
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 22 July 2024
Sec. Microbial Physiology and Metabolism
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1409771
Cyanobacteria have great potential in CO2-based bio-manufacturing and synthetic biological studies. The filamentous cyanobacterium, Leptolyngbya sp. strain BL0902, is comparable to Arthrospira (Spirulina) platensis in commercial-scale cultivation while proving to be more genetically tractable. Here, we report the analyses of the whole genome sequence, gene inactivation/overexpression in the chromosome and deletion of non-essential chromosomal regions in this strain. The genetic manipulations were performed via homologous double recombination using either an antibiotic resistance marker or the CRISPR/Cpf1 editing system for positive selection. A desD-overexpressing strain produced γ-linolenic acid in an open raceway photobioreactor with the productivity of 0.36 g·m−2·d−1. Deletion mutants of predicted patX and hetR, two genes with opposite effects on cell differentiation in heterocyst-forming species, were used to demonstrate an analysis of the relationship between regulatory genes in the non-heterocystous species. Furthermore, a 50.8-kb chromosomal region was successfully deleted in BL0902 with the Cpf1 system. These results supported that BL0902 can be developed into a stable photosynthetic cell factory for synthesizing high value-added products, or used as a model strain for investigating the functions of genes that are unique to filamentous cyanobacteria, and could be systematically modified into a genome-streamlined chassis for synthetic biological purposes.
The goal of synthetic biology is to build biological systems that are able to perform the desired functions for various purposes, such as producing chemicals (Ro et al., 2006) or biofuels (d’Espaux et al., 2015), detecting trace contaminants (Webster et al., 2014), creating new disease models or treatment methods (Ruder et al., 2011; Hutmacher et al., 2015), or studying basic science issues (Davies, 2017). To this end, synthetic biology adopts a bottom up approach to build biological systems with the ideal characteristics or to reconstruct the existing natural systems by learning from the engineering principles of design and construction (Mukherji and van Oudenaarden, 2009). The engineered biological systems are based on the availability of standardized and characterized bioparts, from which biodevices and then complex biosystems are assembled. This would also need a host and a set of models for a predictable outcome (Mukherji and van Oudenaarden, 2009). Therefore, synthetic biology is implemented by making genetic alterations in the host cells, while the host serves as a platform for achieving design goals using various tools and strategies. The basic requirements for a host include rapid and robust growth, complete genome sequence and gene annotation, facile genetic engineering and in-depth research in physiology and genetics (Kim et al., 2017). Initial studies in synthetic biology depended on model heterotrophic organisms such as Escherichia coli and Saccharomyces cerevisiae. With the constant extension of application fields of synthetic biology, the hosts for various purposes would have more specific requirements. For example, when used as a cell factory for production of components or additives of cosmetics, medicines and foods, a host suitable for safe, green and industrializable bio-manufacturing would be required.
Cyanobacteria are a group of oxygen-evolving photosynthetic prokaryotes that utilize the light as the energy and CO2 as the carbon source, and are major participants in the geochemical cycles of carbon, nitrogen and oxygen (Hamilton et al., 2016; Lobus and Kulikovskiy, 2023). There is a great interest in engineering cyanobacteria as hosts for photosynthetic conversion of CO2 into high value-added products. In recent years, some laboratory model strains, for instance, Synechococcus elongatus PCC 7942, Synechococcus sp. PCC 7002 and Synechocystis sp. PCC 6803, have been utilized as hosts for the production of biofuels, polymers, pigments and many other value-added chemicals, such as isoprene (Gao et al., 2016), ethanol (Kopka et al., 2017), PHB (Koch et al., 2020), astaxanthin (Diao et al., 2020), limonene (Lin et al., 2017), squalene (Choi et al., 2017), amino acids (Korosh et al., 2017; Brey et al., 2020), fatty acids (Włodarczyk et al., 2020). These efforts have demonstrated the enormous potential of cyanobacteria as model organisms for carbon-negative synthetic biology (Tan et al., 2022). However, these strains have been limited in commercial applications, due to slow growth (relative to heterotrophic bacteria), poor resistance to adversity, predation by protozoa, etc. Therefore, the development of more robust and industrializable strains to serve as cyanobacteria model organisms is critical for future commercial processes.
There are some cyanobacterial strains with superior growth traits meeting the requirements of commercial production. For example, Arthrospira (Spirulina) platensis is a filamentous cyanobacterium with the characteristics of high safety, high protein content, fast growth, convenient harvest, and excellent environmental adaptability, thus is commercially farmed worldwide as a food source (Ahmad et al., 2023). It has long been hoped to be developed as a host platform for syntheses of proteins and chemicals. A recent report showed that exogenous genes cloned on plasmids were transformed into this cyanobacterium with the aid of companion bacteria, integrated into the chromosome via homologous double crossover and efficiently expressed (Jester et al., 2022). However, the described genetic transformation was not based on colony formation on plates, and the segregation process appeared to be very time consuming. Such a technical bottleneck may limit the development of more sophisticated genetic systems for synthetic biology. An alternative filamentous cyanobacterium suitable for large-scale cultivation, called Leptolyngbya sp. BL0902 (hereafter Leptolyngbya BL0902), was initially isolated from an algal production raceway pond (Taton et al., 2012; Ma et al., 2014). It exhibits a series of superior traits, including fast growth in a wide temperature range (22°C ~ 40°C) and high tolerance to salt, alkalinity and light stresses, and is amenable to conjugal gene transfer (based on colony formation on plates) (Taton et al., 2012).
So far, genetic alterations of Leptolyngbya BL0902 depended on the expression of exogenous genes on RSF1010-derived plasmids (Taton et al., 2012; Ma et al., 2014; Poole et al., 2020). To be established as a model strain, its genome sequence and gene annotation must be published, and efficient genetic manipulations of large/small regions on the chromosome should be demonstrated. In this study, we analyzed the genome sequence of Leptolyngbya BL0902 and performed different types of genetic manipulations with the existing genetic tools and strategies. Our results indicated that this strain could be developed into an excellent model strain for metabolic engineering and synthetic biology studies in cyanobacteria.
Leptolyngbya BL0902 was from Dr. Golden JW (University of California-San Diego). Leptolyngbya BL0902 and derivatives were grown in BG11 in flasks, with manual agitation 3–4 times a day, at 30°C under the illumination of 30 μE·m−2·s−1. For selection with antibiotics, spectinomycin (10 μg/mL) or neomycin (25 μg/mL) was added to liquid or solid media as appropriate.
The growth of Leptolyngbya BL0902 and a 50.8 kb-deletion mutant in Zarrouk medium (3 biological replicates) was compared in vertical column-type photobioreactors (48.5 cm × 2.0 cm) bubbled with air supplemented with 1% CO2 in the light of 100 μE·m−2·s−1. The OD730 value was adjusted to 0.05 at the beginning and measured every 12 h.
Conjugation was performed as described by Taton et al. (2012), using Escherichia coli HB101 containing pRL443 (conjugative plasmid), pRL623 (helper plasmid) and the plasmid to be transferred into Leptolyngbya BL0902 as the donor strain. The helper plasmid is not required for the conjugal transfer of RSF1010-based editing plasmids, but it is not necessary to remove this plasmid from the donor strain.
The 100 L-scale semi-continuous cultivation of Leptolyngbya BL0902 PpsbA -desD was carried out in Zarrouk medium bubbled with 4% CO2 (v/v) at 26°C ~ 29°C, in a 1-m2 open raceway photobioreactor, with the constant illumination of 100 μE·m−2·s−1 from both upper and lower sides (Figure 1). The depth of the culture was kept at 10 cm by replenishing water every day to compensate for the evaporative loss. An electric motor-driven paddle wheel, 0.3 m in diameter, rotating at 20 rpm, was used to propel the circulating movement of the culture. The initial OD730nm of the culture was ~0.1. Starting from the 5th day after the inoculation, cells were harvested once every 2 days; at each harvest time point, 50 L of the culture were collected, and the same volume of fresh medium was supplemented to the culture. The growth of cells was recorded based on the dry weight of biomass per liter in two biological replicates. At each time point, 50 mL of cells taken from the culture were vacuum filtered, washed with 0.5 N HCl and dried at 105°C for 4 h, and the dry weight was measured.
