
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 14 June 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1394775
This article is part of the Research TopicSystems Biology and Antimicrobial Drug ResistanceView all 19 articles
 Zhilang Qiu1,2,3†
Zhilang Qiu1,2,3† Kexin Yuan1,3†
Kexin Yuan1,3† Huijun Cao1†
Huijun Cao1† Sufang Chen2,3
Sufang Chen2,3 Feifei Chen2
Feifei Chen2 Fei Mo1*
Fei Mo1* Guo Guo3,4*
Guo Guo3,4* Jian Peng1,2,3,4*
Jian Peng1,2,3,4*Introduction: Acinetobacter baumannii (A. baumannii) is an important opportunistic pathogen causing nosocomial infection in the clinic. The occurrence rate of antibiotic resistance is increasing year by year, resulting in a highly serious situation of bacterial resistance.
Methods: To better understand the local epidemiology of multidrug-resistant A. baumannii, an investigation was conducted on the antibiotic resistance of different types of A. baumannii and its relationship with the genes of A. baumannii. Furthermore, the molecular mechanism underlying antibiotic resistance in A. baumannii was investigated through transcriptome analysis.
Results: These results showed that a total of 9 STs were detected. It was found that 99% of the strains isolated in the hospital belonged to the same STs, and the clone complex CC208 was widely distributed in various departments and all kinds of samples. Furthermore, these A. baumannii strains showed high resistance to ertapenem, biapenem, meropenem, and imipenem, among which the resistance to ertapenem was the strongest. The detection rate of blaOXA–51 gene in these carbapenem resistance A. baumannii (CRAB) reached 100%; Additionally, the transcriptome results showed that the resistance genes were up-regulated in resistance strains, and these genes involved in biofilm formation, efflux pumps, peptidoglycan biosynthesis, and chaperonin synthesis.
Discussion: These results suggest that the CC208 STs were the main clonal complex, and showed high carbapenem antibiotic resistance. All these resistant strains were distributed in various departments, but most of them were distributed in intensive care units (ICU). The blaOXA–23 was the main antibiotic resistance genotype; In summary, the epidemic trend of clinical A. baumannii in Guiyang, China was analyzed from the molecular level, and the resistance mechanism of A. baumannii to carbapenem antibiotics was analyzed with transcriptome, which provided a theoretical basis for better control of A. baumannii.
The Gram-negative bacterium Acinetobacter baumannii (A. baumannii) is known as one of the most important nosocomial pathogens in healthcare-associated infections, especially with ventilator-associated pneumonia and catheter-associated infections (Liu et al., 2022; Kang et al., 2023). Additionally, A. baumannii has been reported to be associated with high mortality in intensive care units (ICUs), particularly among critically ill patients (Inchai et al., 2015). In 2017, The World Health Organization (WHO) also recently listed ESKAPE pathogens in the list of 12 bacteria against which new antibiotics are urgently needed (Mulani et al., 2019; Wang et al., 2022). Overuse of antibiotics has prompted the emergence of carbapenem resistance A. baumannii (CRAB), in consequence, either horizontal gene transfer or other opportunities, such as altered targets, decreased membrane permeability (Chusri et al., 2014), increased production of degrading enzymes (Lee et al., 2017), overexpression of efflux pumps (Roy et al., 2021), metabolic changes, biofilm formation (Yan and Bassler, 2019; Colquhoun and Rather, 2020; Gedefie et al., 2021), or increased nutrient sequestration mechanisms (Tiwari et al., 2019).
Antimicrobial resistance is a prominent feature that helps A. baumannii survive in an antimicrobial stress environment, causing the microorganism to pose a threat to humans and complicating treatments (Han et al., 2022). Multidrug-resistant A. baumannii, extensively drug-resistant A. baumannii, and pan-drug-resistant A. baumannii nosocomial isolates are now commonly found in hospitals (Ramirez et al., 2020). Additionally, A. baumannii is also an emerging pathogen among the elderly and immunocompromised patients in nursing homes and community hospitals (Mody et al., 2015).
The rise of resistant A. baumannii strains prompted the design and execution of epidemiological investigations of A. baumannii epidemics using various molecular typing methods, including multi-locus enzyme electrophoresis (MLEE), pulsed-field gel electrophoresis (PFGE), and subsequently confirmed by multi-locus sequence typing (MLST) (Montanaro et al., 2016). Among these methods, the MLST has become the gold standard for epidemiological (Ravaioli et al., 2022). Furthermore, a prominent advantage of MLST is the derived nomenclature, and sequence type (ST), which has been rapidly and extensively adopted by the community, thus widening the global collective knowledge of the distribution, dissemination, and biological characteristics of crucial clonal populations (Gaiarsa et al., 2019).
A large number of studies have shown that genes play a vital role in the antibiotic resistance of any bacteria (Roy et al., 2022). It reported that a variety of OXA-type β-lactamase genes were found in CRAB strains (Abouelfetouh et al., 2022). Among these genes, the OXA-23 and OXA-51 were key determinants of carbapenem resistance (Chan et al., 2022). In addition, genes associated with efflux pumps also play an important role in carbapenem antibiotic resistance. It is reported that the ΔaceI mutant strain showed lower resistance to chlorhexidine than the parental strain. This result suggests that aceI plays a key role in antibiotic resistance (Liu et al., 2018). Besides, the gene purF, which regulates the biofilm formation, also plays a fatal role in antibiotic resistance (Dai et al., 2021). In addition, the Chaperonin GroEL/GroES also plays an important role in Escherichia coli antibiotic resistance (Goltermann et al., 2015). However, the mechanism of antibiotic resistance of A. baumannii needs to be further explored.
In the present study, the MLST, antibiotic resistance genes and transcriptome of A. baumannii were analyzed. This study found that a total of 9 ST sequence types were detected, and the CC208 was the main clonal complex in this hospital and showed high antibiotic resistance to carbapenems; The blaOXA–23 gene was the main antibiotic resistance genotypes; The transcriptome result suggests that carbapenem antibiotic resistance of A. baumannii involved biofilm formation, efflux pumps, peptidoglycan biosynthesis, and chaperonin synthesis.
The 199 strains CRAB were isolated from patients in various departments of Affiliated Hospital of Guizhou Medical University, Guizhou, China from 2017 to 2019. Standard A. baumannii ATCC19606, pan-resistant A. baumannii strains are preserved in the Key Laboratory of Modern Pathogenic Biology, School of Basic Medicine, Guizhou Medical University.
The genomic DNA of A. baumannii was extracted using the kit (BIOTEKE, Beijing, China). Seven housekeeping genes were amplified by PCR. The primers used are listed in Table 1. The PCR reactions were performed using PrimeSTAR® Max DNA Polymerase (Takara, Japan) with the following amplification parameters: 95°C for 5 min; 94°C for 30 s, 50°C for 30 s, and 72°C for 40 s, 30 cycles; 72°C for 10 min.
The amplified products of MLST typing of A. baumannii were sequenced, and the sequence results were compared on the MLST comparison website of A. baumannii. The sequencing results were submitted to the website for comparison to obtain allele number and ST type. The Bio Numerics 7.1 software is used for typing data processing, and generating minimum spanning tree (MST).
The minimal inhibitory concentration (MIC) values of A. baumannii isolated from clinical isolates were measured by the micro-dilution broth method. The antibiotics determined were imipenem, meropenem, ertapenem, and biapenem. There were three repeats for each strain and antibiotic. Except for 5 gradients of biapenem, 6 gradients were done. The concentration of the mother liquid before dilution is shown in Table 2. The results were judged by the Institute of Clinical and Laboratory Standards (CLSI2012).
In this study, the blaOA–23 and blaOXA–51 were selected as molecular markers for rapid identification of A. baumannii. The primers are listed in Table 3, and the PCR reaction system is 15 μL, including the premixTaq enzyme 7.5 μL, the primers 1 μL with 10 μM, DNA template 1 μL, and the ddH2O 4.5 μL. Meanwhile, the PCRs were performed at 94°C for 5 min, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C, and a final extension of 10 min at 72°C. The 5 μL PCR products were sampled in 1% agarose gel and 30 min was electrophoretic at a constant voltage of 100V. The photos were taken by WD-9413B gel imaging analyzer.
The AB77 (weak imipenem resistance) and AB98 (strong imipenem resistance) were selected for transcriptome sequencing with three biological repeats per sample. After resuscitation of CRAB77 and CRAB98, respectively, 1 mL solution was absorbed and cultured in 100 mL LB liquid medium. After overnight incubation at 37°C 220 rpm until logarithmic growth, the organisms were collected by centrifugation, and three biological replicates were performed in each group, totaling six samples. The collected bacterial precipitates were immediately liquid nitrogen snap-frozen and stored in the refrigerator at −80°C. The total RNA of these samples was extracted using Trizol reagent (Sigma-Aldrich, United States) according to the specification described method. The RNA concentration and quality were measured using a Nanodrop 2000 micro-spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), and agarose gel electrophoresis. RNA-seq library construction and RNA sequencing were performed by the Meiji Biomedical Technology Co., Ltd. (Shanghai, China).
