- 1Division of Food Microbiology, National Institute of Food and Drug Safety Evaluation, Ministry of Food and Drug Safety, Cheongju, Republic of Korea
- 2Institute of Life Sciences & Resources and Department of Food Science and Biotechnology, Kyung Hee University, Yongin, Republic of Korea
- 3Department of Food and Animal Biotechnology, Research Institute of Agriculture and Life Sciences, Seoul National University, Seoul, Republic of Korea
- 4Department of Agricultural Biotechnology, Seoul National University, Seoul, Republic of Korea
- 5Department of Biotechnology, and Carbohydrate Bioproduct Research Center, Sejong University, Seoul, Republic of Korea
- 6Center for Food and Bioconvergence, Seoul National University, Seoul, Republic of Korea
Introduction: The predominant hybrid pathogenic E. coli, enterohemorrhagic E. coli (EHEC), combines characteristics of Shiga toxin-producing E. coli (STEC) and enteropathogenic E. coli (EPEC), contributing to global outbreaks with severe symptoms including fatal consequences. Since EHEC infection was designated as a notifiable disease in 2000 in South Korea, around 2000 cases have been reported, averaging approximately 90 cases annually.
Aim: In this work, genome-based characteristic analysis and cell-based assay of hybrid STEC/aEPEC strains isolated from livestock feces, animal source foods, and water in South Korea was performed.
Methods: To identify the virulence and antimicrobial resistance genes, determining the phylogenetic position of hybrid STEC/aEPEC strains isolated in South Korea, a combination of real-time PCR and whole-genome sequencing (WGS) was used. Additionally, to assess the virulence of the hybrid strains and compare them with genomic characterization, we performed a cell cytotoxicity and invasion assays.
Results: The hybrid STEC/aEPEC strains harbored stx and eae genes, encoding Shiga toxins and E. coli attachment/effacement related protein of STEC and EPEC, respectively. Furthermore, all hybrid strains harbored plasmid-carried enterohemolysin(ehxCABD), a key virulence factor in prevalent pathogenic E. coli infections, such as diarrheal disease and hemolytic-uremic syndrome (HUS). Genome-wide phylogenetic analysis revealed a close association between all hybrid strains and specific EPEC strains, suggesting the potential acquisition of Stx phages during STEC/aEPEC hybrid formation. Some hybrid strains showed cytotoxic activity against HeLa cells and invasive properties against epithelial cells. Notably, all STEC/aEPEC hybrids with sequence type (ST) 1,034 (n = 11) exhibited higher invasiveness than those with E2348/69. This highlights the importance of investigating potential correlations between STs and virulence characteristics of E. coli hybrid strains.
Conclusion: Through genome-based characterization, we confirmed that the hybrid STEC/aEPEC strains are likely EPEC strains that have acquired STEC virulence genes via phage. Furthermore, our results emphasize the potential increased danger to humans posed by hybrid STEC/aEPEC strains isolated in South Korea, containing both stx and eaeA, compared to STEC or EPEC alone.
1 Introduction
Escherichia coli (E. coli) commonly resides in the gastrointestinal tracts of humans and animals as part of their natural gut flora (Kaper et al., 2004). Although most strains are harmless members of this gut community, certain E. coli variants, including those that have acquired specific virulence traits, can cause diarrheal and extraintestinal illnesses in humans and animals (Kaper et al., 2004; Dobrindt, 2005). Diarrheagenic E. coli (DEC) accounts for approximately 30–40% of acute diarrhea cases in children under 5 years old in developing countries (Dutta et al., 2013; Miliwebsky et al., 2016) and is a significant cause of both isolated instances and outbreaks of diarrhea worldwide (Croxen et al., 2013).
DEC, characterized by distinct combinations of virulence characteristics, are categorized into pathotypes, each with specific host preferences, global occurrence, disease impacts, and transmission methods (Croxen and Finlay, 2010). DEC are categorized into six main pathotypes: enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC). Each E. coli pathotype possesses distinct pathogenic mechanisms and a unique set of virulence factors encoded by specific gene clusters (Jesser and Levy, 2020; Pakbin et al., 2021). These genes related to pathogenicity may control functions such as adhesion, invasion, attachment, iron acquisition, motility, and toxin activity (Kaper et al., 2004; Croxen and Finlay, 2010).
STEC is an E. coli pathotype capable of producing at least one of the Shiga toxins (Stxs), namely Stx1 and Stx2 (Kaper et al., 2004; Croxen et al., 2013). Within each group, numerous subtypes or variants of Stx have been identified and characterized (Burgos and Beutin, 2012). STEC is a globally prevalent pathogen associated with human diseases that primarily causes diarrhea, hemorrhagic colitis (HC), and hemolytic-uremic syndrome (HUS) (Jesser and Levy, 2020; Pakbin et al., 2021). EPEC induces diarrhea and initiates the formation of attaching and effacing (A/E) lesions in the human gut epithelium (Kaper et al., 2004; Croxen et al., 2013). EPEC strains are categorized as either typical or atypical according to whether or not they have the E. coli adherence factor plasmid (EAF) expressing the bfpA gene. This gene is responsible for the production of bundle-forming pili. Typical EPEC (tEPEC) possesses both the locus of enterocyte effacement (LEE) region responsible for attaching and causing effacing lesions (eae), and bfpA. In contrast, atypical EPEC (aEPEC) lacks bfpA (Jesser and Levy, 2020; Pakbin et al., 2021). aEPEC strains exhibit comparable prevalence rates among healthy adults and children with diarrhea, and are also found in asymptomatic adults (Hazen et al., 2016). However, certain aEPEC strains have been linked to diarrhea outbreaks (Vieira et al., 2016).
The Enterohemorrhagic Escherichia coli (EHEC) is commonly used to refer to a subset of STEC strains that simultaneously harbors genes for Stx production and LEE. As STEC strains have the potential to induce diarrhea and hemolytic uremic syndrome (HUS) in human subjects regardless of the presence of LEE, this PAI is an accessory set of virulence genes that enhances STEC pathogenicity. EHEC are considered a pathotype of E. coli and are referred to as such in the scientific literature rather than referring to them as hybrid STEC/aEPEC. However, according to previous studies, there are reports indicating that EPEC/STEC hetero-pathogens consist of atypical EPEC carrying Stx (Croxen et al., 2013; Eichhorn et al., 2015; Gomes et al., 2016; Silva et al., 2019). Especially, Gioia-Di Chiacchio et al. (2018) were the first to report a typical EPEC strain carrying Stx, identified in eight E. coli strains isolated from birds, which simultaneously harbored Stx2, LEE, and BFP.
Globally, emerging hybrid diarrheagenic E. coli strains incorporating genetic markers from diverse pathotypes pose substantial public health concerns (Santos et al., 2020). This emergence is attributed to the horizontal gene transfer (HGT) among diarrheagenic E. coli or the prevalence of E. coli virulence genes typically located on plasmids, enabling transmission via conjugation (Kaper et al., 2004; Dobrindt, 2005; Geurtsen et al., 2022). Hybrid DEC pathotype strains are increasingly reported worldwide. Different hybrid strains, such as EPEC/ETEC, have been identified in India (Dutta et al., 2015), with Germany (Bielaszewska et al., 2011) and Indonesia (Setyarini et al., 2020) reporting STEC/EAEC hybrids. Brazil has documented cases of EPEC/STEC (Gioia-Di Chiacchio et al., 2018) and EPEC/EAEC (de Lira et al., 2021). Sweden has several instances of STEC/ETEC (Bai et al., 2019). Diarrheal disease outbreaks associated with various DEC pathotypes occurred in Germany in 2011 (Buchholz et al., 2011), Japan in 2016 (Furukawa et al., 2018) and 2020 (Kashima et al., 2021), Nottingham, UK in 2014 (Newitt et al., 2016), South Korea in 2018 (Lim et al., 2020), and the United Kingdom in 2020 (Mulchandani et al., 2021). Hybrid strains are characterized by the presence of virulence genes unique to multiple E. coli pathotypes, typically detected via PCR or comprehensive whole-genome sequencing (WGS) (Jesser and Levy, 2020; Lee et al., 2023a,b). Cell-based assays have also been used to assess the virulence potential of hybrid strains (Valiatti et al., 2020).
The purpose of the present study was to characterize the genetic structure and evaluate the virulence potential of hybrid STEC/aEPEC strains isolated from livestock feces, animal source foods, and water in South Korea. This study enhances our understanding of the horizontal transfer of several virulence-related genes via mobile genetic elements (MGEs) and provides information on the virulence potential of hybrid STEC/aEPEC strains. Based on our findings, we highlighted the potential implications of these hybrid E. coli strains for public health, emphasizing the need for comprehensive monitoring to mitigate risks associated with the horizontal transfer of virulence-related genes.
2 Materials and methods
2.1 Bacterial strains and serotyping
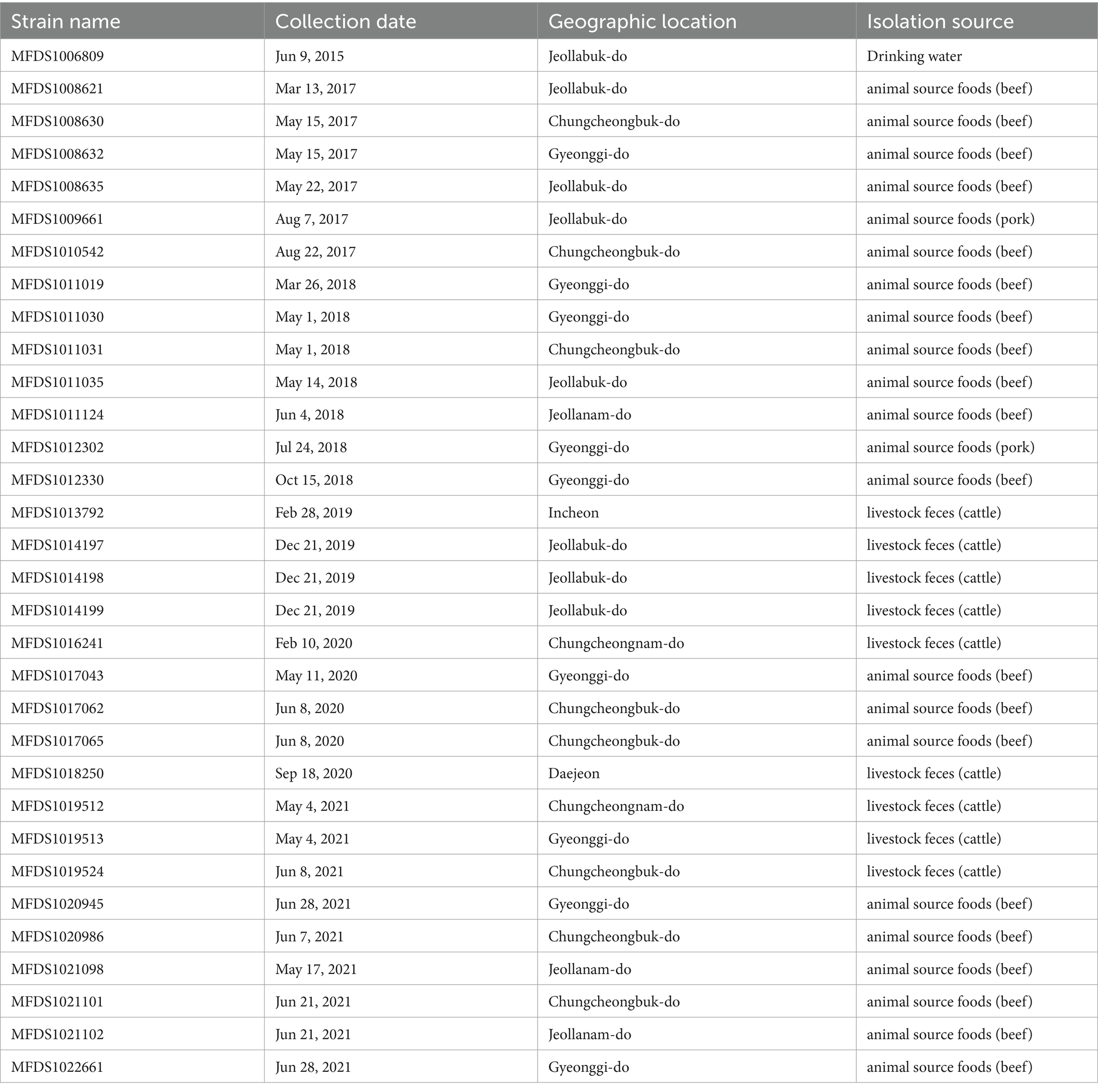
Pathogenic E. coli strains (n = 1,433) were isolated from animal food (beef, pork, chicken, and duck) (n = 961), animal feces (cattle, pigs, and poultry) (n = 358), vegetables such as salad (n = 75), and other (n = 39) during 2015 to 2021 in the Korea and distributed by the Korean Culture Collection for Foodborne Pathogens (Ministry of Food and Drug Safety, Cheongju, Republic of Korea). The 32 hybrid strains used in this study are summarized in Table 1. The strains were identified using a VITEK MS system (BioMérieux Inc., Marcy-l’Etoile, France). Additionally, to identify the serotype, bacteria were tested for agglutination using specific somatic (O1–O181) antisera from the E. coli Reference Laboratory (LREC) in Lugo, Spain. This method helps detect variations in somatic (O) antigens (Guinée et al., 1972, 1981; Blanco et al., 1996).
