- 1Center on Biologics Development and Evaluation, Department of Anatomy and Physiology, College of Veterinary Medicine, Kansas State University, Manhattan, KS, United States
- 2BioApplications Inc., Pohang-si, Republic of Korea
- 3MEDEXX Co., Ltd., Seongnam-si, Republic of Korea
- 4Celltrix Co., Ltd., Seongnam-si, Republic of Korea
- 5Department of Diagnostic Medicine and Pathobiology, College of Veterinary Medicine, Kansas State University, Manhattan, KS, United States
Introduction: African swine fever (ASF) is a lethal and highly contagious transboundary animal disease with the potential for rapid international spread. In the absence of a widely available and definitively proven vaccine, rapid and early detection is critical for ASF control. The quick and user-friendly lateral flow assay (LFA) can easily be performed by following simple instructions and is ideal for on-site use. This study describes the development and validation of two LFAs for the rapid detection of ASF virus (ASFV) in pig serum.
Methods: The highly immunogenic antigens (p30 and p72) of ASFV Georgia 2007/1 (genotype II) were expressed in plants (Nicotiana benthamiana) and were used to immunize BALB/c mice to generate specific monoclonal antibodies (mAbs) against the p30 and p72 proteins. mAbs with the strongest binding ability to each protein were used to develop p30_LFA and p72_LFA for detecting the respective ASFV antigens. The assays were first evaluated using a spike-in test by adding the purified p30 or p72 protein to a serum sample from a healthy donor pig. Further validation of the tests was carried out using serum samples derived from experimentally infected domestic pigs, field domestic pigs, and feral pigs, and the results were compared with those of ASFV real-time PCR.
Results: p30_LFA and p72_LFA showed no cross-reaction with common swine viruses and delivered visual results in 15 min. When testing with serially diluted proteins in swine serum samples, analytical sensitivity reached 10 ng/test for p30_LFA and 20 ng/test for p72_LFA. Using real-time PCR as a reference, both assays demonstrated high sensitivity (84.21% for p30_LFA and 100% for p72_LFA) with experimentally ASFV-infected pig sera. Specificity was 100% for both LFAs using a panel of PBS-inoculated domestic pig sera. Excellent specificity was also shown for field domestic pig sera (100% for p30_LFA and 93% for p72_LFA) and feral pig sera (100% for both LFAs).
Conclusion: The results obtained in this study suggest that p30_LFA and p72_LFA hold promise as rapid, sensitive, user-friendly, and field-deployable tools for ASF control, particularly in settings with limited laboratory resources.
1 Introduction
ASF is a devastating and highly contagious transboundary animal disease with the potential for rapid international spread (Dixon et al., 2020). The causative agent, African swine fever virus (ASFV), is a large, double-stranded DNA virus belonging to the Asfarviridae family. This complex virus genome is approximately 170–194 kilobase pairs (kbp) and contains over 150 open reading frames (ORFs), depending on the strain (Karger et al., 2019; Wang et al., 2021; Li et al., 2022). While warthogs and specific soft ticks act as natural reservoirs for ASFV, harboring the virus with no signs of illness, domestic pigs face a different fate. Infection in domestic pigs triggers a severe and often fatal disease with high mortality rates (Karger et al., 2019; Netherton et al., 2019; Wang Y. et al., 2021; Li et al., 2022). Based on the p72 major capsid p72 protein gene (B646L), 24 ASFV genotypes (I–XXIV) have been identified (Quembo et al., 2018). Genotype II ASFV that emerged in the Caucasus region in 2007 is responsible for the contemporary pandemic. The situation worsened dramatically when ASF reached China in 2018, swiftly spreading across Asia (Le et al., 2019; Berends et al., 2021; Mighell and Ward, 2021). Notably, the disease re-emerged in the Caribbean in 2021, impacting the Dominican Republic and Haiti after approximately four decades of absence (Jean-Pierre et al., 2022; Ramirez-Medina et al., 2022). Currently, ASF remains widespread in sub-Saharan Africa, parts of West Africa, and Sardinia and continues to spread in Europe, Asia, the Pacific, and the Caribbean regions (Shi et al., 2021; Wang et al., 2023).
The lack of effective tools makes prevention and control extremely challenging. Attempts to immunize animals using vaccine formulations prepared by conventional means and comprising infected cell extracts, supernatants of infected pig peripheral blood leukocytes, purified and inactivated virions, infected glutaraldehyde-fixed macrophages, or detergent-treated infected alveolar macrophages failed to induce protective immunity (Escribano et al., 2013; Rock, 2021; Pikalo et al., 2022). Similarly, subunit vaccines targeting specific viral proteins, even in combination, have not provided complete protection (Gaudreault and Richt, 2019; Rock, 2021). Live-attenuated vaccine (LAVs) candidates, though promising, face hurdles related to stable production, safety concerns, and the lack of methods to differentiate between vaccinated and infected animals (DIVAs) (Bosch-Camós et al., 2020; Rock, 2021; Wang T. et al., 2021; Brake, 2022; Truong et al., 2023). There is currently no ASF vaccine commercially available outside of Vietnam. Consequently, control strategies currently rely heavily on strict sanitation measures and the culling of infected and potentially exposed animals (Gallardo et al., 2015; Wang et al., 2023). Therefore, rapid and accurate diagnosis of ASFV infection is urgently required for the prevention, control, and eradication of the disease in affected countries. The OIE-recommended tests for ASFV detection include virus isolation, fluorescent antibody testing, real-time PCR, and conventional PCR (World Organisation for Animal Health (OIE), 2023). However, these methods are time-consuming and require well-equipped laboratories and trained personnel, which can lead to delays in disease diagnosis in remote or underserved areas. Newer molecular tests, such as portable PCR and LAMP assays, offer promise for field use but still require some technical expertise for nucleic acid extraction (Fan et al., 2020; Fu et al., 2021; Yang et al., 2022; Bohorquez et al., 2023).
LFAs offer a compelling solution for detecting pathogens in the field, especially in resource-limited settings. These tests are rapid, inexpensive, and user-friendly, requiring minimal training and equipment. They are ideal for point-of-care testing (POCT), providing results quickly and conveniently. Several lateral flow assays (LFAs) have been developed for detecting various viral and bacterial antigens (Magambo et al., 2014; Carrio et al., 2015; Schramm et al., 2015; Ang et al., 2016; Onyilagha et al., 2022). In this report, we expressed the highly immunogenic antigens (p30 and p72) of ASFV Georgia 2007/1 (genotype II) in the well-established Nicotiana benthamiana plant expression system (Alcaraz et al., 1990; Revilla et al., 2018; Park et al., 2019; Kim et al., 2023; Shin et al., 2023). Plant-based systems offer several advantages for recombinant protein production compared to bacterial or mammalian cell platforms. These include safety, low cost, compatibility with green technologies, minimal contamination risks, and widespread societal acceptance (Lee and Ko, 2017; Islam et al., 2019; Kim et al., 2023). Importantly, the potential for large-scale production, optimized growth conditions, low cultivation costs, and the ability to produce complex proteins further highlight the benefits of plant expression systems for recombinant protein manufacturing (Burnett and Burnett, 2020). By utilizing mAbs targeting plant-expressed ASFV p30 and p72 proteins, we successfully developed two novel LFAs for ASFV detection. These LFAs demonstrate high sensitivity and excellent specificity and are suitable for rapid, user-friendly, and field-deployable ASF surveillance.
2 Materials and methods
2.1 Animals, viruses, and cells
Specific pathogen-free female Balb/c female mice (6 weeks old) were purchased from Orient Bio, Sungnam, Korea. All animal experiments were authorized by the Institutional Animal Care and Use Committee of MEDEXX (IACUC# AEC-20160713-0001). All animal experiments were performed under strict adherence to the IACUC protocol.
Murine myeloma cell line Sp2/0Ag14 was purchased from the American Type Culture Collection (ATCC-CRL-1581, Rockville, MD, United States) and was maintained in RPMI-1640 (Gibco, New York, NY, United States) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Flowery Branch, GA, United States) at 37 °C with 5% CO2.
