
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 06 June 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1414401
This article is part of the Research TopicHorizontal Transfer of Antibiotic Resistance Genes in the Environment: Dynamic, Contributing Factors, and ControlView all 8 articles
 Logan C. Ott1,2
Logan C. Ott1,2 Melha Mellata1,2*
Melha Mellata1,2*The animal gut acts as a potent reservoir for spreading and maintaining conjugative plasmids that confer antimicrobial resistance (AMR), fitness, and virulence attributes. Interventions that inhibit the continued emergence and expansion of AMR and virulent strains in agricultural and clinical environments are greatly desired. This study aims to determine the presence and efficacy of short-chain fatty acids (SCFA) inhibitory effects on the conjugal transfer of AMR plasmids. In vitro broth conjugations were conducted between donor Escherichia coli strains carrying AMP plasmids and the plasmid-less Escherichia coli HS-4 recipient strain. Conjugations were supplemented with ddH2O or SCFAs at 1, 0.1, 0.01, or 0.001 molar final concentration. The addition of SCFAs completely inhibited plasmid transfer at 1 and 0.1 molar and significantly (p < 0.05) reduced transfer at 0.01 molar, regardless of SCFA tested. In explant models for the chicken ceca, either ddH2O or a final concentration of 0.025 M SCFAs were supplemented to the explants infected with donor and recipient E. coli. In every SCFA tested, significant decreases in transconjugant populations compared to ddH2O-treated control samples were observed with minimal effects on donor and recipient populations. Finally, significant reductions in transconjugants for plasmids of each incompatibility type (IncP1ε, IncFIβ, and IncI1) tested were detected. This study demonstrates for the first time the broad inhibition ability of SCFAs on bacterial plasmid transfer and eliminates AMR with minimal effect on bacteria. Implementing interventions that increase the concentrations of SCFAs in the gut may be a viable method to reduce the risk, incidence, and rate of AMR emergence in agricultural and human environments.
The emergence and spread of antimicrobial resistance (AMR) and virulence attributes in bacteria are of immense concern to both animal and human health (Teuber, 1999; Nordmann and Poirel, 2005; Robicsek et al., 2006; Sommer et al., 2009; Penders et al., 2013; Lopatkin et al., 2017; Fukuda et al., 2019; Hedman et al., 2020; Techitnutsarut and Chamchod, 2021). The gut of food animals, such as poultry, acts as a potent reservoir for this spread and storage of plasmids (Jochum et al., 2021). Plasmid spread among bacteria results in the emergence of novel bacterial strains with potential for pathogenicity and often results in resistant infections in both animal agriculture as well as in the clinical environments (Guiyoule et al., 2001; Wang et al., 2003; Nordmann and Poirel, 2005; Hong et al., 2009; Aguado-Urda et al., 2012; Soares et al., 2014; Tang et al., 2017; Dolejska and Papagiannitsis, 2018; Millan, 2018). In humans, this leads to prolonged care and recovery of patients, and increased mortality (Roca et al., 2015; Woolhouse et al., 2015; Hofer, 2019).
Identification of strategies for inhibiting conjugal spread of plasmids among bacteria in the environment and within animal hosts is of dire importance (Ruzin and Novick, 1998; Fernandez-Lopez et al., 2005; Ripoll-Rozada et al., 2016; García-Cazorla et al., 2018; Graf et al., 2018). The use of the diet has been briefly examined for its role in preventing AMR (Liu et al., 2019; Elshaghabee et al., 2021). Preliminary studies have demonstrated using medium and long-chain polyunsaturated fatty acids (PUFAs) to inhibit the conjugal ATPase TrwD. However, no in vivo or host-associated studies have confirmed this function, nor has the mechanism of action been confirmed (Fernandez-Lopez et al., 2005; Ripoll-Rozada et al., 2016; García-Cazorla et al., 2018). Regardless, a limitation to the application of PUFAs as a dietary intervention for gut-mediated plasmid transfer is that medium and long-chain fatty acids (MCFA, LCFA) are readily absorbed in the upper intestinal tract and often do not persist to the gut microbiota to mediate an effect in vivo (Ramírez et al., 2001).
Short-chain fatty acids (SCFAs) have a carbon chain length of no more than six and are potent regulators of the gut environment. Increased concentrations of SCFAs in the gut have been found to select for beneficial microbes, inhibit colonization and persistence of harmful microbes, and SCFAs act as a metabolic source for healthy epithelial cell growth and maintenance (Sakata and Yajima, 1984; Vinolo et al., 2011; Kim et al., 2014; Tan et al., 2014; Baxter et al., 2019; Parada Venegas et al., 2019; Schulthess et al., 2019; Liao et al., 2020). Contrary to MCFAs and LCFAs, SCFAs are the metabolic output of microbial fermentation of indigestible fibers from the host diet and are generated locally in the midgut by microbial constituents (Kim et al., 2014). Supplementation of the diet with nonfermentable fibers is currently understood to lead to increased gut homeostasis and epithelial health through the enrichment of SCFA and SCFA-producing bacteria (Baxter et al., 2019; Mueller et al., 2020). There is evidence for an inhibitory effect on the colonization and persistence of organisms commonly found to host plasmids through modification of the gut environment pH, but the combinatorial role of SCFA inhibition of both colonization and conjugation has not yet been demonstrated (Kim et al., 2014; Baxter et al., 2019; Schulthess et al., 2019; Liao et al., 2020).
Overall, the goal of this study is to determine what effect SCFAs have on the incidence of bacterial plasmid transfer in an in vitro model for the gut environment by testing the impact of the naturally occurring SCFAs from the animal gut, as well as the greater family of SCFAs to determine if the biochemical effects are dependent on fatty acid chain length. Additionally, we tested the broad efficacy of SCFAs against conjugal plasmids of varying genetic types to identify if inhibition is plasmid-specific or conserved across plasmid biology. Here, we demonstrate the apparent inhibition of bacterial plasmid conjugation in an in vitro and an ex vivo avian host co-culture ceca explant. Using traditional broth plasmid conjugation assays, we identified the ability of SCFAs to impact the rate and incidence of plasmid conjugation. Furthermore, we showed the ability of SCFAs to confer an effect in ex vivo host-associated environments, indicating a potential role in vivo.
Unless otherwise stated, all SCFAs used in this study were purchased from VWR International (Radnor, Pennsylvania, United States). RPMI, ADMEM, fetal bovine serum, and L-glutamine were purchased from Thermo Fisher Scientific (Waltham, Massachusetts, United States) and were Gibco brand. SCFAs were dissolved and diluted into sterile High-performance liquid chromatography (HPLC) grade ultrapure water and filter sterilized through 0.22 μM syringe filters before use. SCFAs were stored covered at room temperature between uses. SafMannan® yeast cell wall preparations were provided by Phileo by Lesaffre.
The multidrug-resistant (MDR) Avian Pathogenic Escherichia (E.) coli (APEC) strain APEC O2-211 was used as the primary donor in both in vitro and ex vivo conjugation experiments (Nielsen et al., 2018). APEC harbors one large MDR plasmid, pAPECO2-211A-ColV (197 Kbp), and two small plasmids, pAPEC-O2-211B (4,231 bp) and pAPEC-O2-211C (2,096 bp). The pAPECO2-211A-ColV was the plasmid screened for transfer, and confers resistance to tetracycline, macrolide, fluoroquinolone, carbapenem, cephalosporin, and aminoglycoside antibiotics. Furthermore, pAPEC-O20211A-ColV contains the virulence determinants; ibeA, iutA, iroN, sitA, tsh, fim, fyuA, pap, and vat (Nielsen et al., 2018). Salmonella (S.) enterica Serovar Kentucky strain CVM29188 was isolated from a chicken meat sample and served as a supplemental donor during in vitro conjugation assays (30). CVM29188 contains 3 plasmids, 2 of which carry MDR, including pVCM29188_146 (146 kb, GenBank accession CP001122) that carries resistance genes to aminoglycosides and tetracyclines and pCVM29188_101 (101 kb, GenBank accession CP001121) that carries resistance to cephalosporins and quaternary ammonium compounds. For conjugations with the S. Kentucky strain, the transfer of the pCVM29188_146 plasmid was monitored as with the pAPEC-O2-211A-ColV plasmid.
To limit host effect on conjugation assays, E. coli SP915 strain, an MG1655 derivative with a chromosomal mCherry insertion, was used as the donor for the plasmids pKJK5-GM (tetracycline and trimethoprim resistant), pCVM29188_146 (tetracycline and streptomycin resistant), and pC20-GM (cefotaxime resistant) (Ott et al., 2021). The E. coli HS-4 strain maintained a spontaneous chromosomal resistance to rifampicin and was used as a recipient in all conjugations (Ott et al., 2020, 2021). E. coli HS-4 was initially isolated from a healthy human gut and was a proxy for a commensal animal gut microbe (Rasko et al., 2008). Full information on the strains and plasmids used in this study is reported in Table 1. Fresh bacterial cultures were streaked on MacConkey agar supplemented with antibiotics targeting each plasmid resistance marker (pAPEC-O2-211A-ColV, 15 μg/mL tetracycline; pKJK5-GM, 15 μg/mL tetracycline; pCVM29188_146, 15 μg/mL tetracycline; pC20-GM, 4 μg/mL cefotaxime; HS-4, 100 μg/mL rifampicin) before each experiment.
