
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 10 October 2024
Sec. Infectious Agents and Disease
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1425437
This article is part of the Research Topic Listeria monocytogenes: Do We Know Enough About This Pathogen? View all 7 articles
Listeria monocytogenes is a foodborne pathogen that causes listeriosis in humans, the severity of which depends on multiple factors, including intrinsic characteristics of the affected individuals and the pathogen itself. Additionally, emerging evidence suggests that epigenetic modifications may also modulate host susceptibility to infection. Therefore, different clinical outcomes can be expected, ranging from self-limiting gastroenteritis to severe central nervous system and maternal-neonatal infections, and bacteremia. Furthermore, L. monocytogenes is a genetically and phenotypically diverse species, resulting in a large variation in virulence potential between strains. Multilocus sequence typing (MLST) has been widely used to categorize the clonal structure of bacterial species and to define clonal complexes (CCs) of genetically related isolates. The combination of MLST and epidemiological data allows to distinguish hypervirulent CCs, which are notably more prevalent in clinical cases and typically associated with severe forms of the disease. Conversely, other CCs, termed hypovirulent, are predominantly isolated from food and food processing environments and are associated with the occurrence of listeriosis in immunosuppressed individuals. Reports of genetic traits associated with this diversity have been described. The Food and Agriculture Organization (FAO) is encouraging the search for virulence biomarkers to rapidly identify the main strains of concern to reduce food waste and economical losses. The aim of this review is to comprehensively collect, describe and discuss the methodologies used to discriminate the virulence potential of L. monocytogenes CCs. From the exploration of in vitro and in vivo models to the study of expression of virulence genes, each approach is critically explored to better understand its applicability and efficiency in distinguishing the virulence potential of the pathogen.
Within the genus Listeria, twenty-eight species are recognized; however, only two are considered pathogenic: Listeria ivanovii and Listeria monocytogenes (Raufu et al., 2022; Vázquez-Boland et al., 2001). Both L. ivanovii and L. monocytogenes can cause listeriosis, but the majority of cases are attributed to L. monocytogenes and only a few to L. ivanovii. Although much rarer than those caused by L. monocytogenes and L. ivanovii, L. innocua infections have been reported in humans and ruminants (Favaro et al., 2014; Moura et al., 2019; Perrin et al., 2003; Rocha et al., 2013; Walker et al., 1994). Moura et al. (2019) demonstrated the virulence potential of atypical haemolytic L. innocua strains.
Human listeriosis, primarily caused by the consumption of contaminated food, is a severe illness that can manifest in one of two forms: non-invasive gastrointestinal infection in immunocompetent individuals or invasive listeriosis in risk groups, including pregnant women and newborns, the elderly and immunocompromised individuals (Vázquez-Boland et al., 2001; World Health Organization & Food and Agriculture Organization of the United Nations, 2004). In the invasive form, the pathogen surpasses the blood–brain and placental barriers, resulting in septicaemia, meningitis, spontaneous abortion and stillbirth (Lecuit, 2005). In 2022, the European Union reported 2,738 confirmed cases of listeriosis, which is 50 times fewer cases than the predominant gastrointestinal infection reported in humans, campylobacteriosis. Among the surveyed zoonotic pathogens, L. monocytogenes had the highest rates of hospitalization (96%) and case fatalities (18.1%) (EFSA and ECDC, 2023). These highlights the gravity of this major public health issue in developed nations. In addition to posing a significant public health risk, contamination of foods with this pathogen leads to disruptions in production, distribution, and recalls. As a result, it is receiving considerable attention from the food industry and authorities due to the significant economic losses and food waste involved (Li et al., 2022).
The study of L. monocytogenes bacterial model is of undoubtable importance; however, scientific research cannot be directly performed in humans. The investigation of this foodborne pathogen infection in humans has been mainly through reported clinical cases, epidemiological data, genome analysis and the use of infection models. In addition, L. monocytogenes has relatively low incidence in humans and extended incubation periods can be challenging in listeriosis studies, hindering the identification of causing pathogen and contamination routes (Hoelzer et al., 2012; Vázquez-Boland et al., 2001). Although different methods have evolved to better characterize L. monocytogenes, this species is genetically heterogeneous and different typing methods (discussed below) can be used to subtype this species at different levels. Due to the great variety of typing methods available, comparative analysis between studies can be challenging (Koopmans et al., 2023). Additionally, the study of L. monocytogenes virulence potential can be conducted through different host species used as infection models, however, differences in the selected model, infection dose, incubation time, etc. can be difficult when comparing between studies. Several phenotypic and genotypic tools as well as in vitro and in vivo models have been used to evaluate the uneven virulence potential among distinct strains. Given the diversity of L. monocytogenes studies, it is challenging to define criteria that are universally objective, consistent and applicable. Therefore, in this review we aim to explore and analyse the current methodologies utilized for evaluating differences in the virulence potential among strains of distinct CCs (summarized in Figure 1), giving readers an overview of the available literature.
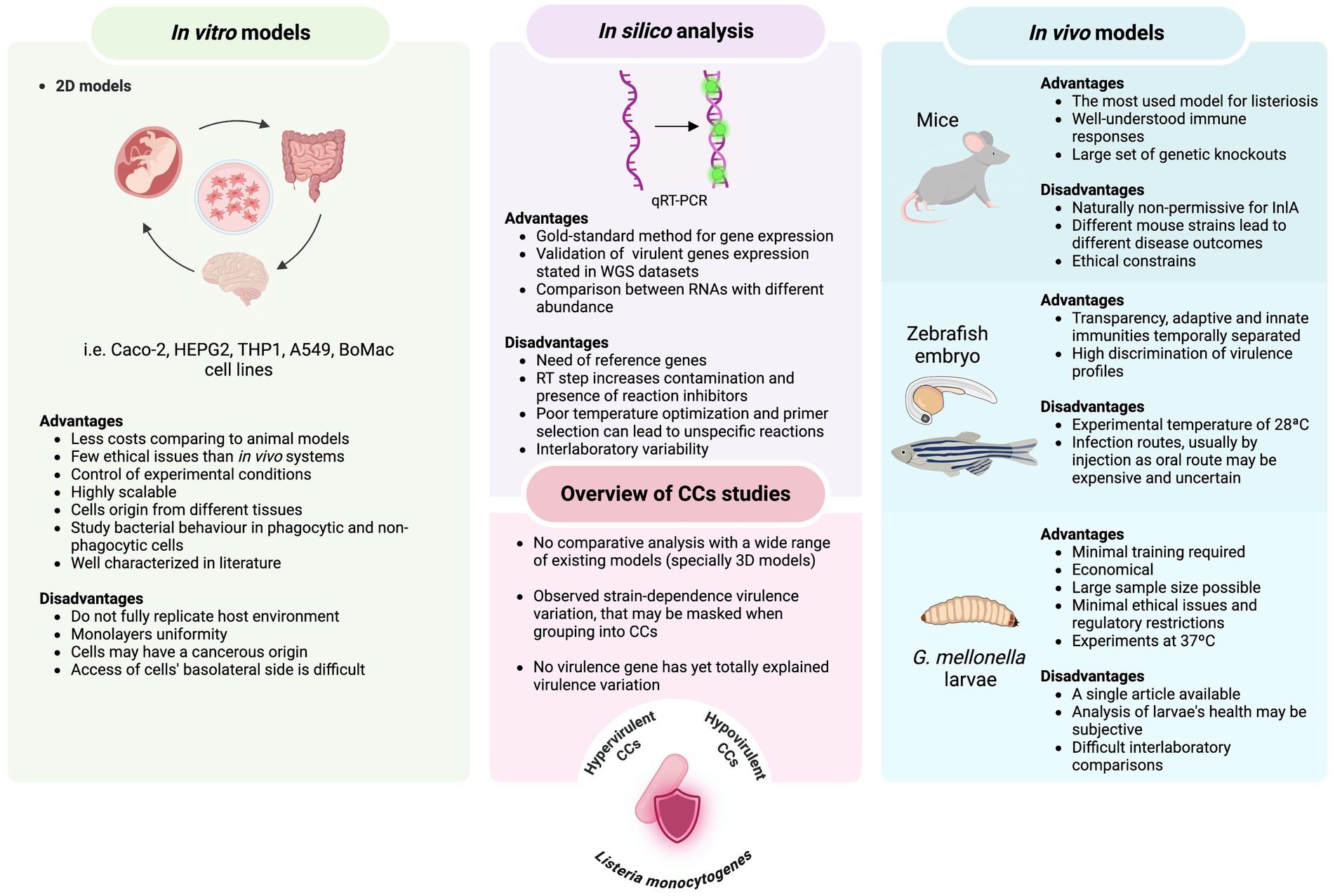
Figure 1. Overview of advantages and disadvantages of infection systems (both in vitro and in vivo models) and molecular approaches used to assess the virulence potential among L. monocytogenes clonal complexes.
Typing of L. monocytogenes has been essential in epidemiological studies of listeriosis, allowing for the establishment of clonal relatedness among collected isolates. Over the decades, the development and implementation of pheno-and geno-typing methods have made it possible to confirm outbreaks, trace sources of contamination and identify transmission routes within the food chain. Additionally, the increasing adoption of standardized typing methods has facilitated the establishment of effective national and international surveillance systems, enabling the monitoring of evolutionary trends and the generation of comparisons across different geographical regions. This has indubitably had a major influence on the responses and strategies of public health systems worldwide. On the other hand, these methods have massively enhanced our perception of the remarkable biodiversity within L. monocytogenes species and their distribution in different environments. The first method, largely employed in epidemiological studies, was based on the serological antigen structure of the bacterium, specifically on the agglutinating activity of somatic (O) and flagellar (H) antigens (Seeliger and Höhne, 1979; Seeliger and Langer, 1989). This method was gradually replaced by more expeditious methods – namely, a gel-based multiplex-polymerase chain reaction (PCR serogroup) that differentiates, between four major serogroups, including the serovars more frequently isolated from food and patients (> 98%, i.e., 1/2a, 1/2b, 1/2c, and 4b): serogroup IVb (comprising serovars 4b, 4d, 4e), serogroup IIa (comprising serovar 1/2a, 3a); IIb (comprising serovars 1/2b, 3b, 7); and serogroup IIc (comprising serovars 1/2c, 3c) (Doumith et al., 2004). Later, a real-time triplex-PCR assay that differentiates these groups was made available (Vitullo et al., 2013). Although many L. monocytogenes serotypes have been discovered, three major serovars (1/2a, 1/2b, and 4b) are responsible for a substantial fraction of listeriosis cases (about 90 to 95% of human infections) (Schiavano et al., 2022).
Several typing methods have been used for multiple purposes, with genotypic methods being particularly highlighted due to their higher discriminatory power (e.g., amplified fragment length polymorphism (AFLP), multilocus variable-number tandem repeat analysis (MLVA) or ribotyping) compared to phenotypic methods. In the specific case of epidemiological studies, pulsed-field gel electrophoresis (PFGE), based on the analysis of DNA restriction patterns, has been considered the “gold standard” technique for typing L. monocytogenes for many years (Graves and Swaminathan, 2001). However, PFGE has some drawbacks, such as the difficulty of standardizing the analysis of fingerprints, which poses a challenge for inter-laboratory and inter-country comparisons. In addition, while it is valuable for assessing genetic relatedness between isolates, pinpointing sources of contamination and identifying outbreaks, it is not sufficient for establishing comprehensive phylogenetic relationships between strains. Sequence-based typing methods such as multilocus sequence typing (MLST) or multi-virulence-locus sequence typing (MLVST) are more appropriate for this purpose (Maiden et al., 1998; Salcedo et al., 2003; Zhang et al., 2004). Currently, MLST is widely used as a reference method to categorize the clonal structure of bacterial species and to define clonal complexes (CCs) of genetically related isolates, i.e., those descended from the same ancestor. In L. monocytogenes, MLST is based on the sequencing of seven housekeeping genes (acbZ, bglA, cat, dapE, dat, ldh, and lhkA), that allow the determination of sequence types (STs) (Bergholz et al., 2018; Orsi et al., 2011; Ragon et al., 2008). Additionally, Ragon et al. (2008) grouped these STs within CCs, with strains sharing at least six out of seven MLST alleles being assigned to the same CC. Currently, the preferred method for epidemiological and phylogenetic studies has shifted to whole genome sequencing (WGS), which has become more accessible to a broader range of laboratories due to technological advances and reduced costs (Gerner-Smidt et al., 2019). Whole genome sequencing of Listeria provides high-resolution data that not only allows phylogenetic relationships between strains to be determined, but also provides in-depth knowledge of the genomic structure of a given strain, including information on specific virulence factors and other genes that contribute to pathogenesis, as well as potential antibiotic resistance prediction (Hurley et al., 2019; Moura et al., 2024).
This species presents a diverse genetic pool and its virulence potential is very heterogeneous, resulting in an uneven capacity of strains to cause disease (Pyz-Łukasik et al., 2022). Currently, this species is divided into four major evolutionary lineages (I-IV), comparable to subspecies (Liu, 2006; Orsi et al., 2008; Rasmussen et al., 1995; Roberts et al., 2006; Ward et al., 2008; Wiedmann et al., 1997); lineage I includes serotypes 1/2b, 3b, 4b, 4d, and 4e and 7, and is significantly overrepresented in human listeriosis cases (Gray et al., 2004; Orsi et al., 2011); lineage II includes serotypes 1/2a, 1/2c, 3a, and 3c, prevalent among isolates from environmental samples, food, and animal listeriosis cases (Nightingale et al., 2005b; Sauders et al., 2006), and contribute significantly to sporadic cases of human listeriosis (Jeffers et al., 2001); and, lineage III and IV include serotypes 4a, 4c and atypical serotype 4b isolates, which are rare and are mainly associated with listeriosis in animals (Liu, 2006). Clonal complexes are grouped within lineages, for example, CC1, CC2, CC4 and CC6 (serotype 4b, lineage I) and CC121 and CC9 (serotypes 1/2a and 1/2c, respectively, lineage II) (Maury et al., 2016). A methodology for cloning L. monocytogenes and assessing potential human infectivity has been patented (WO2017009198A1).
More than one hundred CCs have been reported globally. The predominance of particular CCs is highly heterogeneous among different sources and regions. In 2011, Chenal-Francisque et al. (2011) characterized the genotypic profile of three hundred isolates collected from 42 countries on five distinct continents, and these isolates were distributed within 111 STs, assembled into only 17 CCs. This reinforces the idea that there is an irregular geographical distribution, with a few prevailing CCs (Chenal-Francisque et al., 2011; Wagner et al., 2022). However, these isolates were collected between 1933 and 2007, and it has been established that the distribution of CCs tends to change over time with some CCs, such as CC9, CC121, CC5, and CC6, emerging more recently (Bergholz et al., 2018). Information on STs/CCs associated with listeriosis outbreaks in European countries during the last decade are presented in Table 1. When molecular characterization of outbreak strains was not available from publications, additional information was collected from the Institut Pasteur MLST database.1 CC6 and CC8 were the two main CCs, accounting for 17.4% of the total of 23 outbreaks identified. Many studies across European countries have also reported that some clonal complexes, such as CC1, CC2, CC3, CC4, CC5, CC6, CC8, CC9, CC37, CC121, and CC388 are globally prevalent with some geographical disparities (Domínguez et al., 2023; Félix et al., 2022; Maury et al., 2016; Painset et al., 2019). To better characterize this heterogeneity between strains from different CCs, two independent terms have been established: CCs with a high frequency in human clinical cases are considered hypervirulent; conversely, CCs associated with food, persistence in food manufacturing environments, and with a lower frequency in human listerioses cases are considered hypovirulent (Maury et al., 2016). Therefore, CC1, CC2, CC4, and CC6 (lineage I) are considered to be hypervirulent clones since they are clinically related and mainly infect individuals with low or no comorbidities. Contrarily, strains belonging to CC9 and CC121 (lineage II), recognized as hypovirulent clones, are regularly isolated from food and food processing environments. The latter are often associated with individuals with a compromised immune system (Maury et al., 2016). There is also an intermediate classification for those clones that may be in transition from their host-associated lifestyle due to loss of virulence and acquisition of stress resistance genes (FAO and WHO, 2022). In 2018, Fritsch and co-workers also established three different levels of virulence among CCs and STs for risk characterization: hypovirulence, medium virulence, and hypervirulence. The latter, includes, in addition to the previously mentioned hypervirulent CCs, CC224, ST54, CC101 + 90, ST87, ST451, ST504, CC220, ST388, and CC207 (Fritsch et al., 2018). Hypovirulent CCs include CC9 and CC121, as well as CC11, CC19, CC31, CC193, CC199, CC204, and ST124.
It is important to note that, although hypovirulent CCs such as CC9 and CC121 are mainly associated with food and food processing environments, cases of invasive listeriosis caused by these CCs have also been reported. For instance, CC121 was considered the second most common CC isolated from human clinical cases in Norway and in France (Fagerlund et al., 2022; Maury et al., 2016).
Despite the potential ability to predict the risk of a specific strain of L. monocytogenes causing disease after consumption of contaminated food, most regulatory authorities worldwide take action when any L. monocytogenes is found in ready-to-eat (RTE) food that is capable of supporting growth, regardless of its strain characteristics. This approach is recommended by the Food and Agriculture Organization (FAO) and the World Health Organization (WHO) (2022), although in some countries risk managers are permitted to use information on L. monocytogenes subtypes to guide risk management decisions. However, the FAO and the WHO encourage the search for other virulence markers to predict, based on genetic virulence profiles (CCs characterization) (FAO and WHO, 2022). The discovery of one or multiple biomarkers that would allow to predict the real virulence potential of a given strain, and a clear distinction between hypo-and hypervirulence would be of great value to reassess the risks associated with different L. monocytogenes strains and to develop appropriate policies that neither overstate nor underestimate the risk posed by each strain. Ultimately, this finding would also contribute massively to the reduction of costs associated with the recall and destruction of contaminated food products and to reduced food waste and its social and economic consequences.
The L. monocytogenes infection cycle comprises various steps: adhesion and invasion, lysis and escape from the vacuole, cytosolic multiplication, actin-tails polymerization, spread to neighbouring cells, and rupture of a double-membrane vacuole (Luque-Sastre et al., 2018; Pizarro-Cerdá et al., 2012). Some virulence genes are important for infection, such as, InlA-E-cadherin and/or InlB-C-Met (L. monocytogenes internalins-host receptors) for invasion, listeriolysin O (LLO) and phospholipases A and B (PlcA and PlcB) for both primary and double-layer vacuoles disruption, ActA for actin tail polymerization and intracellular motility (Quereda et al., 2021; Radoshevich and Cossart, 2018).