A virulent VNUA-ASFV-05 L1 strain (genotype II) was isolated from the spleen of a domestic pig with typical acute ASF during an ASF outbreak in Northern Vietnam in 2020 (Truong et al., 2021). It is maintained in BSL-3 laboratories of Kansas State University. This virus was used to make the standards for the Quantitative ASFV Real-Time PCR.
2.2 Porcine serum samples
This study utilized serum samples from domestic pigs and feral pigs in Dr. Jishu Shi’s laboratory at Kansas State University. These samples include the following:
i. ASFV negative pig sera: serum samples from pigs inoculated with phosphate-buffered saline (PBS, pH 7.4, Thermo Scientific, Bridgewater, NJ, United States).
ii. iASFV-infected pig sera: serum samples from pigs infected with the ASFV VNUA-ASFV-05 L1 at different time points: day 0, day 7, and the day of euthanasia.
iii. Other common swine virus-infected pig sera: serum samples from pigs infected with classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (PRV), and bovine viral diarrhea virus (BVDV).
iv. Feral pig sera: serum samples from feral pigs caught in Kansas (collaboration with USDA APHIS Wildlife Services, Kansas Wildlife Services, United States).
Additionally, serum samples from field pigs with no known exposure to ASFV were collected and tested with the presented LFAs in four farms within South Korea.
2.3 Protein expression and generation of monoclonal antibodies
DNA sequences encoding p30 and p72 protein of ASFV Georgia 2007/1 (GenBank accession number FR682468.2) were codon optimized for N. benthamiana. For the purification, six histidine tags or porcine Fc domain were fused at the C-terminus of p30 or p72, respectively. Each fusion gene was additionally fused with the ER signal sequence of binding protein (BiP) and then inserted into the pCAMBIA1300 vector harboring CaMV 35 s promoter and NoS terminator. Expression of each protein in N. benthamiana was performed according to a previously described protocol (Sohn et al., 2023). To confirm the purity of the isolated proteins, samples were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 10% gel. Gels were then stained with Coomassie Brilliant Blue R-250 (BioSolutions, Suwon, South Korea) according to the manufacturer’s instructions.
For the generation of mAbs against p30 and p72 proteins, 100 μL of protein p30 or p72 (2.5 μg/mL) mixed with an equal volume of Freund’s complete adjuvant (Sigma-Aldrich Inc., St. Louis, MO, United States) was used as immunogens to inject (intraperitoneal injection, IP) each of five female Balb/c mice. Two booster injections, each with the same protein dose and equal volume of Freund’s incomplete adjuvant (Sigma-Aldrich Inc., St. Louis, MO, United States), were administered at 2-week intervals. One week after the second booster, blood samples were collected for antibody titer testing. The mouse with the highest antibody titer received a final injection of 2.5 μg protein without adjuvant. Three days later, these mice were euthanized, and their spleen cells were fused with SP2/0Ag14 cells using 50% polyethylene glycol (Sigma-Aldrich Inc., St. Louis, MO, United States) at a 5:1 ratio. Following fusion, cells were resuspended in a HAT-selective medium (Sigma-Aldrich Inc., St. Louis, MO, United States) at a concentration of 105 cells/ml. Then, 100 μL of this cell suspension was added to each well of a 96-well plate and incubated at 37°C with 5% CO2. After 10 days, culture supernatants were screened for the presence of antibodies against p30 or p72 using an indirect ELISA. Positive wells were subjected to multiple rounds of single-cell cloning through limiting dilution until monoclonals were achieved.
2.4 Indirect ELISA
ELISA plates were coated overnight at 4°C with 100 μL/well of protein (1 μg/mL) in PBS (pH 7.4, Thermo Scientific, Bridgewater, NJ, United States). To perform the ELISA assay for screening the hybridomas, the plates were washed three times with PBS containing 0.05% Tween 20, blocked with 300 μL of 5% skimmed milk in PBS, and incubated at 37°C for 1 h. After washing, the plates were incubated with 100 μL/well of primary Ab (culture supernatants) for 1 h. After washing, the plates were incubated with secondary Ab (0.8 mg/mL at 1:10,000 dilution Goat anti-Mouse IgG HRP, 100 μL/well) at 37°C for 1 h. Enzyme assay was performed using TMB (Invitrogen, Carlsbad, CA, United States). The reaction was allowed to occur for 10 min and then stopped with 100 μL/well of 1 M sulfuric acid. The absorbance at 450 nm was measured using a microtiter plate reader.
2.5 Nucleic acid extraction and quantitative ASFV real-time PCR
Viral nucleic acids were extracted using an automated King Fisher™ Duo Prime DNA/RNA extraction system (Thermo Fisher Scientific, Waltham, MA, United States) with MagMAX™ Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, MA, United States), according to the manufacturer’s protocols. Serially diluted Genotype II ASFVs were added to the extraction plate as standards for quantification. ASFV DNA was then detected using Path-ID Multiplex One-Step RT-PCR Kit (Applied Biosystems, Grand Island, NY, United States) on StepOnePlus™ Real-Time PCR System (Applied Biosystems, Grand Island, NY, United States) using previously described primers and probes (Shi et al., 2016). The reaction condition involved a 95°C incubation for 5 min, followed by 45 cycles of denaturation at 95°C for 15 s and a combined annealing and extension step at 60°C for 45 s. Upon completion, standard curves, Ct values, and virus quantities in each sample were recorded. All experiments were performed in duplicate.
2.6 Development and assembly of the lateral flow device
Lateral flow device development and assembly incorporated minor modifications from previously described methods (Sastre et al., 2016). Briefly, a dispenser applied specific capture solutions: goat anti-rabbit IgG (1 mg/mL) for the control line and either anti-p30 mAb 89G6 (2 mg/mL) or anti-p72 mAb 5G11 (2 mg/mL) for the test line, at a rate of 1 μL/cm. The membranes were then dried for over 4 h at a low humidity (below 20%).
Colloidal gold conjugation began by heating 90 mL of distilled water to 100°C. After reaching boiling, 10 mL of 1% (w/v) gold chloride trihydrate and 1 mL of 1% sodium citrate tribasic dihydrate were added and stirred until completely dissolved. The solution turned a final red color and was then stirred for an additional 10 min at room temperature. Following dilution, the solution’s absorbance was measured using a spectrophotometer. Antibody conjugation involved adjusting the pH of 50 mL of colloidal gold solution to 7.2 with 0.1 M potassium carbonate. Either anti-p30 mAb 91G3 (7 μg/mL in PBS) or anti-p72 mAb 7D11 (7 μg/mL in PBS) was slowly added dropwise and stirred for 30 min. Subsequently, 5 mL of 10% (w/v) bovine serum albumin (BSA) was incorporated, and the mixture was stirred for another 30 min. The conjugate was centrifuged (10,000 rpm, 30 min) to separate the liquid (supernatant) from the solid (pellet). The pellet was then resuspended in 1% (w/v) BSA in PBS and stored in a refrigerator.
The conjugation pad was prepared with a solution containing 3% sucrose, 0.1% sodium azide, 1% BSA, and the appropriate antibody-gold conjugate (with an optical density (OD) of 2 at 540 nm) in PBS. Gold-conjugated rabbit IgG (OD 0.7 at 540 nm) was included for testing the control line. This solution was absorbed onto a glass fiber conjugate pad and dried for over 4 h at low humidity (below 20%). Finally, the membrane with capture antibodies, the conjugate pad, and a sample pad were assembled in a plastic housing to create the final LFA kit (Figure 1).