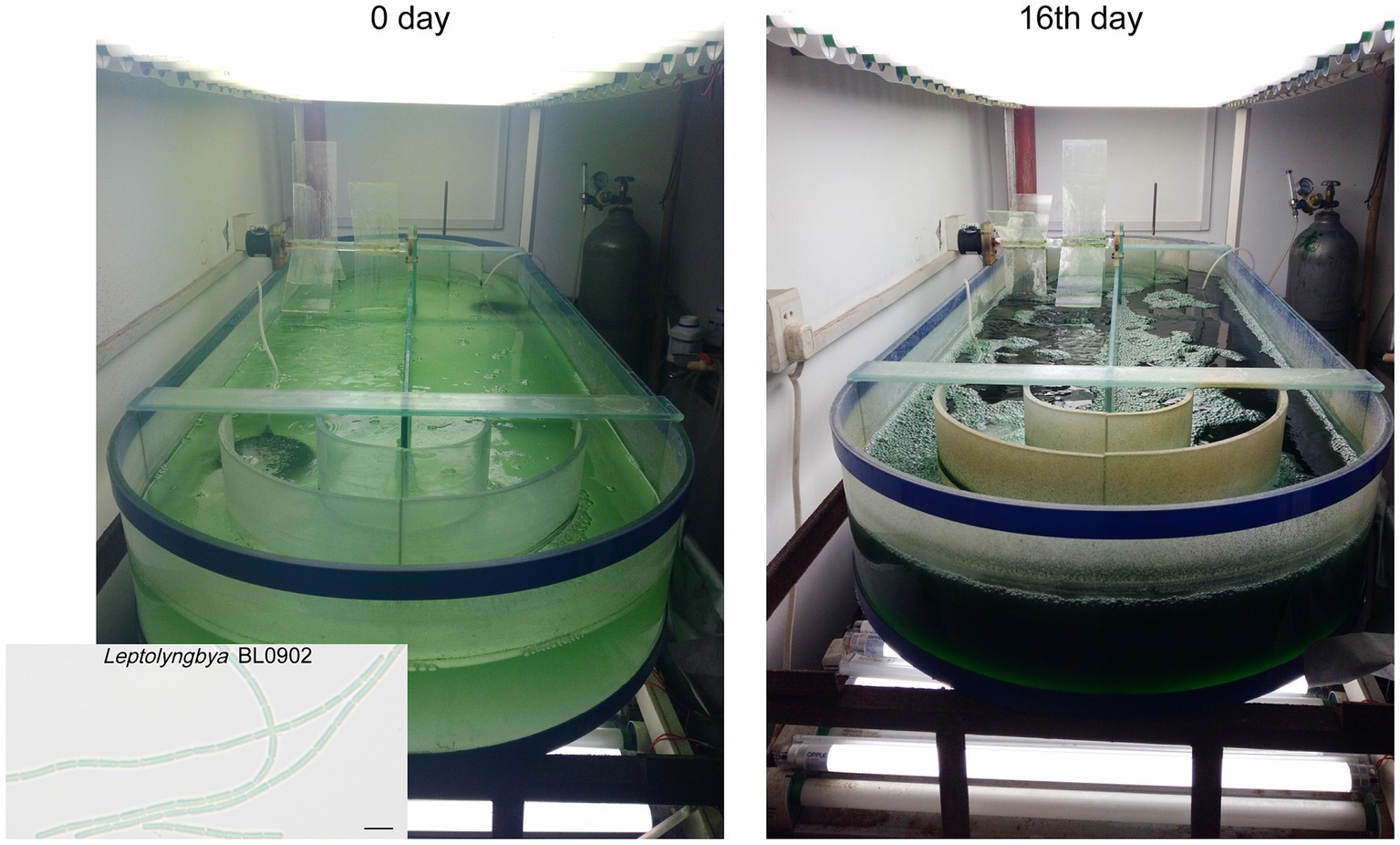
Figure 1. Cultivation of Leptolyngbya BL0902 PpsbA -desD in the open raceway photobioreactor. The scale bar in the photomicrograph at the lower left corner stands for 5 μm.
Fatty acid composition was analyzed as previously described (Laurens et al., 2012) with 3 biological replicates. Lipids were extracted from 10 mg freeze-dried cells and simultaneously transesterified, with sequentially added chloroform/methanol (2:1) and 5% HCl in methanol, and analyzed by GC (Trace Ultra, Thermo Electric) equipped with a DB-23 capillary column (60 m in length, 0.25 mm in diameter, 0.25 μm in film thickness) coupled to a FID detector. For quantification of the GLA content in Leptolyngbya strains, tridecanoic acid (C13:0) was added to the dried cells as an internal standard before extraction and transesterification of lipids.
Molecular cloning manipulations were performed according to standard protocols, but some ligation reactions were performed using ClonExpress Ultra One step Cloning Kit V2 (Vazyme, Nanjing, China) based on in vitro homologous recombination. DNA fragments generated by PCR were confirmed by sequencing after cloning in plasmids. Details of plasmid construction and strain generation are provided in Supplementary Table S1 but briefly described as below.
For interruption of desB with the omega cassette (Prentki and Krisch, 1984) or PpsbA-desD-omega in Leptolyngbya BL0902, pHB6115 and pHB6127 were constructed, with these fragments inserted at the Bal I site of Leptolyngbya desB, cloned into pRL271, a sacB-bearing vector (Cai and Wolk, 1990). The desD gene was generated by PCR using Synechocystis PCC 6803 DNA as the template.
For generation of deletions in the chromosome of Leptolyngbya BL0902, a DNA fragment with the two homologous arms for double crossover recombination was generated by overlap PCR (Horton et al., 1989) and cloned into the Bgl II-BamH I site of a pCpf1 plasmid, and a short dsDNA with the gRNA sequence was cloned into the plasmid replacing the DNA fragment between the two Aar I sites. Such constructed plasmids pHB7912, pHB7913, pHB7914 and pJS2529 were used to generate the ΔhetR, Δ[patX-hetR], ΔpatX and Δ07990-08580 mutants, respectively.
These plasmids were introduced into Leptolyngbya BL0902 by conjugation (Taton et al., 2012), and exconjugants were randomly picked and streaked on plates with appropriate antibiotics, followed by selection in liquid medium. Genomic DNA was extracted from these exconjugants with a quick mini-preparation method (Cai and Wolk, 1990) for PCR examinations.
High-molecular-weight genomic DNA was extracted from Leptolyngbya BL0902 using the cetyltrimethylammonium bromide (CTAB) method (Murray and Thompson, 1980), and RNA was removed by using RNase A. The genome sequence was determined by third generation long read sequencing technology on PacBio platforms (Pacific Biosciences, CA, United States). A total of 283,232 reads were obtained, encompassing 1.7 Gb, with an average read length of 5,990 bp. In addition, Illumina sequencing was performed on Illumina HiSeq 2,500 (Illumina Inc., San Diego, CA, USA) to obtain high quality reads, producing 3 Gb high quality data. The genome was assembled using HGAP3 (Chin et al., 2013) based on PacBio sequencing reads and polished using Illumina reads. Finally, the Leptolyngbya BL0902 genome was assembled into a complete circular chromosome with no gap and five circular plasmids.
Protein-coding genes were predicted by using Glimmer3 (Delcher et al., 2007) with default parameters, and the predicted proteins were annotated by searching against NCBI non-redundant protein (NR),1 Swiss-Prot (Wu et al., 2006), COG (Tatusov et al., 1997), KEGG (Kanehisa and Goto, 2000) and GO (Ashburner et al., 2000) databases. The rRNA sequences were identified using RNAmmer (Lagesen et al., 2007), tRNA genes identified using tRNAscan-SE (Lowe and Eddy, 1997), tandem repeats (> 6 bp) identified using Trf407b.linux in RepeatMasker,2 and regularly interspaced short palindromic repeat (CRISPR) arrays identified using MinCED.3
Leptolyngbya strains grown to OD730nm ~ 0.8 were collected by centrifugation, quickly frozen and stored in liquid nitrogen. Total RNA was extracted from cells using the RNA pre-purification Cell/Bacteria Kit (Tiangen Biotech Co., Ltd., Beijing, China), then DNA was removed with DNase I, and rRNA was removed using the Ribo-off rRNA Depletion Kit V2 (Bacteria) (Vazyme, Nanjing, China). One microgram of total RNA was used to construct RNA libraries after retro-transcription, and 150 nt paired-end sequencing was performed on Illumina Novaseq 6,000.
