
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 13 May 2024
Sec. Antimicrobials, Resistance and Chemotherapy
Volume 15 - 2024 | https://doi.org/10.3389/fmicb.2024.1393266
This article is part of the Research TopicMetagenomic Approach for Exploration of Antimicrobial Resistance in Uncultivated MicrobiotaView all 4 articles
Antimicrobial resistance (AMR) poses a significant global health threat as the silent pandemic. Because of the use of antimicrobials in aquaculture systems, fish farms may be potential reservoirs for the dissemination of antimicrobial resistance genes (ARGs). Treatments with disinfectants have been promoted to reduce the use of antibiotics; however, the effect of these types of treatments on AMR or ARGs is not well known. This study aimed to evaluate the effects of low dose ozone treatments (0.15 mg/L) on ARG dynamics in pond water using metagenomic shotgun sequencing analysis. The results suggested that ozone disinfection can increase the relative abundance of acquired ARGs and intrinsic efflux mediated ARGs found in the resistance nodulation cell division (RND) family. Notably, a co-occurrence of efflux and non-efflux ARGs within the same bacterial genera was also observed, with most of these genera dominating the bacterial population following ozone treatments. These findings suggest that ozone treatments may selectively favor the survival of bacterial genera harboring efflux ARGs, which may also have non-efflux ARGs. This study underscores the importance of considering the potential impacts of disinfection practices on AMR gene dissemination particularly in aquaculture settings where disinfectants are frequently used at low levels. Future endeavors should prioritize the evaluation of these strategies, as they may be associated with an increased risk of AMR in aquatic environments.
The persistence and spread of antimicrobial resistance (AMR) in the environment, is considered a global silent pandemic and one of the leading health issues facing the world (Mendelson et al., 2022). It is estimated that AMR leads to approximately 700,000 deaths annually and the number is projected to rise to 10 million deaths per year in 2050 (Joshua et al., 2022). Additionally, by 2050, AMR could cost $100 trillion in lost economic output (Burki, 2018). Given the use of antibiotics in aquaculture, fish farms have been suggested as potential sources and sinks for antimicrobial resistance (Watts et al., 2017). Addressing AMR associated with aquaculture practices requires a comprehensive approach, including setting standards for antibiotic use, promoting responsible and prudent use of antibiotics, and adopting alternative disease management strategies and biosecurity to prevent disease outbreaks and reduce reliance on these drugs (Bhat and Altinok, 2023).
One of the strategies commonly used in fish farming to decrease the use of antibiotics is treatment with low dose disinfectants (Rico and Van den Brink, 2014). These disinfectants can help decrease bacterial pathogens on the fish or in the water column (Hushangi and Hosseini Shekarabi, 2018), thereby lowering the risk of systemic infections and minimizing antibiotic dependence. Topical disinfectants can be administered in aquaculture systems continuously, as low dose indefinite baths, or at higher dose as short-term dips (Mota et al., 2022). Regardless of the strategy used, the process rarely eliminates all the bacteria in the system. The remaining bacteria have the potential to regrow and drive the microbial community (Pei et al., 2019; Huang et al., 2023).
It has been shown that bacteria that remerge after disinfection may be more resistant not only to the disinfectant used, but also to antibiotics (Schweizer, 2001; McBain et al., 2002; Sharma et al., 2016). The mechanisms for this selection have still to be elucidated, but specific gene mutations, changes in cell envelope permeability, increased expression of efflux pump, and increased ability to form biofilms have been hypothesized (McDonnell and Russell, 1999; Poole, 2002; Rozman et al., 2021). These findings highlight the importance of considering the potential impacts of disinfection on ARGs particularly in aquaculture where disinfection is used frequently. The objective of this study was to evaluate the effect of low dose ozone on the relative abundance of ARGs in pond water using shotgun metagenomic sequencing analysis.
The experiment was conducted in August 2022 at the Au Tau Fisheries Office, Agriculture, Fisheries and Conservation Department in Yuen Long, Hong Kong SAR, PR China. The pond water we used had not been treated with any antibiotics and the last H2O2 (7 mg/L) treatment for algae control was done over a year prior to our study. The original aim of the study was to assess the effect of different ozone delivery systems on the microbial communities. The detailed methods and results of the initial study are published elsewhere (Huang et al., 2023). We used the sequence data from this prior study to subsequently assess the effect of ozone on the relative abundance of ARGs. In brief, pond water was pumped into twelve 75L tanks. There were 4 groups in total, two ozone treatment groups, ozone macrobubbles (O3MB) and ozone nanobubbles (O3NB), and two control groups, air macrobubbles (AirMB) and air nanobubbles (AirNB). All groups were triplicated.
To generate ozone nanobubbles, a nanobubbler (aQua+075MO, AquaPro Solutions Pte Ltd., Singapore) was connected to an ozone generator (DNO-15G, Dino Purification Co., Ltd, China) with an incoming oxygen stream flow rate of approximately 0.3 LPM. Water from the tanks was circulated into the nanobubbler and subsequently reintroduced back into the tanks. A dissolved ozone meter (DOZ-30, Dino Purification Co., Ltd, China) was used to monitor the ozone concentration meanwhile, and the system was stopped when the desired ozone concentration of 0.15 mg/L was attained in the three O3NB tanks. the same ozone concentration of 0.15 mg/L was achieved in O3MB group by attaching the same ozone generator to an air stone to deliver gaseous ozone macrobubbles to the tanks. For the AirMB and AirNB group, air pumps (Dazs model AP-528, Hong Kong) with a flow rate of 1.2 L/min were attached to air stones and the nanobubbler, respectively to provide microbubble and nanobubble aeration, and these were run for the same amount of time as the pumps used in the O3MB and O3NB treatment group, respectively.
Water samples (400 mL) were collected before and 24 h after the different treatments, and ozone was not detected at either of these time points (0 mg/L). Water was filtered, DNA was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen™), and then submitted to Novogene for 150 bp pair-end metagenomic shotgun sequencing. A sequencing depth of 6 Gb (Giga base pairs) was applied for each sample on Illumina HiSeq platform.
After the sequencing, raw reads were cleaned using Readfq (V8).1 The clean data were assembled using MEGAHIT software (v1.0.4-beta) (Li et al., 2015). Fragments shorter than 500 bp in the contigs were removed from the assembly. MetaGeneMark (v2.10) (Gemayel et al., 2022) was then used to predict genes on contigs, and the information with a length less than 100 nt in the prediction results was filtered out. We used the CD-HIT software (v4.5.8)2 with the default setting to create a unique initial gene catalog. Clean data of each sample was aligned to this initial gene catalog to determine the number of reads that matched the genes in our catalog using Bowtie2 (Bowtie2.2.4). Genes with reads ≤2 in each sample were filtered out. The filtered data was used for identification of ARGs and subsequent analysis. Based on the number of reads aligned and the length of the genes, the relative abundance of each gene in each sample was calculated by the following formula (Cotillard et al., 2013; Le Chatelier et al., 2013; Zeller et al., 2014), in which r was the number of gene reads, and L is the length of gene.
Notes: “k” represents a certain gene and “i” was a value from 1 to n, used to cycle through all genes.
The presence of acquired antimicrobial resistance genes was determined using the ResFinder tool (Florensa et al., 2012). Because we did not know the bacterial origin of most of our genes it was sometimes difficult to classify genes as acquired or intrinsic in this database as in cases such as (blaPAO, blaSGM and oleB) the nature of the gene could differ depending on the bacterial origin. In these cases, we classified the gene as acquired because it has been described in the literature as acquired in at least one bacterial species. To find the intrinsic ARGs belonging to the following 4 families, ATP binding cassette (ABC), resistance nodulation cell division (RND), small multidrug resistance (SMR), and major facilitator super (MFS) family, we used the CARD-RGI tool (McArthur et al., 2013). The threshold values for inclusion in our study were, e-value < 1e−9, identity >70%, and alignment length >100 bp. The Comprehensive Antibiotic Resistance Database (CARD) was also used to determine the mechanism of action and the antibiotic class that the genes conferred to.
DIAMOND (Version 0.9.9.110) was utilized to search genes with the e-value threshold of 1e−5 against sequences of bacteria, fungi, archaea, and viruses obtained from the NR database of the National Center for Biotechnology Information (NCBI). We used the LCA algorithm to ensure accurate species annotation. Finally, we identified whether the ten most abundant bacterial genera, which dominated post treatment had either intrinsic or acquired AMR genes associated with efflux pump proteins.