Conjugation reactions between donor strains and the recipient HS-4 strain were assayed through traditional broth conjugation methods using Luria Bertani (LB) broth as the suspension medium as previously (Ott et al., 2020, 2021). Briefly, overnight cultures of logarithmic phase donor and recipient strains were pelleted at 16,000 × G for 2 min, rinsed twice with pre-warmed phosphate-buffered saline (PBS), and the final pellet was then resuspended to OD600nm ~ 1.0. Cultures were mixed 1:1 for a total volume of 180 μL in individual wells of a 96-well tissue culture plate. SCFAs were added at a dilution of 1:10 for final desired concentrations of either 0 and 0.025 M, or 0, 0.001, 0.01, 0.1, and 1 molar, or at physiological concentrations previously observed from the chicken ceca (Walugembe et al., 2015; González-Ortiz et al., 2020). Conjugation mixtures were then incubated for 6 h at the optimal bacterial growth temperature of 37°C. After incubation, conjugation mixtures were immediately serially diluted and plated on MacConkey agar selecting for each bacterial population (donors, 15 μg/mL tetracycline or 4 μg/mL cefotaxime; recipients, 100 μg/mL rifampicin; transconjugants, 15 μg/mL tetracycline or 4 μg/mL cefotaxime and 100 μg/mL rifampicin).
All animal work in this study was reviewed and approved by the Institutional Animal Care Use Committee at Iowa State University under protocol IACUC-21-265. On two separate occasions, three ~2-week-old white leghorn chickens were collected from a local commercial hatchery, CO2 euthanized, and aseptically necropsied. From each bird, 3 cm long segments of each cecum were removed, opened longitudinally, and placed in tissue collection media (RPMI, 50 ug/mL gentamycin, 1% penicillin/streptomycin) and incubated at 40°C for 1 h to clear remaining resident microbiota and remove fecal material. Tissues were then rinsed in sterile PBS to remove residual antibiotics and any remaining fecal material, aseptically cut into 0.5 cm2 segments, and placed into individual wells of a 96-well tissue culture plate.
Complete growth media (ADMEM, 8 mM L-Glutamine, 1% Pen/strep, 10% fetal bovine serum) supplemented with OD600nm ~ 1.0 of donor and recipient bacterial mixture prepared as previously described during in vitro conjugations with complete growth media as the pellet diluent was added. To each well of paired ceca tissue, sterile ddH2O or SCFA were added in 1:10 dilutions for a final concentration of 0.025 M and a final volume of 200 μL as to represent a realistic but increased concentration of SCFAs in the gut compared to physiological conditions. Following incubation at 40°C for 6 h in 5% CO2, supernatant from each tissue co-culture was pipette homogenized and serially diluted in tenfold dilutions and plated on MacConkey agar selecting for either donor (15 ug/mL tetracycline), recipient (100 ug/mL rifampicin), or transconjugants (15 ug/mL tetracycline and 100 ug/mL rifampicin) populations.
Statistical analyses were completed using the GraphPad Prism 6 Software suite. In vitro, ddH2O and SCFA groups were compared using unpaired two-way ANOVA (concentration ~ SCFA). Ex vivo ddH2O and SCFA groups were compared using a paired one-way ANOVA within each bacterial population. p-values (p) less than 0.05 were considered significant. When used, conjugation frequency was calculated as the total population of transconjugants divided by the total population of donors per sample.
In vitro broth plasmid conjugations between E. coli SP915 (pCVM29188_146) and the plasmid-free E. coli HS-4 recipient resulted in significantly decreased transconjugants populations at all propionate treatment levels (29.5, 59, 147.5, and 295 mM), with no transconjugants detected at the 295 mM concentration following enrichment (Figure 1A). Donor populations in the 295 mM propionate treatment group were significantly reduced compared to the control group (Figure 1A). Additionally, the conjugation frequency was significantly decreased for all treatment groups compared to the control (Figure 1B).
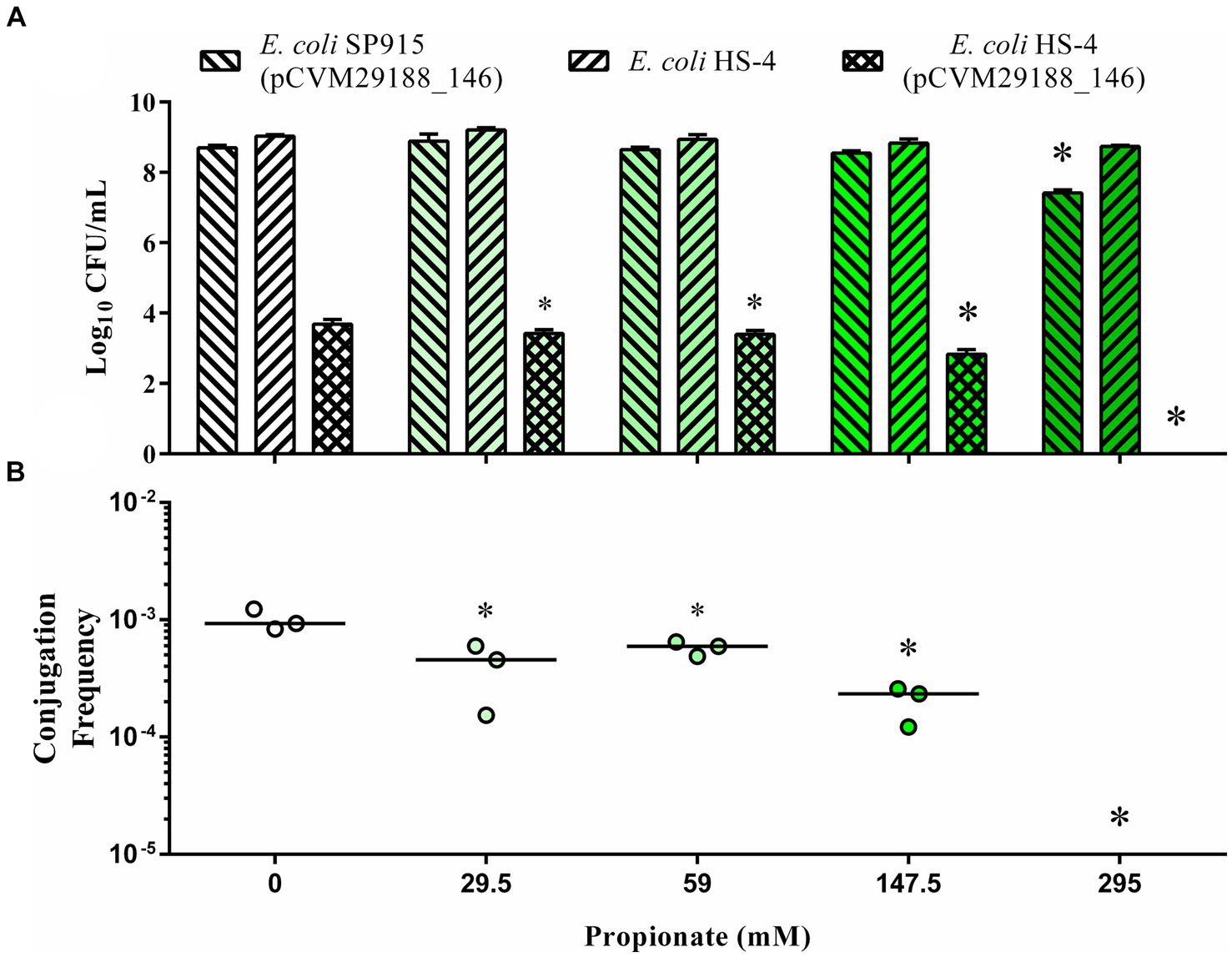
Figure 1. In vitro inhibition of bacterial plasmid transfer by the SCFA propionate in liquid broth. The Log10 CFU/mL of donors (forward slash), recipients (backward slash), and transconjugants (crossed) (A) and conjugation efficiency (B), as calculated by the total transconjugant population divided by the total donor population for each reaction. Bars in (A) represent the mean and standard deviations, while the bars in (B) represent the median. p-values were corrected for multiple comparisons by Dunnet post-hoc hypothesis testing. *p < 0.05, *p < 0.005, *p < 0.0005, *p < 0.00005. p-values less than 0.05 were considered significant.
Conjugation between either S. Kentucky or E. coli APEC-02–211 and the plasmid-free recipient E. coli HS-4 was demonstrated in the presence of acetate, propionate, and butyrate at physiological concentrations from the chicken ceca (Figure 2). Addition of acetate to the conjugation broths led to a significant reduction in conjugation frequency in all groups, with no detectable transconjugants in the 90 or 180 mM and 45, 90, and 180 mM groups for either S. Kentucky or APEC-02 (p < 0.05) (Figures 2A,B). Adding propionate at 2-, 4-, and 8-mM did not result in significant changes in conjugation frequency. However, a negative trend is visible in the S. Kentucky group, and a positive trend is present in the APEC-O2 group (p > 0.05) (Figures 2A,B). Addition of butyrate at 10, 20, or 40 mM demonstrated significant reductions in both donor groups in a dose-dependent response (p < 0.005) except for 10 mM in the APEC-02 group (p > 0.05) (Figures 2A,B).
