- 1School of Environmental and Chemical Engineering, Xi’an Polytechnic University, Xi’an, China
- 2Key Laboratory of Risk Assessment and Control for Environment and Food Safety, Tianjin Institute of Environmental and Operational Medicine, Tianjin, China
- 3College of Oceanography and Ecological Science, Shanghai Ocean University, Shanghai, China
- 4School of Public Health, North China University of Science and Technology, Tangshan, China
- 5Key Laboratory of Karst Geological Resources and Environment, Guizhou University, Guizhou, China
Introduction: Bacterial resistance presents a major challenge to both the ecological environment and human well-being, with persistence playing a key role. Multiple studies were recently undertaken to examine the factors influencing the formation of persisters and the underlying process, with a primary focus on Gram-negative bacteria and Staphylococcus aureus (Gram-positive bacteria). Enterococcus faecalis (E. faecalis) is capable of causing a variety of infectious diseases, but there have been few studies of E. faecalis persisters. Previous studies have shown that the sex pheromone cCF10 secreted by E. faecalis induces conjugative plasmid transfer. However, whether the pheromone cCF10 regulates the persistence of E. faecalis has not been investigated.
Methods: As a result, we investigated the effect and potential molecular mechanism of pheromone cCF10 in regulating the formation of persisters in E. faecalis OG1RF using a persistent bacteria model.
Results and discussion: The metabolically active E. faecalis OG1RF reached a persistence state and temporarily tolerated lethal antibiotic concentrations after 8 h of levofloxacin hydrochloride (20 mg/mL) exposure, exhibiting a persistence rate of 0.109 %. During the growth of E. faecalis OG1RF, biofilm formation was a critical factor contributing to antibiotic persistence, whereas 10 ng/mL cCF10 blocked persister cell formation. Notably, cCF10 mediated the antibiotic persistence of E. faecalis OG1RF via regulating metabolic activity rather than suppressing biofilm formation. The addition of cCF10 stimulated the Opp system and entered bacterial cells, inhibiting (p)ppGpp accumulation, thus maintaining the metabolically active state of bacteria and reducing persister cell generation. These findings offer valuable insights into the formation, as well as the control mechanism of E. faecalis persisters.
1 Introduction
The emergence of antimicrobial resistance (AMR) has resulted in increased morbidity and mortality rates, making it a serious public health concern (Li et al., 2021; Pennino et al., 2023). Worldwide, AMR is reported to be responsible for 700,000 deaths each year (Wang et al., 2019). As a result, AMR has attracted great attention, particularly regarding its formation mechanisms (Cho et al., 2018; Cepas et al., 2019; Shahin et al., 2019; Kaszab et al., 2023) and removal processes (Saha and Mukherjee, 2019; Alt et al., 2023). AMR include intrinsic (Roberts et al., 2021) and acquired resistance (Shahin et al., 2019). Intrinsic resistance is determined by chromosomes, and different bacterial cell structures and chemical compositions make them naturally insensitive to some antibacterial drugs. Acquired resistance can occur via several methods, including genetic mutations (Roberts et al., 2021) and plasmid-mediated horizontal gene transfer (Yang et al., 2023). Efforts to mitigate AMR involve various removal mechanisms, including antibiotic stewardship programs (Pallares et al., 2022), wastewater treatment (Łuczkiewicz et al., 2010) and bioremediation (Apreja et al., 2022). Generally, AMR development is attributed to the spread of antibiotic resistance genes (ARGs). Recently, however, it has been found that persisters not only promote the evolution of AMR but are also a major cause of recalcitrant infections and persistent contamination (Zhang, 2014; Gollan et al., 2019; Liu et al., 2022). Persisters, a subpopulation of bacterial cells that exist in a non-growing and non-responsive state, exhibit phenotypic but not genetic changes (Harms et al., 2016). Bacteria spontaneously enter a persistence state and could survive under various stresses, including antibiotic exposure. A biphasic kill curve revealed that persisters could survive even after exposure to lethal antibiotic concentrations (Balaban et al., 2019) and could resume normal growth after antibiotic removal, thereby recovering antibiotic sensitivity (Fang and Allison, 2023). Although their minimum inhibitory concentration (MIC) may remain constant in the dormant population, persisters could cause treatment failure and disease recurrence (Gollan et al., 2019). For example, Staphylococcus aureus (S. aureus) persisters have been associated with suppurative infection and hospital cross infection (Chang et al., 2020; Yee et al., 2022), and Escherichia coli (E. coli) persisters have been linked with recurrent urinary tract infections (Morales-Espinosa et al., 2016). Furthermore, the presence of persisters means that traditional antibiotic targets are inactive (Li et al., 2018; Bartell et al., 2020; Beam et al., 2021), limiting the accessibility of drugs (Cao et al., 2022) for bacterial infections treatment. Moreover, persistence promotes antibiotic resistance evolution (Windels et al., 2019a,b) and population tolerance development (Van den Bergh et al., 2017). Therefore, preventing persister formation is a crucial AMR control strategy.
Persistence is a defensive strategy bacteria use to resist antibiotics. Hitherto, persister cells have been found in almost all varieties of bacterial populations, including E. coli (Tkachenko et al., 2014; Xu et al., 2021), Pseudomonas aeruginosa (P. aeruginosa; Mlynarcik and Kolar, 2017), and S. aureus (Lechner et al., 2012; Kim et al., 2018), among other prevalent clinical and environmental bacteria (Kaur et al., 2018; Fernandes et al., 2022). Previous research has shown that prolonged exposure to high concentration of antibiotics (Kaur et al., 2018; Paranjape and Shashidhar, 2019), nanomaterials (Wang et al., 2022), and other conventional or innovative antimicrobial agents (Ye et al., 2022) could result in phenotypic persistence. Therefore, in the laboratory researchers used a variety of strategies to provoke persisters, including nutritional restriction, exposure to lethal concentrations of antibiotics, and so on (Svenningsen et al., 2022). These environmental stimuli triggered bacterial persister formation via influencing the membrane potential through an oxidative stress (OS)-reliant mechanism (Wang et al., 2017), regulating bacterial metabolism and toxin-antitoxin (TA; Wagner and Unoson, 2012) modules in E. coli. Furthermore, persister formation involves both DNA damage and repair, as well as SOS response (McCourt et al., 2014; Podlesek and Žgur Bertok, 2020). In the context of Gram-positive bacteria, it is believed that biofilms significantly impact (Liu et al., 2021; Zhang et al., 2022) persister formation in S. aureus and that bacterial biofilms also serve as a protective growth modality (Lebeaux et al., 2014) for acclimation to harsh environments, accounting for almost 80% (Hrynyshyn et al., 2022) of chronic infections. Moreover, Dufour et al. (2022) discovered that quorum sensing (QS) could promote bacterial persistence by regulating the TA system. Based on existing insights into these mechanisms, researchers hypothesized that specific chemicals and small molecules (Liu et al., 2021; Narimisa et al., 2021) might impede persister formation by modulating QS systems (Allegretta et al., 2017), biofilms (Lin et al., 2022; Nguyen et al., 2023), and intracellular ATP levels (Conlon et al., 2016; Aedo et al., 2019) in both Gram-positive and Gram-negative bacteria. The use of single-cell technologies including flow cytometry, microscopy, microfluidic devices, and other technologies in conjunction with each other can enhance our ability to recognize persisters (Balomenos et al., 2017; Hare et al., 2021). Currently, the mechanisms that facilitate or inhibit bacterial persistence formation are relatively constrained, with the specific target yet to be elucidated.
Currently, persister research focuses primarily on E. coli, P. aeruginosa, and Gram-positive bacteria such as S. aureus, whereas E. faecalis persisters, which frequently precipitate nosocomial infections, have received minimal attention (Van Tyne et al., 2019; Kalfopoulou and Huebner, 2020). According to research, E. faecalis possesses a robust cell wall (Canfield et al., 2023), making it inherently resistant (Korir et al., 2019) to various antibiotics. Moreover, it readily acquires resistance via the uptake of resistant plasmids (Tomita et al., 2023; Yang et al., 2023), transposons (Dale et al., 2015), or genetic mutations (García-Solache and Rice, 2019). Due to the severity of E. faecalis resistance, there are few antibiotics available for the clinical treatment of Enterococcus infections. Phenotypic resistance is a characteristic feature of persistent E. faecalis infections. Kaviar et al. (2022) discovered that E. faecalis clinical isolates exhibited a high proportion of persister cells and vancomycin tolerance tendency, further complicating treatment. Therefore, to control their formation and prevent chronic infections, more attention should be paid to enterococcal persisters.
Several studies have proposed strategies to combat bacterial persistence. However, favorable environmental (Wainwright et al., 2021) conditions could revive persister cells. Through single amino acid testing, Yamasaki et al. (2020) confirmed that nutrient signaling molecules could facilitate persister resuscitation. Bacteria-secreted signaling molecules could regulate bacterial behavior (Dunny and Leonard, 1997), and as an essential mode of bacterial communication, they have been demonstrated to regulate persistence processes (Zarkan et al., 2020). Specifically, it has been reported that the fatty acid signaling molecule cis-2-decenoic acid (cis-DA) can resuscitate persister cells to a metabolically active state in E. coli and P. aeruginosa (Marques et al., 2014). These findings suggest that signaling molecules could regulate bacterial metabolism. Therefore, we hypothesized that signaling molecules might sustain bacterial metabolic activity, preventing bacteria from assuming a state of persistence.
The secretion of intercellular signaling molecules among bacteria, a prevalent phenomenon, is crucially involved in bacterial behavior regulation (Mori et al., 1984; Vickerman and Mansfield, 2019). The peptide pheromones secreted by E. faecalis can be sensed and induced by bacteria as signal molecules. There are many similar pheromones such as cCF10, cAM373 and cAD 1 secreted by E. faecalis that regulate expression of conjugative plasmid transfer genes in E. faecalis (Mori et al., 1984; Vickerman and Mansfield, 2019). Of these, pheromone cCF10 has the highest level of attention. The amino acid sequence of the sex pheromone cCF10 is LVTLVFV (Dunny and Leonard, 1997). Pro-C is a secreted lipoprotein comprising a cleaved signal peptide from the ccfA gene product in E. faecalis. Following the initial cleavage, the released 22-amino acid signal peptide was further cleaved by Eep protease to produce C (cCF10) and released into the growth medium (Flannagan and Clewell, 2002; Chandler and Dunny, 2008; Varahan et al., 2014). Generally, cCF10 and its antagonistic signaling molecule, iCF10 (Chen et al., 2017), competitively bind to PrgX (Price et al., 2016) in OG1RF (pCF10). This process initiates prgQ transcription, inducing conjugative plasmid (pCF10) transfer (Chatterjee et al., 2013). Moreover, in the absence of pCF10, cCF10 can also be taken up by OG1RF through the Opp2 system (Segawa et al., 2021). However, whether the pheromone cCF10 regulates the persistence in E. faecalis during this process has not been investigated. Therefore, we proposed that pheromone cCF10 may prevent bacteria from transitioning to a low-energy metabolic state, impeding the formation of persisters in E. faecalis. Herein, we exposed E. faecalis to levofloxacin hydrochloride (LVF) to screen for persisters in vitro experiments and we investigated the impact of pheromone cCF10 on the formation of persister cells in E. faecalis OG1RF, along with exploring its potential molecular mechanism. Furthermore, this study aimed to enhance our understanding of the formation of E. faecalis persisters and provide a novel approach and theory for studying the prevention and treatment of E. faecalis persisters.
2 Materials and methods
2.1 Bacterial strains and culture condition
All the E. faecalis used herein were derived from E. faecalis OG1RF (ATCC 47077). The OG1RF∆ccfA strain was constructed in our lab as outlined in Yang et al. (2023). Supplementary Text S1 presents the details. The bacteria were grown in the brain heart infusion (BHI) medium (Coolaber Science and Technology, China) at 37°C and 150 rpm agitation.
