
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
PERSPECTIVE article
Front. Microbiol. , 12 June 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1415100
This article is part of the Research Topic tRNA and Protein Synthesis in Microorganisms View all 10 articles
Interpretation of the genetic code from triplets of nucleotides to amino acids is fundamental to life. This interpretation is achieved by cellular tRNAs, each reading a triplet codon through its complementary anticodon (positions 34–36) while delivering the amino acid charged to its 3′-end. This amino acid is then incorporated into the growing polypeptide chain during protein synthesis on the ribosome. The quality and versatility of the interpretation is ensured not only by the codon-anticodon pairing, but also by the post-transcriptional modifications at positions 34 and 37 of each tRNA, corresponding to the wobble nucleotide at the first position of the anticodon and the nucleotide on the 3′-side of the anticodon, respectively. How each codon is read by the matching anticodon, and which modifications are required, cannot be readily predicted from the codon-anticodon pairing alone. Here we provide an easily accessible modification pattern that is integrated into the genetic code table. We focus on the Gram-negative bacterium Escherichia coli as a model, which is one of the few organisms whose entire set of tRNA modifications and modification genes is identified and mapped. This work provides an important reference tool that will facilitate research in protein synthesis, which is at the core of the cellular life.
The genetic code is universally conserved, with minor exceptions, as the central mechanism that translates genetic information stored in nucleic acids to amino acid sequences of proteins. This code consists of 64 triplets of nucleotides, of which 61 are sense codons, each specifying the insertion of an amino acid to protein synthesis, while the remaining 3 are non-sense codons, each signaling termination of protein synthesis. Importantly, interpretation of the genetic code is entirely dependent on cellular tRNAs providing matching anticodons to each codon, while delivering the specified amino acid as the building block to protein synthesis. The speed and quality of interpreting each codon is essential for life. This demand is addressed by post-transcriptional modifications in tRNA molecules, most notably at position 34 and at position 37 (Figure 1A). The presence of tRNA modifications at positions 34 and 37 also controls and regulates the codon-anticodon wobble pairing (Agris et al., 2018), thus solving the conundrum that organisms across the tree of life usually have a fewer number of tRNA anticodons relative to 64 codons of the genetic code. Notably, while all native tRNA molecules contain an extensive set of modifications, those at positions 34 and 37 encompass the greatest structural diversity. Some of these modifications are also functionally coupled to tRNA aminoacylation and to maintenance of the translational reading frame (Bjork et al., 1987; Muramatsu et al., 1988; Gamper et al., 2015; Thiaville et al., 2016; Gamper et al., 2021a,b; Masuda et al., 2022). Additionally, tRNA modifications at these two positions can be inducible, responding to cellular stress to facilitate translation of codon-biased stress genes (Endres et al., 2015). Indeed, dysregulation of tRNA modifications at these two positions is linked to various pathologies (Suzuki, 2021). However, despite the importance, how each anticodon is associated with a modification at positions 34 and/or 37 is not obvious from the genetic code and is not readily predictable, as subtle changes of the modification can alter the codon-anticodon pairing. This lack of information has limited researchers from a better understanding of the genetic code. Here, we address this critical gap, by revealing a pattern of tRNA modifications that associates the modifications at positions 34 and 37 of each tRNA to translation of its codon(s). We use this pattern to convey the information of modifications in a systematic organization that can be referenced by users. Notably, while the genetic code is conserved in evolution, the pattern of modifications is specific to each organism, depending on its GC content, codon usage, and dynamics of the tRNA repertoires (Diwan and Agashe, 2018).
