- 1National Engineering Research Center of Wheat and Corn Further Processing, Henan University of Technology, Zhengzhou, Henan, China
- 2College of Biological Engineering, Henan University of Technology, Zhengzhou, Henan, China
- 3School of Food and Pharmacy, Xuchang University, Xuchang, Henan, China
- 4Collaborative Innovation Center of Functional Food by Green Manufacturing, Xuchang, Henan, China
- 5Henan Provincial Engineering Laboratory of Preservation and Breeding of Industrial Microbial Strains, Zhengzhou, Henan, China
- 6School of International Education, Henan University of Technology, Zhengzhou, Henan, China
- 7State Key Laboratory of Crop Stress Adaptation and Improvement, College of Agriculture, Henan University, Kaifeng, Henan, China
Sweet potato residue (SPR) is the by-product of starch extraction from fresh sweet potatoes and is rich in carbohydrates, making it a suitable substrate for bioethanol production. An amylolytic industrial yeast strain with co-expressing α-amylase and glucoamylase genes would combine enzyme production, SPR hydrolysis, and glucose fermentation into a one-step process. This consolidated bioprocessing (CBP) shows great application potential in the economic production of bioethanol. In this study, a convenient heterologous gene integration method was developed. Eight copies of a Talaromyces emersonii α-amylase expression cassette and eight copies of a Saccharomycopsis fibuligera glucoamylase expression cassette were integrated into the genome of industrial diploid Saccharomyces cerevisiae strain 1974. The resulting recombinant strains exhibited clear transparent zones in the iodine starch plates, and SDS-PAGE analysis indicated that α-amylase and glucoamylase were secreted into the culture medium. Enzymatic activity analysis demonstrated that the optimal temperature for α-amylase and glucoamylase was 60–70°C, and the pH optima for α-amylase and glucoamylase was 4.0 and 5.0, respectively. Initially, soluble corn starch with a concentration of 100 g/L was initially used to evaluate the ethanol production capability of recombinant amylolytic S. cerevisiae strains. After 7 days of CBP fermentation, the α-amylase-expressing strain 1974-temA and the glucoamylase-expressing strain 1974-GA produced 33.03 and 28.37 g/L ethanol, respectively. However, the 1974-GA-temA strain, which expressed α-amylase and glucoamylase, produced 42.22 g/L ethanol, corresponding to 70.59% of the theoretical yield. Subsequently, fermentation was conducted using the amylolytic strain 1974-GA-temA without the addition of exogenous α-amylase and glucoamylase, which resulted in the production of 32.15 g/L ethanol with an ethanol yield of 0.30 g/g. The addition of 20% glucoamylase (60 U/g SPR) increased ethanol concentration to 50.55 g/L, corresponding to a theoretical yield of 93.23%, which was comparable to the ethanol production observed with the addition of 100% α-amylase and glucoamylase. The recombinant amylolytic strains constructed in this study will facilitate the advancement of CBP fermentation of SPR for the production of bioethanol.
1 Introduction
Sweet potato is a high-yielding and adaptable food crop, which is widely cultivated in tropical and subtropical regions (Karmee, 2016). The annual output of sweet potatoes can reach approximately 100 million tons, with China accounting for the majority of global production, at approximately 80% (Xia et al., 2019). At present, the sweet potato–processing industry focuses on starch extraction, food production, and microbial fermentation (El Sheikha and Ray, 2016). Sweet potato residue (SPR), which is extracted from fresh sweet potatoes after crushing, washing, and filtering, is primarily composed of starch, cellulose, and pectin (Gou et al., 2023). However, given its low water solubility and high levels of polysaccharides and proteins, SPR is susceptible to mildew and contamination. Rancid SPR releases methane as a residue, which is the second most prevalent greenhouse gas and whose impact on climate change can be more than 25 times that of carbon dioxide (Lu et al., 2013). In general, SPR is used in animal protein feed production, pectin extraction, and dietary fiber extraction (Aziz and Mohsen, 2002; Hao et al., 2014; Pagana et al., 2014; Wang et al., 2016). However, long-term storage of SPR can result in mycotoxin production, which limits its application as animal feed. Meanwhile, the extraction rates for pectin and dietary fibers from SPR are insufficiently high to reach the industrial production scale. Researchers have developed biotechnologies to transform SPR into various biological products through microbial fermentation. This process involves the hydrolysis of starch and cellulose by enzymes, thereby producing monosaccharides and oligosaccharides, which can be fermented to produce butanol and other products (Jin et al., 2022).
Bioethanol is widely regarded as a potential source of energy and an alternative to fossil fuels (Osman et al., 2021). At present, bioethanol is predominantly produced from sugar and starchy food crops, such as sugar cane, corn, and cassava, because of the well-established processing techniques and high ethanol yields associated with these crops (Cripwell et al., 2020). Nevertheless, the development of the corn ethanol industry is constrained by the rising demand for food and animal feed. Furthermore, feedstock represents an important proportion of the overall production costs of bioethanol, necessitating the identification of alternative, low-cost feedstocks (Wang et al., 2019). SPR, a cheap and abundant starchy by-product from the sweet potato–processing industry, shows great application potential in this regard.
Irrespective of the feedstock, there are other substantial costs associated with the current starch-to-ethanol production process (Görgens et al., 2014). These costs are predominantly associated with conventional energy-intensive gelatinization or the addition of exogenous enzyme cocktails for the liquefaction and saccharification of raw starch. The energy requirement for conventional gelatinization accounts for 10%–20% of the fuel value of ethanol in a typical refinery (Lorenzo and Marina, 2010). Moreover, the cost of enzymes represents 8% of the total processing cost (Görgens et al., 2014). Therefore, the production of ethanol from starch can benefit from the combination of all these steps into a single process, which is known as consolidated bioprocessing (CBP). In such a process, genetically engineered ethanologenic yeast strains, such as amylolytic Saccharomyces cerevisiae strains, are required to produce raw starch-degrading enzymes that enable the yeast to simultaneously hydrolyze the starch and ferment the resulting sugars to ethanol (Chandel et al., 2018; Cripwell et al., 2019b).
A number of S. cerevisiae strains with raw starch CBP capabilities have been developed with varying degrees of success (Viktor et al., 2013; Favaro et al., 2015; Sakwa et al., 2018). Most previous studies have focused on the engineering of S. cerevisiae laboratory strains (Cripwell et al., 2019a), which are easy to manipulate and allow for the screening of transformants without antibiotic selection. However, laboratory strains showed lower thermotolerance and decreased glucose fermentation rates when compared with industrial yeast strains (Kong et al., 2018). Furthermore, starch-degrading enzymes genes were always expressed using episomal plasmids rather than integration into the genome, and the selection marker and copy number of the plasmids will affect the growth ability and cellular metabolism of the cell (Karim et al., 2013). Given the aforementioned considerations, Cripwell et al. used the industrial S. cerevisiae yeast strain Ethanol Red as a host, integrating seven copies of the codon-optimized Talaromyces emersonii glucoamylase-encoding gene (temG_Opt) and four copies of the native T. emersonii α-amylase-encoding gene (temA) into the genome via δ-integration. The recombinant industrial amylolytic yeast strain could produce 89.35 g/L ethanol from 200 g/L raw corn starch in a single step after 192 h at 30°C, which represents the highest value to date (Cripwell et al., 2019a).
In this study, the industrial bioethanol-producing diploid S. cerevisiae 1974 was selected as the parental strain. The T. emersonii α-amylase gene (temA) and codon-optimized Saccharomycopsis fibuligera glucoamylase gene (GA), both of which have been proven to have a high starch-hydrolyzing activity were integrated into the specific sites on the S. cerevisiae 1974 genome (Cripwell et al., 2019a; Wang et al., 2021). Based on a quick and easy high-copy number gene integration method that was developed, a recombinant S. cerevisiae strain harboring eight copies of temA and eight copies of GA was constructed. The recombinant yeast strains were assessed for their starch hydrolysis capability using iodine starch plates. In addition, SDS-PAGE analysis was performed to analyze the expression of recombinant proteins in the supernatant fermentation broth. The effect of time, pH, and temperature on the starch-hydrolyzing enzymatic activity was also investigated. Finally, the ability of recombinant strains to ferment soluble starch and SPR to produce ethanol was also evaluated. To the best of our knowledge, this study is the first to utilize a starch-hydrolyzing yeast strain for CBP fermentation of SPR to produce ethanol.
