
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 29 July 2024
Sec. Food Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1428808
 Prerna Saini1,2
Prerna Saini1,2 Repally Ayyanna1†
Repally Ayyanna1† Rishi Kumar1,2
Rishi Kumar1,2 Sayan Kumar Bhowmick1,2
Sayan Kumar Bhowmick1,2 Vinay Bhaskar1
Vinay Bhaskar1 Bappaditya Dey1,2*‡
Bappaditya Dey1,2*‡The accelerated rise in antimicrobial resistance (AMR) poses a significant global health risk, necessitating the exploration of alternative strategies to combat pathogenic infections. Biofilm-related infections that are unresponsive to standard antibiotics often require the use of higher-order antimicrobials with toxic side effects and the potential to disrupt the microbiome. Probiotic therapy, with its diverse benefits and inherent safety, is emerging as a promising approach to prevent and treat various infections, and as an alternative to antibiotic therapy. In this study, we isolated novel probiotic bacteria from the gut of domestic goats (Capra hircus) and evaluated their antimicrobial and anti-biofilm activities against the ‘ESKAPE’ group of pathogens. We performed comprehensive microbiological, biochemical, and molecular characterizations, including analysis of the 16S-rRNA gene V1-V3 region and the 16S-23S ISR region, on 20 caprine gut-derived lactic acid bacteria (LAB). Among these, six selected Lactobacillus isolates demonstrated substantial biofilm formation under anaerobic conditions and exhibited robust cell surface hydrophobicity and autoaggregation, and epithelial cell adhesion properties highlighting their superior enteric colonization capability. Notably, these Lactobacillus isolates exhibited broad-spectrum growth inhibitory and anti-biofilm properties against ‘ESKAPE’ pathogens. Additionally, the Lactobacillus isolates were susceptible to antibiotics listed by the European Food Safety Authority (EFSA) within the prescribed Minimum Inhibitory Concentration limits, suggesting their safety as feed additives. The remarkable probiotic characteristics exhibited by the caprine gut-derived Lactobacillus isolates in this study strongly endorse their potential as compelling alternatives to antibiotics and direct-fed microbial (DFM) feed supplements in the livestock industry, addressing the escalating need for antibiotic-free animal products.
Antimicrobial resistance (AMR) is a growing global public health concern because many pathogens are becoming resistant to standard antibiotics. The ‘ESKAPE’ group of six nosocomial pathogens is leading the priority pathogen list of multidrug-resistant (MDR) and extensively drug-resistant (XDR) bacteria that includes, i.e., Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. (Boucher et al., 2009). These pathogens can escape the bactericidal actions of various antimicrobial agents. Inappropriate use or overuse of antibiotics has resulted in the global emergence and spread of these pathogens, causing outbreaks, community-acquired infections, and transmission Infection-related fatalities caused by drug-resistant (DR) pathogens are expected to account for the largest number of deaths worldwide by 2050 (Shankar, 2016). Biofilm formation is a major mechanism by which DR and MDR-ESKAPE bacteria exhibit a drug resistance phenotype (Mulani et al., 2019). Biofilms protect specialized dormant persister cells that are tolerant to antibiotics, as well as host immune cells, leading to difficult-to-treat recalcitrant infections (Lewis, 2005). Antibiotics are administered alone or in combination to effectively treat these infections. However, with every passing year, the number of antibiotics to treat these infections is declining, predisposing humanity towards a future with fewer antibiotics that will probably become ineffective in the near future (Andersson et al., 2020). Hence, there is a dire need to find safe and natural alternative antibiotic agents, such as probiotics, to treat infections caused by such pathogens (Mulani et al., 2019; da Rosa et al., 2020).
Probiotics are live microorganisms that confer health benefits to the host when administered in adequate amounts and are considered a potential alternative to antibiotics (Hill et al., 2014). Most probiotics belong to lactic acid bacteria (LAB), a group of bacteria that are generally regarded as safe (GRAS), and are the oldest known probiotic to inhibit or treat infections caused by DR pathogens (Mattia and Merker, 2008; Silva et al., 2020). LAB strains belonging to Lactobacillus and Bifidobacterium genera are known to inhibit pathogens by a plethora of mechanisms, including competitive exclusion, adhesion to the intestinal mucosa, host immunomodulation, enhancement of epithelial barrier integrity, and production of organic acids, hydrogen peroxide, bacteriocins, and antimicrobial peptides etc. (Bermudez-Brito et al., 2012). The application of Lactobacillus has shown promise for treating infections caused by ESKAPE bacteria in both animals and humans. For example, topical application of Lactobacillus acidophilus (L. acidophilus) or Limosilactobacillus reuteri (Lm. reuteri) was effective in treating wound infections caused by A. baumanii (Stanbro et al., 2020; Todorov et al., 2023). The application of Limosilactobacillus fermentum (Lm. fermentum) improved the condition of ischemic wounds in rabbits (Jones et al., 2012). Similar activities against skin pathogens, such as E. coli, P. aeruginosa, S. aureus, and Propionibacterium have been reported for Lactiplantibacillus plantarum (Lp. plantarum) ATCC 10241 and Lactobacillus delbrueckii (Lb. delbrueckii) DSMZ 20081 (Fijan et al., 2019; Yilmaz and Turkyilmaz, 2023). Additionally, Lacticaseibacillus rhamnosus (Ls. rhamnosus), Lm. fermentum, L. acidophilus, and Lp. plantarum prevented the adhesion and regrowth of E. faecalis and E. faecium biofilms (Velraeds et al., 1996). Lactobacillus gasseri (L. gasseri) LBM220 isolated from the feces of breastfed infants showed strong antibacterial activity against all six MDR-ESKAPE pathogens (Rastogi et al., 2021). Lactobacillus can inhibit the growth of a variety of livestock pathogens, including bovine mastitis-causing Methicillin Resistant Staphylococcus aureus (MRSA), which also causes skin abscesses and septicemia (Fitzgerald, 2012; Kang et al., 2017). Gram-negative bacillus K. pneumoniae, a common causative agent of clinical mastitis in dairy cattle, is an emerging zoonotic and foodborne pathogen worldwide (Munoz et al., 2006; Darniati et al., 2021). Lp. plantarum CIRM653 impaired K. pneumoniae biofilms independently of its bactericidal effect (Lagrafeuille et al., 2018). Lb. delbrueckii subsp. delbrueckii LDD01 also showed the highest inhibitory effect against K. pneumoniae (Mogna et al., 2016). Pseudomonas aeruginosa is also associated with many diseases in livestock and companion animals, including urinary tract infections in dogs, mastitis in dairy cows, and endometritis in horses (Leitner and Krifucks, 2007; Haenni et al., 2015). Growth inhibition and anti-biofilm effects of L. fermentum against MDR, XDR, and pan-drug-resistant (PDR) P. aeruginosa strains was also reported (Shokri et al., 2018). Notably, the L. acidophilus ATCC 4356 strain is well known for its strong inhibition of biofilm growth against a majority of P. aeruginosa strains (Alexandre et al., 2014; Elbadri et al., 2019). European Food Safety Authority (EFSA) has listed E. faecalis as an opportunistic pathogen in birds, poultry, and reptiles [EFSA Panel on Animal Health and Welfare (AHAW) et al., 2022]. Enterococcus faecalis has also been detected in animals, meat, and meat-based products, as well as in human fecal samples and in patients with bloodstream infections (Hammerum, 2012). Biosurfactants and conditioned media from several probiotic bacteria were found to prevent adhesion and biofilm formation by E. faecalis (Safadi et al., 2022). Lipoteichoic acids from Lp. plantarum have been proven effective in disrupting mature E. faecalis biofilms (Kim et al., 2020). Furthermore, livestock animals are the main reservoir of Shiga toxin-producing E. coli with zoonotic potential, and several Lactobacillus isolates exhibit antagonistic activity against these E. coli strains (Frank and Marth, 1977; Byakika et al., 2019).
Since the antibacterial properties of probiotic Lactobacillus depend on strain specificity, comprehensive characterization and evaluation of the probiotic properties of new or novel Lactobacillus isolates from natural sources are essential before selecting them for preclinical evaluation for their potential use (Ramos et al., 2013; Campana et al., 2017). To date, no study has demonstrated the isolation and inhibitory effects of Caprine gut-derived Lactobacillus against ESKAPE group pathogens. Here, we isolated Lactobacillus from the small intestine of domestic goats, identified them by biochemical and molecular methods, and assessed their probiotic properties, such as acid and bile tolerance, surface properties, epithelial cell adhesion, intrinsic colonization-cum-biofilm formation ability, and antibiotics susceptibility, and finally demonstrated their antagonistic and anti-biofilm effects against ESKAPE group pathogens.
The Lp. plantarum strain (MTCC-2621) used as the positive control strain was procured from the Microbial Type Culture Collection (MTCC), CSIR-Institute of Microbial Technology (IMTECH), Chandigarh, India. Lactobacillus isolates were grown in MRS agar or broth, as applicable. Pathogenic ESKAPE group bacteria were obtained from: E. coli and S. aureus (American Type Culture collection-ATCC, United States), K. pneumonae, A. baumanii, P. aeruginosa (MTCC, Chandigarh, India), and E. faecalis (National Center for Microbial Resources-NCMR, Pune, India). All bacteria were subcultured from glycerol stocks in Luria Bertani (LB) broth at 37°C overnight in a shaker incubator. Frozen glycerol stocks were prepared and stored at −80°C for future use.
Intestinal tissues (jejunum) were collected from goats (n = 11) immediately after slaughter in the Govt. authorized slaughterhouse in chilled phosphate-buffered saline (PBS, pH 7.4), transported on ice to the laboratory, and processed on the same day for bacterial isolation in a BSL-2 facility. Goat intestinal tissue samples were thoroughly washed with 1XPBS to remove intestinal content and debris. Each intestinal tissue sample was dissected into small pieces and collected in screw-capped 2 mL homogenization tubes containing 1.0 mm glass beads. Tissue sections were homogenized twice for 30 s at 4,000 rpm using a bead beater (BeadBug™). Subsequently, 100 μL of the homogenate was inoculated into 10 mL of sterile MRS broth. In parallel, the intestinal content was collected separately in a 1.5 mL tube, and 100 μL of the soup was directly inoculated into 10 mL of MRS broth. All the samples were incubated at 37°C in an orbital shaker incubator at 200 rpm for 24 h. Subsequently, cultures were 10-fold serially diluted up to 1: 106, and 100 μL samples from the three highest dilutions were plated onto MRS agar plates and incubated at 37°C aerobically for 48 h. Single isolated colonies were picked and replated onto MRS agar for colony morphology evaluation and subsequent characterization. Typically, round, creamy-white, shiny colonies with smooth and proper margins were selected based on the morphology description in the Bergey’s Manual of Determinative Bacteriology.
Each colony was screened for colony and bacterial morphology, Gram staining, and catalase activity. Only Gram-positive and catalase-negative rods were characterized further. Various other tests were performed to characterize Lactobacillus, as recommended in Bergey’s Manual of Determinative Bacteriology, including the indole test for the production of indole from tryptophan, the nitrate test for the reduction of nitrate to nitrite, and the Vogues Proskauer (VP) test to determine the presence of acetyl methyl carbinol after glucose fermentation (Bergey, 1994). The carbohydrate fermentation ability of all isolates was assessed using the Himedia Hilacto identification kit (KB020, Himedia), which is a standardized colorimetric identification system based on pH change and substrate utilization for the genus Lactobacillus. The kit contained one strip that included 12 wells, one for esculin and another for catalase, and 10 wells for 10 different carbohydrate sugars, that is, xylose, cellobiose, arabinose, maltose, galactose, mannose, melibiose, raffinose, sucrose, and trehalose. 50 μL of 0.1 OD (A600nm) bacterial inoculum (50 μL) was added to each well of a strip by surface inoculation and incubated at 37°C for 24–48 h. For carbohydrate fermentation and esculin tests, a color change was observed in each well, according to the manufacturer’s protocol. For the catalase test, a loopful of growth was scraped from the surface of the plate and dipped in a clean glass test tube containing 3% of freshly prepared H2O2, and effervescence was observed on the surface of the loop. No effervescence was observed in the case of a negative catalase test.
To identify the genus of the Lactobacillus isolates, colony-PCR was carried out on the genomic DNA extracted from the Lactobacillus isolates through the Triton-X-100 boiling lysis method using previously reported Lactobacillus genus-specific primers Forward-R16-1 and Reverse-LbLMA1 as described previously (Supplementary Table S1) (Dubernet et al., 2002). The optimized PCR cycling conditions included initial denaturation at 94°C for 1 min, followed by 30 cycles consisting of denaturation at 98°C for 5 s, annealing at 55°C for 5 s, and extension at 72°C for 5 s, and 2 min of final extension. The amplified PCR products were analyzed using agarose gel electrophoresis with ethidium bromide staining and visualized under UV light.
For Lactobacillus species identification, a second round of PCR was performed, targeting the 16S-rRNA V1-V3 region and 16S-23S Intergenic Spacer Region (ISR). Two separate primer pair sets were used to amplify ~509 bp and ~ 565 bp, using 16S(8-27)-F and V3(519-536)-R, and 16S(ISR)-F and 23S(ISR)-R primer pairs, respectively (Supplementary Table S1) (Weisburg et al., 1991; Gurtler and Stanisich, 1996; Turner et al., 1999). The first PCR was conducted using sequencing primer set-1 under the following conditions: initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 57°C for 30 s, extension at 72°C for 60 s, and final extension of 7 min. For primer set-2, PCR conditions involved an initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 59°C for 30 s, extension at 72°C for 60 s, and a final extension of 7 min. Amplified PCR products were subjected to agarose gel electrophoresis and ethidium bromide staining. PCR products were subsequently excised from the agarose gel and DNA was extracted using the Qiagen Gel Extraction Kit (#28704) according to the manufacturer’s protocol. The excised amplicons were sequenced by Sanger sequencing, using the forward primers for each PCR described above. The trimmed sequences were subjected to NCBI BLAST analysis using default parameters to identify the closest bacterial species via % sequence cover and percentage identity.
