- 1School of Food and Bioengineering, Xihua University, Chengdu, China
- 2Key Laboratory of Food Microbiology of Sichuan, Xihua University, Chengdu, China
The toxin-antitoxin (TA) system plays a key role in bacteria escaping antibiotic stress with persistence, however, the mechanisms by which persistence is controlled remain poorly understood. Weissella cibaria, a novel probiotic, can enters a persistent state upon encountering ciprofloxacin stress. Conversely, it resumes from the persistence when ciprofloxacin stress is relieved or removed. Here, it was found that PemIK TA system played a role in transitioning between these two states. And the PemIK was consisted of PemK, an endonuclease toxic to mRNA, and antitoxin PemI which neutralized its toxicity. The PemK specifically cleaved the U↓AUU in mRNA encoding enzymes involved in glycolysis, TCA cycle and respiratory chain pathways. This cleavage event subsequently disrupted the crucial cellular processes such as hydrogen transfer, electron transfer, NADH and FADH2 synthesis, ultimately leading to a decrease in ATP levels and an increase in membrane depolarization and persister frequency. Notably, Arg24 was a critical active residue for PemK, its mutation significantly reduced the mRNA cleavage activity and the adverse effects on metabolism. These insights provided a clue to comprehensively understand the mechanism by which PemIK induced the persistence of W. cibaria to escape ciprofloxacin stress, thereby highlighting another novel aspect PemIK respond for antibiotic stress.
1 Introduction
The discovery of antibiotics in the late 1920s marked a significant milestone in the history of combating bacterial infection. However, it is now widely acknowledged that antibiotics are not the panacea they were once thought to be. Rather, antibiotics can and do fail, often with severe consequences (Page and Peti, 2016). This occurs because bacteria acquire resistance mutations to render the antibiotics ineffective or activate their endogenous mechanisms to evade antibiotic stress (Page and Peti, 2016). The former is a major culprit, but it would be overly simplistic to attribute all failures solely to this factor (Maisonneuve and Gerdes, 2014). In fact, when bacteria are treated with antibiotic, it is nearly impossible to kill them completely. Even if they are sensitive to antibiotic, there will always be a small percentage of cells known as persister cells that can withstand the lethal effect of antibiotic (Wilmaerts et al., 2019). Bacterial persistence is a state of dormancy induced by an endogenous mechanism, and unlike drug-resistant mutant, it does not proliferate in the presence of antibiotic, but, randomly in time, switches from a dormant state back to a growing state when antibiotic stress is relieved or removed. Therefore, it is generally accepted that persistence is an important reason for failure of antibiotic prevention or treatment of bacteria (Wilmaerts et al., 2019).
Persister process of bacteria is a survival strategy that allows populations to survive in sudden adverse environments without going extinct completely (Gerdes and Maisonneuve, 2012; Page and Peti, 2016; Wilmaerts et al., 2019). To date, persistence has been observed in nearly all bacterial species, even eukaryotic microorganisms (Wilmaerts et al., 2019). Although persister process is governed by phenotypic switching in isogenic populations, it occurs stochastically and independently of environment, albeit at a higher frequency in adverse conditions (Maisonneuve and Gerdes, 2014; Page and Peti, 2016; Wilmaerts et al., 2019). Numerous researchers have suggested that persister formation depends on the expression level of toxin-antitoxin systems (TAs) in bacteria. These systems encode two components, a stable toxin protein that inhibits cell growth and a labile antitoxin (either RNA or protein) that regulates toxin activity. When toxin protein exceeds a certain threshold in bacterial cell, persistence is induced (Wilmaerts et al., 2019).
PemIK is a novel type II TA system, originally discovered in plasmid pCH91 of Staphylococcus aureus (Bukowski et al., 2013). The toxin PemK dimer, an RNase, specifically cleaves the mRNA in a ribosome-independent manner. The antitoxin PemI neutralizes the toxicity of PemK, thereby facilitating bacterial recovery from persistence (Bukowski et al., 2013; Wilmaerts et al., 2019). In pathogenic bacteria such as S. aureus, Klebsiella pneumonia, Bacillus anthracis and Mycobacterium tuberculosis, free PemK dimers lead to a rapid formation and higher proportion of persister cells (Kim et al., 2022). Although PemK can induce persister cells, it possesses distinct pattern in regulating physiological activities across different organisms. In S. aureus, it comprehensively regulates virulence by cleaving target mRNA encoding proteins involved in virulence synthesis (Bukowski et al., 2013; Kim et al., 2022). However, in Caenorhabditis elegans, it affects fundamental physiological processes including ATP production, lipid synthesis, cytoskeleton organization, stress response, and others (Kim et al., 2022). However, the mechanism by which PemK orchestrates the specific regulation of bacterial transcription, thereby manipulating the transition from growth to persistence, remains elusive.
Weissella cibaria, a novel probiotic and hetero-lactic bacterium, is usually present in various spontaneous fermentation foods. Interestingly, it can survive in various stresses through persistence, including antibiotic residue in raw materials, high acidity and nutrient hunger, and even growth inhibitor. In its persister cells, PemIK responds strongly to the persister formation under ciprofloxacin stress. Although the structure and function of PemIK in pathogenic bacteria, particularly S. aureus, are somewhat known, it remains unknown in W. cibaria due to its species-specific mode of regulation. Therefore, the present study analyzed the structure and function of PemIK in W. cibaria, and investigated the PemK’s adverse effects on basic metabolic processes. Meanwhile, the detoxification effect of PemI on PemK was also determined. The results provided a structural basis for understanding the molecular mechanism by which PemIK induced persistence of W. cibaria to survive in ciprofloxacin stress, and also offered insights into strategies for discovering new agents that could activate or inhibit W. cibaria.
2 Materials and methods
2.1 Bacterial strains and culture conditions
Weissella cibaria CGMCC 1.19376 was cultured in De Man, Rogosa, and Sharpe (MRS) broth at 30°C (Cai et al., 2022). Escherichia coli BL21 was cultivated in Luria-Bertani (LB) liquid me-dium at 37°C.
2.2 Transcription analysis of PemIK
Total RNA was extracted using TRIzol Reagent (ThermoFisher Scientific Inc., Massachusetts, USA). Subsequently, cDNA synthesis was performed using the prime-script reverse transcriptase and random primers according to manufacturer’s instructions (TaKaRa, Beijing, China). Polymerase chain reaction (PCR) was conducted with PemK-F, PemK-R, PemI-F, and PemI-R primers (Supplementary Table 1) for amplifying the PemI and PemK genes. Templates included cDNA, mRNA, and gDNA (genomic DNA of W. cibaria). The promoter and terminator analysis of PemKI were performed by BPROM program (Shahmuradov, 2003).
2.3 Structural analysis of PemK
The primary and secondary structures of PemK were analyzed by Clustal Omega and Jalview,1 respectively. The results were visualized by ESPript 3.0 (Robert and Gouet, 2014). Three-dimensional models of PemK protein were generated using SWISS-MODEL (Waterhouse et al., 2018). The dimeric structure of PemK was submitted along with mRNA containing 5′ UAUU 3′ to the HADDOCK 2.2 webserver (Van Zundert et al., 2016). Subsequently, a model of PemK bound to RNA was generated and visualized by PyMOL (PyMOL Molecular Graphics System, version 2.3.0) (Schiffrin et al., 2020).
2.4 Identification of active residues in PemK
The SNAP22 predicted the mutation of active amino acid in PemK (Bromberg and Rost, 2007). The thermal stability, solvent-accessible surface area and electrostatic potential energy of PemK were analyzed by MUpro,3 CCP44 and APBS plug in PyMOL, respectively.
2.5 Cytotoxicity analysis of PemIK
PemK and its mutant were ligated to pET28a plasmid, while PemI was ligated to pBAD43 plasmid. Subsequently, the recombinant pET28a and pBAD43 were co-transformed into E. coli BL21. Following cultivation to an optical density (OD) 600 nm of 0.2–0.3, PemK and PemI were induced by 1 mM IPTG and 0.2% L-arabinose, respectively. Cytotoxicity of PemK and neutralization of PemK by PemI were assessed through growth curves of E. coli BL21 at OD600nm.
2.6 Regulation of cell membrane potential, endogenous reactive oxygen species (ROS) and intracellular ATP by PemK
Weissella cibaria at 1 × 107 CFU/ml in logarithmic stage was treated with 1/2 MIC ciprofloxacin (16 μg/ml) at 30°C for 1–8 h. Recombinant E. coli BL21 was cultured to logarithmic stage and adjusted to 1 × 107 CFU/ml, followed by induction of PemK using 1 mM IPTG at 37°C for 1–8 h. The cell membrane potential was assessed using the rhodamine 123 fluorescence method (Yu et al., 2022). The endogenous ROS level was analyzed following the method described by Xiong et al. (2023). Intracellular ATP was determined as described by Shan et al. (2017). PBS (pH 7.4) treatment served as control.
2.7 Effect of PemK on expression of metabolic enzymes
The splicing mRNA of PemK were analyzed based on the genome sequence of W. cibaria, focusing on metabolic enzymes in oxidative phosphorylation (OP), glycolysis, and citric acid cycle (TCA). To further validate these impacts, transcriptome analysis was conducted in recombinant E. coli BL21. The total RNA with high-quality from recombinant E. coli BL21 using TRIzol Reagent (ThermoFisher Scientific Inc., MA, USA) was used to construct the RNA-Seq library for sequencing on Illumina Hiseq platform (Hiseq 4000, 150PE) with NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) by Majorbio Technology Co. Ltd (Shanghai, China). The RNA-seq data analysis was conducted as methods described by Wang et al. (2015). The differential expression was calculated by DESeq R package (1.18.0). The transcription with log2 fold change > 1 was defined as the metabolic enzymes affected by PemK.
2.8 Regulation of cell persistence and morphology by PemK
The persister cells were detected using method of biphasic bactericidal curve (Sun et al., 2017). Briefly, W. cibaria was incubated in MRS broth at 30°C to 107 CFU/ml, then ciprofloxacin was added to 1/2 MIC and incubated again for 0, 1, 3, 5, 7 and 9 h. Subsequently, 0.5 ml cultures were transferred into 20 ml MRS broth containing MBC (2,048 μg/ml) ciprofloxacin and incubated at 30°C for 24 h. The 1 ml cultures were harvested by centrifugation at 4,000 rpm for 1 min and washed twice with PBS (pH 7.4). After cell pellets were resuspended and serially diluted 10-fold with PBS (pH 7.4), the dilution cells were inoculated on MRS agar plate to count after 24 h incubation at 30°C. The persister frequency was expressed as the ratio of viable bacteria in treated and untreated group with 1/2 MIC ciprofloxacin. After PemK was induced by IPTG in recombinant E. coli BL21, no-persister cells were killed with MBC ciprofloxacin in LB broth medium. The persister frequency of recombinant E. coli BL21 was carried out following similar procedures described above. The cell morphology was observed in FEI inspect F50 scanning electron microscope (Inspect F50, FEI, Hillsboro, USA).
