- Faculty of Forestry and Wood Sciences, Czech University of Life Sciences Prague, Prague, Czechia
Background: Climate change has recently boosted the severity and frequency of pine bark beetle attacks. The bacterial community associated with these beetles acts as “hidden players,” enhancing their ability to infest and thrive on defense-rich pine trees. There is limited understanding of the environmental acquisition of these hidden players and their life stage-specific association with different pine-feeding bark beetles. There is inadequate knowledge on novel bacterial introduction to pine trees after the beetle infestation. Hence, we conducted the first comparative bacterial metabarcoding study revealing the bacterial communities in the pine trees before and after beetle feeding and in different life stages of two dominant pine-feeding bark beetles, namely Ips sexdentatus and Ips acuminatus. We also evaluated the bacterial association between wild and lab-bred beetles to measure the deviation due to inhabiting a controlled environment.
Results: Significant differences in bacterial amplicon sequence variance (ASVs) abundance existed among different life stages within and between the pine beetles. However, Pseudomonas, Serratia, Pseudoxanthomonas, Taibaiella, and Acinetobacter served as core bacteria. Interestingly, I. sexdentatus larvae correspond to significantly higher bacterial diversity and community richness and evenness compared to other developmental stages, while I. acuminatus adults displayed higher bacterial richness with no significant variation in the diversity and evenness between the life stages. Both wild and lab-bred I. sexdentatus beetles showed a prevalence of the bacterial family Pseudomonadaceae. In addition, wild I. sexdentatus showed dominance of Yersiniaceae, whereas Erwiniaceae was abundant in lab-bred beetles. Alternatively, Acidobacteriaceae, Corynebacteriaceae, and Microbacteriaceae were highly abundant bacterial families in lab-bred, whereas Chitinophagaceae and Microbacteriaceae were highly abundant in wild I. accuminatus. We validated the relative abundances of selected bacterial taxa estimated by metagenomic sequencing with quantitative PCR.
Conclusion: Our study sheds new insights into bacterial associations in pine beetles under the influence of various drivers such as environment, host, and life stages. We documented that lab-breeding considerably influences beetle bacterial community assembly. Furthermore, beetle feeding alters bacteriome at the microhabitat level. Nevertheless, our study revisited pine-feeding bark beetle symbiosis under the influence of different drivers and revealed intriguing insight into bacterial community assembly, facilitating future functional studies.
Introduction
Bark beetles (Coleoptera: Curculionidae: Scolytinae) are economically important forest pests that cause large-scale forest damage across Europe (Biedermann et al., 2019; Singh et al., 2024). The outbreaks of pine bark beetles such as Ips sexdentatus (Börner, 1776) and Ips acuminatus (Gyllenhal, 1827) heavily impact forestry economics, affecting the forest-dependent sector and international wood markets (Montagné-Huck and Brunette, 2018). These outbreaks reduce the forest tree lifespan, decrease carbon uptake, and alter microclimatic situations in the forest, along with recreational and aesthetic values (Abdullah et al., 2018; Dobor et al., 2018). According to recent reports, particularly in the Czech Republic, the severity of pinewood damage by bark beetles increased by over 80,000 m3 in 2019 from about 10,000 m3 in 2009 (Liška et al., 2021). Moreover, climate changes such as increased air temperature, altered precipitation patterns, and increased frequency of drought and heat events have critically stimulated the outbreaks of bark beetles by altering the tree defence physiology (Raffa et al., 2008; Marini et al., 2017). Precisely, drought affects host tree fitness and can stimulate bark-beetle populations to overcome the epidemic threshold and cause an outbreak (McNichol et al., 2022). For instance, high spring temperature stimulates I. acuminatus to infest weakened and vigorous trees (Chinellato et al., 2014). Subsequently, the increasing infestations by I. acuminatus have listed the species among the most-aggressive bark beetle species within Europe (Faccoli et al., 2012; Colombari et al., 2013; Plewa and Mokrzycki, 2017; Liška et al., 2021). Climate change has also stimulated the importance of I. sexdentatus as a forest pest with considerable dispersal competency (Pineau et al., 2017b).
Although I. sexdentatus and I. acuminatus share several ecological characteristics, including host, I. sexdentatus infests the lower part of the bole, whereas I. acuminatus attacks the higher bark canopy (top branches) (Davydenko et al., 2017). Nonetheless, both Ips species must overcome the robust pine defence system to thrive. The robust pine defence system has physical and chemical barriers against biotic stress, including bark beetles (Mumm and Hilker, 2006). Physical barriers include tissues having lignin and suberin polymers that offer protection against degradation, penetration and ingestion, while the chemical barriers involve phenolic compounds and terpenoids (monoterpenes, diterpenes, and sesquiterpenes) with entomotoxic competency. A high concentration of monoterpenes shows ovicidal, repellent, adulticidal, and larvicidal effects against bark beetles (Krokene, 2015). Therefore, the defence barrier in healthy pine trees is a formidable challenge for bark beetles. Alternatively, gut microbial assemblage in bark beetles might promote the competency of bark beetles to exhaust conifer defence (Chakraborty et al., 2020a; Chakraborty et al., 2020b). Bacterial genera such as Pseudomonas, Serratia, and Rahnella associated with Dendroctonus valens (LeConte, 1860) might play an important role in metabolising monoterpenes (Boone et al., 2013; Xu et al., 2016). Similarly, Erwinia typography isolated from Ips typographus (Linnaeus, 1758) is suggested to tolerate a high concentration of myrcene-a plant defensive monoterpene (Skrodenytė-Arbačiauskienė et al., 2012). Furthermore, mountain pine beetle (Dendroctonus ponderosae Hopkins) has been reported to have bacterial mutualists containing genes (i.e., dit genes-diterpene degrading genes) that could be associated with terpene degradation (Adams et al., 2013).
Apart from overcoming the tree defence, a significant challenge for bark beetles is nutrient acquisition, as they primarily thrive on phloem tissues with limited nutrition and complex carbohydrates. Subsequently, the gut microbiome comes to the rescue, helping the beetles to digest and metabolise complex polysaccharides (i.e., lignin from conifers) (Hu et al., 2014) and nutrient acquirement (Morales-Jiménez et al., 2009; Morales-Jiménez et al., 2013). Beetle-associated microbiome extends their symbiosis with the host by producing pheromones, thus facilitating chemical communications (Xu et al., 2015). Furthermore, beetle-bacterial mutualism also plays an important role in defence against pathogens (González-Dominici et al., 2021). However, such microbial contribution to bark beetle survival is limited to only a few beetle genera, such as the red turpentine and mountain pine beetle (Adams et al., 2013; Cheng et al., 2018). Nevertheless, our understanding of bark beetles as holobionts is restricted. Few studies have evaluated microbial acquisition and succession in bark beetles (Ibarra-Juarez et al., 2020; Veselská et al., 2023; Baños-Quintana et al., 2024). In addition, limited studies in pine bark beetles have focused on understanding the effect of environment and metamorphosis that can cause comprehensive changes in the beetle microbiome, resulting in distinct bacterial communities. Moreover, microbial community assembly at the level of bark beetle microhabitat is still lacking. There is limited information on the influence of the host plant microbiome in shaping the beetle bacterial community, particularly in terms of Ips bark beetles (Kolasa et al., 2019; Silver et al., 2021; Chakraborty et al., 2023; Pirttilä et al., 2023; Baños-Quintana et al., 2024). No literature highlights the impact of beetle feeding on the pine-associated microbiome. Subsequently, it is also crucial to assess the microbial assembly in wild and lab-bred beetles to evaluate the influence of the bark beetle breeding facility.
We hypothesised that metamorphosis, microhabitat, and lab-breeding might shape the bacterial community structure in the Ips pine beetles similar to other insects (Chen et al., 2018; González-Serrano et al., 2020; Tian et al., 2022; Chakraborty et al., 2023; Veselská et al., 2023; Baños-Quintana et al., 2024). Hence, we conducted the first comparative metabarcoding study on two pine-feeding beetles (I. sexdentatus, I. acuminatus) to evaluate our hypothesis and respond to the following fundamental questions regarding bark beetle symbiosis: (1) What are the bacterial communities associated with the two pine-feeding Ips beetles across life-stages? (2) What influence does the host microbiome have in shaping the bacterial community assemblage or vice versa? (3) How does lab-breeding impact pine beetle bacterial community assemblage?
Materials and methods
Bark beetle collection and breeding for life-stage specific sample collection
Adult bark beetles (Coleoptera: Curculionidae: Scolytinae) were collected from infested logs (dbh ~20 cm) obtained from different beetle colonies in the Rouchovany forest area (49.0704° N, 16.1076° E) in the Czech Republic in 2020. The two bark beetle species (I.sexdentatus and I.acuminatus) were identified based on the work of Nunberg (Nunberg, 1981) and Pfeffer (Pfeffer, 1955, 1995). The infested logs were brought to the debarking room directly from the forest, where the adult beetles were collected using surface disinfected tweezers and bred to F2 generation in laboratory conditions (temp around 25°C in the day and 19°C at night, RH 60%) on fresh pine logs collected from the same forest area. Over 50 samples from each life stage (larva, pupa, adult) were collected from multiple infested logs in 50 mL plastic conical tubes, snap-frozen with liquid nitrogen, and kept at −80°C for DNA extraction. The gallery wood was collected from the same F2 generation infested logs using surface sterilised blades and kept in plastic tubes with RNAlater solution at −80°C for future use. All the beetle and wood samples were collected at least 10 cm below the edges of the logs to minimise contamination (Table 1). Uninfected, fresh woods with no beetle infestation were taken as control wood (unfed fresh phloem) and were kept in plastic conical tubes with RNAlater solution at −80°C.
Wild beetle collection
Similarly, the infested logs, collected from the same Rouchovany forest area in 2021, were directly brought to the debarking room, where adult I. sexdentatus and I. acuminatus wild beetles were collected using surface sterilised tweezers in 50 mL plastic conical tubes, snap-frozen with liquid nitrogen and kept at −80°C for DNA extraction. The gallery wood samples were collected from the same infested logs in RNAlater solution and stored at −80°C. Similarly, unfed wood samples (fresh phloem tissue) were collected from logs from the same locality without any beetle infestation. Beetle bacteriome variability of individual colonies was not explored in the current study due to the mixing of beetle samples from different colonies. However, such a sampling method can reduce the random variability from the heterologous sampling material. The sample details are provided in Table 1.
DNA extraction
Developmental stage-specific lab-bred and wild-collected samples were randomly selected and disinfected by rinsing the beetles twice with 70% ethanol and subsequently washing them with sterile water. Any sample with apparent infection was discarded. Due to size differences, the sampling amount per replicate for both species differed. One individual per replicate was used for I. sexdentatus, whereas, for I. acuminatus, four larvae/replicate, two pupae/replicate, and five adults/replicate were used. Total DNA from whole beetles was extracted using the MACHEREY-NAGEL NucleoSpin Soil DNA kit with modifications in the manufacturer’s protocol. Similarly, wood microbial DNA was extracted (~120 mg wood/replicate) using QIAGEN DNeasy Plant Mini Kit following the manufacturer’s protocol with modifications. The samples were homogenised under liquid nitrogen, and the lysis was performed for 1 min. The extracted DNA quantity and quality were accessed using a Qubit 2.0 Fluorometer (Thermo Scientific) and 1% agarose gel electrophoresis. High-quality samples (five biological replicates from each life stage for both species and 4 biological replicates for each wood sample) and two negative extraction controls were selected for 16S rRNA gene metabarcoding at Novogene Company, China.
16S amplicon sequencing
16S amplicon sequencing was executed at Novogene, China, using a pre-optimised protocol. Precisely, 1 ng/μl template DNA, bacterial 16S rRNA gene primers (341F-806R) (Klindworth et al., 2013) containing unique barcodes, and Phusion High-Fidelity PCR Master Mix (New England Biolabs) were used to set up PCR reactions. Subsequently, a PCR reaction mixture without template DNA was used as a negative control. PCR products were visualised in 2% agarose gel electrophoresis. The samples with amplification between 450 and 480 bp were selected and mixed in equidensity ratios for gel purification using a QIAGEN Gel extraction kit. NEBNext Ultra DNA Library Pre-Kit for Illumina was used to create sequencing libraries followed by index code ligation. Sequencing libraries were quantitively and qualitatively analysed using Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and Agilent Bioanalyser 2,100 system. An Illumina Novaseq 6,000 platform obtained 250 bp paired-end reads from the sequenced libraries.
