
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Microbiol. , 24 May 2024
Sec. Microorganisms in Vertebrate Digestive Systems
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1396031
This article is part of the Research Topic Rodent model organisms: Therapeutic treatments and drugs interaction with the gut microbiome View all 41 articles
Metformin is of great focus because of its high safety, low side effects, and various effects other than lowering blood sugar, such as anti-inflammation, anti-tumor, and anti-aging. Studies have shown that metformin has a modulating effect on the composition and function of the intestinal microbiota other than acting on the liver. However, the composition of microbiota is complex and varies to some extent between species and individuals, and the experimental design of each study is also different. Multiple factors present a major obstacle to better comprehending the effects of metformin on the gut microbiota. This paper reviews the regulatory effects of metformin on the gut microbiota, such as increasing the abundance of genus Akkermansia, enriching short-chain fatty acids (SCFAs)-producing bacterial genus, and regulating gene expression of certain genera. The intestinal microbiota is a large and vital ecosystem in the human body and is considered to be the equivalent of an “organ” of the human body, which is highly relevant to human health and disease status. There are a lot of evidences that the gut microbiota is responsible for metformin’s widespread effects. However, there are only a few systematic studies on this mechanism, and the specific mechanism is still unclear. This paper aims to summarize the possible mechanism of metformin in relation to gut microbiota.
Metformin is a synthetic derivative of guanidine derived from the guanidine alkaloid of the plant named Galega officinalis L. with significant hypoglycemic effects (Ursini et al., 2018), which was first successfully synthesized by two Irish scientists, Werner and Bell (1922). Since its clinical application in 1957, metformin’s cornerstone status has remained consistent and it is still the first-line preferred treatment for type 2 diabetes mellitus (T2DM) side by side with lifestyle (Montvida et al., 2018; Nicolucci et al., 2019; Ahmad et al., 2020). T2DM is a chronic, low-grade inflammatory disease characterized by elevated blood glucose and is regulated by a combination of genetics and environment. The incidence of T2DM continues to rise worldwide due to people’s bad dietary habits, the decrease in exercise and unhealthy lifestyles. According to the International Diabetes Federation, 1 in 8 adults, about 783 million people, will live with diabetes by 2045, and more than 90% of them will have T2DM (The Idf Diabetes Atlas, 2021). Metformin is widely used because of its low cost, good efficacy and few adverse reactions (Lancet, 1998), but its exact mechanism of action still remains partly unclear.
Now the confirmed mechanism of metformin’s hypoglycemic action is to inhibit hepatic gluconeogenesis and reduce hepatic glucose output through both AMPK-dependent (Song et al., 2001; Shaw et al., 2005; Miller et al., 2013) and AMPK-independent (Madiraju et al., 2014, 2018) pathways. At the same time, metformin also promotes the uptake and utilization of glucose by peripheral tissues. Extensive scientific evidences have also shown that in the gut, metformin can also increase the secretion of glucagon-like peptide-1 (GLP-1) by L cells (Kim et al., 2014; Napolitano et al., 2014; Duca et al., 2015), thereby improving blood glucose homeostasis and reducing the secretion of lipids by intestinal epithelial cells. Therefore, metformin can not only regulate glucose metabolism, but also regulate lipid metabolism. This feature makes it very suitable for obese patients with T2DM. In addition, metformin was also found to have anti-tumor effects.
The process of metformin exerting its extensive effects cannot be separated from the regulation of gut microbiota. According research (Koropatkin and Martens, 2017), when metformin is administered orally, its bioavailability is about 50%, and it is higher compared to intravenous administration. Its action mainly occurs in the intestine, suggesting that intestinal microbiota may be another target of metformin (Wang J. H. et al., 2018). With the widespread use of metformin in clinical practice, the research on it has become increasingly advanced. There have been substantial evidences that metformin has a regulatory effect on the gut microbiota (Zhang et al., 2015; Elbere et al., 2018; Harsch and Konturek, 2018; Brandt et al., 2019), and the process by which metformin exerts its broad effects is also closely correlated with the gut microbiota. Even though this correlation has been verified, its specific mechanisms are not fully understood. In this paper, we comprehensively review the progress of its research.
After oral administration of metformin, it enters the gastrointestinal (GI) tract and is absorbed by the intestinal epithelial cells of the upper small intestine. This absorption in the stomach and large intestine is almost negligible (Graham et al., 2011). Because the hydrophilicity of the drug inhibits its transport process across cell membranes, metformin must rely on transporters to actively transport in and out of cells (Hardie et al., 2012). Intestinal epithelial cells take up metformin through transporters located on the inner surface of the intestinal epithelium such as plasma monoamine transporter protein (PMAT) and organic cation transporter protein 3 (OCT3) (Figure 1). However, some studies have found that metformin is also taken up in the intestine by passive diffusion. But there is no consensus on whether this type of translocation occurs via paracellular or transcellular pathways (Shirasaka et al., 2022). The drug then leaves the intestinal epithelium via organic cation transporter protein 1 (OCT1) and is delivered to the liver via the portal vein. Metformin enters the liver via OCT1 and OCT3, where it suppresses gluconeogenesis. Multi-specificity is one of the most distinguishing features of OCT1 (Meyer and Tzvetkov, 2021). Drugs are not metabolized by the liver, but multidrug and toxin extrusion protein 1 (MATE1) expressed in hepatocytes is involved in the clearance of drugs, which is transported to the kidney through bile or through blood (Gong et al., 2012). Metformin then enters the renal epithelium via organic cation transporter protein 2 (OCT2). The drug is then secreted by MATE1 and MATE2 of the kidney in an unaltered form and is cleared through the urine.
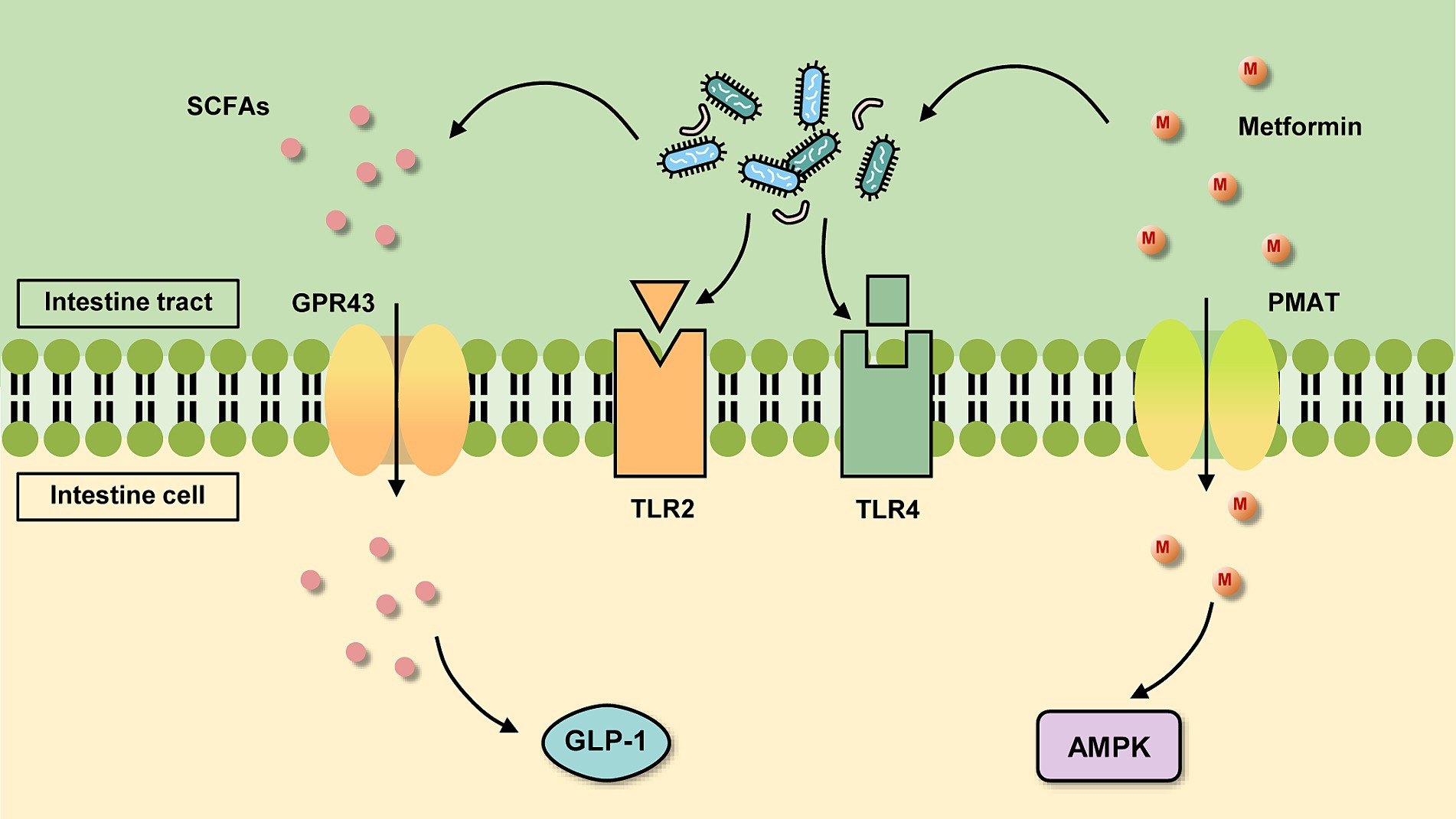
Figure 1. Basic mechanism of metformin action in the intestine. On the one hand, metformin is absorbed by intestinal cells through transporters such as plasma monoamine transporter protein (PMAT) and organic cation transporter protein 3 (OCT3), which activate the AMPK pathway. On the other hand, it can act in the gut on the gut microbiota, which changes and acts on certain receptors or produces certain metabolites for further regulation.