2.2 Real-time PCR-based identification of hybrid strains
Real-time PCR was performed using a PowerCheckTM 20/15 Pathogen Multiplex Real-time PCR kit (Kogen Biotech Co., Ltd., Seoul, Republic of Korea) to detect virulence genes in different DEC pathogens, including VT1 and VT2 (STEC); bfpA and eaeA (EPEC); LT, STh and STp (ETEC); aggR (EAEC); ipaH (EIEC) (Lee et al., 2023a,b).
2.3 Genome sequencing, assembly, and annotation
Genomic DNA was extracted using the MagListo™ 5 M Genomic DNA Extraction Kit for cells and tissues (Bioneer, Daejeon, Korea) following the manufacturer’s instructions. A DNA library was prepared using the Nextera DNA Flex Library Prep Kit (Illumina, San Diego, CA, USA). Short-read sequencing was performed using the Illumina MiSeq sequencing system with the MiSeq Reagent Kit v3 (600-cycle). A hybrid genome assembly approach was employed to ensure a comprehensive genome sequence by integrating additional long-read sequence data obtained from PacBio Sequel (Pacific Bioscience, Menlo Park, CA, USA) or ONT MinION (Oxford Nanopore Technologies, Oxford, UK). A hybrid assembly of short-read sequence data (Illumina MiSeq) and long-read sequence data (PacBio Sequel, ONT MinION) was performed using Unicycler (v0.4.91; default options). The assembled genome was annotated using the Rapid Annotation with the Subsystem Technology toolkit, implemented in the BV-BRC annotation web service (v3.32.13a).
2.4 DNA sequence analysis and bioinformatics
Antimicrobial resistance genes and virulence factors were identified using ResFinder v4.1 and the virulence factor database (Bortolaia et al., 2020; Liu et al., 2022). Pathogenic and toxigenic factors essential for EPEC pathogenesis in the human intestine, including LEE-encoded virulence factors, were identified using the Basic Local Alignment Search Tool (BLAST). RM10042 and RM10466 were used as reference strains for STEC (Parker et al., 2018). And E2348/69 and E110019 were used as reference strains for EPEC (Iguchi et al., 2009; Hazen and Rasko, 2019). Genome data for each strain were acquired from the NCBI database, with the corresponding accession numbers as follows: RM10042 (CP028122), RM10466 (CP028381), E2348/69 (FM180568), and E110019 (CP035751). MGEs were identified using MobileElementFinder2 (Camacho et al., 2009). Whole genome multi-locus sequence typing (wgMLST) was performed using BioNumerics (v8.0; Applied Maths, Sint-Martens-Latem, Belgium), and the analysis included 17,380 loci from E. coli and Shigella. Additionally, SerotypeFinder 2.0 (Joensen et al., 2015), a web-based serotyping tool, was used to predict the antigen profiles of different E. coli strains.
2.5 Prophage prediction and analysis
The PHAge Search Tool Enhanced Release (PHASTER) (Arndt et al., 2016) was used to detect bacteriophage sequences within distinct genome sequences. PHASTER predicted potential prophage regions, classifying them as “intact (scoring above 90),” “questionable (with scores ranging between 70 and 90),” or “incomplete (scoring below 70),” based on the presence of phage-related genes in the identified phage region of the genome containing Stx. Following the identification of these prophage sequences, the RAST toolkit within the BV-BRC genome annotation web service (v3.32.13a) was used for the annotation of virulence genes.
2.6 Identification of plasmid-associated sequences
PlasmidFinder 2.1 was used to investigate the plasmid features of the entire genome (Carattoli et al., 2014). The threshold for identification was 95% and the minimum coverage was set at 60%. Identification was based on the discovery of replicon sequences from various known plasmid incompatibility (Inc) groups, forming the basis for the identification process. Extracted plasmid sequences were annotated to detect virulence genes, including hemolysin-related genes, using Rapid Annotation with Subsystem Technology tools in the BV-BRC annotation web service (v3.32.13a).
2.7 Phylogenetic analysis and population structure analysis
We conducted a comparative genomic analysis of 32 hybrid STEC/aEPEC strains isolated in South Korea and 187 other pathogenic E. coli strains (Lee et al., 2023a,b). To analyze the pan-genome, we used the bacterial pan-genome analysis (BPGA) tool (v1.3) with default parameters. Clustering was performed using the USEARCH tool with 95% sequence identity as the cutoff value. A phylogenetic tree was constructed using the neighbor-joining method and visualized using the Interactive Tree of Life (iTOL) v6. For population structure analysis, RhierBAPs (Tonkin-Hill et al., 2018) were used.
2.8 Cell culture
HeLa cells (ATCC CCL-2) were grown in Dulbecco’s modified Eagle’s medium (Gibco, Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco) and incubated at 37°C for 24 h in a 5% CO2 environment. Cell monolayers were seeded in 96-well tissue culture plates at a density of 5 × 104 cells/well for the cytotoxicity assay and 1 × 105 cells/well for the invasion assay (Horwitz and Latimer, 2000; Shifrin et al., 2002.).
2.9 Cell cytotoxicity and invasion assays
For cytotoxicity assessment, hybrid STEC/aEPEC strains were prepared as previously described (Roberts et al., 2001). The supernatants from 107 CFU of polymyxin B-treated bacteria were obtained and applied to the cell monolayer. Cell cytotoxicity was evaluated by measuring lactate dehydrogenase (LDH) release using the cytotoxicity detection kit (LDH) (Roche, Basel, Switzerland) according to the manufacturer’s instructions. The absorbance was measured at 495 nm using a SpectraMax i3 platform (Molecular Devices, CA, USA). The results were calculated as relative values compared to those of untreated cells. The LDH release (% cytotoxicity) was calculated according to the following equation: ((LDHtreated – LDHuntreated)/(LDHtotallysis − LDHuntreated)) × 100. The experiments were performed in triplicate in duplicate wells. E. coli DH5α and the STEC strain ATCC 43890 were used as the non-cytotoxic control and cytotoxic positive control in the cytotoxicity assessment (Lang et al., 1994; Robinson et al., 2006).
To assess the invasive abilities of hybrid strains, a classical gentamicin protection assay was performed as previously described (Choi et al., 2007), with minor modifications. The cultures of hybrid STEC/aEPEC strains were applied to the cell monolayer at a multiplicity of infection (MOI) of 100. Invasion ability was determined as the percentage of the initial colony-forming units recovered after gentamicin treatment and expressed as relative invasion (%) of the non-invasive control strain, E. coli DH5α, defined as 100%. The experiments were performed twice in duplicate wells. E. coli DH5α and the EPEC strain E2348/69 were used as the non-invasive control and invasive positive control in the cell invasion assay (Rosa et al., 2001; Robinson et al., 2006; Iguchi et al., 2009).
2.10 Statistical analysis
The significance of differences between the bacterial strains was assessed using the GraphPad Prism 5.01 software, by using the Student’s unpaired t-test. The data are represented as means and standard errors of the mean (SEM) from two independent experiments.
3 Results
3.1 Serotyping and sequence type of STEC/aEPEC hybrids
E. coli O and H antigens were identified using specific somatic (O1–O181) antisera and with the sequence based SerotypeFinder 2.0. The sequence type (ST) was determined using the wgMLST database for E. coli and Shigella species. Serotyping and sequencing results for the 32 hybrid STEC/aEPEC strains are summarized in Table 2. A comparison between in vitro and in silico serotyping showed consistent findings across the 32 hybrid STEC/aEPEC strains. Based on in vitro and in silico serotyping, the 32 hybrid E. coli strains belonged to 10 distinct O:H serogroups [O103:H2 (n = 13), O26:H11 (n = 3), O74:H25 (n = 3), O84:H2 (n = 3), O108:H25 (n = 3), O98:H21 (n = 2), O103:H8 (n = 2), O71:H8 (n = 1), O156:H25 (n = 1), and O177:H25 (n = 1)]. The hybrid strains represented seven STs [ST1034 (n = 11), ST591 (n = 5), ST774 (n = 5), ST481 (n = 4), ST1249 (n = 2), ST1621 (n = 2), ST2366 (n = 2), and ST2277 (n = 1)].
3.2 Genome assemblies of STEC/aEPEC hybrids
Thirty-two hybrid STEC/aEPEC strains from South Korea were sequenced; 16 strains had one chromosome and one plasmid, 11 had one chromosome and two plasmids, and five had one chromosome and three plasmids. Genomic characteristics of the hybrid STEC/aEPEC strains are summarized in Table 2. The genome lengths of these strains ranged from 5,314,960 to 5,984,701 bp. In addition, these strains had G + C contents between 50.3 and 50.8%, the number of coding DNA sequences was between 5,437 and 9,866, tRNA genes between 87 and 105, and rRNA genes between 22 and 26.
