- Department of Biology, Santa Clara University, Santa Clara, CA, United States
Most Escherichia coli isolates from humans do not utilize D-sucrose as a substrate for fermentation or growth. Previous work has shown that the Csc pathway allows some E. coli to utilize sucrose for slow growth, and this pathway has been engineered in E. coli W strains to enhance use of sucrose as a feedstock for industrial applications. An alternative sucrose utilization pathway, Scr, was first identified in Klebsiella pneumoniae and has been reported in some E. coli and Salmonella enterica isolates. We show here that the Scr pathway is native to an important subset of E. coli phylogroup B2 lineages that lack the Csc pathway but grow rapidly on sucrose. Laboratory E. coli strains derived from MG1655 (phylogroup A, ST10) are unable to utilize sucrose and lack the scr and csc genes, but a recombinant plasmid-borne scr locus enables rapid growth on and fermentation of sucrose. Genome analyses of Enterobacteriaceae indicate that the scr locus is widespread in other Enterobacteriaceae; including Enterobacter and Klebsiella species, and some Citrobacter and Proteus species. In contrast, the Csc pathway is limited mostly to E. coli, some Shigella species (in which csc loci are rendered non-functional by various mutations), and Citrobacter freundii. The more efficient Scr pathway likely has greater potential than the Csc pathway for bioindustrial applications of E. coli and other Enterobacteriaceae using sucrose as a feedstock.
Introduction
D-Sucrose, a disaccharide of glucose and fructose, is common in the biosphere due to its production in many plant tissues. It is a major product of human agriculture and can be advantageous for microbially-mediated production of alcohols and other industrially-relevant chemicals (Peters et al., 2010). However, the ability to utilize sucrose as a carbon and energy source is not universal among microbes, and is highly variable among Gram-negative bacteria of the family Enterobacteriaceae (Le Bouguénec and Schouler, 2011). For example, isolates of Salmonella and Shigella rarely utilize sucrose, but isolates of Enterobacter and Klebsiella usually do. In our experience, roughly one third of clinical and commensal E. coli isolates ferment sucrose in the API20E rapid identification system (Holmes et al., 1978). We describe here the genomic basis for variability in sucrose utilization in E. coli, Shigella, and other Enterobacteriaceae.
Variability in sucrose utilization within Enterobacteriaceae was initially suggested to be due to carriage of key genes on plasmids, with Wohlhieter et al. (1975) observing that a rare sucrose-utilizing Salmonella isolate could conjugally transfer the phenotype to E. coli, and Palchaudhuri et al. (1977) reporting a sucrose-utilizing E. coli isolate in which the Suc+ trait was apparently carried on a non-conjugal plasmid. An S. typhimurium plasmid, pUR400, conveying the Suc+ phenotype was subsequently characterized (Schmid et al., 1982; García, 1985). Analysis of the plasmid-based Salmonella system (Schmid et al., 1988), along with the homologous chromosomally-encoded Klebsiella pneumoniae system (Sprenger and Lengeler, 1988; Titgemeyer et al., 1996) brought the details of the scr pathway into focus (Figure 1). Sucrose is brought into the periplasm via a sucrose-specific outer membrane porin, ScrY (Hardesty et al., 1991; Schmid et al., 1991; Schülein et al., 1991). From the periplasm, sucrose crosses the cytoplasmic membrane via a PEP-dependent transport (PTS) system, with ScrA comprising the sucrose-specific EIIscr membrane channel component (Lengeler et al., 1982; Schmid et al., 1982). In the cytoplasm, sucrose-6-phosphate is cleaved by sucrose-6-phosphate hydrolase (ScrB) into fructose and glucose-6-phosphate (Schmid et al., 1982). G-6-P is fed into glycolysis, and fructose is phosphorylated to fructose-6-phosphate by fructokinase (ScrK) (Aulkemeyer et al., 1991).
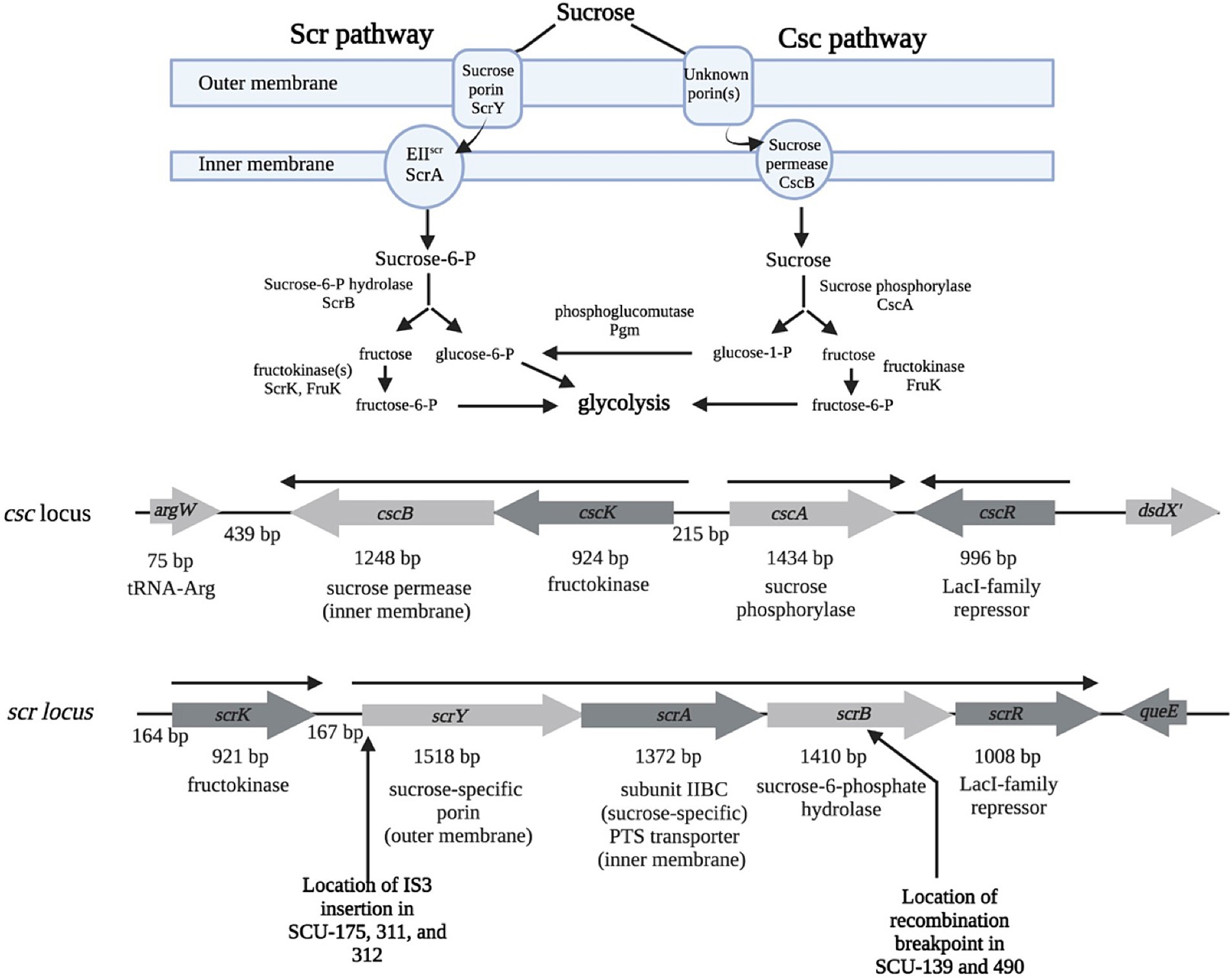
Figure 1. Function and genetic organization of the Scr and Csc pathways for sucrose utilization. Inferred structure and function of the Scr and Csc pathways (reviewed in Reid and Abratt, 2005). Organization of the E. coli genetic loci encoding the Scr and Csc pathways, and the regulators controlling them (cscR and scrR, respectively). The transcriptional organizations of the loci are shown in solid arrows above the genes. Approximate locations of IS3 insertions (scrY) and recombination event (scrB) discussed in Results are indicated below the scr locus. Figure created using BioRender.
As the scr genes were being explored, Alaeddinoglu and Charles (1979) found that sucrose utilization can be a chromosomally-encoded trait in some E. coli, where it was mutually exclusive with D-serine utilization. Bockmann et al. (1992) found that E. coli EC3132 could process sucrose through a pathway they designated Csc (“chromosomally-coded sucrose genes”) (Figure 1; Bockmann et al., 1992). There is no dedicated porin component in the Csc system, so non-specific transport of sucrose into the periplasm via one or more constitutive porin(s) is assumed. Cytoplasmic membrane transport is via the CscB sucrose permease, a non-PTS sucrose-H+ symporter homologous to LacY (Bockmann et al., 1992). In the cytoplasm, sucrose is cleaved by sucrose phosphorylase (CscA) into glucose-1-phosphate and fructose. The csc locus also includes a LacI-type repressor (CscR) that controls expression of the csc genes in response to sucrose; the genes also respond to catabolite repression via cAMP and CAP (Bockmann et al., 1992). As Jahreis et al. (2002) noted, growth of EC3132 on sucrose as the sole carbon and energy source is slow, leading to efforts to increase growth rates by mutation and selection in both EC3132 and E. coli W (Jahreis et al., 2002; Bruschi et al., 2012; Sabri et al., 2013). Enhanced growth rates on sucrose have been achieved via mutations derepressing csc expression by targeting the interaction of CscR with its operator target(s), or by mutationally increasing transport flux through CscB (Jahreis et al., 2002; Sabri et al., 2013).
