
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 14 June 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1416454
This article is part of the Research Topic Global Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and Treatment, Volume III View all 14 articles
 Jingshu Ji1,2†
Jingshu Ji1,2† Yufeng Zhu1,3†
Yufeng Zhu1,3† Feng Zhao1,2
Feng Zhao1,2 Jingjing Zhang1,2
Jingjing Zhang1,2 Bingyan Yao1,2
Bingyan Yao1,2 Mingli Zhu3
Mingli Zhu3 Yunsong Yu4,5,6
Yunsong Yu4,5,6 Jun Zhang1,2*
Jun Zhang1,2* Ying Fu1,2*
Ying Fu1,2*Introduction: The dissemination of carbapenem-resistant Enterobacteriales (CRE) in nosocomial settings is primarily associated with the horizontal transfer of plasmids. However, limited research has focused on the in-host transferability of carbapenem resistance. In this study, ten isolates were collected from gut specimens of five individuals, each hosting two different species, including Escherichia coli, Klebsiella pneumoniae, Klebsiella aerogenes, Enterobacter cloacae, or Citrobacter koseri.
Methods: Species identification and antimicrobial susceptibility were determined by MALDI-TOF MS and broth microdilution method. Carbapenemase genes were detected and localized using PCR, S1-PFGE and southern blot. The transferability of carbapenemase genes between species was investigated through filter mating experiments, and the genetic contexts of the plasmids were analyzed using whole genome sequencing.
Results and discussion: Our results revealed that each of the ten isolates harbored a carbapenemase gene, including blaNDM-5, blaNDM-1, or blaKPC-2, on a plasmid. Five different plasmids were successfully transferred to recipient cells of E. coli, K. pneumoniae or A. baumannii by transconjugation. The genetic contexts of the carbapenemase gene were remarkably similar between the two CRE isolates from each individual. This study highlights the potential for interspecies plasmid transmission in human gut, emphasizing the colonization of CRE as a significant risk factor for the dissemination of carbapenemase genes within the host. These findings underscore the need for appropriate intestinal CRE screening and colonization prevention.
Antibiotic resistance of bacteria has become a global concern, particularly in clinical settings (Laxminarayan et al., 2013). Carbapenems are often considered as a last resort for treating multidrug-resistant bacteria, alongside polymyxin and tigecycline (Brink, 2019). Among the various mechanisms of carbapenem resistance, the production of carbapenemase is a significant factor (Mancuso et al., 2021). The spread of carbapenemase between different pathogens is primarily associated with horizontal gene transfer (HGT) (Partridge et al., 2018; San Millan, 2018). Conjugation, mediated by transferable elements such as plasmids and integrative conjugative elements (ICE), is a major mechanism of HGT, surpassing transduction and transformation (Halary et al., 2010; Partridge et al., 2018). Consequently, the transfer of carbapenemase-producing plasmids between pathogens could exacerbate the antibiotic resistance situation and contribute to the global distribution of carbapenem-resistant Enterobacteriales (CRE).
The human gut serves as a natural reservoir for a diverse microbial population, including important pathogens with varying resistances and relevant genes, making it an ideal environment for interspecies HGT (McInnes et al., 2020). Given that gut microbiota act as a defense against pathogen invasion, disruption of this protection could lead to gut-derived infections in critically ill patients (Wang et al., 2019). The use of broad-spectrum antibiotics is known as an important risk of microbiota disruption and CRE colonization, such as carbapenems, cephalosporins, ciprofloxacin and so on (Kim et al., 2017). As we have known, bacteria need nutrients to grow and colonize in gut, some of its nutrient metabolites even can inhibit pathogen growth (Ng et al., 2013). However, the killing of gut microbiota by broad-spectrum antibiotics, not only enriches the nutrients for CRE to grow, but also reduces the inhibitory metabolites, causing the CRE expansion and colonization (Reed and Theriot, 2021; Yip et al., 2023). Common opportunistic pathogens in the human gut microbiota include Enterobacteriales such as Escherichia coli and Klebsiella pneumoniae. Colonization of multidrug-resistant K. pneumoniae in patients’ gut can lead to transmission in the intensive care unit, with approximately 50% of infections originating from the patients’ own microbiota (Gorrie et al., 2017). Previous studies have reported the transfer of carbapenem-resistant Enterobacteriales between patients, which is significant in healthcare-associated infections (David et al., 2019). However, the in vivo transmission of carbapenem-resistant genes between gut microbes are not fully understood.
Therefore, we conducted this study to assess the colonization of carbapenem-resistant bacteria in patients’ gut in our hospital and to investigate their potential in vivo transmission pathways.
A project was initiated to monitor the colonization of carbapenem-resistant Enterobacterales in the human gut and to assess the risk of secondary infections in hospitalized patients. The project was conducted in accordance with the ethical approval granted by Sir Run Run Shaw Hospital under the reference number 20200831-36, spanning from 2018 to 2021.