2.9 Statistical analysis
All experiments were conducted in triplicate for each sample, and the data was presented as the mean values ± standard deviation, with variations less than 10%. Statistical significance analysis was carried out using analysis of variance (ANOVA) with Tukey’s post-hoc test in SPSS 20.0, and a p-value less than 0.05 was considered statistically significant.
3 Results
3.1 PemIK is a typical II TA stress regulator
Co-localization and co-transcription in an operon are common characteristics of type II-TAs (Zhou et al., 2021). In W. cibaria, the PemI and PemK genes were situated at locus 8,267–8,949 on plasmid PA010 (Supplementary Figure 1), sharing a promoter and terminator in transcription but possessing independent start and stop codons in translation (Figure 1A). Both open reading frames (ORFs) consisted of a frame with 342 bp encoding 114 amino acids followed by a TAA termination codon. Interestingly, a genetic overlap of seven base pairs between them (Figure 1A), which distinguished it from other PemIK structures.
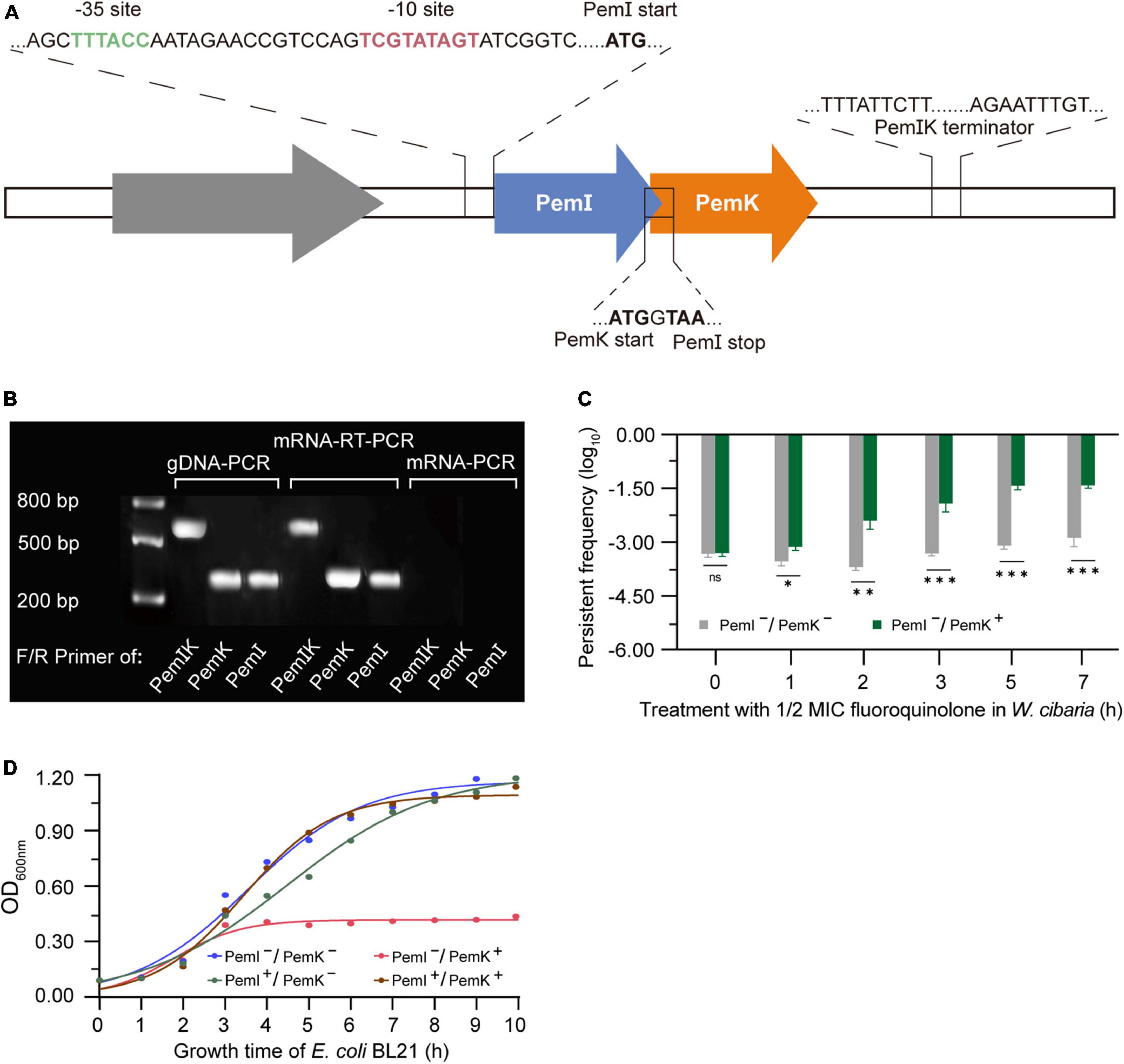
Figure 1. The structural characteristics and pressure response mechanism of PemIK. (A) Operon structure of PemIK of W. cibaria. (B) Validation of co-transcription. (C) Persister frequency of W. cibaria after treatment with 1/2 MIC ciprofloxacin. (D) Growth curve of recombinant E. coli BL21, demonstrating that PemI counteracted the cytotoxic effects of PemK through physical binding. Data were assessed by ANOVA with Tukey’s post-hoc test. The asterisk (*) indicated significant level: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns), P > 0.05.
When W. cibaria was exposed to 1/2 MIC ciprofloxacin, the co-transcription of PemIK was activated (Figure 1B), while PemI underwent degradation by Lon enzyme. Consequently, free PemK exhibited cytotoxicity to W. cibaria, leading to a severe growth inhibition and a significant increase in proportion of persister cells (Figure 1C). It has been demonstrated that PemI could counteract the cytotoxicity of PemK in recombinant E. coli BL21 (Figure 1D). Moreover, it also functioned as a transcription factor by inhibiting their co-transcription. Therefore, once the adverse conditions were alleviated or eliminated, the persistence of W. cibaria resumed its growth.
3.2 PemK is a novel specific endonuclease of mRNA
The toxins of II-type TAs are generally enzymes, particularly RNases (Díaz-Orejas et al., 2017). PemK from W. cibaria shared conserved residues and exhibited similar folds with its homologs, a barrel-shaped pattern of 7 β-sheets and 2 α-helices (Figure 2A). However, there were some differences in their secondary structures (Figure 2B), suggesting that they have similar functions but different mechanisms in controlling mRNA binding. The binding of mRNA typically occurred at the like-gate interfaces, which corresponded to the crevices between β1–β2 and β3–β4 sheets of one monomer and α1 helices of the other monomer in PemK dimers from W. cibaria (Figure 2C). The gate of PemK dimer from W. cibaria differed from that observed in its homologs from S. aureus, E. coli, B. subtilis, and B. anthracis (Figure 2B). Simultaneously, its electrostatic potential surface also displayed variations compared to its homologs (Figures 2D–H), it featured a weak positive charge plaque on its surface. These unique structural differences suggested that its mechanism for binding and cleaving mRNA differed from its homologs.
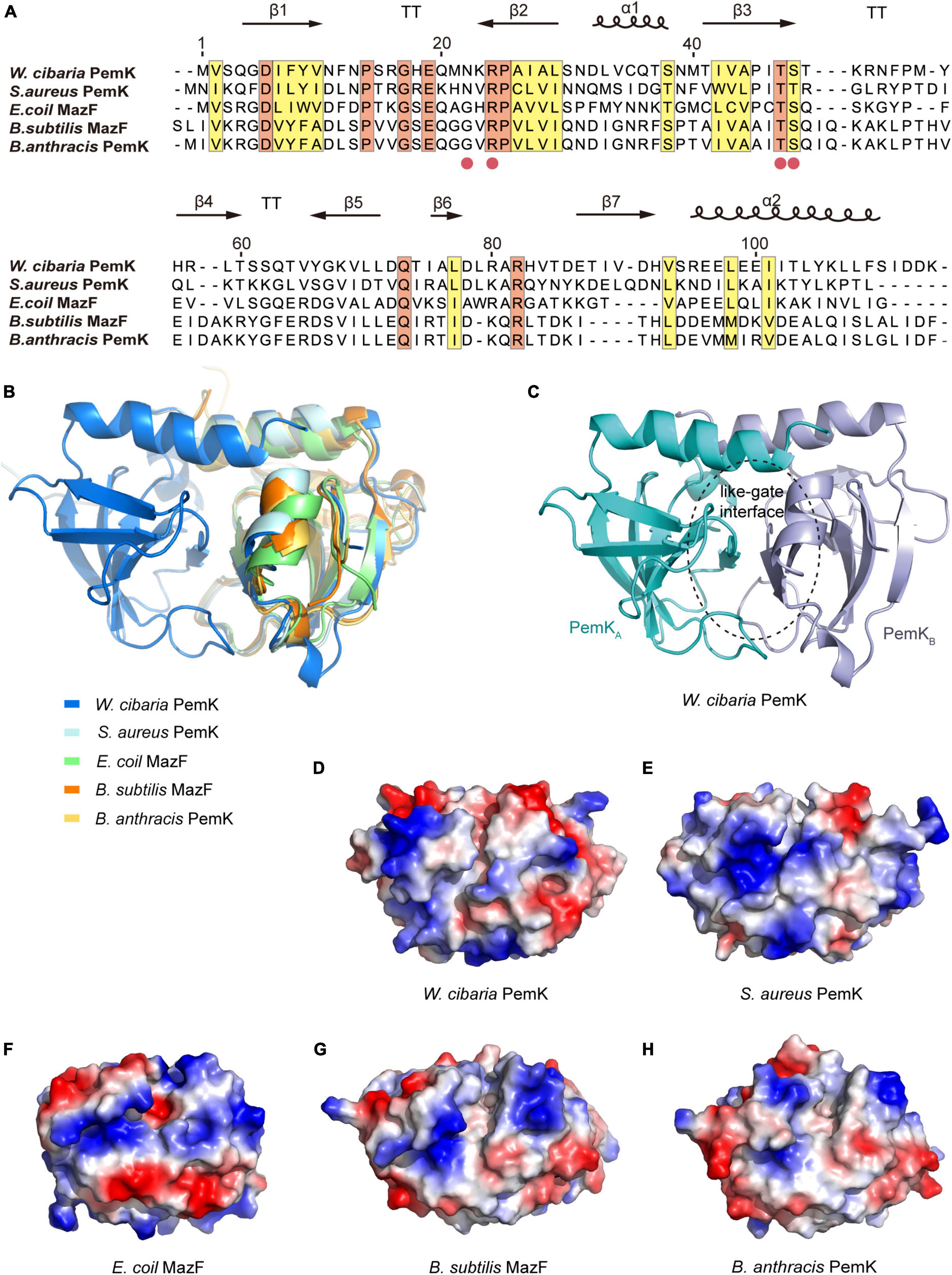
Figure 2. The structural comparison of PemK with its homologs. (A) Sequence alignment of PemK with its homologs. Secondary structures were denoted in the upper region. Potential active site residues are marked as red circles. Conserved and similar residues were shown in orange and yellow boxes, respectively. α: α-helix, β: β-sheet, T: turn. (B) Overlay structure of PemK with other homologs. (C) Overall structure of the PemK dimer. (D–H) Electrostatic surface potential represented the different charge distributions at the gate-like interface regions in PemK and its homologs.