2.2 The biphasic killing curve experiment of Enterococcus faecalis
First, OG1RF cultures in the logarithmic growth phase were initiated by inoculating them into the BHI medium (1:1,000). Following that, cells were grown for 4 h and then exposed to different LVF concentrations. Herein, LVF concentrations were MIC-based. Supplementary Text S2 describes antibiotic type selection and MIC measurement. Bacteria were treated with various antibiotic concentrations, followed by sample collection at different time intervals for enumeration. Subsequently, the cultures were gradient-diluted with a Phosphate-Buffered Solution (PBS) and then grown on BHI agar for counting.
2.3 Detection of the persistence rate during the growth of Enterococcus faecalis
First, OG1RF cultures in the logarithmic growth phase were inoculated into the BHI medium (1:1000) and then cultured at 37°C. At specific culture time points (4, 4.5, 5, 5.5, and 6 h), a sample was removed from the culture medium and challenged with LVF at a final concentration of 20 mg/L. The mixed cultures were then incubated at 37°C for 8 h. Subsequently, the samples were diluted with PBS before inoculating their appropriate dilutions on BHI agar to count the surviving bacteria. Simultaneously, the samples’ bacterial concentrations before LVF treatment was determined. The results were presented as a colony-forming unit (CFU)/mL, and the persister frequency () was determined using the following Formula (1):
where represents the number of bacteria in the culture after antibiotic treatment (CFU/mL), and is the number of bacteria in the culture before antibiotic treatment (CFU/mL).
2.4 Effects of cCF10 on persister formation in Enterococcus faecalis
First, OG1RF cultures in the logarithmic growth phase were inoculated into the BHI medium (1:1,000) and then supplemented with different concentrations of the pheromone cCF10 (8, 10, 12, 14, 16, and 20 ng/mL). After incubating the cultures for 3 h, the bacteria were collected through centrifugation at 8,000 rpm for 3 min. The collected material was re-suspended in the BHI medium, and then exposed with LVF at a final concentration of 20 mg/L before determining the persistence rate as earlier mentioned. We detected the persistence rate of OG1RF∆ccfA to further verify the effect of cCF10 on the formation of E. faecalis persistence. We also examined the effects of different cCF10 concentrations (1, 10, and 20 ng/mL) on the persistence rate of OG1RF∆ccfA. Notably, Kingsray Biotechnology Co., Ltd. (Nanjing, China) synthesized the peptide pheromone used herein, the 7-amino acid sex pheromone cCF10 (C-clumping-inducing peptide, amino acid sequence LVTLVFV; Dunny and Leonard, 1997). To confirm the specificity of cCF10, a site-mutated sequence called cCF10-F (sequence = LVFLVTV) was employed and cCF10-F were synthesized by GenScript (China). The procured pheromones cCF10 and cCF10-F were dissolved in acetonitrile (Maclean’s, China), respectively, and then stored at −20°C in the dark.
2.5 Extraction and detection of the extracellular pheromone cCF10
We extracted the extracellular pheromone per the methodology described by Zhou et al. (2023). Briefly, the collected samples were placed on ice for 20 min and then centrifuged at 6,000 rpm for 10 min at 4°C. The supernatant was then filtered using a 0.22 μm filter (SLGP033RB Millipore, Massachusetts, United States) and mixed with ammonium hydroxide solution and acetonitrile (both from Shanghai Macklin Biochemical Technology Co. Ltd., China) at a ratio of 8:1:1. The resulting mixtures were vortexed at 1,400 rpm for 15 min at room temperature (RT) and then centrifuged for 15 min at 25,000 rpm (ST16R; ThermoFisher Scientific, Massachusetts, United States). Following that, thorough mixing was done with the supernatant and an equivalent amount of an aqueous solution of 10% ammonium hydroxide. The cCF10 elution process was as follows. First, the extraction column (Sep-Pak C18 WAT054945 Waters, Massachusetts, United States) was activated by sequentially flowing 5 mL acetonitrile through it, followed by 5 mL water at a rate of one drop/s. A volume of 30 mL of test solution was passed through the extraction column at a speed of one drop per 3 s. Subsequently, to eliminate water-soluble impurities from the column bed matrix material within the extraction column or those that have been absorbed during activation or sample loading, 2 mL of water was introduced as an eluent. This step was followed by flushing with an aqueous acetonitrile solution (30%), also amounting to 2 mL, to ensure the impurities were properly removed. Finally, the collected eluate was sent to Science Compass (Zhejiang, China) for Liquid Chromatography Mass Spectrometry (LCMS) testing. Supplementary Text S4 presents the detailed detection procedure. Preparation of Scanning Electron Microscope (SEM) samples.
The cell pellet was gently re-suspended in a pre-cooled 2.5% glutaraldehyde fixative for 24 h at 4°C. The fixative was then removed from the sample via centrifugation at 6000 rpm for 5 min, followed by dehydration using an ethanol gradient approach. Ultimately, the bacteria underwent lyophilization with a FDU1200 EYELA freeze-dryer (Tokyo, Japan) and were then imaged using a Sigma 300 SEM (Zeiss, Germany).
2.6 Transcriptome sequencing
Firstly, E. faecalis OG1RF normal bacteria, persisters, and cCF10 treatment persisters were collected as previously described. The bacterial cultures were first collected via centrifugation at 8,000 rpm for 3 min. The collected residues were then cooled in liquid nitrogen for 15 min and stored at −80°C before being sent to Allwegene Technology Co. Ltd. (Beijing, China) for transcriptome sequencing. Supplementary Text S5 describes the specific sequencing methods employed. The transcriptome data (project number PRJCA026007) has been deposited in the China National center for Bioinformation. The URL is https://ngdc.cncb.ac.cn/bioproject/browse/PRJCA026007.
2.7 Measurement of biofilm formation
The OG1RF strain was cultured in a six-well plate, and any unadhered bacteria were gently washed off with PBS. The plates with adherent biofilms were then air-dried at RT and fixed with methanol (1 mL/well) for 15 min. The methanol was discarded, and the plates were air-dried again. A 1% crystal violet staining solution (1 mL/well) was then added to stain the biofilms for 1 h. Subsequently, the PBS was used to carefully wash away the crystal violet staining solution until it became colorless, after which the plates were left to air dry. The OD570 was then determined using a Multifunctional Fluorescent Enzyme Labeler (Spectra Max M5, United States).
2.8 Adenosine triphosphate measurement
First, the bacterial cultures were centrifugally washed with PBS at 6,000 rpm for 5 min and then subjected to ATP measurement per the instructions in the ATP Assay Kit (Beyotime Biotechnology Co. Ltd., China). Chemiluminescence measurements were recorded using MFEL (SpectraMax M5, United States), and the data were normalized to the total amount of protein measured per the instructions in the BCA Protein Concentration Measurement Kit (Beyotime Biotechnology Co. Ltd., China).
2.9 Total RNA extraction and real-time fluorescent quantitative reverse transcription polymerase chain reaction analysis
The samples were transferred to ice for immediate cooling, and total RNA extraction was performed per the instructions in the Gram-Positive Microbes RNA Isolation Kit (Beibei Biotechnology Co, China). Subsequently, reverse transcription polymerase chain reaction (RT-qPCR) was conducted to convert total RNA into cDNA using the first strand cDNA synthesis kit (Tiangen, China) and random primers. Subsequently, the Power Up SYBR Master Mix (ThermoFisher, United States) was used for real-time PCR analysis, along with the CFX96 Real-Time System (Bio-Rad Laboratories Inc., Hercules, United States). The absolute quantification method was utilized for determining the mRNA levels of specific genes related to pheromones, such as those encoding pheromones and responding to them. The 16S rRNA gene was utilized as an internal reference for normalization purposes. The primer sequences for RT-qPCR analysis can be found in Supplementary Table S1, and were designed with the assistance of DNASTAR.
2.10 Statistical analysis
The experiments were independently repeated a minimum of three times. Data analysis was conducted with SPSS 25 software (IBM, Armonk, NY). All data were presented as mean ± standard deviation (SD) and were analyzed using the independent samples t-test or one-way analysis of variance followed by the student–Newman–Keuls test. Results or differences with p < 0.05 were considered statistically significant.
3 Results
3.1 Pheromone cCF10 prevented the formation of persister cells in Enterococcus faecalis
Herein, we first established a persistent bacteria model and subjected E. faecalis strains to a series of persister analyses to examine the effect of pheromone cCF10 on the formation of E. faecalis persister cells (Supplementary Figures S1, S2). According to the “biphasic killing” curve, bacterial populations decreased sharply after antibiotic exposure and plateaued at 8 h (Supplementary Figure S2B). Furthermore, the number of surviving bacteria was almost stable when antibiotic concentrations exceeded 20 mg/L, indicating that the surviving bacteria were persisters (Balaban et al., 2004; Lewis, 2007; Kint et al., 2012; Harms et al., 2016). Based on SEM images (Supplementary Figure S2C), normal-growing E. faecalis cells exhibited a relatively round and full morphology, whereas LVF-exposed cells shriveled, with a large amount of biofilm attached to their surfaces, further indicating that the surviving bacteria were persisters. Extracellular polymers (EPS) is mainly a number of polymer substances, such as polysaccharides, proteins and nucleic acids. Our results showed that more polysaccharides, proteins and nucleic acids were produced around the persisters compared to normal bacteria (Supplementary Figure S3). Unlike dead bacteria, the persistent bacteria were able to resuscitate after the removal of antibiotic pressure. To illustrate this point, we added electron microscope images of the persistent bacteria at different resuscitation times (Supplementary Figure S4). We observed changes in the morphology of bacteria that had been screened for antibiotics when added to fresh media. We found that after 1 h of resuscitation, most of the bacteria in the visual field returned to normal bacterial form. In addition, we also compared the resuscitation curves of the persisters and the sterilization curves of the recovered persisters. We obtained the same growth curve as normal bacteria (Supplementary Figure S5A). Then we exposed the resuscitated to 20 mg/L of levofloxacin hydrochloride for 12 h. We found that the screened bacteria were still sensitive to levofloxacin hydrochloride (Supplementary Figure S5B). Our results indicated that the bacteria we screened were persisters rather than resistant bacteria.
Although bacterial growth was not affected when E. faecalis OGIRF was exposed to 10 and 12 ng/mL cCF10 concentrations (Supplementary Figure S6), the persister rate decreased from 0.109 to 0.050 and 0.047% (Figure 1A). This finding indicated that cCF10 could inhibit the formation of E. faecalis persisters. Interestingly, the persister rate of E. faecalis increased to 0.201%, 0.211%, and 0.205% when pheromone concentrations reached 14, 18, and 20 ng/mL, respectively. It could be attributed to the fact that the excessively high pheromone concentrations exceeded the regulatory range of E. faecalis, interfering with the physiological metabolism of the bacterium.
