
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol. , 31 July 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1401802
This article is part of the Research Topic Global Dissemination and Evolution of Epidemic Multidrug-Resistant Gram-Negative Bacterial Pathogens: Surveillance, Diagnosis and Treatment, Volume III View all 14 articles
 Hong-Xian Chen1,2
Hong-Xian Chen1,2 Fang-Jie Chen1,2
Fang-Jie Chen1,2 Qian-Jin Zhou1,2,3*
Qian-Jin Zhou1,2,3* Shi-Lin Shang2
Shi-Lin Shang2 Biao Tang4
Biao Tang4 Zhong-Jie Xu1,2
Zhong-Jie Xu1,2 Li-Jun Duan2,5
Li-Jun Duan2,5 Jing-Lei Jin2
Jing-Lei Jin2 Gui-Zong Xu1,2
Gui-Zong Xu1,2 Mao-Cang Yan6
Mao-Cang Yan6 Jiong Chen1,2,3*
Jiong Chen1,2,3*Introduction: Aeromonas spp. are ubiquitous inhabitants of ecosystems, and many species are opportunistically pathogenic to humans and animals. Multidrug-resistant (MDR) Aeromonas species have been widely detected in hospitals, urban rivers, livestock, and aquatic animals.
Results: In this study, we identified two Aeromonas isolates, namely Aeromonas veronii 0728Q8Av and Aeromonas caviae 1029Y16Ac, from coastal waters in Zhejiang, China. Both isolates exhibited typical biochemical characteristics and conferred MDR to 11 kinds of antibiotics, remaining susceptible to ceftazidime. Whole-genome sequencing revealed that both isolates harbored multiple antibiotic resistance genes (ARGs) and several mobile genetic elements (MGEs) on the chromosomes, each containing a resistance genomic island (GI), a typical class 1 integron, a transposon, and various insertion sequences (ISs). Most ARGs were situated within the multiple resistance GI, which contained a class 1 integron and a transposon in both Aeromonas isolates. Furthermore, a chromosomal mcr-3.16 gene was identified in A. veronii 0728Q8Av, while a chromosomal mcr-3.3 was found in A. caviae 1029Y16Ac. Both mcr-3 variants were not located within but were distanced from the multidrug resistance GI on the chromosome, flanking by multiple ISs. In addition, a mcr-3-like was found adjacent to mcr-3.16 to form a tandem mcr-3.16-mcr-3-like-dgkA structure; yet, Escherichia coli carrying the recombinants of mcr-3-like did not exhibit resistance to colistin. And an incomplete mcr-3-like was found adjacent to mcr-3.3 in A. caviae 1029Y16Ac, suggesting the possibility that mcr-3 variants originated from Aeromonas species. In vivo bacterial pathogenicity test indicated that A. veronii 0728Q8Av exhibited moderate pathogenicity towards infected ayu, while A. caviae 1029Y16Ac was non-virulent.
Discussion: Thus, both Aeromonas species deserve further attention regarding their antimicrobial resistance and pathogenicity.
Antibiotic resistance genes (ARGs) are considered emerging environmental contaminants due to their potential risks to human and animal health (Zheng et al., 2021). Infections caused by multidrug-resistant (MDR) Gram-negative bacteria (GNB) are increasingly common and pose a paramount therapeutic challenge, raising significant concerns worldwide (Osei Sekyere et al., 2016; Vivas et al., 2019). Colistin is considered one of the few “last-resort” antibiotics against MDR GNB infections (Osei Sekyere et al., 2016; Yin et al., 2017). The emergence and rapid dissemination of the mobile colistin resistance (mcr) genes further compound the challenges in antibiotic treatment of MDR bacteria (Vivas et al., 2019; Shen et al., 2020).
Aeromonas, a Gram-negative bacillus, is widely distributed in the environment, particularly in freshwater and estuarine ecosystems (Janda and Abbott, 2010). Currently, the Aeromonas genus encompasses 36 species, many of which are opportunistically pathogenic to both animals and humans (Janda and Abbott, 2010; Fernández-Bravo and Figueras, 2020). Aeromonas spp. infect humans, causing various gastroenteritis and extra-intestinal diseases (Fernández-Bravo and Figueras, 2020; Carusi et al., 2023). Long recognized as a significant pathogen in aquatic animals, Aeromonas instigates a broad spectrum of opportunistic infections, resulting in severe clinical manifestations such as sepsis, bleeding, ulcers, ascites, and high mortality (Janda and Abbott, 2010; Carusi et al., 2023). Many studies have demonstrated that Aeromonas serves as a MDR carrier and a reservoir of many ARGs like mar genes (Piotrowska and Popowska, 2015; Ling et al., 2017; Eichhorn et al., 2018; Shen et al., 2020).
The coastal waters, being significant areas of human activity, harbor a wealth of ARGs and antibiotic-resistant bacteria (ARB) (Zhu et al., 2017; Zheng et al., 2021). These genes and bacteria originate not only from coastal aquaculture but also from various human activities such as animal husbandry, freshwater aquaculture, clinical medicine, and industrial production (Zhu et al., 2017; Jang et al., 2018; Zheng et al., 2021). They are introduced into coastal water environments through mechanisms such as rainfall and sewage discharge (Shao et al., 2018). Exposure of environmental microorganisms to this mixture stimulates horizontal gene transfer (HGT) events, spreading genetic resistance elements across different microbial strains and increasing microbial abundance and penetration in new hosts (Juhas et al., 2009; Shao et al., 2018). Aeromonas spp. is ubiquitously distributed in coastal waters and mariculture animals, many of which harbors abundant ARGs and mobile genetic elements (MGEs) (Piotrowska and Popowska, 2014; Carusi et al., 2023). Although Aeromonas carrying mcr genes have been repeatedly detected in freshwater animals and water bodies (Eichhorn et al., 2018; Xu et al., 2020; Sakulworakan et al., 2021), the incidence of mobile colistin resistance determinants and their genetic environment in Aeromonas genomes from coastal waters and marine animals have largely been ignored. In this study, we investigated the antibiotic susceptibility of two MDR Aeromonas species isolated from the coastal waters near Wenzhou, Zhejiang Province, China. Through genome sequencing, we examined the genetic profiles for antimicrobial resistance, analyzed the structural characteristics of MDR gene islands on their chromosomes, and investigated the distribution of mcr variants. Additionally, we experimentally tested the ability of the mcr variants to mediate colistin resistance. Furthermore, we assessed the in vivo pathogenicity of the two MDR Aeromonas species in ayu (Plecoglossus altivelis), an important amphidromous economic fish species found in East Asia.
In our previous study, we selected 25 sampling sites in the coaster waters near Wenzhou in the East China Sea and collected surface seawater samples (Jin et al., 2021). We isolated culturable bacteria resistant to sulfonamides, tetracyclines, and quinolones from the seawater samples using antibiotic resistance plates and detected the target ARGs in these isolates using polymerase chain reaction (PCR). We isolated 1,605 bacterial strain with antibiotic resistance phenotypes, among which 51 isolates tested positive for resistance genes related to sulfonamides, tetracyclines, and quinolones, with 43 of them belonging to the genus Aeromonas (Jin et al., 2021). Additionally, we identified 39 MDR strains (Jin et al., 2021). Using PCR primers for the mcr genes, we identified 2 Aeromonas isolates containing the mcr-3 variants (Jin et al., 2021; Xu et al., 2021) (Supplementary Table S1). Preliminary 16S rDNA sequencing suggested that both isolates likely belonged to Aeromonas veronii and Aeromonas caviae. In this study, further identification of both isolates was performed using multilocus phylogenetic analysis (MLPA) based on six genes (gyrB, rpoD, dnaJ, gyrA, dnaX, and atpD) (Martinez-Murcia et al., 2011). Phylogenetic trees were constructed based on the concatenated sequences containing these six genes using the maximum likelihood method in the MEGA 7.1 software package. Subsequently, biochemical tests were conducted on the isolates using biochemical test kits (Tiangen, Beijing, China).
Antimicrobial susceptibility to 12 antibiotics belonging to seven antibiotic classes, including β-lactams [ampicillin (AMP) and ceftazidime (CAZ)], tetracyclines [oxytetracycline (OTC) and tetracycline (TET)], amphenicols [chloramphenicol (CHP) and florfenicol (FFC)], sulfonamides [sulfamethoxazole (SMZ) and trimethoprim (TMP)], quinolones [ciprofloxacin (CIP) and enrofloxacin (ENR)], aminoglycosides [gentamycin (GEN)], and polymyxin [colistin (COL)], was assessed using the disc diffusion method, with modifications derived from the methodology outlined by Huq (2020). Briefly, A. veronii 0728Q8Av and A. caviae 1029Y16Ac were cultured overnight in Mueller Hinton II (cation-adjusted)/CAMHB broth (Solarbio, Beijing, China). A 0.1 mL bacterial culture of each strain was evenly spread on a Mueller–Hinton (MH) agar plate. The sterile paper discs (7 mm in diameter) containing the antibiotics (10 μg/disc for APM, GEN, 30 μg/disc for CAZ, TET, OTC, CHP, FFC, 5 μg/disc for CIP, ENR, TMP, 300 IU/disc for COL, 250 μg/disc for SMZ) were placed on the surface of the inoculated MH agar plates. The plates were then incubated at 28°C, and the inhibition zones were measured after 18 h of incubation.
The antibiotics that induced resistance in either of the tested bacterial strains were selected to calculate the minimum inhibitory concentrations (MICs) against both strains. This was achieved using the broth microdilution method (BMD) recommended by the Clinical and Laboratory Standards Institute (CLSI) (CLSI, 2018, M100-S28). BMD is conducted using Mueller Hinton II (cation-adjusted)/CAMHB broth, a range of 2-fold dilutions of antibiotics (ranging from 0.5 to 256 μg/mL), and a bacterial inoculum density was adjusted equivalent to a 0.5 McFarland standard per well. The MIC breakpoints were interpreted according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST)1 and CLSI guidelines (CLSI, 2018, M100-S28). Escherichia coli ATCC 25922 was used as a quality control.
The genomic DNA of A. veronii 0728Q8Av and A. caviae 1029Y16Ac was extracted using cetyltrimethylammonium bromide method, following the manufacturer’s protocol BGI-PB-TQ-DNA-003A0. Subsequently, a 20 kb fragment library was constructed for each isolate, utilizing high-quality genomic DNA that met the requirements for whole-genome sequencing. Sequencing was carried out on a Pacbio Sequel II platform (BGI, Shenzhen, China). The reads were assembled using Canu (version 1.5). After multiple adjustments to achieve the optimal assembly, base correction, circularization, and plasmid library alignment were performed on the assembled sequences to obtain the final assembly results. Gene prediction was performed with Glimmer (version 3.02). The annotation of genes was accomplished through in-house pipeline on the RAST server2 (Ebu et al., 2023). Acquired antimicrobial resistance genes were predicted using ResFinder 4.13 with a threshold of percent identity set at 90% and a minimum coverage length of 60%. Genomic islands (GIs) were determined using IslandPath-DIMOB4 (Bertelli and Brinkman, 2018). Insertion sequences (ISs) were identified through ISfinder5 (Siguier et al., 2006). Transposons and integrons were predicted using TnCentral6 and Integron Finder,7 respectively (Damas et al., 2022). Comparative analysis of genome sequences was performed using Easyfig with a maximum e-value of 0.001 and identification threshold set at 98% and BRIG (with an identification threshold of 50%) (Tang et al., 2022). Virulence factors were predicted using the Virulence Factor Database (VFDB, http://www.mgc.ac.cn/VFs/) (Liu et al., 2022).
