
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 27 May 2024
Sec. Microbe and Virus Interactions with Plants
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1402653
 Dengfeng Qi1†
Dengfeng Qi1† Qiao Liu1†
Qiao Liu1† Liangping Zou1
Liangping Zou1 Miaoyi Zhang1
Miaoyi Zhang1 Kai Li1
Kai Li1 Yankun Zhao1
Yankun Zhao1 Yufeng Chen1
Yufeng Chen1 Junting Feng1
Junting Feng1 Dengbo Zhou1
Dengbo Zhou1 Yongzan Wei1
Yongzan Wei1 Wei Wang1*
Wei Wang1* Lu Zhang2*
Lu Zhang2* Jianghui Xie1*
Jianghui Xie1*Banana wilt caused by Fusarium oxysporum f. sp. cubense tropical race 4 (Foc TR4) is a devastating fungal disease. Biocontrol strategies hold immense potential for inhibiting the spread of Foc TR4. Here, 30 actinobacteria were isolated from soils and screened for their antagonistic activity against Foc TR4. Strain SCA4-21T was selected due to its strongest antagonistic activity against Foc TR4. Strain SCA4-21T also exhibited strong antagonistic activity against the other eight phytopathogenic fungi. The strain was identified as the genus Streptomyces according to its physiological, biochemical, and phenotypic characteristics. The phylogenetic trees of 16S rRNA sequences demonstrated that strain SCA4-21T formed a subclade with S. iranensis HM 35T and/or S. rapamycinicus NRRL B-5491T with low bootstrap values. Considering that 16S rRNAs did not provide sufficient resolution for species-level identification, the whole genome of strain SCA4-21T was sequenced. Multilocus sequence analysis (MLSA) based on five housekeeping gene alleles (atpD, gyrB, recA, rpoB, and trpB) revealed that strain SCA4-21T clustered into S. hygroscopicus subsp. hygroscopicus NBRC 13472T with 100% of bootstrap value. The analysis of the genome-based phylogeny also approved the results. Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) were 91.26 and 44.30%, respectively, with values below the respective species level threshold of 95 and 70%. Hence, strain SCA 4–21T represented a novel species within the genus Streptomyces, named Streptomyces luomodiensis sp. nov. The type strain is SCA4-21T (=GDMCC4.340T = JCM36555T). By the CAZymes analysis, 348 carbohydrate-active enzymes (CAZymes) were detected, including 15 chitinases and eight β-1,3-glucanases. The fermentation broth of strain SCA4-21T, exhibiting strong antagonistic activity against Foc TR4, demonstrated high activities of chitinase and β-1,3-glucanase, which might be involved in antifungal activity. Our results showed an innovative potential biocontrol agent for managing plant fungal diseases, specifically banana fusarium wilt.
Bananas (Musa spp.) are an important fruit and staple food crop in the tropics and subtropics (Mohandas and Ravishankar, 2016). However, the world banana industry is suffering from the serious harm of Fusarium wilt disease, which is caused by the soil-borne Fusarium oxysporum f. sp. cubense (Foc). Foc race 1 (Foc 1) destroyed the major cultivar of the Gros Michel banana in Central America and the Caribbean during the mid-20th century (Pérez-Vicente et al., 2014). Although the detriment of Foc 1 was overcome by the resistant Cavendish cultivars, a new strain of Foc known as tropical race 4 (Foc TR4) emerged and spread rapidly (Ploetz, 2015). Foc TR4 can infect almost all banana cultivars. It is difficult to control Foc TR4 because of its long survival ability in soil (Ploetz, 2015). Utilizing antagonistic microbes to control Foc TR4 is a promising strategy due to its environmentally friendly and highly effective advantages (Fu et al., 2017).
Actinobacteria are ubiquitously distributed in the soil and natural environment. They are considered an ideal candidate for biocontrol agents (Gonzalez-Franco and Hernandez, 2009; Bhatti et al., 2017). Actinobacteria successfully colonizing the plant rhizosphere provides a biocontrol potential for soil-borne disease (Barka et al., 2015). Streptomyces is the largest genus of the phylum Actinomycetota (Law et al., 2017). They are Gram-positive, aerobic, filamentous bacteria with approximately 70% of G + C content in their genomes (Waksman and Henrici, 1943). The traditional method of DNA–DNA hybridization allowed the classification of prokaryotes, but serious shortcomings limited its application such as a time-consuming procedure and operational feasibility (Gevers et al., 2005). Currently, the identification of Streptomyces primarily relies on the combination of phenotypic and genotypic characteristics. 16S rRNA sequences did not provide a sufficient resolution for the species-level identification. Average nucleotide identity (ANI) provided an efficient method to identify the level of species (Gevers et al., 2005). At the moment of writing, there are 726 Streptomyces species with a validly published and correct name, according to the LPSN–list of Prokaryotic names with Standing in Nomenclature (Parte et al., 2020).1 However, there is still a lack of Streptomyces with strong adaptability and high antimicrobial activity in agricultural practice.
Streptomyces are also massive reservoirs of bioactive substances such as antibiotics, enzymes, and siderophores (Jose et al., 2021). Moreover, they also participate in the cycling of carbon due to their ability to secrete many carbohydrate-active enzymes (CAZymes). These enzymes can facilitate the biodegradation of remains from animals, plants, and fungi (Barka et al., 2015). CAZymes are classified into six major families according to the functional characteristics (Mann et al., 2013). The members of the glycoside hydrolase family contain chitinase and β-1,3-glucanase (Aktuganov et al., 2008; Zacky and Ting, 2013). Chitinase and β-1,3-glucanase are responsible for the antifungal activity of Streptomyces sp. 5–4 (Yun et al., 2022) and Streptomyces griseus (Anitha and Rebeeth, 2009). However, the antifungal mechanism of Streptomyces is still unclear due to the physiological property differences of diverse members.
In this study, 30 actinobacteria were isolated from a dry-hot valley of Huili County, Sichuan Province, China. Their antagonistic activity was screened against Foc TR4. Strain SCA4-21T was further investigated due to its strongest antagonistic activity against Foc TR4. The broad-spectrum antifungal activity of strain SCA4-21T was initially detected against the selected eight phytopathogenic fungi. Subsequently, the taxonomic status of strain SCA4-21T was identified based on its phenotypic and genotypic characteristics. To explore the antifungal mechanism of strain SCA4-21T, CAZymes of its genome were predicted, and the antifungal activity of the fermentation broth from strain SCA4-21T was determined. Additionally, the activities of chitinase and β-1,3-glucanase in the fermentation broth from strain SCA4-21T were tested.
The soil samples were collected from the Luomodi Village in a dry-hot valley of Huili County, Sichuan Province, China. Actinobacteria were isolated using a serial dilution method on the starch-casein medium (SCA, 10 g of soluble starch, 0.3 g of casein, 2.0 g of KNO3, 2.0 g of NaCl, 2.0 g of K2HPO4, 0.05 g of MgSO4⋅7H2O, 0.02 g of CaCO3, 0.01 g of FeSO4⋅H2O, and 18 g of agar in 1 L sterile ddH2O, pH 7.0–7.4). The addition of potassium dichromate (50 mg/L) and nystatin (50 mg/L) was used to suppress the growth of other fungi and bacteria, respectively (Williams et al., 1983). Briefly, soil samples were sieved through a 0.6 mm mesh. A one-gram sample was suspended in 9 mL of sterile water. After incubation at 55°C for 20 min, the soil homogenate was diluted with sterile water into 10−1, 10−2, and 10−3 and poured into the SCA plates. The plates were cultured at 28°C for 7 d, and the colonies with different shapes and colors were picked up and further purified on ISP2 (yeast extract-malt extract agar) (Shirling and Gottlieb, 1966). All the purified isolates were preserved in the ISP2 (yeast malt extract agar) slant agar medium at 4°C and 30% of glycerol (v/v) at −80°C, respectively.
The antifungal activity of actinobacteria was screened against Foc TR4 using the plate confrontation method (Jing et al., 2020). Briefly, these isolates were inoculated at four symmetrical points 2.5 cm away from the center of potato dextrose agar (PDA) plates. After 2 d, the mycelial disk (0.5 cm diameter) of Foc TR4 was inoculated into the center of the PDA plate. The mycelial disk of Foc TR4 only was used as a control. The experiment was set up in triplicate. After incubation at 28°C for 7 d, the diameters of Foc TR4 were measured. The antifungal activity of actinobacteria was calculated according to the following formula: MI = [(C-T)/C] × 100%, where C and T represented the growth diameters of the control and treatment groups, respectively. Strain SCA4-21 (the type strain SCA4-21T) was selected to be investigated further due to the strong antifungal activity against Foc TR4.
The broad-spectrum antagonistic activity of strain SCA4-21T was determined using the plate confrontation method as described above. The selected eight pathogenic fungi include Curvularia fallax (ATCC 34598), Colletotrichum gloeosporioides (Penzig) Penzig et Saccardo (ATCC MYA-456), Colletotrichum gloeosporioides (ACCC 36351), Colletotrichum acutatum (ATCC 56815), Colletotrichum fragariae (ATCC 58718), Colletotrichum gloeosporioides (Penz) Saec (ACCC 36351), Fusarium oxysporum f. sp. cucumerinum Owen (ATCC 204378), and Fusarium graminearum (DSM 21803). The pathogens were kept in the Institute of Tropical Bioscience and Biotechnology, China Academy of Tropical Agricultural Sciences, Haikou, China.