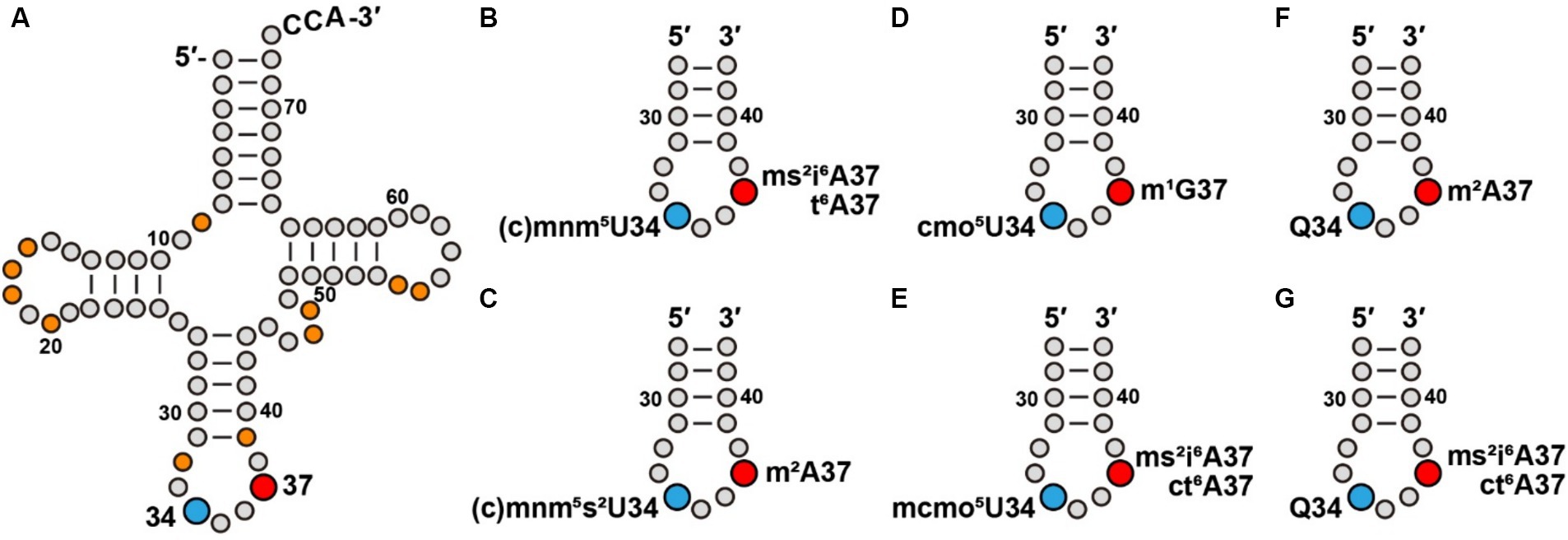
Figure 1. Modifications at tRNA positions 34 and 37 contribute to a modification pattern. (A) Modifications at positions 34 and 37 of E. coli tRNAs, in blue and red, respectively, in the cloverleaf structure that also include other commonly present modifications (in orange), such as s4U8, D16 (D: dihydrouridine), D17, Gm18, D20, Cm/Um32, ψ39 (ψ: pseudouridine), m7G46, acp3U47 (acp3U, 3-amino-3-carboxy-propyl-uridine), m5U54, and ψ55, most of which contribute to stabilization of the tRNA structure. (B) Association of cmnm5U34 and mnm5U34 with ms2i6A37 or t6A37 series in an ASL. (C) Association of cmnm5s2U34 and mnm5s2U34 with m2A37 in an ASL. (D) Association of cmo5U34 with m1G37 in an ASL. (E) Association of mcmo5U34 with ms2i6A37 or with ct6A37 in an ASL. (F) Association of Q34 with m2A37 in an ASL. (G) Association of Q34 with ms2i6A37 or with ct6A37 in an ASL.
We provide the pattern of tRNA modifications for E. coli strain MG1655 – one of the few organisms whose entire set of tRNA modifications is known and confirmed by identification of the associated modification enzyme(s) of each (de Crecy-Lagard and Jaroch, 2021). While some modifications are synthesized by one enzyme, others are synthesized in a pathway involving several enzymes. An example of the former is the N1-methylation of guanosine at position 37 (m1G37) by the TrmD tRNA methyltransferase (Hou et al., 2017), while an example of the latter is cmo5U34 (see the abbreviation below), which requires four enzymes catalyzing the successive conversion of U34 to ho5U34 and then to cmo5U34 (Sakai et al., 2019). The development of the pattern of modifications of E. coli is an important step to better understand the relationship between each codon and its anticodons for accurate interpretation of the genetic code. It also lays the framework for developing the pattern for related organisms and even higher eukaryotes, as the chemical structure of some modifications at position 34 or 37 are conserved (e.g., I34, m1G37, and ms2i6A37) or were evolved to a similar structure by addition of a new chemical moiety (e.g., from t6A37 to ms2t6A37).
Importantly, given the power of the genetic tools available to E. coli, investigation of each modification in the pattern is feasible, even if it is essential for cell viability (Masuda et al., 2019; Masuda and Hou, 2023). Additionally, E. coli is the single model organism that has received substantial advancement of bioengineering to expand the genetic code. Notably, while the genetic code provides the basic framework to synthesize proteins with the canonical 20 amino acids and selenocysteine (see below), efforts are being made to expand this code to incorporate a non-canonical amino acid at a site-specific position to diversify the structure and function of proteins, thus paving the way to protein-based medicines and industrial biocatalysts. Active efforts are aimed to engineer a tRNA to read a non-sense codon or a quadruplet codon for site-specific delivery of a non-canonical amino acid to protein synthesis (Shandell et al., 2021; Gamper et al., 2022; Kim et al., 2024). These efforts inevitably must consider tRNA modifications at positions 34 and 37 to control the quality of the engineered codon-anticodon pairing. For example, in a genetically isolated derivative of E. coli Pro(GGG) tRNA (see abbreviations below) that would be a strong candidate for reading of a quadruplet codon motif such as CCC-C, we show that the presence of m1G37, which disrupts the Watson-Crick (W-C) base pairing of the guanosine, readily compromises this activity (Gamper et al., 2015, 2021a,b), pointing to the need to better engineer this tRNA for genetic code expansion. Thus, the tRNA modification pattern as shown here will serve as a critical reference for consideration to push the frontier forward of genetic code expansion.