The raw paired-end reads were trimmed and quality controlled by fastp version 0.21.0 (Chen et al., 2018) with default parameters, then clean reads (over 3 G for each strain) were separately aligned to reference genome using hisat2 version: 2.0.1-beta (Kim et al., 2015). The featureCounts function from the Subread package (Liao et al., 2013) was used to count reads that mapped to each one of the protein-coding genes. Raw count data was then used as input into DESeq2 v.1.42.0 (Love et al., 2014) for differential expression analyses (3 biological replicates). Differential expression was considered significant if the absolute FoldChange value was >2 or < 0.5 and the false discovery rate (FDR) adjusted p-value was <0.05. After applying a regularized-logarithm transformation to the raw count gene matrix, we calculated the average expression value of each gene across three biological replicates, then standardized the expression of each gene along the samples to generate a standardized matrix and visualized the result as a heatmap using the package ggplot2 v.3.4.4 (Wickham, 2016), in R.
The genome of Leptolyngbya BL0902 (GenBank accession numbers: CP046155-CP046160) is composed of six circular DNA molecules, the chromosome and five plasmids (Table 1), with a total size of 4.71 Mb. The 4.309 Mb-long chromosome is predicted to have 3,981 protein-coding genes, 6 rRNA (two copies of rRNA operons) and 43 tRNA genes; the five plasmids, with sizes of 158.483 kb, 93.510 kb, 78.547 kb, 44.466 kb and 25.969 kb, contain 177, 84, 66, 68 and 27 protein-coding genes, respectively. To our best knowledge, this is the smallest among the Leptolyngbya genomes (Table 2) and those of the closely related genus Nodosilinea (Supplementary Table S2). As seen in some cyanobacteria with rRNA operon variants, such as Anabaena sp. PCC7120 (Iteman et al., 2000), the two 16S-23S internal transcribed spacer regions of Leptolyngbya BL0902 contain either tRNA-Ile or tRNA-Ala gene. In addition, 571 tandem repeats (> 6 bp) and 16 clustered regularly interspaced short palindromic repeat (CRISPR) arrays were identified in the genome (Table 1).
Some cyanobacterial species produce cyanotoxins that pose risks to human and animals, and the syntheses of cyanotoxins are typically dependent upon conserved gene clusters. We searched the genome of Leptolyngbya BL0902 for all the eleven representative cyanotoxin biosynthesis gene clusters (Pearson et al., 2016) but found no similar one. Type II restriction enzymes may cut foreign DNA and greatly reduce gene transfer efficiency in cyanobacteria (Elhai et al., 1997). However, no genes (homologs) for type II restriction enzymes from REBASE (Roberts et al., 2010) were found in the genome of Leptolyngbya BL0902.
Cyanobacteria can be classified into 4 groups according to the types of fatty acid desaturases (Murata and Wada, 1995): (1) those only with DesC that generates a single double bond at position 9 (Δ9, from the C-terminus) on the fatty acid chain in acyl-lipids; (2) those with DesA (Δ12), DesB (Δ15) and DesC; (3) those with DesA, DesC and DesD (Δ6); (4) those with DesA, DesB, DesC and DesD. In the genome of Leptolyngbya BL0902, we found genes encoding DesA (GFS31_05880), DesB (GFS31_39000) and DesC (GFS31_27570), therefore it should be a strain of group 2.
In filamentous cyanobacteria, there is a monophyletic group that form specialized cells termed heterocysts for nitrogen fixation (Wolk et al., 1994). Some non-heterocystous cyanobacteria can also perform nitrogen fixation (Rippka and Waterbury, 1977), and these diazotrophic cyanobacteria all possess the nif (nitrogen fixation) gene cluster (Tsujimoto et al., 2014). Leptolyngbya BL0902 is a non-heterocystous cyanobacterium without the nif gene cluster; however, it possesses genes (GFS31_32400, GFS31_16620-GFS31_16630) similar to hetR (Buikema and Haselkorn, 2001) and hetZ-patU (Zhang et al., 2007), which play central roles in regulation of heterocyst differentiation. In heterocyst-forming cyanobacteria, HetR directly regulates the expression of hetZ via a HetR-binding site, while HetZ regulates the expression of hetR and two genes encoding RG(S/T)GR-containing peptides, namely patS and patX, directly or indirectly via the DIF1 promoter (Du et al., 2020). HetR also activates or inhibits the expression of some other genes, such as hetP and the hetP-like gene alr3234 in the heterocyst-forming cyanobacterium Anabaena sp. PCC 7120 (Hou et al., 2015). The pentapeptide RG(S/T)GR, derived from PatS (Yoon and Golden, 1998), PatX (Elhai and Khudyakov, 2018) and a protein called HetN (Higa et al., 2012), is an inhibitor of HetR. In Leptolyngbya BL0902, patS and hetN are not found, but a protein-encoding gene, GFS31_32390, meets the definition of patX (Elhai and Khudyakov, 2018); of the three ORFs similar to hetR, the one (GFS31_32400) with the highest similarity is located immediately downstream of patX and appears to be co-transcribed with patX from the predicted DIF1 promoter (Supplementary Figure S1).
A gene transfer system based on wide-host-range plasmids has been established in Leptolyngbya BL0902 (Taton et al., 2012; Poole et al., 2020). However, manipulations of genetic loci on the chromosome are required for gene function analyses and generation of more stable cell factories. To this end, we tested integration of genes into the chromosome by homologous double-crossover. In Leptolyngbya BL0902, the three fatty acid desaturases generate double bonds at Δ9, Δ12 and Δ15 positions on C18 fatty acid chains of acyl-lipids, leading to the formation of α linolenic acid (ALA). First, we constructed the plasmid pHB6125 (Supplementary Table S1) for interrupting desB with the omega cassette (Spr) in Leptolyngbya BL0902 (Figures 2A,B). A DNA fragment containing desB was cloned in the plasmid, and the omega cassette was inserted at the Bal I site of desB. sacB on the vector can provide the positive selection for double-crossover recombinants. We planned to introduce the plasmid into Leptolyngbya BL0902 by conjugation to obtain single-crossover recombinants, then to select double-crossover mutants on sucrose-containing plates. However, when we checked 9 randomly picked spectinomycin-resistant exconjugants by PCR examinations, one of them was already double-crossover mutant (ΔdesD), whereas the rest 8 were single-crossovers, therefore the selection on sucrose-containing plates was omitted. Then, we tried to interrupt desB with PpsbA -desD and the omega cassette, so as to replace α-linolenic acid with γ-linolenic acid (GLA, C18 fatty acid with double bonds at Δ6, Δ9 and Δ12) in Leptolyngbya BL0902. PpsbA is a strong promoter from the chloroplast of Amaranthus hybridus (Elhai, 1993), while desD is from Synechocystis PCC 6803. The plasmid pHB6127 was constructed in a structure similar to pHB6125, but a fragment with PpsbA -desD and the omega cassette was inserted into the BalI site of desB. Of 11 randomly picked exconjugants, 10 formed single-crossover recombination between pHB6127 and the chromosome, but one directly formed double-crossover, with PpsbA-desD and the omega cassette inserted within desB, and this desD-overexpressing strain was called PpsbA-desD (Figures 2A,B).