We explore here the genomic basis for sucrose utilization, or lack thereof, in diverse E. coli lineages, in Shigella species, and in related Enterobacteriacea, finding that the Scr and Csc pathways show starkly contrasting evolutionary histories. These insights not only shed light on the evolutionary history of sucrose utilization in this group of bacteria, but in practical terms may encourage more effective engineering of E. coli for bioindustrial applications with sucrose-rich organic materials as feedstock.
Materials and methods
Bacterial strains and media
Commensal E. coli isolates used in this work were isolated from healthy college students, as described in Stephens et al. (2020). Bacteria were routinely cultured on LB broth or agar. Utilization of sugars as sole carbon and energy sources was tested by growth on M9 minimal salts medium, with the designated sugar at a starting concentration of 10 mM. Plates or liquid media containing D-glucose are referred to as “M9G,” while those containing D-sucrose are referred to as “M9S” herein. Cultures were incubated at 37°C (with shaking for liquid cultures). Fermentation was tested by growth in phenol red broth (Hardy Diagnostics) with the designated sugar present at a starting concentration of 0.5%. Cultures were incubated at 37°C with no shaking.
Bioinformatic analysis
The genome sequences of commensal E. coli used herein have been described previously (Stephens et al., 2020). Isolates were assigned to MLST groups using the web-based MLST 2.0 algorithm (Larsen et al., 2012). BLAST/MegaBLAST searches (Altschul et al., 1990) of these genomes, as well as basic molecular biological processes such as PCR primer design, were done locally using the Geneious Prime desktop bioinformatics package (Dotmatics). Searches of the RefSeq database (Pruitt et al., 2007) were done with NCBI BLAST (Johnson et al., 2008). scr genes from the closest known related species were used as queries, to compensate for evolutionary drift between gene sequences from more distant relatives. To identify E. coli genes uniquely associated with sucrose utilization, a subset of commensal E. coli genomes of known sucrose phenotypes were submitted to the Genomes Online Database (GOLD; Mukherjee et al., 2023). The Phylogenetic Profiler tool within the Integrated Microbial Genomes and Microbiomes platform (IMG/G; Joint Genome Institute, Walnut Creek, USA) (Chen et al., 2023) was then used for analysis.
Cloning and expression of scr locus
Genomic DNA was extracted using the NucleoSpin microbial DNA purification kit (Macherey-Nagel). For amplification of the scr genes by polymerase chain reaction (PCR), primer pairs for the target gene regions were designed using Geneious bioinformatics software (Biomatters LTD), and synthesized by Integrated DNA Technologies (Alameda, CA, USA). The entire 6.6 kb scr locus was amplified using primers scrK43F (TCC CGG CAT ATT CAC GTT TCC AC) and scrR6689R (CCG TTT TAC AGG GGC GAT GCA). The scrY gene alone was amplified using primers scrY1058F (ACC GCC TTA CCC CGA CAA CA) and scrA2818R (TTC AGT AAA AGC CTC ACA TCC GT). Genomic DNA from E. coli strain SCU-147 (Stephens et al., 2020) was used as a template for amplification of the scr genes for cloning. Products of appropriate sizes were verified by agarose gel electrophoresis, then prepared for cloning using the Gel and PCR Clean-up kit (Macherey-Nagel). The PCR Cloning Kit (New England Biolabs) was used to clone amplicons into plasmid pMiniT2.0, and ligated DNA was electroporated into E. coli NEB 10-beta C3019H, with selection on LB agar containing 100 μg/mL ampicillin. Four colonies were selected from each ligation and transformation and used for colony PCR. The same primers were used to assess putative recombinant plasmids. Colonies that produced strong product bands of the target size were grown overnight in liquid LB with 100 μl/mL ampicillin, and plasmid DNA was prepared using a commercial kit (Zymo). Plasmid DNA was digested with restriction enzyme EcoRI to verify expected insert size, and candidate clones were further verified by Sanger sequencing (Sequetech, Mountain View, CA, USA) to confirm the expected insert. Plasmid DNA was subsequently used to transform natural commensal E. coli isolates from Stephens et al. (2020), prepared by calcium treatment (Chang et al., 2017). To remove scrR from the scr locus, pMM007 was digested with BamHI, resulting in a 7.7 kb fragment product (pMM008). A partial internal 500 bp fragment of scrY was removed by digestion with EcoRV. The larger linearized plasmid was re-ligated with T4 DNA ligase using quick T4 DNA ligase (New England Biolabs), then transformed into NEB 10-beta C3019H.
Results
Utilization of D-sucrose by commensal Escherichia coli isolates
To explore the genetic and physiological basis for sucrose utilization in E. coli, we employed a collection of diverse commensal E. coli strains with sequenced genomes (Stephens et al., 2020). The collection includes more than 100 representatives of all major E. coli phylogroups (A, B1, B2, C, D, E, and F) (Denamur et al., 2021). Table 1 shows results for a limited subset of 32 isolates, each representing a different MLST group, and each with fully assembled, closed genomes available in GenBank. Broth and agar plate-based assays were used to examine growth using sucrose as sole carbon and energy source. Representative growth curves and images of M9G and M9S plates are shown in Figure 2. The “Suc+” phenotype (Figures 2B,D) was defined here as being able to utilize D-sucrose as sole carbon and energy source in M9 minimal salts medium at a growth rate similar to glucose; under the conditions applied (small test tube cultures grown in a shaking incubator at 37°C), this corresponded to a growth rate μ of 0.8–1.1 generations/h. Roughly 20% of commensal E. coli strains in our collection were Suc+ (6/32 shown in Table 1). A slightly higher fraction (9/32, Table 1) were able to more slowly utilize D-sucrose for growth (Figure 2C and Table 1; isolates with μ = 0.4–0.7 gen h−1 were designated “slow,” isolates with μ = 0.1–0.3 were designated “very slow” growth). The growth rate of colonies on M9S for these isolates was noticeably slower (Figure 2C). The remaining isolates (17/32 in Table 1) were unable to use D-sucrose as the sole carbon and energy source for significant growth (Suc−, Figure 2A).
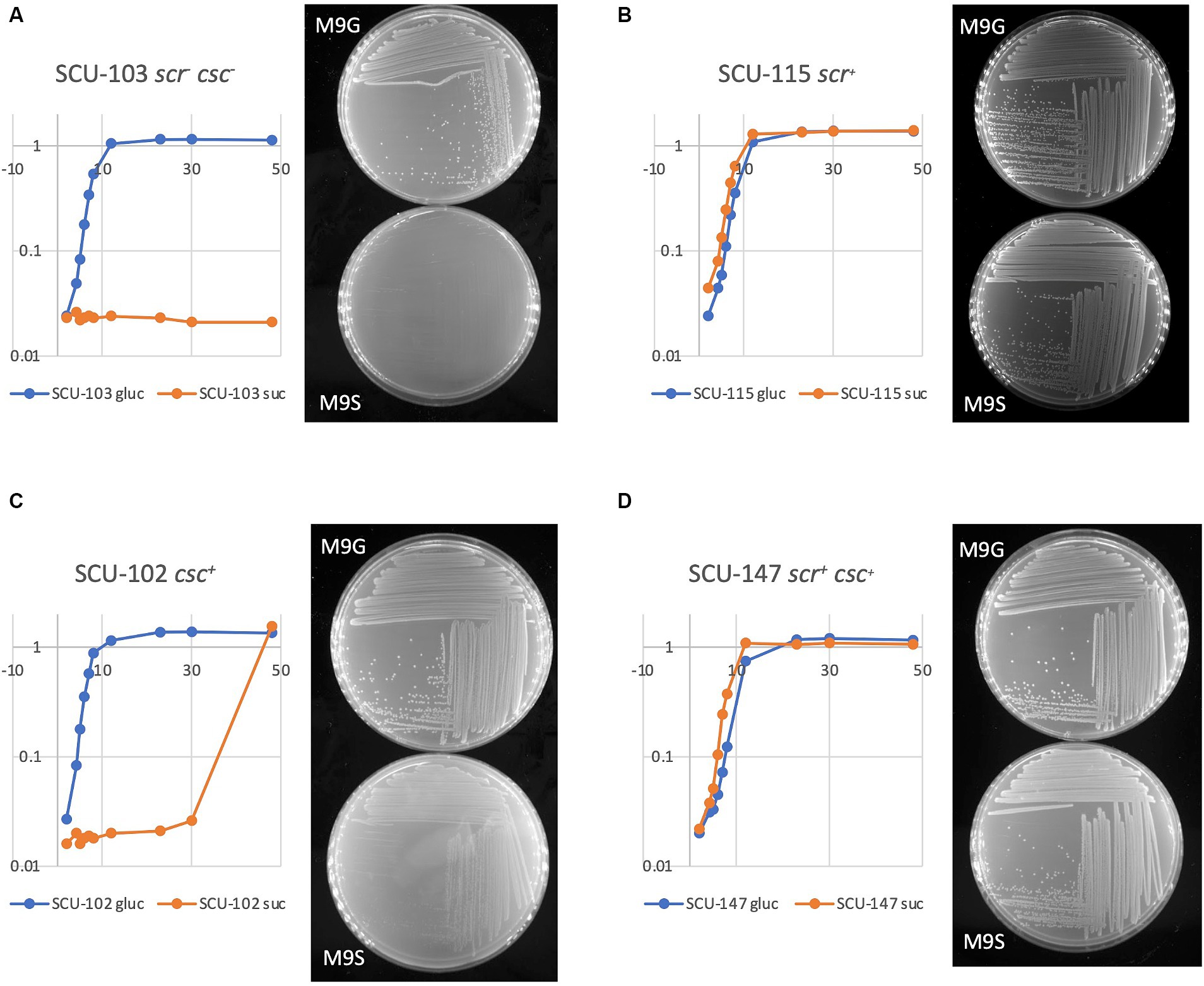
Figure 2. Assays for sucrose utilization in liquid and agar media. Culture growth in M9 minimal salts broth with either glucose or sucrose as sole carbon/energy source was monitored in a spectrophotometer at 600 nm, whom on the left of each panel. M9 agar plates with either glucose (M9G) or sucrose (M9S) were photographed 24 h after streaking.
