
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 19 July 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1426510
Escherichia coli is one of the most important pathogens causing bloodstream infections (BSIs) throughout the world. We sought to characterize the phylogroup classification, major human sequence types (STs), antimicrobial resistance, presence of selected antimicrobial resistance and virulence genes, and genetic diversity of E. coli isolated from patients with BSIs at the University Hospital in Iran. A total of 100 E. coli bloodstream isolates were collected between December 2020 and June 2022. This study used PCR to investigate phylogenetic groups (A, B1, B2, C, D, E, and F), four major STs (ST69, ST73, ST95, and ST131), antibiotic resistance genes (ARGs), virulence-associated genes (VAGs), and pathogenicity islands (PAIs). Antimicrobial susceptibility testing was done by disk diffusion method. Genetic diversity was analyzed by repetitive element sequence-based PCR (REP-PCR). The phylogenetic group B2 (32%) predominated, followed by phylogenetic group E (25%). ST131 (28%) was the most prevalent ST and the majority of these isolates (89.3%) were of serotype O25b. Most of E. coli isolates (75%) were categorized as multidrug resistant (MDR) with high rates of resistance (>55%) to ampicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, cefazolin, and ceftriaxone. The most frequent ARGs were blaTEM (66%), sul1 (57%), and sul2 (51%). The most prevalent VAGs and PAIs were fimH (type 1 fimbriae adhesin; 85%), aer (iucC) (aerobactin; 79%), traT (serum resistance; 77%), iutA (aerobactin siderophore receptor; 69%), and PAI IV536 (75%), respectively. The highest rate of ARGs and VAGs was observed in the ST131 isolates. REP-PCR analysis showed high diversity among the studied isolates. The high prevalence of MDR septicemic E. coli with different types of ARGs, VAGs and genotypes is an extremely worrisome sign of BSIs treatment and poses a major threat for hospitalized patients. Active surveillance, stringent prescribing policies, increasing the awareness of ARGs among clinicians and re-defining the infection control measures are essential to curb the dissemination of these strains.
Escherichia coli is responsible for a wide variety of diseases such as urinary tract infections (UTIs), pneumonia, bloodstream infections (BSIs), and meningitis in humans in both developed and developing countries (Mathers et al., 2015a; Sarowska et al., 2019). E. coli BSIs (EC-BSIs) have been associated with prolonged hospital stays, high mortality, and costs (Chen et al., 2021). Based on phylogenetic group classification, E. coli was classified into 7 groups (A, B1, B2, C, D, E, and F) (Zhao et al., 2021). EC-BSIs commonly belonged to group B2, and to a lesser extent, group D, while groups A and B1 were frequently associated with the commensal and less virulent strains (Cole et al., 2019). Invasive E. coli strains have various virulence-associated genes (VAGs) including adhesins, toxins, siderophores and capsular antigens, which allow to colonize the host and invade the bloodstream (Ciesielczuk et al., 2015). Molecular studies revealed that four clones or sequence types (STs) 69, 73, 95, and 131 were commonly responsible for most E. coli infections in humans (Hojabri et al., 2019b). In a UK study, it was shown that these four STs were responsible for a large proportion of UTIs and BSIs (Doumith et al., 2015). Antibiotic resistance patterns associated with these clones differ markedly: isolates belonging to STs 69, 73, and 95 are mainly susceptible to antibiotics and rarely have resistance to extended-spectrum cephalosporins, while members of ST131 are connected with high rates of cephalosporins or fluoroquinolones resistance (Doumith et al., 2015). Rapid identification of these major STs by molecular techniques can assist clinicians to select appropriate antimicrobials for treatment before conventional susceptibility test results become available (Doumith et al., 2015). Recently, EC-BSIs have been increasing in multiple countries and of greater concern is the emergence of multidrug resistant (MDR) strains, which can cause inappropriate empirical treatment (Gladstone et al., 2020). Different typing techniques have been developed for epidemiological relationships of E. coli (Fratamico et al., 2016). Among polymerase chain reaction (PCR)-based approaches, repetitive extra-genic palindromic elements (REP)-PCR is a rapid, simple, reliable and cost-effective method with high discriminatory power compared to other genotyping techniques (Sabat et al., 2013). Information about the molecular epidemiology of EC-BSIs is limited in Iran. Therefore, the present study aimed to evaluate phylogroup classification, four major human STs, antimicrobial resistance, presence of selected antimicrobial resistance and virulence genes, and also genetic diversity in a teaching hospital of Tehran, Iran.
This cross-sectional study was performed in a 640-beds university-affiliated hospital (Shariati hospital) in Iran between December 2020 and June 2022, using a total of 100 non-duplicate (one isolate per patient) E. coli were isolated from blood samples of patients with clinical symptoms of BSIs. E. coli was identified using colony morphology and biochemical profile by the standard laboratory guidelines (Pathak et al., 2013). The isolates were stored at −70°C in Trypticase Soy Broth (TSB) containing 30% glycerol until further characterization.
Genomic bacterial DNA was extracted from freshly cultured bacteria by boiling method (Iranpour et al., 2015). Major E. coli phylogenetic groups (A, B1, B2, C, D, E, or F) were determined using multiplex PCR (Clermont et al., 2013). The four main E. coli STs (ST69, ST73, ST95, and ST131) were evaluated by PCR as previously described (Doumith et al., 2015). Furthermore, molecular serotyping of ST131 isolates was performed by PCR method to detect two common serogroups, including O16 and O25b (Park et al., 2018).
Antibiotic susceptibility testing was performed using a disk-diffusion method on Mueller-Hinton agar according to Clinical Laboratory Standard Institute (CLSI) 2022 guidelines. The following antimicrobial agents were tested in this study: ampicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, piperacillin-tazobactam, cefazolin, ceftriaxone, cefepime, ceftazidime, gentamicin, ampicillin-sulbactam, imipenem, meropenem, and amikacin. MDR refers an isolate resistant to at least one potentially effective antimicrobial drug in three or more classes (such as penicillins/cephalosporins, fluoroquinolones, carbapenems, and aminoglycosides) (Tam et al., 2010; Zeighami et al., 2019). Extensively drug-resistant (XDR) was defined as those that were resistant to all except one or two antimicrobial classes (Falagas et al., 2010). The resistance score was calculated by the number of resistant antimicrobial agents for each isolate. A score of 1 shows resistant; 0.5, intermediary; and 0, sensitive (Duan et al., 2020).
Molecular determination of ARGs was carried out by PCR based on amplification of genes with specific primers previously described. Isolates were screened for extended-spectrum beta-lactamases (ESBLs) genes (blaCTX-M, blaTEM, and blaSHV) (Neyestanaki et al., 2014), plasmid-mediated quinolone resistance genes (qnrA and qnrB) (Wu et al., 2007), plasmid-mediated trimethoprim resistance gene (dfrA) (Boroumand et al., 2021), plasmid-mediated sulfonamide resistance genes (sul1, sul2, and sul3) (Jiang et al., 2019), carbapenemase genes (blaOXA, blaNDM, blaIMP, blaVIM, blaSPM and blaKPC) (Neyestanaki et al., 2014; Bian et al., 2019), and plasmid-mediated colistin resistance gene (mcr-1) (Peng et al., 2019). Furthermore, blaCTX-M and blaOXA positive isolates were screened for blaOXA-48, blaOXA-181 (Sheikh et al., 2021), blaCTX-M-14 and blaCTX-M-15 (Tawfik et al., 2011). The resistance-gene score was the number of unique ARGs detected for each isolate.
PCR was performed to screen 13 different VAGs, including adhesins (fimH, papC, papG II, papG III, and sfa/foc) (Yun et al., 2014), toxins (cnf-1, cvaC, and hlyC) (Dezfulian et al., 2003), capsules (rfc and kpsMTII) (Chapman et al., 2006), siderophores (aer [iucC] and iutA) (Guiral et al., 2015), serum resistance factor (traT) (Rasoulinasab et al., 2021), and 8 PAIs (PAI I536, PAI II536, PAI III536, PAI IV536, PAI ICFT073, PAI IICFT073, PAI IJ96, and PAI IIJ96) (Rasoulinasab et al., 2021). The virulence-gene score was calculated by the number of the virulence genes detected for each isolate (Merino et al., 2020).
REP-PCR method was performed using a T100™ Thermal Cycler (BioRad, Germany) instrument with primers rep-F (5′-ICGICTTAT CIGGCCTAC-3′) and rep-R (5′-IIIICGICGICATCIGGC-3′). Amplification and electrophoresis were carried out as described previously (Erfanimanesh et al., 2022). In the lack of appropriate software, the band patterns generated by REP-PCR were assessed visually. The binary matrix was built based on the absence or presence of each band (coded 0 or 1, respectively) (Pusparini et al., 2018). Phylogenetic tree based on binary matrix was constructed by the free online service at http://insilico.ehu.eus/dice_upgma/. Isolates were classified together into the same cluster when their similarity coefficient was ≥80%.
Statistical analyses were performed using SPSS software (version 16.0). Comparison between the variables was performed using Pearson’s Chi-square test or Fisher’s exact test. The differences were considered statistically significant if p-value ≤ 0.05. Correlation analyses (Spearman R correlation coefficient) were performed to evaluate the correlation between the variables, demonstrated with correlation heatmaps. Cut off value was considered Spearman R ≥ 0.4 (Rezatofighi et al., 2021). The preparation of heatmaps was done on R software (version 4.3.2).
Phylogenetic analysis showed that 32% of our isolates belonged to phylogenetic group B2, 25% to group E, 13% to group C, 11% to group B1, 9% to group A, 7% to group F, and 3% to group D. Analysis of major E. coli STs indicated that the most prevalent ST was ST131 (28%, n = 28/100), followed by ST69 (17%, n = 17/100) and ST95 (2%, n = 2/100). ST73 was not found. Fifty-three (53%, n = 53/100) isolates were unknown STs. Distribution of STs in different phylogenetic groups was as follow: group B2 (ST131: 78.6%, n = 22; ST95: 100%, n = 2; unknown STs: 15.1%, n = 8); group E (ST69: 64.7%, n = 11; unknown STs: 26.4%, n = 14); group C (ST131: 10.8%, n = 3; ST69: 5.9%, n = 1; unknown STs: 17%, n = 9); group B1 (ST131: 7.1%, n = 2; unknown STs: 17%, n = 9); group A (ST69: 11.8%, n = 2; unknown STs: 13.2%, n = 7); group F (ST69: 11.8%, n = 2; unknown STs: 9.4%, n = 5); group D (ST131: 3.5%, n = 1; ST69: 5.9%, n = 1; unknown STs: 1.9%, n = 1). The majority of ST131 and all ST95 isolates belonged to phylogenetic group B2, while most ST69 isolates belonged to phylogenetic group E. PCR O-typing of 28 ST131 isolates revealed that the vast majority of the isolates (25/28, 89.3%) harbored O25b and 10.7% (3/28) had O16.
