- 1Water Research Institute, CNR-IRSA, National Research Council, Rome, Italy
- 2PhD Program in Evolutionary Biology and Ecology, Department of Biology, University of Rome “Tor Vergata”, Rome, Italy
- 3Water Research Institute, CNR-IRSA, National Research Council, Verbania, Italy
- 4NBFC, National Biodiversity Future Center, Palermo, Italy
- 5Laboratory of Biology of Algae, Department of Biology, University of Rome “Tor Vergata”, Rome, Italy
The plastisphere, a unique microbial biofilm community colonizing plastic debris and microplastics (MPs) in aquatic environments, has attracted increasing attention owing to its ecological and public health implications. This review consolidates current state of knowledge on freshwater plastisphere, focussing on its biodiversity, community assembly, and interactions with environmental factors. Current biomolecular approaches revealed a variety of prokaryotic and eukaryotic taxa associated with plastic surfaces. Despite their ecological importance, the presence of potentially pathogenic bacteria and mobile genetic elements (i.e., antibiotic resistance genes) raises concerns for ecosystem and human health. However, the extent of these risks and their implications remain unclear. Advanced sequencing technologies are promising for elucidating the functions of plastisphere, particularly in plastic biodegradation processes. Overall, this review emphasizes the need for comprehensive studies to understand plastisphere dynamics in freshwater and to support effective management strategies to mitigate the impact of plastic pollution on freshwater resources.
1 Introduction
The term “plastisphere” was coined to describe the unique community of aquatic microbes that colonize plastic debris in marine environments (Zettler et al., 2013). Early studies on marine plastisphere were mainly based on the morphological identification of microorganisms using scanning electron microscopy (SEM), which highlighted the presence of filamentous bacteria and phototrophic eukaryotes (e.g., diatoms) (Carson et al., 2013; Zettler et al., 2013; Oberbeckmann et al., 2014; Amaral-Zettler et al., 2020). The use of biomolecular methods is contributing to improve our understanding of plastisphere biodiversity in freshwater environments revealing the full breadth and complexity of plastic-associated biofilms (Besemer et al., 2012; Besemer, 2015; Di Pippo et al., 2020).
Recent studies have shown that plastisphere is composed of microbial photoautotrophs, heterotrophs, protistan grazers and decomposers, most of which are known as biofilm formers and biofilm-associated microbes (Du et al., 2022; Li W. et al., 2023; Nikolopoulou et al., 2023). Once in water, the plastic debris provides hard surfaces for rapid microbial colonization and a new pelagic habitat for benthonic species (Yang Y. et al., 2020; Dąbrowska, 2021; Haram et al., 2021). Plastics and microplastics (MPs) were reported either as dispersal vehicles for microorganisms of health concern (Di Pippo et al., 2022; Du et al., 2022; Rubin and Zucker, 2022) or as hotspots for horizontal gene transfer, including antibiotic resistance genes (ARGs) (Luo et al., 2023; Chen et al., 2024; Li K. et al., 2024). Furthermore, plastisphere microorganisms were also found to be directly involved in polymer biodegradation (Du et al., 2022; Mishra et al., 2022; Yuan et al., 2022; Li K. et al., 2023).
Much of the current literature has so far demonstrated that several environmental conditions and local factors influence microbial communities developing on plastic surfaces (Hoellein et al., 2014; Oberbeckmann et al., 2018; Amaral-Zettler et al., 2020; Coons et al., 2021; Amaneesh et al., 2023). However, fundamental questions remain unanswered. The role of the polymer type and properties on plastisphere structure and biodiversity is still unclear (Sooriyakumar et al., 2022; Li K. et al., 2023; Miao et al., 2023). In addition, there is a knowledge gap about community assembly over time and on the microbial taxa involved in the different stages of plastisphere succession (Pollet et al., 2018; Amaral-Zettler et al., 2020; Eronen-Rasimus et al., 2022; Forero-López et al., 2022; Wallbank et al., 2022; Miao et al., 2023).
Despite the growing recognition of plastic waste pervasiveness in marine ecosystems and the large body of research focusing on plastic- and microplastic-associated biofilms, the freshwater plastisphere remains relatively understudied. Given the critical importance of quality freshwaters to provide essential services to human health and society, this disparity highlights the need for a more comprehensive understanding of plastic pollution in freshwater ecosystems, which can harbor a complex and diverse array of microorganisms differently sensitive to environmental and anthropogenic pollution (Hoellein et al., 2017; Di Pippo et al., 2020, 2022; Eronen-Rasimus et al., 2022; Nguyen et al., 2023).
This study was entailed to synthetically overview the current knowledge of freshwater plastisphere and the main environmental factors potentially influencing its development and microbial community assembly. In particular, we examined the current understanding of how the plastisphere might affect freshwater ecosystems and human health. We also examined its potential for biodegradation and identified critical aspects that require further investigation.
2 Plastisphere biodiversity, taxon composition, and factors affecting microbial community assembly
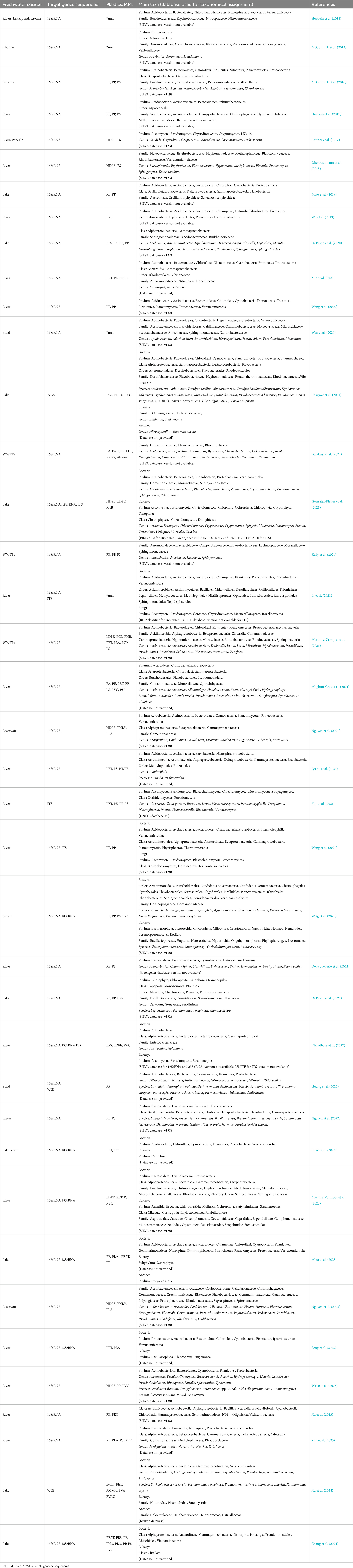
Once dispersed in water, plastics and MPs are rapidly colonized by planktonic microorganisms, which can adhere and grow onto solid surfaces forming complex plastic-associated biofilms whose biodiversity profiles consistently differ from those of the surrounding environments (see Table 1). While there is a shared consensus on the definition of a new plastic-associated micro-ecosystem with distinct microbiota, it is still debated whether the freshwater plastisphere harbors higher or lower biodiversity than planktonic communities and biofilms formed on natural substrates (McCormick et al., 2014, 2016; Hoellein et al., 2017; Arias-Andres et al., 2018; Wu et al., 2019; Wang et al., 2020; Xue et al., 2020; Galafassi et al., 2021; Kelly et al., 2021).
Most of the available information on freshwater plastisphere biodiversity comes from culture-independent methods (Table 1), which allow a comprehensive characterization of plastisphere-associated microbiomes. Among them, the use of high-throughput sequencing methods, both as amplicon sequencing of SSU RNA genes and shotgun metagenomic sequencing, are essential to decipher the taxonomic and functional diversity of samples, thus providing the composition and the metabolic potential of the entire microbial community (Dey et al., 2022; Wani et al., 2023). Recent developments in sequencing techniques have led to sequence very long reads (up to 50 kb) offering multiple cutting-edge options for understanding microbiome structure and functioning (Tedersoo et al., 2021). For example, when applied to amplicon sequencing (e.g., 16S rRNA gene), long-reads can resolve microbial taxonomy at deeper levels rather than short-reads due to the ability to read the entire gene with single nucleotide resolution leading to the identification of sub-species clades or “strains” within the community. Most studies have focused on Bacteria, with very few reports on archaeal and eukaryotic biodiversity (Table 1).
Proteobacteria, Bacteroidetes, Actinobacteria, Firmicutes, Verrucomicrobia, Planctomycetes, and Acidobacteria were the bacterial phyla most frequently detected on plastic particles in freshwaters (Table 1). Plastisphere bacteria are mainly affiliated with Gammaproteobacteria (family Burkolderaceae), Alphaproteobacteria (e.g., families Sphingomonadaceae, Rhodobacteraceae, and Hyphomicrobiaceae), Flavobacteria, and Firmicutes (Table 1). Rhodobacteraceae and Burkolderaceae are recurrent, particularly owing to their role as initial colonizers (Polz et al., 2006; Bhagwat et al., 2021; Di Pippo et al., 2022). Their involvement in different biogeochemical cycles and their mutualisms with eukaryotes suggest an important role in microbial community succession on plastic surfaces (Simon et al., 2017).