Bacteriome data analysis
Data processing and species annotation
The bioinformatic data analysis was performed using a standardised pipeline in QIIME2 (version 2022.2) (Bolyen et al., 2019) as described in our earlier studies (Gupta et al., 2023). The barcodes and primer sequences were removed, and the sample sequences were merged using FLASH (V1.2.11)1 to generate raw Illumina pair-end reads (Magoč and Salzberg, 2011) and then checked for high-quality reads using fastp software. VSEARCH software (Rognes et al., 2016) was used to identify and remove chimeric sequences. The chloroplast and mitochondrial sequences were discarded to facilitate downstream bioinformatic analyses. The amplicon sequence variant (ASV) (Li et al., 2020) abundance table was obtained using the DADA2 module (Callahan et al., 2016). The ASV abundance table or the feature table is a matrix of samples, and the feature (ASV) abundance represents the number of times each feature/ASV was observed in each sample. Sequences that have an abundance lesser than five were discarded. Furthermore, classify-sklearn algorithm, a pre-trained Naive Bayes classifier was used for species annotation with the bacterial SILVA database (version 138.1) (Quast et al., 2012; Bokulich et al., 2018) to obtain individual ASVs with respective species annotation in QIIME2 (version 2022.2) (Bolyen et al., 2019). The core consortium of the bacterial communities in the samples was defined as ASVs present in ≥60% of each beetle sample group to avoid the occurrence of any transient species.
Alpha diversity
Alpha diversity indices like bacterial community richness (Chao1), evenness (Pielou) (Magurran, 1988) and bacterial diversity (Shannon) (Magurran, 1988) were used to analyse bacterial community structure. Kruskal-Wallis-pairwise-group test was performed to test the significance level within the samples. In addition, QIIME2 (version 2022.2) was used to estimate Good’s coverage (sequence depth) (Chao et al., 1988) and observed species from all stage-specific and wood samples that were represented by R software (Version 2.15.3; R Core Team, 2013, Vienna, Austria) (R Core Team, 2013).
Beta diversity
The bacterial diversity variation between different life stages and wood samples for both Ips species was estimated using the UniFrac distance metric (Lozupone et al., 2011) determined in QIIME2 (version 2022.2). Moreover, unweighted UniFrac distance measurement was used for non-metric multi-dimensional scaling (NMDS) analysis illustrated by R software (Oksanen, 2007). The functions ADONIS (analysis and partitioning sum of squares using dissimilarities) and ANOSIM (analysis of similarities) (Clarke, 1993; Anderson, 2001) were used to determine significant differences in life-stage specific and wood bacteriome in QIIME2 (version 2022.2). ADONIS is a non-parametric multivariate variance test analysis that utilises a distance metric (Lozupone et al., 2011) to determine significant differences in the bacterial community among the sample groups (Stat et al., 2013). However, ANOSIM analysis utilises the same distance metric to estimate if the variation among different sample groups is larger than within the sample group (Chapman and Underwood, 1999). Additionally, a permutational multivariate analysis of variance (PERMANOVA) (Anderson, 2001) using Bray Curtis distance was conducted to assess the significance of overall bacterial diversity across different life stages and to evaluate the impact of lab-breeding on the two pine beetles. Furthermore, a t-test was used to determine significantly abundant bacterial species (p < 0.05) in the sample groups (D’Argenio et al., 2014). Similarly, Metastats analysis using false discovery rate (FDR) and multiple hypothesis tests for sparsely-sampled features revealed that the intra-group differed significantly on abundant bacterial species (Paulson et al., 2011). LEfSe (linear discriminant analysis effect size) analysis was performed to obtain significant biomarkers that can help to distinguish two samples in an experimental condition (Segata et al., 2011). These biologically consistent, statistically significant biomarkers derived from LEfSe can disclose metagenomic attributes (taxa/metabolites/genes) to distinguish between the two samples.
Quantitative PCR assay
The relative abundance of selected bacterial taxa was estimated using quantitative PCR assay and correlated with the metagenomic sequencing results. Six individuals for each life stage (larvae/pupae/adults) were pooled per replicate, and four biological replicates were prepared. However, due to the limited availability of I. acuminatus samples, only four replicates of adult beetles were prepared. For the same reason, life stage comparisons for I. acuminatus were not performed using qPCR. Tubulin beta-1 chain (β-Tubulin) was used as reference genes for I. sexdentatus (Sellamuthu et al., 2021), while ribosomal protein (RPL7) and the elongation factor (EF1a) genes were selected for I. accuminatus (unpublished data). Six primer pairs representing different bacterial taxa and one eubacterial primer revealing the total bacterial population were used for qPCR assay (Supplementary Table 5). The specific bacterial primers were selected from previously published studies, while Psedoxanthomonas and Serratia genus-specific primers were designed in-house based on 16S rRNA gene sequences available in NCBI. The in-house designed primers were validated by sequencing the amplified product of the genus-specific primers and confirming it by NCBI blast. In addition, we also checked the primer with other non-specific bacterial cultures. The qPCR was performed with 10 μL of reaction mixture containing 4 μL of gDNA (10 ng/μl), 5 μL SYBR® Green PCR Master Mix (Applied Biosystems), 0.5 μL forward and reverse primer (10 μM). Amplification conditions included initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s and 60°C for 30 s. The relative quantification (RQ) of the selected bacterial population was estimated using the delta–delta Ct method (2−ΔΔCt), where ΔCt was estimated as the difference between the threshold Ct values with specific bacterial primers and the housekeeping reference gene. In this study, the 2−ΔΔCt method reveals the fold change of the bacterial abundance relative to the housekeeping genes. It is worth mentioning here that the relative abundance of a specific bacterial population compared to the total bacterial population often lacks consistency (Navidshad et al., 2012). Hence, the housekeeping genes with stable gene expression were considered to normalise the data and determine the relative bacterial abundance. In the qPCR analysis, the significant difference in relative bacterial abundance between different life stages of I. sexdentatus and I. acuminatus was estimated using the method described by Pekár and Brabec (2016). The best linear model was elected under Akaike’s information criterion (AIC), and goodness of fit and heterogeneity were ensured by plotting residuals of the model against fitted values. Then, the variable was tested using ANOVA, and the difference between the categorical variable levels was compared by treatment contrasts t-test (Crawley, 2012). All analyses were performed in the R 4.3.1 environment (R Core Team, 2013).
Results
Bacteriome structure
Sequencing statistics
The sequencing data from two pine bark beetle species, I. sexdentatus (ISX) and I. acuminatus (IAC), of different populations (lab-bred, wild collection) and life stages (larva, pupa, adult) along with host tissue (control wood, fed/gallery wood) generated 6,393,911 raw reads. A Phred Quality score > 30 was used for quality control. Therefore, a total of 5,812,983 clean reads were obtained (Control wood-374,493; Fed wood-374,084; I. sexdentatus larvae-463,927; I. sexdentatus pupae-440,213; I. sexdentatus adult-446,527; I. acuminatus larvae-463,230; I. acuminatus pupae-446,017; I. acuminatus adult-372,076; I. sexdentatus wild adult-475,613; I. sexdentatus Control wood-364,782; I. sexdentatus Fed wood-381,812; I. acuminatus wild adult-447,180; I. acuminatus Control wood - 387, 312; I. acuminatus Fed wood-375,717) (Supplementary Excel 1).
Bacterial relative abundance at distinct taxonomic levels
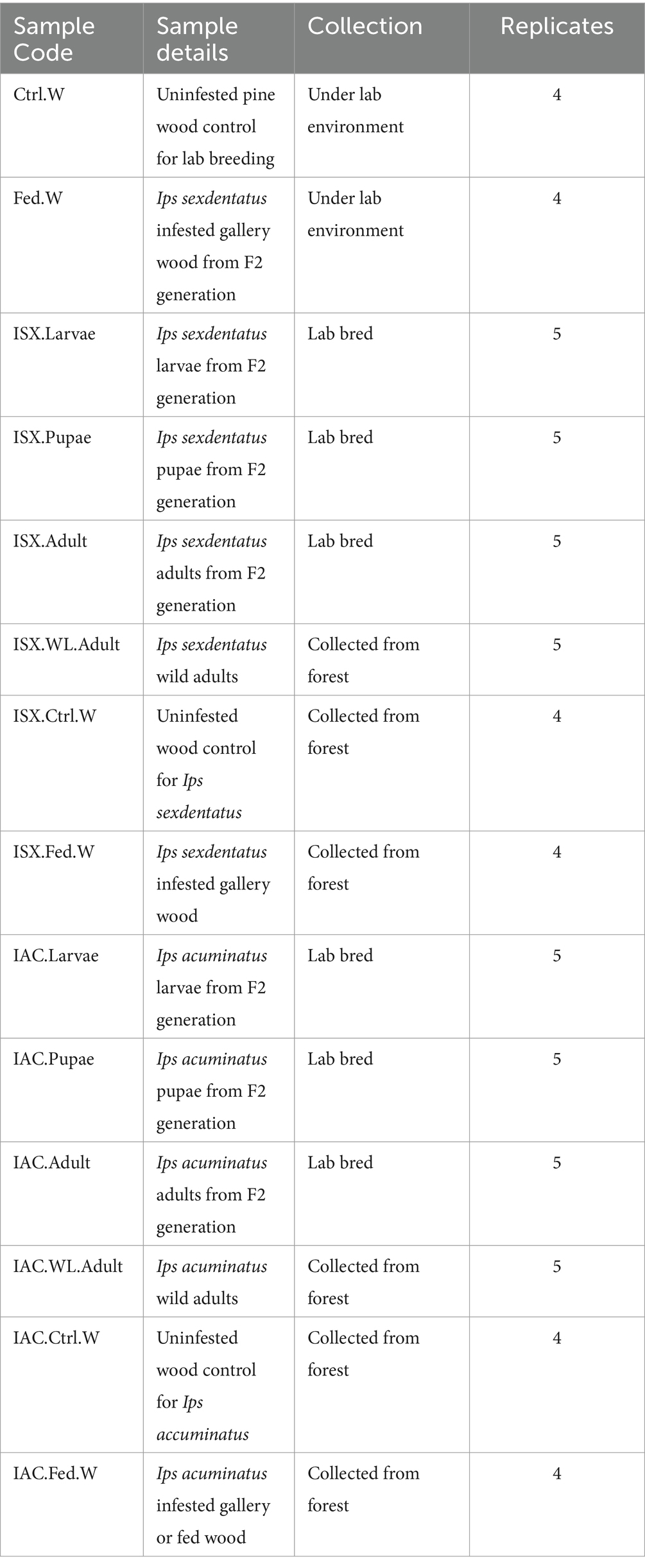
The bacterial sequences obtained from the two beetle species and wood samples generate 4,056 ASVs at a 100% similarity level (Supplementary Excel 2). The Good’s coverage indicator (>98%) and the rarefaction curve indicated sampling comprehensiveness that represented the bacterial communities associated with the beetle and wood samples (Table 2; Supplementary Figure 1). The estimation of the Goods coverage index (0.98) after filtering of sequence read (n > 5) suggests that the majority of the ASVs present in the samples were detected, and approximately only 2% of the ASVs were not covered during the sequencing. The predominant bacterial classes across all samples were Gammaproteobacteria, Alphaproteobacteria, Actinobacteria, Bacteroidia, Bacilli (Figure 1A). The relative abundance of Gammaproteobacteria was higher in I. sexdentatus pupae (0.94 ± 0.03) and adult (0.99 ± 0.002) samples compared to larvae (0.57 ± 0.15) and wild-collected adults (0.63 ± 0.20) samples (Supplementary Excel 2). Similarly, the relative abundance of Gammaproteobacteria was highest in I. acuminatus larvae (0.95 ± 0.004) compared to other stages (larvae-0.84 ± 0.06, pupae-0.79 ± 0.06) and wild-collected adults (0.88 ± 0.02). Furthermore, Gammaproteobacteria [I. sexdentatus Control wood (0.73 ± 0.03), I. sexdentatus Fed wood (0.80 ± 0.01), and I. acuminatus Control wood (0.36 ± 0.02) I. acuminatus Fed wood (0.72 ± 0.05)] was also the dominant class in different phloem wood samples (Supplementary Excel 2). The relative abundance of Alphaproteobacteria was documented as highest in I. sexdentatus wild-collected adults (0.34 ± 0.19) compared to all other samples (Supplementary Excel 2).
