
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 26 July 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1420156
Introduction: Trichoderma species establish symbiotic relationships with plants through both parasitic and mutualistic mechanisms. While some Trichoderma species act as plant pathogenic fungi, others utilize various strategies to protect and enhance plant growth.
Methods: Phylogenetic positions of new species of Trichoderma were determined through multi-gene analysis relying on the internal transcribed spacer (ITS) regions of the ribosomal DNA, the translation elongation factor 1-α (tef1-α) gene, and the RNA polymerase II (rpb2) gene. Additionally, pathogenicity experiments were conducted, and the aggressiveness of each isolate was evaluated based on the area of the cross-section of the infected site.
Results: In this study, 13 Trichoderma species, including 9 known species and 4 new species, namely, T. delicatum, T. robustum, T. perfasciculatum, and T. subulatum were isolated from the diseased tubers of Gastrodia elata in Yunnan, China. Among the known species, T. hamatum had the highest frequency. T. delicatum belonged to the Koningii clade. T. robustum and T. perfasciculatum were assigned to the Virens clade. T. subulatum emerged as a new member of the Spirale clade. Pathogenicity experiments were conducted on the new species T. robustum, T. delicatum, and T. perfasciculatum, as well as the known species T. hamatum, T. atroviride, and T. harzianum. The infective abilities of different Trichoderma species on G. elata varied, indicating that Trichoderma was a pathogenic fungus causing black rot disease in G. elata.
Discussion: This study provided the morphological characteristics of new species and discussed the morphological differences with phylogenetically proximate species, laying the foundation for research aimed at preventing and managing diseases that affect G. elata.
The genus Trichoderma, belonging to the Hypocreales in Ascomycota, is widely distributed in different types of habitats (Migheli et al., 2009). They show significant potential for applications in human societal production. In agriculture, Trichoderma species as a soil conditioner can produce abundant secondary metabolites to promote plant growth and enhance plant stress tolerance (Degenkolb et al., 2008; Pruksakorn et al., 2010; Cheng et al., 2012). In industry, the improved strain Trichoderma reesei QM6a is used as a cellulase-producing strain (Kubicek et al., 2019). However, some Trichoderma spp. are potentially pathogenic fungi that can cause infections in humans. Moreover, several Trichoderma species can secrete toxins that decrease the yield of large edible fungi, negatively impact human health and economic development (Kim et al., 2012; Sandoval–Denis et al., 2014).
Gastrodia elata BI. contains various active components, including G. elata alkaloids and G. elata polysaccharides, which enhance immune function, alleviate pain, and exhibit anti-inflammatory and antioxidant properties (Fan et al., 2013; Chen et al., 2016; Liu and Huang, 2017). A special symbiotic relationship exists between G. elata and Armillariella mellea P. Karst, where G. elata relies on Armillariella mellea P. Karst for nutrient supply (Zeng et al., 2018). However, certain Trichoderma species can parasitize on A. mellea in symbiosis with G. elata, causing A. mellea to die, which consequently affects the growth of G. elata (Wu and Ma, 1998). Additionally, T. viride Pers. and T. hamatum Bainier were reported as the pathogens responsible for causing the black rot disease in G. elata (Zeng et al., 2007; Han et al., 2017). Zhang et al. (2020) investigated the diseases of Gastrodia elata in Xiaocaoba, Yunnan, and Dafang, Guizhou areas and found that the incidence of diseased Gastrodia elata ranged from 6% to 17%, with a yield reduction of 10% to 30%. However, Trichoderma also served as a natural defender of G. elata. Zhang et al. (2020) isolated T. harzianum Rifai from diseased G. elata tissues, ensuring the normal growth of G. elata by controlling the growth of pathogenic fungi in G. elata (Zhang et al., 2020). T. viride also exhibited antagonistic effects against corn stalk rot pathogens on G. elata (Wu and Ma, 1998). Therefore, the connection between G. elata and Trichoderma spp. is significantly important and complex.
Research on Trichoderma species began more than 200 years ago. With the development of sequencing technology, researchers began to incorporate morphological methods into fungal identification (Lieckfeldt and Seifert, 2000; Kullnig–Gradinger et al., 2002). The internal transcribed spacer (ITS), as the molecular barcode for Trichoderma, is the primary sequence used for identification (Lieckfeldt and Seifert, 2000; Chaverri et al., 2001; Kullnig–Gradinger et al., 2002; Samuels et al., 2002). Although the ITS sequence can provide preliminary insights into the classification of Trichoderma species, a single gene cannot accurately depict interspecies relationships. New molecular barcodes have gradually been introduced into the classification, such as the translation elongation factor 1-α (tef1) and the RNA polymerase II (rpb2) gene (Berney et al., 2000; Chaverri et al., 2003; Jaklitsch et al., 2013; Jaklitsch and Voglmayr, 2015). The universality of tef1 and rpb2 makes them ideal identification tools (Berney et al., 2000; Chaverri et al., 2003). The multi-gene analysis supports the exploration of the diversity of Trichoderma species associated with G. elata, which provides a theoretical basis for the prevention of Gastrodia diseases.
In this study, 93 strains were isolated from the diseased tubers of G. elata collected from Yiliang County, Zhaotong City, Yunnan Province. Among these strains, 85 belonged to 9 known species. Four new Trichoderma species belonging to three clades were described and illustrated using multi-gene analysis. Additionally, the pathogenicity of the representative species was tested.
Approximately more than 100 diseased tubers of G. elata were collected from the planting area in Xiaocaoba, Yiliang County, Zhaotong City, Yunnan Province. Some tubers were collected from a planting pond, while others were obtained from markets. The samples were then placed in sterile self-sealing plastic bags, labeled, and transported to the laboratory. These samples were stored at 4°C.
The plant tissues were washed to remove soil and debris, and then they were immersed in 75% ethanol (75 mL of ethanol was diluted with water to obtain a total volume of 100 mL) for 2 min. After rinsing three times with sterile water for 30 s each, the tissues were immersed in 1.5% sodium hypochlorite (1.5 g of sodium hypochlorite was dissolved in 100 mL of water) for 2 min, and then the rinsing step was repeated. Subsequently, the tissues were air-dried. The plant tissues were cut into slices of 5 mm × 5 mm dimensions and were placed on potato dextrose agar (PDA) plates (PDA: 200 g potato, 20 g dextrose, 18 g agar, and 1,000 mL distilled water) at 25°C. After mycelium growth, the strains with morphological characteristics of Trichoderma, such as abundant and fluffy hyphae, and yellow-green or green colonies, were selected for transferring into PDA plates for initial purification culture and identification. The pure and dried cultures were deposited in the herbarium of the Laboratory for Conservation and Utilization of Bio-Resources, Yunnan University, Kunming, Yunnan, China (YMF).
Perforate PDA plates were covered with mycelium using a 5 mm diameter punch. Inoculate circular mycelial plugs were secured onto the PDA medium, cornmeal agar (CMA) plates (CMA: 20 g of cornmeal, 18 g of agar, and 1,000 mL of distilled water), and synthetic nutrient-poor agar (SNA) plates (SNA: 1 g of KH2PO4, 1 g of KNO3, 0.5 g of MgSO4, 0.5 g of KCl, 0.2 g of glucose, 0.2 g of sucrose, 18 g of agar, and 1,000 mL of distilled water). The plates were incubated at 25°C, 30°C, and 35°C under alternating 12 h light and 12 h darkness conditions. The colony radius was recorded after 3 days, and the time taken for complete colony coverage was noted. Microscopic observations of hyphae, conidiophores, phialides, conidia, and other microscopic structures were performed using an Olympus BX51 microscope (Tokyo, Japan) connected to a DP controller digital camera. At least 30 datasets were measured for each structure. Colonies were photographed after 7 days of production, and conidia were photographed after 14 days of production.
The 0.5 g fungal mycelia were collected and transferred into a 1.5 mL microcentrifuge tube with a 0.7–0.8 mL of lysis buffer [7 mol/L Urea, 50 mmol/L Tris-HCl, 62.5 mmol/L NaCl, 10 g/L Sodium Dodecyl Sulfate (SDS)]. The mixture was centrifuged at 12,000 r/min for 5 min, and the aqueous phase was transferred into a new 1.5 mL tube. An equal volume of the DNA extract (phenol/chloroform/isoamyl alcohol, 25:24:1) was added to the homogenates. The mixture was centrifuged at 12,000 r/min for 5 min, and the aqueous phase was transferred into a new 1.5 mL tube. The homogenates containing DNA were re-extracted by adding an equal volume of isopropanol and 1/10 volume of 3 mol/L NaAc. The mixture was placed at −20°C for 20 min and then centrifuged at 12,000 r/min for 5 min. The aqueous phase was then discarded. The DNA pellet was washed twice with 70% ethanol (70 mL of ethanol was diluted with water to obtain a total volume of 100 mL) in order to precipitate the pellet further. Afterward, it was dried and resuspended in 50 μL of H2O for PCR analysis (Sun et al., 2000; Liu et al., 2005). Fragments of ITS, rpb2, and tef1 were amplified with three primer pairs: ITS4 and ITS5 for ITS (White et al., 1990), frpb2-5f and frpb2-7cr for rpb2 (Liu et al., 1999), and EF1-728F (Carbone and Kohn, 1999) and TEF1LLErev (Jaklitsch et al., 2005) for tef1. A 25 μL reaction volume contained 1.0 μL of DNA template, 1.0 μL of each forward and reverse primer, 12.5 μL of 2 × MasterMix (Tiangen Biotech), and 9.5 μL of ddH2O. The PCR thermal cycle programs are shown in Table 1. PCR products were purified using the PCR product purification kit (Biocolor Bioscience and Technology Co., Shanghai, China), and forward and reverse sequencing was carried out on an ABI 3730 XL DNA sequencer (Applied Biosystems, Foster City, California) with the primers used during the PCR amplification. The sequences were deposited in the GenBank database at the National Center for Biotechnology Information (NCBI). The accession numbers are listed in Table 2.