3.3 In silico identification of virulence genes
We conducted virulence gene mapping to identify an array of virulence factors within the STEC/aEPEC hybrid genome (Figure 1). All hybrid STEC/aEPEC strains harbored intimin (eae)- and Shiga toxin (stx)-encoding genes. In addition, numerous virulence determinants contributed to E. coli pathogenesis, including LEE-associated genes, enterotoxins, and additional components such as the LEE-locus-encoded type III secretion system (TTSS), ACE type VI secretion system (T6SS), and LEE-encoded TTSS effectors. Detailed results pertaining to virulence gene mapping of these hybrid genomes are documented in Supplementary Table S1. We also identified LEE-encoding genes associated with EPEC pathogenicity. Figure 2 presents a comparative analysis of 32 hybrid STEC/aEPEC strains, STEC strains (RM10042 (Parker et al., 2018) and RM10466 (Parker et al., 2018)), EPEC strains (E2348/69 (Iguchi et al., 2009) and E110019 (Hazen and Rasko, 2019)), and non-O157 EHEC (STEC/EPEC hybrid) strain (O26:H11 strains 11,368, O103:H2 strain 12,009). Genome data for each strain were acquired from the NCBI database, with the corresponding accession numbers as follows: RM10042 (CP028122), RM10466 (CP028381), E2348/69 (FM180568), E110019 (CP035751), 11,368 (AP010953), and 12,009 (AB253581). In a comparative assessment using EPEC strains E2348/69 and E110019 as positive controls, all hybrid STEC/aEPEC strains harbored the LEE region. Hemolysin plays a crucial role as a virulence factor in the prevalence E. coli UTIs and various extraintestinal locations (Elliott et al., 1998). Plasmid-carried enterohemolysin (ehxA) is a ubiquitous exotoxin produced by E. coli that amplifies the virulence of several clinical infections. To investigate the ehxCABD in hybrid STEC/aEPEC strains, we performed plasmid-related sequence analysis using PlasmidFinder 2.1. The analysis revealed the presence of three plasmid replicon sequences (IncFII, IncFIB, and IncY) associated with the established Inc. groups within the STEC/aEPEC genome, as indicated in Figure 3 and Supplementary Figure S1. Hemolysin-related genes were detected in all plasmids found in the 32 hybrid STEC/aEPEC strains (Figure 3).
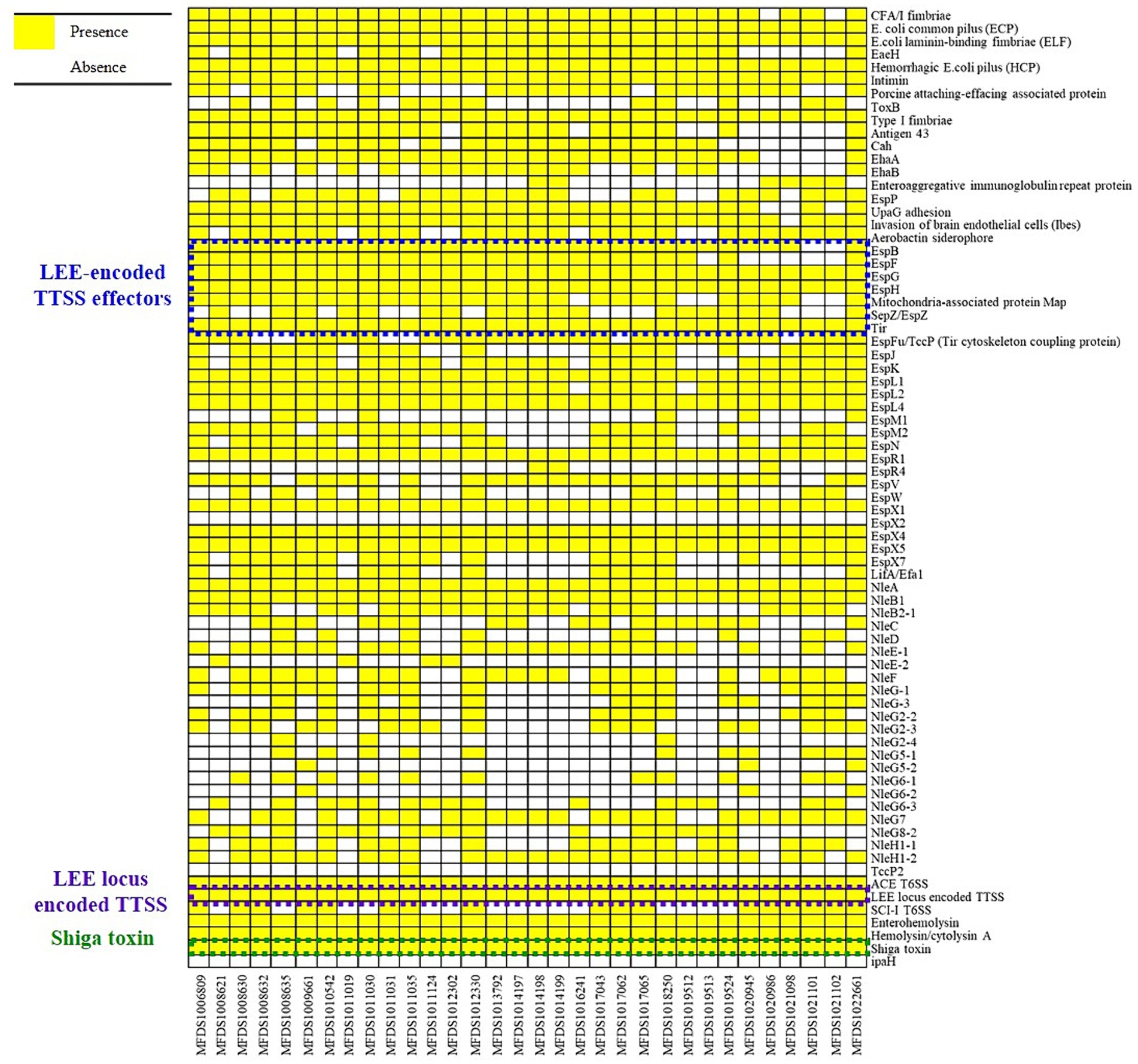
Figure 1. Heatmap of virulence factors across hybrid STEC/aEPEC strains. Heatmap showing the presence/absence of virulence factors (y-axis) within hybrid STEC/aEPEC isolates identified in this study (x-axis). The presence of virulence genes is shown in yellow, and the absence in white. Heatmap produced using the gplot (v3.1.3) package in R (v4.1.3).
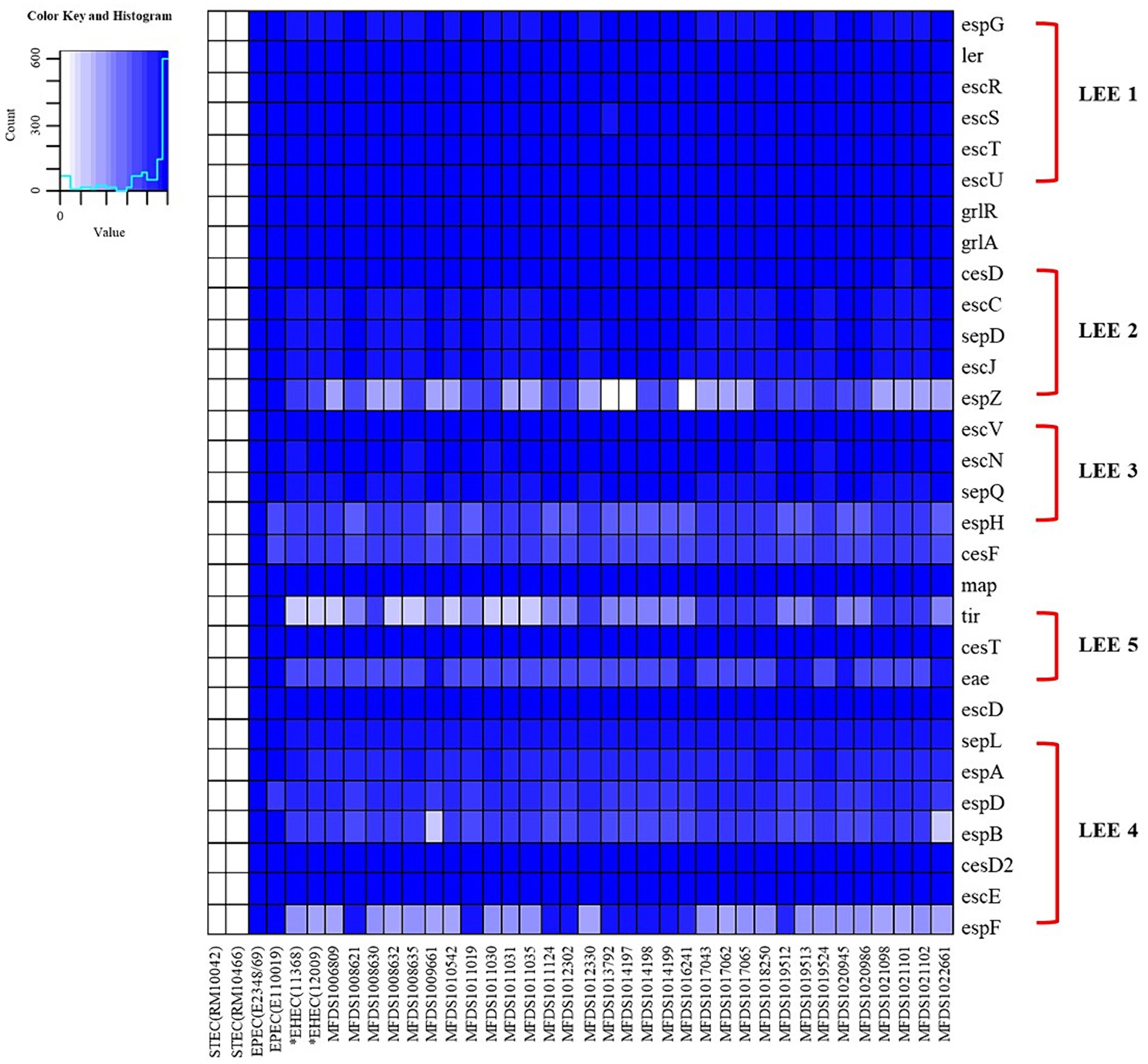
Figure 2. The heatmap of the protein-encoding genes within the LEE region of STEC strains (RM10042 and RM10466), EPEC strains (E2348/69 and E110019), non-O157 EHEC strain (11,368 and 12,009), and STEC/EPEC genomes analyzed in this study. Heatmap showing the presence/absence of protein-encoding genes (y-axis) within hybrid STEC/aEPEC isolates identified in this study (x-axis). The presence of protein-encoding genes within the LEE region is shown in blue, whereas the absence is shown in white, as indicated in the color key. Heatmap produced using the gplot (v3.1.3) package in R (v4.1.3).