CRE species were initially isolated from stool samples using Chromatic CRE selective plates (Liofilchem® Chromatic™, Italy) following standard manufacturing protocols. Colonies grown on the plates were then identified using Matrix Assisted Laser Desorption Ionization Time-of-Flight mass spectrometry (MALDI-TOF MS, bioMérieux, France) and tested for susceptibility to meropenem, imipenem, and ertapenem. Isolates identified as Enterobacterales species and confirmed to be resistant to any of the three carbapenems were subjected to antimicrobial susceptibility testing (AST) using the VITEK2 compact system (bioMérieux, France) for 16 antimicrobial agents (ampicillin; ampicillin/sulbactam; piperacillin/tazobactam; ceftazidime; cefotetan; ceftriaxone; cefepime; imipenem; ertapenem; aztreonam; ciprofloxacin; gentamicin; amikacin; levofloxacin; sulfamethoxazole/trimethoprim; tobramycin), and three others (tigecycline; colistin; ceftazidime/avibactam) were tested using the broth microdilution method as per CLSI M07Ed11 guidelines (CLSI, 2018). The MIC interpretation followed the CLSI M100 (30th edition) (CLSI, 2020) breakpoints, except for tigecycline, for which the breakpoints and interpretive criteria of EUCAST (Version 11.0, 2021) were applied (EUCAST, 2021). Quality control strains included E. coli ATCC25922 and Pseudomonas aeruginosa ATCC 27853.
The presence of carbapenemase in CRE was determined using NG-Test® CARBA 5 (Hardy Diagnostics, France) and confirmed by PCR amplification followed by sequencing (Du et al., 2013). Patients with two CRE species were included, and the CRE isolates were collected for further analysis.
The bacterial isolates were embedded in agarose plugs containing total DNAs, and then treated with S1 nuclease. Plasmids were separated using the CHEF-Mapper XA PFGE system (Bio-Rad, United States) (Hunter et al., 2005). The gel was stained with GelRed (Biotium, United States) and visualized under 254 nm UV light. Subsequently, the DNAs on the gel were transferred to a nylon membrane (Millipore, United States) under pressure, hybridized with digoxigenin-labeled blaNDM or blaKPC probes, and detected using the NBT/BCIP color detection kit (Roche Applied Sciences, Germany) (Chen et al., 2011).
To assess the transferability of plasmids carrying carbapenem-resistant genes through conjugation, a filter mating experiment was conducted between donor cells containing the carbapenem-resistant gene on plasmid and recipient cells of different species. Four recipient isolates from three species were selected: E. coli J53 (Sodium azide resistant, NaN3R), E. coli EC600 (Rifampicin resistant, RifR), Acinetobacter baumannii ATCC17978 (Rifampicin resistant, RifR), and K. pneumoniae ATCC13883 (Rifampicin resistant, RifR) (Supplementary Table S1). Each recipient strain was mated with the donor strain, except when both of them were of the same species. The mating experiment was performed on a 0.22 μm filter membrane cultured on antibiotic-free Mueller-Hinton agar medium. After 18 h of incubation at 37°C, the bacterial mixture was transferred to Mueller-Hinton agar medium supplemented with specific antibiotics as listed in Supplementary Table S1. The transconjugants were then confirmed by PCR for the presence of carbapenemase genes. Antimicrobial susceptibility testing of the transconjugants was conducted using the methods described above.
Genomic DNAs of all the isolates were extracted using the QIAamp DNA MiniKit (Qiagen, New York, United States) according to the manufacturer’s instructions. Subsequently, the DNA samples were sequenced using the Illumina-Hiseq X-10 (Illumina Inc., San Diego, CA) and/or the MinION platform (Nanopore, Oxford, United Kingdom). All the 10 CRE isolates were sequenced by the Illumina-Hiseq X-10 (Illumina Inc., San Diego, CA), while only 6 isolates (Eco13188, Eco20779, Eco20155, Kpn20156, Kpn20795 and Kpn40) were sequenced by the MinION platform (Nanopore, Oxford, UK). The sequenced reads were assembled using LC Genomic Workbench (CLC Bio 10.0) and Unicycler software. Gene prediction and annotation were performed using prokka. Plasmid maps were generated and compared using Proksee1 (Grant et al., 2023). The whole genome sequences were submitted to the Center for Genomic Epidemiology website2 for analysis of plasmid types, virulence genes, and resistance genes. The genome sequences of isolates Eco13188, Eco20779, Eco20155, Kpn20156, Kpn20795 and Kpn40 have been deposited in NCBI under the BioProject number PRJNA809193.
During the course of the project between July and September 2021, five inpatients (referred to as P1 to P5) were included, each of whom harbored two distinct CRE species simultaneously (Table 1). The patients’ ages ranged from 30 to 88 years, with an average age of 60.8 years. They were admitted to the Hematology department (2 patients), Infectious Disease department (2 patients), and Gastroenterology department (1 patient). All patients were afflicted with severe illnesses, such as hematological malignancies, septicemia, and hepatic failure.
A total of 10 CRE isolates were collected and identified. The antimicrobial susceptibility testing of the 10 isolates is summarized in Table 2 (Donor part), indicating resistance to imipenem, ertapenem, ampicillin, ampicillin/sulbactam, and ceftriaxone, while showing susceptibility to tigecycline, colistin, and amikacin. Subsequent testing using NG-Test® CARBA 5 and PCR sequencing confirmed the production of NDM-1 carbapenemase in isolates from P4, NDM-5 carbapenemase in isolates from P1 and P5, and KPC-2 carbapenemase in isolates from P2 and P3 (Table 1).