Subsequently, phylogenetic analysis of the 12 Lactobacillus isolates was performed using either the sequences of the 16S rRNA-V1-V3 region or 16S-23S ISR via hierarchical clustering using the neighbor-joining method. Briefly, both the sequences were aligned using ClustalW (default parameters). The alignment results were then used to determine the best-fit nucleotide-substitutional model. The Kimura 2-parameter and Tamura 3-parameter models were used for the V3 and ISR regions, respectively. The neighbor-joining method (bootstrap test −1,000 replicates) was used to generate the phylogenetic trees. Evolutionary analyses were conducted using MEGA11 software.
The pH and acid tolerance of the samples were assessed under three different conditions (pH 2.0, 3.0, and 4.0) by adjusting the pH of MRS broth. In brief, 96-well microplates were added with 150 μL/well of MRS broth, adjusted to three different pH levels, and inoculated with 1% (v/v) bacterial culture from an overnight broth culture, which had been adjusted to 0.8 OD (A600nm). The microplates were incubated at 37°C for 12 h at 200 rpm in a microplate incubator. Absorbance was read at 600 nm at two-hour intervals for a total of 12 h.
To evaluate the resistance of Lactobacillus isolates to high bile salt conditions, the isolates were cultured in MRS broth containing three different bile salt concentrations: 0.5, 1, and 2% w/v. The same culture method was employed as described above for pH shock, with the exception that standard MRS medium (pH 5.5) was used as the base medium along with the appropriate bile salt concentration.
The cell surface hydrophobicity of Lactobacillus isolates was determined using the microbial adhesion to hydrocarbons (MATH) method as previously described (Vinderola et al., 2004). This method evaluates hydrophobicity by measuring the affinity of microorganisms for organic solvents such as hexane, xylene, or toluene. The Lactobacillus isolates were grown overnight in a shaker incubator at 37°C in MRS broth and then harvested by centrifugation at 8000 rpm at room temperature. The cells were washed twice using 1 × PBS and adjusted to an optical density of 0.8–1.0 at A600 nm (A0). Next, 1 mL of xylene or hexane was added to each suspension and the mixture was vortexed vigorously for 2 min. After 1 h of incubation at room temperature without shaking and phase separation, the aqueous phase was carefully removed and its absorbance (At) was measured. The hydrophobicity percentage was calculated using the following formula: hydrophobicity (%) = (1 − At/A0) × 100.
Auto-aggregation assays were performed as described previously (Zuo et al., 2016). Briefly, Lactobacillus isolates were grown overnight in MRS broth at 37°C and harvested by centrifugation at 8000 rpm at room temperature. The cells were then washed twice with 1 × PBS (pH = 7.4), and the resulting bacterial suspensions’ OD was adjusted to 0.8–1.0 at A600nm (A0). The bacterial suspension was then incubated for 2 h at 37°C without shaking. The upper phase was removed and the OD was measured (At). Finally, the auto-aggregation percentage was determined using the following formula: Auto-aggregation (%) = (OD A0 − OD At /A0) × 100.
The biofilm formation ability of the Lactobacillus isolates was assessed using a previously described protocol with minor modifications (Aoudia et al., 2016). The tests were performed under aerobic and anaerobic conditions. Briefly, Lactobacillus isolates were cultured overnight in MRS broth at 37°C in a shaker incubator using glycerol stock to obtain the primary culture. After this, all isolates were sub-cultured to achieve log-phase growth (0.5–0.6 OD at A600nm). In each well of a 24-well polystyrene plate, 100 μL of the secondary log phase culture was added to 2 mL of MRS medium and incubated at 37°C for 72 h. After incubation, non-adherent bacteria and the culture medium were removed, and the wells were washed twice with sterile distilled water. Plastic-adhered biofilms were fixed with 1 mL of methanol for 15 min, methanol was removed, and the biofilms were air-dried. Finally, the biofilms were stained by using 200 μL of 0.2% crystal violet in distilled water for 10 min, the excess stain was removed with sterile distilled water, and the stain was extracted from the adherent cells using 500 μL of 0.5 M glacial acetic acid. Absorbance was measured using a microplate reader at A570nm.
The adhesion abilities of the Lactobacillus isolate to epithelial cells were measured as described previously by Jacobsen et al. with minor modifications (Jacobsen et al., 1999). Madin-Darby bovine kidney (MDBK) epithelial cells were cultured in DMEM medium (DMEM, Gibco) at 37°C in a humidified atmosphere containing 5% CO2, and were seeded at a density of 3 × 105 cells/mL per well in 6 well plates (Corning Inc., NY, United States). Lactobacillus isolates including Lactobacillus plantarum MTCC-2621 control strain was grown in MRS broth (MRS-Broth, Himedia) to an ODA600nm of 0.8, and were labeled with Carboxyfluorescein succinimidyl ester (CFSE) fluorescent dye as per the manufacturer’s instructions (Thermos scientific). Briefly, 3 × 107 bacterial cells were washed twice with 1x PBS (phosphate-buffered saline, pH 7.4) via centrifugation and resuspended in 250 μL of 1x CFSE dye and incubated in a shaker at 37°C, 200 rpm for 40 min. Post-labelling with CFSE dye, bacterial cells were washed again centrifuged with 1X PBS at 4500 g for 10 min, and resuspended in DMEM (without FBS, Antibiotics and Antimycotic solution). CFSE dye labeled Lactobacillus were used to infect the MDBK cells at an MOI of 1:10. Infected MDBK cells were incubated for 2 h at 37°C in 5% CO2, and subsequently washed three times with Dulbecco’s PBS buffer (DPBS, Gibco) to remove un-adhered bacteria, and adherent bacterial cells were evaluated by fluorescent microscopy (ZEISS, AXIO Observer 7) following DAPI (#62248, ThermoFisher) counter staining. In parallel, bacterial CFU were measured via plating on to MRS-agar plates. For CFU plating cells were detached via addition of 1 mL solution of 0.25% trypsin–EDTA (Sigma, United States) and incubating for 5-min at room temperature (RT). The detached cells were gently aspirated and mixed repeatedly to make homogenous suspension, and was then serially diluted in MRS medium and plated onto MRS agar plates. After incubation for 24 h at 37°C, the colonies were counted. Data were expressed as the percentage of adhesion = (CFU of adhered bacteria per well /bacterial cells initially added) × 100.
Overnight grown Lactobacillus cultures were streaked onto blood agar media containing 5% goat blood and incubated at 37°C for 24 h. Listeria monocytogenes and S. aureus were used as positive controls for α- and β-hemolysis, respectively. Clear and colored zones surrounding the colonies were examined. Clear zones indicated beta hemolysis, greenish zones indicated alpha hemolysis, and the absence of zones indicated no hemolysis or gamma hemolysis.
Antibiotic susceptibility of the Lactobacillus isolates was tested using the broth microdilution method. Nine antibiotics were selected, including β-lactams like ampicillin (0.125–64 mg/L), macrolides like erythromycin (0.0039–2 mg/L), lincosamides like clindamycin (0.0156–8 mg/L), glycopeptides such as vancomycin (0.125–64 mg/L), aminoglycosides like gentamicin (0.031–16 mg/L), kanamycin (0.125–64 mg/L), streptomycin (0.125–64 mg/L), tetracycline (0.125–64 mg/L), and chloramphenicol (0.125–64 mg/L), as recommended by FEEDAP and EFSA panels for human and veterinary importance [EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2012]. Briefly, the isolated Lactobacillus were propagated overnight in MRS broth at 37°C to reach an OD (A600nm) of 1.0. Microtiter test plates containing 2-fold serially diluted antibiotics in Lactobacillus susceptibility medium (LSM-broth) were inoculated at a final inoculum density of 0.001 (A600nm), equivalent to 105 CFU/mL. After aerobic incubation at 37°C for 24 and 48 h, MIC values of each antibiotic were visually evaluated two times at 24 and 48 h as the lowest antibiotic concentrations at which no growth was observed. The susceptibility status of strains was interpreted according to the microbiological cut-off values defined by the EFSA Panel on bacterial Feed Additives and Products or Substances used in Animal Feed in relation to antimicrobial resistance (EFSA Panel on Animal Health and Welfare (AHAW) et al., 2021).
The antimicrobial activity of all Lactobacillus isolates was analyzed against ESKAPE pathogens using an agar well diffusion assay. ESKAPE bacteria were grown overnight to an OD (A600nm) of 1.0, and 10-fold serial dilutions were made in normal saline. 100 μL of the bacterial inoculum was spread onto nutrient agar plates. Wells (10 mm diameter) were made on agar plates using 1 mL sterile tip bottoms. To extract CFS from Lactobacillus isolates, they were grown overnight and centrifuged at 9,000 rpm at 4°C, and supernatants were collected and filter sterilized with a 0.2-micron filter. 100 μL of CFS was added to the wells on each agar plate. To obtain the bacterial cell extract, overnight cultures of Lactobacillus were centrifuged at 9,000 rpm and 4°C, and the supernatant was discarded. Cell pellets were washed with 1x PBS (phosphate buffer saline, pH 7.4), and cells were disrupted by homogenization using a bead-beater using 0.1 mm beads in phosphate buffer; finally, following centrifugation cell lysate was filtered through a 0.2-micron filter and the sterile filtrate was used for the antimicrobial assay. After 24 h of incubation at 37°C under aerobic conditions, zone inhibition (mm) was recorded using the zone of inhibition scale (PW297, Himedia).
The anti-biofilm properties of CFS of Lactobacillus isolates against ESKAPE pathogens were assessed in 24-well plates. Briefly, 500 μL of ESKAPE bacterial culture of 0.3 OD (A600nm) in LB broth was added to each well along with 500 μL of CFS (in MRS media). 1: 1 mixture of L.B broth and MRS was used as a negative control. ESKAPE bacterial culture (0.3 OD) without CFS was used as a positive control. The microtiter plates were incubated at 37°C for 48 h to allow for biofilm formation. Subsequently, biofilm formation was measured as described in the ‘Biofilm formation assay’ section.
GraphPad Prism 9 was used for the preparation of the graphs and to perform the statistical analysis. For comparison of group means, One-Way ANOVA or t-test was performed where ever applicable, and differences were considered significant when p < 0.05. All the results are shown as the mean ± SD unless otherwise described in the corresponding figure legends.
Gastrointestinal (GI) infections in livestock ruminants not only have significant implications for animal health and productivity but also impact public health as a source of zoonotic enteric and other diseases in humans. Resident probiotic bacteria in the small intestine of domestic goats may have evolved niche-specific colonization and host-favoring properties that promote the maintenance of enteric homeostasis and antagonize the growth of pathogens in the gut. To exploit the beneficial properties of such enteric probiotic Lactobacillus, we first collected small intestinal (jejunum) tissues from goats and subjected them to a standard MRS media-based Lactobacillus isolation method (De Man et al., 1960; Dave and Shah, 1996). After several rounds of isolation of potential—Lactobacillus colonies with creamy white color on MRS agar, we selected 54 colonies for further confirmation by Lactobacillus genus-specific PCR using 16S-rRNA gene-specific primers, Forward-R16-1 and Reverse-LbLMA1, as described previously by Dubernet et al. (2002) (Supplementary Table S1). Out of 54 colonies, we selected 20 that showed an expected amplicon of ~250 bp (Supplementary Figure S1A). Furthermore, these 20 isolates were subjected to a second round of PCR and amplicon sequencing targeting 16S-rRNA V1-V3 region and the 16S-23S Intergenic Spacer Region (ISR) for species identification (Weisburg et al., 1991; Gurtler and Stanisich, 1996; Berthier et al., 1998; Turner et al., 1999). Two separate primer pair sets were used to amplify ~509 bp (Supplementary Figure S1B) and ~ 565 bp regions (Supplementary Figure S1C) from the 16S-rRNA-V1-V3 region and 16S-23S ISR using 16S(8–27)-F and V3(519–536)-R and 16S(ISR)-F and 23S(ISR)-R primer pairs, respectively (Supplementary Table S1). Out of the 20 Lactobacillus -like colonies that were subjected to PCR, followed by Sanger sequencing and NCBI BLAST-based nucleotide homology analysis, 12 isolates showed the closest identity to Lactobacillus spp., while the other colonies were found to belong to either Enterococcus spp. or Acetobacter spp. (Table 1). All sequences were submitted to NCBI Bio-project No. PRJNA985412, PRJNA986841, PRJNA986842. Among the 13 Lactobacillus species, 5 were identified as Lp. plantarum, 2 as L. salivarius, 2 as L. crispatus, 2 as L. amylovorous, 1 as L. kitasatonis, and the other as L. jhonsonii (Table 1).