According to Jin et al. (2021), the prevalence of Aeromonas spp. with mcr genes in coastal waters is low (2 out of 51). A. veronii 0728Q8Av and A. caviae 1029Y16Ac show different levels of resistance to colistin and harbor different mcr-3 variants. To assess whether these mcr-3 variants affect resistance to colistin, we conducted functionality assays. To in silico verify mcr-3 variants in both A. veronii 0728Q8Av and A. caviae 1029Y16Ac, the sequences of mcr-3.16, mcr-3.3, and mcr-3-like, was aligned, respectively, referred to sequences of mcr-3 variants downloaded from NCBI.8 And a phylogenetic tree was constructed based on mcr-3 variants’ sequences using the Maximum likelihood method in the MEGA 7.1 software package.
To determine the function of mcr-3 variants, four DNA fragments, corresponding to each open reading frame (ORF) of mcr-3.16, mcr-3-like, mcr-3.16-mcr-3-like, and mcr-3.3 regions along with their 5′- and 3′-flanking regions, respectively, were amplified using the primers Q8MCR3.16-F/Q8MCR3.16-R, Q8MCR-3-like-F/Q8MCR-3-like-R, Q8MCR3.16-F/MCR-3-like-R, and Y16MCR3.3-F/Y16MCR3.3-R, and cloned into pUC19 vector by digestion of BamHI and EcoRI, respectively. The recombinant plasmids were separately transformed into E. coli DH5α. Transformants were screened on LB agar plates containing 50 μg/mL ampicillin.
Meanwhile, the ORFs of mcr-3.16, mcr-3-like, mcr-3.16-mcr-3-like, and mcr-3.3, were amplified using the primers Q8MCR3.16-BMQ-F/Q8MCR3.16-BMQ-R, Q8MCR-3-likeBMQ-F/Q8MCR-3-likeBMQ-R, Q8MCR3.16-BMQ-F/Q8MCR-3-likeBMQ-R, and Y16MCR3.3-BMQ-F/Y16MCR3.3-BMQ-R, and cloned into pET28a vector by digestion of XhoI and EcoRI, respectively. The recombinant plasmids were separately transformed into E. coli BL21. Transformants were screened using 50 μg/mL kanamycin and were induced by addition of IPTG. Colistin susceptibility of E. coli transformed with any of the recombinant plasmids was determined in triplicates.
To determine whether the mcr-3 variants in A. veronii 0728Q8Av and A. caviae 1029Y16Ac are located on the chromosome or plasmid, conjugation assays were conducted as described previously (Sun et al., 2016; Tang et al., 2024). The E. coli J53 was used as the recipient strain, and A. veronii 0728Q8Av or A. caviae 1029Y16Ac was the donor strain. The E. coli ECCNB20-2 carrying mcr-1 was used as a positive control donor (Chang et al., 2020). All bacterial strains were cultured to be in the logarithmic growth phase in LB broth at 37°C. Each of the donor bacteria, that is A. veronii 0728Q8Av, A. caviae 1029Y16Ac, and E. coli ECCNB20-2, was mixed with E. coli J53 in suitable proportion, and the bacterial mixture was transferred to filter paper on the LB agar and cultured for 8 h. Then the filter paper was suspended into a centrifuge tube containing LB broth and serially diluted. Each diluted bacterial solution (10 μL) was cultured onto screening LB agar supplemented with 2 μg/mL colistin and 100 μg/mL NaN3, and with 100 μg/mL NaN3, respectively. Conjugation transfer was determined as described by Tang et al. (2022).
Healthy ayu, weighing 20–25 g, were purchased from a commercial farm in Ningbo, China. Fish were raised in a recirculating system with filtered water at 20–22°C (Zhou et al., 2020). The fish were fed with pelleted dry food once a day and acclimatized to laboratory conditions for 2 weeks before experiments. Then, randomly allocate the fish into 21 tanks, each containing 10 individuals. The fish in each three tanks were intraperitoneally administered 100 μL of bacterial suspension of A. veronii 0728Q8Av and A. caviae 1029Y16Ac, at concentrations of 1 × 107, 1 × 108, and 1 × 109 CFU/mL. Additionally, three tanks of fish were injected sterile PBS in volumes matching those of the experimental groups, serving as negative controls. All fish were kept at 20–22°C for 7 days to observe and record the morbidity and mortality rates. The degree of virulence, expressed as the 50% mean lethal dose (LD50), was calculated using the Bliss method (Finney, 1985). All intervention measures are strictly carried out in accordance with the guiding principles of the Animal Experiment Ethics Committee of Ningbo University (No. 11102).
The complete genome sequences of A. veronii 0728Q8Av and A. caviae 1029Y16Ac were submitted to GenBank under Bioproject accession numbers PRJNA1070378 and PRJNA1070380, respectively.
Jin et al. (2021) identified both mcr-3.16 and mcr-3-like in strain 0728Q8Av and mcr-3.3 in strain 1029Y16Ac by DNA sequencing of PCR products. In this study, MLPA based on six house-keeping genes demonstrated that strain 0728Q8Av clustered together with selected A. veronii strains and strain 1029Y16Ac clustered together with A. caviae strains, which indicated that the two isolates belonged to A. veronii and A. caviae, respectively (Figure 1).
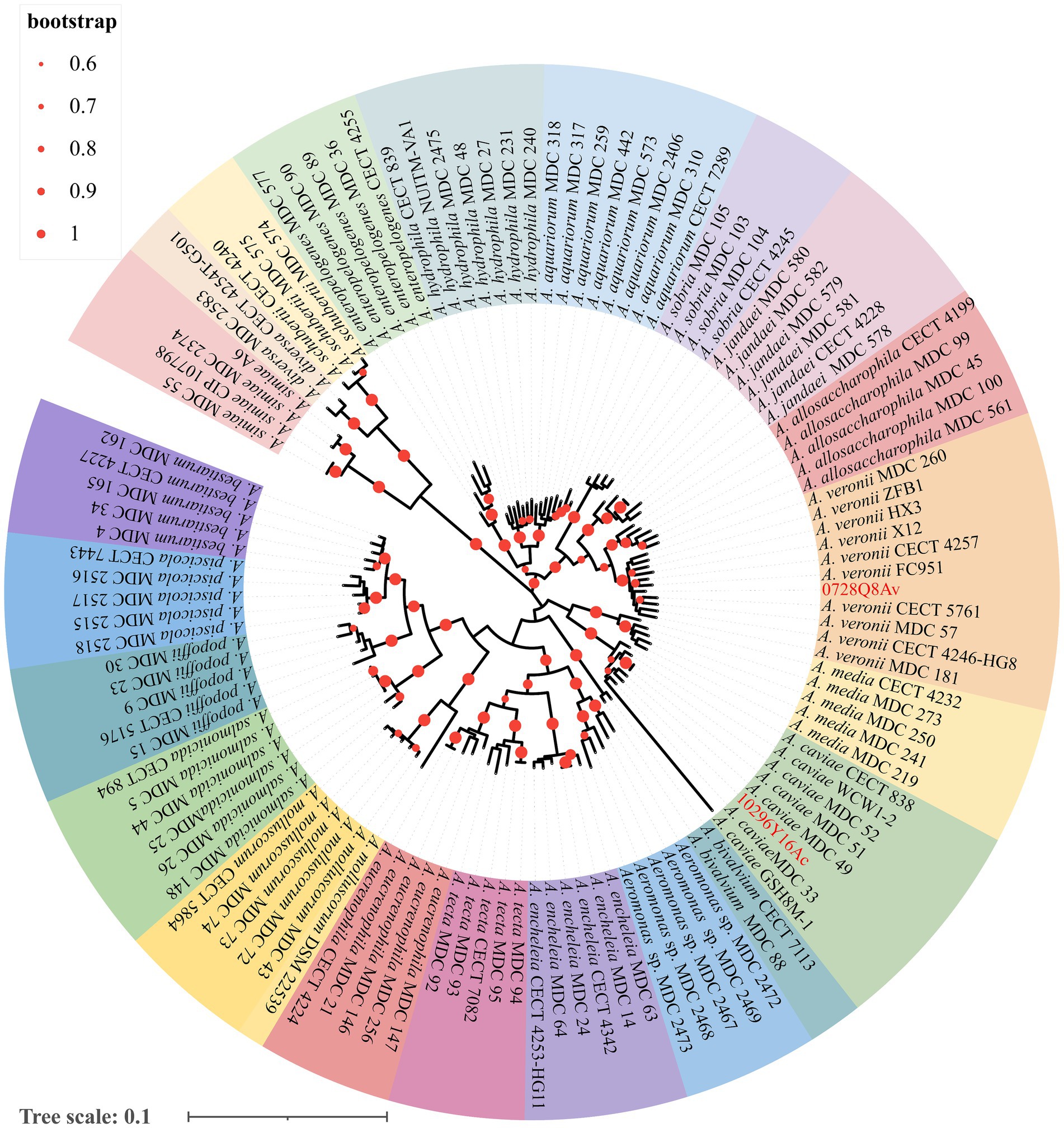
Figure 1. The multilocus phylogenetic tree analysis based on the sequences of six house-keeping genes (gyrB, rpoD, dnaJ, gyrA, dnaX, and atpD) from Aeromonas spp. using the maximum likelihood method. The values at the forks indicate the percentage of bootstrap values (1,000 replicates; shown only when >60%). Scale bar shows the number of substitutions per base. Accession numbers of the sequences used are listed in Supplementary Table S2.
Regarding the biochemical characteristics, both A. veronii 0728Q8Av and A. caviae 1029Y16Ac exhibited numerous identical traits. For example, both isolates demonstrated growth at a concentration of 1–3% sodium chloride (NaCl) (w/v) but were unable to be maintained at >6% NaCl. They were both positive for acid formation when provided with glucose, sucrose, mannose, maltose, OPNG, mannitol, and saligenin, while showing negativity towards xylose, raffinose, and sorbose. Additionally, both bacterial isolates demonstrated the ability to produce oxidase, indole, and arginine dihydrolase (Table 1). However, the two bacterial isolates presented some differing characteristics. Specifically, A. veronii 0728Q8Av, unlike A. caviae 1029Y16Ac, tested positive in the Voges-Proskauer test and exhibited urea hydrolysis. A. caviae 1029Y16Ac could utilize arabinose and lactose, producing acid, whereas A. veronii 0728Q8Av could not (Table 1).
The susceptibility testing showed that A. veronii 0728Q8Av exhibited resistance to COL (with a MIC of 8 μg/mL), OTC (64 μg/mL), TET (32 μg/mL), TMP (32 μg/mL), SMZ (256 μg/mL), AMP (64 μg/mL), CHP (16 μg/mL), and FFC (64 μg/mL), and intermediate to GEN (8 μg/mL), CIP (2 μg/mL), and ENR (2 μg/mL), but remained susceptible to CAZ (4 μg/mL) (Table 2). In contrast, A. caviae 1029Y16Ac exhibited resistance to COL (with a MIC of 4 μg/mL), AMP (32 μg/mL), TET (32 μg/mL), OTC (32 μg/mL), TMP (256 μg/mL), SMZ (256 μg/mL), CIP (4 μg/mL), ENR (4 μg/mL), and intermediate to CHP (8 μg/mL), FFC (16 μg/mL), and GEN (8 μg/mL), but remained susceptible to CAZ (8 μg/mL) (Table 2).
