
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 24 July 2024
Sec. Terrestrial Microbiology
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1411264
This article is part of the Research Topic Soilborne Pathogenic Fungi: Systematics, Pathogenesis and Disease Control View all 8 articles
The diversity of Ganoderma remains largely unexplored, with little information available due to fungiphobia and the morphological plasticity of the genus. To address this gap, an ongoing study aims to collect and identify species with this genus using nuclear ribosomal DNA regions called the “Internal Transcribed Spacer” (ITS1-5.8S-ITS2 = ITS). In this study, a new species, Ganoderma segmentatum sp. nov., was discovered on the dead tree trunk of the medicinal plant, Vachellia nilotica. The species was identified through a combination of morpho-anatomical characteristics and phylogenetic analyses. This new species was closely related to Ganoderma multipileum, G. mizoramense, and G. steyaertanum, with a 99% bootstrap value, forming a distinct branch in the phylogenetic tree. Morphologically, G. segmentatum can be distinguished by its frill-like appearance on the margin of basidiome. Wilt or basal stem rot, a serious disease of trees caused by Ganoderma species and V. nilotica, is brutally affected by this disease, resulting in substantial losses in health and productivity. This Ganoderma species severely damages V. nilotica through deep mycelial penetration in the upper and basal stems of the host species. Pathogenic observational descriptions of G. segmentatum on dead tree trunks showed the exudation of viscous reddish-brown fluid from the basal stem portion, which gradually extended upward. Symptoms of this disease include decay, stem discoloration, leaf drooping, and eventual death, which severely damaged the medicinal tree of V. nilotica.
The members of the genus Ganoderma P. Karst. (Basidiomycota, Polyporales) can be found all over the world and play a diverse role in forest ecosystems (Coetzee et al., 2015; Sun et al., 2022). The tropical diversity of Ganoderma is not completely clear, as shown by the current number of novelties (Wang et al., 2009; Xing et al., 2018; Cabarroi-Hernández et al., 2019; Luangharn et al., 2019). The synonymization in the genus Ganoderma indicates that the present nomenclatural situation is unsatisfactory and confused, and its improvement needs continuous endeavor (Hong and Jung, 2004; Wang et al., 2009; Fryssouli et al., 2020). Ganoderma has been investigated at macro- and micro-levels, and some existing records are purely based on morphological descriptions. The main morpho-anatomical characteristics of Ganoderma species are shelf-like basidiomata, laccate appearance, and a maroon-brown color (Ahmad, 1956; Steyaert, 1972; Irshad et al., 2012; Fakhar-ud-Din and Mukhtar, 2019). Unfortunately, no DNA sequences are available for a few records, and there is an urgent need to use molecular methods to study the Ganoderma species (Umar et al., 2021a).
Molecular phylogeny provides interesting progress in this genus (Volk, 1992; Moncalvo, 2000; Hong and Jung, 2004). Most sequences of ITS rDNA have been analyzed to delimit and resolve the confusion surrounding this species in Ganoderma complexes (Hong and Jung, 2004; Cabarroi-Hernández et al., 2019). The internal transcribed spacer (ITS: ITS1-5.8S-ITS2) shows maximum efficiency in solving the problems regarding species identification among Ganoderma taxon (Fryssouli et al., 2020). ITS presents a clear barcoding gap to sort out the morphological and biological species concepts; therefore, it can be used for identification (Badotti et al., 2017).
Ganoderma, a hazardous white rot fungus present on living trees, possesses cell wall-degrading enzymes to degrade major wood components such as lignin, cellulose, and hemicellulose (Schwarze et al., 2000). Many Ganoderma species are pathogenic, causing butt and stem rot diseases of woody plants and crops (Moncalvo, 2000; Elliott et al., 2001; Sahebi et al., 2017). Saprophytic Ganoderma species are highly ligninolytic in nature due to white rot (Volk, 1992). In natural ecosystems, laccase plays an important role in lignin production and degradation in plants. Laccases have garnered multiple industrial interests, e.g., xenobiotic bioremediation and detoxification of phenolics (Wang et al., 2015). This enzyme has been presented in many Ganoderma species and is associated with diverse biological events, especially in the life cycle of white-rotter's mycelium development, pigmentation, sporulation, and fruiting body formation (Jin et al., 2018). Ganoderma species are emerging as major fungal pathogens. Laccases of these species have been assigned other biological functions, e.g., plant pathogenesis, lignin degradation, and provides understanding of how a fungal saprophyte becomes a dangerous pathogen. It is an intrinsic attribute for an opportunistic pathogen to realize that its regulatory pathways have evolved under environmental rather than host pressure. Nevertheless, its production pathway defines the pathogen's ability to cause host damage because evolution allows for appropriate survival in the environment and pathogenicity in the host plant (Zhu and Williamson, 2004).
Higher plants assimilate laccases in lignin biosynthesis and polymerization (a structural compound), which further facilitate plant development and defense of plant tissues in response to pathogenic species (Arregui et al., 2019; Westrick et al., 2024). Interestingly, Ganoderma, a wood rotter, secretes laccase during lignin degradation, in which terminal phenolic lignin is directly oxidized by laccases (Westrick et al., 2024).
Many diseases are caused by different pathogens in multiple crops and trees, e.g., Ganoderma sp., which leads to basal and upper stem rot (Fee, 2011). The genus Ganoderma significantly minimizes the yield of medicinal plants, and the world economy experiences a huge product loss, if we fail to protect the medicinal plants (Corley and Tinker, 2008). Degradation by Ganoderma slowly leads to the decay of tree trunks, a process where lignin and cellulose are diminished with the passage of time. Trunks of trees die due to the infectious pathogenic action of Ganoderma, where lignin and cellulose of the lower bole or root flare become decayed (Sinclair and Lyon, 2005). Proteins and cell wall-degrading enzymes are biological macromolecules involved in fungal life activities (Wu et al., 2017). Fungal attack directly exposed the spongy cellulose by decomposition of lignin, and white rot appeared on the plant's woody tissues (Paterson et al., 2009). The disease of rot starts with the penetration of fungal mycelia into the tree's roots, which is sporadic toward the shoot, and finally, the trunk stands collapse (Rees et al., 2009).
Vachellia nilotica is a tree of 5–20 m long, fissured bark with a dense spheric crown. The stems and branches are usually dark to black in color. This tree has thin, straight, light gray spines in axillary pairs without thorns. Leaves are bipinnate, with pairs of pinnulae, tomentose, and rachis, with a gland at the bottom of the last pair of pinnulae. Flowers are globulous (1.2–1.5 cm dia.), bright golden-yellow in color, and whorly on 2–3 cm long peduncles located at the ends of branches (Wardill et al., 2005). The present study was conducted to analyze the morpho-anatomical characters of basidiomata and the ITS sequence phylogeny of new Ganoderma specimens, which act as a major pathogen of V. nilotica. This species leads to massive economic losses by threatening the medicinal host plants with basal rot diseases.
The Ganoderma specimens were collected in 2019 from the University of the Punjab, New Campus, Lahore, Pakistan (under the authority letter of the Director, Institute of Botany, University of the Punjab). The average annual rainfall is approximately 607 mm, and the temperature is 24°C at this site. This area is covered by Vachellia nilotica. Basidiomata were found at the bases of the live tree trunks of V. nilotica trees, usually after the onset of the rainy season in early summer. We observed microscopic structures from cross-sections of the dried basidiomata in KOH (5%), stained them with Congo red (1%), and viewed them under an MX4300H compound light microscope (Meiji Techo Co., Ltd., Japan). Drawings were made with the aid of a drawing tube. Data on anatomical features were recorded from at least 30 measurements at a magnification of 1000X. Basidiospore measurements were presented as length × width (Nagy et al., 2010). Basidiospores were measured without taking into account the apiculus when they were not shrunk. The morphological descriptions of the microscopic features are in part following the study by Cabarroi-Hernández et al. (2019). Color terms in parentheses were recorded by following Kornerup and Wanscher (1975).
Genomic DNA was extracted from dried specimens using the modified cetyltrimethylammonium bromide (CTAB) procedure (Doyle and Doyle, 1987). ITS (ITS1+5.8S+ITS2 rDNA) was amplified with polymerase chain reaction (PCR) using the primers ITS1 and ITS2 (White et al., 1990). Reaction mixtures (20 μL) contained 0.5 μL DNA template, 8.5 mL of distilled water, 0.5 μL of each primer, and 10 mL PCR mix [DreamTaqGreen PCR Master Mix (2 X), Fermentas]. PCR cycling conditions were 35 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. Sequencing was conducted using the same PCR primers. New sequences were edited using MEGA v. 10.0 (Kumar et al., 2016) and submitted to GenBank under accession numbers MZ666127 and MZ666128.
