- Department of Biology, University of Regina, Regina, SK, Canada
Members of the bacterial genus Pantoea produce a variety of antimicrobial products that are effective against plant, animal, and human pathogens. To date, little is known about the distribution and evolutionary history of these clusters. We surveyed the public databases for the 12 currently known antibiotic biosynthetic gene clusters found across Pantoea strains to determine their distribution. We show that some clusters, namely pantocin B, PNP-3, and PNP-4 are found strictly in Pantoea, while agglomerin, andrimid, AGA, dapdiamide, herbicolin, PNP-1, PNP-2, PNP-5, and pantocin A, are more broadly distributed in distantly related genera within Vibrionaceae, Pectobacteriaceae, Yersiniaceae, Morganellaceae, and Hafniaceae. We evaluated the evolutionary history of these gene clusters relative to a cpn60-based species tree, considering the flanking regions of each cluster, %GC, and presence of mobile genetic elements, and identified potential occurrences of horizontal gene transfer. Lastly, we also describe the biosynthetic gene cluster of pantocin B in the strain Pantoea agglomerans Eh318 more than 20 years after this antibiotic was first described.
1 Introduction
The genus Pantoea is a diverse group within the Erwiniaceae whose members inhabit a variety of aquatic and terrestrial environments (Walterson and Stavrinides, 2015). Members of Pantoea have also been found in association with various insects, animals, and humans (Walterson and Stavrinides, 2015). As a genus, Pantoea has a variety of unique characteristics and capabilities that make it of interest in both environmental and clinical settings including applications in biocontrol, bioremediation, biosensing, and therapeutics (Walterson and Stavrinides, 2015). Of significance, many Pantoea strains produce natural product antimicrobials effective against human, plant, and/or animal pathogens (Walterson and Stavrinides, 2015). These antimicrobials are primarily synthesized through secondary metabolic pathways via biosynthetic gene clusters (BGCs), which include genes that encode enzymes, as well as genes involved in regulation, export, and resistance. To date, there are at least 22 known unique antimicrobials found in members of Pantoea, which collectively represent 12 different BGCs.
Among the earliest described antimicrobials produced by Pantoea were the tetronate antibiotics, agglomerin A, B, C, and D, which were isolated from Pantoea agglomerans PB-6042 (Shoji et al., 1989). These compounds are synthesized by the same seven gene cluster and differ only in the structure of the hydrocarbon chain found on the acyl group of the compound (Terui et al., 1990; Kanchanabanca et al., 2013). The agglomerins show moderate activity against both Gram-positive and Gram-negative anaerobic bacteria (Shoji et al., 1989). Andrimid, another Pantoea antibiotic, is a pseudopeptide synthesized through a hybrid NRPS-PKS pathway (Jin et al., 2006). Andrimid was first extracted from the broth culture of a symbiont of a brown planthopper in 1987 (Fredenhagen et al., 1987); however, the 21-gene cluster responsible for the synthesis of andrimid was not described until almost 20 years later in P. agglomerans Eh335 (Jin et al., 2006). Andrimid acts by inhibiting the β-subunit of acetyl-CoA carboxylase in fatty acid synthesis, thus inhibiting cell growth (Freiberg et al., 2004; Jin et al., 2006). Andrimid is effective against both Gram-positive and Gram-negative pathogens, including methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus (Needham et al., 1994).
Pantoea also produces phenazine antibiotics, most notably, D-alanylgriseoluteic acid (AGA). AGA was first described in P. agglomerans Eh1087 as inhibiting the growth of the phytopathogen, Erwinia amylovora, the causative agent of fire blight in apples and pears (Kearns and Hale, 1996; Kearns and Mahanty, 1998; Giddens et al., 2002). The spectrum of activity of AGA has since been expanded to include many Gram-positive bacteria including clinical strains of S. pneumoniae and methicillin-resistant S. aureus (Giddens and Bean, 2007). A 16-gene cluster is responsible for AGA biosynthesis and resistance, but also directs the synthesis of at least two phenazine intermediates: phenazine-1,6-dicarboxylic acid and griseoluteic acid (Giddens et al., 2002).
The tripeptide antibiotics, dapdiamide A, B, C, D, and E were first identified from P. agglomerans CU0119 (Dawlaty et al., 2010). These structurally similar compounds share a central L-2,3-diaminopropionic acid fragment, but differ in their two amide-linked side chains (Dawlaty et al., 2010). Dapdiamides A-E are synthesized by a nine-gene cluster (Dawlaty et al., 2010). The dapdiamide cluster is homologous to two other Pantoea antibiotic BGCs: herbicolin I produced by Pantoea vagans C9-1, and 2-amino-3-(oxirane-2,3-dicarboxamido)-propanoyl-valine (APV) produced by P. agglomerans 48b/90 (Völksch and Sammer, 2008; Smits et al., 2010, 2011; Kamber et al., 2012). Herbicolin I and APV are structurally identical to dapdiamide E, although the stereochemistry of these has not been confirmed (Völksch and Sammer, 2008; Smits et al., 2011; Kamber et al., 2012).
Pantoea strains have also been reported to produce the antifungals, herbicolin A and B. These two lipopeptides were first isolated from P. agglomerans A 111 (Winkelmann et al., 1981), and later from P. agglomerans ZJU23 (Xu et al., 2022), which led to the characterization of the BGC responsible for the production of both molecules and their mechanism of action. Herbicolin A is synthesized by a 10-gene cluster and is thought to inhibit sterol-containing fungi through the disruption of ergosterol-containing lipid rafts in fungal cell membranes (Xu et al., 2022). Herbicolin B is an intermediate of herbicolin A and is glycosylated by AcbI in the last steps of synthesis to form herbicolin A (Xu et al., 2022). Herbicolin B is also biologically active against sterol-containing fungi, but less so than herbicolin A (Greiner and Winkelmann, 1991; Xu et al., 2022).
The peptide antibiotics pantocin A and B, produced by P. agglomerans Eh318, were first described as inhibitors of E. amylovora (Wright et al., 2001). Pantocin A acts by inhibiting histidine biosynthesis, and thus, its activity is neutralized if exogenous histidine is available (Wright et al., 2001; Jin et al., 2003a). The BGC for pantocin A has been described as a three-gene operon approximately 2.5 kb in size (Jin et al., 2003b). A small open reading frame (ORF) directly upstream of the cluster, which encodes a 30-amino acid precursor peptide is also required for pantocin A biosynthesis (Jin et al., 2003b). The BGCs of two other previously described Pantoea antibiotics, the microcin MccEh252, produced by P. agglomerans Eh252, and herbicolin O, produced by P. vagans C9-1 share high sequence identity to the pantocin A BGC (Vanneste et al., 2008). Genetic and chemical analysis of herbicolin O has recently confirmed it to be the same antibiotic as pantocin A (Ishimaru et al., 2017). Some discrepancies between the physiochemical characteristics of MccEh252 and pantocin A have been reported; however, these may be due to differences in assaying methodologies (Vanneste et al., 2008). The range of activity of MccEh252 is also indistinguishable from that of pantocin A (Vanneste et al., 2008); thus, it is likely that these two antibiotics are identical. In contrast to pantocin A, pantocin B is arginine reversible and works to inhibit its target by disrupting arginine biosynthesis through inhibition of N-acetylornithine transaminase (Brady et al., 1999). Although the structure and mechanism of action are known for pantocin B (Brady et al., 1999), the BGC has yet to be published.