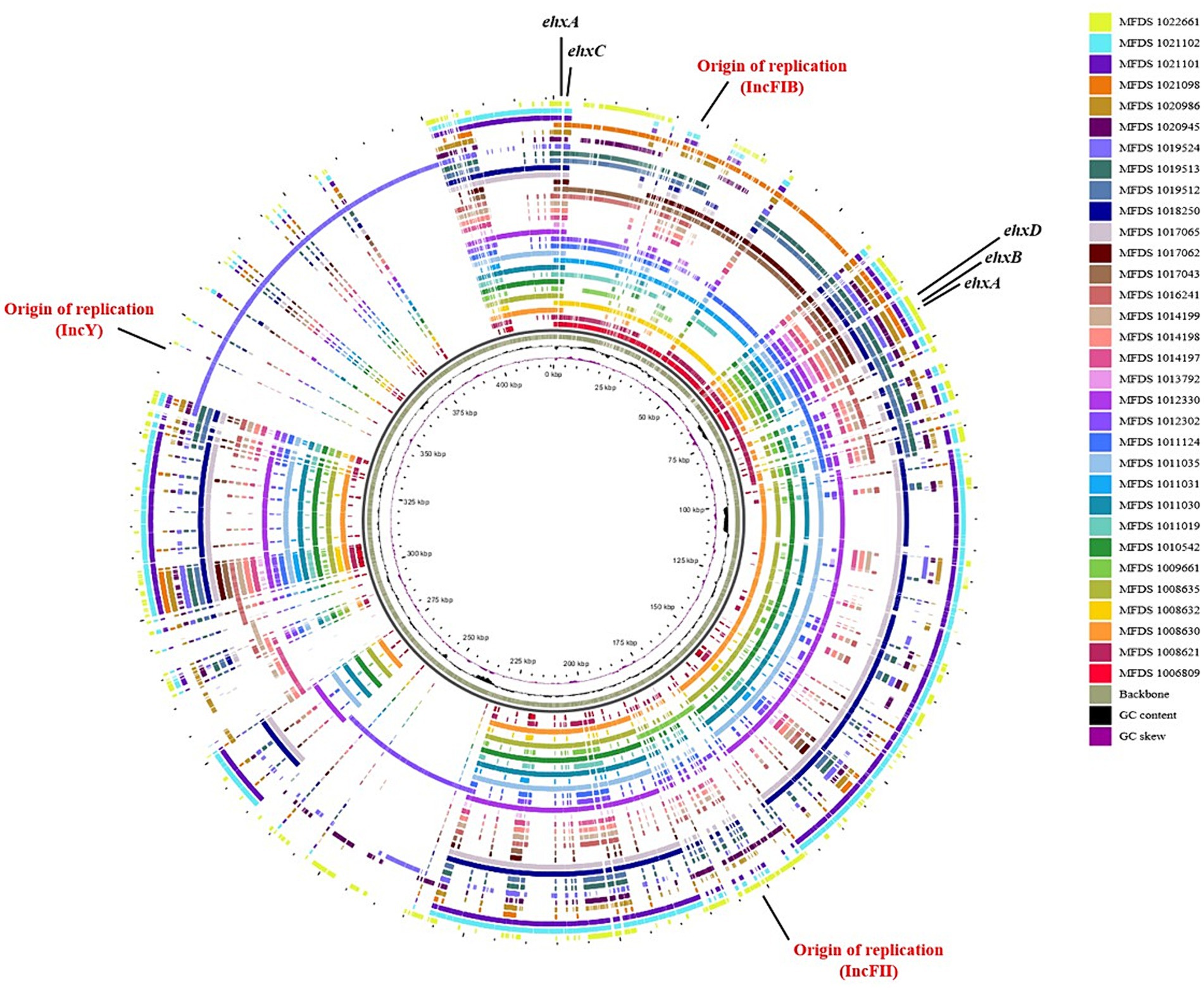
Figure 3. Comparative analysis of hemolysin-encoding plasmids. The pangenome shows the hemolysin-encoding plasmids for the 32 hybrid STEC/aEPEC strains. This pangenome is divided into plasmids IncFI1, IncFIB, IncY, and phage. The graphical circular maps of plasmids were generated using GView.
3.4 In silico identification of antimicrobial resistance genes
ResFinder was used to predict the antimicrobial resistance genes within the hybrid genomes (Figure 4). Only three genomes (MFDS1011030, MFDS1012302, and MFDS1021098) harbored various antimicrobial resistance genes, with streptomycin- and sulfamethoxazole-related antibiotic resistance genes identified as common. Supplementary Table S2 shows the extensive mapping of antimicrobial resistance genes within hybrid E. coli genomes. MFDS1011030 harbored ampicillin resistance genes (TEM-1B), streptomycin resistance genes (aph(3″)-Ib, aph(6)-Id, aph(3′)-la), sulfamethoxazole resistance genes (sul2), and tetracycline resistance genes (tet(A)). MFDS1012302 harbored ampicillin resistance genes (TEM-1B) and streptomycin resistance genes (aph(3″)-Ib, aph(6)-Id), sulfamethoxazole resistance genes (sul2). MFDS1021098 streptomycin resistance genes (aph(3″)-Ib, aph(6)-Id, aph(3′)-la), sulfamethoxazole resistance genes (sul2), and tetracycline resistance genes (tet(B)). In particular, streptomycin resistance genes and sulfamethoxazole genes harbored common among the three STEC/aEPEC hybrids (MFDS1011030, MFDS1012302, and MFDS1021098).
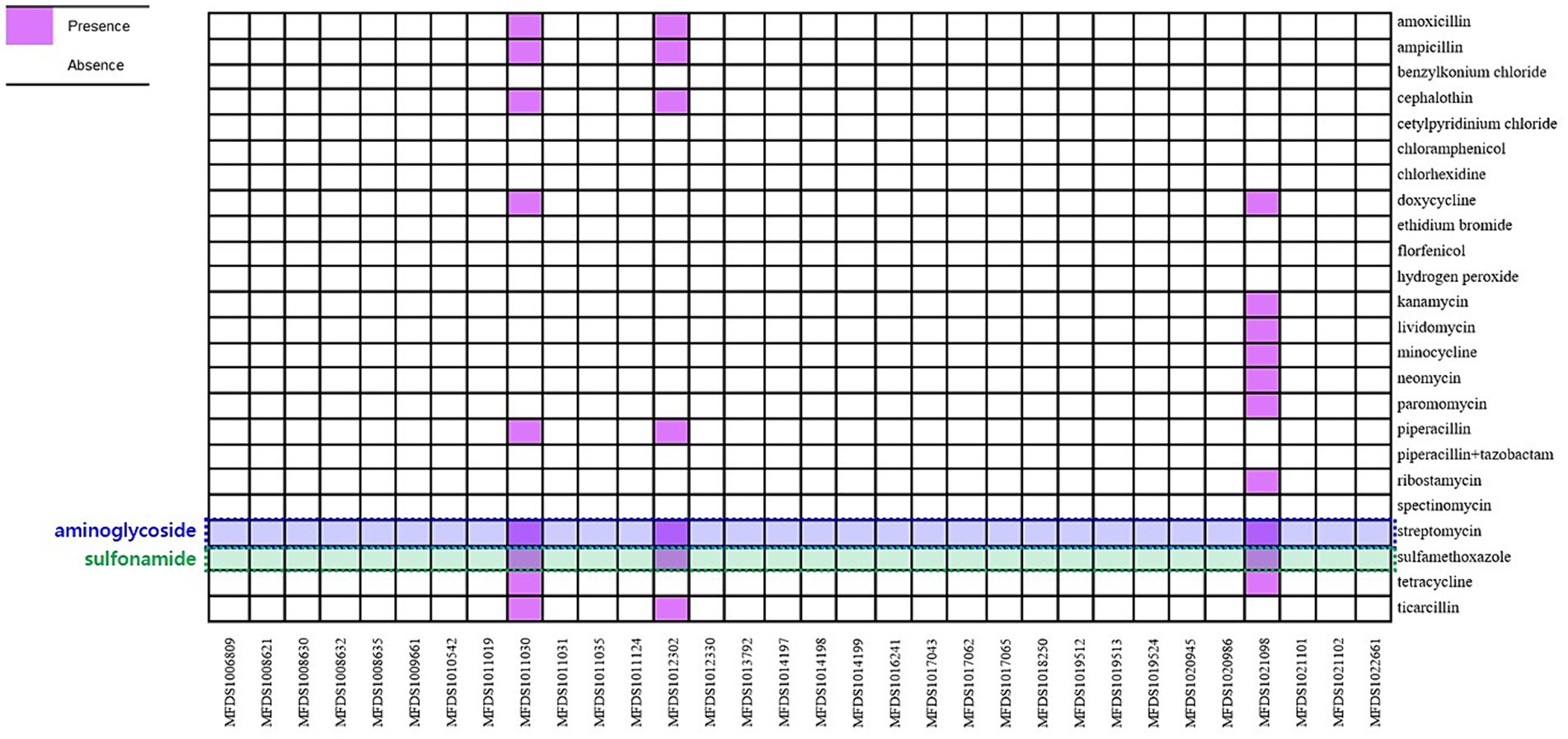
Figure 4. Heatmap of antimicrobial profiles across hybrid STEC/aEPEC strains. Heatmap showing the presence/absence of antimicrobial resistance (y-axis) among the hybrid STEC/aEPEC isolates identified in this study (x-axis). The presence of antimicrobial resistance genes is shown in purple, and the absence in white. Heatmap was generated using the gplot (v3.1.3) package in R software (v4.1.3).
3.5 Phage characterization
To detect the presence of stx-encoding phage in hybrid STEC/aEPEC strains, we conducted a comprehensive identification of bacteriophage sequences using PHASTER. The outcomes of our investigation, involving 32 hybrid STEC/aEPEC strains, are presented in Table 2. Validation of the entire set of stx1 gene sequences was confirmed within the respective “intact” phage sequence region. Notably, the stx1 (stx 1a and stx 1b) gene sequence was detected in most STEC/aEPEC strains (30/32), with only two strains (MFDS1009661 and MFDS1019524) confirmed to possess both stx1 (stx 1a and stx 1b) and stx2 (stx 2a and stx 2b) gene sequences.
3.6 Phylogenetic analysis and population structure analysis
This study aimed to elucidate the genomic relationships between STEC/aEPEC hybrids and other pathogenic E. coli strains. The genomes of 187 isolates, comprising 41 STEC, 46 ETEC, 72 EPEC, 18 EAEC, and 10 EIEC strains, were used for the phylogenetic analysis to determine the genomic relationship between the STEC/aEPEC hybrids and other pathogenic E. coli isolates. The 187 genome datasets included the sequencing results of 160 pathogenic E. coli as well as 32 hybrid STEC/aEPEC genomes, which were deposited in the NCBI database. The genomes analyzed in this study are summarized in Supplementary Table S3. The phylogenetic analysis based on BPGA, which used coding sequences (CDSs) with protein sequences, revealed a close association between all hybrid strains and specific EPEC strains, suggesting the potential acquisition of STEC virulence genes during their emergence (Figure 5A). Furthermore, hierBAPS analysis was based on aligning SNPs from pan-genomes, and it’s utilized to identify genomic similarities among strains through hierarchical clustering of DNA sequence data. The phylogenetic analysis based on hierBAPS revealed their division into six primary sequence clusters (Bayesian analysis of population structure [BAPS] hierarchical level 1), which were further segregated into 28 lineages (BAPS level 2) (Figure 5B). All 32 hybrid strains were closely related to EPEC and were distributed across six groups (two at level 1 and six at level 2) within the total distribution.
