- 1Department of Microbiology, University of Delhi, South Campus, New Delhi, India
- 2King Abdullah University of Science and Technology (KAUST), KAUST Catalysis Center, Thuwal, Makkah, Saudi Arabia
- 3School of Life Sciences, University of Nottingham, Queen’s Medical Centre, Nottingham, United Kingdom
- 4Institute for Experimental Molecular Imaging, University Clinic, RWTH Aachen University, Aachen, Germany
Extremophilic proteins are valuable in various fields, but their expression can be challenging in traditional hosts like Escherichia coli due to misfolding and aggregation. Haloferax volcanii (H. volcanii), a halophilic expression system, offers a solution. This study examined cleavable and non-cleavable purification tags at both the N- and C-termini when fused with the superfolder green fluorescent protein (sfGFP) in H. volcanii. Our findings reveal that an N-terminal 8xHis-tag or Strep-tag®II significantly enhances protein production, purity, and yield in H. volcanii. Further experiments with mCherry and halophilic alcohol dehydrogenase (ADH) showed improved expression and purification yields when the 8xHis-tag or Strep-tag®II was positioned at the C-terminus for mCherry and at the N-terminus for ADH. Co-positioning 8xHis-tag and Twin-Strep-tag® at the N-terminus of sfGFP, mCherry, and ADH yielded significantly enhanced results. These findings highlight the importance of thoughtful purification tag design and selection in H. volcanii, providing valuable insights for improving protein production and purification with the potential to advance biotechnological applications.
Introduction
Extremophiles thrive in extreme physical or geochemical conditions that are inhospitable to most life forms. These environments encompass extreme temperatures, significant variations in salinity, high pressures, the presence of heavy metals, and drastic pH levels (Sysoev et al., 2021). Extremophilic proteins hold a unique position in biotechnology due to their exceptional stability under harsh environmental conditions (Wang et al., 2024). They contribute to sustainable biotechnology on Earth and offer valuable insights into the potential existence of life on other planets (Allers, 2010; Schultz et al., 2023; Wang et al., 2024). The efficient expression and cost-effective purification of recombinant proteins are paramount in today’s pharmaceutical and biotechnology industries (Puetz and Wurm, 2019). While Escherichia coli (E. coli) expression systems are widely adopted for producing heterologous recombinant proteins, they come with inherent limitations, including a limited capacity for forming disulfide bonds, the inability to perform posttranslational modifications, and the absence of an efficient secretion system (Baeshen et al., 2015; Ibrahim et al., 2023). Extremophilic proteins, which are adapted to extremes of temperature, pH, or salinity, often elude successful expression in mesophilic systems like E. coli due to issues of protein misfolding, conformational stress, and the formation of inclusion bodies (Elleuche et al., 2014; Kruglikov et al., 2022). Consequently, there is a pressing demand for alternative expression systems that can effectively accommodate the expression of extremophilic proteins in their native state.
Haloferax volcanii (H. volcanii) offers a solution to these challenges as an extremophilic protein expression host. H. volcanii is an obligate halophilic archaeon, originally isolated from the Dead Sea, which naturally thrives in extreme conditions (Strillinger et al., 2016; Grötzinger et al., 2018; Akal et al., 2019; Sysoev et al., 2021). H. volcanii has been equipped with a suite of microbiological and molecular genetics techniques, including an efficient DNA transformation system, shuttle plasmids, and selectable markers, making it a versatile platform for genetic studies, proteomics, and biotechnology research (Allers, 2010; Strillinger et al., 2016; Haque et al., 2019; Pérez-Arnaiz et al., 2020; Karan et al., 2023). Moreover, H. volcanii grows, compared to other extreme halophiles, under laboratory culture conditions at high density and in a wide range of temperatures (37–55°C) and salt concentrations (1.8–3.5 M NaCl) (Haque et al., 2019, 2020; Sysoev et al., 2021). Due to the fragile nature of the outer S-layer cell walls, H. volcanii cells easily lyse in hypotonic conditions, such as low salt buffer or water, releasing all cellular proteins (Dyall-Smith, 2008). This allows a simplified downstream protein purification with minimal labor and cost, making H. volcanii attractive for biotechnology use (Grötzinger et al., 2018; Akal et al., 2019; Karan et al., 2020, 2023; Alshehri et al., 2022).
The H. volcanii strain H1895, coupled with the pTA1992 plasmid, is a dedicated host with genetic modifications for enhanced protein overexpression and purification (Haque et al., 2020). Several genetic modifications have been made to H. volcanii strain H1895 to optimize it as an expression host for protein overexpression. Notably, biofilm-forming genes HVO_1033 and HVO_1034 have been intentionally deleted. Additionally, naturally histidine-rich proteins PitA and Cdc48d have been replaced with orthologous genes containing fewer histidine residues (Allers et al., 2010; Strillinger et al., 2016; Haque et al., 2019, 2020). These changes serve a dual purpose: they reduce potential contaminants and enable polyhistidine-tag (His-tag) purification, a common method in protein purification. Moreover, H1895 has proven to be an excellent platform for expressing various enzymes and larger molecular structures (Allers, 2010; Strillinger et al., 2016; Haque et al., 2019, 2020; Karan et al., 2023). Furthermore, the lack of biofilm formation extends its utility to bioreactor applications, further underlining its versatility and potential for biotechnological endeavors (Strillinger et al., 2016; Haque et al., 2020).
Functional protein expression and purification are essential for exploring protein structure, function, and biotechnological applications (Du et al., 2022). Therefore, applying appropriate purification tags that enhance solubility and are cleavable in protein production is crucial. These tags help achieve high expression yields, target protein purity, save time, and reduce production costs (Gomari et al., 2020). However, it is important to note that the construction of fusion proteins can often affect the expression level, solubility, stability, and subsequent purification efficiency of recombinant proteins (Yadav et al., 2016; Köppl et al., 2022; Le et al., 2022). Even in well-studied expression systems like E. coli, there is no universal rule for determining the optimal expression conditions, purification tag types, and positions (Köppl et al., 2022). Regarding haloarchaeal proteins, expression levels are typically low, and the procedures for purifying these proteins can be complex and costly (Martínez-Espinosa, 2019). Systematic studies are missing to investigate the effects of purification tag positions and the influence of commonly used affinity and solubility-enhancing tags on expression, solubility, and purification in haloarchaeal expression systems.
This study systematically investigates the impact of various purification tags on protein expression, solubility, and subsequent purification of H. volcanii expressed proteins. Consequently, fusion constructs using the superfolder green fluorescent protein (sfGFP) and a range of common purification tags (polyhistidine-tag or His-tag, Strep-tag®II, Twin-Strep-tag®, a FLAG-tag, and 3x-FLAG-tag), both at the N- and C-termini, with and without cleavage sites were designed (Skerra and Schmidt, 1999; Einhauer and Jungbauer, 2001; Kimple and Sondek, 2004; Pédelacq et al., 2006; Shahravan et al., 2008; Allers et al., 2010; Hermans et al., 2012; Schmidt et al., 2012, 2013; Young et al., 2012; Beznosov et al., 2013; Kimple et al., 2013; Zhao et al., 2013; Djender et al., 2014; Nguyen et al., 2014; Yeliseev et al., 2017; Mishra, 2020; Köppl et al., 2022). Next, the study also investigates the novel four amino acid short C-tag at the C-terminus of sfGFP to diversify the tag library further. Fluorescence measurements of sfGFP enable rapid and efficient expression, solubility, and purification estimations of these fusion proteins (Pédelacq et al., 2006). Furthermore, a comparison was conducted using 8xHis-tag and Strep-tag®II at both the N- and C-termini with the red fluorescent protein mCherry and a previously fully characterized halophilic alcohol dehydrogenase (ADH) from the deep-sea brine pool of the Red Sea (Shaner et al., 2004; Grötzinger et al., 2018; Akal et al., 2019). Additionally, the dual-affinity-tag approach by combining the 8xHis-tag and the Strep-tag®II at the N-terminus of sfGFP, mCherry, and ADH was explored. These investigations unveiled the substantial impact of different tags, tag lengths, and positions on protein expression and purification. The insights gained from this study hold significant promise in addressing the challenges associated with producing and purifying halophilic proteins in H. volcanii.
Materials and methods
Materials
All chemicals and solvents were purchased from Sigma-Aldrich (St. Louis, MO, United States). Water was desalted and purified using a milli-Q® (Merck, Darmstadt, Germany) system. This study used Haloferax volcanii H1895 and the vector pTA1992 (Haque et al., 2019).
Plasmid preparation
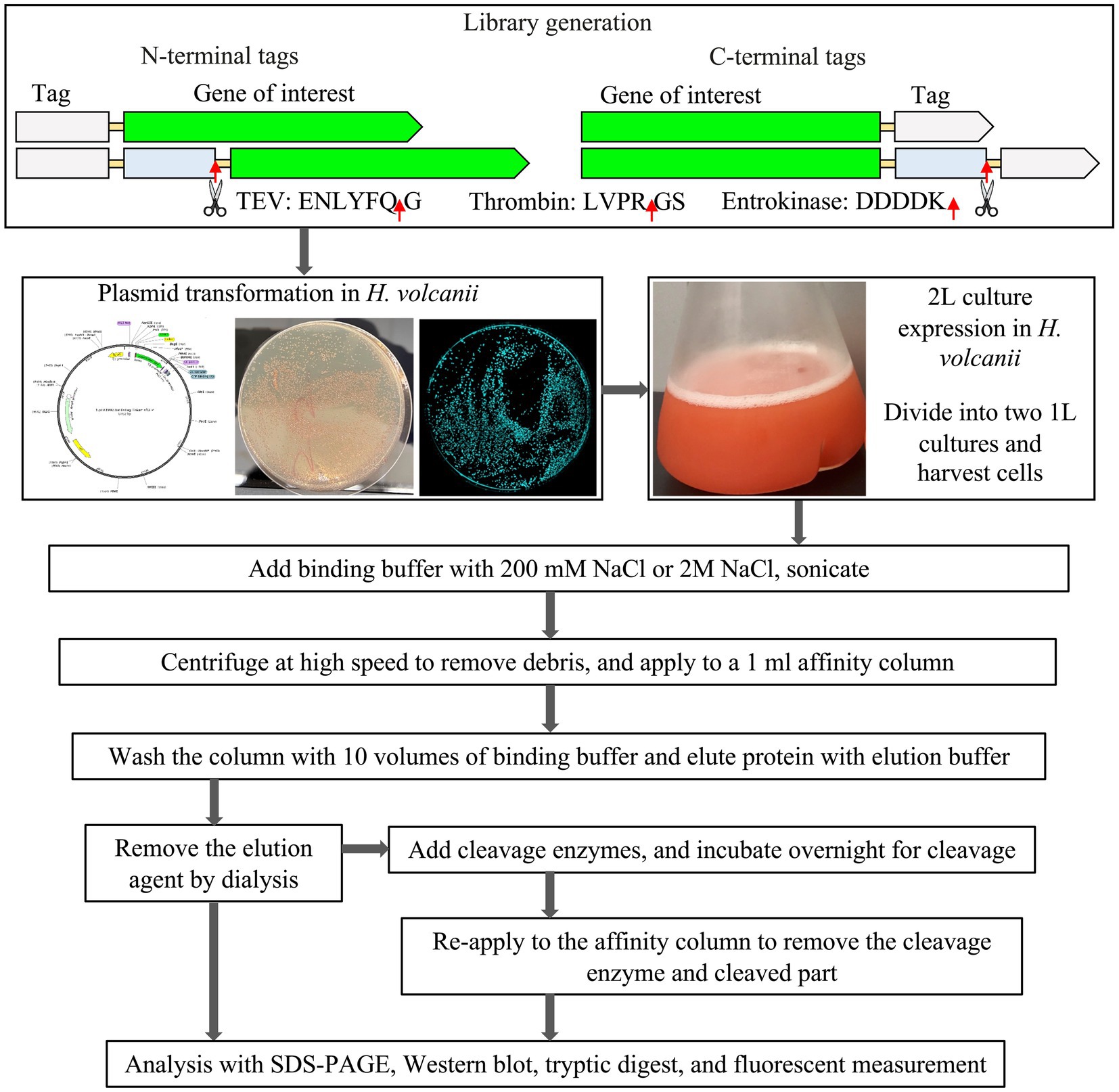
The purification-tag-fusion-sfGFP protein synthetic gene library (GenScript Biotech Corporation, HK), together with the mCherry and ADH constructs were codon-optimized using the java codon adaptation online tool Jcat for Halobacterium sp. (strain NRC-1/ATCC 700922/JCM 11081) (Grote et al., 2005) and cloned into the pTA1921 vector via NdeI and BamHI restriction enzymes (Supplementary Figure S1 and Supplementary Table S1).