All data, including specific acquired ARGs, as well as their associated antibiotic classes, and resistance mechanisms, were graphically represented using box plots and stacked bar plots. The data were expressed as mean ± the standard deviation. To assess significant changes in relative abundance between pre- and 24-h post-treatments within treatment groups, we performed paired t-tests after log-transforming the data when the model residuals did not meet the assumptions of normality and homogeneity of variance. A p-value threshold of less than 0.05 was used to determine statistical significance. We also evaluated the difference in the relative abundance of intrinsic resistance genes for efflux protein families, RND, ABC, SMR and MFS, between pre and 24 h-post treatment in the four treatment groups using the same statistical methods. All statistical analysis were done in R Statistical Software (v4.1.2) (R Core Team, 2021).
A dendrogram was used to visualize the hierarchical cluster analysis, which was performed using Ward’s clustering method with squared Euclidean distance to determine if the acquired ARG profiles between treatments were different (Ward, 1963).
Additionally, we investigated whether acquired or intrinsic ARGs co-existed within the same genera of bacteria for overall and post-ozone treated samples by using a Sankey chart, which linked the relationships between these ARGs and the bacterial genera, while highlighting the interplay of resistance mechanisms. This was done using the “networkD3” package in the R Statistical Software (v4.1.2) (R Core Team, 2021).
We obtained an average of 43,673,320 ± 260,619 paired-end reads per water sample. After quality control, we retained 43,584,916 ± 261,566 paired-end reads per sample, generating a total of 21,792,458 ± 130,783 merged reads. The details of the quality control steps are outlined in Huang et al. (2023). The processed reads were on average assembled into 251,556 contigs per sample with an average length of 1,208 bp (Supplementary Table 1). These reads were used to identify the ARGs in the pre and post air or ozone treated water samples.
Based on shotgun metagenomic analysis, there were 26 different acquired ARGs identified within the predicted open reading frames (ORFs), which were associated with resistance against ten different classes of antibiotics and encompassed five resistance mechanisms (Supplementary Table 2).
Our hierarchical clustering analysis on the acquired ARGs revealed three distinct clusters corresponding to the two ozone treatments and the air treatment groups (controls) (Figure 1). In summary, all pre-treatment samples and 24 h-post air treated samples clustered together (Cluster 1), and the 24h-post treated samples in the two ozone groups clustered separately based on the type of bubbles used to deliver the gas (Cluster2 & 3). The clustering was mainly driven by the increase in relative abundance of five genes oqxB, qepA, oleB, blaSGM and tmexD (Figures 2A–F).
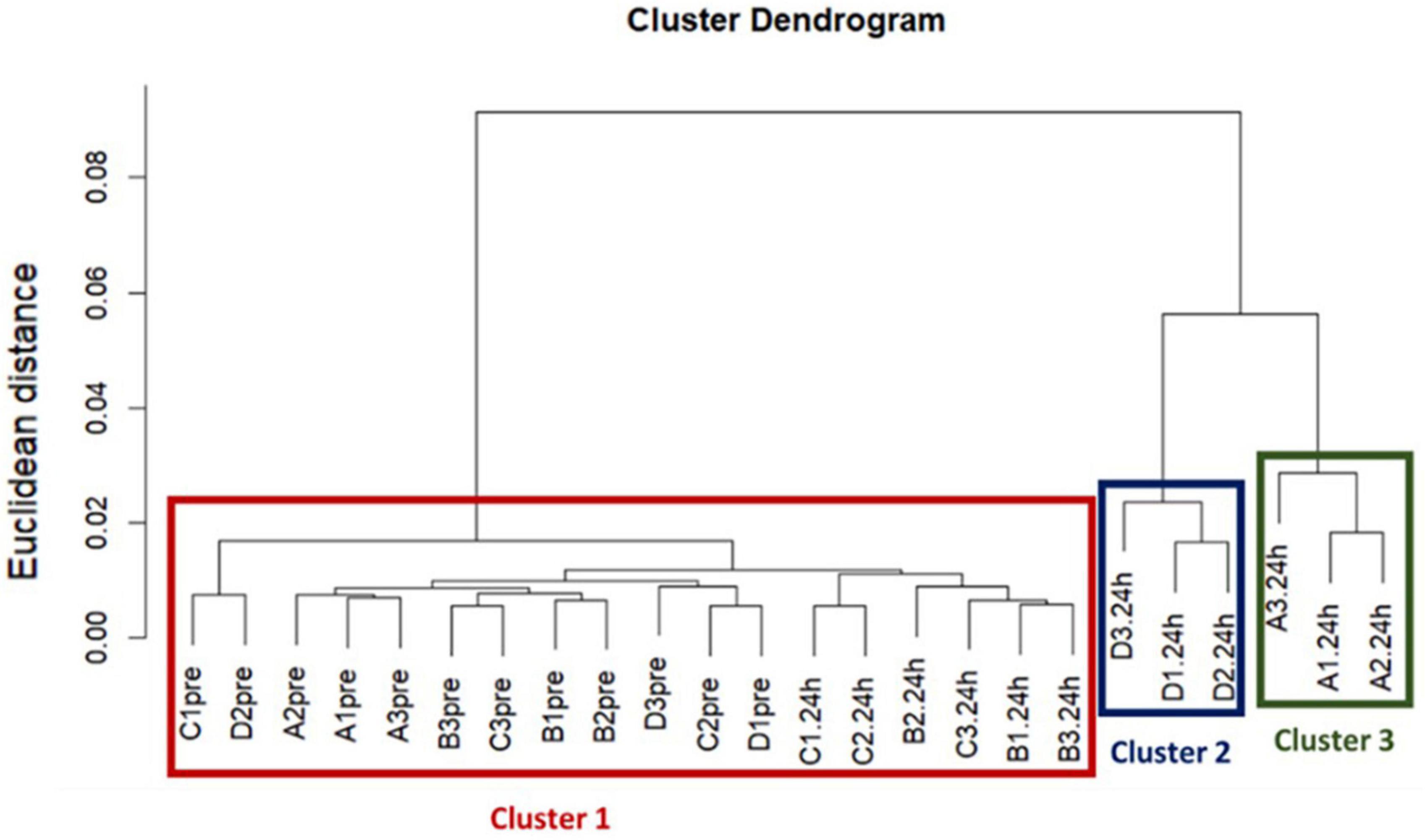
Figure 1. The results of a Hierarchical cluster analysis on acquired resistance genes revealed three distinct clusters corresponding to the two ozone treatments and the control air treatment groups. Treatments are labeled as, A: ozone macrobubble group (O3MB), B: air macrobubble group (AirMB), C: air nanobubble group (AirNB) and D: ozone nanobubble group (O3NB). The number following is the replicate, followed by the sampling time, pre: pretreatment sample, 24 h: sample after 24 h of the treatment. Our analysis showed all pre-treatment samples and 24 h-post control air treatment samples clustered together in the Cluster 1, and the three samples from the 24 h-post ozone nanobubble treated group were categorized into cluster 2. Cluster 3 was comprised by three samples in 24 h-post ozone macrobubble treated group.