Preliminary Basic Local Alignment Search Tool (BLAST) searches using ITS sequences of isolates against NCBI and UNITE databases have identified both the known species and suspected new species. The suspected new species were subjected to a BLAST search from GenBank, and the top three species in terms of similarity were selected as reference strains, preferably including the type strain of each species. The results indicated that the rpb2 similarity failed to meet the ≥99% standard, and the tef1 similarity failed to meet the ≥97% standard (Table 3). The rpb2 and tef1 sequences did not conform to the standards for the known species. This discrepancy indicates that further analysis is required to determine the systematic position of the suspected new species (Cai et al., 2022).
The research revealed that the new species belonged to three clades: the Spirale clade, the Virens clade, and the Koningii clade. We selected all strains reported in recent years within the Spirale clade as reference strains. For the Virens clade, we selected the main species of this clade as reference strains. In addition, the Helicum and Strictipile clades were selected as secondary references because the identification of some strains was ambiguous. For the Koningii clade, strains with a high sequence similarity were selected as reference strains. Moreover, the main strains of the Atroviride clade were selected as reference strains. Protocrea farinosa Berk & Broome (CBS 121551) and P. pallida Ellis & Everh (CBS 299.78) were used as outgroups.
Sequence alignment was performed using ClustalX 1.83. Based on the alignment, the sequence was trimmed to the appropriate length using MEGA11. Appending was performed using BioEdit v.7.0. The three genes were merged into a single sequence for subsequent operations. The combined sequence matrix (Fasta file) contained 2,775 characters from each gene (561 from ITS, 932 from rpb2, and 1,282 from tef1). The alignment data from phylogenetic analyses were deposited in TreeBASE (http://purl.org/phylo/treebase/phylows/study/TB2:S31222).
The systematic position of the new species was determined using Maximum Likelihood (ML) and Bayesian Inference (BI) analyses. The ML analysis was conducted using Iqtree. The command “iqtree -s example.fas -m MF -nt AUTO” was used to select the best model and generate the output. The best model used in this study was TIM2e+R9. Bootstrap values were calculated from 1,000 replicates. After setting the outgroup, Iqtree was run to complete the ML analysis. Bayesian trees were constructed using MrBayes v3.1.2, with the best model chosen through MrModeltest 2.3. The corresponding NEXUS file was prepared, Metropolis-coupled Markov Chain Monte Carlo (MCMC) was initiated for 1,000,000 generations, and every 100 generations were sampled to achieve a final average standard deviation of split frequencies below 0.01. The first 25% of the MCMC samples were discarded, and the remaining data were used to calculate posterior probabilities for Bayesian trees. The trees were visualized in FigTree v1.4, with branches showing Maximum Likelihood Bootstrap Proportions (MLBPs) above 70 and Bayesian Inference Posterior Probabilities (BIPPs) above 0.85.
Healthy, fresh Gastrodia elata tubers were selected for pathogenicity testing of the strains. Three type strains of the new species (YMF 1.08101, YMF 1.08102, and YMF 1.07894) and two type strains of the known species with a high isolation frequency (T. hamatum D5-13-3-2, T. atroviride D5-2-2, and T. harzianum J2-10-2) were selected. The Gastrodia elata tubers were surface-sterilized, according to the method described in Section 2.2, and then dried on a sterile filter paper. Then, each tuber was placed in a Petri dish, measuring 15 cm in diameter and 3 cm in depth, the filter paper sheet was soaked in 10% glycerin at the bottom of the Petri dish to maintain humidity. A U-shaped tube was placed on the filter paper. The healthy G. elata tubers were inoculated by placing mycelial plugs (5 mm in diameter), which were cut from the margins of actively growing colonies, into premade holes that were 4 mm in diameter and 2 mm in depth, with the mycelial side facing down on the tubers. Three replicate tubers were inoculated for each isolate, and non-colonized agar plugs were used as negative controls. The Petri dishes were sealed with a plastic wrap, incubated at 25°C, and were observed periodically. After 12 days, pathogens were isolated and identified from each G. elata tuber with symptomatic lesions. The aggressiveness of each isolate was evaluated based on the area of the cross-section of the infected site on a continuous scale of 0 to 3, in which 0 indicated no visible lesions, 1 indicated the area of disease lesions up to 1.0 cm2, 2 indicated the area of disease lesions from 1.1 to 2.0 cm2, and 3 indicated the area of disease lesions >2.0 cm2.
In this study, 93 strains of Trichoderma were isolated and purified from the diseased tubers of G. elata. Among these strains, 85 were identified as the known species and 8 were defined as potential new species based on the BLAST search results of the ITS sequence.
Phylogenetic analyses inferred from the ITS sequence were conducted to identify the known species, which included nine species, namely, T. hamatum, T. atroviride P. Karst, T. harzianum, T. crassum Bissett, T. lixii Chaverri, T. longipile Bissett, T. spirale Bissett, T. koningii Oudem., and T. scalesiae Samuels & H.C. Evans (Supplementary Figure 1). The highest isolation frequency was observed in T. hamatum, reaching 35.87%. The isolation frequencies of the remaining species were as follows: 18.48% for T. atroviride, 17.39% for T. harzianum, 9.78% for T. crassum, 5.43% for T. lixii, 2.17% for T. longipile, and 1.09% for T. spirale, T. koningii, and T. scalesiae (Supplementary Table 1).
The multi-gene analysis involving ITS, rpb2, and tef1 provided evidence for the identification of the new species. The ML analysis demonstrated a similar tree topology, which was consistent with the results obtained from BI analyses (Figure 1). The tree topology exhibited four new species: T. delicatum (MLBP/BIPP = 100/1.00), T. robustum (MLBP/BIPP = 100/1.00), T. perfasciculatum (MLBP/BIPP = 100/1.00), and T. subulatum (MLBP/BIPP = 100/1.00). T. delicatum belonged to the Koningii clade (MLBP/BIPP = 99/1.00). T. robustum and T. perfasciculatum belonged to the Virens clade (MLBP/BIPP = 85/1.00). T. subulatum was located in the Spirale clade (MLBP/BIPP = 100/1.00). T. delicatum was related to T. dorothopsis A.A. Tomah and J.Z. Zhang, and T. texanum Montoya. T. robustum and T. perfasciculatum were grouped together and associated with T. crassum, T. neocrassum Samuels, and T. virens Arx. T. subulatum was related to T. variegatum Z.F. Yu and T.T. Jing, T. hunanense K. Chen and W.Y. Zhuang, and T. longisporum K. Chen and W.Y. Zhuang.
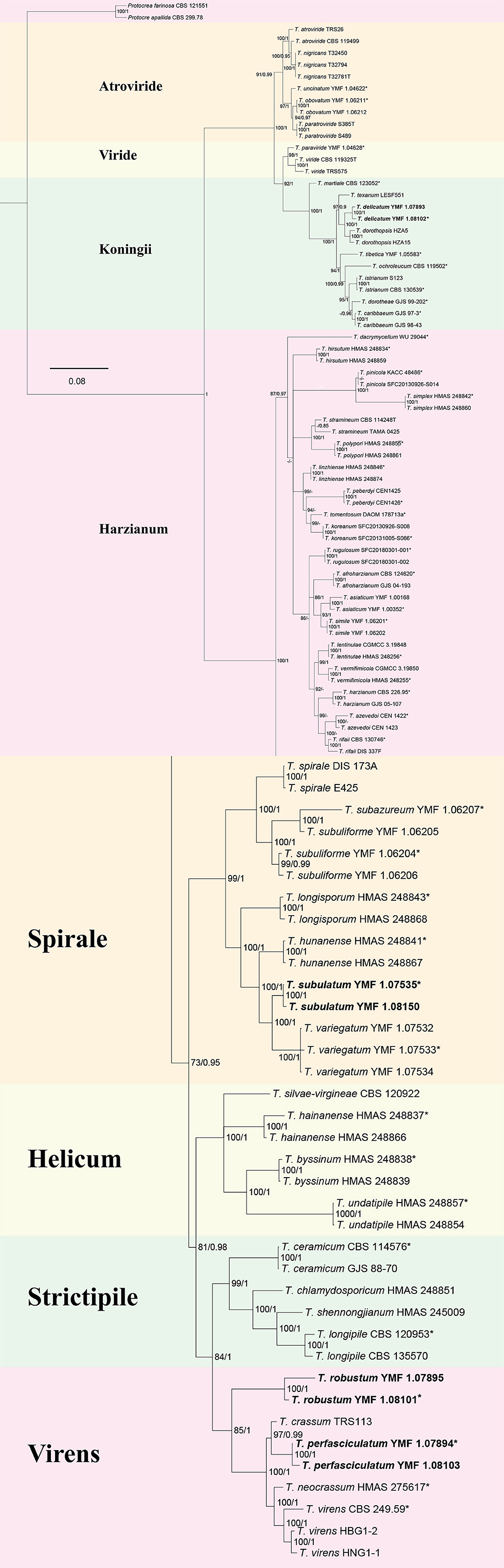
Figure 1. The phylogenetic tree of Trichoderma species based on the combined ITS, tef1, and rpb2 gene sequences constructed using the Maximum Likelihood (ML) analysis and Bayesian inference (BI) analysis. The former value near the nodes represents bootstrap support from ML bootstrap over 70%, and the latter value represents Bayesian posterior probabilities over 0.85. Protocrea farinose CBS 121551 and P. pallida CBS 299.78 were used as outgroups. Bold font indicates newly described species.