2 Materials and methods
2.1 Strains, plasmids, and growth conditions
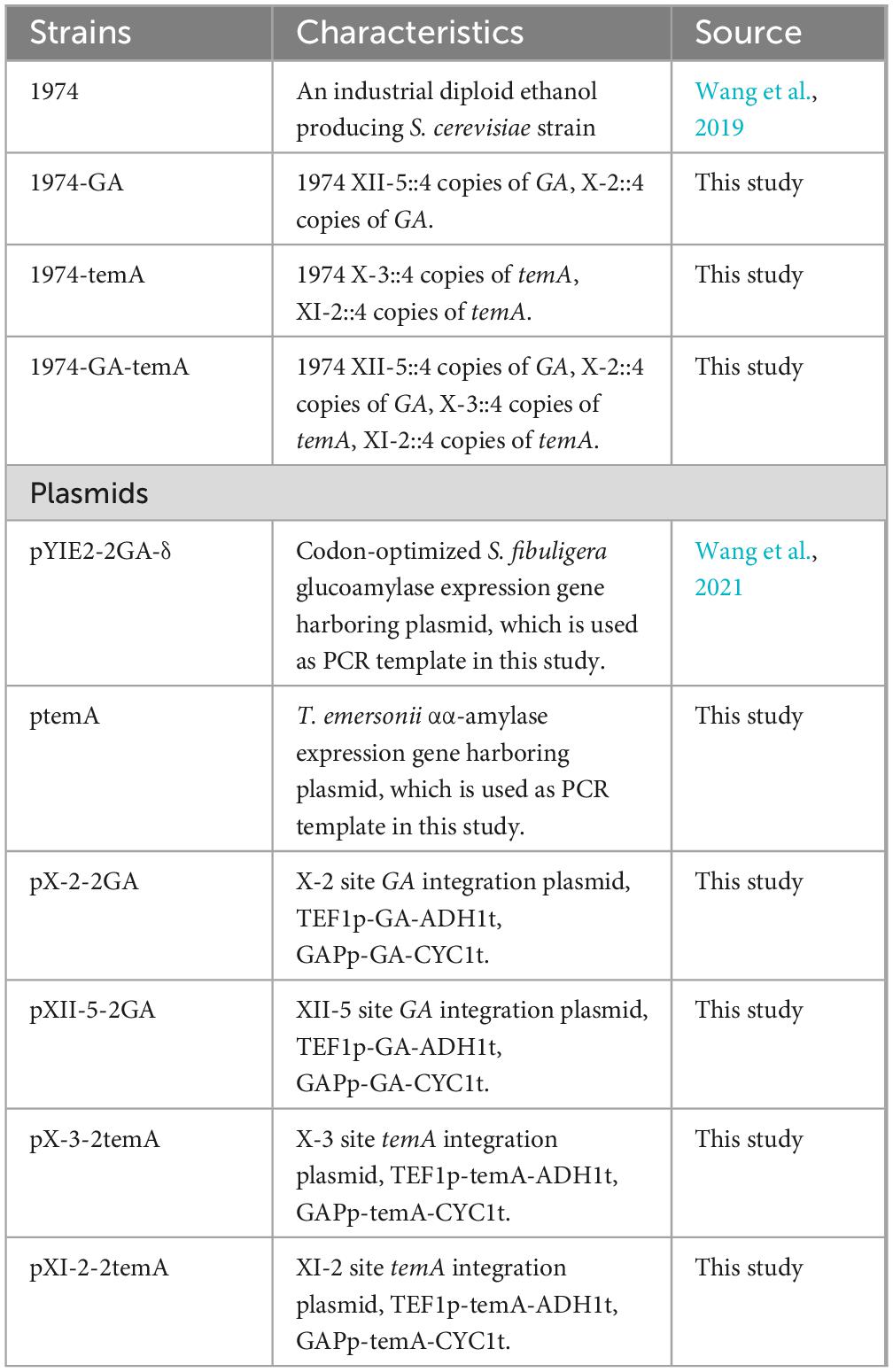
The strains and plasmids used in this study are listed in Table 1. In brief, the industrial bioethanol production diploid S. cerevisiae strain 1974 was used as the original host. All molecular cloning operations were conducted in Escherichia coli strain DH5α. In addition, strain DH5α was cultured in LB medium at 37°C, and 100 μg/mL ampicillin was added if necessary. For routine S. cerevisiae molecular engineering, strains were maintained in a YPD medium. Antibiotics (300 μg/mL G418 or 400 μg/mL hygromycin) was added as required. The composition of the medium used is shown in the supporting information. α-Amylase (Beijing Aoboxing Biotechnology Co., Ltd, 3700 U/g) and glucoamylase (Beijing Aoboxing Biotechnology Co., Ltd,100000 U/g) were used in the CBP fermentation test.
2.2 Plasmid and strain construction
The KOD-plus-neo DNA polymerase (Toyobo, Japan) or KOD FX polymerase (Toyobo, Japan) was used for PCR amplifications. The DNA restriction enzymes for cloning were obtained from Thermo Fisher Scientific (USA). The primers used for plasmid construction are listed in Supplementary Table 1. The codon-optimized S. fibuligera glucoamylase gene (GenBank accession number: MW082635) was amplified from the plasmid pYIE2-2GA-δ. The T. emersonii amylase gene temA was synthesized by GenScript Biotech Corporation (China), and the resulting plasmid was ptemA. Detailed procedures for plasmid and strain construction are described in the supporting information. The map of the plasmids pX-2-2GA, pXII-5-2GA, pX-3-2temA, and pXI-2-2 are shown in Supplementary Figures 1, 2.
2.3 Determination of the copy number of GA and temA genes in recombinant S. cerevisiae strain by colony PCR and real-time PCR
After the transformation of GA or temA gene expression cassette fragments, several transformants grown in the YPD-G418-HYG plate were selected randomly and pretreated with lysis buffer for colony PCR. One primer was located within the GA or temA gene expression cassette, while another primer was located in the chromosomal homology arm. This combination of primers was used to ascertain whether the target genes had been integrated into the specific region of the chromosome. Genomic DNA was extracted from the positive transformants and used as the next PCR template. In addition, the primer pairs located in the up and down regions of the heterologous gene insertion sites were selected, and the strain 1974 was used as a control. If a short fragment was amplified, then only one copy of the chromosome had been inserted by heterologous genes. By contrast, no fragment was amplified if the two chromosomes had been inserted.
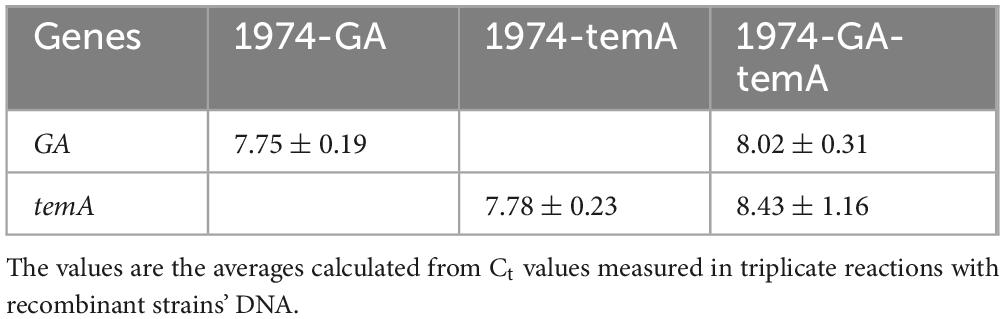
For analysis of the gene copy number, the strains’ genomic DNA was isolated with a TIANamp Yeast DNA Kit (Tiangen Biotech [Beijing] Co., Ltd) according to the manufacturer’s protocol and quantified by NanoDrop ND-1000 (Thermo Fisher Scientific). The fold changes of the gene copy number compared with the reference gene ACT1 were determined using the equation, NTarget / NReference = (2Ct (Reference) /2Ct (Target)) × 2. Note that the copy number of ACT1 is 2 because the strain is a diploid yeast. The primers used for real-time quantitative PCR are listed in Supporting Information Supplementary Table 1.
2.4 Starch-hydrolyzing plate and SDS-PAGE analysis
The recombinant S. cerevisiae strains were inoculated into 3 mL of YPD medium and cultured overnight at 30°C and 240 rpm until reaching an OD600 value of 4.7. Subsequently, 10 μL of the culture supernatant was inoculated into the YPS plate and incubated at 30°C for 72 h. After incubation, the starch-degradation test plate was flooded with approximately 10 mL of 50 mM potassium iodide–iodine solution and incubated for 15 min at room temperature. The excess iodine solution was poured off, and the plates were washed with 1 M NaCl. The formation of a clear hydrolysis zone around the recombinant strains indicated the presence of starch-hydrolyzing enzymatic activity.
The protein content of aliquots (100 μL) of the medium was precipitated with 15% TCA and left on ice for 30 min. Subsequently, centrifugation in a microcentrifuge (10,600 × g, 15 min, 4°C) was performed to remove the supernatants of the TCA-treated samples, after which the pellets were washed with ice-cold acetone. Following a second round of centrifugation in a microcentrifuge (10,600 × g, 15 min, 4°C), the supernatants were discarded, and the pellets were air dried. Then, SDS-PAGE sample buffer was added, and the proteins were separated on 15% SDS-PAGE gels.
2.5 Genetic stability and enzymatic activity analysis
The frozen 1974-GA-temA glycerol stock was streaked onto a YPD plate and cultured at 30°C. Then, the single colony was streaked onto a new YPD plate. This procedure was repeated until 30 serial passages were performed. Colonies from the 10th, 20th, and 30th passages were selected to test the starch hydrolysis activity using the iodine vapor method.