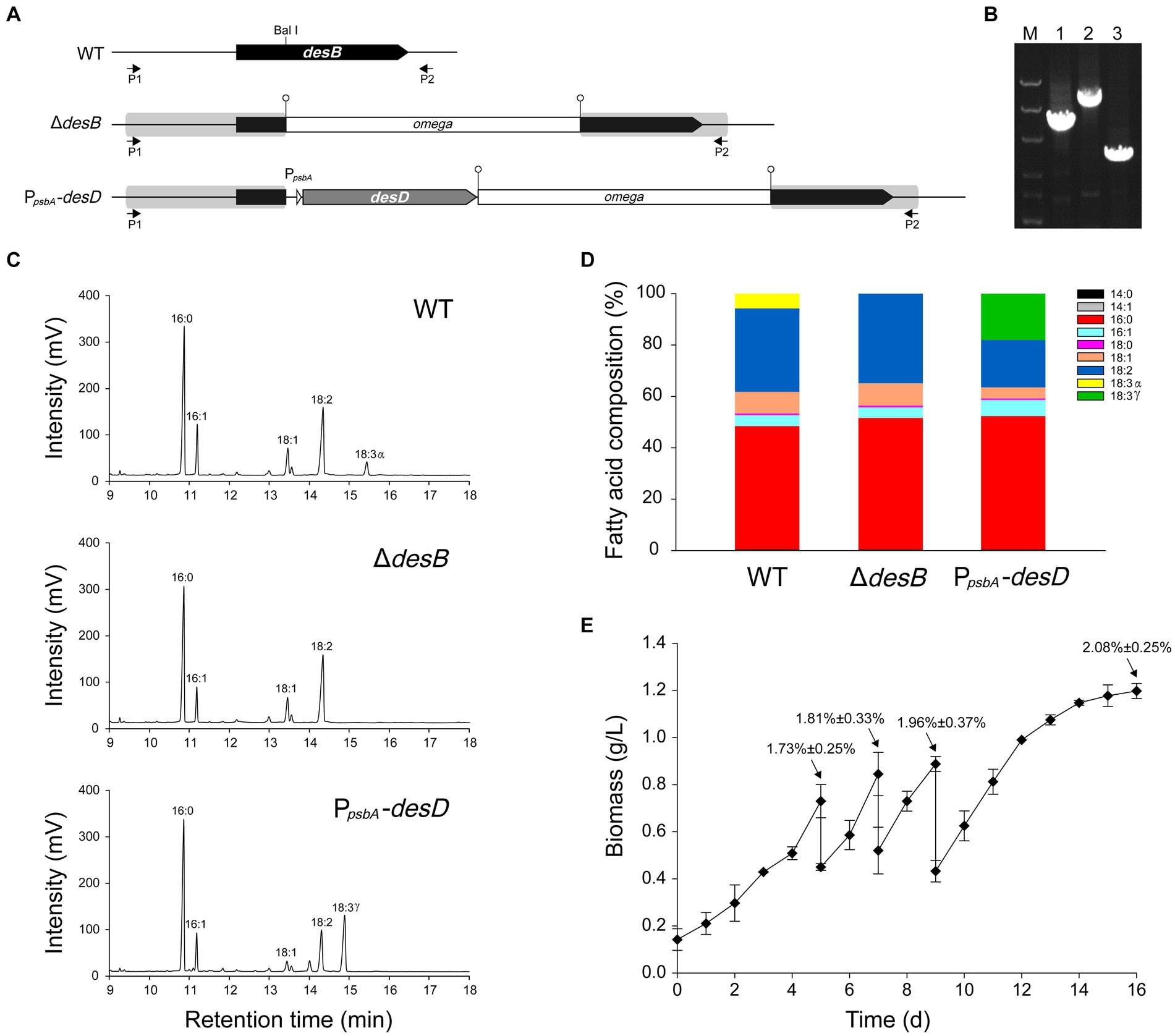
Figure 2. Generation of a Leptolyngbya BL0902 strain that produces GLA as the only C18:3 fatty acid. (A) The structure of the desB region in the wild type (WT) and two mutant strains ΔdesB and PpsbA-desD. PpsbA-desD is the GLA-producing strain. P1 and P2 represent the primers, L-desB-1 and L-desB-2, for PCR examination. (B) PCR examination of the desB region, as shown in the electrophoretogram: lane 1, △desB; lane 2, PpsbA-desD; lane 3, WT. M, dsDNA marker (8 kb, 5 kb, 3 kb, 1.5 kb, 1 kb, 0.5 kb). (C) Gas chromatograms of fatty acid methyl esters prepared from acyl-lipids of Leptolyngbya strains. (D) Fatty acid compositions of Leptolyngbya strains (also see Supplementary Table S3). (E) Growth of the PpsbA-desD strain under semi-continuous cultivation conditions in the open raceway photobioreactor (see Figure 1). Percentages (mean ± SD) indicated at the four time points are GLA contents of cells (w/w).
Gas chromatography analyses of fatty acid composition showed that unlike in the wild type, ALA was no longer formed in the ΔdesB mutant (Figures 2C,D; Supplementary Table S3). In the PpsbA-desD strain, ALA (18:3α) was replaced with GLA (18:3γ), which accounted for 18.02% ± 0.92% of long chain fatty acids. Compared to the fatty acid composition in the wild type, C18:3 increased in this strain at the expense of mono- and di-unsaturated C18 fatty acids (Figure 2D; Supplementary Table S3). The desD-overexpressing strain and GLA content were stable over past 6 years under laboratory conditions. To demonstrate the potential for commercial production, we evaluated the biomass and GLA productivities of this strain under semi-continuous cultivation conditions in a 1-m2 open raceway photobioreactor (Figures 1, 2E). On the 5th, 7th and 9th day, half of the culture was collected for cell harvest, and fresh medium of the same volume was supplemented to the culture. Productivities were calculated based on the increase from the 5th day to the 7th day and that from the 7th day to the 9th day (3 samples × 2). The cultivation was extended to the 16th day. The GLA contents gradually increased at the four time points, reaching ~2% of the biomass (dry weight). Based on two independent cultivation experiments (3 samples × 2 × 2), we calculated that the productivity of biomass was 19.1 ± 4.9 g·m−2·d−1, and that of GLA was 0.36 ± 0.14 g·m−2·d−1 (p < 0.05).
For analyses of gene functions or biotechnological genetic manipulations, markerless deletions or insertions are sometimes required. The CRISPR/Cpf1 editing system is suitable for such purposes (Ungerer and Pakrasi, 2016; Niu et al., 2018). hetR and patX are two genes found in almost all filamentous cyanobacteria, heterocyst-forming or not (Elhai and Khudyakov, 2018). The functions of these two genes in those species that do not form heterocysts remain a mystery. We constructed Cpf1-based editing plasmids pHB7912, pHB7913 and pHB7914 (Supplementary Table S1), for deleting hetR, patX-hetR and patX, respectively. The editing plasmids were introduced into Leptolyngbya BL0902 by conjugation, and exconjugants were checked with PCR using specific primers (Figures 3A–D; Supplementary Table S1). In the resulted mutants ΔhetR, Δ[patX-hetR] and ΔpatX, the predicted DIF1 promoter upstream of patX-hetR (Supplementary Figure S1) remained unchanged. The editing plasmids were then removed from the mutants by positive selection on sucrose-containing plates, and the removal of plasmids was confirmed by PCR examination and antibiotic-resistance assay (data not shown).