The newly generated sequences were used to search for the most similar sequences deposited in GenBank using the BLASTn tool. Sequences were aligned using the MAFFT online version (Katoh et al., 2019) and manually adjusted using MEGA v. 10.0 (Tamura et al., 2011). A phylogenetic analysis was conducted via maximum likelihood (ML) and Bayesian inference (BI). ML analyses were performed using RAxML-HPC Blackbox version 8.2.10 (Stamatakis, 2014) implemented on the CIPRES Science Gateway (Miller et al., 2010), with an estimated proportion of invariable sites GTRGAMMA+I and branch support evaluated by 1,000 bootstrap replicates. Bayesian inference was carried out in MrBayes v. 3.2.2 (Ronquist et al., 2012). The best substitution model for tree reconstruction was estimated by both the Akaike information criterion and the Bayesian information criterion jModelTest 2.0 (Darriba et al., 2012). Four Markov chain Monte Carlo chains were run simultaneously, starting from random trees for 2,000,000 generations and sampling every 100th generation. After discarding the first 25% of trees as burn-in phase, a consensus Bayesian tree and Bayesian posterior probabilities (BPP) were determined based on all remaining trees. A BPP above 0.90 was considered a significant value. Trees were visualized and further edited in Tree Graph 2 (Stöver and Müller, 2010).
Malt Extract Agar (MEA) media was prepared by the addition (g/L) of MEA 7, Agar 10, K2HPO4 0.5, KH2PO4 0.5, MgSO4·7H2O 0.5, Peptone 2.5, and Glucose 15 at pH 5.0. Culture media was sterilized (for 25 min at 121°C) and then autoclaved. The autoclaved MEA was augmented with 0.02% guaiacol for typical laccase (phenolic) and 0.04% veratryl alcohol (non-phenolic) for atypical laccase. Chromogen was added to evaluate the atypical and the typical laccase-producing abilities of Ganoderma mycelium. The replicates were incubated at 30°C for 5 days.
Submerged culture broth was designed for peak absorption of laccase in shake flasks. Submerged nutrients broth (g/L), composed of yeast extract 5 g, starch 1 g, MgSO4·7H2O 0.5 g, NaCl 0.5 g, FeSO4·7H2O 0.5 g, CaCl2 0.5 g, and ZnSO4 0.02 g, were taken in shake flasks for the growth of mycelium and incubated at 27± 2°C in static condition for 5 days. The pH range of 8.0 to 11 for blue laccase and 4 to 12 for white laccase was selected. Afterward, absorbance was monitored at 280, 330, 470, and 605 nm (3 min) using a UV-Vis spectrophotometer (Xing et al., 2018). Three replicates were designed for accuracy in the results.
The collected data from various parameters was analyzed and represented by means ± standard deviation (SD). Statistical analysis was calculated using SPSS 18.0 software.
The two ITS rDNA sequences (MZ666127 and MZ666128) of the new species had 0.5% site differences. The phylogenetic tree of G. segmentatum sp. nov. was closely matrixed to G. multipileum, G. mizoramense, and G. steyaertanum B. J. Smith and K. Sivasithamparam with a bootstrap value of 99%. The ITS dataset included 98 nucleotide sequences of Ganoderma species, and one sequence of Amauroderma (A. rude KF372587) was chosen as an outgroup in this tree. There were 697 bp in the dataset. The nucleotide frequencies were A = 22.21%, T/U = 29.35%, C = 23.56%, and G = 24.87%, and the maximum log likelihood was InL-4430,914. The best model, GTR+G+I, selected by jModelTest, was used for BI. Both ML and BI analyses resulted in trees of identical topology, and the differences mostly supported the nodes. The ML tree is shown in Figure 1.
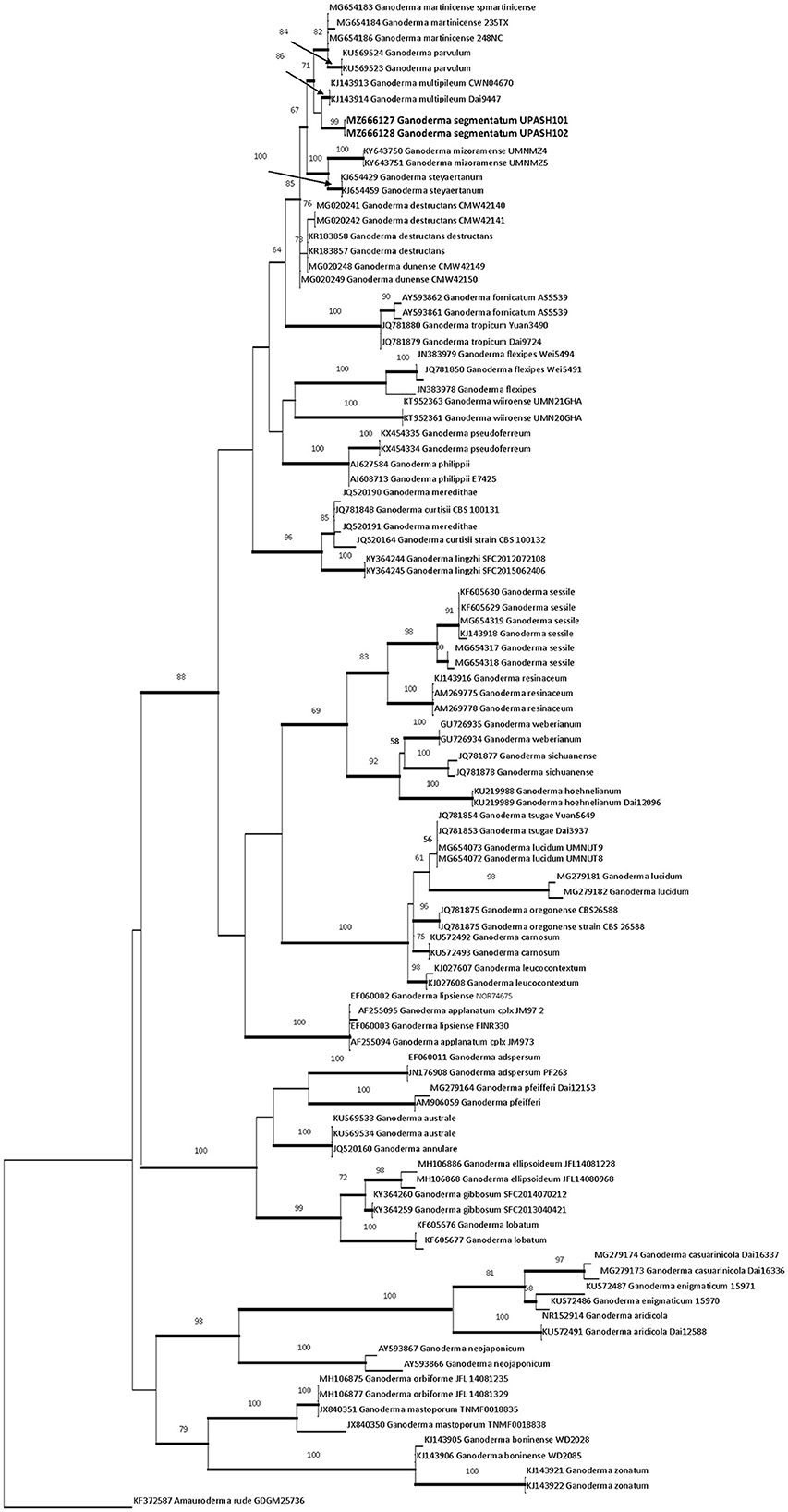
Figure 1. RaXmL tree reconstructed based on ITS rDNA sequences showing the phylogenetic placement of G. segmentatum sp. nov. Maximum likelihood bootstrap (MLB) values higher than 50% (based on 1,000 replicates) are displayed at the nodes, and thick lines represent Bayesian posterior probabilities (BPP) >0.95. The tree is rooted in Amauroderma rude (KF372587). The sequences in bold are from the new species.
Ganoderma segmentatum A. Umar (Figures 2–4)

Figure 2. G. segmentatum sp. nov. (A) basidiome upper surface with a frill-like margin and laccate appearance. (B) milky white lower pore surface. (C) side view of the basidiome with thin margins. (D) caccate dark maroon brown stipe. (E) contextum in layers form and longitudinal tubes below the contextum [scale bars: (A–D) = 3 cm, (E) =0.5 mm].