To date, about 150 different types of modifications have been identified in all classes of RNAs, including tRNAs, mRNA, rRNAs, and other RNAs (Boccaletto et al., 2018). Focusing on tRNAs specifically, we analyzed the entire set of 47 unique sequences of E. coli MG1655 and compiled all the modifications found at positions 34 and 37 (Bjork and Hagervall, 2014), followed by literature searches to update the information (Keseler et al., 2021). We developed the tRNA modification pattern by showing each codon of the genetic code next to its anticodon(s), and by showing each anticodon with the modifications at positions 34 and 37. Modifications at position 34 include: mnm5U (5-methyl-amino-methyl-uridine), cmnm5Um (5-carboxy-methyl-aminomethyl-2′-O-methyl uridine), Cm (2′-O-methyl cytidine), cmo5U (uridine 5-oxyacetic acid), ac4C (N4-acetyl-cytidine), mcmo5U (5-(methoxy)-carbonyl methoxy-uridine), mcmo5Um (5-(methoxy)-carbonyl methoxy-2′-O-methyl-uridine), cmnm5s2U (5-carboxy-methyl-amino-methyl-2-thiouridine), mnm5s2U (5-methyl-amino-methyl-2-thiouridine), I (inosine), L (lysidine), Q (queuosine), and gluQ (glutamyl-queuosine). Conversely, modifications at position 37 include: ms2i6A (2-methylthio-N6-isopentenyl-adenosine), i6A (N6-isopentenyl-adenosine), m1G (N1-methyl-guanosine), m2A (2-methyl-adenosine), ct6A (cyclic N6-threonyl-carbamoyl-adenosine), m6t6A (N6-methyl-N6-threonyl-carbamoyl-adenosine), and m6A (N6-methyl-adenosine). Below we describe each modified nucleotide by the abbreviated form and follow it with the number 34 or 37, indicating its position in the tRNA sequence framework. We also cite each tRNA by the three-letter code of the amino-acid identity, followed by the modified anticodon in parentheses. Thus, Arg(ICG) means that it is tRNAArg with the anticodon that is modified from ACG to ICG in the native tRNA.
We integrate the tRNA modification pattern into the genetic code table to provide an easy access to the association of each codon with the matching anticodon(s) (Figure 2). At each codon, we show directly in the table the modification at position 34 of the anticodon, which is responsible for wobble base pairing with the codon. We include the tRNA gene that expresses each anticodon in parentheses to facilitate cross-referencing with published work. Although specific patterns of modifications at position 34 were reported previously for all three domains of life (Grosjean et al., 2010), we describe below a few trends for bacterial tRNAs.
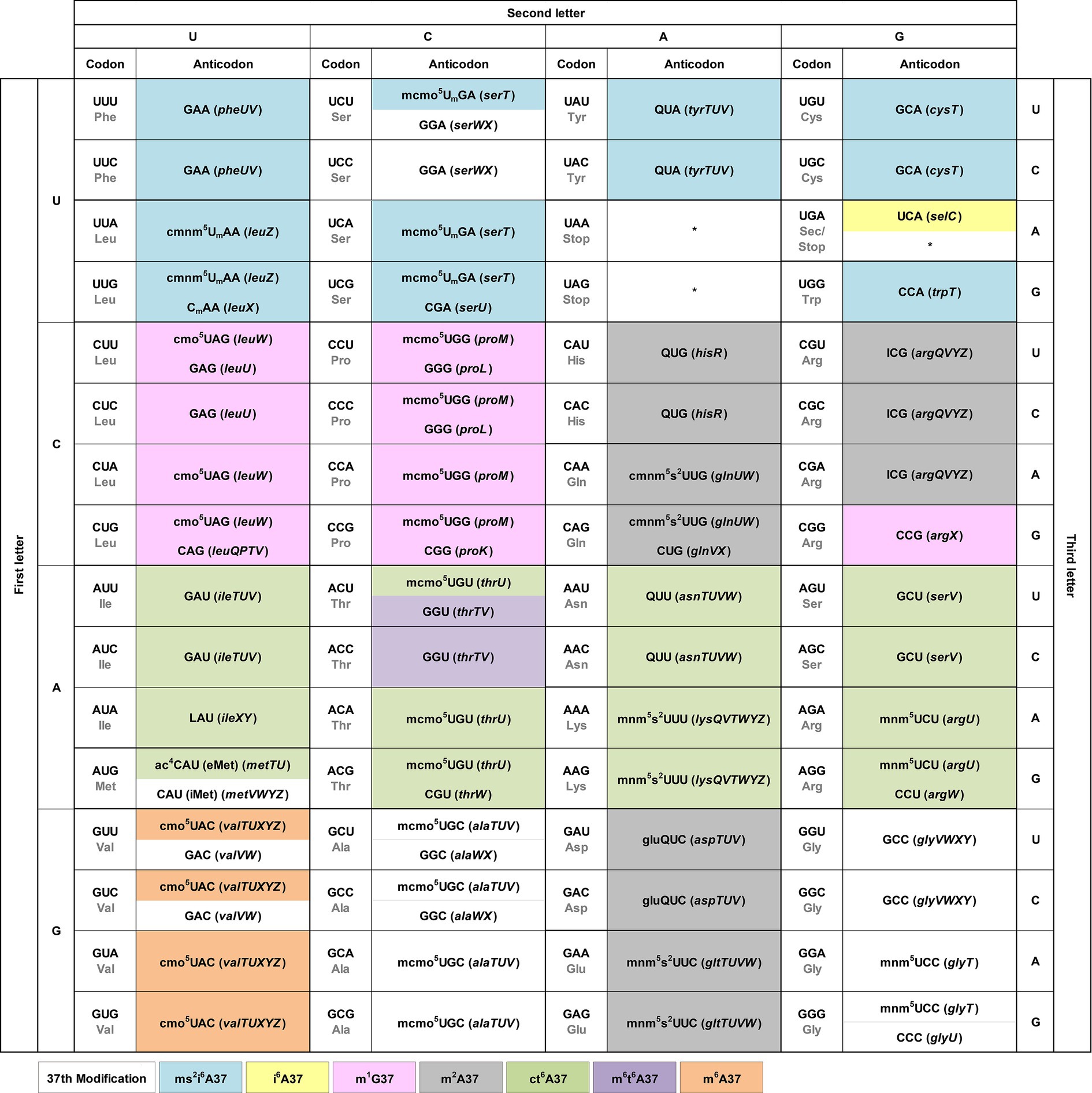
Figure 2. The modification pattern of E. coli tRNAs. Each sense codon of the genetic code is shown with its anticodon(s) that can form W-C or wobble base pairs. Each anticodon is shown with the identified modifications at position 34 directly in the genetic code table, while the modifications at position 37 shown by coloring. The associated tRNA gene that expresses each anticodon is shown in italic in parentheses. Each non-sense codon is marked with a * symbol.