PemK specifically recognizes and cleaves the tetrad U↓AHU (where H is C, A, or U) sequence in target mRNAs (Zhang et al., 2004; Zorzini et al., 2016; Cho et al., 2017). In S. aureus, Arg25 and Thr48 had been confirmed as key active residues for PemK (Kim et al., 2022). In W. cibaria, the mRNA’s U↓AHU (where H is U) sequence bound to the like-gate interfaces of PemK dimer through four hydrogen bonds and four van der Waals contacts (Figures 3A, B). Specifically, Asn22 and Arg24 formed hydrogen bonds with the phosphate group between U↓A, while Thr47 and Ser48 bound to the adenine base of U↓A through hydrogen bonds (Figure 3B). However, among these residues, Asn22 was not highly conserved in homologs based on sequence alignment (Figure 2A), and its substitution with any amino acid had little impact on PemK’s function (Figure 3C). Even when Asn22 was mutated to Trp in situ, there were no changes observed in the electrostatic potential surface of its gate interface (Figures 3D, E). Therefore, Arg24, Thr47 and Ser48 might be important residues influencing mRNA-binding and catalytic activity.
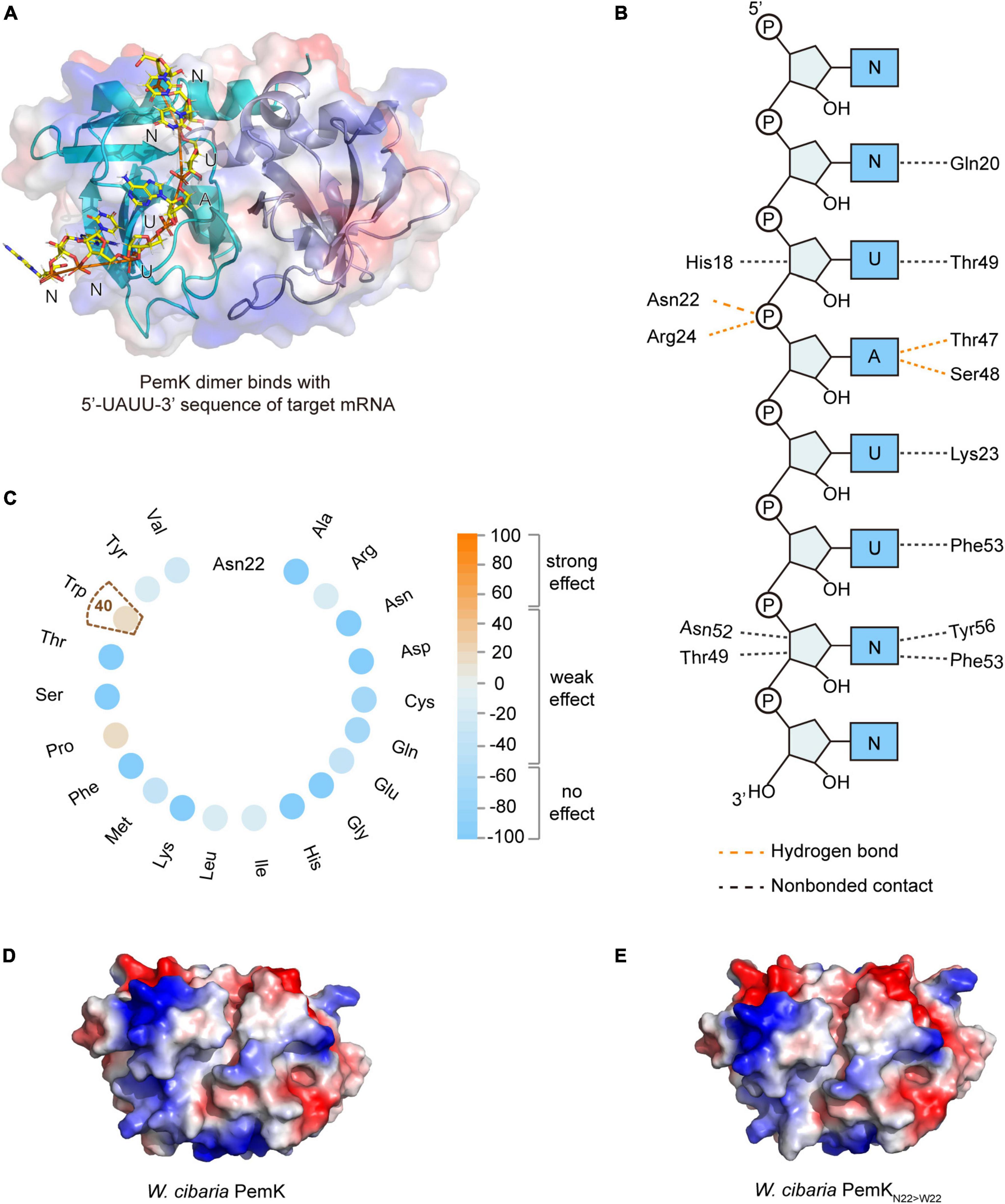
Figure 3. The mRNA binding and cleavage of PemK. (A) Model of PemK-mRNA complex depicted the stick model of PemK dimer and target mRNA. (B) NUCPLOT diagram of contacts between mRNA and PemK. Orange and black dotted lines represented hydrogen bonds and non-bonded contacts, respectively. (C) SNAP2 predicted the impact scores on the function of PemK after Asn22 (N22) was mutated to 20 amino acids, respectively. (D) Electrostatic surface potential of PemK. (E) Electrostatic surface potential of PemKN22>W22 that Asn22 was mutated to Trp22 (W22).
3.3 Arg24 is crucial for the RNase activity of PemK
The function of protein is often closely related to its structure and charge distribution (Gorham et al., 2011). To gain more detailed insights into the active site of PemK, mutations were predicted in situ at Arg24, Thr47, and Ser48 residues (Figure 4A). Interestingly, substitution of Arg24 by most amino acid strongly affected PemK function, resulting in a score ranging from 52 to 91 (Figure 4A and Supplementary Table 2). While substitution of Ser48 had weak or no effects with a score below 65 (Figure 4A). The impact of Thr47 substitution was generally less pronounced than that of Arg24, a score below 80 (Figure 4A). In PemK-mRNA complex, Arg24 bound to the phosphate group in U↓A through a hydrogen bond (Figure 3B). In S. aureus PemK and B. subtilis MazF, the amino acid residues, which bound to the phosphate group between U↓A via hydrogen bonding, were considered active sites involved in cleaving the tetrad U↓AUU sequence (Bukowski et al., 2013). Notably, Arg24 of PemK from W. cibaria corresponded well to Arg25 and Arg26 (Figure 2A), an important active residue in S. aureus PemK and B. subtilis MazF. So it might be a crucial active site for PemK to cleave the 3′,5′-phosphodiester bond of target mRNA.
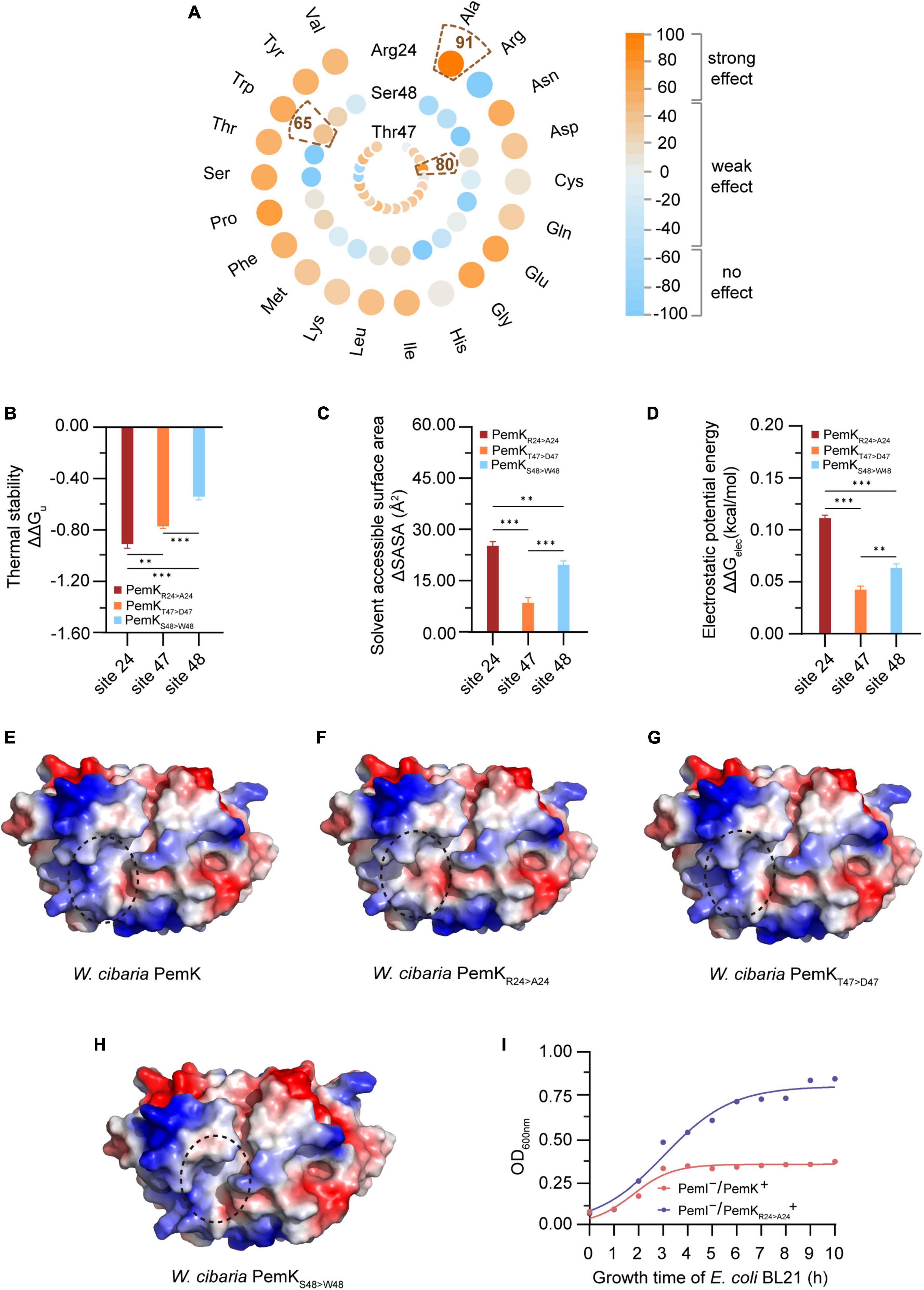
Figure 4. The properties of PemK after mutation of Arg24, Thr47 and Ser48. (A) SNAP2 predicted the impact scores on the function of PemK after Arg24, Thr47 and Ser48 were mutated to 20 amino acids, respectively. (B–D) Differences of thermal stability, solvent accessible surface area, electrostatic potential energy of PemKR24>A24 (Arg24 was mutated to Ala24), PemKT47>D47 (Thr47 was mutated to Asp47) and PemKS48>W48 (Ser48 was mutated to Trp48), compared with the wild-type PemK. (E–H) Electrostatic surface potential of PemK, PemKR24>A24, PemKT47>D47, and PemKS48>W48. The change in electrical potential energy at the point mutation locations were highlighted in dotted circles. (I) Growth curve of recombinant E. coli BL21 after induction of PemK and PemKR24>A24 expression. Data were assessed by ANOVA with Tukey’s post-hoc test. The asterisk (*) indicated significant level: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns), P > 0.05.