It is well known that the pharmacodynamic property of metformin is anti-diabetic, but there is growing evidence that it has a number of extra-hypoglycemic effects. Firstly, metformin has been shown to improve lipid metabolism (He, 2020) and therefore has a favorable effect on body weight (Day et al., 2019; Zhang et al., 2023), blood lipids (Wulffelé et al., 2004), and cardiovascular risk (Després, 2003) associated with T2DM. Secondly, the role of metformin that has received the most attention is its effect on tumor tissue, not only in terms of treatment, but also in terms of prevention (Coyle et al., 2016; Cejuela et al., 2022). In addition, many studies have emphasized the anti-inflammatory (Pålsson-McDermott and O’Neill, 2020) and anti-aging (Soukas et al., 2019) effects of metformin. Current research done on the potential beneficial effects of metformin has extended to treating diabetic nephropathy (Han et al., 2021), metabolic syndrome (Ladeiras-Lopes et al., 2015), and polycystic ovary syndrome (Peng et al., 2023). There are even recent studies that have found metformin to have protective and therapeutic effects against COVID-19 (Bramante et al., 2021, 2023).
The gut microbiota is a fairly complex ecosystem. Luckey (1972) found in 1972 that the number of human intestinal microbiota can reach 1014 species, and this data has been accepted and quoted by most scholars after publication. The amount of genetic material in the intestinal microbiota exceeds the human genome by more than 100 times (Bäckhed et al., 2005). The intestinal microbial community is diverse and dominated by five main bacterial phyla: Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria and Verrucomicrobia. Among these five phyla, the phylum Bacteroidetes and the phylum Firmicutes accounted for more than 90% of the total population. Most of the bacteria under the phylum Bacteroidetes belong to the genera Bacteroides and Prevotella. And bacteria under the phylum Firmicutes that dominate the gut microbiota include the genera Clostridium, Eubacterium and Ruminococcus (Eckburg et al., 2005). The proportion of various microbiota is different depending on the site of the GI tract. The upper GI tract was mainly enriched with Firmicutes, Proteobacteria and Lactobacillaceae, while the microbiota of lower GI tract was mainly composed of Bacteroidetes, Firmicutes, and A.muciniphila (Scheithauer et al., 2016; Meijnikman et al., 2018). In addition to taxonomic classification, the human microbiota can be categorized into three different enterotypes: Bacteroides, Prevotella or Ruminococcus. Bacteroides-dominated enterotype is characterized by saccharolytic and proteolytic activities involved in synthesis of riboflavin, biotin, ascorbate and pantothenate. The enterotype of Prevotella-dominated plays the role of mucin glycoprotein degraders involved in the synthesis of thiamine and folate. The last enterotype is dominated by Ruminococcus, characterized by membrane transportation of sugars and mucin degrading activities (Arumugam et al., 2011). A study from Taiwan categorized enterotypes into Bacteroides, Prevotella and Enterobacteriaceae and claimed that Enterobacteriaceae may be a new subtype in Asian populations (Liang et al., 2017). However, the concept of enterotypes has been debated due to the high degree of variability observed in the gut microbiota between individuals and the fact that many of the data show that there are not three completely discrete clusters (Knights et al., 2014). It is indisputable that regardless of enterotypes, some members of the microbial population act as a “core microbiota,” while others are more of a “flexible pool.” The “core microbiota” is composed of host-adapted microbes reproducibly included in the gut microbiota made up of different environmental combinations and determined by genetic factors. While the “flexible pool” is usually obtained from water, food and various components of the environment, which contributes to the adaptation of the host (Shapira, 2016).
Human cells coexist with bacteria for a long time, and there is a complex process of material and energy exchange between them. Intestinal microbiota plays a crucial role in many key metabolic processes of human body, such as SCFAs, amino acids, bile acids and vitamin synthesis (Montandon and Jornayvaz, 2017). There is no doubt that the gut microbiome is extremely relevant to human health and disease status. In many disease states, such as metabolic syndrome, inflammatory bowel disease (IBD), cardiovascular diseases, malignant tumors, etc., the structure and function of intestinal microbiota are obviously different from those of healthy bowel microbiota (Tilg and Moschen, 2014; Shin et al., 2017; Harsch and Konturek, 2018; Eibl and Rozengurt, 2019). Regulating the structure and function of intestinal microflora can improve the disease status of the body to a certain extent. So, targeting the gut microbiome may provide new therapeutic thoughts for some intractable diseases. In particular, the role of gut microbiota in T2DM and obesity has attracted the attention of many researchers (Boulangé et al., 2016; Pedersen et al., 2016; Cani, 2018; Chobot et al., 2018; Zhao et al., 2018). Gut bacterial therapies that improve insulin sensitivity by altering the composition of the gut microbiota have become a new therapeutic modality.
Study has shown that changes in modern lifestyles have led to a decrease in the diversity of the gut microbiota in many developed populations (Clemente et al., 2015). This reduced diversity may promote the development of metabolic disorders. Individuals with low gut microbiota abundance have been found to be susceptible to obesity, insulin resistance and dyslipidemia (le Chatelier et al., 2013). Lactobacillus, Prevotella, Bacteroides, Desulfovibrio, and Oxalobacter spp. are decreasing in the gut microbiota of low-gene-count patients compared with high-gene-count patients. Functional changes in the microbiota of low-gene-count patients primarily consist of a decrease in butyrate-producing bacteria and an increase in the ratio of Akkermansia to Ruminococcus gnavus, which lead to decreased methane production potential, decreased hydrogen production potential, enhanced mucus degradation, and increased peroxidase activity (le Chatelier et al., 2013). The metabolic disturbances resulting from this imbalance of anti-inflammatory and pro-inflammatory bacterial species puts people at increased risk of suffering from T2DM (Ouchi et al., 2011). Approximately 86% of patients with T2DM are overweight or obese, and obesity is considered the greatest risk factor for T2DM (Daousi et al., 2006; Singer-Englar et al., 2019). A number of studies have reported significant changes in the gut microbiota of patients with obesity and T2DM compared to healthy adults (Qin et al., 2012; Karlsson et al., 2013; Tims et al., 2013; Forslund et al., 2015; Dao et al., 2016; le Roy et al., 2018), manifested in an overall increase in the abundance of Bacteroides and Prevotella copri as well as a decrease in the abundance of Akkermansia, Roseburia and Ruminococcus.
Metformin comes from Galega officinalis L., and later the function of lowering blood glucose has been found in animals, and more and more studies on its hypoglycemic effect have been conducted, and the effect has been furtherly confirmed. There has also been a great deal of research on its mechanisms. Stepensky et al. (2002) stated that metformin was better able to lower blood glucose by intestinal administration. Subsequently, researchers boldly hypothesized that metformin acted through the gut. Duca et al. (2015) found that the hypoglycemic effect of metformin was reduced by blocking the cAMP pathway in the gut, and speculated that the target of metformin was mainly in the gut. Buse et al. (2016) confirmed again that the gut is the main locus of hypoglycemic effects of metformin through comparable plasma levels of metformin. Meanwhile, due to its safety and few side effects, metformin has been widely studied, and it has been found that its effect is not only limited to reducing hyperglycemia, but also delaying the development of diabetes complications (Scheen and Paquot, 2013; Yaribeygi et al., 2019; American Diabetes Association, 2020). Therefore, people have found that it has many extra-hypoglycemic effects, such as anti-inflammatory, anti-tumor, anti-aging and so on. Among them, the anti-inflammatory effect has attracted the most attention, because in many diseased states, besides specific symptoms the body is generally in a state of chronic inflammation.
The mechanism of metformin has been studied endlessly, but the mechanism has not yet been fully clarified. In recent years, with advances in gene sequencing technology, more and more research has focused on the effects of metformin on the gut microbiota. It has been possible to confirm the association of metformin with certain intestinal microbiota. Most animal studies (Lee et al., 2018; Wang Z. et al., 2018; Zheng et al., 2018; Brandt et al., 2019; Ji et al., 2019; Ahmadi et al., 2020; Chung et al., 2020; Ryan et al., 2020) used C57BL/6 mice to construct a high-fat diet (HFD)-induced diabetes model followed by metformin treatment. Some studies have also used db/db mice (Chen et al., 2018; Zhang et al., 2019) or KKAy mice (Gao et al., 2018). Although the structure of the intestinal microbiota is very complex and the experimental design varies among studies, such as the different timing of HFD induction, distinctions in the duration of metformin treatment, and variations in mouse strains, but the impact of metformin on gut microbiota derived from these studies are nearly consistent. For example, the proportions of phyla Bacteroidetes (Gao et al., 2018; Ahmadi et al., 2020; Ryan et al., 2020) and Verrucomicrobia (Gao et al., 2018; Lee et al., 2018; Ryan et al., 2020) and genera Akkermansia (Chen et al., 2018; Gao et al., 2018; Lee et al., 2018; Zheng et al., 2018; Ji et al., 2019; Zhang et al., 2019; Ryan et al., 2020) and Bacteroides (Chen et al., 2018; Gao et al., 2018; Lee et al., 2018; Ryan et al., 2020) were significantly increased in the metformin treatment group. Similar results were also manifested in the rat model as in the mouse model (Zhang et al., 2015; Cui et al., 2019). Details are shown in Table 1.