The median age of the 100 patients was 67 years old (range 0–86 years). Thirty-eight (38%) patients were female (mean age 61 years, range 16–87 years), and 62 (62%) patients were male (mean age 54 years, range 0–86 years). Sixty-nine (69%) were aged over 50 years and one (1%) of patients was aged less than 1 year. Most (60.7%, n = 17) ST131 isolates were predominant in females, followed by unknown STs (30.2%, n = 16) and ST69 (29.4%, n = 5) (p = 0.01); whereas, ST95 (100%, n = 2) isolates predominated in males, followed by ST69 (70.6%, n = 12), unknown STs (69.8%, n = 37), and ST131 (39.3%, n = 11) (p = 0.01). Elderly patients over 50 years were more frequent; however, a significant association was not observed with the patterns of ST distribution (p = 0.1).
Antimicrobial susceptibility testing results revealed that more than 55% of our isolates were resistant to ampicillin (89%), trimethoprim-sulfamethoxazole (75%), ciprofloxacin (71%), cefazolin (64%), and ceftriaxone (58%). In contrast, the low resistance rates were observed for carbapenems (4%) and amikacin (2%). The frequencies of MDR and XDR isolates were 75 and 5%, respectively (Table 1). Concerning the antibiotic susceptibility of ST131 and non-ST131 isolates, ST131 had higher resistance rates than non-ST131 isolates and these differences were significant for cephalosporins, gentamicin, and amikacin (p < 0.05). Moreover, most ST131 isolates (92.9%) were MDR.
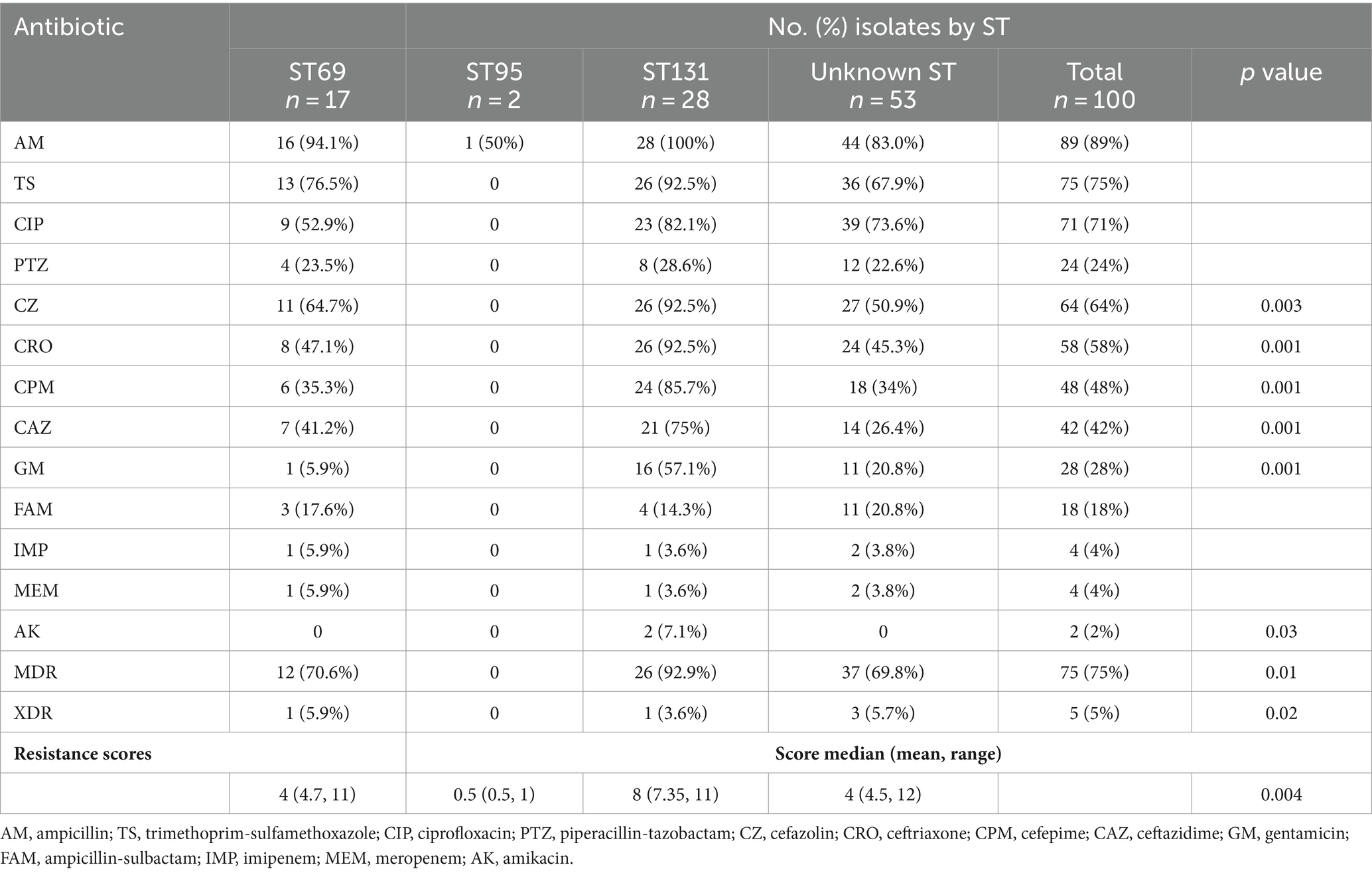
Table 1. Antimicrobial resistance based on major sequence types (STs) among Escherichia coli isolates.
The overall prevalence of individual ARGs ranged from 0% (blaKPC, blaIMP, blaSPM, qnrA, and mcr-1) to 66% (blaTEM) (Table 2). More than half of the isolates harbored blaTEM, sul1, and sul2 genes. blaCTX-M, dfrA, sul1, blaOXA, and blaVIM genes were more frequent in ST131 isolates, while blaTEM, qnrB and blaNDM genes were more common in ST69 isolates. Additionally, resistance-gene scores of the ST131 isolates were slightly higher than other non-ST131 isolates.
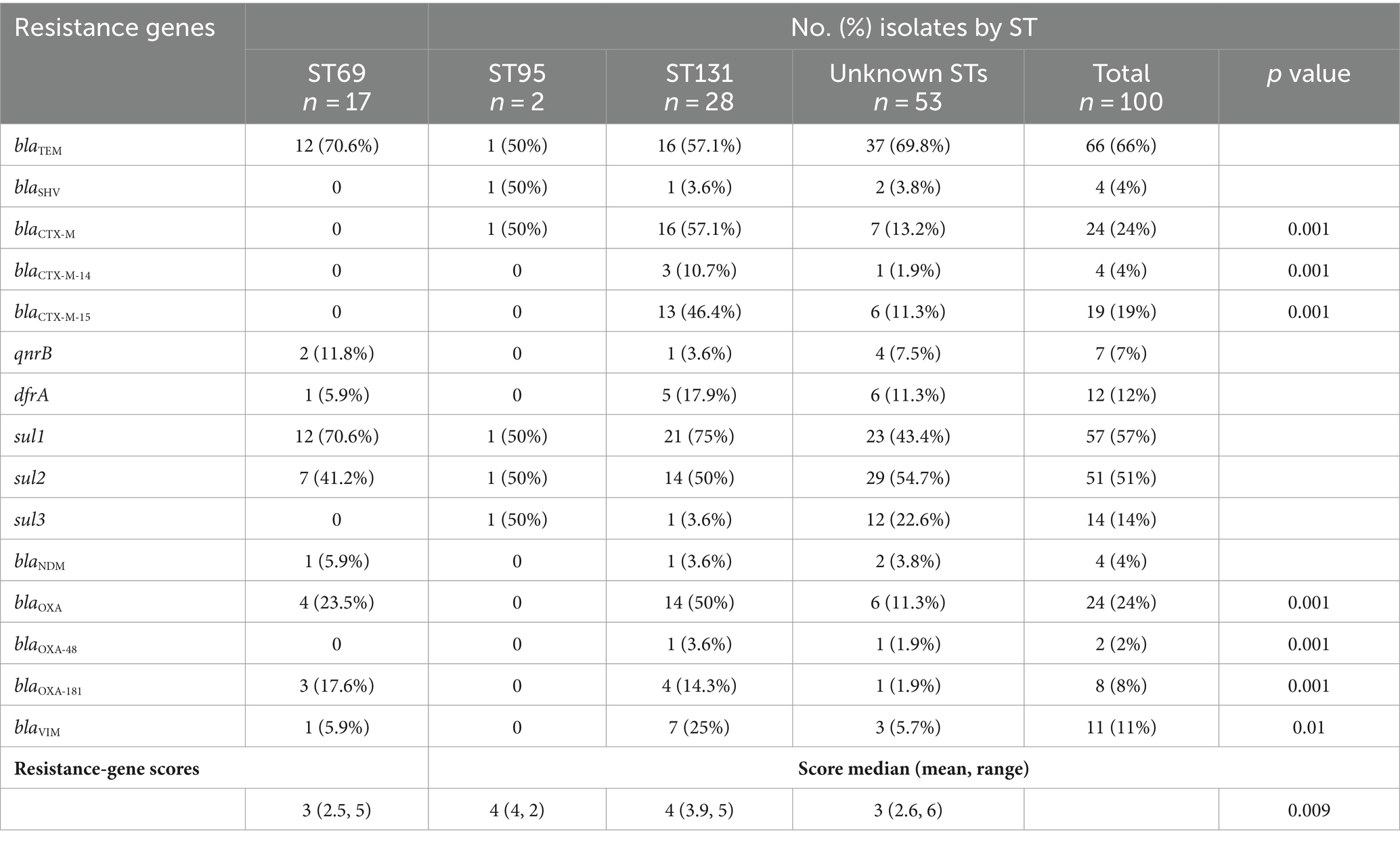
Table 2. Distribution of resistance genes among major sequence types (STs) of Escherichia coli isolates.
We evaluated association between the ARGs and AMR phenotype by Spearman’s rank correlation coefficient. As demonstrated in the correlation matrix in Figure 1, no strong positive association was observed between them in the present study. While, it was found that there is a weak positive association between the blaTEM gene and resistance to piperacillin-tazobactam. We also found a weak positive association between the resistance genes, including sul3 with sul1 and sul2 with blaOXA. Furthermore, analysis of the co-resistance phenomenon showed a strong positive association between the following antibiotics: (cefazolin with ampicillin, ceftriaxone, cefepime, ceftazidime, and piperacillin-tazobactam); (ceftriaxone with cefepime, ceftazidime, and piperacillin-tazobactam); (cefepime with ceftazidime, gentamicin, and piperacillin-tazobactam); (ceftazidime with gentamicin, ampicillin-sulbactam, and piperacillin-tazobactam); (piperacillin-tazobactam with ampicillin-sulbactam); (ampicillin-sulbactam with imipenem and meropenem).