Trichoderma delicatum Z.F. Yu & C.W. Ye, sp. nov. Figure 2
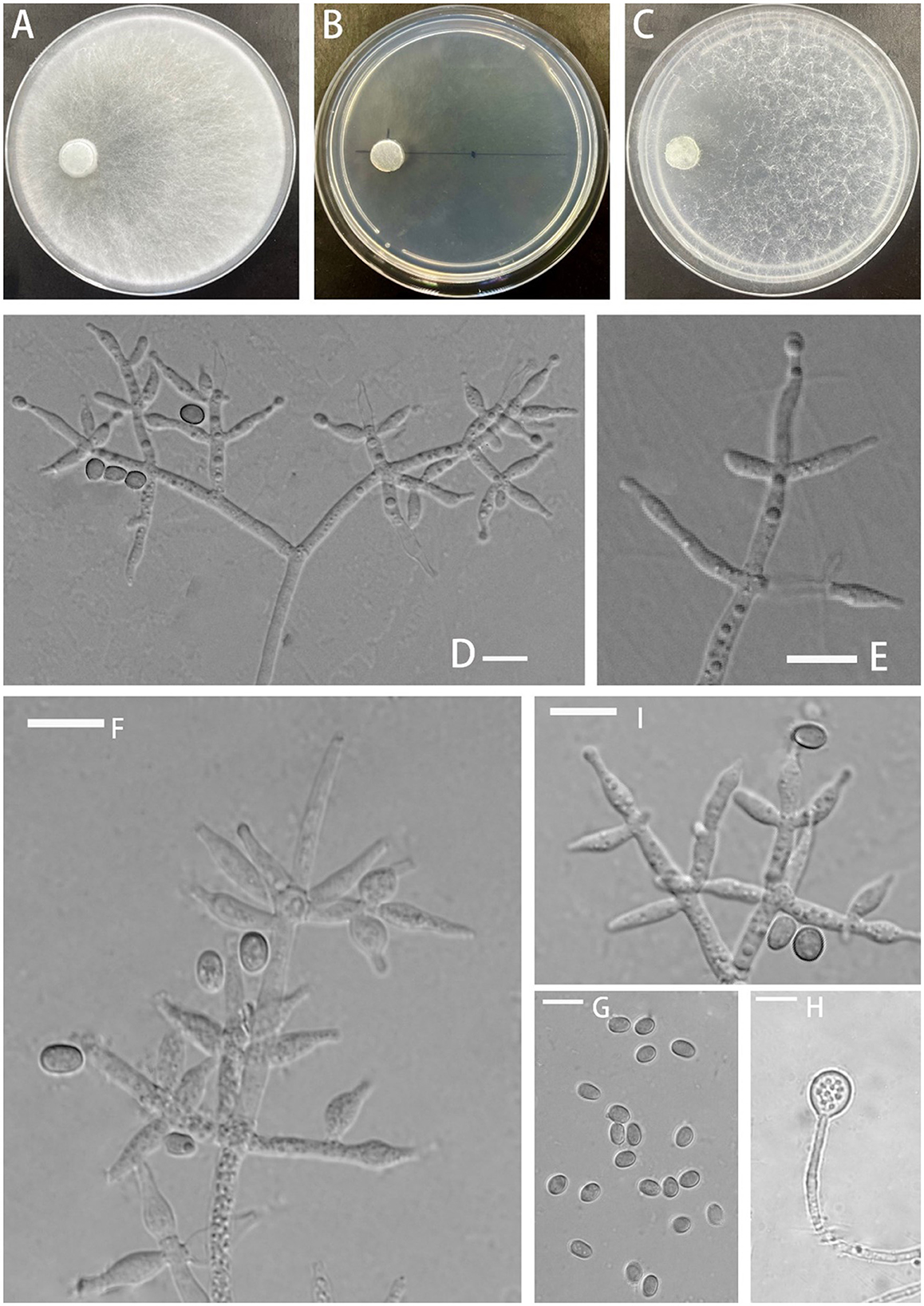
Figure 2. Morphology of Trichoderma delicatum (YMF 1.08102). (A–C) cultures on PDA plates, 7d; CMA plates, 7d; SNA plates, 7d; 25°C; (D–F, I) conidiophores and phialides; (G) conidia; and (H) chlamydospores. Scale bars: 10 μm (D–I).
MycoBank No: 853529
Etymology: Latin, delicatum, meaning nice, elegant, fine, slender and referring to conidiophores and conidiogenous cells.
Description: The sexual morph of the species is unknown. The asexual morph features conidiophores that are tree-shaped, with a prominent main axis. The branches are irregularly arranged around the main axis, usually including secondary branches. These branches are oriented randomly. Phialide are either ampulliform or narrowly vase-shaped, measuring (−10)10.4–13.0(15.3–) × (−3.4)4.1–4.8(5.4–) μm with a l/w ratio of (−2)2.3–3(3.9–). They are (−2.1)2.4–3.2(3.6–) μm wide at the base and are widest around the middle. Oblong or ellipsoid conidia are green and smooth, measuring (−5.0)5.2–6.0(6.5–) × (−3.3)3.7–4.6(5.4–) μm, with a l/w ratio of (−1.0)1.2–1.5(1.7–). Chlamydospores were observed growing at the tip of hyphae, they are spherical, measuring 8.1–8.9 × 7.3–9.3(−8.6) μm, with a l/w ratio of 1.0–1.2.
Culture characteristics: The optimum temperature for growth was 25°C.
Colony radius on the PDA plate after 72 h: 56 mm at 25°C, 53 mm at 30°C, and 25 mm at 35°C, covering the plate after 3.2 days at 25°C. There was a similar growth rate on the PDA plate at 25°C and 30°C. The colony slightly stratified and radial, with abundant aerial hyphae. Thea peripheral hyphae are more numerous than those in center, creating a plush, white appearance. The conidiation time is long. No distinct odor noted.
Colony radius on the CMA plate after 72 h: 26 mm at 25°C, 25 mm at 30°C, and 9 mm at 35°C, covering the plate after 7 days at 25°C. Colony hyaline, aerial hyphae lacking. Conidiation formed after 5.5 days. A little blister-like structures randomly distributed. No distinct odor noted.
Colony radius on the SNA plate after 72 h: 40 mm at 25°C, 29 mm at 30°C, and 20 mm at 35°C, covering the plate after 5.5 days at 25°C. Colony transparent, no zonate, aerial hyphae sparse, abundant at margin, arachnoid. Conidiation started after 5 days, the white mycelium gradually turned green. No obvious odor noted.
Materials examined: China, Yunnan Province, Zhaotong City, Yiliang County, Xiaocaoba Town, on diseased Gastrodia elata, 25 Oct 2021, T.T. Jing (holotype YMF 1.08102). lbid. (cultures: YMF 1.07893).
Notes: Phylogenetically, T. delicatum is associated with T. dorothopsis and T. texanum. T. delicatum can be distinguished based on the size of the spores. The spores of T. dorothopsis and T. texanum are spherical or nearly spherical, with dimensions ranging between 3 and 4 μm (T. dorothopsis: (−3.2)3.3–3.9(4.2–) × (−2.9)3.1–3.5(3.6–) μm, T. texanum: 1–4 × 1.8–3.1 μm) (Quimi et al., 2016; Ali et al., 2020). In contrast, the spores of T. delicatum are ellipsoidal, with a l/w ratio of 1.2–1.5. T. delicatum exhibits the fastest growth rate at 25°C, with growth restricted at 30°C and 35°C. T. dorothopsis maintains an optimal growth rate at 27–30°C (Ali et al., 2020). The preferred growth temperature for T. texanum on the PDA plate is 25°C, while on CMA and the SNA plate, it is 30°C (Quimi et al., 2016). Sporulation of T. delicatum is challenging to observe on the PDA plate, and it takes approximately 5.5 days to observe sporulation on the CMA plate. In comparison, T. dorothopsis shows sporulation after 4 days on the PDA plate (Ali et al., 2020). Additionally, T. texanum exhibits pigment production on the PDA plate, but the phenomenon is not observed in T. delicatum and T. dorothopsis (Quimi et al., 2016; Ali et al., 2020). Chlamydospores are observable in T. delicatum on the PDA plate, and this structure is slightly larger than that of T. texanum (Quimi et al., 2016).