However, the results in humans are not entirely consistent with those in animals. This may be due to the significant differences in gut microbial diversity between humans and animals (Peng et al., 2013). Moreover, clinical trials in patients with diabetes are affected by many complex factors, such as diet, race, comorbidities, and drug combinations. An analysis of a T2DM cohort based on metagenomic sequencing and validated in an independent cohort found that metformin-treated patients had an increase in Escherichia coli and a decrease in Intestinibacter, and that the abundance of several intestinal microbial genera was more similar to that of normal control levels, particularly Akkermansia (Forslund et al., 2015). Moreover, A double-blind study of patients with T2DM treated with metformin led to significant changes in the relative abundance of more than 80 bacterial species compared with placebo after 4 months, in which an increase in Escherichia and Akkermansia muciniphila and a decrease in Intestinibacter were also observed, whereas most of the changes with placebo occurred in phylum Firmicutes and phylum Proteobacteria (Wu et al., 2017). Similar results were found in several studies on the effects of metformin on the gut microbiota of patients with obesity or T2DM (de la Cuesta-Zuluaga et al., 2017; Hiel et al., 2020), and even a study on metformin-induced changes in the gut microbiota of healthy young men likewise observed an increase in Escherichia/Shigella and a reduction in Intestinibacter (Bryrup et al., 2019). Details are shown in Table 1.
Nevertheless, the alterations in the gut microbiota of diabetic individuals by metformin treatment show consistent results in certain bacteria, in both humans and rodents. This was almost always demonstrated by enrichment of mucin-degrading A. muciniphila and SCFA-producing bacteria such as Bifidobacterium bifidum and Butyrivibrio (de la Cuesta-Zuluaga et al., 2017). However, this discrepancy between the human and animal studies suggests that we should still focus our researches in clinical setting and interpreting the mechanisms of action of the human gut microbiota. However, the reported results on the effect of metformin on human gut microbiota diversity are inconsistent. In patients with newly diagnosed T2DM, Tong et al. (2018) reported an increase in gut microbiota diversity with metformin based on Chao1 enrichment estimates, whereas Sun et al. (2018) observed a slight decrease in alpha diversity. In studies of healthy individuals, Bryrup et al. (2019) found no significant changes in gut microbial richness and diversity after metformin treatment. However, Elbere et al. (2018) reported a significant decrease in gut microbiota diversity after 24 h of metformin administration using the Shannon index. Overall, the therapy of metformin was associated with changes in the abundance of specific bacterial genera in the human gut microbiota, but the effects on gut microbiota diversity were variable, which highlights the need for further research to understand the potential mechanisms and clinical significance.
Akkermansia muciniphila, a strictly anaerobic, endospore-free ovoid gut bacterium, is true of the phylum Verrucomicrobia (Derrien et al., 2004). It mainly colonizes the outer mucus layer of the GI tract, and uses the mucin of the GI tract as a carbon and nitrogen source to maintain its growth. The dynamic balance between the consumption of mucin and the production of mucin by goblet cells is obtained to maintain the stability of the mucus layer (Sicard et al., 2017). And the abundance of A. muciniphila is obviously reduced in patients with obesity, T2DM, and cardiovascular disease. At the same time, a large number of studies have shown that the content of mucin-degrading bacteria A. muciniphila in the intestinal microbiota is significantly increased after the administration of metformin (Hur and Lee, 2015; de la Cuesta-Zuluaga et al., 2017; Wu et al., 2017; Lee et al., 2018; Verdura et al., 2019), which can make it reach 20% of the total microbiome (de la Cuesta-Zuluaga et al., 2017; Cani, 2018). Moreover, some studies have verified that metformin directly promotes the growth of this bacterium in vitro (Wu et al., 2017).
SCFAs (such as acetic acid, propionic acid and butyric acid) are usually metabolites produced by various SCFAs-producing bacteria in the gut that metabolizes carbohydrates such as dietary fiber (Rau et al., 2018), and they are essential for the health of the gut, the body and even the brain. SCFAs can affect brain function by directly or indirectly regulating the gut-brain axis through immune, vagal, endocrine and other humoral pathways (Dalile et al., 2019). They also suppress appetite as well as regulate energy homeostasis by stimulating the secretion of hormones such as GLP-1 from intestinal L cells (De Silva and Bloom, 2012). There is evidence that metformin can increase the abundance of microbiota which can all produce SCFAs such as Butyrivibrio, Bifidobacterium, Megasphaera, Prevotella, and so on (de la Cuesta-Zuluaga et al., 2017).
Lactobacillus is a class of anaerobic or partly anaerobic, non-bacteriophage bacteria belonging to the phylum Firmicutes. The bacteria of this genus have a strong ability to break down sugar, and the main end product is lactic acid, which can protect the intestinal barrier and reduce the inflammatory response. Metformin has been found to increase the abundance of Lactobacillus in rodents with HFD (Zhou et al., 2016; Bauer et al., 2018). In recent studies, this trend has also been observed in certain clinical studies in patients with T2DM (Xu et al., 2022).
One study Forbes et al. (2018) indicated that the gut microbiota of patients with immune-mediated inflammatory diseases such as IBD, ankylosing spondylitis, rheumatoid arthritis and systemic lupus erythematosus differed sharply from normal controls. For example, the abundance of Intestinibacter is sensibly higher in the gut of patients with Crohn’s disease (CD). In addition, Bacteroides fragilis is the only bacterium that has been proved to beget abscess formation, has a potent virulence factor, and is the most common anaerobic pathogen (Wexler, 2007). Other bacteria associated with the disease are Clostridioides difficile [here we use its new nomenclature according to Zheng et al. (2020)], an anaerobic enteric pathogen that can cause severe diarrhea and lead to death (Sandhu and Mcbride, 2018). Metformin declines the number of disease-associated bacteria such as Intestinibacter spp., Bacteroides fragilis, and Clostridioides difficile (de la Cuesta-Zuluaga et al., 2017; Elbere et al., 2018; Bryrup et al., 2019).
The GI mucosal barrier with important physiological roles is a three-dimensional protective structure consisting of mechanical, chemical, immune and biological barriers. Many of the mechanisms of metformin action revolve around stabilization of the GI mucosal barrier. An emphatic increase in the expression of two markers of mucin levels, MUC2 and MUC5 genes, was observed after metformin treatment (Kyriachenko et al., 2019). This suggests that the increased abundance of A. muciniphila after metformin administration may be due to the fact that metformin increases the number of goblet cells, providing more substrate for the growth of A. muciniphila. Moreover, A. muciniphila was discovered to improve HFD-induced intestinal hyperpermeability and also affect intestinal barrier function by the way that upregulates the expression of tight junction and closure proteins (Li et al., 2016). In this way, it decreases the entry of pro-inflammatory lipopolysaccharides (LPS) into the circulation, thus reducing inflammation in the organism. Additionally, Everard et al. (2013). measured the concentration of endocannabinoids (ECs) in the gut of the mice treated with A. muciniphila and found that A. muciniphila also raised the release of ECs. Increased endocannabinoids inhibit monoacylglycerol lipase (Alhouayek et al., 2011), thereby reducing systemic inflammation, altering intestinal peptide secretion and increasing intestinal mucosal barrier thickness. It is well known that T2DM is a metabolic and inflammatory disease characterized by deteriorating islet function and elevated levels of inflammatory cytokines. Therefore, therapies targeting inflammation can also recover glycemic control in T2DM patients. In a study, the treatment with three different strains of A. muciniphila was found to reverse the low-grade chronic inflammatory state in HFD mice to a certain extent, including a significant increase in the gene expression of inflammatory and immunosuppressive factor interleukin (IL)-10, as well as a decrease in the mRNA levels of tumor necrosis factor-α (TNF-α), Monocyte chemoattractant protein-1, and Toll-like receptor 2 (TLR2) (Yang et al., 2020). In terms of alleviating inflammation, A. muciniphila also modulates Forkhead box protein 3 (Foxp3) in mouse adipose tissue (Choi et al., 2021), resulting in a diminution of regulatory T cells (Tregs), and anti-inflammatory while indirectly improving glucose homeostasis. In addition, a specific membrane protein, Amuc_1100, exists on the outer membrane of A. muciniphila, and many studies have reported that it induces the production of specific cytokines by binding to TLR2 and activating the downstream pathway of TLR2 (Figure 1), which in turn improves the host’s immune homeostasis and intestinal mucosal barrier function (Plovier et al., 2017; Song et al., 2023; Wang et al., 2024), and then improves the blood glucose level. The protein retains its active conformation and functions even after pasteurization (Plovier et al., 2017). Amuc_1100 protein also has the function of lowering blood lipids, which can reduce the content of low-density lipoprotein (LDL) and cholesterol (Lee et al., 2018). Plus, vesicles secreted by A. muciniphila reduce the expression of Toll-like receptor 4 (TLR4) (Figure 1), which impacts the nuclear factor-κB (NF-κB) pathway and thus lessens the secretion of the pro-inflammatory factors IL-6, IL-8 (Ottman et al., 2017; Ashrafian et al., 2019).
What’s more, metformin can increase the abundance of Bifidobacterium which can promote the growth of gastric mucin, thereby stabilizing the GI mucosa. Cani et al. (2009) identified that after increasing the quantity of Bifidobacterium in the intestine, the content of proglucagon mRNA was higher, which promoted the secretion of glucagon-derived peptides. During the course, glucagon-like peptide-2 (GLP-2) enhanced intestinal epithelial cells proliferation and reduced gut permeability, thereby stabilizing the intestinal mucosal barrier.
For the gut, SCFAs have important roles in regulating intestinal microbiota balance, maintaining electrolyte balance and improving intestinal immune function. Administration of metformin increases the abundance of SCFAs-producing microbiota (described previously, see Chapter 4.3), thereby increasing the production of SCFAs. The produced SCFAs are partially absorbed rapidly by colonocytes via monocarboxylate transporter protein (MCT), then undergo a series of oxidative reactions, and finally provide energy to the cells in the form of ATP (Dalile et al., 2019). The SCFAs not metabolized in colonocytes enter the portal circulation of the liver through the basolateral membrane and provide substrates for energy metabolism in hepatocytes. SCFAs are also implicated in the biosynthesis of glucose, fatty acids and cholesterol in hepatocytes. After the above metabolism, only a small fraction of SCFAs remains to reach the whole body through the blood circulation (Dalile et al., 2019).
