- 1Department of Pedology and Agricultural Durable, UFR STRM, FHB University, Abidjan, Côte d'Ivoire
- 2Center National of Research Agronomic-CNRA, Man, Côte d'Ivoire
The impact of chemical fertilizers on soil microbial communities is well acknowledged. This study assesses the influence of various phosphorus sources on soil bacterial composition, abundance, and Phosphorus Cycle Gene Abundance. Three phosphorus sources (natural phosphate rock, triple super phosphate (TSP), and chemical fertilizer NPK) were field tested following two rice cultivation cycles. Soil samples were subsequently collected and analyzed for bacterial groups and phosphorus cycle genes. Results indicated that the bacterial community composition remained consistent, comprising five main phyla: Firmicutes, Actinobacteria, Proteobacteria, Halobacterota, and Chloroflexia, regardless of fertilizer type. NPK fertilizer significantly reduced the relative abundance of Chloroflexia by 19% and Firmicutes by 16.4%, while increasing Actinobacteria and Proteobacteria by 27.5 and 58.8%, respectively. TSP fertilizer increased Actinobacteria by 27.1% and Halobacterota by 24.8%, but reduced Chloroflexia by 8.6%, Firmicutes by 12.6%, and Proteobacteria by 0.6%. Phosphate rock application resulted in reductions of Chloroflexia by 27.1%, Halobacterota by 22.9%, and Firmicutes by 6.2%, alongside increases in Actinobacteria by 46.6% and Proteobacteria by 23.8%. Combined application of TSP, NPK, and phosphate rock led to increases in Proteobacteria (24–40%) and Actinobacteria (13–39%), and decreases in Chloroflexia (5.2–22%) and Firmicutes (6–12.3%) compared to the control (T0). While the different phosphorus sources did not alter the composition of phosphorus cycle genes, they did modulate their abundance. NPK fertilizer did not significantly affect ppK genes (57–59%) but reduced gcd (100 to 69%), 3-phytase (74 to 34%), appA (91 to 63%), and phoD (83 to 67%). Phosphate rock reduced appA and gcd by 27 and 15%, respectively, while increasing 3-phytase by 19%. TSP decreased ppK and phoD by 42 and 40%, respectively, and gcd and appA by 34 and 56%, respectively. Combined fertilizers reduced appA (49 to 34%), 3-phytase (10 to 0%), and gcd (27 to 6%), while increasing ppK (72 to 100%). Among tested phosphorus sources, natural phosphate rock was best, causing moderate changes in bacterial composition and phosphorus genes, supporting balanced soil microbial activity. These findings highlight the complex interactions between fertilizers and soil microbial communities, underscoring the need for tailored fertilization strategies to maintain soil health and optimize agricultural productivity.
Introduction
Phosphorus (P) is essential for plant growth, but its accessibility in acidic tropical soils is often limited (Kpan et al., 2023). To overcome this challenge, chemical fertilizers, whether organic or inorganic, are widely employed to enhance phosphorus availability and boost crop yields (Sahrawat et al., 2001; Koné et al., 2023; Kpan et al., 2023). The application of mineral and organic fertilizers has proven effective in improving soil fertility and crop productivity, serving as primary nutrient sources for arable soils (Wang et al., 2017; Wu et al., 2021; Kpan et al., 2023). For instance, Islam et al. (2015) reveal the positive impact of Nitrogen (N) fertilizer on wheat grain yield, while Das et al. (2023) emphasize the stabilizing effect of long-term manure application on soil organic matter and overall soil health. Nitrogen treatments were found to influence soil pH, cation exchange capacity (CEC), and phosphorus levels (Setu, 2022; Wang et al., 2023; Zeng et al., 2024).
Microbes in agricultural soil play pivotal roles in nutrient cycling, participating in processes such as organic matter decomposition and the biogeochemical cycling of elements (C, N, P, S) (Morris and Blackwood, 2015). They contribute essential nutrients for crop growth, and alterations in soil nutrient levels post-fertilization significantly influence microbial biomass, composition, and diversity. The sensitivity of soil microbial biomass and diversity to variations in soil nutrients, pH, and organic matter content has been well-documented (Wang et al., 2017). Furthermore, the growth and activities of bacteria and fungi exhibit adaptability in response to crop yield and the chemical, physical, and biological properties of the soil (Kou et al., 2023; Zeng et al., 2024).
Several studies have documented shifts in soil microbial communities following fertilizer application (Sivojiene et al., 2021; Wang et al., 2022). For example, Sivojiene et al. (2021) observed an increase in microbial biomass and diversity with organic fertilizations, while Wang et al. (2022) noted the impact of different fertilization treatments on the bacterial community. Ren et al. (2018) and (2020) highlighted that adequate nitrogen and carbon from organic fertilizer supplementation promote soil microbial growth, but excess phosphorus from fertilization reduces microbial community diversity (Liu et al., 2022). The application of fertilizers, particularly chemical ones, can lead to various adverse effects on soil properties and indirectly impact its microbial ecosystem (Cheng et al., 2020). Continuous application of chemical fertilizers can cause soil acidification, negatively affecting soil structure and fertility (Pahalvi et al., 2021). Imbalanced use of chemical fertilizers can alter soil pH, increase pest attacks, and exacerbate acidification (Krasilnikov et al., 2022). Consequently, acidification can lead to the loss of essential nutrients, decreased soil pH, and reduced availability of micronutrients, ultimately resulting in poorer crop performance and soil degradation (Ozlu and Kumar, 2018).
Fertilizers can also alter the composition and functioning of microbial communities (Kai et al., 2020). They affect soil microorganisms indirectly by changing soil properties or directly through the addition of nutrients (Pan et al., 2020; Yan et al., 2021). Research has shown that prolonged use of chemical fertilizers can significantly reduce soil bacterial diversity, primarily due to a decrease in soil pH levels (Xu et al., 2020; Yang et al., 2020). While some research suggests that organic fertilizer application can increase soil microbial diversity compared to chemical fertilizers (Sun et al., 2015; Kai et al., 2020), other studies indicate the opposite (Hu et al., 2018; Xu et al., 2020; Yang et al., 2020). According to Zhang et al. (2022), while moderate fertilizer use may enhance microbial activity and diversity, excessive application can lead to a decline in beneficial microbes and an increase in pathogenic species. Additionally, the use of combined inorganic and organic fertilization in rice field soils across South China was found to change the abundance of soil microbial communities (Liu et al., 2020). To mitigate the negative impacts of intensive fertilizer use, there is a pressing need for comprehensive and sustainable soil management practices. This has led to a growing interest in eco-friendly and cost-effective technologies, including the utilization of phosphate rock (PR), a low-cost natural resource recognized as a sustainable alternative for agriculture (Vassilev et al., 2001; Reddy et al., 2002).
In this context, our study aims to elucidate the impact of three phosphorus sources—natural phosphorus rock of Morocco (PRM), Triple Super Phosphate (TSP), and NPK 15/15/15—on the composition of soil bacterial groups and the genes involved in the phosphorus cycle.
Materials and methods
Study sites and soil sampling
Our research was conducted in the western region of Ivory Coast from 2019 to 2021, specifically in a rice field situated at the National Center of Agricultural Research (CNRA) station (7° 18′57” N; 7° 27′19” W). The experiment was conducted during the main cropping season, characterized by a mono-modal rainfall pattern. The area receives an average annual rainfall of 1771 mm (Koné et al., 2010). The rainy season extends from February to October, with peak rainfall occurring between March and June. The climate is warm and humid, with mean annual maximum and minimum temperatures of 32°C and 17°C, respectively, and an average relative humidity of 79%.
Before initiating the experiments, soil samples were systematically collected at a depth of 0–20 cm from various locations within the plot to ensure comprehensive coverage and representativeness. These individual subsamples were then combined into a composite sample, which was subsequently sieved (2 mm) and divided into two parts. The first part underwent thorough physico-chemical analysis, while the second part was stored at −4°C for microbiological analysis. The soil’s characterization before experimentation is presented in Table 1.
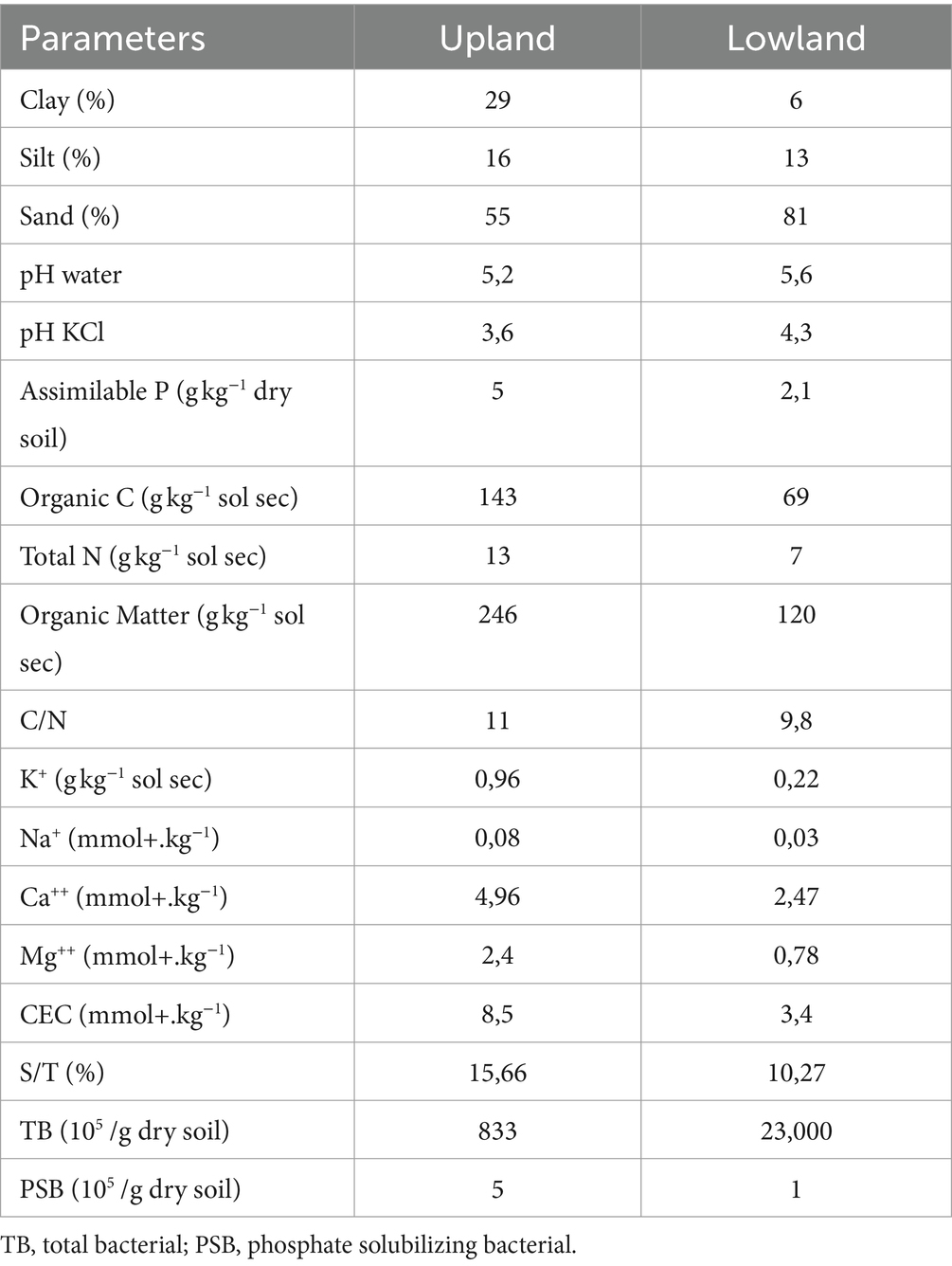
Table 1. Physico-chemical and microbiology characteristics of the paddy soils at 0–20 cm depth before experimentation.