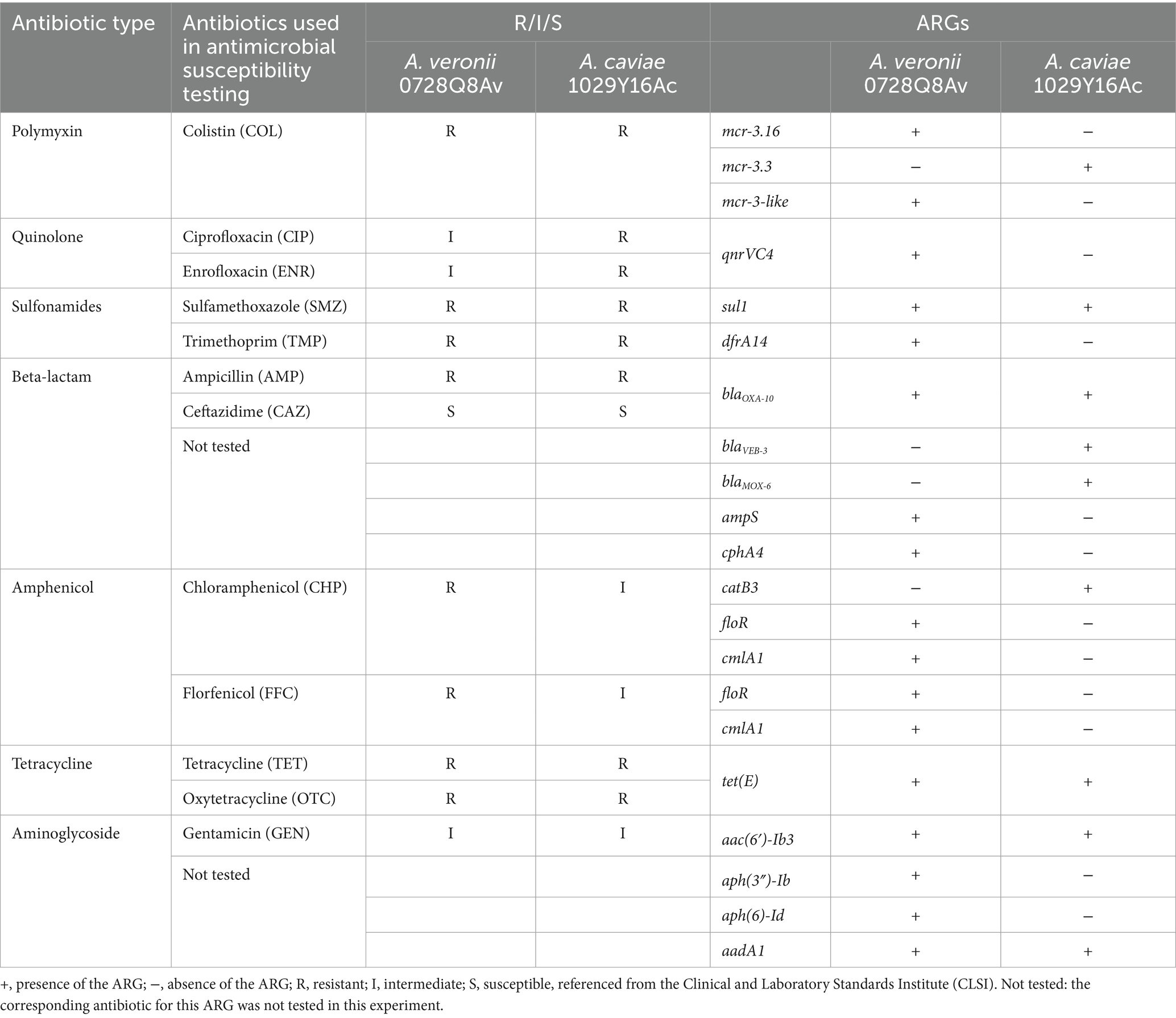
Table 2. Antibiotic resistance phenotype and antibiotic resistance genotypes of both A. veronii 0728Q8Av and A. caviae 1029Y16Ac.
A. veronii 0728Q8Av harbored one circular chromosomal genome of 4,752,946 bp with G + C content of 58.5% and no plasmid was found. A. caviae 1029Y16Ac harbored one circular chromosomal genome of 4,779,291 bp with G + C content of 60.86% and two plasmids, 1029Y16AcP1 (21,050 bp) and 1029Y16AcP2 (8,340 bp).
Acquired ARGs were identified by Resfinder 4.1 analysis. The chromosomal genome of A. veronii 0728Q8Av contained eight types of acquired ARGs indicating resistance to aminoglycoside [aac(6′)-Ib3, aph(3″)-Ib, aph(6)-Id, aadA1], polymyxin (mcr-3.16, mcr-3-like), quinolone (qnrVC4), sulfonamides (sul1, dfrA14), beta-lactam (blaOXA-10, ampS, cphA4), tetracycline [tet(E)], amphenicol (floR, cmlA1), and quaternary ammonium compound (qacE∆1). The genome contained 15 GIs and almost all the ARGs were located on the GI1 of A. veronii 0728Q8Av (AvGI1), except for mcr-3.16, mcr-3-like, ampS and cphA4 (Supplementary Figure S1A; Supplementary Table S3). The chromosomal genome of A. caviae 1029Y16Ac contained seven types of acquired ARGs indicating resistance to aminoglycoside [aac(6′)-Ib3, aadA1], polymyxin (mcr-3.3), sulfonamides (sul1), beta-lactam (blaVEB-3, blaMOX-6, blaOXA-10), tetracycline [tet(E)], amphenicol (catB3) and quaternary ammonium compound (qacE∆1). No ARGs were identified on plasmid 1029Y16AcP1 and 1029Y16AcP2. The chromosomal genome presented 23 GIs, and almost all the ARGs were located on the GI11 of A. caviae 1029Y16Ac (AcGI11), except for mcr-3.3, blaMOX-6, and tet(E) (Table 2; Supplementary Figure S1B; Supplementary Table S3).
The virulence factors’ prediction suggested that virulence factors were significantly different between A. veronii 0728Q8Av and A. caviae 1029Y16Ac (Table 3). The Tap type IV pili of both strains exhibited similar structural features. However, A. veronii 0728Q8Av possessed 16 genes related to MSHA type IV pili, whereas A. caviae 1029Y16Ac had only 3. And A. veronii 0728Q8Av had type I and Flp type IV pili, whereas A. caviae1029Y16Ac did not. Flagella analysis revealed that both strains had polar flagella. However, A. veronii 0728Q8Av possessed 54 genes related to polar flagella, whereas A. caviae 1029Y16Ac had only 7. Both stains had a type II secretion system (T2SS) containing 14 genes. Toxin analysis revealed that both strains contained Hemolysin and Thermostable hemolysin (TH). Interestingly, A. veronii 0728Q8Av had Aerolysin, but A. caviae 1029Y16Ac did not.
Blast analysis revealed that both mcr-3.16 (1623-bp of ORF) of A. veronii 0728Q8Av and mcr-3.3 (1623-bp of ORF) of A. caviae 1029Y16Ac exhibited 100% identity to previous reported mcr-3.16 (WP-111273845.1) and mcr-3.3 (WP-099982814.1). MCR-3-like of A. veronii 0728Q8Av provided a 145-aa substitution by 29 amino acids at the carbon terminus, while a single amino acid difference occurred at the 273rd residue within the 396 amino acid residues at the Nitrogen terminus, compared to reported MCR-3-like (MF495680) (Figure 2). The phylogenetic tree unveiled a close clustering of both MCR-3.16 and MCR-3.3 with other predetermined MCR-3 variants. Additionally, the MCR-3-like variant in this study exhibited clustering alongside the one reported in A. veronii isolated from retail chicken (MF495680) (Supplementary Figure S2).
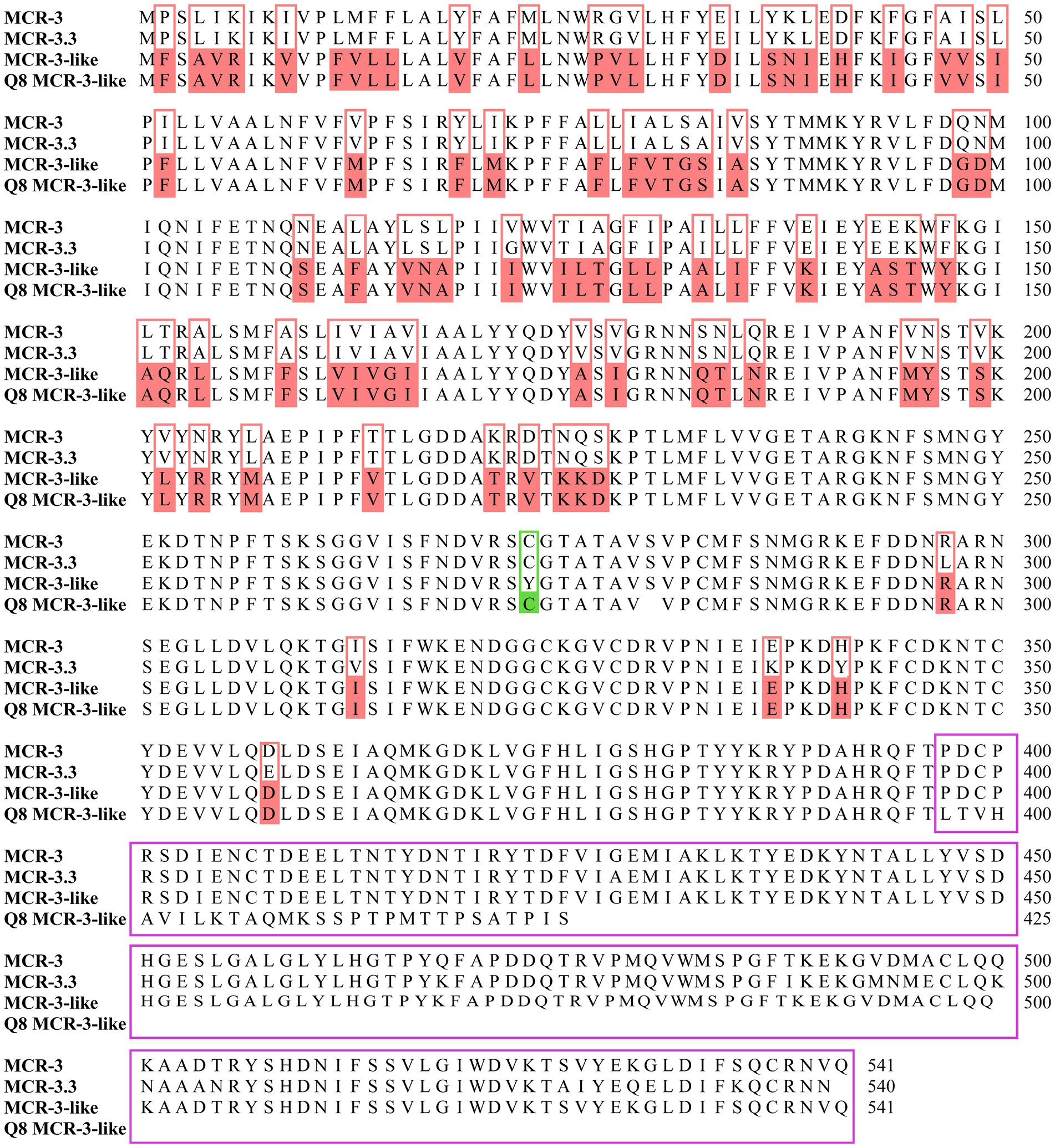
Figure 2. Alignment of the deduced amino acid sequences of Q8 MCR-3-like (A. veronii 0728Q8Av, PRJNA1070378) with previously reported MCR-3 (E. coli pWJ1, WP-039026394.1), MCR-3.3 (Aeromonas veronii 172, WP-099982814.1) and MCR-3-like (Aeromonas veronii 172, MF495680). The amino acid residues of Q8 MCR-3-like identical to MCR-3-like but differing from MCR-3 and MCR-3.3 were highlighted in pink boxes. The 273rd amino acid residue at the N-terminus of Q8 MCR-3-like differed from that of MCR-3-like but matched those of MCR-3 and MCR-3.3, highlighted in a green box. A 145-aa substitution by 29 amino acids at the C-terminus of Q8 MCR-3-like, compared to MCR-3-like, was indicated by a purple box.