Genetic basis of sucrose utilization
To identify genes associated with the Suc+ phenotype, fully assembled and annotated genomes of four Suc+ isolates (SCU-115, phylogroup B2, MLST type ST1155; SCU-147, B2/ST1193; SCU-182, B2/ST131; SCU-387, B2/ST14) were compared to the genomes of eight phylogenetically diverse Suc− isolates (SCU-103, A/ST10; SCU-152, B1/ST54; SCU-108 B2/ST95; SCU-112 B2/ST73; SCU-321, D/ST69; SCU-106, E/ST3695; SCU-120 F/ST648, and SCU-301 F/ST967) using the Phylogenetic Profiler tool of the IMG/G platform. Just five genes (scrA, scrB, scrK, scrR, and scrY) were present in the genomes of all four Suc+ isolates (minimum 80% identity) and absent in all of the genomes of the eight Suc− isolates. The scr genes are clustered in two adjacent transcriptional units, scrK and scrYABR (Cowan et al., 1991). The 6.7 kb scr locus was highly conserved, with ≥99% identity among the four Suc+ isolates.
A recombinant plasmid-borne scr locus enables growth on sucrose
To test the sufficiency of the scr locus for sucrose utilization, the region was amplified by PCR and cloned into plasmid vector pMiniT2.0 (Figure 3). The recombinant scr+ plasmid construct pMM_027 was introduced into the E. coli NEB10-beta strain, a derivative of the standard DH10B cloning strain, which is in turn derived from E. coli MG1655 (Durfee et al., 2008), a phylogroup A/ST10 strain. Phylogroup A E. coli in our collection were universally unable to use sucrose, and lack the scr or csc loci. Neither the parental NEB10-beta strain or the recombinant NEB10-beta/pMM_027 strain were able to grown on M9 minimal medium with glucose or sucrose as sole carbon and energy source, presumably due to auxotrophies resulting from the complex genotype of NEB10b. They were, however, able to ferment glucose in phenol-red broth. NEB10-beta/pMM_027 gained the ability to rapidly ferment sucrose in phenol red broth. When pMM_027 was moved into SCU-113 (B1, ST5974, scr− csc−), a prototrophic E. coli commensal isolate, the resulting strain was able to both ferment sucrose and utilize sucrose as sole carbon and energy source on M9 medium, albeit more slowly than a native Suc+ B2 strain. These results indicate that the scr locus is sufficient to convey the Suc+ phenotype.
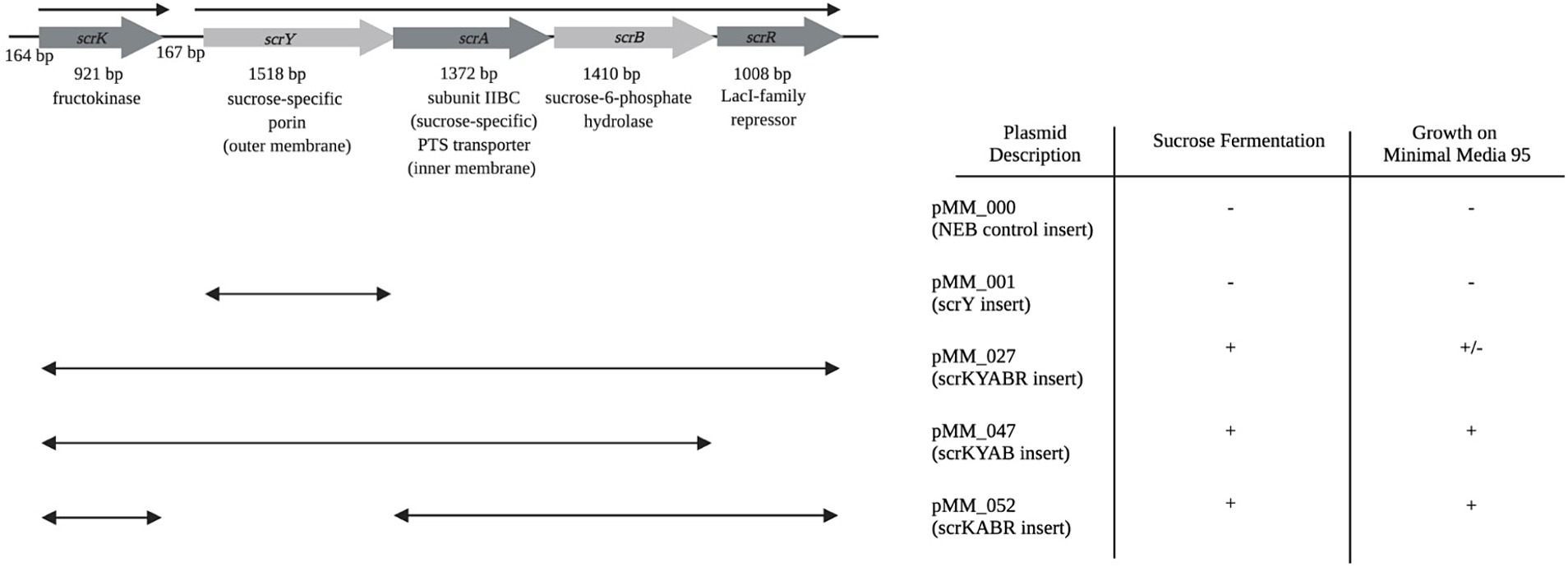
Figure 3. Utilization of sucrose conferred by recombinant plasmids with scr genes. Life side of the figure diagrams inserts in recombinant plasmids (named on the right side) containing the entire scr operon, or fragments thereof. Plasmids were introduced into E. coli SCU-113 (B1, ST5974, scr− csc−). In the table, “+” indicates that fast fermentation/growth was observed, “+/−” indicates that slower fermentation and growth was observed, and “−” indicates that no fermentation/growth was observed.
Removal of the putative scrR transcriptional regulator from the recombinant construct (pMM_047) accelerated growth and fermentation of sucrose, as did deletion of the porin-encoding scrY gene (pMM_052). The scrY deletion removed the scrYABR operon promoter, but we speculate that it is possible that read-through transcription from scrK in the deletion construct provided sufficient expression of scrA and scrB to allow sucrose fermentation and growth.
Phylogenetic distribution of scr locus
The sequenced commensal E. coli genomes in our collection were queried via BLAST for the scr locus (Table 1). The Suc+ phenotype and scr are largely associated with a branch of the B2 clade that includes the ST1193 and ST131 MLST groups, known for their extraintestinal virulence (Pitout et al., 2022). Overall, scr was found in nearly half of B2 isolates.
The presence of the scr locus correlated well with the Suc+ phenotype, as the majority of scr-containing isolates were Suc+ (6/9 in Table 1, and 20/24 in our larger sequenced E. coli collection). Three unusual scr-containing isolates (SCU-175, 311, 312) from outside the B2 clade showed a Suc− phenotype when incubated on M9S plates or liquid media for up to 1 day. However, isolated colonies did appear over time on these plates (SCU-175 is shown as an example in Figure 4). Genome sequence analysis revealed identical 1.9 kb Is3 insertions located 12 bp into the scrY gene (Figure 1). This insertion is expected to knock out transport by ScrY, the sucrose-specific porin of the Scr system (Hardesty et al., 1991; Schmid et al., 1991). The pMM_052 recombinant construct discussed above showed that scrY is not strictly necessary for the Suc+ phenotype, but the Is3 insertion in scrY may also have negative polar effects on expression of the downstream scrA, scrB, and scrR genes. GenBank searches identified four other examples of this precise configuration in E. coli F/ST62 isolates from around the world, but the Is3 insertion is not universal in F/ST62 isolates. In fact, the closely-related SCU-114 isolate in our collection has an intact scr locus and is Suc+ (data not shown). As shown in Figure 4, the isolated colonies selected from prolonged incubation of scrY::Is3 strains on M9 sucrose media stably gain the ability to utilize sucrose, as would be expected if the Is3 element has been excised.
