- 1Hellenic Agricultural Organization - DIMITRA (ELGO - DIMITRA), Institute of Plant Breeding and Genetic Resources, Thessaloniki, Greece
- 2Centre for Crop and Disease Management, Department of Molecular and Life Sciences, Curtin University, Perth, WA, Australia
- 3Laboratory of Plant Pathology, Department of Agriculture, International Hellenic University, Thessaloniki, Greece
- 4Laboratory of Plant Pathology, Department of Crop Science, Agricultural University of Athens, Athens, Greece
Septoria pistaciarum, a causal agent of Septoria leaf spot disease of pistachio, is a fungal pathogen that causes substantial losses in the cultivation, worldwide. This study describes the first pan-genome-based survey of this phytopathogen—comprising a total of 27 isolates, with 9 isolates each from 3 regional units of Greece (Pieria, Larissa and Fthiotida). The reference isolate (SPF8) assembled into a total of 43.1 Mb, with 38.6% contained within AT-rich regions of approximately 37.5% G:C. The genomes of the 27 isolates exhibited on average 42% gene-coding and 20% repetitive regions. The genomes of isolates from the southern Fthiotida region appeared to more diverged from each other than the other regions based on SNP-derived trees, and also contained isolates similar to both the Pieria and Larissa regions. In contrast, isolates of the Pieria and Larissa were less diverse and distinct from one another. Asexual reproduction appeared to be typical, with no MAT1-2 locus detected in any isolate. Genome-based prediction of infection mode indicated hemibiotrophic and saprotrophic adaptations, consistent with its long latent phase. Gene prediction and orthology clustering generated a pan-genome-wide gene set of 21,174 loci. A total of 59 ortholog groups were predicted to contain candidate effector proteins, with 36 (61%) of these either having homologs to known effectors from other species or could be assigned predicted functions from matches to conserved domains. Overall, effector prediction suggests that S. pistaciarum employs a combination of defensive effectors with roles in suppression of host defenses, and offensive effectors with a range of cytotoxic activities. Some effector-like ortholog groups presented as divergent versions of the same protein, suggesting region-specific adaptations may have occurred. These findings provide insights and future research directions in uncovering the pathogenesis and population dynamics of S. pistaciarum toward the efficient management of Septoria leaf spot of pistachio.
1 Introduction
Pistachio (Pistacia vera L.) is an important crop in various regions around the world including the Mediterranean basin (Drais et al., 2023). The global economic value of pistachios is estimated to be over $1 billion annually, with the top five producers being the United States, Iran, Turkey, Syria and Greece (Mateos et al., 2022). Septoria pistaciarum—the causal agent of Septoria blight of pistachios, is an important pathogen reported across all pistachio-growing regions, including: the United States (Young and Michailides, 1989), Middle-East (Akgul et al., 2011), Central Asia (Ahmad et al., 2011) and the Mediterranean (Eskalen et al., 2001; Gusella et al., 2021; Lopez-Moral et al., 2022; Drais et al., 2023).
Septoria pistaciarum infects leaves, shoots, and nuts of pistachio trees, causing a variety of symptoms, including leaf spots, shoot blight, and nut rot (Crous et al., 2013) and can significantly reduce pistachio quality and yield (Drais et al., 2023). In leaves the symptoms of the disease appear on both sides from spring until the end of summer, where the pathogen produces conidia from pycnidial conidiomata that dispersed initiating new infections (Gusella et al., 2021). Pistachio trees may prematurely defoliate due to severe inoculum pressure and favorable environmental conditions (Drais et al., 2023). In turn, the bearing shoots may be also affected along with the he physiological processes of assimilation of carbohydrates necessary for bud differentiation (Drais et al., 2023). The host range of S. pistaciarum appears to be limited to pistachio trees, but the pathogen can survive in infected plant debris on the ground, in fallen leaves, or infected nursery stock (Chitzandis, 1956). Although other closely related species of the Mycosphaerellaceae also infect pistachio, S. pistaciarum is morphologically distinctive with angular leafspots confined by leaf-veins (Crous et al., 2013). Spread can occur by wind-blown spores which can disperse in long distances (Gusella et al., 2021), or by contact with infected plant material (Chitzandis, 1956).
As various aspects of the pathogen life cycle and epidemiology are still unclear, the management of this pathogen with commercial fungicides is quite a challenging task. Early studies report the good efficacy of copper and triazole fungicides against Septoria leaf spot of pistachios in USA (Call and Matheron, 1994) and Turkey (Çat, 2022). In Greece the standard farming practice includes the application of various copper formulations until flowering and then a combination of fungicides from the classes of demethylation inhibitors (DMIs), quinone outside inhibitors (QoIs), fourth generation succinate dehydrogenase inhibitors (SDHIs) and anilinopyrimidines (APs) at 14-day intervals for at least 2 months. Detecting latent infections within the pistachio tissues could be a crucial step in limiting the number of field sprays, as well as in ensuring a sustainable use of effective fungicides through an accurate spray program (Gusella et al., 2021).
Genomics has been extensively applied to some plant-pathogenic fungal taxa (Aylward et al., 2017), and has been a useful tool enabling bioinformatic inference and molecular biology methods to validate their pathogenicity mechanisms (Jones et al., 2018). In this context, S. pistaciarum has not yet received much attention, although it is related to other hemibiotrophic pathogens with comparatively extensive pan-genomic resources, including: Zymoseptoria tritici (Testa et al., 2015); Pseudocercospora fijiensis (Arango Isaza et al., 2016); Ramularia collo-cygni (McGrann et al., 2016; Stam et al., 2018; Sjokvist et al., 2019) and; Cercospora spp. (Chand et al., 2015; Orner et al., 2015; Albu et al., 2017; Vaghefi et al., 2017; Wingfield et al., 2017; Zeng et al., 2017a,b; Sautua et al., 2019; Gu et al., 2020; Lin et al., 2022; Cheng et al., 2023; Yang et al., 2023). A previous phylogenetic study based on 5 conserved loci—which distinguished S. pistaciarum from other pistachio-infecting Mycosphaerellaceae spp. (Crous et al., 2013)—amounts to the current extent of genomic study of this organism. This study represents the first analysis of the pan-genome of a local Mediterranean population of S. pistaciarum. These resources will enable the study of the genomic features driving the pathogenicity and biological complexity of this foliar pathogen, with new insights into its population dynamics and pan-genomic structure.
2 Methods
2.1 Origin of Septoria pistaciarum isolates and DNA extraction
During the 2022 growing period, S. pistaciarum isolates were obtained in June from pistachio leaves showing leaf spot symptoms (cv. Aeginis) across three distinct sites located in the areas of Amfikleia, Fthiotida (38.629963 N, 22.681427E; code FTH), Kiparissia, Larissa (39.5138684 N, 22.5733432E; code LAR) and Kitros, Pieria (40.3785945 N, 22.607257E; code PIE). The trees were over 20 years old and trained under the standard open vase system to an average height of 10 m. A standard fungicide program based on the fungicides boscalid + pyraclostrobin (Signum 26,7/6,7 WG; BASF), fluxapyroxad (Sercadis 30 SC; BASF), pyrimethanil (Scala 40 SC; BASF), dodine (Syllit 544 SC; UPL), and copper (Cuprofix Ultra 40 WG; ADAMA) was applied on all orchards in spring. Each diseased leaf sample was transferred to the lab in a separate moist polyethylene bag to prevent cross-contamination and stored at room temperature for 72 h to induce the formation of cirrhi in the pycnidia. From each sample a single-spore isolate was obtained by slight touching a flamed wire loop onto a freshly formatted cirrhus of conidia from one spot per leaf randomly picked up. A sparse pycnidiospore suspension in 0.5 mL distilled water was prepared and spread onto Potato Dextrose Agar (PDA) in Petri dishes amended with 100 mg/L of streptomycin sulfate. After 48 h of incubation at 24°C in the dark, individual germinated single-spore conidia were transferred in glass test tubes with potato dextrose agar and stored until use (Figure 1). Pycnidiospores were hyaline, curved, with obtuse ends, having 1 to 5 septa (Chitzandis, 1956) typically of S. pistaciarum. In total, 27 isolates were obtained (with nine isolates representing each region; Table 1) and they were maintained in PDA tubes at 4°C. Because of the very slow growth of the hyphae, in order to obtain larger colonies in plates for further analysis each representative plate was seeded with 0.4 mL of a conidial suspension from each isolate (Dhingra and Sinclair, 1985). After 4 weeks of incubation in the dark, fungal DNA was extracted from conidiomata (Figure 1) of all isolates using the Quick-DNA™ Fungal/Bacterial Miniprep Kit (Zymo Research).

Figure 1. Morphological features of Septoria pistaciarum: (A) Germinated single-spore conidia on PDA after 48 h of incubation at 24°C in the dark. (B,C) Representative isolates SPF8 (B) and SPF9 (C) with grown on PDA after 4 weeks of incubation at 24°C in the dark; (D,E) Symptoms of Septoria leaf spot observed in pistachio leaves infected by SPF8 (D) and SPF9 (E).