The raw data was filtered to remove contaminated and low-quality sequences from connectors and to obtain clean reads. Bowtie2-2.5.0 was used to map the genome of the filtered sequence (Langmead and Salzberg, 2012). The reference genome of A. baumannii (ATCC 19606) was downloaded from GenBank (NZ_CP045110.1). The gene expression levels were calculated based on TPM (transcript per million) (Zhao et al., 2021). Differential expression analysis between CRAB77 and CRAB98 was performed using the DEGSeq2 R package. The significance of the DEGs was determined with a P-value < 0.05 found by DEseq2. Transcripts with log 2 (Fold Change) > 1 and P-value < 0.05 were considered significant differential expressions. KOBAS software was used to test the statistical enrichment of differentially expressed genes in KEGG pathways. Significantly enriched KEGG pathways was identified by a P-value < 0.05.
qRT-PCR was performed to confirm the DEGs identified using RNA-seq. The primer sequences of candidate genes and a housekeeping gene (16S rRNA) were designed using Primer Premier 5 (Table 4). The first strand of cDNA was synthesized by PrimeScript TMRT reagent Kit with gDNA Eraser (TaKaRa, Beijing, China) according to the manufacturer’s instructions. The qRT-PCR was carried out with a PikoReal 96 real-time PCR system (Thermofisher Scientific) using PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Waltham, MA, USA). The sense and anti-sense primers were designed by Primer Premier 5 and they were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). Sequences of the primers used in this study were shown in detail in Table 4. The reactions were performed with three biological and technical replicates per sample. The data were analyzed using the 2–ΔCt method (Schmittgen and Livak, 2008).
Statistical analysis was performed using SPSS21.0. All experiments were performed in triplicate. Statistical differences between groups were assessed by the two-tailed Student’s t-test and one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test or by the nonparametric Mann–Whitney U test and Kruskal-Wallis test, followed by Duncan’s multiple-comparison test. A P-value < 0.05 was considered statistically significant.
The sequences of these 7 housekeeping genes in 199 A. baumannii strains were submitted to the MLST database of the Pasteur Institute1 to investigate whether there were base substitutions or deletions. The sequences obtained from housekeeping gene sequencing of each strain were submitted to the database, and the corresponding alleles of each sequence were queried, to obtain the ST model of each strain. The results showed that there were 9 different sequence types (Table 5), among which ST136, ST191, and ST208 had high frequency in the hospital.
The minimum spanning tree showed that the ST types of the 199 strains are relatively concentrated, and all of them originated from ST191 (Figure 1). In addition, there is only gpi difference between ST136/ST195/ST368/ST369/ST208/ST540 and ST191. While there are two alleles difference between ST457 and ST191, the genetic relationship is close. It is reported that the mutation rate and recombination rate of gpi in the MLST typing scheme of A. baumannii are higher than those of other housekeeping genes, and it is easy to mutate in the process of evolution. It is found that a similar situation in the study of A. baumannii typing, suggests that A. baumannii MLST scheme has a better resolution on the housekeeping gene gpi. Among the 199 strains of CRAB, there is only one ST, which is widely distributed in various departments. Among these departments, the ICU region is the most distributed. However, the ST2206 does not belong to these ST, and there are six allele differences between ST2206 and ST191, and the ST2206 is isolated in the comprehensive ICU region.
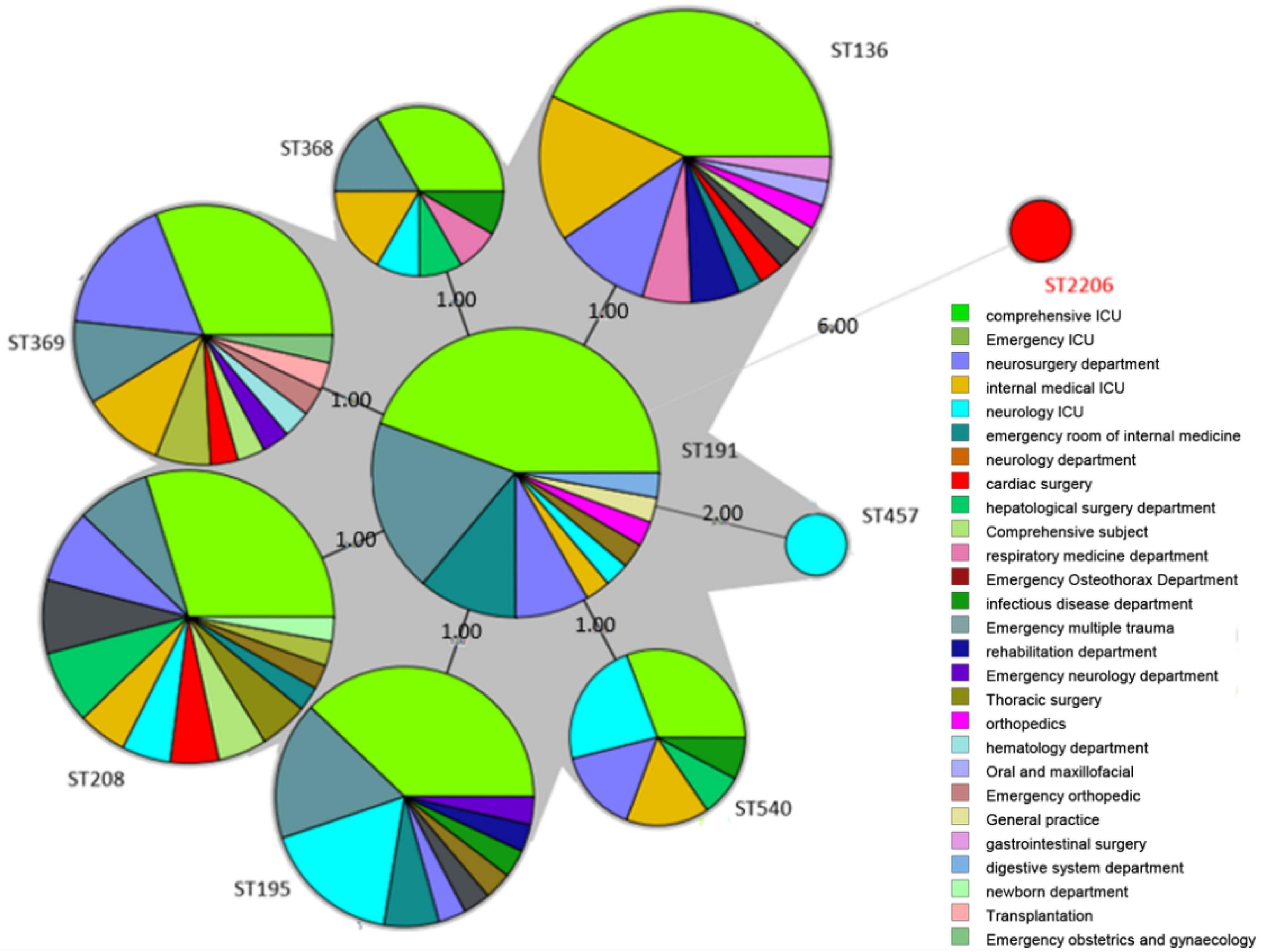
Figure 1. Minimum spanning tree (MST) of 199 A. baumannii strains. The size of the circle in MST represents the number of the same ST genotypes, and the black solid line indicates that there are 3 or fewer differences in housekeeping genes between the two ST genotypes. The thicker the black solid line, the smaller the difference in housekeeping genes between the two ST genotypes. The dotted line indicates that there are 4 or more differences in housekeeping genes between the two ST genotypes. The denser the dotted line, the greater the difference in housekeeping genes between ST genotypes.
To understand the classification position of these identified strains in the entire A. baumannii database, the ST of all strains were placed in the A. baumannii ST database. The results showed that the ST136 / ST195 / ST368 / ST369 / ST208 / ST540 / ST191 belongs to CC208, and the ST2206 distant relatives (Figure 2).
