
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 12 June 2024
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1407324
 Liu-dan Liang1,2,3,4
Liu-dan Liang1,2,3,4 Sheng Li2,5
Sheng Li2,5 Mei-jin Huang2,3
Mei-jin Huang2,3 Hui-xin Peng2,6
Hui-xin Peng2,6 Zi-jun Lu2,5
Zi-jun Lu2,5 Zhuo-hua Zhang1,2,3
Zhuo-hua Zhang1,2,3 Li-ye Su2,5
Li-ye Su2,5 Suren R. Sooranna6,7*
Suren R. Sooranna6,7* Yan Liu2,4*†
Yan Liu2,4*† Zhao-he Huang1,2,5*†
Zhao-he Huang1,2,5*†Background: Some recent observational studies have shown that gut microbiota composition is associated with puerperal sepsis (PS) and no causal effect have been attributed to this. The aim of this study was to determine a causal association between gut microbiota and PS by using a two-sample Mendelian randomization (MR) analysis.
Methods: This study performed MR analysis on the publicly accessible genome-wide association study (GWAS) summary level data in order to explore the causal effects between gut microbiota and PS. Gut microbiota GWAS (n = 18,340) were obtained from the MiBioGen study and GWAS-summary-level data for PS were obtained from the UK Biobank (PS, 3,940 cases; controls, 202,267 cases). Identification of single nucleotide polymorphisms associated with each feature were identified based on a significance threshold of p < 1.0 × 10–5. The inverse variance weighted (IVW) parameter was used as the primary method for MR and it was supplemented by other methods. Additionally, a set of sensitivity analytical methods, including the MR-Egger intercept, Mendelian randomized polymorphism residual and outlier, Cochran’s Q and the leave-one-out tests were carried out to assess the robustness of our findings.
Results: Our study found 3 species of gut microbiota, Lachnospiraceae FCS020, Lachnospiraceae NK4A136, and Ruminococcaceae NK4A214, to be associated with PS. The IVW method indicated an approximately 19% decreased risk of PS per standard deviation increase with Lachnospiraceae FCS020 (OR = 0.81; 95% CI 0.66–1.00, p = 0.047). A similar trend was also found with Lachnospiraceae NK4A136 (OR = 0.80; 95% CI 0.66–0.97, p = 0.024). However, Ruminococcaceae NK4A214 was positively associated with the risk of PS (OR = 1.33, 95% CI: 1.07–1.67, p = 0.011).
Conclusion: This two-sample MR study firstly found suggestive evidence of beneficial and detrimental causal associations of gut microbiota on the risk of PS. This may provide valuable insights into the pathogenesis of microbiota-mediated PS and potential strategies for its prevention and treatment.
Maternal sepsis is a life-threatening condition that leads to significant maternal morbidity and mortality, posing a tremendous socio-economic and healthcare burden worldwide. Data from the World Health Organization estimate that sepsis is responsible for 11.0–30.9% of total maternal deaths, ranking fifth among the causes of maternal mortality, with puerperal sepsis (PS) accounting for a significant portion of this burden (Say et al., 2014; Chebbo et al., 2016; Ngonzi et al., 2016; Padilla and Palanisamy, 2017; Parfitt and Hering, 2018; Plante et al., 2019). Hensley et al. (2019) analyzed data from 57.8% of the U.S. population and found that the incidence rate of maternal sepsis was 0.04%. In recent years, the incidence of maternal sepsis have been increasing. Kendle et al. (2019) found an annual increase of 6.6% in the incidence rate of delivery-associated sepsis in the United States, as well as an elevated fatality rate (20.6%) (Kuriya et al., 2016; Creanga, 2018). Furthermore, PS also increased the risk of postpartum anxiety and depression in mothers, and this not only interfered with family bonding but it also had a negative impact on breastfeeding of newborn infants (Groer and Morgan, 2007; Best et al., 2019). Hence, there is an urgent need to identify therapeutic as well as preventive targets for PS.
As the largest reservoir of bacteria and endotoxins in the body, the intestine is regarded as the “engine” of sepsis and multiple organ dysfunction syndrome (MODS), as well as participating in the metabolism of matter and energy (Anand and Mande, 2022). Under normal circumstances, the gut microbiota plays a beneficial role in human physiological processes and immune homeostasis, which is crucial for maintaining human health (O'Hara and Shanahan, 2006; Clarke et al., 2014). Gut microbiota dysbiosis has been demonstrated to increase the susceptibility to sepsis and these microorganisms may be a significant conductor for the pathogenesis of sepsis (Yang et al., 2021). In particular, in critically ill patients, it has been shown that the homeostasis of gut microbiota was disrupted, leading to abnormal changes in the type, number, proportion and location of gut microbes (McDonald et al., 2016). The interaction between disease and gut microbiota disorders can lead to clinical deterioration and the development of MODS (Adelman et al., 2020). During pregnancy, the gut microbiota undergoes profound changes, and imbalances in the gut microbiota composition are often associated with adverse pregnancy outcomes (Qi et al., 2021). There is increasing evidence supporting the relationship between gut microbiome dysbiosis and PS (Winther et al., 2020). Pregnancy hormones, the immune system and gut microbiome can interact with each other and changes in bacterial composition may be conducive to maintaining homeostasis and adapting to the physiological changes that accompanies pregnancy. These changes can also make women more vulnerable during pregnancy and postpartum, especially with respect to infectious diseases (Fuhler, 2020). Therefore, a better understanding of the changes in maternal gut microbiota may help to control the risk of PS.
Randomized controlled trials (RCTs) are an ideal method for inferring causality in epidemiological studies, but due to ethical limitations, the practice of RCTs remains challenging (Hariton and Locascio, 2018). Mendelian randomization (MR) is an alternative method which can effectively apply the data obtained from existing genome-wide association studies (GWAS) and we can use the genetic variation as an instrumental variable (IV) to explore the causal relationship between risk factors and outcomes (Hemani et al., 2018). According to Mendelian laws of inheritance, parental alleles are randomly assigned to offspring, a process equivalent to randomization in RCTs. In theory, genetic variations are not affected by common confounding factors such as the environment after birth. In addition, genetic variations predate exposures and outcomes, ruling out the problem of reverse causation and therefore, their use as IVs to analyze the causal relationship between exposures and outcomes is suitable for epidemiological studies.
At present, the role of gut microbiota in the risk of PS has not been evaluated within the MR framework. Therefore, this study is the first attempt to employ a two-sample MR approach to explore the causal relationship between genetically predicted gut microbiota and PS risk using aggregated statistics from GWAS obtained data.
