
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 19 June 2024
Sec. Extreme Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1427806
This article is part of the Research TopicProkaryotic Microbes in Arid Regions: Distribution, Environmental Adaptation, Biogeochemical Cycling, and CultivationView all 9 articles
Altitude and ultraviolet (UV) radiation may affect the community composition and distribution of microorganisms in soil ecosystems. In this study, 49 soil samples from 10 locations were collected from different elevations on the eastern Pamir Plateau and analyzed for soil microbial community structure and function using high-throughput sequencing. The results showed that soil samples from different elevations of the eastern Pamir Plateau contained 6834 OTUs in 26 phyla and 399 genera. The dominant phyla common to different elevations were Actinobacteria, Proteobacteria, Bacteroidota, Acidobacteriota, and Gemmatimonadota. The dominant genera were Rubrobacter, Sphingomonas, Nocardioides, and Solirubrobacter. Species richness increased slightly with elevation, and there were significant differences in community composition between the elevations. Elevation and UV exposure are important factors that drive changes in bacterial communities. The results of the KEGG pathway showed that drug resistance, antineoplastic, aging, replication, and repair were enhanced and then slightly decreased with increasing elevation. Bacterial communities at different elevations were rich in radiation-resistant microorganisms, and the main genera were Rubrobacter, Sphingomonas, Nocardioides, Pontibacter, and Streptomyces. The findings have shown the composition and distribution of bacterial communities at different elevations on the Eastern Pamir Plateau. Potentially radiation tolerant microbial species were also examined. The results are of considerable importance for the succession of bacterial microorganisms in the plateau region, the study of radiation tolerant bacterial germplasm resources, and the application of biofunctionality.
Plateau ecosystems are among the most important terrestrial ecosystems. Their climate, vegetation, and soil properties vary widely over short spatial distances and altitudinal gradients (McCain, 2010). Francos et al. (2021) found that elevation and total nitrogen (TN) are two important factors affecting organic matter (OM) in surface and subsurface soil layers. For areas above 3000–4000 m elevation, fractional vegetation cover has the strongest response to precipitation and temperature (Zhang et al., 2019). Owing to low temperatures and soil aridity or salinization in plateau areas, such habitats can reduce soil productivity or disrupt soil system function. A specific pattern of soil microbial diversity and taxonomic composition is formed. In this context, environmental stress plays a key role in soil microbial community changes (Khomutovska et al., 2021; Ji et al., 2022). Elevation has multiple response mechanisms in plateau ecosystems. These relationships may reflect the different feedback mechanisms and processes of ecosystems in response to environmental gradients (Bagousse-Pinguet et al., 2019).
The Pamir Plateau is one of the world’s top ten plateaus with massive mountains and precipitous terrain. It is the center of the concentrated development of alpine glaciers and is an intense natural environment with extremely arid and cold climates. This climatic region harbors a wide range of microbial resources (Yang and Yan, 2008; Yang et al., 2018; Barandun et al., 2021; Zhang et al., 2021). To date, domestic and international research on microorganisms in the Pamir region has focused on the glaciers, rocky substrates, and mountainous deserts. Khomutovska et al. (2017, 2020) studied endolithic microorganisms from the cold desert ecosystems of Eastern Pamir. Their community composition and structure were influenced by the microhabitat structure and associated microenvironmental conditions. Li et al. (2015) showed that plateau glacier habitats are nutrient-poor and a wide range of their microorganisms are better suited to oligotrophic survival environments. However, to date, there have been relatively few studies conducted on the composition of bacterial communities and potential radiation-tolerant bacteria in soil samples collected from different elevations in the East Pamir region.
The Pamir Plateau is exposed to high solar radiation. The highest solar radiation intensity in the region is 943.2 kW h/m2 in summer with the lowest value being 301.6 kW h/m2 in winter (Safaraliev et al., 2020). This is an extreme habitat that may contain abundant extreme microbial resources that can be used. Microbes that can survive high radiation doses are known as radiation-tolerant microorganisms. They are mainly found in desert soils, plateau lakes, radiation-polluted areas, and Antarctica (Fredrickson et al., 2004; Paulino-Lima et al., 2016; Portero et al., 2019; Monsalves et al., 2020). They have extreme resistance to DNA-damaging environments, that is, ionizing radiation and UV rays, mutagens, and desiccation environments (Narasimha and Basu, 2021). They possess highly efficient and precise DNA repair systems (Agapov and Kulbachinskiy, 2015) and antioxidant systems (Qi et al., 2020). Developing such microbial resources and studying their DNA damage repair mechanisms are of considerable importance for exploring the molecular mechanisms of DNA repair. They also play a substantial role in promoting the development of DNA technology. Meanwhile, the research and development of radiation-resistant microorganisms have a range of considerable potential applications in fields such as environmental engineering (Shang et al., 2021), medicine (Singh and Gabani, 2011; Gabani and Singh, 2013), and agriculture (Pan et al., 2009).
To date, relatively few studies have been conducted on the composition of bacterial communities at different elevations on the Pamir Plateau. Although there has been an increase in research on radiation-tolerant microorganisms, the use of radiation-tolerant microbial resources remains limited. The specific habitat of the Pamir Plateau is likely to have abundant radiation-tolerant microorganisms. In this study, we used high-throughput sequencing technology to determine the composition and distribution of bacteria at different elevations in East Pamir. We also examined the correlation between environmental factors and species preferences and investigated typical strains of radiation-tolerant bacteria; this would lay the foundation for the acquisition and use of radiation-tolerant resources.
This study was conducted on the eastern Pamir Plateau (38°–41°N, 73°–76°E), China. Samples were collected at three elevations, with 49 soil samples from 10 sampling locations selected based on the geomorphology of the sampling sites (Table 1). Among them, the low elevation range was 1400–2400 m with an average elevation of 2231 m, including TH (Tokaji Aketedu County, shade), TS (Tokaji Aketedu County, sunny), and WQ (Wuqia County). The middle elevation range was 2400–3400 m with an average elevation of 3146 m, including G2 (Gaizi Bridge 2), G1 (Gaizi Bridge 1), B (Baisha Lake), and S7 (Subash Seventh Bridge). The high elevation range was 3400–4400 m with an average elevation of 3829 m including WH (Wakhan Corridor), MT (204 Mustag Iceberg Base), and SB (Subash Daban). Information on the elevation, temperature, moisture, coordinates, and UV radiation dose was collected at each location. After selecting a sample point, parallel samples were randomly selected at four points within a 50–100 m radius. To collect the soil samples, a surface layer of approximately 2 cm was removed, and soil samples were collected at a depth of 0–20 cm. Approximately 1 kg of soil was collected for each sample, placed in a self-sealing bag, and stored in the laboratory at 4°C. Parallel soil samples were mixed from each sampling site. Roots, stones, and other debris were removed, before 200 g of the samples were weighed for air-drying, They were then sent to the Testing Center of the Xinjiang Academy of Agricultural Sciences, Xinjiang Academy of Agricultural Sciences (Xinjiang, China) for physicochemical property analysis.
A standard soil test series (NY/T 1121) was conducted. Organic matter (OM) was determined using the K2Cr2O7 oxidation method. The total nitrogen (TN) was measured using the Kjeldahl method. Available nitrogen (AN) was determined using the sulfate extraction method. Available phosphorus (AP) was detected using the hydrochloric acid–ammonium fluoride extraction–molybdenum antimony colorimetric method. Available potassium (AK) was detected using the ammonium acetate extraction–flame photometric method. Soluble salt (Salt) was detected using the mass method. The pH was determined using a potentiometric method.
Total soil DNA was extracted from a 0.5 g soil sample using the PowerSoil DNA Isolation Kit (Tiangen Biochemical, Beijing, China), according to the manufacturer’s instructions. DNA quality and quantity were assessed using a UVZ2550 spectrophotometer (Shimadzu Corporation, Kyoto, Japan) and 1% agarose gel electrophoresis, respectively. The samples were sent to Novogene Bioinformatics (Beijing) for amplification and sequencing of the V3–V4 regions of the 16S rRNA gene with standard Illumina 16S primers (338F:5′-ACTCCTACGGGGAGGCAGCA-3′,806R:5′-GGACTACHVGGGGTWTCTAAT-3′). Before amplification, tag sequences were added to the 5′ of downstream primers to distinguish between different samples.