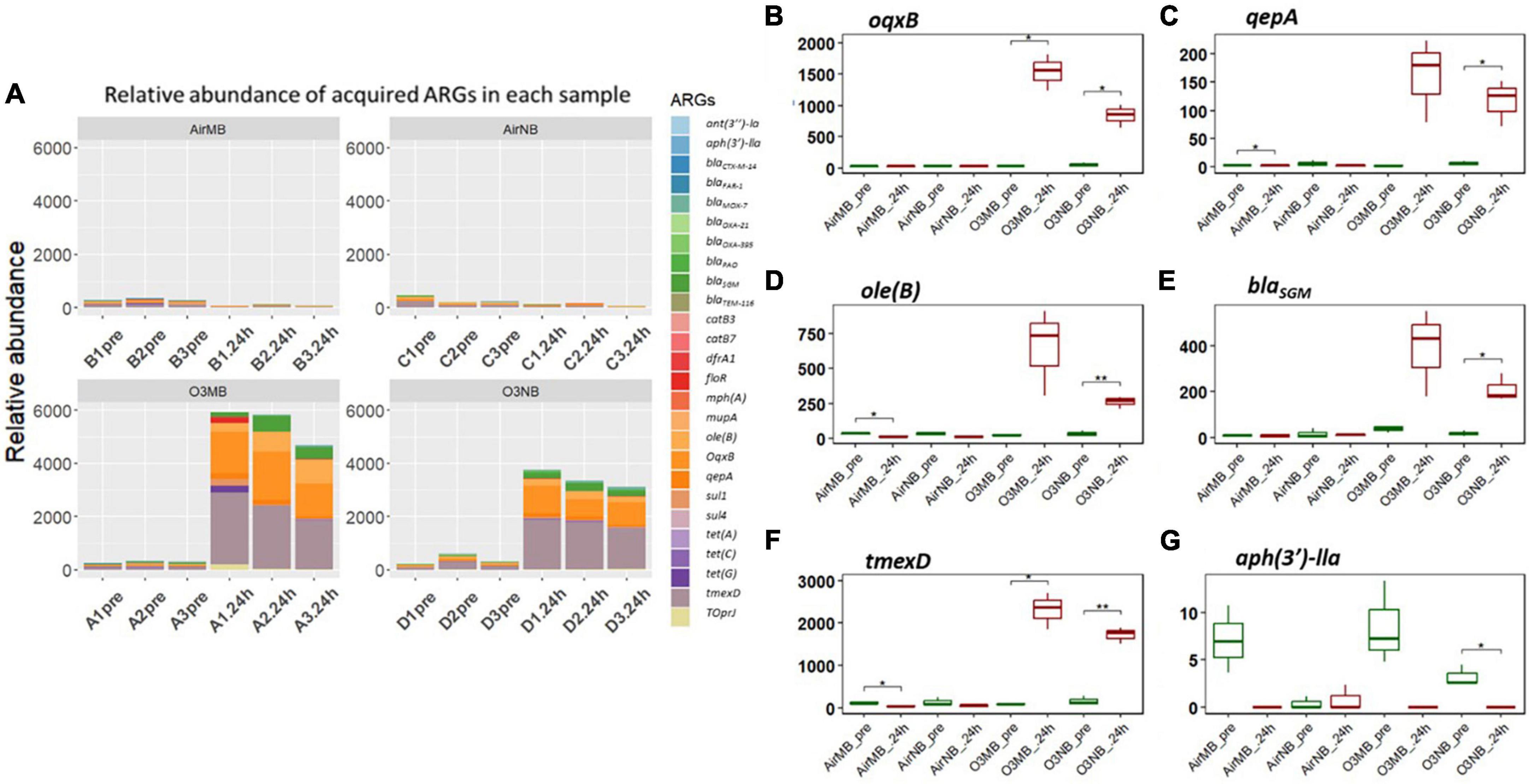
Figure 2. (A) Relative abundance of acquired ARGs of each sample across four treatments; (B–G) Relative abundance of specific ARGs before (pre) and 24 h after (24 h) air or ozone treatments. Significant difference in the relative abundance of ARGs between pre and post treatment samples were determined using a Paired T-test and is indicated with *. Only one ARG showed a significant decrease. Y-axis represents relative abundance of each gene. X-axis represents sample name, in which A is ozone macrobubble group (O3MB), B is air macrobubble group (AirMB), C is air nanobubble group (AirNB) and D is ozone nanobubble group (O3NB). The number following is the replicate, followed by the sampling time, pre, pretreatment sample; 24 h, sample after 24 h of the treatment.
In contrast, the aph(3′)-IIa gene showed a significant decrease following ozone nanobubble (O3NB) treatment compared to the air treatment groups (Figure 2G). Although there was a trend suggesting the ozone macrobubble (O3MB) treated water also had a reduction in this particular gene compared to the O3NB group, this difference was not statistically significant (Figure 2G).
Our analysis of the acquired ARGs categorized by the class of antibiotics they affect, suggested a notably increase in the relative abundance of genes associated with resistance to the following classes of drugs: fluoroquinolones, macrolides, and beta-lactam antibiotics in the 24 h-post ozone treated groups compared to the controls (Figures 3A–C). In addition, the genes associated with resistance to multidrugs accounted for over 70% of ARGs after treatment with ozone (Figures 3D, G). While there was an observable increase in trimethoprim-resistance genes in the ozone treated groups, the difference between pre- and 24 h-post treatment was not statistically significant (Figure 3E). Conversely, there was a decrease in the genes encoding aminoglycoside-resistance after ozone treatments; however, this decrease was only significant in the O3NB group (Figure 3F).
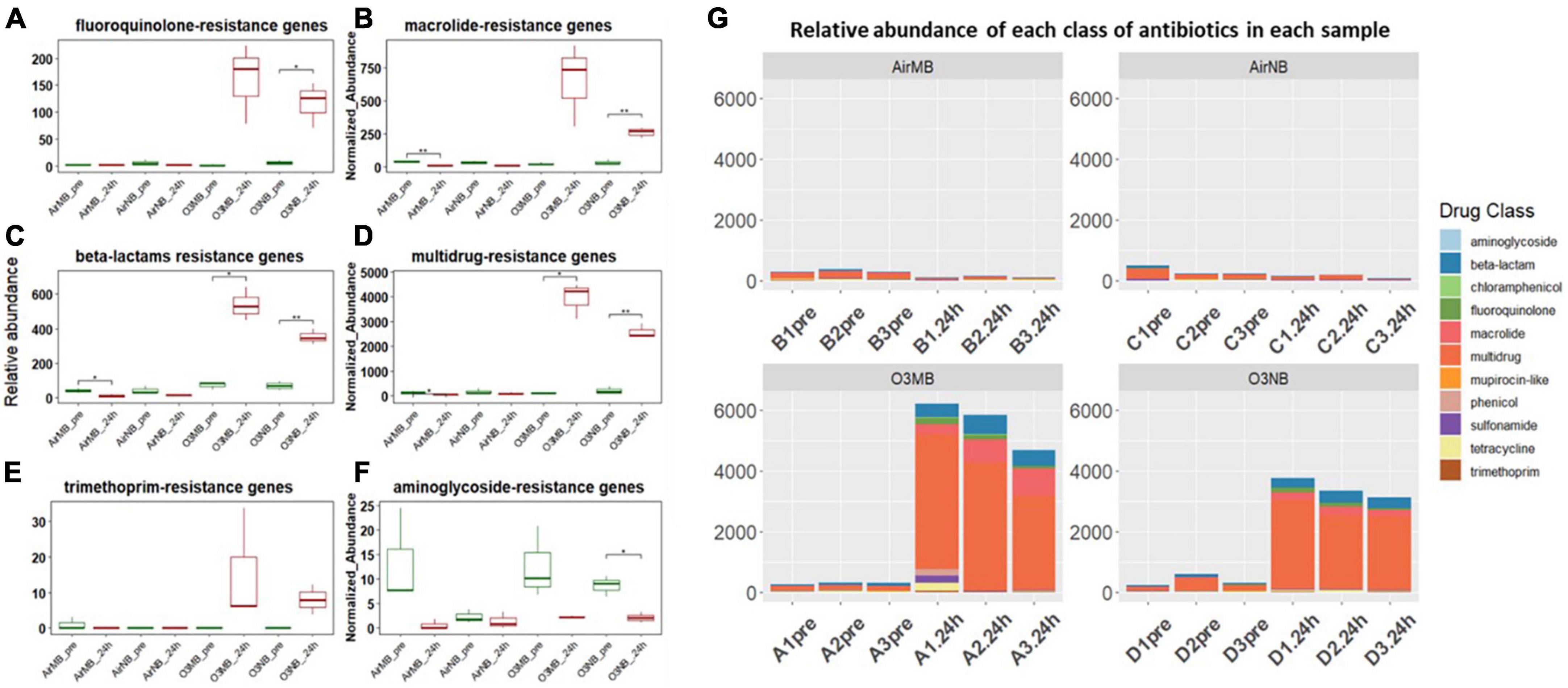
Figure 3. (A–F) Relative abundance of ARGs grouped by antibiotic classes before (pre) and 24 h after (24 h) air or ozone treatments. Significant difference in the relative abundance of genes in different antibiotic classes between pre and post treatment samples were determined using a Paired T-test and is indicated with *. Only one class showed a in significant decrease; (G) Normalized abundance of antibiotics classes of each sample across four treatments, which showed ARGs associated with multidrug resistance increased and dominated in the community after the ozone treatments. Y-axis represents relative abundance of each antibiotic class. X-axis represents sample name, in which A is ozone macrobubble group (O3MB), B is air macrobubble group (AirMB), C is air nanobubble group (AirNB) and D is ozone nanobubble group (O3NB). The number following is the replicate, followed by the sampling time, pre, pretreatment sample; 24 h, sample after 24 h of the treatment.
The acquired ARGs identified in our study were also categorized into five primary resistance mechanisms, which included antibiotic inactivation, antibiotic efflux systems, antibiotic target alternation, antibiotic target protection, and antibiotic target replacement (Figures 4A–E and Supplementary Table 2). When we grouped the genes by their mechanisms of action, we observed significant increases in the relative abundance of ARGs encoding for the antibiotic efflux systems, antibiotic inactivation, and antibiotic target protection following the ozone treatments (Figures 4A–C, F). In contrast, the genes associated with antibiotic target alteration, initially present at low levels, exhibited a significant decrease in relative abundance after all treatments (Figure 4D).
