- 1Department of Public Health, Shihezi University School of Medicine, Shihezi, Xinjiang, China
- 2Key Laboratory for Prevention and Control of Emerging Infectious Diseases and Public Health Security, The Xinjiang Production and Construction Corps, Shihezi, Xinjiang, China
- 3Department of Public Health and Key Laboratory of Xinjiang Endemic and Ethnic Diseases of the Ministry of Education, School of Medicine, Shihezi University, Shihezi, Xinjiang, China
Background: Type 1 diabetes mellitus (T1DM) is a chronic metabolic disease that seriously jeopardizes human physical and mental health and reduces quality of life. Intestinal flora is one of the critical areas of exploration in T1DM research.
Objective: This study aims to explore the research hotspot and development trend of T1DM and intestinal flora to provide research direction and ideas for researchers.
Methods: We used the Web of Science (WOS) Core Collection and searched up to 18 November 2023, for articles on studies of the correlation between T1DM and intestinal flora. CiteSpace, VOSviewers and R package “bibliometrix” were used to conduct this bibliometric analysis.
Results: Eventually, 534 documents met the requirements to be included, and as of 18 November 2023, there was an upward trend in the number of publications in the field, with a significant increase in the number of articles published after 2020. In summary, F Susan Wong (UK) was the author with the most publications (21), the USA was the country with the most publications (198), and the State University System of Florida (the United States) was the institution with the most publications (32). The keywords that appeared more frequently were T cells, fecal transplants, and short-chain fatty acids. The results of keywords with the most robust citation bursts suggest that Faecalibacterium prausnitzii and butyrate may become a focus of future research.
Conclusion: In the future, intestinal flora will remain a research focus in T1DM. Future research can start from Faecalibacterium prausnitzii and combine T cells, fecal bacteria transplantation, and short-chain fatty acids to explore the mechanism by which intestinal flora affects blood glucose in patients with T1DM, which may provide new ideas for the prevention and treatment of T1DM.
1 Introduction
Type 1 diabetes mellitus (T1DM) is a metabolic disorders caused by autoimmunity leading to a lack of insulin secretion from pancreatic beta cells (Zhang et al., 2022a). According to data published by the International Diabetes Federation (IDF) in 2021, approximately 1.2 million people worldwide with T1DM were adolescents and children (Saeedi et al., 2019). The low age and poor cognition of most T1DM patients at the onset of the disease make glycemic control very difficult and complication rates high, seriously jeopardizing the lives and health of patients (Bruno et al., 2013; Katsarou et al., 2017; Li et al., 2022; Riveline et al., 2022). The pathogenesis of T1DM is genetic, and non-genetic factors play an increasingly important role in it (Christovich and Luo, 2022; Zhang et al., 2022c). In recent years, there has been a growing interest in non-genetic factors in intestinal flora changes such as the pathogenesis of T1DM (Del Chierico et al., 2022). The intestinal flora of patients with T1DM differs from that of healthy individuals. For instance, the Firmicutes to Bacteroidetes ratio significantly decreased, with the quantity of Bacteroidetes significantly increasing for healthy children (Murri et al., 2013). Intestinal flora affected host metabolism and immune network activity, ultimately influencing diabetes development (Musso et al., 2011). In addition, intestinal flora also plays an important role in the complications of T1DM. T1DM patients with depression have a higher abundance of Megasphaera than those without depression (Liu et al., 2024). Moreover, the metabolites of intestinal flora also play an important role in T1DM. The tryptophan metabolites mediated by intestinal flora are an important marker for distinguishing between rapid and chronic progressors of T1DM (Lamichhane et al., 2023). The phenolic acids produced by beneficial bacteria have high antioxidant, anti-diabetic, and anti-inflammatory properties, which can accelerate T cell differentiation, increase the number of Treg, and inhibit the release of inflammatory cytokines in macrophages (Hu et al., 2023). Most patients with type 1 diabetes do not have a family history. Low genetic risk does not rule out the development of clinical type 1 diabetes. Ruminococcus is a powerful determinant for distinguishing control infants from future type 1 diabetes infants in a random forest analysis (Bélteky et al., 2023). Current research has found that the results of T1DM and intestinal flora in terms of α and β diversity are inconsistent (Traversi et al., 2022; Flasbeck et al., 2023). In addition, current research focuses on bacteria, with few studies exploring fungi and viruses. Coxsackievirus B infection is considered to be a modulator of beta cell autoimmunity, which may be developed into a vaccine to reduce the incidence of T1DM (Traversi et al., 2022). Therefore, exploring the intestinal flora is essential for studying the mechanism of T1DM. In the past, many scholars focused on exploring the mechanisms and therapeutic means for the occurrence of T1DM with the intestinal flora, including intestinal permeability. Many cutting-edge technological approaches, such as macro-genomics and metabolomics, have also been applied to this field (Stewart et al., 2018). However, the specific mechanisms between T1DM and intestinal flora and new research hotspots remain to be explored.
Through statistical techniques, bibliometric analysis allows for analyzing research trends (Zhu et al., 2021). The methodology has been created and applied to various industries, including studies in analytical chemistry, environmental studies, and epidemiology (Fu et al., 2022; Yang et al., 2022; Zhong et al., 2022). Currently, we found only one bibliometric study on T1DM and intestinal flora. Zhang et al.’s (2022b) study only selected the literature on diabetes and intestinal flora correlation studies conducted between 2011 and 2021, with a collection timeframe of only ten years and a discussion focusing on Type 2 Diabetes Mellitus. Therefore, this study conducted a bibliometric analysis based on the records of literature published up to 18 November 2023, including the year, keywords, authors, country, institution, and co-citations of the relevant research publications, to identify the research hotspots and trends in the field, and to provide a reference for future research and applications in the field of T1DM and intestinal flora.
2 Materials and methods
2.1 Data source
The original data for this study were obtained from the Web of Science (WOS) Core Collection, searched up to 18 November 2023, for literature published during the period.
2.2 Data search strategy
To obtain as comprehensive data as possible, this study was searched with the help of the Medical Subject Headings (MeSHs), using T1DM AND intestinal flora and its upstream and downstream terms as the subject terms according to the search rules of Web of Science. In the Supplementary material, specific search tactics were given.