The unfolding free energy (ΔGu) is an important indicator of protein thermostability, and the ΔΔGu (ΔΔGu = ΔGmut–u—ΔGwt–u) can characterize the change in the thermostability caused by site-directed mutations (Verstraeten et al., 2015). The substituting in situ of Arg24, Thr47 and Ser48 decreased the thermostability of PemK, with substituting of Arg24 with Ala being more pronounced than the others, ΔΔGu = −0.91 (Figure 4B). In addition, the solvent accessible surface area (SASA) also characterized the instability caused by mutations (Figure 4C). A larger SASA value indicates a higher solvent-accessible surface area of protein, which is associated with lower stability. Three site-directed mutations in situ resulted in an increase in SASA of PemK, in which the ΔSASAArg24>Ala24 (25.18) > ΔSASASer48>Trp48 (19.64) > ΔSASAThr47>Asp47 (8.47) (Figure 4C).
The mutations may alter the charge distribution of protein’s active site or ligand-binding site, thereby impacting its affinity and specificity for the target molecules (Gorham et al., 2011). In W. cibaria, substituting Arg24 with Ala significantly altered the charge distribution on electrostatic potential surface of PemK dimers (Figures 4E, F), particularly at the like-gate interfaces where a transition occurred from positive charge to weak negative charge. Moreover, substituting Arg24 with Ala led to a more pronounced destabilization of PemK compared to Thr47 and Ser48 mutations, as indicated by ΔΔGelec (ΔΔGelec = ΔGmut–elec—ΔGwt–elec): ΔΔGelec–Arg24>Ala24 (0.11) > ΔΔGelec–Ser48>Trp48 (0.06) > ΔΔGelec–Thr47>Asp47 (0.04) (Figures 4D, G, H). Additionally, it was observed that PemKArg24>Ala24 significantly reduced its cellar toxicity, without a significant inhibition on the recombinant E. coli BL21 (Figure 4I). These findings suggested that Arg24 was more likely to affect PemK’s ability to cleave U↓AUU sequence, thereby influencing cell regulation in response to adverse external environments.
3.4 PemKArg24>Ala24 mitigates the impairment of respiratory chain
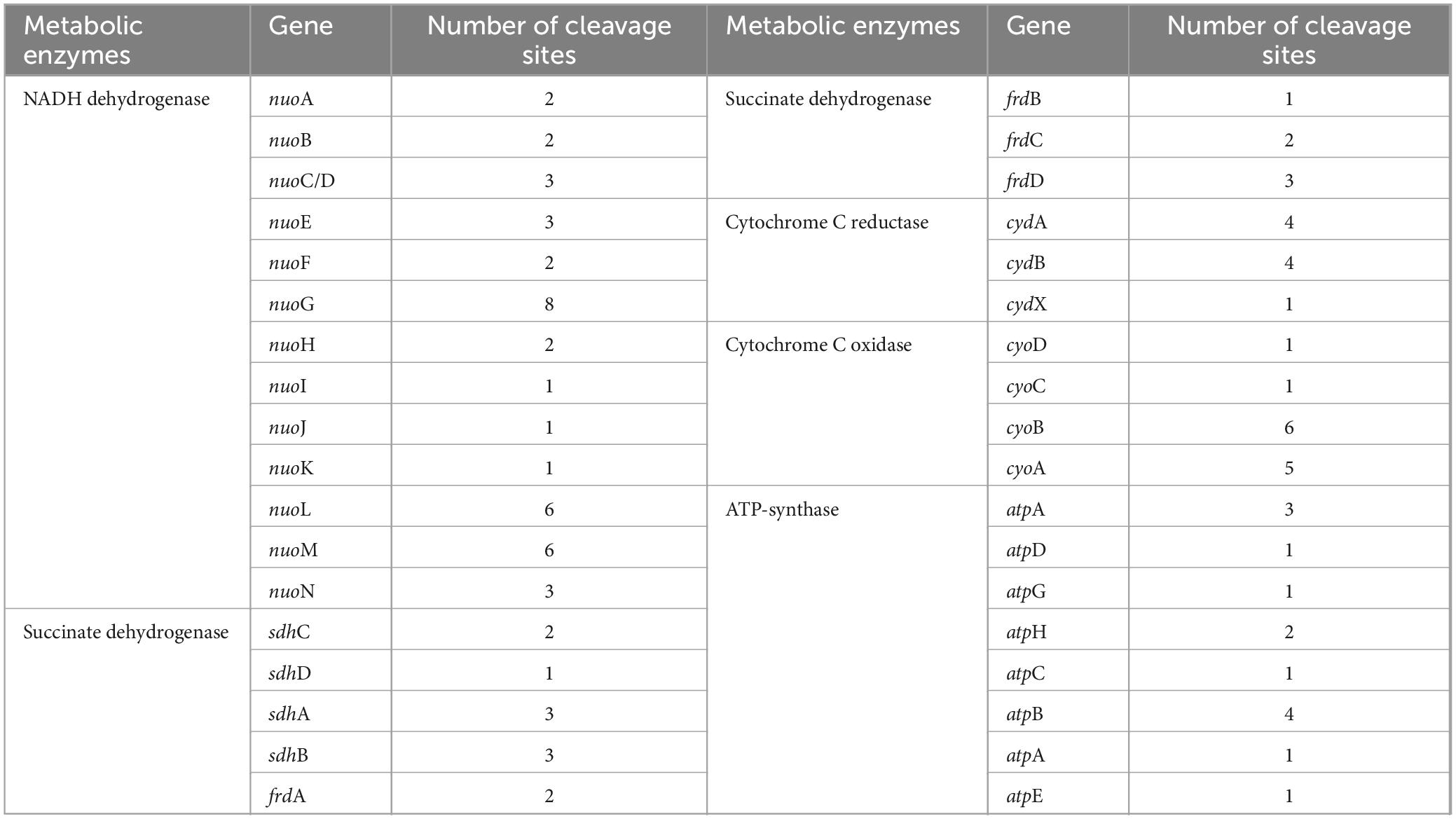
Persistence has been associated with reduced levels of cellular ATP (Verstraeten et al., 2015; Wilmaerts et al., 2019). The respiratory chain is the main process to generate cellular ATP, which is composed of a series of hydrogen transfer and electron transfer reactions mediated by complexes I to IV and ATP-synthase on cell plasma membrane (Figure 5A). In W. cibaria, PemK cleaved the target mRNAs of complexes I–IV and ATP-synthase, including 13 genes in complex I, 8 genes of complex II, 3 genes of complex III, 4 genes of complex IV, and 8 genes of ATP-synthase (Table 1 and Supplementary Table 3). In recombinant E. coli BL21, Ala substitution for Arg24 impaired PemK’s ability to cleave these target mRNAs, especially nuoE in complex I, sdhD and frdA in comolex II, atpG and atpF in ATP-synthase (Figures 5B–F), thus reducing the adverse effects of PemK on respiratory chain.
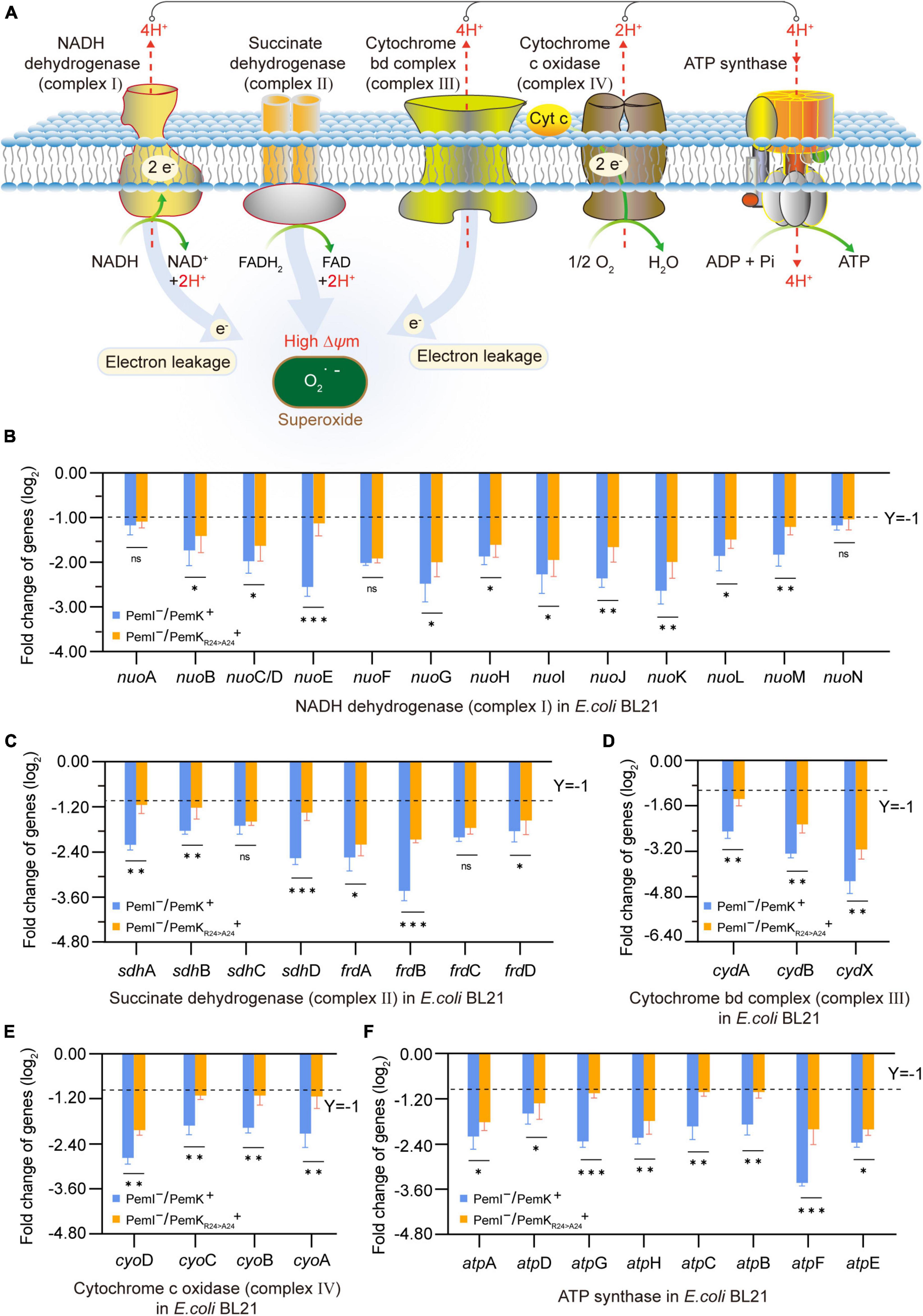
Figure 5. The effects on the respiratory chain before and after PemK mutation. (A) Schematic of respiratory chain. (B–F) Effects of PemK and PemKR24>A24 on gene transcription of complexes I–IV and ATP-synthase in recombinant E. coli BL21. Data were assessed by ANOVA with Tukey’s post-hoc test. The asterisk (*) indicated significant level: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns), P > 0.05.