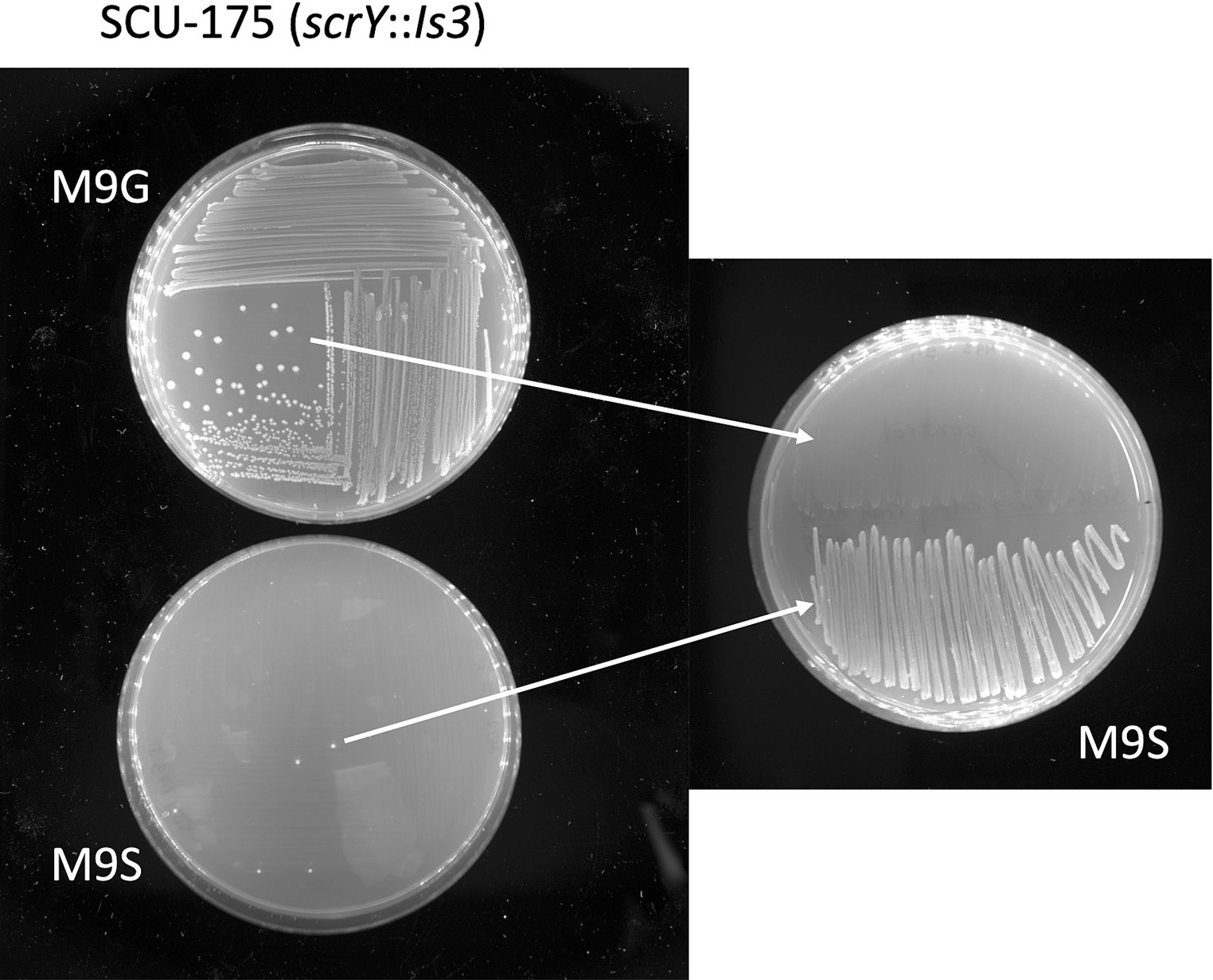
Figure 4. Rare spontaneous conversion of Suc− SCU-175 (scrY::Is3) to Suc+ phenotype. The left panel shows that SCU-175 showed no growth on M9S plates after 48 h, with the exception of rare spontaneous colonies. When those colonies were restreaked on M9S plates (right panel, bottom half of plate) they were Suc+, in contrast to the parental strain streaked on the same plate as a control (right panel, top half).
Some scr+ isolates examined were not fully Suc+; for example, SCU-479 grew slowly, and SCU-176 grew very slowly on M9 sucrose. The basis for these phenotypes is not yet known, although we also noted that SCU-176 grew very slowly on glucose as well, so the growth phenotype may not be attributable specifically to a sugar utilization pathway.
Among 79 isolate genomes in our collection initially determined to lack scr (using the SCU-147 scr locus as a BLAST query), 54 isolates were Suc−, 26 were Sucslow, and two were Suc+: SCU-391 (A, ST216) and SCU-490 (B1, ST6496). Analysis of the SCU-391 and SCU-490 genomes did not reveal hits above 80% identity to the SCU-147 scr locus. However, further inspection showed that these genomes contained a more distantly related scr locus (~67% identity with SCU-147 scr over 6.7 kb). The SCU-391 scr locus closely matches SCU-490 for the scrKYA genes, but they diverge at codon 229 of scrB, continuing into scrR (Figure 1). The scrKYA genes of SCU-391/490 were used as queries to search GenBank, identifying closely related loci (>95% identity, >95% query coverage) in more than 200 E. coli RefSeq genomes (0.8% of the total). Strikingly similar (>99.7% identity, 100% query coverage) scr loci were found in 230/494 (47%) C. freundii RefSeq genomes, and in the genomes of several other non-freundii Citrobacter, Enterobacter, and Klebsiella isolates in GenBank. The divergent SCU-391 scrBR region was found in genomes of distinct C. freundii isolates. It therefore seems likely that C. freundii or a closely related species was the ancestral source of the scr loci in SCU-391 and 490, via independent transfers to A and B1 E. coli. Phenotypic analysis indicated that these scr loci are fully functional in E. coli; indeed, growth curve analysis of SCU-391 found that it grew more rapidly on sucrose than glucose (μsuc = 1.15 gen h−1 vs. μgluc = 0.85 gen h−1) and that it’s growth rate on sucrose is higher than B2 isolates with the native scr locus (μsuc = 0.8–1.1 gen h−1).
In E. coli SCU-147 and nearly all of the other scr+ E. coli B2 and F isolates in our collection with fully closed genomes, the scr locus is positioned on the chromosome adjacent to the queE gene. As noted by Díez-Villaseñor et al. (2010), in scr− E. coli strains this is the location of the CRISPR2.2 array of CRISPR2 repeats, also known as iap repeats (Treviño-Quintanilla et al., 2007). The queE-CRISPR2.2 region is roughly 25 kb from the region encoding the CRISPR-CAS-E system in E. coli with the CAS-E genes. The 6.7 kb scr locus replaces approximately 0.5 kb of non-homologous DNA adjacent to queE. No IS or transposable elements flanking scr were noted that might mechanistically account for the insertion. In SCU-391 the C. freundii-like scr locus was located nearly on the opposite side of the chromosome, adjacent to a prophage genome resembling Shigella SFII. It appears likely that the C. freundii-like scr locus has been horizontally transferred among Gram-negative enterobacterial species. Indeed, GenBank searches suggest that many of these C. freundii-like scr loci in E. coli genomes reside on plasmids (data not shown), the chromosomal location in SCU-391 notwithstanding.
For a more historic view of the scr locus in E. coli, genomes of strains in the ECOR collection (Ochman and Selander, 1984) were examined. This collection was assembled to reflect the genetic diversity of E. coli as understood in the mid- to late 20th century. The scr locus was present in 8/72 of the ECOR collection genomes (11%), lower than the 21% in our contemporary commensal collection, which is significantly richer in B2 isolates. The scr locus is found in 4/16 (25%) of the ECOR B2 strains and 4/6 (67%) of the F strains (all intact scr loci). No other versions of the scr genes, such as the C. freundii-like scr, were detected in the ECOR genomes.
Shigella species have emerged evolutionarily multiple times from within the broader E. coli lineage (The et al., 2016), but clinical diagnostic databases such as API20E indicate that Shigella isolates rarely if ever ferment sucrose. The presence of the scr locus in Shigella genomes was queried in the RefSeq genome database. The scr genes were very rare, being present in less than 1% of sequenced S. sonnei and S. flexneri genomes, and in no S. dysenteriae or S. boydii genomes.