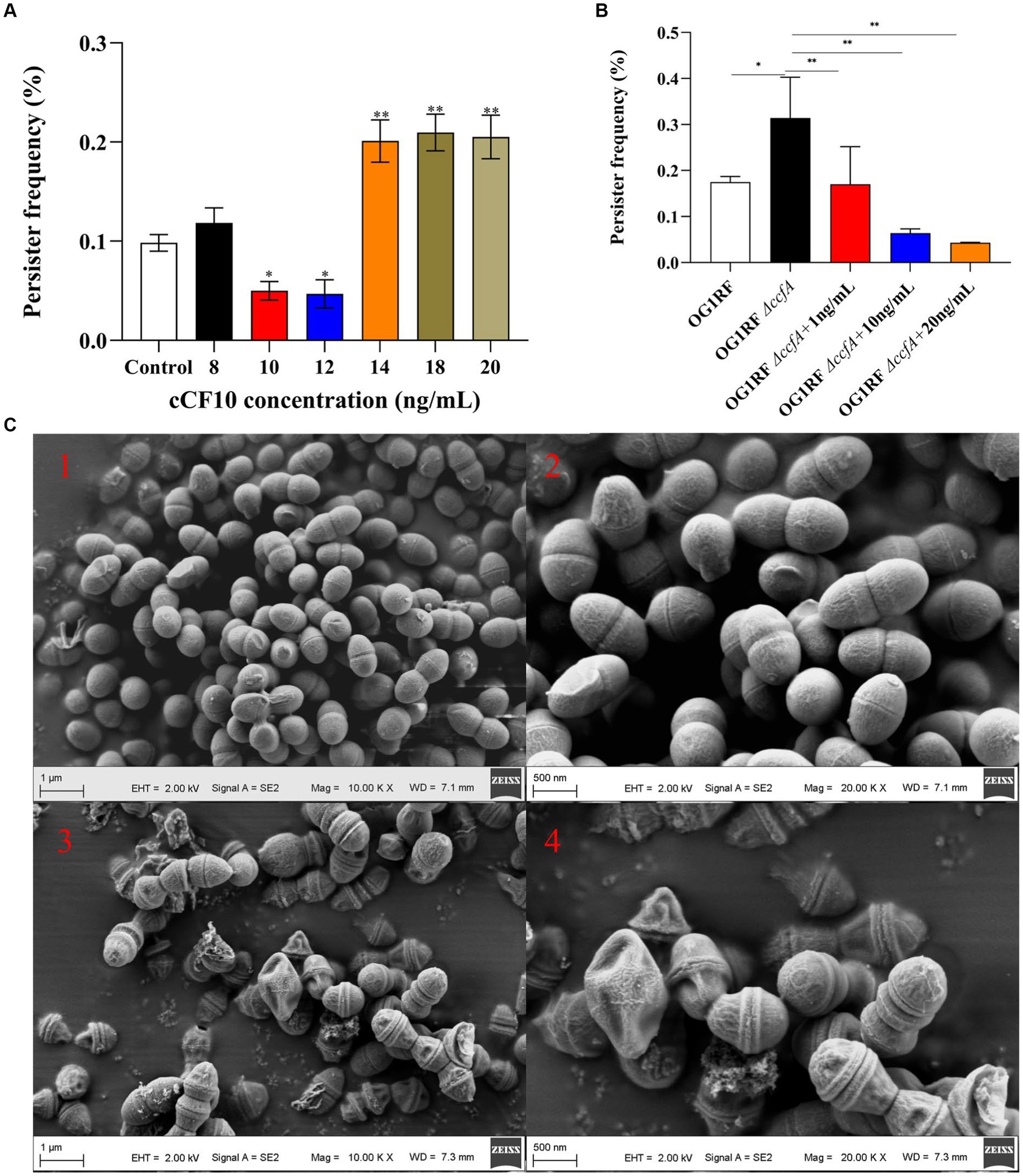
Figure 1. Pheromone cCF10 inhibited the formation of persister cells. (A) The effects of different concentrations of cCF10 on persister rate in OG1RF. (B) Influence of different concentrations of cCF10 on persister rate in OG1RF∆ccfA. The results represent the mean ± standard deviation (SD) of three biological replicates. (C) Representative SEM images of showing the morphology of bacteria after LVF treatment in the OG1RF∆ccfA (1–2) and the control group (3–4). Significant differences between groups are presented with *p < 0.05 and **p < 0.01.
We also detected the persister rate of a mutant OG1RF∆ccfA strain to verify the effect of cCF10 on the formation of E. faecalis persistence. According to the results, the persister rate of the mutant strain increased to 0.31%. On the other hand, adding exogenous cCF10 at 10 and 20 ng/mL concentrations reduced the persister rate to 0.064% and 0.043%, respectively (Figure 1B), further indicating that the pheromone cCF10 could inhibit the persistence of E. faecalis within a certain range. To confirm the specificity of cCF10, we employed a site-mutated sequence called cCF10-F. Our results indicated that unlike cCF10, cCF10-F did not impact the formation of OG1RF persisters (Supplementary Figure S7). Based on SEM images (Figure 1C), normal-growing OG1RF∆ccfA cells exhibited a rounded and plump morphology, whereas LVF-exposed cells had a shriveled appearance, indicating that the surviving bacteria were persisters.
3.2 Accumulation of biofilm played a crucial role in the generation of Enterococcus faecalis persisters
Herein, the persisters were screened with 20 mg/L LVF at different culture times to evaluate the changes in persister rate during the OG1RF growth stages. The bacteria reached the logarithmic phase at 4 h, at which the persister rate was 0.11%, and plateaued at 6 h at which the persister rate was 7.68%. Furthermore, the persister generation rate gradually increased with the growth of OG1RF, exhibiting 0.25%, 0.72%, and 2.48% persister rates after growth for 4.5, 5, and 5.5 h, respectively (Figure 2A). To explore the potential mechanisms, we evaluated the biofilm content of OG1RF during the growth process. The results (Figure 2B) revealed OG1RF biofilm accumulation during the growth process (the OD570 changed from 0.204 to 1.289). Notably, although there was a concomitant increase in pheromone content as the bacterial population proliferated (Figure 2C), this heightened pheromone concentration did not cause a decline in the persister rate. Furthermore, the biofilm exerted its primary influence during this period. Additionally, we measured biofilm content in E. faecalis to investigate the potential impact of pheromones on biofilm formation. According to the results, bacteria accumulated a large amount of biofilm after persistence (OD570 increased from 0.442 to 0.600 to 0.601), and cCF10 addition did not alter biofilm accumulation (Supplementary Figure S8). To support this idea, we added the results of different concentrations (8, 10, 12, 14, 18, and 20 ng/mL) of cCF10 on OG1RF biofilm formation. Our results suggested that pheromones do not affect the formation of bacterial biofilms compared to control group (Supplementary Figure S9).
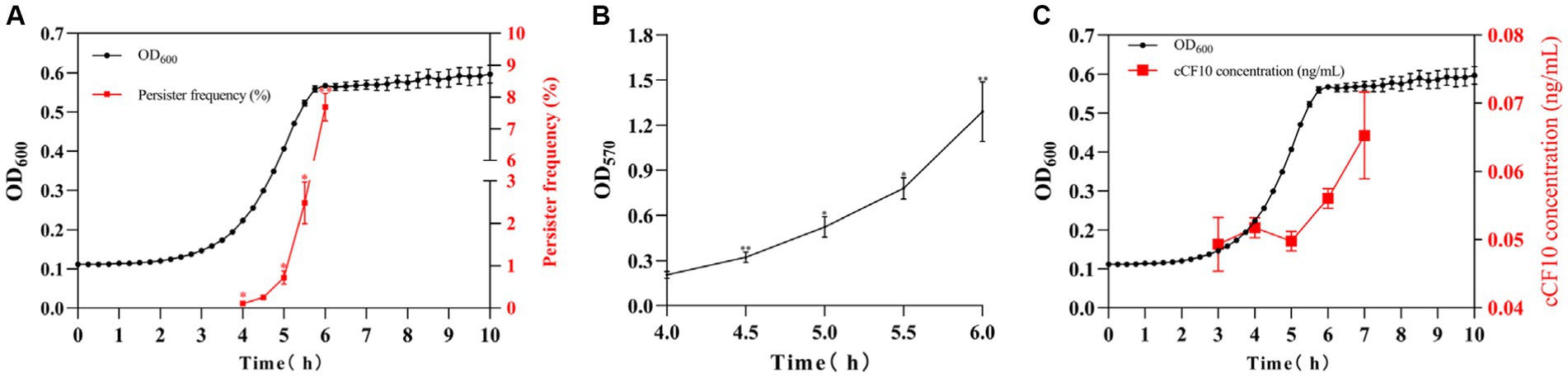
Figure 2. The formation of the persistence depends on biofilm accumulation in Enterococcus faecalis. Changes in persister rates (A), biofilm accumulation (B), and cCF10 concentration (C) at different growth periods in OG1RF. Significant differences between groups were calculated using repeated measures variance and are expressed as *p < 0.05 and **p < 0.01. The results represent the mean ± SD of three biological replicates.
3.3 Gene regulatory pathways involved in cCF10 inhibited Enterococcus faecalis persistence
Herein, we examined transcript level changes to further determine the molecular mechanisms of E. faecalis persistence and the mechanism by which cCF10 inhibits OG1RF persistence. We described the process of transcriptomics analysis in detail in Supplementary Text S5. Persisters had 2,065 differential genes compared to normal bacteria (Figure 3A). On the other hand, the cCF10 treatment group had 231 differential genes compared to the persisters group (Figure 3B). The top 20 enriched pathways are shown in order of q-value from smallest to largest. DEGs were mainly concentrated in metabolic pathways in normal and persisters groups (Figure 3C). On the other hand, in the persisters and cCF10 treatment groups, the results showed that the DEGs were mainly enriched in ABC transfer system (Figure 3D). Furthermore, in the persisters group, the expression levels of genes involved in DNA replication (Figure 3E) and ATP synthesis (Figure 3F) were decreased compared to normal bacteria. Among genes involved in DNA replication, danB2 exhibited the most significant changes (Figure 3E), with reductions to 0.89 and 0.90 times that of the control group in the persister and cCF10 treatment groups, respectively. On the other hand, among the genes associated with ATP synthesis, atpB (Figure 3F) had the most significant alterations, with reductions to 0.79-fold and 0.80-fold that of the control group in the persister and cCF10 treatment groups, respectively. In addition, to further verify whether cCF10 could alter the energy metabolism of OG1RF and prevent bacterial persistence, we focused on changes in gene expression levels associated with glycolysis and tricarboxylic acid (TCA) cycling. Clearly, the expression levels of genes related to metabolism decreased after the bacteria entered the persistence state. In cCF10 treatment group, the expression levels of genes (gatC, gatA, gatB, galK, nifJ, lpd, aceF, and pdhA) were all increased. Of these, galk showed the most significant change with a 1.07-fold increase compared to the persisters group (Figures 3K,L). Furthermore, phoU expression (Figure 3G) in the persister and cCF10 treatment groups were 0.89-fold and 0.90-fold that of the control group, respectively. These findings indicate a notable phoU downregulation following bacterial persistence. Furthermore, recA, which is associated with SOS response, was upregulated by 1.19-fold (in the persisters group) and 1.17-fold (in the cCF10 treatment group) relative to the control group (Figure 3H). The Opp2 system serves as the conduit for cCF10 secretion and uptake in OG1RF (Segawa et al., 2021). In the persisters group, the expression levels of genes involved in Opp2 system were decreased compared to normal bacteria (Figure 3I). However, no changes in Opp2 system gene expression were found in cCF10 treatment group compared to persisters (Figure 3I). In order to further verify this result, we carried out laboratory verification in 3.4. Our results have shown that the accumulation of biofilm played a crucial role in the generation of E. faecalis persisters. Although transcriptomic results showed that the expression of several genes (gelE, ccpA, eno, can and OG1RF_12096) associated with biofilm formation was mostly reduced in the persistent bacteria compared with normal bacteria (Figure 3J). However, no changes in biofilm formation genes expression were found in cCF10 treatment group compared to persisters. In addition, we added different concentrations of cCF10 during the growth of OG1RF in order to further verify the effect of cCF10 on OG1RF biofilm formation. Consistently, the addition of cCF10 did not alter the formation of OG1RF biofilms (Supplementary Figure S9).