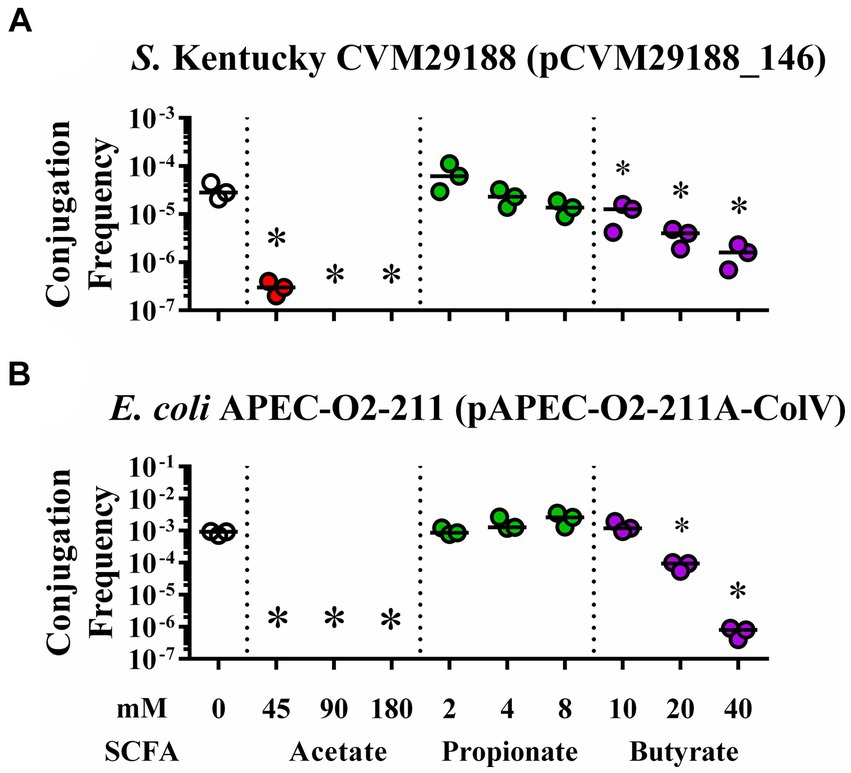
Figure 2. In vitro inhibition of bacterial plasmid transfer by SCFAs at gut physiological concentrations in liquid broth. Conjugation efficiency of plasmids pCVM29188_146 (A) and pAPEC-02–211A-ColV (B) during in vitro exposure to physiological levels of SCFAs from the chicken ceca. Bacteria were incubated with ddH2O (clear), acetate (red), propionate (green), or butyrate (purple) at 0, 45, 90, 180; 0, 2, 4, 8; and 0, 10, 20, or 40 mM, respectively. Bars represent the median. Comparisons between control and varying concentrations of SCFAs were completed by one-way ANOVA within each SCFA group. Ps were corrected for multiple comparisons by Dunnet hypothesis testing. *p < 0.05, *p < 0.005, *p < 0.0005, *p < 0.00005. p-values less than 0.05 were considered significant.
To explore the role of SCFAs on the incidence of plasmid conjugation, liquid in vitro plasmid conjugation assays between E. coli APEC-02–211 and E. coli HS-4 were conducted in the presence of 0, 0.001, 0.01, 0.1, and 1 M of either formate, acetate, propionate, butyrate, isobutyrate, valerate, isovalerate, or 2-methyl butyrate (Figure 3). Additions of each SCFA at 0.1 M or 1 M resulted in complete reductions in transconjugant populations, with no transconjugants detected following enrichment (p < 0.00005). Additionally, 0.01 M concentrations of either acetate, propionate, butyrate, valerate, or isovalerate resulted in significant decreases in transconjugant populations compared to the ddH2O treatment control (p < 0.005) (Figure 3). The 0.001 mM of both formate and butyrate treatments resulted in small but significant increases in total transconjugant populations (p < 0.05). No other significant differences between the 0.001 M SCFAs and the control were observed.
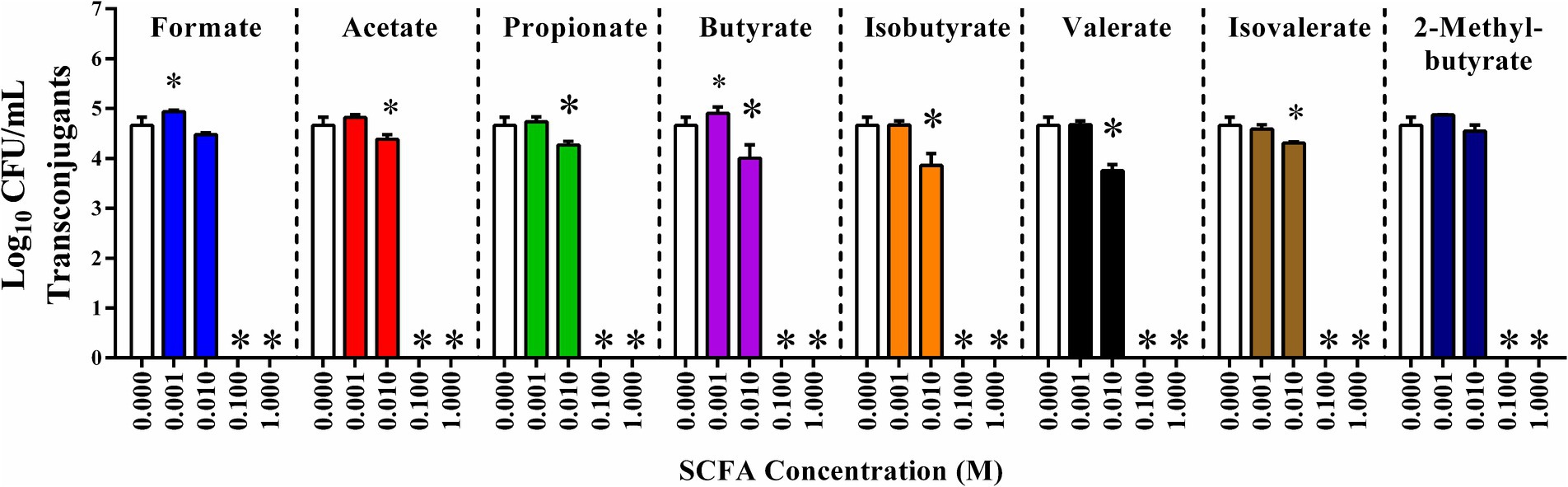
Figure 3. In vitro inhibition of IncF plasmid transfer by SCFAs in liquid broth. Conjugation reaction mixture supplemented with 0 (ddH2O), or 0.001, 0.01, 0.1, or 1 molar of each SCFA: formate (light blue), acetate (red), propionate (green), butyrate (purple), isobutyrate (orange), valerate (black), isovalerate (brown), and 2-methylbutyrate (dark blue). Bars represent the mean of three replicates, and error bars represent the standard deviation above and below the mean. Comparisons between ddH2O treatments and varying concentrations of SCFAs were completed by one-way ANOVA within each SCFA group. Ps were corrected for multiple comparisons by Dunnet hypothesis testing. *p < 0.05, *p < 0.005, *p < 0.0005, *p < 0.00005. p-values less than 0.05 were considered significant.
Chicken ceca explants were harvested and used in co-culture conjugation experiments to determine if the reduction in conjugation was observable in host-associated environments. The total donor (Figure 4A), recipient (Figure 4B), and transconjugant populations (Figure 4C) were, respectively, enumerated from co-cultures in the presence or absence of supplemental SCFAs at physiological concentrations (Figure 4). The addition of SCFAs individually to explant cultures resulted in slight but insignificant increases in donor APEC-02-211 populations in the formate, acetate, propionate, and isobutyrate treatment groups (Figure 4A). However, the increase in the donor population in the valerate group compared to the ddH2O control was significant (p < 0.0005). A non-significant decrease in the donor population was further observed in the butyrate, isovalerate, and 2-methylbutyrate groups (Figure 4A).
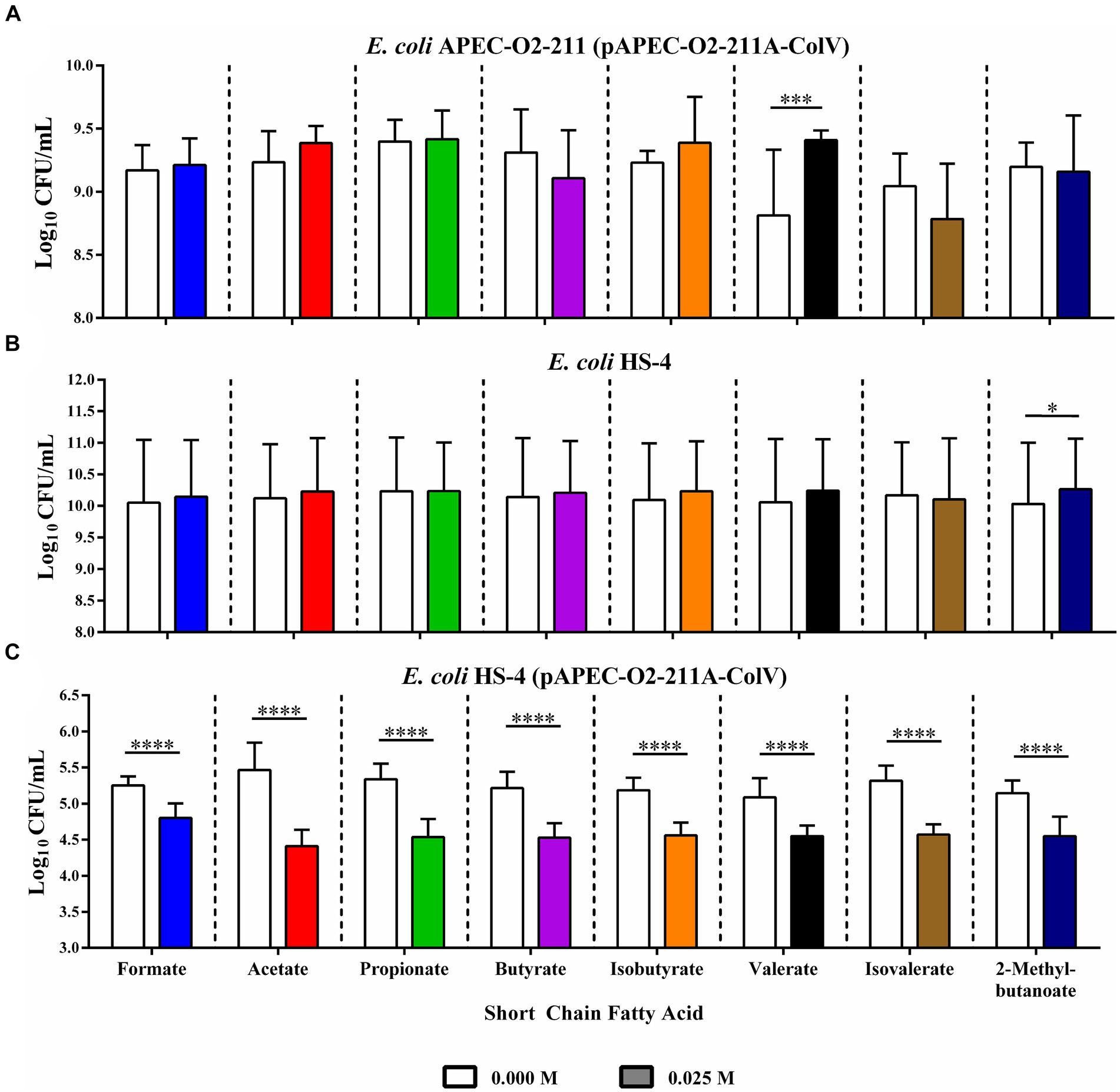
Figure 4. Ex vivo inhibition of IncF conjugation by SCFAs in chicken cecal explants. Bacterial enumeration of donors (A), recipients (B), and transconjugants (C) from cecal explant co-cultures. Bars represent the mean of three replicates, and error bars represent the standard deviation above and below the mean. Comparisons between ddH2O and SCFA treatments were completed by paired two-way ANOVA, and the Šidák correction for multiple comparisons was applied. *p < 0.05, *p < 0.005, *p < 0.0005, *p < 0.00005. p-values less than 0.05 were considered significant.