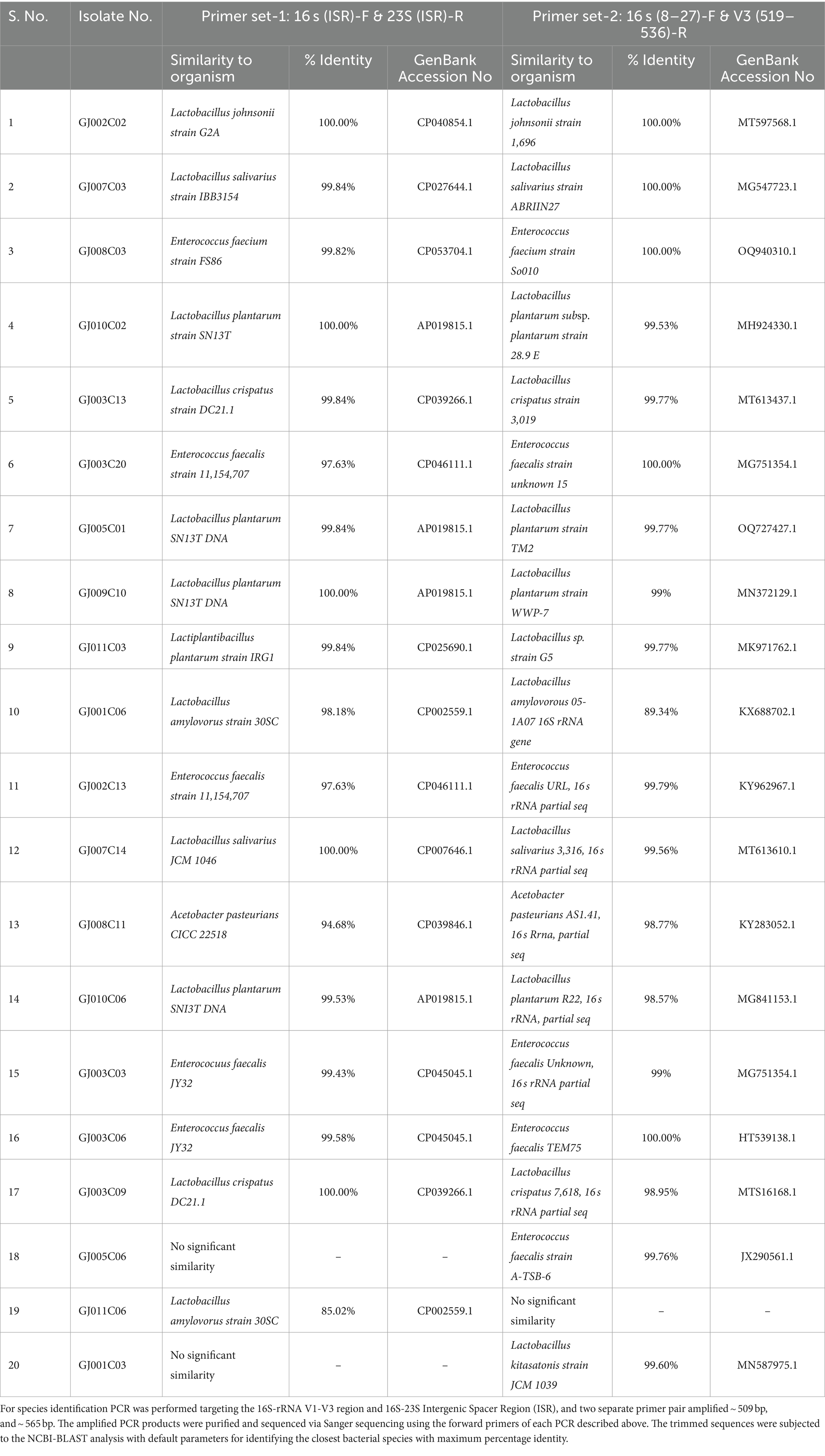
Table 1. 16 s-rRNA gene V1-V3 region and 16 s-23 s ISR amplicon sequencing and identification of Lactobacillus isolates.
Subsequently, phylogenetic analysis of the 12 Lactobacillus isolates using either of the sequences of the 16S rRNA-V1-V3 region (Figure 1A) or 16S-23S ISR (Figure 1B) via hierarchical clustering using the neighbor-joining method exhibited close evolutionary relationships and relatedness among the caprine gut-derived Lactobacillus isolates. As shown in the dendrograms in Figure 1, isolates belonging to the same species of Lactobacillus were mapped to be closely related by both methods. Moreover, isolates from the same animal exhibited the closest phylogenetic relationship.
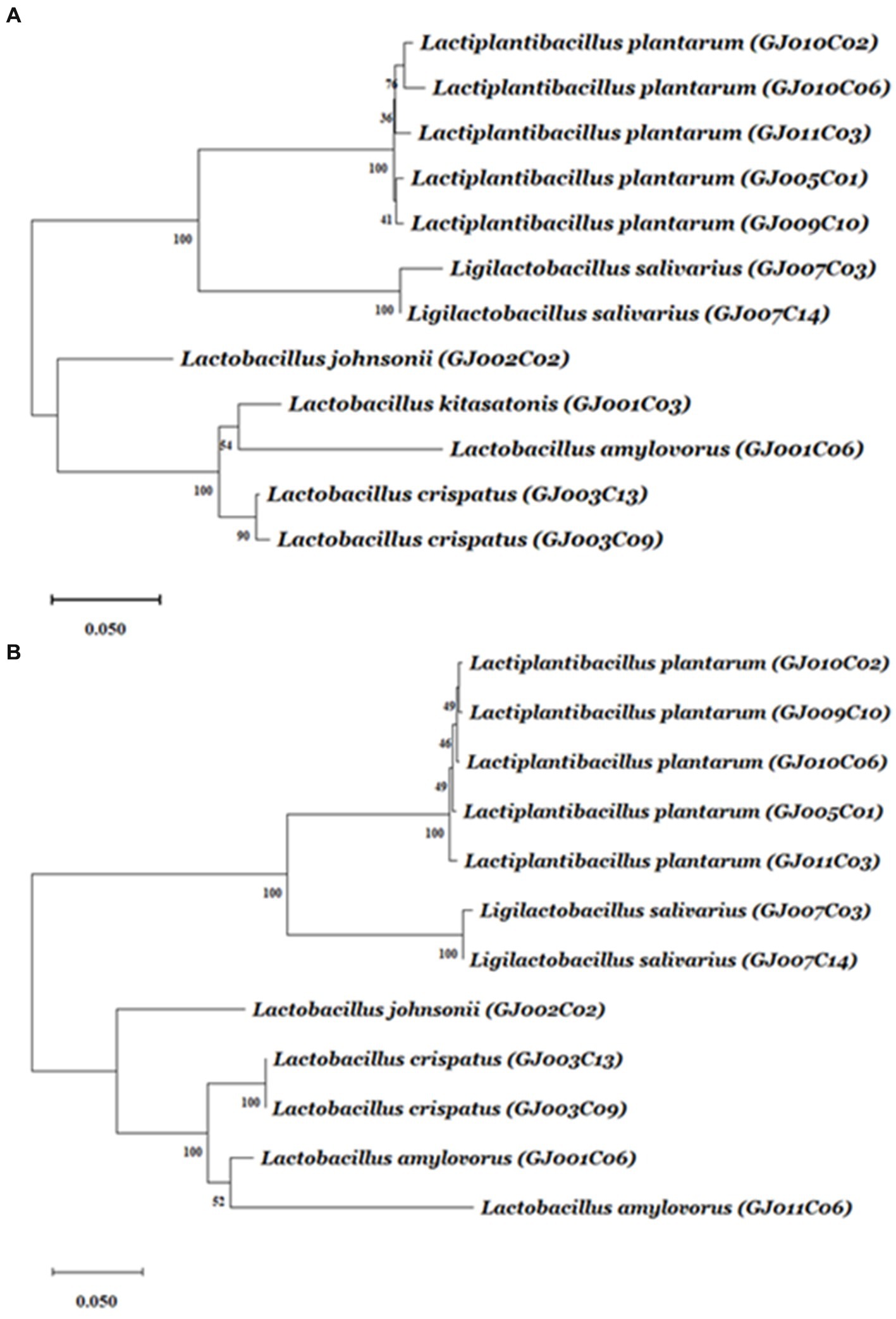
Figure 1. The dendrogram tree of Lactobacillus isolates based on 16S rRNA gene V1-V3 region and 16S-23S ISR sequences. The amplicons from the 16S rRNA gene V1-V3 region and 16S-23S ISR were sequenced using the primers 16S(8-27)-F and 16S(ISR)-F, respectively. From each isolate, either of the sequences was aligned using ClustalW with default parameters. The alignment results were then used to find out best fitted nucleotide-substitutional model. The Kimura 2-parameter model and Tamura 3-parameter model are used for the V1-V3 region, and ISR region, respectively. The neighbor-joining method (bootstrap test −1,000 replicates) was used to generate the hierarchical clustering-based dendrogram trees based on (A) 16S rRNA gene V1-V3 region, and (B) 16S-23S ISR region. A unit of the distance scale represents the percentage of differences between two sequences. Evolutionary analyses were conducted using MEGA11 software.
Furthermore, 12 selected Lactobacillus isolates were subjected to a series of biochemical tests to meet the criteria for Lactobacillus species, as described in Bergey’s Manual of Determinative Bacteriology (Bergey, 1994). These include Gram staining, catalase test, nitrate reduction (NR) test, indole test, and Vogues–Proskauer (VP) tests. All 12 isolates showed gram-positive staining, of which six were identified as catalase-negative, while the remaining six were catalase-positive. All 12 Lactobacillus isolates showed negative results for the indole test, nitrate test, and VP test (except one showing weak + VP test) (Table 2). These features of the Lactobacillus isolates corroborate the standard characteristics of Lactobacillus spp. Further, 6 catalase negative Lactobacillus isolates were selected for further characterization by carbohydrate fermentation test.
Probiotic Lactobacillus with complex sugar fermentation capability is important for their use in ruminant feed because ruminants rely on microbial fermentation of complex sugars in their digestive process (Matthews et al., 2019). Here, we assessed the carbohydrate fermentation ability of these six Lactobacillus isolates on 10 different carbohydrate sugars, namely, monosaccharides (Xylose, Arabinose, Galactose, Mannose), disaccharides (Cellobiose, Maltose, Melibiose, Sucrose, Trehalose), and trisaccharide (Raffinose) using a Hilacto™ Identification kit for Lactobacillus which also includes catalase and esculin tests. All six Lactobacillus isolates utilized sugars at different rates of hydrolysis, indicating varied carbohydrate fermentation capabilities (Table 3; Supplementary Figure S2). Furthermore, the Lactobacillus isolates displayed a positive esculin test, which is a typical feature of Lactobacillus spp. confirming their ability to break down complex coumarin glycoside-esculin into glucose and esculetin in the presence of β-glucosidase or esculinase enzyme (Chuard and Reller, 1998).
The ability to tolerate harsh conditions, such as low pH or acidic environment in the stomach, and sustenance in the high bile salt concentration in the initial part of the small intestine are the major determinants for the survival of probiotic bacteria in the gastrointestinal tract (Chou and Weimer, 1999). Each of the six selected Lactobacillus isolates displayed different degrees of sensitivity towards pH and bile salt stress, as evidenced by their differential growth patterns monitored over a period of 12 h at different pH (2.0, 3.0, 4.0, and 6.5) and bile salt (0.5, 1.0, and 2.0%)-containing media. In a non-acidic environment at pH 6.5, four of the Lactobacillus isolates exhibited typical sigmoidal bacterial growth, except the GJ010C06 and GJ003C09 isolates that displayed longer lag phases with minimal growth (Figure 2A). At pH 4.0, half of the Lactobacillus isolates exhibited moderate growth, while the other half displayed reduced or stalled growth (Figure 2B). Nevertheless, all the isolates, including GJ010C06 and GJ003C09, survived acid stress at lower pH values of 2.0, 3.0, while the GJ011C03 strain exhibited the highest growth in these low pH environments (Figures 2C,D).
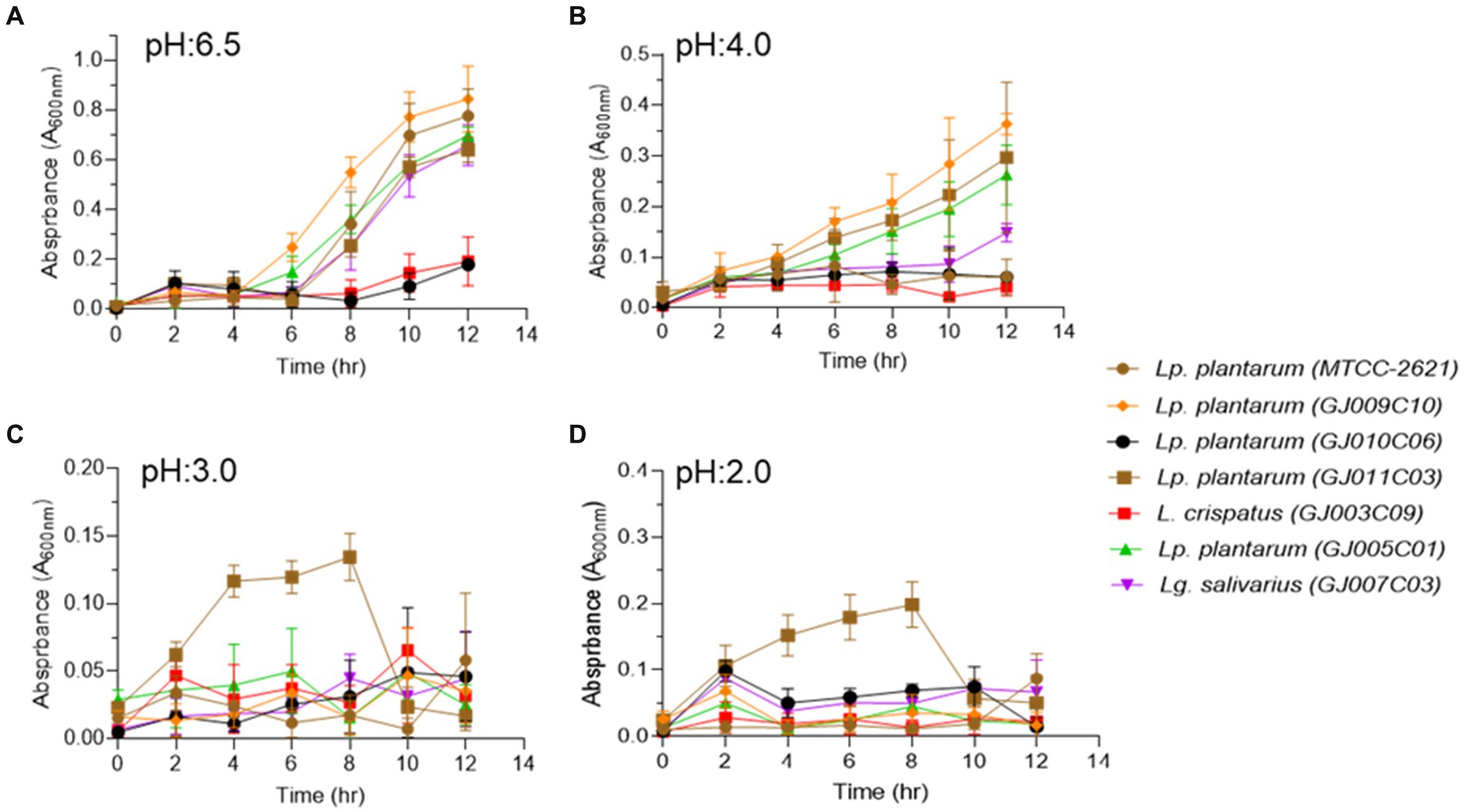
Figure 2. Profiling of the ability of Lactobacillus isolates to pH stress. Tolerance of Lactobacillus isolates to lower pH were assessed in three different conditions in addition to the MRS media (pH 6.5). Absorbance (A600nm) was taken at an interval of 2 h and till 12 h post-inoculation. Each connecting point in the line graphs depicts the mean ± SD of the absorbance values at each time point indicating the growth of Lactobacillus isolates at (A) pH 6.5, (B) pH 4.0, (C) pH 3.0, and (D) pH 2.0 conditions. Lp. plantarum (MTCC-2621) was used as positive control.