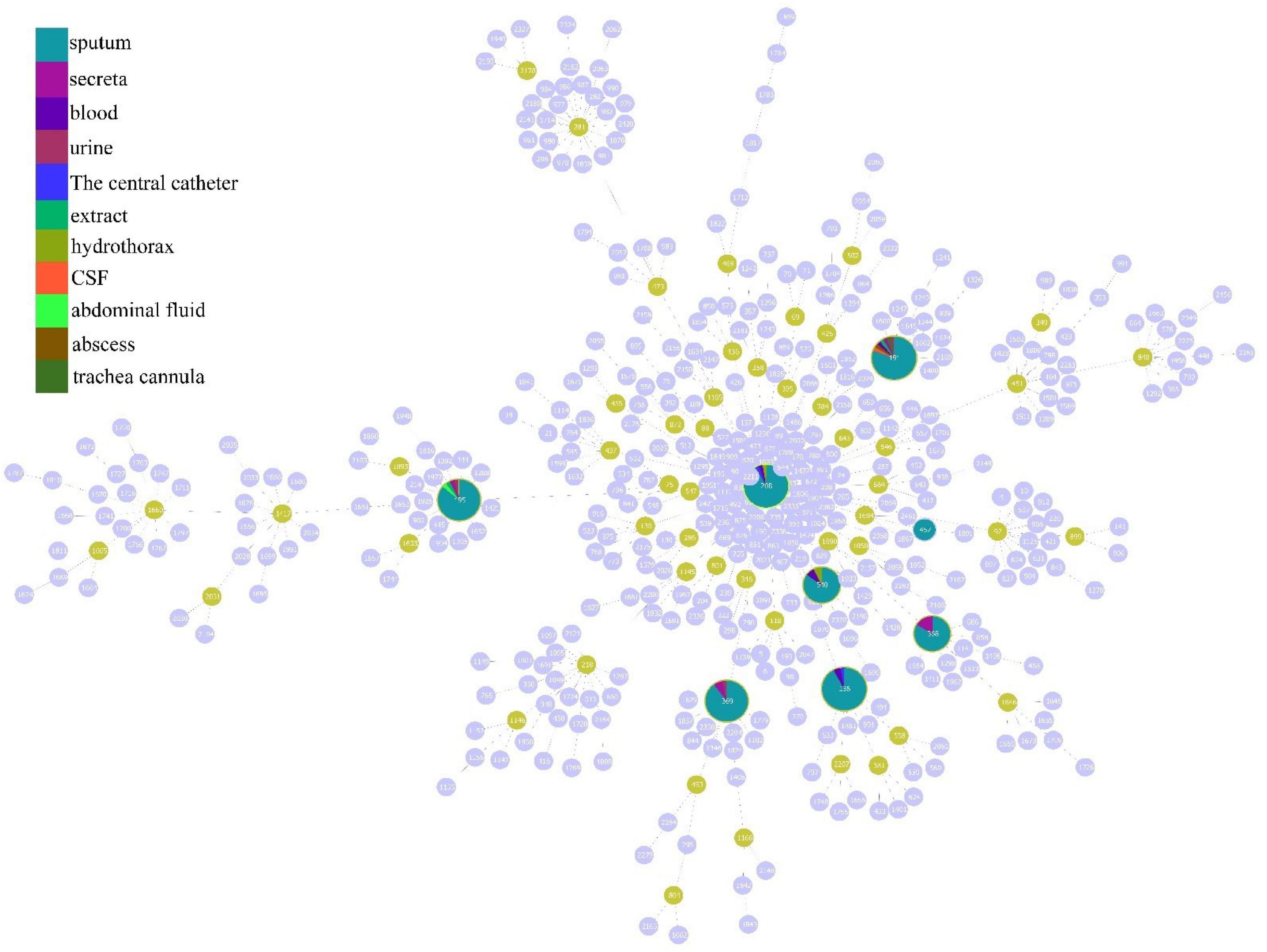
Figure 2. Minimum spanning tree analysis of CRAB isolates based on multilocus sequence typing (MLST) data. Each circle represents an independent sequence type (ST). The size of each circle corresponds to a different number of isolates, with larger sizes representing higher isolate quantity. The lines connecting the circles indicate the relationship between different STs. Different types of lines represent a difference in one allele (solid lines), two alleles (dashed lines), and three or more alleles (dotted lines).
According to the analysis of the source distribution of 199 CRAB strains, the results showed that most of these clinical strains came from sputum, and a total of 171 strains of each ST type were detected in these sputum samples, while the distribution was less in other samples (Figure 3A). Based on the analysis of sputum samples in various departments, these CRAB were found in all clinical sputum samples in many departments, while there were few types of CRAB samples in the remaining departments (Figure 3B). This result implies that the ST is widely prevalent in all departments in the hospital, and it is necessary to focus on detecting the presence of CRAB in sputum samples to timely control the spread of CRAB.
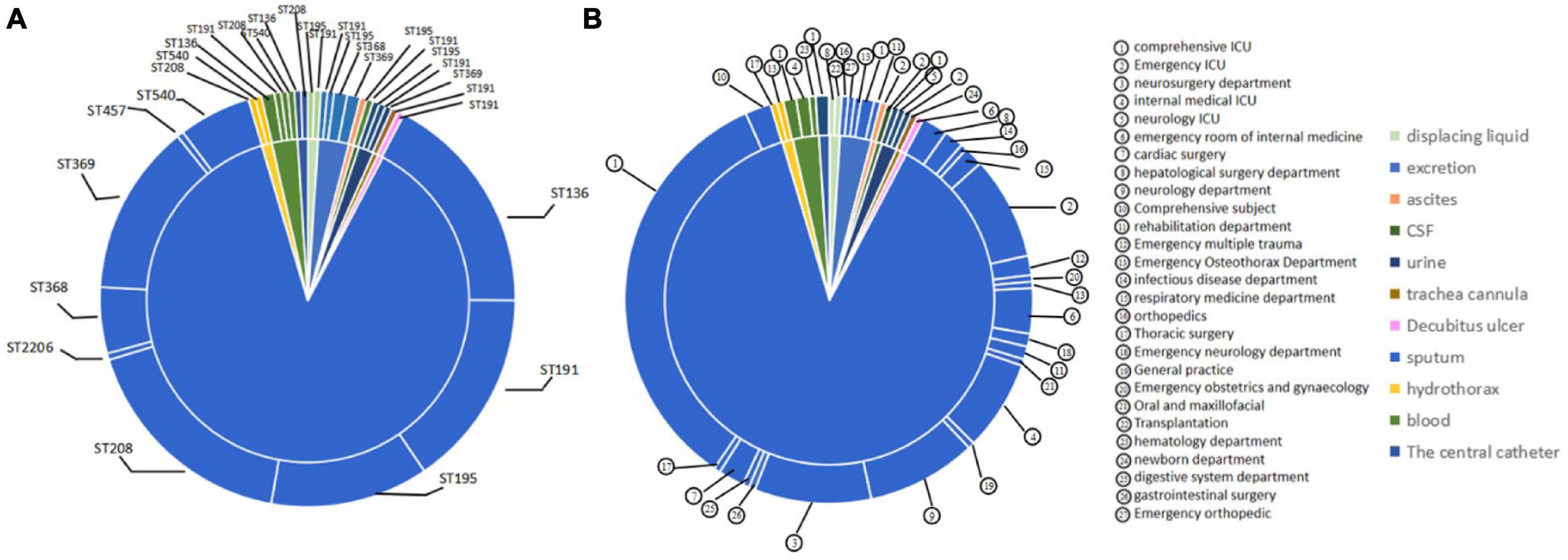
Figure 3. Molecular epidemiological characteristics of strains from different tissue and clinical departments. (A) The source of different typing strains. (B) The distribution of CRAB strains in sputum in each department.
The result of MIC values for the 199 A. baumannii strains showed that the MIC values for biapenem were concentrated at 128 μg/mL, accounting for 44.5%; the MIC value of meropenem was concentrated at 128 μg/mL, accounting for 39%; the MIC value for ertapenem was concentrated at 512 μg/mL, accounting for 39.5%; as well as the MIC values for imipenem were concentrated at 64 μg/mL, accounting for 51% (Figure 4 and Supplementary Table 1). These results indicate that all experimental strains were highly resistant to the four carbapenems antibiotics, among which the experimental strains had the strongest resistance to ertapenem.
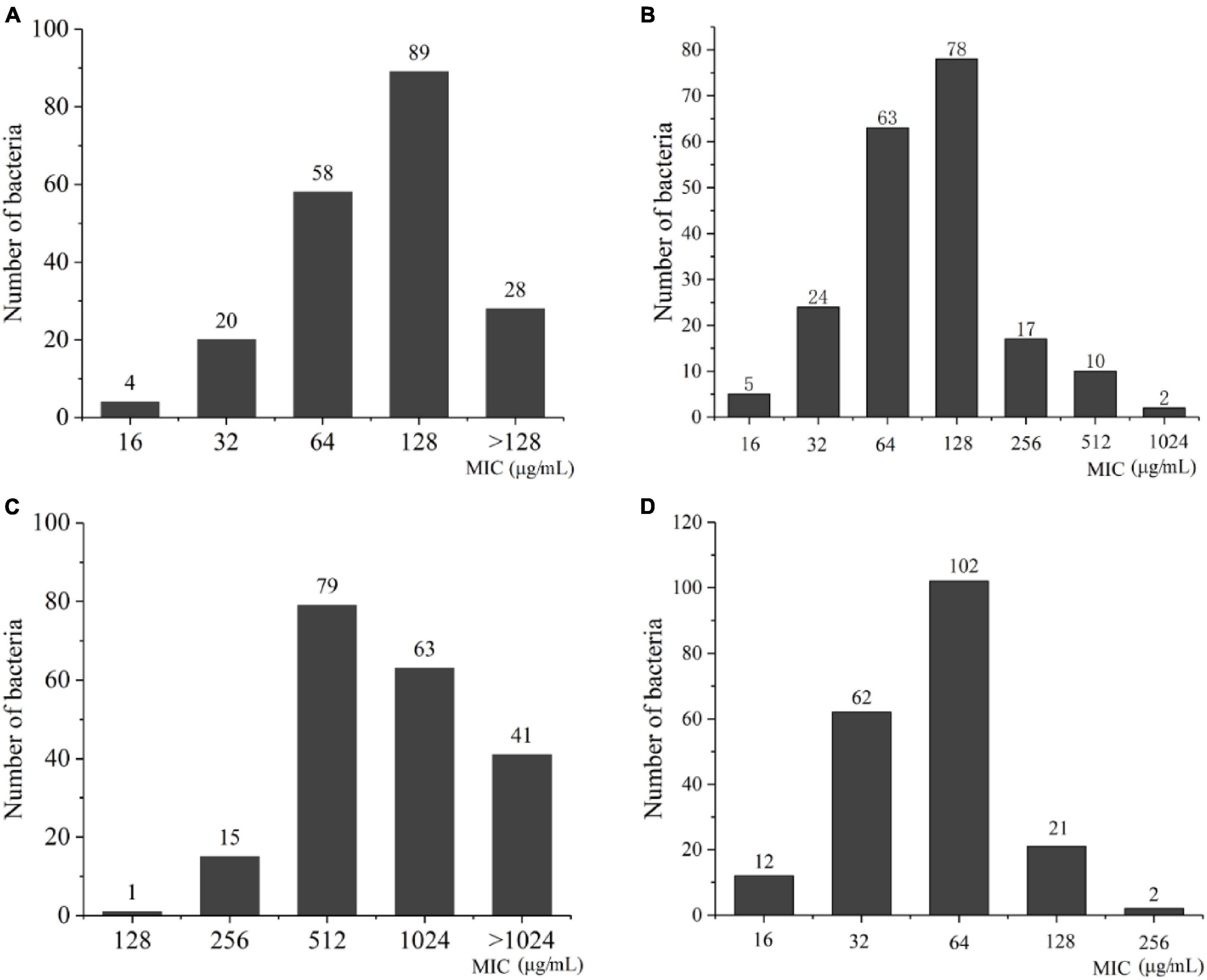
Figure 4. The distribution of MIC value in A. baumannii. (A) Biapenem; (B) meropenem; (C) ertapenem; (D) imipenem.