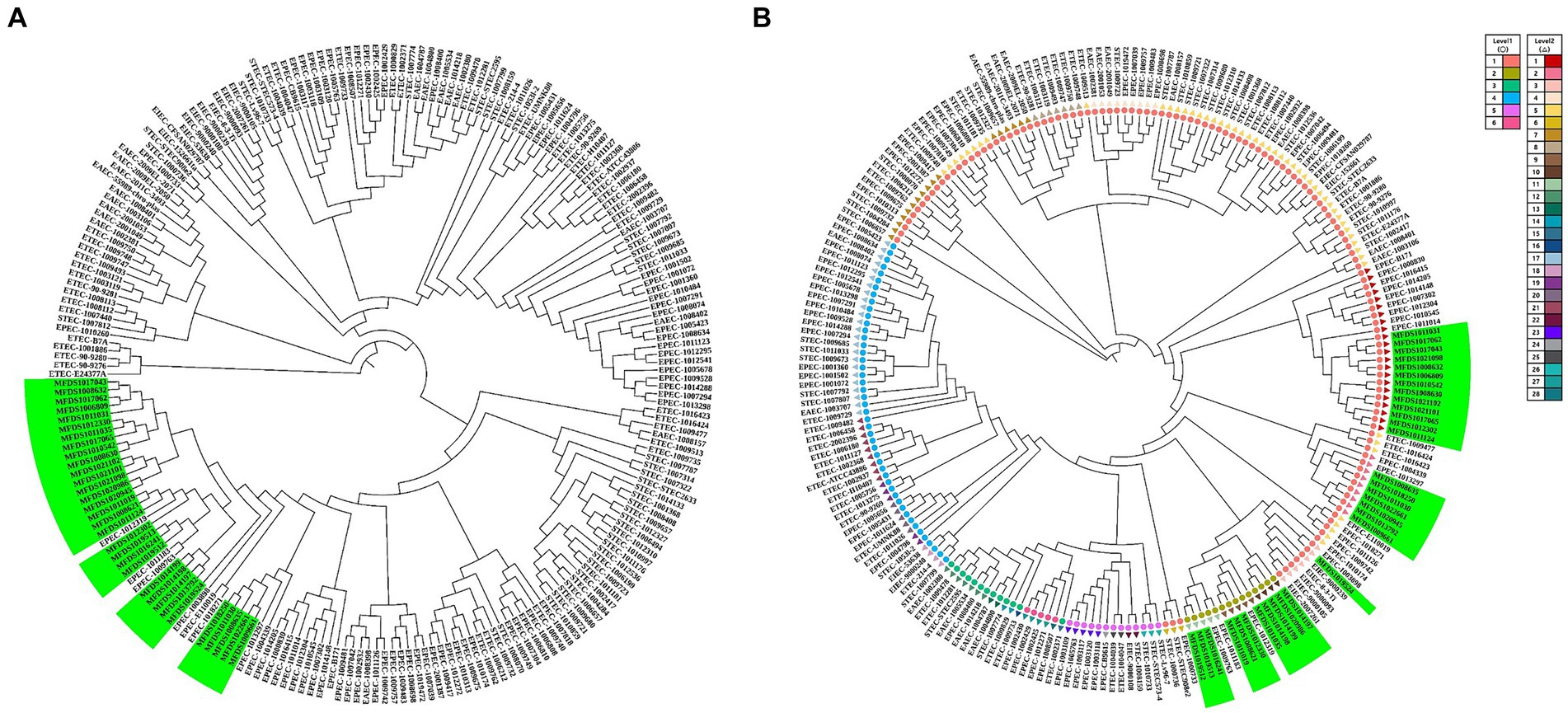
Figure 5. Phylogenetic and population structure analysis of hybrid STEC/aEPEC strains. (A) Green bars represent the 32 hybrid STEC/aEPEC strains that are closely related to specific EPEC strains. (B) Sequence clusters (1 to 6) are indicated in the outer colored dot, which are further divided into 28 lineages (inner ring). The green bar represents 32 STEC/EPEC hybrid strains isolated identified in this study. Among the identified strains, 32 hybrid strains closely related to EPEC were divided into six groups (two level 1, six level 2).
3.7 Virulence of STEC/aEPEC hybrids
As indicated in Figure 1, all STEC/aEPEC hybrids contained both stx- and LEE-encoding genes, suggesting the potential for enhanced virulence in a distinct set of pathotypes. Therefore, we evaluated the cytotoxic activities induced by Stx and the invasive abilities mediated by A/E formation in STEC/aEPEC hybrid strains (Figure 6). The cytotoxicity of the hybrid strains was determined by measuring the levels of cytoplasmic LDH released from HeLa cells. (Figure 6A). All hybrid strains and the STEC reference strain, ATCC 43890, were cytotoxic to HeLa cells (Maldonado et al., 2005), varying from 14% (MFDS1013792) to 51% (MFDS1021101). Among the 32 STEC/aEPEC hybrid strains, six (MFDS1019512, MFDS1019513, MFDS1019524, MFDS1022101, MFDS1021102, and MFDS1022661) exhibited higher cytotoxicity than the STEC strain ATCC 43890, although the differences were not statistically significant (p > 0.05, Student’s t-test). Notably, MFDS1009661 also exhibited high cytotoxic activity, similar to that of the STEC strain ATCC 43890.
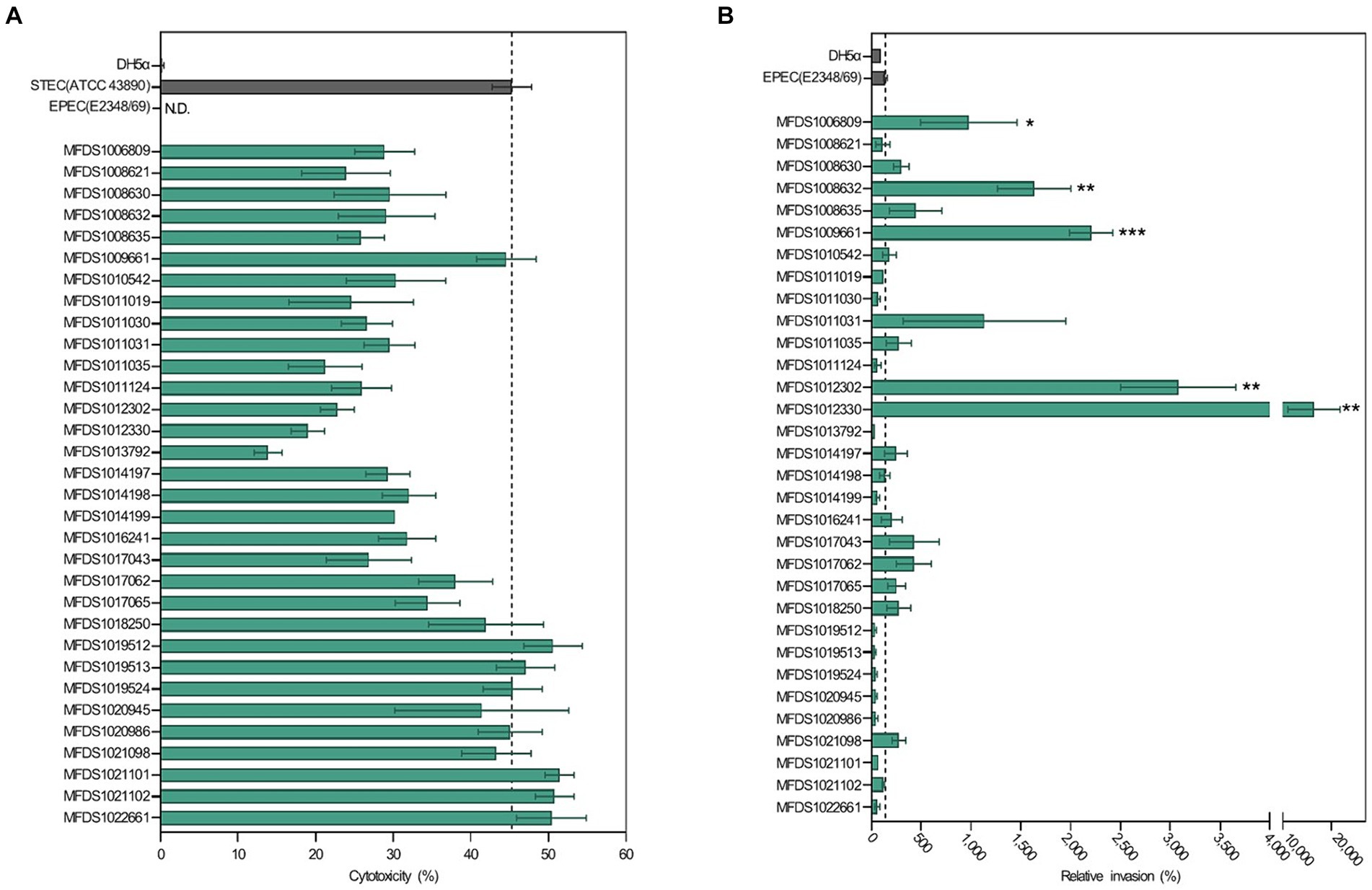
Figure 6. Assessment of cytotoxicity and invasive ability of STEC/aEPEC strains. (A) Cytotoxicity (%) of STEC/aEPEC hybrid strains was measured via LDH release from HeLa cells. Error bars represent S.E.M. from two independent experiments. ATCC 43890 and E2348_69 were used as reference strains for STEC and EPEC, respectively. N.D., Not detected. (B) The invasive ability of STEC/aEPEC hybrid strains was expressed as the relative invasion (%) of the negative control strain E. coli DH5α, defined as 100%. Error bars represent S.E.M. from two independent experiments. Statistical analyses were performed using Student’s t-test. *p < 0.05, **p < 0.01, ***p < =0.005.
The invasive abilities of hybrid STEC/aEPEC strains were also examined and compared to those of the EPEC strain E2348/69 (142%) and E. coli DH5α (100%), respectively (Figure 6B). Notably, five hybrid strains MFDS1006809 (979%, p = 0.0481), MFDS1008632 (1,635%, p = 0.0028), MFDS1009661 (2,207%, p = 0.0001), MFDS1012302 (3,080%, p = 0.0012), and MFDS1012330 (16,401%, p = 0.0078) exhibited significantly higher invasive ability than the control EPEC strain E2348/69 (142%). Moreover, MFDS1017062 and MFDS1021098 showed higher invasive abilities (428 and 280%, respectively) and a greater cytotoxicity potential (38 and 43%, respectively) than the control EPEC strain E2348/69 (142%).
4 Discussion
STEC and EPEC, both foodborne pathogens, can cause severe consequences such as potentially fatal infant diarrhea and hemolytic uremic syndrome, respectively. As STEC strains have the potential to induce diarrhea and hemolytic uremic syndrome (HUS) in human subjects regardless of the presence of LEE, this PAI is an accessory set of virulence genes that enhances STEC pathogenicity. The STEC/EPEC pathotype has been linked to numerous global outbreaks, severe symptoms, and, in some cases, fatal outcomes. Here, we first report 32 hybrid STEC/aEPEC strains identified in South Korea between 2015 and 2021, aiming to profile their virulence and associated genes for deeper exploration of their phylogenetic relationships with other pathogenic E. coli strains. Additionally, the genotypic profiles of AMR were analyzed. Furthermore, a cell-based assay was used to evaluate the virulence potential of hybrid STEC/aEPEC strains.