The two-sample MR study design enables the estimation of the causal impacts of the exposure on the outcome by utilizing GWAS summary statistics from two distinct studies. The flowchart of our study is outlined in Figure 1. This approach relies on three core assumptions: (1) The genetic instruments should exhibit a robust correlation with the exposure, (2) the influence of the genetic instruments on the outcome should be solely mediated through the exposure and (3) the genetic instruments must be unrelated to any known factors that may confound the association between the exposure and the outcome. All methods carried out in this study were in accordance with relevant guidelines and regulations. This study has been approved by the Ethics Committee of the Affiliated Hospital of Youjiang Medical University for Nationalities (No. YFY-LL-2023-169).
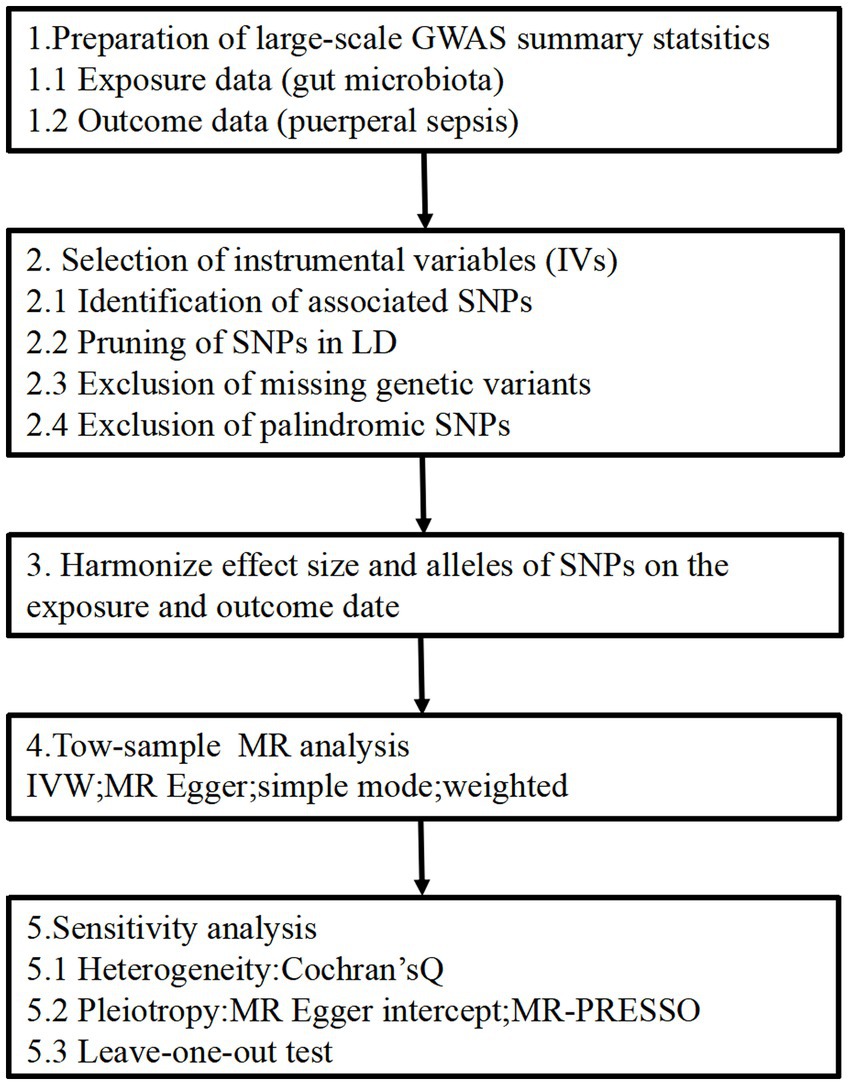
Figure 1. Study design of the MR study on the associations of gut microbiota and puerperal sepsis. SNPs, single nucleotide polymorphisms; LD, linkage disequilibrium; IVW, inverse variance weighted; MR, Mendelian randomization; MR-PRESSO, Mendelian randomized polymorphism residual and outlier.
The genetic summary-level data for gut microbiota was derived from the largest meta-analysis conducted by the MiBioGen consortium, which encompassed 24 cohorts and a total of 18,340 individuals. 72.3% of the participants (n = 13,266) in this study were of European descent (Kurilshikov et al., 2021). This research group targeted three distinct regions of the 16S rRNA gene (specifically V4, V3-V4, and V1-V2) to analyze the microbial composition and the taxonomic classification was conducted through direct taxonomic binning. During the microbiota quantitative trait loci (mbQTL) mapping analysis for each cohort, only the bacterial taxa that were detected in more than 10% of the samples were considered. This selection criterion was applied to evaluate the impact of host genetic variants on the abundance levels of these specific bacterial taxa within the gut microbiota. After excluding 12 unknown genera, a total of 119 others that passed the taxon inclusion cutoffs were included in this study. More detailed descriptions on genetic variants for gut microbiota can be found in the source publication (Kurilshikov et al., 2021).
For the risk of PS, we acquired GWAS summary data from the FinnGen study (R9) (Kurki et al., 2023). This ongoing project, integrates the germline genotype data from Finnish biobanks with health record data obtained from Finnish health registries, providing information on clinically defined outcomes. The dataset included 3,940 cases with PS and 202,267 controls for use in our analysis.
IVs can be used to control for confounding and measurement errors in observational studies because they allow for the possibility of making causal inferences with observed and unobserved data in epidemiological studies. A series of quality control steps was used in the selection of IVs for each feature in this study and these included: (1) Identification of associated single nucleotide polymorphisms (SNPs): SNPs associated with each feature were identified based on a significance threshold of p < 1.0 × 10−5. (2) Pruning of SNPs in linkage disequilibrium (LD): SNPs within a 500-kilobase window and with a LD threshold of r2 < 0.1 were removed. This process was based on the 1,000 Genomes Project reference panel for European populations (Phase 3). Among the remaining SNPs, those with the lowest p-values from the GWAS were retained. (3) Exclusion of missing genetic variants: genetic variants that were not present in the outcome of the GWAS dataset were excluded from the analysis. This was to avoid their replacement by proxy variants. (4) Exclusion of palindromic SNPs, which are SNPs with alleles consisting of a base as well as its complementary base, were excluded. This step ensured that the effects of the SNPs on the exposures corresponded to the same allele as their effects on the outcomes. We estimated the value for each MR test conducted and this represented the proportion of variance in the phenotype explained by the genetic variants. Additionally, we calculated the mean F-statistical value as a measure of the strength of the IV used (Burgess and Thompson, 2011; Papadimitriou et al., 2020; Ni et al., 2021).
