). The expression of OsAREB8 showed no noticeable difference between the mock- and PGPR-treated plants under water stress (Figure 7E).
Analysis of the expression of ABA-independent DRGs displayed that at the early stage of water stress, the CACC109- and CACC119-treated plants showed significantly higher OsWRKY47 expression than the mock-treated plants. However, at the middle stage of water stress, all plants exhibited similar expression levels of OsWRKY47. At the late stage, OsWRKY47 expression was significantly upregulated in the PGPR-treated plants compared with the mock-treated plants (Figure 8A). At the early and middle stages of water stress, the expression of OsDREB2A was similar in both the PGPR-treated and mock-treated plants. However, in the late stage, the CACC109- and CACC119-treated plants showed significantly higher levels of OsDREB2A expression compared to the mock-treated plants (Figure 8B). The expression levels of both splice variants of OsDREB2B, OsDREB2B1 and OsDREB2B2, were higher in the CACC109-treated plants than in the CACC119- and mock-treated plants at the early stage of water stress. At the middle stage, the PGPR- and mock-treated plants exhibited comparable expression levels of OsDREB2B1 and OsDREB2B2. However, at the late stage, the expression levels of OsDREB2B1 and OsDREB2B2 were significantly higher in the PGPR-treated plants than in the mock-treated plants, which displayed similar levels to the non-stressed plants (Figures 8C,D). No significant difference in OsERD1 expression was found between the mock- and PGPR-treated plants under water stress (Figure 8E).
These results indicated that CACC109 and CACC119 enhanced the drought tolerance in rice plants by upregulating the expression of DRGs via ABA-dependent and -independent pathways.
3.8 Genome analysis of CACC109 (PRJNA932602) and CACC119 (PRJNA1049983)
Whole-genome sequencing of B. megaterium CACC109 and CACC119 yielded 672 and 621 Mb of data with approximately 34× and 5× coverage, respectively. Sequencing reads with a Phred score greater than Q30 were used for de novo assembly using PacBio sequencing data. The Microbial Assembly resulted in draft genomes of ~5.3 and 5.7 Mb (5,309,578 and 5,703,136 bp) with 4 and 10 contigs with N50 of 5,083,237 and 4,735,530 bp, respectively. The average GC contents of the CACC109 and CACC119 genomes were 38.1 and 37.8%, respectively. Annotation of the CACC109 and CACC119 draft genomes resulted in 5,423 and 5,787 CDSs with median lengths of 714 and 702 bp, 42 and 49 rRNAs, and 130 and 187 tRNAs, respectively, as predicted by Prodigal 2.6.2 (Supplementary Table S3). These genomic features were visualized in circular maps of the B. megaterium CACC109 and CACC119 genomes (Supplementary Figure S2).
We deciphered genes enhancing plant growth and abiotic stress resilience from the entire genomes of CACC109 and CACC119. The presence of genes associated with phosphate solubilization, nitrogen fixation, and IAA production highlighted their potential as PGPs. In addition, the detection of genes related to siderophore production and ACC deaminase activity underscores their ability to thrive under adverse environmental conditions and improve plant tolerance to abiotic stresses (Supplementary Table S4). The identification of genes associated with motility and chemotaxis further indicated that CACC109 and CACC119 are plant-associated bacteria capable of navigating to plant roots in response to plant-secreted substances and nutrients (Supplementary Table S5).
3.9 Effects of PGPR on soluble organic components in soil
To investigate whether the PGPR affect the balance of soluble organic components in soil, we compared the levels of P2O5, Ca2+, and K+ in soils treated with mock, CACC109, or CACC119. Under water stress, P2O5 content was significantly higher in the CACC109- and CACC119-treated soils than in the mock-treated soils. Furthermore, the application of both PGPR significantly increased the K+ and Ca2+ contents in the soils by 22.9 and 31.4%, respectively, compared with the mock treatment (Supplementary Figure S3). These results indicated that the PGPR CACC109 and CACC119 played a role in altering the balance of soluble organic components in soils under drought.
4 Discussion
Drought is an abiotic stress that can severely affect the growth and production of crops, including rice, and the agricultural demand for food security (Zhang et al., 2018). Many strategies have been developed to increase the resilience of plants to abiotic stresses, allowing them to thrive in challenging environments (Ashry et al., 2022). Among these, inoculation with PGPR is effective in increasing crop productivity in drought-stressed environments (Rodríguez-Salazar et al., 2009). The enhancement of plant growth by PGPR can be attributed to several mechanisms, including the synthesis of PGP hormones and other PGP activities (Glick, 1995). Over the past two decades, various soil microorganisms have been used to alleviate abiotic stresses in plants. PGPR mitigate the effects of drought on crops (Niu et al., 2018; Jochum et al., 2019; Batool et al., 2020). However, the selection of bacteria with abiotic stress tolerance and in vitro PGP properties is crucial to alleviate abiotic stress in plants (Jinal et al., 2021).
In the present study, 31 isolates were obtained from ginseng rhizosphere and field soils, of which 28 were identified as Bacillus spp. Among the isolates, B. megaterium CACC109 and CACC119 showed comparatively better osmotic stress tolerance (Figure 1A). Both strains exhibited EPS and siderophore production (Pallai, 2005; Naseem et al., 2018), nitrogen fixation (Cattelan et al., 1999), IAA synthesis (Loper and Schroth, 1986), and ACC deaminase activity (Glick, 2014) (Figures 1C–H). Notably, CACC109 showed additional advantages, including higher ACC deaminase activity, phosphate solubilization, and EPS production under osmotic stress, enhancing its ability to promote plant growth and alleviate abiotic stress. Phosphate-solubilizing microorganisms act as biofertilizers (Sivasakthi et al., 2013) that increase phosphorus availability and crop yield (Shi et al., 2014). Nitrogen-fixing Bacillus species are promising in agriculture because they facilitate plant nutrient uptake (Novo et al., 2018). The IAA produced by PGPR promotes plant growth and nutrient absorption (Loper and Schroth, 1986; Venturi and Keel, 2016; Gowtham et al., 2017). Microbial EPS production under stress conditions enhances bacterial survival, indicating drought tolerance (Sandhya et al., 2009). Specifically, PGPR produce EPS composed of complex organic macromolecules, including polysaccharides, proteins, and uronic acid. These compounds protect microbial cells by stabilizing their membrane structure against adverse environmental conditions such as drought, high salinity and extreme temperatures (Ojuederie and Babalola, 2017). Siderophores are critical for microbial survival and facilitate iron binding and uptake, particularly under conditions of low iron availability. Higher siderophore levels correlate with improved drought tolerance in plants (Andrews et al., 2003; Arzanesh et al., 2011). ACC deaminase activity in many PGPR helps reduce ET levels in plants, mitigating growth inhibition under stress and thereby promoting plant growth (Glick, 2014).