U34 is consistently modified to one of two series. The cmnm5U34 series includes cmnm5Um34 and mnm5s2U34 [e.g., Arg(mnm5UCU), Gln(cmnm5s2UUG), Glu(mnm5s2UUC), Gly(mnm5UCC), Leu(cmnm5UmAA), and Lys(mnm5s2UUU)], while the cmo5U34 series includes mcmo5U34 [e.g., Ala(mcmo5UGC), Pro(mcmo5UGG), Thr(mcmo5UGU), and Ser(mcmo5UmGA)]. These two series share the common 5-carboxyl group modification of uridine. The only exception without any modification of U34 is in the anticodon of Sec(UCA) (Sec: selenocysteine), which inserts Sec to an internal UGA codon in mRNAs using a different decoding mechanism that specifies the site of insertion by a 3′-stem-loop structure and requires dedicated accessory factors (Commans and Bock, 1999). This U34, however, is modified in human Sec(mcm5UCA) (Songe-Moller et al., 2010). By contrast, the modification status of C34 is more diverse, which can be unmodified as in Arg(CCG), Arg(CCU), Gln(CUG), Gly(CCC), Leu(CAG), iMet(CAU) (the initiator tRNAMet), Pro(CGG), Thr(CGU), and Trp(CCA), or modified as in Leu(CmAA) or in eMet(ac4CAU) (the elongator tRNAMet). An important modification is L34 in Ile(LAU), which is the single determinant that reads the Ile codon AUA and discriminates against the Met codon AUG, while also serving as the major determinant that prevents the tRNA from mis-aminoacylation with Met (Muramatsu et al., 1988; Suzuki and Miyauchi, 2010). Similarly, A34 is consistently modified to I34, without exception, which expands the capacity of I34 to base-pair with A/C/U. This A-to-I modification is found in the single tRNA Arg(ICG). In contrast, G34 is mostly unmodified, except when it is in the GUX sequence, where it is modified to Q34 as in Tyr(QUA), His(QUG), Asn(QUU), and in Asp(gluQUC). The modification to Q34 equalizes the quality of base pairing with C/U (Morris et al., 1999), thus conferring a similar strength and efficiency of decoding of the two related codons.
We show the modification at position 37 on the 3′-side of each anticodon using different colors (Figure 2). The nucleotide at position 37 is predominantly a purine, whose modification can provide a stacking interaction with the nucleotide at position 36 of the anticodon to stabilize the structure of the anticodon-stem loop (ASL) (Konevega et al., 2004). This stabilization is necessary to neutralize differences of the ASL sequence for docking into the ribosome decoding site with a uniform speed and quality during protein synthesis (Konevega et al., 2004; Seelam Prabhakar et al., 2021). It is also necessary to assist the extensive conformational rearrangement of the ASL during the ribosome translocation to move down to the next codon (Nguyen et al., 2019). Indeed, deficiency of the modification at position 37 can cause ribosome stalling and +1-frameshifting (Masuda et al., 2021), indicating the importance of the modification in translation of the genetic code. Analysis of the modification pattern at position 37 in relationship with the nucleotide at position 36 identifies the following trends.
