
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Microbiol. , 24 May 2024
Sec. Virology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1415698
The Picornaviridae is a family of icosahedral viruses with single-stranded, highly diverse positive-sense RNA genomes. Virions consist of a capsid, without envelope, surrounding a core of RNA genome. A typical genome of picornavirus harbors a well-conserved and highly structured RNA element known as the internal ribosome entry site (IRES), functionally essential for viral replication and protein translation. Based on differences in their structures and mechanisms of action, picornaviral IRESs have been categorized into five types: type I, II, III, IV, and V. Compared with the type IV IRES, the others not only are structurally complicated, but also involve multiple initiation factors for triggering protein translation. The type IV IRES, often referred to as hepatitis C virus (HCV)-like IRES due to its structural resemblance to the HCV IRES, exhibits a simpler and more compact structure than those of the other four. The increasing identification of picornaviruses with the type IV IRES suggests that this IRES type seems to reveal strong retention and adaptation in terms of viral evolution. Here, we systematically reviewed structural features and biological functions of the type IV IRES in picornaviruses. A comprehensive understanding of the roles of type IV IRESs will contribute to elucidating the replication mechanism and pathogenesis of picornaviruses.
The family Picornaviridae comprises a large group of RNA viruses, including several significant human and animal pathogens (Zell, 2018). Since the discovery of the first picornavirus, foot-and mouth disease virus in 1898, this family has expanded to include at least 158 recognized species divided into 68 genera (Ng et al., 2015; Zell et al., 2021). These viruses have single positive-stranded RNA genomes ranging from 6.7 to 10.1 kilonucleotides in length (Andino et al., 2023). Although picornaviruses express different proteins, their RNA genomes share a similar organization (Yang et al., 2002). The picornaviral genome codes for a single polyprotein, subsequently cleaved into multiple mature structural and nonstructural proteins by virus-encoded proteinases (Zell, 2018). The 5′ end of picornaviral genome lacks a cap structure present in eukaryotic mRNAs. Instead, it is covalently linked to a virus-encoded protein, VPg, serving as a primer for RNA synthesis (Martinez-Salas et al., 2015). Immediately following VPg, there is a long and highly structured 5′ untranslated region (5’ UTR) containing the internal ribosome entry site (IRES), a cis-acting element essential for the synthesis of viral polyprotein (Daijogo and Semler, 2011).
The IRES element is an RNA fragment capable of folding into a complex structure, enabling itself to interact with one or more components of the canonical translation apparatus in a specific, cap-independent manner (Fernández-Miragall and Martínez-Salas, 2007; Firth and Brierley, 2012; Holmes and Semler, 2019). These interactions facilitate the recruitment of ribosomes or ribosomal pre-initiation complexes to an internal site in mRNA following infection with RNA viruses. By bypassing the conventional process of translation initiation, IRES allows for the continuous synthesis of viral proteins, even in the presence of the host’s translational shutdown of its own proteins (Jackson et al., 2010; Yu et al., 2011b; Firth and Brierley, 2012; Arhab et al., 2020). Consequently, RNA viruses can effectively evade antiviral defenses in hosts.
Due to their diversities in structure and function, picornaviral IRES elements have been classified into five distinct types (Martinez-Salas et al., 2017). The type IV IRES is specifically characterized by its compact structure and minimal reliance on eukaryotic initiation factors (eIFs) (Sweeney et al., 2014). Moreover, the wide distribution of type IV IRESs across numerous genera in the Picornaviridae family (Arhab et al., 2020) has aroused widespread interest among virologists. Here, we systematically reviewed the type IV IRES, mainly involved in its structure, function and impact on the initiation of polyprotein translation.
IRESs were originally found in picornaviruses, such as poliovirus (PV) (Pelletier and Sonenberg, 1988) and encephalomyocarditis virus (EMCV) (Jang et al., 1988). Subsequently, the presence of IRESs has been identified in many other viruses and cellular mRNAs (Yang and Wang, 2019). In recent years, with in-depth analysis on their structure- and action-related mechanisms, picornavirus IRESs have been classified into five types, designated type I, II, III, IV, and V. The type IV and V are also referred to as hepatitis C virus (HCV)-like IRES and aichivirus (AV)-like IRES, respectively (Sweeney et al., 2014). Each type of IRES element contains specific RNA secondary structures that vary across IRES types. These structures can exclusively recognize and bind specific factors, such as the 40S ribosomal subunit, a subset of eIFs, and several RNA-binding proteins (RBPs). Through these specific interactions, one IRES is able to recruit directly the ribosome to a viral start codon, facilitating the pathway of translation initiation, distinct from that of the cap-binding initiation mode (Lozano and Martínez-Salas, 2015).
Type I, II, and III IRESs, exemplified by PV, foot-and-mouth disease virus (FMDV) and hepatitis A virus (HAV), respectively, utilize nearly all canonical translation initiation factors, as well as non-canonical ones known as IRES trans-acting factors (ITAFs) (Pacheco and Martinez-Salas, 2010), to enhance the IRES activity (Hanson et al., 2012). Although initiation factors, like eIF4E and intact eIF4G, are superfluous for type I and II IRESs, these factors are essential for the function of type III IRESs (Ali et al., 2001; Sadahiro et al., 2018). Similarly, the type V IRES found in AV involves multiple canonical eIFs during its process of translation initiation (Yu et al., 2011b; Sweeney et al., 2012). Distinct from other IRES categories, type IV elements demonstrate a diminished dependence on eIFs and eliminate the requirement for ITAFs in assembling 48S complexes (Pisarev et al., 2004; Mailliot and Martin, 2018). These IRES-mediated translation mechanisms are primarily associated with two models (Komar and Hatzoglou, 2015): the first one, whereby the 40S ribosome positions itself in the vicinity of AUG, subsequently scanning to locate the AUG (Figure 1A), and the second one, in which the 40S ribosomal subunit directly localizes to the AUG (Figures 1B–D).
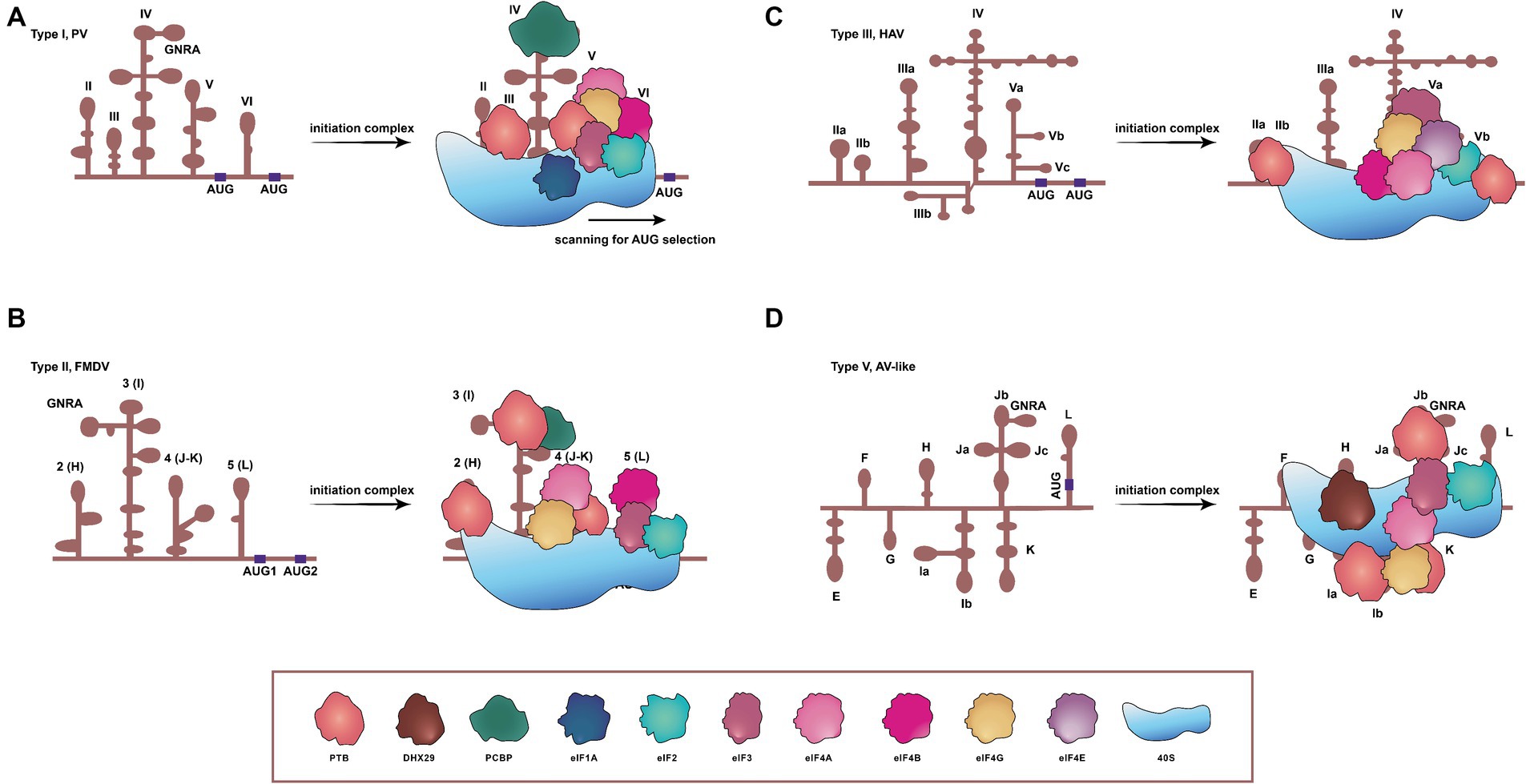
Figure 1. Schematic representations of secondary structures, and requirements of translation initiation factors for four IRESs in picornaviruses. (A) Type I IRES, exemplified by PV, comprises five principal structural domains (dII to dVI). It involves all translation initiation factors and many ITAFs to initiate translation, concerning a scanning process. (B) FMDV contains the type II IRES, arranged in modular domains (H to L). The type II IRES does not scan the mRNA, and instead, the 48S-like complex is recruited directly to the start codon. (C) The type III IRES has been found only in HAV, and the protein translation mediated by this type depends on eIF4E. (D) The type V IRES is present in AV and harbors eight domains. Distinct from all previously characterized IRESs, the one of AV exhibits an absolute dependency on DHX29, necessitated by the entrapment of its AUG within a stable hairpin structure. DHX29: DExH-box protein DHX29; eIF1A, eIF2, eIF3, eIF4A, eIF4B, eIF4E, and eIF4G: eukaryotic initiation factor 1A, 2, 3, 4A, 4B, 4E, and 4G. PTB: pyrimidine tract-binding protein; PCBP: poly(C)-binding protein; 40S: 40S small ribosomal subunit.
