- 1State Key Laboratory of Microbial Metabolism, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
- 2Department of Respiratory and Critical Care Medicine, Shanghai Pulmonary Hospital, Shanghai, China
- 3Shanghai Public Health Clinical Center, Shanghai Institute of Infectious Diseases and Biosecurity, Fudan University, Shanghai, China
- 4Department of Laboratory Medicine, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, China
Introduction: The unique dormancy of Mycobacterium tuberculosis plays a significant role in the major clinical treatment challenge of tuberculosis, such as its long treatment cycle, antibiotic resistance, immune escape, and high latent infection rate.
Methods: To determine the function of MtrA, the only essential response regulator, one strategy was developed to establish its regulatory network according to high-quality genome-wide binding sites.
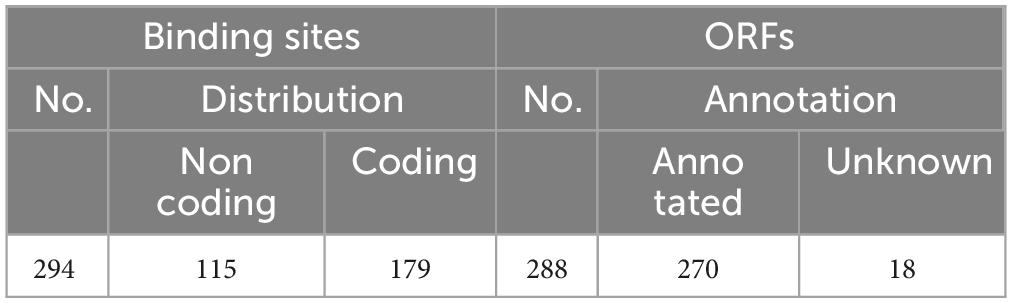
Results and discussion: The complex modulation mechanisms were implied by the strong bias distribution of MtrA binding sites in the noncoding regions, and 32.7% of the binding sites were located inside the target genes. The functions of 288 potential MtrA target genes predicted according to 294 confirmed binding sites were highly diverse, and DNA replication and damage repair, lipid metabolism, cell wall component biosynthesis, cell wall assembly, and cell division were the predominant pathways. Among the 53 pathways shared between dormancy/resuscitation and persistence, which accounted for 81.5% and 93.0% of the total number of pathways, respectively, MtrA regulatory genes were identified not only in 73.6% of their mutual pathways, but also in 75.4% of the pathways related to dormancy/resuscitation and persistence respectively. These results suggested the pivotal roles of MtrA in regulating dormancy/resuscitation and the apparent relationship between dormancy/resuscitation and persistence. Furthermore, the finding that 32.6% of the MtrA regulons were essential in vivo and/or in vitro for M. tuberculosis provided new insight into its indispensability. The findings mentioned above indicated that MtrA is a novel promising therapeutic target for tuberculosis treatment since the crucial function of MtrA may be a point of weakness for M. tuberculosis.
1 Introduction
Tuberculosis (TB) caused by Mycobacterium tuberculosis infection remains a serious worldwide health concern in terms of infection incidence and associated mortality (Kone et al., 2022), despite the significant improvement in the situation; for example, the global mortality of TB considerably reduced by 47% from 1990 to 2015 through the global efforts of many countries, including China (Wang et al., 2014; World Health Organization, 2015). However, the incidence of tuberculosis is on the rise again; for instance, its global incidence rate in 2021 increased by 3.6% compared with that in 2020 (Ding et al., 2020; World Health Organization, 2022). Although approximately 70% of new tuberculosis cases are diagnosed predominantly in developing countries, especially in regions such as Southeast Asia and Africa, which have become heavy burdens for these countries (World Health Organization, 2022), the incidence rates in developed nations are increasing significantly as well, especially for vulnerable populations, such as children and AIDS patients (Dheda et al., 2016; Tomà et al., 2017).
Since the metabolism of dormant bacilli is considerably suppressed (Shleeva et al., 2015; Jain et al., 2016), DNA replication (Gopinath et al., 2015) and cell division (Muñoz-Elías and McKinney, 2006; Iona et al., 2016) are arrested, and even cell wall composition and structures are altered (Bacon et al., 2014), dormancy/resuscitation could significantly improve the survival of bacilli under attack by the host immune system (Wayne and Sohaskey, 2001), such as macrophages (Kundu and Basu, 2021), tolerance to antibiotics (Hu et al., 2003; Nuermberger et al., 2004; Gengenbacher and Kaufmann, 2012) and recurrence (Gengenbacher and Kaufmann, 2012). Therefore, dormancy/resuscitation is the primary cause of the challenges associated with the clinical treatment of tuberculosis (Gupta et al., 2018), such as prolonged treatment cycles (Zhang, 2014), poor treatment efficacy (Wayne and Hayes, 1996), and difficulties in disease control (Gupta et al., 2018).
Among the approximately 400 dormancy/resuscitation genes identified thus far (Kundu and Basu, 2021), genes involved in DNA replication (Nandi et al., 2019); translation (Nandi et al., 2019); fatty acid and mycolic acid biosynthesis (Salina et al., 2014); primary metabolism (Betts et al., 2002); and cell wall biosynthesis (Iona et al., 2016) were downregulated, while genes involved in fatty acid degradation (Muñoz-Elías and McKinney, 2005; Del Portillo et al., 2018); the transporter systems of molybdate, phosphate and other nutrients (Betts et al., 2002; Muñoz-Elías and McKinney, 2006); iron storage proteins (Voskuil et al., 2004); mycobactin synthesis (Bacon et al., 2004); and toxin/antitoxin systems (Aguilar-Ayala et al., 2017; Hudock et al., 2017) were upregulated in dormant bacilli. Correspondingly, during resuscitation, genes involved in aerobic respiration (Salina et al., 2014), DNA replication (Nandi et al., 2019), expression regulation (Rodríguez et al., 2014; Nandi et al., 2019), cell division (Iona et al., 2016), mycolic acid and lipoarabinomannan (LAM) synthesis, phthiocerol mycocerosate (PDIM) and sulfolipid (Kundu and Basu, 2021), and ribosome biosynthesis were upregulated, in addition to five essential resuscitation promoting factors (Rpfs) (Iona et al., 2016). Then, the structure of the peptidoglycan (PG) layer was reconstituted by PG lytic transglycosylases to meet the metabolic requirements of cell growth and division (Downing et al., 2005; Tufariello et al., 2006; Kondratieva et al., 2011; Gengenbacher and Kaufmann, 2012). Therefore, neither dormancy nor resuscitation can occur on wheels without global transcriptional regulator(s).
DosR-DosS (or DevR-DevS), which is thought to be a potential regulator of dormancy/resuscitation (Leistikow et al., 2010; Schubert et al., 2015) because it responds to hypoxia (Sherman et al., 2001), NO (Voskuil et al., 2003) and CO (Kumar et al., 2008). However, DosR is dispensable for M. tuberculosis, and no more than 50 target genes have been identified thus far (Park et al., 2003); moreover, the survival rate of the DosR knockout mutant was only slightly lower than that of the wild type under hypoxic conditions (Rustad et al., 2008, 2009; Taneja et al., 2010).
Currently, dormancy and persistence, two highly similar reversible states of low metabolism and growth arrest, are mainly distinguished by growth-related features (Kell et al., 2015); for example, dormant tubercle bacilli must undergo resuscitation under specific conditions before being cultured in vitro (Kell et al., 1998), whereas persistent cells can be cultured in vitro immediately (Joshi et al., 2021). Research on their discrepancies and associations could undoubtedly make significant contributions to the understanding of M. tuberculosis (Zhang, 2004; Lipworth et al., 2016).
MtrA, the response regulator of MtrA/B, which is the only essential two-component system of M. tuberculosis (Zahrt and Deretic, 2000), was revealed to be one of five essential genes of M. tuberculosis under all in vivo (Sassetti and Rubin, 2003; Rengarajan et al., 2005) and in vitro (Griffin et al., 2011; DeJesus et al., 2017; Minato et al., 2019) conditions by the transposon mutagenesis strategy. It is highly conserved in the Mycobacterium genus, even for M. leprae, which has lost most of its sigma factors and two-component systems (Tyagi and Sharma, 2004). MtrA has been shown to be associated with cell division, cell wall synthesis, antibiotic resistance, cell morphology and osmoprotection not only in M. tuberculosis (Gorla et al., 2018; Peterson et al., 2023) but also in M. smegmatis (Gorla et al., 2018), M. avium (Cangelosi et al., 2006), M. leprae (Marques et al., 1998) and even other actinobacterial species (Brocker et al., 2011).
MtrA not only binds to oriC in M. tuberculosis (Rajagopalan et al., 2010) but also regulates essential resuscitation-promoting factors (rpfA, rpfB, and rpfC) (Sharma et al., 2015; Chatterjee et al., 2018), dnaA (DNA replication) (Li et al., 2010), and wag31 (cell division) (Gorla et al., 2018). Furthermore, the intracellular survival rate of M. tuberculosis inside THP-1 cells was significantly reduced after MtrA was overexpressed (Fol et al., 2006), and slightly knocking down the expression of MtrA could considerably sensitize M. tuberculosis to vancomycin, bedaquiline, isoniazid and rifampicin (Peterson et al., 2023) and inhibit its cell division and growth (Peterson et al., 2021, 2023). Additionally, whiB4, an oxidative stress regulator (Chatterjee et al., 2018), and fbpB (Ag85B), which are responsible for cell adhesion (Rajagopalan et al., 2010), are regulatory targets of MtrA (Chatterjee et al., 2018). The above reports strongly suggested an association between MtrA and dormancy/resuscitation.
To date, the number of M. tuberculosis MtrA target genes reported ranged from 45 (Chatterjee et al., 2018) to 279 (Minch et al., 2015; Gorla et al., 2018), and only 15 genes were supported by all three studies utilizing chromatin immunoprecipitation sequencing (ChIP-seq), the mainstream research technique currently exploring the regulons of transcription factors, due to its high false positive and negative rates and unsatisfactory reproducibility (Park, 2009; Lihu and Holban, 2015). This study explored the roles and functions of M. tuberculosis MtrA by revealing its regulons according to verified genome-wide binding sites.
2 Materials and methods
2.1 Reagents
The majority of reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2 Bacterial strains, culture and transformation
The bacterial strains used in this study, M. tuberculosis H37Rv, M. smegmatis MC2-155 and Escherichia coli Top10 and BL21 (DE3), were stocked in our laboratory.
E. coli was incubated with LB media at 37°C and 220 RPM. M. tuberculosis H37Rv and M. smegmatis MC2-155 were cultured in Middlebrook 7H9 broth supplemented with 10% OADC and 0.05% Tween 80 at 37°C and 200 RPM or cultured on 7H10 supplemented with 50 μg/ml kanamycin when needed.
The preparation and transformation of electrocompetent M. smegmatis cells were performed as previously reported (Jacobs et al., 1991).
2.3 Construction of plasmids
The eGFP fragment from the plasmid pUC-eGFP was PCR amplified using the primers EGFP-F (5′- gctaggatccATGGTGAGCAAGGGCGAGG -3′) and EGFP-R (5′- ctacgtcgacTTACTTGTACAGCTCGTCCATGCC -3′), which were subsequently ligated with pMV261 to construct pMV261-eGFP after both fragments were double digested by BamHI and SalI.