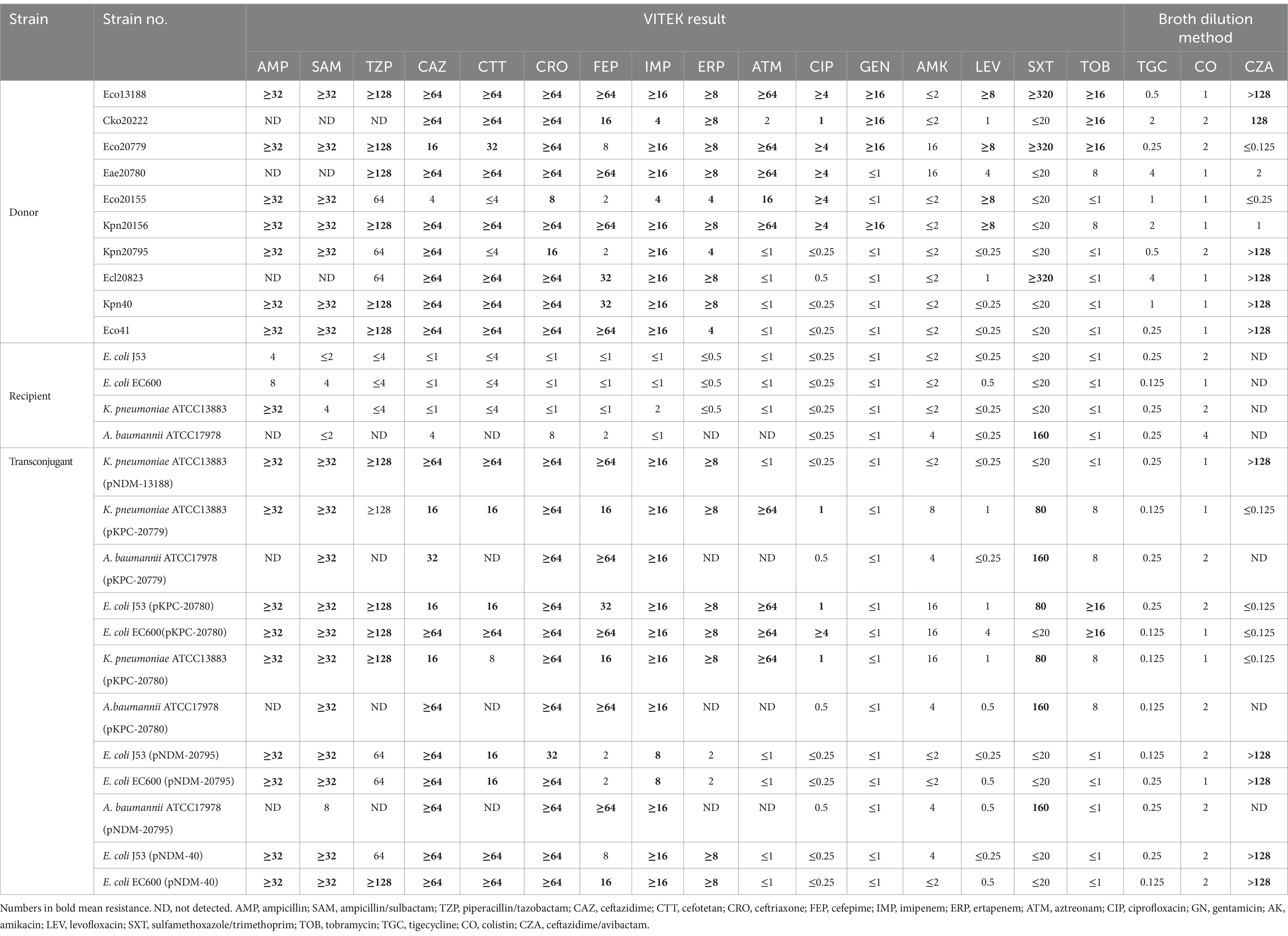
Table 2. Antimicrobial susceptibility testing of both donors, recipients and transconjugants (μg/mL).
As depicted in Figure 1, the six CRE isolates from P1 (Eco13188 & Cko20222), P4 (Kpn20795 & Ecl20823), and P5 (Kpn40 & Eco41) exhibited a positive signal for the blaNDM probe, confirming the presence of blaNDM gene on plasmids. The six blaNDM-carrying plasmids had nearly the same size within the range of 33.3 kb to 54.7 kb. The remaining four CRE isolates from P2 (Eco20779 & Eae20780) and P3 (Eco20155 & Kpn20156) showed a blaKPC signal, indicating the carriage of blaKPC gene on plasmids. The two blaKPC-carrying plasmids of P2 had the same size within the range of 54.7 kb ~78.2 kb, while the other two plasmids of P3 had obvious size differences.