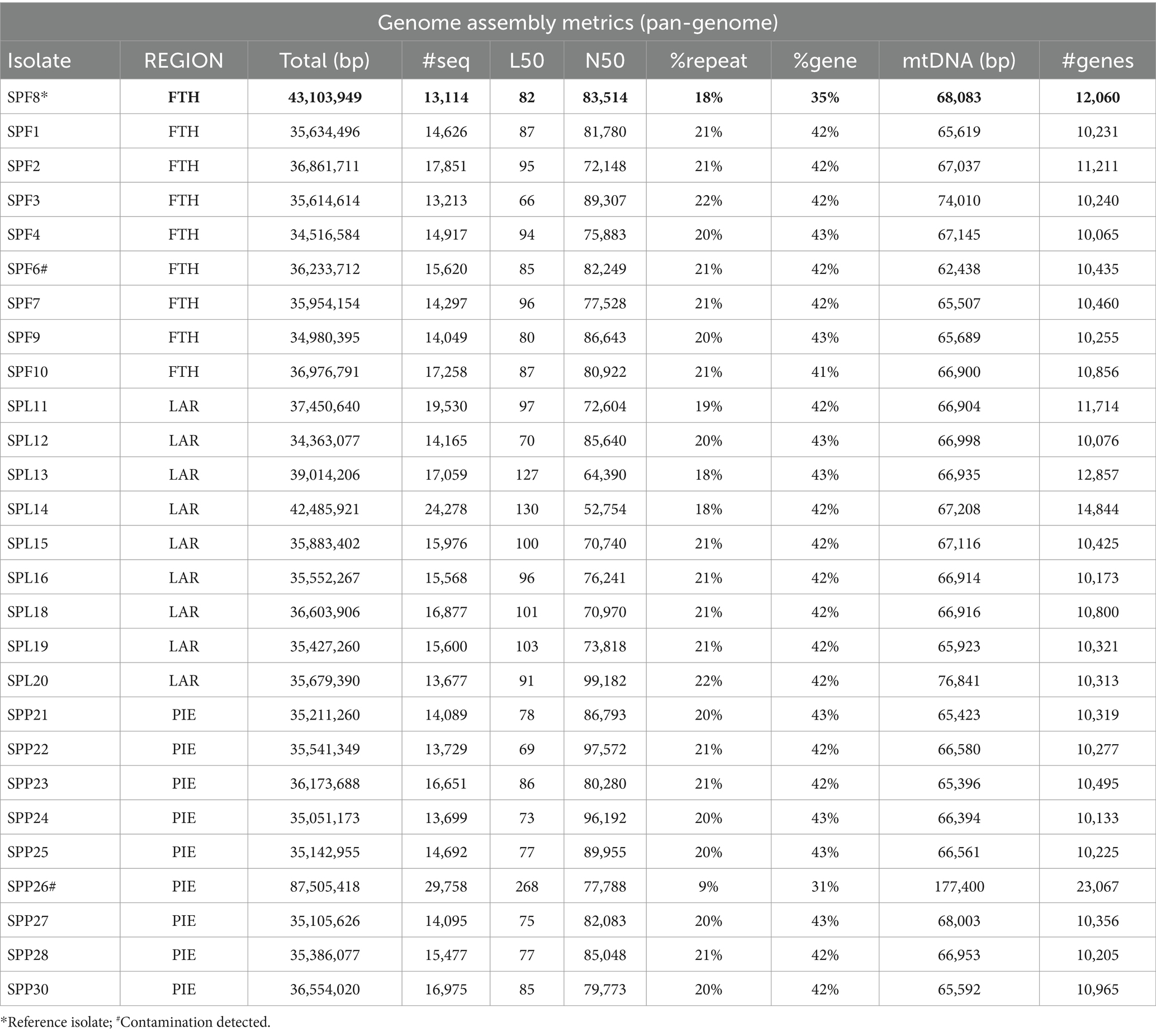
Table 1. Summary of genome assembly metrics for 27 isolates of Septoria pisticiarum isolated across 3 sub-regions of Greece (FTH, Fthiotida; LAR, Larisa; PIE, Pieria).
2.2 Genome assembly
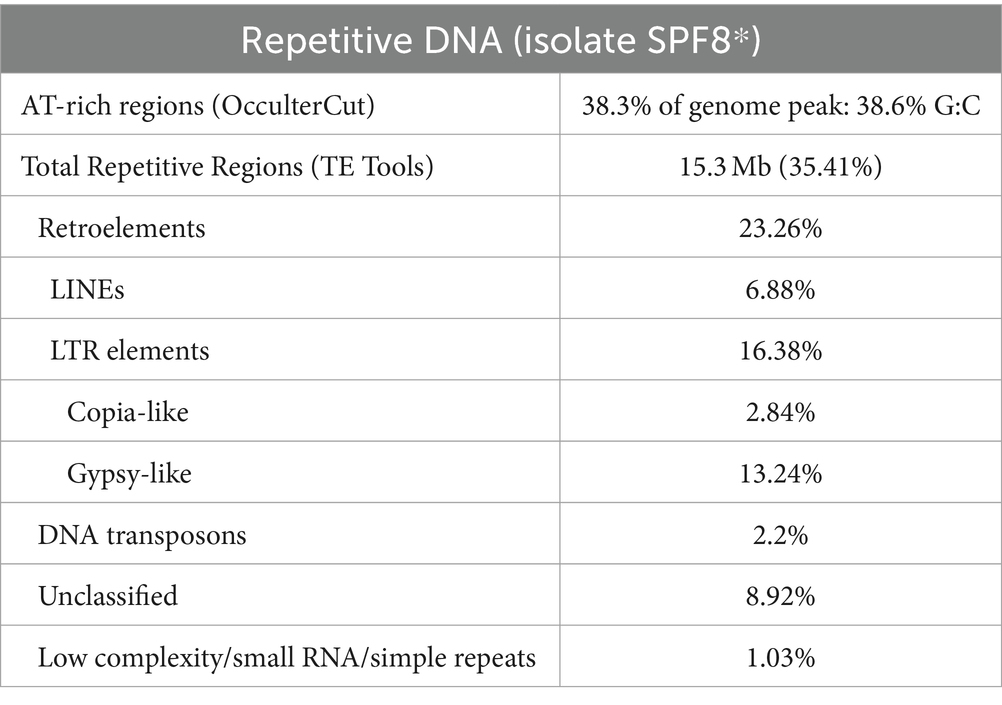
Fungal DNA from all isolates was randomly fragmented into 350 bp inserts, genomic libraries were constructed using the Novogene NGS DNA Library Prep Set (Cat No.PT004) and sequenced by a PE150 sequencing strategy. Illumina reads were trimmed by cutadapt v3.7 (-a AAGTCGGAGGCCAAGCGGTCTTAGGAAGACAA-A AAGTCGGATCGTAGCCATGTCGTTCTGTGAGCCAAGGAGTTG --minimum-length = 50 -- max-n = 3 -n2 -q30; Martin, 2011). All isolates were assembled by SPAdes v3.15.4 (--only_assembler –cov-cutoff auto; Bankevich et al., 2012). Assembly quality and gene set completeness was predicted via BUSCO v5.4.6 (Simao et al., 2015). Initial genome assembly metrics were assessed for contiguity and BUSCO completeness (Supplementary Data Sheet 1), from which the “SPF8” isolate was selected as a reference isolate. Reads for isolate ‘SPF8’ were merged and gap-filled with BBMerge (Bushnell et al., 2017) then de novo assembled with SPAdes as above. Genome assemblies of all isolates were further scaffolded were possible using pairwise comparisons vs. other isolates with ragtag v2.1.0 (scaffold: –remove-small -f 75 -r, merge; Alonge et al., 2022). Mitochondrial genomic DNA (mtDNA) was assembled via MitoZ v3.6 (Meng et al., 2019), and assembled mtDNA contigs were appended to the genome assemblies, while previously-assembled sequences matching mtDNA contigs were removed. Contamination checks for each assembly was performed via BLAST v2.12.0 (blastn -max_target_seqs 5 -evalue 1e-100 -perc_identity 95; Altschul et al., 1990) to the UNITE database (Abarenkov et al., 2010). Overall G:C content and AT-rich compartments were predicted with OcculterCut v1.1 using the reference isolate SPF8 genome assembly (Testa et al., 2016).
2.3 Prediction of genomic repeats, genes and functions
Repetitive sequences were predicted using Dfam TE Tools 1.88 (Lerat et al., 2016). Gene prediction was performed across all isolates in 2 rounds. In the first round, funannotate v1.8.15 (predict; max_intronlen 1000; Palmer and Stajich, 2020) was used to predict genes for all isolates. In order to provide further evidence for the accurate support of gene loci and their exon boundaries, transcriptomic data was obtained for the isolate SPF8. Thus, total mRNA was extracted in triplicate from the mycelial phase of this isolate grown on PDA medium from a fresh two-weeks-old culture. The Quick-RNA™ Fungal/Bacterial Miniprep kit (Zymo Research) was employed, and RNAseq reads were generated through Illumina platform (PE150 Novaseq 6000) using the Novogene NGS RNA Library Prep Set (PT042). RNAseq alignment vs. the reference isolate was performed with HiSAT2 v2.2.1 (max-intronlen 5000; dta; Kim et al., 2019), converted to GFF via Stringtie v2.2.1 (Levy Karin et al., 2020) and provided as input to funnannotate. Funannotate-predicted proteomes were clustered into orthogroups (including singletons) with ProteinOrtho v6.3.1 (selfblast; singles; Lechner et al., 2011). A representative pan-genome proteome dataset was selected from the longest member of each orthogroup, and used as input to MetaEuk Release 6-a5d39d9 (--easy-predict –max-intron 500; Levy Karin et al., 2020) for a second round of gene prediction in all isolates, and orthogroups with MetaEuk matches were retained as a representative ‘pan-genome’ proteome set. Functional annotations were predicted across the pan-genome proteome dataset with InterProScan v5.63-95.0 (Jones et al., 2014). Secretion and effector-like properties were predicted using Predector v1.2.7 (Jones et al., 2021). Trophic niche (i.e., biotrophy, necrotrophy, etc.) was predicted via CATAStrophy v0.1.0 (Hane et al., 2020; using HMMER 3.3) vs. dbCAN v10. Mating type genes were screened vs. a representative dataset (Wilken et al., 2017) using MetaEuk (as above). Secondary metabolite synthesis gene clusters were predicted in the reference isolate assembly (SPF8) with antiSMASH v6.1.1 (Blin et al., 2021). Fungicide-resistance mutations were predicted with the fungicide-resistance allele screening tool (FRAST) (Oliver et al., 2024).