Considering all the above information, a detailed investigation regarding the putative virulence markers linked to both hyper-and hypovirulence is still ongoing, and some interesting findings have been reported. Regarding the core genome, inlA is normally present and expressed as a full-length form within clinical isolates (Lecuit et al., 2001). Premature stop codons mutations (PMSCs) have been found in the inlA gene, resulting in a truncated non-functional internalin in food isolates. In some studies, these PMSCs have been found amid strains from hypovirulent CCs, such as CC9 and CC121, and thus it is hypothesized that in some way, the lower virulence potential of these strains can be justified by the InlA truncation, leading to a reduced capacity to cross the intestinal barrier (Jacquet et al., 2004; Lachtara et al., 2022; Moura et al., 2016). The significant role of InlA-mediated crossing of L. monocytogenes through the intestinal barrier has been described. However, some studies have showed that the inoculation of ΔinlA mutants still resulted in L. monocytogenes infection (Bierne et al., 2002; Bou Ghanem et al., 2012; Disson et al., 2008). Therefore, it was hypothesized that L. monocytogenes employes alternative routes to cross the intestinal barrier. Besides the M cell-mediated translocation in Peyer’s patches, Drolia et al. (2018, 2024) showed that the linkage between Listeria adhesion protein (LAP) and its surface receptor Hsp60 promotes cell disruption by using the cell innate system, consequently leading to bacterial translocation (Drolia et al., 2024; Drolia et al., 2018). These studies have shown that L. monocytogenes can cross the intestine through InlA-independent routes, which could explain the isolation of strains belonging to hypovirulent CCs (normally associated with the production of truncated inlA) in clinical cases. However, to our knowledge no comparative studies have investigated LAP or other InlA-independent invasion factors as putative candidate to distinguish hyper-or hypovirulent strains of L. monocytogenes.
All strains of L. monocytogenes carry the Listeria Pathogenicity Island 1 (LIPI-1), which clusters several fundamental genes for L. monocytogenes pathogenicity (Moura et al., 2016; Vázquez-Boland et al., 2001). These include the hly gene, which encodes a hemolysin – LLO – that provides the capacity to lyse erythrocytes. As mentioned above, this toxin can form a pore and allow the bacteria to escape from the internalization vacuole; thus, this virulence factor is detrimental to the virulence of L. monocytogenes. Another important virulence factor is PrfA, known as the main regulator of virulence genes in L. monocytogenes, such as the prfA, actA and hly genes. However, some studies have reported the existence of non-hemolytic L. monocytogenes strains, belonging to both lineages I and II, that have mutations in either the prfA or hly genes, and consequently a lower virulence potential (Maury et al., 2017).
Regarding the accessory genome, the pathogenicity island LIPI-3 carries eight genes. Listeriolysin S (LLS) encoded by llsA, functions as a bacteriocin with the capacity to modify the composition of the intestinal microbiota by eliminating or hindering the growth of neighbouring bacteria. This virulence cluster is often present within lineage I isolates, especially those from CC1, CC2 and CC6 – constituting a potential marker of hypervirulence (Cotter et al., 2008; Moura et al., 2016; Quereda et al., 2016). Additionally, Maury et al. (2016) identified a novel virulence cluster termed LIPI-4, which aggregates six genes that encode a cellobiose family phosphotransferase system (PTS). This gene cluster is strongly associated with strains of CC4, which are highly relevant to human brain and placental infections (Maury et al., 2016; Moura et al., 2016). Furthermore, it was thought that this pathogenic island was exclusively related to CC4 strains, but isolates from CC87 in China also displayed this locus (Wang et al., 2019; Zhang et al., 2020). These findings suggest that this could be a putative marker of hypervirulence, although it was found that this island was also present in L. innocua – a non-pathogenic species – and thus its role in hypervirulence is still controversial, reinforcing the need for further studies. Another intriguing gene is lmo2776, which acts as a bacteriocin and plays an important role in modulating the intestinal microbiome, mainly targeting Prevotella copri – a common gut commensal that has the capacity to modify the intestinal mucus layer and potentially intensify gut infection. The critical aspect is its significant presence in lineage I strains compared to its low frequency in lineage II strains. Curiously, deletion of lmo2776 resulted in a better spread of the bacteria to the liver and spleen – the primary target organs of L. monocytogenes after crossing the intestinal barrier. This can be explained by the capacity of L. monocytogenes to discriminate between P. copri, preventing exorbitant inflammation and leading to longer periods of infection (Rolhion et al., 2019).
In virulence studies, both pathogen characteristics and host physiology and anatomy must be considered, as microbial infections result from interactions between pathogens, hosts and the surrounding environment (Prescott, 2022). Preclinical trials using in vivo and in vitro biological models, have provided valuable insights into host-pathogen interactions (Anju et al., 2020; Khan et al., 2018). In vivo systems, used for various purposes from drug development to investigating physiological processes, complement in vitro studies by providing a more comprehensive understanding of biological responses. However, neither system alone is sufficient to make absolute predictions (Khan et al., 2018). Some of these models will be detailed in the following sections (Table 2).
In order to improve human health research, both mammalian and non-mammalian models are used due to ethical constraints with experiments involving humans (World Medical Association, 2013). The broad host range of L. monocytogenes allows the use of various animal models, such as Drosophila melanogaster (fly), Galleria mellonella (moth), Caenorhabditis elegans (nematode), Mus musculus (mouse), Cavia porcellus (guinea pig), Oryctolagus cuniculus (rabbit), among others (Anju et al., 2020; Prescott, 2022) – some of which will be discussed further. Animal models offer advantages that make them invaluable for human health research: they have identical biological processes, anatomical similarities (especially in vertebrates animals) – which are difficult to replicate in in vitro systems – compatible diseases such as cancer and diabetes, short life cycle and some can be easily genetically transformed to acquire some fundamental characteristics to express the disease phenotype (Kiani et al., 2022). Additionally, in vivo models are essential because they possess some unique characteristics when compared to in vitro models, for instance, the immunity associated with commensals and the intestinal mucosa throughout infection (Eng and Pearson, 2021). Depending on the final objective of the study, several aspects must be considered when selecting the ideal animal model: (1) the pathogen should have a similar tissue and cell affinity as in humans; (2) it should reveal the identical observable disease outcome and immunopathological harm; and (3) it should be susceptible to genetic manipulation (Lecuit, 2007). In addition to animal features, to study L. monocytogenes virulence, understanding listeriosis pathophysiology is crucial to select the adequate animal model. As already mentioned, L. monocytogenes can cross the intestinal, blood–brain and placental barriers. Therefore, pregnant, non-pregnant and geriatric animal models have been used in the study of L. monocytogenes pathogenesis, this was exhaustively described by Hoelzer et al. (2012). Animal models have played crucial roles in the characterization the virulence of L. monocytogenes. Generally, insightful data about the different pathways of bacterial translocation through host’s defensive barriers, the exploitation of host’s immunity to improve disease, performance of dose-dependent assays, the complex host immune responses to infection, the species specificity, virulence factors and strains virulence potential have been emerged from animal models studies (Hoelzer et al., 2012; Koopmans et al., 2023; Lecuit, 2007; Maury et al., 2016; Tsai et al., 2013). To our knowledge, an ideal animal model for listeriosis has not been established. Continuous new insights into animal physiology have increase the possibilities for infection systems, yet no single animal model completely aggregates the desirable characteristics to study human listeriosis. Therefore, the selection of an in vivo system is according to the specific objectives of the research being conducted.
Although animal models bring unquestionable insights into the study of infectious diseases, their extensive and indiscriminate use is strongly condemned by the European commission. This authority bases its policy on the Three R’s principle (Replacement, Reduction and Refinement), which aims to replace the use of animals with non-animal strategies, to use a reduced number of animals per experiment without compromising the ultimate aim of the research, and to improve practices that contribute to the welfare of animals from birth to death. When animal replacement is not possible, the use of animals must follow strict guidelines set out in EU Directive 2010/63/EU (European Parliament, 2010; Zuang et al., 2024).
The establishment of Robert Koch postulates to determine the etiological agent of an infectious disease, marked the inception of using mammalian species, phylogenetically related to humans, as healthy susceptible models (Kaito et al., 2020; Short and MacInnes, 2022).
Listeria monocytogenes is a ubiquitous microorganism, which enables it to infect a wide range of animals (Kammoun et al., 2022). However, in addition to humans, it mainly causes disease in ruminants, which, in an immediate and logical thought, should be the primary models to study listeriosis. However, this brings up many limitations. Thus, mice are the standard in vivo model to study listeriosis due to their size, ease of breeding and reproduction, rapid acclimation to confinement and an equivalent physiology when compared to humans (Lecuit, 2007). Commonly, mice are intravenously infected with the pathogen, and the role of some virulence factors, such as ActA and LLO, have emanated from this technique (Disson et al., 2009). Although mice are widely used in L. monocytogenes studies, the efficacy of oral infection is low due to the species-specific associated of with mammalian cells. As mentioned above, InlA binds to the E-cad receptor, which is a specific linkage for each species, depending on its 16th amino-acid type. Permissive species, such as guinea pigs, rabbits, humans and gerbils, have a proline in this position while non-permissive species have a glutamic acid – mice and rats have the glutamic acid and, consequently do not allow InlA binding (Lecuit et al., 1999). On the other hand, InlB naturally binds to C-Met in mice, humans and gerbils (Khelef et al., 2006). Theoretically, animals that naturally possess the imperative requirements to be bound to L. monocytogenes internalins, such as ruminants, non-human primates, and gerbils, should be selected to study listeriosis (Kammoun et al., 2022). Nonetheless, the ethical hurdles do not allow their wide application, so humanized mice have surged to overcome this limitation (Disson et al., 2008; Lecuit et al., 2001). Additionally, a “murinized” L. monocytogenes strain was developed to interact more closely with mouse E-cadherin. This modification involved altering the inlA gene in the L. monocytogenes EGDe strain to successfully infect wild-type mice (Wollert et al., 2007). Although this species-specific limitation was overcome, it was further discovered that the altered InlA was able to interact with both E-cadherin and N-cadherin in mice, luminally accessible in goblet and M-cells respectively, leading the bacteria to target both cells, increasing gut inflammation and consequently, hindering the capacity of L. monocytogenes to spread in the host (Tsai et al., 2013).
Currently there has not been a published comparative analysis of clonal complexes and their virulence in some animal models, such as gerbils, non-human primates, guinea pigs or rats. Although gerbils are permissive to both receptors, their use in listeriosis studies is limited. This may be related to the decreased sensitive to oral infection with L. monocytogenes when compared to other models, insufficient characterization when compared to mice and guinea pigs, absence of genetic models, and limited specific reagents and antibodies. The guinea pig infection model is advantageous in maternal-fetal studies as its placenta is the most comparable to human placenta among all rodents and has equivalent placental tropism. However, its narrow use may be related not only to species-specificity but also due to different disease symptoms from human listeriosis, with weak central nervous system tropism. Guinea pigs also present long gestation periods compared to mice, lack of gene deletion and transgenic models, and their larger size is more costly, limiting the number of animals per experiment. Despite the similarities of rats to mice, this infection model has shown low susceptibility to infection, requiring high infection doses to provoke disease. The use of non-human primates, as expected, is limited due to extended gestation periods, reduced number of available animals per study, and limited gene libraries compared to mice. Additionally, all these models are most costly when compared to mice (Cossart, 2011; D'Orazio, 2014; Eallonardo and Freitag, 2024; Hoelzer et al., 2012; Khelef et al., 2006; Roulo et al., 2014; Yan et al., 2023).
Considering this, very few articles have employed mice to investigate this phylogenetic association, either directly or indirectly (Domínguez et al., 2023; Maudet et al., 2022; Soni et al., 2017). Although the objective was not to compare strains from different CCs, Soni et al. (2017) inoculated three strains from CC1 in mice and observed a varying disease-causing capacity. One strain did not kill any mouse, while the other two presented 60 and 100% relative virulence. This highlights that although CCs are a more thorough classification, strains within a single CC can exhibit different virulence potentials. They also observed that the three strains harboured the major virulence genes, with the strain showing the lower pathogenicity presenting mutations in crucial virulence factors, such as listeriolysin O. However, no conclusion has been reached as to which mutation or genes better explain this unequal pathogenicity between phylogenetically close strains (Soni et al., 2017). Furthermore, in 2019, a large outbreak of listeriosis occurred in the Andalusian region, causing 207 cases, which was later associated with the strain ST388 from CC388 (Ministerio de Sanidad Consumo y Bienestar Social de España, 2019). Domínguez and her colleagues proceeded to investigate the virulence potential of this strain by comparing it with other strains from hypervirulent CCs (CC1 and CC4). In vivo infection assays were performed, and mice were infected intravenously with four strains (reference ATCC® 19115™, CC1, CC4, and CC388 strains). The results showed no significant differences between the CC388 strain and the other hypervirulent strains, as CC4 and CC388 isolates exhibited identical infection and spread ability (Domínguez et al., 2023). Maudet et al. (2022) selected strains from CC1, CC4, and CC6 – previously characterized as highly neuroinvasive CCs (Maury et al., 2016) – to perform infection assays in humanized KIE16P mice. The comparative analyses performed between these hypervirulent CCs and EGDe strain (CC9), corroborated the increased capacity of hypervirulent CCs to invade mice brains. Additionally, gene expression assays showed that hypervirulent strains presented upregulated levels of the inlAB operon, when compared to EGDe. Throughout these experiments different ∆inlB mutant strains were constructed to validate its relevance in the neuroinvasion capacity of L. monocytogenes. Despite the reduced neuroinvasion levels of EGDe when compared to CC4 strains, this study showed that whether using hypovirulent or hypervirulent strains, the inlB gene deletion reduced bacterial loads in the brain, confirming the need of overexpressing the inlB gene in L. monocytogenes neuroinvasiveness. Furthermore, the authors reported that InlB has immunosuppressive properties that are crucial to protect infected cells from host immune responses, resulting in an increase of infected monocytes’ lifespan and L. monocytogenes propagation to the brain. Additionally, as hypervirulent strains exhibit overexpression of inlB and are mainly associated with infections in immunocompetent individuals, this article highlights the need to continuously study hypervirulent CCs to improve our perspective regarding the bacterial factors employed in L. monocytogenes infection mechanism. Altogether, these finding showed that regardless of some reports suggesting strain-dependence in L. monocytogenes virulence studies, strains from hypervirulent CCs confer a significant concern to human health with distinct virulence factors that allows them to evade the host immune system. Moreover, mice models have proven to be a reliable tool to study L. monocytogenes infection cycle in mammals and we believe they will continue to be useful in future works.
Although mammalian models are the paradigm for studying host-pathogen interactions, they still present many obstacles, such as ethical issues due to animal welfare, high costs, adequate facilities and differentiated training requirements. Therefore, alternative models are needed for in vivo experiments that are less costly, easier to manipulate, with a short life cycle and are ethically acceptable. The complexity and relevance of these models lie between the sophisticated humanized mice and the simplicity of in vitro approaches (Lecuit, 2007). A variety of invertebrate and vertebrate models have been used to study the virulence potential of pathogens and the host immune response (Ahlawat and Sharma, 2022; Mylonakis et al., 2007). In L. monocytogenes studies, we highlight the use of G. mellonella larvae, Drosophila melanogaster, Caenorhabditis elegans and Danio rerio (zebrafish), which have given valuable insights in the study of listeriosis. For instance, C. elegans model was previously used to evaluate the effects and toxicity of antimicrobial or antibiofilm substances in host-pathogen interactions and study nitrogen metabolism of L. monocytogenes after nematodes gut colonization (Kern et al., 2016; Muthulakshmi et al., 2022; Silva et al., 2015; Sivaranjani et al., 2016). The pathogenesis of L. monocytogenes has also been explored in D. melanogaster model, focusing in host immune system modulation, fly’s metabolism alterations upon infection, association between the bacterial growth dynamics and host’s genotypes (Chambers et al., 2012a,b; Hotson and Schneider, 2015; Mansfield et al., 2003; Taillebourg et al., 2014). Besides the widely use of both C. elegans and D. melanogaster models in the study of pathogenic bacteria, to our knowledge there are no comparative studies between L. monocytogenes CCs. In the fly model this can be related to the fact that the favourable temperature to flies is between 22 and 25°C, however, listeriosis studies are mainly conducted at 30–37°C and its inadequacy to distinguish between avirulent and virulent Listeria spp. after bacterial injection into the flies thorax (Jensen et al., 2007). On the other hand, C. elegans limited use may be associated to the fact that the deletion of some bacterial virulence genes (i.e., ActA) did not affect nematode’s death and that the C. elegans intestine architecture may be different from mammals, since neither cell junction during cell extrusion or in goblet cells lumen are common in this nematode (Balla and Troemel, 2013; Thomsen et al., 2006). Considering this, we focused on both zebrafish and waxworms to give an overview about the study of virulence potential between L. monocytogenes strains/CCs in non-mammals.
In the past, it was thought that insects were not a good in vivo model to study microorganisms that cause disease in humans since they are not phylogenetically close. However, they share a few physiological aspects with humans. Human pathogens present an analogous virulence capacity in humans and insects, with similar virulence factors involved (Mansfield et al., 2003; Martinez et al., 2017; Mukherjee et al., 2010; Tsai et al., 2016). In addition, the pathogen follows similar infection cycle steps in both hosts. Consequently, insects have evolved some defence mechanisms that are shared between mammals and insect hosts, for instance, the innate immune system with physical and phagocytic barriers, that have a homologous function (Kemp and Massey, 2007; Peterson et al., 2008). However, insects lack the capacity to develop an adaptive immune response, which is a common feature in vertebrates (Ahlawat and Sharma, 2022; Tsai et al., 2016). Hence, insects as host models have been a convenient alternative to mammals for infectious disease research.
Experiments with Galleria mellonella larvae have been carried out for some time, with increasing interest in recent year as a potential surrogate model to explore pathogen infections (Dinh et al., 2021). Besides being small, cheap, short life cycle, easy to maintain and to obtain in large numbers, it is also adapted to temperatures from 25°C to 37°C – the optimum growth temperature for the vast majority of human pathogens (Dinh et al., 2021; Mylonakis et al., 2005). The wax worm is selected stage to be utilized as a model, with infection normally occurring by injection, which requires minimal training (Singkum et al., 2019; Tsai et al., 2016). Its whole genome has recently been sequenced, enabling the search for further novel insights (Lange et al., 2018). Moreover, these insects possess a relatively advanced innate immune system, comprising two main components – the cellular and humoral immune response. The primer is composed of hemocytes – phagocytic cells that prevail in the hemolymph, and they are also capable of encapsulation and nodulation of pathogens. The humoral response results from the production of lytic enzymes, antimicrobial peptides (AMPs), opsonins and melanin upon microbial exposure (Boman and Hultmark, 1987; Kavanagh and Reeves, 2004; Pereira et al., 2018). It has been reported that G. mellonella larvae infected with L. monocytogenes are prone to produce AMPs such as galiomycin, lysozyme, gallerimycin, insect metalloproteinase inhibitor (IMPI) and cecropin D (Mukherjee et al., 2011; Mukherjee et al., 2010). Beside the analysis of host’s immune modulation through hemocytes enumeration and variations in AMPs expression, the melanization, survival capacity, development of cocoon, motion ability can be evaluated in infected larvae with L. monocytogenes (Kavanagh and Sheehan, 2018).