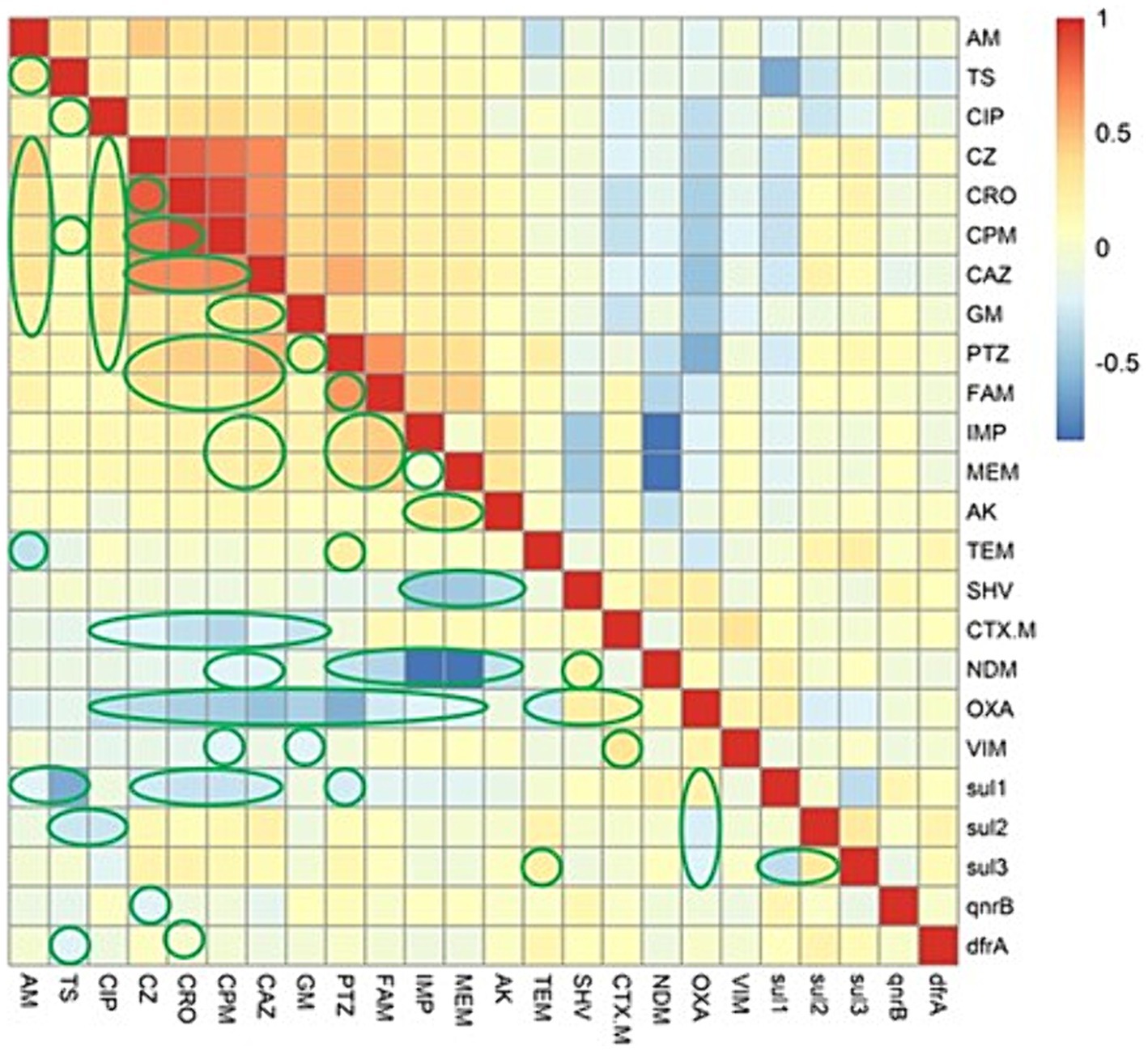
Figure 1. The heatmap demonstrates the spearman correlation among various antimicrobial agents and the antimicrobial resistance genes in Escherichia coli causing bloodstream infections. The statistically significant associations (p < 0.05) are indicated by closed green lines in the heatmap cells. The red color represents the positive correlation, the darker red denotes a stronger positive correlation coefficient, the blue color represents negative correlation, and the darker blue denotes a stronger negative correlation coefficient. AM, ampicillin; TS, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; PTZ, piperacillin-tazobactam; CZ, cefazolin; CRO, ceftriaxone; CPM, cefepime; CAZ, ceftazidime; GM, gentamicin; FAM, ampicillin-sulbactam; IMP, imipenem; MEM, meropenem; AK, amikacin.
The overall prevalence of VAGs ranged from 0% (papG III) to 85% (fimH). Five virulence genes (fimH, aer [iucC], traT, iutA, and kpsMT II) were found in ≥56% of the isolates. In contrast, four VAGs (hlyC, cnf-1, cvaC, and sfa/foc) were found in less than 16% of the isolates (Table 3). In this study, the most predominant PAI marker was PAI IV536 (75%), followed by PAI IICFT073 (30%), and PAI ICFT073 (28%), whereas three PAIs (PAI IIJ96, PAI II536 and PAI I536) were found in less than 13% of the isolates. Two PAIs (PAI IJ96 and PAI III536) were not detected in any isolate. A number of VAGs (papC, papG II, cnf-1, hlyC, kpsMTII, aer [iucC], iutA, and traT) and PAIs (other than PAI IV536) were observed significantly more frequently in ST131 isolates compared with non-ST131 isolates. Statistical analysis showed that VAGs were more prevalent among phylogroup B2 isolates (mean: 9.6, median: 8.0), followed by group D (mean: 7.3, median: 7.0), group E (mean: 6.2, median: 6.0), group C (mean: 4.9, median: 5.0), group F (mean: 4.1, median: 5.0), group B1 (mean: 4.4, median: 4.0), and group A (mean: 3.2, median: 3.0) (p < 0.001).
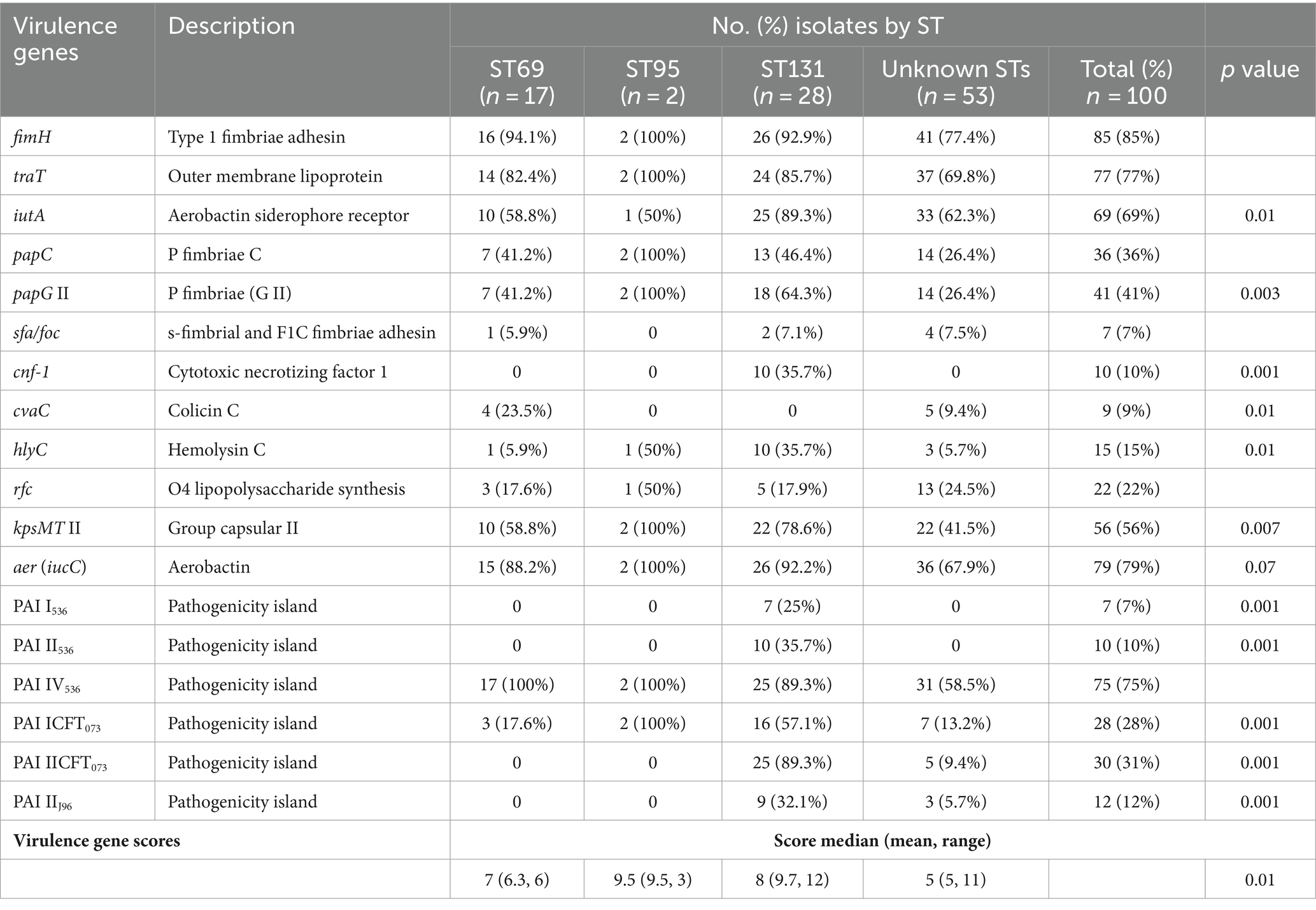
Table 3. Distribution of virulence genes among major sequence types (STs) of Escherichia coli isolates.
We performed Spearman’s rank correlation coefficient analysis to evaluate the association between the virulence determinants in EC-BSIs. On the basis of Figure 2, Spearman correlation analysis displayed strong positive associations among the following genes: (fimH with PAI IIJ96); (iutA with aer [iucC]); (papC with papG II, hlyC, cnf-1, and PAI II536); (papG II with cnf-1, papC and hlyC); (cnf-1 with hlyC, PAI I536, PAI II536, PAI ICFT073, PAI IICFT073, and PAI IIJ96); (hlyC with PAI I536, PAI II536, PAI ICFT073, and PAI IIJ96); (PAI I536 with PAI II536, PAI ICFT073, PAI IICFT073, and PAI IIJ96); (PAI II536 with PAI ICFT073, PAI IICFT073, and PAI IIJ96); (PAI ICFT073 with PAI IICFT073, and PAI IIJ96).