All analyses were conducted using the TwoSampleMR package (version 0.5.6) in R, specifically version 4.2.1. Our primary analysis utilized the inverse-variance weighted (IVW) approach, which offers the highest statistical power (Burgess et al., 2016). To address potential violations of MR assumptions resulting from different pleiotropy scenarios, we performed multiple sensitivity analyses, including the MR-Egger, weighted median, simple mode and weighted mode (Hartwig et al., 2017). Previous studies have suggested that relying solely on p-value thresholds to determine “significance” was not ideal (Sterne and Davey, 2001; Li et al., 2017). Therefore, we employed three criteria to establish a reliable causal association: (1) The MR-IVW estimate for each feature should surpassed the p-value threshold, (2) The MR estimates must consistently demonstrate the same direction of effect across all sensitivity analyses, and (3) Limited evidence of horizontal pleiotropy should be observed through either the MR-Egger intercept test or the Mendelian Randomization Pleiotropy Residual Sum and Outlier (MR-PRESSO) global test (Bowden et al., 2015; Verbanck et al., 2018). Furthermore, we also assessed the heterogeneity of IVs using the Cochran’s Q-statistical test and performed a leave-one SNP-out analysis in order to ensure that the results were not biased by any individual SNP (Burgess et al., 2017).
A total 1,269 IVs were screened for 119 bacterial genera from the MiBioGen consortium. These IVs explained 0.34–3.33% of the variance and that the F-statistics for them were all greater than 10 (ranging from 20.46 to 28.66). These parameters signified the robustness and strength of the IVs employed in this study. This indicated the absence of any significant weak instrument bias in the results we obtained, making the findings more reliable and acceptable (Supplementary Table S1).
We identified three species of gut microbiota, Lachnospiraceae FCS020, Lachnospiraceae NK4A136, and Ruminococcaceae NK4A214, related to PS according to the criteria outlined above in which the IVW method demonstrated a significant difference (p < 0.05), and the other methods indicated consistent directional effects. The IVW method indicated an approximately 19% decreased risk of PS per standard deviation (SD) increase in Lachnospiraceae FCS020 (OR = 0.81; 95% CI 0.66–1.00, p = 0.047). A similar trend was also found with Lachnospiraceae NK4A136 (OR = 0.80; 95% CI 0.66–0.97, p = 0.024). However, the IVW method also showed that Ruminococcaceae NK4A214 was positively associated with a risk of PS (OR = 1.33, 95% CI: 1.07–1.67, p = 0.011) (Table 1; Figure 2; Supplementary Table S2).
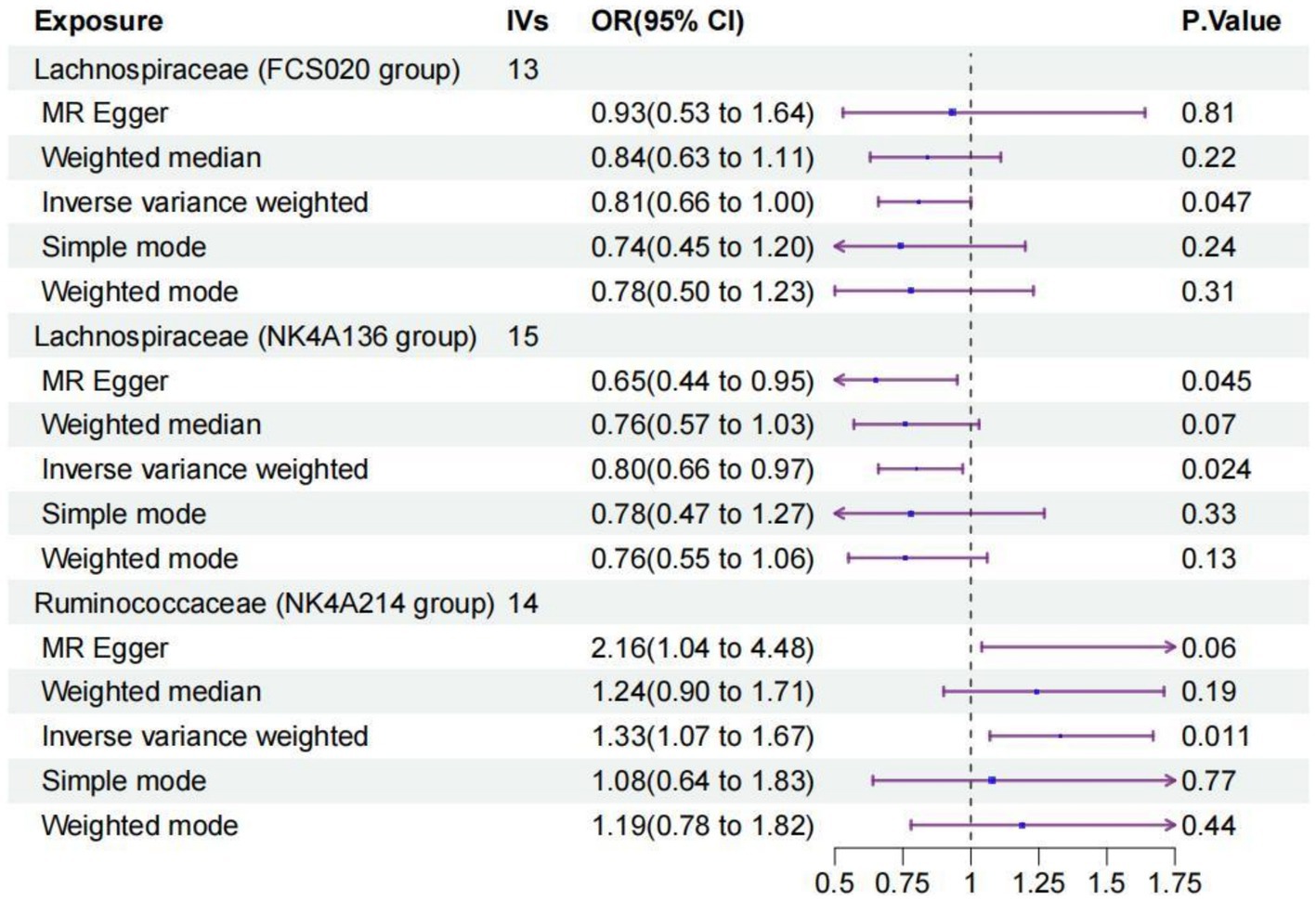
Figure 2. Forest plot of the causal relationships estimated for three gut microbiota and puerperal sepsis by using five MR methods including IVW, MR-Egger, weighted median, simple mode and weighted mode. The blue dots and purple bars represent the MR and the 95% confidence intervals of IMR analysis estimates, respectively. The OR > 1 indicates increased risk while <1 indicates decreased risk. MR, Mendelian randomization; CI, confidence interval; OR, odds ratio; IVW, inverse-variance weighted.