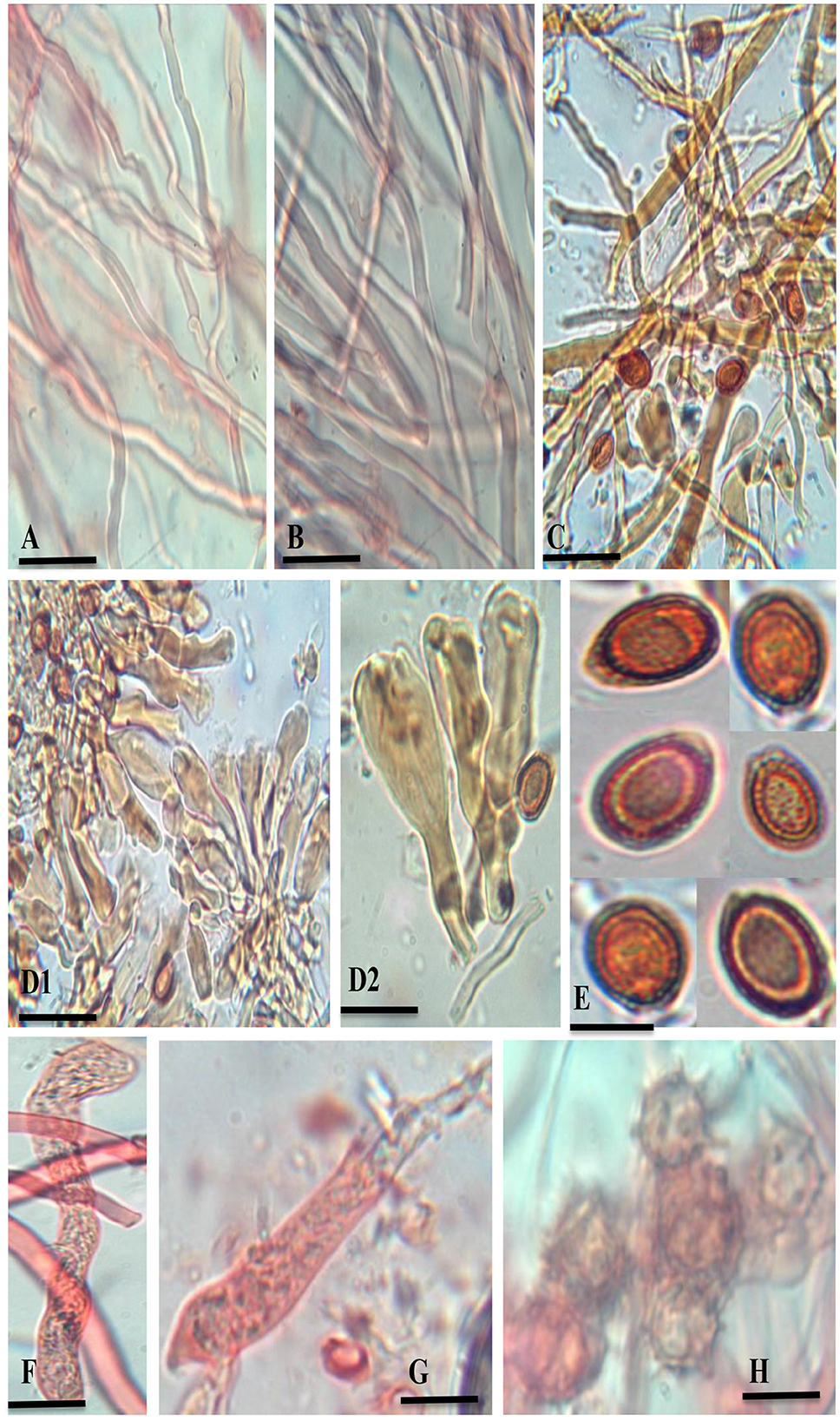
Figure 3. G. segmentatum sp. nov. (A) generative hyphae (clamp connections indicated by an arrow). (B) skeletal hyphae. (C) binding hyphae with multiple branches. (D1, D2) crustohymeniderm cells/cuticle cells taken from basidiomata. (E) broadly ellipsoid, bitunicate, and coarsely echinulated basidiospores. (F, G) basidia. (H) chlamydospores [scale bars: (A–C) =5 μm, (D–G) =10 μm].
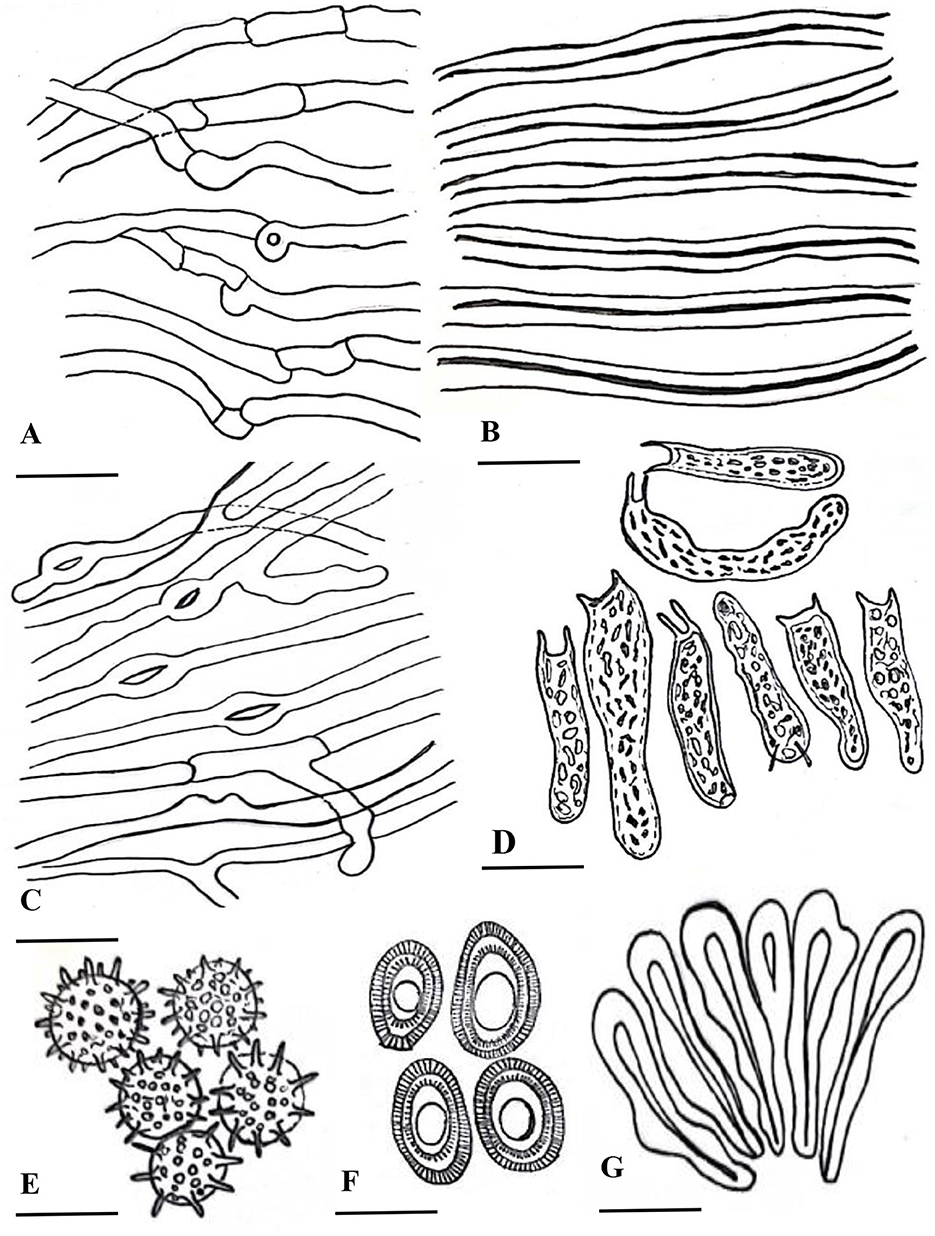
Figure 4. Line drawing of anatomical characters of G. segmentatum. (A) cenerative hyphae. (B) skeletal hyphae. (C) binding hyphae. (D) basidia. (E) chlamydospores. (F) basidiospores. (G) crustohymeniderm cells [scale bars: (A–C) =5 μm, (D–G) = 10 μm].
MycoBank: MB840865
Ganoderma segmentatum is morphologically characterized by annual, dimidiate, stipitate, and brick-red-colored laccate basidiome with a thin, light brown margin. A context without resinous melanoid bands was observed in this species.
Holotype: PAKISTAN. Punjab Province: Lahore, New Campus, University of Punjab (31.4981°N 73.3044°E), elevation 217 m a.s.l., attached to the trunk of the living tree of Vachellia nilotica, 22 June 2018, Aisha Umar (holotype UPASH101). GenBank: ITS = MZ666127.
Etymology: The species epithet “segmentatum” indicates the frill-like appearance at the pileus margin.