The type I IRES in picornaviruses is composed of five principal domains, designated dII to dVI (Martinez-Salas et al., 2015). Translation initiation on PV IRES entails the scanning by 43S ribosomal preinitiation complexes, and engages a set of eIFs (eIF2, eIF3, eIF4A, eIF4G, eIF4B, and eIF1A) and a single ITAF, identified as the poly(C)-binding protein 2 (PCBP2), as shown in Figure 1A (Sweeney et al., 2014; Andreev et al., 2022). In instance of conventional pathways of translation initiation inaccessible, these viruses can employ an alternative IRES-independent transition mechanism, which depends on eIF2A/eIF2D and utilizes a non-AUG codon for initiation. This alternative pathway seems to facilitate ongoing translation and viral genome replication in the presence of activated intrinsic defenses against viruses (Kim et al., 2023).
Similar to type I IRESs, type II IRESs, such as those of EMCV and FMDV, also comprise five major domains, designated H to L (Yu et al., 2011a). Translation initiation mediated by IRESs of EMCV and FMDV involves the specific binding of eIF4G and eIF4A to the Y-shaped J-K domain, an interaction dependent on a conserved sequence/structural motif at the apex of domain J (Figure 1B) (López de Quinto and Martínez-Salas, 2000; Yu et al., 2011a; Sweeney et al., 2012; Imai et al., 2016). In EMCV, the IRES structure can be remodeled by eIF4G/eIF4A to promote EMCV-IRES/40S interactions, and rearrange the coding region to accommodate correctly the AUG in the ribosomal mRNA cleft. The 43S complex can then undergo structural rearrangement due to both AUG/tRNA recognition and close contact between the ribosome and IRES. This would allow the 60S to join the complex and to initiate translation (Chamond et al., 2014). Type II IRESs therefore can function dependent neither on the eIF4E nor on factors implicated in ribosomal scanning, such as eIF1 and eIF1A (Sweeney et al., 2012).
The secondary structure of the HAV IRES element comprises six structural domains, and this IRES element is much longer than types I and II (Brown et al., 1991; Francisco-Velilla et al., 2022). HAV IRES requires the intact eIF4G to initiate translation, unlike types I and II, both of which require only the C-terminal two-thirds fragment of eIF4G (Francisco-Velilla et al., 2022). The absence of a cap structure at the 5′ end of picornaviral genome indicates that the involvement of the cap-binding initiation factor eIF4E is unnecessary for the translation initiation of picornaviruses. However, eIF4E is essential in HAV for the activation of IRES-mediated translation (Figure 1C) (Redondo et al., 2012). Thus, HAV is unable to shut down the protein synthesis in hosts by a similar mechanism as those of other picornaviruses, and its IRES is inefficient, probably due to its unfair competition for the cellular translation machinery and tRNAs (Pinto et al., 2018, 2021). To ameliorate the tRNA competition, HAV has evolved a highly biased and deoptimized codon usage with respect to its hosts (Pinto et al., 2007; Costafreda et al., 2014; Pinto et al., 2018). During translation, the eIF4F complex recognizes the IRES element and, subsequently, the 43S preinitiation complex and the 60S large ribosomal subunits are directly recruited onto HAV mRNA to synthesize the viral polyprotein (Sadahiro et al., 2018).
The AV IRES consists of eight domains, designated E to L. In vitro reconstitution of initiation on the AV IRES reveals that it shares some characteristics with type I and II IRESs, suggesting that it appears to be a chimera between type I and type II (Yu et al., 2011b; Sweeney et al., 2012). The formation of 48S complex on the AV IRES requires eIF2, eIF3, eIF4A, the eIF4A-interacting central domain of eIF4G (eIF4Gm) and the DExH-box protein DHX29. This process is strongly stimulated by the pyrimidine tract-binding protein (Figure 1D). The lack of dependence on eIF1 and eIF1A, which robustly stimulate scanning and monitor the fidelity of AUG selection during this process, implies that the AUG can be directly recognized and bound by the 43S complex on the AV IRES, hence bypassing the need for scanning (Yu et al., 2011b).
The translation initiation mediated by the type IV IRES occurs through direct binding of the IRES element to the 40S ribosomal subunit. The IRES–40S complex positions the AUG at the P site and forms a 48S complex with eIF2/GTP/initiator tRNA (Hashem et al., 2013), stabilized by the binding of eIF3 (Ji et al., 2004). Following two steps of GTP hydrolysis, eIF2 is released, and the 60S ribosomal subunit associates under the action of eIF5B, resulting in the formation of an active 80S ribosome. This 80S ribosome subsequently initiates the synthesis of viral polyprotein (Figure 2) (Ji et al., 2004; Johnson et al., 2017; Niepmann and Gerresheim, 2020).
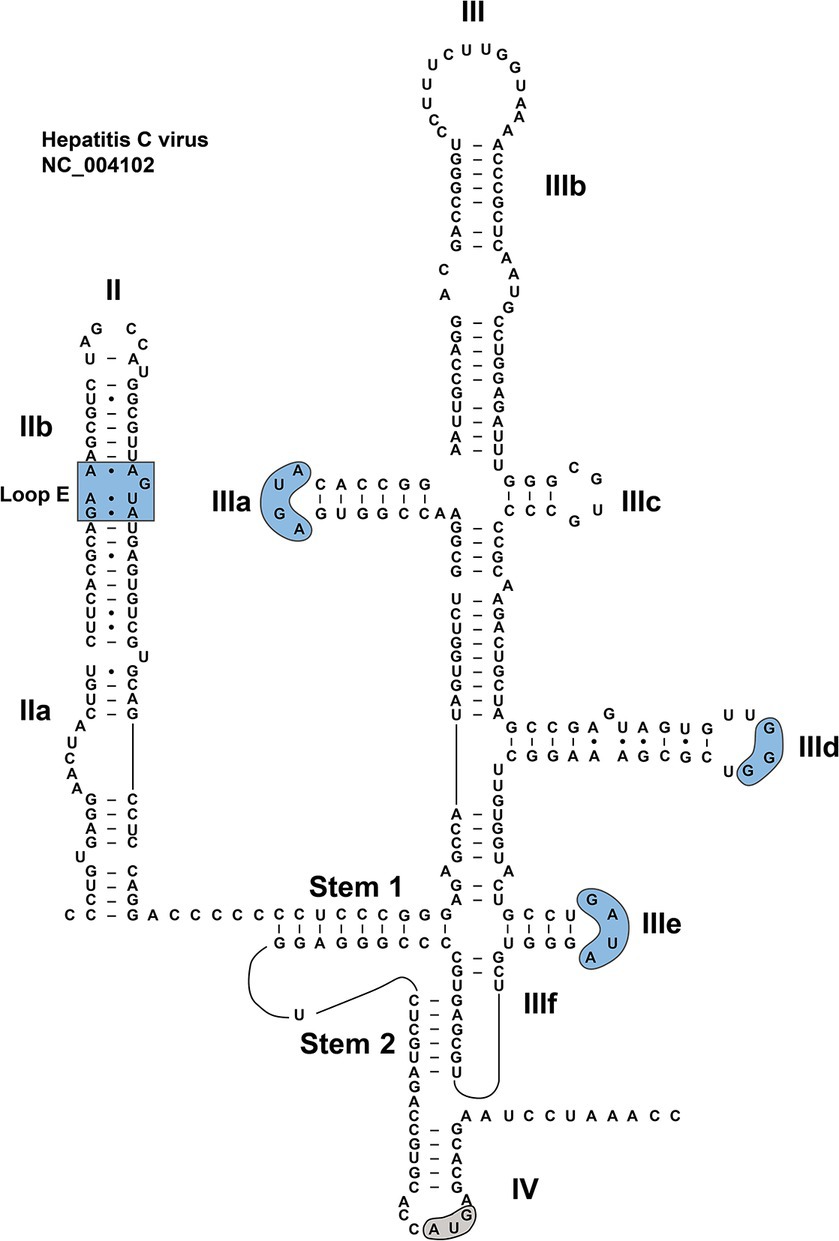
Figure 2. RNA sequence and secondary structure of HCV IRES. The conserved motifs in dII and dII are indicated by blue shades. The start codon is indicated by a gray shade.
The relationship between RNA structure and biological function of distinct IRESs has been intensively analyzed by various experimental methods (Malnou et al., 2002; Bailey and Tapprich, 2007; Serrano et al., 2009), providing insights into the initiation mechanism of viral protein translation. Since the secondary and tertiary structures of type IV IRES are very different from those of the other four types, the type IV IRES is systematically reviewed here.
The type IV IRES was initially identified in HCV, bovine viral diarrhea virus (BVDV) and classical swine fever virus (CSFV) (Arhab et al., 2020). This specific IRES type is subsequently found in approximately 25 distinct genera in the Picornaviridae family (Table 1) (Arhab et al., 2020). In the context of the Picornaviridae family, the porcine teschovirus-1 (PTV), belonging to the Teschovirus genus, stands as one of the earliest documented instances (Pisarev et al., 2004). Although the major portion of the PTV-1 genome had been determined prior to 2002, its 5′-terminal sequence had been neither cloned nor characterized (Kaku et al., 2002). Kaku and colleagues subsequently demonstrated the presence of a functional IRES element in the PTV 5’ UTR by constructing and analyzing plasmids that expressed bicistronic mRNAs. Further tests confirmed that the activity of this element was unaffected by the co-expression of an enterovirus 2A protease and the cleavage of eIF4G. It could also function effectively in an RRL (rabbit reticulocyte lysate) in vitro translation system (Kaku et al., 2002). Thus, the biological properties of the PTV IRES are most similar to those of EMCV and FMDV IRES elements, but the computer-predicted secondary structure of the PTV IRES shows no apparent resemblance to those of cardiovirus and aphthovirus IRES elements. The PTV IRES is consequently classified into a new category of IRES (Kaku et al., 2002).
Following the definition of the type IV IRES, this group further determined its boundaries and identified the requirements for the formation of 48S preinitiation complexes through toeprinting assays. It was found that the formation of 48S preinitiation complex on the PTV-1 IRES had no need of the initiation factors eIF1, eIF1A, eIF3, eIF4B, and eIF4F (Pisarev et al., 2004). It only involves purified 40S ribosomal subunits plus the ternary complex of eIF2, Met-tRNA, and GTP, although this process is enhanced in the presence of eIF3. Indeed, the PTV-1 IRES can form a binary complex with 40S subunits alone (Chard et al., 2006a,b). These data demonstrate that the PTV-1 IRES has properties completely different from those of other IRES elements in picornaviruses, but highly resembles to the HCV IRES (Pisarev et al., 2004). Therefore, this unique class of type IV IRES is also referred to as HCV-like IRES.