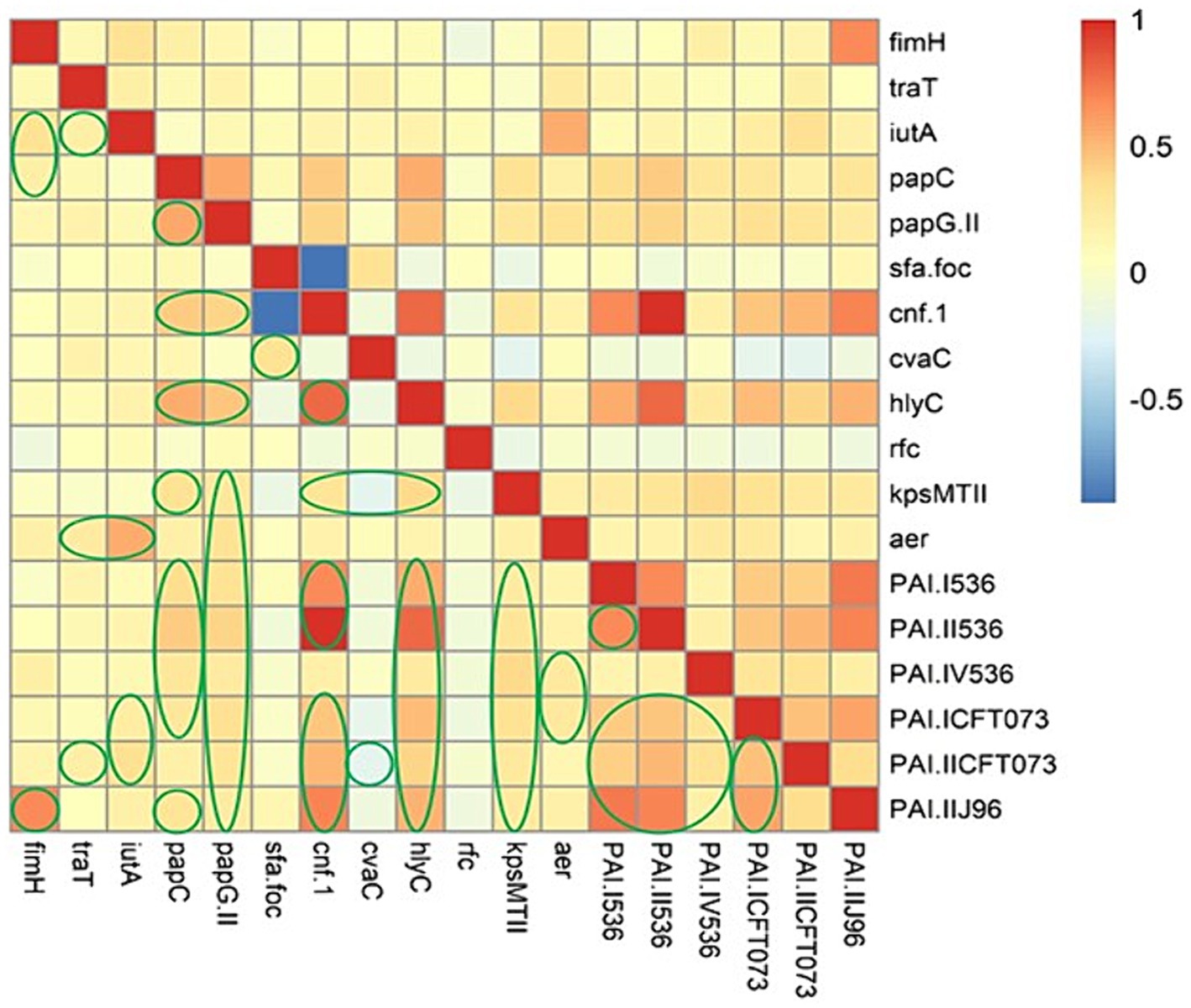
Figure 2. The heatmap illustrates the spearman correlation between different virulence markers in Escherichia coli causing bloodstream infections. The statistically significant associations (p < 0.05) are indicated by closed green lines in the heatmap cells. The red color represents the positive correlation, the darker red denotes a stronger positive correlation coefficient, the blue color represents negative correlation, and the darker blue denotes a stronger negative correlation coefficient.
In this regard, we were not observed any strong positive association between the AMR phenotype and virulence factors (Figure 3); however, a weak positive association was observed as follow: (ampicillin-sulbactam with fimH, iutA, kpsMTII, iucC, and PAI IV536); (imipenem with fimH, and iutA); (meropenem with fimH, and iutA). Strong positive associations were detected between the presence of some ARGs and VAGs, including: (blaCTX-M with kpsMTII, PAI ICFT073, and PAI IICFT073); (blaOXA with cnf-1, hlyC, PAI I536, PAI II536, and PAI IIJ96).
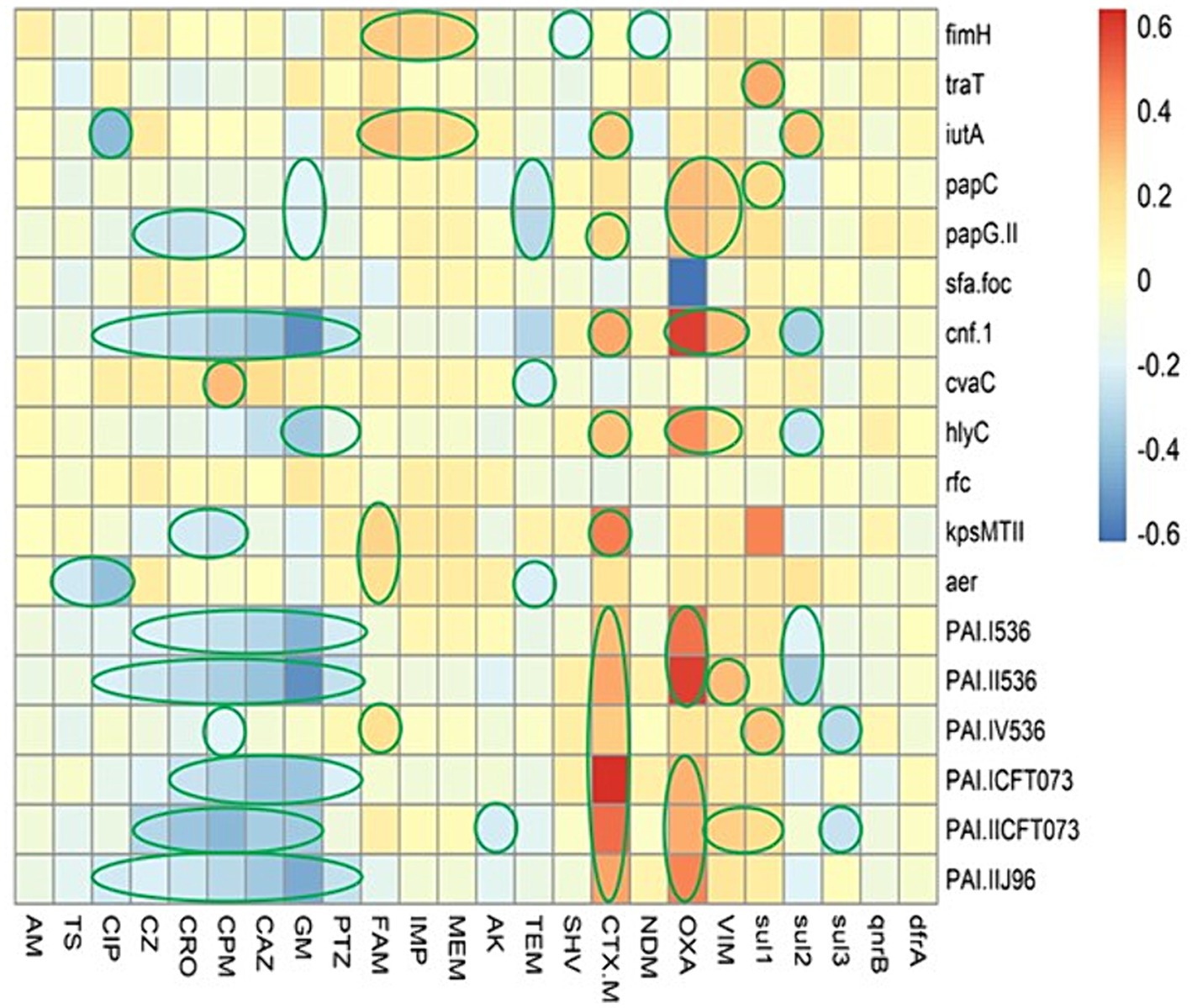
Figure 3. The heatmap demonstrates the spearman correlation between various virulence markers and antimicrobial resistance phenotype in Escherichia coli causing bloodstream infections. The statistically significant associations (p < 0.05) are indicated by closed green lines in the heatmap cells. The red color represents the positive correlation, the darker red denotes a stronger positive correlation coefficient, the blue color represents negative correlation, and the darker blue denotes a stronger negative correlation coefficient. AM, ampicillin; TS, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; PTZ, piperacillin-tazobactam; CZ, cefazolin; CRO, ceftriaxone; CPM, cefepime; CAZ, ceftazidime; GM, gentamicin; FAM, ampicillin-sulbactam; IMP, imipenem; MEM, meropenem; AK, amikacin.
REP-PCR technique produced varying amplification products from about 100 to about 3,000 bp with different bands (between 1 and 13 bands) for each isolate. According to Figure 4, REP-PCR analysis displayed 91 distinct genetic patterns which clustered the isolates into 33 REP types (named as C1-C33). No band was observed in one isolate. C14 was the most abundant cluster, comprising 13.14% (13/99) of the isolates, followed by C6 (10.1%, 10/99). Other clusters included <10 isolates. Comparison of isolates in the same cluster showed that most isolates were different in relation to the gene distributions and antimicrobial resistance profile. Furthermore, REP-PCR showed that the isolates belonging to the same STs, exhibited a wide range of genetic diversity.