Strain SCA4-21T was inoculated on eight media including ISP2, ISP3 (oatmeal agar), ISP4 (inorganic salts-starch agar), ISP5 (glycerol-asparagine agar), ISP6 (peptone yeast-iron agar), ISP7 (tyrosine agar), and Gause No. 1 agar and cultured at 28°C for 7 d (Shirling and Gottlieb, 1966). The culture characteristics were observed such as the growth of aerial and substrate hyphae and the production of soluble pigment. The spore chain and spore morphology of strain SCA4-21T were observed by a scanning electron microscope (SEM, Zeiss ∑IGMA, Germany) after 7 and 21 d of growth on ISP2 medium according to our previous method (Qi et al., 2021). Mycelial blocks (5 mm diameter) were cut from plates of strain SCA4-21T using a sterile scalpel. The samples were fixed with 2.5% of glutaraldehyde at 4°C overnight and were rinsed twice with phosphoric acid buffer solution (PBS, 0.1 mol/L, pH 7.0). The materials were dehydrated in a graded ethanol solution (70, 80, 90, 95, and 100%) for 2 min, respectively. The specimens were soaked in fresh t-butyl alcohol overnight. Dehydrated samples were freeze-dried and coated with a film of gold–palladium alloy under vacuum for SEM.
The physiological characteristics of strain SCA4-21T were determined such as the utilization of nitrogen and carbon and the tolerance to NaCl (0–11%, w/v), temperature (16–50°C), and pH (4–10). We also measured its biochemical features, including the production of urease, degradation of Tweens 20, 40, and 60, gelatin liquefaction, starch hydrolysis, nitrate reduction, siderophore, and antibiotic sensitivity test (Qi et al., 2021).
Strain SCA2-4 T was inoculated in the ISP2 liquid culture medium and shaken at 180 rpm and 28°C for 5 d. The biomass was collected by centrifugation at 12000 rpm for 5 min. The precipitate was freeze-dried. The cellular fatty acids were determined by gas chromatography (Agilent 6,890) using the Sherlock Microbial Identification System (MIS) software (Sasser, 1990). The composition of cell wall diaminopimelic acid (DAP) was analyzed according to the method of Hasegawa et al. (1983). Respiration quinone was performed as described by Minnikin et al. (1984).
Strain SCA4-21T was cultured in ISP2 liquid medium at 28°C for 3 d. Mycelia of strain SCA4-21T were collected by centrifugation and frozen in nitrogen. The whole genome was sequenced using the Illumina Hiseq + PacBio sequencing platform (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China). Raw reads were trimmed and filtered using the Trimmomatic (v 0.36) software (Bolger et al., 2014). The clean reads were assembled using the Unicycler (v 0.4.8) software (Wick et al., 2017). The protein-coding genes were predicted using the Glimmer (v3.02) (Delcher et al., 2007), Prodigal (v 2.6.3) (Hyatt et al., 2010), and GeneMarkS (v4.3) (Besemer and Borodovsky, 2005). Genome annotation was performed by the Prokaryotic Genome Annotation Pipeline of NCBI (Tatusova et al., 2016). CAZymes were first predicted using the online CAZy v 6 software (Cantarel et al., 2009).
Strain SCA4-21T was inoculated in the ISP2 liquid medium and shaken at 180 rpm and 28°C for 3 d. DNA was extracted using the bacterial genome rapid DNA kit (RN43, Aidlab Co., Ltd., Beijing, China). The 16S rRNA sequence was amplified using a pair of the common primers 27F (5’-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5’-GGTTACCTTGTTACGACTT-3′) (Wang et al., 2015). The PCR reaction system was set according to Wang et al. (2022). After sequencing, the multiple alignment of the 16S rRNA sequence was carried out using EzBioCloud (Hall, 1999)2 and CLUSTALW within BioEdit 7.0.5.3 (Thompson et al., 1994). Phylogenetic trees were constructed using neighbor-joining, maximum parsimony, and maximum likelihood methods in MEGA7.0 (Kumar et al., 2016). Multilocus sequence analysis (MLSA) was performed based on five housekeeping genes including atpD, gyrB, recA, rpoB, and trpB (Rong and Huang, 2010). These housekeeping genes of strain SCA4-21T were extracted from the sequenced genome. The homolog sequences of five housekeeping genes from other species were downloaded from GenBank.3 The phylogenetic trees were constructed using the above method. The evolutionary distance was calculated by using the Kimura two-parameter (K2P) model for nucleotide sequences (Kimura, 1980). Based on the alignment of the whole genomes with other type strains of Streptomyces, the phylogenomic tree was constructed using the Type (Strain) Genome Server (TyGS) with default parameters (Meier-Kolthoff and Göker, 2019).4 The GenBank accession numbers of these housekeeping genes and genomes are listed in Supplementary Table S1.
To further identify the species of strain SCA4-21T, the overall genome-related indexes (OGRIs), such as average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH), between strain SCA4-21T and its closely related species were calculated by the online OrthoANI (Yoon et al., 2017)5 and the Genome-to-Genome Distance Calculator (GGDC) version 3.0 (Meier-Kolthoff et al., 2022),6 respectively.
Antifungal activity of fermentation broth of Streptomyces sp. SCA4-21T was determined by the agar well diffusion method (Magaldi et al., 2004). Briefly, strain SCA4-21T was inoculated into 100 mL of liquid YE medium and cultured at 28°C and 180 rpm for 7 d. Culture supernatant was harvested from the culture media of strain SCA4-21T by centrifugation at 2000 rpm for 20 min, followed by filtration through a 0.22-μm sterile filter (Millipore, Bedford, MA, United States). A total of 100 μL of sterile supernatant was aliquoted into each symmetrical well 2.5 cm away from the center of the PDA plate. The mycelial disk (5 mm diameter) of Foc TR4 was placed on the center of the PDA plate. Sterilized H2O was used as a control. Three replicates were set for each experiment. After incubating at 28°C for 5–7 d, the antifungal activity of fermentation broth was calculated using the formula MI = [(C-T)/C] × 100%, where C and T expressed the growth diameters of the control and treatment groups, respectively.
The activities of CAZymes, including chitinase and glucanase, were measured. Briefly, a 5-mm block of strain SCA4-21T was inoculated into 100 mL of liquid YE medium and shaken at 28°C and 180 rpm for 7 d. Culture filtrates were collected from the culture media of strain SCA4-21T by centrifugation at 2000 rpm for 20 min. The activities of chitinase and β-1,3-glucanase were determined using the Chitinase Activity Assay Kit (Beijing Solarbio Science & Technology Co., Ltd.) and the microorganism β-1,3-glucanase ELISA Kit (Jiangsu Meibiao Biotechnology Co., Ltd.), respectively. The amount of chitinase that breaks down chitin to produce 1 μg of N-acetylglucosamine per milliliter of sample solution per minute at 37°C is defined as one unit of chitinase activity. One unit of β- 1,3-glucanase activity is expressed as the amount of enzyme that converts 1 μmol of substrate into product per milliliter of sample solution per minute at 37°C. All experiments were repeated three times.
A total of 30 actinobacteria were isolated from soil samples in the dry-hot valley and designated as SCA4-1 to SCA4-30 (Figure 1A). The antifungal activities of these isolates against Foc TR4 were analyzed using the plate confrontation method. Seven strains showed antifungal activity against Foc TR4. Among them, SCA4-21T (the type strain SCA4-21T) has the highest percentage of mycelial inhibition (74.22 ± 1.23) %, followed by SCA4-6 (54.95 ± 0.07) %, SCA4-14 (52.62 ± 0.19)%, SCA4-1 (51.31 ± 0.11)%, SCA4-17 (46.46 ± 0.07)%, SCA4-8 (43.23 ± 0.17)%, and SCA4-27 (28.69 ± 0.13) %. Therefore, strain SCA4-21T was selected for the following experiment (Figure 1B).
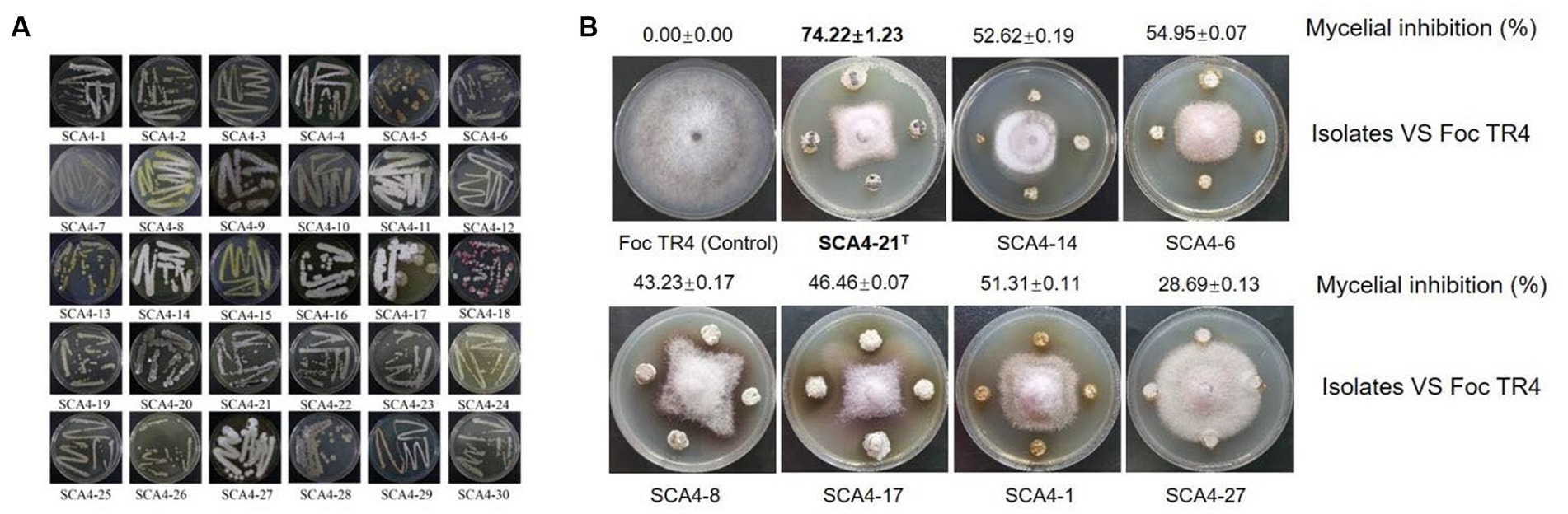
Figure 1. Isolation of actinobacteria and screening of their antagonistic activity against Foc TR4 (A) Thirty actinobacteria were isolated from wheat rhizosphere soil samples. (B) Seven actinobacteria exhibited antifungal activity against Foc TR4.