Quality filtering of downstream data from the Illumina MiSeq sequencing platforms was performed using QIIME software (version 1.9.1). Using UCLUST (version 1.1.579) software to cluster all the clean reads from all the samples, the sequences were clustered into operational taxonomic units (OTUs) by default with 97% identity. Species annotation analysis was performed using the Mothur method with the SSUrRNA database of SILVA132 with the threshold set at 0.8–1. Shannon, Simpson, Chao1, and other diversity indices were calculated using QIIME software. Redundancy analysis (RDA) was used to evaluate the correlation between the community structure and environmental variables using CANOCO (version 5.0, Microcomputer Power, Ithaca, N.Y., USA). Principal co-ordination analysis (PCoA) is a multivariate statistical technique used to explore and visualize the similarities or differences among samples and was performed using Wekemo BioIncloud (Gao et al., 2024). Co-occurrence network was constructed to predict interactions among members within bacterial communities. The frequencies of co-occurrence association (with | rho| > 0.7, p < 0.001) presented were recorded to reveal the association persistency among communities. Spearman’s rho statistic is used to estimate correlation with function cor.test in “stats” packagee (R Development Core Team, 2016). The co-occurrence networks were visualized with “igraph” package (Csardi and Nepusz, 2006). Network characteristics were determined using functions in “bipartite” package (Dormann et al., 2009). PUCRUSt2 predicts gene by link the ASV/OTU sequences to references genomes in PUCRUSt2 database. PUCRUSt2 database contains KEGG annotation informations of these reference genomes. PUCRUSt2 database meas built-in database of PICRUSt2 software (Douglas et al., 2020). We identified importance of species (variable) points by the randomforest R (Version 3.2.1). Cross-validation was performed by the “rfcv” function for selecting appropriate features. The “varImp-Plot” function was used to show the importance of features in the classification. Community composition maps were plotted using OriginPro 2021 software. IBM SPSS Statistics software for Windows (version 27.0, IBM Corp., Armonk, N.Y., USA) was used to analyze the differences between treatments. p value < 0.05 was considered significant.
Based on the sequencing results from 16S rRNA gene amplicons, the eastern Pamir Plateau soil samples from different elevations contained 6834 OTUs belonging to 26 phyla, 71 classes, 142 orders, 212 families, and 399 genera. Unidentified species were highly abundant. The bacterial communities at different elevations on the Pamir Plateau were significantly different at different taxonomic levels. Across all the samples, the explanation for a large proportion of the phyla was as follows. Actinobacteriota (56.21–28.45%) was the predominant phylum, followed by Proteobacteria (10.12–21.59%), Bacteroidota (7.04–9.73%), Acidobacteriota (4.30–12.72%), and Gemmatimonadota (5.55–8.06%). The top ten dominant phyla in terms of abundance were the same at both low and middle elevations. However, there were differences in relative abundance, with only Myxococcota being more abundant than Firmicutes at high altitudes, representing a difference between the low and middle elevations. In the three elevation ranges, the Actinobacteriota abundance decreased with increasing elevation. Meanwhile, the abundances of Proteobacteria, Bacteroidetes, Gemmatimonadota, and Acidobacteria increased with increasing elevation (Figure 1). This indicates that the four phyla with increased abundance are better suited for higher elevation habitats.
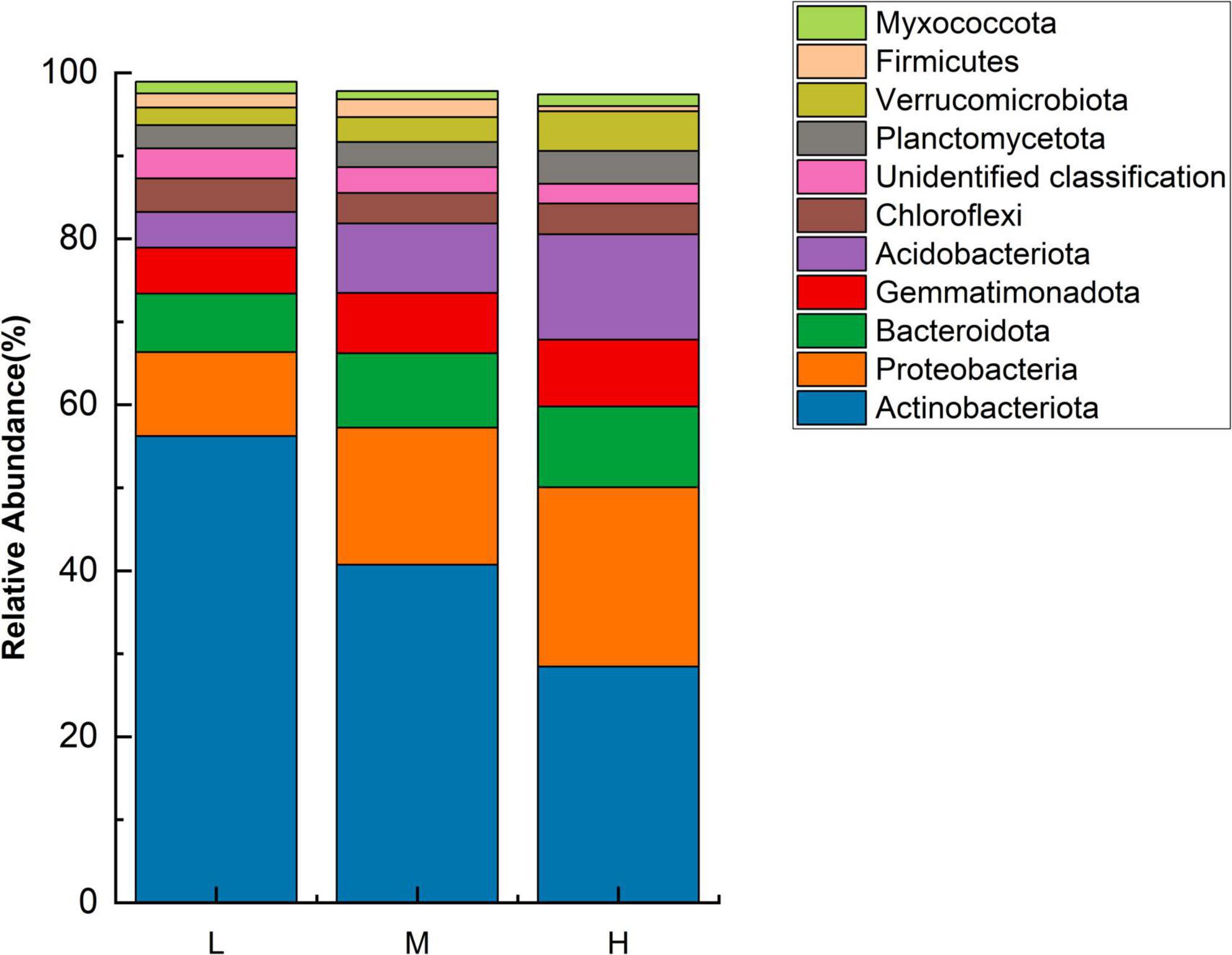
Figure 1. Relative abundance of major bacterial phyla at different elevations (n = 3) in the Eastern Pamir region.
At the genus level, the highest abundances of unidentified genera were observed at all three elevations. The dominant genera identified were Rubrobacter, Sphingomonas, Nocardioides, and Solirubrobacter. There were differences in the dominant genera existed at different elevations. The low elevation dominant genera were Rubrobacter (4.43%), Nocardioides (1.47%), and Glycomyces (1.46%). The medium elevation dominant genera were Rubrobacter (3.46%), Sphingomonas (1.44%), and Pontibacter (1.44%). At the higher elevation, the dominant genera were Sphingomonas (2.15%), Subgroup 10 (1.25%), and Solirubrobacter (1.02%). With increasing elevation, the relative abundances of Rubrobacter and Nocardioides decreased, Sphingomonas increased, and Solirubrobacter remained stable (Figure 2).
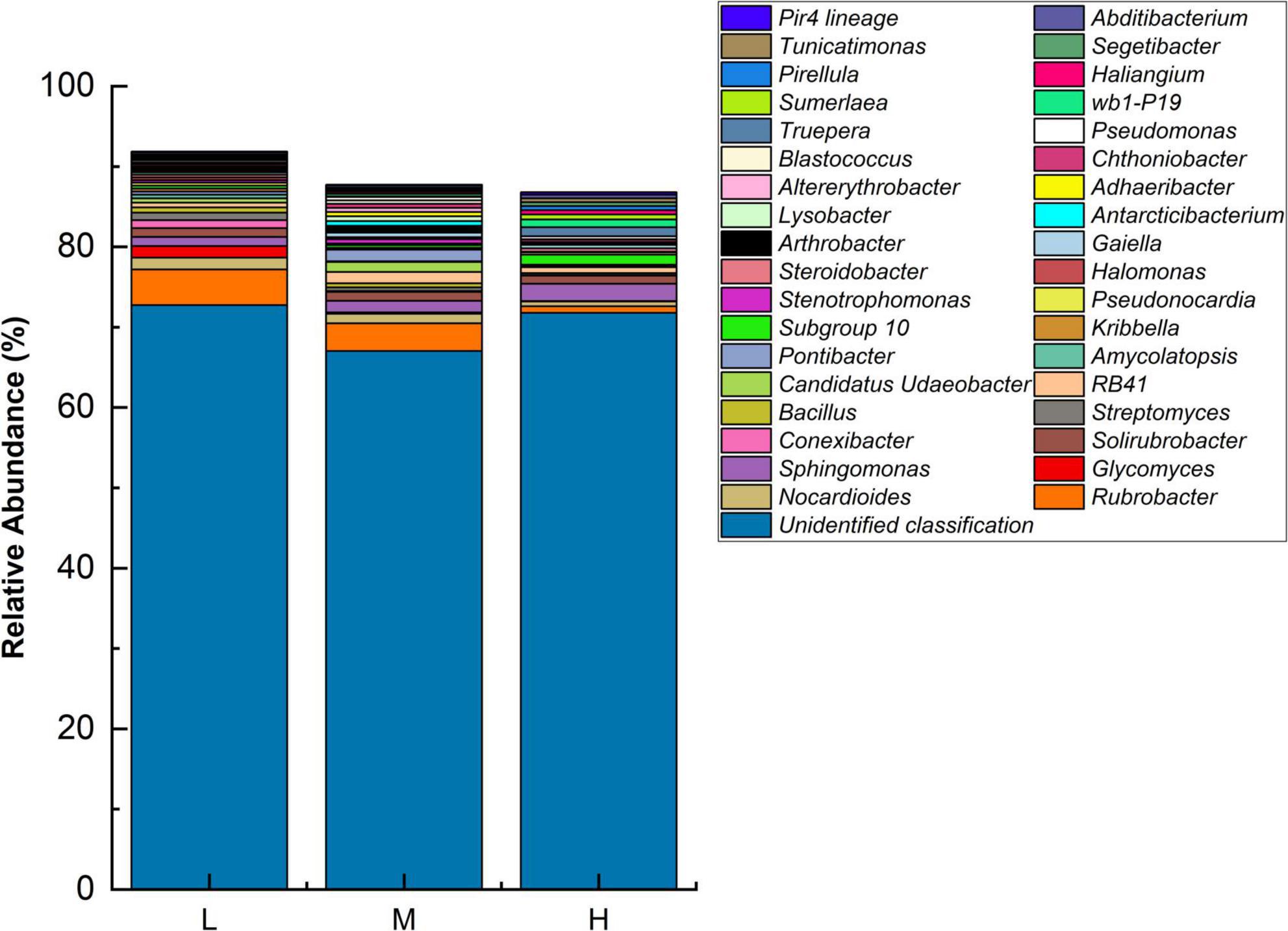
Figure 2. Relative abundance of major bacterial genera at different elevations (n = 3) in the Eastern Pamir region.