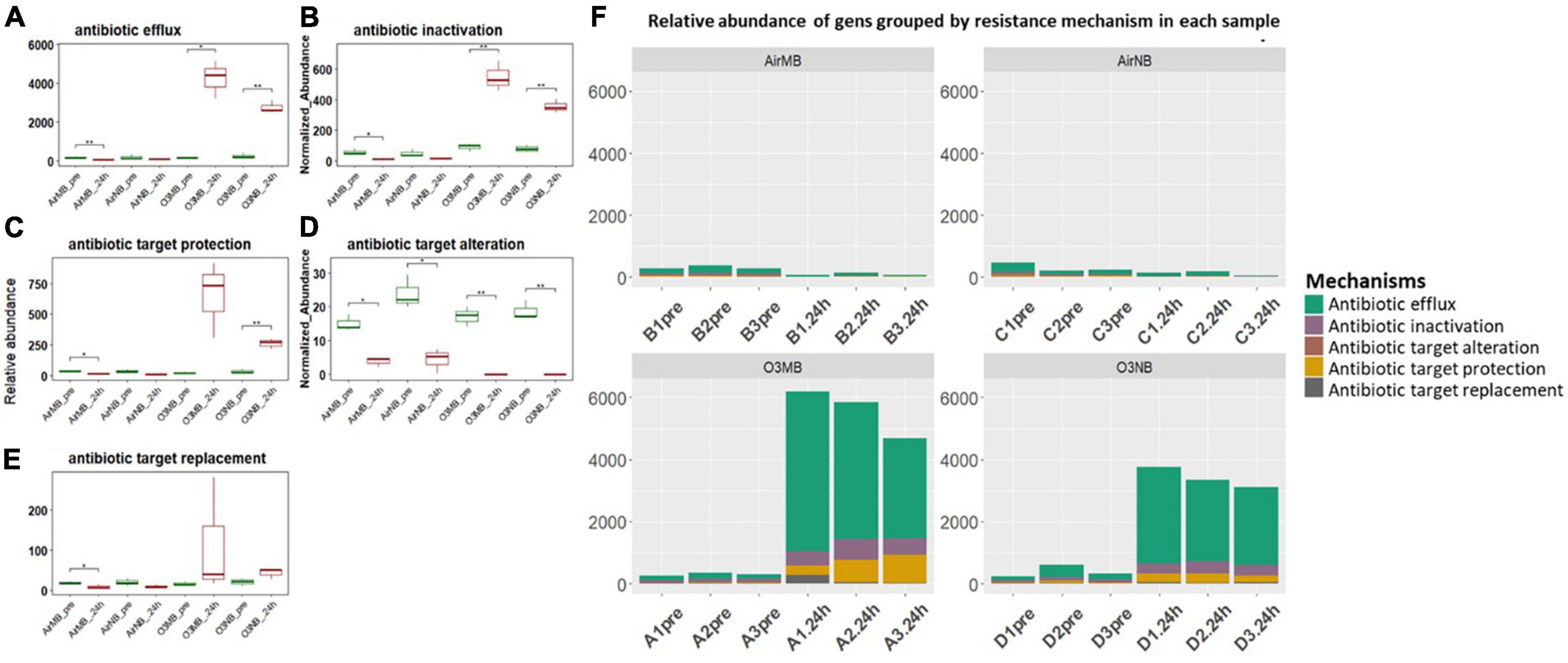
Figure 4. (A–E) Relative abundance of ARGs grouped by resistance mechanisms before (pre) and 24 h after (24 h) air or ozone treatments. Significant difference in the relative abundance of genes in different resistance mechanisms between pre and post treatment samples were determined using a Paired T-test and is indicated with *. Antibiotic target alteration showed a significant decrease after treatments in all groups; (F) Relative abundance of genes grouped by resistance mechanism in each sample across four treatments, which showed ARGs associated with antibiotic efflux mechanism increased and dominated in the community after the ozone treatments. Y-axis represents relative abundance of genes grouped by mechanism. X-axis represents sample name, in which A is ozone macrobubble group (O3MB), B is air macrobubble group (AirMB), C is air nanobubble group (AirNB) and D is ozone nanobubble group (O3NB). The number following is the replicate, followed by the sampling time, pre, pretreatment sample; 24 h, sample after 24 h of the treatment.
The intrinsic efflux ARGs, which were identified using the CARD-RGI were initially more abundant in all samples compared to the acquired ARGs. Similar to the change in relative abundance of acquired efflux ARGs, the intrinsic efflux ARGs from the RND family were significantly higher in ozone treated samples compared to air treated samples 24 h after the treatment (Figure 5), with the adeF gene becoming the most abundant intrinsic efflux ARGs in RND family in the water samples post ozone treatments (Figure 6). However, the intrinsic efflux ARGs in the ABC family significantly decline after ozone treatments, which was driven mainly by mel, tlrC, and otrC (Figure 5).
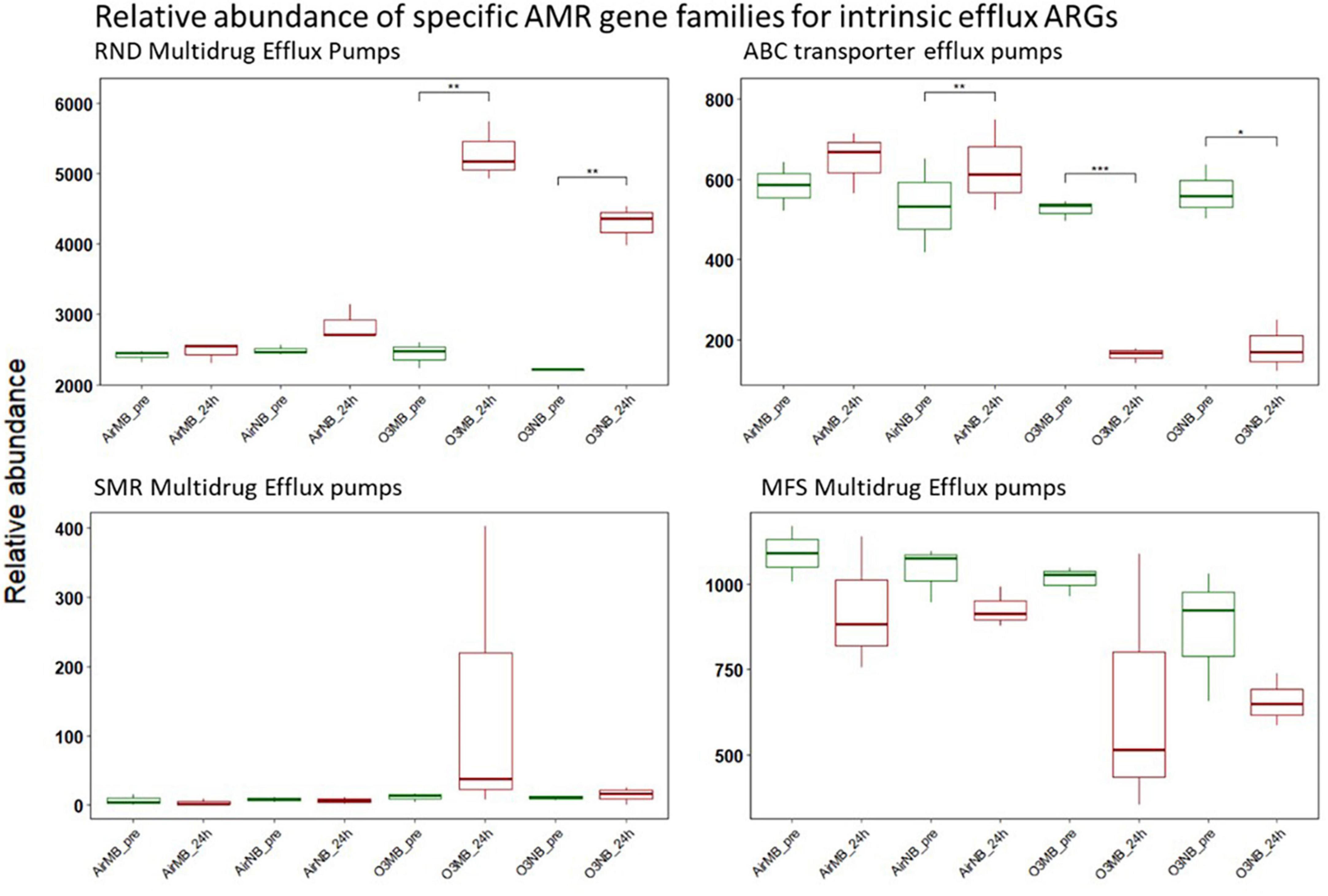
Figure 5. Relative abundance of specific AMR gene families for intrinsic efflux ARGs before (pre) and 24 h after (24 h) air or ozone treatments. Significant difference in the relative abundance of genes in different families between pre and post treatment samples were determined using a Paired T-test and is indicated with *. Only genes in the RND family showed a significant increase in relative abundance after ozone treatments, while ABC family decreased significantly after ozone treatments. Y-axis represents relative abundance of genes grouped by mechanism. X-axis represents sample name, in which AirMB is air macrobubble group, AirNB is air nanobubble group, O3MB is ozone macrobubble group and O3NB is ozone nanobubble group. The number following is the replicate, followed by the sampling time, pre, pretreatment sample; 24 h, sample after 24 h of the treatment.