For these three gut microbiota species, no heterogeneity was found by using either the IVW method or the MR-Egger regression, and the p-values for Cochran’s Q test were all >0.05 (Supplementary Table S3). The MR Egger regression intercepts were near zero and these revealed little evidence of horizontal pleiotropy in the IVs (Supplementary Table S4). Using the MR-PRESSO global outlier test, no evidence for outliers was observed (Supplementary Table S5). The scatter plots did not demonstrate any potential outliers in the IVs for these three gut microbiota species (Figure 3). In the leave-one SNP-out analyses, the risk estimates of all gut microbiota genera generally remained consistent after eliminating each SNP at a time, suggesting that these were no identified leverage points with high influence (Figure 4). The full results of the MR estimates for the 119 gut microbiota genera on PS are presented in Supplementary Tables S2–S4.
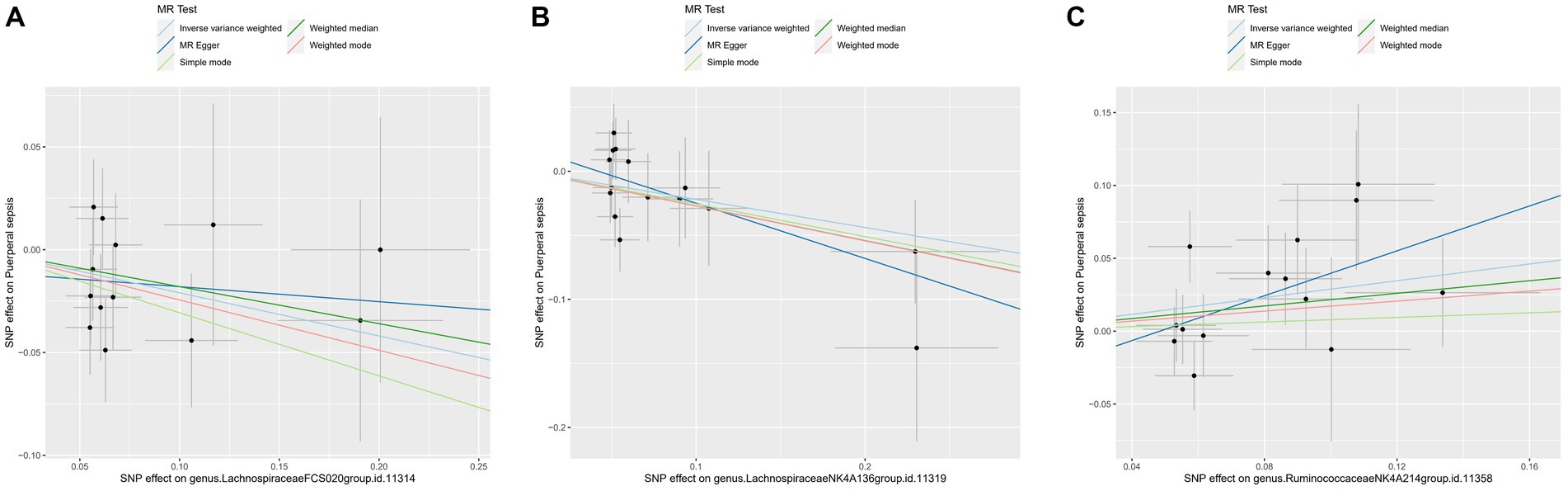
Figure 3. The scatter plot of MR estimates for (A) Lachnospiraceae FCS020, (B) Lachnospiraceae NK4A136 and (C) Ruminococcaceae NK4A214. The light blue, dark blue, light green, dark green, and red lines represent the association between microbiota (exposure) and puerperal sepsis (outcome) when estimated by the IVW method, MR-Egger regression, simple mode, weighted median and weighted mode, respectively. The vertical and horizontal black lines around each point represent the 95% confidence interval for each polymorphism with respect to associations with exposure and outcome, respectively. SNP, single nucleotide polymorphism; MR, Mendelian randomization; IVW, inverse-variance weighted.
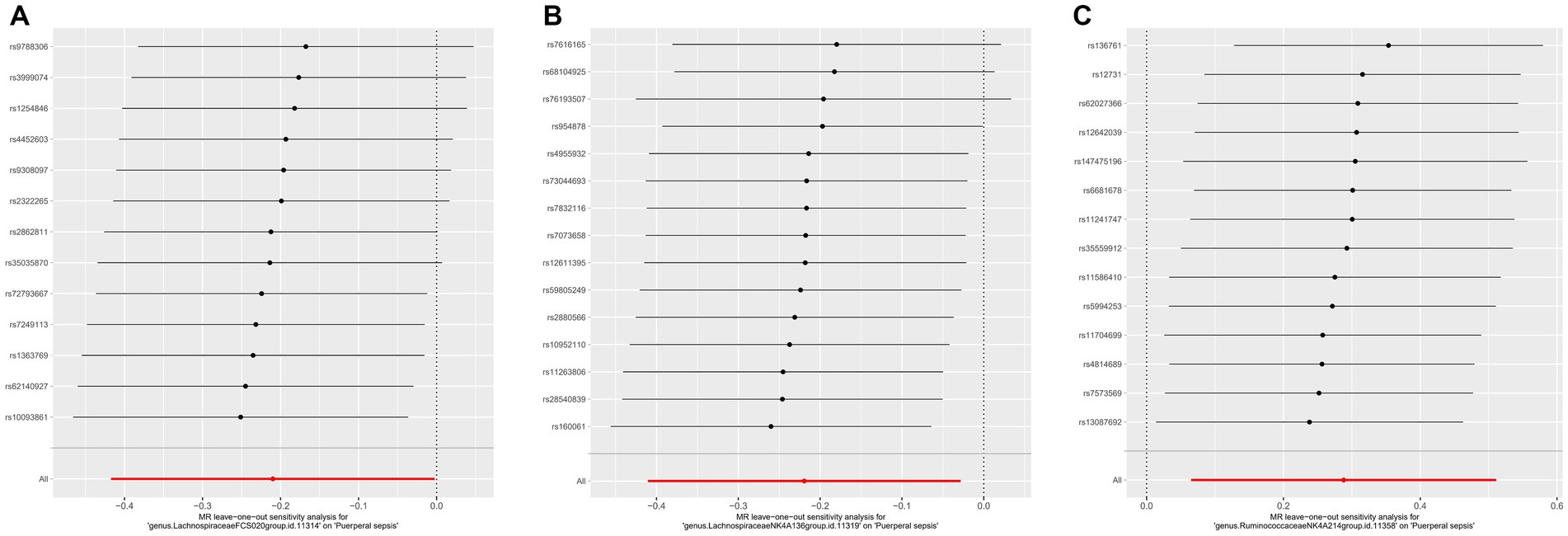
Figure 4. The leave one out analysis of MR estimates for (A) Lachnospiraceae FCS020, (B) Lachnospiraceae NK4A136 and (C) Ruminococcaceae NK4A214. The black dots represent the causal estimate for the association between a specific exposure and target after discarding each SNP in turn. The red dots represent the overall causal estimate by using the random-effect IVW. The horizontal lines denote 95% confidence intervals. SNP, single nucleotide polymorphism; MR, Mendelian randomization; IVW, inverse-variance weighted.