Plant material
Two rice varieties, WITA 9 and IDSA 10, were thoughtfully selected from the National Center of Agricultural Research (CNRA) of Ivory Coast, taking into consideration the specific ecologies of the plot. The rice variety WITA 9 is also called Nimba. It was developed in 1984 by the International Institute of Tropical Agriculture (IIAT) by mixing the variety HI 2042-178-1 and the variety CT19 is an improved variety which was chosen mainly for its short cycle (90 days). It’s average yield of 6 t ha−1 and its potential yield of 10 t ha−1. The seed of WITA 9 provided in National Center of Agricultural Research (CNRA) in Man. The rice variety IDSA 10, also known as Fafa, was provided by the National Center for Agronomic Research (CNRA) in Man. Resulting from a cross between IRAT 112 and Iguape Cateto, it is suited to uplands and slopes and has a short growth cycle of 105 days. Its potential grain yield is 4.8 t/ha. However, in agricultural practice in Côte d’Ivoire, the average harvest is 2.5 t/ha, which can vary depending on the agroecology. This variety is widely adopted in the country.
Phosphorus sources material
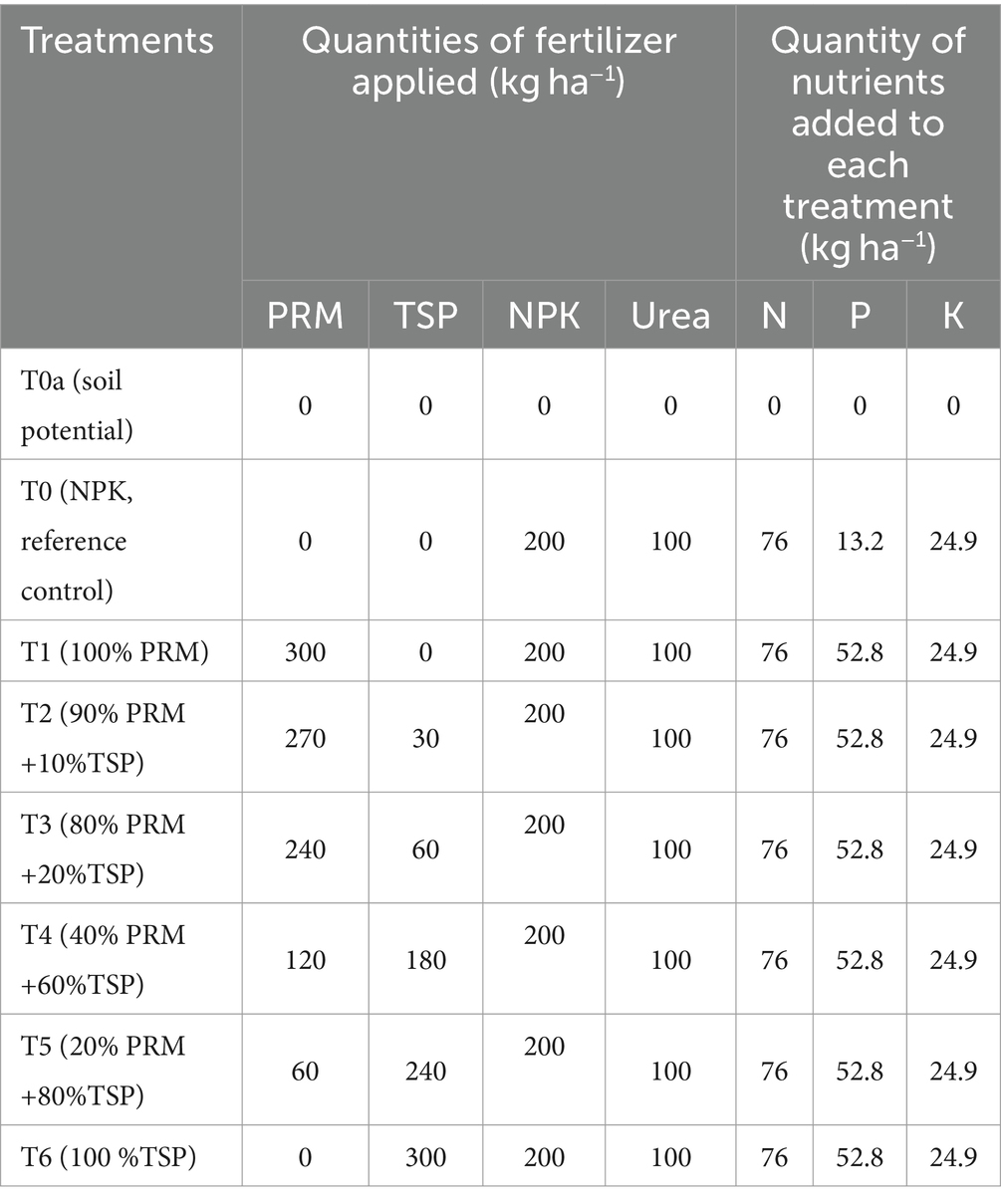
Three different phosphorus sources were employed in this study. The first is a natural phosphorus rock, specifically Morocco phosphate rock (PR), boasting a P2O5 content of 30% with a solubility of 3% in water (Table 2). The second is a chemical phosphorus fertilizer, Triple Superphosphate (TSP), also containing 45% P2O5. The third phosphorus source is NPK fertilizer (15/15/15) was incorporated into the study to ensure a recommended dose of NPK for rice and was applied at a dose of 200 kg NPK ha−1. Both fertilizers (Morocco phosphate rock (PR) and Triple Superphosphate (TSP)) were generously supplied by the Office Cherifien of Phosphate (OCP) and was applied at a dose of 90 kg P2O5 ha−1 or 300 kg TSP or PR per hectare. Another additional nutrient, such as a nitrogen fertilizer in the form of Urea (46% N) was employed at a dose of 100 kg Urea ha−1. The Table 3 gives the composition of the different treatments applied.

Table 2. Elemental chemical composition of Moroccan phosphate rock (MPR) analyzed in 100 g MPR samples.
Trial design
The experimental setup used was a randomized complete block design (RCBD), with a single application of treatments in the first cycle. The experiment was conducted on a 500 m2 plot using a randomized complete block design, with 100 m2 blocks serving as replications. Five replication was considered in each Block design. These blocks were then subdivided into 8 microplots of 25 m2 each, with each microplot representing a distinct treatment. Different treatments based of different phosphorus sources (PR, TSP, NPK) were applied, at a dose of 90 kg P2O5 ha−1 or 300 kg TSP or PR per hectare, only at the beginning of the first cycle in the plots, excluding the control treatment (T0). Additionally, 100 kg ha−1 of 46% Urea was applied, with 50 kg ha−1 at the tillering stage and 50 kg ha−1 at the at panicle stage. For upland plots, after applied the treatment, seeds were directly sown at a rate of four seeds per hole. After germination, thinning was carried out to leave two plants per hole before tillering. To avoid competition between the rice and weeds, manual weeding was performed as needed. No insecticides or fungicides were applied to the plots.
For lowland plots, two days before sowing, the 15 day-old rice plants, NPK 15-15-15 (200 kg ha−1) and the different treatments were applied, as background fertilizer for each plot except for the absolute control. The transplanting was carried out at a rate of 02 plants per pocket.
Microbiological characterization soil samples were collected by treatment at the end of the second cropping cycle.
Quantification and analysis of microbial diversity in rice plot soils
The analysis of the bacterial communities present in soils was carried out using molecular biology based on the extraction of total DNA from the indigenous bacterial at ecology laboratory of the Institute of research of development in agroenvironment (IRDA) in Quebec, Canada (Quebec, QC Canada).
Extraction, amplification, and sequencing of total bacterial DNA from soils
DNA from soil samples was extracted from 0.5 g of soil using the FastDNA Spin kit for Soil (MP Biomedicals, Solon, OH, United States), according to manufacturer’s instructions at the Laboratory of Microbial Ecology of the IRDA (Québec, QC Canada) with three replicates per sample. Extracted DNA was stored at −20°C for amplification and sequencing.
Bacterial DNA amplification focused on the V4 regions of prokaryotic 16S rRNA, employing primer sequences specific to the V4 regions of the SSU rRNA gene (515F and 806R) following the methods outlined by Apprill et al. (2015) and Parada et al. (2016) (Table 4). The chosen approach involved a dual-index, two-step PCR designed for Illumina MiSeq high-throughput sequencing. Consequently, DNA was amplified with primers 515F/806RB and sequenced to evaluate amplification bias and sequencing error rate as described (Tekeu et al., 2023). Paired-end sequencing (2 × 300 base pairs) occurred on Illumina MiSeq at the genomic analysis platform at the Institute of Research and Development in Agroenvironment (IRDA) in Quebec, Canada (Quebec, QC Canada).