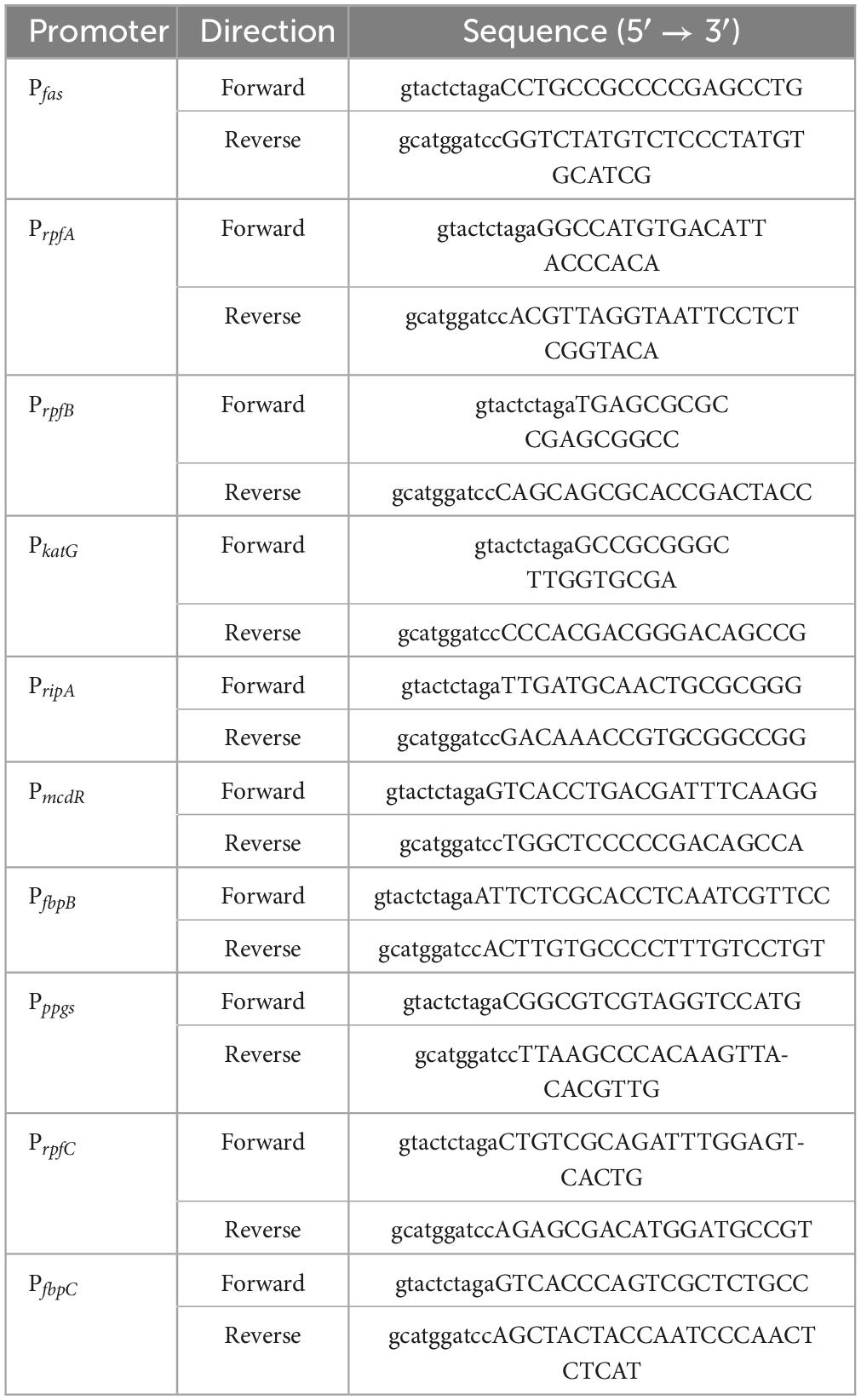
PCR-amplified promoters of MtrA target genes, such as fas, were ligated with XbaI and BamHI double-digested pMV261-eGFP to replace the hsp60 promoter (398 bp) to yield pMV261-Pfas-eGFP (Figure 1A). All of the primers used are listed in Table 1.
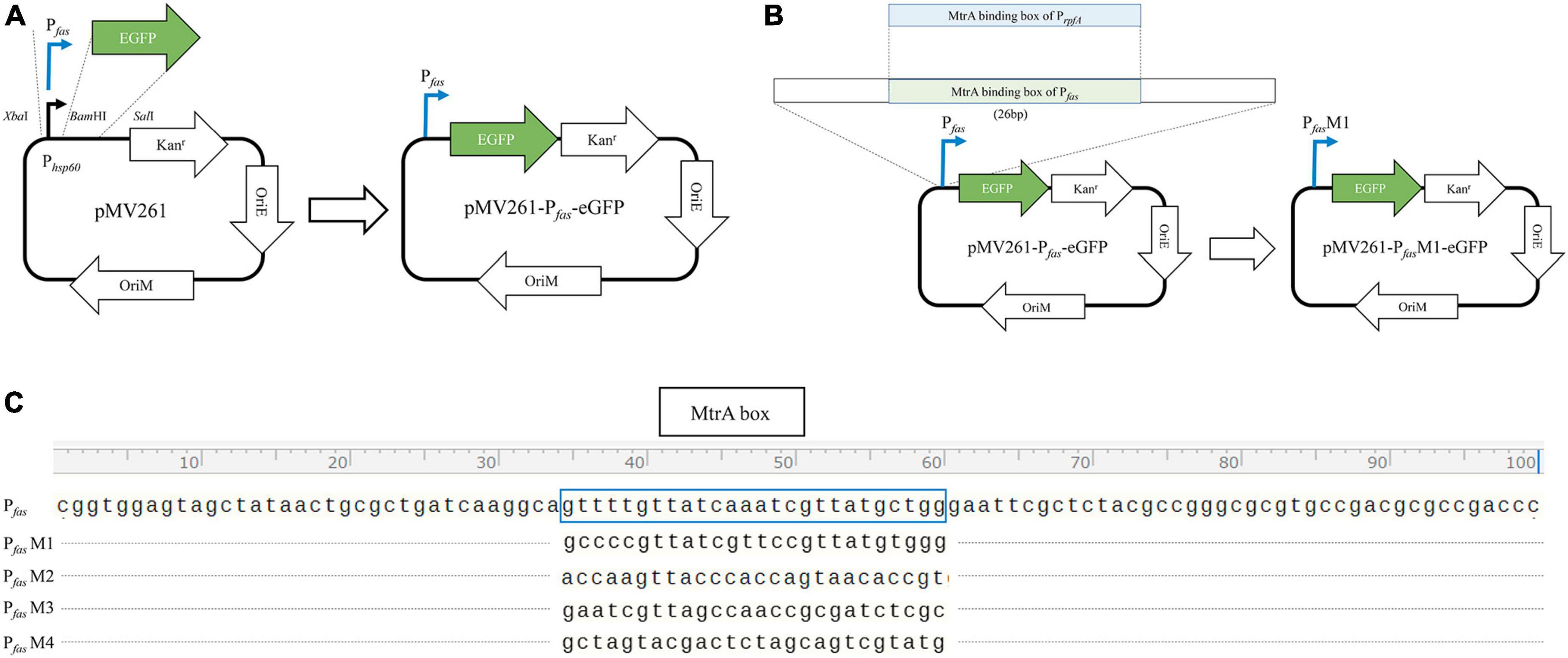
Figure 1. Diagram of the construction of the reporter plasmid. (A) Construction diagram of pMV261-pfas-eGFP. (B) Diagram of MtrA binding box replacement. (C) The MtrA binding site (boxed) of fas (KD = 4.62E-08 M) was replaced with those of rpfA (KD = 1.35E-07 M, PfasM1), fadE5 (KD = 1.42E-05 M, PfasM2), fbpC (KD = 9.85E-03 M, PfasM3) and carA (KD > 0.01 M, PfasM4).
The MtrA-binding fragment (26 bp) from rpfA, fadE5, fbpC, and carA(no binding) with a 15 nt single-stranded homologous arm on each side was recombined with PCR-amplified pMV261-Pfas-eGFP (pRPfas-F:5′- tgccttgatcagcgcagttatagc -3′, and pRPfas–R: 5′- gaattcgctctacgccgggc -3′) using Exnase II (Vazyme, Nanjing, China) to replace the binding fragment of Pfas (Figures 1B, C). All constructs were used for subsequent experiments after sequencing verification.
2.4 Verification of promoter activity in vivo
Bacterial cells of M. smegmatis confirmed transformants growing to OD600 = 0.6 were harvested by centrifugation (5000 × g, 10 min at 4°C), washed and resuspended in PBS. Then, the concentration of the cells (diluted 10-fold) was adjusted to OD600 = 0.7. The eGFP expression of cells was assayed by measuring the fluorescence using a multifunction microplate reader (Tecan Infinite F200, λex: 488 nm, λem: 507 nm) in a flat bottom 96-well black plate (Falcon).
2.5 MtrA expression, purification and binding affinity assay
MtrA expression, purification and binding affinity assays by biolayer interferometry (BLI) were conducted following previous reports (Memon et al., 2023).
2.6 Genome-wide mining for MtrA binding sites
Fifty-two potential ca. 200-bp binding fragments of MtrA retrieved according to the ChIP-seq data (Minch et al., 2015; Chatterjee et al., 2018; Gorla et al., 2018) were amplified using M. tuberculosis H37Rv genomic DNA as a template, primers were designed using Primer3 (Untergasser et al., 2012), and the sequences are listed in Supplementary Table 1.
The sequences verified by EMSA (Supplementary Figure 1) were further analyzed by MEME (Bailey and Elkan, 1994) to identify conserved motifs. Then, the minimum length of the binding fragment was determined utilizing BLI to assay the affinity of the sequentially truncated fragments (Supplementary Table 2). The position weight matrix (PWM) was established by MEME (Bailey and Elkan, 1994) by analyzing twenty-six high-affinity (KD < 5.77E-06 M) 26-bp fragments. Then the genome of M. tuberculosis H37Rv (AL123456) was comprehensively searched for potential binding sites (q-value ≤ 0.5) by FIMO (Grant et al., 2011) utilizing the established PWM. All potential binding sites identified were further verified by BLI, and all fragments were synthesized by Tsingke Biotechnology (Shanghai, China). The consensus sequence of MtrA binding fragments was generated by WebLogo (Crooks et al., 2004).
2.7 Gene annotation and gene enrichment analysis
The genes closest to binding sites were annotated by Tuberculist (Lew et al., 2011), TBDB (Galagan et al., 2010) and BioCyc (Karp et al., 2019). The protein families were categorized by COG (Galperin et al., 2021), and the orthology clusters were determined by BlastKOALA (Kanehisa et al., 2016). Gene enrichment (the proportion of MtrA target genes within specific pathway) was analyzed using KOBAS-i (Bu et al., 2021); the node size represents the P value, and the edge represents a correlation value greater than 0.35.