Archaea are likely to represent a minor component of the plastic-associated microbial community (<0.1% of total amplicon sequences) (Mughini-Gras et al., 2021), showing a lower diversity compared to Bacteria.
Microscopy observations indicated that freshwater eukaryotic microbes of various trophic levels may represent a significant portion of plastisphere biodiversity (Carson et al., 2013; Oberbeckmann et al., 2014; Bryant et al., 2016; Masó et al., 2016), although high-throughput sequencing data are still limited (Kettner et al., 2017; Bhagwat et al., 2021; González-Pleiter et al., 2021; Li et al., 2021; Wang et al., 2021; Weig et al., 2021; Xue et al., 2021; Chaudhary et al., 2022; Di Pippo et al., 2022; Li W. et al., 2023; Martínez-Campos et al., 2023; Miao et al., 2023; Song et al., 2023; Xu et al., 2024; Zhang et al., 2024). Different taxa of primary producers (Chlorophyta, Charophyta, Bacillariophyta), primary/secondary consumers (Peritrichia, Oligotrichia), mixotrophs (Dinophyceaea), saprotrophic/parasitic fungi (Cryptomycota, Peronosperales, Oomycetes), and metazoan consumers were retrieved (Table 1). In recent studies various conventional and bio-based plastic polymers (e.g., postconsumer plastic, “raw” plastic from known manufacturing sources) were used to assess microbial plastic colonization in freshwaters under different field and laboratory settings (Table 1). Regardless of the varying tested conditions, different environmental, spatial, and temporal factors (e.g., redox potential, salinity, nutrient concentration, geographical location, anthropogenic influence, seasonality) appeared to drive the microbial composition and assembly of the freshwater plastisphere (Zhang et al., 2004; Kettner et al., 2017; Bhagwat et al., 2021; González-Pleiter et al., 2021; Li et al., 2021; Weig et al., 2021; Xue et al., 2021; Di Pippo et al., 2022; Martínez-Campos et al., 2023; Miao et al., 2023; Xu et al., 2024). The role of polymer types and properties is unclear and currently under debate (Jacquin et al., 2019; Bhagwat et al., 2021; Coons et al., 2021; Delacuvellerie et al., 2021; Mughini-Gras et al., 2021; Weig et al., 2021; Sooriyakumar et al., 2022; Wang et al., 2022; Li K. et al., 2023; Miao et al., 2023), since only few studies have reported that specific plastic polymers can select different communities (McCormick et al., 2016; Di Pippo et al., 2020; Delacuvellerie et al., 2021; Li et al., 2021; Martínez-Campos et al., 2021; Mughini-Gras et al., 2021). Surface properties such as roughness, topography, and electrostatic charge are known to influence freshwater bacterial attachment and biofilm assembly (Rummel et al., 2017; Nguyen et al., 2021). Microbial colonization during the early developmental stages and the subsequent microbial biofilm maturation are directly influenced by the chemical, physical, mechanical, and morphological properties of the polymer substrata (Dexter, 1979; Rummel et al., 2017; Kreve and Reis, 2021; Nguyen et al., 2021; Zheng et al., 2021; Jia, 2022). On the one hand, the presence of plastic additives (e.g., plasticizers, flame retardants, colorants) can promote hydrophobicity and alter the original properties of the bare solid surfaces (Karlsson et al., 1988; De Tender et al., 2015; Bhagwat et al., 2021). On the other hand, environmental aging, weathering, and photo-oxidation can reduce the surface hydrophobicity, thus promoting microbial colonization (Gong et al., 2019; Bao et al., 2022). The surface colonization processes involve a succession of microorganisms that contribute to the establishment of a stable biofilm consortium. By producing extracellular polymeric substances (EPS), pioneer microorganisms can facilitate their attachment to surfaces, but also provide a suitable carbon source for other microbial species (Yang Y. et al., 2020). Consequently, early colonizers can be outcompeted by other taxa with increasing duration of exposure to environmental conditions (e.g., incubation time in water), thus leading to converging community composition over time on different materials (Pinto et al., 2019; Yang Y. et al., 2020; Nguyen et al., 2021; Chaudhary et al., 2022; Martínez-Campos et al., 2023; Miao et al., 2023; Xu et al., 2023).
3 Ecosystem and human health-related issues: occurrence of plastic-associated pathogens and genetic elements of health concern
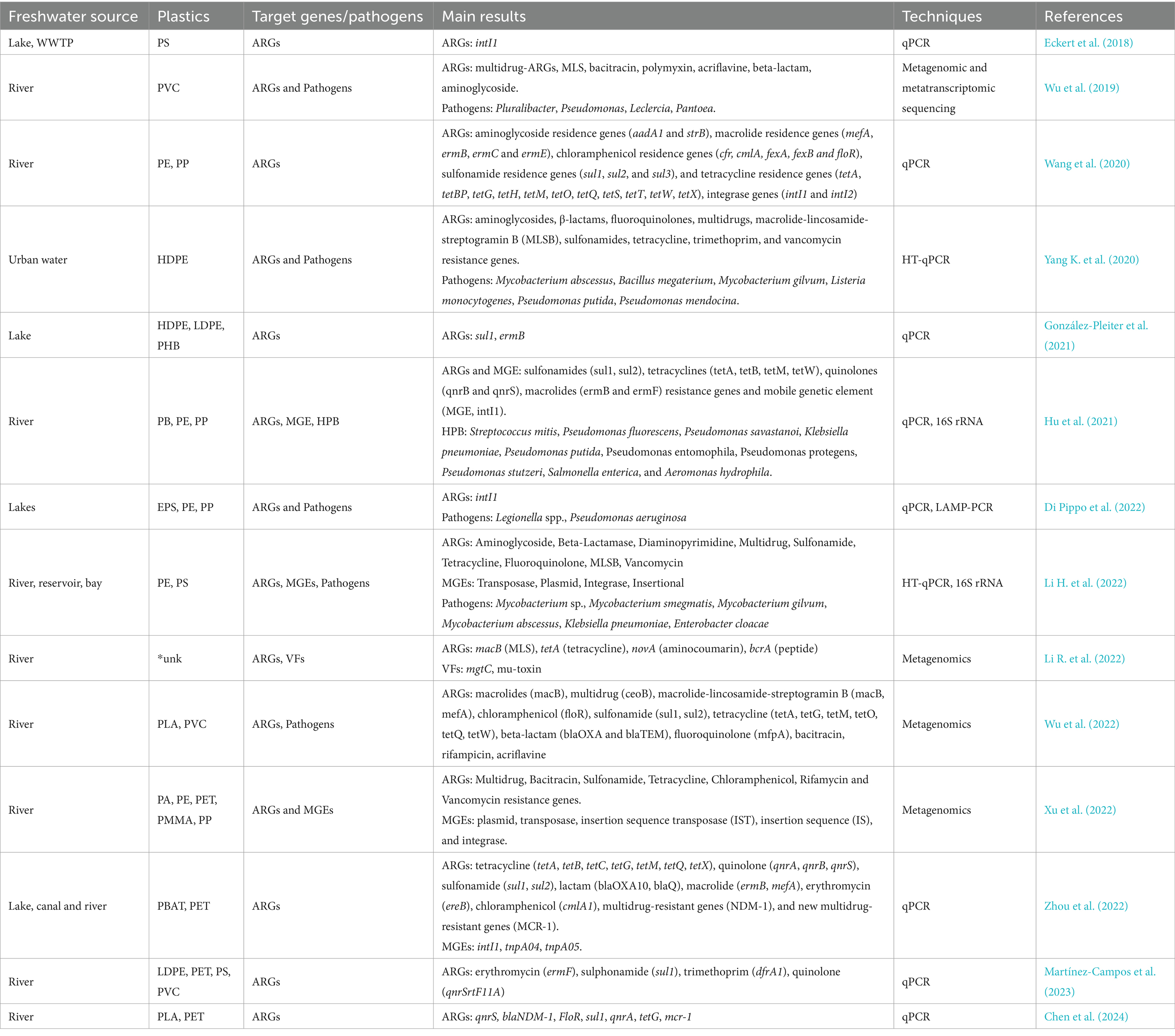
Plastic-associated microbiological elements of health concern are rarely monitored in freshwater ecosystems, despite their fundamental services provided to human health and society (e.g., drinking water supply, agricultural/industrial activities, recreational activities). Plastic debris and associated biofilms have been reported to represent newly available ecological niches that facilitate the accumulation of various harmful microbes. Recent studies have highlighted the presence of potentially pathogenic bacteria in freshwater plastisphere communities, including members of the genera Vibrio, Pseudomonas, Acinetobacter, Arcobacter, Bacillus, Aquabacterium, Mycobacterium, Aeromonas, Tenacibaculum, Escherichia, Klebsiella, and Legionella (Kirstein et al., 2016; McCormick et al., 2016). These bacteria can pose a significant risk to aquatic life and human health by causing infections, skin irritation, and even systemic diseases. In addition to bacterial pathogens, the plastisphere can also harbor eukaryotic microorganisms that can have a potential negative impact. Potentially toxic microalgae and potentially pathogenic fungi (i.e., Chytridiomycota and Cryptomycota species) were found on plastic debris, raising concerns about its role in promoting harmful algal blooms and the spread of water-borne fungal diseases (Barros and Seena, 2021; Di Pippo et al., 2022). More recently, several studies on plastic and MP dispersal in freshwaters showed the co-presence of pathogens and Mobile Genetic Elements (MGEs), including ARGs, thus suggesting a higher probability of antibiotic resistance acquisition mediated by the plastisphere (Junaid et al., 2022; Table 2). Considering the worldwide spread of MPs in the environment, ARGs presence on MPs may exacerbate risk for human to acquire ARGs and-or resistant microorganisms of health concern. Indeed, some studies have revealed that marine microorganisms can uptake MPs from the water environment transferring in the food chain and more recently has been shown that ARGs can transfer through the trophic level into the food chain (Zhu et al., 2019; Figure 1).