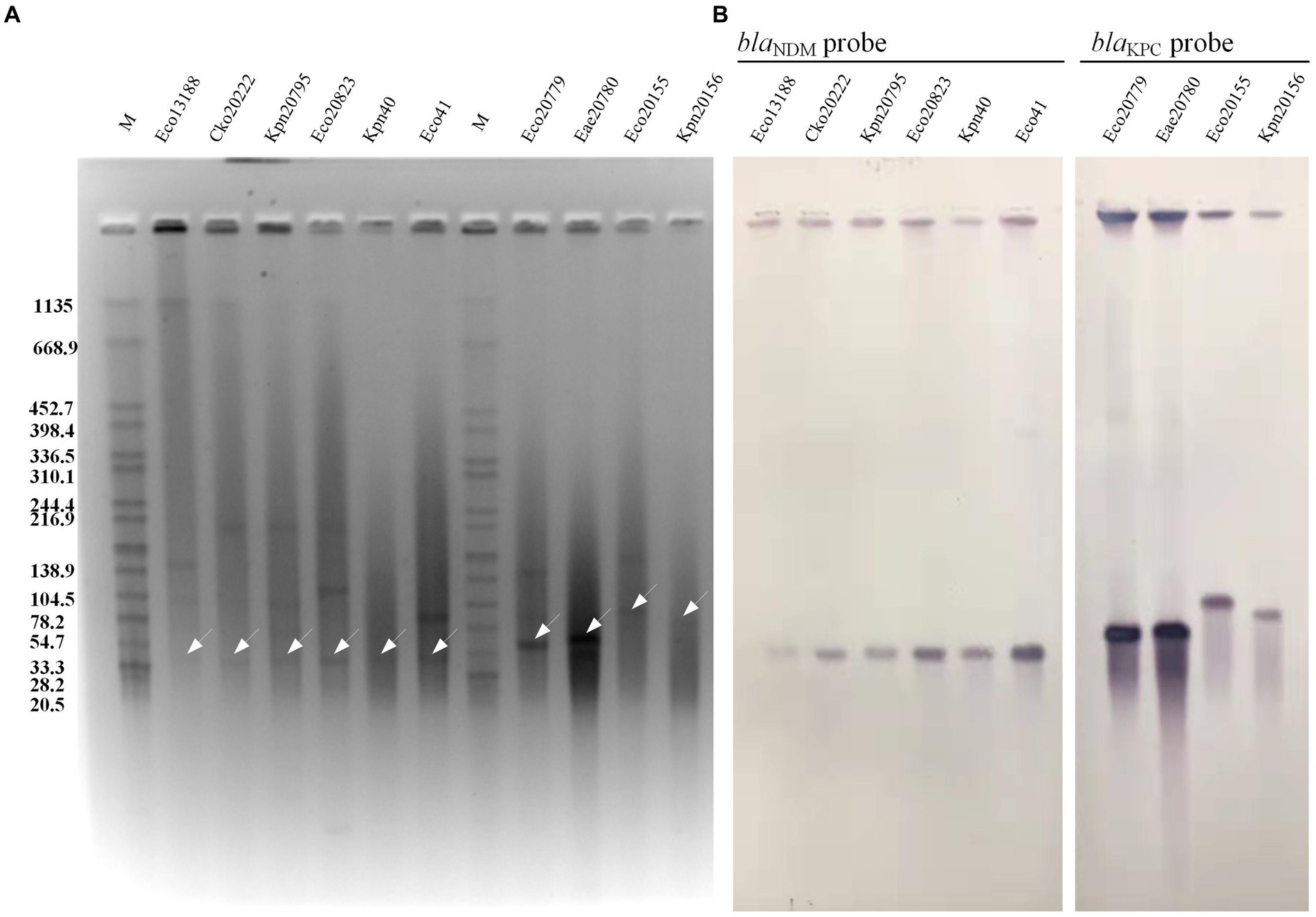
Figure 1. S1 PFGE and Southern blot hybridization of 10 CRE isolates. (A) S1 PFGE of the 10 CRE isolates. (B) Southern blot hybridization of probe blaNDM and blaKPC. The white arrows showed the positive bands of plasmid hybridized with blaNDM probe or blaKPC probe. M, the Salmonella serotype Braenderup strain H9812 was used as a molecular marker.
The subsequent filter mating experiment showed that blaKPC-carrying isolates Eco20779 (E. coli) and Eae20780 (K. aerogenes) from P2 successfully transconjugated plasmids to recipients of other species, while Eco20155 (E. coli) and Kpn20156 (K. pneumoniae) from P3 failed to transfer plasmids to the selected recipients (Table 2, Transconjugant part). Specifically, Eco20779 and Eae20780 both transferred their KPC plasmids to K. pneumoniae ATCC13883 and A. baumannii ATCC17978, while Eae20780 transferred to E. coli J53 and E. coli EC600 in addition. The blaNDM-carrying isolates Kpn20795 (K. pneumoniae) from P4 successfully transferred its NDM plasmid to E. coli J53, E. coli EC600, and A. baumannii ATCC17978. And Kpn40 (K. pneumoniae) from P5 successfully transferred to E. coli J53, E. coli EC600. Eco13188 (E. coli) from P1 successfully transferred to K. pneumoniae ATCC13883.
Consequently, a total of 12 transconjugants were obtained from 5 CRE isolates, each from one patient. And all of the 12 transconjugants were resistant to imipenem and most β-lactams (Table 2, Transconjugant part), which were consistent with the phenotypes of the donors. S1-PFGE and southern blot analysis of the 12 transconjugants and donors confirmed the presence of blaNDM or blaKPC-carrying plasmids (Supplementary Figure S1). Although not all the 10 CRE isolates successfully transferred the plasmid, the results still provide evidence that plasmids-carrying carbapenemases can be transferred interspecies.
Whole genome sequencing was conducted to investigate the genetic background of CRE isolates, and information of six complete genomes was shown in Table 3. Each genome contained sequences of chromosome and several plasmids, and the carbapenemase-carrying plasmids belonged to different plasmid incompatibility (Inc) types, mainly IncX3 and IncN.