Galleria mellonella has been utilized as an infection model to study the virulence potential of L. monocytogenes through comparative studies with different Listeria species or comparisons between L. monocytogenes serotypes (Martinez et al., 2017; Mukherjee et al., 2010; Pan et al., 2024; Rakic Martinez et al., 2020). Mukherjee and co-workers explored the ability of this insect model to discriminate between non-pathogenic and pathogenic Listeria species. When injected with 106 CFU/larva, strains belonging to non-pathogenic species, such as L. innocua and L. seeligeri were observed to have a lower infection capacity than the L. monocytogenes EGD-e strain; and, although L. ivanovii caused a significant but slightly higher mortality than the non-pathogenic species, it presented a reduced pathogenicity efficiency compared to L. monocytogenes (Mukherjee et al., 2010). These results were corroborated by Martinez et al. (2017), who observed that, at the same inoculum level, the L. monocytogenes LS1209 reference strain displayed a LT50 (lethal time to kill 50% of larvae) 4 to 6 times lower than the non-pathogenic Listeria strains (Martinez et al., 2017).
The wax model was used to test the virulence potential of L. monocytogenes strains of different serotypes. The serotype 4b strain, commonly associated with clinical cases, expressed the highest larvae killing rate and was more pathogenic than the serotype 1/2a strain, usually related to food isolates. Other serotypes tested, 4a, 4c and 4d, also showed a lower pathogenic potential (Mukherjee et al., 2010). However, the study conducted by Martinez et al. (2017) showed that strains from different serotypes (1/2a, 4b, 1/2b) resulted in similar larvae mortality and identical LT50 at 24 h when administered at 106 CFU/larva (Martinez et al., 2017). This lack of correlation between serotypes and virulence potential was in clear contrast to the findings of the former study, highlighting the importance of considering potential confounding. These may include differences in the dose of bacteria injected (106 CFU/larva in the first study, whereas three different concentrations – 106 CFU/larva, 105 CFU/larva and 104 CFU/larva – were used in the second) and the parameters analysed (Mukherjee et al. monitored the % survival along 7 days, whereas Martinez et al. focused on LT50 at 24 h and % mortality – not specifying its progression over the infection period). Nonetheless, it was concluded in both studies that the virulence potential of L. monocytogenes is dose and strain dependent, so these different results could be explained by the use of different L. monocytogenes strains. Another factor that could externally influence on the observed results is the larvae’s diet, since no information was available on the rearing of the larvae used in the Martinez et al. research. Previous studies have shown the importance of the diet in the larvae development, health, hemolymph volume and hemocyte concentration, which subsequently affect the immune response of G. mellonella (Jorjão et al., 2018; Kwadha et al., 2017). It has also been published that the diet of worms has an important impact in microbiological studies (Banville et al., 2012; Jorjão et al., 2018). Hence, standardization of diets could reduce external biases on results allowing for interlaboratory comparisons.
To date, virulence evaluation of different L. monocytogenes CCs using G. mellonella has only been performed by Cardenas-Alvarez et al. (2019). This insect model was used to compare the pathogenic potential of CC1, CC6, CC7, CC9, CC14, CC37, and CC204 strains. Briefly, differences were observed between strains from different CCs, with strains from the putatively hypervirulent CCs, CC1, and CC14, causing a reduced average survival rate (33.2 and 29.1%, respectively). Oppositely, isolates from CC9, widely accepted as hypovirulent CC, presented the highest survival rate (53.5%). In addition, the remaining CCs (6, 7, 37 and 204) showed an intermediate range of survival rates from 40 to 50%. Another parameter evaluated was the LD50 value (median lethal dose) – calculated from the colonies counted on plates and the number of larvae killed per day – lower values were observed for CC14, meaning that fewer cells of the pathogen are needed to kill G. mellonella. Cytotoxicity was also evaluated by measuring the level of lactate dehydrogenase (LDH), which is a signal of cell damage after bacterial infection. CC14 strains caused significantly less cytotoxicity than other CCs (CC6 and CC7). A positive correlation was found between LD50 and cytotoxicity, therefore CC14, strains by having a reduced LD50, also caused less injuries to host cells, which is hypothesized to be a defence mechanism to escape the host immune system and successfully spread (Cardenas-Alvarez et al., 2019). Considering these results, G. mellonella as an infection model, besides the capacity to differentiate non-pathogenic from the pathogenic Listeria species, has the potential to distinguish between virulent and attenuated L. monocytogenes strains from different CCs, validating its ability to discriminate the virulence potential of L. monocytogenes.
The non-mammalian vertebrate Danio rerio, known as the zebrafish, is an in vivo model that has been gradually catching the attention of researchers for the study of infectious diseases as it meets the ideal features of vertebrate and mammalian models (Shan et al., 2015). As other models already described in this review, zebrafish is more easily applicable and economically and ethically acceptable than most mammalian models. Being a vertebrate, its morphological and genetic similarities with humans are more pronounced than with invertebrate (Pont and Blanc-Potard, 2021). In addition to the zebrafish’s large clutch dimension, ex-utero growth and small size, its transparency makes zebrafish a distinctive mode, allowing observation of the early stages of growth and enabling the real-time observation of bacterial infections (Pont and Blanc-Potard, 2021; Shan et al., 2015; van der Sar et al., 2004). Another interesting peculiarity is that the innate and the adaptive immune systems are temporally separated, where the primer acts singularly during early weeks while the latter is perceived just during the 4–6 weeks post-fertilization (Herbomel et al., 1999; Herbomel et al., 2001; Lam et al., 2004; Trede et al., 2004).
The use of this vertebrate model in the study of host-pathogen interactions began in 1999, when Philippe Herbomel et al., reported that primitive macrophages – which evolve during the embryo’s development and subsequently give rise to hematopoietic stem cells – develop in the zebrafish embryos at 22 h post-fertilization (Herbomel et al., 1999). Therefore, zebrafish have been used to explore host-pathogen interactions and provide new insights into the capacity of L. monocytogenes to cause disease in this in vivo model (Levraud et al., 2009; Shan et al., 2015; Zakrzewski et al., 2020). The different stages of development of zebrafish are used for research and have their advantages, but infection assays have only been performed in zebrafish’s embryos to study the association of clonal complexes with hyper-and hypovirulence of L. monocytogenes strains, thus, our review will merely focus on this developmental stage. Among these, Hurley et al. (2019) made use of L. monocytogenes strains collected from three meat and vegetable processing facilities over 4 years. Genome analysis on these isolates reported distinct virulence genotypes and grouped them into hypervirulent, hypovirulent and unknown virulence groups (Hurley et al., 2019). This classification was slightly different from that previously described by Maury et al. (2016), as the isolates commonly associated with clinical cases (strains from CC1, CC2 and CC6) were underrepresented among the isolates collected. Therefore, hypervirulent strains were selected based on the presence of additional virulence factors such as listeriolysin S from LIPI-3 or LIPI-4. Selected hypovirulent strains (CC121, CC9, CC31) harboured PMSC mutation in the inlA gene and some of them had a deletion on the actA gene, which is associated with a decrease in intracellular spread. Isolates with integral virulence factors or with minimal mutations in some genes were classified as having unknown virulence capacity (CC3). Zebrafish embryos infected with putatively hypervirulent strains presented only a 3% survival rate, followed by zebrafish embryos infected with isolates of unknown virulence (20% survival rate), while hypovirulent strains caused a higher survival rate of 53–83%, requiring 72 h post-infection to cause this decline. Using the zebrafish infection model, Hurley et al. (2019) were able to discriminate the different virulence phenotypes and confirm the previous virulence genotypes obtained by WGS (Hurley et al., 2019). Muchaamba et al. (2022) also performed infection assays using the zebrafish embryo model, comparing the virulence potential of L. monocytogenes strains by lineage, serotype, and clonal complex. When the strains were grouped by CC, the researchers observed virulence discrepancies by CC and strain-specific intra-clonal complex. Embryos infected with CCs that are generally considered hypervirulent showed higher mortality than isolates from CC9 or CC8. Within some CCs, such as CC1 and CC9, strain-dependent virulence variation was observed – three CC1 strains required more than 24 h post-infection to cause 100% mortality, and while two CC9 strains exhibited no virulence, the other three CC9 strains presented variable levels of virulence. The conclusion of the in vivo assays was that the virulence potential of this pathogen varies with genotype, serotype and strain (Muchaamba et al., 2022). Therefore, both studies confirmed the previous categorization of hypervirulent and hypovirulent L. monocytogenes CCs using the zebrafish embryo infection model. This underscores the model’s relevance as an in vivo tool for further elucidating the virulence phenotypes of L. monocytogenes strains.
The in vitro systems represent alternative processes to study bacterial virulence as they mimic the infectious mechanism, allowing, for example, screening of pathogen gene expression and how the deletion of some genes affects the behaviour of strains in physiological environments mimicking in vivo conditions. In vitro assays are based on the assumption that pathogens, such as L. monocytogenes, have the ability to infect hosts by attachment, invasion, multiplication and subsequent dissemination in either phagocytic or non-phagocytic cells through the production of virulence factors (Liu et al., 2007). Although these systems do not precisely replicate the full features of the host-pathogen interaction, as infectious agents may encounter unfavourable conditions and the host immune system, when compared to in vivo models they are less expensive, less time consuming and less ethically demanding, allowing large-scale experiments. Additionally, the ability to control experimental conditions allows to unravel favouring factors in disease. Therefore, their use is recommended for preliminary studies to find new virulence factors, after which in vivo models can be used on a limited scale to confirm the results (Lehr, 2002; McCoy et al., 2024; Chiang et al., 1999). For these reasons many different in vitro models have been developed. The standard in vitro system, that has been used for decades, is the 2D monolayer culture of immortalized human cells. More recently, in a bioengineering context, there has been an increase in the use of different systems based on in vitro and ex vivo models, such as organoids and 3D cell cultures, to improve the monolayer model (Taebnia et al., 2023). The choice of an appropriate in vitro model should focus on the definite biological issue, for example, cell lines are more adequate to study precise interaction processes of pathogens. The addition of unneeded complexity can be disadvantageous, shrouding relevant host-pathogen interactions (McCoy et al., 2024). Therefore, many tissue culture experiments to study adhesion, invasion, cell to cell spread in different cell lines, survival in macrophages, evaluation of cytotoxicity and pathogens activity upon different host environmental conditions (e.g., pH and temperature) have been reported to describe and determine novel virulence concepts of bacteria (Conte et al., 1994; Hasebe et al., 2017; Wagner et al., 2022). In L. monocytogenes, in vitro models have been used to investigate host-pathogen interactions at either an intestinal, cerebral or placental level. These models brought significant knowledge regarding the L. monocytogenes intracellular cycle, invasion at cell extrusion sites, the role of putative virulent genes in cell invasion, required internalins (InlA, InlB and InlP) to placental invasion, L. monocytogenes bacteriocins in intestinal commensals, pathogen’s routes to invade the brain and other related aspects (Banović et al., 2020; Cabanes et al., 2004; Cabanes et al., 2005; Lamond and Freitag, 2018; Pentecost et al., 2006; Rolhion et al., 2019).
In 1948 the first cell line based on subcutaneous mouse tissues was developed. Thenceforth, various mammalian cell lines have been developed and used as the primary in vitro model to investigate infectious diseases, since they mimic host defence mechanisms (Magdalena, 2017). In L. monocytogenes, the human colorectal adenocarcinoma cell line Caco-2 is one of the most popular cell models that replicate the intestinal barrier, along with HT-29, Henle-407, HeLa, and many other cell lines (Liu et al., 2007; Pizarro-Cerdá et al., 2012). Different cell lines used in listeriosis studies were represented in Table 3.
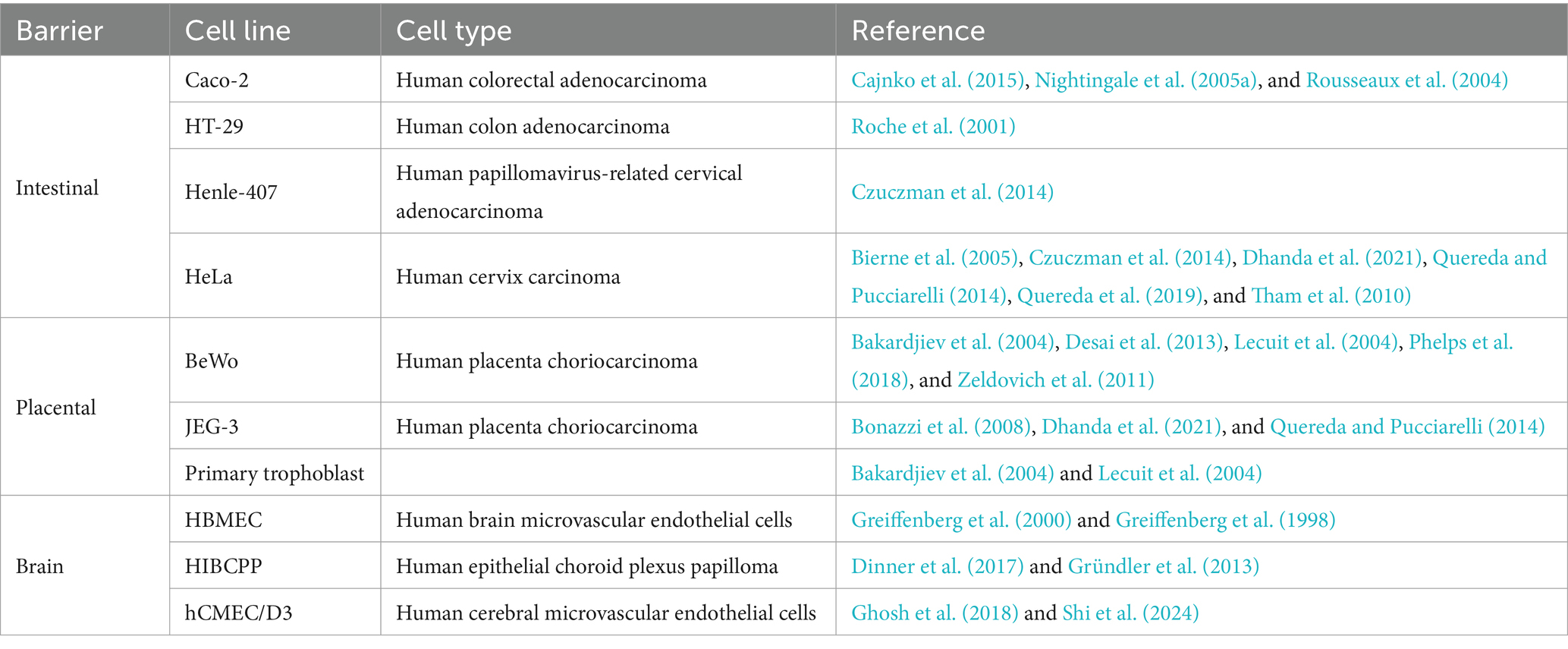
Table 3. Examples of cell lines used to study human listeriosis, representing the intestinal, placental and brain barriers.
The main limitation of cell models is their uniformity, not truly mimicking environment and morphology of epithelial tissues where a panoply of distinct cells can be found (Hidalgo, 1996; Pearce et al., 2018). One way to overcome this limitation is to co-culture different cell lines. However, to our knowledge, this strategy has not been commonly used to study the virulence potential of L. monocytogenes (Laparra and Sanz, 2009; Wikman-Larhed and Artursson, 1995). Another limitation of these cell models is their cancer origin, which makes it difficult to extrapolate the data since they may not reflect the actual physiological context. Additionally, the static conditions in which these monolayers are performed lead to very rapid bacteria overgrowth, thus compromising the duration of the culture and the search for new insights about the interaction between the host and its microbiome (Rodriguez, 2018; Taebnia et al., 2023).
Summing up, these in vitro cell models have been widely used to study the virulence potential of L. monocytogenes and have contributed to expand the current knowledge of the virulence mechanism of this bacterium. Consequently, this research has led to the development of strategies to control the dissemination of listeriosis. To date, very few studies have used these models to differentiate between strains from different CCs (Domínguez et al., 2023; Møretrø et al., 2024; Schiavano et al., 2022; Wagner et al., 2022). Schiavano et al. (2022) performed adhesion and invasion assays using the Caco-2 cell line in order to do a comparative analysis between human clinical isolates and food isolates. They observed that two out of three clinical strains (from CC1 and CC101) expressed a relatively high adhesion regarding to L. innocua. However, the invasion efficiency was not significantly higher than that of the non-pathogenic strain. On the other hand, the food isolates showed a variable adhesion capacity, with strains from CC7, CC121 and CC1 showing significantly higher values than L. innocua.
For invasiveness, strains from CC7 and CC1 displayed significant higher capacity than L. innocua. No correlation was found between adhesion and invasion for food-derived strains. Curiously, one clinical strain (566 strain) did not show high levels of both adhesion and invasion. However, this strain belongs to CC31, which has been reported to be isolated more frequently from food than from humans. CC31 is not considered to be a hypervirulent clonal complex, as corroborated by its low invasive ability. Cases of human listeriosis caused by strains of this CC may be justified by the compromised immune system of the host (Schiavano et al., 2022). In contrast, the clinical strain tested from CC1, considered to be a common hypervirulent CC, showed an unexpectedly reduced invasion although a high level of adhesion was detected. Regarding the food isolates, the two strains with higher invasiveness belong to CC1 and CC7, which are usually associated with human cases but have also been reported in food. CC1 is highly associated with dairy and cattle products while CC7 has already been described as an intermediate CC (Lüth et al., 2020a). As expected, strains from CC9 and CC121 showed a low invasion ability. As both hypo-and hypervirulent CCs with a reduced invasion capacity were found in clinical cases, we can conclude that the state of the host immune system is very important since it may facilitate the occurrence of listeriosis. In addition, hypervirulent CCs can be found in foods, emphasizing their threat for individuals (Schiavano et al., 2022).