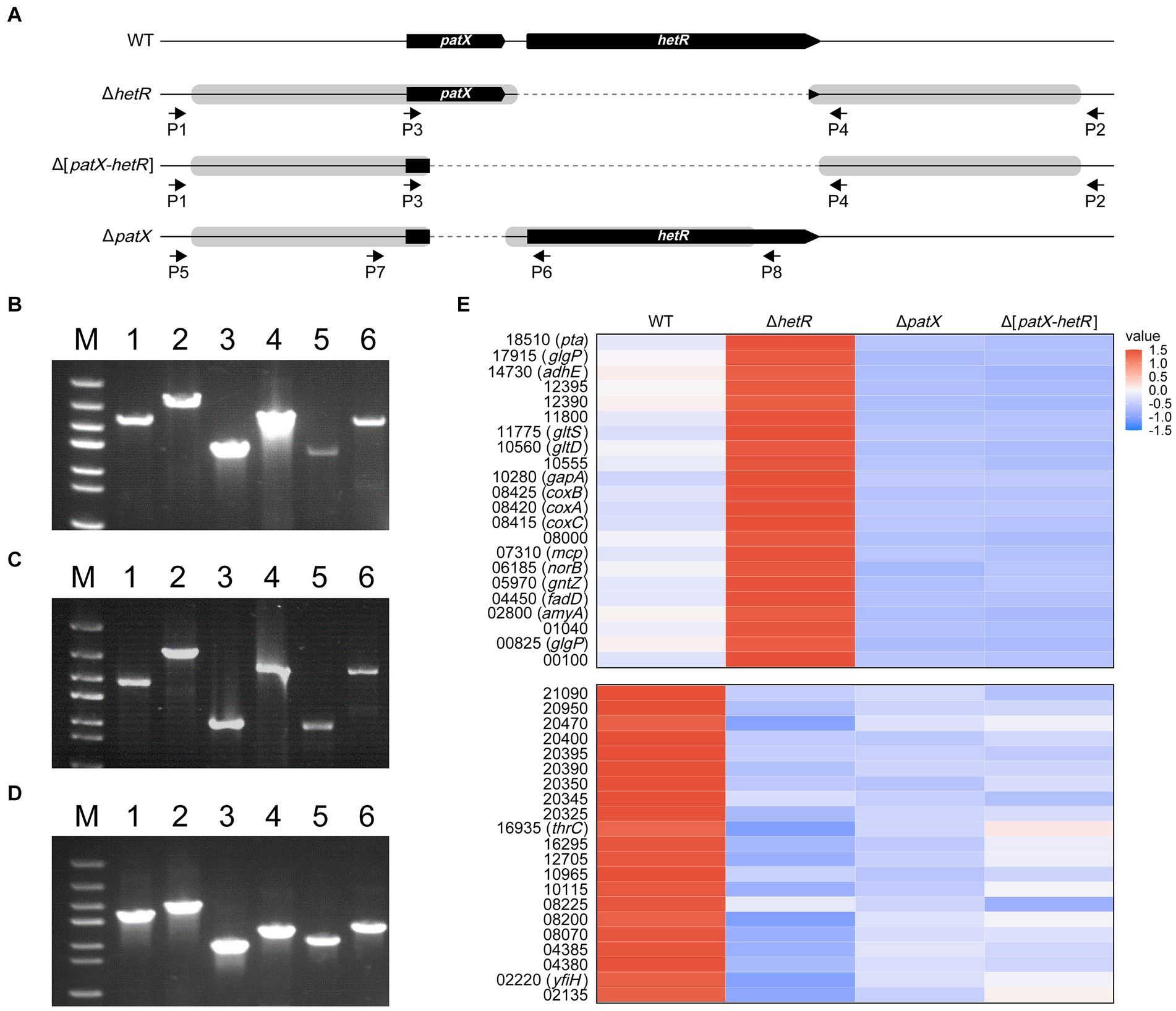
Figure 3. Markerless deletions at the patX-hetR region of Leptolyngbya BL0902. (A) The structure of the patX-hetR region in the WT and mutant strains ΔhetR, Δ[patX-hetR] and ΔpatX. P1, P2, P3, P4, P5, P6, P7 and P8 represent the primers Ch-0902-hetR-F, Ch-0902-hetR-R, Ch-patX-hetR-F-in, Ch-hetR-R-in, Ch-patX-F1, Ch-patX-rev, Ch-patX-For and Ch-patX-R1, respectively. (B–D) Electrophoretograms of PCR products for examination of the mutants (lanes 1, 3, 5), with the wild type as the control (lanes 2, 4, 6). Primers used in B and C (ΔhetR, Δ[patX-hetR]): lanes 1 and 2, P1/P2; lanes 3 and 4, P1/P4; lanes 5 and 6, P3/P2. Primers used in D (ΔpatX): lanes 1 and 2, P5/P8; lanes 3 and 4, P5/P6; lanes 5 and 6, P7/P8. M, dsDNA marker (5 kb, 3 kb, 2 kb, 1.5 kb, 1 kb, 0.8 kb, 0.5 kb). (E) A partial heatmap showing two types of differential expression patterns in the three mutants and the wild type. The full heatmap is shown in Supplementary Figure S2, based on the standardized expression levels of the genes identified in RNA-seq analyses (Supplementary Table S4). The blue shades designate decreasing levels, and red shades increasing levels. The color intensity denotes the standardized expression, as indicated by the scale bar.
In heterocyst-forming cyanobacteria, PatX is thought to be one of the precursors for the RG(S/T)GT-containing inhibitor of HetR (Elhai and Khudyakov, 2018; Khudyakov et al., 2020). We wondered whether PatX counteracts the regulatory effects of HetR, either activating or inhibitory, on gene expression in Leptolyngbya BL0902. Transcriptomic analyses were performed to compare the transcriptional profiles of the WT and mutants. The results showed that there were 993 genes differentially expressed in ΔhetR vs the wild type, 428 in ΔpatX, 477 in Δ[patX-hetR], including genes involved in photosynthesis, respiration, nitrogen metabolism, etc. Of these genes, 126 were up- or down-regulated in all these mutants compared to the wild type. According to the differential expression patterns in mutants compared to the wild type, these 126 genes can be classified into 5 groups (Supplementary Figure S2, with 2 groups shown in Figure 3E as examples). It appeared that HetR downregulates genes in the first and second groups but upregulates those in the third, fourth and fifth groups. Only in groups 2 and 3, PatX showed opposite effects on gene expression compared to HetR. If PatX acts as a (pre)inhibitor of HetR, the effect of patX on gene expression should be dependent upon that of hetR. However, in groups 2 and 3, Δ[patX-hetR] was similar to ΔpatX but contrary to ΔhetR in differential expression. Apparently, the effects of PatX on gene expression are basically independent of HetR in Leptolyngbya BL0902.
Deletions of large non-essential regions in the chromosome are required for genome streamlining in cyanobacteria, which would reduce the genomic complexity and may improve some productive traits (Hou et al., 2023; Sengupta et al., 2024). By BLAST searching against the essential genes experimentally identified in Synechococcus elongatus PCC 7942 (Rubin et al., 2015), we obtained a list of genes that are probably essential in Leptolyngbya BL0902. Then we tried to delete some chromosomal regions without these genes, for example, a 50.8-kb region (chromosomal bp 825,418–876,218), extending from GFS31_07990 to GFS31_08580. The Cpf1-based editing plasmid pJS2529 (Supplementary Table S1) was constructed and transferred into Leptolyngbya BL0902 by conjugation. Of 20 randomly picked exconjugants, 6 exhibited complete deletion of the 50.8-kb region. The complete segregation of the resulted mutant, Δ07990-08580, was confirmed by PCR using 3 pairs of primers (Figure 4). When grown in Zarrouk medium in column photobioreactors with aeration, Δ07990-08580 showed a slightly reduced growth rate compared to that of the wild type (Figure 4), but this does not compromise the feasibility of genome streamlining in Leptolyngbya BL0902.