To molecularly characterize all hybrid STEC/aEPEC strains, a combination of real-time PCR and serotyping was used. WGS analysis of these hybrids corroborated the findings related to the presence of virulence factors, serotyping, and antimicrobial susceptibility. All 32 STEC/EPEC hybrid strains confirmed the presence of genes encoding Shiga toxin 1 and/or 2 (stx1a, stx1b and/or stx2a, stx2b) and E. coli attaching and effacing lesions (eae). Stxs, the major virulence factors of STEC, are broadly classified into two types: stx1 and stx2. Furthermore, in the Stx1 group, stx1a is found in major STEC strains causing HC and HUS in humans, whereas stx1c and stx1d are rarely associated with human infections. In the Stx2 group, stx2a, stx2c, and stx2d were reported to be more virulent in human infections compared to isolates carrying stx2b, stx2e, stx2f, and stx2g (Burgos and Beutin, 2012; Probert et al., 2014; Zhi et al., 2019; Bai et al., 2021). Epidemiological data indicate a stronger association between Stx2-producing E. coli strains and HUS than strains producing Stx1 (Basu et al., 2016). However, the E. coli O157:H7 strain, producing stx1, and not stx2, was isolated from a Korean patient with HUS (Kim et al., 1998). Most hybrid STEC/aEPEC strains isolated from livestock feces, animal source foods, and water in South Korea harbored stx1. EPEC strains were categorized as typical or atypical based on the presence or absence of the E. coli adherence factor plasmid (EAF) housing bfpA, which encodes the bundle-forming pili (BFP). tEPEC includes the LEE region, including eaeA for attaching and effacing lesions, and bfpA. In contrast, aEPEC lacks bfpA. aEPEC causes endemic, epidemic, and persistent diarrhea in both humans and farm animals (Trabulsi et al., 2002), with certain cases linked to acute illness in humans (Abba et al., 2009). Our investigation confirmed that the STEC/EPEC hybrid strains identified in South Korea belonged to the aEPEC category and were distinguished by the absence of a gene encoding BFP.
All hybrid STEC/aEPEC strains encompassed chromosomally encoded virulence genes characteristic of EPEC, such as intimin and TTSS within the LEE region, and additional effectors not linked to LEE. The hallmark of an EPEC infection is the development of A/E lesions within the intestinal epithelium (Cepeda-Molero et al., 2017). These lesions result from genes situated on the LEE pathogenicity island, which encodes adhesin intimin, a TTSS, and six effectors, including the pivotal translocated intimin receptor (Tir) (Cepeda-Molero et al., 2017). In additionally, the chromosome of STEC/aEPEC contains T6SS which along with TTSS are important for cell–cell interaction and virulence factors. Previous studies have shown that the prevalence of the T6SS gene cluster is significantly higher in pathogenic E. coli strains compared to non-pathogenic E. coli strains. Pathogenic strains of E. coli have been identified with multiple T6SSs, some carrying more than one T6SS cluster, while others possess a single T6SS cluster but with multiple copies of hcp and vgrG genes. This highlights the functional role of the T6SS system in enhancing host pathogenicity.
The hybrid STEC/aEPEC strains in South Korea carry a phage-encoded STEC virulence gene (stx) responsible for Stx production. Additionally, phylogenetic tree analysis revealed close relationships between all hybrids and specific EPEC strains. These findings indicated that STEC/aEPEC hybrids potentially originated from EPEC strains that acquired STEC virulence genes via phage transmission. Moreover, our investigation revealed that all the hybrid strains possessed plasmid-carried enterohemolysin genes (ehxCABD). Enterohemolysin (Ehx) is a pore-forming toxin in enterohemorrhagic Escherichia coli that increases virulence in various clinical infections and associated with the presence of shiga toxin (Schwidder et al., 2019). This toxin is encoded on plasmids as part of an operon comprised of four genes: ehxA (coding for functional toxin), ehxC (managing post-translational modification of EhxA), ehxB and ehxD (responsible for the transport of EhxA through the inner membrane) (Kolenda et al., 2021).
The hybrids represented diverse serotypes [O103:H2 (n = 13), O26:H11 (n = 3), O74:H25 (n = 3), O84:H2 (n = 3), O108:H25 (n = 3), O98:H21 (n = 2), O103:H8 (n = 2), O71:H8 (n = 1), O156:H25 (n = 1), and O177:H25 (n = 1)]. The O157:H7 serotype strains are the primary human pathogens among enterohemorrhagic E. coli (EHEC) strains; however, other non-O157 serotypes (O26, O91, O103, O111, O121, and O145) have also exhibited the capacity to cause severe illnesses (Allerberger et al., 2003; Karmali et al., 2003; Kaper et al., 2004; Johnson et al., 2006; Messens et al., 2015; Soysal et al., 2016). Moreover, incidents of EHEC O80:H2 triggering lethal complications in HUS have recently emerged in France (Mariani-Kurkdjian et al., 2014; Soysal et al., 2016), the Netherlands (Wijnsma et al., 2017), and Switzerland (Nüesch-Inderbinen et al., 2018).
The cytotoxicity of Shiga toxin-producing E. coli is linked to the presence of stx (Maldonado et al., 2005). Polymyxin B is effective in extracting intracellular proteins, including heat-labile enterotoxins and Shiga-like toxins, from the periplasmic space of gram-negative bacteria (Evans et al., 1974; Karmali et al., 1985). Using the Polymyxin B extraction method for bacterial preparations, we examined the toxic potential of Stxs in STEC/aEPEC hybrids. All hybrid strains harboring stx and the STEC reference strain ATCC 43890 were cytotoxic to HeLa cells, whereas the EPEC reference strain E2348/69 and non-cytotoxic control E. coli DH5α showed no cytotoxicity. Notably, hybrid strains (MFDS1009661 and MFDS1019524) harboring both stx1 and stx2 were highly cytotoxic (45%) (Table 2), without discernible differences in cytotoxicity from the STEC strain ATCC 43890 (p > 0.05, Student’s t-test). Some hybrid strains lacking the stx2 gene sequence also exhibited high cytotoxicity to HeLa cells, suggesting the involvement of other unidentified virulence genes in the cytotoxicity of hybrid STEC/aEPEC strains.
EPEC adherence occurs through three stages of adherence to epithelial cells: initial adherence (i), signal transduction (ii), and intimate attachment (iii) (Donnenberg et al., 1997). In particular, the EPEC adherence factor (EAF) plasmid confers localized adherence on the epithelial cells called the attaching and effacing (A/E) lesions by expressing BFP, which makes differentiation between tEPEC and aEPEC (Nataro and Kaper, 1998; Trabulsi et al., 2002). It is also distinct from the invasion mechanism of enteroinvasive E. coli (EIEC), which is mediated by the invasion plasmid (pINV) encoding genetic determinants for epithelial cell invasion, including the invasion plasmid antigen gene H (ipaH) (Sethabutr et al., 1993). The results of virulence gene mapping for 32 STEC/aEPEC strains demonstrated that ipaH sequences were not detected in the hybrid genomes (Figure 1). Considering the lack of EAF plasmid in aEPEC strains, the absence of ipaH suggests that the invasive potential of STEC/aEPEC hybrids may be derived from mechanisms different from those of tEPEC or EIEC. Instead, it was reported that some aEPEC strains possessing the intimin-encoding gene (eae) exhibited invasive potential in human intestinal cells in vitro with varying invasion efficiencies (Hernandes et al., 2008; Yamamoto et al., 2009). Despite the absence of the genes responsible for tEPEC and EIEC invasion, the presence of intimin within the genomes of 32 STEC/aEPEC hybrids may support the possibility of highly invasive strains. However, further analyses are still needed to evaluate the ability of hybrid strains to invade the epithelial cells in vivo. As the STEC strains are inefficient in invading epithelial cells (Oelschlaeger et al., 1994), invasive ability distinguishes EPEC from STEC types. The five hybrid strains were highly invasive to HeLa cells compared to the EPEC strain E2348/69. Notably, all five hybrid strains were related to EPEC in the phylogenetic analysis, suggesting a correlation between the phylogenetic and virulence profiles of the STEC/aEPEC hybrids. Moreover, all ST 1034 (n = 11) hybrid STEC/aEPEC strains displayed higher invasive potential than the EPEC strain E2348/69, including the highly invasive and cytotoxic hybrid strains MFDS1017062 and MFDS1021098. In particular, these genomes harbored two T6SS gene clusters, each containing the 13 core T6SS genes (TssJ, TssL, TssM, TssK, TssF, TssG, TssE, TssA, TssB, TssC, ClpV, Hcp, VgrG) necessary for the proper functioning of a T6SS. These results suggested the need to explore possible correlations between STs and virulence profiles in hybrid E. coli strains.
This study investigated the transfer of virulence genes in hybrid STEC/aEPEC strains identified in South Korea, emphasizing the efficacy of WGS as a robust method for scrutinizing bacterial genomes, particularly in detecting regions containing MGEs like phages and plasmids. Antibiotic resistance was confirmed in only three hybrid STEC/aEPEC strains (MFDS1011030, MFDS1012302, and MFDS1021098), displaying multiple drug resistance. Moreover, five hybrid strains (MFDS1006809, MFDS1008632, MFDS1009661, MFDS1012302, and MFDS1012330) showed cytotoxic activity against HeLa cells and significantly higher invasive ability than the EPEC strain E2348/69. These genomic and virulence findings substantially contribute to our understanding of hybrid STEC/aEPEC strains and pave the way for further investigations of E. coli pathogenicity. Furthermore, this study observes the emergence of MDR hybrid E. coli strains with high virulence potential.
5 Conclusion
This study documented the virulence markers and antimicrobial resistance profiles of hybrid STEC/aEPEC strains isolated in South Korea. Comprehensive genome-wide phylogenetic analysis demonstrated close genetic relationships between all hybrids and specific EPEC strains. Furthermore, the genes encoding stx1 or stx2 within these hybrid strains were harbored by phages. Additionally, some hybrid strains exhibited cytotoxicity and invasive abilities in human epithelial HeLa cells. Notably, all hybrid STEC/aEPEC strains that belonged to ST 1034 (n = 11) had a higher invasive potential than the EPEC strain E2348/69, including the two highly invasive and highly cytotoxic hybrid strains MFDS1017062 and MFDS1021098. Our results emphasize the potential increased danger to humans posed by hybrid STEC/aEPEC strains isolated in South Korea, containing both stx and eaeA, compared to STEC or EPEC alone. Furthermore, these genomic and virulence findings considerably contribute to our understanding of hybrid STEC/aEPEC strains and encourage further investigations into E. coli pathogenicity.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
WL: Data curation, Methodology, Writing – original draft. JH: Data curation, Methodology, Writing – review & editing. JC: Data curation, Methodology, Writing – review & editing. YJ: Data curation, Methodology, Writing – review & editing. EK: Methodology, Writing – review & editing. EA: Methodology, Writing – review & editing. SeK: Writing – review & editing. HS: Writing – review & editing. SR: Writing – review & editing. SoK: Conceptualization, Supervision, Writing – review & editing. H-YK: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the Ministry of Food and Drug Safety (grant number 23194MFDS017). The findings and conclusions of this study are our own and do not necessarily represent the views of the Ministry of Food and Drug Safety.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1398262/full#supplementary-material
Footnotes
References
Abba, K., Sinfield, R., Hart, C. A., and Garner, P. (2009). Pathogens associated with persistent diarrhoea in children in low and middle income countries: systematic review. BMC Infect. Dis. 9:88. doi: 10.1186/1471-2334-9-88
Allerberger, F., Friedrich, A. W., Grif, K., Dierich, M. P., Dornbusch, H. J., Mache, C. J., et al. (2003). Hemolytic-uremic syndrome associated with enterohemorrhagic Escherichia coli O26:H infection and consumption of unpasteurized cow’s milk. Int. J. Infect. Dis. 7, 42–45. doi: 10.1016/s1201-9712(03)90041-5
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bai, X., Scheutz, F., Dahlgren, H. M., Hedenström, I., and Jernberg, C. (2021). Characterization of clinical Escherichia coli strains producing a novel Shiga toxin 2 subtype in Sweden and Denmark. Microorganisms 9:2374. doi: 10.3390/microorganisms9112374
Bai, X., Zhang, J., Ambikan, A., Jernberg, C., Ehricht, R., Scheutz, F., et al. (2019). Molecular characterization and comparative genomics of clinical hybrid Shiga toxin-producing and enterotoxigenic Escherichia coli (STEC/ETEC) strains in Sweden. Sci. Rep. 9:5619. doi: 10.1038/s41598-019-42122-z
Basu, D., Li, X. P., Kahn, J. N., May, K. L., Kahn, P. C., and Tumer, N. E. (2016). The A1 subunit of Shiga toxin 2 has higher affinity for ribosomes and higher catalytic activity than the A1 subunit of Shiga toxin 1. Infect. Immun. 84, 149–161. doi: 10.1128/IAI.00994-15
Bielaszewska, M., Mellmann, A., Zhang, W., Köck, R., Fruth, A., Bauwens, A., et al. (2011). Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. Lancet Infect. Dis. 11, 671–676. doi: 10.1016/S1473-3099(11)70165-7
Blanco, J. E., Blanco, M., Blanco, J., Mora, A., Balaguer, L., Mouriño, M., et al. (1996). O serogroups, biotypes, and eae genes in Escherichia coli strains isolated from diarrheic and healthy rabbits. J. Clin. Microbiol. 34, 3101–3107. doi: 10.1128/jcm.34.12.3101-3107.1996
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Buchholz, U., Bernard, H., Werber, D., Böhmer, M. M., Remschmidt, C., Wilking, H., et al. (2011). German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 365, 1763–1770. doi: 10.1056/NEJMoa1106482
Burgos, Y., and Beutin, L. (2012). “Evaluation of an immuno-chromatographic detection system for Shiga toxins and the E. coli O157 antigen” in Trends in Immunolabelled and related techniques. ed. E. Abuelzein (Berlin: InTech), 29–40.