2.3 Data extraction and collection
The type of literature included in this study only had research articles and reviews, and the search language was limited to English. The screening of this study was carried out independently by two researchers simultaneously. A total of 2,134 records were searched, and the titles, abstracts, and keywords were read to determine whether the content had a clear association with “T1DM” or “intestinal flora” and then manually filtered. If these were insufficient to determine whether there was a clear correlation between “T1DM and intestinal flora,” the full text was reviewed. The two researchers retrieved 547 and 517 articles after de-duplication and exclusion of literature that did not meet the inclusion criteria. The results of the inclusion analyses were exchanged, and the third researcher decided to include those articles that were difficult to have due to disagreement. In the end, 534 articles met the inclusion criteria. The flow chart of the study is shown in Supplementary Figure 1.
2.4 Bibliometric analysis
The 534 documents that met the criteria were selected for the study, exported and downloaded as Plain text files, Tab delimited files and BibTeX, and the output journals and authors were identified using VOSviewers (1.6.18) to construct a relevant network of major co-cited journals and authors. Keywords, countries, institutions, and journals were extracted and organized according to the frequency of occurrence using CiteSpace (6.1.R3), and co-occurrence mapping was generated. The R package “bibliometrix” (version 4.3.1) was applied to visualize the geographical location and cooperation of countries. Annual articles and co-citations were managed using Microsoft Excel (2019) to collect data and analyze publication trends. Ethical approval was not applicable for the present study.
3 Results
3.1 Publication outputs and trends
The current study included 534 on T1DM and intestinal flora up to 18 November 2023. As shown in Figure 1, the number of publications was low from 2004 to 2010, all of which were ten or less. From 2010 onward, the number of articles in this field gradually increased, and studies related to intestinal flora and T1DM received widespread attention. The number of citations in this field in the WoSCC peaked in 2022.
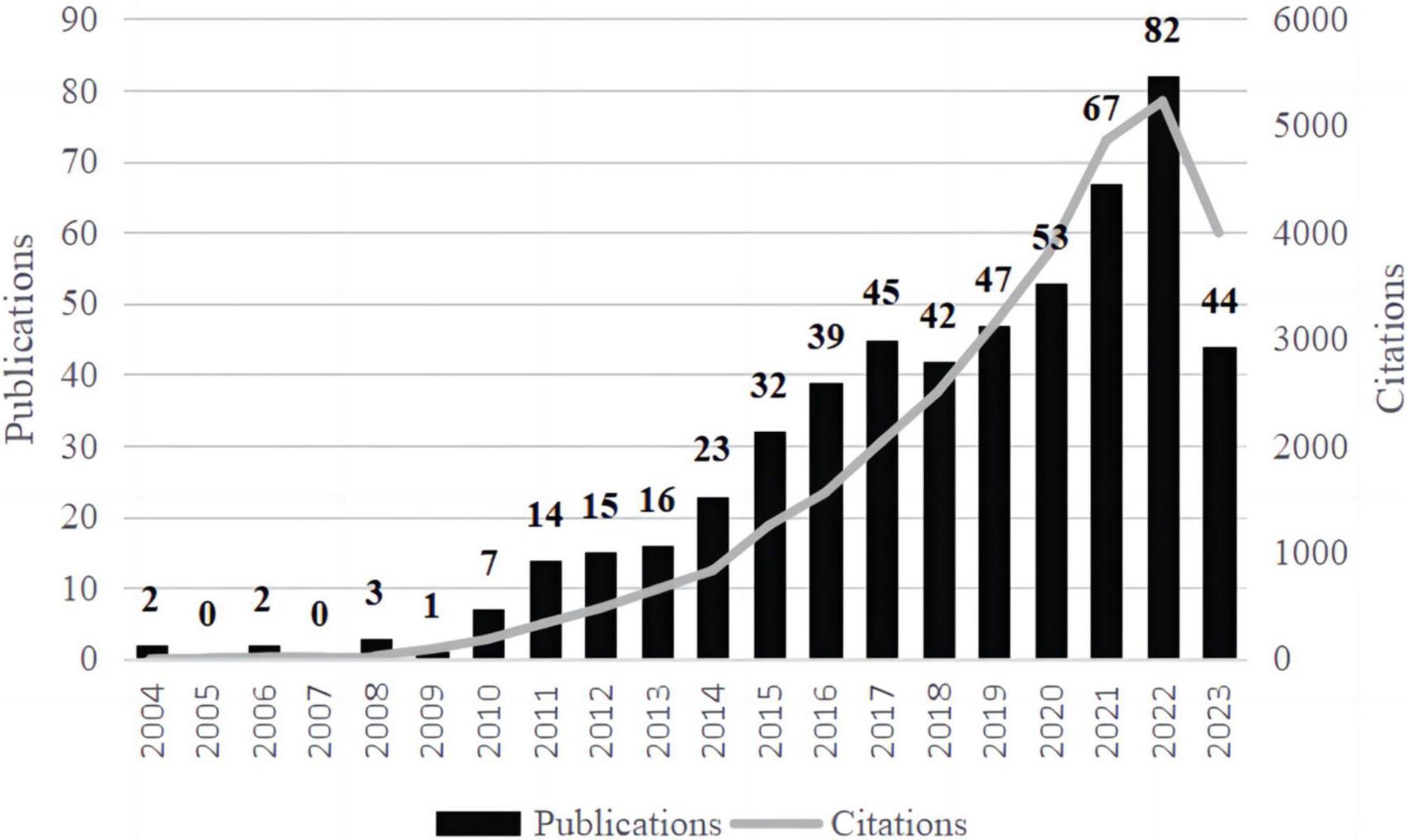
Figure 1. Trends in posting volume and citation frequency annually. The broken line represented the paper’s citation frequency, and the bar graph represented the number of publications of the article.