Compared with the control group, strain SCA4-21T exhibited a strong broad-spectrum antifungal activity. The highest inhibition rates were observed in C. fallax (86.63 ± 1.43)% and C. gloeosporioides (Penzig) Penzig et Saccardo (84.96 ± 1.43)%, respectively (Figure 2), followed by C. gloeosporioides (78.73 ± 1.35)%, F. graminearum Sehw (78.63 ± 1.33)%, F. oxysporum f. sp. cucumerinum (75.21 ± 1.25)%, C. fragariae (74.78 ± 1.15)%, C. acutatum (73.04 ± 1.08)%, and C. gloeosporioides (Penz) Saec (70.05 ± 1.15)%.
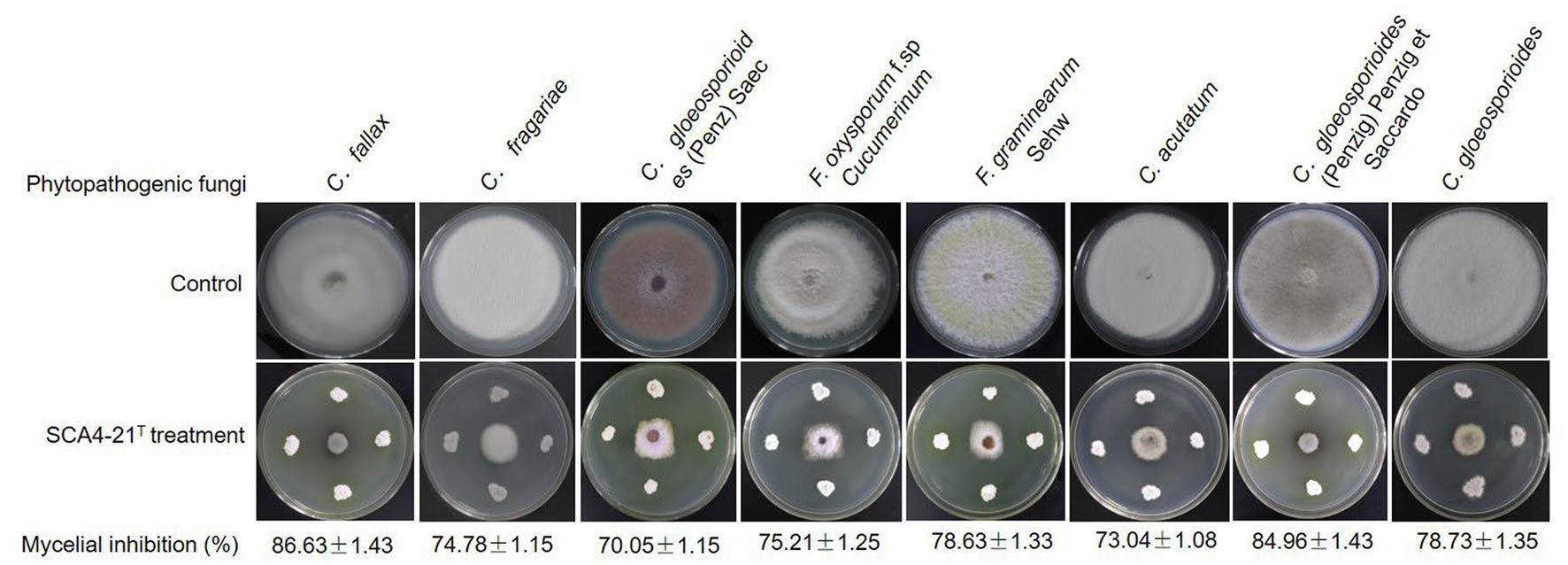
Figure 2. Strain SCA4-21T exhibited broad-spectrum antifungal activity against eight other Phytopathogenic fungi.
Strain SCA4-21T was an aerobic and Gram-positive bacterium. It could grow well on the ISP2, PDA, and Gause’s no.1 plates, followed by ISP5-7. The colors of colonies on different media exhibited white, cream, pure white, gray white, and silver gray. Strain SCA4-21T produced light-brown and light-yellow soluble pigment only in ISP7 and PDA plates, respectively (Figure 3A and Table 1). Morphological observations of strain SCA4-21T on ISP2 plates revealed the presence of branched aerial mycelia, a spiral spore chain, and cylindrical spores with a shrinkage surface. On 7-day-old plates, abundant aerial mycelia and spore chains were observed, while on 21-day-old plates, a large number of mature spores detached from spore chains were observed (Figure 3B). On ISP3, PDA, and Gause’s no.1 plates, these spirals gradually merged into dark masses of spores as they aged. This phenomenon was frequently observed in the members of Streptomyces hygroscopicus and Streptomyces iranensis groups (Shirling and Gottlieb, 1972; Hamedi et al., 2010). The cultural and morphological characteristics of strain SCA4-21T were consistent with those of the genus Streptomyces.
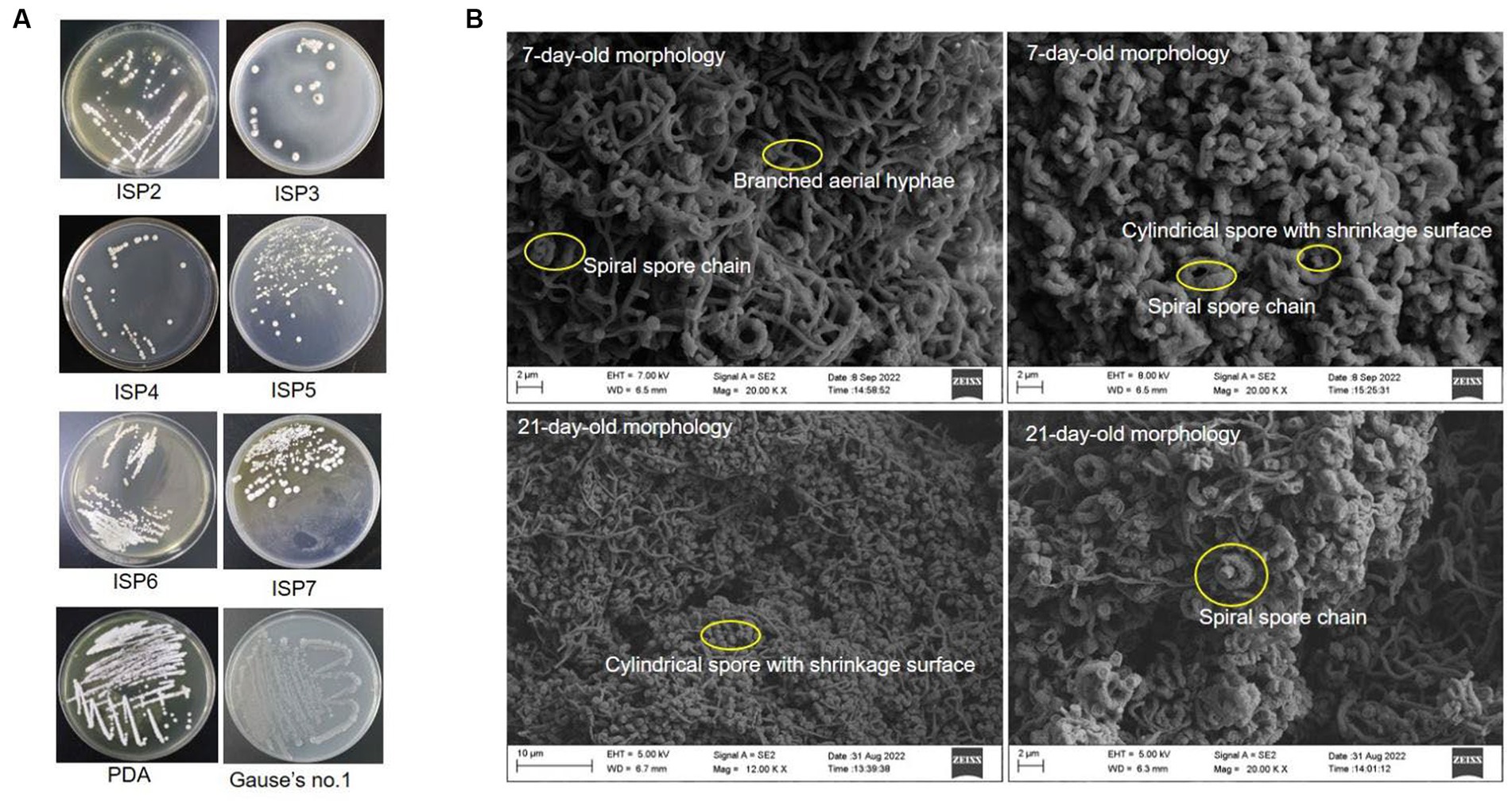
Figure 3. Cultural and morphological characteristics of strain SCA4-21T (A) Cultural characteristics of strain SCA4-21T cultured on eight different media, including ISP2, ISP3, ISP4, ISP5, ISP6, ISP7, PDA, and Gause’s no.1 medium. (B) Morphological characteristics of strain SCA4-21T cultured on ISP2 plates for 7d and 14 d at 28°C.