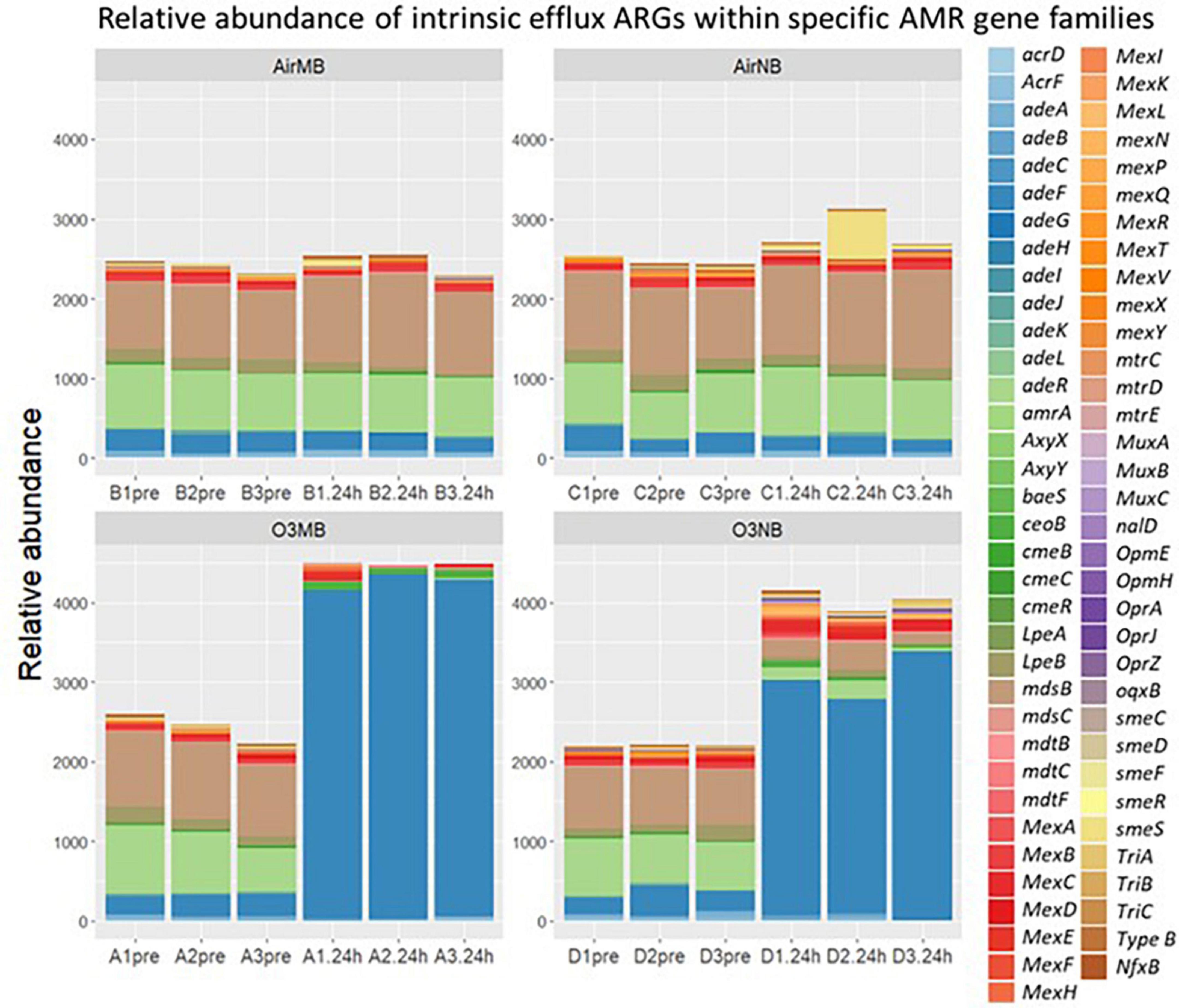
Figure 6. Relative abundance of intrinsic efflux ARGs within specific AMR gene families including ATP binding cassette (ABC), resistance nodulation cell division (RND), small multidrug resistance (SMR), and major facilitator super (MFS) family detected by CARD-RGI identifier before (pre) and 24 h after (24 h) air or ozone treatments delivered by macrobubbles or nanobubbles, which showed adeF gene became the most abundant intrinsic efflux ARGs in the water samples 24 h after the ozone treatments. Y-axis represents relative abundance of genes. X-axis represents sample name, in which A is ozone macrobubble group (O3MB), B is air macrobubble group (AirMB), C is air nanobubble group (AirNB) and D is ozone nanobubble group (O3NB). The number following is the replicate, followed by the sampling time, pre, pretreatment sample; 24 h, sample after 24 h of the treatment.
Of the acquired ARGs identified in our study, only 9.63% were classified to 33 of the 2,552 bacterial genera present in our samples (Figure 7). The other acquired ARGs could not be linked to specific bacteria in our sample, though this could be because there was insufficient information in our database to classify their origin. Further, 44 genera of bacteria in our samples had genes belonging to the four intrinsic efflux pump families. The relative abundance of the bacterial genera associated with (acquired or intrinsic) ARGs increased post ozone treatments, while bacteria genera associated with ARGs did not increase in relative abundance after the air treatment samples (Figure 8).

Figure 7. Sankey diagram used to investigate whether acquired ARGs or intrinsic efflux ARGs co-existed within the same known genera of bacteria 24 h after air treatment ed samples (A) and ozone treatment (B), while highlighting the interplay of resistance mechanisms. Regardless of the treatment efflux pump ARGs were linked to almost all bacterial genera with the exception of 2 or 3, suggesting ozone treatment did not alter the co-existence pattern of ARGs.