3 Results
3.1 Promoters were functional in vivo, and 288 target ORFs were predicted according to 294 binding sites identified
The conserved motif of MtrA binding fragments were identified using MEME by analyzing the sequences of 52 binding fragments verified by EMSA (Supplementary Figure 1) from 76 reported ones supported by three independent ChIP-seq experiments (Minch et al., 2015; Chatterjee et al., 2018; Gorla et al., 2018). After the minimum length of binding fragment of MtrA was determined to be 26 bp through assaying affinity (BLI) of MtrA binding with the sequentially truncated fragments. 294 binding sites were confirmed to be bound by MtrA (KD < 1.00E-3 M) (Supplementary Table 1) from 355 potential binding sites (Supplementary Data) predicted by FIMO utilizing the position weight matrix (PWM) established by MEME using 26 sequences of high-affinity (KD < 5.77E-06 M) 26-bp fragments from the whole genome of M. tuberculosis H37Rv.The conserved MtrA motif generated from sequences of 294 binding sites consisting of two 7-bp degenerate direct repeats of t/c-G/t-T/a-n-a-C/T-c (MtrA binding box) with 4-bp spacer (Supplementary Figure 2), was significant similar with the results based on DNaseI footprinting of oriC and PfbpB (Rajagopalan et al., 2010) and ChIP-seq analysis (Minch et al., 2015; Gorla et al., 2018) with some differences in the abundance of bases at specific locations, for example, the proportion of 7th T and 9th A in the fragment was significantly higher than that of reports (Rajagopalan et al., 2010; Gorla et al., 2018).
As shown in Figure 2, 10 randomly selected promoters of MtrA target genes could drive the expression of the reporter gene individually (Figure 2A); furthermore, the sequence of the 26-bp fragment of the MtrA binding site significantly influenced the expression of the reporter gene (Figure 2B). According to 294 binding sites confirmed, 288 ORFs were assigned as potential MtrA target genes, accounting for 7.2% of the total number of M. tuberculosis H37Rv ORFs. A total of 39.1% of the binding sites were found in noncoding regions, including oriC, and six ORFs had two binding sites identified to be closest to them (Table 2, Supplementary Data).
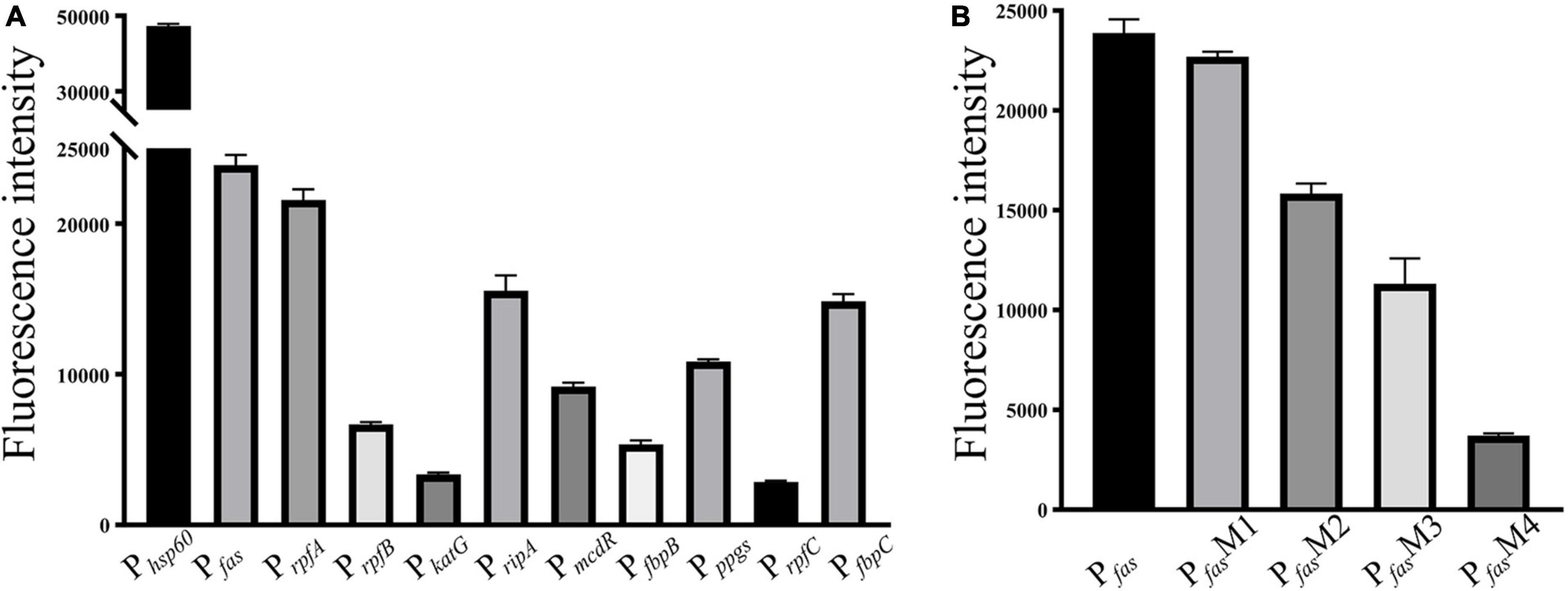
Figure 2. Promoter in vivo activity assay. (A) Selected promoters of MtrA target genes. (B) Effect of replacement of the MtrA binding box in the fas promoter.
As shown in Figure 3, the percentages of binding sites in noncoding and coding regions whose distances to their target genes were shorter than 200 bp were 83.9% and 48.6%, respectively. A total of 65.6% of the binding sites (193) were located in regions upstream of target genes, and the rest were in the target genes (Table 2, Supplementary Data). Among the 193 binding sites located in regions upstream of target genes, 57.0% were in intergenic regions, and 43.0% were in genic regions of the preceding gene.
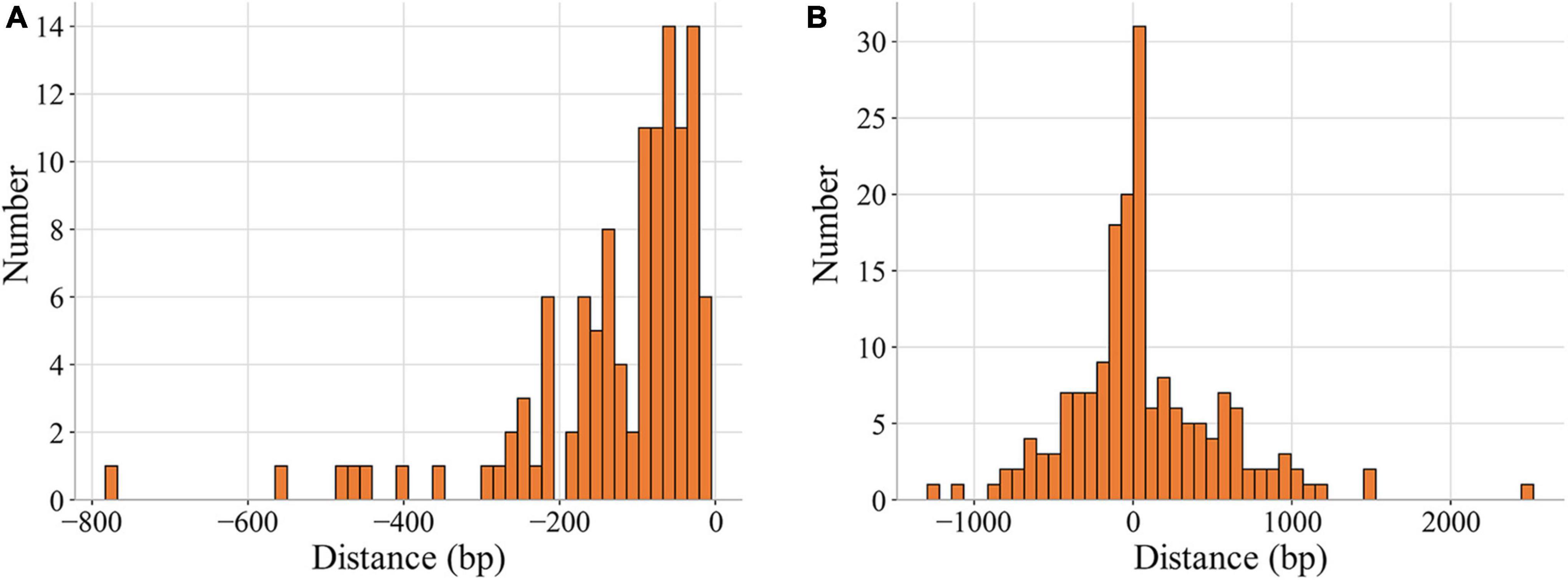
Figure 3. Distribution of the distances of MtrA binding sites to target genes. The distance represents the number of base pairs between the first nucleotide of the binding site and the start codon (0 on the X axis) of the target gene (A) noncoding region. (B) Coding region.
A total of 127 distinct KEGG ortholog groups (KOs) were identified in 270 annotated ORFs (93.8%), and most KO categories contained only 1 or 2 genes, except for K06994, which comprised 6 ORFs coding for RND superfamily drug exporters. The functions of MtrA target genes are highly diverse and include various enzymes, PE/PPE family proteins, toxin-antitoxin (TA) systems, transport/secretion systems, transcriptional regulation factors, protein kinases, etc., and even MtrA itself (Supplementary Table 3).