The alpha diversity indices for bacterial communities in the soil samples from different elevations were analyzed. This included the Chao1 observed_features index for species richness, faith_pd for phylogenetic diversity, and the Shannon and Simpson indices for species diversity. The species richness and diversity indices were the lowest in the low elevation samples, significantly different from those in the middle and high elevations (Table 2). In contrast, diversity was highest at higher elevations but was not significantly different from species diversity at mid-elevations. Species diversity and richness indices increased with increasing elevation, indicating that higher elevation areas had higher species richness and diversity.
Bacterial Alpha diversity and taxonomic composition analyses were performed using the community clustering approach of principal coordination analysis (PCoA). Unweighted UniFrac Bray–Curtis PCoA analysis showed that 31.4% of the total variation was explained by axis 1 and 21.4% by axis 2 (Figure 3). The low- and high-evaluation samples were significantly separated spatially, indicating a low similarity and high specificity of the community composition between the two samples. Middle elevation samples overlapped with all other samples, suggesting a similar species community composition between low and middle elevations, and between middle and high elevations.
The core microbiome has been defined in different ways (Sun et al., 2023). Turnbaugh et al. (2007, 2009) first proposed the core microbiome concept as “all taxa that are common to the microbiomes in all or the vast majority of habitats”. Delgado-Baquerizo et al. (2018) and Shade and Stopnisek (2019) defined core microbiome based on abundance-occupancy distributions that include highly abundant and ubiquitous taxa. In conclusion, core members is crucial to understanding the assembly mechanism of communities and identifying functionally important community members.
Based on a 97% similarity level, groups were set up at elevation intervals to obtain the OTUs data for each group of samples. The results of the Venn diagram for 6834 OTUs from 48 soil samples from the Pamir Plateau region are shown in Figure 4. There were 2966 identical OTUs at the three elevations, which accounted for 43.4% of the total OTUs. This indicated the presence of a core microbiome in the soil samples from different elevations. Among the total 2966 OTUs, the OTUs with higher relative abundance, that is, a relative abundance greater than 1%, were annotated as Sphingomonadaceae_B_OTU_17858, Rubrobacter, and Micrococcaceae_B_OTU_1232. These results suggest that these taxa have adapted well to the Pamir Plateau ecosystem and can survive in soil environments at different elevations.
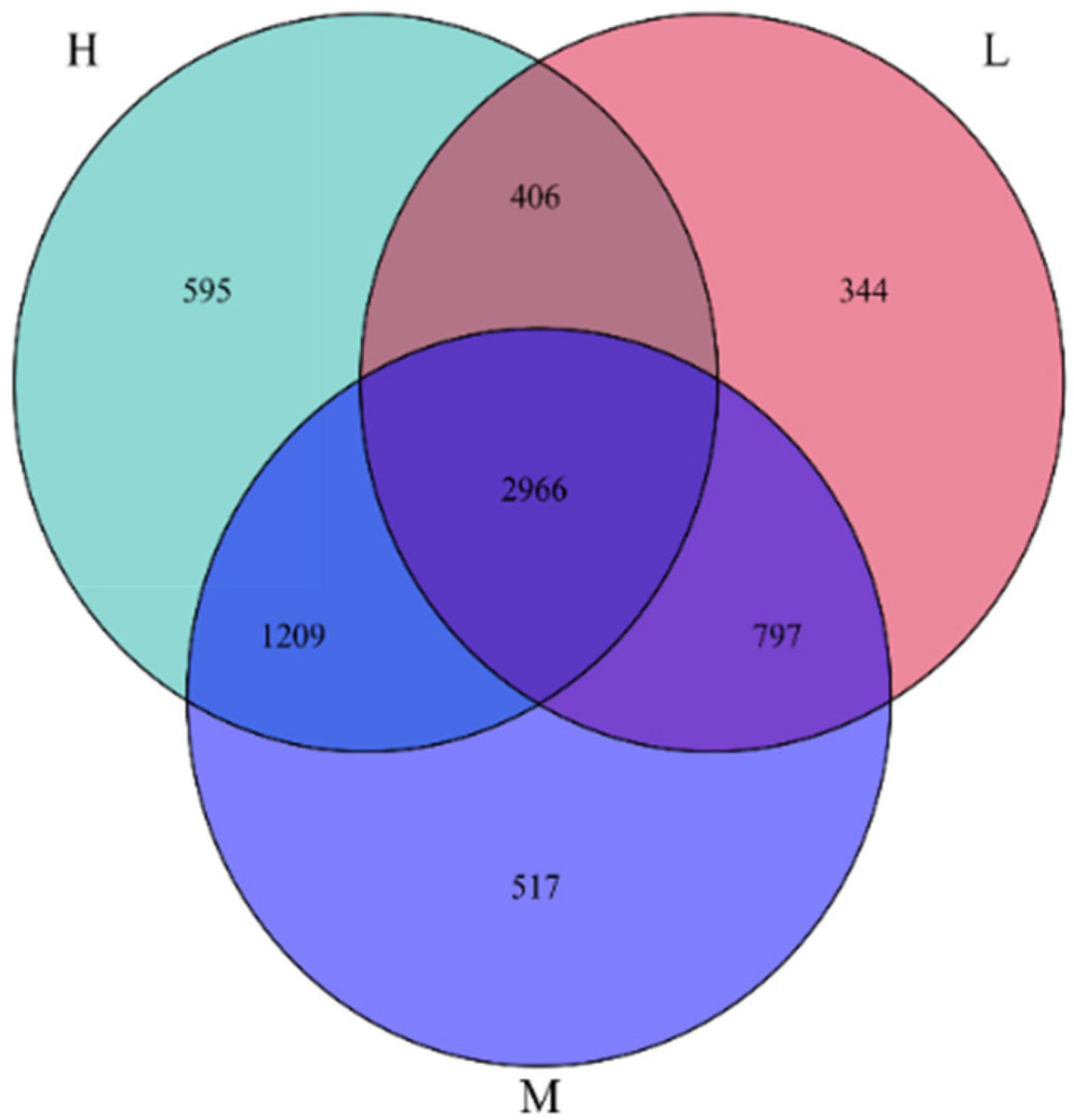
Figure 4. Venn diagram of the number of OTUs of samples at different elevation in the eastern Pamir Plateau
There were substantial differences in the OTUs at different elevations. The numbers of OTUs specific to low, medium, and high elevations were 344, 517, and 595, respectively, accounting for 5.03, 7.57, and 8.71% of the total OTUs, respectively. This indicates that there were specific microbial communities in the soil samples at different elevations with a minor percentage. However, they were sensitive to elevation changes.
The soil physicochemical properties were analyzed at three different elevations, and the results are shown in Table 3. The UV intensity increased with elevation, but the change was not significant between middle and high elevations. The soil temperature decreased from 25.7°C to 17.53°C with elevation. The OM increased from 4.63 g kg–1 to 12.03 g kg–1 with elevation. Soil AP content and pH increased with elevation. AN increased, and then decreased with altitude. The Salt initially increased and then decreased. TN content was significantly higher at high elevations than at low and middle elevations. Soil moisture and available potassium (AK) did not change significantly. Overall, the soil samples from different elevations in the eastern Pamir Plateau were nutrient-poor, arid, low-temperature alkaline soils.
Collected environmental factor information and measured physicochemical characteristics of soil samples from the three elevations were correlated with the top 10 bacterial phyla in abundance (Figure 5). The RDA showed that 78.71% of the total variation was explained by Axis 1 and 5.45% by Axis 2. We found that UV (Contribution % = 60.9, p = 0.022) was the most important factor driving changes in the bacterial community. Elevation (p = 0.022), OM (p = 0.044), TN (p = 0.022), AK (p = 0.022), and Salt (p = 0.022) were the relevant influencing factors.