With U36-ending anticodons, the modification at position 37 is mostly ct6A37 and in one case it is m6t6A37 [in Thr(GGU)]. With C36-ending anticodons, the nucleotide at position 37 is usually not modified, except in Val(cmo5UAC), where it is modified to m6A37, and in Asp(gluQUC) and Glu(QUC), where it is modified to m2A37. With A36-ending anticodons, the modification at position 37 is usually ms2i6A37, except in one case Sec(UCA), where it is modified to just i6A37. Notably, with G36-ending anticodons, the modification at position 37 is m1G37 or m2A37, where the former is in Leu(cmo5UAG), Leu(GAG), Leu(CAG), Pro(mcmo5UGG), Pro(GGG), and Pro(CGG), and in Arg(CGG), while the latter is in His(QUC), Gln(cmnm5s2UUG), and in Arg(ICG). The nucleotide coupling between G36 and m1G37 has an enzymatic rationale, where the modification enzyme TrmD for m1G37 in bacteria indeed requires G36 as a recognition element to perform the modification reaction (Hou et al., 2017). This enzymatic rationale is likely to hold for other 36–37 nucleotide pairs, although experimental validation is necessary.
Given the importance of the modifications at both positions 34 and 37, we explore the possibility that some of them may have co-evolved with an inter-dependent relationship in a concept known as the “modification circuitry” (Han and Phizicky, 2018; Barraud and Tisne, 2019). Indeed, the presence of the m1G37 modification in E. coli Pro(mcmo5UGG) is the prerequisite for the methoxyl addition to convert cmo5U34 to mcmo5U34 (Masuda et al., 2018), while the presence of ms2i6A37 in E. coli Leu(CmAA) and Leu(cmnm5UmAA) is the prerequisite for 2′-O-methylation in Cm34 and Um34, respectively (Zhou et al., 2015). Similarly, the formation of Cm32 and Cm34 in the ASL of yeast Phe(GAA) is the prerequisite to convert m1G37 to yW37 (wybutosine) (Guy et al., 2012). To further evaluate the concept of the modification circuitry in the E. coli modification pattern, we analyzed the co-existence of the modifications at positions 34 and 37. Notably, we emphasize that co-existence only provides an implicit for circuitry, which requires experimental validation.
The list of a potential modification circuitry is complex and idiosyncratic, indicating that it is strongly dependent on individual ASL sequences, as well as the presence of other modifications. Here, we provide a framework for the potential circuitry of modifications at positions 34 and 37. We note that the cmnm5 and mnm5U34 series of modifications are often associated with i6A37 and t6A37 series of modifications generated by one of the more complex enzymatic pathways (Figure 1B). For example, cmnm5U34 and mnm5U34 co-exist with ms2i6A37 in Leu(cmnm5UmAA) and with ct6A37 in Arg(mnm5UCU), respectively. However, addition of the 2-thio (s2) group to form cmnm5s2U34 or mnm5s2U34 simplifies the modification at position 37 to usually a single methyl group (Figure 1C). This is shown in the association of cmnm5s2U34 with m2A37 in Gln(cmnm5s2UUG) and the association of mnm5s2U34 with m2A37 in Glu(mnm5s2UUC). An exception is the association of mnm5s2U34 with ct6A37 in Lys(mnm5s2UUU), where both modifications are generated from a multi-enzyme pathway. In contrast, while the cmo5U34 modification is generally associated with the simple structure of m1G37 (Figure 1D), such as in Leu(cmo5UAG), the addition of one methyl group to form mcmo5U34 requires a more complex structure at position 37 (Figure 1E), such as ms2i6A37 in Ser(mcmo5UGA) and ct6A37 in Thr(mcmo5UGU). An exception is that the mcmo5U34 modification remains associated with m1G37 in Pro(mcmo5UGG).
In contrast, the pairing of Q34 is diverse. While it is paired with the simple structure of m2A37 in His(QUG) and in Asp(gluQUC) (Figure 1F), it is paired with the complex structure of ms2i6A37 in Tyr(QUA) and the complex structure of ct6A37 in Asn(QUU) (Figure 1G).