3.2 Analysis of journals output
Studies on the correlation between T1DM and intestinal flora were published in 242 journals. In the accompanying Supplementary Figure 2, The dual-map overlay of journals was superimposed to show the distribution of topics. Supplementary Figure 3 depicted the association between journals and time. The journals that have been active in publishing articles in this field in the last five years are Frontiers in Immunology (IF: 7.3), International Journal of Molecular Sciences (IF: 5.6), and so on, which were in the top ten journals in terms of the number of articles issued. The top ten journals regarding the number of reports issued were shown in Table 1, all of which were in Journal Citation Report (JCR)1 Q1 and Q2. The journal with the highest impact factor is Diabetologia (IF: 8.2), followed by Diabetes (IF: 7.7).
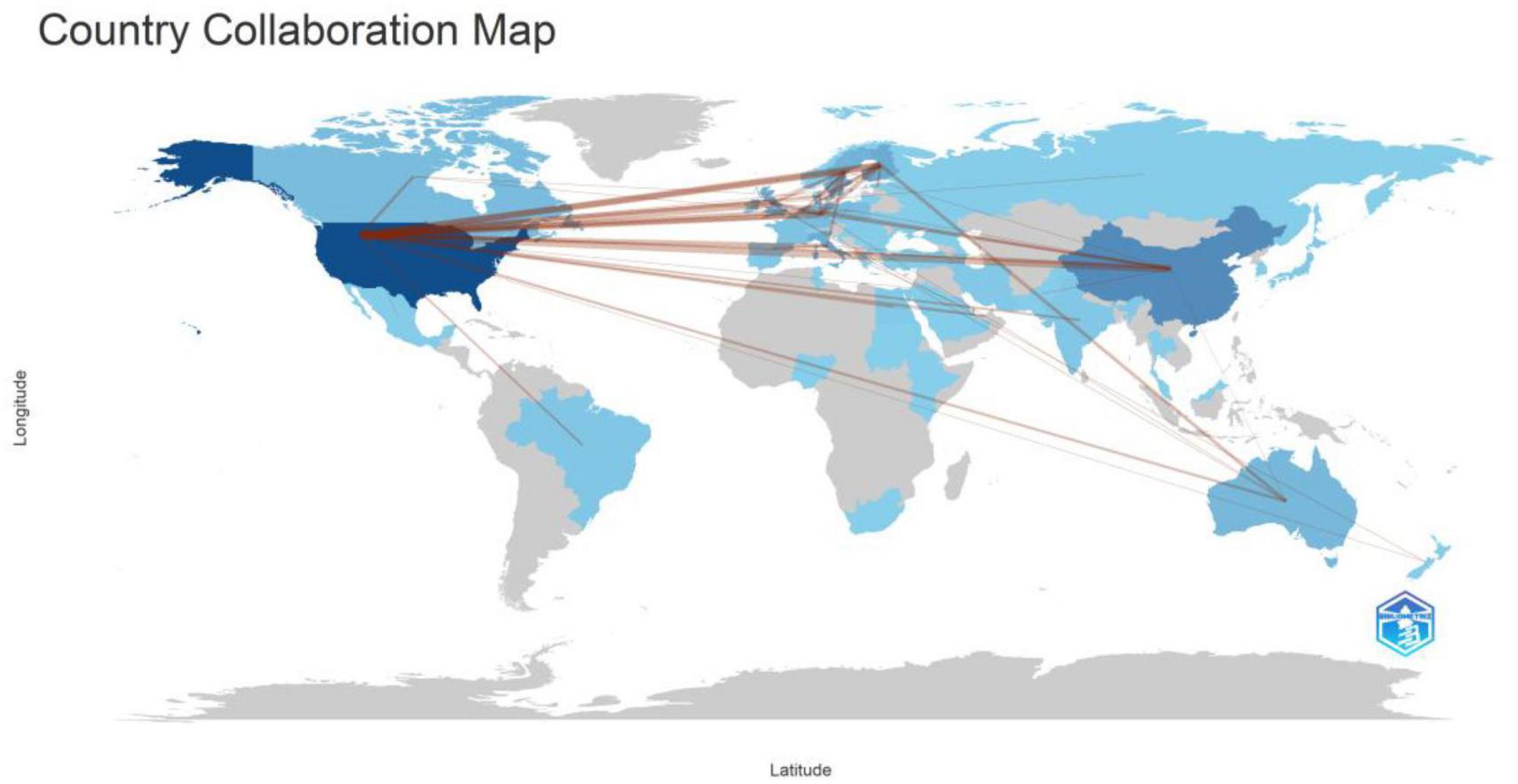
3.3 Analysis of country cooperation
Studies on T1DM and intestinal flora were conducted in 206 countries or regions. We filtered and visualized these country states and constructed a collaborative network based on the number of publications and relationships in each country (Figure 2). Figure 3A depicted the partnership between individual countries, with the ability of governments to collaborate increasing with increasing centrality. Supplementary Table 1 listed the ten countries with the most published articles. The United States ranks first with 198 pieces, followed by China and Finland, with over 50 publications.