2.4 Comparative genomics
Gene ortholog group presence-absence variation (PAV) across the S. pistaciarum pan-genome was determined based on matches to a pan-genome-wide representative proteome dataset using MetaEuk (see above). Genome sequencing reads were aligned to the SPF8 assembly with BWA v0.7.17-r1198-dirty (Li and Durbin, 2009). Genome sequencing reads were processed fastqToSam to generate raw ubam inputs, and passed through MarkIlluminaAdapters, MarkDuplicates, and combined with BWA alignmed bam data with MergeBamAlignment (-CREATE_INDEX true -ADD_MATE_CIGAR true) to generate alignmed uban inputs for variant calling. Variant calling was performed with GATK v4.2.6.1 (HaplotypeCaller -ERC GVCF –minimum-mapping-quality 20 –min-base-quality-score 20 -G StandardAnnotation -G AS_StandardAnnotation -G StandardHCAnnotation; McKenna et al., 2010). Bi-allellic single nucleotide polymorphisms (SNPs) were filtered and randomly selected for 1 per 5 kb with BCFTools v1.15 (+prune -w 5,000 bp -n1 -N rand; Danecek et al., 2021), and used to generate an unrooted distance-based tree via IQTree v2.2.2.7 (1,000 iterations, -bb 1000 -alrt 1000; Minh et al., 2020). The tree was visualized alongside selected and summarized predicted protein function data (mating types, effector-like proteins) with iTOL (mid-point root; Letunic and Bork, 2021).
3 Results and discussion
3.1 Assessment of genome quality and features
The reference isolate (SPF8) assembled into a total of 43.1 Mb. All isolate assemblies exhibited ~70-120X sequencing depth (ex. SPP26), an average N50 of 80.4 kb, an average contig number of 9,084, and an average of 42% and 20% gene-coding and repetitive regions, respectively (Table 2) with LTR retrotransposons being the most common repeat type (Table 2; Supplementary Data Sheet 1). The genome metrics above were comparable to pan-genome datasets from the sister species Zymoseptoria tritici, which had an average core chromosome coverage of ~30X, an average N50 of 84.7 kb and average contig number of 1,088 (Feurtey et al., 2023). CATAStrophy (Hane et al., 2020) predictions for S. pistaciarum indicated non-haustorial hemibiotrophy (Table 3), which may correspond with prior reports of a 2 week latent phase (Crous et al., 2013). OcculterCut analysis of G:C content of the reference isolate assembly (Figure 2B) also indicated a bi-modal G:C distribution typical of hemibiotrophic fungi (Testa et al., 2016), with the AT-rich peak at approximately 38.6% G:C containing over a third (38.3%) of the genome length. Blastn vs. UNITE (Abarenkov et al., 2010) indicated >99.25% identity to the ITS region for all isolates to Mycosphaerella pistaciarum, however isolate 6 was also contaminated with plant DNA, and isolate 26 was contaminated with a Basidiomycete matching Tomentella fuscograulosa [NUC: UDB028526] (Supplementary Data Sheet 2). Despite varying levels of contamination in these two assemblies, the pan-genome survey approach of this study (Figures 2C,D) was capable of focusing on genome features relevant to the S. pistaciarum population as a whole, as genes specific only to isolates 6 and 26 were not the subject of further study. Assessment of the reference assembly via BUSCO indicated 98.7% completeness relative to the capnodiales_odb10 dataset (2020-08-05, genomes:13, BUSCOs:3578), with 3738/3786 BUSCOs detected completely (Supplementary Data Sheet 3).
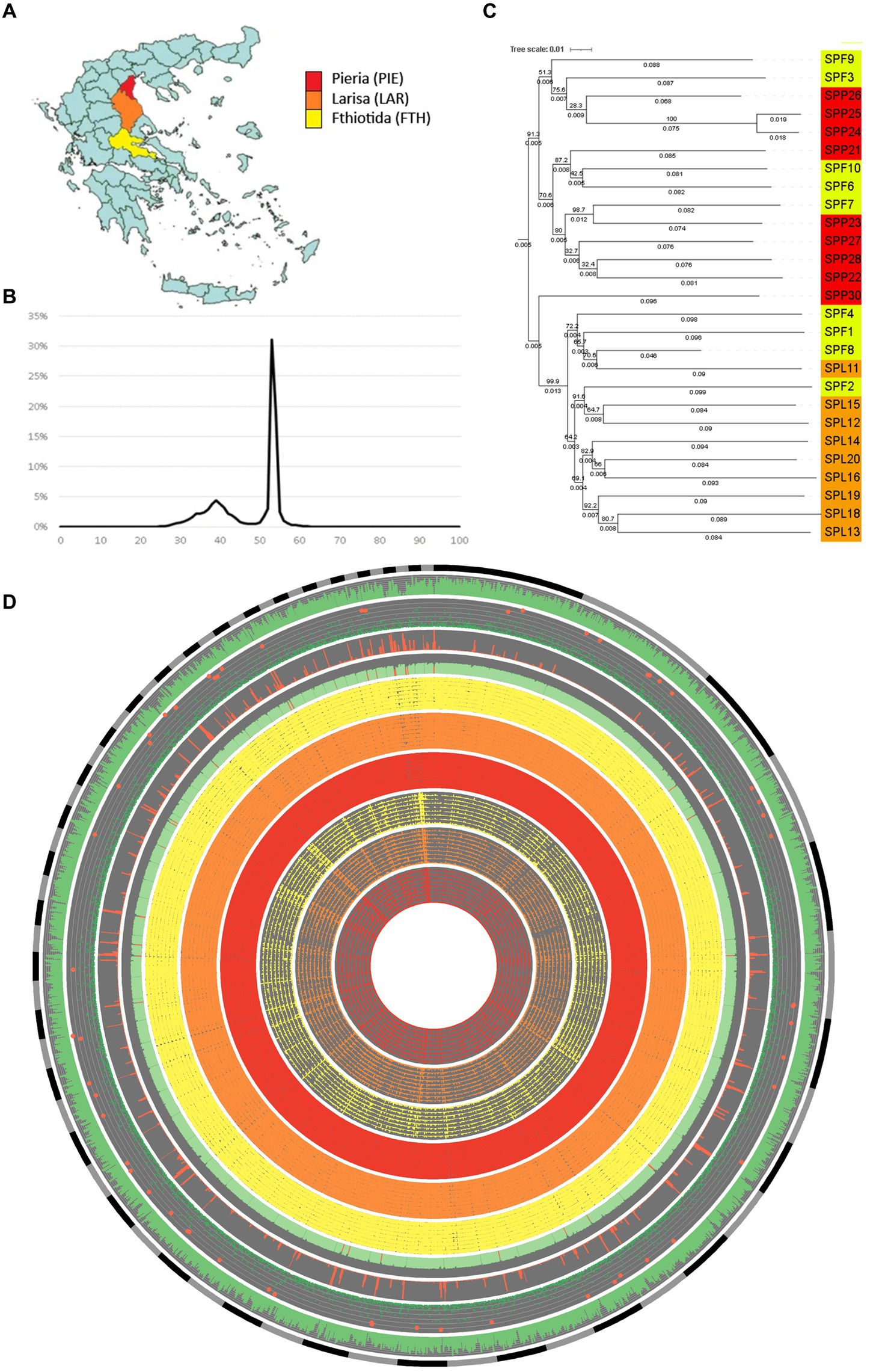
Figure 2. Summary of 27 Septoria pistaciarum isolates sampled across Greece and their genomic features. (A) Nine isolates each were sampled from 3 regions bordering the Aegean Sea: Pieria (PIE-red); Larisa (LAR-orange); Fthiotida (FTH-yellow). (B) G:C content (x-axis) summarized by the relative proportion of the reference isolate (SPF8) genome assembly (y-axis) indicated bi-modal G:C distribution with a significant proportion of the genome organized into AT-rich compartments (C) Midpoint-rooted distance-based tree of the 27 isolates based on SNP variant calling relative to the reference isolate. (D) A circular plot of genomic features relative to the reference isolate assembly (excluding sequences < 100 kb), with co-centric rings representing (in inward order): sequence length (gray and black); gene density (% coverage/10 kb, green); Predector effector-likelihood scores (−3 to 3, green, with scores >2 highlighted red); repeat density (% coverage/10 kb, red); G:C content (%G:C/10 kb, green, with <40% highlighted red); Homology to alternate isolates (%match coverage/10 kb, FTH = yellow, LAR = orange, PIE = red); Mutation density (from 1 to 500 SNPs/10 kb, FTH = yellow, LAR = orange, PIE = red).