Cleavage of mRNA caused the mRNA to break before translation, thus it blocked the synthesis of complexes I–IV and ATPase, resulting in the failure of H+ and e– transfer reactions in respiratory chain, which reduced the membrane potential and ATP production of W. cibaria (Figures 6A, B). Alterations in membrane potential caused membrane depolarization (Figure 6C), so the heightened fluorescence intensity corresponded to a more lower membrane potential (Figure 6A). In recombinant E. coli BL21, PemK significantly reduced the membrane potential and increased the membrane depolarization, while Ala substitution for Arg24 significantly alleviated the degree of membrane potential reduction and membrane depolarization, with an alleviation of 7.8–19.6% (Figure 6D). The membrane potential generated a proton motive force (PMF), which promotes the movement of H+ across cell membrane and ultimately produces ATP via ATP synthase. Therefore, the changes in membrane potential directly affected the production of ATP in W. cibaria, generally reducing the amount of ATP by 20–40% (Figure 6B). Additionally, blocked electron transport obstructed the reduction of O2, leading to the accumulation of excess ROS in W. cibaria to 1.50–2.30 times of normal levels (Figure 6E). Similarly, Ala substitution for Arg24 had also moderated the adverse effects on the ATP production and reduced ROS accumulation in recombinant E. coli BL21, with ATP content increased by 20.8–276.60% and ROS accumulation reduced by 7.8–15.6%, respectively (Figures 6F–K). These insights implied that Arg24 played an important role in regulation of respiratory chain by PemK.
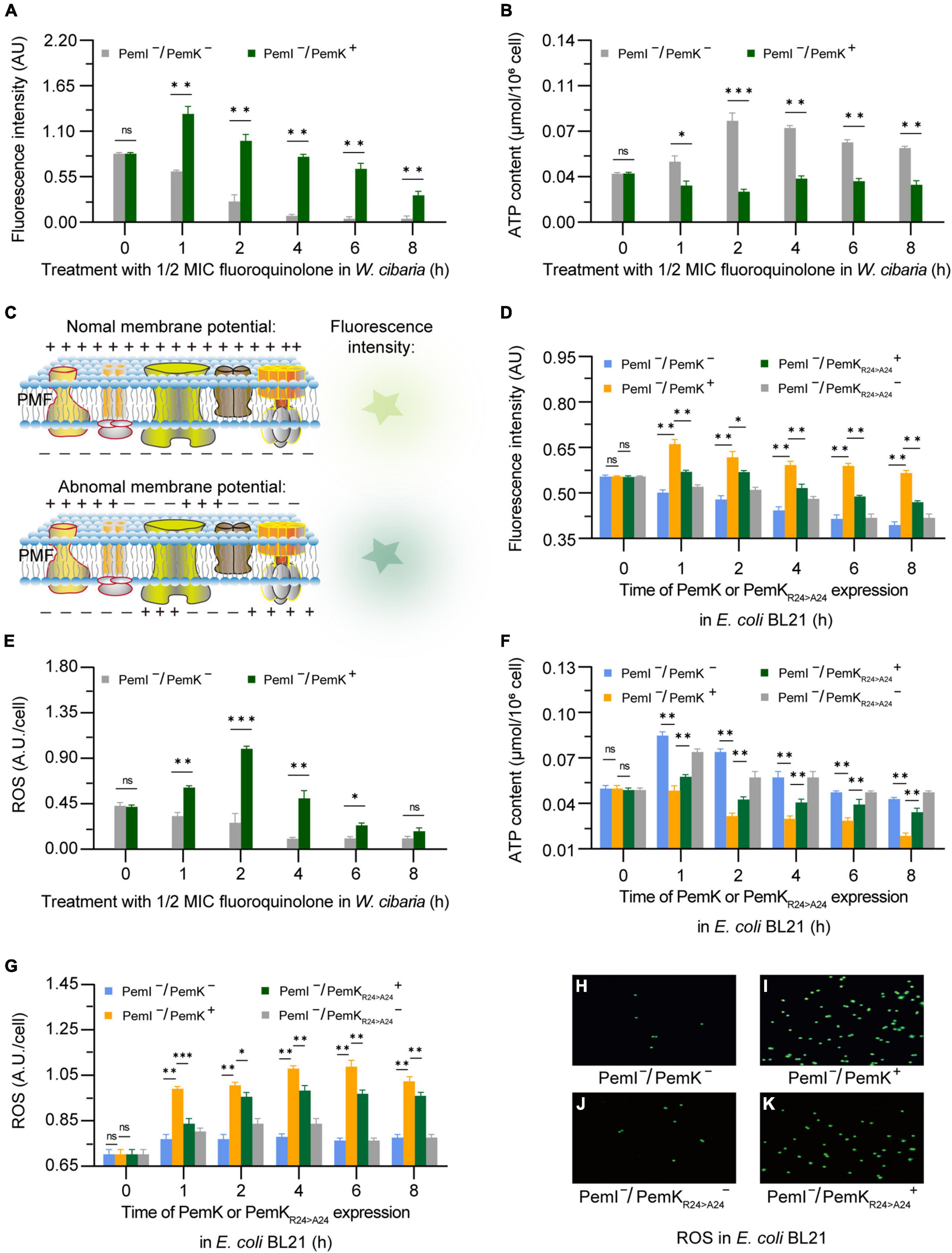
Figure 6. The alterations in membrane potential, ATP, and ROS caused by PemK and PemKR24>A24 in W. cibaria and recombinant E. coli BL21, respectively. (A,B,E) Membrane potential, ATP, and ROS of W. cibaria after treatment with 1/2 MIC ciprofloxacin. (C) Schematic of membrane potential. (D,F,G) Membrane potential, ATP, and ROS of recombinant E. coli BL21 after induction of PemK and PemKR24>A24 protein expression. (H–K) Fluorescence of intracellular ROS in recombinant E. coli BL21. Data were assessed by ANOVA with Tukey’s post-hoc test. The asterisk (*) indicated significant level: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns), P > 0.05.
3.5 PemKArg24>Ala24 alleviates the adverse effects on NADH and FADH2 synthesis
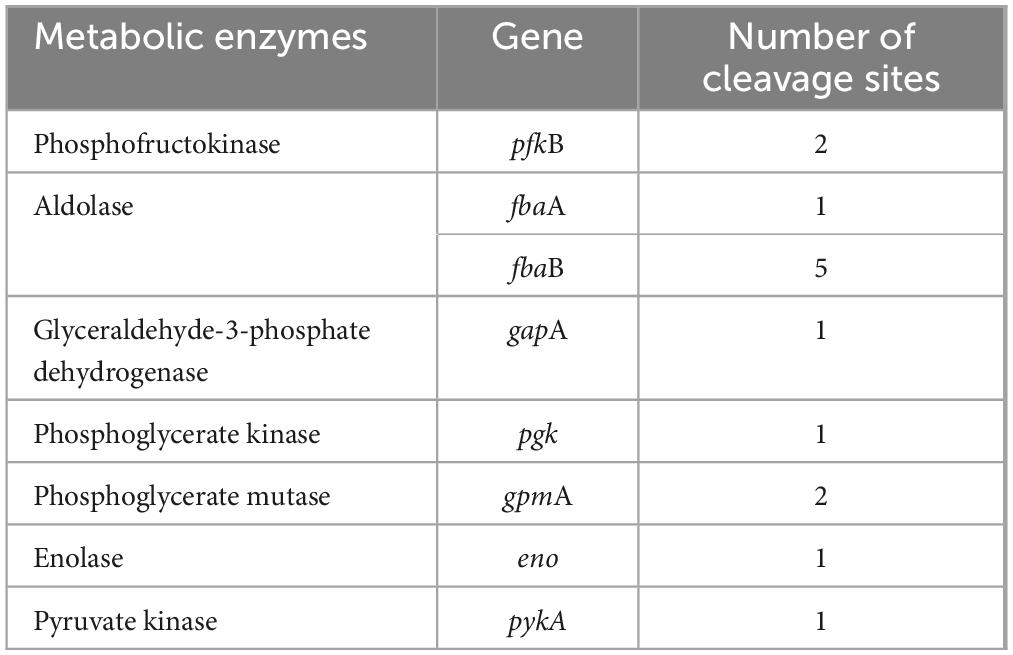
NADH and FADH2 are the important reducing agents to supply H+ for cells via the respiratory chain pathway, while glycolysis and TCA cycle serve as their primary sources. Glycolysis catalyzes a glucose into two pyruvates, accompanied by production of 2 NADH. This process involves ten enzyme-catalyzed reactions (Chandel, 2021), and any dysfunction of them can lead to disorders in NADH synthesis (Figure 7A). In glycolysis of W. cibaria, PemK only targeted the mRNA of one or two subunits of phosphofructokinase, aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, phosphoglycerate mutase, enolase and pyruvate kinase (Table 2 and Supplementary Table 4).