In this study, we performed a two-sample MR analysis in order to evaluate the causal relationship between gut microbiota and PS. We used the summary statistics on gut microbiota from the largest GWAS meta-analysis conducted by the MiBioGen Consortium and the summary statistics on PS from the FinnGen Consortium R9 published data. We found a causal relationship between the risk of PS and three species of gut microbiota, namely Lachnospiraceae FCS020, Lachnospiraceae NK4A136, and Ruminococcaceae NK4A214. The increased abundance of Lachnospiraceae FCS020 and Lachnospiraceae NK4A136 was negatively associated with the risk of PS, while Ruminococcaceae NK4A214 appeared to be a risk factor for the disease.
The human intestine has a complex ecosystem of bacteria, accounting for about 80% of the total microbiota. Human gut microbiota consists of numerous microorganisms, but the most abundant species belong to the phyla, Firmicutes (synonym Bacillota) and Bacteroidetes (synonym Bacteroidota) (Ramakrishna, 2013; Akash et al., 2019; Firmino et al., 2020), fulfilling an irreplaceable role in maintaining host health. Firmicutes possess anti-inflammatory properties and can alleviate the progression of inflammatory bowel disease. Bacteroidetes perform metabolic conversions such as degradation of proteins and complex sugar polymers which are essential for the host. The perinatal period is a unique period during which a woman’s physiological status (including hormone levels, metabolism and immunity) undergoes significant alternations (Newbern and Freemark, 2011). As hormones can impact bacterial growth, changes in hormonal levels may affect gut microbiota during pregnancy (Neuman and Koren, 2017). Moreover, substantial changes in host hormones levels, metabolism and immunity before and after delivery may be accompanied by changes in gut microbiota (Salzman et al., 2010; Vijay-Kumar et al., 2010). The gut microbiome during pregnancy is characterized by a decrease in abundance and an increase in inter-subject diversity. The relative abundance of Actinobacteria, Proteobacteria, and Coprobacilli increases on average from the first to the third trimesters of pregnancy, and these effects persist until the perinatal period (Koren et al., 2012). No substantial differences in maternal gut microbiome have been found when comparing the third trimester with the first few months postpartum (Koren et al., 2012; Jost et al., 2014).
Sepsis is defined as a life-threatening organ dysfunction caused by a dysfunctional host response to infection (Singer et al., 2016). An increasing body of evidence now suggests that gut microbiota play an important role in the pathophysiology of sepsis and dysbiosis of these organisms been associated with numerous sepsis-associated pathogenic conditions (Dickson, 2016; Klingensmith and Coopersmith, 2016). The spectrum of gut microbiota can also potentially be used as a diagnostic and prognostic marker for sepsis (Liu et al., 2021). Specific physiological changes during pregnancy can alter the gut microbiome, thus increasing the risk of sepsis-like infections.
Our study identified 3 specific bacterial species having a causal relationship with the risk of PS. Among them, Lachnospiraceae FCS020 and Lachnospiraceae NK4A136 are important members of Lachnospiraceae. Lachnospiraceae and Ruminococcaceae belong to the phylum Firmicutes and are the most common bacterial groups found in the human gut. The bacteria which belong to the Lachnospiraceae family are anaerobic and they are abundant in a healthy gut. They can produce short-chain-fatty acids (SCFA) (Vacca et al., 2020) by converting primary into secondary bile acids. SCFA can enhance the integrity of the intestinal epithelial barrier and inhibit inflammation, which is important for maintaining the gastro-intestinal health (Hu et al., 2019). Sorbara et al. (2020) found a protective effect of Lachnospiraceae on metabolic health during sepsis. Consistent with these findings, we found an increased abundance of Lachnospiraceae FCS020 and Lachnospiraceae NK4A136 can reduce the risk of PS. However, Muratsu et al. (2022) showed that the Lachnospiraceae family increased in the subacute stage (day 7 after caecal ligation and puncture) in a mouse model of sepsis (Muratsu et al., 2022), which was inconsistent with our findings. A study on the component flux of the gut microbiome and the risk of gram-negative bloodstream infections found that Ruminococcaceae is associated with a reduced risk of gram-negative dominance (Stoma et al., 2021). The proportion of Ruminococcaceae in healthy patients was higher than that in sepsis patients (Antharam et al., 2013). However, we found that Ruminococcaceae NK4A214 may be a risk factor for PS. Ruminococcaceae are known to have a beneficial effect on gut barrier function (Madsen, 2001). In addition, some species are also depleted in Crohn’s disease and ulcerative colitis and they have also been shown to have anti-inflammatory properties (Sokol et al., 2008).
At present, the changes in gut microbiota in PS patients have not been reported. These inconsistent results may be explained by the considerable inter-species and intra-species diversity in the gut microbiota, which may have had a significant impact on PS. Standardized and specific gut microbiota classification systems are essential for subsequent mechanistic studies and clinical guidance. Other studies that have explored the role of gut microbiota in sepsis and other related conditions. For example, a recent study looked at the correlation between gut microbiota, C- reactive protein and sepsis, and suggested that this protein has a potential role as a mediator in facilitating the impact of some bacterial species on sepsis (Zhang et al., 2023). Microbiota may also cause an imbalance in the lung and intestinal immunity by causing bacterial translocation through the TLR4 and NF-kB pathway resulting in inflammation of lung tissues (Sun et al., 2022).