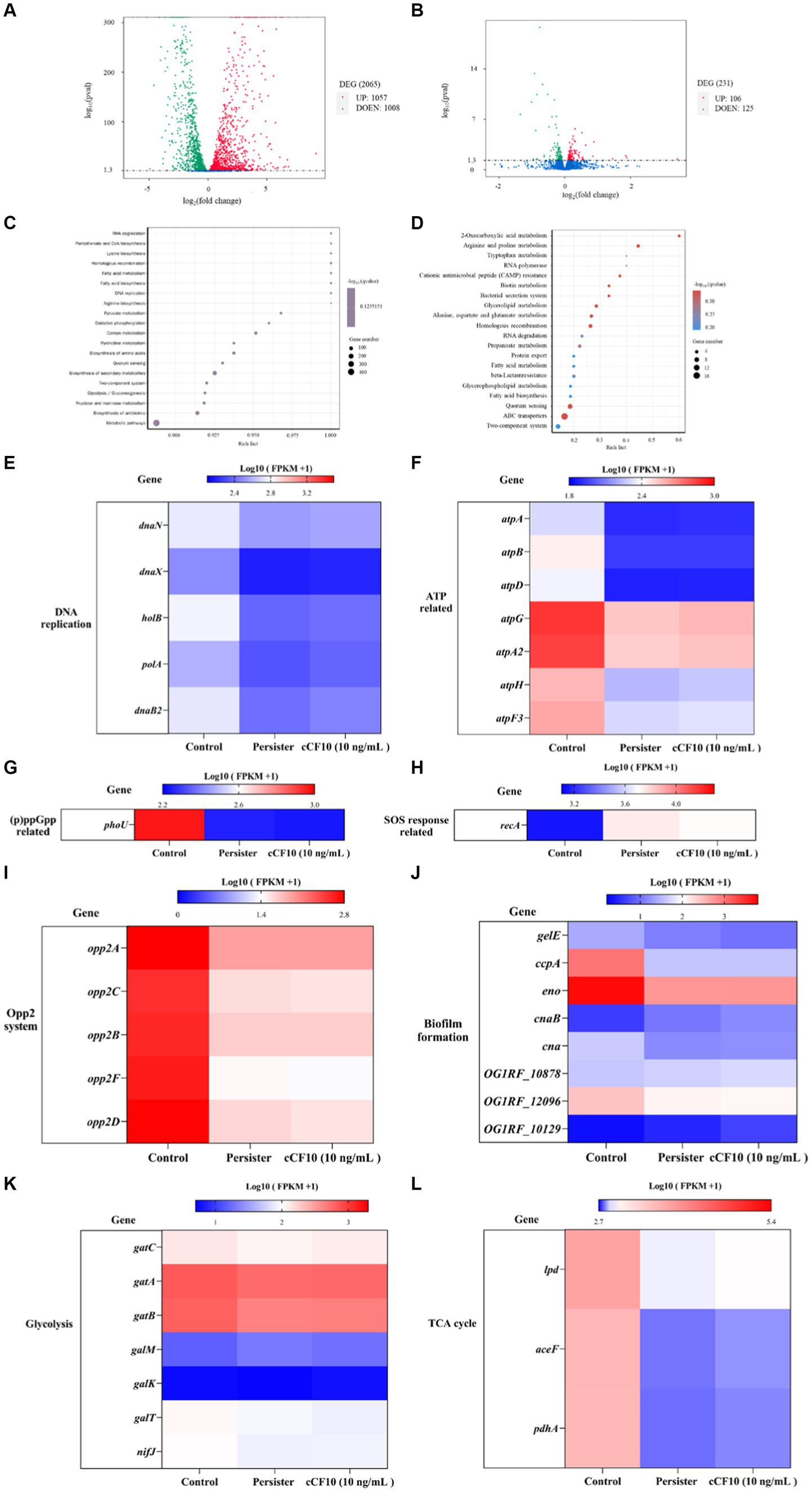
Figure 3. Alterations in the abundance of key genes associated with the persistence. (A) A volcano map of differential genes in the normal bacteria and persister cells. DEG, differentially expressed genes. (B) A volcano map of differential genes in the persister cells and 10 ng/mL cCF10 treatment groups. Red dots represent up regulation, and green dots indicate down-regulated. KEGG pathway enriched analysis. (C) The enriched pathway involved in target genes were showed in the normal bacteria and persister cells. (D) The enriched pathway involved in target genes were showed in the persister cells and 10 ng/mL cCF10 treatment groups. KEGG, Kyoto Encyclopedia of Genes and Genomes. Rich factor refers to the ratio of the number of differentially expressed genes enriched to the number of annotated genes in the pathway. Q-value (value range 0–1) is the p-value after correction by multiple hypothesis testing. Changes in the expression of genes related to DNA replication (E), ATP (F), (p)ppGpp (G), SOS response (H), Opp2 system (I), biofilm formation (J), glycolysis (K) and TCA cycle (L) in different groups (N = 4). TCA, tricarboxylic acid cycle.
3.4 cCF10 prevented bacterial persistence by motivating the Opp2 system and hampering (p)ppGpp accumulation
The Opp system serves as the conduit for pheromone secretion and uptake (Segawa et al., 2021). Segawa et al. (2021) discovered that pheromone uptake in E. faecalis OG1RF was primarily dependent on the Opp2 system. The OppA lipoprotein is a well-conserved protein known for its ability to bind peptides. It interacts with an ABC transporter system consisting of two channel-forming proteins (OppB and OppC) and two membrane-associated ATPases (OppD and OppF; Segawa et al., 2021). Herein, we found that the expression of genes related to the Opp2 system decreased after bacteria entered the persistence phase (Figure 4), indicating that the pheromone-binding ability of E. faecalis persisters was reduced and that the uptake pathway was partially blocked. However, opp2A expression (Figure 4A) increased by 1.61-fold (10 ng/mL of cCF10) and 1.11-fold (14 ng/mL of cCF10) relative to the persisters group, suggesting that the cCF10-binding ability was improved. Similar changes were observed in opp2B, opp2C, opp2D, and opp2F. The expression of opp2B (Figure 4B) increased, respectively, by 1.25-fold (10 ng/mL of cCF10) and 1.50-fold (14 ng/mL of cCF10) relative to the persisters group, suggesting that the cCF10-binding ability was improved. The expression of opp2C (Figure 4C) increased, respectively, by 1.90-fold (10 ng/mL of cCF10) and 1.62-fold (14 ng/mL of cCF10) relative to the persisters group, suggesting that the cCF10-binding ability was improved. The expression of opp2D (Figure 4D) increased, respectively, by 1.53-fold (10 ng/mL of cCF10) and 1.66-fold (14 ng/mL of cCF10) relative to the persisters group, suggesting that the cCF10-binding ability was improved. The expression of opp2F (Figure 4E) increased, respectively, by 2.43-fold (10 ng/mL of cCF10) and 3.11-fold (14 ng/mL of cCF10) relative to the persisters group, suggesting that the cCF10-binding ability was improved. These findings suggested that cCF10 could induce the Opp2 system and enter bacterial cells.
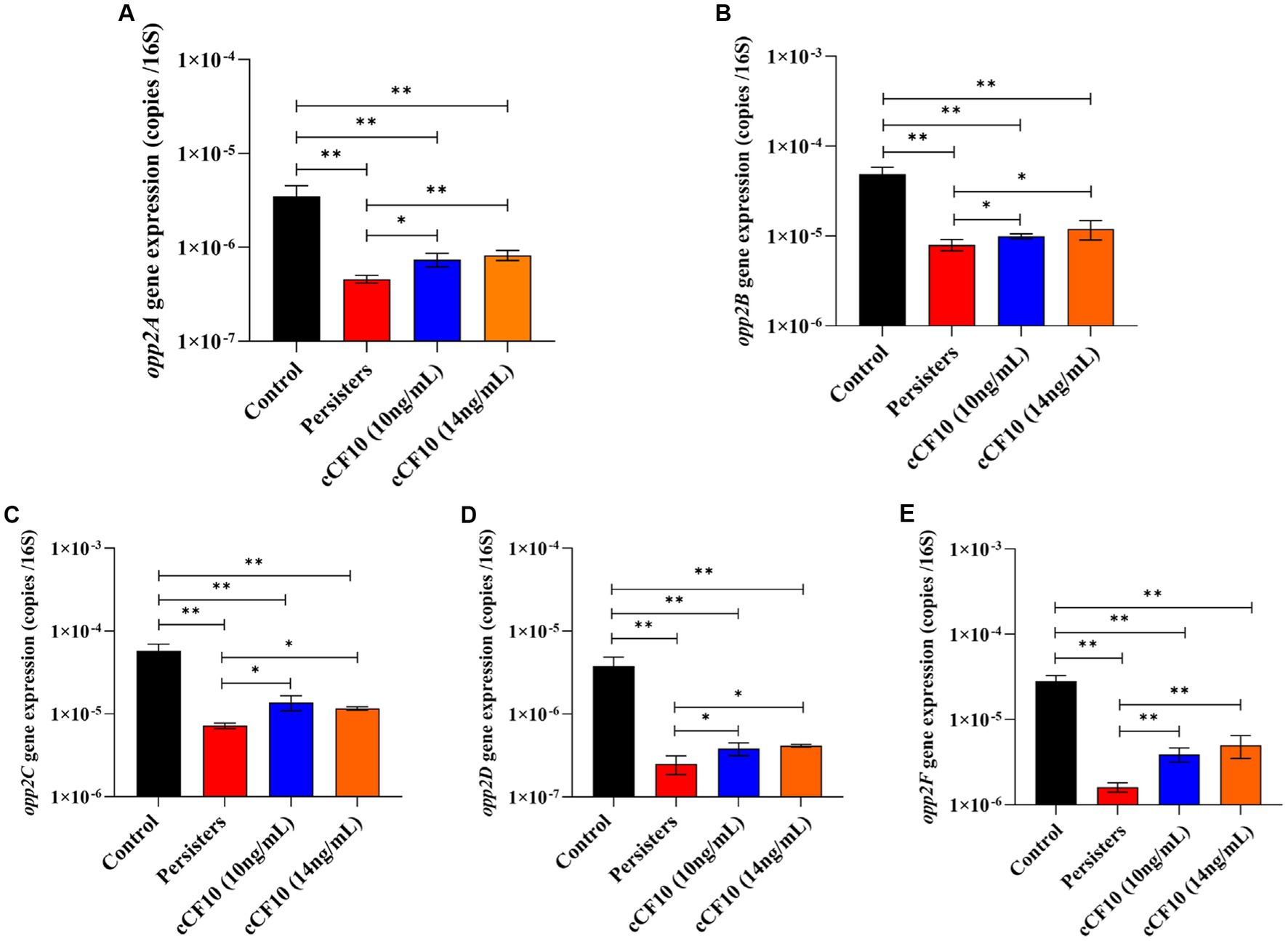
Figure 4. Exogenous cCF10 induces Opp system and enters cells in Enterococcus faecalis. Expression of the related genes of Opp system including opp2A (A), opp2B (B), opp2C (C), opp2D (D), and opp2F (E) in different treatment groups. Significant differences between groups were calculated using analysis of variance and presented by *p < 0.05 and **p < 0.01. The results represent the mean ± SD of three biological replicates.
(p)ppGpp is a signaling molecule that bacteria produce when they encounter challenging conditions, such as exposure to antibiotics. The build-up of (p)ppGpp can ultimately interfere with the movement of protons and inhibit ATP synthesis, leading to bacterial dormancy (Shang et al., 2020). Alterations in (p)ppGpp production negatively affect bacterial stress survival and virulence in E. faecalis, and (p)ppGpp directly inhibits the activity of enzymes involved in GTP biosynthesis (Gaca et al., 2013, 2015). According to research, RelA (synthase) and SpoT (hydrolase) maintain a steady state of (p)ppGpp (Hobbs and Boraston, 2019). At the same time, phoU was reported to affect the intracellular level of (p)ppGpp (Shang et al., 2020). Herein, relA (Figure 5A) and spoT (Figure 5B) expression decreased after the formation of persisters, and no significant changes were observed in the cCF10 treatment group. Furthermore, phoU expression (Figure 5C) in the cCF10 treatment group was 4.5 × 10−5 copies/16 s RNA, a 3.21-fold increase compared to the persisters group. These findings indicate that cCF10 could inhibit persistence by suppressing (p)ppGpp production in E. faecalis.