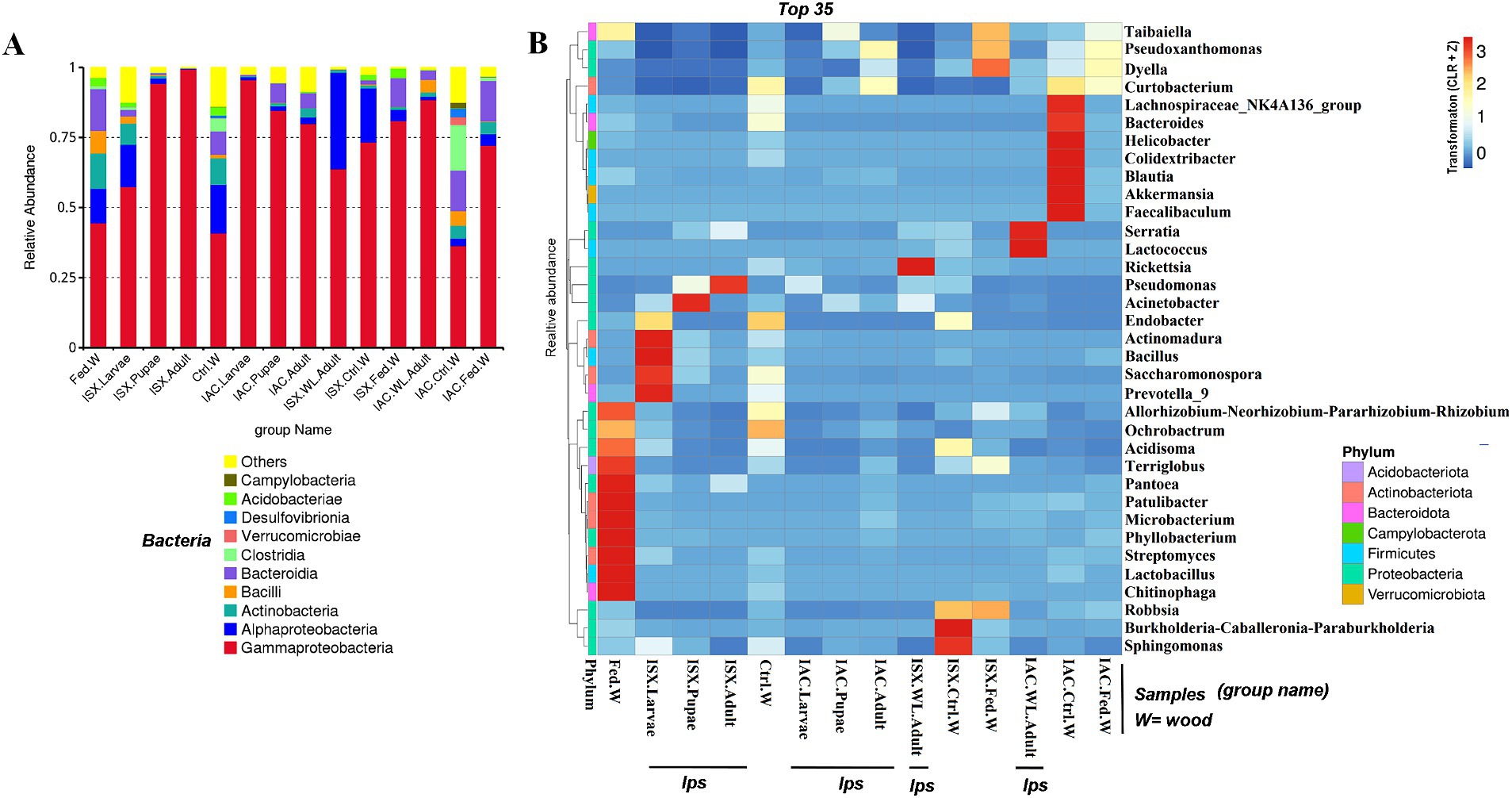
Figure 1. Bacterial diversity in lab-bred (life-stages), wild-type adult beetles and wood samples. (A) The bar plot represents the relative abundance of bacteriome at the class level (top 10). (B) Heatmap depicting the relative abundance of 35 dominant bacterial genera among lab-bred (life-stages), wild-type adult beetles and wood samples. The relative ASV abundance is represented by a colour gradient where the darker colour indicates higher abundance, whereas the lighter colour indicates low abundance for a specific bacterial genus (ISX-Ips sexdentatus, IAC-Ips acuminatus).
Life stage-specific bacteriome
Bacterial associations in I. sexdentatus (ISX)
Our study revealed that the ASV distribution of the different life stages of both pine beetles comprises a pool of diverse bacterial populations. I. sexdentatus larvae, pupae, and adult stages contained 16, 5, and 3 unique ASVs, respectively (Figure 2A). Moreover, nine common ASVs were present in all life stages of I. sexdentatus, constituting the core bacteriome. However, it is essential to mention that each ASV may not represent an individual species. The I. sexdentatus core consortium consisted of bacterial genera, including Pseudomonas, Pseudoxanthomonas, Sphingomonas, Acinetobacter, and members belonging to the bacterial family Erwiniaceae (Supplementary Excel 3). The heatmap revealed the high abundance of Pseudomonas in adults, whereas Acinetobacter, Saccharomonospora, and Rickettsia were dominant in pupae, larvae, and wild adults, respectively (Figure 1B). The alpha diversity analysis showed that I. sexdentatus adult beetles had substantially lower bacterial richness (Chao1 63.32 ± 12.32) than the other developmental stages (Chao1, larvae-396.9 ± 101.78, p < 0.01 and pupae-181.37 ± 56.39, p < 0.01) (Table 2; Supplementary Figure 2). However, I. sexdentatus larvae represented significantly higher bacterial diversity (Shannon-4.99 ± 1.32) compared to the adult (Shannon-2.34 ± 0.38, p < 0.01) (Table 2; Supplementary Figure 2). Similarly, the bacterial community evenness was significantly higher in I. sexdentatus larvae (Pielou-0.61 ± 0.14) compared to adults (Pielou-0.43 ± 0.07) (p < 0.05) (Table 2; Supplementary Figure 2). The overall bacterial diversity showed significant differences while comparing different developmental stages of I. sexdentatus (Permanova analysis, Pseudo F statistics = 5.399, p = 0.00003). Subsequently, Metastat analysis revealed the differential abundance of top bacterial genera between different developmental stages (Table 3). Among the bacterial genera present, Pseudomonas was the most dominant bacterial genus in all the three life stages of I. sexdentatus beetles, with differences in their relative abundance. LEfSe represented the key bacterial biomarkers in I. sexdentatus life stage-specific bacterial populations (Figure 2D; Supplementary Figure 3A; Supplementary Table 3). I. sexdentatus pupae documented the bacterial families, including Enterobacteriaceae and Moraxellaceae, as biomarkers (Figure 2D). In contrast, Yersiniaceae and Pseudomonadaceae were the biomarkers of I. sexdentatus adults. Similarly, I. sexdentatus larvae represented the class Alphaproteobacteria as a distinct biomarker (Supplementary Table 3).
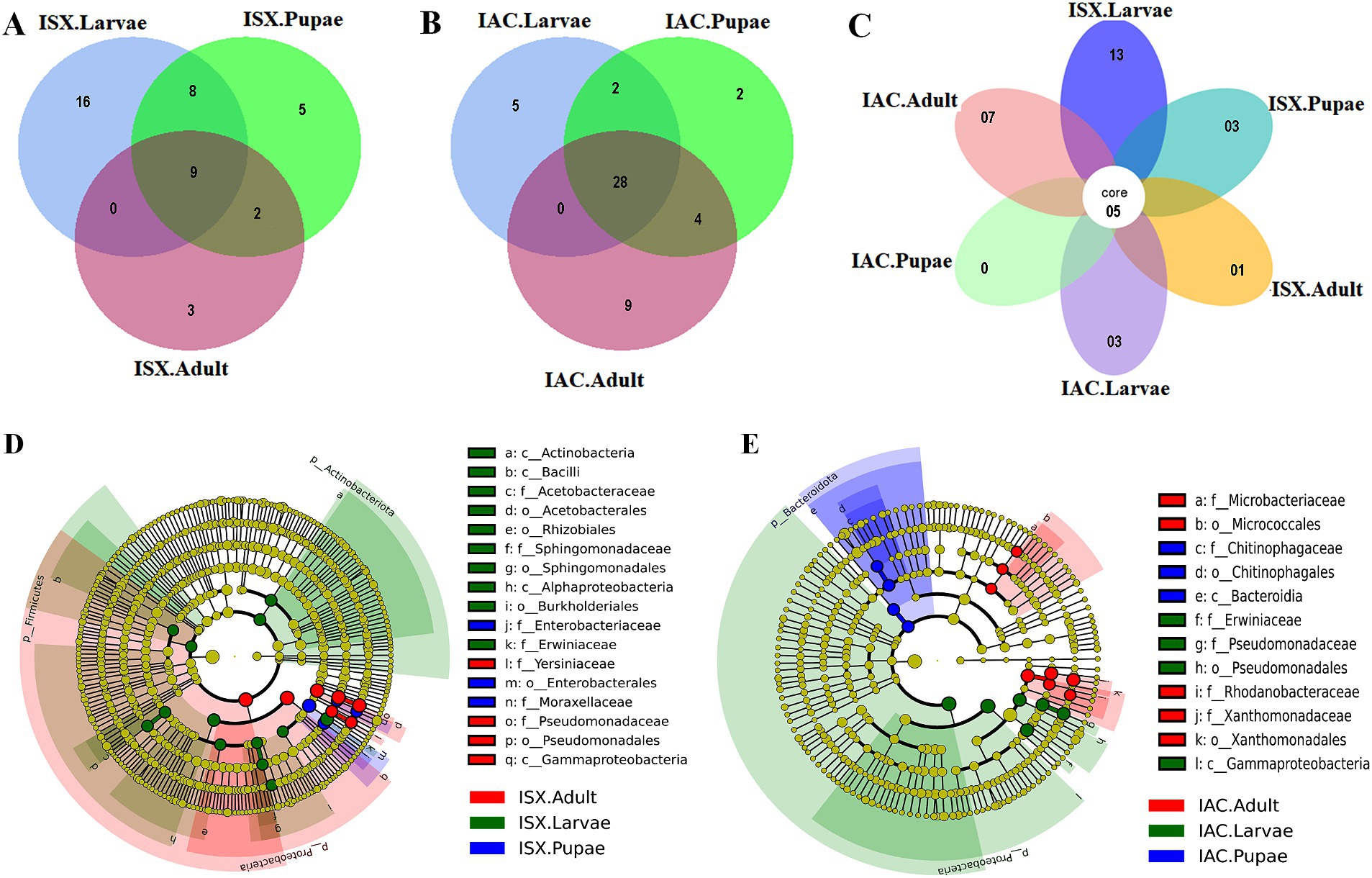
Figure 2. Core bacteriome. (A) Venn diagram illustrating bacterial ASV distribution in I. sexdentatus life stages (ISX.Larvae, ISX. Pupae, ISX.Adult). (B) Venn diagram depicting bacterial ASV distribution in I. acuminatus life stages (IAC.Larvae, IAC.Pupae, IAC.Adult). The shaded regions indicate the common areas between the sample groups. (C) Flower diagram representing the core ASVs across the life stages of two pine beetles. (D) Cladogram illustrating the results from LEfSe analysis revealing the biologically consistent, statistically significant bacterial biomarkers across different life stages of I. sexdentatus (ISX). (E) Cladogram representing significantly distinct bacterial biomarkers across I. acuminatus (IAC) life stages. Distinct taxonomic level (phylum to genus) is denoted in the circle from inward to outward. The different coloured nodes (red, green, and blue) represent bacterial species that play a significant role in different life stages across two different beetles (ISX and IAC), whereas yellowish-green circles represent non-significant bacterial species. Specific bacterial biomarkers are denoted by letters above the circles. The size of the nodes represents the relative abundance of the bacterial species at a particular taxon (ISX-Ips sexdentatus, IAC-Ips acuminatus).

Table 3. Metastat analysis representing the top 10 differently abundant bacterial genera across different life stages of Ips pine beetles.
Bacterial associations in I. acuminatus (IAC)
The core bacteriome in I. acuminatus comprised 28 ASVs that were categorised into 10 families including Erwiniaceae, Pseudomonadaceae, Xanthomonadaceae, and Rhodanobacteraceae being the most dominant bacterial families (Figure 2B; Supplementary Excel 4). Comparing the developmental stages, Pseudomonas showed a high abundance in I. acuminatus larvae, while Taibaiella, Sphingomonas, and Curtobacterium were dominant in the pupal stage (Figure 1B). Furthermore, the alpha diversity indices revealed higher bacterial richness in I. acuminatus adults (Chao1, 251.42 ± 51.88) compared to larvae (Chao1, 117.92 ± 13.59) (p < 0.01) (Table 2; Supplementary Figure 2). However, no stage-specific differences in bacterial diversity and evenness were observed in I. acuminatus beetles (Table 2). Similar to I. sexdentatus, significant differences in the bacterial diversity were observed between I. acuminatus life stages (Permanova analysis Pseudo F statistics = 5.789, p = 0.00008). Metastat analysis revealed a significantly high abundance of Pseudomonas in I. acuminatus larvae, while Arachidicoccus, Dyella and Burkholderia-Caballeronia-Paraburkholderia were prevalent in adults and pupae (Table 3). Furthermore, LEfSe analysis corresponds to the bacterial biomarkers in I. acuminatus life stages (Figure 2E; Supplementary Figure 3B; Supplementary Table 3). I. acuminatus pupal biomarkers were categorised into class Bacteroidia (Figure 2E; Supplementary Table 3), while members from the phyla Proteobacteria (class Gammaproteobacteria, family Erwiniaceae, Pseudomonadaceae) were represented as the biomarkers in I. acuminatus larvae. Similarly, I. acuminatus adults documented family-Rhodanobactericeae, Xanthomonadeceae and Microbactericeae as predominant biomarkers (Supplementary Table 3).