After SCFAs are absorbed in the colon, it works further via action on the G protein-coupled receptors GPR43 and GPR41 (Figure 1). SCFAs stimulate the proliferation of colonic epithelial L cells and are bound to GPR43 above them, increasing intracytoplasmic calcium ion and cyclic adenosine cAMP concentrations and increasing GLP-1 secretion (Chambers et al., 2018; Rodriguez et al., 2018) which inhibits glucose uptake by intestinal wall cells. Activation of GPR41 increases the secretion of intestinal peptide YY (PYY) and decreases the secretion of glucose-dependent insulin-releasing polypeptide (GIP) and growth hormone-releasing peptide, thereby reducing glucose uptake (Hu et al., 2018). GPR43 is distributed in adipose tissue, bone marrow, spleen, pancreas, peripheral blood mononuclear cells, small intestine, and mammary gland, while GPR41 is distributed in various tissues, especially in immune cells. Hence, SCFAs also have beneficial effects on peripheral tissues, controlling substrate metabolism and optimizing systemic insulin sensitivity. For example, in immune cells, SCFAs can inhibit inflammation by binding to GPR41 and GPR43 (Allin et al., 2015; Thorburn et al., 2015). SCFAs also induce browning of white adipose tissue, boost glucose tolerance, control blood glucose levels, and exert anti-obesity and anti-diabetic effects (He et al., 2020).
Moreover, SCFAs are inhibitors of histone deacetylase (HDAC). SCFAs inhibit HDAC of Treg, in turn affecting Treg production (Arpaia et al., 2013; Furusawa et al., 2013). Also, the inhibition of HDAC by SCFAs can further modulate the function of intestinal macrophages (Chang et al., 2014) and dendritic cells (Trompette et al., 2014; Meijnikman et al., 2018) and downregulate LPS-induced pro-inflammatory mediators, such as NO, IL-6, and IL-12, resulting in anti-inflammatory effects. There is also evidence that SCFAs decline the release of TNF-α from neutrophils (Lee et al., 2019), which is also one of the typical pro-inflammatory cytokines that trigger subclinical inflammation.
Notably, SCFAs also play a crucial role in maintaining the Intestinal mucosal barrier integrity. SCFAs can increase transcription of mucin genes (described previously, see Chapter 4.1), then upgrading intestinal barrier function (Willemsen et al., 2003; Gaudier et al., 2004). At the same time, the increased A. muciniphila would continue to degrade mucin and produce more SCFAs. A prospective study by Dao et al. (2016) verified the relationship. This study spotted that individuals with higher abundance of A. muciniphila had a better metabolic profile, and the concentration of SCFAs in such populations was positively correlated with A. muciniphila abundance.
Bile acids are a class of amphiphilic metabolites produced by hepatocytes and secreted by the bile ducts into the intestine, which have an important emulsifying effect on fat ingested by the body during digestion. The substantial increase in plasma bile acid concentrations (primary, secondary, total and unconjugated bile acid concentrations) with metformin may be explained by the fact that metformin increased the abundance of probiotic bacteria, especially Lactobacillus, and thus their bile salt hydrolase (BSH) gene expression. The BSH gene, a gene encoding the production of BSH by the intestinal microbiota, is associated with obesity and T2DM was significantly correlated (Jia et al., 2020). An essential part of cholesterol regulation by gut microbes is achieved by the hydrolysis of cross-linked bile salts by BSH, thus reducing serum cholesterol levels. The de-cross-linked bile acids have low water solubility and are readily secreted into the feces. Due to the loss of fecal bile acids and the decrease in bile salts, the liver increases the conversion of cholesterol to bile salts to maintain bile salt metabolic balance. It has been demonstrated that oral administration of BSH-containing Lactobacillus can reduce serum cholesterol levels in animals and humans, especially Limosilactobacillus fermentum [here we use its new nomenclature according to Naghmouchi et al. (2020), Zheng et al. (2020), Çiftci and Tuna (2021), Ng et al. (2022)]. However, no studies have revealed a correlation between the presence of specific bacteria and bile acid concentrations after metformin treatment (Wu et al., 2017).
Subsequently, secondary bile acids, key signaling molecules produced by bile acids and gut microbiota metabolism, are bound to membrane receptors (Takeda G protein-coupled receptor 5, TGR5/G protein-coupled bile acid receptor 1, GPBAR1) and nuclear receptors (Farnesoid X Receptor, FXR), which stimulates colonic secretion of GLP-1 and PYY. It plays a role in promoting proliferation and inhibiting apoptosis in pancreatic 𝛽-cells, and directly scales insulin secretion up. Another study (Sun et al., 2020) demonstrated that serum fibroblast growth factor 19 (FGF19) and bile acid levels were simultaneously increased in patients with T2DM early in life. This may be due to the fact that bile acids entering the intestine stimulate intestinal secretion and expression of the gut-derived hormone FGF19 via bile acid-FXR signaling (Mulla et al., 2019), which stimulates hepatic glycogen synthesis in an insulin non-dependent manner, restrains hepatic gluconeogenesis, and exerts hypoglycemic effects. FGF19 also dramatically promotes glucose tolerance (Zhang et al., 2019).
There are other pathways for FXR signaling activation. Sun et al. (2018) showed that metformin treatment altered the metabolism of folate and methionine in mice, which in turn inhibited the growth of B. fragilis. The reduction of B. fragilis increased the level of Glycoursodeoxycholic acid in the gut, then led to the activation of intestinal FXR signaling. Several studies have implied that FXR activation relieves intestinal inflammatory state in IBD patients, as well as that colonic inflammation in patients with CD and in rodent models of colitis is linked to diminished expression of FXR mRNA (Vavassori et al., 2009). During inflammation in the liver, there is a sustained inhibition of bile salt export pump expression and reduced FXR expression. Hepatic transporter protein function is then decreased, which leads to both an increase in hepatic bile acid chelation and persistent inflammation. Activation of FXR stabilizes the nuclear co-repressor (NcoR) at the NF-κB response element on the IL-1β promoter, while decreasing the expression of TNF-α, IL-1β, IL-2, IL-6 and IFN-γ mRNA, thereby reducing the extent of inflammation (Vavassori et al., 2009).
In addition to regulating the gene expression of BSH of probiotic bacteria (described previously, see Chapter 4.3), metformin also modulates the protein-coding genes of another microbiota. Wu et al. (2017) discovered that the abundance of E. coli, Bifidobacterium adolescentic, and A. muciniphila was plainly increased in the intestine after metformin administration. They further proceeded transcriptomic analysis of fecal samples and found that some of the protein-coding genes of A. muciniphila and B. wadsworthia were dramatically regulated by metformin, and most of them encoded metalloproteins or metal transporter proteins. Metal ions (e.g., Ca2+, Mn2+, Zn2+, and Mg2+) play an important role in maintaining the structure and homeostasis of various proteins in microorganisms. And these proteins affect the metabolism of microorganisms (Murphy et al., 1981). Several studies have shown that metallothionein has a negative regulatory role in all kinds of organ tissues, various types (including LPS-related, allergic and oxidative) of inflammation (Inoue et al., 2009; Inoue and Takano, 2013). It has also been indicated that metformin is able to bind to metal ions, thus causing changes in chemical structure (Logie et al., 2012). However, further metabolomic and proteomic studies are still needed to investigate the mechanisms of relevant microbial metabolism-host interactions.
Metformin was found to reduce the number of C. difficile in the intestine in several studies (Elbere et al., 2018; Bryrup et al., 2019). C. difficile can release toxin A and toxin B (Popoff, 2018), which induce the production of pro-inflammatory cytokines and inflammatory mediators in a variety of cell lines (Zhang and Feng, 2016). And toxin A is able to trigger IL-8 secretion by human intestinal cells (He et al., 2002). It has also been reported that prostaglandin E2 (PGE2) secretion and cyclooxygenase-2 (COX-2) expression were significantly increased in colon cells exposed to toxin A either in vivo or in vitro (Kim et al., 2007). Therefore, metformin is able to decrease the release of pro-inflammatory factors by a reduction in the number of opportunistic pathogenic bacteria.
Metformin has been used globally for more than 60 years as the drug of choice for the treatment of T2DM. In clinical applications, in addition to treating T2DM, metformin has been found to be effective in certain diseases, such as obesity, cardiovascular disease, IBD, and other inflammation-related diseases. In addition, metformin has received much attention for its anti-tumor and anti-aging effects. Even recent literature provides evidence that metformin slows the development of COVID-19. Some people may experience side effects after taking metformin, with the main symptoms being gastrointestinal distress and, in rare cases, lactic acidosis (Sanchez-Rangel and Inzucchi, 2017). However, the mechanisms involved, whether therapeutic or side effects, are not fully understood. Previously, it was believed that metformin targets the liver and exerts its hypoglycemic effect through AMPK-dependent and non-AMPK-dependent pathways. However, this is not sufficient to explain the mechanism by which metformin achieves its broad therapeutic effects. A number of studies have provided evidence that the gut is the site of metformin’s wide-ranging effects, and found that gut microbiota may be an important “target” in this regard. However, differences in study subjects and experimental designs, especially the heterogeneity of the gut microbiota itself, have made it difficult to study the modulatory effects of metformin on the gut microbiota. The results of changes in the diversity and abundance of the human gut microbiota after metformin treatment are not consistent. Changes in the abundance of certain microbiota were also not identical across studies, and it is even possible to draw opposite conclusions. This highlights the importance of the need for further research to understand the underlying mechanisms of these changes. Nonetheless, metformin use has been associated with changes in the abundance of specific bacterial genera in the gut microbiota, and this has centered on the enrichment of A. muciniphila and bacteria that can produce SCFAs. By enriching these bacteria to stabilize the GI mucosal barrier as well as to regulate bile acid metabolism, metformin exerts a role in regulating glucose metabolism and lipid metabolism, which in turn maintains glucose homeostasis and improves the inflammatory state. In addition, with the increased interest of researchers in multi-omics studies, transcriptomic and metabolomic studies on the gut microbiota have increased. It was gradually found that metformin can also regulate the expression of certain genes in the gut microbiota, such as BSH genes and metalloproteins genes. However, the intestinal microbiota is very complex and it is challenging to study the mechanism of action of metformin. The mechanism by which metformin acts by regulating the gut microbiota needs to be further clarified.