Here we provide an easily accessible modification pattern of E. coli tRNAs that is integrated into the genetic code. In this modification pattern, each anticodon that reads the corresponding codon is shown with the modifications at positions 34 and 37. These modifications are likely selected during evolution to serve two functions – to facilitate accurate and diverse reading of each codon and to neutralize differences among various codon-anticodon pairings to ensure a uniform speed and quality of decoding across all codons, which is important for protein homeostasis (Ranjan and Rodnina, 2016). As these modifications are contained within the associated tRNA, which is charged with the specified amino acid for the codon, they constitute the necessary mechanism to facilitate interpretation of the genetic code. Despite the diverse structures of these modifications and their complex distribution in the genetic code, we have elucidated two principles for how each anticodon is associated with modifications to facilitate interpretation of the codon. These principles provide the basis to guide the users to search for more detailed information.
First, we find that modifications of complex structures are usually associated with AU-rich codons, while modifications of a simple structure or no modification are associated with GC-rich codons. Indeed, U34 is almost always modified with chemical structures that are synthesized by a multi-enzyme pathway and A34 is exclusively modified to I34, whereas C34 is not necessarily modified, while G34 is modified to Q34 when it is followed by a uridine on the 3′-side of the anticodon. At position 37, which stabilizes the stacking interaction with the nucleotide at position 36, the modification at 37 is usually a complex structure when the preceding nucleotide is U36 or A36. In contrast, the nucleotide at position 37 is not modified when the preceding nucleotide is C36, while the nucleotide at position 37 is modified to the simple structure of m1G37 when the preceding nucleotide is G36. In one of the most AU-rich cases, Lys(mnm5s2UUU) is modified with mnm5s2U34 and with ct6A37, both generated from a complex pathway.
Second, we suggest the possibility of a diverse, but potentially important, circuitry relationship between modifications at positions 34 and 37. Specifically, the addition of a simple chemical moiety at position 34 can change the profile of the modifications at position 37. This change is likely sequence-dependent to optimize each ASL to dock to the ribosome decoding site, which is structured by the 16S rRNA that is also extensively modified (Demirci et al., 2010; Kimura and Suzuki, 2010). While further experimental validation is necessary to validate the potential modification circuitry at positions 34 and 37 of each ASL, additional consideration of the circuitry should include the potential base pairing interactions at positions 31–39 and at 32–38, as they can also modulate the quality of the codon-anticodon pairing (Grosjean and Westhof, 2016).
Notably, several bacterial pathogens lack specific tRNA modifications, emphasizing that a more detailed study of these modifications will be important to human health. For example, our homology search suggests that Pseudomonas aeruginosa and Acinetobacter baumannii lack the gene for TmcA to synthesize ac4C34 on Met(CAU) and the gene for TrmM to synthesize m6A37 on Val(UAC). Additionally, our homology search suggests that A. baumannii lack the genes for CmoA and CmoB to synthesize cmo5U34, which would be required to expand wobble pairing in several tRNAs. This raises the question of how these bacteria species achieve the quality and diversity of anticodon-codon pairing to remain viable. Additionally, Mycobacterium bovis alters the level of cmo5U34 to adapt to hypoxia (Chionh et al., 2016) and P. aeruginosa alters the level of Cm/Um/Am32 to adapt to oxidative stress (Jaroensuk et al., 2016), reinforcing the notion that tRNA modifications play an essential role in stress response of bacterial pathogens.
In summary, the development of the modification pattern for E. coli tRNAs provides an important reference that will facilitate researchers to investigate the origin and evolution of the genetic code, the interpretation of the code during protein synthesis, and the exploration of the code for bioengineering purposes. It will also provide a rewarding path to investigate the potentially even more complex landscape of modifications in higher eukaryotes.
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