3.2 Genes related to cell wall biogenesis, lipid metabolism, cell division, DNA replication and damage repair were the main targets of MtrA
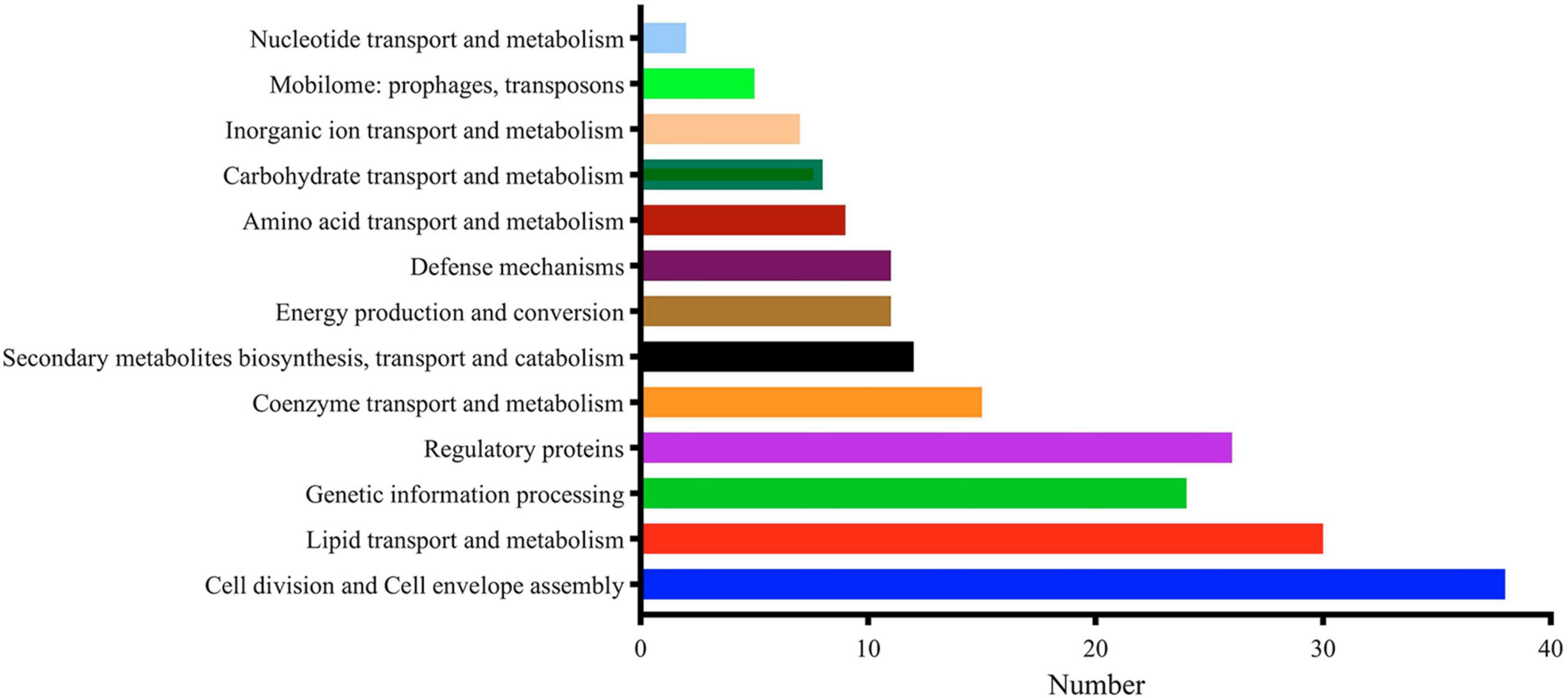
MtrA target genes involved in the initiation and elongation of chromosome replication (dnaA, dnaB, holB, dnaN), four of nine M. tuberculosis DNA repair systems, such as translesion synthesis and base excision repair (dnaE2, nth, mpg and ligD); cell wall synthesis, modification and maintenance of cell morphology (murJ, ldtA, ponA2, rpfA-E, aftC, fas, ppgS, fbpA-C, etc.); LAMs, PDIMs and other important cell wall lipid synthesis (pimE, mptC, capA, fadD28-29, ppsE, etc.); and divisome formation, cell separation, cell division regulation and other cell division processes (sepF, ftsW, wag31, cwlM, ami1, ripA) accounted for 17.01% of the total number of regulons (Supplementary Table 3). As shown in Figure 4, regulatory factors (26) were also the primary targets of MtrA, including 8 transcription factors from the WhiB (whiB1, whiB3, whiB4 and whiB7) and sigma factor families (sigB, sigD, sigE and sigH), three members of the Ser/Thr protein kinase family (pknA, pknF, and pknH) (Supplementary Table 3), cell division and cell envelope assembly (38), lipid transport and metabolism (30) and genetic information processing (24).
3.3 32.6% of MtrA target genes were essential genes
Among the MtrA regulons, 94 (32.6%) genes were essential genes reported (Sassetti and Rubin, 2003; Rengarajan et al., 2005; Griffin et al., 2011; DeJesus et al., 2017; Minato et al., 2019), including 25 in vivo and 74 in vitro essential genes (Table 3). Interestingly, among the MtrA target genesthat were essential under any one of five in vivo or in vitro experimental conditions, only 4 genes were indispensable in both murine spleens (Sassetti and Rubin, 2003) and macrophages (Rengarajan et al., 2005), accounting for 28.6% and 26.7%, respectively, of the total number of essential genes. However, 40 genes were essential for all three in vitro experimental conditions, accounting for 76.9%, 81.6% and 60.6%, respectively, of the total number of essential genes (Supplementary Table 3). however, only MtrA was indispensable under all in vivo and in vitro experimental conditions (Supplementary Table 3).
3.4 The primary carbohydrate, lipid, nucleotide and amino acid metabolic pathways were the main regulatory targets of MtrA
Among the 60 pathways in which MtrA target genes were found, primary metabolism pathways were predominant (83.3%), including 11 carbohydrate metabolism pathways, 7 lipid metabolism pathways, 2 nucleotide metabolism pathways, 11 amino acid metabolism pathways, and 3 glycan biosynthesis and metabolism pathways, followed by genetic information processing pathways (11.7%), including 4 replication and repair pathways, 2 translation pathways and 1 folding, sorting and degradation pathway. The remaining pathways were mainly associated with environmental information processing and cellular process pathways (Table 4). Furthermore, the gene enrichment ratios of the LAM biosynthesis (38.5%) and arabinogalactan biosynthesis (37.5%) pathways were approximately two times greater than those of the other pathways (Figure 5).
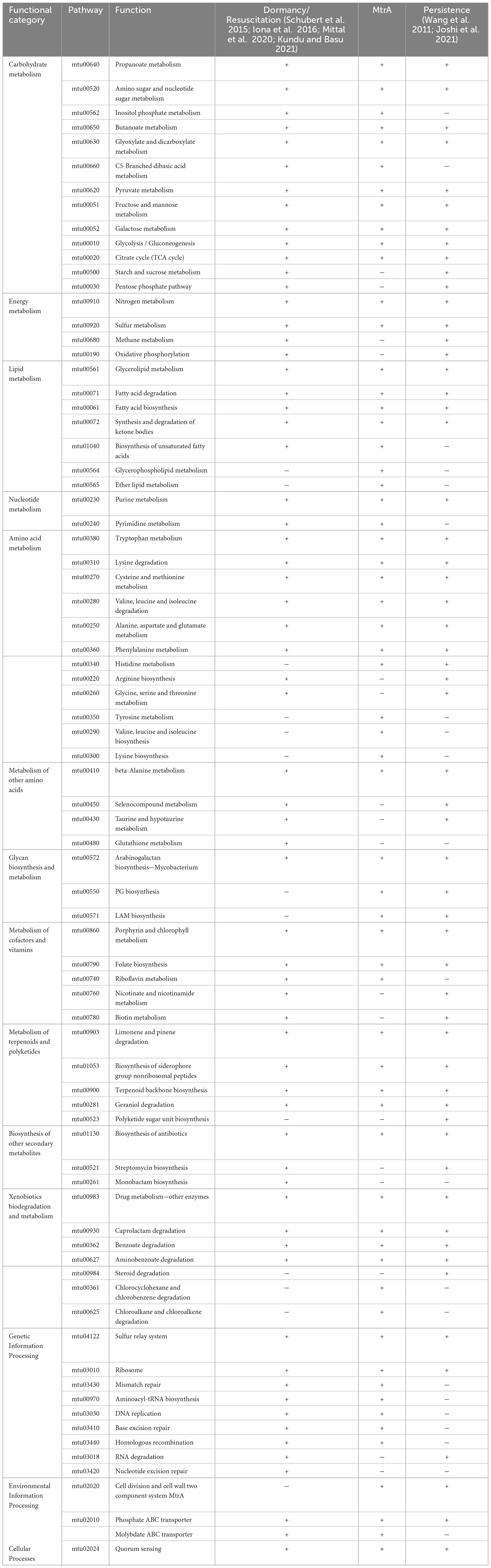
Table 4. Comparison among MtrA target pathways, dormancy/resuscitation pathways and persistence pathways.
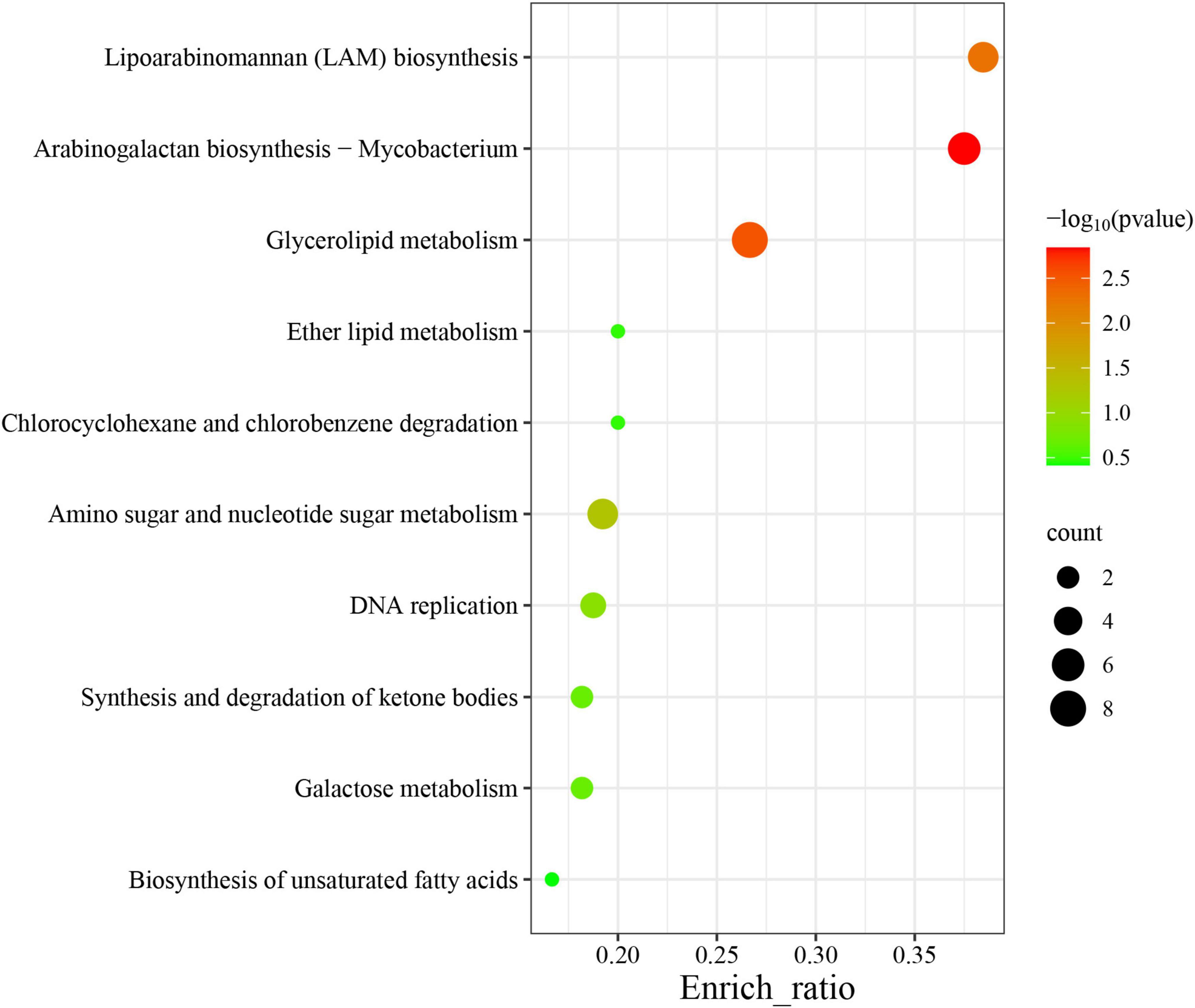
Figure 5. Top 10 enriched pathways of MtrA target genes. The abscissa label represents the enrichment ratio of pathways, which is calculated as the input gene number/background gene number.