Likewise, the recipient populations were slightly, but insignificantly, increased in formate, acetate, butyrate, isobutyrate, valerate, and 2-methyl butyrate treatment groups, with the increase in recipient population in the 2-methyl butyrate treatment group being statistically significant (p < 0.05) (Figure 4B). The addition of isovalerate resulted in a slight, but insignificant, decrease in the total recipient population.
Finally, the addition of any of the eight SCFAs at 0.025 mM resulted in a large and significant reduction in total transconjugant populations when compared to the corresponding ddH2O-treated control groups (p < 0.00005) (Figure 4C).
To ascertain whether the inhibitive properties of SCFAs occurred consistently with plasmids of various incompatibility types, in vitro conjugations between E. coli SP915 carrying either pKJK5-GM (IncP1ε), pCVM29188_146 (IncFIβ), or pC20-GM (IncI1) and the plasmid-less HS-4 recipient were conducted in the presence or absence of 0.025 M of the gut associated SCFAs acetate, propionate, or butyrate (Figure 5). The addition of each of the SCFAs tested to conjugations between E. coli SP915 containing the pKJK5-GM broad host range plasmid did not result in significant changes in donor or recipient populations compared to the ddH2O treated control group (Figure 5A). However, total transconjugant populations were significantly decreased (p < 0.00005) in each of the SCFA treatment groups when compared to the ddH2O treatment control group. Furthermore, the butyrate treatment resulted in significantly fewer transconjugants compared to the propionate (p < 0.005) but not compared to the acetate.
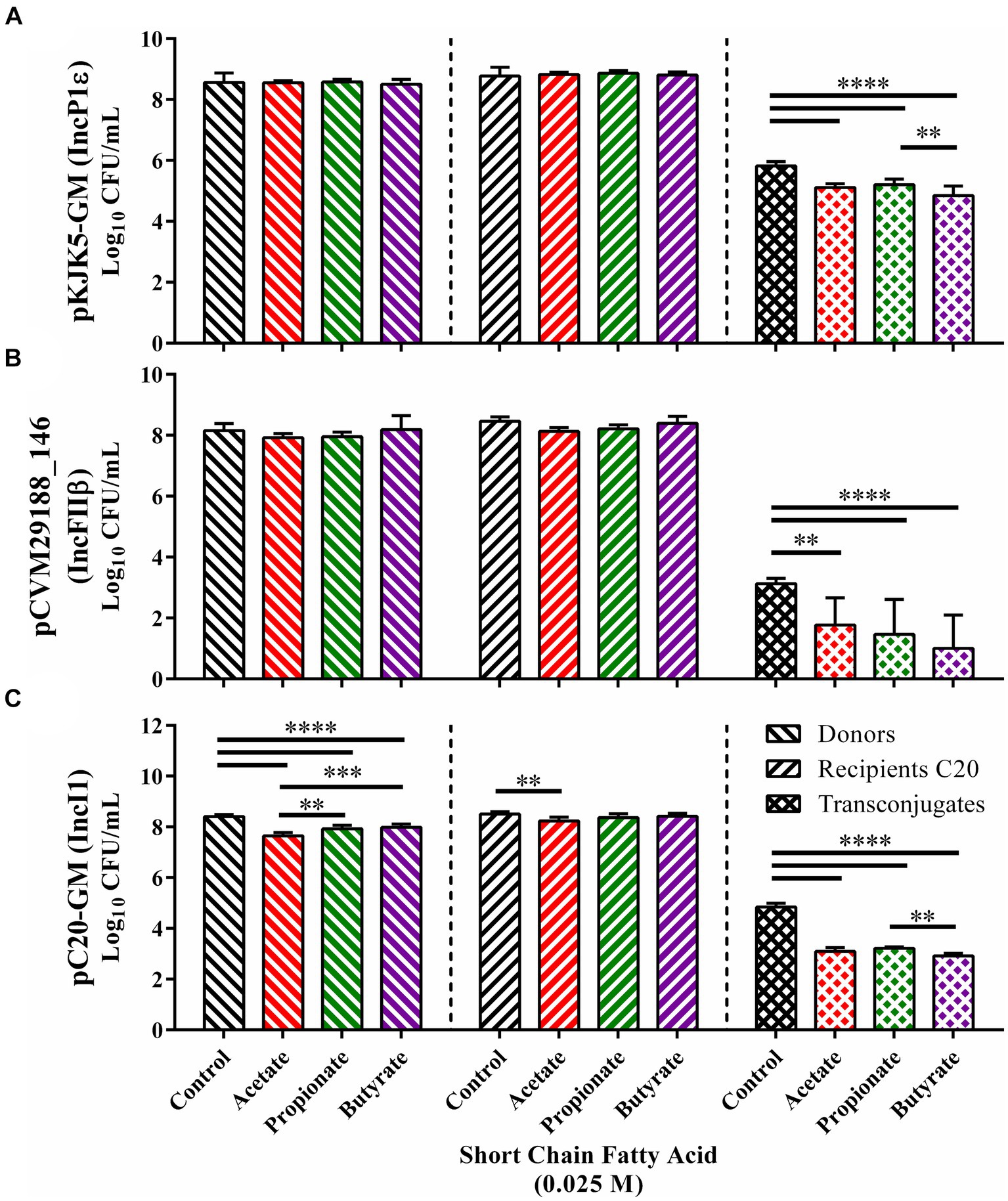
Figure 5. In vitro inhibition of the transfer of multiple plasmid incompatibility groups in liquid broth. Conjugation in the presence of acetate (red), propionate (green), and butyrate (purple) for E. coli SP915 (pCVM29188_146) (A), E. coli SP915 (pKJK5-GM) (B), and E. coli SP915 (pC20-GM) (C). Comparisons between ddH2O and SCFA treatments were completed by paired two-way ANOVA, and the Šidák correction for multiple comparisons was applied. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005. p-values less than 0.05 were considered significant.
In plasmids conjugations between E. coli SP915 carrying the pCVM29188_146 narrow host range plasmid and HS-4, again, no significant differences between donor and recipient populations and the ddH2O treated controls were observed (Figure 5B). However, significant reductions in the transconjugant populations were observed in all three SCFA groups tested with the most significant decreases being in propionate and butyrate (p < 0.00005) and a lesser but significant (p < 005) reduction in the acetate treatment group when compared to the ddH2O treatment group (Figure 5B).
Finally, plasmid conjugations between E. coli SP915 carrying the narrow host range plasmid pC20-GM and the plasmid-free HS-4 resulted in significant (p < 0.00005) reductions in donors for each of the three SCFAs acetate, propionate, butyrate tested with decreasing reductions from acetate to propionate (p < 0.005), and acetate to butyrate (p < 0.0005) (Figure 5C). Additionally, a significant decrease in recipients was observed in the acetate group but not in the propionate nor butyrate treatment groups. Again, total transconjugant populations were significantly decreased (p < 0.00005) in each SCFA treatment group compared to the ddH2O treatment control group. Furthermore, the butyrate treatment group resulted in significantly fewer transconjugants compared to propionate (p < 0.005) but not compared to acetate (Figure 5C).
The misuse of antibiotics in both agricultural and clinical settings has resulted in the rapid evolution and dissemination of AMR and virulence genes (Threlfall et al., 2000; Lowy, 2003; Nordmann and Poirel, 2005; Robicsek et al., 2006; Roca et al., 2015). This dissemination is empowered through the use and transfer of extrachromosomal plasmid DNA (Dolejska and Papagiannitsis, 2018; Millan, 2018; Redondo-Salvo et al., 2020). The transfer of this DNA through bacterial conjugation enables the direct transfer and rapid expression of whole AMR and virulence genes (Licht et al., 1999; Grohmann et al., 2003; Mellata et al., 2010; Virolle et al., 2020). This phenomenon contributes to the rapid emergence and spread of novel MDR pathogens not only in human healthcare but also in agricultural environments (Dowson et al., 1989; Guiyoule et al., 2001; Wang et al., 2003; Skyberg et al., 2006; Hong et al., 2009; Mellata et al., 2010, 2012; Aguado-Urda et al., 2012; Soares et al., 2014; Card et al., 2017; Lacey et al., 2017; Millan, 2018; Mobasseri et al., 2019). This is significant due to the transfer of bacterial pathogens from food products, such as poultry meat, to humans having been frequently observed (Fricke et al., 2009; Mellata, 2013; Mitchell et al., 2015; Liu et al., 2018; Mellata et al., 2018).