When bile salts were added to the growth medium at three different concentrations (0.5, 1, and 2% w/v), all selected Lactobacillus isolates exhibited suppressed growth compared with the standard growth media (Figure 3A). However, they were able to endure the salt stress for up to 12 h in medium containing 0.5% bile salt (Figure 3B). In media containing 1.0 and 2.0% bile salt, after an initial normal growth until 4 h post-inoculation, the Lactobacillus isolates displayed a sharp reduction in culture absorbance, followed by a plateau phase of growth (Figures 3C,D). Among the six Lactobacillus isolates, GJ009C10, GJ010C06, and GJ011C03 maintained higher culture absorbance levels than the other three Lactobacillus isolates.
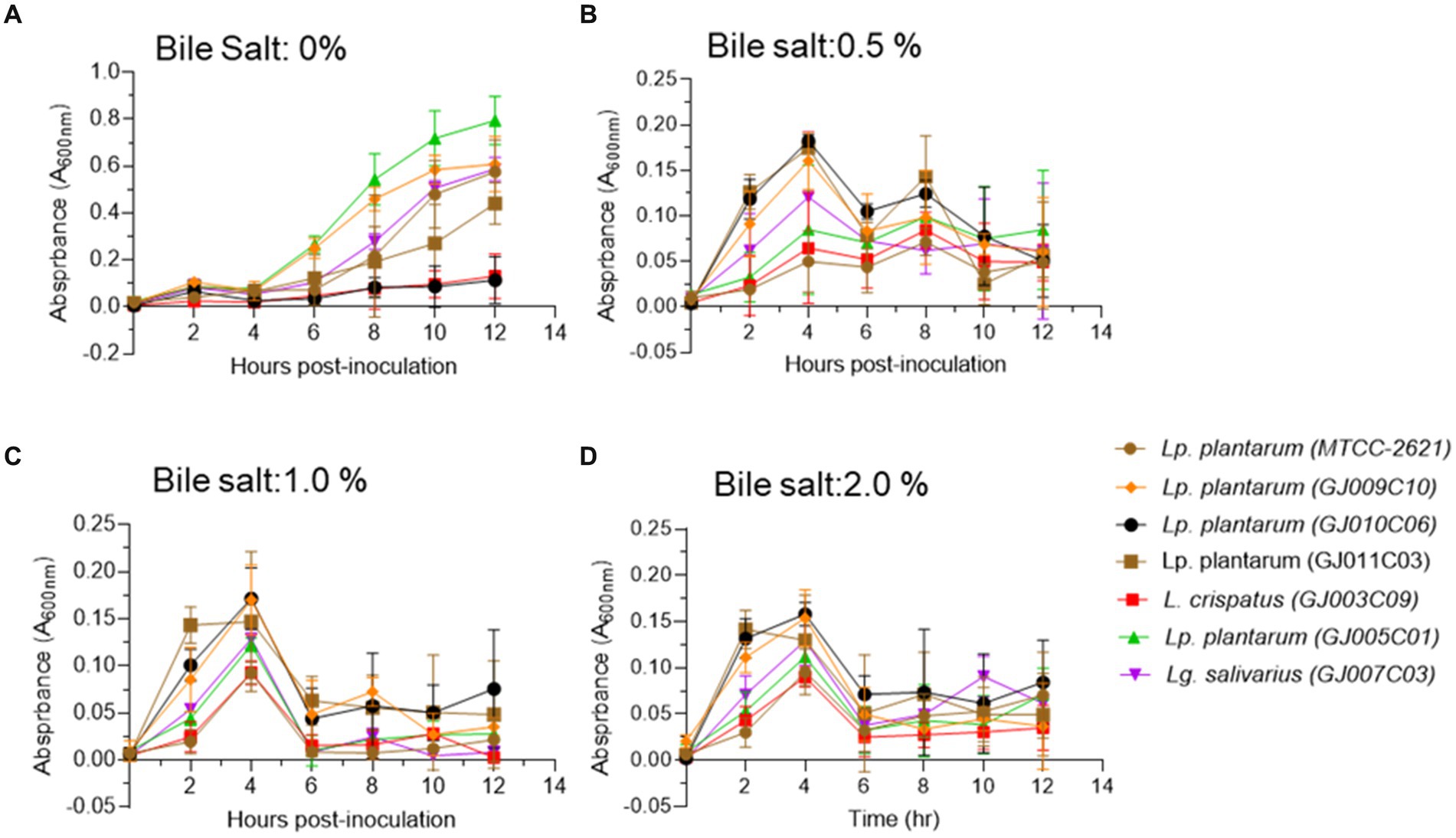
Figure 3. Bile tolerance profile of Lactobacillus isolates. Bile salt tolerance of Lactobacillus isolates was assessed in three different concentrations in addition to the standard MRS media. Absorbance (A600nm) was taken at an interval of 2 h and till 12 h post-inoculation. Each connecting point in the line graphs depicts the mean ± SD of the absorbance values at each time point indicating the growth of Lactobacillus isolates in (A) 0% bile salt, (B) 0.5%, (C) 1%, and (D) 2% w/v conditions. Lp. plantarum (MTCC-2621) was used as positive control.
Probiotic bacteria need to adhere to the mucus in the gut to survive, and their ability to do so is crucial for competition with harmful bacteria (Tuomola et al., 2001; Ohland and MacNaughton, 2010). Adhesion to the intestinal wall involves both nonspecific and specific interactions facilitated by different cell components (Magnusson et al., 1985; Krausova et al., 2019). Cell surface hydrophobicity, measured by the microbial adhesion to hydrocarbon method (MATH), is an important factor for adhesion capacity and is considered a pre-test for epithelial cell adhesion ability (Vinderola et al., 2004). Similarly, auto-aggregation is a process in which bacteria physically interact with each other with the help of cell surface components, such as proteins, carbohydrates, and lipoteichoic acid. Auto-aggregation of probiotics is necessary for adherence to the gut lining and acts as a barrier against undesirable bacteria (Tuo et al., 2013). These characteristics provide advantages for probiotics to colonize the gut. Hydrophobicity was assessed using two organic compounds: Hexane (Figure 4A) and Xylene (Figure 4B). All of the Lactobacillus isolates exhibited significant adhesion rates (% hydrophobicity) of over 60%. Lactobacillus isolates GJ009C10 and GJ005C01 exhibited >80% adhesion rates. Although both organic solvent-based techniques showed a comparable trend in % Hydrophobicity, the xylene-based method yielded higher values than the hexane-based method. The selected Lactobacillus isolates also displayed a recommended range of 60 to 80% auto-aggregation percentage, with GJ007C03 and GJ010C06 as top performers (Figure 4C). Interestingly, all the Lactobacillus isolates exhibited comparatively higher % Hydrophobicity and % Autoaggregation compared to the Lp. plantarum (MTCC-2621) control strain.
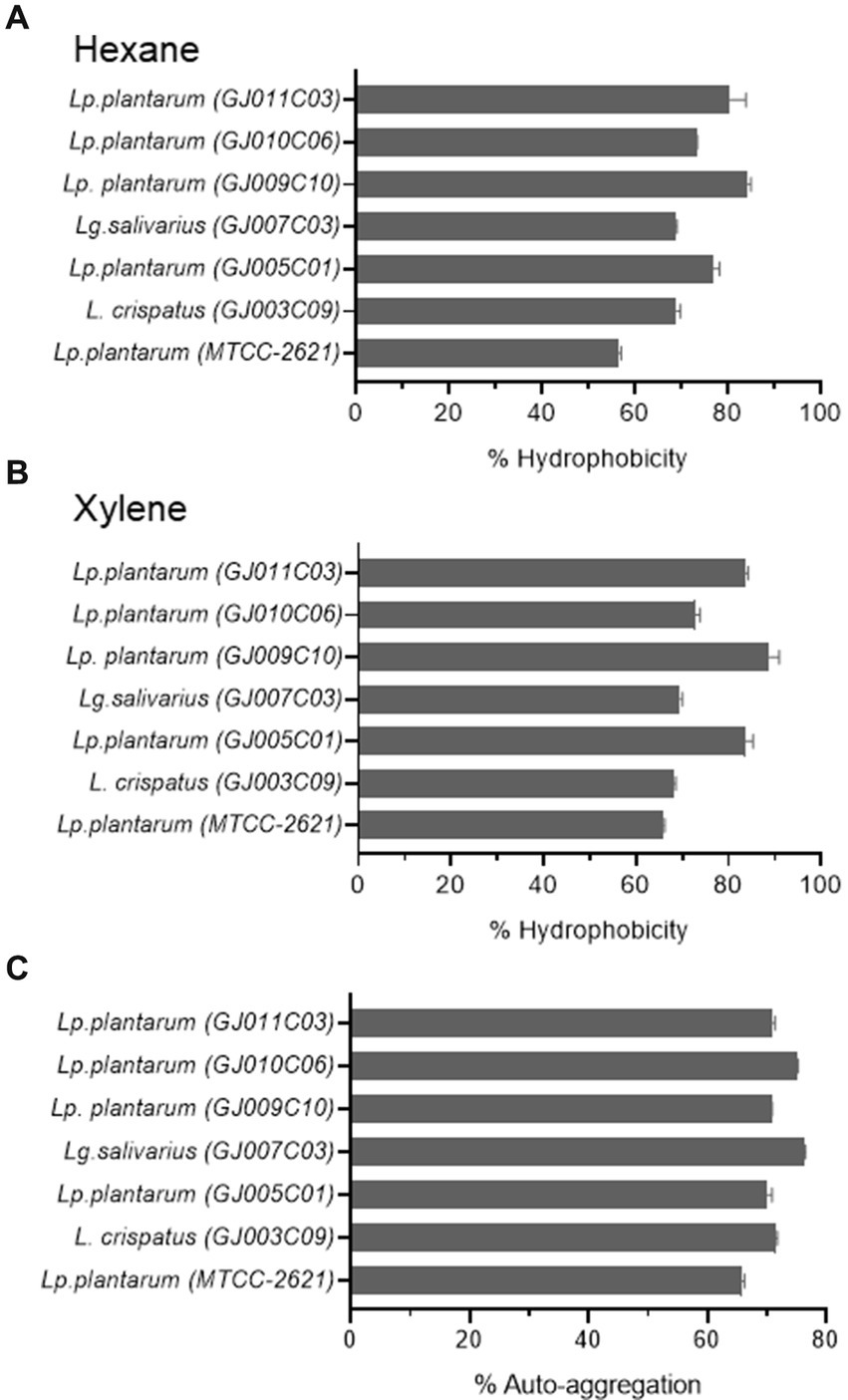
Figure 4. Cell surface properties of selected Lactobacillus isolates. The figure depicts the (A,B) % Hydrophobicity and (C) % Autoaggregation of Lactobacillus isolates. The % hydrophobicity was measured using the microbial adhesion to hydrocarbons (MATH) using (A) hexane, and (B) xylene as organic solvents as described in the Materials & methods section. (C) Autoaggregation is measured by evaluating the physical clumping ability of the Lactobacillus isolates in PBS solution as described earlier. The data is the Mean ± SD of triplicate experiments. Lp. plantarum (MTCC-2621) was used as positive control.
A promising approach for the control of pathogenic bacterial biofilms is the use of probiotics to colonize epithelial surfaces and counteract the proliferation of other bacterial species via competitive exclusion (Salas-Jara et al., 2016). Probiotic biofilms can promote their self-colonization and longer persistence in the intestinal mucosa, and Lactobacillus species produce more robust biofilms than other species (Kubota et al., 2008). Here, we tested the biofilm formation ability of Lactobacillus isolates under both aerobic (Figure 5A) and anaerobic conditions (Figure 5B) as they are facultative-anaerobic in nature, and both conditions prevail in the gastrointestinal tract. While all selected Lactobacillus isolates formed biofilms under both aerobic and anaerobic conditions, a significantly greater mass of biofilms was formed under anaerobic conditions. The most abundant biofilm-forming Lactobacillus isolates under both aerobic and anaerobic conditions were GJ005C01, GJ009C10, GJ003C09, and GJ011C03.