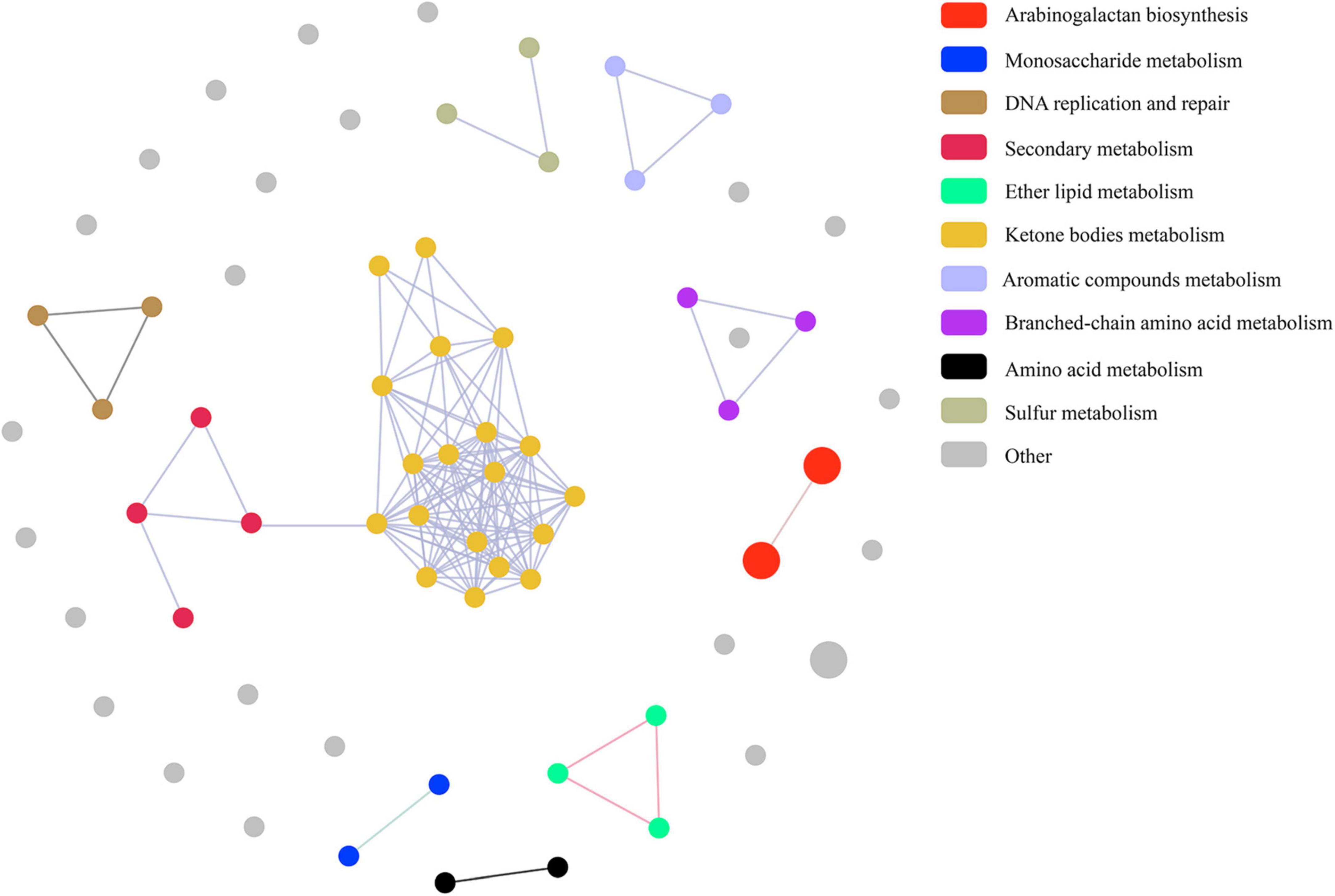
Pathway correlation analysis revealed that 37 MtrA target pathways comprised 10 distinct functional clusters, including arabinogalactan biosynthesis pathways (cluster 1), monosaccharide metabolism pathways (cluster 2), and DNA replication and repair pathways (cluster 3), and the remaining 23 pathways were orphan pathways (Figure 6). Although the majority of clusters were composed of 2-4 pathways, there were 19 pathways in cluster 6, including 5 carbon metabolism pathways and 4 amino acid metabolism pathways. Furthermore, the average number of genes per pathway in cluster 1 was 7, while that in the remaining clusters ranged from 1 to 4 (Figure 6). Interestingly, 59 MtrA regulons were reported to be relevant to dormancy/resuscitation, accounting for 13.9% of the total number of target genes of MtrA, including seven well-documented genes involved in dormancy/resuscitation, such as rpfA, rpfB, rpfC and dnaA (Supplementary Table 3).
3.5 MtrA target genes were identified in 75.4% pathways related to dormancy/resuscitation and persistence
Among 65 dormancy/resuscitation pathways predicted according to 425 dormancy/resuscitation genes reported thus far, such as carbohydrate metabolism, energy metabolism, lipid metabolism, replication and repair, etc., MtrA target genes were identified in 49 (75.4%) pathways, including 11 carbohydrate metabolism, 6 amino acid metabolism, 5 lipid metabolism, and 7 genetic information processing pathways (Table 4).
Among 57 persistence pathways predicted based on 159 reported persistence genes (Wang et al., 2011; Joshi et al., 2021), such as ripA, fbpC and other well-documented genes, MtrA target genes were identified in 43 pathways (75.4%), including 9 carbohydrate, 7 amino acid and 4 lipid metabolism pathways etc. (Table 4).
4 Discussion
Identifying target genes of prokaryotic global transcription factors that are crucial for revealing their functions remains a challenging task.
Among the 355 binding sites predicted in the genome of M. tuberculosis H37Rv using this study strategy, 82.8% (294) could be bound by MtrA with affinities (KDs) ranging from 10–8 to 10–3 M. According to the position of the binding sites in the genome, 288 target genes were predicted, including 17 out of the 18 genes regulated by MtrA reported thus far, excluding rpoB (Fol et al., 2006; Rajagopalan et al., 2010; Plocinska et al., 2012; Sharma et al., 2015; Chatterjee et al., 2018; Gorla et al., 2018).
For the MtrA target genes reported by three ChIP-seq studies (Minch et al., 2015; Chatterjee et al., 2018; Gorla et al., 2018), those supported by all three independent ChIP-seq studies were 100% consistent with our results; however, those supported by any two or one showed 65.6% and 32.9% overlap with our findings, respectively, which demonstrated the good performance of our strategy in identifying target genes of bacterial global transcription factors.
Since 7 bp motif used by Li et al. to mine for MtrA target genes was generated from one promoter (dnaA) through DNaseI footprinting and SPR analysis, only 14 of their 155 predicted MtrA target genes of M. tuberculosis were overlapped with our results, and for 264 potential MtrA target genes of M. smegmatis, the expression of less than 50% (12 out of 26 genes) of the genes assayed was significantly changed when MtrA was knocked down in M. smegmatis (Li et al., 2010). The aforementioned results further demonstrated that the attainment of successful genome-wide binding site identification for global transcription factors heavily relied on the acquisition of high-quality binding motif(s), which could be achieved only by analyzing a substantial number of promoters.
Since the coding region accounted for 90.7% of the genome of M. tuberculosis H37Rv (Lew et al., 2011), the probability of MtrA binding sites in the noncoding regions was 4.2-fold greater than the theoretical value, which indicated a strong bias in their distribution. Moreover, 32.7% of binding sites were found inside the target genes, suggesting complex regulatory mechanisms and functions since this kind of binding site can influence transcription, splicing and translation (Hua et al., 2022), and many target genes are associated with virulence, drug resistance, etc. (Galagan et al., 2013a,b; Stephanie et al., 2022).
As Supplementary Table 3 shows, the regulatory genes suggested that MtrA was involved in DNA replication (dnaA, oriC) (Ditse et al., 2017), DNA damage repair (dnaE2, mku, ligD) (Singh, 2017), cell division and cell wall assembly (wag31, ripA and fbpB) (Rajagopalan et al., 2010; Peterson et al., 2023) and even associated with virulence through stress adaptation, such as nutrient deprivation (Betts et al., 2002), oxidation (Narayan et al., 2007), acidity (Rodrigue et al., 2006), and antibiotics (Alam et al., 2009) (whiB1, whiB3, whiB4, whiB7, sigB, sigD, sigE, sigH, pknA, pknF, pknH, etc.).
A total of 32.6% (94) of the MtrA regulons were essential, and MtrA itself was essential under both in vivo and three in vitro experimental conditions (Sassetti and Rubin, 2003; Rengarajan et al., 2005; Griffin et al., 2011; DeJesus et al., 2017; Minato et al., 2019), which not only provided new explanations and insight into its indispensability but also demonstrated that the dynamic and precise regulation of MtrA was crucial in all stages of disease, from infection to survival and growth inside host cells.
The number of in vivo MtrA regulatory essential genes was significantly less than that in vitro, and the consistency of in vivo essential genes under different conditions was much lower than that in vitro (Table 3). These findings suggested that the indispensability of genes was significantly contingent upon the growing conditions and environment of tubercle bacilli and provided further evidence to support the hypothesis that M. tuberculosis evolved into a highly specific human pathogen (Gagneux, 2018; Pepperell, 2022).
The above findings not only indicated that the regulon study of global transcription factors through genome-wide binding profile investigations could comprehensively reveal their functions but also raised one question: why has M. tuberculosis MtrA evolved to regulate such large number of genes with diverse functions?
Several crucial genes involved in dormancy/resuscitation (Downing et al., 2005; Tufariello et al., 2006; Kondratieva et al., 2011) are regulated by MtrA, including resuscitation-promoting factors (rpfA, rpfB, and rpfC) (Sharma et al., 2015; Chatterjee et al., 2018), the replication initiation protein dnaA (Fol et al., 2006), the cell division protein wag31 (Plocinska et al., 2012), the mycolyltransferase fbpB (Rajagopalan et al., 2010) and transcription factors such as sigD (Gorla et al., 2018) and whiB3 (Gorla et al., 2018). After more dormancy/resuscitation-related genes were added to the regulatory list of MtrA in this study, MtrA regulatory genes were found to be involved in 75.4% of the dormancy/resuscitation pathways predicted according to the genes reported thus far (Schubert et al., 2015; Iona et al., 2016; Mittal et al., 2020; Kundu and Basu, 2021), from carbohydrate metabolism, energy metabolism, lipid metabolism, DNA replication and repair pathways to virulence pathways, such as ldtA, ldtC (Tolufashe et al., 2020), ppgS, rv3779 (Draper et al., 1997; Bhamidi et al., 2008; Skovierová et al., 2010; Wheat et al., 2015), capA, and mptC (Maeda et al., 2003; Kang et al., 2005), which respond to the long-term survival of tubercle bacilli inside macrophages by inhibiting phagolysosome maturation and infecting dendritic cells (Table 4, Supplementary Table 3).
Modulating dormancy/resuscitation is the most reasonable and plausible explanation for why MtrA regulating approximately 300 genes involved in DNA replication, DNA damage repair, cell division, stress tolerance to the primary metabolism of carbohydrates, lipids, amino acids, etc.