Protein expression in Haloferax volcanii
The construct containing vectors was transformed into the Haloferax volcanii H1895 using the PEG/EDTA method (Dyall-Smith, 2008). H. volcanii and derivatives were cultured in the Hv-YPC medium at 45°C with shaking for 24–36 h, as previously described (Strillinger et al., 2016; Grötzinger et al., 2018; Akal et al., 2019; Karan et al., 2020; Alshehri et al., 2022). For solid media, 2% (w/v) agar was added. Stock cultures were maintained in glycerol at −80°C. For short-term use, cultures were maintained on stock plates.
Estimation of expression level and solubility
The expression level of the sfGFP or mCherry constructs was evaluated by measuring the fluorescence (ex 485 nm/em 507 nm) or mCherry (ex 587 nm/em 610 nm) of 24 h grown cell culture/OD600 with three different colonies. The expression level of the ADH constructs was evaluated by determining the final protein concentration after purification using NanoDrop absorption at 280 nm. The results were transformed into relative expression, with the highest fluorescence (for sfGFP N-ter Strep-tag, mCherry C-ter 8xHis-tag, and ADH N-ter 8xHis-tag) set to 100%.
The solubility of sfGFP and mCherry was evaluated by comparing the fluorescence after sonication, before and after centrifugation. For ADH, which lacks fluorescence, solubility was evaluated using SDS-PAGE and Western blotting to compare the soluble fraction against the pellet.
Cell harvesting and lysis
One liter of cell culture was grown until an OD600 of 1.0–1.5. Cells were harvested by centrifugation (4,000 × g, 4°C, 45 min) in a Legend XFR Centrifuge (Thermo Scientific, Waltham, United States) and disrupted in binding buffer (50 mM sodium phosphate buffer, pH 7.4 containing 200 mM or 2 M NaCl) containing Pierce™ Protease Inhibitor EDTA-free tablet (Thermo Scientific) using a sonicator (Model Q500, QSONICA, Newtown, CT, United States) with a 1.9 cm probe. Cell debris was removed by centrifugation (24,000 × g, 4°C, 45 min) in a Multifuge X1R Centrifuge (Thermo Scientific).
Purification of constructs
All constructs were purified using the ÄKTAprime plus chromatography system (GE Healthcare Life Sciences, Piscataway, NJ, United States) according to the purification tag in the binding buffer (50 mM sodium phosphate, pH 7.4, 200 mM or 2 M NaCl).
Purification of His-tag fused constructs
The polyHis and SUMO fused constructs were purified on a 1 mL HiTrap Ni2+ chelating column (GE Healthcare Life Sciences, Piscataway, NJ, United States). The supernatant was loaded at a 1.0 mL/min flow rate onto the column pre-equilibrated with a binding buffer (50 mM sodium phosphate, pH 7.4, 200 mM, or 2 M NaCl) containing 20 mM imidazole. After washing the column with binding buffer, the protein was eluted in one step with His-elution buffer (binding buffer containing 250 mM imidazole).
Purification of Strep-tag®II fused constructs
The Strep-tag®II and Twin-Strep-tag® tag fused constructs were purified on a 1 mL Strep-Tactin®XT 4Flow (GenScript Biotech Corporation, HK). The supernatant was loaded at a 1.0 mL/min flow rate onto the column pre-equilibrated with binding buffer (50 mM sodium phosphate, pH 7.4, 200 mM, or 2 M NaCl). After washing the column with binding buffer, the protein was eluted in one step with a strep-elution buffer (binding buffer containing 50 mM biotin).
Purification of FLAG-tag fused constructs
The FLAG-tag and 3X FLAG-tag fused constructs were purified on a 1 mL Anti-DYKDDDDK G1 Affinity Resin column (GenScript Biotech Corporation, HK). The supernatant was loaded at a 1.0 mL/min flow rate onto the column pre-equilibrated with binding buffer (50 mM sodium phosphate, pH 7.4, 200 mM, or 2 M NaCl). After washing the column with binding buffer, the protein was eluted in a one-step FLAG-elution buffer (100 mM Tris/HCl buffer, pH 12.0, 200 mM, or 2 M NaCl). To purify the FLAG/3xFLAG-tag fused constructs, MonoRab™ Anti-DYKDDDDK Magnetic Beads (Cat No. L00835, GenScript) were tested as well. The cell lysate was added to the pre-washed beads with bead binding buffer (50 mM sodium phosphate, pH 7.4, 200 mM or 2 M NaCl) and incubated on a shaker at 4°C for 2 h. After magnetic separation, the supernatant was removed, and the magnetic beads were washed with the bead binding buffer three times. Finally, the DYKDDDDK fused construct bound to the beads was eluted by bead elution buffer (100 mM Tris/HCl buffer, pH 12.0).
Purification of C-tag fused constructs
The C-tag fused constructs were purified on a 1 mL CaptureSelect™ C-tagXL column (Thermo Scientific, Waltham, United States). The supernatant was loaded at a 1.0 mL/min flow rate onto the column pre-equilibrated with binding buffer (50 mM sodium phosphate, pH 7.4, 200 mM, or 2 M NaCl). The protein was eluted in one step with a C-tag-elution buffer (50 mM sodium phosphate, pH 7.4, 2 M MgCl2).
Supplementary steps for purification and analysis
The purified fractions were combined, further purified, and concentrated with Amicon® Ultra-4 Centrifugal Filter Units, 10 kDa. The protein was then dialyzed against dialysis buffer (20 mM sodium phosphate, pH 7.4, 200 mM, or 2 M NaCl). Protein concentration was determined using NanoDrop absorption at 280 nm.
Note: The purification procedures of each construct were not optimized (e.g., elution gradient, wash steps) to compare better the purity of sfGFP, respectively, mCherry and ADH, constructs for each purification tag.
Protein cleavage
For fusion constructs containing cleavable tags, the cleavage of the purification tags was performed overnight at 4°C in dialysis buffer (50 mM Tris-HCl, 1 mM DTT, pH 8.0) containing 200 mM NaCl or 2 M NaCl, with the respective SUMO-Protease, containing a His-tag, (ratio of 1:10 w/w) and TEV-Protease, containing a His-tag, (200 U for 1 mg of fusion protein), and then reapplied to the respective affinity column to remove the cleaved purification tag. The SUMO- and TEV-Protease were removed by the HiTrap Ni2+ chelating column (GE Healthcare Life Sciences, Piscataway, NJ, United States).
The FLAG tag contains an internal enterokinase (EK) cleavage site, which allows the removal of the affinity tag after purification. The eluted fractions were incubated with Enterokinase, containing a His-tag (Z03376-1, Genscript, 200 U for 1 mg of fusion protein) to remove the FLAG-tag. The Enterokinase was removed by the HiTrap Ni2+ chelating column (GE Healthcare Life Sciences, Piscataway, NJ, United States).
SDS-PAGE and western blotting analysis
Western blotting analysis was performed as previously described (DasSarma et al., 2013; Andar et al., 2017; Karan et al., 2020). The SDS-PAGE analysis was performed using Novex® Tris-glycine gels (4–20%, Invitrogen, Carlsbad, CA, United States). Proteins were then transferred to 0.45 μm nitrocellulose membranes (Millipore Corp., Boston, MA). Membranes were blocked for 15 min in Pierce Fast Blocking Buffer (Thermo Fisher Scientific) and then incubated in Anti-GFP Polyclonal Antibody (Thermo Fisher Scientific) at 1:2000 dilution, followed by three washing steps with TBST, then incubated with Goat anti-Rabbit IgG (H + L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Invitrogen, Catalog # A-11008).
The purity of the proteins was evaluated using SDS-PAGE stained with Coomassie brilliant blue and quantified with ImageJ software1 (Schneider et al., 2012). Purity was specified as a percentage of the total protein content, with the intensity of the target protein band compared to the total protein intensity on each lane of the gel.
Tryptic digest and LC-MS/MS analysis
The identification of corresponding peptides was performed by LC-MS/MS analysis. 10 μg of the sample was digested with trypsin using the FASP protocol (Wiśniewski et al., 2009). Peptides were measured using an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, United States) and analyzed by MASCOT v2.3 (Matrix Sciences Ltd., United Kingdom).
Results and discussion
Experimental design for systematic screening
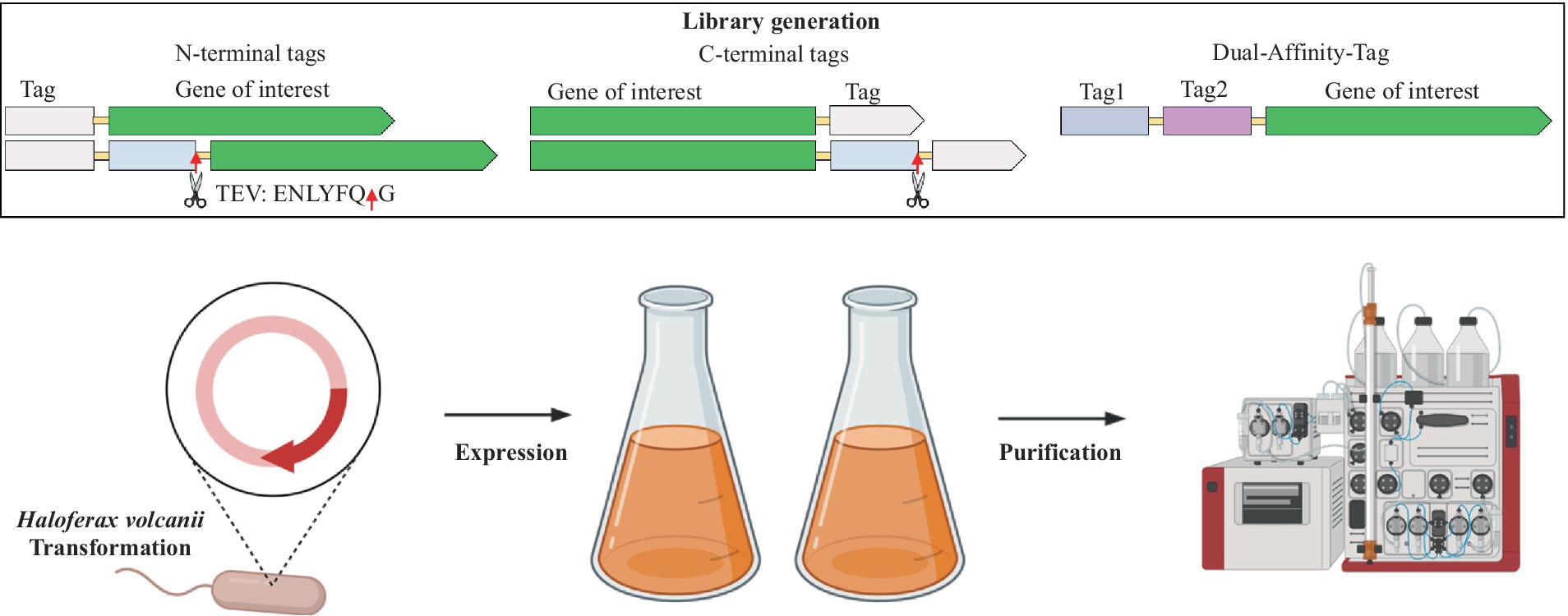
In the study, H. volcanii strain H1895 as the host, in combination with the pTA1992 plasmid that features the exceptionally strong constitutive p.syn promoter (Haque et al., 2019, 2020) was employed. Next, a versatile purification-tag-fusion-protein library using the sfGFP protein as a reporter (Pédelacq et al., 2006), was designed and constructed. Within this library, a range of purification tags, including 6xHis, 8xHis, Strep-tag®II, Twin-Strep-tag®, FLAG, and 3x-FLAG, strategically placed at both the N-terminus and C-terminus of sfGFP (Figure 1) were introduced.
His-tags are the most used and cost-effective affinity tags to purify recombinant proteins using an immobilized metal affinity chromatography (IMAC) column (Porath et al., 1975). Notably, protein activity is rarely affected by polyhistidine affinity tags because of their relatively small size and charge (Block et al., 2009). In the context of H. volcanii strains where naturally histidine-rich proteins PitA and Cdc48d have been replaced with orthologous genes containing fewer histidine residue, the 6xHis-tag has been widely adopted for recombinant protein purification (Grötzinger et al., 2018; Haque et al., 2019, 2020; Karan et al., 2020). The Small Ubiquitin-like Modifier (SUMO)-tag, which incorporated either a 6xHis-tag or an 8xHis-tag for expression and purification (Butt et al., 2005) was used for the library design. Strep-tag®II, an 8-amino acid peptide tag (WSHPQFEK), exhibits a strong affinity for Strep-Tactin® (or Strep-Tactin®XT), a specially engineered streptavidin under physiological buffer conditions (Skerra and Schmidt, 1999; Schmidt et al., 2013; Yeliseev et al., 2017). This interaction allows for the rapid one-step purification of nearly any recombinant protein at high purity and functionality. Moreover, the Strep-tag®II can be found as a repetitive motif, generating a Twin-Strep-tag®. The Twin-Strep-tag®, characterized by its relatively higher affinity for Strep-Tactin® matrices, offers enhanced purification capabilities for recombinant proteins.