With the emergence and spread of MDR plasmids, a new emphasis on non-antibiotic therapeutics for managing the microbiome associated with humans and animals has been developed (Fernandez-Lopez et al., 2005; Solis-Cruz et al., 2020; Cabezn et al., 2017; Lopatkin et al., 2017; Zhang et al., 2019; Li et al., 2021). Inhibition of bacterial plasmid transfer, as opposed to indiscriminately eliminating bacterial hosts, is a desirable approach to preventing the spread of MDR plasmids. This targeted approach avoids directly remodeling the gut microbiota through the depletion of potential reservoir strains such as the recipient populations. The resident strains are often beneficial or neutral in the gut and act as an alternate or temporary host for plasmid DNA (Salyers et al., 2004; Huddleston, 2014; Lerner et al., 2017). A plasmid specific approach is different from using antigen-based vaccination or even the use of antibiotics themselves, both of which remodel the gut microbiota by both discriminatory and nondiscriminatory depletion of microbiota members (Midtvedt et al., 1986; Candon et al., 2015; Mir et al., 2019; Zhang et al., 2019; Guevarra et al., 2021; Techitnutsarut and Chamchod, 2021). Alternative approaches to address this shift in focus have been regularly reported and examined for their ability to regulate the microbial populations in these environments and are periodically reviewed (Cohen, 2009; McDougall et al., 2009; Ghouri et al., 2018; Du et al., 2021; Sadowska et al., 2021; Wisener et al., 2021).
On such approach involves the use of dietary additives, such as the PUFAs linoleic acid and alpha-linolenic acid, which have been briefly demonstrated as conjugation inhibitors (COINs) (Li et al., 2021). However, the application of this novel dietary intervention has not yet been shown in host-associated conditions. Additionally, using medium to long-chain PUFAs offers limitations for practical use, as in the animal host, they are rapidly assimilated in the small intestine (Krogdahl, 1985; Ramírez et al., 2001). An alternative to the use of medium to long-chain PUFAs is the use of the microbial secreted SCFA dietary metabolites, as they are generated by the gut resident microbiota in the caecum of fermentative animals and in the colon of non-fermentative animals such as humans (Tan et al., 2014). The SCFAs have potent effects on the host gut and gut microbiota that are categorized as beneficial (Sakata and Yajima, 1984; Vinolo et al., 2011; Kim et al., 2014; Tan et al., 2014; Baxter et al., 2019; Parada Venegas et al., 2019; Liao et al., 2020). Unlike medium to long-chain PUFA, SCFAs escape rapid absorption in the small intestine as non-digestible intermediates, making them ideal candidates for directed regulation of the microbiota in the context of the gut (Krogdahl, 1985; Ramírez et al., 2001).
Propionate, a SCFA with a three-carbon chain length, is used as an antifungal and bacteriostatic preservative in foods, such as in bread and cheese, as well as in technical applications, such as in the media recipes used for the maintenance of Drosophila (Suhr et al., 2004; Guynot et al., 2005; Marin et al., 2010; BDSC, 2019). Additionally, propionate is one of the three SCFAs regularly found in the gut (i.e., acetate, propionate, and butyrate). Propionate is currently proposed to act on fungi by crossing the cell wall and interacting with succinyl-CoA, an intermediate of the metabolite catabolism pathway, to form propionyl-CoA at significantly increased levels. Subsequently, increased concentrations of propionyl-CoA inhibit the catabolism or anabolism of metabolites and prevent metabolic activity in fungi (Brock et al., 2000).
Initial conjugations with SCFAs were conducted as part of our previous work with the Drosophila animal model and included concentrations typical of those found in Drosophila media (Figure 1) (Ott et al., 2021). The minimal concentration for antimicrobial and antifungal effects of propionate have been previously studied and are inherently not novel (Maruyama and Kitamura, 1985; Brock et al., 2000; Guynot et al., 2005; Marin et al., 2010; Langfeld et al., 2021). Minimal inhibitory concentrations of propionate on E. coli and fungal growth were reported to be ~25 and 50 mM, respectively, levels which were similar to or lower than those found in Drosophila media (59 mM) (Brock et al., 2000; Guynot et al., 2005; Langfeld et al., 2021). While the concentrations we used for our initial in vitro conjugations were similar in scale, they were still greater than those traditionally identified in the gut of humans and other animals under physiological conditions (Sakata and Yajima, 1984; Liao et al., 2020; Mueller et al., 2020).
Unlike in fungi, the currently proposed mechanism of action of propionate in bacteria is like that of PUFAs, where medium-chain PUFAs are expected to bind to and directly inhibit the TrwD ATPase protein itself (Ripoll-Rozada et al., 2016; García-Cazorla et al., 2018). The contribution of the carboxylic acid functional group may be responsible for the conserved inhibitory effects of both SCFAs and longer-chain PUFAs in inhibiting bacterial plasmid conjugation. Structural analysis of the binding of SCFAs to conjugal proteins is also underway to determine if that mechanism may also provide a combinatory effect here. However, there is additional evidence that the import of propionate occurs in a protonated form and is rapidly deprotonated upon entry into the cell, increasing cytoplasmic pH and significantly increasing proton concentration (Maruyama and Kitamura, 1985; Langfeld et al., 2021). The increase in intracellular hydrogen ions may be critical to inhibiting the use of the proton gradient in producing ATP through membrane-bound ATP synthase (Langfeld et al., 2021). Inhibition of ATP synthase using the proton gradient may be essential in the functional transfer of plasmid DNA in vitro and even in vivo, as plasmid DNA processing, loading, and transfer are all dependent on the hydrolysis of cytoplasmic ATP (Willetts and Wilkins, 1984). However, the increase in cytoplasmic pH and the effect on the incidence or rate of plasmid conjugation are yet to be determined for SCFAs.
Poultry may be a primary source of MDR bacteria, and the transfer of such bacteria to humans (Al-Ghamdi et al., 1999; van den Bogaard et al., 2001; Fricke et al., 2009; Bortolaia et al., 2016; Hedman et al., 2020; Jochum et al., 2021). In this study, we first tested in vitro bacterial conjugations using the physiological concentration of SCFAs from the chicken ceca for the three main gut SCFAs, including acetate, propionate, and butyrate (Figure 2). We observed significant reductions in the frequency of plasmid conjugation for two MDR plasmids with significance to the poultry industry (pCVM29188_146 and pAPEC_02–211A-ColV). However, in these conjugations, the physiological concentration of propionate does not appear to be great enough to significantly reduce conjugation frequency as it is for acetate or butyrate. This indicates that at physiological levels of 2, 4, and 8 mM, propionate does not play a significant role in regulating conjugation in broth and likely does not at typical levels within the ceca of chickens (Dunkley et al., 2007; Józefiak et al., 2010; Sunkara et al., 2011). Acetate and butyrate, conversely, significantly reduced conjugation in vitro, as indicated by the significant reduction in conjugation frequency at all concentrations tested (Figure 2).
While the gut SCFAs acetate, propionate, and butyrate may have potential as COINs, it is not immediately clear if this is a conserved characteristic of all SCFAs or limited to those that regularly occur in the gut. To address this uncertainty, we repeated in vitro conjugations in the presence of all eight standard SCFAs at concentrations ranging between 1 and 0.001 M final concentrations (Figure 3). The complete depletion of transconjugant populations for all SCFAs at concentrations of SCFA of 0.1 and 1 M may be attributed to the intense shift in the pH of the media. It may make these extreme concentrations impractical or physiologically incompatible with the gut environment, so these specific results may not reflect the practical application of these concentrations in vivo.
We did, however, observe a significant reduction in the total transconjugant population in six of the eight SCFAs tested: acetate, propionate, butyrate, valerate, and isovalerate, with a trending decrease in the other two, formate and 2-methyl butyrate at a final concentration of 0.01 M. This concentration is partially between the previously reported minimal growth inhibition value and the physiological concentrations previously tested and found in the cecum of chickens (Dunkley et al., 2007; Józefiak et al., 2010; Sunkara et al., 2011). These results indicate that the greater family of SCFAs share a physiochemical factor that mediates the inhibitory effect observed in these studies, and further experimentation is required to identify and potentially optimize this approach. Furthermore, these data indicate that elevating the physiological concentrations of SCFAs in the gut may be a practical way to increase the observed reduction in transconjugants in vivo.
To mitigate the complexities of the in vivo gut environment and to overcome the limitations of in vitro models’ ability to account for complex host factors, we repeated conjugation assays in a chicken ceca explant model (Ott and Mellata, 2022). Chicken ceca explants were collected from two-week-old chickens from a commercial farm and used as substrate in broth conjugation reactions using donor and recipient populations in the presence of ddH2O or 0.025 M of each SCFA (Figure 4). To eliminate the possibility that pH changes due to the addition of SCFAs, not SCFAs themselves, may have caused the significant changes observed in our experiments, ex vivo explant conjugations were completed in a pH buffered solution, which mitigates changes in pH.
Similarly to our in vitro data, we observed little to no effect on the donor and recipient populations for each SCFA tested, with the exemption of a small but significant increase in donors in the valerate treatment group, and a small but significant rise in recipients in the 2-methylbutyrate treatment group. This data indicates that each of the eight SCFAs may have potential as a COIN in host-associated environments. Our data warrant future follow up in vivo studies to further examine the role of SCFA in regulating the incidence and rate of bacterial plasmid transfer in the gut of living hosts with intact resident microbiota.
In addition to the role of host association in microbial activity, the biology of bacterial plasmid transfer also relies on the plasmids’ genetic content (Gama et al., 2017; Virolle et al., 2020; Ott et al., 2021). As for the plasmids used in this study, each encodes their own transfer machinery, enabling them to be individually sufficient for both replication and transfer within the donor cell. However, the specific proteins and genes involved in the replication and transfer of each plasmid vary (Shintani et al., 2015). The genetic differences dictate the breadth of possible conjugal recipients and may lead to differential responses to COINs. To determine if SCFAs offer a COIN effect on a plasmid specific basis, we tested three plasmids of both broad and narrow host range types for conjugation under exposure to SCFAs. We observed a universal reduction in total transconjugant populations for each plasmid type tested in vitro (IncP1ε, IncFIβ, and IncI1) (Figure 5). These data indicate that SCFAs do have a universal mechanism of action for inhibiting plasmid conjugation outside of targeting specific genes. This may be different than what was previously observed with medium-chain PUFAs or may be additive and provide a synergistic outcome (Li et al., 2021).