For quantitative amylase analysis, single clones or frozen glycerol stock cultures of 1974, 1974-GA, and 1974-GA-temA were inoculated into 3 mL of YPD medium in test tubes and grown overnight at 30°C and 240 rpm. A total of 200 μL of the cultures were transferred to 50 mL of fresh YPD medium. After 72 h of incubation, the cultures were centrifuged at 12,000 rpm for 10 min. The resulting supernatant was utilized for the analysis of starch-hydrolyzing enzymatic activity. The reaction mixture, comprising 450 μL of 1% soluble starch in 0.1 M citrate–phosphate buffer (pH 4.5) and 50 μL of enzyme solution, was incubated at 40°C for 30 min. Then, the reaction was terminated by heating for 10 min in boiling water. A control sample was incubated for 10 min in boiling water to inactivate the enzyme and incubated under the same conditions. The reducing sugar content was measured using the dinitrosalicylic acid method (Gou et al., 2023). One unit of enzymatic activity was defined as the amount of enzyme that produced 1 μmol of reducing sugar (equivalent to 1 μmol glucose) per minute under the abovementioned standard assay conditions. A standard curve of the glucose is shown in Supplementary Figure 4.
2.6 Fermentation studies
For soluble starch fermentation, the recombinant strains were inoculated into 6 mL of YPD medium in test tubes and grown overnight at 30°C and 240 rpm, the OD600 of the strain was about 6.0. The cells were harvested by centrifugation at 12,000 rpm, washed two times with sterile water, suspended using YPS medium and inoculated into 50 mL of YPS medium in anaerobic flasks at an initial inoculation volume of 10% (v/v). During fermentation, samples were taken at specific time intervals for OD600 readings and metabolite analysis.
SPR was provided by Luoyang Feed Factory of Henan Dongfang Zhengda Co., Ltd., China. The dried SPR was pulverized, passed through a 60-mesh sieve, and stored at room temperature. The dried SPR is composed of 51.94% ± 0.55% starch, 18.22% ± 0.47% cellulose, 3.38% ± 0.02% hemicellulose, 1.66% ± 1.31% pectin, 2.92% ± 0.03% protein, and 2.23% ± 0.01% ash (Gou et al., 2023). For SPR fermentation, the recombinant S. cerevisiae strain 1974-GA-temA was inoculated into 3 mL of YPD medium in test tubes and grown overnight at 30°C and 240 rpm. A total of 200 μL of the cultures was transferred to 25 mL of YPD medium in flasks to repeat the procedure. After aerobic growth, cells were harvested by centrifugation at 12,000 rpm, washed two times with sterile water, and inoculated into the SPR medium in a 250 mL anaerobic flask at an inoculation volume of 10% (v/v). The SPR medium was prepared by weighing 10 g of SPR powder and mixing it with deionized water in a 1:6 solid-to-liquid ratio. The pH was set to 5.0 with 1M HCl or 1 M NaOH. For α-amylase and glucoamylase addition test, a total of 0.13 g of α-amylase (48 U/g SPR) and 0.03 g of glucoamylase (300 U/g SPR) were used to supplement the CBP fermentation of strains, which were designated as 100% dosage. Samples were taken at specific time intervals for OD600 readings and metabolite analysis during fermentation. The OD600 values, as well as glucose and ethanol concentrations, were determined in accordance with a previous report (Gou et al., 2023).
3 Results and discussion
3.1 High copy number of α-amylase and glucoamylase integration strain construction
In this study, the industrial diploid S. cerevisiae strain 1974 was selected as the expression host due to its excellent fermentation capacity in the bioethanol production field (Wang et al., 2021). Two copies of the codon-optimized S. fibuligera glucoamylase expression gene were cloned into plasmids pX-2-2GA and plasmid pXII-5-2GA, respectively. Two copies of the T. emersonii α-amylase expression gene were also cloned into the plasmids pX-3-2temA and pXI-2-2temA. GA and temA genes were expressed under the control of the strong promoter and terminator combinations PENO1-TENO1 and PADH1-TPDH1. In preventing the potential loss of the GA and temA gene pairs during mitotic recombination, the two copies were assembled in a tail-to-tail manner (Figure 1A). The NotI-linearized GA expression cassette was integrated into the chromosome XII-5 and X-2 sites of the 1974 genome and verified using diagnostic PCR. In parallel, the NotI-linearized temA expression cassette was integrated into the chromosome X-3 and XI-2 sites of the 1974 genome and verified using diagnostic PCR (Figure 1B).
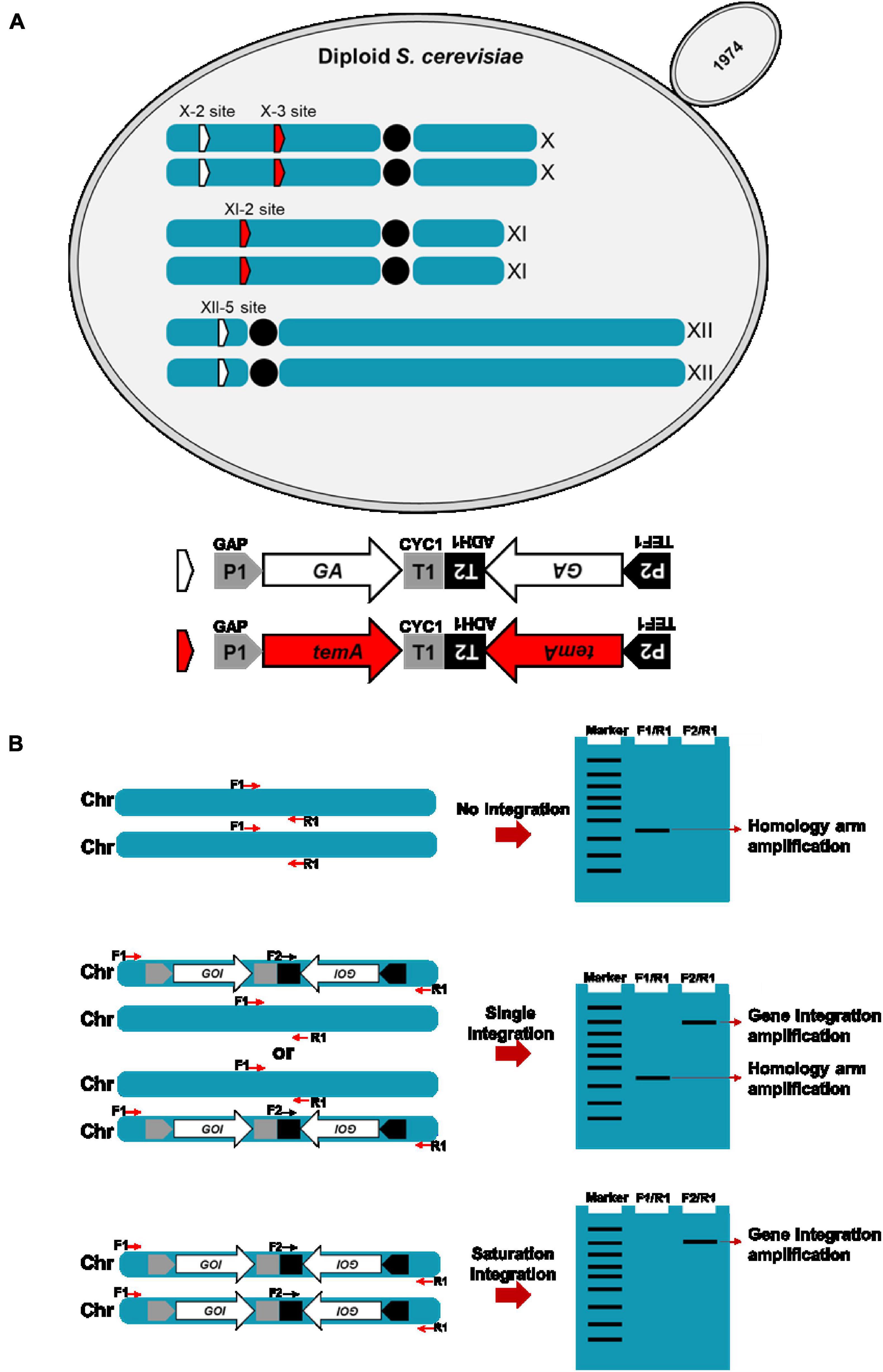
Figure 1. Schematic representation of the construction of the αα-amylase- and glucoamylase-producing S. cerevisiae strain (A) and gene copy number verification by PCR (B). (A) The GA expression cassettes were integrated at chromosome XII-5 and X-2 sites. temA expression cassettes were integrated at chromosome X-3 and XI-2 sites. P1, GAP promoter. T1, CYC1 terminator. P2, TEF1 promoter. T2, ADH1 terminator. (B) Sketch map of gene copy number determination by using conventional PCR.