3.2 Local geographic distribution of the Greek Septoria pistaciarum pan-genome
Based on analysis of SNP-based distance trees the two regions—Pieria (north) and Larissa (central)—were distinct, whereas Fthiotida (south) was similar to both Pieria and Larissa regions (Figures 2A,C). While the geographic range and number of isolates sampled in this study is limited, based on the available data, we speculate that various events of long-distance dispersal (Golan and Pringle, 2017) caused by anthropogenic influences, spore dispersal, and infected nursery stock may be the cause of this clustering of isolates from the Fthiotida region with a relatively higher genetic divergence than their expected geographic distance. Such a clustering, where the populations were not strictly clustered based on their geographical origins, has also been reported in Zymoseptoria tritici populations (Mekonnen et al., 2020; Chedli et al., 2022). It may also indicate that these isolates belong to a population that might has experienced range expansions across the other two regions, which has resulted in admixture complemented by gene flow (Rogers and Rogers, 1999), gradually reducing the isolation by distance (Mills et al., 2007) of Fthiotida isolates. On the other hand, the extensive clustering into geographically-restricted lineages of isolates from Larissa and Pieria regions may indicate that these populations experienced a geographic isolation leading to local adaptation (Bazzicalupo, 2022). Future expansion of pan-genomic resources with a broader sampling to include surrounding regions or a globally-representative set of isolates of S. pistaciarum, might be able to confirm if isolates of the southern Fthiotida region represent a “wilder” population, relative those of the genetically narrower Pieria and Larissa regions. At present it is not clear whether pathways of movement through neighboring regions that were not sampled may have contributed to the transportation history of these isolates. Assemblies for all isolates possessed two versions of MAT1-1 type mating type genes, homologous to MAT-1-5/COX13 [ABS19615] and MAT1-1-6/APN2 [ABS19616] (Wilken et al., 2017; Supplementary Data Sheet 4). No MAT1-2 homolog was detected, suggesting limited potential for meiotic recombination across these three regions.
3.3 Potential secondary metabolite production
AntiSMASH prediction of secondary metabolite synthesis gene clusters (SMCs) in the reference isolate (SPF8) assembly, indicated homologs of 4 T1PKS (polyketide synthase) regions potentially encoding toxins similar to cercosporin (Chen et al., 2007), fusarubin/oxyjavanicin (Chowdhury et al., 2017), aspyridone A (Bergmann et al., 2007), fumonisin (Kamle et al., 2019), as well as melanin which may have a role in strengthening the pathogen cell wall (Jacobson, 2000). Also predicted were 15 non-ribosomal peptide synthase (NRPS)-like clusters potentially encoding the cytotoxic cyclic peptide serinocyclin (Krasnoff et al., 2007) and other unknown metabolites, and 5 terpene sythase clusters (Supplementary Data Sheet 5). Accurate prediction of secondary metabolite biosynthesis products remains challenging and requires further validation, however the clusters for serinocyclin and melanin were highly conserved. Overall, the predicted SMC profile of S. pistaciarum was similar to that previously reported for the relatively closely-related species Zymoseptoria tritici (Hassani et al., 2022), however S. pistaciarum had more NRPS SMCs whereas Z. tritici had more PKS SMCs (Supplementary Data Sheet 5).
3.4 Potential fungicide resistance
Although testing of differential fungicide efficacy and resistance across S. pistaciarum isolates was beyond the scope of this current study, it was possible to predict potential fungicide resistance mutations from pan-genomic data. Analysis of amino acid changes in the products of known fungicide resistance loci (Mair et al., 2016) revealed several potential resistance adaptations, inferred from reported resistance associated with equivalent mutations in other fungal species (Supplementary Data Sheet 6; Oliver et al., 2024). The CYP51A protein of all isolates contained amino acid residues corresponding to P216L, M220I, and H147Y of Aspergillus fumigatus CYP51A [NCBI: AF338659], which may confer azole resistance (Howard et al., 2009). Some isolates had mutations in CYP51B corresponding to deletions at Q287 and G412, and a mutation at S208T (relative to Zymoseptoria tritici [NCBI: AY253234]), that may be involved in DMI resistance (Stammler and Semar, 2011), CytB loci were not well represented across this pan-genome with data for only two isolates, however for these no intron/intein mutations were detected, and one deletion mutation corresponded to site D203 (relative to Z. tritici [NCBI: AY247413]), which in Plasmopora viticola, was involved in resistance to cyazofamid (Mounkoro et al., 2019). Beta-tubulin exhibited mutations corresponding to E198A and M257L (relative to Aspergillus nidulans [NCBI: M17519]), which may confer benzamidazole resistance (Leroux et al., 2002). A single isolate had a mutation in the OS1 protein corresponding to A350S of Botrytis cinerea [NCBI: AF435964] which may confer resistance to Fludioxonil (Ren et al., 2016). Overall, the mutations detected above and their corresponding resistance phenotypes inferred from related fungal species may reflect the broad application of boscalid + pyraclostrobin, fluxapyroxad, pyrimethanil, dodine and copper that was applied to all isolates in this study.
3.5 Prediction of pathogenicity effector genes
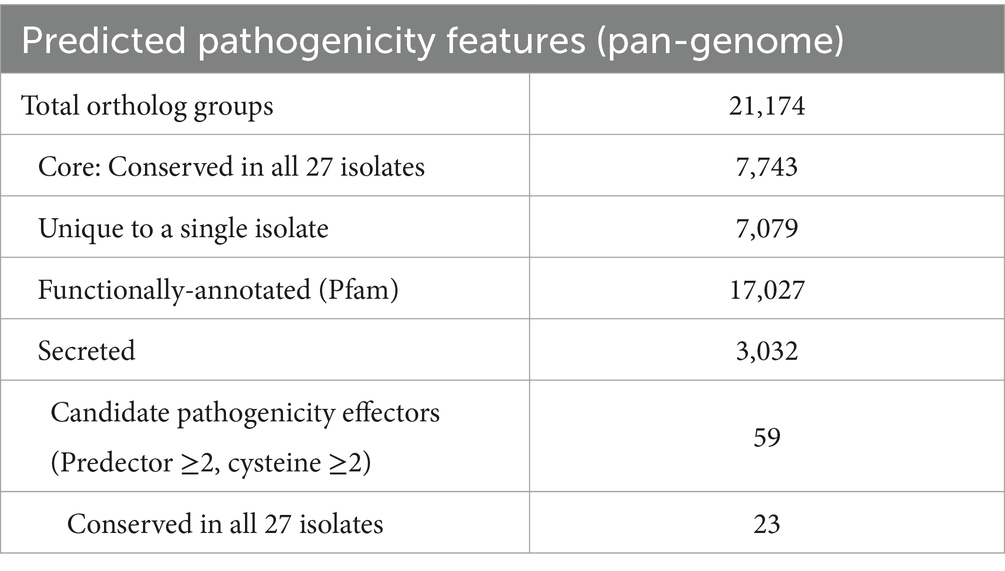
Comparative analysis of orthologous groups across the pan-genome indicated 21,174 ortholog groups (Supplementary Data Sheet 7), with isolates encoding an average of 11.2 K genes each, and 6,805 (32%) core groups containing a single member that was present in all isolates (Supplementary Data Sheet 8). Candidate secreted effector-like protein (CSEP) prediction across the representative pan-genome orthologous gene set, which in an effort to highlight high-priority effector candidates, was filtered for: predicted secretion (Predector—any method), Predector Score ≥ 2, and cysteine residues ≥ 2, which resulted in 59 CSEP-orthogroups (Figure 3) (Supplementary Data Sheet 8). Of these, notable CSEPs with conserved functional domains and/or with homology to confirmed effectors of other pathogen species (Supplementary Data Sheets 9, 10) are summarized below according to their speculative roles at various phases of S. pistaciarum infection. Overall, the predicted CSEP set indicated that S. pistaciarum may employ a combination of defensive effectors with roles in suppression of host defenses, and offensive effectors with a range of cytotoxic activities. Notably, some effector-like ortholog groups may have presented as divergent versions of the same protein (Table 4; Supplementary Data Sheet 8), suggesting region-specific adaptations may have occurred. There were 23/59 highly-conserved CSEP orthogroups, common to all 27 isolates, with another 5 similarly conserved sets of groups (WSC, PBP, DLH, ZtNip2 and Cutinase, Table 4) presenting as separate orthogroups that may be highly divergent versions of the same ortholog.