Figure 5 showed elevation was significantly and positively correlated with Proteobacteria, Bacteroidetes, Gemmatimonadota, Verrucomicrobiota, Myxococcus, Acidobacteriota, and Planctomycetota. UV radiation was significantly and positively correlated with Proteobacteria, Bacteroidetes, Gemmatimonadota, Verrucomicrobiota, Myxococcus, and Acidobacteria. OM was significantly and negatively correlated with Firmicutes, Bacteroidetes, and unidentified classifications. TN was negatively correlated with Firmicutes and Bacteroidetes. Temperature was negatively correlated with Firmicutes, Bacteroidetes, and Proteobacteria. AK was negatively correlated with Verrucomicrobiota, Planctomycetota, Myxococcus, and Acidobacteria. Planctomycetota, Chloroflexi, Actinobacteria, unidentified classification, and Firmicutes. Salt was negatively correlated with Chloroflexi, Actinobacteria, and a unidentified classification, whereas Firmicutes was positively correlated. The low elevation samples had the highest correlation with Salt, while the high elevation samples were positively correlated with factors including elevation and OM.
Network co-occurrence showed the ecological relationships among species in the microbial communities. Screening the top 100 OTUs for species abundance with a correlation coefficient of 0.8 and P = 0.05 (Figure 6), the results showed that positive correlations arose among 11 OTUs belonging to three phyla, namely, Proteobacteria, Actinobacteria, and Gemmatimonadota. OTUs with the highest betweenness and closeness centralities were annotated as Sphingomonas, Gemmatimonadaceae_B_OTU_8822, and Solirubrobacter. These OTUs were located on the lines of other nodes. These changes resulted in relevant OTU variations. Therefore, OTUs are the core microorganisms in the bacterial co-occurrence network, and changes in them caused significant changes in microbial communities.
The random forest importance of species (variable) points showed that the genera Glycomyces, Streptomyces, and Chthoniobacter had the highest importance of species (variable) points across soil samples (Figure 7).
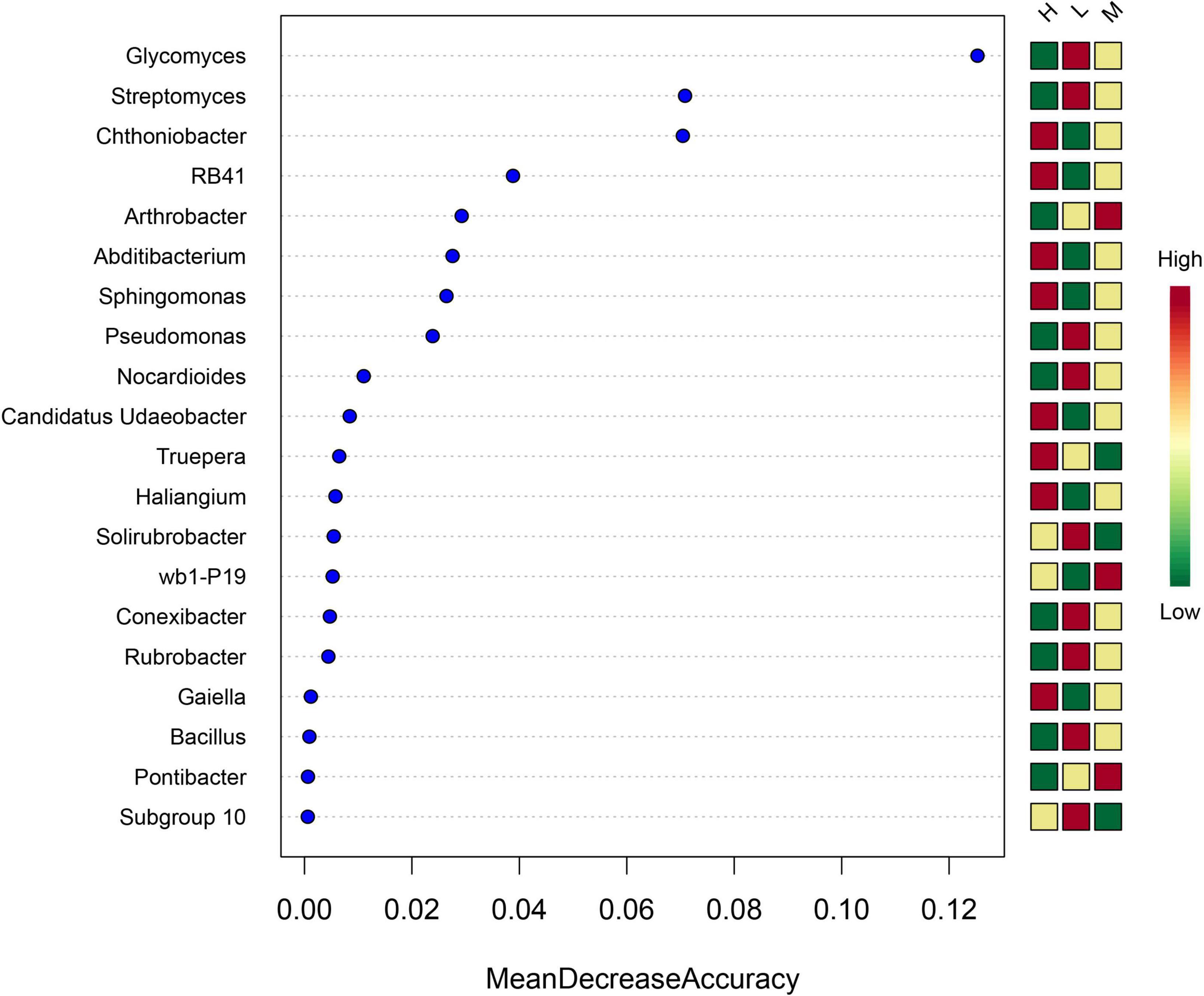
Figure 7. Importance of species (variable) points for the top 20 soil bacterial genera as affected by elevation.
Functional prediction of microbial communities using the PICRUSt2 database reflects microbial community function to a certain extent. A total of 10 major functional classifications were obtained on the KEGG secondary classification (Figure 8A). The top three were aging, drug resistance, antineoplastics, replication, and repair. Most of these pathways were enhanced at mid-elevations and reduced at higher elevations but were still stronger than at lower elevations. At Level 3 KEGG, the top three pathways were procyanidin biosynthesis, insect hormone biosynthesis, and adipocytokine signaling, which were inhibited at high elevations. Platinum drug resistance pathways increased with elevation. Other pathways increased at mid-altitudes, with no significant difference between low and high elevations (Figure 8A).
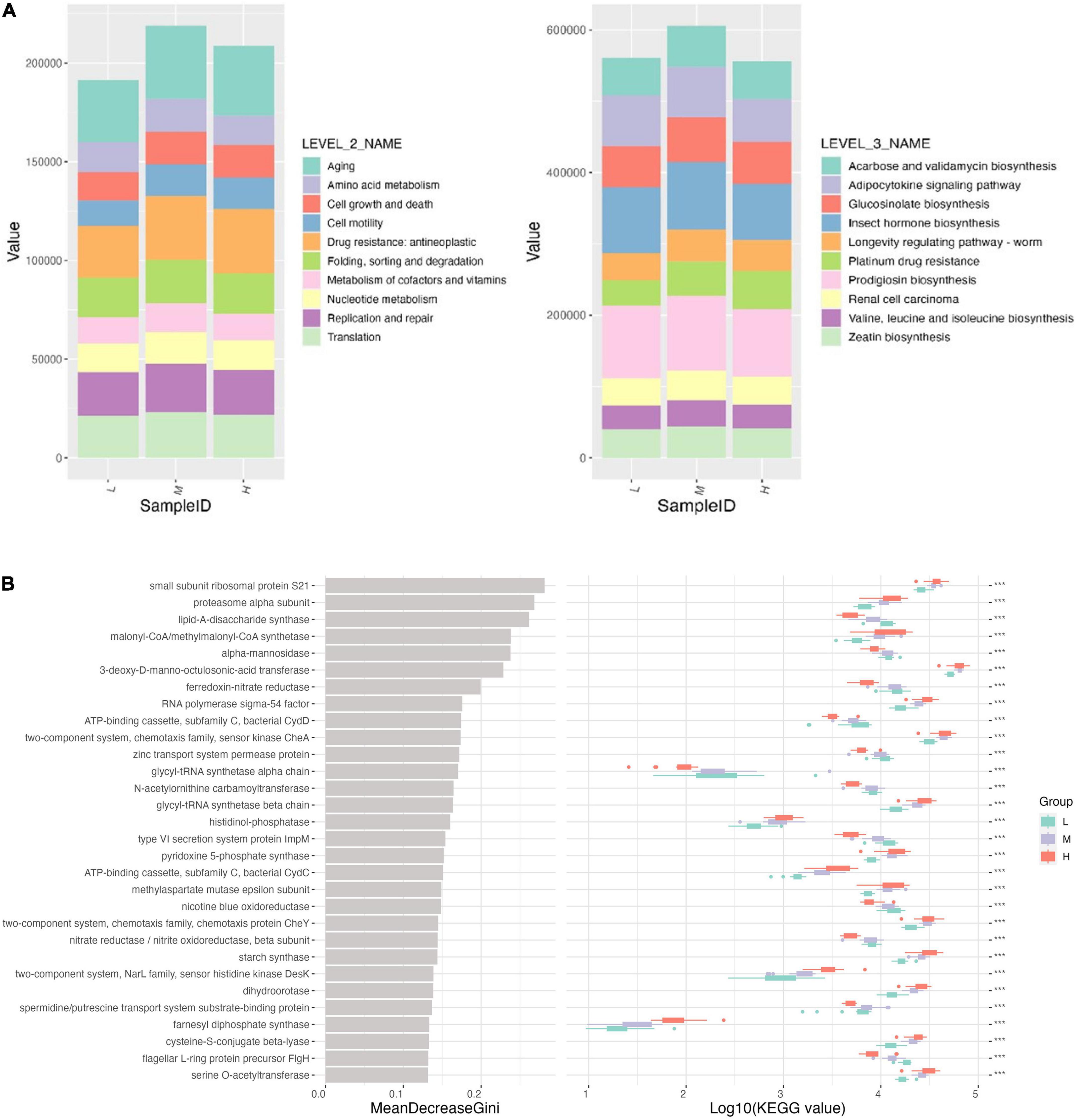
Figure 8. (A) Function profiling showing divergences in rare and abundant bacterial functions at KEGG module levels 2 and 3. (B) Functional pathway importance of the top 30 pathways affected by elevation.