Interestingly, the “unique” IRES was subsequently found in the Sapelovirus (Krumbholz et al., 2002; Oberste et al., 2003; Li et al., 2011; Son et al., 2014) and Anativirus genera (Tseng and Tsai, 2007). Functional assays demonstrated that these IRES elements remained still active, when eIF4G was cleaved and when the activity of eIF4A was blocked (Chard et al., 2006a). On the basis of the sequence information and the structure prediction, the type IV IRES has also been demonstrated to exist in the Kobuvirus (Reuter et al., 2009), Aalivirus (Wang et al., 2014), Tremovirus (Hellen and de Breyne, 2007; Bakhshesh et al., 2008), Avihepatovirus (Tseng et al., 2007; Pan et al., 2012), Pasivirus (Sauvage et al., 2012; Yu et al., 2013; Asnani et al., 2015), and Senecavirus genera (Willcocks et al., 2011).
In addition to the aforementioned picornaviruses that mainly infect pigs and poultry, the others harboring the type IV IRES have also been found in a wide range of vertebrates from fishes to mammals. For example, seal picornavirus type 1 (SePV-1) was first identified in marine mammals in 2007, and then classified into the genus Aquamavirus (Kapoor et al., 2008). The second member of this genus is bear picornavirus 1 (BePV-1), another novel picornavirus isolated from black bear. Sequence analyses of SePV-1 and BePV-1 revealed that both of them had the type IV IRES (Wang et al., 2019b). In 2011, three novel picornaviruses, bat picornavirus 1 (BPV-1), BPV-2, and BPV-3, were identified in bats. Both BPV-1 and BPV-2 were characterized by the presence of the type IV IRES, whereas the BPV-3 harbored the type I IRES (Lau et al., 2011; Zell, 2018). More picornaviruses have been recently identified in bat species, including Hepatovirus (Drexler et al., 2015), Diresapivirus (Wu et al., 2016), Crohivirus and Kunsagivirus (Asnani et al., 2015; Yinda et al., 2017) genera, all of which are found to possess the type IV IRES.
A novel feline picornavirus, FePV in the genus Felipivirus, was identified in stray cats in Hong Kong. FePV is closely related both to members in the genus Sapelovirus and to the unclassified BPV-3, but with the difference that FePV possesses the type IV IRES instead of the type I IRES found in BPV-3 (Lau et al., 2012). Feline sakobuvirus A, the second picornavirus with the type IV IRES isolated from cats, does not belong to the same genus as that of FePV, but exhibits a closer relation to members of the genus Kobuvirus (Ng et al., 2014). Type IV-containing picornaviruses are also recognized in other mammals, including rats (genus Parechovirus) (Firth et al., 2014), ferret (genus Parechovirus) (Smits et al., 2013), marmot (genus Mosavirus) (Luo et al., 2018) and baboons (genus Kunsagivirus) (Buechler et al., 2017).
The genus Megrivirus, recognized to contain one of the longest picornaviral genomes currently described, comprises five species, named A to E. The type IV IRES is present in the 5’ UTR region of all species, with the sole exception of species D (Honkavuori et al., 2011; Phan et al., 2013; Lau et al., 2014; Liao et al., 2014; Boros et al., 2014a; Kim et al., 2015; Wang et al., 2017; Zell, 2018; Yinda et al., 2019; Haji Zamani et al., 2023). This type of virus is additionally found in migrant bird (genus Kunsagivirus) (Boros et al., 2013), pigeon (genus Colbovirus) (Kofstad and Jonassen, 2011), quail (genus Phacovirus) (Pankovics et al., 2012), goose (genus Ludopivirus) (Boros et al., 2018), duck (Pink-eared duck picornavirus), red-crowned crane (genus Grusopivirus) (Wang Y. et al., 2019; Arhab et al., 2020), lorikeet (genus Grusopivirus) (Wang et al., 2019a), as well as other species, such as amphibians, reptiles (Ng et al., 2015; Arhab et al., 2020) and fishes (Table 1) (Barbknecht et al., 2014; Lange et al., 2014; Phelps et al., 2014; Asnani et al., 2015). Most known type IV-containing picornaviruses are found in mammals and birds, whereas this type of picornavirus has also been identified in other vertebrates in recent years. These findings further emphasize the relative ubiquity and diversity of the type IV IRES.
A great deal of works have been devoted to studies on the HCV IRES (Lukavsky, 2009; Berry et al., 2011; Perard et al., 2013; Yamamoto et al., 2015; Yokoyama et al., 2019; Brown et al., 2022), which is of critical importance for understanding the biological mechanisms of the type IV IRES. The structure of HCV IRES can serve as an ideal model for unveiling secondary structures of type IV IRESs, especially considering the similar roles these elements play in the synthesis of viral proteins within their individual viruses. Although a significant portion of the currently identified HCV-like IRES structures are based on the model of HCV IRES for research, each of them has its own specific structure and function. The arrangement of type IV IRES is relatively simple, mainly composed of secondary stem–loop structures and tertiary structures (Asnani et al., 2015; Lee et al., 2017). A typical HCV IRES is approximately 340 nt in length, containing many stem–loop structures, known as dII, dIII, and dIV, a pseudoknot (PK), and a helical structure that links dII with dIII and dIV (Kalliampakou et al., 2002). DII is a 70-nt-long flexible hairpin, composed of two subdomains, IIa and IIb: the former harboring an asymmetric internal loop; the latter including an internal loop E motif and an apical hairpin (Figure 3). These structural features are relatively conserved among closely related HCV-like IRESs in members of the Flaviviridae family (Locker et al., 2007; Lukavsky, 2009).
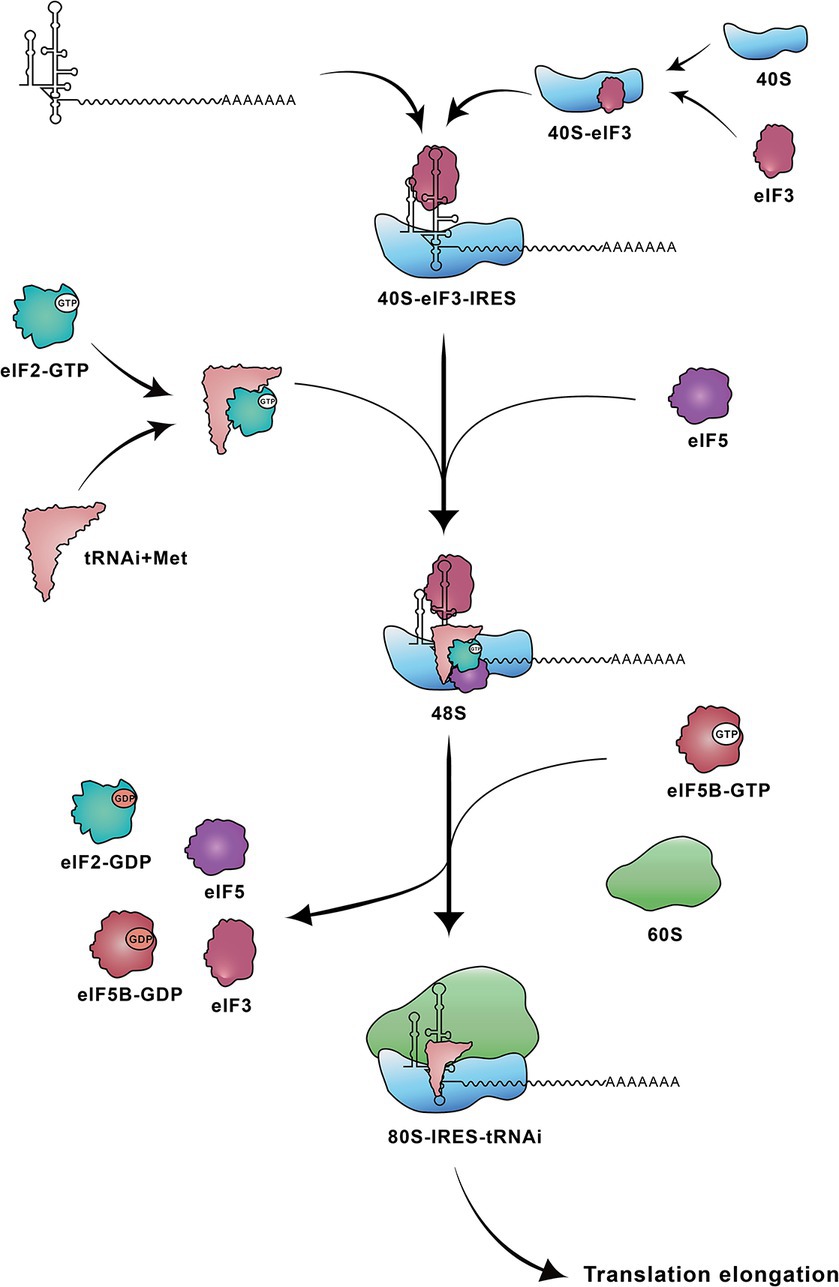
Figure 3. Steps involved in translation initiation mediated by HCV IRES. The HCV IRES first binds to a 40S-eIF3 complex. Subsequently the ternary 40S-IRES-eIF3 complex acquires the eIF2-tRNAiMet-GTP and eIF5. Following two steps of GTP hydrolysis, the 60S ribosomal subunit joins to form the elongation-competent 80S ribosome. 40S: 40S small ribosomal subunit; 40S-eIF3: 40S ribosomal subunit-eukaryotic initiation factor 3; 48S: 48S complex; 60S: 60S large ribosomal subunit; 80S-IRES-tRNAi: 80S-IRES-initiator tRNA; eIF2-GTP: eIF2-guanosine triphosphate; eIF2-GDP: eIF2-guanosine diphosphate; eIF5B-GTP: eIF5B-guanosine triphosphate; eIF5B-GDP: eIF5B-guanosine diphosphate; tRNAi+Met: tRNAi-methionine.