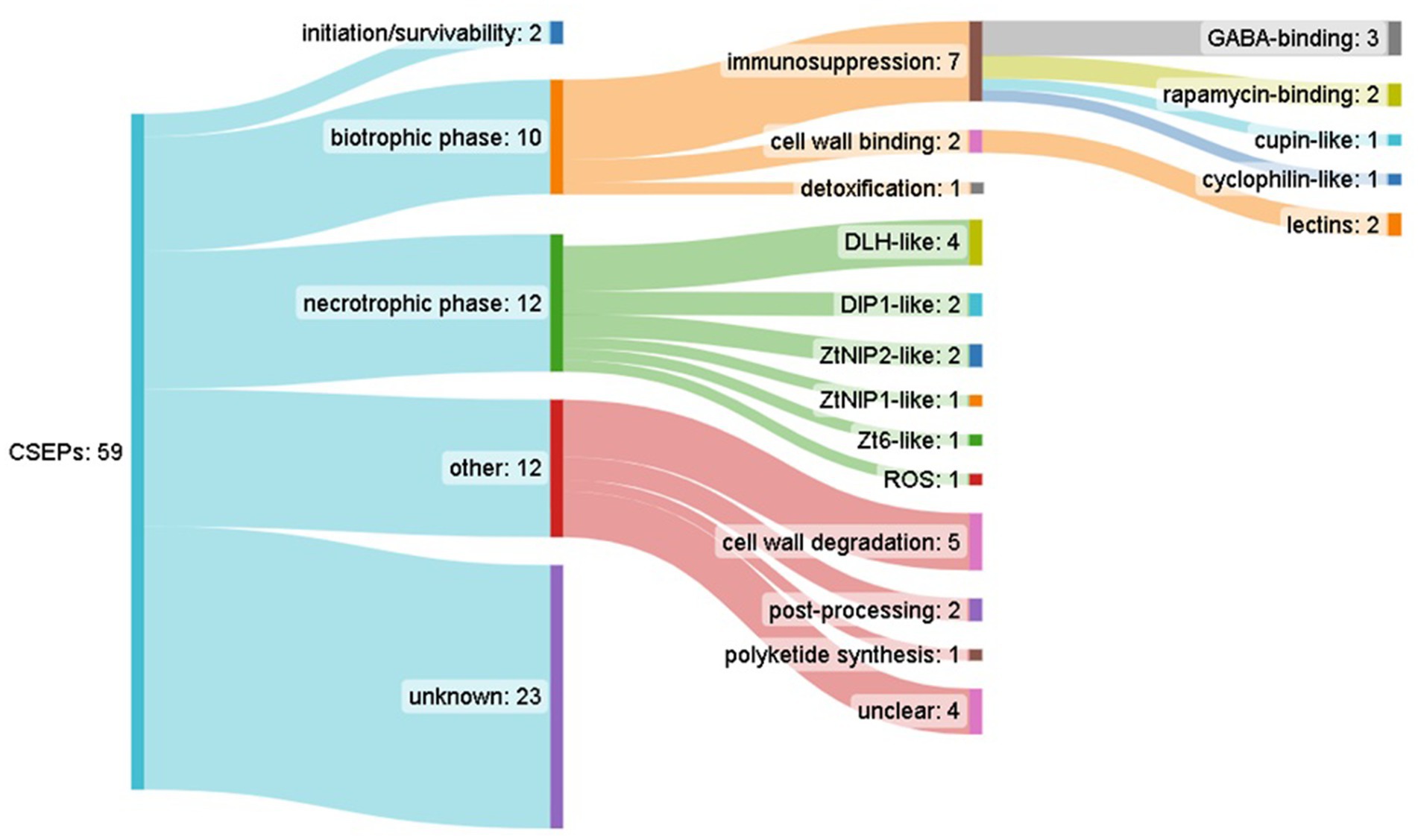
Figure 3. Summary of candidate secreted effector-like protein (CSEP) predictions across Septoria pistaciarum pan-genome-derived orthogroups, and their predicted functional annotations.
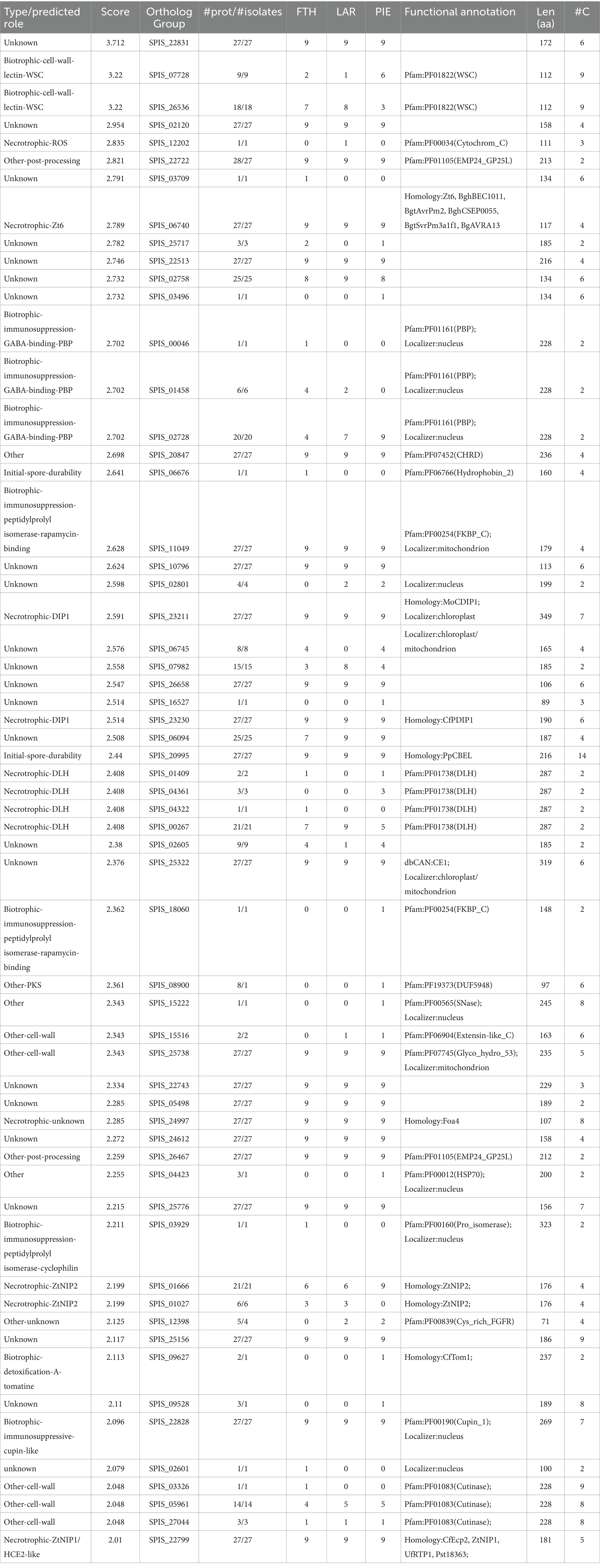
Table 4. Summary of candidate secreted effector-like protein (CSEP) predictions across Septoria pistaciarum pan-genome-derived orthogroups, and their predicted functional annotations.
3.5.1 Spore survival prior to infection
Two CSEP orthogroups were categorized as having potential roles in spore durability and/or promoting initial colonization prior to infection. All 27 isolates possessed a PpCBEL lectin homolog (orthogroup SPIS_20995), which may promote colonization of leaf surface without directly causing virulence and may be recognized as a PAMP leading to HR (Gaulin et al., 2006). A single isolate (#4 of the FTH region) had a copy of orthogroup SPIS_03709, matching the Hydrophobin_2 domain. Other members of this family include the cerato-ulmins, a class of hydrophobins best studied in Dutch Elm Disease (Gallo et al., 2023). These CSEPs may promote infection rates over time by increasing spore resistance to dessication.
3.5.2 Latent/biotrophic phase
There were 10 CSEP orthogroups which were categorized as having potential roles in the biotrophic/latent phase of infection, 7 of which appear to have roles in suppression of host defenses. Three orthogroups (SPIS_00046, SPIS_01458, SPIS_02728) matched to the PBP domain (periplasmic binding proteins), which may bind Gamma-aminobutyric acid (GABA). These 3 orthogroups are likely diverged from a common ortholog, as counts were distributed across the 27 isolates (1, 6, and 20 respectively), with SPIS_00046 specific to the FTH region, SPIS_01458 occurring in both FTH and LAR, and SPIS_02728 occurring across all 3 regions. The PBP-domain CSEPs could potentially interfere with GABA-regulated aspects of host metabolism and defense, as increased GABA concentration increases photosynthesis, reduces ROS production, regulates stomatal opening and increases biotic stress tolerance (Li et al., 2021). There were 2 CSEP orthogroups (SPIS_11049, SPIS_18060) which matched to the FKPBP_C domain, the former present in all 27 isolates, and the latter specific to a single isolate (#26—PIE). These are FK506-binding proteins—or FKBP-type peptidyl-prolyl cis-trans isomerases—which are functionally related to cyclophilins/immunophilins, and are receptors for ‘rapalog’-type immunosuppressant molecules including rapamycin, FK506, and cyclosporins. The best-studied example, rapamycin, has antifungal activity (Singh et al., 1979; Cruz et al., 1999), can increase tolerance to abiotic stresses (Dong et al., 2018), and can increase mitochondrial respiration and ROS production in host cells (Villa-Cuesta et al., 2014). Notably the CSEP SPIS_11049, common to all 27 isolates, was predicted by localizer to target the mitochondrion. Another CSEP SPIS_03929, only in isolate #3 of FTH, also matched to a cyclophilin-like domain with predicted nuclear localization. There were 2 very high-ranking (Predector score = 3.2) CSEP orthogroups (SPIS_07728, SPIS_26536) which matched the WSC domain, which are beta-glucan-binding lectins (Wawra et al., 2019) which have been reported to alter cell wall composition and suppress host PTI (Wawra et al., 2016). The two WSC-type CSEPs were another divergent set, with 9 and 18 isolates belonging to these groups respectively, with the former more prevalent in the Pieria region and the latter more prevalent in the FTH and LAR regions. There was also a cupin-like CSEP (SPIS_22828) common to all 27 isolates, localized to the nucleus, which may also have an immunosuppressive role (Yan et al., 2022).
3.5.3 Necrotrophic phase
Aside from CSEP orthogroups that appear to support the biotrophic phase, there were several with putative necrotrophic effector functions. Four orthogroups (SPIS_00267, SPIS_01409, SPIS_04361, SPIS_04322) appear to be part of a larger divergent group (21, 2, 3, and 1 isolates, respectively) matching dienelactone hydrolases with the DLH domain. These may be involved in chlorocatechol degradation, may be required for virulence and potentially laterally-transferred between plant pathogen species (Gardiner et al., 2012). The less common orthogroups did not occur in the LAR region. Two CSEP groups (SPIS_23211, SPIS_23230) were both present in all 27 isolates, and matched (defense-inducing-protein) DIP1 pectate lyases, which can induce host HR, and increase hydrogen peroxide and alkanisation in host cells (Ashwin et al., 2018). There were several CSEP orthogroups which matched well-studied effectors of the closely-related wheat pathogen Zymoseptoria tritici.