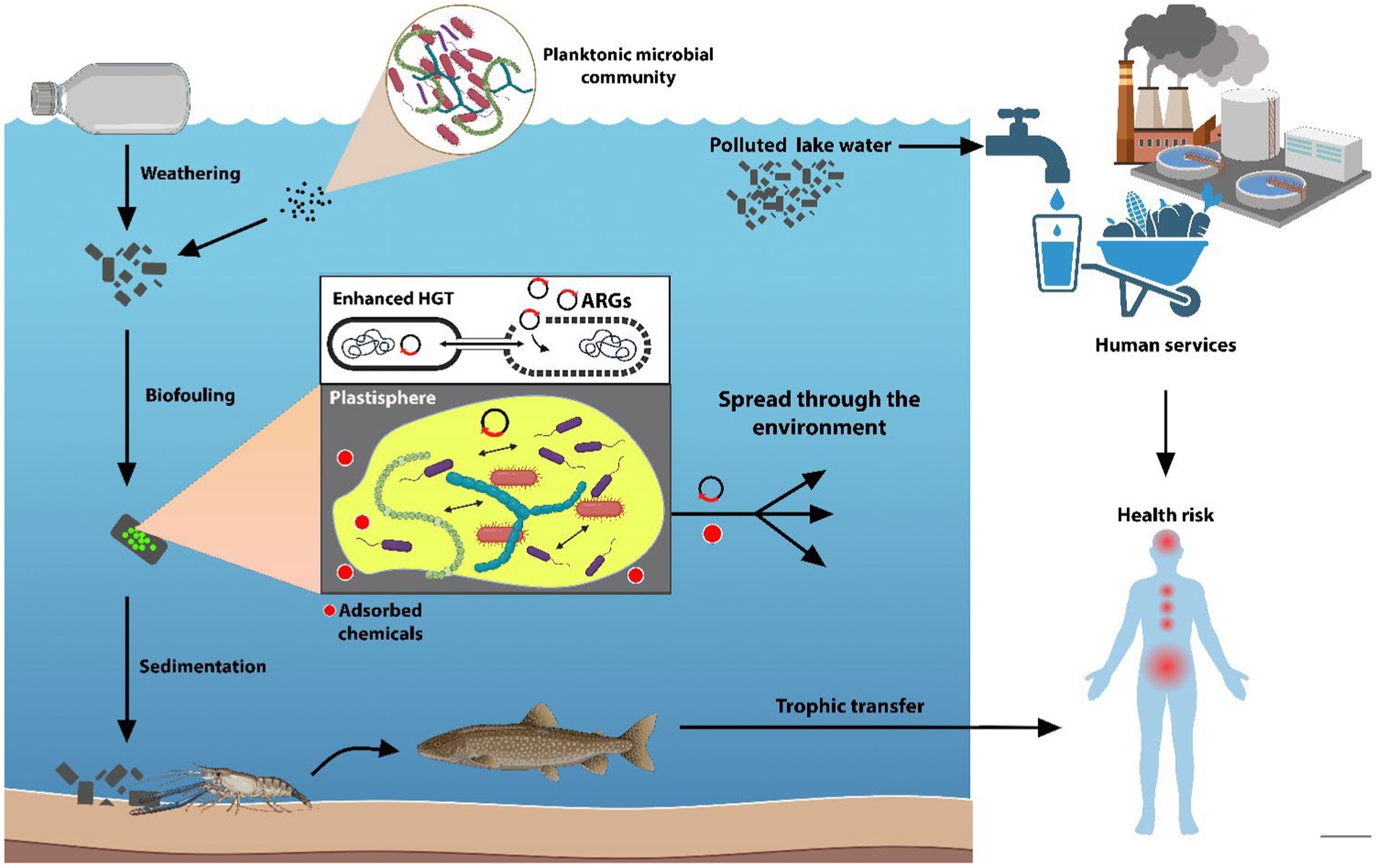
Figure 1. Pathways through which plastisphere and plastics may affect ecosystems and human health. Plastics released in freshwater as vectors for both microorganisms and chemicals adsorbed on their surfaces. Once in water, plastics are affected by abiotic factors leading to their fragmentation into smaller particles, that are colonized by the planktonic microbial community (biofouling) with an increase of density of the particles, causing their sedimentation through the water column. The close relationship between the biofilm microorganisms lead to an enhanced horizontal gene transfer (HGT) with the spreading of the ARGs through the plastisphere members. As ubiquitous pollutants, plastic debris travel throughout the environment leading to the spread of antibiotic resistance and adsorbed chemicals into freshwater, finally reaching services provided to human health and society. The persistent plastic debris can also enter the trophic chain. (Images created using BioRender.com.)
ARG enrichment in plastic-associated biofilms is promoted by the proximity and close contact between bacterial cells that facilitate horizontal gene transfer, contributing to long-distance dispersal and long-term persistence of ARGs in the environment (Di Pippo et al., 2022; Du et al., 2022; Rubin and Zucker, 2022; Luo et al., 2023; Chen et al., 2024; Li et al., 2024; Figure 1). Moreover, the presence of plastic-adsorbed xenobiotics and metals might enhance ARG occurrence through co-selection processes (Abe et al., 2020; Junaid et al., 2022; Michaelis and Grohmann, 2023).
Unlike in marine environments, studies in freshwater showed limited plastisphere enrichment of ARGs compared to the surrounding waters and natural substrata (Wu et al., 2019; Wang et al., 2020; Xu et al., 2022), with no differences in ARGs and MGEs observed between plastisphere and natural biofilms (Hu et al., 2021). Recent studies on freshwater plastisphere have mainly analyzed the differences in ARG abundance and diversity by comparing (i) surface waters and other natural surfaces (González-Pleiter et al., 2021; Xu et al., 2022; Martínez-Campos et al., 2023), (ii) biodegradable and non-biodegradable plastics (Zhou Q. et al., 2022), (iii) different stages of biofilm development at different contamination levels (Table 2). Although clearly showing the worldwide spread of plastic-associated pathogens and ARGs, the information available on freshwater plastisphere is still limited to properly evaluate human health risks (Manaia, 2017; Zhang et al., 2021).
Notably, the identification of potential pathogens is at the genus level, which does not provide direct evidence of the pathogen’s occurrence, infectivity or virulence (Liu et al., 2022). Furthermore, quantitative PCR-based methods are limited to known functional genes and may miss novel or uncharacterized ARGs (Li et al., 2015). Combined omics approaches can provide detailed information on the collection of ARGs within the entire microbial community, namely the resistome (Bengtsson-Palme et al., 2018). However, due to the large diversity of ARGs and their incomplete coverage by the applied monitoring methods, the plastic-associated ARGs profiles are hardly comparable between different studies.
Recent publications showed how long-reads sequencing can help to overcome these technical limitations (Zhang et al., 2022). Long-read sequencing techniques improve the quality and completeness of metagenome-assembled genomes allowing to reduce errors and improving the accuracy of ARG identification and characterization (Table 2).
4 The role of plastisphere in plastic biodegradation processes
Once plastic items are transported through the aquatic environment, abiotic factors can cause changes in their mechanical and physico-chemical properties (Luo et al., 2022) and plastisphere microorganisms can modify MP surface properties by degrading additives, secreting MP-modifying/degrading enzymes. The plastisphere-mediated biodegradation of plastic debris and MPs is a complex multifaceted process in which polymers are first bio-fragmented through the secretion of extracellular enzymes. In the subsequent assimilation phase, the small and water-soluble molecules produced during the depolymerization of plastics are transported through the cell membrane.
Once inside the cell, plastic-derived molecules can be used as a carbon source to produce biomass and energy before being mineralized to CO2/CH4 and H2O (Tiwari et al., 2020; Yuan et al., 2020; Zeenat et al., 2021; Priya et al., 2022; Zhou Y. et al., 2022; Sun et al., 2023).
Plastic-degrading enzymes and microorganisms have been identified using culture-based approaches in which selected strains, isolated from environmental samples, are grown and screened for plastic-degrading activity under laboratory conditions (Mierzwa-Hersztek et al., 2019; Mohanan et al., 2020; Tiwari et al., 2020, 2022; Yuan et al., 2020; Amobonye et al., 2021; Nguyen et al., 2021; Othman et al., 2021; Priya et al., 2022; Zhou Y. et al., 2022). Microorganisms exhibiting plastic-degrading activity are typically isolated and enriched in a plastic-containing medium, while polymer-degrading enzymes are conventionally identified using a combination of biochemical and biomolecular approaches (Viljakainen and Hug, 2021; Herbert et al., 2022; Tiwari et al., 2022; Zhu et al., 2022; Tournier et al., 2023). To date, various genes and enzymes have been found to be associated with the plastisphere, including PETase (Polyethylene terephthalatease), MHETase (Mono (2-hydroxyethyl) terephthalate hydrolase), cutinases, lipases, oxidoreductases, laccases, peroxidases, and esterases (Tournier et al., 2023). In addition, several genes and enzymes enabling the breakdown of aromatic compounds into simpler and less toxic forms have been found in the plastisphere, mostly including dioxygenases (Seo et al., 2009).