BGCs responsible for the production of Pantoea Natural Products (PNP) 1 through 5 have also been described, but little is known about their structures or modes of action (Walterson et al., 2014; Robinson et al., 2020; Williams et al., 2020; Williams and Stavrinides, 2020; Kirk and Stavrinides, 2023). PNP-1 from Pantoea ananatis BRT175 is encoded by an eight-gene cluster and is effective against E. amylovora and select Pantoea strains (Walterson et al., 2014). After the discovery of the PNP-1 gene cluster, the structure of PNP-1 was determined to be 4-formylaminooxyvinylglycine, a non-proteogenic amino acid belonging to the class of antibiotics known as the oxyvinylglycines (Okrent et al., 2018). The cluster for PNP-2, identified in the cystic fibrosis isolate P. agglomerans TX10 is composed of six genes (Robinson et al., 2020). The resulting antibiotic is broad-spectrum, with the ability to inhibit both Gram-positive and Gram-negative bacteria including Enterobacter, Escherichia, Klebsiella, Kosakonia, Pseudocitrobacter, Salmonella, Staphylococcus, and Streptococcus (Robinson et al., 2020). PNP-3 is also broad-spectrum, with the ability to inhibit drug-resistant strains of Pseudomonas aeruginosa and Acinetobacter baumannii in addition to Klebsiella spp., E. coli, Enterobacter spp., S. aureus, and Streptococcus mutans (Williams and Stavrinides, 2020). PNP-3 is encoded by an eight-gene cluster, and was found to be produced by P. agglomerans strains 3581 and SN01080 (Williams and Stavrinides, 2020).
The BGC for PNP-4 found in P. agglomerans B025670 consists of 14 genes and was identified using comparative genomic approaches (Williams et al., 2020). PNP-4 is effective against Enterobacter spp., E. amylovora, E. coli, Kosakonia spp., Pseudocitrobacter spp., and Salmonella Typhimurium, including some MDR strains (Williams et al., 2020). Lastly, the cluster for PNP-5, found in the clinical isolate P. agglomerans 20KB447973, is composed of 10 genes and shares similarity to previously described gene clusters for the dithiolopyrrolone antibiotic, holomycin, found in Streptomyces clavuligerus, Yersinia ruckeri, and Photobacterium galatheae (Li and Walsh, 2010; Qin et al., 2013; Sheng-Da et al., 2021). PNP-5 has not yet been confirmed to be structurally identical to holomycin. PNP-5 shows activity against a variety of Gram-positive and Gram-negative bacteria including Citrobacter spp., E. hormaechei, Enterobacter spp., E. amylovora, E. coli, Klebsiella spp., L. lactis, Salmonella Typhimurium, and S. mutans (Kirk and Stavrinides, 2023).
In this study, we determined the distribution of these 12 Pantoea antibiotic BGCs. We then created phylogenies to establish the evolutionary relationships between homologous clusters from different species and combined this with an analysis of the flanking regions to help assess the potential evolutionary history of each cluster. We show that some antibiotic gene clusters may represent more recent acquisitions, while other clusters that appear widely distributed among Pantoea strains may be older. We also delineate and describe the sequence of the pantocin B gene cluster from the strain P. agglomerans Eh318.
2 Materials and methods
2.1 Sequences and BLAST
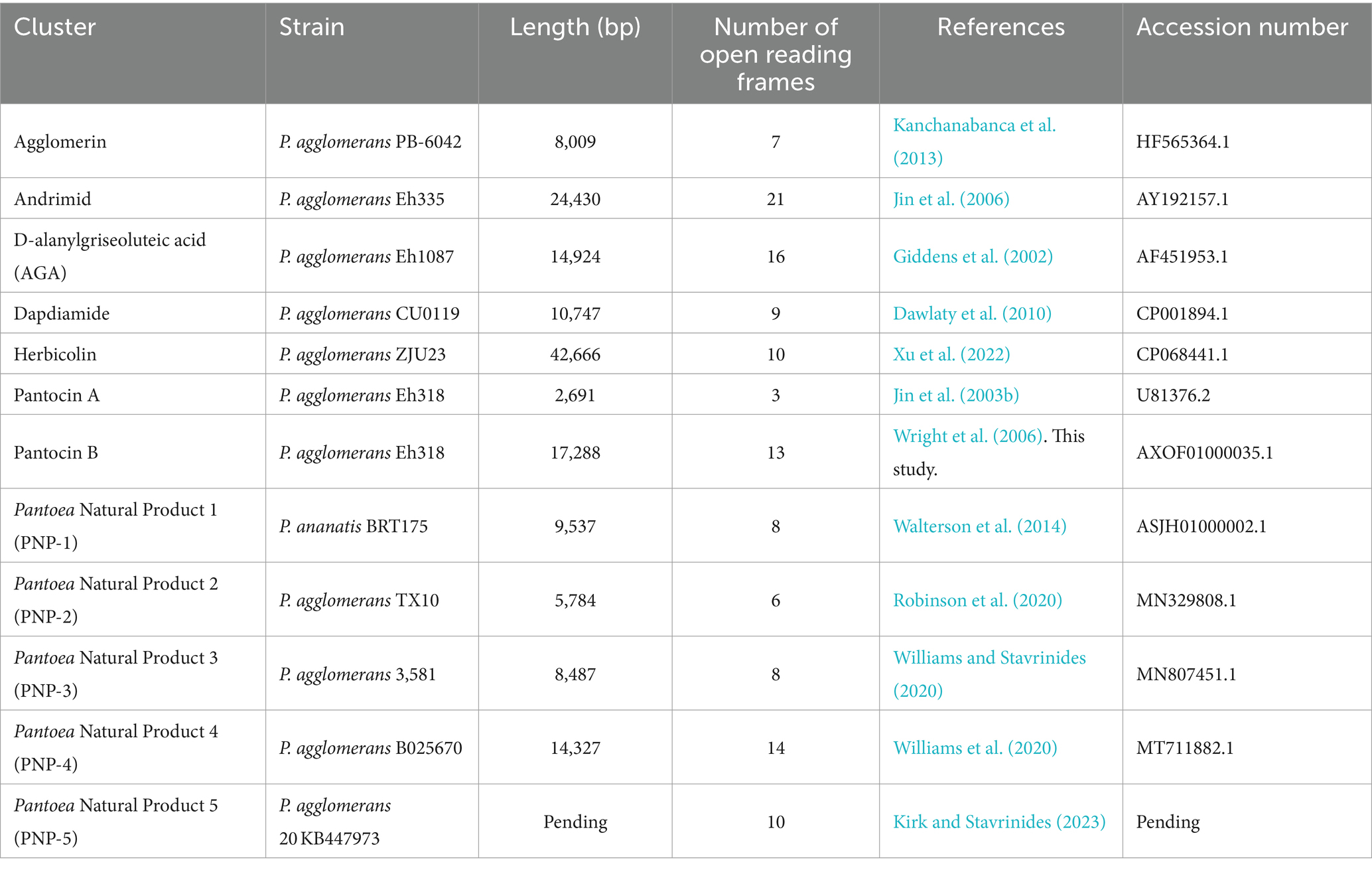
The distributions of 12 antibiotic BGCs identified in Pantoea were assessed: agglomerin, andrimid, AGA, dapdiamide, herbicolin, PNP-1, PNP-2, PNP-3, PNP-4, PNP-5, pantocin A, and pantocin B (Table 1). BGCs were BLASTed against the non-redundant (nr) and whole-genome shotgun contigs (wgs) databases at NCBI1 using default blastn parameters (Supplementary Table 1). Standalone BLAST was also used for some incomplete and draft genomes using a word size of 11 with all other parameters set to default. Candidate BGCs that did not contain all of the genes found in the query cluster, or whose genes were not entirely syntenic relative to the query cluster were excluded. Also excluded were candidate BGCs that diverged by more than 60% nucleotide identity across the length of the query cluster. BGCs that met our thresholds, but were found across contigs in draft genomes were included in our analyses. The 5 kb flanking each end of each cluster was also extracted and ORFs were predicted using GeneMark.hmm with Heuristic Models (Besemer and Borodovsky, 1999). Clinker (Gilchrist and Chooi, 2021) was used to visualize clusters, and to identify homologs between clusters. Parameters were set to default, with a 30% amino acid identity cut-off for this analysis.