YW: Investigation, Writing – original draft. XJ: Writing – original draft. BC: Writing – review & editing, Conceptualization, Supervision.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This article was supported by the CAMS Innovation Fund for Medical Sciences (2019-I2M-5-055); Natural Science Foundation of China (82304360).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, E., Sargeant, J. A., Zaccardi, F., Khunti, K., Webb, D., and Davies, M. (2020). Where does metformin stand in modern Day Management of Type 2 diabetes? Pharmaceuticals (Basel) 13:427. doi: 10.3390/ph13120427
Ahmadi, S., Razazan, A., Nagpal, R., Jain, S., Wang, B., Mishra, S. P., et al. (2020). Metformin reduces aging-related leaky gut and improves cognitive function by beneficially modulating gut microbiome/goblet cell/mucin Axis. J. Gerontol. A Biol. Sci. Med. Sci. 75, e9–e21. doi: 10.1093/gerona/glaa056
Alhouayek, M., Lambert, D. M., Delzenne, N. M., Cani, P. D., and Muccioli, G. G. (2011). Increasing endogenous 2-arachidonoylglycerol levels counteracts colitis and related systemic inflammation. FASEB J. 25, 2711–2721. doi: 10.1096/fj.10-176602
Allin, K. H., Nielsen, T., and Pedersen, O. (2015). Mechanisms in endocrinology: gut microbiota in patients with type 2 diabetes mellitus. Eur. J. Endocrinol. 172, R167–R177. doi: 10.1530/EJE-14-0874
American Diabetes Association (2020). Pharmacologic approaches to glycemic treatment: standards of medical Care in Diabetes—2020. Diabetes Care 43, S98–s110. doi: 10.2337/dc20-S009
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Arumugam, M., Raes, J., Pelletier, E., le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Ashrafian, F., Behrouzi, A., Shahriary, A., Ahmadi Badi, S., Davari, M., Khatami, S., et al. (2019). Comparative study of effect of Akkermansia muciniphila and its extracellular vesicles on toll-like receptors and tight junction. Gastroenterol. Hepatol. Bed Bench 12, 163–168
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science 307, 1915–1920. doi: 10.1126/science.1104816
Bauer, P. V., Duca, F. A., Waise, T. M. Z., Rasmussen, B. A., Abraham, M. A., Dranse, H. J., et al. (2018). Metformin alters upper small intestinal microbiota that impact a glucose-Sglt1-sensing Glucoregulatory pathway. Cell Metab. 27, 101–17.e5. doi: 10.1016/j.cmet.2017.09.019
Boulangé, C. L., Neves, A. L., Chilloux, J., Nicholson, J. K., and Dumas, M. E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med. 8:42. doi: 10.1186/s13073-016-0303-2
Bramante, C. T., Buse, J. B., Liebovitz, D. M., Nicklas, J. M., Puskarich, M. A., Cohen, K., et al. (2023). Outpatient treatment of Covid-19 and incidence of post-Covid-19 condition over 10 months (Covid-out): a multicentre, randomised, quadruple-blind, parallel-group, phase 3 trial. Lancet Infect. Dis. 23, 1119–1129. doi: 10.1016/S1473-3099(23)00299-2
Bramante, C. T., Ingraham, N. E., Murray, T. A., Marmor, S., Hovertsen, S., Gronski, J., et al. (2021). Metformin and risk of mortality in patients hospitalised with Covid-19: a retrospective cohort analysis. Lancet Healthy Longev 2, e34–e41. doi: 10.1016/S2666-7568(20)30033-7
Brandt, A., Hernández-Arriaga, A., Kehm, R., Sánchez, V., Jin, C. J., Nier, A., et al. (2019). Metformin attenuates the onset of non-alcoholic fatty liver disease and affects intestinal microbiota and barrier in small intestine. Sci. Rep. 9:6668. doi: 10.1038/s41598-019-43228-0
Bryrup, T., Thomsen, C. W., Kern, T., Allin, K. H., Brandslund, I., Jørgensen, N. R., et al. (2019). Metformin-induced changes of the gut microbiota in healthy young men: results of a non-blinded, one-armed intervention study. Diabetologia 62, 1024–1035. doi: 10.1007/s00125-019-4848-7
Buse, J. B., DeFronzo, R. A., Rosenstock, J., Kim, T., Burns, C., Skare, S., et al. (2016). The primary glucose-lowering effect of metformin resides in the gut, not the circulation: results from short-term pharmacokinetic and 12-week dose-ranging studies. Diabetes Care 39, 198–205. doi: 10.2337/dc15-0488
Cani, P. D. (2018). Human gut microbiome: hopes, threats and promises. Gut 67, 1716–1725. doi: 10.1136/gutjnl-2018-316723
Cani, P. D., Possemiers, S., van de Wiele, T., Guiot, Y., Everard, A., Rottier, O., et al. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving Glp-2-driven improvement of gut permeability. Gut 58, 1091–1103. doi: 10.1136/gut.2008.165886
Cejuela, M., Martin-Castillo, B., Menendez, J. A., and Pernas, S. (2022). Metformin and breast Cancer: where are we now? Int. J. Mol. Sci. 23:2705. doi: 10.3390/ijms23052705
Chambers, E. S., Preston, T., Frost, G., and Morrison, D. J. (2018). Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr. Nutr. Rep. 7, 198–206. doi: 10.1007/s13668-018-0248-8
Chang, P. V., Hao, L., Offermanns, S., and Medzhitov, R. (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 111, 2247–2252. doi: 10.1073/pnas.1322269111
Chen, C., You, L. J., Huang, Q., Fu, X., Zhang, B., Liu, R. H., et al. (2018). Modulation of gut microbiota by mulberry fruit polysaccharide treatment of obese diabetic db/db mice. Food Funct. 9, 3732–3742. doi: 10.1039/C7FO01346A
Chobot, A., Górowska-Kowolik, K., Sokołowska, M., and Jarosz-Chobot, P. (2018). Obesity and diabetes-not only a simple link between two epidemics. Diabetes Metab. Res. Rev. 34:e3042. doi: 10.1002/dmrr.3042
Choi, Y., Bose, S., Seo, J., Shin, J. H., Lee, D., Kim, Y., et al. (2021). Effects of live and pasteurized forms of Akkermansia from the human gut on obesity and metabolic dysregulation. Microorganisms 9:2039. doi: 10.3390/microorganisms9102039
Chung, E., Elmassry, M. M., Kottapalli, P., Kottapalli, K. R., Kaur, G., Dufour, J. M., et al. (2020). Metabolic benefits of annatto-extracted tocotrienol on glucose homeostasis, inflammation, and gut microbiome. Nutr. Res. 77, 97–107. doi: 10.1016/j.nutres.2020.04.001
Çiftci, G., and Tuna, E. (2021). Effects of cholesterol and Lactobacillus acidophilus on testicular function. Clin. Exp. Reprod. Med. 48, 229–235. doi: 10.5653/cerm.2020.04322
Clemente, J. C., Pehrsson, E. C., Blaser, M. J., Sandhu, K., Gao, Z., Wang, B., et al. (2015). The microbiome of uncontacted Amerindians. Sci. Adv. 1:e1500183. doi: 10.1126/sciadv.1500183
Coyle, C., Cafferty, F. H., Vale, C., and Langley, R. E. (2016). Metformin as an adjuvant treatment for cancer: a systematic review and meta-analysis. Ann. Oncol. 27, 2184–2195. doi: 10.1093/annonc/mdw410
Cui, H. X., Zhang, L. S., Luo, Y., Yuan, K., Huang, Z. Y., and Guo, Y. (2019). A purified Anthraquinone-glycoside preparation from rhubarb ameliorates type 2 diabetes mellitus by modulating the gut microbiota and reducing inflammation. Front. Microbiol. 10:1423. doi: 10.3389/fmicb.2019.01423
Dalile, B., van Oudenhove, L., Vervliet, B., and Verbeke, K. (2019). The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478. doi: 10.1038/s41575-019-0157-3
Dao, M. C., Everard, A., Aron-Wisnewsky, J., Sokolovska, N., Prifti, E., Verger, E. O., et al. (2016). Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65, 426–436. doi: 10.1136/gutjnl-2014-308778
Daousi, C., Casson, I. F., Gill, G. V., MacFarlane, I. A., Wilding, J. P. H., and Pinkney, J. H. (2006). Prevalence of obesity in type 2 diabetes in secondary care: association with cardiovascular risk factors. Postgrad. Med. J. 82, 280–284. doi: 10.1136/pmj.2005.039032
Day, E. A., Ford, R. J., Smith, B. K., Mohammadi-Shemirani, P., Morrow, M. R., Gutgesell, R. M., et al. (2019). Metformin-induced increases in Gdf15 are important for suppressing appetite and promoting weight loss. Nat. Metab. 1, 1202–1208. doi: 10.1038/s42255-019-0146-4
de la Cuesta-Zuluaga, J., Mueller, N. T., Corrales-Agudelo, V., Velásquez-Mejía, E. P., Carmona, J. A., Abad, J. M., et al. (2017). Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care 40, 54–62. doi: 10.2337/dc16-1324
De Silva, A., and Bloom, S. R. (2012). Gut hormones and appetite control: a focus on Pyy and Glp-1 as therapeutic targets in obesity. Gut. Liver 6, 10–20. doi: 10.5009/gnl.2012.6.1.10
Derrien, M., Vaughan, E. E., Plugge, C. M., and de Vos, W. M. (2004). Akkermansia muciniphila gen. Nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476. doi: 10.1099/ijs.0.02873-0
Després, J. P. (2003). Potential contribution of metformin to the management of cardiovascular disease risk in patients with abdominal obesity, the metabolic syndrome and type 2 diabetes. Diabetes Metab. 29:6s53-61. doi: 10.1016/S1262-3636(03)72788-8
Duca, F. A., Côté, C. D., Rasmussen, B. A., Zadeh-Tahmasebi, M., Rutter, G. A., Filippi, B. M., et al. (2015). Metformin activates a duodenal Ampk-dependent pathway to lower hepatic glucose production in rats. Nat. Med. 21, 506–511. doi: 10.1038/nm.3787