The FLAG-tag is a small, highly soluble tag consisting of a hydrophilic octapeptide epitope (DYKDDDDK) with an enterokinase-cleavage site (DDDDK). This tag’s hydrophilic nature positions it primarily on the surface of the recombinant protein, increasing accessibility to resins and minimizing potential adverse effects on the fusion protein (Hopp et al., 1988; Schmidt et al., 2012). Similarly, the 3xFLAG-tag is a robust epitope tag (DYKDHDG-DYKDHDI-DYKDDDDK) was fused with sfGFP at the N- and C-termini.
A new class of purification tags, the four-peptide (E-P-E-A) short C-tag, which exhibits a strong affinity for a camelid antibody fragment when expressed at the C-terminus of a protein was also explored (Djender et al., 2014).
We also scrutinized the effect of positioning the TEV cleavage site at the N-terminus and C-terminus of the sfGFP fusion constructs (Supplementary Table S1 and Supplementary Figure S1). A total of 26 distinct sfGFP constructs were studied, each evaluated for expression through fluorescence measurements (Supplementary Figure S2), further corroborated by Western blot analysis (Supplementary Figure S3). The different sfGFP constructs were compared for their relative expression level, solubility, and purification profile (Supplementary Figure S4). Tryptic digest analysis was additionally performed to confirm the identity of the fusion proteins (Supplementary Figure S5). The focus was primarily on purification at low salt (200 mM NaCl) and high salt concentrations (2 M NaCl). This comprehensive evaluation aimed to determine the most suitable purification tag for the native purification of extremophilic halophilic proteins, capitalizing on the suitability of H. volcanii as the expression host.
The initial systematic screening library was expanded to examine the applicability of the sfGFP results to the red fluorescent protein mCherry (Shaner et al., 2004) and a previously fully characterized halophilic alcohol dehydrogenase (ADH) from the deep-sea brine pool of the Red Sea (Grötzinger et al., 2018) (Supplementary Figure S7). These investigations revealed the significant impact of different tags, tag lengths, and positions on protein expression and purification. This resulted in a total of 37 screened constructs.
Expression and purification of sfGFP fusion proteins
The corresponding overview of the expression and purification of the sfGFP constructs is presented in the order of the different studied affinity purification tag systems.
Polyhistidine-tag (His-tag)
The experimental design explored the efficacy of both a 6xHis-tag and an 8xHis-tag at the N- and C-termini of sfGFP, both with and without a TEV cleavage site. Furthermore, the N-terminal solubility-enhancing and cleavable Small Ubiquitin-like Modifier (SUMO)-tag, which incorporated either a 6xHis-tag or an 8xHis-tag for expression and purification (Butt et al., 2005), was used. All the constructs underwent purification and cleavage, where applicable, under varying salt concentrations, ranging from low (200 mM NaCl) to high (2 M NaCl) in a 50 mM sodium phosphate buffer, pH 7.4. The expression level, solubility, purity, and final yield of the fusion sfGFP constructs are presented in Table 1 and Figure 1.
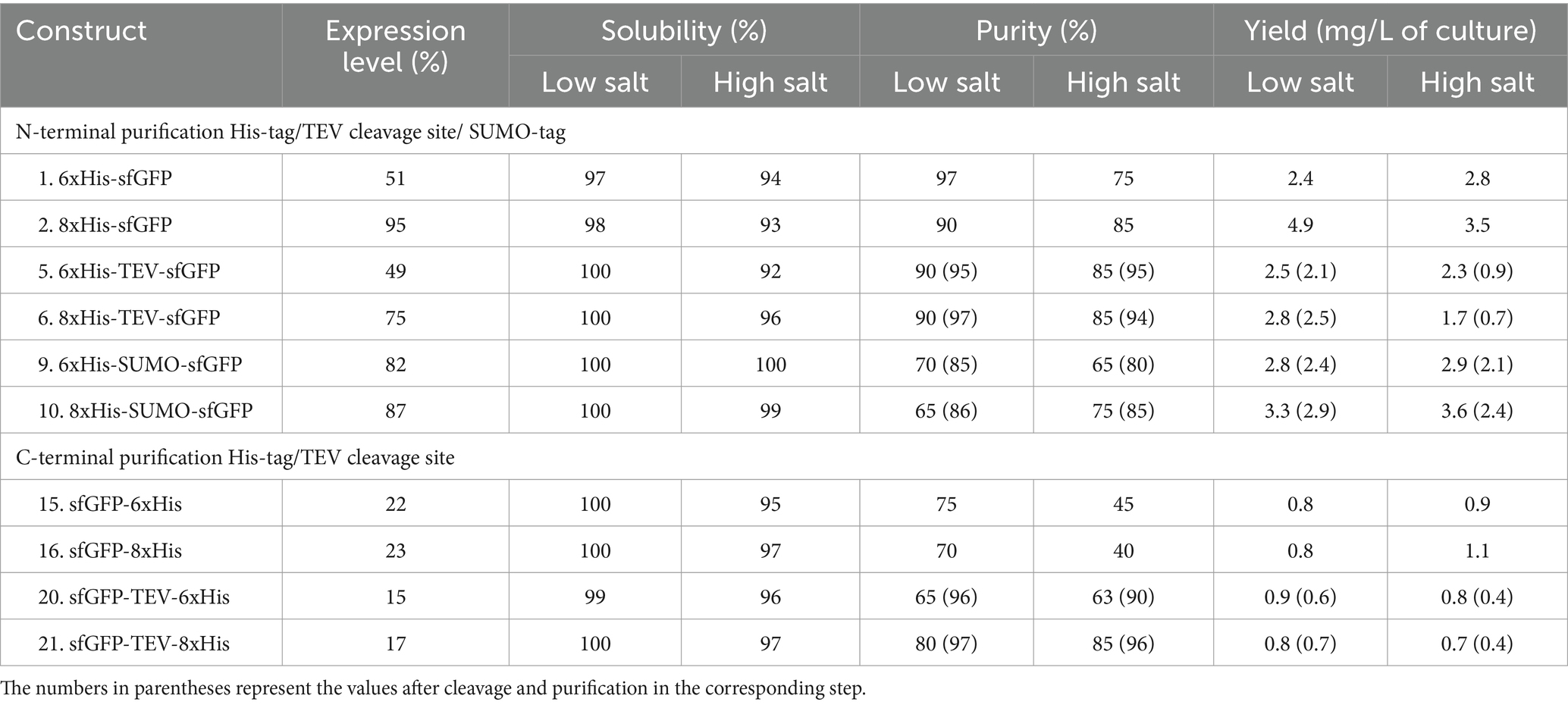
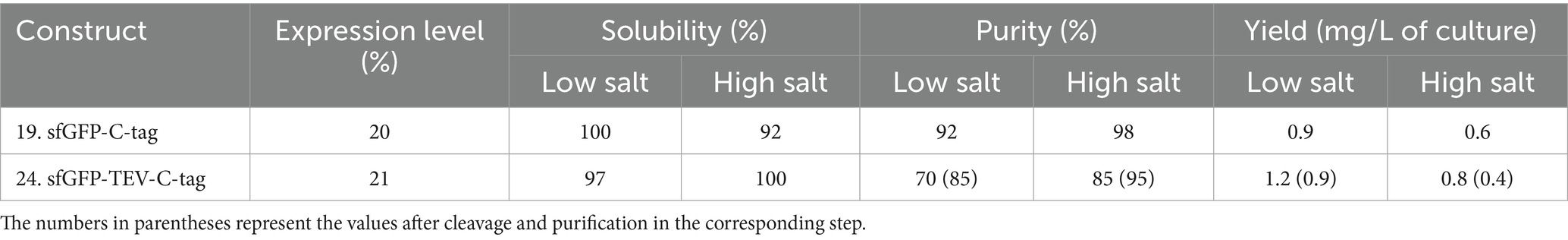
Table 1. Relative expression, solubility, purity, and yield of sfGFP expressed with His-tag constructs.
N-terminal vs. C-terminal and 6xHis- vs. 8xHis-tag
The expression of sfGFP is significantly influenced by the choice of tag and its location (Yilmaz and Arslanyolu, 2015). Notably, the highest level of sfGFP expression was achieved with the N-terminal 8xHis-tag (Table 1). Surprisingly, the introduction of the N-terminal 8xHis-SUMO-tag did not significantly enhance expression yield, and the addition of a cleavable TEV cleavage site had a noticeable impact on expression levels. Conversely, sfGFP expression with C-terminal His-tags demonstrated significantly lower levels, up to one-fifth, compared to the N-terminal 8xHis-tag. While the expression level of sfGFP varied regarding the position and length of the tag, it’s noteworthy that these variations did not substantially impact solubility. All constructs exhibited consistently high solubility levels ranging from 92 to 100%, irrespective of salt concentration. The purity of the sfGFP fusion proteins displayed notable variation concerning His-tag length, position, and salt concentration in the purification buffer. The 8xHis-tag consistently outperformed all other His-tag constructs, resulting in high purity and yield in high and low salt buffers. This outcome aligns with expectations, as the increased number of histidine residues in the tag leads to more efficient binding on IMAC columns. Furthermore, it allows for more rigorous washes, enhancing the overall purity of the protein.
Sumo-tag vs. TEV cleavage site and the impact of salt
A comparative assessment of the solubility-enhancing and cleavable SUMO-tag versus the TEV cleavage site yielded valuable insights into the performance of these constructs. The SUMO-tag exhibited robust performance, outperforming the cleavable TEV cleavage site. The SUMO-tag was pivotal in enhancing sfGFP expression and ensuring high purity, irrespective of the buffering composition. sfGFP constructs with the TEV cleavage site also displayed strong expression, achieving even higher purity levels. However, the TEV cleavage site did not cleave efficiently at high salt concentrations (Figure 2). This limitation resulted in approximately 40–50% final yields compared to the uncleaved constructs. Of note, the sfGFP construct featuring a C-terminal 6xHis-tag with a TEV cleavage site exhibited the lowest production level among all the tested His-tagged constructs.
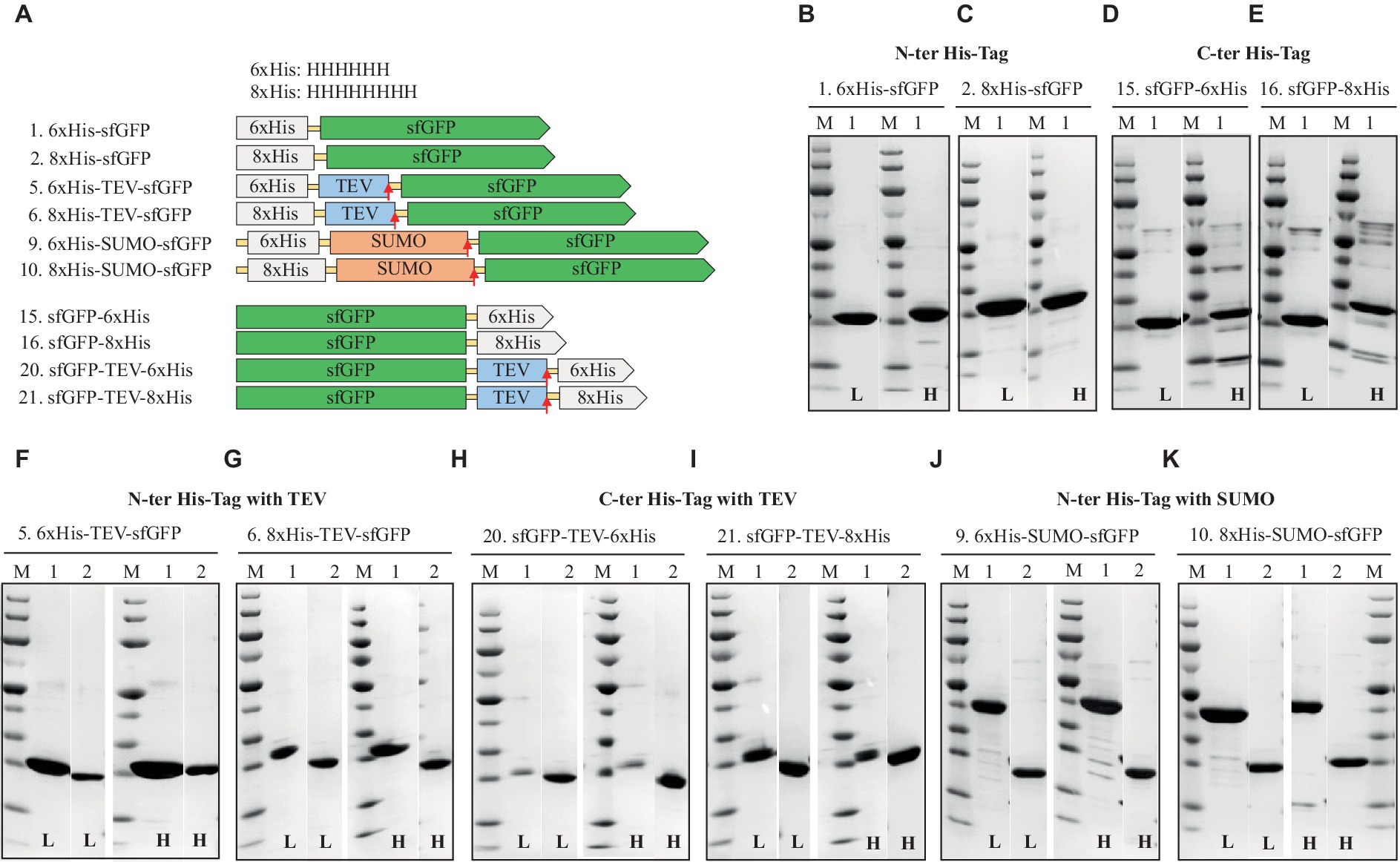
Figure 2. Schematic representation of His-tag constructs (A) and SDS-PAGE analysis of the purification of the sfGFP with N-terminal 6xHis (B), 8xHis (C), and C-terminal 6xHis (D) 8xHis (E), N-terminal His-tag with cleavable tag, TEV, 6xHis, (F) 8xHis (G), and C-terminal His-tag with TEV cleavage site, 6xHis (H), 8xHis (I), N-terminal His-tag with solubility and cleavable SUMO-tag with 6xHis (J), or 8xHis (K). Lane M: molecular weight marker; Lane 1: Elution from HisTrap affinity column using the binding buffer containing 250 mM imidazole; Lane 2: Flow-through from HisTrap affinity column loaded after TEV-/SUMO-Protease cleavage. L: Low salt (200 mM NaCl); H: High salt (2 M NaCl) in the binding, washing, and elution buffers.