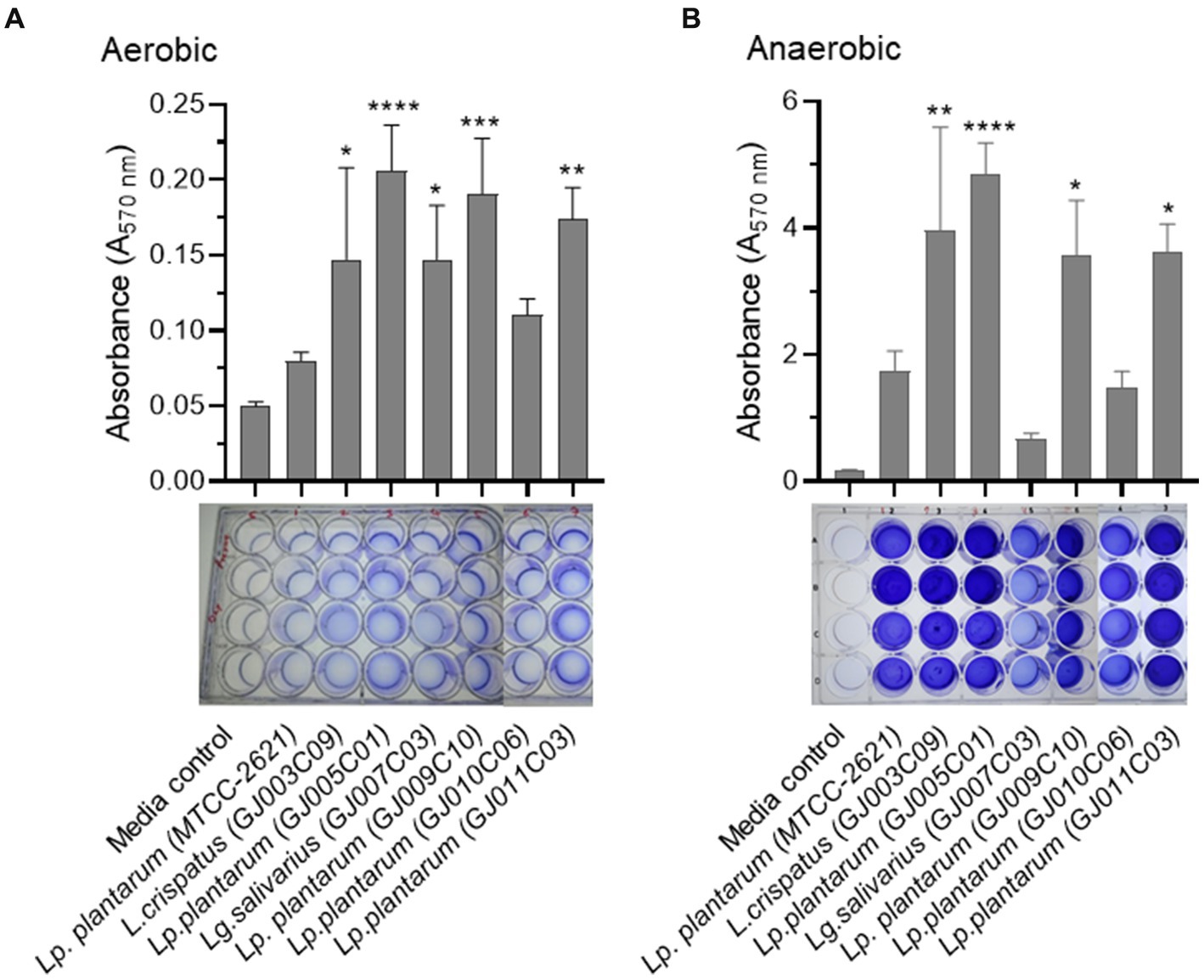
Figure 5. Biofilm formation by Lactobacillus isolates in aerobic and anaerobic environments. The biofilm formation ability of Lactobacillus isolates was assessed using both (A) aerobic, and (B) anaerobic growth conditions. The assay was performed in 24-well polystyrene plates. Lactobacillus isolates were cultured in static conditions in MRS media at 37°C for 72 h. Subsequently, quantitative measurement of biofilm was performed using the standard crystal violet-based assay. The bar diagrams depict the Mean ± SD of the absorbance (A570nm) values from triplicate wells. The experiment was performed twice. One-way ANOVA was performed to compare the mean differences of each Lactobacillus isolate compared to the Lp. plantarum (MTCC-2621) control strain. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. All Lactobacillus isolates produced more biofilm in the anaerobic conditions compared to the aerobic environment.
The adhesion ability of Lactobacillus isolates onto the MDBK cells was determined by fluorescence microscopy and CFU counting. Fluorescence microscopy showed CFSE labeled green fluorescent bacteria bound to cell surface, however variations in the adhesion was apparent across the Lactobacillus isolates (Figures 6A–H). CFU counts of the adhered bacteria (Supplementary Figure S3) and derivation of the % adhesion ability (Figure 6I) supported the microscopic findings. The percentage of adhesion varied from 45.5% (GJ010C06) to 74.4% (GJ011C03) among the Lactobacillus isolates. Lactobacillus isolates were then categorized based on their % adhesion as described previously (Jacobsen et al., 1999): strongly adhesive (>70%, GJ011C03 and GJ003C09), moderately adhesive (50–70%, GJ009C10, GJ007C03 and GJ005C01), and weakly adhesive (<50%, GJ010C06). The % adhesion ability of GJ003C09, GJ009C10, GJ011C03, and GJ005C01 were statistically similar to the MTCC control strain, while GJ007C03 and GJ010C06 showed significantly low % adhesion (p < 0.05 and p < 0.01, respectively).
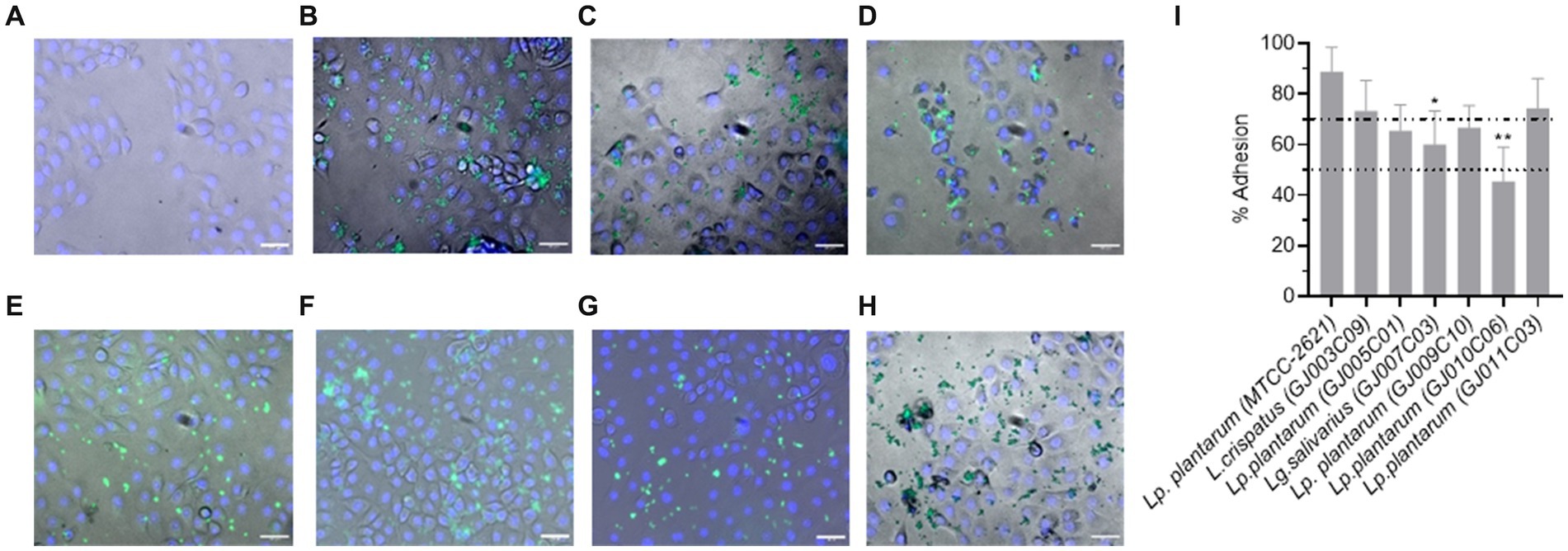
Figure 6. Cell adhesion ability of Lactobacillus isolates. Representative fluorescence microscopic images of MDBK cells after 2-h of incubation with fluorescently labeled probiotic isolates at an MOI of 1:10. (A). MDBK cells only (Negative control), (B–H) MDBK cells incubated with (B) MTCC-2621 (Positive control), (C) GJ003C09, (D) GJ005C01, (E) GJ007C03, (F) GJ011C03, (G) GJ009C10, and (H) GJ010C06. Scale bars: 100 μm for 20X objectives. (I) CFU of all the Lactobacillus isolates was measured by bacteriological plating from triplicate wells and % adhesion was calculated and presented as a bar diagram depicting mean ± SD. The top and bottom dotted lines represent the boundaries between the strong and moderate, and moderate and low adherent Lactobacillus isolates. ANOVA test was performed to compare the mean of Lactobacillus isolates to that of Lp. plantarum (MTCC-2621) control strain. *p < 0.05; **p < 0.01.
While Lactobacillus belong to the GRAS category of bacteria and are typically safe for consumption, newly isolated LAB strains must be proven to be devoid of any toxic effects, such as hemolytic properties, which can lead to serious health problems including anemia and kidney damage (Naidu et al., 2012; Tanaka et al., 2023). To ensure that the caprine gut-derived novel Lactobacillus isolates were safe, we performed hemolytic profiling for α-hemolysis (Supplementary Figure S4A) and β-hemolysis (Supplementary Figure S4B), wherein L. monocytogenes and S. aureus were used as positive controls for α- and β-hemolysis, respectively (Mogrovejo et al., 2020). None of the selected caprine gut-derived Lactobacillus isolates showed a hemolytic effect, highlighting their safety for consumption (Supplementary Figure S4C).
Antimicrobial resistance is a significant safety concern when evaluating the use of Lactobacillus as feed additive and therapeutic [European Food Safety Authority (EFSA), 2005]. The food chain has been identified as a major route of drug-resistant bacterial transmission between animals and humans, highlighting the need to monitor the safety of Lactobacillus used in animal nutrition (Brashears et al., 2005). Here, we screened the caprine gut-derived Lactobacillus isolates against nine antibiotics, including ampicillin (0.125–64 mg/L), erythromycin (0.0039–2 mg/L), clindamycin (0.0156–8 mg/L), vancomycin (0.125–64 mg/L), gentamicin (0.031–16 mg/L), kanamycin (0.125–64 mg/L), streptomycin (0.125–64 mg/L), tetracycline (0.125–64 mg/L), and chloramphenicol (0.125–64 mg/L) using the broth dilution-based minimum inhibitory concentration (MIC) method in LSM media using the microbiological breakpoint (BP), or cut-off values recommended by the FEEDAP document, and EFSA [EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2012]. The MIC for all LAB isolates were measured using a U-bottom microplate-based growth assay followed by visual observation at 24 h post-inoculation (Table 4). All isolated Lactobacillus strains were found to be susceptible to nine of the antibiotics tested, except vancomycin, and the MIC was below the epidemiological cutoff suggested by EFSA [EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2012].
ESKAPE pathogens are major causes of infection in animals and humans; however, the incidence of antibiotic failure against these pathogens is a critical public health problem (Manyi-Loh et al., 2018). These bacteria form biofilms, which make them resistant to antibiotics and immune cells. Probiotic Lactobacillus are known to inhibit the growth of pathogens via the production of antimicrobial substances such as organic acids, hydrogen peroxide, bacteriocins, and antimicrobial peptides (Plaza-Diaz et al., 2019). Antagonistic activity against such pathogens is a prerequisite for potential probiotics. Therefore, we screened the antagonistic activity against ESKAPE pathogen of the Lactobacillus isolates in two sets of experiments. First, the antagonistic growth potential of lactobacillus cell-free supernatant (CFS) and cell lysate was evaluated by an agar well diffusion assay (Supplementary Figure S5 and Figure 7), and second, anti-biofilm efficacy of CFS was evaluated by measuring their ability to prevent biofilm formation in a microplate-based assay (Figure 8). In the agar well diffusion assay, Lactobacillus isolates GJ009C10, GJ011C03, GJ005C01, and GJ007C03 demonstrated considerable levels of inhibition against the majority of ESKAPE pathogens in the case of both CFS (Figure 7A) and cell lysate (Figure 7B). In contrast, growth inhibition was relatively lesser for the isolates GJ003C09 and GJ010C06. Notably, CFS and cell lysate from all the Lactobacillus isolates exhibited highest inhibition of A. baumannii, among the ESKAPE pathogens. The zone inhibition data showed similar trends between CFS and cell lysate treatments (Figure 7).
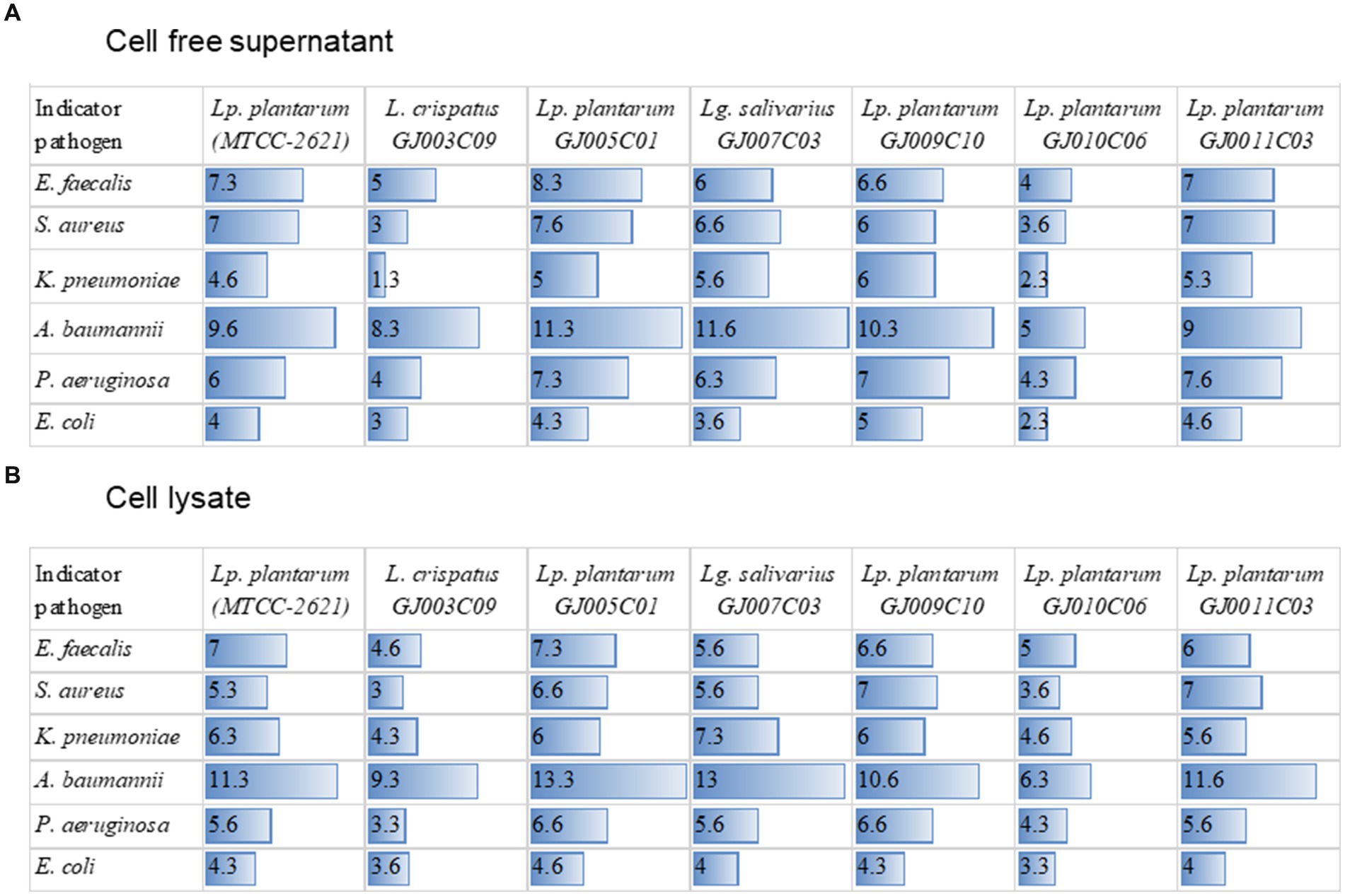
Figure 7. Zone of inhibition of ESKAPE pathogen growth by cell-free supernatant and cell-lysates of LAB isolates. The figure depicts the zone of inhibition radius from agar well diffusion assay. The zone of inhibition was recorded with the zone of inhibition scale (PW297, Himedia) following 24 h of incubation after the addition of sterile-filtered (A) cell-free culture supernatants, and (B) cell lysate of Lactobacillus isolates to the agar-wells. The horizontal bar depicts the Mean (values embedded) of the zone of inhibition radius of three experiments (mm) excluding the diameter of the agar well; Lp. plantarum (MTCC-2621) used as a positive control.