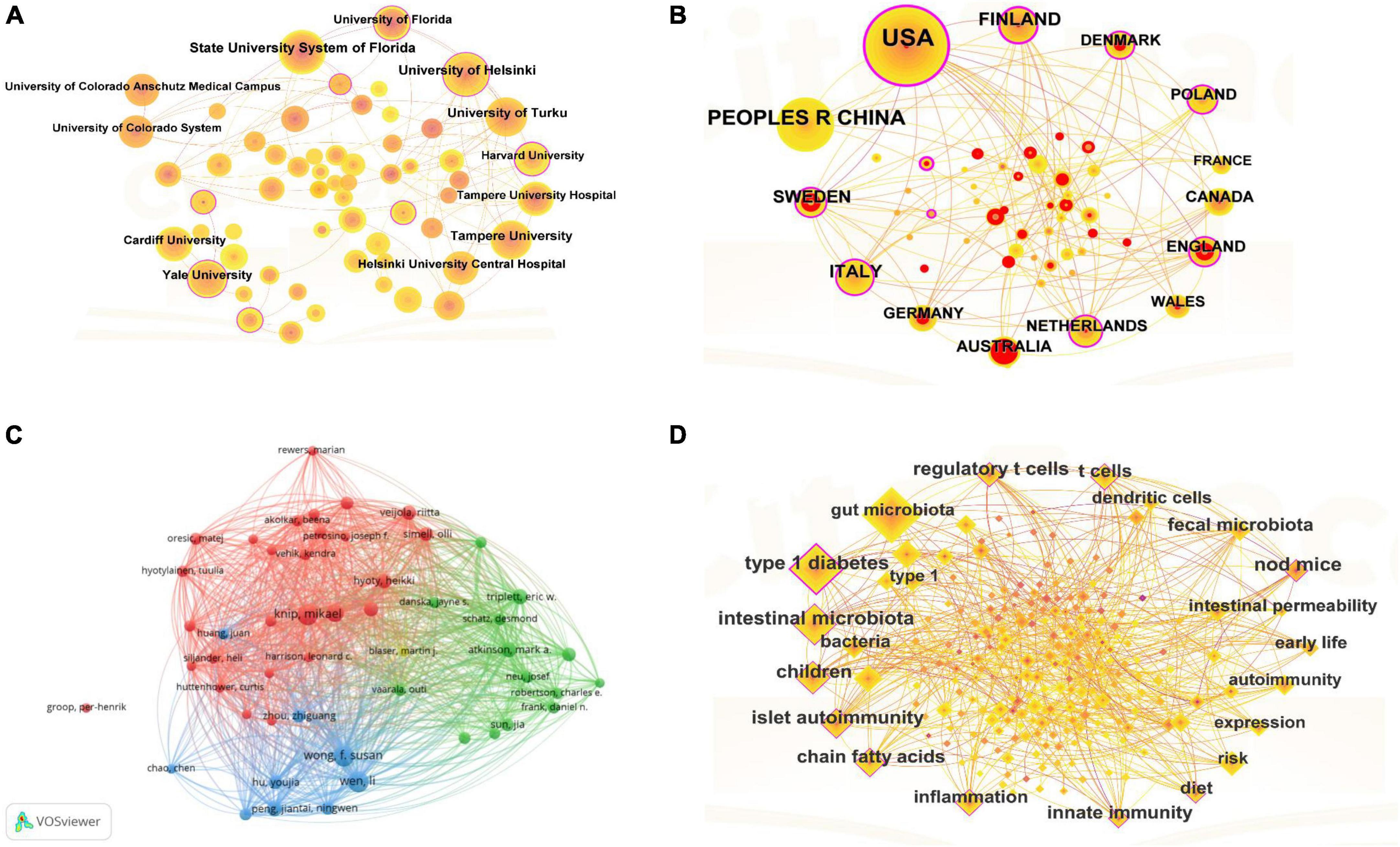
Figure 3. (A) Each country was represented as a node, and the node size was proportional to the sum of publications. Different nodes were given different colors according to the color gradient in the lower right corner. (B) In this map, a node represented an institution, and the size of each node represented its relative quantity of research output. Each line represented the strength of the cooperation relationship between the two institutions, and strength value was displayed between the lines. (C) Each author was represented as a node, and the node size was proportional to the sum of citations. A link between two nodes indicated a co-citation relationship. The distance between nodes indicated the relatedness, and a smaller distance implied a higher relatedness and would be assigned to one cluster with the same colors. (D) Each keyword represented a node; the node size was proportional to the number of documents, and the line between the two nodes represented the connection between the keywords.
3.4 Analysis of institutional cooperation
There were 2,081 institutions involved in research on T1DM and intestinal flora, and the partnerships between institutions were depicted in Figure 3B. The top five institutions in terms of the number of publications were State University System of Florida (32), University of Helsinki (30), University of Turku (26), Tampere University (24) and Yale University (24). Notably, no Chinese institutions were among the top five regarding publications. High-productivity institutions led institutional collaborations, and each institution was well-communicated and closely connected with Yale University or the State University System of Florida and other universities, respectively.
3.5 Analysis of author collaboration network
There were 2,796 authors involved in the T1DM and intestinal flora studies. Figure 3C described the authors’ co-citations using VOSviewers (1.6.18). In the visualization map, the authors formed their collaborative teams, and the different colored clusters reflected the collaborative relationship between the authors. In the red group, Knip, Mikael and Lernmark, Ake were the members of the “The Environmental Determinants of Diabetes in Young People” (TEDDY). They have worked primarily with children and have co-authored 75 articles and published in highly rated journals such as the Jama-Journal of the American Medical Association (IF = 120.7) (Aronsson et al., 2019). T1DM genetically susceptible children, higher gluten intake in the first five years was associated with increased risk of celiac disease autoimmunity and celiac disease. In the blue cluster, Wong, F. Susan and Wen, Li have co-authored 106 publications, mostly in mice, and the two have published an article in Nature proposing that the interaction between gut microbes and the innate immune system was a critical epigenetic factor that modifies susceptibility to T1DM (Pearson et al., 2023). The green group needed more collaboration between authors and more publications.
3.6 Analysis of co-cited references
Supplementary Table 2 listed the 15 most cited articles. Among them, in terms of research methodology, four pieces used single-omics analyses, such as macro-genomics and metabolomics, and five parts used multiple-omics analyses. In terms of study design, three teams designed case-control studies, and two teams designed cohort studies with sample sizes of 33 and 222, respectively, all in Northern Europe. Two other groups, part of the TEDDY study, created cohort and case-control studies of 903 and 783 children from six clinical centers in four countries (Finland, Germany, Sweden, and the USA).
3.7 Analysis of keywords
Figure 3D showed that 20 keywords appeared more than 30 times in this field. In terms of frequency, in addition to the keywords related to the subject terms “intestinal flora” and “T1DM,” the keywords with a frequency of more than 30 and had a high degree of centrality included “T cells,” “short-chain fatty acids,” and “fecal transplants,” and the rest of the keywords were closely related.
Supplementary Table 3 showed the top 35 terms with the most robust citation bursts in the field, butyrate, and Faecalibacterium prausnitzii appearing in 2021 and continuing to this day.
4 Discussion
In this study, we conducted a systematic bibliometric assessment of the literature on correlation studies between T1DM and intestinal flora from 18 November 2023.