In the present study, we evaluated the effects of the selected PGPR strains, CACC109 and CACC119, on seed germination, seedling growth, and drought stress in rice plants. Both strains improved rice seed germination and root development under osmotic stress induced by PEG 6000 and mannitol (Figure 2) possibly by enhancing EPS production, which mitigated damage and promoted metabolic processes in the seeds (Bakhshandeh et al., 2020). Similarly, a previous study reported a 50% increase in germination rate under stress conditions with the application of EPS-producing bacterial strains (Tewari and Arora, 2014). CACC109 and CACC119 also significantly improved the drought tolerance of rice plants (Figure 4). Notably, CACC109 showed superior EPS production to CACC119 under high osmotic stress (Figure 1C), which is consistent with the observed phenotype. Bacteria can survive under stressful conditions by producing EPS, which increases water retention, regulates the diffusion of organic carbon sources, and allows irreversible attachment and colonization of roots by forming a network of fibrillar material that binds bacteria to the root surface (Chenu and Roberson, 1996; Bashan and Holguin, 1997). EPS are hydrated compounds containing 97% water in the matrix, protecting both bacteria and plants from desiccation. Plants treated with EPS-producing bacteria exhibit enhanced water retention and drought stress tolerance (Bensalim et al., 1998). Bacterial EPS can form a biofilm on the root surface, ultimately enhancing soil structure and supporting plant development while mitigating the impact of drought stress (Sandhya et al., 2009). EPS produced by various bacterial strains enhance soil permeability by improving soil aggregation and maintaining a higher water potential and better nutrient uptake around roots, resulting in increased plant growth and drought tolerance (Upadhyay et al., 2012; Vejan et al., 2016). Niu et al. (2018) reported that several PGPR strains, including Enterobacter hormaechei, Pseudomonas fluorescens, and Pseudomonas migulae, produce EPS and enhance seed germination and seedling growth under drought conditions. The inoculation of sunflower rhizosphere with the EPS-producing rhizobial strain YAS34 under drought conditions significantly increases the ratio of root-adhering soil per root tissue (Alami et al., 2000). Vurukonda et al. (2016) found that inoculating maize seeds with EPS-producing bacterial strains Alcaligenes faecalis, Pseudomonas aeruginosa, and Proteus penneri increases soil moisture and relative water contents, shoot and root length, leaf area, and plant biomass under drought conditions. The results of present study suggest that EPS-producing CACC109 and CACC119 are promising candidates for enhancing plant stress tolerance and promoting seedling establishment under adverse environmental conditions.
The CACC109- and CACC119-treated rice plants had significantly higher shoot and root lengths, leaf widths, plant weights, and chlorophyll contents than the control plants (Figures 3, 4). Notably, CACC109 treatment substantially increased root biomass due to axial and lateral root development, whereas CACC119 exerted a similar but less pronounced effect (Supplementary Figure S1). This result is consistent with the growth-promoting effects of PGPR reported in previous studies. For example, Bacillus aryabhattai AB211 from maize (Bhattacharyya et al., 2017), Bacillus aryabhattai SRB02 from soybean roots (Park et al., 2017), Bacillus sp. BCLRB2 in saline soil (Hadj Brahim et al., 2019), Bacillus subtilis EA-CB0575 from banana roots (Franco-Sierra et al., 2020), and Bacillus sp. D5 from saffron corms (Magotra et al., 2021) significantly improve plant growth through various mechanisms, such as synthesis of hormones, facilitation of phosphate uptake, and production of beneficial compounds. Our results highlight the potential benefits of nutrient uptake and enhanced adaptability to adverse abiotic stress conditions in rice plants.
Drought decreases plant growth as essential nutrients and solutes are redirected toward stress-related processes (Brestic et al., 2018). Nonetheless, bacterial inoculation enhances plant growth, resulting in significant increases in root and shoot lengths and leaf area (Lin et al., 2020). Different PGPR strains enhance the growth of various plants under abiotic stresses, such as drought and salinity (Kumari et al., 2016; Creus et al., 2020; Mishra et al., 2020; Muhammad Usman Aslam et al., 2020). In the present study, the application of CACC109 and CACC119 significantly improved the shoot length, leaf width, plant weight, chlorophyll content, and proline production in the rice plants under drought, ultimately increasing the drought tolerance in the PGPR-treated plants compared with the mock-treated plants (Figures 3, 4).
Drought can affect the chlorophyll levels in plant leaves and consequently decrease photosynthesis (Liu et al., 2019; Khayatnezhad and Gholamin, 2021). It can significantly reduce photosynthetic efficiency by altering the leaf area, affecting chlorophyll synthesis, increasing lipid peroxidation, and promoting chlorophyll degradation (Khayatnezhad and Gholamin, 2021). Consequently, plants capable of maintaining higher chlorophyll levels under drought stress may exhibit greater tolerance to water shortage and sustain increased growth and biomass production. This hypothesis is supported by the higher total chlorophyll contents and water retention in the CACC109- and CACC119-treated plants than in the mock-treated plants under drought.
The accumulation of osmolytes such as proline, sugars, and proteins in plants is a key indicator of drought tolerance because it facilitates osmotic adjustment and prevents water loss (Vaishnav and Choudhary, 2019; Karimzadeh et al., 2021). In the present study, proline production and proline biosynthesis gene expression significantly increased in the plants treated with CACC109 and CACC119 under drought (Figure 5). One of the defense strategies of PGPR is to improve the tolerance of plants to drought stress, indicating the substantial contribution of CACC109 and CACC119 in alleviating drought in plants by enhancing their defense mechanisms.
Endogenous signaling molecules such as ROS play crucial roles in plant growth, development, and stress adaptation, acting as double-edged swords because of their dual nature as signaling molecules and potential toxins, especially under drought (Miller et al., 2010; Steffens, 2014). Although excessive ROS accumulation can be detrimental to plant cells, plants can enhance drought tolerance by modulating ROS levels through the activities of various antioxidant enzymes, such as SOD, CAT, POD, and GPX (Blokhina, 2003; Miller et al., 2010; Fang et al., 2015; Nadarajah, 2020). Elevated expression of ROS detoxification genes improves plant drought tolerance by mitigating the deleterious effects of oxidative stress on plant metabolism (Ning et al., 2010). In the present study, the CACC109-treated plants had significantly higher expression of antioxidant-related genes OsSOD, OsAPX, OsCAT, OsPOD, and OsGPX under drought, whereas the CACC119-treated plants had notably higher expression of OsCAT and OsPOD compared with the mock-treated plants (Figure 6). These findings suggest that CACC109 and CACC119 influence cellular ROS homeostasis and potentially improve drought tolerance in rice plants by regulating oxidative stress levels. In particular, the relative expression trend of these genes varies over time. Specifically, the relative expression of OsCAT and OsPOD was higher at the late stage of water stress, whereas that of OsAPX, OsSOD, and OsGPX was lower at the same stage in the plants inoculated with CACC109 (Figure 6). This differential expression of antioxidant genes can be attributed to several factors. The enzymes APX, GPX, CAT, and POD utilize different mechanisms to scavenge hydrogen peroxide (H2O2) and manage oxidative stress in plants. Ascorbate peroxidase requires an ascorbate and glutathione (GSH) regeneration system, known as the ascorbate-glutathione cycle. In the first reaction of this cycle, catalyzed by APX, H2O2 and ascorbate produce H2O and monodehydroascorbate (Mittler, 2002). Similarly, GPX reduces H2O2 to H2O but uses GSH directly as the reducing agent (Noctor et al., 2014). By contrast, CAT directly converts H2O2 to H2O and 1/2 O2 and is primarily involved in detoxifying H2O2 rather than regulating it as a signaling molecule in plants (Apel and Hirt, 2004). Meanwhile, peroxidase can utilize various substrates to reduce H2O2, often employing phenolic compounds as electron donors, and is involved in a wide range of physiological processes (Gill and Tuteja, 2010). Changes in the balance of these scavenging enzymes trigger compensatory mechanisms. For instance, a decrease in CAT activity upregulates APX and GPX to maintain ROS homeostasis. Similarly, POD activity may increase in response to stress conditions to compensate for changes in the levels or activity of other antioxidant enzymes, thus contributing to the overall ROS detoxification and stress adaptation (Apel and Hirt, 2004). Inoculation with CACC109 could influence the expression of ROS detoxification genes by modulating plant hormone balance, signaling pathways, or enhancing drought stress-specific response mechanisms. This phenomenon could result in a more targeted activation of OsCAT and OsPOD while downregulating others such as OsAPX, OsSOD, and OsGPX in the late stage. These observed differential expressions likely reflect the strategic and dynamic response of plants to effectively manage ROS levels. Further studies involving detailed temporal gene expression analysis and the role of CACC109 inoculation may provide deeper insights into these regulatory mechanisms.