Description: “Basidiomata annual, solitary or in group, rigid, dull laccate, stipitate, convex, consistency corky-woody; Pileus 17–18 × 13–13.5 cm, applanate, non-imbricate, dimidiate to flabelliform, laccate, plano convex, hard, shiny upper surface, rugose to verrucose, thin crusted, conspicuously concentrically sulcate to groovy, particularly with frill-like appearance near the margin and around the whole basidiomata; purplish red (14A8), earth colored (5F3), brick red; Stipe 5.5–5.5 × 3.5–3.7 cm, pleuropode, acentric, stout, cylindrical to flat dorso-lateral, dark brown (8F7–6F8) thus darker than the pileus, woody hard, strongly laccate with a thick crust; Margin 0.3–0.5 mm thick, obtuse, entire, few undulations, usually lighter than the rest of the pileus, dull yellow (4A8) to brown (5E8) when fresh, and later cream (4A3) to orange (5A4) in dry condition; Stipe 5.5–5.5 × 3.5–3.7 cm, pleuropode, acentric, stout, cylindrical to flat dorso-lateral, dark brown (8F7–6F8) thus darker than the pileus, woody hard, strongly laccate with a thick crust; Pore surface at first cream (4A3) becomes brown (6E8) to dark brown (6F8) when old, pores almost invisible to the naked eye, subcircular to circular, dissepiments entire and thick, sterile margin concolorous; Tubes 1.1–1.2 cm deep, non-stratified, grayish brown (6E3) with earth-colored (5F3) walls, strongly contrasting with the pore mouth and the context; Context: 0.9–1.2 cm thick, dry, azonate in both pileus and stipe, loose fibrils, rusty brown (6E8) to dark brown (6F8), hard, non-melanoid, separated from the crust by a yellowish orange (4A7) thin line; Hyphal system trimitic: generative hyphae hyaline, thin-walled, septate, with clamp connection, abundant in the dissepiments; somatic hyphae composed of skeletal and binding hyphae; skeletal hyphae thick-walled to solid in the trama; frequently branched, yellowish (2A3), dichotomous to arboriform with thick stalks gradually tapering in the context; and cuticle yellowish (2A3) to brownish (6D6), with thick branches shorter than in the trama; binding hyphae thick-walled, light brown (5D5), randomly bulbous (from middle or near the branch), branched slender, and acutes; Pileipellis: composed of a palisade of vertical club-shaped to broadly clavate, pale yellow, yellowish-brown (5E8) to pale dark brownish (6E8) apices, 38.5–55.7 × 12.2–14.4 μm, smooth, thick-walled to solid, with scant lumen, largely stalked with peduncles continuing as hyaline thick-walled hyphae; Basidia 10.5–20.2 × 2.2–2.7 μm, fusiform to clavate, mostly two-spored with acute sterigmata, thin-walled, content with fine to coarse granular oily content; Basidiospores 8.5–9.6 × 5.2–6.7 μm (average L = 8.84 μm, W = 5.78 μm, Q = L/W = 1.52), broadly ellipsoid, bitunicate, exospore smooth, endospore coarsely echinulate, with turgid vesicular appendix, guttulated, spore print light brown (6D8); Chlamydospores 2.4–2.9 × 2.3–2.6 μm, numerous, round, thick-walled, ornamented with long pillars, hyaline, yellowish (2A3) to reddish brown (8E7).
Another specimen examined: PAKISTAN. PUNJAB PROVINCE: Lahore, New Campus, University of Punjab (31.4981° N 73.3044° E), elevation 217 m a.s.l., attached to a dead tree trunk of Vachellia nilotica, 25 July 2019, Aisha Umar (isotype UPASH102). GenBank: ITS = MZ666128.
Ganoderma segmentatum is reproduced by vegetative mycelia and sexually by spores. Basidiospores are believed to be the source of the inoculum. Spores germinated and formed the monokaryotic vegetative mycelia, which are saprophytically grown in the plant trunk. Dikaryotic mycelium was seen via migration and the nuclear exchange of two different hyphae. The dikaryotic mycelium of Ganoderma species caused a deep and speedy invasion within the plant host. The dikaryotic mycelia formed the large basidiomata under suitable environmental conditions. Basidiomata is a multicellular reproductive body where karyogamy and meiotic spores lead to the completion of the sexual phase (Figure 5). The tetrapolar mating system in sexual reproduction promoted the diversity of genetic content in the same plantation of the genus Ganoderma, leading to population dynamics. This is the primary cause of inefficient disease management. Basal rot management is a big challenge, because the infective dikaryotic mycelium continued the process of penetration into a healthy tree trunk and produced new basidiospores.
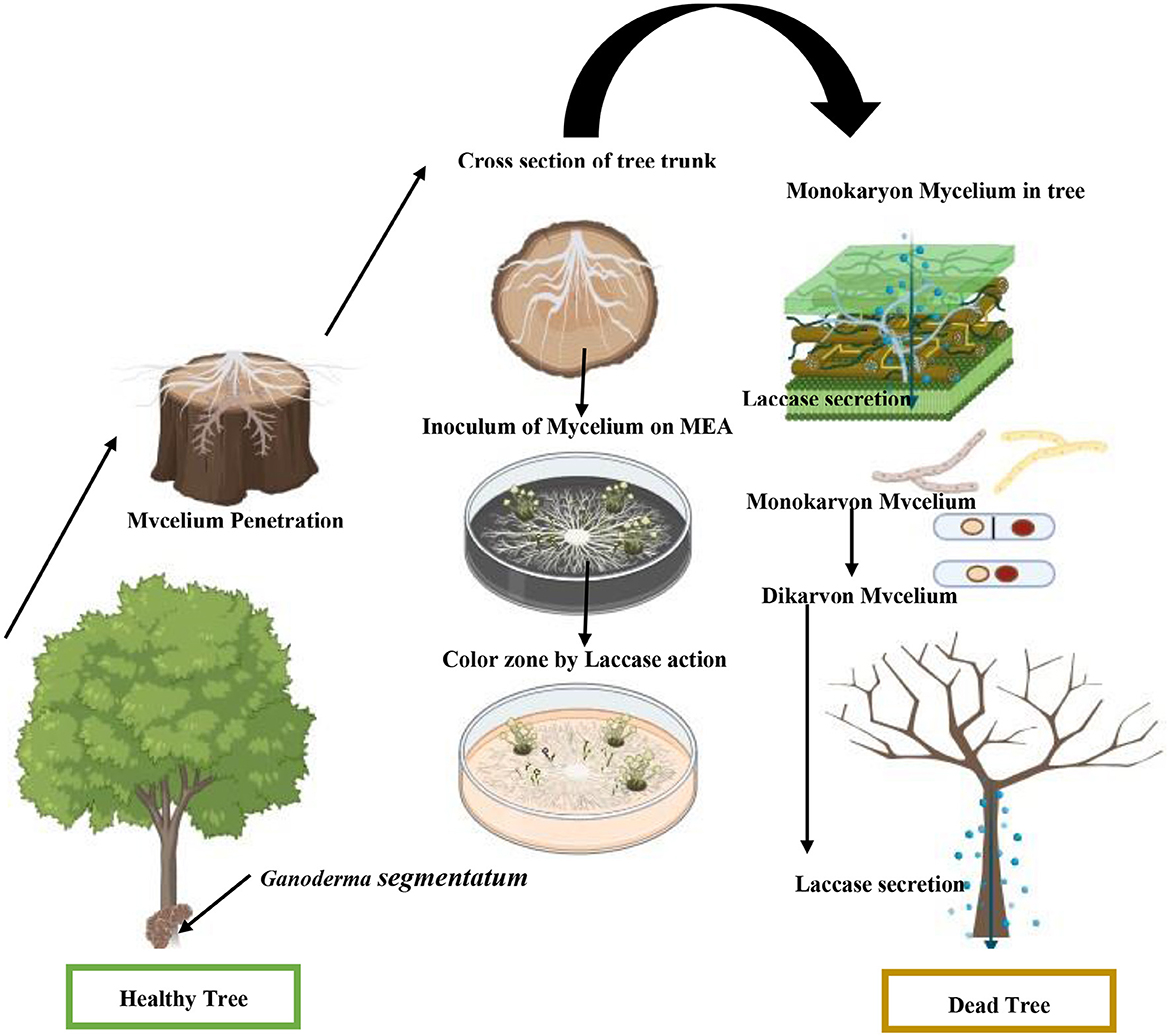
Figure 5. Schematic presentation of stem root causes the decay of V. nilotica by G. segmentatum sp. nov.
Colonization of G. segmentatum with V. nilotica was observed through contact between healthy and diseased roots. The colonization started when the Ganoderma species found a wound for penetration (Figure 5). G. segmentatum initially exhibited the symptoms of reddish-brown viscous fluid exudated from the basal stem portion, which gradually extended upward in direction. In severe cases, the basal portion of the stem decayed completely, and sporophores appeared at the base of the trunk. Brutally rot roots, decay and discoloration of the stem, drooping of all leaves, browning of branches, and death of the plant are the characteristic symptoms of this disease.
The growing patches of BSR infection covered the healthy tree species with the passage of time. The specialized reproductive basidiospores of Ganoderma play an important role in the maintenance of the sexual cycle. Ganoderma segmentatum is characterized by broadly ellipsoid, bitunicate, smooth exospores, endospores that are coarsely echinulated, turgid vesicular appendix, and guttulated basidiospores typically grown in the trunk. Similarly, other white-rot fungal species degrade the wood lignin component during pathogenic action. Basidiomata on the basal stem badly infected the Vachellia nilotica tree. Symptoms of basal stem rot disease on V. nilotica were the collapsing of lower leaves and leaves hanging downward from the point of attachment, and finally, the trunk falling down due to stem decay. Ganoderma segmentatum, as a pathogen, released cell wall-degrading laccase to soften and loosen the host cell wall made up of lignin. The fungal and its laccase facilitated the softening and deep penetration of needle-like microhyphae in the stem of V. nilotica. Microhyphae with extracellular matrix degraded the cellulose components, leading to minute cell wall cracks. This pathogenic interaction of mycelium with living cells and tissues also facilitated the continuous supply of nutrients. Microscopic examination of infected V. nilotica revealed the biotrophic nutrition provided by G. segmentatum during colonization. Needle-like fungal structures puncture the healthy plant cells. Embedded thick and blackish-gray lines were also observed in the host tree.