Genomic analysis of plasmids revealed the possible horizontal gene transfer (HGT) approach of carbapenemases (Figure 2). The four blaNDM-5-carrying plasmids from Eco13188, Cko20222, Kpn40 and Eco41 were almost identical in size, and had nearly the same surrounding context. Two blaNDM-1-carrying plasmids from Kpn20795 and Ecl20823 were almost identical in size and surrounding context of blaNDM-1, varying in some IS elements. The context of blaNDM-1-carrying plasmids were similar to the blaNDM-5 plasmid. The blaKPC-2-harboring plasmids from Eco20779 and Eae20780 were not the very same as the plasmid sequences of Eae20780 were assembled to several small contigs, while the main context and the surrounding context of blaKPC-2 was overlapped. Besides blaKPC-2, the plasmids also contained blaCTX-M-3 and blaTEM-1, flanked by tnpR gene and Tn3 family transposase, constituting a classical Tn3 transposon. The other two blaKPC-2-harboring plasmids from Eco20155 and Kpn20156 varied in sizes, but they shared a similar context of flanking region by blaKPC-2, which was a ca.10 kb IS26 associated HGT element. The main difference between the two elements was an addition of ISKpn28 fragment downstream ISKpn27 in plasmid of Eco20155. The carbapenemase gene contexts suggested the horizontal gene transfer capacity of carbapenemase-carrying plasmids.
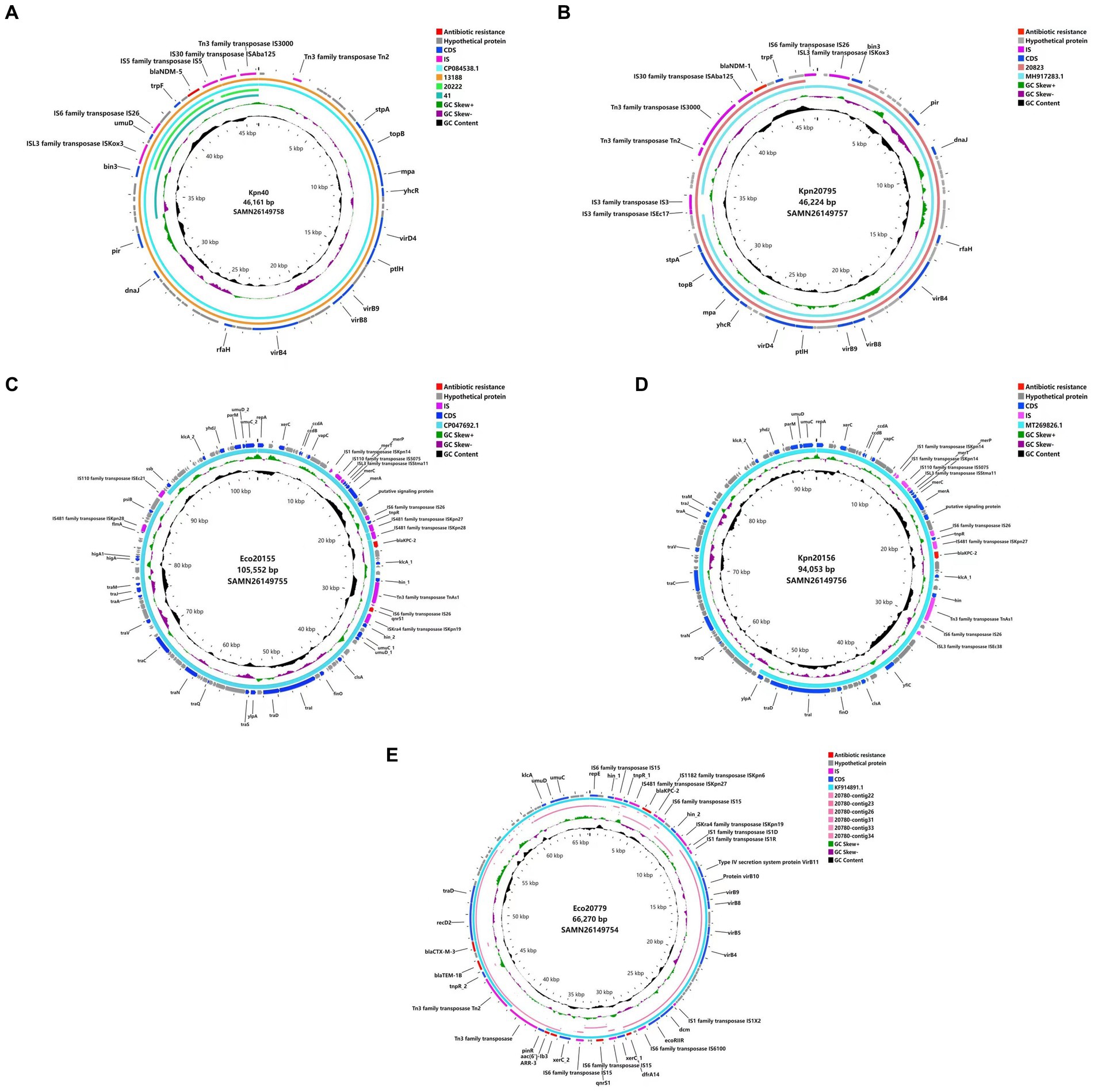
Figure 2. Plasmid comparison based on WGS. (A) WGS of Kpn40 plasmid versus reference sequence CP084538.1, Eco13188, Cko20222, and Eco41. (B) WGS of Kpn20795 plasmid versus reference sequence MH917283.1, and Ecl20823. (C) WGS of Eco20155 plasmid versus reference sequence CP047692.1. (D) WGS of Kpn20156 plasmid versus reference sequence MT269826.1. (E) WGS of Eco20779 plasmid versus reference sequence KF914891.1, and Eae20780 contig22,23,26,31,33,34. Different colors represent various genetic types.  , antibiotic resistance;
, antibiotic resistance;  , hypothetical protein;
, hypothetical protein;  , IS;
, IS;  , CDS;
, CDS;  , reference sequence;
, reference sequence;  , GC Skew+;
, GC Skew+;  , GC Skew−;
, GC Skew−;  , GC content;
, GC content; 




 , Eae20780 contig22, 23, 26, 31, 33, 34;
, Eae20780 contig22, 23, 26, 31, 33, 34;  , Ecl20823;
, Ecl20823;  , Eco13188;
, Eco13188;  , Cko20222;
, Cko20222;  , Eco41.