A single orthogroup present in all 27 isolates (SPIS_22799) matched the necrosis-inducing effector ZtNIP1, and a divergent set of 2 orthogroups (SPIS_01666, SPIS_01027—present in 21 and 6 isolates respectively) matched the chlorosis-inducing effector ZtNIP2 (Ben M'Barek et al., 2015; Zhang et al., 2019). Another CSEP group (SPIS_06740) present in all 27 isolates matched the necrosis-inducing Zt6 ribonuclease, which may also have a dual-role in cytotoxicity against other microbes (Kettles et al., 2018). Other notable CSEP groups included a cytochrome C homolog (SPIS_12202) only present in 1 isolate (Sep_pis-14/LAR) and a Foa4 effector candidate homolog present in all isolates (SPIS_24997; Tintor et al., 2020).
3.5.4 Other CSEPs with indeterminate roles
Among CSEP groups not assigned to the above categories, there were two groups (SPIS_25738 and SPIS_246467) which were present in all isolates, matching GH53 glycosyl hydrolases and EMP24/GP25L/P24/GOLD family proteins. The latter may have a role in transporting proteins from the endoplasmic reticulum in order to bind coat proteins to cytoplasmic domains, and effectors such PITC_013620 have this function (Li et al., 2022). There were also 3 groups matching CE5 Cutinase domains (SPIS_03326, SPIS_05961, SPIS_27044—from 1, 14 and 3 isolates respectively) which may have a role cell wall degradation during early infection.
4 Conclusion
We present these S. pistaciarum pan-genome resources as a foundational resource for pistachio disease surveillance and future effector gene discovery. Overall, these pan-genome-based analyses indicate that S. pistaciarum employs a combination of directly offensive effector proteins, as well as host-defense suppression during its relatively long biotrophic latent phase. A small set of toxic secondary metabolites and high-confidence effector candidate proteins have been generated, with indication of the relative conservation and/or regional-specificity of diverged sequence variants, which may help to direct focus to the study of the role in pathogenicity of these candidates in future validation studies. This study also demonstrates how previously under-studied patho-systems can now be rapidly surveyed using a combination of low-cost pan-genomic sequencing and the transfer of recent bioinformatic approaches and increasingly-informative pathogenicity-relevant datasets developed across other model fungal pathogen species.
Data availability statement
The genome sequencing and transcriptome datasets generated for this study can be found in the NCBI database under BioProject: PRJNA1115914.
Author contributions
AZ: Conceptualization, Data curation, Formal Analysis, Investigation, Software, Supervision, Validation, Writing – original draft, Writing – review & editing. AB: Investigation, Writing – original draft. NG: Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MH: Data curation, Formal Analysis, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MC: Investigation, Visualization, Writing – original draft, Writing – review & editing. DIT: Writing – original draft, Writing – review & editing. EP: Writing – original draft, Writing – review & editing. JH: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was undertaken with the assistance of resources and services from the Pawsey Supercomputing Centre and the National Computational Infrastructure (NCI), which are supported by the Australian Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1396760/full#supplementary-material
SUPPLEMENTARY DATA SHEET 1 | Summary of genome assembly, gene and repetitive DNA metrics, for 27 genome assemblies of Septoria pistaciarum.
SUPPLEMENTARY DATA SHEET 2 | Summary of contamination checks with BlastN versus UNITE, for 27 genome assemblies of Septoria pistaciarum.
SUPPLEMENTARY DATA SHEET 3 | BUSCO gene completeness report versus the Septoria pistaciarum reference isolate SPF8 genome assembly.
SUPPLEMENTARY DATA SHEET 4 | Screening results for mating type loci across 27 genome assemblies of Septoria pistaciarum.
SUPPLEMENTARY DATA SHEET 5 | Secondary metabolite synthesis clusters predicted by AntiSMASH in the Septoria pistaciarum SPF8 reference isolate.
SUPPLEMENTARY DATA SHEET 6 | Fungicide resistance mutations predicted by FRAST, for 27 genome assemblies of Septoria pistaciarum.
SUPPLEMENTARY DATA SHEET 7 | Representative protein sequences for ortholog groups predicted in the Septoria pistaciarum pan-genome.
SUPPLEMENTARY DATA SHEET 8 | Presence of ortholog groups across the 27 isolates of the Septoria pistaciarum pan-genome.
SUPPLEMENTARY DATA SHEET 9 | Functional annotation and other pathogenicity metadata predicted by Predector, for pan-genome ortholog groups of Septoria pistaciarum.
SUPPLEMENTARY DATA SHEET 10 | Protein feature locations of functional annotations predicted by Predector, for pan-genome ortholog groups of Septoria pistaciarum.
References
Abarenkov, K., Henrik Nilsson, R., Larsson, K. H., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi--recent updates and future perspectives. New Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Ahmad, S., Khan, N., and Ashraf, S. (2011). First report of leaf blight caused by Septoria pistaciarum on Pistacia vera in India. J. Plant Pathol. 93:72. doi: 10.5555/20123315613
Akgul, H., Yılmazkaya, D., and Ergul, C. C. (2011). New microfungi records on pistachio (Pistacia vera L.) from Gaziantep province of Turkey. Afr. J. Biotechnol. 10, 14439–14442. doi: 10.5897/AJB11.1952
Albu, S., Sharma, S., Bluhm, B. H., Price, P. P., Schneider, R. W., and Doyle, V. P. (2017). Draft genome sequence of Cercospora cf. sigesbeckiae, a causal agent of Cercospora leaf blight on soybean. Genome Announc. 5:17. doi: 10.1128/genomeA.00708-17
Alonge, M., Lebeigle, L., Kirsche, M., Jenike, K., Ou, S., Aganezov, S., et al. (2022). Automated assembly scaffolding using RagTag elevates a new tomato system for high-throughput genome editing. Genome Biol. 23:258. doi: 10.1186/s13059-022-02823-7
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Arango Isaza, R. E., Diaz-Trujillo, C., Dhillon, B., Aerts, A., Carlier, J., Crane, C. F., et al. (2016). Combating a global threat to a clonal crop: Banana black Sigatoka pathogen Pseudocercospora fijiensis (synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genet. 12:e1005876. doi: 10.1371/journal.pgen.1005876
Ashwin, N. M. R., Barnabas, L., Ramesh Sundar, A., Malathi, P., Viswanathan, R., Masi, A., et al. (2018). CfPDIP1, a novel secreted protein of Colletotrichum falcatum, elicits defense responses in sugarcane and triggers hypersensitive response in tobacco. Appl. Microbiol. Biotechnol. 102, 6001–6021. doi: 10.1007/s00253-018-9009-2
Aylward, J., Steenkamp, E. T., Dreyer, L. L., Roets, F., Wingfield, B. D., and Wingfield, M. J. (2017). A plant pathology perspective of fungal genome sequencing. IMA Fungus 8, 1–15. doi: 10.5598/imafungus.2017.08.01.01
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bazzicalupo, A. (2022). Local adaptation in fungi. FEMS Microbiol. Rev. 46:fuac026. doi: 10.1093/femsre/fuac026
Ben M'barek, S., Cordewener, J. H., Tabib Ghaffary, S. M., Van Der Lee, T. A., Liu, Z., Mirzadi Gohari, A., et al. (2015). FPLC and liquid-chromatography mass spectrometry identify candidate necrosis-inducing proteins from culture filtrates of the fungal wheat pathogen Zymoseptoria tritici. Fungal Genet. Biol. 79, 54–62. doi: 10.1016/j.fgb.2015.03.015
Bergmann, S., Schumann, J., Scherlach, K., Lange, C., Brakhage, A. A., and Hertweck, C. (2007). Genomics-driven discovery of PKS-NRPS hybrid metabolites from aspergillus nidulans. Nat. Chem. Biol. 3, 213–217. doi: 10.1038/nchembio869
Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., Van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Bushnell, B., Rood, J., and Singer, E. (2017). BBMerge–accurate paired shotgun read merging via overlap. PLoS One 12:e0185056. doi: 10.1371/journal.pone.0185056
Call, R. E., and Matheron, M. E. (1994). Control of Septoria leaf spot of pistachio (Pistacia Vera). HortScience 29, 1408f–1409f. doi: 10.21273/HORTSCI.29.12.1408f
Çat, A. (2022). “Current status of Septoria leaf spot disease in pistachio (Pistacia vera L.)” in Forest and agricultural studies from different perspectives. ed. A. B. M. D. Ulusan (Lithuania: SRA Academic Publishing), 133–146.
Chand, R., Pal, C., Singh, V., Kumar, M., Singh, V. K., and Chowdappa, P. (2015). Draft genome sequence of Cercospora canescens: a leaf spot causing pathogen. Curr Sci India 109, 2103–2110. doi: 10.18520/cs/v109/i11/2103-2110
Chedli, R. B. H., Aouini, L., M’barek, S. B., Bahri, B. A., Verstappen, E., Kema Gerrit, H., et al. (2022). Genetic diversity and population structure of Zymoseptoria tritici on bread wheat in Tunisia using SSR markers. Eur. J. Plant Pathol. 163, 429–440. doi: 10.1007/s10658-022-02486-x
Chen, H., Lee, M. H., Daub, M. E., and Chung, K. R. (2007). Molecular analysis of the cercosporin biosynthetic gene cluster in Cercospora nicotianae. Mol. Microbiol. 64, 755–770. doi: 10.1111/j.1365-2958.2007.05689.x
Cheng, Z., Lv, X., Duan, C., Zhu, H., Wang, J., Xu, Z., et al. (2023). Pathogenicity variation in two genomes of Cercospora species causing gray leaf spot in maize. Mol. Plant Microbe Interact. 36, 14–25. doi: 10.1094/MPMI-06-22-0138-R
Chitzandis, A. (1956). Species of Septoria on the leaves of Pistacia vera L. and their perfect states. Ann Inst Phytopathol Benaki 10, 29–44.