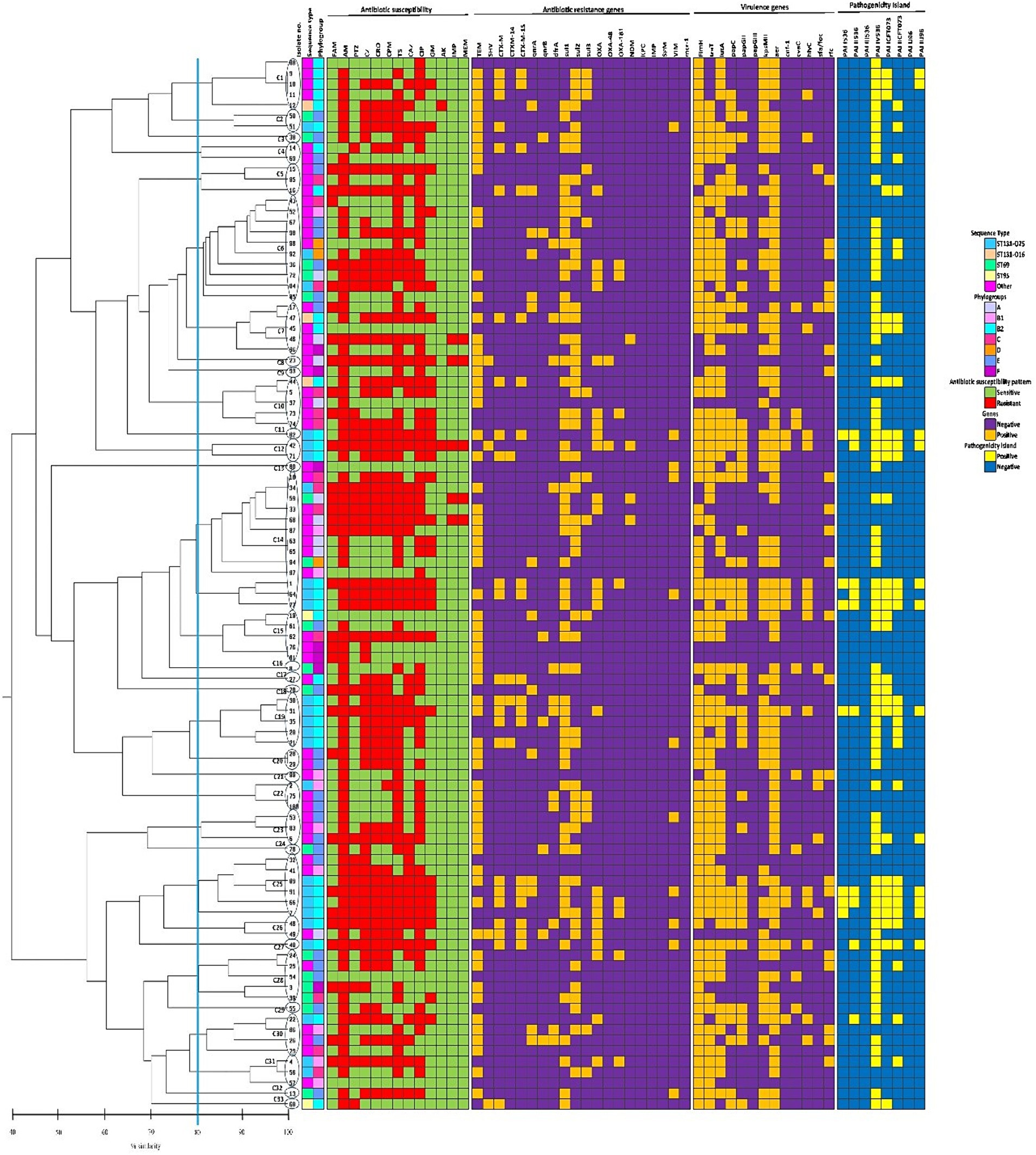
Figure 4. The phenotypic and genotypic characteristics of 99 Escherichia coli bloodstream infection isolates included in the present study. The left side of the figure is the REP-PCR dendrogram and the right side is the heat map of isolates based on their antimicrobial resistance patterns, the distribution of antibiotic resistance genes, virulence-associated genes, and pathogenicity islands which are color-coded feature categories. The vertical line displays the 80% similarity cut-off value. Based on a similarity index ≥80%, 33 clusters were found. Numbers at the terminal branches are strain name. Phylogenetic tree generated with DNA banding patterns based on REP primers using unweighted-pair group method with arithmetic mean cluster analysis. AM, ampicillin; TS, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; PTZ, piperacillin-tazobactam; CZ, cefazolin; CRO, ceftriaxone; CPM, cefepime; CAZ, ceftazidime; GM, gentamicin; FAM, ampicillin-sulbactam; IMP, imipenem; MEM, meropenem; AK, amikacin.
In the current study, we investigated 100 clinical E. coli isolates from BSIs for phylogenetic group distributions, four major STs, virulence/resistance genes as well as the genetic relatedness. In our study, the phylogenetic classification indicated that phylogenetic groups B2 and E were the predominant groups. In 2023, Li et al. analyzed 60 E. coli bloodstream isolates in China and found that phylogroups B2 and D were the most prevalent groups (Li et al., 2023). In 2019, Daga et al. studied 48 E. coli bloodstream isolates in Brazil and observed that phylogroups B2 and B1 were the most common one (Daga et al., 2019). Previous studies have shown that phylogroup A was the most predominant among uropathogenic E. coli (UPEC) isolates in Russia and Egypt (Khairy et al., 2019). In this study, PCR assay for detection of four major E. coli STs revealed that ST131 (28%) was the predominant clone. Based on recent studies, the prevalence of ST131 varies from 12.5% to nearly 30% among human clinical isolates (Banerjee and Johnson, 2014). In a previous study in a single hospital in Semnan (Iran), Hojabri et al. reported that 29.8% of E. coli strains isolated from urine belonged to ST131 (Hojabri et al., 2017). A previous study conducted in China during 2020 to 2021 revealed that ST131 (26.7%) was the most common clone in blood samples (Li et al., 2023). In Korean study, Park et al. showed that ST131 (11.9%) predominated among E. coli bloodstream isolates (Park et al., 2018). Similar to previous studies, we observed that O25b was the predominant serotype among the studied ST131 isolates (Hojabri et al., 2017; Namaei et al., 2017; Zhong et al., 2019). In contrast, Zhong et al. reported that O16 was the most prevalent serotype (75%) among ST131 isolates (Zhong et al., 2015). As mentioned earlier, the distribution of phylogenetic groups, STs and serotypes varies between different countries and these discrepancies may be due to the geographical variation, origin of isolates, differences in the sample size and host populations (Zhang et al., 2002). Regarding antibiotic resistance, high rates of resistance were observed against ampicillin, trimethoprim-sulfamethoxazole, ciprofloxacin, cefazolin, and ceftriaxone. Furthermore, resistance patterns of tested isolates have shown that 75 and 5% were MDR and XDR, respectively. Similar trends have been reported in Iran; where, Hojabri et al. have observed that 90.8% of E. coli isolates collected from different clinical samples were MDR (Hojabri et al., 2020). On the other hand, Ny et al. have indicated that the MDR rate varies from 1.7% (Latvia) to 26.9% (Russia) among six European countries (Ny et al., 2019). A possible explanation for this variation is that inappropriate and excessive drug usage is still common in Iran, whereas in developed countries policies have been implemented to limit the use of antibiotics. In the current study, amikacin and carbapenems were the most effective antibiotic against E. coli than other tested antibiotics, which is in agreement with previous studies (Mazumder et al., 2020; Li et al., 2023). We also statistically compared antibiotic resistance characteristics of isolates between STs. It should be noted that antibiotic resistance of ST131 isolates was significantly higher than ST95 and ST73 isolates. This is not surprising, because E. coli ST131 is a pandemic MDR clone and is associated with a large proportion of antibiotic-resistant E. coli infections in various clinical settings (Riley, 2014; Mathers et al., 2015b). In this study, we detected different resistance genes, and the most were blaTEM (66%), sul1 (57%), sul2 (51%), blaCTX-M (24%), and blaOXA (24%). The prevalence rate of ARGs varies widely between countries. In Palestine, Tayh et al. reported that sul (28.6%), blaTEM (17%) and blaOXA (2.4%) were the most prevalent ARGs among E. coli strains isolated from urine samples (Tayh et al., 2023). In a Chinese study, 18.3, 13.3 and 13.3% of the E. coli isolates harbored blaCTX-M-15, blaTEM and blaOXA-1 genes, respectively (Li et al., 2023). In Bangladesh, Mazumder et al. observed that blaCTX-M-15 (52%), blaTEM (20) and blaNDM-1 (5%) were the most prevalent ESBL and carbapenemase genes among E. coli isolates (Mazumder et al., 2020). The observed variation may be attributed to the different patterns of use of antibiotics, dissemination of specific clones harboring multiple ARGs and the number of isolates studied. In current study, we found that the highest resistance score belonged to the ST131 isolates and these isolates carried multiple ARGs which could potentially complicate the medical treatment of BSIs. Notably, we observed a common phenomenon that genotypic and phenotypic resistances were not entirely consistent. In this study, of the eleven isolates harboring blaVIM carbapenemase, only four isolates were resistant to imipenem and meropenem, while the remaining seven isolate were susceptible. This might be due to the mutations, unusual expression of ARGs or the expression of ARGs has not reached a level that can lead to drug resistance (Yu et al., 2020; Zhang et al., 2021). Generally, the antibiotic resistance phenotype relies on the carriage and expression of ARGs. The associations between AMR phenotypes and ARGs varied in this study. Mostly, negative associations were more common between different AMR and ARGs. But, we found some weak positive associations among AMR and ARGs. Positive correlations between AMR and ARGs might be attributed to the selective pressure exerted by antimicrobial agents on bacteria, leading to increased expression of ARGs or the simultaneous colocation of ARGs on single mobile genetic elements (MGEs), such as integrons, transposons, or plasmids (Birkegård et al., 2017). Nonetheless, the correlation reported in this study is based only on statistical data, and more investigations are needed to clarify the underlying mechanisms of this correlation. In contrast to the findings in our study, Rahman et al. reported that the number of ARGs is strongly associated with the phenotypic AMR in UPEC isolates (Rahman et al., 2022). Rosengren et al. reported that the agreement between AMR and ARGs varied from 33 to 85% (Rosengren et al., 2009). We found various VAGs and PAIs, of which fimH (85%), iucC (79%), traT (77%), PAI IV536 (75%), iutA (69%), and kpsMTII (56%) were the most prevalent. It seems that the high prevalence of these VAGs appears to be good indicators for EC-BSIs. While papG III, hlyC, cnf-1, cvaC, and sfa/foc genes, which were observed at low frequencies in this study, are considered to be key factors for UTIs caused by UPEC isolates (Bingen-Bidois et al., 2002; Doye et al., 2002; Whelan et al., 2023). Our findings are in agreement with the results reported by Daga et al., showing the fimH (95.8%), traT (77.1%), PAI IV536 (77.1%), iutA (64.3%), kpsMTII (45.8%) and to be considerably more prevalent in the septicemic E. coli strains (Daga et al., 2019). The current study, along with other studies from different part of the world (Cooke et al., 2010; Blanco et al., 2013; Johnson et al., 2014), shows that these virulence factors are essential for the development of E. coli extraintestinal infections and could be considered as vaccine candidates. Our results also revealed that the ST131 bloodstream isolates exhibited a higher virulence score than non-ST131 isolates, indicating higher virulence power and therefore the possible reason for their high prevalence among our isolates. In this study we observed that phylogroups B2 and D had high level of VFs score. On the other hand, isolates belonged to phylogroups A and B1 exhibited low level of VFs score. In agreement with this finding, other studies in the China (Li et al., 2023), Ethiopia (Dadi et al., 2020), and Pakistan (Ali et al., 2019) have shown that E. coli strains belong to phylogroups B2 and D possessed more VFs than the other phylogroups. However, in a study from Egypt, Khairy et al. exhibited that VFs were more common in phylogroup A (Khairy et al., 2019). This discrepancy may be related to the geographical differences and type of infections. Association between VAGs and PAIs were also analyzed. The strongest positive associations were observed between some VAGs and PAIs, suggesting the genetic linkages between them. Concerning the relationship between VAGs and AMR, two hypotheses have been postulated (Erjavec et al., 2007; Rezatofighi et al., 2021). First, resistant strains can be more virulent because mobile genetic elements (MGEs) including transposons, integrons, and plasmids may contain VAGs and AMR genes simultaneously. On the other hand, some investigators claim that AMR strains are not more virulent than susceptible ones because the evolution of AMR and virulence may not have occurred simultaneously (Erjavec et al., 2007; Rezatofighi et al., 2021). In the current study, no relationship was observed between VAGs and resistance phenotype. Only weak positive associations were found between some VAGs and resistance phenotype. The relationship between VAGs and resistance phenotypes is a complex phenomenon and influenced by numerous factors, such as geographical locations, specific hosts, and type of antimicrobial drugs used, methods used to identify VAGs and evaluate antibiotic resistance, genetic background of the organisms, types of VAGs and antimicrobials, chromosomal mutations, sample type, and colocation of both virulence and resistance genes on single MGEs (Boerlin et al., 2005; Liu et al., 2017; Neamati et al., 2020; Pan et al., 2020; Rezatofighi et al., 2021). In this study, we observed that resistance and virulence genes were randomly distributed among our isolates, suggesting various E. coli STs, each with different profiles of resistance and virulence genes, have the potential to cause BSIs. Comparison of ARGs and virulence factors of ST131 and non- ST131 exhibited increased frequencies of six ARGs (blaCTX-M, blaCTX-M-14, blaCTX-M-15, blaOXA, blaOXA-48, and blaVIM) and ten virulence genes (iutA, papG II, cnf-1, hlyC, kpsMT II, PAI I536, PAI II536, PAI ICFT073, PAI IICFT073, and PAI IIJ96) in ST131 (p < 0.05). However, cvaC and blaOXA-181 were significantly higher in ST69 (p < 0.05). Our results are in line with previous studies that reported a higher prevalence of certain genes in ST69 and ST131 strains (Hojabri et al., 2019b; Hung et al., 2019). Molecular typing is a valuable tool in clinical epidemiology for determining the identical or closely related strains, rule out or confirm clonal outbreaks and sources of infection (Sabat et al., 2013). In our study, REP-PCR differentiated 100 E. coli strains into 91 genotypes, suggesting that isolates were clonally unrelated and dissemination of isolates was not due to a clonal outbreak. Additionally, we found that isolates belonging to the same clusters revealed the distinct ARGs and VAGs profiles, suggesting that the horizontal transmission and rearrangement of DNA may have occurred (Aibinu et al., 2012). REP-PCR method also demonstrated that our isolates were genetically diverse and highly heterogeneous. This may be related to genetic variation in our isolates. In a previous study in northeast of Iran, Fallah et al. differentiated the 85 diarrhoeagenic E. coli strains into 84 genotypes using REP-PCR (Fallah et al., 2021). Similarly, Ateba and Mbewe found that REP-PCR has more discriminatory power in differentiating E. coli strains (Ateba and Mbewe, 2013). However, Foley et al. reported that this method was a limited technique to differentiate among the strains (Foley et al., 2004). In this study, REP-PCR method demonstrated good discriminatory ability in discriminating E. coli isolates belonging to same STs. Similar findings have been previously reported (Can et al., 2016; Hojabri et al., 2019a; Lienard et al., 2021; Shahbazi et al., 2023). Although REP-PCR is simple, rapid and cost-effective method, its reproducibility is low and depends on quality of bacterial DNA and the PCR conditions (Foxman et al., 2005). With the purpose of obtaining more reliable and stronger results, sequence-based typing approaches such as multilocus sequence typing (MLST) and whole genome sequencing (WGS) have been developed (Foxman et al., 2005; Vanstokstraeten et al., 2023). MLST is based on the sequencing of conserved housekeeping genes and provides portable, reproducible and an unambiguous data. WGS provides a vast amount of information contained in the genome sequence of bacterial pathogens (Vanstokstraeten et al., 2023); however, the use of WGS in Iran is limited, due to lack of funding and cost, specialized equipment, awareness and education. Moreover, many researchers, physicians and hospitals lack timely access to WGS, constrained by a shortage of specialists as well as clinical workflow and operational challenges. Consequently, conventional methods such as PCR and REP-PCR, along with other cost-effective typing methods, are still used more extensively in Iran. It should be emphasized that the current study has several limitations, including lack of demographics and clinical features of the patients (all isolates studied were from a single hospital in Tehran), antibiotic prescribing data, the small sample size and the lack of other molecular typing methods such as MLST and WGS for further genotypic characterization of these isolates.
This study demonstrated that high prevalence of MDR septicemic E. coli strains with different types of ARGs, VAGs and genotypes is an extremely worrisome sign of BSIs treatment, increases the risk of therapy failure and consequently poses a major threat for hospitalized patients. Active surveillance, stringent prescribing policies, increasing the awareness of ARGs among clinicians and re-defining the infection control measures are essential to curb the dissemination of these strains.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
The studies involving humans were approved by Ethics Committee of Tehran University of Medical Sciences (ethical approval reference number IR.TUMS.MEDICINE.REC.1401.412). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants' legal guardians/next of kin.
SaH: Investigation, Methodology, Writing – original draft. ShH: Formal analysis, Software, Writing – review & editing. FJ: Resources, Writing – review & editing. ME: Writing – review & editing. RB: Supervision, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research has been supported by Tehran University of Medical Sciences and Health Services. Study grant no 1401-02-410/58630.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aibinu, I., Odugbemi, T., Koenig, W., and Ghebremedhin, B. (2012). Sequence type ST131 and ST10 complex (ST617) predominant among CTX-M-15-producing Escherichia coli isolates from Nigeria. Clin. Microbiol. Infect. 18, E49–E51. doi: 10.1111/j.1469-0691.2011.03730.x
Ali, I., Rafaque, Z., Ahmed, I., Tariq, F., Graham, S. E., Salzman, E., et al. (2019). Phylogeny, sequence-typing and virulence profile of uropathogenic Escherichia coli (UPEC) strains from Pakistan. BMC Infect. Dis. 19:620. doi: 10.1186/s12879-019-4258-y
Ateba, C. N., and Mbewe, M. (2013). Determination of the genetic similarities of fingerprints from Escherichia coli O157: H7 isolated from different sources in the north West Province, South Africa using ISR, BOXAIR and REP-PCR analysis. Microbiol. Res. 168, 438–446. doi: 10.1016/j.micres.2013.02.003
Banerjee, R., and Johnson, J. R. (2014). A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 58, 4997–5004. doi: 10.1128/AAC.02824-14
Bian, F., Yao, M., Fu, H., Yuan, G., Wu, S., and Sun, Y. (2019). Resistance characteristics of CTX-M type Shigella flexneri in China. Biosci. Rep. 39:BSR20191741. doi: 10.1042/BSR20191741
Bingen-Bidois, M., Clermont, O., Bonacorsi, S., Terki, M., Brahimi, N., Loukil, C., et al. (2002). Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70, 3216–3226. doi: 10.1128/IAI.70.6.3216-3226.2002
Birkegård, A. C., Halasa, T., Græsbøll, K., Clasen, J., Folkesson, A., and Toft, N. (2017). Association between selected antimicrobial resistance genes and antimicrobial exposure in Danish pig farms. Sci. Rep. 7:9683. doi: 10.1038/s41598-017-10092-9
Blanco, J., Mora, A., Mamani, R., López, C., Blanco, M., Dahbi, G., et al. (2013). Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b: H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J. Clin. Microbiol. 51, 3358–3367. doi: 10.1128/JCM.01555-13
Boerlin, P., Travis, R., Gyles, C. L., Reid-Smith, R., Heather Lim, N. J., Nicholson, V., et al. (2005). Antimicrobial resistance and virulence genes of Escherichia coli isolates from swine in Ontario. Appl. Environ. Microbiol. 71, 6753–6761. doi: 10.1128/AEM.71.11.6753-6761.2005
Boroumand, M., Naghmachi, M., and Ghatee, M. A. (2021). Detection of phylogenetic groups and drug resistance genes of Escherichia coli causing urinary tract infection in Southwest Iran. Jundishapur J. Microbiol. 14:e112547. doi: 10.5812/jjm.112547
Can, F., Kurt-Azap, Ö., İspir, P., Nurtop, E., Seref, C., Loçlar, İ., et al. (2016). The clinical impact of ST131 H30-Rx subclone in urinary tract infections due to multidrug-resistant Escherichia coli. J. Glob. Antimicrob. Resist. 4, 49–52. doi: 10.1016/j.jgar.2015.10.006
Chapman, T. A., Wu, X.-Y., Barchia, I., Bettelheim, K. A., Driesen, S., Trott, D., et al. (2006). Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl. Environ. Microbiol. 72, 4782–4795. doi: 10.1128/AEM.02885-05
Chen, F., Lv, T., Xiao, Y., Chen, A., Xiao, Y., and Chen, Y. (2021). Clinical characteristics of patients and whole genome sequencing-based surveillance of Escherichia coli community-onset bloodstream infections at a non-tertiary hospital in China. Front. Microbiol. 12:748471. doi: 10.3389/fmicb.2021.748471