Comparing bacteriome of two pine-feeding Ips beetles
Comparing the two beetle species, I. acuminatus and I. sexdentatus, 5 ASVs were shared across the developmental stages in both species (Figure 2C) that were assigned to 3 families including Xanthomonadaceae, Erwiniaceae, and Sphingomonadaceae (Supplementary Excel 5). Furthermore, the adult and larval stages of both species (I. sexdentatus and I. acuminatus) showed a significant difference (p < 0.01) in bacterial richness. For instance, in I. sexdentatus beetles, the larvae have higher bacterial richness, but in I. acuminatus, it was adult beetles. Although there was not much variation in bacterial diversity and evenness within the pupal and the larval stages between the two beetle species (I. acuminatus and I. sexdentatus), I. acuminatus adult showed significantly higher bacterial diversity (Shannon-3.73 ± 0.66) compared to I. sexdentatus adult (Shannon-2.34 ± 0.38, p < 0.05) (Table 2; Supplementary Figure 2). Additionally, NMDS using unweighted UniFrac distances revealed the differences between I. sexdentatus and I. acuminatus bacterial communities by hierarchically clustering different life stages and wood samples (Figure 3A). The larval and adult beetle-associated bacteria in I. acuminatus and I. sexdentatus had significant differences within and among them. Similarly, ADONIS and ANOSIM analysis revealed significant differences between the developmental stages of the two pine-feeding beetles (Supplementary Tables 1, 2).
Figure 3. Impact of lab breeding on bacterial association. (A) Non-metric Multi-Dimensional Scaling (NMDS) based on unweighted UniFrac distance matrix represents bacterial diversity variation in different life stages of two beetles along with the wood samples. (B) Cladogram representing significant bacterial biomarkers among I. sexdentatus, I. acuminatus wild type and lab-bred beetles (ISX-Ips sexdentatus, IAC-Ips acuminatus). Distinct taxonomic level (phylum to genus) is denoted in the circle from inward to outward. The different coloured nodes (red, green, blue and purple) represent bacterial species that play a significant role in wild-collected adults and lab-reared adults in two different beetles (ISX and IAC), whereas yellowish-green circles represent non-significant bacterial species. Specific bacterial biomarkers are denoted by letters above the circles. The size of the nodes represents the relative abundance of the bacterial species at a particular taxon (ISX-Ips sexdentatus, IAC-Ips acuminatus).
Impact of beetle lab breeding on bacterial assemblage
I. sexdentatus: wild vs. lab-bred
According to our study, the lab-bred adult population (F2 generation) and the wild beetle population have considerable differences in ASV composition. The I. sexdentatus wild and lab-bred adults had 7, 1 unique ASVs, and 22 shared ASVs (Supplementary Excel 6). The core bacterial consortium accounted for 6 families, including Xanthomonadaceae, Erwiniaceae, Yersiniaceae, and Pseudomonadaceae as the dominant families (Supplementary Figure 5; Supplementary Excel 6), while Pseudomonas was the highly abundant genera. Alpha diversity comparisons revealed that in Ips sexdentatus samples, the lab-bred adult beetles (Chao1-63.32 ± 12.32) had lower bacterial richness than wild adults (Chao1-138.87 ± 12.60, p < 0.05), while no significant variation was observed in the bacterial diversity and community evenness (Table 2; Supplementary Figure 2). However, lab-breeding showed a significant influence on the bacterial beta diversity in I. sexdentatus adults (Permanova analysis, Pseudo F statistics = 4.816, p = 0.00794) Metastat analysis revealed significant differences in bacterial abundance between the lab-bred and wild-collected beetles (Table 3). Additionally, the lab-bred and wild-collected beetles possess distinct bacterial markers (Figure 3B; Supplementary Figure 3C). For instance, LEfSe analysis revealed the members of the class Gammaproteobacteria, family-Pseudomonadaceae as the biomarkers of lab-bred adults, while class Alphaproteobacteria, family-Erwiniaceae, and Rickettsiaceae was represented as the biomarker of wild-collected adults (Figure 3B; Supplementary Table 4).
I. acuminatus: wild vs. lab-bred
The lab-bred and wild-collected I. acuminatus adult beetles comprised 30 common bacterial ASVs belonging to Xanthomonadaceae, Erwiniaceae, Pseudomonadaceae, Rhodanobacteraceae, Chitinophagaceae, and Microbacteriaceae (Supplementary Excel 6). The heatmap indicated that Curtobacterium, Dyella, and Pseudoxanthomonas dominated lab-bred adults, while Taibaiella, Lactococcus, and Serratia were prevalent in wild I. acuminatus beetles (Figure 1B). In contrast to I. sexdentatus, the alpha diversity analysis revealed that lab-bred I. acuminatus adult (Chao1 251.42 ± 51.88) had higher bacterial richness compared to wild adults (123.57 ± 19.99) (p < 0.05) (Table 2; Supplementary Figure 2). However, no significant differences in bacterial diversity and evenness were observed (Table 2; Supplementary Figure 2). Beta diversity analysis revealed significant differences between the bacterial communities in the lab-bred and wild-collected I. acuminatus adults (Permanova analysis, Pseudo F statistics = 6.389, p = 0.00794). LEfSe analysis represented biomarkers in I. acuminatus adult belonging to different phyla-Actinobacterioda (class-Actinobacteria; order-Micrococcales; family-Microbacteriaceae), Bacteroidota (class-Bacteroidia; order-Chitinophagales; family-Chitinophagaceae), Proteobacteria (family-Rhodanobactericeae, Xanthomonadeceae) (Figure 3B; Supplementary Figure 3C; Supplementary Table 4). I. acuminatus wild adults contained biomarkers belonging to two phyla-Proteobacteria (order-Enterobacterales; family-Yersiniaceae) and Firmicutes (class Bacilli; order-Lactobacillales; family-Streptococcaceae). Nevertheless, the wild adult beetles for both Ips species were clustered separately from lab-bred adults in the NMDS plot, indicating laboratory breeding impact on beetle bacteriome (Figure 3A). The ADONIS and ANOSIM results also demonstrate significant differences between the bacterial communities associated with wild and lab-bred beetles (Supplementary Tables 1, 2).
Host contribution in shaping beetle bacteriome
Comparing the I. sexdentatus wild beetles with their control and fed woods revealed a consortium of 20 shared ASVs (Figure 4A; Supplementary Excel 7). The shared bacteriome documented a high abundance of bacterial families Pseudomonadaceae, Yersiniaceae, and Erwiniaceae, while Pseudomonas is the dominant genus (Supplementary Excel 7). The unfed pine wood (I. sexdentatus Control wood) from the forest documented a high abundance of bacterial genera-Burkholderia_Caballeronia_Paraburkholderia whereas Pseudoxanthomonas, Robbsia were dominant in gallery wood of the wild I. sexdentatus (Figure 1B).
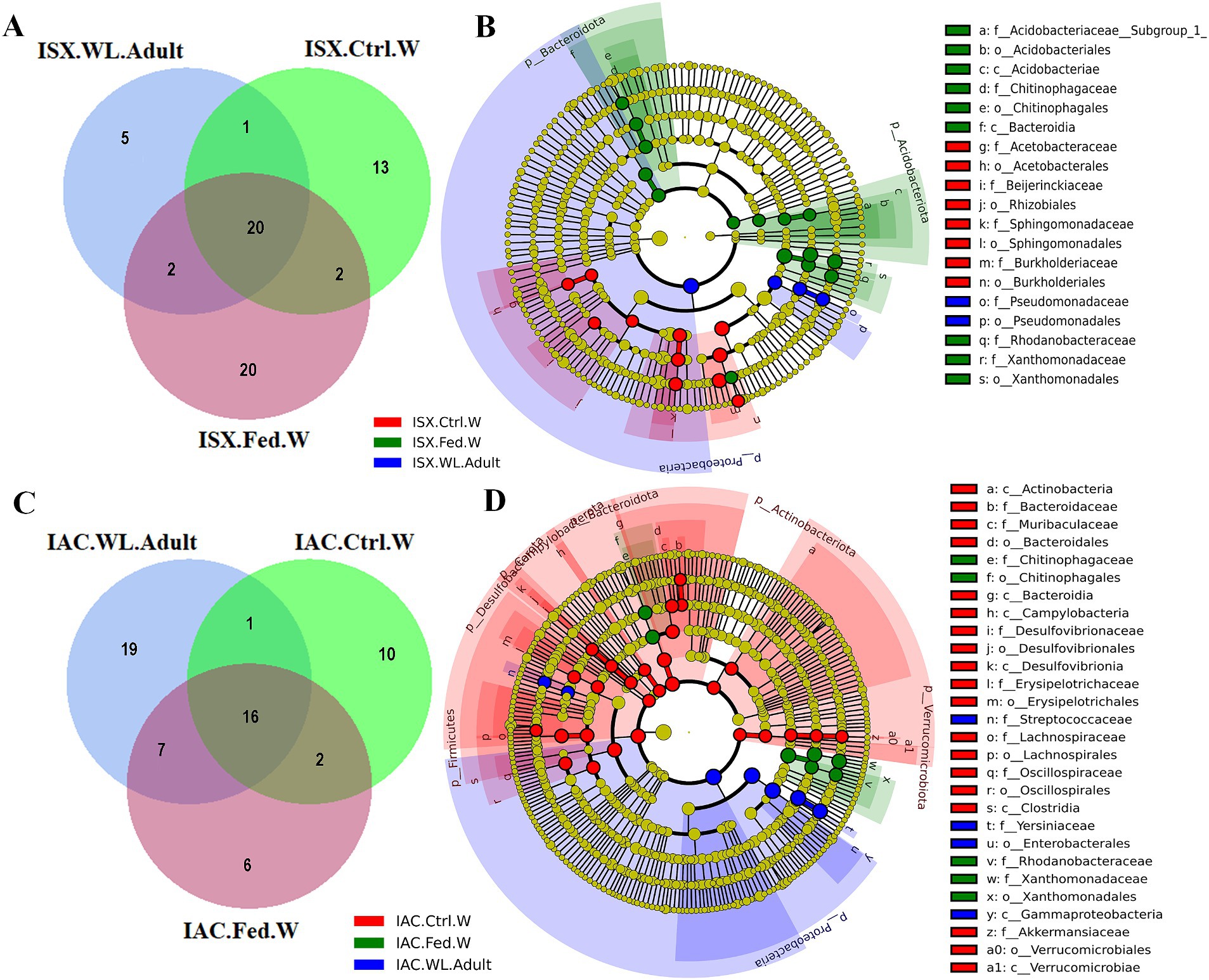
Figure 4. Host contribution in shaping beetle bacteriome. (A) Venn diagram showing the bacterial ASVs contribution of wood bacteriome in shaping I. sexdentatus bacteriome. (B) Cladogram representing significant bacterial biomarkers in I. sexdentatus wild-adult beetle and different wood types (control wood and fed wood). (C) Venn diagram showing the bacterial ASVs contribution of wood bacteriome in shaping I. acuminatus bacteriome. (D) Cladogram representing significant bacterial biomarkers among I. acuminatus wild adult beetle and different wood types (control wood and fed wood) (ISX-Ips sexdentatus, IAC-Ips acuminatus).
Similarly, considering the I. acuminatus wild beetles with their control and fed wood documented 16 shared ASVs belonging to bacterial families Xanthomonadaceae, Erwiniaceae, Rhodanobacteraceae, Chitinophagaceae, Burkholderiaceae, Acetobacteraceae (Figure 4C; Supplementary Excel 8). The heatmap revealed the prevalence of Pseudoxanthomonas, Taibaiella, Dyella in gallery wood of the wild I. acuminatus, which in turn had a lower abundance in unfed control pine wood (I. acuminatus Control wood), indicating an alteration in pine wood bacterial assemblage after beetle feeding (Figure 1B).
Several biomarkers were found while comparing the wild adult and wood bacterial communities. For instance, Proteobacteria was a significant biomarker with high abundance in the wild adult population (I. sexdentatus and I. acuminatus wild adult) (Figures 4B,D; Supplementary Figures 4A,B; Supplementary Table 3). Other biomarkers such as bacterial orders-Burkholderiales (family-Burkholderiaceae) and Pseudomonadales showed low abundance in I. sexdentatus fed wood, while an increase in the relative abundance of bacterial biomarkers belonging to bacterial family Chitinophagaceae, Rhodanobacteraceae, Xanthomonadaceae were observed compared to the control wood (Supplementary Table 3). Subsequently, biomarkers in I. sexdentatus wild adults were categorised into the family-Pseudomonadaceae, order-Pseudomonadales, and the biomarkers for I. acuminatus wild adults were classified into the family-Streptococcaceae, Yersiniaceae, order-Enterobacterales, class-Gammaproteobacteria. Concomitantly, in I. acuminatus control wood samples Bacterodiota, Dessulfobacteriota, Verrucomicrobiota were the significant biomarkers predominated by families like Bacterodiaceae, Desulfovibrionaceae, Akkermansiaceae. However, in the fed wood samples from two beetle species (I. sexdentatus and I. acuminatus fed wood), Proteobacteria, Acidobacteriota, and Bacterodiota were the predominant phyla. Similarly, there were differences at other taxa levels. LefSe analysis revealed that I. sexdentatus fed-wood biomarkers belonged to families such as Acidobacteriaceae _(Subgroup_1), Chitinophagaceae, Rhodanobacteraceae, and Xanthomonadaceae (Figure 4B; Supplementary Table 3). Similarly, Chitinophagaceae, Rhodanobacteraceae, and Xanthomonadaceae were the prevalent biomarkers in I. acuminatus fed wood (Figure 4D; Supplementary Table 3). Investigating the functional relevance of these biomarkers in different sample groups will be intriguing.