The changes observed in the levels of Lachnospiraceae FCS020, Lachnospiraceae NK4A136, and Ruminococcaceae NK4A214 in this study with PS may potentially involve interactions with the microbiomes from other epithelial sites of the body, which are known to impact human physiology, both in health and in disease (Ogunrinola et al., 2020). In particular, the microbiome directly associated with the reproductive tract (Rizzo et al., 2021), urinary tract (Kim and Lee, 2023) and skin (Byrd et al., 2018) can have more direct and localized effects on specific diseases. Lactobacilli dominate the microbiome of the human reproductive tract and the maintenance of a healthy microbiota can protect the host from pathogens, increase reproductive potential and reduce the rates of adverse pregnancy outcomes (Rizzo et al., 2021; Wang et al., 2024). These types of studies suggest that our observed changes may be due to modifications in the gut microbiome as it tries to either overcome or ameliorate the onset of PS. However, further speculations are beyond the scope of this study.
The present study has several advantages. Firstly, the causal relationship between gut microbiota and PS was determined by MR analysis, thereby eliminating confounding factors and avoiding the effect of reverse causation on possible causal inference. Secondly, the genetic variation in the gut microbiota was obtained from the largest available GWAS meta-analysis, ensuring the strength of the IVs in the MR analysis. In addition, we detected and corrected for the effect of horizontal pleiotropy by using MR-Egger regression and MR-PRESSO analyses. However, our study also had some limitations. Firstly, the SNPs based on the genome-wide statistical significance threshold (5 × 10−8) were too limited, so we included a genome-wide significance level (1 × 10−5) for this study. Secondly, the GWAS data in this study were mainly from European populations, with only a small portion of gut microbiome data from other races, thus potentially leading to partial bias. In addition, our study was based on the 16S rRNA gene and the broad classification of PS and these are accompanied by their own limitations until more detailed GWAS data become available. Thirdly, we could not rule out possible diet-gene and gene–environment interactions in our analysis and these may have influenced the observed results we obtained. We would also like to perform longitudinal observational studies in future in order to provide insights on how changes in gut microbiota over time correlate with the incidence of PS. The goal of these studies will lead to clinical trials on modifying gut microbiota (e.g., through probiotics and/or diet) to directly test the effectiveness of such interventions in future prevention and management of PS.
In this study we used two large publicly accessible genomic databases to study the link gut microbes and a type of sepsis. Based on the statistical methods used, we found evidence for some beneficial as well as detrimental effects of gut microbiota on the risk of PS. This may provide insights on the pathogenesis of microbe-mediated PS and lead to potential prevention and remedies for this condition.
This two-sample MR study found a causal relationship between gut microbiota and PS, providing ideas for further research on the pathogenesis of this disease. In addition, the data suggest that PS may be mediated by gut microbiota. This study provides valuable insights into the pathogenesis of microbiota-mediated PS and suggestions for potential novel targets for prevention and treatment strategies of this condition. Further RCTs are needed in order to further elucidate the role of gut microbiota in PS and the specific mechanisms involved.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
This study was conducted after approval by the Affiliated Hospital of Youjiang Medical University for Nationalities (No. YFY-LL-2023-169). All the methods used were carried out in accordance with relevant guidelines and regulations.
L-dL: Conceptualization, Data curation, Formal analysis, Writing – original draft. SL: Formal analysis, Writing – original draft. M-jH: Formal analysis, Writing – original draft. H-xP: Formal analysis, Writing – original draft. Z-jL: Formal analysis, Writing – original draft. Z-hZ: Formal analysis, Writing – original draft. L-yS: Formal analysis, Writing – original draft. SS: Conceptualization, Writing – review & editing. YL: Conceptualization, Formal analysis, Funding acquisition, Methodology, Writing – review & editing. Z-hH: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Fund (No. 82260883), the 2020 Guangxi Science and Technology Plan project (No. 2020GXNSFAA297102), High-level talents of the Affiliated Hospital of Youjiang Medical College for Nationalities (No. R20212603), and the project of Guangxi Zhuang Autonomous Region Health Commission (Nos. GXZYL20220312 and Z20201440).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1407324/full#supplementary-material
Adelman, M. W., Woodworth, M. H., Langelier, C., Busch, L. M., Kempker, J. A., Kraft, C. S., et al. (2020). The gut microbiome's role in the development, maintenance, and outcomes of sepsis. Crit. Care 24:278. doi: 10.1186/s13054-020-02989-1
Akash, M. S. H., Fiayyaz, F., Rehman, K., Sabir, S., and Rasool, M. H. (2019). Gut microbiota and metabolic disorders: advances in therapeutic interventions. Crit. Rev. Immunol. 39, 223–237. doi: 10.1615/CritRevImmunol.2019030614
Anand, S., and Mande, S. S. (2022). Host-microbiome interactions: gut-liver axis and its connection with other organs. NPJ Biofilms Microbiomes 8:89. doi: 10.1038/s41522-022-00352-6