The growth of strain SCA4-21T was observed at pH 6.0 to 9.0 (optimal at 7), at temperature 16–46°C(optimal at 28°C), and with NaCl of 0 to 3% (w/v) (optimal at 2%). It could degrade starch, urea, and Tween-20 while producing melanoid pigment and siderophores (Figure 4A). It could neither degrade gelatin, Tween 40, Tween 60, nitrate, and cellulose nor produce H2S (Supplementary Table S2). The strain demonstrated resistance to 10 antibiotics, namely furazolidone, compound sulfamethoxazole, polymyxin B, vancomycin, erythromycin, minocycline, kanamycin, gentamicin, ceftriaxone, and cefuroxime. However, it was sensitive to 11 antibiotics, namely, clindamycin, chloramphenicol, penicillin, cefepime, cefotaxime, cefamandole, midecamycin, carbenicillin, ampicillin, benzylpenicillin, and piperacillin (Figure 4B; Supplementary Table S2). In addition, as shown in Supplementary Table S3, the isolate was able to utilize D-mannose, D-trehalose, sorbitol, D-fructose, lactose, mannitol, and xylan as carbon sources but could not use L-arabinose and raffinose. Strain SCA4-21T used L-phenylalanine, L-asparagine, L-methionine, L-valine, L-histidine, L-tryptophan, D-cellobiose, L-hydroxyproline, and L- cysteine but did not utilize anhydrous inositol, L-glutamic acid, L-tyrosine, and arginine.
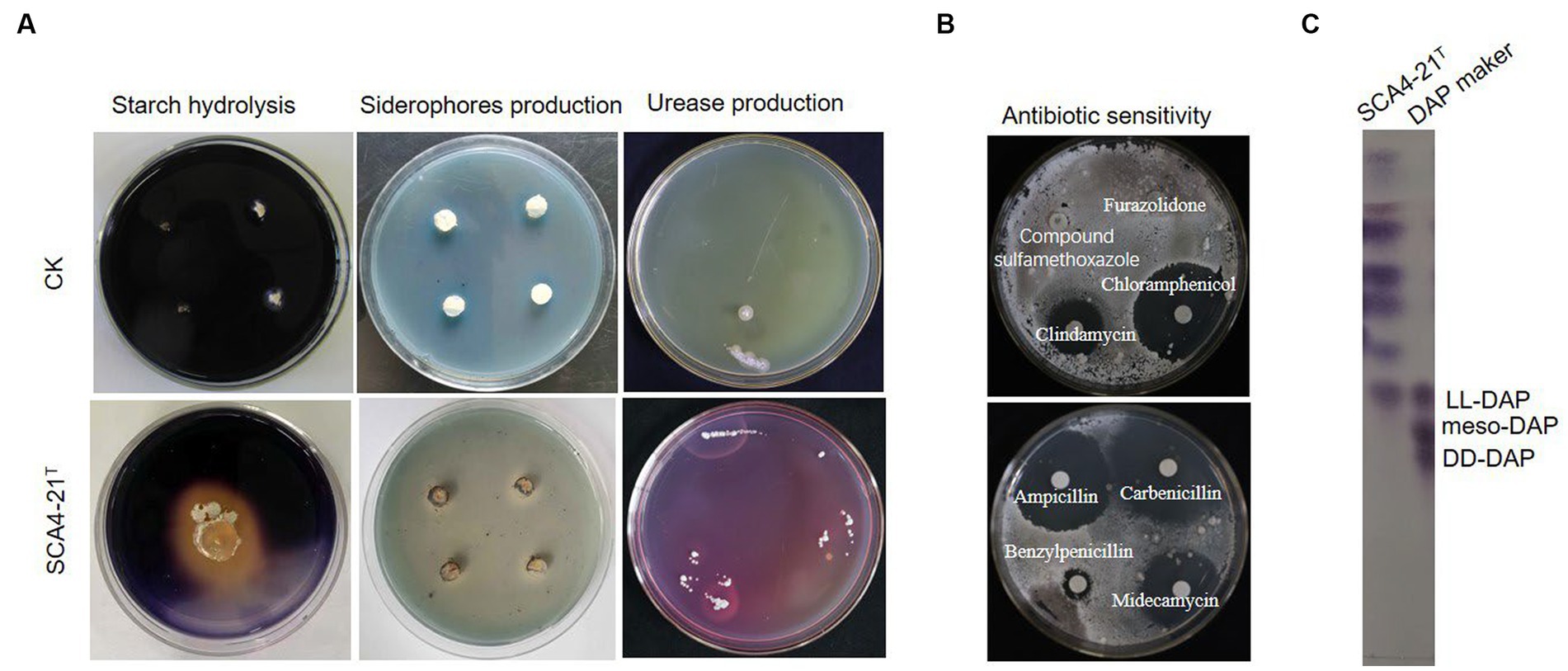
Figure 4. Physiological and chemotaxonomic characteristics of strain SCA4-21T (A) Compared with the control strain, strain SCA4-21T could produce amylase, siderophores, and urease. (B) Antibiotic sensitivity experiment. The strain was resistant to some antibiotics such as furazolidone, compound sulfamethoxazole, but sensitive to other antibiotics like clindamycin, chloramphenicol, carbenicillin, midecamycin, ampicillin, and benzylpenicillin. (C) Composition analysis of DAP in cell wall. The cell wall of strain SCA4-21T mainly contained LL-DAP.
The chemotaxonomic characteristics of strain SCA4-21T are listed in Table 2. The dominant menaquinones were MK9 (H8) (65.50%) and MK10(H2) (34.50%) (Supplementary Figure S1). The dominant fatty-acid compositions of the isolate consisted of anteiso-C15:0 (34.78%) and C16:0 (19.89%). The presence of LL-DAP in the peptidoglycan of the cell wall was considered one of the effective methods for the identification of the genus Streptomyces (Kieser et al., 2000). The cell wall of strain SCA4-21T largely contained LL-diaminopimelic acid (LL-DAP) (Figure 4C), which helped in the rapid identification of this strain as Streptomyces.
The complete genome of strain SCA4-21T was sequenced and assembled into a circular chromosome of 10,044,493 bp with 71.48% of GC content (Figure 5A). Genomic features of strain SCA4-21T are listed in Table 3. The genome contained 8,246 protein-coding genes (CDS) with 8,847,075 bp of gene total length, 64 tRNA genes, 18 rRNA genes, 134 sRNA genes, 1,436 repeat genes, 80 CRISPR-Cas genes, and 6 insert sequences. Functional analysis revealed that 7,548, 3,460, and 4,391 genes were annotated to KEGG, COG, and GO categories, respectively (Figures 5B,C). In COG function classification, most of the predicted CDS related to metabolism (47.4%), followed by cellular processes and signaling (20.2%) and information storage and processing (19.7%). A total of 12.6% of CDS are poorly characterized (Supplementary Table S4).
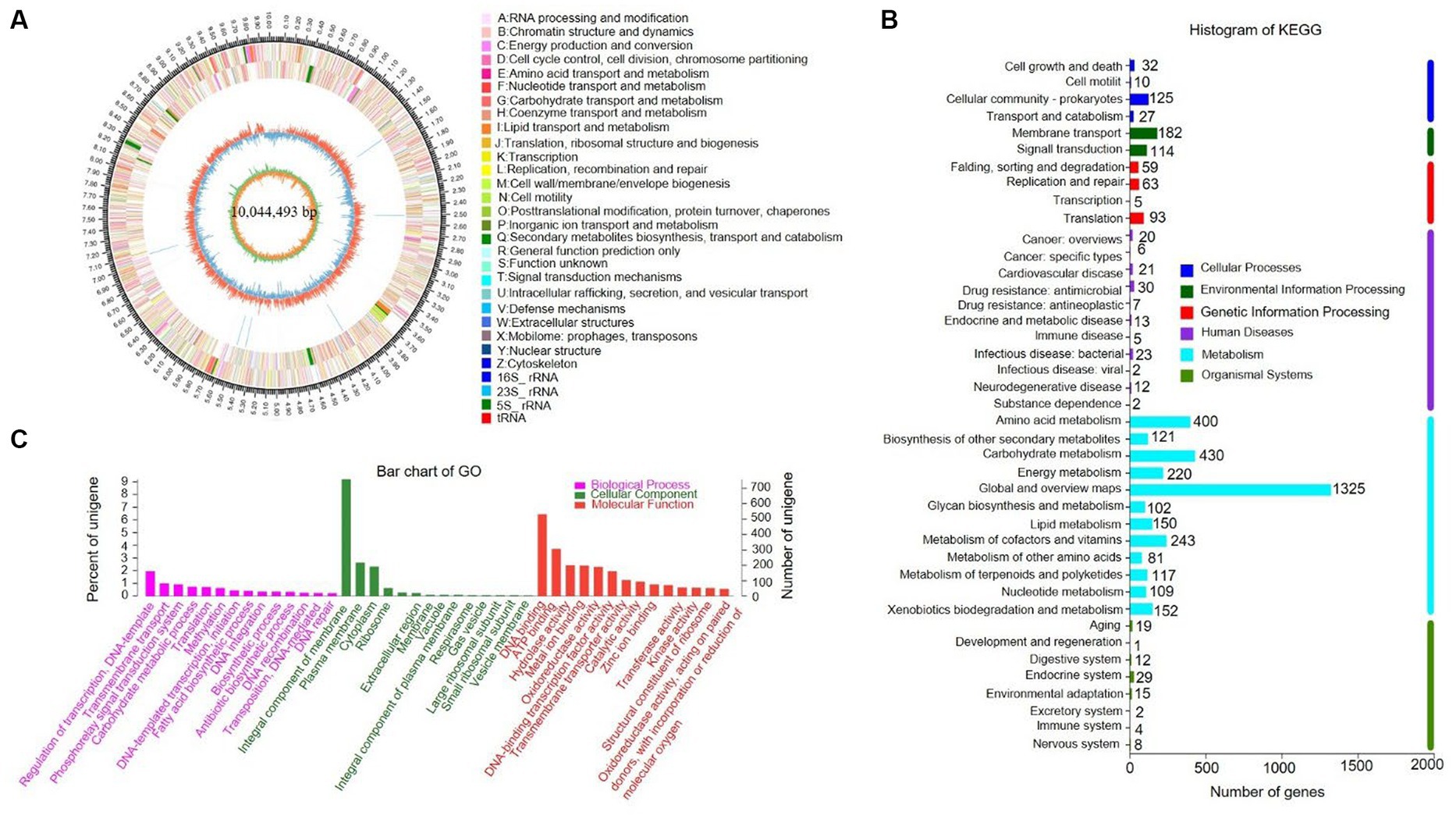
Figure 5. Genome information and function annotation of strain SCA4-21T (A) Circular map. From the outside to the middle, ring 1 was the mark of genome size. Rings 2 and 3 represent CDS on forward/reverse strand. Different colors indicate the functional categories of CDS. Ring 4 is tRNA and rRNA. Ring 5 is the G + C content. The outward red portion indicates that the GC content of this region is higher than the average GC content of the whole genome. The inward blue part indicates that the GC content of this region is lower than the average GC content of the whole genome, followed by G + C skew in ring 6. (B) The KEGG pathway annotation. (C) The GO annotation.