More recently, the focus has shifted from studying specific strains or enzymes to examining the plastisphere community as a whole “degradation unit” (Jacquin et al., 2019; Yuan et al., 2020; Eronen-Rasimus et al., 2022; Joshi et al., 2022; Taipale et al., 2022; Cai et al., 2023; Maheswaran et al., 2023; Miao et al., 2023; Niu et al., 2023; Vaksmaa et al., 2023; Yu et al., 2023; Zhu et al., 2023). Various key microbial taxa can cooperate and show enzymatic potential for polymer biodegradation (Li K. et al., 2023; Li W. et al., 2023; Miao et al., 2023), also promoting changes in surface material properties, including chemical composition, morphology roughness, formation of holes and cracks, and weight loss. Such microbial driven surface features were assessed by scanning electron microscopy (SEM), Fourier-transform infrared spectroscopy (FTIR), atomic force microscopy, contact angle analysis, calorimetry, and mechanical testing (Chen et al., 2020; Denaro et al., 2020; Kosiorowska et al., 2022; Li J. et al., 2023; Sun et al., 2023). Several studies have shown that the presence of taxa associated with the biodegradation process is strictly connected to environmental factors such as light exposure (including UV radiation), heat, humidity, absorbed chemicals, pH, and oxygen levels, depending on the specific sampling sites (Tiwari et al., 2020; Yuan et al., 2020; Martínez-Campos et al., 2021; Vincent et al., 2022; Li W. et al., 2023). However, although mediated by biofilm composition and development (Miao et al., 2021; for further details see Sun et al., 2023), plastic polymer type and properties as surface morphology, topography, hydrophobicity, electric charge distribution, molecular weight, mobility, crystallinity, types of functional groups, additives, and plasticizers were likely the main abiotic factors affecting the overall biodegradation process (Tiwari et al., 2020; Yuan et al., 2020; Song et al., 2023).
Few studies currently available report inconsistent results on the direct involvement of plastisphere in biodegradation processes. The metabolic potential to hydrolyse and use the plastic polymers as carbon sources was not convincingly demonstrated, while plastic materials were mostly used as adhesion surfaces by opportunistic aquatic microbes (Oberbeckmann et al., 2021; Di Pippo et al., 2023). Further investigations are thus needed to provide a deeper understanding of plastisphere role in plastic biodegradation.
Advanced culture-independent approaches based on sequencing technologies are accelerating discoveries in this field. Although still in their infancy, “plastic-omics” (Viljakainen and Hug, 2021) are emerging as important tools for understanding the functional potential of the plastisphere, providing important insights into the identification of potentially degrading bacterial taxa, the factors influencing their enrichment, and the plastic degrading genes/enzymes, and thus a holistic understanding of the plastic degradation process (Viljakainen and Hug, 2021; Malik et al., 2023). Metatranscriptomics can be a powerful approach to reveal the gene expression profiles and transcriptional activity of microorganisms associated with plastic surfaces, elucidating metabolic pathways and gene regulatory networks involved in plastic biodegradation (Gilbert et al., 2008; Kirstein et al., 2016; Xu et al., 2019; Yang et al., 2019; Lu et al., 2020).
5 Conclusion
This review paper sheds light on the intricate relationship between plastic pollution and microbial communities in freshwater ecosystems, specifically focusing on the freshwater plastisphere. While molecular methods have expanded our understanding of plastisphere biodiversity, fundamental questions regarding the influence of the polymer type and properties and environmental factors on plastisphere structure, biodiversity and on community assembly remain unanswered. The presence of potentially pathogenic microbes and genetic elements of concern within the plastisphere raises important implications for ecosystem and human health. However, the extent of these risks and their impacts are still not fully elucidated, necessitating further research efforts. Advanced sequencing technologies offer promising avenues for uncovering the functional potential of the plastisphere, including its role in plastic biodegradation processes. Overall, the findings underscore the urgent need for comprehensive investigations into freshwater plastisphere dynamics, which are crucial for informing effective management strategies to mitigate the environmental and health impacts of plastic pollution in freshwater ecosystems.
Author contributions
VB: Writing – review & editing, Writing – original draft. SG: Conceptualization, Writing – review & editing, Writing – original draft. CL: Writing – review & editing, Writing – original draft. SC: Writing – review & editing. SA: Conceptualization, Writing – review & editing. RC: Writing – review & editing. BM: Writing – review & editing, Writing – original draft. SR: Conceptualization, Writing – review & editing. FP: Conceptualization, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the project “National Biodiversity Future Center–NBFC” (code CN_00000033, CUP B83C22002930006) funded under the National Recovery and Resilience Plan (NRRP) by the Italian Ministry of University and Research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abe, K., Nomura, N., and Suzuki, S. (2020). Biofilms: hot spots of horizontal gene transfer (HGT) in aquatic environments, with a focus on a new HGT mechanism. FEMS Microbiol. Ecol. 96:fiaa 031. doi: 10.1093/femsec/fiaa031
Amaneesh, C., Anna Balan, S., Silpa, P. S., Kim, J. W., Greeshma, K., Aswathi Mohan, A., et al. (2023). Gross negligence: impacts of microplastics and plastic leachates on phytoplankton community and ecosystem dynamics. Environ. Sci. Technol. 57, 5–24. doi: 10.1021/acs.est.2c05817
Amaral-Zettler, L. A., Zettler, E. R., and Mincer, T. J. (2020). Ecology of the plastisphere. Nat. Rev. Microbiol. 18, 139–151. doi: 10.1038/s41579-019-0308-0
Amobonye, A., Bhagwat, P., Singh, S., and Pillai, S. (2021). Plastic biodegradation: frontline microbes and their enzymes. Sci. Total Environ. 759:143536. doi: 10.1016/j.scitotenv.2020.143536
Arias-Andres, M., Kettner, M. T., Miki, T., and Grossart, H.-P. (2018). Microplastics: new substrates for heterotrophic activity contribute to altering organic matter cycles in aquatic ecosystems. Sci. Total Environ. 635, 1152–1159. doi: 10.1016/j.scitotenv.2018.04.199
Bao, R., Pu, J., Xie, C., Mehmood, T., Chen, W., Gao, L., et al. (2022). Aging of biodegradable blended plastic generates microplastics and attached bacterial communities in air and aqueous environments. J. Hazard. Mater. 434:128891. doi: 10.1016/j.jhazmat.2022.128891
Barros, J., and Seena, S. (2021). Plastisphere in freshwaters: an emerging concern. Environ. Pollut. 290:118123. doi: 10.1016/j.envpol.2021.118123
Bengtsson-Palme, J., Kristiansson, E., and Larsson, D. G. J. (2018). Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol. Rev. 42:fux 053. doi: 10.1093/femsre/fux053
Besemer, K. (2015). Biodiversity, community structure and function of biofilms in stream ecosystems. Res. Microbiol. 166, 774–781. doi: 10.1016/j.resmic.2015.05.006
Besemer, K., Peter, H., Logue, J. B., Langenheder, S., Lindström, E. S., Tranvik, L. J., et al. (2012). Unraveling assembly of stream biofilm communities. ISME J. 6, 1459–1468. doi: 10.1038/ismej.2011.205
Bhagwat, G., Zhu, Q., O’Connor, W., Subashchandrabose, S., Grainge, I., Knight, R., et al. (2021). Exploring the composition and functions of plastic microbiome using whole-genome sequencing. Environ. Sci. Technol. 55, 4899–4913. doi: 10.1021/acs.est.0c07952
Bryant, J. A., Clemente, T. M., Viviani, D. A., Fong, A. A., Thomas, K. A., Kemp, P., et al. (2016). Diversity and activity of communities inhabiting plastic debris in the North Pacific gyre. mSystems 1, e00024–e00016. doi: 10.1128/mSystems.00024-16