, Eco41.
Colonization of CRE in the gut is an asymptomatic infection (Nordmann and Poirel, 2019) that poses a potential risk to immunocompromised patients, such as those with hematological malignancies or undergoing chemotherapy for cancer (Tancrede and Andremont, 1985; Trecarichi et al., 2012; Shimasaki et al., 2019). This can lead to bacteremia through bacterial translocation, contributing to high mortality rates in these patient populations. In our previous screening project, carbapenem-resistant K. pneumoniae (CRKP) and carbapenem-resistant E. coli (CREC) were the predominant species in patients’ gut, and the co-existence of two CRE species in one patient was uncommon. In this study, we isolated 5 pairs of co-existent CRE species from 5 severely ill patients. During their treatment, the frequent use of carbapenem, tigecycline, or piperacillin/tazobactam posed a high risk for CRE proliferation due to microbiota dysbiosis (Harris et al., 2021). Studies have shown that antibiotic administration can lead to a significant increase in CRKP populations, owing to microbiota dysbiosis induced by antibiotic stress (Rooney et al., 2019). Under antibiotic stress, gut microbiota dysbiosis makes CRE take a competitive advantage and obtain a large number of nutrients, which promotes CRE growth and colonization (Rivera-Chavez et al., 2017). Also, the metabolites of normal intestinal flora can inhibit the growth of pathogens, while the destruction of intestinal flora reduces the metabolites, which ultimately leads to the massive growth and colonization of pathogens (Jacobson et al., 2018; McDonald et al., 2018; Sorbara et al., 2019). Therefore, it is necessary to screen for intestinal CRE in immunocompromised patients, evaluate the risk of CRE colonization, and maintain the stability of intestinal flora, both during the treatment and upon admission.
Plasmids are mobile genetic elements that play a crucial role in horizontal gene transfer. Conjugation is the primary mechanism through which plasmids facilitate HGT, with approximately 14% of plasmids being conjugative (Smillie et al., 2010). Previous research has predominantly focused on demonstrating plasmid-mediated transfer of antibiotic resistance genes in vitro. However, our study provides evidence of in vivo HGT of plasmids carrying antibiotic resistance genes.
As previously mentioned, we collected 10 CRE isolates from 5 patients, with each patient harboring 2 species. Through genome sequencing and conjugation experiments, we confirmed that blaKPC and blaNDM were located on plasmids. The 6 blaNDM-carrying plasmids from patients P1, P4, and P5 were all IncX3 type conjugative plasmids containing a T4SS system, which enhances the efficiency of plasmid conjugation (Smillie et al., 2010; Costa et al., 2021). Furthermore, the genomic context of plasmids from patients P1 and P5 (blaNDM-5) and P4 (blaNDM-1) were largely similar, differing only in a few IS insertions. Liu et al. reported that the backbone of IncX3 plasmids has remained highly conserved over 10 years, while the genetic context of blaNDM has diversified into 5 subtypes, indicating the potent ability of blaNDM genetic context (IS and transposon structures) to spread across different species (Liu et al., 2021). A recent study also confirmed that an IncX3 plasmid carrying blaNDM was present in 47 different isolates of Enterobacteriales species in Hong Kong (Wang et al., 2018).
The IncX3 plasmid, which typically has a narrow spectrum of Enterobacteriales, was found to carry the blaNDM-1 gene in P4-Kpn20795, a strain of K. pneumoniae. This plasmid was successfully transferred to A. baumannii ATCC17978, indicating the potential for horizontal gene transfer between different species. The presence of ISAba125 in the context of blaNDM suggests its involvement in the mobilization of blaNDM, a phenomenon that has been observed in Acinetobacter spp. as well (Nigro et al., 2011). Additionally, the blaNDM-5-carrying isolates from P1 were collected in a time sequence, with P1-Cko20222 being collected 3 days later than P1-Eco13188. This time interval indicates the possibility of in vivo transmission from E. coli to C. koseri, highlighting the occurrence of interspecies plasmid transfer within the gut. A similar in vivo transfer was reported by H. L. Nielsen et al., where a blaNDM-1-carrying IncN2 plasmid was identified in Salmonella Kottbus from the fecal sample of an individual, and was also found in E. coli and Citrobacter freundii in the same patient, suggesting a transfer between species (Nielsen et al., 2021).
The blaKPC plasmids from P2, identified as IncN type, were found to be identical and capable of conjugating to both K. pneumoniae ATCC13883 and A. baumannii ATCC17978, demonstrating their ability for interspecies transfer. Previous studies have also provided evidence of interspecies transfer of blaKPC-carrying plasmids, such as the transmission of KPC-2 carbapenemase between C. freundii, K. pneumoniae, E. coli, and Morganella morganii during a patient’s long-term hospital stay (Polcarova et al., 2019). The IncN type plasmids are known for their broad-host range and ability to carry various resistance genes, including extended-spectrum β-lactams, quinolones, and aminoglycosides (Rozwandowicz et al., 2018). In addition to blaKPC, the P2 plasmids also carried blaCTX-M-3 and blaTEM-1, with blaCTX-M often being associated with IncN plasmids in E. coli. On the other hand, the blaKPC plasmids from P3 did not belong to any Inc. type according to PlasmidFinder analysis and failed in conjugation. Despite this, a manual inspection of the P3 plasmids revealed the presence of the repA gene and IncF plasmid conjugative transfer associated tra genes, although this could not explain the failure of conjugation.