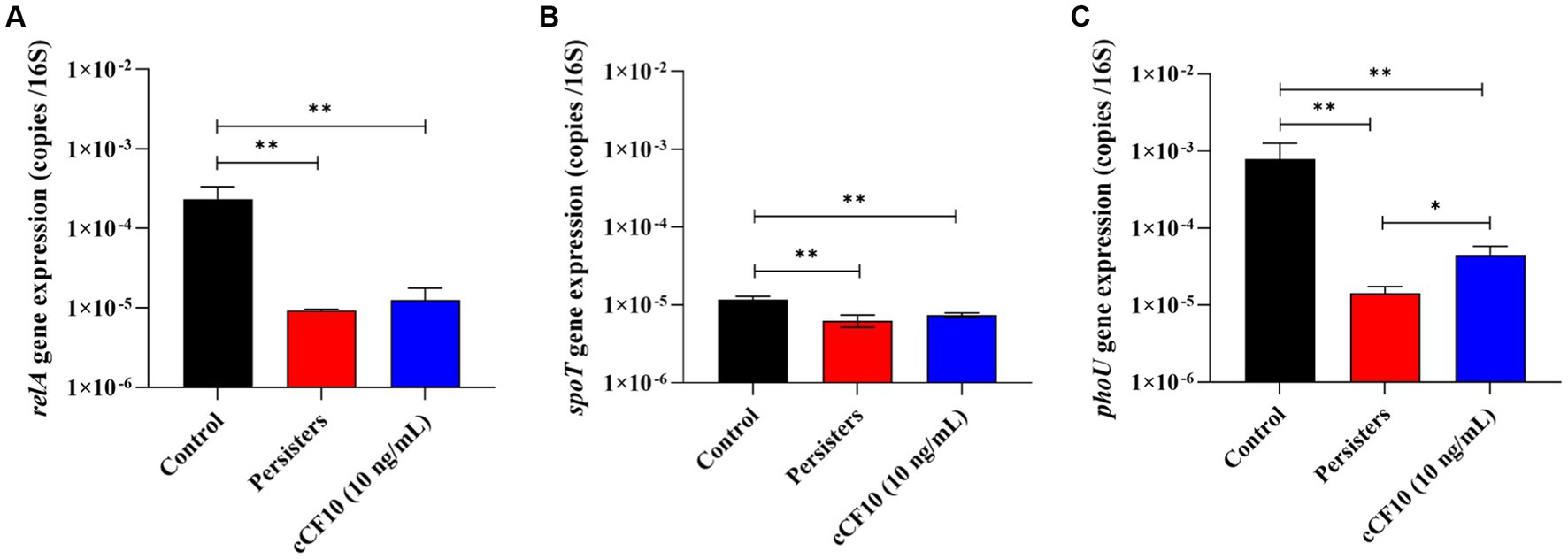
Figure 5. cCF10 inhibits (p)ppGpp accumulation. The expression of the (p)ppGpp encoding genes relA (A) and spoT (B) in different treatment groups. The expression of phoU, a gene encoding the regulatory protein PhoU of the phosphate transport system, in different treatment groups (C). Significant differences between groups were obtained using analysis of variance and presented with *p < 0.05 and **p < 0.01. The results represent the mean ± SD of three biological replicates.
3.5 cCF10 reduces persister cell generation via maintaining the metabolically active state of bacteria
Repressed metabolic activity is a distinctive feature of bacterial persistence (Amato et al., 2014). Herein, When OG1RF entered the persistence state, ATP level decreased from 1.91 μmol/ mg protein to 0.20 μmol/ mg protein. We also observed that compared to the persisters group, 10 ng/mL cCF10 treatment group significantly promoted ATP synthesis (about 1.16-fold). However, the ATP content decreased by ~0.91 times relative to that of the persisters group after adding 14 ng/mL of cCF10 (Figure 6A). Additionally, we assessed the expression levels of genes involved in energy metabolism, including ATP synthesis and DNA replication to further investigate the effect of cCF10 in preventing the formation of persister cells. As expected, atpB (Figure 6B) and atpD (Figure 6C) were downregulated after persistence (~0.073-fold and 0.069-fold of normal bacteria, respectively). After cCF10 exposure, atpB and atpD were upregulated by 2.82-fold and 1.29-fold, respectively, compared to the persisters group, further implying that cCF10 could inhibit persistence by increasing ATP levels. Similarly, danB (Figure 6D), danE (Figure 6E), and recG (Figure 6F) expression levels in persister cells were 0.15, 0.18, and 0.11 times those in normal bacteria, respectively. Notably, cCF10 upregulated danE (Figure 6E) from 7.9 × 10−6 to 1.3 × 10−5 copies/16 s RNA (~1.6-fold increase) compared to persisters. These findings illustrated that cCF10 reduces the generation of persister cells by maintaining the metabolic activity of E. faecalis OG1RF.
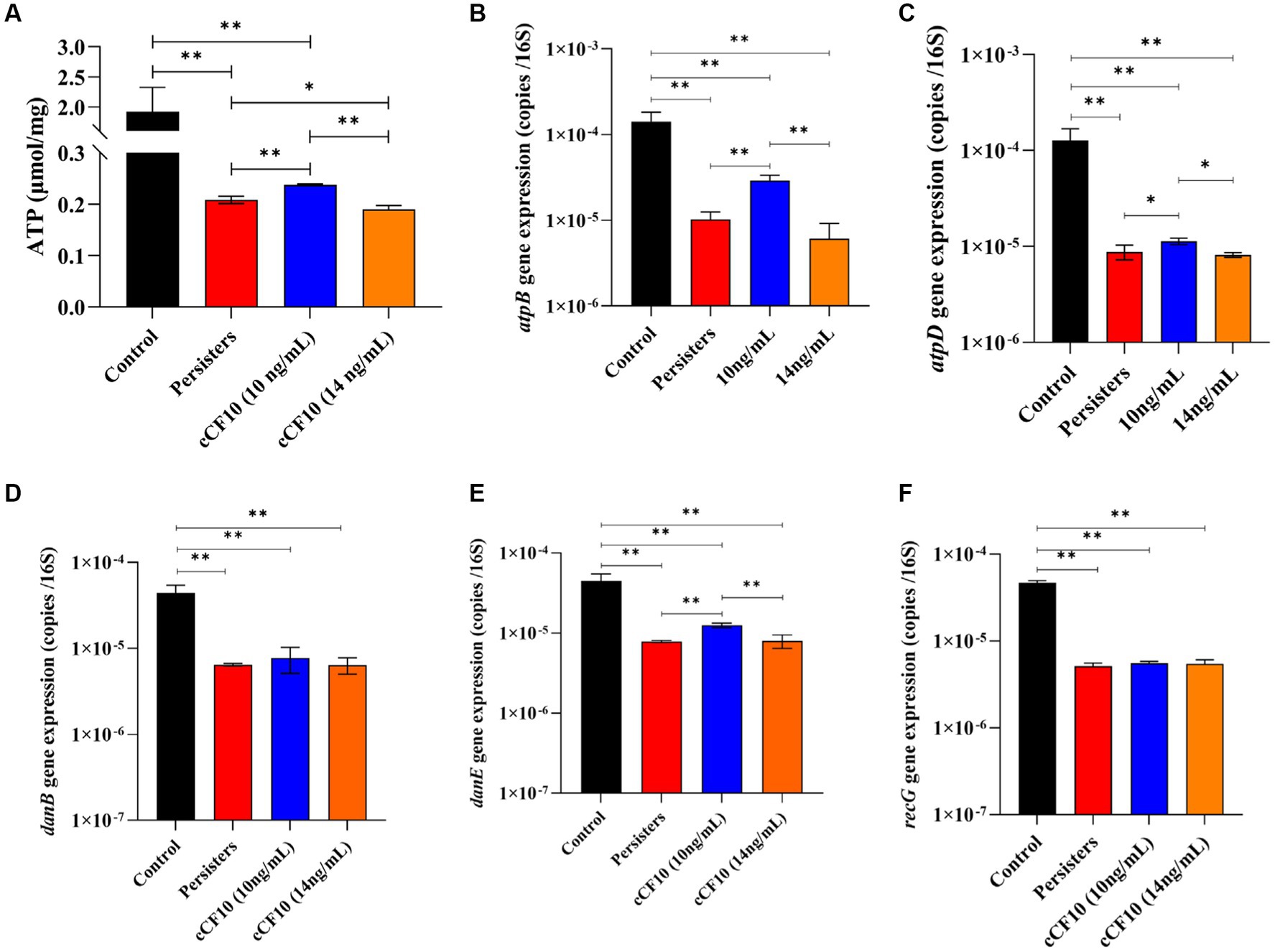
Figure 6. cCF10 maintains the metabolically active state of Enterococcus faecalis. Changes in ATP synthesis in the indicated treatment groups (A). The expression of the ATP related genes atpB (B) and atpD (C) in different groups. The expression of danB (D), danE (E), and recG (F) genes related to DNA replication in the indicated groups. Significant differences between groups were analyzed using analysis of variance and are presented with *p < 0.05 and **p < 0.01. The results represent the mean ± SD of three biological replicates.
4 Discussion
Persisters are key in the recurrence of chronic infections (Harms et al., 2016), exerting a significant influence on disease persistence and treatment outcomes. Persister cells have been the focus of extensive research in recent times. Persisters, specifically Enterococci, the second most Gram-positive pathogen (Kalfopoulou and Huebner, 2020), are a common cause of chronic recurrent infections such as chronic endocarditis. Although Enterococci-initiated nosocomial infections may potentially be associated with bacterial persistence (Conlon, 2014), there are few studies on Enterococci persisters. Herein, we investigated the processes and mechanisms underlying the formation of E. faecalis persistence using an E. faecalis persistence model. The persistence rate of E. faecalis OG1RF was as high as 0.109%, ~10-fold greater than that of Gram-negative bacteria such as E. coli (Wang et al., 2022). Our findings indicated that Enterococci exhibited a heightened propensity for persister cell formation, warranting widespread attention. According to research, Enterococci biofilms exhibit inherent antimicrobial tolerance, posing a significant challenge to managing infections (Ch’ng et al., 2019) and potentially contributing to persistence. This study revealed that biofilm accumulation promoted persister formation in E. faecalis (Figure 2). Consistently, Kaviar et al. (2022) reported that the expression of biofilm-associated genes (esp., agg, and gelE) were higher among persisters compared to non-persister E. faecalis isolates. Therefore, biofilm enrichment serves as a pivotal step in E. faecalis persistence formation.
Numerous studies have proposed strategies for combating bacterial resistance (Allegretta et al., 2017; Baldry et al., 2020; Liu et al., 2020, 2021; Wainwright et al., 2021). Among them, enhancing the effectiveness of conventional antibiotics has been reported as a viable approach, and recent studies have extensively documented different categories of metabolites, as well as adjuncts for augmenting antibiotic potency (Liu et al., 2019; Lv et al., 2022). Certain compounds, including n-butanol and cajaninstilbene acid derivatives (Liu et al., 2019; Lv et al., 2022), have demonstrated therapeutic efficacy against persistent bacteria. Nevertheless, these compounds could cause cellular toxicity. Facilitating persister resuscitation is also considered an effective treatment against bacterial persistence (Defraine et al., 2018). Specific signaling molecules involved in nutritional (Dunny and Leonard, 1997) and fatty acid signaling (Marques et al., 2014) can modulate bacterial metabolism, facilitating persister resuscitation. For example, Davies and Marques (2009) demonstrated that cis-DA, a fatty acid signaling molecule, could induce the dissipation of already-formed biofilms and inhibit biofilm development. Marques et al. (2014) reported that cis-DA could induce a bacterial transition from a persistent to a highly antimicrobial-sensitive state. Compared to those treated with ciprofloxacin alone, the presence of cis-DA resulted in a significant reduction of 1 to 2 logarithmic orders in the population of persister cells for both P. aeruginosa and E. coli (Marques et al., 2014). Similarly, our findings demonstrated that cCF10, existing as short peptides in Enterococci, regulated the formation of E. faecalis persister cells in a concentration-dependent manner. The persister rate decreased from 0.109 to 0.050 and 0.047% when E. faecalis OGIRF was exposed to cCF10 concentrations of 10 and 12 ng/mL (Figure 1A).