Chowdhury, N. S., Sohrab, M. H., Rana, M. S., Hasan, C. M., Jamshidi, S., and Rahman, K. M. (2017). Cytotoxic naphthoquinone and Azaanthraquinone derivatives from an endophytic fusarium solani. J. Nat. Prod. 80, 1173–1177. doi: 10.1021/acs.jnatprod.6b00610
Crous, P. W., Quaedvlieg, W., Sarpkaya, K., Can, C., and Erkilic, A. (2013). Septoria-like pathogens causing leaf and fruit spot of pistachio. IMA Fungus 4, 187–199. doi: 10.5598/imafungus.2013.04.02.04
Cruz, M. C., Cavallo, L. M., GöRlach, J. M., Cox, G., Perfect, J. R., Cardenas, M. E., et al. (1999). Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol. Cell. Biol. 19, 4101–4112. doi: 10.1128/MCB.19.6.4101
Danecek, P., Bonfield, J. K., Liddle, J., Marshall, J., Ohan, V., Pollard, M. O., et al. (2021). Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008
Dhingra, O., and Sinclair, J. (1985). Basic plant pathology methods. Boca Raton, Florida, USA: CRC Press, Inc.
Dong, Q., Mao, K., Duan, D., Zhao, S., Wang, Y., Wang, Q., et al. (2018). Genome-wide analyses of genes encoding FK506-binding proteins reveal their involvement in abiotic stress responses in apple. BMC Genomics 19, 1–16. doi: 10.1186/s12864-018-5097-8
Drais, M. I., Gusella, G., Mazzaglia, A., and Polizzi, G. (2023). A quantitative PCR assay for the detection and quantification of Septoria pistaciarum, the causal agent of pistachio leaf spot in Italy. PLoS One 18:e0286130. doi: 10.1371/journal.pone.0286130
Eskalen, A., Küsek, M., Danıstı, L., and Karada, S. (2001). Fungal diseases in pistachio trees in East-Mediterranean and southeast Anatolian regions. Cah Options Mediterr 56, 261–264.
Feurtey, A., Lorrain, C., Mcdonald, M. C., Milgate, A., Solomon, P. S., Warren, R., et al. (2023). A thousand-genome panel retraces the global spread and adaptation of a major fungal crop pathogen. Nat. Commun. 14:1059. doi: 10.1038/s41467-023-36674-y
Gallo, M., Luti, S., Baroni, F., Baccelli, I., Cilli, E. M., Cicchi, C., et al. (2023). Plant defense elicitation by the Hydrophobin Cerato-Ulmin and correlation with its structural features. Int. J. Mol. Sci. 24:2251. doi: 10.3390/ijms24032251
Gardiner, D. M., Mcdonald, M. C., Covarelli, L., Solomon, P. S., Rusu, A. G., Marshall, M., et al. (2012). Comparative pathogenomics reveals horizontally acquired novel virulence genes in fungi infecting cereal hosts. PLoS Pathog. 8:e1002952. doi: 10.1371/journal.ppat.1002952
Gaulin, E., Drame, N., Lafitte, C., Torto-Alalibo, T., Martinez, Y., Ameline-Torregrosa, C., et al. (2006). Cellulose binding domains of a Phytophthora cell wall protein are novel pathogen-associated molecular patterns. Plant Cell 18, 1766–1777. doi: 10.1105/tpc.105.038687
Golan, J. J., and Pringle, A. (2017). Long-distance dispersal of Fungi. Microbiol Spectr 5:2016. doi: 10.1128/microbiolspec.FUNK-0047-2016
Gu, X., Ding, J., Liu, W., Yang, X., Yao, L., Gao, X., et al. (2020). Comparative genomics and association analysis identifies virulence genes of Cercospora sojina in soybean. BMC Genomics 21:172. doi: 10.1186/s12864-020-6581-5
Gusella, G., Aiello, D., Michailides, T. J., and Polizzi, G. (2021). Update of pistachio leaf spot caused by Septoria pistaciarum in light of new taxonomic advances in Italy. Fungal Biol. 125, 962–970. doi: 10.1016/j.funbio.2021.08.006
Hane, J. K., Paxman, J., Jones, D. A., Oliver, R. P., and De Wit, P. (2020). “CATAStrophy,” a genome-informed trophic classification of filamentous plant pathogens–how many different types of filamentous plant pathogens are there? Front. Microbiol. 10:3088. doi: 10.3389/fmicb.2019.03088
Hassani, M. A., Oppong-Danquah, E., Feurtey, A., Tasdemir, D., and Stukenbrock, E. H. (2022). Differential regulation and production of secondary metabolites among isolates of the fungal wheat pathogen Zymoseptoria tritici. Appl. Environ. Microbiol. 88, e02296–e02221. doi: 10.1128/aem.02296-21
Howard, S. J., Cerar, D., Anderson, M. J., Albarrag, A., Fisher, M. C., Pasqualotto, A. C., et al. (2009). Frequency and evolution of azole resistance in aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15, 1068–1076. doi: 10.3201/eid1507.090043
Jacobson, E. S. (2000). Pathogenic roles for fungal melanins. Clin. Microbiol. Rev. 13, 708–717. doi: 10.1128/CMR.13.4.708
Jones, D. A., Bertazzoni, S., Turo, C. J., Syme, R. A., and Hane, J. K. (2018). Bioinformatic prediction of plant-pathogenicity effector proteins of fungi. Curr. Opin. Microbiol. 46, 43–49. doi: 10.1016/j.mib.2018.01.017
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Jones, D. A., Rozano, L., Debler, J. W., Mancera, R. L., Moolhuijzen, P. M., and Hane, J. K. (2021). An automated and combinative method for the predictive ranking of candidate effector proteins of fungal plant pathogens. Sci. Rep. 11:19731. doi: 10.1038/s41598-021-99363-0
Kamle, M., Mahato, D. K., Devi, S., Lee, K. E., Kang, S. G., and Kumar, P. (2019). Fumonisins: impact on agriculture, food, and human health and their management strategies. Toxins 11:328. doi: 10.3390/toxins11060328
Kettles, G. J., Bayon, C., Sparks, C. A., Canning, G., Kanyuka, K., and Rudd, J. J. (2018). Characterization of an antimicrobial and phytotoxic ribonuclease secreted by the fungal wheat pathogen Zymoseptoria tritici. New Phytol. 217, 320–331. doi: 10.1111/nph.14786
Kim, D., Paggi, J. M., Park, C., Bennett, C., and Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915. doi: 10.1038/s41587-019-0201-4
Krasnoff, S. B., Keresztes, I., Gillilan, R. E., Szebenyi, D. M., Donzelli, B. G., Churchill, A. C., et al. (2007). Serinocyclins a and B, cyclic heptapeptides from Metarhizium anisopliae. J. Nat. Prod. 70, 1919–1924. doi: 10.1021/np070407i
Lechner, M., Findeiss, S., Steiner, L., Marz, M., Stadler, P. F., and Prohaska, S. J. (2011). Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. doi: 10.1186/1471-2105-12-124
Lerat, E., Fablet, M., Modolo, L., Lopez-Maestre, H., and Vieira, C. (2016). TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Res. 45, gkw953–e17. doi: 10.1093/nar/gkw953
Leroux, P., Fritz, R., Debieu, D., Albertini, C., Lanen, C., Bach, J., et al. (2002). Mechanisms of resistance to fungicides in field strains of Botrytis cinerea. Pest Manag. Sci. 58, 876–888. doi: 10.1002/ps.566
Letunic, I., and Bork, P. (2021). Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Levy Karin, E., Mirdita, M., and Soding, J. (2020). MetaEuk-sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome 8:48. doi: 10.1186/s40168-020-00808-x
Li, L., Dou, N., Zhang, H., and Wu, C. (2021). The versatile GABA in plants. Plant Signal. Behav. 16:1862565. doi: 10.1080/15592324.2020.1862565
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, X., Yang, S., Zhang, M., Yang, Y., and Peng, L. (2022). Identification of pathogenicity-related effector proteins and the role of Piwsc1 in the virulence of Penicillium italicum on Citrus fruits. J Fungi 8:646. doi: 10.3390/jof8060646
Lin, Z., Wang, W., and Chen, J. (2022). Genome sequence resource for Cercospora rodmanii J1, a potential biological control agent for water hyacinth. Phytopathology 112, 2462–2465. doi: 10.1094/PHYTO-04-22-0119-A
Lopez-Moral, A., Agusti-Brisach, C., Raya, M. D. C., Lovera, M., Trapero, C., Arquero, O., et al. (2022). Etiology of Septoria leaf spot of pistachio in southern Spain. Plant Dis. 106, 406–417. doi: 10.1094/PDIS-02-21-0331-RE
Mair, W., Lopez-Ruiz, F., Stammler, G., Clark, W., Burnett, F., Hollomon, D., et al. (2016). Proposal for a unified nomenclature for target-site mutations associated with resistance to fungicides. Pest Manag. Sci. 72, 1449–1459. doi: 10.1002/ps.4301
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12. doi: 10.14806/ej.17.1.200
Mateos, R., Salvador, M. D., Fregapane, G., and Goya, L. (2022). Why should pistachio be a regular food in our diet? Nutrients 14:3207. doi: 10.3390/nu14153207
Mcgrann, G. R., Andongabo, A., Sjokvist, E., Trivedi, U., Dussart, F., Kaczmarek, M., et al. (2016). The genome of the emerging barley pathogen Ramularia collo-cygni. BMC Genomics 17:584. doi: 10.1186/s12864-016-2928-3
Mckenna, A., Hanna, M., Banks, E., Sivachenko, A., Cibulskis, K., Kernytsky, A., et al. (2010). The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. doi: 10.1101/gr.107524.110
Mekonnen, T., Haileselassie, T., Goodwin, S. B., and Tesfayea, K. (2020). Genetic diversity and population structure of Zymoseptoria tritici in Ethiopia as revealed by microsatellite markers. Fungal Genet. Biol. 141:103413. doi: 10.1016/j.fgb.2020.103413
Meng, G., Li, Y., Yang, C., and Liu, S. (2019). MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47:e63. doi: 10.1093/nar/gkz173
Mills, S., Lunt, D. H., and Gomez, A. (2007). Global isolation by distance despite strong regional phylogeography in a small metazoan. BMC Evol. Biol. 7:225. doi: 10.1186/1471-2148-7-225
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mounkoro, P., Michel, T., Benhachemi, R., Surpateanu, G., Iorga, B. I., Fisher, N., et al. (2019). Mitochondrial complex III qi-site inhibitor resistance mutations found in laboratory selected mutants and field isolates. Pest Manag. Sci. 75, 2107–2114. doi: 10.1002/ps.5264
Oliver, R. P., Hane, J. K., Mair, W., and Lopez-Ruiz, F. (2024). The 2023 update of target site mutations associated with resistance to fungicides and a web-tool to assist label designations. J Plant Dis Protect, 1–6. doi: 10.1007/s41348-024-00872-7
Orner, V. A., Cantonwine, E. G., Wang, X. M., Abouelleil, A., Bochicchio, J., Nusbaum, C., et al. (2015). Draft genome sequence of Cercospora arachidicola, causal agent of early leaf spot in peanuts. Genome Announc. 3:15. doi: 10.1128/genomeA.01281-15
Palmer, J., and Stajich, J. (2020). Funannotate v1. 8.1: Eukaryotic genome annotation. 4054262. https://zenodo.org/records/4054262
Ren, W., Shao, W., Han, X., Zhou, M., and Chen, C. (2016). Molecular and biochemical characterization of laboratory and field mutants of Botrytis cinerea resistant to Fludioxonil. Plant Dis. 100, 1414–1423. doi: 10.1094/PDIS-11-15-1290-RE
Rogers, S. O., and Rogers, M. A. (1999). “Gene flow in fungi” in Structure and dynamics of fungal populations. Population and community biology series. ed. J. Worall, vol. 25 (Dordrecht: Springer), 97–121.