Our study models the transfer of both broad and narrow host range MDR plasmids from both pathogenic and commensal E. coli to a representative commensal E. coli from the human gut, which then served as both a reservoir and potential pathobiont strain with potential virulence and pathogenic capacity (Salyers et al., 2004; Sommer et al., 2009; Penders et al., 2013; Rolain, 2013). The IncP pKJK5 plasmid is a MDR plasmid that maintains determinants of resistance to both tetracycline and sulfasalazine class antibiotics (Table 1) (Klümper et al., 2015). Likewise, both pCVM29188_146 and pAPEC-O2-211A-ColV are narrow host range (IncF) plasmids that maintain multiple AMR and virulence genes and are associated with food production hosts, such as chickens (Table 1) (Fricke et al., 2009; Nielsen et al., 2018). The ability of AMR and virulent plasmids from broad host range, like IncP plasmids, to transfer to a wide range of recipient organisms, including Gram-positive and Gram-negative bacteria of multiple genera and species (Klümper et al., 2015), could lead to diverse severe infections and antibiotic treatment failure. Thus, treatment like ours using SCFAs that significantly reduce and even eliminate plasmid transfer would highly benefit human health by preventing complications from infections and even death.
Overall, this study demonstrates SCFAs as a potential universal conjugation inhibitor during in vitro broth conjugations and chicken ceca explant co-culture experiments. While this does not directly demonstrate the in vivo action of SCFAs as COINs, it may hint at their role in the gut as COINs and identify them as potential novel therapeutic or dietary interventions to mitigate the emergence and spread of MDR strains in the gut. Continued studies on both the complete mechanism of action of SCFA COIN and in vivo application of SCFAs are ongoing.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was approved by Iowa State University, Institutional Animal Care Use Committee Protocol IACUC-21-265. The study was conducted in accordance with the local legislation and institutional requirements.
LCO: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. MM: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work is supported by the United States Department of Agriculture (USDA) National Institute of Food and Agriculture AFRI Predoctoral Fellowship IOW05679 to LCO and USDA Hatch project IOW05700-NC1202 and IOW04202 to MM.
The authors would like to thank Mark Engelken and Julia Marks for their contribution to conducting conjugation assay experiments associated with this study.
LCO and MM are listed as inventors on a pending patent covering research presented in this paper.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aguado-Urda, M., Gibello, A., Blanco, M. M., López-Campos, G. H., Cutuli, M. T., and Fernández-Garayzábal, J. F. (2012). Characterization of plasmids in a human clinical strain of Lactococcus garvieae. PLoS One 7:e40119. doi: 10.1371/journal.pone.0040119
Al-Ghamdi, M. S., El-Morsy, F., Al-Mustafa, Z. H., Al-Ramadhan, M., and Hanif, M. (1999). Antibiotic resistance of Escherichia coli isolated from poultry workers, patients and chicken in the Eastern Province of Saudi Arabia. Trop. Med. Int. Health 4, 278–283. doi: 10.1046/J.1365-3156.1999.00392.X
Anjum, M., Madsen, J. S., Espinosa-Gongora, C., Jana, B., Wiese, M., Nielsen, D. S., et al. (2018). A culture-independent method for studying transfer of IncI1 plasmids from wild-type Escherichia coli in complex microbial communities. J. Microbiol. Methods 152, 18–26. doi: 10.1016/J.MIMET.2018.07.009
Baxter, N. T., Schmidt, A. W., Venkataraman, A., Kim, K. S., Waldron, C., and Schmidt, T. M. (2019). Dynamics of human gut microbiota and short-chain fatty acids in response to dietary interventions with three fermentable fibers. MBio 10, 10:e02566-18. doi: 10.1128/mBio.02566-18
BDSC (2019). Bloomington Drosophila Stock Center: BDSC Cornmeal Food. Available at: https://bdsc.indiana.edu/information/recipes/index.html (Accessed May 19, 2021).
Bortolaia, V., Espinosa-Gongora, C., and Guardabassi, L. (2016). Human health risks associated with antimicrobial-resistant enterococci and Staphylococcus aureus on poultry meat. Clin. Microbiol. Infect. 22, 130–140. doi: 10.1016/J.CMI.2015.12.003
Brock, M., Fischer, R., Linder, D., and Buckel, W. (2000). Methylcitrate synthase from aspergillus nidulans: implications for propionate as an antifungal agent. Mol. Microbiol. 35, 961–973. doi: 10.1046/J.1365-2958.2000.01737.X
Cabezn, E., de la Cruz, F., and Arechaga, I. (2017). Conjugation inhibitors and their potential use to prevent dissemination of antibiotic resistance genes in Bacteria. Front. Microbiol. 8, 1–7. doi: 10.3389/fmicb.2017.02329
Candon, S., Perez-Arroyo, A., Marquet, C., Valette, F., Foray, A.-P., Pelletier, B., et al. (2015). Antibiotics in early life Alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes. PLoS One 10:e0125448. doi: 10.1371/journal.pone.0125448
Card, R. M., Cawthraw, S. A., Nunez-Garcia, J., Ellis, R. J., Kay, G., Pallen, M. J., et al. (2017). An in vitro chicken gut model demonstrates transfer of a multidrug resistance plasmid from Salmonella to commensal Escherichia coli. mBio 8, 1–15. doi: 10.1128/mBio.00777-17
Cohen, J. (2009). Non-antibiotic strategies for Sepsis. Clin. Microbiol. Infect. 15, 302–307. doi: 10.1111/J.1469-0691.2009.02753.X
Dolejska, M., and Papagiannitsis, C. C. (2018). Plasmid-mediated resistance is going wild. Plasmid 99, 99–111. doi: 10.1016/j.plasmid.2018.09.010
Dowson, C. G., Hutchison, A., Brannigan, J. A., George, R. C., Hansman, D., Linares, J., et al. (1989). Horizontal transfer of penicillin-binding protein genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. 86, 8842–8846. doi: 10.1073/pnas.86.22.8842
Du, K., Bereswill, S., and Heimesaat, M. M. (2021). A literature survey on antimicrobial and immune-modulatory effects of butyrate revealing non-antibiotic approaches to tackle bacterial infections. Eur. J. Microbiol. Immunol. (Bp) 11, 1–9. doi: 10.1556/1886.2021.00001
Dunkley, K. D., Dunkley, C. S., Njongmeta, N. L., Callaway, T. R., Hume, M. E., Kubena, L. F., et al. (2007). Comparison of in vitro fermentation and molecular microbial profiles of high-fiber feed substrates incubated with chicken Cecal Inocula. Poult. Sci. 86, 801–810. doi: 10.1093/ps/86.5.801
Elshaghabee, F. M. F., Rokana, N., Elshaghabee, F. M. F., and Rokana, N. (2021). “Dietary management by probiotics, prebiotics and synbiotics for the prevention of antimicrobial resistance” in Sustainable Agriculture Reviews, vol. 49.
Fernandez-Lopez, R., Machón, C., Longshaw, C. M., Martin, S., Molin, S., Zechner, E. L., et al. (2005). Unsaturated fatty acids are inhibitors of bacterial conjugation. Microbiology (NY) 151, 3517–3526. doi: 10.1099/mic.0.28216-0
Fricke, W. F., McDermott, P. F., Mammel, M. K., Zhao, S., Johnson, T. J., Rasko, D. A., et al. (2009). Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica Serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75, 5963–5971. doi: 10.1128/AEM.00786-09
Fukuda, A., Usui, M., Okamura, M., Dong-Liang, H., and Tamura, Y. (2019). Role of flies in the maintenance of antimicrobial resistance in farm environments. Microb. Drug Resist. 25, 127–132. doi: 10.1089/mdr.2017.0371
Gama, J. A., Zilhão, R., and Dionisio, F. (2017). Conjugation efficiency depends on intra and intercellular interactions between distinct plasmids: plasmids promote the immigration of other plasmids but repress co-colonizing plasmids. Plasmid 93, 6–16. doi: 10.1016/J.PLASMID.2017.08.003
García-Cazorla, Y., Getino, M., Sanabria-Ríos, D. J., Carballeira, N. M., De La Cruz, F., Arechaga, I., et al. (2018). Conjugation inhibitors compete with palmitic acid for binding to the conjugative traffic ATPase TrwD, providing a mechanism to inhibit bacterial conjugation. J. Biol. Chem. 293, 16923–16930. doi: 10.1074/jbc.RA118.004716
Ghouri, F., Hollywood, A., and Ryan, K. (2018). A systematic review of non-antibiotic measures for the prevention of urinary tract infections in pregnancy. BMC Pregnancy Childbirth 18, 1–10. doi: 10.1186/S12884-018-1732-2/TABLES/6
González-Ortiz, G., Olukosi, O. A., Jurgens, G., Apajalahti, J., and Bedford, M. R. (2020). Short-chain fatty acids and ceca microbiota profiles in broilers and turkeys in response to diets supplemented with phytase at varying concentrations, with or without xylanase. Poult. Sci. 99, 2068–2077. doi: 10.1016/J.PSJ.2019.11.051
Graf, F. E., Palm, M., Warringer, J., and Farewell, A. (2018). Inhibiting conjugation as a tool in the fight against antibiotic resistance. Drug Dev. Res. 80, 19–23. doi: 10.1002/ddr.21457