The laccase in this study was detected in the preliminary test. The engraved mycelium was taken exactly below side of the fruiting body grown on the tree trunk of V. nilotica. Trunk-penetrated mycelia were inoculated on an MEA plate augmented with veratryl alcohol. White laccase transformed the veratryl alcohol into a brown color by oxidation (Figure 6A). Blue laccase formed the reddish-brown oxidation zone on the agar plate after oxidation of guaiacol, indicating the ability of Ganoderma sp. to release the laccase (Figure 6B). It is well known that blue laccases generally react with organic substrates in the presence of mediators (guaiacol), while yellow laccases perform the same act without any mediator molecules. The laccase of Ganoderma species oxidizes the non-phenolic lignin compounds, which cannot be oxidized by the laccase alone. Thus, the oxidation of lignin was dependent on the presence of primary laccase substrates (mediators). In this study, typical blue laccase exhibited a reddish-brown color by reaction with chromogen, while white or yellow laccase did not exhibit any color with guaiacol.
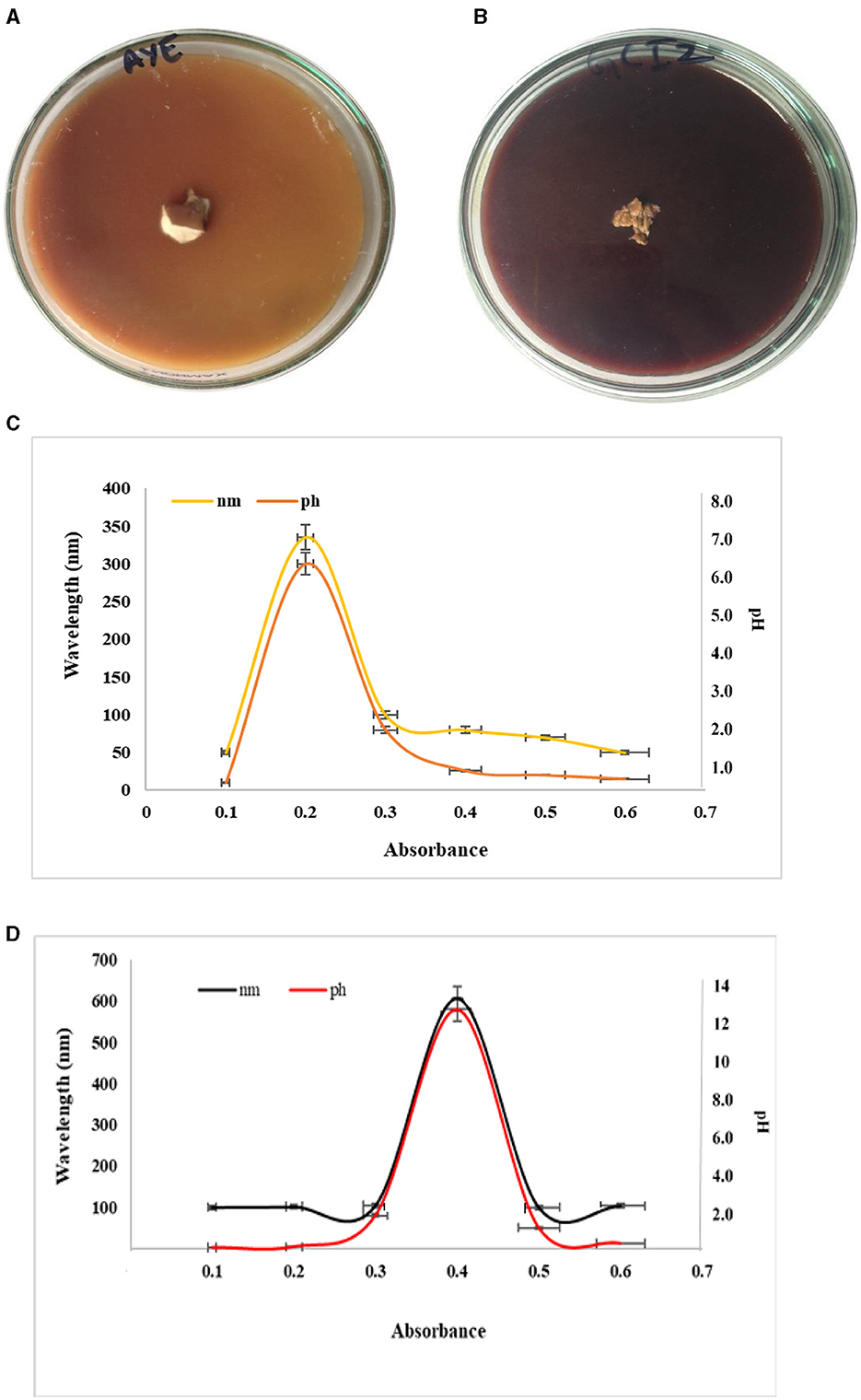
Figure 6. Preliminary plate test for laccase (A) veratryl alcohol, (B) guaiacol, (C) absorbance of laccase, and (D) laccase at a particular pH.
Laccase secreted by mycelium taken from the lower side of the fruiting body exhibited a peak at 340 nm at a pH of 6.5 (Figure 6C). The absorption peak was 600 nm at a pH of 12 for laccase in submerged broth (Figure 6D). The results indicated from the peak that laccase broke down the trunk and caused basal stem rot via the production of white laccase. The peak observed for laccase in the submerged broth indicated blue laccase.
White/yellow laccase had an absorption peak at 330 nm to 400 nm, but no peak was observed at 605 to 610 nm. Yellow laccase can be reduced artificially into blue laccase but it does not exhibit absorption at 600 nm. Conversion of laccase from blue to yellow occurred by the reduction of the type I Cu site via aromatic products or the binding of specific amino acids to the enzyme polypeptide formed during lignin degradation. The reason behind the colorlessness of white laccase is the change in the valence state of the copper ion (Cu2+). White laccase is also considered a member of the laccase family because its primary structure is identical to the known laccase, which uses oxygen as an oxidative agent.
In the ITS phylogeny, the new species clustered together with G. multipileum, G. martinicense, G. mizoramense, and G. destructans M.P.A. Coetzee, Marinc, and M. J. Wingf and G. steyaertanum in a highly supported clade (99%). The global meta-analysis of ITS rDNA Ganoderma sequences (Fryssouli et al., 2020) resolved all five species with a maximum supported (72% ML, 0.97 BPP) lineage. Fryssouli et al. (2020) used the threshold of interspecific values for ITS similarities (≤ 98%), and genetic distance (≥ 0.015) effectively applied to the 21 putatively new phylospecies. The widely adopted thresholds for separating the species in Basidiomycota are < 97% to 98% for ITS sequence similarity, while p-values are > 0.010–0.020 for genetic distances (Zervakis et al., 2019). Therefore, ITS-based phylogenies are helpful in addressing taxonomic issues (in conjunction with other criteria like distinct morpho-anatomical characters).
The present study introduced G. segmentatum as a new member of the genus Ganoderma. It also substantiated the reliability of our ITS phylogeny in resolving Ganoderma species in this clade. The six species in the lineage can be roughly characterized by the geographical origin of their specimens. The specimens of G. destructans originated from South Africa (Darriba et al., 2012; Coetzee et al., 2015). G. martinicense is reported from the USA, Mexico, Cuba, Martinique, Colombia, Brazil, and Argentina (Darriba et al., 2012). G. steyaertanum is distributed in Indonesia and Australia, while G. multipileum is described as including the specimens formerly assigned to tropical Asian Ganoderma lucidum (Stöver and Müller, 2010). As shown by the internal clade resolution, G. multipileum had a strong phylogenetic relationship (86% ML, 0.95 BPP) with G. segmentatum, which was resolved in the sister position. Both species form a sister subclade with G. martinicense. Based on present knowledge, G. multipileum and G. segmentatum have the same geographical origin, especially G. multipileum, which is known from Pakistan. Besides the phylogenetic separation, the two species can be well differentiated by their morphological features. Basidiomata of both species are usually laterally stipitate, while in G. segmentatum, they can be concrete, with pilei often growing together from the lower pilei in G. multipileum (Wang et al., 2009). A group of laccate pilei in G. multipileum overlap, while our new species is less laccate and solitary, appearing on a stem. The basidiomata is maroon-brown in G. multipileum and brick-red in G. segmentatum. In addition, the imbricate and concrete basidiomata of G. multipileum may reach a large size (up to 36 cm long and 54 cm wide) (Wang et al., 2009). Moreover, the pileial surface of G. multipileum showed a range of colors from orange-yellow (4A7) to orange-red (8A8) to brown-red (8C8) and was radially striate (Wang et al., 2009) compared to the sulcate concentrically zonate margin of G. segmentatum.