2.2 Phylogenetic analysis
The nucleotide sequences of homologous gene clusters together with their intergenic regions were aligned with MUSCLE (Madeira et al., 2019) using default parameters. Maximum-likelihood trees for each gene cluster set were constructed with MEGAX (Stecher et al., 2020) using best-fit models (Supplementary Table 2) with 1,000 bootstrap replicates. Maximum-likelihood species trees were generated using available cpn60 sequences for representative strains.
3 Results
3.1 Agglomerin
Homologs of the agglomerin cluster described from P. agglomerans PB-6042 were identified in a total of 181 strains, 178 of which belonged to the genus Dickeya (Table 2; Supplementary Tables 3, 4). No homologs were identified in any other strain of Pantoea. Neither the genome nor any barcoding regions for P. agglomerans PB-6042 are currently available to confirm the identity of this strain; consequently, the agglomerin cluster was not analyzed further.
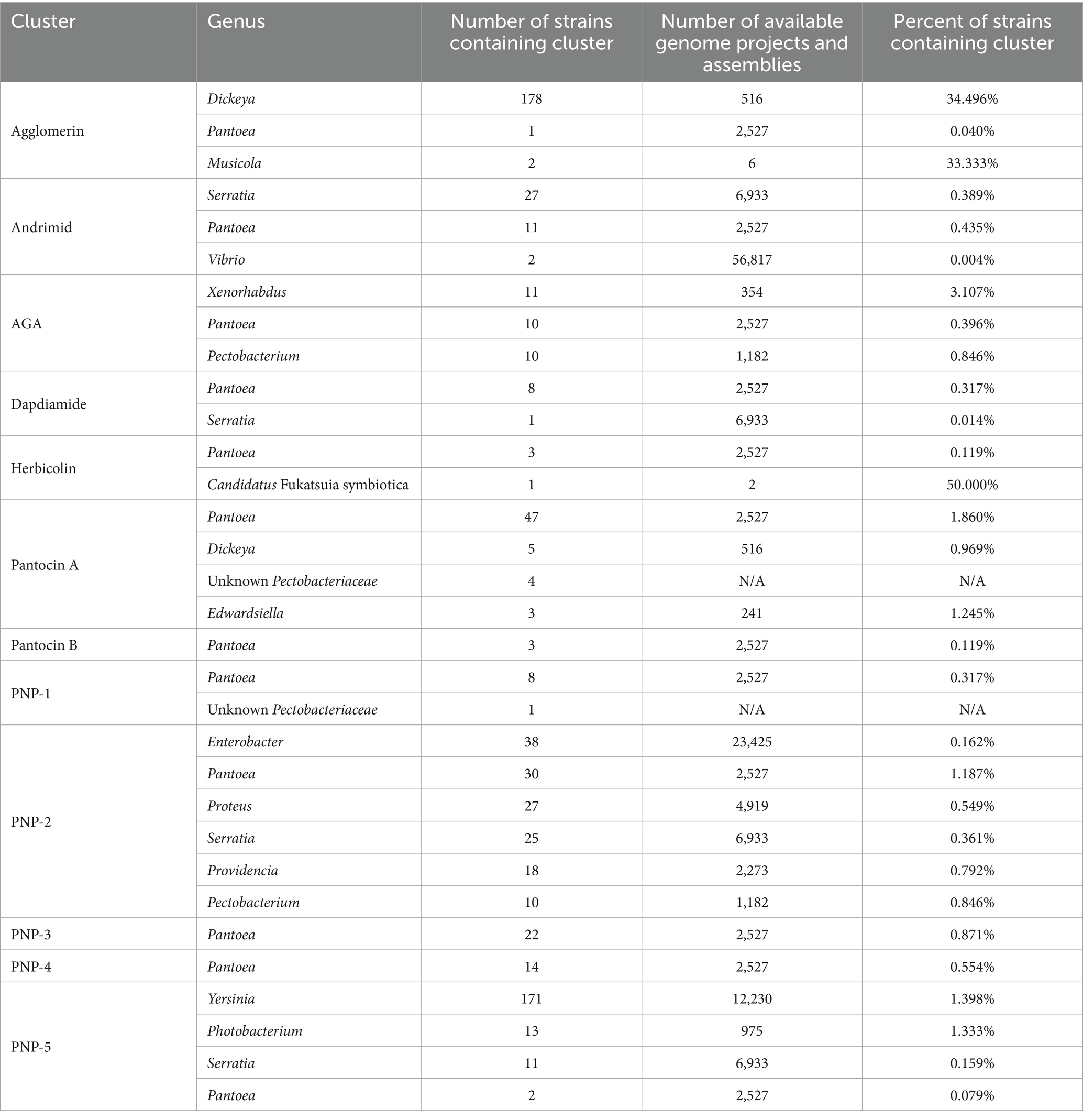
Table 2. Distribution of Pantoea antibiotic biosynthetic gene clusters across representative genera.
3.2 Andrimid
The andrimid BGC was identified in 11 genomes from the nr database and 29 from the wgs (Supplementary Table 3). Of these 40 BGCs, 11 were found in Pantoea: six within P. ananatis, two within P. stewartii, one within P. agglomerans, and two within uncharacterized Pantoea strains (Supplementary Table 4). Homologs of the andrimid cluster were also found in members of Serratia and Vibrio (Supplementary Table 4). Serratia had the greatest number of andrimid homologs out of the three representative genera (Supplementary Table 4); however, there were almost three times the number of Serratia genome projects and assemblies available as compared to Pantoea (Table 2). A phylogenetic analysis of the andrimid cluster revealed two distinct lineages within each clade (Supplementary Figure 1A). The first lineage contained all Pantoea and Vibrio strains as well as some Serratia representatives, while the other contained only a subset of the Serratia representatives (Supplementary Figure 1A). The cpn60 tree shows the expected relationships between representative taxa, with each species forming its own unique lineage (Supplementary Figure 1B).
Predicted ORFs flanking andrimid clusters were largely conserved within genera although there were some exceptions (Supplementary Figure 1C). The flanking region of the P. agglomerans Eh355 cluster was not conserved relative to the other Pantoea clusters, and there was also significant intra-species variation in the flanking regions of S. plymuthica andrimid clusters (Supplementary Figure 1C). One predicted ORF encoding a LysR-family transcriptional regulator was found upstream of all Pantoea and Serratia andrimid clusters, but not upstream of the Vibrio BGCs (Supplementary Figure 1C; orange arrow). Similarly, directly upstream of the andrimid cluster, a hypothetical protein-encoding gene was predicted in both Vibrio strains, most Serratia, and P. agglomerans Eh335 (Supplementary Figure 1C; green arrow). Interestingly, the last gene in the published andrimid cluster, admU, a predicted transposase, was only found in the original strain, P. agglomerans Eh355 (Supplementary Figure 1C). An analysis of the %GC of the andrimid cluster from P. agglomerans Eh335 revealed a GC content of 46.87%, while the genome of P. agglomerans Eh335 had a GC content of 54.56% (Table 3).