Quantitative PCR
The qPCR system utilized primers eub338/eub518 (Fierer et al., 2005) to detect total bacteria. Detection was conducted in duplicate on a CFX96 instrument (Biorad, Hercules, CA, United States) using SYBR green qPCR mix (Qiagen, Toronto, ON, Canada), following the procedure outlined in Tekeu et al. (2023). The detection system was designed within a detection range spanning 4 LOG. This method served to quantify the relative abundance of bacteria in the soil samples, expressed as the number of targeted sequences (Amplified Units) per gram of dry soil (AU g−1 dry soil). Subsequently, these values underwent normalization, and a log transformation was applied.
Predictive functions of phosphate-solubilizing bacteria
Phosphate-solubilizing bacteria are characterized by several biochemical activities, including acid production and enzyme. These functions are determined from taxonomic data obtained by analyzing the diversity of prokaryotes. As part of this study, several genes involved in phosphate solubilization by bacteria or that could serve as biomarkers were identified and mentioned as method described by Wan et al. (2020) and Wu et al. (2022).
Bioinformatics and biostatistics processing
Sequence analysis and grouping into sequence taxonomic units was performed on IRDA’s LEM bioinformatics platform and involved various processing strategies Qiime2 (Bolyen et al., 2019) and R (R Core Team project, 2014), including quality validation steps, reference bases and indices for measuring microbial richness, and comparative measures of microbial diversity. These were grouped into OTUs based on 97% sequence similarity. Taxon assignment was performed using the SILVA version 138 reference database (Quast et al., 2012), which was also used for the analysis of bacterial diversity. For the analysis of predictive functions related to phosphate solubilization, the data obtained from the taxonomic analysis were processed using the Picrust2 approach (Douglas et al., 2019) and 5 marker genes related to this functionality were filtered.
Statistical analysis
Statistical models were developed using the lm function from the agricolae package in R software version 4.3.2 (R Core Team, 2023), with its RStudio interface. The data are presented as mean ± standard deviation. Statistical differences were considered significant for p < 0.05. The significant difference between treatments was analyzed by Newman–Keuls test in one-way ANOVA and multiple comparisons of means (p < 0.05) were conducted using the Newman–Keuls test to identify homogenous groups at the 5% probability threshold.
Results and discussion
Results
Treatment effect on the relative abundance of bacterial community composition
The addition of different phosphate amendment does not alter the soil indigenous bacterial community composition in the two rice plots studied, with the presence of five (5) phyla Firmicutes, Actinobacteria, Proteobacteria, Halobacterota, and Chloroflexia (Figure 1), regardless of the treatments applied and the ecological characteristics of the plots (upland/lowland).
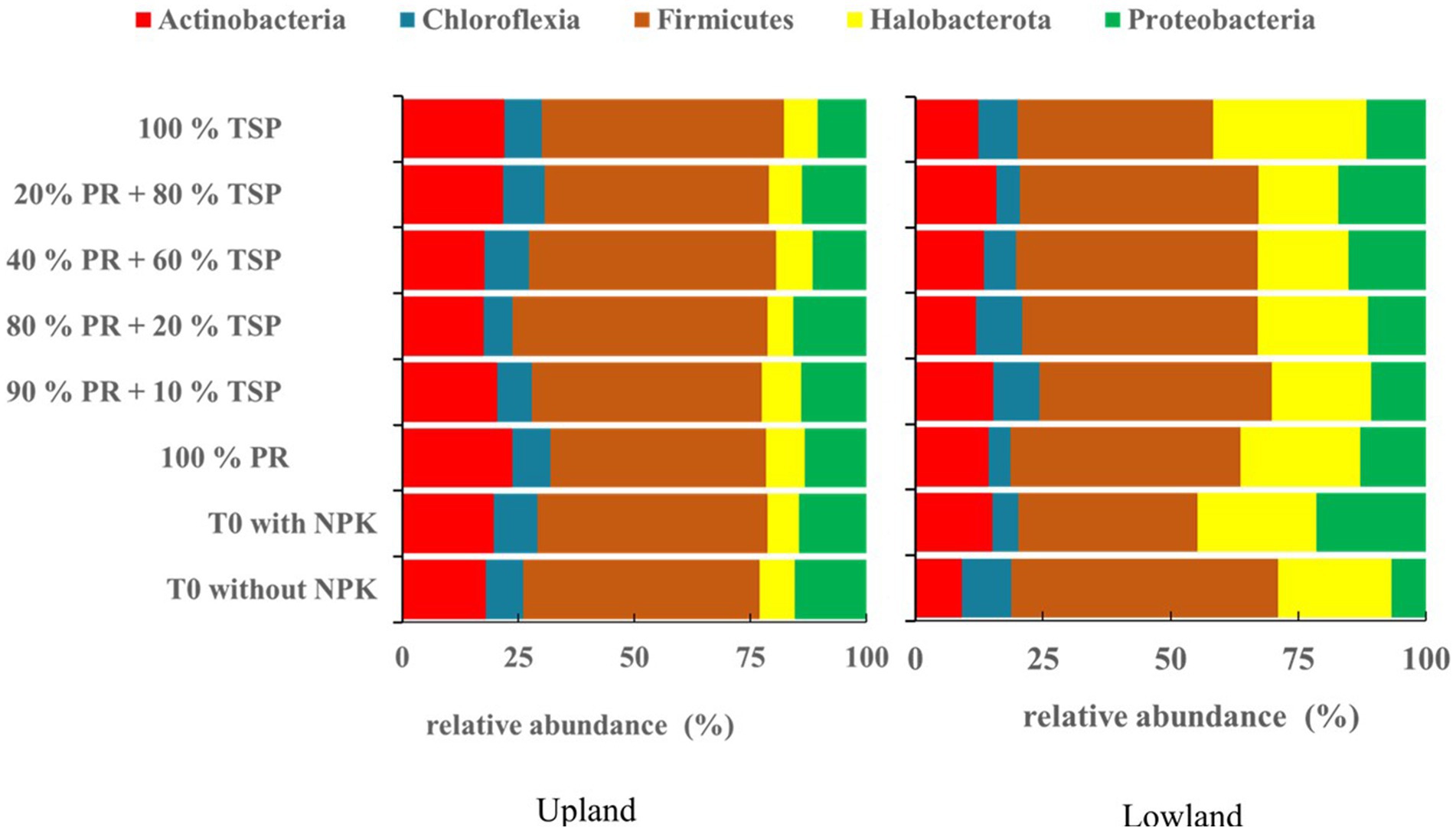
Figure 1. The relative abundance (%) of bacterial community composition among the primary prokaryotic groups detected in upland and lowland habitats (groups greater than 1%) under different treatments.
The Firmicutes phylum (35 to 53.25%) is the most dominant, followed by the Halobacterota phylum (6.7 to 30%), Actinobacteria phylum (9 to 23.8%), and Proteobacteria phylum (5 to 21%) in each phylum (Figure 1). While, the Chloroflexia phylum (4 to 9.58%) appears least abundant phylum in the both rice plots (Figure 1).
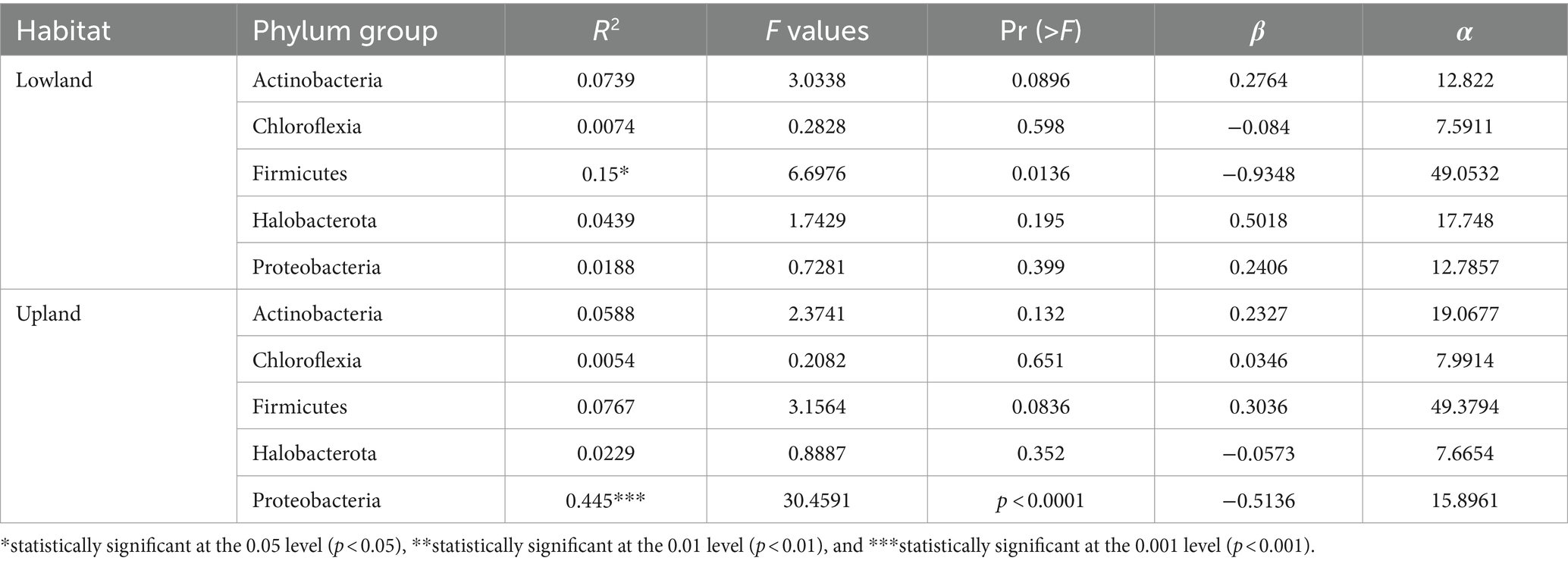
Our results demonstrate that the relative abundance of different phyla varied between the two ecological plot types studied, irrespective of the treatment applied. Specifically, Actinobacteria, Chloroflexia, Firmicutes and Proteobacteria phlyla exhibited significantly higher relative abundances in upland plots compared to lowland plots (Figure 2 and Table 5). Conversely, for Halobacterota phylum, their relative abundance was higher in lowland plots than in upland plots (Figure 2 and Table 5).