To determine the function in mediating colistin resistance of mcr-3 variants, 2,685 bp, 1,920 bp, 4,008 bp, and 2,787 bp DNA fragments, corresponding to the ORFs of mcr-3.16, mcr-3-like, mcr-3.16-mcr-3-like, and mcr-3.3 along with their 5′- and 3′-flanking regions, respectively, were amplified to construct the recombinants pUC19-mcr-3.16, pUC19-mcr-3-like, pUC19-mcr-3.16-mcr-3-like, and pUC19-mcr-3.3, and transformed into E. coli DH5α. The results showed that transformants containing pUC19-mcr-3.16, pUC19-mcr-3.16-mcr-3-like, and pUC19-mcr-3.3, had colistin MICs of 4 μg/mL, 4 μg/mL, and 2 μg/mL, respectively, which were 8-, 8- and 4-fold higher than the MIC of DH5α containing pUC19 alone (0.5 μg/mL). Transformants containing pUC19-mcr-3-like had a colistin MIC of 0.5 μg/mL, similar to DH5α containing pUC19 alone (Table 4). Meanwhile, the ORFs of the three mcr-3 variants were cloned to construct the recombinants pET28a-mcr-3.16, pET28a-mcr-3-like, pET28a-mcr-3.16-mcr-3-like, and pET28a-mcr-3.3. The results showed that the transformants containing pET28a-mcr-3.16, pET28a-mcr-3.16-mcr-3-like, and pET28a-mcr-3.3, had colistin MICs of 16 μg/mL, which were 2-fold higher than the MIC of BL21 containing pET28a alone (8 μg/mL). Transformants containing pET28a-mcr-3-like had a colistin MIC of 8 μg/mL, similar to BL21 containing pET28a alone (Table 4).

Table 4. Colistin susceptibility profiles of A. veronii 0728Q8Av and A. caviae 1029Y16Ac harboring mcr-3.16 and mcr-3-like, and mcr-3.3, respectively.
A segment consisting of mcr-3.16-mcr-3-like-dgkA was observed in A. veronii 0728Q8Av, which was identical to that in A. salmonicida Z5-5 (GCA_003265515.1), a foodborne isolate from chicken meat in China. A similar genetic composition pattern mcr-3.3-mcr-3-like-dgkA was identified in many other A. veronii isolates (MF495680, CP040717, GCA_003265545.1, and GCA_003265585.1). The mcr-3-like gene in A. veronii 0728Q8Av was located 66 bp downstream of the mcr-3.16 gene, and the dgkA gene was located 118 bp downstream of mcr-3-like, which were also found in A. veronii 172 (MF495680), A. veronii Z2-7 (GCA_003265545.1) and A. veronii ZJ12-3 (GCA_003265585.1). In comparison, a 66 bp intergenic region sequence existed between mcr-3.16 (mcr-3.3) and mcr-3-like in A. salmonicida Z5-5 (GCA_003265515.1) and A. veronii HX3 (CP040717), while the intergenic region sequence between mcr-3-like and dgkA spaned 206 nucleotides. Similar to A. salmonicida Z5-5, the 5′ flanking region of the segment mcr-3.16-mcr-3-like-dgkA in A. veronii 0728Q8Av contained an insertion sequence ISKpn3. However, in A. salmonicida Z5-5, two hypothetical proteins were inserted between mcr-3.16 and ISKpn3, distinguishing it from A. veronii 0728Q8Av. Additionally, an insertion sequence ISAs29 could be found adjacent to the dgkA gene in the 3′ flanking region of mcr-3.16-mcr-3-like-dgkA (Figure 3).
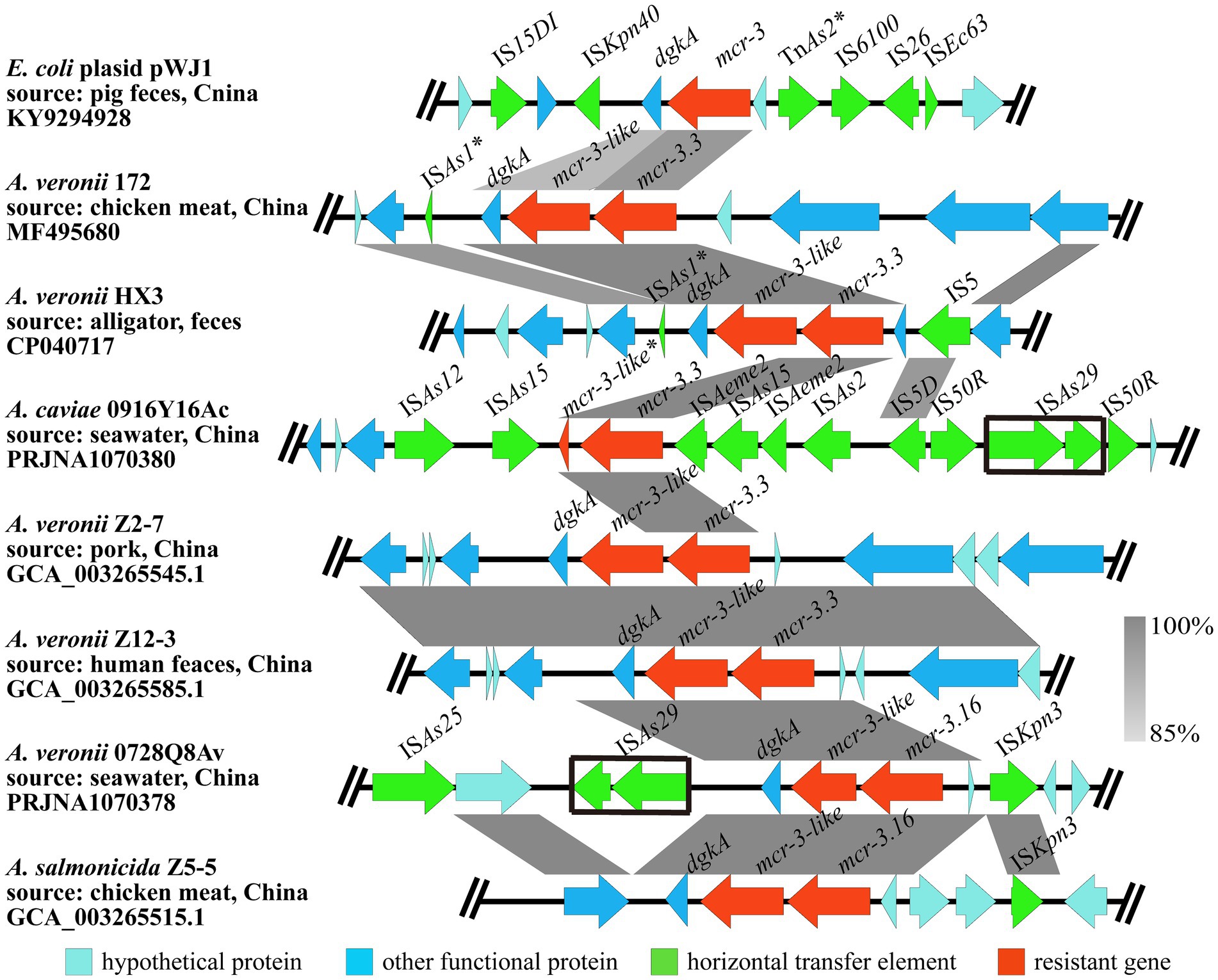
Figure 3. The genetic context of mcr-3.16 and mcr-3.3 in the A. veronii 0728Q8Av and A. caviae 1029Y16Ac, respectively. The arrows indicate the direction of gene transcription. The grayscale intensity indicates the sequence similarity between two linked regions. The black box indicates the identical insertion sequence ISAS29. *Representing incomplete sequences.
A segment consisting of mcr-3.3-mcr-3-like was discovered in A. caviae 1029Y16Ac. But the mcr-3-like gene was incomplete and only had 116 amino acids, which showed 100% identity to the Nitrogen terminus of the MCR-3-like (MF495680). Compared to other Aeromonas spp., it seemed that the 3′ end of the mcr-3-like and its downstream dgkA gene were completely absent in A. caviae 1029Y16Ac. None ORFs but 10 ISs, namely ISAs12, ISAs15, ISAs2, ISAs29, IS5D, IS50R, and ISAeme2, were observed at the flanking regions of the segment mcr-3.3-mcr-3-like in A. caviae 1029Y16Ac (Figure 3). No transconjugants were observed on the colistin-containing screening LB agar plates coated with bacterial culture from the conjugation assay using A. veronii 0728Q8Av or A. caviae 1029Y16Ac. This indicated that the mcr-3 variants, that is mcr-3.16 and mcr-3-like in A. veronii 0728Q8Av and mcr-3.3 in A. caviae 1029Y16Ac, were not transferred into E. coli J53 through conjugation, suggesting that these mcr-3 variants were located on the chromosomes of both A. veronii 0728Q8Av and A. caviae 1029Y16Ac (Supplementary Figure S3).
The results showed that gene island AvGI1 in A. veronii 0728Q8Av harbored 12 ARGs, including tet(E), aph (3″)-Ib, aph(6)-Id, floR, qnrVC4, cmlA1, blaOXA-10, aac(6′)-Ib3, aadA1, dfrA14, truncated qacE (qacE∆1), and sul, which potentially mediated the resistance against tetracyclines, amphenicol, aminoglycosides, quinolones, beta lactams, quaternary ammonium compounds, and sulfonamides antibiotics (Figure 4A). Meanwhile, AvGI1 contained five complete ISs (ISAs1, ISVsa3, IS26, IS6100, and IS3000), one incomplete IS (IS15DI), and one transposon Tn5393. In addition, AvGI1 had a typical class 1 integron containing the integrase intI1 localized at the attI1 site and the genes qacE∆1 and sul1 conferring resistance to quaternary ammonium compounds and sulfonamides, respectively. The gene cassette array featured four attC sites, partitioning it into four gene boxes: qnrVC4, orf-clmlA, blaOXA-10-aac (6′)-Ib3, and orf-aadA1 (Figure 4A).
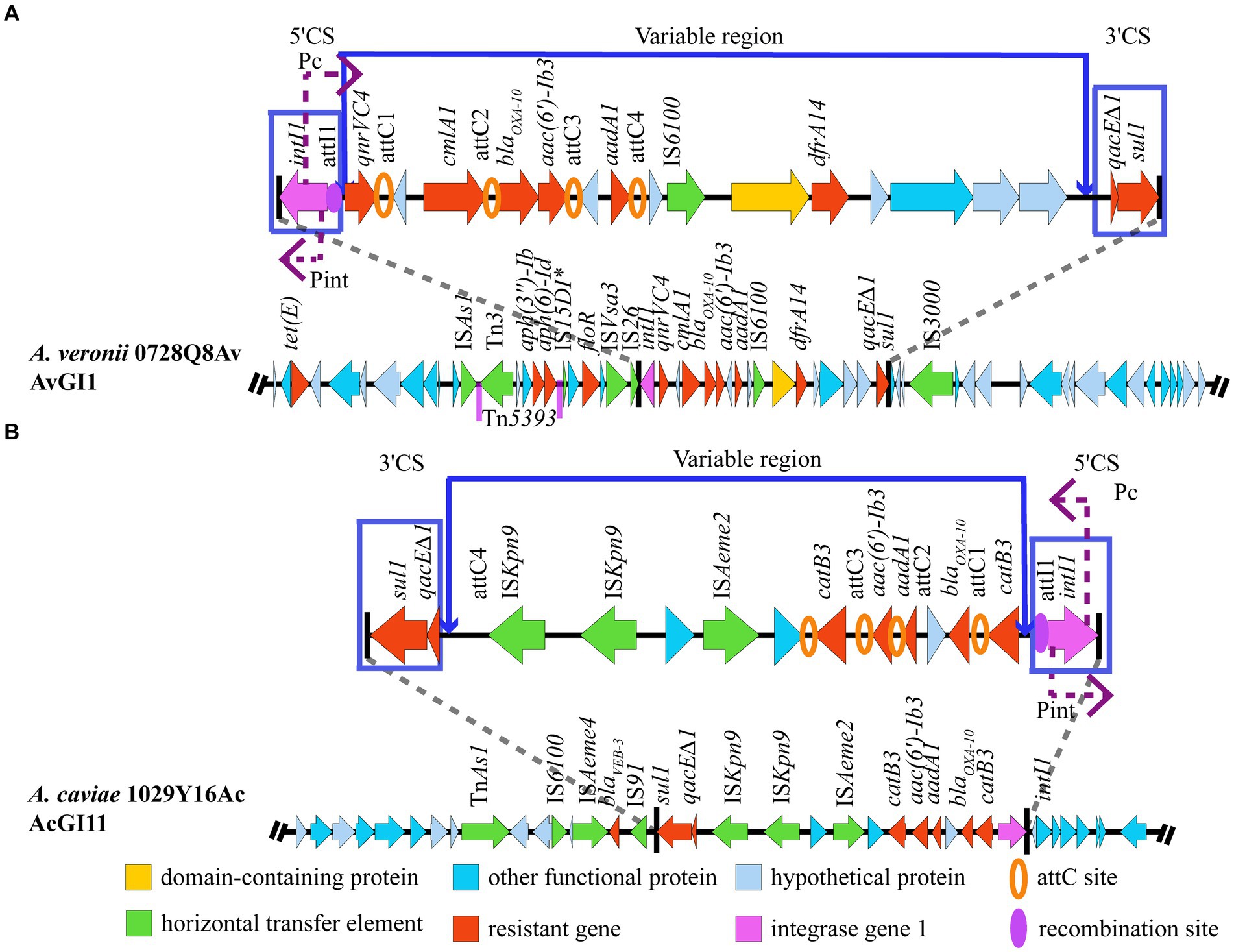
Figure 4. Structure of the GIs harboring MDR genes. (A) AvGI1 in A. veronii 0728Q8Av. (B) AcGI11 in A. caviae 1029Y16Ac. *Representing incomplete sequences.