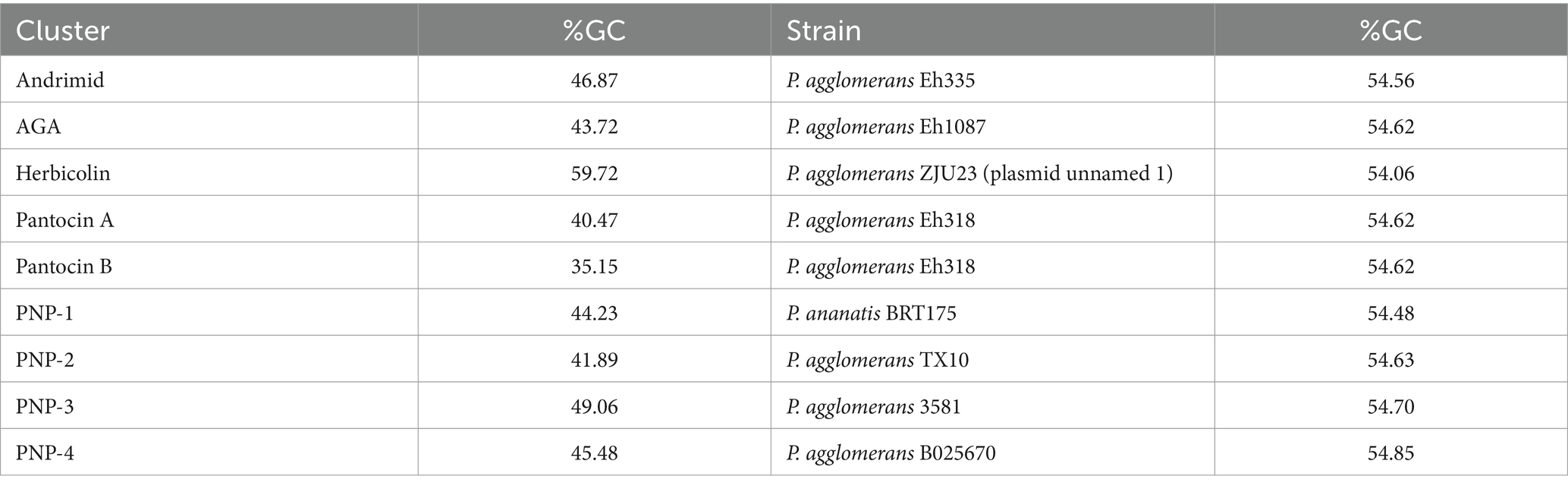
Table 3. %GC content of Pantoea antibiotic biosynthetic gene clusters and their host genomes/plasmids.
3.3 D-alanylgriseoluteic acid
AGA cluster homologs were identified in 10 P. agglomerans strains, 10 Pectobacterium strains and 11 Xenorhabdus strains for a total of 31 AGA-containing representatives (Supplementary Tables 3, 4). AGA clusters were identified in a relatively small proportion of sequenced Pantoea genomes (0.4%) whereas it was distributed across 3% of available Xenorhabdus genomes (Table 2). The phylogeny of the AGA clusters formed three distinct lineages corresponding to the three genera, and this phylogeny was congruent with the cpn60 tree (Supplementary Figures 2A,B). An analysis of the flanking regions of all Pantoea strains containing the AGA cluster revealed extensive conservation, except for potential frameshifts in the second predicted ORF upstream of the clusters and the third predicted ORF downstream of the clusters of P. agglomerans 190, CFSAN047154, CFSAN047153, Pa39-23, and Pa39-21 (Supplementary Figure 2C). Very few flanking regions of Pectobacterium AGA clusters were available, but those that could be analyzed exhibited little conservation between representatives (Supplementary Figure 2C). Flanking regions of BGCs identified in Xenorhabdus showed conservation within, but not across the two representative species, with Xenorhabdus sp. SF857 flanking regions found to be highly divergent in comparison to other Xenorhabdus strains (Supplementary Figure 2C). Interestingly, an approximately 500 bp ORF predicted to encode a DUF2165 family protein was identified downstream of the cluster across all representatives in all three genera, suggesting it may be important for AGA biosynthesis (Supplementary Figure 2C; purple arrow). Notably, the %GC of the P. agglomerans Eh1087 AGA cluster was over 10% lower than that of the genome (Table 3).
3.4 Dapdiamide
The dapdiamide cluster was found almost exclusively in P. agglomerans, with seven representatives carrying the cluster (Supplementary Tables 3, 4). Homologous clusters were also identified in both P. vagans and Serratia inhibens (Supplementary Table 4). A phylogenetic analysis of the dapdiamide clusters showed all Pantoea representatives grouping together (Supplementary Figure 3A), although P. vagans C9-1 nested within the agglomerans clusters (Supplementary Figures 3A,B). Further, P. agglomerans C410P1 and SI1_M5 formed sister taxa in the gene cluster tree, but not in the cpn60 tree (Supplementary Figure 3B). The flanking regions surrounding the dapdiamide clusters showed the least amount of conservation in comparison to other antibiotic BGCs (Supplementary Figure 3C). Only one ORF predicted to encode a thioredoxin domain-containing protein directly downstream of the Pantoea clusters was found to be conserved, except in P. agglomerans DAPP-PG734 (Supplementary Figure 3C; green arrow). There were no shared ORFs between the flanking regions of Pantoea and the single Serratia representative.
3.5 Herbicolin
The sequence of the herbicolin cluster was obtained from the P. agglomerans ZJU23 genome (Xu et al., 2022). Our open-reading frame prediction resulted in two additional ORFs between acbB and acbC not previously described (Supplementary Figure 4C; Xu et al., 2022). These additional ORFs appeared to be the result of frameshift mutations in acbC, which may be sequencing artifacts. A search of the databases for this cluster identified it in three Pantoea strains and one Candidatus Fukatsuia symbiotica strain (Supplementary Tables 3, 4). It is unclear how widely distributed this cluster is across Candidatus F. symbiotica strains as there were only two genomes available; however, the herbicolin cluster appeared narrowly distributed across Pantoea (Table 2). Two P. agglomerans clusters were more closely related to each other than to the other two Pantoea representatives, which was consistent with the cpn60 species tree (Supplementary Figures 4A,B). The ORFS upstream of the four BGCs were not conserved, although some of the genes downstream were conserved across the three Pantoea BGCs (Supplementary Figure 4C). The herbicolin cluster appears to be plasmid-encoded in P. agglomerans ZJU23, P. agglomerans 9Rz4 and Candidatus F. symbiotica 5D. The P. agglomerans ZJU23 herbicolin cluster had a 59.72% GC content, while its host plasmid had a GC content of 54.06% (Table 3).
3.6 Pantocin A
Fifty-nine strains were identified as carrying the pantocin A cluster, 47 of which were members of Pantoea and the remaining 12 were strains of Dickeya chrysanthemi, Edwardsiella hoshinae, and various members of the Pectobacteriaceae (Supplementary Table 4). Among members of Pantoea, the pantocin A cluster was most prevalent in P. agglomerans and P. ananatis, but it is also present in strains of P. brenneri, P. vagans, P. stewartii, and Pantoea spp. (Supplementary Table 4). The representative pantocin A clusters grouped into four distinct lineages that correspond to each genus, with the only exception being D. chrysanthemi Ech1591, which clustered with Pantoea strains instead of other Dickeya (Supplementary Figure 5A). The Pantoea lineage is subdivided into two well-supported lineages, one containing only P. ananatis and two P. stewartii strains, and the other all other representative Pantoea strains including one subset of P. ananatis strains (Supplementary Figure 5A). The antibiotic cluster tree was found to be mostly incongruent with the corresponding cpn60 tree at the individual gene cluster level, although highly congruent at the genus level (Supplementary Figures 5A,B).
A comparison of the clusters identified in Edwardsiella, Dickeya, and the members of the Pectobacteriaceae showed that their flanking regions were mostly conserved, in contrast to those of Pantoea, which were variable (Supplementary Figure 5C). One ORF predicted to encode a hypothetical protein directly upstream of the cluster was found to be conserved across most Pantoea strains except for P. agglomerans Eh318, 540Y, Pa39-3, 20TX10122, Pa39-1, TX10, and PA4 (Supplementary Figure 5C; purple arrow). A predicted ABC transporter ATP-binding protein found upstream of the clusters of P. ananatis strains PNA_18_8S, PNA_18_9S, and PNA_18_10S was also identified upstream of the clusters of Pectobacteriaceae strains C52, C80, and CE70 and (Supplementary Figure 5C). The %GC content of P. agglomerans Eh318 was calculated to be 54.62%, while the pantocin A cluster from this strain was significantly lower at 40.47% GC (Table 3).