To verify the integration of foreign genes, two pairs of primers were designed for PCR. One pair is located on the homology arms surrounding the integration site (F1/R1), whereas the other pair is located within the integrated gene and downstream homology arms (F2/R1, Figure 1B). Initially, the F2/R1 primer pair is used to determine whether the foreign gene is integrated at a specific site. The presence of a band indicates that the foreign gene expression cassette has been integrated into at least one chromosome. Subsequently, the F1/R1 primers are used to confirm these transformants. If a short fragment (homologous arm region) is amplified, then only one chromosome has integrated the foreign gene expression cassette (single integration). Conversely, the absence of the amplification of a short fragment indicates that both chromosomes have integrated the foreign gene expression cassette (saturation integration), resulting in four copies (Figure 1B).
A high expression level of exogenous genes in S. cerevisiae is often necessary to overexpress pathway enzymes leading to bioethanol production (Myburgh et al., 2019; Wang et al., 2019). However, the common methods used for identifying gene copy numbers, such as real-time PCR or genome sequencing, are time consuming and laborious. Based on the site-specific gene copy number identification method developed in this study, several transformants were selected randomly to determine the gene copy number after GA and temA expression cassette transformation. Figures 2A, B demonstrate that the GA expression cassette was successfully integrated at the X-2 and XII-5 sites in colony 1 to colony 5 except for colony 6, which was integrated only at the XII-5 site. No band was amplified using the primer pairs F1/R1 (Figures 2C, D), indicating that both chromosomes have integrated the GA expression cassette, resulting in eight copies. Using the same method, six colonies were randomly selected to verify the integration of the temA expression cassette (Figures 2E–H). Colonies 1 and 2 exhibited saturation integration for the temA expression cassette. To construct stain 1974-GA-temA, the temA expression cassette was transformed into strain 1974-GA (Supplementary Figure 3), and strain-harbored newly integrated eight copies of the temA expression cassette were selected (Figures 2I–L). Copy numbers for integrated genes in each genome were also determined by using real-time PCR. As shown in Table 2, GA copy number in strain 1974-GA was 7.75, temA copy number in strain 1974-temA was 7.78, GA and temA copy number in strain 1974-GA-temA were 8.02 and 8.43, respectively. The copy number determined by real-time PCR was consistent with the colony PCR method.
3.2 Functional secretion of recombinant α-amylase and glucoamylase in recombinant S. cerevisiae strains
Following the cultivation of recombinant S. cerevisiae strains in YPS medium for 3 days, iodine staining was conducted using iodine particles (Cripwell et al., 2019b). The results demonstrated that no transparent circle was formed around the control strain 1974, whereas a transparent circle was observed around the recombinant strains (Figure 3A), indicating their ability to grow with starch as the sole carbon source. The diameter of the transparent circle was 1.33 cm for the 1974-GA strain, 2.26 cm for the 1974-temA strain, and 2.68 ± 0.011 cm for the 1974-GA-temA strain. This result indicates that α-amylase has a stronger ability to hydrolyze starch than glucoamylase. The recombinant S. cerevisiae strain co-expressing α-amylase and glucoamylase exhibited the strongest ability to hydrolyze starch (Figure 3A).
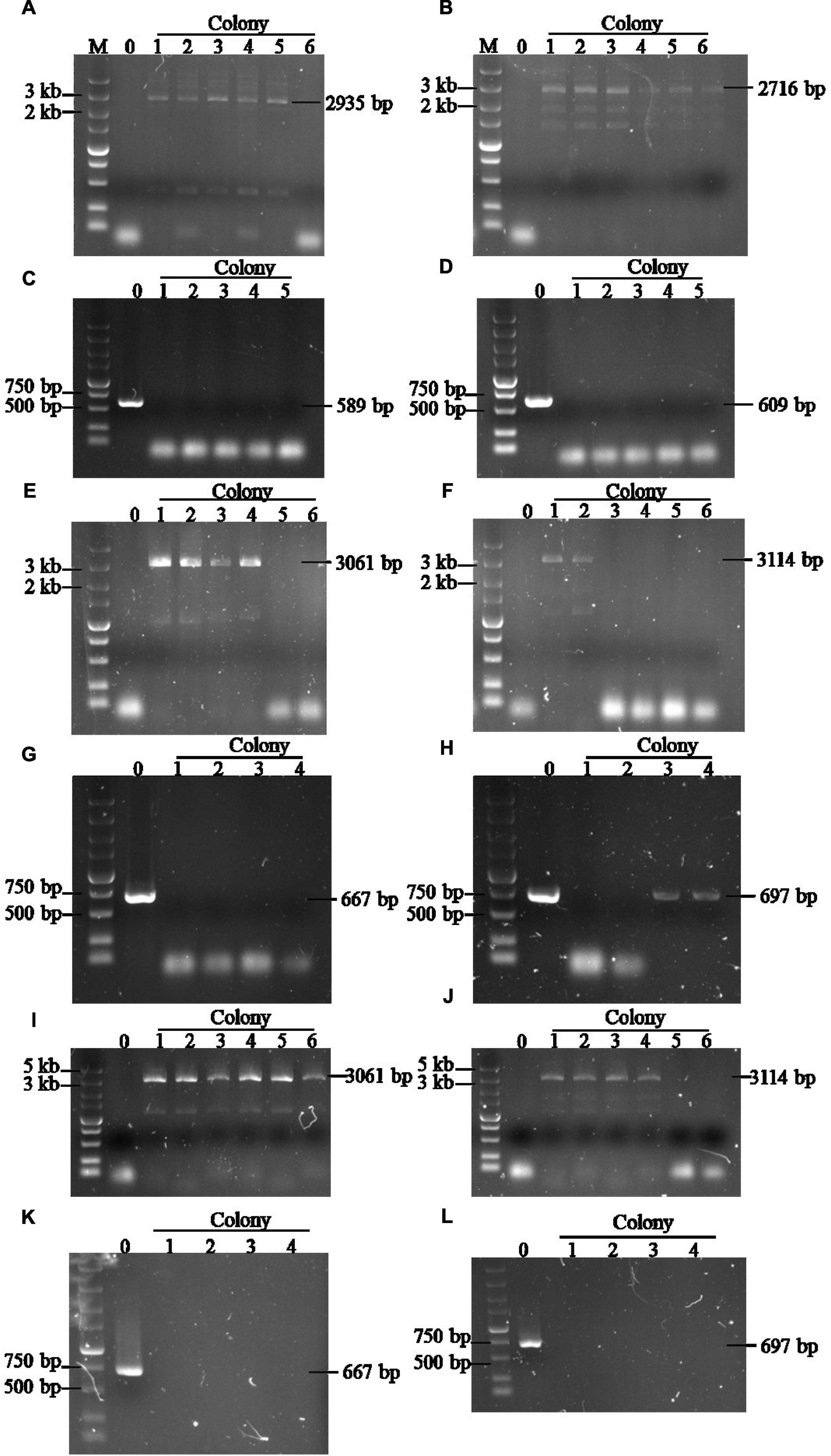
Figure 2. Agarose gel electrophoretic analysis of the GA copy number in strain 1974-GA (A–D), temA copy number in strain 1974-temA (E–H), and GA and temA copy number in strain 1974-GA-temA (I–L). (A,B) Verification of GA integration at X-2 and XII-5 sites using F2/R1 primers (based on 1974 strain). (C,D) Verification of GA integration at X-2 and XII-5 sites using F1/R1 primers, respectively (based on 1974 strain). (E,F) Verification of temA integration at X-3 and XI-2 sites using F2/R1 primers, respectively (based on 1974 strain). (G,H) Verification of temA integration at X-3 and XI-2 sites using F1/R1 primers, respectively (based on 1974 strain). (I,J) Verification of temA integration at X-3 and XI-2 sites using F2/R1 primers, respectively (based on 1974-GA strain). (K,L) Verification of temA integration at X-3 and XI-2 sites using F1/R1 primers, respectively (based on 1974-GA strain). Number zero refers to the control strain 1974. The original gel of Figures 2A, B was presented as Supplementary Figure 5. The original gel of Figure 2C was presented as Supplementary Figure 6. The original gel of Figure 2D was presented as Supplementary Figure 7. The original gel of Figures 2E, F was presented as Supplementary Figure 8. The original gel of Figure 2G was presented as Supplementary Figure 9. The original gel of Figure 2H was presented as Supplementary Figure 10. The original gel of Figures 2I, J was presented as Supplementary Figure 11. The original gel of Figure 2K was presented as Supplementary Figure 12. The original gel of Figure 2L was presented as Supplementary Figure 13.