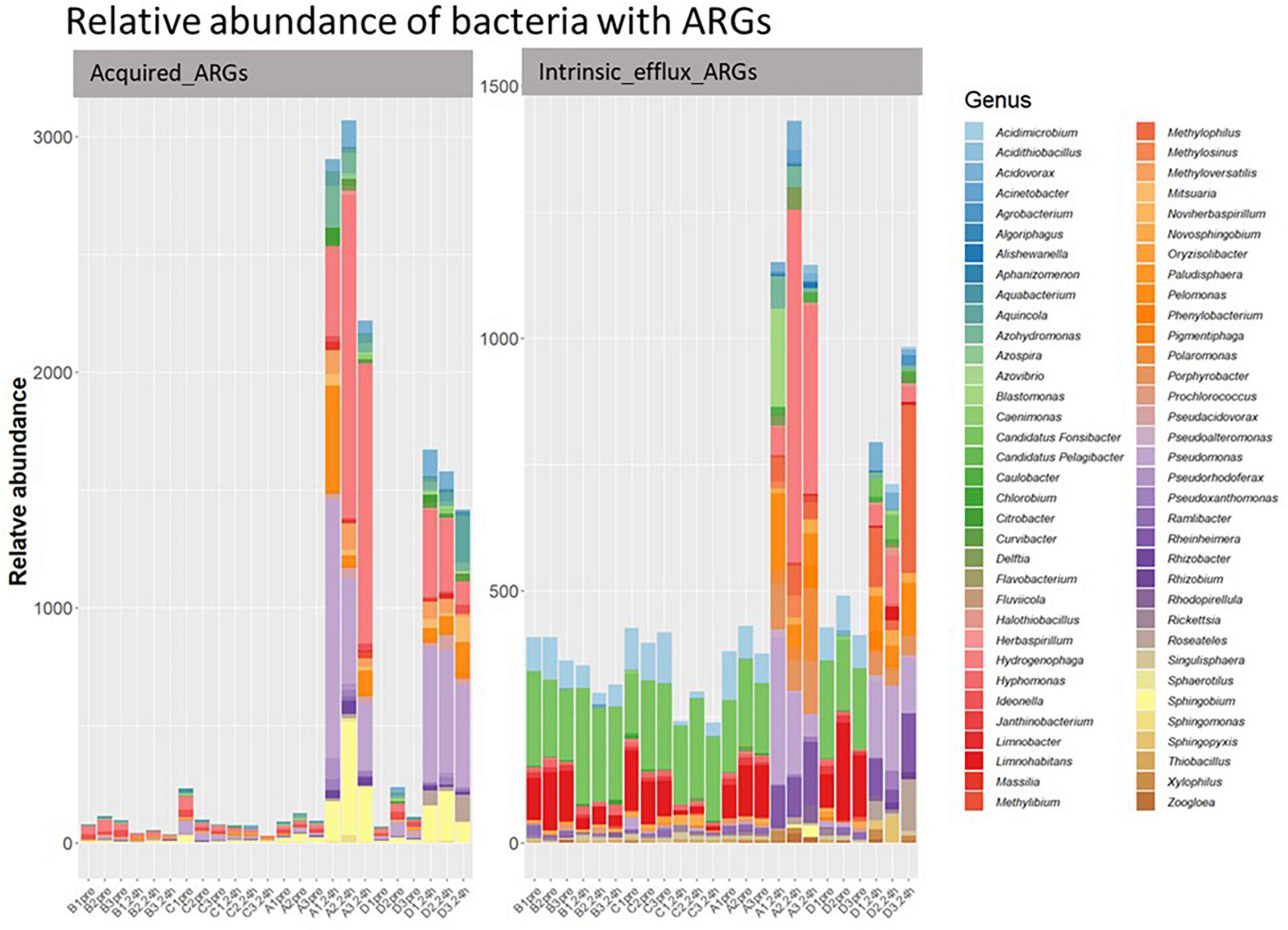
Figure 8. The relative abundance of classified bacterial genera with acquired efflux ARGs (left) and intrinsic efflux ARGs (right) in all samples. Y-axis represents relative abundance of bacteria with ARGs. X-axis represents sample name, in which A is ozone macrobubble group; B is air macrobubble group; C is air nanobubble group and D is ozone nanobubble group. The number following is the replicate, followed by the sampling time, pre, pretreatment sample; 24 h, sample after 24 h of the treatment.
Most of the 33 genera in our study had at least one gene associated with acquired efflux pumps (Figure 7). For example, the tmexD, oqxB, qepA, and toprJ, four efflux pump genes, were present in 21, 13, 5, and 4 bacterial genera (Figure 7), respectively, and all of these genes increased after ozone treatment (Figure 2). Interestingly, of the 10 most abundant bacterial genera (Supplementary Figure 1) in our post-ozone treated samples, all had at least eight efflux ARGs (Supplementary Table 3). In contrast, the number of genera in the 24 h-post air treated groups that had efflux pump ARGs was at most 6 (Supplementary Table 3).
It is worth noting that after the ozone treatments, a number of specific non-efflux ARGs of importance (i.e. beta-lactam-resistance genes (blaOXA–395, blaMOX–7 and blaPAO), chloramphenicol resistance genes (catB7) remained in the bacterial population (Figure 7B). However, these genes were always in genera that also had efflux pump genes with the few exceptions of Citrobacter and Sphingomonas.
Our shotgun metagenomic analysis of pond water treated with low dose ozone (0.15 mg/L) indicated that a one-time treatment, irrespective of how this gas was delivered (nanobubbles or macrobubbles), played a role in influencing the composition and relative abundance of antimicrobial resistant genes (ARGs) 24 h after treatments. Our findings suggested an increase in intrinsic efflux and acquired ARGs. Five out of 33 acquired ARGs detected in our samples increased significantly in relative abundance 24 h after a low dose ozone treatment. These 5 genes were associated with antibiotic efflux, antibiotic activation, and antibiotic target protection mechanisms; however, the efflux pump genes appeared to be more prominent in the ten most abundant bacterial genera post treatment. Other researchers have also reported similar ARG changes in bacterial communities exposed to different disinfectants, such as chlorine, hydrogen peroxide, quaternary ammonium, peracetic acid, chlorhexidine diacetate and trisodium phosphate (Su and D’Souza, 2012; Capita et al., 2013; Biswal et al., 2014; Molina-González et al., 2014; Gadea et al., 2016). Interestingly, we found an increase in relative abundance for a few specific non-efflux ARGs after ozone treatments, but closer inspection revealed that they usually co-existed in bacterial genera associated with intrinsic or acquired efflux ARGs.
The increased relative abundance of both intrinsic efflux and acquired ARGs associated with an efflux pump mechanism after treatment with ozone was somewhat expected. Many of these genes play a crucial role in the survival of bacteria, particularly Gram-negative bacteria, by expelling toxic substances, including antibiotics and other compounds such as those generated during oxidative stress (Nelson, 2002; Huwaitat et al., 2016; Masi et al., 2017). In fact, many researchers have found both acquired efflux pump genes such as qepA, tet(G) and oqxB, and the intrinsic efflux genes in the RND family to be up-regulated in response to different stressors, suggesting they play an important role in bacterial survival (Shi et al., 2013; Tandukar et al., 2013; Xu et al., 2017; Gu et al., 2020; Hu et al., 2023; McCarlie et al., 2023). Therefore, it is not surprising that we detected an increase in the relative abundance of genes associated with the efflux pump mechanism after exposure to low dose ozone, which is associated with oxidative stress (Pell et al., 1997).
It is important to better understand the selective pressure that ozone exerts on bacterial communities at different doses used in aquaculture particularly low dose treatments that may not kill all bacteria (Iakovides et al., 2019, 2021; Huang et al., 2023). Our study suggests at lower ozone concentrations, which are used to treat fish, bacteria are not completely eliminated (Huang et al., 2023), and the surviving bacteria may be more resistant to ozone. This shift in population could have additional consequences on other types of treatments, particularly antibiotic therapies as the genes suspected of conferring ozone resistance may also bestow resistance to antibiotics.
Besides an increase in ARGs associated with efflux pumps, we also observed a significant increase in the relative abundance of a few non-efflux genes either involved in antibiotic target protection or antibiotic inactivation after our ozone treatments. Previous studies also found an increase in antibiotic specific genes in wastewater after treatment of with 0.9 ± 0.1 g ozone per 1 g DOC. In one study, there was an increase in the abundance of vanA and blaVIM genes, encoding vancomycin and carbapenem resistance (Alexander et al., 2016). Another study found after an ozone treatment, the surviving R. ornithinolytica bacterial population had an increase in the blaCTX–M–62 gene (Azuma et al., 2022). It is possible genes that conferred ozone resistance in our study (i.e., efflux pump genes) co-occurred with AMR specific (i.e., non-efflux pump) genes, which could therefore lead to their co-selection under ozone exposure. Co-selection of genes under different environment stressors has been reported by others (Ding et al., 2019). In our study, there was evidence for this hypothesis as we found several non-efflux pump ARG often co-occurring within the same genera of bacteria as acquired efflux ARGs (i.e., OqxB) or the intrinsic RND efflux genes (i.e., adeF). In fact, all but two of the ten most abundant bacteria that survived and colonized our samples after an ozone treatment had either intrinsic RND efflux genes or acquired efflux ARGs. Interestingly, these two genera Sphingomonas and Citrobacter, are known to form biofilms. This protective mechanism protects bacteria against disinfectants (Donlan and Costerton, 2002; Gulati and Ghosh, 2017) and may explain their survival post ozone treatment.
Notably, a few ARGs such as dfrA1, floR and ant(3″)-Ia, which increased in relative abundance after 24 h after ozone treatments are known to be located on mobile genetic elements (MGEs) (Zhang et al., 2017; Sánchez-Osuna et al., 2020; Raza et al., 2022; Zhu et al., 2022). If the increased ARGs observed in our study post treatment were present within MGEs, the enrichment of those ARGs could lead to nonpathogenic microbial species acting as an ecological reservoir for pathogenic bacteria (Peterson and Kaur, 2018). Bacteria can acquire new ARGs through horizontal gene transfer from plasmids, transposons, or other MGEs originating from other bacteria or the environment (Jian et al., 2021).