The most distinguishing features are the size and shape of the basidiospore in the genus Ganoderma (Steyaert, 1972; Kirk et al., 2008; Torres-Torres and Guzmán-Dávalos, 2012; Umar et al., 2021b). The spore size of G. multipileum is larger (8.0–13.2 × 5.5–7.4 μm) than that of our species. On an ITS basis, 26 additional nucleotides are present in G. segmentatum sp. nov. and differentiated by 10 nucleotides from G. multipileum. Therefore, according to the nucleotide basis, G. multipileum is different from our species. Ganoderma martinicense Welti and Courtec. basidiospores are larger (9.5–12 × 5–7 μm) with a golden yellow pileal surface (Welti and Courtecuisse, 2010), contrary to G. segmentatum. One additional nucleotide is present in G. martinicense but absent in our species. Almost seven nucleotides are different in our new species of G. martinicense. The spores of Ganoderma parvulum are longer (8–10 × 5–6 μm) than those of G. segmentatum identified in Pakistan (De Lima Júnior et al., 2014).
G. parvulum is characterized by a pale ochraceous context with resinaceous streaks (Murrill, 1902), while the context is light chocolate brown without resinaceous streaks in G. segmentatum. G. parvulum has two additional nucleotides, while two gaps are present in our species. Both species are differentiated by 17 nucleotides. The basidiomes of Ganoderma destructans and G. steyaertanum exhibited a few similarities, but the spores of G. destructans are slightly larger than those of G. steyaertanum (7.3–12.7 × 5.0–9.5 μm). G. steyaertanum possesses laccate dimidiate basidiocarp, occasionally sessile, umbonate that is dark to chestnut (dark brown) in color, yellowish-white margin, and pale yellow to grayish orange pores (Stöver and Müller, 2010). These characteristics are contrary to those of this new Ganoderma species. G. segmentatum sp. nov. is differentiated from G. steyaertanum by eight nucleotides, while our new species possesses one additional nucleotide and two gaps in nucleotide sequence. Basidiospores of G. destructans are larger (11–14 × 7–9 μm, av. 12.3 × 8.0 μm) than our new species. The pileus surface of G. destructans is covered by white to creamy soft non-poroid tissue of hymenophore, which turns brown when this species becomes old (Coetzee et al., 2015). At the molecular level, analysis of ITS sequence alignments revealed that three additional and five different nucleotides are present in G. segmentatum sp. nov., while they are absent in G. destructans.
The combinatorial mating system of sexual reproduction promoted diversity in the genus Ganoderma. The genetic divergence in the genetic pool was higher, especially in isolates of different geographical origins. Different isolates and strains exhibit a different degree of aggressiveness and patience toward biological control (Kok et al., 2013; Midot et al., 2019; Wong et al., 2021).
In this study, upper and basal stem rot severely affected the Vachellia nilotica plant species. This rot is the primary cause of an inefficient disease control system. Studies indicated that there is a need of compatible partners for Ganoderma species to “mate and initiate” the sexual cycle (heterothallism). Monokaryons possess two “unlinked mating” loci, which initiate the mating process to complete sexual reproduction (ruled by the tetrapolar mating system) (Ramzi et al., 2019). Ganoderma, as a pathogen, releases small cell wall-degrading enzymes, e.g., cellulase, manganese peroxidase, polygalacturonase, and laccase, which soften the host cell wall (Tan et al., 2018). Hyphae of G. boninense colonized the oil palm root by secreting trace amounts of degrading enzymes (polygalacturonase and laccase) (Dhillon et al., 2021). These enzymes degenerate the integrity of the polysaccharides (host cell wall) (Ho et al., 2019). This process starts with the production of multiple response molecules (reactive oxygen species, phytoalexins, and pathogenesis-related proteins) to trigger the alterations in the cell wall. Currently, little study is available on the proteins or enzymes involved in host penetration. Hyphal mating ensures the survival and continuation of the genetic variety of many fungal pathogenic species. Variation in mating facilitates the species in adoption of changed environmental conditions (Morrow and Fraser, 2009). Different studies have reported that Ganoderma is a common disease-causing agent. This species gradually leads to the death and decline of trees (Glen et al., 2009; Elshafie et al., 2013; Bhadra, 2014; Coetzee et al., 2015). The pathogenic action of G. adspersum on young Tilia species and Aesculus sp. was reported in Italy by Nicolotti et al. (2009). Post-inoculation, after 2 years, make the decay columns by drilling the method of inoculation. In New York, Pirone (1957) also performed the pathogenicity tests via agar plugs or fruiting body (pieces) of G. lucidum placed by drilling into the holes of the lower trunk in young Acer platanoides, and symptoms of infection were also observed. Elliott and Broschat (2005) considered Ganoderma zonatum a destructive palm pathogen, similarly, infections or diseases were observed in palms according to Elliott and Uchida (2024). A few Ganoderma species are host-specific and found on certain groups of hosts (palms, conifers, and hardwoods) (Gilbertson and Ryvarden, 1986); For example, Ganoderma meredithiae attacked pines, while Ganoderma curtisii made connections with hardwoods (Elliott and Broschat, 2005; Adaskaveg et al., 2011).
Non-phenolic substrates are oxidized by white/yellow laccases, which are required in the case of blue laccases. Yellow laccase was detected by converting veratryl alcohol into veratraldehyde (Chaurasia et al., 2013a,b). Veratryl alcohol is a secondary metabolite synthesized de novo by white-rot fungi and its low amount enhance the laccase activity (Jensen et al., 1994). Veratryl alcohol, a non-phenolic lignin compound, can be oxidized by the laccase of Coriolus versicolor.
In the reported literature on submerged culture, fungi secreted blue laccases, while secreting yellow laccases when grown on lignin-containing solid substrates. Yellow laccases are better biocatalysts than blue ones. Yellow laccase was produced in Pleurotus ostreatus, G. fornicatum, Panus tigrinus, Phlebia radiate, P. tremellosa, Sclerotinia sclerotiorum (Daroch et al., 2014; Agrawal et al., 2018), G. lucidum, Scytalidium thermophilum (Ben Younes and Sayadi, 2011), Pycnoporus sanguineus (Dantán-González et al., 2008), Cerrena unicolor (Michniewicz et al., 2008), Pycnoporus cinnabarinus (Schliephake et al., 2000), and Myrothecium verrucaria (Zhao et al., 2012). The blue laccase was identified by Trametes versicolor, T. trogii, T. villosa, Rigidoporus lignosus, and Coriolus versicolor (Levin et al., 2010). Phellinus linteuis MTCC-1175 exhibited a peak at 610 nm (Chaurasia et al., 2013a,b), while yellow laccase showed a characteristic spectrum in Coriolopsis floccosa MTCC-1177 (Chaurasia et al., 2013a,b).
It was previously observed that the yellow laccases were obtained from cultures grown on a solid-state medium, while the blue forms isolated from cultures grown on a liquid medium without lignin (Daroch et al., 2014). The authors suggested that the yellow laccase can be fabricated by the modification of blue laccase via 1. low-molecular-weight lignin decomposition products; 2. glycosylation; 3. turnover-dependent oxidation of the active site; 4. amino acids; and 5. copper ligands. This mediator can be obtained from the culture medium, from which the enzyme was isolated (Dantán-González et al., 2008; Ben Younes and Sayadi, 2011; Agrawal et al., 2018).
The morpho-anatomical and molecular study of Ganoderma segmentatum indicated that it is a new species matrixed separately in the clade of Ganoderma species with a strong bootstrap value (99%). Ganoderma is a globally distributed genus comprising species associated with forest ecology and medicinal trees. The Ganoderma species are facultative parasites on living, dead, or rotting trees. They also cause the white rot of hardwoods by decomposing cellulose, lignin, and polysaccharides. The decay of roots and lower trunk or stem flares leads to hazardous tree conditions and tree failures, resulting in serious damage to property and life.
It is clear that Ganoderma are ecologically indispensable, but their pathogenic nature causes tree diseases. Although this new species is a major contributor to the genus Ganoderma and its pathogenic relationship with host plants has brutally damaged the V. nilotica.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
AU: Writing – original draft, Writing – review & editing. WY: Writing – original draft. JL: Writing – review & editing. FA: Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was partly supported by the Chongqing Municipal Education Commission (KJQN201900533, KJQN202300518). This research was funded by Research Supporting Project Number (RSP2024R364), King Saud University, Riyadh, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor IA declared a past co-authorship with the author AU.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Adaskaveg, J., Blanchette, R., and Gilbertson, R. (2011). Decay of date palm wood by white-rot and brown-rot fungi. Canad. J. Bot. 69, 615–629. doi: 10.1139/b91-083
Agrawal, K., Chaturvedi, V., and Verma, P. (2018). Fungal laccase discovered but yet undiscovered. Bioresour. Bioproc. 5:4. doi: 10.1186/s40643-018-0190-z
Ahmad, S. (1956). Fungi of Pakistan, mon. I. Biological Society of Pakistan. Lahore: Laboratories, Government College, 126.