Alpha diversity analysis revealed higher bacterial diversity in control wood samples compared to fed wood. However, no significant difference was observed between wood samples (Table 2; Supplementary Figure 2). In contrast, the I. acuminatus fed/gallery wood samples collected from the forest demonstrated significantly different lower bacterial community evenness (I. acuminatus Fed wood-0.64 ± 0.03) than its respective control wood samples (I. acuminatus Control wood 0.84 ± 0.01) (p < 0.05) (Table 2; Supplementary Figure 2). Such findings indicate the enrichment of bacterial species in the feeding gallery after beetle feeding.
Consequently, the NMDS plot represented I. acuminatus wild adults, fed wood, and control wood into distinct clusters (Figure 3A). However, no such clustering was observed in the case of wild-collected I. sexdentatus samples. In contrast, comparing lab-bred and wild-collected wood samples (control wood and fed wood) for the I. sexdentatus revealed a distinct bacterial population, suggesting the influence of environment and beetle feeding as drivers in shaping the host microbiome (Figure 3A; Supplementary Excel 9; Supplementary Tables 1, 2). A schematic diagram containing the top five bacterial families across the developmental stages of two pine beetles and their respective wood and their putative role in the beetle holobiont is illustrated as a summary figure (Figure 5).
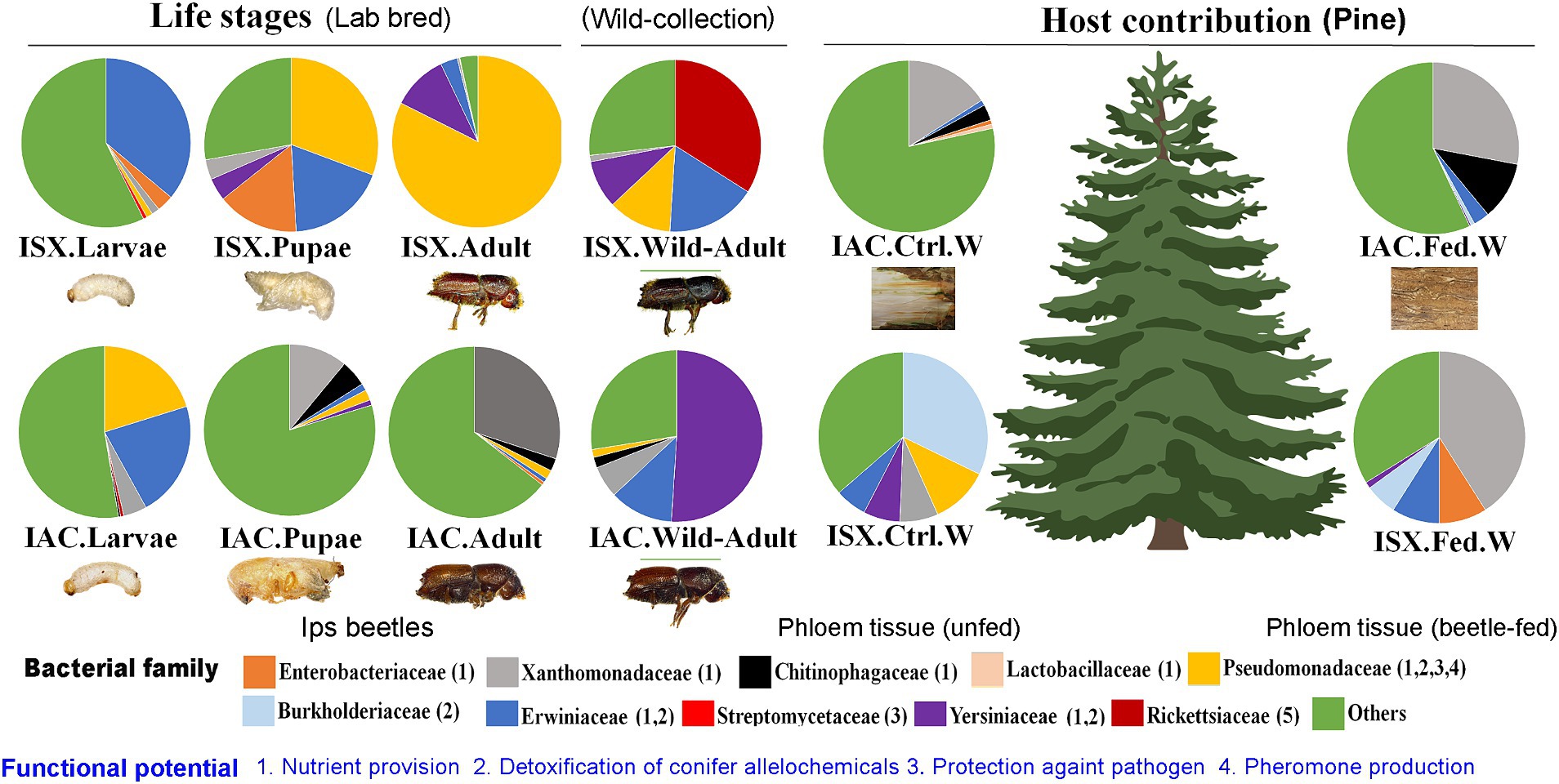
Figure 5. Schematic diagram containing the top five bacterial families at each developmental stage of two pine beetles (ISX-Ips sexdentatus, IAC-I. acuminatus) and their respective wood samples (control wood, gallery wood).
Relative bacterial abundance using quantitative PCR assay
I. sexdentatus life stages
The qPCR assay revealed a relatively high abundance of total bacterial population (eubacterial primers) in I. sexdentatus adults compared to all other life stages (Supplementary Figure 6A; Supplementary Excel 10). Precisely, there is a difference between the life stages of I. sexdentatus (ANOVA; df = 2, p < 0.01). However, the relative bacterial abundance between larval and pupal stages is marginally varied (contrast t-test; p = 0.099); adult bacterial assemblage was significantly different from larvae (contrast t-test; p < 0.01) but slightly diverse from pupae (contrast t-test; p = 0.054). The relative abundance of Bacteroidetes (phylum), Enterobacteriaceae (family), Pseudoxanthomonas, Pseudomonas, and Serratia (genus) varied within the life stages of I. sexdentatus (Figures 6A,B,D–F). No abundance difference was observed for Firmicutes (Figure 6C).
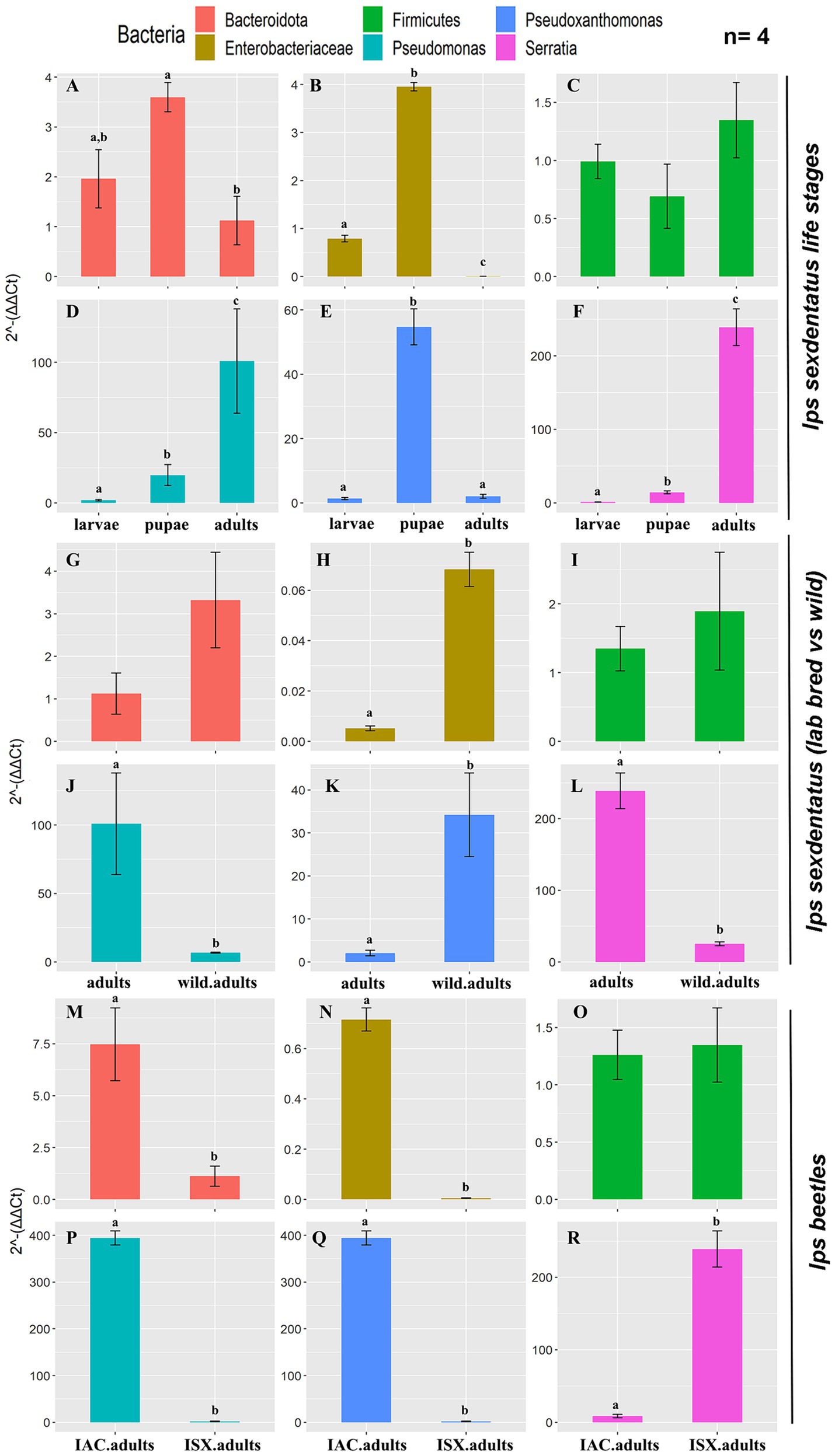
Figure 6. Quantitative PCR assay representing the relative abundance of selected bacterial taxa present in the two pine-feeding beetles. The 2−ΔΔCt revealed the fold change of the bacterial abundance relative to the stable reference genes (ISX-β-Tubulin; IAC-EF1a, RPL7). The data indicates the mean fold change of bacterial abundance within the beetles (n = 4). The data was tested using ANOVA, and the difference between the categorical variable levels was compared using the treatment contrasts t-test. Different letters indicate the statistical significance, p < 0.01. (A–F) I. sexdentatus life stage comparisons; (G–L) I. sexdentatus wild vs. lab-bred adults; (M–R) I. sexdentatus vs. I. accuminatus lab-bred adults. The statistical analysis results were presented in Supplementary Excel 10.
Wild adults vs. lab-bred adults
Similarly, the overall difference in bacterial abundance (eubacterial primers) between wild and breeding adult beetles was non-significant (ANOVA; df = 1, p = 0.076) (Supplementary Figure 6B; Supplementary Excel 10). However, the lab-bred I. sexdentatus adults showed a higher abundance of Pseudomonas and Serratia than wild adults (Figures 6J,L). Meanwhile, Enterobacteriaceae and Pseudoxanthomonas are more abundant in wild beetles (Figures 6H,K). No differences were observed in the relative abundance of Bacteroidetes and Firmicutes (Figures 6G,I).
I. sexdentatus vs. I. acuminatus
Comparing the two pine beetles, adults of I. acuminatus had significantly higher relative abundance than I. sexdentatus (ANOVA; df = 1, p < 0.001) (Supplementary Figure 6C; Supplementary Excel 10). The relative abundance of Bacteroidetes (phylum), Enterobacteriaceae (family), Pseudomonas, and Pseudoxanthomonas (genus) was significantly higher in I. acuminatus adults (Figures 6M,N,P,Q). However, Serratia (genus) abundance was greater in I. sexdentatus adults (Figure 6R). No significant differences in the relative abundance of Firmicutes were observed for any comparisons (Figures 6C,I,O).