The EzBioCloud analysis of the 16S rRNA gene sequence (1,524 bp) of strain SCA4-21T revealed that the strain shared the highest similarity to S. iranensis HM 35T (99.45%), S. rapamycinicus NRRL B-5491T (99.35%), and S. hygroscopicus subsp. hygroscopicus NBRC 13472T (99.17%). The phylogenetic trees based on the 16S rRNA sequences were constructed using neighbor-joining (NJ), maximum parsimony (MP), and maximum likelihood (ML) methods and are shown in Figure 6A; Supplementary Figures S2, S3, respectively. NJ and MP trees revealed that strain SCA4-21T formed a subclade with S. iranensis HM 35T with low bootstrap values, indicating that the strain belongs to the genus Streptomyces (Figure 6A; Supplementary Figure S2). However, the ML tree showed that strain SCA4-21T formed a subclade with S. iranensis HM 35T and S. rapamycinicus NRRL B-5491T with a bootstrap value of 63% (Supplementary Figure S3). The above results indicate that it is difficult to distinguish this strain from its closely related species using the 16S rRNA gene.
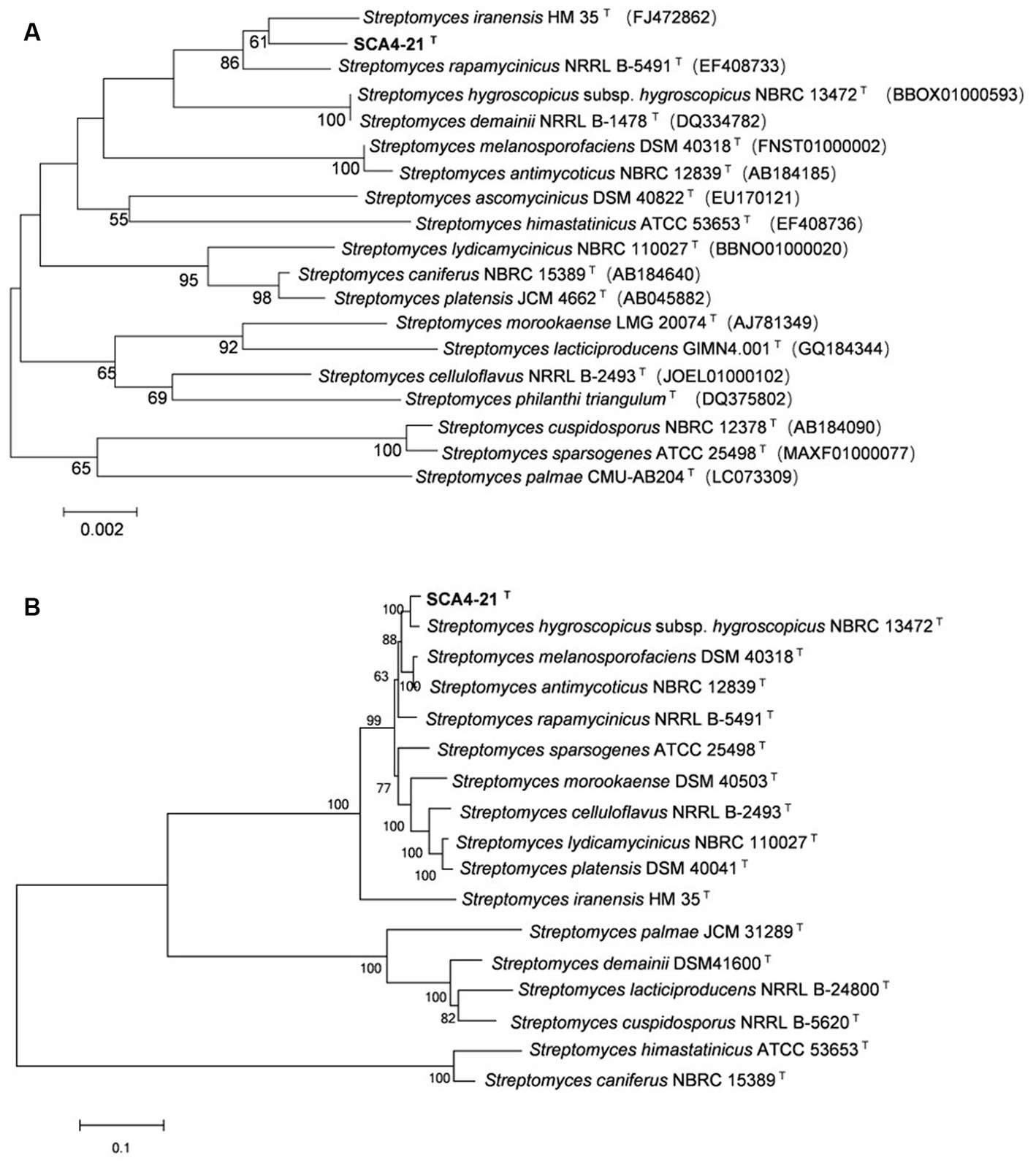
Figure 6. The phylogenetic trees of strain SCA4-21T (A) The phylogenetic tree based on the complete 16S rRNA sequences (1,524 bp) was constructed using neighbor-joining method. Accession numbers of the selected genes were listed in brackets. (B) The phylogenetic tree based on concatenated five housekeeping genes (atpD, gyrB, recA, rpoB, and trpB) (9,381 bp) was constructed using neighbor-joining method. Strain names and accession numbers are listed in Supplementary Table S1. Bootstrap percentages (based on 1,000 replications) were shown at branching points. Bar, 0.002 or 0.1 substitutions per nucleotide position.
Therefore, to assign strain SCA4-21T to species, MLSA was performed using five housekeeping genes: atpD, gyrB, recA, rpoB, and trpB. The phylogenetic trees using all three algorithms showed that SCA4-21T formed a well-delineated subclade with Streptomyces hygroscopicae subsp. hygroscopicus NBRC 1372T with bootstrap values of 100% (Figure 6B; Supplementary Figures S4, S5). Therefore, five housekeeping genes, namely, atpD, gyrB, recA, rpoB, and trpB, could distinguish closely related Streptomyces species. The MLSA distance values ranged from 0.021 to 1.092 (Table 4), which is much higher than the threshold of 0.007 for the delineation of bacterial species (Rong and Huang, 2012). The results were also approved by the analysis of the genome-based phylogeny (Supplementary Figure S6). The above data indicated that strain SCA4-21T represents a novel species of Streptomyces.
The ANI and dDDH values were calculated between strain SCA4-21T and closely related species. The species were selected based on the results of the phylogenetic trees constructed using the 16S rRNA gene and five housekeeping genes. Strain SCA4-21T showed the highest ANI and dDDH values with Streptomyces hygroscopicus subsp. hygroscopicus NBRC 13472T, which were 91.26 and 48%, respectively (Table 5). These values were below the novel species threshold of 95% (ANI) and 70% (dDDH) (Meier-Kolthoff et al., 2022; Riesco and Trujillo, 2024). These findings further supported that strain SCA4-21T belongs to a novel species in the genus Streptomyces.
To predict enzyme genes related to fungal cell wall degradation, CAZymes of strain SCA4-21T were annotated (Figure 7A). The results demonstrated that the genome of strain SCA4-21T encoded 348 CAZymes including 36 auxiliary activities (AAs), 6 carbohydrate-binding modules (CBMs), 74 carbohydrate esterases (CEs), 155 glycoside hydrolases (GHs), 65 glycosyltransferases (GTs), and 12 polysaccharide lyases (PLs). The proportion of CAZymes was 4.22% in strain SCA4-21T genome (Figure 7B). Family and gene ID of strain SCA4-21T CAZymes are listed in Supplementary Table S5. AAs were divided into nine families, namely AA1, AA2, AA3, AA3_2, AA4, AA5, AA6, AA7, and AA10. CBMs contained four families: CBM2, CBM13, CBM35, and CBM66. CEs included 11 families: CE1, CE2, EC3, EC4, EC7, EC8, EC9, EC10, EC12, EC14, and EC15. GTs were classified into 15 families: GT1, GT2_Glycos_transf_2, GT2_Glyco_tranf_2_3, GT4, GT5, GT9, GT20, GT28, GT35, GT39, GT41, GT76, GT81, GT83, and GT87. PLs were divided into 11 families: PL1, PL1_5, PL1_6, PL3_4, PL7_3, PL8, PL9, PL9_3, PL11, PL26, and PL31. GHs were classified into 80 families: GH1, GH2, GH3, GH4, GH5_1, GH5_8, GH5_18, GH5_19, GH5_40, GH5_43, GH6, GH8, GH9, GH10, GH11, GH12, GH13_3, GH13_9, GH13_10, GH13_11, GH13_13, GH13_16, GH13_26, GH13_30, GH13_32, GH15, GH16, GH18, GH19, GH20, GH23, GH25, GH26, GH27, GH30_5, GH30_7, GH31, GH33, GH35, GH36, GH42, GH43, GH43_3, GH43_5, 1 GH43_10, GH43_24, GH43_26, GH43_34, GH44, GH46, GH48, GH51, GH54, GH55, GH62, GH63, GH64, GH65, GH67, GH74, GH76, GH77, GH78, GH85, GH87, GH89, GH92, GH93, GH95, GH106, GH109, GH113, GH114, GH127, GH128, GH135, GH145, GH146, GH152, and GH154. Fifteen Chitinase (EC 3.2.1.14) were identified in GH18, GH19, GH23, and GH48 families. Additionally, eight β-1,3-glucanases (EC 3.2.1.39) were found in GH16, GH55, GH64, and GH128 families (Figure 6D). Chitinase and β-1,3-glucanases are often found to degrade the cell walls of plant pathogenic fungi (Fan et al., 2002; Arora et al., 2007). Therefore, we suggest that chitinase and β-1,3-glucanases could be responsible for the antifungal activity of strain SCA4-21T.