Beyond E. coli and Shigella, sucrose utilization is variable within the family Enterobacteriaceae. Data on sucrose fermentation from the API 20E database was used as a surrogate phenotypic assay for comparison with genome-based surveys of the scr locus in the RefSeq database. Results are shown in Table 2 for the scrA locus. The presence of scrA was correlated with sucrose utilization by species. For example, less than 1% of Salmonella isolates ferment sucrose, and in the Salmonella enterica RefSeq database of over 10,000 genomes, only eight contained the entire scr locus (minimum 80% coverage, 80% nucleotide identity). A rare exception to the exclusive role of the Scr pathway in sucrose utilization was seen in Citrobacter freundii. Isolates of this species near universally ferment sucrose, but only half of the C. freundii genomes in RefSeq contain the scr genes; however, as described below, the alternative csc locus is ubiquitous in C. freundii. Other Citrobacter species rarely or never utilize sucrose, and the scr and csc genes are concomitantly scarce. Conversely, Enterobacter and Klebsiella species almost always ferment sucrose, and nearly all of those genomes in the RefSeq collection contain the scr locus. Proteus vulgaris isolates almost all ferment sucrose and contain the scr locus, but P. mirabilis isolates rarely do either. Moving beyond the Enterobacteriaceae to related families in the order Enterobacterales, the scr locus is near universal in Erwinia amylovora, Serratia marcescens, and Yersinia enterocolitica, but Y. pestis and Y. pseudotuberculosis rarely ferment sucrose or contain scr genes.
Identifying the Scr pathway in a wider range of species offers the opportunity to examine evolutionary conservation of the proteins and regulatory components. On the protein side, the ScrA PTS transporter subunit (456 amino acids) is the most highly conserved across species, ranging from 91% amino acid identity between the E. coli and C. freundii proteins, to 81% between the E. coli and Yersinia enterocolitica proteins. The outer-membrane-spanning ScrY porin, the largest of the gene products at 505 amino acids, retains 64% identity between the E. coli and Y. enterocolitica proteins. The least conserved of the five scr gene products is the ScrB fructose-6-phosphate hydrolase enzyme (467 aa), with only 55% conservation between the E. coli and Y. enterocolitica proteins.
Comparison of non-coding DNA sequences with homologous functions can allow identification of conserved regulatory elements across species. Expression of scrYAB is highly inducible by sucrose in vivo, with regulation dependent on ScrR, a member of the LacI family of transcriptional regulators. Cowan et al. (1991) and Jahreis and Lengeler (1993) characterized the scrY promoter region of pUR400, identifying two 14 bp inverted repeats immediately upstream and downstream of the scrY −35 and −10 promoter elements, and providing mutational evidence that these serve as operators for ScrR binding. Jahreis and Lengeler (1993) further showed that, although sucrose induces expression from the scrY promoter in vivo, ScrR bound to D-fructose or fructose-1-phosphate (products of sucrose degradation) is released from operator DNA to allow RNA polymerase to initiate transcription. Figure 5 shows scrY promoter regions from eight species aligned to determine the extent to which the Scr regulatory sequences are conserved across species. The ScrR operator sites are indeed highly conserved sequences in the promoter region. The putative operators are positioned immediately upstream and downstream of the −35 and −10 regions of the promoter, and the downstream operator overlaps directly with the expected transcription start site.
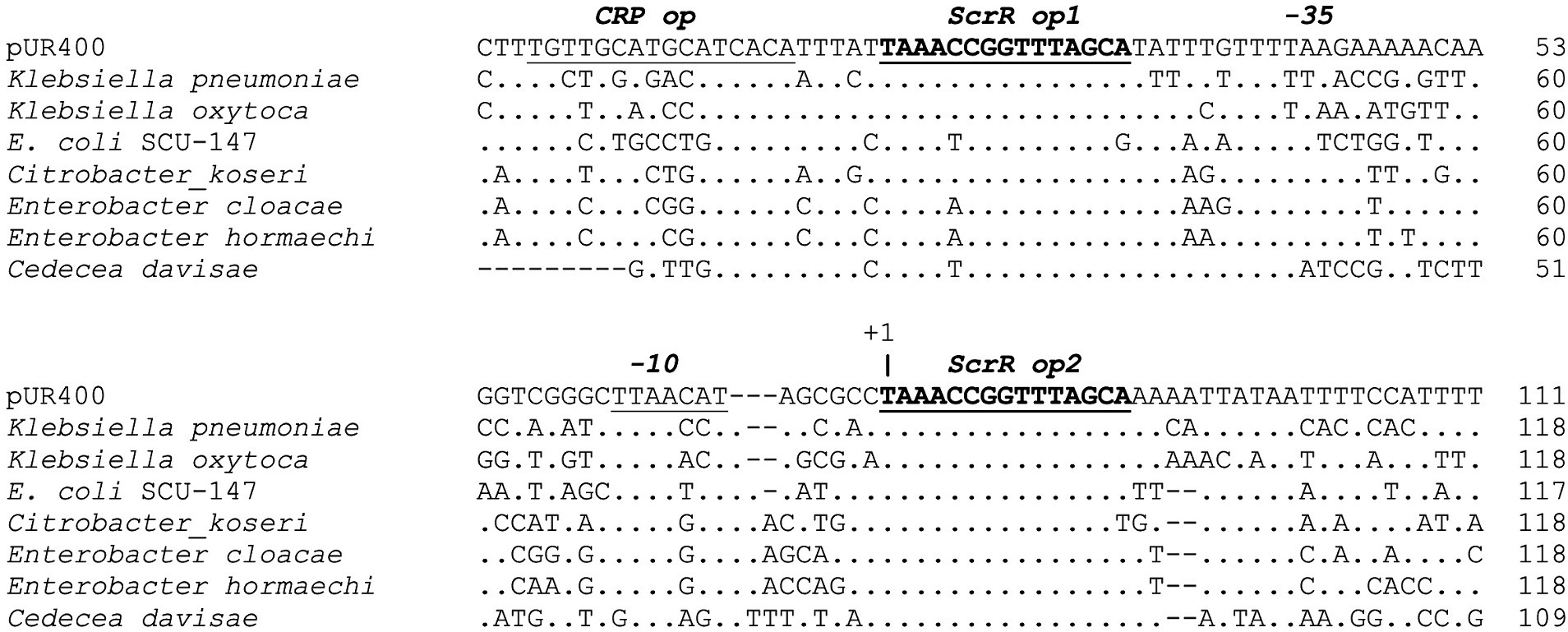
Figure 5. Conservation of the scrY promoter and regulatory region. DNA sequences upstream of the scrY start codon were aligned using the Geneious alignment algorithm. An identical base at a position is indicated by “.”, and gaps by “−”. Annotations above the aligned sequences are based on Cowan et al. (1991) and Jahreis and Lengeler (1993).
Jahreis and Lengeler (1993) also identified a CRP operator positioned further upstream of the pUR400 scrY promoter and demonstrated catabolite repression of this promoter. As seen in Figure 5, the putative CRP operator is not well conserved across species. CRP-mediated catabolite repression of scr gene expression therefore may not be conserved in other Enterobacteriaceae, but this has not been tested experimentally.
Analysis of csc locus
Most of the literature on sucrose utilization in E. coli focuses on the csc locus. In our collection of sequenced commensal E. coli, seven Suc+ isolates contained both the scr and csc loci, and 15 Suc+ isolates contained only the scr locus, again demonstrating that it is the scr locus that is critical for rapid sucrose utilization. In the absence of the scr locus, the presence of the csc genes was most strongly associated with the Sucslow phenotype, often with an extended lag phase (see Figure 2C for an example). No csc+ scr− isolates were Suc+, but 23 out of 29 were scored as either “slow” or “very slow” for growth on M9 sucrose, including 2 “slow” and 5 “very slow” isolates shown in Table 1. These results are consistent with the literature describing csc-dependent sucrose utilization in E. coli to be “unusually slow” (Jahreis et al., 2002). Five csc+ scr− isolates [SCU-171 (B2/ST657), 152 (B1/ST54), 483 (B1/ST10955), 106 (B1/ST3695), and 316 (E/ST57)] were completely unable to utilize sucrose for growth. SCU-106 has a 10 bp insertion in the cscB coding region, with an accompanying frameshift, but the other isolates have no obvious genetic defect in the csc locus to explain the growth deficiencies.
In our contemporary collection, the csc locus was found in 37% of isolates. The csc locus was universal in phylogroup D and E isolates, nearly universal in B1 isolates, uncommon in B2 isolates, and completely absent in A and F isolates. In the historical ECOR collection, the csc locus was present in 25/72 ECOR isolates (35%), similar in frequency to our contemporary collection. The csc genes were present in nearly all ECOR B1, D, and E strains (23/27), but rare in B2 strains (2/16), and absent in A and F strains (29 in aggregate).
csc resides at a recombination hotspot on the E. coli chromosome in close proximity to the argW tRNA gene (Jahreis et al., 2002). Moritz and Welch (2006) examined utilization of D-serine by diarrheagenic and uropathogenic E. coli and found that D-serine and (slow) sucrose utilization were mutually exclusive, consistent with earlier observations by Alaeddinoglu and Charles (1979). The cscRAKB and dsdCXA loci typically reside at roughly the same chromosomal location, though there is more than one possible configuration of each locus, perhaps because this is also a frequent target for integration of lambdoid phage. We observed diverse arrangements in this genomic region in our collection, with dsdC often retained even when the csc locus was present. In a few strains, both the csc and dsd loci were present in their entirety, but more often isolates had one functional locus or the other, as seen by Moritz and Welch (2006).