Cai, Z., Li, M., Zhu, Z., Wang, X., Huang, Y., Li, T., et al. (2023). Biological degradation of plastics and microplastics: a recent perspective on associated mechanisms and influencing factors. Microorganisms 11:1661. doi: 10.3390/microorganisms11071661
Carson, H. S., Nerheim, M. S., Carroll, K. A., and Eriksen, M. (2013). The plastic-associated microorganisms of the North Pacific gyre. Mar. Pollut. Bull. 75, 126–132. doi: 10.1016/j.marpolbul.2013.07.054
Chaudhary, A., Dunn, S. T., Kelly, J., and Hoellein, T. J. (2022). Plastic microbiome development in a freshwater ecosystem. Sci. Total Environ. 848:157697. doi: 10.1016/j.scitotenv.2022.157697
Chen, Y., Yan, Z., Zhou, Y., Zhang, Y., Jiang, R., Wang, M., et al. (2024). Dynamic evolution of antibiotic resistance genes in plastisphere in the vertical profile of urban rivers. Water Res. 249:120946. doi: 10.1016/j.watres.2023.120946
Chen, Z., Zhao, W., Xing, R., Xie, S., Yang, X., Cui, P., et al. (2020). Enhanced in situ biodegradation of microplastics in sewage sludge using hyperthermophilic composting technology. J. Hazard. Mater. 384:121271. doi: 10.1016/j.jhazmat.2019.121271
Coons, A. K., Busch, K., Lenz, M., Hentschel, U., and Borchert, E. (2021). Biogeography rather than substrate type determines bacterial colonization dynamics of marine plastics. Peer J 9:e12135. doi: 10.7717/peerj.12135
Dąbrowska, A. (2021). A roadmap for a Plastisphere. Mar. Pollut. Bull. 167:112322. doi: 10.1016/j.marpolbul.2021.112322
Delacuvellerie, A., Ballerini, T., Frère, L., Matallana-Surget, S., Dumontet, B., and Wattiez, R. (2022). From rivers to marine environments: a constantly evolving microbial community within the plastisphere. Mar. Pollut. Bull. 179:113660. doi: 10.1016/j.marpolbul.2022.113660
Delacuvellerie, A., Benali, S., Cyriaque, V., Moins, S., Raquez, J.-M., Gobert, S., et al. (2021). Microbial biofilm composition and polymer degradation of compostable and non-compostable plastics immersed in the marine environment. J. Hazard. Mater. 419:126526. doi: 10.1016/j.jhazmat.2021.126526
Denaro, R., Aulenta, F., Crisafi, F., Di Pippo, F., Cruz Viggi, C., Matturro, B., et al. (2020). Marine hydrocarbon-degrading bacteria breakdown poly (ethylene terephthalate) (PET). Sci. Total Environ. 749:141608. doi: 10.1016/j.scitotenv.2020.141608
De Tender, C. A., Devriese, L. I., Haegeman, A., Maes, S., Ruttink, T., and Dawyndt, P. (2015). Bacterial community profiling of plastic litter in the Belgian part of the North Sea. Environ. Sci. Technol. 49, 9629–9638. doi: 10.1021/acs.est.5b01093
Dexter, S. C. (1979). Influence of substratum critical surface tension on bacterial adhesion—in situ studies. J. Colloid Interface Sci. 70, 346–354. doi: 10.1016/0021-9797(79)90038-9
Dey, S., Rout, A. K., Behera, B. K., and Ghosh, K. (2022). Plastisphere community assemblage of aquatic environment: plastic-microbe interaction, role in degradation and characterization technologies. Environ. Microbiome 17:32. doi: 10.1186/s40793-022-00430-4
Di Pippo, F., Bocci, V., Amalfitano, S., Crognale, S., Levantesi, C., Pietrelli, L., et al. (2023). Microbial colonization patterns and biodegradation of petrochemical and biodegradable plastics in lake waters: insights from a field experiment. Front. Microbiol. 14:441. doi: 10.3389/fmicb.2023.1290441
Di Pippo, F., Crognale, S., Levantesi, C., Vitanza, L., Sighicelli, M., Pietrelli, L., et al. (2022). Plastisphere in lake waters: microbial diversity, biofilm structure, and potential implications for freshwater ecosystems. Environ. Pollut. 310:119876. doi: 10.1016/j.envpol.2022.119876
Di Pippo, F., Venezia, C., Sighicelli, M., Pietrelli, L., Di Vito, S., Nuglio, S., et al. (2020). Microplastic-associated biofilms in lentic Italian ecosystems. Water Res. 187:116429. doi: 10.1016/j.watres.2020.116429
Du, Y., Liu, X., Dong, X., and Yin, Z. (2022). A review on marine plastisphere: biodiversity, formation, and role in degradation. Comput. Struct. Biotechnol. J. 20, 975–988. doi: 10.1016/j.csbj.2022.02.008
Eckert, E. M., Di Cesare, A., Kettner, M. T., Arias-Andres, M., Fontaneto, D., Grossart, H.-P., et al. (2018). Microplastics increase impact of treated wastewater on freshwater microbial community. Environ. Pollut. 234, 495–502. doi: 10.1016/j.envpol.2017.11.070
Eronen-Rasimus, E. L., Näkki, P. P., and Kaartokallio, H. P. (2022). Degradation rates and bacterial community compositions vary among commonly used bioplastic materials in a brackish marine environment. Environ. Sci. Technol. 56, 15760–15769. doi: 10.1021/acs.est.2c06280
Forero-López, A. D., Brugnoni, L. I., Abasto, B., Rimondino, G. N., Lassalle, V. L., Ardusso, M. G., et al. (2022). Plastisphere on microplastics: in situ assays in an estuarine environment. J. Hazard. Mater. 440:129737. doi: 10.1016/j.jhazmat.2022.129737
Galafassi, S., Sabatino, R., Sathicq, M. B., Eckert, E. M., Fontaneto, D., Dalla Fontana, G., et al. (2021). Contribution of microplastic particles to the spread of resistances and pathogenic bacteria in treated wastewaters. Water Res. 201:117368. doi: 10.1016/j.watres.2021.117368
Gilbert, J. A., Field, D., Huang, Y., Edwards, R., Li, W., Gilna, P., et al. (2008). Detection of large numbers of novel sequences in the Metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042. doi: 10.1371/journal.pone.0003042
Gong, M., Yang, G., Zhuang, L., and Zeng, E. Y. (2019). Microbial biofilm formation and community structure on low-density polyethylene microparticles in lake water microcosms. Environ. Pollut. 252, 94–102. doi: 10.1016/j.envpol.2019.05.090
González-Pleiter, M., Velázquez, D., Casero, M. C., Tytgat, B., Verleyen, E., Leganés, F., et al. (2021). Microbial colonizers of microplastics in an Arctic freshwater lake. Sci. Total Environ. 795:148640. doi: 10.1016/j.scitotenv.2021.148640
Haram, L. E., Carlton, J. T., Centurioni, L., Crowley, M., Hafner, J., Maximenko, N., et al. (2021). Emergence of a neopelagic community through the establishment of coastal species on the high seas. Nat. Commun. 12:6885. doi: 10.1038/s41467-021-27188-6
Herbert, J., Beckett, A. H., and Robson, S. C. (2022). A review of cross-disciplinary approaches for the identification of novel industrially relevant plastic-degrading enzymes. Sustain. For. 14:898. doi: 10.3390/su142315898
Hoellein, T. J., McCormick, A. R., Hittie, J., London, M. G., Scott, J. W., and Kelly, J. J. (2017). Longitudinal patterns of microplastic concentration and bacterial assemblages in surface and benthic habitats of an urban river. Freshw. Sci. 36, 491–507. doi: 10.1086/693012
Hoellein, T., Rojas, M., Pink, A., Gasior, J., and Kelly, J. (2014). Anthropogenic litter in urban freshwater ecosystems: distribution and microbial interactions. PLoS One 9:e98485. doi: 10.1371/journal.pone.0098485
Huang, J.-N., Wen, B., Miao, L., Liu, X., Li, Z.-J., Ma, T.-F., et al. (2022). Microplastics drive nitrification by enriching functional microorganisms in aquaculture pond waters. Chemosphere 309:136646. doi: 10.1016/j.chemosphere.2022.136646
Hu, H., Jin, D., Yang, Y., Zhang, J., Ma, C., and Qiu, Z. (2021). Distinct profile of bacterial community and antibiotic resistance genes on microplastics in Ganjiang River at the watershed level. Environ. Res. 200:111363. doi: 10.1016/j.envres.2021.111363
Jacquin, J., Cheng, J., Odobel, C., Pandin, C., Conan, P., Pujo-Pay, M., et al. (2019). Microbial ecotoxicology of marine plastic debris: a review on colonization and biodegradation by the “Plastisphere”. Front. Microbiol. 10:865. doi: 10.3389/fmicb.2019.00865
Jia, Z. (2022). “Antifouling strategies-interference with bacterial adhesion” in Focus on bacterial biofilms. eds. T. Das and A. C. Di (London: Intech Open)
Joshi, G., Goswami, P., Verma, P., Prakash, G., Simon, P., Vinithkumar, N. V., et al. (2022). Unraveling the plastic degradation potentials of the plastisphere-associated marine bacterial consortium as a key player for the low-density polyethylene degradation. J. Hazard. Mater. 425:128005. doi: 10.1016/j.jhazmat.2021.128005
Junaid, M., Liu, S., Liao, H., Liu, X., Wu, Y., and Wang, J. (2022). Wastewater plastisphere enhances antibiotic resistant elements, bacterial pathogens, and toxicological impacts in the environment. Sci. Total Environ. 841:156805. doi: 10.1016/j.scitotenv.2022.156805