DIII consists of several subdomains (IIIa to IIIf). The basal part of dIII contains a 4-way junction, which includes a predicted PK (IIIf) and a small stem–loop (IIIe) (Melcher et al., 2003; Lukavsky, 2009). In the middle of the dIII, the dIIId forms part of a three-way junction. The apical region of dIII contains a 4-way junction (Figure 3), involved in a central dIII stem, as well as dIIIa, dIIIb and dIIIc (Koirala et al., 2020). Certain motifs in the HCV dIII also exhibit a high degree of conservation in some viruses, such as in CSFV and BVDV. For example, dIIIa and dIIIe contain AGUA and GA[U/C]A tetraloops, respectively. The dIIId features a G-rich loop, with at least three consecutive G residues, in the hairpin (Fletcher and Jackson, 2002; Lukavsky, 2009). However, the sequence of highly conserved dIIIa loop is unimportant for the maintenance of full IRES activity (Fletcher and Jackson, 2002). The dIV is the last subdomain in the type IV IRES, and harbors the viral start codon (Berry et al., 2011; Tanaka et al., 2018).
Over 40 structures associated with the type IV IRES have currently been identified or predicted. Representative models of type IV IRESs are schematically shown in Supplementary Figure S1. These IRESs are 180–420 nucleotides in length (Asnani et al., 2015), exhibiting the significant diversity in their structures, whereas all these IRES structures contain a crucial, highly conserved core region, including the PK as well as subdomains IIId, IIIe, and IIIf (Arhab et al., 2020). The core region is functionally essential for the translation initiation. The dIII is typically composed of four to seven subdomains in the type IV IRES, whereas the number of subdomains varies greatly among different picornaviruses. The simplest one is derived from Guangxi changeable lizard picornavirus 2, only having four subdomains: IIIa, IIId, IIIe, and IIIf. The most complex ones, like members in Crohivirus, Parechovirus, Tropivirus, Senecavirus, Ludopivirus, and Tremovirus genera, are composed of seven subdomains: IIIa to IIIf. In addition to the complete complement of characteristic subdomains and motifs, these complex IRESs also bear an extra dIIIa2 (Ludopivirus genus) (Boros et al., 2018), dIIId2 (Senecavirus, Parechovirus, Crohivirus, and Tropivirus genera) or dIIIb2 (Tremovirus genus) (Hellen and de Breyne, 2007; Arhab et al., 2020).
Several conserved sequences exist in dIII, including the apical GGG motif in dIIId and a GA[U/C]A tetraloop in dIIIe. An “AGUA” loop at the top of dIIIa, similar to that in HCV IRES, is only found in the genera Pasivirus, Tremovirus, Tropivirus, Senecavirus, Crohivirus, Parechovirus, and Pasivirus (Hellen and de Breyne, 2007; Asnani et al., 2015; Arhab et al., 2020). The GGG motif at the top of IIId loop was found to be a GGGGG pentaloop in a homologous position in simian sapelovirus (Hellen and de Breyne, 2007). In some cases, the G-rich sequence has four consecutive G residues in some picornaviruses, including in carp picornavirus 1, fathead minnow picornavirus, BPV-1, Ia io picornavirus 1, California sea lion sapelovirus 1, grusopivirus, FePV, tortoise rafivirus A1 and crohivirus B (Asnani et al., 2015; Arhab et al., 2020). However, only two G residues are identified in pink-eared duck picornavirus and pemapivirus (Arhab et al., 2020). The dIIIe, consisting of an apical GA[U/C]A tetraloop and a 4-bp stem, is conserved in almost all type IV IRESs, whereas members of the species Megrivirus B have a GAUC loop, and Aalivirus has a 3-bp stem and 5-nt apical loop (Wang et al., 2014; Asnani et al., 2015).
Significant sequence variations are observed in the apical region of dIII. For example, in Kobuvirus, Kunsagivirus, Limnipivirus, and Rafivirus genera, the apical region of dIII may be severely truncated, possibly lacking one (genera Kunsagivirus, Limnipivirus, Sapelovirus, and Teschovirus) or two (genus Aalivirus) subdomains (Arhab et al., 2020). Additionally, subdomains IIIa and IIIc may also be disposed in a staggered configuration rather than forming a 4-way junction. Subdomain IIIb may be greatly elongated, as observed in the Megrivirus, Phacovirus, and Colbovirus genera, which are topped by a 20-nt-long “8”-like structure (Asnani et al., 2015; Boros et al., 2016). Such a structure is conserved in avian-origin and seal picornaviruses, and may be crucial for the viral translation (Pankovics et al., 2012; Boros et al., 2014b).
The dII of type IV IRES, with a length range of 21–120 nt, is shorter or longer than that of the HCV IRES. Compared to the dIII, the dII shows the great variability. In Teschovirus, Tropivirus, and Tremovirus genera, the dII contains a small branching hairpin (dIIb) (Arhab et al., 2020). Despite the high variability of dII in the HCV IRES, its apical hairpin loop, internal loop E motif and basal internal loops are conserved among HCV, and closely related to those of HCV-like IRESs (Lukavsky, 2009). Notably, dIV in HCV is not unique to it. This special structure is also found in some picornaviral genera (e.g., Colbovirus, Megrivirus, Sapelovirus, Crohivirus, Pasivirus, and Limnipivirus) containing the type IV IRES. The dIV in these genera shows the lower stability than the homologous region in HCV (Asnani et al., 2015).
Collectively, the type IV IRES contains a highly conserved core region, which is crucial for the translation initiation. Surrounding this core, other regions, e.g., dII, vary significantly among IRESs from different species. This combination of structural conservation and variability may be related to the function and regulation of type IV IRES.
The heterogeneity of type IV IRES structures exemplifies biological variety, and equips viruses with adaptive mechanisms to thrive in various host environments (Martinez-Salas et al., 2017). Each IRES configuration is likely tailored to particular environmental and biological demands, facilitating viral proliferation and transmission across diverse hosts. Cellular proteins binding to viral IRES are critical for orchestrating the life cycle of viruses. For example, certain cellular proteins can enhance the activity of IRES, while others may suppress its function (Liu et al., 2020; Embarc-Buh et al., 2021; Lopez-Ulloa et al., 2022; Marques et al., 2022). During this process, the secondary and tertiary structures of IRES constitute the foundation for the interaction with various cellular factors. Perturbation of specific conserved motifs may compromise IRES structural integrity, potentially impeding viral replication.
The flexibility of secondary structure and the uniqueness of primary sequence within dIII contribute to its role as the most active region in RNA–protein interactions (Barría et al., 2009; El Awady et al., 2009; Lukavsky, 2009; Johnson et al., 2017). The dIIIa, dIIIb, and dIIIc of HCV are primarily involved in the binding of eIF3 and the 40S subunit (Kieft et al., 2001; Rijnbrand et al., 2004). Elements equivalent to HCV dIIIa and dIIIc are less conserved in picornaviral IRESs, possibly implying that these domains are less critical for their functional activities. Nevertheless, the possibility that these two domains still have other functions in picornaviral IRESs cannot be wholly dismissed, and these functions may be achieved by the evolution of additional sequences (Chard et al., 2006b). The interaction between dIIIb and eIF3 affects a key process of active ribosomal assembly, during which the binding of eIF3 to dIIIb will exert effects on both eIF3 and eIF2 stabilities in the formation of preinitiation complexes (Easton et al., 2009). Deletion mutations at this site reduce initiator tRNA deposition, leading to further compromise of the 80S complex assembly (Ji et al., 2004). Although the “8”-like structure is present in the apical region of dIIIb in certain instances, a comprehensive research on its precise mechanism of action remains to be conducted further. A limited number of studies (Boros et al., 2014b) have documented the impact of deletions or mutations in this structure on IRES functionality. However, given its prevalent occurrence in avian picornaviruses, the structure can hold unique significance.
The GGG motif in the dIIId loop is a major determinant of ribosome binding dIIId (Jubin et al., 2000; Friis et al., 2012; Niepmann et al., 2018), capable of base-pairing with CCC nucleotides in the apical loop of ES7 of 18S ribosomal RNAs (Hashem et al., 2013; Lattimer et al., 2019). The dIIId apical loop contains a GG motif in the genus Pemapivirus, indicating that this dinucleotide sequence may be adequate for ribosomal binding (Arhab et al., 2020). Mutations in this sequence can reduce the binding affinity of the IRES for the 40S subunit, thereby hindering the assembly of the 48S complex (Ji et al., 2004; de Breyne et al., 2008; Willcocks et al., 2011; Pan et al., 2012). Certain viruses possess two dIIId: dIIId1 and dIIId2. The latter exhibits the variability in function across various viruses. For instance, dIIId2 in senecavirus A (SVA), albeit unessential for IRES-mediated translation as evidenced by its non-impact on eIF3 and 40S recruitment, 48S complexes and 80S ribosomes assembly, plays a crucial role in viral infectivity (Willcocks et al., 2011). The absence of dIIId2 would interfere with the assembly of 80S ribosome in both BDV- and CSFV-infected cells, consequently impeding the translation initiation (Willcocks et al., 2017).
The GA[U/C]A tetraloop observed in the dIIIe region of HCV-like IRES diverges from the standard GNRA tetraloop that is characteristic of type I and II IRES elements (López de Quinto and Martínez-Salas, 1997; Psaridi et al., 1999; Robertson et al., 1999; Asnani et al., 2016; Mailliot and Martin, 2018). Despite minor variations in the tetraloop bases across IRES types, their nucleotide sequences are all essential for IRES function. Nucleotide insertions, mutations or deletions in this motif may lead to IRES inactivation, thereby affecting the conformation and stability of IRES by modulating the interaction of RNA–RNA tertiary structures (Bhattacharyya and Das, 2006; Fernandez-Miragall et al., 2006; Bakhshesh et al., 2008; Pan et al., 2012). For example, the apical part of dIII would be reorganized in the FMDV IRES after a single nucleotide substitution in the GNRA motif to GUAG, or its substitution by a UNCG motif (Fernandez-Miragall and Martinez-Salas, 2003). Change in each of the four nucleotide positions in the GAUA loop of HCV IRES severely impairs the IRES activity (Psaridi et al., 1999), and this hairpin loop may enhance the interaction between the IIId and the 40S subunit (Lukavsky et al., 2000; Angulo et al., 2016).
Type IV IRES features a single PK structure, in contrast to three PKs in dicistroviruses (Abaeva et al., 2023). The structure is formed via base pairing between loop nucleotides of IIIf and other single-stranded regions, showing two base-paired stems, stem I and II (Liu et al., 2021a). The IRES activity is dependent both on the primary sequence and on the secondary structure in both stem regions. Any mutation, if responsible for disruption of the secondary structure of stem region, mutations on both sides of the stem sequence without disturbing the overall conformation, or insertion (or deletion) mutations in the stem region, would invariably compromise the IRES function (Chard et al., 2006b; Berry et al., 2010; Willcocks et al., 2011; Pan et al., 2012; Wang et al., 2021; Liu et al., 2021b). Furthermore, sequence alterations within the PK region of SVA stem II can potentially exert an impact on virion assembly (Liu et al., 2021a). The function of the PK is contingent upon its intact structure. When the PK participates in the binding of the 40S subunit, its primary role is associated with positioning the start codon to the ribosomal P site (Kieft et al., 2001; Chard et al., 2006b; Lukavsky, 2009). An optimal distance is also maintained between the PK and the start codon. Either insertion or deletion of nucleotides in this sequence would interfere with viral replication to some extent, possibly attributed to deleterious effects on the AUG reaching the P site (Berry et al., 2010; Liu et al., 2021c).