In conclusion, our study has demonstrated that the blaKPC and blaNDM carbapenemases of the 10 CRE isolates are located on plasmids and can be transferred interspecies through horizontal gene transfer in human intestinal environment. CRE intestinal colonization brings the risk of carbapenem resistance genes spreading in the host, so appropriate intestinal CRE screening and colonization prevention are necessary.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA809193.
This work was approved by Ethics Committee of Sir Run Run Shaw Hospital (no. 20200831-36).
JJ: Writing – review & editing, Writing – original draft. YZ: Writing – review & editing, Project administration, Methodology. FZ: Writing – review & editing, Project administration, Methodology. JiZ: Writing – review & editing, Formal Analysis. BY: Writing – review & editing, Project administration, Methodology. MZ: Writing – review & editing, Formal Analysis. YY: Writing – review & editing, Conceptualization. JuZ: Writing – review & editing, Conceptualization. YF: Writing – review & editing, Conceptualization.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China (no. 82072344), Medical Health Science and Technology Project of Zhejiang Province (2023KY974), Research Fund for Applications of High-throughput Sequencing Technology in Infectious Diseases (no. MTP2022B019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1416454/full#supplementary-material
Brink, A. J. (2019). Epidemiology of carbapenem-resistant gram-negative infections globally. Curr. Opin. Infect. Dis. 32, 609–616. doi: 10.1097/QCO.0000000000000608
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob. Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
CLSI (2018). Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. CLSI Standard M07. 11th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 30th Edn. Wayne, PA, Clinical and Laboratory Standards Institute.
Costa, T. R. D., Harb, L., Khara, P., Zeng, L., Hu, B., and Christie, P. J. (2021). Type IV secretion systems: advances in structure, function, and activation. Mol. Microbiol. 115, 436–452. doi: 10.1111/mmi.14670
David, S., Reuter, S., Harris, S. R., Glasner, C., Feltwell, T., Argimon, S., et al. (2019). Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat. Microbiol. 4, 1919–1929. doi: 10.1038/s41564-019-0492-8
Du, X. X., Wang, J. F., Fu, Y., Zhao, F., Chen, Y., Wang, H. P., et al. (2013). Genetic characteristics of blaNDM-1-positive plasmid in Citrobacter freundii isolate separated from a clinical infectious patient. J. Med. Microbiol. 62, 1332–1337. doi: 10.1099/jmm.0.057091-0
EUCAST (2021). The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 11.0. Available at: http://www.eucast.org.
Gorrie, C. L., Mirceta, M., Wick, R. R., Edwards, D. J., Thomson, N. R., Strugnell, R. A., et al. (2017). Gastrointestinal carriage is a major reservoir of Klebsiella pneumoniae infection in intensive care patients. Clin. Infect. Dis. 65, 208–215. doi: 10.1093/cid/cix270
Grant, J. R., Enns, E., Marinier, E., Mandal, A., Herman, E. K., Chen, C.-Y., et al. (2023). Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51, W484–W492. doi: 10.1093/nar/gkad326
Halary, S., Leigh, J. W., Cheaib, B., Lopez, P., and Bapteste, E. (2010). Network analyses structure genetic diversity in independent genetic worlds. Proc. Natl. Acad. Sci. USA 107, 127–132. doi: 10.1073/pnas.0908978107
Harris, H. C., Buckley, A. M., Spittal, W., Ewin, D., Clark, E., Altringham, J., et al. (2021). The effect of intestinal microbiota dysbiosis on growth and detection of carbapenemase-producing Enterobacterales within an in vitro gut model. J. Hosp. Infect. 113, 1–9. doi: 10.1016/j.jhin.2021.04.014
Hunter, S. B., Vauterin, P., Lambert-Fair, M. A., Van Duyne, M. S., Kubota, K., Graves, L., et al. (2005). Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J. Clin. Microbiol. 43, 1045–1050. doi: 10.1128/JCM.43.3.1045-1050.2005
Jacobson, A., Lam, L., Rajendram, M., Tamburini, F., Honeycutt, J., Pham, T., et al. (2018). A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe 24, 296–307.e7. doi: 10.1016/j.chom.2018.07.002
Kim, S., Covington, A., and Pamer, E. G. (2017). The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol. Rev. 279, 90–105. doi: 10.1111/imr.12563
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K., Wertheim, H. F., Sumpradit, N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098. doi: 10.1016/S1473-3099(13)70318-9
Liu, B., Guo, Y., Liu, N., Wang, J., Li, F., Yao, L., et al. (2021). In silico evolution and comparative genomic analysis of IncX3 plasmids isolated from China over ten years. Front. Microbiol. 12:725391. doi: 10.3389/fmicb.2021.725391
Mancuso, G., Midiri, A., Gerace, E., and Biondo, C. (2021). Bacterial antibiotic resistance: the Most critical pathogens. Pathogens 10:1310. doi: 10.3390/pathogens10101310