Antharam, V. C., Li, E. C., Ishmael, A., Sharma, A., Mai, V., Rand, K. H., et al. (2013). Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51, 2884–2892. doi: 10.1128/JCM.00845-13
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Best, E., Parnell, P., Couturier, J., Barbut, F., le Bozec, A., Arnoldo, L., et al. (2019). Corrigendum to “environmental contamination by bacteria in hospital washrooms according to hand-drying method: a multi-centre study”. J. Hosp. Infect. 101:487. doi: 10.1016/j.jhin.2019.01.014
Bowden, J., Davey Smith, G., and Burgess, S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080
Burgess, S., Bowden, J., Fall, T., Ingelsson, E., and Thompson, S. G. (2017). Sensitivity analyses for robust causal inference from Mendelian randomization analyses with multiple genetic variants. Epidemiology 28, 30–42. doi: 10.1097/EDE.0000000000000559
Burgess, S., Dudbridge, F., and Thompson, S. G. (2016). Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat. Med. 35, 1880–1906. doi: 10.1002/sim.6835
Burgess, S., and Thompson, S. G. (2011). Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 40, 755–764. doi: 10.1093/ije/dyr036
Byrd, A. L., Belkaid, Y., and Segre, J. A. (2018). The human skin microbiome. Nat. Rev. Microbiol. 16, 143–155. doi: 10.1038/nrmicro.2017.157
Chebbo, A., Tan, S., Kassis, C., Tamura, L., and Carlson, R. W. (2016). Maternal Sepsis and septic shock. Crit. Care Clin. 32, 119–135. doi: 10.1016/j.ccc.2015.08.010
Clarke, G., Stilling, R. M., Kennedy, P. J., Stanton, C., Cryan, J. F., and Dinan, T. G. (2014). Minireview: gut microbiota: the neglected endocrine organ. Mol. Endocrinol. (Baltimore, MD) 28, 1221–1238. doi: 10.1210/me.2014-1108
Creanga, A. A. (2018). Maternal mortality in the United States: a review of contemporary data and their limitations. Clin. Obstet. Gynecol. 61, 296–306. doi: 10.1097/GRF.0000000000000362
Dickson, R. P. (2016). The microbiome and critical illness. Lancet Respir. Med. 315, 801–872. doi: 10.1016/S2213-2600(15)00427-0
Firmino, F. C., Porcellato, D., Cox, M., Suen, G., Broadbent, J. R., and Steele, J. L. (2020). Characterization of microbial communities in ethanol biorefineries. J. Ind. Microbiol. Biotechnol. 47, 183–195. doi: 10.1007/s10295-019-02254-7
Fuhler, G. M. (2020). The immune system and microbiome in pregnancy. Best Pract. Res. Clin. Gastroenterol. 44-45:101671. doi: 10.1016/j.bpg.2020.101671
Groer, M. W., and Morgan, K. (2007). Immune, health and endocrine characteristics of depressed postpartum mothers. Psychoneuroendocrinology 32, 133–139. doi: 10.1016/j.psyneuen.2006.11.007
Hariton, E., and Locascio, J. J. (2018). Randomised controlled trials - the gold standard for effectiveness research: study design: randomised controlled trials. BJOG 125:1716. doi: 10.1111/1471-0528.15199
Hartwig, F. P., Davey Smith, G., and Bowden, J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998. doi: 10.1093/ije/dyx102
Hemani, G., Zheng, J., Elsworth, B., Wade, K. H., Haberland, V., Baird, D., et al. (2018). The MR-base platform supports systematic causal inference across the human phenome. eLife 7:e34408. doi: 10.7554/eLife.34408
Hensley, M. K., Bauer, M. E., Admon, L. K., and Prescott, H. C. (2019). Incidence of maternal Sepsis and Sepsis-related maternal deaths in the United States. JAMA 322, 890–892. doi: 10.1001/jama.2019.9818
Hu, S., Wang, J., Xu, Y., Yang, H., Wang, J., Xue, C., et al. (2019). Anti-inflammation effects of fucosylated chondroitin sulphate from Acaudina molpadioides by altering gut microbiota in obese mice. Food Funct. 10, 1736–1746. doi: 10.1039/c8fo02364f
Jost, T., Lacroix, C., Braegger, C., and Chassard, C. (2014). Stability of the maternal gut microbiota during late pregnancy and early lactation. Curr. Microbiol. 68, 419–427. doi: 10.1007/s00284-013-0491-6
Kendle, A. M., Salemi, J. L., Tanner, J. P., and Louis, J. M. (2019). Delivery-associated sepsis: trends in prevalence and mortality. Am. J. Obstet. Gynecol. 220, 391.e1–391.e16. doi: 10.1016/j.ajog.2019.02.002
Kim, D. S., and Lee, J. W. (2023). Urinary tract infection and microbiome. Diagnostics (Basel) 13:1921. doi: 10.3390/diagnostics13111921
Klingensmith, N. J., and Coopersmith, C. M. (2016). The gut as the Motor of Multiple Organ Dysfunction in critical illness. Crit. Care Clin. 32, 203–212. doi: 10.1016/j.ccc.2015.11.004
Koren, O., Goodrich, J. K., Cullender, T. C., Spor, A., Laitinen, K., Kling Bäckhed, H., et al. (2012). Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell 150, 470–480. doi: 10.1016/j.cell.2012.07.008
Kurilshikov, A., Medina-Gomez, C., Bacigalupe, R., Radjabzadeh, D., Wang, J., Demirkan, A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1
Kuriya, A., Piedimonte, S., Spence, A. R., Czuzoj-Shulman, N., Kezouh, A., and Abenhaim, H. A. (2016). Incidence and causes of maternal mortality in the USA. J. Obstet. Gynaecol. Res. 42, 661–668. doi: 10.1111/jog.12954
Kurki, M. I., Karjalainen, J., Palta, P., Sipilä, T. P., Kristiansson, K., Donner, K. M., et al. (2023). FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518. doi: 10.1038/s41586-022-05473-8
Li, X., Meng, X., Timofeeva, M., Tzoulaki, I., Tsilidis, K. K., Ioannidis, P. A., et al. (2017). Serum uric acid levels and multiple health outcomes: umbrella review of evidence from observational studies, randomised controlled trials, and Mendelian randomisation studies. BMJ (Clinical Research ed) 357:j2376. doi: 10.1136/bmj.j2376
Liu, J., Wang, M., Chen, W., Ma, J., Peng, Y., Zhang, M., et al. (2021). Altered gut microbiota taxonomic compositions of patients with Sepsis in a pediatric intensive care unit. Front. Pediatr. 9:645060. doi: 10.3389/fped.2021.645060
Madsen, K. L. (2001). Inflammatory bowel disease: lessons from the IL-10 gene-deficient mouse. Clin. Invest. Med. 24, 250–257.