Ciesielczuk, H., Betts, J., Phee, L., Doumith, M., Hope, R., Woodford, N., et al. (2015). Comparative virulence of urinary and bloodstream isolates of extra-intestinal pathogenic Escherichia coli in a galleria mellonella model. Virulence 6, 145–151. doi: 10.4161/21505594.2014.988095
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Cole, B. K., Ilikj, M., McCloskey, C. B., and Chavez-Bueno, S. (2019). Antibiotic resistance and molecular characterization of bacteremia Escherichia coli isolates from newborns in the United States. PLoS One 14:e0219352. doi: 10.1371/journal.pone.0219352
Cooke, N. M., Smith, S. G., Kelleher, M., and Rogers, T. R. (2010). Major differences exist in frequencies of virulence factors and multidrug resistance between community and nosocomial Escherichia coli bloodstream isolates. J. Clin. Microbiol. 48, 1099–1104. doi: 10.1128/JCM.02017-09
Dadi, B. R., Abebe, T., Zhang, L., Mihret, A., Abebe, W., and Amogne, W. (2020). Distribution of virulence genes and phylogenetics of uropathogenic Escherichia coli among urinary tract infection patients in Addis Ababa, Ethiopia. BMC. Infect. Dis. 20:108. doi: 10.1186/s12879-020-4844-z
Daga, A. P., Koga, V. L., Soncini, J. G. M., de Matos, C. M., Perugini, M. R. E., Pelisson, M., et al. (2019). Escherichia coli bloodstream infections in patients at a university hospital: virulence factors and clinical characteristics. Front. Cell. Infect. Microbiol. 9:191. doi: 10.3389/fcimb.2019.00191
Dezfulian, H., Batisson, I., Fairbrother, J. M., Lau, P. C., Nassar, A., Szatmari, G., et al. (2003). Presence and characterization of extraintestinal pathogenic Escherichia coli virulence genes in F165-positive E. coli strains isolated from diseased calves and pigs. J. Clin. Microbiol. 41, 1375–1385. doi: 10.1128/JCM.41.4.1375-1385.2003
Doumith, M., Day, M., Ciesielczuk, H., Hope, R., Underwood, A., Reynolds, R., et al. (2015). Rapid identification of major Escherichia coli sequence types causing urinary tract and bloodstream infections. J. Clin. Microbiol. 53, 160–166. doi: 10.1128/JCM.02562-14
Doye, A., Mettouchi, A., Bossis, G., Clément, R., Buisson-Touati, C., Flatau, G., et al. (2002). CNF1 exploits the ubiquitin-proteasome machinery to restrict rho GTPase activation for bacterial host cell invasion. Cell 111, 553–564. doi: 10.1016/s0092-8674(02)01132-7
Duan, Y., Cao, Y., and Luo, Y. (2020). Antibiotic resistance and virulence of extraintestinal pathogenic Escherichia coli (ExPEC) vary according to molecular types. Front. Microbiol. 11:598305. doi: 10.3389/fmicb.2020.598305
Erfanimanesh, S., Emaneini, M., Modaresi, M. R., Feizabadi, M. M., Halimi, S., Beigverdi, R., et al. (2022). Distribution and characteristics of bacteria isolated from cystic fibrosis patients with pulmonary exacerbation. Can. J. Infect. Dis. Med. Microbiol. 2022, 1–13. doi: 10.1155/2022/5831139
Erjavec, M. S., Rijavec, M., Križan-Hergouth, V., Fruth, A., and Žgur-Bertok, D. (2007). Chloramphenicol-and tetracycline-resistant uropathogenic Escherichia coli (UPEC) exhibit reduced virulence potential. Int. J. Antimicrob. Agents 30, 436–442. doi: 10.1016/j.ijantimicag.2007.06.025
Falagas, M. E., Maraki, S., Karageorgopoulos, D. E., Kastoris, A. C., Mavromanolakis, E., and Samonis, G. (2010). Antimicrobial susceptibility of multidrug-resistant (MDR) and extensively drug-resistant (XDR) Enterobacteriaceae isolates to fosfomycin. Int. J. Antimicrob. Agents 35, 240–243. doi: 10.1016/j.ijantimicag.2009.10.019
Fallah, N., Rad, M., Ghazvini, K., Ghaemi, M., and Jamshidi, A. (2021). Molecular typing and prevalence of extended-spectrum β-lactamase genes in diarrhoeagenic Escherichia coli strains isolated from foods and humans in Mashhad, Iran. J. Appl. Microbiol. 131, 2033–2048. doi: 10.1111/jam.15062
Foley, S. L., Simjee, S., Meng, J., White, D. G., Mcdermott, P. F., and Zhao, S. (2004). Evaluation of molecular typing methods for Escherichia coli O157: H7 isolates from cattle, food, and humans. J. Food Prot. 67, 651–657. doi: 10.4315/0362-028x-67.4.651
Foxman, B., Zhang, L., Koopman, J. S., Manning, S. D., and Marrs, C. F. (2005). Choosing an appropriate bacterial typing technique for epidemiologic studies. Epidemiol. Perspect. Innov. 2:10. doi: 10.1186/1742-5573-2-10
Fratamico, P. M., DebRoy, C., Liu, Y., Needleman, D. S., Baranzoni, G. M., and Feng, P. (2016). Advances in molecular serotyping and subtyping of Escherichia coli. Front. Microbiol. 7:195060. doi: 10.3389/fmicb.2016.00644
Gladstone, R. A., McNally, A., Pöntinen, A. K., Tonkin-Hill, G., Lees, J. A., Skytén, K., et al. (2020). Emergence and dissemination of antimicrobial resistance in Escherichia coli causing bloodstream infections in Norway in 2002-17: a nationwide, longitudinal, microbial population genomic study. Lancet Microbe 2, e331–e341. doi: 10.1016/S2666-5247(21)00031-8
Guiral, E., Sáez-López, E., Bosch, J., Goncé, A., López, M., Sanz, S., et al. (2015). Antimicrobial resistance and virulence characterization among Escherichia coli clinical isolates causing severe obstetric infections in pregnant women. J. Clin. Microbiol. 53, 1745–1747. doi: 10.1128/JCM.00487-15
Hojabri, Z., Darabi, N., Arab, M., Saffari, F., and Pajand, O. (2019a). Clonal diversity, virulence genes content and subclone status of Escherichia coli sequence type 131: comparative analysis of E. coli ST131 and non-ST131 isolates from Iran. BMC Microbiol. 19:117. doi: 10.1186/s12866-019-1493-8
Hojabri, Z., Darabi, N., Mirmohammadkhani, M., Rahimi, H., Hemmati, R., Saeedi, Z., et al. (2020). Expansion of a subset within the C2 subclade of Escherichia coli sequence type 131 (ST131) is driving the increasing rates of aminoglycoside resistance. Open Forum Infect. Dis. 7:ofaa410. doi: 10.1093/ofid/ofaa410
Hojabri, Z., Mirmohammadkhani, M., Darabi, N., Arab, M., and Pajand, O. (2019b). Characterization of antibiotic-susceptibility patterns and virulence genes of five major sequence types of Escherichia coli isolates cultured from extraintestinal specimens: a 1-year surveillance study from Iran. Infect. Drug. Resist. 12, 893–903. doi: 10.2147/IDR.S199759
Hojabri, Z., Mirmohammadkhani, M., Kamali, F., Ghassemi, K., Taghavipour, S., and Pajand, O. (2017). Molecular epidemiology of Escherichia coli sequence type 131 and its H30/H30-Rx subclones recovered from extra-intestinal infections: first report of OXA-48 producing ST131 clone from Iran. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1859–1866. doi: 10.1007/s10096-017-3021-9
Hung, W. T., Cheng, M. F., Tseng, F. C., Chen, Y. S., Shin-Jung Lee, S., Chang, T. H., et al. (2019). Bloodstream infection with extended-spectrum beta-lactamase-producing Escherichia coli: the role of virulence genes. J. Microbiol. Immunol. Infect. 52, 947–955. doi: 10.1016/j.jmii.2019.03.005
Iranpour, D., Hassanpour, M., Ansari, H., Tajbakhsh, S., Khamisipour, G., and Najafi, A. (2015). Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. Biomed. Res. Int. 2015, 1–7. doi: 10.1155/2015/846219
Jiang, H., Cheng, H., Liang, Y., Yu, S., Yu, T., Fang, J., et al. (2019). Diverse mobile genetic elements and conjugal transferability of sulfonamide resistance genes (sul1, sul2, and sul3) in Escherichia coli isolates from Penaeus vannamei and pork from large markets in Zhejiang, China. Front. Microbiol. 10:1787. doi: 10.3389/fmicb.2019.01787
Johnson, J. R., Clermont, O., Johnston, B., Clabots, C., Tchesnokova, V., Sokurenko, E., et al. (2014). Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J. Clin. Microbiol. 52, 1358–1365. doi: 10.1128/JCM.03502-13
Khairy, R. M., Mohamed, E. S., Abdel Ghany, H. M., and Abdelrahim, S. S. (2019). Phylogenic classification and virulence genes profiles of uropathogenic E. coli and diarrhegenic E. coli strains isolated from community acquired infections. PLoS One 14:e0222441. doi: 10.1371/journal.pone.0222441
Li, R., Xu, H., Tang, H., Shen, J., and Xu, Y. (2023). The characteristics of extended-Spectrum β-lactamases (ESBLs)-producing Escherichia coli in bloodstream infection. Infect. Drug. Resist. 16, 2043–2060. doi: 10.2147/IDR.S400170
Lienard, A., Hosny, M., Jneid, J., Schuldiner, S., Cellier, N., Sotto, A., et al. (2021). Escherichia coli isolated from diabetic foot osteomyelitis: clonal diversity, resistance profile, virulence potential, and genome adaptation. Microorganisms 9:380. doi: 10.3390/microorganisms9020380
Liu, X., Liu, H., Li, Y., and Hao, C. (2017). Association between virulence profile and fluoroquinolone resistance in Escherichia coli isolated from dogs and cats in China. J. Infect. Dev. Ctries. 11, 306–313. doi: 10.3855/jidc.8583
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015a). Escherichia coli ST131: the quintessential example of an international multiresistant high-risk clone. Adv. Appl. Microbiol. 90, 109–154. doi: 10.1016/bs.aambs.2014.09.002
Mathers, A. J., Peirano, G., and Pitout, J. D. (2015b). The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin. Microbiol. Rev. 28, 565–591. doi: 10.1128/CMR.00116-14
Mazumder, R., Abdullah, A., Ahmed, D., and Hussain, A. (2020). High prevalence of Bla(CTX-M-15) gene among extended-Spectrum β-lactamase-producing Escherichia coli isolates causing extraintestinal infections in Bangladesh. Antibiotics (Basel) 9:796. doi: 10.3390/antibiotics9110796
Merino, I., Porter, S. B., Johnston, B., Clabots, C., Thuras, P., Ruiz-Garbajosa, P., et al. (2020). Molecularly defined extraintestinal pathogenic Escherichia coli status predicts virulence in a murine sepsis model better than does virotype, individual virulence genes, or clonal subset among E. coli ST131 isolates. Virulence 11, 327–336. doi: 10.1080/21505594.2020.1747799