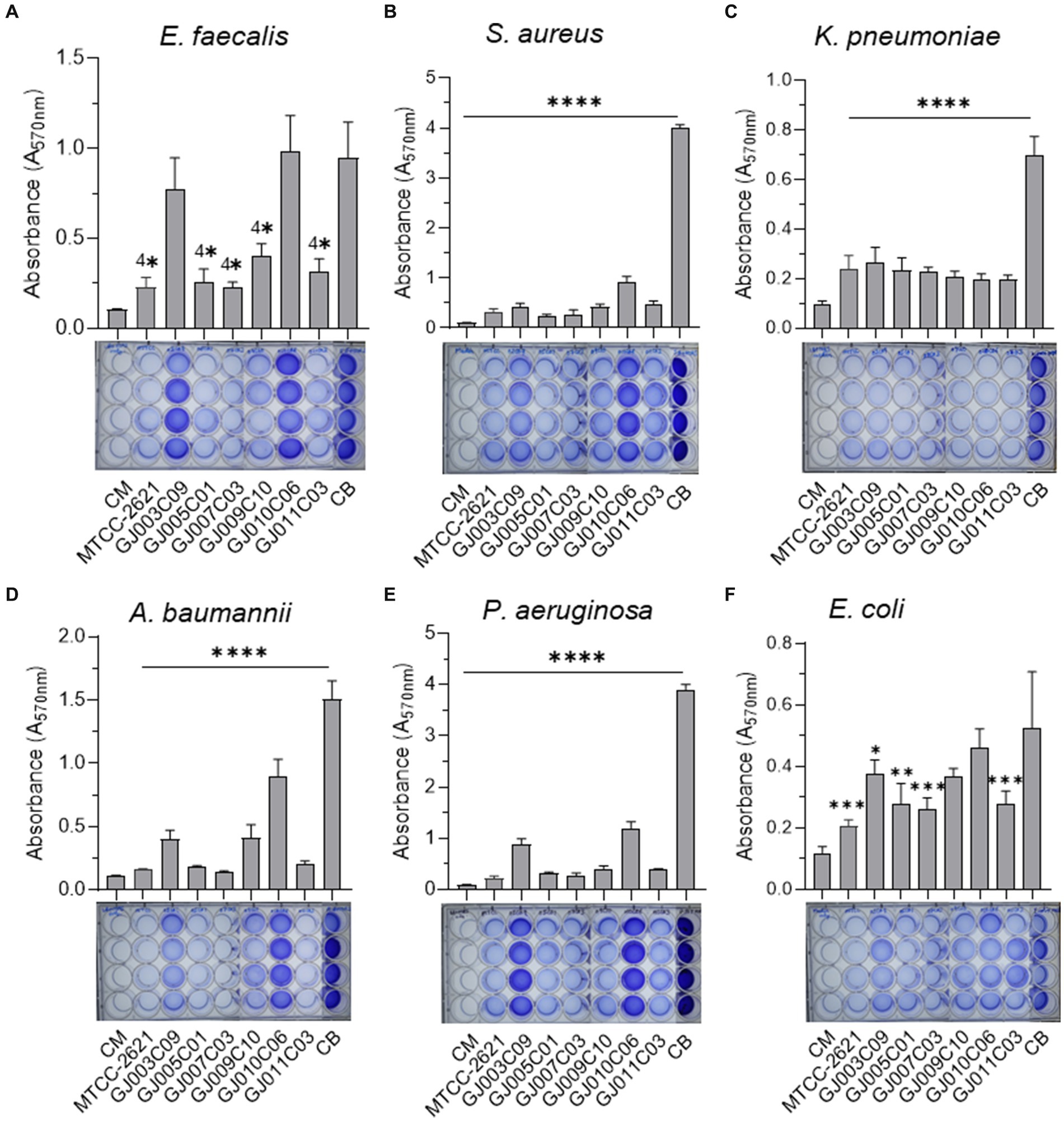
Figure 8. Lactobacillus isolates prevent biofilm formation by ESKAPE pathogens. The biofilm inhibition ability of Lactobacillus isolates was assessed using Cell-free culture filtrate (CSF). The assay was performed in 24-well polystyrene plates. ESKAPE pathogens [(A) E. faecalis, (B) S. aureus, (C) K. pneumoniae, (D) A. baumannii, (E) P. aeruginosa, and (F) E. coli] were cultured either in the presence or absence of Lactobacillus-CSF and incubated in static conditions at 37 ˚C for 48 hours. Subsequently, quantitative measurement of biofilm was performed using the standard crystal violet-based assay. The bar diagrams depict the Mean ± SD of the absorbance (A570nm) values from triplicate wells. The experiment was performed twice. One-way ANOVA was performed to compare the mean differences of each condition compared to the base value without any CSF (negative control biofilm, CB). *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. All Lactobacillus -CSF exhibited various degrees of biofilm inhibition on ESKAPE bacteria. Lp plantarum (MTCC-2621) used as a positive control.
Cell-free supernatants of Lactobacillus isolates exhibited anti-biofilm activity against ESKAPE pathogens (Figure 8). All Lactobacillus culture-free supernatants exhibited highly significant inhibition of biofilm formation by E. faecalis, S. aureus, K. pneumoniae, A. baumannii, and P. aeruginosa, although there was a certain degree of variation in their efficiency of inhibition (Figures 8A–E). In the case of E. coli biofilms, while the degree of inhibition was lower than that observed against the other five bacteria, the Lactobacillus—CFS of GJ007C03 and GJ011C03 showed considerable inhibition, followed by moderate biofilm inhibition observed with CFS of GJ003C01 and GJ009C10 (Figure 8F). Of the six Lactobacillus isolates, the CSF of four isolates, specifically GJ009C10, GJ011C03, GJ005C01, and GJ007C03, significantly (p < 0.01) reduced biofilm formation by ESKAPE pathogens.
The use of probiotics in animals has gained significant interest in recent years because of their numerous health benefits and market demands. While most commercial probiotics used in livestock are of human or dairy origin, probiotics from a similar host origin as the target host are preferred because of their greater evolutionary adaptability to the host’s gastrointestinal environment and superior biological activity (Dowarah et al., 2018). Probiotics isolated from animal intestines possess distinct characteristics, such as higher resistance to bile salts and low pH levels, as well as enhanced intestinal adherence abilities compared to dairy-based probiotics (Sornplang and Piyadeatsoontorn, 2016; Bazireh et al., 2020). Additionally, there is a growing need to develop species-specific probiotics capable of combating DR and MDR pathogens to improve the health and performance of livestock animals (Ripamonti et al., 2011; Blajman et al., 2015). Previous studies have reported the isolation and characterization of probiotic bacterial species from goat milk and feces (Setyawardani et al., 2011; Andrada et al., 2022); however, there are currently no reports on Lactobacillus species specifically derived from the jejunum intestinal compartment. Thus, this study aimed to isolate and characterize Lactobacillus species from goat small intestine jejunum, focusing on their morphological, biochemical, molecular, functional, and antagonistic properties against the proliferation and biofilm formation of the ESKAPE group of pathogens. The selection of probiotic bacteria was based on various criteria outlined by the Food and Agriculture Organization (FAO) and World Health Organization (WHO) (Bajagai et al., 2016). These criteria include bacterial origin, species, strain characterization, functional aspects (gastric acid and bile tolerance, carbohydrate utilization, and antimicrobial activity), antibiotic resistance, surface properties, and biosafety assessment (Bajagai et al., 2016). Here, out of 54 morphologically similar prospective Lactobacillus colonies that grew on MRS agar from the goat-jejunum tissue homogenates, following multi-layered and multi-parametric identification via molecular, microbiological, and biochemical methods, only six catalase negative isolates were shortlisted for further assessment of beneficial probiotic properties. Catalase-negative probiotics are considered superior as they can survive and thrive in the low-oxygen environment of the digestive system, and are also less likely to cause harmful changes in the gut microbiota by producing reactive oxygen species (ROS), which can damage the intestinal lining and trigger inflammation. These six isolates were gram-positive, bacillus, positive for esculin hydrolysis, and negative for indole production, nitrate utilization, and Voges Proskauer test. They could ferment complex carbohydrates and showed >99% sequence similarity with 16S rRNA gene and the 16S-23S ISR region for Lactobacillus species.
Probiotic candidates must tolerate low pH and bile salts to survive in the gastrointestinal tract (Chou and Weimer, 1999). Bacteria must survive the low pH of <2.0 in the stomach in the case of simple stomach animals, and < 2.5, in the abomasum in the case of ruminants, and remain viable for at least ~4 h before reaching the intestines. Furthermore, free bile acid that is synthesized in the liver conjugates with glycine or taurine, generating conjugated bile salts and releasing them in the duodenum (Urdaneta and Casadesús, 2017). These bile salts exhibit antimicrobial activity by damaging the bacterial cell walls and inducing DNA damage (Begley et al., 2005). To cope with bile salts, bacteria in the gut must possess an intrinsic resistance mechanisms (Ruiz et al., 2013). Certain probiotic strains, including Lactobacillus spp., have specific proteins devoted to the efflux of bile salts and protons, modifying sugar metabolism, and preventing the misfolding of proteins (Ruiz et al., 2013). In this study, the survival of the different Lactobacillus isolates varied under acidic and bile salt conditions. Some strains showed considerable survival at pH 2.0 and 3.0, while others demonstrated tolerance to different concentrations of bile salts, suggesting a strain-specific pattern. Notably, maximum survival or tolerance to acidity was observed in the case of GJ011C06 at a lower pH, and GJ009C10, GJ010C06, and GJ011C03 tolerated a higher bile salt environment than the other three Lactobacillus isolates.
To increase the chances of survival and colonization in the gastrointestinal tract, probiotic bacteria must adhere to the intestinal epithelium (Tuo et al., 2013). Both auto-aggregation and surface hydrophobicity testing serve as pre-tests for evaluating the adhesion capacity of probiotic bacteria to the epithelial cells. Various bacterial structures and components, such as pili, fimbriae, adhesins, mucus-binding proteins, fibronectin-binding proteins, surface layer proteins, lipoteichoic acid, and exopolysaccharides enable epithelial colonization (Krausova et al., 2019). This attachment serves a protective role by competing with intestinal pathways for host cell-binding sites. All the Lactobacillus isolates in our study exhibited hydrophobicity values ranging from 60 to 80% using both the Xylene and Hexane methods. Similar hydrophobicity values were reported in several earlier studies for Lactobacillus isolates (Gomaa, 2013), and Lactobacillus with >40% hydrophobicity were previously selected as supplement for swine feed (Dowarah et al., 2018). Furthermore, the Lactobacillus isolates exhibited significant auto-aggregation ranging from 60–80%, indicating their potential for colonization and adhesion in the gastrointestinal tract.
Adherence to epithelial cells and mucous has long been considered one of the selection criteria for probiotic microorganisms. It is an important requirement for both colonization and persistence inside gastrointestinal tract. Our findings showed that, Lactobacillus isolates GJ011C03 (74.4%) and GJ03C09 (73.3%) were strongly adhesive to MDBK cells, while moderate adhesion was observed for GJ009C10, GJ007C03 and GJ005C01, and least adhesion (45.5%) was observed for isolate GJ010C06. Varied levels of adhesion to mammalian cells were reported by several previous studies (Wang et al., 2009; Grover et al., 2011; Pringsulaka et al., 2015) Variations among adhesion can be seen between isolates as probiotic properties are generally strain specific within same species (Jacobsen et al., 1999). Although, the in vitro assays used for assessing the adherence potential of probiotic strains may not Truly mimic the gut environment, it provides valuable clue in short listing the potential probiotic strains for further validation in the animal models and livestock for their utility as direct-fed microbials.
Microbial communities, such as biofilms, when attached to a substratum or to each other, are effective in controlling biofilm formation by other bacterial species (Elbadri et al., 2019). Lactobacillus spp. are known to form robust biofilms compared to other bacteria (Kubota et al., 2008). Probiotic biofilms can enhance colonization, prolong persistence in the intestinal mucosa, and regulate pathogenic biofilms (Lagrafeuille et al., 2018). In this study, the Lactobacillus isolates exhibited varied levels of biofilm formation under both aerobic and anaerobic conditions; however, they had significantly higher levels of biofilm formation under anaerobic conditions, and the top three biofilm-forming Lactobacillus isolates were GJ005C01, GJ009C10, and GJ011C03. In addition to the anti-pathobiont effect and increased colonization due to enhanced biofilm formation by probiotic Lactobacillus in the gut under anaerobic conditions, it may exert other beneficial effects. These include (i) enhanced nutrient absorption: biofilms provide a protective layer and retain nutrients that may be lost in the conventional gastrointestinal tract, allowing better nutrient absorption by the probiotic bacteria; (ii) defense against antibiotics, which enables the probiotic bacteria to survive and flourish even in the presence of antibiotics, thereby maintaining their beneficial effects on the gut during antibiotic therapy; and (iii) biofilm-mediated communication: biofilms are known to facilitate communication among bacteria, allowing for better coordination of metabolic and physiological functions and enabling the probiotic bacteria to function more efficiently for better gut homeostasis (Barzegari et al., 2020).