Furthermore, among the 53 mutual pathways between dormancy/resuscitation and persistence, which accounted for 81.5% and 93.0% of the total number of pathways, respectively, energy metabolism, lipid metabolism, amino acid metabolism, environmental information processing, and MtrA target genes were identified in 73.6% of the pathways (39) (Table 4).
As Table 4 shows, the cellular processes and representative pathways of latent tuberculosis, such as fatty acid degradation (25), glyoxylate shunting (102) and gluconeogenesis (103), were exactly identical between dormancy/resuscitation and persistence, and they were all regulated by MtrA. Approximately 80.0% of the pathways related to xenobiotic biodegradation and metabolism, terpenoid and polyketide metabolism, and lipid and carbohydrate metabolism were shared between dormancy/resuscitation and persistence, and more than 80.0% of these pathways were regulated by MtrA. The fact that dormancy/resuscitation shares the majority of its pathways mentioned above with persistence provides an explanation for why they are significantly similar in their cellular processes and physiological states (Kell et al., 2015).
However, for pathways related to genetic information processing, glycan biosynthesis and metabolism, only approximately 30% of the pathways were identical between dormancy/resuscitation and persistence (Table 4). Interestingly, the pathways crucial for resuscitation, such as DNA replication, homologous recombination repair, mismatch repair, nucleotide excision repair and RNA degradation (Gengenbacher and Kaufmann, 2012), were identified only in dormancy/resuscitation, and all of these pathways are regulated by MtrA. These results indicated that dormancy/resuscitation and persistence are unique pathways; for example, rpfA-E are essential for reconstituting the structure of the PG layer to meet the metabolic requirements of cell growth and division (Downing et al., 2005; Tufariello et al., 2006; Kondratieva et al., 2011; Gengenbacher and Kaufmann, 2012), and several genes involved in DNA repair pathways are crucial for repairing DNA damage accumulated during long periods of deep growth arrest (Salina and Makarov, 2022) when tubercle bacilli reinitiate growth during the process of resuscitation.
Additionally, MtrA plays an important role in tuberculosis (mtu05152), β-lactam resistance (mtu01501) and vancomycin resistance (mtu01502) since its binding sites were identified in the promoter region of several crucial genes in the PG biosynthesis pathway (murA, murJ, ldtA, ldtC, ponA2, PBP-lipo and dacB1, etc.) and LAM biosynthesis pathway (pimE, mptB mptC, and capA) (Supplementary Table 3).
For similar studies on M. tuberculosis MtrA employing ChIP-seq technique, 216 and 155 target genes were identified from exponential and stationary phases of M. tuberculosis by expressing phosphorylation competent MtrAY102C mutant protein (function independent of MtrB)(Gorla et al., 2018), and 93 mutual genes were shared by two phases, while only 89 (Minch et al., 2015) and 45 (Chatterjee et al., 2018) target genes were reported when wildtype MtrA was expressed. Since less than 1% purified MtrA expressed by E. coli was phosphorylated at positions like D13, D56 (Fol et al., 2006; Rajagopalan et al., 2010; Al Zayer et al., 2011), our previous work demonstrated that unphosphorylated MtrA could bind with its target DNA fragments after its dimerization was facilitated by the substrate DNA regardless of affinity (Memon et al., 2023). However, the dimerization of other members from OmpR subfamily, such as KdpE (Narayanan et al., 2014), PhoP (He et al., 2016) and PmrA (Lou et al., 2015), was facilitated by phosphorylation on regulatory domain (Gao and Stock, 2010; Barbieri et al., 2013). Interestingly, we recently found that phosphorylation could significantly increase (Rv1796, Rv1908c) or reduce (Rv2524c) the affinity of MtrA to its target fragment (data not show). The aforementioned results suggests that the affinity alterations of phosphorylated and unphosphorylated MtrA to its target fragments in fragment-specific manner are likely crucial for M. tuberculosis to adapt to diverse environmental conditions and stresses through dynamically adjusting the profile of MtrA target genes.
In summary, we should pay utmost attention to MtrA, as its potentially indispensable roles in tuberculosis and antibiotic resistance, especially in dormancy/resuscitation and persistence, indicate that it may be the Achilles heel of M. tuberculosis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
Author contributions
XF: Writing−review and editing, Writing−original draft, Investigation, Formal analysis. XW: Writing−review and editing, Supervision. AM: Investigation, Writing−review and editing. X-YF: Investigation, Writing−review and editing, Resources. QS: Writing−review and editing, Formal analysis. HC: Supervision, Writing−review and editing, Formal analysis. YY: Writing−review and editing, Supervision. ZD: Supervision, Conceptualization, Writing−review and editing. JM: Investigation, Writing−review and editing, Supervision. WM: Writing−original draft, Conceptualization, Writing−review and editing, Supervision, Resources, Funding acquisition.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of the article. This study was supported by grants from the National Key R&D Program of China (2018YFE0102400), the State Key Laboratory of Microbial Metabolism (no. 2011DA105494) and the Shanghai Science and Technology Development Foundation (AX-2212).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1415554/full#supplementary-material
Supplementary Table 1 | Sequences of primers used for EMSA.
Supplementary Table 2 | Effect of the fragment length of Rv0867c on MtrA binding affinity.
Supplementary Table 3 | Summary of MtrA regulons.
Supplementary Figure 1 | EMSA analysis of MtrA binding to selected promoters of target genes.
Supplementary Figure 2 | MtrA logo diagram of the motif.
Supplementary Data | MtrA binding sites.
References
Aguilar-Ayala, D. A., Tilleman, L., Van Nieuwerburgh, F., Deforce, D., Palomino, J. C., Vandamme, P., et al. (2017). The transcriptome of Mycobacterium tuberculosis in a lipid-rich dormancy model through RNAseq analysis. Sci. Rep. 7:17665. doi: 10.1038/s41598-017-17751-x
Al Zayer, M., Stankowska, D., Dziedzic, R., Sarva, K., Madiraju, M. V., and Rajagopalan, M. (2011). Mycobacterium tuberculosis mtrA merodiploid strains with point mutations in the signal-receiving domain of MtrA exhibit growth defects in nutrient broth. Plasmid 65, 210–218. doi: 10.1016/j.plasmid.2011.01.002
Alam, M. S., Garg, S. K., and Agrawal, P. (2009). Studies on structural and functional divergence among seven WhiB proteins of Mycobacterium tuberculosis H37Rv. Febs J. 276, 76–93. doi: 10.1111/j.1742-4658.2008.06755.x
Bacon, J., Alderwick, L. J., Allnutt, J. A., Gabasova, E., Watson, R., Hatch, K. A., et al. (2014). Non-replicating Mycobacterium tuberculosis elicits a reduced infectivity profile with corresponding modifications to the cell wall and extracellular matrix. PLoS One 9:e87329. doi: 10.1371/journal.pone.0087329
Bacon, J., James, B. W., Wernisch, L., Williams, A., Morley, K. A., Hatch, G. J., et al. (2004). The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis 84, 205–217. doi: 10.1016/j.tube.2003.12.011
Bailey, T. L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36.
Barbieri, C. M., Wu, T., and Stock, A. M. (2013). Comprehensive analysis of OmpR phosphorylation, dimerization, and DNA binding supports a canonical model for activation. J. Mol. Biol. 425, 1612–1626. doi: 10.1016/j.jmb.2013.02.003
Betts, J. C., Lukey, P. T., Robb, L. C., McAdam, R. A., and Duncan, K. (2002). Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43, 717–731. doi: 10.1046/j.1365-2958.2002.02779.x
Bhamidi, S., Scherman, M. S., Rithner, C. D., Prenni, J. E., Chatterjee, D., Khoo, K. H., et al. (2008). The identification and location of succinyl residues and the characterization of the interior arabinan region allow for a model of the complete primary structure of Mycobacterium tuberculosis mycolyl arabinogalactan. J. Biol. Chem. 283, 12992–13000. doi: 10.1074/jbc.M800222200
Brocker, M., Mack, C., and Bott, M. (2011). Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J. Bacteriol. 193, 1237–1249. doi: 10.1128/jb.01032-10
Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S., He, Z., et al. (2021). KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, W317–W325. doi: 10.1093/nar/gkab447
Cangelosi, G. A., Do, J. S., Freeman, R., Bennett, J. G., Semret, M., and Behr, M. A. (2006). The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 50, 461–468. doi: 10.1128/aac.50.2.461-468.2006
Chatterjee, A., Sharma, A. K., Mahatha, A. C., Banerjee, S. K., Kumar, M., Saha, S., et al. (2018). Global mapping of MtrA-binding sites links MtrA to regulation of its targets in Mycobacterium tuberculosis. Microbiology 164, 99–110. doi: 10.1099/mic.0.000585
Crooks, G. E., Hon, G., Chandonia, J. M., and Brenner, S. E. (2004). WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190. doi: 10.1101/gr.849004
DeJesus, M. A., Gerrick, E. R., Xu, W., Park, S. W., Long, J. E., Boutte, C. C., et al. (2017). Comprehensive essentiality analysis of the Mycobacterium tuberculosis Genome via saturating transposon mutagenesis. mBio 8:e02133-16. doi: 10.1128/mBio.02133-16
Del Portillo, P., García-Morales, L., Menéndez, M. C., Anzola, J. M., Rodríguez, J. G., Helguera-Repetto, A. C., et al. (2018). Hypoxia is not a main stress when Mycobacterium tuberculosis is in a dormancy-like long-chain fatty acid environment. Front. Cell Infect. Microbiol. 8:449. doi: 10.3389/fcimb.2018.00449
Dheda, K., Barry, C. E., and Maartens, G. (2016). Tuberculosis. Lancet 387, 1211–1226. doi: 10.1016/s0140-6736(15)00151-8
Ding, C., Wang, S., Shangguan, Y., Feng, X., Guo, W., Shi, P., et al. (2020). Epidemic trends of tuberculosis in China from 1990 to 2017: evidence from the global burden of disease study. Infect. Drug Resist. 13, 1663–1672. doi: 10.2147/idr.S249698
Ditse, Z., Lamers, M. H., and Warner, D. F. (2017). DNA replication in Mycobacterium tuberculosis. Microbiol. Spectr. 5, 581–606. doi: 10.1128/microbiolspec.TBTB2-0027-2016
Downing, K. J., Mischenko, V. V., Shleeva, M. O., Young, D. I., Young, M., Kaprelyants, A. S., et al. (2005). Mutants of Mycobacterium tuberculosis lacking three of the five like genes are defective for growth in vivo and for resuscitation in vitro. Infect. Immun. 73, 3038–3043. doi: 10.1128/iai.73.5.3038-3043.2005
Draper, P., Khoo, K. H., Chatterjee, D., Dell, A., and Morris, H. R. (1997). Galactosamine in walls of slow-growing mycobacteria. Biochem. J. 327, 519–525. doi: 10.1042/bj3270519
Fol, M., Chauhan, A., Nair, N. K., Maloney, E., Moomey, M., Jagannath, C., et al. (2006). Modulation of Mycobacterium tuberculosis proliferation by MtrA, an essential two-component response regulator. Mol. Microbiol. 60, 643–657. doi: 10.1111/j.1365-2958.2006.05137.x