Wagner and his colleagues used Caco-2 cells to evaluate the invasion capacity of strains from thirteen different CCs. Strains from three CCs (CC5, CC9, and CC14) were not able to invade these cells, with only CC14 strains encoding the complete functional inlA gene. Strains from CC403 and CC415 were the most invasive, while strains from CC3, CC8 and CC121 were significantly less invasive. Among strains of these CCs with attenuated invasion, CC14 strains comprised all major virulent factors but still showed inefficiency at invading Caco-2 cells. Therefore, invasion and intracellular dissemination assays in Caco-2 and HEPG2 cells, both with an epithelial-like morphology, were performed with two strains of this CC. In Caco-2 cells, both CC14 strains showed a significantly reduced invasion capacity when compared to EGDe. However, only one CC14 strain showed a significantly lower invasion in HEPG2 cells. Conversely, intracellular multiplication in Caco-2 cells was significantly increased for both CC14 strains, while only one CC14 strain showed a greater intracellular spread in HEPG2 cells compared to EGDe (Wagner et al., 2022). Therefore, the virulence result of CC14 may be confusing when compared with previous findings, such as those in section 4.1.2., where it was characterized as a hypervirulent CC (Cardenas-Alvarez et al., 2019). These differences could not only be related to methodology used in the G. mellonella infection assays, where L. monocytogenes was injected directly into the hemolymph, bypassing the intestinal barrier, and thus not utilising the inlA and inlB genes, but also two distinct models (in vitro and in vivo) were used, highlighting the fact that different complexity could bring variable outcomes. Additionally, in Wagner’s study although the inlA and inlB genes were present in the genetic profile of selected CC14 strains, their expression was significantly reduced compared to L. monocytogenes EGDe. This reduction in expression could possibly be explained by a point mutation in the promoter region (Wagner et al., 2022). These findings of Wagner et al. (2022), point up the drawback of using solely WGS data to define virulence potentials, since presence of a virulence gene does not necessarily lead to gene expression. Besides the different approaches used, these models showed some similarities regarding the high capacity of CC14 strains to spread within the selected models.
More recently, researchers investigated the 2019 outbreak in Andalusian, Spain, caused by a strain of CC388. Adhesion and invasion in vitro assays were performed on A549 cell line to compare the virulence potential of this outbreak strain with strains belonging to CC1, CC4 and a reference strain from CC2 (ATCC 19115). They observed a higher adhesion of the CC388 strain compared to CC1 and the reference strains, and lower adhesion compared to CC4 strain, although no significant differences were reported. A similar pattern was observed in invasion assays, with significant differences between strains of CC1, ATCC 19115 and CC388 versus CC4. It was therefore concluded that the CC388 strain had an equal or greater virulence potential compared to other strains from hypervirulent CCs (Domínguez et al., 2023).
Concluding, 2D monolayer models have significantly advanced L. monocytogenes research, providing insight into key virulence properties and host-pathogen interactions. However, variations in results between different models require careful analysis. Therefore, their use must be complemented by molecular biology approaches to elucidate any unexpected phenotypic differences found by in vitro methods.
Macrophages play a crucial role in the innate immune response, using their skilled phagocytic activity to fight infection. They recognise pathogens through pattern recognition receptors (PRRs) that bind to microbial-associated molecular patterns (MAMPs) (Kumar, 2020) such as DNA, RNA, lipopolysaccharides, and lipoproteins, thereby activating the host immune response (Fitzgerald and Kagan, 2020). This interaction triggers signalling pathways that culminate in the secretion of cytokines and the process of phagocytosis. Once engulfed by macrophages, pathogens are entrapped in acidic phagosomes where antimicrobial molecules can be found (Levin et al., 2016). However, certain microbes have evolved mechanisms to evade the host’s immune defences and proliferate intracellularly, rendering macrophages ineffective in protecting against such pathogens (Galli and Saleh, 2021).
Listeria monocytogenes is able to survive within macrophages, as demonstrated by Tilney and Portnoy in 1989, who elucidated its mechanism of infection within these immune cells (Tilney and Portnoy, 1989). Considering this, different macrophage cell lines have been used to simulate the host barriers following bacterial intestinal invasion (Liu et al., 2007). Besides the employment of epithelial-like cells (Caco-2 and HEPG2) to evaluate L. monocytogenes virulence potential, Wagner et al. (2022) also used the human macrophage-like THP-1 cell line for this purpose. As CC14 strains were unable to invade Caco-2 cells, despite the presence of all key virulence genes, two CC14 isolates were used to invade and multiply intracellularly within macrophage cells. Although no significant differences in invasion capacity were observed between two CC14 strains and EGDe, intracellular multiplication was significantly increased in THP1 for both CC14 strains (Wagner et al., 2022). In 2016, Dreyer et al. conducted a study to investigate putative L. monocytogenes strain-associated virulence in isolates from the farm environment and diseased animals, focusing on rhombencephalitis cases where ST1 (CC1) was overrepresented. The in vitro assays were conducted on a bovine macrophage cell line (BoMac) and it was observed that STs associated with encephalitic infections (ST1, ST4 (CC4) and ST412 (CC412 – lineage II)) were able to invade and replicate more efficiently than those from the farm environment. Additionally, none of the isolates presented truncated InlA, which is commonly associated with virulence attenuation. Thus, although ST412 isolates from lineage II accounted for only 7% of rhombencephalitis cases (which is not a statistically significant association between ST and clinical outcome), they presented an increased virulence potential, highlighting the fact that these clinical-associated characteristics are not exclusive to lineage I isolates, raising awareness of the potential risk other CCs’. In addition, the inlA gene is not the only biomarker for differential virulence (Dreyer et al., 2016).
Another study conducted in 2017 aimed to test the relevance of certain virulence genes (inlJ1, inlF and lls) in the hypervirulence capacity of CC1 strains. The L. monocytogenes CC1 parental strain and its respective deletion gene mutants were compared to the EGDe (CC9) strain in different cell culture models, including macrophages. The BoMac cell line was used to mimic the intracellular phagosome environment. Despite both strains infecting all cell models, the CC1 isolate exhibited higher invasiveness than EGDe in some cell lines. For example, invasion into BoMac cells was 2.2 times higher for the strain from the hypervirulent CC. Moreover, the CC1 strain showed a significantly greater number of infection foci in BoMac cells, indicating an enhanced capacity to spread intercellularly and corroborating its stronger internalization phenotype. However, the intracellular multiplication in all cell lines was not significantly different between the two strains. Listeria monocytogenes exhibits a cell-specific interaction, as evidenced by the differential infection capacity observed between these two strains in specific cell types, including macrophages (Rupp et al., 2017).
In a more recent study, macrophages were used for a different comparative analysis of CCs beyond those previously mentioned, using various cell models for investigating invasion capacity, intracellular spread, and multiplication associated with potential PMSC mutations in InlA. Chalenko et al. (2022) found that InlB could modify the invasion and proliferation capacity of L. monocytogenes within macrophages. Consequently, they proposed to investigate whether the interaction between InlB and cell receptors would affect the intracellular infection cycle of the pathogen in these immune cells. Interestingly, this study explored the phylogenetically determined diversity of InlB to understand its impact on pathogen-cell interactions. The first step was to investigate the effective interaction between different InlB isoforms found in lineage I and II of L. monocytogenes strains and their two target receptors, using three distinct receptor-binding domains of InlB (idInlB) – idInlBCC1, idInlBCC7 and idInlBCC9 – representing different virulence potentials. The study of the interaction of idInlB-human receptors (c-Met and gC1qR) was possible by the measurement of dissociation constants using Microscale Thermoforesis technology. The results showed that the interaction between idInlB variants and human receptors differs in terms of binding strength. idInlBcc1 showed stronger binding to C-Met receptor compared to idInlBcc7 and idInlBcc9, while idInlBcc9 presented weaker binding to the gC1qR receptor, with no significance differences observed between idInlBCC1 and idInlBCC7 variants. Based on the EGDeΔinlB strain (lacking the inlB gene), isogenic L. monocytogenes strains were also constructed, namely LmInlBCC1, LmInlBCC7 and LmInlBCC9. These strains harboured full-length internalin B isoforms that differed only in the idInlB domain. When human M1 macrophages were infected with these strains, different multiplication capacities were observed, with LmInlBCC1 showing a significantly higher proliferation capacity compared to the other. However, no significant differences were observed regarding the cell uptake. Altogether, these results suggest that phylogenetic differences in InlB affect the ability of L. monocytogenes to interfere with macrophage activity. In particular, it was suggested that InlBCC1 efficiently overcomes immune barriers of these cells, which is consistent with the high occurrence of CC1 observed in epidemiological data (Chalenko et al., 2023).
Despite the unquestionable use of 2D models in studying microbe-host interactions, these models are limited in mimicking real features, such as peristaltic movements, transitions between different intestinal cells, interactions with the intestinal microbiota, and their inability to be maintained for long periods (Taebnia et al., 2023). These gaps in accurately reproducing the function and structure of the human intestinal epithelium, limit the value of 2D models. Therefore, researchers have developed more comprehensive and complex models, that do not replace monolayer models, but rather complement them by incorporating physiological components or simulating infectious disease scenarios that are difficult to assess using simpler culture methods (Taebnia et al., 2023). In the 21st century, steam cells started to be cultivated to generate organoids, which have effectively bridged the gap between traditional 2D monolayer cultures and ex vivo models (Han et al., 2021; Sato et al., 2009; Taebnia et al., 2023).
In 2014, an important finding allowed the development of a reproducible method known as directed differentiation, which enables the specialization of leucine-rich repeat containing G protein-coupled receptor 5 (LGR5+) stem cells into various cell types, including goblet cells, enterocytes, stem cells, Paneth cells and enteroendocrine cells (Yin et al., 2014). These findings hold great promise for studying host-pathogen interactions and for investigating the responses and properties of specific cell types, such as the barrier role. Directed differentiation cannot be overstated in light of previous research showing that L. monocytogenes can surpass the intestinal barrier not only through enterocytes but also through M cells and goblet cells (Corr et al., 2006; Nikitas et al., 2011). Currently, organoids can be derived from cells of different species and consists of either differentiated cells, stem cells or a combination of thereof (Davies, 2018). The organoids have their apical side facing the lumen (central position), while basolateral crypt regions are directed to the outside (budding structure), mimicking the real intestinal epithelium. This reversed polarity is a challenge for microbiological research, as it is difficult to access to the lumen intestinal organoids (Huang et al., 2021). To overcome this obstacle, microinjection, mechanical dissociation techniques or “apical-out organoids” can be used (Co et al., 2019; Huang et al., 2021; Karve et al., 2017).
Many studies have used organoids to investigate host-pathogen interactions involving various microorganisms, including L. monocytogenes (Co et al., 2019; Huang et al., 2021; Kim et al., 2021; Roodsant et al., 2020; Zhou et al., 2022a,c). In the research of listeriosis, different types of intestinal (i.e., fetal human intestinal organoids, adult human intestinal basal-out and apical-out organoids, adult and young murine small intestinal organoids) and brain (i.e., organotypic brain slices) organoids have been used (Co et al., 2019; Guldimann et al., 2012; Hentschel et al., 2021; Huang et al., 2021; Kim et al., 2021). However, to our knowledge the use of placental organoids in the study of L. monocytogenes have not yet been reported (Yan et al., 2023).
Additionally, presently, the comparative analysis of different virulent L. monocytogenes strains using organoids has only been performed by Zhou and colleagues. However, the main objective of the authors was the study of protein changes in the host epithelium. Organoids were infected with two different strains – a “virulent strain” (serotype 1/2a) and a “low virulent strain” (serotype 4a) – and quantitative proteomic analysis of the infected mice organoids was performed (Zhou et al., 2022b). In general, it was shown that both strains were able to reduce the host’s energy metabolism, stimulate the host’s immune response and increase the expression of proteins related to adhesion and invasion, as expected. Although some differences were found between the two strains, while the virulent strain significantly activated the ferroptosis pathway – known as a cell death pathway – the attenuated strain exhibited a higher activation of the complement system, which has a crucial function in innate immune responses. Notably, both strains down-regulated nucleotide-binding oligomerization domain 2 (NOD2), a receptor in the NOD-like receptor signalling pathway, which is crucial for innate immune responses and can recognise pathogens through muramyl dipeptide (MDP) (Liu et al., 2023; Zhou et al., 2022b). This down regulation could potentially hamper the multiplication and protection of intestinal stem cells. So, it was expected that organoids infected with L. monocytogenes would show upregulation of NOD2. However, another study showed that germ-free mice expressed reduced levels of NOD2, which subsequently increased when gut commensals were added. This observation could potentially explain the reduced expression of NOD2 in organoids as they lack gut-associated bacteria (Petnicki-Ocwieja et al., 2009). Overall, it was concluded that the immune activity and biological functions were identical in the two different strains, although some differential expression of different proteins within the pathway were observe (Zhou et al., 2022b). Considering this, the use of organoids for L. monocytogenes studies have been reported. However, to the best of our knowledge, no article on CC-associated virulence has been published. Thus, further research on the use of organoids as infection models for this pathogen is needed to explore their viability for comparative analysis of differentially virulent strains.
The success of polymerase chain reaction (PCR) is largely due to its capacity to amplify minimal amounts of genetic material, to millions of copies in a very short time (Farrell, 2010; Mullis et al., 1986). Technical improvements have led to the quantification of gene expression through techniques such as quantitative reverse transcriptase PCR (qRT-PCR), which enables the detection and quantification of RNA products (Farrell, 2010; Zhu et al., 2020). Although qRT-PCR is considered the gold-standard for mRNA quantification, some drawbacks limit its use. Among these we highlight the additional reverse transcription (RT) step, which can introduce contamination and inhibitions, common issues in the quantification of the actual cDNA present in the sample. In addition, relative quantification requires well established reference genes that are not affected by experimental conditions, and inadequate optimization of primers and annealing temperatures can lead to non-specific sequence targeting. To address these limitations, other molecular approaches such as digital PCR (dPCR) have been developed, which allow absolute quantification without the need for reference genes and calibration curves and with a reduced probability of contamination, thus improving interlaboratory comparability (Baettig et al., 2023; Cao et al., 2020; Lei et al., 2021; Witte et al., 2016). The use of dPCR for gene expression analysis increasing, although its application is primarily limited to the detection and quantification of L. monocytogenes cells and biofilms (Chen et al., 2017; Grudlewska-Buda et al., 2020; Ricchi et al., 2017; Witte et al., 2016). Gene expression profiling techniques have greatly improved our understanding of how pathogens’ modulate gene transcription after interacting after interacting with the host environment, strategically recruiting their genome during the infection life cycle. Consequently, genes that exhibit differential expression during infections have captured the attention of researchers, providing insight into which virulence genes are essential for microbial pathogenicity (Shelburne and Musser, 2004).
To date, no studies using dPCR in virulence potential analysis among CCs have been reported. RT-qPCR technology is the only that has been used to study how L. monocytogenes strains from distinct CCs differentially express stress response genes in diverse contexts (da Silva et al., 2021; Guerreiro et al., 2022; Wambui et al., 2020; Wu et al., 2022). However, only a few research papers reporting the use of this technique to analyse differential gene expression of virulence genes associated with CCs have been published (Rupp et al., 2017; Wagner et al., 2022). Rupp et al. (2017), aimed to understand the hypervirulence of CC1, hypothesizing that it could be due to additional virulence features or genetic variation within previously studied virulence genes. They focused on three specific genes – alleles of inlJ1 and inlF and lls gene – two of which show some differences from lineage II strains, while the last is commonly found in lineage I CCs. Therefore, a strain from CC1 and EGDe strain (CC9) were used for intracellular (Caco-2 and BoMac cell lines) and extracellular (BHI broth) infection assays, where the gene expression of the CC1 strain was further analysed. These three virulence genes were expressed in the CC1 strain under both conditions, but the resulting PCR bands were less intense than the control genes (such as actA and rrs (16S)). However, no comparative analysis of gene expression between these two strains was explored. This study concluded that the CC1 strain was able to invade cells more effectively and exhibited an enhanced intracellular spread compared to EGDe. Nevertheless, the selected virulence genes were not found to correlate with these phenotypes, despite their strong association with the CC1 strains (Rupp et al., 2017).
Wagner and colleagues worked on the genotypic and phenotypic characterization of L. monocytogenes strains from the meat and salmon processing industry in Norway (Wagner et al., 2022). In vitro assays in Caco-2 cells and WGS analysis showed that CC14 strains lacked invasion capacity while carrying the full-length inlA gene. This interesting result led to a further analysis of the invasion and internalization capacity of CC14 strains using different cell lines (Caco-2, HEPG2 and THP1 cells) and the subsequent gene transcription analysis. EGDe was used as a reference. Under the conditions of Caco-2 cells infection, there were significant differences in gene expression between CC14 strains and EGDe – the expression of inlA and inlB genes was decreased in CC14 isolates, the other virulence genes (actA, hly and prfA) were not differentially expressed when compared to the EGDe strain. qRT-PCR was also used to study gene expression after the reconstruction of inlA in isolates from CC9 and CC121 that carried PMSCs mutations. Under Caco-2 cells growth conditions, the researchers observed no significant differences in the expression of inlA, inlB and prfA between the wildtype (WT) strains and their respective mutants. However, significant differences in the inlA expression were observed in both CC9 WT and one of its mutants compared to EGDe. The CC121 inlA reconstructed mutants showed no significant differences in gene expression when compared to EGDe, indicating successful gene reconstruction (Wagner et al., 2022). This study highlights the need for analysis of gene expression, since strains from CC14, although harbouring the full-length inlA gene, showed reduced expression of this virulence gene. We can conclude that the qRT-PCR technique is an essential tool to obtain further information about the virulence potential of L. monocytogenes strains.
Characterization of the virulence potential within L. monocytogenes strains is essential for effective risk assessment and to reduce the human and economic losses associated with listeriosis. Multi-locus sequence typing, combined with epidemiological data, has facilitated the identification of more or less virulent CCs, helping researchers to refine their understanding of the infection patterns of this pathogen. However, despite advances in CC characterization, the specific virulence markers that confer distinct disease-causing capacities remain poorly understood.