IM: Conceptualization, Investigation, Visualization, Writing – review & editing, Data curation. Y-MH: Conceptualization, Funding acquisition, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing, Project administration.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the NIH grant award R35GM134931 to Y-MH. We are grateful for the support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agris, P. F., Eruysal, E. R., Narendran, A., Vare, V. Y. P., Vangaveti, S., and Ranganathan, S. V. (2018). Celebrating wobble decoding: half a century and still much is new. RNA Biol. 15, 537–553. doi: 10.1080/15476286.2017.1356562
Barraud, P., and Tisne, C. (2019). To be or not to be modified: miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life 71, 1126–1140. doi: 10.1002/iub.2041
Bjork, G. R., Ericson, J. U., Gustafsson, C. E., Hagervall, T. G., Jonsson, Y. H., and Wikstrom, P. M. (1987). Transfer RNA modification. Annu. Rev. Biochem. 56, 263–285. doi: 10.1146/annurev.bi.56.070187.001403
Bjork, G. R., and Hagervall, T. G. (2014). Transfer RNA modification: presence, synthesis, and function. EcoSal Plus 6. doi: 10.1128/ecosalplus.esp-0007-2013
Boccaletto, P., Machnicka, M. A., Purta, E., Piatkowski, P., Baginski, B., Wirecki, T. K., et al. (2018). MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 46, D303–D307. doi: 10.1093/nar/gkx1030
Chionh, Y. H., McBee, M., Babu, I. R., Hia, F., Lin, W., Zhao, W., et al. (2016). tRNA-mediated codon-biased translation in mycobacterial hypoxic persistence. Nat. Commun. 7:13302. doi: 10.1038/ncomms13302
Commans, S., and Bock, A. (1999). Selenocysteine inserting tRNAs: an overview. FEMS Microbiol. Rev. 23, 335–351. doi: 10.1111/j.1574-6976.1999.tb00403.x
de Crecy-Lagard, V., and Jaroch, M. (2021). Functions of bacterial tRNA modifications: from ubiquity to diversity. Trends Microbiol. 29, 41–53. doi: 10.1016/j.tim.2020.06.010
Demirci, H., Ft, M., Belardinelli, R., Kelley, A. C., Ramakrishnan, V., Gregory, S. T., et al. (2010). Modification of 16S ribosomal RNA by the KsgA methyltransferase restructures the 30S subunit to optimize ribosome function. RNA 16, 2319–2324. doi: 10.1261/rna.2357210
Diwan, G. D., and Agashe, D. (2018). Wobbling forth and drifting Back: the evolutionary history and impact of bacterial tRNA modifications. Mol. Biol. Evol. 35, 2046–2059. doi: 10.1093/molbev/msy110
Endres, L., Dedon, P. C., and Begley, T. J. (2015). Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 12, 603–614. doi: 10.1080/15476286.2015.1031947
Gamper, H., Li, H., Masuda, I., Miklos Robkis, D., Christian, T., Conn, A. B., et al. (2021a). Insights into genome recoding from the mechanism of a classic +1-frameshifting tRNA. Nat. Commun. 12:328. doi: 10.1038/s41467-020-20373-z
Gamper, H., Mao, Y., Masuda, I., McGuigan, H., Blaha, G., Wang, Y., et al. (2021b). Twice exploration of tRNA +1 frameshifting in an elongation cycle of protein synthesis. Nucleic Acids Res. 49, 10046–10060. doi: 10.1093/nar/gkab734
Gamper, H. B., Masuda, I., Frenkel-Morgenstern, M., and Hou, Y. M. (2015). Maintenance of protein synthesis reading frame by EF-P and m(1)G37-tRNA. Nat. Commun. 6:7226. doi: 10.1038/ncomms8226
Gamper, H., Masuda, I., and Hou, Y. M. (2022). Genome expansion by tRNA +1 frameshifting at quadruplet codons. J. Mol. Biol. 434:167440. doi: 10.1016/j.jmb.2021.167440
Grosjean, H., de Crecy-Lagard, V., and Marck, C. (2010). Deciphering synonymous codons in the three domains of life: co-evolution with specific tRNA modification enzymes. FEBS Lett. 584, 252–264. doi: 10.1016/j.febslet.2009.11.052
Grosjean, H., and Westhof, E. (2016). An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res. 44, 8020–8040. doi: 10.1093/nar/gkw608
Guy, M. P., Podyma, B. M., Preston, M. A., Shaheen, H. H., Krivos, K. L., Limbach, P. A., et al. (2012). Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 18, 1921–1933. doi: 10.1261/rna.035287.112
Han, L., and Phizicky, E. M. (2018). A rationale for tRNA modification circuits in the anticodon loop. RNA 24, 1277–1284. doi: 10.1261/rna.067736.118
Hou, Y. M., Matsubara, R., Takase, R., Masuda, I., and Sulkowska, J. I. (2017). TrmD: a methyl transferase for tRNA methylation with m(1)G37. Enzyme 41, 89–115. doi: 10.1016/bs.enz.2017.03.003
Jaroensuk, J., Atichartpongkul, S., Chionh, Y. H., Wong, Y. H., Liew, C. W., McBee, M. E., et al. (2016). Methylation at position 32 of tRNA catalyzed by TrmJ alters oxidative stress response in Pseudomonas aeruginosa. Nucleic Acids Res. 44, 10834–10848. doi: 10.1093/nar/gkw870
Keseler, I. M., Gama-Castro, S., Mackie, A., Billington, R., Bonavides-Martinez, C., Caspi, R., et al. (2021). The EcoCyc database in 2021. Front. Microbiol. 12:711077. doi: 10.3389/fmicb.2021.711077