Interestingly, we observed a gradual increase in the persister rate of E. faecalis during growth, which was accompanied by an upward trend in extracellular cCF10 pheromone concentration. This phenomenon could be attributed to the fact that the extracellular pheromone concentrations ranged from 0.04 to 0.08 ng/mL (Figure 2), whereas the inhibitory effects against E. faecalis persistence were observed when the additional cCF10 concentration reached 10 ng/mL (Figure 1), indicating that bacterial persistence could only be suppressed at a specific pheromone concentration threshold. Similarly, in our previous studies on conjugative transfer of plasmid, we also found that cCF10 induced the active state of bacteria in a certain range and then induced the transfer of plasmid, whereas higher concentrations of cCF10 inhibited the process (Yang et al., 2022). It may be because the high concentration of cCF10 affects the normal metabolism of bacteria. Besides, our results also indicated that the growth of OG1RF was inhibited by 40 ng/mL of cCF10 (Supplementary Figure S6). Concentrations of cCF10 below 40 ng/mL above 10 ng/mL affected OG1RF metabolism, although not OG1RF growth. Therefore, it is that elevated cCF10 levels disrupt bacterial metabolism and lead to bacterial persistence. Our findings also confirmed that biofilm content increased gradually during the growth process and that the gradual augmentation of biofilm content increased the persistence rates (Perez et al., 2014). However, elevated levels of cCF10 alone could not reverse this phenomenon. Consequently, we elucidated the two modes that impact the persistence process of E. faecalis, specifically the regulation of biofilm formation and pheromone effects. This deduction offers valuable insights into future efforts to combat E. faecalis persisters.
Subsequently, we investigated the mechanism by which cCF10 regulates the formation of persisters. Our results confirmed that the Opp system was suppressed during persistence, indicating a partial closure of the pheromone intake pathway in E. faecalis. Moreover, the results further confirmed that cCF10 modulated the expression of Opp2 system (Figure 4), implying that cCF10 may induce the Opp2 system. Peptide uptake in numerous bacterial species is predominantly facilitated by oligopeptide permease systems, which consists of a set of five protein constituents (Dunny, 2013). The OppA lipoprotein is a well-conserved protein known for its ability to bind peptides. It interacts with an ABC transporter system consisting of two channel-forming proteins (OppB and OppC) and two membrane-associated ATPases (OppD and OppF; Berntsson et al., 2012; Moraes et al., 2014). Together, this system facilitates the binding and transport of a wide range of peptides across different species (Moraes et al., 2014). Exogenous pheromone cCF10 is transported into the cell via the Opp system during ATP hydrolysis (Berntsson et al., 2012; Dunny, 2013). Segawa et al. (2021) indicated that the pheromone import in E. faecalis OG1RF was dependent on a functional Opp2 system. The Opp transport protein system, besides serving as a signaling molecule for intercellular communication in Gram-positive bacteria, also can capture extracellular nutrients (Moraes et al., 2014). Therefore, we thought that activation of the Opp system by cCF10 sustained cellular energy metabolism, thus inhibiting bacterial persistence. Formation of the persistence is induced by growth stagnation (Manuse et al., 2021). Bekale et al. (2023) reported that the persistence of E. coli might be regulated by ATP levels. Notably, accumulation of (p)ppGpp stimulated the production of toxin, thereby causing membrane potential collapse and ATP depletion, leading to bacterial dormancy (Pan et al., 2023). Analysis of the transcriptome results showed that cCF10 affected the expression of energy metabolism and (p)ppGpp related genes in E. faecalis (Figure 3). Two enzymes RelA (synthase) and SpoT (hydrolase) maintain a steady state of (p)ppGpp (Hobbs and Boraston, 2019). Simultaneously, inactivation of phoU can enhance (p)ppGpp accumulation (Shang et al., 2020). This study revealed that exposure to cCF10 upregulated the expression of phoU, thereby inhibiting (p)ppGpp accumulation (Figure 5). Further, accumulation of (p)ppGpp inhibited bacterial energy metabolism, thereby inducing bacterial dormancy (Pan et al., 2023). Our results demonstrated that cCF10 promoted the expression of atpB and atpD (Figure 6), which encode ATP synthetases.
Therefore, the present results indicate that the exogenous cCF10 entered the cell through the Opp channel, promoting ATP synthesis and sustaining bacterial energy metabolism. In general, the diminution of energy metabolism levels in bacteria induces bacterial persistence, as the majority of antibiotics exert their bactericidal effects by disrupting active, energy-dependent targets. In addition, the primary mechanism of action of the LVF employed in our study entails the suppression of bacterial DNA gyrase activity. Our study further confirmed that the external pheromone cCF10 enhanced the expression of dnaE (Figure 6), which encodes DNA polymerase III. This enzyme regulates the DNA replication by facilitating accurate and swift synthesis of DNA strands. The results suggested that cCF10 enhanced DNA replication, thereby augmenting the susceptibility of E. faecalis to the bactericidal effect of LVF. Consequently, cCF10 induced Opp system and entered bacterial cells to inhibit (p)ppGpp accumulation which maintained the metabolically active state of bacteria and reduced the generation of persister cells.
5 Conclusion
In conclusion, this study found that during the growth of E. faecalis OG1RF, accumulation of biofilm contributed to the development of antibiotic persistence. Moreover, we found for the first time that cCF10 prevented the formation of persister cells at certain concentrations. Surprisingly, cCF10 mediated the antibiotic persistence of E. faecalis OG1RF by altering the metabolic activity rather than inhibiting biofilm formation. Addition of cCF10 improved the Opp system and entered bacterial cells to suppress the accumulation of (p)ppGpp. This maintained the metabolically active state of bacteria and reduced the formation of persister cells. These results provide valuable insights and expand our understanding of the formation and control mechanism of persisters in E. faecalis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
LZ: Writing – original draft, Writing – review & editing, Investigation, Formal analysis. XY: Writing – review & editing, Writing – original draft. XF: Formal analysis, Writing – review & editing. PY: Formal analysis, Writing – review & editing. XL: Formal analysis, Writing – review & editing. FW: Formal analysis, Writing – review & editing. ZS: Resources, Writing – review & editing. JW: Resources, Writing – review & editing. FS: Supervision, Writing – review & editing, Resources. ZQ: Supervision, Writing – review & editing, Resources.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by National Natural Science Foundation of China (grant no. 42177414), the Shaanxi Provincial Science and Technology Key Project (no. 2023-YBNY-167), and the Xi’an Municipal Science and Technology Project (no. 22GXFW0040).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1408701/full#supplementary-material
References
Aedo, S. J., Orman, M. A., and Brynildsen, M. P. (2019). Stationary phase persister formation in Escherichia coli can be suppressed by piperacillin and PBP3 inhibition. BMC Microbiol. 19:140. doi: 10.1186/s12866-019-1506-7
Allegretta, G., Maurer, C. K., Eberhard, J., Maura, D., Hartmann, R. W., Rahme, L., et al. (2017). In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Front. Microbiol. 8:924. doi: 10.3389/fmicb.2017.00924
Alt, L. M., Flater, J. S., Howe, A., Moorman, T. B., Schulte, L. A., and Soupir, M. L. (2023). Prairie strips remove swine manure associated antimicrobial resistance genes and bacteria from runoff. Agric. Ecosyst. Environ. 349:108469. doi: 10.1016/j.agee.2023.108469
Amato, S. M., Fazen, C. H., Henry, T. C., Mok, W. W., Orman, M. A., Sandvik, E. L., et al. (2014). The role of metabolism in bacterial persistence. Front. Microbiol. 5:70. doi: 10.3389/fmicb.2014.00070
Apreja, M., Sharma, A., Balda, S., Kataria, K., Capalash, N., and Sharma, P. (2022). Antibiotic residues in environment: antimicrobial resistance development, ecological risks, and bioremediation. Environ. Sci. Pollut. Res. Int. 29, 3355–3371. doi: 10.1007/s11356-021-17374-w
Balaban, N. Q., Helaine, S., Lewis, K., Ackermann, M., Aldridge, B., Andersson, D. I., et al. (2019). Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 17, 441–448. doi: 10.1038/s41579-019-0196-3
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L., and Leibler, S. (2004). Bacterial persistence as a phenotypic switch. Science 305, 1622–1625. doi: 10.1126/science.1099390
Baldry, M., Bojer, M. S., Najarzadeh, Z., Vestergaard, M., Meyer, R. L., Otzen, D. E., et al. (2020). Phenol-soluble Modulins modulate Persister cell formation in Staphylococcus aureus. Front. Microbiol. 11:573253. doi: 10.3389/fmicb.2020.573253
Balomenos, A. D., Tsakanikas, P., Aspridou, Z., Tampakaki, A. P., Koutsoumanis, K. P., and Manolakos, E. S. J. B. S. B. (2017). Image analysis driven single-cell analytics for systems microbiology. BMC Syst. Biol. 11:43. doi: 10.1186/s12918-017-0399-z
Bartell, J. A., Cameron, D. R., Mojsoska, B., Haagensen, J. A. J., Pressler, T., Sommer, L. M., et al. (2020). Bacterial persisters in long-term infection: emergence and fitness in a complex host environment. PLoS Pathog. 16:e1009112. doi: 10.1371/journal.ppat.1009112
Beam, J. E., Wagner, N. J., Shook, J. C., Bahnson, E. S. M., Fowler, V. G. Jr., Rowe, S. E., et al. (2021). Macrophage-produced Peroxynitrite induces antibiotic tolerance and supersedes intrinsic mechanisms of Persister formation. Infect. Immun. 89:e0028621. doi: 10.1128/iai.00286-21
Bekale, L. A., Sharma, D., Bacacao, B., Chen, J., and Maria, P. L. S. (2023). Eradication of bacterial persister cells by leveraging their low metabolic activity using adenosine triphosphate coated gold nanoclusters. Nano Today 51:101895. doi: 10.1016/j.nantod.2023.101895
Berntsson, R. P., Schuurman-Wolters, G. K., Dunny, G., Slotboom, D. J., and Poolman, B. (2012). Structure and mode of peptide binding of pheromone receptor PrgZ. J. Biol. Chem. 287, 37165–37170. doi: 10.1074/jbc.M112.386334
Canfield, G. S., Chatterjee, A., Espinosa, J., Mangalea, M. R., Sheriff, E. K., Keidan, M., et al. (2023). Lytic bacteriophages facilitate antibiotic sensitization of Enterococcus faecium. Antimicrob. Agents Chemother. 65:21. doi: 10.1128/aac.00143-21
Cao, Z., Chen, X., Chen, J., Xia, A., Bacacao, B., Tran, J., et al. (2022). Gold nanocluster adjuvant enables the eradication of persister cells by antibiotics and abolishes the emergence of resistance. Nanoscale 14, 10016–10032. doi: 10.1039/d2nr01003h
Cepas, V., López, Y., Muñoz, E., Rolo, D., Ardanuy, C., Martí, S., et al. (2019). Relationship between biofilm formation and antimicrobial resistance in gram-negative Bacteria. Microb. Drug Resist. 25, 72–79. doi: 10.1089/mdr.2018.0027
Ch’ng, J.-H., Chong, K. K. L., Lam, L. N., Wong, J. J., and Kline, K. A. (2019). Biofilm-associated infection by enterococci. Nat. Rev. Microbiol. 17, 82–94. doi: 10.1038/s41579-018-0107-z
Chandler, J. R., and Dunny, G. M. (2008). Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J. Bacteriol. 190, 1172–1183. doi: 10.1128/jb.01327-07
Chang, J., Lee, R. E., and Lee, W. (2020). A pursuit of Staphylococcus aureus continues: a role of persister cells. Arch. Pharm. Res. 43, 630–638. doi: 10.1007/s12272-020-01246-x
Chatterjee, A., Cook, L. C. C., Shu, C.-C., Chen, Y., Manias, D. A., Ramkrishna, D., et al. (2013). Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer. Proc. Natl. Acad. Sci. 110, 7086–7090. doi: 10.1073/pnas.1212256110
Chen, Y., Bandyopadhyay, A., Kozlowicz, B. K., Haemig, H. A. H., Tai, A., Hu, W. S., et al. (2017). Mechanisms of peptide sex pheromone regulation of conjugation in Enterococcus faecalis. Microbiol Open 6:492. doi: 10.1002/mbo3.492
Cho, H., Kwon, K., Kim, S., Park, Y., and Koo, S. (2018). Association between biofilm formation and antimicrobial resistance in Carbapenem-resistant Pseudomonas Aeruginosa. Ann. Clin. Lab. Sci. 48, 363–368.