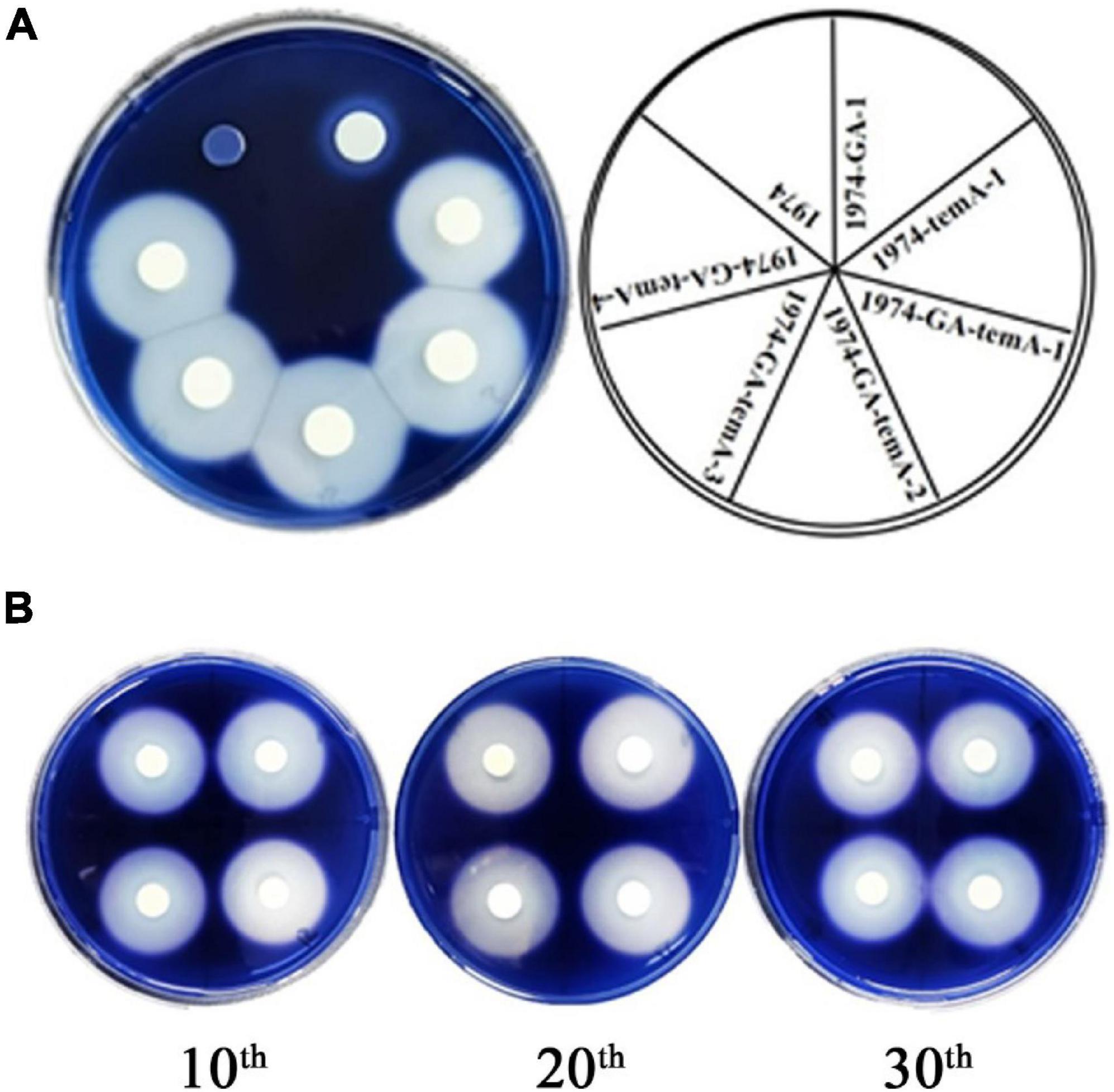
Figure 3. Hydrolysis zones indicate the production of starch-degrading enzymes (A) and genetic stability evolution (B).
The genetic stability of the strain 1974-GA-temA was evaluated by performing 30 serial passages on non-selective YPD plates. The average diameter of the transparent circle for the 10th, 20th, and 30th generations of the 1974-GA-temA strain was 2.68, 2.66, and 2.66 cm, respectively (Figure 3B). The consistent diameters observed after 30 generations indicate that the starch hydrolysis ability of the strain 1974-GA-temA has not changed. This result is due to the fact that the α-amylase and glucoamylase genes were integrated into specific sites on the S. cerevisiae chromosome, rather than expressed in plasmid form or integrated at the δ sites, both of which are prone to lose the copy number of foreign genes (Stepchenkova et al., 2023). Again we tested the GA and temA copy number in strain 1974-GA-temA after serial passages on non-selective YPD plates by using real-time PCR, eight copies of GA and eight copies of temA remained present in the strain (Supplementary Table 2). The results of the strain genetic stability testing indicate that the strain 1974-GA-temA has potential for industrial application.
α-Amylase and glucoamylase should be secreted into the medium to hydrolyze starch into glucose, which is then utilized by S. cerevisiae. SDS-PAGE analysis of the supernatant of fermentation broth was performed to identify the expression of α-amylase and glucoamylase directly. Based on the amino acid sequences, the predicted molecular weights of the unglycosylated GA and temA protein were 57.4 and 68.3 kDa, respectively. SDS-PAGE analysis of the supernatant indicated prominent bands for strains expressing GA, with the molecular size of the band being consistent with the predicted GA (Figure 4A). The strain 1974-temA exhibited clear protein bands between 55 and 72 kDa, whereas the strain 1974-GA-temA displayed the GA and temA protein bands (Figure 4A). The SDS-PAGE result indicated that α-amylase and glucoamylase were expressed and secreted into the medium.
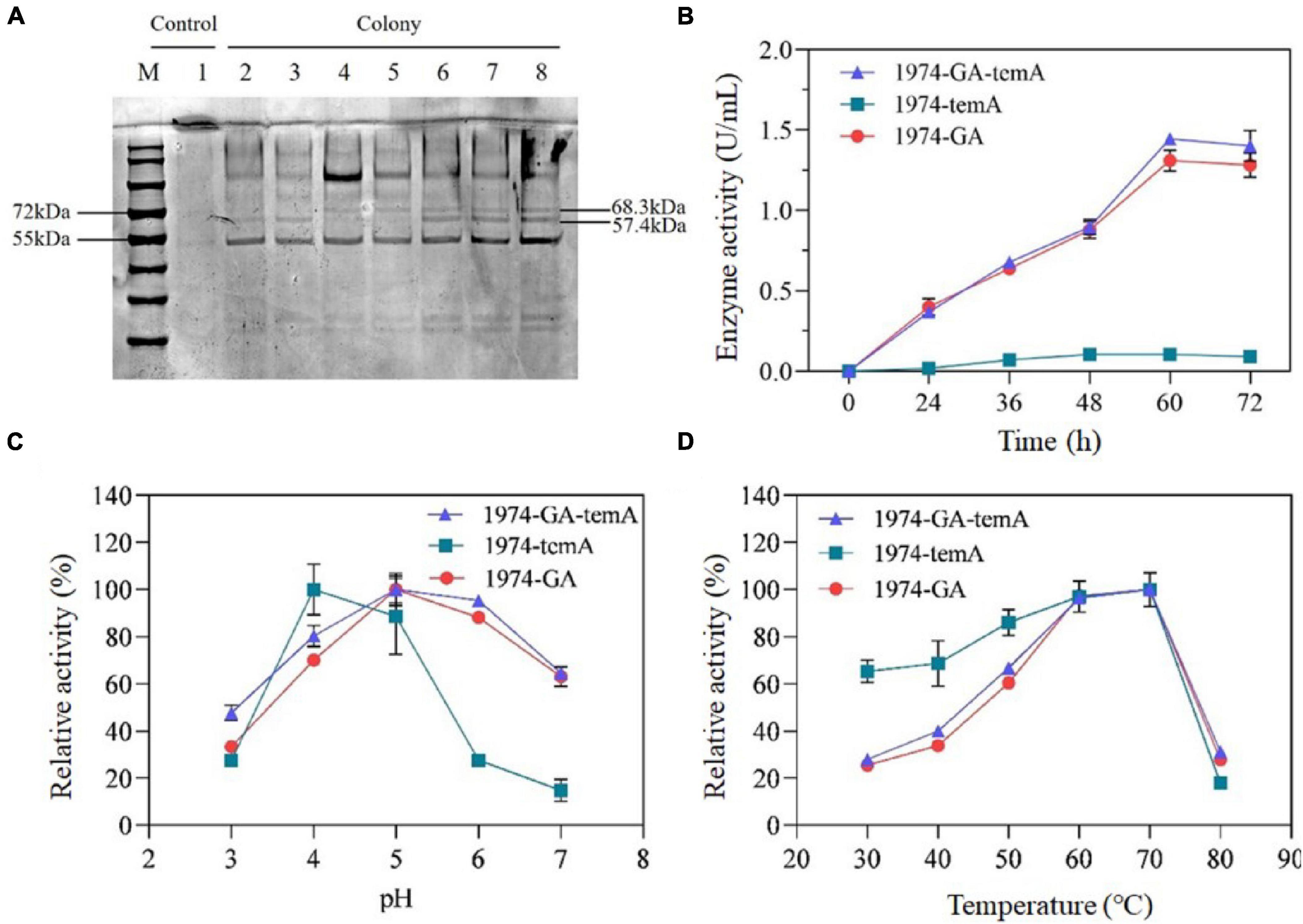
Figure 4. SDS-PAGE analysis of the expression of αα-amylase and glucoamylase (A) and enzymatic activity assays. Lane 1, 1974. Lane 2 to lane 3, 1974-GA. Lane 4 to lane 5, 1974-temA. Lane 6 to lane 8, 1974-GA-temA. Volumetric enzymatic activity profiles over time for the strain 1974-GA-temA (B). Effect of pH (C) and temperature (D) on the enzymatic activity of recombinant enzymes. The original gel of Figure 4A was presented as Supplementary Figure 14. Data points represent the average of the results from three independent experiments.