Overall, the N-terminal 8xHis-tag proved the best performer, offering a simple, rapid, and highly efficient one-step purification method for sfGFP. This approach employed cost-effective materials and delivered approximately double the final yield compared to the 6xHis-tag and nearly five times the yield compared to the C-terminal 6xHis- or 8xHis-tag constructs (Table 1).
The 6xHis-tag remains the most favored purification tag for H. volcanii due to its compatibility with high-salt media (Haque et al., 2019). However, the results, which contradict earlier studies on N-terminal His-tagged GFP that indicated adverse effects on protein production, emphasize the ongoing challenge of selecting the optimal position and length for the His-tag within various expression systems (Nguyen et al., 2014; Phan et al., 2015; Tian et al., 2019; Le et al., 2022). This study underscores the importance of making informed choices regarding His-tag design to maximize expression levels and purification efficiency.
Strep-tag®II and twin-Strep-tag®
Next, the sfGFP constructs fused with Strep-tag®II and Twin-Strep-tag® were investigated.
N-terminal vs. C-terminal and Strep-tag®II vs. twin-Strep-tag®
The results of the sfGFP constructs fused with Strep-tag®II are summarized in Table 2 and Figure 3. Expression levels of sfGFP varied based on the position of the Strep-tag®II and Twin-Strep-tag®. Like the His-tag, the N-terminal Strep-tag®II yielded the highest sfGFP expression, surpassing the C-terminal Strep-tag®II or Twin-Strep-tag® by up to fivefold. Notably, while C-terminal Strep-tag®II has been utilized for protein production in H. volcanii and proven compatible with high salt concentrations, the study reveals that N-terminal Strep-tag®II results in approximately five times higher expression, suggesting that C-terminal usage in H. volcanii should be reconsidered (Braun et al., 2019). Intriguingly, Twin-Strep-tag®, which generally exhibits a higher affinity for Strep-Tactin® and greater tolerance to salts and detergents in buffers than Strep-tag®II (Yeliseev et al., 2017), displayed an unexpected reduction in sfGFP expression to about one-third when compared to N-terminal Strep-tag®II. The introduction of Strep-tags (Strep-tag®II or Twin-Strep-tag®) appeared to have no significant effect on the solubility of the fusion proteins, regardless of salt conditions. Furthermore, these tags enabled high-purity purification processes (Table 2 and Figure 2).
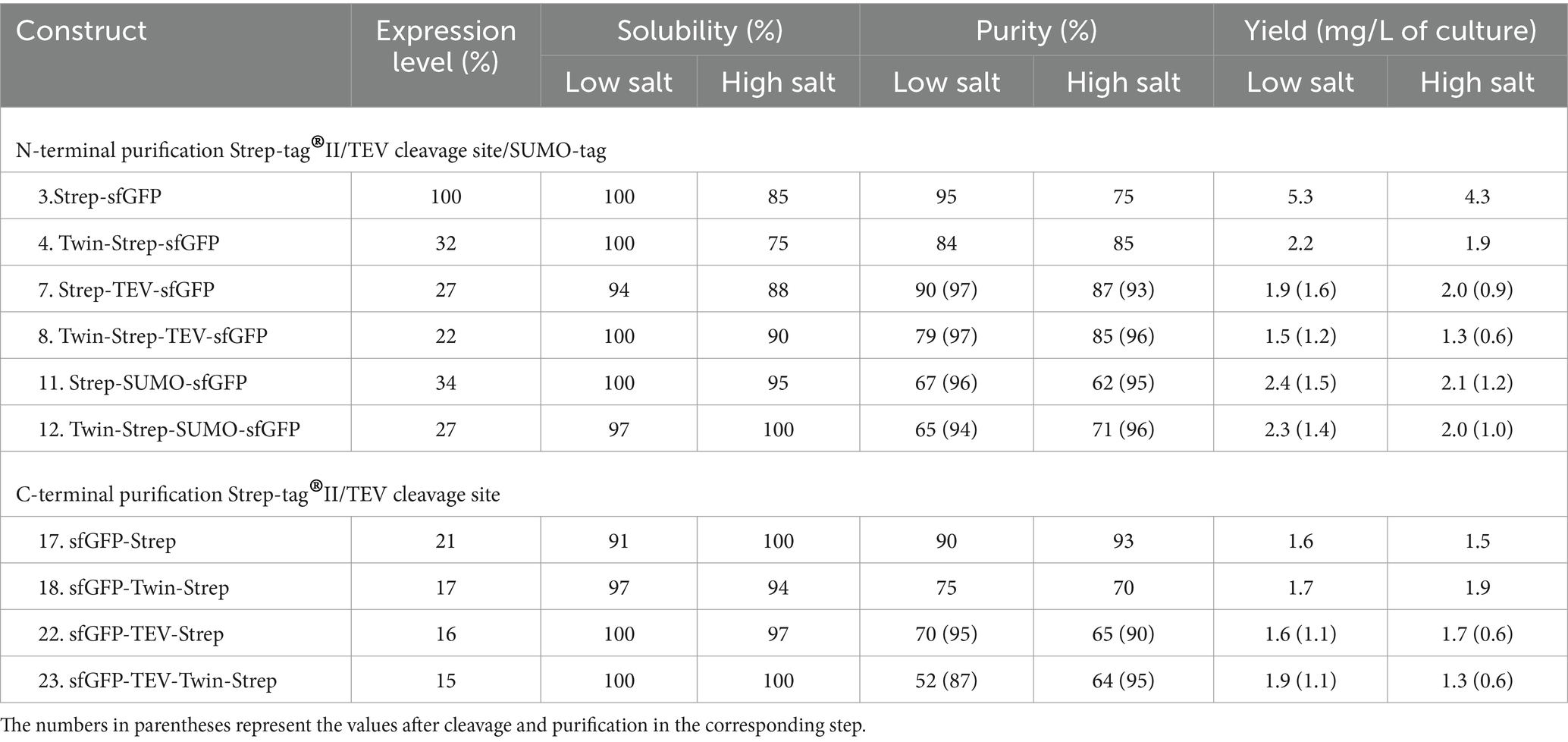
Table 2. Relative expression, solubility, and purity of sfGFP expressed with Strep-tag®II and Twin-Strep-tag® constructs.
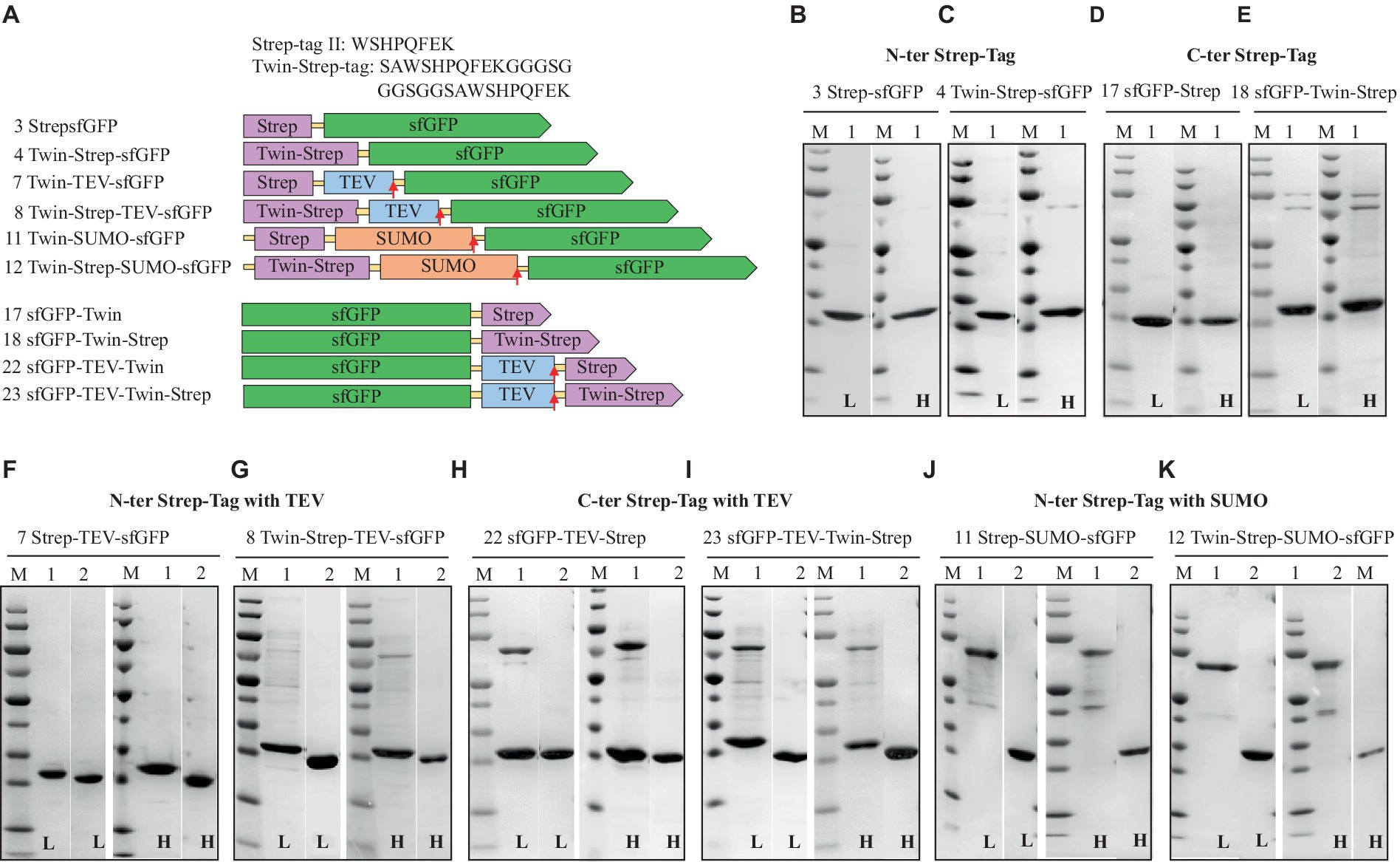
Figure 3. Schematic representation of Strep-tag (Strep-tag®II and Twin-Strep-tag®) constructs (A) and SDS-PAGE analysis of the purification of the sfGFP with N-Terminal Strep (B), Twin-Strep-tag® (C), and C-terminal Strep (D) Twin-Strep-tag®s (E). N-terminal Strep with TEV cleavage site, Strep, (F) Twin-Strep-tag® (G), and C-terminal Strep-tag®II with TEV cleavage site, Strep (H), Twin-Strep-tag®s (I), N-terminal Strep-tag®II with solubility and cleavable SUMO-Strep-tag (J), and Twin-Strep-tag® (K). Lane M: molecular weight marker; Lane 1: Elution from Strep-Tactin affinity column using the binding buffer containing 50 mM biotin; Lane 2: Flow-through from HisTrap and Strep-Tactin affinity column loaded after TEV-/SUMO-Protease cleavage. L: Low salt (200 mM NaCl); H: High salt (2 M NaCl) in the binding, washing, and elution buffers.