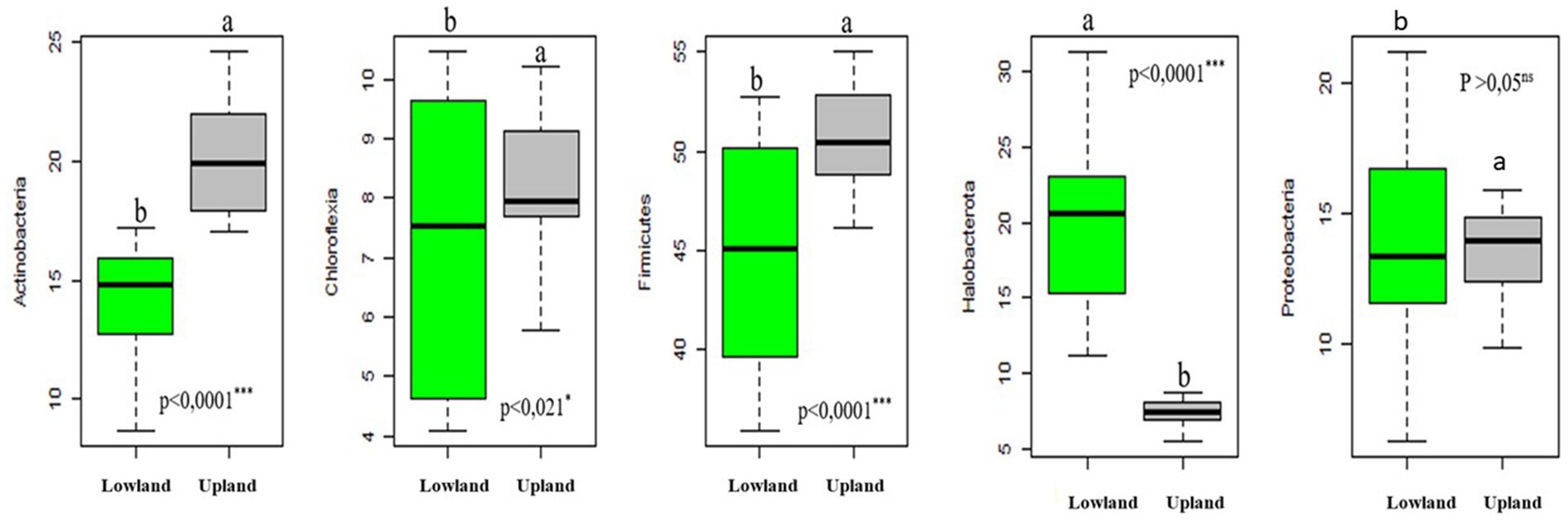
Figure 2. Boxplot showing the relative abundance bacterial groups (phyla) for two-habitats plots studied (upland and lowland) (groups greater than 1%). The thick horizontal line represents the median of the distribution. Different letters indicate differences among medians according to the Tukey test (p < 0.001).
Indeed, the addition of soluble NPK fertilizer decreases the relative abundance of the Chloroflexia phylum from 9% under T0a to 5% under T0 (with NPK), and of the Firmicutes phylum from 52.3% under T0a to 36.7% under T0 (Figure 3), regardless of plot habitat. As for the Actinobacteria and Proteobacteria phyla, there is a significant notable increase in relative abundance, respectively, from 9% under T0a to 19.6% under T0, and from 6% under T0a without NPK to 20.9% under T0 (Figure 3). Additionally, it is observed that the addition of NPK does not affect the relative abundance of Halobacterota compared to the control T0a (Figure 3).
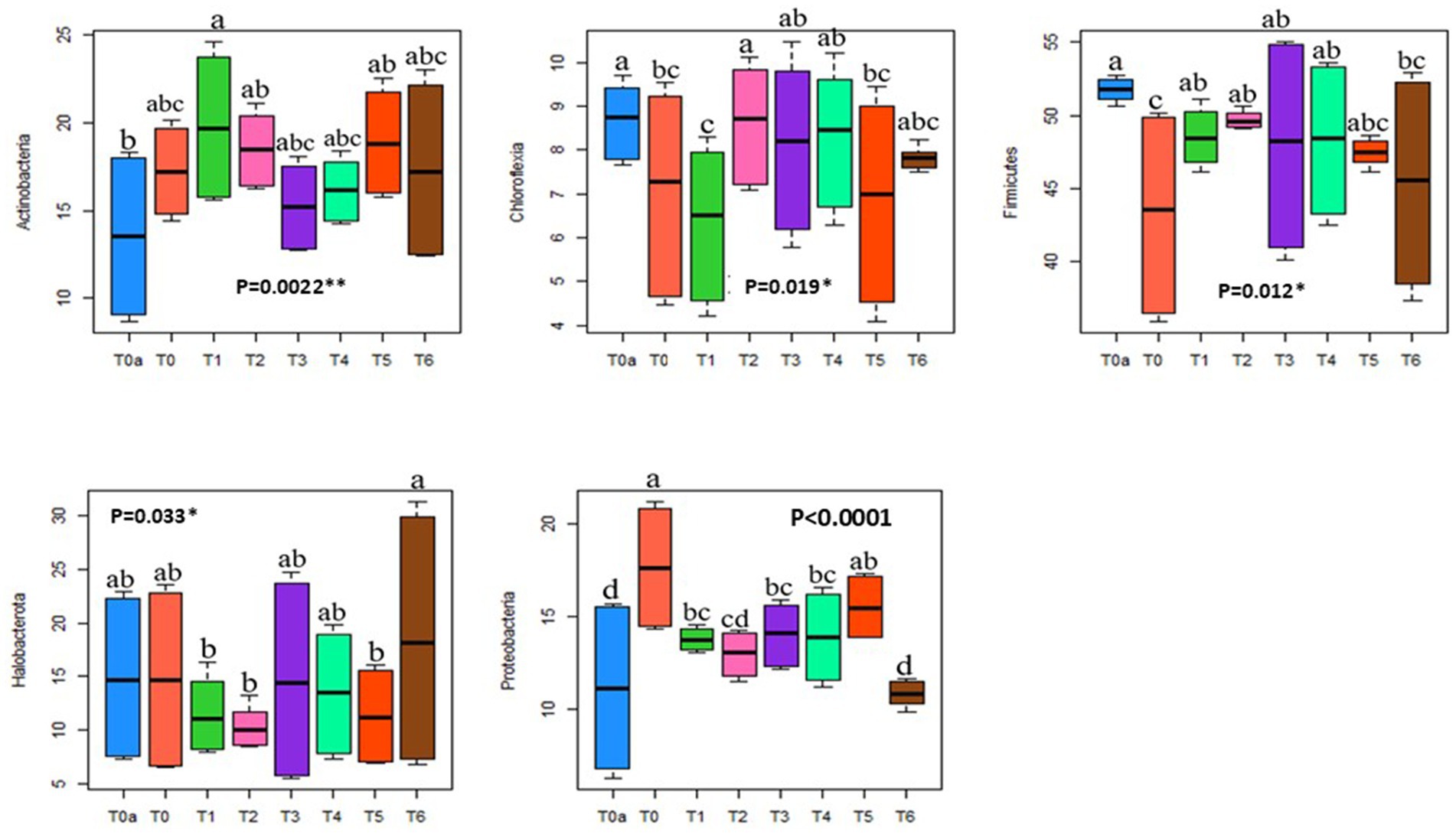
Figure 3. Boxplot showing the relative abundance of bacterial phyla among the primary prokaryotic groups detected (greater than 1%) under different treatments. The thick horizontal line represents the median abundance of each phylum within each treatment. Blue indicates T0a (soil potential without NPK, PRM, and TSP), orange indicates T0 (reference control with NPK), dark green indicates T1 (100% PRM), pink indicates T2 (90% PRM + 10% TSP), violet indicates T3 (80% PRM + 20% TSP), light green indicates T4 (40% PRM + 60% TSP), red indicates T5 (20% PRM + 80% TSP), and brown indicates T6 (0% PRM + 100% TSP).
Our findings demonstrate that amending the plot with phosphate rock from Morocco (100% PRM, T1) significantly alters the relative abundance of the five detected phyla compared to the unamended plot (T0a). The presence of phosphate rock (PRM) significantly reduces the relative abundance of the Chloroflexia (from 8.7 to 6.31%), Halobacterota (from 14.9 to 11.5%), and Firmicutes (from 51.8 to 48.6%) phyla, while increasing the relative abundance of Actinobacteria (from13.5 to 19.8%) and Proteobacteria (from 11.1 to 13.8%), compared to the control treatment (T0a) (Figure 3).
On the other hand, when plots were amended with a combination of phosphate rock and triple superphosphate (TSP), particularly in treatments T3 (80% PRM + 20% TSP) and T4 (40% PRM + 60% TSP), resulted in a reduction in the relative abundance of Chloroflexia (from 8.65 to 8%), and Firmicutes (from 51.8 to 47.92%), alongside an elevation in the relative abundance of Proteobacteria (from 11.1 to 14%) and Actinobacteria (from 13.5 to 16.2%), compared to the untreated plot (T0a) (Figure 3). Additionally, the relative abundance of Halobacterota remained relatively constant at 14.93 to 13.5% compared to the unamended plot (T0a).
Furthermore, when plots received only Triple Super Phosphate (TSP) as the phosphate amendment, compared to the control treatment (T0a), significant modifications were observed in the relative abundance of the five detected phyla. The presence of TSP alone decreased the relative abundance of Chloroflexia (from 8.65 to 7.8%) and Firmicutes (from 51.8 to 45.4%), while increasing the relative abundance of Actinobacteria (from 13.5 to 17.4%), as well as Halobacterota (from 14.93 to 18.6%), compared to the unamended plot (T0a) (Figure 3). There was no significant effect on the relative abundance of Proteobacteria remained relatively constant at 11.13 to 10.8% compared to the unamended plot (T0a).