According to the ST type classification of 199 CRAB strains, MIC values of these 199 CRAB strains against ertapenem were calculated. The results showed that ST136, ST191, ST195, and ST208 were more resistant to ertapenem than ST368, ST369, ST457, and ST540. However, the ST2206, which is distantly related to STs strain, also had a MIC value of 1,024 μg/mL. This result indicates that ST2206 with strong antibiotic resistance (Table 6).
In addition, according to the antibiotic resistance detection of these 199 strains to biapenem, the results showed that the MIC values of ST types concentrated at 128 μg/mL. The result showed that these ST types have the same resistance to biapenem. However, the ST2206 showed lower resistance to biapenem than other ST types (Table 7).
According to the detection of the resistance for these 199 CRAB strains to meropenem, it was found that the MIC values of STs to meropenem in these 199 CRAB strains were higher, while the ST2206 showed low resistance to meropenem (Table 8).
Based on the detection of the resistance STs to imipenem in 199 CRAB strains, the results showed that MIC values were concentrated in 32 μg/mL-128 μg/mL, indicating strong resistance. Moreover, the ST2206 also showed resistance to imipenem, but the resistance was low, with a MIC value of 32 μg/mL (Table 9). According to this data, it was be defined weakly resistant strain with MIC values ≤ 32 to imipenem, while strains with MIC value > 32 was defined as strongly resistant strain. By this definition, ST136 and ST208, have quite different resistance to imipenem; In this study, weakly resistant strains accounted for 18% in ST136, and 47.22% were weakly resistant strains in ST208. This result indicates that different STs may have different resistance mechanisms to imipenem.
To understand the distribution of antibiotic resistance genes, the blaOXA–23, and blaOXA–51 were detected in 199 CRAB, standard A. baumannii (ATCC19606), pan-resistant A. baumannii (PRAB) and SJZ24 by using PCR amplification. The results showed that the target sequence of blaOXA–23 (Figure 5A) and blaOXA–51 (Figure 5B) genes were found in these strains. The BLAST analysis was conducted after sequencing these target bands, and it was found that these target bands were all target antibiotic resistance genes. Statistical data showed that the detection rate of the blaOXA–23 gene and blaOXA–51 gene in CRAB was 92.5 and 100%, respectively (Figure 5C). These results suggest that the class D carbapenemases genes are widely distributed in these 199 CRAB strains, which may be one of the reasons for its high antibiotic resistance to carbapenems antibiotics.
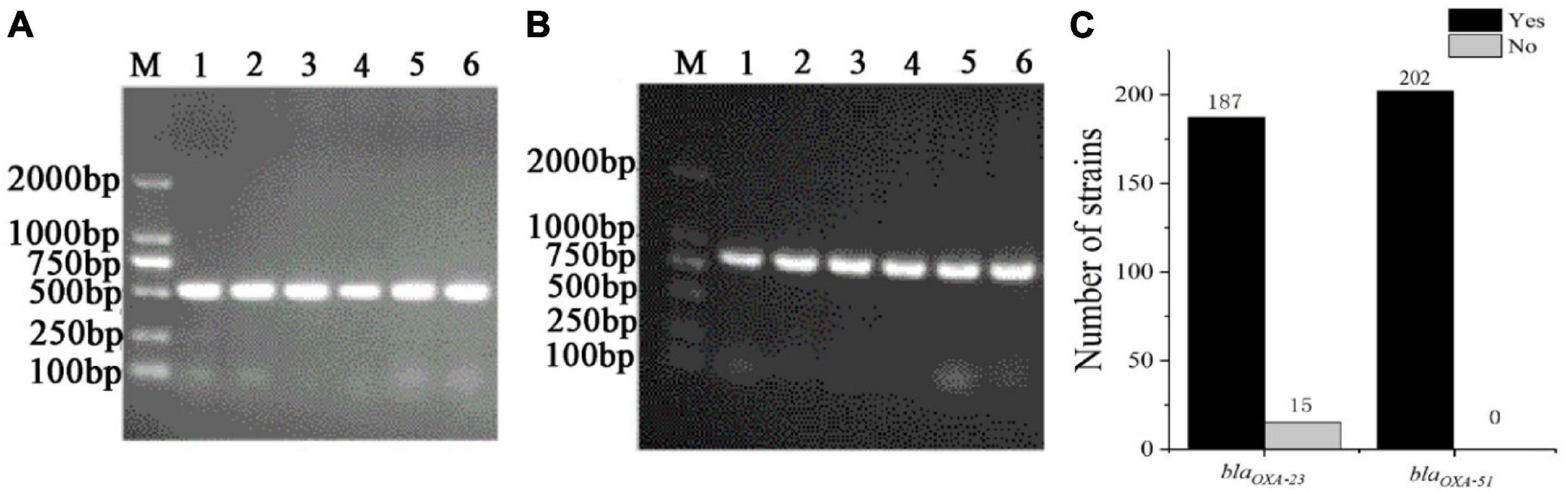
Figure 5. The PCR amplification of CRAB (part of the sample) resistance genes and the statistics of positive tests of antibiotic resistance genes. (A) blaOXA–23; (B) blaOXA–51; (C) Statistics of positive tests of antibiotic resistance genes; M, DNA maker; Yes, the number of positive strains; no, the number of negative strains.
To understand the resistance mechanisms of different STs to imipenem, the transcriptome sequencing analysis between ST208 (AB77) and ST136 (AB98) was performed in this study. The results showed that a sum of 159.60 million reads was generated and each sample provided an average production of 26.60 million. A mean of 26.60 million clean reads was obtained from each library with an adequate read ratio of 94.63% after removing adaptor sequences, N-containing reads, and low-quality. A mean of 20.44 million CDS-mapped reads was obtained from each library with an adequate read ratio of 80.11% (Supplementary Table 2). A total of 3,829 genes were mapped from all the samples. The resistance strain and sensitive strain gene expression levels were analyzed, and 390 differentially expressed genes (DEGs) were recognized (Supplementary Table 3). Among such DEGs, the resistance strain had 256 DEGs up-regulated and 134 DEGs down-regulated (Figure 6A). It is worth pointing out that some genes related to antibiotic resistance, efflux pump, and biofilm formation were up-regulated in the resistance strain (Figure 6B). The increased expression level of these genes may increase the antibiotic resistance of A. baumannii.
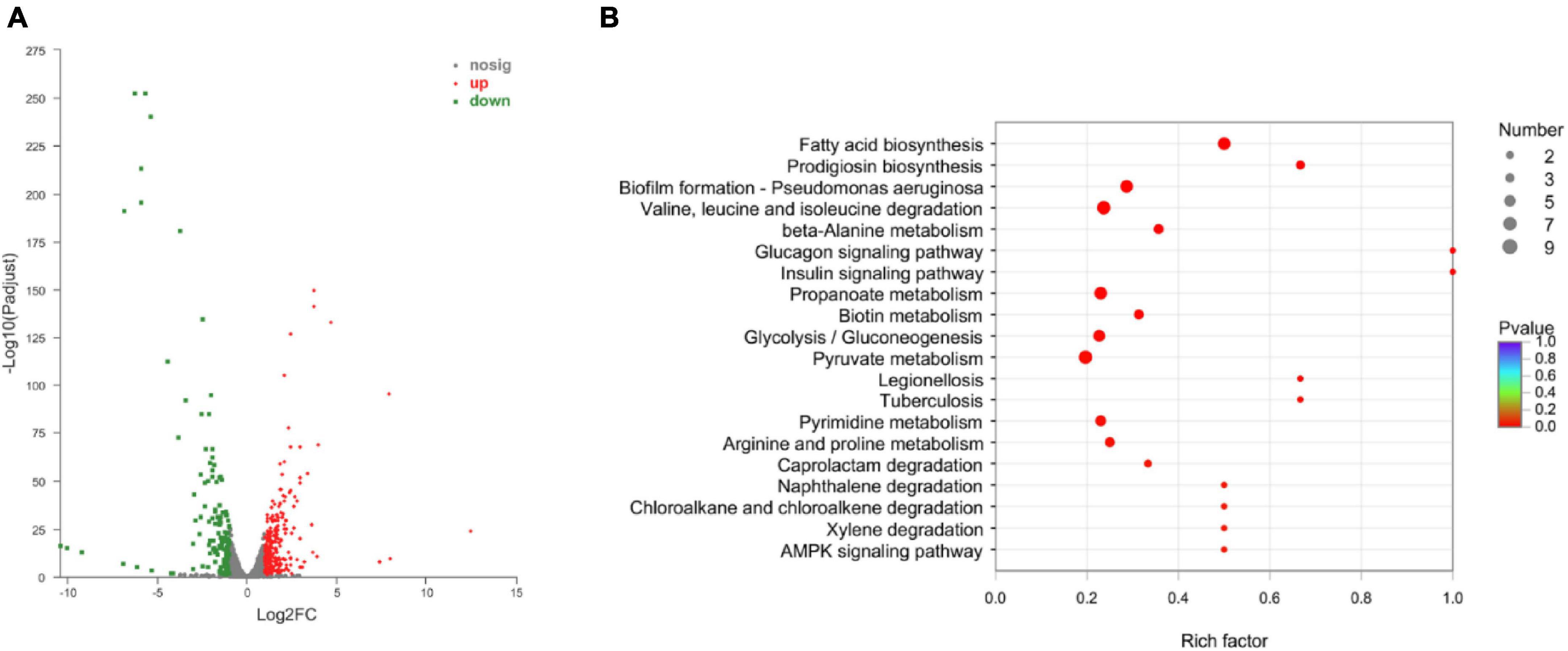
Figure 6. The Volcano diagram of differentially expressed genes between resistance strain and sensitive strain (A) and DEGs involved in KEGG pathway enrichment analysis in comparison with resistance strain and sensitive strain (B). In (A), The x-axis represents the multiple differential expressions; the y-axis represents significance. In (B), The x-axis represents the Rich factor, GeneRatio = Term Candidate Gene Number/Term Gene Number. The y-axis represents KEGG Pathway. The size of the bubble is proportional to the number of genes in the KEGG Pathway. And the color represents the P-value of enrichment. The deeper the color, the smaller the P-value.