Trichoderma robustum Z.F. Yu & C.W. Ye, sp. nov. Figure 3
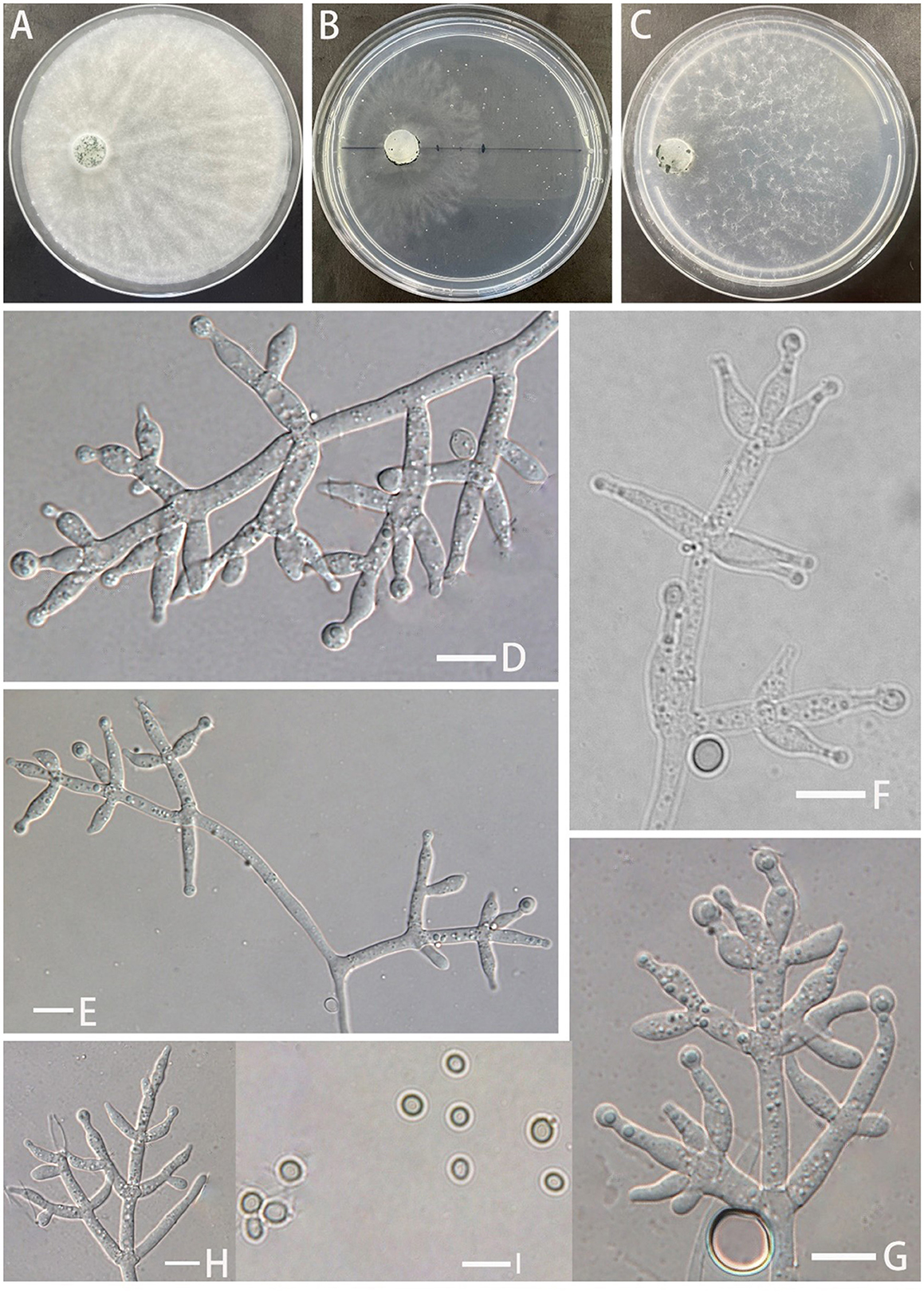
Figure 3. Morphology of Trichoderma robustum (YMF 1.08101). (A–C) cultures on PDA plates, 7d; CMA plates, 7d; SNA plates, 7d; (D–H) conidiophores and phialides; and (I) conidia. Scale bars: 10 μm (D–I).
MycoBank No: 853530
Etymology: Latin, robustum, referring to having or exhibiting strength or vigorous conidiophores and conidiogenous cells.
Description: Sexual morph: Unknown. Asexual morph: Conidiophores with a main axis, mostly secondary branches, tertiary branches also observed, branches asymmetrically distributed on both sides of the main axis toward the tip. Pedicels ampulliform or flask–shaped, 2–3 whorls arising from the ends of the branches, (−8.7)10.1–13.7(15.2–) × (2.5–)2.9–3.5(−3.8) μm, l/w ratio (−2.5)3.0–4.2(5.2–), (−1.9)2.1–2.6(2.8–) μm wide at the base, widest around the middle. Conidia ovate or suborbicular, green, smooth (−4.1)4.3–4.6(4.8–) × (−3.7)4.0–4.3(4.5–) μm, l/w ratio 1.0–1.1(1.3–). No chlamydospores were observed.
Culture characteristics: Optimum temperature for growth 25°C.
Colony radius on the PDA plate after 72 h: 36 mm at 25°C, 12 mm at 30°C, and 9 mm at 35°C, covering the plate after 3.8 days at 25°C. Colony slightly stratified, radial, aerial hyphae abundant, plush, and white. The colony producing a green dotted conidiation area from the center outward. No distinct odor noted.
Colony radius on the CMA plate after 72 h: 16 mm at 25°C, 9 mm at 30°C, and 7 mm at 35°C. Colony translucent, aerial hyphae nearly lacking, slow growth, fuzzy stratification. Conidiation starting after 6 days. No distinct odor noted.
Colony radius on the SNA plate after 72 h: 10 mm at 25°C, 8 mm at 30°C, and 5 mm at 35°C, covering the plate after 3.8 days at 25°C. Colony translucent, no ring band, aerial hyphae sparse, flocculent. Conidiation starting after 4 days, green blister-like dots scattered around the plate. No distinct odor noted.
Materials examined: China, Yunnan Province, Zhaotong City, Yiliang County, Xiaocaoba Town, on diseased Gastrodia elata, 25 Oct 2021, T.T. Jing (holotype YMF 1.08101). lbid. (cultures: YMF 1.07895).
Notes: T. robustum is phylogenetically associated with T. crassum, T. virens, and T. neocrassum. Morphologically, T. robustum exhibits a prominent main axis with asymmetrical branches distributed on both sides of the main axis, but the branches display uniform intervals. Two to three whorls of phialides arise at the end of each branch, resembling T. crassum (2–4 whorls) (Chaverri et al., 2003). The sizes of the phialides in T. robustum are comparable to those of T. crassum (13.5–15.7 × 4.3–4.6 μm) and are larger than those of T. virens (7.9–9.9 × 3.8–4.5 μm) and T. neocrassum (7–22 × 2.5–4 μm). Sporulation of T. robustum is not observed on PDA plates. In comparison to T. crassum, T. virens, and T. neocrassum, T. robustum exhibits a significantly slower growth rate (Chaverri et al., 2001, 2003; Zhang and Zhuang, 2019). T. robustum is similar to T. crissum, which cannot grow at 35°C. However, T. robustum shows faster growth on PDA plates than on SNA plates, while the opposite is observed for T. crassum (Chaverri et al., 2003). T. robustum cannot produce pigments, which distinguishes it from T. crassum, T. virens, and T. neocrassum. The spores of T. robustum, unlike those of T. crassum and T. neocrassum, are round or nearly round, resembling those of T. virens (Chaverri et al., 2001, 2003; Zhang and Zhuang, 2019).
Trichoderma perfasciculatum Z.F. Yu & C.W. Ye, sp. nov. Figure 4
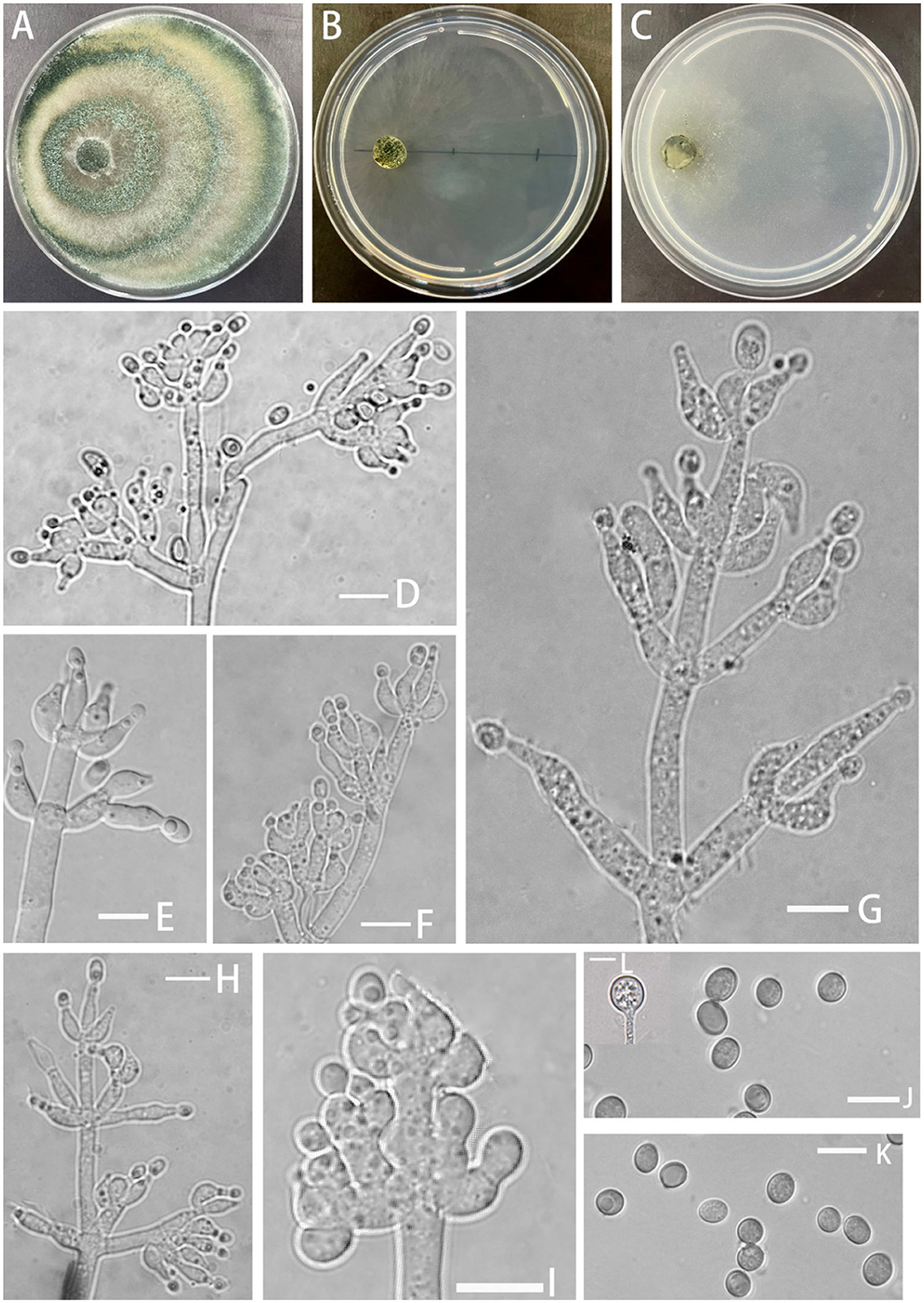
Figure 4. Morphology of Trichoderma perfasciculatum (YMF 1.07894). (A–C) cultures on PDA plates, 7d; CMA plates, 7d; SNA plates, 7d; (D–I) conidiophores and phialides; (J, K) conidia; and (L) chlamydospores. Scale bars: 10 μm (D–L).