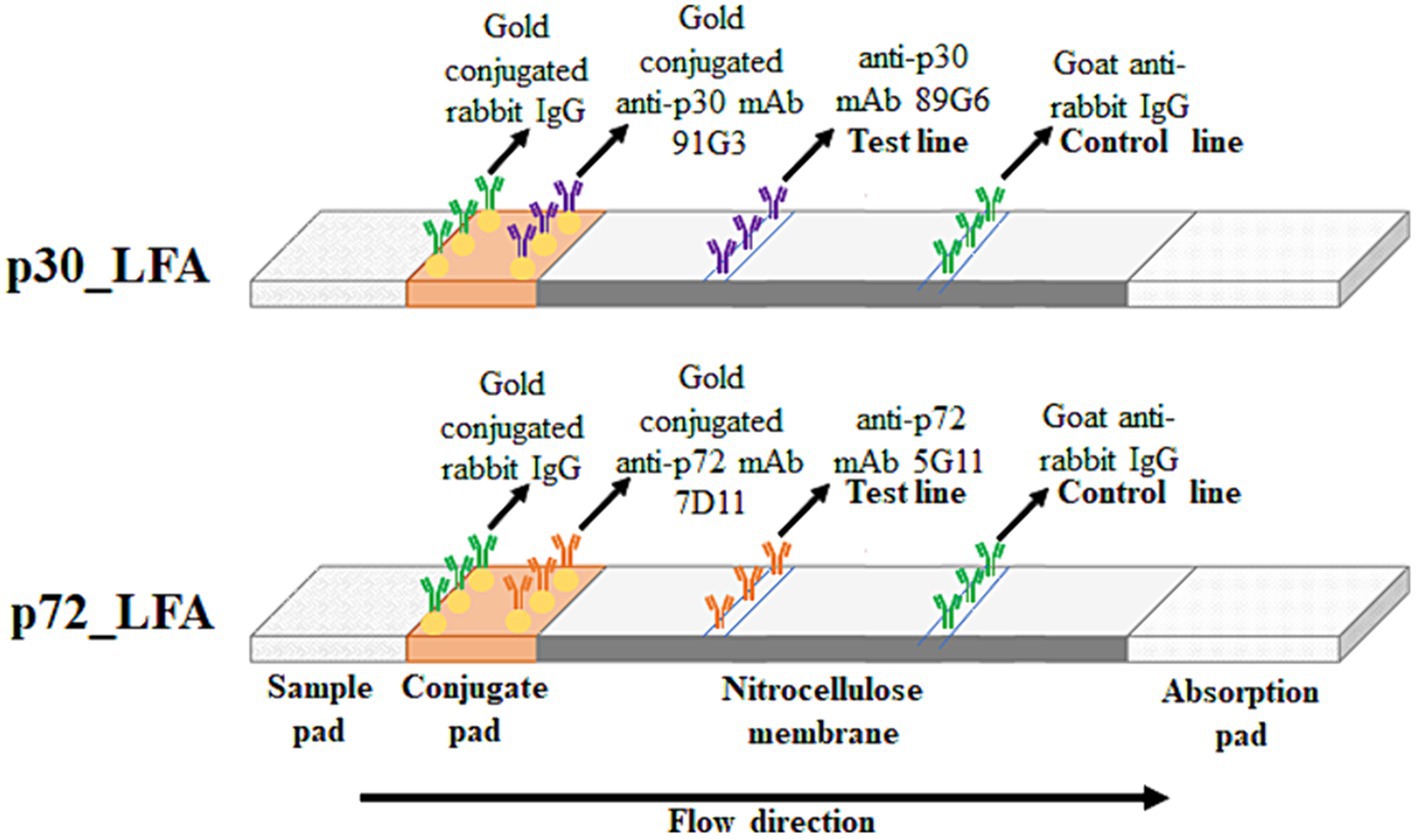
Figure 1. Schematic diagram of p30_LFA and p72_LFA. The sample (pig serum or plasma) migrates through the conjugate pad and the nitrocellulose membrane by capillarity. In the presence of ASFV, the p30/p72 protein is captured first by the gold-conjugated mAb91G3/mAb7D11, forming an antibody–antigen immune complex. This immune complex then reacts with the immobilized mAb89G6/mAb5G11 on the membrane, making the test line visible along with the control line (gold-conjugated rabbit IgG captured by goat anti-rabbit IgG). In the case of a negative test, only a control line appears.
2.7 Test procedure
The test is designed for use with porcine plasma or serum samples without dilution. Samples should be brought to room temperature (15–30°C) prior to test. A measure of 120 μL of the samples is applied to the sample pad. The sample will flow along the result window by capillarity. The results are interpreted 15 min after adding the sample. In the presence of ASFV, the test line (red/pink) is visible along with the red/pink control line. In the case of a negative test, only a red/pink control line appears. The control line must appear always. Otherwise, the test has to be considered invalid and needs to be repeated with a new LFA cassette.
2.8 Statistical analysis
For this study, quantitative ASFV real-time PCR was regarded as the standard (reference) for pathogen detection. Sensitivity and specificity analyses were carried out by the web-based MedCalc statistical software.1
3 Results
3.1 Development of the lateral flow assay
The ASFV proteins p30 and p72 were successfully expressed and purified from N. benthamiana as expected protein size (Figure 2). To establish the LFA test, we selected mAbs with the strongest binding ability to each plant-derived protein. A specific mAb for p72 (mAb 5G11) was used as the capture reagent on the test line for the p72_LFA. Similarly, a high-affinity mAb for p30 (mAb 89G6) was used on the test line in the p30_LFA. For increased sensitivity, we designed the LFA to avoid diluting the sample. Pig serum or plasma can be directly applied to the designated well on the device. After optimization, we established a user-friendly testing procedure:
i. Add 120 μL of the sample directly to the sample well (or use the provided dropper to add four drops).
ii. Wait for 15 min.
iii. Interpret the results in the designated window.
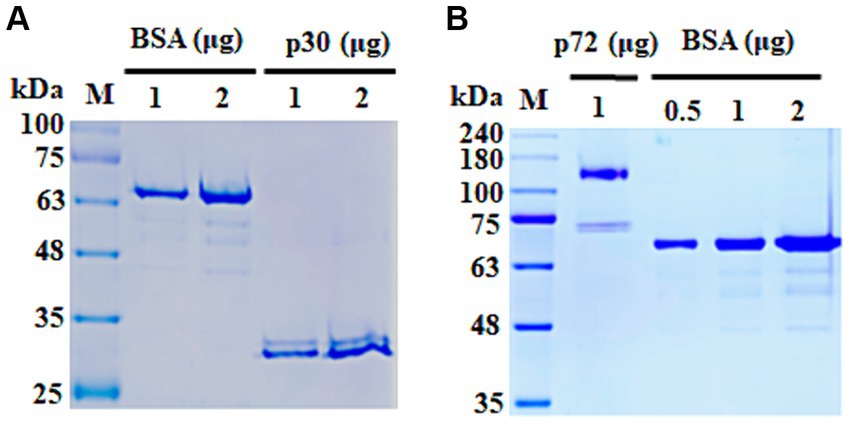
Figure 2. SDS-PAGE analysis of purified ASFV proteins. (A) Purified p30 protein. (B) Purified p72 protein. Bovine serum albumin (BSA) was used as a loading control. kDa, kilo Dalton.
The appearance of a pink test line and a pink control line indicates a positive result, while only the pink control line indicates a negative result (Figure 3).