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
Eibl, G., and Rozengurt, E. (2019). Kras, yap, and obesity in pancreatic cancer: a signaling network with multiple loops. Semin. Cancer Biol. 54, 50–62. doi: 10.1016/j.semcancer.2017.10.007
Elbere, I., Kalnina, I., Silamikelis, I., Konrade, I., Zaharenko, L., Sekace, K., et al. (2018). Association of metformin administration with gut microbiome dysbiosis in healthy volunteers. PLoS One 13:e0204317. doi: 10.1371/journal.pone.0204317
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Forbes, J. D., Chen, C. Y., Knox, N. C., Marrie, R. A., el-Gabalawy, H., de Kievit, T., et al. (2018). A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 6:221. doi: 10.1186/s40168-018-0603-4
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Gao, K., Yang, R., Zhang, J., Wang, Z., Jia, C., Zhang, F., et al. (2018). Effects of Qijian mixture on type 2 diabetes assessed by metabonomics, gut microbiota and network pharmacology. Pharmacol. Res. 130, 93–109. doi: 10.1016/j.phrs.2018.01.011
Gaudier, E., Jarry, A., Blottière, H. M., de Coppet, P., Buisine, M. P., Aubert, J. P., et al. (2004). Butyrate specifically modulates Muc gene expression in intestinal epithelial goblet cells deprived of glucose. Am. J. Physiol. Gastrointest. Liver Physiol. 287, G1168–G1174. doi: 10.1152/ajpgi.00219.2004
Gong, L., Goswami, S., Giacomini, K. M., Altman, R. B., and Klein, T. E. (2012). Metformin pathways: pharmacokinetics and pharmacodynamics. Pharmacogenet. Genomics 22, 820–827. doi: 10.1097/FPC.0b013e3283559b22
Graham, G. G., Punt, J., Arora, M., Day, R. O., Doogue, M. P., Duong, J. K., et al. (2011). Clinical pharmacokinetics of metformin. Clin. Pharmacokinet. 50, 81–98. doi: 10.2165/11534750-000000000-00000
Han, Y. C., Tang, S. Q., Liu, Y. T., Li, A. M., Zhan, M., Yang, M., et al. (2021). Ampk agonist alleviate renal tubulointerstitial fibrosis via activating mitophagy in high fat and streptozotocin induced diabetic mice. Cell Death Dis. 12:925. doi: 10.1038/s41419-021-04184-8
Hardie, D. G., Ross, F. A., and Hawley, S. A. (2012). Amp-activated protein kinase: a target for drugs both ancient and modern. Chem. Biol. 19, 1222–1236. doi: 10.1016/j.chembiol.2012.08.019
Harsch, I. A., and Konturek, P. C. (2018). The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into "old" diseases. Med. Sci. 6:32. doi: 10.3390/medsci6020032
He, L. (2020). Metformin and systemic metabolism. Trends Pharmacol. Sci. 41, 868–881. doi: 10.1016/j.tips.2020.09.001
He, D., Sougioultzis, S., Hagen, S., Liu, J., Keates, S., Keates, A. C., et al. (2002). Clostridium difficile toxin a triggers human colonocyte Il-8 release via mitochondrial oxygen radical generation. Gastroenterology 122, 1048–1057. doi: 10.1053/gast.2002.32386
He, J., Zhang, P., Shen, L., Niu, L., Tan, Y., Chen, L., et al. (2020). Short-chain fatty acids and their association with signalling pathways in inflammation, glucose and lipid metabolism. Int. J. Mol. Sci. 21:6356. doi: 10.3390/ijms21176356
Hiel, S., Gianfrancesco, M. A., Rodriguez, J., Portheault, D., Leyrolle, Q., Bindels, L. B., et al. (2020). Link between gut microbiota and health outcomes in inulin -treated obese patients: lessons from the Food4Gut multicenter randomized placebo-controlled trial. Clin. Nutr. 39, 3618–3628. doi: 10.1016/j.clnu.2020.04.005
Hu, J., Lin, S., Zheng, B., and Cheung, P. C. K. (2018). Short-chain fatty acids in control of energy metabolism. Crit. Rev. Food Sci. Nutr. 58, 1243–1249. doi: 10.1080/10408398.2016.1245650
Hur, K. Y., and Lee, M. S. (2015). New mechanisms of metformin action: focusing on mitochondria and the gut. J. Diab. Invest. 6, 600–609. doi: 10.1111/jdi.12328
Inoue, K., and Takano, H. (2013). Metallothionein as a negative regulator of pulmonary inflammation. Curr. Pharm. Biotechnol. 14, 414–419. doi: 10.2174/1389201011314040005
Inoue, K., Takano, H., Shimada, A., and Satoh, M. (2009). Metallothionein as an anti-inflammatory mediator. Mediat. Inflamm. 2009:101659, 1–7. doi: 10.1155/2009/101659
Ji, S., Wang, L., and Li, L. (2019). Effect of metformin on short-term high-fat diet-induced weight gain and anxiety-like behavior and the gut microbiota. Front. Endocrinol. (Lausanne) 10:704. doi: 10.3389/fendo.2019.00704
Jia, B., Park, D., Hahn, Y., and Jeon, C. O. (2020). Metagenomic analysis of the human microbiome reveals the association between the abundance of gut bile salt hydrolases and host health. Gut Microbes 11, 1300–1313. doi: 10.1080/19490976.2020.1748261
Karlsson, F. H., Tremaroli, V., Nookaew, I., Bergström, G., Behre, C. J., Fagerberg, B., et al. (2013). Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103. doi: 10.1038/nature12198
Kim, M. H., Jee, J. H., Park, S., Lee, M. S., Kim, K. W., and Lee, M. K. (2014). Metformin enhances glucagon-like peptide 1 via cooperation between insulin and Wnt signaling. J. Endocrinol. 220, 117–128. doi: 10.1530/JOE-13-0381
Kim, H., Rhee, S. H., Pothoulakis, C., and LaMont, J. T. (2007). Inflammation and apoptosis in Clostridium difficile enteritis is mediated by Pge2 up-regulation of Fas ligand. Gastroenterology 133, 875–886. doi: 10.1053/j.gastro.2007.06.063
Knights, D., Ward, T. L., McKinlay, C. E., Miller, H., Gonzalez, A., McDonald, D., et al. (2014). Rethinking "enterotypes". Cell Host Microbe 16, 433–437. doi: 10.1016/j.chom.2014.09.013
Koropatkin, N. M., and Martens, E. C. (2017). Meds modify microbiome, mediating their effects. Cell Metab. 26, 456–457. doi: 10.1016/j.cmet.2017.08.022
Kyriachenko, Y., Falalyeyeva, T., Korotkyi, O., Molochek, N., and Kobyliak, N. (2019). Crosstalk between gut microbiota and antidiabetic drug action. World J. Diabetes 10, 154–168. doi: 10.4239/wjd.v10.i3.154
Ladeiras-Lopes, R., Fontes-Carvalho, R., Bettencourt, N., Sampaio, F., Gama, V., and Leite-Moreira, A. (2015). Novel therapeutic targets of metformin: metabolic syndrome and cardiovascular disease. Expert Opin. Ther. Targets 19, 869–877. doi: 10.1517/14728222.2015.1025051
Lancet (1998). Effect of intensive blood-glucose control with metformin on complications in overweight patients with type 2 diabetes (Ukpds 34). Uk prospective diabetes study (Ukpds) group. Lancet 352, 854–865. doi: 10.1016/S0140-6736(98)07037-8
le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
le Roy, C. I., Beaumont, M., Jackson, M. A., Steves, C. J., Spector, T. D., and Bell, J. T. (2018). Heritable components of the human fecal microbiome are associated with visceral fat. Gut Microbes 9, 61–67. doi: 10.1080/19490976.2017.1356556
Lee, S. E., Choi, Y., Jun, J. E., Lee, Y. B., Jin, S. M., Hur, K. Y., et al. (2019). Additional effect of dietary Fiber in patients with type 2 diabetes mellitus using metformin and sulfonylurea: An open-label, pilot trial. Diabetes Metab. J. 43, 422–431. doi: 10.4093/dmj.2018.0090
Lee, H., Lee, Y., Kim, J., An, J., Lee, S., Kong, H., et al. (2018). Modulation of the gut microbiota by metformin improves metabolic profiles in aged obese mice. Gut Microbes 9, 155–165. doi: 10.1080/19490976.2017.1405209
Li, J., Lin, S., Vanhoutte, P. M., Woo, C. W., and Xu, A. (2016). Akkermansia Muciniphila protects against atherosclerosis by preventing metabolic Endotoxemia-induced inflammation in Apoe−/− mice. Circulation 133, 2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645
Liang, C., Tseng, H. C., Chen, H. M., Wang, W. C., Chiu, C. M., Chang, J. Y., et al. (2017). Diversity and enterotype in gut bacterial community of adults in Taiwan. BMC Genomics 18:932. doi: 10.1186/s12864-016-3261-6
Logie, L., Harthill, J., Patel, K., Bacon, S., Hamilton, D. L., Macrae, K., et al. (2012). Cellular responses to the metal-binding properties of metformin. Diabetes 61, 1423–1433. doi: 10.2337/db11-0961
Luckey, T. D. (1972). Introduction to intestinal microecology. Am. J. Clin. Nutr. 25, 1292–1294. doi: 10.1093/ajcn/25.12.1292
Madiraju, A. K., Erion, D. M., Rahimi, Y., Zhang, X. M., Braddock, D. T., Albright, R. A., et al. (2014). Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510, 542–546. doi: 10.1038/nature13270
Madiraju, A. K., Qiu, Y., Perry, R. J., Rahimi, Y., Zhang, X. M., Zhang, D., et al. (2018). Metformin inhibits gluconeogenesis via a redox-dependent mechanism in vivo. Nat. Med. 24, 1384–1394. doi: 10.1038/s41591-018-0125-4
Meijnikman, A. S., Gerdes, V. E., Nieuwdorp, M., and Herrema, H. (2018). Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr. Rev. 39, 133–153. doi: 10.1210/er.2017-00192
Forslund, K., Hildebrand, F., Nielsen, T., Falony, G., le Chatelier, E., et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266. doi: 10.1038/nature15766