Treatment effect on phosphorus cycle genes
Whatever the habitat of the rice plots (upland/lowland) and treatment applied, five (5) phosphorus cycle genes were identified in the soils. These were (Table 6):
• Enzymes involved in inorganic phosphate solubilization (gcd [EC:1.1.5.2] quinoprotein glucose dehydrogenase and ppk [EC:2.7.4.1] polyphosphate kinase).
• Enzymes involved in organic phosphate mineralization (3-phytase [EC:3.1.3.8], appA [EC:3.1.3.26 3.1.3.2] 4-phytase/acid phosphatase and phoD [EC:3.1.3.1] alkaline phosphatase D).
In our study, among the five phosphorus cycle genes identified in the soil plots, the phoD and ppk genes were found to have the highest relative abundance in upland soils, regardless of the applied treatment. However, in lowland soils, the phoD, ppk, and gcd genes were identified as having the highest relative abundance (Figure 4).
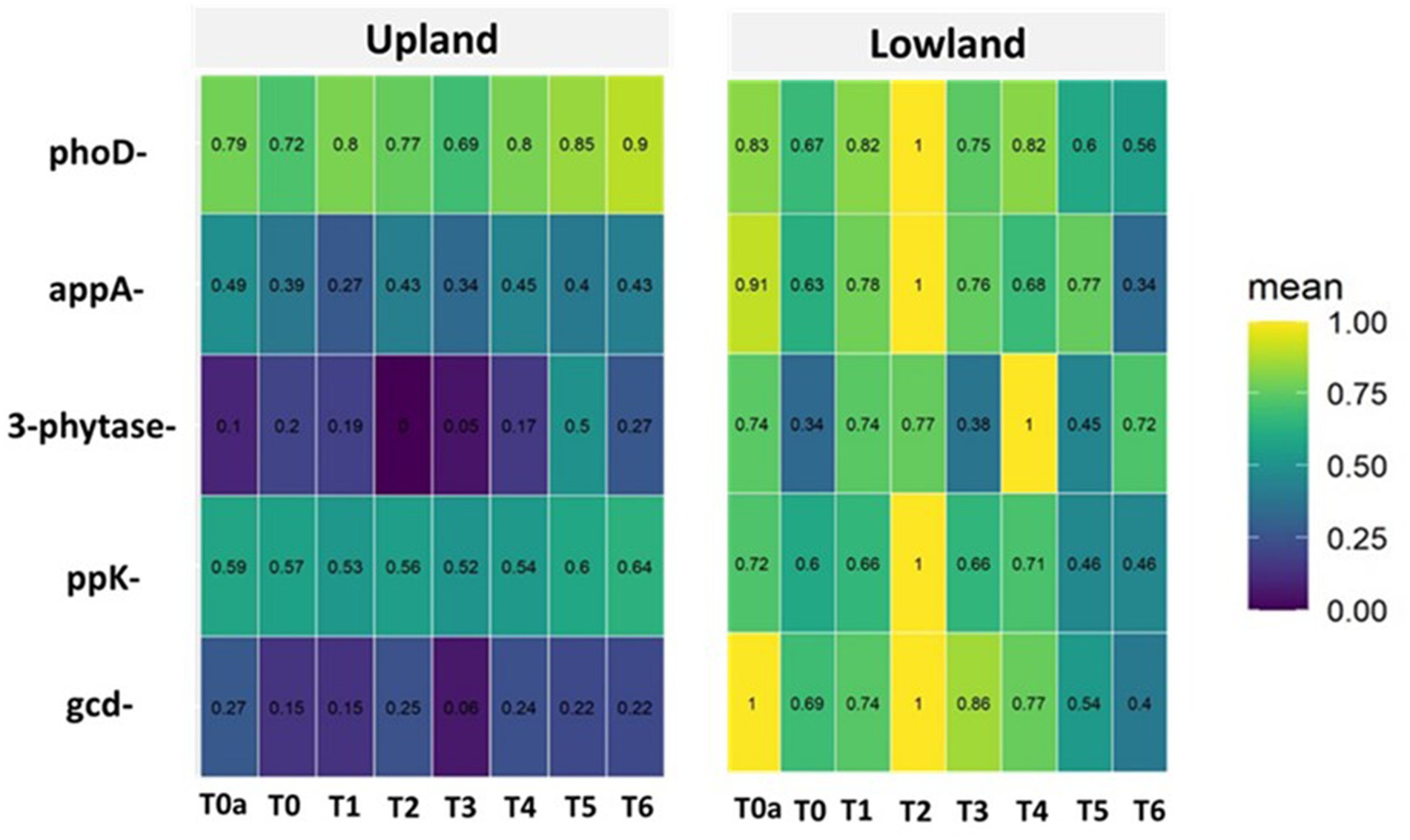
Figure 4. Abundance of phosphorus cycle genes identified in upland and lowland habitat. Treatments: T0a (absolute control without NPK), T0 (Control with NPK), T1 (100% RP); T2 (90% RP + 10% TSP), T3 (80% RP + 20% TSP), T4 (60% RP + 40% TSP), T5 (20% RP + 80% TSP), T6 (100% TSP).
The application of treatments has influenced the abundance of phosphorus cycle genes identified.
Indeed, when compared to soil potential (T0a, without NPK, control), it is evident that the addition of soluble NPK fertilizer decreases in upland, the relative abundance of appA gene (from 49% under T0a to 39%) under T0 (with NPK), and of gcd gene (from 27% under T0a to 15%) under T0 (Figure 4). As for 3-phytase gene, there is a significant notable increase in relative abundance, from 10% under T0a to 20% under T0 (Figure 3). Additionally, it is observed that the addition of NPK does not affect the relative abundance of phoD (79 to 72%) and ppK (59 to 57%) genes compared to the control T0a (Figure 4). In lowland, there is a significant notable decrease in relative abundance of gcd (100 to 69%), 3-phytase (74 to 34%), appA (91 to 63%) and phoD (83 to 67%).
Our findings demonstrate that amending the plot with phosphate rock from Morocco (100% PRM, T1) significantly reduces the abundance of appA (49 to 27%) and gcd (27 to 15%) genes, compared to the unamended plot (T0a), in upland plot, while increasing the relative abundance of 3-phytase (10 to 19%). Additionally, it is observed that the presence of PRM does not affect ppk gene (59 to 53%), and phoD (79 to 80%), compared to the control treatment (T0a) (Figure 4). In lowland, the presence of PRM significantly reduces the abundance of appA (91 to 78%), ppk (72 to 66%), and gcd (100 to 74%) genes, compared to the unamended plot (T0a). There was no significant effect of PRM on the abundance of phoD and 3-phytase genes remained relatively constant, respectively, at 83 to 82% and at 74 to 74%, compared to the unamended plot (T0a).
On the other hand, when plots were amended with a combination of phosphate rock and triple superphosphate (TSP), particularly in treatments T3 (80% PRM + 20% TSP) and T4 (40% PRM + 60% TSP), resulted in upland a reduction in the abundance of appA (49to 34%), 3-phytase (10 to 0%), gcd (27 to 6%), alongside there was no significant effect of PRM on the abundance of phoD and ppk genes compared to the unamended plot (T0a) (Figure 4). In lowland, the combination of phosphate rock and triple superphosphate (TSP) significantly reduces the abundance of appA (91 to 68%), phoD (83 to 75%), and gcd (100 to 77%) genes, compared to the unamended plot (T0a), while increasing the relative abundance of 3-phytase (74 to 100%) and ppK (72 to 100%) genes (Figure 4).
Furthermore, when plots received only Triple Super Phosphate (TSP) as the phosphate amendment, compared to the control treatment (T0a), significant modifications were observed in the abundance of phosphorus genes. In lowland plot, the presence of TSP alone decreased the abundance of ppk (72 to 42%), gcd (100 to 40%), appA (91 to 34%), and phoD (83 to 56%), genes, while there was no significant effect on the abundance of 3-phytase (74 to 72%). In upland plot, the presence of TSP alone increased the abundance of ppk (59 to 64%), phoD (79 to 90%) and 3-phytase (10 to 27%) genes, while there was no significant effect on the abundance of gcd (27 to 22%) and appA (49 to 43%) genes (Figure 4).
Discussion
In agricultural soil, in order to mitigate the limited availability of phosphorus nutrients, agriculturists commonly resort to artificial supplementation through both inorganic and organic fertilization methods. Soil fertilization has been shown to improve bacterial diversity and abundance in the soil (Sun et al., 2015).
Impact of plot habitat on composition and abundance bacterial communities
The investigation carried out on rice field soils from both upland and lowland plots at the Man Research Station revealed the presence of five (05) phyla bacterial (Firmicutes, Actinobacteria, Proteobacteria, Halobacterota, Chloroflexi) in plots of both ecologies, indicating stability in bacterial communities composition regardless ecological differences. However, variations in the relative abundance of phyla were observed depending on the plot habitat, likely attributable to divergent agricultural practices employed in these two ecological settings. Numerous studies indicate that various soil management practices strongly affect the soil microbiota by enhancing bacterial community structure and simultaneously improving soil health (Khmelevtsova et al., 2022; Su et al., 2022; Singh et al., 2023; Górska et al., 2024). In fact, in lowland areas, the cultivation technique is based on incorporating organic residues into the soil before transplanting, while on the upland, it involves burning and then direct seeding. This finding is consistent with previous studies by of Szoboszlay et al. (2017) and Li et al. (2018), which highlight the adverse effects of certain agricultural practices, particularly plowing, on fungi presence by disrupting hyphae. Consequently, numerous prior investigations have underscored the profound impact of agricultural practices on microbial communities (Geisler, 2009), with evidence indicating influence on the abundance of denitrifying bacteria groups (Assémien et al., 2017). This study suggests that the composition of bacterial communities in soils remains relatively stable across different ecologies, yet modifications in the relative abundance of phyla occur, as proposed in the works of Vian et al. (2009).
Effect of diverse phosphorus sources on the composition and abundance soil bacterial groups
The study investigated how different phosphorus sources, such as natural phosphate rock from Morocco (PRM) and chemical fertilizers (Triple super phosphate (TSP) and NPK fertilizers), impact soil bacterial groups. Intriguingly, despite the various treatments applied, the composition of bacterial communities remained unaltered, comprising five main phyla: Firmicutes, Actinobacteria, Proteobacteria, Halobacterota, and Chloroflexia.