McDonald, D., Ackermann, G., Khailova, L., Baird, C., Heyland, D., Kozar, R., et al. (2016). Extreme dysbiosis of the microbiome in critical illness. mSphere 1:e00199-16. doi: 10.1128/mSphere.00199-16
Muratsu, A., Ikeda, M., Shimizu, K., Kameoka, S., Motooka, D., Nakamura, S., et al. (2022). Dynamic change of fecal microbiota and metabolomics in a polymicrobial murine sepsis model. Acute Med. Surg. 9:e770. doi: 10.1002/ams2.770
Neuman, H., and Koren, O. (2017). The pregnancy microbiome. Nestle Nutr. Inst. Workshop Ser. 88, 1–9. doi: 10.1159/000455207
Newbern, D., and Freemark, M. (2011). Placental hormones and the control of maternal metabolism and fetal growth. Curr. Opin. Endocrinol. Diabetes Obes. 18, 409–416. doi: 10.1097/MED.0b013e32834c800d
Ngonzi, J., Tornes, Y. F., Mukasa, P. K., Salongo, W., Kabakyenga, J., Sezalio, M., et al. (2016). Puerperal sepsis, the leading cause of maternal deaths at a tertiary university teaching Hospital in Uganda. BMC Pregnancy Childbirth 16:207. doi: 10.1186/s12884-016-0986-9
Ni, J. J., Xu, Q., Yan, S. S., Han, B. X., Zhang, H., Wei, X. T., et al. (2021). Gut microbiota and psychiatric disorders: a two-sample Mendelian randomization study. Front. Microbiol. 12:737197. doi: 10.3389/fmicb.2021.737197
Ogunrinola, G. A., Oyewale, J. O., Oshamika, O. O., and Olasehinde, G. I. (2020). The human microbiome and its impacts on health. Int. J. Microbiol. 2020, 8045646–8045647. doi: 10.1155/2020/8045646
O'Hara, A. M., and Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. doi: 10.1038/sj.embor.7400731
Padilla, C., and Palanisamy, A. (2017). Managing maternal Sepsis: early warning criteria to ECMO. Clin. Obstet. Gynecol. 60, 418–424. doi: 10.1097/GRF.0000000000000269
Papadimitriou, N., Dimou, N., Tsilidis, K. K., Banbury, B., Martin, R. M., Lewis, S. J., et al. (2020). Physical activity and risks of breast and colorectal cancer: a Mendelian randomisation analysis. Nat. Commun. 11:597. doi: 10.1038/s41467-020-14389-8
Parfitt, S. E., and Hering, S. L. (2018). Recognition and Management of Sepsis in the obstetric patient. AACN Adv Crit Care 29, 303–315. doi: 10.4037/aacnacc2018171
Plante, L. A., Pacheco, L. D., and Louis, J. M. (2019). SMFM consult series #47: Sepsis during pregnancy and the puerperium. Am. J. Obstet. Gynecol. 220, B2–b10. doi: 10.1016/j.ajog.2019.01.216
Qi, X., Yun, C., Pang, Y., and Qiao, J. (2021). The impact of the gut microbiota on the reproductive and metabolic endocrine system. Gut Microbes 13, 1–21. doi: 10.1080/19490976.2021.1894070
Ramakrishna, B. S. (2013). Role of the gut microbiota in human nutrition and metabolism. J. Gastroenterol. Hepatol. 28, 9–17. doi: 10.1111/jgh.12294
Rizzo, A. E., Gordon, J. C., Berard, A. R., Burgener, A. D., and Avril, S. (2021). The female reproductive tract microbiome-implications for gynecologic cancers and personalized medicine. J. Pers. Med. 11:546. doi: 10.3390/jpm11060546
Salzman, N. H., Hung, K., Haribhai, D., Chu, H., Karlsson-Sjöberg, J., Amir, E., et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11, 76–82. doi: 10.1038/ni.1825
Say, L., Chou, D., Gemmill, A., Tunçalp, Ö., Moller, A. B., Daniels, J., et al. (2014). Global causes of maternal death: a WHO systematic analysis. Lancet Glob. Health 2, e323–e333. doi: 10.1016/S2214-109X(14)70227-X
Singer, M., Deutschman, C. S., Seymour, C. W., Shankar-Hari, M., Annane, D., Bauer, M., et al. (2016). The third international consensus definitions for Sepsis and septic shock (Sepsis-3). JAMA 315, 801–810. doi: 10.1001/jama.2016.0287
Sokol, H., Lay, C., Seksik, P., and Tannock, G. W. (2008). Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm. Bowel Dis. 14, 858–867. doi: 10.1002/ibd.20392
Sorbara, M. T., Littmann, E. R., Fontana, E., Moody, T. U., Kohout, C. E., Gjonbalaj, M., et al. (2020). Functional and genomic variation between human-derived isolates of Lachnospiraceae reveals inter- and intra-species diversity. Cell Host Microbe 28, 134–146.e4. doi: 10.1016/j.chom.2020.05.005
Sterne, J. A., and Davey, S. G. (2001). Sifting the evidence-what's wrong with significance tests? BMJ (Clinical Research ed) 322, 226–231. doi: 10.1136/bmj.322.7280.226
Stoma, I., Littmann, E. R., Peled, J. U., Giralt, S., van den Brink, M. R. M., Pamer, E. G., et al. (2021). Compositional flux within the intestinal microbiota and risk for bloodstream infection with gram-negative Bacteria. Clin. Infect. Dis. 73, e4627–e4635. doi: 10.1093/cid/ciaa068
Sun, T., Wang, L., and Zhang, H. (2022). intestinal microbiota in sepsis. Intensive Care Res. 2, 1–7. doi: 10.1007/s44231-022-00001-8
Vacca, M., Celano, G., Calabrese, F. M., Portincasa, P., Gobbetti, M., and De Angelis, M. (2020). The controversial role of human gut Lachnospiraceae. Microorganisms 8:573. doi: 10.3390/microorganisms8040573
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Vijay-Kumar, M., Aitken, J. D., Carvalho, F. A., Cullender, T. C., Mwangi, S., Srinivasan, S., et al. (2010). Metabolic syndrome and altered gut microbiota in mice lacking toll-like receptor 5. Science (New York, N.Y.) 328, 228–231. doi: 10.1126/science.1179721
Wang, N., Chen, L., Yi, K., Zhang, B., Li, C., and Zhou, X. (2024). The effects of microbiota on reproductive health: a review. Crit. Rev. Food Sci. Nutr. 64, 1–22. doi: 10.1080/10408398.2022.2117784
Winther, A., Axelsson, P. B., Clausen, T. D., and Løkkegaard, E. (2020). Prophylactic antibiotics in caesarean delivery before or after cord clamping - protecting the mother at the expense of the infant's microbiota? BJOG 127, 203–206. doi: 10.1111/1471-0528.15960
Yang, X. J., Liu, D., Ren, H. Y., Zhang, X. Y., Zhang, J., and Yang, X. J. (2021). Effects of sepsis and its treatment measures on intestinal flora structure in critical care patients. World J. Gastroenterol. 27, 2376–2393. doi: 10.3748/wjg.v27.i19.2376
Keywords: causal relationship, gut microbiota, Lachnospiraceae, puerperal sepsis, Mendelian randomization, Ruminococcaceae
Citation: Liang L-d, Li S, Huang M-j, Peng H-x, Lu Z-j, Zhang Z-h, Su L-y, Sooranna SR, Liu Y and Huang Z-h (2024) Causal relationship between gut microbiota and puerperal sepsis: a 2-sample Mendelian randomization study. Front. Microbiol. 15:1407324. doi: 10.3389/fmicb.2024.1407324
Received: 26 March 2024; Accepted: 21 May 2024;
Published: 12 June 2024.
Edited by:
Munis Dundar, Erciyes University, TürkiyeReviewed by:
Zhengrui Li, Shanghai Jiao Tong University, ChinaCopyright © 2024 Liang, Li, Huang, Peng, Lu, Zhang, Su, Sooranna, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suren R. Sooranna, cy5zb29yYW5uYUBpbXBlcmlhbC5hYy51aw==; Yan Liu, NjA1MDEyMjAzQHFxLmNvbQ==; Zhao-he Huang, bGxkMjAxOTQ0QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.