In Random Forest, the small subunit ribosomal protein S21, proteasome alpha subunit, and lipid-A-disaccharide synthase were the most important for maintaining the stability of the bacterial community at different elevations (Figure 8B). Meanwhile, each pathway differed significantly among the elevations.
Based on the results of previous studies, we selected 55 genera belonging to five bacterial phyla that have been reported to have radiation resistance potential (Figure 9A). Among them, Actinobacteria had high diversity with 24 genera, followed by Proteobacteria with 18 genera, Firmicutes with six genera, Bacteroidetes with five genera, Deinococcus–Thermus with two genera, and Deinococcus and Truepera. These two genera include bacteria that are resistant to harsh environments, such as Deinococcus radiodurans, which is a radiation-resistant microorganism.
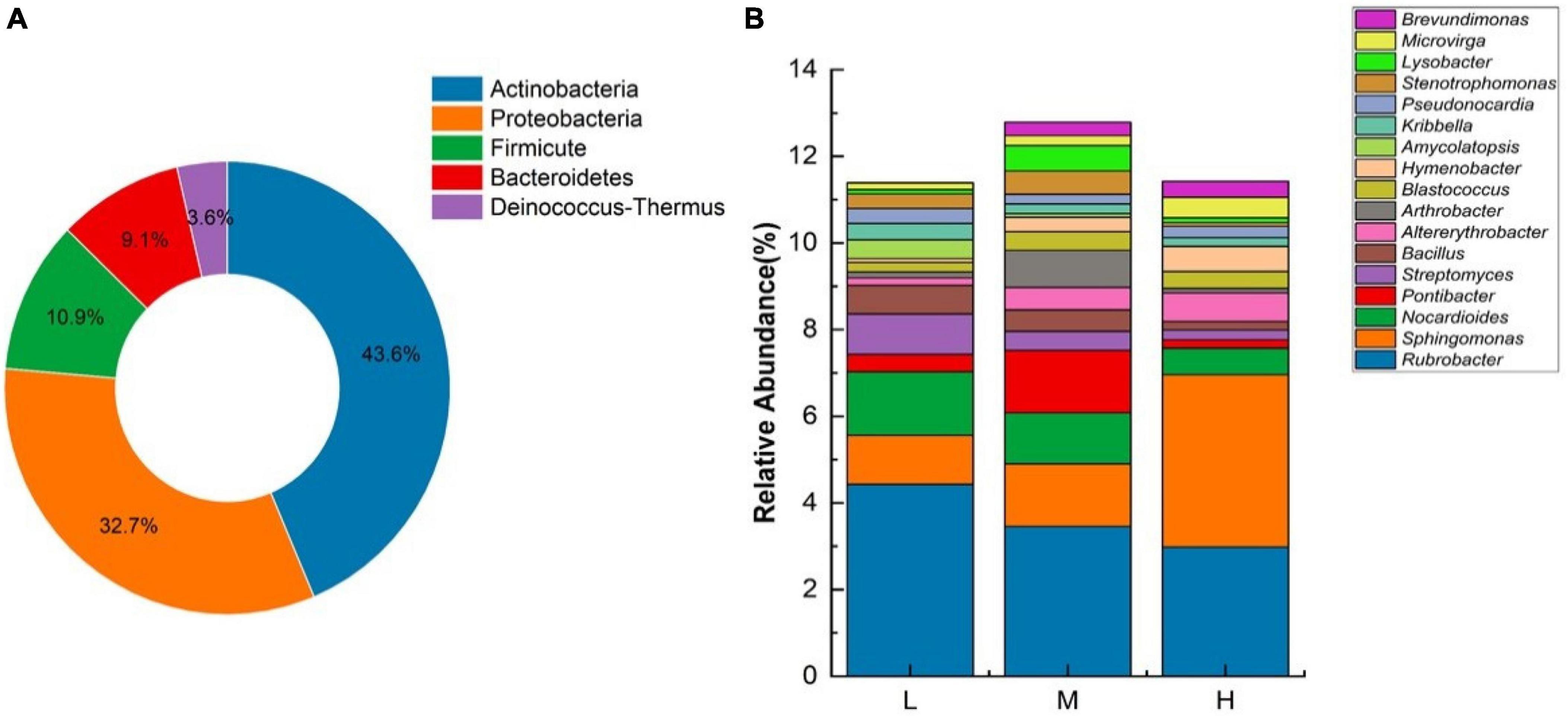
Figure 9. (A) Radiation-resistant bacterial community composition at different elevations on the Eastern Pamir Plateau based on the phyla level. (B) Radiation-resistant bacterial community composition at different elevations of the Eastern Pamir Plateau based on genus level.
The top 10 potentially radiation-tolerant microorganisms distributed at each elevation were selected and mapped to their relative abundance (Figure 9B). The top five genera in terms of abundance of potential radiation-tolerant microorganisms were Rubrobacter, Sphingomonas, Nocardioides, Pontibacter, and Streptomyces. Species abundance varied greatly between elevations. At low elevations, Rubrobacter, Nocardioides, and Sphingomonas are the dominant genera. At the middle elevations, the dominant genera were Rubrobacter, Sphingomonas, and Pontibacter. At high altitudes, the dominant genera were Sphingomonas and Rubrobacter. The abundance of Rubrobacter, Nocardioides, and Streptomyces genera showed a decreasing trend at 4.43–2.98%, 1.47–0.61%, and 0.94–0.22%, respectively. The abundance of Sphingomonas genera showed an increasing trend (1.13–3.98%) among different elevations. The Pontibacter relative abundance initially increased, and then decreased. Deinococcus and Truepera were also detected at different elevations with abundances ranging from 0.0018–0.0039% to 0.10–0.16%, respectively. Because the dominant genera in the samples from different elevations were potentially radiation-resistant, this region contains rich resources for radiation-resistant microorganisms.
A Venn diagram showed that the distribution of OTUs differed across habitats (Figure 10). A total of 472 OTUs sequences were obtained, of which 201 were common among the three elevations, accounting for 42.58% of the total. The numbers of OTUs specific to the low, medium, and high elevations were 35 (7.42%), 50 (10.59%), and 40 (8.47%), respectively. This indicated that the diversity of radiation tolerant species was greater at the middle elevations.
The correlation of community composition across sites was analyzed using Unfaire Bray–Curtis PCoA (Figure 11). Principal components 1 (PCoA1) and 2 (PCoA2) explained 17.4% and 13.8% of the radiation-tolerant bacteria in the soil samples, respectively. The samples were spatially overlapping, indicating that the radiation-resistant species were widely distributed among the three altitude zones. Only some strains were distributed at low, middle, and high elevations, and most radiation-resistant microorganisms survived more stably at different elevations, forming a stable core system.