The csc locus was examined in Shigella sonnei, S. flexneri, S. dysenteriae, and S. boydii genomes, and was found to be defective in nearly all cases. Figure 4 compares the wild-type E. coli csc locus with loci commonly observed in the genomes of Shigella species. The cscA and cdcR genes were often present and intact, but the cscK (fructokinase) and cscB (sucrose permease) genes were usually interrupted with IS1, IS3, or IS4 elements, or missing altogether. An absence of scr loci, combined with disruption of csc loci, may explain why utilization/fermentation of sucrose is rare or absent in Shigella isolates. Shigella genomes rarely contain the dsd locus either; only 4% of nearly 2,200 Shigella genomes in the RefSeq database contained a full-length, uninterrupted dsd locus.
Querying GenBank with the entire E. coli csc locus (Ec_csc hereafter), only four examples were found in Escherichia genomes other than E. coli, four examples in coliphage genomes, four in Citrobacter farmeri genomes, and one each in a Citrobacter telavivensis genome and an Enterobacter hormaechei genome. These may have arisen from horizontal gene transfer from E. coli. Further inspection also revealed that Citrobacter freundii genomes contain a csc locus (Cf_csc hereafter) that falls below the detection threshold with the Ec_csc query in Blastn. Cf_csc is clearly homologous to Ec_csc when the amino acid sequences of the encoded proteins are compared. The arrangement of genes in Cf_csc is altered (Figure 4), with cscA and cscR each independently inverted. Nearly all (95%) of C. freundii RefSeq genomes contain a complete, uninterrupted (and likely functional) csc locus with this arrangement. Querying GenBank with Cf_csc identified several homologous loci, possibly also resulting from horizontal gene transfer, in Citrobacter portucalensis, Citrobacter youngae, and Kluyvera ascorbata genomes.
As with scr, identifying the csc locus in multiple species allowed comparison of the promoter regions to search for potentially conserved regulatory elements. In E. coli, expression of cscA is inducible by sucrose in vivo and dependent on CscR, which like ScrR is a LacI family member. Jahreis et al. (2002) identified two copies of a 12 bp inverted repeat in the intergenic region between cscA and cscK; one of these putative operators was the site of a mutation that upregulated expression of cscA. The promoter regions upstream of the cscA gene in five species were aligned and compared (Figure 6). The inverted repeats identified as potential CscR operators were indeed conserved between E. coli and the other species. Assuming a similar functional organization to the E. coli cscA promoter, these operators flank either side of the −35 and −10 regions of the promoter, with the downstream operator overlapping the transcriptional start site. Jahreis et al. (2002) also identified a putative Crp-cAMP operator, conventionally positioned adjacent to the −35 region of the cscA promoter and demonstrated catabolite repression of the Csc pathway. Extensive conservation of the putative Crp operator is seen between species (Figure 7).
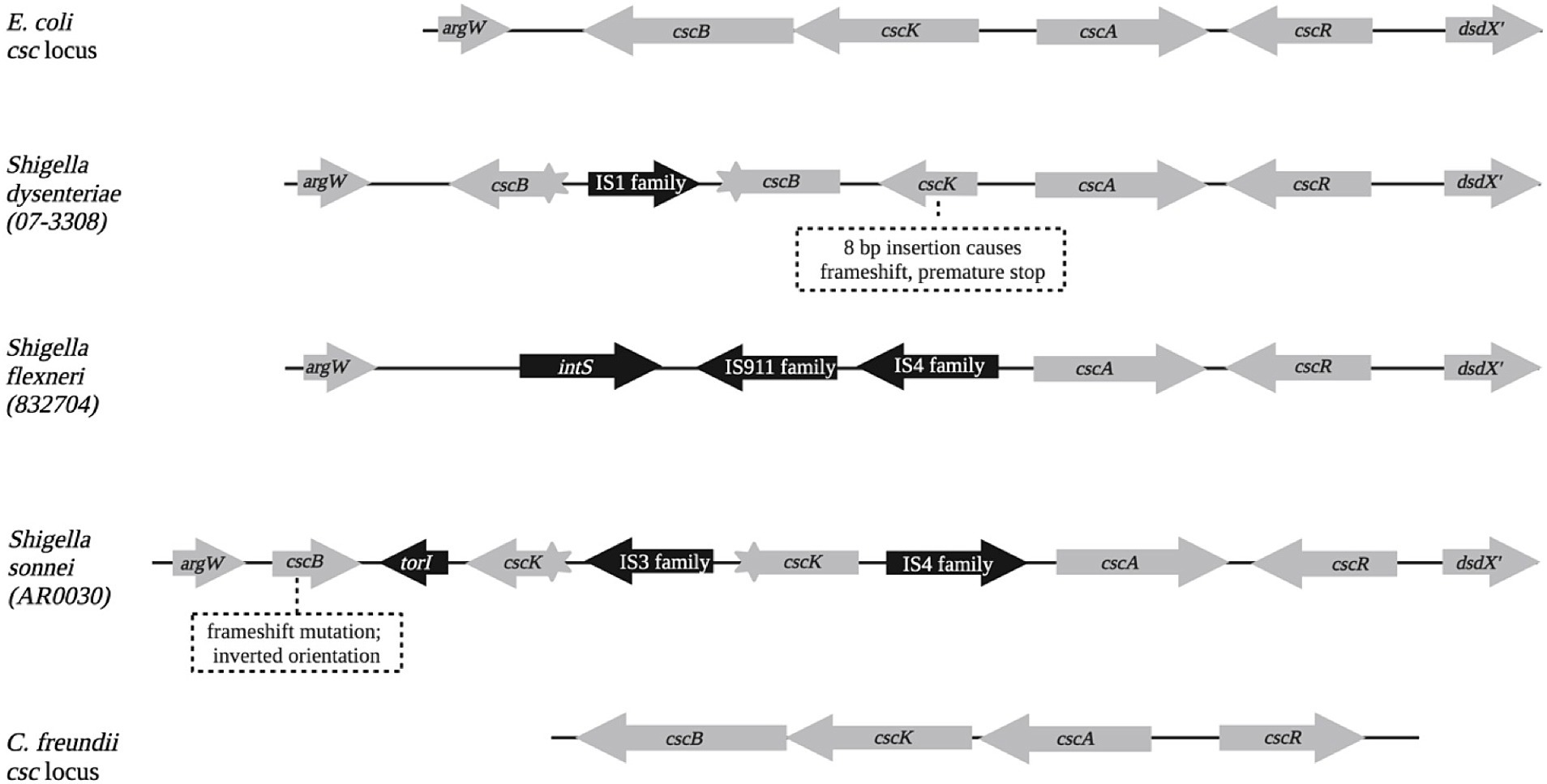
Figure 6. Representative structures of E. coli, Shigella, and C. freundii csc loci. Shigella species and strain names are shown on the left side. Note that in the C. freundii locus, the cscA and cscR genes are in the same relative locations, but their orientations are each flipped.