This study had several limitations that should be acknowledged. Firstly, our results were based on only one pond. Different pond water sources could be included in future studies to increase the external validity of the findings. Secondly, we used shotgun metagenomic analysis which only provided us with short-read sequencing. This is particularly problematic for determining the origin of genes within the mixed bacterial community. Despite attempts to address this limitation by employing read assembly techniques (Li et al., 2022), we could not determine the source of many ARGs (i.e., bacterial genomic or MGEs). Another limitation is that the classifiers we used to determine whether genes were intrinsic or acquired were not consistent across all bacterial genera. There were some genes (i.e., blaPAO, blaSGM, oleB) that are likely intrinsic for some bacteria and considered acquired for others, therefore, it was difficult for us to categorize these as we did not know the bacterial origin of all the genes. Despite this limitation, of the genes we classified as acquired (n = 33), there were only a few which could have been intrinsic depending on the origin (Li et al., 2022). Finally, it is important to note that our reporting of relative changes in certain genes between samples, was achieved by normalizing the reads to the total sequence reads, therefore we could only assess relative abundance. This measure does not necessarily translate to phenotypic antibiotic resistance or absolute abundance. To investigate the absolute abundance of ARGs, quantitative polymerase chain reaction (qPCR) analyses would be required, and this was beyond the scope of this study.
In this study, we provide valuable insights into the effects of low-dose ozone treatments on ARG dynamics in aquaculture pond water. The results indicate that low dose ozone disinfection, delivered through macrobubbles or nanobubbles, can lead to an increase in the relative abundance of ARGs. We saw the increase mainly with the efflux pumps, whether acquired or naturally present in the bacteria. We also have evidence to suggest co-selection of other resistance genes. This study emphasizes the importance of considering the potential impacts of disinfection on ARGs, particularly in aquaculture settings where disinfectants are frequently used at low levels.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/1100576.
QH: Writing – review and editing, Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. PB: Writing – review and editing, Methodology, Formal analysis. PN: Writing – review and editing, Investigation. JZ: Writing – review and editing, Investigation. WC: Writing – review and editing, Investigation. SS-H: Writing – review and editing, Writing – original draft, Supervision, Project administration, Investigation, Funding acquisition.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the International Development Research Centre (grant number 9240005).
We would like to thank our funding agency International Development Research Centre (IDRC) and the students who helped with the initial water treatment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1393266/full#supplementary-material
Supplementary Figure 1 | Relative abundance of the top 10 genera present in the microbial communities before (pre) and 24 h after (24 h) air or ozone treatments delivered by macrobubbles or nanobubbles. Y-axis represents accumulated proportion of the genera. X-axis represents sample name, in which A is ozone macrobubble group, B is air macrobubble group, C is air nanobubble group and D is ozone nanobubble group.
ABC, ATP binding cassette; AMR, antimicrobial resistance; ARGs, antimicrobial resistance genes; AirMB, air macrobubble; AirNB, air nanobubble; CARD, comprehensive antibiotic resistance database; MFS, major facilitator super; NCBI, national center for biotechnology information; O3MB, ozone macrobubble; O3NB, ozone nanobubble; ORFs, open reading frames; RND, resistance nodulation cell division; SMR, small multidrug resistance.
Alexander, J., Knopp, G., Dötsch, A., Wieland, A., and Schwartz, T. (2016). Ozone treatment of conditioned wastewater selects antibiotic resistance genes, opportunistic bacteria, and induce strong population shifts. Sci. Total Environ. 559, 103–112. doi: 10.1016/j.scitotenv.2016.03.154
Azuma, T., Katagiri, M., Sekizuka, T., Kuroda, M., and Watanabe, M. (2022). Inactivation of bacteria and residual antimicrobials in hospital wastewater by ozone treatment. Antibiotics 11:862. doi: 10.3390/antibiotics11070862
Bhat, R. A. H., and Altinok, I. (2023). Antimicrobial Resistance (AMR) and alternative strategies for combating AMR in aquaculture. Turk. J. Fish. Aquat. Sci. 23:11. doi: 10.4194/TRJFAS24068
Biswal, B., Khairallah, R., Bibi, K., Mazza, A., Gehr, R., Masson, L., et al. (2014). Impact of UV and peracetic acid disinfection on the prevalence of virulence and antimicrobial resistance genes in uropathogenic Escherichia coli in wastewater effluents. Appl. Environ. Microbiol. 80, 3656–3666. doi: 10.1128/AEM.00418-14
Burki, T. K. (2018). Superbugs: An arms race against bacteria. Lancet Respir. Med. 6:668. doi: 10.1016/S2213-2600(18)30271-6
Capita, R., Álvarez-Fernández, E., Fernández-Buelta, E., Manteca, J., and Alonso-Calleja, C. (2013). Decontamination treatments can increase the prevalence of resistance to antibiotics of Escherichia coli naturally present on poultry. Food Microbiol. 34, 112–117. doi: 10.1016/j.fm.2012.11.011
Cotillard, A., Kennedy, S., Kong, L., Prifti, E., Pons, N., Le Chatelier, E., et al. (2013). Dietary intervention impact on gut microbial gene richness. Nature 500, 585–588. doi: 10.1038/nature12480
Ding, J., An, X., Lassen, S., Wang, H., Zhu, D., and Ke, X. (2019). Heavy metal-induced co-selection of antibiotic resistance genes in the gut microbiota of collembolans. Sci Total Environ. 683, 210–215. doi: 10.1016/j.scitotenv.2019.05.302
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: Survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/cmr.15.2.167-193.2002
Florensa, A., Kaas, R., Clausen, P., Aytan-Aktug, D., and Aarestrup, F. (2012). ResFinder–an open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb Genom 8:748. doi: 10.1099/mgen.0.000748
Gadea, R., Fuentes, M., Pulido, R., Gálvez, A., and Ortega, E. (2016). Effects of exposure to quaternary-ammonium-based biocides on antimicrobial susceptibility and tolerance to physical stresses in bacteria from organic foods. Food Microbiol. 63, 58–71. doi: 10.1016/j.fm.2016.10.037
Gemayel, K., Lomsadze, A., and Borodovsky, M. (2022). MetaGeneMark-2: Improved gene prediction in metagenomes. BioRxiv [Preprint]. doi: 10.1101/2022.07.25.500264
Gu, Y., Wang, S., Huang, L., Sa, W., Li, J., Huang, J., et al. (2020). Development of resistance in Escherichia coli ATCC25922 under exposure of sub-inhibitory concentration of olaquindox. Antibiotics 9:791. doi: 10.3390/antibiotics9110791
Gulati, P., and Ghosh, M. (2017). Biofilm forming ability of Sphingomonas paucimobilis isolated from community drinking water systems on plumbing materials used in water distribution. J. Water Health 15, 942–954. doi: 10.2166/wh.2017.294
Hu, Z., Yang, L., Liu, Z., Han, J., Zhao, Y., Jin, Y., et al. (2023). Excessive disinfection aggravated the environmental prevalence of antimicrobial resistance during COVID-19 pandemic. Sci. Total Environ. 882:163598. doi: 10.1016/j.scitotenv.2023.163598
Huang, Q., Ng, P., Marques, A., Cheng, T., Man, K., Zu, L., et al. (2023). Effect of ozone nanobubbles on the microbial ecology of pond water and safety for jade perch (Scortum barcoo). Aqua 576:739866. doi: 10.1016/j.aquaculture.2023.739866
Hushangi, R., and Hosseini Shekarabi, S. P. (2018). Effect of a peracetic acid-based disinfectant on growth, hematology and histology of juvenile rainbow trout (Oncorhynchus mykiss). Fishes 3:10. doi: 10.3390/fishes3010010
Huwaitat, R., McCloskey, A., Gilmore, B., and Laverty, G. (2016). Potential strategies for the eradication of multidrug-resistant Gram-negative bacterial infections. Fut. Microbiol. 11, 955–972. doi: 10.2217/fmb-2016-0035