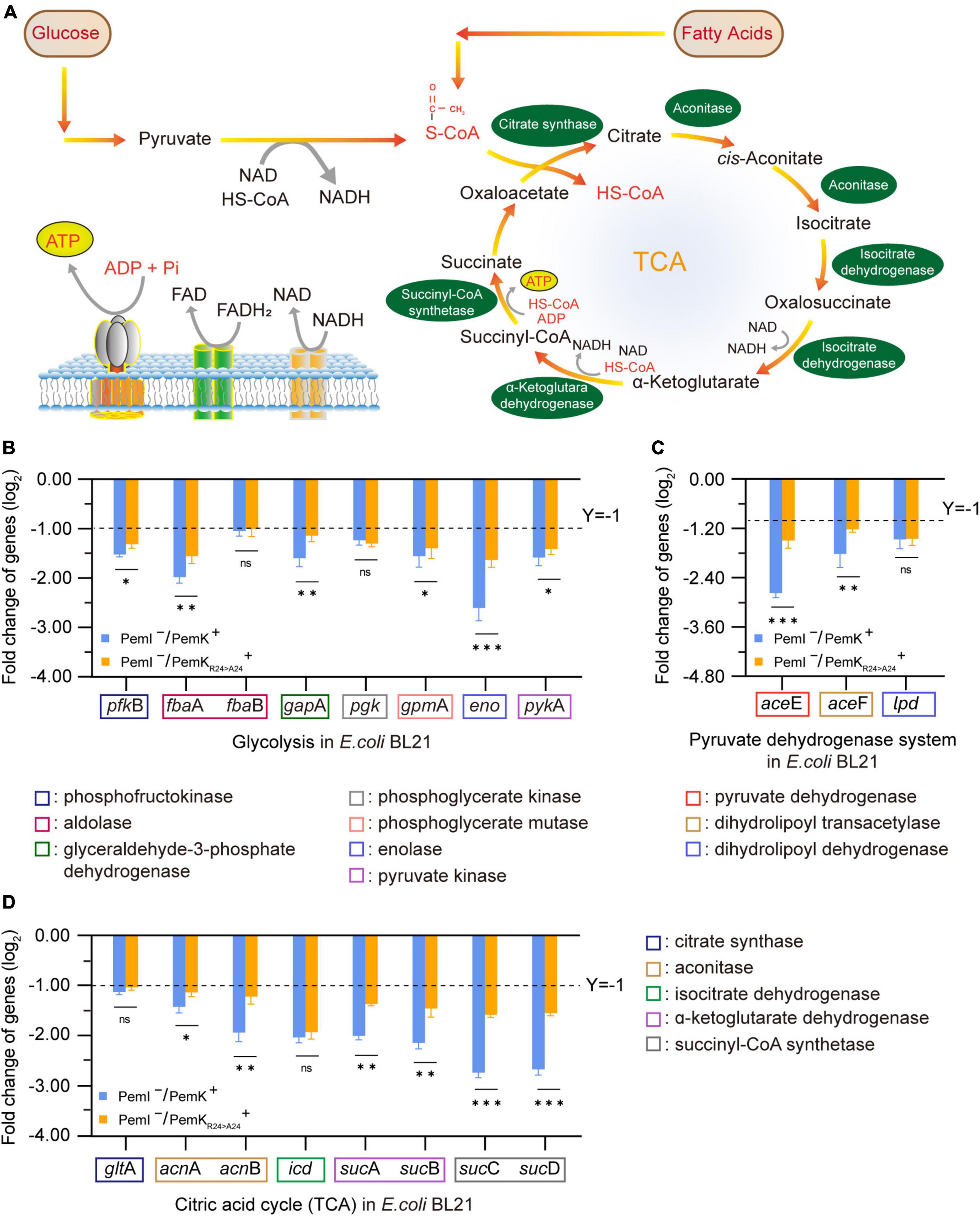
Figure 7. The effects on NADH and FADH2 synthesis before and after PemK mutation in recombinant E. coli BL21. (A) Schematic of glycolysis and TCA cycle. (B) Effects of PemK and PemKR24>A24 on the gene transcription of glycolysis. (C) Effects of PemK and PemKR24>A24 on the gene transcription of pyruvate dehydrogenase system. (D) Effects of PemK and PemKR24>A24 on the gene transcription of TCA cycle. Data were assessed by ANOVA with Tukey’s post-hoc test. The asterisk (*) indicated significant level: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns), P > 0.05.
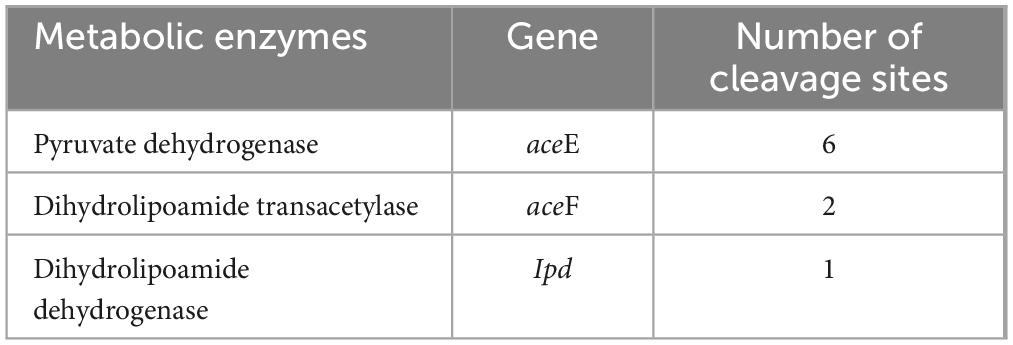
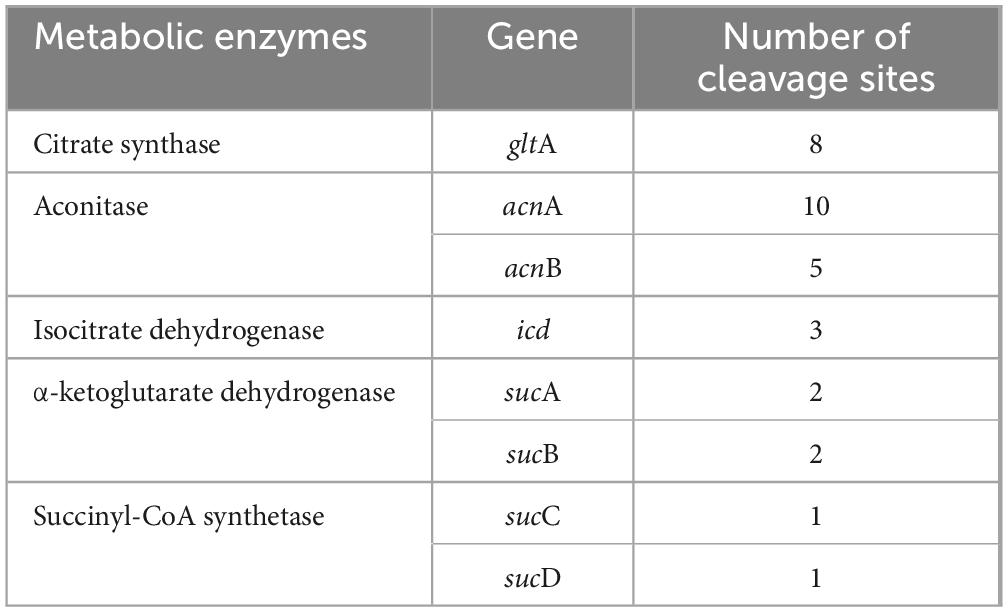
Acetyl-CoA serves as the bridge between glycolysis and TCA cycle, generated by the pyruvate dehydrogenase system consisting of pyruvate dehydrogenase, dihydrolipoamide transacetylase, and dihydrolipoamide dehydrogenase. In W. cibaria, PemK also cleaved their mRNA (Table 3 and Supplementary Table 5). Acetyl-CoA enters TCA to undergo a series of enzymatic reactions into CO2 and H2O while producing 3 NADH and 1 FADH2. However, PemK only targeted the mRNA of one or two subunit of citrate synthase, aconitase, isocitrate dehydrogenase, α-ketoglutarate dehydrogenase and succinyl-CoA synthetase in W. cibaria (Table 4 and Supplementary Table 6). In recombinant E. coli BL21, PemK significantly impaired the mRNA of these target enzymes involved in glycolysis, pyruvate oxidative decarboxylation and TCA cycle by −1.05–2.98 log2 folds (Figures 7B–D). However, Ala substitution for Arg24 effectively alleviated these adverse impacts except for phosphoglycerate kinase, dihydrolipoamide dehydrogenase, citrate synthase and isocitrate dehydrogenase. Therefore, Arg24 was a crucial site for PemK to disrupt the NADH and FADH2 synthesis in W. cibaria.
3.6 PemKArg24>Ala24 attenuates growth inhibition and persister frequency
Induction of persistence appears to be the primary physiological function of TAs in bacteria, and stimulating the expression of type II toxins can significantly enhance the persister frequency (Maisonneuve and Gerdes, 2014; Merfa et al., 2016). In recombinant E. coli BL21, induction of PemK resulted in significant growth inhibition, with OD600nm maintained at 0.29–0.31 (Figure 8A). Although the persister frequency also increased from −3.5 log10 to −2.58 log10 during growth of E. coli BL21 with PemK–/PemI–, PemK expression dramatically elevated the persister frequency of E. coli BL21 with PemI–/PemK+ up to −1.82 log10 to −0.24 log10 (Figure 8B). Substitution of Arg24 with Ala24 significantly weakened the growth inhibition and induced persister frequency of E. coli BL21 with PemI–/PemKArg24>Ala24+, decreasing it to −2.82 to −2.24 log10 (Figure 8B).