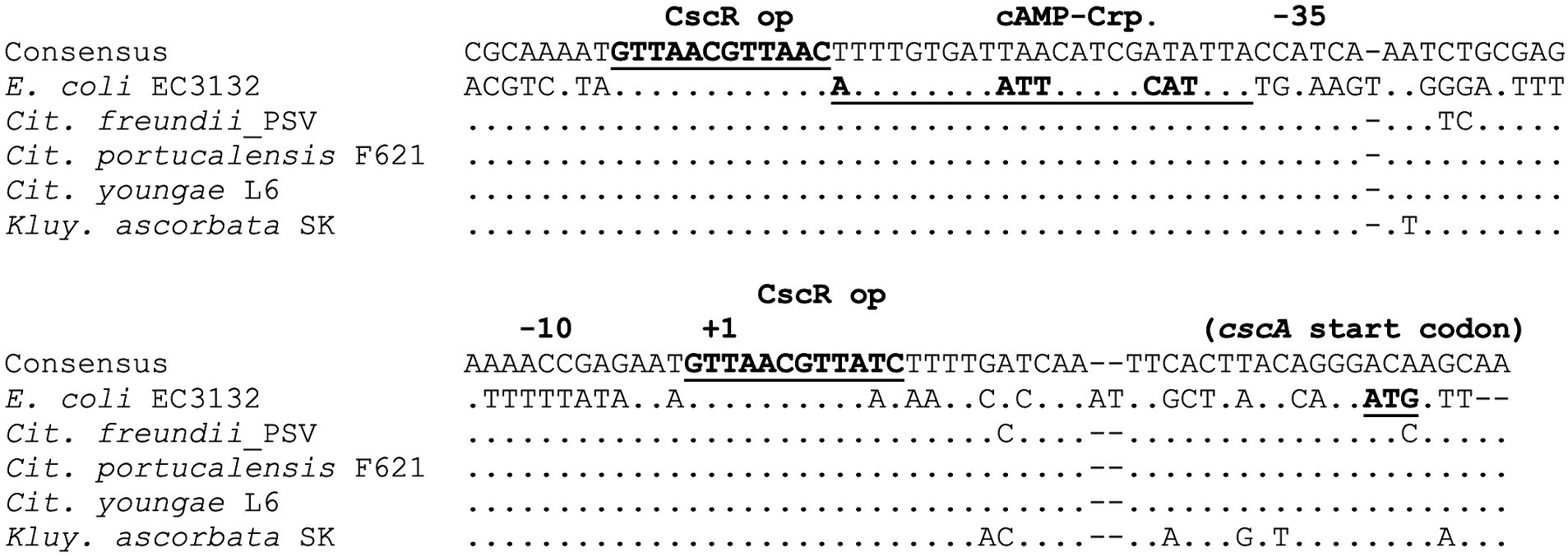
Figure 7. Conservation of the cscA promoter and regulatory region. DNA sequences upstream of the cscA start codon were aligned using the Geneious alignment algorithm. Identity with the consensus sequence is indicated by “.”, and gaps by “−”. Annotations above the aligned sequences are based on Jahreis et al. (2002).
Discussion
The results presented here support several key findings: (1) a native Scr pathway allows many E. coli strains to grow rapidly on sucrose, while the Csc pathway supports slower growth; (2) the Scr and Csc pathways are relatively common and independent in E. coli, and sometimes coexist; (3) the Scr and Csc pathways are generally chromosomally encoded, but are occasionally mobilized by non-chromosomal elements; and (4) the Scr pathway is widespread but not universal in Enterobacteriaceae, while the Csc pathway is largely confined to E. coli, Shigella (where it is non-functional), and Citrobacter species.
Because the E. coli K-12 strains (phylogroup A, ST10) that have been the dominant laboratory models lack both the scr and csc loci and thus do not utilize sucrose, exploration of sucrose metabolism in E. coli has been limited. Prior to this work there were only two reports describing chromosomally-encoded scr loci in E. coli, in uropathogenic phylogroup B2 ST92 isolate E. coli 536 (Brzuszkiewicz et al., 2006), and in enterohemorrhagic phylogroup F isolates (Treviño-Quintanilla et al., 2007). Phylogroup F represents a small fraction of the human-associated E. coli in most studies, but B2 strains are more common (e.g., Marin et al., 2022), and B2 isolates carrying scr represented nearly 20% of the E. coli in our commensal collection, and an even larger fraction among UTI isolates (data not shown).
Some (rare) non-B2 E. coli isolates appear to contain scr due to horizontal gene transfer (HGT), such as three isolates we identified containing scr loci mobilized from Citrobacter freundii. In SCU-391, Cf-scr is surrounded by prophage genes, suggesting that scr could have moved by phage transduction. Other HGT possibilities not seen in our collection include mobilization of scr by recombination onto large plasmids, as with the pUR400 plasmid found in the original example of scr mobilization from Salmonella into E. coli. Another possibility is a conjugative transposon, as with CTnscr94 in Salmonella senftenburg (Hochhut et al., 1997). Nevertheless, Smith and Parsell (1975), showed that only three out of 152 sucrose-using E. coli strains could transmit the phenotype by conjugation, so scr movement by plasmids or conjugative transposons is probably not common in E. coli.
The Scr pathway is present in both Gram-negative and Gram-positive bacteria (Reid and Abratt, 2005), and is widespread in the family Enterobacteriaceae (Table 2). An upper bound for the evolutionary age of a functioning bacterial Scr system may be the evolution in higher plants of the capacity to synthesize sucrose as a photosynthesis-derived product for carbon and energy storage (Ruan, 2014). Variability across bacterial lineages may reflect erratic selection pressure due to the variability of sucrose abundance in various niches. An association with plants may be more likely to favor this capacity; the gram-negative phytopathogen Erwinia amylovora, for example, requires the scr locus as a key virulence factor (Bogs and Geider, 2000).
The csc locus was the first set of E. coli genes experimentally associated with sucrose utilization (Bockmann et al., 1992). Previous genomic analyses showed that csc was mutually exclusive with the dsd locus for D-serine utilization, which generally resides at the same location on the chromosome. Some authors have speculated that the dsd locus is selected for in uropathogenic E. coli, as D-serine (but not sucrose) is present in significant concentrations in urine and may serve as both signal and growth substrate (Roesch et al., 2003). Conversely, intestinal pathogenic E. coli may not encounter D-serine, but may be selected for utilization of carbohydrates such as sucrose. That rationale must be reconsidered, as the scr locus allows E. coli isolates to utilize sucrose regardless of whether the csc genes are present. Furthermore, we see no significant difference in the frequency of the dsd, csc, and scr genes between the genomes of commensal and uropathogenic E. coli in our lab (data not shown).
The appearance of the csc locus in E. coli must have been prior to divergence of contemporary Shigella species, as many Shigella genomes have retained the csc locus, albeit in non-functional form. Ubiquitous destruction of the csc locus, eliminating the ability to utilize sucrose, suggests that this phenotype has been actively selected against in the pathogenic context that shaped Shigella evolution. However, a functional dsd locus has not replaced csc in Shigellae, suggesting that metabolism of D-serine is likewise not advantageous for these species.
In conclusion, the Scr pathway is highly effective in supporting rapid growth of E. coli with sucrose as a substrate. Little effort toward physiological analysis and/or genetic manipulation of the E. coli Scr pathway has been applied to optimize it for bioindustrial applications, particularly relative to what has been done with the Csc pathway. Wider recognition of the significance of the Scr pathway in E. coli and other Enterobacteriaceae could encourage research on exploitation of sucrose as a feedstock for bacterially-based processes.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
CS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Writing – original draft, Writing – review & editing. MM: Investigation, Writing – original draft, Writing – review & editing. VL: Investigation, Visualization, Writing – original draft, Writing – review & editing. JJ: Investigation, Visualization, Writing – original draft, Writing – review & editing. AM: Data curation, Investigation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Institutes of Health grant R15AI130816-01A1, and by Santa Clara University.
Acknowledgments
The authors thank Alex King and Eric Jedel for their early contributions to this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alaeddinoglu, N. G., and Charles, H. P. (1979). Transfer of a gene for sucrose utilization into Escherichia coli K12, and consequent failure of expression of genes for D-serine utilization. Microbiology 110, 47–59.
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Aulkemeyer, P., Ebner, R., Heilenmann, G., Jahreis, K., Schmid, K., Wrieden, S., et al. (1991). Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol. Microbiol. 5, 2913–2922. doi: 10.1111/j.1365-2958.1991.tb01851.x
Bockmann, J., Heuel, H., and Lengeler, J. W. (1992). Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol. Gen. Genet. MGG 235, 22–32. doi: 10.1007/BF00286177
Bogs, J., and Geider, K. (2000). Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J. Bacteriol. 182, 5351–5358. doi: 10.1128/JB.182.19.5351-5358.2000
Bruschi, M., Boyes, S. J., Sugiarto, H., Nielsen, L. K., and Vickers, C. E. (2012). A transferable sucrose utilization approach for non-sucrose-utilizing Escherichia coli strains. Biotechnol. Adv. 30, 1001–1010. doi: 10.1016/j.biotechadv.2011.08.019
Brzuszkiewicz, E., Brüggemann, H., Liesegang, H., Emmerth, M., Ölschläger, T., Nagy, G., et al. (2006). How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103, 12879–12884. doi: 10.1073/pnas.0603038103
Chang, A. Y., Chau, V., Landas, J. A., and Pang, Y. (2017). Preparation of calcium competent Escherichia coli and heat-shock transformation. JEMI Methods 1, 22–25.