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). Blast+: architecture and applications. BMC Bioinformatics 10, 1–9. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cepeda-Molero, M., Berger, C. N., Walsham, A. D. S., Ellis, S. J., Wemyss-Holden, S., Schüller, S., et al. (2017). Attaching and effacing (a/E) lesion formation by enteropathogenic E. coli on human intestinal mucosa is dependent on non-LEE effectors. PLoS Pathog. 13:e1006706. doi: 10.1371/journal.ppat.1006706
Choi, J., Shin, D., and Ryu, S. (2007). Implication of quorum sensing in Salmonella enterica serovar typhimurium virulence: the luxS gene is necessary for expression of genes in pathogenicity island 1. Infect. Immun. 75, 4885–4890. doi: 10.1128/IAI.01942-06
Croxen, M. A., and Finlay, B. B. (2010). Molecular mechanisms of Escherichia coli pathogenicity. Nat. Rev. Microbiol. 8, 26–38. doi: 10.1038/nrmicro2265
Croxen, M. A., Law, R. J., Scholz, R., Keeney, K. M., Wlodarska, M., and Finlay, B. B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. doi: 10.1128/CMR.00022-13
de Lira, D. R. P., Cavalcanti, A. M. F., Pinheiro, S. R. S., Orsi, H., Dos Santos, L. F., and Hernandes, R. T. (2021). Identification of a hybrid atypical enteropathogenic and enteroaggregative Escherichia coli (aEPEC/EAEC) clone of serotype O3:H2 associated with a diarrheal outbreak in Brazil. Braz. J. Microbiol. 52, 2075–2079. doi: 10.1007/s42770-021-00580-6
Dobrindt, U. (2005). (Patho-)genomics of Escherichia coli. Int. J. Med. Microbiol. 295, 357–371. doi: 10.1016/j.ijmm.2005.07.009
Donnenberg, M. S., Kaper, J. B., and Finlay, B. B. (1997). Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 5, 109–114. doi: 10.1016/S0966-842X(97)01000-7
Dutta, S., Guin, S., Ghosh, S., Pazhani, G. P., Rajendran, K., Bhattacharya, M. K., et al. (2013). Trends in the prevalence of diarrheagenic Escherichia coli among hospitalized diarrheal patients in Kolkata, India. PLoS One 8:e56068. doi: 10.1371/journal.pone.0056068
Dutta, S., Pazhani, G. P., Nataro, J. P., and Ramamurthy, T. (2015). Heterogenic virulence in a diarrheagenic Escherichia coli: evidence for an EPEC expressing heat-labile toxin of ETEC. Int. J. Med. Microbiol. 305, 47–54. doi: 10.1016/j.ijmm.2014.10.006
Eichhorn, I., Heidemanns, K., Semmler, T., Kinnemann, B., Mellmann, A., Harmsen, D., et al. (2015). Highly virulent non-O157 enterohemorrhagic Escherichia coli (EHEC) serotypes reflect similar phylogenetic lineages, providing new insights into the evolution of EHEC. Appl. Environ. Microbiol. 81, 7041–7047. doi: 10.1128/AEM.01921-15
Elliott, S. J., Srinivas, S., Albert, M. J., Alam, K., Robins-Browne, R. M., Gunzburg, S. T., et al. (1998). Characterization of the roles of hemolysin and other toxins in enteropathy caused by alpha-hemolytic Escherichia coli linked to human diarrhea. Infect. Immun. 66, 2040–2051. doi: 10.1128/IAI.66.5.2040-2051.1998
Evans, D. J. Jr., Evans, D. G., and Gorbach, S. L. (1974). Polymyxin B-induced release of low-molecular-weight, heat-labile enterotoxin from Escherichia coli. Infect. Immun. 10, 1010–1017. doi: 10.1128/iai.10.5.1010-1017.1974
Furukawa, I., Suzuki, M., Masaoka, T., Nakajima, N., Mitani, E., Tasaka, M., et al. (2018). Outbreak of enterohemorrhagic Escherichia coli O157:H7 infection associated with minced meat cutlets consumption in Kanagawa, Japan. J. Infect. Dis. 71, 436–441. doi: 10.7883/yoken.JJID.2017.495
Geurtsen, J., de Been, M., Weerdenburg, E., Zomer, A., McNally, A., and Poolman, J. (2022). Genomics and pathotypes of the many faces of Escherichia coli. FEMS Microbiol. Rev. 46:fuac031. doi: 10.1093/femsre/fuac031
Gioia-Di Chiacchio, R. M., Cunha, M. P. V., de Sá, L. R. M., Davies, Y. M., Pereira, C. B. P., Martins, F. H., et al. (2018). Novel hybrid of typical enteropathogenic Escherichia coli and Shiga-toxin-producing E. coli (tEPEC/STEC) emerging from pet birds. Front. Microbiol. 9:2975. doi: 10.3389/fmicb.2018.02975
Gomes, T. A. T., Elias, W. P., Scaletsky, I. C. A., Guth, B. E. C., Rodrigues, J. F., Piazza, R. M. F., et al. (2016). Diarrheagenic Escherichia coli. Brazilian. J. Microbiol. 47, 3–30. doi: 10.1016/j.bjm.2016.10.015
Guinée, P. A. M., Agterberg, C. M., and Jansen, W. H. (1972). Escherichia coli O antigen typing by means of a mechanized microtechnique. Appl. Microbiol. 24, 127–131. doi: 10.1128/am.24.1.127-131.1972
Guinée, P. A. M., Jansen, W. H., Wadström, T., and Sellwood, R. (1981). Escherichia coli associated with neonatal diarrhoea in piglets and calves. In: P. W. Leeuwde and P. A. M. Guinée (eds) Laboratory diagnosis in neonatal calf and pig diarrhoea. Dordrecht: Springer.
Hazen, T. H., Donnenberg, M. S., Panchalingam, S., Antonio, M., Hossain, A., Mandomando, I., et al. (2016). Genomic diversity of EPEC associated with clinical presentations of differing severity. Nat. Microbiol. 1:15014. doi: 10.1038/nmicrobiol.2015.14
Hazen, T. H., and Rasko, D. A. (2019). The complete genome of the atypical enteropathogenic Escherichia coli archetype isolate E110019 highlights a role for plasmids in dissemination of the type III secreted effector EspT. Infect. Immun. 87:e00412-19. doi: 10.1128/IAI.00412-19
Hernandes, R. T., Silva, R. M., Carneiro, S. M., Salvador, F. A., Fernandes, M. C. C., Padovan, A. C. B., et al. (2008). The localized adherence pattern of an atypical enteropathogenic Escherichia coli is mediated by intimin omicron and unexpectedly promotes HeLa cell invasion. Cell. Microbiol. 10, 415–425. doi: 10.1111/j.1462-5822.2007.01054.x
Horwitz, W., and Latimer, G. W. (2000). Official methods of analysis of AOAC international. Gaithersburg: AOAC International.
Iguchi, A., Thomson, N. R., Ogura, Y., Saunders, D., Ooka, T., Henderson, I. R., et al. (2009). Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J. Bacteriol. 191, 347–354. doi: 10.1128/JB.01238-08
Jesser, K. J., and Levy, K. (2020). Updates on defining and detecting diarrheagenic Escherichia coli pathotypes. Curr. Opin. Infect. Dis. 33, 372–380. doi: 10.1097/QCO.0000000000000665
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Johnson, K. E., Thorpe, C. M., and Sears, C. L. (2006). The emerging clinical importance of non-O157 Shiga toxin-producing Escherichia coli. Clin. Infect. Dis. 43, 1587–1595. doi: 10.1086/509573
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Karmali, M. A., Mascarenhas, M., Shen, S., Ziebell, K., Johnson, S., Reid-Smith, R., et al. (2003). Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. J. Clin. Microbiol. 41, 4930–4940. doi: 10.1128/JCM.41.11.4930-4940.2003
Karmali, M. A., Petric, M., Lim, C., Cheung, R., and Arbus, G. S. (1985). Sensitive method for detecting low numbers of verotoxin-producing Escherichia coli in mixed cultures by use of colony sweeps and polymyxin extraction of verotoxin. J. Clin. Microbiol. 22, 614–619. doi: 10.1128/jcm.22.4.614-619.1985
Kashima, K., Sato, M., Osaka, Y., Sakakida, N., Kando, S., Ohtsuka, K., et al. (2021). An outbreak of food poisoning due to Escherichia coli serotype O7:H4 carrying astA for enteroaggregative E. coli heat-stable enterotoxin1 (EAST1). Epidemiol. Infect. 149:e244. doi: 10.1017/S0950268821002338
Kim, Y. B., Okuda, J., Matsumoto, C., Morigaki, T., Asai, N., Watanabe, H., et al. (1998). Isolation of an Escherichia coli O157:H7 strain producing Shiga toxin 1 but not Shiga toxin 2 from a patient with hemolytic uremic syndrome in Korea. FEMS Microbiol. Lett. 166, 43–48. doi: 10.1111/j.1574-6968.1998.tb13181.x
Kolenda, R., Sidorczuk, K., Noszka, M., Aleksandrowicz, A., Khan, M. M., Burdukiewicz, M., et al. (2021). Genome placement of alpha-haemolysin cluster is associated with alpha-haemolysin sequence variation, adhesin and iron acquisition factor profile of Escherichia coli. Microb. Genom. 7:000743. doi: 10.1099/mgen.0.000743
Lang, A. L., Tsai, Y. L., Mayer, C. L., Patton, K. C., and Palmer, C. J. (1994). Multiplex PCR for detection of the heat-labile toxin gene and Shiga-like toxin I and II genes in Escherichia coli isolated from natural waters. Appl. Environ. Microbiol. 60, 3145–3149. doi: 10.1128/aem.60.9.3145-3149.1994
Lee, W., Kim, M. H., Sung, S., Kim, E., An, E. S., Kim, S. H., et al. (2023a). Genome-based characterization of hybrid Shiga toxin-producing and enterotoxigenic Escherichia coli (STEC/ETEC) strains isolated in South Korea, 2016–2020. Microorganisms 11:1285. doi: 10.3390/microorganisms11051285
Lee, W., Sung, S., Ha, J., Kim, E., An, E. S., Kim, S. H., et al. (2023b). Molecular and genomic analysis of the virulence factors and potential transmission of hybrid enteropathogenic and enterotoxigenic Escherichia coli (EPEC/ETEC) strains isolated in South Korea. Int. J. Mol. Sci. 24:12729. doi: 10.3390/ijms241612729
Lim, M. A., Kim, J. Y., Acharya, D., Bajgain, B. B., Park, J. H., Yoo, S. J., et al. (2020). A diarrhoeagenic Enteropathogenic Escherichia coli (EPEC) infection outbreak that occurred among elementary school children in Gyeongsangbuk-Do Province of South Korea was associated with consumption of water-contaminated food items. Int. J. Environ. Res. Public Health 17:3149. doi: 10.3390/ijerph17093149
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Maldonado, Y., Fiser, J. C., Nakatsu, C. H., and Bhunia, A. K. (2005). Cytotoxicity potential and genotypic characterization of Escherichia coli isolates from environmental and food sources. Appl. Environ. Microbiol. 71, 1890–1898. doi: 10.1128/AEM.71.4.1890-1898.2005
Mariani-Kurkdjian, P., Lemaître, C., Bidet, P., Perez, D., Boggini, L., Kwon, T., et al. (2014). Haemolytic-uraemic syndrome with bacteraemia caused by a new hybrid Escherichia coli pathotype. New Microbes New Infect. 2, 127–131. doi: 10.1002/nmi2.49
Messens, W., Bolton, D., Frankel, G., Liebana, E., McLauchlin, J., Morabito, S., et al. (2015). Defining pathogenic verocytotoxin-producing Escherichia coli (VTEC) from cases of human infection in the European Union, 2007–2010. Epidemiol. Infect. 143, 1652–1661. doi: 10.1017/S095026881400137X
Miliwebsky, E., Schelotto, F., Varela, G., Luz, D., Chinen, I., and Piazza, R. M. (2016). “Human diarrheal infections: diagnosis of diarrheagenic Escherichia coli Pathotypes” in Escherichia coli in the Americas. ed. A. G. Torres (Switzerland: Springer), 343–369.