Our findings revealed that the application of soluble NPK alone did not alter the composition of bacterial indigenous groups in the studied soils. Regardless of the treatments, the presence of all five phyla remained unchanged. However, compared to the control treatment (T0a, without NPK), the relative abundance of certain phyla in the soil varied in response to NPK application. Specifically, it reduced the relative abundance of Chloroflexia by 19% and Firmicutes by 16.4%, while increasing the relative abundance of Actinobacteria and Proteobacteria by 27.5 and 58.8%, respectively. This finding contrasts with the study by Ma et al. (2018), which showed low abundance levels of Proteobacteria under NPK treatment. Notably, the application of soluble NPK fertilizer alone seemed to affect the relative abundance of soil bacterial groups but not their diversity, as demonstrated by Zhou et al. (2021), which observed a decrease in certain soil bacterial classes under both low and high NPK application. Our results align with several previous studies that reported an increase in the abundance and activity of ammonium-oxidizing bacteria (AOB) in response to NPK fertilizer addition (Simonin et al., 2015; Assémien et al., 2017). According to Rao et al. (2021), this is because NPK fertilization only affects the quantity of soil organic matter rather than its diversity, resulting in minimal impact on the functional structure of the soil.
Similarly, the application of phosphate rock from Morocco (100% PRM, T1) alone resulted in variable changes in the relative abundance of phyla. Specifically, reductions of 27.1% for Chloroflexia, 22.9% for Halobacterota, and 6.2% for Firmicutes were observed, alongside increases of 46.6% for Actinobacteria and 23.8% for Proteobacteria. This highlights the differing impacts of natural phosphate rock on soil bacterial community distribution. Previous studies have shown that the application of rock phosphate improves the phosphorus (P) content in soil, causing significant changes in the soil microorganism community (He et al., 1997; Khoshru et al., 2023; Muhammad Bello et al., 2023). This enhancement in phosphorus availability can lead to shifts in microbial populations, fostering a more diverse and active soil microbiome, as demonstrated by Silva et al. (2017). Furthermore, the variation in the relative abundances of different bacterial groups identified could be linked to soil phosphorus forms, as suggested by Wu et al. (2022).
The use of chemical triple superphosphate (TSP) fertilizer led to increases of 27.1% for Actinobacteria and 24.8% for Halobacterota, while reducing the relative abundance of Chloroflexia by 8.6%, Firmicutes by 12.6%, and Proteobacteria by 0.6%. These findings align with Trabelsi et al. (2017), who observed an increase in Actinobacteria abundance in TSP-treated soil. In contrast, previous studies like Silva et al. (2017) noted a decline in Proteobacteria and Chloroflexi under chemical TSP fertilization compared to untreated soil. Additionally, Peine et al. (2019) demonstrated that TSP application alone boosted microbial biomass and subsequent phosphorus uptake in the soil compared to unfertilized controls.
Additionally, our results revealed that the combination of chemical fertilizers (TSP, NPK) and natural phosphate rock resulted in an increase in the abundance of Proteobacteria (24 to 40%) and Actinobacteria (13 to 39%), and a decrease in the relative abundance of Chloroflexia (5.2 to 22%) and Firmicutes (6 to 12.3%) compared to the control treatment (T0). This study demonstrates that the combination of phosphate rock and chemical fertilizers, such as triple superphosphate modifies bacterial abundance, either increasing or decreasing it (Silva et al., 2017). Similar effect has been observed by Kou et al. (2023); which noted significantly changed the microbial community structure, increasing microbial activity (microbial biomass and soil respiration) when N was combined to P fertilizers. Our results indicate that when chemical fertilizers are combined with phosphate rock, their effects are mitigated, particularly on Firmicutes and Halobacterota. This result does not differ from those of Lori et al. (2023) and Wang et al. (2018), who reported that bacterial α-diversity was unaffected by fertilization intensity, while their community structure changed consistently.
These findings illuminate the intricate relationship between soluble NPK, TSP fertilizers, and natural phosphate rock and their impacts on the relative abundance of bacterial groups in soils. They highlight that each of these phosphorus sources exerts distinct effects on soil bacterial communities. Furthermore, the response of these bacterial groups to phosphate amendments appears to be intricately linked to their inherent characteristics and resilience, as suggested by Kumar et al. (2013). This study affirmed that in soil, the resistance and resilience of microbial populations vary according to their characteristics and nature. For instance, fungi and actinomycetes exhibit greater resistance and resilience compared to other groups of organisms.
These findings imply that soil bacteria possess varying degrees of adaptability and sensitivity to changes induced by different phosphorus sources. This complexity underscores the intricate nature of soil microbial ecology and emphasizes the importance of gaining a nuanced understanding of how soil bacteria interact with environmental factors, including phosphorus availability and soil pH, to uphold ecosystem balance and function.
Effect of phosphate amendments on the abundance of phosphorus cycle genes in soils
The analysis of phosphorus cycle genes involved in phosphate rock dissolution within rice field plots revealed the presence of five key phosphorus cycle genes (phoD, appA, ppk, gcd, 3-phytase), regardless of plot habitat. Our findings indicate that these diverse P-cycle genes are exclusively attributed to bacterial populations. This corroborates the observations of Siles et al. (2022), who highlight that 3-phytase and phoD genes are present in archaea, fungi, and bacteria, while the appA gene is solely found in bacteria. Similarly, Li et al. (2018) and Enebe and Babalola (2021), assert that gcd and ppk genes originate from bacteria. Specifically, the ppk gene serves as the primary enzyme catalyzing polyphosphate synthesis in bacteria and is essential for bacterial mobility (Zhu et al., 2005), while the gcd gene plays a pivotal role in microbial phosphorus metabolism (Li et al., 2018). Our results showed the abundance of P-cycle gene is higher in lowland than upland habitat, probably due to soil environment or characteristic as suggested by Li et al. (2022).
Inputs from various phosphorus sources do not exert influence on the composition of phosphorus-cycle genes in rice field soil. Nevertheless, the presence of these diverse P-cycle genes can enhance the dissolution of phosphate rocks or maintain P solubility in the soil. Interestingly, the application of 90 kg P2O5 ha−1 only in the first year of cultivation did not affect the composition of soil P-cycle genes, in contrast to findings by Liu et al. (2017) and (2022), who observed modifications in the composition of inorganic P-solubilization and organic P-mineralization genes in soil due to long-term high-P inputs (200 kg P2O5 ha−1 year−1).
Furthermore, regardless of the applied treatment, the phoD and ppk genes, are the significantly most abundant genes in these soils. The abundance of these P-cycle genes indicates that microorganisms in the rice field soils of the Man research station have a strong potential to mineralize organic phosphates in the soil through alkaline phosphatases (phoD) and acid phosphatases (appA, 3-phytases) (Li et al., 2018; Dai et al., 2020) and to solubilize inorganic phosphates through quinoprotein glucose dehydrogenases (gcd) and polyphosphate kinases (ppk) (Siles et al., 2022).
However, our findings revealed that the application of soluble NPK alone did not alter the composition of phosphorus cycle genes in the studied soils but modified their abundance by decreasing the abundance of inorganic P-solubilization (ppk and gcd) and organic P-mineralization genes (3-phytase, appA, and phoD).
Moreover, the input of natural phosphate rock from Morocco (100% PRM) as phosphate amendment significantly reduces the abundance of appA and gcd genes, have various effect on the abundance of ppK genes, according on soil characteristic, and have no significant effect on 3-phytase and phoD genes compared to the unamended plot (T0a).
The exclusive use of Triple Super Phosphate (TSP) as the phosphate amendment resulted in notable changes in the abundance of phosphorus genes. Specifically, there was a significant decrease in the abundance of ppk, gcd, appA, and phoD genes. In contrast, the abundance of 3-phytase showed no significant effect, exhibiting both increases and decreases in abundance of ppk, phoD, and 3-phytase genes, depending on the soil characteristic (Liu et al., 2023).
However, when plots were amended with a combination of phosphate rock and triple superphosphate (TSP), there was a reduction in the abundance of appA, 3-phytase, and gcd genes and an enhancement in the abundance of 3-phytase and ppk genes, alongside varying effects of PRM on the abundance of phoD depending on soil characteristic.
It appears that the nature of phosphorus sources does not alter the composition of phosphorus cycle genes but significantly modifies the abundance of P-cycle genes, depending on soil characteristics or environmental factors. This variation is attributed to the nature of the amendment and also to environment factors.
Conclusion
The analysis of bacterial diversity in paddy field soils reveals the presence of various bacterial groups, including the Firmicutes phylum, which are known for their ability to solubilize and mineralize organic and inorganic phosphates. Additionally, the identification of five different genes involved in the phosphorus cycle could contribute to the effective dissolution of applied phosphate amendments. This study highlights variable changes in the abundance of bacterial communities and phosphorus cycle genes depending on both the phosphorus source in the amendments and environmental factors. However, combining natural phosphate rock with chemical fertilizers appears to mitigate this impact on soil microorganism abundance.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: Github (https://github.com/BONGOUA/BONGOUA-DEVISME--10.3389-fmicb.2024.1409559/commit/9f0f392b9b9dbc92d33b6f55c0630e406e9eee1f) and NCBI BioSample (https://www.ncbi.nlm.nih.gov/biosample/), accession numbers SAMN43473894- SAMN43473925.