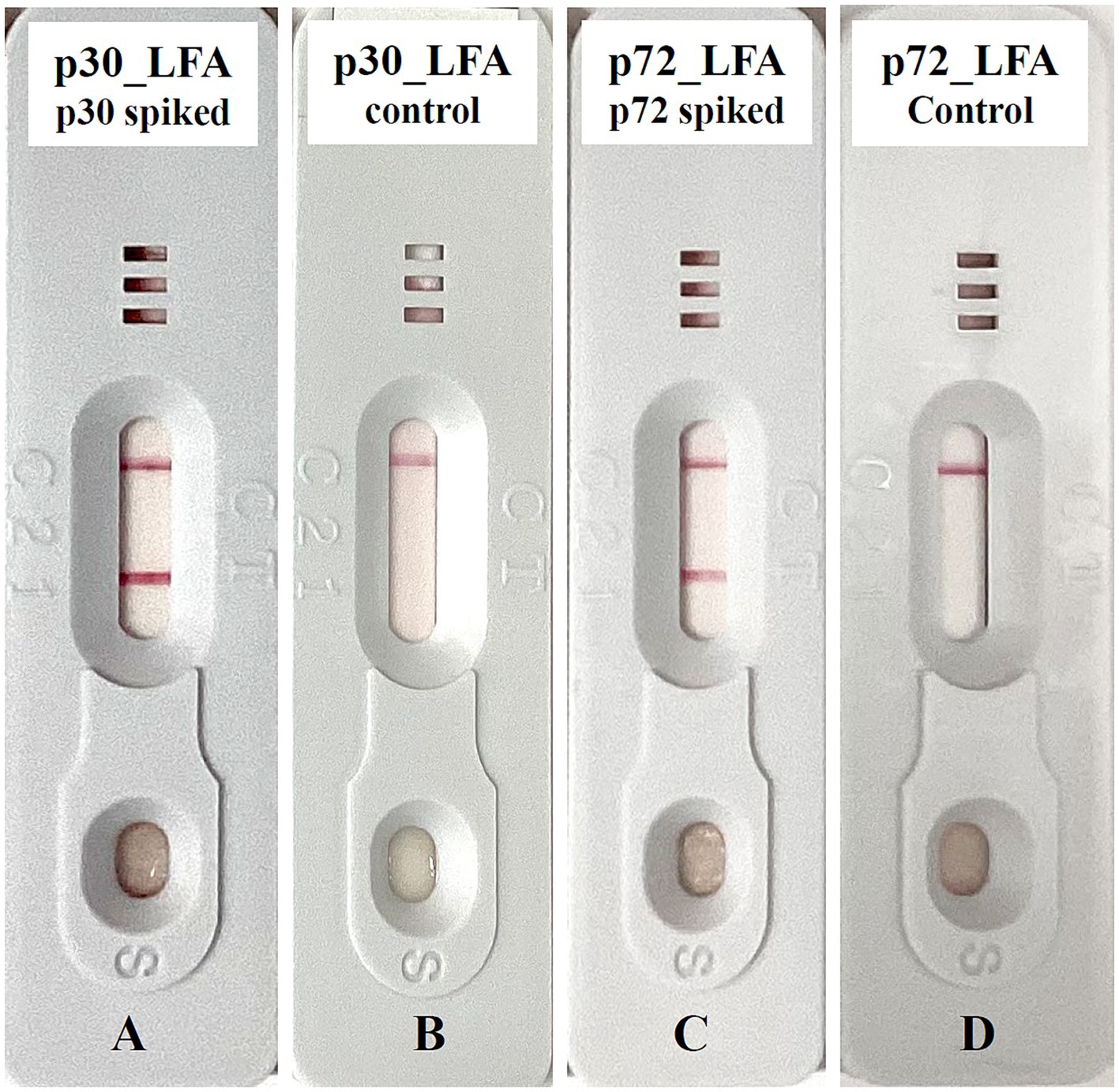
Figure 3. Exemplary lateral flow device for ASFV detection. (A) p30_LFA positive: test line and control line are detected. (B) p30_LFA is negative: only the control line is detected. (C) p72_LFA is positive: test line and control line are detected. (D) p72_LFA is negative: only the control line is detected.
3.2 Analytical specificity and sensitivity of p30_LFA and p72_LFA
To confirm that our p30_LFA and p72_LFA tests only detect ASFV and do not react to other viruses, we tested them with various serum sample categories. These categories include serum samples from pigs injected with PBS (n = 30), serum samples from pigs infected with CSFV (n = 12), serum samples from pigs infected with PRRSV (n = 12), serum samples from pigs infected with PRV (n = 10), and serum samples from pigs infected with BVDV (n = 2). The results are encouraging (Table 1). Both p30_LFA and p72_LFA tests are negative for all samples (100% specificity). This indicates that the tests are highly specific for ASFV and do not react with other common swine viruses.
In order to evaluate the analytical sensitivity, spike-in tests were carried out. We added known amounts of purified ASFV p30 and p72 proteins to serum samples from a healthy donor pig. We then progressively diluted these samples by two-fold serial dilutions. These diluted samples were then analyzed using both p30_LFA and p72_LFA. The p30_LFA test can detect ASFV p30 protein as low as 10 nanograms per test (ng/test) (Figure 4A). The p72_LFA test can detect ASFV p72 protein down to a concentration of 20 ng/test (Figure 4B).
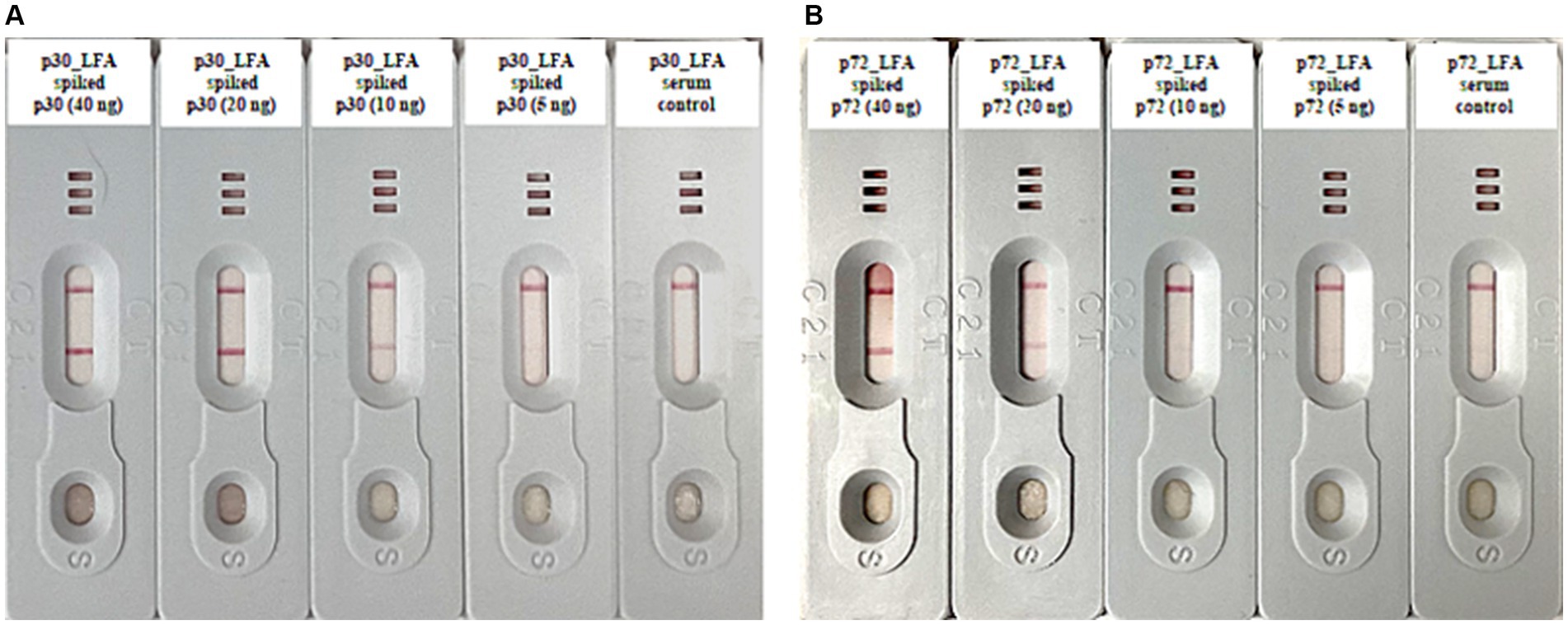
Figure 4. Analytical sensitivity tests of p30_LFA and p72_LFA. (A) Two-fold serial dilutions of purified ASFV p30 protein (40, 20, 10, 5, and 0 ng) in serum samples (from healthy donor pig) tested by p30_LFA. (B) Two-fold serial dilutions of purified ASFV p72 protein (40, 20, 10, 5, and 0 ng) in serum samples (from healthy donor pig) tested by p72_LFA.