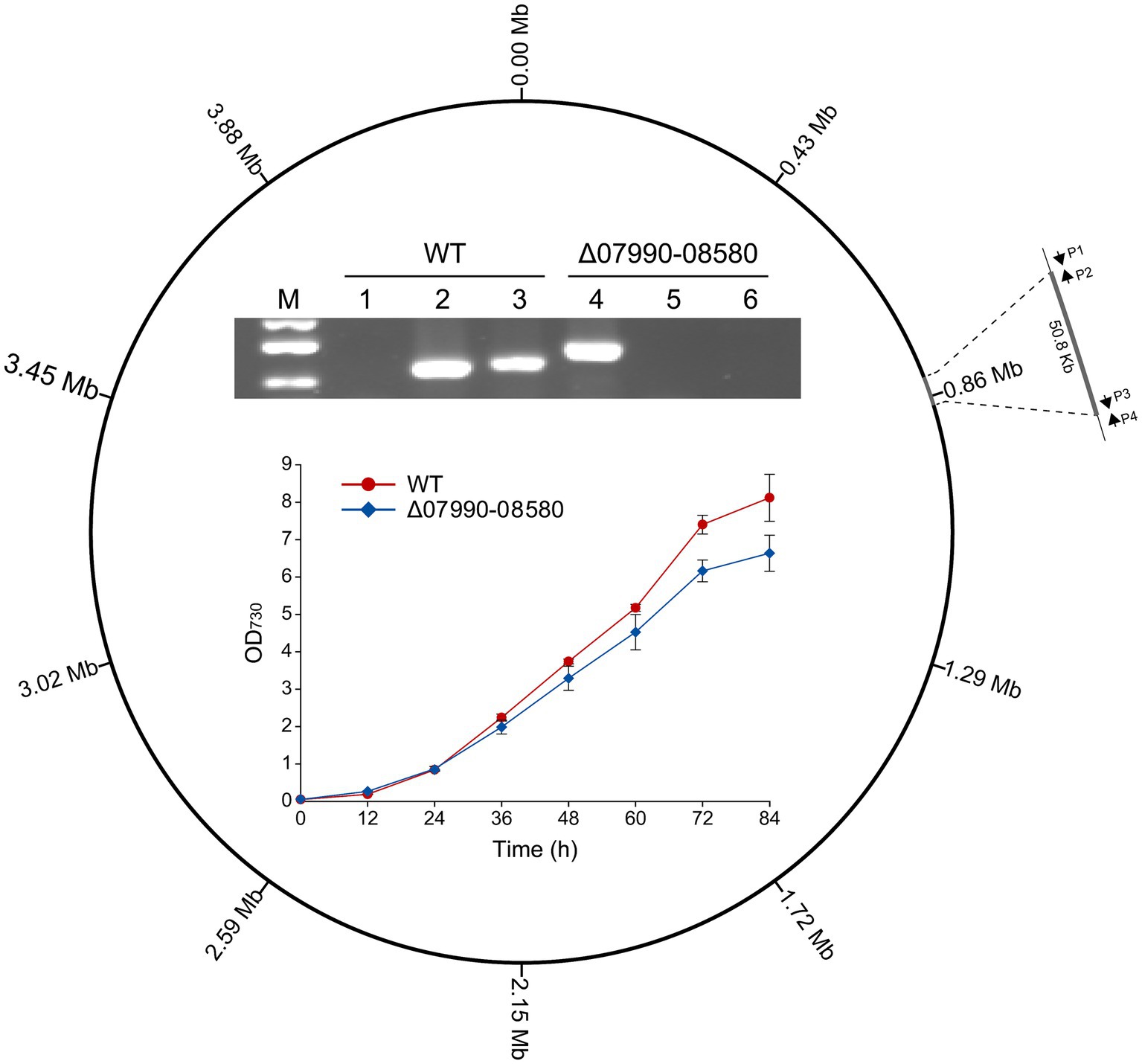
Figure 4. Deletion of a 50.8-kb region in the chromosome of Leptolyngbya BL0902, generating the Δ07990-08580 mutant. P1, P2, P3 and P4 represent the primers BL0783-0842-F, BL0783-0842-R′, BL0783-0842-F′ and BL0783-0842-R. The electrophoretogram shows the result of PCR examination of the mutant: lanes 1 and 4, using primers P1/P4; lanes 2 and 5, P1/P2; lanes 3 and 6, P3/P4. M, dsDNA marker (0.75 kb, 0.5 kb and 0.25 kb). The growth of the mutant and the wild type was compared in Zarrouk medium in column-type photobioreactors.
For biological research in cyanobacteria, there are some frequently used model strains, such as unicellular species Synechocystis sp. PCC 6803, Synechococcus sp. PCC 7942 and Synechococcus sp. PCC 7002, and heterocyst-forming filamentous species Anabaena/Nostoc sp. PCC 7120 and A. variabilis ATCC 29413. However, no filamentous cyanobacteria that do not form heterocysts have been widely used as a model strain. This is probably due to two reasons: (1) lack of a focus question that needs to be addressed with this type of cyanobacteria; (2) lack of a strain that is suitable for different types of genetic manipulations. However, with the development of synthetic biology in cyanobacteria, it is increasingly realized that a non-heterocystous filamentous model strain is necessary, because a filamentous strain, if it is tolerant to a high concentration of sodium bicarbonate and grows quickly (for example, in Zarrouk medium), is more suitable for commercial-scale cultivation and low-cost harvest than unicellular species. On the other hand, for the sake of basic research, to find out how heterocysts originated or how genes involved in heterocyst differentiation originated, our efforts must be extended to filamentous species that do not form heterocysts. Leptolyngbya BL0902 is such a filamentous cyanobacterium, with the potential to be developed into a model strain. In the whole genome sequence of this strain, we found no genes involved in cyanotoxin biosynthesis and no genes encoding type II restriction enzymes. Therefore, it could be a safe host for production of nutrients, food/cosmetics additives or constituents of medicine, and the existing genetic tools and strategies may be directly used in manipulations of its genomic DNA.
Genetic manipulations in filamentous cyanobacteria usually depend on conjugal transfer of plasmids from E. coli to recipient cells (Elhai et al., 1997) and single/double-crossover recombination between the homologous sequence(s) on the plasmid and the target genomic DNA. Double-crossover mutants are often generated employing positive selection strategies, such as the use of a sacB gene on a non-replicable vector (Cai and Wolk, 1990) or a CRISPR/Cpf1 system on a replicative plasmid (Ungerer and Pakrasi, 2016). The former strategy is expected to generate the single-crossover mutant at the first step, then from the single cross-over mutant, double crossovers are selected based on the lethal effect of sacB on cyanobacterial cells grown on sucrose-containing plates. However, in Leptolyngbya BL0902, we found that about 1/10 of exconjugants generated at the first step were already double-crossover mutants, therefore the second step was omitted. The mechanism for the high ratio of double-crossover mutants in the exconjugants remains to be elucidated. One possibility is that a substantial proportion of the plasmid was linearized after transfer into Leptolyngbya cells. Apparently, this strategy is only suitable for insertion of a DNA fragment with an antibiotic-resistance marker into the target sequence; for generation of markerless deletions in the chromosome of Leptolyngbya BL0902, the CRISPR/Cpf1 system would be much more efficient.
Based on the genetic manipulation system, we generated a Leptolyngbya strain that produced GLA as the only C18:3 fatty acid and four markerless deletion strains. The GLA-producing strain was semi-continuously cultivated with an open raceway photobioreactor, and the result demonstrated that value-added chemicals could be stably produced in Leptolyngbya BL0902 in a way potentially for commercial production (Figures 1, 2). Of the four deletion mutants, three were used to analyze the relationship of hetR and patX in non-heterocystous filamentous cyanobacterium. Although PatX contributes to the inhibition of HetR in Anabaena PCC 7120, the transcriptomic analysis of the differential expression between ΔhetR, Δ[patX-hetR], ΔpatX mutants and the wild type of Leptolyngbya BL0902 did not show a similar effect of PatX on HetR (Figure 3). Probably, PatX acts as an independent regulator in Leptolyngbya. In addition to deletions in the patX-hetR region, we also generated a mutant in which a 50.8-kb nonessential region was deleted (Figure 4). Generation of the large fragment deletion in the chromosome further lends support to our proposal that Leptolyngbya BL0902 could be used as a model strain for synthetic biological studies in filamentous cyanobacteria. The affected growth of the large-fragment deletion mutant (or multiple-deletion mutants afterwards) may be restored by using a hypermutation system (Sun et al., 2023).
In the future, Leptolyngbya BL0902 may be further developed into ready-to-use ‘plug and play’ chassis cells. First, the genome can be extensively streamlined. In bacterial genomes, there are many nonessential regions that are disposable under favorable conditions (Martínez-García and de Lorenzo, 2016; Kurokawa and Ying, 2019). Elimination of these regions can lower genomic complexity and improve the predictability and operability of genetic engineering. Second, different types of regulatory modules may be integrated into the genome. Such gene expression platforms would allow desired products to be synthesized under specific conditions or at the stationary growth phase, so as to alleviate the contradiction between cell propagation and product accumulation. In addition to the applied purposes, gene function analyses in Leptolyngbya BL0902 may contribute to the studies on the origin of heterocysts, and reconstruction of functional modules of heterocyst differentiation in this strain would greatly consolidate research models proposed for heterocyst formation and patterning.
The Leptolyngbya sp. BL0902 genome sequences with annotations have been deposited in the NCBI GenBank under accession numbers CP046155.1 (chromosome), CP046156.1 (plasmid 1), CP046157.1 (plasmid 2), CP046158.1 (plasmid 3), CP046159.1 (plasmid 4) and CP046160.1 (plasmid 5). RNA-Seq raw sequence data are available at the NCBI BioProject repository under identification no. PRJNA1087288.