Kim, Y., Cho, S., Kim, J. C., and Park, H. S. (2024). tRNA engineering strategies for genetic code expansion. Front. Genet. 15:1373250. doi: 10.3389/fgene.2024.1373250
Kimura, S., and Suzuki, T. (2010). Fine-tuning of the ribosomal decoding center by conserved methyl-modifications in the Escherichia coli 16S rRNA. Nucleic Acids Res. 38, 1341–1352. doi: 10.1093/nar/gkp1073
Konevega, A. L., Soboleva, N. G., Makhno, V. I., Semenkov, Y. P., Wintermeyer, W., Rodnina, M. V., et al. (2004). Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal a site by stacking and Mg2+−dependent interactions. RNA 10, 90–101. doi: 10.1261/rna.5142404
Masuda, I., and Hou, Y. M. (2023). Protocol to identify the core gene supported by an essential gene in E. coli bacteria using a genome-wide suppressor screen. STAR Protoc. 4:102196. doi: 10.1016/j.xpro.2023.102196
Masuda, I., Hwang, J. Y., Christian, T., Maharjan, S., Mohammad, F., Gamper, H., et al. (2021). Loss of N(1)-methylation of G37 in tRNA induces ribosome stalling and reprograms gene expression. eLife 10:e70619. doi: 10.7554/eLife.70619
Masuda, I., Matsubara, R., Christian, T., Rojas, E. R., Yadavalli, S. S., Zhang, L., et al. (2019). tRNA methylation is a global determinant of bacterial multi-drug resistance. Cell Syst. 8, 302–314. doi: 10.1016/j.cels.2019.03.008
Masuda, I., Takase, R., Matsubara, R., Paulines, M. J., Gamper, H., Limbach, P. A., et al. (2018). Selective terminal methylation of a tRNA wobble base. Nucleic Acids Res. 46:e37. doi: 10.1093/nar/gky013
Masuda, I., Yamaki, Y., Detroja, R., Tagore, S., Moore, H., Maharjan, S., et al. (2022). tRNA methylation resolves codon usage bias at the limit of cell viability. Cell Rep. 41:111539. doi: 10.1016/j.celrep.2022.111539
Morris, R. C., Brown, K. G., and Elliott, M. S. (1999). The effect of queuosine on tRNA structure and function. J. Biomol. Struct. Dyn. 16, 757–774. doi: 10.1080/07391102.1999.10508291
Muramatsu, T., Nishikawa, K., Nemoto, F., Kuchino, Y., Nishimura, S., Miyazawa, T., et al. (1988). Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336, 179–181. doi: 10.1038/336179a0
Nguyen, H. A., Hoffer, E. D., and Dunham, C. M. (2019). Importance of a tRNA anticodon loop modification and a conserved, non-canonical anticodon stem pairing in tRNACGGProfor decoding. J. Biol. Chem. 294, 5281–5291. doi: 10.1074/jbc.RA119.007410
Ranjan, N., and Rodnina, M. V. (2016). tRNA wobble modifications and protein homeostasis. Translation 4:e1143076. doi: 10.1080/21690731.2016.1143076
Sakai, Y., Kimura, S., and Suzuki, T. (2019). Dual pathways of tRNA hydroxylation ensure efficient translation by expanding decoding capability. Nat. Commun. 10:2858. doi: 10.1038/s41467-019-10750-8
Seelam Prabhakar, P., Takyi, N. A., and Wetmore, S. D. (2021). Posttranscriptional modifications at the 37th position in the anticodon stem-loop of tRNA: structural insights from MD simulations. RNA 27, 202–220. doi: 10.1261/rna.078097.120
Shandell, M. A., Tan, Z., and Cornish, V. W. (2021). Genetic code expansion: a brief history and perspective. Biochemistry 60, 3455–3469. doi: 10.1021/acs.biochem.1c00286
Songe-Moller, L., van den Born, E., Leihne, V., Vagbo, C. B., Kristoffersen, T., Krokan, H. E., et al. (2010). Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol. Cell. Biol. 30, 1814–1827. doi: 10.1128/MCB.01602-09
Suzuki, T. (2021). The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392. doi: 10.1038/s41580-021-00342-0
Suzuki, T., and Miyauchi, K. (2010). Discovery and characterization of tRNAIle lysidine synthetase (TilS). FEBS Lett. 584, 272–277. doi: 10.1016/j.febslet.2009.11.085
Thiaville, P. C., Legendre, R., Rojas-Benitez, D., Baudin-Baillieu, A., Hatin, I., Chalancon, G., et al. (2016). Global translational impacts of the loss of the tRNA modification t(6)a in yeast. Microb Cell. 3, 29–45. doi: 10.15698/mic2016.01.473
Keywords: tRNA anticodon, position 34 of the anticodon, position 37 on the 3′-side of the anticodon, post-transcriptional modifications, modification circuitry
Citation: Masuda I and Hou Y-M (2024) A tRNA modification pattern that facilitates interpretation of the genetic code. Front. Microbiol. 15:1415100. doi: 10.3389/fmicb.2024.1415100
Received: 09 April 2024; Accepted: 31 May 2024;
Published: 12 June 2024.
Edited by:
Matthieu G. Gagnon, University of Texas Medical Branch at Galveston, United StatesReviewed by:
Valerie De Crecy-Lagard, University of Florida, United StatesCopyright © 2024 Masuda and Hou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ya-Ming Hou, eWEtbWluZy5ob3VAamVmZmVyc29uLmVkdQ==
†ORCID: Isao Masuda, https://orcid.org/0000-0001-9385-4424
Ya-Ming Hou, https://orcid.org/0000-0001-6546-2597
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.