Among these DEGs, 19 genes may be involved the carbapenem resistance, including biofilm formation, efflux pump, peptidoglycan biosynthesis, chaperonin synthesis, and drug transporter. In the biofilm formation process, there were pilus assembly protein (Pap, FQU82_RS03605), urease accessory protein (UreD, FQU82_RS05390), OmpA family protein (FQU82_RS07900), NADP-specific glutamate dehydrogenase (gdhA), phosphoenolpyruvate–protein phosphotransferase (pstP), and amido phosphoribosyl transferase (purF, FQU82_RS13115). In addition, there were 5 genes involved in efflux pump, including LysR family transcriptional regulator (FQU82_RS08650), PACE efflux transporter (FQU82_RS09170), chlorhexidine efflux PACE transporter (aceI, FQU82_RS12055), MacB family efflux pump subunit (FQU82_RS03105), MacA family efflux pump subunit (FQU82_RS03110). Meanwhile, there were 3 genes may contribute to peptidoglycan biosynthesis, including D-alanyl-D-alanine carboxypeptidase PBP5/6 (dacC, FQU82_RS14285), endocytic transglycosylase (MltG, FQU82_RS14770), and UDP-glucose 4-epimerase (GalE, FQU82_RS00890). Additionally, there were 3 chaperonins, including DnaK, GroEL, and GroES may be involved in antibiotic resistance. It is worth mentioning that the MFS transporter and ABC transporter substrate-binding protein may participate in drug transport (Table 10).
To confirm findings of gene expression obtained from transcriptome data, 6 DEGs concerned with antibiotic resistance were chosen for qRT-PCR. These DEGs are mainly involved in biofilm formation, efflux pump, peptidoglycan biosynthesis, and chaperonin synthesis. As shown in, the 6 DEGs had a very similar expression pattern based on the transcriptome data and qRT-PCR, which indicates the trustworthiness of the transcriptomic analysis (Figure 7).
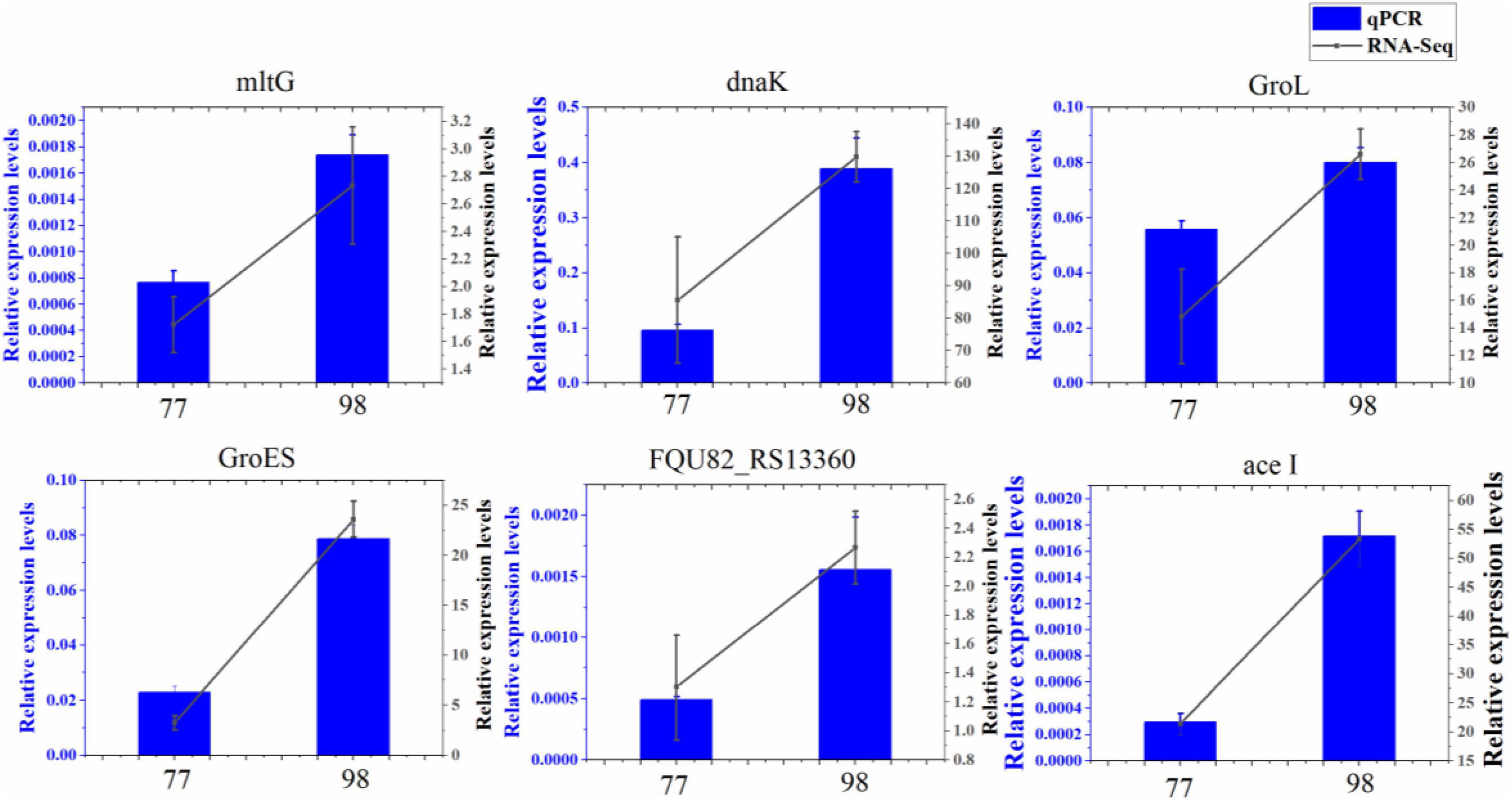
Figure 7. Relative expression levels of 6 DEGs in 77 and 98. MltG, endolytic transglycosylase MltG; dnaK, molecular chaperone DnaK; GroL, chaperonin GroEL; GroES, co-chaperone GroES; FQU82_RS13360, ABC transporter substrate-binding protein; ace I, chlorhexidine efflux PACE transporter AceI.
A. baumannii is mainly related to nosocomial infections globally and is an oxybiotic Gram-negative pathogen that results in infections of the skin, urinary tract, bloodstream, and other soft tissues (Lee et al., 2017). In recent years, A. baumannii have become a significant cause of critical healthcare-associated diseases and infections (Lima et al., 2019). Among these β-lactam antibiotics, the carbapenems are considered the most effective class with the most extensive spectrum of antimicrobial activity (O’Donnell et al., 2021). Hence, carbapenem antibiotics are also often used in the treatment of A. baumannii infection (Nguyen and Joshi, 2021). However, with the use of carbapenem antibiotics, some carbapenem-resistant bacteria, including A. baumannii, Pseudomonas aeruginosa, and Stenotrophomonas maltophilia were appeared (Nordmann and Poirel, 2019). It is reported that A. baumannii infections occur in critically ill patients in the ICU setting and account for up to 20% of infections in ICUs worldwide (Lee et al., 2017). To better understand the carbapenem resistance A. baumannii to be able to develop a good treatment plan. In this study, 199 CRAB strains were used as materials to study their typing. Among the 199 CRAB strains, two strains were used to explore the mechanism of antibiotic resistance.
According to the MLST analysis of the 199 CRAB strains, it was found that the ST136 and ST208 were the most STs of CRAB, which have strong resistance to the four kinds of carbapenem antibiotics. As in previous studies, the ST136 and ST208 belong to CC92, which is the most widely and largest disseminated complex worldwide (Alcantar-Curiel et al., 2019; Lima et al., 2019). Additionally, ST208 has become the main sequence type of A. baumannii infection outbreak in western China (Qu et al., 2016; Gao et al., 2022). Furthermore, along with the global phenomenon, CC92 has become the dominant circulating strain in children (Kang et al., 2023). In addition, the present study showed that the ST195, ST191, and ST369 were divided into the same group, while the time of emergence of these two STs was different. It is worth noting that the virulence of ST369 is higher than that of ST191 (Jiang et al., 2021). Moreover, the resistance of ST191 to carbapenem is also very serious (Tables 6–9). Therefore, it is extremely important to control the spread of ST136, ST191, ST208, ST195, ST369.