MycoBank No: 853531
Etymology: Latin, perfasciculatum, per- meaning very, completely + -fasciculatum referring to the conidiogenous cells growing and arranging in the fascicles.
Description: Sexual morph: unknown. Asexual morph: Conidiophores penicillium-like, mostly asymmetrically arranged, main axis conspicuous, with secondary branches, oriented toward the main axis, 2–4 bottlenecks in whorls, clustered. Pedicels ampulla-shaped or narrow ampulla-shaped, partly curved (−10)10.4–13.0(15.3–) × (−3.4)4.1–4.8(5.4–) μm, l/w ratio (−2)2.3–3(3.9–), (−2.1)2.4–3.2(3.6–) μm wide at the base, widest at middle. Conidia ellipsoid, green, and smooth (−5.0)5.2–6.0(6.5–) × (−3.3)3.7–4.6(5.4–) μm, l/w ratio 1.2–1.5. Chlamydospore noted, rounded, terminal, 8.1–8.9 × 7.3–9.3 μm, l/w ratio 1.0–1.2.
Culture characteristics: Optimum temperature for growth 25°C.
Colony radius on the PDA plate after 72 h: 54 mm at 25°C, 39 mm at 30°C, and 12 mm at 35°C, covering the plate after 3.8 days at 25°C. Colony dense, white, turning green after conidiation on the 3rd day, producing spores at a fast rate, with obvious circular annular bands, aerial hyphae abundant. No obvious odor noted. Yellow pigment noted.
Colony radius on the CMA plate after 72 h: 25 mm at 25°C, 11 mm at 30°C, and 11 mm at 35°C, covering the plate after 8 days. Colonies were hyaline, radial, mycelium was lacking, slow growing, and stratification not obvious. No distinct odor noted, no chlamydospore observed.
Colony radius on the SNA plate after 72 h: 34 mm at 25°C, 24 mm at 30°C, and 14 mm at 35°C, covering the plate after 4 days at 25°C. Colonies translucent, radial, no ring band, aerial hyphae loose, conidiation starting after 6 days. No obvious odor noted.
Materials examined: China, Yunnan Province, Zhaotong City, Yiliang County, Xiaocaoba Town, on diseased Gastrodia elata, 25 Oct 2021, T.T. Jing (holotype YMF 1.07895). lbid. (cultures: YMF 1.08103).
Notes: From a systematic perspective, T. perfasciculatum is closely related to T. crassum and associated with T. virens and T. neocrassum. The growth rate of T. perfasciculatum on PDA plates at 25°C is significantly higher than that of T. crassum (35–45 μm), T. virens (approximately 28μm), and T. neocrassum (32–34 μm) within the same clade (Chaverri et al., 2001, 2003; Zhang and Zhuang, 2019). T. perfasciculatum and T. crassum cannot grow at 35°C, while T. virens exhibits the opposite behavior. However, the growth rate of T. perfasciculatum on SNA plates is lower than that on PDA plates, and T. crassum and T. virens present stronger adaptation to SNA plates (Chaverri et al., 2001, 2003). Circular annular bands can be observed in T. perfasciculatum and T. neocrassum (Zhang and Zhuang, 2019). All four species, T. perfasciculatum, T. crassum, T. virens, and T. neocrassum, can produce yellow pigments. The phialides of T. perfasciculatum and T. crassum are more irregular than those of T. virens, but all exhibit 2–4 whorls arising from the apex. The phialides of T. crassum (l/w radio 3–3.6) are narrower than those of T. perfasciculatum. T. perfasciculatum, T. crassum (3.7–5.3 × 2.6–3.7 μm, l/w radio 1.4), and T. neocrassum (4.5–8 × 3.5–5(7–) μm, l/w radio 1.1–1.6) produce ellipsoidal spores, while the spores of T. virens (4.2–4.9 × 3.8–4.2 μm) are smaller and more rounded (Chaverri et al., 2001, 2003; Zhang and Zhuang, 2019).
Trichoderma subulatum Z.F. Yu & T.T. Jing, sp. nov. Figure 5
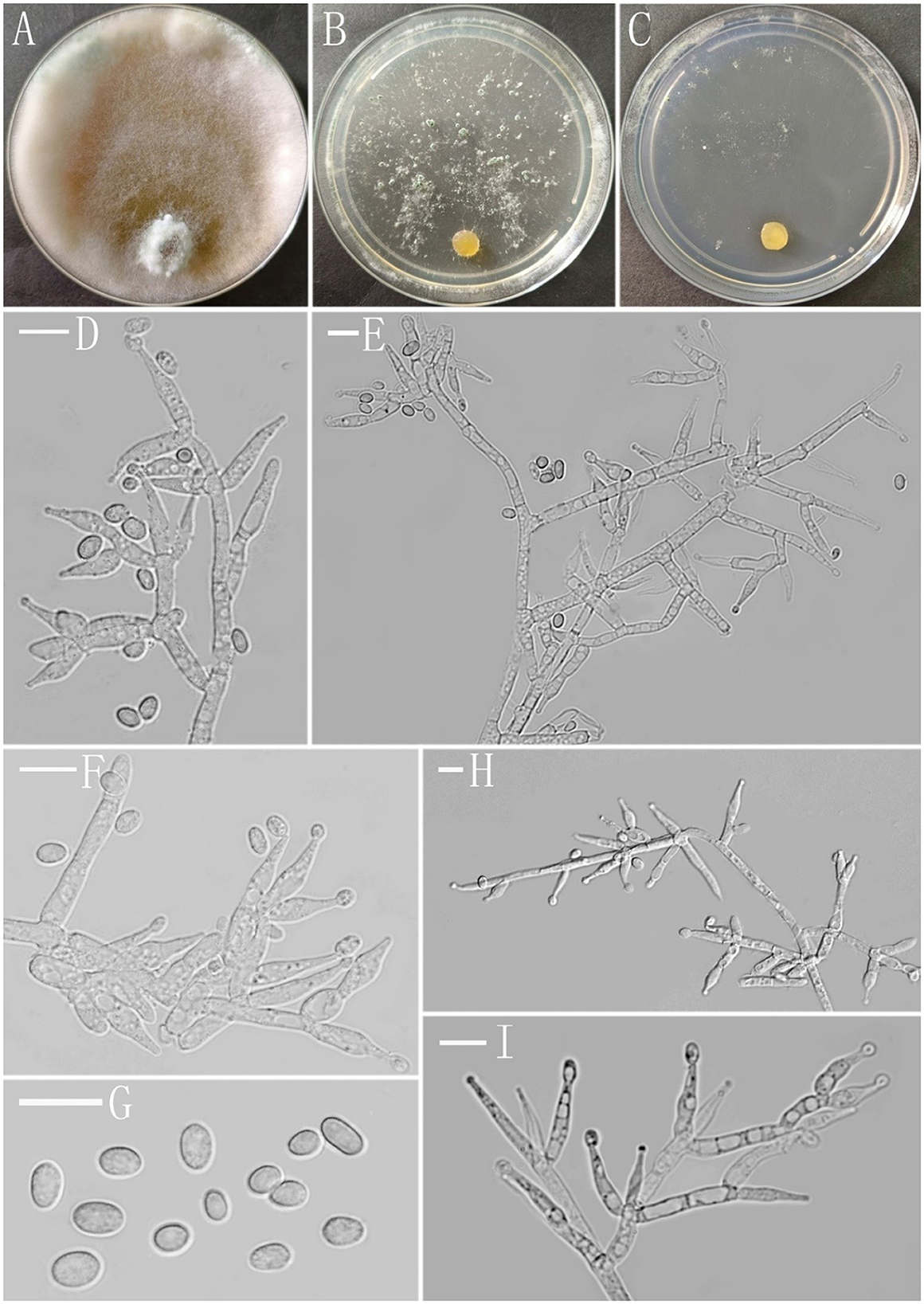
Figure 5. Morphology of Trichoderma subulatum (YMF 1.07835). (A–C) cultures on PDA plates, 7d; SNA plates, 7d; CMA plates, 7d; (D–F, H, I) conidiophores and phialides; and (G) conidia. Scale bars: 10 μm (D–I).
MycoBank No: 853532
Etymology: Latin, subulatum, referring to subulate phialides.