Conlon, B. P. (2014). Staphylococcus aureus chronic and relapsing infections: evidence of a role for persister cells: an investigation of persister cells, their formation and their role in S. aureus disease. Bioessays 36, 991–996. doi: 10.1002/bies.201400080
Conlon, B. P., Rowe, S. E., Gandt, A. B., Nuxoll, A. S., Donegan, N. P., Zalis, E. A., et al. (2016). Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1:16051. doi: 10.1038/nmicrobiol.2016.51
Dale, J., Cagnazzo, J., Phan, C., Barnes, A., and Dunny, G. (2015). Multiple roles for Enterococcus faecalis glycosyltransferases in biofilm-associated antibiotic resistance, cell envelope integrity, and conjugative transfer. Antimicrob. Agents Chemother. 59, 4094–4105. doi: 10.1128/AAC.00344-15
Davies, D. G., and Marques, C. N. (2009). A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 191, 1393–1403. doi: 10.1128/jb.01214-08
Defraine, V., Fauvart, M., and Michiels, J. (2018). Fighting bacterial persistence: current and emerging anti-persister strategies and therapeutics. Drug Resist. Updat. 38, 12–26. doi: 10.1016/j.drup.2018.03.002
Dufour, D., Zhao, H., Gong, S. G., and Lévesque, C. M. (2022). A DNA-damage inducible gene promotes the formation of antibiotic Persisters in response to the quorum sensing signaling peptide in Streptococcus mutans. Genes 13:1434. doi: 10.3390/genes13081434
Dunny, G. M. (2013). Enterococcal sex pheromones: signaling, social behavior, and evolution. Annu. Rev. Genet. 47, 457–482. doi: 10.1146/annurev-genet-111212-133449
Dunny, G. M., and Leonard, B. A. (1997). Cell-cell communication in gram-positive bacteria. Annu. Rev. Microbiol. 51, 527–564. doi: 10.1146/annurev.micro.51.1.527
Fang, X., and Allison, K. R. (2023). Resuscitation dynamics reveal persister partitioning after antibiotic treatment. Mol. Syst. Biol. 19:e11320. doi: 10.15252/msb.202211320
Fernandes, S., Gomes, I. B., Sousa, S. F., and Simões, M. (2022). Antimicrobial susceptibility of Persister biofilm cells of Bacillus cereus and Pseudomonas fluorescens. Microorganisms 10:160. doi: 10.3390/microorganisms10010160
Flannagan, S. E., and Clewell, D. B. (2002). Identification and characterization of genes encoding sex pheromone cAM373 activity in Enterococcus faecalis and Staphylococcus aureus. Mol. Microbiol. 44, 803–817. doi: 10.1046/j.1365-2958.2002.02922.x
Gaca, A. O., Colomer-Winter, C., and Lemos, J. A. (2015). Many means to a common end: the intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J. Bacteriol. 197, 1146–1156. doi: 10.1128/JB.02577-14
Gaca, A. O., Kajfasz, J. K., Miller, J. H., Liu, K., Wang, J. D., Abranches, J., et al. (2013). Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. MBio 4, e00646–e00613. doi: 10.1128/mBio.00646-13
García-Solache, M., and Rice, L. B. (2019). The Enterococcus: a model of adaptability to its environment. Clin. Microbiol. Rev. 32:18. doi: 10.1128/cmr.00058-18
Gollan, B., Grabe, G., Michaux, C., and Helaine, S. (2019). Bacterial Persisters and infection: past, present, and progressing. Annu. Rev. Microbiol. 73, 359–385. doi: 10.1146/annurev-micro-020518-115650
Hare, P. J., LaGree, T. J., Byrd, B. A., DeMarco, A. M., and Mok, W. W. K. (2021). Single-cell technologies to study phenotypic heterogeneity and bacterial Persisters. Microorganisms 9:2277. doi: 10.3390/microorganisms9112277
Harms, A., Maisonneuve, E., and Gerdes, K. (2016). Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:4268. doi: 10.1126/science.aaf4268
Hobbs, J. K., and Boraston, A. B. (2019). (p)ppGpp and the stringent response: an emerging threat to antibiotic therapy. ACS Infect Dis 5, 1505–1517. doi: 10.1021/acsinfecdis.9b00204
Hrynyshyn, A., Simões, M., and Borges, A. (2022). Biofilms in surgical site infections: recent advances and novel prevention and eradication strategies. Antibiotics 11:69. doi: 10.3390/antibiotics11010069
Kalfopoulou, E., and Huebner, J. (2020). Advances and prospects in vaccine development against enterococci. Cells 9:397. doi: 10.3390/cells9112397
Kaszab, E., Laczkó, L., Kardos, G., and Bányai, K. (2023). Antimicrobial resistance genes and associated mobile genetic elements in Lactobacillales from various sources. Front. Microbiol. 14:1281473. doi: 10.3389/fmicb.2023.1281473
Kaur, A., Sharma, P., and Capalash, N. (2018). Curcumin alleviates persistence of Acinetobacter baumannii against colistin. Sci. Rep. 8:11029. doi: 10.1038/s41598-018-29291-z
Kaviar, V. H., Khoshnood, S., Asadollahi, P., Kalani, B. S., Maleki, A., Yarahmadi, S., et al. (2022). Survey on phenotypic resistance in Enterococcus faecalis: comparison between the expression of biofilm-associated genes in Enterococcus faecalis persister and non-persister cells. Mol. Biol. Rep. 49, 971–979. doi: 10.1007/s11033-021-06915-8
Kim, W., Hendricks, G. L., Tori, K., Fuchs, B. B., and Mylonakis, E. (2018). Strategies against methicillin-resistant Staphylococcus aureus persisters. Future Med. Chem. 10, 779–794. doi: 10.4155/fmc-2017-0199
Kint, C. I., Verstraeten, N., Fauvart, M., and Michiels, J. (2012). New-found fundamentals of bacterial persistence. Trends Microbiol. 20, 577–585. doi: 10.1016/j.tim.2012.08.009
Korir, M. L., Dale, J. L., and Dunny, G. M. (2019). Role of epaQ, a previously uncharacterized Enterococcus faecalis gene, in biofilm development and antimicrobial resistance. J. Bacteriol. 201:19. doi: 10.1128/jb.00078-19
Lebeaux, D., Ghigo, J. M., and Beloin, C. (2014). Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 78, 510–543. doi: 10.1128/mmbr.00013-14
Lechner, S., Lewis, K., and Bertram, R. (2012). Staphylococcus aureus Persisters tolerant to bactericidal antibiotics. J. Mol. Microbiol. Biotechnol. 22, 235–244. doi: 10.1159/000342449
Lewis, K. (2007). Persister cells, dormancy and infectious disease. Nat. Rev. Microbiol. 5, 48–56. doi: 10.1038/nrmicro1557
Li, Z., Shi, L., Wang, B., Wei, X., Zhang, J., Guo, T., et al. (2021). In vitro assessment of antimicrobial resistance dissemination dynamics during multidrug-resistant-bacterium invasion events by using a continuous-culture device. Appl. Environ. Microbiol. 87:20. doi: 10.1128/aem.02659-20
Li, Y., Zhang, L., Zhou, Y., Zhang, Z., and Zhang, X. (2018). Survival of bactericidal antibiotic treatment by tolerant persister cells of Klebsiella pneumoniae. J. Med. Microbiol. 67, 273–281. doi: 10.1099/jmm.0.000680
Lin, J. S., Bekale, L. A., Molchanova, N., Nielsen, J. E., Wright, M., Bacacao, B., et al. (2022). Anti-persister and anti-biofilm activity of self-assembled antimicrobial Peptoid ellipsoidal micelles. ACS Infect Dis 8, 1823–1830. doi: 10.1021/acsinfecdis.2c00288
Liu, Y., Li, R., Xiao, X., and Wang, Z. (2019). Bacterial metabolism-inspired molecules to modulate antibiotic efficacy. J. Antimicrob. Chemother. 74, 3409–3417. doi: 10.1093/jac/dkz230
Liu, Y., She, P., Xu, L., Chen, L., Li, Y., Liu, S., et al. (2021). Antimicrobial, Antibiofilm, and anti-persister activities of Penfluridol against Staphylococcus aureus. Front. Microbiol. 12:727692. doi: 10.3389/fmicb.2021.727692
Liu, X., Tang, R., Li, H., Wang, L., and Wan, C. (2022). The physiological and ecological properties of bacterial persisters discovered from municipal sewage sludge and the potential risk. Environ. Res. 205:112481. doi: 10.1016/j.envres.2021.112481
Liu, X., Wang, C., Yan, B., Lyu, L., Takiff, H. E., and Gao, Q. (2020). The potassium transporter KdpA affects persister formation by regulating ATP levels in Mycobacterium marinum. Emerg Microbes Infect 9, 129–139. doi: 10.1080/22221751.2019.1710090
Łuczkiewicz, A., Jankowska, K., Fudala-Książek, S., and Olańczuk-Neyman, K. (2010). Antimicrobial resistance of fecal indicators in municipal wastewater treatment plant. Water Res. 44, 5089–5097. doi: 10.1016/j.watres.2010.08.007
Lv, B., Bian, M., Huang, X., Sun, F., Gao, Y., Wang, Y., et al. (2022). N-butanol potentiates subinhibitory aminoglycosides against bacterial Persisters and multidrug-resistant MRSA by rapidly enhancing antibiotic uptake. ACS Infect Dis 8, 373–386. doi: 10.1021/acsinfecdis.1c00559
Manuse, S., Shan, Y., Canas-Duarte, S. J., Bakshi, S., Sun, W. S., Mori, H., et al. (2021). Bacterial persisters are a stochastically formed subpopulation of low-energy cells. PLoS Biol. 19:e3001194. doi: 10.1371/journal.pbio.3001194
Marques, C. N. H., Morozov, A., Planzos, P., and Zelaya, H. M. (2014). The fatty acid signaling molecule cis-2-Decenoic acid increases metabolic activity and reverts Persister cells to an antimicrobial-susceptible state. Appl. Environ. Microbiol. 80, 6976–6991. doi: 10.1128/AEM.01576-14
McCourt, J., O’Halloran, D. P., McCarthy, H., O’Gara, J. P., and Geoghegan, J. A. (2014). Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol. Lett. 353, 157–164. doi: 10.1111/1574-6968.12424
Mlynarcik, P., and Kolar, M. (2017). Starvation- and antibiotics-induced formation of persister cells in Pseudomonas aeruginosa. Biomed Pap Med Fac Univ Palacky Olomouc Czech 161, 58–67. doi: 10.5507/bp.2016.057
Moraes, P. M., Seyffert, N., Silva, W. M., Castro, T. L., Silva, R. F., Lima, D. D., et al. (2014). Characterization of the Opp peptide transporter of Corynebacterium pseudotuberculosis and its role in virulence and pathogenicity. Biomed. Res. Int. 2014:489782, 1–7. doi: 10.1155/2014/489782
Morales-Espinosa, R., Hernandez-Castro, R., Delgado, G., Mendez, J. L., Navarro, A., Manjarrez, A., et al. (2016). UPEC strain characterization isolated from Mexican patients with recurrent urinary infections. J. Infect. Dev. Ctries. 10, 317–328. doi: 10.3855/jidc.6652