The gene island AcGI11 in A. caviae 1029Y16Ac harbored 7 ARGs, including blaVEB-3, sul1, qacE∆1, aac (6′)-Ib3, aadA1, blaOXA-10 and catB3, which potentially mediated the resistance against beta lactams, sulfonamides, quaternary ammonium compounds, amphenicol, and aminoglycosides (Figure 4B). Meanwhile, AcGI11 contained six complete ISs, including IS6100, ISAeme2, ISAeme4, two copies of ISKpn9, and IS91, and one transposon TnAs1. Similarly, AcGI11 had the typical class 1 integron and the gene cassette array featured four attC sites, partitioning it into four gene boxes: catB3, blaOXA-10-orf-aadA1, aac(6′)-Ib3, and catB3 (Figure 4B).
Blast analysis indicated that three genomic segments were similar to AvGI1 (query coverage >72%, identities >90%), that is A. hydrophila NUITM-VA1 (AP025277), A. caviae WCW1-2 (CP039832) and A. simaiae A6 (CP040449), while no genomic segment was similar to AcGI11. Multiple sequence alignment revealed a high similarity in the class 1 integron, including the ORFs and ISs within the gene cassettes, between A. veronii 0728Q8Av (PRJNA1070380) and A. hydrophila NUITM-VA1 (AP025277), as well as A. caviae WCW1-2 (CP039832). The transposon Tn5393 and the insertion sequence IS3000 were observed upstream and downstream of the class 1 integrons in A. veronii 0728Q8Av, A. caviae WCW1-2 (CP039832), and A. simaiae A6 (CP040449). But, A. veronii 0728Q8Av contained an additional segment, IS15DI-orf-floR-orf-ISVsa3-IS26, located between Tn5393 and the class 1 integron. A. hydrophila NUITM-VA1 (AP025277) did not form the transposon Tn5393 (Figure 5).
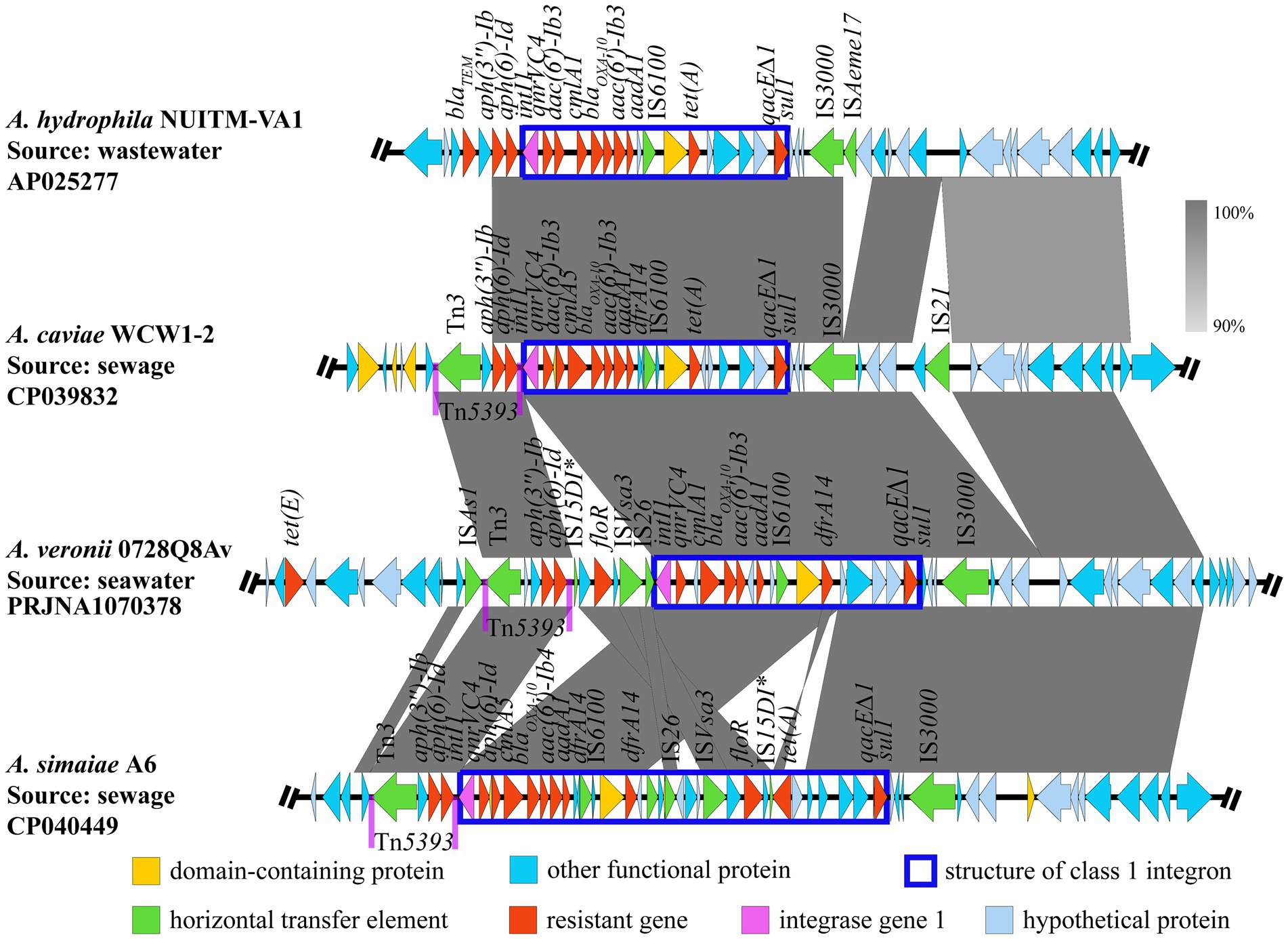
Figure 5. Comparative analysis of genetic environments of AvGI1. The arrows indicate the direction of gene transcription. The grayscale intensity indicates the sequence similarity between two linked regions. *Representing incomplete sequences.
The mortality of fish infected with the A. veronii 0728Q8Av and A. caviae 1029Y16Ac isolate are summarized in Table 5. Using the calculated method reported by Mittal et al. (1980), the LD50 value in a 7-day period obtained from A. veronii 0728Q8Av was measured at 2.15 × 107 CFU/mL and was categorized as virulent, while the LD50 of A. caviae 1029Y16Ac was calculated at 1.35 × 109 CFU/mL and was non-virulent (Table 5).
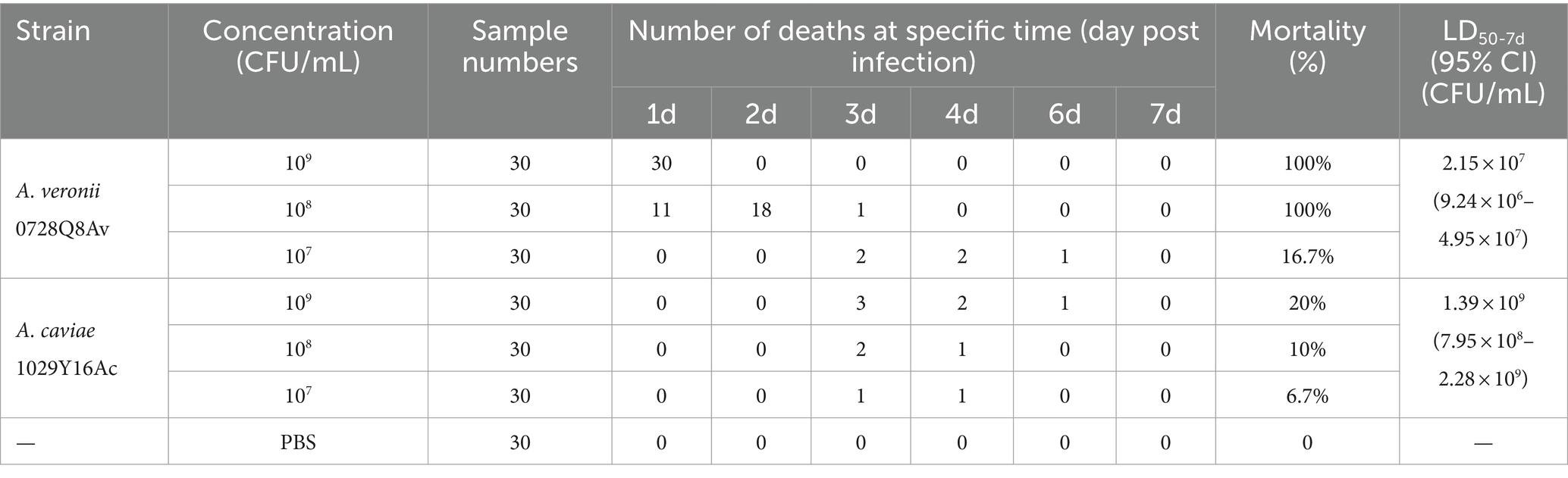
Table 5. Determination of median lethal dosage (LD50) of A. veronii 0728Q8Av and A. caviae 1029Y16Ac.
Aeromonas species isolated from coastal waters are prone to developing MDR, contributing to the interchange of ARGs between land-derived and marine environments (Zhu et al., 2017; Jang et al., 2018; Zheng et al., 2021). In this study, we pinpointed two MDR Aeromonas species, that is A. veronii 0728Q8Av and A. caviae 1029Y16Ac, which exhibited distinct biochemical characteristics as previously documented (Abbott et al., 2003). Both isolates exhibited resistance to colistin and harbored different mcr-3 variants. All resistance elements, including ARGs and MGEs, were located on their chromosomes, irrespective of the presence of plasmids. The mcr-3 variants were not clustered with other ARGs but were independently located on the chromosomal DNA, flanked by several ISs. In addition, A. veronii 0728Q8Av, but not A. caviae 1029Y16Ac, exhibits significant pathogenicity to ayu (Plecoglossus altivelis).