3.7 Pantocin B
At the time of our survey, the pantocin B gene cluster had not yet been fully described. To identify the BGC in the P. agglomerans Eh318 genome, we used a Perl script to search the available genome sequence for restriction site patterns that were previously reported (Wright et al., 2006). A single ~25 kb region with a nearly identical restriction site pattern was identified, which also matched the cluster size (~17 kb) and number of ORFs (13) that had been predicted previously (Supplementary Figure 6; Wright et al., 2006). Predicted protein products of ORFs in the pantocin B cluster based on BLASTx searches can be found in Supplementary Figure 6. Analysis of the distribution of the pantocin B gene cluster identified only two strains in P. ananatis that carry it in addition to P. agglomerans Eh318 (Supplementary Tables 3, 4; Supplementary Figure 7). The flanking regions of the cluster in the two P. ananatis strains were consistent with one another, although it is predicted that these strains may be clonal (Supplementary Figure 7C). The %GC content of the pantocin B cluster in P. agglomerans Eh318 was calculated to be almost 20% lower than that of the Eh318 genome (Table 3).
3.8 PNP-1
Eight homologous PNP-1 clusters were identified within P. ananatis and P. stewartii subsp. indologenes strains, along with one cluster in a strain belonging to a member of the Pectobacteriaceae (Supplementary Tables 3, 4). Clusters identified in P. stewartii formed a separate monophyletic group from those of P. ananatis, which was consistent with the relationship between these two species as shown in the cpn60 tree (Supplementary Figures 8A,B). The Pectobacteriaceae bacterium CE90 cluster also formed its own unique lineage in the cluster phylogeny, consistent with the species tree (Supplementary Figures 8A,B). The flanking regions of the PNP-1 clusters were conserved across Pantoea, with some divergence seen upstream of the clusters between P. ananatis and P. stewartii subsp. indologenes strains, with some of the P. ananatis strains appearing to be clonal (i.e., PANS 99–36, PANS_99_36, and 99–36; PANS_99_25 and 99–25) (Supplementary Figures 8A–C). Flanking regions of Pantoea strains and the single Pectobacteriaceae representative were not conserved (Supplementary Figure 8C). Additionally, the %GC content of the PNP-1 cluster in P. ananatis BRT175 is approximately 10% lower than that of the BRT175 genome (Table 3).
3.9 PNP-2
The PNP-2 cluster was found to be broadly distributed, with homologs identified in 148 strains (Supplementary Tables 3, 4). The majority of Pantoea representatives containing the PNP-2 cluster were P. agglomerans, with others belonging to the species P. vagans and P. pleuroti, as well as unclassified Pantoea spp. (Supplementary Table 4). Almost half of the strains containing the PNP-2 cluster were non-Pantoea strains and included those from the genera Providencia, Proteus, Pectobacterium, Enterobacter, and Serratia (Supplementary Table 4). Proportionally, however, PNP-2 was found to be most broadly distributed within Pantoea (Table 2). Of note, homologs of pnp2E, a predicted ferredoxin reductase, in Proteus strains were truncated, with only the first portion of the gene being conserved (Supplementary Figure 9C).
A phylogenetic analysis of the PNP-2 BGC showed that clusters from each genus formed their own distinct lineages, with a few exceptions (Supplementary Figure 9A). First, P. vagans Mg1, P. vagans UBA6298, and Pantoea sp. S62 formed their own lineage separate from other Pantoea representatives, which was consistent with the cpn60 tree (Supplementary Figures 9A,B). Second, several E. asburiae strains along with P. agglomerans T6 and SI1_M5 formed their own lineage separate from both genera (Supplementary Figure 9A). In contrast, these strains grouped with their respective species groups in the cpn60 tree (Supplementary Figure 9B).
The flanking regions of the PNP-2 clusters identified in Proteus and Providencia were conserved in all representative strains, while the flanking regions of the Enterobacter clusters were conserved across all strains except for the six E. asburiae strains that were similar to each other (Supplementary Figure 9C). Similarly, the majority of Pectobacterium strains shared flanking regions, except for Pe. colocasium LJ1 and Pectobacterium sp. F1-1, which were more similar to each other, and Pe. carotovorum 251, which was unique (Supplementary Figure 9C). The most diversity in flanking regions was seen in Serratia and Pantoea (Supplementary Figure 9C). Additionally, the %GC of the PNP-2 cluster in P. agglomerans TX10 was significantly lower (41.89%) than that of the genome (Table 3).
3.10 PNP-3
The PNP-3 cluster was found only in members of Pantoea (Supplementary Tables 3, 4). Of the 22 strains, most were P. agglomerans and P. vagans, along with some strains identified as only Pantoea spp. (Supplementary Tables 3, 4). The PNP-3 cluster phylogeny formed three main lineages (Supplementary Figure 10A). The first lineage contained only P. agglomerans, the second contained P. agglomerans strains along with P. vagans C9-1, and the third contained only Pantoea spp. (Supplementary Figure 10A). Although P. agglomerans strains were not fully resolved in the cpn60 tree, there was incongruence between the two trees, particularly within the second lineage, which grouped five P. agglomerans strains with the single P. vagans strain (Supplementary Figures 10A,B). Flanking regions of PNP-3 clusters showed high variability despite all representatives belonging to three Pantoea species (Supplementary Figure 10C). ORFs predicted to encode hypothetical proteins were found downstream of the cluster in all but four representatives (Supplementary Figure 10C; pink arrow). There were also several frameshift mutations identified within the PNP-3 cluster in several strains: P. agglomerans 553Y, 540Y, Pa31-4, Pa39-1, Pa39-7, Pa39-21, Pa39-23, P. vagans C9-1, Pantoea sp. M_6, M_8, and M_10 (Supplementary Figure 10C). The %GC of the P. agglomerans 3581 PNP-3 cluster is 49.06%, while that of the genome is 54.70% (Table 3).
3.11 PNP-4
The PNP-4 cluster was also restricted to Pantoea, but was distributed across multiple representatives of P. dispersa, P. deleyi, P. ananatis, P. agglomerans, and Pantoea spp. for a total of 14 representatives (Supplementary Tables 3, 4). Three distinct monophyletic groups emerged in the PNP-4 cluster tree (Supplementary Figure 11A), including one that comprised P. agglomerans strains as well as Pantoea spp. and P. deleyi, and the two others P. dispersa and P. ananatis, exclusively (Supplementary Figure 11A). The cpn60 tree closely mirrored the cluster tree, except P. deleyi LMG24200, which formed its own lineage in the cpn60 tree (Supplementary Figure 11B).
Flanking regions of PNP-4 clusters fell into four main patterns. The first group was composed of clusters from P. deleyi and P. agglomerans strains, and the second group comprised Pantoea sp. EKM21T and EKM22T, which had upstream genes that shared some similarity with the first group (Supplementary Figure 11C). P. agglomerans CFBP1316 was basal to these two groups and showed the same upstream ORF pattern as group one, but matched the downstream pattern of group 2 (Supplementary Figure 11C). Two P. ananatis strains made up group 3, while both P. dispersa strains as well as P. agglomerans M1657A made up group 4 (Supplementary Figure 11C). Neither group 3 nor group 4 shared flanking regions with the first two groups (Supplementary Figure 11C). The GC content of the P. agglomerans B025670 genome was approximately 10% higher than that of the PNP-4 cluster from this strain (45.48%) (Table 3).