Ribosomal translocation is a crucial stage in the protein synthesis, requiring the mRNA template to move so that new codons are positioned within the A site for decoding (Zhou et al., 2014; Milicevic et al., 2024). The conformational shift induced by HCV dII in the 40S subunit affects the translocation process (Brown et al., 2022). Despite the spatial separation between dII and dIII, the asymmetrical interior loop in dIIa facilitates the formation of bent dII conformation (Lukavsky et al., 2003). This bending permits the apical region of dIIb to spatially contact the 40S ribosomal subunit bound to dIII, not only facilitating the adoption of a specific HCV RNA configuration in the decoding groove, but also potentially assisting in the proper selection of start codon (Lukavsky et al., 2003; Filbin and Kieft, 2011; Johnson et al., 2017; Brown et al., 2022).
The hairpin loop and loop E motif in dIIb can promote both eIF5-induced GTP hydrolysis and eIF2/GDP release from the initiation complex (Locker et al., 2007; Martinez-Salas et al., 2008; Barría et al., 2009). Both deletion and mutation of the dII region would lead to a significant reduction in the IRES function (Friis et al., 2012; Filbin et al., 2013). The majority of functional studies are currently involved in the dII of HCV IRES. The dII in the type IV IRES shows considerable variation among picornaviruses, some of which contain neither a loop E motif nor an asymmetric inner loop. In certain viruses, their dII regions simply show a stem-loop structure formed via a 21-nt-long motif (Asnani et al., 2015). This simplified dII may exert functions distinct from those of HCV through synergistic actions with other IRES domains, or interaction with specific host factors, reflecting the evolutionary plasticity of viral IRES in both structure and function.
To summarize, IRES dIII primarily engages in the binding of eIF3 and 40S ribosomal subunits, and in the precise localization of the start codon. The dII is mainly involved in 80S ribosome assembly, and in the following initiation steps. The attachment of 40S ribosomal subunit or eIF3 to IRES is not confined to an individual dIII subdomain. Rather, the higher-order structures formed between multiple IRES domains, along with their exposed conserved sequences and structural motifs, collectively provide multiple regulatory interfaces for recruitment and precise localization of these molecules. Therefore, maintaining the structural integrity of the IRES is functionally crucial for viral replication. Any mutation, which disrupts the IRES structure or alters its conserved sequences, will lead to IRES inactivation, thereby affecting the translation process of the viral proteins.
The first reports concerning IRES elements unequivocally demonstrated the cap-independent initiation of translation in PV and EMCV genomes. There has been substantial progress in recent 20 years in uncovering structures and functions of picornaviral IRESs. The type IV IRES can fold into a compact tertiary structure, making its own key elements interact with the small ribosomal subunit through a cluster of specific ribosomal proteins for triggering the translation of viral polyprotein. Unfortunately, most valuable reports associated with the type IV IRES are based on the HCV, rather than on picornaviruses. There is still much to be learned about the interaction of type IV IRES with host factors or even other RNA elements. Nowadays, characterizing its high-order organization of RNA structure, in conjunction with the recognition of IRES–protein interaction network, is the greatest challenge to unveil how these specialized RNA structures function during viral replication.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
YL: Writing – original draft. LZ: Writing – original draft. LW: Writing – original draft. JL: Software, Writing – original draft. YZ: Software, Writing – review & editing. FL: Conceptualization, Writing – review & editing. QW: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (Grant No. 32273000), the Qingdao Demonstration Project for People-benefit from Science and Techniques (Grant No. 23-2-8-xdny-14-nsh; 24-2-8-xdny-4-nsh), and the National Program of Undergraduate Innovation and Entrepreneurship (Grant No. 202310435039).
LZ was employed by Shandong New Hope Liuhe Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1415698/full#supplementary-material
Supplementary Figure S1 | Representative structures of type IV IRESs potentially forming in picornaviral genomes. The conserved motifs in DII and DIII are indicated by blue shades. The start codon is indicated by a gray shade.
Abaeva, I. S., Young, C., Warsaba, R., Khan, N., Tran, L. V., Jan, E., et al. (2023). The structure and mechanism of action of a distinct class of dicistrovirus intergenic region IRESs. Nucleic Acids Res. 51, 9294–9313. doi: 10.1093/nar/gkad569
Ali, I. K., McKendrick, L., Morley, S. J., and Jackson, R. J. (2001). Activity of the hepatitis a virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75, 7854–7863. doi: 10.1128/jvi.75.17.7854-7863.2001
Andino, R., Kirkegaard, K., Macadam, A., Racaniello, V. R., and Rosenfeld, A. B. (2023). The Picornaviridae family: knowledge gaps, animal models, countermeasures, and prototype pathogens. J. Infect. Dis. 228, S427–S445. doi: 10.1093/infdis/jiac426
Andreev, D. E., Niepmann, M., and Shatsky, I. N. (2022). Elusive trans-acting factors which operate with type I (poliovirus-like) IRES elements. Int. J. Mol. Sci. 23:15497. doi: 10.3390/ijms232415497
Angulo, J., Ulryck, N., Deforges, J., Chamond, N., Lopez-Lastra, M., Masquida, B., et al. (2016). LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 44, 1309–1325. doi: 10.1093/nar/gkv1325
Arhab, Y., Bulakhov, A. G., Pestova, T. V., and Hellen, C. U. T. (2020). Dissemination of internal ribosomal entry sites (IRES) between viruses by horizontal gene transfer. Viruses 12:612. doi: 10.3390/v12060612
Asnani, M., Kumar, P., and Hellen, C. U. (2015). Widespread distribution and structural diversity of type IV IRESs in members of Picornaviridae. Virology 478, 61–74. doi: 10.1016/j.virol.2015.02.016
Asnani, M., Pestova, T. V., and Hellen, C. U. (2016). PCBP2 enables the cadicivirus IRES to exploit the function of a conserved GRNA tetraloop to enhance ribosomal initiation complex formation. Nucleic Acids Res. 44, gkw609–gkw9917. doi: 10.1093/nar/gkw609
Bailey, J. M., and Tapprich, W. E. (2007). Structure of the 5′ nontranslated region of the coxsackievirus b3 genome: chemical modification and comparative sequence analysis. J. Virol. 81, 650–668. doi: 10.1128/JVI.01327-06
Bakhshesh, M., Groppelli, E., Willcocks, M. M., Royall, E., Belsham, G. J., and Roberts, L. O. (2008). The picornavirus avian encephalomyelitis virus possesses a hepatitis C virus-like internal ribosome entry site element. J. Virol. 82, 1993–2003. doi: 10.1128/JVI.01957-07
Barbknecht, M., Sepsenwol, S., Leis, E., Tuttle-Lau, M., Gaikowski, M., Knowles, N. J., et al. (2014). Characterization of a new picornavirus isolated from the freshwater fish Lepomis macrochirus. J. Gen. Virol. 95, 601–613. doi: 10.1099/vir.0.061960-0
Barría, M. I., González, A., Vera-Otarola, J., León, U., Vollrath, V., Marsac, D., et al. (2009). Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role in cap-independent translation. Nucleic Acids Res. 37, 957–971. doi: 10.1093/nar/gkn1022
Berry, K. E., Waghray, S., and Doudna, J. A. (2010). The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA 16, 1559–1569. doi: 10.1261/rna.2197210
Berry, K. E., Waghray, S., Mortimer, S. A., Bai, Y., and Doudna, J. A. (2011). Crystal structure of the HCV IRES central domain reveals strategy for start-codon positioning. Structure 19, 1456–1466. doi: 10.1016/j.str.2011.08.002
Bhattacharyya, S., and Das, S. (2006). An apical GAGA loop within 5' UTR of the coxsackievirus B3 RNA maintains structural organization of the IRES element required for efficient ribosome entry. RNA Biol. 3, 60–68. doi: 10.4161/rna.3.2.2990
Boros, A., Kiss, T., Kiss, O., Pankovics, P., Kapusinszky, B., Delwart, E., et al. (2013). Genetic characterization of a novel picornavirus distantly related to the marine mammal-infecting aquamaviruses in a long-distance migrant bird species, European roller (Coracias garrulus). J. Gen. Virol. 94, 2029–2035. doi: 10.1099/vir.0.054676-0
Boros, A., Pankovics, P., Adonyi, A., Fenyvesi, H., Day, J. M., Phan, T. G., et al. (2016). A diarrheic chicken simultaneously co-infected with multiple picornaviruses: complete genome analysis of avian picornaviruses representing up to six genera. Virology 489, 63–74. doi: 10.1016/j.virol.2015.12.002
Boros, A., Pankovics, P., Knowles, N. J., Nemes, C., Delwart, E., and Reuter, G. (2014a). Comparative complete genome analysis of chicken and Turkey megriviruses (family picornaviridae): long 3′ untranslated regions with a potential second open reading frame and evidence for possible recombination. J. Virol. 88, 6434–6443. doi: 10.1128/JVI.03807-13
Boros, A., Pankovics, P., and Reuter, G. (2014b). Avian picornaviruses: molecular evolution, genome diversity and unusual genome features of a rapidly expanding group of viruses in birds. Infect. Genet. Evol. 28, 151–166. doi: 10.1016/j.meegid.2014.09.027