The enzymatic activities of the recombinant strains are shown in Figure 4B. After 60 h of fermentation, the enzymatic activity stabilized. The α-amylase enzymatic activity reached 0.12 U/mL ± 0.01 U/mL, whereas the glucoamylase enzymatic activity reached 1.36 U/mL ± 0.07 U/mL. α-Amylase primarily hydrolyzes starch into dextrin and oligosaccharides, whereas glucoamylase primarily hydrolyzes dextrin and oligosaccharides into glucose (Cripwell et al., 2020). Consequently, the glucose released from starch by the enzyme of the strain 1974-GA is higher than that in the fermentation broth of the strain 1974-temA, resulting in lower α-amylase activity compared with glucoamylase activity. The strain 1974-GA-temA can simultaneously secrete α-amylase and glucoamylase. The fermentation broth of the strain 1974-GA-temA exhibits the highest glucose content, with enzymatic activity reaching 1.47 U/mL ± 0.10 U/mL (Figure 4B). The extracellular amylase activity of strain 1974-GA-temA was comparable with the strain ER T1 published in the previous report (Cripwell et al., 2019a).
As the combination of T. emersonii-derived α-amylase and S. fibuligera-derived glucoamylase has not been reported, the optimal pH and temperature for enzymatic activity were determined. A series of pH buffer systems (pH 3.0, pH 4.0, pH 5.0, pH 6.0, and pH 7.0) and temperature values (30°C, 40°C, 50°C, 60°C, 70°C, and 80°C) was tested (Figures 4C, D). The enzymatic activity of the recombinant enzyme was quantified by exhibiting the highest enzymatic activity designated as 100% relative enzymatic activity. The results indicated that the optimal pH for α-amylase was approximately 4.0, whereas that for glucoamylase was around 5.0 (Figure 4C). Furthermore, at a pH of 5.0, the optimal hydrolysis temperature for α-amylase and glucoamylase is approximately 60°C to 70°C, indicating that these enzymes exhibit relatively high hydrolysis temperatures (Figure 4D). α-Amylase and glucoamylase from different microbial sources have different optimum pH and temperature. For example, the optimal pH for Aspergillus tubingensis derived α-amylase and glucoamylase are 4.0, while the optimum temperatures are 60 and 70°C, respectively (Viktor et al., 2013). The combinations of α-amylase and glucoamylase from T. emersonii exhibited excellent raw starch hydrolyzed capability (Cripwell et al., 2019a), however, the optima pH and temperature for T. emersonii derived glucoamylase remains unclear.
3.3 Ethanol production from soluble starch with recombinant S. cerevisiae strains
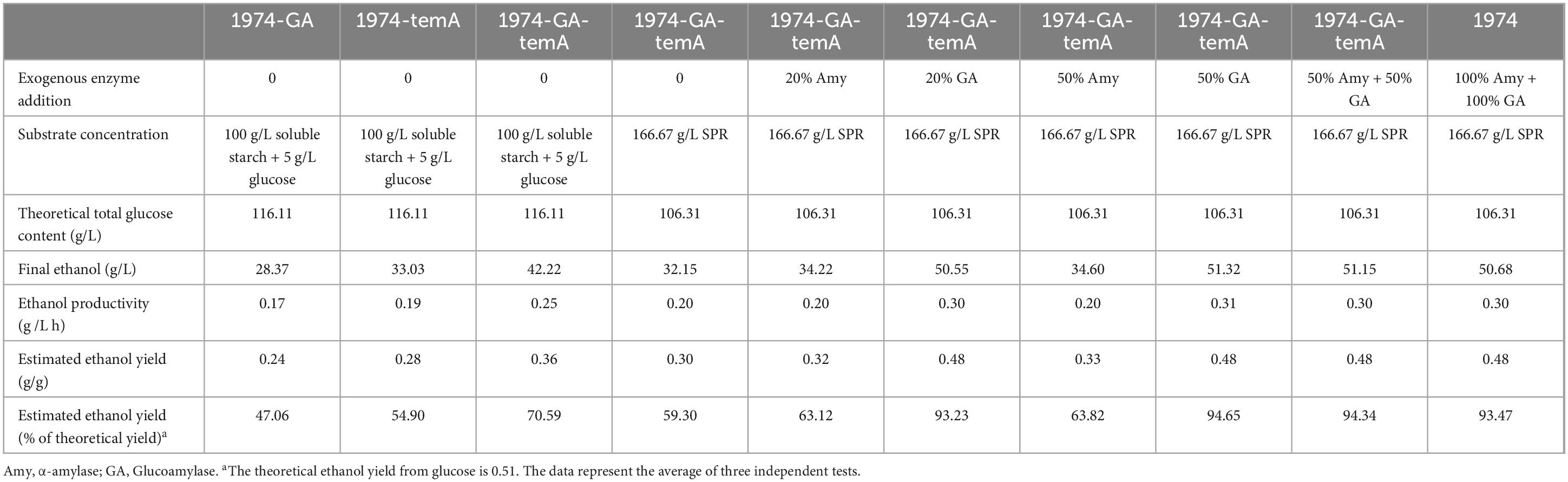
The CBP simulation was initially conducted under fermentative conditions with recombinant S. cerevisiae strains utilizing 100 g/L soluble starch and 5 g/L glucose as the initial carbon source for cells. Samples were taken every 24 h to measure glucose and ethanol levels, and the iodine–starch reaction was used to determine the completion of starch hydrolysis. The strain 1974-GA-temA produced 42.2 g/L ± 0.52 g/L ethanol after 5 days of fermentation (Figure 5 and Table 3), corresponding to 70.59% of the theoretical yield. The strains 1974-GA and 1974-temA exhibited incomplete starch hydrolysis for 9 days and produced 28.4 g/L ± 0.57 g/L and 33.0 g/L ± 0.97 g/L ethanol, respectively (Figure 5). Although all three engineering strains can utilize soluble starch to produce ethanol, the strains 1974-GA and 1974-temA exhibited a weaker ability to convert starch. Therefore, complete hydrolysis of starch requires the synergistic action of α-amylase and glucoamylase.
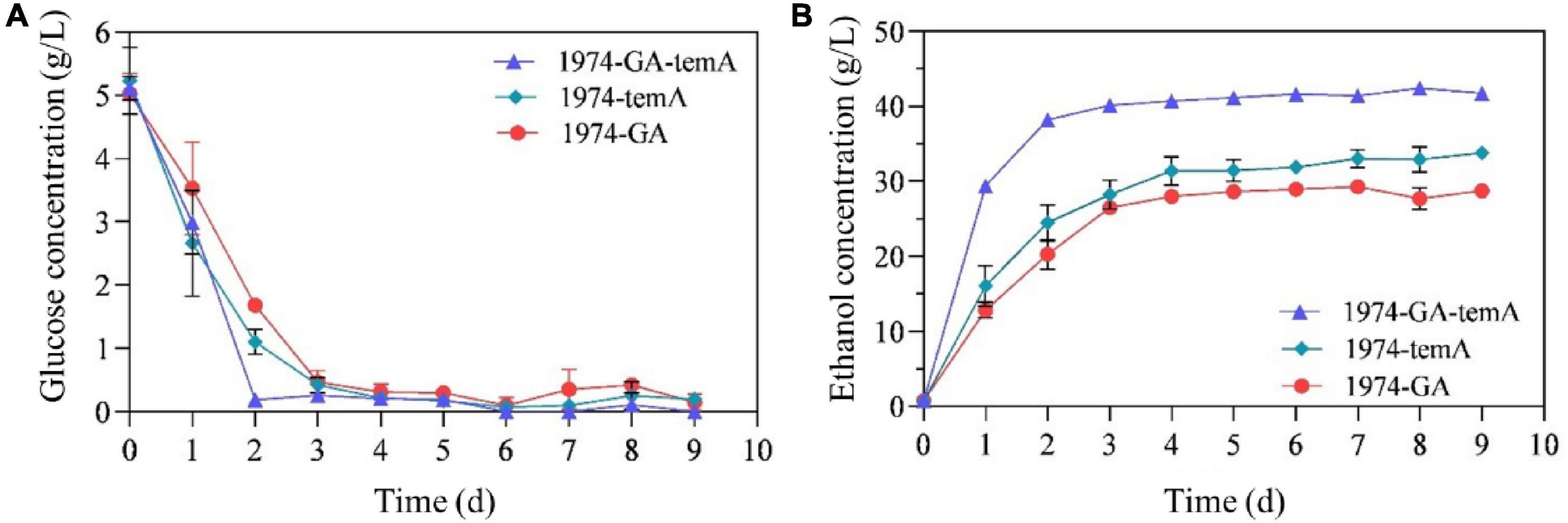
Figure 5. Fermentation of soluble starch by recombinant S. cerevisiae strain. (A) Residual glucose concentration during fermentation. (B) Ethanol concentration during fermentation. Data points represent the average of the results from three independent fermentation experiments.