3.12 PNP-5
The PNP-5 cluster was found to be distributed across 197 strains (Supplementary Tables 3, 4). Only two Pantoea species carry the cluster, Pantoea sp. 1.19 and P. agglomerans 20KB447973; however, previous work has reported that Pantoea sp. 1.19 may be misidentified (Kirk and Stavrinides, 2023). The remaining 195 clusters are found in members of Serratia, Photobacterium, and Yersinia (Supplementary Table 4). Yersinia strains accounted for 171 of the total representatives (Supplementary Table 4), which represented approximately 1.4% of all available Yersinia genomes (Table 2). The gene cluster tree is largely congruent to that of the species tree with respect to the monophyly of the representative genera; however, there were noticeable differences between the Serratia clades of the two trees (Supplementary Figures 12A,B).
The predicted ORFs in the flanking regions of PNP-5 clusters were conserved within genera, except Serratia sp. DD3, which did not show similarity to other Serratia representatives (Supplementary Figure 12C). Photobacterium strains showed the most diversity in flanking regions (Supplementary Figure 12C). Across most strains in Photobacterium, predicted ORFs for a LysR substrate-binding domain-containing protein and a DUF1127 domain-containing protein were found directly downstream of the cluster (Supplementary Figure 12C; green arrows). All Photobacterium strains also contained an additional gene within the PNP-5 cluster encoding a metallophosphoesterase (Supplementary Figure 12C; green arrow).
4 Discussion
We assessed the distribution of 12 known Pantoea antibiotic BGCs across public databases, and found that some clusters, namely pantocin B, PNP-3, and PNP-4 were found only in a relatively small subset of Pantoea strains. All other clusters had more broad distributions, encompassing members of Pantoea as well as other genera in the Enterobacteriaceae, Pectobacteriaceae, Yersiniaceae, Morganellaceae, and Hafniaceae. The andrimid and PNP-5 clusters also had representatives in the Vibrionaceae. In general, the andrimid, dapdiamide, pantocin A, PNP-1, and PNP-2 BGCs were more represented across Pantoea genomes than in any other genus, while the reverse was true for the AGA and PNP-5 BGCs (Table 2). This could suggest that the metabolites encoded by BGCs that are more prevalent across a diversity of Pantoea strains play a more central role in the general ecology of the group. Secondary metabolite BGCs are often reported to be reflective of ecotype or phylotype, with many gene clusters being species-specific (Jensen et al., 2007; Penn et al., 2009; Soucy et al., 2015; Xu et al., 2019). Still, inter-species and even inter-kingdom horizontal transfer of BGCs can occur (Vior et al., 2018; Kominek et al., 2019).
Our results are consistent with previous reports that many strains carry more than one BGC. P. agglomerans Pa31-3, Pa39-3, and Pa39-1 possess the BGCs for both pantocin A and PNP-3, P. agglomerans DAPP-PG734 has the BGCs for dapdiamide and PNP-4, and P. agglomerans strain 4 has the BGCs for dapdiamide, PNP-2 and PNP-3 (Sulja et al., 2022). Our previous work reported that P. agglomerans TX10 had clusters for both PNP-2 and pantocin A (Robinson et al., 2020) while P. agglomerans 3581 had the pantocin A cluster in addition to the PNP-3 cluster (Williams and Stavrinides, 2020). Our analysis also supported previous reports that P. vagans C9-1 has the BGCs for pantocin A, dapdiamide, and PNP-3 (Smits et al., 2011; Williams and Stavrinides, 2020; Sulja et al., 2022) and P. agglomerans Eh318 has the BGCs for pantocin A and B (Wright et al., 2001). In addition to these examples, we found several other strains that carry multiple BGCs. P. agglomerans CFSAN047153 and CFSAN047154 as well as Pectobacterium sp. F1-1 have both the AGA and PNP-2 clusters while P. agglomerans 39–23 and 39–7 have the AGA and PNP-3 clusters. P. agglomerans 540Y has both the Pantocin A and PNP-3 clusters, P. agglomerans Sl1_M5 has the dapdiamide and PNP-2 clusters, and P. agglomerans 9Rz4 has both the PNP-3 cluster as well as the herbicolin cluster. P. agglomerans VRA_MhP_f has both the PNP-2 and PNP-3 clusters. Pectobacteriaceae bacterium CE90 also contains two clusters: pantocin A and PNP-1. While our survey provides a glimpse into the distribution of these BGCs across strains, whether all these gene clusters are expressed and produce a bioactive metabolite remains unclear.
Our survey only focused on BGCs that shared significant sequence identity to the original reference cluster. For example, the PNP-1 cluster as described in P. ananatis BRT175 is composed of eight genes and produces 4-formylaminooxyvinylglycine, whereas the gene cluster in P. fluorescens that produces 4-formylaminooxyvinylglycine is composed of 11 genes (Walterson et al., 2014; Okrent et al., 2018). Experimental validation was needed to establish that these two BGCs produce a similar metabolite; consequently, we opted to use more stringent criteria and thresholds for this analysis. Expanding the survey to include other BGCs that share most or all of the genes of the reference BGCs would provide unique insight into the step-wise evolution of these BGCs.
Given the roles of natural product BGCs in competition and niche-specific adaptation, they are often found associated with mobile genetic elements (Ziemert et al., 2014; Vior et al., 2018). Potential horizontal gene transfer (HGT) events can be identified by comparing the evolutionary history of the BGCs to that of the host strains, with congruence indicating vertical transmission (Matter et al., 2009; Ziemert et al., 2014). For our analysis we used the barcoding gene cpn60, which can efficiently establish phylogenetic relationships between bacteria at the species level (Verbeke et al., 2011). We also constructed phylogenies using entire antimicrobial gene clusters, which helps to offset any strong phylogenetic signal from individual genes that may be evolving at different rates (Ziemert et al., 2014). Several instances of potential HGT were seen when comparing these trees to their sister cpn60 trees. For example, in the dapdiamide cluster tree, P. agglomerans DAPP-PG734 did not group with other P. agglomerans strains as seen in the cpn60 tree (Supplementary Figures 3A,B), suggesting the cluster has a unique evolutionary history in comparison to host strain. Analysis of the flanking regions of P. agglomerans DAPP-PG734 supports evolution through horizontal transfer, as it does not share homologous genes with any other Pantoea representatives (Supplementary Figure 3C). The flanking regions of the cluster in DAPP-PG734 also contain remnants of transposase genes, which are often associated with HGT events. In addition, the dapdiamide clusters found in P. vagans C9-1 and P. agglomerans C410P1, DAPP-PG734 and Sl1_M5 are predicted to be plasmid-encoded, and in some cases are adjacent to plasmid-associated genes such as the conjugation-associated gene, traI (Supplementary Figure 3C; dark blue arrow). The association of BGCs with mobile genetic elements can also be seen in strains carrying the andrimid, pantocin A, PNP-2, PNP-3, and PNP-4 clusters.
Although only some BGCs were directly associated with mobile genetic elements, our %GC analysis indicated that most BGCs have 5–20% lower %GC relative to their parent genome or plasmid, with the exception of herbicolin BGC, which has a %GC approximately 5% higher than the genome (Table 3). The herbicolin cluster is found in Candidatus F. symbiotica 5D, a symbiont of pea aphids (Patel et al., 2019), many of which provide their hosts with antifungal defenses (Łukasik et al., 2013). Given the narrow distribution of this cluster, it is interesting that these two relatively distant species share this cluster. As previously suggested, HGT may have occurred between Pantoea and Candidatus F. symbiotica (Xu et al., 2022), which is the most parsimonious explanation given the current data. Similarly, HGT may account for the PNP-1 cluster being common to both Pantoea and the unknown Pectobacteriaceae strain, as no other closely related strains have been identified that carry the cluster.
Our survey provides a comprehensive assessment of the distribution and potential evolutionary histories of known Pantoea antibiotic BGCs to date. We show these clusters have very different distributions, ranging from more restricted to broadly distributed across distantly related families. Some of these distributions, coupled with the presence of mobile genetic elements and analyses of %GC are suggestive of HGT. An understanding of the roles of the metabolites produced by these BGCs will provide much needed insight into the ecology and evolution of Pantoea and other closely related taxa.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
AK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. JS: Conceptualization, Formal analysis, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The authors gratefully acknowledge financial support from the Natural Sciences and Engineering Research Council of Canada (RGPIN-02863-2021). AK was supported by the Canadian Institutes of Health Research‘s Graduate Scholarships -Master’s award and the University of Regina Faculty of Graduate Studies and Research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1416674/full#supplementary-material
Footnotes
1. ^ncbi.nlm.nih.gov; accessed on October 20, 2023.