This review highlights the limited use of different infection models to study the virulence potential of L. monocytogenes CC. While some models, such as G. mellonella larvae, have not been extensively studied, results suggest variability in results depending on the model used. Some studies successfully distinguish hyper-and hypovirulent CCs, while others show inconsistent results, suggesting a strain-dependent characterization of infection risk within CCs. The strain-dependent nature of L. monocytogenes virulence potential, together with highly variable host susceptibility and evidence that virulence may be influenced by the food matrix, limits the usefulness of subtypes. Grouping them into Clonal Complexes (CCs) may mask relevant differences in their pathogenicity. Additionally, we highlight the fact that a standard infection model for studying the virulence potential of L. monocytogenes CCs has not yet been established. This lack of standardization difficult comparisons between dies, as variations in experimental conditions, infection doses and techniques may lead to different virulence outcomes. Despite these challenges, the use of different approaches to validate previous findings is recommended, and improvements to existing models, mainly 3D systems, are ongoing.
The presence or absence of virulence genes alone is not sufficient to determine pathogenicity, highlighting the multifactorial nature of listeriosis. Consequently, the achievement of a zero risk infection remains elusive and requires a comprehensive understanding of the host immunity and pathogen virulence machinery. Despite ongoing efforts, a reliable virulence biomarker capable of differentiating attenuated strains has yet to be identified, underscoring the importance of continued research and refinement of infection models to advance our understanding and control of listeriosis. To conclude, we would like to highlight a recent study that used artificial intelligence to predict the virulence potential of L. monocytogenes at the subspecies level using WGS datasets. The use of the pan-genome showed the best predictive results, pointing up the possible value of pan-genome related genes in virulence. Although the authors acknowledge some of the limitations of their models, such as the difficulty of generalizing data based on WGS from only three countries, which rises concerns about the application of these predictive machine learning models in other regions (Gmeiner et al., 2024), these findings show that innovative technologies are being explored for future risk assessment. Additionally, besides the limitations of the current use of subtype classification for characterizing L. monocytogenes virulence, their use cannot be ruled out as more is learned about risk variability. In the future, standardized risk assessment models may serve as valuable tools for the food industry in risk management upon L. monocytogenes contamination.
MS: Writing – review & editing, Writing – original draft, Methodology, Investigation, Conceptualization. RM: Writing – review & editing, Visualization, Supervision, Project administration, Funding acquisition. VF: Conceptualization, Writing – review & editing, Visualization, Supervision. PT: Writing – review & editing, Visualization, Validation, Supervision, Resources, Project administration, Funding acquisition.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Funds from FCT-Fundação para a Ciência e a Tecnologia through project GenoPhenoTraits4Persitence-Genomic and phenotypic traits contributing to persistence of Listeria monocytogenes in food processing environment (PTDC/BAA-AGR/4194/2021; https://doi.org/10.54499/PTDC/BAA-AGR/4194/2021).
We would also like to thank the scientific collaboration under the FCT project UIDB/50016/2020.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahlawat, S., and Sharma, K. K. (2022). Lepidopteran insects: emerging model organisms to study infection by enteropathogens. Folia Microbiol. 68, 181–196. doi: 10.1007/s12223-022-01014-y
Anju, V. T., Siddhardha, B., and Dyavaiah, M. (2020). Animal models to understand host–pathogen interactions. 1st Edn. Singapore: Springer.
Baettig, C. G., Zirngibl, M., Smith, K. F., Lear, G., and Tremblay, L. A. (2023). Comparison between droplet digital PCR and reverse transcription-quantitative PCR methods to measure ecotoxicology biomarkers. Mar. Pollut. Bull. 190:114829. doi: 10.1016/j.marpolbul.2023.114829
Bakardjiev, A. I., Stacy, B. A., Fisher, S. J., and Portnoy, D. A. (2004). Listeriosis in the pregnant guinea pig: a model of vertical transmission. Infect. Immun. 72, 489–497. doi: 10.1128/iai.72.1.489-497.2004
Balla, K. M., and Troemel, E. R. (2013). Caenorhabditis elegans as a model for intracellular pathogen infection. Cell. Microbiol. 15, 1313–1322. doi: 10.1111/cmi.12152
Banović, F., Schroten, H., and Schwerk, C. (2020). Potential Roles and Functions of Listerial Virulence Factors during Brain Entry. Toxins 12:297. doi: 10.3390/toxins12050297
Banville, N., Browne, N., and Kavanagh, K. (2012). Effect of nutrient deprivation on the susceptibility of Galleria mellonella larvae to infection. Virulence 3, 497–503. doi: 10.4161/viru.21972
Bergholz, T. M., Shah, M. K., Burall, L. S., Rakic-Martinez, M., and Datta, A. R. (2018). Genomic and phenotypic diversity of Listeria monocytogenes clonal complexes associated with human listeriosis. Appl. Microbiol. Biotechnol. 102, 3475–3485. doi: 10.1007/s00253-018-8852-5
Bierne, H., Mazmanian, S. K., Trost, M., Pucciarelli, M. G., Liu, G., Dehoux, P., et al. (2002). Inactivation of the srtA gene in Listeria monocytogenes inhibits anchoring of surface proteins and affects virulence. Mol. Microbiol. 43, 869–881. doi: 10.1046/j.1365-2958.2002.02798.x
Bierne, H., Miki, H., Innocenti, M., Scita, G., Gertler, F. B., Takenawa, T., et al. (2005). WASP-related proteins, Abi1 and Ena/VASP are required for Listeria invasion induced by the Met receptor. J. Cell Sci. 118, 1537–1547. doi: 10.1242/jcs.02285
Boman, H. G., and Hultmark, D. (1987). Cell-free immunity in insects. Ann. Rev. Microbiol. 41, 103–126. doi: 10.1146/annurev.mi.41.100187.000535
Bonazzi, M., Veiga, E., Pizarro-Cerdá, J., and Cossart, P. (2008). Successive post-translational modifications of E-cadherin are required for InlA-mediated internalization of Listeria monocytogenes. Cell. Microbiol. 10, 2208–2222. doi: 10.1111/j.1462-5822.2008.01200.x
Bou Ghanem, E. N., Jones, G. S., Myers-Morales, T., Patil, P. D., Hidayatullah, A. N., and D'Orazio, S. E. F. (2012). InlA Promotes Dissemination of Listeria monocytogenes to the Mesenteric Lymph Nodes during Food Borne Infection of Mice. PLoS Pathog. 8:e1003015. doi: 10.1371/journal.ppat.1003015
Cabal, A., Allerberger, F., Huhulescu, S., Kornschober, C., Springer, B., Schlagenhaufen, C., et al. (2019). Listeriosis outbreak likely due to contaminated liver pâté consumed in a tavern, Austria, December 2018. Eur. Secur. 24:1900274. doi: 10.2807/1560-7917.ES.2019.24.39.1900274
Cabanes, D., Dussurget, O., Dehoux, P., and Cossart, P. (2004). Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51, 1601–1614. doi: 10.1111/j.1365-2958.2003.03945.x
Cabanes, D., Sousa, S., Cebriá, A., Lecuit, M., García-del Portillo, F., and Cossart, P. (2005). Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. EMBO J. 24, 2827–2838. doi: 10.1038/sj.emboj.7600750
Cajnko, M. M., Marušić, M., Kisovec, M., Rojko, N., Benčina, M., Caserman, S., et al. (2015). Listeriolysin O Affects the Permeability of Caco-2 Monolayer in a Pore-Dependent and Ca2+-Independent Manner. PLoS One 10:e0130471. doi: 10.1371/journal.pone.0130471
Cao, Y., Yu, M., Dong, G., Chen, B., and Zhang, B. (2020). Digital PCR as an Emerging Tool for Monitoring of Microbial Biodegradation. Molecules 25:706. doi: 10.3390/molecules25030706
Cardenas-Alvarez, M. X., Townsend Ramsett, M. K., Malekmohammadi, S., and Bergholz, T. M. (2019). Evidence of hypervirulence in Listeria monocytogenes clonal complex 14. J. Med. Microbiol. 68, 1677–1685. doi: 10.1099/jmm.0.001076
Chalenko, Y. M., Abdulkadieva, M. M., Safarova, P. V., Kalinin, E. V., Slonova, D. A., and Yermolaeva, S. A. (2022). InlB protein secreted by Listeria monocytogenes controls the pathogen interaction with macrophages. Bull. RSMU 3, 5–10. doi: 10.24075/brsmu.2022.034
Chalenko, Y. M., Slonova, D. A., Kechko, O. I., Kalinin, E. V., Mitkevich, V. A., and Ermolaeva, S. A. (2023). Natural isoforms of Listeria monocytogenes virulence factor Inlb differ in c-Met binding efficiency and differently affect uptake and survival Listeria in macrophage. Int. J. Mol. Sci. 24, 1–15. doi: 10.3390/ijms24087256
Chambers, M. C., Lightfield, K. L., and Schneider, D. S. (2012a). How the fly balances its ability to combat different pathogens. PLoS Pathog. 8:e1002970. doi: 10.1371/journal.ppat.1002970
Chambers, M. C., Song, K. H., and Schneider, D. S. (2012b). Listeria monocytogenes infection causes metabolic shifts in Drosophila melanogaster. PLoS One 7:e50679. doi: 10.1371/journal.pone.0050679
Chen, Y., Gonzalez-Escalona, N., Hammack, T. S., Allard, M. W., Strain, E. A., and Brown, E. W. (2016). Core genome multilocus sequence typing for Identification of Globally Distributed Clonal Groups and Differentiation of Outbreak Strains of Listeria monocytogenes. Appl. Environ. Microbiol. 82, 6258–6272. doi: 10.1128/aem.01532-16
Chen, J. Q., Healey, S., Regan, P., Laksanalamai, P., and Hu, Z. L. (2017). PCR-based methodologies for detection and characterization of Listeria monocytogenes and Listeria ivanovii in foods and environmental sources. Food Sci. Human Wellness 6, 39–59. doi: 10.1016/j.fshw.2017.03.001
Chenal-Francisque, V., Lopez, J., Cantinelli, T., Caro, V., Tran, C., Leclercq, A., et al. (2011). Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 17, 1110–1112. doi: 10.3201/eid/1706.101778
Chiang, S. L., Mekalanos, J. J., and Holden, D. W. (1999). In vivo genetic analysis of bacterial virulence. Ann. Rev. Microbiol. 53, 129–154. doi: 10.1146/annurev.micro.53.1.129
Co, J. Y., Margalef-Català, M., Li, X., Mah, A. T., Kuo, C. J., Monack, D. M., et al. (2019). Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 26, 2509–2520.e4. doi: 10.1016/j.celrep.2019.01.108
Comandatore, F., Corbella, M., Andreoli, G., Scaltriti, E., Aguzzi, M., Gaiarsa, S., et al. (2017). Genomic characterization helps dissecting an outbreak of listeriosis in northern Italy. PLoS Curr. 9:ecurrents.outbreaks.633fd8994e9f06f31b3494567c7e504c. doi: 10.1371/currents.outbreaks.633fd8994e9f06f31b3494567c7e504c
Conte, M. P., Longhi, C., Petrone, G., Polidoro, M., Valenti, P., and Seganti, L. (1994). Listeria monocytogenes infection of Caco-2 cells: role of growth temperature. Res. Microbiol. 145, 677–682. doi: 10.1016/0923-2508(94)90039-6
Corr, S., Hill, C., and Gahan, C. G. (2006). An in vitro cell-culture model demonstrates internalin-and hemolysin-independent translocation of Listeria monocytogenes across M cells. Microb. Pathog. 41, 241–250. doi: 10.1016/j.micpath.2006.08.003
Cossart, P. (2011). Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 108, 19484–19491. doi: 10.1073/pnas.1112371108
Cotter, P. D., Draper, L. A., Lawton, E. M., Daly, K. M., Groeger, D. S., Casey, P. G., et al. (2008). Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 4, e1000144–e1000110. doi: 10.1371/journal.ppat.1000144
Czuczman, M. A., Fattouh, R., van Rijn, J. M., Canadien, V., Osborne, S., Muise, A. M., et al. (2014). Listeria monocytogenes exploits efferocytosis to promote cell-to-cell spread. Nature 509, 230–234. doi: 10.1038/nature13168
da Silva, D. A. L., de Melo Tavares, R., Camargo, A. C., Yamatogi, R. S., De Martinis, E. C. P., and Nero, L. A. (2021). Biofilm growth by Listeria monocytogenes on stainless steel and expression of biofilm-related genes under stressing conditions. World J. Microbiol. Biotechnol. 37:119. doi: 10.1007/s11274-021-03092-5
Davies, J. A. (2018). “Chapter 1- Organoids and mini-organs: Introduction, history, and potential” in Organoids and mini-organs. eds. J. A. Davies and M. L. Lawrence (Cambridge, Massachusetts: Academic Press), 3–23.
Desai, R. W., Amosu, M., and Smith, M. A. (2013). Invasion of Listeria monocytogenes strains in a human trophoblast cell line (BeWo). Birth defects research part a-clinical and molecular teratology, 97, 305
Dhanda, A. S., Vogl, A. W., Ness, F., Innocenti, M., and Guttman, J. A. (2021). mDia1 Assembles a Linear F-Actin Coat at Membrane Invaginations To Drive Listeria monocytogenes Cell-to-Cell Spreading. MBio 12, e02939–e02921. doi: 10.1128/mBio.02939-21
Dinh, H., Semenec, L., Kumar, S. S., Short, F. L., and Cain, A. K. (2021). Microbiology's next top model: Galleria in the molecular age. Pathog. Dis. 79:ftab006. doi: 10.1093/femspd/ftab006
Dinner, S., Kaltschmidt, J., Stump-Guthier, C., Hetjens, S., Ishikawa, H., Tenenbaum, T., et al. (2017). Mitogen-activated protein kinases are required for effective infection of human choroid plexus epithelial cells by Listeria monocytogenes. Microbes Infect. 19, 18–33. doi: 10.1016/j.micinf.2016.09.003
Disson, O., Grayo, S., Huillet, E., Nikitas, G., Langa-Vives, F., Dussurget, O., et al. (2008). Conjugated action of two species-specific invasion proteins for fetoplacental listeriosis. Nature 455, 1114–1118. doi: 10.1038/nature07303
Disson, O., Nikitas, G., Grayo, S., Dussurget, O., Cossart, P., and Lecuit, M. (2009). Modeling human listeriosis in natural and genetically engineered animals. Nat. Protoc. 4, 799–810. doi: 10.1038/nprot.2009.66
Domínguez, A. V., Ledesma, M. C., Domínguez, C. I., Cisneros, J. M., Lepe, J. A., and Smani, Y. (2023). In vitro and in vivo virulence study of Listeria monocytogenes isolated from the Andalusian outbreak in 2019. Trop. Med. Infect. Dis. 8:58. doi: 10.3390/tropicalmed8010058
D'Orazio, S. E. (2014). Animal models for oral transmission of Listeria monocytogenes. Front. Cell. Infect. Microbiol. 4:15. doi: 10.3389/fcimb.2014.00015
Doumith, M., Buchrieser, C., Glaser, P., Jacquet, C., and Martin, P. (2004). Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42, 3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004
Dreyer, M., Aguilar-Bultet, L., Rupp, S., Guldimann, C., Stephan, R., Schock, A., et al. (2016). Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 6:36419. doi: 10.1038/srep36419
Drolia, R., Bryant, D. B., Tenguria, S., Jules-Culver, Z. A., Thind, J., Amelunke, B., et al. (2024). Listeria adhesion protein orchestrates caveolae-mediated apical junctional remodeling of epithelial barrier for Listeria monocytogenes translocation. MBio 15:e0282123. doi: 10.1128/mbio.02821-23
Drolia, R., Tenguria, S., Durkes, A. C., Turner, J. R., and Bhunia, A. K. (2018). Listeria Adhesion Protein Induces Intestinal Epithelial Barrier Dysfunction for Bacterial Translocation. Cell Host Microbe 23, 470–484.e7. doi: 10.1016/j.chom.2018.03.004
Eallonardo, S. J., and Freitag, N. E. (2024). Crossing the Barrier: A Comparative Study of Listeria monocytogenes and Treponema pallidum in Placental Invasion. Cells 13:88. doi: 10.3390/cells13010088
ECDC and EFSA (2019). Multi-country outbreak of Listeria monocytogenes clonal complex 8 infections linked to consumption of cold-smoked fish products. EFSA Support. Publ. 16:1665E. doi: 10.2903/sp.efsa.2019.EN-1665
EFSA and ECDC (2018). Multi-country outbreak of Listeria monocytogenes serogroup IVb, multi-locus sequence type 6, infections linked to frozen corn and possibly to other frozen vegetables – first update. EFSA Support. Publ. 15:1448E. doi: 10.2903/sp.efsa.2018.EN-1448
EFSA and ECDC (2023). The European Union One Health 2022 Zoonoses Report. EFSA J. 21:e8442. doi: 10.2903/j.efsa.2023.8442
EFSAKoutsoumanis, K., Alvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., Chemaly, M., et al. (2020). The public health risk posed by Listeria monocytogenes in frozen fruit and vegetables including herbs, blanched during processing. EFSA J. 18:e06092. doi: 10.2903/j.efsa.2020.6092
Eng, V. V., and Pearson, J. S. (2021). In vivo studies on Citrobacter rodentium and host cell death pathways. Curr. Opin. Microbiol. 64, 60–67. doi: 10.1016/j.mib.2021.09.005
European Parliament Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes, (2010). Available at: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX:32010L0063 (Accessed August 20, 2024).
Fagerlund, A., Idland, L., Heir, E., Møretrø, T., Aspholm, M., Lindbäck, T., et al. (2022). Whole-genome sequencing analysis of Listeria monocytogenes from rural, urban, and farm environments in Norway: genetic diversity, persistence, and relation to clinical and food isolates. Appl. Environ. Microbiol. 88, e0213621–e0202121. doi: 10.1128/aem.02136-21
FAO and WHO. (2022). Listeria monocytogenes in ready-to-eat (RTE) foods: attribution, characterization and monitoring – Meeting report (Microbiological risk assessment), Issue. Rome, Italy: FAO and WHO.
Farrell, R. E. (2010). “Chapter 18- RT-PCR A Science and an Art Form” in RNA methodologies. 4th ed (Cambridge, Massachusetts: Academic Press), 385–448.