Discussion
Conifers have evolved a formidable defence against pests and pathogens (Klepzig et al., 1996; Krokene, 2015). In contrast, pests, including bark beetles and their associated microbiota, can successfully invade host trees by compromising host defence (Cheng et al., 2018). The insect microbiota is often influenced by the diet, sex, life stages, and the environment and facilitates the expansion of the ecological and evolutionary potential of their hosts (Yun et al., 2014; Lange et al., 2023). Recently, there has been growing interest in understanding the role of bark beetle-microbial association in shaping the plasticity of beetles. For instance, a recent study on Eurasian spruce bark beetles (ESBB), I. typographus, reported the developmental stage and the geo-location as drivers in shaping the beetle-microbial association (Chakraborty et al., 2023; Veselská et al., 2023; Baños-Quintana et al., 2024; Moussa et al., 2024). The gut bacterial dynamics in the pine beetle Dendroctonus rhizophagus revealed that the presence of persistent bacterial communities across the life stages of the beetle might be essential for ensuring certain physiological functions for the host (Briones-Roblero et al., 2017). However, information on the contribution of life stage and environment on the pine-feeding Ips bark beetles, I. sexdentatus, and I. acuminatus (Coleoptera: Curculionidae) is lacking. Hence, the present comparative study is focused on the influence of life stage and environment on the bacterial communities associated with the two pine-feeding Ips bark beetles. It is also important to mention that as our study focused on different life stages of the beetles, we did not separately evaluate sex-specific microbial assemblage in adult beetles. The sex-specific variation in the microbial association and their functional relevance will be an exciting avenue for future investigation. However, a recent gut proteomics study on ESBB failed to find any significant sex-specific protein expression difference in male and female adult beetles (Ashraf et al., 2023), suggesting conservation in gene expression. It will be interesting to see if such conservations exist in the case of microbial associations or not.
The bark beetle larvae spend their entire life gregariously feeding and developing under the bark. The larvae acquire microbial communities during feeding that mainly aid nutrient acquisition and detoxify plant defensive compounds (Peral-Aranega et al., 2020; Liu et al., 2022). During metamorphosis, beetles undergo complete structural changes from larvae to adults via the non-feeding pupal stage, which might lead to the gain or loss of certain microbiota (Sela et al., 2020). The compartmentalisation of the internal structures and organogenesis during metamorphosis leads to different physiological conditions, including redox potential, oxygen concentrations, and pH changes influencing the distribution and survival of the insect microbiota (Callegari et al., 2021). Recent findings by Peral-Aranega et al. (2023) revealed that the bacterial diversity in I. typographus reduced in the pupal stage compared to the larvae and regained in the adult stages, which was also observed with I. acuminatus beetles in our current study. However, there is no such trend in I. sexdentatus beetles. In I. sexdentatus beetles, bacterial richness and diversity gradually decreased after each life stage. Interestingly, adult beetles perform multiple duties on maturation, including feeding, host finding, and reproduction. Such responsibilities of the adult beetles can be associated with the maintenance and selectivity of their symbionts (Lemoine et al., 2020). Nevertheless, such possibilities need further experimental validation.
Our results demonstrated that the bacterial richness and diversity varied across the same developmental stages between the two Ips beetles. For instance, a high bacterial richness and diversity in I. acuminatus (IAC) adults was observed compared to I. sexdentatus (ISX). This interspecies variation in the bacterial community might be associated with their variable preference for pine trees. Although both I. sexdentatus and I. acuminatus feed on the same pine trees, they have particular preferences for pine trees. I. acuminatus beetles can attack healthy trees, whereas I. sexdentatus beetles are considered a secondary pest that prefers weakened and stressed pine trees to colonise (Wermelinger et al., 2008; Pineau et al., 2017a). In addition, I. acuminatus beetles infest young pine stands and plantations more aggressively than I. sexdentatus (Davydenko et al., 2017). Therefore, both Ips species might have differential resistance to host chemical defence that can influence bacterial partner selection and maintenance. Also, I. acuminatus beetles might require a wide range of bacterial species to deal with pine allelochemicals, which might explain the higher bacterial diversity in I. acuminatus adults compared to I. sexdentatus adults. In addition, I. acuminatus adult (wild) feeds on the upper canopy of the pine tree where the bark is thin, whereas I. sexdentatus adult (wild) beetles feed on the lower part of the bole where the bark is thick (Davydenko et al., 2017). Such habitat specialisation can be reflected by the differences in the bacterial diversity and richness between the two bark beetle species feeding on the same host. Recent research documented specific bark beetle fungal symbiotic system-mediated adaptation to feeding on different parts of the same conifer (spruce) hosts (Bai et al., 2024). Therefore, it can be assumed that microclimatic conditions can influence the symbiont association, richness and diversity in bark beetles. Nonetheless, such interpretations need to be further investigated in these beetles.
Moreover, for bark beetles and many other species, the competency of an organism to associate with other microorganisms can allow the species to thrive under challenging scenarios like climatic fluctuations and resources (Lemoine et al., 2020). Therefore, the fungus-feeding nature of I. acuminatus larvae might have some influence on its bacterial population dynamics. In addition, I. acuminatus also possess oral mycetangium for fungal transmission, which is rare to other Ips beetles, making them phylogenetically and morphologically distinct (Papek et al., 2024). The fungal species can be pathogenic to the host tree, while on the contrary, they may play nutritional roles in the I. acuminatus larvae (Villari et al., 2012) and might also interact with the bacterial communities residing in the developing larvae (Zhang et al., 2018). This could explain the lower bacterial richness in fungus-feeding I. acuminatus larvae but a higher bacterial diversity in I. sexdentatus larvae. However, dedicated investigations are needed to confirm the ecological relevance of such observations.
Laboratory adaptation and breeding conditions are essential factors that can affect bacterial associations unpredictably (Augustinos et al., 2019; Baños-Quintana et al., 2024). Compared to laboratory-bred beetles, broader and continuous environmental challenges in wild-collected beetles can encourage highly diversified and rich bacterial assemblage in wild-collected beetles. Furthermore, the bottleneck effect and high selective pressure are the key drivers in laboratory populations that often reduce symbiont load (Augustinos et al., 2019). Such observations may result in I. sexdentatus adults (wild) having higher bacterial diversity and richness than lab-bred adults (Table 2). However, no such trend was observed in I. acuminatus. Hence, dedicated investigations are needed to understand the ecological relevance of such observation. It is worth mentioning that the beetles were collected in different years; hence, the observed variation can also be due to feeding on different wood under different environmental conditions. A comparison of lab-bred (control wood, fed wood) and wild-collected (I. sexdentatus control and fed wood) wood samples for the same species (I. sexdentatus) has revealed that each wood sample poses a distinct bacterial population (Supplementary Excel 9) endorsing our prediction.
Although both pine beetles have distinct bacterial communities influenced by microclimatic conditions, canopy preference, metamorphosis, and feeding behaviour, our study revealed a core bacteriome across all developmental stages of two pine beetles (Figure 2C). Nevertheless, the bacterial composition did not vary, but there were distinct variations in the relative abundance of the bacterial genera. The core bacterial communities within the developmental stages of the two pine-feeding beetles were dominated by bacterial genera belonging to Erwiniaceae and Pseudomonaceae. A similar observation was reported in other beetles (Cambronero-Heinrichs et al., 2023; Peral-Aranega et al., 2023; Veselská et al., 2023). Our results documented the prevalence of Pseudomonas in the pine-feeding larvae. Pseudomonas is one of the most prominent bacterial genera isolated from different bark beetle species and across life stages (Saati-Santamaría et al., 2021), suggesting their pivotal role in the survival of the beetles. For example, the bacterial strains P. bohemica and P. typographi that were consistently present in I. typographus are reported to produce lytic enzymes and antifungal compounds (Peral-Aranega et al., 2023). Other abundant bacterial genera, including Serratia, Pseudoxanthomonas, Sphingmonas, Acinetobacter were present throughout the developmental stages of the two beetles, I. acuminatus and I. sexdentatus. The persistence of such bacterial strains across the life stages indicates their pivotal role in bark beetle holobiont (Cardoza et al., 2009; Fang et al., 2020; Ge et al., 2021; Peral-Aranega et al., 2023). Different strains of Pseudomonas, Serratia, Pseudoxanthomonas have been shown to be associated with lignocellulose degradation (Kumar et al., 2015; García-Fraile, 2018; Saati-Santamaría et al., 2021), indicating their role in the provision of nutrients to the beetles. For instance, Pseudomonas putida and P. azotoformans isolated from Dendroctonus rhizophagus were shown to have cellulolytic, amylolytic, xylanolytic, lipolytic, and esterase activity (Briones-Roblero et al., 2017). Our findings revealed a high abundance of Serratia in adults and the pupal stage of the pine beetles. Such observation was corroborated by Fang et al. (2020) where Serratia was dominant in adults and pupal stages of Ips typographus (Fang et al., 2020). Bacterial strains Serratia marcescens, and Pseudomonas mandelii, isolated from Dendroctonus ponderosae adults, have been shown to degrade plant defense chemicals (monoterpenes) (Boone et al., 2013). Additionally, bacterial strains from the core consortium, including Pseudomonas, Acinetobacter, and members of the Erwiniaceae family in the two pine beetle species, were reported to produce siderophores, aiding in competition by limiting the availability of iron in the environment (Peral-Aranega et al., 2023). Such results can provide some working hypothesis to follow up in future with individual stains isolated from these beetles via culture dependent methods.
Our study also reveals beetle-mediated alterations in wood bacterial assemblage, similar to insect herbivores that reshape the native plant leaf microbiome (Humphrey and Whiteman, 2020). However, the degree of overlap or distinctiveness between insect and host (wood) microbiome remains ambiguous (Pirttilä et al., 2023). Precisely, higher sharing between fed wood and beetle samples followed by lower bacterial diversity in fed wood samples than in control wood can be due to several reasons. It can be presumed that bacterial association in both beetle species is influenced by the horizontal transfer of bacteria from the pine host. Further studies are required to comprehend the ecological relevance of beetle-mediated tree bacteriome alteration.
Limitation
The sample size in our study is relatively small, with fewer individuals per replicate, which may have reduced our ability to eliminate transient species while characterising the core microbiome. Secondly, we collected samples from one forest location in the Czech Republic, and that geographical distribution might introduce a potential confounder that could influence the composition of the pine beetle bacteriome, leading to some biases in the results. Moreover, 16S rRNA amplicon sequencing restricted our taxonomic identification to the genus level, potentially missing crucial functional associations at the species or strain levels. Nonetheless, our study offers valuable hypotheses for delving into pine bark beetle symbiosis in future.
Conclusion
Our comparative metagenomic study reported the impact of life stage and lab-breeding on the bacterial communities in two pine-feeding beetles. The core bacterial genera, including Pseudomonas, Pseudoxanthomonas, and Acinetobacter were dominant in the pine beetles. The bacterial diversity and richness varied significantly across life stages, wild collected, and laboratory populations. I. sexdentatus larvae represented significantly higher bacterial diversity and community richness and evenness compared to other developmental stages, while I. acuminatus adults demonstrated higher bacterial richness with no significant variation in the bacterial diversity and richness between the life stages. Lab-bred and wild beetles showed distinct bacterial diversity and richness. Beetle feeding substantially influenced the host bacteriome at the feeding galleries. Further downstream studies to characterise the key highly abundant and transient bacterial species will provide new ecological insights into pine bark beetle symbiosis, particularly concerning their metabolic capacities, interactions with other symbionts, and roles in the detoxification of conifer allelochemicals. Essential bacterial partners can also be used for microbiome-based sustainable pest management practises as carriers for dsRNA against bark beetles (Whitten et al., 2016; Qadri et al., 2020; Joga et al., 2021; Gupta et al., 2023).
Data availability statement
The bacteriome dataset in the study is available under NCBI Bio-project PRJNA854390.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
AK: Writing – original draft, Visualization, Validation, Software, Methodology, Investigation, Formal analysis, Data curation. AC: Writing – original draft, Supervision, Software, Resources, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. RM: Writing – review & editing, Visualization, Validation, Software, Formal analysis. JS: Methodology, Writing – review & editing, Visualization, Resources. AR: Writing – review & editing, Visualization, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. AR, AC, and AK financed by EVA 4.0,” No. CZ.02.1.01/0.0/0.0/16 019/0000803 supported by OP RDE. The project is funded by “EXTEMIT-K,” No. CZ.02.1.01/0.0/0.0/15_003/0000433 financed by OP RDE. AK was also supported by the Internal Grant Agency FLD (IGA 2021–2022; project number-A_21_05). AC and AR are also supported by the “Excellent Team Grant (2023–2024)” from FLD, CZU.