Karlsson, S., Ljungquist, O., and Albertsson, A.-C. (1988). Biodegradation of polyethylene and the influence of surfactants. Polym. Degrad. Stab. 21, 237–250. doi: 10.1016/0141-3910(88)90030-4
Kelly, J. J., London, M. G., McCormick, A. R., Rojas, M., Scott, J. W., and Hoellein, T. J. (2021). Wastewater treatment alters microbial colonization of microplastics. PLoS One 16:e0244443. doi: 10.1371/journal.pone.0244443
Kettner, M. T., Rojas-Jimenez, K., Oberbeckmann, S., Labrenz, M., and Grossart, H.-P. (2017). Microplastics alter composition of fungal communities in aquatic ecosystems. Environ. Microbiol. 19, 4447–4459. doi: 10.1111/1462-2920.13891
Kirstein, I. V., Kirmizi, S., Wichels, A., Garin-Fernandez, A., Erler, R., Löder, M., et al. (2016). Dangerous hitchhikers? Evidence for potentially pathogenic Vibrio spp. on microplastic particles. Mar. Environ. Res. 120, 1–8. doi: 10.1016/j.marenvres.2016.07.004
Kosiorowska, K. E., Biniarz, P., Dobrowolski, A., Leluk, K., and Mirończuk, A. M. (2022). Metabolic engineering of Yarrowia lipolytica for poly (ethylene terephthalate) degradation. Sci. Total Environ. 831:154841. doi: 10.1016/j.scitotenv.2022.154841
Kreve, S., and Reis, A. C. D. (2021). Bacterial adhesion to biomaterials: what regulates this attachment? A review. Jpn. Dent. Sci. Rev. 57, 85–96. doi: 10.1016/j.jdsr.2021.05.003
Li, B., Yang, Y., Ma, L., Ju, F., Guo, F., Tiedje, J. M., et al. (2015). Metagenomic and network analysis reveal wide distribution and co-occurrence of environmental antibiotic resistance genes. ISME J. 9, 2490–2502. doi: 10.1038/ismej.2015.59
Li, C., Wang, L., Ji, S., Chang, M., Wang, L., Gan, Y., et al. (2021). The ecology of the plastisphere: Microbial composition, function, assembly, and network in the freshwater and seawater ecosystems. Water Res. 202:117428. doi: 10.1016/j.watres.2021.117428
Li, H., Luo, Q., Zhao, S., Zhao, P., Yang, X., Huang, Q., et al. (2022). Watershed urbanization enhances the enrichment of pathogenic bacteria and antibiotic resistance genes on microplastics in the water environment. Environ. Pollut. 313:120185. doi: 10.1016/j.envpol.2022.120185
Li, J., Zhang, T., Shan, X., Zheng, W., Zhang, Z., Ouyang, Z., et al. (2023). Abandoned disposable masks become hot substrates for plastisphere, whether in soil, atmosphere or water. J. Hazard. Mater. 452:131321. doi: 10.1016/j.jhazmat.2023.131321
Li, K., Jia, W., Xu, L., Zhang, M., and Huang, Y. (2023). The plastisphere of biodegradable and conventional microplastics from residues exhibit distinct microbial structure, network and function in plastic-mulching farmland. J. Hazard. Mater. 442:130011. doi: 10.1016/j.jhazmat.2022.130011
Li, K., Xu, L., Bai, X., Zhang, G., Zhang, M., and Huang, Y. (2024). Potential environmental risks of field bio/non-degradable microplastic from mulching residues in farmland: evidence from metagenomic analysis of plastisphere. J. Hazard. Mater. 465:133428. doi: 10.1016/j.jhazmat.2024.133428
Li, R., Zhu, L., Wang, Y., and Zhu, Y.-G. (2022). Metagenomic insights into environmental risk of field microplastics in an urban river. Water Res. 223:119018. doi: 10.1016/j.watres.2022.119018
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Li, W., Miao, L., Adyel, T. M., Wu, J., Yu, Y., and Hou, J. (2023). Characterization of dynamic plastisphere and their underlying effects on the aging of biodegradable and traditional plastics in freshwater ecosystems. J. Hazard. Mater. 446:130714. doi: 10.1016/j.jhazmat.2022.130714
Luo, G., Liang, B., Cui, H., Kang, Y., Zhou, X., Tao, Y., et al. (2023). Determining the contribution of Micro/Nanoplastics to antimicrobial resistance: challenges and perspectives. Environ. Sci. Technol. 57, 12137–12152. doi: 10.1021/acs.est.3c01128
Luo, H., Liu, C., He, D., Xu, J., Sun, J., Li, J., et al. (2022). Environmental behaviors of microplastics in aquatic systems: A systematic review on degradation, adsorption, toxicity and biofilm under aging conditions. Journal of Hazardous Materials, 423, 126915. doi: 10.1016/j.jhazmat.2021.126915
Lu, R., Wu, X., Wan, Z., Li, Y., Zuo, L., Qin, J., et al. (2020). Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virol. Sin. 35, 344–347. doi: 10.1007/s12250-020-00218-1
Maheswaran, B., Al-Ansari, M., Al-Humaid, L., Sebastin Raj, J., Kim, W., Karmegam, N., et al. (2023). In vivo degradation of polyethylene terephthalate using microbial isolates from plastic polluted environment. Chemosphere 310:136757. doi: 10.1016/j.chemosphere.2022.136757
Malik, N., Lakhawat, S. S., Kumar, V., Sharma, V., Bhatti, J. S., and Sharma, P. K. (2023). Recent advances in the omics-based assessment of microbial consortia in the plastisphere environment: deciphering the dynamic role of hidden players. Process Saf. Environ. Prot. 176, 207–225. doi: 10.1016/j.psep.2023.06.013
Manaia, C. M. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 25, 173–181. doi: 10.1016/j.tim.2016.11.014
Martínez-Campos, S., González-Pleiter, M., Fernández-Piñas, F., Rosal, R., and Leganés, F. (2021). Early and differential bacterial colonization on microplastics deployed into the effluents of wastewater treatment plants. Sci. Total Environ. 757:143832. doi: 10.1016/j.scitotenv.2020.143832
Martínez-Campos, S., González-Pleiter, M., Rico, A., Schell, T., Vighi, M., Fernández-Piñas, F., et al. (2023). Time-course biofilm formation and presence of antibiotic resistance genes on everyday plastic items deployed in river waters. J. Hazard. Mater. 443:130271. doi: 10.1016/j.jhazmat.2022.130271
Masó, M., Fortuño, J. M., De Juan, S., and Demestre, M. (2016). Microfouling communities from pelagic and benthic marine plastic debris sampled across Mediterranean coastal waters. Sci. Mar. 80, 117–127. doi: 10.3989/scimar.04281.10A
McCormick, A., Hoellein, T. J., Mason, S. A., Schluep, J., and Kelly, J. J. (2014). Microplastic is an abundant and distinct microbial habitat in an Urban River. Environ. Sci. Technol. 48, 11863–11871. doi: 10.1021/es503610r
McCormick, A. R., Hoellein, T. J., London, M. G., Hittie, J., Scott, J. W., and Kelly, J. J. (2016). Microplastic in surface waters of urban rivers: concentration, sources, and associated bacterial assemblages. Ecosphere 7:1556. doi: 10.1002/ecs2.1556
Miao, L., Li, W., Adyel, T. M., Yao, Y., Deng, Y., Wu, J., et al. (2023). Spatio-temporal succession of microbial communities in plastisphere and their potentials for plastic degradation in freshwater ecosystems. Water Res. 229:119406. doi: 10.1016/j.watres.2022.119406
Miao, L., Wang, P., Hou, J., Yao, Y., Liu, Z., Liu, S., et al. (2019). Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 650, 2395–2402. doi: 10.1016/j.scitotenv.2018.09.378
Miao, L., Yu, Y., Adyel, T. M., Wang, C., Liu, Z., Liu, S., et al. (2021). Distinct microbial metabolic activities of biofilms colonizing microplastics in three freshwater ecosystems. J. Hazard. Mater. 403:123577. doi: 10.1016/j.jhazmat.2020.123577
Michaelis, C., and Grohmann, E. (2023). Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiotics 12:328. doi: 10.3390/antibiotics12020328
Mierzwa-Hersztek, M., Gondek, K., and Kopeć, M. (2019). Degradation of polyethylene and biocomponent-derived polymer materials: an overview. J. Polym. Environ. 27, 600–611. doi: 10.1007/s10924-019-01368-4
Mishra, S., Swain, S., Sahoo, M., Mishra, S., and Das, A. P. (2022). Microbial colonization and degradation of microplastics in aquatic ecosystem: a review. Geomicrobiol J. 39, 259–269. doi: 10.1080/01490451.2021.1983670
Mohanan, N., Montazer, Z., Sharma, P. K., and Levin, D. B. (2020). Microbial and enzymatic degradation of synthetic plastics. Front. Microbiol. 11:580709. doi: 10.3389/fmicb.2020.580709
Mughini-Gras, L., Van Der Plaats, R. Q. J., Van Der Wielen, P. W. J. J., Bauerlein, P. S., and De Roda Husman, A. M. (2021). Riverine microplastic and microbial community compositions: a field study in the Netherlands. Water Res. 192:116852. doi: 10.1016/j.watres.2021.116852