4.1 Global publication and journal trends
Changes in the number of academic publications were an essential indicator of the development trend of a field (Peng et al., 2020). With the year-on-year increase in publications and co-citations, we have clarified that studying the correlation between T1DM and intestinal flora has become a research hotspot. The number of literature in the field will increase further with the deepening of the molecular mechanism of T1DM and intestinal flora.
The research on T1DM and intestinal flora was extensive and published in 242 journals. Supplementary Figure 2 showed that literature published in Molecular, Biology, and Genetics journals were cited in the same journals and Medicine, Medical, and Clinical journals with a higher. The high frequency implied that the current research on intestinal flora and T1DM mainly focused on basic medical research. Supplementary Figure 3 showed that articles in this field can be published in journals in multiple areas, such as GUT, MICROBIOME, and NUTRIENTS. Prospective scholars can prioritize research outputs from journals with the highest number of publications and the top three impact factors (Diabetologia, Diabetes, Frontiers in Immunology) to gain access to their cutting-edge information.
4.2 Countries, institutions, and authors
Combining Figures 2, 3A, B, There was much optimistic cooperation between the USA, European countries, and China. Among the top five countries in terms of the number of publications, the United States and China have the highest number of publications, and they also have the top five diabetics in the world. The top five institutions in the number of publications are universities in developed countries (Finland and the United States). However, there was no Chinese institution among the top five institutions. This suggested that research on the correlation between T1DM and the intestinal flora required a certain degree of academic atmosphere and financial support. Combined with Figure 3C, the team leaders of each cluster have published in highly rated journals, which researchers can read for easy reference and learning of cutting-edge content. Although the authors within each collection worked closely together and produced a lot of collaborative outputs, the collaboration between team leaders was not strong, and it was expected that researchers could strengthen their cooperation in the future to promote the development of research in the field of T1DM and intestinal flora.
The top-ranked issuing organizations, countries, and authors were more closely linked to each other, suggesting that collaboration would lead to more productive outputs. Meanwhile, analyzing the dynamics of intestinal flora required extensive cohort studies. Individual country studies include the Type 1 Diabetes Prediction and Prevention (DIPP) Study (Nanto-Salonen et al., 2008) and the All Babies in South-East Sweden (ABIS) study (Russell et al., 2019). Multinational studies in several countries include the “The Environmental Determinants of Diabetes in Young People” (TEDDY) study (Stewart et al., 2018; Vatanen et al., 2018), one of the most extensive infant microbiome datasets to date, of which Knip, Mikael, and Lernmark, Ake, who were members of the Red Cluster of the Authors’ Collaborative Network and whose results have also been published in highly rated journals. Collaboration between different countries can collect samples from other regions, integrating different geographic environments and living habits and exploring their effects on the composition of the intestinal flora (Abela and Fava, 2019). However, no extensive birth cohort studies have been conducted in Asian countries such as China or African and South American countries.
4.3 References co-citation analysis
The top fifteen cited literature in Supplementary Table 2 were published in journals with high-impact factors. Cohort and case-control studies were the main research methods, and both had advantages and disadvantages. Case-control studies were less costly, but the sample was poorly represented, and care needed to be taken to control the matching of dietary habits, duration of breastfeeding, mode of delivery, and physiological factors such as age, gender, ethnicity, and geographic factors (Murri et al., 2013). Cohort studies were costly and proved to loss of visits. Notably, some researchers combined the design of cohort studies with case-control study methodology and published them in Nature, providing a new idea to explore the relationship between dynamic changes in intestinal flora and T1DM (Stewart et al., 2018).
Researchers were currently using single-omics, such as 16S rRNA sequencing and Metagenome sequencing, for more analytical studies of intestinal flora. 16S rRNA analytical methods usually do not provide a functional genetic composition of the bacterial community. In contrast, Metagenome sequencing studies were particularly well suited to characterize the microbial assemblage at a functional level (Hamady and Knight, 2009). Metabolomics analyses can help researchers understand cellular metabolism and reveal the microbiome’s impact on human health and disease (Joice et al., 2014; Wishart, 2016). The limitation was that metabolomics only provides static information on metabolite abundance. However, researchers have made less use of multi-omics, and little literature has been published on applying lipidomics and proteomics in intestinal flora related to T1DM, which is expected to be developed and used by researchers.
4.4 Hot trends and hot changes
Combined with the keyword analysis results in Figure 3D, T cells appeared most frequently (excluding the subject term). Several specific bacteria have been shown to influence T1DM by modulating the phenotypic differentiation of T cells (Krych et al., 2015). Clostridial clusters IV and XIVa, Bacteroides fragilis, and Lactobacillus can generate regulatory T cells (Atarashi et al., 2013). However, the dominant microorganisms associated with T1DM Acetatifactor, Coprococcus_2, or Lachnoclostridium and T cells have not been published and need further investigation in future work. Gazali et al. (2020) used the most extensive and rigorous age-matched cohort study analyzed to demonstrate that peripheral blood MAIT cell alterations were associated with the clinical onset of T1DM. However, their research found reduced expression of β7 integrins on regular CD3 T cells in children with T1DM. Others have not verified this result, suggesting a defect in mucosal T-cell homing in children with T1DM, which should be investigated in more detail in the future (Gazali et al., 2020).
In addition, we found that fecal microbiota transplantation (FMT) has increased in research fervor in recent years. A recently published long-term cohort study showed that FMT had a favorable short- and long-term safety profile without increasing the risk of infection transmission and new-onset disease (Tian et al., 2022). A longitudinal randomized controlled study published by de Groot et al. (2021) showed that when FMT was administered to patients with a recent diagnosis of TIDM, FMT prevented the decline in endogenous insulin secretion in patients after 12 months (de Groot et al., 2021). This was the first report that FMT can affect residual β-cells in vivo in patients with new-onset T1DM, and future researchers can continue to add in this direction. However, the ability of the newly established intestinal flora to remain homeostatic for an extended period after performing FMT and its ability to match rather than exacerbate the immune response highly were issues that researchers needed to be concerned about when conducting experiments (Halaweish et al., 2022; Zhang et al., 2022c).