Favaro, M., Sarmati, L., Sancesario, G., and Fontana, C. (2014). First case of Listeria innocua meningitis in a patient on steroids and eternecept. JMM Case Rep. 1:e003103. doi: 10.1099/jmmcr.0.003103
Félix, B., Sevellec, Y., Palma, F., Douarre, P. E., Felten, A., Radomski, N., et al. (2022). A European-wide dataset to uncover adaptive traits of Listeria monocytogenes to diverse ecological niches. Sci. Data 9, 1–12. doi: 10.1038/s41597-022-01278-6
Fernández-Martínez, N. F., Ruiz-Montero, R., Briones, E., Baños, E., García San Miguel Rodríguez-Alarcón, L., Chaves, J. A., et al. (2022). Listeriosis outbreak caused by contaminated stuffed pork, Andalusia, Spain, July to October 2019. Eur. Secur. 27:2200279. doi: 10.2807/1560-7917.ES.2022.27.43.2200279
Ferreira, V., Magalhães, R., Almeida, G., Cabanes, D., Fritzenwanker, M., Chakraborty, T., et al. (2018). Genome sequence of Listeria monocytogenes 2542, a serotype 4b strain from a cheese-related outbreak in Portugal. Genome Announc. 6:e00540-18. doi: 10.1128/genomeA.00540-18
Fischer, M. A., Thürmer, A., Flieger, A., and Halbedel, S. (2021). Complete genome sequences of three clinical Listeria monocytogenes sequence type 8 strains from recent german listeriosis outbreaks. Microbiol. Resour. Announc. 10:e00303-21. doi: 10.1128/MRA.00303-21
Fitzgerald, K. A., and Kagan, J. C. (2020). Toll-like receptors and the control of immunity. Cell 180, 1044–1066. doi: 10.1016/j.cell.2020.02.041
Fretz, R., Pichler, J., Sagel, U., Much, P., Ruppitsch, W., Pietzka, A. T., et al. (2010a). Update: multinational listeriosis outbreak due to “Quargel”, a sour milk curd cheese, caused by two different L. monocytogenes serotype 1/2a strains, 2009-2010. Eur. Secur. 15:19543. doi: 10.2807/ese.15.16.19543-en
Fretz, R., Sagel, U., Ruppitsch, W., Pietzka, A. T., Stöger, A., Huhulescu, S., et al. (2010b). Listeriosis outbreak caused by acid curd cheese “Quargel”, Austria and Germany 2009. Eur. Secur. 15:19477. doi: 10.2807/ese.15.05.19477-en
Fritsch, L., Guillier, L., and Augustin, J.-C. (2018). Next generation quantitative microbiological risk assessment: Refinement of the cold smoked salmon-related listeriosis risk model by integrating genomic data. Microb. Risk Anal. 10, 20–27. doi: 10.1016/j.mran.2018.06.003
Galli, G., and Saleh, M. (2021). Immunometabolism of macrophages in bacterial infections. Front. Cell. Infect. Microbiol. 10:607650. doi: 10.3389/fcimb.2020.607650
Gerner-Smidt, P., Besser, J., Concepción-Acevedo, J., Folster, J. P., Huffman, J., Joseph, L. A., et al. (2019). Whole genome sequencing: bridging one-health surveillance of foodborne diseases. Front. Public Health 7:172. doi: 10.3389/fpubh.2019.00172
Ghosh, P., Halvorsen, E. M., Ammendolia, D. A., Mor-Vaknin, N., O’Riordan, M. X. D., Brumell, J. H., et al. (2018). Invasion of the Brain by Listeria monocytogenes Is Mediated by InlF and Host Cell Vimentin. MBio 9:e00160-18. doi: 10.1128/mBio.00160-18
Gillesberg, S., Ethelberg, S., Björkman, J. T., Jensen, T., Sørensen, G., Kvistholm Jensen, A., et al. (2016). Two listeria outbreaks caused by smoked fish consumption—using whole-genome sequencing for outbreak investigations. Clin. Microbiol. Infect. 22, 620–624. doi: 10.1016/j.cmi.2016.04.017
Gmeiner, A., Njage, P. M. K., Hansen, L. T., Aarestrup, F. M., and Leekitcharoenphon, P. (2024). Predicting Listeria monocytogenes virulence potential using whole genome sequencing and machine learning. Int. J. Food Microbiol. 410:110491. doi: 10.1016/j.ijfoodmicro.2023.110491
Graves, L. M., and Swaminathan, B. (2001). PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 65, 55–62. doi: 10.1016/S0168-1605(00)00501-8
Gray, M. J., Zadoks, R. N., Fortes, E. D., Dogan, B., Cai, S., Chen, Y., et al. (2004). Listeria monocytogenes isolates from foods and humans form distinct but overlapping populations. Appl. Environ. Microbiol. 70, 5833–5841. doi: 10.1128/AEM.70.10.5833-5841.2004
Greiffenberg, L., Goebel, W., Kim, K. S., Daniels, J., and Kuhn, M. (2000). Interaction of Listeria monocytogenes with Human Brain Microvascular Endothelial Cells: an Electron Microscopic Study. Infect. Immun. 68, 3275–3279. doi: 10.1128/iai.68.6.3275-3279.2000
Greiffenberg, L., Goebel, W., Kim, K. S., Weiglein, I., Bubert, A., Engelbrecht, F., et al. (1998). Interaction of Listeria monocytogenes with Human Brain Microvascular Endothelial Cells: InlB-Dependent Invasion, Long-Term Intracellular Growth, and Spread from Macrophages to Endothelial Cells. Infect. Immun. 66, 5260–5267. doi: 10.1128/iai.66.11.5260-5267.1998
Grudlewska-Buda, K., Skowron, K., and Gospodarek-Komkowska, E. (2020). Comparison of the intensity of biofilm formation by Listeria monocytogenes using classical culture-based method and digital droplet PCR. AMB Express 10:75. doi: 10.1186/s13568-020-01007-5
Gründler, T., Quednau, N., Stump, C., Orian-Rousseau, V., Ishikawa, H., Wolburg, H., et al. (2013). The surface proteins InlA and InlB are interdependently required for polar basolateral invasion by Listeria monocytogenes in a human model of the blood–cerebrospinal fluid barrier. Microbes Infect. 15, 291–301. doi: 10.1016/j.micinf.2012.12.005
Guerreiro, D. N., Wu, J., McDermott, E., Garmyn, D., Dockery, P., Boyd, A., et al. (2022). In vitro evolution of Listeria monocytogenes reveals selective pressure for loss of SigB and AgrA function at different incubation temperatures. Appl. Environ. Microbiol. 88, e0033022–e0000322. doi: 10.1128/aem.00330-22
Guldimann, C., Lejeune, B., Hofer, S., Leib, S. L., Frey, J., Zurbriggen, A., et al. (2012). Ruminant organotypic brain-slice cultures as a model for the investigation of CNS listeriosis. Int. J. Exp. Pathol. 93, 259–268. doi: 10.1111/j.1365-2613.2012.00821.x
Halbedel, S., Sperle, I., Lachmann, R., Kleta, S., Fischer, M. A., Wamp, S., et al. (2023). Large multicountry outbreak of invasive listeriosis by a Listeria monocytogenes ST394 clone linked to smoked rainbow trout, 2020 to 2021. Microbiol. Spectr. 11:e0352022. doi: 10.1128/spectrum.03520-22
Halbedel, S., Wilking, H., Holzer, A., Kleta, S., Fischer, M. A., Lüth, S., et al. (2020). Large nationwide outbreak of invasive listeriosis associated with blood sausage, Germany, 2018-2019. Emerg. Infect. Dis. 26, 1456–1464. doi: 10.3201/eid2607.200225
Han, X., Mslati, M. A., Davies, E., Chen, Y., Allaire, J. M., and Vallance, B. A. (2021). Creating a more perfect union: Modeling intestinal bacteria-epithelial interactions using organoids. Cell. Mol. Gastroenterol. Hepatol. 12, 769–782. doi: 10.1016/j.jcmgh.2021.04.010
Harter, E., Lassnig, C., Wagner, E. M., Zaiser, A., Wagner, M., and Rychli, K. (2019). The novel internalins InlP1 and InlP4 and the internalin-like protein InlP3 enhance the pathogenicity of Listeria monocytogenes. Front. Microbiol. 10:1644. doi: 10.3389/fmicb.2019.01644
Hasebe, R., Nakao, R., Ohnuma, A., Yamasaki, T., Sawa, H., Takai, S., et al. (2017). Listeria monocytogenes serotype 4b strains replicate in monocytes/macrophages more than the other serotypes. J. Vet. Med. Sci. 79, 962–969. doi: 10.1292/jvms.16-0575
Hentschel, V., Arnold, F., Seufferlein, T., Azoitei, N., Kleger, A., and Müller, M. (2021). Enteropathogenic Infections: Organoids Go Bacterial. Stem Cells Int. 2021, 8847804–8847814. doi: 10.1155/2021/8847804
Herbomel, P., Thisse, B., and Thisse, C. (1999). Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126, 3735–3745. doi: 10.1242/dev.126.17.3735
Herbomel, P., Thisse, B., and Thisse, C. (2001). Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev. Biol. 238, 274–288. doi: 10.1006/dbio.2001.0393
Hidalgo, I. J. (1996). Cultured intestinal epithelial cell models. Pharm. Biotechnol. 8, 35–50. doi: 10.1007/978-1-4899-1863-5_3
Hoelzer, K., Pouillot, R., and Dennis, S. (2012). Animal models of listeriosis: a comparative review of the current state of the art and lessons learned. Vet. Res. 43:18. doi: 10.1186/1297-9716-43-18
Hotson, A. G., and Schneider, D. S. (2015). Drosophila melanogaster Natural Variation Affects Growth Dynamics of Infecting Listeria monocytogenes. G3 (Bethesda) 5, 2593–2600. doi: 10.1534/g3.115.022558
Huang, J., Zhou, C., Zhou, G., Li, H., and Ye, K. (2021). Effect of Listeria monocytogenes on intestinal stem cells in the co-culture model of small intestinal organoids. Microb. Pathog. 153:104776. doi: 10.1016/j.micpath.2021.104776
Hurley, D., Luque-Sastre, L., Parker, C. T., Huynh, S., Eshwar, A. K., Nguyen, S. V., et al. (2019). Whole-genome sequencing-based characterization of 100 Listeria monocytogenes isolates collected from food processing environments over a four-year period. mSphere 4, e00252–e00219. doi: 10.1128/msphere.00252-19
Jacquet, C., Doumith, M., Gordon, J. I., Martin, P. M. V., Cossart, P., and Lecuit, M. (2004). A Molecular Marker for Evaluating the Pathogenic Potential of Foodborne Listeria monocytogenes. J. Infect. Dis. 189, 2094–2100. doi: 10.1086/420853
Jeffers, G. T., Bruce, J. L., McDonough, P. L., Scarlett, J., Boor, K. J., and Wiedmann, M. (2001). Comparative genetic characterization of Listeria monocytogenes isolates from human and animal listeriosis cases. Microbiology 147, 1095–1104. doi: 10.1099/00221287-147-5-1095
Jensen, A. K., Björkman, J. T., Ethelberg, S., Kiil, K., Kemp, M., and Nielsen, E. M. (2016). Molecular typing and epidemiology of human listeriosis cases, Denmark, 2002-2012. Emerg. Infect. Dis. 22, 625–633. doi: 10.3201/eid2204.150998
Jensen, R. L., Pedersen, K. S., Loeschcke, V., Ingmer, H., and Leisner, J. J. (2007). Limitations in the use of Drosophila melanogaster as a model host for gram-positive bacterial infection. Lett. Appl. Microbiol. 44, 218–223. doi: 10.1111/j.1472-765X.2006.02040.x
Jorjão, A. L., Oliveira, L. D., Scorzoni, L., Figueiredo-Godoi, L. M. A., Cristina, A. P. M., Jorge, A. O. C., et al. (2018). From moths to caterpillars: Ideal conditions for Galleria mellonella rearing for in vivo microbiological studies. Virulence 9, 383–389. doi: 10.1080/21505594.2017.1397871
Kaito, C., Murakami, K., Imai, L., and Furuta, K. (2020). Animal infection models using non-mammals. Microbiol. Immunol. 64, 585–592. doi: 10.1111/1348-0421.12834
Kammoun, H., Kim, M., Hafner, L., Gaillard, J., Disson, O., and Lecuit, M. (2022). Listeriosis, a model infection to study host-pathogen interactions in vivo. Curr. Opin. Microbiol. 66, 11–20. doi: 10.1016/j.mib.2021.11.015
Karve, S. S., Pradhan, S., Ward, D. V., and Weiss, A. A. (2017). Intestinal organoids model human responses to infection by commensal and Shiga toxin producing Escherichia coli. PLoS One 12, e0178966–e0178920. doi: 10.1371/journal.pone.0178966
Kavanagh, K., and Reeves, E. P. (2004). Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28, 101–112. doi: 10.1016/j.femsre.2003.09.002
Kavanagh, K., and Sheehan, G. (2018). The use of Galleria mellonella larvae to identify novel antimicrobial agents against fungal species of medical interest. J. Fungi 4:113. doi: 10.3390/jof4030113
Kemp, M. W., and Massey, R. C. (2007). The use of insect models to study human pathogens. Drug Discov. Today Dis. Model. 4, 105–110. doi: 10.1016/j.ddmod.2007.06.007
Kern, T., Kutzner, E., Eisenreich, W., and Fuchs, T. M. (2016). Pathogen–nematode interaction: Nitrogen supply of Listeria monocytogenes during growth in Caenorhabditis elegans. Environ. Microbiol. Rep. 8, 20–29. doi: 10.1111/1758-2229.12344
Khan, A., Waqar, K., Shafique, A., Irfan, R., and Gul, A. (2018). “Chapter 18- In vitro and in vivo animal models: the engineering towards understanding human diseases and therapeutic interventions” in Omics technologies and bio-engineering (pp. 431–448). eds. D. Barh and V. Azevedo (Cambridge, Massachusetts: Academic Press).
Khelef, N., Lecuit, M., Bierne, H., and Cossart, P. (2006). Species specificity of the Listeria monocytogenes InlB protein. Cell. Microbiol. 8, 457–470. doi: 10.1111/j.1462-5822.2005.00634.x
Kiani, A. K., Pheby, D., Henehan, G., Brown, R., Sieving, P., Sykora, P., et al. (2022). Ethical considerations regarding animal experimentation. J. Prev. Med. Hyg. 63, e255–e266. doi: 10.15167/2421-4248/jpmh2022.63.2S3.2768
Kim, M., Fevre, C., Lavina, M., Disson, O., and Lecuit, M. (2021). Live imaging reveals Listeria hijacking of E-cadherin recycling as it crosses the intestinal barrier. Curr. Biol. 31, 1037–1047.e4. doi: 10.1016/j.cub.2020.11.041
Kleta, S., Hammerl, J. A., Dieckmann, R., Malorny, B., Borowiak, M., Halbedel, S., et al. (2017). Molecular tracing to find source of protracted invasive listeriosis outbreak, southern Germany, 2012-2016. Emerg. Infect. Dis. 23, 1680–1683. doi: 10.3201/eid2310.161623
Koopmans, M. M., Brouwer, M. C., Vázquez-Boland, J. A., and Beek, D. V. D. (2023). Human Listeriosis. Clin. Microbiol. Rev. 36, e0006019–e0000019. doi: 10.1128/cmr.00060-19
Kumar, V. (2020). Phagocytosis: phenotypically simple yet a mechanistically complex process. Int. Rev. Immunol. 39, 118–150. doi: 10.1080/08830185.2020.1732958
Kvistholm, J., Nielsen, E., Björkman, J., Jensen, T., Müller, L., Persson, S., et al. (2016). Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin. Infect. Dis. 63, 64–70. doi: 10.1093/cid/ciw192
Kwadha, C. A., Ong'amo, G. O., Ndegwa, P. N., Raina, S. K., and Fombong, A. T. (2017). The biology and control of the greater wax moth, Galleria mellonella. Insects 8:61. doi: 10.3390/insects8020061
Lachmann, R., Halbedel, S., Adler, M., Becker, N., Allerberger, F., Holzer, A., et al. (2021). Nationwide outbreak of invasive listeriosis associated with consumption of meat products in health care facilities, Germany, 2014–2019. Clin. Microbiol. Infect. 27, 1035.e1–1035.e5. doi: 10.1016/j.cmi.2020.09.020
Lachtara, B., Wieczorek, K., and Osek, J. (2022). Genetic diversity and relationships of Listeria monocytogenes serogroup IIa isolated in Poland. Microorganisms 10:532. doi: 10.3390/microorganisms10030532
Lam, S. H., Chua, H. L., Gong, Z., Lam, T. J., and Sin, Y. M. (2004). Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev. Comp. Immunol. 28, 9–28. doi: 10.1016/S0145-305X(03)00103-4
Lamond, N. M., and Freitag, N. E. (2018). Vertical Transmission of Listeria monocytogenes: Probing the Balance between Protection from Pathogens and Fetal Tolerance. Pathogens 7:52. doi: 10.3390/pathogens7020052
Lange, A., Beier, S., Huson, D. H., Parusel, R., Iglauer, F., and Frick, J.-S. (2018). Genome sequence of Galleria mellonella (greater wax moth). Genome Announc. 6, e01220–e01217. doi: 10.1128/genomeA.01220-17
Laparra, J. M., and Sanz, Y. (2009). Comparison of in vitro models to study bacterial adhesion to the intestinal epithelium. Lett. Appl. Microbiol. 49, 695–701. doi: 10.1111/j.1472-765X.2009.02729.x
Lecuit, M. (2005). Understanding how Listeria monocytogenes targets and crosses host barriers. Clin. Microbiol. Infect. 11, 430–436. doi: 10.1111/j.1469-0691.2005.01146.x
Lecuit, M. (2007). Human listeriosis and animal models. Microbes Infect. 9, 1216–1225. doi: 10.1016/j.micinf.2007.05.009
Lecuit, M., Dramsi, S., Gottardi, C., Fedor-Chaiken, M., Gumbiner, B., and Cossart, P. (1999). A single amino acid in E-cadherin responsible for host specificity towards the human pathogen Listeria monocytogenes. EMBO J. 18, 3956–3963. doi: 10.1093/emboj/18.14.3956
Lecuit, M., Nelson, D. M., Smith, S. D., Khun, H., Huerre, M., Vacher-Lavenu, M.-C., et al. (2004). Targeting and crossing of the human maternofetal barrier by Listeria monocytogenes: Role of internalin interaction with trophoblast E-cadherin. Proc. Natl. Acad. Sci. 101, 6152–6157. doi: 10.1073/pnas.0401434101
Lecuit, M., Vandormael-Pournin, S., Lefort, J., Huerre, M., Gounon, P., Dupuy, C., et al. (2001). A transgenic model for listeriosis: role of internalin in crossing the intestinal barrier. Science 292, 1722–1725. doi: 10.1126/science.1059852
Lehr, C.-M. (ed.). (2002). “In vitro test systems for drug absorption and delivery” in Cell culture models of biological barriers. 1st ed. (London: CRC Press).