Nguyen, H. T., Choi, W., Kim, E.-J., and Cho, K. (2022). Microbial community niches on microplastics and prioritized environmental factors under various urban riverine conditions. Sci. Total Environ. 849:157781. doi: 10.1016/j.scitotenv.2022.157781
Nguyen, N. H. A., El-Temsah, Y. S., Cambier, S., Calusinska, M., Hrabak, P., Pouzar, M., et al. (2021). Attached and planktonic bacterial communities on bio-based plastic granules and micro-debris in seawater and freshwater. Sci. Total Environ. 785:147413. doi: 10.1016/j.scitotenv.2021.147413
Nguyen, N. H. A., Marlita, M., El-Temsah, Y. S., Hrabak, P., Riha, J., and Sevcu, A. (2023). Early stage biofilm formation on bio-based microplastics in a freshwater reservoir. Sci. Total Environ. 858:159569. doi: 10.1016/j.scitotenv.2022.159569
Nikolopoulou, I., Piperagkas, O., Moschos, S., and Karayanni, H. (2023). Bacteria release from microplastics into new aquatic environments. Diversity 15:115. doi: 10.3390/d15010115
Niu, L., Chen, Y., Li, Y., Wang, Y., Shen, J., Wang, L., et al. (2023). Diversity, abundance and distribution characteristics of potential polyethylene and polypropylene microplastic degradation bacterial communities in the urban river. Water Res. 232:119704. doi: 10.1016/j.watres.2023.119704
Oberbeckmann, S., Bartosik, D., Huang, S., Werner, J., Hirschfeld, C., Wibberg, D., et al. (2021). Genomic and proteomic profiles of biofilms on microplastics are decoupled from artificial surface properties. Environ. Microbiol. 23, 3099–3115. doi: 10.1111/1462-2920.15531
Oberbeckmann, S., Kreikemeyer, B., and Labrenz, M. (2018). Environmental factors support the formation of specific bacterial assemblages on microplastics. Front. Microbiol. 8:2709. doi: 10.3389/fmicb.2017.02709
Oberbeckmann, S., Loeder, M. G. J., Gerdts, G., and Osborn, A. M. (2014). Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in northern European waters. FEMS Microbiol. Ecol. 90, 478–492. doi: 10.1111/1574-6941.12409
Othman, A. R., Hasan, H. A., Muhamad, M. H., Ismail, N. I., and Abdullah, S. R. S. (2021). Microbial degradation of microplastics by enzymatic processes: a review. Environ. Chem. Lett. 19, 3057–3073. doi: 10.1007/s10311-021-01197-9
Pinto, M., Langer, T. M., Hüffer, T., Hofmann, T., and Herndl, G. J. (2019). The composition of bacterial communities associated with plastic biofilms differs between different polymers and stages of biofilm succession. PLoS One 14:e0217165. doi: 10.1371/journal.pone.0217165
Pollet, T., Berdjeb, L., Garnier, C., Durrieu, G., Poupon, C. L., Misson, B., et al. (2018). Prokaryotic community successions and interactions in marine biofilms: the key role of Flavobacteriia. FEMS Microbiol. Ecol. 94:83. doi: 10.1093/femsec/fiy083
Polz, M. F., Hunt, D. E., Preheim, S. P., and Weinreich, D. M. (2006). Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. B Biol. Sci. 361, 2009–2021. doi: 10.1098/rstb.2006.1928
Priya, A., Dutta, K., and Daverey, A. (2022). A comprehensive biotechnological and molecular insight into plastic degradation by microbial community. J. Chem. Technol. Biotechnol. 97, 381–390. doi: 10.1002/jctb.6675
Qiang, L., Cheng, J., Mirzoyan, S., Kerkhof, L. J., and Häggblom, M. M. (2021). Characterization of microplastic-associated biofilm development along a freshwater-estuarine gradient. Environ. Sci. Technol. 55, 16402–16412. doi: 10.1021/acs.est.1c04108
Rubin, A. E., and Zucker, I. (2022). Interactions of microplastics and organic compounds in aquatic environments: a case study of augmented joint toxicity. Chemosphere 289:133212. doi: 10.1016/j.chemosphere.2021.133212
Rummel, C. D., Jahnke, A., Gorokhova, E., Kühnel, D., and Schmitt-Jansen, M. (2017). Impacts of biofilm formation on the fate and potential effects of microplastic in the aquatic environment. Environ. Sci. Technol. Lett. 4, 258–267. doi: 10.1021/acs.estlett.7b00164
Seo, J.-S., Keum, Y.-S., and Li, Q. (2009). Bacterial degradation of aromatic compounds. Int. J. Environ. Res. Public Health 6, 278–309. doi: 10.3390/ijerph6010278
Simon, M., Scheuner, C., Meier-Kolthoff, J. P., Brinkhoff, T., Wagner-Döbler, I., Ulbrich, M., et al. (2017). Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 11, 1483–1499. doi: 10.1038/ismej.2016.198
Song, X., Ding, J., Tian, W., Xu, H., Zou, H., and Wang, Z. (2023). Effects of plastisphere on phosphorus availability in freshwater system: critical roles of polymer type and colonizing habitat. Sci. Total Environ. 870:161990. doi: 10.1016/j.scitotenv.2023.161990
Sooriyakumar, P., Bolan, N., Kumar, M., Singh, L., Yu, Y., Li, Y., et al. (2022). Biofilm formation and its implications on the properties and fate of microplastics in aquatic environments: a review. J. Hazard. Mater. Adv. 6:100077. doi: 10.1016/j.hazadv.2022.100077
Sun, X.-L., Xiang, H., Xiong, H.-Q., Fang, Y.-C., and Wang, Y. (2023). Bioremediation of microplastics in freshwater environments: a systematic review of biofilm culture, degradation mechanisms, and analytical methods. Sci. Total Environ. 863:160953. doi: 10.1016/j.scitotenv.2022.160953
Taipale, S. J., Vesamäki, J., Kautonen, P., Kukkonen, J. V. K., Biasi, C., Nissinen, R., et al. (2022). Biodegradation of microplastic in freshwaters: a long-lasting process affected by the lake microbiome. Environ. Microbiol. 25, 2669–2680. doi: 10.1111/1462-2920.16177
Tedersoo, L., Albertsen, M., Anslan, S., and Callahan, B. (2021). Perspectives and benefits of high-throughput long-read sequencing in microbial ecology. Appl. Environ. Microbiol. 87:e0062621. doi: 10.1128/AEM.00626-21
Tiwari, N., Bansal, M., Santhiya, D., and Sharma, J. G. (2022). Insights into microbial diversity on plastisphere by multi-omics. Arch. Microbiol. 204:216. doi: 10.1007/s00203-022-02806-z
Tiwari, N., Santhiya, D., and Sharma, J. G. (2020). Microbial remediation of micro-nano plastics: current knowledge and future trends. Environ. Pollut. 265:115044. doi: 10.1016/j.envpol.2020.115044
Tournier, V., Duquesne, S., Guillamot, F., Cramail, H., Taton, D., Marty, A., et al. (2023). Enzymes’ power for plastics degradation. Chem. Rev. 123, 5612–5701. doi: 10.1021/acs.chemrev.2c00644
Vaksmaa, A., Polerecky, L., Dombrowski, N., Kienhuis, M. V. M., Posthuma, I., Gerritse, J., et al. (2023). Polyethylene degradation and assimilation by the marine yeast Rhodotorula mucilaginosa. ISME Commun. 3:68. doi: 10.1038/s43705-023-00267-z
Viljakainen, V. R., and Hug, L. A. (2021). New approaches for the characterization of plastic-associated microbial communities and the discovery of plastic-degrading microorganisms and enzymes. Comput. Struct. Biotechnol. J. 19, 6191–6200. doi: 10.1016/j.csbj.2021.11.023
Vincent, A. E. S., Chaudhary, A., Kelly, J. J., and Hoellein, T. J. (2022). Biofilm assemblage and activity on plastic in urban streams at a continental scale: site characteristics are more important than substrate type. Sci. Total Environ. 835:155398. doi: 10.1016/j.scitotenv.2022.155398
Wallbank, J. A., Lear, G., Kingsbury, J. M., Weaver, L., Doake, F., Smith, D. A., et al. (2022). Into the Plastisphere, where only the generalists thrive: early insights in Plastisphere microbial community succession. Front. Mar. Sci. 9:841142. doi: 10.3389/fmars.2022.841142
Wang, C., Wang, L., Ok, Y. S., Tsang, D. C. W., and Hou, D. (2022). Soil plastisphere: exploration methods, influencing factors, and ecological insights. J. Hazard. Mater. 430:128503. doi: 10.1016/j.jhazmat.2022.128503
Wang, J., Chu, L., Wojnárovits, L., and Takács, E. (2020). Occurrence and fate of antibiotics, antibiotic resistant genes (ARGs) and antibiotic resistant bacteria (ARB) in municipal wastewater treatment plant: an overview. Sci. Total Environ. 744:140997. doi: 10.1016/j.scitotenv.2020.140997
Wang, L., Tong, J., Li, Y., Zhu, J., Zhang, W., Niu, L., et al. (2021). Bacterial and fungal assemblages and functions associated with biofilms differ between diverse types of plastic debris in a freshwater system. Environ. Res. 196:110371. doi: 10.1016/j.envres.2020.110371