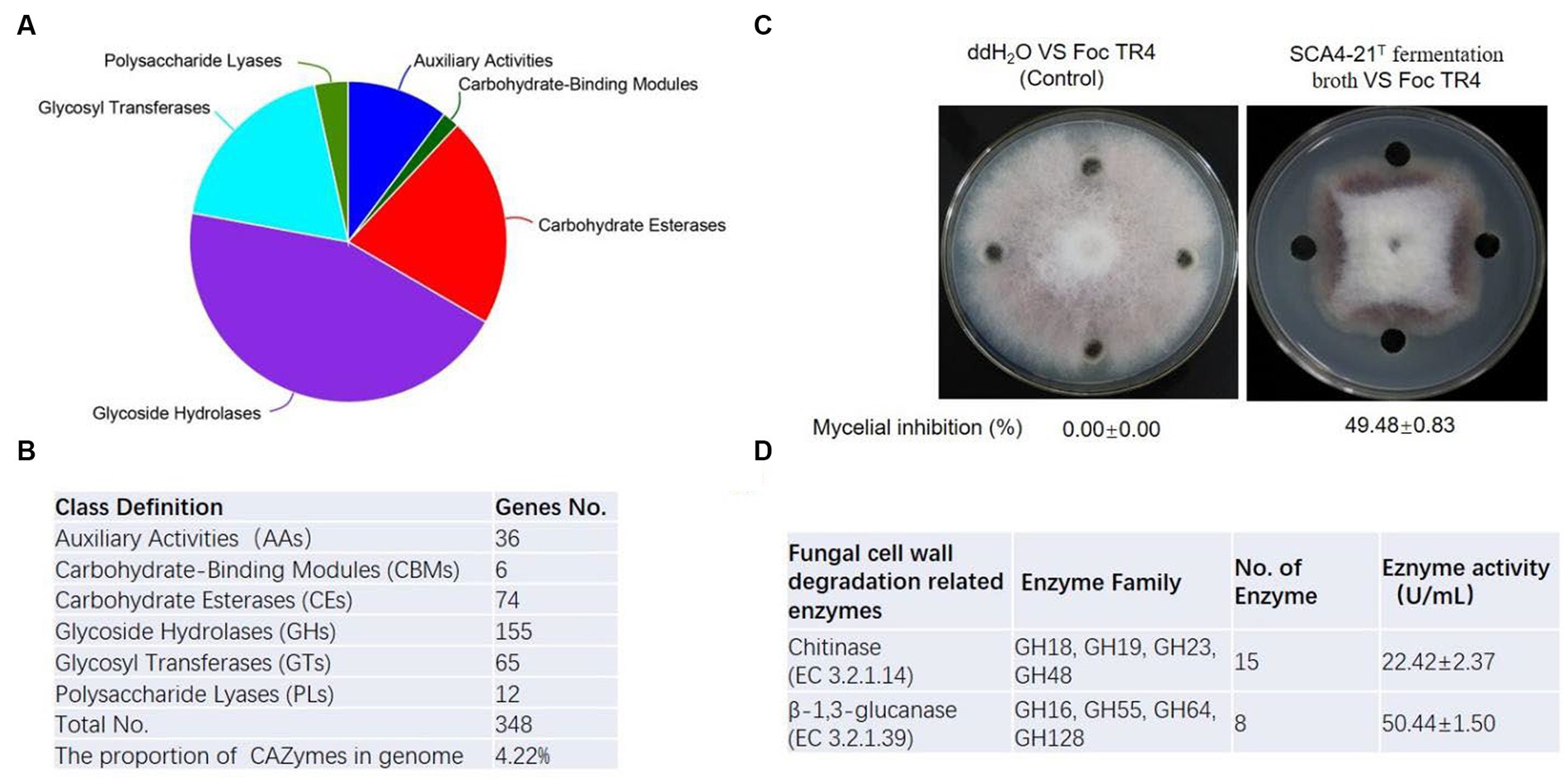
Figure 7. CAZymes analysis of strain SCA4-21T (A) Pie chart of CAZymes (B) The number of genes in different families of CAZymes. (C) Determination of antifungal activity in fermentation broth of SCA4-21T. (D) Evaluation of chitinase and β-1,3-glucanase activity in fermentation broth of SCA4-21T.
To further analyze the antifungal activity of the fermentation broth from strain SCA4-21T, the inhibition percentage against Foc TR4 mycelia was measured. By determination, the inhibition rate was 49.48 ± 0.83% (Figure 7C). The activities of chitinase and β-1,3-glucanase were 22.42 ± 2.37 U/mL and 50.44 ± 1.50 U/mL in the fermentation broth, respectively (Figure 6D), suggesting that two enzymes could be contributed to the antifungal activity against Foc TR4.
Streptomyces luomodiensis (luo.mo.di.en’sis. N.L. masc./fem. Adj. luomodiensis of Luomodi, a village in China, referring to the place where the type strain was first isolated).
Gram-positive, aerobic, and non-motile actinobacteria forming branched substrate and aerial mycelia, which differentiate into spiral spore chains. Spores are cylindrical with a shrinkage surface. Abundant growth occurs on all ISP, PDA, and Gause’s no.1 plates, and the colonies are white, pure or gray white, silver gray, and cream. Growth appears at 20–46°C with an optimum of 28°C, at pH 6–9 with an optimum of 7.0, and 0–3% (w/v) NaCl concentration with an optimum of 2%. Soluble pigment forms on ISP7 and PDA media. They degrade urea, Tween 20, and starch while producing melanoid pigment and siderophores. They neither degrade gelatin, Tween 40 and 60, nitrate, or cellulose nor produce H2S. D-mannose, D-trehalose, sorbitol, D-fructose, lactose, mannitol, and xylan are utilized as sole source of carbon and L-phenylalanine, L-asparagine, L-methionine, L-valine, L-histidine, L-tryptophan, D-cellobiose, L-hydroxyproline, and L- cysteine are utilized as sole source of nitrogen. The predominant menaquinones are MK9 (H8) and MK10 (H2). Major fatty acids (>10.0%) are anteiso-C15:0 and C16:0. Cell wall hydrolysates contain LL-diaminopimelic acid (LL-DAP), but they do not contain meso-DAP or DD-DAP.
The type strain, SCA4-21T (=GDMCC4.340T = JCM36555T), was isolated from the soil of Luomodi Village, Huili County, Sichuan Province, China. The genome of the type strain is characterized by a size of 10,044,493 bp and a G + C content of 71.48 mol%. Raw reads of strain SCA4-21T were deposited to the SRA database with the accession number SRP303585, and 16S rRNA gene and complete genome sequence were submitted to GenBank with accession numbers OQ352835 and CP117522, respectively.
Banana Fusarium wilt caused by Foc TR4 is a destructive fungal disease. More than 80% of bananas are susceptible to Foc TR4 (Pérez-Vicente et al., 2014). The utilization of antagonistic microorganisms for controlling banana fusarium wilt has received increasing attention due to its safety and efficacy. To enhance the effectiveness of biological control, it is crucial to screen broad-spectrum and novel antagonistic microorganisms as biocontrol agents (Heydan and Pessarakh, 2010). Unique environments often harbor an abundance of novel microorganisms (Bull, 2010). Here, 30 actinobacteria displaying diverse morphologies were isolated from the dry-hot valley of Huili County (Figure 1A). Among them, seven actinobacteria exhibited antagonistic activity against Foc TR4 (Figure 1B). Especially, strain SCA4-21T showed the most antifungal activity against Foc TR4 and other eight phytopathogenic fungi (Figure 2B).
Fungal cell walls are mainly composed of polysaccharides, accounting for 80–90% of their dry weight. CAZymes are the most important enzymes for polysaccharide degradation. The functional genes encoding CAZymes participate in fungal cell wall degradation. CAZymes analysis demonstrates that the genome of strain SCA4-21T contains 348 CAZymes, accounting for 4.22% of total genes (Figures 7A,B). In most bacterial genomes, CAZymes typically comprise less than 2% of total genes (Mann et al., 2013). The genome of strain SCA4-21T exhibited a high percentage of CAZymes, suggesting its strong potential for degrading fungal cell walls. Specifically, the GH family of CAZymes in strain SCA4-21T genome contained 15 chitinase genes and eight β-1,3-glucanase genes (Figure 7D). These enzymes are responsible for catalyzing the hydrolysis of chitin and β-1,3-glucan, which are the major structural components of fungal cell walls. Accumulated data indicate that chitinases and β-1,3-glucanases produced by biocontrol microbes and plants can inhibit fungal growth (Li et al., 2021; Ruangwong et al., 2022). The study conducted by Arora et al. (2007) showed that the chitinase and β-1,3-glucanase produced by fluorescent pseudomonads can inhibit the growth of Rhizoctonia solani. Aktuganov et al. (2008) demonstrated that two enzymes chitinase and β-1,3-glucanase purified from Paenibacillus ehimensis IB-X-b have wide-range antifungal activity by degrading fungal mycelia cell walls. Anitha and Rebeeth (2009) reported that chitinase extracted from Streptomyces griseus can inhibit soil-borne plant pathogens such as Fusarium oxysporum, Alternaria alternata, Rhizoctonia solani, and Fusarium solani. Chitinase and β-1,3-glucanase produced by Streptomyces cavourensis SY224 contribute to the biocontrol of anthracnose in pepper (Lee et al., 2012). In the present study, the fermentation broth of strain SCA4-21T has high levels of chitinase and β-1,3-glucanase activities (Figure 7C). These findings suggest that these two enzymes could play a crucial role in the antifungal activity of strain SCA4-21T against Foc TR4.