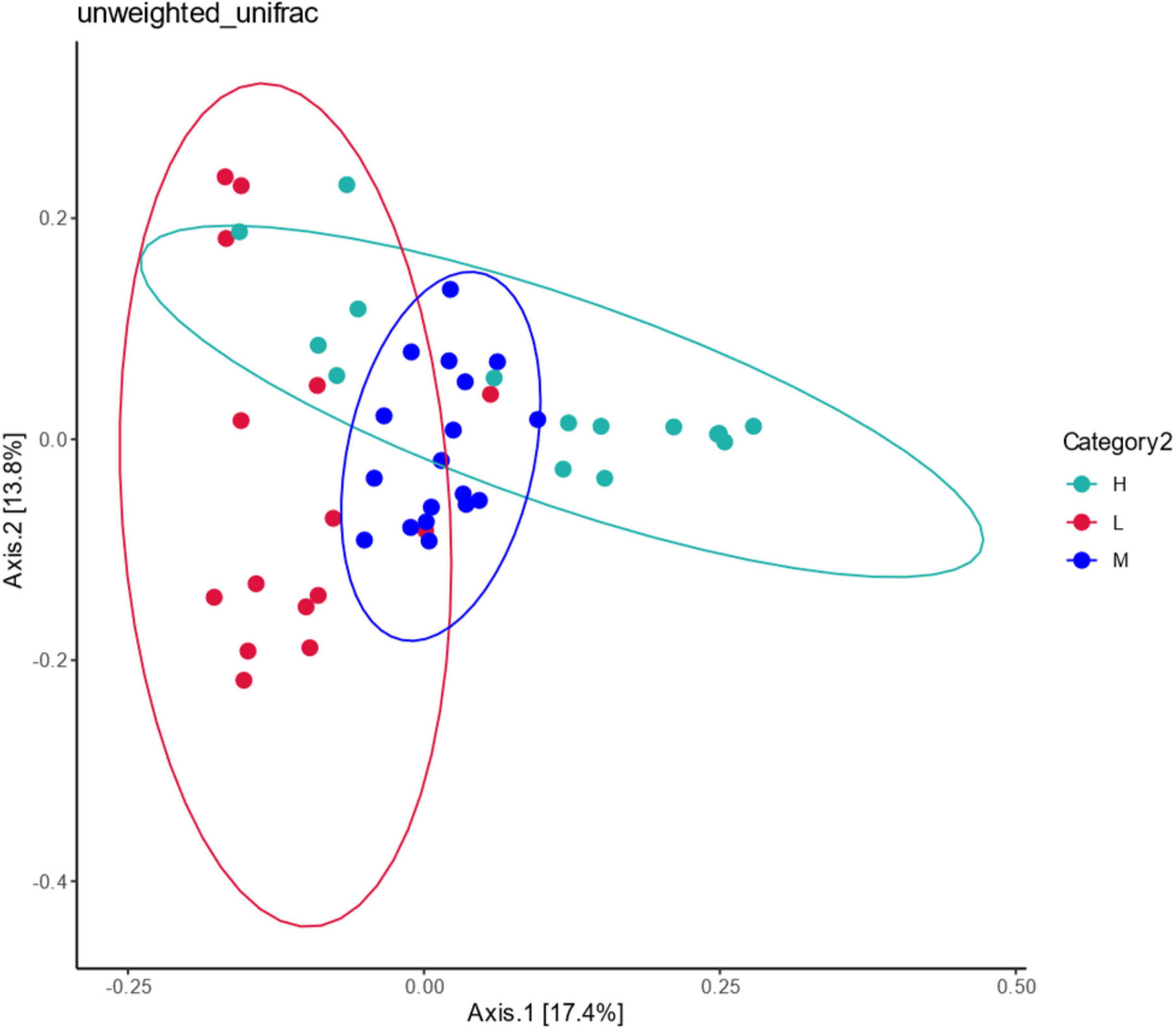
Figure 11. Principal coordination analysis of potentially radiation-tolerant bacteria at different elevations in East Pamir.
Microorganisms are the main drivers of biochemical cycles, and their community structure influences ecosystem stability. Plateau regions have limited soil nutrients, fragile ecosystems, and harsh environments. The Pamir Plateau is located in central Asia, with a total glacial coverage of approximately 12,260 km2 (Yao et al., 2012). It mainly has cold mountainous deserts. Low vegetation cover, scarce precipitation, and poor soil nutrient concentration and water content result in an extremely arid environment (Mętrak et al., 2015, 2017). To date, prior studies on microorganisms in plateau regions have focused on the Tibetan, Loess, and Mongolian Plateaus. These studies have involved variations in soil enzyme activities and microbial communities along elevation gradients (Fan et al., 2021), reflections of functional microorganisms in soil to natural factors such as nitrogen deposition and precipitation (Xu et al., 2021; Li et al., 2023), or different responses of soil microorganisms from different plant communities to changes in environmental stresses (Xu et al., 2023). Relatively few studies have been conducted on the distribution of bacterial communities in East Pamir.
Under the influence of environmental conditions, soil microorganisms adapt to the environment by regulating their community structure (Zeng et al., 2021). Zhang et al. (2021) studied bacterial diversity in desert habitats in the Tibetan Plateau region at an average altitude of 2800 m, where Proteobacteria, Actinobacteria, Bacteroidetes, and Chloroflexi were the dominant phyla. Cui et al. (2019) studied the microbial diversity of alpine ecosystems in the Tibetan Plateau at 2800–3500 m and found that the dominant phyla included Acidobacteria and Proteobacteria, whereas Actinobacteria, Bacteroidetes, and Gemmatimonadota were the dominant bacterial phyla. In this study, Actinobacteria was the dominant phylum, followed by Proteobacteria, Bacteroidetes. Acidobacteria and Gemmatimonadota were the secondary dominant phyla, which is in line with the results of Cui et al. (2019) and Zhang et al. (2021), with the same results but with differences in species abundance. Rubrobacter, Sphingomonas, Nocardioides, and Solirubrobacter were the dominant genera at the genus level. There were differences in the dominant genera among different elevations, which may be mainly because of the ecology of the sampling area, geographic location, climate change, and anthropogenic activities (Mou et al., 2020).
There are three main patterns of relationships among species diversity, community structure, and elevation: monotonically decreasing (Ding et al., 2023), humping (Nottingham et al., 2018), and U-shaped (Shen et al., 2020). Many studies have shown that Alpha diversity decreases with elevation, whereas others have reported the greatest diversity at mid-elevations, or insignificant elevation patterns (Yao et al., 2017). Ogwu et al. (2019) found that microbial abundance increased with elevation in a study on alpine microorganisms in Japan. Shen et al. (2013) found that soil bacterial diversity did not vary with elevation in the Changbai Mountain region. In contrast, the present study found differences in bacterial community diversity at different elevation, with an increasing trend with increasing elevation. Although the data described vary, mechanisms driving the different elevation patterns have not been explicitly discussed. Yao et al. (2017) revealed that the elevational patterns of dominant soil microbial taxa along Changbai mountain slope were shaped more by vegetation and soil nutrient status. This suggests that discussion about soil community structure and species diversity at different elevations also needs to incorporate the effects of environmental factors.
Elevation and soil are the main factors driving soil microbial communities (Tang et al., 2020). Bacterial communities respond to environmental factors in a complex manner, with pH usually being the main influence (Griffiths et al., 2011; Bahram et al., 2018). In addition, factors such as soil temperature, soluble salt, nitrogen, Organic matter, and precipitation also affect the composition of soil bacterial communities. Zhao et al. (2022) studied soil characteristics and microbial communities along an altitudinal gradient (1700–2300 m) in a Huashan pine forest in the Qinling Mountains and found that in addition to altitudinal factors, pH, soil temperature, OM, and AK were important factors influencing microbial communities. Wang et al. (2022) found that differences in bacterial community composition at the OTU level in soils and lake sediments from the lakeshore of Kekexili with a mean elevation of 4600 m were mainly determined by the mean annual temperature, salinity, total organic carbon, and TN content. This finding is similar to that of the present study. The environmental factors in this study were significantly distributed at different elevations and had a significant influence on microbial community composition. In study of the effects of UV on microorganisms, Natarajan et al. (2008) found the melanin content of Aspergillus niger spores on south slopes with strong UV radiation was three times higher than on shaded north slopes (south slopes have 2–8 times more sunlight than north slopes) in the “Evolution Canyon”, Mount Carmel, Israel. In this study, pH did not show a significant correlation to community composition. However, UV, OM, temp, TN, AK, and Salt were environmental factors affecting the bacterial community in the Pamir region and were significantly correlated with the major bacterial phyla. Low altitude samples showed the strongest correlation with Salt, whereas high elevation samples showed a positive correlation with factors such as elevation and OM.
Environmental factors at low elevations are highly variable due to human activities. Meanwhile, environmental constraints at high elevations, such as low temperatures, dryness, and high radiation intensity, lead to fragile ecosystems (Nottingham et al., 2018; Portero et al., 2019). The co-occurrence network among microorganisms reflects the function and stability of a biome (Ma et al., 2020). The co-occurrence network was used to analyze the key interactions between microorganisms and their response to the environment, examine the fundamentals of microbial communities in different soil environments and identify key hypothetical taxa in the community. In this study, 11 OTUs in the co-occurrence network belonged to three phyla, Proteobacteria, Actinobacteria, and Gemmatimonadota, of which Sphingomonas, Gemmatimonadaceae_B_OTU_8822, Solirubrobacter were in a central position as core microorganisms in the symbiotic network. Gene function prediction revealed differences in gene pathways between soil samples from neighboring elevations. Random forest results also showed that the small subunit ribosomal protein S21, proteasome α-subunit, and lipid-A-disaccharide synthase were the most important factors in maintaining the stability of the bacterial community between altitudes.
Since 1956, when DR was isolated from canned meat by Andersen et al. (1956), which deteriorated after radiation sterilization (Andersen et al., 1956), radiation-resistant microbial resources have attracted the attention of researchers. More than 1500 strains of radiation-resistant microbial strains have been identified. The isolation sources are mainly deserts, radiation-contaminated areas, hot springs, and marine sediments, which are seldom found in highland areas. In this study, we predicted and screened out species with a potential radiation tolerance function based on the currently reported genera of radiation tolerant bacteria and revealed their distribution at different elevations. The results showed that Rubrobacter, Sphingomonas, Nocardioides, Pontibacter and Streptomyces were the dominant genera among the potentially radiation-tolerant species in the Pamir Plateau, while Rubrobacter, Sphingomonas and Nocardioides were the dominant genera in the Pamir Plateau at different altitudes. This suggests this region is rich in potential radiation tolerant microbial resources. In the bacterial co-occurrence network, the genera Rubrobacter and Sphingomonadaceae, which are highly abundant, have been reported to be radiation tolerant. Therefore, it is assumed that the strains from this habitat also have a high potential for radiation tolerance.