Namaei, M. H., Yousefi, M., Ziaee, M., Salehabadi, A., Ghannadkafi, M., Amini, E., et al. (2017). First report of prevalence of CTX-M-15-producing Escherichia coli O25b/ST131 from Iran. Microb. Drug Resist. 23, 879–884. doi: 10.1089/mdr.2016.0272
Neamati, F., Khorshidi, A., Moniri, R., and Hosseini Tafreshi, S. A. (2020). Molecular epidemiology of antimicrobial resistance of uropathogenic Escherichia coli isolates from patients with urinary tract infections in a tertiary teaching hospital in Iran. Microb. Drug Resist. 26, 60–70. doi: 10.1089/mdr.2019.0184
Neyestanaki, D. K., Mirsalehian, A., Rezagholizadeh, F., Jabalameli, F., Taherikalani, M., and Emaneini, M. (2014). Determination of extended spectrum beta-lactamases, metallo-beta-lactamases and AmpC-beta-lactamases among carbapenem resistant Pseudomonas aeruginosa isolated from burn patients. Burns 40, 1556–1561. doi: 10.1016/j.burns.2014.02.010
Ny, S., Edquist, P., Dumpis, U., Gröndahl-Yli-Hannuksela, K., Hermes, J., Kling, A.-M., et al. (2019). Antimicrobial resistance of Escherichia coli isolates from outpatient urinary tract infections in women in six European countries including Russia. J. Glob. Antimicrob. Resist. 17, 25–34. doi: 10.1016/j.jgar.2018.11.004
Pan, Y., Zeng, J., Li, L., Yang, J., Tang, Z., Xiong, W., et al. (2020). Coexistence of antibiotic resistance genes and virulence factors deciphered by large-scale complete genome analysis. mSystems 5:e00821-19. doi: 10.1128/mSystems
Park, J. Y., Yun, K. W., Choi, E. H., and Lee, H. J. (2018). Prevalence and characteristics of sequence type 131 Escherichia coli isolated from children with bacteremia in 2000–2015. Microb. Drug Resist. 24, 1552–1558. doi: 10.1089/mdr.2017.0224
Pathak, A., Chandran, S. P., Mahadik, K., Macaden, R., and Lundborg, C. S. (2013). Frequency and factors associated with carriage of multi-drug resistant commensal Escherichia coli among women attending antenatal clinics in Central India. BMC Infect. Dis. 13, 1–9. doi: 10.1186/1471-2334-13-199
Peng, Z., Li, X., Hu, Z., Li, Z., Lv, Y., Lei, M., et al. (2019). Characteristics of carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-1 and MCR-1 from pig farms in China. Microorganisms 7:482. doi: 10.3390/microorganisms7110482
Pusparini, N., Waturangi, D. E., Usia, T., and Nikastri, E. (2018). Genetic diversity of Escherichia coli isolated from ice cube production sites. BMC. Res. Notes 11, 659–658. doi: 10.1186/s13104-018-3737-3
Rahman, M. M., Hossain, M. M. K., Rubaya, R., Halder, J., Karim, M. E., Bhuiya, A. A., et al. (2022). Association of Antibiotic Resistance Traits in Uropathogenic Escherichia coli (UPEC) isolates. Can. J. Infect. Dis. Med. Microbiol. 2022, 1–9. doi: 10.1155/2022/4251486
Rasoulinasab, M., Shahcheraghi, F., Feizabadi, M. M., Nikmanesh, B., Hajihasani, A., Sabeti, S., et al. (2021). Distribution of pathogenicity island markers and H-antigen types of Escherichia coli O25b/ST131 isolates from patients with urinary tract infection in Iran. Microb. Drug Resist. 27, 369–382. doi: 10.1089/mdr.2019.0485
Rezatofighi, S. E., Najafifar, A., Askari Badouei, M., Peighambari, S. M., and Soltani, M. (2021). An integrated perspective on virulence-associated genes (VAGs), antimicrobial resistance (AMR), and phylogenetic clusters of pathogenic and non-pathogenic avian Escherichia coli. Front. Vet. Sci. 8:758124. doi: 10.3389/fvets.2021.758124
Riley, L. (2014). Pandemic lineages of extraintestinal pathogenic Escherichia coli. Clin. Microbiol. Infect. 20, 380–390. doi: 10.1111/1469-0691.12646
Rosengren, L. B., Waldner, C. L., and Reid-Smith, R. J. (2009). Associations between antimicrobial resistance phenotypes, antimicrobial resistance genes, and virulence genes of fecal Escherichia coli isolates from healthy grow-finish pigs. Appl. Environ. Microbiol. 75, 1373–1380. doi: 10.1128/AEM.01253-08
Sabat, A., Budimir, A., Nashev, D., Sá-Leão, R., Van Dijl, J., Laurent, F., et al. (2013). Overview of molecular typing methods for outbreak detection and epidemiological surveillance. Euro Surveill. 18:20380. doi: 10.2807/ese.18.04.20380-en
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., Bugla-Ploskonska, G., et al. (2019). Virulence factors, prevalence and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut Pathog. 11, 1–16. doi: 10.1186/s13099-019-0290-0
Shahbazi, R., Salmanzadeh-Ahrabi, S., Aslani, M. M., Alebouyeh, M., Falahi, J., and Nikbin, V. S. (2023). The genotypic and phenotypic characteristics contributing to high virulence and antibiotics resistance in Escherichia coli O25-B2-ST131 in comparison to non-O25-B2-ST131. BMC Pediatr. 23:59. doi: 10.1186/s12887-023-03866-w
Sheikh, A. F., Khoshnood, S., Saki, M., Shahi, F., Seyed-Mohammadi, S., Abdi, M., et al. (2021). Prevalence of carbapenemases and ESBL encoding genes among K. pneumoniae isolates obtained from an educational hospital in Ahvaz, southwestern Iran. Gene Rep. 23:101128. doi: 10.1016/j.genrep.2021.101128
Tam, V. H., Chang, K.-T., Abdelraouf, K., Brioso, C. G., Ameka, M., McCaskey, L. A., et al. (2010). Prevalence, resistance mechanisms, and susceptibility of multidrug-resistant bloodstream isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 54, 1160–1164. doi: 10.1128/AAC.01446-09
Tawfik, A. F., Alswailem, A. M., Shibl, A. M., and Al-Agamy, M. H. (2011). Prevalence and genetic characteristics of TEM, SHV, and CTX-M in clinical Klebsiella pneumoniae isolates from Saudi Arabia. Microb. Drug Resist. 17, 383–388. doi: 10.1089/mdr.2011.0011
Tayh, G., Elmanama, A., Selmi, R., and Slama, K. B. (2023). Antibiotic resistance profile and molecular characterization of extraintestinal pathogenic Escherichia coli (ExPEC) from human clinical samples in gaza strip, Palestine. Lett. Appl. Microbiol. 76:ovac033. doi: 10.1093/lambio/ovac033
Vanstokstraeten, R., Piérard, D., Crombé, F., De Geyter, D., Wybo, I., Muyldermans, A., et al. (2023). Genotypic resistance determined by whole genome sequencing versus phenotypic resistance in 234 Escherichia coli isolates. Sci. Rep. 13:449. doi: 10.1038/s41598-023-27723-z
Whelan, S., Lucey, B., and Finn, K. (2023). Uropathogenic Escherichia coli (UPEC)-associated urinary tract infections: the molecular basis for challenges to effective treatment. Microorganisms 11:2169. doi: 10.3390/microorganisms11092169
Wu, J.-J., Ko, W.-C., Tsai, S.-H., and Yan, J.-J. (2007). Prevalence of plasmid-mediated quinolone resistance determinants QnrA, QnrB, and QnrS among clinical isolates of Enterobacter cloacae in a Taiwanese hospital. Antimicrob. Agents Chemother. 51, 1223–1227. doi: 10.1128/AAC.01195-06
Yu, Z., Wang, J., Ho, H., Wang, Y., Huang, S., and Han, R. (2020). Prevalence and antimicrobial-resistance phenotypes and genotypes of Escherichia coli isolated from raw milk samples from mastitis cases in four regions of China. J. Glob. Antimicrob. Resist. 22, 94–101. doi: 10.1016/j.jgar.2019.12.016
Yun, K. W., Kim, H. Y., Park, H. K., Kim, W., and Lim, I. S. (2014). Virulence factors of uropathogenic Escherichia coli of urinary tract infections and asymptomatic bacteriuria in children. J. Microbiol. Immunol. Infect. 47, 455–461. doi: 10.1016/j.jmii.2013.07.010
Zeighami, H., Valadkhani, F., Shapouri, R., Samadi, E., and Haghi, F. (2019). Virulence characteristics of multidrug resistant biofilm forming Acinetobacter baumannii isolated from intensive care unit patients. BMC Infect. Dis. 19:629. doi: 10.1186/s12879-019-4272-0
Zhang, S., Chen, S., Rehman, M. U., Yang, H., Yang, Z., Wang, M., et al. (2021). Distribution and association of antimicrobial resistance and virulence traits in Escherichia coli isolates from healthy waterfowls in Hainan, China. Ecotoxicol. Environ. Saf. 220:112317. doi: 10.1016/j.ecoenv.2021.112317
Zhang, L., Foxman, B., and Marrs, C. (2002). Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. J. Clin. Microbiol. 40, 3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002
Zhao, X., Lv, Y., Adam, F. E. A., Wang, X., and Yang, Z. (2021). Comparison of antimicrobial resistance, virulence genes, phylogroups, and biofilm formation of Escherichia coli isolated from intensive farming and free-range sheep. Front. Microbiol. 12:699927. doi: 10.3389/fmicb.2021.699927
Zhong, Y.-M., Liu, W.-E., Liang, X.-H., Li, Y.-M., Jian, Z.-J., and Hawkey, P. M. (2015). Emergence and spread of O16-ST131 and O25b-ST131 clones among faecal CTX-M-producing Escherichia coli in healthy individuals in Hunan Province, China. J. Antimicrob. Chemother. 70, 2223–2227. doi: 10.1093/jac/dkv114
Keywords: Eshcerichia coli, bloodstream infections, phylogenetic group, sequence type, virulence, antimicrobial resistance, REP-PCR
Citation: Hemati S, Halimi S, Jabalameli F, Emaneini M and Beigverdi R (2024) Phylogenetic group, antibiotic resistance, virulence gene, and genetic diversity of Escherichia coli causing bloodstream infections in Iran. Front. Microbiol. 15:1426510. doi: 10.3389/fmicb.2024.1426510
Received: 01 May 2024; Accepted: 09 July 2024;
Published: 19 July 2024.
Edited by:
Abdelazeem Algammal, Suez Canal University, EgyptReviewed by:
Amit Ranjan, Columbia University, United StatesCopyright © 2024 Hemati, Halimi, Jabalameli, Emaneini and Beigverdi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reza Beigverdi, ci1iZWlndmVyZGlAdHVtcy5hYy5pcg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.