Also appearing at a higher frequency in the keywords were short-chain fatty acids (SCFAs). SCFAs were produced by the fermentation of dietary fiber by intestinal flora, which can suppress the immune response and control the symptoms of T1DM (Wahlstrom et al., 2016). Recent animal studies have shown that altered metabolism of intestinal SCFAs plays a causal role in T1DM (Desai et al., 2016; Marino et al., 2017). Butyrate was one of the main types of SCFAs produced in the colon. Yuan et al. (2022) found in an independent cohort that the microbiota of children with new-onset TIDM was characterized by reduced butyrate production at the species, gene, and metabolite levels. In streptozotocin-induced TIDM mice, butyrate was protective of islet structure and function. Another short-term cohort study had shown that targeting dietary SCFAs may be a mechanism to alter the immune profile, promote immune tolerance, and improve glycemic control for the treatment of T1DM (Bell et al., 2022). However, it has also been demonstrated that oral butyrate treatment in patients with T1DM resulted in a significant decrease in fecal SCFA levels but no change in residual β-cell function, which was inconsistent with findings in the mouse model of T1DM (de Groot et al., 2020). This may be related to the method of administration of butyrate, differences in administration and dosage, differences in pathophysiology between humans and mice, and other aspects. The discrepancy in the results of this study needs to be examined in subsequent studies.
Shilo et al. confirmed that fecal bacterium prausnitzii was one of the most commonly altered intestinal bacterial species in patients with T1DM (Jamshidi et al., 2019; Shilo et al., 2022). A study showed a negative correlation between Faecalibacterium abundance and glycated hemoglobin compared to a healthy population (Rouland et al., 2022). The exact causal mechanism between the two has yet to be reported. The intestinal mucosal changes in T1DM may be related to intestinal inflammation, which may be due to the low abundance of Faecalibacterium prausnitzii and other butyrate-producing bacteria (Louis and Flint, 2009). Experimental studies by investigators were needed to determine whether using Faecalibacterium prausnitzii as a probiotic had therapeutic or preventive value for intestinal inflammation in patients with T1DM.
5 Conclusion
This study used CiteSpace (6.1.R3), VOSviewers (1.6.18) and The R package “bibliometrix” (version 4.3.1) to evaluate 534 literatures as of 18 November 2023 on research on T1DM and intestinal flora. Research on T1DM and intestinal flora was increasing, with 206 countries, 2,081 institutions, and 2,796 authors conducting research in this field. Longitudinal studies with long-term follow-up observations are recommended in this field of research to explore the relationship between T cells, fecal bacterial transplantation, and short-chain fatty acids and to explore the therapeutic value of Faecalibacterium prausnitzii, which may provide new ideas for the prevention and treatment of T1DM.
Data availability statement
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
XC: Writing – original draft, Writing – review & editing. ZW: Writing – original draft, Writing – review & editing. YZ: Writing – review & editing. LD: Writing – review & editing. YC: Writing – review & editing. HH: Writing – review & editing. XS: Writing – review & editing. YL: Methodology, Project administration, Supervision, Writing – review & editing. HW: Methodology, Project administration, Supervision, Writing – review & editing. LZ: Methodology, Project administration, Supervision, Writing – review & editing. JH: Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (No. 81560551), Shihezi University High-Level Talent Research Launch Project (No. RCZK202367), and the Shihezi University Innovation Outstanding Young Talents Program (Natural Science) (No. CXPY202004).
Acknowledgments
We thank Professor JH for reviewing the bibliometric methodology employed in this edition. We are also grateful to our collaborators from the Department of Public Health and Key Laboratory for Prevention and Control of Emerging Infectious Diseases and Public Health Security and Department of Public Health and Key Laboratory of Xinjiang Endemic and Ethnic Diseases of the Ministry of Education for their involvement in the full-text reviews.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1403514/full#supplementary-material
Footnotes
References
Abela, A. G., and Fava, S. (2019). Incidence of type 1 diabetes and distance from the sea: A descriptive epidemiological study. J. Diabetes 11, 345–347. doi: 10.1111/1753-0407.12862
Aronsson, C. A., Lee, H. S., Segerstad, E. M. H., Uusitalo, U., Yang, J. M., Koletzko, S., et al. (2019). Association of gluten intake during the first 5 years of life with incidence of celiac disease autoimmunity and celiac disease among children at increased risk. JAMA 322, 514–523. doi: 10.1001/jama.2019.10329
Atarashi, K., Tanoue, T., Oshima, K., Suda, W., Nagano, Y., Nishikawa, H., et al. (2013). T-reg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232. doi: 10.1038/nature12331
Bell, K. J., Saad, S., Tillett, B. J., and Helen, M. (2022). Metabolite-based dietary supplementation in human type 1 diabetes is associated with microbiota and immune modulation. Microbiome 10:9. doi: 10.1186/s40168-021-01193-9
Bélteky, M., Milletich, P. L., Ahrens, A. P., Triplett, E. W., and Ludvigsson, J. (2023). Infant gut microbiome composition correlated with type 1 diabetes acquisition in the general population: The ABIS study. Diabetologia 66, 1116–1128. doi: 10.1007/s00125-023-05895-7