3.3 Validate p30_LFA and p72_LFA with experimental samples
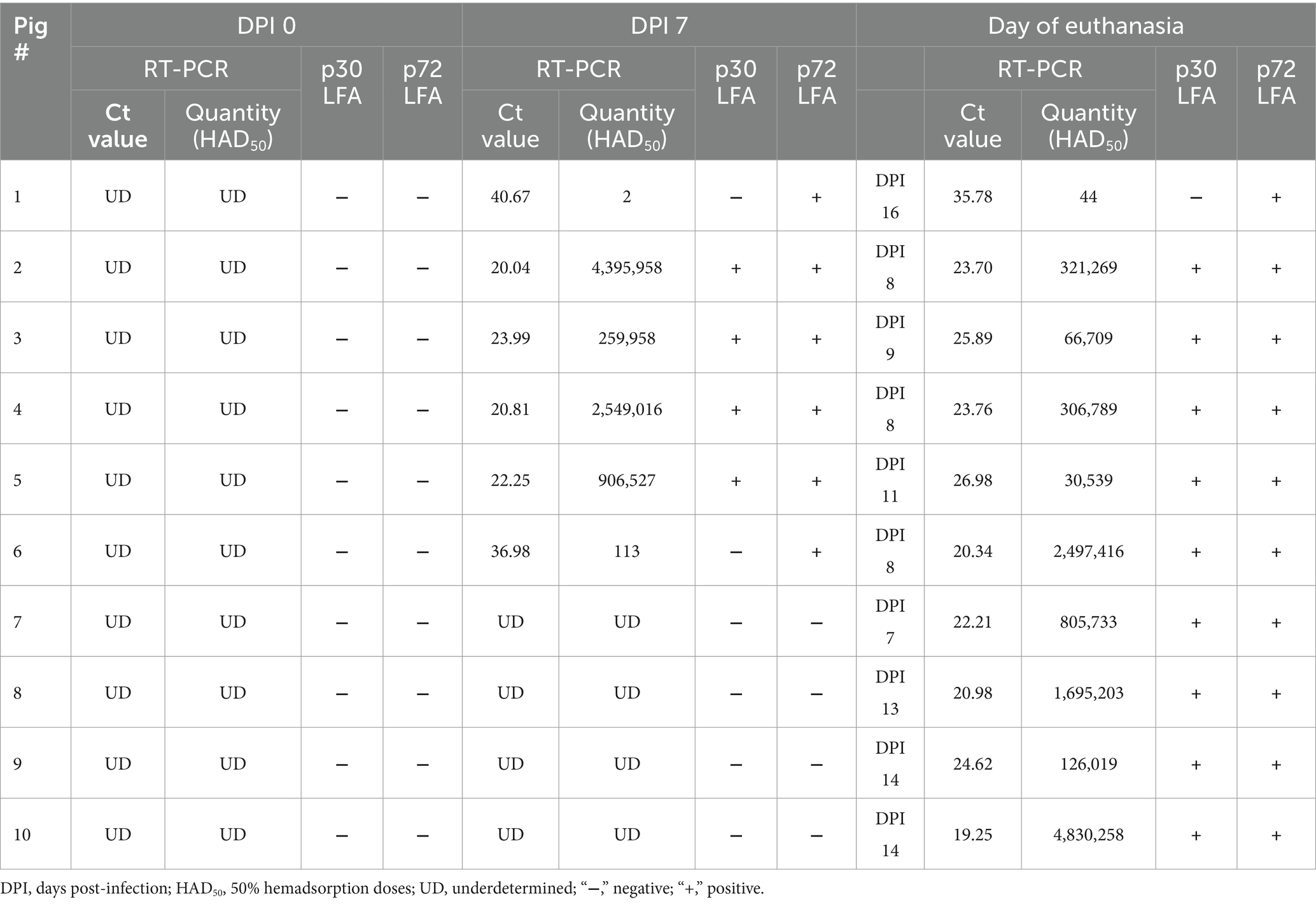
To further evaluate p30_LFA and p72_LFA tests, we used serum samples collected at 0-day post-infection (DPI), 7 DPI, and the day of euthanasia from 10 pigs infected with ASFV VNUA-ASFV-05 L1 (genotype II). This virus was isolated from a domestic pig during an ASF outbreak in Northern Vietnam in 2020 and caused typical clinical signs of acute ASF (Truong et al., 2021). Six pigs tested positive for ASFV using quantitative ASFV real-time PCR at 7 DPI and all pigs tested positive on the day they were euthanized (between 8 and 16 DPI). The amount of ASFV in their serum samples (viral load) varied, ranging from a high level (Ct values of 19.25, ASFV quantity 4,830,258 HAD50) to a very low level (Ct values of 40.67, ASFV quantity 2 HAD50) (Table 2).
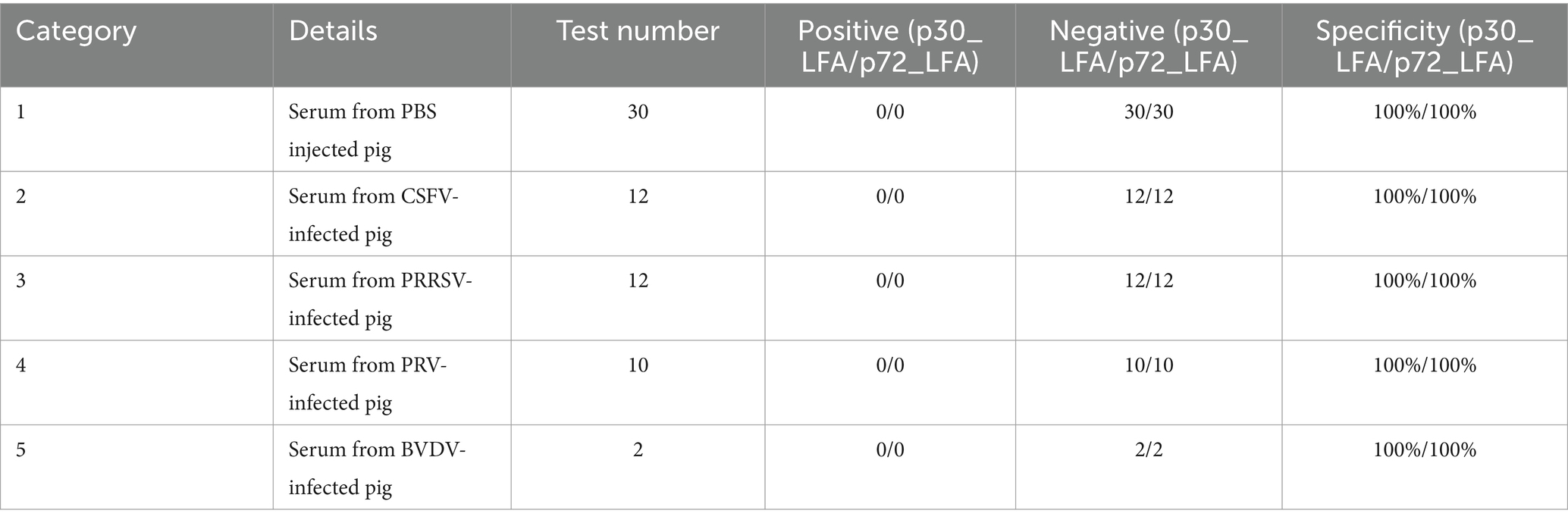
Table 2. Comparing results of p30_LFA and p72_LFA tests with the quantitative ASFV real-time PCR using serum samples from ASFV VNUA-ASFV-05 L1 (genotype II) experimentally infected pigs.
The results showed the following:
i. For p72_LFA: All serum samples that were tested positive by real-time PCR were also tested positive on the p72_LFA test. This indicates that the p72_LFA test has 100% sensitivity (95% confidence interval: 79.41 to 100%) for detecting ASFV in these samples.
ii. For p30_LFA: Three samples (pig #1 at 7 DPI and 16 DPI, pig#6 at 7 DPI) with very low viral loads (Ct values of 40.67, 35.78, and 36.98, respectively) tested negative for p30_LFA. This indicates that the p30_LFA test has 84.21% sensitivity (95% confidence interval: 60.42 to 96.62%) for detecting ASFV in these samples.