Imipenem is a carbapenem antibiotic commonly used in clinical treatment and plays an important role in preventing A. baumannii infection (Khaled et al., 2021). A large number of studies have shown that A. baumannii ST136 is highly resistant to imipenem (Firoozeh et al., 2023; You et al., 2023). In the present study, ST136 also showed strong resistance to imipenem, while ST208 showed weak resistance to imipenem (Table 9). This phenomenon may be caused by differences in gene expression at different STs. Transcriptome results showed that these two STs A. baumannii were significantly differentially expressed in biofilm formation, efflux pump, peptidoglycan biosynthesis, and chaperone biosynthesis. These results suggest that the resistance ability of ST136 to imipenem is caused by the differential gene expression of these pathways.
As we all know, bacterial resistance to antibiotics involves many factors, such as the formation of biofilm, which can increase antibiotic resistance thousands of times (Guo et al., 2022). Additionally, the efflux pump, the chaperonins, and peptidoglycan biosynthesis also play important roles in antibiotic resistance (Grishin et al., 2020; Stevens et al., 2020; Barnabas et al., 2022). During the biofilm formation, the pilus assembly protein plays an important role in the formation of fimbriae (Sanchez et al., 2017; Shanmugasundarasamy et al., 2022). In this study, the pilus assembly protein was up-regulated in the antibiotic resistance strains (Table 6). In addition, the ureD (Maisuria et al., 2015), ptsP (Abisado et al., 2021), and purF (Li et al., 2021) also contribute to biofilm formation, which may play an important in the process of antibiotic resistance. In the meanwhile, a ureD, and ptsP were also up-regulated in this study (Table 10). These results suggest that biofilm formation plays an important role in carbapenem antibiotic resistance.
To date, different categories of efflux pumps have been identified in A. baumannii, including RND-family (resistance-nodulation-division), MFS (major facilitator superfamily), MATE (multidrug and toxic compound extrusion), SMR (small multi antibiotic resistance), PACE family and ABC (ATP-binding cassette) superfamilies are the main components of the efflux pump (Blair et al., 2014; Li et al., 2015; Hassan et al., 2018). These efflux pumps play a key role in antibiotic resistance (Roy et al., 2022). In this study, the MFS transporter, and PACE efflux transporter were up-regulated (Table 10), which may improve the ability of carbapenem antibiotic resistance. Most importantly, LysR, a transcriptional regulator, also indirectly regulates the efflux pump (Fu et al., 2020; Lu et al., 2021), which is also significantly up-regulated in this study.
Besides, the peptidoglycan also contributes to antibiotic resistance (Kvesić et al., 2022). It is reported that the deposition of Pseudomonas aeruginosa with the interference of peptidoglycan circulation in the lungs of mice was significantly reduced, and the mortality caused by systemic infection was significantly reduced (Torrens et al., 2019). Moreover, the peptidoglycan synthesis related gene dacC was more frequently found in populations with lower relative fitness across the measured growth parameters (Barbosa et al., 2017). In the meanwhile, the dacC in the antibiotic resistance strain was upregulated (Table 10), indicating that dacC may also be involved in antibiotic resistance.
As in previous studies, chaperone proteins also play an important role in antibiotic resistance (Chiang et al., 2022). For instance, in Escherichia coli, the Chaperonin GroEL/GroES over-expression can promote aminoglycoside resistance and reduce drug susceptibilities (Goltermann et al., 2015). Moreover, the GroEL can increase the antibiotic resistance of Klebsiella pneumoniae by promoting the refolding of misfolded proteins (Ye et al., 2021). It is worth mentioning that the chaperone GroEL/GroES of A. baumannii has also been reported to play an important role in antimicrobial resistance (Abirami et al., 2023). In addition, the DnaK system supports rifampicin resistance (Fay et al., 2021). In this study, the chaperone proteins GroEL, GroES and DnaK are up-regulated in the antibiotic-resistance strain. These results suggest that chaperones may also play a crucial role in the process of carbapenem antibiotic resistance (Figure 8).
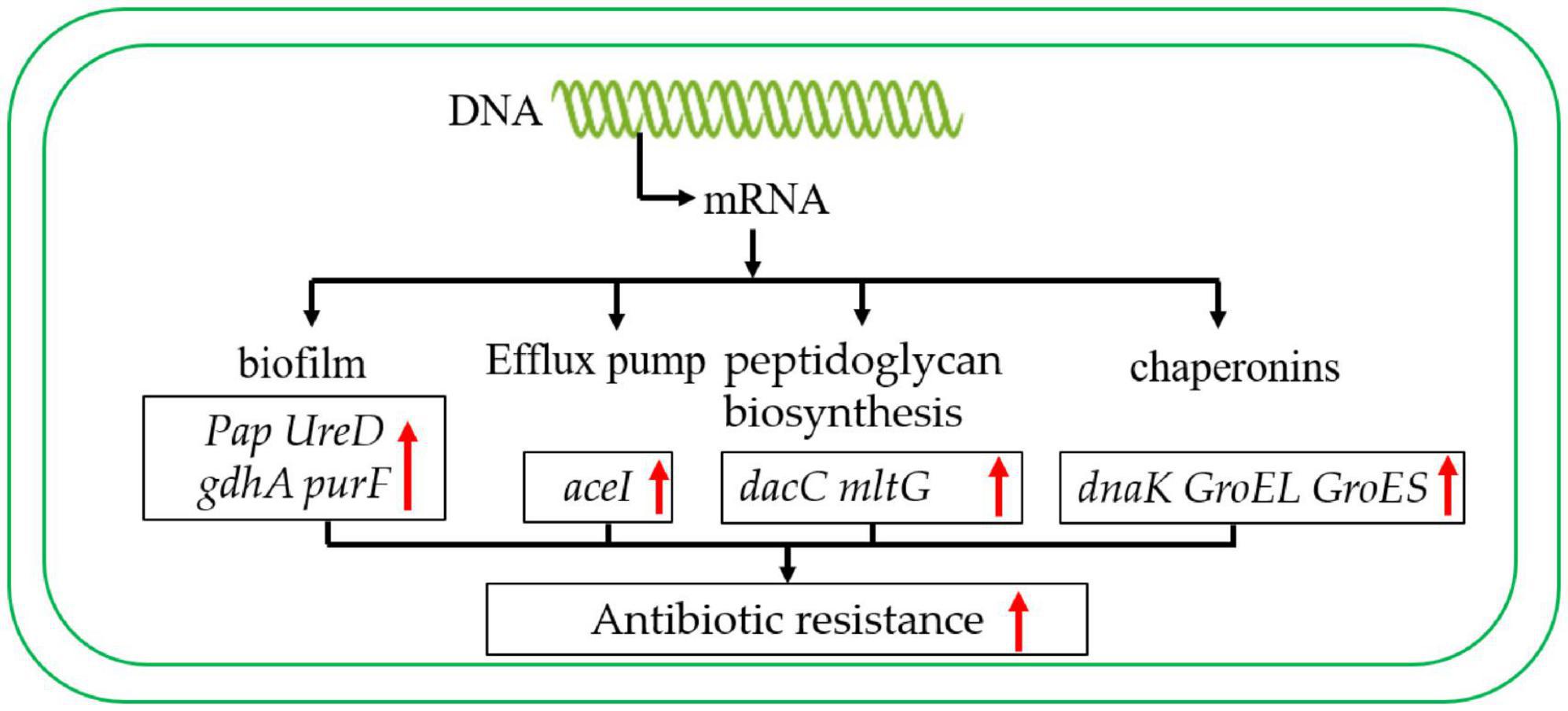
Figure 8. Carbapenem-resistant A. baumannii antibiotic resistance regulation model. The red arrow indicates that the expression level was up-regulated.
In summary, these results showed that the main STs of CRAB were ST136, ST191, ST208, ST195, ST369, and it was mainly distributed in ICU. The analysis of the antibiotic resistance mechanism suggested that the antibiotic resistance mechanism of A. baumannii was mainly due to the expression of biofilm formation and efflux pump-related genes. In addition, to peptidoglycan biosynthesis, chaperonin synthesis also plays an important role. This study provides a theoretical basis for better control of CRAB.
In this study, there were 9 ST sequence types were detected, and the CC208 was the main clonal complex in this hospital and showed high carbapenems resistance; as well as the blaOXA–23 gene was the main carbapenems resistance genotypes; the results of transcriptome data showed that the mechanism of enhanced carbapenems resistance of A. baumannii may be due to the enhanced biofilm-forming ability, efflux pump expression, peptidoglycan biosynthesis, and molecular chaperone protein expression. Therefore, we need to strengthen the research and development of new antibiotics to provide an effective way to control carbapenem-resistant A. baumannii. This study lays the foundation for the detection and control of carbapenem A. baumannii. In summary, the epidemic trend of clinical A. baumannii in Guiyang, China was analyzed from the molecular level, and the antibiotic resistance mechanism of A. baumannii to carbapenem antibiotics was analyzed with transcriptome, which provided a theoretical basis for better control of A. baumannii.
The original contributions presented in this study are included in this article/Supplementary material, further inquiries can be directed to the corresponding authors.