References
Besemer, J., and Borodovsky, M. (1999). Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27, 3911–3920. doi: 10.1093/nar/27.19.3911
Brady, S. F., Wright, S. A., Lee, J. C., Sutton, A. E., Zumoff, C. H., Wodzinski, R. S., et al. (1999). Pantocin B, an antibiotic from Erwinia herbicola discovered by heterologous expression of cloned genes. J. Am. Chem. Soc. 121, 11912–11913. doi: 10.1021/ja992790m
Dawlaty, J., Zhang, X., Fischbach, M. A., and Clardy, J. (2010). Dapdiamides, tripeptide antibiotics formed by unconventional amide ligases. J. Nat. Prod. 73, 441–446. doi: 10.1021/np900685z
Fredenhagen, A., Tamura, S. Y., Kenny, P. T. M., Komura, H., Naya, Y., Nakanishi, K., et al. (1987). Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J. Am. Chem. Soc. 109, 4409–4411. doi: 10.1021/ja00248a055
Freiberg, C., Brunner, N. A., Schiffer, G., Lampe, T., Pohlmann, J., Brands, M., et al. (2004). Identification and characterization of the first class of potent bacterial acetyl-CoA aarboxylase inhibitors with antibacterial activity. J. Biol. Chem. 279, 26066–26073. doi: 10.1074/jbc.M402989200
Giddens, S. R., and Bean, D. C. (2007). Investigations into the in vitro antimicrobial activity and mode of action of the phenazine antibiotic D-alanylgriseoluteic acid. Int. J. Antimicrob. Agents 29, 93–97. doi: 10.1016/j.ijantimicag.2006.08.028
Giddens, S. R., Feng, Y., and Mahanty, H. K. (2002). Characterization of a novel phenazine antibiotic gene cluster in Erwinia herbicola Eh1087. Mol. Microbiol. 45, 769–783. doi: 10.1046/J.1365-2958.2002.03048.X
Gilchrist, C. L. M., and Chooi, Y.-H. (2021). Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics 37, 2473–2475. doi: 10.1093/bioinformatics/btab007
Greiner, M., and Winkelmann, G. (1991). Fermentation and isolation of herbicolin a, a peptide antibiotic produced by Erwinia herbicola strain a 111. Appl. Microbiol. Biotechnol. 34, 565–569. doi: 10.1007/BF00167899
Ishimaru, C. A., Lansdell, T. A., Clardy, J., Duffy, B., and Smits, T. H. M. (2017). The histidine-reversible antibiotic herbicolin O produced by Pantoea vagans C9-1 is pantocin A. J. Plant Pathol. 99, 91–97. doi: 10.4454/JPP.V99I0.3910
Jensen, P. R., Williams, P. G., Oh, D. C., Zeigler, L., and Fenical, W. (2007). Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl. Environ. Microbiol. 73, 1146–1152. doi: 10.1128/AEM.01891-06
Jin, M., Fischbach, M. A., and Clardy, J. (2006). A biosynthetic gene cluster for the acetyl-CoA aarboxylase inhibitor andrimid. J. Am. Chem. Soc. 128, 10660–10661. doi: 10.1021/ja063194c
Jin, M., Liu, L., Wright, S. A., Beer, S. V., and Clardy, J. (2003a). Structural and functional analysis of pantocin a: an antibiotic from Pantoea agglomerans discovered by heterologous expression of cloned genes. Angew. Chemie Int. Ed. 42, 2898–2901. doi: 10.1002/anie.200351053
Jin, M., Wright, S. A., Beer, S. V., and Clardy, J. (2003b). The biosynthetic gene cluster of pantocin A provides insights into biosynthesis and a tool for screening. Angew. Chemie Int. Ed. 42, 2902–2905. doi: 10.1002/ANIE.200351054
Kamber, T., Lansdell, T. A., Stockwell, V. O., Ishimaru, C. A., Smits, T. H., and Duffy, B. (2012). Characterization of the biosynthetic operon for the antibacterial peptide herbicolin in Pantoea vagans biocontrol strain C9-1 and incidence in Pantoea species. Appl. Environ. Microbiol. 78, 4412–4419. doi: 10.1128/AEM.07351-11
Kanchanabanca, C., Tao, W., Hong, H., Liu, Y., Hahn, F., Samborskyy, M., et al. (2013). Unusual acetylation–elimination in the formation of tetronate antibiotics. Angew. Chemie 125, 5897–5900. doi: 10.1002/ANGE.201301680
Kearns, L. P., and Hale, C. N. (1996). Partial characterization of an inhibitory strain of Erwinia herbicola with potential as a biocontrol agent for Erwinia amylovora, the fire blight pathogen. J. Appl. Bacteriol. 81, 369–374. doi: 10.1111/J.1365-2672.1996.TB03521.X
Kearns, L. P., and Mahanty, H. K. K. (1998). Antibiotic production by Erwinia herbicola Eh1087: its role in inhibition of Erwinia amylovora and partial characterization of antibiotic biosynthesis genes. Appl. Environ. Microbiol. 64, 1837–1844. doi: 10.1128/AEM.64.5.1837-1844.1998
Kirk, A., and Stavrinides, J. (2023). A replica plating method for efficient, high-throughput screening of antibiotic gene clusters in bacteria uncovers a holomycin-like cluster in the clinical isolate, Pantoea agglomerans 20KB447973. J. Microbiol. Methods 213:106822. doi: 10.1016/j.mimet.2023.106822
Kominek, J., Doering, D. T., Opulente, D. A., Shen, X.-X., Zhou, X., DeVirgilio, J., et al. (2019). Eukaryotic acquisition of a bacterial operon. Cell 176, 1356–1366.e10. doi: 10.1016/j.cell.2019.01.034
Li, B., and Walsh, C. T. (2010). Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc. Natl. Acad. Sci. 107, 19731–19735. doi: 10.1073/pnas.1014140107
Łukasik, P., van Asch, M., Guo, H., Ferrari, J., Charles, J., and Godfray, H. (2013). Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 16, 214–218. doi: 10.1111/ele.12031
Madeira, F., Park, Y. M., Lee, J., Buso, N., Gur, T., Madhusoodanan, N., et al. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47, W636–W641. doi: 10.1093/nar/gkz268
Matter, A. M., Hoot, S. B., Anderson, P. D., Neves, S. S., and Cheng, Y.-Q. (2009). Valinomycin biosynthetic gene cluster in Streptomyces: conservation, ecology and evolution. PLoS One 4:e7194. doi: 10.1371/journal.pone.0007194
Needham, J., Kelly, M. T., Ishige, M., and Andersen, R. J. (1994). Andrimid and moiramides A-C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens: structure elucidation and biosynthesis. J. Org. Chem. 59, 2058–2063. doi: 10.1021/jo00087a020
Okrent, R. A., Trippe, K. M., Manning, V. A., and Walsh, C. M. (2018). Detection of 4-formylaminooxyvinylglycine in culture filtrates of Pseudomonas fluorescens WH6 and Pantoea ananatis BRT175 by laser ablation electrospray ionization-mass spectrometry. PLoS One 13:e0200481. doi: 10.1371/journal.pone.0200481
Patel, V., Chevignon, G., Manzano-Marín, A., Brandt, J. W., Strand, M. R., Russell, J. A., et al. (2019). Cultivation-assisted genome of Candidatus Fukatsuia symbiotica; the enigmatic “X-type” symbiont of aphids. Genome Biol. Evol. 11, 3510–3522. doi: 10.1093/gbe/evz252
Penn, K., Jenkins, C., Nett, M., Udwary, D. W., Gontang, E. A., McGlinchey, R. P., et al. (2009). Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J. 3, 1193–1203. doi: 10.1038/ismej.2009.58