Plants have evolved numerous defense mechanisms against drought, among which transcription factors (TFs) play a crucial role in regulating the expression of stress-related genes. Members of the basic leucine zipper (bZIP) family are important TFs responsible for ABA signaling and plant responses to drought (Joo et al., 2021). OsbZIP23 is a key regulator of drought and salinity tolerance in rice (Xiang et al., 2008). AREBs, which belong to the group A subfamily of bZIP TFs, play key roles in the ABA signaling pathway in rice. They regulate the expression of stress-responsive genes, especially under drought and salinity, thus contributing to the stress tolerance mechanisms in rice (Yoshida et al., 2010). NAC, one of the largest families of TFs in plants, is involved in the regulation of growth and stress responses (Nakashima et al., 2012; Shao et al., 2015). OsNAC066 positively regulates oxidative and drought stress tolerance, as demonstrated by functional analyses of overexpression and RNAi transgenic rice plants (Yuan et al., 2019). WRKY TFs play an important role in regulating transcriptional reprogramming associated with tolerance to multiple abiotic stresses in plants (Chen et al., 2012). OsWRKY47 is a positive regulator of drought response, and OsWRKY47-overexpressing rice plants exhibit greater drought tolerance than wild-type plants (Raineri et al., 2015). DREB TFs play critical roles in improving abiotic stress tolerance, mostly in an ABA-independent manner (Dubouzet et al., 2003; Lata and Prasad, 2011). In rice, OsDREB2A expression is induced by water stress, resulting in increased drought tolerance (Cui et al., 2011). The OsDREB2B transcript exhibits functional and non-functional forms under drought, with alternative splicing induced by its pre-mRNA, thereby enhancing drought tolerance (Matsukura et al., 2010). These findings highlight the critical role of OsDREB2 in regulating drought tolerance in rice plants.
In the present study, we investigated the molecular mechanisms underlying PGPR-induced drought tolerance by examining the expression of DRGs. CACC109 and CACC119 positively influenced the response of rice plants to drought. Under well-watered conditions, the mock- and PGPR-treated plants showed no significant difference in DRG expression (Figures 7, 8). However, under water stress, the PGPR- and mock-treated plants showed significantly higher expression of DRGs than the control plants. OsZIP23 expression peaked in the CACC109-treated plants. OsDREB2A expression was significantly higher in both CACC-treated plants than in the mock-treated plants at the late drought stage. The expression of OsDREB2B1 and OsDREB2B2 peaked in the CACC109-treated plants during water stress. The highest expression levels of OsNAC066 were observed in the CACC109-treated plants. The expression levels of OsAREB1 and OsAREB2 were significantly higher in both CACC-treated plants than in the mock-treated plants at the middle and late stages of water stress. However, OsAREB8 expression was similar or significantly lower in the CACC-treated plants at these same stages. The AREB family members, though functionally related, can have different regulatory controls and promoter regions responding to specific signals. The inoculation with CACC could interact differently with the signaling pathways involving OsAREB8 compared to OsAREB1 and OsAREB2. This could result in a distinct modulation of OsAREB8 expression, possibly due to specific hormonal changes or stress signals induced by CACC treatment. Both PGPR-treated plants exhibited significantly higher OsWRKY47 expression at the early and late stages of water stress. Moreover, the expression of OsDREB2B1 and OsDREB2B2 was significantly upregulated in the CACC109-treated plants during the early stage and in both PGPR-treated plants at the late stage of drought. These results suggest that CACC109 and CACC119 enhance drought tolerance in rice via ABA-dependent and -independent pathways, as reflected by the altered expression of the abovementioned DRGs. Taken together, the results of our study highlight the beneficial effects of PGPR CACC109 and CACC119 on plant response to drought and provide novel insights into the molecular mechanisms underlying PGPR-induced drought tolerance in rice.
The whole genome analysis of CACC109 and CACC119 reveals a suite of genes that underscore their potential as effective PGPR, particularly under stress conditions. The genomes of these strains encode genes associated with key physiological traits that enhance plant growth and stress resilience. For instance, the presence of genes related to phosphate solubilization, nitrogen fixation, and IAA production highlights their ability to improve nutrient availability and hormonal balance, which are crucial for plant growth and stress adaptation (Supplementary Table S4) (Lugtenberg and Kamilova, 2009). Moreover, the detection of genes involved in siderophore production and ACC deaminase activity points to their role in mitigating the adverse effects of abiotic stresses. Siderophores can help plants acquire iron under limited conditions, while ACC deaminase can reduce the levels of ET, a hormone that often exacerbates stress responses (Hayat et al., 2010; Singh et al., 2022). The presence of genes related to motility and chemotaxis further indicates that these bacteria can navigate toward plant roots, enhancing their colonization and interaction with plant roots in response to plant-secreted signals and nutrient gradients (Supplementary Table S5) (Feng et al., 2021). These genomic insights are instrumental in understanding how CACC109 and CACC119 can positively impact rice growth under drought stress. The array of beneficial traits encoded in their genomes supports their potential as effective PGPR, capable of not only promoting growth but also enhancing stress resilience. Future research should focus on validating these genomic predictions in practical field settings, particularly under varied stress conditions, to fully elucidate their roles and optimize their application for crop improvement.
The PGPR have long been recognized for their beneficial effects on plant growth under optimal conditions. However, their efficacy can be complex when plants face different types of stress. Recent studies suggest that while PGPR can significantly enhance plant growth, their benefits might be context-dependent, especially under stress conditions. For instance, under deficit stresses such as drought, the growth-promoting effects of PGPR might be counterproductive as they could divert the plant’s energy away from crucial stress responses, leading to potential negative outcomes (Glick, 2012; Vurukonda et al., 2016). Conversely, under cumulative stresses, such as salinity, PGPR might help alleviate some of the stress impacts through mechanisms such as dilution effects, which can help reduce the severity of stress impacts on plant growth (Kumar et al., 2023). In the present study, the strains CACC109 and CACC119 demonstrated positive effects on rice plants under drought stress (Figures 3, 4). This finding is consistent with the notion that some PGPR strains can enhance plant resilience to specific stress conditions. However, the response of these strains to cumulative stresses, including combinations of drought and salinity, remains unexplored. Further research is warranted to understand how these PGPR strains perform under such combined stresses. This would provide a comprehensive view of their functional capabilities and help differentiate between mere growth promotion and genuine alleviation of stress-induced growth inhibition, offering deeper insights into the interplay between PGPR and environmental stressors.