HG: Funding acquisition, Investigation, Methodology, Writing – review & editing. YW: Funding acquisition, Investigation, Writing – original draft. ZH: Investigation, Writing – original draft. FY: Investigation, Writing – original draft. XH: Validation, Visualization, Writing – review & editing. DN: Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. XX: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key R & D Program of China (2021YFA0909700), the Wuhan Science and Technology Key Projects (2023020302020708) and the Key Research Projects of Central China Normal University (CCNU24JCPT018).
The authors are indebted to Yiming Li, for her assistance in comparing the growth of the wild type and the 50.8 kb-deletion mutant of Leptolyngbya BL0902.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1409771/full#supplementary-material
Ahmad, A. M. R., Intikhab, A., Zafar, S., Farooq, U., Shah, H. B. U., Akram, S., et al. (2023). Spirulina, an FDA-approved functional food: worth the hype? Cell. Mol. 69, 137–144. doi: 10.14715/cmb/2022.69.1.24
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Brey, L. F., Włodarczyk, A. J., Bang Thøfner, J. F., Burow, M., Crocoll, C., Nielsen, I., et al. (2020). Metabolic engineering of Synechocystis sp. PCC 6803 for the production of aromatic amino acids and derived phenylpropanoids. Metab. Eng. 57, 129–139. doi: 10.1016/j.ymben.2019.11.002
Buikema, W. J., and Haselkorn, R. (2001). Expression of the Anabaena hetR gene from a copper-regulated promoter leads to heterocyst differentiation under repressing conditions. Proc. Natl. Acad. Sci. USA 98, 2729–2734. doi: 10.1073/pnas.051624898
Cai, Y. P., and Wolk, C. P. (1990). Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172, 3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chin, C. S., Alexander, D. H., Marks, P., Klammer, A. A., Drake, J., Heiner, C., et al. (2013). Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10, 563–569. doi: 10.1038/nmeth.2474
Choi, S. Y., Wang, J. Y., Kwak, H. S., Lee, S. M., Um, Y., Kim, Y., et al. (2017). Improvement of squalene production from CO2 in Synechococcus elongatus PCC 7942 by metabolic engineering and scalable production in a photobioreactor. ACS Synth. Biol. 6, 1289–1295. doi: 10.1021/acssynbio.7b00083
d’Espaux, L., Mendez-Perez, D., Li, R., and Keasling, J. D. (2015). Synthetic biology for microbial production of lipid-based biofuels. Curr. Opin. Chem. Biol. 29, 58–65. doi: 10.1016/j.cbpa.2015.09.009
Davies, J. A. (2017). Using synthetic biology to explore principles of development. Development 144, 1146–1158. doi: 10.1242/dev.144196
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Diao, J., Song, X., Zhang, L., Cui, J., Chen, L., and Zhang, W. (2020). Tailoring cyanobacteria as a new platform for highly efficient synthesis of astaxanthin. Metab. Eng. 61, 275–287. doi: 10.1016/j.ymben.2020.07.003
Du, Y., Zhang, H., Wang, H., Wang, S., Lei, Q., Li, C., et al. (2020). Expression from DIF1-motif promoters of hetR and patS is dependent on HetZ and modulated by PatU3 during heterocyst differentiation. PLoS One 15:e0232383. doi: 10.1371/journal.pone.0232383
Elhai, J. (1993). Strong and regulated promoters in the cyanobacterium Anabaena PCC 7120. FEMS Microbiol. Lett. 114, 179–184. doi: 10.1111/j.1574-6968.1993.tb06570.x
Elhai, J., and Khudyakov, I. (2018). Ancient association of cyanobacterial multicellularity with the regulator HetR and an RGSGR pentapeptide-containing protein (PatX). Mol. Microbiol. 110, 931–954. doi: 10.1111/mmi.14003
Elhai, J., Vepritskiy, A., Muro-Pastor, A. M., Flores, E., and Wolk, C. P. (1997). Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179, 1998–2005. doi: 10.1128/jb.179.6.1998-2005
Gao, X., Gao, F., Liu, D., Zhang, H., Nie, X., and Yang, C. (2016). Engineering the methylerythritol phosphate pathway in cyanobacteria for photosynthetic isoprene production from CO2. Energy Environ. Sci. 9, 1400–1411. doi: 10.1039/C5EE03102H
Hamilton, T. L., Bryant, D. A., and Macalady, J. L. (2016). The role of biology in planetary evolution: cyanobacterial primary production in low-oxygen Proterozoic oceans. Environ. Microbiol. 18, 325–340. doi: 10.1111/1462-2920.13118
Higa, K. C., Rajagopalan, R., Risser, D. D., Rivers, O. S., Tom, S. K., Videau, P., et al. (2012). The RGSGR amino acid motif of the intercellular signaling protein, HetN, is required for patterning of heterocysts in Anabaena sp. strain PCC 7120. Mol. Microbiol. 83, 682–693. doi: 10.1111/j.1365-2958.2011.07949.x
Horton, R. M., Hunt, H. D., Ho, S. N., Pullen, J. K., and Pease, L. R. (1989). Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68. doi: 10.1016/0378-1119(89)90359-4
Hou, F., Ke, Z., Xu, Y., Wang, Y., Zhu, G., Gao, H., et al. (2023). Systematic large fragment deletions in the genome of Synechococcus elongatus and the consequent changes in transcriptomic profiles. Genes 14:1091. doi: 10.3390/genes14051091
Hou, S., Zhou, F., Peng, S., Gao, H., and Xu, X. (2015). The HetR-binding site that activates expression of patA in vegetative cells is required for normal heterocyst patterning in Anabaena sp. PCC 7120. Science Bull. 60, 192–201. doi: 10.1007/s11434-014-0724-5
Hutmacher, D. W., Holzapfel, B. M., De-Juan-Pardo, E. M., Pereira, B. A., Ellem, S. J., Loessner, D., et al. (2015). Convergence of regenerative medicine and synthetic biology to develop standardized and validated models of human diseases with clinical relevance. Curr. Opin. Biotechnol. 35, 127–132. doi: 10.1016/j.copbio.2015.06.001
Iteman, I., Rippka, R., Tandeau De Marsac, N., and Herdman, M. (2000). Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146, 1275–1286. doi: 10.1099/00221287-146-6-1275
Jester, B. W., Zhao, H., Gewe, M., Adame, T., Perruzza, L., Bolick, D. T., et al. (2022). Development of Spirulina for the manufacture and oral delivery of protein therapeutics. Nat. Biotechnol. 40, 956–964. doi: 10.1038/s41587-022-01249-7
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. doi: 10.1093/nar/28.1.27
Khudyakov, I., Gladkov, G., and Elhai, J. (2020). Inactivation of three RG(S/T)GR pentapeptide-containing negative regulators of HetR results in lethal differentiation of Anabaena PCC 7120. Life (Basel) 10:326. doi: 10.3390/life10120326
Kim, D., Langmead, B., and Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kim, W. J., Lee, S.-M., Um, Y., Sim, S. J., and Woo, H. M. (2017). Development of SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942. Front. Plant Sci. 8, 1–9. doi: 10.3389/fpls.2017.00293
Koch, M., Bruckmoser, J., Scholl, J., Hauf, W., Rieger, B., and Forchhammer, K. (2020). Maximizing PHB content in Synechocystis sp. PCC 6803: a new metabolic engineering strategy based on the regulator PirC. Microb. Cell Factories 19:231. doi: 10.1186/s12934-020-01491-1
Kopka, J., Schmidt, S., Dethloff, F., Pade, N., Berendt, S., Schottkowski, M., et al. (2017). Systems analysis of ethanol production in the genetically engineered cyanobacterium Synechococcus sp. PCC 7002. Biotechnol. Biofuels 10:56. doi: 10.1186/s13068-017-0741-0
Korosh, T. C., Markley, A. L., Clark, R. L., McGinley, L. L., McMahon, K. D., and Pfleger, B. F. (2017). Engineering photosynthetic production of L-lysine. Metab. Eng. 44, 273–283. doi: 10.1016/j.ymben.2017.10.010