Chen, I. M. A., Chu, K., Palaniappan, K., Ratner, A., Huang, J., Huntemann, M., et al. (2023). The IMG/M data management and analysis system v. 7: content updates and new features. Nucleic Acids Res. 51, D723–D732. doi: 10.1093/nar/gkac976
Cowan, P. J., Nagesha, H., Leonard, L., Howard, J. L., and Pittard, A. J. (1991). Characterization of the major promoter for the plasmid-encoded sucrose genes scrY, scrA, and scrB. J. Bacteriol. 173, 7464–7470. doi: 10.1128/jb.173.23.7464-7470.1991
Denamur, E., Clermont, O., Bonacorsi, S., and Gordon, D. (2021). The population genetics of pathogenic Escherichia coli. Nat. Rev. Microbiol. 19, 37–54. doi: 10.1038/s41579-020-0416-x
Díez-Villaseñor, C., Almendros, C., García-Martínez, J., and Mojica, F. J. (2010). Diversity of CRISPR loci in Escherichia coli. Microbiology 156, 1351–1361. doi: 10.1099/mic.0.036046-0
Durfee, T., Nelson, R., Baldwin, S., Plunkett, G. III, Burland, V., Mau, B., et al. (2008). The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J. Bacteriol. 190, 2597–2606. doi: 10.1128/JB.01695-07
García, J. L. (1985). Cloning in Escherichia coli and molecular analysis of the sucrose system of the Salmonella plasmid SCR-53. Mol. Gen. Genet. MGG 201, 575–577. doi: 10.1007/BF00331358
Hardesty, C., Ferran, C., and DiRienzo, J. M. (1991). Plasmid-mediated sucrose metabolism in Escherichia coli: characterization of scrY, the structural gene for a phosphoenolpyruvate-dependent sucrose phosphotransferase system outer membrane porin. J. Bacteriol. 173, 449–456. doi: 10.1128/jb.173.2.449-456.1991
Hochhut, B., Jahreis, K., Lengeler, J. W., and Schmid, K. (1997). CTnscr94, a conjugative transposon found in enterobacteria. J. Bacteriol. 179, 2097–2102. doi: 10.1128/jb.179.7.2097-2102.1997
Holmes, B., Willcox, W. R., and Lapage, S. P. (1978). Identification of Enterobacteriaceae by the API 20E system. J. Clin. Pathol. 31, 22–30. doi: 10.1136/jcp.31.1.22
Jahreis, K., Bentler, L., Bockmann, J., Hans, S., Meyer, A., Siepelmeyer, J., et al. (2002). Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J. Bacteriol. 184, 5307–5316. doi: 10.1128/JB.184.19.5307-5316.2002
Jahreis, K., and Lengeler, J. W. (1993). Molecular analysis of two ScrR repressors and of a ScrR–FruR hybrid repressor for sucrose and D-fructose specific regulons from enteric bacteria. Mol. Microbiol. 9, 195–209. doi: 10.1111/j.1365-2958.1993.tb01681.x
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Le Bouguénec, C., and Schouler, C. (2011). Sugar metabolism, an additional virulence factor in enterobacteria. Int. J. Med. Microbiol. 301, 1–6. doi: 10.1016/j.ijmm.2010.04.021
Lengeler, J. W., Mayer, R. J., and Schmid, K. (1982). Phosphoenolpyruvate-dependent phosphotransferase system enzyme III and plasmid-encoded sucrose transport in Escherichia coli K-12. J. Bacteriol. 151, 468–471. doi: 10.1128/jb.151.1.468-471.1982
Marin, J., Clermont, O., Royer, G., Mercier-Darty, M., Decousser, J. W., Tenaillon, O., et al. (2022). The population genomics of increased virulence and antibiotic resistance in human commensal Escherichia coli over 30 years in France. Appl. Environ. Microbiol. 88, e00664–e00622.
Moritz, R. L., and Welch, R. A. (2006). The Escherichia coli argW-dsdCXA genetic island is highly variable, and E. coli K1 strains commonly possess two copies of dsdCXA. J. Clin. Microbiol. 44, 4038–4048. doi: 10.1128/JCM.01172-06
Mukherjee, S., Stamatis, D., Li, C. T., Ovchinnikova, G., Bertsch, J., Sundaramurthi, J. C., et al. (2023). Twenty-five years of Genomes OnLine Database (GOLD): data updates and new features in v. 9. Nucleic Acids Res. 51, D957–D963. doi: 10.1093/nar/gkac974
Ochman, H., and Selander, R. K. (1984). Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157, 690–693. doi: 10.1128/jb.157.2.690-693.1984
Palchaudhuri, S., Rahn, S., Santos, D. S., and Maas, W. K. (1977). Characterization of plasmids in a sucrose-fermenting strain of Escherichia coli. J. Bacteriol. 130, 1402–1403. doi: 10.1128/jb.130.3.1402-1403.1977
Peters, S., Rose, T., and Moser, M. (2010). Sucrose: a prospering and sustainable organic raw material. Top. Curr. Chem. 294, 1–23. doi: 10.1007/128_2010_58
Pitout, J. D., Peirano, G., Chen, L., DeVinney, R., and Matsumura, Y. (2022). Escherichia coli ST1193: following in the footsteps of E. coli ST131. Antimicrob. Agents Chemother. 66, e00511–e00522.
Pruitt, K. D., Tatusova, T., and Maglott, D. R. (2007). NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35, D61–D65. doi: 10.1093/nar/gkl842
Reid, S. J., and Abratt, V. R. (2005). Sucrose utilisation in bacteria: genetic organisation and regulation. Appl. Microbiol. Biotechnol. 67, 312–321. doi: 10.1007/s00253-004-1885-y
Roesch, P. L., Redford, P., Batchelet, S., Moritz, R. L., Pellett, S., Haugen, B. J., et al. (2003). Uropathogenic Escherichia coli use D-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 49, 55–67. doi: 10.1046/j.1365-2958.2003.03543.x
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65, 33–67. doi: 10.1146/annurev-arplant-050213-040251
Sabri, S., Nielsen, L. K., and Vickers, C. E. (2013). Molecular control of sucrose utilization in Escherichia coli W, an efficient sucrose-utilizing strain. Appl. Environ. Microbiol. 79, 478–487. doi: 10.1128/AEM.02544-12
Schmid, K., Ebner, R., Altenbuchner, J., Schmitt, R., and Lengeler, J. W. (1988). Plasmid-mediated sucrose metabolism in Escherichia coli K12: mapping of the scr genes of pUR400. Mol. Microbiol. 2, 1–8. doi: 10.1111/j.1365-2958.1988.tb00001.x
Schmid, K., Ebner, R., Jahreis, K., Lengeler, J. W., and Titgemeyer, F. (1991). A sugar-specific porin, ScrY, is involved in sucrose uptake in enteric bacteria. Mol. Microbiol. 5, 941–950. doi: 10.1111/j.1365-2958.1991.tb00769.x
Schmid, K., Schupfner, M., and Schmitt, R. (1982). Plasmid-mediated uptake and metabolism of sucrose by Escherichia coli K-12. J. Bacteriol. 151, 68–76. doi: 10.1128/jb.151.1.68-76.1982
Schülein, K., Schmid, K., and Benzl, R. (1991). The sugar-specific outer membrane channel ScrY contains functional characteristics of general diffusion pores and substrate-specific porins. Mol. Microbiol. 5, 2233–2241. doi: 10.1111/j.1365-2958.1991.tb02153.x
Smith, H. W., and Parsell, Z. (1975). Transmissible substrate-utilizing ability in enterobacteria. Microbiology 87, 129–140.
Sprenger, G. A., and Lengeler, J. W. (1988). Analysis of sucrose catabolism in Klebsiella pneumoniae and in Scr+ derivatives of Escherichia coli K12. Microbiology 134, 1635–1644. doi: 10.1099/00221287-134-6-1635
Stephens, C., Arismendi, T., Wright, M., Hartman, A., Gonzalez, A., Gill, M., et al. (2020). F plasmids are the major carriers of antibiotic resistance genes in human-associated commensal Escherichia coli. MSphere 5, 10–1128.
The, H. C., Thanh, D. P., Holt, K. E., Thomson, N. R., and Baker, S. (2016). The genomic signatures of Shigella evolution, adaptation and geographical spread. Nat. Rev. Microbiol. 14, 235–250. doi: 10.1038/nrmicro.2016.10
Titgemeyer, F., Jahreis, K., Ebner, R., and Lengeler, J. W. (1996). Molecular analysis of the scrA and scrB genes from Klebsiella pneumoniae and plasmid pUR400, which encode the sucrose transport protein enzyme II Scr of the phosphotransferase system and a sucrose-6-phosphate invertase. Mol. Gen. Genet. MGG 250, 197–206.
Treviño-Quintanilla, L. G., Escalante, A., Caro, A. D., Martínez, A., González, R., Puente, J. L., et al. (2007). The phosphotransferase system-dependent sucrose utilization regulon in enteropathogenic Escherichia coli strains is located in a variable chromosomal region containing iap sequences. Microbial Physiol. 13, 117–125. doi: 10.1159/000103603
Keywords: Escherichia coli, sucrose, evolutionary genomics, metabolism, Scr, Csc
Citation: Stephens C, Martinez M, Leonardi V, Jaing J and Miller A (2024) The Scr and Csc pathways for sucrose utilization co-exist in E. coli, but only the Scr pathway is widespread in other Enterobacteriaceae. Front. Microbiol. 15:1409295. doi: 10.3389/fmicb.2024.1409295
Edited by:
Harold J. Schreier, University of Maryland, Baltimore County, United StatesReviewed by:
Vineet Kumar, The University of Texas at Austin, United StatesDong-Woo Lee, Yonsei University, Republic of Korea
Copyright © 2024 Stephens, Martinez, Leonardi, Jaing and Miller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Craig Stephens, Y3N0ZXBoZW5zQHNjdS5lZHU=
 Craig Stephens
Craig Stephens Mireille Martinez
Mireille Martinez