Lei, S., Chen, S., and Zhong, Q. (2021). Digital PCR for accurate quantification of pathogens: Principles, applications, challenges and future prospects. Int. J. Biol. Macromol. 184, 750–759. doi: 10.1016/j.ijbiomac.2021.06.132
Levin, R., Grinstein, S., and Canton, J. (2016). The life cycle of phagosomes: formation, maturation, and resolution. Immunol. Rev. 273, 156–179. doi: 10.1111/imr.12439
Levraud, J.-P., Disson, O., Kissa, K., Bonne, I., Cossart, P., Herbomel, P., et al. (2009). Real-time observation of Listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect. Immun. 77, 3651–3660. doi: 10.1128/iai.00408-09
Li, Y., Gan, Z., Zhou, X., and Chen, Z. (2022). Accurate classification of Listeria species by MALDI-TOF mass spectrometry incorporating denoising autoencoder and machine learning. J. Microbiol. Methods 192:106378. doi: 10.1016/j.mimet.2021.106378
Liu, D. (2006). Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J. Med. Microbiol. 55, 645–659. doi: 10.1099/jmm.0.46495-0
Liu, D., Lawrence, M. L., Ainsworth, A. J., and Austin, F. W. (2007). Toward an improved laboratory definition of Listeria monocytogenes virulence. Int. J. Food Microbiol. 118, 101–115. doi: 10.1016/j.ijfoodmicro.2007.07.045
Liu, B., Zhu, X., Zhang, N., Zhang, H., Li, H., Qi, Y., et al. (2023). Direct ferrous sulfate exposure facilitates the VBNC state formation rather than ferroptosis in Listeria monocytogenes. Microbiol. Res. 269:127304. doi: 10.1016/j.micres.2023.127304
Luque-Sastre, L., Arroyo, C., Fox, E. M., McMahon, B. J., Bai, L., Li, F., et al. (2018). Antimicrobial Resistance in Listeria Species. Microbiol. Spectr. 6, 10–1128. doi: 10.1128/microbiolspec.ARBA-0031-2017
Lüth, S., Halbedel, S., Rosner, B., Wilking, H., Holzer, A., Roedel, A., et al. (2020). Backtracking and forward checking of human listeriosis clusters identified a multiclonal outbreak linked to Listeria monocytogenes in meat products of a single producer. Emerg. Microbes. Infect. 9, 1600–1608. doi: 10.1080/22221751.2020.1784044
Mäesaar, M., Mamede, R., Elias, T., and Roasto, M. (2021). Retrospective use of whole-genome sequencing expands the multicountry outbreak cluster of Listeria monocytogenes ST1247. Int. J. Genom. 2021:6636138. doi: 10.1155/2021/6636138
Mäesaar, M., and Roasto, M. (2020). Draft genome sequence of a multicountry outbreak-related Listeria monocytogenes sequence type 1247 Strain, VLTRLM2013. Microbiol. Resour. Announc. 9:e00698-20. doi: 10.1128/mra.00698-20
Magalhaes, R., Almeida, G., Ferreira, V., Santos, I., Silva, J., Mendes, M. M., et al. (2015). Cheese-related listeriosis outbreak, Portugal, March 2009 to February 2012. Euro Surveill. 20:21104. doi: 10.2807/1560-7917.es2015.20.17.21104
Magdalena, J.-S. (2017). “History of cell culture” in New insights into cell culture technology. ed. G. S. J. Thatha (Rijeka, Croatia: Intech Open), Ch. 1.
Maiden, M. C. J., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Mansfield, B. E., Dionne, M. S., Schneider, D. S., and Freitag, N. E. (2003). Exploration of host–pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5, 901–911. doi: 10.1046/j.1462-5822.2003.00329.x
Martinez, M. R., Wiedmann, M., Ferguson, M., and Datta, A. R. (2017). Assessment of Listeria monocytogenes virulence in the Galleria mellonella insect larvae model. PLoS One 12, e0184557–e0184517. doi: 10.1371/journal.pone.0184557
Maudet, C., Kheloufi, M., Levallois, S., Gaillard, J., Huang, L., Gaultier, C., et al. (2022). Bacterial inhibition of Fas-mediated killing promotes neuroinvasion and persistence. Nature 603, 900–906. doi: 10.1038/s41586-022-04505-7
Maurella, C., Gallina, S., Ru, G., Adriano, D., Bellio, A., Bianchi, D. M., et al. (2018). Outbreak of febrile gastroenteritis caused by Listeria monocytogenes 1/2a in sliced cold beef ham, Italy, May 2016. Eur. Secur. 23, 17–00155. doi: 10.2807/1560-7917.ES.2018.23.10.17-00155
Maury, M. M., Chenal-Francisque, V., Bracq-Dieye, H., Han, L., Leclercq, A., Vales, G., et al. (2017). Spontaneous loss of virulence in natural populations of Listeria monocytogenes. Infect. Immun. 85, e00541–e00517. doi: 10.1128/iai.00541-17
Maury, M. M., Tsai, Y. H., Charlier, C., Touchon, M., Chenal-Francisque, V., Leclercq, A., et al. (2016). Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 48, 308–313. doi: 10.1038/ng.3501
McCoy, R., Oldroyd, S., Yang, W., Wang, K., Hoven, D., Bulmer, D., et al. (2024). In Vitro Models for Investigating Intestinal Host-Pathogen Interactions. Adv. Sci. 11:2306727. doi: 10.1002/advs.202306727
McLauchlin, J., Aird, H., Amar, C., Barker, C., Dallman, T., Lai, S., et al. (2021). An outbreak of human listeriosis associated with frozen sweet corn consumption: Investigations in the UK. Int. J. Food Microbiol. 338:108994. doi: 10.1016/j.ijfoodmicro.2020.108994
Ministerio de Sanidad Consumo y Bienestar Social de España. (2019). Informe de Fin de Seguimiento Del Brote de Listeriosis. Available at: https://www.sanidad.gob.es/profesionales/saludPublica/ccayes/alertasActual/listeriosis/docs/Informe_cierre_Listeriosis_20190927.pdf (accessed May 28, 2019).
Møretrø, T., Wagner, E., Heir, E., Langsrud, S., and Fagerlund, A. (2024). Genomic analysis of Listeria monocytogenes CC7 associated with clinical infections and persistence in the food industry. Int. J. Food Microbiol. 410:110482. doi: 10.1016/j.ijfoodmicro.2023.110482
Moura, A., Criscuolo, A., Pouseele, H., Maury, M. M., Leclercq, A., Tarr, C., et al. (2016). Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2:16185. doi: 10.1038/nmicrobiol.2016.185
Moura, A., Disson, O., Lavina, M., Thouvenot, P., Huang, L., Leclercq, A., et al. (2019). Atypical Hemolytic Listeria innocua Isolates Are Virulent, albeit Less than Listeria monocytogenes. Infect. Immun. 87:e00758-18. doi: 10.1128/IAI.00758-18
Moura, A., Leclercq, A., Vales, G., Tessaud-Rita, N., Bracq-Dieye, H., Thouvenot, P., et al. (2024). Phenotypic and genotypic antimicrobial resistance of Listeria monocytogenes: an observational study in France. Lancet Reg. Health Eur. 37:100800. doi: 10.1016/j.lanepe.2023.100800
Muchaamba, F., Eshwar, A. K., Stevens, M. J. A., Stephan, R., and Tasara, T. (2022). Different shades of Listeria monocytogenes: strain, serotype, and lineage-based variability in virulence and stress tolerance profiles. Front. Microbiol. 12:792162. doi: 10.3389/fmicb.2021.792162
Mukherjee, K., Abu Mraheil, M., Silva, S., Müller, D., Cemic, F., Hemberger, J., et al. (2011). Anti-Listeria activities of Galleria mellonella hemolymph proteins. Appl. Environ. Microbiol. 77, 4237–4240. doi: 10.1128/aem.02435-10
Mukherjee, K., Altincicek, B., Hain, T., Domann, E., Vilcinskas, A., and Chakraborty, T. (2010). Galleria mellonella as a model system for studying Listeria pathogenesis. Appl. Environ. Microbiol. 76, 310–317. doi: 10.1128/aem.01301-09
Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., and Erlich, H. (1986). Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb. Symp. Quant. Biol. 51, 263–273. doi: 10.1101/sqb.1986.051.01.032
Muthulakshmi, L., Suganya, K., Murugan, M., Annaraj, J., Duraipandiyan, V., Al Farraj, D. A., et al. (2022). Antibiofilm efficacy of novel biogenic silver nanoparticles from Terminalia catappa against food-borne Listeria monocytogenes ATCC 15,313 and mechanisms investigation in-vivo and in-vitro. J. King Saud Univ. Sci. 34:102083. doi: 10.1016/j.jksus.2022.102083
Mylonakis, E., Casadevall, A., and Ausubel, F. M. (2007). Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 3, e101–e107. doi: 10.1371/journal.ppat.0030101
Mylonakis, E., Moreno, R., El Khoury, J. B., Idnurm, A., Heitman, J., Calderwood, S. B., et al. (2005). Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect. Immun. 73, 3842–3850. doi: 10.1128/iai.73.7.3842-3850.2005
Nightingale, K. K., Windham, K., Martin, K. E., Yeung, M., and Wiedmann, M. (2005a). Select Listeria monocytogenes subtypes commonly found in foods carry distinct nonsense mutations in inlA, leading to expression of truncated and secreted internalin A, and are associated with a reduced invasion phenotype for human intestinal epithelial cells. Appl. Environ. Microbiol. 71, 8764–8772. doi: 10.1128/aem.71.12.8764-8772.2005
Nightingale, K. K., Windham, K., and Wiedmann, M. (2005b). Evolution and molecular phylogeny of Listeria monocytogenes isolated from human and animal listeriosis cases and foods. J. Bacteriol. 187, 5537–5551. doi: 10.1128/jb.187.16.5537-5551.2005
Nikitas, G., Deschamps, C., Disson, O., Niault, T., Cossart, P., and Lecuit, M. (2011). Transcytosis of Listeria monocytogenes across the intestinal barrier upon specific targeting of goblet cell accessible E-cadherin. J. Exp. Med. 208, 2263–2277. doi: 10.1084/jem.20110560
Nüesch-Inderbinen, M., Bloemberg, G. V., Müller, A., Stevens, M. J. A., Cernela, N., Kollöffel, B., et al. (2021). Listeriosis caused by persistence of Listeria monocytogenes serotype 4b sequence type 6 in cheese production environment. Emerg. Infect. Dis. 27, 284–288. doi: 10.3201/eid2701.203266
Orsi, R. H., Bakker, H. C. D., and Wiedmann, M. (2011). Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 301, 79–96. doi: 10.1016/j.ijmm.2010.05.002
Orsi, R. H., Sun, Q., and Wiedmann, M. (2008). Genome-wide analyses reveal lineage specific contributions of positive selection and recombination to the evolution of Listeria monocytogenes. BMC Evol. Biol. 8:233. doi: 10.1186/1471-2148-8-233
Painset, A., Björkman, J. T., Kiil, K., Guillier, L., Mariet, J. F., Félix, B., et al. (2019). LiSEQ – whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genom. 5:e000257. doi: 10.1099/mgen.0.000257
Pan, X., Shen, J., Hong, Y., Wu, Y., Guo, D., Zhao, L., et al. (2024). Comparative analysis of growth, survival, and virulence characteristics of Listeria monocytogenes isolated from imported meat. Microorganisms 12:345. doi: 10.3390/microorganisms12020345
Pearce, S. C., Coia, H. G., Karl, J. P., Pantoja-Feliciano, I. G., Zachos, N. C., and Racicot, K. (2018). Intestinal in vitro and ex vivo models to study host-microbiome interactions and acute stressors. Front. Physiol. 9:1584. doi: 10.3389/fphys.2018.01584
Pentecost, M., Otto, G., Theriot, J. A., and Amieva, M. R. (2006). Listeria monocytogenes Invades the Epithelial Junctions at Sites of Cell Extrusion. PLoS Pathog. 2:e3. doi: 10.1371/journal.ppat.0020003
Pereira, T. C., de Barros, P. P., Fugisaki, L. R. O., Rossoni, R. D., Ribeiro, F. C., de Menezes, R. T., et al. (2018). Recent advances in the use of Galleria mellonella model to study immune responses against human pathogens. J. Fungi 4:128. doi: 10.3390/jof4040128
Perrin, M., Bemer, M., and Delamare, C. (2003). Fatal Case of Listeria innocua Bacteremia. J. Clin. Microbiol. 41, 5308–5309. doi: 10.1128/jcm.41.11.5308-5309.2003
Peterson, R. T., Nass, R., Boyd, W. A., Freedman, J. H., Dong, K., and Narahashi, T. (2008). Use of non-mammalian alternative models for neurotoxicological study. Neurotoxicology 29, 546–555. doi: 10.1016/j.neuro.2008.04.006
Petnicki-Ocwieja, T., Hrncir, T., Liu, Y.-J., Biswas, A., Hudcovic, T., Tlaskalova-Hogenova, H., et al. (2009). Nod2 is required for the regulation of commensal microbiota in the intestine. Proc. Natl. Acad. Sci. 106, 15813–15818. doi: 10.1073/pnas.0907722106
Phelps, C. C., Vadia, S., Arnett, E., Tan, Y., Zhang, X., Pathak-Sharma, S., et al. (2018). Relative Roles of Listeriolysin O, InlA, and InlB in Listeria monocytogenes Uptake by Host Cells. Infect. Immun. 86:e00555-18. doi: 10.1128/iai.00555-18
Pietzka, A., Allerberger, F., Murer, A., Lennkh, A., Stöger, A., Cabal Rosel, A., et al. (2019). Whole genome sequencing based surveillance of L. monocytogenes for early detection and investigations of listeriosis outbreaks. Front. Public Health 7:139. doi: 10.3389/fpubh.2019.00139
Pizarro-Cerdá, J., Kühbacher, A., and Cossart, P. (2012). Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb. Perspect. Med. 2:a010009. doi: 10.1101/cshperspect.a010009
Pont, S., and Blanc-Potard, A.-B. (2021). Zebrafish embryo infection model to investigate Pseudomonas aeruginosa interaction with innate immunity and validate new therapeutics. Front. Cell. Infect. Microbiol. 11:745851. doi: 10.3389/fcimb.2021.745851
Prescott, J. F. (2022). “Themes in bacterial pathogenesis” in Pathogenesis of bacterial infections in animals (Hoboken, New Jersey: John Wiley & Sons, Ltd.), 1–14.
Pyz-Łukasik, R., Paszkiewicz, W., Kiełbus, M., Ziomek, M., Gondek, M., Domaradzki, P., et al. (2022). Genetic diversity and potential virulence of Listeria monocytogenes isolates originating from polish artisanal cheeses. Food Secur. 11:2805. doi: 10.3390/foods11182805
Quereda, J. J., Dussurget, O., Nahori, M.-A., Ghozlane, A., Volant, S., Dillies, M.-A., et al. (2016). Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. 113, 5706–5711. doi: 10.1073/pnas.1523899113
Quereda, J. J., Morón-García, A., Palacios-Gorba, C., Dessaux, C., García-del Portillo, F., Pucciarelli, M. G., et al. (2021). Pathogenicity and virulence of Listeria monocytogenes: A trip from environmental to medical microbiology. Virulence 12, 2509–2545. doi: 10.1080/21505594.2021.1975526
Quereda, J. J., and Pucciarelli, M. G. (2014). Deletion of the membrane protein Lmo0412 increases the virulence of Listeria monocytogenes. Microbes Infect. 16, 623–632. doi: 10.1016/j.micinf.2014.07.002
Quereda, J. J., Rodríguez-Gómez, I. M., Meza-Torres, J., Gómez-Laguna, J., Nahori, M. A., Dussurget, O., et al. (2019). Reassessing the role of internalin B in Listeria monocytogenes virulence using the epidemic strain F2365. Clin. Microbiol. Infect. 25, 252.e1–252.e4. doi: 10.1016/j.cmi.2018.08.022
Radoshevich, L., and Cossart, P. (2018). Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat. Rev. Microbiol. 16, 32–46. doi: 10.1038/nrmicro.2017.126
Ragon, M., Wirth, T., Hollandt, F., Lavenir, R., Lecuit, M., Le Monnier, A., et al. (2008). A New Perspective on Listeria monocytogenes Evolution. PLoS Pathog. 4, e1000146–e1000114. doi: 10.1371/journal.ppat.1000146
Rakic Martinez, M., Ferguson, M., and Datta, A. R. (2020). Virulence assessment of Listeria monocytogenes grown in different foods using a Galleria mellonella model. PLoS One 15:e0232485. doi: 10.1371/journal.pone.0232485
Rasmussen, O. F., Skouboe, P., Dons, L., Rossen, L., and Olsen, J. E. (1995). Listeria monocytogenes exists in at least three evolutionary lines: evidence from flagellin, invasive associated protein and listeriolysin O genes. Microbiology 141, 2053–2061. doi: 10.1099/13500872-141-9-2053
Raufu, I. A., Moura, A., Vales, G., Ahmed, O. A., Aremu, A., Thouvenot, P., et al. (2022). Listeria ilorinensis sp. nov., isolated from cow milk cheese in Nigeria. Int. J. Syst. Evol. Microbiol. 72:005437. doi: 10.1099/ijsem.0.005437
Ricchi, M., Bertasio, C., Boniotti, M. B., Vicari, N., Russo, S., Tilola, M., et al. (2017). Comparison among the Quantification of Bacterial Pathogens by qPCR, dPCR, and Cultural Methods. Front. Microbiol. 8:1174. doi: 10.3389/fmicb.2017.01174
Roberts, A., Nightingale, K., Jeffers, G., Fortes, E., Kongo, J. M., and Wiedmann, M. (2006). Genetic and phenotypic characterization of Listeria monocytogenes lineage III. Microbiology 152, 685–693. doi: 10.1099/mic.0.28503-0
Rocha, P. R. D. A., Dalmasso, A., Grattarola, C., Casalone, C., Del Piero, F., Bottero, M. T., et al. (2013). Atypical cerebral listeriosis associated with Listeria innocua in a beef bull. Res. Vet. Sci. 94, 111–114. doi: 10.1016/j.rvsc.2012.07.017
Roche, S. M., Velge, P., Bottreau, E., Durier, C., Marquet-van der Mee, N., and Pardon, P. (2001). Assessment of the virulence of Listeria monocytogenes: agreement between a plaque-forming assay with HT-29 cells and infection of immunocompetent mice. Int. J. Food Microbiol. 68, 33–44. doi: 10.1016/S0168-1605(01)00460-3
Rodriguez, A. G. (2018). The applicability of in vitro models of the intestinal barrier for the risk assessment of engineered nanomaterials used as food additives [Doctoral Dissertation, Universitat Autònoma de Barcelona].