Qin, Z., Baker, A. T., Raab, A., Huang, S., Wang, T., Yu, Y., et al. (2013). The fish pathogen Yersinia ruckeri produces holomycin and uses an RNA methyltransferase for self-resistance. J. Biol. Chem. 288, 14688–14697. doi: 10.1074/jbc.M112.448415
Robinson, L. J., Verrett, J. N., Sorout, N., and Stavrinides, J. (2020). A broad-spectrum antibacterial natural product from the cystic fibrosis isolate, Pantoea agglomerans Tx10. Microbiol. Res. 237:126479. doi: 10.1016/J.MICRES.2020.126479
Sheng-Da, Z., Thomas, I., Louise, L. L., Ostenfeld, L. T., and Lone, G. (2021). Holomycin, an antibiotic secondary metabolite, is required for biofilm formation by the native producer Photobacterium galatheae S2753. Appl. Environ. Microbiol. 87, e00169–e00121. doi: 10.1128/AEM.00169-21
Shoji, J. I., Sakazaki, R., Hattori, T., Matsumoto, K., Uotani, N., and Yoshida, T. (1989). Isolation and characterization of agglomerins A, B, C and D. J. Antibiot. 42, 1729–1733. doi: 10.7164/antibiotics.42.1729
Smits, T. H. M., Rezzonico, F., Kamber, T., Blom, J., Goesmann, A., Ishimaru, C. A., et al. (2011). Metabolic versatility and antibacterial metabolite biosynthesis are distinguishing genomic features of the fire blight antagonist Pantoea vagans C9-1. PLoS One 6:e22247. doi: 10.1371/journal.pone.0022247
Smits, T. H. M., Rezzonico, F., Kamber, T., Goesmann, A., Ishimaru, C. A., Stockwell, V. O., et al. (2010). Genome sequence of the biocontrol agent Pantoea vagans strain C9-1. J. Bacteriol. 192, 6486–6487. doi: 10.1128/jb.01122-10
Soucy, S. M., Huang, J., and Gogarten, J. P. (2015). Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 16, 472–482. doi: 10.1038/nrg3962
Stecher, G., Tamura, K., and Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 37, 1237–1239. doi: 10.1093/molbev/msz312
Sulja, A., Pothier, J. F., Blom, J., Moretti, C., Buonaurio, R., Rezzonico, F., et al. (2022). Comparative genomics to examine the endophytic potential of Pantoea agglomerans DAPP-PG 734. BMC Genomics 23:742. doi: 10.1186/s12864-022-08966-y
Terui, Y., Sakazaki, R., and Shoji, J. I. (1990). Structures of agglomerins. J. Antibiot. (Tokyo) 43, 1245–1253. doi: 10.7164/antibiotics.43.1245
Vanneste, J. L., Yu, J., Cornish, D. A., Vanneste, J. L., Yu, J., and Cornish, D. A. (2008). Presence of genes homologous to those necessary for synthesis of microcin MccEh252 in strains of Pantoea agglomerans. Acta Hortic. 793, 391–396. doi: 10.17660/ActaHortic.2008.793.58
Verbeke, T. J., Sparling, R., Hill, J. E., Links, M. G., Levin, D., and Dumonceaux, T. J. (2011). Predicting relatedness of bacterial genomes using the chaperonin-60 universal target (cpn60 UT): application to Thermoanaerobacter species. Syst. Appl. Microbiol. 34, 171–179. doi: 10.1016/j.syapm.2010.11.019
Vior, N. M., Lacret, R., Chandra, G., Dorai-Raj, S., Trick, M., and Truman, A. W. (2018). Discovery and biosynthesis of the antibiotic bicyclomycin in distantly related bacterial classes. Appl. Environ. Microbiol. 84, e02828–e02817. doi: 10.1128/AEM.02828-17
Völksch, B., and Sammer, U. (2008). “Characterization of the inhibitory strain Pantoea sp. 48b/90 with potential as a biocontrol agent for bacterial plant pathogens” in Pseudomonas syringae pathovars and related pathogens – identification, epidemiology and genomics. eds. M. Fatmi, A. Collmer, N. S. Iacobellis, J. W. Mansfield, J. Murillo, and N. W. Schaad, et al. (Dordrecht: Springer), 111–116.
Walterson, A. M., Smith, D. D. N., and Stavrinides, J. (2014). Identification of a Pantoea biosynthetic cluster that directs the synthesis of an antimicrobial natural product. PLoS One 9:e96208. doi: 10.1371/JOURNAL.PONE.0096208
Walterson, A. M., and Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 39, 968–984. doi: 10.1093/FEMSRE/FUV027
Williams, A. N., Sorout, N., Cameron, A. J., and Stavrinides, J. (2020). The integration of genome mining, comparative genomics, and functional genetics forbiosynthetic gene cluster identification. Front. Genet. 11:1543. doi: 10.3389/FGENE.2020.600116
Williams, A. N., and Stavrinides, J. (2020). Pantoea natural product 3 is encoded by an eight-gene biosynthetic gene cluster and exhibits antimicrobial activity against multi-drug resistant Acinetobacter baumannii and Pseudomonas aeruginosa. Microbiol. Res. 234:126412. doi: 10.1016/J.MICRES.2020.126412
Winkelmann, G., Glück, C., Lupp, R., and Jung, G. (1981). “Herbicolins - new peptide antibiotics from Enterobacteria” in Structure and activity of natural peptides. eds. W. Voelter and G. Weitzel (Berlin: De Gruyter), 237–252.
Wright, S. A. I., Jin, M., Clardy, J., and Beer, S. V. (2006). The biosynthetic genes of pantocin A and pantocin B of Pantoea agglomerans Eh318. Acta Hortic. 704, 313–320. doi: 10.17660/ActaHortic.2006.704.45
Wright, S. A. I., Zumoff, C. H., Schneider, L., and Beer, S. V. (2001). Pantoea agglomerans strain-EH318 produces two antibiotics that inhibit Erwinia amylovora in vitro. Appl. Environ. Microbiol. 67, 284–292. doi: 10.1128/AEM.67.1.284-292.2001
Xu, L., Ye, K.-X., Dai, W.-H., Sun, C., Xu, L.-H., and Han, B.-N. (2019). Comparative genomic insights into secondary metabolism biosynthetic gene cluster distributions of marine Streptomyces. Mar. Drugs 17:498. doi: 10.3390/md17090498
Xu, S., Yong-Xin, L., Cernava, T., Wang, H., Zhou, Y., Xia, T., et al. (2022). Fusarium fruiting body microbiome member Pantoea agglomerans inhibits fungal pathogenesis by targeting lipid rafts. Nat. Microbiol. 7, 831–843. doi: 10.1038/s41564-022-01131-x
Keywords: Pantoea, bacterial natural products, biosynthetic gene clusters, horizontal gene transfer, phylogenetics, comparative genomics, pantocin B, antibiotics
Citation: Kirk A and Stavrinides J (2024) Distribution and comparative genomic analysis of antimicrobial gene clusters found in Pantoea. Front. Microbiol. 15:1416674. doi: 10.3389/fmicb.2024.1416674
Edited by:
Miklos Fuzi, Independent Researcher, Seattle, WA, United StatesReviewed by:
Baikui Wang, University of Chinese Academy of Sciences, ChinaCyril Bontemps, Université de Lorraine, France
Copyright © 2024 Kirk and Stavrinides. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John Stavrinides, am9obi5zdGF2cmluaWRlc0B1cmVnaW5hLmNh
 Ashlyn Kirk
Ashlyn Kirk John Stavrinides
John Stavrinides