While this study offers valuable insights into the potential of CACC109 and CACC119 as PGPR for promoting plant growth and enhancing drought tolerance in rice plants, the limitations of the present study should be acknowledged. Firstly, the study focused only on rice plants, and the efficacy of these PGPR strains may vary across different plant species or even across cultivars of the same species. Additionally, the experimental conditions may not accurately replicate real agricultural environments, potentially affecting the generalizability of the findings. Moreover, the mechanisms underlying the observed effects of the PGPR strains have not been fully elucidated, indicating the need for further research to comprehensively understand their mode of action. Furthermore, the long-term effects of PGPR inoculation on soil health and ecosystem dynamics were not investigated in this study, underscoring the necessity for future studies to assess the broader ecological implications of PGPR use in agriculture.
5 Conclusion
This study aimed to identify novel PGPR strains that can promote plant growth and alleviate drought stress. B. megaterium strains CACC109 and CACC119 displayed various PGP activities. They significantly enhanced rice seed germination under osmotic stress and promoted root growth under stressed and non-stressed conditions. Additionally, these strains improved overall plant growth parameters, such as shoot and root lengths and weights, and leaf widths, while also enhancing plant physiological characteristics, such as chlorophyll levels and osmolyte production. Notably, the rice plants treated with CACC109 and CACC119 exhibited enhanced drought tolerance, as evidenced by their higher survival rates, increased chlorophyll and proline contents, reduced water loss rates, and upregulated expression of antioxidant-related and drought-responsive genes. Overall, this study provides valuable insights into the beneficial effects of PGPR on plant growth and their response to environmental stressors, offering promising avenues for increasing crop productivity and mitigating the impacts of climate change on agriculture.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
SL: Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Validation, Visualization, Writing – original draft, Writing – review & editing, Resources. J-AK: Formal analysis, Investigation, Methodology, Validation, Resources, Writing – review & editing. JS: Investigation, Writing – review & editing. SC: Investigation, Writing – review & editing. GJ: Writing – review & editing. YK: Funding acquisition, Supervision, Validation, Writing – review & editing, Conceptualization, Resources.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was supported by Technology commercialization support project Ministry of Agriculture, Food and Rural Affairs, Republic of Korea (122022–3).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1430546/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Effects of CACC109 and CACC119 on rice shoot growth and root development. (A) Comparison of shoot growth and root development in 9-week-old mock-, CACC109-, and CACC119-treated rice plants grown under well-watered conditions. (B) Magnification of the root parts.
SUPPLEMENTARY FIGURE S2 | Circular map of the genome and genome features circles of B. megaterium CACC109 (A) and CACC119 (B). From the outside to the inside, the outer black circle represents the scale line in Mbps; the second gray band represents contigs; the third and fourth circles represent the annotated reference genes (specifically, CDSs) found in the forward and reverse strands, respectively; the fifth circle displays only rRNA and tRNA found in the genome; the sixth circle displays the GC skew metric. The genomic mean GC-skew value is used as the baseline, relative to which higher-than-average values are displayed in green, whereas lower-than-average values are displayed in red; the seventh circle displays the GC ratio metric. GC ratio also uses the genomic mean GC ratio value as its baseline, with higher-than-average values in blue and lower-than-average values in yellow.
SUPPLEMENTARY FIGURE S3 | Effects of CACC109 and CACC119 on soluble organic components. Comparison of soil nutrients (P2O5, K+, and Ca2+) between mock and PGPR (CACC109 and CACC119)-treated soils in pots. Data are presented as the mean ± SD of at least three independent samples. Different letters indicate significant differences (P < 0.05, Student’s t-test). The experiment was performed three times.
Footnotes
References
Ahmad, B., Sahar, N., Nausheen, M., and Shumaila, B. (2013). Isolation and characterization of cellulolytic nitrogen fixing Azotobacter species from wheat rhizosphere of Khyber Pakhtunkhwa. World Appl. Sci. J. 27, 51–60. doi: 10.5829/idosi.wasj.2013.27.01.81120
Alami, Y., Achouak, W., Marol, C., and Heulin, T. (2000). Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl. Environ. Microbiol. 66, 3393–3398. doi: 10.1128/AEM.66.8.3393-3398.2000
Ammar, M. H., Anwar, F., el-Harty, E. H., Migdadi, H. M., Abdel-Khalik, S. M., al-Faifi, S. A., et al. (2015). Physiological and yield responses of Faba bean (Vicia faba L.) to drought stress in managed and open field environments. J. Agron. Crop Sci. 201, 280–287. doi: 10.1111/jac.12112
Andrews, S. C., Robinson, A. K., and Rodríguez-Quiñones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Anjum, S. A., Wang, L. C., Farooq, M., Hussain, M., Xue, L. L., and Zou, C. M. (2011). Brassinolide application improves the drought tolerance in maize through modulation of enzymatic antioxidants and Leaf gas exchange. J. Agron. Crop Sci. 197, 177–185. doi: 10.1111/j.1439-037X.2010.00459.x
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Arzanesh, M. H., Alikhani, H. A., Khavazi, K., Rahimian, H. A., and Miransari, M. (2011). Wheat (Triticum aestivum L.) growth enhancement by Azospirillum sp. under drought stress. World J. Microbiol. Biotechnol. 27, 197–205. doi: 10.1007/s11274-010-0444-1
Ashry, N. M., Alaidaroos, B. A., Mohamed, S. A., Badr, O. A. M., El-Saadony, M. T., and Esmael, A. (2022). Utilization of drought-tolerant bacterial strains isolated from harsh soils as a plant growth-promoting rhizobacteria (PGPR). Saudi J. Biol. Sci. 29, 1760–1769. doi: 10.1016/j.sjbs.2021.10.054
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting Rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9:1473. doi: 10.3389/fpls.2018.01473
Bakhshandeh, E., Gholamhosseini, M., Yaghoubian, Y., and Pirdashti, H. (2020). Plant growth promoting microorganisms can improve germination, seedling growth and potassium uptake of soybean under drought and salt stress. Plant Growth Regul. 90, 123–136. doi: 10.1007/s10725-019-00556-5
Barnawal, D., Singh, R., and Singh, R. P. (2019). “Role of plant growth promoting Rhizobacteria in drought tolerance” in PGPR amelioration in sustainable agriculture (Elsevier), 107–128.