Arregui, L., Ayala, M., Gómez-Gil, X., Gutiérrez-Soto, G., Hernández-Luna, C. E., Herrera De Los Santos, M., et al. (2019). Laccases: structure, function, and potential application in water bioremediation. Microb. Cell Factor. 18, 1–33. doi: 10.1186/s12934-019-1248-0
Badotti, F., de Oliveira, F. S., Garcia, C. F., Vaz, A. B. M., Fonseca, P. L. C., Nahum, L. A., et al. (2017). Effectiveness of ITS and sub-regions as DNA barcode markers for the identification of Basidiomycota (Fungi). BMC Microbiol. 17:42. doi: 10.1186/s12866-017-0958-x
Ben Younes, S., and Sayadi, S. (2011). Purification and characterization of a novel trimeric and thermotolerant laccase produced from the ascomycete Scytalidium thermophilum strain. J. Mol. Catal. B: Enzymatic 73, 35–42. doi: 10.1016/j.molcatb.2011.07.014
Bhadra, M. (2014). Ganoderma association with the mortality of Acacia auriculiformis, susceptibility to different hosts and its controls. J. Plant Pathol. Microbiol. 05:238. doi: 10.4172/2157-7471.1000238
Cabarroi-Hernández, M., Villalobos-Arámbula, A. R., Torres-Torres, M. G., Decock, C., and Guzmán-Dávalos, L. (2019). The Ganoderma weberianum-resinaceum lineage: multilocus phylogenetic analysis and morphology confirm G. mexicanum and G. parvulum in the Neotropics. MycoKeys 59, 95–131. doi: 10.3897/mycokeys.59.33182
Chaurasia, P. K., Yadav, A., Yadav, R. S., and Yadava, S. (2013a). Purification and characterization of laccase from Coriolopsis floccosa MTCC-1177 and its use in the selective oxidation of aromatic methyl group to aldehyde without mediators. J. Chem. Sci. 125, 1395–1403. doi: 10.1007/s12039-013-0525-4
Chaurasia, P. K., Yadav, A., Yadav, S. S., and Yadava, S. (2013b). Purification and characterization of laccase secreted by Phellinus linteus MTCC-1175 and its role in the selective oxidation of aromatic methyl group. Prikl. Biokhim. Mikrobiol. 49, 592–599. doi: 10.1134/S000368381306006
Coetzee, M. P. A., Marincowitz, S., Muthelo, V. G., and Wingfield, M. J. (2015). Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus 6, 249–256. doi: 10.5598/imafungus.2015.06.01.16
Dantán-González, E., Vite-Vallejo, O., Martínez-Anaya, C., Méndez-Sánchez, M., González, M. C., Palomares, L. A., et al. (2008). Production of two novel laccase isoforms by a thermotolerant strain of Pycnoporus sanguineus isolated from an oil-polluted tropical habitat. Int. Microbiol. 11, 163–169. doi: 10.2436/20.1501.01.56
Daroch, M., Houghton, C. A., Moore, J. K., Wilkinson, M. C., Carnell, A. J., Bates, A. D., et al. (2014). Glycosylated yellow laccases of the basidiomycete Stropharia aeruginosa. Enzyme Microb. Technol. 58–59, 1–7. doi: 10.1016/j.enzmictec.2014.02.003
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. (2012). jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9, 772–772. doi: 10.1038/nmeth.2109
De Lima Júnior, N. C., Baptista Gibertone, T., and Malosso, E. (2014). Delimitation of some neotropical laccate Ganoderma (Ganodermataceae): molecular phylogeny and morphology. Rev. Biol. Trop. 62, 1197–1208. doi: 10.15517/rbt.v62i3.12380
Dhillon, B., Hamelin, R. C., and Rollins, J. A. (2021). Transcriptional profile of oil palm pathogen, Ganoderma boninense, reveals activation of lignin degradation machinery and possible evasion of host immune response. BMC Genom. 22, 326. doi: 10.1186/s12864-021-07644-9
Doyle, J. J., and Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19, 11–15
Elliott, M. L., and Broschat, T. K. (2005). Ganoderma Butt Rot of Palms. Gainesville: University of Florida IFAS Extension Bulletin, 5.
Elliott, M. L., Broschat, T. K., Elliott, M. L., and Broschat, T. K. (2001). Observations and pathogenicity experiments on Ganoderma zonatum in Florida. Palms 45, 62–72.
Elliott, M. L., and Uchida, J. Y. (2024). Diseases and disorders of ornamental palms. Phytopathol News. doi: 10.1094/APSnetFeature-2004-0304
Elshafie, A. E., Al-Bahry, S. N., El-Nagerabi, S. A. F., and Al-Kindi, K. K. (2013). New record of Ganoderma colossum associated with Sclerocarya birrea dieback. Australas. Plant Dis. Notes 8, 85–87. doi: 10.1007/s13314-013-0102-1
Fakhar-ud-Din, and Mukhtar, T. (2019). Morphological characterization of Ganoderma species from Murree hills of Pakistan. Plant Protect. 3:0128. doi: 10.33804/pp.003.02.0128
Fee, C. G. (2011). Management of Ganoderma Diseases in Oil Palm Plantations. Kuala Lumpur: Sustainable Agriculture-an insight into Ganoderma, 325–339.
Fryssouli, V., Zervakis, G. I., Polemis, E., and Typas, M. A. (2020). A global meta-analysis of ITS rDNA sequences from material belonging to the genus Ganoderma (Basidiomycota, Polyporales) including new data from selected taxa. MycoKeys 75, 71–143. doi: 10.3897/mycokeys.75.59872
Gilbertson, R., and Ryvarden, L. (1986). North American Polypores: Volume I &II. Oslo, Norway: Fungi Flora.
Glen, M., Bougher, N. L., Francis, A. A., Nigg, S. Q., Lee, S. S., Irianto, R., et al. (2009). Ganoderma and Amauroderma species associated with root-rot disease of Acacia mangium plantation trees in Indonesia and Malaysia. Australas. Plant Path. 38, 345–356. doi: 10.1071/AP09008
Ho, C. L., Tan, Y. C., Yeoh, K. A., Lee, W. K., Ghazali, A. K., Yee, W. Y., et al. (2019). Leaf transcriptome of oil palm (Elaeis guineensis Jacq.) infected by Ganoderma boninense. Trees. 33, 943–950. doi: 10.1007/s00468-019-01830-9
Hong, S. G., and Jung, H. S. (2004). Phylogenetic analysis of Ganoderma based on nearly complete mitochondrial small-subunit ribosomal DNA sequences. Mycologia 96, 742–755. doi: 10.1080/15572536.2005.11832922
Irshad, M., Anwar, Z., Gulfraz, M., Butt, H. I., Ejaz, A., and Nawaz, H. (2012). Purification and characterization of α-amylase from Ganoderma tsuage growing in waste bread medium. Afr. J. Biotechnol. 11, 8288–8294. doi: 10.5897/AJB11.3643
Jensen, K. A., Evans, K. M., Kirk, T. K., and Hammel, K. E. (1994). Biosynthetic pathway for veratryl alcohol in the ligninolytic fungus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 60, 709–714. doi: 10.1128/aem.60.2.709-714.1994
Jin, W., Li, J., Feng, H., You, S., Zhang, L., Norvienyeku, J., et al. (2018). Importance of a laccase gene (Lcc1) in the development of Ganoderma tsugae. Int. J. Mol Sci. 19, 471. doi: 10.3390/ijms19020471
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformat. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kirk, P. M., Cannon, P. F., Minter, D. W., and Stalpers, J. A. (2008). Dictionary of the Fungi.10th edn. Wallingford: CAB International.
Kok, S. M., Goh, Y. K., Jiat, T., Goh, K., Wei Chee, W., and Goh, Y. K. (2013). In vitro growth of Ganoderma boninense isolates on novel palm extract medium and virulence on oil palm (Elaeis guineensis) seedlings. Malays. J. Microbiol. 9, 33–42. doi: 10.21161/mjm.45212
Kornerup, A., and Wanscher, J. H. (1975). Taschenlexikon der Farben, 1400 Farbnuancen und 600 Farbnamen. Zürich-Göttingen: Musterschmidt-verlag, 242.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Levin, L., Melignani, E., and Ramos, A. M. (2010). Effect of nitrogen sources and vitamins on ligninolytic enzyme production by some white-rot fungi. Dye decolorization by selected culture filtrates. Bioresour. Technol. 101, 4554–4563. doi: 10.1016/j.biortech.2010.01.102
Luangharn, T., Karunarathna, S. C., Mortimer, P. E., Hyde, K. D., and Xu, J. (2019). Additions to the knowledge of Ganoderma in Thailand: Ganoderma casuarinicola, a new record; and Ganoderma thailandicum sp. nov. MycoKeys 59, 47–65. doi: 10.3897/mycokeys.59.36823
Michniewicz, A., Ledakowicz, S., Ullrich, R., and Hofrichter, M. (2008). Kinetics of the enzymatic decolorization of textile dyes by laccase from Cerrena unicolor. Dyes Pigm. 77, 295–302. doi: 10.1016/j.dyepig.2007.05.015
Midot, F., Lau, S. Y. L., Wong, W. C., Tung, H. J., Yap, M. L., Lo, M. L., et al. (2019). Genetic diversity and demographic history of Ganoderma boninense in oil palm plantations of Sarawak, Malaysia inferred from ITS regions. Microorganisms 7:464. doi: 10.3390/microorganisms7100464
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). “Creating the CIPRES Science Gateway for inference of large phylogenetic trees,” in 2010 Gateway Computing Environments Workshop (GCE) (San Diego, CA: Supercomputer Center), 1–8.