Author contributions
AB-D: Formal analysis, Supervision, Writing – review & editing, Writing – original draft. SK: Methodology, Writing – original draft. K-KK: Supervision, Writing – review & editing. BL: Methodology, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
We would like to sincerely thank the “Office Chérifien du Phosphate” (OCP-Africa) for the financial support and the National Center for Agronomic Research (NCAR) of Man and Gagnoa for the technical support provided to the realization of the ASORPRI research project. We also thank each farmer who lent his plot to carry out the field trials.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Apprill, A., McNally, S., Parsons, R., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Assémien, F. L., Pommier, T., Gonnety, J. T., Gervaix, J., and Le Roux, X. (2017). Adaptation of soil nitrifiers to very low nitrogen level jeopardizes the efficiency of chemical fertilization in west african moist savannas. Sci. Rep. 7:10275. doi: 10.1038/s41598-017-10185-5
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Cheng, H., Yuan, M., Duan, Q., Sun, R., Shen, Y., Yu, Q., et al. (2020). Influence of phosphorus fertilization patterns on the bacterial community in upland farmland. Ind. Crop. Prod. 155:112761. doi: 10.1016/j.indcrop.2020.112761
Dai, Z., Liu, G., Chen, H., Chen, C., Wang, J., Ai, S., et al. (2020). Long-term nutrient inputs shift soil microbial functional profiles of phosphorus cycling in diverse agroecosystems. ISME J. 14, 757–770. doi: 10.1038/s41396-019-0567-9
Das, S., Liptzin, D., and Maharjan, B. (2023). Long-term manure application improves soil health and stabilizes carbon in continuous maize production system. Geoderma 430:116338. doi: 10.1016/j.geoderma.2023.116338
Douglas, G. M., Maffei, V. J., Zaneveld, J., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2019). PICRUSt2: an improved and extensible approach for metagenome inference. BioRxiv 672295. doi: 10.1101/672295
Enebe, M. C., and Babalola, O. (2021). The influence of soil fertilization on the distribution and diversity of phosphorus cycling genes and microbes Community of Maize Rhizosphere Using Shotgun Metagenomics. Genes 12:1022. doi: 10.3390/genes12071022
Fierer, N., Jackson, J. A., Vilgalys, R., and Jackson, R. B. (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71, 4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005
Geisler, O. (2009). Etude de l’impact des pratiques agricoles sur les capacités fonctionnelles des communautés microbiennes en lien avec le recyclage de la matière organique. Thèse de doctorat, université de Lorraine, 49
Górska, E. B., Stępień, W., Hewelke, E., Lata, J.-C., Gworek, B., Gozdowski, D., et al. (2024). Response of soil microbiota to various soil management practices in 100-year-old agriculture field and identification of potential bacterial ecological indicator. Ecol. Indic. 158:111545. doi: 10.1016/j.ecolind.2024.111545
He, Z. L., Baligar, V. C., Martens, D. C., and Ritchey, K. D. (1997). Effect of phosphate rock, lime and cellulose on soil microbial biomass in acidic forest soil and its significance in carbon cycling. Biology Fertility Soils 24, 329–334.
Hu, X. J., Liu, J. J., Zhu, P., Wei, D., Jin, J., Liu, X., et al. (2018). Long-term manure addition reduces diversity and changes community structure of diazotrophs in a neutral black soil of Northeast China. J. Soil Sediment 18, 2053–2062. doi: 10.1007/s11368-018-1975-6
Islam, M. R., Hossain, M. B., Siddique, A. B., Rahman, M. T., and Malika, M. (2015). Contribution of green manure incorporation in combination with nitrogen fertilizer in rice production. SAARC J. Agric. 12, 134–142. doi: 10.3329/sja.v12i2.21925
Kai, T., Kumano, M., and Tamaki, M. (2020). A study on Rice growth and soil environments in Paddy fields using different organic and chemical fertilizers. J. Agric. Chem. Environ. 9, 331–342. doi: 10.4236/jacen.2020.94024
Khmelevtsova, L. E., Sazykin, I. S., Azhogina, T. N., and Sazykina, M. A. (2022). Influence of agricultural practices on bacterial Community of Cultivated Soils. Agriculture 12:371. doi: 10.3390/agriculture12030371
Khoshru, B., Alireza, F. N., Debasis, M., Manju, C., Younes, R. D., Gökhan, B., et al. (2023). Rock phosphate solubilizing potential of soil microorganisms: advances in sustainable crop production. Bacteria 2, 98–115. doi: 10.3390/bacteria2020008
Koné, B., Bongoua-Devisme, A. J., Hien, M. P., Coulibaly, K. D., and Koné, A. (2023). Combined effect of Morocco rock phosphate and chemical fertilizer in low-land Rice production in Guinea savanna zone of Côte D’Ivoire: replenishment of degraded Fluvisol for boosting Rice production. J. Waste Manag. Recycl. Technol. 1, 2–7. doi: 10.47363/JWMRT/2023(1)111
Koné, B., Ettien, J.B., Amadji, G.L., Diatta, S., and Camara, M., (2010). Effects of phosphate fertilizers of different origins on rainfed rice production on acidic soils in semi-mountainous forest areas in tropical climates Case of hyperdystric ferralsols under fallows in Ivory Coast. 22, 55–63
Kou, Z., Li, C., Chang, S., Miao, Y., Zhang, W., Li, Q., et al. (2023). Effects of nitrogen and phosphorus additions on soil microbial community structure and ecological processes in the farmland of Chinese loess plateau. J. Aride Land 15, 960–974. doi: 10.1007/s40333-023-0023-6
Kpan, W. H., Bongoua-D, A. J., Kouadio, K.-K. H., Kone, B., and Bahan, F. M. L. (2023). Response of lowland rice to phosphate amendments in three acidics agroecological zones of Côte d'Ivoire: man-Gagnoa-Bouaké. Int. J. Environ. Agric. Biotechnol. 8, 135–144. doi: 10.22161/ijeab.85.18
Krasilnikov, P., Taboada, M. A., and Amanullah, (2022). Fertilizer use, soil health and agricultural sustainability. Agriculture 12:462. doi: 10.3390/agriculture12040462
Kumar, S., Patra, A. K., Singh, D., Purakayastha, T. J., Rosin, K. G., and Kumar, M. (2013). Balanced fertilization along with farmyard manures enhances abundance of microbial groups and their resistance and resilience against heat stress in a semi-arid Inceptisol. Commun. Soil Sci. Plant Anal. 44, 2299–2313. doi: 10.1080/00103624.2013.803562
Li, Y., Wang, J., He, L., Xu, X., Wang, J., Ren, C., et al. (2022). Different mechanisms driving increasing abundance of microbial phosphorus cycling gene groups along an elevational gradient. iScience 25:105170. doi: 10.1016/j.isci.2022.105170
Li, J., Wu, X., Gebremikael, M. T., Wu, H., Cai, D., Wang, B., et al. (2018). Response of soil organic carbon fractions, microbial community composition and carbon mineralization to high-input fertilizer practices under an intensive agricultural system. PLoS One 13:e0195144. doi: 10.1371/journal.pone.0195144
Liu, L., Gao, Z., Yang, Y. G., Gao, Y., Mahmood, M., Jiao, H., et al. (2023). Long-term high-P fertilizer input shifts soil P cycle genes and microorganism communities in dryland wheat production systems. Agric. Ecosyst. Environ. 342, 108226–108809. doi: 10.1016/j.agee.2022.108226
Liu, H., Li, S., Qiang, R., Lu, E., Li, C., Zhang, J., et al. (2022). Response of soil microbial community structure to phosphate fertilizer reduction and combinations of microbial fertilizer. Front. Environ. Sci. 10:899727. doi: 10.3389/fenvs.2022.899727
Liu, L., Li, C., Zhu, S., Xu, Y., Li, H., Zheng, X., et al. (2020). Combined application of organic and inorganic nitrogen fertilizers affects soil prokaryotic communities compositions. Agronomy 10:132. doi: 10.3390/agronomy10010132
Liu, S., Meng, J., Jiang, L., Yang, X., Lan, Y., Cheng, X., et al. (2017). Rice husk biochar impacts soil phosphorous availability, phosphatase activities and bacterial community characteristics in three different soil types. Appl. Soil Ecol. 116, 12–22. doi: 10.1016/j.apsoil.2017.03.020
Lori, M., Hartmann, M., Kundel, D., Mayer, J., Mueller, R. C., Mäder, P., et al. (2023). Soil microbial communities are sensitive to differences in fertilization intensity in organic and conventional farming systems. FEMS Microbiol. Ecol. 99:fiad046. doi: 10.1093/femsec/fiad046
Ma, M., Zhou, J., Ongena, M., Liu, W., Wei, D., Zhao, B., et al. (2018). Effect of long-term fertilization strategies on bacterial community composition in a 35-year field experiment of Chinese Mollisols. AMB Expr 8:20. doi: 10.1186/s13568-018-0549-8
Morris, S. J., and Blackwood, C. B. (2015). The Ecology of the soil biota and their function, Soil microbiology, ecology and biochemistry, 4th edn, Ed. E. A Eldor . Cambridge, MA: Academic Press, 273–309
Muhammad Bello, Z., Muhammad, S., Aliyu Aliero, A., Rabani, A., and Aliyu Dabai, I. (2023). Phosphate solubilization improvement for plant uptake from phosphate rock and phosphate solubilizing microbes consortium: impact on food security. Food Security Challeng. Appr. 72:107029. doi: 10.5772/intechopen.107029
Ozlu, E., and Kumar, S. (2018). Response of soil organic carbon, pH, electrical conductivity, and water stable aggregates to long-term annual manure and inorganic fertilizer. Soil Sci. Soc. Am. J. 82, 1243–1251. doi: 10.2136/sssaj2018.02.0082
Pahalvi, H.N., Rafiya, L., Rashid, S., Nisar, B., and Kamili, A.N. (2021), Chemical fertilizers and their impact on soil health. Microbiota and biofertilizers ; G.H. Dar, R.A. Bhat, M.A. Mehmood, and K.R. Hakeem, Eds.; Springer: Cham, Switzerland, 2, 1–20
Pan, H., Chen, M. M., Feng, H. J., Wei, M., Song, F. P., Lou, Y., et al. (2020). Organic and inorganic fertilizers respectively drive bacterial and fungal community compositions in a fluvo-aquic soil in northern China. Soil Tillage Res. 198:104540. doi: 10.1016/j.still.2019.104540