Bashan, Y., and Holguin, G. (1997). Azospirillum – plant relationships: environmental and physiological advances (1990–1996). Can. J. Microbiol. 43, 103–121. doi: 10.1139/m97-015
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Batool, T., Ali, S., Seleiman, M. F., Naveed, N. H., Ali, A., Ahmed, K., et al. (2020). Plant growth promoting rhizobacteria alleviates drought stress in potato in response to suppressive oxidative stress and antioxidant enzymes activities. Sci. Rep. 10:16975. doi: 10.1038/s41598-020-73489-z
Bensalim, S., Nowak, J., and Asiedu, S. K. (1998). A plant growth promoting rhizobacterium and temperature effects on performance of 18 clones of potato. Am. J. Pot Res 75, 145–152. doi: 10.1007/BF02895849
Bhandari, U., Gajurel, A., Khadka, B., Thapa, I., Chand, I., Bhatta, D., et al. (2023). Morpho-physiological and biochemical response of rice (Oryza sativa L.) to drought stress: a review. Heliyon 9:e13744. doi: 10.1016/j.heliyon.2023.e13744
Bhattacharyya, C., Bakshi, U., Mallick, I., Mukherji, S., Bera, B., and Ghosh, A. (2017). Genome-guided insights into the plant growth promotion capabilities of the physiologically versatile Bacillus aryabhattai strain AB211. Front. Microbiol. 8:411. doi: 10.3389/fmicb.2017.00411
Bland, C., Ramsey, T. L., Sabree, F., Lowe, M., Brown, K., Kyrpides, N. C., et al. (2007). CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformat. 8:209. doi: 10.1186/1471-2105-8-209
Blokhina, O. (2003). Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann. Bot. 91, 179–194. doi: 10.1093/aob/mcf118
Brestic, M., Zivcak, M., Hauptvogel, P., Misheva, S., Kocheva, K., Yang, X., et al. (2018). Wheat plant selection for high yields entailed improvement of leaf anatomical and biochemical traits including tolerance to non-optimal temperature conditions. Photosynth. Res. 136, 245–255. doi: 10.1007/s11120-018-0486-z
Cattelan, A. J., Hartel, P. G., and Fuhrmann, J. J. (1999). Screening for plant growth–promoting Rhizobacteria to promote early soybean growth. Soil Sci. Soc Amer J 63, 1670–1680. doi: 10.2136/sssaj1999.6361670x
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chenu, C., and Roberson, E. B. (1996). Diffusion of glucose in microbial extracellular polysaccharide as affected by water potential. Soil Biol. Biochem. 28, 877–884. doi: 10.1016/0038-0717(96)00070-3
Cherif-Silini, H., Silini, A., Yahiaoui, B., Ouzari, I., and Boudabous, A. (2016). Phylogenetic and plant-growth-promoting characteristics of Bacillus isolated from the wheat rhizosphere. Ann. Microbiol. 66, 1087–1097. doi: 10.1007/s13213-016-1194-6
Choi, K., Choi, J., Lee, P. A., Roy, N., Khan, R., Lee, H. J., et al. (2020). Alteration of bacterial wilt resistance in tomato plant by microbiota transplant. Front. Plant Sci. 11:1186. doi: 10.3389/fpls.2020.01186
Claeys, H., Van Landeghem, S., Dubois, M., Maleux, K., and Inzé, D. (2014). What is stress? Dose-response effects in commonly used in vitro stress assays. Plant Physiol. 165, 519–527. doi: 10.1104/pp.113.234641
Cohen, A. C., Bottini, R., Pontin, M., Berli, F. J., Moreno, D., Boccanlandro, H., et al. (2015). Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 153, 79–90. doi: 10.1111/ppl.12221
Conesa, M. R., Rosa, J. M. De La, Domingo, R., Bañon, S., and Pérez-Pastor, A. (2016). Changes induced by water stress on water relations, stomatal behaviour and morphology of table grapes (cv. Crimson seedless) grown in pots. Sci. Hortic. 202, 9–16. doi: 10.1016/j.scienta.2016.02.002
Creus, C. M., Sueldo, R. J., and Barassi, C. A. (2004). Water relations and yield in Azospirillum-inoculated wheat exposed to drought in the field. Can. J. Bot. 82, 273–281. doi: 10.1139/b03-119
Creus, C. M., Sueldo, R. J., Barassi, C. A., Taamalli, W., Abdelly, C., and Djébali, N. (2020). Auxin and proline producing rhizobacteria mitigate salt-induced growth inhibition of barley plants by enhancing water and nutrient status. S. Afr. J. Bot. 128, 209–217. doi: 10.1016/j.sajb.2019.10.023
Cui, M., Zhang, W., Zhang, Q., Xu, Z., Zhu, Z., Duan, F., et al. (2011). Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 49, 1384–1391. doi: 10.1016/j.plaphy.2011.09.012
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., and Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
Devarajan, A. K., Muthukrishanan, G., Truu, J., Truu, M., Ostonen, I., Kizhaeral, S., et al. (2021). The foliar application of rice phyllosphere bacteria induces drought-stress tolerance in Oryza sativa (L.). Plan. Theory 10:387. doi: 10.3390/plants10020387
Ding, K., Tabuchi, Y., and Makino, T. (2022). Effect of steam-processing of the Panax ginseng root on its inducible activity on granulocyte-colony stimulating factor secretion in intestinal epithelial cells in vitro. J. Ethnopharmacol. 287:114927. doi: 10.1016/j.jep.2021.114927
Dubouzet, J. G., Sakuma, Y., Ito, Y., Kasuga, M., Dubouzet, E. G., Miura, S., et al. (2003). OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 33, 751–763. doi: 10.1046/j.1365-313X.2003.01661.x
Durairaj, K., Velmurugan, P., Park, J.-H., Chang, W.-S., Park, Y.-J., Senthilkumar, P., et al. (2018). An investigation of biocontrol activity Pseudomonas and Bacillus strains against Panax ginseng root rot fungal phytopathogens. Biol. Control 125, 138–146. doi: 10.1016/j.biocontrol.2018.05.021
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2007). PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinformat. 8:18. doi: 10.1186/1471-2105-8-18
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Fang, Y., Liao, K., Du, H., Xu, Y., Song, H., Li, X., et al. (2015). A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 66, 6803–6817. doi: 10.1093/jxb/erv386
FAO (2017). The future of food and agriculture: Trends and challenges. Rome: Food and Agriculture Organization of the United Nations.
Feng, H., Fu, R., Hou, X., Lv, Y., Zhang, N., Liu, Y., et al. (2021). Chemotaxis of beneficial Rhizobacteria to root exudates: the first step towards root-microbe rhizosphere interactions. IJMS 22:6655. doi: 10.3390/ijms22136655
Ferreira, M. J., Silva, H., and Cunha, A. (2019). Siderophore-producing Rhizobacteria as a promising tool for empowering plants to cope with Iron limitation in saline soils: a review. Pedosphere 29, 409–420. doi: 10.1016/S1002-0160(19)60810-6
Fleischer, H., Vorberg, E., Thurow, K., Warkentin, M., and Behrend, D. (2014). Determination of calcium and phosphor in bones using microwave digestion and ICP-MS: Comparison of manual and automated methods using ICP-MS. in 5th IMEKO TC19 Symposium on environmental instrumentation and measurement, 23–24 September 201. Chemnitz, Germany. 94–99.