Bruno, G., Spadea, T., Picariello, R., Gruden, G., Barutta, F., Cerutti, F., et al. (2013). Early life socioeconomic indicators and risk of type 1 diabetes in children and young adults. J. Pediatr. 162:600. doi: 10.1016/j.jpeds.2012.09.010
Christovich, A., and Luo, X. M. (2022). Gut microbiota, leaky gut, and autoimmune diseases. Front. Immunol. 13:946248. doi: 10.3389/fimmu.2022.946248
de Groot, P. F., Nikolic, T., Imangaliyev, S., Bekkering, S., Duinkerken, G., Keij, F. M., et al. (2020). Oral butyrate does not affect innate immunity and islet autoimmunity in individuals with longstanding type 1 diabetes: A randomised controlled trial. Diabetologia 63, 597–610. doi: 10.1007/s00125-019-05073-8
de Groot, P., Nikolic, T., Pellegrini, S., Sordi, V., Imangaliyev, S., Rampanelli, E., et al. (2021). Faecal microbiota transplantation halts progression of human new-onset type 1 diabetes in a randomised controlled trial. Gut 70, 92–105. doi: 10.1136/gutjnl-2020-322630
Del Chierico, F., Rapini, N., Deodati, A., Matteoli, M. C., Cianfarani, S., and Putignani, L. (2022). Pathophysiology of type 1 diabetes and gut microbiota role. Int. J. Mol. Sci. 23:650. doi: 10.3390/ijms232314650
Desai, M. S., Seekatz, A. M., Koropatkin, N. M., Kamada, N., Hickey, C. A., Wolter, M., et al. (2016). A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167:1339. doi: 10.1016/j.cell.2016.10.043
Flasbeck, V., Hirsch, J., Petrak, F., Meier, J. J., Herpertz, S., Gatermann, S., et al. (2023). Microbiome composition and central serotonergic activity in patients with depression and type 1 diabetes. Eur. Arch. Psychiatry Clin. Neurosci. doi: 10.1007/s00406-023-01694-8
Fu, L., Mao, S., Chen, F., Zhao, S., Su, W., Lai, G., et al. (2022). Graphene-based electrochemical sensors for antibiotic detection in water, food and soil: A scientometric analysis in CiteSpace (2011-2021). Chemosphere 297:127. doi: 10.1016/j.chemosphere.2022.134127
Gazali, A. M., Schroderus, A. M., Nanto-Salonen, K., Rintamaki, R., Pihlajamaki, J., Knip, M., et al. (2020). Mucosal-associated invariant T cell alterations during the development of human type 1 diabetes. Diabetologia 63, 2396–2409. doi: 10.1007/s00125-020-05257-7
Halaweish, H. F., Boatman, S., and Staley, C. (2022). Encapsulated fecal microbiota transplantation: Development, efficacy, and clinical application. Front. Cell. Infect. Micrbiol. 12:17. doi: 10.3389/fcimb.2022.826114
Hamady, M., and Knight, R. (2009). Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 19, 1141–1152. doi: 10.1101/gr.085464.108
Hu, J., Ding, J., Li, X., Li, J., Zheng, T., Xie, L., et al. (2023). Distinct signatures of gut microbiota and metabolites in different types of diabetes: A population-based cross-sectional study. EClinicalMedicine 62:102132. doi: 10.1016/j.eclinm.2023.102132
Jamshidi, P., Hasanzadeh, S., Tahvildari, A., Farsi, Y., Arbabi, M., Mota, J. F., et al. (2019). Is there any association between gut microbiota and type 1 diabetes? A systematic review. Gut Pathog. 11:7. doi: 10.1186/s13099-019-0332-7
Joice, R., Yasuda, K., Shafquat, A., Morgan, X. C., and Huttenhower, C. (2014). Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 20, 731–741. doi: 10.1016/j.cmet.2014.10.003
Katsarou, A., Gudbjornsdottir, S., Rawshani, A., Dabelea, D., Bonifacio, E., Anderson, B. J., et al. (2017). Type 1 diabetes mellitus. Nat. Rev. Dis. Prim. 3:17. doi: 10.1038/nrdp.2017.16
Krych, L., Nielsen, D. S., Hansen, A. K., and Hansen, C. H. F. (2015). Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-gamma level in NOD Mice. Gut Microbes 6, 101–109. doi: 10.1080/19490976.2015.1011876
Lamichhane, S., Sen, P., Dickens, A. M., Kråkström, M., Ilonen, J., Lempainen, J., et al. (2023). Circulating metabolic signatures of rapid and slow progression to type 1 diabetes in islet autoantibody-positive children. Front. Endocrinol. 14:1211015. doi: 10.3389/fendo.2023.1211015
Li, G. H., Huang, K., Dong, G. P., Zhang, J. W., Gong, C. X., Luo, F. H., et al. (2022). Clinical incidence and characteristics of newly diagnosed type 1 diabetes in chinese children and adolescents: A nationwide registry study of 34 medical centers. Front. Pediatr. 10:888370. doi: 10.3389/fped.2022.888370
Liu, Z., Yue, T., Zheng, X., Luo, S., Xu, W., Yan, J., et al. (2024). Microbial and metabolomic profiles of type 1 diabetes with depression: A case-control study. J. Diabetes 16:e13542. doi: 10.1111/1753-0407.13542
Louis, P., and Flint, H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x
Marino, E., Richards, J. L., McLeod, K. H., Stanley, D., Yap, Y. A., Knight, J., et al. (2017). Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat. Immunol. 18, 552–562. doi: 10.1038/ni.3713
Murri, M., Leiva, I., Gomez-Zumaquero, J. M., Tinahones, F. J., Cardona, F., Soriguer, F., et al. (2013). Gut microbiota in children with type 1 diabetes differs from that in healthy children: A case-control study. BMC Med. 11:46. doi: 10.1186/1741-7015-11-46
Musso, G., Gambino, R., and Cassader, M. (2011). Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu. Rev. Med. 62, 361–380. doi: 10.1146/annurev-med-012510-175505
Nanto-Salonen, K., Kupila, A., Simell, S., Siljander, H., Salonsaari, T., Hekkala, A., et al. (2008). Nasal insulin to prevent type 1 diabetes in children with HLA genotypes and autoantibodies conferring increased risk of disease: A double-blind, randomised controlled trial. Lancet 372, 1746–1755. doi: 10.1016/S0140-6736(08)61309-4