ZQ: Funding acquisition, Methodology, Resources, Writing – original draft. KY: Methodology, Software, Writing – original draft. HC: Methodology, Software, Writing – original draft. SC: Methodology, Software, Writing – review & editing. FC: Writing – review & editing. FM: Funding acquisition, Writing – review & editing: GG: Funding acquisition, Writing – review & editing. JP: Funding acquisition, Resources, Writing – review & editing.
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (82002180, 81660347), Characteristic Key Laboratory of Guizhou Province [2021] 313, Guizhou Provincial Natural Science Foundation (ZK [2022] Key Program 039), the Science and Technology Fund Project of Guizhou Provincial Health Commission (gzwkj2023-567), and the High-Level Talent Initiation Project of Guizhou Medical University (J[2022]023).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1394775/full#supplementary-material
Abirami, G., Alexpandi, R., Sudhin, S., Durgadevi, R., Roshni, P. S., Kumar, P., et al. (2023). Pyrogallol downregulates the expression of virulence-associated proteins in Acinetobacter baumannii and showing anti-infection activity by improving non-specific immune response in zebrafish model. Int. J. Biol. Macromol. 226, 853–869. doi: 10.1016/j.ijbiomac.2022.12.045
Abisado, R. G., Kimbrough, J. H., McKee, B. M., Craddock, V. D., Smalley, N. E., Dandekar, A. A., et al. (2021). Tobramycin adaptation enhances policing of social cheaters in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 87:e0002921. doi: 10.1128/AEM.00029-21
Abouelfetouh, A., Mattock, J., Turner, D., Li, E., and Evans, B. A. (2022). Diversity of carbapenem-resistant Acinetobacter baumannii and bacteriophage-mediated spread of the Oxa23 carbapenemase. Microb. Genom. 8:752. doi: 10.1099/mgen.0.000752
Alcantar-Curiel, M. D., Rosales-Reyes, R., Jarillo-Quijada, M. D., et al. (2019). Carbapenem-resistant Acinetobacter baumannii in three tertiary care hospitals in Mexico: Virulence profiles, innate immune response and clonal dissemination. Front. Microbiol. 10:2116. doi: 10.3389/fmicb.2019.02116
Barbosa, C., Trebosc, V., Kemmer, C., Gayosso-Vázquez, C., Fernández-Vázquez, J. L., Toledano-Tableros, J. E., et al. (2017). Alternative evolutionary paths to bacterial antibiotic resistance cause distinct collateral effects. Mol. Biol. Evol. 34, 2229–2244. doi: 10.1093/molbev/msx158
Barnabas, V., Kashyap, A., Raja, R., Newar, K., Rai, D., Dixit, N. M., et al. (2022). the extent of antimicrobial resistance due to efflux pump regulation. ACS Infect. Dis. 8, 2374–2388. doi: 10.1021/acsinfecdis.2c00460
Blair, J. M., Richmond, G. E., and Piddock, L. J. (2014). Multidrug efflux pumps in gram-negative bacteria and their role in antibiotic resistance. Future Microbiol. 9, 1165–1177.
Chan, K. W., Liu, C. Y., Wong, H. Y., Chan, W. C., Wong, K. Y., and Chen, S. (2022). Specific amino acid substitutions in OXA-51-type β-lactamase enhance catalytic activity to a level comparable to carbapenemase OXA-23 and OXA-24/40. Int. J. Mol. Sci. 23:4496.
Chiang, M.-H., Chang, F.-J., Kesavan, D. K., Vasudevan, A., Xu, H., Lan, K. L., et al. (2022). Proteomic network of antibiotic-induced outer membrane vesicles released by extensively drug-resistant Elizabethkingia anophelis. Microbiol. Spectr. 10:e0026222. doi: 10.1128/spectrum.00262-22
Chusri, S., Na-Phatthalung, P., Siriyong, T., Paosen, S., and Voravuthikunchai, S. P. (2014). Holarrhena antidysenterica as a resistance modifying agent against Acinetobacter baumannii: Its effects on bacterial outer membrane permeability and efflux pumps. Microbiol. Res. 169, 417–424. doi: 10.1016/j.micres.2013.09.004
Colquhoun, J. M., and Rather, P. N. (2020). insights into mechanisms of biofilm formation in Acinetobacter baumannii and Implications for uropathogenesis. Front. Cell Infect. Microbiol. 10:253. doi: 10.3389/fcimb.2020.00253
Dai, W., Zhang, Y., Zhang, J., Xue, C., Yan, J., Li, X., et al. (2021). Analysis of antibiotic-induced drug resistance of Salmonella enteritidis and its biofilm formation mechanism. Bioengineered 12, 10254–10263. doi: 10.1080/21655979.2021.1988251
Fay, A., Philip, J., Saha, P., Hendrickson, R. C., Glickman, M. S., and Burns-Huang, K. (2021). The DnaK chaperone system buffers the fitness cost of antibiotic resistance mutations in mycobacteria. mBio 12:21. doi: 10.1128/mBio.00123-21
Firoozeh, F., Nikibakhsh, M., Badmasti, F., Zibaei, M., and Nikbin, V. S. (2023). Clonal relatedness of carbapenem-resistant Acinetobacter baumannii: High prevalence of ST136pas in a burn center. Ann. Clin. Microbiol. Antimicrob. 22:9. doi: 10.1186/s12941-023-00589-9
Fu, Y., Zhang, L., Wang, G., Lin, Y., Ramanathan, S., Yang, G., et al. (2020). The LysR-type transcriptional regulator YeeY plays important roles in the regulatory of furazolidone resistance in Aeromonas hydrophila. Front. Microbiol. 11:577376. doi: 10.3389/fmicb.2020.577376
Gaiarsa, S., Batisti Biffignandi, G., Esposito, E. P., Castelli, M., Jolley, K. A., Brisse, S., et al. (2019). Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) schemes. Front. Microbiol. 10:930. doi: 10.3389/fmicb.2019.00930
Gao, Y., Li, H., Chen, H., Zhang, J., Wang, R., Wang, Z., et al. (2022). Origin, phylogeny, and transmission of the epidemic clone ST208 of carbapenem-resistant Acinetobacter baumannii on a global scale. Microbiol. Spectr. 10:e0260421. doi: 10.1128/spectrum.02604-21
Gedefie, A., Demsis, W., Ashagrie, M., Kassa, Y., Tesfaye, M., Tilahun, M., et al. (2021). Acinetobacter baumannii biofilm formation and its role in disease pathogenesis: A review. Infect. Drug Resist. 14, 3711–3719. doi: 10.2147/IDR.S332051
Goltermann, L., Sarusie, M. V., and Bentin, T. (2015). Chaperonin GroEL/GroES over-expression promotes aminoglycoside resistance and reduces drug susceptibilities in Escherichia coli Following exposure to Sublethal aminoglycoside doses. Front. Microbiol. 6:1572. doi: 10.3389/fmicb.2015.01572
Grishin, A. V., Karyagina, A. S., Vasina, D. V., Vasina, I. V., Gushchin, V. A., and Lunin, V. G. (2020). Resistance to peptidoglycan-degrading enzymes. Crit. Rev. Microbiol. 46, 703–726. doi: 10.1080/1040841x.2020.1825333
Guo, H., Tong, Y., Cheng, J., Abbas, Z., Li, Z., Wang, J., et al. (2022). Biofilm and small colony variants–an update on Staphylococcus aureus strategies toward drug resistance. Int. J. Mol. Sci. 23:1241. doi: 10.3390/ijms23031241
Han, L., Gao, Y., Liu, Y., Yao, S., Zhong, S., Zhang, S., et al. (2022). An outer membrane protein YiaD contributes to adaptive resistance of Meropenem in Acinetobacter baumannii. Microbiol. Spectr. 10:e0017322. doi: 10.1128/spectrum.00173-22
Hassan, K. A., Liu, Q., Elbourne, L. D. H., Ahmad, I., Sharples, D., Naidu, V., et al. (2018). Pacing across the membrane: The novel PACE family of efflux pumps is widespread in Gram-negative pathogens. Res. Microbiol. 169, 450–454. doi: 10.1016/j.resmic.2018.01.001
Inchai, J., Pothirat, C., Bumroongkit, C., Limsukon, A., Khositsakulchai, W., and Liwsrisakun, C. (2015). Prognostic factors associated with mortality of drug-resistant Acinetobacter baumannii ventilator-associated pneumonia. J. Intensive Care 3:9. doi: 10.1186/s40560-015-0077-4
Jiang, M., Chen, X., Liu, S., Zhang, Z., Li, N., Dong, C., et al. (2021). Epidemiological analysis of multidrug-resistant Acinetobacter baumannii isolates in a tertiary hospital over a 12-year period in China. Front. Public Health 9:707435. doi: 10.3389/fpubh.2021.707435
Kang, H. M., Yun, K. W., and Choi, E. H. (2023). Molecular epidemiology of Acinetobacter baumannii complex causing invasive infections in Korean children during 2001-2020. Ann. Clin. Microbiol. Antimicrob. 22:32. doi: 10.1186/s12941-023-00581-3
Khaled, J. M., Alharbi, N. S., Siddiqi, M. Z., Alobaidi, A. S., Nauman, K., Alahmedi, S., et al. (2021). A synergic action of colistin, imipenem, and silver nanoparticles against pandrug-resistant Acinetobacter baumannii isolated from patients. J. Infect. Public Health 14, 1679–1685. doi: 10.1016/j.jiph.2021.09.015
Kvesić, M., Kalinić, H., Dželalija, M., Šamanić, I., Andričević, R., and Maravić, A. (2022). Microbiome and antibiotic resistance profiling in submarine effluent-receiving coastal waters in Croatia. Environ. Pollut. 292:118282. doi: 10.1016/j.envpol.2021.118282
Langmead, B., and Salzberg, S. L. (2012) Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359.