Foyer, C. H., Rasool, B., Davey, J. W., and Hancock, R. D. (2016). Cross-tolerance to biotic and abiotic stresses in plants: a focus on resistance to aphid infestation. J. Exp. Bot. 67, 2025–2037. doi: 10.1093/jxb/erw079
Franco-Sierra, N. D., Posada, L. F., Santa-María, G., Romero-Tabarez, M., Villegas-Escobar, V., and Álvarez, J. C. (2020). Bacillus subtilis EA-CB0575 genome reveals clues for plant growth promotion and potential for sustainable agriculture. Funct. Integr. Genomics 20, 575–589. doi: 10.1007/s10142-020-00736-x
Fukagawa, N. K., and Ziska, L. H. (2019). Rice: importance for global nutrition. J. Nutr. Sci. Vitaminol. 65, S2–S3. doi: 10.3177/jnsv.65.S2
García, J. E., Maroniche, G., Creus, C., Suárez-Rodríguez, R., Ramirez-Trujillo, J. A., and Groppa, M. D. (2017). In vitro PGPR properties and osmotic tolerance of different Azospirillum native strains and their effects on growth of maize under drought stress. Microbiol. Res. 202, 21–29. doi: 10.1016/j.micres.2017.04.007
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Glick, B. R. (1995). The enhancement of plant growth by free living Bacteria. Can. J. Microbiol. 41, 109–117. doi: 10.1139/m95-015
Glick, B. R. (2012). Plant growth-promoting Bacteria: mechanisms and applications. Scientifica 2012, 1–15. doi: 10.6064/2012/963401
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Glick, B. R., Penrose, D. M., and Li, J. (1998). A model for the lowering of plant ethylene concentrations by plant growth-promoting Bacteria. J. Theor. Biol. 190, 63–68. doi: 10.1006/jtbi.1997.0532
Gowtham, H. G., Duraivadivel, P., Hariprasad, P., and Niranjana, S. R. (2017). A novel split-pot bioassay to screen indole acetic acid producing rhizobacteria for the improvement of plant growth in tomato [Solanum lycopersicum L.]. Sci. Hortic. 224, 351–357. doi: 10.1016/j.scienta.2017.06.017
Hadj Brahim, A., Lobna, D., Mouna, J., Asmahen, A., Houda, H., Manel, B. A., et al. (2019). Bacillus sp. BCLRB2: an efficient diazotrophic halotolerant PGPB strain, in Proceedings of MOL2NET 2018, International Conference on Multidisciplinary Sciences, 4th, 6126
Hardoim, P. R., Overbeek, L. S. Van, Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/mmbr.00050-14
Haworth, M., Elliott-Kingston, C., and McElwain, J. C. (2013). Co-ordination of physiological and morphological responses of stomata to elevated [CO2] in vascular plants. Oecologia 171, 71–82. doi: 10.1007/s00442-012-2406-9
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117-1
Hayat, J. P. S., Hayat, Q., Alyemeni, M. N., Wani, A. S., and Ahmad, A. (2012). Role of proline under changing environments. Plant Signal. Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Hyatt, D., Chen, G.-L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformat. 11:119. doi: 10.1186/1471-2105-11-119
Islam, S., Akanda, A. M., Prova, A., Islam, M. T., and Hossain, M. M. (2016). Isolation and identification of plant growth promoting Rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front. Microbiol. 6:1360. doi: 10.3389/fmicb.2015.01360
Ji, L., Nasir, F., Tian, L., Chang, J., Sun, Y., Zhang, J., et al. (2021). Outbreaks of root rot disease in different aged American ginseng plants are associated with field microbial dynamics. Front. Microbiol. 12:676880. doi: 10.3389/fmicb.2021.676880
Jinal, H. N., Gopi, K., Kumar, K., and Amaresan, N. (2021). Effect of zinc-resistant Lysinibacillus species inoculation on growth, physiological properties, and zinc uptake in maize (Zea mays L.). Environ. Sci. Pollut. Res. 28, 6540–6548. doi: 10.1007/s11356-020-10998-4
Jochum, M. D., McWilliams, K. L., Borrego, E. J., Kolomiets, M. V., Niu, G., Pierson, E. A., et al. (2019). Bioprospecting plant growth-promoting Rhizobacteria that mitigate drought stress in grasses. Front. Microbiol. 10:2106. doi: 10.3389/fmicb.2019.02106
Joo, H., Baek, W., Lim, C. W., and Lee, S. C. (2021). Post-translational modifications of bZIP transcription factors in Abscisic acid signaling and drought responses. Curr. Genomics 22, 4–15. doi: 10.2174/1389202921999201130112116
Kanehisa, M., Goto, S., Sato, Y., Kawashima, M., Furumichi, M., and Tanabe, M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucl. Acids Res. 42, D199–D205. doi: 10.1093/nar/gkt1076
Karimzadeh, J., Alikhani, H. A., Etesami, H., and Pourbabaei, A. A. (2021). Improved phosphorus uptake by wheat plant (Triticum aestivum L.) with rhizosphere fluorescent pseudomonads strains under water-deficit stress. J. Plant Growth Regul. 40, 162–178. doi: 10.1007/s00344-020-10087-3
Kaushal, M. (2019). “Portraying rhizobacterial mechanisms in drought tolerance” in PGPR amelioration in sustainable agriculture. eds. A. K. Singh, A. Kumar, and P. K. Singh (San Diego, CA: Elsevier, Inc., Science & Technology/Academic Press), 195–216.
Khayatnezhad, M., and Gholamin, R. (2021). The effect of drought stress on the superoxide dismutase and chlorophyll content in durum wheat genotypes. Adva. Life Sci. 8, 119–123.
Kim, J.-H. (2018). Pharmacological and medical applications of Panax ginseng and ginsenosides: a review for use in cardiovascular diseases. J. Ginseng Res. 42, 264–269. doi: 10.1016/j.jgr.2017.10.004
Kumar, A. (2017). Development and characterization of Rice genotypes for water use efficiency and drought resistance. Ph.D. Dissertation. Fayetteville, AR, USA: University of Arkansas.
Kumar, A., Behera, I., Langthasa, M., and PrakashNaroju, S. (2023). Effect of plant growth-promoting rhizobacteria on alleviating salinity stress in plants: a review. J. Plant Nutr. 46, 2525–2550. doi: 10.1080/01904167.2022.2155548
Kumari, S., Vaishnav, A., Jain, S., Varma, A., and Choudhary, D. K. (2016). Induced drought tolerance through wild and mutant bacterial strain Pseudomonas simiae in mung bean (Vigna radiata L.). World J. Microbiol. Biotechnol. 32:4. doi: 10.1007/s11274-015-1974-3
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Li, Z., Fu, J., Zhou, R., and Wang, D. (2018). Effects of phenolic acids from ginseng rhizosphere on soil fungi structure, richness and diversity in consecutive monoculturing of ginseng. Saudi J. Biol. Sci. 25, 1788–1794. doi: 10.1016/j.sjbs.2018.07.007
Li, H., Guo, Q., Jing, Y., Liu, Z., Zheng, Z., Sun, Y., et al. (2020). Application of Streptomyces pactum Act12 enhances drought resistance in wheat. J. Plant Growth Regul. 39, 122–132. doi: 10.1007/s00344-019-09968-z
Lin, Y., Watts, D. B., Kloepper, J. W., Feng, Y., and Torbert, H. A. (2020). Influence of plant growth-promoting Rhizobacteria on corn growth under drought stress. Commun. Soil Sci. Plant Anal. 51, 250–264. doi: 10.1080/00103624.2019.1705329
Liu, B., Liang, J., Tang, G., Wang, X., Liu, F., and Zhao, D. (2019). Drought stress affects on growth, water use efficiency, gas exchange and chlorophyll fluorescence of Juglans rootstocks. Sci. Hortic. 250, 230–235. doi: 10.1016/j.scienta.2019.02.056