Boros, A., Pankovics, P., Simmonds, P., Kiss, T., Phan, T. G., Delwart, E., et al. (2018). Genomic analysis of a novel picornavirus from a migratory waterfowl, greater white-fronted goose (Anser albifrons). Arch. Virol. 163, 1087–1090. doi: 10.1007/s00705-017-3696-3
Brown, Z. P., Abaeva, I. S., De, S., Hellen, C. U. T., Pestova, T. V., and Frank, J. (2022). Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES. EMBO J. 41:e110581. doi: 10.15252/embj.2022110581
Brown, E. A., Day, S. P., Jansen, R. W., and Lemon, S. M. (1991). The 5′ nontranslated region of hepatitis a virus RNA: secondary structure and elements required for translation in vitro. J. Virol. 65, 5828–5838. doi: 10.1128/JVI.65.11.5828-5838.1991
Buechler, C. R., Bailey, A. L., Lauck, M., Heffron, A., Johnson, J. C., Campos Lawson, C., et al. (2017). Genome sequence of a novel Kunsagivirus (Picornaviridae: Kunsagivirus) from a wild baboon (Papio cynocephalus). Genome Announc. 5, e00261–e00217. doi: 10.1128/genomeA.00261-17
Chamond, N., Deforges, J., Ulryck, N., and Sargueil, B. (2014). 40S recruitment in the absence of eIF4G/4A by EMCV IRES refines the model for translation initiation on the archetype of type II IRESs. Nucleic Acids Res. 42, 10373–10384. doi: 10.1093/nar/gku720
Chard, L. S., Bordeleau, M. E., Pelletier, J., Tanaka, J., and Belsham, G. J. (2006a). Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J. Gen. Virol. 87, 927–936. doi: 10.1099/vir.0.81546-0
Chard, L. S., Kaku, Y., Jones, B., Nayak, A., and Belsham, G. J. (2006b). Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 80, 1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006
Costafreda, M. I., Perez-Rodriguez, F. J., D'Andrea, L., Guix, S., Ribes, E., Bosch, A., et al. (2014). Hepatitis a virus adaptation to cellular shutoff is driven by dynamic adjustments of codon usage and results in the selection of populations with altered capsids. J. Virol. 88, 5029–5041. doi: 10.1128/JVI.00087-14
Daijogo, S., and Semler, B. L. (2011). Mechanistic intersections between picornavirus translation and RNA replication. Adv. Virus Res. 80, 1–24. doi: 10.1016/B978-0-12-385987-7.00001-4
de Breyne, S., Yu, Y., Pestova, T. V., and Hellen, C. U. (2008). Factor requirements for translation initiation on the simian picornavirus internal ribosomal entry site. RNA 14, 367–380. doi: 10.1261/rna.696508
Drexler, J. F., Corman, V. M., Lukashev, A. N., van den Brand, J. M., Gmyl, A. P., Brunink, S., et al. (2015). Evolutionary origins of hepatitis a virus in small mammals. Proc. Natl. Acad. Sci. USA 112, 15190–15195. doi: 10.1073/pnas.1516992112
Easton, L. E., Locker, N., and Lukavsky, P. J. (2009). Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 37, 5537–5549. doi: 10.1093/nar/gkp588
El Awady, M. K., Azzazy, H. M., Fahmy, A. M., Shawky, S. M., Badreldin, N. G., Yossef, S. S., et al. (2009). Positional effect of mutations in 5'UTR of hepatitis C virus 4a on patients' response to therapy. World J. Gastroenterol. 15, 1480–1486. doi: 10.3748/wjg.15.1480
Embarc-Buh, A., Francisco-Velilla, R., and Martinez-Salas, E. (2021). RNA-binding proteins at the host-pathogen Interface targeting viral regulatory elements. Viruses 13:952. doi: 10.3390/v13060952
Fernandez-Miragall, O., and Martinez-Salas, E. (2003). Structural organization of a viral IRES depends on the integrity of the GNRA motif. RNA 9, 1333–1344. doi: 10.1261/rna.5950603
Fernández-Miragall, O., and Martínez-Salas, E. (2007). In vivo footprint of a picornavirus internal ribosome entry site reveals differences in accessibility to specific RNA structural elements. J. Gen. Virol. 88, 3053–3062. doi: 10.1099/vir.0.83218-0
Fernandez-Miragall, O., Ramos, R., Ramajo, J., and Martinez-Salas, E. (2006). Evidence of reciprocal tertiary interactions between conserved motifs involved in organizing RNA structure essential for internal initiation of translation. RNA 12, 223–234. doi: 10.1261/rna.2153206
Filbin, M. E., and Kieft, J. S. (2011). HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA 17, 1258–1273. doi: 10.1261/rna.2594011
Filbin, M. E., Vollmar, B. S., Shi, D., Gonen, T., and Kieft, J. S. (2013). HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 20, 150–158. doi: 10.1038/nsmb.2465
Firth, C., Bhat, M., Firth, M. A., Williams, S. H., Frye, M. J., Simmonds, P., et al. (2014). Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in new York City. mBio 5, e01933–e01914. doi: 10.1128/mBio.01933-14
Firth, A. E., and Brierley, I. (2012). Non-canonical translation in RNA viruses. J. Gen. Virol. 93, 1385–1409. doi: 10.1099/vir.0.042499-0
Fletcher, S. P., and Jackson, R. J. (2002). Pestivirus internal ribosome entry site (IRES) structure and function: elements in the 5′ untranslated region important for IRES function. J. Virol. 76, 5024–5033. doi: 10.1128/jvi.76.10.5024-5033.2002
Francisco-Velilla, R., Embarc-Buh, A., Abellan, S., and Martinez-Salas, E. (2022). Picornavirus translation strategies. FEBS Open Bio 12, 1125–1141. doi: 10.1002/2211-5463.13400
Friis, M. B., Rasmussen, T. B., and Belsham, G. J. (2012). Modulation of translation initiation efficiency in classical swine fever virus. J. Virol. 86, 8681–8692. doi: 10.1128/jvi.00346-12
Haji Zamani, N., Hosseini, H., Ziafati Kafi, Z., Sadri, N., Hojabr Rajeoni, A., Esmaeelzadeh Dizaji, R., et al. (2023). Whole-genome characterization of avian picornaviruses from diarrheic broiler chickens co-infected with multiple picornaviruses in Iran. Virus Genes 59, 79–90. doi: 10.1007/s11262-022-01938-0
Hanson, P. J., Zhang, H. M., Hemida, M. G., Ye, X., Qiu, Y., and Yang, D. (2012). IRES-dependent translational control during virus-induced endoplasmic reticulum stress and apoptosis. Front. Microbiol. 3:92. doi: 10.3389/fmicb.2012.00092
Hashem, Y., des Georges, A., Dhote, V., Langlois, R., Liao, H. Y., Grassucci, R. A., et al. (2013). Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503, 539–543. doi: 10.1038/nature12658
Hellen, C. U., and de Breyne, S. (2007). A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 81, 5850–5863. doi: 10.1128/JVI.02403-06
Holmes, A. C., and Semler, B. L. (2019). Picornaviruses and RNA metabolism: local and global effects of infection. J. Virol. 93, e02088–e02017. doi: 10.1128/JVI.02088-17
Honkavuori, K. S., Shivaprasad, H. L., Briese, T., Street, C., Hirschberg, D. L., Hutchison, S. K., et al. (2011). Novel picornavirus in Turkey poults with hepatitis, California, USA. Emerg. Infect. Dis. 17, 480–487. doi: 10.3201/eid1703.101410
Imai, S., Kumar, P., Hellen, C. U., D'Souza, V. M., and Wagner, G. (2016). An accurately preorganized IRES RNA structure enables eIF4G capture for initiation of viral translation. Nat. Struct. Mol. Biol. 23, 859–864. doi: 10.1038/nsmb.3280
Jackson, R. J., Hellen, C. U., and Pestova, T. V. (2010). The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. doi: 10.1038/nrm2838
Jang, S. K., Kräusslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C., and Wimmer, E. (1988). A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988
Ji, H., Fraser, C. S., Yu, Y., Leary, J., and Doudna, J. A. (2004). Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl. Acad. Sci. USA 101, 16990–16995. doi: 10.1073/pnas.0407402101
Johnson, A. G., Grosely, R., Petrov, A. N., and Puglisi, J. D. (2017). Dynamics of IRES-mediated translation. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 372:20160177. doi: 10.1098/rstb.2016.0177
Jubin, R., Vantuno, N. E., Kieft, J. S., Murray, M. G., Doudna, J. A., Lau, J. Y., et al. (2000). Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 74, 10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000
Kaku, Y., Chard, L. S., Inoue, T., and Belsham, G. J. (2002). Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 talfan. J. Virol. 76, 11721–11728. doi: 10.1128/jvi.76.22.11721-11728.2002
Kalliampakou, K. I., Psaridi-Linardaki, L., and Mavromara, P. (2002). Mutational analysis of the apical region of domain II of the HCV IRES. FEBS Lett. 511, 79–84. doi: 10.1016/s0014-5793(01)03300-2
Kapoor, A., Victoria, J., Simmonds, P., Wang, C., Shafer, R. W., Nims, R., et al. (2008). A highly divergent picornavirus in a marine mammal. J. Virol. 82, 311–320. doi: 10.1128/JVI.01240-07
Kieft, J. S., Zhou, K., Jubin, R., and Doudna, J. A. (2001). Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7, 194–206. doi: 10.1017/s1355838201001790
Kim, H., Aponte-Diaz, D., Sotoudegan, M. S., Shengjuler, D., Arnold, J. J., and Cameron, C. E. (2023). The enterovirus genome can be translated in an IRES-independent manner that requires the initiation factors eIF2A/eIF2D. PLoS Biol. 21:e3001693. doi: 10.1371/journal.pbio.3001693
Kim, H. R., Yoon, S. J., Lee, H. S., and Kwon, Y. K. (2015). Identification of a picornavirus from chickens with transmissible viral proventriculitis using metagenomic analysis. Arch. Virol. 160, 701–709. doi: 10.1007/s00705-014-2325-7
Kofstad, T., and Jonassen, C. M. (2011). Screening of feral and wood pigeons for viruses harbouring a conserved mobile viral element: characterization of novel Astroviruses and picornaviruses. PLoS One 6:e25964. doi: 10.1371/journal.pone.0025964