Kurokawa, M., and Ying, B. W. (2019). Experimental challenges for reduced genomes: the cell model Escherichia coli. Microorganisms 8:3. doi: 10.3390/microorganisms8010003
Lagesen, K., Hallin, P., Rødland, E. A., Staerfeldt, H. H., Rognes, T., and Ussery, D. W. (2007). RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35, 3100–3108. doi: 10.1093/nar/gkm160
Laurens, L. M., Quinn, M., Van Wychen, S., Templeton, D. W., and Wolfrum, E. J. (2012). Accurate and reliable quantification of total microalgal fuel potential as fatty acid methyl esters by in situ transesterification. Anal. Bioanal. Chem. 403, 167–178. doi: 10.1007/s00216-012-5814-0
Liao, Y., Smyth, G. K., and Shi, W. (2013). The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41:e108. doi: 10.1093/nar/gkt214
Lin, P. C., Saha, R., Zhang, F., and Pakrasi, H. B. (2017). Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep. 7:17503. doi: 10.1038/s41598-017-17831-y
Lobus, N. V., and Kulikovskiy, M. S. (2023). The co-evolution aspects of the biogeochemical role of phytoplankton in aquatic ecosystems: a review. Biology (Basel). 12:92. doi: 10.3390/biology12010092
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lowe, T. M., and Eddy, S. R. (1997). tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25, 955–964. doi: 10.1093/nar/25.5.955
Ma, A. T., Schmidt, C. M., and Golden, J. W. (2014). Regulation of gene expression in diverse cyanobacterial species by using theophylline-responsive riboswitches. Appl. Environ. Microbiol. 80, 6704–6713. doi: 10.1128/AEM.01697-14
Martínez-García, E., and de Lorenzo, V. (2016). The quest for the minimal bacterial genome. Curr. Opin. Biotechnol. 42, 216–224. doi: 10.1016/j.copbio.2016.09.001
Mukherji, S., and van Oudenaarden, A. (2009). Synthetic biology: understanding biological design from synthetic circuits. Nat. Rev. Genet. 10, 859–871. doi: 10.1038/nrg2697
Murata, N., and Wada, H. (1995). Acyl-lipid desaturases and their importance in the tolerance and acclimatization to cold of cyanobacteria. Biochem. J. 308, 1–8. doi: 10.1042/bj3080001
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Niu, T. C., Lin, G. M., Xie, L. R., Wang, Z. Q., Xing, W. Y., Zhang, J. Y., et al. (2018). Expanding the potential of CRISPR-Cpf1-based genome editing technology in the cyanobacterium Anabaena PCC 7120. ACS Synth. Biol. 8, 170–180. doi: 10.1021/acssynbio.8b00437
Pearson, L. A., Dittmann, E., Mazmouza, R., Ongley, S. A., D’Agostino, P. M., and Neilan, B. A. (2016). The genetics, biosynthesis and regulation of toxic specialized metabolites of cyanobacteria. Harmful Algae 54, 98–111. doi: 10.1016/j.hal.2015.11.002
Poole, L. B., Parsonage, D., Sergeant, S., Miller, L. R., Lee, J., Furdui, C. M., et al. (2020). Acyl-lipid desaturases and Vipp1 cooperate in cyanobacteria to produce novel omega-3 PUFA-containing glycolipids. Biotechnol. Biofuels 13:83. doi: 10.1186/s13068-020-01719-7
Prentki, P., and Krisch, H. M. (1984). In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29, 303–313. doi: 10.1016/0378-1119(84)90059-3
Rippka, R., and Waterbury, J. B. (1977). The synthesis of nitrogenase by nonheterocystous cyanobacteria. FEMS Microbiol. Lett. 2, 83–86. doi: 10.1111/j.1574-6968.1977.tb00913.x
Ro, D. K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. doi: 10.1038/nature04640
Roberts, R. J., Vincze, T., Posfai, J., and Macelis, D. (2010). REBASE-a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38, D234–D236. doi: 10.1093/nar/gkp874
Rubin, B. E., Wetmore, K. M., Price, M. N., Diamond, S., Shultzaberger, R. K., Lowe, L. C., et al. (2015). The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. USA 112, 6634–6643. doi: 10.1073/pnas.1519220112
Ruder, W. C., Lu, T., and Collins, J. J. (2011). Synthetic biology moving into the clinic. Science 333, 1248–1252. doi: 10.1126/science.1206843
Sengupta, A., Bandyopadhyay, A., Sarkar, D., Hendry, J. I., Schubert, M. G., Liu, D., et al. (2024). Genome streamlining to improve performance of a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. MBio 15:e0353023. doi: 10.1128/mbio.03530-23
Sun, H., Luan, G., Ma, Y., Lou, W., Chen, R., Feng, D., et al. (2023). Engineered hypermutation adapts cyanobacterial photosynthesis to combined high light and high temperature stress. Nat. Commun. 14:1238. doi: 10.1038/s41467-023-36964-5
Tan, C., Xu, P., and Tao, F. (2022). Carbon-negative synthetic biology: challenges and emerging trends of cyanobacterial technology. Trends Biotechnol. 40, 1488–1502. doi: 10.1016/j.tibtech.2022.09.012
Taton, A., Lis, E., Adin, D. M., Dong, G., Cookson, S., Kay, S. A., et al. (2012). Gene transfer in Leptolyngbya sp. strain BL0902, a cyanobacterium suitable for production of biomass and bioproducts. PLoS One 7:e30901. doi: 10.1371/journal.pone.0030901
Tatusov, R. L., Koonin, E. V., and Lipman, D. J. (1997). A genomic perspective on protein families. Science 278, 631–637. doi: 10.1126/science.278.5338.631
Tsujimoto, R., Kamiya, N., and Fujita, Y. (2014). Transcriptional regulators ChlR and CnfR are essential for diazotrophic growth in nonheterocystous cyanobacteria. Proc. Natl. Acad. Sci. USA 111, 6762–6767. doi: 10.1073/pnas.1323570111
Ungerer, J., and Pakrasi, H. B. (2016). Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria. Sci. Rep. 6:39681. doi: 10.1038/srep39681
Webster, D. P., TerAvest, M. A., Doud, D. F. R., Chakravorty, A., Holmes, E. C., Radens, C. M., et al. (2014). An arsenic-specific biosensor with genetically engineered Shewanella oneidensis in a bioelectrochemical system. Biosens. Bioelectron. 62, 320–324. doi: 10.1016/j.bios.2014.07.003
Włodarczyk, A., Selão, T. T., Norling, B., and Nixon, P. J. (2020). Newly discovered Synechococcus sp. PCC 11901 is a robust cyanobacterial strain for high biomass production. Commun. Biol. 3:215. doi: 10.1038/s42003-020-0910-8
Wolk, C. P., Ernst, A., and Elhai, J. (1994). “Heterocyst metabolism and development” in The molecular biology of Cyanobacteria. ed. D. Bryant (Dordrecht, Netherland: Kluwer Academic Publishers), 769–823.
Wu, C. H., Apweiler, R., Bairoch, A., Natale, D. A., Barker, W. C., Boeckmann, B., et al. (2006). The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 34, D187–D191. doi: 10.1093/nar/gkj161
Yoon, H. S., and Golden, J. W. (1998). Heterocyst pattern formation controlled by a diffusible peptide. Science 282, 935–938. doi: 10.1126/science.282.5390.935
Keywords: synthetic biology, model organism, γ-linolenic acid, patX-hetR , filamentous cyanobacteria
Citation: Gao H, Wang Y, Huang Z, Yu F, Hu X, Ning D and Xu X (2024) Development of Leptolyngbya sp. BL0902 into a model organism for synthetic biological research in filamentous cyanobacteria. Front. Microbiol. 15:1409771. doi: 10.3389/fmicb.2024.1409771
Received: 01 April 2024; Accepted: 10 July 2024;
Published: 22 July 2024.
Edited by:
Weiwen Zhang, Tianjin University, ChinaReviewed by:
Takashi Osanai, Meiji University, JapanCopyright © 2024 Gao, Wang, Huang, Yu, Hu, Ning and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Degang Ning, bmluZ2RlZ2FuZ0BpaGIuYWMuY24=; Xudong Xu, eHV4dWRvbmdAY2NudS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.