Loper, J. E., and Schroth, M. N. (1986). Influence of bacterial sources of Indole-3-acetic acid on root elongation of sugar beet. Phytopathology 76, 386–389. doi: 10.1094/Phyto-76-386
Lugtenberg, B., and Kamilova, F. (2009). Plant-growth-promoting Rhizobacteria. Ann. Rev. Microbiol. 63, 541–556. doi: 10.1146/annurev.micro.62.081307.162918
Magotra, S., Bhagat, N., Ambardar, S., Ali, T., Hurek, B. R., Hurek, T., et al. (2021). Field evaluation of PGP Bacillus sp. strain D5 native to Crocus sativus, in traditional and non traditional areas, and mining of PGP genes from its genome. Sci. Rep. 11:5454. doi: 10.1038/s41598-021-84585-z
Mahmood, T., Khalid, S., Abdullah, M., Ahmed, Z., Shah, M. K. N., Ghafoor, A., et al. (2019). Insights into drought stress signaling in plants and the molecular genetic basis of cotton drought tolerance. Cells 9:105. doi: 10.3390/cells9010105
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Gen. Genomics. 283, 185–196. doi: 10.1007/s00438-009-0506-y
Mayak, S., Tirosh, T., and Glick, B. R. (2004). Plant growth-promoting bacteria that confer resistance to water stress in tomatoes and peppers. Plant Sci. 166, 525–530. doi: 10.1016/j.plantsci.2003.10.025
Meudt, W. J., and Gaines, T. P. (1967). Studies on the oxidation of Indole-3-acetic acid by peroxidase enzymes. I. Colorimetric determination of Indole-3-acetic acid oxidation products. Plant Physiol. 42, 1395–1399. doi: 10.1104/pp.42.10.1395
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signalling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mishra, S. K., Khan, M. H., Misra, S., Dixit, V. K., Gupta, S., Tiwari, S., et al. (2020). Drought tolerant Ochrobactrum sp. inoculation performs multiple roles in maintaining the homeostasis in Zea mays L. subjected to deficit water stress. Plant Physiol. Biochem. 150, 1–14. doi: 10.1016/j.plaphy.2020.02.025
Misra, S., and Chauhan, P. S. (2020). ACC deaminase-producing rhizosphere competent Bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. Three Biotech 10:119. doi: 10.1007/s13205-020-2104-y
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Muhammad Usman Aslam, S. A., Raza, M. A. S., Saleem, M. F., Waqas, M., Iqbal, R., and Haider, I. (2020). Improving strategic growth stage-based drought tolerance in quinoa by Rhizobacterial inoculation. Commun. Soil Sci. Plant Anal. 51, 853–868. doi: 10.1080/00103624.2020.1744634
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. IJMS 21:5208. doi: 10.3390/ijms21155208
Nagarajkumar, M., Bhaskaran, R., and Velazhahan, R. (2004). Involvement of secondary metabolites and extracellular lytic enzymes produced by Pseudomonas fluorescens in inhibition of Rhizoctonia solani, the rice sheath blight pathogen. Microbiol. Res. 159, 73–81. doi: 10.1016/j.micres.2004.01.005
Nakabayashi, R., and Saito, K. (2015). Integrated metabolomics for abiotic stress responses in plants. Curr. Opin. Plant Biol. 24, 10–16. doi: 10.1016/j.pbi.2015.01.003
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophy. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Naseem, H., Ahsan, M., Shahid, M. A., and Khan, N. (2018). Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J. Basic Microbiol. 58, 1009–1022. doi: 10.1002/jobm.201800309
Nawrocki, E. P., Burge, S. W., Bateman, A., Daub, J., Eberhardt, R. Y., Eddy, S. R., et al. (2015). Rfam 12.0: updates to the RNA families database. Nucleic Acids Res. 43, D130–D137. doi: 10.1093/nar/gku1063
Ning, J., Li, X., Hicks, L. M., and Xiong, L. (2010). A Raf-like MAPKKK gene DSM1 mediates drought resistance through reactive oxygen species scavenging in Rice. Plant Physiol. 152, 876–890. doi: 10.1104/pp.109.149856
Niu, X., Song, L., Xiao, Y., and Ge, W. (2018). Drought-tolerant plant growth-promoting Rhizobacteria associated with foxtail millet in a semi-arid agroecosystem and their potential in alleviating drought stress. Front. Microbiol. 8:2580. doi: 10.3389/fmicb.2017.02580
Noctor, G., Mhamdi, A., and Foyer, C. H. (2014). The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 164, 1636–1648. doi: 10.1104/pp.113.233478
Novo, L. A. B., Castro, P. M. L., Alvarenga, P., and Da Silva, E. F. (2018). “Plant growth–promoting Rhizobacteria-assisted phytoremediation of mine soils” in Bio-geotechnologies for mine site rehabilitation (Amsterdam, Netherlands: Elsevier), 281–295.
Ojuederie, O. B., and Babalola, O. O. (2017). Microbial and plant-assisted bioremediation of heavy metal polluted environments: a review. Int. J. Environ. Res. Public Health 14:504. doi: 10.3390/ijerph14121504
Olsen, S. R., Cole, C. V., and Watanabe, F. S. (1954). Estimation of available phosphorus in soils by extraction with sodium bicarbonate. Washington DC: US Government Printing Office.
Omara, A. E.-D., Hafez, E. M., Osman, H. S., Rashwan, E., El-Said, M. A. A., Alharbi, K., et al. (2022). Collaborative impact of compost and beneficial Rhizobacteria on soil properties, physiological attributes, and productivity of wheat subjected to deficit irrigation in salt affected soil. Plan. Theory 11:877. doi: 10.3390/plants11070877
Overbeek, R. (2005). The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 33, 5691–5702. doi: 10.1093/nar/gki866
Pallai, R. (2005). Effect of plant growth-promoting rhizobacteria on canola (Brassica napus L.) and lentil (Lens culinaris Medik) plants. Department of Applied Microbiology and Food Science, University of Saskatchewan, Saskatoon, Canada.
Park, Y.-G., Mun, B.-G., Kang, S.-M., Hussain, A., Shahzad, R., Seo, C.-W., et al. (2017). Bacillus aryabhattai SRB02 tolerates oxidative and nitrosative stress and promotes the growth of soybean by modulating the production of phytohormones. PLoS One 12:e0173203. doi: 10.1371/journal.pone.0173203
Punja, Z. K., Wan, A., Rahman, M., Goswami, R. S., Barasubiye, T., Seifert, K. A., et al. (2008). Growth, population dynamics, and diversity of Fusarium equiseti in ginseng fields. Eur. J. Plant Pathol. 121, 173–184. doi: 10.1007/s10658-007-9261-2
Raineri, J., Wang, S., Peleg, Z., Blumwald, E., and Chan, R. L. (2015). The rice transcription factor OsWRKY47 is a positive regulator of the response to water deficit stress. Plant Mol. Biol. 88, 401–413. doi: 10.1007/s11103-015-0329-7
Richardson, A. E., and Simpson, R. J. (2011). Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol. 156, 989–996. doi: 10.1104/pp.111.175448
Rodríguez-Salazar, J., Suárez, R., Caballero-Mellado, J., and Iturriaga, G. (2009). Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett. 296, 52–59. doi: 10.1111/j.1574-6968.2009.01614.x
Ryu, C.-M., Farag, M. A., Hu, C.-H., Reddy, M. S., Wei, H.-X., Paré, P. W., et al. (2003). Bacterial volatiles promote growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 4927–4932. doi: 10.1073/pnas.0730845100
Saakre, M., Baburao, T. M., Salim, A. P., Ffancies, R. M., Achuthan, V. P., Thomas, G., et al. (2017). Identification and characterization of genes responsible for drought tolerance in Rice mediated by Pseudomonas fluorescens. Rice Sci. 24, 291–298. doi: 10.1016/j.rsci.2017.04.005