Sumo vs. TEV cleavage site and the impact of salt
Next, experiments to assess the removal of Strep-tags using protease cleavage sites (TEV and SUMO) positioned between the sfGFP protein and the Strep-tags were conducted. Subsequently, a purification step was employed to separate the sfGFP protein from the cleaved Strep-tags. Consequently, the findings indicate that both Strep-tag®II and Twin-Strep-tag®, when fused with SUMO or TEV cleavage sites, could be efficiently removed through protease cleavage (SUMO- or TEV-Protease).
Among all the tested tags fused to the N-terminus of sfGFP, Strep-tag®II consistently yielded the highest levels of sfGFP expression.
Flag-tag
Next, the expression and solubility of sfGFP fused with the FLAG-tag and 3xFLAG-tag, in combination with the SUMO-tag or TEV cleavage site (Einhauer and Jungbauer, 2001; Schmidt et al., 2012; Beznosov et al., 2013) were assessed.
N-terminal vs. C-terminal FLAG-tag and 3xFLAG-tag
Comparable to His and Strep-tagged sfGFP findings, N-terminal FLAG, and 3xFLAG-tags exhibited higher expression levels (50 and 75%, respectively) than C-terminal tags (Table 3). Interestingly, N-terminal 3xFLAG-fused sfGFP demonstrated expression levels 1.5 to 3 times superior to N-terminal FLAG-tag, C-terminal FLAG, or 3xFLAG-tagged sfGFP. Additionally, all constructs displayed high solubility, exceeding 90%, regardless of whether the FLAG or 3xFLAG was positioned at the N-terminus or C-terminus of the sfGFP protein.
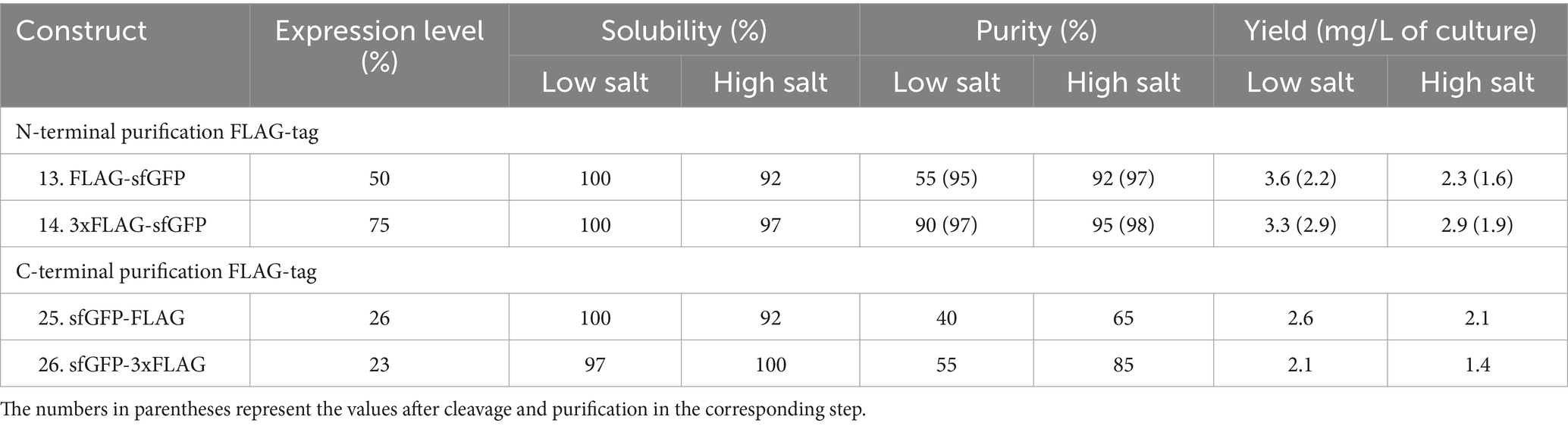
Table 3. Relative expression, solubility, and purity of sfGFP expressed with FLAG-tag and 3xFLAG-tag.
Cleaving the FLAG-tag/3xFLAG-tag and the impact of salt
The Enterokinase efficiently removed the FLAG-tag even at higher salt concentrations, which led to a relatively lower yield (Figure 4). All FLAG-tagged expressions resulted in highly pure sfGFP with no visible contaminating proteins on SDS-PAGE (Figure 4). To purify FLAG and 3xFLAG-tagged sfGFP from sonicated H. volcanii cell culture, the anti-DYKDDDDK magnetic beads (GenScript) were also tested. Remarkably, we were able to streamline the purification process by eliminating centrifugation or filtration steps and still achieved high purification (data not shown).
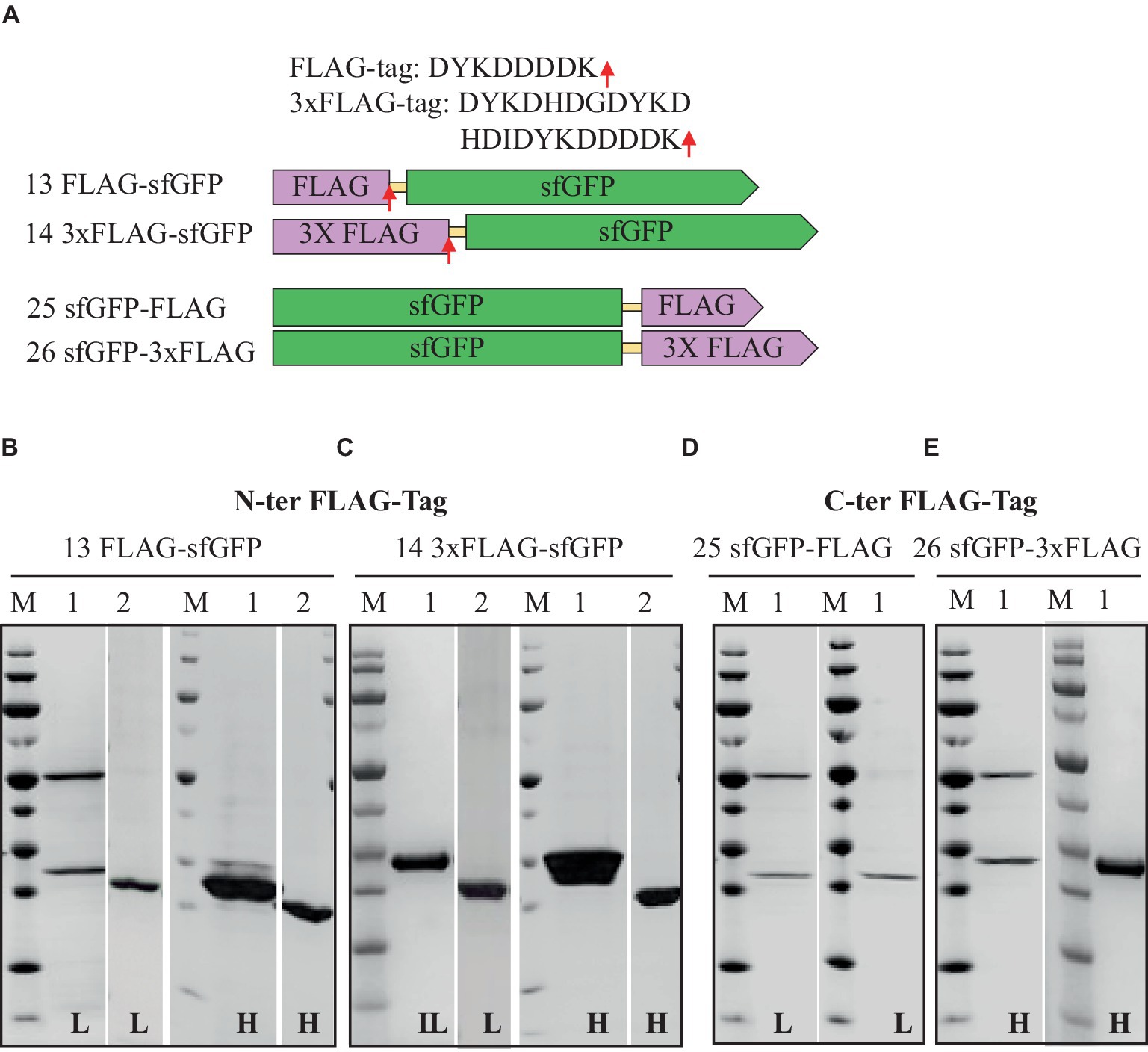
Figure 4. Schematic representation of FLAG-tag constructs (A) and SDS-PAGE analysis of the purification of the sfGFP with N-Terminal FLAG (B), 3xFLAG (C), and C-terminal FLAG (D) 3xFLAG with TEV cleavage site (E). Lane M: molecular weight marker; Lane 1: Elution from anti-FLAG affinity column using 100 mM Tris/HCl buffer, pH 12.0; Lane 2: Flow-through from HisTrap affinity column loaded after TEV-Protease cleavage. L: Low salt (200 mM NaCl); H: High salt (2 M NaCl) in the binding, washing, and elution buffers.
While FLAG-tags have previously been employed to purify recombinant proteins in various expression systems, including bacteria, baculovirus, mammalian cells, and yeast, there has been limited information regarding their use in halophilic systems (Einhauer and Jungbauer, 2001; Mishra, 2020). FLAG peptide has been tagged with modified archaellin genes (flaA1, flaA2, and flaB2) in haloarchaea Halobacterium salinarum cells and utilized for the identification of proteins within the archaellum (Beznosov et al., 2013). However, no reports existed on FLAG-tagged protein expression and purification in the halophilic context until this study. These findings suggest that the FLAG and 3xFLAG-tags suit protein expression and purification in halophilic systems. The FLAG-tag carries several advantages over His and Strep tags. Like the 6xHis-, 8xHis-, and Strep-tag®II, the FLAG-tag comprises a small hydrophilic peptide (8 amino acids), unlikely to impact protein folding or function. Furthermore, it can be easily removed with Enterokinase. However, it is worth noting that Enterokinase, while versatile, is sensitive to high salt concentrations compared to other proteases like the SUMO-Protease, limiting its application in the extremophilic environment (Shahravan et al., 2008).
C-tag
Next, the C-tag constructs were examined, and the results detailing the expression level, solubility, purity, and final yield of the fusion sfGFP constructs are outlined in Table 4 and Figure 5. The C-tag-binding resin facilitated the purification of pure proteins in both low-salt and high-salt buffers. Notably, under the 2 M NaCl purification buffer, the protein achieved high purity at 98%, albeit with a lower yield. However, for the 200 mM NaCl purification buffer, the purity of sfGFP could be significantly improved by column washing with 1 M NaCl (Supplementary Figure S6). Moreover, including a TEV cleavage site aided in removing the C-tag, resulting in higher purity, although this came at the expense of yield, particularly in high-salt conditions due to the reduced activity of TEV-Protease in high-salt environments (Parks et al., 1995; Waugh, 2011).
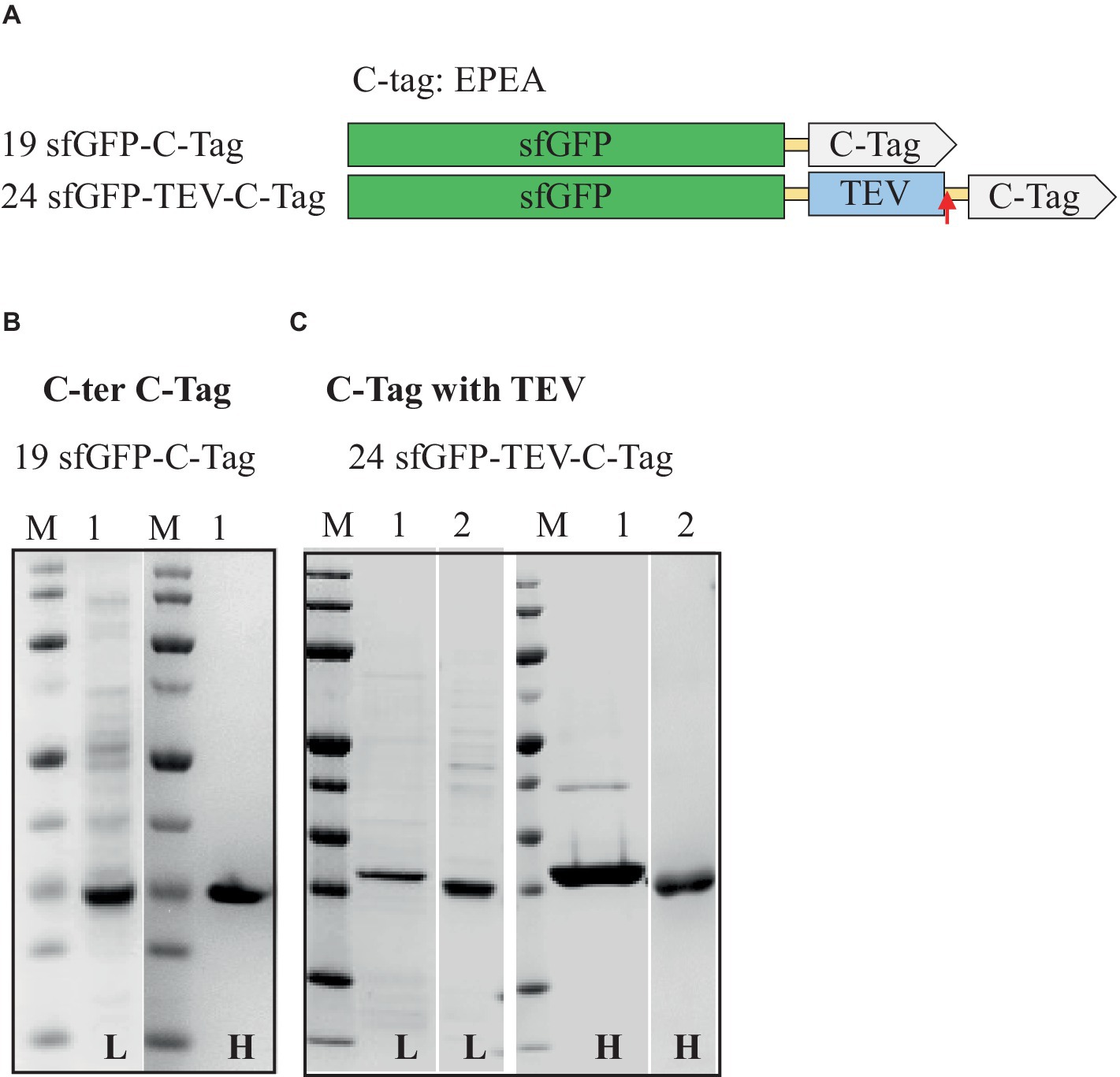
Figure 5. Schematic representation of C-tag constructs (A) and SDS-PAGE analysis of the purification of the sfGFP with C-tag (B) and C-tag with TEV cleavage site (C). Lane M: molecular weight marker; Lane 1: Elution from C-tag XL affinity column using 2 M MgCl2; Lane 2: Flow-through from HisTrap affinity column loaded after TEV-Protease cleavage. L: Low salt (200 mM NaCl); H: High salt (2 M NaCl) in the binding and washing buffers.