Mulchandani, R., Brehmer, C., Butt, S., Vishram, B., Harrison, M., Marchant, E., et al. (2021). Outbreak of Shiga toxin-producing Escherichia coli O157 linked with consumption of a fast-food product containing imported cucumbers, United Kingdom, august 2020. Int. J. Infect. Dis. 110, S62–S68. doi: 10.1016/j.ijid.2021.04.001
Nataro, J. P., and Kaper, J. B. (1998). Diarrheagenic escherichia coli. Clin. Microbiol. Rev. 11, 142–201. doi: 10.1128/cmr.11.1.142
Newitt, S., MacGregor, V., Robbins, V., Bayliss, L., Chattaway, M. A., Dallman, T., et al. (2016). Two linked enteroinvasive Escherichia coli outbreaks, Nottingham, UK, June 2014. Emerg. Infect. Dis. 22, 1178–1184. doi: 10.3201/eid2207.152080
Nüesch-Inderbinen, M., Cernela, N., Wüthrich, D., Egli, A., and Stephan, R. (2018). Genetic characterization of Shiga toxin producing Escherichia coli belonging to the emerging hybrid pathotype O80:H2 isolated from humans 2010–2017 in Switzerland. Int. J. Med. Microbiol. 308, 534–538. doi: 10.1016/j.ijmm.2018.05.007
Oelschlaeger, T. A., Barrett, T. J., and Kopecko, D. J. (1994). Some structures and processes of human epithelial cells involved in uptake of enterohemorrhagic Escherichia coli O157:H7 strains. Infect. Immun. 62, 5142–5150. doi: 10.1128/iai.62.11.5142-5150.1994
Pakbin, B., Brück, W. M., and Rossen, J. W. A. (2021). Virulence factors of enteric pathogenic Escherichia coli: a review. Int. J. Mol. Sci. 22:9922. doi: 10.3390/ijms22189922
Parker, C. T., Cooper, K. K., Huynh, S., Smith, T. P., Bono, J. L., and Cooley, M. (2018). Genome sequences of eight Shiga toxin-producing Escherichia coli strains isolated from a produce-growing region in California. Microbiol. Resour. Announc. 7:10. doi: 10.1128/MRA.00807-18
Probert, W. S., McQuaid, C., and Schrader, K. (2014). Isolation and identification of an Enterobacter cloacae strain producing a novel subtype of Shiga toxin type 1. J. Clin. Microbiol. 52, 2346–2351. doi: 10.1128/jcm.00338-14
Roberts, P. H., Davis, K. C., Garstka, W. R., and Bhunia, A. K. (2001). Lactate dehydrogenase release assay from Vero cells to distinguish verotoxin producing Escherichia coli from non-verotoxin producing strains. J. Microbiol. Methods 43, 171–181. doi: 10.1016/s0167-7012(00)00222-0
Robinson, C. M., Sinclair, J. F., Smith, M. J., and O’Brien, A. D. (2006). Shiga toxin of enterohemorrhagic Escherichia coli type O157:H7 promotes intestinal colonization. Proc. Natl. Acad. Sci. USA 103, 9667–9672. doi: 10.1073/pnas.0602359103
Rosa, A. C., Vieira, M. A., Tibana, A., Gomes, T. A., and Andrade, J. R. (2001). Interactions of Escherichia coli strains of non-EPEC serogroups that carry eae and lack the EAF and stx gene sequences with undifferentiated and differentiated intestinal human Caco-2 cells. FEMS Microbiol. Lett. 200, 117–122. doi: 10.1111/j.1574-6968.2001.tb10702.x
Santos, A. C. M., Santos, F. F., Silva, R. M., and Gomes, T. A. T. (2020). Diversity of hybrid- and hetero-pathogenic Escherichia coli and their potential implication in more severe diseases. Front. Cell. Infect. Microbiol. 10:339. doi: 10.3389/fcimb.2020.00339
Schwidder, M., Heinisch, L., and Schmidt, H. (2019). Genetics, toxicity, and distribution of Enterohemorrhagic Escherichia coli Hemolysin. Toxins 11:502. doi: 10.3390/toxins11090502
Sethabutr, O., Venkatesan, M., Murphy, G. S., Eampokalap, B., Hoge, C. W., and Echeverria, P. (1993). Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J. Infect. Dis. 167, 458–461. doi: 10.1093/infdis/167.2.458
Setyarini, W., Raharjo, D., Arizandy, R. Y., Pamoengkas, Z., Sudarmo, S. M., Athiyyah, A. F., et al. (2020). Molecular detection of a new pathotype enteroaggregative haemorrhagic Escherichia coli (EAHEC) in Indonesia, 2015. Infect. Dis. Rep. 12:8745. doi: 10.4081/idr.2020.8745
Shifrin, Y., Kirschner, J., Geiger, B., and Rosenshine, I. (2002). Enteropathogenic Escherichia coli induces modification of the focal adhesions of infected host cells. Cell. Microbiol. 4, 235–243. doi: 10.1046/j.1462-5822.2002.00188.x
Silva, M., Santos, A., Rocha, L., Caetano, B., Mitsunari, T., Santos, L., et al. (2019). Development and validation of Shiga toxin-producing Escherichia coli immunodiagnostic assay. Microorganisms 7:276. doi: 10.3390/microorganisms7090276
Soysal, N., Mariani-Kurkdjian, P., Smail, Y., Liguori, S., Gouali, M., Loukiadis, E., et al. (2016). Enterohemorrhagic Escherichia coli hybrid pathotype O80:H2 as a new therapeutic challenge. Emerg. Infect. Dis. 22, 1604–1612. doi: 10.3201/eid2209.160304
Tonkin-Hill, G., Lees, J. A., Bentley, S. D., Frost, S. D. W., and Corander, J. (2018). RhierBAPS: an R implementation of the population clustering algorithm hierBAPS. Wellcome Open Res. 3:93. doi: 10.12688/wellcomeopenres.14694.1
Trabulsi, L. R., Keller, R., and Tardelli Gomes, T. A. (2002). Typical and atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8, 508–513. doi: 10.3201/eid0805.010385
Valiatti, T. B., Santos, F. F., Santos, A. C. M., Nascimento, J. A. S., Silva, R. M., Carvalho, E., et al. (2020). Genetic and virulence characteristics of a hybrid atypical enteropathogenic and uropathogenic Escherichia coli (aEPEC/UPEC) strain. Front. Cell. Infect. Microbiol. 10:492. doi: 10.3389/fcimb.2020.00492
Vieira, M. A., Dos Santos, L. F., Dias, R. C. B., Camargo, C. H., Pinheiro, S. R. S., Gomes, T. A. T., et al. (2016). Atypical enteropathogenic Escherichia coli as aetiologic agents of sporadic and outbreak-associated diarrhoea in Brazil. J. Med. Microbiol. 65, 998–1006. doi: 10.1099/jmm.0.000313
Wijnsma, K. L., Schijvens, A. M., Rossen, J. W. A., Kooistra-Smid, A. M. D. M., Schreuder, M. F., and van de Kar, N. C. A. J. (2017). Unusual severe case of hemolytic uremic syndrome due to Shiga toxin 2d-producing E. coli O80:H2. Pediatr. Nephrol. 32, 1263–1268. doi: 10.1007/s00467-017-3642-3
Yamamoto, D., Hernandes, R. T., Blanco, M., Greune, L., Schmidt, M. A., Carneiro, S. M., et al. (2009). Invasiveness as a putative additional virulence mechanism of some atypical Enteropathogenic Escherichia coli strains with different uncommon intimin types. BMC Microbiol. 9, 1–10. doi: 10.1186/1471-2180-9-146
Keywords: Shiga toxin-producing E. coli, enteropathogenic E. coli, whole genome sequencing, virulence factor, bacteriophages, cytotoxicity
Citation: Lee W, Ha J, Choi J, Jung Y, Kim E, An ES, Kim SH, Shin H, Ryu S, Kim SH and Kim H-Y (2024) Genetic and virulence characteristics of hybrid Shiga toxin-producing and atypical enteropathogenic Escherichia coli strains isolated in South Korea. Front. Microbiol. 15:1398262. doi: 10.3389/fmicb.2024.1398262
Edited by:
Adrian Canizalez-Roman, Autonomous University of Sinaloa, MexicoReviewed by:
Uriel Alberto Angulo-Zamudio, Professor Autonomous University of Sinaloa Culiacán, MexicoSusan Leonard, United States Food and Drug Administration, United States
Copyright © 2024 Lee, Ha, Choi, Jung, Kim, An, Kim, Shin, Ryu, Kim and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Soon Han Kim, bGFtYm5kb2dAa29yZWEua3I=; Hae-Yeong Kim, aHlraW1Aa2h1LmFjLmty
 Woojung Lee
Woojung Lee Jina Ha1
Jina Ha1 Eiseul Kim
Eiseul Kim Hakdong Shin
Hakdong Shin Sangryeol Ryu
Sangryeol Ryu Hae-Yeong Kim
Hae-Yeong Kim