Pearson, J. A., Peng, J., Huang, J., Yu, X. Q., Tai, N. W., Hu, Y. J., et al. (2023). NLRP6 deficiency expands a novel CD103(+) B cell population that confers immune tolerance in NOD mice. Front. Immunol. 14:1147925. doi: 10.3389/fimmu.2023.1147925
Peng, C., He, M., Cutrona, S. L., Kiefe, C. I., Liu, F., and Wang, Z. (2020). Theme trends and knowledge structure on mobile health apps: Bibliometric analysis. Jmir Mhealth Uhealth 8:13. doi: 10.2196/18212
Riveline, J. P., Verges, B., Detournay, B., Picard, S., Benhamou, P. Y., Bismuth, E., et al. (2022). Design of a prospective, longitudinal cohort of people living with type 1 diabetes exploring factors associated with the residual cardiovascular risk and other diabetes-related complications: The SFDT1 study. Diabetes Metab. 48:8. doi: 10.1016/j.diabet.2021.101306
Rouland, M., Beaudoin, L., Rouxel, O., Bertrand, L., Cagninacci, L., Saffarian, A., et al. (2022). Gut mucosa alterations and loss of segmented filamentous bacteria in type 1 diabetes are associated with inflammation rather than hyperglycaemia. Gut 71, 296–308. doi: 10.1136/gutjnl-2020-323664
Russell, J. T., Roesch, L. F. W., Ordberg, M., Ilonen, J., Atkinson, M. A., Schatz, D. A., et al. (2019). Genetic risk for autoimmunity is associated with distinct changes in the human gut microbiome. Nature 10:3621. doi: 10.1038/s41467-019-11460-x
Saeedi, P. I, Petersohn, P., Salpea, B., Malanda, S., Karuranga, N., Unwin, S., et al. (2019). Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the international diabetes federation diabetes atlas, 9th edition. Diabetes Res. Clin. Pract. 157:107843. doi: 10.1016/j.diabres.2019.107843
Shilo, S., Godneva, A., Rachmiel, M., Korem, T., Bussi, Y., Kolobkov, D., et al. (2022). The gut microbiome of adults with type 1 diabetes and its association with the host glycemic control. Diabetes Care 45, 555–563. doi: 10.2337/dc21-1656
Stewart, C. J., Ajami, N. J., O’Brien, J. L., Hutchinson, D. S., Smith, D. P., Wong, M. C., et al. (2018). Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature 562:583. doi: 10.1038/s41586-018-0617-x
Tian, H. L., Zhang, S. Y., Qin, H. L., Li, N., and Chen, Q. Y. (2022). Long-term safety of faecal microbiota transplantation for gastrointestinal diseases in China. Lancet 7, 702–703. doi: 10.1016/S2468-1253(22)00170-4
Traversi, D., Scaioli, G., Rabbone, I., Carletto, G., Ferro, A., Franchitti, E., et al. (2022). Gut microbiota, behavior, and nutrition after type 1 diabetes diagnosis: A longitudinal study for supporting data in the metabolic control. Front. Nutr. 9:968068. doi: 10.3389/fnut.2022.968068
Vatanen, T., Franzosa, E. A., Schwager, R., Tripathi, S., Arthur, T. D., Vehik, K., et al. (2018). The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562:589. doi: 10.1038/s41586-018-0620-2
Wahlstrom, A., Sayin, S. I., Marschall, H., and Backhed, F. (2016). Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50. doi: 10.1016/j.cmet.2016.05.005
Wishart, D. (2016). Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 15, 473–484. doi: 10.1038/nrd.2016.32
Yang, L., Fang, X., and Zhu, J. (2022). Citizen environmental behavior from the perspective of psychological distance based on a visual analysis of bibliometrics and scientific knowledge mapping. Front. Psychol. 12:766907. doi: 10.3389/fpsyg.2021.766907
Yuan, X., Wang, R., Han, B., Sun, C., Chen, R., Wei, H., et al. (2022). Functional and metabolic alterations of gut microbiota in children with new-onset type 1 diabetes. Nat. Commun. 13:6356. doi: 10.1038/s41467-022-33656-4
Zhang, S., Deng, F. Y., Chen, J. X., Chen, F. W., Wu, Z. Z., Li, L. P., et al. (2022c). Fecal microbiota transplantation treatment of autoimmune-mediated type 1 diabetes: A systematic review. Front. Cell. Infect. Micrbiol. 12:1075201. doi: 10.3389/fcimb.2022.1075201
Zhang, L., Zhang, H. C., Xie, Q., Xiong, S., Jin, F. C., Zhou, F., et al. (2022b). A bibliometric study of global trends in diabetes and gut flora research from 2011 to 2021. Front. Endocrinol. 13:990133. doi: 10.3389/fendo.2022.990133
Zhang, L., Jonscher, K. R., Zhang, Z. Y., Xiong, Y., Mueller, R. S., Friedman, J. E., et al. (2022a). Islet autoantibody seroconversion in type-1 diabetes is associated with metagenome-assembled genomes in infant gut microbiomes. Nat. Commun. 13:14. doi: 10.1038/s41467-022-31227-1
Zhong, D., Li, Y., Huang, Y., Hong, X., Li, J., and Jin, R. (2022). Molecular mechanisms of exercise on cancer: A bibliometrics study and visualization analysis via CiteSpace. Front. Mol. Biosci. 8:797902. doi: 10.3389/fmolb.2021.797902
Keywords: T1DM, intestinal flora, bibliometrics, CiteSpace, VOSviewers
Citation: Cui X, Wu Z, Zhou Y, Deng L, Chen Y, Huang H, Sun X, Li Y, Wang H, Zhang L and He J (2024) A bibliometric study of global trends in T1DM and intestinal flora research. Front. Microbiol. 15:1403514. doi: 10.3389/fmicb.2024.1403514
Received: 19 March 2024; Accepted: 12 June 2024;
Published: 04 July 2024.
Edited by:
Moez Rhimi, INRAE Centre Jouy-en-Josas, FranceReviewed by:
Kailash Singh, Uppsala University, SwedenJuan Hernandez, INRA Ecole Nationale Vétérinaire, Agroalimentaire et de l’alimentation de Nantes-Atlantique (Oniris), France
Copyright © 2024 Cui, Wu, Zhou, Deng, Chen, Huang, Sun, Li, Wang, Zhang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jia He, aGVqaWExMjMuc2hpaGV6aUAxNjMuY29t
†These authors share first authorship
 Xinxin Cui
Xinxin Cui Zhen Wu
Zhen Wu Yangbo Zhou1
Yangbo Zhou1 Xiangbin Sun
Xiangbin Sun Yu Li
Yu Li Haixia Wang
Haixia Wang Li Zhang
Li Zhang Jia He
Jia He