Meyer, M. J., and Tzvetkov, M. V. (2021). Oct1 Polyspecificity-friend or foe? Front. Pharmacol. 12:698153. doi: 10.3389/fphar.2021.698153
Miller, R. A., Chu, Q., Xie, J., Foretz, M., Viollet, B., and Birnbaum, M. J. (2013). Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic amp. Nature 494, 256–260. doi: 10.1038/nature11808
Montandon, S. A., and Jornayvaz, F. R. (2017). Effects of antidiabetic drugs on gut microbiota composition. Genes (Basel) 8:250. doi: 10.3390/genes8100250
Montvida, O., Shaw, J., Atherton, J. J., Stringer, F., and Paul, S. K. (2018). Long-term trends in Antidiabetes drug usage in the U.S.: Real-world evidence in patients newly diagnosed with type 2 diabetes. Diabetes Care 41, 69–78. doi: 10.2337/dc17-1414
Mulla, C. M., Goldfine, A. B., Dreyfuss, J. M., Houten, S., Pan, H., Pober, D. M., et al. (2019). Plasma Fgf-19 levels are increased in patients with post-bariatric hypoglycemia. Obes. Surg. 29, 2092–2099. doi: 10.1007/s11695-019-03845-0
Murphy, G., McGuire, M. B., Russell, R. G., and Reynolds, J. J. (1981). Characterization of collagenase, other metallo-proteinases and an inhibitor (Timp) produced by human synovium and cartilage in culture. Clin. Sci. (Lond.) 61, 711–716. doi: 10.1042/cs0610711
Naghmouchi, K., Belguesmia, Y., Bendali, F., Spano, G., Seal, B. S., and Drider, D. (2020). Lactobacillus fermentum: a bacterial species with potential for food preservation and biomedical applications. Crit. Rev. Food Sci. Nutr. 60, 3387–3399. doi: 10.1080/10408398.2019.1688250
Napolitano, A., Miller, S., Nicholls, A. W., Baker, D., van Horn, S., Thomas, E., et al. (2014). Novel gut-based pharmacology of metformin in patients with type 2 diabetes mellitus. PLoS One 9:e100778. doi: 10.1371/journal.pone.0100778
Ng, S. C., Xu, Z., Mak, J. W. Y., Yang, K., Liu, Q., Zuo, T., et al. (2022). Microbiota engraftment after faecal microbiota transplantation in obese subjects with type 2 diabetes: a 24-week, double-blind, randomised controlled trial. Gut 71, 716–723. doi: 10.1136/gutjnl-2020-323617
Nicolucci, A., Charbonnel, B., Gomes, M. B., Khunti, K., Kosiborod, M., Shestakova, M. V., et al. (2019). Treatment patterns and associated factors in 14 668 people with type 2 diabetes initiating a second-line therapy: results from the global discover study programme. Diabetes Obes. Metab. 21, 2474–2485. doi: 10.1111/dom.13830
Ottman, N., Reunanen, J., Meijerink, M., Pietilä, T. E., Kainulainen, V., Klievink, J., et al. (2017). Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One 12:e0173004. doi: 10.1371/journal.pone.0173004
Ouchi, N., Parker, J. L., Lugus, J. J., and Walsh, K. (2011). Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 11, 85–97. doi: 10.1038/nri2921
Pålsson-McDermott, E. M., and O’Neill, L. A. J. (2020). Targeting immunometabolism as an anti-inflammatory strategy. Cell Res. 30, 300–314. doi: 10.1038/s41422-020-0291-z
Pedersen, H. K., Gudmundsdottir, V., Nielsen, H. B., Hyotylainen, T., Nielsen, T., Jensen, B. A. H., et al. (2016). Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381. doi: 10.1038/nature18646
Peng, Q., Chen, X., Liang, X., Ouyang, J., Wang, Q., Ren, S., et al. (2023). Metformin improves polycystic ovary syndrome in mice by inhibiting ovarian ferroptosis. Front. Endocrinol. (Lausanne) 14:1070264. doi: 10.3389/fendo.2023.1070264
Peng, X., Yu, K. Q., Deng, G. H., Jiang, Y. X., Wang, Y., Zhang, G. X., et al. (2013). Comparison of direct boiling method with commercial kits for extracting fecal microbiome Dna by Illumina sequencing of 16S rrna tags. J. Microbiol. Methods 95, 455–462. doi: 10.1016/j.mimet.2013.07.015
Plovier, H., Everard, A., Druart, C., Depommier, C., van Hul, M., Geurts, L., et al. (2017). A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113. doi: 10.1038/nm.4236
Popoff, M. R. (2018). Clostridium difficile and Clostridium sordellii toxins, proinflammatory versus anti-inflammatory response. Toxicon 149, 54–64. doi: 10.1016/j.toxicon.2017.11.003
Qin, J., Li, Y., Cai, Z., Li, S., Zhu, J., Zhang, F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490, 55–60. doi: 10.1038/nature11450
Rau, M., Rehman, A., Dittrich, M., Groen, A. K., Hermanns, H. M., Seyfried, F., et al. (2018). Fecal Scfas and Scfa-producing bacteria in gut microbiome of human Nafld as a putative link to systemic T-cell activation and advanced disease. United European Gastroenterol J 6, 1496–1507. doi: 10.1177/2050640618804444
Rodriguez, J., Hiel, S., and Delzenne, N. M. (2018). Metformin: old friend, new ways of action-implication of the gut microbiome? Curr. Opin. Clin. Nutr. Metab. Care 21, 294–301. doi: 10.1097/MCO.0000000000000468
Ryan, P. M., Patterson, E., Carafa, I., Mandal, R., Wishart, D. S., Dinan, T. G., et al. (2020). Metformin and dipeptidyl Peptidase-4 inhibitor differentially modulate the intestinal microbiota and plasma metabolome of metabolically dysfunctional mice. Can. J. Diabetes 44, 146–55.e2. doi: 10.1016/j.jcjd.2019.05.008
Sanchez-Rangel, E., and Inzucchi, S. E. (2017). Metformin: clinical use in type 2 diabetes. Diabetologia 60, 1586–1593. doi: 10.1007/s00125-017-4336-x
Sandhu, B. K., and Mcbride, S. M. (2018). Clostridioides difficile. Trends Microbiol. 26, 1049–1050. doi: 10.1016/j.tim.2018.09.004
Scheen, A. J., and Paquot, N. (2013). Metformin revisited: a critical review of the benefit-risk balance in at-risk patients with type 2 diabetes. Diabetes Metab. 39, 179–190. doi: 10.1016/j.diabet.2013.02.006
Scheithauer, T. P., Dallinga-Thie, G. M., de Vos, W. M., Nieuwdorp, M., and van Raalte, D. H. (2016). Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol. Metab. 5, 759–770. doi: 10.1016/j.molmet.2016.06.002
Shapira, M. (2016). Gut microbiotas and host evolution: scaling up Symbiosis. Trends Ecol. Evol. 31, 539–549. doi: 10.1016/j.tree.2016.03.006
Shaw, R. J., Lamia, K. A., Vasquez, D., Koo, S. H., Bardeesy, N., DePinho, R. A., et al. (2005). The kinase Lkb1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science 310, 1642–1646. doi: 10.1126/science.1120781
Shin, N. R., Bose, S., Wang, J. H., Ansari, A. Z., Lim, S. K., Chin, Y. W., et al. (2017). Flos Lonicera combined with metformin ameliorates Hepatosteatosis and glucose intolerance in association with gut microbiota modulation. Front. Microbiol. 8:2271. doi: 10.3389/fmicb.2017.02271
Shirasaka, Y., Seki, M., Hatakeyama, M., Kurokawa, Y., Uchiyama, H., Takemura, M., et al. (2022). Multiple transport mechanisms involved in the intestinal absorption of metformin: impact on the nonlinear absorption kinetics. J. Pharm. Sci. 111, 1531–1541. doi: 10.1016/j.xphs.2022.01.008
Sicard, J. F., le Bihan, G., Vogeleer, P., Jacques, M., and Harel, J. (2017). Interactions of intestinal Bacteria with components of the intestinal mucus. Front. Cell. Infect. Microbiol. 7:387. doi: 10.3389/fcimb.2017.00387
Singer-Englar, T., Barlow, G., and Mathur, R. (2019). Obesity, diabetes, and the gut microbiome: an updated review. Expert Rev. Gastroenterol. Hepatol. 13, 3–15. doi: 10.1080/17474124.2019.1543023
Song, S., Andrikopoulos, S., Filippis, C., Thorburn, A. W., Khan, D., and Proietto, J. (2001). Mechanism of fat-induced hepatic gluconeogenesis: effect of metformin. Am. J. Physiol. Endocrinol. Metab. 281, E275–E282. doi: 10.1152/ajpendo.2001.281.2.E275
Song, Z., Chen, J., Ji, Y., Yang, Q., Chen, Y., Wang, F., et al. (2023). Amuc attenuates high-fat diet-induced metabolic disorders linked to the regulation of fatty acid metabolism, bile acid metabolism, and the gut microbiota in mice. Int. J. Biol. Macromol. 242:124650. doi: 10.1016/j.ijbiomac.2023.124650
Soukas, A. A., Hao, H., and Wu, L. (2019). Metformin as anti-aging therapy: is it for everyone? Trends Endocrinol. Metab. 30, 745–755. doi: 10.1016/j.tem.2019.07.015
Stepensky, D., Friedman, M., Raz, I., and Hoffman, A. (2002). Pharmacokinetic-pharmacodynamic analysis of the glucose-lowering effect of metformin in diabetic rats reveals first-pass pharmacodynamic effect. Drug Metab. Dispos. 30, 861–868. doi: 10.1124/dmd.30.8.861
Sun, L., Xie, C., Wang, G., Wu, Y., Wu, Q., Wang, X., et al. (2018). Gut microbiota and intestinal Fxr mediate the clinical benefits of metformin. Nat. Med. 24, 1919–1929. doi: 10.1038/s41591-018-0222-4
Sun, Y., Zhu, M., Zhao, H., Ni, X., Chang, R., Su, J., et al. (2020). Serum fibroblast growth factor 19 and Total bile acid concentrations are potential biomarkers of hepatocellular carcinoma in patients with type 2 diabetes mellitus. Biomed. Res. Int. 2020, 1–9. doi: 10.1155/2020/1751989
The Idf Diabetes Atlas (2021). Available at: https://idf.org/about-diabetes/diabetes-facts-figures/.