Lactobacillus can acquire and spread antibiotic resistance genes through horizontal gene transfer (van Reenen and Dicks, 2011). Hence, antibiotic resistance testing is crucial when using new probiotic isolates as feed supplements, because of the potential risk of transferring antibiotic resistance to pathogenic bacteria and reducing the efficacy of antibiotics. According to EU regulatory frameworks, any bacterial strain carrying an acquired gene conferring AMR or strains with the unknown genetic nature of demonstrated resistance to antimicrobial agents should not be used as a feed additive because of the risk of horizontal spread (Stefańska et al., 2021). In this study, all caprine gut-probiotic Lactobacillus isolates were found to be susceptible to all the antibiotics listed in the EFSA panel, except vancomycin. Although vancomycin did not have breakpoints suggested by EFSA for L. plantarum obligate-, facultative-, heterofermentative, and homofermentative species, most of the Lactobacillus isolates showed intrinsic resistance to it [Tynkkynen et al., 1998; EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2012]. Furthermore, screening for streptomycin MIC wasn’t essential for homofermentative Lactobacillus spp. [EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), 2012], as an added caution, we evaluated the MIC for streptomycin, and were found to be below the cut-off values provided by EFSA. These findings on the selected Lactobacillus isolates, in addition to their non-hemolytic nature, indicate their potential use as feed supplements to promote animal health and prevent unwarranted emergence of antimicrobial resistance.
All six ESKAPE groups of pathogens commonly infect animals, with Enterobacter being the most common, followed by K. pneumoniae, P. aeruginosa, S. aureus, E. faecium, and A. baumannii. The major challenges of AMR in these pathogens are the multimodal antibiotic subversion mechanisms that have evolved, with biofilm formation being one of the leading causes. Therefore, agents that prevent biofilm formation and disperse preformed biofilms are associated with therapeutic benefits in animals. Components of the CFS of lactobacilli, such as exopolysaccharides and biosurfactants, may inhibit biofilm formation, as previously reported against MDR pathogens (Kaur et al., 2018; Rezaei et al., 2021). The Anti-biofilm effect of CFS of L. rhamnosus and Lp. plantarum is well-known against food pathogens, such as P. aeruginosa and L. monocytogenes (Rezaei et al., 2021). Probiotic Lactobacillus spp. have also shown inhibition of biofilm formation as well as a reduction in gene expression involved in the quorum sensing pathway in Streptococcus mutans (Wasfi et al., 2018). The Anti-biofilm effects of cell-free supernatants of L. pentosus and Lp. plantarum have also been reported against B. cereus and P. aeruginosa (Khiralla et al., 2015). In this study, the CFS of all six Lactobacillus isolates from goat small intestine inhibited biofilm formation by ESKAPE pathogens, with variation in inhibition ability among the strains. The present findings are in agreement with a previous report on Lactobacillus spp. isolated from the GI tract of other animal species (Dowarah et al., 2018).
In conclusion, this study has significantly advanced our understanding of the probiotic potential of Lactobacillus spp. isolated from the caprine gut, revealing not only their resilience under gastrointestinal-like stress but also their remarkable antimicrobial and biofilm inhibitory capacities. The standout isolates, GJ005C01, GJ007C03, GJ009C10, and GJ011C03, have shown exceptional promise, offering viable alternatives to traditional antibiotics in the livestock farming. These findings underscore the transformative potential of these probiotics in enhancing livestock health, with far-reaching implications for the food and pharmaceutical sectors. By harnessing the power of these probiotics, we can potentially mitigate the risks associated with emergence of antibiotic resistance, ushering in a new era of sustainable and effective disease management strategies in both agricultural and clinical settings.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
All experiments were reviewed and approved by the Institutional Biological Safety Committee (IBSC, approval no. IBSC/2022/NIAB/BD/02) of the National Institute of Animal Biotechnology, Hyderabad. All procedures were performed in accordance with the relevant guidelines and regulations laid down by DBT-RCGM, Govt. of India. The requirement of the Institutional Animal Ethics Committee (IAEC) was waived because no animal experiments were involved, and only authorized slaughterhouse goat tissue samples were used for the isolation of probiotics.
PS: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing, Conceptualization. RA: Data curation, Investigation, Methodology, Writing – review & editing. RK: Data curation, Investigation, Methodology, Software, Writing – review & editing. SB: Formal analysis, Visualization, Writing – original draft, Writing – review & editing. VB: Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. BD: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The financial support as intramural grant from the National Institute of Animal Biotechnology is gratefully acknowledged. Junior and Senior Research Fellowships to PS, RA, and RK were provided by the Council of Scientific and Industrial Research (CSIR), Indian Council of Medical Research (ICMR), and Department of Biotechnology (DBT), Govt. of India, respectively. Junior Research Fellowship to SB and VB were provided by the University Grant Commission (UGC), and DBT, Govt. of India, respectively.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1428808/full#supplementary-material
Alexandre, Y., le Berre, R., Barbier, G., and le Blay, G. (2014). Screening of Lactobacillus spp. for the prevention of Pseudomonas aeruginosa pulmonary infections. BMC Microbiol. 14:107. doi: 10.1186/1471-2180-14-107
Andersson, D. I., Balaban, N. Q., Baquero, F., Courvalin, P., Glaser, P., Gophna, U., et al. (2020). Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 44, 171–188. doi: 10.1093/femsre/fuaa001
Andrada, E., Mechoud, M. A., Abeijón-Mukdsi, M. C., Chagra Dib, E. P., Cerviño, S., Perez Chaia, A., et al. (2022). Ferulic acid esterase producing Lactobacillus johnsonii from goat feces as corn silage inoculants. Microorganisms 10:1732. doi: 10.3390/microorganisms10091732
Aoudia, N., Rieu, A., Briandet, R., Deschamps, J., Chluba, J., Jego, G., et al. (2016). Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol. 53, 51–59. doi: 10.1016/j.fm.2015.04.009
Bajagai, Y. S., Klieve, A. V., Dart, P. J., and Bryden, W. (2016). Probiotics in animal nutrition: production, impacts and regulation. FAO. Animal Production and Health : ed. Harinder P. S. Makkar.
Barzegari, A., Kheyrolahzadeh, K., Hosseiniyan Khatibi, S. M., Sharifi, S., Memar, M. Y., and Zununi Vahed, S. (2020). The battle of probiotics and their derivatives against biofilms. Infect. Drug Resist. 13, 659–672. doi: 10.2147/IDR.S232982
Bazireh, H., Shariati, P., Azimzadeh Jamalkandi, S., Ahmadi, A., and Boroumand, M. A. (2020). Isolation of novel probiotic Lactobacillus and Enterococcus strains from human salivary and fecal sources. Front. Microbiol. 11:597946. doi: 10.3389/fmicb.2020.597946
Begley, M., Gahan, C. G., and Hill, C. (2005). The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651. doi: 10.1016/j.femsre.2004.09.003
Bergey, D. H. (1994). Bergey's manual of determinative bacteriology : Lippincott Williams & Wilkins, Baltimore.
Bermudez-Brito, M., Plaza-Díaz, J., Muñoz-Quezada, S., Gómez-Llorente, C., and Gil, A. (2012). Probiotic mechanisms of action. Ann. Nutr. Metab. 61, 160–174. doi: 10.1159/000342079
Berthier, R., Jacquier-Sarlin, M., Schweitzer, A., Block, M. R., and Molla, A. (1998). Adhesion of mature polyploid megakaryocytes to fibronectin is mediated by beta 1 integrins and leads to cell damage. Exp. Cell Res. 242, 315–327. doi: 10.1006/excr.1998.4119
Blajman, J., Gaziano, C., Zbrun, M. V., Soto, L., Astesana, D., Berisvil, A., et al. (2015). In vitro and in vivo screening of native lactic acid bacteria toward their selection as a probiotic in broiler chickens. Res. Vet. Sci. 101, 50–56. doi: 10.1016/j.rvsc.2015.05.017
Boucher, H. W., Talbot, G. H., Bradley, J. S., Edwards, J. E., Gilbert, D., Rice, L. B., et al. (2009). Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 48, 1–12. doi: 10.1086/595011
Brashears, M. M., Amezquita, A., and Jaroni, D. (2005). Lactic acid bacteria and their uses in animal feeding to improve food safety. Adv. Food Nutr. Res. 50, 1–31. doi: 10.1016/S1043-4526(05)50001-9
Byakika, S., Mukisa, I. M., Mugab, R., and Muyanja, C. (2019). Antimicrobial activity of lactic acid bacteria starters against acid tolerant, antibiotic resistant, and potentially virulent E. coli isolated from a fermented Sorghum-millet beverage. Int. J. Microbiol. :2013539. doi: 10.1155/2019/2013539
Campana, R., van Hemert, S., and Baffone, W. (2017). Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut pathogens 9, 1–12. doi: 10.1186/s13099-017-0162-4
Chou, L.-S., and Weimer, B. (1999). Isolation and characterization of acid-and bile-tolerant isolates from strains of Lactobacillus acidophilus. J. Dairy Sci. 82, 23–31. doi: 10.3168/jds.S0022-0302(99)75204-5
Chuard, C., and Reller, L. B. (1998). Bile-esculin test for presumptive identification of enterococci and streptococci: effects of bile concentration, inoculation technique, and incubation time. J. Clin. Microbiol. 36, 1135–1136. doi: 10.1128/JCM.36.4.1135-1136.1998
da Rosa, T. F., Coelho, S. S., Foletto, V. S., Bottega, A., Serafin, M. B., Machado, C. S., et al. (2020). Alternatives for the treatment of infections caused by ESKAPE pathogens. J. Clin. Pharm. Ther. 45, 863–873. doi: 10.1111/jcpt.13149
Darniati, D., Setiyaningsih, S., Agungpriyono, D. R., and Handharyani, E. (2021). First evidence of Klebsiella pneumoniae infection in Aceh cattle: Pathomorphology and antigenic distribution in the lungs. Vet World 14, 1007–1013. doi: 10.14202/vetworld.2021.1007-1013
Dave, R., and Shah, N. (1996). Evaluation of media for selective enumeration of Streptococcus thermophilus, Lactobacillus delbrueckii ssp. bulgaricus, Lactobacillus acidophilus, and bifidobacteria. J. Dairy Sci. 79, 1529–1536. doi: 10.3168/jds.S0022-0302(96)76513-X
De Man, J., Rogosa, D., and Sharpe, M. E. (1960). A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23, 130–135.
Dowarah, R., Verma, A. K., Agarwal, N., Singh, P., and Singh, B. R. (2018). Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS One 13:e0192978. doi: 10.1371/journal.pone.0192978
Dubernet, S., Desmasures, N., and Guéguen, M. (2002). A PCR-based method for identification of lactobacilli at the genus level. FEMS Microbiol. Lett. 214, 271–275. doi: 10.1111/j.1574-6968.2002.tb11358.x
EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2012). Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J. 10:2740. doi: 10.2903/j.efsa.2012.2740
EFSA Panel on Animal Health and Welfare (AHAW)Nielsen, S. S., Alvarez, J., Bicout, D. J., Calistri, P., Canali, E., et al. (2021). Research priorities to fill knowledge gaps in wild boar management measures that could improve the control of African swine fever in wild boar populations. EFSA J. 19:e06689. doi: 10.2903/j.efsa.2021.6716
EFSA Panel on Animal Health and Welfare (AHAW)Nielsen, S. S., Bicout, D. J., Calistri, P., Canali, E., Drewe, J. A., et al. (2022). Assessment of listing and categorisation of animal diseases within the framework of the animal health law (regulation (EU) no 2016/429): antimicrobial-resistant Enterococcus faecalis in poultry. EFSA J. 20:e07126. doi: 10.2903/j.efsa.2022.7126
Elbadri, A. S., Fathi, M. S., Hamid, A. E. A., and ElGendy, H. A. (2019). The effect of Lactobacillus acidophilus as a probiotic against Pseudomonas aeruginosa growth and biofilm formation. Nov. Res. Microbiol. J. 3, 428–439. doi: 10.21608/nrmj.2019.44951
European Food Safety Authority (EFSA) (2005). Opinion of the scientific panel on additives and products or substances used in animal feed (FEEDAP) on the updating of the criteria used in the assessment of bacteria for resistance to antibiotics of human or veterinary importance. EFSA J. 3:223. doi: 10.2903/j.efsa.2005.223
Fijan, S., Frauwallner, A., Langerholc, T., Krebs, B., ter Haar (née Younes), J. A., Heschl, A., et al. (2019). Efficacy of using probiotics with antagonistic activity against pathogens of wound infections: an integrative review of literature. Biomed. Res. Int. 2019, 1–21. doi: 10.1155/2019/7585486
Fitzgerald, J. R. (2012). Livestock-associated Staphylococcus aureus: origin, evolution and public health threat. Trends Microbiol. 20, 192–198. doi: 10.1016/j.tim.2012.01.006
Frank, J. F., and Marth, E. H. (1977). Inhibition of Enteropathogenic Escherichia coli by Homofermentative lactic acid Bacteria in Skimmilk. J. Food Prot. 40, 754–759. doi: 10.4315/0362-028X-40.11.754
Gomaa, E. Z. (2013). Antimicrobial and anti-adhesive properties of biosurfactant produced by lactobacilli isolates, biofilm formation and aggregation ability. J. Gen. Appl. Microbiol. 59, 425–436. doi: 10.2323/jgam.59.425
Grover, S., Rajput, Y. S., Duary, R. K., and Batish, V. K. (2011). Assessing the adhesion of putative indigenous probiotic lactobacilli to human colonic epithelial cells. Indian J. Med. Res. 134, 664–671. doi: 10.4103/0971-5916.90992
Gurtler, V., and Stanisich, V. A. (1996). New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology (Reading) 142, 3–16. doi: 10.1099/13500872-142-1-3
Haenni, M., Hocquet, D., Ponsin, C., Cholley, P., Guyeux, C., Madec, J. Y., et al. (2015). Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet. Res. 11:9. doi: 10.1186/s12917-015-0324-x
Hammerum, A. M. (2012). Enterococci of animal origin and their significance for public health. Clin. Microbiol. Infect. 18, 619–625. doi: 10.1111/j.1469-0691.2012.03829.x
Hill, C., Guarner, F., Reid, G., Gibson, G. R., Merenstein, D. J., Pot, B., et al. (2014). Expert consensus document: the international scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514. doi: 10.1038/nrgastro.2014.66
Jacobsen, C. N., Rosenfeldt Nielsen, V., Hayford, A. E., Møller, P. L., Michaelsen, K. F., Pærregaard, A., et al. (1999). Screening of probiotic activities of forty-seven strains of Lactobacillus spp. by in vitro techniques and evaluation of the colonization ability of five selected strains in humans. Appl. Environ. Microbiol. 65, 4949–4956. doi: 10.1128/AEM.65.11.4949-4956.1999
Jones, M., Ganopolsky, J. G., Labbé, A., Gilardino, M., Wahl, C., Martoni, C., et al. (2012). Novel nitric oxide producing probiotic wound healing patch: preparation and in vivo analysis in a New Zealand white rabbit model of ischaemic and infected wounds. Int. Wound J. 9, 330–343. doi: 10.1111/j.1742-481X.2011.00889.x
Kang, M. S., Lim, H. S., Oh, J. S., Lim, Y. J., Wuertz-Kozak, K., Harro, J. M., et al. (2017). Antimicrobial activity of Lactobacillus salivarius and Lactobacillus fermentum against Staphylococcus aureus. Pathog Dis 75, 1–10. doi: 10.1093/femspd/ftx0009
Kaur, S., Sharma, P., Kalia, N., Singh, J., and Kaur, S. (2018). Anti-biofilm properties of the fecal probiotic lactobacilli against Vibrio spp. Front. Cell. Infect. Microbiol. 8:120. doi: 10.3389/fcimb.2018.00120
Khiralla, G. M., Mohamed, E., Farag, A., and Elhariry, H. (2015). Antibiofilm effect of Lactobacillus pentosus and Lactobacillus plantarum cell-free supernatants against some bacterial pathogens. J. Biotech Res. 6:86.