Antimicrobial resistance in Aeromonas is rapidly escalating worldwide, and resistance elements have become increasingly complex. For example, Aeromonas spp. resistance to 3rd-generation cephalosporin and producing broad-spectrum carbapenemase KPC-24 have been isolated globally (Bhaskar et al., 2015; Yang et al., 2022). Aeromonas demonstrates notable activity in MDR, exhibiting widespread resistance to various classes of antibiotics including β-lactam, aminoglycosides, fluoroquinolones, tetracyclines, macrolides, sulfonamides, polymyxins, and phenicols, particularly in wastewater (Figueira et al., 2011; Carusi et al., 2023; Neil et al., 2024). However, few reports have highlighted the contribution of Aeromonas to the development and dissemination of antimicrobial resistance elements in coastal aquatic environments (Gambino et al., 2022; Liang et al., 2024). In the present study, the two Aeromonas species exhibited broad MDR profiles, with sulfamethoxazole, oxytetracycline, ampicillin and florfenicol being the least effective to A. veronii 0728Q8Av and sulfamethoxazole and trimethoprim being the least effective to A. caviae 1029Y16Ac. Correspondingly, various ARGs could be found in the chromosomal genomes of both Aeromonas strains, such as mcr-3.16, qnrVC4, sul1, dfrA14, blaOXA-10, ampS, cphA4, tet(E), floR, and cmlA1 in A. veronii 0728Q8Av, and mcr-3.3, sul1, blaVEB-3, blaMOX-6, blaOXA-10, tet(E), and catB3 in A. caviae 1029Y16Ac. If only considering ARGs as determinants of antibiotic resistance, sul1 exhibits highly potent resistance to sulfamethoxazole (with an MIC value of 256 μg/mL). No ARGs coding for trimethoprim-resistant dihydrofolate reductases (DHFRs) was found in the genome of A. caviae 1029Y16Ac, indicating other resistance mechanisms probably responsible for trimethoprim. Interestingly, both strains remained susceptible to ceftazidime, which is supported by their resistance gene profiles.
MGEs are the major contributors to the horizontal transfer of ARGs among bacteria (Juhas et al., 2009; Piotrowska and Popowska, 2015; shao et al., 2018). Aeromonas spp. can adeptly utilize these elements to develop its MDR (Piotrowska and Popowska, 2015). Unlike Enterobacteriaceae, which primarily transfer ARGs via plasmids, the extra-chromosomal DNA (Rozwandowicz et al., 2018), Aeromonas spp. also use plasmids for horizontal gene transfer (Piotrowska and Popowska, 2015). However, a significant proportion of their ARGs are located on the chromosome and are mainly transferred through integrons, ISs, and transposons (L’Abée-Lund and Sørum, 2001; Schmidt et al., 2001; Sakulworakan et al., 2021). ISs (e.g., IS26, ISPa12, ISKpn8,) and transposons (e.g., Tn3, Tn21, Tn1213) frequently involved in the formation of diverse genetic units characterized by variable regions containing various ARGs, often segmented by conservative and unique DNA structures, or forming multiple gene cassettes (Piotrowska and Popowska, 2015). Moreover, novel multiple drug-resistant MGEs are continually being identified in Aeromonas spp., complicating the formation and transmission of multidrug resistance further (Piotrowska and Popowska, 2015; Carusi et al., 2023). In this study, two multidrug resistance GIs, AvGI1 and AcGI11, were identified in A. veronii 0728Q8Av and A. caviae 1029Y16Ac, respectively. Both AvGI1 and AcGI11 harbored a class 1 integron which is considered the predominant type in Aeromonas (Piotrowska and Popowska, 2015). AvGI1 contained more ARGs than AcGI11. Apart from sul1-qacE∆1 (a common structural combination found at 3′ end of the typical class 1 integron) (Deng et al., 2015), aadA1, and blaOXA-10, the ARGs in both GIs were largely distinct (Figure 4). AVGI1 contained a Tn3 family transposase Tn5393, whereas AcGI11 harbored another Tn3 family transposase TnAs1, and both GIs also possessed several ISs (Figure 4). Tn5393, located tandemly upstream of the class 1 integron in AvGI1, is likely involved in the mobility and stability of two ARGs, that is aph (3″)-Ib and aph (6)-Id, conferring resistance to aminoglycosides. Although TnAs1, along with ISs like IS6100 (belonging to the IS6 family members) and IS91, were found downstream of the 3′ flanking region of the integron in AcGI11, all the ARGs located within the class 1 integron. Furthermore, the ARGs were carried by the gene cassette array, and two idle ISKpn9s were tandemly adjacent to the sul1-qacE∆1 structure within the class 1 integron in AcGI11 (Figure 4). IS26 was identified in the 5′ flanking region of the class 1 integron in AvGI1, forming the IS26-intI structure (Figure 4). However, IS3000 was identified in the 3′ flanking region of the class 1 integron in AvGI1, forming an IS26-integron-IS3000 unit rather than the IS26-integron-IS26 structure reported in the literature (Shang et al., 2021; Ma et al., 2023; Wang et al., 2023). Nevertheless, the horizontal transfer of IS26-integron-IS3000 remains to be explored, although IS26-orf-IS3000 could serve as a vehicle for ARG transmission (Doi et al., 2014). AvGI1 exhibited high sequence similarity with several Aeromonas strains isolated from wastewater and sewage in China, including A. hydrophila NUITM-VA1 (AP025277), A. caviae WCW1-2 sewage (CP039832), and A. simaiae A6 (CP040449) (Chen C. et al., 2019; Chen Q. et al., 2019; Dao et al., 2022) (Figure 5). The composition and arrangement of ARGs carried by class 1 integron in AvGI1, together with aph(3″)-Ib and aph(6)-Id, closely resembled those observed in both A. hydrophila NUITM-VA1 and A. caviae WCW1-2. The tet(E) gene located not within but upstream of the class 1 integron in AvGI1, which was the only significant difference compare to A. hydrophila NUITM-VA1 and A. caviae WCW1-2. Compare to A. hydrophila NUITM-VA1, Tn5393 probably enhanced the transmission of both ARGs in A. caviae WCW1-2 and A. veronii 0728Q8Av. Additionally, the Tn5393-aph(3″)-Ib-aph(6)-Id unit was also found tandemly located upstream of the class 1 integron in A. simaiae A6. Interestingly, an IS15DI-floR-ISVsa3-IS26 unit was also found within the class 1 integron in A. simaiae A6 was present as a reverse insertion between Tn5393 and class 1 integron in AvGI1 (Figure 5). These results indicated the transposon Tn5393, combined with the IS IS3000, could serve as a vehicle for the transmission of class 1 integron, reflecting the plasticity of MGEs in ARGs transmission among bacteria.
Colistin resistance in Aeromonas has been extensively documented, with various mcr variants being detected (Ling et al., 2017; Yin et al., 2017; Eichhorn et al., 2018; Shen et al., 2018). Among these, mcr-3 variants were detected most frequently in Aeromonas spp., including mcr-3.13, mcr-3.14, mcr-3.15, mcr-3.16, mcr-3.17, mcr-3.18, mcr-3.3, mcr-3.6, mcr-3.7, mcr-3.8, mcr-3.9, and mcr-5 (Ling et al., 2017; Yin et al., 2017; Eichhorn et al., 2018; Ma et al., 2018; Shen et al., 2018). The mcr-3-mcr-3-like segment was originally reported in A. veronni 172 isolated from chicken meat (Ling et al., 2017). Subsequently, the mcr-3.6-mcr-3-like, mcr-3.8-mcr-3-like, and mcr-3.9-mcr-3-like segments were reported in A. allosaccharophila, A. jandaei, and A. hydrophila, respectively (Eichhorn et al., 2018). In this study, we identified a mcr-3.16-mcr-3-like segment in A. veronii 0728Q8Av and a mcr-3.3-mcr-3-like (incomplete) segment in A. caviae 1029Y16Ac. E. coli transformants carrying pUC19-mcr-3.16 or pUC19-mcr-3.16-mcr-3-like showed an 8-fold higher colistin MIC compared to transformants containing pUC19 alone, while transformants containing pET28a-mcr-3.16 or pET28a-mcr-3.16-mcr-3-like had a 2-fold higher colistin MIC than those containing pET28a alone. These results indicated that it is not mcr-3-like but mcr-3.16 capable of mediating resistance to colistin in A. veronii 0728Q8Av, which was consistent with previous reports (Yin et al., 2017; Shen et al., 2018). In contrast, transformants carrying pUC19-mcr-3.3 showed 4-fold higher colistin MIC than pUC19 alone, and those containing pET28a-mcr-3.3 had a 2-fold higher colistin MIC than pET28a alone, which indicated that mcr-3.3 could mediate resistance to colistin in A. caviae 1029Y16Ac (Ling et al., 2017; Shen et al., 2018). An incomplete mcr-3-like fragment, predictively coding partial N-terminal transmembrane domain of phosphoethanolamine transferase, was found downstream and adjacent to mcr-3.3 A. caviae 1029Y16Ac. This incompleteness of the phosphoethanolamine transferase mutant suggests the likelihood that mcr-3 originated from Aeromonas species (Shen et al., 2018). Additionally, a dgkA gene encoding diacylglycerol kinase was identified downstream and adjacent to mcr-3-like in A. veronii 0728Q8Av, forming a mcr-3.16-mcr-3-like-dgkA segment. This positioning of dgkA was also found in several diploid analogs of mcr-3 variants, such as mcr-3.16-mcr-3-like in A. salmonicida Z5-5, mcr-3.3-mcr-3-like in A. veronii Z12-3, A. veronii Z2-7, A. veronii HX3, and A. veronii 172 (Figure 3). Further exploration is needed to determine whether this contributes to an improvement for mcr-3-like in resistance to colistin. Definitely, an ISKpn40-mcr-3-dgkA-ISKpn40 segment frequently occurred in various bacteria, such as Klebsiella pneumoniae, E. coli, and Salmonella enterica (Xiang et al., 2018; Hadjadj et al., 2019; Sia et al., 2020). Gallardo et al. (2020) demonstrated that dgkA, closely situated to mcr-3 variants, could marginally enhance resistance to colistin. This enhancement may stem from its ability to compensate for alterations in phospholipid metabolism functions induced by LPS modification through colistin resistance determinants (Gallardo et al., 2020).
After the ban on colistin as a growth promoter in several countries for example China, Japan, and Thailand, both the colistin residue concentrations and mcr variants (especially mcr-1) in different environments have reportedly decreased (Wang et al., 2020; Rhouma et al., 2023). This has alleviated the predicament of Aeromonas in the spread of mcr genes. However, the emergence of MDR in treating Aeromonas infections remains a concern. In this study, the two Aeromonas strains exhibited similar resistance profiles, showing resistance to commonly used antibiotics such as tetracycline, oxytetracycline, florfenicol, ampicillin, sulfamethoxazole, and trimethoprim (Table 2). Both strains remained sensitive to ceftazidime and lacked common ARGs for other antibiotics, such as the tetX family conferring resistance to tetracycline and blaNDM conferring resistance to carbapenem, indicating that therapeutic options are still available. Nevertheless, both strains were found to carry the blaOXA-10 gene encoding ESBL, and A. caviae 1029Y16Ac harbored the blaVEB-3 gene, highlighting the need to monitor the spread of these strains.
A. caviae and A. veronii, along with A. dhakensis, A. hydrophila and A. salmonicida, are the most frequently detected Aeromonas species causing diseases in both humans and animals (Janda and Abbott, 2010; Fernández-Bravo and Figueras, 2020). As opportunistic pathogens, their pathogenicity is restricted by various factors, including genetic virulence factors, temperature, and host immune status (Fernández-Bravo and Figueras, 2020; Carusi et al., 2023). In this study, according to a virulence assessment method (Mittal et al., 1980), A. veronii 0728Q8Av demonstrated moderate virulence towards the tested ayu, while A. caviae 1029Y16Ac showed no virulence. Additionally, a 100% mortality rate was observed within 2 days post-infection when fish were injected with A. veronii 0728Q8Av at concentrations ranging from 108 to 109 CFU/mL, indicating a short incubation period for A. veronii 0728Q8Av infection (Chen F. et al., 2019). Compared to A. caviae 1029Y16Ac, more virulence factors related to pili (such as Type I, Flp type IV, and MSHA type IV) and polar flagella were identified in A. veronii 0728Q8Av, which probably enhanced the bacteria adhesion and persistence thus to promote its pathogenicity (Kirov et al., 2004; Boyd et al., 2008; Dacanay et al., 2010). Moreover, although the genomes of both A. veronii 0728Q8Av and A. caviae 1029Y16Ac contained genes encoding toxins hemolysin and thermostable hemolysin, only A. veronii 0728Q8Av additionally harbored aerolysin, a pore-forming toxin (Ran et al., 2018). This may be associated with the pathogenicity observed in A. veronii 0728Q8Av.