Importantly, both the p30_LFA and p72_LFA tests did not show any false positives, meaning they have 100% specificity for detecting ASFV in these samples.
3.4 Validate p30_LFA and p72_LFA with field samples
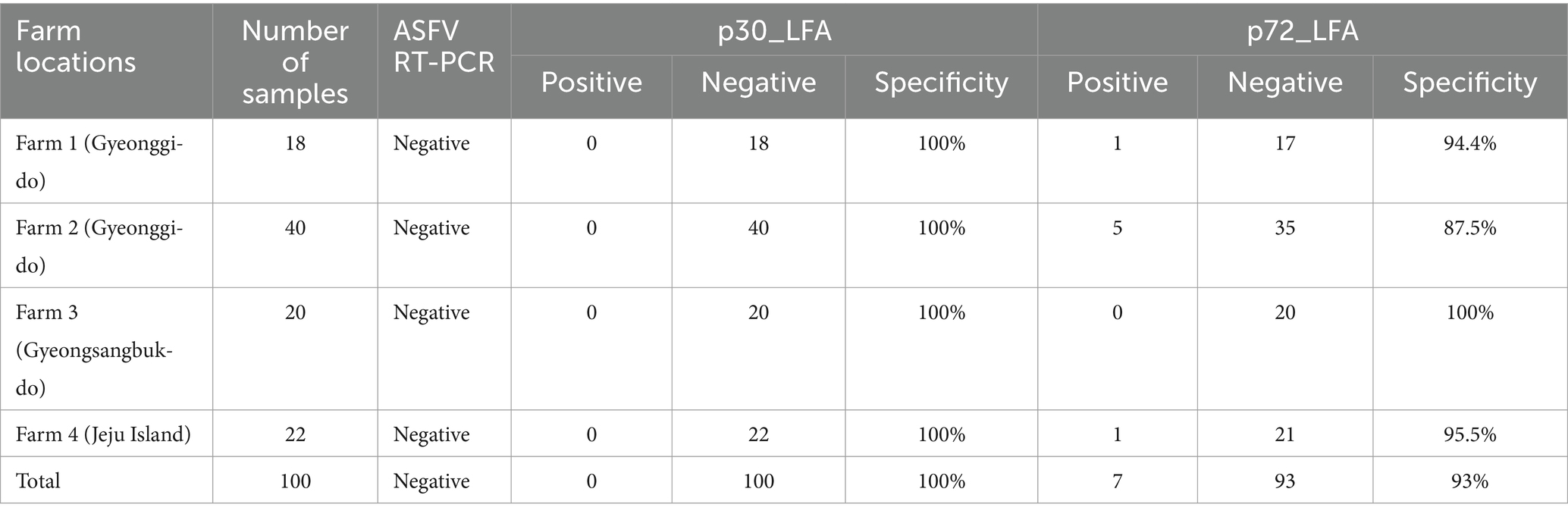
To validate the performance of p30_LFA and p72_LFA with field samples, we collected serum samples from domestic pigs (n = 100) across four farms in different geographical regions of South Korea (Table 3). These farms had no history of exposure to ASFV, and all samples tested negative for ASFV using an ASFV real-time PCR assay.
The p30_LFA test results are ideal, showing negative results for all 100 serum samples, demonstrating 100% specificity. This means the p30_LFA accurately identified pigs without ASFV. However, the p72_LFA test produced unexpectedly positive results. Despite these farms being ASFV-free, seven samples reacted with the p72_LFA test. This translates to a specificity of 93% for the p72-LFA test.
In addition, we tested serum samples from feral pigs (n = 6) with p30_LFA and p72_LFA. Both p30_LFA and p72_LFA showed negative results for these samples, demonstrating 100% specificity.
4 Discussion
While highly sensitive and specific molecular tests such as real-time PCR exist for ASFV detection (Fan et al., 2020; Fu et al., 2021; Yang et al., 2022; Bohorquez et al., 2023; World Organisation for Animal Health (OIE), 2023), these assays can be expensive, require specialized training, and are limited to laboratory use. LFA offers a promising alternative. It is cost-effective, portable, requires no additional equipment, can easily be performed outside the laboratory, and provides results within minutes. These features make LFA a valuable POCT. A large number of such assays have been applied as efficient tests for the on-site analysis of biomarkers, such as proteins, small molecules, and nucleic acids, from a variety of different biological samples, including serum, blood, urine, saliva, and many other types (Magambo et al., 2014; Carrio et al., 2015; Schramm et al., 2015; Omidfar et al., 2023). A key limitation of LFAs is their generally lower sensitivity compared to molecular assays such as real-time PCR, which can lead to false-negative results (Onyilagha et al., 2022). However, this limitation might be less critical for ASFV detection. Pigs infected with virulent ASFV strains, such as the contemporary pandemic genotype II ASFV strain, develop high levels of the virus in their blood within a few days after infection. These virulent strains have a short incubation (2–3 days), followed by early clinical signs such as fever within 3–5 days. The fever in ASFV-infected animals coincides with viremia (the number of viruses in the blood), which quickly peaks (within 1–2 days of fever) up to 109 HAD50/ml (Dixon et al., 2020; Onyilagha et al., 2022). The high viral load during early infection makes LFA a viable option for detecting ASFV antigens in the early stage of ASFV infection.
This study focused on developing rapid and highly sensitive LFA for detecting the infection of ASFV in pigs. We developed p30_LFA, which is designed to detect the early viral structural protein p30 (encoded by the CP204L gene), expressed as early as 2–4 h post-infection and throughout the infection cycle (Revilla et al., 2018; Omidfar et al., 2023). In the meantime, we developed p72_LFA, which is designed to detect the p72 capsid protein (encoded by the B646L gene), the main structural protein of ASFV, accounting for approximately 33% of the total viral mass (Alcaraz et al., 1990; García-Escudero et al., 1998). Our tests achieved high analytical sensitivity in the spike-in test. p30_LFA detects as low as 10 ng of p30 protein per test (Figure 4A). p72_LFA detects down to 20 ng of p72 protein per test (Figure 4B). Both p30_LFA and p72_LFA showed no cross-reactions with other tested viruses (CSFV, PRRSV, PRV, and BVDV) (Table 1).
Compared to real-time PCR using samples from pigs experimentally infected with ASFV, our p30_LFA and p72_LFA assays demonstrated promising sensitivity and specificity. The p30_LFA detected ASFV in 84.21% of positive samples, while the p72_LFA achieved 100% sensitivity. Both assays exhibited 100% specificity, meaning no false positives were observed (Table 2). This is a significant improvement over previously reported ASFV LFAs, which only detected 68% of real-time PCR-positive samples and required sample dilution before testing (Sastre et al., 2016). Our assays achieve high sensitivity (low false negatives) due to two key factors. First, we utilize high-affinity mAbs in p30_LFA and p72_LFA. Second, these assays are designed for direct use with porcine plasma or serum samples, eliminating the need for dilution steps. Since serum samples were used, p72_LFA is expected to be more sensitive than p30_LFA. The p30_LFA targets the inner core shell protein of ASFV, while the p72_LFA targets the outer capsid protein of ASFV, granting it direct access to assay capture antibodies (Alcaraz et al., 1990; Revilla et al., 2018). To further improve p30_LFA sensitivity, we are incorporating an additional sample lysis step to liberate inner antigens. We will report these results in the near future.
Field testing using serum samples from South Korean farms revealed high specificities of 100 and 93% for p30_LFA and p72_LFA, respectively (Table 3). Seven unexpected false positives were observed with p72_LFA out of 100 field samples. These discrepancies might be due to farm-specific factors, such as underlying disease conditions or vaccination protocols, warranting further investigation. Further assessment of the specificity and sensitivity of p72_LFA under field conditions, as well as the exploration of strategies to enhance its diagnostic accuracy, are planned for future studies. Encouragingly, both LFAs exhibited 100% specificity with feral pig serum, suggesting they could be a reliable tool for identifying ASFV infection in wild pigs. These findings highlight the potential of p30_LFA and p72_LFA as field-deployable tools for identifying ASFV-infected farms and monitoring ASFV in wild pigs, particularly in areas with limited access to molecular assays or centralized laboratories.