Kim, A. R., Kang, M., Yoo, Y. J., Yun, C. H., Perinpanayagam, H., Kum, K. Y., et al. (2020). Lactobacillus plantarum lipoteichoic acid disrupts mature Enterococcus faecalis biofilm. J. Microbiol. 58, 314–319. doi: 10.1007/s12275-020-9518-4
Krausova, G., Hyrslova, I., and Hynstova, I. (2019). In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 5:100. doi: 10.3390/fermentation5040100
Kubota, H., Senda, S., Nomura, N., Tokuda, H., and Uchiyama, H. (2008). Biofilm formation by lactic acid Bacteria and resistance to environmental stress. J. Biosci. Bioeng. 106, 381–386. doi: 10.1263/jbb.106.381
Lagrafeuille, R., Miquel, S., Balestrino, D., Vareille-Delarbre, M., Chain, F., Langella, P., et al. (2018). Opposing effect of Lactobacillus on in vitro Klebsiella pneumoniae in biofilm and in an in vivo intestinal colonisation model. Benef Microbes 9, 87–100. doi: 10.3920/BM2017.0002
Leitner, G., and Krifucks, O. (2007). Pseudomonas aeruginosa mastitis outbreaks in sheep and goat flocks: antibody production and vaccination in a mouse model. Vet. Immunol. Immunopathol. 119, 198–203. doi: 10.1016/j.vetimm.2007.05.007
Lewis, K. (2005). Persister cells and the riddle of biofilm survival. Biochem. Mosc. 70, 267–274. doi: 10.1007/s10541-005-0111-6
Magnusson, K.-E., Dahlgren, C., Maluszyńska, G., Kihlström, E., Skogh, T., Stendahl, O., et al. (1985). Non-specific and specific recognition mechanisms of bacterial and mammalian cell membranes. J. Dispers. Sci. Technol. 6, 69–89. doi: 10.1080/01932698508943934
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/molecules23040795
Matthews, C., Crispie, F., Lewis, E., Reid, M., O’Toole, P. W., and Cotter, P. D. (2019). The rumen microbiome: a crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 10, 115–132. doi: 10.1080/19490976.2018.1505176
Mattia, A., and Merker, R. (2008). Regulation of probiotic substances as ingredients in foods: premarket approval or “generally recognized as safe” notification. Clin. Infect. Dis. 46, S115–S118. doi: 10.1086/523329
Mogna, L., Nicola, S., Deidda, F., Amoruso, A., del Piano, M., Mogna, G., et al. (2016). In vitro inhibition of Klebsiella pneumoniae by Lactobacillus delbrueckii Subsp. delbrueckii LDD01 (DSM 22106). J. Clin. Gastroenterol. 50, S136–S139. doi: 10.1097/MCG.0000000000000680
Mogrovejo, D. C., Perini, L., Gostinčar, C., Sepčić, K., Turk, M., Ambrožič-Avguštin, J., et al. (2020). Prevalence of antimicrobial resistance and hemolytic phenotypes in Culturable Arctic Bacteria. Front. Microbiol. 11:570. doi: 10.3389/fmicb.2020.00570
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: a review. Front. Microbiol. 10:539. doi: 10.3389/fmicb.2019.00539
Munoz, M. A., Ahlström, C., Rauch, B. J., and Zadoks, R. N. (2006). Fecal shedding of Klebsiella pneumoniae by dairy cows. J. Dairy Sci. 89, 3425–3430. doi: 10.3168/jds.S0022-0302(06)72379-7
Naidu, K. S. B., Adam, J. K., and Govender, P. (2012). The use of probiotics and safety concerns: A review. African journal of microbiology research. 6, 6871–6877. doi: 10.5897/AJMR12.1281
Ohland, C. L., and MacNaughton, W. K. (2010). Probiotic bacteria and intestinal epithelial barrier function. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G807–G819. doi: 10.1152/ajpgi.00243.2009
Plaza-Diaz, J., Ruiz-Ojeda, F. J., Gil-Campos, M., and Gil, A. (2019). Mechanisms of action of probiotics. Adv. Nutr. 10, S49–S66. doi: 10.1093/advances/nmy063
Pringsulaka, O., Rueangyotchanthana, K., Suwannasai, N., Watanapokasin, R., Amnueysit, P., Sunthornthummas, S., et al. (2015). In vitro screening of lactic acid bacteria for multi-strain probiotics. Livest. Sci. 174, 66–73. doi: 10.1016/j.livsci.2015.01.016
Ramos, C. L., Thorsen, L., Schwan, R. F., and Jespersen, L. (2013). Strain-specific probiotics properties of Lactobacillus fermentum, Lactobacillus plantarum and Lactobacillus brevis isolates from Brazilian food products. Food Microbiol. 36, 22–29. doi: 10.1016/j.fm.2013.03.010
Rastogi, S., Mittal, V., and Singh, A. (2021). Selection of potential probiotic Bacteria from exclusively breastfed infant faeces with antagonistic activity against multidrug-resistant ESKAPE pathogens. Probiotics Antimicrob. Proteins 13, 739–750. doi: 10.1007/s12602-020-09724-w
Rezaei, Z., Khanzadi, S., and Salari, A. (2021). Biofilm formation and antagonistic activity of Lacticaseibacillus rhamnosus (PTCC1712) and Lactiplantibacillus plantarum (PTCC1745). AMB Express 11, 1–7. doi: 10.1186/s13568-021-01320-7
Ripamonti, B., Agazzi, A., Bersani, C., de Dea, P., Pecorini, C., Pirani, S., et al. (2011). Screening of species-specific lactic acid bacteria for veal calves multi-strain probiotic adjuncts. Anaerobe 17, 97–105. doi: 10.1016/j.anaerobe.2011.05.001
Ruiz, L., Margolles, A., and Sánchez, B. (2013). Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front. Microbiol. 4:396. doi: 10.3389/fmicb.2013.00396
Safadi, S., Maan, H., Kolodkin-Gal, I., Tsesis, I., and Rosen, E. (2022). The products of probiotic Bacteria effectively treat persistent Enterococcus faecalis biofilms. Pharmaceutics 14:751. doi: 10.3390/pharmaceutics14040751
Salas-Jara, M. J., Ilabaca, A., Vega, M., and García, A. (2016). Biofilm forming Lactobacillus: new challenges for the development of probiotics. Microorganisms 4:35. doi: 10.3390/microorganisms4030035
Setyawardani, T., Maheswari, R., Palupi, N., and Rahayu, W. (2011). Identification and characterization of probiotic lactic acid bacteria isolated from indigenous goat milk. Anim. Prod. 13, 57–63.
Shankar, P. R. (2016). Book review: tackling drug-resistant infections globally. Arch. Pharm. Pract. 7, 110–111. doi: 10.4103/2045-080X.186181
Shokri, D., Khorasgani, M. R., Mohkam, M., Fatemi, S. M., Ghasemi, Y., and Taheri-Kafrani, A. (2018). The inhibition effect of lactobacilli against growth and biofilm formation of Pseudomonas aeruginosa. Probiotics Antimicrob. Proteins 10, 34–42. doi: 10.1007/s12602-017-9267-9
Silva, D. R., Sardi, J. C. O., Pitangui, N. S., Roque, S. M., Silva, A. C. B., and Rosalen, P. L. (2020). Probiotics as an alternative antimicrobial therapy: current reality and future directions. J. Funct. Foods 73:104080. doi: 10.1016/j.jff.2020.104080
Sornplang, P., and Piyadeatsoontorn, S. (2016). Probiotic isolates from unconventional sources: a review. J Anim Sci Technol 58:26. doi: 10.1186/s40781-016-0108-2
Stanbro, J., Park, J. M., Bond, M., Stockelman, M. G., Simons, M. P., and Watters, C. (2020). Topical delivery of lactobacillus culture supernatant increases survival and wound resolution in traumatic Acinetobacter baumannii infections. Probiotics Antimicrob Proteins 12, 809–818. doi: 10.1007/s12602-019-09603-z
Stefańska, I., Kwiecień, E., Jóźwiak-Piasecka, K., Garbowska, M., Binek, M., and Rzewuska, M. (2021). Antimicrobial susceptibility of lactic acid bacteria strains of potential use as feed additives-the basic safety and usefulness criterion. Front. Vet. Sci. 8:687071. doi: 10.3389/fvets.2021.687071
Tanaka, Y., Aryantini, N. P. D., Yamasaki, E., Saito, M., Tsukigase, Y., Nakatsuka, H., et al. (2023). In vitro probiotic characterization and safety assessment of lactic acid Bacteria isolated from raw Milk of Japanese-Saanen goat (Capra hircus). Animals 13:7. doi: 10.3390/ani13010007
Todorov, S. D., Baretto Penna, A. L., Venema, K., Holzapfel, W. H., and Chikindas, M. L. (2023). Recommendations for the use of standardised abbreviations for the former Lactobacillus genera, reclassified in the year 2020. Benef. Microbes 15, 1–4. doi: 10.1163/18762891-20230114
Tuo, Y., Yu, H., Ai, L., Wu, Z., Guo, B., and Chen, W. (2013). Aggregation and adhesion properties of 22 Lactobacillus strains. J. Dairy Sci. 96, 4252–4257. doi: 10.3168/jds.2013-6547
Tuomola, E., Crittenden, R., Playne, M., Isolauri, E., and Salminen, S. (2001). Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr. 73, 393s–398s. doi: 10.1093/ajcn/73.2.393s
Turner, S., Pryer, K. M., Miao, V. P. W., and Palmer, J. D. (1999). Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 46, 327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x
Tynkkynen, S., Singh, K. V., and Varmanen, P. (1998). Vancomycin resistance factor of Lactobacillus rhamnosus GG in relation to enterococcal vancomycin resistance (van) genes. Int. J. Food Microbiol. 41, 195–204. doi: 10.1016/S0168-1605(98)00051-8
Urdaneta, V., and Casadesús, J. (2017). Interactions between Bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4, 1–13. doi: 10.3389/fmed.2017.00163
van Reenen, C. A., and Dicks, L. M. (2011). Horizontal gene transfer amongst probiotic lactic acid bacteria and other intestinal microbiota: what are the possibilities? A review. Arch. Microbiol. 193, 157–168. doi: 10.1007/s00203-010-0668-3
Velraeds, M. M., van der Mei, H. C., Reid, G., and Busscher, H. J. (1996). Inhibition of initial adhesion of uropathogenic Enterococcus faecalis by biosurfactants from Lactobacillus isolates. Appl. Environ. Microbiol. 62, 1958–1963. doi: 10.1128/aem.62.6.1958-1963.1996
Vinderola, C. G., Medici, M., and Perdigón, G. (2004). Relationship between interaction sites in the gut, hydrophobicity, mucosal immunomodulating capacities and cell wall protein profiles in indigenous and exogenous bacteria. J. Appl. Microbiol. 96, 230–243. doi: 10.1046/j.1365-2672.2004.02158.x
Wang, B., Li, J., Li, Q., Zhang, H., and Li, N. (2009). Isolation of adhesive strains and evaluation of the colonization and immune response by Lactobacillus plantarum L2 in the rat gastrointestinal tract. Int. J. Food Microbiol. 132, 59–66. doi: 10.1016/j.ijfoodmicro.2009.03.016
Wasfi, R., Abd el-Rahman, O. A., Zafer, M. M., and Ashour, H. M. (2018). Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell. Mol. Med. 22, 1972–1983. doi: 10.1111/jcmm.13496
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Yilmaz, O., and Turkyilmaz, S. (2023). Investigation of the potential probiotic effects of lactic acid bacteria and cell-free supernatants against important pathogens leading to wound infections. Minerva Biotechnol Biomol Res. 35, 41–53. doi: 10.23736/S2724-542X.22.02935-2
Keywords: probiotics, Lactobacillus, ESKAPE pathogens, biofilm, antimicrobial resistance
Citation: Saini P, Ayyanna R, Kumar R, Bhowmick SK, Bhaskar V and Dey B (2024) Restriction of growth and biofilm formation of ESKAPE pathogens by caprine gut-derived probiotic bacteria. Front. Microbiol. 15:1428808. doi: 10.3389/fmicb.2024.1428808
Received: 07 May 2024; Accepted: 05 July 2024;
Published: 29 July 2024.
Edited by:
Massimo Iorizzo, University of Molise, ItalyReviewed by:
Mapitsi Thantsha, University of Pretoria, South AfricaCopyright © 2024 Saini, Ayyanna, Kumar, Bhowmick, Bhaskar and Dey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bappaditya Dey, YmRleUBuaWFiLm9yZy5pbg==
†Present address: Repally Ayyanna, The University of Alabama in Huntsville, Chemical & Material Engineering, Huntsville, AL, United States
‡ORCID: Bappaditya Dey, https://orcid.org/0000-0003-2728-4683
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.