Lee, C. R., Lee, J. H., Park, M., Park, K. S., Bae, I. K., Kim, Y. B., et al. (2017). Biology of Acinetobacter baumannii: Pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front. Cell Infect. Microbiol. 7:55. doi: 10.3389/fcimb.2017.00055
Li, L., Li, Y., Zhu, F., Li, Y., Zhu, F., Cheung, A. L., et al. (2021). new mechanistic insights into purine biosynthesis with second messenger c-di-AMP in relation to biofilm-related persistent methicillin-resistant Staphylococcus aureus infections. mBio 12:e0208121. doi: 10.1128/mBio.02081-21
Li, X. Z., Plesiat, P., and Nikaido, H. (2015). The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 28, 337–418. doi: 10.1128/CMR.00117-14
Lima, W. G., Silva Alves, G. C., Sanches, C., Antunes Fernandes, S. O., and de Paiva, M. C. (2019). Carbapenem-resistant Acinetobacter baumannii in patients with burn injury: A systematic review and meta-analysis. Burns 45, 1495–1508. doi: 10.1016/j.burns.2019.07.006
Liu, M., Hernandez-Morales, A., Clark, J., Le, T., Biswas, B., Bishop-Lilly, K. A., et al. (2022). Comparative genomics of Acinetobacter baumannii and therapeutic bacteriophages from a patient undergoing phage therapy. Nat. Commun. 13:3776. doi: 10.1038/s41467-022-31455-5
Liu, Q., Hassan, K. A., Ashwood, H. E., Gamage, H. K. A. H., Li, L., Mabbutt, B. C., et al. (2018). Regulation of the aceI multidrug efflux pump gene in Acinetobacter baumannii. J. Antimicrob. Chemother. 73, 1492–1500. doi: 10.1093/jac/dky034
Lu, H. F., Tsai, Y. C., Li, L. H., Lin, Y. T., and Yang, T. C. (2021). Role of AzoR, a LysR-type transcriptional regulator, in SmeVWX pump-mediated antibiotic resistance in Stenotrophomonas maltophilia. J. Antimicrob. Chemother. 76, 2285–2293. doi: 10.1093/jac/dkab203
Maisuria, V. B., Hosseinidoust, Z., and Tufenkji, N. (2015). Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Appl. Environ. Microbiol. 81, 3782–3792. doi: 10.1128/AEM.00239-15
Mody, L., Gibson, K. E., Horcher, A., Prenovost, K., McNamara, S. E., Foxman, B., et al. (2015). Prevalence of and risk factors for multidrug-resistant Acinetobacter baumannii colonization among high-risk nursing home residents. Infect. Control Hosp. Epidemiol. 36, 1155–1162. doi: 10.1017/ice.2015.143
Montanaro, L., Ravaioli, S., and Ruppitsch, W. (2016). Molecular characterization of a prevalent ribocluster of methicillin-sensitive Staphylococcus aureus from orthopedic implant infections. correspondence with MLST CC30. Front. Cell Infect. Microbiol. 6:8. doi: 10.3389/fcimb.2016.00008
Mulani, M. S., Kamble, E. E., Kumkar, S. N., Tawre, M. S., and Pardesi, K. R. (2019). Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front. Microbiol. 10:539. doi: 10.3389/fmicb.2019.00539
Nguyen, M., and Joshi, S. G. (2021). Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: A scientific review. J. Appl. Microbiol. 131, 2715–2738. doi: 10.1111/jam.15130
Nordmann, P., and Poirel, L. (2019). Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 69, S521–S528. doi: 10.1093/cid/ciz824
O’Donnell, J. N., Putra, V., and Lodise, T. P. (2021). Treatment of patients with serious infections due to carbapenem-resistant Acinetobacter baumannii: How viable are the current options? Pharmacotherapy 41, 762–780. doi: 10.1002/phar.2607
Qu, J., Du, Y., Yu, R., and Lu, X. (2016). The first outbreak caused by Acinetobacter baumannii ST208 and ST195 in China. Biomed. Res. Int. 2016:9254907. doi: 10.1155/2016/9254907
Ramirez, M. S., Bonomo, R. A., and Tolmasky, M. E. (2020). Carbapenemases: Transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 10:720. doi: 10.3390/biom10050720
Ravaioli, S., Campoccia, D., Ruppitsch, W., Allerberger, F., Poggi, A., Chisari, E., et al. (2022). Comparison of automated ribotyping, spa typing, and MLST in 108 clinical isolates of Staphylococcus aureus from orthopedic infections. Int. J. Mol. Sci. 23:1660. doi: 10.3390/ijms23031660
Roy, S., Chatterjee, S., Bhattacharjee, A., Chattopadhyay, P., Saha, B., Dutta, S., et al. (2021). Overexpression of efflux pumps, mutations in the pumps’ regulators, chromosomal mutations, and AAC(6’)-Ib-cr are associated with fluoroquinolone resistance in diverse sequence types of neonatal septicaemic Acinetobacter baumannii: A 7-year single center study. Front. Microbiol. 12:602724. doi: 10.3389/fmicb.2021.602724
Roy, S., Chowdhury, G., Mukhopadhyay, A. K., Dutta, S., and Basu, S. (2022). Convergence of biofilm formation and antibiotic resistance in Acinetobacter baumannii infection. Front. Med. 9:793615. doi: 10.3389/fmed.2022.793615
Sanchez, B. C., Chang, C., Wu, C., Tran, B., and Ton-That, H. (2017). Electron transport chain is biochemically linked to pilus assembly required for polymicrobial interactions and biofilm formation in the gram-positive Actinobacterium Actinomyces oris. mBio 8, e399–e317. doi: 10.1128/mBio.00399-17
Schmittgen, T. D., and Livak, K. J. (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 73, 1101–1108. doi: 10.1038/nprot.2008.73
Shanmugasundarasamy, T., Karaiyagowder Govindarajan, D., and Kandaswamy, K. (2022). A review on pilus assembly mechanisms in gram-positive and Gram-negative bacteria. Cell Surf. 8:100077. doi: 10.1016/j.tcsw.2022.100077
Stevens, M., Howe, C., Ray, A. M., Washburn, A., Chitre, S., Sivinski, J., et al. (2020). Analogs of nitrofuran antibiotics are potent GroEL/ES inhibitor pro-drugs. Bioorg. Med. Chem. 28:115710. doi: 10.1016/j.bmc.2020.115710
Tiwari, V., Rajeswari, M. R., and Tiwari, M. (2019). Proteomic analysis of iron-regulated membrane proteins identify FhuE receptor as a target to inhibit siderophore-mediated iron acquisition in Acinetobacter baumannii. Int. J. Biol. Macromol. 125, 1156–1167. doi: 10.1016/j.ijbiomac.2018.12.173
Torrens, G., Sanchez-Diener, I., Jordana-Lluch, E., Barceló, I. M., Zamorano, L., Juan, C., et al. (2019). In vivo validation of peptidoglycan recycling as a target to disable AmpC-mediated resistance and reduce virulence enhancing the cell-wall-targeting immunity. J. Infect. Dis. 220, 1729–1737. doi: 10.1093/infdis/jiz377
Wang, Y., Yang, J., Sun, X., Li, M., Zhang, P., Zhu, Z., et al. (2022). CRISPR-Cas in Acinetobacter baumannii contributes to antibiotic susceptibility by targeting endogenous AbaI. Microbiol. Spectr. 10, e0082922. doi: 10.1128/spectrum.00829-22
Yan, J., and Bassler, B. L. (2019). Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe 26, 15–21. doi: 10.1016/j.chom.2019.06.002
Ye, C., Li, W., Yang, Y., Liu, Q., Li, S., Zheng, P., et al. (2021). Inappropriate use of antibiotics exacerbates inflammation through OMV-induced pyroptosis in MDR Klebsiella pneumoniae infection. Cell Rep. 36:109750. doi: 10.1016/j.celrep.2021.109750
You, Q., Du, X., Hu, N., Zhang, Y., Zhang, N., Wang, F., et al. (2023). Local characteristics of molecular epidemiolgy of Acinetobacter baumannii in Jilin province (northeast China). BMC Microbiol. 23:19. doi: 10.1186/s12866-023-02761-9
Keywords: carbapenem resistance A. baumannii, MLST typing, antibiotic resistance, antibiotic resistance mechanism, transcriptome
Citation: Qiu Z, Yuan K, Cao H, Chen S, Chen F, Mo F, Guo G and Peng J (2024) Cross-talk of MLST and transcriptome unveiling antibiotic resistance mechanism of carbapenem resistance Acinetobacter baumannii clinical strains isolated in Guiyang, China. Front. Microbiol. 15:1394775. doi: 10.3389/fmicb.2024.1394775
Received: 02 March 2024; Accepted: 23 May 2024;
Published: 14 June 2024.
Edited by:
Vijay Soni, NewYork-Presbyterian, United StatesReviewed by:
Chuan Wang, University of South China, ChinaCopyright © 2024 Qiu, Yuan, Cao, Chen, Chen, Mo, Guo and Peng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jian Peng, amlhbnBlbmdAZ21jLmVkdS5jbg==; Guo Guo, Z3VvZ3VvQGdtYy5lZHUuY24=; Fei Mo, bW9mZWkwMzEyQGdtYy5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.