Description: Sexual morph: Unknown. Asexual morph: Conidiophores asymmetry, often with a main axis, mostly irregularly branched, the terminus bearing phialides, branches often tended toward the conidiophore terminus, rebranching one to three times, the distance between branches relatively large, each branch terminating in a whorl of 2–3 phialides. Phialides subulate often asymmetric, straight or slightly curved, constricted below the tip forming a narrow neck, solitary or in whorls of 2–3(−4), (−7.4)9.4–15.7(16.4–) × (−3.0)3.4–5.1(5.9–) μm, l/w ratio (−1.7)2.1–4.4(4.9–), 2.0–3.3(−3.6) μm wide at the base, widest around the middle. Conidia ellipsoidal to oblong, less globose, green, smooth, (−3.6)4.0–6.1(6.4–) × (−2.7)2.9–3.4(3.6–) μm, l/w ratio (−1.2)1.5–1.9(2.1–).
Culture characteristics: Optimum temperature for growth 25°C. Very fast growing colony.
Colony radius on PDA plates after 72 h: 46–47 mm at 25°C, 40–42 mm at 30°C, and 16 mm after 9 days at 35°C, covering the plate after 4 days at 25°C. Colony dense, zonate indistinctly, aerial hyphae abundant, cotton-like, more abundant near the original inoculum and around the periphery of the colony. Conidiation starting after 6 days, formed numerous on aerial hyphae, green. Chlamydospores common, subglobose to globose, smooth, terminal and intercalary, 5.7–10.4 × 5.7–9.2 μm. A distinct odor noted, and yellow pigment noted.
Colony radius on the CMA plate after 72 h: 27–30 mm at 25°C, 25–27 mm at 30°C, and 5 mm after 9 days at 35°C, covering the plate after 6 days at 25°C. Colony homogenous, hyaline, radial, not zonate, aerial hyphae sparse. Conidiation starting after 7 days in pustules, not common, appearing at the colony margin, hemispherical, first white, turning grayish green after 8 days. No chlamydospores observed. A distinct odor noted, and light yellow pigment noted.
Colony radius on the SNA plate after 72 h: 33–35 mm at 25°C, 30–32 mm at 30°C, and 12 mm after 9 days at 35°C, covering the plate at 25°C after 7 days. Colony hyaline, indistinctly zonate, mycelium loose, aerial hyphae floccus. Conidiation formed in pustules after 4 days, pustules appearing around the periphery of the colony, hemispherical, first white, turning deep green after 5 days. No chlamydospores observed. A distinct odor noted, light yellow pigment noted.
Materials examined: China, Yunnan Province, Zhaotong City, Yiliang County, Xiaocaoba Town, on diseased Gastrodia elata, 25 Oct 2021, T.T. Jing (holotype YMF 1.07535). lbid. (cultures: YMF 1.08150).
Notes: Phylogenetically, T. subulatum is closely related to T. hunanense and T. variegatum in the Spirale clade. Compared with the new species, T. hunanense produces narrower phialides (3.1–3.9 μm) and shorter and wider conidia [(−3.6)4.2–5.6(7.5–) × 3.1–3.9 μm], forming distinct zonate of pustules on SNA plates (Chen and Zhuang, 2017). T. variegatum differs in its longer and wider conidia [(−4.3)4.7–7.4(8.6–) × (−2.7)3.0–4.1(4.3–) μm] and exhibits a lower growth rate on CMA plates (20–22 mm), PDA plates (30–32 mm), and SNA plates (18–22 mm) (Ye et al., 2023). T. subulatum, T. hunanense, and T. variegatum produce yellow pigment in PDA plates; however, yellow pigment can also be observed in CMA and SNA plates from T. variegatum. T. subulatum produces a distinct odor in culture, which is not noted in T. hunanense and T. variegatum (Chen and Zhuang, 2017; Ye et al., 2023).
After 12 days, the inoculated tubers exhibited varying degrees of infection, while all uninoculated controls remained healthy (Figure 6). The black rot lesions were confined to the area around the inoculation site and did not spread to the area around the uninoculated site. The cross-sections of the inoculation sites on the G. elata tubers also demonstrated infection. Dark brown and dry rot areas formed on the superficial skin of the G. elata tubers near the inoculation points. The strains that were reisolated from symptomatic tissues were morphologically and phylogenetically identical to the test strains.
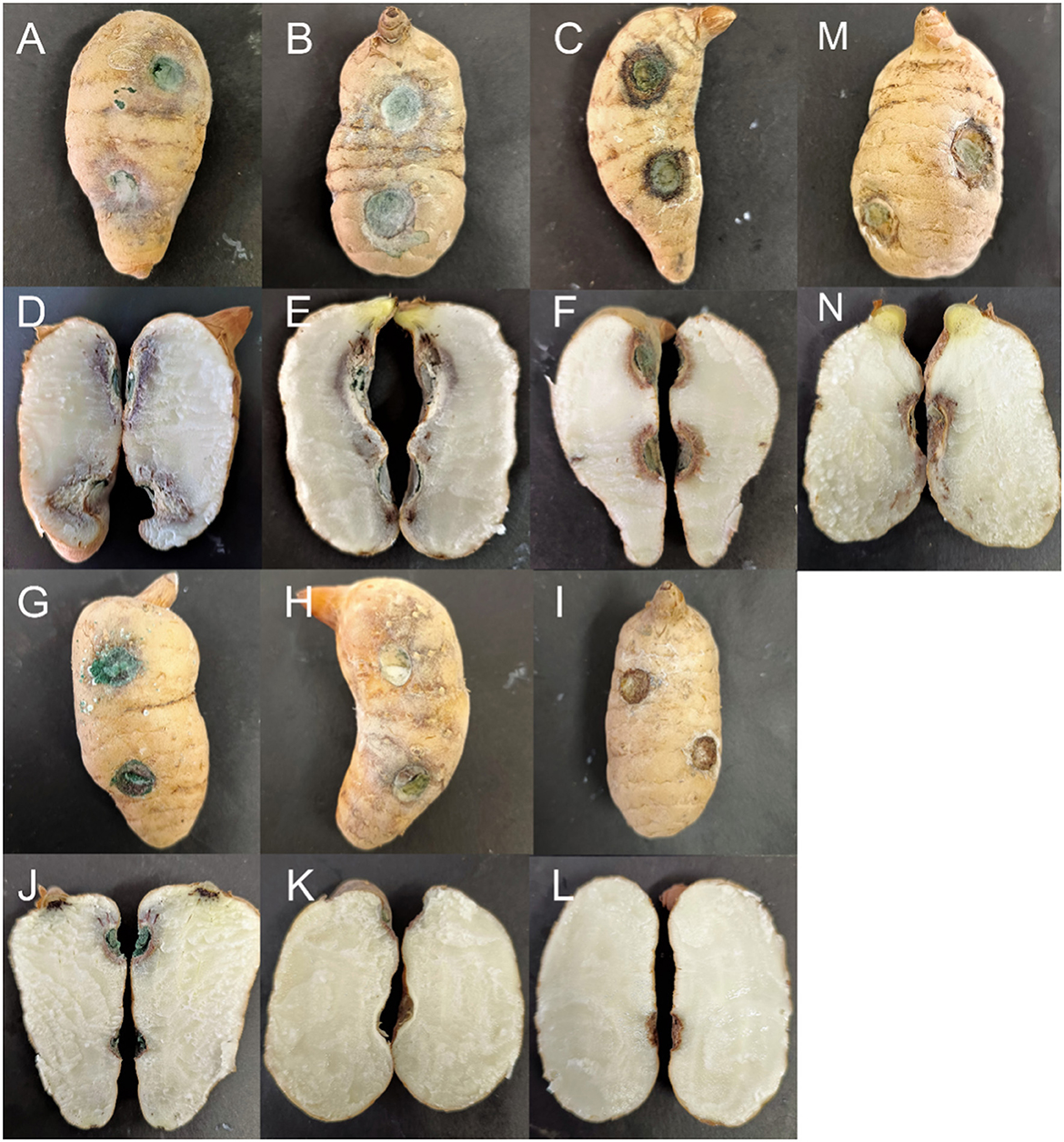
Figure 6. Symptoms of Gastrodia elata after its inoculation with testing strains. (A, D) Trichoderma robustum YMF 1.08101; (B, E) Trichoderma perfasciculatum YMF 1.07894; (C, F) Trichoderma hamatum D5-13-3-2; (G, J) Trichoderma atroviride D5-2-2; (H, K) Trichoderma delicatum YMF 1.08102; (I, L) Control; and (M, N) Trichoderma harzianum J2-10-2.
The results indicated that the infection abilities of different Trichoderma species varied. T. robustum (YMF 1.08101) and T. perfasciculatum (YMF 1.07894) caused the most severe symptoms, spreading from the inoculation site into the interior of the Gastrodia elata. The border between the diseased and healthy tissues appeared purple, and the completely diseased areas near the cortex appeared white and tofu-like. The aggressiveness of these two species reached a scale of 3. T. hamatum (D5-13-3-2) appeared moderately pathogenic. The epidermis around the inoculation site darkened, and the mycelium invaded the cortex to form brown spots. The aggressiveness reached a scale of 2. T. atroviride (D5-2-2) and T. harzianum (J2-10-2) demonstrated weak infection ability, with lesions spreading only to the superficial layers. The aggressiveness reached a scale of 1. G. elata inoculated with T. delicatum (YMF 1.08102) exhibited no obvious symptoms either on the epidermis or internally. The aggressiveness reached a scale of 0 (Table 4).