Sandhya, V., Shaik, Z. A., Grover, M., Reddy, G., and Venkateswarlu, B. (2009). Alleviation of drought stress effects in sunflower seedlings by the exopolysaccharides producing Pseudomonas putida strain GAP-P45. Biol. Fertil. Soils 46, 17–26. doi: 10.1007/s00374-009-0401-z
Schattner, P., Brooks, A. N., and Lowe, T. M. (2005). The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33, W686–W689. doi: 10.1093/nar/gki366
Sen, S., Ghosh, D., and Mohapatra, S. (2018). Modulation of polyamine biosynthesis in Arabidopsis thaliana by a drought mitigating Pseudomonas putida strain. Plant Physiol. Biochem. 129, 180–188. doi: 10.1016/j.plaphy.2018.05.034
Shahbandeh, M. (2024). World rice acreage from 2010 to 2022 (in million hectares) : Statista. Available at: https://www.statista.com/statistics/271969/world-rice-acreage-since-2008/
Shao, H., Wang, H., and Tang, X. (2015). NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front. Plant Sci. 6:902. doi: 10.3389/fpls.2015.00902
Sharma, P., Jha, A. B., Dubey, R. S., and Pessarakli, M. (2012). Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Bot. 2012:217037, 1–26. doi: 10.1155/2012/217037
Sharma, A., and Johri, B. N. (2003). Growth promoting influence of siderophore-producing Pseudomonas strains GRP3A and PRS9 in maize (Zea mays L.) under iron limiting conditions. Microbiol. Res. 158, 243–248. doi: 10.1078/0944-5013-00197
Shi, F., Yin, Z., Jiang, H., and Fan, B. (2014). Screening, identification of P-dissolving fungus P83 strain and its effects on phosphate solubilization and plant growth promotion. Wei Sheng Wu Xue Bao 54, 1333–1343
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Singh, P., Chauhan, P. K., Upadhyay, S. K., Singh, R. K., Dwivedi, P., Wang, J., et al. (2022). Mechanistic insights and potential use of siderophores producing microbes in rhizosphere for mitigation of stress in plants grown in degraded Land. Front. Microbiol. 13:898979. doi: 10.3389/fmicb.2022.898979
Sivasakthi, S., Kanchana, D., Usharani, G., and Saranraj, P. (2013). Production of plant growth promoting substance by Pseudomonas fluorescens and Bacillus subtilis isolates from Paddy rhizosphere soil of Cuddalore District, Tamil Nadu, India. Int. J. Microbiol. Res. 4, 227–233.Available at: https://api.semanticscholar.org/CorpusID:1419152
Steffens, B. (2014). The role of ethylene and ROS in salinity, heavy metal, and flooding responses in rice. Front. Plant Sci. 5:685. doi: 10.3389/fpls.2014.00685
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tewari, S., and Arora, N. K. (2014). Multifunctional exopolysaccharides from Pseudomonas aeruginosa PF23 involved in plant growth stimulation, biocontrol and stress amelioration in sunflower under saline conditions. Curr. Microbiol. 69, 484–494. doi: 10.1007/s00284-014-0612-x
The UniProt Consortium (2015). UniProt: a hub for protein information. Nucleic Acids Research. 43, D204–D212. doi: 10.1093/nar/gku989
Tiepo, A. N., Hertel, M. F., Rocha, S. S., Calzavara, A. K., Oliveira, A. L. M. D., Pimenta, J. A., et al. (2018). Enhanced drought tolerance in seedlings of Neotropical tree species inoculated with plant growth-promoting bacteria. Plant Physiol. Biochem. 130, 277–288. doi: 10.1016/j.plaphy.2018.07.021
Tiwari, P., Srivastava, D., Chauhan, A. S., Indoliya, Y., Singh, P. K., Tiwari, S., et al. (2021). Root system architecture, physiological analysis and dynamic transcriptomics unravel the drought-responsive traits in rice genotypes. Ecotoxicol. Environ. Saf. 207:111252. doi: 10.1016/j.ecoenv.2020.111252
Upadhyay, S. K., Singh, J. S., Saxena, A. K., and Singh, D. P. (2012). Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol. 14, 605–611. doi: 10.1111/j.1438-8677.2011.00533.x
Vaishnav, A., and Choudhary, D. K. (2019). Regulation of drought-responsive gene expression in Glycine max L. Merrill is mediated through Pseudomonas simiae strain AU. J. Plant Growth Regul. 38, 333–342. doi: 10.1007/s00344-018-9846-3
Vargas, L., Santa Brígida, A. B., Mota Filho, J. P., De Carvalho, T. G., Rojas, C. A., Vaneechoutte, D., et al. (2014). Drought tolerance conferred to sugarcane by association with Gluconacetobacter diazotrophicus: a transcriptomic view of hormone pathways. PLoS One 9:e114744. doi: 10.1371/journal.pone.0114744
Vejan, P., Abdullah, R., Khadiran, T., Ismail, S., and Nasrulhaq Boyce, A. (2016). Role of plant growth promoting Rhizobacteria in agricultural sustainability—a review. Molecules 21:573. doi: 10.3390/molecules21050573
Venturi, V., and Keel, C. (2016). Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. doi: 10.1016/j.tplants.2016.01.005
Vurukonda, S. S. K. P., Vardharajula, S., Shrivastava, M., and SkZ, A. (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184, 13–24. doi: 10.1016/j.micres.2015.12.003
Woo, O.-G., Kim, H., Kim, J.-S., Keum, H. L., Lee, K.-C., Sul, W. J., et al. (2020). Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiol. Biochem. 148, 359–367. doi: 10.1016/j.plaphy.2020.01.032
Xiang, Y., Tang, N., Du, H., Ye, H., and Xiong, L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring Abscisic acid sensitivity and salinity and drought tolerance in Rice. Plant Physiol. 148, 1938–1952. doi: 10.1104/pp.108.128199
Yang, X., Lu, M., Wang, Y., Wang, Y., Liu, Z., and Chen, S. (2021). Response mechanism of plants to drought stress. Horticulturae 7:50. doi: 10.3390/horticulturae7030050
Yoshida, T., Fujita, Y., Sayama, H., Kidokoro, S., Maruyama, K., Mizoi, J., et al. (2010). AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 61, 672–685. doi: 10.1111/j.1365-313X.2009.04092.x
Yuan, X., Wang, H., Cai, J., Bi, Y., Li, D., and Song, F. (2019). Rice NAC transcription factor ONAC066 functions as a positive regulator of drought and oxidative stress response. BMC Plant Biol. 19:278. doi: 10.1186/s12870-019-1883-y
Keywords: Bacillus megaterium, drought, plant growth-promoting rhizobacteria, rice, stress-responsive gene
Citation: Lee S, Kim J-A, Song J, Choe S, Jang G and Kim Y (2024) Plant growth-promoting rhizobacterium Bacillus megaterium modulates the expression of antioxidant-related and drought-responsive genes to protect rice (Oryza sativa L.) from drought. Front. Microbiol. 15:1430546. doi: 10.3389/fmicb.2024.1430546
Edited by:
Debasis Mitra, Graphic Era University, IndiaReviewed by:
Nagaraju Yalavarthi, Central Silk Board, IndiaNivetha Nagarajan, Sea6 Energy Pvt. Ltd., India
Sumera Yasmin, National Institute for Biotechnology and Genetic Engineering, Pakistan
Copyright © 2024 Lee, Kim, Song, Choe, Jang and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangseon Kim, eWFuZ3Nlb25AY2lhbG0ub3Iua3I=
†ORCID: Jung-Ae Kim, https://orcid.org/0000-0002-0694-477X
Jeongsup Song, https://orcid.org/0000-0003-0916-5613
Seonbong Choe, https://orcid.org/0009-0004-1257-1588
Geupil Jang, https://orcid.org/0000-0002-6231-828X
 Sanghun Lee
Sanghun Lee Jung-Ae Kim1†
Jung-Ae Kim1† Yangseon Kim
Yangseon Kim