Among all the affinity tags tested, the smallest among them, the C-tag comprising just four amino acids, proved to be a rapid and efficient tool for purification. The C-tag offers several advantages over more established tags such as His, Strep, and FLAG. It is the smallest affinity purification tag with high binding affinity and selectivity. Furthermore, its limited impact on protein folding and functionality due to its small size makes it an attractive choice for applications in protein purification (Hermans et al., 2012; Djender et al., 2014; Jin et al., 2017).
The systematic screening of the different purification tags and tag positions with sfGFP revealed a significant enhancement in protein production, purity, and yield in H. volcanii when an 8xHis-tag or Strep-tag®II was placed at the N-terminus of sfGFP. This enhancement is potentially linked to favorable folding kinetics of newly synthesized proteins and increased accessibility of these tags during affinity purification, which may facilitate higher yield and purity. To explore whether this trend was unique to the expression system or exclusive to sfGFP, the investigation was extended to include the red fluorescent protein mCherry and a previously fully characterized halophilic alcohol dehydrogenase (ADH) from the deep-sea brine pool of the Red Sea (Shaner et al., 2004; Grötzinger et al., 2018).
Transferring the expression and purification results from sfGFP to mCherry and ADH
The 8xHis-tag and Strep-tag®II were both positioned at the N- and C-termini to investigate the impact of tag position on mCherry and the ADH (Supplementary Table S2 and Supplementary Figure S1). Surprisingly, we discovered that mCherry demonstrated improved expression levels when the 8xHis-tag, or Strep-tag®II, was located at the C-terminus of the protein. Conversely, ADH exhibited more efficient expression when the 8xHis-tag or Strep-tag®II was situated at the N-terminus of the respective proteins (Table 5 and Figure 6). Although these proteins shared similar relative solubility levels, they exhibited divergent expression patterns, emphasizing the nuanced nature of tag effects in different protein contexts.
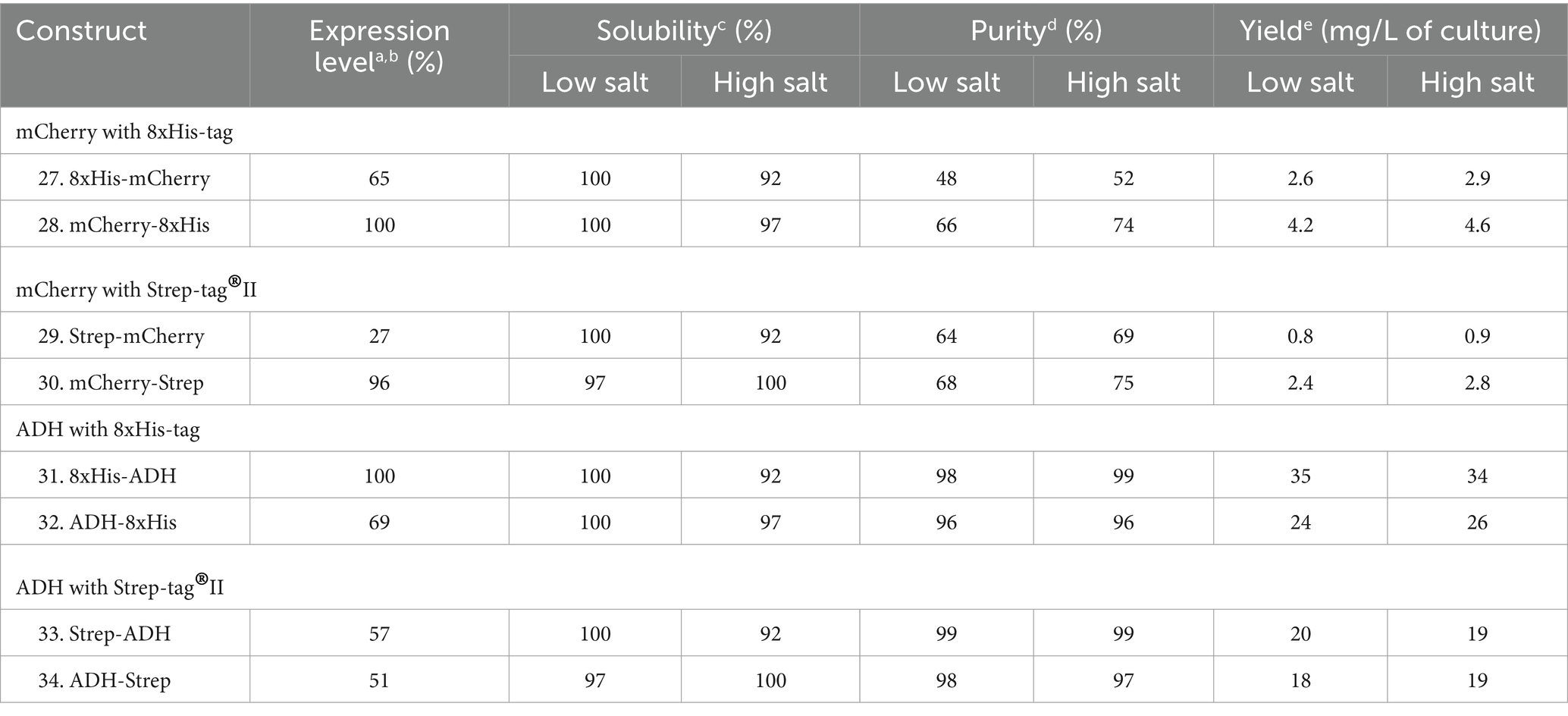
Table 5. Relative expression, solubility, and purity of mCherry and ADH expressed with N- and C-terminal 8xHis-tag and Strep-tag®II.
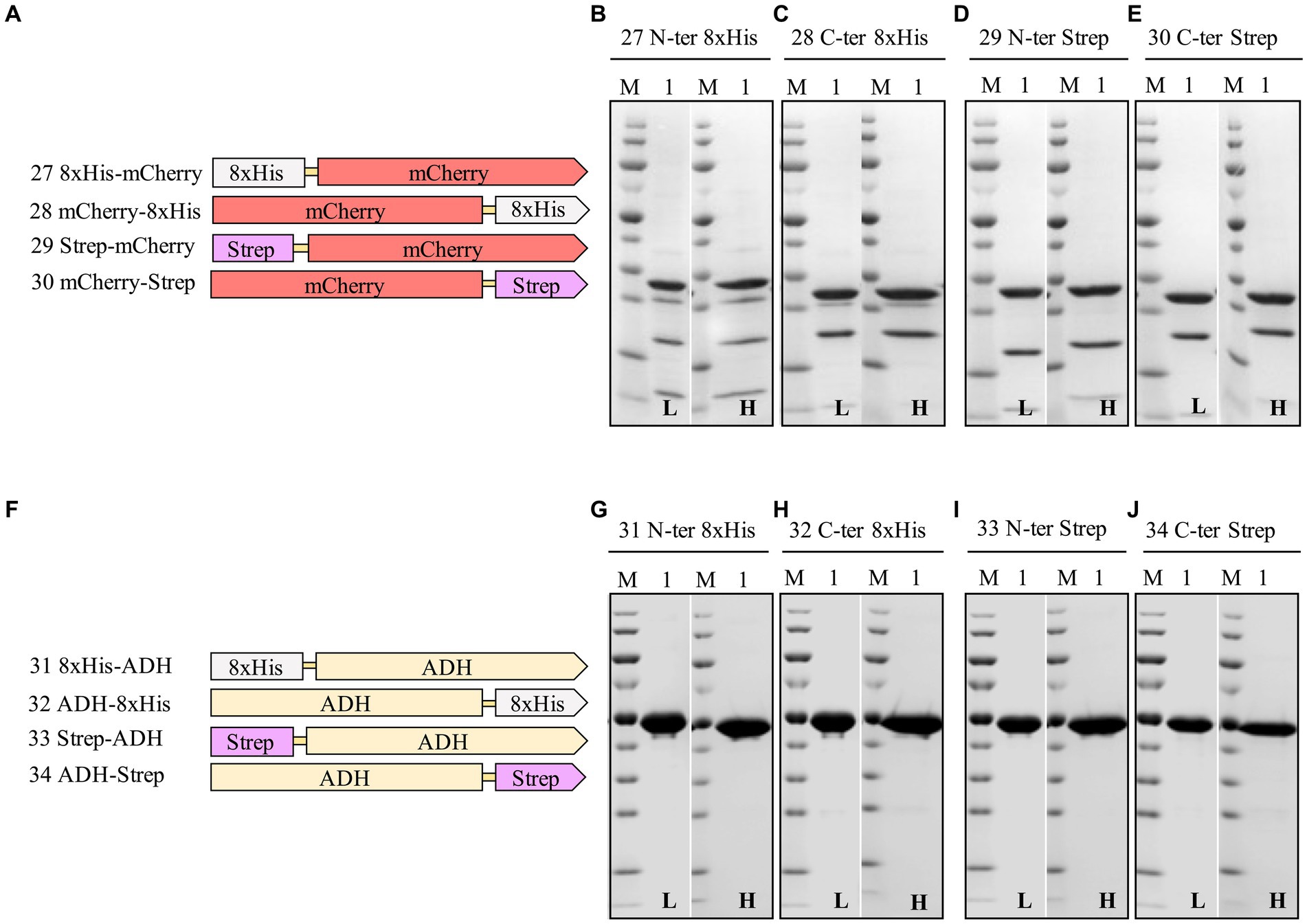
Figure 6. Schematic representation of N- and C-terminal 8xHis-tag and Strep-tag constructs of mCherry (A), and ADH (F), and SDS-PAGE analysis of the purification of the mCherry (B–E), ADH (G–J) Lane M: molecular weight marker; Lane 1: Elution from affinity column. L: Low salt (200 mM NaCl); H: High salt (2 M NaCl) in the binding and washing buffers.
These findings underscore the critical role of purification tags, their positions, and lengths in determining fusion protein expression, purification, and yields. Moreover, this observation emphasizes the necessity of exploring various fusion strategies to optimize expression and purification efficiencies. We considered applying a dual-affinity-tag approach to create a more general purification tag for H. volcanii.
Impact of combining 8xHis-tag and twin-Strep-tag® at the N-terminus on the expression and purification of sfGFP, mCherry, and ADH: a dual-affinity-tag approach
Assessing different tag combinations, positions, and lengths for a novel protein can be resource-intensive and time-consuming (Bernier et al., 2018). To address this challenge, we applied a dual-affinity-tag approach by combining 8xHis-tag and Twin-Strep-tag® tags at the N-terminus of sfGFP, mCherry, and ADH (Supplementary Table S3 and Supplementary Figure S1). In the purification process first a HiTrap Ni2+ chelating column was used to exploit the 8xHis-tag, followed by a Strep-Tactin®XT 4Flow column to utilize the Twin-Strep-tag®, achieving high-purity protein. Surprisingly, this dual-affinity-tag approach consistently outperformed all previously tested tag configurations regarding expression level and purification yields (Table 6 and Figure 7). Moreover, the dual-affinity-tag seems to be a universal solution for protein expression and purification in H. volcanii since all tested proteins were notably expressed in higher amounts and yielded higher overall protein content.