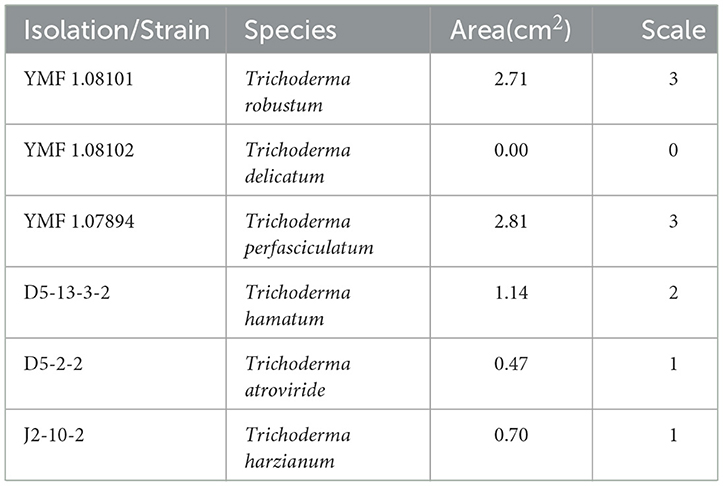
Table 4. Pathogenicity test results of Trichoderma strains isolated from Gastrodia elata affected by the black rot disease.
Since Wu and Ma (1998) reported that Trichoderma could affect the growth of Gastrodia elata by parasitizing Armillariella mellea, T. viride, and T. hamatum were successively reported as pathogenic factors of G. elata (Zeng et al., 2007; Han et al., 2017; Zhang et al., 2020). Previously, we also reported that T. albidum and T. variegatum were isolated from the rot tubers of G. elata (Ye et al., 2023). In this study, we reported that 13 species of Trichoderma, including T. atorviride, T. crissum, and others, were connected with the black rot disease of G. elata. Trichoderma demonstrated the highest separation frequency of 36.61%. However, it was not detected in an analysis of the association between the brown rot tuber of G. elata and soil microbial communities by high—throughput sequencing, in which Ilyonectria P. Chaverri & Salgado was the first dominant genus. The genus Ilyonectria ranked third with a frequency of isolation of 17.72% (Tang et al., 2020), which may be attributed to variations in methods or regions.
The four new species belonged to the Koningii clade, the Virens clade, and the Spirale clade. The Koningii clade was first proposed as an “aggregate” and was later proposed by researchers as a subclade in the Trichoderma sect. (Rifai, 1969; Bissett, 1991). Lieckfeldt et al. (1998) introduced the T. koningii aggregate species on SNA plates, which were characterized by long, narrow spores and slow growth rates. Samuels observed that species morphologically similar to T. koningii clustered on a branch in the phylogenetic tree, distinct from T. viride and T. atroviride (Samuels et al., 2006). Jaklitsch and Voglmayr (2015) named the Viride clade, corresponding to the previously defined Trichoderma sect. Qin and Zhuang (2016) reported seven new Trichoderma species, and the authors assigned them to different subclades under the Viride clade: Hamatum, Koningii, Neorufum, Rogersonii, Viride, and Viridescent. Zheng et al. (2021) identified two new species belonging to the Koningii clade. The spores of T. delicatum were narrower than those of T. dorothopsis and T. texanum. T. delicatum exhibited a slower growth rate on SNA plates. T. texanum exhibited pigment production on PDA plates, but the phenomenon was not observed in T. delicatum and T. dorothopsis.
The Virens clade and the Harzianum clade were previously demonstrated to be closely related. However, researchers now classify the Harzianum/Virens clade as two independent clades (Saravanakumar et al., 2016; Przylucka et al., 2017; Carmen et al., 2021). The representative species in the Virens clade is T. virens. Subsequent studies placed T. crassum and T. neocrassum into the Virens clade, expanding the diversity (Chaverri et al., 2003; Zhang and Zhuang, 2019). Later, Zheng et al. (2021) added T. inaequilaterale Z.F. Yu & Y.F. Lv to the Virens clade. Species in this clade generally exhibit faster growth rates and produce gliocladium-like conidiophores and green, smooth spores (Chaverri et al., 2003). The phialides of T. robustum and T. perfasciculatum are consistent with those of T. crassum and T. virens, displaying 2–3(−4) whorls arising at the end of the conidiophores (Chaverri et al., 2003). T. perfasciculatum exhibits the fastest growth rate in the Virens clade. Neither T. robustum nor T. perfasciculatum can grow at 35°C, which is consistent with T. crassum.
The Spirale clade was initially proposed by Chen and Zhuang (2017). Previous studies showed that T. spirale was a distinct terminal branch, but identification using only tef1 differed from that using rpb2 and tef1. Chaverri et al. (2003) found that T. spirale belonged to the Strictipile clade, while Jaklitsch (2009) found that it belonged to the Polysporum clade. Chen and Zhuang (2017) utilized multi-gene analysis composed of ITS, tef1, and rpb2 to discover that T. hunanense, T. longisporum, and T. spirale were closely related under statistical support. Zheng et al. (2021) later added T. subuliforme Z.F. Yu and Y.F. Lv, as well as T. subazureum Z.F. Yu and Y.F. Lv, into the Spirale clade. Recently, T. variegatum was discovered and grouped into the Spirale clade. Members of the Spirale clade generally form hairy pustules, produce yellow pigments in culture, and have more or less oblong conidia (Chen and Zhuang, 2017), which are typical characteristics of T. subulatum. This is the second time that Trichoderma in the Spirale clade has been isolated from the rot tuber of Gastrodia elata.
The fungal diseases affecting G. elata were diverse. Besides Trichoderma spp., other pathogens include Fusarium oxysporum, Poliota praecox and Botrytis cinerea (Zeng et al., 2007), and Ilyonectria spp. (Zhang et al., 2020; Qiao et al., 2024). All these studies only reported on pathogens from diseased G. elata and not on the interaction between pathogens and G. elata. This may be due to the limited distribution of G. elata. The cultivation and production of G. elata are mainly concentrated in Asian countries, such as China and Korea. In China, Yunnan and Guizhou are the main production areas. Pathogenicity tests revealed differences in the pathogenic capabilities of different Trichoderma species. T. robustum, T. perfasciculatum, T. hamatum, T. atroviride, and T. harzianum showed pathogenic effects on G. elata, indicating that Trichoderma is indeed a pathogen that causes black rot disease in G. elata. However, further research is required to understand how Trichoderma causes disease in G. elata and whether changes in the fungal community structure of the rhizosphere soil can affect the normal growth of G. elata. This study proposed a connection between Gastrodia elata, Trichoderma, and black rot disease. It highlighted the impact of Trichoderma on Gastrodia elata and provided a theoretical basis for future research.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
CY: Methodology, Formal analysis, Validation, Writing – review & editing, Writing – original draft. YY: Investigation, Writing – original draft. WL: Investigation, Writing – original draft. TJ: Investigation, Writing – original draft. MM: Conceptualization, Funding acquisition, Writing – review & editing. MQ: Conceptualization, Writing – review & editing. ZY: Conceptualization, Resources, Writing – review & editing
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financed by the National Natural Science Foundation Program of PR China (32170017, 32370017), the National Key R & D Program of China (2022YFD1400700), and the Scientific Research and Innovation Project of Postgraduate Students in the Academic Degree of Yunnan University (KC-23236624).