We are presently conducting additional field validation studies using samples collected from both domestic and wild pigs to validate these promising findings. The validations will encompass the quality of the samples and environmental variables. Field samples primarily consist of carcasses from animals discovered either deceased or harvested, a crucial aspect to consider during LFA sample analysis. Factors including cold temperatures have the potential to decelerate reactions, potentially leading to skewed results. Windy conditions could introduce dust or debris that might disrupt the testing process. We will investigate the specific influence of these variables on the p30-LFA and p72-LFA assays and report the findings soon.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee of MEDEXX. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LW: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. JK: Data curation, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. HK: Investigation, Writing – original draft, Writing – review & editing. H-JP: Investigation, Writing – original draft, Writing – review & editing. M-JL: Investigation, Writing – original draft, Writing – review & editing. S-HH: Investigation, Writing – original draft, Writing – review & editing. C-WS: Investigation, Writing – original draft, Writing – review & editing. RM: Investigation, Writing – original draft, Writing – review & editing. YL: Investigation, Writing – original draft, Writing – review & editing. AC: Investigation, Writing – original draft, Writing – review & editing. JR: Investigation, Writing – original draft, Writing – review & editing. FM-F: Investigation, Writing – original draft, Writing – review & editing. E-JS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. JS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the USDA NIFA Award #2022–67015-36516, National Bio and Agro-Defense Facility Transition Fund, USDA NIFA Hatch-Multistate project (grant no.: 1021491), USDA ARS Non-Assistance Cooperative Agreements (grant nos.: 58-8064-8-011, 58-8064-9-007, 58-3020-9-020, and 59-0208-9-222), USDA NIFA Award #2022–67015-36516, USDA NIFA Subaward #25–6226–0633-002, National Pork Board Grant (grant no.: 18-059), Department of Homeland Security (grant no.: 70RSAT19CB0000027), and Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET), funded by Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (grant no.: RS-2022-IP122009).
Acknowledgments
The authors would like to thank the Comparative Medicine staff and staff of the Biosecurity Research Institute at Kansas State University for their technical help.
Conflict of interest
JK, HK, and E-JS were employed by the BioApplications Inc. H-JP and M-JL were employed by the MEDEXX Co., Ltd. S-HH and C-WS were employed by the Celltrix Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://www.medcalc.org/calc/diagnostic_test.php, accessed on 21 June 2024.
References
Alcaraz, C., De Diego, M., Pastor, M. J., and Escribano, J. M. (1990). Comparison of a radioimmunoprecipitation assay to immunoblotting and ELISA for detection of antibody to African swine fever virus. J. Vet. Diagn. Invest. 2, 191–196. doi: 10.1177/104063879000200307
Ang, S. H., Rambeli, M., Thevarajah, T. M., Alias, Y. B., and Khor, S. M. (2016). Quantitative, single-step dual measurement of hemoglobin A1c and total hemoglobin in human whole blood using a gold sandwich immunochromatographic assay for personalized medicine. Biosens. Bioelectron. 78, 187–193. doi: 10.1016/j.bios.2015.11.045
Berends, J., da Costa, B., Jong, J., Cooper, T. L., Dizyee, K., Morais, O., et al. (2021). Investigating the socio-economic and livelihoods impacts of African swine fever in Timor-Leste: an application of spatial group model building. Front. Vet. Sci. 8:687708. doi: 10.3389/fvets.2021.687708
Bohorquez, J. A., Lanka, S., Rosell, R., Pérez-Simó, M., Alberch, M., Rodriguez, F., et al. (2023). Efficient detection of African swine fever virus using minimal equipment through a LAMP PCR method. Front. Cell. Infect. Microbiol. 13:1114772. doi: 10.3389/fcimb.2023.1114772
Bosch-Camós, L., López, E., and Rodriguez, F. (2020). African swine fever vaccines: a promising work still in progress. Porcine Health Manag. 6:17. doi: 10.1186/s40813-020-00154-2
Brake, D. A. (2022). African swine fever modified live vaccine candidates: transitioning from discovery to product development through harmonized standards and guidelines. Viruses 14:2619. doi: 10.3390/v14122619
Burnett, M. J. B., and Burnett, A. C. (2020). Therapeutic recombinant protein production in plants: challenges and opportunities. Plants People Planet. 2, 121–132. doi: 10.1002/ppp3.10073
Carrio, A., Sampedro, C., Sanchez-Lopez, J. L., Pimienta, M., and Campoy, P. (2015). Automated low-cost smartphone-based lateral flow saliva test reader for drugs-of-abuse detection. Sensors 15, 29569–29593. doi: 10.3390/s151129569
Dixon, L. K., Stahl, K., Jori, F., Vial, L., and Pfeiffer, D. U. (2020). African swine fever epidemiology and control. Annu. Rev. Anim. Biosci. 8, 221–246. doi: 10.1146/annurev-animal-021419-083741
Escribano, J. M., Galindo, I., and Alonso, C. (2013). Antibody-mediated neutralization of African swine fever virus: myths and facts. Virus Res. 173, 101–109. doi: 10.1016/j.virusres.2012.10.012
Fan, X., Li, L., Zhao, Y., Liu, Y., Liu, C., Wang, Q., et al. (2020). Clinical validation of two recombinase-based isothermal amplification assays (RPA/RAA) for the rapid detection of African swine fever virus. Front. Microbiol. 11:1696. doi: 10.3389/fmicb.2020.01696
Fu, J., Zhang, Y., Cai, G., Meng, G., and Shi, S. (2021). Rapid and sensitive RPA-Cas12a-fluorescence assay for point-of-care detection of African swine fever virus. PLoS One 16:e0254815. doi: 10.1371/journal.pone.0254815
Gallardo, C., Nieto, R., Soler, A., Pelayo, V., Fernández-Pinero, J., Markowska-Daniel, I., et al. (2015). Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in eastern European Union countries: how to improve surveillance and control programs. J. Clin. Microbiol. 53, 2555–2565. doi: 10.1128/JCM.00857-15
García-Escudero, R., Andrés, G., Almazán, F., and Viñuela, E. (1998). Inducible gene expression from African swine fever virus recombinants: analysis of the major capsid protein p72. J. Virol. 72, 3185–3195. doi: 10.1128/JVI.72.4.3185-3195.1998
Gaudreault, N. N., and Richt, J. A. (2019). Subunit vaccine approaches for African swine fever virus. Vaccines 7:56. doi: 10.3390/vaccines7020056
Islam, M. R., Kwak, J. W., Lee, J. S., Hong, S. W., Khan, M. R. I., Lee, Y., et al. (2019). Cost-effective production of tag-less recombinant protein in Nicotiana benthamiana. Plant Biotechnol. J. 17, 1094–1105. doi: 10.1111/pbi.13040
Jean-Pierre, R. P., Hagerman, A. D., and Rich, K. M. (2022). An analysis of African swine fever consequences on rural economies and smallholder swine producers in Haiti. Front. Vet. Sci. 9:960344. doi: 10.3389/fvets.2022.960344
Karger, A., Pérez-Núñez, D., Urquiza, J., Hinojar, P., Alonso, C., Freitas, F. B., et al. (2019). An update on African swine fever virology. Viruses 11:864. doi: 10.3390/v11090864