Acknowledgments
We appreciate the helpful feedback provided by the reviewers and handling editor.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1400894/full#supplementary-material
Footnotes
References
Abdullah, H., Darvishzadeh, R., Skidmore, A. K., Groen, T. A., and Heurich, M. (2018). European spruce bark beetle (Ips typographus, L.) green attack affects foliar reflectance and biochemical properties. Int. J. Appl. Earth Obs. Geoinf. 64, 199–209. doi: 10.1016/j.jag.2017.09.009
Adams, A. S., Aylward, F. O., Adams, S. M., Erbilgin, N., Aukema, B. H., Currie, C. R., et al. (2013). Mountain pine beetles colonizing historical and naive host trees are associated with a bacterial community highly enriched in genes contributing to terpene metabolism. Appl. Environ. Microbiol. 79, 3468–3475. doi: 10.1128/AEM.00068-13
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Ashraf, M. Z., Mogilicherla, K., Sellamuthu, G., Siino, V., Levander, F., and Roy, A. (2023). Comparative gut proteomics study revealing adaptive physiology of Eurasian spruce bark beetle, Ips typographus (Coleoptera: Scolytinae). Front. Plant Sci. 14:1157455. doi: 10.3389/fpls.2023.1157455
Augustinos, A. A., Tsiamis, G., Cáceres, C., Abd-Alla, A. M., and Bourtzis, K. (2019). Taxonomy, diet, and developmental stage contribute to the structuring of gut-associated bacterial communities in tephritid pest species. Front. Microbiol. 10:2004. doi: 10.3389/fmicb.2019.02004
Bai, Z., Fang, J., Yu, C., Zhang, S., Liu, F., Han, F., et al. (2024). Divergent response of two bark beetle–fungal symbiotic systems to host monoterpenes reflects niche partitioning strategies. J. Chem. Ecol. 12, 1–12. doi: 10.1007/s10886-024-01535-5
Baños-Quintana, A. P., Gershenzon, J., and Kaltenpoth, M. (2024). The Eurasian spruce bark beetle Ips typographus shapes the microbial communities of its offspring and the gallery environment. Front. Microbiol. 15:1367127. doi: 10.3389/fmicb.2024.1367127
Biedermann, P. H., Müller, J., Grégoire, J.-C., Gruppe, A., Hagge, J., Hammerbacher, A., et al. (2019). Bark beetle population dynamics in the Anthropocene: challenges and solutions. Trends Ecol. Evol. 34, 914–924. doi: 10.1016/j.tree.2019.06.002
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 1–17. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Boone, C. K., Keefover-Ring, K., Mapes, A. C., Adams, A. S., Bohlmann, J., and Raffa, K. F. (2013). Bacteria associated with a tree-killing insect reduce concentrations of plant defense compounds. J. Chem. Ecol. 39, 1003–1006. doi: 10.1007/s10886-013-0313-0
Briones-Roblero, C. I., Hernández-García, J. A., Gonzalez-Escobedo, R., Soto-Robles, L. V., Rivera-Orduña, F. N., and Zúñiga, G. (2017). Structure and dynamics of the gut bacterial microbiota of the bark beetle, Dendroctonus rhizophagus (Curculionidae: Scolytinae) across their life stages. PloS one 12:e0175470. doi: 10.1371/journal.pone.0175470
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callegari, M., Crotti, E., Fusi, M., Marasco, R., Gonella, E., De Noni, I., et al. (2021). Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. NPJ Biofilms Microbiomes 7:42. doi: 10.1038/s41522-021-00212-9
Cambronero-Heinrichs, J. C., Battisti, A., Biedermann, P. H., Cavaletto, G., Castro-Gutierrez, V., Favaro, L., et al. (2023). Erwiniaceae bacteria play defensive and nutritional roles in two widespread ambrosia beetles. FEMS Microbiol. Ecol. 99:fiad144. doi: 10.1093/femsec/fiad144
Cardoza, Y. J., Vasanthakumar, A., Suazo, A., and Raffa, K. F. (2009). Survey and phylogenetic analysis of culturable microbes in the oral secretions of three bark beetle species. Entomol. Exp. Appl. 131, 138–147. doi: 10.1111/j.1570-7458.2009.00844.x
Chakraborty, A., Ashraf, M. Z., Modlinger, R., Synek, J., Schlyter, F., and Roy, A. (2020a). Unravelling the gut bacteriome of Ips (Coleoptera: Curculionidae: Scolytinae): identifying core bacterial assemblage and their ecological relevance. Sci. Rep. 10:18572. doi: 10.1038/s41598-020-75203-5
Chakraborty, A., Modlinger, R., Ashraf, M. Z., Synek, J., Schlyter, F., and Roy, A. (2020b). Core mycobiome and their ecological relevance in the gut of five Ips bark beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 11:568853. doi: 10.3389/fmicb.2020.568853
Chakraborty, A., Purohit, A., Khara, A., Modlinger, R., and Roy, A. (2023). Life-stage and geographic location determine the microbial assemblage in Eurasian spruce bark beetle, Ips typographus L. (Coleoptera: Curculionidae). Front. Forests Global Change 6:1176160. doi: 10.3389/ffgc.2023.1176160
Chao, A., Lee, S.-M., and Chen, T.-C. (1988). A generalized Good's nonparametric coverage estimator. Chin. J. Mathematic. 1, 189–199.
Chapman, M., and Underwood, A. (1999). Ecological patterns in multivariate assemblages: information and interpretation of negative values in ANOSIM tests. Mar. Ecol. Prog. Ser. 180, 257–265. doi: 10.3354/meps180257
Chen, B., Du, K., Sun, C., Vimalanathan, A., Liang, X., Li, Y., et al. (2018). Gut bacterial and fungal communities of the domesticated silkworm (Bombyx mori) and wild mulberry-feeding relatives. ISME J. 12, 2252–2262. doi: 10.1038/s41396-018-0174-1
Cheng, C., Wickham, J. D., Chen, L., Xu, D., Lu, M., and Sun, J. (2018). Bacterial microbiota protect an invasive bark beetle from a pine defensive compound. Microbiome 6, 1–16. doi: 10.1186/s40168-018-0518-0
Chinellato, F., Battisti, A., Finozzi, V., and Faccoli, M. (2014). Better today but worse tomorrow: how warm summers affect breeding performance of a scots pine pest. Agrochimica Int. J. Plant Chem. Soil Sci. Plant Nutr. 58, 133–145.
Clarke, K. R. (1993). Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18, 117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x
Colombari, F., Schroeder, M. L., Battisti, A., and Faccoli, M. (2013). Spatio-temporal dynamics of an Ips acuminatus outbreak and implications for management. Agric. For. Entomol. 15, 34–42. doi: 10.1111/j.1461-9563.2012.00589.x
D’Argenio, V., Casaburi, G., Precone, V., and Salvatore, F. (2014). Comparative metagenomic analysis of human gut microbiome composition using two different bioinformatic pipelines. BioMed Res. Int. 2014:325340. doi: 10.1155/2014/325340
Davydenko, K., Vasaitis, R., and Menkis, A. (2017). Fungi associated with Ips acuminatus (Coleoptera: Curculionidae) in Ukraine with a special emphasis on pathogenicity of ophiostomatoid species. Eur. J. Entomol. 114, 77–85. doi: 10.14411/eje.2017.011
Dobor, L., Hlásny, T., Rammer, W., Barka, I., Trombik, J., Pavlenda, P., et al. (2018). Post-disturbance recovery of forest carbon in a temperate forest landscape under climate change. Agric. For. Meteorol. 263, 308–322. doi: 10.1016/j.agrformet.2018.08.028
Faccoli, M., Finozzi, V., and Colombari, F. (2012). Effectiveness of different trapping protocols for outbreak management of the engraver pine beetle Ips acuminatus (Curculionidae, Scolytinae). Int. J. Pest Manag. 58, 267–273. doi: 10.1080/09670874.2011.642824
Fang, J.-X., Zhang, S.-F., Liu, F., Zhang, X., Zhang, F.-B., Guo, X.-B., et al. (2020). Differences in gut bacterial communities of Ips typographus (Coleoptera: curculionidae) induced by enantiomer-specific α-pinene. Environ. Entomol. 49, 1198–1205. doi: 10.1093/ee/nvaa098
García-Fraile, P. (2018). Roles of bacteria in the bark beetle holobiont–how do they shape this forest pest? Ann. Appl. Biol. 172, 111–125. doi: 10.1111/aab.12406
Ge, S.-X., Shi, F.-M., Pei, J.-H., Hou, Z.-H., Zong, S.-X., and Ren, L.-L. (2021). Gut bacteria associated with Monochamus saltuarius (Coleoptera: Cerambycidae) and their possible roles in host plant adaptations. Front. Microbiol. 12:687211. doi: 10.3389/fmicb.2021.687211
González-Dominici, L. I., Saati-Santamaría, Z., and García-Fraile, P. (2021). Genome analysis and genomic comparison of the novel species Arthrobacter ipsi reveal its potential protective role in its bark beetle host. Microb. Ecol. 81, 471–482. doi: 10.1007/s00248-020-01593-8
González-Serrano, F., Pérez-Cobas, A. E., Rosas, T., Baixeras, J., Latorre, A., and Moya, A. (2020). The gut microbiota composition of the moth Brithys crini reflects insect metamorphosis. Microb. Ecol. 79, 960–970. doi: 10.1007/s00248-019-01460-1
Gupta, S., Chakraborty, A., and Roy, A. (2023). Prospects for deploying microbes against tree-killing beetles (Coleoptera) in Anthropocene. Front. Forests Global Change 6:1182834. doi: 10.3389/ffgc.2023.1182834
Hu, X., Yu, J., Wang, C., and Chen, H. (2014). Cellulolytic bacteria associated with the gut of Dendroctonus armandi larvae (Coleoptera: Curculionidae: Scolytinae). Forests 5, 455–465. doi: 10.3390/f5030455
Humphrey, P. T., and Whiteman, N. K. (2020). Insect herbivory reshapes a native leaf microbiome. Nat. Ecol. Evol. 4, 221–229. doi: 10.1038/s41559-019-1085-x
Ibarra-Juarez, L. A., Burton, M., Biedermann, P., Cruz, L., Desgarennes, D., Ibarra-Laclette, E., et al. (2020). Evidence for succession and putative metabolic roles of fungi and bacteria in the farming mutualism of the ambrosia beetle Xyleborus affinis. Msystems 5, e00541–e00520. doi: 10.1128/mSystems.00541-20
Joga, M. R., Mogilicherla, K., Smagghe, G., and Roy, A. (2021). RNA interference-based forest protection products (FPPs) against wood-boring coleopterans: Hope or hype? Front. Plant Sci. 12:733608. doi: 10.3389/fpls.2021.733608
Klepzig, K. D., Smalley, E. B., and Raffa, K. F. (1996). Combined chemical defenses against an insect-fungal complex. J. Chem. Ecol. 22, 1367–1388. doi: 10.1007/BF02027719
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41:808. doi: 10.1093/nar/gks808
Kolasa, M., Ścibior, R., Mazur, M. A., Kubisz, D., Dudek, K., and Kajtoch, Ł. (2019). How hosts taxonomy, trophy, and endosymbionts shape microbiome diversity in beetles. Microb. Ecol. 78, 995–1013. doi: 10.1007/s00248-019-01358-y
Krokene, P. (2015). “Conifer defense and resistance to bark beetles” in Bark beetles. eds. K. F. Raffa and J. C. Gregoire (Amsterdam: Elsevier), 177–207.
Kumar, M., Revathi, K., and Khanna, S. (2015). Biodegradation of cellulosic and lignocellulosic waste by Pseudoxanthomonas sp R-28. Carbohydr. Polym. 134, 761–766. doi: 10.1016/j.carbpol.2015.08.072
Lange, C., Boyer, S., Bezemer, T. M., Lefort, M.-C., Dhami, M. K., Biggs, E., et al. (2023). Impact of intraspecific variation in insect microbiomes on host phenotype and evolution. ISME J. 17, 1798–1807. doi: 10.1038/s41396-023-01500-2
Lemoine, M. M., Engl, T., and Kaltenpoth, M. (2020). Microbial symbionts expanding or constraining abiotic niche space in insects. Curr. Opin. Insect Sci. 39, 14–20. doi: 10.1016/j.cois.2020.01.003
Li, M., Shao, D., Zhou, J., Gu, J., Qin, J., Chen, W., et al. (2020). Signatures within esophageal microbiota with progression of esophageal squamous cell carcinoma. Chin. J. Cancer Res. 32, 755–767. doi: 10.21147/j.issn.1000-9604.2020.06.09
Liška, J., Knížek, M., and Véle, A. (2021). Evaluation of insect pest occurrence in areas of calamitous mortality of scots pine. Lesnicky Casopis 67, 85–90. doi: 10.2478/forj-2021-0006
Liu, F., Ye, F., Cheng, C., Kang, Z., Kou, H., and Sun, J. (2022). Symbiotic microbes aid host adaptation by metabolizing a deterrent host pine carbohydrate d-pinitol in a beetle-fungus invasive complex. Sci. Adv. 8:eadd5051. doi: 10.1126/sciadv.add5051
Lozupone, C., Lladser, M. E., Knights, D., Stombaugh, J., and Knight, R. (2011). UniFrac: an effective distance metric for microbial community comparison. ISME J. 5, 169–172. doi: 10.1038/ismej.2010.133
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Magurran, A. E. (1988). Ecological diversity and its measurement. Princeton, NJ: Princeton University Press.