This work acknowledges the strain provided by the Microbial Library of the Germplasm Bank of Wild Species from Southwest China.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1420156/full#supplementary-material
Ali, A., Iman, L.i, and B. Zhang, J. (2020). A new species of Trichoderma and gliotoxin role: A new observation in enhancing biocontrol potential of T. virens against Phytophthora capsici on chili pepper. Biol. Control 145:104261. doi: 10.1016/j.biocontrol.2020.104261
Berney, C., Pawlowski, J., and Louisette, Z. (2000). Elongation factor 1–alpha sequences do not support an early divergence of the acoela. Mol. Biol. Evol. 17, 1032–1039. doi: 10.1093/oxfordjournals.molbev.a026384
Bissett, J. (1991). A revision of the genus Trichoderma. II. infrageneric classification. Canadian J. Botany 69, 2357–2372. doi: 10.1139/b91-297
Cai, F., Dou, K., Wang, P., Komal, C., Chen, J., and Druzhinina, I. (2022). “The current state of Trichoderma taxonomy and species identification,” in Advances in Trichoderma biology for agricultural applications (Cham: Springer International Publishing), 3–35. doi: 10.1007/978-3-030-91650-3_1
Carbone, I., and Kohn, L. (1999). A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553. doi: 10.2307/3761358
Carmen, D.el, Evans, H., Abreu, L., Macedo, D., Ndacnou, M., Kifle, B., and Barreto, R. (2021). New species and records of Trichoderma isolated as mycoparasites and endophytes from cultivated and wild coffee in Africa. Sci. Rep. 11:5671. doi: 10.1038/s41598-021-84111-1
Chaverri, P., Castlebury, L., Overton, B., and Samuels, G. (2003). Hypocrea/Trichoderma: species with conidiophore elongations and green conidia. Mycologia 95:1100. doi: 10.2307/3761915
Chaverri, P., Samuels, G., and Stewart, E. (2001). Hypocrea virens sp. nov., the Teleomorph of Trichoderma virens. Mycologia 93:1113. doi: 10.2307/3761672
Chen, J., Tian, S., Shu, X., Du, H., Li, N., and Wang, J. (2016). Extraction, characterization and immunological activity of polysaccharides from Rhizoma gastrodiae. Int. J. Mol. Sci. 17, 1011–1011. doi: 10.3390/ijms17071011
Chen, K., and Zhuang, W. (2017). Discovery from a large–scaled survey of Trichoderma in soil of China. Sci. Rep. 7:9090. doi: 10.1038/s41598-017-07807-3
Cheng, C., Yang, C., and Peng, K. (2012). Antagonism of Trichoderma harzianum ETS 323 on botrytis cinerea mycelium in culture conditions. Phytopathology 102, 1054–1063. doi: 10.1094/PHYTO-11-11-0315
Degenkolb, T., Hans, V., Kristian, F., Samuels, G., and Brueckner, H. (2008). Recent advances and future prospects in peptaibiotics, hydrophobin, and mycotoxin research, and their importance for chemotaxonomy of trichoderma and hypocrea. ChemInform 5, 671–680. doi: 10.1002/cbdv.200890064
Fan, Q., Chen, C., Lin, Y., Zhang, C., Liu, B., and Zhao, S. (2013). Fourier transform infrared (FT–IR) spectroscopy for discrimination of Rhizoma gastrodiae (Tianma) from different producing areas. J. Mol. Struct. 1051, 66–71. doi: 10.1016/j.molstruc.2013.07.039
Han, M., Mi, N., Lee, H., and Park, E. (2017). First report of soft rot associated with Trichoderma hamatum in Gastrodia elata. Plant Dis. 101:1048. doi: 10.1094/PDIS-11-16-1630-PDN
Jaklitsch, W. (2009). European species of Hypocrea Part I. The green–spored species. Stud. Mycol. 63, 1–91. doi: 10.3114/sim.2009.63.01
Jaklitsch, W., Komon, M., Kubicek, C., and Druzhinina, I. (2005). Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97, 1365–1378. doi: 10.1080/15572536.2006.11832743
Jaklitsch, W., Samuels, G., Adnan, I., and Hermann, V. (2013). Disentangling the Trichoderma viridescens complex. Persoonia 31, 112–146. doi: 10.3767/003158513X672234
Jaklitsch, W., and Voglmayr, H. (2015). Biodiversity of Trichoderma (Hypocreaceae) in southern Europe and macaronesia. Stud. Mycol. 80, 1–87. doi: 10.1016/j.simyco.2014.11.001
Kim, C., Park, M., Kim, S., Maekawa, N., and Yu, S. (2012). Identification of Trichoderma, a competitor of shiitake mushroom (Lentinula edodes), and competition between Lentinula edodes and Trichoderma species in Korea. Plant Pathol. J. 28, 137–148. doi: 10.5423/PPJ.2012.28.2.137
Kubicek, C., Steindorff, A., Chenthamara, K., Manganiello, G., Henrissat, B., Zhang, J., et al. (2019). Evolution and comparative genomics of the most common Trichoderma species. BMC Genom. 20, 1–24. doi: 10.1186/s12864-019-5680-7
Kullnig–Gradinger, C., Szakacs, G., and Kubicek, C. (2002). Phylogeny and evolution of the genus Trichoderma: a multigene approach. Mycol. Res. 106, 757–767. doi: 10.1017/S0953756202006172
Lieckfeldt, E., Samuels, G., Börner, T., and Gams, W. (1998). Trichoderma koningii: neotypification and Hypocrea teleomorph. Canad. J. Botany 76, 1507–1522. doi: 10.1139/b98-090
Lieckfeldt, E., and Seifert, K. (2000). An evaluation of the use of ITS sequences in taxonomy of the Hypocreales. Stud. Mycol. 45, 35–44.
Liu, S., Jin, P., and Dai, F. (2005). A rapid and simple extraction method for plant pathogenic fungi. Acta Phytopathol. Sinica 35, 362–365.
Liu, Y., and Huang, G. (2017). The chemical composition, pharmacological effects, clinical applications and market analysis of Gastrodia Elata. Pharm. Chem. J. 51, 211–215. doi: 10.1007/s11094-017-1584-5
Liu, Y., Whelen, S., and Hall, B. (1999). Phylogenetic relationships among Ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Migheli, Q., Balmas, V., Komoñ–Zelazowska, M., Scherm, B., Fiori, S., Kopchinskiy, A., et al. (2009). Soils of a Mediterranean hot spot of biodiversity and endemism (Sardinia, Tyrrhenian Islands) are inhabited by pan–European, invasive species of Hypocrea/Trichoderma. Environ. Microbiol. 11, 35–46. doi: 10.1111/j.1462-2920.2008.01736.x
Pruksakorn, P., Arai, M., Kotoku, N., Vilchèze, C., Baughn, A., Moodley, P., et al. (2010). Trichoderins, novel aminolipopeptides from a marine sponge–derived Trichoderma sp., are active against dormant mycobacteria. Bioorg. Med. Chem. Lett. 20, 3658–3663. doi: 10.1016/j.bmcl.2010.04.100
Przylucka, A., Akcapinar, G., Chenthamara, K., Cai, F., Grujic, M., Karpenko, J., et al. (2017). HFB7 – A novel orphan hydrophobin of the Harzianum and Virens clades of Trichoderma, is involved in response to biotic and abiotic stresses. Fungal Genet. Biol. 102, 63–76. doi: 10.1016/j.fgb.2017.01.002
Qiao, M., Jing, T., Wan, Y., and Yu, Z. (2024). Analyses of multi-locus sequences and morphological features reveal Ilyonectria species associated with black rot disease of Gastrodia elata. Plant Dis. 108, 382–397. doi: 10.1094/PDIS-01-22-0001-RE
Qin, W., and Zhuang, W. (2016). Seven wood–inhabiting new species of the genus Trichoderma (Fungi, Ascomycota) in Viride clade. Sci. Rep. 6:27074. doi: 10.1038/srep27074
Quimi, V., Meirelles, L., Chaverri, P., and Rodrigues, A. (2016). Unraveling Trichoderma species in the attine ant environment: description of three new taxa. Antonie van Leeuwenhoek 109, 633–651. doi: 10.1007/s10482-016-0666-9
Samuels, G., Dodd, S., Gams, W., Castlebury, L., and Petrini, O. (2002). Trichoderma species associated with the green mold epidemic of commercially grown Agaricus bisporus. Mycologia 94:146. doi: 10.2307/3761854
Samuels, G., Dodd, S., Lu, B., Petrini, O., Schroers, H., and Druzhinina, I. (2006). The Trichoderma koningii aggregate species. Stud. Mycol. 56, 67–133. doi: 10.3114/sim.2006.56.03
Sandoval–Denis, M., Sutton, D., Cano–Lira, J., Gene, J., Fothergill, A., Wiederhold, N., et al. (2014). Phylogeny of the clinically relevant species of the emerging fungus Trichoderma and their antifungal susceptibilities. J. Clin. Microbiol. 52, 2112–2125. doi: 10.1128/JCM.00429-14
Saravanakumar, K., Yu, C., Dou, K., Wang, M., Li, Y., and Chen, J. (2016). Biodiversity of Trichoderma community in the tidal flats and wetland of southeastern China. PLoS ONE 11:e0168020. doi: 10.1371/journal.pone.0168020
Sun, Y., Zhang, W., Li, F., Guo, Y., Liu, T., and Huang, H. (2000). Identification and genetic mapping of four novel genes that regulate leaf development in Arabidopsis. Cell Res. 10, 325–335. doi: 10.1038/sj.cr.7290059
Tang, X., Zhang, J., Teng, L., Zhou, T., Yuan, Q., Jiang, W., et al. (2020). Association study between rhizoma Gastrodia elata brown–rot and soil microbial communities. Chin. J. Exp. Tradit. Med. Form. 24, 153–160. doi: 10.13422/j.cnki.syfjx.20202083
White, T., Bruns, T., Lee, S., Taylor, W. T., Lee, S., and Shawe-Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications. PCR Protocols 1, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wu, D., and Ma, J. (1998). A Parasitical trichoderma viride in Armillariella mellea depended on by Gastrodia elata. J. Hubei Agric. Coll. 18, 112–114.
Ye, C., Jing, T., Sha, Y., Mo, M. H., and Yu, Z. (2023). Two new Trichoderma species (Hypocreales, Hypocreaceae) isolated from decaying tubers of Gastrodia elata. MycoKeys 99, 187–207. doi: 10.3897/mycokeys.99.109404
Zeng, L., Yuan, J., Li, D., Yuan, C., Xian, F., and Xia, J. (2007). Investigation and comprehensive management of pest and disease types in the authentic Chinese medicinal herb tianma (Gastrodia elata) GAP base in Guizhou. Guizhou Agric. Sci. 35:3. doi: 10.13346/j.mycosystema.170197
Zeng, X., Yang, J., Ling, H., Guo, S., and Zhang, Y. (2018). RNA–Seq analysis of Gastrodia elata seed germination infected with Mycena dendrobii. Mycosystema 37, 52–63.
Zhang, J., Xin, T., Xiao, C., Xu, J., Yuan, Q., Wang, X., et al. (2020). Investigation, analysis and identification of disease of Gastrodia elata. Zhongguo Zhong yao za zhi 45, 478–484. doi: 10.19540/j.cnki.cjcmm.20191204.108
Zhang, Y., and Zhuang, W. (2019). Epitypifications of three Trichoderma species and new Chinese records of the genus. Mycosystema 38, 23–38. doi: 10.13346/j.mycosystema.180224
Keywords: taxonomy, Trichoderma, plant disease, Gastrodia elata, multi-gene phylogeny
Citation: Ye C, You Y, Li W, Jing T, Mo M, Qiao M and Yu Z (2024) Diversity of Trichoderma species associated with the black rot disease of Gastrodia elata, including four new species. Front. Microbiol. 15:1420156. doi: 10.3389/fmicb.2024.1420156
Received: 22 April 2024; Accepted: 06 June 2024;
Published: 26 July 2024.
Edited by:
Fred O. Asiegbu, University of Helsinki, FinlandReviewed by:
Jing Si, Beijing Forestry University, ChinaCopyright © 2024 Ye, You, Li, Jing, Mo, Qiao and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zefen Yu, emZ5dTIwMjFAMTYzLmNvbQ==; Min Qiao, cWlhb21pbmdAeW51LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.