A complete database of 16S rRNA sequences is available for the type strains of prokaryotic species, allowing rapid identification of strains at the genus level. Nonetheless, the phylogeny of the 16S rRNA gene is difficult to distinguish closely related species because of the high level of sequence conservation (Gevers et al., 2005; Guo et al., 2008; Richter and Rosselló-Móra, 2009). The gene clusters of atpD, gyrB, recA, rpoB, and trpB have a higher level of sequence divergence than that of 16S rRNA genes. MLSA based on these five housekeeping genes has been used as an alternative phylogenetic marker in species discrimination of Streptomyces (Rong et al., 2009; Huang et al., 2019; Qi et al., 2021, 2022). In our present study, strain SCA4-21T exhibited the highest similarity to S. iranensis HM 35T (99.45%), S. rapamycinicus NRRL B-5491T (99.35%), and S. hygroscopicus subsp. hygroscopicus NBRC 13472T (99.17%). However, the 16S rRNA phylogenetic analysis revealed that strain SCA4-21T clustered with S. iranensis HM 35T in the NJ and MP trees, with bootstrap values of 61 and 36%, respectively (Figure 6A; Supplementary Figure S2), while it clustered with S. iranensis HM 35T and S. rapamycinicus NRRL B-5491T in the ML tree (Supplementary Figure S3). Consequently, the phylogeny of the 16S rRNA gene was unable to effectively differentiate strain SCA4-21T from its closely related species. Therefore, MLSA of this strain based on five housekeeping genes (atpD, gyrB, recA, rpoB, and trpB) was performed. The NJ, MP, and ML phylogenetic trees all consistently indicated that strain SCA4-21T had the highest bootstrap values and formed a well-delineated subclade with S. hygroscopicus subsp. hygroscopicus NBRC 13472T (Figure 6B; Supplementary Figures S4, S5). The phylogenomic tree of strain SCA4-21T by comparing it to the type strains of Streptomyces further supported the above findings (Supplementary Figure S6). Therefore, strain SCA4-21T is closely related to S. hygroscopicus subsp. hygroscopicus NBRC 13472T.
After the identification of most closest species, the similarity alignment of the genome is required to assign the isolate to a known or unknown species (Gevers et al., 2005). When it comes to identifying prokaryotic species, the traditional DNA–DNA hybridization (DDH) is time-consuming and labor-intensive, with low inter-laboratory reproducibility (Richter and Rosselló-Móra, 2009). With the rapid development of sequencing technology, it has become possible to directly use the alignment of genome sequences to classify the species. The overall genome-related indexes (OGRI), such as ANI and dDDH, have been widely used to replace the traditional DDH to identify different species (Richter and Rosselló-Móra, 2009; Zhang et al., 2019; Meier-Kolthoff et al., 2022). The ANI and dDDH values for accepted species boundary are around 95–96 and 70%, respectively (Riesco and Trujillo, 2024). In this study, ANI and dDDH values were calculated between strain SCA4-21T and the adjacent seven strains in the phylogenetic trees of the 16S rRNA gene or MLSA. The genome of strain SCA4-21T showed 91.26% of ANI and 44.3% of dDDH with S. hygroscopicus subsp. hygroscopicus NBRC 13472T (Table 5). Both values are below the respective threshold values, indicating that strain SCA4-21T represents a novel species of Streptomyces. These findings offer a novel potential biocontrol agent for the management of plant fungal diseases.
A total of 30 actinobacteria were isolated and screened for their antagonistic activity against Foc TR4. Strain SCA4-21T exhibited a strong antifungal activity. Based on the morphological, physiological, biochemical, and chemotaxonomic characteristics, strain SCA4-21T was identified as genus Streptomyces. The genome of strain SCA4-21T was subjected to complete genome sequencing and assembly, resulting in a 10,044,493 base pair chromosome. This chromosome encodes 8,246 protein-coding genes, with a GC content of 71.48%. CAZymes analysis revealed the presence of 348 carbohydrate-active enzymes, which accounted for 4.22% of the total genes. These included 15 chitinases and 8 β-1, 3-glucanases related to fungal cell wall degradation. The phylogenetic trees of 16S rRNA do not distinguish the close species Streptomyces. MLSA showed that strain SCA4-21T clustered with S. hygroscopicus subsp. hygroscopicus NBRC 13472T with 100% of bootstrap value. The MLSA distance between strain SCA4-21T and S. hygroscopicus subsp. hygroscopicus NBRC 13472T is 0.021, significantly exceeding the threshold of 0.007 for the delineation of bacterial species, thereby suggesting that strain SCA4-21T represents a novel species. The taxonomic status of strain SCA4-21T was also supported by the phylogenomic tree. ANI and dDDH values are calculated to further verify the conclusion. The strain is named after Streptomyces luomodiensis sp. nov. The type strain is SCA4-21T (=GDMCC4.340T = JCM 34963T).
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
DQ: Conceptualization, Methodology, Writing – review & editing. QL: Formal analysis, Investigation, Writing – original draft. LiZ: Methodology, Writing – original draft. MZ: Methodology, Writing – original draft. KL: Writing – original draft, Formal analysis. YZ: Writing – original draft, Formal analysis. YC: Visualization, Writing – original draft. JF: Visualization, Writing – original draft. DZ: Writing – original draft, Resources. YW: Writing – original draft, Formal analysis. WW: Funding acquisition, Project administration, Writing – review & editing. LuZ: Writing – review & editing, Resources. JX: Funding acquisition, Project administration, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Natural Science Foundation of Hainan (322QN417, 322RC660, 321MS066, and 321RC543), the Central Public-interest Scientific Institution Basal Research Fund (1630052022006; CATASCXTD202309), the Hainan Science and Technology Association young talent lifting project (QCQTXM202203), and Chinese Academy of Tropical Agricultural Sciences for Science and Technology Innovation Team of National Tropical Agricultural Science Center (CATASCXTD202312).
We are grateful to Zhufeng Gao and Tianyu Li for their help in this work.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1402653/full#supplementary-material
1. ^ https://www.bacterio.net/
2. ^ https://www.ezbiocloud.net/
3. ^ https://www.ncbi.nlm.nih.gov/
Aktuganov, G., Melentjev, A., Galimzianova, N., Khalikova, E., Korpela, T., and Susi, P. (2008). Wide-range antifungal antagonism of Paenibacillus ehimensis IB-x-b and its dependence on chitinase and β-1,3-glucanase production. Can. J. Microbiol. 54, 577–587. doi: 10.1139/W08-043
Anitha, A., and Rebeeth, M. (2009). In vitro antifungal activity of Streptomyces griseus against phytopathogenic fungi of tomato field. Acad J Plant Sci. 2, 119–123,
Arora, N. K., Kim, M. J., Kang, S. C., and Maheshwari, D. K. (2007). Role of chitinase and beta-1,3-glucanase activities produced by a fluorescent pseudomonad and in vitro inhibition of Phytophthora capsici and Rhizoctonia solani. Can. J. Microbiol. 53, 207–212. doi: 10.1139/w06-119
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2015). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/MMBR.00019-15
Besemer, J., and Borodovsky, M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454. doi: 10.1093/nar/gki487
Bhatti, A. A., Haq, S., and Bhat, R. A. (2017). Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 111, 458–467. doi: 10.1016/j.micpath.2017.09.036
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bull, A. T. (2010). “Actinobacteria of the extremobiosphere” in Extremophiles handbook. eds. K. Horikoshi, G. Antranikian, A. T. Bull, F. Robb, and K. Stelter (Berlin: Springer-Verlag GmbH), 1203–1240.
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics 23, 673–679. doi: 10.1093/bioinformatics/btm009
Fan, Q., Tian, S., Liu, H., and Xu, Y. (2002). Production of p-1,3-glucanase and chitinase of two biocontrol agents and their possible modes of action. Chin. Sci. Bull. 47, 292–296. doi: 10.1360/02tb9070
Fu, L., Penton, C. R., Ruan, Y., Shen, Z., Xue, C., Li, R., et al. (2017). Inducing the rhizosphere microbiome by biofertilizer application to suppress banana Fusarium wilt disease. Soil Biol. Biochem. 104, 39–48. doi: 10.1016/j.soilbio.2016.10.008
Gevers, D., Cohan, F. M., Lawrence, J. G., Spratt, B. G., Coenye, T., Feil, E. J., et al. (2005). Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 3, 733–739. doi: 10.1038/nrmicro1236
Gonzalez-Franco, A. C., and Hernandez, L. R. (2009). Actinomycetes as biological control agents of phytopathogenic fungi. Tecnociencia Chihuahua 3, 62–73,
Guo, Y., Zheng, W., Rong, X., and Huang, Y. (2008). A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 58, 149–159. doi: 10.1099/ijs.0.65224-0
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 734, 95–98. doi: 10.1021/bk-1999-0734.ch008
Hamedi, J., Mohammadipanah, F., Klenk, H. P., Pötter, G., Schumann, P., Spröer, C., et al. (2010). Streptomyces iranensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 60, 1504–1509. doi: 10.1099/ijs.0.015339-0
Hasegawa, T., Takizawa, M., and Tanida, S. (1983). A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322. doi: 10.2323/jgam.29.319
Heydan, A., and Pessarakh, M. (2010). A review on biological control of fungal plant pathogens using microbial antagonists. J. Biol. Sci. 10, 273–290. doi: 10.3923/jbs.2010.273.290
Huang, X., Kong, F., Zhou, S., Huang, D., Zheng, J., and Zhu, W. (2019). Streptomyces tirandamycinicus sp. nov. a novel marine sponge-derived actinobacterium with antibacterial potential against Streptococcus agalactiae. Front. Microbiol. 10:482. doi: 10.3389/fmicb.2019.00482
Hyatt, D., Chen, G. L., LoCascio, P. F., Land, M. L., Larimer, F. W., and Hauser, L. J. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119
Jing, T., Zhou, D., Zhang, M., Yun, T., Qi, D., Wei, Y., et al. (2020). Newly isolated Streptomyces sp. JBS5-6 as a potential biocontrol agent to control banana Fusarium wilt: genome sequencing and secondary metabolite cluster profiles. Front. Microbiol. 11:3036. doi: 10.3389/fmicb.2020.602591
Jose, P. A., Maharshi, A., and Jha, B. (2021). Actinobacteria in natural products research: Progress and prospects. Microbiol. Res. 246:126708. doi: 10.1016/j.micres.2021.126708
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., Hopwood, D. A., Charter, K., et al. (2000). Practical streptomyces genetics. Norwich: John Innes Foundation, 2–33.