Gagneux, S. (2018). Ecology and evolution of Mycobacterium tuberculosis. Nat. Rev. Microbiol. 16, 202–213. doi: 10.1038/nrmicro.2018.8
Galagan, J., Lyubetskaya, A., and Gomes, A. (2013a). ChIP-Seq and the complexity of bacterial transcriptional regulation. Curr. Top. Microbiol. Immunol. 363, 43–68. doi: 10.1007/82_2012_257
Galagan, J. E., Minch, K., Peterson, M., Lyubetskaya, A., Azizi, E., Sweet, L., et al. (2013b). The Mycobacterium tuberculosis regulatory network and hypoxia. Nature 499, 178–183. doi: 10.1038/nature12337
Galagan, J. E., Sisk, P., Stolte, C., Weiner, B., Koehrsen, M., Wymore, F., et al. (2010). TB database 2010: overview and update. Tuberculosis 90, 225–235. doi: 10.1016/j.tube.2010.03.010
Galperin, M. Y., Wolf, Y. I., Makarova, K. S., Vera Alvarez, R., Landsman, D., and Koonin, E. V. (2021). COG database update: focus on microbial diversity, model organisms, and widespread pathogens. Nucleic Acids Res. 49, D274–D281. doi: 10.1093/nar/gkaa1018
Gao, R., and Stock, A. M. (2010). Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr. Opin. Microbiol. 13, 160–167. doi: 10.1016/j.mib.2009.12.009
Gengenbacher, M., and Kaufmann, S. H. (2012). Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532. doi: 10.1111/j.1574-6976.2012.00331.x
Gopinath, V., Raghunandanan, S., Gomez, R. L., Jose, L., Surendran, A., Ramachandran, R., et al. (2015). Profiling the Proteome of Mycobacterium tuberculosis during dormancy and reactivation. Mol. Cell Proteomics 14, 2160–2176. doi: 10.1074/mcp.M115.051151
Gorla, P., Plocinska, R., Sarva, K., Satsangi, A. T., Pandeeti, E., Donnelly, R., et al. (2018). MtrA response regulator controls cell division and cell wall metabolism and affects susceptibility of mycobacteria to the first line antituberculosis drugs. Front. Microbiol. 9:2839. doi: 10.3389/fmicb.2018.02839
Grant, C. E., Bailey, T. L., and Noble, W. S. (2011). FIMO: scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018. doi: 10.1093/bioinformatics/btr064
Griffin, J. E., Gawronski, J. D., Dejesus, M. A., Ioerger, T. R., Akerley, B. J., and Sassetti, C. M. (2011). High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 7:e1002251. doi: 10.1371/journal.ppat.1002251
Gupta, V. K., Kumar, M. M., Singh, D., Bisht, D., and Sharma, S. (2018). Drug targets in dormant Mycobacterium tuberculosis: can the conquest against tuberculosis become a reality? Infect. Dis. 50, 81–94. doi: 10.1080/23744235.2017.1377346
He, X., Wang, L., and Wang, S. (2016). Structural basis of DNA sequence recognition by the response regulator PhoP in Mycobacterium tuberculosis. Sci. Rep. 6:24442. doi: 10.1038/srep24442
Hu, Y., Coates, A. R., and Mitchison, D. A. (2003). Sterilizing activities of fluoroquinolones against rifampin-tolerant populations of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47, 653–657. doi: 10.1128/aac.47.2.653-657.2003
Hua, C., Huang, J., Wang, T., Sun, Y., Liu, J., Huang, L., et al. (2022). Bacterial transcription factors bind to coding regions and regulate internal cryptic promoters. mBio 13:e0164322. doi: 10.1128/mbio.01643-22
Hudock, T. A., Foreman, T. W., Bandyopadhyay, N., Gautam, U. S., Veatch, A. V., LoBato, D. N., et al. (2017). Hypoxia sensing and persistence genes are expressed during the intragranulomatous survival of Mycobacterium tuberculosis. Am. J. Respir. Cell. Mol. Biol. 56, 637–647. doi: 10.1165/rcmb.2016-0239OC
Iona, E., Pardini, M., Mustazzolu, A., Piccaro, G., Nisini, R., Fattorini, L., et al. (2016). Mycobacterium tuberculosis gene expression at different stages of hypoxia-induced dormancy and upon resuscitation. J. Microbiol. 54, 565–572. doi: 10.1007/s12275-016-6150-4
Jacobs, W. R., Kalpana, G. V., Cirillo, J. D., Pascopella, L., Snapper, S. B., Udani, R. A., et al. (1991). Genetic systems for mycobacteria. Methods Enzymol. 204, 537–555. doi: 10.1016/0076-6879(91)04027-l
Jain, P., Weinrick, B. C., Kalivoda, E. J., Yang, H., Munsamy, V., Vilcheze, C., et al. (2016). Dual-Reporter Mycobacteriophages (Φ2DRMs) reveal preexisting Mycobacterium tuberculosis persistent cells in human sputum. mBio 7, e01023–16. doi: 10.1128/mBio.01023-16
Joshi, H., Kandari, D., and Bhatnagar, R. (2021). Insights into the molecular determinants involved in Mycobacterium tuberculosis persistence and their therapeutic implications. Virulence 12, 2721–2749. doi: 10.1080/21505594.2021.1990660
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Kang, P. B., Azad, A. K., Torrelles, J. B., Kaufman, T. M., Beharka, A., Tibesar, E., et al. (2005). The human macrophage mannose receptor directs Mycobacterium tuberculosis lipoarabinomannan-mediated phagosome biogenesis. J. Exp. Med. 202, 987–999. doi: 10.1084/jem.20051239
Karp, P. D., Billington, R., Caspi, R., Fulcher, C. A., Latendresse, M., Kothari, A., et al. (2019). The BioCyc collection of microbial genomes and metabolic pathways. Brief. Bioinform. 20, 1085–1093. doi: 10.1093/bib/bbx085
Kell, D., Potgieter, M., and Pretorius, E. (2015). Individuality, phenotypic differentiation, dormancy and ‘persistence’ in culturable bacterial systems: commonalities shared by environmental, laboratory, and clinical microbiology. F1000Res 4:179. doi: 10.12688/f1000research.6709.2
Kell, D. B., Kaprelyants, A. S., Weichart, D. H., Harwood, C. R., and Barer, M. R. (1998). Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Van Leeuwenhoek 73, 169–187. doi: 10.1023/a:1000664013047
Kondratieva, T., Rubakova, E., Kana, B. D., Biketov, S., Potapov, V., Kaprelyants, A., et al. (2011). Mycobacterium tuberculosis attenuated by multiple deletions of rpf genes effectively protects mice against TB infection. Tuberculosis 91, 219–223. doi: 10.1016/j.tube.2011.01.005
Kone, B., Somboro, A. M., Kone, M., Holl, J. L., Baya, B., Dabitao, D., et al. (2022). Molecular epidemiology and genetic diversity of Mycobacterium tuberculosis complex in referral health centers of Bamako, Mali: What is new? Int. J. Infect. Dis. 117, 204–211. doi: 10.1016/j.ijid.2022.01.061
Kumar, A., Deshane, J. S., Crossman, D. K., Bolisetty, S., Yan, B. S., Kramnik, I., et al. (2008). Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283, 18032–18039. doi: 10.1074/jbc.M802274200
Kundu, M., and Basu, J. (2021). Applications of transcriptomics and proteomics for understanding dormancy and resuscitation in Mycobacterium tuberculosis. Front. Microbiol. 12:642487. doi: 10.3389/fmicb.2021.642487
Leistikow, R. L., Morton, R. A., Bartek, I. L., Frimpong, I., Wagner, K., and Voskuil, M. I. (2010). The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192, 1662–1670. doi: 10.1128/jb.00926-09
Lew, J. M., Kapopoulou, A., Jones, L. M., and Cole, S. T. (2011). TubercuList–10 years after. Tuberculosis 91, 1–7. doi: 10.1016/j.tube.2010.09.008
Li, Y., Zeng, J., Zhang, H., and He, Z. G. (2010). The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol. 10:242. doi: 10.1186/1471-2180-10-242
Lihu, A., and Holban, Ş (2015). A review of ensemble methods for de novo motif discovery in ChIP-Seq data. Brief. Bioinform. 16, 964–973. doi: 10.1093/bib/bbv022
Lipworth, S., Hammond, R. J. H., Baron, V. O., Hu, Y., Coates, A., and Gillespie, S. H. (2016). Defining dormancy in mycobacterial disease. Tuberculosis 99, 131–142. doi: 10.1016/j.tube.2016.05.006
Lou, Y. C., Weng, T. H., Li, Y. C., Kao, Y. F., Lin, W. F., Peng, H. L., et al. (2015). Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA. Nat. Commun. 6:8838. doi: 10.1038/ncomms9838
Maeda, N., Nigou, J., Herrmann, J. L., Jackson, M., Amara, A., Lagrange, P. H., et al. (2003). The cell surface receptor DC-SIGN discriminates between Mycobacterium species through selective recognition of the mannose caps on lipoarabinomannan. J. Biol. Chem. 278, 5513–5516. doi: 10.1074/jbc.C200586200
Marques, M. A., Chitale, S., Brennan, P. J., and Pessolani, M. C. (1998). Mapping and identification of the major cell wall-associated components of Mycobacterium leprae. Infect. Immun. 66, 2625–2631. doi: 10.1128/iai.66.6.2625-2631.1998
Memon, A. A., Fu, X., Fan, X. Y., Xu, L., Xiao, J., Rahman, M. U., et al. (2023). Substrate DNA promoting binding of Mycobacterium tuberculosis MtrA by facilitating dimerization and interpretation of affinity by minor groove width. Microorganisms 11:2505. doi: 10.3390/microorganisms11102505
Minato, Y., Gohl, D. M., Thiede, J. M., Chacón, J. M., Harcombe, W. R., Maruyama, F., et al. (2019). Genomewide assessment of mycobacterium tuberculosis conditionally essential metabolic pathways. mSystems 4, e00070–19. doi: 10.1128/mSystems.00070-19
Minch, K. J., Rustad, T. R., Peterson, E. J., Winkler, J., Reiss, D. J., Ma, S., et al. (2015). The DNA-binding network of Mycobacterium tuberculosis. Nat. Commun. 6:5829. doi: 10.1038/ncomms6829
Mittal, P., Sinha, R., Kumar, A., Singh, P., Ngasainao, M. R., Singh, A., et al. (2020). Focusing on DNA repair and damage tolerance mechanisms in Mycobacterium tuberculosis: an emerging therapeutic theme. Curr. Top. Med. Chem. 20, 390–408. doi: 10.2174/1568026620666200110114322
Muñoz-Elías, E. J., and McKinney, J. D. (2005). Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat. Med. 11, 638–644. doi: 10.1038/nm1252
Muñoz-Elías, E. J., and McKinney, J. D. (2006). Carbon metabolism of intracellular bacteria. Cell Microbiol. 8, 10–22. doi: 10.1111/j.1462-5822.2005.00648.x
Nandi, M., Sikri, K., Chaudhary, N., Mande, S. C., Sharma, R. D., and Tyagi, J. S. (2019). Multiple transcription factors co-regulate the Mycobacterium tuberculosis adaptation response to vitamin C. BMC Genomics 20:887. doi: 10.1186/s12864-019-6190-3
Narayan, A., Sachdeva, P., Sharma, K., Saini, A. K., Tyagi, A. K., and Singh, Y. (2007). Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol. Genomics 29, 66–75. doi: 10.1152/physiolgenomics.00221.2006
Narayanan, A., Kumar, S., Evrard, A. N., Paul, L. N., and Yernool, D. A. (2014). An asymmetric heterodomain interface stabilizes a response regulator-DNA complex. Nat. Commun. 5:3282. doi: 10.1038/ncomms4282
Nuermberger, E., Bishai, W. R., and Grosset, J. H. (2004). Latent tuberculosis infection. Semin. Respir. Crit. Care Med. 25, 317–336. doi: 10.1055/s-2004-829504
Park, H. D., Guinn, K. M., Harrell, M. I., Liao, R., Voskuil, M. I., Tompa, M., et al. (2003). Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843. doi: 10.1046/j.1365-2958.2003.03474.x
Park, P. J. (2009). ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 10, 669–680. doi: 10.1038/nrg2641
Pepperell, C. S. (2022). Evolution of tuberculosis pathogenesis. Annu. Rev. Microbiol. 76, 661–680. doi: 10.1146/annurev-micro-121321-093031
Peterson, E. J. R., Brooks, A. N., Reiss, D. J., Kaur, A., and Baliga, N. S. (2021). MtrA Regulation of Essential Peptidoglycan Cleavage in Mycobacterium tuberculosis during Infection. New York: Cold Spring Harbor Laboratory.