Thorburn, A. N., McKenzie, C. I., Shen, S., Stanley, D., Macia, L., Mason, L. J., et al. (2015). Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 6:7320. doi: 10.1038/ncomms8320
Tilg, H., and Moschen, A. R. (2014). Microbiota and diabetes: an evolving relationship. Gut 63, 1513–1521. doi: 10.1136/gutjnl-2014-306928
Tims, S., Derom, C., Jonkers, D. M., Vlietinck, R., Saris, W. H., Kleerebezem, M., et al. (2013). Microbiota conservation and Bmi signatures in adult monozygotic twins. ISME J. 7, 707–717. doi: 10.1038/ismej.2012.146
Tong, X., Xu, J., Lian, F., Yu, X., Zhao, Y., Xu, L., et al. (2018). Structural alteration of gut microbiota during the amelioration of human type 2 diabetes with hyperlipidemia by metformin and a traditional Chinese herbal formula: a multicenter, randomized, open label clinical trial. MBio 9:e02392–17. doi: 10.1128/mBio.02392-17
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Ursini, F., Russo, E., Pellino, G., D’Angelo, S., Chiaravalloti, A., de Sarro, G., et al. (2018). Metformin and autoimmunity: a "new Deal" of an old drug. Front. Immunol. 9:1236. doi: 10.3389/fimmu.2018.01236
Vavassori, P., Mencarelli, A., Renga, B., Distrutti, E., and Fiorucci, S. (2009). The bile acid receptor Fxr is a modulator of intestinal innate immunity. J. Immunol. 183, 6251–6261. doi: 10.4049/jimmunol.0803978
Verdura, S., Cuyàs, E., Martin-Castillo, B., and Menendez, J. A. (2019). Metformin as an archetype immuno-metabolic adjuvant for cancer immunotherapy. Onco Targets Ther 8:e1633235. doi: 10.1080/2162402X.2019.1633235
Wang, J. H., Bose, S., Shin, N. R., Chin, Y. W., Choi, Y. H., and Kim, H. (2018). Pharmaceutical impact of Houttuynia Cordata and metformin combination on high-fat-diet-induced metabolic disorders: link to intestinal microbiota and metabolic Endotoxemia. Front. Endocrinol. (Lausanne) 9:620. doi: 10.3389/fendo.2018.00620
Wang, L. J., Jin, Y. L., Pei, W. L., Li, J. C., Zhang, R. L., Wang, J. J., et al. (2024). Amuc_1100 pretreatment alleviates acute pancreatitis in a mouse model through regulating gut microbiota and inhibiting inflammatory infiltration. Acta Pharmacol. Sin. 45, 570–580. doi: 10.1038/s41401-023-01186-4
Wang, Z., Saha, S., van Horn, S., Thomas, E., Traini, C., Sathe, G., et al. (2018). Gut microbiome differences between metformin- and liraglutide-treated T2dm subjects. Endocrinol. Diab. Metab. 1:e00009. doi: 10.1002/edm2.9
Werner, E., and Bell, J. (1922). CCXIV.—The preparation of methylguanidine, and of ββ-dimethylguanidine by the interaction of dicyanodiamide, and methylammonium and dimethylammonium chlorides respectively. J. Chem. Soc., Trans. 121, 1790–1794. doi: 10.1039/ct9222101790
Wexler, H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Willemsen, L. E., Koetsier, M. A., van Deventer, S., and van Tol, E. (2003). Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut 52, 1442–1447. doi: 10.1136/gut.52.10.1442
Wu, H., Esteve, E., Tremaroli, V., Khan, M. T., Caesar, R., Mannerås-Holm, L., et al. (2017). Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat. Med. 23, 850–858. doi: 10.1038/nm.4345
Wulffelé, M. G., Kooy, A., de Zeeuw, D., Stehouwer, C. D. A., and Gansevoort, R. T. (2004). The effect of metformin on blood pressure, plasma cholesterol and triglycerides in type 2 diabetes mellitus: a systematic review. J. Intern. Med. 256, 1–14. doi: 10.1111/j.1365-2796.2004.01328.x
Xu, Y., Zheng, S., Jiang, S., Chen, J., Zhu, X., and Zhang, Y. (2022). The effect of Chinese herbal formulas combined with metformin on modulating the gut microbiota in the amelioration of type 2 diabetes mellitus: a systematic review and meta-analysis. Front. Endocrinol. (Lausanne) 13:927959. doi: 10.3389/fendo.2022.927959
Yang, M., Bose, S., Lim, S., Seo, J. G., Shin, J. H., Lee, D., et al. (2020). Beneficial effects of newly isolated Akkermansia muciniphila strains from the human gut on obesity and metabolic dysregulation. Microorganisms 8:1413. doi: 10.3390/microorganisms8091413
Yaribeygi, H., Farrokhi, F. R., Butler, A. E., and Sahebkar, A. (2019). Insulin resistance: review of the underlying molecular mechanisms. J. Cell. Physiol. 234, 8152–8161. doi: 10.1002/jcp.27603
Zhang, S. Y., Bruce, K., Danaei, Z., Li, R. J. W., Barros, D. R., Kuah, R., et al. (2023). Metformin triggers a kidney Gdf15-dependent area postrema axis to regulate food intake and body weight. Cell Metab. 35, 875–86.e5. doi: 10.1016/j.cmet.2023.03.014
Zhang, Y., and Feng, H. (2016). Pathogenic effects of glucosyltransferase from Clostridium difficile toxins. Pathog. Dis. 74:ftw024. doi: 10.1093/femspd/ftw024
Zhang, J., Li, H., Bai, N., Xu, Y., Song, Q., Zhang, L., et al. (2019). Decrease of Fgf19 contributes to the increase of fasting glucose in human in an insulin-independent manner. J. Endocrinol. Investig. 42, 1019–1027. doi: 10.1007/s40618-019-01018-5
Zhang, W., Xu, J. H., Yu, T., and Chen, Q. K. (2019). Effects of berberine and metformin on intestinal inflammation and gut microbiome composition in db/db mice. Biomed. Pharmacother. 118:109131. doi: 10.1016/j.biopha.2019.109131
Zhang, X., Zhao, Y., Xu, J., Xue, Z., Zhang, M., Pang, X., et al. (2015). Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 5:14405. doi: 10.1038/srep14405
Zhao, L., Zhang, F., Ding, X., Wu, G., Lam, Y. Y., Wang, X., et al. (2018). Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156. doi: 10.1126/science.aao5774
Zheng, J., Li, H., Zhang, X., Jiang, M., Luo, C., Lu, Z., et al. (2018). Prebiotic Mannan-oligosaccharides augment the hypoglycemic effects of metformin in correlation with modulating gut microbiota. J. Agric. Food Chem. 66, 5821–5831. doi: 10.1021/acs.jafc.8b00829
Zheng, J., Wittouck, S., Salvetti, E., Franz, C. M. A. P., Harris, H. M. B., Mattarelli, P., et al. (2020). A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 70, 2782–2858. doi: 10.1099/ijsem.0.004107
Keywords: metformin, intestinal microbiota, gastrointestinal mucosal barrier, SCFAs, bile acid metabolism
Citation: Wang Y, Jia X and Cong B (2024) Advances in the mechanism of metformin with wide-ranging effects on regulation of the intestinal microbiota. Front. Microbiol. 15:1396031. doi: 10.3389/fmicb.2024.1396031
Received: 05 March 2024; Accepted: 29 April 2024;
Published: 24 May 2024.
Edited by:
Jiajia Song, Southwest University, ChinaReviewed by:
Rosa Del Campo, Ramón y Cajal Institute for Health Research, SpainCopyright © 2024 Wang, Jia and Cong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Cong, Y29uZzY0MDZAaGVibXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.