Kimura, M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. doi: 10.1007/BF01731581
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Law, J. W., Ser, H. L., Khan, T. M., Chuah, L. H., Pusparajah, P., Chan, K. G., et al. (2017). The potential of Streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae). Front. Microbiol. 17:3. doi: 10.3389/fmicb.2017.0000
Lee, S. Y., Tindwa, H., Lee, Y. S., Naing, K. W., Hong, S. H., Nam, Y., et al. (2012). Biocontrol of anthracnose in pepper using chitinase, beta-1,3 glucanase, and 2-furancarboxaldehyde produced by Streptomyces cavourensis SY224. J. Microbiol. Biotechnol. 22, 1359–1366. doi: 10.4014/jmb.1203.02056
Li, W., Long, Y., Mo, F., Shu, R., Yin, X., Wu, X., et al. (2021). Antifungal activity and biocontrol mechanism of Fusicolla violacea J-1 against soft rot in kiwifruit caused by Alternaria alternata. J. Fungi (Basel) 7:937. doi: 10.3390/jof7110937
Magaldi, S., Mata-Essayag, S., Hartung de Capriles, C., Perez, C., Colella, M. T., Olaizola, C., et al. (2004). Well diffusion for antifungal susceptibility testing. Int. J. Infect. Dis. 8, 39–45. doi: 10.1016/j.ijid.2003.03.002
Mann, A. J., Hahnke, R. L., Huang, S., Werner, J., Xing, P., Barbeyron, T., et al. (2013). The genome of the alga-associated marine Flavobacterium Formosa agariphila KMM 3901T reveals a broad potential for degradation of algal polysaccharides. Appl. Environ. Microbiol. 79, 6813–6822. doi: 10.1128/AEM.01937-13
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Meier-Kolthoff, J. P., Sardà Carbasse, J., Peinado-Olarte, R. L., and Göker, M. (2022). TYGS and LPSN: a database tandem for fast and reliable genome-based classification and nomenclature of prokaryotes. Nucleic Acid Res. 50, D801–D807. doi: 10.1093/nar/gkab902
Minnikin, D. E., O’Donnell, A. G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., et al. (1984). An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241. doi: 10.1016/0167-7012(84)90018-6
Mohandas, S., and Ravishankar, K. V. (2016). Banana: Genomics and transgenic approaches for genetic improvement. Singapore: Springer, 211–226.
Parte, A. C., Sardà Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Göker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Pérez-Vicente, L., Dita, M., and Martinez, D. L. P. E. (2014). Technical manual prevention and diagnostic of fusarium wilt (Panama disease) of banana caused by Fusarium oxysporum f. sp. cubense tropical race 4 (TR4). Rome: FAO, 4–13.
Ploetz, R. C. (2015). Management of Fusarium wilt of banana: a review with special reference to tropical race 4. Crop Prot. 73, 7–15. doi: 10.1016/j.cropro.2015.01.007
Qi, D., Zou, L., Zhou, D., Zhang, M., Wei, Y., Li, K., et al. (2022). Biocontrol potential and antifungal mechanism of a novel Streptomyces sichuanensis against Fusarium oxysporum f. sp. cubense tropical race 4 in vitro and in vivo. Appl. Microbiol. Biotechnol. 106, 1633–1649. doi: 10.1007/s00253-022-11788-3
Qi, D., Zou, L., Zhou, D., Zhang, M., Wei, Y., Zhang, L., et al. (2021). Identification and antifungal mechanism of a novel actinobacterium Streptomyces huiliensis sp. nov. against Fusarium oxysporum f. sp. cubense tropical race 4 of banana. Front. Microbiol. 12:722661. doi: 10.3389/fmicb.2021.722661
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U. S. A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Riesco, R., and Trujillo, M. E. (2024). Update on the proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 74:006300. doi: 10.1099/ijsem.0.006300
Rong, X., Guo, Y., and Huang, Y. (2009). Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst. Appl. Microbiol. 32, 314–322. doi: 10.1016/j.syapm.2009.05.003
Rong, X., and Huang, Y. (2010). Taxonomic evaluation of the Streptomyces griseus clade using multilocus sequence analysis and DNA-DNA hybridization, with proposal to combine 29 species and three subspecies as 11 genomic species. Int. J. Syst. Evol. Microbiol. 60, 696–703. doi: 10.1099/ijs.0.012419-0
Rong, X., and Huang, Y. (2012). Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 35, 7–18. doi: 10.1016/j.syapm.2011.10.004
Ruangwong, O. U., Kunasakdakul, K., Chankaew, S., Pitija, K., and Sunpapao, A. (2022). A rhizobacterium, Streptomyces albulus Z1-04-02, displays antifungal activity against sclerotium rot in mungbean. Plants (Basel) 11:2607. doi: 10.3390/plants11192607
Sasser, M. (1990). Identification of Bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. Newark, DE: MIDI Inc.
Shirling, E. B., and Gottlieb, D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Evol. Microbiol. 16, 313–340. doi: 10.1099/00207713-16-3-313
Shirling, E. B., and Gottlieb, D. (1972). Comparative description of type strains of Streptomyces: V. Additional Descriptions. Int J Syst Bacteriol. 22, 265–394. doi: 10.1099/00207713-22-4-265
Tatusova, T., DiCuccio, M., Badretdin, A., Chetvernin, V., Nawrocki, E. P., Zaslavsky, L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624. doi: 10.1093/nar/gkw569
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Waksman, S. A., and Henrici, A. T. (1943). The nomenclature and classification of the actinomycetes. J. Bacteriol. 46, 337–341. doi: 10.1128/jb.46.4.337-341.1943
Wang, J., Cai, B., Li, K., Zhao, Y., Li, C., Liu, S., et al. (2022). Biological control of fusarium oxysporum f. sp. cubense tropical race 4 in banana plantlets using newly isolated Streptomyces sp. WHL7 from marine soft coral. Plant Dis. 106, 254–259. doi: 10.1094/PDIS-06-21-1275-RE
Wang, L., Xing, M., Di, R., and Luo, Y. (2015). Isolation, identification and antifungal activities of Streptomyces aureoverticillatus HN6. J. Plant Pathol. Microb. 2, 233–241. doi: 10.4172/2157-7471.1000281
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Williams, S. T., Goodfellow, M., Alderson, G., Wellington, E. M. H., Sneath, P. H. A., and Sackin, M. J. (1983). Numerical classification of streptomyces and related genera. J. Gen. Microbiol. 129, 1743–1813. doi: 10.1099/00221287-129-6-1743
Yoon, S. H., Ha, S. M., Lim, J., Kwon, S., and Chun, J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110, 1281–1286. doi: 10.1007/s10482-017-0844-4
Yun, T., Jing, T., Zhou, D., Zhang, M., Zhao, Y., Li, K., et al. (2022). Potential biological control of endophytic Streptomyces sp. 5-4 against Fusarium wilt of banana caused by Fusarium oxysporum f. sp. cubense tropical race 4. Phytopathology 112, 1877–1885. doi: 10.1094/PHYTO-11-21-0464-R
Zacky, F. A., and Ting, A. S. Y. (2013). Investigating the bioactivity of cells and cell-free extracts of Streptomyces griseus towards Fusarium oxysporum f. sp. cubense race 4. Biol. Control 66, 204–208. doi: 10.1016/j.biocontrol.2013.06.001
Keywords: Streptomyces, novel species, antifungal activity, taxonomic identification, banana Fusarium disease, antagonistic mechanism
Citation: Qi D, Liu Q, Zou L, Zhang M, Li K, Zhao Y, Chen Y, Feng J, Zhou D, Wei Y, Wang W, Zhang L and Xie J (2024) Taxonomic identification and antagonistic activity of Streptomyces luomodiensis sp. nov. against phytopathogenic fungi. Front. Microbiol. 15:1402653. doi: 10.3389/fmicb.2024.1402653
Received: 19 March 2024; Accepted: 09 May 2024;
Published: 27 May 2024.
Edited by:
James T. Tambong, Agriculture and Agri-Food Canada, CanadaReviewed by:
Raul Riesco Jarrin, University of Salamanca, SpainCopyright © 2024 Qi, Liu, Zou, Zhang, Li, Zhao, Chen, Feng, Zhou, Wei, Wang, Zhang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianghui Xie, eGllamlhbmdodWlAaXRiYi5vcmcuY24=; Lu Zhang, bHV6aGFuZ3Rlc3RAaGFpbm51LmVkdS5jbg==; Wei Wang, d2FuZ3dlaXN5c0BhaGF1LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.