A study of bacterial communities in soil samples from different elevations of the Pamir Plateau showed that Actinobacteria was the dominant phylum, whereas Proteobacteria, Bacteroidetes, Acidobacteria, and Gemmatimonadota were the subdominant phyla. The genera were dominated by Rubrobacter, Sphingomonas, Nocardioides, and Solirubrobacter. Species diversity increased slightly with elevation, whereas community specificity increased with elevation. Elevation and UV radiation were the main factors affecting the bacterial communities. Under different environmental conditions at different elevations, soil bacteria can adapt to their environment by altering KEGG pathways. The soil samples from different elevations were rich in typical radiation-tolerant resources. The main genera were Rubrobacter, Sphingomonas, Nocardioides, Pontibacter, and Streptomyces. The composition of radiation-tolerant bacterial communities varied significantly among different elevations. The results of this study have shown the composition and distribution of bacterial communities at different elevations on the Pamir Plateau. It is of considerable importance to examine the potential genera of radiation-resistant microorganisms. This will be important for the succession of bacterial microorganisms on the plateau and for the study of radiation-resistant microbial germplasm resources and their biofunctional applications.
The datasets presented in this study have been deposited in the Sequence Read Archive of the National Center for Biotechnology Information (SRA, NCBI, www.ncbi.nlm.nih.gov/sra) under the BioProject IDs PRJNA1032247. The ASV tables, fasta sequences, and taxonomy data for bacteria were uploaded to Figshare repository, resulting in https://doi.org/10.6084/m9.figshare.25435474.
JZ: Conceptualization, Funding acquisition, Investigation, Writing–original draft, Writing–review and editing. H-NW: Formal analysis, Writing–original draft. Q-YT: Data curation, Investigation, Writing–review and editing. M-YG: Investigation, Supervision, Writing–review and editing. Z-DZ: Conceptualization, Investigation, Writing–review and editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of the article. This research was funded by the Project of Fund for Stable Support to Agricultural Sci-Tech Renovation (xjnkywdzc-2023005), National Natural Science Foundation of China (32360033), Special Incubation Project of Science and Technology Renovation of Xinjiang Academy of Agricultural Sciences (xjkcpy-2022004).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agapov, A. A., and Kulbachinskiy, A. V. (2015). Mechanisms of stress resistance and gene regulation in the radioresistant bacterium Deinococcus radiodurans. Biochemistry 80, 1201–1216. doi: 10.1134/S0006297915100016
Andersen, A. W., Nordan, H. C., Cain, R. F., Parrish, G., and Duggan, D. (1956). Studies on a radio-resistant micrococcus. Isolation, morphology, cultural characteristics, and resistance to gamma radiation. Food Technol. 10, 575–578.
Bagousse-Pinguet, Y., Soliveres, S., Gross, N., Torices, R., Berdugo, M., and Maestre, F. T. (2019). Phylogenetic, functional, and taxonomic richness have both positive and negative effects on ecosystem multifunctionality. Proc. Natl. Acad. Sci. U.S.A. 116, 8419–8424. doi: 10.1073/pnas.1815727116
Bahram, M., Hildebrand, F., Forslund, S. K., Anderson, J. L., Soudzilovskaia, N. A., Bodegom, P. M., et al. (2018). Structure and function of the global topsoil microbiome. Nature 560:233. doi: 10.1038/s41586-018-0386-6
Barandun, M., Pohl, E., Naegeli, K., McNabb, R., Huss, M., Berthier, E., et al. (2021). Hot Spots of glacier mass balance variability in central Asia. Geophys. Res. Lett. 48:e2020GL092084. doi: 10.1029/2020GL092084
Csardi, G., and Nepusz, T. (2006). The igraph software package for complex network research. InterJ. Complex Syst. 1695, 1–9.
Cui, Y., Bing, H., Fang, L., Wu, Y., Yu, J., Shen, G., et al. (2019). Diversity patterns of the rhizosphere and bulk soil microbial communities along an altitudinal gradient in an alpine ecosystem of the eastern Tibetan Plateau. Geoderma 338, 118–127. doi: 10.1016/j.geoderma.2018.11.047
Delgado-Baquerizo, M., Oliverio, A. M., Brewer, T. E., Benavent-Gonzalez, A., Eldridge, D. J., Bardgett, R. D., et al. (2018). A global atlas of the dominant bacteria found in soil. Science 359, 320–325. doi: 10.1126/science.aap9516
Ding, B., Feng, L., Ba, S., Jiang, X., Liu, G., and Liu, W. (2023). Temperature drives elevational diversity patterns of different types of organisms in Qinghai-Tibetan Plateau wetlands. iScience 26:107252. doi: 10.1016/j.isci.2023.107252
Dormann, C. F., Fründ, J., Blüthgen, N., and Gruber, B. (2009). Indices, graphs and null models: Analyzing bipartite ecological networks. Open Ecol. J. 2, 7–24. doi: 10.2174/1874213000902010007
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Fan, S. Y., Sun, H., Yang, J. Y., Qin, J. H., Shen, D. J., and Chen, Y. X. (2021). Variations in soil enzyme activities and microbial communities along an altitudinal gradient on the eastern qinghai–Tibetan plateau. Forests 12, 681–681. doi: 10.3390/f12060681
Francos, M., Sánchez, G. C., Girona, G. A., and Fernández, G. V. (2021). Influence of topography on sediment dynamics and soil chemical properties in a Mediterranean forest historically affected by wildfires: NE Iberian Peninsula. Environ. Earth Sci. 80:436. doi: 10.1007/s12665-021-09731-2
Fredrickson, J. K., Zachara, J. M., Balkwill, D. L., Kennedy, D., Kostandarithes, H. M., Daly, M. J., et al. (2004). Geomicrobiology of high-level nuclear waste-contaminated vadose sediments at the Hanford site, Washington state. Appl. Environ. Microbiol. 70, 4230–4241. doi: 10.1128/AEM.70.7.4230-4241.2004
Gabani, P., and Singh, O. V. (2013). Radiation-resistant extremophiles and their potential in biotechnology and therapeutics. Appl. Microbiol. Biotechnol. 97, 993–1004. doi: 10.1007/s00253-012-4642-7
Gao, Y. Y., Zhang, G. X., Jiang, S. Y., and Liu, Y. X. (2024). Wekemo Bioincloud: A user-friendly platform for meta-omics data analyses. iMeta 3:e175. doi: 10.1002/imt2.175
Griffiths, R. I., Thomson, B. C., James, P., Bell, T., Bailey, M., and Whiteley, A. S. (2011). The bacterial biogeography of British soils. Environ. Microbiol. 13, 1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x
Ji, M., Kong, W., Jia, H., Delgado-Baquerizo, M., Zhou, T., Liu, X., et al. (2022). Polar soils exhibit distinct patterns in microbial diversity and dominant phylotypes. Soil Biol. Biochem. 166:108550. doi: 10.1016/j.soilbio.2022.108550
Khomutovska, N., de los Ríos, A., and Jasser, I. (2020). Diversity and colonization strategies of endolithic cyanobacteria in the cold mountain desert of Pamir. Microorganisms 9:6. doi: 10.3390/microorganisms9010006
Khomutovska, N., de los Ríos, A., Syczewski, M. D., and Jasser, I. (2021). Connectivity of edaphic and endolithic microbial niches in cold mountain desert of eastern Pamir (Tajikistan). Biology 10:314. doi: 10.3390/biology10040314
Khomutovska, N., Jerzak, M., Kostrzewska-Szlakowska, I., Kwiatowski, J., Suska-Malawska, M., Syczewski, M., et al. (2017). Life in extreme habitats: Diversity of endolithic microorganisms from cold desert ecosystems of eastern pamir. Polish J. Ecol. 65, 303–319. doi: 10.3161/15052249PJE2017.65.4.001
Li, M., Wang, J., and Saw, G. (2015). Culturable bacterial diversity in snow, ice and meltwater of the Yangbark Glacier, Muztag Ata. J. Glaciol. Geocryol. 37, 1634–1641.