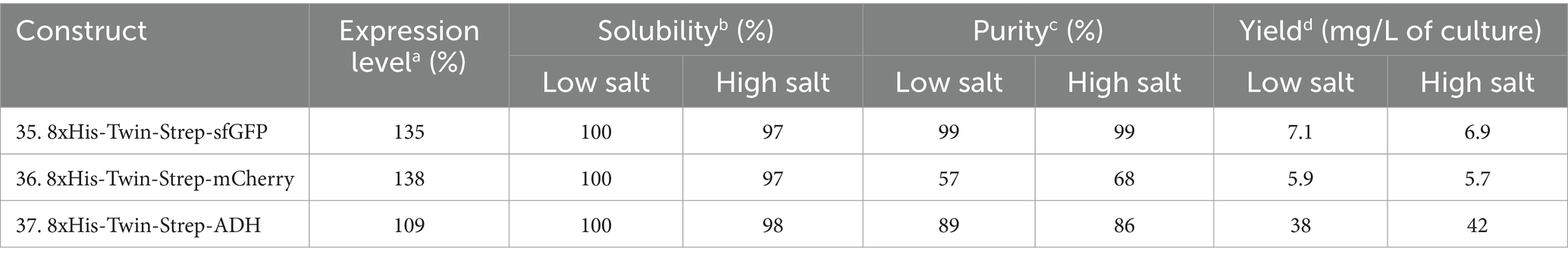
Table 6. Relative expression, solubility, and purity of sfGFP, mCherry, and ADH expressed with N-terminal dual-affinity-tag consisting of 8xHis-tag and Twin-Strep-tag®.
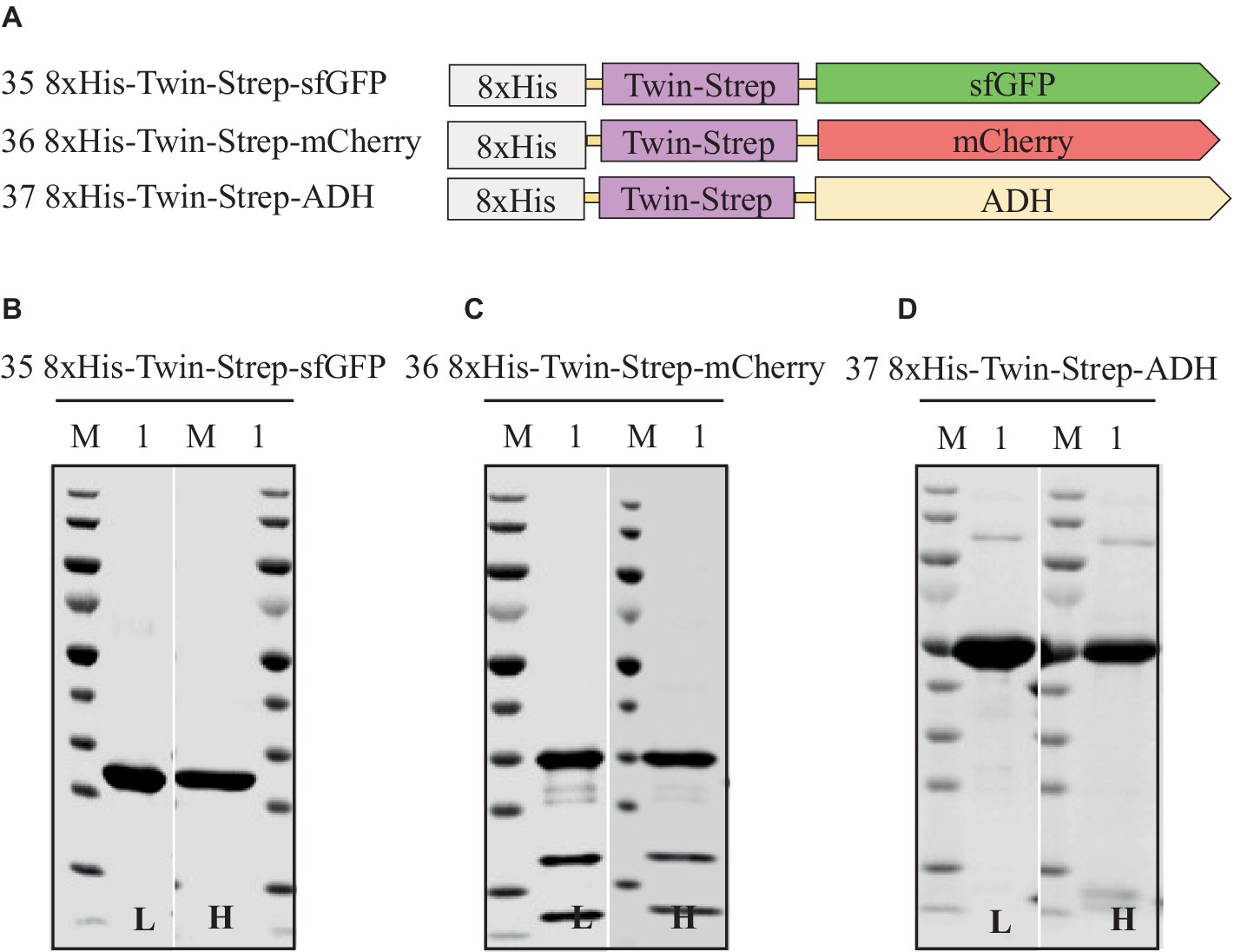
Figure 7. Schematic representation of N-terminal dual-affinity-tag consisting of 8xHis-tag and Twin-Strep-tag® constructs of sfGFP, mCherry, and ADH (A) and SDS-PAGE analysis of the purification of the sfGFP (B), mCherry (C), ADH (D). Lane M: molecular weight marker; Lane 1: Elution from affinity column. L: Low salt (200 mM NaCl); H: High salt (2 M NaCl) in the binding and washing buffers.
The dual-affinity-tag approach is a method that streamlines the purification of recombinant proteins, yielding homogeneous preparations of the proteins of interest in H. volcanii. This is achieved by enabling two consecutive affinity chromatography steps, efficiently removing contaminating proteins.
Conclusion
In conclusion, the study demonstrates the efficient production and purification of sfGFP proteins utilizing various affinity tags within the Haloferax volcanii expression system. The choice of tag and its position significantly impacts the levels of protein production and purification, with the dual-affinity-tag configuration of the 8xHis-tag and Strep-tag®II at the N-terminus generally providing optimal results. This approach has proven effective not just for sfGFP but also for mCherry and ADH, highlighting the importance of tag configuration over mere expression levels.
The options for cleaving the purification tag at high salt concentrations are limited due to the salt sensitivity of commonly tested proteases, including enterokinase and Tobacco Etch Virus (TEV) protease. Other frequently used proteases, such as human rhinovirus 3C protease (HRV 3C) and thrombin, also exhibit salt sensitivity (Jenny et al., 2003; Dušeková et al., 2022). However, salt-insensitive alternatives like SUMO protease and Factor Xa exist (Jenny et al., 2003). Additionally, emerging alternatives, such as the Plum Pox Virus (PPV) NIa protease, demonstrate reduced salt sensitivity, presenting viable options for such conditions (Zheng et al., 2008).
By expanding the library with mCherry and a halophilic alcohol dehydrogenase, we observed that optimal tag positioning varies depending on the protein, with C-terminal tags more effective for mCherry and N-terminal tags better suited for ADH. This underscores the nuanced approach needed in tag placement and protease selection to accommodate different protein characteristics.
As the investigation advance, incorporating proteins with inherently poor solubility will be crucial to further test and validate the adaptability of the suggested tagging strategies. This future work will enhance the general understanding of the expression system’s capabilities, providing a more comprehensive foundation for biotechnological applications in extremophilic organisms.
In summary, the study not only reinforces the significance of strategic tag design and placement but also the critical role of compatible protease selection in the successful production and purification of proteins in extremophilic systems like Haloferax volcanii. By focusing on these elements, researchers can optimize protein yield and purity, thereby improving the overall efficiency of their expression and purification protocols.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
RK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. DR: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Validation, Writing – review & editing. TA: Resources, Validation, Writing – review & editing. MR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The research reported in this publication was supported by funding from the King Abdullah University of Science and Technology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1403623/full#supplementary-material
Abbreviations
sfGFP, superfolder green fluorescent protein; His6 or his8 tag, hexa or octa histidine-tag, halophilic alcohol dehydrogenase (ADH); IMAC, immobilized metal affinity chromatography; SDS-PAGE, sodium dodecyl sulfate-polyacrylamide gel electrophoresis; SUMO, small ubiquitin-like modifier; TEV, tobacco etch virus; Haloferax volcanii, H. volcanii; Escherichia coli, E. coli
Footnotes
References
Akal, A. L., Karan, R., Hohl, A., Alam, I., Vogler, M., Grötzinger, S. W., et al. (2019). A polyextremophilic alcohol dehydrogenase from the Atlantis II deep Red Sea brine pool. FEBS Open Bio 9, 194–205. doi: 10.1002/2211-5463.12557
Allers, T. (2010). Overexpression and purification of halophilic proteins in Haloferax volcanii. Bioeng Bugs 1, 290–292. doi: 10.4161/bbug.1.4.11794
Allers, T., Barak, S., Liddell, S., Wardell, K., and Mevarech, M. (2010). Improved strains and plasmid vectors for conditional overexpression of His-tagged proteins in Haloferax volcanii. Appl. Environ. Microbiol. 76, 1759–1769. doi: 10.1128/AEM.02670-09
Alshehri, S., Karan, R., Ghalayini, S., Kahin, K., Khan, Z., Renn, D., et al. (2022). Air-loaded gas vesicle nanoparticles promote cell growth in three-dimensional bioprinted tissue constructs. Int J Bioprint 8:489. doi: 10.18063/ijb.v8i3.489
Andar, A. U., Karan, R., Pecher, W. T., DasSarma, P., Hedrich, W. D., Stinchcomb, A. L., et al. (2017). Microneedle-assisted skin permeation by nontoxic Bioengineerable gas vesicle nanoparticles. Mol. Pharm. 14, 953–958. doi: 10.1021/acs.molpharmaceut.6b00859
Baeshen, M. N., Al-Hejin, A. M., Bora, R. S., Ahmed, M. M. M., Ramadan, H. A. I., Saini, K. S., et al. (2015). Production of biopharmaceuticals in E. coli: current scenario and future perspectives. J. Microbiol. Biotechnol. 25, 953–962. doi: 10.4014/jmb.1412.12079
Bernier, S. C., Cantin, L., and Salesse, C. (2018). Systematic analysis of the expression, solubility and purification of a passenger protein in fusion with different tags. Protein Expr. Purif. 152, 92–106. doi: 10.1016/j.pep.2018.07.007
Beznosov, S., Pyatibratov, M., Veluri, P., Mitra, S., and Fedorov, O. (2013). A way to identify archaellins in Halobacterium salinarum archaella by FLAG-tagging. Open Life Sci 8, 828–834. doi: 10.2478/s11535-013-0202-0
Block, H., Maertens, B., Spriestersbach, A., Brinker, N., Kubicek, J., Fabis, R., et al. (2009). Immobilized-metal affinity chromatography (IMAC): a review. Methods Enzymol. 463, 439–473. doi: 10.1016/S0076-6879(09)63027-5
Braun, F., Thomalla, L., van der Does, C., Quax, T. E. F., Allers, T., Kaever, V., et al. (2019). Cyclic nucleotides in archaea: cyclic di-AMP in the archaeon Haloferax volcanii and its putative role. Microbiology 8:e00829. doi: 10.1002/mbo3.829
Butt, T. R., Edavettal, S. C., Hall, J. P., and Mattern, M. R. (2005). SUMO fusion technology for difficult-to-express proteins. Protein Expr. Purif. 43, 1–9. doi: 10.1016/j.pep.2005.03.016
DasSarma, S., Karan, R., DasSarma, P., Barnes, S., Ekulona, F., and Smith, B. (2013). An improved genetic system for bioengineering buoyant gas vesicle nanoparticles from haloarchaea. BMC Biotechnol. 13:112. doi: 10.1186/1472-6750-13-112
Djender, S., Beugnet, A., Schneider, A., and De Marco, A. (2014). The biotechnological applications of recombinant single-domain antibodies are optimized by the C-terminal fusion to the EPEA sequence (C tag). Antibodies 3, 182–191. doi: 10.3390/antib3020182
Du, M., Hou, Z., Liu, L., Xuan, Y., Chen, X., Fan, L., et al. (2022). Progress, applications, challenges and prospects of protein purification technology. Front. Bioeng. Biotechnol. 10:28691. doi: 10.3389/fbioe.2022.1028691
Dušeková, E., Berta, M., Sedláková, D., Řeha, D., Dzurillová, V., Shaposhnikova, A., et al. (2022). Specific anion effect on properties of HRV 3C protease. Biophys. Chem. 287:106825. doi: 10.1016/j.bpc.2022.106825
Dyall-Smith, M. (2008). The Halohandbook: Protocols for haloarchaeal genetics. Haloarchaeal Genetics Laboratory: Melbourne, VI, Australia, 14.