The MDR Aeromonas are ubiquitously distributed among humans, animals, and their environments. Numerous Aeromonas species carrying various mcr variants, which confer resistance to colistin, have been detected in hospitals, urban rivers, livestock, and aquatic animals. However, their presence in coastal waters remains relatively underreported. In this study, we identified two MDR Aeromonas strains, namely A. veronii 0728Q8Av and A. caviae 1029Y16Ac, from coastal waters in Zhejiang, China. Both Aeromonas isolates exhibited significant resistance to 11 kinds of antibiotics and remained susceptible to ceftazidime, a 3rd-generation cephalosporin antibiotic. And both isolates harbored multiple ARGs located on their chromosomes, with the majority concentrated within in a resistance GI, respectively. Both islands harbored typical class 1 integrons. Notably, both isolates carried ARGs mediating colistin resistance, namely mcr-3.16 on A. veronii 0728Q8Av and mcr-3.3 on A. caviae 1029Y16Ac. Both mcr-3 variants were located on the chromosome, distanced from the multidrug resistance GIs, flanking by multiple ISs. Additionally, a mcr-3-like was identified in the genome of A. veronii 0728Q8Av, forming a tandem mcr-3.16-mcr-3-like-dgkA structure. However, the mcr-3-like recombinants did not confer colistin resistance in E. coli. Furthermore, an incomplete mcr-3-like was found adjacent to mcr-3.3 in the genome of A. caviae 1029Y16Ac, suggesting the likelihood that mcr-3 originated from Aeromonas species. Additionally, we demonstrated that A. veronii 0728Q8Av exhibited pathogenicity towards infected ayu. These findings indicated the presence of terrestrial MDR Aeromonas species in the coastal waters of China, posing a potential threat to the aquaculture, necessitating the development of more effective strategies to mitigate the spread of antibiotic resistance.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1070378, https://www.ncbi.nlm.nih.gov/genbank/, PRJNA1070380.
The animal study was approved by the Animal Experiment Ethics Committee of Ningbo University. The study was conducted in accordance with the local legislation and institutional requirements.
H-XC: Visualization, Investigation, Writing – original draft. F-JC: Investigation, Visualization, Writing – original draft. Q-JZ: Visualization, Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. S-LS: Investigation, Visualization, Writing – original draft. BT: Investigation, Visualization, Writing – original draft. Z-JX: Investigation, Visualization, Writing – original draft. L-JD: Investigation, Visualization, Writing – original draft. J-LJ: Investigation, Visualization, Writing – original draft. G-ZX: Investigation, Visualization, Writing – original draft. M-CY: Resources, Writing – original draft. JC: Funding acquisition, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The work was supported by National Natural Science Foundation of China (42276110), the Program of Science and Technology Department of Zhejiang Province (LGN22C010001), the Key Research and Development Project of Zhejiang Province (2021C02059), Natural Science Foundation of Ningbo City, China (2021J061), the Program of Science and Technology Department of Ningbo City (2022S210), State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products (2021DG700024-ZZ2102), One Health Interdisciplinary Research Project, Ningbo University (HZ202201), the Program of Zhejiang Agriculture and Rural Affairs (2023SNJF062), Zhejiang Key Laboratory of Exploitation and Preservation of Coastal Bio-Resource (J2022001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1401802/full#supplementary-material
2. ^http://rast.theseed.org/FIG/rast.cgi
3. ^https://cge.cbs.dtu.dk/services/ResFinder/
4. ^http://www.pathogenomics.sfu.ca/islandviewer
5. ^https://www-is.biotoul.fr/
6. ^https://tncentral.ncc.unesp.br/
Abbott, S. L., Cheung, W. K. W., and Janda, J. M. (2003). The genus Aeromonas: biochemical characteristics, atypical reactions, and phenotypic identification schemes. J. Clin. Microbiol. 41, 2348–2357. doi: 10.1128/JCM.41.6.2348-2357.2003
Bertelli, C., and Brinkman, F. S. L. (2018). Improved genomic island predictions with IslandPath-DIMOB. Bioinformatics 34, 2161–2167. doi: 10.1093/bioinformatics/bty095
Bhaskar, M., Dinoop, K. P., and Mandal, J. (2015). Characterization of ceftriaxone-resistant Aeromonas spp. isolates from stool samples of both children and adults in southern India. J. Health Popul. Nutr. 33:26. doi: 10.1186/s41043-015-0036-7
Boyd, J. M., Dacanay, A., Knickle, L. C., Touhami, A., Brown, L. L., Jericho, M. H., et al. (2008). Contribution of type IV pili to the virulence of Aeromonas salmonicida subsp. salmonicida in Atlantic salmon (Salmo salar L.). Infect. Immun. 76, 1445–1455. doi: 10.1128/IAI.01019-07
Carusi, J., Kabuki, D. Y., de Seixas Pereira, P. M., and Cabral, L. (2023). Aeromonas spp. in drinking water and food: occurrence, virulence potential and antimicrobial resistance. Food Res. Int. 175:113710. doi: 10.1016/j.foodres.2023.113710
Chang, J., Tang, B., Chen, Y., Xia, X., Qian, M., and Yang, H. (2020, 2020). Two IncHI2 plasmid-mediated colistin-resistant Escherichia coli strains from the broiler chicken supply chain in Zhejiang province, China. J. Food Prot. 83, 1402–1410. doi: 10.4315/JFP-20-041
Chen, C., Chen, L., Zhang, Y., Cui, C. Y., Wu, X. T., He, Q., et al. (2019). Detection of chromosome-mediated tet(X4)-carrying Aeromonas caviae in a sewage sample from a chicken farm. J. Antimicrob. Chemother. 74, 3628–3630. doi: 10.1093/jac/dkz387
Chen, F., Sun, J., Han, Z., Yang, X., Xian, J. A., and Shi, H. (2019). Isolation, identification and characteristics of Aeromonas veronii from diseased crucian carp (Carassius auratus gibelio). Front. Microbiol. 10:2742. doi: 10.3389/fmicb.2019.02742
Chen, Q., Zhou, W., Shen, K., Li, K., Xu, T., Bao, Q., et al. (2019). OXA-830, a novel chromosomally encoded extended-spectrum class D β-lactamase in Aeromonas simiae. Front. Microbiol. 10:2732. doi: 10.3389/fmicb.2019.02732
Dacanay, A., Boyd, J. M., Fast, M. D., Knickle, L. C., and Reith, M. E. (2010). Aeromonas salmonicida type I pilus system contributes to host colonization but not invasion. Dis. Aquat. Org. 88, 199–206. doi: 10.3354/dao02157
Damas, M. S. F., Ferreira, R. L., Campanini, E. B., Soares, G. G., Campos, L. C., Laprega, P. M., et al. (2022). Whole genome sequencing of the multidrug-resistant Chryseobacterium indologenes isolated from a patient in Brazil. Front. Med. 9:931279. doi: 10.3389/fmed.2022.931379
Dao, T. D., Yano, H., Takemura, T., Hirabayashi, A., Trang, L. T., Tran, H. H., et al. (2022). Strand-biased circularizing integrative elements spread tmexCD-toprJ gene clusters encoding RND-type multidrug efflux pumps by repeated transpositions. bioRxiv. Available at: https://doi.org/10.1101/2022.09.22.508988. [Epub ahead of preprint]
Deng, Y., Bao, X., Ji, L., Chen, L., Liu, J., Miao, J., et al. (2015). Resistance integrons: class 1, 2 and 3 integrons. Ann. Clin. Microbiol. Antimicrob. 14:45. doi: 10.1186/s12941-015-0100-6
Doi, Y., Hazen, T. H., Boitano, M., Tsai, Y. C., Clark, T. A., Korlach, J., et al. (2014). Whole-genome assembly of Klebsiella pneumoniae coproducing NDM-1 and OXA-232 carbapenemases using single-molecule, real-time sequencing. Antimicrob. Agents Chemother. 58, 5947–5953. doi: 10.1128/AAC.03180-14
Ebu, S. M., Ray, L., Panda, A. N., and Gouda, S. K. (2023). De novo assembly and comparative genome analysis for polyhydroxyalkanoates-producing Bacillus sp. BNPI-92 strain. J. Genet. Eng. Biotechnol. 21:132. doi: 10.1186/s43141-023-00578-7
Eichhorn, I., Feudi, C., Wang, Y., Kaspar, H., Feßler, A. T., Lübke-Becker, A., et al. (2018). Identification of novel variants of the colistin resistance gene mcr-3 in Aeromonas spp. from the national resistance monitoring programme GERM-Vet and from diagnostic submissions. J. Antimicrob. Chemother. 73, 1217–1221. doi: 10.1093/jac/dkx538
Fernández-Bravo, A., and Figueras, M. J. (2020). An update on the genus Aeromonas: taxonomy, epidemiology, and pathogenicity. Microorganisms 8:129. doi: 10.3390/microorganisms8010129
Figueira, V., Vaz-Moreira, I., Silva, M., and Manaia, C. M. (2011). Diversity and antibiotic resistance of Aeromonas spp. in drinking and waste water treatment plants. Water Res. 45, 5599–5611. doi: 10.1016/j.watres.2011.08.021
Finney, D. J. (1985). The median lethal dose and its estimation. Arch. Toxicol. 56, 215–218. doi: 10.1007/BF00295156
Gallardo, A., Ugarte-Ruiz, M., Hernández, M., Miguela-Villoldo, P., Rodríguez-Lázaro, D., Domínguez, L., et al. (2020). Involvement of hpap2 and dgka genes in colistin resistance mediated by mcr determinants. Antibiotics 9:531. doi: 10.3390/antibiotics9090531
Gambino, D., Savoca, D., Sucato, A., Gargano, V., Gentile, A., Pantano, L., et al. (2022). Occurrence of antibiotic resistance in the Mediterranean Sea. Antibiotics 11:332. doi: 10.3390/antibiotics11030332
Hadjadj, L., Baron, S. A., Olaitan, A. O., Morand, S., and Rolain, J. M. (2019). Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front. Microbiol. 10:2720. doi: 10.3389/fmicb.2019.02720
Huq, M. A. (2020). Green synthesis of silver nanoparticles using Pseudoduganella eburnea MAHUQ-39 and their antimicrobial mechanisms investigation against drug resistant human pathogens. Int. J. Mol. Sci. 21:1510. doi: 10.3390/ijms21041510
Janda, J. M., and Abbott, S. L. (2010). The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 23, 35–73. doi: 10.1128/CMR.00039-09
Jang, H. M., Kim, Y. B., Choi, S., Lee, Y., Shin, S. G., Unno, T., et al. (2018). Prevalence of antibiotic resistance genes from effluent of coastal aquaculture, South Korea. Environ. Pollut. 233, 1049–1057. doi: 10.1016/j.envpol.2017.10.006
Jin, J. L., Zhou, Q. J., Shao, X. B., Wang, Y. H., Chen, J., Yan, M. C., et al. (2021). Investigation on resistance genes against four types of common antibiotics in Wenzhou waters of the East China Sea. Oceanol. Limnol. Sin. 52, 947–959. doi: 10.11693/hyhz20210200040