Grohmann, E., Muth, G., and Espinosa, M. (2003). Conjugative plasmid transfer in gram-positive Bacteria. Microbiol. Mol. Biol. Rev. 67, 277–301. doi: 10.1128/mmbr.67.2.277-301.2003
Guevarra, R. B., Cho, J. H., Cho, J. H., Lee, J. H., Kim, H., Kim, S., et al. (2021). Oral vaccination against Lawsonia intracellularis changes the intestinal microbiome in weaned piglets. Animals 11:2082. doi: 10.3390/ANI11072082
Guiyoule, A., Gerbaud, G., Buchrieser, C., Galimand, M., Rahalison, L., Chanteau, S., et al. (2001). Transferable plasmid-mediated resistance to streptomycin in clinical isolate of Yersinia pestis. Emerg. Infect. Dis. 7, 43–48. doi: 10.3201/eid0701.010106
Guynot, M. E., Ramos, A. J., Sanchis, V., and Marín, S. (2005). Study of benzoate, propionate, and sorbate salts as Mould spoilage inhibitors on intermediate moisture bakery products of low pH (4.5–5.5). Int. J. Food Microbiol. 101, 161–168. doi: 10.1016/J.IJFOODMICRO.2004.11.003
Hedman, H. D., Vasco, K. A., and Zhang, L. (2020). A review of antimicrobial resistance in poultry farming within low-resource settings. Animals 10, 1–39. doi: 10.3390/ani10081264
Hofer, U. (2019). The cost of antimicrobial resistance. Nat. Rev. Microbiol. 17:3. doi: 10.1038/s41579-018-0125-x
Hong, B. K., Wang, M., Chi, H. P., Kim, E. C., Jacoby, G. A., and Hooper, D. C. (2009). oqxAB encoding a multidrug efflux pump in human clinical isolates of Enterobacteriaceae. Antimicrob. Agents Chemother. 53, 3582–3584. doi: 10.1128/AAC.01574-08
Huddleston, J. R. (2014). Horizontal gene transfer in the human gastrointestinal tract: potential spread of antibiotic resistance genes. Infect. Drug Resist. 7, 167–176. doi: 10.2147/IDR.S48820
Jochum, J. M., Redweik, G. A. J., Ott, L. C., and Mellata, M. (2021). Bacteria broadly-resistant to last resort antibiotics detected in commercial chicken farms. Microorganisms 9, 1–16. doi: 10.3390/microorganisms9010141
Józefiak, D., Rutkowski, A., Kaczmarek, S., Jensen, B. B., Engberg, R. M., and Højberg, O. (2010). Effect of β-Glucanase and xylanase supplementation of barley- and Rye-based diets on Caecal microbiota of broiler chickens. Br. Poult. Sci. 51, 546–557. doi: 10.1080/00071668.2010.507243
Kim, C. H., Park, J., and Kim, M. (2014). Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw. 14, 277–288. doi: 10.4110/in.2014.14.6.277
Klümper, U., Riber, L., Dechesne, A., Sannazzarro, A., Hansen, L. H., Sørensen, S. J., et al. (2015). Broad host range plasmids can invade an unexpectedly diverse fraction of a soil bacterial community. ISME J. 9, 934–945. doi: 10.1038/ismej.2014.191
Krogdahl, A. (1985). Digestion and absorption of lipids in poultry. J. Nutr. 115, 675–685. doi: 10.1093/jn/115.5.675
Lacey, J. A., Keyburn, A. L., Ford, M. E., Portela, R. W., Johanesen, P. A., Lyras, D., et al. (2017). Conjugation-mediated horizontal gene transfer of Clostridium perfringens plasmids in the chicken gastrointestinal tract results in the formation of new virulent strains. Appl. Environ. Microbiol. 83, 1–13. doi: 10.1128/AEM.01814-17
Langfeld, L. Q., Du, K., Bereswill, S., and Heimesaat, M. M. (2021). A review of the antimicrobial and immune-modulatory properties of the gut microbiota derived short chain fatty acid propionate – what is new? Eur. J. Microbiol. Immunol. (Bp) 11, 50–56. doi: 10.1556/1886.2021.00005
Lerner, A., Matthias, T., and Aminov, R. (2017). Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 8:27. doi: 10.3389/fimmu.2017.01630
Li, G., Xia, L. J., Zhou, S. Y., Wang, X. R., Cui, C. Y., He, Y. Z., et al. (2021). Linoleic acid and α-linolenic acid inhibit conjugative transfer of an IncX4 plasmid carrying mcr-1. J. Appl. Microbiol. 130, 1893–1901. doi: 10.1111/jam.14885
Liao, X., Shao, Y., Sun, G., Yang, Y., Zhang, L., Guo, Y., et al. (2020). The relationship among gut microbiota, short-chain fatty acids, and intestinal morphology of growing and healthy broilers. Poult. Sci. 99, 5883–5895. doi: 10.1016/j.psj.2020.08.033
Licht, T. R., Christensen, B. B., Krogfelt, K. A., and Molin, S. (1999). Plasmid transfer in the animal intestine and other dynamic bacterial populations: the role of community structure and environment. Microbiology (NY) 145 (Pt 9), 2615–2622. doi: 10.1099/00221287-145-9-2615
Liu, C. M., Stegger, M., Aziz, M., Johnson, T. J., Waits, K., Nordstrom, L., et al. (2018). Escherichia coli ST131- H 22 as a foodborne Uropathogen. MBio 9:e00470-18. doi: 10.1128/mBio.00470-18
Liu, J., Taft, D. H., Maldonado-Gomez, M. X., Johnson, D., Treiber, M. L., Lemay, D. G., et al. (2019). The fecal Resistome of dairy cattle is associated with diet during nursing. Nat. Commun. 10, 1–15. doi: 10.1038/s41467-019-12111-x
Lopatkin, A. J., Meredith, H. R., Srimani, J. K., Pfeiffer, C., Durrett, R., and You, L. (2017). Persistence and reversal of plasmid-mediated antibiotic resistance. Nat. Commun. 8:1689. doi: 10.1038/s41467-017-01532-1
Lowy, F. D. (2003). Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 111, 1265–1273. doi: 10.1172/JCI18535
Marin, S., Sanchis, V., Sanz, D., Castel, I., Ramos, A. J., Canela, R., et al. (2010). Control of growth and Fumonisin B1 production by fusarium verticillioides and fusarium proliferatum isolates in moist maize with propionate preservatives 16, 555–563. doi: 10.1080/026520399283696
Maruyama, K., and Kitamura, H. (1985). Mechanisms of growth inhibition by propionate and restoration of the growth by sodium bicarbonate or acetate in Rhodopseudomonas sphaeroides S. J. Biochem. 98, 819–824. doi: 10.1093/oxfordjournals.jbchem.a135340
McDougall, S., Parker, K. I., Heuer, C., and Compton, C. W. R. (2009). A review of prevention and control of heifer mastitis via non-antibiotic strategies. Vet. Microbiol. 134, 177–185. doi: 10.1016/J.VETMIC.2008.09.026
Mellata, M. (2013). Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends. Foodborne Pathog. Dis. 10, 916–932. doi: 10.1089/fpd.2013.1533
Mellata, M., Ameiss, K., Mo, H., and Curtiss, R. I. I. I. (2010). Characterization of the contribution to virulence of three large plasmids of avian pathogenic Escherichia coli x7122 (O78:K80:H9). Infect. Immun. 78, 1528–1541. doi: 10.1128/IAI.00981-09
Mellata, M., Johnson, J. R., and Curtiss, R. (2018). Escherichia coli isolates from commercial chicken meat and eggs cause sepsis, meningitis and urinary tract infection in rodent models of human infections. Zoonoses Public Health 65, 103–113. doi: 10.1111/ZPH.12376
Mellata, M., Maddux, J. T., Nam, T., Thomson, N., Hauser, H., Stevens, M. P., et al. (2012). New insights into the bacterial fitness-associated mechanisms revealed by the characterization of large plasmids of an avian pathogenic E. coli. PLoS One 7:e29481. doi: 10.1371/journal.pone.0029481
Midtvedt, T., Carlstedt-Duke, B., Høverstad, T., Lingaas, E., Norin, E., Saxerholt, H., et al. (1986). Influence of Peroral antibiotics upon the Biotransformatory activity of the intestinal microflora in healthy subjects. Eur. J. Clin. Investig. 16, 11–17. doi: 10.1111/j.1365-2362.1986.tb01300.x
Millan, A. S. (2018). Evolution of plasmid-mediated antibiotic resistance in the clinical context. Trends Microbiol. 26, 978–985. doi: 10.1016/j.tim.2018.06.007
Mir, R. A., Schaut, R. G., Allen, H. K., Looft, T., Loving, C. L., Kudva, I. T., et al. (2019). Cattle intestinal microbiota shifts following Escherichia coli O157:H7 vaccination and colonization. PLoS One 14:e0226099. doi: 10.1371/JOURNAL.PONE.0226099
Mitchell, N. M., Johnson, J. R., Johnston, B., Curtiss, R. III, and Mellata, M. (2015). Zoonotic potential of Escherichia coli isolates from retail chicken meat products and eggs. Appl. Environ. Microbiol. 81, 1177–1187. doi: 10.1128/AEM.03524-14
Mobasseri, G., JuTeh, C. S., Ooi, P. T., and Thong, K. L. (2019). The emergence of Colistin-resistant Klebsiella pneumoniae strains from swine in Malaysia. J. Glob. Antimicrob. Resist. 17, 227–232. doi: 10.1016/j.jgar.2018.12.015
Mueller, N. T., Zhang, M., Juraschek, S. P., Miller, E. R., and Appel, L. J. (2020). Effects of high-Fiber diets enriched with carbohydrate, protein, or unsaturated fat on circulating short chain fatty acids: results from the OmniHeart randomized trial. Am. J. Clin. Nutr. 111, 545–554. doi: 10.1093/ajcn/nqz322
Nielsen, D. W., Mangiamele, P., Ricker, N., Barbieri, N. L., Allen, H. K., Nolan, L. K., et al. (2018). Complete genome sequence of avian pathogenic Escherichia coli strain APEC O2-211. Microbiol. Resour. Announc. 7:e01046-18. doi: 10.1128/MRA.01046-18
Nordmann, P., and Poirel, L. (2005). Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56, 463–469. doi: 10.1093/jac/dki245