Moncalvo, J. M. (2000). “Systematics of Ganoderma,” in Ganoderma Diseases of Perennial Crops, 23–45.
Morrow, C. A., and Fraser, J. A. (2009). Sexual reproduction and dimorphism in the pathogenic basidiomycetes. FEMS Yeast Res. 9, 161–177. doi: 10.1111/j.1567-1364.2008.00475.x
Murrill, W. A. (1902). The Polyporaceae of North America. I. The genus Ganoderma. Bull. Torrey Bot. Club 29, 599–608.
Nagy, G. L., Vágvölgyi, C., and Papp, T. (2010). Type studies and nomenclatural revisions in Parasola (Psathyrel laceae) and related taxa. Mycotaxon 112, 103–141. doi: 10.5248/112.103
Nicolotti, G., Gonthier, P., Guglielmo, F., and Garbelotto, M. M. (2009). A biomolecular method for the detection of wood decay fungi: a focus on tree stability assessment. J. Arboric. 35:14. doi: 10.48044/jauf.2009.004
Paterson, R. R. M., Moen, S., and Lima, N. (2009). The feasibility of producing oil palm with altered lignin content to control Ganoderma disease. J. Phytopathol. 157, 649–656. doi: 10.1111/j.1439-0434.2009.01553.x
Pirone, P. P. (1957). Ganoderma lucidum, a parasite of shade trees. Bull. Torrey Bot. Club 84, 424–428. doi: 10.2307/2482973
Ramzi, A. B., Che Me, M. L., Ruslan, U. S., Baharum, S. N., and Nor Muhammad, N. A. (2019). Insight into plant cell wall degradation and pathogenesis of Ganoderma boninense via comparative genome analysis. PeerJ 7:e8065. doi: 10.7717/peerj.8065
Rees, R. W., Flood, J., Hasan, Y., Potter, U., and Cooper, R. M. (2009). Basal stem rot of oil palm (Elaeis guineensis); mode of root infection and lower stem invasion by Ganoderma boninense. Plant Pathol. 58, 982–989. doi: 10.1111/j.1365-3059.2009.02100.x
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Sahebi, M., Hanafi, M. M., van Wijnen, A. J., Akmar, A. S. N., Azizi, P., Idris, A. S., et al. (2017). Profiling secondary metabolites of plant defence mechanisms and oil palm in response to Ganoderma boninense attack. Int. Biodeter. Biodegrad. 122, 151–164. doi: 10.1016/j.ibiod.2017.04.016
Schliephake, K., Mainwaring, D. E., Lonergan, G. T., Jones, I. K., and Baker, W. L. (2000). Transformation and degradation of the disazo dye Chicago Sky Blue by a purified laccase from Pycnoporus cinnabarinus. Enzyme Microb. Technol. 27, 100–107. doi: 10.1016/S0141-0229(00)00181-2
Sinclair, W. A., and Lyon, H. H. (2005). Diseases of Trees and Shrubs. Ithaca, NY: Comstock Publishing Associates.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steyaert, R. L. (1972). Species of Ganoderma and related genera mainly of the Bogor and Leiden Herbaria. Persoonia 7, 55–118.
Stöver, B. C., and Müller, K. F. (2010). TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformat. 11, 7. doi: 10.1186/1471-2105-11-7
Sun, Y.-F., Lebreton, A., Xing, J.-H., Fang, Y.-X., Si, J., Morin, E., et al. (2022). Phylogenomics and comparative genomics highlight specific genetic features in Ganoderma Species. J Fungi. 8:311. doi: 10.3390/jof8030311
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tan, J. S., Lee, Y. P., Sulaiman, S., Camus-Kulandaivelu, L., Klopp, C., Mercière, M., et al. (2018). The route to the development of basal stem rot resistance in oil palm (Elaeis guineensis) via the discovery of lignin degradation process in the pathogen Ganoderma boninense. Acta Hortic. 359–370. doi: 10.17660/ActaHortic.2018.1205.42
Torres-Torres, M. G., and Guzmán-Dávalos, L. (2012). The morphology of Ganoderma species with a laccate surface. Mycotaxon 119, 201–216. doi: 10.5248/119.201
Umar, A., Ahmed, S., and Bashir, H. (2021a). Ganoderma leucocontextum, a new record from Pakistan. Mycotaxon 136, 529–539. doi: 10.5248/136.529
Umar, A., Ahmed, S., and Gafforov, Y. (2021b). Ganoderma pakistanicum sp. nov. (Ganodermataceae, Basidiomycota) from Pakistan. Nova Hedwigia 531–543. doi: 10.1127/nova_hedwigia/2021/0656
Volk, T. J. (1992). Review of genera of polypores: nomenclature and taxonomy: synopsis fungorum 5. Mycologia 84, 950–951. doi: 10.2307/3760304
Wang, D.-M., Wu, S.-H., Su, C.-H., Peng, J.-T., Shih, Y.-H., and Chen, L.-C. (2009). Ganoderma multipileum, the correct name for “G. lucidum” in tropical Asia. Bot. Stud. 50, 451–458.
Wang, W., Liu, F., Jiang, Y., Wu, G., Guo, L., Chen, R., et al. (2015). The multigene family of fungal laccases and their expression in the white rot basidiomycete Flammulina velutipes. Gene 563, 142–149. doi: 10.1016/j.gene.2015.03.020
Wardill, T. J., Graham, G. C., Zalucki, M., Palmer, W. A., Playford, J., and Scott, K. D. (2005). The importance of species identity in the biocontrol process: identifying the subspecies of Acacia nilotica (Leguminosae: Mimosoideae) by genetic distance and the implications for biological control. J. Biogeogr. 32, 2145–2159. doi: 10.1111/j.1365-2699.2005.01348.x
Welti, S., and Courtecuisse, R. (2010). The Ganodermataceae in the French West Indies (Guadeloupe and Martinique). Fungal Diver. 43, 103–126. doi: 10.1007/s13225-010-0036-2
Westrick, N. M., Dominguez, E. G., Bondy, M., Hull, C. M., Smith, D. L., and Kabbage, M. (2024). A single laccase acts as a key component of environmental sensing in a broad host range fungal pathogen. Commun Biol. 7:348. doi: 10.1038/s42003-024-06034-7
White, T., Bruns, T., Lee, S., Taylor, J., Innis, M., Gelfand, D., et al. (1990). “Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics,” in Pcr Protocols: a Guide to Methods and Applications, 315–322.
Wong, W. C., Tung, H. J., Fadhilah, M. N., Midot, F., Lau, S. Y. L., Melling, L., et al. (2021). Genetic diversity and gene flow amongst admixed populations of Ganoderma boninense, causal agent of basal stem rot in African oil palm (Elaeis guineensis Jacq.) in Sarawak (Malaysia), Peninsular Malaysia, and Sumatra (Indonesia). Mycologia 113, 902–917. doi: 10.1080/00275514.2021.1884815
Wu, S., Ren, X., Li, Y., Guo, W., Lei, X., Yao, J., et al. (2017). Effect of dietary Astragalus Polysaccharide supplements on testicular miRNA expression profiles and enzymatic changes of breeder cocks. Sci. Rep. 7:38864. doi: 10.1038/srep38864
Xing, J.-H., Sun, Y.-F., Han, Y.-L., Cui, B.-K., and Dai, Y.-C. (2018). Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. MycoKeys 7, 93–108. doi: 10.3897/mycokeys.34.22593
Zervakis, G. I., Venturella, G., Fryssouli, V., Inglese, P., Polemis, E., and Gargano, M. L. (2019). Pleurotus opuntiae revisited - an insight to the phylogeny of dimitic Pleurotus species with emphasis on the P. djamor complex. Fungal Biol. 123, 188–199. doi: 10.1016/j.funbio.2018.12.005
Zhao, D., Zhang, X., Cui, D., and Zhao, M. (2012). Characterisation of a novel white laccase from the deuteromycete fungus Myrothecium verrucaria NF-05 and its decolourisation of dyes. PLoS ONE 7:e38817. doi: 10.1371/journal.pone.0038817
Keywords: laccate fungi, paleotropic species, basal rot, ITS rDNA, Ganodermataceae, laccase
Citation: Umar A, Yuan W, Lu J and Ameen F (2024) Fungal–plant interaction: a pathogenic relationship between Ganoderma segmentatum sp. nov. and Vachellia nilotica. Front. Microbiol. 15:1411264. doi: 10.3389/fmicb.2024.1411264
Received: 02 April 2024; Accepted: 09 July 2024;
Published: 24 July 2024.
Edited by:
Iftikhar Ali, Columbia University, United StatesReviewed by:
Yijuan Ding, Southwest University, ChinaCopyright © 2024 Umar, Yuan, Lu and Ameen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aisha Umar, YXNoLmRyODhAZ21haWwuY29t; Junxing Lu, anVueGx1QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.