Rolhion, N., Chassaing, B., Nahori, M.-A., de Bodt, J., Moura, A., Lecuit, M., et al. (2019). A Listeria monocytogenes bacteriocin can target the commensal Prevotella copri and modulate intestinal infection. Cell Host Microbe 26, 691–701.e5. doi: 10.1016/j.chom.2019.10.016
Roodsant, T., Navis, M., Aknouch, I., Renes, I. B., van Elburg, R. M., Pajkrt, D., et al. (2020). A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front. Cell. Infect. Microbiol. 10:272. doi: 10.3389/fcimb.2020.00272
Roulo, R. M., Fishburn, J. D., Amosu, M., Etchison, A. R., and Smith, M. A. (2014). Dose Response of Listeria monocytogenes Invasion, Fetal Morbidity, and Fetal Mortality after Oral Challenge in Pregnant and Nonpregnant Mongolian Gerbils. Infect. Immun. 82, 4834–4841. doi: 10.1128/iai.01514-14
Rousseaux, S., Olier, M., Lemaître, J. P., Piveteau, P., and Guzzo, J. (2004). Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 70, 2180–2185. doi: 10.1128/aem.70.4.2180-2185.2004
Rupp, S., Bärtschi, M., Frey, J., and Oevermann, A. (2017). Hyperinvasiveness and increased intercellular spread of Listeria monocytogenes sequence type 1 are independent of listeriolysin S, internalin F and internalin J1. J. Med. Microbiol. 66, 1053–1062. doi: 10.1099/jmm.0.000529
Ruppitsch, W., Prager, R., Halbedel, S., Hyden, P., Pietzka, A., Huhulescu, S., et al. (2015). Ongoing outbreak of invasive listeriosis, Germany, 2012 to 2015. Eur. Secur. 20:30094. doi: 10.2807/1560-7917.ES.2015.20.50.30094
Salcedo, C., Arreaza, L., Alcalá, B., De La Fuente, L., and Vázquez, J. A. (2003). Development of a multilocus sequence typing method for analysis of Listeria monocytogenes clones. J. Clin. Microbiol. 41, 757–762. doi: 10.1128/jcm.41.2.757-762.2003
Sato, T., Vries, R. G., Snippert, H. J., van de Wetering, M., Barker, N., Stange, D. E., et al. (2009). Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265. doi: 10.1038/nature07935
Sauders, B. D., Durak, M. Z., Fortes, E., Windham, K., Schukken, Y., Lembo, A. J., et al. (2006). Molecular characterization of Listeria monocytogenes from natural and urban environments. J. Food Prot. 69, 93–105. doi: 10.4315/0362-028X-69.1.93
Schiavano, G. F., Ateba, C. N., Petruzzelli, A., Mele, V., Amagliani, G., Guidi, F., et al. (2022). Whole-genome sequencing characterization of virulence profiles of Listeria monocytogenes food and human isolates and in vitro adhesion/invasion assessment. Microorganisms 10:62. doi: 10.3390/microorganisms10010062
Schjørring, S., Gillesberg Lassen, S., Jensen, T., Moura, A., Kjeldgaard, J. S., Müller, L., et al. (2017). Cross-border outbreak of listeriosis caused by cold-smoked salmon, revealed by integrated surveillance and whole genome sequencing (WGS), Denmark and France, 2015 to 2017. Eur. Secur. 22, 17–00762. doi: 10.2807/1560-7917.ES.2017.22.50.17-00762
Schmid, D., Allerberger, F., Huhulescu, S., Pietzka, A., Amar, C., Kleta, S., et al. (2014). Whole genome sequencing as a tool to investigate a cluster of seven cases of listeriosis in Austria and Germany, 2011-2013. Clin. Microbiol. Infect. 20, 431–436. doi: 10.1111/1469-0691.12638
Seeliger, H. P. R., and Höhne, K. (1979). “Chapter II serotyping of Listeria monocytogenes and related species” in Methods in Microbiology. eds. T. Bergan and J. R. Norris, vol. 13 (Cambridge, Massachusetts: Academic Press), 31–49.
Seeliger, H. P. R., and Langer, B. (1989). Serological analysis of the genus Listeria. Its values and limitations. Int. J. Food Microbiol. 8, 245–248. doi: 10.1016/0168-1605(89)90020-2
Shan, Y., Fang, C., Cheng, C., Wang, Y., Peng, J., and Fang, W. (2015). Immersion infection of germ-free zebrafish with Listeria monocytogenes induces transient expression of innate immune response genes. Front. Microbiol. 6:373. doi: 10.3389/fmicb.2015.00373
Shelburne, S. A., and Musser, J. M. (2004). Virulence gene expression in vivo. Curr. Opin. Microbiol. 7, 283–289. doi: 10.1016/j.mib.2004.04.013
Shi, W., Zhang, Q., Li, H., Du, D., Ma, X., Wang, J., et al. (2024). Biofilm Formation, Motility, and Virulence of Listeria monocytogenes Are Reduced by Deletion of the Gene lmo0159, a Novel Listerial LPXTG Surface Protein. Microorganisms 12:1354. doi: 10.3390/microorganisms12071354
Short, F. L., and MacInnes, J. I. (2022). “Experimental approaches to understanding pathogenesis” in Pathogenesis of bacterial Infections in animals. eds. J. F. Prescott, A. N. Rycroft, J. D. Boyce, J. I. MacInnes, F. Van Immerseel, and J. A. Vázquez-Boland (Hoboken, New Jersey: John Wiley & Sons, Ltd.), 57–78.
Silva, A., Genovés, S., Martorell, P., Zanini, S. F., Rodrigo, D., and Martinez, A. (2015). Sublethal injury and virulence changes in Listeria monocytogenes and Listeria innocua treated with antimicrobials carvacrol and citral. Food Microbiol. 50, 5–11. doi: 10.1016/j.fm.2015.02.016
Singkum, P., Suwanmanee, S., Pumeesat, P., and Luplertlop, N. (2019). A powerful in vivo alternative model in scientific research: Galleria mellonella. Acta Microbiol. Immunol. Hung. 66, 31–55. doi: 10.1556/030.66.2019.001
Sivaranjani, M., Gowrishankar, S., Kamaladevi, A., Pandian, S. K., Balamurugan, K., and Ravi, A. V. (2016). Morin inhibits biofilm production and reduces the virulence of Listeria monocytogenes — An in vitro and in vivo approach. Int. J. Food Microbiol. 237, 73–82. doi: 10.1016/j.ijfoodmicro.2016.08.021
Smith, B., Larsson, J. T., Lisby, M., Müller, L., Madsen, S. B., Engberg, J., et al. (2011). Outbreak of listeriosis caused by infected beef meat from a meals-on-wheels delivery in Denmark 2009. Clin. Microbiol. Infect. 17, 50–52. doi: 10.1111/j.1469-0691.2010.03200.x
Soni, D. K., Ghosh, A., Chikara, S. K., Singh, K. M., Joshi, C. G., and Dubey, S. K. (2017). Comparative whole genome analysis of Listeria monocytogenes 4b strains reveals least genome diversification irrespective of their niche specificity. Gene Rep. 8, 61–68. doi: 10.1016/j.genrep.2017.05.007
Stephan, R., Althaus, D., Kiefer, S., Lehner, A., Hatz, C., Schmutz, C., et al. (2015). Foodborne transmission of Listeria monocytogenes via ready-to-eat salad: A nationwide outbreak in Switzerland, 2013–2014. Food Control 57, 14–17. doi: 10.1016/j.foodcont.2015.03.034
Taebnia, N., Römling, U., and Lauschke, V. M. (2023). In vitro and ex vivo modeling of enteric bacterial infections. Gut Microbes 15:2158034. doi: 10.1080/19490976.2022.2158034
Taillebourg, E., Schneider, D. S., and Fauvarque, M.-O. (2014). The Drosophila Deubiquitinating Enzyme dUSP36 Acts in the Hemocytes for Tolerance to Listeria monocytogenes Infections. J. Innate Immun. 6, 632–638. doi: 10.1159/000360293
Tasara, T., Ebner, R., Klumpp, J., and Stephan, R. (2015). Complete Genome Sequence of Listeria monocytogenes N2306, a Strain Associated with the 2013-2014 Listeriosis Outbreak in Switzerland. Genome Announc. 3:e00553-15. doi: 10.1128/genomea.00553-15
Tham, T. N., Gouin, E., Rubinstein, E., Boucheix, C., Cossart, P., and Pizarro-Cerda, J. (2010). Tetraspanin CD81 Is Required for Listeria monocytogenes Invasion. Infect. Immun. 78, 204–209. doi: 10.1128/iai.00661-09
Thomsen, L. E., Slutz, S. S., Tan, M. W., and Ingmer, H. (2006). Caenorhabditis elegans is a model host for Listeria monocytogenes. Appl. Environ. Microbiol. 72, 1700–1701. doi: 10.1128/aem.72.2.1700-1701.2006
Tilney, L. G., and Portnoy, D. A. (1989). Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109, 1597–1608. doi: 10.1083/jcb.109.4.1597
Trede, N. S., Langenau, D. M., Traver, D., Look, A. T., and Zon, L. I. (2004). The use of zebrafish to understand immunity. Immunity 20, 367–379. doi: 10.1016/S1074-7613(04)00084-6
Tsai, Y.-H., Disson, O., Bierne, H., and Lecuit, M. (2013). Murinization of Internalin Extends Its Receptor Repertoire, Altering Listeria monocytogenes Cell Tropism and Host Responses. PLoS Pathog. 9, e1003381–e1003316. doi: 10.1371/journal.ppat.1003381
Tsai, C. J., Loh, J. M., and Proft, T. (2016). Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 7, 214–229. doi: 10.1080/21505594.2015.1135289
van der Sar, A. M., Appelmelk, B. J., Vandenbroucke-Grauls, C. M., and Bitter, W. (2004). A star with stripes: zebrafish as an infection model. Trends Microbiol. 12, 451–457. doi: 10.1016/j.tim.2004.08.001
Vázquez-Boland, J. A., Kuhn, M., Berche, P., Chakraborty, T., Domínguez-Bernal, G., Goebel, W., et al. (2001). Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14, 584–640. doi: 10.1128/cmr.14.3.584-640.2001
Vitullo, M., Grant, K. A., Sammarco, M. L., Tamburro, M., Ripabelli, G., and Amar, C. F. (2013). Real-time PCRs assay for serogrouping Listeria monocytogenes and differentiation from other Listeria spp. Mol. Cell. Probes 27, 68–70. doi: 10.1016/j.mcp.2012.10.001
Wagner, E., Fagerlund, A., Thalguter, S., Jensen, M. R., Heir, E., Møretrø, T., et al. (2022). Deciphering the virulence potential of Listeria monocytogenes in the Norwegian meat and salmon processing industry by combining whole genome sequencing and in vitro data. Int. J. Food Microbiol. 383:109962. doi: 10.1016/j.ijfoodmicro.2022.109962
Walker, J. K., Morgan, J. H., McLauchlin, J., Grant, K. A., and Shallcross, J. A. (1994). Listeria innocua isolated from a case of ovine meningoencephalitis. Vet. Microbiol. 42, 245–253. doi: 10.1016/0378-1135(94)90023-X
Wambui, J., Eshwar, A. K., Aalto-Araneda, M., Pöntinen, A., Stevens, M. J. A., Njage, P. M. K., et al. (2020). The Analysis of Field Strains Isolated From Food, Animal and Clinical Sources Uncovers Natural Mutations in Listeria monocytogenes Nisin Resistance Genes. Front. Microbiol. 11:549531. doi: 10.3389/fmicb.2020.549531
Wang, Y., Luo, L., Li, Q., Wang, H., Wang, Y., Sun, H., et al. (2019). Genomic dissection of the most prevalent Listeria monocytogenes clone, sequence type ST87, in China. BMC Genomics 20:1014. doi: 10.1186/s12864-019-6399-1
Ward, T. J., Ducey, T. F., Usgaard, T., Dunn, K. A., and Bielawski, J. P. (2008). Multilocus Genotyping Assays for Single Nucleotide Polymorphism-Based Subtyping of Listeria monocytogenes Isolates. Appl. Environ. Microbiol. 74, 7629–7642. doi: 10.1128/AEM.01127-08
Wiedmann, M., Bruce, J. L., Keating, C., Johnson, A. E., McDonough, P. L., and Batt, C. A. (1997). Ribotypes and virulence gene polymorphisms suggest three distinct Listeria monocytogenes lineages with differences in pathogenic potential. Infect. Immun. 65, 2707–2716. doi: 10.1128/iai.65.7.2707-2716.1997
Wikman-Larhed, A., and Artursson, P. (1995). Co-cultures of human intestinal goblet (HT29-H) and absorptive (Caco-2) cells for studies of drug and peptide absorption. Eur. J. Pharm. Sci. 3, 171–183. doi: 10.1016/0928-0987(95)00007-Z
Wilking, H., Lachmann, R., Holzer, A., Halbedel, S., Flieger, A., and Stark, K. (2021). Ongoing high incidence and case-fatality rates for invasive listeriosis, Germany, 2010-2019. Emerg. Infect. Dis. 27, 2485–2488. doi: 10.3201/eid2709.210068
Witte, A. K., Fister, S., Mester, P., Schoder, D., and Rossmanith, P. (2016). Evaluation of the performance of quantitative detection of the Listeria monocytogenes prfA locus with droplet digital PCR. Anal. Bioanal. Chem. 408, 7583–7593. doi: 10.1007/s00216-016-9861-9
Wollert, T., Pasche, B., Rochon, M., Deppenmeier, S., van den Heuvel, J., Gruber, A. D., et al. (2007). Extending the Host Range of Listeria monocytogenes by Rational Protein Design. Cell 129, 891–902. doi: 10.1016/j.cell.2007.03.049
World Health Organization & Food and Agriculture Organization of the United Nations. (2004). Risk assessment of Listeria monocytogenes in ready-to-eat foods: technical report (9241562625). (Microbiological Risk Assessment Series); no. 5, Issue. F. a. A. Organization. Available at: https://apps.who.int/iris/handle/10665/42875 (Accessed August 20, 2024).
World Medical Association (2013). World Medical Association Declaration of Helsinki: Ethical Principles for Medical Research Involving Human Subjects. JAMA 310, 2191–2194. doi: 10.1001/jama.2013.281053
Wu, J., NicAogáin, K., McAuliffe, O., Jordan, K., and O’Byrne, C. (2022). Phylogenetic and Phenotypic Analyses of a Collection of Food and Clinical Listeria monocytogenes Isolates Reveal Loss of Function of Sigma B from Several Clonal Complexes. Appl. Environ. Microbiol. 88, e0005122–e0000022. doi: 10.1128/aem.00051-22
Yan, H., Wu, M., Dong, Q., and Li, Z. (2023). Advances in in vitro and in vivo models for Listeria monocytogenes placental infection. Chin. J. Biotechnol. 39, 3985–4003. doi: 10.13345/j.cjb.230263
Yde, M., Naranjo, M., Mattheus, W., Stragier, P., Pochet, B., Beulens, K., et al. (2012). Usefulness of the European Epidemic Intelligence Information System in the management of an outbreak of listeriosis, Belgium, 2011. Eur. Secur. 17:20279. doi: 10.2807/ese.17.38.20279-en
Yin, X., Farin, H. F., van Es, J. H., Clevers, H., Langer, R., and Karp, J. M. (2014). Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat. Methods 11, 106–112. doi: 10.1038/nmeth.2737
Zakrzewski, A. J., Chajęcka-Wierzchowska, W., Zadernowska, A., and Podlasz, P. (2020). Virulence Characterization of Listeria monocytogenes, Listeria innocua, and Listeria welshimeri Isolated from Fish and Shrimp Using In Vivo Early Zebrafish Larvae Models and Molecular Study. Pathogens 9:1028. doi: 10.3390/pathogens9121028
Zeldovich, V. B., Robbins, J. R., Kapidzic, M., Lauer, P., and Bakardjiev, A. I. (2011). Invasive extravillous trophoblasts restrict intracellular growth and spread of Listeria monocytogenes. PLoS Pathog. 7:e1002005. doi: 10.1371/journal.ppat.1002005
Zhang, H., Chen, W., Wang, J., Xu, B., Liu, H., Dong, Q., et al. (2020). 10-Year Molecular Surveillance of Listeria monocytogenes Using Whole-Genome Sequencing in Shanghai, China, 2009–2019. Front. Microbiol. 11:551020. doi: 10.3389/fmicb.2020.551020
Zhang, W., Jayarao, B. M., and Knabel, S. J. (2004). Multi-Virulence-Locus Sequence Typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70, 913–920. doi: 10.1128/AEM.70.2.913-920.2004
Zhou, C., Zhang, Y., Bassey, A., Huang, J., Zou, Y., and Ye, K. (2022a). Expansion of Intestinal Secretory Cell Population Induced by Listeria monocytogenes Infection: Accompanied With the Inhibition of NOTCH Pathway. Front. Cell. Infect. Microbiol. 12:793335. doi: 10.3389/fcimb.2022.793335
Zhou, C., Zou, Y., Huang, J., Zhao, Z., Zhang, Y., Wei, Y., et al. (2022b). TMT-Based Quantitative Proteomic Analysis of Intestinal Organoids Infected by Listeria monocytogenes Strains with Different Virulence. Int. J. Mol. Sci. 23:6231. doi: 10.3390/ijms23116231
Zhou, C., Zou, Y., Zhang, Y., Teng, S., and Ye, K. (2022c). Involvement of CCN1 Protein and TLR2/4 Signaling Pathways in Intestinal Epithelial Cells Response to Listeria monocytogenes. Int. J. Mol. Sci. 23:2739. doi: 10.3390/ijms23052739
Zhu, H., Zhang, H., Xu, Y., Laššáková, S., Korabečná, M., and Neužil, P. (2020). PCR past, present and future. Biotechniques 69, 317–325. doi: 10.2144/btn-2020-0057
Keywords: listeriosis, virulence, risk assessment, CC, infection
Citation: Sousa M, Magalhães R, Ferreira V and Teixeira P (2024) Current methodologies available to evaluate the virulence potential among Listeria monocytogenes clonal complexes. Front. Microbiol. 15:1425437. doi: 10.3389/fmicb.2024.1425437
Received: 29 April 2024; Accepted: 26 September 2024;
Published: 10 October 2024.
Edited by:
Jens Andre Hammerl, Bundesinstitut für Risikobewertung, GermanyReviewed by:
Rishi Drolia, Old Dominion University, United StatesCopyright © 2024 Sousa, Magalhães, Ferreira and Teixeira. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Paula Teixeira, cGN0ZWl4ZWlyYUB1Y3AucHQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.