Koirala, D., Lewicka, A., Koldobskaya, Y., Huang, H., and Piccirilli, J. A. (2020). Synthetic antibody binding to a Preorganized RNA domain of hepatitis C virus internal ribosome entry site inhibits translation. ACS Chem. Biol. 15, 205–216. doi: 10.1021/acschembio.9b00785
Komar, A. A., and Hatzoglou, M. (2015). Exploring internal ribosome entry sites as therapeutic targets. Front. Oncol. 5:233. doi: 10.3389/fonc.2015.00233
Krumbholz, A., Dauber, M., Henke, A., Birch-Hirschfeld, E., Knowles, N. J., Stelzner, A., et al. (2002). Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J. Virol. 76, 5813–5821. doi: 10.1128/jvi.76.11.5813-5821.2002
Lange, J., Groth, M., Fichtner, D., Granzow, H., Keller, B., Walther, M., et al. (2014). Virus isolate from carp: genetic characterization reveals a novel picornavirus with two aphthovirus 2A-like sequences. J. Gen. Virol. 95, 80–90. doi: 10.1099/vir.0.058172-0
Lattimer, J., Stewart, H., Locker, N., Tuplin, A., Stonehouse, N. J., and Harris, M. (2019). Structure-function analysis of the equine hepacivirus 5′ untranslated region highlights the conservation of translational mechanisms across the hepaciviruses. J. Gen. Virol. 100, 1501–1514. doi: 10.1099/jgv.0.001316
Lau, S. K., Woo, P. C., Lai, K. K., Huang, Y., Yip, C. C., Shek, C. T., et al. (2011). Complete genome analysis of three novel picornaviruses from diverse bat species. J. Virol. 85, 8819–8828. doi: 10.1128/JVI.02364-10
Lau, S. K., Woo, P. C., Yip, C. C., Choi, G. K., Wu, Y., Bai, R., et al. (2012). Identification of a novel feline picornavirus from the domestic cat. J. Virol. 86, 395–405. doi: 10.1128/JVI.06253-11
Lau, S. K. P., Woo, P. C. Y., Yip, C. C. Y., Li, K. S. M., Fan, R. Y. Y., Bai, R., et al. (2014). Chickens host diverse picornaviruses originated from potential interspecies transmission with recombination. J. Gen. Virol. 95, 1929–1944. doi: 10.1099/vir.0.066597-0
Lee, K. M., Chen, C. J., and Shih, S. R. (2017). Regulation mechanisms of viral IRES-driven translation. Trends Microbiol. 25, 546–561. doi: 10.1016/j.tim.2017.01.010
Li, L., Shan, T., Wang, C., Côté, C., Kolman, J., Onions, D., et al. (2011). The fecal viral flora of California Sea lions. J. Virol. 85, 9909–9917. doi: 10.1128/jvi.05026-11
Liao, Q., Zheng, L., Yuan, Y., Shi, J., and Zhang, D. (2014). Genomic characterization of a novel picornavirus in Pekin ducks. Vet. Microbiol. 172, 78–91. doi: 10.1016/j.vetmic.2014.05.002
Liu, F., Wang, N., Huang, Y., Wang, Q., and Shan, H. (2021a). Stem II-disrupting pseudoknot does not abolish ability of Senecavirus a IRES to initiate protein expression, but inhibits recovery of virus from cDNA clone. Vet. Microbiol. 255:109024. doi: 10.1016/j.vetmic.2021.109024
Liu, F., Wang, N., Wang, Q., and Shan, H. (2021b). Motif mutations in pseudoknot stem I upstream of start codon in Senecavirus a genome: impacts on activity of viral IRES and on rescue of recombinant virus. Vet. Microbiol. 262:109223. doi: 10.1016/j.vetmic.2021.109223
Liu, F., Wang, Q., Wang, N., and Shan, H. (2021c). Impacts of single nucleotide deletions from the 3′ end of Senecavirus a 5′ untranslated region on activity of viral IRES and on rescue of recombinant virus. Virology 563, 126–133. doi: 10.1016/j.virol.2021.09.002
Liu, W., Yang, D., Sun, C., Wang, H., Zhao, B., Zhou, G., et al. (2020). hnRNP K is a novel internal ribosomal entry site-transacting factor that negatively regulates foot-and-mouth disease virus translation and replication and is antagonized by viral 3C protease. J. Virol. 94, e00803–e00820. doi: 10.1128/jvi.00803-20
Locker, N., Easton, L. E., and Lukavsky, P. J. (2007). HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26, 795–805. doi: 10.1038/sj.emboj.7601549
López de Quinto, S., and Martínez-Salas, E. (1997). Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 71, 4171–4175. doi: 10.1128/jvi.71.5.4171-4175.1997
López de Quinto, S., and Martínez-Salas, E. (2000). Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6, 1380–1392. doi: 10.1017/s1355838200000753
Lopez-Ulloa, B., Fuentes, Y., Pizarro-Ortega, M. S., and Lopez-Lastra, M. (2022). RNA-binding proteins as regulators of internal initiation of viral mRNA translation. Viruses 14:188. doi: 10.3390/v14020188
Lozano, G., and Martínez-Salas, E. (2015). Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 12, 113–120. doi: 10.1016/j.coviro.2015.04.008
Lukavsky, P. J. (2009). Structure and function of HCV IRES domains. Virus Res. 139, 166–171. doi: 10.1016/j.virusres.2008.06.004
Lukavsky, P. J., Kim, I., Otto, G. A., and Puglisi, J. D. (2003). Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 10, 1033–1038. doi: 10.1038/nsb1004
Lukavsky, P. J., Otto, G. A., Lancaster, A. M., Sarnow, P., and Puglisi, J. D. (2000). Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7, 1105–1110. doi: 10.1038/81951
Luo, X. L., Lu, S., Jin, D., Yang, J., Wu, S. S., and Xu, J. (2018). Marmota himalayana in the Qinghai-Tibetan plateau as a special host for bi-segmented and unsegmented picobirnaviruses. Emerg Microbes Infect 7:20. doi: 10.1038/s41426-018-0020-6
Mailliot, J., and Martin, F. (2018). Viral internal ribosomal entry sites: four classes for one goal. Wiley Interdiscip Rev RNA 9:e1458. doi: 10.1002/wrna.1458
Malnou, C. E., Poyry, T. A., Jackson, R. J., and Kean, K. M. (2002). Poliovirus internal ribosome entry segment structure alterations that specifically affect function in neuronal cells: molecular genetic analysis. J. Virol. 76, 10617–10626. doi: 10.1128/jvi.76.21.10617-10626.2002
Marques, R., Lacerda, R., and Romao, L. (2022). Internal ribosome entry site (IRES)-mediated translation and its potential for novel mRNA-based therapy development. Biomedicines 10:1865. doi: 10.3390/biomedicines10081865
Martinez-Salas, E., Francisco-Velilla, R., Fernandez-Chamorro, J., and Embarek, A. M. (2017). Insights into structural and mechanistic features of viral IRES elements. Front. Microbiol. 8:2629. doi: 10.3389/fmicb.2017.02629
Martinez-Salas, E., Francisco-Velilla, R., Fernandez-Chamorro, J., Lozano, G., and Diaz-Toledano, R. (2015). Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res. 206, 62–73. doi: 10.1016/j.virusres.2015.01.012
Martinez-Salas, E., Pacheco, A., Serrano, P., and Fernandez, N. (2008). New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 89, 611–626. doi: 10.1099/vir.0.83426-0
Melcher, S. E., Wilson, T. J., and Lilley, D. M. (2003). The dynamic nature of the four-way junction of the hepatitis C virus IRES. RNA 9, 809–820. doi: 10.1261/rna.5130703
Milicevic, N., Jenner, L., Myasnikov, A., Yusupov, M., and Yusupova, G. (2024). mRNA reading frame maintenance during eukaryotic ribosome translocation. Nature 625, 393–400. doi: 10.1038/s41586-023-06780-4
Ng, T. F., Mesquita, J. R., Nascimento, M. S., Kondov, N. O., Wong, W., Reuter, G., et al. (2014). Feline fecal virome reveals novel and prevalent enteric viruses. Vet. Microbiol. 171, 102–111. doi: 10.1016/j.vetmic.2014.04.005
Ng, T. F., Wellehan, J. F., Coleman, J. K., Kondov, N. O., Deng, X., Waltzek, T. B., et al. (2015). A tortoise-infecting picornavirus expands the host range of the family Picornaviridae. Arch. Virol. 160, 1319–1323. doi: 10.1007/s00705-015-2366-6
Niepmann, M., and Gerresheim, G. K. (2020). Hepatitis C virus translation regulation. Int. J. Mol. Sci. 21:2328. doi: 10.3390/ijms21072328
Niepmann, M., Shalamova, L. A., Gerresheim, G. K., and Rossbach, O. (2018). Signals involved in regulation of hepatitis C virus RNA genome translation and replication. Front. Microbiol. 9:395. doi: 10.3389/fmicb.2018.00395
Oberste, M. S., Maher, K., and Pallansch, M. A. (2003). Genomic evidence that simian virus 2 and six other simian picornaviruses represent a new genus in Picornaviridae. Virology 314, 283–293. doi: 10.1016/s0042-6822(03)00420-3
Pacheco, A., and Martinez-Salas, E. (2010). Insights into the biology of IRES elements through riboproteomic approaches. J. Biomed. Biotechnol. 2010:458927. doi: 10.1155/2010/458927
Pan, M., Yang, X., Zhou, L., Ge, X., Guo, X., Liu, J., et al. (2012). Duck hepatitis a virus possesses a distinct type IV internal ribosome entry site element of picornavirus. J. Virol. 86, 1129–1144. doi: 10.1128/JVI.00306-11
Pankovics, P., Boros, A., and Reuter, G. (2012). Novel picornavirus in domesticated common quail (Coturnix coturnix) in Hungary. Arch. Virol. 157, 525–530. doi: 10.1007/s00705-011-1192-8
Pelletier, J., and Sonenberg, N. (1988). Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. doi: 10.1038/334320a0
Perard, J., Leyrat, C., Baudin, F., Drouet, E., and Jamin, M. (2013). Structure of the full-length HCV IRES in solution. Nat. Commun. 4:1612. doi: 10.1038/ncomms2611
Phan, T. G., Vo, N. P., Boros, A., Pankovics, P., Reuter, G., Li, O. T., et al. (2013). The viruses of wild pigeon droppings. PLoS One 8:e72787. doi: 10.1371/journal.pone.0072787
Phelps, N. B., Mor, S. K., Armien, A. G., Batts, W., Goodwin, A. E., Hopper, L., et al. (2014). Isolation and molecular characterization of a novel picornavirus from baitfish in the USA. PLoS One 9:e87593. doi: 10.1371/journal.pone.0087593
Pinto, R. M., Aragones, L., Costafreda, M. I., Ribes, E., and Bosch, A. (2007). Codon usage and replicative strategies of hepatitis a virus. Virus Res. 127, 158–163. doi: 10.1016/j.virusres.2007.04.010