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Peine, M., Vitow, N., Grafe, M., Baum, C., Zicker, T., Eichler-Löbermann, B., et al. (2019). Effect of triple superphosphate and biowaste compost on mycorrhizal colonization and enzymatic P mobilization under maize in a long-term field experiment. J. Plant Nutr. Soil Sci. 182, 167–174. doi: 10.1002/jpln.201800499
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, 590–596. doi: 10.1093/nar/gks1219
Rao, D., Meng, F., Yan, X., Zhang, M., Yao, X., Kim, K. S., et al. (2021). Changes in soil microbial activity, bacterial community composition and function in a long-term continuous soybean cropping system after corn insertion and fertilization. Front. Microbiol. 12:638326. doi: 10.3389/fmicb.2021.638326
R Core Team . (2014). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
R Core Team . (2023). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/ was added in the main manuscript
Reddy, M. S., Kumar, S., Babita, K., and Reddy, M. S. (2002). Biosolubilization of poorly soluble rock phosphates by aspergillus tubingensis and aspergillus Niger. Bioresour. Technol. 84, 187–189. doi: 10.1016/S0960-8524(02)00040-8
Ren, B., Hu, Y., Chen, B., Zhang, Y., Thiele, J., Shi, R., et al. (2018). Soil pH and plant diversity shape soil bacterial community structure in the active layer across the latitudinal gradients in continuous permafrost region of northeastern China. Sci. Rep. 8:5619. doi: 10.1038/s41598-018-24040-8
Ren, N., Wang, Y., Ye, Y., Zhao, Y., Huang, Y., Fu, W., et al. (2020). Effects of continuous nitrogen fertilizer application on the diversity and composition of rhizosphere soil Bacteria. Front. Microbiol. 11:1948. doi: 10.3389/fmicb.2020.01948
Sahrawat, K. L., Jones, M., Diatta, S., and Adam, A. (2001). Response of upland Rice to fertilizer phosphorus and its residual value in an Ultisol. Commun. Soil Sci. Plant Anal. 32, 2457–2468. doi: 10.1081/CSS-120000384
Setu, H. (2022). Effect of phosphorus and potassium fertilizers application on soil chemical characteristics and their accumulation in potato plant tissues. Appl. Environ. Soil Sci. 2022:1. doi: 10.1155/2022/5342170
Siles, J. A., Starke, R., Martinovic, T., Fernandes, M. L. P., Orgiazzi, A., and Bastida, F. (2022). Distribution of phosphorus cycling genes across land uses and microbial taxonomic groups based on metagenome and genome mining. Soil Biol. Biochem. 174:108826. doi: 10.1016/j.soilbio.2022.108826
Silva, U. C., Medeiros, J. D., Leite, L. R., Morais, D. K., Cuadros-Orellana, S., Oliveira, C. A., et al. (2017). Long-term rock phosphate fertilization impacts the microbial communities of maize rhizosphere. Front. Microbiol. 8:1266. doi: 10.3389/fmicb.2017.01266
Simonin, M., Le Roux, X., Poly, F., Lerondelle, C., Hungate, B. A., Nunan, N., et al. (2015). Coupling between and among ammonia oxidizers and nitrite oxidizers in grassland mesocosms submitted to elevated CO2 and nitrogen supply. Microb. Ecol. 70, 809–818. doi: 10.1007/s00248-015-0604-9
Singh, I., Hussain, M., Manjunath, G., Chandra, N., and Ravikanth, G. (2023). Regenerative agriculture augments bacterial community structure for a healthier soil and agriculture. Front. Agron. 5:1134514. doi: 10.3389/fagro.2023.1134514
Sivojiene, D., Kacergius, A., Baksiene, E., Maseviciene, A., and Zickiene, L. (2021). The influence of organic fertilizers on the abundance of soil microorganism communities, agrochemical indicators, and yield in east lithuanian light soils. Plan. Theory 10:2648. doi: 10.3390/plants10122648
Su, Y., Hu, Y., Zi, H., Chen, Y., Deng, X., Hu, B., et al. (2022). Contrasting assembly mechanisms and drivers of soil rare and abundant bacterial communities in 22-year continuous and non-continuous cropping systems. Sci. Rep. 12:3264. doi: 10.1038/s41598-022-07285-2
Sun, R., Zhang, X.-X., Guo, X., Wang, D., and Chu, H. (2015). Bacterial diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 88, 9–18. doi: 10.1016/j.soilbio.2015.05.007
Szoboszlay, M., Dohrmann, A. B., Poeplau, C., Don, A., and Tebbe, C. C. (2017). Impact of land-use change and soil organic carbon quality on microbial diversity in soils across Europe. FEMS Microbiol. Ecol. 93:fix146. doi: 10.1093/femsec/fix146
Tekeu, H., Jeanne, T., D’Astous-Pagé, J., and Hogue, R. (2023). Artificial network inference analysis reveals the impact of biostimulant on bacterial communities in fumigated soil for potato production against common scab. Front. Soil Sci. 3, 1–19. doi: 10.3389/fsoil.2023.1208909
Trabelsi, D., Cherni, A., Zineb, A. B., Dhane, S. F., and Mhamdi, R. (2017). Fertilization of Phaseolus vulgaris with the Tunisian rock phosphate affects richness and structure of rhizosphere bacterial communities. Appl. Soil Ecol. 114, 1–8. doi: 10.1016/j.apsoil.2016.11.014
Vassilev, N., Vassileva, M., Fenice, M., and Federici, F. (2001). Immobilized cell technology applied in solubilization of insoluble inorganic (rock) phosphates and P plant acquisition. Bioresour. Technol. 79, 263–271. doi: 10.1016/S0960-8524(01)00017-7
Vian, J. F., Peigné, J., Chaussod, R., and Roger-Estrade, J. (2009). Effects of four tillage systems on soil structure and soil microbial biomass in organic farming. Soil Use Manag. 25, 1–10. doi: 10.1111/j.1475-2743.2008.00176.x
Wan, W., Qin, Y., Wu, H., Zuo, W., He, H., Tan, J., et al. (2020). Isolation and characterization of phosphorus solubilizing bacteria with multiple phosphorus sources utilizing capability and their potential for lead immobilization in soil. Front. Microbiol. 11:752. doi: 10.3389/fmicb.2020.00752
Wang, R., Bicharanloo, B., Hou, E., Jiang, Y., and Dijkstra, F. A. (2022). Phosphorus supply increases nitrogen transformation rates and retention in soil: a global meta-analysis. Earth's Future 10:e2021EF002479. doi: 10.1029/2021EF002479
Wang, Q., Wang, C., Yu, W., Turak, A., Chen, D., Huang, Y., et al. (2018). Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese fir plantations. Front. Microbiol. 9:1543. doi: 10.3389/fmicb.2018.01543
Wang, R., Wang, Y., Zhang, Z., Pan, H., Lan, L., Huang, R., et al. (2023). Effects of exponential N application on soil exchangeable base cations and the growth and nutrient contents of clonal Chinese fir seedlings. Plan. Theory 12:851. doi: 10.3390/plants12040851
Wang, C., Zheng, M., Song, W., Wen, S., Wang, B., Zhu, C., et al. (2017). Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol. Biochem. 113, 240–249. doi: 10.1016/j.soilbio.2017.06.019
Wu, Q., Chen, D., Zhou, W., Zhang, X., and Ao, J. (2022). Long-term fertilization has different impacts on bacterial communities and phosphorus forms in sugarcane rhizosphere and bulk soils under low-P stress. Front. Plant Sci. 13:1019042. doi: 10.3389/fpls.2022.1019042
Wu, X., Cui, Z., Peng, J., Zhang, F., and Liesack, W. (2022). Genome-resolved metagenomics identifies the particular genetic traits of phosphate-solubilizing bacteria in agricultural soil. ISME Commun. 2:17. doi: 10.1038/s43705-022-00100-z
Wu, J., Sha, C., Wang, M., Ye, C., Li, P., and Huang, S. (2021). Effect of organic fertilizer on soil Bacteria in maize fields. Land 10:328. doi: 10.3390/land10030328
Xu, Q. C., Ling, N., Chen, H., Duan, Y. H., Wang, S., Shen, Q., et al. (2020). Long-term chemical-only fertilization induces a diversity decline and deep selection on the soil bacteria. Am. Soc. Microbiol. mSystems 5, 337–320. doi: 10.1128/msystems.00337-20
Yan, T. T., Xue, J. H., Zhou, Z. D., and Wu, Y. B. (2021). Biochar-based fertilizer amendments improve the soil microbial community structure in a karst mountainous area. Sci. Total Environ. 794:148757. doi: 10.1016/j.scitotenv.2021.148757
Yang, Y. R., Li, X. G., Liu, J. G., Zhou, Z. G., Zhang, T. L., and Wang, X. X. (2020). Fungal community structure in relation to manure rate in red soil in southern China. Appl. Soil Ecol. 147:103442. doi: 10.1016/j.apsoil.2019.103442
Zeng, P., Zhao, Q., Hu, J., Zhang, X., Mao, B., Qy, S., et al. (2024). Nitrogen addition has divergent effects on phosphorus fractions in four types of soils. Ecol. Process. 13:43. doi: 10.1186/s13717-024-00523-7
Zhang, S., Li, X., Chen, K., Shi, J., Wang, Y., Luo, P., et al. (2022). Long-term fertilization altered microbial community structure in an aeolian sandy soil in Northeast China. Front. Microbiol. 13:979759. doi: 10.3389/fmicb.2022.979759
Zhou, J., Kong, Y., Zhao, W., Wei, G., Wang, Q., Ma, L., et al. (2021). Comparison of bacterial and archaeal communities in two fertilizer doses and soil compartments under continuous cultivation system of garlic. PLoS One 16:e0250571. doi: 10.1371/journal.pone.0250571
Keywords: NPK fertilizers, TSP, natural phosphate rock, bacteria phylum, chemical fertilizer
Citation: Bongoua-Devisme AJ, Kouakou SAa, Kouadio K-KH and Lemonou Michael BF (2024) Assessing the influence of diverse phosphorus sources on bacterial communities and the abundance of phosphorus cycle genes in acidic paddy soils. Front. Microbiol. 15:1409559. doi: 10.3389/fmicb.2024.1409559
Edited by:
Arnab Majumdar, Imperial College London, United KingdomReviewed by:
Poonam Yadav, Banaras Hindu University, IndiaTin Mar Lynn, Ministry of Education, Myanmar, Myanmar
Debojyoti Moulick, Independent Researcher, Kolkata, India
Copyright © 2024 Bongoua-Devisme, Kouakou, Kouadio and Lemonou Michael. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Affi Jeanne Bongoua-Devisme, Ym9uZ291YV9qZWFubmVAeWFob28uZnI=
 Affi Jeanne Bongoua-Devisme1*
Affi Jeanne Bongoua-Devisme1*