Juhas, M., Van Der Meer, J. R., Gaillard, M., Harding, R. M., Hood, D. W., and Crook, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kirov, S. M., Castrisios, M., and Shaw, J. G. (2004). Aeromonas flagella (polar and lateral) are enterocyte adhesins that contribute to biofilm formation on surfaces. Infect. Immun. 72, 1939–1945. doi: 10.1128/IAI.72.4.1939-1945.2004
L’Abée-Lund, T. M., and Sørum, H. (2001). Class 1 integrons mediate antibiotic resistance in the fish pathogen Aeromonas salmonicida worldwide. Microb. Drug Resist. 7, 263–272. doi: 10.1089/10766290152652819
Liang, H., Huang, J., Tao, Y., Klümper, U., Berendonk, T. U., Zhou, K., et al. (2024). Investigating the antibiotic resistance genes and their potential risks in the megacity water environment: a case study of Shenzhen Bay Basin, China. J. Hazard. Mater. 465:133536. doi: 10.1016/j.jhazmat.2024.133536
Ling, Z., Yin, W., Li, H., Zhang, Q., Wang, X., Wang, Z., et al. (2017). Chromosome-mediated mcr-3 variants in Aeromonas veronii from chicken meat. Antimicrob. Agents Chemother. 61:e01272. doi: 10.1128/AAC.01272-17
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Ma, L., Qu, Y., Wang, W., and Wang, D. (2023). Characterization of Klebsiella pneumoniae carrying the blaNDM-1 gene in IncX3 plasmids and the rare In1765 in an IncFIB-IncHI1B plasmid. Front. Cell. Infect. Microbiol. 13:1324846. doi: 10.3389/fcimb.2023.1324846
Ma, S., Sun, C., Hulth, A., Li, J., Nilsson, L. E., Zhou, Y., et al. (2018). Mobile colistin resistance gene mcr-5 in porcine Aeromonas hydrophila. J. Antimicrob. Chemother. 73, 1777–1780. doi: 10.1093/jac/dky110
Martinez-Murcia, A. J., Monera, A., Saavedra, M. J., Oncina, R., Lopez-Alvarez, M., Lara, E., et al. (2011). Multilocus phylogenetic analysis of the genus Aeromonas. Syst. Appl. Microbiol. 34, 189–199. doi: 10.1016/j.syapm.2010.11.014
Mittal, K. R., Lalonde, G., Leblanc, D., Olivier, G., and Lallier, R. (1980). Aeromonas hydrophila in rainbow trout: relation between virulence and surface characteristics. Can. J. Microbiol. 26, 1501–1503. doi: 10.1139/m80-248
Neil, B., Cheney, G. L., Rosenzweig, J. A., Sha, J., and Chopra, A. K. (2024). Antimicrobial resistance in aeromonads and new therapies targeting quorum sensing. Appl. Microbiol. Biotechnol. 108:205. doi: 10.1007/s00253-024-13055-z
Osei Sekyere, J., Govinden, U., Bester, L. A., and Essack, S. Y. (2016). Colistin and tigecycline resistance in carbapenemase-producing Gram-negative bacteria: emerging resistance mechanisms and detection methods. J. Appl. Microbiol. 121, 601–617. doi: 10.1111/jam.13169
Piotrowska, M., and Popowska, M. (2014). The prevalence of antibiotic resistance genes among Aeromonas species in aquatic environments. Ann. Microbiol. 64, 921–934. doi: 10.1007/s13213-014-0911-2
Piotrowska, M., and Popowska, M. (2015). Insight into the mobilome of Aeromonas strains. Front. Microbiol. 6:494. doi: 10.3389/fmicb.2015.00494
Ran, C., Qin, C., Xie, M., Zhang, J., Li, J., Xie, Y., et al. (2018). Aeromonas veronii and aerolysin are important for the pathogenesis of motile aeromonad septicemia in cyprinid fish. Environ. Microbiol. 20, 3442–3456. doi: 10.1111/1462-2920.14390
Rhouma, M., Madec, J. Y., and Laxminarayan, R. (2023). Colistin: from the shadows to a One Health approach for addressing antimicrobial resistance. Int. J. Antimicrob. Agents 61:106713. doi: 10.1016/j.ijantimicag.2023.106713
Rozwandowicz, M., Brouwer, M. S. M., Fischer, J., Wagenaar, J. A., Gonzalez-Zorn, B., Guerra, B., et al. (2018). Plasmids carrying antimicrobial resistance genes in Enterobacteriaceae. J. Antimicrob. Chemother. 73, 1121–1137. doi: 10.1093/jac/dkx488
Sakulworakan, R., Chokmangmeepisarn, P., Dinh-Hung, N., Sivaramasamy, E., Hirono, I., Chuanchuen, R., et al. (2021). Insight into whole genome of Aeromonas veronii isolated from freshwater fish by resistome analysis reveal extensively antibiotic resistant traits. Front. Microbiol. 12:733668. doi: 10.3389/fmicb.2021.733668
Schmidt, A. S., Bruun, M. S., Larsen, J. L., and Dalsgaard, I. (2001). Characterization of class 1 integrons associated with R-plasmids in clinical Aeromonas salmonicida isolates from various geographical areas. J. Antimicrob. Chemother. 47, 735–743. doi: 10.1093/jac/47.6.735
Shang, D., Zhao, H., Xu, X., Arunachalam, K., Chang, J., Bai, L., et al. (2021). Conjugative IncHI2 plasmid harboring novel class 1 integron mediated dissemination of multidrug resistance genes in Salmonella typhimurium. Food Control 122:107810. doi: 10.1016/j.foodcont.2020.107810
Shao, S., Hu, Y., Cheng, J., and Chen, Y. (2018). Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment. Crit. Rev. Biotechnol. 38, 1195–1208. doi: 10.1080/07388551.2018.1471038
Shen, Y., Xu, C., Sun, Q., Schwarz, S., Ou, Y., Yang, L., et al. (2018). Prevalence and genetic analysis of mcr-3-positive Aeromonas species from humans, retail meat, and environmental water samples. Antimicrob. Agents Chemother. 62:e00404. doi: 10.1128/AAC.00404-18
Shen, Y., Zhang, R., Schwarz, S., Wu, C., Shen, J., Walsh, T. R., et al. (2020). Farm animals and aquaculture: significant reservoirs of mobile colistin resistance genes. Environ. Microbiol. 22, 2469–2484. doi: 10.1111/1462-2920.14961
Sia, C. M., Greig, D. R., Day, M., Hartman, H., Painset, A., Doumith, M., et al. (2020). The characterization of mobile colistin resistance (mcr) genes among 33 000 Salmonella enterica genomes from routine public health surveillance in England. Microb. Genom. 6:e000331. doi: 10.1099/mgen.0.000331
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sun, J., Yang, R. S., Zhang, Q., Feng, Y., Fang, L. X., Xia, J., et al. (2016). Co-transfer of blaNDM-5 and mcr-1 by an IncX3-X4 hybrid plasmid in Escherichia coli. Nat. Microbiol. 1:16176. doi: 10.1038/nmicrobiol.2016.176
Tang, B., Chang, J., Chen, Y., Lin, J., Xiao, X., Xia, X., et al. (2022). Escherichia fergusonii, an underrated repository for antimicrobial resistance in food animals. Microbiol. Spectr. 10:e0161721. doi: 10.1128/spectrum.01617-21
Tang, B., Zhao, H., Li, J., Liu, N., Huang, Y., Wang, J., et al. (2024). Detection of clinical Serratia marcescens isolates carrying blaKPC-2 in a hospital in China. Heliyon 10:e29702. doi: 10.1016/j.heliyon.2024.e29702
Vivas, R., Barbosa, A. A. T., Dolabela, S. S., and Jain, S. (2019). Multidrug-resistant bacteria and alternative methods to control them: an overview. Microb. Drug Resist. 25, 890–908. doi: 10.1089/mdr.2018.0319
Wang, Y., Xu, C., Zhang, R., Chen, Y., Shen, Y., Hu, F., et al. (2020). Changes in colistin resistance and mcr-1 abundance in Escherichia coli of animal and human origins following the ban of colistin-positive additives in China: an epidemiological comparative study. Lancet Infect. Dis. 20, 1161–1171. doi: 10.1016/S1473-3099(20)30149-3
Wang, L., Zhu, M., Yan, C., Zhang, Y., He, X., Wu, L., et al. (2023). Class 1 integrons and multiple mobile genetic elements in clinical isolates of the Klebsiella pneumoniae complex from a tertiary hospital in eastern China. Front. Microbiol. 14:985102. doi: 10.3389/fmicb.2023.985102
Xiang, R., Liu, B. H., Zhang, A. Y., Lei, C. W., Ye, X. L., Yang, Y. X., et al. (2018). Colocation of the polymyxin resistance gene mcr-1 and a variant of mcr-3 on a plasmid in an Escherichia coli isolate from a chicken farm. Antimicrob. Agents Chemother. 62, e00501–e00518. doi: 10.1128/AAC.00501-18
Xu, C., Lv, Z., Shen, Y., Liu, D., Fu, Y., Zhou, L., et al. (2020). Metagenomic insights into differences in environmental resistome profiles between integrated and monoculture aquaculture farms in China. Environ. Int. 144:106005. doi: 10.1016/j.envint.2020.106005
Xu, T., Zhang, C., Ji, Y., Song, J., Liu, Y., Guo, Y., et al. (2021). Identification of mcr-10 carried by self-transmissible plasmids and chromosome in Enterobacter roggenkampii strains isolated from hospital sewage water. Environ. Pollut. 268:115706. doi: 10.1016/j.envpol.2020.115706
Yang, C., Guo, M., Yang, H., Wen, Y., Zhu, Z., Wang, T., et al. (2022). blaKPC-24-harboring Aeromonas veronii from the hospital sewage samples in China. Microbiol. Spectr. 10:e0055522. doi: 10.1128/spectrum.00555-22
Yin, W., Li, H., Shen, Y., Liu, Z., Wang, S., Shen, Z., et al. (2017). Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio 8:e00543. doi: 10.1128/mBio.00543-17
Zheng, D., Yin, G., Liu, M., Chen, C., Jiang, Y., Hou, L., et al. (2021). A systematic review of antibiotics and antibiotic resistance genes in estuarine and coastal environments. Sci. Total Environ. 777:146009. doi: 10.1016/j.scitotenv.2021.146009
Zhou, Y., Zhou, Q. J., Qiao, Y., Chen, J., and Li, M. Y. (2020). The host defense peptide β-defensin confers protection against Vibrio anguillarum in ayu, Plecoglossus altivelis. Dev. Comp. Immunol. 103:103511. doi: 10.1016/j.dci.2019.103511
Keywords: Aeromonas, antibiotic resistance genes, mobile genetic elements, mcr-3, pathogenicity, coastal water
Citation: Chen H-X, Chen F-J, Zhou Q-J, Shang S-L, Tang B, Xu Z-J, Duan L-J, Jin J-L, Xu G-Z, Yan M-C and Chen J (2024) Two colistin resistance-producing Aeromonas strains, isolated from coastal waters in Zhejiang, China: characteristics, multi-drug resistance and pathogenicity. Front. Microbiol. 15:1401802. doi: 10.3389/fmicb.2024.1401802
Received: 16 March 2024; Accepted: 18 July 2024;
Published: 31 July 2024.
Edited by:
Fang He, Zhejiang Provincial People’s Hospital, ChinaReviewed by:
Fengxia Yang, Ministry of Agriculture and Rural Affairs, ChinaCopyright © 2024 Chen, Chen, Zhou, Shang, Tang, Xu, Duan, Jin, Xu, Yan and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qian-Jin Zhou, bXVtdTIzMjVAMTYzLmNvbQ==; emhvdXFpYW5qaW5AbmJ1LmVkdS5jbg==; Jiong Chen, amNoZW4xOTc1QDE2My5jb20=; Y2hlbmppb25nQG5idS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.