Li, Y. M., Xiong, L. L., Yu, H., Zeng, K., Wei, Y. L., Li, H. Y., et al. (2023). Function and distribution of nitrogen-cycling microbial communities in the Napahai plateau wetland. Arch. Microbiol. 205:357. doi: 10.1007/s00203-023-03695-6
Ma, B., Wang, Y., Ye, S., Liu, S., Stirling, E., Gilbert, J., et al. (2020). Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 8:82. doi: 10.1186/s40168-020-00857-2
McCain, C. M. (2010). Could temperature and water availability drive elevational species richness patterns? A global case study for bats. Glob. Ecol. Biogeogr. 16, 1–13. doi: 10.1111/j.1466-8238.2006.00263.x
Mętrak, M., Chachulski, Ł, Navruzshoev, D., Pawlikowski, P., Rojan, E., Sulwiński, M., et al. (2017). Nature’s Patchwork: How water sources and soil salinity determine the distribution and structure of halophytic plant communities in arid environments of the Eastern Pamir. PLoS One 12:e0174496. doi: 10.1371/journal.pone.0174496
Mętrak, M., Sulwiński, M., Chachulski, Ł, Wilk, M. B., and Suskamalawska, M. (2015). “Creeping environmental problems in the Pamir mountains: Landscape conditions, climate change, wise use and threats,” in Proceedings of the climate change impacts on high-altitude ecosystems, (Berlin: Springer Science and Business Media LLC), 665–694. doi: 10.1007/978-3-319-12859-7_28
Monsalves, M. T., Ollivet-Besson, G. P., Amenabar, M. J., and Blamey, J. M. (2020). Isolation of a psychrotolerant and UV-C-resistant bacterium from elephant island, antarctica with a highly thermoactive and thermostable catalase. Microorganisms 8:95. doi: 10.3390/microorganisms8010095
Mou, X. M., Wu, Y. N., Niu, Z. Q., Jia, B., Guan, Z. H., Chen, J., et al. (2020). Soil phosphorus accumulation changes with decreasing temperature along a 2300 m altitude gradient. Agric. Ecosyst. Environ. 301:107050. doi: 10.1016/j.agee.2020.107050
Narasimha, A., and Basu, B. (2021). New insights into the activation of radiation desiccation response regulon in Deinococcus radiodurans. J. Biosci. 46:10. doi: 10.1007/s12038-020-00123-5
Natarajan, S., Isabella, G., Alex, B., Kazumasa, W., Shosuke, I., and Eviatar, N. (2008). Adaptive melanin response of the soil fungus Aspergillus niger to UV radiation stress at “evolution canyon”, Mount Carmel, Israel. PLoS One 3:e2993. doi: 10.1371/journal.pone.0002993
Nottingham, A. T., Fierer, N., Turner, B. L., Whitaker, J., Ostle, N. J., McNamara, N. P., et al. (2018). Microbes follow Humboldt: Temperature drives plant and soil microbial diversity patterns from the Amazon to the Andes. Ecology 99, 2455–2466. doi: 10.1002/ecy.2482
Ogwu, M. C., Takahashi, K., Dong, K., Song, H. K., Moroenyane, I., Waldman, B., et al. (2019). Fungal elevational Rapoport pattern from a high mountain in Japan. Sci. Rep. 9:6570. doi: 10.1038/s41598-019-43025-9
Pan, J., Wang, J., Zhou, Z., Yan, Y., Zhang, W., Lu, W., et al. (2009). IrrE, a global regulator of extreme radiation resistance in Deinococcus radiodurans, enhances salt tolerance in Escherichia coli and Brassica napus. PLoS One 4:e4422. doi: 10.1371/journal.pone.0004422
Paulino-Lima, I. G., Fujishima, K., Navarrete, J. U., Galante, D., Rodrigues, F., Azua-Bustos, A., et al. (2016). Extremely high UV-C radiation resistant microorganisms from desert environments with different manganese concentrations. J. Photochem. Photobiol. B Biol. 163, 327–336. doi: 10.1016/j.jphotobiol.2016.08.017
Portero, L. R., Alonso-Reyes, D. G., Zannier, F., Vazquez, M. P., Farías, M. E., Gärtner, W., et al. (2019). Photolyases and cryptochromes in UV-resistant bacteria from high-altitude Andean lakes. Photochem. Photobiol. 95, 315–330. doi: 10.1111/php.13061
Qi, H. Z., Wang, W. Z., He, J. Y., Ma, Y., Xiao, F. Z., and He, S. Y. (2020). Antioxidative system of Deinococcus radiodurans. Res. Microbiol. 171, 45–54. doi: 10.1016/j.resmic.2019.11.002
R Development Core Team (2016). R: A language and environment for statistical computing. Vienna: R Development Core Team.
Safaraliev, M. K., Odinaev, I. N., Ahyoev, J. S., Rasulzoda, K. N., and Otashbekov, R. A. (2020). Energy potential estimation of the region’s solar radiation using a solar tracker. Appl. Solar Energ. 4, 270–275. doi: 10.3103/S0003701X20040118
Shade, A., and Stopnisek, N. (2019). Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr. Opin. Microbiol. 49, 50–58. doi: 10.1016/j.mib.2019.09.008
Shang, D., Chen, Q., Jiang, M., Wang, B., Xie, Z., Yu, N., et al. (2021). Colonized extremophile Deinococcus radiodurans alleviates toxicity of cadmium and lead by suppressing heavy metal accumulation and improving antioxidant system in rice. Environ. Pollut. 284:117127. doi: 10.1016/j.envpol.2021.117127
Shen, C. C., Xiong, J. B., Zhang, H. Y., Feng, Y. Z., Lin, X. G., Li, X. Y., et al. (2013). Soil pH drives the spatial distribution of bacterial communities along elevation on Changbai Mountain. Soil Biol. Biochem. 57, 204–211. doi: 10.1016/j.soilbio.2012.07.013
Shen, C., Gunina, A., Luo, Y., Wang, J., He, J. Z., Kuzyakov, Y., et al. (2020). Contrasting patterns and drivers of soil bacterial and fungal diversity across a mountain gradient. Environ. Microbiol. 22, 3287–3301. doi: 10.1111/1462-2920.15090
Singh, O. V., and Gabani, P. (2011). Extremophiles: Radiation resistance microbial reserves and therapeutic implications. Appl. Microbiol. 110, 851–861. doi: 10.1111/j.1365-2672.2011.04971.x
Sun, X., Sharon, O., and Sharon, A. (2023). Distinct features based on partitioning of the endophytic fungi of cereals and other grasses. Microbiol. Spectr. 11, e00611–e00623. doi: 10.1128/spectrum.00611-23
Tang, M., Li, L., Wang, X., You, J., Li, J., and Chen, X. (2020). Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai Mountain. Sci. Rep. 10:12442. doi: 10.1038/s41598-020-69441-w
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Wang, X., Ren, Y., Yu, Z., Shen, G., Cheng, H., and Tao, S. (2022). Effects of environmental factors on the distribution of microbial communities across soils and lake sediments in the Hoh Xil Nature Reserve of the Qinghai-Tibetan Plateau. Sci. Total Environ. 838:156148. doi: 10.1016/j.scitotenv.2022.156148
Xu, M. J., Li, T. T., Liu, W., Ding, J. J., Gao, L. L., Han, X. G., et al. (2021). Sensitivity of soil nitrifying and denitrifying microorganisms to nitrogen deposition on the Qinghai–Tibetan plateau. Ann. Microbiol. 71:6. doi: 10.1186/s13213-020-01619-z
Xu, Y. T., Sun, R., Yan, W. M., and Zhong, Y. Q. W. (2023). Divergent response of soil microbes to environmental stress change under different plant communities in the Loess Plateau. Catena 230:107240. doi: 10.1016/j.catena.2023.107240
Yang, S. P., and Yan, P. (2008). Study on floristic phytogepgraphy of seed plants in the Pamirs of China. J. Xinyang Norm. Univers. Nat. Sci. Edn. 4, 543–546.
Yang, S. P., Jiang, J., and Yan, P. (2018). Eco-geographic distribution of rare and endangered plants and endemism in the Pamirs Region of China. J. Arid Land Resourc. Environ. 32, 115–120.
Yao, F., Yang, S., Wang, Z. R., Wang, X., Ye, J., Wang, X. G., et al. (2017). Microbial taxa distribution is associated with ecological trophic cascades along an elevation gradient. Front. Microbiol. 8:2071. doi: 10.3389/fmicb.2017.02071
Yao, T., Thompson, L., Yang, W., Yu, W., Gao, Y., Guo, X., et al. (2012). Different glacier status with atmospheric circulations in Tibetan Plateau and surroundings. Nat. Clim. Change 2, 663–667. doi: 10.1038/nclimate1580
Zeng, Q. C., Liu, D., and An, S. S. (2021). Decoupled diversity patterns in microbial geographic distributions on the arid area (the Loess Plateau). Catena 196:104922. doi: 10.1016/j.catena.2020.104922
Zhang, W., Bahadur, A., Zhang, G., Nasir, F., Zhang, B., Wu, X., et al. (2021). Bacterial diversity and community composition distribution in cold-desert habitats of Qinghai-Tibet Plateau, China. Microorganisms 9:262. doi: 10.3390/microorganisms9020262
Zhang, Y., Xu, G., Li, P., Li, Z., Wang, Y., Wang, B., et al. (2019). Vegetation change and its relationship with climate factors and elevation on the Tibetan Plateau. Int. J. Environ. Res. Public Health 16:4709. doi: 10.3390/ijerph16234709
Keywords: bacterial communities, elevation gradient, radiation resistance, environmental adaptation, Pamir plateau
Citation: Zhu J, Wang H-N, Tang Q-Y, Gu M-Y and Zhang Z-D (2024) Composition and distribution of bacterial communities and potential radiation-resistant bacteria at different elevations in the eastern Pamirs. Front. Microbiol. 15:1427806. doi: 10.3389/fmicb.2024.1427806
Received: 04 May 2024; Accepted: 03 June 2024;
Published: 19 June 2024.
Edited by:
Zhendong Yang, Chengdu University, ChinaCopyright © 2024 Zhu, Wang, Tang, Gu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhi-Dong Zhang, emhhbmd6aGVlZG9uZ0Bzb2h1LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.