Previously, the enzymatic hydrolysates of SPR were used as a fermentation substrate to produce ethanol (Gou et al., 2023). Herein, SPR was directly used as the fermentation substrate. However, in the absence of α-amylase or glucoamylase, the strain 1974-GA-temA exhibited incomplete starch hydrolysis after 10 days (Figure 6), which could be attributed to the fermentation temperature not being optimal for the recombinant enzyme, resulting in a slow starch hydrolysis rate. Despite the advantages of CBP, recombinant proteins still need to be produced by the yeast strain before substrate hydrolysis can accelerate at the start of fermentation. Therefore, a portion of exogenous α-amylase or glucoamylase was supplemented. The results demonstrated that after adding exogenous α-amylase and glucoamylase to the initial strain 1974 for synchronous saccharification and fermentation for 8 days, the starch in the SPR was completely hydrolyzed (Figure 6 and Table 3). This reaction resulted in a final ethanol production of 50.68 g/L ± 0.07 g/L, with an ethanol yield of 0.48 g/g.
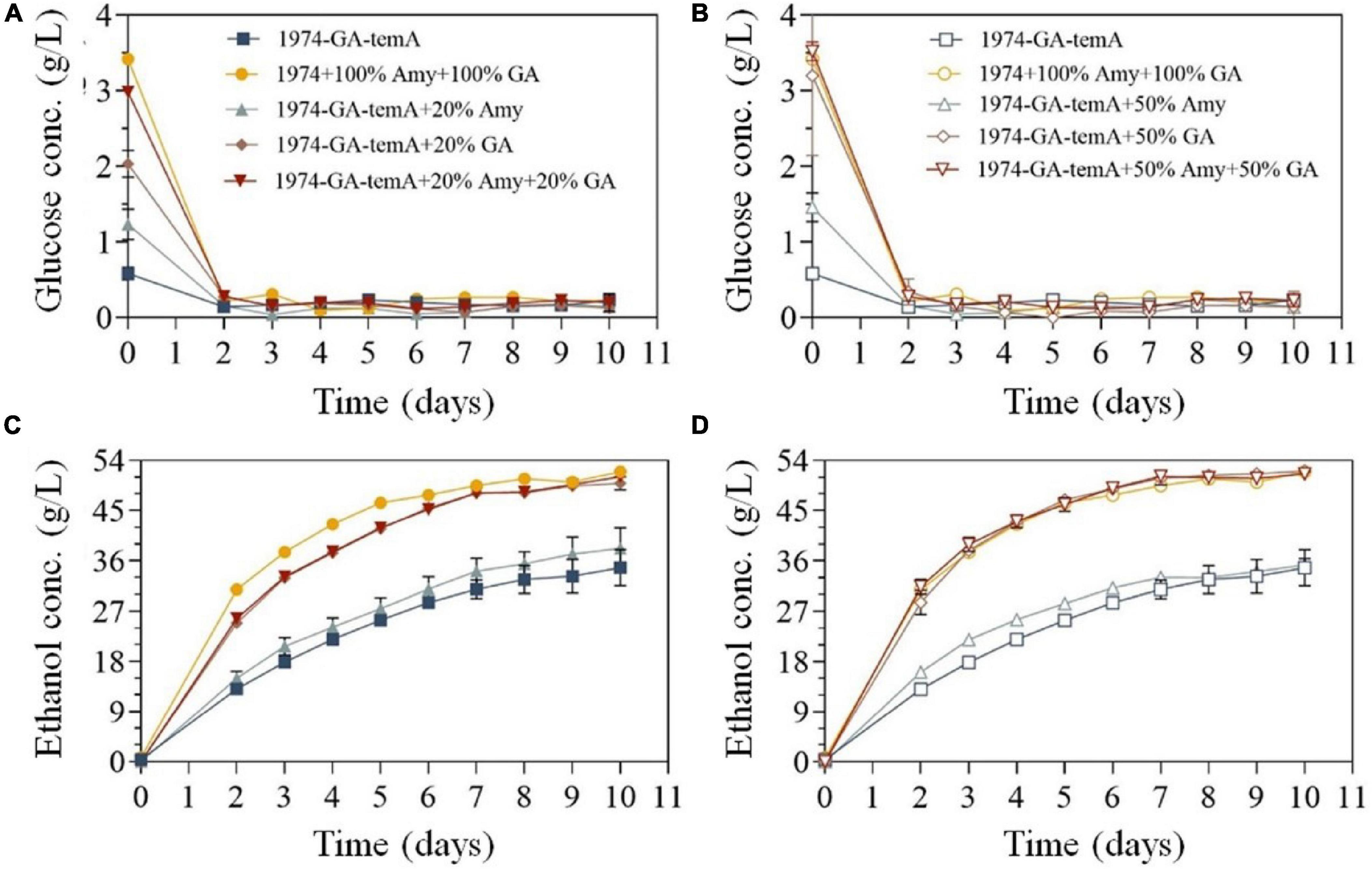
Figure 6. S. cerevisiae strains 1974 and 1974-GA-temA during fermentation in 250 mL fermentation flasks with SPR. The residual glucose concentrations (A,B) and ethanol concentrations (C,D) were monitored throughout fermentation. A total of 0.13 g of αα-amylase (3700 U/g) and 0.03 g of glucoamylase (100000 U/g) were used to supplement the CBP fermentation of strains, which were designated as 100% dosage. Data are the mean of three repeats showing standard deviation.
When the S. cerevisiae strain 1974-GA-temA was supplemented with exogenous α-amylase or glucoamylase, the ethanol production improved compared with that without exogenous α-amylase and/or glucoamylase amylase was added. Nevertheless, although exogenous α-amylase was added, the strain 1974-GA-temA cannot completely hydrolyze the starch after 10 days of fermentation. The results shown in Figure 6 demonstrate that the S. cerevisiae strain 1974-GA-temA can ferment starch after 10 days. This fermentation was achieved by adding either 20% exogenous glucoamylase or 20% exogenous α-amylase and 20% exogenous glucoamylase. In both cases, complete hydrolysis of starch was achieved. Furthermore, when 20% or 50% exogenous glucoamylase was supplemented, the starch was completely hydrolyzed on the 7th to 8th day of fermentation. In this case, the ethanol yield (50.55 g/L ± 0.13 g/L and 51.32 g/L ± 0.15 g/L, respectively) was comparable to the yields obtained when 100% α-amylase or 100% glucoamylase were added in the S. cerevisiae strain 1974 (Table 3). These findings indicate that exogenous glucoamylase exert a more pronounced influence on the fermentation of raw meal by the S. cerevisiae strain 1974-GA-temA. Considering its economic benefits, only 20% exogenous glucoamylase should be supplemented during the fermentation of SPR. Throughout the SPR fermentation cycle, the glucose in the culture medium was maintained at a very low level (Figure 6), indicating that the strain was metabolically active and could quickly utilize the decomposed glucose. In previous reports, as the fermentation time increased, the glucose content in the culture medium increased, indicating incomplete metabolism of the strain (Cripwell et al., 2019a,b). The results demonstrated that strain 1974-GA-temA constructed in this study showed great potential in SPR CBP bioethanol fermentation.
4 Conclusion
In this study, recombinant industrial ethanol-producing diploid S. cerevisiae strains with eight copies of α-amylase and eight copies of glucoamylase genes integration were successfully constructed. In vitro enzymatic activity analysis demonstrated that the optimal temperature for α-amylase and glucoamylase was 70°C, and the optimal pH for α-amylase and glucoamylase was 4.0 and 5.0, respectively. CBP fermentation of SPR with the recombinant amylolytic strain 1974-GA-temA demonstrated that the strain 1974-GA-temA produced 32.15 g/L ethanol in the absence of exogenous α-amylase and glucoamylase, corresponding to 59.30% of the theoretical yield. When 20% glucoamylase was added, ethanol production increased to 50.55 g/L, corresponding to 93.23% of the theoretical yield. This result is comparable to the parental strain 1974 fermented with 100% α-amylase and glucoamylase. Therefore, the yeast strain constructed in this study can be regarded as a potential CBP yeast for the commercial production of ethanol from SPR substrate.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
XW: Writing–review and editing, Writing–original draft, Supervision, Investigation, Funding acquisition, Conceptualization. NG: Writing–review and editing, Writing–original draft, Visualization, Methodology. JH: Writing–original draft, Methodology, Data curation. CG: Writing–original draft, Methodology. XX: Writing–original draft, Methodology. HZ: Methodology, Writing–review and editing. AL: Writing–review and editing, Funding acquisition. JH: Supervision, Funding acquisition, Writing–review and editing. MH: Writing–review and editing, Project administration. NL: Writing–review and editing, Writing–original draft, Project administration, Funding acquisition, Conceptualization.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of the article. This study was funded by the Open Project Program of the National Engineering Research Center of Wheat and Corn Further Processing, Henan University of Technology (Grant Number NL2022015), Major Science and Technology Projects in Henan Province (Grant Number 231100110300), Key Research and Development Project of Henan Province (Grant Number 231111310700), Science and Technology Foundation of Henan Province (Grant Number 232102310302), and Open Competition Research Projects of Xuchang University (Grant Number 2022JBGS06).