Marini, L., Økland, B., Jönsson, A. M., Bentz, B., Carroll, A., Forster, B., et al. (2017). Climate drivers of bark beetle outbreak dynamics in Norway spruce forests. Ecography 40, 1426–1435. doi: 10.1111/ecog.02769
McNichol, B. H., Clarke, S. R., Faccoli, M., Montes, C. R., Nowak, J. T., Reeve, J. D., et al. (2022). “Relationships between drought, coniferous tree physiology, and Ips bark beetles under climatic changes” in Bark beetle management, ecology, and climate change. eds. R. W. Hofstetter and K. Gandhi (Amsterdam: Elsevier), 153–194.
Montagné-Huck, C., and Brunette, M. (2018). Economic analysis of natural forest disturbances: a century of research. J. For. Econ. 32, 42–71. doi: 10.1016/j.jfe.2018.03.002
Morales-Jiménez, J., Vera-Ponce de León, A., García-Domínguez, A., Martínez-Romero, E., Zúñiga, G., and Hernández-Rodríguez, C. (2013). Nitrogen-fixing and uricolytic bacteria associated with the gut of Dendroctonus rhizophagus and Dendroctonus valens (Curculionidae: Scolytinae). Microb. Ecol. 66, 200–210. doi: 10.1007/s00248-013-0206-3
Morales-Jiménez, J., Zúñiga, G., Villa-Tanaca, L., and Hernández-Rodríguez, C. (2009). Bacterial community and nitrogen fixation in the red turpentine beetle, Dendroctonus valens LeConte (Coleoptera: Curculionidae: Scolytinae). Microb. Ecol. 58, 879–891. doi: 10.1007/s00248-009-9548-2
Moussa, A., Nones, S., Vannucchi, P. E., Shahzad, G.-I.-R., Dittmer, J., Corretto, E., et al. (2024). The bacterial community of the European spruce bark beetle in space and time. Entomol. General. 44, 211–222. doi: 10.1127/entomologia/2023/2114
Mumm, R., and Hilker, M. (2006). Direct and indirect chemical defence of pine against folivorous insects. Trends Plant Sci. 11, 351–358. doi: 10.1016/j.tplants.2006.05.007
Navidshad, B., Liang, J. B., and Jahromi, M. F. (2012). Correlation coefficients between different methods of expressing bacterial quantification using real time PCR. Int. J. Mol. Sci. 13, 2119–2132. doi: 10.3390/ijms13022119
Nunberg, M. (1981). Klucze do rozpoznawania owadów Polski [Keys for the identification of Polish Insects]. Czȩść XIX. Chrzzşzcze–Coleoptera, Korniki–Scolytidae, Wyrynniki–Platypodidae. PWN, Warszawa-Wroclaw. Zeszyt, 99–100.
Oksanen, J. (2007). Vegan: Community ecology package. R package version 1.8–5. Available at: http://www.cran.r-project.org (Accessed June 14, 2024).
Papek, E., Ritzer, E., Biedermann, P. H., Cognato, A. I., Baier, P., Hoch, G., et al. (2024). The pine bark beetle Ips acuminatus: an ecological perspective on life-history traits promoting outbreaks. J. Pest. Sci. 97, 1093–1122. doi: 10.1007/s10340-024-01765-2
Paulson, J. N., Pop, M., and Bravo, H. C. (2011). Metastats: an improved statistical method for analysis of metagenomic data. Genome Biol. 12 (Suppl 1), 17. doi: 10.1186/gb-2011-12-s1-p17
Pekár, S., and Brabec, M. (2016). Modern analysis of biological data: Generalized linear models in R. Brno: Masarykova Univerzita.
Peral-Aranega, E., Saati-Santamaría, Z., Ayuso-Calles, M., Kostovčík, M., Veselská, T., Švec, K., et al. (2023). New insight into the bark beetle ips typographus bacteriome reveals unexplored diversity potentially beneficial to the host. Environ. Microb. 18:53. doi: 10.1186/s40793-023-00510-z
Peral-Aranega, E., Saati-Santamaría, Z., Kolařik, M., Rivas, R., and García-Fraile, P. (2020). Bacteria belonging to Pseudomonas typographi sp. nov. from the bark beetle Ips typographus have genomic potential to aid in the host ecology. Insects 11:593. doi: 10.3390/insects11090593
Pfeffer, A. (1955). Fauna ČSR. Svazek 6: Kůrovci-Scolytoidea. Praha: Brouci-Coleoptera Nakladatelství Československé akadmie věd.
Pfeffer, A. (1995). Zentral-und westpaläarktische Borken-und Kernkäfer:Coloptera: Scolytidae, Platypodidae. Basel: Pro Entomologia.
Pineau, X., Bourguignon, M., Jactel, H., Lieutier, F., and Sallé, A. (2017a). Pyrrhic victory for bark beetles: successful standing tree colonization triggers strong intraspecific competition for offspring of Ips sexdentatus. For. Ecol. Manag. 399, 188–196. doi: 10.1016/j.foreco.2017.05.044
Pineau, X., David, G., Peter, Z., Sallé, A., Baude, M., Lieutier, F., et al. (2017b). Effect of temperature on the reproductive success, developmental rate and brood characteristics of I ps sexdentatus (B oern.). Agric. For. Entomol. 19, 23–33. doi: 10.1111/afe.12177
Pirttilä, A., Brusila, V., Koskimäki, J., Wäli, P., Ruotsalainen, A., Mutanen, M., et al. (2023). Exchange of microbiomes in plant-insect herbivore interactions. MBio 14, e03210–e03222. doi: 10.1128/mbio.03210-22
Plewa, R., and Mokrzycki, T. (2017). Occurrence, biology, and economic importance of the sharp-dentated bark beetle Ips acuminatus (Gyllenhal, 1827) (Coleoptera, Curculionidae, Scolytinae) in Poland. Sylwan 161, 619–629. doi: 10.26202/sylwan.2017077
Qadri, M., Short, S., Gast, K., Hernandez, J., and Wong, A. C.-N. (2020). Microbiome innovation in agriculture: development of microbial based tools for insect pest management. Front. Sust. Food Syst. 4:547751. doi: 10.3389/fsufs.2020.547751
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
R Core Team (2013). R: A language and environment for statistical computing (version 2.15. 3) [computer software] R foundation for statistical computing. Vienna: R Core Team.
Raffa, K. F., Aukema, B. H., Bentz, B. J., Carroll, A. L., Hicke, J. A., Turner, M. G., et al. (2008). Cross-scale drivers of natural disturbances prone to anthropogenic amplification: the dynamics of bark beetle eruptions. Bioscience 58, 501–517. doi: 10.1641/B580607
Rognes, T., Flouri, T., Nichols, B., Quince, C., and Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. doi: 10.7717/peerj.2584
Saati-Santamaría, Z., Rivas, R., Kolařik, M., and García-Fraile, P. (2021). A new perspective of Pseudomonas—host interactions: distribution and potential ecological functions of the genus Pseudomonas within the bark beetle Holobiont. Biology 10:164. doi: 10.3390/biology10020164
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60–R18. doi: 10.1186/gb-2011-12-6-r60
Sela, R., Laviad-Shitrit, S., and Halpern, M. (2020). Changes in microbiota composition along the metamorphosis developmental stages of Chironomus transvaalensis. Front. Microbiol. 11:586678. doi: 10.3389/fmicb.2020.586678
Sellamuthu, G., Amin, S., Bílý, J., Synek, J., Modlinger, R., Sen, M. K., et al. (2021). Reference gene selection for normalizing gene expression in Ips sexdentatus (Coleoptera: Curculionidae: Scolytinae) under different experimental conditions. Front. Physiol. 12:752768. doi: 10.3389/fphys.2021.752768
Silver, A., Perez, S., Gee, M., Xu, B., Garg, S., Will, K., et al. (2021). Persistence of the ground beetle (Coleoptera: Carabidae) microbiome to diet manipulation. PLoS One 16:e0241529. doi: 10.1371/journal.pone.0241529
Singh, V. V., Naseer, A., Mogilicherla, K., Trubin, A., Zabihi, K., Roy, A., et al. (2024). Understanding bark beetle outbreaks: exploring the impact of changing temperature regimes, droughts, forest structure, and prospects for future forest pest management. Rev. Environ. Sci. Biotechnol. 23, 257–290. doi: 10.1007/s11157-024-09692-5
Skrodenytė-Arbačiauskienė, V., Radžiutė, S., Stunžėnas, V., and Būda, V. (2012). Erwiniatypographi sp. nov., isolated from bark beetle (Ips typographus) gut. Int. J. Syst. Evol. Microbiol. 62, 942–948. doi: 10.1099/ijs.0.030304-0
Stat, M., Pochon, X., Franklin, E. C., Bruno, J. F., Casey, K. S., Selig, E. R., et al. (2013). The distribution of the thermally tolerant symbiont lineage (Symbiodinium clade D) in corals from Hawaii: correlations with host and the history of ocean thermal stress. Ecol. Evol. 3, 1317–1329. doi: 10.1002/ece3.556
Tian, H., Zhao, L., Koski, T.-M., and Sun, J. (2022). Microhabitat governs the microbiota of the pinewood nematode and its vector beetle: implication for the prevalence of pine wilt disease. Microbiol. Spectr. 10, e00783–e00722. doi: 10.1128/spectrum.00783-22
Veselská, T., Švec, K., Kostovčík, M., Peral-Aranega, E., Garcia-Fraile, P., Křížková, B., et al. (2023). Proportions of taxa belonging to the gut core microbiome change throughout the life cycle and season of the bark beetle Ips typographus. FEMS Microbiol. Ecol. 99:fiad072. doi: 10.1093/femsec/fiad072
Villari, C., Battisti, A., Chakraborty, S., Michelozzi, M., Bonello, P., and Faccoli, M. (2012). Nutritional and pathogenic fungi associated with the pine engraver beetle trigger comparable defenses in scots pine. Tree Physiol. 32, 867–879. doi: 10.1093/treephys/tps056
Wermelinger, B., Rigling, A., Schneider Mathis, D., and Dobbertin, M. (2008). Assessing the role of bark-and wood-boring insects in the decline of scots pine (Pinus sylvestris) in the Swiss Rhone valley. Ecol. Entomol. 33, 239–249. doi: 10.1111/j.1365-2311.2007.00960.x
Whitten, M. M., Facey, P. D., Del Sol, R., Fernández-Martínez, L. T., Evans, M. C., Mitchell, J. J., et al. (2016). Symbiont-mediated RNA interference in insects. Proc. R. Soc. B Biol. Sci. 283:20160042. doi: 10.1098/rspb.2016.0042
Xu, L., Lou, Q., Cheng, C., Lu, M., and Sun, J. (2015). Gut-associated bacteria of Dendroctonus valens and their involvement in verbenone production. Microb. Ecol. 70, 1012–1023. doi: 10.1007/s00248-015-0625-4
Xu, L. T., Lu, M., and Sun, J. H. (2016). Invasive bark beetle-associated microbes degrade a host defensive monoterpene. Insect Sci. 23, 183–190. doi: 10.1111/1744-7917.12255
Yun, J.-H., Roh, S. W., Whon, T. W., Jung, M.-J., Kim, M.-S., Park, D.-S., et al. (2014). Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80, 5254–5264. doi: 10.1128/AEM.01226-14
Keywords: Ips acuminatus, Ips sexdentatus, core bacteriome, microhabitat, holobiont, amplicon sequence variances (ASVs)
Citation: Khara A, Chakraborty A, Modlinger R, Synek J and Roy A (2024) Comparative metagenomic study unveils new insights on bacterial communities in two pine-feeding Ips beetles (Coleoptera: Curculionidae: Scolytinae). Front. Microbiol. 15:1400894. doi: 10.3389/fmicb.2024.1400894
Edited by:
Kim Yrjälä, University of Helsinki, FinlandReviewed by:
Zaki Saati Santamaría, University of Salamanca, SpainCesar Hugo Hernández-Rodríguez, Instituto Politécnico Nacional, Mexico
Copyright © 2024 Khara, Chakraborty, Modlinger, Synek and Roy. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amrita Chakraborty, Q2hha3JhYm9ydHlAZmxkLmN6dS5jeg==; Amit Roy, Um95QGZsZC5jenUuY3o=
 Arunabha Khara
Arunabha Khara Amrita Chakraborty
Amrita Chakraborty Roman Modlinger
Roman Modlinger Jiří Synek
Jiří Synek Amit Roy
Amit Roy