Ott, L. C., Engelken, M., Scott, S. M., McNeill, E. M., and Mellata, M. (2021). Drosophila model for gut-mediated horizontal transfer of narrow- and broad-host-range plasmids. mSphere 6:6. doi: 10.1128/mSphere.00698-21
Ott, L. C., and Mellata, M. (2022). Models for gut-mediated horizontal gene transfer by bacterial plasmid conjugation. Front. Microbiol. 13:2371. doi: 10.3389/FMICB.2022.891548
Ott, L. C., Stromberg, Z. R., Redweik, G. A. J., Wannemuehler, M. J., and Mellata, M. (2020). Mouse genetic background affects transfer of an antibiotic resistance plasmid in the gastrointestinal tract. mSphere 5, 1–13. doi: 10.1128/mSphere.00847-19
Parada Venegas, D., de la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Penders, J., Stobberingh, E. E., Savelkoul, P. H. M., and Wolffs, P. F. G. (2013). The human microbiome as a reservoir of antimicrobial resistance. Front. Microbiol. 4:87. doi: 10.3389/fmicb.2013.00087
Popowska, M., and Krawczyk-Balska, A. (2013). Broad-host-range IncP-1 plasmids and their resistance potential. Front. Microbiol. 4:44. doi: 10.3389/fmicb.2013.00044
Ramírez, M., Amate, L., and Gil, A. (2001). Absorption and distribution of dietary fatty acids from different sources. Early Hum. Dev. 65, S95–S101. doi: 10.1016/S0378-3782(01)00211-0
Rasko, D. A., Rosovitz, M. J., Myers, G. S. A. A., Mongodin, E. F., Fricke, W. F., Gajer, P., et al. (2008). The Pangenome structure of Escherichia coli: comparative genomic analysis of E. coli; commensal and pathogenic isolates. J. Bacteriol. 190, 6881–6893. doi: 10.1128/JB.00619-08
Redondo-Salvo, S., Fernández-López, R., Ruiz, R., Vielva, L., de Toro, M., Rocha, E. P. C., et al. (2020). Pathways for horizontal gene transfer in Bacteria revealed by a global map of their plasmids. Nat. Commun. 11:3602. doi: 10.1038/s41467-020-17278-2
Ripoll-Rozada, J., Garca-Cazorla, Y., Getino, M., Machn, C., Sanabria-Ros, D., de la Cruz, F., et al. (2016). Type IV traffic ATPase TrwD as molecular target to inhibit bacterial conjugation. Mol. Microbiol. 100, 912–921. doi: 10.1111/mmi.13359
Robicsek, A., Jacoby, G. A., and Hooper, D. C. (2006). The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6, 629–640. doi: 10.1016/S1473-3099(06)70599-0
Roca, I., Akova, M., Baquero, F., Carlet, J., Cavaleri, M., Coenen, S., et al. (2015). The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 6, 22–29. doi: 10.1016/j.nmni.2015.02.007
Rolain, J.-M. (2013). Food and human gut as reservoirs of transferable antibiotic resistance encoding genes. Front. Microbiol. 4:173. doi: 10.3389/fmicb.2013.00173
Ruzin, A., and Novick, R. P. (1998). Glycerol Monolaurate inhibits induction of vancomycin resistance in Enterococcus faecalis. J. Bacteriol. 180, 182–185. doi: 10.1128/jb.180.1.182-185.1998
Sadowska, J. M., Genoud, K. J., Kelly, D. J., and O’Brien, F. J. (2021). Bone biomaterials for overcoming antimicrobial resistance: advances in non-antibiotic antimicrobial approaches for regeneration of infected osseous tissue. Mater. Today 46, 136–154. doi: 10.1016/J.MATTOD.2020.12.018
Sakata, T., and Yajima, T. (1984). Influence of short chain fatty acids on the epithelial cell division of digestive tract. Q. J. Exp. Physiol. 69, 639–648. doi: 10.1113/expphysiol.1984.sp002850
Salyers, A., Gupta, A., and Wang, Y. (2004). Human intestinal Bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416. doi: 10.1016/j.tim.2004.07.004
Schulthess, J., Pandey, S., Capitani, M., Rue-Albrecht, K. C., Arnold, I., Franchini, F., et al. (2019). The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–445.e7. doi: 10.1016/j.immuni.2018.12.018
Shintani, M., Sanchez, Z. K., and Kimbara, K. (2015). Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front. Microbiol. 6:242. doi: 10.3389/FMICB.2015.00242/ABSTRACT
Skyberg, J. A., Johnson, T. J., Johnson, J. R., Clabots, C., Logue, C. M., and Nolan, L. K. (2006). Acquisition of avian pathogenic Escherichia coli plasmids by a commensal Escherichia coli isolate enhances its abilities to kill chicken embryos, grow in human urine, and colonize the murine kidney. Infect. Immun. 74, 6287–6292. doi: 10.1128/IAI.00363-06
Soares, T. C. S., Paes, A. C., Megid, J., Ribolla, P. E. M., dos Santos Paduan, K., and Gottschalk, M. (2014). Antimicrobial susceptibility of Streptococcus suis isolated from clinically healthy swine in Brazil. Can. J. Vet. Res. 78, 145–149
Solis-Cruz, B., Hernandez-Patlan, D. M., Hargis, B. M., and Tellez, G. (2020). Use of Prebiotics as an Alternative to Antibiotic Growth Promoters in the Poultry Industry [Internet]. Prebiotics and Probiotics - Potential Benefits in Nutrition and Health. in IntechOpen. doi: 10.5772/intechopen.89053
Sommer, M. O. A., Dantas, G., and Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131. doi: 10.1126/science.1176950
Suhr, K. I., and Nielsen, P. V. (2004). Effect of weak acid preservatives on growth of bakery product spoilage Fungi at different water activities and pH values. Int. J. Food Microbiol. 95, 67–78. doi: 10.1016/J.IJFOODMICRO.2004.02.004
Sunkara, L. T., Achanta, M., Schreiber, N. B., Bommineni, Y. R., Dai, G., Jiang, W., et al. (2011). Butyrate enhances disease resistance of chickens by inducing antimicrobial host defense peptide gene expression. PLoS One 6:e27225. doi: 10.1371/journal.pone.0027225
Tan, J., McKenzie, C., Potamitis, M., Thorburn, A. N., Mackay, C. R., and Macia, L. (2014). The Role of Short-Chain Fatty Acids in Health and Disease. Advances in Immunology [Internet]. 121, 91–119. Available from: https://www.sciencedirect.com/science/article/pii/B9780128001004000039?via%3Dihub
Tang, Y., Dai, L., Sahin, O., Wu, Z., Liu, M., and Zhang, Q. (2017). Emergence of a plasmid-borne multidrug resistance gene cfr(C) in foodborne pathogen Campylobacter. J. Antimicrob. Chemother. 72, 1581–1588. doi: 10.1093/jac/dkx023
Techitnutsarut, P., and Chamchod, F. (2021). Modeling bacterial resistance to antibiotics: bacterial conjugation and drug effects. Adv. Differ. Equat. 2021:290. doi: 10.1186/s13662-021-03423-8
Teuber, M. (1999). Spread of antibiotic resistance with food-borne pathogens. Cell. Mol. Life Sci. 56, 755–763. doi: 10.1007/s000180050022
Threlfall, E. J., Ward, L. R., Frost, J. A., and Willshaw, G. A. (2000). The emergence and spread of antibiotic resistance in food-borne Bacteria. Int. J. Food Microbiol. 62, 1–5. doi: 10.1016/S0168-1605(00)00351-2
van den Bogaard, A. E., London, N., Driessen, C., and Stobberingh, E. E. (2001). Antibiotic resistance of Faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemother. 47, 763–771. doi: 10.1093/jac/47.6.763
Vinolo, M. A. R., Rodrigues, H. G., Nachbar, R. T., and Curi, R. (2011). Regulation of inflammation by short chain fatty acids. Nutrients 3, 858–876. doi: 10.3390/nu3100858
Virolle, C., Goldlust, K., Djermoun, S., Bigot, S., and Lesterlin, C. (2020). Plasmid transfer by conjugation in gram-negative bacteria: from the cellular to the community level. Genes (Basel) 11, 1–33. doi: 10.3390/genes11111239
Walugembe, M., Hsieh, J. C. F., Koszewski, N. J., Lamont, S. J., Persia, M. E., and Rothschild, M. F. (2015). Effects of dietary fiber on cecal short-chain fatty acid and cecal microbiota of broiler and laying-hen chicks. Poult. Sci. 94, 2351–2359. doi: 10.3382/ps/pev242
Wang, M., Tran, J., and Jacoby, G. (2003). Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrobial Agents 47, 2242–2248. doi: 10.1128/AAC.47.7.2242-2248.2003
Willetts, N., and Wilkins, B. (1984). Processing of plasmid DNA during bacterial conjugation. Microbiol. Rev. 48, 24–41. doi: 10.1128/MR.48.1.24-41.1984
Wisener, L. V., Sargeant, J. M., O’Sullivan, T. L., O’Connor, A. M., McEwen, S. A., Reist, M., et al. (2021). Non-antibiotic approaches for disease prevention and control in nursery pigs: A scoping review. Front. Vet. Sci. 8:227. doi: 10.3389/FVETS.2021.620347/BIBTEX
Woolhouse, M., Ward, M., Van Bunnik, B., and Farrar, J. (2015). Antimicrobial resistance in humans, livestock and the wider environment. Philos. Trans. R. Soc. B Biol. Sci. 370:20140083. doi: 10.1098/rstb.2014.0083
Keywords: horizontal gene transfer, short-chain fatty acids, plasmid transfer inhibition, antimicrobial resistance, bacterial plasmid conjugation
Citation: Ott LC and Mellata M (2024) Short-chain fatty acids inhibit bacterial plasmid transfer through conjugation in vitro and in ex vivo chicken tissue explants. Front. Microbiol. 15:1414401. doi: 10.3389/fmicb.2024.1414401
Received: 08 April 2024; Accepted: 22 May 2024;
Published: 06 June 2024.
Edited by:
Umer Zeeshan Ijaz, University of Glasgow, United KingdomReviewed by:
Chang-Wei Lei, Sichuan University, ChinaCopyright © 2024 Ott and Mellata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Melha Mellata, bW1lbGxhdGFAaWFzdGF0ZS5lZHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.