Kim, C. M., Kim, D. M., Bang, M. S., Seo, J. W., Kim, D. Y., Yun, N. R., et al. (2023). Efficacy of plant-made human recombinant ACE2 against COVID-19 in a Golden Syrian Hamster model. Viruses 15:964. doi: 10.3390/v15040964
Le, V. P., Jeong, D. G., Yoon, S. W., Kwon, H. M., Trinh, T. B. N., Nguyen, T. L., et al. (2019). Outbreak of African swine fever, Vietnam, 2019. Emerg. Infect. Dis. 25, 1433–1435. doi: 10.3201/eid2507.190303
Lee, J. H., and Ko, K. (2017). Production of recombinant anti-Cancer vaccines in plants. Biomol. Ther. 25, 345–353. doi: 10.4062/biomolther.2016.126
Li, Z., Chen, W., Qiu, Z., Li, Y., Fan, J., Wu, K., et al. (2022). African swine fever virus: a review. Life 12:1255. doi: 10.3390/life12081255
Magambo, K. A., Kalluvya, S. E., Kapoor, S. W., Seni, J., Chofle, A. A., Fitzgerald, D. W., et al. (2014). Utility of urine and serum lateral flow assays to determine the prevalence and predictors of cryptococcal antigenemia in HIV-positive outpatients beginning antiretroviral therapy in Mwanza, Tanzania. J. Int. AIDS Soc. 17:19040. doi: 10.7448/IAS.17.1.19040
Mighell, E., and Ward, M. P. (2021). African swine fever spread across Asia, 2018-2019. Transbound. Emerg. Dis. 68, 2722–2732. doi: 10.1111/tbed.14039
Netherton, C. L., Connell, S., Benfield, C. T. O., and Dixon, L. K. (2019). The genetics of life and death: virus-host interactions underpinning resistance to African swine fever, a viral hemorrhagic disease. Front. Genet. 10:402. doi: 10.3389/fgene.2019.00402
Omidfar, K., Riahi, F., and Kashanian, S. (2023). Lateral flow assay: a summary of recent Progress for improving assay performance. Biosensors 13:837. doi: 10.3390/bios13090837
Onyilagha, C., Nguyen, K., Luka, P. D., Hussaini, U., Adedeji, A., Odoom, T., et al. (2022). Evaluation of a lateral flow assay for rapid detection of African swine fever virus in multiple sample types. Pathogens 11:138. doi: 10.3390/pathogens11020138
Park, Y., An, D. J., Choe, S., Lee, Y., Park, M., Park, S., et al. (2019). Development of recombinant protein-based vaccine against classical swine fever virus in pigs using transgenic Nicotiana benthamiana. Front. Plant Sci. 10:624. doi: 10.3389/fpls.2019.00624
Pikalo, J., Porfiri, L., Akimkin, V., Roszyk, H., Pannhorst, K., Kangethe, R. T., et al. (2022). Vaccination with a gamma irradiation-inactivated African swine fever virus is safe but does not protect against a challenge. Front. Immunol. 13:832264. doi: 10.3389/fimmu.2022.832264
Quembo, C. J., Jori, F., Vosloo, W., and Heath, L. (2018). Genetic characterization of African swine fever virus isolates from soft ticks at the wildlife/domestic interface in Mozambique and identification of a novel genotype. Transbound. Emerg. Dis. 65, 420–431. doi: 10.1111/tbed.12700
Ramirez-Medina, E., O'Donnell, V., Silva, E., Espinoza, N., Velazquez-Salinas, L., Moran, K., et al. (2022). Experimental infection of domestic pigs with an African swine fever virus field strain isolated in 2021 from the Dominican Republic. Viruses 14:1090. doi: 10.3390/v14051090
Revilla, Y., Pérez-Núñez, D., and Richt, J. A. (2018). African swine fever virus biology and vaccine approaches. Adv. Virus Res. 100, 41–74. doi: 10.1016/bs.aivir.2017.10.002
Rock, D. L. (2021). Thoughts on African swine fever vaccines. Viruses 13:943. doi: 10.3390/v13050943
Sastre, P., Gallardo, C., Monedero, A., Ruiz, T., Arias, M., Sanz, A., et al. (2016). Development of a novel lateral flow assay for detection of African swine fever in blood. BMC Vet. Res. 12:206. doi: 10.1186/s12917-016-0831-4
Schramm, E. C., Staten, N. R., Zhang, Z., Bruce, S. S., Kellner, C., Atkinson, J. P., et al. (2015). A quantitative lateral flow assay to detect complement activation in blood. Anal. Biochem. 477, 78–85. doi: 10.1016/j.ab.2015.01.024
Shi, X., Liu, X., Wang, Q., Das, A., Ma, G., Xu, L., et al. (2016). A multiplex real-time PCR panel assay for simultaneous detection and differentiation of 12 common swine viruses. J. Virol. Methods 236, 258–265. doi: 10.1016/j.jviromet.2016.08.005
Shi, J., Wang, L., and McVey, D. S. (2021). Of pigs and men: the best-laid plans for prevention and control of swine fevers. Anim. Front. 11, 6–13. doi: 10.1093/af/vfaa052
Shin, M., Kang, H., Shin, K. R., Lee, R., Kim, K., Min, K., et al. (2023). Plant-expressed Zika virus envelope protein elicited protective immunity against the Zika virus in immunocompetent mice. Sci. Rep. 13:22955. doi: 10.1038/s41598-023-47428-7
Sohn, E. J., Kang, H., Min, K., Park, M., Kim, J. H., Seo, H. W., et al. (2023). A plant-derived maternal vaccine against porcine epidemic diarrhea protects piglets through maternally derived immunity. Vaccines 11:965. doi: 10.3390/vaccines11050965
Truong, Q. L., Nguyen, T. L., Nguyen, T. H., Shi, J., Vu, H. L. X., Lai, T. L. H., et al. (2021). Genome sequence of a virulent African swine fever virus isolated in 2020 from a domestic pig in northern Vietnam. Microbiol. Resour. Announc. 10, 10–1128. doi: 10.1128/MRA.00193-21
Truong, Q. L., Wang, L., Nguyen, T. A., Nguyen, H. T., Tran, S. D., Vu, A. T., et al. (2023). A cell-adapted live-attenuated vaccine candidate protects pigs against the homologous strain VNUA-ASFV-05L1, a representative strain of the contemporary pandemic African swine fever virus. Viruses 15:2089. doi: 10.3390/v15102089
Wang, L., Ganges, L., Dixon, L. K., Bu, Z., Zhao, D., Truong, Q. L., et al. (2023). 2023 international African swine fever workshop: critical issues that need to be addressed for ASF control. Viruses 16:4. doi: 10.3390/v16010004
Wang, Y., Kang, W., Yang, W., Zhang, J., Li, D., and Zheng, H. (2021). Structure of African swine fever virus and associated molecular mechanisms underlying infection and immunosuppression: a review. Front. Immunol. 12:715582. doi: 10.3389/fimmu.2021.715582
Wang, T., Luo, R., Sun, Y., and Qiu, H. J. (2021). Current efforts towards safe and effective live attenuated vaccines against African swine fever: challenges and prospects. Infect. Dis. Poverty 10:137. doi: 10.1186/s40249-021-00920-6
World Organisation for Animal Health (OIE) . (2023). African swine fever. https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.09.01_ASF.pdf (Accessed April 15, 2024).
Keywords: African swine fever, lateral flow assay, rapid, sensitive, development
Citation: Wang L, Kim J, Kang H, Park H-J, Lee M-J, Hong S-H, Seo C-W, Madera R, Li Y, Craig A, Retallick J, Matias-Ferreyra F, Sohn E-J and Shi J (2024) Development and evaluation of two rapid lateral flow assays for on-site detection of African swine fever virus. Front. Microbiol. 15:1429808. doi: 10.3389/fmicb.2024.1429808
Edited by:
Ke Liu, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Fangfeng Yuan, Massachusetts Institute of Technology, United StatesDenis V. Kolbasov, Federal Research Center of Virology and Microbiology, Russia
Keke Wu, South China Agricultural University, China
Copyright © 2024 Wang, Kim, Kang, Park, Lee, Hong, Seo, Madera, Li, Craig, Retallick, Matias-Ferreyra, Sohn and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihua Wang, bGlodWFAdmV0Lmstc3RhdGUuZWR1; Eun-Ju Sohn, ZWpzb2huQGJpb2FwcC5jby5rcg==; Jishu Shi, anNoaUB2ZXQuay1zdGF0ZS5lZHU=
 Lihua Wang
Lihua Wang Juhun Kim2
Juhun Kim2 Franco Matias-Ferreyra
Franco Matias-Ferreyra