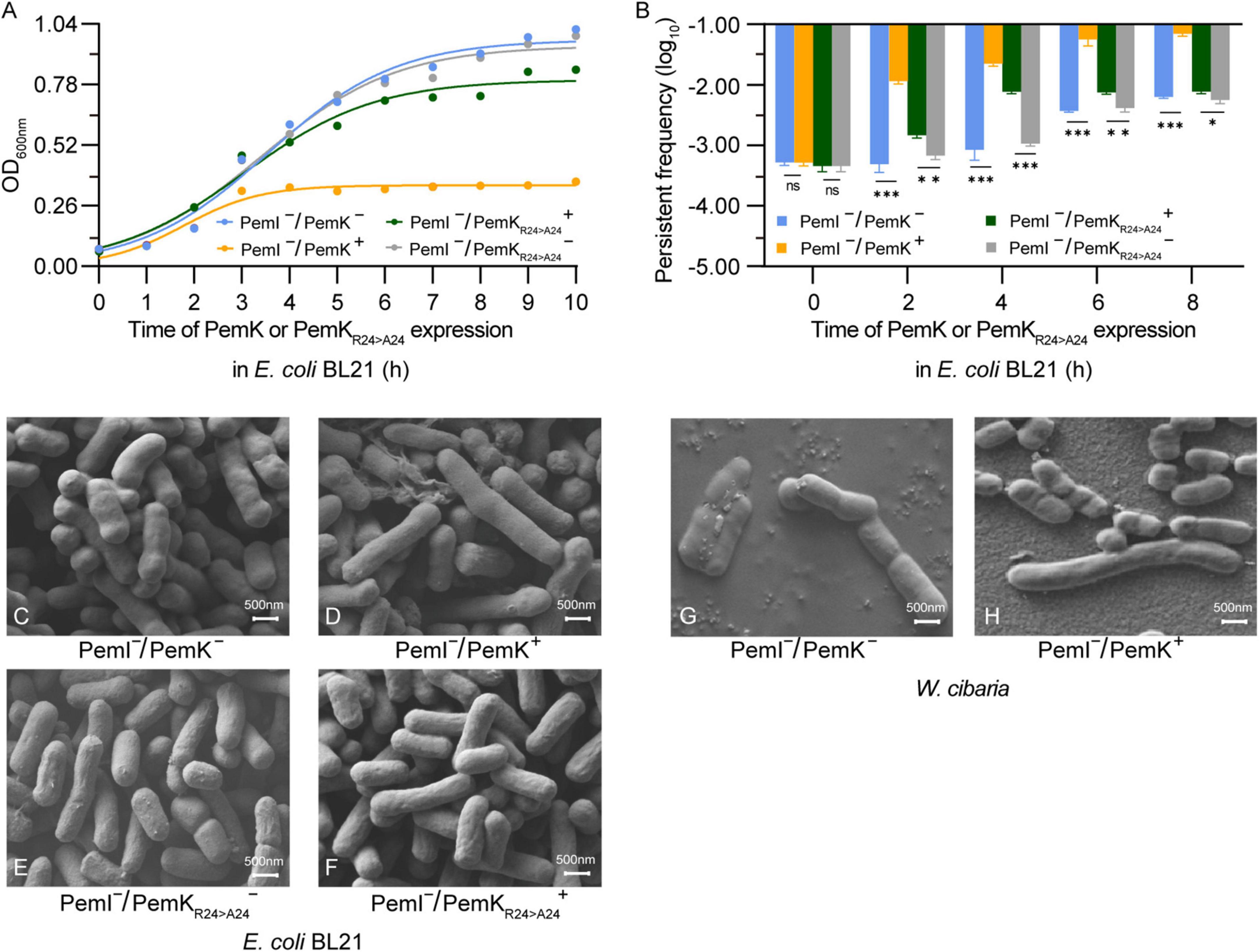
Figure 8. Persister frequency and cell morphology after induction of PemK and PemKR24>A24 expression. (A) Growth curve of recombinant E. coli BL21. (B) Persister frequency of recombinant E. coli BL21. (C–F) Cell morphology of E. coli BL21. (G,H) Cell morphology before and after induction of PemK expression in W. cibaria. Data were assessed by ANOVA with Tukey’s post-hoc test. The asterisk (*) indicated significant level: *P < 0.05, **P < 0.01, ***P < 0.001, not significant (ns), P > 0.05.
II-type toxin expression does not necessarily lead to changes in morphology of persister cells, but when it does, it usually involves abnormal elongation, filamentation, and even a transition from spiral to round coccoid (Cho et al., 2017; El Mortaji et al., 2020; Yu et al., 2023). In recombinant E. coli BL21, PemK expression significantly prolonged the morphology of some cells, increasing from 0.75–1.5 μm to 1.5–2.5 μm. This change was also observed in W. cibaria, but with more pronounced effects from 0.75–1.25 to 0.75–3.5 μm. However, the mutation PemKArg24>Ala24 did not induce significant alterations of E. coli BL21 (Figures 8C–H). It was generally believed that ectopic expression of a single toxin gene resulted in a phenotype similar to naturally occurring persister bacteria (Merfa et al., 2016; Page and Peti, 2016). Therefore, PemK have prolonged the persister cell of W. cibaria through the same mechanism as E. coli BL21.
4 Discussion
The mechanism underlying TA-mediated persistence remains incompletely understood, although it is generally believed that the direct cause is the shutdown or disruption of major cellular processes induced by TA toxins (Verstraeten et al., 2015; Wilmaerts et al., 2019). However, experimental confirmation of this understanding has been scarce. Our presented results revealed the molecular mechanism through which PemK induced persistence of W. cibaria by disrupting crucial cellular processes such as hydrogen transfer, electron transfer, NADH and FADH2 synthesis (Figures 3, 5, 7). This disruption occurred via mRNA fragmentation of targeting metabolic enzymes involved in glycolysis, TCA cycle and respiratory chain pathways, ultimately leading to reduced ATP levels and membrane depolarization (Figures 6A–C). These findings provided valuable insights into how W. cibaria evaded ciprofloxacin stress, that was, when faced with ciprofloxacin stress, it entered the persistence, conversely, once the stress was alleviated, the persistence was awakened.
Bacterial persistence is a programmed phenotypic transformation with a genetic basis (Gerdes and Maisonneuve, 2012; Wilmaerts et al., 2019). Currently, it is widely accepted that TAs play a crucial role in bacterial persistence (Page and Peti, 2016). Our findings further supported this consensus, as PemKI’s mRNA was one of the most up-regulated transcripts during the persistence of W. cibaria under ciprofloxacin stress (Figure 1C). Additionally, ectopic expression of PemK significantly increased the persistence frequency of recombinant E. coli BL21 (Figure 8B). In W. cibaria, the PemKI module consisted of PemK and its cognate antitoxin PemI (Figures 1A, B). Although PemK shared conserved residues and similar structure with homologs, its gate crevice and charge distribution differed from those homologs due to amino acid variations. Notably, the gate crevice of PemK closely resembled that of S. aureus, but differed significantly from E. coli, B. subtilis and B. anthracis (Figure 2). These unique characteristics determined its binding specificity for mRNA and physiological functions.
The significant function of PemK and its homologues is to catalyze the cleavage of bacterial mRNA with specific sequences (Bukowski et al., 2013; Kim et al., 2022), and their catalytic activities are closely related to the type and location of amino acid residues involved in mRNA binding and cleavage. In E. coli, MazF specifically cleaves the P-O5′ phosphoester bond between G↓ACA or G/UA↓CA of mRNA through transphosphorylation, with Arg29 and Thr52 likely serving as catalytic residues (Zorzini et al., 2016). MazF from B. subtilis cleaves mRNAs with U↓ACAU, where Arg25 and Thr48 are crucial for its activity, while Gln50 and Arg71 have minimal effect on activity (Simanshu et al., 2013). For S. aureus, Glu20, Arg25, Thr48, and Arg84 are key residues involved in binding and cleavage of mRNA with U↓AUU. Among them, Arg25 and Thr48 may be responsible for mRNA cleavage, while Arg84 maintains the “closed form” conformation of PemK dimer despite not directly interacting with mRNA. Conserved Arg and Thr residues located in β2 and β3 sheet, respectively, are generally considered as the active residues for catalysis in PemK homologues, where Arg acts as both a general acid/base catalyst, while Thr stabilizes transition states during reactions. In W. cibaria, Arg24 bound the phosphate group between U↓A, its mutation significantly affected the conformation and charge distribution of its gate crevice, as well as the thermostability, SASA and instability of PemK, thus significantly reducing the catalytic activity of PemK (Figures 3B, 4–8). Interestingly, substitution at Thr48 with any amino acid did not obviously affect any properties described above for PemK (Figure 4A), most likely due to fact that it bound the adenine between U↓A rather than phosphate group in target mRNA sequence (Figure 3B). Therefore, the active residues of PemK from W. cibaria differed significantly from those found in its homologs, where only Arg24 was the crucial active residue.
A decrease in intracellular ATP and/or low PMF across the membrane are characteristic indicators of persister formation resulting from TA-toxin attacking on conservative metabolic processes (Yu et al., 2023). Various small toxins, such as those in the Hok family, abrogate the PMF and inhibit ATP synthesis by depolarizing bacterial membranes (Harms et al., 2018). However, there is currently limited knowledge regarding the detailed mechanism through which TA-toxins target ATP and PMF. In W. cibaria, ATP synthesis primarily occurs through substrate level phosphorylation in glycolysis and TCA cycle, as well as oxidative phosphorylation in the respiratory chain. The former serves as an auxiliary pathway for energy acquisition, with only 3 ATP production. However, PemK inhibited their ATP synthesis, more importantly, it reduced the supply of NADH + H+ and FADH2 for oxidative phosphorylation by cleaving mRNA of enzymes in metabolic framework (Figures 5–7). The oxidative phosphorylation is the primary pathway for ATP production, generating 10 ATP molecules along with the efflux of 10 H+ through complexes I, III, and IV, and the influx of 4 H+ through ATP-synthase. Consequently, in W. cibaria, PemK cleaved target mRNAs encoding complexes I, III and IV to result in impaired H+ efflux from inner-membrane, subsequently reduced the membrane potential and abolished PMF (Figures 5, 6A, C). Simultaneously, PemK targeted complexes I–IV and ATP synthase to block the electron transfer and H+ influx which ultimately disrupts ATP synthesis (Figures 6B, F). In bacteria, disruption of ATP synthesis appears to be a promising mechanism for TA-toxins as most antibiotics corrode active targets by energy-dependent processes. Therefore, the disruption of ATP synthesis leading to cell depolarization was likely to be the primary physiological function of PemK inducing W. cibaria persistence to evade ciprofloxacin stress.
Recently, it has been documented that ATP content was the decisive factor in whether exponentially growing cells transition into persister cells. However, this correlation was not observed in S. aureus (Kamphuis et al., 2006; Somerville and Proctor, 2009). Under ciprofloxacin stress, PemK significantly augmented the persister frequency of W. cibaria, from 0.03% up to 0.6% (Figure 1C), achieved through disruption of ATP synthesis pathway and depolarization of cell membrane, which has been further validated in recombinant E. coli BL21 (Figures 6A–D, F). Moreover, when Ala replaced Arg24, PemKArg24>Ala24 considerably weakened its cytotoxicity to ATP synthesis pathway and membrane potential of recombinant E. coli BL21, resulting in a decrease in persister frequency from −1.82 to −1.24 log10 to −2.82 to −2.24 log10 (Figures 6D, 8A). These ectopic results further corroborated the physiological mechanism underlying PemK’s involvement in the persister formation of W. cibaria, while also highlighted the significance of Arg24 for maintaining PemK’s activity.