Wani, A. K., Akhtar, N., Naqash, N., Rahayu, F., Djajadi, D., Chopra, C., et al. (2023). Discovering untapped microbial communities through metagenomics for microplastic remediation: recent advances, challenges, and way forward. Environ. Sci. Pollut. Res. 30, 81450–81473. doi: 10.1007/s11356-023-25192-5
Weig, A. R., Löder, M. G. J., Ramsperger, A. F. R. M., and Laforsch, C. (2021). In situ prokaryotic and eukaryotic communities on microplastic particles in a small headwater stream in Germany. Front. Microbiol. 12:660024. doi: 10.3389/fmicb.2021.660024
Wen, B., Liu, J.-H., Zhang, Y., Zhang, H.-R., Gao, J.-Z., and Chen, Z.-Z. (2020). Community structure and functional diversity of the plastisphere in aquaculture waters: does plastic color matter? Sci. Total Environ. 740:140082. doi: 10.1016/j.scitotenv.2020.140082
Witsø, I. L., Basson, A., Vinje, H., Llarena, A.-K., Bringas, C. S., Aspholm, M., et al. (2023). Freshwater plastispheres as a vector for foodborne bacteria and viruses. Environ. Microbiol. 25, 2864–2881. doi: 10.1111/1462-2920.16536
Wu, X., Liu, Z., Li, M., Bartlam, M., and Wang, Y. (2022). Integrated metagenomic and metatranscriptomic analysis reveals actively expressed antibiotic resistomes in the plastisphere. J. Hazard. Mater. 430:128418. doi: 10.1016/j.jhazmat.2022.128418
Wu, X., Pan, J., Li, M., Li, Y., Bartlam, M., and Wang, Y. (2019). Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 165:114979. doi: 10.1016/j.watres.2019.114979
Xu, C., Hu, C., Lu, J., Yang, T., Shen, C., Li, F., et al. (2024). Lake plastisphere as a new biotope in the Anthropocene: potential pathogen colonization and distinct microbial functionality. J. Hazard. Mater. 461:132693. doi: 10.1016/j.jhazmat.2023.132693
Xu, C., Lu, J., Shen, C., Wang, J., and Li, F. (2022). Deciphering the mechanisms shaping the plastisphere antibiotic resistome on riverine microplastics. Water Res. 225:119192. doi: 10.1016/j.watres.2022.119192
Xue, N., Fang, Q., Pan, X., and Zhang, D. (2021). Distinct fungal plastisphere across different river functional zones: a watershed scale study. Sci. Total Environ. 752:141879. doi: 10.1016/j.scitotenv.2020.141879
Xue, N., Wang, L., Li, W., Wang, S., Pan, X., and Zhang, D. (2020). Increased inheritance of structure and function of bacterial communities and pathogen propagation in plastisphere along a river with increasing antibiotics pollution gradient. Environ. Pollut. 265:114641. doi: 10.1016/j.envpol.2020.114641
Xu, L., Li, K., Zhang, M., Guo, J., Jia, W., Bai, X., et al. (2023). Plastic substrate and residual time of microplastics in the urban river shape the composition and structure of bacterial communities in plastisphere. J. Environ. Manag. 345:118710. doi: 10.1016/j.jenvman.2023.118710
Xu, X., Wang, S., Gao, F., Li, J., Zheng, L., Sun, C., et al. (2019). Marine microplastic-associated bacterial community succession in response to geography, exposure time, and plastic type in China’s coastal seawaters. Mar. Pollut. Bull. 145, 278–286. doi: 10.1016/j.marpolbul.2019.05.036
Yang, K., Chen, Q.-L., Chen, M.-L., Li, H.-Z., Liao, H., Pu, Q., et al. (2020). Temporal dynamics of antibiotic Resistome in the Plastisphere during microbial colonization. Environ. Sci. Technol. 54, 11322–11332. doi: 10.1021/acs.est.0c04292
Yang, Y., Liu, G., Song, W., Ye, C., Lin, H., Li, Z., et al. (2019). Plastics in the marine environment are reservoirs for antibiotic and metal resistance genes. Environ. Int. 123, 79–86. doi: 10.1016/j.envint.2018.11.061
Yang, Y., Liu, W., Zhang, Z., Grossart, H.-P., and Gadd, G. M. (2020). Microplastics provide new microbial niches in aquatic environments. Appl. Microbiol. Biotechnol. 104, 6501–6511. doi: 10.1007/s00253-020-10704-x
Yuan, J., Ma, J., Sun, Y., Zhou, T., Zhao, Y., and Yu, F. (2020). Microbial degradation and other environmental aspects of microplastics/plastics. Sci. Total Environ. 715:136968. doi: 10.1016/j.scitotenv.2020.136968
Yuan, Z., Nag, R., and Cummins, E. (2022). Human health concerns regarding microplastics in the aquatic environment—from marine to food systems. Sci. Total Environ. 823:153730. doi: 10.1016/j.scitotenv.2022.153730
Yu, Y., Miao, L., Adyel, T. M., Waldschläger, K., Wu, J., and Hou, J. (2023). Aquatic plastisphere: interactions between plastics and biofilms. Environ. Pollut. 322:121196. doi: 10.1016/j.envpol.2023.121196
Zeenat, E., Elahi, A., Bukhari, D. A., Shamim, S., and Rehman, A. (2021). Plastics degradation by microbes: a sustainable approach. J. King Saud Univ. Sci. 33:101538. doi: 10.1016/j.jksus.2021.101538
Zettler, E. R., Mincer, T. J., and Amaral-Zettler, L. A. (2013). Life in the “Plastisphere”: microbial communities on plastic marine debris. Environ. Sci. Technol. 47, 7137–7146. doi: 10.1021/es401288x
Zhang, A.-N., Gaston, J. M., Dai, C. L., Zhao, S., Poyet, M., Groussin, M., et al. (2021). An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat. Commun. 12:4765. doi: 10.1038/s41467-021-25096-3
Zhang, W., Bhagwat, G., Palanisami, T., Liang, S., Wan, W., and Yang, Y. (2024). Lacustrine plastisphere: distinct succession and assembly processes of prokaryotic and eukaryotic communities and role of site, time, and polymer types. Water Res. 248:120875. doi: 10.1016/j.watres.2023.120875
Zhang, X. M., Elkoun, S., Ajji, A., and Huneault, M. A. (2004). Oriented structure and anisotropy properties of polymer blown films: HDPE, LLDPE and LDPE. Polymer 45, 217–229. doi: 10.1016/j.polymer.2003.10.057
Zhang, Z., Zhang, Q., Wang, T., Xu, N., Lu, T., Hong, W., et al. (2022). Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 13:1553. doi: 10.1038/s41467-022-29283-8
Zheng, S., Bawazir, M., Dhall, A., Kim, H.-E., He, L., Heo, J., et al. (2021). Implication of surface properties, bacterial motility, and hydrodynamic conditions on bacterial surface sensing and their initial adhesion. Front. Bioeng. Biotechnol. 9:643772. doi: 10.3389/fbioe.2021.643722
Zhou, Q., Zhang, J., Zhang, M., Wang, X., Zhang, D., and Pan, X. (2022). Persistent versus transient, and conventional plastic versus biodegradable plastic? —two key questions about microplastic-water exchange of antibiotic resistance genes. Water Res. 222:118899. doi: 10.1016/j.watres.2022.118899
Zhou, Y., Kumar, M., Sarsaiya, S., Sirohi, R., Awasthi, S. K., Sindhu, R., et al. (2022). Challenges and opportunities in bioremediation of micro-nano plastics: a review. Sci. Total Environ. 802:149823. doi: 10.1016/j.scitotenv.2021.149823
Zhu, B., Wang, D., and Wei, N. (2022). Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 40, 22–37. doi: 10.1016/j.tibtech.2021.02.008
Zhu, L., Wang, H., Chen, B., Sun, X., Qu, K., and Xia, B. (2019). Microplastic ingestion in deep-sea fish from the South China Sea. Science of The Total Environment, 677, 493–501. doi: 10.1016/j.scitotenv.2019.04.380
Zhu, M., Qi, X., Yuan, Y., Zhou, H., Rong, X., Dang, Z., et al. (2023). Deciphering the distinct successional patterns and potential roles of abundant and rare microbial taxa of urban riverine plastisphere. J. Hazard. Mater. 450:131080. doi: 10.1016/j.jhazmat.2023.131080
Glossary
Keywords: freshwater plastisphere, biodiversity, antibiotic resistance genes, pathogenic bacteria, plastic biodegradation
Citation: Bocci V, Galafassi S, Levantesi C, Crognale S, Amalfitano S, Congestri R, Matturro B, Rossetti S and Di Pippo F (2024) Freshwater plastisphere: a review on biodiversity, risks, and biodegradation potential with implications for the aquatic ecosystem health. Front. Microbiol. 15:1395401. doi: 10.3389/fmicb.2024.1395401
Edited by:
Tanja Shabarova, Academy of Sciences of the Czech Republic (ASCR), CzechiaReviewed by:
Lei Ren, Guangdong Ocean University, ChinaCopyright © 2024 Bocci, Galafassi, Levantesi, Crognale, Amalfitano, Congestri, Matturro, Rossetti and Di Pippo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesca Di Pippo, ZnJhbmNlc2NhLmRpcGlwcG9AaXJzYS5jbnIuaXQ=
 Valerio Bocci
Valerio Bocci Silvia Galafassi
Silvia Galafassi Caterina Levantesi
Caterina Levantesi Simona Crognale
Simona Crognale Stefano Amalfitano
Stefano Amalfitano Roberta Congestri
Roberta Congestri Bruna Matturro
Bruna Matturro Simona Rossetti
Simona Rossetti Francesca Di Pippo
Francesca Di Pippo