Peterson, E. J. R., Brooks, A. N., Reiss, D. J., Kaur, A., Do, J., Pan, M., et al. (2023). MtrA modulates Mycobacterium tuberculosis cell division in host microenvironments to mediate intrinsic resistance and drug tolerance. Cell Rep. 42:112875. doi: 10.1016/j.celrep.2023.112875
Plocinska, R., Purushotham, G., Sarva, K., Vadrevu, I. S., Pandeeti, E. V., Arora, N., et al. (2012). Septal localization of the Mycobacterium tuberculosis MtrB sensor kinase promotes MtrA regulon expression. J. Biol. Chem. 287, 23887–23899. doi: 10.1074/jbc.M112.346544
Rajagopalan, M., Dziedzic, R., Al Zayer, M., Stankowska, D., Ouimet, M. C., Bastedo, D. P., et al. (2010). Mycobacterium tuberculosis origin of replication and the promoter for immunodominant secreted antigen 85B are the targets of MtrA, the essential response regulator. J. Biol. Chem. 285, 15816–15827. doi: 10.1074/jbc.M109.040097
Rengarajan, J., Bloom, B. R., and Rubin, E. J. (2005). Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc. Natl. Acad. Sci. U S A. 102, 8327–8332. doi: 10.1073/pnas.0503272102
Rodrigue, S., Provvedi, R., Jacques, P. E., Gaudreau, L., and Manganelli, R. (2006). The sigma factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 30, 926–941. doi: 10.1111/j.1574-6976.2006.00040.x
Rodríguez, J. G., Hernández, A. C., Helguera-Repetto, C., Aguilar Ayala, D., Guadarrama-Medina, R., Anzóla, J. M., et al. (2014). Global adaptation to a lipid environment triggers the dormancy-related phenotype of Mycobacterium tuberculosis. mBio 5, e01125–14. doi: 10.1128/mBio.01125-14
Rustad, T. R., Harrell, M. I., Liao, R., and Sherman, D. R. (2008). The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502. doi: 10.1371/journal.pone.0001502
Rustad, T. R., Sherrid, A. M., Minch, K. J., and Sherman, D. R. (2009). Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 11, 1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x
Salina, E. G., and Makarov, V. (2022). Mycobacterium tuberculosis dormancy: how to fight a hidden danger. Microorganisms 10:2334. doi: 10.3390/microorganisms10122334
Salina, E. G., Waddell, S. J., Hoffmann, N., Rosenkrands, I., Butcher, P. D., and Kaprelyants, A. S. (2014). Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biol. 4:140106. doi: 10.1098/rsob.140106
Sassetti, C. M., and Rubin, E. J. (2003). Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U S A. 100, 12989–12994. doi: 10.1073/pnas.2134250100
Schubert, O. T., Ludwig, C., Kogadeeva, M., Zimmermann, M., Rosenberger, G., Gengenbacher, M., et al. (2015). Absolute proteome composition and dynamics during dormancy and resuscitation of Mycobacterium tuberculosis. Cell Host Microbe 18, 96–108. doi: 10.1016/j.chom.2015.06.001
Sharma, A. K., Chatterjee, A., Gupta, S., Banerjee, R., Mandal, S., Mukhopadhyay, J., et al. (2015). MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology 161, 1271–1281. doi: 10.1099/mic.0.000087
Sherman, D. R., Voskuil, M., Schnappinger, D., Liao, R., Harrell, M. I., and Schoolnik, G. K. (2001). Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc. Natl. Acad. Sci. U S A. 98, 7534–7539. doi: 10.1073/pnas.121172498
Shleeva, M., Kondratieva, T., Rubakova, E., Vostroknutova, G., Kaprelyants, A., and Apt, A. (2015). Reactivation of dormant “non-culturable” Mycobacterium tuberculosis developed in vitro after injection in mice: both the dormancy depth and host genetics influence the outcome. Microb. Pathog. 78, 63–66. doi: 10.1016/j.micpath.2014.11.016
Singh, A. (2017). Guardians of the mycobacterial genome: a review on DNA repair systems in Mycobacterium tuberculosis. Microbiology 163, 1740–1758. doi: 10.1099/mic.0.000578
Skovierová, H., Larrouy-Maumus, G., Pham, H., Belanová, M., Barilone, N., Dasgupta, A., et al. (2010). Biosynthetic origin of the galactosamine substituent of Arabinogalactan in Mycobacterium tuberculosis. J. Biol. Chem. 285, 41348–41355. doi: 10.1074/jbc.M110.188110
Stephanie, F., Tambunan, U. S. F., and Siahaan, T. J. (2022). M. tuberculosis transcription machinery: a review on the mycobacterial RNA polymerase and drug discovery efforts. Life 12:1774. doi: 10.3390/life12111774
Taneja, N. K., Dhingra, S., Mittal, A., Naresh, M., and Tyagi, J. S. (2010). Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5:e10860. doi: 10.1371/journal.pone.0010860
Tolufashe, G. F., Sabe, V. T., Ibeji, C. U., Ntombela, T., Govender, T., Maguire, G. E. M., et al. (2020). Structure and function of L,D- and D,D-transpeptidase family enzymes from Mycobacterium tuberculosis. Curr. Med. Chem. 27, 3250–3267. doi: 10.2174/0929867326666181203150231
Tomà, P., Lancella, L., Menchini, L., Lombardi, R., Secinaro, A., and Villani, A. (2017). Radiological patterns of childhood thoracic tuberculosis in a developed country: a single institution’s experience on 217/255 cases. La Radiol. Med. 122, 22–34. doi: 10.1007/s11547-016-0683-9
Tufariello, J. M., Mi, K., Xu, J., Manabe, Y. C., Kesavan, A. K., Drumm, J., et al. (2006). Deletion of the Mycobacterium tuberculosis resuscitation-promoting factor Rv1009 gene results in delayed reactivation from chronic tuberculosis. Infect. Immun. 74, 2985–2995. doi: 10.1128/iai.74.5.2985-2995.2006
Tyagi, J. S., and Sharma, D. (2004). Signal transduction systems of mycobacteria with special reference to M. tuberculosis. Curr. Sci. 86, 93–102.
Untergasser, A., Cutcutache, I., Koressaar, T., Ye, J., Faircloth, B. C., Remm, M., et al. (2012). Primer3–new capabilities and interfaces. Nucleic Acids Res. 40:e115. doi: 10.1093/nar/gks596
Voskuil, M. I., Schnappinger, D., Visconti, K. C., Harrell, M. I., Dolganov, G. M., Sherman, D. R., et al. (2003). Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713. doi: 10.1084/jem.20030205
Voskuil, M. I., Visconti, K. C., and Schoolnik, G. K. (2004). Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis 84, 218–227. doi: 10.1016/j.tube.2004.02.003
Wang, L., Zhang, H., Ruan, Y., Chin, D. P., Xia, Y., Cheng, S., et al. (2014). Tuberculosis prevalence in China, 1990-2010; a longitudinal analysis of national survey data. Lancet 383, 2057–2064. doi: 10.1016/s0140-6736(13)62639-2
Wang, X., Wang, H., and Xie, J. (2011). Genes and regulatory networks involved in persistence of Mycobacterium tuberculosis. Sci. China Life Sci. 54, 300–310. doi: 10.1007/s11427-011-4134-5
Wayne, L. G., and Hayes, L. G. (1996). An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64, 2062–2069. doi: 10.1128/iai.64.6.2062-2069.1996
Wayne, L. G., and Sohaskey, C. D. (2001). Nonreplicating persistence of mycobacterium tuberculosis. Annu. Rev. Microbiol. 55, 139–163. doi: 10.1146/annurev.micro.55.1.139
Wheat, W. H., Dhouib, R., Angala, S. K., Larrouy-Maumus, G., Dobos, K., Nigou, J., et al. (2015). The presence of a galactosamine substituent on the arabinogalactan of Mycobacterium tuberculosis abrogates full maturation of human peripheral blood monocyte-derived dendritic cells and increases secretion of IL-10. Tuberculosis 95, 476–489. doi: 10.1016/j.tube.2015.04.002
World Health Organization (2015). Global Tuberculosis Report 2015. Geneva: World Health Organization.
World Health Organization (2022). Global Tuberculosis Report 2022. Geneva: World Health Organization.
Zahrt, T. C., and Deretic, V. (2000). An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182, 3832–3838. doi: 10.1128/jb.182.13.3832-3838.2000
Zhang, Y. (2004). Persistent and dormant tubercle bacilli and latent tuberculosis. Front. Biosci. 9:1136–1156. doi: 10.2741/1291
Keywords: Mycobacterium tuberculosis, MtrA, regulatory network, dormancy, resuscitation, persistence
Citation: Fu X, Wan X, Memon AA, Fan X-Y, Sun Q, Chen H, Yao Y, Deng Z, Ma J and Ma W (2024) Regulatory role of Mycobacterium tuberculosis MtrA on dormancy/resuscitation revealed by a novel target gene-mining strategy. Front. Microbiol. 15:1415554. doi: 10.3389/fmicb.2024.1415554
Received: 10 April 2024; Accepted: 28 May 2024;
Published: 17 June 2024.
Edited by:
Amanda J. Martinot, Tufts University, United StatesReviewed by:
Thomas R. Ioerger, Texas A and M University, United StatesKenan C. Murphy, University of Massachusetts Medical School, United States
Copyright © 2024 Fu, Wan, Memon, Fan, Sun, Chen, Yao, Deng, Ma and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ma, d21hQHNqdHUuZWR1LmNu; Jian Ma, bWFqaWFuXzcyM0BzaW5hLmNvbQ==
 Xiang Fu1
Xiang Fu1 Xiao-Yong Fan
Xiao-Yong Fan Haifeng Chen
Haifeng Chen Yufeng Yao
Yufeng Yao Wei Ma
Wei Ma