Mori, M., Sakagami, Y., Narita, M., Isogai, A., Fujino, M., Kitada, C., et al. (1984). Isolation and structure of the bacterial sex pheromone, cAD1, that induces plasmid transfer in Streptococcus faecalis. FEBS Lett. 178, 97–100. doi: 10.1016/0014-5793(84)81248-x
Narimisa, N., Sadeghi Kalani, B., Mohammadzadeh, R., and Masjedian Jazi, F. (2021). Combination of antibiotics-Nisin reduces the formation of Persister cell in Listeria monocytogenes. Microb. Drug Resist. 27, 137–144. doi: 10.1089/mdr.2020.0019
Nguyen, L., DeVico, B., Mannan, M., Chang, M., Rada Santacruz, C., Siragusa, C., et al. (2023). Tea tree essential oil kills Escherichia coli and Staphylococcus epidermidis Persisters. Biomol. Ther. 13:1404. doi: 10.3390/biom13091404
Pallares, C., Hernández-Gómez, C., Appel, T. M., Escandón, K., Reyes, S., Salcedo, S., et al. (2022). Impact of antimicrobial stewardship programs on antibiotic consumption and antimicrobial resistance in four Colombian healthcare institutions. BMC Infect. Dis. 22:420. doi: 10.1186/s12879-022-07410-6
Pan, X., Liu, W., Du, Q., Zhang, H., and Han, D. (2023). Recent advances in bacterial persistence mechanisms. Int. J. Mol. Sci. 24:4311. doi: 10.3390/ijms241814311
Paranjape, S. S., and Shashidhar, R. (2019). Comparison of starvation-induced Persister cells with antibiotic-induced Persister cells. Curr. Microbiol. 76, 1495–1502. doi: 10.1007/s00284-019-01777-7
Pennino, F., Maccauro, M. L., Sorrentino, M., Gioia, M., Riello, S., Messineo, G., et al. (2023). Insights from a cross-sectional study on knowledge, attitudes and behaviors concerning antibiotic use in a large metropolitan area: implications for public health and policy interventions. Antibiotics 12:1476. doi: 10.3390/antibiotics12101476
Perez, A. C., Pang, B., King, L. B., Tan, L., Murrah, K. A., Reimche, J. L., et al. (2014). Residence of Streptococcus pneumoniae and Moraxella catarrhalis within polymicrobial biofilm promotes antibiotic resistance and bacterial persistence in vivo. Pathog Dis 70, 280–288. doi: 10.1111/2049-632x.12129
Podlesek, Z., and Žgur Bertok, D. (2020). The DNA damage inducible SOS response is a key player in the generation of bacterial Persister cells and population wide tolerance. Front. Microbiol. 11:1785. doi: 10.3389/fmicb.2020.01785
Price, V. J., Huo, W., Sharifi, A., and Palmer, K. L. (2016). CRISPR-Cas and restriction-modification act additively against conjugative antibiotic resistance plasmid transfer in Enterococcus faecalis. mSphere 1:16. doi: 10.1128/mSphere.00064-16
Roberts, M. G., Burgess, S., Toombs-Ruane, L. J., Benschop, J., Marshall, J. C., and French, N. P. (2021). Combining mutation and horizontal gene transfer in a within-host model of antibiotic resistance. Math. Biosci. 339:108656. doi: 10.1016/j.mbs.2021.108656
Saha, D., and Mukherjee, R. (2019). Ameliorating the antimicrobial resistance crisis: phage therapy. IUBMB Life 71, 781–790. doi: 10.1002/iub.2010
Segawa, T., Johnson, C. M., Berntsson, R. P., and Dunny, G. M. (2021). Two ABC transport systems carry out peptide uptake in Enterococcus faecalis: their roles in growth and in uptake of sex pheromones. Mol. Microbiol. 116, 459–469. doi: 10.1111/mmi.14725
Shahin, K., Bouzari, M., Wang, R., and Khorasgani, M. R. (2019). Distribution of antimicrobial resistance genes and integrons among Shigella spp. isolated from water sources. J Glob Antimicrob Resist 19, 122–128. doi: 10.1016/j.jgar.2019.04.020
Shang, Y., Wang, X., Chen, Z., Lyu, Z., Lin, Z., Zheng, J., et al. (2020). Staphylococcus aureus PhoU homologs regulate Persister formation and virulence. Front. Microbiol. 11:865. doi: 10.3389/fmicb.2020.00865
Svenningsen, M. S., Svenningsen, S. L., Sørensen, M. A., and Mitarai, N. (2022). Existence of log-phase Escherichia coli persisters and lasting memory of a starvation pulse. Life Sci Alliance 5:e202101076. doi: 10.26508/lsa.202101076
Tkachenko, A., Kashevarova, N., Shumkov, M., and Karavaeva, E. (2014). Polyamines upregulate the formation of E. coli persister cells through the modulation of rpoS expression. FEBS J. 281:666.
Tomita, H., Lu, J. J., and Ike, Y. (2023). High incidence of multiple-drug-resistant pheromone-responsive plasmids and transmissions of VanA-type vancomycin-resistant Enterococcus faecalis between livestock and humans in Taiwan. Antibiotics 12:1668. doi: 10.3390/antibiotics12121668
Van den Bergh, B., Fauvart, M., and Michiels, J. (2017). Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 41, 219–251. doi: 10.1093/femsre/fux0001
Van Tyne, D., Manson, A. L., Huycke, M. M., Karanicolas, J., Earl, A. M., and Gilmore, M. S. (2019). Impact of antibiotic treatment and host innate immune pressure on enterococcal adaptation in the human bloodstream. Sci. Transl. Med. 11:8418. doi: 10.1126/scitranslmed.aat8418
Varahan, S., Harms, N., Gilmore, M. S., Tomich, J. M., and Hancock, L. E. (2014). An ABC transporter is required for secretion of peptide sex pheromones in Enterococcus faecalis. MBio 5, e01726–e01714. doi: 10.1128/mBio.01726-14
Vickerman, M. M., and Mansfield, J. M. (2019). Streptococcal peptides that signal Enterococcus faecalis cells carrying the pheromone-responsive conjugative plasmid pAM373. Mol. Oral Microbiol. 34, 254–262. doi: 10.1111/omi.12271
Wagner, E. G. H., and Unoson, C. (2012). The toxin-antitoxin system tisB-istR1. RNA Biol. 9, 1513–1519. doi: 10.4161/rna.22578
Wainwright, J., Hobbs, G., and Nakouti, I. (2021). Persister cells: formation, resuscitation and combative therapies. Arch. Microbiol. 203, 5899–5906. doi: 10.1007/s00203-021-02585-z
Wang, T., El Meouche, I., and Dunlop, M. J. (2017). Bacterial persistence induced by salicylate via reactive oxygen species. Sci. Rep. 7:43839. doi: 10.1038/srep43839
Wang, Y., Lu, J., Mao, L., Li, J., Yuan, Z., Bond, P. L., et al. (2019). Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera. ISME J. 13, 509–522. doi: 10.1038/s41396-018-0275-x
Wang, S., Zhao, C., Xue, B., Li, C., Zhang, X., Yang, X., et al. (2022). Nanoalumina triggers the antibiotic persistence of Escherichia coli through quorum sensing regulators lrsF and qseB. J. Hazard. Mater. 436:129198. doi: 10.1016/j.jhazmat.2022.129198
Windels, E. M., Michiels, J. E., Fauvart, M., Wenseleers, T., Van den Bergh, B., and Michiels, J. (2019a). Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. ISME J. 13, 1239–1251. doi: 10.1038/s41396-019-0344-9
Windels, E. M., Michiels, J. E., Van den Bergh, B., Fauvart, M., and Michiels, J. (2019b). Antibiotics: combatting tolerance to stop resistance. mBio 10:19. doi: 10.1128/mBio.02095-19
Xu, Y., Liu, S., Zhang, Y., and Zhang, W. (2021). DNA adenine methylation is involved in persister formation in E. coli. Microbiol. Res. 246:126709. doi: 10.1016/j.micres.2021.126709
Yamasaki, R., Song, S., Benedik, M. J., and Wood, T. K. (2020). Persister cells resuscitate using membrane sensors that activate chemotaxis, lower cAMP levels, and revive ribosomes. iScience 23:100792. doi: 10.1016/j.isci.2019.100792
Yang, X., Niu, Y., Yang, Y., Zhou, H., Li, J., Fu, X., et al. (2023). Pheromone effect of estradiol regulates the conjugative transfer of pCF10 carrying antibiotic resistance genes. J. Hazard. Mater. 451:131087. doi: 10.1016/j.jhazmat.2023.131087
Yang, Y., Yang, X., Zhou, H., Niu, Y., Li, J., Fu, X., et al. (2022). Bisphenols promote the pheromone-responsive plasmid-mediated conjugative transfer of antibiotic resistance genes in Enterococcus faecalis. Environ. Sci. Technol. 56, 17653–17662. doi: 10.1021/acs.est.2c05349
Ye, M. S., Chen, H. L., Liu, C. X., Ren, A. J., Yang, H. W., and Wang, S. S. (2022). Caspofungin at sub-inhibitory concentration promotes the formation of Candida albicans persister cells. J. Appl. Microbiol. 133, 2466–2473. doi: 10.1111/jam.15718
Yee, R., Yuan, Y., Tarff, A., Brayton, C., Gour, N., Feng, J., et al. (2022). Eradication of Staphylococcus aureus biofilm infection by Persister drug combination. Antibiotics 11:1278. doi: 10.3390/antibiotics11101278
Zarkan, A., Matuszewska, M., Trigg, S. B., Zhang, M., Belgami, D., Croft, C., et al. (2020). Inhibition of indole production increases the activity of quinolone antibiotics against E. coli persisters. Sci. Rep. 10:11742. doi: 10.1038/s41598-020-68693-w
Zhang, Y. (2014). Persisters, persistent infections and the yin-Yang model. Emerg Microbes Infect 3, e3–e10. doi: 10.1038/emi.2014.3
Zhang, S., Qu, X., Jiao, J., Tang, H., Wang, M., Wang, Y., et al. (2022). Felodipine enhances aminoglycosides efficacy against implant infections caused by methicillin-resistant Staphylococcus aureus, persisters and biofilms. Bioactive Materials 14, 272–289. doi: 10.1016/j.bioactmat.2021.11.019
Keywords: persister, Enterococcus faecalis, pheromone cCF10, biofilm, (p)ppGpp
Citation: Zhu L, Yang X, Fu X, Yang P, Lin X, Wang F, Shen Z, Wang J, Sun F and Qiu Z (2024) Pheromone cCF10 inhibits the antibiotic persistence of Enterococcus faecalis by modulating energy metabolism. Front. Microbiol. 15:1408701. doi: 10.3389/fmicb.2024.1408701
Edited by:
You-Hee Cho, CHA University, Republic of KoreaReviewed by:
Amrita Panda, Kumaun University, IndiaLynn Ernest Hancock, University of Kansas, United States
Nikolaos Strepis, Erasmus Medical Center, Netherlands
Copyright © 2024 Zhu, Yang, Fu, Yang, Lin, Wang, Shen, Wang, Sun and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Feilong Sun, c3VuZmVpbG9uZ0B4cHUuZWR1LmNu; Zhigang Qiu, emhpZ2FuZ3FpdTk5QGdtYWlsLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Li Zhu1,2†
Li Zhu1,2† Xiaobo Yang
Xiaobo Yang Zhiqiang Shen
Zhiqiang Shen Jingfeng Wang
Jingfeng Wang Feilong Sun
Feilong Sun Zhigang Qiu
Zhigang Qiu