Einhauer, A., and Jungbauer, A. (2001). The FLAG™ peptide, a versatile fusion tag for the purification of recombinant proteins. J. Biochem. Biophys. Methods 49, 455–465. doi: 10.1016/S0165-022X(01)00213-5
Elleuche, S., Schröder, C., Sahm, K., and Antranikian, G. (2014). Extremozymes—biocatalysts with unique properties from extremophilic microorganisms. Curr. Opin. Biotechnol. 29, 116–123. doi: 10.1016/j.copbio.2014.04.003
Gomari, M. M., Saraygord-Afshari, N., Farsimadan, M., Rostami, N., Aghamiri, S., and Farajollahi, M. M. (2020). Opportunities and challenges of the tag-assisted protein purification techniques: applications in the pharmaceutical industry. Biotechnol. Adv. 45:107653. doi: 10.1016/j.biotechadv.2020.107653
Grote, A., Hiller, K., Scheer, M., Munch, R., Nortemann, B., Hempel, D. C., et al. (2005). JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, W526–W531. doi: 10.1093/nar/gki376
Grötzinger, S. W., Karan, R., Strillinger, E., Bader, S., Frank, A., Al Rowaihi, I. S., et al. (2018). Identification and experimental characterization of an extremophilic brine pool alcohol dehydrogenase from single amplified genomes. ACS Chem. Biol. 13, 161–170. doi: 10.1021/acschembio.7b00792
Haque, R. U., Paradisi, F., and Allers, T. (2019). Haloferax volcanii as immobilised whole cell biocatalyst: new applications for halophilic systems. Appl. Microbiol. Biotechnol. 103, 3807–3817. doi: 10.1007/s00253-019-09725-y
Haque, R. U., Paradisi, F., and Allers, T. (2020). Haloferax volcanii for biotechnology applications: challenges, current state and perspectives. Appl. Microbiol. Biotechnol. 104, 1371–1382. doi: 10.1007/s00253-019-10314-2
Hermans, P., Clasen, R., and Detmers, F. (2012). Reinventing affinity tags: innovative technology designed for routine purification of C-terminal EPEA-tagged recombinant proteins. Gen. Eng. Biotechnol. News 32, 48–49. doi: 10.1089/gen.32.17.20
Hopp, T. P., Prickett, K. S., Price, V. L., Libby, R. T., March, C. J., Pat Cerretti, D., et al. (1988). A short polypeptide marker sequence useful for recombinant protein identification and purification. Bio/Technology 6, 1204–1210. doi: 10.1038/nbt1088-1204
Ibrahim, T. N. B. T., Abas, A. B., and Razak, N. F. A. (2023). Escherichia coli cell factory for synthesis of biomolecules. Biomanuf. Sustain. Prod. Biomol. 7, 141–163. doi: 10.1007/978-981-19-7911-8_7
Jenny, R. J., Mann, K. G., and Lundblad, R. L. (2003). A critical review of the methods for cleavage of fusion proteins with thrombin and factor Xa. Protein Expr. Purif. 31, 1–11. doi: 10.1016/S1046-5928(03)00168-2
Jin, J., Hjerrild, K. A., Silk, S. E., Brown, R. E., Labbé, G. M., Marshall, J. M., et al. (2017). Accelerating the clinical development of protein-based vaccines for malaria by efficient purification using a four amino acid C-terminal C-tag. Int. J. Parasitol. 47, 435–446. doi: 10.1016/j.ijpara.2016.12.001
Karan, R., Mathew, S., Muhammad, R., Bautista, D. B., Vogler, M., Eppinger, J., et al. (2020). Understanding high-salt and cold adaptation of a polyextremophilic enzyme. Microorganisms 8:1594. doi: 10.3390/microorganisms8101594
Karan, R., Renn, D., Nozue, S., Zhao, L., Habuchi, S., Allers, T., et al. (2023). Bioengineering of air-filled protein nanoparticles by genetic and chemical functionalization. J. Nanobiotechnol. 21:108. doi: 10.1186/s12951-023-01866-7
Kimple, M. E., Brill, A. L., and Pasker, R. L. (2013). Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 73, 9.9.1–9.9.23. doi: 10.1002/0471140864.ps0909s73
Kimple, M. E., and Sondek, J. (2004). Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 36, 9.9.1–9.9.19. doi: 10.1002/0471140864.ps0909s36
Köppl, C., Lingg, N., Fischer, A., Kröß, C., Loibl, J., Buchinger, W., et al. (2022). Fusion tag design influences soluble recombinant protein production in Escherichia coli. Int. J. Mol. Sci. 23:7678. doi: 10.3390/ijms23147678
Kruglikov, A., Wei, Y., and Xia, X. (2022). Proteins from thermophilic Thermus thermophilus often do not fold correctly in a mesophilic expression system such as Escherichia coli. ACS Omega 7, 37797–37806. doi: 10.1021/acsomega.2c04786
Le, N. T. P., Phan, T. T. P., Phan, H. T. T., Truong, T. T. T., Schumann, W., and Nguyen, H. D. (2022). Influence of N-terminal His-tags on the production of recombinant proteins in the cytoplasm of Bacillus subtilis. Biotechnol. Rep. 35:e00754. doi: 10.1016/j.btre.2022.e00754
Martínez-Espinosa, R. M. (2019). Heterologous and homologous expression of proteins from haloarchaea: denitrification as case of study. Int. J. Mol. Sci. 21:82. doi: 10.3390/ijms21010082
Mishra, V. (2020). Affinity tags for protein purification. Curr. Protein Pept. Sci. 21, 821–830. doi: 10.2174/1389203721666200606220109
Nguyen, N. H., Phan, T. T. P., Tran, T. L., and Nguyen, H. D. (2014). Investigating the expression of GFP fused with HIS-TAG at N-or Cterminus using plasmid pHT253 and pHT254 in Bacillus subtilis. VNUHCM J. Sci. Technol. Dev. 17, 5–11. doi: 10.32508/stdj.v17i4.1550
Parks, T. D., Howard, E. D., Wolpert, T. J., Arp, D. J., and Dougherty, W. G. (1995). Expression and purification of a recombinant tobacco etch virus NIa proteinase: biochemical analyses of the full-length and a naturally occurring truncated proteinase form. Virology 210, 194–201. doi: 10.1006/viro.1995.1331
Pédelacq, J.-D., Cabantous, S., Tran, T., Terwilliger, T. C., and Waldo, G. S. (2006). Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24, 79–88. doi: 10.1038/nbt1172
Pérez-Arnaiz, P., Dattani, A., Smith, V., and Allers, T. (2020). Haloferax volcanii—a model archaeon for studying DNA replication and repair. Open Biol. 10:293. doi: 10.1098/rsob.200293
Phan, T. T. P., Tran, L. T., Schumann, W., and Nguyen, H. D. (2015). Development of P grac 100-based expression vectors allowing high protein production levels in Bacillus subtilis and relatively low basal expression in Escherichia coli. Microb. Cell Factories 14, 1–9. doi: 10.1186/s12934-015-0255-z
Porath, J., Carlsson, J., Olsson, I., and Belfrage, G. (1975). Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 258, 598–599. doi: 10.1038/258598a0
Puetz, J., and Wurm, F. M. (2019). Recombinant proteins for industrial versus pharmaceutical purposes: a review of process and pricing. PRO 7:476. doi: 10.3390/pr7080476
Schmidt, T. G. M., Batz, L., Bonet, L., Carl, U., Holzapfel, G., Kiem, K., et al. (2013). Development of the twin-Strep-tag® and its application for purification of recombinant proteins from cell culture supernatants. Protein Expr. Purif. 92, 54–61. doi: 10.1016/j.pep.2013.08.021
Schmidt, P. M., Sparrow, L. G., Attwood, R. M., Xiao, X., Adams, T. E., and McKimm-Breschkin, J. L. (2012). Taking down the FLAG! How insect cell expression challenges an established tag-system. PLoS One 7:e37779. doi: 10.1371/journal.pone.0037779
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schultz, J., dos Santos, A., Patel, N., and Rosado, A. S. (2023). Life on the edge: bioprospecting extremophiles for astrobiology. J Indian Inst Sci 103, 721–737. doi: 10.1007/s41745-023-00382-9
Shahravan, S. H., Qu, X., Chan, I. S., and Shin, J. A. (2008). Enhancing the specificity of the enterokinase cleavage reaction to promote efficient cleavage of a fusion tag. Protein Expr. Purif. 59, 314–319. doi: 10.1016/j.pep.2008.02.015
Shaner, N. C., Campbell, R. E., Steinbach, P. A., Giepmans, B. N. G., Palmer, A. E., and Tsien, R. Y. (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572. doi: 10.1038/nbt1037
Skerra, A., and Schmidt, T. G. M. (1999). Applications of a peptide ligand for streptavidin: the Strep-tag. Biomol. Eng. 16, 79–86. doi: 10.1016/S1050-3862(99)00033-9
Strillinger, E., Grötzinger, S. W., Allers, T., Eppinger, J., and Weuster-Botz, D. (2016). Production of halophilic proteins using Haloferax volcanii H1895 in a stirred-tank bioreactor. Appl. Microbiol. Biotechnol. 100, 1183–1195. doi: 10.1007/s00253-015-7007-1
Sysoev, M., Grötzinger, S. W., Renn, D., Eppinger, J., Rueping, M., and Karan, R. (2021). Bioprospecting of novel extremozymes from prokaryotes—the advent of culture-independent methods. Front. Microbiol. 12:630013. doi: 10.3389/fmicb.2021.630013
Tian, R., Liu, Y., Chen, J., Li, J., Liu, L., Du, G., et al. (2019). Synthetic N-terminal coding sequences for fine-tuning gene expression and metabolic engineering in Bacillus subtilis. Metab. Eng. 55, 131–141. doi: 10.1016/j.ymben.2019.07.001
Wang, Y., Qian, J., Shi, T., Wang, Y., Ding, Q., and Ye, C. (2024). Application of extremophile cell factories in industrial biotechnology. Enzym. Microb. Technol. 175:110407. doi: 10.1016/j.enzmictec.2024.110407
Waugh, D. S. (2011). An overview of enzymatic reagents for the removal of affinity tags. Protein Expr. Purif. 80, 283–293. doi: 10.1016/j.pep.2011.08.005
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362. doi: 10.1038/nmeth.1322
Yadav, D. K., Yadav, N., Yadav, S., Haque, S., and Tuteja, N. (2016). An insight into fusion technology aiding efficient recombinant protein production for functional proteomics. Arch. Biochem. Biophys. 612, 57–77. doi: 10.1016/j.abb.2016.10.012
Yeliseev, A., Zoubak, L., and Schmidt, T. G. M. (2017). Application of Strep-Tactin XT for affinity purification of twin-Strep-tagged CB(2), a G protein-coupled cannabinoid receptor. Protein Expr. Purif. 131, 109–118. doi: 10.1016/j.pep.2016.11.006
Yilmaz, G., and Arslanyolu, M. (2015). Efficient expression of codon-adapted affinity tagged super folder green fluorescent protein for synchronous protein localization and affinity purification studies in Tetrahymena thermophila. BMC Biotechnol. 15:22. doi: 10.1186/s12896-015-0137-9
Young, C. L., Britton, Z. T., and Robinson, A. S. (2012). Recombinant protein expression and purification: a comprehensive review of affinity tags and microbial applications. Biotechnol. J. 7, 620–634. doi: 10.1002/biot.201100155
Zhao, X., Li, G., and Liang, S. (2013). Several affinity tags commonly used in chromatographic purification. J. Anal. Methods Chem. 2013, 1–8. doi: 10.1155/2013/581093
Keywords: halophiles, archaea, extremophiles, purification tags, His-tag, Strep-tag®II, Haloferax volcanii, bioengineering
Citation: Karan R, Renn D, Allers T and Rueping M (2024) A systematic analysis of affinity tags in the haloarchaeal expression system, Haloferax volcanii for protein purification. Front. Microbiol. 15:1403623. doi: 10.3389/fmicb.2024.1403623
Edited by:
Mohammad Ali Amoozegar, University of Tehran, IranReviewed by:
Satya P. Singh, Saurashtra University, IndiaAdele Williamson, University of Waikato, New Zealand
Copyright © 2024 Karan, Renn, Allers and Rueping. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ram Karan, cmFta2FyYW5Ac291dGguZHUuYWMuaW4=; Dominik Renn, ZG9taW5pay5yZW5uQGthdXN0LmVkdS5zYQ==
 Ram Karan
Ram Karan Dominik Renn
Dominik Renn Thorsten Allers
Thorsten Allers Magnus Rueping
Magnus Rueping