McDonald, J. A. K., Mullish, B. H., Pechlivanis, A., Liu, Z., Brignardello, J., Kao, D., et al. (2018). Inhibiting growth of Clostridioides difficile by restoring Valerate, produced by the intestinal microbiota. Gastroenterology 155, 1495–1507.e15. doi: 10.1053/j.gastro.2018.07.014
McInnes, R. S., McCallum, G. E., Lamberte, L. E., and van Schaik, W. (2020). Horizontal transfer of antibiotic resistance genes in the human gut microbiome. Curr. Opin. Microbiol. 53, 35–43. doi: 10.1016/j.mib.2020.02.002
Ng, K. M., Ferreyra, J. A., Higginbottom, S. K., Lynch, J. B., Kashyap, P. C., Gopinath, S., et al. (2013). Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99. doi: 10.1038/nature12503
Nielsen, H. L., Thomsen, P. K., Litrup, E., Torpdahl, M., Overballe-Petersen, S., Hansen, F., et al. (2021). A case of blaNDM-1-positive Salmonella Kottbus, Denmark, November 2020. Euro Surveill. 26:569. doi: 10.2807/1560-7917.ES.2021.26.26.2100569
Nigro, S. J., Post, V., and Hall, R. M. (2011). Aminoglycoside resistance in multiply antibiotic-resistant Acinetobacter baumannii belonging to global clone 2 from Australian hospitals. J. Antimicrob. Chemother. 66, 1504–1509. doi: 10.1093/jac/dkr163
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of Carbapenem resistance in gram-negative Bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Polcarova, P., Hobzova, L., Kukla, R., Skorepa, P., Smetana, J., Sosovickova, R., et al. (2019). In-vivo interspecies transmission of carbapenemase KPC in a long-term treated female patient. Epidemiol. Mikrobiol. Imunol. 68, 99–102.
Reed, A. D., and Theriot, C. M. (2021). Contribution of inhibitory metabolites and competition for nutrients to colonization resistance against Clostridioides difficile by commensal Clostridium. Microorganisms 9:371. doi: 10.3390/microorganisms9020371
Rivera-Chavez, F., Lopez, C. A., and Baumler, A. J. (2017). Oxygen as a driver of gut dysbiosis. Free Radic. Biol. Med. 105, 93–101. doi: 10.1016/j.freeradbiomed.2016.09.022
Rooney, C. M., Sheppard, A. E., Clark, E., Davies, K., Hubbard, A. T. M., Sebra, R., et al. (2019). Dissemination of multiple carbapenem resistance genes in an in vitro gut model simulating the human colon. J. Antimicrob. Chemother. 74, 1876–1883. doi: 10.1093/jac/dkz106
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
San Millan, A. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26, 978–985. doi: 10.1016/j.tim.2018.06.007
Shimasaki, T., Seekatz, A., Bassis, C., Rhee, Y., Yelin, R. D., Fogg, L., et al. (2019). Increased relative abundance of Klebsiella pneumoniae Carbapenemase-producing Klebsiella pneumoniae within the gut microbiota is associated with risk of bloodstream infection in long-term acute care hospital patients. Clin. Infect. Dis. 68, 2053–2059. doi: 10.1093/cid/ciy796
Smillie, C., Garcillan-Barcia, M. P., Francia, M. V., Rocha, E. P., and de la Cruz, F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. doi: 10.1128/MMBR.00020-10
Sorbara, M. T., Dubin, K., Littmann, E. R., Moody, T. U., Fontana, E., Seok, R., et al. (2019). Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J. Exp. Med. 216, 84–98. doi: 10.1084/jem.20181639
Tancrede, C. H., and Andremont, A. O. (1985). Bacterial translocation and gram-negative bacteremia in patients with hematological malignancies. J. Infect. Dis. 152, 99–103. doi: 10.1093/infdis/152.1.99
Trecarichi, E. M., Cauda, R., and Tumbarello, M. (2012). Detecting risk and predicting patient mortality in patients with extended-spectrum beta-lactamase-producing Enterobacteriaceae bloodstream infections. Future Microbiol. 7, 1173–1189. doi: 10.2217/fmb.12.100
Wang, C., Li, Q., and Ren, J. (2019). Microbiota-immune interaction in the pathogenesis of gut-derived infection. Front. Immunol. 10:1873. doi: 10.3389/fimmu.2019.01873
Wang, Y., Tong, M. K., Chow, K. H., Cheng, V. C., Tse, C. W., Wu, A. K., et al. (2018). Occurrence of highly conjugative IncX3 epidemic plasmid carrying Bla NDM in Enterobacteriaceae isolates in geographically widespread areas. Front. Microbiol. 9:2272. doi: 10.3389/fmicb.2018.02272
Keywords: carbapenem-resistant Enterobacteriales, KPC, NDM, horizontal gene transfer, interspecies, host gut
Citation: Ji J, Zhu Y, Zhao F, Zhang J, Yao B, Zhu M, Yu Y, Zhang J and Fu Y (2024) Co-colonization of different species harboring KPC or NDM carbapenemase in the same host gut: insight of resistance evolution by horizontal gene transfer. Front. Microbiol. 15:1416454. doi: 10.3389/fmicb.2024.1416454
Received: 12 April 2024; Accepted: 03 June 2024;
Published: 14 June 2024.
Edited by:
Fang He, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Ruichao Li, Yangzhou University, ChinaCopyright © 2024 Ji, Zhu, Zhao, Zhang, Yao, Zhu, Yu, Zhang and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Fu, eWluZy5mdUB6anUuZWR1LmNu; Jun Zhang, amFtZXN6aGFuZzIwMDBAemp1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.