Acknowledgments
We would like to thank Professor Sheng Yang from the CAS Center for Excellence in Molecular Plant Sciences for kindly providing strain 1974.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1419293/full#supplementary-material
Abbreviations
CBP, consolidated bioprocessing; GA, glucoamylase; temA: α-amylase.
References
Aziz, N., and Mohsen, G. (2002). Bioconversion of acid- and gamma-ray-treated sweet potato residue to microbial protein by mixed cultures. J. Ind. Microbiol. Biotechnol. 29, 264–267. doi: 10.1038/sj.jim.7000297
Chandel, A., Garlapati, V., Singh, A., Antunes, F., and da Silva, S. (2018). The path forward for lignocellulose biorefineries: Bottlenecks, solutions, and perspective on commercialization. Bioresour. Technol. 264, 370–381. doi: 10.1016/j.biortech.2018.06.004
Cripwell, R., Favaro, L., Viljoen-Bloom, M., and van Zyl, W. (2020). Consolidated bioprocessing of raw starch to ethanol by Saccharomyces cerevisiae: Achievements and challenges. Biotechnol. Adv. 42:107579. doi: 10.1016/j.biotechadv.2020.107579
Cripwell, R., Rose, S., Viljoen-Bloom, M., and van Zyl, W. (2019b). Improved raw starch amylase production by Saccharomyces cerevisiae using codon optimisation strategies. FEMS Yeast Res. 19:127. doi: 10.1093/femsyr/foy127
Cripwell, R., Rose, S., Favaro, L., and van Zyl, W. (2019a). Construction of industrial Saccharomyces cerevisiae strains for the efficient consolidated bioprocessing of raw starch. Biotechnol. Biofuels 12:201. doi: 10.1186/s13068-019-1541-5
El Sheikha, A., and Ray, R. (2016). Potential impacts of bioprocessing of sweet potato: Review. Crit. Rev. Food Sci. Nutr. 57, 455–471. doi: 10.1080/10408398.2014.960909
Favaro, L., Viktor, M., Rose, S., Viljoen-Bloom, M., van Zyl, W., Basaglia, M., et al. (2015). Consolidated bioprocessing of starchy substrates into ethanol by industrial Saccharomyces cerevisiae strains secreting fungal amylases. Biotechnol. Bioeng. 112, 1751–1760. doi: 10.1002/bit.25591
Görgens, J., Bressler, D., and van Rensburg, E. (2014). Engineering Saccharomyces cerevisiae for direct conversion of raw, uncooked or granular starch to ethanol. Crit. Rev. Biotechnol. 35, 369–391. doi: 10.3109/07388551.2014.888048
Gou, C. C., Wang, X., Yu, Y., Huang, J., Wang, X., and Hui, M. (2023). One-step enzymatic hydrolysis of sweet potato residue after gelatinization for bioethanol production by Saccharomyces cerevisiae. Biomass Conversion Biorefinery 14, 15853–15862.
Hao, Z. H., Wang, D. J., Chen, H., Sun, J., and Xu, Y. (2014). Sweet potato starch residue as starting material to prepare polyacrylonitrile adsorbent via SI-SET-LRP. J. Agric. Food Chem. 62, 1765–1770. doi: 10.1021/jf4048397
Jin, Y. L., Ding, F., Wang, J., Yi, Z., Gao, Y., Yang, L., et al. (2022). One-step conversion of sweet potato waste to butanol via fermentation by Clostridium acetobutylicum. Biomass Conversion Biorefinery 14, 13087–13098.
Karim, A., Curran, K., and Alper, H. (2013). Characterization of plasmid burden and copy number in Saccharomyces cerevisiae for optimization of metabolic engineering applications. FEMS Yeast Res. 13, 107–116. doi: 10.1111/1567-1364.12016
Karmee, S. K. (2016). Liquid biofuels from food waste: Current trends, prospect and limitation. Renew. Sustain. Energy Rev. 53, 945–953.
Kong, I., Turner, T., Kim, H., Kim, S., and Jin, Y. (2018). Phenotypic evaluation and characterization of 21 industrial Saccharomyces cerevisiae yeast strains. FEMS Yeast Res. 18:1. doi: 10.1093/femsyr/foy001
Lorenzo, F., and Marina, B. (2010). Engineering amylolytic yeasts for industrial bioethanol production. Chem. Eng. Trans. 20, 97–102.
Lu, H. J., Gui, Y., Zheng, L., and Liu, X. (2013). Morphological, crystalline, thermal and physicochemical properties of cellulose nanocrystals obtained from sweet potato residue. Food Res. Int. 50, 121–128.
Myburgh, M. W., Cripwell, R. A., Favaro, L., and van Zyl, W. H. (2019). Application of industrial amylolytic yeast strains for the production of bioethanol from broken rice. Bioresour. Technol. 294:122222. doi: 10.1016/j.biortech.2019.122222
Osman, A. I., Qasim, U., Jamil, F., Al-Muhtaseb, A. H., Jrai, A. A., Al-Riyami, M., et al. (2021). Bioethanol and biodiesel: Bibliometric mapping, policies and future needs. Renew. Sustain. Energy Rev. 152:111677.
Pagana, I., Morawicki, R., and Hager, T. J. (2014). Lactic acid production using waste generated from sweet potato processing. Int. J. Food Sci. Technol. 49, 641–649.
Sakwa, L., Cripwell, R., Rose, S., and Viljoen-Bloom, M. (2018). Consolidated bioprocessing of raw starch with Saccharomyces cerevisiae strains expressing fungal alpha-amylase and glucoamylase combinations. FEMS Yeast Res. 18:85. doi: 10.1093/femsyr/foy085
Stepchenkova, E., Zadorsky, S., Shumega, A., and Aksenova, A. (2023). Practical approaches for the yeast Saccharomyces cerevisiae genome modification. Int. J. Mol. Sci. 24:11960. doi: 10.3390/ijms241511960
Viktor, M., Rose, S., van Zyl, W., and Viljoen-Bloom, M. (2013). Raw starch conversion by Saccharomyces cerevisiae expressing Aspergillus tubingensis amylases. Biotechnol. Biofuels 6:167. doi: 10.1186/1754-6834-6-167
Wang, F., Jiang, Y., Guo, W., Niu, K., Zhang, R., Hou, S., et al. (2016). An environmentally friendly and productive process for bioethanol production from potato waste. Biotechnol. Biofuels 9:50. doi: 10.1186/s13068-016-0464-7
Wang, X., Liao, B., Li, Z., Liu, G., Diao, L., Qian, F., et al. (2021). Reducing glucoamylase usage for commercial-scale ethanol production from starch using glucoamylase expressing Saccharomyces cerevisiae. Bioresour Bioprocess. 8, 20. doi: 10.1186/s40643-021-00375-5
Wang, X., Yang, J., Yang, S., and Jiang, Y. (2019). Unraveling the genetic basis of fast l-arabinose consumption on top of recombinant xylose-fermenting Saccharomyces cerevisiae. Biotechnol. Bioeng. 116, 283–293. doi: 10.1002/bit.26827
Keywords: sweet potato residue fermentation, bioethanol production, recombinant amylolytic Saccharomyces cerevisiae strain construction, without exogenous enzyme addition, consolidated bioprocessing
Citation: Wang X, Guo N, Hu J, Gou C, Xie X, Zheng H, Liao A, Huang J, Hui M and Liu N (2024) Construction of an amylolytic Saccharomyces cerevisiae strain with high copies of α-amylase and glucoamylase genes integration for bioethanol production from sweet potato residue. Front. Microbiol. 15:1419293. doi: 10.3389/fmicb.2024.1419293
Received: 18 April 2024; Accepted: 19 July 2024;
Published: 06 August 2024.
Edited by:
Zhiqiang Wen, Nanjing Normal University, ChinaReviewed by:
Riaan Den Haan, University of the Western Cape, South AfricaFu-Li Li, Chinese Academy of Sciences (CAS), China
Copyright © 2024 Wang, Guo, Hu, Gou, Xie, Zheng, Liao, Huang, Hui and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin Wang, d2FuZ3hpbkBoYXV0LmVkdS5jbg==; Na Liu, bGl1bmEzNDU2QDE2My5jb20=
 Xin Wang
Xin Wang Na Guo
Na Guo Jingting Hu2
Jingting Hu2 Haobo Zheng
Haobo Zheng Aimei Liao
Aimei Liao Na Liu
Na Liu