Iakovides, I., Manoli, K., Karaolia, P., Michael-Kordatou, I., Manaia, C., and Fatta-Kassinos, D. (2021). Reduction of antibiotic resistance determinants in urban wastewater by ozone: Emphasis on the impact of wastewater matrix towards the inactivation kinetics, toxicity and bacterial regrowth. J. Hazard Mater. 420:126527. doi: 10.1016/j.jhazmat.2021.126527
Iakovides, I., Michael-Kordatou, I., Moreira, N., Ribeiro, A., Fernandes, T., Pereira, M., et al. (2019). Continuous ozonation of urban wastewater: Removal of antibiotics, antibiotic-resistant Escherichia coli and antibiotic resistance genes and phytotoxicity. Water Res. 159, 333–347. doi: 10.1016/j.watres.2019.05.025
Jian, Z., Zeng, L., Xu, T., Sun, S., Yan, S., Yang, L., et al. (2021). Antibiotic resistance genes in bacteria: Occurrence, spread, and control. J Basic Microbiol. 61, 1049–1070. doi: 10.1002/jobm.202100201
Joshua, I., Bobai, M., and Woje, C. S. (2022). “Managing antimicrobial resistance beyond the hospital antimicrobial stewardship: The role of one health,” in The Global Antimicrobial Resistance Epidemic-Innovative Approaches and Cutting-Edge Solutions, ed. T. Guillermo (London: IntechOpen), doi: 10.5772/intechopen.104170
Le Chatelier, E., Nielsen, T., Qin, J., Prifti, E., Hildebrand, F., Falony, G., et al. (2013). Richness of human gut microbiome correlates with metabolic markers. Nature 500, 541–546. doi: 10.1038/nature12506
Li, D., Liu, C., Luo, R., Sadakane, K., and Lam, T. (2015). MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, L., Xiao, Y., Olsen, R. H., Wang, C., Meng, H., and Shi, L. (2022). Short-and long-read metagenomics insight into the genetic contexts and hosts of mobile antibiotic resistome in Chinese swine farms. Sci. Total Environ. 827:154352. doi: 10.1016/j.scitotenv.2022.154352
Masi, M., Réfregiers, M., and Pos, K. (2017). Mechanisms of envelope permeability and antibiotic influx and efflux in Gram-negative bacteria. Nat. Microbiol. 2, 1–7. doi: 10.1038/nmicrobiol.2017.1
McArthur, A., Waglechner, N., Nizam, F., Yan, A., Azad, M., Baylay, A., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 57, 3348–3357. doi: 10.1128/aac.00419-13
McBain, A., Rickard, A., and Gilbert, P. (2002). Possible implications of biocide accumulation in the environment on the prevalence of bacterial antibiotic resistance. J. Ind. Microbiol. Biotechnol. 29, 326–330. doi: 10.1038/sj.jim.7000324
McCarlie, S., du Preez, L., Hernandez, J., Boucher, C., and Bragg, R. (2023). Transcriptomic signature of bacteria exposed to benzalkonium chloride. Res. Microbiol. [Online ahead of print]. doi: 10.1016/j.resmic.2023.104151
McDonnell, G., and Russell, A. D. (1999). Antiseptics and disinfectants: Activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179. doi: 10.1128/cmr.12.1.147
Mendelson, M., Sharland, M., and Mpundu, M. (2022). Antibiotic resistance: Calling time on the ‘silent pandemic’. JAC-AMR 4:dlac016. doi: 10.1093/jacamr/dlac016
Molina-González, D., Alonso-Calleja, C., Alonso-Hernando, A., and Capita, R. (2014). Effect of sub-lethal concentrations of biocides on the susceptibility to antibiotics of multi-drug resistant Salmonella enterica strains. Food Control 40, 329–334. doi: 10.1016/j.foodcont.2013.11.046
Mota, V., Eggen, M., and Lazado, C. (2022). Acute dose-response exposure of a peracetic acid-based disinfectant to Atlantic salmon parr reared in recirculating aquaculture systems. Aqua 554:738142. doi: 10.1016/j.aquaculture.2022.738142
Nelson, M. (2002). Modulation of antibiotic efflux in bacteria. Curr. Med. Chem. 1, 35–54. doi: 10.2174/1568012023355054
Pei, L., Ke, Y., Zhao, H., Liu, W., Wang, L., Jia, C., et al. (2019). Regulatory effect of Garidisan on dysbiosis of the gut microbiota in the mouse model of ulcerative colitis induced by dextran sulfate sodium. BMC Complement. Altern. Med. 19:329. doi: 10.1186/s12906-019-2750-y
Pell, E., Schlagnhaufer, C., and Arteca, R. (1997). Ozone-induced oxidative stress: Mechanisms of action and reaction. Physiol. Plant 100, 264–273. doi: 10.1111/j.1399-3054.1997.tb04782.x
Peterson, E., and Kaur, P. (2018). Antibiotic resistance mechanisms in bacteria: Relationships between resistance determinants of antibiotic producers, environmental bacteria, and clinical pathogens. Front. Microbiol. 9:2928. doi: 10.3389/fmicb.2018.02928
Poole, K. (2002). Mechanisms of bacterial biocide and antibiotic resistance. J. Appl. Microbiol. 92, 55S–64S.
R Core Team (2021). R: A Language and Environment for Statistical Computing (4.1.2 ed.). Vienna: R Foundation for Statistical Computing.
Raza, S., Shin, H., Hur, H., and Unno, T. (2022). Higher abundance of core antimicrobial resistant genes in effluent from wastewater treatment plants. Water Res. 208:117882. doi: 10.1016/j.watres.2021.117882
Rico, A., and Van den Brink, P. (2014). Probabilistic risk assessment of veterinary medicines applied to four major aquaculture species produced in Asia. Sci. Total Environ. 468, 630–641. doi: 10.1016/j.scitotenv.2013.08.063
Rozman, U., Pušnik, M., Kmetec, S., Duh, D., and Šostar Turk, S. (2021). Reduced susceptibility and increased resistance of bacteria against disinfectants: A systematic review. Microorganisms 9:2550. doi: 10.3390/microorganisms9122550
Sánchez-Osuna, M., Cortés, P., Llagostera, M., Barbé, J., and Erill, I. (2020). Recurrent mobilization of ancestral and novel variants of the chromosomal di-hydrofolate reductase gene drives the emergence of clinical resistance to trimethoprim. bioRxiv [Preprint]. doi: 10.1101/2020.07.31.230557
Schweizer, H. P. (2001). Triclosan: A widely used biocide and its link to antibiotics. FEMS Microbiol. Lett. 202, 1–7. doi: 10.1111/j.1574-6968.2001.tb10772.x
Sharma, V., Johnson, N., Cizmas, L., McDonald, T., and Kim, H. (2016). A review of the influence of treatment strategies on antibiotic resistant bacteria and antibiotic resistance genes. Chemosphere 150, 702–714. doi: 10.1016/j.chemosphere.2015.12.084
Shi, P., Jia, S., Zhang, X., Zhang, T., Cheng, S., and Li, A. (2013). Metagenomic insights into chlorination effects on microbial antibiotic resistance in drinking water. Water Res. 47, 111–120. doi: 10.1016/j.watres.2012.09.046
Su, X., and D’Souza, D. H. (2012). Reduction of Salmonella typhimurium and Listeria monocytogenes on produce by trisodium phosphate. Food Sci. Technol. 45, 221–225. doi: 10.1016/j.lwt.2011.08.010
Tandukar, M., Oh, S., Tezel, U., Konstantinidis, K., and Pavlostathis, S. (2013). Long-term exposure to benzalkonium chloride disinfectants results in change of microbial community structure and increased antimicrobial resistance. Environ. Sci. Technol. 47, 9730–9738. doi: 10.1021/es401507k
Ward, J. Jr. (1963). Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 58, 236–244. doi: 10.1080/01621459.1963.10500845
Watts, J., Schreier, H., Lanska, L., and Hale, M. (2017). The rising tide of antimicrobial resistance in aquaculture: Sources, sinks and solutions. Mar. Drugs 15:158. doi: 10.3390/md15060158
Xu, Y., Xu, J., Mao, D., and Luo, Y. (2017). Effect of the selective pressure of sub-lethal level of heavy metals on the fate and distribution of ARGs in the catchment scale. Environ. Pollut. 220, 900–908. doi: 10.1016/j.envpol.2016.10.074
Zeller, G., Tap, J., Voigt, A., Sunagawa, S., Kultima, J., Costea, P., et al. (2014). Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 10:766. doi: 10.15252/msb.20145645
Zhang, G., Leclercq, S., Tian, J., Wang, C., Yahara, K., Ai, G., et al. (2017). A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT (3″)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 13:e1006602. doi: 10.1371/journal.pgen.1006602
Keywords: ozone treatments, disinfection, metagenomic shotgun sequencing, antimicrobial resistance genes, efflux mediated ARGs
Citation: Huang Q, Butaye P, Ng PH, Zhang J, Cai W and St-Hilaire S (2024) Impact of low-dose ozone nanobubble treatments on antimicrobial resistance genes in pond water. Front. Microbiol. 15:1393266. doi: 10.3389/fmicb.2024.1393266
Received: 28 February 2024; Accepted: 22 April 2024;
Published: 13 May 2024.
Edited by:
Karsten Becker, University Medicine Greifswald, GermanyReviewed by:
Bijay Khajanchi, United States Food and Drug Administration, United StatesCopyright © 2024 Huang, Butaye, Ng, Zhang, Cai and St-Hilaire. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sophie St-Hilaire, c3N0aGlsYWlAY2l0eXUuZWR1Lmhr
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.