The significance of TA system in bacterial persistence renders it a natural target for the discovery of innovative strategies that can either trigger the persistence or awaken the persister cell. Our study provided some crucial structural and functional insights into PemK of W. cibaria, serving as a fundamental basis for uncovering this novel approach. However, the pivotal question remains whether interference with mRNA and physiological function by ciprofloxacin-induced PemK characterizes universal regulatory on W. cibaria under diverse stresses, which currently lacks clarity and necessitates further investigation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in this article/Supplementary material.
Author contributions
H-YZ: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing – original draft. W-LX: Conceptualization, Funding acquisition, Resources, Supervision, Writing – review & editing. TC: Data curation, Formal analysis, Writing – original draft. MZ: Data curation, Investigation, Methodology, Writing – original draft. H-YW: Formal analysis, Validation, Writing – original draft.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the Natural Science Foundations of Sichuan, China, grant number 2018JY0045, National Natural Science Foundations of China, grant number 31571935, and Open Foundation of State Key Laboratory of Environmental Criteria and Risk Assessment, China, grant number SKLECRA 20240618.
Acknowledgments
We are grateful to Keo Lee for advice on transcription experiments, statistical analysis, and stimulating discussions.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1402319/full#supplementary-material
Footnotes
- ^ http://www.jalview.org
- ^ https://rostlab.org/services/snap/
- ^ https://www.ics.uci.edu/∼baldig/mutation.html
- ^ http://www.ccp4.ac.uk/
References
Bromberg, Y., and Rost, B. (2007). SNAP: Predict effect of non-synonymous polymorphisms on function. Nucleic Acids Res. 35, 3823–3835. doi: 10.1093/nar/gkm238
Bukowski, M., Lyzen, R., Helbin, W. M., Bonar, E., Szalewska-Palasz, A., Wegrzyn, G., et al. (2013). A regulatory role for Staphylococcus aureus toxin–antitoxin system PemIKSa. Nat. Commun. 4:2012. doi: 10.1038/ncomms3012
Cai, T., Zhao, Q., Xiang, W., Zhu, L., Rao, Y., and Tang, J. (2022). HigBA toxin–antitoxin system of Weissella cibaria is involved in response to the bile salt stress. J. Sci. Food Agric. 102, 6749–6756. doi: 10.1002/jsfa.12042
Chandel, N. S. (2021). Glycolysis. Cold Spring Harb. Perspect. Biol. 13:a040535. doi: 10.1101/cshperspect.a040535
Cho, J., Carr, A. N., Whitworth, L., Johnson, B., and Wilson, K. S. (2017). MazEF toxin-antitoxin proteins alter Escherichia coli cell morphology and infrastructure during persister formation and regrowth. Microbiology 163, 308–321. doi: 10.1099/mic.0.000436
Díaz-Orejas, R., Espinosa, M., and Yeo, C. C. (2017). The importance of the expendable: Toxin–antitoxin genes in plasmids and chromosomes. Front. Microbiol. 8:1479. doi: 10.3389/fmicb.2017.01479
El Mortaji, L., Tejada-Arranz, A., Rifflet, A., Boneca, I. G., Pehau-Arnaudet, G., Radicella, J. P., et al. (2020). A peptide of a type I toxin-antitoxin system induces Helicobacter pylori morphological transformation from spiral shape to coccoids. Proc. Natl. Acad. Sci. U. S. A. 117, 31398–31409. doi: 10.1073/pnas.2016195117
Gerdes, K., and Maisonneuve, E. (2012). Bacterial persistence and toxin-antitoxin loci. Annu. Rev. Microbiol. 66, 103–123. doi: 10.1146/annurev-micro-092611-150159
Gorham, R. D., Kieslich, C. A., and Morikis, D. (2011). Electrostatic clustering and free energy calculations provide a foundation for protein design and optimization. Ann. Biomed. Eng. 39, 1252–1263. doi: 10.1007/s10439-010-0226-9
Harms, A., Brodersen, D. E., Mitarai, N., and Gerdes, K. (2018). Toxins, targets, and triggers: An overview of toxin-antitoxin biology. Mol. Cell 70, 768–784. doi: 10.1016/j.molcel.2018.01.003
Kamphuis, M. B., Bonvin, A. M. J. J., Monti, M. C., Lemonnier, M., Muñoz-Gómez, A., Van Den Heuvel, R. H. H., et al. (2006). Model for RNA binding and the catalytic site of the RNase kid of the bacterial parD toxin–antitoxin system. J. Mol. Biol. 357, 115–126. doi: 10.1016/j.jmb.2005.12.033
Kim, D. H., Kang, S. M., Baek, S. M., Yoon, H. J., Jang, D. M., Kim, H. S., et al. (2022). Role of PemI in the Staphylococcus aureus PemIK toxin–antitoxin complex: PemI controls PemK by acting as a PemK loop mimic. Nucleic Acids Res. 50, 2319–2333. doi: 10.1093/nar/gkab1288
Maisonneuve, E., and Gerdes, K. (2014). Molecular mechanisms underlying bacterial persisters. Cell 157, 539–548. doi: 10.1016/j.cell.2014.02.050
Merfa, M. V., Niza, B., Takita, M. A., and De Souza, A. A. (2016). The MqsRA toxin-antitoxin system from Xylella fastidiosa plays a key role in bacterial fitness, pathogenicity, and persister cell formation. Front. Microbiol. 7:904. doi: 10.3389/fmicb.2016.00904
Page, R., and Peti, W. (2016). Toxin-antitoxin systems in bacterial growth arrest and persistence. Nat. Chem. Biol. 12, 208–214. doi: 10.1038/nchembio.2044
Robert, X., and Gouet, P. (2014). Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W336. doi: 10.1093/nar/gku316
Schiffrin, B., Radford, S. E., Brockwell, D. J., and Calabrese, A. N. (2020). PYXLINKVIEWER: A flexible tool for visualization of protein chemical crosslinking data within the PYMOL molecular graphics system. Protein Sci. 29, 1851–1857. doi: 10.1002/pro.3902
Shahmuradov, I. A. (2003). PlantProm: A database of plant promoter sequences. Nucleic Acids Res. 31, 114–117. doi: 10.1093/nar/gkg041
Shan, Y., Brown Gandt, A., Rowe, S. E., Deisinger, J. P., Conlon, B. P., and Lewis, K. (2017). ATP-dependent persister formation in Escherichia coli. mBio 8:e02267-16. doi: 10.1128/mbio.02267-16
Simanshu, D. K., Yamaguchi, Y., Park, J. H., Inouye, M., and Patel, D. J. (2013). Structural basis of mRNA recognition and cleavage by toxin MazF and its regulation by antitoxin MazE in Bacillus subtilis. Mol. Cell 52, 447–458. doi: 10.1016/j.molcel.2013.09.006
Somerville, G. A., and Proctor, R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73, 233–248. doi: 10.1128/mmbr.00005-09
Sun, C., Guo, Y., Tang, K., Wen, Z., Li, B., Zeng, Z., et al. (2017). MqsR/MqsA toxin/antitoxin system regulates persistence and biofilm formation in Pseudomonas putida KT2440. Front. Microbiol. 8:840. doi: 10.3389/fmicb.2017.00840
Van Zundert, G. C. P., Rodrigues, J. P. G. L. M., Trellet, M., Schmitz, C., Kastritis, P. L., Karaca, E., et al. (2016). The HADDOCK2.2 Web Server: User-fiendly integrative modeling of biomolecular complexes. J. Mol. Biol. 428, 720–725. doi: 10.1016/j.jmb.2015.09.014
Verstraeten, N., Knapen, W. J., Kint, C. I., Liebens, V., Van den Bergh, B., Dewachter, L., et al. (2015). Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol. Cell 59, 9–21. doi: 10.1016/j.molcel.2015.05.011
Wang, L., Zhang, J., Cao, Z., Wang, Y., Gao, Q., Zhang, J., et al. (2015). Inhibition of oxidative phosphorylation for enhancing citric acid production by Aspergillus niger. Microb. Cell Fact. 14:7. doi: 10.1186/s12934-015-0190-z
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303. doi: 10.1093/nar/gky427
Wilmaerts, D., Windels, E. M., Verstraeten, N., and Michiels, J. (2019). General mechanisms leading to persister formation and awakening. Trends Genet. 35, 401–411. doi: 10.1016/j.tig.2019.03.007
Xiong, H. B., Zhou, X. H., Xiang, W. L., Huang, M., Lin, Z. X., Tang, J., et al. (2023). Integrated transcriptome reveals that d-limonene inhibits Candida tropicalis by disrupting metabolism. LWT Food Sci. Technol. 176:114535. doi: 10.1016/j.lwt.2023.114535
Yu, H., Lin, Z. X., Xiang, W. L., Huang, M., Tang, J., Lu, Y., et al. (2022). Antifungal activity and mechanism of d-limonene against foodborne opportunistic pathogen Candida tropicalis. LWT Food Sci. Technol. 159:113144. doi: 10.1016/j.lwt.2022.113144
Yu, Z., Goodall, E. C. A., Henderson, I. R., and Guo, J. (2023). Plasmids can shift bacterial morphological response against antibiotic stress. Adv. Sci. 10:2203260. doi: 10.1002/advs.202203260
Zhang, J., Zhang, Y., Zhu, L., Suzuki, M., and Inouye, M. (2004). Interference of mRNA function by sequence-specific endoribonuclease PemK. J. Biol. Chem. 279, 20678–20684. doi: 10.1074/jbc.M314284200
Zhou, J., Li, S., Li, H., Jin, Y., Bai, F., Cheng, Z., et al. (2021). Identification of a toxin–antitoxin system that contributes to persister formation by reducing NAD in Pseudomonas aeruginosa. Microorganisms 9:753. doi: 10.3390/microorganisms9040753
Keywords: Weissella cibaria, toxin-antitoxin system, PemIK module, ciprofloxacin stress, persister cell, metabolic processes
Citation: Zhu H-Y, Xiang W-L, Cai T, Zhang M and Wang H-Y (2024) PemK’s Arg24 is a crucial residue for PemIK toxin–antitoxin system to induce the persistence of Weissella cibaria against ciprofloxacin stress. Front. Microbiol. 15:1402319. doi: 10.3389/fmicb.2024.1402319
Received: 17 March 2024; Accepted: 15 April 2024;
Published: 14 May 2024.
Edited by:
Vijay Soni, NewYork-Presbyterian, United StatesReviewed by:
Yaxin Li, Cornell University, United StatesSmriti Parashar, University of California, San Diego, United States
Tripti Nair, University of Southern California, United States
Copyright © 2024 Zhu, Xiang, Cai, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Liang Xiang, YmlvdW5pY29tQG1haWwueGh1LmVkdS5jbg==
†These authors share first authorship
 Hao-Yu Zhu
Hao-Yu Zhu Wen-Liang Xiang
Wen-Liang Xiang Ting Cai1,2
Ting Cai1,2