Pinto, R. M., Perez-Rodriguez, F. J., Costafreda, M. I., Chavarria-Miro, G., Guix, S., Ribes, E., et al. (2021). Pathogenicity and virulence of hepatitis a virus. Virulence 12, 1174–1185. doi: 10.1080/21505594.2021.1910442
Pinto, R. M., Perez-Rodriguez, F. J., D'Andrea, L., de Castellarnau, M., Guix, S., and Bosch, A. (2018). Hepatitis a virus codon usage: implications for translation kinetics and capsid folding. Cold Spring Harb. Perspect. Med. 8:a031781. doi: 10.1101/cshperspect.a031781
Pisarev, A. V., Chard, L. S., Kaku, Y., Johns, H. L., Shatsky, I. N., and Belsham, G. J. (2004). Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 78, 4487–4497. doi: 10.1128/jvi.78.9.4487-4497.2004
Psaridi, L., Georgopoulou, U., Varaklioti, A., and Mavromara, P. (1999). Mutational analysis of a conserved tetraloop in the 5′ untranslated region of hepatitis C virus identifies a novel RNA element essential for the internal ribosome entry site function. FEBS Lett. 453, 49–53. doi: 10.1016/s0014-5793(99)00662-6
Redondo, N., Sanz, M. A., Steinberger, J., Skern, T., Kusov, Y., and Carrasco, L. (2012). Translation directed by hepatitis a virus IRES in the absence of active eIF4F complex and eIF2. PLoS One 7:e52065. doi: 10.1371/journal.pone.0052065
Reuter, G., Boldizsar, A., and Pankovics, P. (2009). Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch. Virol. 154, 101–108. doi: 10.1007/s00705-008-0288-2
Rijnbrand, R., Thiviyanathan, V., Kaluarachchi, K., Lemon, S. M., and Gorenstein, D. G. (2004). Mutational and structural analysis of stem-loop IIIC of the hepatitis C virus and GB virus B internal ribosome entry sites. J. Mol. Biol. 343, 805–817. doi: 10.1016/j.jmb.2004.08.095
Robertson, M. E., Seamons, R. A., and Belsham, G. J. (1999). A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5, 1167–1179. doi: 10.1017/s1355838299990301
Sadahiro, A., Fukao, A., Kosaka, M., Funakami, Y., Takizawa, N., Takeuchi, O., et al. (2018). Translation of hepatitis a virus IRES is upregulated by a hepatic cell-specific factor. Front. Genet. 9:307. doi: 10.3389/fgene.2018.00307
Sauvage, V., Ar Gouilh, M., Cheval, J., Muth, E., Pariente, K., Burguiere, A., et al. (2012). A member of a new Picornaviridae genus is shed in pig feces. J. Virol. 86, 10036–10046. doi: 10.1128/JVI.00046-12
Serrano, P., Ramajo, J., and Martinez-Salas, E. (2009). Rescue of internal initiation of translation by RNA complementation provides evidence for a distribution of functions between individual IRES domains. Virology 388, 221–229. doi: 10.1016/j.virol.2009.03.021
Smits, S. L., Raj, V. S., Oduber, M. D., Schapendonk, C. M., Bodewes, R., Provacia, L., et al. (2013). Metagenomic analysis of the ferret fecal viral flora. PLoS One 8:e71595. doi: 10.1371/journal.pone.0071595
Son, K. Y., Kim, D. S., Kwon, J., Choi, J. S., Kang, M. I., Belsham, G. J., et al. (2014). Full-length genomic analysis of Korean porcine Sapelovirus strains. PLoS One 9:e107860. doi: 10.1371/journal.pone.0107860
Sweeney, T. R., Abaeva, I. S., Pestova, T. V., and Hellen, C. U. (2014). The mechanism of translation initiation on type 1 picornavirus IRESs. EMBO J. 33, 76–92. doi: 10.1002/embj.201386124
Sweeney, T. R., Dhote, V., Yu, Y., and Hellen, C. U. (2012). A distinct class of internal ribosomal entry site in members of the Kobuvirus and proposed Salivirus and Paraturdivirus genera of the Picornaviridae. J. Virol. 86, 1468–1486. doi: 10.1128/JVI.05862-11
Tanaka, T., Otoguro, T., Yamashita, A., Kasai, H., Fukuhara, T., Matsuura, Y., et al. (2018). Roles of the 5' untranslated region of nonprimate Hepacivirus in translation initiation and viral replication. J. Virol. 92, e01997–e01917. doi: 10.1128/JVI.01997-17
Tseng, C. H., Knowles, N. J., and Tsai, H. J. (2007). Molecular analysis of duck hepatitis virus type 1 indicates that it should be assigned to a new genus. Virus Res. 123, 190–203. doi: 10.1016/j.virusres.2006.09.007
Tseng, C. H., and Tsai, H. J. (2007). Sequence analysis of a duck picornavirus isolate indicates that it together with porcine enterovirus type 8 and simian picornavirus type 2 should be assigned to a new picornavirus genus. Virus Res. 129, 104–114. doi: 10.1016/j.virusres.2007.06.023
Wang, F., Liang, T., Liu, N., Ning, K., Yu, K., and Zhang, D. (2017). Genetic characterization of two novel megriviruses in geese. J. Gen. Virol. 98, 607–611. doi: 10.1099/jgv.0.000720
Wang, X., Liu, N., Wang, F., Ning, K., Li, Y., and Zhang, D. (2014). Genetic characterization of a novel duck-origin picornavirus with six 2A proteins. J. Gen. Virol. 95, 1289–1296. doi: 10.1099/vir.0.063313-0
Wang, N., Wang, H., Shi, J., Li, C., Liu, X., Fan, J., et al. (2021). The stem-loop I of Senecavirus a IRES is essential for cap-independent translation activity and virus recovery. Viruses 13:2159. doi: 10.3390/v13112159
Wang, Y., Yang, S., Liu, D., Zhou, C., Li, W., Lin, Y., et al. (2019). The fecal virome of red-crowned cranes. Arch. Virol. 164, 3–16. doi: 10.1007/s00705-018-4037-x
Wang, H., Yang, S., Shan, T., Wang, X., Deng, X., Delwart, E., et al. (2019a). A novel picornavirus in feces of a rainbow lorikeet (Trichoglossus moluccanus) shows a close relationship to members of the genus Avihepatovirus. Arch. Virol. 164, 1911–1914. doi: 10.1007/s00705-019-04246-5
Wang, H., Zhang, W., Yang, S., Kong, N., Yu, H., Zheng, H., et al. (2019b). Asian black bear (Ursus thibetanus) picornavirus related to seal aquamavirus a. Arch. Virol. 164, 653–656. doi: 10.1007/s00705-018-4101-6
Willcocks, M. M., Locker, N., Gomwalk, Z., Royall, E., Bakhshesh, M., Belsham, G. J., et al. (2011). Structural features of the Seneca Valley virus internal ribosome entry site (IRES) element: a picornavirus with a pestivirus-like IRES. J. Virol. 85, 4452–4461. doi: 10.1128/JVI.01107-10
Willcocks, M. M., Zaini, S., Chamond, N., Ulryck, N., Allouche, D., Rajagopalan, N., et al. (2017). Distinct roles for the IIId2 sub-domain in pestivirus and picornavirus internal ribosome entry sites. Nucleic Acids Res. 45, 13016–13028. doi: 10.1093/nar/gkx991
Wu, Z., Yang, L., Ren, X., He, G., Zhang, J., Yang, J., et al. (2016). Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 10, 609–620. doi: 10.1038/ismej.2015.138
Yamamoto, H., Collier, M., Loerke, J., Ismer, J., Schmidt, A., Hilal, T., et al. (2015). Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. EMBO J. 34, 3042–3058. doi: 10.15252/embj.201592469
Yang, Y., Rijnbrand, R., McKnight, K. L., Wimmer, E., Paul, A., Martin, A., et al. (2002). Sequence requirements for viral RNA replication and VPg uridylylation directed by the internal cis-acting replication element (cre) of human rhinovirus type 14. J. Virol. 76, 7485–7494. doi: 10.1128/jvi.76.15.7485-7494.2002
Yang, Y., and Wang, Z. (2019). IRES-mediated cap-independent translation, a path leading to hidden proteome. J. Mol. Cell Biol. 11, 911–919. doi: 10.1093/jmcb/mjz091
Yinda, C. K., Esefeld, J., Peter, H. U., Matthijnssens, J., and Zell, R. (2019). Penguin megrivirus, a novel picornavirus from an Adelie penguin (Pygoscelis adeliae). Arch. Virol. 164, 2887–2890. doi: 10.1007/s00705-019-04404-9
Yinda, C. K., Zell, R., Deboutte, W., Zeller, M., Conceicao-Neto, N., Heylen, E., et al. (2017). Highly diverse population of Picornaviridae and other members of the Picornavirales in Cameroonian fruit bats. BMC Genomics 18:249. doi: 10.1186/s12864-017-3632-7
Yokoyama, T., Machida, K., Iwasaki, W., Shigeta, T., Nishimoto, M., Takahashi, M., et al. (2019). HCV IRES captures an actively translating 80S ribosome. Mol. Cell 74, 1205–1214.e8. doi: 10.1016/j.molcel.2019.04.022
Yu, Y., Abaeva, I. S., Marintchev, A., Pestova, T. V., and Hellen, C. U. (2011a). Common conformational changes induced in type 2 picornavirus IRESs by cognate trans-acting factors. Nucleic Acids Res. 39, 4851–4865. doi: 10.1093/nar/gkr045
Yu, J. M., Li, X. Y., Ao, Y. Y., Li, L. L., Liu, N., Li, J. S., et al. (2013). Identification of a novel picornavirus in healthy piglets and seroepidemiological evidence of its presence in humans. PLoS One 8:e70137. doi: 10.1371/journal.pone.0070137
Yu, Y., Sweeney, T. R., Kafasla, P., Jackson, R. J., Pestova, T. V., and Hellen, C. U. (2011b). The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J. 30, 4423–4436. doi: 10.1038/emboj.2011.306
Zell, R. (2018). Picornaviridae-the ever-growing virus family. Arch. Virol. 163, 299–317. doi: 10.1007/s00705-017-3614-8
Zell, R., Knowles, N. J., and Simmonds, P. (2021). A proposed division of the family Picornaviridae into subfamilies based on phylogenetic relationships and functional genomic organization. Arch. Virol. 166, 2927–2935. doi: 10.1007/s00705-021-05178-9
Keywords: picornavirus, IRES, type IV IRES, stem-loop structure, ribosome, host factor, translation
Citation: Li Y, Zhang L, Wang L, Li J, Zhao Y, Liu F and Wang Q (2024) Structure and function of type IV IRES in picornaviruses: a systematic review. Front. Microbiol. 15:1415698. doi: 10.3389/fmicb.2024.1415698
Received: 11 April 2024; Accepted: 13 May 2024;
Published: 24 May 2024.
Edited by:
Jianjun Zhao, Heilongjiang Bayi Agricultural University, ChinaReviewed by:
Bo Wan, Henan Agricultural University, ChinaCopyright © 2024 Li, Zhang, Wang, Li, Zhao, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fuxiao Liu, bGF1ZGF3bkAxMjYuY29t; Qianqian Wang, d3FxMTA3MzIzQDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.