Sautua, F. J., Gonzalez, S. A., Doyle, V. P., Berretta, M. F., Gordo, M., Scandiani, M. M., et al. (2019). Draft genome sequence data of Cercospora kikuchii, a causal agent of Cercospora leaf blight and purple seed stain of soybeans. Data Brief 27:104693. doi: 10.1016/j.dib.2019.104693
Simao, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V., and Zdobnov, E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi: 10.1093/bioinformatics/btv351
Singh, K., Sun, S., and Vezina, C. (1979). Rapamycin (AY-22,989), a new antifungal antibiotic. IV. Mechanism of action. J. Antibiot. 32, 630–645. doi: 10.7164/antibiotics.32.630
Sjokvist, E., Lemcke, R., Kamble, M., Turner, F., Blaxter, M., Havis, N. H. D., et al. (2019). Dissection of Ramularia leaf spot disease by integrated analysis of barley and Ramularia collo-cygni transcriptome responses. Mol. Plant Microbe Interact. 32, 176–193. doi: 10.1094/MPMI-05-18-0113-R
Stam, R., Münsterkötter, M., Pophaly, S. D., Fokkens, L., Sghyer, H., Güldener, U., et al. (2018). A new reference genome shows the one-speed genome structure of the barley pathogen Ramularia collo-cygni. Genome Biol. Evol. 10, 3243–3249. doi: 10.1093/gbe/evy240
Stammler, G., and Semar, M. (2011). Sensitivity of Mycosphaerella graminicola (anamorph: Septoria tritici) to DMI fungicides across Europe and impact on field performance. EPPO Bull 41, 149–155. doi: 10.1111/j.1365-2338.2011.02454.x
Testa, A., Oliver, R., and Hane, J. (2015). Overview of genomic and bioinformatic resources for Zymoseptoria tritici. Fungal Genet. Biol. 79, 13–16. doi: 10.1016/j.fgb.2015.04.011
Testa, A. C., Oliver, R. P., and Hane, J. K. (2016). OcculterCut: a comprehensive survey of AT-rich regions in fungal genomes. Genome Biol. Evol. 8, 2044–2064. doi: 10.1093/gbe/evw121
Tintor, N., Paauw, M., Rep, M., and Takken, F. L. (2020). The root-invading pathogen fusarium oxysporum targets pattern-triggered immunity using both cytoplasmic and apoplastic effectors. New Phytol. 227, 1479–1492. doi: 10.1111/nph.16618
Vaghefi, N., Kikkert, J. R., Bolton, M. D., Hanson, L. E., Secor, G. A., and Pethybridge, S. J. (2017). De novo genome assembly of Cercospora beticola for microsatellite marker development and validation. Fungal Ecol. 26, 125–134. doi: 10.1016/j.funeco.2017.01.006
Villa-Cuesta, E., Holmbeck, M. A., and Rand, D. M. (2014). Rapamycin increases mitochondrial efficiency by mtDNA-dependent reprogramming of mitochondrial metabolism in Drosophila. J. Cell Sci. 127, 2282–2290. doi: 10.1242/jcs.142026
Wawra, S., Fesel, P., Widmer, H., Neumann, U., Lahrmann, U., Becker, S., et al. (2019). FGB1 and WSC3 are in planta-induced beta-glucan-binding fungal lectins with different functions. New Phytol. 222, 1493–1506. doi: 10.1111/nph.15711
Wawra, S., Fesel, P., Widmer, H., Timm, M., Seibel, J., Leson, L., et al. (2016). The fungal-specific beta-glucan-binding lectin FGB1 alters cell-wall composition and suppresses glucan-triggered immunity in plants. Nat. Commun. 7:13188. doi: 10.1038/ncomms13188
Wilken, P. M., Steenkamp, E. T., Wingfield, M. J., De Beer, Z. W., and Wingfield, B. D. (2017). Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biol. Rev. 31, 199–211. doi: 10.1016/j.fbr.2017.05.003
Wingfield, B. D., Berger, D. K., Steenkamp, E. T., Lim, H. J., Duong, T. A., Bluhm, B. H., et al. (2017). IMA genome-F 8: draft genome of Cercospora zeina, fusarium pininemorale, Hawksworthiomyces lignivorus, Huntiella decipiens and Ophiostoma ips. IMA Fungus 8, 385–396. doi: 10.5598/imafungus.2017.08.02.10
Yan, T., Zhou, X., Li, J., Li, G., Zhao, Y., Wang, H., et al. (2022). FoCupin1, a Cupin_1 domain-containing protein, is necessary for the virulence of fusarium oxysporum f. sp. cubense tropical race 4. Front. Microbiol. 13:1001540. doi: 10.3389/fmicb.2022.1001540
Yang, L., Gao, G., Gao, W., Zhang, C., Wu, F., Huo, J., et al. (2023). Complete genome sequence resource for Cercospora apii causing leaf spot of celery. Mol. Plant Microbe Interact. 36, 670–673. doi: 10.1094/MPMI-04-23-0039-A
Young, D., and Michailides, T. (1989). 1st report of Septoria leaf-spot of pistachio in Arizona. Plant Dis. 73:775. doi: 10.1094/PD-73-0775E
Zeng, F., Lian, X., Zhang, G., Yu, X., Bradley, C. A., and Ming, R. (2017a). A comparative genome analysis of Cercospora sojina with other members of the pathogen genus Mycosphaerella on different plant hosts. Genom Data 13, 54–63. doi: 10.1016/j.gdata.2017.07.007
Zeng, F., Wang, C., Zhang, G., Wei, J., Bradley, C. A., and Ming, R. (2017b). Draft genome sequence of Cercospora sojina isolate S9, a fungus causing frogeye leaf spot (FLS) disease of soybean. Genom Data 12, 79–80. doi: 10.1016/j.gdata.2017.02.014
Keywords: Septoria pistaciarum, pistachio, Mycosphaerellaceae, plant-pathogen, pathogenicity effectors
Citation: Zambounis A, Boutsika A, Gray N, Hossain M, Chatzidimopoulos M, Tsitsigiannis DI, Paplomatas E and Hane J (2024) Pan-genome survey of Septoria pistaciarum, causal agent of Septoria leaf spot of pistachios, across three Aegean sub-regions of Greece. Front. Microbiol. 15:1396760. doi: 10.3389/fmicb.2024.1396760
Edited by:
Levente Kiss, University of Southern Queensland, AustraliaReviewed by:
Niloofar Vaghefi, The University of Melbourne, AustraliaMuhammad Zahid Mumtaz, The University of Lahore, Pakistan
Stefan Kusch, RWTH Aachen University, Germany
Copyright © 2024 Zambounis, Boutsika, Gray, Hossain, Chatzidimopoulos, Tsitsigiannis, Paplomatas and Hane. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Antonios Zambounis, azampounis@elgo.gr; James Hane, james.hane@curtin.edu.au
 Antonios Zambounis
Antonios Zambounis Anastasia Boutsika
Anastasia Boutsika Naomi Gray2
Naomi Gray2 Mohitul Hossain
Mohitul Hossain Dimitrios I. Tsitsigiannis
Dimitrios I. Tsitsigiannis James Hane
James Hane